Abstract
Hantavirus causes two important rodent-borne viral zoonoses, hemorrhagic fever with renal syndrome (HFRS) in Eurasia and hantavirus pulmonary syndrome (HPS) in North and South America. Twenty-four species that represent sero- and genotypes have been registered within the genus Hantavirus by the International Committee on Taxonomy of Viruses (ICTV). Among the viral proteins, nucleocapsid (N) protein possesses an immunodominant antigen. The antigenicitiy of N protein is conserved compared with that of envelope glycoproteins. Therefore, N protein has been used for serological diagnoses and seroepidemiological studies. An understanding of the antigenic properties of N protein is important for the interpretation of results from serological tests using N antigen. N protein consists of about 430 amino acids and possesses various epitopes. The N-terminal quarter of N protein bears linear and immunodominant epitopes. However, a serotype-specific and multimerization-dependent antigenic site was found in the C-terminal half of N protein. In this paper, the structure, function, and antigenicity of N protein are reviewed.
Keywords: nucleocapsid, monoclonal antibody, epitope
1. Introduction
Species of the genus Hantavirus are classified in the family Bunyaviridae. Rodents, shrews, and bats are persistently infected with hantaviruses, whereas humans are infected accidentally, occasionally causing severe illness. Among rodent-borne hantaviruses, the causative agents of two important zoonoses are known: hemorrhagic fever with renal syndrome (HFRS) and Hantavirus pulmonary syndrome (HPS) [1,2].
Like other viral members of the Bunyaviridae family, hantaviruses are enveloped RNA viruses that contain three-segmented negative-sense RNAs, designated S, M, and L based on the molecular weight of their virion. The S, M, and L RNA segments encode nucleocapsid protein (N), envelope glycoproteins (Gn and Gc), and RNA-dependent RNA polymerase (L) protein, respectively [3]. Nonstructural (NS) protein was detected in the S genome segment of some hantaviruses as a candidate interferon antagonist [4,5,6]. Among the structural proteins, N protein is the most abundant in the hantavirus virion and accumulates in the cytoplasm of infected cells. Since N protein is immunodominant, diverse recombinant N proteins produced by various expression systems have been applied as diagnostic antigens to detect hantavirus-specific antibody (US patent number: 5614193) [7,8,9,10].
In addition to its value as a diagnostic antigen, N protein exhibits RNA-binding activity [11,12,13] and multimerization [14], which are crucial functions for the encapsidation of the viral genome in the virion [15]. Furthermore, N protein has been suggested to play an important role in the initiation of transcription and translation of the hantavirus genome. Thus, it is a multifunctional protein that contributes to not only virus encapsulation and assembly but also to the translation and transcription of genomic RNA to complete the viral lifecycle.
Each species of Hantavirus appears to have a single predominant rodent, shrew, mole, or bat species that serves as its natural reservoir [16,17,18,19,20], probably because of the co-evolution of hantaviruses with their animal reservoirs. Rodent-borne hantaviruses comprise three large groups according to their host rodents: Murinae-, Arvicolinae-, and Sigmodontinae/Neotominae-associated. Due to the close association between rodent species and hantaviruses, their geographical distributions are the same. Therefore, Murinae- and Arvicolinae-associated hantaviruses, which are distributed in Eurasia, are called Old World hantaviruses. Alternatively, Sigmodontinae/Neotominae-associated hantaviruses are distributed in North and South American countries and are called New World hantaviruses [21]. Causative agents of HFRS and HPS are exclusive to rodents. Murinae-associated hantaviruses include causative agents of HFRS: Hantaan virus (HTNV), Seoul virus (SEOV), and Dobrava virus (DOBV). Thailand virus (THAIV), which is carried by Bandicota indica, is suspected of being pathogenic to humans in Asia [22]. Arvicolinae-associated hantaviruses are distributed throughout Eurasia, and Puumala virus (PUUV) is a causative agent of HFRS that has been called nephropathia epidemica (NE) in Northern Europe. Numerous Arvicorinae rodent-borne hantaviruses have been reported from both the Old World and New World. Among them, only PUUV is known to be pathogenic. The group of Sigmodontinae/Neotominae-associated hantaviruses, Sin Nombre virus (SNV), Andes virus (ANDV), Laguna Negra virus (LANV), and variable HPS-related hantaviruses were found in North and South America [23,24]. Hantavirus N protein shares a common antigenic site with each group.
Here, we review the antigenic properties of epitopes on hantavirus N protein, particularly in relation to the structure, virus species and function.
2. Hantavirus N Protein
2.1. Antigenic Profiling of N Protein Using Monoclonal Antibodies (MAbs) and Polyclonal Antibodies
The initial research using MAbs against N protein of HTNV, SEOV, and PUUV was reported by Ruo and coauthors [25]. Most clones against HTNV and SEOV were cross-reactive against two viruses but not cross-reactive with PUUV. Also, one serotype-specific clone against HTNV was established. Yoshimatsu et al. confirmed the binding region of MAbs produced by Ruo et al. All clones excluding the serotype-specific form were found to bind with the N-terminal part of N [26]. On the other hand, Lundkvist and coauthors reported MAbs against PUUV [27]. All clones against PUUV exhibited unique epitopes distinct from HTNV and SEOV. Among eleven clones, six were PUUV‑specific. The clone 3H9 was a PUUV-specific antibody and its epitope was determined as amino acids (aa) 251 to 260 (VKPGTPAQEI) using the pepscan assay [28]. However, epitopes of other MAbs were not determined with the same assay [28]. Elgh et al. showed that most PUUV‑specific MAbs bind to 100 aa of the N terminus of PUUV N [29]. MAbs against N protein of New World hantaviruses and against ANDV and Carizale virus (CARV) [30,31] were reported. In those studies, group-common, genus-common, and serotype-specific epitopes were found within the N-terminal region of N protein. MAbs against the N-terminal 120 aa of N of PUUV showed cross‑reactivity against New World hantaviruses, and these antibodies were useful for detecting antigens in immunohistochemical assays [32]. These results obtained by using MAbs indicate that the N terminus of hantavirus N protein is immunodominant.
Thottapalayam virus (TPMV) is a prototype virus of shrew-borne hantavirus and is the most distinct form of rodent-borne hantaviruses [33]. Shigel et al. reported MAbs against TPMV [34]. All four clones were directed against the N-terminal region. These results indicate that, even in TPMV, the N terminus of N protein is immunodominant. Furthermore, the N-terminal region showed strong reactivity in a Western blot assay on staining by MAbs and polyclonal antisera. This indicated that the epitope found in the N terminus is linear and immundominant [34]. Similarly, antibody epitopes induced in HFRS and HPS patient sera and rodent-borne hantavirus-infected animals using a synthetic peptide antigen and/or truncated antigens were investigated. The results indicated that linear and immunodominant epitopes of N protein are also present in the N-terminal region even in these polyclonal antibodies [28,34,35,36,37,38,39]. Based on the results of epitope-mapping studies with MAb observations, MAbs against N protein were mostly produced against the N-terminal region of N protein of Old World, New World, and shrew-borne hantaviruses. These results reveal that the basic structure of N protein may be common among viruses within the genus Hantavirus.
2.2. Deduced Structure of the N-Terminal Region of N Protein
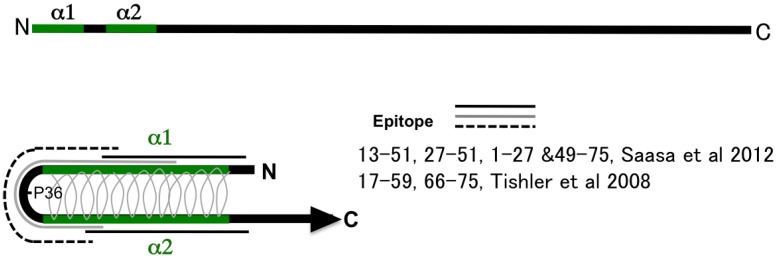
As shown in Figure 1, secondary structure prediction based on the deduced aa sequence, two α-helices in the N-terminal region (α1 and α2), was reported by Alfadhli et al. [40,41]. At present, the intramolecular coiled-coil structure is a model of the N-terminal region favored by several researchers [42,43]. A coiled-coil structure of aa 1 to 74 based on crystal structure analysis [42] and NMR [44] was reported. Therefore, aa 1-74 produced two long helices (α1 and α2) that intertwine into a coiled-coil domain [44]. A schema of 100 aa of the N terminus of N protein is presented in the figure. The aa 1-74 produced the antiparallel coiled-coil structure of α1 and α2 helices shown in Figure 1A. The conserved proline at the 36th position was a vertex of the structure [42,44]. Based on the model, Tischler and Saasa proposed an antigenic model of the N-terminal region [30,31]. They found several overlapping epitopes in the N-terminal region by employing a competitive binding assay of MAbs. These MAbs showed various antigen-binding profiles including group-common, genus-common, and serotype-specific profiles. However, the N-terminal region seems to have only group-common epitopes and not to have serotype-specific or genus-common epitopes in actual infection. The N-terminal 100 amino acid antigens showed a group-common binding profile and low cross-reactivities with patient sera. Sera from HFRS patients infected with PUUV showed extremely low or no cross-reactivities against HTNV and DOBV antigens and slight cross-reactivity against SNV antigen [38,39]. The N-terminal part of N of ANDV and PUUV showed low cross-reactivities between Sigmodontinae/Neotominae-borne and Microtinae-borne hantaviruses [45]. These observations indicate that the antigenic region shown with black and solid lines (1-27 & 49-75) mainly induced antibodies in patient and animal sera during actual infection. The mutant N protein lacking aa 1-35 (α1-deletion mutant) completely lost its antigenicity [26]. Using MAb binding to the N-terminal 100 aa of HTNV N protein (such as GBO4, ECO2, and ECO1), it was not possible to determine their epitopes by a peptide-scanning assay employing a previously described peptide‑scanning assay [46]. These results also indicate that the coiled-coil structure of the N terminus was the main structure involved in the antigenicity of N.
Figure 1.
Schema of the structure of the N terminus of hantavirus N and epitope mapping. Two α helices α1 and α2, of the N-terminal region are shown [42]. The model of the intramolecular antiparallel coiled-coil structure has gained much support. Regarding the structure, several epitopes were reported as genus-common, type-common, or a type‑specific epitopes [30,31]. The turn of this structure was proline at the 36th position.
2.3. Deduced Structure of the Central Part of N Protein
As shown in Figure 2, another report also showed that the central region of N protein was responsible for RNA-binding activity [12,13]. N protein may bind to hantaviral genomic RNA selectively rather than to general RNA; however, this has yet to be confirmed [11,47,48]. Yoshimatsu and coauthors reported that Region I (aa 100-125) was highly conserved among hantaviruses, and they identified an important region assisting in N-N homotypic interaction [49]. A variable region of N (aa 230-302) was identified in the central part of N protein (175-218) [50,51]. This region may be involved in the serotype-specific epitope of N protein.
Figure 2.

Schema of the central region of hantavirus N. Region 1 (aa 100-12) assists in N‑N homotypic interaction [49]. RNA-binding regions involving aa 175-218 [12] and 100 aa of the C terminus [11] are shown.
2.4. Deduced Structure of the C-Terminal Region of N Protein
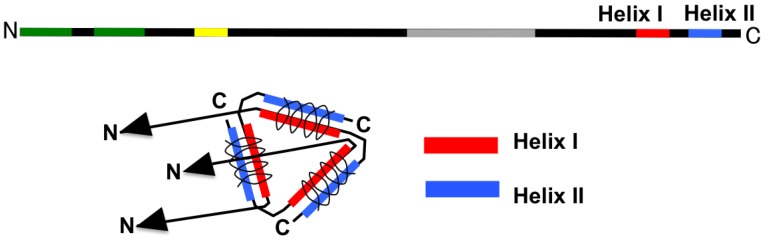
Two additional alpha-helical structures in the C-terminal region, Helix I and Helix II, were predicted by secondary structure analysis by Kaukinen et al. [52]. As shown in Figure 3, Helix I and Helix II contribute to the interaction with other N proteins through the intermolecular coiled-coil structure [52]. With deletion of Helix II from entire N, homotypic interaction did not occur in a yeast two-hybrid assay. On the other hand, N lacking Helix II showed interaction with entire N in the same assay [49]. These results also supported intermolecular interaction between Helix I and Helix II.
Figure 3.
Interaction of the hantavirus N by using the C-terminal part. The coiled coil of Helix I (around aa 381-384) and that of Helix II (around aa 413-414) are parallel, with α helices aligned in the same direction [52]. Three molecules formed trimerized N throughout the three intermolecular coiled-coil structures.
2.5. Deduced Structure of Trimerized N Protein
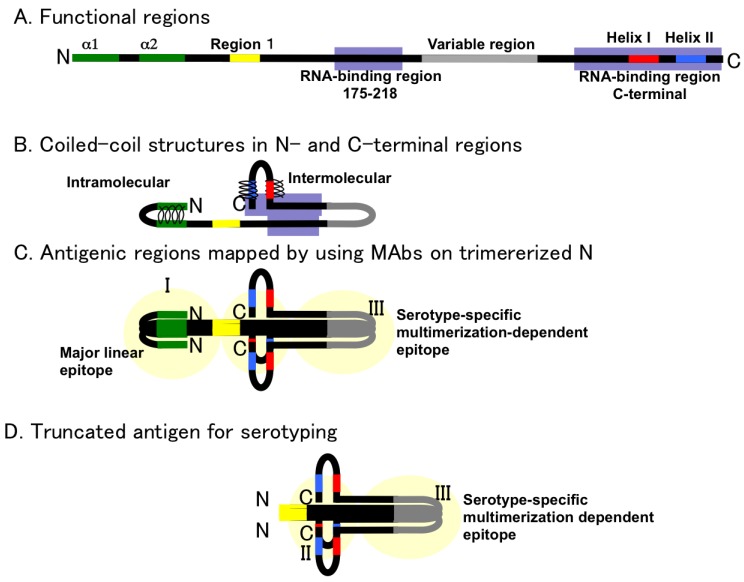
Mir and coauthors reported that N protein formed a trimer [53]. Based on their observations, a trimerized N protein is presented in Figure 4. In Figure 4A,B, a schema of N protein including coiled-coil interactions in the N and C terminal regions is presented. From epitope mapping of N protein using the competitive binding assay of MAbs, N- and C-terminal regions were in close proximity. Further, a serotype-specific epitope on N protein, which was expected to be located in the central region of N, was found as a projection from the N terminus [26]. According to these observations, an outline of N protein is proposed. In this schema, two RNA-binding regions are in close proximity. These two regions of N protein may cooperatively contribute to RNA binding. In Figure 4C, a schema of trimerized N is presented [54]. Antigenic regions previously designated as I, II, and III [26] were overlaid on the trimerized model. Yoshimatsu et al. showed that antigenic regions I and III were immunodominant in actual infection. MAb binding to regions I and III competed with patient sera and infected rodent sera. The antigenicity of region I was reproducible by both E. coli and a baculovirus expression system. On the other hand, the antigenicity of region III was reproducible only by the baculovirus expression system [26]. Two HTNV-specific MAb clones, BDO1 [25] and C24B4 [26], were able to bind to N protein expressed by the baculovirus vector. However, recombinant N protein expressed by E. coli [26] and the yeast expression system (unpublished observation) showed low binding activity with these HTNV-specific MAbs. To express serotype-specific epitopes among hantavirus N, the selection of an appropriate expression system is important. In Figure 4D, the concept of serotyping antigen based on truncation of N antigen is shown. Morii et al. proposed serotyping antigens designed with deletion of group-common and major linear epitopes from N antigen [55].
Figure 4.
A schema of trimerized hantavirus N. A. Functional regions of N were plotted on the primary structure of hantavirus N shown in Figure 1, Figure 2, and Figure 3. B. Interactions in the N-terminal and C-terminal part of hantavirus N. C. Antigenic regions mapped by poly- and monoclonal antibodies: From the competitive binding assay, two major antigenic regions were found in HTNV N. One was the N terminus (antigenic region I) and the other was the C terminus (antigenic region III). The central region was not a major antigenic site. Serotype-specific epitopes were found in the edge region and as discontinuous epitopes [26]. D. Concept of serotyping antigen based on truncation of N antigen. By deletion of group-common and major linear epitopes, serotyping antigens were designed [55].
2.6. Variety of Trimerization of N Protein Depending on the Viruses and Vectors
Multimerization of N is involved in the antigenicity of N. Most of the serotype-specific epitopes appear to be multimerization-dependent [49]. Authentic hantavirus N proteins in the virion should be multimerized. Native HTNV N proteins in inoculated Vero E6 cells were also detected as multimerized N proteins by competitive sandwich ELISA, as previously described [49]. However, the entire N protein of HTNV expressed in an E. coli vector system as described previously was detected as a monomer [26]. On the other hand, recombinant N proteins of SNV, HTNV, and SEOV showed an N-N interaction in yeast and mammalian two-hybrid assays [40,49]. These results indicate that the multimerization of recombinant N protein varied depending on the expression system. On the other hand, recombinant N proteins using the baculovirus expression system were more complicated. As shown in Table 1, all of the recombinant entire N proteins of SNV, ANDV, and LNV were monomeric. Hantavirus N proteins expressed in the baculovirus vector were detected as multimers. Although 155-429 aa of SEOV and DOBV N proteins were detected as multimers, 155-429 aa of HTNV were detected as a monomer. These results indicate that the region of N protein required for multimerization varies among hantaviruses in the baculovirus expression system. They also suggest that the reproducibility of N protein multimerization is dependent on the virus species. Furthermore, as shown in the results for New World hantaviruses except BCCV in Table 1, N-terminal region of the N protein inhibited N-N homotypic interaction.
Table 1.
Homotypic interactions of recombinant and truncated N proteins.
| Region (aa) | HTNV | SEOV | DOBV | THAIV | PUUV | SNV | ANDV | LANV | BCCV | CARV |
|---|---|---|---|---|---|---|---|---|---|---|
| 1-429 (Entire) |
M* | M* | M | M | M | S/M* | S | S | M | S |
| 50-429 | M* | M* | M | M | M | S | S* | S* | M | ND |
| 100-429 | M* | M* | M | ND | M | M | M | M | M | M |
| 155-429 | S | M | M | ND | M | S | ND | ND | ND | ND |
Multimerization of recombinant and truncated N proteins expressed by the baculovirus vector were examined by competitive binding assay [49,51,56]. M, multimerized N; S, monomeric N; ND, not done. * Furthermore, interactions between entire and/or truncated N proteins were confirmed using the yeast or mammalian two-hybrid assay [49]. BCCV, Black Creek Canal virus.
2.7. Association of Cellular Components with N
Interactions among hantavirus N proteins were reported as above. In addition to homotypic interaction of N, cellular proteins associated with N were reported. Although interactions of N protein and small ubiquitin-like modifier-1 (SUMO-1) and its E3 ligase Ubc9 and PIAS were reported, the region responsible for the interaction was the C-terminal region of N [57,58]. Because Nedd4-like ubiquitin ligase E3 was found to be associated with the budding of viral particles from the cellular membrane in the case of Lassa virus, filoviruses, and retroviruses [59,60,61,62], contribution of sumoylation-related enzymes to the budding process of hantavirus virus particles was expected. The binding of N and SUMO-1-related protein seemed to be multimerization-dependent [57]. Mir et al. also showed that multimerized N bound to the viral RNA panhandle [53]. These observations suggested that multimerized N might recruit a novel association with a cellular component. On the other hand, Cheng et al. reported interaction of N with the ribosomal protein L19 [63,64], and Ramanathan et al. showed N traffic in microtubules to the ER-Golgi intermediate compartment (ERGIC) [65,66]. Although regions responsible for N protein binding with L19 or ERGIC remain unclear, it is thought that these interactions participate in viral particle formation. An indirect interaction between N protein and the actin filament or Gn and Gc proteins may also be associated with the assembly of viral particles [67,68,69,70].
3. Conclusions
Hantavirus N protein is a major antigenic protein. Therefore, it has been important for serological diagnosis. At first glance, it appears simple. However, it has various epitopes such as genus-common, group-common, and serotype-specific epitopes. In addition to the cross-reactivity, both linear and discontinuous epitopes were found from analyses using monoclonal and polyclonal antibodies. Furthermore, the structure that constituted those epitopes has become clear. The N-terminus of N, which was constituted by an antiparallel coiled-coil structure, was found to be immunodominant. On the other hand, the C-terminal half of N, constituted by a parallel intermolecular coiled-coil structure, possessed serotype-specific and multimerization-dependent epitopes. The novel structure of N protein after multimerization might add a novel association with cellular proteins and/or RNA derived from host cells. Finally, reproducibility of epitopes of recombinant antigens is dependent on the expression system and viral species. By understanding the relation of the structure and antigenicity of N protein, a better diagnostic system will be constructed and interpretation of the results of serological tests will be more correct and informative.
Acknowledgments
This study was supported in part by JSPS KAKENHI Grant Number and the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases, MEXT, Japan. This work was also supported in part by Grants-in-Aid for Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labour and Welfare, Japan.
Conflicts of Interest
The authors declare no conflict of interest.
References and Notes
- 1.Schmaljohn C.S., Hasty S.E., Dalrymple J.M., LeDuc J.W., Lee H.W., von Bonsdorff C.H., Brummer-Korvenkontio M., Vaheri A., Tsai T.F., Regnery H.L., et al. Antigenic and genetic properties of viruses linked to hemorrhagic fever with renal syndrome. Science. 1985;227:1041–1044. doi: 10.1126/science.2858126. [DOI] [PubMed] [Google Scholar]
- 2.Nichol S.T., Spiropoulou C.F., Morzunov S., Rollin P.E., Ksiazek T.G., Feldmann H., Sanchez A., Childs J., Zaki S., Peters C.J. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 3.Schmaljohn C.S., Hasty S.E., Harrison S.A., Dalrymple J.M. Characterization hantaan virions, the prototype virus of hemorrhagic fever with renal syndrome. J. Infect. Dis. 1983;148:1005–1011. doi: 10.1093/infdis/148.6.1005. [DOI] [PubMed] [Google Scholar]
- 4.Jaaskelainen K.M., Kaukinen P., Minskaya E.S., Plyusnina A., Vapalahti O., Elliott R.M., Weber F., Vaheri A., Plyusnin A. Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J. Med. Virol. 2007;79:1527–1536. doi: 10.1002/jmv.20948. [DOI] [PubMed] [Google Scholar]
- 5.Virtanen J.O., Jaaskelainen K.M., Djupsjobacka J., Vaheri A., Plyusnin A. Tula hantavirus NSs protein accumulates in the perinuclear area in infected and transfected cells. Arch. Virol. 2010;155:117–121. doi: 10.1007/s00705-009-0546-y. [DOI] [PubMed] [Google Scholar]
- 6.Vera-Otarola J., Solis L., Soto-Rifo R., Ricci E.P., Pino K., Tischler N.D., Ohlmann T., Darlix J.L., Lopez-Lastra M. The Andes hantavirus NSs protein is expressed from the viral small mRNA by a leaky scanning mechanism. J. Virol. 2012;86:2176–2187. doi: 10.1128/JVI.06223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmaljohn C.S., Sugiyama K., Schmaljohn A.L., Bishop D.H. Baculovirus expression of the small genome segment of Hantaan virus and potential use of the expressed nucleocapsid protein as a diagnostic antigen. J. Gen. Virol. 1988;69:777–786. doi: 10.1099/0022-1317-69-4-777. [DOI] [PubMed] [Google Scholar]
- 8.Vapalahti O., Lundkvist A., Kallio-Kokko H., Paukku K., Julkunen I., Lankinen H., Vaheri A. Antigenic properties and diagnostic potential of puumala virus nucleocapsid protein expressed in insect cells. J. Clin. Microbiol. 1996;34:119–125. doi: 10.1128/jcm.34.1.119-125.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulrich R., Lundkvist A., Meisel H., Koletzki D., Sjolander K.B., Gelderblom H.R., Borisova G., Schnitzler P., Darai G., Kruger D.H. Chimaeric HBV core particles carrying a defined segment of Puumala hantavirus nucleocapsid protein evoke protectiveimmunity In an animal model. Vaccine. 1998;16:272–280. doi: 10.1016/S0264-410X(97)00172-2. [DOI] [PubMed] [Google Scholar]
- 10.Brus Sjolander K., Golovljova I., Plyusnin A., Lundkvist A. Diagnostic potential of puumala virus nucleocapsid protein expressed in Drosophila melanogaster cells. J. Clin. Microbiol. 2000;38:2324–2329. doi: 10.1128/jcm.38.6.2324-2329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gott P., Stohwasser R., Schnitzler P., Darai G., Bautz E.K. RNA binding of recombinant nucleocapsid proteins of hantaviruses. Virology. 1993;194:332–337. doi: 10.1006/viro.1993.1263. [DOI] [PubMed] [Google Scholar]
- 12.Xu X., Severson W., Villegas N., Schmaljohn C.S., Jonsson C.B. The RNA binding domain of the hantaan virus N protein maps to a central, conserved region. J. Virol. 2002;76:3301–3308. doi: 10.1128/JVI.76.7.3301-3308.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Severson W., Xu X., Kuhn M., Senutovitch N., Thokala M., Ferron F., Longhi S., Canard B., Jonsson C.B. Essential amino acids of the hantaan virus N protein in its interaction with RNA. J. Virol. 2005;79:10032–10039. doi: 10.1128/JVI.79.15.10032-10039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alminaite A., Backstrom V., Vaheri A., Plyusnin A. Oligomerization of hantaviral nucleocapsid protein: charged residues in the N-terminal coiled-coil domain contribute to intermolecular interactions. J. Gen. Virol. 2008;89:2167–2174. doi: 10.1099/vir.0.2008/004044-0. [DOI] [PubMed] [Google Scholar]
- 15.Kaukinen P., Vaheri A., Plyusnin A. Hantavirus nucleocapsid protein: A multifunctional molecule with both housekeeping and ambassadorial duties. Arch. Virol. 2005;150:1693–1713. doi: 10.1007/s00705-005-0555-4. [DOI] [PubMed] [Google Scholar]
- 16.Vaheri A., Strandin T., Hepojoki J., Sironen T., Henttonen H., Makela S., Mustonen J. Uncovering the mysteries of hantavirus infections. Nat. Rev. Microbiol. 2013;11:539–550. doi: 10.1038/nrmicro3066. [DOI] [PubMed] [Google Scholar]
- 17.Hughes A.L., Friedman R. Evolutionary diversification of protein-coding genes of hantaviruses. Mol. Biol. Evol. 2000;17:1558–1568. doi: 10.1093/oxfordjournals.molbev.a026254. [DOI] [PubMed] [Google Scholar]
- 18.Kang H.J., Bennett S.N., Sumibcay L., Arai S., Hope A.G., Mocz G., Song J.W., Cook J.A., Yanagihara R. Evolutionary insights from a genetically divergent hantavirus harbored by the European common mole (Talpa europaea) PLoS One. 2009;4:e6149. doi: 10.1371/journal.pone.0006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arai S., Nguyen S.T., Boldgiv B., Fukui D., Araki K., Dang C.N., Ohdachi S.D., Nguyen N.X., Pham T.D., Boldbaatar B., et al. Novel bat-borne hantavirus, Vietnam. Emerg. Infect. Dis. 2013;19:1159–1161. doi: 10.3201/eid1907.121549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss S., Witkowski P.T., Auste B., Nowak K., Weber N., Fahr J., Mombouli J.V., Wolfe N.D., Drexler J.F., Drosten C., et al. Hantavirus in bat, Sierra Leone. Emerg. Infect. Dis. 2012;18:159–161. doi: 10.3201/eid1801.111026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young J.C., Mills J.N., Enria D.A., Dolan N.E., Khan A.S., Ksiazek T.G. New World hantaviruses. Br. Med. Bull. 1998;54:659–673. doi: 10.1093/oxfordjournals.bmb.a011718. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura I., Yoshimatsu K., Lee B.H., Okumura M., Taruishi M., Araki K., Kariwa H., Takashima I., Arikawa J. Development of a serotyping ELISA system for Thailand virus infection. Arch. Virol. 2008;153:1537–1542. doi: 10.1007/s00705-008-0128-4. [DOI] [PubMed] [Google Scholar]
- 23.Mills J.N., Amman B.R., Glass G.E. Ecology of Hantaviruses and Their Hosts in North America. Vector Borne Zoonotic Dis. 2010;10:563–574. doi: 10.1089/vbz.2009.0018. [DOI] [PubMed] [Google Scholar]
- 24.Palma R.E., Polop J.J., Owen R.D., Mills J.N. Ecology of rodent-associated hantaviruses in the Southern Cone of South America: Argentina, Chile, Paraguay, and Uruguay. J. Wildl. Dis. 2012;48:267–281. doi: 10.7589/0090-3558-48.2.267. [DOI] [PubMed] [Google Scholar]
- 25.Ruo S.L., Sanchez A., Elliott L.H., Brammer L.S., McCormick J.B., Fisher-Hoch S.P. Monoclonal antibodies to three strains of hantaviruses: Hantaan, R22, and Puumala. Arch. Virol. 1991;119:1–11. doi: 10.1007/BF01314318. [DOI] [PubMed] [Google Scholar]
- 26.Yoshimatsu K., Arikawa J., Tamura M., Yoshida R., Lundkvist A., Niklasson B., Kariwa H., Azuma I. Characterization of the nucleocapsid protein of hantaan virus strain 76–118 using monoclonal antibodies. J. Gen. Virol. 1996;77:695–704. doi: 10.1099/0022-1317-77-4-695. [DOI] [PubMed] [Google Scholar]
- 27.Lundkvist A., Fatouros A., Niklasson B. Antigenic variation of European haemorrhagic fever with renal syndrome virus strains characterized using bank vole monoclonal antibodies. J. Gen. Virol. 1991;72:2097–2103. doi: 10.1099/0022-1317-72-9-2097. [DOI] [PubMed] [Google Scholar]
- 28.Lundkvist A., Bjorsten S., Niklasson B., Ahlborg N. Mapping of B-cell determinants in the nucleocapsid protein of Puumala virus: definition of epitopes specific for acute immunoglobulin G recognition in humans. Clin. Diagn. Lab. Immunol. 1995;2:82–86. doi: 10.1128/cdli.2.1.82-86.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elgh F., Lundkvist A., Alexeyev O.A., Wadell G., Juto P. A major antigenic domain for the human humoral response to Puumala virus nucleocapsid protein is located at the amino-terminus. J. Virol. Methods. 1996;59:161–172. doi: 10.1016/0166-0934(96)02042-3. [DOI] [PubMed] [Google Scholar]
- 30.Tischler N.D., Rosemblatt M., Valenzuela P.D. Characterization of cross-reactive and serotype-specific epitopes on the nucleocapsid proteins of hantaviruses. Virus. Res. 2008;135:1–9. doi: 10.1016/j.virusres.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Saasa N., Yoshida H., Shimizu K., Sanchez-Hernandez C., Romero-Almaraz M.D., Koma T., Sanada T., Seto T., Yoshii K., Ramos C., et al. The N-terminus of the Montano virus nucleocapsid protein possesses broadly cross-reactive conformation-dependent epitopes conserved in rodent-borne hantaviruses. Virology. 2012;428:48–57. doi: 10.1016/j.virol.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Kucinskaite-Kodze I., Petraityte-Burneikiene R., Zvirbliene A., Hjelle B., Medina R.A., Gedvilaite A., Razanskiene A., Schmidt-Chanasit J., Mertens M., Padula P., et al. Characterization of monoclonal antibodies against hantavirus nucleocapsid protein and their use for immunohistochemistry on rodent and human samples. Arch. Virol. 2011;156:443–456. doi: 10.1007/s00705-010-0879-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song J.W., Baek L.J., Schmaljohn C.S., Yanagihara R. Thottapalayam virus, a prototype shrewborne hantavirus. Emerg. Infect. Dis. 2007;13:980–985. doi: 10.3201/eid1307.070031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlegel M., Tegshduuren E., Yoshimatsu K., Petraityte R., Sasnauskas K., Hammerschmidt B., Friedrich R., Mertens M., Groschup M.H., Arai S., et al. Novel serological tools for detection of Thottapalayam virus, a Soricomorpha-borne hantavirus. Arch. Virol. 2012;157:2179–2187. doi: 10.1007/s00705-012-1405-9. [DOI] [PubMed] [Google Scholar]
- 35.Yamada T., Hjelle B., Lanzi R., Morris C., Anderson B., Jenison S. Antibody responses to Four Corners hantavirus infections in the deer mouse (Peromyscus maniculatus): Identification of an immunodominant region of the viral nucleocapsid protein. J. Virol. 1995;69:1939–1943. doi: 10.1128/jvi.69.3.1939-1943.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gott P., Zoller L., Darai G., Bautz E.K.F. A Major Antigenic Domain Of Hantaviruses Is Located On the Aminoproximal Site Of the Viral Nucleocapsid Protein. Virus Genes. 1997;14:31–40. doi: 10.1023/A:1007983306341. [DOI] [PubMed] [Google Scholar]
- 37.Lundkvist A., Meisel H., Koletzki D., Lankinen H., Cifire F., Geldmacher A., Sibold C., Gott P., Vaheri A., Kruger D.H., et al. Mapping of B-cell epitopes in the nucleocapsid protein of Puumala hantavirus. Viral. Immunol. 2002;15:177–192. doi: 10.1089/088282402317340323. [DOI] [PubMed] [Google Scholar]
- 38.Elgh F., Lundkvist A., Alexeyev O.A., Stenlund H., Avsic-Zupanc T., Hjelle B., Lee H.W., Smith K.J., Vainionpaa R., Wiger D., et al. Serological diagnosis of hantavirus infections by an enzyme-linked immunosorbent assay based on detection of immunoglobulin G and M responses to recombinant nucleocapsid proteins of five viral serotypes. J. Clin. Microbiol. 1997;35:1122–1130. doi: 10.1128/jcm.35.5.1122-1130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elgh F., Linderholm M., Wadell G., Tarnvik A., Juto P. Development of humoral cross-reactivity to the nucleocapsid protein of heterologous hantaviruses in nephropathia epidemica. FEMS Immunol. Med. Microbiol. 1998;22:309–315. doi: 10.1111/j.1574-695X.1998.tb01220.x. [DOI] [PubMed] [Google Scholar]
- 40.Alfadhli A., Love Z., Arvidson B., Seeds J., Willey J., Barklis E. Hantavirus nucleocapsid protein oligomerization. J. Virol. 2001;75:2019–2023. doi: 10.1128/JVI.75.4.2019-2023.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alfadhli A., Steel E., Finlay L., Bachinger H.P., Barklis E. Hantavirus nucleocapsid protein coiled-coil domains. J. Biol. Chem. 2002;277:27103–27108. doi: 10.1074/jbc.M203395200. [DOI] [PubMed] [Google Scholar]
- 42.Boudko S.P., Kuhn R.J., Rossmann M.G. The coiled-coil domain structure of the Sin Nombre virus nucleocapsid protein. J. Mol. Biol. 2007;366:1538–1544. doi: 10.1016/j.jmb.2006.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alminaite A., Halttunen V., Kumar V., Vaheri A., Holm L., Plyusnin A. Oligomerization of hantavirus nucleocapsid protein: analysis of the N-terminal coiled-coil domain. J. Virol. 2006;80:9073–9081. doi: 10.1128/JVI.00515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Boudreaux D.M., Estrada D.F., Egan C.W., St Jeor S.C., De Guzman R.N. NMR structure of the N-terminal coiled coil domain of the Andes hantavirus nucleocapsid protein. J. Biol. Chem. 2008;283:28297–28304. doi: 10.1074/jbc.M804869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amada T., Yoshimatsu K., Koma T., Shimizu K., Gamage C.D., Shiokawa K., Nishio S., Ahlm C., Arikawa J. Development of an immunochromatography strip test based on truncated nucleocapsid antigens of three representative hantaviruses. Virol. J. 2014 doi: 10.1186/1743-422X-11-87. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okumura M., Yoshimatsu K., Araki K., Lee B.H., Asano A., Agui T., Arikawa J. Epitope analysis of monoclonal antibody E5/G6, which binds to a linear epitope in the nucleocapsid protein of hantaviruses. Arch. Virol. 2004;149:2427–2434. doi: 10.1007/s00705-004-0393-9. [DOI] [PubMed] [Google Scholar]
- 47.Mir M.A., Brown B., Hjelle B., Duran W.A., Panganiban A.T. Hantavirus N protein exhibits genus-specific recognition of the viral RNA panhandle. J. Virol. 2006;80:11283–11292. doi: 10.1128/JVI.00820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mir M.A., Sheema S., Haseeb A., Haque A. Hantavirus nucleocapsid protein has distinct m7G cap- and RNA-binding sites. J. Biol. Chem. 2010;285:11357–11368. doi: 10.1074/jbc.M110.102459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshimatsu K., Lee B.H., Araki K., Morimatsu M., Ogino M., Ebihara H., Arikawa J. The multimerization of hantavirus nucleocapsid protein depends on type-specific epitopes. J. Virol. 2003;77:943–952. doi: 10.1128/JVI.77.2.943-952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang M., Rossi C., Schmaljohn C.S. Expression of non-conserved regions of the S genome segments of three hantaviruses: evaluation of the expressed polypeptides for diagnosis of haemorrhagic fever with renal syndrome. J. Gen. Virol. 1993;4:1115–1124. doi: 10.1099/0022-1317-74-6-1115. [DOI] [PubMed] [Google Scholar]
- 51.Koma T., Yoshimatsu K., Taruishi M., Miyashita D., Endo R., Shimizu K., Yasuda S.P., Amada T., Seto T., Murata R., et al. Development of a serotyping enzyme-linked immunosorbent assay system based on recombinant truncated hantavirus nucleocapsid proteins for New World hantavirus infection. J. Virol. Methods. 2012;185:74–81. doi: 10.1016/j.jviromet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 52.Kaukinen P., Kumar V., Tulimaki K., Engelhardt P., Vaheri A., Plyusnin A. Oligomerization of Hantavirus N protein: C-terminal alpha-helices interact to form a shared hydrophobic space. J. Virol. 2004;78:13669–13677. doi: 10.1128/JVI.78.24.13669-13677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mir M.A., Panganiban A.T. Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J. Virol. 2004;78:8281–8288. doi: 10.1128/JVI.78.15.8281-8288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaukinen P., Vaheri A., Plyusnin A. Mapping of the regions involved in homotypic interactions of Tula hantavirus N protein. J. Virol. 2003;77:10910–10916. doi: 10.1128/JVI.77.20.10910-10916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morii M., Yoshimatsu K., Arikawa J., Zhou G., Kariwa H., Takashima I. Antigenic characterization of Hantaan and Seoul virus nucleocapsid proteins expressed by recombinant baculovirus: application of a truncated protein, lacking an antigenic region common to the two viruses, as a serotyping antigen. J. Clin. Microbiol. 1998;36:2514–2521. doi: 10.1128/jcm.36.9.2514-2521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koma T., Yoshimatsu K., Pini N., Safronetz D., Taruishi M., Levis S., Endo R., Shimizu K., Yasuda S.P., Ebihara H., et al. Truncated hantavirus nucleocapsid proteins for serotyping Sin Nombre, Andes, and Laguna Negra hantavirus infections in humans and rodents. J. Clin. Microbiol. 2010;48:1635–1642. doi: 10.1128/JCM.00072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee B.-H., Yoshimatsu K., Maeda A., Ochiai K., Morimatsu M., Araki K., Ogino M., Morikawa S., Arikawa J. Association of the nucleocapsid protein of the Seoul and Hantaan hantaviruses with small ubiquitin-like modifier-1-related molecules. Virus Research. 2003;98:83–91. doi: 10.1016/j.virusres.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Kaukinen P., Vaheri A., Plyusnin A. Non-covalent interaction between nucleocapsid protein of Tula hantavirus and small ubiquitin-related modifier-1, SUMO-1. Virus Res. 2003;92:37–45. doi: 10.1016/S0168-1702(02)00312-X. [DOI] [PubMed] [Google Scholar]
- 59.Sakurai A., Yasuda J., Takano H., Tanaka Y., Hatakeyama M., Shida H. Regulation of human T-cell leukemia virus type 1 (HTLV-1) budding by ubiquitin ligase Nedd4. Microbes Infect. 2004;6:150–156. doi: 10.1016/j.micinf.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 60.Yasuda J., Hunter E., Nakao M., Shida H. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 2002;3:636–640. doi: 10.1093/embo-reports/kvf132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yasuda J., Nakao M., Kawaoka Y., Shida H. Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 2003;77:9987–9992. doi: 10.1128/JVI.77.18.9987-9992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Urata S., Noda T., Kawaoka Y., Yokosawa H., Yasuda J. Cellular factors required for Lassa virus budding. J. Virol. 2006;80:4191–4195. doi: 10.1128/JVI.80.8.4191-4195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng E., Haque A., Rimmer M.A., Hussein I.T., Sheema S., Little A., Mir M.A. Characterization of the Interaction between hantavirus nucleocapsid protein (N) and ribosomal protein S19 (RPS19) J. Biol. Chem. 2011;286:11814–11824. doi: 10.1074/jbc.M110.210179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haque A., Mir M.A. Interaction of hantavirus nucleocapsid protein with ribosomal protein S19. J. Virol. 2010;84:12450–12453. doi: 10.1128/JVI.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramanathan H.N., Jonsson C.B. New and Old World hantaviruses differentially utilize host cytoskeletal components during their life cycles. Virology. 2008;374:138–150. doi: 10.1016/j.virol.2007.12.030. [DOI] [PubMed] [Google Scholar]
- 66.Ramanathan H.N., Chung D.H., Plane S.J., Sztul E., Chu Y.K., Guttieri M.C., McDowell M., Ali G., Jonsson C.B. Dynein-dependent transport of the hantaan virus nucleocapsid protein to the endoplasmic reticulum-Golgi intermediate compartment. J. Virol. 2007;81:8634–8647. doi: 10.1128/JVI.00418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ravkov E.V., Nichol S.T., Peters C.J., Compans R.W. Role Of Actin Microfilaments In Black Creek Canal Virus Morphogenesis. J. Virol. 1998;72:2865–2870. doi: 10.1128/jvi.72.4.2865-2870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hepojoki J., Strandin T., Wang H., Vapalahti O., Vaheri A., Lankinen H. Cytoplasmic tails of hantavirus glycoproteins interact with the nucleocapsid protein. J. Gen. Virol. 2010;91:2341–2350. doi: 10.1099/vir.0.021006-0. [DOI] [PubMed] [Google Scholar]
- 69.Wang H., Alminaite A., Vaheri A., Plyusnin A. Interaction between hantaviral nucleocapsid protein and the cytoplasmic tail of surface glycoprotein Gn. Virus Res. 2010;151:205–212. doi: 10.1016/j.virusres.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 70.Shimizu K., Yoshimatsu K., Koma T., Arikawa J. Role of Nucleocapsid Protein of Hantaviruses in Intracellular Traffic of Viral Glycoproteins. Virus Res. 2013;178:349–356. doi: 10.1016/j.virusres.2013.09.022. [DOI] [PubMed] [Google Scholar]