Abstract
Somatic cells can be reprogrammed to induced pluripotent stem cells (iPSCs) by defined factors. However, substantial cell numbers subjected to iPSC induction stray from the main reprogramming route and are immortalized as partial iPSCs. These partial iPSCs can become genuine iPSCs by exposure to the ground state condition. However, such conversion is only possible for mouse partial iPSCs, and it is not applicable to human cells. Moreover, the molecular basis of this conversion is completely unknown. Therefore, we performed genome-wide screening with a piggyBac vector to identify genes involved in conversion from partial to genuine iPSCs. This screening led to identification of Cnot2, one of the core components of the Ccr4-Not complex. Subsequent analyses revealed that other core components, Cnot1 and Cnot3, also contributed to the conversion. Thus, our data have uncovered a novel role of core components of the Ccr4-Not complex as regulators of transition from partial to genuine iPSCs.
Introduction
Somatic cells such as skin cells can be reprogrammed to pluripotent cells by forced expression of defined factors [1]. However, the generation of induced pluripotent stem cells (iPSCs) by this method is very slow and inefficient, making it difficult to investigate the underlying molecular mechanisms of pluripotency acquisition [2–6]. Partial iPSCs are an unwanted by-product generated from cells subjected to iPSC induction [6–8]. A recent investigation [5] has suggested that this partial iPSC state is an unnecessary step toward becoming genuine iPSCs. However, substantial proportions of somatic cells stray from the main reprogramming route and become partial iPSCs. DNA microarray analyses have revealed that the expression profiles of partial iPSCs from different laboratories show an extremely high similarity with each other [5,6]. These analyses suggest that the partial iPSC state is reasonably homogeneous rather than a generic term for cells that fail to complete iPSC induction. The high similarity among expression profiles is probably because of the numerous strict criteria that define partial iPSCs. These criteria include an embryonic stem cell (ESC)-like morphology, positivity for the SSEA-1 marker and alkaline phosphatase activity, maintenance of exogenous reprogramming factor expression, rapid and stable propagation, and expression of some but not all pluripotency markers, such as Fbx15 and FGF-4 genes. Because of the intrinsic character of stable and rapid cell proliferation, partial iPSCs occasionally become the most prevalent subpopulation among cells subjected to iPSC induction. Although partial iPSCs efficiently convert to iPSCs by exposure to a ground state or its related condition (2i) using MAPK and GSK3β inhibitors [8], the genes and pathways involved in this transition are largely unknown except for the involvement of endogenous expression of principal core pluripotency genes, such as Oct3/4 and Nanog.
Here, we performed genome-wide screening with a piggyBac vector to identify genes that participate in transition from partial to genuine iPSCs. Our screening identified the gene encoding Cnot2, one of the core components of the Ccr4-Not complex [9,10]. Our knockdown studies suggested that all of the core components of the complex, that is, Cnot1, Cnot2, and Cnot3, as well as Trim28, which shares genomic binding sites with Cnot3 [11], equally contribute to this transition. Analyses of genes with altered expression as a result of the forced expression in partial iPSCs indicated that the major role of these factors is suppression of gene expression associated with developmental processes. Thus, our screening identified core components of the Ccr4-Not complex and Trim28 as new players that contribute to the transition of abnormally reprogrammed partial iPSCs to genuine iPSCs.
Materials and Methods
iPSC induction
Mouse embryonic fibroblasts (MEFs) were prepared from 13.5 dpc embryos carrying a GFP reporter gene whose expression faithfully recapitulated Nanog gene expression [12]. iPSC induction was performed as described by Takahashi and Yamanaka [1]. iPSC induction shown in Supplementary Fig. S6 was conducted with retroviruses carrying Cnot2 and/or Trim28 in addition to those carrying the four reprogramming factors (Oct3/4, Sox2, Klf4, and c-Myc).
Establishment of partial iPSC lines
To isolate partial iPSC lines, MEFs bearing a Nanog-GFP reporter gene were subjected to iPSC induction. Colonies with ESC colony-like morphology at 3 or 4 weeks post-iPSC induction were individually recovered and maintained on feeder cells. Among them, clones exhibiting strong DsRed, but not GFP, fluorescence were chosen as candidates for partial iPSC clones. After elimination of clones that showed spontaneous conversion to genuine iPSCs, two clones designated as 2B1 and 5C5 were selected based on specific criteria. These criteria included high expression of retroviral genes and almost complete lack of expression of principal pluripotency marker genes because of strong DNA methylation. Finally, these clones were subjected to single cell sorting to ensure clonality.
iPSC culture and expression of exogenous genes by retroviral infection
Partial and genuine iPSCs were cultured in the standard mouse ESC medium supplemented with fetal bovine serum and leukemia inhibitory factor (LIF) on feeder cells unless indicated otherwise. To convert the majority of partial iPSCs to genuine iPSCs, partial iPSCs were subjected to the 2i (MAPK and GSK3β inhibitors) + LIF condition with knockout serum replacement. For genome-wide screening, partial iPSCs were cultured in standard mouse ESC medium or under the 2i condition with serum. Cnot1, Cnot2, Cnot3, Trim28, and Nanog were expressed individually or in combination in partial iPSCs by retroviral infection. For individual expression of genes, G418 was used for stable selection. Even for combinatorial expression of multiple genes, only a single gene was expressed for each infection in which appropriate antibiotics, such as hygromycin and zeocin, were used for selection.
Reagents and antibodies
The following reagents were used at the indicated concentrations unless stated otherwise: 1.5 μg/mL puromycin (Sigma); 600 μg/mL hygromycin, 300 μg/mL Zeocin (Invitrogen); 400 μg/mL G418 (Sigma); 1 μM PD0325901 (Axon Medchem); 3 μM CHIR99021 (Axon Medchem); anti-Cnot1 (14276-1-AP), anti-Cnot2 (10313-1-AP) (ProteinTech); anti-Cnot3 (H0004849-MO1) (Abnova); anti-Trim28 (KAP1) (ab10483) (Abcam); anti-β-actin (sc-47778) (Santa Cruz Biotechnology).
Western blotting
For western blotting, proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene difluoride membrane, and then probed with the indicated primary antibodies and appropriate secondary antibodies conjugated to horseradish peroxidase. Specific protein bands were detected by an enhanced chemiluminescence system (GE Healthcare).
Single cell sorting
To ensure clonality, partial iPSCs (2B1 and 5C5) and their derivative iPSCs were subjected to single cell sorting by fluorescence-activated cell sorting (FACS) of GFP−/DsRed+ and GFP+/DsRed− cells. Among the cell clones, cells exhibiting continuous expansion without a change in reporter gene expression were recovered as stable cell lines.
Production of retroviruses for shRNA-mediated knockdown
Retroviral vectors for shRNAs against the expression of Cnot1, Cnot2, Cnot3, Trim28, and luciferase were produced by subcloning the following oligonucleotides together with their complementary sequences into pLKO.1-puro vectors:
Cnot1 KD:
5′-CCGGTGGTTAGGAATGATCACATTACTCGAGTAATGTGATCATTCCTAACCATTTTTG-3′;
Cnot2 KD:
5′-CCGGCTCTTAGCTGCGGTAGAACTTCAAGAGAGTTCTACCGCAGCTAAGAGTTTTTG-3′;
Cnot3 KD:
5′-CCGGGATAAGAAGAGAGGCCGATTTCAAGAGAATCGGCCTCTCTTCTTATCTTTTTG-3′;
Trim28 KD:
5′-CCGGGGACTACAATCTGATTGTTATCTCGAGATAACAATCAGATTGTAGTCCTTTTTG-3′;
Lucifease KD:
5′-CCGGCTTACGCTGAGTACTTCGATTCAAGAGATCGAAGTACTCAGCGTAAGTTTTTG-3′.
U6-shRNA-puro fragments were recovered after NotI and NsiI double digestion and then transferred to pMX-based vectors.
To produce viruses carrying a specific shRNA expression unit, vectors bearing the above sequences were individually transfected into Plat-E cells [13] by lipofection. Partial iPSCs (2B1) were infected with the generated retroviruses in the presence of 8 μg/mL polybrene, and then selected with 1.5 μg/mL puromycin.
PiggyBac-mediated expression of Cnot3 and/or Trim28 in partial iPSCs (2B1)
Cnot3-IRES-hygromycin and Trim28-IRES-neomycin resistance genes were individually subcloned into piggyBac vectors carrying the constitutively active chicken actin gene promoter [14]. These plasmid vectors were introduced into partial iPSCs (2B1) individually or in combination together with the piggyBac transposase expression vector by lipofection. Then, stable cell lines were obtained by selection with G418 and/or hygromycin.
Bisulfite sequencing
Bisulfite sequencing of genomic DNA was performed using the same primers described by Kanatsu-Shinohara et al. [15] for the Oct3/4 gene promoter and those described by Mikkelsen et al. [7] for Nanog, Utf1, Dppa5, and Rex1 gene promoters.
Microarray and bioinformatics analyses
In vitro-transcribed biotin-labeled cRNA was synthesized according to the Affymetrix guidelines. Labeled samples were hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 arrays following the manufacturer's instructions. Microarray expression data were background-subtracted and normalized using the robust multiarray analysis method [16] with R-package 2.11.1 and Bioconductor 2.6 [17]. Spotfire X.X (TIBCO) was used for hierarchical clustering. Gene ontology (GO) analysis was performed using DAVID 6.7 software [18,19].
Accession number
DNA microarray data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE44339.
Results
Isolation of partial iPSC lines
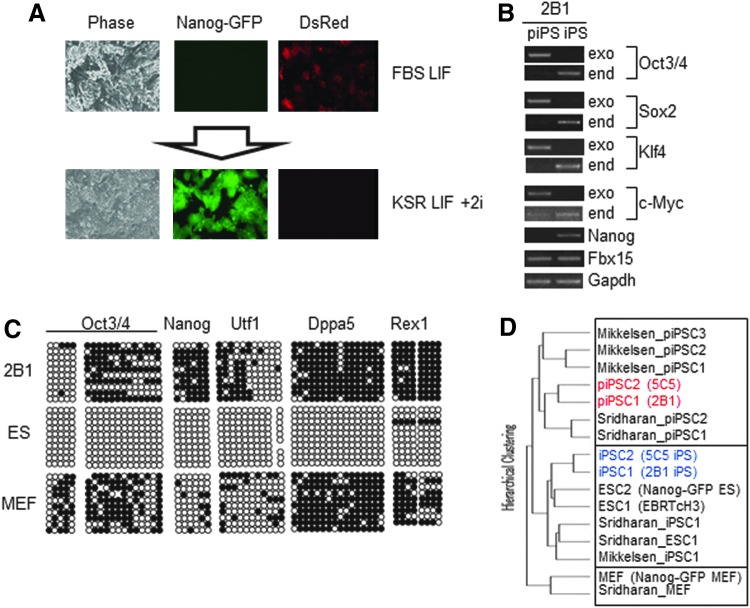
As the first step for identification of genes that participate in the transition from partial to genuine iPSCs, we generated our own partial iPSC lines. MEFs from embryos carrying a Nanog-GFP reporter gene were used for iPSC induction [12]. To induce iPSCs, the MEFs were infected with a retrovirus carrying the DsRed reporter gene together with the four reprogramming factors (Oct3/4, Sox2, Klf4, and c-Myc) to monitor infection efficiency and as a marker to distinguish between partial and genuine iPSCs. At 3 or 4 weeks post-iPSC induction, colonies with ESC-like morphology were randomly picked up and transferred to 24-well plates. During expansion of the picked up colonies, two cell clones termed 2B1 (Fig. 1A) and 5C5 (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd), which exhibited strong DsRed expression but no GFP fluorescence, were selected as candidates for partial iPSCs. Next, the two cell clones were subjected to single cell sorting to ensure the clonality. Similar to previously reported partial iPSCs [8], the 2B1 clone showed efficient conversion to genuine iPSCs by exposure to the 2i condition, which was determined by the loss of retroviral DsRed gene expression and induction of GFP expression (Fig. 1A). We found that the 5C5 clone also showed the same switch in reporter gene expression (Supplementary Fig. S1). Next, we monitored the expression of exogenous and endogenous reprogramming factor genes in the 2B1 clone. When maintained under empirical culture conditions, we found high expression levels of exogenous genes, but very low expression levels of endogenous pluripotency genes (Oct3/4, Sox2, and Klf4). However, upon exposure to the 2i condition, the expression of the exogenous pluripotency markers was replaced with the endogenous gene expression (Fig. 1B). Similar to other partial iPSC clones [7], bisulfite sequencing analyses indicated extensive methylation of pluripotency marker gene promoters in the 2B1 clone (Fig. 1C). To assess the similarities to previously generated partial iPSCs by other groups [6,7], we performed DNA microarray analyses to compare the gene expression profiles of our iPSCs with those of other partial iPSCs. We found that all of the cell lines designated as partial iPSCs, including the 2B1 and 5C5 clones, showed highly similar expression profiles that were clearly different from those of genuine iPSCs and MEFs (Fig 1D). These results confirmed that the two clones (2B1 and 5C5) had characteristics that met all of the criteria needed for designation as partial iPSCs.
FIG. 1.
Generation of partial induced pluripotent stem cell (iPSC) clones. (A) Bright field and fluorescence images of partial iPSC clone 2B1 (upper panels) and those subjected to the 2i condition with MAPK and GSK3β inhibitors (lower panels). (B) Reverse transcription-PCR analyses of the expression of endogenous (end) and exogenous (exo) reprogramming factor genes and other indicated embryonic stem cell (ESC) marker genes in partial iPSCs (2B1) and those exposed to the 2i condition for 8 days. For the latter cells, GFP-positive cells were sorted and then expanded before the analyses. (C) Bisulfite sequencing analyses of pluripotency marker genes in partial iPSC clone 2B1. The analyses were also conducted with mouse embryonic fibroblasts (MEFs) and ESCs as references. (D) Hierarchical clustering of whole-genome transcripts in partial iPSC clones 2B1 and 5C5, and iPSCs generated from them under the 2i condition together with those from different laboratories [6,7]. Partial iPSC clones 2B1 and 5C5 and their derivative iPSCs are indicated with red and blue letters, respectively. Tree branch heights are proportional to the distances between samples.
Identification of the Cnot2 gene as a regulator participating in the transition from partial to genuine iPSCs
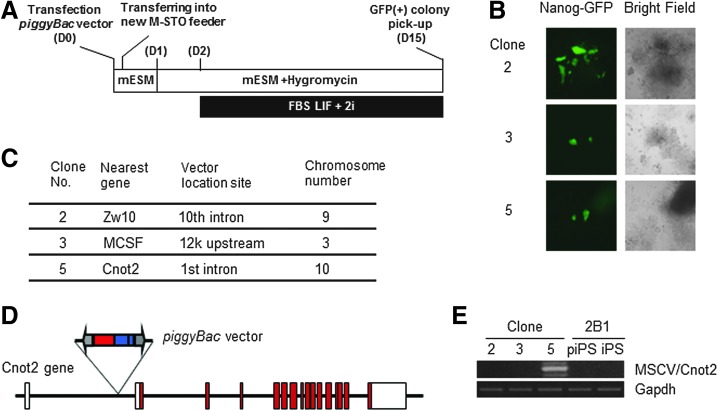
To identify genes involved in the conversion from partial to genuine iPSCs, we introduced a piggyBac gene-trap vector containing the murine stem cell virus enhancer/promoter [20] into the two partial iPSC clones bearing the Nanog-GFP transgene. The cells were cultured under the conventional LIF + serum mouse ESC medium condition, and the vector was allowed to move throughout their genomes by transposase-mediated transposition. However, several trials did not produce any clones expressing the GFP reporter gene that was under the control of the Nanog gene promoter. We assumed that these failures were attributed to the requirement for the activation of more than two critical genes for the transition, which could not be achieved by the employed method. However, we considered that changing the culture condition may overcome this problem. Although kinase inhibitors (2i) are usually used together with LIF under a serum-free condition [8,21,22], an equivalent 2i + LIF condition with serum did not induce conversion of partial iPSCs to GFP-positive iPSCs (Supplementary Fig. S1A). Therefore, we used the 2i + LIF condition with serum as a starting condition for the screen (Fig. 2A). After screening, we obtained three GFP-positive iPSC clones (Fig. 2B). Next, genomic DNAs from these clones were recovered to determine the genomic integration sites of the vector. Figure 2C shows the integration site of the vector in each clone. Among the three clones, we chose clone 5 for the following reasons. First, piggyBac vector insertion in clone 5 did not disrupt the coding region of the Cnot2 gene because of the presence of the initiating methionine codon in the second exon (Fig. 2D). More importantly, reverse transcription-PCR analyses revealed that exogenous reprogramming factor genes were almost completely silenced in clone 5, but not in clones 2 or 3 (Supplementary Fig. S2B). Figure 2E shows the presence of Cnot2 mRNA fused with the sequence from the piggyBac vector in clone 5.
FIG. 2.
Identification of the Cnot2 gene by screening with the piggyBac vector as a positive regulator of conversion from partial to genuine iPSCs. (A) Protocol for the piggyBac-mediated activation screen. A piggyBac vector bearing the murine stem cell virus (MSCV) enhancer/promoter [20] and a transposase expression vector were introduced into partial iPSC clone 2B1 by lipofection. Changes of the culture conditions including hygromycin selection (600 μg/mL) were performed as indicated. (B) Bright field and fluorescence images of three GFP-positive clones yielded by the piggyBac-mediated activation screen. (C) Integration sites of the vector in the three GFP-positive clones. Splinkerette PCR to determine the integration sites of the vector was performed as described by Guo and Smith [20]. (D) Diagram of insertion of the piggyBac vector in the first intron of the Cnot2 gene. Open and solid red boxes represent noncoding and coding exons, respectively. Blue and red boxes in the piggyBac vector indicate the hygromycin-resistance gene and MSCV enhancer/promoter, respectively, while gray arrows represent the 5′ and 3′ long terminal repeats of the vector. (E) Identification of the Cnot2 transcript carrying a part of the 3′ long terminal repeat sequence of the piggyBac vector.
Evidence of the involvement of Ccr4-Not complex components in the transition from partial to genuine iPSCs
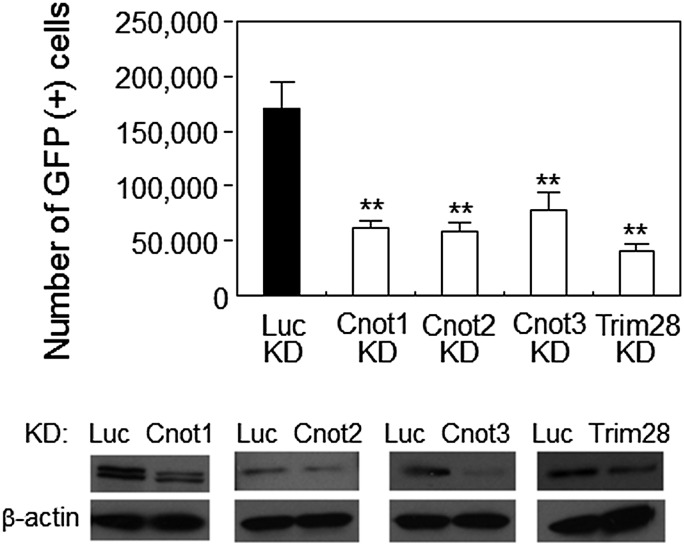
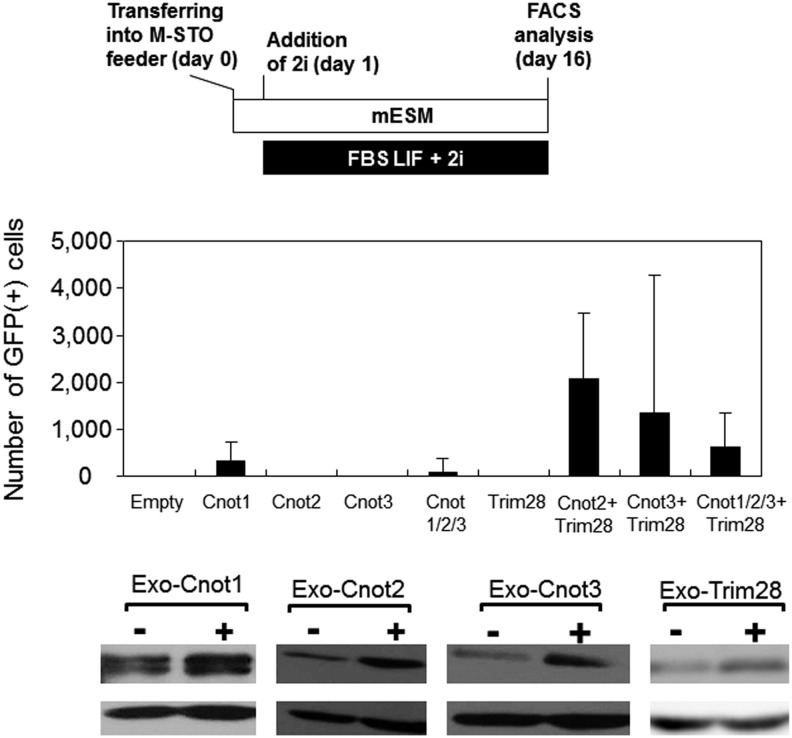
To confirm the participation of Cnot2 in the transition from partial to genuine iPSCs, we first performed knockdown experiments of the Cnot2 gene. Consequently, the reduction of Cnot2 expression significantly lowered the conversion efficiency to genuine iPSCs under the 2i + LIF condition (Fig. 3A). Cnot2 is one of the subunits of the Ccr4-Not complex in which Cnot1, 2, and 3 comprise the core of the complex. It has been demonstrated by chromatin immunoprecipitation analyses that Cnot3 shares genomic binding sites with Trim28 [11,23]. Therefore, we examined whether these factors also contributed to the transition from partial to genuine iPSCs. As a result, the knockdown of Cnot1, Cnot3, or Trim28 expression equally lowered the conversion efficiency to that of Cnot2 knockdown (Fig. 3). Next, we examined the effect of forced expression of these genes on the conversion of partial iPSCs to genuine iPSCs. None of these factors, even in combination, were able to exert a significant effect on the conversion (Fig. 4). However, we observed some positive effects using the combination of Cnot2 or Cnot3 with Trim28, although these effects were extremely subtle compared with those obtained under the regular 2i condition (comparison of GFP-positive cell numbers in Figs. 3 and 4). Although we obtained unexpected results from the above overexpression experiments, we examined whether GFP-positive cells generated by forced expression of Cnot2 and Trim28 could expand stably as an iPSC clone. To this end, we cultured partial iPSCs overexpressing Cnot2 and Trim28 for 2 weeks at a subclonal density and identified a GFP-positive/DsRed-negative clone (Supplementary Fig. S3A). After picking up the clone and transferring to a new dish, GFP-positive cells were expanded and then sorted by FACS. These sorted cells propagated robustly, while maintaining a high level of GFP expression that faithfully represented endogenous Nanog gene expression (Supplementary Fig. S3B). Quantitative PCR (Supplementary Fig. S3C) and western blot (Supplementary Fig. S3D) analyses showed that these cells were equivalent to iPSCs obtained by exposure of partial iPSCs (2B1) to the KSR-LIF + 2i condition in terms of their expression levels of endogenous pluripotency marker genes. Even with successful establishment of the iPSC clone, the extremely low efficiency of transition from partial to genuine iPSCs by forced expression of Cnots and/or Trim28 (Fig. 4) was not compatible with the fact that piggyBac gene trap vector-mediated activation of the Cnot2 gene sufficiently supported conversion from partial to genuine iPSCs (Fig. 2). A possible cause of this discrepancy may lie in the different methods used for the expression of exogenous genes between the two systems in which retroviral-mediated expression would be subjected to severe silencing, whereas piggyBac-mediated expression would persist even after conversion to genuine iPSCs. Therefore, we chose the piggyBac system to express Cnot3 and/or Trim28. However, the vast majority of partial iPSCs remained positive and negative for DsRed and GFP, respectively, even after expression of the two factors individually or in combination (Supplementary Fig. S4). Therefore, we assumed that partial iPSCs converted to genuine iPSCs by piggyBac-mediated activation of the Cnot2 gene were rather special cells that were much more competent for conversion to genuine iPSCs compared with that of the vast majority of other partial iPSCs (see Discussion for details). Another unexpected result shown in Fig. 4 was that either Cnot2 or Cnot3 together with Trim28 had a stronger effect on the conversion from partial to genuine iPSCs compared with that obtained by simultaneous expression of Cnot1, Cnot2, Cnot3, and Trim28. To gain an insight into the underlying molecular basis of this finding, we compared the expression levels of Cnot2 and Cnot3 under various conditions. As a result, we detected large amounts of these proteins under forced expression of Cnot1, Cnot2, and Cnot3 (Supplementary Fig. S5). This result implies that Cnot1, Cnot2, and Cnot3 proteins are stabilized in partial iPSCs by constituting the core of the Ccr4-Not complex. However, we do not know at present why or how construction of the core of the Ccr4-Not complex led to the decline in efficiency of conversion from partial to genuine iPSCs (see Discussion for more details). Next, we examined whether Cnot2 and/or Trim28 expression elevated the efficiency of reprogramming MEFs. To this end, we added retroviruses carrying Cnot2 or Trim28 individually or in combination together with the retrovirus derived-reprogramming factors (Oct3/4, Sox2, klf4, and c-Myc) for iPSC induction of MEFs. However, the additional factors did not elevate, but rather lowered the reprogramming efficiency as assessed by counting alkaline phosphatase-positive colonies (Supplementary Fig. S6). This observation indicated that forced expression of Cnot2 and/or Trim28 during the early stage of iPSC induction affects the reprogramming process negatively, and the promoting effect of these factors becomes evident only at the later stage of induction. An alternative possibility is that these factors promote the reprogramming process rather restrictively in the transition from partial to genuine iPSCs. These possibilities remain to be addressed at present.
FIG. 3.
Effect of knockdown of Cnot1, Cnot2, Cnot3, or Trim28 in partial iPSCs on the production of Nanog-GFP-positive iPSCs. Partial iPSCs (2B1) were infected with a retrovirus expressing a puromycin-resistance gene and shRNA targeting the indicated genes. Transfectants were enriched by puromycin selection (1.5 μg/mL) and then exposed to the 2i condition for iPSC conversion. GFP-positive cells were quantified by flow cytometry. Data are the mean±standard deviation (n=3). **P<0.01. Lower panels show western blots examining the efficiency of knockdown.
FIG. 4.
Effect of overexpression of Cnot1, Cnot2, Cnot3, and Trim28 on the production of Nanog-GFP-positive cells from partial iPSCs. Partial iPSCs (2B1) cultured in the empirical mouse ESC medium were subjected to the overexpression of Cnot1, Cnot2, Cnot3, and Trim28 individually or in the combinations as indicated. After stable integration, MAPK and GSK3β inhibitors (2i) were added to the empirical mouse ESC medium containing leukemia inhibitory factor and serum. Data are the mean±standard deviation (n=6). GFP-positive cells were quantified as described in Fig. 3. Lower panels show western blots confirming forced expression of Cnot1, Cnot2, Cnot3, and Trim28 that were overexpressed individually.
Effect of expression of the core components of the Ccr4-Not complex on the global gene expression profile of partial iPSCs
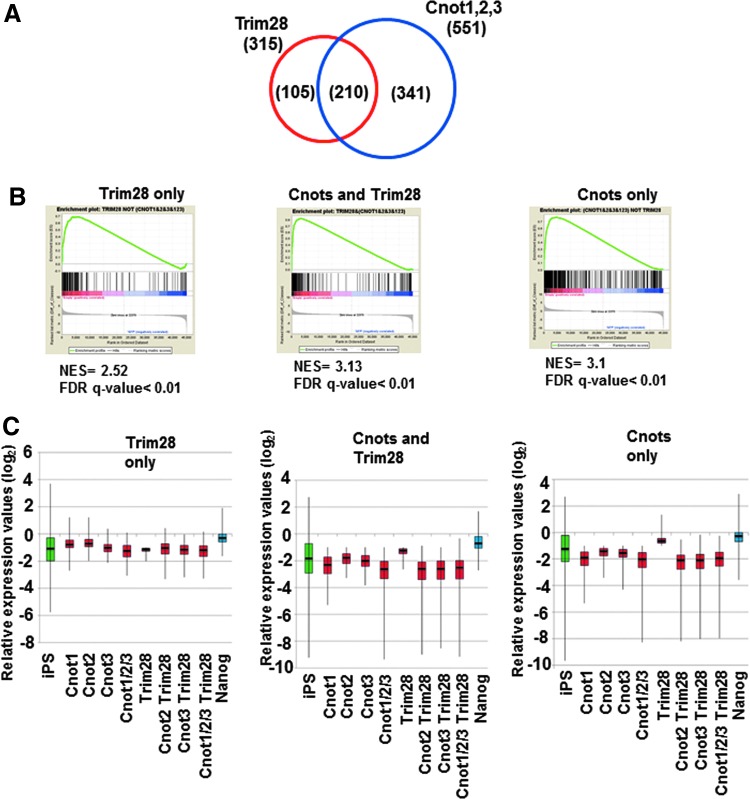
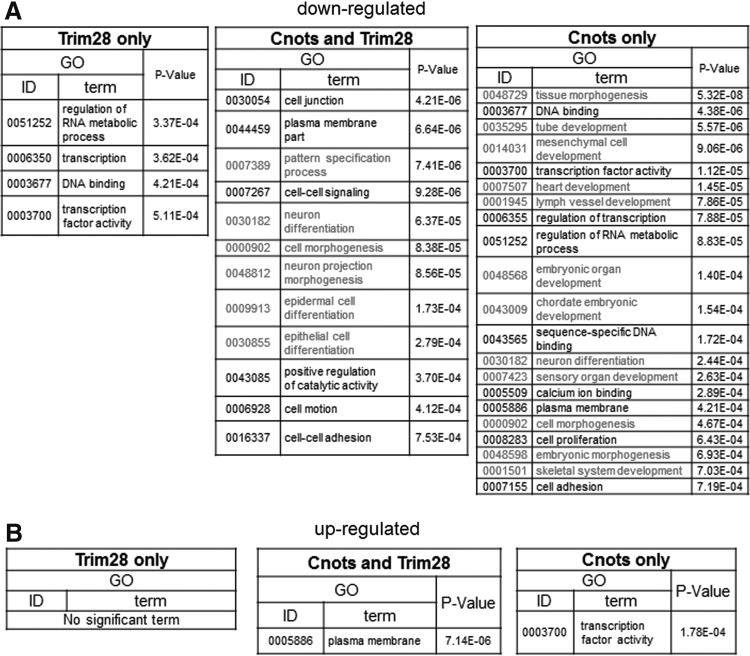
Although there was only a marginal effect of the combinatorial expression of Cnot2 and other factors on the transition from partial to genuine iPSCs, we examined global changes of gene expression caused by forced expression of these factors in partial iPSCs by microarray analyses. We performed these analyses because, although these factors were apparently insufficient to convert the majority of partial iPSCs to genuine iPSCs, they may be sufficient for the modulation of at least some aspects of partial iPSCs to make them somewhat closer to genuine iPSCs. RNA was recovered from the stable cell lines in which Cnot1, Cnot2, Cnot3, and Trim28 were overexpressed individually or in combinations. Because these cells were cultured without any kinase inhibitors (empirical mouse ESC culture condition) during antibiotic selection and expansion, no cells were positive for Nanog-GFP. First, we characterized downregulated genes. We found that genes downregulated by expression of Cnot1, Cnot2, or Cnot3, or simultaneous expression of all three in partial iPSCs were significantly overlapped (Supplementary Fig. S3A). Indeed, 551 genes were commonly downregulated by more than twofold by these factors. The 551 genes were then divided into three groups consisting of genes commonly regulated by Trim28 and core components of the Ccr4-Not complex, and those regulated specifically by either one (Fig. 5A and Supplementary Table S1). We first examined whether these genes showed relatively lower levels of expression in iPSCs compared with those in partial iPSCs by gene set enrichment analyses (GSEA). These analyses revealed that, with all three sets, most, if not all, genes showed a strong tendency of lower expression in genuine iPSCs compared with that in partial iPSCs (Fig. 5B). These results supported the idea that downregulation of these genes by forced expression of the core components of the Ccr4-Not complex and/or Trim28 represents a step in the process required for transition from partial to genuine iPSCs. Analyses of the expression of these genes with box plots also provided results consistent with this notion (Fig. 5C). Notably, the three gene sets did not alter their average expression levels significantly by forced expression of Nanog that is essential for conversion from partial to genuine iPSCs [24]. These results imply that the roles of Ccr4-Not/Trim28 and Nanog are rather distinct in the conversion. To correlate the downregulation of these genes with overall molecular function, we next conducted GO classification analyses. GO terms related to development and morphogenesis were significantly enriched in genes downregulated specifically by core components of the Ccr4-Not complex and those downregulated commonly with Trim28 (Fig. 6A). This finding indicates that the major role of the core components of the Ccr4-Not complex with or without Trim28 is suppression of genes related to developmental processes. We also found that GO terms related to transcription were closed up among genes downregulated specifically by Trim 28. Next, the upregulated genes (Supplementary Figs. S7B and S8, and Supplementary Table S1) were subjected to the same analyses. However, unlike the downregulated genes, GSEA and comparison of average gene expression levels revealed that genes upregulated by core components of the Ccr4-Not complex and/or Trim28 in partial iPSCs did not show any prominent expression in genuine iPSCs compared with that in partial iPSCs (Supplementary Fig. S8B, C). This result strongly opposed the idea that upregulation of these genes affected conversion of partial iPSCs to genuine iPSCs. Accordingly, GO classification analyses yielded only a few significant terms (37 terms among downregulated genes vs. two terms among upregulated genes) (Fig. 6B). This observation suggests that the genes upregulated by Cnots and/or Trim28 in partial iPSCs are not well-ordered gene sets for specific biological outcomes, but rather disordered gatherings. Therefore, the alteration of expression levels of these genes may represent indirect regulation by Cnots and/or Trim28. Taken together, our data suggest that core components of the Ccr4-Not complex may participate in the transition from partial to genuine iPSCs by repressing rather than activating gene expression and that the major targets of regulation are genes related to developmental and morphogenetic processes.
FIG. 5.
Characterization of genes downregulated by Ccr4-Not complex components and/or Trim28. (A) Venn diagram illustrating three groups of genes downregulated by Ccr4-Not complex components and Trim28 or specifically by either one in partial iPSCs (2B1). (B) The three groups of genes in A were individually subjected to gene set enrichment analyses to compare the expression profiles between partial and genuine iPSCs. (C) Box plots showing the expression levels of the three groups of genes in A using data from partial iPSCs (2B1) in which an empty vector had been introduced as a reference. Whiskers represent the upper and lower limits of the range. Boxes represent the first and third quartiles, and the bold line represents the median.
FIG. 6.
Gene ontology (GO) terms associated with gene sets showing differential expression caused by forced expression of Ccr4-Not complex components and/or Trim28. (A) Prominent GO terms (P<10−3) among genes downregulated by Ccr4-Not complex components and/or Trim28. Only one GO term was selected in cases where more than two GO terms clustering in the same tree were denoted by the analyses. (B) GO terms (P<10−3) among genes upregulated by Ccr4-Not complex components and/or Trim28. Only one GO term was selected from terms clustering in the same tree as described in A.
Discussion
It was originally thought that the partial iPSC state is an absolute barrier that somatic cells must overcome for conversion to genuine iPSCs [6,7]. However, recent investigation by Polo et al. [5] revealed that partial iPSCs do not resemble any of the intermediate states of cells en route to becoming genuine iPSCs, suggesting that these cells have strayed from the normal reprogramming route at an early point and become immortalized. Therefore, to raise the efficiency of iPSC induction by avoidance of partial iPSC generation, two methods can be considered to prevent straying from the normal reprogramming route to partial iPSCs or correct partial iPSCs back to the normal reprogramming route. The use of L-Myc instead of c-Myc as one of the reprogramming factors appears to correspond to the former type of invention [25]. Glis1 has been also shown to effectively lower the probability of partial iPSC generation [26]. Despite these rather effective former-type inventions, no efficient method for conversion of partial to genuine iPSCs has been devised, except for exposure to the 2i condition. However, our genome-wide screening with the piggyBac vector identified the Cnot2 gene encoding one of the core components of the Ccr4-Not complex as a candidate for supporting such conversion. Subsequent knockdown studies revealed that not only Cnot2 but also other core components of the Ccr4-Not complex, that is, Cnot1 and Cnot3, were also crucially involved in the transition. However, forced expression of either one of these genes or their combinatorial expression including that of Trim28 showed no or only a marginal effect corresponding to about 1% of the effect obtained under the 2i condition. These results were not compatible with the fact that piggyBac vector-mediated activation of the Cnot2 gene was sufficient for the production of genuine iPSCs from partial iPSCs. One possible explanation for this discrepancy is that partial iPSCs converted to genuine iPSCs during the functional screening were rare among the partial iPSC population, and factors other than Cnot2 required for the transition were almost fully expressed, making the cells highly competent for conversion to genuine iPSCs. Another unexpected finding was that expression of either Cnot2 or Cnot3 alone supported more efficient transition from partial to genuine iPSCs in collaboration with Trim28 compared with that of simultaneous expression of Cnot1, Cnot2, Cnot3, and Trim28. Compared with individual expression of either Cnot2 or Cnot3, our western blot data revealed that larger amounts of Cnot2 and Cnot3 when all three core components of the Ccr4-Not complex, that is, Cnot1, Cnot2, and Cnot3, were expressed together. The most likely explanation for this finding is that Cnot2 and Cnot3 are more stable in the Ccr4-Not core complex compared with that of their free forms. If this is indeed the case, our results indicate that there is an inverse relationship between the formation of the Ccr4-Not core complex and the promotion of the transition from partial to genuine iPSCs. More specifically, our data suggest that Cnot2 and Cnot3 may be able to support the transition only in their free forms. Although we do not have any evidence to support this hypothesis at present, we assume that generation of Cnot2 or Cnot3 mutants that are defective for complex formation may resolve this issue. Our microarray data and subsequent GO classification analyses suggested that the major role of the core components of the Ccr4-Not complex with or without Trim28 in the transition of partial to genuine iPSCs is to suppress the expression of genes related to developmental processes. Currently, we do not know how these core components perform this function. It is known that the expression of developmental genes is largely repressed in ESCs by a specialized chromosomal architecture termed as the poised bivalent chromatin structure that harbors both transcriptionally active (histone 3-lysine 4 trimethylation) and repressive (histone 3-lysine 27 trimethylation) epigenetic marks [27–30]. Interestingly, a substantial number of genes that should be regulated by the bivalent chromatin structure in ESCs do not bear a complete bivalent architecture in partial iPSCs [7]. Moreover, Cnot3 and Trim28 are substantially colocalized with bivalent genes in ESCs [11,23]. Therefore, it is tempting to speculate that expression of the core components of the Ccr4-Not complex and/or Trim28 participates in construction of the bivalent chromatin structure in partial iPSCs, leading to the downregulation of developmental genes. The roles of the core components of the Ccr4-Not complex in suppressing the expression of developmental genes have been demonstrated previously with ESCs [31] in which impairment of the function of the complex leads to differentiation of ESCs toward trophectodermal cell lineages. Because our data did not indicate any specificity of the genes involved in differentiation toward trophectodermal cells, it is assumed that the common role of the complex is to suppress development/differentiation-associated genes, but sets of genes subjected to the repression are cell type specific.
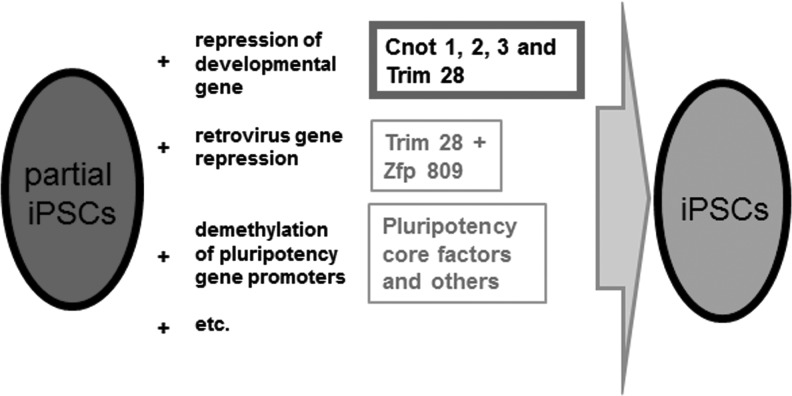
In summary, as depicted in Fig. 7, we demonstrate here that core components of the Ccr4-Not complex with or without Trim28 are strongly involved in the transition of partial to genuine iPSCs mainly through the suppression of genes related to development. We hope that these findings will provide a clue to fully understanding the entire process of this transition at the molecular level and eventually lead to increasing the induction efficiency and quality of iPSCs.
FIG. 7.
Summary of requirements for transition from partial to genuine iPSCs. To convert partial iPSCs to genuine iPSCs, at least three major events must occur, that is, repression of genes related to development/differentiation, silencing of retroviral DNAs [32–34], and acquisition of endogenous pluripotency gene expression [6–8]. Our present study indicates that Ccr4-Not complex components and Trim28 at least in part contribute to one of these events, namely repression of developmental genes.
Supplementary Material
Acknowledgments
We thank Dr. Austin Smith for the piggyBac vector and Tomoko Okuda for technical assistance. This study was performed as a part of the Core Research for Evolutional Science and Technology (CREST) Agency. This study was supported in part by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, and primarily by a Grant-in-Aid for the Support Project of Strategic Research Center in Private Universities to the Saitama Medical University Research Center for Genomic Medicine. A.O. is a recipient of grants from the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant numbers 25293082 and 25670147).
Author Disclosure Statement
The authors declare that they have no conflict of interest.
References
- 1.Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- 2.Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H. and Jaenisch R. (2008). Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2:151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A. and Jaenisch R. (2012). Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150:1209–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D. and Wrana JL. (2012). A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell 11:769–782 [DOI] [PubMed] [Google Scholar]
- 5.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, et al. (2012). A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151:1617–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q. and Plath K. (2009). Role of the murine reprogramming factors in the induction of pluripotency. Cell 136:364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES. and Meissner A. (2008). Dissecting direct reprogramming through integrative genomic analysis. Nature 454:49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW. and Smith A. (2008). Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol 6:e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartlam M. and Yamamoto T. (2010). The structural basis for deadenylation by the CCR4-NOT complex. Protein Cell 1:443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collart MA. and Panasenko OO. (2012). The Ccr4—not complex. Gene 492:42–53 [DOI] [PubMed] [Google Scholar]
- 11.Hu G, Kim J, Xu Q, Leng Y, Orkin SH. and Elledge SJ. (2009). A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 23:837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okita K, Ichisaka T. and Yamanaka S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448:313–317 [DOI] [PubMed] [Google Scholar]
- 13.Morita S, Kojima T. and Kitamura T. (2000). Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 7:1063–1066 [DOI] [PubMed] [Google Scholar]
- 14.Yusa K, Rad R, Takeda J. and Bradley A. (2009). Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods 6:363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanatsu-Shinohara M, Lee J, Inoue K, Ogonuki N, Miki H, Toyokuni S, Ikawa M, Nakamura T, Ogura A. and Shinohara T. (2008). Pluripotency of a single spermatogonial stem cell in mice. Biol Reprod 78:681–687 [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B. and Speed TP. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT. and Lempicki RA. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57 [DOI] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT. and Lempicki RA. (2009). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo G. and Smith A. (2010). A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development 137:3185–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P. and Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453:519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A. and Stunnenberg HG. (2012). The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149:590–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB. and Orkin SH. (2010). A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143:313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I. and Smith A. (2009). Nanog is the gateway to the pluripotent ground state. Cell 138:722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagawa M, Takizawa N, Narita M, Ichisaka T. and Yamanaka S. (2010). Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci U S A 107:14152–14157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maekawa M, Yamaguchi K, Nakamura T, Shibukawa R, Kodanaka I, Ichisaka T, Kawamura Y, Mochizuki H, Goshima N. and Yamanaka S. (2011). Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature 474:225–229 [DOI] [PubMed] [Google Scholar]
- 27.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326 [DOI] [PubMed] [Google Scholar]
- 28.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–353 [DOI] [PubMed] [Google Scholar]
- 29.Jia J, Zheng X, Hu G, Cui K, Zhang J, Zhang A, Jiang H, Lu B, Yates J, 3rd., et al. (2012). Regulation of pluripotency and self-renewal of ESCs through epigenetic-threshold modulation and mRNA pruning. Cell 151:576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, et al. (2008). Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4:e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng X, Dumitru R, Lackford BL, Freudenberg JM, Singh AP, Archer TK, Jothi R. and Hu G. (2012). Cnot1, Cnot2, and Cnot3 maintain mouse and human ESC identity and inhibit extraembryonic differentiation. Stem Cells 30:910–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolf D. and Goff SP. (2007). TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131:46–57 [DOI] [PubMed] [Google Scholar]
- 33.Wolf D, Cammas F, Losson R. and Goff SP. (2008). Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J Virol 82:4675–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolf D. and Goff SP. (2009). Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458:1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.