Abstract
Acetic acid bacteria (AAB) play an important role during cocoa fermentation, as their main product, acetate, is a major driver for the development of the desired cocoa flavors. Here, we investigated the specialized metabolism of these bacteria under cocoa pulp fermentation-simulating conditions. A carefully designed combination of parallel 13C isotope labeling experiments allowed the elucidation of intracellular fluxes in the complex environment of cocoa pulp, when lactate and ethanol were included as primary substrates among undefined ingredients. We demonstrate that AAB exhibit a functionally separated metabolism during coconsumption of two-carbon and three-carbon substrates. Acetate is almost exclusively derived from ethanol, while lactate serves for the formation of acetoin and biomass building blocks. Although this is suboptimal for cellular energetics, this allows maximized growth and conversion rates. The functional separation results from a lack of phosphoenolpyruvate carboxykinase and malic enzymes, typically present in bacteria to interconnect metabolism. In fact, gluconeogenesis is driven by pyruvate phosphate dikinase. Consequently, a balanced ratio of lactate and ethanol is important for the optimum performance of AAB. As lactate and ethanol are individually supplied by lactic acid bacteria and yeasts during the initial phase of cocoa fermentation, respectively, this underlines the importance of a well-balanced microbial consortium for a successful fermentation process. Indeed, AAB performed the best and produced the largest amounts of acetate in mixed culture experiments when lactic acid bacteria and yeasts were both present.
INTRODUCTION
Acetic acid bacteria (AAB) play an important role in cocoa fermentation (1). During fermentation of the pulp that surrounds the cocoa beans, they form acetate. Acetate then diffuses into the beans (2–4), where it initiates a cascade of chemical and biochemical reactions leading to precursor molecules for cocoa flavor (2, 5, 6). Potential substrates for AAB are lactate and ethanol, which are individually produced by lactic acid bacteria (LAB; mainly Lactobacillus fermentum) and yeasts (diverse yeasts such as Saccharomyces cerevisiae, Hanseniaspora opuntiae, and Candida krusei), respectively, during the fermentation process (6–12). Hereby, the degradation of lactate by AAB is desired, since the remaining lactate may provide an off flavor in the final cocoa product (11, 13, 14). In recent years, AAB have been extensively analyzed for their contribution to cocoa fermentation. Obviously, the most prevalent AAB species is Acetobacter pasteurianus (13, 15–17). In addition, Acetobacter ghanensis and Acetobacter senegalensis are found during spontaneous cocoa bean fermentation (9, 13, 17, 18). Further studies provided first insights into the basic microbiological properties of such strains and macroscopic dynamics during cocoa pulp fermentation (12, 15, 18–20). At this point, it appears to be relevant to resolve the metabolic contribution of AAB in greater detail. The basic knowledge that we share indicates a unique metabolism among members of the genus Acetobacter, including a nonfunctional Embden-Meyerhof-Parnas (EMP) pathway and the generation of energy from the incomplete oxidation of ethanol into acetate (21, 22). A few species are able to catabolize lactate (23–26). A pathway from lactate to acetate as the final product has been proposed; however, the role of some of its enzymes remains unclear (26). In general, little is known about the in vivo contribution of these carbon core pathways to the cellular metabolism of AAB. In particular, their lifestyle in complex environments, such as cocoa pulp, is largely unknown. In the past decade, 13C metabolic flux analysis has emerged as a routine approach to study individual microbes grown on single carbon substrates at the level of molecular carbon fluxes (in vivo reaction rates) (27, 28). Without resolving the details, advanced experimental designs have, meanwhile, provided fluxes for complex microbial systems, including mixed populations (29) and nutrient mixtures (30).
Here, we applied 13C-based metabolic flux analysis to study the metabolism of Acetobacter during cocoa pulp fermentation. Acetobacter pasteurianus NCC 316 and A. ghanensis DSM 18895 were selected as representative strains of two major species of AAB naturally occurring in cocoa pulp (23, 31). A validated setup (12) unraveled the metabolic fluxes in AAB under cocoa pulp fermentation-simulating conditions. As a central finding, the concerted use of lactate and ethanol enables optimum fluxes with regard to growth and energy metabolism. Considering recent insights into the extracellular and molecular fluxes of LAB during cocoa fermentation (12, 20, 23, 30), this suggests that the efficiency of AAB depends on compositional traits provided by LAB and yeasts during the initial fermentation process so that the overall success of cocoa fermentation requires a well-balanced microbial consortium.
MATERIALS AND METHODS
Strains and maintenance.
Acetobacter pasteurianus NCC 316, Lactobacillus fermentum NCC 575, and Saccharomyces cerevisiae NYSC 2 were obtained from the Nestlé Culture Collection (Lausanne, Switzerland). A. pasteurianus NCC 316 and L. fermentum NCC 575 were previously isolated from cheese (1978) and from coffee (1984), respectively. S. cerevisiae NYSC 2 is the commonly used laboratory strain X2180-1B (MATα gal2 SUC2 mal CUPJ). Acetobacter ghanensis DSM 18895 was obtained from the German Culture Collection (DSMZ, Braunschweig, Germany). Escherichia coli K-12 was used as a reference strain for enzyme activity assays and was supplied by the Coli Genetic Stock Center (CGSC; New Haven, CT). A. pasteurianus NCC 316 and A. ghanensis DSM 18895 were stored in mannitol-yeast extract-peptone (MYP) medium (32). L. fermentum NCC 575 was maintained in lactobacillus deMan-Rogosa-Sharpe (MRS) broth (32). S. cerevisiae NYSC 2 was stored in yeast malt (YM) medium (32). E. coli K-12 was maintained in LB medium (synonymously called lysogeny broth or Luria-Bertani broth) (33). All frozen stocks were stored at −80°C and contained 15% (vol/vol) glycerol.
Cultivation.
Cells of A. pasteurianus NCC 316 and A. ghanensis DSM 18895 were reactivated on MYP agar at 30°C for 48 h before use. For precultures and the main cultivation of Acetobacter species, cocoa pulp simulation medium for acetic acid bacteria (PSM-AAB) was used (12). It contained the following per liter: 10 g ethanol, 7.2 g sodium lactate, 10 g yeast extract (Roth, Karlsruhe, Germany), 5 g soy peptone (Roth), and 1 ml Tween 80 (Sigma-Aldrich, Taufkirchen, Germany). The initial pH of the medium was 4.5. In 13C tracer experiments, lactate and ethanol were replaced by equimolar amounts of 98% [U-13C]sodium lactate (Cambridge Isotopes, Andover, MA) or 98% [3-13C]sodium lactate (Cambridge Isotopes) and 99% [U-13C]ethanol (Sigma-Aldrich), respectively. Precultures and the main cultures of L. fermentum NCC 575 and S. cerevisiae NYSC 2 were performed in cocoa pulp simulation medium for lactic acid bacteria (PSM-LAB) (12), which was prepared as described previously (30). For all experiments, cells from agar plates previously obtained from frozen stocks (Acetobacter) or frozen stocks (L. fermentum and S. cerevisiae) were used to inoculate the first preculture (37°C, 12 to 24 h). Cells were transferred to the second preculture, which was incubated at 37°C. Cells were then harvested in the mid-exponential growth phase, washed (6,000 × g, 5 min, 4°C) with a peptone mixture (10 g liter−1 yeast extract [Roth], 5 g liter−1 soy peptone [Roth]), and used to inoculate the main culture to an initial optical density at 600 nm (OD600) of 0.01. All cultivations of A. pasteurianus NCC 316 and A. ghanensis DSM 18895 were performed in disposable baffled shake flasks (250 ml; PreSens Precision Sensing GmbH, Regensburg, Germany) on a rotary shaker at 280 rpm (Multitron II; Infors, Bottmingen, Switzerland). Dissolved oxygen was monitored using a shake flask reader (SFR; PreSens Precision Sensing GmbH). To account for the potential evaporation of volatile compounds, their loss was monitored in control experiments without cells, in which the cultures were incubated under the fermentation conditions described above, and evaporation rates were used to correct formation and consumption rates. Cultivations with L. fermentum NCC 575 and S. cerevisiae NYSC 2 were conducted in nonbaffled 250-ml shake flasks at 37°C and a rotation speed of 50 rpm.
In order to study the entire cocoa pulp fermentation, PSM-LAB was inoculated with either L. fermentum, S. cerevisiae, or a mixture of both microorganisms and then incubated for 24 h. Subsequently, the fermentation broth was filtered (pore size, 0.22 μm; filter top 250; TPP Techno Plastic Products, Trasadingen, Switzerland), the pH was adjusted to 4.5, and the medium was further incubated with A. pasteurianus NCC 316.
Quantification of substrates and products.
The concentrations of organic acids (lactate, acetate, pyruvate, citrate, and α-ketoglutarate), acetoin, and ethanol in the culture supernatants were determined by high-pressure liquid chromatography (Elite-LaChrom; Hitachi, West Chester, PA) on a chromatograph equipped with an Aminex HPX-87H column (300 by 7.8 mm; Bio-Rad, Hercules, CA) as the stationary phase, with 12.5 mM H2SO4 used as the mobile phase at a flow rate of 0.5 ml min−1 and a column temperature of 45°C. Quantification of acetoin was carried out via UV detection at 290 nm. For detection of all other substances, a refractive index detector was used. Samples were diluted 1:10 with deionized water prior to analysis.
The OD600 was monitored to determine cell growth (Libra S11; Biochrome, Cambridge, United Kingdom). Cell dry weight (CDW) was determined gravimetrically after filtration of culture broth (cellulose acetate; pore size, 0.2 μm; Sartorius, Göttingen, Germany) and subsequent drying of the filters at 105°C until the weight was constant. The correlation factors between OD600 and CDW (three biological replicates each) were as follows: CDW = 0.356 × OD600 (g liter−1) for A. pasteurianus NCC 316 and CDW = 0.240 × OD600 (g liter−1) for A. ghanensis DSM 18995.
GC/MS analysis of 13C labeling pattern of amino acids and of secreted acetoin and acetate.
For analysis of 13C labeling patterns, samples were taken from the first exponential growth phase of the AAB. The mass isotopomer distributions of proteinogenic amino acids were analyzed by gas chromatography/mass spectrometry (GC/MS) in the selective ion monitoring mode (34). The labeling pattern of acetate in the culture supernatants was determined by GC/MS as described previously (30). To determine the labeling pattern of acetoin, a novel method was adapted from a protocol for multifunctional carbonyl compounds (35). Briefly, 500 μl of culture supernatant was mixed with 50 μl of a 0.5% (w/vol) aqueous O-(2,3,4,5,6-pentafluorobenzyl)hydroxylamine hydrochloride (PFBHA) solution, and the mixture was incubated for 30 min at 80°C. Subsequently, PFBHA derivatives were extracted with methyl-t-butylether (MTBE). Excess PFBHA was removed by back-extraction with 0.1 M HCl (500 μl). To eliminate traces of water in the MTBE extracts, dry sodium sulfate powder was added. The remaining water in the organic phase was trapped into clumps of sodium sulfate. After removal of the particles by centrifugation for a few seconds (Sprout minicentrifuge; Heathrow Scientific LLC, Vernon Hills, IL), the samples were derivatized with 50 μl of bis-(trimethylsilyl)trifluoroacetamide (BSTFA) at 80°C for 30 min. Subsequently, PFBHA-BSTFA derivatives were analyzed by GC/MS (7890A; 5975C quadrupole detector; Agilent Technologies, Santa Clara, CA). The oven program was as follows: 60°C for 2 min; ramp at 15°C min−1; final temperature, 325°C. Samples were analyzed in selective ion monitoring mode at m/z 340 to 344 (C1 to C4) and m/z 312 to 314 (C3 to C4). For method validation, a 0.05% (wt/vol) solution of naturally labeled acetoin was treated and analyzed as described above.
Enzyme activity assays.
For enzyme activity assays, cells from the first exponential growth phases were harvested by centrifugation (25,800 × g, 4°C, 10 min), washed with 100 mM Tris-HCl (pH 7.5, 10 mM MgCl2, 0.75 mM dithiothreitol), and resuspended in 10 ml of the same buffer. Disruption was performed mechanically (with 100-μm-diameter silica glass beads and disruption 2 times for 20 s each time at 6.0 m s−1; FastPrep-24; MP Biomedicals, Santa Ana, CA). Cell debris was removed by centrifugation at 17,000 × g for 5 min at 4°C, and the protein content in the crude extract was determined by the Bradford method (36) using a reagent solution (Rothi-Quant, Roth) with bovine serum albumin as the standard. Enzyme activity assays were performed at room temperature in a microplate reader (Varioskan Flash; Thermo Scientific, Waltham, MA). Phosphoenolpyruvate (PEP) carboxykinase (PEPCK; EC 4.1.1.32) and phosphoenolpyruvate carboxylase (PEPC; EC 4.1.1.31) assays were performed as described previously (37). Malic enzyme (ME; EC 1.1.1.40) activity was determined as described previously (38). As a positive control, an isocitrate dehydrogenase assay was used (39). Enzyme activities were calculated from the change in the absorbance. Molar extinction coefficients were as follows: ε340 = 6.22 mM−1 cm−1 for NADH + H+ and NADPH + H+ and ε300 = 0.6 mM−1 cm−1 for oxaloacetate.
Metabolic network.
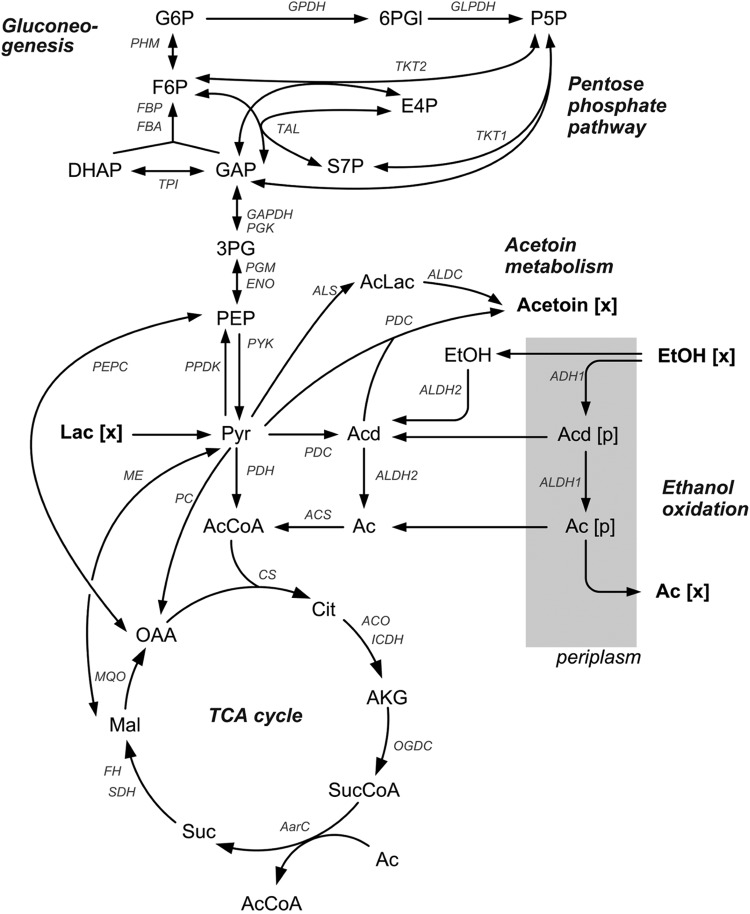
The metabolic pathway network of Acetobacter (Fig. 1) was derived from the genomic repertoire of A. pasteurianus IFO 3283 and A. pasteurianus 386B, which have recently been sequenced (21, 40). A. pasteurianus possesses membrane-bound, pyrroloquinoline quinone (PQQ)-dependent alcohol and aldehyde dehydrogenases for oxidation of ethanol into acetate, as well as genes encoding intracellular alcohol dehydrogenase and aldehyde dehydrogenase (see Table S1 in the supplemental material) (21, 40–43). Lactate is metabolized via lactate dehydrogenase and further metabolized into acetate (26). It has been reported that A. pasteurianus strains accumulate acetoin, when incubated with lactate (23–25). Indeed, two different pathways for acetoin formation are annotated. Furthermore, there is genetic evidence for a modified tricarboxylic acid (TCA) cycle in which succinyl coenzyme A (succinyl-CoA) synthetase and malate dehydrogenase are replaced by succinyl-CoA:acetate CoA transferase (A. pasteurianus IFO 3283 gene annotations APA01_00320, APA01_00310, and APA386B_2589) and malate:quinone oxidoreductase (A. pasteurianus IFO 3283 gene annotations APA01_01110, APA01_11550, and APA386B_2675) (21, 40). This modified pathway has been described previously (44). The glyoxylate pathway and the Entner-Doudoroff (ED) pathway are not encoded in the genomes of A. pasteurianus IFO 3283 and A. pasteurianus 386B, respectively (21, 40). There are predicted genes for PEPC in the genome sequences of A. pasteurianus IFO 3283 and A. pasteurianus 386B (21, 40). Additionally, the activity of PEPCK is predicted putatively (APA01_16650) (40); however, since there is no experimental evidence, it was not considered in the network. Due to the absence of phosphofructokinase, the EMP pathway is nonfunctional (21).
FIG 1.
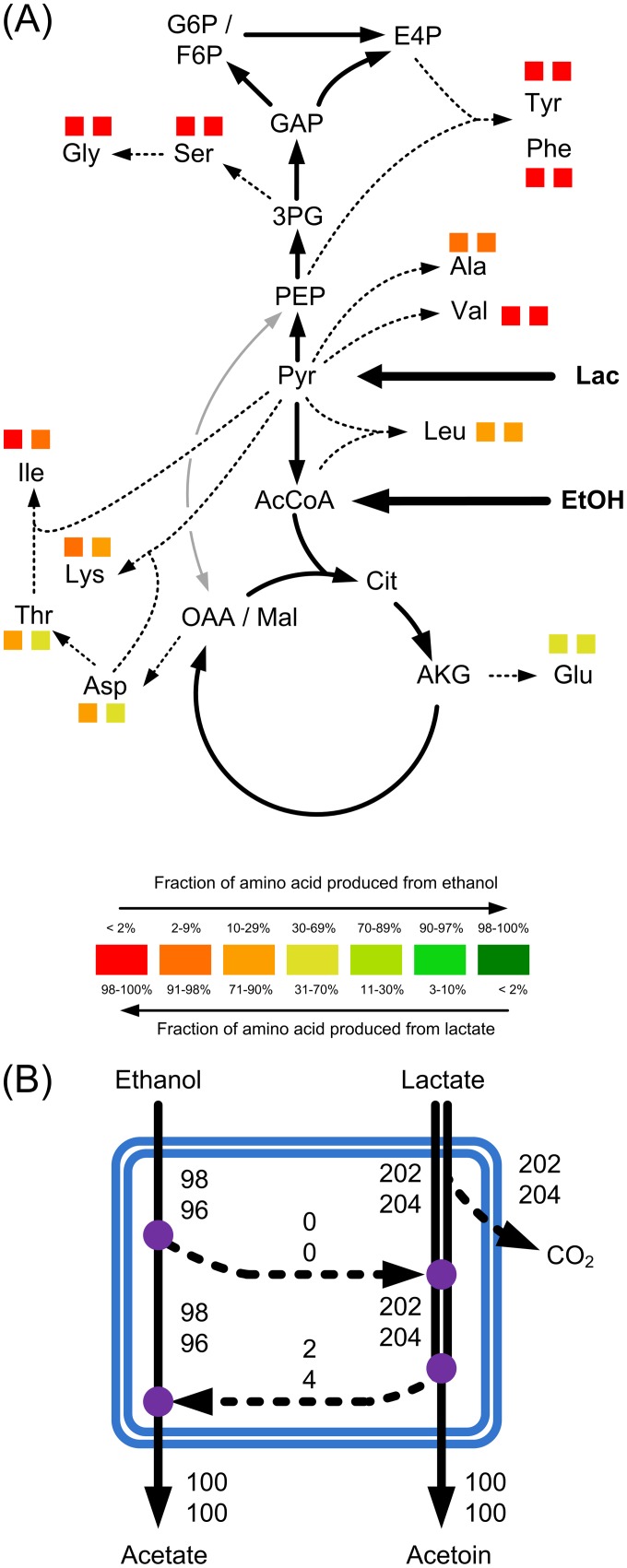
Metabolic network of the central carbon metabolism of Acetobacter pasteurianus. The enzymatic reactions and their corresponding genes and the definitions of the abbreviations for the enzymes are given in Table S1 in the supplemental material. G6P, glucose-6-phosphate; 6PGl, 6-phosphogluconate; P5P, pentose-5-phosphate; F6P, fructose-6-phosphate; E4P, erythrose-4-phosphate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; S7P, sucrose-7-phosphate; 3PG, 3-phosphoglycerate; AcLac, acetolactate; EtOH, ethanol; Acd, acetoin dehydrogenase; AcCoA, acetyl coenzyme A; OGDC, alpha-ketoglutarate dehydrogenase; AarC, succinyl-CoA:acetate CoA transferase; Ac, acetate; Cit, citrate; AKG, α-ketoglutarate; SucCoa, succinyl-CoA; Suc, succinyl; Mal, malate; OAA, oxaloacetate.
Estimation of metabolic fluxes.
Metabolic flux distributions were estimated with the OpenFLUX (v2.1) program (45). Simulated mass isotopomer data were corrected for the natural occurrence of isotopes in derivatization residues and noncarbon atoms (46), whereby the natural abundance of isotopes reflected values from laboratory chemicals (47). For statistical evaluation of the obtained fluxes, Monte Carlo analysis was performed with 100 individual parameter estimations, assuming Gaussian noise for the experimental data (48).
RESULTS
Computer-based experimental design predicts optimum combinations of 13C-labeled lactate and ethanol to resolve metabolic fluxes in Acetobacter.
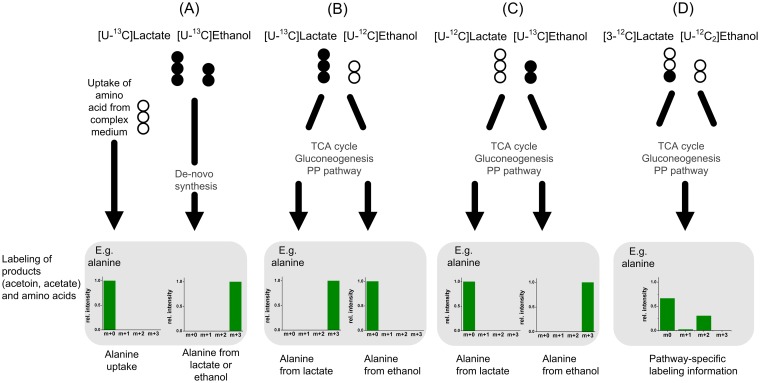
In the medium that represented cocoa pulp, lactate and ethanol were the primary substrates among complex ingredients. In order to elucidate metabolic fluxes in AAB growing on this complex nutrient mixture, a strategy of combined isotope experiments was developed (Fig. 2). Of central importance was the identification of suitable tracer substrates to resolve the fluxes of interest. In a set of computer simulations with various combinations of 13C-labeled lactate and ethanol, excellent resolution of metabolic fluxes was obtained by a combination of [U-13C]lactate, [U-13C]ethanol, and [3-13C]lactate. The relative contribution of de novo synthesis and uptake from the medium to the amino acid supply could be quantified using a mixture of [U13C]lactate and [U13C]ethanol, as deduced previously (49). The individual contribution of lactate and ethanol to central metabolism was elucidated by applying mixtures of one universally labeled substrate and one naturally labeled substrate in two separate labeling experiments. For a more detailed resolution of pathway fluxes, a mixture of [3-13C]lactate and naturally labeled ethanol was found to be adequate. Taken together, metabolic flux analysis of the acetic acid bacteria integrated labeling information from four parallel tracer studies (Fig. 2).
FIG 2.
Labeling strategy used to elucidate metabolic fluxes of Acetobacter pasteurianus and Acetobacter ghanensis DSM 18895 in cocoa pulp simulation medium using specifically enriched isotopic tracer substances and mass spectral labeling analysis of proteinogenic amino acids and extracellular products. (A) Theoretical 13C-labeled mass isotopomer distribution of alanine obtained by de novo synthesis from the labeled substrates ([U-13C]lactate and [U-13C]ethanol) or amino acid uptake (12C). (B and C) Contribution of lactate and ethanol to formation of amino acid precursors. (D) Elucidation of specific metabolic routes of [3-13C]lactate utilization. Black circles, 13C-labeled carbon atoms; white circles, unlabeled carbon (12C); PP, pentose phosphate; Rel. intensity, relative intensity.
Quantification of the labeling profile of acetoin.
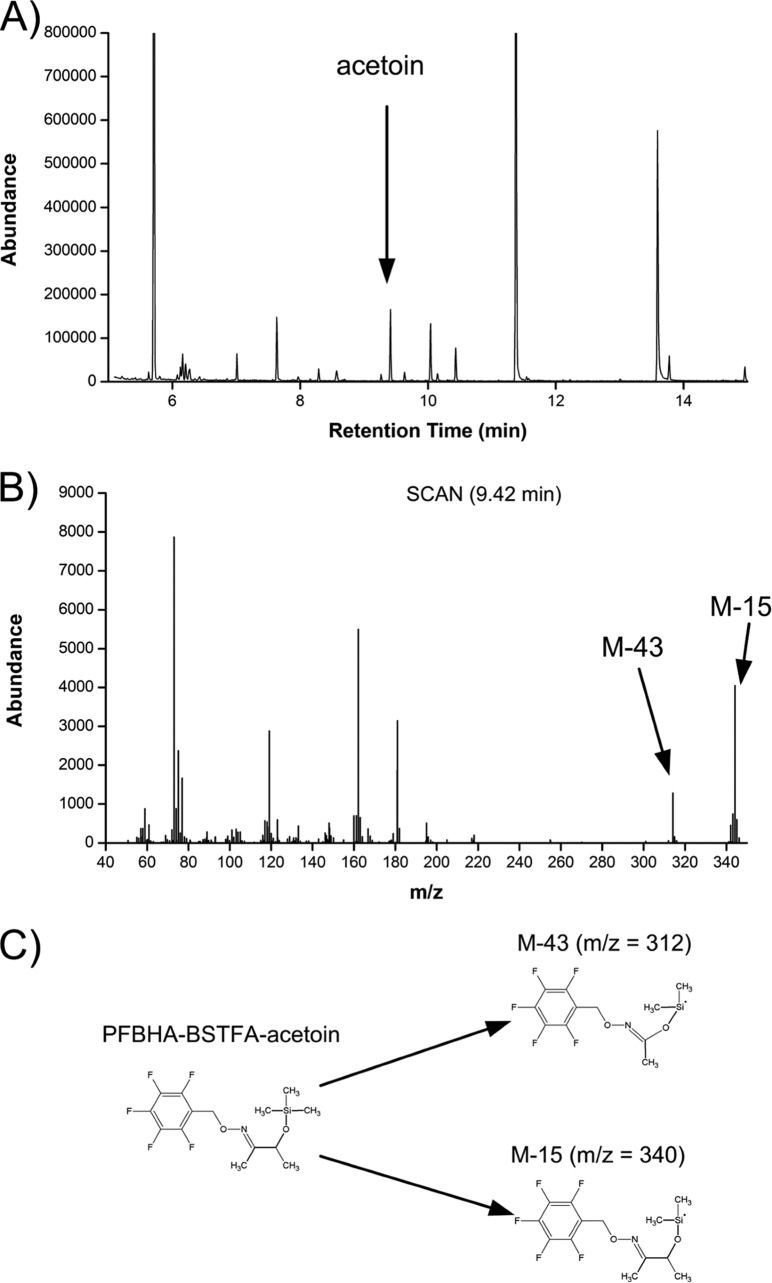
Previously developed protocols allow precise labeling analysis of proteinogenic amino acids (50) and of secreted acetate (30) from isotope experiments. In order to provide the full picture, a method for labeling analysis of secreted acetoin, another typical product of acetic acid bacteria, had to be developed. Tests with different reagents and incubation conditions (data not shown) finally provided a stable PFBHA-BSTFA derivative of acetoin that eluted as a separate peak after 9.4 min, as shown in the GC/MS chromatogram (Fig. 3A). The accuracy of the measurement was validated by analyzing naturally and universally 13C-labeled acetoin. The resulting mass isotopomer distributions perfectly matched the theoretical values derived from the natural isotope abundance (47) (Table 1). The potential isobaric overlay of acetoin ion clusters with the sample matrix was excluded by comparative analysis of acetoin from the supernatant of a culture of A. pasteurianus NCC 316 grown on naturally labeled substrates (Table 1). Accordingly, the developed method was appropriate to quantify the labeling of acetoin from isotope experiments in cocoa pulp simulation medium.
FIG 3.
GC/MS analysis of the 13C labeling pattern of acetoin in culture supernatant. (A) Total ion current of the sample after derivatization with PFBHA and BSTFA, with acetoin eluting after 9.42 min; (B) the corresponding mass spectrum of PFBHA-BSTFA-acetoin; (C) chemical structure of PFBHA-BSTFA-acetoin and the corresponding fragments M-15 and M-43.
TABLE 1.
Mass isotopomer distribution of acetoin determined by GC/MSa
| Fragment | Mass isotopomer fraction | Mean mass isotopomer distribution ± SD |
||
|---|---|---|---|---|
| Theoretical | Unlabeled standard | Unlabeled sampleb | ||
| M-15 | m + 0 | 0.794 | 0.792 ± 0.000 | 0.791 ± 0.002 |
| m + 1 | 0.157 | 0.156 ± 0.000 | 0.157 ± 0.000 | |
| m + 2 | 0.044 | 0.044 ± 0.000 | 0.044 ± 0.000 | |
| m + 3 | 0.005 | 0.006 ± 0.000 | 0.006 ± 0.000 | |
| m + 4 | 0.000 | 0.003 ± 0.000 | 0.002 ± 0.002 | |
| M-43 | m + 0 | 0.811 | 0.806 ± 0.000 | 0.807 ± 0.000 |
| m + 1 | 0.142 | 0.150 ± 0.000 | 0.151 ± 0.000 | |
| m + 2 | 0.041 | 0.043 ± 0.000 | 0.042 ± 0.000 | |
The data comprise the means and standard deviations from duplicate measurements for standard and culture samples and theoretical values inferred from natural isotopic enrichment in nonlabeled substances (47).
Supernatant of A. pasteurianus NCC 316 grown in cocoa pulp simulation medium.
Physiology of A. ghanensis DSM 18895 and A. pasteurianus NCC 316 growth.
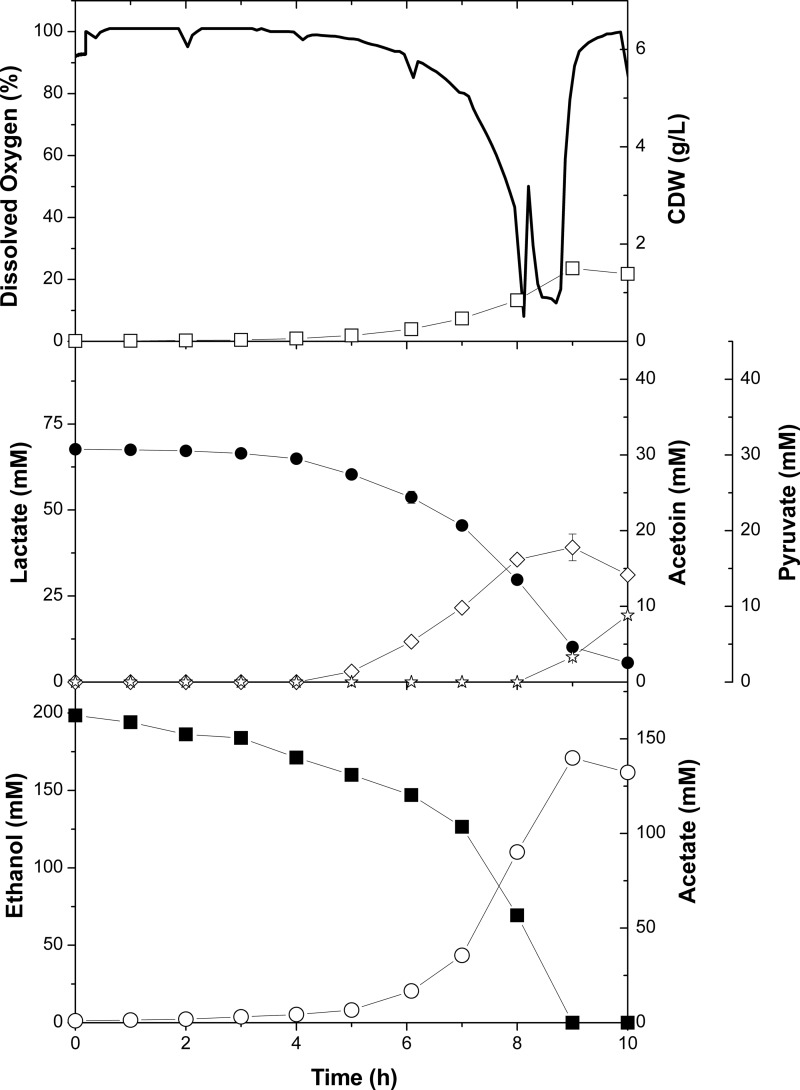
A. pasteurianus NCC 316 coconsumed lactate and ethanol within 8 h of fermentation (Fig. 4). During the first growth phase, acetate was the main product, with a total amount of 140 mM. Additionally, acetoin (18 mM) and pyruvate (13 mM) were produced. The biomass reached a concentration of 1.4 g CDW liter−1, and the pH remained relatively stable at 4.3. The fluxes of substrate uptake and product formation during this phase are listed in Table 2. The concentration of dissolved oxygen (DO) decreased and reached a minimal level of 8% after 8 h of cultivation, which ensures fully aerobic conditions during the process. With depletion of lactate and ethanol, the dissolved oxygen level sharply increased. A. pasteurianus NCC 316 then revealed an intermediate lag phase (see Fig. S1 in the supplemental material). After 20 h, cells started to metabolize acetate, indicated by the decrease of DO. Further on, the biomass increased to a maximum of 2.7 g liter−1 and the final pH after 54 h of cultivation was 8.0.
FIG 4.
Profile of the first growth phase of A. pasteurianus NCC 316 grown in cocoa pulp simulation medium. The data are means from biological duplicates. Black circles, lactate; white circles, acetate; white diamonds, acetoin; black squares, ethanol; white squares, CDW; white stars, pyruvate; black line, dissolved oxygen.
TABLE 2.
Extracellular fluxes of substrates and products and corresponding standard deviations determined from biological duplicates
| Substrate | Mean extracellular flux ± SDa [mmol (g CDW)−1] |
|---|---|
| Lactate | 41.3 ± 1.5 |
| Ethanol | 106.0 ± 2.2 |
| Acetate | 94.0 ± 3.0 |
| Acetoin | 14.3 ± 1.3 |
| Pyruvate | 0 |
The data reflect average fluxes from the first exponential growth phase of A. pasteurianus NCC 316. Pyruvate was not formed during the exponential growth phase.
A. ghanensis DSM 18895 exhibited fermentation characteristics similar to those of A. pasteurianus NCC 316 (see Fig. S2 in the supplemental material). During the first phase of fermentation, lactate and ethanol were coutilized. The concentration of acetate at the end of this growth phase (9 h) was 139 mM, while the DO concentration reached a minimum of 28%, the biomass concentration was 1.0 g liter−1, and the pH was 4.4. Additionally, 12 mM acetoin, but no significant amounts of pyruvate, were formed. While ethanol had been exhausted completely when growth stopped, considerable amounts of lactate (22 mM) remained in the medium and were not metabolized further during the transient lag phase. During a second growth phase, lactate and acetate were then metabolized and the biomass concentration reached a maximum of 1.9 g liter−1. The final pH was 8.5 after 54 h of fermentation.
Overall, both AAB species preferred a combination of lactate and ethanol for their metabolism. However, in the absence of lactate and ethanol, cells could reutilize acetate.
Metabolic origin of proteinogenic amino acids.
In a first set of isotope experiments, cells were grown on fully 13C-labeled lactate or ethanol, together with nonlabeled complex nutrients. According to the experimental design, the 13C labeling pattern of amino acids from cell protein directly provided information on their metabolic origin, i.e., the relative contribution of either lactate or ethanol to the de novo biosynthesis of amino acids during the first exponential growth phase. GC/MS analysis of proteinogenic amino acids of A. ghanensis DSM 18895 and A. pasteurianus NCC 316 revealed a clear picture (see Table S5 in the supplemental material). Aromatic amino acids and amino acids of the serine family were exclusively produced from lactate. These amino acids were naturally labeled when [U-13C]ethanol was present in the fermentation medium but were enriched in 13C when [U-13C]lactate was used. Likewise, amino acids of the pyruvate family were also almost exclusively derived from lactate. This was also observed for lysine and isoleucine, which predominantly originated from lactate. Ethanol contributed to only a small set of biosynthetic pathways. Amino acids stemming from the TCA cycle, including compounds that belong to the aspartate and the glutamate families, were synthesized from both lactate and ethanol.
Contribution of lactate and ethanol to the major fermentation products of A. pasteurianus NCC 316 and A. ghanensis DSM 18895.
The same isotope experiments were next inspected for the contribution of lactate and ethanol to the major products: acetate and acetoin. For this purpose, labeling of the latter was quantified from culture supernatants that were derived from cultures with cocoa pulp simulation medium (first exponential growth phase) containing [U-13C]lactate or [U-13C]ethanol as a tracer substrate (see Tables S7 and S8 in the supplemental material). Taken together, both microorganisms produced most of the acetate (96 to 98%) from ethanol (Fig. 5B). Analogously, acetoin resulted from the metabolism of lactate. Together with the picture from amino acid biosynthesis, this indicated a surprising separation of metabolism. Both strains exhibited two functional pathway modules related to the utilization of lactate and ethanol with only weak exchange fluxes.
FIG 5.
(A) Relative contribution of lactate and ethanol to biosynthesis of amino acids. Squares, fractions originating from lactate and ethanol that were found in the proteinogenic amino acids of A. pasteurianus NCC 316 (left) and A. ghanensis DSM 18895 (right) during the first growth phase in cocoa pulp simulation medium. The full data are given in Table S5 in the supplemental material. (B) Relative contribution (percent) of lactate and ethanol to formation of acetate and acetoin in A. pasteurianus NCC 316 (upper values) and A. ghanensis DSM 18895 (lower values). Violet circles, nodes between the modules of lactate and ethanol metabolism.
Enzyme repertoire of A. pasteurianus NCC 316 and A. ghanensis DSM 18895.
The obviously poor interconnection of the metabolism of lactate and ethanol raised questions about the enzymatic setup around the PEP–pyruvate–acetyl-CoA node, where the two carbon sources are channeled into the core metabolism (Fig. 1). A. pasteurianus NCC 316 and A. ghanensis DSM 18895 tested positive for isocitrate dehydrogenase, an enzyme of the oxidative TCA cycle (Table 3). In both strains, PEP carboxylase was active as anaplerotic enzyme. PEP carboxykinase and ME, enzymes of gluconeogenesis, however, were not expressed (Table 3).
TABLE 3.
Enzymatic repertoire of A. pasteurianus NCC 316 and A. ghanensis DSM 18895 during the first growth phase in cocoa pulp simulation medium
| Enzyme/reaction | Enzyme activity ± SD [U (g CDW)−1] |
|
|---|---|---|
| A. pasteurianus NCC 316 | A. ghanensis DSM 18895 | |
| Isocitrate dehydrogenase (positive control) | 66.9 ± 1.3 | 51.9 ± 5.3 |
| Malic enzyme | ||
| Decarboxylation | NDa | ND |
| Carboxylation | ND | ND |
| PEP carboxylase | 20.3 ± 4.3 | 65.2 ± 15.3 |
| PEP carboxykinase | ||
| Decarboxylation | ND | ND |
| Carboxylation | ND | ND |
ND, not detected [limit of detection, <1 U (g CDW)−1].
Impact of the microbial community on metabolite profiles in simulated cocoa pulp fermentations.
A set of experiments investigated the performance of AAB during an entire two-step cocoa fermentation process. The study combined three different setups for the first phase (only L. fermentum, only S. cerevisiae, L. fermentum plus S. cerevisiae) with a second fermentation phase, in which the setup was inoculated with A. pasteurianus NCC 316. In all cases, A. pasteurianus NCC 316 was grown on a filtrate of prefermented medium from one of the first-phase incubations. Large amounts of lactate and acetate, but not ethanol, were obtained when L. fermentum NCC 575 was used as a starter in the first phase of simulated cocoa pulp fermentation (Table 4). Citrate, which was part of the simulation medium, was largely metabolized by the lactic acid bacteria. The second fermentation phase, in which A. pasteurianus NCC 316 was used as the starter strain, resulted in a significant reduction of lactate, together with the consumption of acetate, whereas the low levels of citrate remained unchanged. A completely different picture was observed when the medium simulating cocoa pulp was incubated with S. cerevisiae NYSC 2. Here, extremely large amounts of ethanol were formed, whereas only low levels of lactate and acetate resulted. The concentration of citrate remained almost unchanged (12). This mixture did not appear to be optimal for A. pasteurianus NCC 316. The AAB only partially utilized ethanol and formed only a small amount of acetate (20 mM). The best performance of A. pasteurianus NCC 316, visualized by the largest amounts of acetate, was observed when L. fermentum and S. cerevisiae were cocultivated during the initial step of fermentation. After the second phase of fermentation, a total amount of 121 mM acetate was observed as the sole product, while citrate, lactate, and ethanol were almost completely consumed.
TABLE 4.
Formation of organic acids and ethanol during simulated two-phase cocoa pulp fermentation
| Organism(s) | Phasea | Mean concn ± SD (mM) |
|||
|---|---|---|---|---|---|
| Citrate | Lactate | Acetate | Ethanol | ||
| L. fermentum NCC 575 | First | 1.3 ± 0.2 | 91.1 ± 1.1 | 106.6 ± 2.0 | NDb |
| Second | 1.0 ± 0.2 | 21.4 ± 1.3 | 76.0 ± 2.2 | ND | |
| S. cerevisiae NYSC 2 | First | 47.0 ± 0.4 | 0.9 ± 0.1 | 7.5 ± 1.3 | 360.9 ± 14.5 |
| Second | 47.1 ± 1.1 | 0.7 ± 0.1 | 21.0 ± 5.7 | 239.2 ± 7.0 | |
| L. fermentum NCC 575 + S. cerevisiae NYSC 2c | First | 0.2 ± 0.2 | 104.1 ± 0.2 | 117.2 ± 3.5 | 75.4 ± 6.9 |
| Second | 0.1 ± 0.1 | 5.8 ± 6.6 | 121.5 ± 15.6 | ND | |
For the first phase, PSM-LAB was inoculated with L. fermentum NCC 575 and/or S. cerevisiae NYSC 2. The cell-free supernatant of each fermentation broth, obtained after incubation for 24 h, was inoculated with A. pasteurianus NCC 316 and incubated for another 12 h (second phase). The concentrations obtained after each of these cultivations are presented.
ND, not detected (limit of detection, <0.2 mM).
Cocultivation of both microorganisms.
DISCUSSION
Acetic acid bacteria exhibit a functionally separated metabolism during coconsumption of two-carbon and three-carbon substrates.
Both Acetobacter species synthesized acetate almost exclusively from ethanol, while only 2 to 4% originated from lactate (Fig. 5B). This was contrary to expectations, because it has previously been suggested that ethanol as well as lactate is converted by A. pasteurianus 386B into acetate (12), but is in line with recent indications from metabolite balancing (23). Based on 13C labeling, our results clearly show that lactate is poorly oxidized into acetate and, vice versa, that ethanol does not contribute to acetoin formation. The latter is completely derived from lactate via pyruvate as a precursor (Fig. 1). Furthermore, our results suggest that the pyruvate decarboxylase pathway is not involved in acetoin formation, since this pathway would use acetaldehyde, produced during assimilation of ethanol (51). Instead, acetoin is synthesized solely via α-acetolactate. The absence of pyruvate decarboxylase activity (low activity) likely seems to be responsible for the low contribution of lactate to acetate formation.
From an energetic perspective, complete oxidation of lactate via the TCA cycle seems optimal (4 mol ATP per mole of lactate) (Table 5). In contrast, the conversion of lactate into acetoin leads to low levels of ATP formation. Further experiments showed that lactate-grown A. pasteurianus NCC 316 indeed shifts from acetoin formation to complete oxidation of lactate into CO2 when ethanol is omitted from the medium (data not shown). We conclude that the TCA cycle is suppressed in the presence of ethanol (52) and that acetoin is an overflow metabolite of pyruvate. Nevertheless, acetoin formation may be advantageous for cellular homoeostasis, as this compound is less toxic than the organic acid. In this regard, acetoin formation could be favorable over acetate production. Analogously, the oxidation of ethanol into acetate and the formation of acetoin from lactate seem to be inefficient from an energetics point of view (Table 5). However, the cells metabolized both products in a second growth phase (see Fig. S1 in the supplemental material) without a significant loss of energy compared to that from the total oxidation of lactate and ethanol (Table 5). Additionally, complete oxidation of lactate via acetate as an overflow metabolite was less efficient (2 mol of ATP per mole of lactate) than complete oxidation of lactate via acetoin (3 mol of ATP per mole of lactate). All of these findings together indicate that A. pasteurianus NCC 316 and A. ghanensis DSM 18895 have adapted their metabolism to a strategy which aims toward the optimization of speed rather than efficiency (53).
TABLE 5.
Energetic efficiency of different metabolic modes for consumption of lactate, ethanol, acetate, and acetoin
| Mode of metabolism | No. of moles formed |
Key enzyme(s), pathway [references(s)] | ||
|---|---|---|---|---|
| NADH + H+ | ATP (GTP) | ATP equivalentsa | ||
| Ethanol-acetate | 2 | 0 | 1 | Membrane-bound dehydrogenase (56, 57) |
| Ethanol-CO2 | 6 | −1c | 2 | Acetyl-CoA synthetase, TCA (22) |
| Acetate-CO2c | 4 | −1c | 1 | Acetyl-CoA synthetase, TCA (22) |
| Lactate-acetate | 2 | 0 | 1 | Pyruvate decarboxylase (73) |
| Lactate-CO2 | 6 | 1 | 4 | Pyruvate dehydrogenase, TCA (22) |
| Lactate-acetoin | 1 | 0 | 0.5 | Acetolactate synthase (21, 40) |
| Acetoinb-CO2 | 10 | 0 | 5 | Acetoin dehydrogenase and acetyl-CoA synthetase, TCA (21, 40) |
As a central finding, our data demonstrate a strict separation of two-carbon (ethanol, acetaldehyde, acetate) and three-carbon (lactate, pyruvate, phosphoenolpyruvate) metabolism in AAB. For example, this becomes obvious from the biosynthesis of amino acids. These are synthesized from precursors of the TCA cycle, from the pentose phosphate pathway, and from gluconeogenesis (Fig. 1 and 5A). Interestingly, all amino acids stemming from intermediates upstream of pyruvate are produced exclusively from lactate during the first growth phase. This confirms that carbon from ethanol does not enter this upper part of metabolism. As an exception to the otherwise strongly separated functional modules of lactate and ethanol metabolism, a minor fraction of lactate-derived carbon replenishes the TCA cycle. Such a separated metabolism is rarely observed in prokaryotic cells and, rather, is a property of eukaryotes, which possess cellular compartments (54). One rare example of a bacterium with a disconnected metabolism is L. fermentum (30, 55). Here, glucose is an energy source, while fructose is entirely used as an electron acceptor for NADH + H+. Thus, separation of metabolism seems to be a property of bacteria with a rather specialized metabolism. AAB are specialized to use membrane-bound oxidation systems for acetate production (41, 56, 57). These systems are directly coupled to respiratory chains, located at the cytoplasmic membrane, and allow oxidation of external substrates in a rather simple manner (41). Since the reaction takes place in the periplasm, no further transport across the membrane into the cytoplasm is required. Thus, although this incomplete oxidation is apparently inefficient (Table 5), it enables bacteria to generate energy with a small set of enzymes. In contrast, channeling of ethanol or acetate into the central metabolism via acetyl-CoA synthetase costs two extra moles of ATP (22, 52, 53). Additionally, the glyoxylate shunt is missing in A. pasteurianus (21, 40) and A. ghanensis (31). As a consequence, ethanol cannot serve as an efficient source for precursors that drain into biomass, such as oxaloacetate, PEP, and pyruvate. Analogously, it was previously shown that A. ghanensis DSM 18895 and A. pasteurianus NCC 316 did not grow on ethanol with ammonium as the only nitrogen source (31). However, due to the direct drain of intermediates from the TCA cycle to biomass, amino acids are partly derived from ethanol (Fig. 5A). Although, as discussed above, a part of the ethanol replenishes the TCA cycle, gluconeogenic intermediates are not derived from ethanol.
Functional separation of two-carbon and three-carbon compound metabolism in acetic acid bacteria under cocoa pulp fermentation-simulating conditions is created by a lack of phosphoenolpyruvate carboxykinase and malic enzyme.
The link between the metabolism of two-carbon compounds, such as ethanol, and that of three-carbon compounds through gluconeogenesis is cataplerotic reactions of PEPCK and ME (Fig. 1). These reactions convert intermediates from the TCA cycle (oxaloacetate, malate) into PEP, which in turn enters gluconeogenesis.
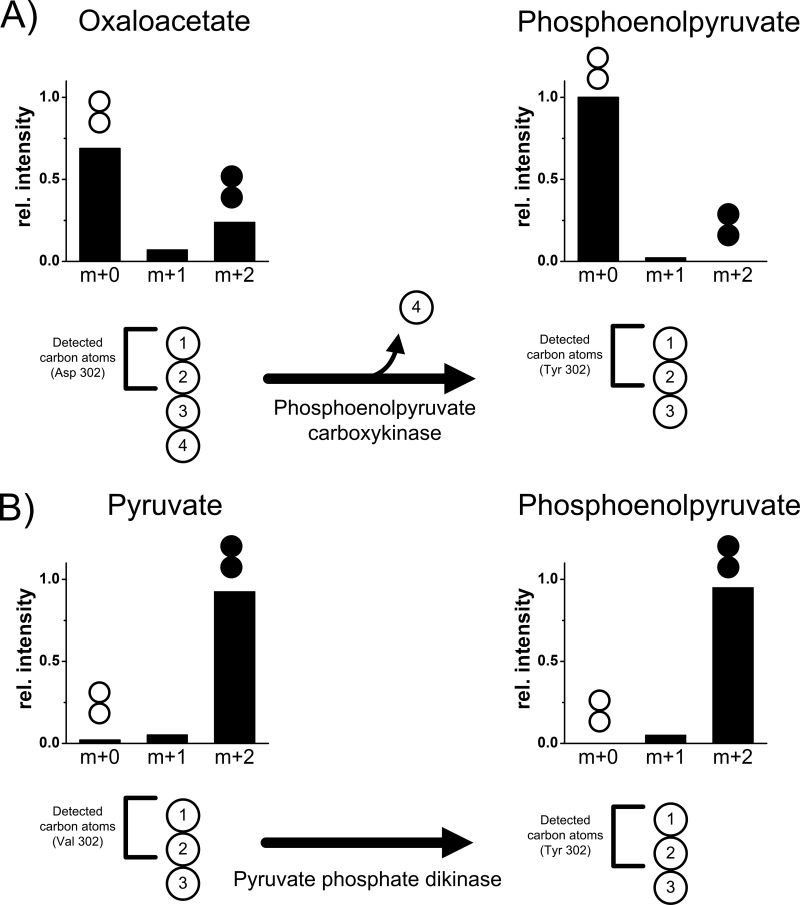
A deeper inspection of the labeling patterns provides direct evidence of their in vivo significance. In the case of a back-flux from oxaloacetate to PEP, one would expect equal 13C enrichment in carbons C-1 and C-2 of oxaloacetate and in the corresponding carbons of PEP. In contrast to significant enrichment in the oxaloacetate pool of A. pasteurianus NCC 316, the PEP pool is completely nonlabeled (Fig. 6A). We conclude that the flux through PEPCK is zero. In agreement with that conclusion, cells did not exhibit activity for this enzyme (Table 3). These results suggest that, in fact, the genome of A. pasteurianus NCC 316 lacks PEPCK. Also, ME is not active, so the possibility of a contribution of TCA cycle metabolites to gluconeogenesis back-flux can be excluded. In fact, the upper part of metabolism is directly supplied from lactate, e.g., for biosynthesis of amino acids of the serine and the aromatic families (see Table S5 in the supplemental material). In this regard, Acetobacter species can make use of pyruvate phosphate dikinase, which converts pyruvate directly into PEP (58, 59). Notably, this reaction requires 2 moles of ATP. Figure 6B shows that the 13C labeling patterns of pyruvate and PEP from isotope experiments are virtually identical. Similarly, Acetobacter aceti lacks oxaloacetate decarboxylation during growth on three-carbon substrates, while pyruvate phosphate dikinase is active (58, 60). Thus, pyruvate phosphate dikinase seems to play an important role in gluconeogenesis in A. pasteurianus NCC 316 and A. ghanensis DSM 18895. Analogously to A. aceti (61), anaplerotic phosphoenolpyruvate carboxylase (PEPC) was present in A. pasteurianus NCC 316 and A. ghanensis DSM 18895 (Table 3).
FIG 6.
Biosynthesis of phosphoenolpyruvate from lactate or ethanol via phosphoenolpyruvate carboxykinase (A) and pyruvate phosphate dikinase (B) by A. pasteurianus NCC 316. (A) Carbon transition in the reaction of phosphoenolpyruvate carboxykinase and experimental labeling profiles of oxaloacetate and phosphoenolpyruvate resulting from growth on a mixture of [12C3]lactate and [13C2]ethanol. Experimental data are derived from the fragments of aspartate and tyrosine at m/z 302 (Asp 302, Tyr 302) containing carbon atoms C-1 and C-2, as described in the supplemental material. (B) Carbon transition in the reaction of pyruvate phosphate dikinase and experimental labeling profiles of pyruvate and phosphoenolpyruvate resulting from growth on a mixture of [13C3]lactate and [12C2]ethanol. Experimental data are derived from the fragments of valine and tyrosine at m/z 302 (Val 302, Tyr 302) containing carbon atoms C-1 and C-2, as described in the supplemental material.
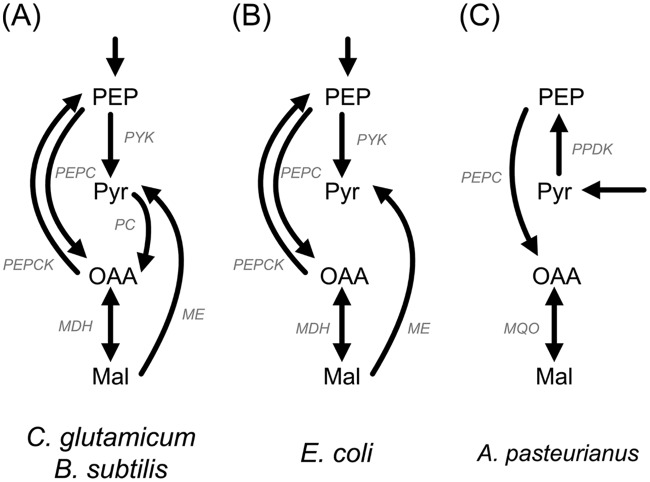
Different enzymes form the metabolic link between glycolysis, gluconeogenesis, and the TCA cycle (Fig. 7). In general, this node is flexible to adjust metabolism in response to anabolic and energetic demands under a given condition (27). Additionally, in some bacteria, carbon flux control through the PEP-pyruvate-oxaloacetate node is complex; e.g., pyruvate carboxylase (PC) and ME are simultaneously active during growth on glucose (62, 63). In contrast, A. pasteurianus NCC 316 and A. ghanensis DSM 18895 use a rather reduced enzymatic setup. For instance, to form PEP from pyruvate, Corynebacterium glutamicum uses PC and PEPCK (Fig. 7). In this set of reactions, 2 moles of ATP is consumed in order to produce 1 mole of PEP. Although the genes for these enzymes are annotated in the genomes of A. pasteurianus IFO 3283_01 and 386B (see Table S1 in the supplemental material), this mode is not used by A. pasteurianus NCC 316 or A. ghanensis DSM 18895. This seems reasonable, because in the presence of an active pyruvate phosphate dikinase, a lack of PEPCK prevents futile cycling. Due to this reduced enzymatic setup, however, there is no interconnection between three-carbon and two-carbon substances in these microorganisms, at least not under the conditions studied. Switching the regulation of malic enzyme expression could be the key event required to switch to different metabolic modes, which are probably needed in other environments.
FIG 7.
Phosphoenolpyruvate-pyruvate-oxaloacetate node in Corynebacterium glutamicum and Bacillus subtilis (A), Escherichia coli (B), and A. pasteurianus (C). Abbreviations for the enzymes are given in Table S1 the supplemental material. MDH, malate dehydrogenase.
The full picture: metabolic flux distribution of A. pasteurianus.
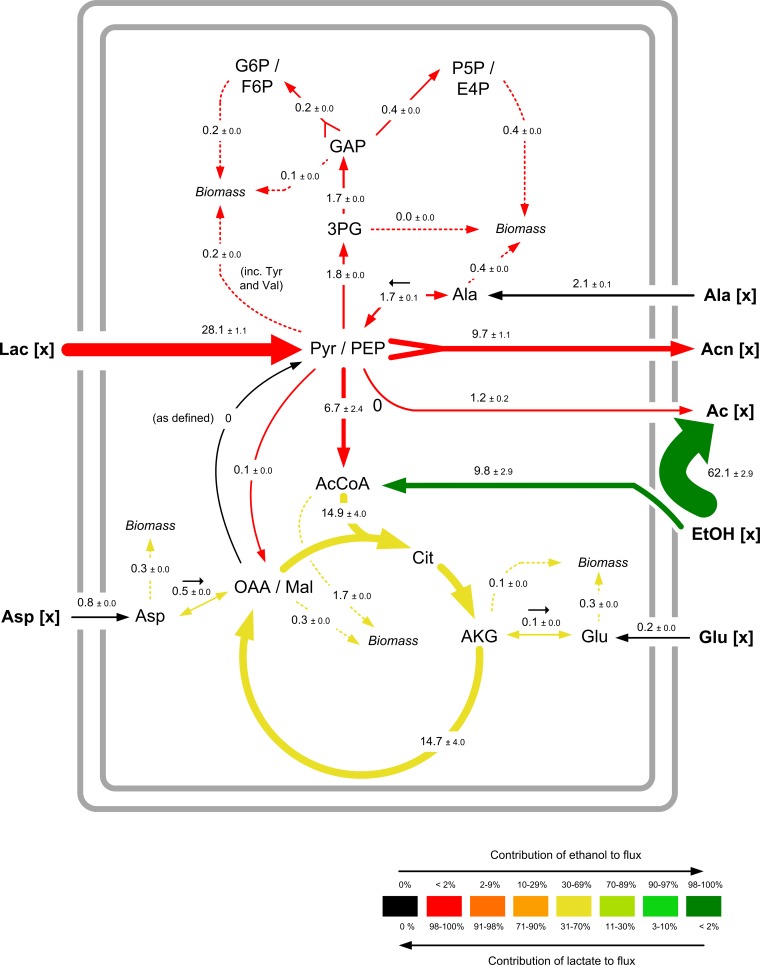
Information on the metabolic repertoire from genome annotation (Fig. 1) and enzyme measurements (Table 3), together with experimental data on growth and product formation (Table 2) and the 13C labeling patterns from the parallel isotope studies (see Table S12 in the supplemental material), has now been integrated. On the basis of the experimental findings, the metabolic network could be condensed into a model (see Table S11 in the supplemental material) that allows high-resolution flux estimations (more detailed information is available in the Appendix). The resulting metabolic flux distribution is shown in Fig. 8. Ethanol is almost completely converted into acetate. Because the corresponding dehydrogenases are directly linked to the respiratory chain in A. pasteurianus (41, 56), 1 molecule of ethanol oxidized into acetate accounts for two electron pairs. With a P/O ratio of 0.5 (53), this corresponds to 62 mmol of ATP per 100 mmol of total substrate influx and 53% of total ATP formation. The TCA cycle contributed to ~40% of total ATP formation, which is composed of ~17% ATP from ethanol metabolism and ~23% ATP from lactate metabolism. Lactate conversion into acetoin is responsible for 8% of the ATP pool. Thus, ethanol is the major energy source of A. pasteurianus NCC 316 under conditions of cocoa fermentation. In contrast, lactate is the main source for the pyruvate pool, which in turn supplies the cell with precursors for biomass compounds, such as amino acids. However, the major part of pyruvate (64% of the total pyruvate/PEP pool) is directed toward the acetoin formation pathway(s). As a result, carbon flux through the TCA cycle is only 15% of the total carbon uptake. This might be due to suppression of the TCA cycle in the presence of ethanol (52). Considering the fully aerobic growth conditions, this suggests that acetoin is formed as an overflow metabolite. Additionally, the decreased activity of the TCA cycle prevents a catabolic breakdown of acetate.
FIG 8.
Metabolic fluxes of A. pasteurianus NCC 316 during the first growth phase in cocoa pulp simulation medium. The data are given as relative fluxes, normalized to the cumulative uptake flux of lactate and ethanol (see Tables S10 and S11 in the supplemental material). The relative flux intensity is illustrated by the arrow thickness, and the individual contribution of lactate and ethanol to the fluxes is indicated by the arrow color. Solid arrows correspond to central metabolic fluxes, and dashed arrows correspond to the drain of precursors into biomass. The net direction of a reversible reaction is indicated by a small black arrow. The fluxes were derived via metabolite and isotopomer balancing recruiting extracellular fluxes (see Table S11 in the supplemental material) and the 13C labeling pattern of proteinogenic amino acids and acetate. Simulated and experimental mass isotopomer distributions are presented in Fig. S3 in the supplemental material. In addition to the net fluxes (in large type), 90% confidence intervals from Monte Carlo analysis are shown (in smaller type). [x], extracellular substrates and products.
In addition to lactate and ethanol as the main energy and carbon sources, respectively, some amino acids played a superior role in the central metabolism of A. pasteurianus NCC 316 (Fig. 8). A small portion of alanine is catabolized and supports the pyruvate pool. Obviously, aspartate serves as a source of oxaloacetate and replenishes the TCA cycle, replacing anaplerosis via PEPC. Glutamate is involved in biosynthetic reactions as a donor of amino groups (64); therefore, there is a steady interconversion between glutamate and its keto acid, α-ketoglutarate. Although alanine, aspartate, and glutamate are metabolized, the biosynthesis of these amino acids is active, suggesting that the involved biosynthetic reactions are constitutive in A. pasteurianus NCC 316.
Bacteria require NADPH + H+ for many biosynthetic processes. The cofactor is typically formed through the pentose phosphate pathway (glucose-6-phosphate dehydrogenase) and the TCA cycle (isocitrate dehydrogenase) (65–67). Since the pentose phosphate pathway is poorly active in A. pasteurianus NCC 316, we wondered how A. pasteurianus covers its biosynthetic demand for NADPH + H+. Considering the cellular composition (68) and the incorporation of amino acids from the extracellular environment, the biosynthetic NADPH + H+ requirement equals approximately 7 mmol/g CDW. Due to the low biomass yield (Table 2), this corresponds to only 5% of total substrate uptake and 7% of ethanol conversion. In contrast, E. coli requires 16 to 17 mmol NADPH + H+ per g CDW, which corresponds to 140% of the glucose uptake flux (69). Through isocitrate dehydrogenase, the TCA cycle provides A. pasteurianus NCC 316 with 22 mmol of NADPH + H+ per g CDW (Fig. 8). Additionally, A. pasteurianus is able to produce NAD(P)H + H+ via soluble alcohol and aldehyde dehydrogenases (21, 51). Among species of Acetobacter, only a soluble aldehyde dehydrogenase of Acetobacter rancens (a later synonym of A. pasteurianus), which requires NADP+, has been characterized (70). Furthermore, an NAD+-dependent soluble alcohol dehydrogenase was identified in A. pasteurianus (51). This indicates that 15 mmol of NADPH + H+ per g CDW is also formed from the intracellular metabolic breakdown of ethanol. Hence, the biosynthetic NADPH demand of A. pasteurianus NCC 316 is fully covered; in fact, there seems to be an apparent excess of NADPH + H+. A proton-translocating nicotinamide nucleotide transhydrogenase might be involved in NADPH oxidation (21).
Taken together, the metabolic flux distribution highlights specific roles of lactate and ethanol in the central metabolism of A. pasteurianus NCC 316 during cocoa pulp fermentation. In this constellation, the main role of ethanol is to generate metabolic energy via acetate production. While lactate is also an energy source, it additionally supports growth by forming precursors, particularly via gluconeogenesis and the pentose phosphate pathway. Thereby, acetate production is intensified, which is one of the main purposes of cocoa fermentation. Consequently, a balanced ratio of lactate and ethanol is important for the success of the fermentation process.
Species interactions determine the quality of cocoa fermentation.
The formation of adequate amounts of acetate is critical for well-fermented cocoa beans (1, 2). In the simulated triculture cocoa pulp fermentation, the largest amount of acetate was obtained when L. fermentum NCC 575 and S. cerevisiae NYSC 2 were cocultivated in the first phase of fermentation. LAB produce lactic acid, which supports the growth of A. pasteurianus NCC 316. Ethanol supplied by yeasts is required for the synthesis of acetate by the acetic acid bacteria and suppression of acetate-degrading enzymes. Furthermore, LAB reduce the amount of citrate and AAB degrade lactate, which positively affects the flavor of the cocoa beans (14). When the cultivation is performed with only L. fermentum NCC 575 in the first phase of cultivation, A. pasteurianus NCC 316 even degrades acetate. For this reason, high levels of ethanol, produced during the initial phase of cocoa pulp fermentation, seem to be important in order to repress the breakdown of acetate. Similarly, small amounts of acetate are produced when only yeast is present in the first phase of simulated cocoa fermentation, since AAB form little biomass on ethanol (53, 71). These findings clearly show that the pathway activities of LAB, AAB, and yeast are important for the successful fermentation of cocoa pulp (11, 23, 72).
APPENDIX
Metabolic model for flux calculations.
The metabolic model for flux estimations was derived from the genomic repertoire (Fig. 1) and experimental findings of this study. Extracellular fluxes are given in Table 2. Since PEPCK and ME were found to be inactive, these reactions were not included. Furthermore, the gluconeogenesis and pentose phosphate pathway were lumped together, since these pathways could not be distinguished in this complex environment. Additionally, gluconeogenesis, the pentose phosphate pathway, and the TCA cycle were considered to be irreversible. Because acetoin was produced exclusively from lactate, the acetoin synthesis pathway was simplified. Pyruvate and PEP were pooled, due to the potentially parallel activity of PEPC and pyruvate carboxylase. Additionally, malate and oxaloacetate as well as erythrose-4-phosphate and pentose-5-phosphate were lumped together, respectively. The drain of precursors to biomass was derived from the bacterial cell composition (68). Additionally, the degrees of de novo synthesis of amino acids were considered, as determined from the experiments. The degrees of synthesis of some amino acids (arginine, cysteine, methionine, and proline) were not determined directly but deduced from those of other amino acids of the same families. The contribution of histidine to the biomass was negligible (68). The mass isotopomer distributions of the [M-57] fragments of alanine, valine, glutamate, and aspartate, the [M-95] fragment of glutamate, and the fragments at m/z 302 of valine and aspartate, as well as the labeling of acetate, were used to fit the model. For this reason, the corresponding uptake fluxes of the amino acids were determined from the model fit. Additionally, the deamination of alanine, aspartate, and glutamate was considered in the metabolic model.
Identical flux distributions, obtained from diverse parameter optimization runs with multiple initialization values, indicated that a global optimum was found. The mean squared error between experimental and simulated mass isotopomer distributions was 4.2 × 10−4, indicating an excellent fit. From Monte Carlo simulations, an average error of the resulting metabolic fluxes of 0.88 was obtained.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stéphane Duboux and Christof Gysler for their support in providing strains of the Nestlé Culture Collection. We further appreciate the technical assistance of Elena Kempf, Sandra Hübner, and Martin Wewers.
Footnotes
Published ahead of print 16 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01048-14.
REFERENCES
- 1.Biehl B, Passern D, Sagemann W. 1982. Effect of acetic acid on subcellular structures of cocoa bean cotyledons. J. Sci. Food Agric. 33:1101–1109. 10.1002/jsfa.2740331107 [DOI] [Google Scholar]
- 2.de Brito ES, García NHP, Gallão MI, Cortelazzo AL, Fevereiro PS, Braga MR. 2001. Structural and chemical changes in cocoa (Theobroma cacao L) during fermentation, drying and roasting. J. Sci. Food Agric. 81:281–288. [DOI] [Google Scholar]
- 3.Quesnel VC. 1965. Agents inducing the death of cacao seeds during fermentation. J. Sci. Food Agric. 16:441–447. 10.1002/jsfa.2740160804 [DOI] [Google Scholar]
- 4.Forsyth WGC, Quesnel VC. 1963. The mechanism of cacao curing. Adv. Enzymol. Relat. Areas Mol. Biol. 25:457–492 [DOI] [PubMed] [Google Scholar]
- 5.Camu N, de Winter T, Addo SK, Takrama JS, Bernaert H, de Vuyst L. 2008. Fermentation of cocoa beans: influence of microbial activities and polyphenol concentrations on the flavour of chocolate. J. Sci. Food Agric. 88:2288–2297. 10.1002/jsfa.3349 [DOI] [Google Scholar]
- 6.Schwan RF, Wheals AE. 2004. The microbiology of cocoa fermentation and its role in chocolate quality. Crit. Rev. Food Sci. Nutr. 44:205–221. 10.1080/10408690490464104 [DOI] [PubMed] [Google Scholar]
- 7.Papalexandratou Z, de Vuyst L. 2011. Assessment of the yeast species composition of cocoa bean fermentations in different cocoa-producing regions using denaturing gradient gel electrophoresis. FEMS Yeast Res. 11:564–574. 10.1111/j.1567-1364.2011.00747.x [DOI] [PubMed] [Google Scholar]
- 8.Daniel H, Vrancken G, Takrama JF, Camu N, de Vos P, de Vuyst L. 2009. Yeast diversity of Ghanaian cocoa bean heap fermentations. FEMS Yeast Res. 9:774–783. 10.1111/j.1567-1364.2009.00520.x [DOI] [PubMed] [Google Scholar]
- 9.Papalexandratou Z, Lefeber T, Bahrim B, Lee OS, Daniel H, de Vuyst L. 2013. Hanseniaspora opuntiae, Saccharomyces cerevisiae, Lactobacillus fermentum, and Acetobacter pasteurianus predominate during well-performed Malaysian cocoa bean box fermentations, underlining the importance of these microbial species for a successful cocoa bean fermentation process. Food Microbiol. 35:73–85. 10.1016/j.fm.2013.02.015 [DOI] [PubMed] [Google Scholar]
- 10.Jespersen L, Nielsen D, Honholt S, Jakobsen M. 2005. Occurrence and diversity of yeasts involved in fermentation of West African cocoa beans. FEMS Yeast Res. 5:441–453. 10.1016/j.femsyr.2004.11.002 [DOI] [PubMed] [Google Scholar]
- 11.Lefeber T, Papalexandratou Z, Gobert W, Camu N, de Vuyst L. 2012. On-farm implementation of a starter culture for improved cocoa bean fermentation and its influence on the flavour of chocolates produced thereof. Food Microbiol. 30:379–392. 10.1016/j.fm.2011.12.021 [DOI] [PubMed] [Google Scholar]
- 12.Lefeber T, Janssens M, Camu N, de Vuyst L. 2010. Kinetic analysis of strains of lactic acid bacteria and acetic acid bacteria in cocoa pulp simulation media toward development of a starter culture for cocoa bean fermentation. Appl. Environ. Microbiol. 76:7708–7716. 10.1128/AEM.01206-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camu N, de Winter T, Verbrugghe K, Cleenwerck I, Vandamme P, Takrama JS, Vancanneyt M, de Vuyst L. 2007. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl. Environ. Microbiol. 73:1809–1824. 10.1128/AEM.02189-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holm CS, Aston JW, Douglas K. 1993. The effects of the organic acids in cocoa on the flavour of chocolate. J. Sci. Food Agric. 61:65–71. 10.1002/jsfa.2740610111 [DOI] [Google Scholar]
- 15.Papalexandratou Z, Falony G, Romanens E, Jimenez JC, Amores F, Daniel H, de Vuyst L. 2011. Species diversity, community dynamics, and metabolite kinetics of the microbiota associated with traditional Ecuadorian spontaneous cocoa bean fermentations. Appl. Environ. Microbiol. 77:7698–7714. 10.1128/AEM.05523-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papalexandratou Z, Vrancken G, de Bruyne K, Vandamme P, de Vuyst L. 2011. Spontaneous organic cocoa bean box fermentations in Brazil are characterized by a restricted species diversity of lactic acid bacteria and acetic acid bacteria. Food Microbiol. 28:1326–1338. 10.1016/j.fm.2011.06.003 [DOI] [PubMed] [Google Scholar]
- 17.Camu N, Gonzalez A, de Winter T, van Schoor A, de Bruyne K, Vandamme P, Takrama JS, Addo SK, de Vuyst L. 2008. Influence of turning and environmental contamination on the dynamics of populations of lactic acid and acetic acid bacteria involved in spontaneous cocoa bean heap fermentation in Ghana. Appl. Environ. Microbiol. 74:86–98. 10.1128/AEM.01512-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefeber T, Gobert W, Vrancken G, Camu N, de Vuyst L. 2011. Dynamics and species diversity of communities of lactic acid bacteria and acetic acid bacteria during spontaneous cocoa bean fermentation in vessels. Food Microbiol. 28:457–464. 10.1016/j.fm.2010.10.010 [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Armisen T, Papalexandratou Z, Hendryckx H, Camu N, Vrancken G, Vuyst L, Cornelis P. 2010. Diversity of the total bacterial community associated with Ghanaian and Brazilian cocoa bean fermentation samples as revealed by a 16 S rRNA gene clone library. Appl. Microbiol. Biotechnol. 87:2281–2292. 10.1007/s00253-010-2698-9 [DOI] [PubMed] [Google Scholar]
- 20.Lefeber T, Janssens M, Moens F, Gobert W, de Vuyst L. 2011. Interesting starter culture strains for controlled cocoa bean fermentation revealed by simulated cocoa pulp fermentations of cocoa-specific lactic acid bacteria. Appl. Environ. Microbiol. 77:6694–6698. 10.1128/AEM.00594-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Illeghems K, de Vuyst L, Weckx S. 2013. Complete genome sequence and comparative analysis of Acetobacter pasteurianus 386B, a strain well-adapted to the cocoa bean fermentation ecosystem. BMC Genomics 14:526. 10.1186/1471-2164-14-526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakurai K, Arai H, Ishii M, Igarashi Y. 2011. Transcriptome response to different carbon sources in Acetobacter aceti. Microbiology 157:899–910. 10.1099/mic.0.045906-0 [DOI] [PubMed] [Google Scholar]
- 23.Moens F, Lefeber T, de Vuyst L. 2014. Oxidation of metabolites highlights the microbial interactions and role of Acetobacter pasteurianus during cocoa bean fermentation. Appl. Environ. Microbiol. 80:1848–1857. 10.1128/AEM.03344-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Ley J, Schell J. 1962. Lactate and pyruvate catabolism in acetic acid bacteria. J. Gen. Microbiol. 29:589–601. 10.1099/00221287-29-4-589 [DOI] [PubMed] [Google Scholar]
- 25.de Ley J. 1959. On the formation of acetoin by Acetobacter. J. Gen. Microbiol. 21:352–365. 10.1099/00221287-21-2-352 [DOI] [PubMed] [Google Scholar]
- 26.Chandra Raj K, Ingram L, Maupin-Furlow J. 2001. Pyruvate decarboxylase: a key enzyme for the oxidative metabolism of lactic acid by Acetobacter pasteurianus. Arch. Microbiol. 176:443–451. 10.1007/s002030100348 [DOI] [PubMed] [Google Scholar]
- 27.Sauer U, Eikmanns BJ. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29:765–794. 10.1016/j.femsre.2004.11.002 [DOI] [PubMed] [Google Scholar]
- 28.Kohlstedt M, Becker J, Wittmann C. 2010. Metabolic fluxes and beyond—systems biology understanding and engineering of microbial metabolism. Appl. Microbiol. Biotechnol. 88:1065–1075. 10.1007/s00253-010-2854-2 [DOI] [PubMed] [Google Scholar]
- 29.Sauer U, Lasko DR, Fiaux J, Hochuli M, Glaser R, Szyperski T, Wüthrich K, Bailey JE. 1999. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J. Bacteriol. 181:6679–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adler P, Bolten CJ, Dohnt K, Hansen CE, Wittmann C. 2013. Core fluxome and metafluxome of lactic acid bacteria under simulated cocoa pulp fermentation conditions. Appl. Environ. Microbiol. 79:5670–5681. 10.1128/AEM.01483-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cleenwerck I, Camu N, Engelbeen K, de Winter T, Vandemeulebroecke K, de Vos P, de Vuyst L. 2007. Acetobacter ghanensis sp. nov., a novel acetic acid bacterium isolated from traditional heap fermentations of Ghanaian cocoa beans. Int. J. Syst. Evol. Microbiol. 57:1647–1652. 10.1099/ijs.0.64840-0 [DOI] [PubMed] [Google Scholar]
- 32.Atlas RM. 2006. Handbook of microbiological media for the examination of food, 2nd ed. CRC Press LLC, Boca Raton, FL [Google Scholar]
- 33.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wittmann C, Hans M, Heinzle E. 2002. In vivo analysis of intracellular amino acid labelings by GC/MS. Anal. Biochem. 307:379–382. 10.1016/S0003-2697(02)00030-1 [DOI] [PubMed] [Google Scholar]
- 35.Spaulding R, Charles M. 2002. Comparison of methods for extraction, storage, and silylation of pentafluorobenzyl derivatives of carbonyl compounds and multi-functional carbonyl compounds. Anal. Bioanal. Chem. 372:808–816. 10.1007/s00216-002-1252-8 [DOI] [PubMed] [Google Scholar]
- 36.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 37.Jetten MSM, Sinskey AJ. 1993. Characterization of phosphoenolpyruvate carboxykinase from Corynebacterium glutamicum. FEMS Microbiol. Lett. 111:183–188. 10.1111/j.1574-6968.1993.tb06383.x [DOI] [Google Scholar]
- 38.Gourdon P, Baucher M, Lindley ND, Guyonvarch A. 2000. Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl. Environ. Microbiol. 66:2981–2987. 10.1128/AEM.66.7.2981-2987.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becker J, Klopprogge C, Schröder H, Wittmann C. 2009. Metabolic engineering of the tricarboxylic acid cycle for improved lysine production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 75:7866–7869. 10.1128/AEM.01942-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azuma Y, Hosoyama A, Matsutani M, Furuya N, Horikawa H, Harada T, Hirakawa H, Kuhara S, Matsushita K, Fujita N, Shirai M. 2009. Whole-genome analyses reveal genetic instability of Acetobacter pasteurianus. Nucleic Acids Res. 37:5768–5783. 10.1093/nar/gkp612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsushita K, Toyama H, Adachi O. 1994. Respiratory chains and bioenergetics of acetic acid bacteria, p 248 In Rose A, Tempest D. (ed), Advances in microbial physiology. Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 42.Kondo K, Beppu T, Horinouchi S. 1995. Cloning, sequencing, and characterization of the gene encoding the smallest subunit of the three-component membrane-bound alcohol dehydrogenase from Acetobacter pasteurianus. J. Bacteriol. 177:5048–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanchanarach W, Theeragool G, Yakushi T, Toyama H, Adachi O, Matsushita K. 2010. Characterization of thermotolerant Acetobacter pasteurianus strains and their quinoprotein alcohol dehydrogenases. Appl. Microbiol. Biotechnol. 85:741–751. 10.1007/s00253-009-2203-5 [DOI] [PubMed] [Google Scholar]
- 44.Matsutani M, Hirakawa H, Yakushi T, Matsushita K. 2011. Genome-wide phylogenetic analysis of Gluconobacter, Acetobacter, and Gluconacetobacter. FEMS Microbiol. Lett. 315:122–128. 10.1111/j.1574-6968.2010.02180.x [DOI] [PubMed] [Google Scholar]
- 45.Quek L, Wittmann C, Nielsen LK, Krömer JO. 2009. OpenFLUX: efficient modelling software for 13C-based metabolic flux analysis. Microb. Cell Fact. 8:25. 10.1186/1475-2859-8-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Winden WA, Wittmann C, Heinzle E, Heijnen JJ. 2002. Correcting mass isotopomer distributions for naturally occurring isotopes. Biotechnol. Bioeng. 80:477–479. 10.1002/bit.10393 [DOI] [PubMed] [Google Scholar]
- 47.Rosman KJR, Taylor PDP. 1998. Isotopic compositions of the elements 1997. Pure Appl. Chem. 70:217–235. 10.1351/pac199870010217 [DOI] [Google Scholar]
- 48.Wittmann C, Heinzle E. 2002. Genealogy profiling through strain improvement by using metabolic network analysis: metabolic flux genealogy of several generations of lysine-producing corynebacteria. Appl. Environ. Microbiol. 68:5843–5859. 10.1128/AEM.68.12.5843-5859.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wittmann C, Heinzle E. 2005. Metabolic activity profiling by 13C tracer experiments and mass spectrometry in Corynebacterium glutamicum. In Barredo JL. (ed), Methods in biotechnology. Humana Press, Totowa, NJ [Google Scholar]
- 50.Wittmann C, Krömer JO, Kiefer P, Binz T, Heinzle E. 2004. Impact of the cold shock phenomenon on quantification of intracellular metabolites in bacteria. Anal. Biochem. 327:135–139. 10.1016/j.ab.2004.01.002 [DOI] [PubMed] [Google Scholar]
- 51.Chinnawirotpisan P, Theeragool G, Limtong S, Toyama H, Adachi OO, Matsushita K. 2003. Quinoprotein alcohol dehydrogenase is involved in catabolic acetate production, while NAD-dependent alcohol dehydrogenase in ethanol assimilation in Acetobacter pasteurianus SKU1108. J. Biosci. Bioeng. 96:564–571. 10.1016/S1389-1723(04)70150-4 [DOI] [PubMed] [Google Scholar]
- 52.Saeki A, Matsushita K, Takeno S, Taniguchi M, Toyama H, Theeragool G, Lotong N, Adachi O. 1999. Enzymes responsible for acetate oxidation by acetic acid bacteria. Biosci. Biotechnol. Biochem. 63:2102–2109. 10.1271/bbb.63.2102 [DOI] [PubMed] [Google Scholar]
- 53.Luttik M, van Spanning R, Schipper D, van Dijken JP, Pronk JT. 1997. The low biomass yields of the acetic acid bacterium Acetobacter pasteurianus are due to a low stoichiometry of respiration-coupled proton translocation. Appl. Environ. Microbiol. 63:3345–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bowsher CG, Tobin AK. 2001. Compartmentation of metabolism within mitochondria and plastids. J. Exp. Bot. 52:513–527. 10.1093/jexbot/52.356.513 [DOI] [PubMed] [Google Scholar]
- 55.Aarnikunnas J, von Weymarn N, Rönnholm K, Leisola M, Palva A. 2003. Metabolic engineering of Lactobacillus fermentum for production of mannitol and pure l-lactic acid or pyruvate. Biotechnol. Bioeng. 82:653–663. 10.1002/bit.10615 [DOI] [PubMed] [Google Scholar]
- 56.Yakushi T, Matsushita K. 2010. Alcohol dehydrogenase of acetic acid bacteria: structure, mode of action, and applications in biotechnology. Appl. Microbiol. Biotechnol. 86:1257–1265. 10.1007/s00253-010-2529-z [DOI] [PubMed] [Google Scholar]
- 57.Adachi O, Miyagawa E, Shinagawam E, Matsushita K, Ameyana M. 1978. Purification and properties of particulate alcohol dehydrogenase from Acetobacter aceti. Agric. Biol. Chem. 42:2331–2340. 10.1271/bbb1961.42.2331 [DOI] [Google Scholar]
- 58.Schwitzguebel J, Ettlinger L. 1979. Pyruvate, orthophosphate dikinase from Acetobacter aceti. Arch. Microbiol. 122:103–108. 10.1007/BF00408052 [DOI] [Google Scholar]
- 59.Benziman M, Eizen N. 1971. Pyruvate-phosphate dikinase and the control of gluconeogenesis in Acetobacter xylinum. J. Biol. Chem. 246:57–61 [PubMed] [Google Scholar]
- 60.Schwitzguébel J, Ettlinger L. 1980. Oxaloacetate decarboxylase from Acetobacter aceti. Arch. Microbiol. 124:63–68. 10.1007/BF00407029 [DOI] [Google Scholar]
- 61.Schwitzguébel J, Ettlinger L. 1979. Phosphoenolpyruvate carboxylase from Acetobacter aceti. Arch. Microbiol. 122:109–115. 10.1007/BF00408053 [DOI] [Google Scholar]
- 62.Petersen S, de Graaf AA, Eggeling L, Möllney M, Wiechert W, Sahm H. 2000. In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J. Biol. Chem. 275:35932–35941. 10.1074/jbc.M908728199 [DOI] [PubMed] [Google Scholar]
- 63.Dauner M, Storni T, Sauer U. 2001. Bacillus subtilis metabolism and energetics in carbon-limited and excess-carbon chemostat culture. J. Bacteriol. 183:7308–7317. 10.1128/JB.183.24.7308-7317.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stephanopoulos G, Aristidou ANJ. 1998. Metabolic engineering. Principles and methodologies, 1st ed. Academic Press, San Diego, CA [Google Scholar]
- 65.Csonka LN, Fraenkel DG. 1977. Pathways of NADPH formation in Escherichia coli. J. Biol. Chem. 252:3382–3391 [PubMed] [Google Scholar]
- 66.Rühl M, Le Coq D, Aymerich S, Sauer U. 2012. 13C-flux analysis reveals NADPH-balancing transhydrogenation cycles in stationary phase of nitrogen-starving Bacillus subtilis. J. Biol. Chem. 287:27959–27970. 10.1074/jbc.M112.366492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Becker J, Klopprogge C, Zelder O, Heinzle E, Wittmann C. 2005. Amplified expression of fructose 1,6-bisphosphatase in Corynebacterium glutamicum increases in vivo flux through the pentose phosphate pathway and lysine production on different carbon sources. Appl. Environ. Microbiol. 71:8587–8596. 10.1128/AEM.71.12.8587-8596.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ingraham JL, Maaloe O, Neidhardt FC. 1983. Growth of the bacterial cell. Sinauer Associates, Sunderland, MA [Google Scholar]
- 69.Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E. 2004. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279:6613–6619. 10.1074/jbc.M311657200 [DOI] [PubMed] [Google Scholar]
- 70.Hommel R, Kurth J, Kleber H. 1988. NADP+-dependent aldehyde dehydrogenase from Acetobacter rancens CCM 1774: purification and properties. J. Basic Microbiol. 28:25–33. 10.1002/jobm.3620280105 [DOI] [Google Scholar]
- 71.Russell JB, Cook GM. 1995. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev. 59:48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ho VTT, Zhao J, Fleet G. 2014. Yeasts are essential for cocoa bean fermentation. Int. J. Food Microbiol. 174:72–87. 10.1016/j.ijfoodmicro.2013.12.014 [DOI] [PubMed] [Google Scholar]
- 73.Chandra Raj K, Ingram L, Maupin-Furlow J. 2001. Pyruvate decarboxylase: a key enzyme for the oxidative metabolism of lactic acid by Acetobacter pasteurianus. Arch. Microbiol. 176:443–451. 10.1007/s002030100348 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.