This work shows that the post-Golgi trafficking of PIP2;7 involves an interaction with SYP61 and SYP121 and that SYP61 and SYP121 colocalize and are physically associated in a SNARE complex. These findings suggest that SNAREs, and possibly a SYP61/SYP121 SNARE complex, play an important role in the regulation of the transport of the plasma membrane aquaporin.
Abstract
Plant plasma membrane intrinsic proteins (PIPs) are aquaporins that facilitate the passive movement of water and small neutral solutes through biological membranes. Here, we report that post-Golgi trafficking of PIP2;7 in Arabidopsis thaliana involves specific interactions with two syntaxin proteins, namely, the Qc-SNARE SYP61 and the Qa-SNARE SYP121, that the proper delivery of PIP2;7 to the plasma membrane depends on the activity of the two SNAREs, and that the SNAREs colocalize and physically interact. These findings are indicative of an important role for SYP61 and SYP121, possibly forming a SNARE complex. Our data support a model in which direct interactions between specific SNARE proteins and PIP aquaporins modulate their post-Golgi trafficking and thus contribute to the fine-tuning of the water permeability of the plasma membrane.
INTRODUCTION
To cope with daily variations in water supply and availability, plants need to adapt their water balance quickly to situations ranging from moderate to severe water shortage episodes to short flooding periods. Such adaptations are achieved partly via the regulation of water channels known as plasma membrane intrinsic proteins (PIPs) (Chaumont et al., 2005; Maurel et al., 2008). To control their abundance and activity in the plasma membrane, PIP aquaporins are tightly regulated at multiple and interconnected levels: transcription, translation, or via posttranslational modifications affecting their trafficking, gating, and degradation (reviewed in Chaumont et al., 2005; Maurel et al., 2008; Hachez et al., 2013; Chaumont and Tyerman, 2014).
The management of the PIP intracellular localization appears to be an important manner by which plant cells modulate the plasma membrane water permeability (reviewed in Hachez et al., 2013). PIPs reach their final destination via the secretory pathway, trafficking from the endoplasmic reticulum (ER) via the Golgi apparatus to the plasma membrane. The ER-to-Golgi trafficking of PIPs is dependent on hetero-oligomerization of PIP1 and PIP2 proteins, the presence of a diacidic ER export motif in several PIP2s, or ubiquitylation (Zelazny et al., 2007, 2009; Lee et al., 2009; Sorieul et al., 2011). PIPs en route to the plasma membrane transit through the trans-Golgi network (TGN) that derives from the most trans-Golgi cisternae. This compartment plays a central role in PIP protein sorting, at the crossroads of secretion and endocytotic pathways. Proteins secreted from the TGN are transported to either the plasma membrane or the lytic or storage vacuoles. Plasma membrane-derived vesicles also merge back at this compartment level, which is assimilated to an early endosome (EE) in plant cells, to be either recycled back to the plasma membrane (constitutive cycling) or further degraded in lytic vacuoles (Li et al., 2011; Besserer et al., 2012; Luu et al., 2012).
Tight regulation of vesicle fusion events is essential to allow specific protein sorting and relies on the action of soluble N-ethylmaleimide-sensitive factor (NSF) protein attachment protein (SNAP) receptor (SNARE) proteins and their associated regulators (NSF and SNAP proteins). SNARE proteins are structurally classified into Q and R groups, based on the presence of a glutamine (Q) or arginine (R) residue in the SNARE domain (Fasshauer et al., 1998). Q-SNAREs can be further divided into three subgroups, Qa, Qb, and Qc. As each organelle in the endomembrane system contains a particular set of SNAREs, the ability to form coiled-coil interactions between specific pairs of Q- and R-SNAREs is thought to provide the specificity of vesicle sorting (Sanderfoot and Raikhel, 1999; Paumet et al., 2004; El Kasmi et al., 2013). Syntaxins are defined as Q-SNAREs with a C-terminal transmembrane domain. In Arabidopsis thaliana, 24 syntaxins have been identified that cluster in eight subgroups (Sanderfoot et al., 2000). Among the plasma membrane-resident syntaxins, SYNTAXIN OF PLANTS121 (SYP121) is known to mediate the trafficking of vesicles between the Golgi complex and the plasma membrane (Geelen et al., 2002). Its function may be disrupted by overexpression of a dominant-negative cytosolic (so-called Sp2) fragment (Tyrrell et al., 2007). Interestingly, besides the regulation of the plasma membrane protein trafficking, SYP121 also determines the gating of the K+ TRANSPORTER1 (AKT1)/K+ RECTIFYING CHANNEL1 (KC1) K+ channel complex through a direct interaction involving an FxRF motif located within the first 12 residues of the protein (Honsbein et al., 2009; Grefen et al., 2010). Such a direct interaction between the maize (Zea mays) SYP121 and aquaporin PIP2;5 was also found to regulate PIP2;5 trafficking to and activity in the plasma membrane (Besserer et al., 2012). Therefore, SYP121 might act as a molecular governor, coordinating the plasma membrane trafficking of ion or water channels in parallel with their gating (Grefen and Blatt, 2008; Honsbein et al., 2011; Besserer et al., 2012), thereby playing an important role in osmotic adjustment during cell expansion or environmental stresses.
The TGN is another SNARE-enriched endomembrane system. At least seven SNAREs are known to be more or less closely associated to this compartment (Uemura et al., 2004, 2012), probably because of its central role in vesicle sorting at the junction of endocytotic and exocytotic paths. Arabidopsis SYP61 is a TGN-localized syntaxin that is part of a protein complex that includes other SNAREs (VESICLE TRANSPORT V-SNARE12 [VTI12] and SYP41) (Drakakaki et al., 2012). It has been implicated in osmotic stress responses (Zhu et al., 2002) and might be linked to trafficking components to and from the prevacuolar compartment. Proteomics analysis of the TGN subcompartment highlighted that SYP61 might play a role in exocytotic trafficking to the plasma membrane, as supported by the nature of the identified proteins (Drakakaki et al., 2012). Altogether, these data point to a potential involvement of SYP61 in transport mechanisms in response to (a)biotic stresses, possibly as an integrated plant response similar to that described for SYP121: Whereas SYP61 vesicles are normally located at the TGN, they might be secreted in response to (a)biotic stresses as a defense/adaptation mechanism (Drakakaki et al., 2012).
As part of our efforts to elucidate the mechanisms regulating PIP2;7 activity and subcellular trafficking in Arabidopsis, we investigated the role of two SNAREs, SYP61 and SYP121. A mass spectrometry-based proteomic approach in maize identified SYP61 as a candidate interactor of the PIPs and the osmotic stress–sensitive1 (osm1; T-DNA insertion in SYP61) Arabidopsis mutant displayed a strong sensitivity to osmotic stress (Zhu et al., 2002), hence suggesting a function for SYP61 in PIP routing and activity. In addition, SYP121, a known PIP interactor (Besserer et al., 2012), colocalized and copurified with SYP61 in Arabidopsis (Drakakaki et al., 2012). This led us to test whether SYP121 and SYP61 might form a SNARE complex potentially involved in PIP trafficking. Our results show that the post-Golgi traffic of PIP2;7 is mediated by both SYP61 and SYP121, which, together, form a previously unknown SNARE complex. These data demonstrate that proper SNARE activity is required for the modulation of the water membrane permeability of plant cells.
RESULTS
PIP2;7 Is Highly Expressed in Elongating Cells in Both Root and Shoot
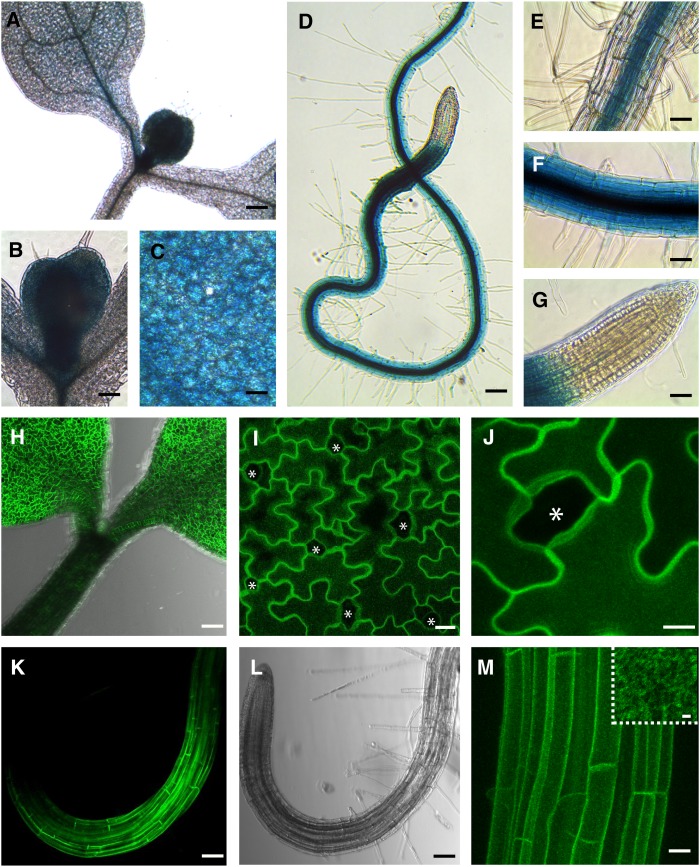
The aquaporin PIP2;7 has been reported to be highly expressed in all plant organs and to be an active water channel when expressed in Xenopus laevis oocytes (Weig et al., 1997; Alexandersson et al., 2005; Prado et al., 2013). To investigate more precisely its expression pattern, we created transcriptional (GUS-GFP [green fluorescent protein]) and translational (Venus-PIP2;7) reporter constructs driven by the native 2-kb-long PIP2;7 promoter (see Methods). Three independent Arabidopsis lines were analyzed for each construct (15 plants/construct) and yielded similar results in term of expression pattern. In aerial parts, β-glucuronidase (GUS) staining was stronger in cotyledons and leaf primordia than in the hypocotyl (Figures 1A to 1C and 1H) and emerging leaf primordia were more strongly labeled than cotyledons (Figures 1A and 1B). In the leaf mesophyll, GUS activity had a diffuse and patchy pattern, with weak staining in the leaf veins, as recently reported (Prado et al., 2013) (Figure 1A). The same trend was observed at the protein level for the PIP2;7 translational reporter (Figure 1H). In cotyledons, Venus-PIP2;7 fusion proteins were found in pavement cells but were not detected in stomatal lineage cells (meristemoids, guard mother cells, and guard cells) (Figures 1I and 1J).
Figure 1.
Analysis of PIP2;7 Transcriptional and Translational Reporters.
(A) to (G) pPIP2;7:GUS transcriptional reporter in Arabidopsis seedlings 7 d postgermination.
(D) to (G) GUS staining of the primary root.
(H) to (J) PIP2;7 translational reporter (pPIP2;7:Venus-PIP2;7) in 7-d-old seedlings showing a labeling of the cotyledons and a weaker signal in the hypocotyl epidermis (H). Note the absence of signal in stomata (asterisks) ([I] and [J]).
(K) and (L) PIP2;7 translational reporter in the primary root.
(M) Root epidermal cells showing plasma membrane labeling by Venus-PIP2;7 proteins that appear not to be homogeneously distributed in the membrane (inset).
(H) to (J), (L), and (M) are three-dimensional reconstitutions by maximum projection of stacked confocal images. Bars = 100 µm in (A), (D), (H), (K), and (L), 50 µm in (B), (C), and (E) to (G), 20 µm in (I) and (M), 10 µm in (J), and 2 µm in inset in (M).
In roots, GUS staining was not detected in the cap and meristem (Figures 1D and 1G), appeared gradually at the end of the meristematic zone (Figure 1F) with a clear peak in the root elongation zone where every cell type was strongly stained (Figures 1D and 1F), and then slightly diminished in older root tissues (Figures 1D and 1E). In the root hair zone, the PIP2;7 promoter activity gradually decreased in the cortex, while remaining high in the stele tissue. In more mature tissues, the GUS signal was strong in the stele, but faint in the cortex (Figures 1D and 1E). The same trend was observed for the PIP2;7 translational reporter in roots (Figures 1K and 1M). Similarly to what was reported for PIP2;1 (Li et al., 2011), the fluorescently tagged PIP2;7 was found distributed in what seemed to be plasma membrane domains (Figures 1H to 1M), although additional analysis using variable-angle evanescent wave microscopy and fluorescence correlation spectroscopy would be needed to ascertain that claim.
The analysis of the PIP2;7 expression pattern in both aerial parts and roots of developing seedlings suggests that it is particularly involved in cell elongation processes and occurs at places where water movement regulation is crucial, such as the root hair zone, stele tissue, and emerging leaf primordia.
PIP2;7 Colocalizes with SYP61 and SYP121
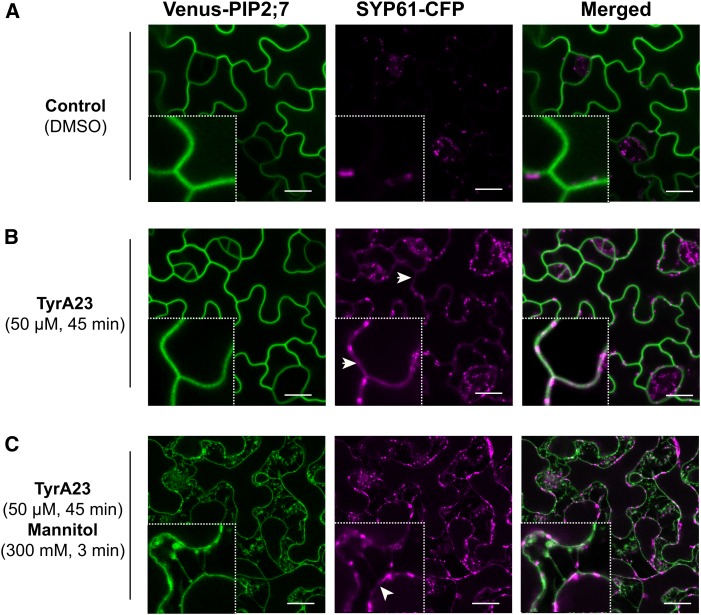
The PIP trafficking to the plasma membrane involves vesicle fusion events along the secretory pathway that are mediated by SNARE proteins. A mass spectrometry-based proteomic approach identified the TGN/EE-localized syntaxin SYP61 as a putative interactor of maize PIPs (Supplemental Figure 1 and Supplemental Methods). To determine whether PIP2;7 might interact with SYP61 in Arabidopsis, colocalization analysis was performed (Figure 2). In cotyledon pavement cells of double transgenic Arabidopsis lines coexpressing pPIP2;7:Venus-PIP2;7 and pSYP61:SYP61-CFP (cyan fluorescent protein), only a weak colocalization between Venus-PIP2;7 and SYP61-CFP (Pearson’s coefficient of 0.166 ± 0.020, mean ± se, n = 15 images, three images/plant) was detected (Figure 2A). Although a vast majority of the SYP61 protein pool was found in TGN/EE (Drakakaki et al., 2012), we hypothesized that SYP61 cycles between this compartment and the plasma membrane and is rapidly internalized by endocytosis. This hypothesis was supported by data showing that BFA treatment triggered an accumulation of SYP61 in brefeldin A bodies and at the plasma membrane (Drakakaki et al., 2012). Upon treatment with tyrphostin A23 (TyrA23; 50 µM, 45 min), a known inhibitor of clathrin-mediated endocytosis in plants (Fujimoto et al., 2010; Barberon et al., 2011; Van Damme et al., 2011), a pool of SYP61-CFP was detected in the plasma membrane where it strongly colocalized with Venus-PIP2;7 (Pearson’s coefficient of 0.583 ± 0.023; mean ± se, n = 15 images, three images/plant) (Figure 2B). This colocalization in the plasma membrane was further confirmed by plasmolysis (300 mM mannitol, 3 min) following the TyrA23 treatment (Figure 2C). The effect of TyrA23 on SYP61-CFP subcellular localization suggests that this protein, whose steady state pool is found in the TGN/EE, might constitutively cycle between the TGN/EE and the plasma membrane.
Figure 2.
Colocalization of PIP2;7 with SYP61 in the Cotyledon Epidermis.
(A) and (B) Venus-PIP2;7 and SYP61-CFP translational reporters in cotyledon epidermal cells under control conditions (A) or upon TyrA23 treatment (50 µM, 45 min) (B). While Venus-PIP2;7 labels the plasma membrane, SYP61-CFP is mostly found in the TGN/EE (insets: 4× magnification) in control conditions (A), whereas a clear overlap of the signals is observed in the plasma membrane upon TyrA23 treatment (B) where a partial colocalization is visible (as highlighted by white arrowheads; insets: 4× magnification).
(C) Colocalization of Venus-PIP2;7 and SYP61-CFP proteins upon TyrA23 treatment (50 µM, 45 min) followed by an osmotic stress (300 mM Mannitol, 3 min). This treatment triggered plasmolysis of the cells. SYP61-CFP still colocalized with Venus-PIP2;7 in the plasmolyzed plasma membrane (insets: 4× magnification). A small but significant difference (P < 0.05) in SYP61-CFP mean fluorescence intensity was detected in individual TGN/EE structures between control conditions (839 ± 9 A.U.; mean ± se, n = 994 endosomes from 11 images) and after TyrA23 treatment (883 ± 6 A.U.; mean ± se, n = 1604 endosomes from 15 images). Bars = 15 µm.
The syntaxin SYP121 had been previously shown to mediate transport of PIP2;5 in maize cells (Besserer et al., 2012). When coexpressed in Arabidopsis under the control of the 35S promoter, Venus-SYP121 and CFP-PIP2;7 strongly colocalized in the plasma membrane of cotyledon pavement cells before and after plasmolysis (Supplemental Figure 2) with an average Pearson’s coefficient of 0.712 ± 0.054 (mean ± se, n = 15 images, three images/plant).
PIP2;7 Physically Interacts with SYP61 and SYP121
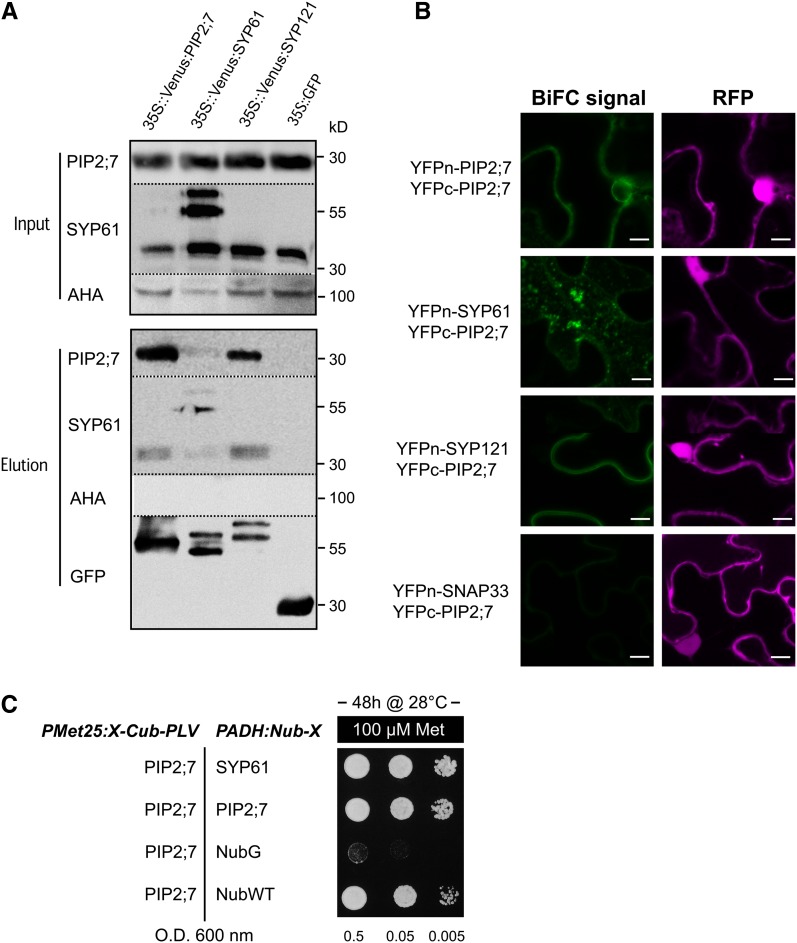
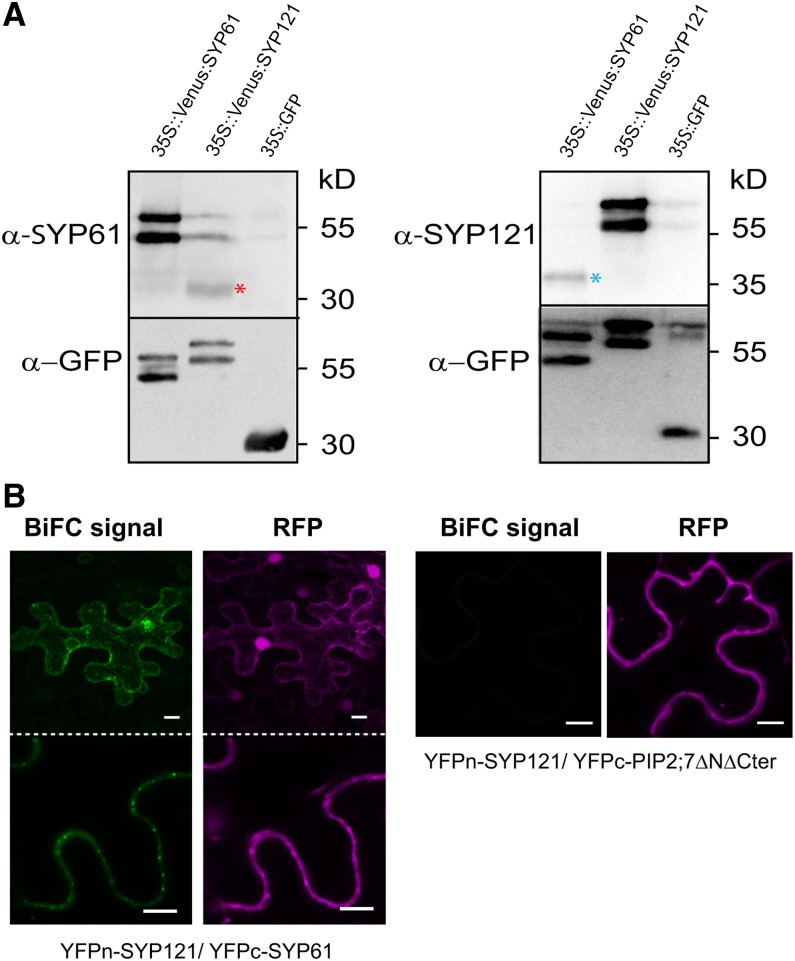
To determine whether PIP2;7 physically interacts with SYP61 or SYP121, we prepared total protein extracts from stable Arabidopsis transgenic lines expressing p35S:Venus-PIP2;7, p35S:Venus-SYP61, p35S:Venus-SYP121, or p35S:GFP (negative control) and performed pull-down assays with anti-GFP columns (see Methods) that resulted in the purification of Venus-tagged or GFP-tagged proteins (Figure 3A). The presence of coeluted endogenous PIP2;7 or SYP61 in the elution fraction was checked by immunodetection with antibodies raised against PIP2;7 or SYP61 (Sanderfoot et al., 2001a) (see Methods). A signal at ∼30 kD corresponding to PIP2;7 monomers was detected in elution fractions from lines overexpressing Venus-PIP2;7 (Figure 3A). Such interaction between endogenous PIP2;7 and Venus-PIP2;7 was expected because aquaporins assemble as tetramers in the membrane and PIPs are able to form disulfide bond-linked dimers (Bienert et al., 2012). The interaction between SYP61 and PIP2;7 was tested in a similar manner. A faint, but significant, signal for endogenous PIP2;7 proteins was recorded in the elution fraction from the Venus-SYP61-overexpressing line (Figure 3A); reciprocally, a significant signal for endogenous SYP61 proteins (∼35 kD) was recorded as well in the elution fraction from the Venus-PIP2;7-overexpressing line (Figure 3A). Coelution of SYP121 and PIP2;7 was also demonstrated by the presence of an on average 10-fold stronger PIP2;7 signal than that in Venus-SYP61-expressing lines in the elution fraction from the Venus-SYP121 line (Figure 3A). Interestingly, endogenous SYP61 proteins were also detected in the elution fraction of Venus-SYP121 (see below). No signal for the plasma membrane H+-ATPases was detected in any of the elution fractions, whereas these proteins were present in all input fractions, demonstrating a specific coelution between PIP2;7 and SYP61 or SYP121. No PIP2;7 signal was observed in elution fractions from lines expressing the soluble GFP, which further confirmed the quality of our coimmunoprecipitation assays.
Figure 3.
Interaction between PIP2;7, SYP61, and SYP121.
(A) GFP trap assay. The presence of PIP2;7, SYP61, and H+-ATPase (AHA) proteins is detected in input fractions (total protein extracts) from homozygous lines expressing Venus-PIP2;7, Venus-SYP61, Venus-SYP121, and soluble GFP (negative control) with PIP2;7, SYP61 and H+-ATPase antibodies (see Methods). The endogenous PIP2;7 signal (band at ∼30 kD) is detected in elution fractions of Venus-PIP2;7, Venus-SYP61, and Venus-SYP121. The endogenous SYP61 signal (band at ∼35 kD) is clearly detected in elution fractions of Venus-PIP2;7 and Venus-SYP121 lines, while a fainter signal is also detected at 35 kD in the Venus-SYP61 line but could originate from degradation of the Venus-SYP61 fusion protein. Note the absence of PIP2;7 and SYP61 signals in the elution fraction of the soluble GFP-negative control. No AHA signal is detected in the elution fraction. These experiments were repeated 5 times with independent biological replicates and representative blots are shown.
(B) BiFC signals for the YFPn-PIP2;7/YFPc-PIP2;7, YFPn-SYP61/YFPc-PIP2;7, YFPn-SYP121/YFPc-PIP2;7, and YFPn-SNAP33/YFPc-PIP2;7 pairs. BiFC signals (YFP) are in green, and soluble RFP signals are depicted in magenta and serve as transfection controls. No signal is detected for the YFPn-SNAP33/YFPc-PIP2;7 pair. Bars = 10 µm.
(C) SUS assays. Yeast coexpressing the Met-repressible bait construct PIP2;7-Cub-PLV and the prey constructs NubG-SYP61 or NubG-PIP2;7 (NubG = negative; NubWT = positive control) were dropped in a dilution series (OD 0.5, 0.05, and 0.005) onto synthetic media containing 100 µM methionine to repress expression of the bait. Yeast growth was recorded after incubation for 48 h. Yeast strain coexpressing the PIP2;7-Cub-PLV/NubG-SYP61 grows similarly to the strain coexpressing the PIP2;7-Cub-PLV/NubG-PIP2;7 pair, while no growth was observed for the negative control (see Methods for details). These experiments were repeated three times with independent biological replicates.
Physical interaction between coeluted proteins was further validated with bimolecular fluorescence complementation (BiFC) assays, performed by transient expression in leaf epidermal cells of tobacco (Nicotiana tabacum) (Figure 3B). PIP2;7 and SYP61 or SYP121 were introduced in the pBiFCt-2in1 vector (Grefen and Blatt, 2012), which allows the coexpression of the half yellow fluorescent protein (YFP) constructs together with a soluble red fluorescent protein (RFP) serving as a transfection control (see Methods). As expected, YFP signal at the plasma membrane was detected for the N-terminal yellow fluorescent protein (YFPn) and C-terminal YFP (YFPc) pair YFPn-PIP2;7/YFPc-PIP2;7 (Figure 3B). For the YFPn-SYP61/YFPc-PIP2;7 pair, BiFC signals were predominantly observed in small vesicular structures (Figure 3B) and to a lesser extent in the plasma membrane. A plasma membrane-localized YFP signal was recorded for the YFPn-SYP121/YFPc-PIP2;7 pair, a result similar to that for PIP2;5/SYP121 of maize (Besserer et al., 2012). SNAP33, a plasma membrane-localized SNARE (Karnik et al., 2013), was used as a negative control for the PIP2;7 interaction. No BiFC signal was detected when the YFPn-SNAP33/YFPc-PIP2;7 pair was expressed, confirming the interaction-dependent specificity of the YFP signal observed for the other pairs.
Additionally, the physical interaction between PIP2;7 and SYP61 was confirmed using a split ubiquitin (SUS) assay (Figure 3C) as previously described (Honsbein et al., 2009, 2011; Grefen et al., 2010; Besserer et al., 2012). SUS constructs were built to test the interaction between NubG-SYP61 (prey) and PIP2;7-Cub-PLV (bait) fusion proteins. NubG-PIP2;7 fusion protein was used as positive interaction control, while NubG and NubWT fragments served as negative and positive Cub interaction controls, respectively. Yeast growth was observed for all protein couples, except the negative control (NubG/PIP2;7-Cub-PLV pair), suggesting that PIP2;7 and SYP61 physically interact. A similar SUS approach was also successfully used to confirm the interaction between PIP2;7 and SYP121 (Supplemental Figure 3). Taken together, these data demonstrate a direct interaction between PIP2;7 and SYP61 or SYP121.
SYP61 and SYP121 Form a SNARE Complex
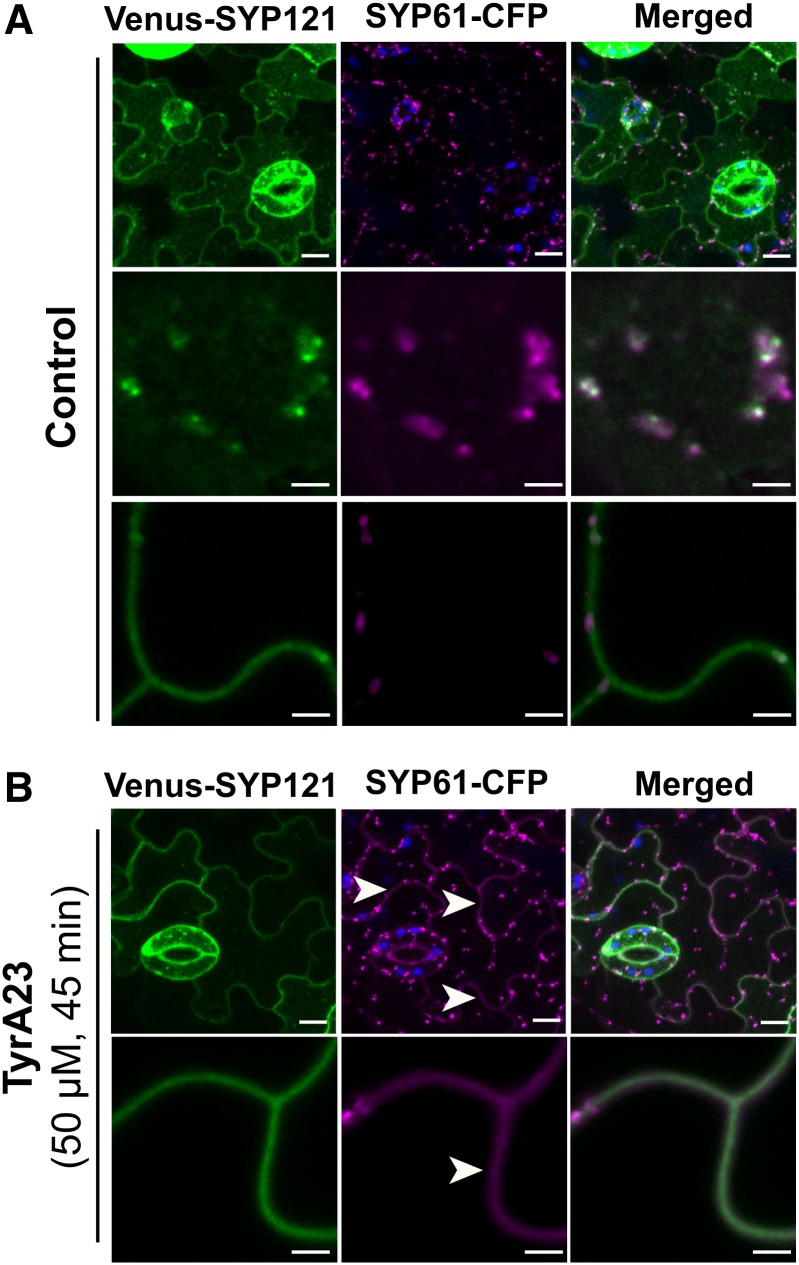
As SYP121 was previously identified in the SYP61 proteome (Drakakaki et al., 2012), we investigated whether these two syntaxins directly interact and possibly form a SNARE complex. In cotyledon epidermal cells, SYP61 predominantly localized in TGN/EEs, whereas SYP121 strongly labeled both the plasma membrane and TGN/EEs (Figure 4A). Whereas colocalization was barely detected at the plasma membrane in control conditions, most of the SYP61 and SYP121 proteins colocalized in the TGN/EE compartments (Figure 4A). Notably, within a particular TGN/EE structure, SYP61-CFP proteins labeled more endosomes than Venus-SYP121 (Figure 4A, top and middle panels); however, the colabeled endosomes moved in a coordinated fashion (Supplemental Movie 1). Within the endosomal compartments of cotyledon pavement cells, Pearson’s coefficients indicated a moderate to strong colocalization of the SYP61-CFP-stained structures with the Venus-SYP121-labeled ones (Pearson’s coefficient range: 0.392 to 0.741). Of the intracellular SYP61-CFP signals, 76% colocalized with the Venus-SYP121 fluorescence and, reciprocally, 73% of the Venus-SYP121 intracellular structures with the SYP61-CFP signals. Statistical analysis of endosomal pools of SYP121 and SYP61 was performed on a total of 45 cells coming from 15 seedlings from the same cotransformed line. Similar colocalization data were observed in root epidermal cells (Pearson’s coefficient range: 0.351 to 0.713) (Supplemental Figure 4).
Figure 4.
Colocalization of SYP61 and SYP121.
Colocalization of SYP61-CFP and Venus-SYP121 expressed from pSYP61:SYP61-CFP and p35S:Venus-SYP121 constructs in cotyledon epidermal cells under control conditions (A) or upon TyrA23 treatment (50 µM, 45 min) (B). In control conditions, SYP61-CFP and Venus-SYP121 mostly colocalized in an endomembrane compartment (TGN/EE) where a partial overlap of Venus-SYP121 and SYP61-CFP signals is detected ([A], middle panels), while SYP61-CFP proteins are barely detected in the plasma membrane ([A], bottom panels). Upon TyrA23 treatment, SYP61-CFP colocalization is also found at the plasma membrane (white arrowheads) ([B], bottom panels) In (A) and (B), images in upper panels are maximum projections of three-dimensional reconstructions. Chloroplast autofluorescence appears in blue. Bars = 10 µm in upper panels and 3 µm in lower panels.
Similarly to the SYP61/PIP2;7 colocalization, we used TyrA23 to block clathrin-mediated endocytosis and assess the impact of such treatment on the colocalization of SYP61 with SYP121. TyrA23 treatment triggered accumulation of SYP61 in the plasma membrane (Figures 2B to 2C) where it strongly colocalized with SYP121 (Figure 4B). The whole-cell Pearson’s coefficient significantly (P < 0.05) increased from 0.357 ± 0.013 in absence of TyrA23 to 0.601 ± 0.027 (mean ± se, n = 15 images, three images/plant) after treatment, mostly due to the accumulation of SYP61-CFP and Venus-SYP121 proteins in the plasma membrane.
GFP pull-down assays revealed the presence of endogenous SYP121 in the elution fraction of the Venus-SYP61 line (Figure 5A). Reciprocally, endogenous SYP61 proteins were detected in the elution fraction from the Venus-SYP121 line (Figures 2A and 5A). These reciprocal pull-downs show that SYP61 and SYP121 coeluted. No signal for SYP61 or SYP121 was detected in the elution fraction of the soluble GFP line. Physical interaction between SYP121 and SYP61 was further validated by BiFC assay performed in tobacco epidermal cells (Figure 5B). As negative interaction control for SYP121, we used a mutated version of PIP2;7 (PIP2;7ΔNΔCter) in which amino acid residues 1 to 39 (cytosolic N terminus) and 263 to 280 (cytosolic C terminus) were deleted. This protein fused to GFP was localized in the plasma membrane and intracellular structures probably corresponding to the ER and Golgi apparatus (Supplemental Figure 5). While YFPn-SYP121/YFPc-PIP2;7ΔNΔCter pair did not yield any fluorescent signal, specific BiFC signals were detected for the YFPn-SYP121/YFPc-SYP61 pair in an endosomal compartment (Figure 5B), confirming that both syntaxins interact in vivo as a presumably part of a SNARE complex.
Figure 5.
Interaction between SYP61 and SYP121.
(A) Coelution of SYP61 and SYP121 by GFP pull-down assay (see Methods for details). Note the specific presence of endogenous SYP61 proteins in the elution fraction of p35S:Venus-SYP121 line (red asterisk), while endogenous SYP121 proteins are detected in the elution fraction of p35S:Venus-SYP61 line (blue asterisk). A total of three independent experiments (biological replicates) were performed and representative blots are shown.
(B) BiFC signals for the YFPn-SYP121/YFPc-SYP61 pair and YFPn-SYP121/YFPc-PIP2;7ΔNΔCter. BiFC signals (YFP) are in green, and soluble RFP signals are depicted in magenta and serve as transfection controls. Bars = 10 µm.
PIP2;7 Trafficking Depends on Functional SYP61
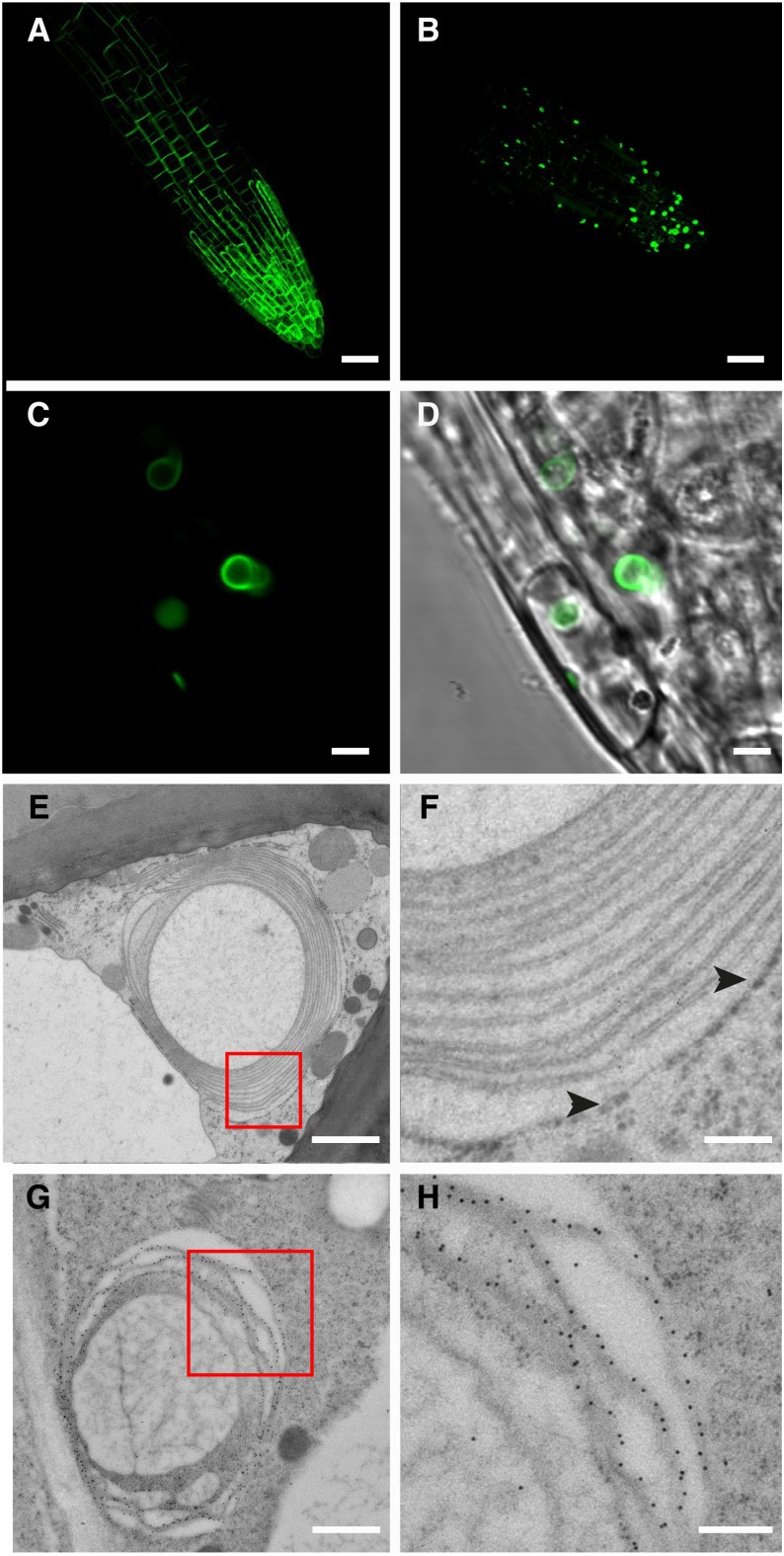
We tested whether alteration of SYP61 function affected the trafficking of PIP2;7. First, we investigated the expression of PIP2;7 in the osm1 mutant background. The osm1 line possesses a T-DNA insertion in the first exon of SYP61 that results in gene disruption and accumulation of truncated SYP61 transcripts (Zhu et al., 2002) and may therefore not be considered as null. However, the observed phenotypes, namely, increased sensitivity to both ionic (NaCl) and nonionic (mannitol) osmotic stress, increased root branching pattern, and faster wilting when grown with limited soil moisture, are rescued by transformation with the wild-type SYP61 allele (Zhu et al., 2002). Venus-PIP2;7 overexpression in the osm1 mutant background resulted in a serious decrease in PIP2;7 amount in the plasma membrane and its abnormal accumulation in globular or lenticular structures with an average diameter of 2.5 µm (Figure 6; Supplemental Figure 6). This observation was confirmed by in situ immunodetection experiments with anti-GFP antibodies that showed accumulation of Venus-PIP2;7 in these globular/lenticular structures (Figures 6B to 6D), in contrast to the auxin transporter PIN2 that labeled only the plasma membrane of the osm1 mutant (Supplemental Figure 7). These abnormal structures were observed in cells from the roots and aerial parts (hypocotyl, cotyledons, and leaf primordia) (Supplemental Figures 6D to 6F).
Figure 6.
Subcellular Localization of Overexpressed Venus-PIP2;7 in osm1.
(A) and (B) Overexpression of Venus-PIP2;7 in C24 (A) and osm1 backgrounds (B). While Venus-PIP2;7 strongly labels the plasma membrane in C24 (A), most of the Venus-PIP2;7 protein pool is trapped in OSER structures in osm1 background (B).
(C) and (D) Close-up view of (B).
(E) and (F) OSER structure as seen via transmission electron microscopy. Note that the membrane stacks are derived from smooth ER. Ribosomes are only found around the most external ER stack ([F], black arrowheads).
(G) and (H) Gold immunolabeling revealing Venus-PIP2;7 in stacked smooth ER membranes.
Bars = 20 µm in (A) and (B), 3 µm in (C) and (D), 1 µm in (E) and (G), and 0.2 µm in (F) and (H).
Transmission electron microscopy analysis of root cap cells from these plants and fine cytological analysis of the samples revealed that these structures corresponded to ER-derived stacked membrane arrays, designated Organized Smooth ER (OSER; also known as karmellae), that sometimes occur in response to elevated levels of specific OSER-inducing proteins (Figures 6E and 6F) (Snapp et al., 2003). These structures were not observed in the absence of Venus-PIP2;7 overexpression in osm1 background. Gold immunolabeling with anti-GFP antibodies showed that Venus-PIP2;7 was closely associated to these OSER membrane stacks (Figures 6G to 6H). Such artifactual OSER structures were formed only in the osm1 line and not in the C24 or Columbia wild-type lines upon Venus-PIP2;7 overexpression, hinting at an accumulation of PIP2;7 in the secretory pathway (ER) due to a transport alteration linked to the syp61 mutation. Interestingly, expression of the pSYP61:SYP61-CFP construct in the osm1 background rescued the plasma membrane localization of Venus-PIP2;7 proteins, demonstrating that formation of OSER structures originates from a deficiency in SYP61 activity upon overexpression of Venus-PIP2;7 (Supplemental Figures 6G to 6I).
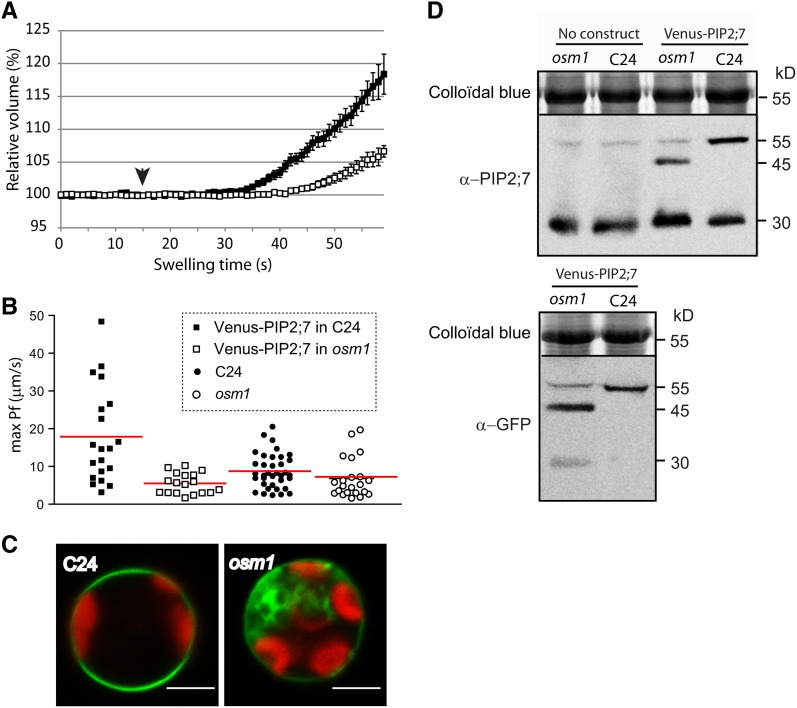
To determine whether alterations of the Venus-PIP2;7 plasma membrane delivery due to altered SYP61 activity correlated with a decrease in the membrane permeability coefficient (Pf), we compared the swelling rate of leaf mesophyll protoplasts originating from the osm1 line and their C24 control accession, both stably transformed with a p35S:Venus:PIP2;7 construct (one homozygous line per genetic background) (Figure 7). The Pf of protoplasts from nontransformed C24 and osm1 backgrounds were also investigated to highlight the contribution of Venus-PIP2;7 to overall water channel activity. A significant difference (P < 0.05) was observed between the Pf values of Venus-PIP2;7-expressing osm1 protoplasts and those of the Venus-PIP2;7-expressing wild-type protoplasts. The Pf values of Venus-PIP2;7-expressing osm1 protoplasts were indeed 69% lower than those of the Venus-PIP2;7-expressing wild-type protoplasts but were not significantly different (P < 0.05) from nontransformed C24 and osm1 backgrounds, indicating that Venus-PIP2;7 trapping into OSER structures prevented the proteins to properly mediate the plasma membrane water permeability. We checked the Venus-PIP2;7 protein levels by immunoblots using both anti-PIP2;7 and anti-GFP antibodies to discriminate between Venus-PIP2;7 and endogenous PIP2;7 dimers that migrate at the same position on the gel (∼55 kD). Endogenous PIP2;7 dimers were only weakly detected in non-transformed C24 and osm1 backgrounds (Figure 7D). An additional band with a molecular mass of ∼45 kD was observed in the osm1 background overexpressing Venus-PIP2;7, probably corresponding to a truncated version of Venus-PIP2;7 (Figure 7D). Free GFP (∼30 kD) was also detected with the anti-GFP antibodies in osm1. A likely explanation would be that Venus-PIP2;7 proteins are partly degraded and cleaved in the osm1 background, presumably as an indirect consequence of their trapping in OSER structures.
Figure 7.
Effect of the osm1 Mutation on Venus-PIP2;7-Mediated Membrane Water Permeability and on the Venus-PIP2;7 Subcellular Distribution in Mesophyll Protoplasts.
(A) Time-lapse analysis of the relative volume change upon a 45-s hypoosmotic challenge of Arabidopsis mesophyll protoplasts overexpressing Venus-PIP2;7 in a C24 background (black squares; n = 20) or in an osm1 background (white squares; n = 18). Cells originated from three independent replicates. Solutions were switched to hypotonic medium at 15 s (black arrowhead). Cells swelled more rapidly in the C24 than in the osm1 backgrounds.
(B) Individual max Pf values plotted for the following lines: 35S:Venus-PIP2;7 in C24 background (black squares), 35S:Venus-PIP2;7 in osm1 background (white squares), nontransformed C24 (black circles), and osm1 lines (white circles). The red line indicates the respective average max Pf value of each data set. The mean max Pf value was significantly higher (P < 0.001) in the 35S:Venus-PIP2;7/C24 line than in the others.
(C) Representative mesophyll protoplasts expressing Venus-PIP2;7 in C24 or osm1 background. Protoplasts were prepared from homozygous lines for the transgene. Note the intracellular accumulation of Venus-PIP2;7 in osm1 background. Bars = 10 µm.
(D) Immunoblot analysis of C24 and osm1 lines expressing Venus-PIP2;7. While endogenous PIP2;7 proteins are detected in similar amounts in both genetic backgrounds (monomeric and dimeric form) using α-PIP2;7 antibody, an additional ∼45-kD band is observed in the osm1 line. Comparison between the size of Venus-PIP2;7 in C24 versus osm1 backgrounds using α-GFP antibody suggests that Venus-PIP2;7 proteins are cleaved in osm1. Blots are representative of the profiles of four independent biological replicates.
Altered SYP121 Activity Affects PIP2;7 Trafficking and Cell Membrane Permeability
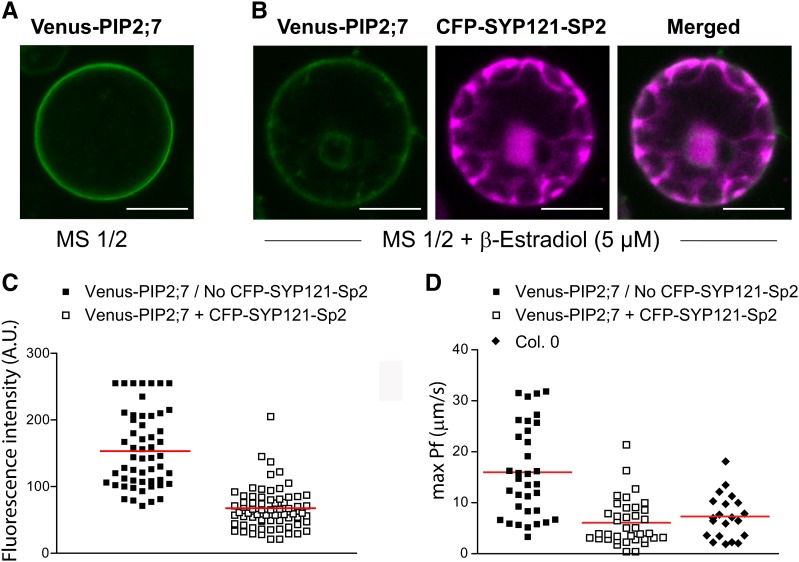
Transient expression assays in maize protoplasts had indicated that the monomeric (m)CFP-SYP121-Sp2 fragment reduced the plasma membrane delivery of PIP2;5 (Besserer et al., 2012). We also previously reported that the Pf values of protoplasts isolated from the syp121-1 Arabidopsis mutated line (Collins et al., 2003) were lower than those of the wild type (Besserer et al., 2012). To further investigate whether the truncated syntaxin interfered with the proper trafficking of stably expressed PIPs, we prepared a genetic construct in which the DNA encoding mCFP-SYP121-Sp2 was placed under the 35S promoter or under a β-estradiol-inducible promoter (see Methods). Transgenic seedlings coexpressing Venus-PIP2;7 and the inducible mCFP-SYP121-Sp2 fragment were obtained, germinated in presence of 5 µM β-estradiol and analyzed at 7 d postgermination. Plants from the same line growing on regular medium to prevent induction of mCFP-SYP121-Sp2 were used as controls. Interestingly, induction of mCFP-SYP121-Sp2 resulted in a 57% lower Venus-PIP2;7 signal intensity in the plasma membrane than in control protoplasts (Figures 8A to 8C). Similarly to what was reported by Besserer et al. (2012), the Venus signal intensity in the whole protoplast was slightly lower but not significantly different in cells coexpressing Venus-PIP2;7 and mCFP-AtSYP121-Sp2 (Venus signal intensity: 32.6 ± 0.8 arbitrary units [A.U.]; mean ± se; n = 116 protoplasts) compared with protoplasts expressing Venus-PIP2;7 alone (Venus signal intensity: 34.9 ± 0.9 A.U.; mean ± se; n = 183 protoplasts). Coexpression of Venus-PIP2;7 and mCFP-SYP121-Sp2 significantly (P < 0.0001) reduced (64%) the Pf values of mesophyll cell protoplasts when compared with cells expressing Venus-PIP2;7 alone (Figure 8D).
Figure 8.
Effect of the CFP-SYP121-Sp2 Fragment on Venus-PIP2;7 Subcellular Distribution and Membrane Water Permeability in Mesophyll Protoplasts.
(A) and (B) Venus-PIP2;7 localization under control conditions (no β-estradiol treatment; [A]) or upon coexpression with the CFP-SYP121-Sp2 dominant-negative fragment (B). The Venus-PIP2;7 signal intensity is reduced in the cell periphery upon coexpression with the CFP-SYP121-Sp2 construct. Bars = 10 µm.
(C) Venus-PIP2;7 fluorescence intensity measurement in the cell periphery without (n = 54) or after induction of CFP-SYP121-Sp2 (n = 67). The peripheral Venus-PIP2;7 signal intensity is significantly (P < 0.001) lower (57%) upon coexpression with CFP-SYP121-Sp2 than that under noninducing conditions.
(D) Individual max Pf values plotted for the following lines: 35S:Venus-PIP2;7 in Columbia in absence of CFP-SYP121-Sp2 protein induction (black squares) or under SP2 inducing conditions (white squares). The Pf of wild-type Columbia mesophyll protoplasts is displayed as well.
The red lines indicate the respective average max Pf value of each data set. The mean max Pf value was significantly (P < 0.0001) reduced to wild-type levels upon coexpression of the SYP121-Sp2 dominant-negative mutant. Overexpression of Venus-PIP2;7 significantly increases (P < 0.001) the cell Pf compared with wild-type levels.
Such an inhibitory effect on PIP plasma membrane trafficking was also observed in intact root cells for Venus-PIP2;7 and was also found for YFP-PIP1;4 or RFP-PIP2;1 after coexpression with mCFP-SYP121-Sp2 (Supplemental Figure 8). The mechanism is therefore not PIP2;7 specific but seems to be rather general for PIP proteins. The overexpression of the SYP121-Sp2 fragment also affected the plasma membrane delivery of the leucine-rich repeat receptor kinase BRASSINOSTEROID INSENSITIVE1 (BRI1) fused to GFP (Friedrichsen et al., 2000), of which the signal was depleted from the plasma membrane and the overall fluorescence intensity was reduced (Supplemental Figure 9). By contrast, PIN2-GFP was not affected and was seemingly transported to the plasma membrane via a SYP121-independent route (Supplemental Figure 9) as for the trafficking of H+-ATPase to the plasma membrane (Sutter et al., 2006). Taken together, these data demonstrate that SYP121-mediated plasma membrane delivery is not restricted to PIP and K+-channels but does not constitute a general mechanism for plasma membrane-localized proteins.
DISCUSSION
PIP aquaporins facilitate the water movement through the plasma membrane and therefore are key proteins that regulate their water permeability. However, to reach the plasma membrane, PIPs have to traffic through the secretory pathway, a process that requires vesicle fusions between different compartments and specific SNARE complex activities (Pratelli et al., 2004). Here, we demonstrated that the post-Golgi trafficking of PIP2;7 involves an interaction with SYP61 and SYP121 and that the proper subcellular localization of PIP2;7 depends on the correct activity of these syntaxins. We also show that SYP61 and SYP121 colocalize and are physically associated in a SNARE complex. These findings suggest that SNAREs, and possibly a SYP61/SYP121 SNARE complex, play an important role in the regulation of the transport of the plasma membrane aquaporin.
The Syntaxin SYP61 Is Part of a SNARE Complex That Mediates the PIP2;7 Post-Golgi Trafficking
Colocalization, immunoprecipitation, BiFC, and SUS experiments showed that SYP61 and PIP2;7 physically interact at the level of the plasma membrane and in the TGN/EE. However, the exact role of SYP61 in modulating the PIP2;7 post-Golgi trafficking is still unknown: It might influence either the PIP anterograde transport from the TGN/EE to the plasma membrane, as suggested by the proteomics analysis of the SYP61 compartment that identified several plasma membrane-resident cargos (Drakakaki et al., 2012), or the retrograde transport from the plasma membrane to the TGN/EE. In fact, depending on its association with specific SNARE proteins, SYP61 might be involved in both anterograde and retrograde transport between the TGN/EE and the plasma membrane. Interestingly, upon treatment with the endocytosis inhibitor TyrA23, SYP61 accumulated in the plasma membrane where it colocalized with PIP2;7 and SYP121, indicating its cycling between TGN/EE and plasma membrane (Figure 2C). These data are similar to what was recently reported for IRON-REGULATED TRANSPORTER1 (IRT1), whose accumulation in the plasma membrane could be detected only after TyrA23 treatment due to inhibition of its constitutive cycling between the PM and TGN/EE (Barberon et al., 2011). Inhibition of endocytosis by TyrA23 also seemed to slightly affect the overall abundance/stability of SYP61-CFP as a small (5% compared with control condition) but statistically significant (P < 0.05) accumulation of SYP61-CFP could be noted in TGN/EE following TyrA23 treatment.
Null mutations of key TGN-localized syntaxins are usually lethal (Sanderfoot et al., 2001b). The only available known SYP61 mutant is osm1, which is not considered a null mutant because of the aberrant SYP61 transcripts it produces (Zhu et al., 2002). However, because the observed phenotypes are rescued by the wild-type SYP61 allele, its use, as a first line of study, can be very informative regarding the role of SYP61 (Zhu et al., 2002). Overexpression of Venus-PIP2;7 in osm1 induced the formation of karmellae, whorls, and crystalloid OSER structures that have been reported in various cells, tissues, and organisms, including plants, fungi, and mammals under physiological conditions or by overproduction of resident ER transmembrane proteins (reviewed in Snapp et al., 2003). A “zipper mechanism” was proposed to explain the OSER biogenesis, in which the cytoplasmic domains of OSER-inducing proteins on opposing membranes bind tightly to each other and stabilize the membranes together. This model predicts that OSER-inducing proteins that reside within these structures are tightly bound and trapped. Interestingly, even when OSER structures are known artifacts of overproduced membrane proteins, accumulation of Venus-PIP2;7 in these structures was observed only in the osm1, but not in the C24 wild-type background overexpressing Venus-PIP2;7, and disappeared when the osm1 line was complemented with the pSYP61:SYP61-CFP construct, demonstrating that the altered SYP61 activity negatively affects PIP2;7 secretion and might lead to ER accumulation and, thereby, to OSER formation. The fate of the proteins accumulated in such structures is still unclear: They could simply be stored there without further degradation to mitigate their cell toxicity; alternatively, OSER compartments might facilitate bulk disposal of overproduced proteins by an autophagy pathway. However, some data obtained in mammalian cells support that OSER structures are not subjected to autophagic degradation (Korkhov, 2009), even if disruption of genes essential to autophagy leads to accumulation of OSER membranes (Komatsu et al., 2005). Interestingly, in plant cells, SYP61 forms a complex with VTI12 (Zouhar et al., 2009) for which there is evidence for a role in autophagy (Surpin et al., 2003), raising the hypothesis that OSER structures observed in Venus-PIP2;7-expressing osm1 plants might be linked to a SYP61-related autophagy defect. As null mutants for SYP61 are not available, conditional or cell-type-specific silencing of SYP61 via inducible artificial microRNA expression might help clarify the physiological role of SYP61. In addition, as SYP61 is part of a TGN-based complex, including SYP41, VTI12, and YKT61 proteins (Zouhar et al., 2009), it would be worth checking how mutations in other members of this complex affect PIP anterograde trafficking, constitutive cycling, and/or degradation.
SYP121 Regulates PIP2;7 Trafficking and the Osmotic Water Permeability of Membranes
Coexpression of CFP-PIP2;7 with the soluble SYP121-Sp2 dominant-negative fragment significantly reduced its plasma membrane abundance and severely affected the Pf of mesophyll protoplasts. This Pf inhibition originated from PIP2;7 activity depletion in the plasma membrane due to its intracellular accumulation at the ER level, a mechanism similar to that observed upon transient coexpression of PIP2;5 and SYP121-Sp2 in maize protoplasts (Besserer et al., 2012). These data confirmed the key role of SYP121 in the anterograde post-Golgi trafficking of PIP proteins. It is striking that the SNARE SYP121 regulates transport and activity of both plasma membrane K+ channels and aquaporins, transporters involved in the regulation of cellular water homeostasis (Sutter et al., 2006; Honsbein et al., 2009; Grefen et al., 2010; Besserer et al., 2012). At this stage, we cannot exclude that SYP121 simply bridges both proteins, resulting in the formation of an enhanced PIP/K+-channel complex. The occurrence of such a complex that allows SYP121-mediated coregulation of these channels/transporters might play a key role in the control of physiological processes in which coordination of ion and water fluxes is crucial, as is the case for cell expansion or turgor regulation (Besserer et al., 2012). Therefore, the interplay between PIPs, K+ channels, and SYP121 needs to be further investigated and could pave the way to exciting discoveries in the field of plant osmoregulation.
A SYP121/SYP61 SNARE Complex Might Regulate the PIP2;7 Subcellular Routing
Proteomics analysis identified SYP121 in the SYP61-defined compartment (Drakakaki et al., 2012), and these two syntaxins had been colocalized in Arabidopsis protoplasts, implying that SYP61 is involved in anterograde transport of the SYP121 SNARE complex to the plasma membrane. SYP121 forms the SYP121-SNAP33-VESICLE-ASSOCIATED MEMBRANE PROTEIN721 (VAMP721)/VAMP722 tertiary SNARE complex at the plasma membrane through interaction with the SNAP33 adaptor and the two functionally redundant VAMP72 R-SNAREs (Collins et al., 2003; Karnik et al., 2013). Similarly, SYP61 is known to establish such a tertiary complex with SYP41 and VTI12 and to act at the TGN/EE (Zouhar et al., 2009; Drakakaki et al., 2012). We confirmed previously published results showing the colocalization of the two syntaxins in the TGN/EE (Drakakaki et al., 2012) and demonstrated by BiFC and pull-down assays that SYP61 interacted directly with SYP121 in vivo. A functional SNARE complex mediates the fusion of vesicles and consists of a tetrameric assembly of Qa-, Qb-, Qc-, and R-SNARE domains (Fukuda et al., 2000). After vesicle tethering to its target compartment, the vesicle-localized R-SNARE interacts with a preformed Q-SNARE complex, made of Qa-, Qb-, and Qc-SNAREs (endosomal type) or Qa- and Qb,c-SNAREs (plasma membrane type) to form a trans-SNARE complex (El Kasmi et al., 2013). Different associations of SNAREs within these complexes confer target selectivity to membrane fusion events.
This background raises the possibility of two nonexclusive interpretations. First, as SYP121 and SYP61 belong to the Qa- and Qc-SNARE family, respectively, our data support the existence of an endosomal type SNARE complex, in which SYP121 and SYP61 would physically interact to mediate a vesicular fusion between the TGN/EE and the plasma membrane (Figure 9). Given the nature of the SYP61 proteome and these data, we postulate that the SYP61/SYP121 SNARE complex could interact directly with the PIP2;7 protein and mediate its transport between the TGN/EE and plasma membrane. It is still unclear whether this SYP-mediated PIP transport regulation affects the anterograde route to the plasma membrane or their retrograde movement. Such a SNARE association probably also regulates transport of other plasma membrane-resident proteins as supported by the proteomics data of the SYP61 compartment (Drakakaki et al., 2012). The second interpretation could be that SYP121 might also function independently of SYP61 in a different SNARE complex, so that only a fraction of the SYP121 pool would interact with SYP61. This is supported by several facts. First, the steady state pool of SYP121 in the plasma membrane far exceeded that of SYP61, as revealed by the colocalization of both syntaxins and immunoprecipitation assays: SYP61 appeared to only transiently reside in the plasma membrane without accumulating there (Figure 4). Second, at the TGN level, the SYP61 and SYP121 distributions overlapped only partially both in terms of population of tagged endosomes (SYP61 labeled more endosomes than SYP121) (Figure 4A; Supplemental Figure 4) as well as of spreading within a particular TGN structure. Indeed, whereas SYP61 and SYP121 colocalized somewhat at the TGN, both fluorescently tagged syntaxins colabeled only some TGN subdomains (Figure 4A; Supplemental Figure 4 and Supplemental Movie 1). This observation suggests a partition of the SYP121 protein pool in two distinct SNARE complexes: an endosomal type in which SYP121/SYP61 would form a Q-SNARE complex and the previously described SYP61-independent plasma membrane type (Collins et al., 2003), involved in anterograde transport. Such involvement of a Qa-SNARE isoform in distinct SNARE complexes has recently been demonstrated for KNOLLE (SYP111), in which two distinct types of KNOLLE complexes jointly mediated the membrane fusion in cytokinesis of Arabidopsis (El Kasmi et al., 2013). Regardless of the alternative yet not mutually exclusive interpretations, the two SNAREs contribute in a synergistic fashion to influence the traffic of PIPs. The nature of the Qb-SNARE(s) in these complexes as well as the interacting R-SNARE(s) need to be identified. Such models open the way to new research directions bridging membrane protein transport and cellular homeostasis regulation.
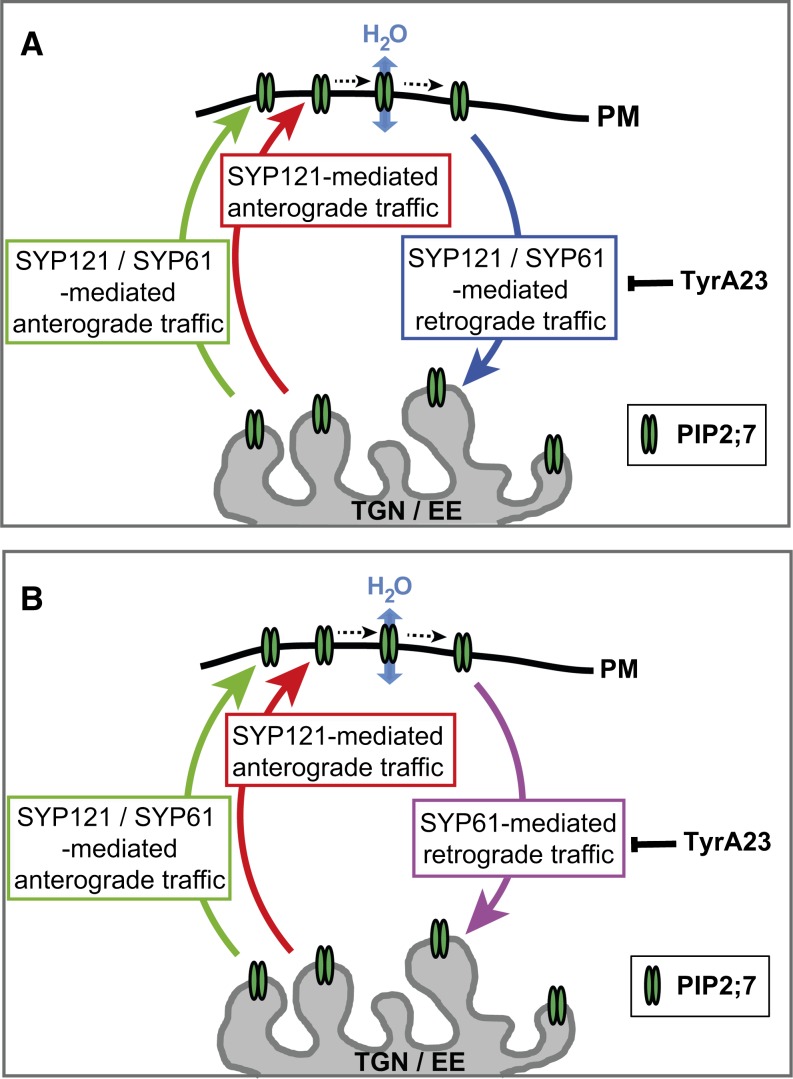
Figure 9.
SYP-Mediated Post-Golgi Trafficking of PIP2;7.
After exiting the trans-Golgi cisternae, PIP2;7 localizes in the TGN compartment that hosts most of the steady state SYP61 protein pool as well as a fraction of that of SYP121. However, this latter isoform is more abundant in the plasma membrane. Our model postulates that PIP2;7-containing vesicles budding from the TGN can fuse with the plasma membrane via a SYP121-mediated membrane fusion event (red arrow; Besserer et al., 2012) or a similar SYP61/SYP121-mediated mechanism (green arrow), as SYP61 quickly cycles between the TGN/EE and the plasma membrane where it might be associated with SYP121 as a transient trans-SNARE complex. Plasma membrane-derived PIP2;7-containing endosomes can merge back to the TGN/EE via SYP61/SYP121-mediated membrane fusion mechanism ([A], blue arrow) or via SYP61-mediated fusion ([B], purple arrow). TyrA23, an inhibitor of CCV-mediated endocytosis, was shown to trigger plasma membrane accumulation of SYP61 and SYP121.
METHODS
Genetic Constructs
The full-length cDNAs encoding PIP2;7, SYP121, and SYP61 were amplified by PCR from total cDNA (extracted from 7-d-old seedlings) and directionally subcloned with a uracil excision-based improved high-throughput USER cloning technique (Nour-Eldin et al., 2006) into the USER-compatible plant expression vectors pCAMBIA2300 35Su Nterm mYFP and pCAMBIA2300 35Su Nterm mCFP (Bienert et al., 2011). Mutated versions of SYP61 and SYP121 were also created by truncating the full-length cDNA after the nucleotides 660 and 864, respectively, to create Sp2 fragments that were cloned according to the same procedure and plasmids mentioned above.
The 2-in-1 BiFC vectors (Grefen and Blatt, 2012) were used to carry out BiFC and produce SYP121, SYP61, PIP2;7, and SNAP33 fused at their N-terminal end to the split YFP. For the YFPn-SYP61/YFPc-PIP2;7 BiFC constructs, SYP61 and PIP2;7 cDNAs were amplified from total cDNA and subcloned in pDONR221P3P2 and pDONR221P1P4 entry vectors, respectively, with the BP clonase II enzyme kit (Invitrogen). Transfer of the Gateway cassette from these two entry vectors to the pBIFCt-2in1-NN destination vector (Grefen and Blatt, 2012) was performed through LR clonase II-mediated recombination according to the manufacturer’s recommendations. The 2-in-1 BiFC constructs for the YFPn-SYP121/YFPc-PIP2;7 and YFPn-SNAP33/YFPc-PIP2;7 pairs were built the same way. For the YFPn-SYP121/YFPc-SYP61 BiFC assay, SYP121 and SYP61 cDNAs were subcloned in pDONR221P3P2 and pDONR221P1P4 entry vectors prior to their integration in the pBIFCt-2in1-NN destination vector. Entry vectors and BiFC constructs for the mutated version of PIP2;7 (PIP2;7ΔNΔCter), where amino acid residues 1 to 39 (cytosolic N terminus) and 263 to 280 (cytosolic C terminus were deleted) were prepared and cloned the same way as PIP2;7.
For the SUS assay, PIP2;7, SYP61, and SYP121 cDNAs were subcloned in pDONR221P1P2 prior to their integration in the SUS destination vectors. The Split-ubiquitin vectors pMetYC-DEST and pNX35-DEST (Grefen et al., 2009; Grefen and Blatt, 2012) were used to produce the Met-repressible bait construct PIP2;7-Cub-PLV and the prey constructs NubG-SYP61, NubG-SYP121, or NubG-PIP2;7, respectively. The NubWT fragment was obtained from the pNubWT-Xgate vector (Grefen et al., 2009).
The β-estradiol-inducible mCFP-SYP121-Sp2 and mYFP-SYP61-Sp2 constructs were obtained by PCR amplification of the corresponding template from the previously built 35S constructs with D-TOPO cloning-compatible primers. PCR fragments were recombined into a pENTR vector with the pENTR Directional TOPO cloning kit (Invitrogen) and further recombined into the pMDC7 destination vector (Curtis and Grossniklaus, 2003) by LR cloning (Invitrogen). The PIP2;7 translational reporter construct was prepared by PCR amplification of the 2.0-kb PIP2;7 promoter region with BP cloning-compatible primers from a genomic DNA extract and by inserting the PCR product into pDONR P4-P1R (Invitrogen) via BP cloning. Similarly, PIP2;7 cDNA was amplified by PCR from a plasmid cDNA template with BP cloning-compatible primers and introduced into pDONR P2RP3 (Invitrogen). The PIP2;7 promoter in pDONR P4-P1R, PIP2;7 cDNA in pDONR P2RP3, and Venus cDNA in pDONR 221 (Mylle et al., 2013) were recombined with MultiSite Gateway technology (Invitrogen) with the destination vector pK7m34GW (Karimi et al., 2002). Plasmid extractions for DNA manipulation and sequencing were prepared with the mini-prep purification kit Nucleobond (Clontech) according to the manufacturer’s protocol. All constructs in the destination vectors were sequenced to verify that correct fragments were cloned in frame and subsequently introduced into Agrobacterium tumefaciens strain GV3101 (for stable Arabidopsis thaliana transformation) or AGL1 (for transient Nicotiana tabacum transformation). Tobacco plants were transiently transformed by classical leaf infiltration, whereas Arabidopsis plants were stably transformed by floral dipping (Clough and Bent, 1998).
Primer sequences used to build the different constructs are listed in Supplemental Table 1.
Plant Material
Seeds were surface-sterilized and sown on half-strength Murashige and Skoog agar plates. After 2 d at 4°C, the plates were transferred to a growth chamber with a 16-h-light/8-h-dark regime for 7 d. Markers, mutants, and previously published transgenic lines are osm1 (Zhu et al., 2002), pSYP61:SYP61-CFP (Robert et al., 2008), pBRI1:BRI1:GFP (Friedrichsen et al., 2000), pPIN2:PIN2:GFP (Xu and Scheres, 2005).
Confocal Microscopy
Plant materials were imaged according to standard procedures on a Zeiss LSM710 confocal microscope equipped with a spectral detector. In each experiment in which the fluorescence intensity between the control and mutated lines needed to be compared, calibration of the laser beam intensity, gain, and offset parameters were achieved on cells expressing the fluorescent reporter in appropriate control (Columbia-0 or C24) backgrounds. The same parameters were used in images acquired on mutated backgrounds or backgrounds in which constructs were coexpressed, allowing a subsequent calculation and comparison of the plasma membrane fluorescence intensity by means of the Zen 2009 software (Carl Zeiss MicroImaging). The fluorescence intensity in the plasma membrane of protoplasts and intracellular compartments was quantified with an in-house developed macro for the ImageJ software as described (Besserer et al., 2012). The BiFC assay was done in tobacco epidermal cells transiently transformed by Agrobacterium infiltration (Batoko et al., 2000). Samples were analyzed 3 d after infiltration to allow sufficient time for protein production. In cells exhibiting a fluorescence signal after excitation at 514 nm, an emission spectrum was determined to validate the signal specificity. Imaris software (BitPlane) was used to provide a quantitative measurement of the colocalization of SYP61-CFP with Venus-PIP2;7 or Venus-SYP121. A Pearson’s correlation coefficient was calculated by the software for each channel from images acquired by sequential scanning. Background correction values were automatically adjusted for all images. The Pearson coefficients were calculated from 15 independent images. The degree of colocalization from the Pearson’s coefficient values was categorized based on a previously published description (Zinchuk et al., 2013). Quantification of SYP61-CFP mean fluorescence intensity in TGN/EE structures was also performed using this software. The effects of induction and presence of mCFP-SYP121-Sp2 on the subcellular localization and abundance of Venus-PIP2;7 proteins in protoplasts were investigated with microscope settings similar between Sp2-inducing and control conditions. Sixty-seven and 54 cells were analyzed for SP2-inducing or noninducing conditions, respectively.
Electron Microscopy
Root tips of 5-d-old seedlings expressing the p35S:Venus:PIP2;7 construct in C24 or osm1 backgrounds were excised, immersed in 20% (w/v) BSA, and frozen immediately in a high-pressure freezer EM PACT (Leica Microsystems). Immunolocalization freeze-substitution steps were performed as described (Tanaka et al., 2009).
Immunoprecipitation Assay
To prepare a protein extract suitable for immunoprecipitation, 500 mg of seedling tissue was used. Whole seedlings were ground in liquid nitrogen and thawed in extraction buffer (250 mM sucrose, 20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 10 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1% Nonidet P-40, and 0.1% sodium deoxycholate) supplemented by one tablet of phosphoSTOP phosphatase inhibitor cocktail and one tablet of complete protease inhibitor cocktail (Roche). Ground samples were incubated on ice for 15 min and centrifuged at 10,000g for 15 min at 4°C. The supernatant was collected and the centrifugation step was repeated once. The concentration of extracted proteins was determined by the Bradford assay and adjusted to 5 mg/mL. Protein extracts (3.5 mg) were mixed to 15 μL anti-GFP beads (Chromotek) in Micro Bio-Spin columns (Bio-Rad) and incubated with gentle mixing for 1 h at 4°C. The beads were pelleted by centrifugation at 2,800g for 10 s and the flow-through was discarded. The beads were washed five times with 700 μL extraction buffer. At the end of the process, bound proteins were eluted from the beads by adding 100 μL Laemmli buffer and boiling for 5 min at 95°C. Proteins (45 µL/well) were electrophoresed by SDS-PAGE, transferred to a nitrocellulose membrane, and immunodetected with antibodies directed against GFP (Duby et al., 2001), plasma membrane H+-ATPase (Morsomme et al., 1996), SYP61 (Sanderfoot et al., 2001a), SYP121 (Tyrrell et al., 2007), or PIP2;7 (Agrisera). Stripping of the polyvinylidene fluoride membranes was performed with 0.5 n NaOH treatment for 5 min, followed by three washes with milliQ water and 30 min blocking of the membrane in blocking buffer (PBS, 3% milk powder, and 0.5% Tween 20) prior to incubation with a different antibody.
SUS Assay
Electroporation-competent THY.AP4 yeast strain was cotransformed with the Nub and Cub constructs of interest. Yeast colonies coexpressing the bait and prey constructs were recovered 48 h after transfer to selective media (CSM, -Leu-, Trp-) (Grefen et al., 2009). Growth assays were performed as follows. Yeast coexpressing the Met-repressible bait construct PIP2;7-Cub-PLV and the prey constructs NubG-PIP2;7, NubG-SYP61, NubG-SYP121, NubG (negative control), or NubWT (positive control) were dropped in a dilution series (O.D. 0.5, 0.05, and 0.005) onto synthetic (CSM, -Leu-, Trp-, Ade-, His-, Met-) medium containing 100 µM methionine to repress expression of the bait. Yeast growth was recorded after incubation for 48 h at 30°C.
Isolation of Arabidopsis Protoplasts and Protoplast Swelling Assay
Protoplasts from 7-d-old Arabidopsis aerial parts were isolated from ∼50 seedlings per line as described (Ramahaleo et al., 1999). Protoplasts originated from the nontransformed C24, Columbia, and osm1 backgrounds, the C24 and osm1 lines expressing the p35S:Venus-PIP2;7 construct, or a Columbia line coexpressing the p35S:Venus-PIP2;7 and the β-estradiol-inducible CFP-SYP121-SP2 construct. The solutions used for Arabidopsis protoplast swelling assays and experiments were as described (Postaire et al., 2010; Moshelion et al., 2004; Volkov et al., 2007). Maximum Pf values were calculated, plotted on a frequency diagram, and used in statistical analysis as described (Besserer et al., 2012). Three biological replicates were performed for each set of experiments, using the same homozygous lines (one line per genetic background).
Statistical Analysis of the Data
Statistical analysis was conducted using GraphPad Prism software, version 3.00, to determine the significance of the presented data.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At-PIP2;7, At4g35100; At-SYP121, At3g11820 ; At-SYP61, At1g28490; At-AKT1, At2g26650; At-KC1, At4g32650; At-VTI12, At1g26670; At-SYP41, At5g26980; At-SNAP33, At5g61210; At-PIN2, At5g57090; At-PIP1;4, At4g00430; At-PIP2;1, At3g53420; At-BRI1, At4g39400; At-IRT1, At4g19690; At-VAMP721, At1g04750; At-VAMP722, At2g33120; At-SYP111, At1g08560; At-YKT61, At5g58060; Zm-SYP121, NP_001150776; and Zm-PIP2;5, AF130975.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification of Zm-SYP61 as Putative Interactor of Zm-PIP2;6 by Affinity Chromatography Coupled to MALDI/TOF-TOF Analysis.
Supplemental Figure 2. Colocalization of SYP121 and PIP27.
Supplemental Figure 3. Mating-Based Split-Ubiquitin Assays Demonstrating PIP2;7 and SYP121 Interaction.
Supplemental Figure 4. Colocalization of SYP61 and SYP121 Expressed from pSYP61:SYP61-CFP and p35S:Venus-SYP121 Constructs in Elongated Primary Root Cells.
Supplemental Figure 5. Subcellular Localization of Transiently Expressed GFP-PIP2;7ΔNΔCter Proteins in Tobacco Epidermal Cells.
Supplemental Figure 6. Mistargeting of Overexpressed Venus-AtPIP2;7 in osm1 Compared with Wild-Type Backgrounds and Phenotype Complementation.
Supplemental Figure 7. Immunolocalization of Venus-PIP2;7 and PIN2 Proteins in the osm1 Background.
Supplemental Figure 8. Effect of the SYP121-Sp2 Fragment on the Subcellular Localization of PIP Fusion Proteins.
Supplemental Figure 9. Effect of SYP121-Sp2 Fragments on PIN2 and BRI1 Subcellular Localization.
Supplemental Methods. Identification of Proteins Interacting with Zm-PIP2;6.
Supplemental Movie 1. Movement of SYP61-CFP/Venus-SYP121 Colabeled Endosomes.
Supplemental Table 1. PCR Primers Used to Build the Different Genetic Constructs.
Supplementary Material
Acknowledgments
We thank Georgia Drakakaki and Henri Batoko for helpful discussions, Tomohiro Uemura for the Venus-SYP61-overexpressing line, Jianhua Zhu for the osm1 line, Jiří Friml for anti-GFP and anti-PIN2 antibodies, Natasha Raikhel for anti-SYP61 antibodies, the Nottingham Arabidopsis Stock Center for seeds, and the ABRC for DNA stocks and plasmids used in this study. Confocal microscopy was carried out at the Université catholique de Louvain imaging platform IMABIOL with advice from Abdelmounaim Errachid. Electron microscopy was performed at the DMBR-PSB TEM core facility in VIB. We thank Martine De Cock for English editing of the article. This work was supported by the Belgian National Fund for Scientific Research (FNRS), the Interuniversity Attraction Poles Programme-Belgian Science Policy (Grant IAP7/29), the “Communauté française de Belgique-Actions de Recherches Concertées,” the Francqui Foundation, and the Bauchau Award. C.H. was a FNRS postdoctoral researcher and was supported for 6 months by a Francqui-Inter-Community Postdoctoral fellowship.
AUTHOR CONTRIBUTIONS
C.H. designed and performed the research, analyzed data, and wrote the article. T.L., H.D., D.C., and H.R. performed research. R.D.R. provided technical assistance. C.G. designed experiments. M.R.B. analyzed data. D.I. supervised part of the study. E.R. participated in the design and supervision of the research and analyzed data. F.C. designed and supervised the whole project, analyzed data, and wrote the article. All authors contributed to the final article.
Glossary
- ER
endoplasmic reticulum
- TGN
trans-Golgi network
- EE
early endosome
- TyrA23
tyrphostin A23
- BiFC
bimolecular fluorescence complementation (
- SUS
split ubiquitin
- A.U.
absorbance units
Footnotes
The online version of this article contains Web-only data.
References
- Alexandersson E., Fraysse L., Sjövall-Larsen S., Gustavsson S., Fellert M., Karlsson M., Johanson U., Kjellbom P. (2005). Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 59: 469–484. [DOI] [PubMed] [Google Scholar]
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H., Zheng H.-Q., Hawes C., Moore I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A., Burnotte E., Bienert G.P., Chevalier A.S., Errachid A., Grefen C., Blatt M.R., Chaumont F. (2012). Selective regulation of maize plasma membrane aquaporin trafficking and activity by the SNARE SYP121. Plant Cell 24: 3463–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert G.P., Bienert M.D., Jahn T.P., Boutry M., Chaumont F. (2011). Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J. 66: 306–317. [DOI] [PubMed] [Google Scholar]
- Bienert G.P., Cavez D., Besserer A., Berny M.C., Gilis D., Rooman M., Chaumont F. (2012). A conserved cysteine residue is involved in disulfide bond formation between plant plasma membrane aquaporin monomers. Biochem. J. 445: 101–111. [DOI] [PubMed] [Google Scholar]
- Chaumont F., Moshelion M., Daniels M.J. (2005). Regulation of plant aquaporin activity. Biol. Cell 97: 749–764. [DOI] [PubMed] [Google Scholar]
- Chaumont F., Tyerman S.D. (2014). Aquaporins: highly regulated channels controlling plant water relations. Plant Physiol. 164: 1600–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Collins N.C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J.-L., Hückelhoven R., Stein M., Freialdenhoven A., Somerville S.C., Schulze-Lefert P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G., van de Ven W., Pan S., Miao Y., Wang J., Keinath N.F., Weatherly B., Jiang L., Schumacher K., Hicks G., Raikhel N. (2012). Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 22: 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duby G., Oufattole M., Boutry M. (2001). Hydrophobic residues within the predicted N-terminal amphiphilic α-helix of a plant mitochondrial targeting presequence play a major role in in vivo import. Plant J. 27: 539–549. [DOI] [PubMed] [Google Scholar]
- El Kasmi F., Krause C., Hiller U., Stierhof Y.-D., Mayer U., Conner L., Kong L., Reichardt I., Sanderfoot A.A., Jürgens G. (2013). SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol. Biol. Cell 24: 1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D., Sutton R.B., Brunger A.T., Jahn R. (1998). Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA 95: 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D.M., Joazeiro C.A.P., Li J., Hunter T., Chory J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M., Arimura S., Ueda T., Takanashi H., Hayashi Y., Nakano A., Tsutsumi N. (2010). Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc. Natl. Acad. Sci. USA 107: 6094–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R., McNew J.A., Weber T., Parlati F., Engel T., Nickel W., Rothman J.E., Söllner T.H. (2000). Functional architecture of an intracellular membrane t-SNARE. Nature 407: 198–202. [DOI] [PubMed] [Google Scholar]
- Geelen D., Leyman B., Batoko H., Di Sansebastiano G.P., Moore I., Blatt M.R. (2002). The abscisic acid-related SNARE homolog NtSyr1 contributes to secretion and growth: evidence from competition with its cytosolic domain. Plant Cell 14: 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Blatt M.R. (2008). SNAREs—molecular governors in signalling and development. Curr. Opin. Plant Biol. 11: 600–609. [DOI] [PubMed] [Google Scholar]
- Grefen C., Blatt M.R. (2012). A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC). Biotechniques 53: 311–314. [DOI] [PubMed] [Google Scholar]
- Grefen C., Chen Z., Honsbein A., Donald N., Hills A., Blatt M.R. (2010). A novel motif essential for SNARE interaction with the K+ channel KC1 and channel gating in Arabidopsis. Plant Cell 22: 3076–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Obrdlik P., Harter K. (2009). The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS). Methods Mol. Biol. 479: 217–233. [DOI] [PubMed] [Google Scholar]
- Hachez C., Besserer A., Chevalier A.S., Chaumont F. (2013). Insights into plant plasma membrane aquaporin trafficking. Trends Plant Sci. 18: 344–352. [DOI] [PubMed] [Google Scholar]
- Honsbein A., Blatt M.R., Grefen C. (2011). A molecular framework for coupling cellular volume and osmotic solute transport control. J. Exp. Bot. 62: 2363–2370. [DOI] [PubMed] [Google Scholar]
- Honsbein A., Sokolovski S., Grefen C., Campanoni P., Pratelli R., Paneque M., Chen Z., Johansson I., Blatt M.R. (2009). A tripartite SNARE-K+ channel complex mediates in channel-dependent K+ nutrition in Arabidopsis. Plant Cell 21: 2859–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Karnik R., Grefen C., Bayne R., Honsbein A., Köhler T., Kioumourtzoglou D., Williams M., Bryant N.J., Blatt M.R. (2013). Arabidopsis Sec1/Munc18 protein SEC11 is a competitive and dynamic modulator of SNARE binding and SYP121-dependent vesicle traffic. Plant Cell 25: 1368–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., et al. (2005). Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkhov V.M. (2009). GFP-LC3 labels organised smooth endoplasmic reticulum membranes independently of autophagy. J. Cell. Biochem. 107: 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Cho S.K., Son O., Xu Z., Hwang I., Kim W.T. (2009). Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang X., Yang Y., Li R., He Q., Fang X., Luu D.-T., Maurel C., Lin J. (2011). Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23: 3780–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu D.-T., Martinière A., Sorieul M., Runions J., Maurel C. (2012). Fluorescence recovery after photobleaching reveals high cycling dynamics of plasma membrane aquaporins in Arabidopsis roots under salt stress. Plant J. 69: 894–905. [DOI] [PubMed] [Google Scholar]
- Maurel C., Verdoucq L., Luu D.-T., Santoni V. (2008). Plant aquaporins: membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 59: 595–624. [DOI] [PubMed] [Google Scholar]
- Morsomme P., de Kerchove d’Exaerde A., De Meester S., Thinès D., Goffeau A., Boutry M. (1996). Single point mutations in various domains of a plant plasma membrane H(+)-ATPase expressed in Saccharomyces cerevisiae increase H(+)-pumping and permit yeast growth at low pH. EMBO J. 15: 5513–5526. [PMC free article] [PubMed] [Google Scholar]
- Moshelion M., Moran N., Chaumont F. (2004). Dynamic changes in the osmotic water permeability of protoplast plasma membrane. Plant Physiol. 135: 2301–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylle E., Codreanu M.-C., Boruc J., Russinova E. (2013). Emission spectra profiling of fluorescent proteins in living plant cells. Plant Methods 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin H.H., Hansen B.G., Nørholm M.H.H., Jensen J.K., Halkier B.A. (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumet F., Rahimian V., Rothman J.E. (2004). The specificity of SNARE-dependent fusion is encoded in the SNARE motif. Proc. Natl. Acad. Sci. USA 101: 3376–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postaire O., Tournaire-Roux C., Grondin A., Boursiac Y., Morillon R., Schäffner A.R., Maurel C. (2010). A PIP1 aquaporin contributes to hydrostatic pressure-induced water transport in both the root and rosette of Arabidopsis. Plant Physiol. 152: 1418–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado K., Boursiac Y., Tournaire-Roux C., Monneuse J.-M., Postaire O., Da Ines O., Schäffner A.R., Hem S., Santoni V., Maurel C. (2013). Regulation of Arabidopsis leaf hydraulics involves light-dependent phosphorylation of aquaporins in veins. Plant Cell 25: 1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R., Sutter J.-U., Blatt M.R. (2004). A new catch in the SNARE. Trends Plant Sci. 9: 187–195. [DOI] [PubMed] [Google Scholar]
- Ramahaleo T., Morillon R., Alexandre J., Lassalles J.-P. (1999). Osmotic water permeability of isolated protoplasts. Modifications during development. Plant Physiol. 119: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S., Chary S.N., Drakakaki G., Li S., Yang Z., Raikhel N.V., Hicks G.R. (2008). Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl. Acad. Sci. USA 105: 8464–8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A.A., Raikhel N.V. (1999). The specificity of vesicle trafficking: coat proteins and SNAREs. Plant Cell 11: 629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A.A., Assaad F.F., Raikhel N.V. (2000). The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 124: 1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A.A., Kovaleva V., Bassham D.C., Raikhel N.V. (2001a). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell 12: 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A.A., Pilgrim M., Adam L., Raikhel N.V. (2001b). Disruption of individual members of Arabidopsis syntaxin gene families indicates each has essential functions. Plant Cell 13: 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E.L., Hegde R.S., Francolini M., Lombardo F., Colombo S., Pedrazzini E., Borgese N., Lippincott-Schwartz J. (2003). Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163: 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorieul M., Santoni V., Maurel C., Luu D.-T. (2011). Mechanisms and effects of retention of over-expressed aquaporin AtPIP2;1 in the endoplasmic reticulum. Traffic 12: 473–482. [DOI] [PubMed] [Google Scholar]
- Surpin M., Zheng H., Morita M.T., Saito C., Avila E., Blakeslee J.J., Bandyopadhyay A., Kovaleva V., Carter D., Murphy A., Tasaka M., Raikhel N. (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15: 2885–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter J.-U., Campanoni P., Tyrrell M., Blatt M.R. (2006). Selective mobility and sensitivity to SNAREs is exhibited by the Arabidopsis KAT1 K+ channel at the plasma membrane. Plant Cell 18: 935–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19: 391–397. [DOI] [PubMed] [Google Scholar]
- Tyrrell M., Campanoni P., Sutter J.-U., Pratelli R., Paneque M., Sokolovski S., Blatt M.R. (2007). Selective targeting of plasma membrane and tonoplast traffic by inhibitory (dominant-negative) SNARE fragments. Plant J. 51: 1099–1115. [DOI] [PubMed] [Google Scholar]
- Uemura T., Kim H., Saito C., Ebine K., Ueda T., Schulze-Lefert P., Nakano A. (2012). Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc. Natl. Acad. Sci. USA 109: 1784–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R.L., Nakano A., Takeyasu K., Sato M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29: 49–65. [DOI] [PubMed] [Google Scholar]
- Van Damme D., Gadeyne A., Vanstraelen M., Inzé D., Van Montagu M.C., De Jaeger G., Russinova E., Geelen D. (2011). Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc. Natl. Acad. Sci. USA 108: 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov V., Hachez C., Moshelion M., Draye X., Chaumont F., Fricke W. (2007). Water permeability differs between growing and non-growing barley leaf tissues. J. Exp. Bot. 58: 377–390. [DOI] [PubMed] [Google Scholar]
- Weig A., Deswarte C., Chrispeels M.J. (1997). The major intrinsic protein family of Arabidopsis has 23 members that form three distinct groups with functional aquaporins in each group. Plant Physiol. 114: 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005). Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny E., Borst J.W., Muylaert M., Batoko H., Hemminga M.A., Chaumont F. (2007). FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc. Natl. Acad. Sci. USA 104: 12359–12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny E., Miecielica U., Borst J.W., Hemminga M.A., Chaumont F. (2009). An N-terminal diacidic motif is required for the trafficking of maize aquaporins ZmPIP2;4 and ZmPIP2;5 to the plasma membrane. Plant J. 57: 346–355. [DOI] [PubMed] [Google Scholar]
- Zhu J., Gong Z., Zhang C., Song C.P., Damsz B., Inan G., Koiwa H., Zhu J.K., Hasegawa P.M., Bressan R.A. (2002). OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell 14: 3009–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinchuk V., Wu Y., Grossenbacher-Zinchuk O. (2013). Bridging the gap between qualitative and quantitative colocalization results in fluorescence microscopy studies. Sci. Rep. 3: 1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhar J., Rojo E., Bassham D.C. (2009). AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol. 149: 1668–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.