Abstract
Recent genomic analyses of pathologically-defined tumor types identify “within-a-tissue” disease subtypes. However, the extent to which genomic signatures are shared across tissues is still unclear. We performed an integrative analysis using five genome-wide platforms and one proteomic platform on 3,527 specimens from 12 cancer types, revealing a unified classification into 11 major subtypes. Five subtypes were nearly identical to their tissue-of-origin counterparts, but several distinct cancer types were found to converge into common subtypes. Lung squamous, head & neck, and a subset of bladder cancers coalesced into one subtype typified by TP53 alterations, TP63 amplifications, and high expression of immune and proliferation pathway genes. Of note, bladder cancers split into three pan-cancer subtypes. The multi-platform classification, while correlated with tissue-of-origin, provides independent information for predicting clinical outcomes. All datasets are available for data-mining from a unified resource to support further biological discoveries and insights into novel therapeutic strategies.
INTRODUCTION
Cancers are typically classified using pathologic criteria that rely heavily on the tissue site of origin. However, large-scale genomics projects are now producing detailed molecular characterizations of thousands of tumors, making a systematic molecular-based taxonomy of cancer possible. Indeed, The Cancer Genome Atlas (TCGA) Research Network has reported integrated genome-wide studies of ten distinct malignancies: glioblastoma multiforme (GBM) (The_Cancer_Genome_Atlas_Network, 2008), serous ovarian carcinoma (OV) (The_Cancer_Genome_Atlas_Network, 2011), colon (COAD) and rectal (READ) adenocarcinomas (The_Cancer_Genome_Atlas_Network, 2012b), lung squamous cell carcinoma (LUSC) (The_Cancer_Genome_Atlas_Network, 2012a), breast cancer (BRCA) (The_Cancer_Genome_Atlas_Network, 2012c), acute myelogenous leukemia (AML) (The_Cancer_Genome_Atlas_Network, 2013b), endometrial cancer (UCEC) (Kandoth et al., 2013b), and renal cell carcinoma (KIRC) (The_Cancer_Genome_Atlas_Network, 2013a), and bladder urothelial adenocarcinoma (The_Cancer_Genome_Atlas_Network, 2014). Those studies have shown that each single-tissue cancer type can be further divided into three to four molecular subtypes. The sub-classification is based on recurrent genetic and epigenetic alterations that converge on common pathways (e.g. p53 and/or Rb checkpoint loss; RTK/RAS/MEK or RTK/PI3K/AKT activation). Meaningful differences in clinical behavior are often correlated with the single-tissue tumor types and, in a few cases, single-tissue subtype identification has led to therapies that target the driving subtype-specific molecular alteration(s). EGFR-mutant lung adenocarcinomas and ERBB2-amplified breast cancer are two well-established examples.
To move toward a molecular taxonomy, we investigated whether tissue-of-origin categories split into sub-types based upon multi-platform genomic analyses, and also extend the analysis in the other direction to look for possible convergence. We looked to see what molecular alterations are shared across cancers arising from different tissues and if previously recognized disease subtypes in fact span multiple tissues of origin. With those questions in mind, we performed a multi-platform integrative analysis of thousands of cancers from 12 tumor types in The Cancer Genome Atlas (TCGA) project. Using data from multiple assay platforms, we tested the hypothesis that molecular signatures provide a distinct taxonomy relative to the currently used tissue-of-origin based classification. At the center of our results is the identification of 11 “integrated subtypes”. Consistent with the histological classification, tissue-of-origin features provided the dominant signal(s) for identification of most subtypes, irrespective of genomic analysis platform or combination thereof. However, approximately 10% of cases were reclassified by the molecular taxonomy, with the newly defined integrated subtypes providing a significant increase in the accuracy for the prediction of clinical outcomes.
RESULTS
Samples, Data Types, and Genomic Platforms
To identify a multi-tissue, molecular signature-based classification of cancer objectively, we first characterized each of the individual tumor types using six different “omic” platforms. The diverse tumor set called “Pan-Cancer-12,” is composed of 12 different malignancies. It comprises 3,527 cases assayed by at least four of the six possible data types routinely generated by TCGA: whole-exome DNA sequence (Illumina HiSeq and GAII), DNA copy number variation (Affymetrix 6.0 microarrays), DNA methylation (Illumina 450,000-feature microarrays), genome-wide mRNA levels (Illumina mRNA-seq), microRNA levels (Illumina microRNA-seq), and protein levels for 131 proteins and/or phosphorylated proteins (Reverse Phase Protein Arrays; RPPA). The 12 tumor types include the ten TCGA Network published data sets listed above and two additional tumor types for which manuscripts have been submitted: lung adenocarcinoma (LUAD) and head & neck squamous cell carcinoma (HNSC). This is the most comprehensive and diverse collection of tumors analyzed by systematic genomic methods to date.
We performed sample-wise clustering to derive subtypes based on six different data types separately: DNA copy number, DNA methylation, mRNA expression, microRNA expression, protein expression, and somatic point mutation (see Supplemental Extended Experimental Procedures and Analyses, Section 1). The classification results from each single-platform analysis produced sets of 8 to 20 groups of samples that each showed high correlation with tissue of origin (Figures S1A–F) and were highly comparable with each other (Figure S2A). For example, patterns of copy number change varied across tissue types, and subtyping of the tumors based on copy number alterations revealed a significant correlation with tissue (p < 6×10−6, Chi-square test).
Integrated Platform Analysis (Cluster of Cluster Assignments)
To identify disease subtypes on a more comprehensive basis than could be done using any single type of data, we developed an integrated subtype classification for all of the tumor samples in the Pan-Cancer-12 collection based on five of the data types, excluding somatic mutations. To do so, the results of the single platform analyses were provided as input to a second-level cluster analysis using a method we refer to as Cluster-Of-Cluster-Assignments (COCA), which was originally developed to define subclasses in the TCGA breast cancer cohort (The_Cancer_Genome_Atlas_Network, 2012c). The algorithm takes as input the binary vectors that represent each of the platform-specific cluster-groups and re-clusters the samples according to those vectors (see Supplemental Text Section 2). One advantage of the method is that data across platforms are combined without the need for normalization steps prior to clustering. In addition, each platform influences the final integrated result with weight proportional to the number of distinct subtypes reproducibly found by Consensus Clustering. Thus, “large” platforms (e.g. 450,000 DNA methylation probes) with orders of magnitude more features than “small” platforms (e.g. 131 RPPA antibodies) do not dominate the solution.
In addition to the COCA classification, we used two additional, independent methods to derive Pan-Cancer-12 subtypes based on integrated data: (i) an algorithm called SuperCluster (Kandoth et al., 2013b) (Figure S2B) and (ii) clustering based on inferred pathway activities from PARADIGM (Vaske et al., 2010), which integrates gene expression and DNA copy number data with a set of predefined pathways to infer the degree of activity of 17,365 pathway features such as proteins, complexes, and cellular processes (Figure S2C). Both SuperCluster and PARADIGM produced classifications that were highly concordant with the COCA subtypes (Figure S2D). Given recent promising results that use gene networks (as opposed to the sparsely populated single-mutation space) to cluster samples based on somatic DNA variants (Hofree et al., 2013), we calculated a mutation-based clustering after first associating genes with pathways and then identifying clusters based on mutated pathways (Figure S1F; Supplemental Data File S1). Including those clusters in the identification of COCA subtypes produced highly similar results to COCA subtypes that did not use the mutation-based clusters (Figure S2D). Thus, we focus here on the COCA results obtained without the mutations, as those five other platform-based classifications required no prior biological knowledge.
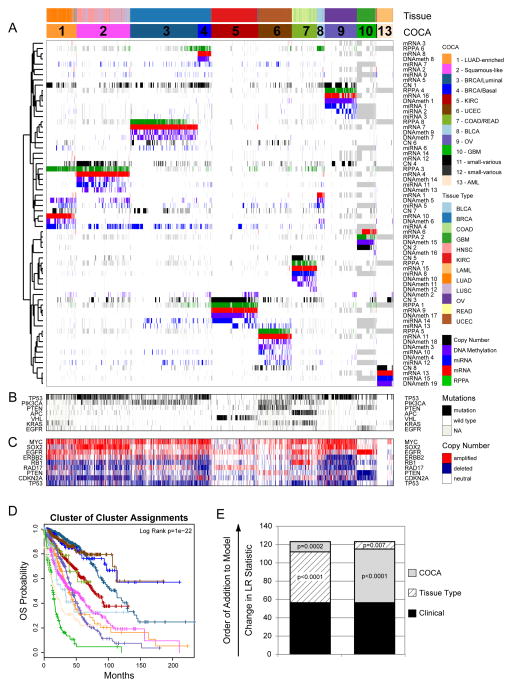
The COCA algorithm identified thirteen clusters of samples, 11 of which included more than ten samples (Table S1). The two small clusters (n=3 and 6) are noted (Table 1), but were excluded from further analyses. We refer to the remaining sample groups by cluster number and a short descriptive mnemonic (Table 1). Of the 11 COCA-integrated subtypes, five show simple, near one-to-one relationships with tissue site of origin: C5-KIRC, C6-UCEC, C9-OV, C10-GBM and C13-LAML (Figure 1A). A sixth COCA type, C1-LUAD-enriched, is predominantly composed (258/306) of non-small cell lung (NSCLC) adenocarcinoma samples (LUAD). The second major constituent of the C1-LUAD-enriched group is a set of NSCLC squamous samples (28/306). Upon re-review of the frozen or formalin fixed sections, 11/28 lung squamous samples that cluster with the C1-LUAD-enriched group did not have squamous features and were reclassified as lung adenocarcinoma (Travis et al., 2011). NSCLCs are often difficult to classify based on histology alone (Grilley-Olson et al., 2013). That difficulty poses an important clinical challenge since histology is used to guide the selection of chemotherapy (Scagliotti et al., 2008) and to select patients for further mutational analysis (e.g., EGFR mutation and ALK fusion testing in non-squamous NSCLC). However, the challenge can be addressed by genomic analysis based on distinct differences in mutation spectrum (Table S2A) and distinct gene expression patterns (Figure S1A). Two clear subtypes of NSCLC (C1-LUAD-enriched and C2-Squamous-like, see discussion below) are identified by COCA.
Table 1. The 12 pathological disease types (rows) and their relationship to the thirteen integrated subtypes defined by the Cluster-of-Cluster-Assignments (COCA) method (see also Supplemental Table S1).
The name of each COCA subtype (top row) includes a cluster number (1 to 13) and a text designation for mnemonic purposes. Two of the subtypes (numbers 11 and 12) were eliminated from further analysis because they included < 10 samples (3 and 6 samples, respectively). Hence, the text focuses on 11 subtypes, not 13.
| Handle | C1-LUAD-enriched | C2-Squamous-like | C3-BRCA/Luminal | C4-BRCA/Basal | C5-KIRC | C6-UCEC | C7-COAD/READ | C8-BLCA | C9-OV | C10-GBM | C11-small-various | C12-small-various | C13-AML | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BLCA | 10 | 31 | 0 | 0 | 1 | 0 | 0 | 74 | 0 | 1 | 1 | 2 | 0 | 120 |
| BRCA | 2 | 1 | 688 | 135 | 5 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 834 |
| COAD | 0 | 0 | 0 | 0 | 0 | 0 | 182 | 0 | 0 | 0 | 0 | 0 | 0 | 182 |
| GBM | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 190 | 0 | 0 | 0 | 195 |
| HNSC | 1 | 302 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 305 |
| KIRC | 1 | 0 | 0 | 0 | 470 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 475 |
| LAML | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 161 | 161 |
| LUAD | 258 | 6 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 270 |
| LUSC | 28 | 206 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 238 |
| OV | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 327 | 0 | 0 | 0 | 0 | 329 |
| READ | 0 | 0 | 0 | 0 | 0 | 0 | 73 | 0 | 0 | 0 | 0 | 0 | 0 | 73 |
| UCEC | 2 | 0 | 0 | 0 | 0 | 340 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 345 |
| Totals | 306 | 546 | 688 | 137 | 479 | 341 | 256 | 79 | 327 | 197 | 3 | 6 | 162 | 3527 |
Figure 1. Integrated Cluster-Of-Cluster Assignments analysis reveals 11 major subtypes (see also Supplemental Figures S1-3 and Data Files S1-3).
A) Integration of subtype classifications from 5 “omic” platforms resulted in the identification of 11 major groups/subtypes from 12 pathologically defined cancer types. The groups are identified by number and color in the second bar, with the tissue of origin specified in the top bar. The matrix of individual “omic” platform type classification/subtype schemes was clustered, and each data type is represented by a different color: copy number=black, DNA methylation=purple, miRNA=blue, mRNA=red and RPPA=green. B) Mutation status for each of 10 Significantly Mutated Genes coded as: wild-type=white, mutant=red, missing data=gray. C) Copy number status for each of 9 important genes: amplified=red, deleted=blue, copy number neutral=white and missing data=gray. The color-coding schema is shown to the right. D) Overall survival (OS) of COCA subtypes by Kaplan-Meier plot. COCA subtypes are highly correlated with overall survival outcomes. E) The log-likelihood ratio (LR) statistic was estimated as we added clinical variables, COCA subtype, or tissue type information to a cox proportional hazards model. Clinical variables included age at diagnosis, tumor size, node status and metastasis status. The change in LR statistic as features were added to the model was assessed for significance by chi-square analysis. The set of samples was limited to the set of tumor types that did not have a one-to-one relationship with a COCA subtype: BLCA, BRCA, COAD, HNSC, LUAD, LUSC, and READ in COCA clusters COCA1 – LUAD-enriched, COCA2-Squamous, COCA3-BRCA/Luminal, COCA4-BRCA/Basal-like, COCA7-COAD/READ and COCA8-BLCA. First bar “A” shows results of adding tissue-of-origin to clinical variables already part of the model, followed by a variable representing the COCA subtyping; bar “B” shows results when COCA is first added on to clinical variables, and then tissue-type is added. In each case the increase in the ability to predict OS was in terms of the LR.
For the other five tissue types, the patterns are more complex. Either a given tissue splits into multiple COCA groups (divergence) or multiple tissue types coalesce into a single COCA group (convergence). A simple example of convergence previously described for TCGA data is the merging of colon (COAD) and rectal (READ) tumors into a single COCA group (The_Cancer_Genome_Atlas_Network, 2012b). The expression features shared by colon and rectal samples were noted in the TCGA Network paper on the two cancer types, but we extend those findings through use of the multi-platform clustering approach (Figure 1, Table 1).
Breast cancers (BRCA) exhibit a pattern of divergence in which two main groups of samples are distinctly identifiable. One group (C3-BRCA/Luminal) contains essentially all of the Luminal (estrogen receptor-positive) (594/597) and HER2-positive tumors (66/66), whereas the other (C4-BRCA/Basal) contains 131/139 of the Breast Basal-like tumors. Although it has previously been appreciated that Basal-like breast cancers (the majority subset of Triple-Negative Breast Cancers) form a distinct subtype (Prat et al., 2013; The_Cancer_Genome_Atlas_Network, 2012c), the findings here provide a more refined, quantitative picture of the extent of difference from Luminal and Basal-like breast cancers. Whereas tissue-of-origin is the dominant signal for combined data on almost all of the other cancer types in the Pan-Cancer-12 collection, Breast Basal-like cancers are as different from Luminal/ER+ breast cancers as they are from cancers of the lung (Figure 1). The data from the present study strongly reinforce the idea that Basal-like breast cancers constitute a unique disease entity.
The remaining three tissue types (HNSC, LUSC and BLCA) provide examples of both divergence and convergence in COCA subtyping (Figure 1 and Table 1). The strongest pattern of convergence is observed for the vast majority of HNSC (301/304), LUSC (206/238) and some of the BLCA (31/120) tumors; they cluster together in a large COCA group (C2-Squamous-like), perhaps reflecting similar cell-type-of-origin or smoking as an etiologic factor. BLCA tumors also exhibit a divergence pattern, distributing predominantly into three distinct groups: 31 BLCA in the C2-Squamous-like group, 10 in the C1-LUAD-enriched group, and 74 in the bladder-only group, C8-BLCA. Five other BLCA samples cluster in four different COCA groups.
Clinical importance of the COCA subtypes
To investigate the clinical relevance of the COCA subtypes, we performed Kaplan-Meier Survival analysis on the Pan-Cancer-12 data set. The results indicate that tissue-of-origin (Figure S3A) and COCA subtype (Figure 1D) are both prognostic and each provides independent information (Figure 1E). Additionally, the two most commonly mutated genes in the overall dataset, TP53 (41%) and PIK3CA (20%), are prognostic, even across different tumor types, as are previously defined genomic signatures of cell proliferation rate (Nielsen et al., 2010) and mutated TP53 gene expression-based signature (Troester et al., 2006) (Figure S3B–F).
We next asked whether prognostic information is provided by the COCA subtypes after accounting for known clinical and tissue-of-origin features. We performed a Multivariate Cox proportional hazards analysis to predict outcomes across the dataset. The analysis was limited to the COCA subtypes that did not have a one-to-one relationship with tissue-of-origin tumor type (COCA1-LUAD enriched, COCA2-Squamous, COCA3-Breast/luminal, COCA4-Breast/Basal, COCA7-COAD/READ, and COCA8-BLCA). In the model we included clinical features such as tumor size, node status, metastasis status, and age at diagnosis, as well as tissue-of-origin. We performed a likelihood ratio test conditioning first on the clinical variables; when either tissue-of-origin or COCA subtype was added to the model, a large increase in the predictive fit of the model was observed, beyond what one would get with the clinical information alone (Figure 1E). That observation supports the classical model in which tissues-of-origin provides strong predictions of outcome. Next, we asked whether the COCA subtypes add additional independent information for predicting survival beyond the combination of tissue-of-origin and clinical features. Indeed, we observed a significant increase in statistical likelihood when COCA is added to a multivariate model that already includes the clinical and tissue-based information (P < 0.0002; Chi-square test; Figure 1E). Thus, while the COCA classification differs from tissue-of-origin based classification in only ~10% of all samples, the difference does provide important molecular information that reflects tumor biology and is associated with clinical outcome.
Genomic Determinants of the Integrated COCA Subtypes
We next identified the major genomic determinants of the COCA subtypes, including somatic mutations and DNA copy number changes. For single nucleotide variants, we analyzed a Pan-Cancer-12 list of 127 Significantly Mutated Genes (SMGs) obtained by MuSiC analysis (Kandoth et al., 2013a). Only three of the genes are mutated at a frequency ≧10% (TP53, PIK3CA and PTEN), and 11 additional are mutated at ≥5% frequency (Table S2A). We also include a list of 291 High-Confidence Cancer Drivers (HCDs) from Pan-Cancer-12 analysis (Tamborero et al., 2013), identified by a combination of five complementary methods to identify signals of positive selection in the mutational pattern of genes across tumors.
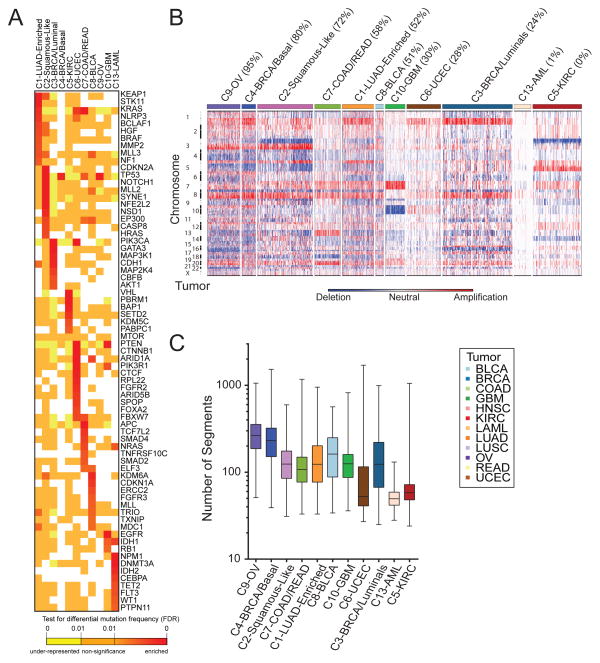
A large number of correlations between COCA subtypes and somatic mutations were found (Figure 2A, Figure S4D, Supplemental Data File S2). Somatic mutations clearly distinguish the C1-LUAD-enriched group from the C2-Squamous-like group. KEAP1 and STK11 are preferentially mutated in C1-LUAD-enriched tumors, whereas CDKN2A, NOTCH1, MLL2 and NFE2L2, among others, are preferentially mutated in C2-Squamous-like (Figure 2A). A similarly distinct set of SMGs was seen for the C3-BRCA/Luminal and C4-BRCA/Basal groups; only two genes are shared (TP53 and PIK3CA), and they show different mutation frequencies (Table S2A). Since the somatic mutation results were not used in any way to determine the COCA subtypes, they provide independent evidence that distinctly different genetic events underlie the subtypes. A protein-protein interaction network analysis of mutations associated with the COCA subtypes obtained using a new version of the HotNet algorithm (Vandin et al., 2012) provides an overview of the genomic determinants of the COCA subtypes (Figure S4E).
Figure 2. Genomic determinants of the Integrative COCA Subtypes (see also Supplemental Figure S4 and Tables S2-3).
A. Genes from the high-confidence list of drivers (Tamborero et al., 2013) found to be mutated at a different rate within one COCA subtype compared outside it based on a two-tailed Fisher’s exact test. Mutation frequency enrichment, red to orange; genes with mutations equaling the background rate, yellow; genes with no observed mutations in a subtype, white. Displayed are top-ranked genes in terms of significant mutation enrichment (FDR<1%) in at least one COCA subtype. B. Somatic copy number alterations (SCNAs) in Integrative Clusters. SCNAs in tumors (horizontal axis) are plotted along chromosomal locations (vertical axis). The heatmap shows the presence of amplifications (red) and deletions (blue) throughout the genome. The color strip along the top indicates integrative COCA cluster membership; the number in parentheses indicates % of samples in a COCA subtype with TP53 mutation. COCA subtypes are ordered from highest TP53 mutant percentage to lowest. C. Range of copy number segments in tumors within each Integrative Cluster. The box and whisker plots show the middle quartiles and the minimum and maximum number of segments in each cluster group.
The degree of genomic instability was a major determinant of subtype, as revealed in copy number variation (CNV) data (Figure 2B). The C9-OV, C4-BRCA/Basal and C1-LUAD-enriched subtypes showed the most marked genomic instability, as assessed by average number of copy number segments per subtype (Figure 2C), whereas AML and UCEC showed the least. Numerous COCA subtype-associated alterations implicated specific regions, arm-level copy number changes (Figure S4A) and/or focal regions of copy number alteration (Figure S4B). Of note were a number of previously described tissue type-specific and subtype-specific alterations, including Chr7 gain and Chr10 loss in GBM (The_Cancer_Genome_Atlas_Network, 2008), 3p loss and 5q gain in kidney (The_Cancer_Genome_Atlas_Network, 2013a), 4q and 5q loss in Breast Basal-like cancers (The_Cancer_Genome_Atlas_Network, 2012c) and 3p loss and 3q gain in Lung Squamous tumors (The_Cancer_Genome_Atlas_Network, 2012a). Of note, the latter were seen in most C2-Squamous-like tumors, regardless of tissue of origin.
Expression-based Determinants of the Integrated Subtypes
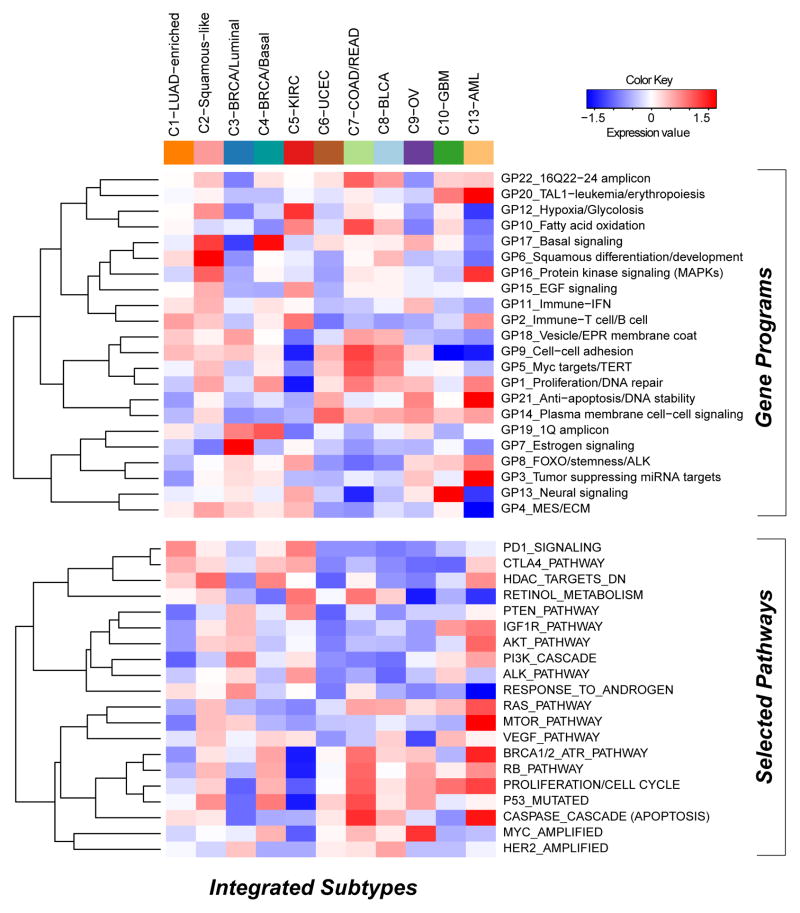
We next sought to identify gene expression modules characteristic of each COCA subtype. First, we started with 6,898 sets of gene signatures documented to be co-expressed, co-amplified, or to function together. From these, we identified gene programs as those whose genes have mRNA-seq signatures of high mutual correlation across the Pan-Cancer-12 dataset. After applying a bimodality filter and Weighted Gene Correlation Network-based clustering, 22 non-redundant gene programs were identified (Supplemental Table S4A, Figure S5A, Experimental Procedures and Analyses, Section 5, and Supplemental Data File S5). Linear classification with the 22 gene programs reconstituted the 11 integrated subtypes with 90% accuracy (Figure S5A; Table S4B). To view the expression-based determinants of the integrated subtypes we plotted the average expression level of each gene program within each COCA cluster (Figure 3). As expected, the gene programs GP6-squamous differentiation/development, GP13-neural signaling and GP20-TAL-1-leukemia/erythropoiesis were the most highly expressed in the C2-Squamous-like, C10-GBM and C13-LAML subtypes, respectively. As well, GP7_Estrogen signaling was highest in the C3-BRCA/luminal cases, whereas GP17_basal signaling had its highest levels in the C4-BRCA/Basal cases. Activated pathway characteristics found by enrichment and sub-network analyses based on PARADIGM inferences, many of which were consistent with the gene program analysis, are summarized in Table S4A (see Supplemental Extended Procedures and Analyses).
Figure 3. Subtype-specific patterns of gene-program and selected pathway expression characterizing each Pan-Cancer-12 COCA subtype (see also Supplemental Figures S5-6, Table S4, and Data File S5).
The heat map shows integrative subtypes in numerical order. Gene programs (top) and pathway signatures from PARADIGM (bottom) were clustered separately from each other. Red-blue intensities reflect the means of the scores (red=high, white=average, blue=low).
Gene expression programs and PARADIGM pathways carry clinically relevant information beyond tissue-of-origin as evidenced by a multivariate Cox model of survival with COCA subtype as a covariate (see Table S4E). Squamous differentiation/development (GP6), proliferation/cell cycle, and estrogen signaling (GP7) were significant predictors in the model. Intriguingly, GP7, along with fatty acid oxidation (GP10), tumor suppressing miRNA targets (GP3) and the PTEN/MTOR signaling program, were found to be significantly associated with patient outcome in kidney cancer using a Cox proportional hazards survival analysis (Figure S5D; Table S4F). In common with the C3-BRCA/Luminal subtype cases, higher levels of estrogen signaling (GP7) were also associated with better prognosis for C5-KIRC cancers. Consistent with the higher frequency of elevated HER2 protein levels in bladder, colorectal and serous endometrial cancers (Akbani et al., 2014), the HER2-amplified gene signature appeared elevated in the C8-BLCA, C7-COAD/READ and C6-UCEC subtypes, as well as the C3-BRCA/Luminal subtype which contains all HER2-positive breast cancers. Predictors independent of disease stage included basal signaling (GP17), associated with decreased overall survival, and the immune-related PARADIGM pathways PD1_signaling and CTLA4_pathway, both of which were associated with increased overall survival. These immune-related signatures may reflect varying amounts of lymphocyte infiltrate in the tumors as has been estimated by DNA methylation-based analysis of the Pan-Cancer-12 dataset (Figure S5E). In any case, these immune cell-associated gene programs may be pertinent to emerging treatment strategies based on immune modulation. Overall, despite uneven clinical information and follow-up across the many different Pan-Cancer types, expression-based determinants of the integrated subtypes were sufficiently informative to identify pathway-based features of prognostic value that transcend tissue-of-origin cancer types.
Multiple-Platform Determinants of the Integrated Subtypes
To gain insight into the genetic and epigenetic determinants that characterize each of the COCA subtypes, we calculated differential gene scores derived from each of the separate six platforms (see Supplemental Extended Experimental Procedures and Analysis Section 4) as well as PARADIGM pathway features. All differential activities were mapped to individual genes so that thematic pathways could be identified (see Supplemental Data File S2). Copy number data were summarized at the gene level using GISTIC 2.0 and t-tests for every gene were performed for each COCA subtype. DNA Methylation probes were associated with any gene that fell in the +/1500bp region surrounding gene transcriptional start sites. Genes with differential mRNA expression were identified using a SAM analysis on the RSEM values. Genes with differentially expressed protein products were determined by running a t-test on the 131 protein forms represented on the RPPA data. For mutations, a Fisher’s exact test on the frequency within a COCA subtype compared to outside the subtype was performed for all of a set of 291 high-confidence driver genes (Tamborero et al., 2013). Differentially expressed miRNAs for each COCA subtype were identified using a Wilcoxon rank-sum test based on the miRNA-Seq data. Genes were then identified as those predicted to be targeted by a differentially expressed miRNA that was also anti-correlated across the Pan-Cancer-12 dataset.
Three approaches were used to summarize the unique features of the COCA subtypes. First, Gene Set Enrichment Analysis (GSEA) was run on the single-platform gene-based results and then clustered for visual inspection to elucidate distinctive pathways (Figure S6A). Second, a supervised Elastic Net approach was used to classify the COCA subtypes with 95% accuracy in cross-validation and the predictive features were collected (Figure S6B; Supplemental Data File S3). Third, ‘regulatory hubs’ from PARADIGM with more than 15 downstream targets and found to be differentially activated within a COCA subtype relative to other subtypes were collected (Table S5A). All three approaches revealed that each platform detects different pathways and features with respect to both COCA subtypes and data platforms. The identified discriminating features of the COCA subtypes confirm several expectations: 1) C3-BRCA/Luminal was defined by protein and gene signatures for ER and GATA3 determined by the Elastic Net model as well as by PIK3CA-related signaling revealed by copy number variation-based and mutation-based GSEA, 2) C5-KIRC was defined by multiple features of hypoxia found by mutation- and mRNA-Seq-based GSEA as well as predictive Elastic Net features, and 3) C7-COAD/READ was in part defined by APC mutations.
Convergence of the Squamous-like Subtype
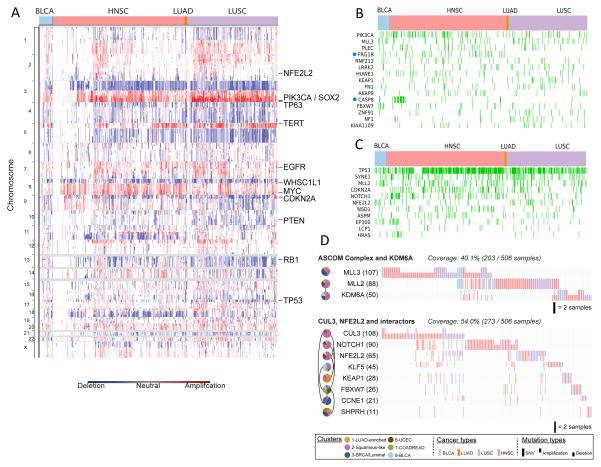
A striking finding of the integrative subtype analysis was the coalescence of four distinct tumor types (LUSC, HNSC, some BLCA and a very few LUAD) into the single C2-Squamous-like subtype. We investigated the genomic- and pathway-based determinants of the subtype. The three main tumor types included shared loss of 3p and increased TP63, PIK3CA and SOX2 gene copies within a characteristic 3q amplicon (Figure 4A). Those regions are well known in LUSC (The_Cancer_Genome_Atlas_Network, 2012a) and HNSC (Bhattacharya et al., 2011; Walter et al., 2013), and the results here extend that observation to include a subgroup of BLCA cases. In addition, the C2-Squamous-like subtype tends to show amplification of MYC and loss of CDKN2A, RB1 and TP53. TP53 mutation is frequent (72%), followed by a dramatic drop-off in mutation frequency to MLL2 (20%), PIK3CA (19%), CDKN2A (18%), NOTCH1 (16%), NFE2L2 (10%) and MALAT1 (6%), the only other genes mutated at ≥10% frequency (Table S2A). Of potential interest in the C2-Squamous-like group, tumors without TP53 mutations show a higher density of PIK3CA mutations (Figure 1), consistent with recent evidence linking PI3K activation and wild-type TP53 inactivation in HNSC (Herzog et al., 2013). Putative driver analysis identified several genes (PIK3CA, MLL3 and KEAP1) frequently mutated in the C2-Squamous-like group but also in other COCA subtypes (Figure 4B). Of these, FRG1B and CASP8 were found to be significantly more associated with HNSC by Fisher’s exact test. Putative driver analysis also revealed a number of genes with higher mutation frequencies in the C2-Squamous-like subtype than in any other subtype: TP53, SYNE1, MLL2, CDKN2A, NOTCH1, NFE2L2 and EP300, among others (Figure 4C; Figure S7A).
Figure 4. Genomic determinants of the C2-Squamous-like COCA subtype (see also Supplemental Figure S7 and Table S5).
A) SCNAs for the C2-Squamous-like subtype are shown, highlighting the importance of 3q26 gains across the different tissue-of-origin samples. B. Selected genes from 291 high-confidence driver (HCD) genes (Tamborero et al., 2013) mutated in > 5% of C2-Squamous-like samples and comparable in frequency in other subtypes. Samples with protein-affecting mutations in those genes are shown in green. C. HCD genes (as in panel B) with mutation frequency significantly higher in C2-Squamous-like tumors relative to others (stated at p<0.01 according to Fisher’s exact test with FDR correction). The method used corrections for imbalance in the number of samples from different tissues (see Supplemental Text Section 8). D. Two sub-networks of mutated pathways identified by an updated HotNet algorithm analysis using HINT interactions (see Supplemental Text) as mutated in at least 20% of the samples of the C2-Squamous-like subtype (cluster 2). Pie charts indicate interactions among the proteins in each subnetwork. Each gene (node) is colored by wedges whose size indicates the relative proportion of the gene’s mutations that are in samples from each integrated subtype. To the right of the pie charts is a gene-by-sample mutation matrix representing the mutation status of each gene across all Squamous-like samples. Full ticks represent SNVs, downticks represent deletions and upticks represent amplifications. The color of each tick indicates tissue-of-origin type, with gray indicating no mutation in the corresponding sample.
An extension of the HotNet algorithm (Vandin et al., 2012) was run on all genes mutated in ≥ 2% of any one subtype in conjunction with the HINT physical protein-protein interaction network (Supplemental Extended Experimental Procedures and Analyses, Section 4; Table S3). HotNet identified four sub-networks of mutated genes characteristic of the C2-Squamous-like subtype (Table S3B). The largest, most frequently mutated sub-network (91.7% of C2-Squamous-like samples) includes many well-known cancer genes and tumor suppressors, including TP53, CDKN2A and PTEN. The second most mutated sub-network (59.9%) consists of NFE2L2, CUL3, and KEAP1, CCNE1, FBXW7, and NOTCH1. NFE2L2, CUL3 and KEAP1 are well known regulators of oxidative stress. The third most mutated sub-network (37.1% of Squamous samples) includes the ASCOM complex (MLL2 and MLL3) and the putative ASCOM-interacting protein KDM6A. These proteins are involved in histone modifications that promote transcription. In addition, consistent with previous reports on collective motility in squamous cell carcinomas (Friedl and Gilmour, 2009), RAC and RHO signaling are also elevated in the C2-Squamous-like subtype based on PARADIGM analysis (Table S4A, Figure S7B).
Molecular Features Common to the Squamous, Breast Basal, and Ovarian Subtypes
Past work highlighted transcriptional similarities between the Breast Basal-like subtype and LUSC (Chung et al., 2002), as well as Breast Basal-like and Serous Ovarian cancers (The_Cancer_Genome_Atlas_Network, 2012c). We therefore asked if those subtypes share additional characteristics. The C9-OV (94%), C4-BRCA/Basal (80%) and C2-Squamous-like (72%) subtypes have the highest frequencies of TP53 mutation. All three show a very high frequency of copy number changes (Figure 2C), and all are significantly enriched with amplifications of 3q26 and 8q24/cMYC and losses of chromosomes 4q, 5q, 8p, and 18q (Figure 2B). The COCA subtypes share features common to a pan-cancer cluster identified by a parallel analysis of the transcriptional profiles of these same tumors (Martinez et al., 2014), which was found to be associated with genomic loss of CDKN2A (p16ARF), increased numbers of DNA double strand breaks, high expression of cyclin B1, and upregulation of proliferation genes.
Consistent with our previous TCGA report noting the similarities between Breast Basal-like and Serous Ovarian cancers (The_Cancer_Genome_Atlas_Network, 2012c), the copy number profiles of the integrative subtypes place the C4-BRCA/basal subtype closest to the C9-OV subtype (Figure S4C); both are also near a cluster tree branch that contains C2-Squamous-like and C8-BLCA. All six of those subtypes show TP53 mutation and large-scale copy number changes.
Pathway commonalities between the C4-BRCA/basal and C9-OV subtypes (Table S5B) largely recapitulate previous finding using PARADIGM analysis that both subtypes show activation of cMYC and FOXM1/proliferation signaling (The_Cancer_Genome_Atlas_Network, 2012c). However, HIF1A signaling in those subtypes, despite previously being reported as high, appears less active in this Pan-Cancer context, probably due to the presence of other cancer types with clearly elevated HIF1A signaling (e.g. C5-KIRC). In terms of gene programs, C2-Squamous-like tumors show high expression of the basal signaling gene program (GP17), at levels comparable with those in the C4-BRCA/Basal tumors (Figure 3). In addition, both subtypes show up-regulation of the proliferation/DNA synthesis gene program (GP1), as well as signatures of TP53 mutation, MYC targets/TERT, VEGF signaling and activation of the PD1 and CTLA4 immune co-stimulatory pathways (Table S4A, Figure 3). Indeed, principal components analysis showed that C2-Squamous-like and C4-BRCA/Basal tumors are the most similar COCA subtypes with regard to gene program/drug pathway expression (Figure S5B).
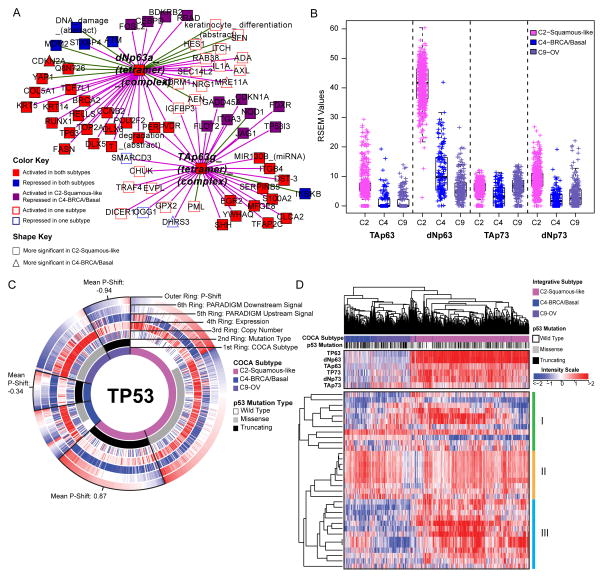
In line with those findings, a systematic search for PARADIGM pathway commonalities between the C2-Squamous-like and C4-BRCA/Basal tumors through the definition of a ‘basalness score’ (The_Cancer_Genome_Atlas_Network, 2012c) reveals shared activation of proliferation- and immune-related pathways. TP63 network dysregulation is apparent in HNSC and LUSC (Figure S7C, Table S5), as found previously (The_Cancer_Genome_Atlas_Network, 2012a; Walter et al., 2013). It has also been associated with normal basal stem/progenitor cell function in other organs (e.g. breast, urogenital tract) (Crum and McKeon, 2010). However, closer scrutiny of the network neighborhood surrounding the TAp63g and dNp63a complexes reveals that TP63 activation is more significant in the C2-Squamous-like tumors than it is in the C4-BRCA/Basals, and it involves a larger number of TP63 network targets (Figure 5A). Indeed, TP63 expression levels, in particular expression of the oncogenic ΔNp63 isoform, are significantly higher in the C2-Squamous-like subtype than in the C4-BRCA/Basal tumors (Figure 5B). Notably, we did not see TP63 network activity or increased expression in the C9-OV subtype (Table S4A and Figure 6B).
Figure 5. Comparison of molecular characteristics of C2-Squamous-like, C4-BRCA/Basal and C9-OV (ovarian) subtypes reveals differences in TP63 and TP53 signaling (see also Supplemental Figure S7, Table S5, and Data File S4).
A) Relative significance of TP63 network activation within the C2-Squamous-like and C4-BRCA/Basal subtypes. The network neighbors surrounding the TAp63γ and ΔNp63α tetramer complexes that show significant activation (or inactivation) within the C2-Squamous-like and/or C4-BRCA/Basal subtypes relative to all other cases were visualized using Cytoscape (Shannon et al., 2003). Node shape reflects relative significance in the one-versus-all comparison (square: more significant in C2-Squamous-like, triangle: more significant in C4-BRCA/Basal). Node color indicates relative activity (red: activated in C2 and C4, blue: inactivated in C2 and C4, purple: activated in C2 but inactivated in C4, white: activated or inactivated in only one subtype). B) Box plot of isoform-specific levels of TP63 and TP73 within three of the TP53-frequently mutated COCA subtypes (C2-Squamous-like, C4-BRCA/Basal, and C9-OV). C) CircleMap of PARADIGM-Shift differences associated with TP53 mutations within the C2, C4 and C9 COCA subtypes. Samples were ordered first by integrative subtype membership (innermost ring), then by TP53 mutation status (second ring), and finally by P-Shift (outer ring, indicating TP53 activity). The GISTIC score (indicating CNV), mRNA expression level, PARADIGM upstream and downstream activities are shown in the third, fourth, fifth and sixth rings, respectively. Red-blue color intensity reflects magnitude (red: positive, blue: negative). TP53-truncating mutants are highlighted (black outlined wedge), and the mean P-shift scores of the truncating mutants are shown. Negative P-Shift scores (outer ring blue) predict loss of function (LOF). D) Unsupervised clustering of C2-Squamous-like, C4-BRCA/basal, and C9-OV cancers based on the expression patterns of 33 published TP53-related gene signatures. Sample subtype assignment (pink: C2-Squamous-like, blue: C4-BRCA/basal, purple: C9-OV) and TP53 mutation status (wild type: white, truncating: black, missense: grey) are indicated in the column color bar. Heatmap red-blue color intensity reflects magnitude (red: positive, white, average: blue: negative). See Supplemental Data File S4 (syn2491513) for complete list.
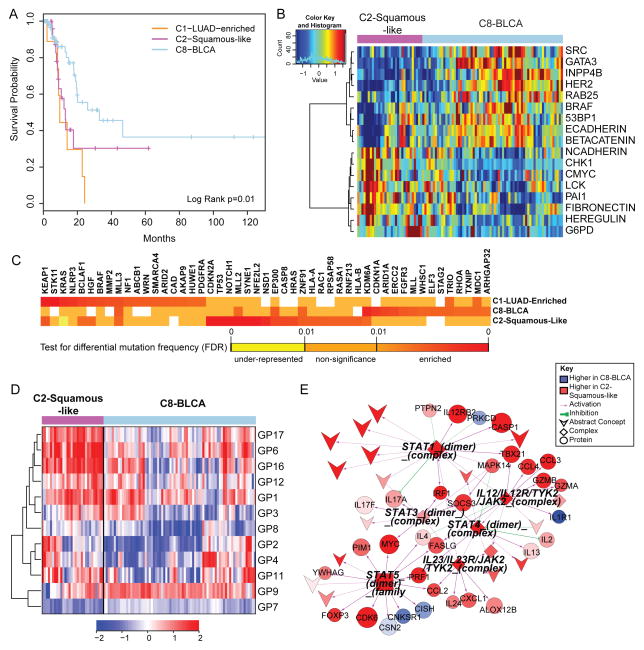
Figure 6. Divergence of the bladder cancer samples across multiple COCA subtypes (see also Supplemental Figure S8 and Table S6).
A) Kaplan-Meier survival analysis of bladder cancers within the C1-LUAD-enriched, C2-Squamous-like, and C8-BLCA subtypes. B) Heatmap of 17 proteins expressed at significantly different levels within the C2-Squamous-like relative to the C8-BLCA bladder cancer samples. Samples are arranged along the column by subtype (pink: C2, light blue: C8); and protein data are ordered along the rows by clustering. Rainbow color scale reflects magnitude (red: high, green: average, blue: low). C) HCD genes with differential mutation frequencies among the bladder samples clustered in COCA subtypes C1, C2 and C8. Differential frequencies reflect frequencies within, relative to frequencies outside of, the COCA subtype. D) Heatmap of 11 gene programs showing significant differential expression between the C2 and C8 bladder cancers. Samples are arranged along the column by subtype (pink: C2, light blue: C8), and gene programs are ordered along the rows by clustering. Red-blue color scale reflects magnitude (red: high, blue: low). E) PARADIGM sub-network of immune-related pathway biomarkers activated in C2 bladder cancers relative to the C8 subtype. Red-blue color scale represents relative activation (red: higher in C2, blue: higher in C8). Node size reflects relative significance, and node shape denotes feature type (diamond: multi-protein complex, inverted v: cellular process, circle: genes, square: gene family). Color of an edge reflects type of interaction within the PARADIGM SuperPathway (purple arrows: activation, green T: inhibition).
High TP53 mutation rates characterize several tumor types including those represented by the COCA subtypes C4-BRCA/Basal, C9-OV, and C2-Squamous-like (Table S2A). Surprisingly, our pathway and gene program analysis reveal a pattern of TP53 compensation in the C2-Squamous-like tumors that distinguishes them from these other subtypes with high TP53 mutation rates. First, the C2-Squamous-like tumors do not exhibit significant loss of PARADIGM-inferred TP53 activity (Table S4A) and PARADIGM-SHIFT analysis (Ng et al., 2012) predicts loss-of-function of TP53-truncating mutations (observed in 43% of C4-BRCA/Basal, 38% of C9-OV and 30% of C2-Squamous-like cases) at a significantly higher degree in the C4-BRCA/Basal and C9-OV subtypes compared to the C2-Squamous-like subtype (Figure 5C). Second, the copy number data when aligned with TP53 missense and truncating mutations, reveals more loss of heterozygosity (LOH) in the C9-OV and C4-BRCA/Basal than in the C2-Squamous-like samples. The apparent higher TP53-pathway activity in C2-Squamous-like tumors may be related to the expression of isoforms of related family members TP63 and/or TP73 (Figure 5B), which may compensate for TP53 mutation in the C2-Squamous-like tumors as revealed by PARADIGM-Shift analysis (Figure 5C), and as supported by functional experimental data in HNSC lines and tumors (Lu et al., 2011). In HNSC, the function of TP63/73 in growth of HNSC is modulated in the presence of inflammatory factor TNF-α and cREL. Third, the transcriptional targets of TP53 shared with TP63/73 appear to be more highly expressed in the C2-Squamous-like subtype than in the C9-OV or C4-BRCA/Basal subtype (Figure S7D). Indeed, hierarchical clustering of 33 TP53-related gene signatures subsets the C2-Squamous-like, C4-BRCA/Basal and C9-OV tumors predominantly by subtype (left side dendrogram sub-tree: 99% C4-BRCA/basal/C9-OV; right-side dendrogram sub-tree: 98% C2-Squamous-like) (Figure 5D). However, with the exception of the C4-BRCA/Basal-like subtype, the levels of TP53 activity were not predictive of overall survival when restricted to the analysis within a subtype. For the C4-BRCA/Basal case, the PARADIGM-Shift scores do provide a moderate predictive degree when only the TP53 truncating mutants are considered (P<0.05). Interestingly, TP63/73 compensatory function has been linked to cisplatin chemo-sensitivity and survival in BRCA1-related triple negative breast cancers (Leong et al., 2007). These studies show the potential for p63/73 compensatory function for mutated or suppressed p53 in HNSCC and breast cancer, which has potential implications for targeted and standard therapy across these malignancies. These data indicate that TP53/63/73 downstream activities are of potentially broader significance among the C2-Squamous-like, C9-OV and C4-BRCA/Basal subtypes, with similarly high TP53 mutation rates.
Divergence of Bladder Cancer Subtypes
Despite a relatively small sample size (n=120), bladder cancer was one of the most diverse of the tumor types, with samples clustering into 7 of the 11 major COCA subtypes (Table S6). The majority of the samples fell into three main COCA groups: 10 in C1–LUAD-enriched, 31 in C2–Squamous-like and 74 in C8–BLCA. Correlation with histology showed that the bladder samples in the C2-Squamous-like group did, indeed, have evidence of squamous features, although most in that subtype had less than 50% squamous differentiation upon review by a team of 5 urological pathologists. The genomic classifications are consistent with evidence for diverse squamous, adenocarcinoma and other variant histologies in bladder carcinoma (Willis et al., 2013). Because it is one of the most diverse tissue-of-origin tumor types in the Pan-Cancer-12 set, we looked at survival differences among the three main COCA groups of bladder cancers. Samples in the C2-Squamous-like and C1-LUAD-enriched groups showed significantly worse overall survival than those in the C8-BLCA group (Figure 6A; Figure S8B). The same distinction held in proteomics-only analyses (Akbani et al., 2014), consistent with the worse overall survival of the other tumor types (LUAD, LUSC, and HNSC) that predominate in the C1-LUAD-enriched and C2-Squamous-like subtypes.
We focused on the two larger subsets (C2-Squamous-like and C8-BLCA) of bladder cancers, performing single-platform and integrated-platform comparisons. There are significant differences in copy number (Figure S4A), protein expression (Figure 6B), mutations (Figure 6C), gene programs (Figure 6D) and PARADIGM pathway networks (Figure 6E; Figure S8A). There is also a significant difference in 3p arm-level events; the C2-Squamous-like subset shows the characteristic squamous-like pattern of 3p loss, whereas the C8-BLCA subtype does not (Figure 2B). Consistent with findings from the Pan-Cancer proteomics analysis (Akbani et al., 2014), higher HER2 and Rab25 protein levels are observed in the majority of the C8-BLCA cases relative to the C2-Squamous-like bladder cases (Figure 6B). Conversely, markers of epithelial-to-mesenchymal transition (EMT) such as low E-cadherin, high fibronectin, and high N-cadherin expression are apparent in the C2-Squamous-like bladder cancers (Figure 6B). Both gene program and PARADIGM analyses reveal differences in immune cell signatures; the bladder C2-Squamous-like samples show higher levels of immune cell-associated signatures (Figure 6D–E). That difference, which has also been noted for lung squamous (The_Cancer_Genome_Atlas_Network, 2012a) and breast Basal-like cancers (Prat et al., 2010), could contribute to differences in outcome and suggest therapeutic targets.
DISCUSSION
This integrated multi-platform analysis of 12 cancer types provides independent and clinically relevant prognostic information above and beyond tumor stage and primary tissue-of-origin. Based on this study, one in ten cancer patients would be classified differently by this new molecular taxonomy versus our current tissue-of-origin tumor classification system. With respect to its therapeutic relevance, this proportion of potentially misclassified tumors is comparable to the rate of EGFR mutations in unselected non-small cell lung cancers (Lynch et al., 2004; Paez et al., 2004) and ERBB2 amplifications among all breast cancers (The_Cancer_Genome_Atlas_Network, 2012c). If used to guide therapeutic decisions, this reclassification would affect a significant number of patients to be considered for non-standard treatment regimens. In addition to identifying several new genomic and pathway insights between and within tissue-of-origin tumor types, this TCGA study provides a public resource compendium of individual and integrated datasets from six different “omic” platforms, comprehensively characterizing >3,500 tumors and enabling researchers to explore new questions and analytical approaches that will perpetuate this discovery process.
It is possible that each COCA subtype reflects tumors arising from distinct cell types. In this new taxonomy, cancers of non-epithelial origin (e.g. neural, muscle, connective tissue) appear most different from epithelial tumors based on virtually all molecular platforms. The next most marked difference is apparent between epithelial cancers arising from basal layer-like cells (C2-Squamous-like and C4-BRCA/Basal) and those with secretory functions (C1-LUAD-enriched and C3-BRCA/Luminal). Molecular commonalities within a COCA subtype suggest common oncogenic pathways. The C2-Squamous-like cancers likely arise from a cellular subtype shared between environmentally exposed epithelial surfaces (e.g. oral cavity, lungs, and bladder); and malignancies from this cellular subtype possess a characteristic set of dysregulated genomic features, including SOX2 and ΔNp63 high expression (by 3q26-29 amplification) with TP53 mutation. Although some of these pathway features have previously been reported for normal squamous tissue development and homeostasis (Crum and McKeon, 2010) and in squamous cell carcinomas of specific organ sites (Maier et al., 2011; Yang et al., 2011), they have not previously emerged collectively as a broad subtype-defining phenotype from an integrated genomic analysis of thousands of different tumors. Cancers in the C2-Squamous-like subtype appear most similar to those in the C4-BRCA/Basal subtype, which in turn show pathway similarities to those in the C9-Ovarian. While all three COCA subtypes exhibit comparably high TP53 mutation frequencies and expression of the GP17_Basal signaling gene program, the C2-Squamous-like cancers are distinguished from all others by their significantly higher TP63 and TP73 expression, both short (ΔNp63, ΔNp73) and long (TAp63, TAp73) isoforms, which may partially compensate TP53 mutation in this COCA subtype.
In this integrated analysis, bladder cancers (BLCA) emerged as the most heterogeneous of all Pan-Cancer-12 malignancies, with multiple samples falling into primarily three different integrated subtypes (C1-LUAD-enriched-like, C2-Squamous-like and C8-BLCA). The clinical relevance of Pan-Cancer integrated subtyping is apparent in this divergent cohort of tumors. Survival is dependent on subtype membership, with the C2-Squamous-like BLCA cases showing earlier mortality than the more common C8-BLCA cases (Figure 7A). Those BLCA cases in the C2-Squamous-like subtype display immune features common to the C2-Squamous-like subtype, that are pertinent to two areas of current interest among bladder cancer researchers: i) epidemiologic and experimental evidence that chronic cystitis and recurrent bladder inflections (or other physical irritants) capable of inducing squamous metaplasia can predispose to squamous cancer of the bladder; and ii) the observation that early-stage bladder cancers are often responsive to intravesicular T-cell induction by Bacillus-Calmette-Guerin (BCG) anti-TB vaccination. These findings strongly support a COCA subtype specific approach to post resection surveillance, adjuvant therapy and management of metastatic disease for bladder cancer patients.
Our results suggest that “cell-of-origin” rather than pathway-based features dominate the molecular taxonomy of diverse tumor types. There are several possible explanations for this observation. First and foremost, there are hundreds to thousands of features (mRNAs, proteins, microRNAs) with cell-type specific expression patterns, whereas pathways tend to regulate a much smaller subset of components – tens to hundreds of genes and their products. Secondly, pathways are often used in a cell type-specific manner (e.g. APC-pathway in colon/rectum); therefore, pathway-based features are likely subsumed by a cell-of-origin-based classification. Further research is needed to uncover pathway dependencies within a cell-of-origin context, of which many such relationships already exist (i.e. EGFR in LUAD, BRAF in Melanoma, etc).
In closing, the refined molecular taxonomy we describe builds on centuries of pathology and genetic research. The datasets and results have been collected into a unified resource on Synapse to support integrative bioinformatics analysis. To support navigation through these findings, the results have been made available through several portals including the UCSC Genome Browser, Gitools, and MD Anderson’s Next Generation Heatmaps (see the Supplemental Extended Procedures and Analyses). New methods to mine these data will enable “subtracting away” the dominant cell-of-origin signals to reveal information about pathway signaling and tumor microenvironments (e.g. stromal and immune components). This new taxonomy provides independent prognostic information above and beyond stage and tissue-of-origin, and further investigations may provide novel pathway-based insights with clues for personalizing therapy. Follow-up studies are needed to validate the findings reported here, and additional samples and tumor types will extend the integrated analysis. However, this initial PanCancer-12 analysis lays the groundwork for a richer classification of tumors into molecularly defined subtypes unlike all prior cancer classification systems.
EXPERIMENTAL PROCEDURES
Data for the complete set of 5,074 TCGA samples were obtained for the December 22, 2012 Pan-Cancer-12 data freeze from the Sage Bionetworks repository, Synapse. All data is made available through the Synapse website (https://www.synapse.org) and referenced with Synapse identifiers denoted as synN, where N provides a unique identifier within the Synapse system. All relevant result files relevant for subtyping and downstream analyses are available from syn2468297.
Mutation data and predicted driver genes
Single nucleotide variant calls for all samples in each of the 12 different tumor types were obtained from the official data freeze for each individual data type. Briefly, mutation calls were obtained from the separate TCGA working groups and processed to de-duplicate and re-annotate them using the ENSEMBLE version 69 transcript database. The combined mutation annotation format (MAF) file is available from the Synapse resource.
127 Significantly mutated genes (SMGs) were identified in the entire sample set as those mutated more frequently than the background model according to MuSiC analysis as described in (Kandoth et al., 2013a). The SMG analysis was also performed by running MuSiC restricted to each COCA subtype. Lastly, genes whose mutations predominantly occur within a given COCA subtype were identified by using the list of high-confidence drivers retrieved by the combined analysis of several signals of positive selection, as described in (Tamborero et al., 2013).
Cluster of Cluster Assignments (COCA)
Subtypes derived from each of the six platforms – mRNA-Seq, miRNA-Seq, reverse-phase protein arrays (RPPA), structural copy number alterations (SCNA), DNA methylation, and somatic mutations – were calculated as described in the Supplemental Extended Procedures and Analyses section. Subtype calls for each of the six platforms were used to identify relationships among the different COCA subtypes and coded into a series of indicator variables for each subtype. The binary matrix was then used in the ConsensusClusterPlus R-package (Wilkerson and Hayes, 2010) to identify patterns of relationship among the samples. ConsensusClusterPlus was run with 80% sample resampling and 1000 iterations of hierarchical clustering based on a Pearson correlation distance metric. More information on integrative subtyping analysis can be found in Supplemental Extended Procedures and Analyses, Section 2. The integrated COCA subtypes are available on the Synapse resource.
Survival Analysis for Pan-Cancer-12 and Squamous Bladder Samples
Overall survival was calculated for samples using information from the enrollment and follow-up forms available at the DCC and downloaded on 6/17/2013. Kaplan-Meier survival plots were generated with the package Survival in R. A log-rank test was used to assess significance.
Supplementary Material
Acknowledgments
We thank the thousands of patients and their loved ones who contributed materially, emotionally, and intellectually to this study. We thank DA Wheeler and ML Meyerson for scientific review of the work and MP Schroeder for help setting up Gitools. We thank KR Shaw, BA Ozenberger, HJ Sofia, CM Hutter, and JC Zenklusen for administrative support. This work was supported by grants from the Chapman Foundation and Dell Foundation to JNW, a MD Anderson Physician Scientist Award to LAB, a Burroughs Welcome Career Award at the Scientific Interface to BJR, and support from the following grants from the United States National Institutes of Health: K08 CA137153, K08 CA176561, P50 CA083639, R01 CA071468, R01 HG005690, R01 HG007069, R01CA180006, R21 CA155679, U01 CA168394, U24 CA143858, U24 CA143867-05, U24 CA143883, U24 CA143848, U24 CA143858, U24 CA143866, U54 CA112970, U24 CA143799, U24 CA143835, U24 CA143840, U24 CA143843, U24 CA143845, U24 CA143848, U24 CA143858, U24 CA143866, U24 CA143867, U24 CA143882, U24 CA143883, U24 CA144025, U54 HG003273, U54 HG003067, U54 HG003079, ZIA-DC-000073, ZIA-DC-000074, and P30 CA016672 for the MD Anderson CCSG Functional Proteomics Core.
AUTHOR CONTRIBUTIONS
KAH, CY, DMW, ADC, LVV, NLB, PWL, LD, GBM, CCB, CMP, and JMS conceived and designed the experiments. KAH, CY, DMW, ADC, DT, MDL, BN, VU, RA, HS, and BJR developed key methodology. RA, PWL, LAB, and GBM acquired data sets. KAH, CY, DMW, ADC, DT, AC, SN, MDL, BN, MDM, VU, CK, RA, HS, NLB, PWL, BJR, LD, AGR, LAB, GBM, JNW, CVW, ZC, EAC, CCB, CMP, and JMS analyzed and interpreted data. JZ, LO, AAM, NLB, RA, JNW, NLB, and JMS created data resources. KAH, DY, DMW, ADC, DT, SN, MDL, BN, VU, RA, BJR, LAB, GBM, JNW, CVW, ZC, EAC, CCB, CMP, JMS drafted the manuscript or provided critical revisions. JZ, CCB, CMP, and JMS led and coordinated the project.
CONSORTIA
The members of The Cancer Genome Atlas Research Network are Rachel Abbott, Scott Abbott, B. Arman Aksoy, Kenneth Aldape, Adrian Ally, Samirkumar Amin, Dimitris Anastassiou, J.Todd Auman, Keith A Baggerly, Miruna Balasundaram, Saianand Balu, Stephen B. Baylin, Stephen C. Benz, Benjamin P. Berman, Brady Bernard, Ami S. Bhatt, Inanc Birol, Aaron D. Black, Tom Bodenheimer, Moiz S. Bootwalla, Jay Bowen, Ryan Bressler, Christopher A. Bristow, Angela N. Brooks, Bradley Broom, Elizabeth Buda, Robert Burton, Yaron S.N. Butterfield, Daniel Carlin, Scott L. Carter, Tod D. Casasent, Kyle Chang, Stephen Chanock, Lynda Chin, Dong-Yeon Cho, Juok Cho, Eric Chuah, Hye-Jung E. Chun, Kristian Cibulskis, Giovanni Ciriello, James Cleland, Melisssa Cline, Brian Craft, Chad J. Creighton, Ludmila Danilova, Tanja Davidsen, Caleb Davis, Nathan D. Dees, Kim Delehaunty, John A. Demchok, Noreen Dhalla, Daniel DiCara, Huyen Dinh, Jason R. Dobson, Deepti Dodda, HarshaVardhan Doddapaneni, Lawrence Donehower, David J. Dooling, Gideon Dresdner, Jennifer Drummond, Andrea Eakin, Mary Edgerton, Jim M. Eldred, Greg Eley, Kyle Ellrott, Cheng Fan, Suzanne Fei, Ina Felau, Scott Frazer, Samuel S Freeman, Jessica Frick, Catrina C. Fronick, Lucinda L. Fulton, Robert Fulton, Stacey B. Gabriel, Jianjiong Gao, Julie M. Gastier-Foster, Nils Gehlenborg, Myra George, Gad Getz, Richard Gibbs, Mary Goldman, Abel Gonzalez-Perez, Benjamin Gross, Ranabir Guin, Preethi Gunaratne, Angela Hadjipanayis, Mark P. Hamilton, Stanley R. Hamilton, Leng Han, Yi Han, Hollie A. Harper, Psalm Haseley, David Haussler, D. Neil Hayes, David I. Heiman, Elena Helman, Carmen Helsel, Shelley M. Herbrich, James G. Herman, Toshinori Hinoue, Carrie Hirst, Martin Hirst, Robert A. Holt, Alan P. Hoyle, Lisa Iype, Anders Jacobsen, Stuart R. Jeffreys, Mark A. Jensen, Corbin D. Jones, Steven J.M. Jones, Zhenlin Ju, Joonil Jung, Andre Kahles, Ari Kahn, Joelle Kalicki-Veizer, Divya Kalra, Krishna-Latha Kanchi, David W. Kane, Hoon Kim, Jaegil Kim, Theo Knijnenburg, Daniel C. Koboldt, Christie Kovar, Roger Kramer, Richard Kreisberg, Raju Kucherlapati, Marc Ladanyi, Eric S. Lander, David E. Larson, Michael S. Lawrence, Darlene Lee, Eunjung Lee, Semin Lee, William Lee, Kjong-Van Lehmann, Kalle Leinonen, Kristen M. Leraas, Seth Lerner, Douglas A. Levine, Lora Lewis, Timothy J. Ley, Haiyan I. Li, Jun Li, Wei Li, Han Liang, Tara M. Lichtenberg, Jake Lin, Ling Lin, Pei Lin, Wenbin Liu, Yingchun Liu, Yuexin Liu, Philip L. Lorenzi, Charles Lu, Yiling Lu, Lovelace J. Luquette, Singer Ma, Vincent J. Magrini, Harshad S. Mahadeshwar, Elaine R. Mardis, Adam Margolin, Marco A. Marra, Michael Mayo, Cynthia McAllister, Sean E. McGuire, Joshua F. McMichael, James Melott, Shaowu Meng, Matthew Meyerson, Piotr A. Mieczkowski, Christopher A. Miller, Martin L. Miller, Michael Miller, Richard A. Moore, Margaret Morgan, Donna Morton, Lisle E. Mose, Andrew J. Mungall, Donna Muzny, Lam Nguyen, Michael S. Noble, Houtan Noushmehr, Michelle O’Laughlin, Akinyemi I. Ojesina, Tai-Hsien Ou Yang, Brad Ozenberger, Angeliki Pantazi, Michael Parfenov, Peter J. Park, Joel S. Parker, Evan Paull, Chandra Sekhar Pedamallu, Todd Pihl, Craig Pohl, David Pot, Alexei Protopopov, Teresa Przytycka, Amie Radenbaugh, Nilsa C. Ramirez, Ricardo Ramirez, Gunnar Rätsch, Jeffrey Reid, Xiaojia Ren, Boris Reva, Sheila M. Reynolds, Suhn K. Rhie, Jeffrey Roach, Hector Rovira, Michael Ryan, Gordon Saksena, Sofie Salama, Chris Sander, Netty Santoso, Jacqueline E. Schein, Heather Schmidt, Nikolaus Schultz, Steven E. Schumacher, Jonathan Seidman, Yasin Senbabaoglu, Sahil Seth, Samantha Sharpe, Ronglai Shen, Margi Sheth, Yan Shi, Ilya Shmulevich, Grace O. Silva, Janae V. Simons, Rileen Sinha, Payal Sipahimalani, Scott M. Smith, Heidi J. Sofia, Artem Sokolov, Mathew G. Soloway, Xingzhi Song, Carrie Sougnez, Paul Spellman, Louis Staudt, Chip Stewart, Petar Stojanov, Xiaoping Su, S. Onur Sumer, Yichao Sun, Teresa Swatloski, Barbara Tabak, Angela Tam, Donghui Tan, Jiabin Tang, Roy Tarnuzzer, Barry S. Taylor, Nina Thiessen, Vesteinn Thorsson, Timothy Triche Jr., David J. Van Den Berg, Fabio Vandin, Richard J. Varhol, Charles J. Vaske, Umadevi Veluvolu, Roeland Verhaak, Doug Voet, Jason Walker, John W. Wallis, Peter Waltman, Yunhu Wan, Min Wang, Wenyi Wang, Zhining Wang, Scot Waring, Nils Weinhold, Daniel J. Weisenberger, Michael C. Wendl, David Wheeler, Matthew D. Wilkerson, Richard K. Wilson, Lisa Wise, Andrew Wong, Chang-Jiun Wu, Chia-Chin Wu, Hsin-Ta Wu, Junyuan Wu, Todd Wylie, Liu Xi, Ruibin Xi, Zheng Xia, Andrew W. Xu, Da Yang, Liming Yang, Lixing Yang, Yang Yang, Jun Yao, Rong Yao, Kai Ye, Kosuke Yoshihara, Yuan Yuan, Alfred K. Yung, Travis Zack, Dong Zeng, Jean Claude Zenklusen, Hailei Zhang, Jianhua Zhang, Nianxiang Zhang, Qunyuan Zhang, Wei Zhang, Wei Zhao, Siyuan Zheng, Jing Zhu, Erik Zmuda, Lihua Zou
References
- Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, Liu W, Yang JY, Yoshihara K, Li J, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nature communications. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Roy R, Snijders AM, Hamilton G, Paquette J, Tokuyasu T, Bengtsson H, Jordan RC, Olshen AB, Pinkel D, et al. Two distinct routes to oral cancer differing in genome instability and risk for cervical node metastasis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:7024–7034. doi: 10.1158/1078-0432.CCR-11-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CH, Bernard PS, Perou CM. Molecular portraits and the family tree of cancer. Nature genetics. 2002;32(Suppl):533–540. doi: 10.1038/ng1038. [DOI] [PubMed] [Google Scholar]
- Crum CP, McKeon FD. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annual review of pathology. 2010;5:349–371. doi: 10.1146/annurev-pathol-121808-102117. [DOI] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nature reviews Molecular cell biology. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- Grilley-Olson JE, Hayes DN, Moore DT, Leslie KO, Wilkerson MD, Qaqish BF, Hayward MC, Cabanski CR, Yin X, Socinski MA, et al. Validation of interobserver agreement in lung cancer assessment: hematoxylin-eosin diagnostic reproducibility for non-small cell lung cancer: the 2004 World Health Organization classification and therapeutically relevant subsets. Archives of pathology & laboratory medicine. 2013;137:32–40. doi: 10.5858/arpa.2012-0033-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog A, Bian Y, Vander Broek R, Hall B, Coupar J, Cheng H, Sowers AL, Cook JD, Mitchell JB, Chen Z, et al. PI3K/mTOR inhibitor PF-04691502 antitumor activity is enhanced with induction of wild-type TP53 in human xenograft and murine knockout models of head and neck cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:3808–3819. doi: 10.1158/1078-0432.CCR-12-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofree M, Shen JP, Carter H, Gross A, Ideker T. Network-based stratification of tumor mutations. Nature methods. 2013 doi: 10.1038/nmeth.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013a;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013b;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. The Journal of clinical investigation. 2007;117:1370–1380. doi: 10.1172/JCI30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Yang X, Duggal P, Allen CT, Yan B, Cohen J, Nottingham L, Romano RA, Sinha S, King KE, et al. TNF-alpha promotes c-REL/DeltaNp63alpha interaction and TAp73 dissociation from key genes that mediate growth arrest and apoptosis in head and neck cancer. Cancer research. 2011;71:6867–6877. doi: 10.1158/0008-5472.CAN-11-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Maier S, Wilbertz T, Braun M, Scheble V, Reischl M, Mikut R, Menon R, Nikolov P, Petersen K, Beschorner C, et al. SOX2 amplification is a common event in squamous cell carcinomas of different organ sites. Human pathology. 2011;42:1078–1088. doi: 10.1016/j.humpath.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Martinez E, Yoshihara K, Kim H, Mills GB, Trevino V, Verhaak RG. Comparison of gene expression patterns across twelve tumor types identifies a cancer supercluster characterized by TP53 mutations and cell cycle defects. Oncogene. 2014 doi: 10.1038/onc.2014.216. To appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Collisson EA, Sokolov A, Goldstein T, Gonzalez-Perez A, Lopez-Bigas N, Benz C, Haussler D, Stuart JM. PARADIGM-SHIFT predicts the function of mutations in multiple cancers using pathway impact analysis. Bioinformatics. 2012;28:i640–i646. doi: 10.1093/bioinformatics/bts402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, Davies SR, Snider J, Stijleman IJ, Reed J, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. The oncologist. 2013;18:123–133. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast cancer research: BCR. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scagliotti GV, Parikh P, von Pawel J, Biesma B, Vansteenkiste J, Manegold C, Serwatowski P, Gatzemeier U, Digumarti R, Zukin M, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26:3543–3551. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborero D, Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Kandoth C, Reimand J, Lawrence MS, Getz G, Bader GD, Ding L, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Scientific reports. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_Cancer_Genome_Atlas_Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_Cancer_Genome_Atlas_Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_Cancer_Genome_Atlas_Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012a;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_Cancer_Genome_Atlas_Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012b;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_Cancer_Genome_Atlas_Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012c;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_Cancer_Genome_Atlas_Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013a;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_Cancer_Genome_Atlas_Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine. 2013b;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_Cancer_Genome_Atlas_Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troester MA, Herschkowitz JI, Oh DS, He X, Hoadley KA, Barbier CS, Perou CM. Gene expression patterns associated with p53 status in breast cancer. BMC cancer. 2006;6:276. doi: 10.1186/1471-2407-6-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandin F, Clay P, Upfal E, Raphael BJ. Discovery of mutated subnetworks associated with clinical data in cancer. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing; 2012. pp. 55–66. [PubMed] [Google Scholar]
- Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, Haussler D, Stuart JM. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter V, Yin X, Wilkerson MD, Cabanski CR, Zhao N, Du Y, Ang MK, Hayward MC, Salazar AH, Hoadley KA, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PloS one. 2013;8:e56823. doi: 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DL, Porten SP, Kamat AM. Should histologic variants alter definitive treatment of bladder cancer? Current opinion in urology. 2013;23:435–443. doi: 10.1097/MOU.0b013e328363e415. [DOI] [PubMed] [Google Scholar]
- Yang X, Lu H, Yan B, Romano RA, Bian Y, Friedman J, Duggal P, Allen C, Chuang R, Ehsanian R, et al. DeltaNp63 versatilely regulates a Broad NF-kappaB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Cancer research. 2011;71:3688–3700. doi: 10.1158/0008-5472.CAN-10-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.