SUMMARY
Many microbial phyla that are widely distributed in open environments have few or no representatives within animal-associated microbiota. Among them, the Chloroflexi comprises taxonomically and physiologically diverse lineages adapted to a wide range of aquatic and terrestrial habitats. A distinct group of uncultured chloroflexi related to free-living anaerobic Anaerolineae inhabits the mammalian gastrointestinal tract and includes low-abundance human oral bacteria that appear to proliferate in periodontitis. Using a single-cell genomics approach we obtained the first draft genomic reconstruction for these organisms and compared their inferred metabolic potential with free-living chloroflexi. Genomic data suggest that oral chloroflexi are anaerobic heterotrophs, encoding abundant carbohydrate transport and metabolism functionalities, similar to those seen in environmental Anaerolineae isolates. The presence of genes for a unique phosphotransferase system and N-acetylglucosamine metabolism suggests an important ecological niche for oral chloroflexi in scavenging material from lysed bacterial cells and the human tissue. The inferred ability to produce sialic acid for cell membrane decoration may enable them to evade the host defense system and colonize the subgingival space. As with other low-abundance but persistent members of the microbiota, discerning community and host factors that influence the proliferation of oral chloroflexi may help understand the emergence of oral pathogens and the microbiota dynamics in health and disease states.
Introduction
The human body is colonized by thousands of distinct types of bacteria that interact with each other, have co-evolved with and are specialized to distinct host niches, with expanding implications for health and disease (Clemente et al., 2012; Human Microbiome Project, 2012; Morgan et al., 2013). Although representatives of approximately 30 microbial phyla have been detected in human microbiota samples based on small subunit (SSU) rRNA gene sequences, a small subset (e.g. Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Fusobacteria) comprise more than 90% of the species richness and abundance across the general human populations (Zhou et al., 2013). Others (e.g. Verrucomicrobia, Spirochaetes, Deferibacteres, Synergistes, Chloroflexi, candidate phyla TM7 and SR1), even though usually present at low abundance (a few percent or less), have been consistently detected in most individuals and include cosmopolitan species in healthy adults or become elevated in disease (Paster et al., 2001; Derrien et al., 2008; Griffen et al., 2011; Wade, 2012; Abusleme et al., 2013).
A deep branching bacterial lineage, the Chloroflexi is a physiologically diverse and ubiquitous group of organisms found in a wide range of aquatic and terrestrial environments. Originally referred to as “green non-sulfur bacteria”, Chloroflexi formally comprises of filamentous anoxygenic photoautotrophs (Chloroflexales), aerobic organotrophs (Herpetosiphonales) and thermophilic chemoheterotrophs (Thermomicrobia) (Hugenholtz et al., 1998; Garrity and Holt, 2001; Hugenholtz and Stackebrandt, 2004). Four other classes subsequently added to this group include the Dehalococcoidetes (anaerobic dehalogenic reducers of chlorinated hydrocarbons), and the heterotrophic Ktedonobacteria, Anaerolineae and Caldilineae identified in wastewater treatment systems and micro-aerobic or anoxic environments (Yamada et al., 2006; Yamada and Sekiguchi, 2009; Yabe et al., 2010). Based on recent comparative genomics and phylogenetic analyses, it has been proposed that those added classes may represent related but distinct phyla, all potentially constituting a Chloroflexi “superphylum” (Gupta et al., 2013).
Chloroflexi represent a low abundance (<1%) but consistent component of human oral and skin microbiota and have also been detected in the gut of other mammals (Ley et al., 2008b; Dewhirst et al., 2010; Griffen et al., 2011; Zhou et al., 2013). A recent study also pointed to their proliferation in the subgingival pockets of patients with periodontitis, with a marked increase in prevalence between healthy (<10%) and diseased individuals (80–90%) (Abusleme et al., 2013), but a role for these bacteria in disease etiology has not yet been established. No study has been dedicated to diversity analysis of mammalian-associated chloroflexi and, because no cultured isolates or genomic information are available, their physiology, mechanisms of host adaptation and pathogenic potential are unknown.
Single-cell genomics has emerged as a powerful approach to gain insights into the physiological potential, evolutionary history and interspecies interactions of uncultured bacteria and archaea, both in open environments and as part of the human microbiota (Marcy et al., 2007; Yoon et al., 2011; Lasken, 2012; Pamp et al., 2012; Campbell et al., 2013a; Campbell et al., 2013b; Podar et al., 2013; Rinke et al., 2013). Here, based on flow cytometry-based bacterial cell isolation and single-cell genomics we present the first genomic dataset for a member of the human oral chloroflexi and its inferred physiological potentials. We also analyzed the diversity and distribution of the different lineages representing this phylum in the healthy human microbiota using SSU rRNA sequences generated under the Human Microbiome Project (Human Microbiome Project, 2012).
Diversity and abundance of the Chloroflexi in the healthy human microbiota
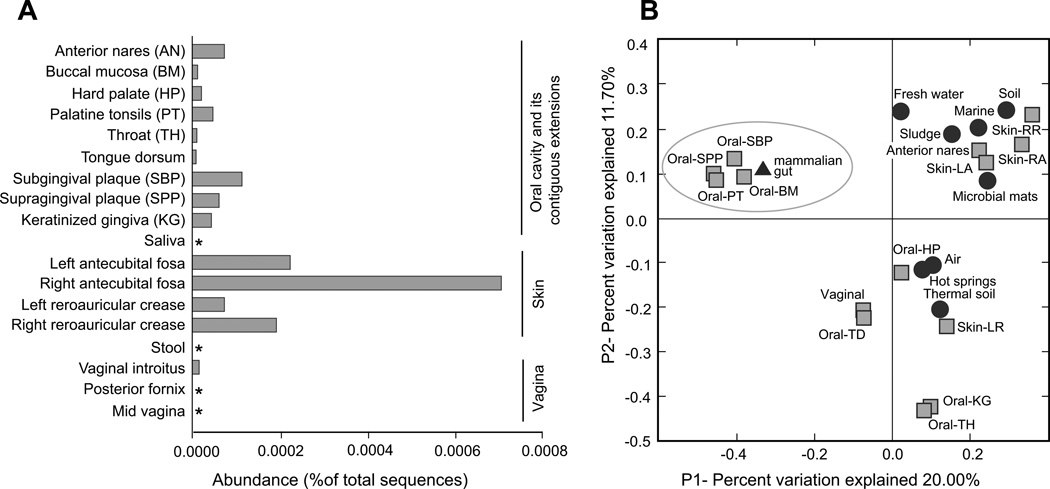
Based on the SSU rRNA dataset generated under the Human Microbiome Project (Methe et al., 2012; Zhou et al., 2013), chloroflexi represented a minor component of the overall healthy human microbiota (408 sequences out of ~24 million, i.e. 0.002%), Most of the chloroflexi sequences (75.3%) originated from skin samples, followed by 24.0% from the oral cavity and its contiguous extensions, and 0.7% were of vaginal origin (Figure 1A). Skin chloroflexi exhibited high diversity and formed 63 operational taxonomic units (OTUs) at 97% nucleotide sequence identity. Chloroflexi from the oral cavity and its contiguous extensions were less diverse and grouped into 7 OTUs with non-unique sequences at 97% identity (Supplementary figure S1). In the Human Oral Microbiome Database (HOMD) (Dewhirst et al., 2010) Chloroflexi are represented by a single phylotype, “oral taxon 439” (de Lillo et al., 2006), affiliated with heterotrophic Anaerolineae (Figure 2) and present at low abundance (0.003%) in healthy individuals. Its relative abundance increases in subjects with periodontitis (de Lillo et al., 2006; Abusleme et al., 2013).
Figure 1.
Diversity of human-associated members of the Chloroflexi. (A) Abundance and distribution of chloroflexi rRNA sequences in the body sites sampled under the NIH-HMP project (asterisks indicate no chloroflexi sequences found). (B) PCoA based on unweighted Unifrac distances between human-associated and environmental chloroflexi. The circled cluster highlights the distinct oral and mammalian gut phylotypes.
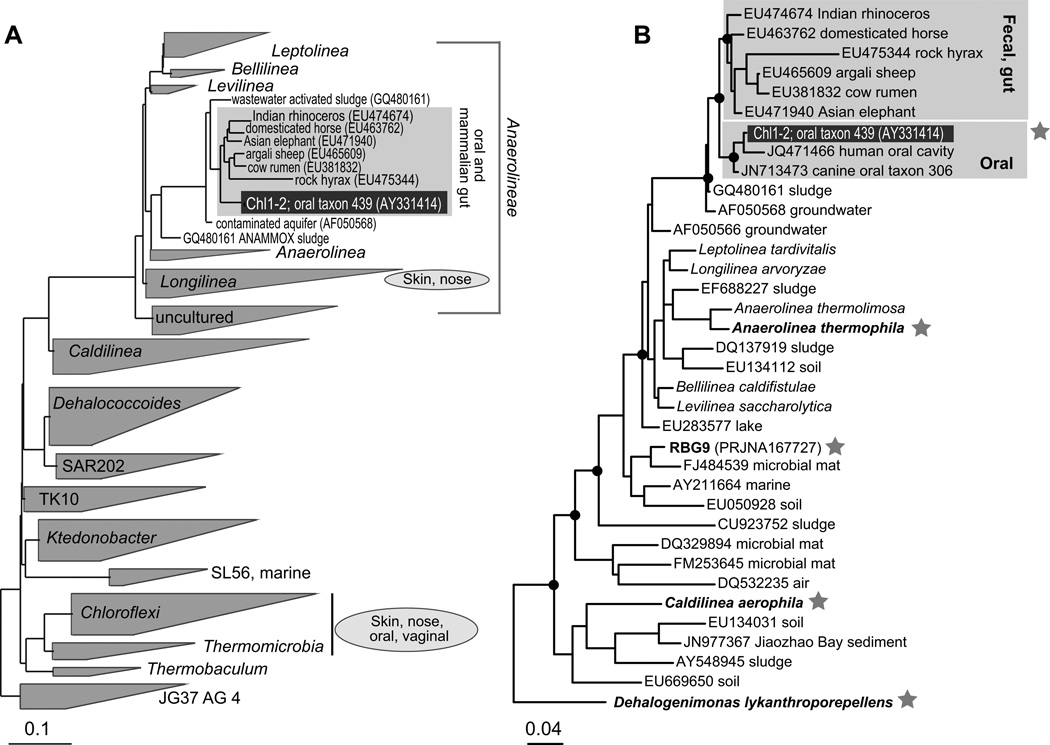
Figure 2.
Phylogenetic diversity of the Chloroflexi. (A). Classes and equivalent level taxa based on ARB and Silva database (Ludwig et al., 2004; Quast et al., 2013). The animal-associated clade that encompasses oral subgingival lineages and their close relatives is expanded. The general class-level placing of HMP sequences is based on a maximum likelihood phylogenetic analysis that included all available sequences. (B) Maximum likelihood phylogeny of the Anaerolineae. A star indicates organisms for which genomic data is available. Major nodes supported by bootstrap values >50 are indicated by filled dots. D. lykanthroporepellens (Dehalococcoides) was used as an outgroup. For environmental sequences only the GenBank accession number and the environment type are listed. The scale bar indicates inferred number of substitutions per nucleotide position.
A principal coordinates analysis (PCoA) of unweighted UniFrac distances grouped skin, nose and oral hard palate sequences with sequences from different open environments (Figure 1B). The close nucleotide similarity between chloroflexi found on relatively exposed habitats of the human body and diverse aquatic, soil and air phylotypes suggests that some might not be stable members of the human microbiota but occasional hitchhikers. Two apparently distinct clusters grouped keratinized gingiva (12 sequences), throat (2 sequences), tongue (1 sequence) and vaginal (3 sequences), but those phylotypes are quite rare in the sampled human population and are phylogenetically diverse. However, one distinct cluster consisted of the largest OTU (58 sequences) identified in the HMP dataset, oral chloroflexi reported in other studies (e.g. “oral taxon 439” (Dewhirst et al., 2010; Dewhirst et al., 2012), chloroflexi from animal gut (Ley et al., 2008b; Kong et al., 2010)(Figure 1B) and formed a phylogenetically distinct clade within the Anaerolineae (Figure 2).
Oral chloroflexi single amplified genomes (SAGs)
Two oral chloroflexi were identified in a library of over 2000 subgingival bacterial single-cell amplified genomic DNAs. The two SAGs (referred to as Chl1 and Chl2) originated from distinct individuals with periodontitis. Based on SSU rRNA genes, Chl1 and Chl2 belong to the class Anaerolineae and form part of a clade that is distinct from its closest relative with a sequenced genome, the free-living thermophilic anaerobe Anaerolinea thermophila (Figure 2). The SSU rRNAs of Chl1 and Chl2 were identical, shared 99.7% identity to the previously recognized “oral taxon 439”, and represent the distinct mammalian oral and gut chloroflexi cluster discussed above.
After abundance normalization of the sequence reads, assembly and contamination removal, Chl1 and Chl2 were comprised of 1.1 Mbp and 1.2 Mbp of DNA sequence, with a G+C content of 53%. Analysis of average nucleotide identity (ANI) revealed an identity of 98.3% between Chl1 and Chl2, based on ~37% overlap of the genomes. Due to this high similarity and the low diversity of oral subgingival chloroflexi revealed by prior studies, we treated these SAGs as members of a single operational taxonomic unit (OTU), the previously recognized uncultured “oral taxon 439” (de Lillo et al., 2006; Dewhirst et al., 2010). All downstream metabolic analyses were performed on a combined assembly of the two datasets, and the organism and its genomic information will be referred to as Chl1-2 throughout the paper. After merging, Chl1-2 consisted of 1.8 Mbp of DNA sequence, with a G+C content of 53% and coded for 1774 proteins and 42 tRNAs and rRNAs. Based on the presence of conserved single copy genes, the estimated genome size for this oral representative of Chloroflexi is approximately 2.7 Mb, with an estimated 67% of the genome present in the Chl1-2 dataset.
Metabolic inferences and comparative genomics
Based on SSU rRNA and protein sequence similarity, Anaerolinea thermophila is the closest sequenced relative to oral taxon 439 (Chl1-2), with 23% of the predicted proteins as top homologues. A genome distance-based tree also revealed a similar relationship (Supplementary figure S2). Many aspects of metabolism are likely to be shared between these lineages, as both encode a rich repertoire of genes for fermentative carbohydrate metabolism. Cultured Anaerolineae have been shown to be strictly anaerobic fermentative chemo-organotrophs, and utilization of carbohydrates has been shown in the laboratory and observed in wastewater sludge granules (Yamada and Sekiguchi, 2009). A related organism (RBG-9) recently uncovered based on metagenomic data from a subsurface environment was predicted to be capable of aerobic sugar respiration in addition to anaerobic fermentation of sugars and amino acids (Hug et al., 2013). Interestingly, related organisms potentially capable of anoxygenic photrophy were also identified in thermal mats from Yellowstone, which indicates that, metabolically, the Anaerolineae can be quite versatile (Klatt et al., 2011).
Glycolysis/gluconeogenesis pathways are present in A. thermophila, RBG-9 and Chl1-2 although genes for the enzymes needed to convert glucose to fructose-6-P are missing in the oral dataset, possibly due to the incomplete genome. Genes encoding enzymes of the pentose phosphate and carbon fixation pathways were also identified as well as for the potential transport and catabolism of a variety of pentoses, hexoses, and disaccharides.
Evidence of energy production capabilities in Chl1-2 includes several subunits of the ATP synthase complex but no cytochrome-encoding genes were found. All Chloroflexi sequenced to date encode an NADH:quinone oxidoreductase (complex I) for electron transport. Although Chl1-2 does contain two genes similar to complex I subunits, these genes match more closely to hymA and hymB, genes that encode subunits responsible for electron transport in an iron-only hydrogenase (Graentzdoerffer et al., 2003). The hymAB-like genes are adjacent to an iron-only hydrogenase catalytic unit gene (hymC). These genes were originally found in association with a formate dehydrogenase, but no evidence of this complex was found in Chl1-2. Not surprisingly, no genes indicating potential for photoautotrophy were found. Because of the incomplete nature of the genome, definitive assertion of the respiratory and fermentative potential for the oral chloroflexi will require cultivation or complete genome sequencing.
To explore for specific functional adaptations of Chl1-2 in a host-associated, oral environment, the presence/absence of clusters of orthologous groups (COGs) was used to compare it with its closest relative, A. thermophila, and other available complete genomes for cultured Chloroflexi. Comparisons to A. thermophila revealed 606 COGs shared by both organisms and 182 COGs unique to Chl1-2.
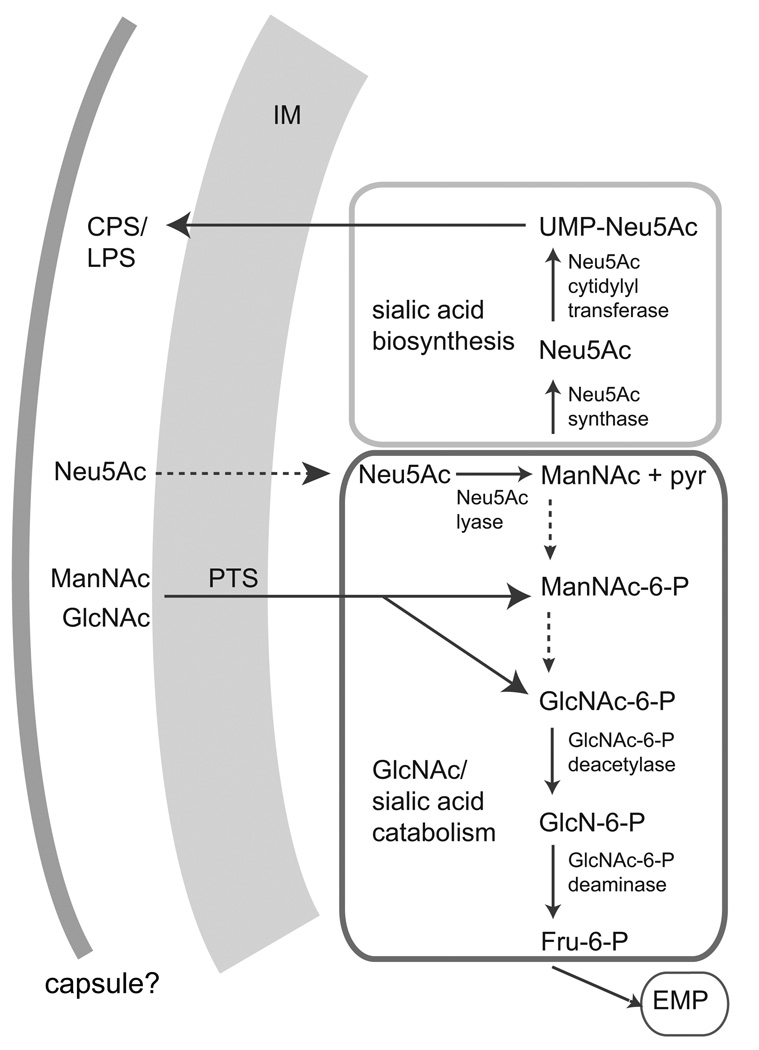
In addition to shared genes, Chl1-2 encodes a unique set of carbohydrate metabolism genes not seen in any other Chloroflexi genomes, including a phosphotransferase system (PTS) that has an ability to transfer a wide range of sugars including mannose, glucose, fructose, N-acetylglucosamine (GlcNAc), glucosamine and N-acetylmannosamine (ManNAc) (Plumbridge and Vimr, 1999). Chl1-2 also contains homologs of GlcNAc-6-phosphate (GlcNAc-6-P) deacetylase and GlcN-6-P deaminase, enzymes involved in the catabolism pathway of GlcNac-6-phosphate to fructose-6-phosphate (Plumbridge and Vimr, 1999). The resulting fructose-6-phosphate could then feed into glycolysis. Together, these genes could encode a pathway for the metabolism of GlcNAc, a major component of bacterial cell walls (Figure 3). The presence of this pathway would support previous in situ reports that members of the subphylum I Chloroflexi are capable of utilizing GlcNAc (Kindaichi et al., 2004; Miura et al., 2007). The Chloroflexi members appear to retrieve GlcNAc from other lysed cells in the environment, as seen with several microautoradiography (MAR)-FISH studies (Okabe et al., 2005; Miura and Okabe, 2008; Zang et al., 2008).
Figure 3.
Genomic highlights for the oral taxon 439, Chl1-2. Inferred N-acetylglucosamine (GlcNAc) and sialic acid N-acetylneuraminate (Neu5Ac) metabolism in oral chloroflexi. Enzymes for which coding genes were not found in the Chl1-2 dataset are indicated by dotted arrows. IM, inner membrane; CPS, capsular polysaccharide; LPS, lipopolysaccharide; pyr, pyruvate; PTS, mannose-type phosphotransferase system; ManNAc, N-acetylmannosamine; GlcN-6-P, glucosamine-6-phosphate; Fru-6-P, fructose-6-phosphate; EMP, Embden-Meyerhof-Parnas glycolytic pathway.
In addition, Chl1-2 encodes a homolog of N-acetylneuraminate (Neu5Ac) lyase, which can convert the sialic acid Neu5Ac into ManNAc and pyruvate (Figure 3). Although no specific transporters were found in the partial genome, the presence of Neu5Ac lyase suggests that Chl1-2 may acquire and catabolize sialic acid from the environment. This ability would be useful in the oral environment, as most mammalian cell surfaces have abundant sialic acid decoration of glycoproteins and glycolipids (Plumbridge and Vimr, 1999). Sialic acid appears to function as a growth factor for some pathogenic oral bacteria, such as Tannerella forsythia (Stafford et al., 2012). Furthermore, Chl1-2 encodes Neu5Ac synthase and Neu5Ac cytidylyltransferase, proteins necessary for the production of the sialic acid (Figure 3). Sialic acid biosynthesis genes are also present in A. thermophila but were not found in available sequencing data from other cultured Chloroflexi. Although it is unclear what the role of this pathway may be in A. thermophila, it is possible that the production of sialic acid in the oral environment helps Chl1-2 evade immune response by mimicking the exterior of host cells (Stafford et al., 2012).
Genes encoding lipopolysaccharide (LPS) biosynthetic enzymes and the production of galactose and rhamnose type O-antigens were identified in Chl1-2. Together with sialic acid, those could be incorporated in LPS and capsular polysaccharides to mimic the surface of epithelial cells (Stafford et al., 2012). Genes encoding proteins involved in export of LPS to the cell surface (Frosch and Muller, 1993), a protein similar to CapD (Lin et al., 1994) and poly-gamma-glutamate biosynthesis protein CapA were also found. The presence of a capsule would benefit Chl1-2 in the oral environment by providing adherence and resistance to both specific and nonspecific host immune systems (Roberts, 1996). Chl1-2 also encodes efflux transporters for multidrug and toxic compound extrusion (MATE) of lantibiotics, xenobiotics, bacitracin and lipid A, HlyD-like proteins and major facilitator superfamily (MFS) proteins necessary for type I secretion and nitroimidazole antibiotic resistance. Genes involved in protection from oxidative stress (methionine sulfoxide reductase, glutathione peroxidase, rubrerythrin and rubredoxin) were also identified, several of them absent in A. thermophila. Chl1-2 also encodes a Clp protease that has been shown to be important for stress response in a number of host-associated bacteria (Capestany et al., 2008; Loughlin et al., 2009; Lourdault et al., 2011). Another common feature of oral associated bacteria is their enhanced ability to acquire iron, which is bound to human lactoferrin and of limited availability (Wang et al., 2012). In that respect, Chl1-2 encodes ferric iron-siderophore transporter proteins as well as a hemolysin III gene. Competence proteins similar to ComEA and ComEC were also present, along with a competence-specific regulator. This suggests that Chl1-2 may be naturally competent, which would facilitate horizontal genes acquisition, a process that has been shown to be rampant in the human microbiota (Smillie et al., 2011).
Conclusions
Vertebrate species harbor characteristic microbiota that, through co-evolution with their hosts, have adapted to distinct eco-physiologies (Ley et al., 2008c; Ley et al., 2008a). Resident microbiota is acquired and selected from the environment after birth, and is shaped and maintained in a dynamic state through complex inter-microbial interactions and interactions with the host. The majority of mammalian-associated microbial species represent a handful of bacterial phyla that display a high level of host-specific diversity (Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria), but many others have been identified at lower abundance and in distinct body niches (Methe et al., 2012; Zhou et al., 2013), some still being known only based on sequence data (Marcy et al., 2007; Campbell et al., 2013b; Di Rienzi et al., 2013). Through single-cell genomics we provided insights into an uncultured representative of the phylum Chloroflexi that belongs to a distinct group of mammalian-adapted Anaerolineae, related to free-living anaerobic fermenters that inhabit a wide range of terrestrial and aquatic environments. Small but exclusively host-adapted clades of bacteria such as the oral chloroflexi revealed here, oral SR1 bacteria (Campbell et al., 2013b) and the gut “‘Melainabacteria” (Ley et al., 2005; Di Rienzi et al., 2013), may indicate that these bacteria colonized mammals relatively early on but did not undergo rampant diversification like other lineages (e.g. Bacteroidetes, Firmicutes) perhaps due to physiological constraints or competition. Like other bacteria adapted to a host environment, oral hloroflexi appear to have acquired specific physiological traits that enable them to inhabit the human body. Inferred glucosamine uptake may be linked to the abundance of cell wall components from surrounding lysed bacterial and epithelial cells. The increased frequency of chloroflexi in periodontitis may thus be linked to elevated bacterial turnover and tissue breakdown in deep subgingival pockets (Abusleme et al., 2013). At the same time, capsular polysaccharides could allow these bacteria cell to mimic the exterior of host cells and enable immune evasion. Overall, the genomic characteristics of subgingival cloroflexi portray organisms similar to their free-living relatives, yet displaying niche-specific adaptations for survival in a host environment. Such inferences may help to design specific laboratory cultivation approaches for these organisms.
Experimental procedures
Diversity analysis of human-associated chloroflexi
Small subunit ribosomal RNA (SSU rRNA) gene sequences for representatives of all major Chloroflexi groups were obtained from GenBank and the Silva database (Quast et al., 2013). We specifically searched for and downloaded Chloroflexi-classified sequences from samples of animal origin. Sequences assigned to Chloroflexi from the Human Microbiome Project dataset (Human Microbiome Project, 2012) were identified and extracted as previously described (Zhou et al., 2013). Overall relationships between the major Chloroflexi taxa and affiliation of human and animal sequences were generated in ARB (Ludwig et al., 2004). Sequence alignments were also generated and edited in Geneious 5.6 (Drummond et al., 2011) followed by maximum likelihood phylogenetic reconstruction by RAxML (Stamatakis, 2006) under a general time reversible model with parameters estimated from the data. Diversity estimation at different sequence divergence levels and principal coordinate analyses based on unweighted Unifrac distances using human, animal-associated and 148 chloroflexi sequences from a variety of open environments were performed in Mothur (Schloss et al., 2009) and QIIME (Caporaso et al., 2010).
Single-cell sorting and genomic amplification
Human subjects enrollment and sample collection protocols were approved by the Ohio State University Institutional Review Board and by the Oak Ridge Site-Wide Institutional Review Board. Signed informed written consent was obtained from all human subjects that provided samples for this study. Subgingival microbiota samples from individual donors diagnosed with periodontitis were collected, fixed and used for single cell sorting and multiple displacement amplification (MDA) as previously described (Campbell et al., 2013a; Campbell et al., 2013b). Among over 2000 taxonomically assigned single amplified genomes (SAGs), two SAGs originating from different donors were identified to represent members of phylum Chloroflexi and were selected for genomic characterization.
SAG sequencing, assembly and annotation
The two Chloroflexi SAGs (Chl1 and Chl2) were sequenced using a Illumina HiSeq paired end sequencing (100 bp each direction) approach at the Hudson Alpha Institute for Biotechnology (Huntsville, AL, USA). A total of 225 million (Chl1) and 45 million (Chl2) reads were generated. Quality read filtering, normalization and assembly were performed for each of the two SAGs as we described previously (Swan et al., 2011; Campbell et al., 2013a). Scaffolding information was used to close gaps between some of the contigs using PCR and Sanger sequencing. Assembled genomic data was loaded into IMG (http://img.jgi.doe.gov) for gene prediction, annotation and for comparative genomic analyses (Markowitz et al., 2012). Several contigs representing potential contamination with non-chloroflexi sequences based on blastp against Genbank, differential GC content and tetramer frequency profiles (Woyke et al., 2009), were identified and removed. Average nucleotide identity (ANI) between common contigs of Chl1 and Chl2 was calculated using JSpecies (Richter and Rossello-Mora, 2009). A combined, hybrid assembly (Chl1-2) was generated by comparing the two datasets using BLASTCLUST (Altschul et al., 1990) (S = 95, L = 0.7) and Mauve (Darling et al., 2004). Regions that exhibited more than 95% identity over at least 1 kb were merged, with sequence of the hybrid assembly chosen based on the longest contig between Chl1 and Chl2 in that region. Genome size estimation was based on frequency analysis using a set of 138 conserved single copy genes (CSCG) from 1516 finished bacterial genomes from IMG, as described in (Campbell et al., 2013b). The sequence data for the individual and combined SAGs has been deposited in GenBank under BioProject accession numbers PRJNA194441-194443 and is also available in IMG (http://img.jgi.doe.gov).
Supplementary Material
Acknowledgements
This research was supported by grant R01 HG004857 from the National Human Genome Research Institute (NHGRI) of the National Institutes of Health (NIH) to M.P. and by grant 1R56DE021567 from the National Institute for Dental and Cranial Research (NIDCR) of the NIH to M.P., A.G. and E.L. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy. P.S. and T.W. were supported by the U.S. Department of Energy Joint Genome Institute and by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. We thank Steve Allman, Sarah Kauffman, Zamin Yang and Alexander Sczyrba for laboratory and bioinformatics assistance.
Footnotes
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher’s web site:
Fig. S1. Distribution and relationship between Chloroflexi OTUs from oral and contiguous sites
Fig. S2. Hierarchical clustering of representative Chloroflexi genomes based on COG composition
References
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Campbell AG, Campbell JH, Schwientek P, Woyke T, Sczyrba A, Allman S, et al. Multiple single-cell genomes provide insight into functions of uncultured Deltaproteobacteria in the human oral cavity. PLoS One. 2013a;8:e59361. doi: 10.1371/journal.pone.0059361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JH, O'Donoghue P, Campbell AG, Schwientek P, Sczyrba A, Woyke T, et al. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci U S A. 2013b;110:5540–5545. doi: 10.1073/pnas.1303090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capestany CA, Tribble GD, Maeda K, Demuth DR, Lamont RJ. Role of the Clp system in stress tolerance, Biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J Bacteriol. 2008;190:1436–1446. doi: 10.1128/JB.01632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J, Kuczynski J, Stombaugh J, Bittinger K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Research. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lillo A, Ashley FP, Palmer RM, Munson MA, Kyriacou L, Weightman AJ, Wade WG. Novel subgingival bacterial phylotypes detected using multiple universal polymerase chain reaction primer sets. Oral Microbiol Immunol. 2006;21:61–68. doi: 10.1111/j.1399-302X.2005.00255.x. [DOI] [PubMed] [Google Scholar]
- Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ Microbiol. 2008;74:1646–1648. doi: 10.1128/AEM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu W-H, et al. The Human Oral Microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Klein EA, Thompson EC, Blanton JM, Chen T, Milella L, et al. The canine oral microbiome. PLoS One. 2012;7:e36067. doi: 10.1371/journal.pone.0036067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife. 2013;2:e01102. doi: 10.7554/eLife.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, et al. Geneious Pro (v5.6.3) 2011 Available at http://www.geneious.com. [Google Scholar]
- Frosch M, Muller A. Phospholipid substitution of capsular polysaccharides and mechanisms of capsule formation in Neisseria meningitidis. Mol Microbiol. 1993;8:483–493. doi: 10.1111/j.1365-2958.1993.tb01592.x. [DOI] [PubMed] [Google Scholar]
- Garrity G, Holt J. Phylum BVI, Chloroflexi phy. nov. In: Boone D, Castenholz R, editors. Bergey’s manual of systematic bacteriology : The Archaea and the deeply branching and phototrophic bacteria. New York: Springer; 2001. pp. 447–450. [Google Scholar]
- Graentzdoerffer A, Rauh D, Pich A, Andreesen JR. Molecular and biochemical characterization of two tungsten- and selenium-containing formate dehydrogenases from Eubacterium acidaminophilum that are associated with components of an iron-only hydrogenase. Archi Microbiol. 2003;179:116–130. doi: 10.1007/s00203-002-0508-1. [DOI] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2011;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Chander P, George S. Phylogenetic framework and molecular signatures for the class Chloroflexi and its different clades; proposal for division of the class Chloroflexia class. nov. [corrected] into the suborder Chloroflexineae subord. nov., consisting of the emended family Oscillochloridaceae and the family Chloroflexaceae fam. nov., and the suborder Roseiflexineae subord. nov., containing the family Roseiflexaceae fam. nov. Antonie Van Leeuwenhoek. 2013;103:99–119. doi: 10.1007/s10482-012-9790-3. [DOI] [PubMed] [Google Scholar]
- Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, Frischkorn KR, et al. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome. 2013;1:22. doi: 10.1186/2049-2618-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Stackebrandt E. Reclassification of Sphaerobacter thermophilus from the subclass Sphaerobacteridae in the phylum Actinobacteria to the class Thermomicrobia (emended description) in the phylum Chloroflexi (emended description) Int J Syst Evol Microbiol. 2004;54:2049–2051. doi: 10.1099/ijs.0.03028-0. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindaichi T, Ito T, Okabe S. Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by microautoradiography-fluorescence in situ hybridization. Appl EnvironMicrobiol. 2004;70:1641–1650. doi: 10.1128/AEM.70.3.1641-1650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt CG, Wood JM, Rusch DB, Bateson MM, Hamamura N, Heidelberg JF, et al. Community ecology of hot spring cyanobacterial mats: predominant populations and their functional potential. ISME J. 2011;5:1262–1278. doi: 10.1038/ismej.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong YH, Teather R, Forster R. Composition, spatial distribution, and diversity of the bacterial communities in the rumen of cows fed different forages. FEMS Microbiology Ecology. 2010;74:612–622. doi: 10.1111/j.1574-6941.2010.00977.x. [DOI] [PubMed] [Google Scholar]
- Lasken RS. Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol. 2012;10:631–640. doi: 10.1038/nrmicro2857. [DOI] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008a;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008b;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WS, Cunneen T, Lee CY. Sequences, analysis and molecular characterization of genes required for the biosynthesis of type-1 capsular polysaccharide in Staphylococcus aureus. J Bacteriol. 1994;176:7005–7016. doi: 10.1128/jb.176.22.7005-7016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin MF, Arandhara V, Okolie C, Aldsworth TG, Jenks PJ. Helicobacter pylori mutants defective in the clpP ATP-dependant protease and the chaperone clpA display reduced macrophage and murine survival. Microbial Pathogenesis. 2009;46:53–57. doi: 10.1016/j.micpath.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Lourdault K, Cerqueira GM, Wunder EA, Picardeau M. Inactivation of clpB in the pathogen Leptospira interrogans reduces virulence and resistance to stress conditions. Infection and Immunity. 2011;79:3711–3717. doi: 10.1128/IAI.05168-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, et al. Dissecting biological "dark matter" with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methe B, Nelson KE, Pop M, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura Y, Okabe S. Quantification of cell specific uptake activity of microbial products by uncultured Chloroflexi by microautoradiography combined with fluorescence in situ hybridization. Environ Sci Technol. 2008;42:7380–7386. doi: 10.1021/es800566e. [DOI] [PubMed] [Google Scholar]
- Miura Y, Watanabe Y, Okabe S. Significance of Chloroflexi in performance of submerged membrane Bioreactors (MBR) treating municipal wastewater. Environ Sci Technol. 2007;41:7787–7794. doi: 10.1021/es071263x. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Segata N, Huttenhower C. Biodiversity and functional genomics in the human microbiome. Trends Genet. 2013;29:51–58. doi: 10.1016/j.tig.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S, Kindaichi T, Ito T. Fate of 14C-labeled microbial products derived from nitrifying bacteria in autotrophic nitrifying biofilms. Appl EnvironMicrobiol. 2005;71:3987–3994. doi: 10.1128/AEM.71.7.3987-3994.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp SJ, Harrington ED, Quake SR, Relman DA, Blainey PC. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB) Genome Res. 2012;22:1107–1119. doi: 10.1101/gr.131482.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J, Vimr E. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J Bacteriol. 1999;181:47–54. doi: 10.1128/jb.181.1.47-54.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Makarova KS, Graham DE, Wolf YI, Koonin EV, Reysenbach AL. Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park. Biol Direct. 2013;8:9. doi: 10.1186/1745-6150-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Roberts IS. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annual Review of Microbiology. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. ApplEnvironMicrobiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–244. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- Stafford G, Roy S, Honma K, Sharma A. Sialic acid, periodontal pathogens and Tannerella forsythia: stick around and enjoy the feast! Molecular Oral Microbiology. 2012;27:11–22. doi: 10.1111/j.2041-1014.2011.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science. 2011;333:1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2012 doi: 10.1016/j.phrs.2012.11.006. S1043-6618(12)00227-7. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Wang RK, Kaplan A, Guo LH, Shi WY, Zhou XD, Lux R, He XS. The influence of iron availability on human salivary microbial community composition. Microbial Ecology. 2012;64:152–161. doi: 10.1007/s00248-012-0013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, Xie G, Copeland A, Gonzalez JM, Han C, Kiss H, et al. Assembling the marine metagenome, one cell at a time. PLoS One. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe S, Aiba Y, Sakai Y, Hazaka M, Yokota A. Thermosporothrix hazakensis gen. nov., sp. nov., isolated from compost, description of Thermosporotrichaceae fam. nov. within the class Ktedonobacteria Cavaletti et al. 2007 and emended description of the class Ktedonobacteria. Int J Syst Evol Microbiol. 2010;60:1794–1801. doi: 10.1099/ijs.0.018069-0. [DOI] [PubMed] [Google Scholar]
- Yamada T, Sekiguchi Y. Cultivation of uncultured chloroflexi subphyla: significance and ecophysiology of formerly uncultured chloroflexi 'subphylum i' with natural and biotechnological relevance. Microbes Environ. 2009;24:205–216. doi: 10.1264/jsme2.me09151s. [DOI] [PubMed] [Google Scholar]
- Yamada T, Sekiguchi Y, Hanada S, Imachi H, Ohashi A, Harada H, Kamagata Y. Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int J Syst Evol Microbiol. 2006;56:1331–1340. doi: 10.1099/ijs.0.64169-0. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Price DC, Stepanauskas R, Rajah VD, Sieracki ME, Wilson WH, et al. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science. 2011;332:714–717. doi: 10.1126/science.1203163. [DOI] [PubMed] [Google Scholar]
- Zang K, Kurisu F, Kasuga I, Furumai H, Yagi O. Analysis of the phylogenetic diversity of estrone-degrading bacteria in activated sewage sludge using microautoradiography-fluorescence in situ hybridization. Syst ApplMicrobiol. 2008;31:206–214. doi: 10.1016/j.syapm.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Gao H, Mihindukulasuriya KA, Rosa PS, Wylie KM, Vishnivetskaya T, et al. Biogeography of the ecosystems of the healthy human body. Genome Biol. 2013;14:R1. doi: 10.1186/gb-2013-14-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.