LEA proteins accumulate in plant seeds prior to maturation drying, and some have been shown to protect membranes from desiccation. This work demonstrates the subcellular distribution of each of 51 Arabidopsis LEA proteins and suggests protection against desiccation or cold stress is tailored for each cellular compartment.
Abstract
Late embryogenesis abundant (LEA) proteins are hydrophilic, mostly intrinsically disordered proteins, which play major roles in desiccation tolerance. In Arabidopsis thaliana, 51 genes encoding LEA proteins clustered into nine families have been inventoried. To increase our understanding of the yet enigmatic functions of these gene families, we report the subcellular location of each protein. Experimental data highlight the limits of in silico predictions for analysis of subcellular localization. Thirty-six LEA proteins localized to the cytosol, with most being able to diffuse into the nucleus. Three proteins were exclusively localized in plastids or mitochondria, while two others were found dually targeted to these organelles. Targeting cleavage sites could be determined for five of these proteins. Three proteins were found to be endoplasmic reticulum (ER) residents, two were vacuolar, and two were secreted. A single protein was identified in pexophagosomes. While most LEA protein families have a unique subcellular localization, members of the LEA_4 family are widely distributed (cytosol, mitochondria, plastid, ER, and pexophagosome) but share the presence of the class A α-helix motif. They are thus expected to establish interactions with various cellular membranes under stress conditions. The broad subcellular distribution of LEA proteins highlights the requirement for each cellular compartment to be provided with protective mechanisms to cope with desiccation or cold stress.
INTRODUCTION
During late development, before they enter the desiccation phase, plant seeds accumulate a class of polypeptides known as LEA (late embryogenesis abundant) proteins, which share common properties such as low sequence complexity, repeat motifs, high hydrophilicity, and often a lack of ordered structure in the native state (Tunnacliffe and Wise, 2007; Shih et al., 2008; Tunnacliffe et al., 2010). Although there is a great diversity in LEA proteins, most can be grouped into eight families in the PFAM database (Finn et al., 2010) according to their primary sequences: dehydrin, LEA_1, LEA_2, LEA_3, LEA_4, LEA_5, LEA_6, and seed maturation protein (SMP). The pertinence of such classification was reinforced and extended by computational analysis of the physico-chemical properties of more than 700 LEA protein sequences deposited in LEAPdb, a publicly available database (Hunault and Jaspard, 2010), which allowed a more detailed organization of the proteins into 12 nonredundant classes (Jaspard et al., 2012). Interestingly, LEA proteins have also been discovered in various species or phyla of invertebrates (bdelloid rotifers, nematodes, tardigrades, and arthropods), which are all anhydrobiotes (Tunnacliffe and Wise, 2007). In these organisms, as in the case of plant seeds, LEA proteins accumulate prior to desiccation, which is a very strong argument for a role of these proteins in desiccation tolerance.
LEA genes are highly represented in plant genomes with, for instance, 51 LEA genes in Arabidopsis thaliana (Bies-Ethève et al., 2008; Hundertmark and Hincha, 2008). According to these studies, 47 of the corresponding LEA proteins are distributed within the eight PFAM families. Two proteins were considered to be members of the LEA_4 family by Hundertmark and Hincha (2008), who also included two other proteins without PFAM classification in a group which we will also refer to as AtM.
Quantitative RT-PCR analysis showed that most Arabidopsis LEA genes were highly expressed in seeds (Hundertmark and Hincha, 2008), in agreement with the accumulation of LEA proteins (Gallardo et al., 2002). Interestingly, 22 LEA genes were found to be highly expressed at the transcript level in vegetative tissues, in particular under dehydration or exposure to low temperature (Hundertmark and Hincha, 2008). Indeed, several LEA proteins are known as cold responsive proteins, such as COLD RESPONSIVE15A (COR15A) in Arabidopsis (Steponkus et al., 1998).
No enzymatic activity has been reported so far for LEA proteins, which are more likely expected to play a role in stress tolerance as protectants of biomolecules and membranes. As intrinsically disordered proteins, they are proposed to have a broad impact in the abiotic stress response in plants (Sun et al., 2013). Individual LEA proteins were shown in vitro to protect enzymes from desiccation and/or freezing induced aggregation (Goyal et al., 2005b; Grelet et al., 2005; Reyes et al., 2005; Pouchkina-Stantcheva et al., 2007; Nakayama et al., 2008; Sharon et al., 2009; Boucher et al., 2010). Such protection of polypeptides might result from chaperone-type effects, implying direct interaction of LEA proteins with clients to prevent aggregation (Kovacs et al., 2008), or from space-filling by LEA proteins, which reduces the collision rates between aggregating proteins, the so-called molecular shield function (Chakrabortee et al., 2012). LEA proteins from the dehydrin family have been shown to bind membranes through their canonical K-segment, a Lys-rich peptide (Koag et al., 2003, 2009). The phosphorylation-dependent binding of the K-segment to phospholipid vesicles was later shown to lower membrane lipid phase transition, in agreement with a role of dehydrin in cold tolerance (Eriksson et al., 2011). A pea (Pisum sativum) seed mitochondrial LEA protein of the LEA_4 family was shown to protect membranes from desiccation (Tolleter et al., 2007, 2010). This intrinsically disordered protein, located in the matrix space, folds during dehydration into a class A amphipathic α-helix form that inserts laterally in the membrane to afford protection in the dry state. LEA proteins have also been shown to sequester calcium (Alsheikh et al., 2005), metal ions (Svensson et al., 2000; Kruger et al., 2002), and reactive oxygen species (Hara et al., 2004), thus potentially contributing to dehydration and cold stress tolerance. They have also been shown to contribute to the glassy state, which is formed by nonreducing sugars such as sucrose or trehalose upon desiccation in the cytoplasm of most of anhydrobiotes (Wolkers et al., 2001; Shih et al., 2004; Shimizu et al., 2010).
Ectopic expression of individual LEA proteins in various organisms has also provided support for a function in stress protection, although the effects are often moderate (for review, see Tunnacliffe and Wise, 2007). For instance, a brown mustard (Brassica juncea) dehydrin overexpressed in yeast (Saccharomyces cerevisiae) or tobacco (Nicotiana tabacum) increased salt and cold tolerance in both organisms (Xu et al., 2008). Another wheat (Triticum aestivum) LEA protein was shown to increase cold tolerance when overexpressed in Arabidopsis (NDong et al., 2002). Recently, a dramatic effect was reported for a brine shrimp (Artemia franciscana) mitochondrial LEA protein expressed in cultured human hepatoma cells (Li et al., 2012b). Indeed, membrane integrity of transgenic cells was preserved after a rapid and uniform cell desiccation, and with the combination of intracellular loading of trehalose before drying, cells maintained almost normal proliferation rates after rehydration.
The diversity of LEA proteins in eukaryotes raises the question of their subcellular localization, and although LEA proteins are predicted to populate most cellular compartments, relatively few have been experimentally localized (Tunnacliffe and Wise, 2007). LEA proteins have been found mostly in the cytosol (Mundy and Chua, 1988; Franz et al., 1989; Roberts et al., 1993; Goyal et al., 2005a) and in chloroplasts (Iturriaga et al., 1992; Lin and Thomashow, 1992; NDong et al., 2002). LEA proteins have also been identified in mitochondria from pea (Bardel et al., 2002; Grelet et al., 2005) and from the brine shrimp (Menze et al., 2009). LEA proteins have also been localized in Arabidopsis peroxisomes (Cutler et al., 2000) and in the endoplasmic reticulum (ER) (Ukaji et al., 2001). In the bdelloid rotifer Adineta ricciae, two LEA proteins were localized in the ER and shown to be distributed to vesicles and partially secreted (Tripathi et al., 2012). Several LEA proteins have been shown to accumulate in the nucleus (Borrell et al., 2002) or to be translocated from the cytosol to the nucleus upon phosphorylation (Jensen et al., 1998; Riera et al., 2004).
For the model plant Arabidopsis, with 51 LEA genes, information about the subcellular localization of the corresponding proteins can be mined in the SUBA (Arabidopsis Subcellular Database) (Heazlewood et al., 2007) and MASCP Gator (Joshi et al., 2011) databases. Experimental information derived from subcellular proteomics or fluorescent protein translational fusions is only available for 16 of the Arabidopsis LEA proteins, and among those, four proteins display several different localizations.
Considering LEA protein diversity, and their major, yet still enigmatic, functions with respect to desiccation tolerance, and possibly other stress responses, establishing the cellular location of each protein is critical to our understanding of their precise biological function. Here, we examined the subcellular localization of the 51 Arabidopsis LEA proteins using a combination of bioinformatic analysis and experimental localization of translational protein fusions. The data revealed their wide distribution and the potential role of the largest family in the protection of the various cellular membranes.
RESULTS
This work aimed to resolve the subcellular localization of the LEA proteins encoded by the 51 LEA genes identified by Hundertmark and Hincha (2008). For the sake of clarity, we will use their nomenclature, which numbers genes successively on the genome. Table 1 shows the correspondence with the AGI code of the protein, alternative names of the proteins, and their classification using PFAM and LEAPdb. For LEA genes with two gene models in The Arabidopsis Information Resource (http://www.arabidopsis.org/), we selected the protein from the gene model with the highest number of ESTs.
Table 1. Arabidopsis LEA Protein Nomenclature and Classification.
| LEA | Gene Model AGI Code | Other Names | Hundertmark and Hincha (2008) Classification | Bies-Ethève et al. (2008) Classification | PFAM Classification | LEAPDB Classification | Previous Subcellular Localizationsa |
|---|---|---|---|---|---|---|---|
| 1 | At1g01470.1 | LEA14 | LEA_2 | Group 7 | PF03168 | Class 7 | C (1) |
| 2 | At1g02820.1 | LEA3 | LEA_3 | Group 6 | PF03242 | Class 9 | |
| 3 | At1g03120.1 | SMP | Group 5 | PF04927 | Class 11 | ||
| 4 | At1g20440.1 | COR47/RD17 | Dehydrin | Group 2 | PF00257 | Class 2 | C (2), N (1), Pm (1), P (2) |
| 5 | At1g20450.1 | ERD10/LTI45/LTI29 | Dehydrin | Group 2 | PF00257 | Class 2 | C (2), Pm (2), P (1) |
| 6 | At1g32560.1 | LEA4-1 | LEA_1 | Group 4 | PF03760 | Class 10 | |
| 7 | At1g52690.1 | AtLEA3-3 | LEA_4 | Group 3 | PF02987 | Class 6 | |
| 8 | At1g54410.1 | Dehydrin | PF00257 | Class 3 | C (1), Pm (2), P (1) | ||
| 9 | At1g72100.1 | LEA_4 | Group 3 | PF02987 | Class 6 | ||
| 10 | At1g76180.1 | ERD14 | Dehydrin | Group 2 | PF00257 | Class 1 | Perox (1), C (1), Pm (3), P (4), V (1) |
| 11 | At2g03740.1 | LEA_4 | PF02987 | Class 6 | |||
| 12 | At2g03850.1 | LEA_4 | PF02987 | Class 6 | Pm (1) | ||
| 13 | At2g18340.1 | LEA_4 | Group 3 | PF02987 | Class 6 | Pm (1) | |
| 14 | At2g21490.1 | Dehydrin | Group 2 | PF00257 | Class 1 | ||
| 15 | At2g23110.1 | PvLEA18 | Group 8 | PF10714 | Class 12 | ||
| 16 | At2g23120.1 | PvLEA18 | Group 8 | PF10714 | Class 12 | Pm (1) | |
| 17 | At2g33690.1 | PvLEA18 | Group 8 | PF10714 | Class 12 | ||
| 18 | At2g35300.1 | LEA4-2 | LEA_1 | Group 4 | PF03760 | Class 10 | |
| 19 | At2g36640.1 | ECP63 | LEA_4 | Group 3 | PF02987 | Class 6 | |
| 20 | At2g40170.1 | EM6 | LEA_5 | Group 1 | PF00477 | Class 5 | |
| 21 | At2g41260.1 | M17 | AtM | Group 9 | |||
| 22 | At2g41280.1 | M10 | AtM | Group 9 | |||
| 23 | At2g42530.1 | COR15B | LEA_4 | Class 6 | P (4) | ||
| 24 | At2g42540.2 | COR15A | LEA_4 | Class 6 | P (6) | ||
| 25 | At2g42560.1 | LEA_4 | Group 3 | PF02987 | Class 6 | ||
| 26 | At2g44060.1 | LEA_2 | Group 7 | PF03168 | Class 8 | C (1), G (1), Pm (4) | |
| 27 | At2g46140.1 | LEA_2 | Group 7 | PF03168 | Class 7 | C (1), Pm (2) | |
| 28 | At3g02480.1 | LEA_4 | Group 3 | PF02987 | Class 6 | ||
| 29 | At3g15670.1 | AtLEA3-4/LEA76 | LEA_4 | Group 3 | PF02987 | Class 6 | N (1) |
| 30 | At3g17520.1 | LEA_4 | Group 4 | PF02987 | Class 6 | ||
| 31 | At3g22490.1 | RAB28 | SMP | Group 5 | PF04927 | Class 11 | N (1) |
| 32 | At3g22500.1 | ECP31 | SMP | Group 5 | PF04927 | Class 11 | |
| 33 | At3g50970.1 | XERO2/LTI30 | Dehydrin | Group 2 | PF00257 | Class 4 | |
| 34 | At3g50980.1 | XERO1 | Dehydrin | Group 2 | PF00257 | Class 1 | |
| 35 | At3g51810.1 | EM1 | LEA_5 | Group 1 | PF00477 | Class 5 | |
| 36 | At3g53040.1 | LEA_4 | Group 3 | PF02987 | Class 6 | ||
| 37 | At3g53770.1 | LEA_3 | Group 6 | PF03242 | Class 9 | ||
| 38 | At4g02380.1 | SAG21/AtLEA5 | LEA_3 | Group 6 | PF03242 | Class 9 | |
| 39 | At4g13230.1 | LEA_4 | PF02987 | Class 6 | |||
| 40 | At4g13560.1 | LEA_4 | Group 3 | PF02987 | Class 6 | ||
| 41 | At4g15910.1 | DI21 | LEA_3 | Group 6 | PF03242 | Class 9 | |
| 42 | At4g21020.1 | LEA_4 | Group 4 | PF02987 | Class 6 | ||
| 43 | At4g36600.1 | LEA_4 | Group 3 | PF02987 | Class 6 | ||
| 44 | At4g38410.1 | Dehydrin | Group 2 | PF00257 | Class 2 | ||
| 45 | At4g39130.1 | Dehydrin | Group 2 | PF00257 | Class 4 | ||
| 46 | At5g06760.1 | LEA4-5 | LEA_1 | Group 4 | PF03760 | Class 10 | Perox (1) |
| 47 | At5g27980.1 | SMP | Group 5 | PF04927 | Class 11 | ||
| 48 | At5g44310.2 | LEA_4 | Group 3 | PF02987 | Class 6 | ||
| 49 | At5g53260.1 | SMP | Group 5 | PF04927 | Class 11 | ||
| 50 | At5g53270.1 | SMP | Group 5 | PF04927 | Class 11 | ||
| 51 | At5g66400.1 | RAB18/AtDI8 | Dehydrin | Group 2 | PF00257 | Class 1 |
Data obtained from SUBA database, corresponding to either GFP or tandem mass spectrometry experiments. Numbers indicate the amount of studies showing same localization. C, cytosol; N, nucleus; Pm, plasma membrane; P, plastid; Perox, peroxisome; V, vacuole; G, Golgi apparatus.
Bioinformatics Subcellular Predictions
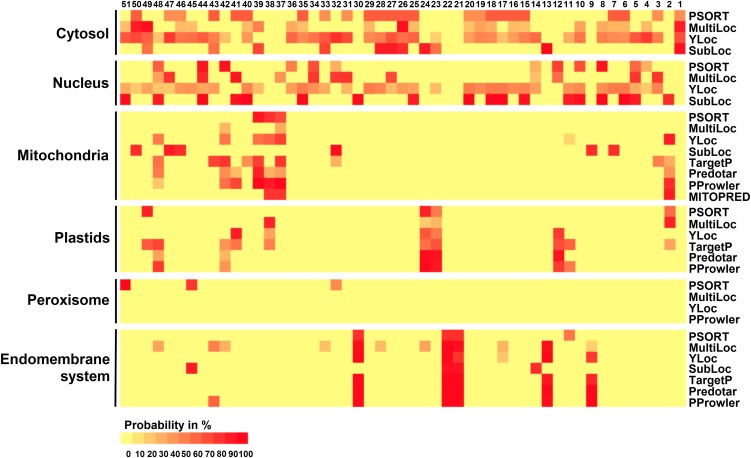
A bioinformatics approach was developed in order to predict the localization of the 51 proteins. Among the several dozen subcellular prediction programs available on the Internet (see http://www.psort.org/), we selected a panel of eight that were able to provide localization probabilities for one or several compartments, including plastid, and thus trained for plant proteins. Two other programs (BaCelLo and WoLF PSORT), which offer predictions for multiple compartments, but without scores, were included because of their reliability (Casadio et al., 2008). All the results obtained with the first eight programs giving prediction scores have been gathered in Figure 1 as a heat map and organized by compartments. The results suggest a rather high representation of LEA proteins in the cytosolic and nuclear compartments, but reveal a lot of heterogeneity in the predictions. If we apply an arbitrary cutoff by considering as valid only predictions for which more than half of the algorithms give scores equal to or over 50% (i.e., three out of the four programs used for these compartments), the heat map appears less scattered (Supplemental Figure 1). The predictions by BaCelLo and WoLF PSORT, which do not provide scores, match poorly with the other predictors (Supplemental Figure 1). In fact, the programs were unanimous in giving high scores for a single localization for only two out of 51 proteins, LEA21 and LEA22, which are expected to populate the endomembrane system (ES). In summary, this bioinformatics analysis, for which up to 10 algorithms were applied to the 51 polypeptides, highlights the difficulty in predicting LEA protein localization with high confidence.
Figure 1.
Heat Map of Subcellular Localization Predictions for the 51 Arabidopsis LEA Proteins.
The heat map illustrates the probability, given by the different programs indicated on the right, of LEA protein targeting to various cellular compartments. LEA proteins are numbered from 1 to 51, and the color coding represents the targeting probability from 0 (yellow) to 100% (red).
Expression of Fluorescent LEA Protein Fusions in Protoplasts, Seedlings, and Plants
In order to acquire experimental data about the subcellular distribution of the 51 LEA proteins, the corresponding coding sequences were genetically fused with green or red fluorescent proteins (GFP and RFP) in vectors designed for transient transgene expression in Arabidopsis leaf protoplasts, which were then observed by laser scanning confocal microscopy. GFP constructs were designed as both N- or C-terminal fusions to each LEA protein. Subcellular compartments markers were used in coexpression experiments to validate the different locations when appropriate and to confirm some peculiar locations. In addition, six constructs were also expressed in seedlings, using a transient Agrobacterium tumefaciens–mediated transformation of Arabidopsis, to examine their location in a tissue context, and transgenic lines were generated to confirm the localization of 11 proteins.
Cytosol and Nucleus
For the majority of LEA proteins (36), both the LEA-GFP and the corresponding GFP-LEA protein fusions were routinely observed to give a diffuse fluorescent pattern typical of a cytosolic location, clearly excluding chloroplasts and vacuoles. Since it is difficult to determine with Arabidopsis protoplast whether cytosolic protein fusions also enter the nucleus, like GFP alone, all of these LEA-GFP constructs were coexpressed with a nuclear localization signal (NLS)-RFP, which is exclusively targeted to the nucleus. Figure 2 shows merged images of the colocalization of NLS-RFP and LEA-GFP fluorescence for a series of four typical LEA proteins exhibiting an exclusive cytosolic localization. The images in Figure 2 show also another series of four LEA proteins displaying a dual localization in the cytosol and nucleus. Among these 36 LEA protein fusions, seven were found exclusively cytosolic, while the others were confirmed with a dual cytosolic-nuclear localization (Table 2). Images captured in separate channels and using the inverted (GFP-LEA) fusion for all these constructs are shown in Supplemental Figure 2. Dual cytosolic-nuclear localization could result from passive diffusion of the fusion proteins into the nucleus or from the presence of degraded LEA-GFP having lost the NLS but still remaining fluorescent. Plotting the repartition of GFP constructs between cytosolic or cytosolic-nuclear localizations according to their molecular mass clearly reveals that dually localized proteins have smaller molecular masses than the cytosolic constructs, which are excluded from the nucleus (Supplemental Figure 3). These data fit well with the classical estimation of the nuclear exclusion limit around 60 kD (Dingwall and Laskey, 1986), and passive diffusion in the nucleus of highly expressed cytosolic LEA-GFP would explain their dual cytosolic-nuclear localization. The integrity of the constructs was verified by protein gel blotting of protoplast protein extracts using an antibody directed against GFP (Supplemental Figure 4). Although some degradation products can be detected, a major band with a SDS-PAGE apparent molecular mass close to, or a few kilodaltons higher than, the theoretical size of the protein fusions is always observed (Supplemental Table 1). Such increased apparent molecular mass on SDS-PAGE gels could be attributed to the disordered character of LEA polypeptides, which leads to reduced binding of SDS (Tompa, 2002). In particular, two cytosolic-nuclear constructs (LEA31-GFP and LEA32-GFP) with respective theoretical masses of 55.370 and 55.405 kD (Supplemental Figure 3) approaching the nuclear exclusion limit displayed strong signals with a very low level of degradation products (Supplemental Figure 4). This indicates that these intact constructs still diffuse passively in the nucleus. Conversely, LEA10-GFP, which has the lower theoretical molecular mass (49.412 kD) of the cytosolic protein group, i.e., below the nuclear exclusion limit, does not diffuse into the nucleus. However, it displays a much higher apparent molecular mass of 63 kD (Supplemental Figure 4 and Supplemental Table 1), and its structural features might prevent passive diffusion through nuclear pores. Taken together, these experiments indicate that 36 LEA-GFP constructs likely accumulate in the cytosol, while 29 of them are able to diffuse passively in the nucleus.
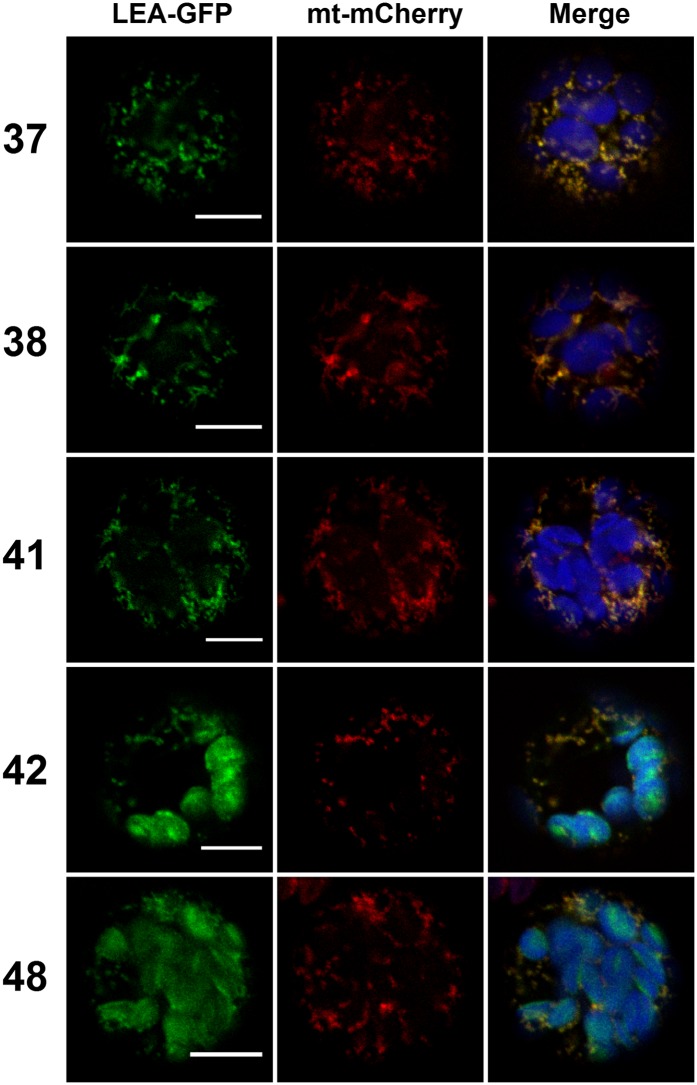
Figure 2.
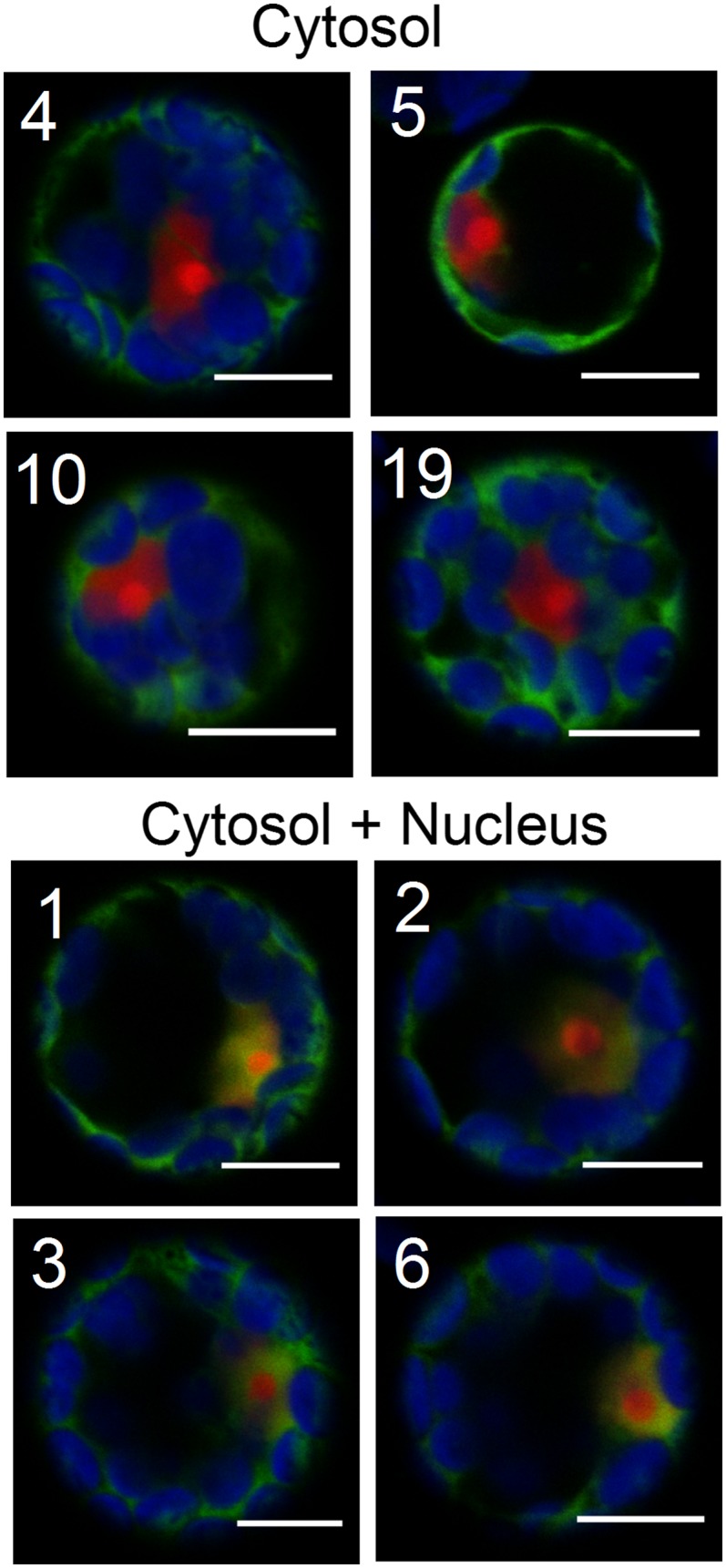
Transient Expression of Cytosolic LEA Protein in Arabidopsis Protoplasts.
Representative examples of LEA-GFP fusion proteins located either in the cytosol only or both in the cytosol and the nucleus were coexpressed with a RFP nuclear marker in Arabidopsis mesophyll protoplasts. Numbers refer to the corresponding LEA proteins. Green, GFP; red, RFP; blue, chlorophyll. The other LEA proteins with similar locations are illustrated in Supplemental Figure 2. Bars = 10 µm.
Table 2. Cytosolic and Nuclear LEA Proteins.
| Localization | LEA Protein |
|---|---|
| Cytosol | 4, 5, 10, 19, 25, 26, 36 |
| Cytosol and nucleus | 1, 2, 3, 6, 7, 8, 14, 15, 16, 17, 18, 20, 27, 28, 29, 31, 32, 33, 34, 35, 39, 40, 44, 45, 46, 47, 49, 50, 51 |
Mitochondria and Plastid
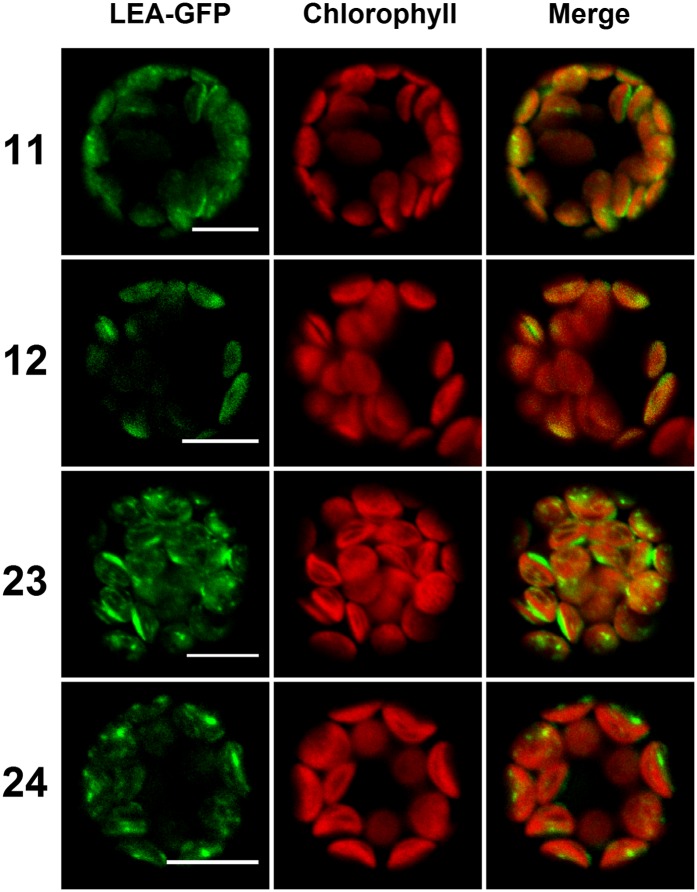
Nine LEA-GFP fusions were found to be targeted to mitochondria, plastid, or to both organelles. LEA37, 38, and 41 fusions displayed a typical mitochondrial matrix pattern illustrated by the perfect colocalization between the GFP signal and the mitochondrial mt-mCherry marker (Figure 3). Interestingly, two fusion proteins (LEA42 and LEA48) were found to be dually targeted to mitochondria and plastids (Figure 3). As a control, GFP-LEA fusions, in which GFP should mask the mitochondrial targeting peptide, were observed in the cytosol as a diffuse signal (Supplemental Figure 5). Figure 4 illustrates the localization of four LEA-GFP proteins (LEA11, 12, 23, and 24) that were found exclusively in plastids, where the autofluorescence of the chlorophyll was used as a marker. Like in the case of the mitochondrial LEA proteins, masking the N terminus targeting peptides (GFP-LEA fusions) resulted in diffuse cytosolic localizations (Supplemental Figure 5). For the five LEA protein fusion targeted to mitochondria or dual targeted to plastid and mitochondria, we established transgenic lines. Confocal examination confirmed the dual targeting of LEA42 and LEA48 and the exclusive mitochondrial localization of LEA 37, 38, and 41 (Supplemental Figure 6).
Figure 3.
Transient Expression of Mitochondrial LEA Proteins in Arabidopsis Protoplasts.
LEA-GFP fusion proteins localized in mitochondria or both in mitochondria and plastids were coexpressed with a mCherry mitochondrial marker (mt-mCherry) in Arabidopsis mesophyll protoplasts. In the merged images, chlorophyll autofluorescence is also shown (in blue). Numbers refer to the corresponding LEA proteins. Green, GFP; red, mCherry; blue, chlorophyll. Bars = 10 µm.
Figure 4.
Transient Expression of Plastid LEA Proteins in Arabidopsis Protoplasts.
LEA-GFP fusion proteins localized in plastids were expressed in Arabidopsis mesophyll protoplasts, with chlorophyll as a natural plastid marker. Numbers refer to the corresponding LEA proteins. Green, GFP; red, chlorophyll. Bars = 10 µm.
For the fusion proteins localized in plastid or dual targeted, the verification of fusion protein expression by protein gel blot analysis of protoplasts (Supplemental Figure 4) revealed major bands with SDS-PAGE apparent molecular masses lower than theoretical sizes of the protein fusions (Supplemental Table 1). These few kilodaltons of differences would be in agreement with the cleavage of targeting peptides during translocation. In the case of the three proteins (LEA37, 38, and 41) exclusively targeted to mitochondria, the differences between theoretical masses of the fusion proteins and their apparent size in protein gel blots were not convincing with respect to the cleavage of targeting peptides. According to protein gel blots, LEA37 fusion was found slightly smaller (0.5 kD), LEA41 almost equal, and LEA38 slightly bigger (1 kD) than their respective theoretical size (Supplemental Figure 4 and Supplemental Table 1). However, because of the small size of these LEA proteins (10 to 14 kD), one cannot rule out the possibility that these proteins would be imported with a very short targeting peptide or even without any cleavable sequence.
ER, Vacuole, and Secretion
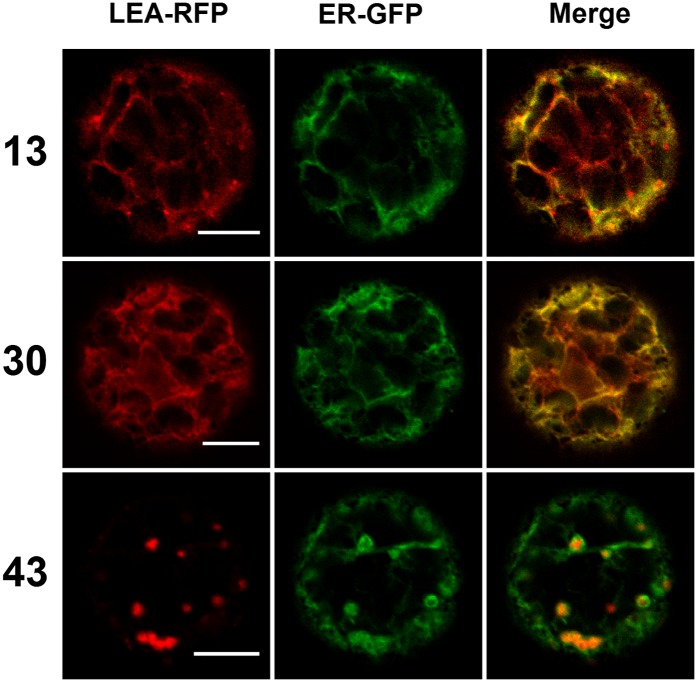
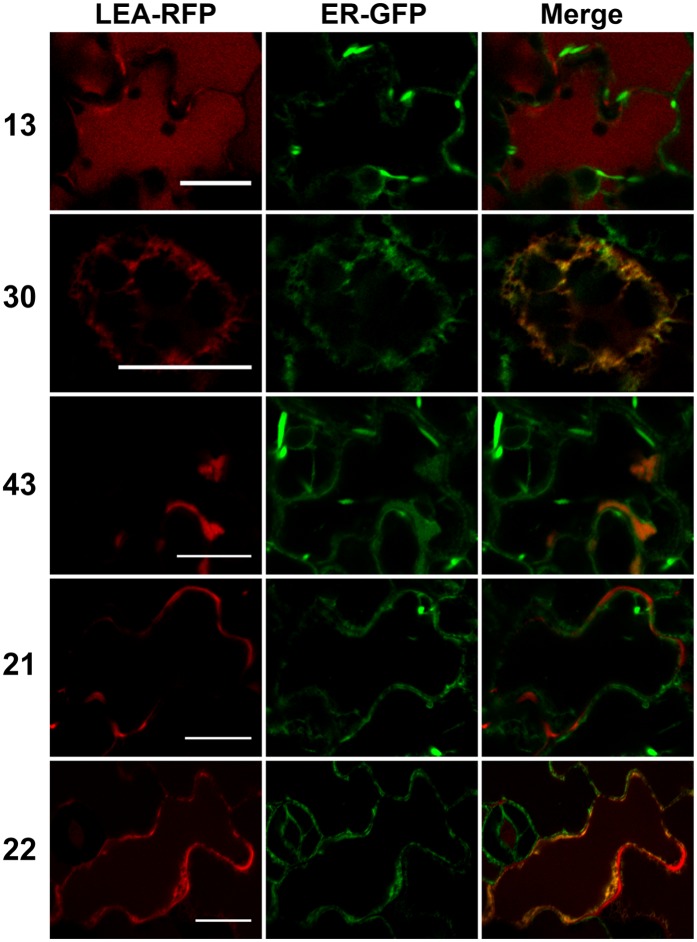
Three LEA-RFP fusion proteins were found to populate the ER in transformed protoplasts, as illustrated in Figure 5. LEA13 and LEA30 signals appeared to clearly colocalize with a GFP-ER marker. When LEA30-RFP was transiently expressed in a seedling system, in order to examine subcellular distribution in intact tissues, the protein was also found to colocalize with the ER marker (Figure 5). In the case of LEA13-RFP, bright red dots, distinct but close to the ER, were observed in protoplasts, suggesting an additional localization in Golgi. However, we could not observe colocalization with a Golgi marker (Supplemental Figure 7). When expressed in seedlings, LEA13-RFP was surprisingly found to accumulate in the vacuole (Figure 6), which could be the final destination of the protein in the secretory pathway. In order to gain more information about the targeting of these proteins, we established transgenic lines, which confirmed the vacuolar localization for LEA13-RFP and showed that LEA30-RFP was localized both in the ER, as in protoplasts and seedlings, but also in the vacuole (Supplemental Figure 8). In protoplasts, LEA43-RFP was found to accumulate specifically in several ovoid regions of the ER (Figure 5). In transiently transformed seedlings, LEA43-RFP accumulated in large and rather diffuse areas of the ER (Figure 6), possibly corresponding to ER sheets, but not in areas known as fusiform bodies (Hawes et al., 2001). In the transgenic lines overexpressing LEA43-RFP, the protein was found to reside in the ER, like in protoplasts and seedlings (Supplemental Figure 8).
Figure 5.
Transient Expression in Arabidopsis Protoplasts of LEA Proteins Located in the ER.
LEA-RFP fusion proteins localized in the ER were coexpressed with an ER constitutive marker (ER-GFP) in Arabidopsis mesophyll protoplasts. Numbers refer to the corresponding LEA proteins. Green, GFP; red, RFP. Bars = 10 µm.
Figure 6.
Transient Expression in Arabidopsis Seedlings of LEA Proteins Located in the ER, Vacuole, or Secreted.
Seedling cotyledon cells were transiently transformed using Agrobacterium infiltration. LEA-RFP fusion proteins were coexpressed with an ER marker (ER-GFP). Numbers refer to the corresponding LEA proteins. Green, GFP; red, RFP. Bars = 20 µm.
Constructs with GFP fused upstream of LEA proteins, in order to mask a putative N terminus signal peptide, were transformed into protoplasts. As expected, all three GFP-LEA fusions were found to reside in the cytosol with a diffuse pattern (Supplemental Figure 9).
To check the integrity of the fusion proteins, protoplasts were transformed with these LEA-GFP constructs and subjected to protein gel blotting. The three proteins fusions were clearly detected with apparent molecular masses close to their theoretical mass (Supplemental Figure 4 and Supplemental Table 1). However, considering the size of the fusions proteins (61 to 78 kD), it is difficult to reach any conclusion regarding the cleavage or lack of a putative signal peptide.
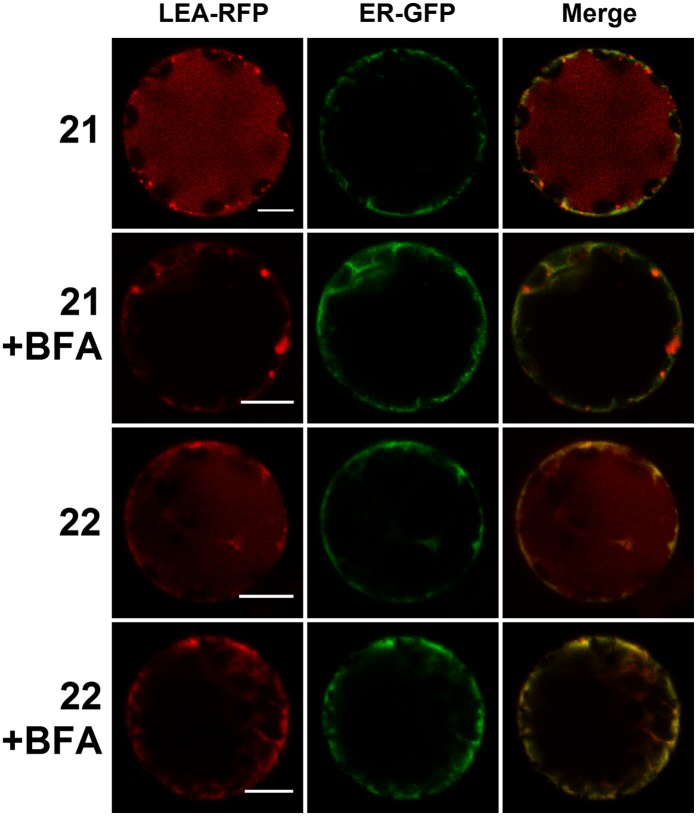
Two LEA fusions (LEA21-RFP and LEA22-RFP) were identified as vacuolar proteins in protoplasts expressing the ER-GFP marker (Figure 7). LEA21-RFP was essentially found in the vacuole (Figure 7), with additional bright spots that could possibly correspond to Golgi (Supplemental Figure 10). LEA22-RFP was observed to accumulate both in the vacuole and in the ER (Figure 7). To confirm the targeting of these two proteins to the vacuole via the secretory pathway, a treatment with Brefeldin A (BFA) was performed to block ER-Golgi transport (Nebenführ et al., 2002) (Figure 7). In BFA-treated protoplasts, LEA21-RFP clearly did not reach the vacuole and was instead found to accumulate in the ER and in large vesicles (Figure 7). These could possibly correspond to ER-Golgi transport vesicles stacking next to the ER exit site and unable to reach the BFA impaired cis-Golgi (Nebenführ et al., 2002). Vacuole accumulation of LEA22-RFP was also prevented by BFA, and the protein instead remained mostly associated with ER (Figure 7). This is in agreement with their ER and vacuolar localization, with BFA blocking the secretion to the vacuole without hampering ER accumulation. To check the integrity of the fusion proteins, protoplasts were transformed with the respective LEA-GFP constructs. In agreement with the fact that GFP targeted to vacuole is readily degraded (Tamura et al., 2003), the corresponding LEA-GFP constructs were not detected in the vacuole of transformed protoplasts, but only as bright spots for LEA21-GFP and ER-type network for LEA22-GFP, although fluorescence was very weak for that construct (Supplemental Figure 11). In protein gel blots, main bands with an SDS-PAGE apparent molecular mass in the range of the theoretical size of the protein fusions could nevertheless be detected for the two LEA proteins (Supplemental Figure 4 and Supplemental Table 1).
Figure 7.
Effect of BFA on ER- or Vacuole-Targeted LEA Proteins Transiently Expressed in Arabidopsis Protoplasts.
LEA-RFP fusion proteins localized in the ER or the vacuole were coexpressed with an ER marker (ER-GFP) in Arabidopsis mesophyll protoplasts. When indicated, transient expression was performed in the presence of BFA (+BFA). Numbers refer to the corresponding LEA proteins. Green, GFP; red, RFP. Bars = 10 µm.
The two LEA-RFP fusion proteins were also transiently expressed in seedlings. Both LEA21 and LEA22 fusions were found to accumulate in diffuse areas in the intercellular space surrounding transformed cells and hence are likely secreted (Figure 6). While LEA21-RFP appears exclusively secreted in these conditions, LEA22-RFP was also observed in the ER and the vacuole. GFP-LEA controls were found localized in the cytosol with a diffuse fluorescence, which suggests that putative N terminus signal peptides would be masked (Supplemental Figure 9). In leaf cells from a transgenic lines expressing LEA21-RFP, the protein was found to be clearly secreted in the intercellular space, while fluorescence was also observed in vacuole (Supplemental Figure 8). In the case of transgenic lines expressing LEA22-RFP, the fusion protein was found to accumulate in the vacuole, but not in the ER (Supplemental Figure 8).
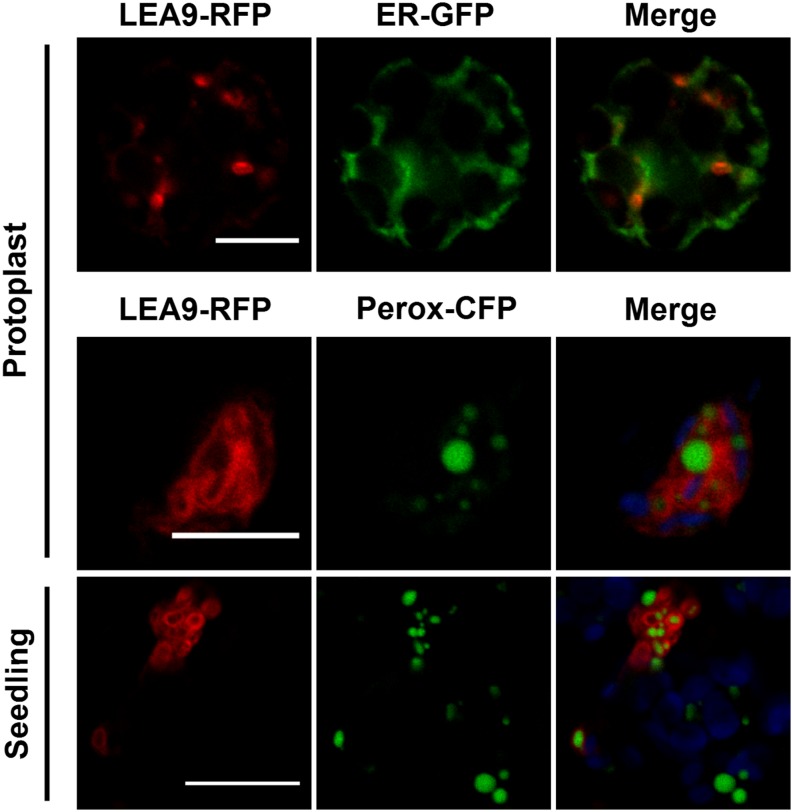
Pexophagosome
Interestingly, when expressed in protoplasts, LEA9-RFP displayed an unusual pattern that could not be attributed to any classical compartment (cytosol, nucleus, ER, Golgi, vacuole, peroxisome, mitochondria, and plastid). The protein accumulates in peculiar spherical and apparently hollow structures (Figure 8). When LEA9-RFP was expressed in cells expressing an ER-GFP marker, these structures appeared to neighbor or partially overlap the ER, suggesting a close association (Figure 8). The coexpression of LEA9-RFP with a peroxisomal cyan fluorescent protein (CFP) marker revealed that the LEA9 fusion populated structures that engulfed most peroxisomes (Figure 8). These unusual structures could therefore correspond to pexophagosomes, which are organelles involved in the recycling of peroxisomes (van Zutphen et al., 2008). Analysis of the integrity of the fusion protein (Supplemental Figure 4) in protoplasts indicates that the apparent molecular mass corresponds to the theoretical size of the protein fusions (Supplemental Table 1).
Figure 8.
Transient Expression of a Pexophagosome-Targeted LEA Protein in Arabidopsis Protoplasts or Seedlings.
LEA9-RFP fusion protein localized in pexophagosomes was coexpressed with an ER marker (ER-GFP) or a peroxisome marker (Perox-CFP) in Arabidopsis mesophyll protoplasts or in Arabidopsis seedling cotyledons using Agrobacterium. Green, GFP or CFP; red, RFP; blue, chlorophyll. Bars = 10 µm for protoplasts and 20 µm for seedlings.
In order to confirm this location, LEA9-RFP was expressed in seedlings where it displayed a pattern typical of pexophagosomes (Figure 8). Transgenic lines expressing LEA9-RFP and a peroxisome marker were established, but few cells were found to accumulate LEA9-RFP. However, the protein always displayed a similar pattern, with association with peroxisomes (Supplemental Figure 8). Finally, a construct with an opposite orientation (GFP-LEA9) expressed in protoplasts displayed similar images to those of its LEA-RFP counterpart (Supplemental Figure 9), suggesting that targeting information was not masked by GFP and, hence, not located in the N or C terminus of the protein.
Targeting Peptide Cleavage Sites for Mitochondrial and Plastid LEA Proteins
Proteins targeted to mitochondria and plastids generally carry an N-terminal targeting peptide, which is cleaved during the import process (Teixeira and Glaser, 2013). As a result, the mature proteins released inside the organelle are usually shorter than their cytosolically synthesized precursors. To facilitate the proper structural and functional characterization of organellar proteins, it is critical to determine the targeting peptide cleavage sites. We previously described a method to experimentally determine the targeting peptide cleavage site of C-terminal GFP fusion proteins imported into mitochondria or chloroplasts during transient expression in protoplasts (Candat et al., 2013). In this approach, protoplasts expressing LEA-GFP proteins are lysed and fusion proteins are immune-captured with GFP antibodies bound to magnetic microbeads. Purified protein-GFP fusions are then separated by electrophoresis and subjected to Edman microsequencing to determine their N terminus. Table 3 displays the targeting peptide cleavage sites we were able to determine for LEA proteins identified in plastids or mitochondria. The cleavage sites for LEA23-GFP and LEA24-GFP, which are localized in plastids, were determined previously (Candat et al., 2013). In the case of LEA11 and LEA12, which are two paralogous proteins targeted to plastids (Figure 4), the cleavage site could be only be determined for LEA11-GFP (Table 3). The level of expression of the LEA12-GFP construct in protoplasts was too low for successful N terminus microsequencing. However, alignment of LEA11 and LEA12 polypeptide sequences, shown in Supplemental Figure 12, indicates a high conservation of the cleavage site regions SWV(S/P)(A/T)↓AVKG(A/D)GNS (arrow indicates cleavage site). It is therefore highly probable that cleavage of the LEA12 precursor occurs at the proposed site. In the case of LEA42 and LEA48, which are dually targeted to plastids and mitochondria (Figure 3), a unique cleavage site was identified (Table 3). This suggests that these proteins should carry an ambiguous transit peptide targeting the precursors to both mitochondria and plastids. We were unfortunately unable, so far, to determine a targeting peptide cleavage site for the three LEA proteins (LEA37, 38, and 41) identified in mitochondria (Figure 3). As in the case of LEA12, their relatively low level of expression in protoplasts hampers the analysis.
Table 3. Determination of the Mature Protein Sequence Using Purification of Protoplasts Synthesized Organelle Proteins.
| LEA Protein | Localization | Experimental Determination of the Mature Protein N-Terminal End (Edman) |
|---|---|---|
| 11 | Plastid | 50-51 AVKGAG |
| 12 | Plastid | ND |
| 23a | Plastid | 51-52 VKSDGN |
| 24a | Plastid | 49-50 AAKGDG |
| 42 | Mitochondria and plastid | 40-41 TSVSQN |
| 48 | Mitochondria and plastid | 41-42 SSVNHS |
ND, not determined.
Determined previously (Candat et al., 2013).
DISCUSSION
LEA proteins are mostly intrinsically disordered proteins expected to play major roles in abiotic stress tolerance in anhydrobiotes. In plants, several dozen genes encode LEA proteins that can be clustered into several families according to their sequence properties. The aim of this research was to provide a subcellular map of the distribution of LEA proteins in the model plant Arabidopsis, for which 51 LEA genes were inventoried (Hundertmark and Hincha, 2008). Because of the high number of gene fusion constructs to investigate (and both N- and C-terminal fusions), a leaf mesophyll protoplast transient assay was selected (Yoo et al., 2007). Although observations performed with mesophyll protoplasts are not representative of all cell types and tissues, they provide a robust system to analyze the intrinsic routing information carried by polypeptides, especially in a comparative manner as in this study. In addition, for most LEA proteins not displaying a cytosolic localization in protoplasts, complementary approaches of Agrobacterium-mediated transient expression in seedlings or stable transgenic lines were used.
Comparison between Subcellular Localization Predictions and Experimental Data
The subcellular localization of more than 50 related proteins, as experimentally determined in this work, represents an interesting data set to examine the performance of different targeting prediction algorithms. A schematic overview of the relative performance of eight different software packages is presented in Supplemental Figure 13. The bar graph illustrates the proportion of correct and erroneous predictions by the different programs for ES, mitochondria, plastids, and cytosolic compartments. For the latter, we considered that all proteins dually localized in the cytosol and nucleus were actually cytosolic and only diffused passively into the nucleus. Since we could not demonstrate exclusive nuclear localization for any construct, all the many nucleus targeting predictions were false and were not included in the graphical analysis. Clearly, the predictions for ES were more accurate than for other compartments, with only three erroneous predictions out of a total of 34, with five programs yielding a high success rate (at least five of the six proteins trafficking in ER) including 100% success achieved by PProwler (Supplemental Figure 13). Such efficiency is likely due to the fact that signal peptides drive the entry of almost all proteins in the secretory pathway in eukaryotes and prokaryotes and thus share common features that facilitate the design of prediction algorithms (Nielsen et al., 1997; Hegde and Bernstein, 2006). At first glance, predictions for cytosolic localization seem robust, with six erroneous predictions alongside 50 correct predictions by four algorithms (Supplemental Figure 13). However, individual predictions appeared more heterogeneous than in the case of SE, and the best success rate was only 67% for YLoc and much lower for the other programs. In the case of plastid and mitochondrial compartments, while the success rate was generally good, the proportion of erroneous predictions was high (Supplemental Figure 13). This certainly reflects the insufficient knowledge about the targeting peptide information for these organelles, as well as the similarity in their import machineries and the increasing number of proteins shown to be dual targeted (Carrie et al., 2009). Overall, PProwler was the most efficient predictor, followed by YLoc and TargetP. However, this conclusion only applies to our data set of proteins, which are highly hydrophilic, largely disordered, and composed of a majority of cytosolic proteins. Nevertheless, this study confirms that assigning protein localization is clearly strengthened by the use of multiple programs, as was demonstrated by analysis of the plant mitochondrial proteome (Heazlewood et al., 2004).
Subcellular Distribution and Classification of LEA Proteins
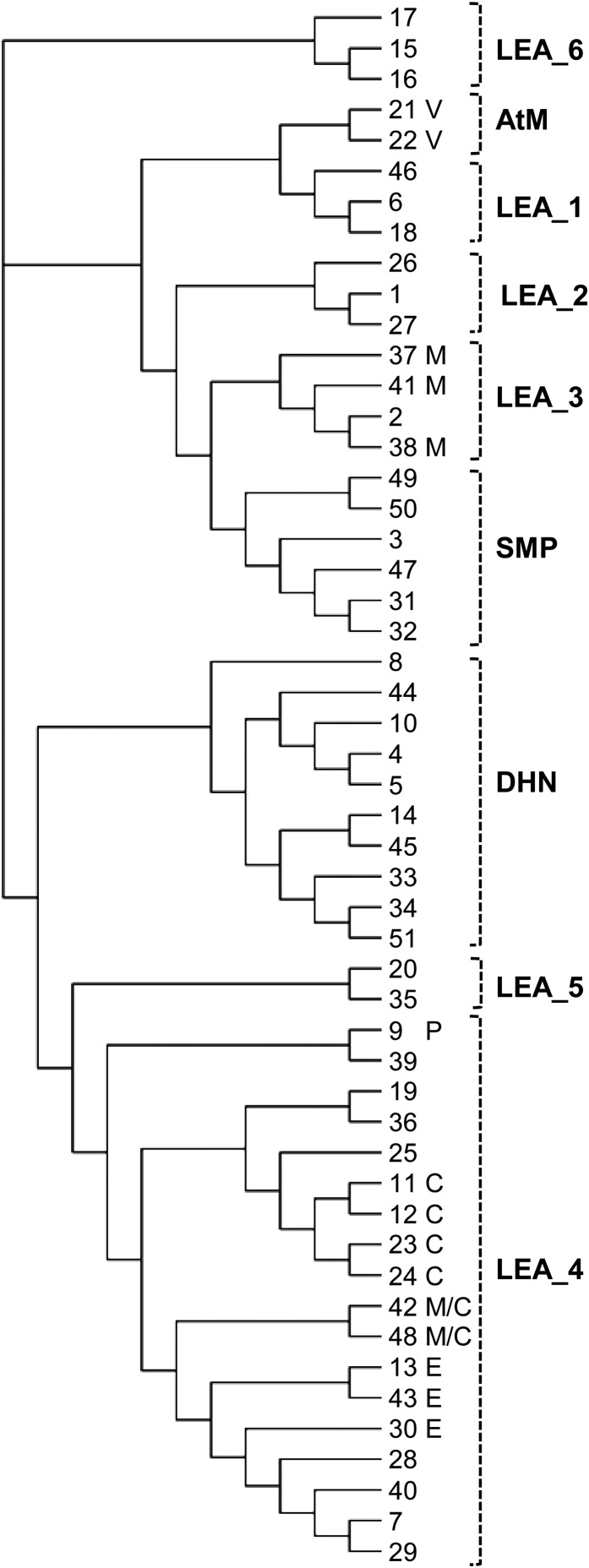
The experimental data enables an examination of the subcellular distribution of the Arabidopsis LEA proteome with respect to the structural classification of the proteins into several families, which likely have different evolutionary histories. Figure 9 shows a cladogram of the 51 LEA proteins incorporating their PFAM classification and their subcellular location. Among the nine families, six (LEA_1, LEA_2, LEA5, LEA6, dehydrin, and SMP) display a cytosolic (or cytosolic and nuclear) localization for all their members (up to 10 in the case of the Dehydrin family). The two members of the AtM family share localization within the secretory pathway, while three out of four members of the LEA_3 family are mitochondrial, the last being cytosolic (Figure 9). In contrast to the rather homogenous location of members of those families, the 18 members of the LEA_4 family, which is the largest family, displayed multiple localizations (cytosol, plastid, mitochondria, ER, and pexophagosome). This overall analysis clearly highlights a link between protein family and location, with the members of eight families generally displaying a single common location while the ninth family is distinguished by members having targeted to diverse locations.
Figure 9.
Cladogram Illustrating the Repartition of Arabidopsis LEA Protein Subcellular Distribution in Each Family.
The rectangular cladogram was built with Dendroscope software (see Methods) using a multiple alignment of the 51 LEA proteins, which are indicated by their number. The subcellular localization is indicated by letters on the right. C, chloroplast; E, ER; M, mitochondria; P, pexophagosome; V, vacuole. All proteins without indication are cytosolic. The brackets on the right indicate the different LEA protein families. DHN, dehydrin.
Cytosolic and Nuclear LEA Proteins
The vast majority (36, distributed in six families) of Arabidopsis LEA protein appeared cytosolic and able to diffuse passively in the nucleus. It is noteworthy that we could not observe any LEA fusion proteins specifically targeted to the nucleus, although LEA31 was previously assigned a nuclear localization using GFP constructs (Borrell et al., 2002). However, from the examination of the published images of Arabidopsis RESPONSIVE TO ABSCISIC ACID28 (Atrab28 and LEA31) stably expressed as a GFP fusion in Arabidopsis (Borrell et al., 2002), the fusion protein appears in fact to be distributed in both the cytosol and nucleus. Three other proteins (LEA4, LEA5, and LEA8) were previously described as cytosolic using GFP fusions (Cutler et al., 2000; Abu-Abied et al., 2006), which we could confirm. Two others (LEA10 and LEA46) are tentatively identified as peroxisomal according to the SUBA database, based on the results of random GFP-cDNA expression in Arabidopsis (Cutler et al., 2000). However, such results should be treated with caution due to the high proportion of non-native coding sequences (i.e., out-of-frame cDNAs) in this large-scale analysis, which resulted in peroxisomal-like localization. Two proteins (LEA7 and LEA29) that appeared cytosolic in our experiments were proposed to reside mainly in the ER, based on GFP fusions expressed in protoplasts (Zhao et al., 2011). However, using colocalization with the ER-GFP marker (Supplemental Figure 14), we could not find any evidence for an ER location for LEA7 or LEA29 GFP fusions.
The increasing amount of subcellular proteomic data that are available in public databases (e.g., MASCP Gator and SUBA) allows a comparison of our results with those obtained using other approaches. Among the LEA set of cytosolic proteins, six (LEA1, 4, 5, 10, 26, and 27) were identified in the cytosolic proteome of Arabidopsis cell suspensions (Ito et al., 2011). Five of them (LEA4, 5, 10, 26, and 27), as well as two others (LEA8 and LEA16), were identified in the plasma membrane proteome of cell cultures or leaves (Alexandersson et al., 2004; Benschop et al., 2007; Marmagne et al., 2007; Mitra et al., 2009; Keinath et al., 2010; Elmore et al., 2012; Li et al., 2012a; Nikolovski et al., 2012). Five proteins (LEA4, 5, 8, 10, and 33) belonging to the dehydrin family could interact with the plasma membrane since LEA5, LEA10, and LEA33 have been shown to bind anionic phospholipid vesicles (Kovacs et al., 2008; Eriksson et al., 2011). Moreover, dehydrins from maize (Zea mays) and the salt-tolerant Thellungiella salsuginea have also been shown to bind phospholipid vesicles (Koag et al., 2003, 2009; Rahman et al., 2010, 2013). Interestingly, four dehydrins (LEA4, 5, 8, and 10) with a clear cytosolic localization in our experiments were also identified in chloroplast proteome (Kleffmann et al., 2004; Zybailov et al., 2008; Ferro et al., 2010; Kong et al., 2011) or in the tonoplast in the case of LEA10 (Whiteman et al., 2008). This suggests interaction of these cytosolic dehydrins with other cellular membranes. LEA26, another cytosolic protein of the LEA_2 family identified in cytosolic and plasma membrane proteomes (see above), was also identified in the Golgi proteome (Parsons et al., 2012). This protein, for which little information apart from its downregulation following cadmium exposure of Arabidopsis cell culture (Sarry et al., 2006) is available, could possibly also interact with membrane. Finally, LEA29, whose localization is debated above, was identified in the nuclear proteome of Arabidopsis leaves (Bae et al., 2003), which agrees well with its capacity to diffuse in the nucleus.
Overall, comparison between our data and that arising from subcellular proteomics indicates discrepancies for several proteins that are in fact easily explicable: for example, localization in the nucleus as a result of diffusion rather than targeting or association with various compartments as a result of simple contamination or membrane binding. Also, the lack of detection of many LEA proteins in subcellular proteomics is likely be due to the absence of transcription or low expression of the corresponding LEA genes.
AtM LEA Proteins Are Vacuolar or Secreted
Two related LEA proteins (LEA21 and LEA22) could be specific to Brassicaceae and were previously classified in the AtM family (Hundertmark and Hincha, 2008). In our study, the two AtM LEA fusions LEA21- and LEA22-RFP appeared clearly in the ES but displayed various localizations (ER, vacuole, and secretion) depending on the expression system, with, for instance, excretion being undetectable with protoplasts. It is worth noting that these two proteins were only detected in seeds (Supplemental Figure 15) in the course a comprehensive organ-specific proteomics analysis in Arabidopsis (Baerenfaller et al., 2011). This suggests a role for these LEA proteins, with respect to their ER and vacuolar localization, in the biogenesis and degradation of protein storage vacuoles (Otegui et al., 2006). If the proteins are actually secreted in seeds, they would be expected to have a role as extracellular chaperones in dehydration tolerance within the intercellular spaces, as was suggested for two LEA proteins from an anhydrobiotic bdelloid rotifer (Tripathi et al., 2012).
Mitochondrial and Cytosolic LEA Proteins in the LEA_3 Family
Three of the four LEA proteins (LEA37, LEA38, and LEA41) comprising the LEA_3 family were exclusively targeted to mitochondria, which was confirmed also in transgenic lines, while the remaining protein (LEA2) was ascribed a cytosolic localization. Interestingly, LEA2 and LEA38 are two paralogous proteins (Hundertmark and Hincha, 2008), which are targeted to different locations. Although the two sequences are very similar, the location differences could be explained by a deletion of six amino acids (36-ValSerSerGlyGlyArg-43) in LEA2, equating to a region prior to the putative cleavage site of LEA38 (Supplemental Figure 16). LEA38 was recently identified in mitochondria, which is consistent with our observations, and proposed to play major roles in plant development as well as abiotic and biotic stress responses (Salleh et al., 2012). Since the protein was previously shown to improve oxidative stress tolerance when expressed in yeast (Mowla et al., 2006), an interaction of LEA38 with enzymes involved in mitochondrial reactive oxygen species metabolism and signaling was suggested to mediate the pleiotropic effects observed in overexpressor or antisense Arabidopsis lines (Salleh et al., 2012). The two other mitochondrial proteins (LEA37 and 41), as well as the cytosolic LEA2, share similar sequence properties, which would suggest analogous molecular functions (albeit in a different compartment for LEA2).
In the organ-specific proteomics mapping, while LEA2 and LEA41 were detected exclusively in flowers, and LEA38 in flower and roots, LEA 37 was not detected in any compartment (Supplemental Figure 15). Although the levels of expression of these genes at the transcript level were very low in the publicly available data (EFP browser: http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), this does not preclude an increase under particular stress conditions.
Multilocalization within the LEA_4 Family
A striking feature of the LEA_4 family in Arabidopsis is the diversity of subcellular locations observed for the 18 corresponding LEA-GFP or LEA-RFP proteins. While eight proteins appeared cytosolic, four were identified in chloroplasts, two in mitochondria and chloroplasts, three in the ER, and one in structures identified as pexophagosomes. Among the four protein fusions targeted to chloroplast in this study, LEA23 was previously identified in chloroplast proteomic analyses (Froehlich et al., 2003; Peltier et al., 2006; Zybailov et al., 2008; Ferro et al., 2010) and LEA24 was shown to be plastidic by transient expression in protoplasts (Candat et al., 2013). However, LEA12, which shows a clear chloroplastic location as a GFP fusion, was previously ascribed a plasma membrane location (Mitra et al., 2009), which could possibly result from contamination by the cytosolic precursor. Organ-specific proteomics indicates that LEA11 and LEA12 are only detected in flowers, while LEA23 and LEA24 are abundant in flowers, leaves, and siliques, but are not detected in seeds (Supplemental Figure 15). These data do not support a general role for plastid LEA proteins in the desiccation tolerance of seeds. However, these proteins could be expressed at higher abundances under stress conditions. Accordingly, LEA23 and LEA24 are known as cold-regulated proteins (COR15B and COR15A) involved in the stabilization of chloroplast membranes or enzymes during freezing (Steponkus et al., 1998; Nakayama et al., 2008; Thalhammer et al., 2010). Two paralogous LEA proteins (LEA42 and LEA48) appeared dual targeted to mitochondria and chloroplasts, both in protoplasts and transgenic lines. They are likely orthologous to LEAM, previously identified in mitochondria from pea seeds (Bardel et al., 2002; Grelet et al., 2005) and from brine shrimp encysted embryos (Menze et al., 2009). Since LEAM-GFP was not found to be targeted in the chloroplast in transformed pea protoplasts (Grelet et al., 2005), dual targeting of these LEA proteins to mitochondria and plastids might not be a general property in plants. It is expected that LEA42 and LEA48 play roles similar to that of LEAM with respect to membrane protection during desiccation (Tolleter et al., 2007), which is in agreement with their seed-specific expression illustrated by their organ-specific proteomics (Supplemental Figure 15) or transcript abundance (EFP browser: http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi).
Three proteins (LEA13, LEA30, and LEA43) were found to engage in the ES pathway. LEA13 was previously detected in a plasma membrane proteomics analysis (Mitra et al., 2009), which may be consistent with transit through the ES since vacuole targeting and secretion through the plasma membrane are correlated. For these three proteins, which reside or transit through the ER, putative retention signals resembling the canonical KDEL sequence were detected in the C terminus (LEA13: YAEL; LEA30: DAEL; LEA43: SAEL). However, ER retention signals can be very specific since a single substitution can abolish their function; moreover, they can be passenger dependent (Denecke et al., 1992; Vitale and Denecke, 1999). Therefore, a more detailed analysis of these putative signals, in particular for LEA13, including the design of constructs with a central fluorescent protein, would be required to ascertain their function. According to the organ-specific proteomics analysis, while these ER proteins can be detected in flowers, leaves, or pollen, LEA30 and LEA43 are especially abundant in seeds, suggesting a role in desiccation tolerance (Supplemental Figure 15).
LEA9, which displays a seed-specific expression pattern (Supplemental Figure 15), was the sole LEA protein that was tentatively identified in pexophagosomes. This raises the question of the existence of pexophagosomes in seeds. Since glyoxysomes and peroxisomes are required for the conversion of lipid reserves during seed germination and the transition to photoautotrophy in Arabidopsis (Graham, 2008), pexophagosomes could have an important role with respect to the biogenesis and recycling of these organelles. LEA9 could therefore contribute to the protection of such structures in the dry state. Since the peroxisome was the only major organelle for which we could not identify the targeting of a LEA protein, this suggests that the organelle could be less susceptible to stress-induced damage or that such damage would not have immediate consequences for cell survival. Peroxisomes do not synthesize phospholipids, and although biogenesis of the organelle is enigmatic, the peroxisomal membrane is expected to derive from the ER (Heiland and Erdmann, 2005). It is also postulated that de novo biogenesis from protoperoxisomes could be a fast process (Lazarow, 2003). It is thus possible that damaged peroxisomes would be rapidly recycled by the pexophagosome, in which LEA9 protein was identified, to allow either elimination of the damaged organelle or possible repair of their membrane by interaction with ER membrane.
Systematic Occurrence of Class A α-Helix Motif in the LEA_4 Family
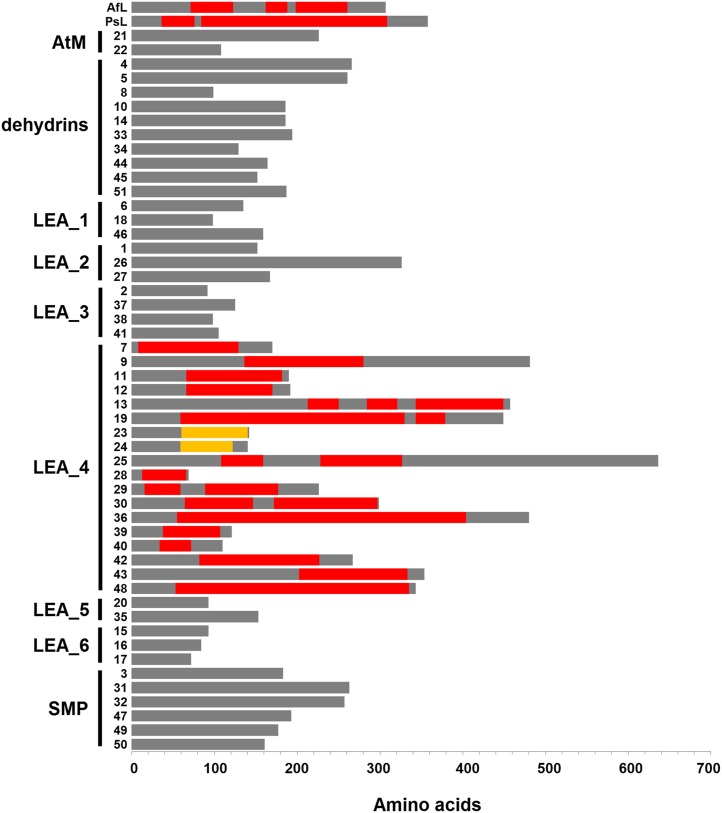
As explained above, LEA42 and LEA48 are putative orthologs of LEAM, a pea mitochondrial protein that was previously shown to fold during dehydration into a peculiar amphipathic helical form, the so-called class A α-helix, allowing the protein to immerse laterally within the inner layer of the inner membrane, reinforcing the membrane in the dry state (Grelet et al., 2005; Tolleter et al., 2007, 2010). In the class A α-helix motif, negatively charged residues form a stripe along the axis of the helix, in the middle of the hydrophilic face, which is flanked on each side by two stripes of positively charged residues (Supplemental Figure 17). This allows the helix to interact laterally with membranes by exposing the hydrophobic face of the helix toward the fatty acid core of the membrane, while positively charged residues interact with phosphate groups of phospholipids, allowing the negatively charged residues to interact with polar heads of positively charged phospholipids (Tolleter et al., 2010). Expectedly, the class A α-helix motif found in pea LEAM protein was clearly detected in their putative homologs LEA42 and LEA48, but also in the case of several other LEA proteins located in different compartments (cytosol, ER, plastid, mitochondria, and pexophagosome), as shown in Supplemental Figure 17.
We decided therefore to scan all Arabidopsis LEA proteins for the occurrence of class A α-helix motifs. A computational analysis was performed using the screening module of the HeliQuest Web server (Gautier et al., 2008) that was adjusted in order to detect protein segments with an α-helix structure sharing sequence parameters and physico-chemical features typical of the LEAM protein class A α-helix motifs (see Methods for details). The results showed that such a structural feature was only shared by proteins from the LEA_4 family (Figure 10). Among the 18 proteins in the family, only two of them (LEA23 and LEA24) displayed amphipathic α-helix motifs resembling those of the class A type, but not canonical, and thus considered here as class A-like (Supplemental Figure 18). In particular, the distribution of charges is not as regular as in the case of other proteins of the LEA_4 family. Interestingly, these two paralogous proteins were included in the LEA_4 group by Hundertmark and Hincha (2008) and in our study because they cluster with other LEA_4 proteins, although they do not match significantly with a PFAM domain (e-value of 1.2 for LEA23; no match for LEA24). It is therefore not surprising that these two proteins do not share structural properties such as the class A α-helix motif with true members of the LEA_4 family. We propose that all LEA_4 proteins exhibiting the class A (and class A-like) α-helix motif would interact and protect various cellular membranes during dehydration, as was shown for LEAM expressed in pea seed mitochondria (Tolleter et al., 2007, 2010). This hypothesis is supported by the fact that LEA_4 proteins are distributed in many subcellular compartments, which would allow a variety of cellular membranes to benefit from protection by LEA proteins during dehydration resulting from desiccation or freezing. LEA7 was indeed shown to interact in vitro with synthetic membranes in the dry state (Popova et al., 2011). Since LEA7 was found to be a cytosolic protein, it could be involved, like the other cytosolic LEA_4 proteins, in the protection of the plasma membrane or organellar membranes, acting on the cytosol exposed side of the membranes. LEA_4 proteins targeted to organelles would provide protection to the corresponding membranes (e.g., inner mitochondrial membrane, inner membrane of the plastid envelope, thylakoid membrane, ER membrane, tonoplast, and pexophagosome). In the case of plastids, it is questionable whether the interactions of class A α-helix motif would be as favorable as in the case of other cell membranes due to the high proportion of galactolipids, which could not easily interact with the positive charges of the proteins because they lack the necessary phosphate group. Indeed, the lipid composition of the envelope inner membrane of chloroplasts, etioplasts, or proplastids comprises only 10% phospholipid and that of thylakoids is even less, at 8%. Whether this would be sufficient to anchor LEA_4 proteins during dehydration is not known. It is interesting that the two plastid proteins with class A-like motifs (LEA23 and LEA24) were shown to protect membranes in vitro (Thalhammer et al., 2010). Interactions of proteins through such amphipathic α-helix were considered biologically relevant because they were favored by lipid compositions mimicking plastid membranes (Thalhammer et al., 2010). Therefore, plastids could harbor a combination of LEA proteins with either class A (LEA11, 12, 42, and 48) or class A-like (LEA23, 24) α-helix motif, which could protect the galactolipid-enriched plastid membranes. Interestingly, in a recent analysis of the degradome of metacaspase 9 (a distant relative of the metazoan caspases) in 2-d-old Arabidopsis seedlings, seven LEA_4 proteins were identified as potential substrates of the processing enzyme (Tsiatsiani et al., 2013). Among these, four proteins (LEA25, 28, 29, and 36) are cytosolic, while two display dual location in mitochondria and plastids (LEA42 and LEA48) and one resides in the ER (LEA30). Since the metacaspase is located in the cytosol (Tsiatsiani et al., 2013), this suggests that the three organellar proteins would be processed before import or in the course of organelle and protein turnover since LEA proteins progressively disappear during early germination and growth. Finally, among the eight PFAM LEA families, the LEA_4 family is the most widely distributed, with representatives in plants, metazoans, fungi, heterolobosea, stramenopiles, and bacteria, and it is possible that most of these proteins harbor the class A α-helix motif and thus could play a role in stress protection of biological membranes.
Figure 10.
Computational Analysis of the Presence of Class A α-Helix Motif in LEA Proteins.
Polypeptide sequences were scanned with a 36-residue sliding window using the scanning module of HeliQuest Web server that was adjusted in order to retrieve typical class A α-helix motifs (shown in red/black) as found in pea LEAM protein (PsL) or its putative homolog from A. franciscana (AfL). Arabidopsis LEA proteins are indicated with their corresponding number and are grouped according to the eight families. Segments shown in orange/light gray correspond to noncanonical class A (class A-like) α-helix motifs.
[See online article for color version of this figure.]
Overall, this analysis of the full complement of LEA proteins within a eukaryotic organism reveals their wide subcellular distribution, with members identified in almost every cellular compartment. Given the current view that all LEA proteins function in stress tolerance, it follows that each compartment requires the presence of LEA protein in certain situations, e.g., dehydration, freezing, cold, or oxidative stress. The predominant cytosolic location agrees with the central position of this compartment, which interacts physically and functionally with all organelles. Cytosolic LEA proteins could therefore be involved in stress protection not only within the cytosol itself, but at the level of membranes delimiting the organelles. The only major organelle for which we could not identify the presence of a LEA protein was the peroxisome, which suggests their rapid biogenesis and recycling through pexophagy could compensate for stress-induced damage.
The complexity of the LEA proteome in plants, which arose from whole-genome duplication and endoreplication events, has been retained along evolution, and the apparent redundancy of LEA proteins in most subcellular compartments likely reflects the versatility of their function in plant life.
METHODS
Plant Culture, Transformation, and Subcellular Localization of Fluorescent Proteins
Arabidopsis thaliana plants (Columbia-0 ecotype) or transgenic lines were grown in Jiffy7 pots (Jiffy Products International) in a growth chamber (23°C, 75% RH, 16 h light with an intensity of 80 to 100 µmol⋅m−2⋅s−1). Transgenic lines expressing ER-GFP or Perox-CFP were obtained from the ABRC (Nelson et al., 2007). Arabidopsis mesophyll protoplasts were isolated according to a procedure adapted from Yoo et al. (2007) as described by Candat et al. (2013). BFA assays were performed after overnight incubation of transformed protoplasts. Protoplasts were treated with BFA at 10 µg/mL for 1.5 h in the dark prior to observations. Transient Arabidopsis seedling transformation was performed according to the procedure from Marion et al. (2008), with some modifications. Sterilized Arabidopsis seeds were sown on MS 1% agar medium (Murashige and Skoog basal medium, Sigma-Aldrich; Agar HP696 CIV, Kalys) in six-well culture plates (CytoOne reference CC7672-7506) and stratified for 2 d at 4°C. Seedlings were grown for 4 d (same growth conditions as above) prior to transformation. After overnight growth at 28°C, Agrobacterium tumefaciens cells (C58 strain) carrying appropriate expression vectors were collected and resuspended at OD600 = 1 in 2 mL of Murashige and Skoog medium supplemented with 200 µM acetosyringone and 0.05% Tween 20 (Sigma-Aldrich). Seedlings were covered by the Agrobacterium suspension for 30 min in the dark. Excess medium was subsequently removed and the plates were transferred to a growth room for 3 d (same growth conditions as above). For a selection of 11 LEA proteins (LEA9, 13, 21, 22, 30, 37, 38, 41, 42, 43, and 48), as well as for the mito-mCherry, transgenic lines expressing fusion proteins were also established by Agrobacterium floral dip transformation (Clough and Bent, 1998) of wild type or lines already expressing organellar markers for ER (YFP-HDEL; Teh and Moore, 2007), peroxisomes (YFP-SKL; Mathur et al., 2002), or mitochondria (mito-mRFP1). Plants from 3 to 21 independent transgenic lines at the T1 stage were selected for microscopy. Subcellular localization of FP-tagged proteins was examined in protoplasts, cotyledons, or leaves using a Nikon A1 laser scanning confocal microscope and NIS-element software (Nikon). GFP, RFP, and chlorophyll were excited with 488-, 561-, or 638-nm laser lines, respectively, with an emission band of 500 to 550 nm for GFP detection, 570 to 620 nm for RFP detection, and 662 to 737 nm for chlorophyll autofluorescence. Experiments using CFP and YFP constructs were performed using the same settings as above for GFP and those with mCherry with the settings for RFP detection. For protoplasts, an average of 50 protoplasts were observed for each experiment (two to five independent protoplast isolation and transformation), with a transformation rate averaging to 25%. In all cases, the subcellular pattern of fluorescent protein was homogeneous, and the images shown in the figures are representative of average protoplasts from a total of 32 transformed protoplasts. Quantitative data are available in Supplemental Table 2 for LEA-FP constructs.
Vector Construction
Plasmid vectors containing full-length LEA coding sequences were obtained from the ABRC or from GenScript, or were kindly provided by Dirk Hincha (Hundertmark and Hincha, 2008) (Supplemental Table 3). Plasmids purified from bacterial strains were used as template to amplify coding sequences by PCR. The Phusion proofreading polymerase was used for amplification following the PCR conditions recommended by the manufacturer (Fermentas Thermo Scientific). The different primers used for each coding sequence amplification are listed in Supplemental Table 4. The PCR product corresponding to the full coding sequence was cloned first in the pENTR/D-TOPO cloning vector (Invitrogen) and then recombined into the appropriate expression vector (p2GWF7,0; p2FGW7,0; p2GWR7.0; pK7RWG2,0) obtained from Plant System Biology (Ghent University) using the LR clonase II kit (Invitrogen). Each coding sequence was cloned up and downstream from the FP sequence in the expression plasmid in order to produce a C- or N-terminal FP-fused protein, respectively. Cloning and expression plasmids were amplified in Escherichia coli One shot DG1 cells (Invitrogen) and purified from an overnight culture using the NucleoSpin plasmid purification kit (Macherey-Nagel). Constructs in binary expression vectors were transformed in Agrobacterium strain C58 for transient or stable transformation.
To build the mito-mCherry line, cDNA encoding the mitochondrial presequence from Nicotiana plumbaginifolia β-ATPase (nucleotides 387 to 666) (Chaumont et al., 1994) was first PCR-amplified using primers containing 5′-BamHI and 3′-SpeI restriction enzyme sites. pRSETb mCherry (Shaner et al., 2004) was obtained from the Tsien Laboratory (University of California, San Diego, CA). The mCherry sequence was PCR-amplified using a 5′ primer containing a 5′-SpeI site and a 3′ primer containing a 3′-SacI site and a stop codon. The presequence cDNA and the mCherry gene were ligated into the BamHI and SacI sites within pBIN121 downstream of the cauliflower mosaic virus 35S promoter in place of the β-glucuronidase gene to create pBIN β-ATPase-mCherry. Agrobacterium was transformed with this vector using standard techniques. The mito-mRFP1 line was generated by transformation with pDCL-mito-mRFP1. The pDCL-X-mRFP1 Gateway destination vector is a modified pMDC43 (Curtis and Grossniklaus, 2003) in which the coding sequence for mGFP6 was replaced by that for mRFP1 (Campbell et al., 2002) via pMDC7 and pMDC24. The pDCL-mito-mRFP1 vector was the product of recombination between pDCL-X-mRFP1 and a pENTR vector containing cDNA encoding the N. plumbaginifolia β-ATPase presequences as described above.
Analysis of GFP Fusion Proteins
The purification and the analysis of GFP-tagged proteins were performed as described (Candat et al., 2013). The samples were separated by SDS-PAGE on a 12% acrylamide gel. They were then electroblotted onto Immobilon polyvinylidene fluoride membrane (Millipore Merck) at 100 V for 1 h, in 10 mM CAPS, pH 11, and 10% (v/v) methanol. For protein gel blot analysis, the membrane was rinsed with deionized water, then blocked for 1 h in TBS buffer (50 mM Tris HCl, pH 7.4, and 150 mM NaCl) containing 1.5% (v/v) Tween 20. The membrane was then incubated for 1 h with a monoclonal anti-GFP antibody conjugated to horseradish peroxidase (Miltenyi Biotec) diluted 1:5000 in TBS buffer containing 0.05% (v/v) Tween 20. The membrane was then washed three times for 10 min with the same buffer. Immunodetection was performed by incubating the membrane for 1 min in 100 mM Tris, pH 8.5, containing 0.4 mM para-hydroxy coumarin acid, 2.5 mM luminol, and 5.43 mM H2O2. Chemiluminescence was monitored by a molecular imager Chemidoc TM XRS system (Bio-Rad).
Bioinformatics Analyses
Subcellular localizations were predicted by the following freely available online programs: PSORT (http://psort.hgc.jp/form.html; Nakai and Horton, 1999), MultiLoc (http://abi.inf.uni-tuebingen.de/Services/MultiLoc; Höglund et al., 2006), YLoc (http://abi.inf.uni-tuebingen.de/Services/YLoc/webloc.cgi; Briesemeister et al., 2010), SubLoc v1.0 (Hua and Sun, 2001), TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/; Emanuelsson et al., 2000), Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html; Small et al., 2004), Protein Prowler v1.2 (http://bioinf.scmb.uq.edu.au/pprowler_webapp_1-2/; Bodén and Hawkins, 2005), and Mitopred (Guda et al., 2004). Default settings were used and plant proteins data sets were selected when available. Predictions were represented as heat map using the Heatmap Builder 1.0 program (King et al., 2005; http://ashleylab.stanford.edu/tools/tools-scripts.html). Helical wheel analyses were performed using the screening module of HeliQuest Web server (Gautier et al., 2008). This module allows the user to screen databases in order to find sequences that have the general physico-chemical features of a target α-helix sequence. We used a personal LEA protein database comprising the 51 Arabidopsis LEA proteins, the mitochondrial pea (Pisum sativum) LEAM protein (accession CAF32327), and its ortholog from Artemia franciscana (accession ACM16586). The module was modified to incorporate new parameters specific to LEAM protein that exhibits class A α-helix motifs (Tolleter et al., 2007). We used a sliding 36-residue window to scan each of the protein sequence for the presence of segments exhibiting class A α-helix motifs. A protein was selected if at least one of its segments fulfilled a set of sequence parameters and physico-chemical criteria comparable to the class A α-helix segments in LEAM protein. Segments containing at least one cysteine (possibly engaged in a disulfide bridge) and/or a proline (a helix-breaker) were excluded. The mean hydrophobicity (H) and the mean hydrophobic moment (µH) (Eisenberg et al., 1982), calculated with the Fauchere-Pliska hydrophobicity scale (Fauchere and Pliska, 1983), the sum (NSTG) of serine, threonine, and glycine residues, the minimum number (Npol) of uncharged polar residues (Ser, Thr, Asn, and Gln), the minimum number of alanine (NA), the number (Ncha) of charged residues at pH 7.4 (Asp, Glu, Lys, and Arg), and the net charge (z) of the sequence must be within the following ranges: −0.4 ≤ H ≤ 0.05; 0.2 ≤ µH ≤ 0.4 ; NSTG ≥ 2; Npol ≥ 22 ; NA ≥ 3; 10 ≤ Ncha ≤ 20; –2 ≥ z ≥ 1. The HeliQuest server generated helical projections for each positive segment that were visually examined to validate the class A α-helix predictions.
Phylogenetic Analysis
A rectangular cladogram of the 51 protein sequences was obtained using the Dendroscope 3.1.0 software (Huson and Scornavacca, 2012) and a multiple alignment generated with ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The alignment (Supplemental Data Set 1) was generated using Gonnet protein weight matrix (open gap = 10, gap extension = 0.20, gap distances = 5, no end gaps, no iteration, neighbor-joining clustering).
Accession Numbers
Accession numbers from this article can be found in Supplemental Table 3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Heat Map of Subcellular Localization Predictions of the 51 Arabidopsis LEA Proteins.
Supplemental Figure 2. Transient Expression of Cytosolic LEA Protein in Arabidopsis Protoplasts.
Supplemental Figure 3. Diffusion of Cytosolic LEA-GFP Proteins in the Nucleus.
Supplemental Figure 4. Protein Gel Blot Analysis of LEA Protein Expressed in Protoplasts.
Supplemental Figure 5. Cytosolic Localization of GFP-LEA Fusions for Proteins Localized in Mitochondria or Plastid When Expressed as LEA-GFP Fusions (See Figures 3 and 4).
Supplemental Figure 6. Leaf Cells from Arabidopsis Transgenic Lines Expressing LEA Proteins Targeted to Mitochondria or Dual Targeted to Mitochondria and Plastid.
Supplemental Figure 7. LEA13-RFP Does Not Colocalize with Golgi in Arabidopsis Protoplasts.
Supplemental Figure 8. Arabidopsis Leaf Cells from Transgenic Lines Expressing LEA Proteins Targeted to the Secretory Pathway or the Pexophagosome.
Supplemental Figure 9. Transient Expression in Arabidopsis Protoplasts of GFP-LEA Fusions for Proteins Which Are Targeted to the Secretory Pathway as LEA-GFP Fusions.
Supplemental Figure 10. Vacuolar and Golgi Localization of LEA 21 in Arabidopsis Protoplasts.
Supplemental Figure 11. Localization of LEA21-GFP and LEA22-GFP in Arabidopsis Protoplasts.
Supplemental Figure 12. LEA11 and LEA12 Sequence Alignment.
Supplemental Figure 13. Estimation of Prediction Software Efficiency.
Supplemental Figure 14. Cytosolic Localization of LEA7 and LEA29 in Arabidopsis Protoplasts.
Supplemental Figure 15. Proteomic Quantification of LEA Proteins in Different Organs of Arabidopsis.
Supplemental Figure 16. LEA2 and LEA38 Sequence Alignment.
Supplemental Figure 17. Modeling of Class A α-Helix Motif of LEA Proteins.
Supplemental Figure 18. Modeling of Class A-Like α-Helix Motif of LEA23 and LEA24.
Supplemental Table 1. Theoretical and Apparent Molecular Masses of LEA and Fusion Proteins.
Supplemental Table 2. Quantitative Data about Transiently Transformed Protoplasts.
Supplemental Table 3. Arabidopsis LEA Templates Used for PCR Amplification.
Supplemental Table 4. Primers Used for the Construction of LEA Gene Fusions and Markers.
Supplemental Data Set 1. ClustalW2 Alignment of the 51 LEA Proteins.
Supplementary Material
Acknowledgments
This research was supported by a grant from ANR Blanc MITOZEN (ANR-12-BSV8-0021-01). We thank Pauline Poupart (IRHS Angers) and Jean-Pierre Andrieu (IBS, Grenoble) for N-terminal sequence determination and Marjorie Juchaux and Fabienne Simonneau (IMAC QUASAV, Angers) for assistance with the confocal microscope.
AUTHOR CONTRIBUTIONS
D.M., M.-H.A.-M., and D.C.L. designed and supervised the research. A.C., M.N., G.P., D.C.L., M.-H.A.-M., and D.M. performed experiments. R.G. performed the Heliquest analysis. D.M. and A.C. wrote the article, and all authors contributed to the editing.
Glossary
- ER
endoplasmic reticulum
- ES
endomembrane system
- BFA
Brefeldin A
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abu-Abied M., Golomb L., Belausov E., Huang S., Geiger B., Kam Z., Staiger C.J., Sadot E. (2006). Identification of plant cytoskeleton-interacting proteins by screening for actin stress fiber association in mammalian fibroblasts. Plant J. 48: 367–379. [DOI] [PubMed] [Google Scholar]
- Alexandersson E., Saalbach G., Larsson C., Kjellbom P. (2004). Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol. 45: 1543–1556. [DOI] [PubMed] [Google Scholar]
- Alsheikh M.K., Svensson J.T., Randall S.K. (2005). Phosphorylation regulated ion-binding is a property shared by the acidic subclass dehydrins. Plant Cell Environ. 28: 1114–1122. [Google Scholar]
- Bae M.S., Cho E.J., Choi E.Y., Park O.K. (2003). Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 36: 652–663. [DOI] [PubMed] [Google Scholar]
- Baerenfaller K., Hirsch-Hoffmann M., Svozil J., Hull R., Russenberger D., Bischof S., Lu Q., Gruissem W., Baginsky S. (2011). pep2pro: a new tool for comprehensive proteome data analysis to reveal information about organ-specific proteomes in Arabidopsis thaliana. Integr. Biol. (Camb.) 3: 225–237. [DOI] [PubMed] [Google Scholar]
- Bardel J., Louwagie M., Jaquinod M., Jourdain A., Luche S., Rabilloud T., Macherel D., Garin J., Bourguignon J. (2002). A survey of the plant mitochondrial proteome in relation to development. Proteomics 2: 880–898. [DOI] [PubMed] [Google Scholar]
- Benschop J.J., Mohammed S., O’Flaherty M., Heck A.J.R., Slijper M., Menke F.L.H. (2007). Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6: 1198–1214. [DOI] [PubMed] [Google Scholar]
- Bies-Ethève N., Gaubier-Comella P., Debures A., Lasserre E., Jobet E., Raynal M., Cooke R., Delseny M. (2008). Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 67: 107–124. [DOI] [PubMed] [Google Scholar]
- Bodén M., Hawkins J. (2005). Prediction of subcellular localization using sequence-biased recurrent networks. Bioinformatics 21: 2279–2286. [DOI] [PubMed] [Google Scholar]
- Borrell A., Cutanda M.C., Lumbreras V., Pujal J., Goday A., Culiáñez-Macià F.A., Pagès M. (2002). Arabidopsis thaliana atrab28: a nuclear targeted protein related to germination and toxic cation tolerance. Plant Mol. Biol. 50: 249–259. [DOI] [PubMed] [Google Scholar]
- Boucher V., Buitink J., Lin X., Boudet J., Hoekstra F.A., Hundertmark M., Renard D., Leprince O. (2010). MtPM25 is an atypical hydrophobic late embryogenesis-abundant protein that dissociates cold and desiccation-aggregated proteins. Plant Cell Environ. 33: 418–430. [DOI] [PubMed] [Google Scholar]
- Briesemeister S., Rahnenführer J., Kohlbacher O. (2010). YLoc—an interpretable web server for predicting subcellular localization. Nucleic Acids Res. 38: W497-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99: 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candat A., Poupart P., Andrieu J.-P., Chevrollier A., Reynier P., Rogniaux H., Avelange-Macherel M.-H., Macherel D. (2013). Experimental determination of organelle targeting-peptide cleavage sites using transient expression of green fluorescent protein translational fusions. Anal. Biochem. 434: 44–51. [DOI] [PubMed] [Google Scholar]
- Carrie C., Giraud E., Whelan J. (2009). Protein transport in organelles: Dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 276: 1187–1195. [DOI] [PubMed] [Google Scholar]
- Casadio R., Martelli P.L., Pierleoni A. (2008). The prediction of protein subcellular localization from sequence: a shortcut to functional genome annotation. Brief. Funct. Genomics Proteomics 7: 63–73. [DOI] [PubMed] [Google Scholar]
- Chakrabortee S., Tripathi R., Watson M., Schierle G.S., Kurniawan D.P., Kaminski C.F., Wise M.J., Tunnacliffe A. (2012). Intrinsically disordered proteins as molecular shields. Mol. Biosyst. 8: 210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F., Silva Filho Mde.C., Thomas D., Leterme S., Boutry M. (1994). Truncated presequences of mitochondrial F1-ATPase beta subunit from Nicotiana plumbaginifolia transport CAT and GUS proteins into mitochondria of transgenic tobacco. Plant Mol. Biol. 24: 631–641. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J., De Rycke R., Botterman J. (1992). Plant and mammalian sorting signals for protein retention in the endoplasmic reticulum contain a conserved epitope. EMBO J. 11: 2345–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Laskey R.A. (1986). Protein import into the cell nucleus. Annu. Rev. Cell Biol. 2: 367–390. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R.M., Terwilliger T.C. (1982). The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299: 371–374. [DOI] [PubMed] [Google Scholar]
- Elmore J.M., Liu J., Smith B., Phinney B., Coaker G. (2012). Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol. Cell. Proteomics 11: 014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Eriksson S.K., Kutzer M., Procek J., Gröbner G., Harryson P. (2011). Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell 23: 2391–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauchere J., Pliska V. (1983). Hydrophobic parameters of amino acid side-chains from the partitioning of N-acetyl aminoacid amide. Eur. J. Med. Chem. 8: 369–375. [Google Scholar]
- Ferro M., et al. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9: 1063–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38: D211–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz G., Hatzopoulos P., Jones T.J., Krauss M., Sung Z.R. (1989). Molecular and genetic analysis of an embryonic gene, DC 8, from Daucus carota L. Mol. Gen. Genet. 218: 143–151. [DOI] [PubMed] [Google Scholar]
- Froehlich J.E., Wilkerson C.G., Ray W.K., McAndrew R.S., Osteryoung K.W., Gage D.A., Phinney B.S. (2003). Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J. Proteome Res. 2: 413–425. [DOI] [PubMed] [Google Scholar]
- Gallardo K., Job C., Groot S.P.C., Puype M., Demol H., Vandekerckhove J., Job D. (2002). Proteomics of Arabidopsis seed germination. A comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol. 129: 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier R., Douguet D., Antonny B., Drin G. (2008). HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics 24: 2101–2102. [DOI] [PubMed] [Google Scholar]
- Goyal K., Pinelli C., Maslen S.L., Rastogi R.K., Stephens E., Tunnacliffe A. (2005a). Dehydration-regulated processing of late embryogenesis abundant protein in a desiccation-tolerant nematode. FEBS Lett. 579: 4093–4098. [DOI] [PubMed] [Google Scholar]
- Goyal K., Walton L.J., Tunnacliffe A. (2005b). LEA proteins prevent protein aggregation due to water stress. Biochem. J. 388: 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham I.A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59: 115–142. [DOI] [PubMed] [Google Scholar]
- Grelet J., Benamar A., Teyssier E., Avelange-Macherel M.H., Grunwald D., Macherel D. (2005). Identification in pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiol. 137: 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda C., Guda P., Fahy E., Subramaniam S. (2004). MITOPRED: a web server for the prediction of mitochondrial proteins. Nucleic Acids Res. 32: W372-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M., Fujinaga M., Kuboi T. (2004). Radical scavenging activity and oxidative modification of citrus dehydrin. Plant Physiol. Biochem. 42: 657–662. [DOI] [PubMed] [Google Scholar]
- Hawes C., Saint-Jore C., Martin B., Zheng H.Q. (2001). ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP! Trends Plant Sci. 6: 245–246. [DOI] [PubMed] [Google Scholar]
- Heazlewood J.L., Tonti-Filippini J.S., Gout A.M., Day D.A., Whelan J., Millar A.H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood J.L., Verboom R.E., Tonti-Filippini J., Small I., Millar A.H. (2007). SUBA: The Arabidopsis subcellular database. Nucleic Acids Res. 35: D213–D218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde R.S., Bernstein H.D. (2006). The surprising complexity of signal sequences. Trends Biochem. Sci. 31: 563–571. [DOI] [PubMed] [Google Scholar]
- Heiland I., Erdmann R. (2005). Biogenesis of peroxisomes. Topogenesis of the peroxisomal membrane and matrix proteins. FEBS J. 272: 2362–2372. [DOI] [PubMed] [Google Scholar]
- Höglund A., Dönnes P., Blum T., Adolph H.W., Kohlbacher O. (2006). MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics 22: 1158–1165. [DOI] [PubMed] [Google Scholar]
- Hua S., Sun Z. (2001). Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17: 721–728. [DOI] [PubMed] [Google Scholar]
- Hunault G., Jaspard E. (2010). LEAPdb: a database for the late embryogenesis abundant proteins. BMC Genomics 11: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M., Hincha D.K. (2008). LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Scornavacca C. (2012). Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Ito J., Batth T.S., Petzold C.J., Redding-Johanson A.M., Mukhopadhyay A., Verboom R., Meyer E.H., Millar A.H., Heazlewood J.L. (2011). Analysis of the Arabidopsis cytosolic proteome highlights subcellular partitioning of central plant metabolism. J. Proteome Res. 10: 1571–1582. [DOI] [PubMed] [Google Scholar]
- Iturriaga G., Schneider K., Salamini F., Bartels D. (1992). Expression of desiccation-related proteins from the resurrection plant Craterostigma plantagineum in transgenic tobacco. Plant Mol. Biol. 20: 555–558. [DOI] [PubMed] [Google Scholar]
- Jaspard E., Macherel D., Hunault G. (2012). Computational and statistical analyses of amino acid usage and physico-chemical properties of the twelve late embryogenesis abundant protein classes. PLoS ONE 7: e36968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen A.B., Goday A., Figueras M., Jessop A.C., Pagès M. (1998). Phosphorylation mediates the nuclear targeting of the maize Rab17 protein. Plant J. 13: 691–697. [DOI] [PubMed] [Google Scholar]
- Joshi H.J., et al. (2011). MASCP Gator: an aggregation portal for the visualization of Arabidopsis proteomics data. Plant Physiol. 155: 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath N.F., Kierszniowska S., Lorek J., Bourdais G., Kessler S.A., Shimosato-Asano H., Grossniklaus U., Schulze W.X., Robatzek S., Panstruga R. (2010). PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 285: 39140–39149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J.Y., et al. (2005). Pathway analysis of coronary atherosclerosis. Physiol. Genomics 23: 103–118. [DOI] [PubMed] [Google Scholar]
- Kleffmann T., Russenberger D., von Zychlinski A., Christopher W., Sjölander K., Gruissem W., Baginsky S. (2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14: 354–362. [DOI] [PubMed] [Google Scholar]
- Koag M.C., Fenton R.D., Wilkens S., Close T.J. (2003). The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 131: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koag M.C., Wilkens S., Fenton R.D., Resnik J., Vo E., Close T.J. (2009). The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 150: 1503–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R.P.W., Siu S.O., Lee S.S.M., Lo C., Chu I.K. (2011). Development of online high-/low-pH reversed-phase-reversed-phase two-dimensional liquid chromatography for shotgun proteomics: a reversed-phase-strong cation exchange-reversed-phase approach. J. Chromatogr. A 1218: 3681–3688. [DOI] [PubMed] [Google Scholar]
- Kovacs D., Kalmar E., Torok Z., Tompa P. (2008). Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 147: 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger C., Berkowitz O., Stephan U.W., Hell R. (2002). A metal-binding member of the late embryogenesis abundant protein family transports iron in the phloem of Ricinus communis L. J. Biol. Chem. 277: 25062–25069. [DOI] [PubMed] [Google Scholar]
- Lazarow P.B. (2003). Peroxisome biogenesis: advances and conundrums. Curr. Opin. Cell Biol. 15: 489–497. [DOI] [PubMed] [Google Scholar]
- Li B., Takahashi D., Kawamura Y., Uemura M. (2012a). Comparison of plasma membrane proteomic changes of Arabidopsis suspension-cultured cells (T87 Line) after cold and ABA treatment in association with freezing tolerance development. Plant Cell Physiol. 53: 543–554. [DOI] [PubMed] [Google Scholar]
- Li S., Chakraborty N., Borcar A., Menze M.A., Toner M., Hand S.C. (2012b). Late embryogenesis abundant proteins protect human hepatoma cells during acute desiccation. Proc. Natl. Acad. Sci. USA 109: 20859–20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Thomashow M.F. (1992). DNA Sequence analysis of a complementary DNA for cold-regulated Arabidopsis gene COR15 and characterization of the COR 15 polypeptide. Plant Physiol. 99: 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion J., Bach L., Bellec Y., Meyer C., Gissot L., Faure J.D. (2008). Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 56: 169–179. [DOI] [PubMed] [Google Scholar]
- Marmagne A., Ferro M., Meinnel T., Bruley C., Kuhn L., Garin J., Barbier-Brygoo H., Ephritikhine G. (2007). A high content in lipid-modified peripheral proteins and integral receptor kinases features in the arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 6: 1980–1996. [DOI] [PubMed] [Google Scholar]
- Mathur J., Mathur N., Hülskamp M. (2002). Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol. 128: 1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menze M.A., Boswell L., Toner M., Hand S.C. (2009). Occurrence of mitochondria-targeted Late Embryogenesis Abundant (LEA) gene in animals increases organelle resistance to water stress. J. Biol. Chem. 284: 10714–10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S.K., Walters B.T., Clouse S.D., Goshe M.B. (2009). An efficient organic solvent based extraction method for the proteomic analysis of Arabidopsis plasma membranes. J. Proteome Res. 8: 2752–2767. [DOI] [PubMed] [Google Scholar]
- Mowla S.B., Cuypers A., Driscoll S.P., Kiddle G., Thomson J., Foyer C.H., Theodoulou F.L. (2006). Yeast complementation reveals a role for an Arabidopsis thaliana late embryogenesis abundant (LEA)-like protein in oxidative stress tolerance. Plant J. 48: 743–756. [DOI] [PubMed] [Google Scholar]
- Mundy J., Chua N.H. (1988). Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J. 7: 2279–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K., Horton P. (1999). PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24: 34–36. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Okawa K., Kakizaki T., Inaba T. (2008). Evaluation of the protective activities of a late embryogenesis abundant (LEA) related protein, Cor15am, during various stresses in vitro. Biosci. Biotechnol. Biochem. 72: 1642–1645. [DOI] [PubMed] [Google Scholar]
- NDong C., Danyluk J., Wilson K.E., Pocock T., Huner N.P., Sarhan F. (2002). Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular characterization and functional analyses. Plant Physiol. 129: 1368–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A., Ritzenthaler C., Robinson D.G. (2002). Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol. 130: 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1–6. [DOI] [PubMed] [Google Scholar]
- Nikolovski N., Rubtsov D., Segura M.P., Miles G.P., Stevens T.J., Dunkley T.P.J., Munro S., Lilley K.S., Dupree P. (2012). Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol. 160: 1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M.S., Herder R., Schulze J., Jung R., Staehelin L.A. (2006). The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18: 2567–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons H.T., et al. (2012). Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 159: 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J.B., Cai Y., Sun Q., Zabrouskov V., Giacomelli L., Rudella A., Ytterberg A.J., Rutschow H., van Wijk K.J. (2006). The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics 5: 114–133. [DOI] [PubMed] [Google Scholar]
- Popova A.V., Hundertmark M., Seckler R., Hincha D.K. (2011). Structural transitions in the intrinsically disordered plant dehydration stress protein LEA7 upon drying are modulated by the presence of membranes. Biochim. Biophys. Acta 1808: 1879–1887. [DOI] [PubMed] [Google Scholar]
- Pouchkina-Stantcheva N.N., McGee B.M., Boschetti C., Tolleter D., Chakrabortee S., Popova A.V., Meersman F., Macherel D., Hincha D.K., Tunnacliffe A. (2007). Functional divergence of former alleles in an ancient asexual invertebrate. Science 318: 268–271. [DOI] [PubMed] [Google Scholar]
- Rahman L.N., Chen L., Nazim S., Bamm V.V., Yaish M.W., Moffatt B.A., Dutcher J.R., Harauz G. (2010). Interactions of intrinsically disordered Thellungiella salsuginea dehydrins TsDHN-1 and TsDHN-2 with membranes - synergistic effects of lipid composition and temperature on secondary structure. Biochem. Cell Biol. 88: 791–807. [DOI] [PubMed] [Google Scholar]
- Rahman L.N., McKay F., Giuliani M., Quirk A., Moffatt B.A., Harauz G., Dutcher J.R. (2013). Interactions of Thellungiella salsuginea dehydrins TsDHN-1 and TsDHN-2 with membranes at cold and ambient temperatures-surface morphology and single-molecule force measurements show phase separation, and reveal tertiary and quaternary associations. Biochim. Biophys. Acta 1828: 967–980. [DOI] [PubMed] [Google Scholar]
- Reyes J.L., Rodrigo M.J., Colmenero-Flores J.M., Gil J.V., Garay-Arroyo A., Campos F., Salamini F., Bartels D., Covarrubias A.A. (2005). Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant Cell Environ. 28: 709–718. [Google Scholar]
- Riera M., Figueras M., López C., Goday A., Pagès M. (2004). Protein kinase CK2 modulates developmental functions of the abscisic acid responsive protein Rab17 from maize. Proc. Natl. Acad. Sci. USA 101: 9879–9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J.K., DeSimone N.A., Lingle W.L., Dure L., III (1993). Cellular concentrations and uniformity of cell-type accumulation of two LEA proteins in cotton embryos. Plant Cell 5: 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salleh F.M., Evans K., Goodall B., Machin H., Mowla S.B., Mur L.A.J., Runions J., Theodoulou F.L., Foyer C.H., Rogers H.J. (2012). A novel function for a redox-related LEA protein (SAG21/AtLEA5) in root development and biotic stress responses. Plant Cell Environ. 35: 418–429. [DOI] [PubMed] [Google Scholar]
- Sarry J.E., et al. (2006). The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics 6: 2180–2198. [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N.G., Palmer A.E., Tsien R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572. [DOI] [PubMed] [Google Scholar]
- Sharon M.A., Kozarova A., Clegg J.S., Vacratsis P.O., Warner A.H. (2009). Characterization of a group 1 late embryogenesis abundant protein in encysted embryos of the brine shrimp Artemia franciscana. Biochem. Cell Biol. 87: 415–430. [DOI] [PubMed] [Google Scholar]
- Shih M.D., Hoekstra F.A., Hsing Y.I.C. (2008). Late embryogenesis abundant proteins. Adv. Bot. Res. 48: 211–255. [Google Scholar]
- Shih M.D., Lin S.C., Hsieh J.S., Tsou C.H., Chow T.Y., Lin T.P., Hsing Y.I.C. (2004). Gene cloning and characterization of a soybean (Glycine max L.) LEA protein, GmPM16. Plant Mol. Biol. 56: 689–703. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Kanamori Y., Furuki T., Kikawada T., Okuda T., Takahashi T., Mihara H., Sakurai M. (2010). Desiccation-induced structuralization and glass formation of group 3 late embryogenesis abundant protein model peptides. Biochemistry 49: 1093–1104. [DOI] [PubMed] [Google Scholar]
- Small I., Peeters N., Legeai F., Lurin C. (2004). Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590. [DOI] [PubMed] [Google Scholar]
- Steponkus P.L., Uemura M., Joseph R.A., Gilmour S.J., Thomashow M.F. (1998). Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95: 14570–14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Rikkerink E.H.A., Jones W.T., Uversky V.N. (2013). Multifarious roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell 25: 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson J., Palva E.T., Welin B. (2000). Purification of recombinant Arabidopsis thaliana dehydrins by metal ion affinity chromatography. Protein Expr. Purif. 20: 169–178. [DOI] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S.I., Watanabe M., Nishimura M., Hara-Nishimura I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35: 545–555. [DOI] [PubMed] [Google Scholar]
- Teh O.K., Moore I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448: 493–496. [DOI] [PubMed] [Google Scholar]
- Teixeira P.F., Glaser E. (2013). Processing peptidases in mitochondria and chloroplasts. Biochim. Biophys. Acta 1833: 360–370. [DOI] [PubMed] [Google Scholar]
- Thalhammer A., Hundertmark M., Popova A.V., Seckler R., Hincha D.K. (2010). Interaction of two intrinsically disordered plant stress proteins (COR15A and COR15B) with lipid membranes in the dry state. Biochim. Biophys. Acta 1798: 1812–1820. [DOI] [PubMed] [Google Scholar]
- Tolleter D., Hincha D.K., Macherel D. (2010). A mitochondrial late embryogenesis abundant protein stabilizes model membranes in the dry state. Biochim. Biophys. Acta 1798: 1926–1933. [DOI] [PubMed] [Google Scholar]
- Tolleter D., Jaquinod M., Mangavel C., Passirani C., Saulnier P., Manon S., Teyssier E., Payet N., Avelange-Macherel M.H., Macherel D. (2007). Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. Plant Cell 19: 1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. (2002). Intrinsically unstructured proteins. Trends Biochem. Sci. 27: 527–533. [DOI] [PubMed] [Google Scholar]
- Tripathi R., Boschetti C., McGee B., Tunnacliffe A. (2012). Trafficking of bdelloid rotifer late embryogenesis abundant proteins. J. Exp. Biol. 215: 2786–2794. [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L., Timmerman E., De Bock P.J., Vercammen D., Stael S., van de Cotte B., Staes A., Goethals M., Beunens T., Van Damme P., Gevaert K., Van Breusegem F. (2013). The Arabidopsis metacaspase9 degradome. Plant Cell 25: 2831–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe, A., Hincha, D.K., Leprince, O., and Macherel, D. (2010). LEA proteins: versatility of form and function. In Sleeping Beauties: Dormancy and Resistance in Harsh Environments, E. Lubzens, J. Cerda, and M. Clark eds (Berlin: Springer), pp. 91–108. [Google Scholar]
- Tunnacliffe A., Wise M.J. (2007). The continuing conundrum of the LEA proteins. Naturwissenschaften 94: 791–812. [DOI] [PubMed] [Google Scholar]
- Ukaji N., Kuwabara C., Takezawa D., Arakawa K., Fujikawa S. (2001). Cold acclimation-induced WAP27 localized in endoplasmic reticulum in cortical parenchyma cells of mulberry tree was homologous to group 3 late-embryogenesis abundant proteins. Plant Physiol. 126: 1588–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutphen T., van der Klei I.J., Kiel J.A.K.W. (2008). Pexophagy in Hansenula polymorpha. Methods Enzymol. 451: 197–215. [DOI] [PubMed] [Google Scholar]
- Vitale A., Denecke J. (1999). The endoplasmic reticulum-gateway of the secretory pathway. Plant Cell 11: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman S.A., Serazetdinova L., Jones A.M.E., Sanders D., Rathjen J., Peck S.C., Maathuis F.J. (2008). Identification of novel proteins and phosphorylation sites in a tonoplast enriched membrane fraction of Arabidopsis thaliana. Proteomics 8: 3536–3547. [DOI] [PubMed] [Google Scholar]
- Wolkers W.F., McCready S., Brandt W.F., Lindsey G.G., Hoekstra F.A. (2001). Isolation and characterization of a D-7 LEA protein from pollen that stabilizes glasses in vitro. Biochim. Biophys. Acta 1544: 196–206. [DOI] [PubMed] [Google Scholar]
- Xu J., Zhang Y.X., Guan Z.Q., Wei W., Han L., Chai T.Y. (2008). Expression and function of two dehydrins under environmental stresses in Brassica juncea L. Mol. Breed. 21: 431–438. [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhao P., Liu F., Ma M., Gong J., Wang Q., Jia P., Zheng G., Liu H. (2011). Overexpression of AtLEA3-3 confers resistance to cold stress in Escherichia coli and provides enhanced osmotic stress tolerance and ABA sensitivity in Arabidopsis thaliana. Mol. Biol. (Mosk.) 45: 851–862. [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.