Abstract
Hirschmanniella oryzae is the most common plant-parasitic nematode in flooded rice cultivation systems. These migratory animals penetrate the plant roots and feed on the root cells, creating large cavities, extensive root necrosis and rotting. The objective of this study was to investigate the systemic response of the rice plant upon root infection by this nematode. RNA sequencing was applied on the above-ground parts of the rice plants at 3 and 7 days post inoculation. The data revealed significant modifications in the primary metabolism of the plant shoot, with a general suppression of for instance chlorophyll biosynthesis, the brassinosteroid pathway, and amino acid production. In the secondary metabolism, we detected a repression of the isoprenoid and shikimate pathways. These molecular changes can have dramatic consequences for the growth and yield of the rice plants, and could potentially change their susceptibility to above-ground pathogens and pests.
Introduction
Rice (Oryza sativa L.) is one of the most important staple foods in the world, feeding more than 50% of the human population. One of the most damaging pathogens, with major impact on rice yield, is the migratory endoparasitic nematode Hirschmanniella oryzae. Hirschmanniella oryzae is present in the soil and irrigation water of the majority of rice-growing areas, mainly where rice is grown under submerged conditions [1], [2]. These nematodes penetrate plant roots and move through the cortex of the root, producing cavities and channels and eventually necrosis. Roots invaded by H. oryzae exhibit discoloration, deterioration and rotting [1], while heavily infested plants also show above-ground symptoms like stunting and up to 60% reduced tillering [1], [2].
In comparison with the existing knowledge on the infection process of dicots by sedentary endoparasitic nematodes [3], far less is known about the interaction between monocot plants and nematodes or plant interactions with migratory endoparasitic nematode species. Plants generally respond to nematode infection by differential expression of genes involved in stress and defense responses, cell wall alteration, metabolism and nutrient allocation, and signal transduction [3]. Our previous transcriptome analysis on the local response of rice roots upon infection with the H. oryzae revealed that at 3 days after inoculation the infected roots accumulated mRNA of many biotic stress-related genes, oxidative stress and cell death pathways [4]. Concurrently the normal primary metabolism of the rice roots appeared impaired. Particularly the jasmonate and ethylene pathways, which are known to be involved in wound responses [5], were strongly induced upon migratory endoparasitic nematode infection in rice roots, although some suppression of specific defense responses was observed at later time points [4]. Next to the accumulating evidence on gene expression and metabolite changes in locally infected tissues, research has also provided evidence on the nematode-induced systemic changes in pathogenesis-related protein levels [6], [7], [8], primary metabolites [9], and hormone-related defense pathways [10]. Below-ground feeding organisms such as insects, nematodes, root pathogens, and ectomycorrhizal fungi are known to influence the concentration of above-ground plant defense compounds including terpenoids, glucosinolates, and phenolics [11], [12], [13], [14], leading to an impact on the susceptibility of the above-ground tissues to subsequent pathogen attack (reviewed in [15]). However, the molecular mechanisms and associated genes driving such a systemic response remain barely investigated.
Therefore, to gain deeper insight into the systemic transcriptional changes in rice after H. oryzae infection we have performed mRNA-Seq on the shoots of rice root nematode infected plants. The observations were independently validated using qRT-PCR and biochemical analyses. This research reveals significant modifications in the metabolism of the plant, with a general suppression of chlorophyll biosynthesis and primary metabolic processes involved in plant growth.
Materials and Methods
Infection
Oryza sativa cv. ‘Nipponbare’ (GSOR-100, USDA) was germinated on wet filter paper for 6 days at 30°C and then transferred to SAP-substrate (Sand and Absorbent Polymer; [16]) and further grown at 26°C under a 16 h/8 h light-regime. Hirschmanniella oryzae infected rice roots were collected in rice fields in Myanmar and sent to Ghent University by Zin Thu Zar Maung (Plant Protection Division, Yangon, Myanmar). This sampling was done at Padan Village, Hlaingtharyar Region in Yangon Division, Myanmar. As this did not involve endangered or protected species, no specific permissions were required. The nematode-infected rice roots were imported in Belgium, issued by permit BE/LOA/2012/033. All further experiments were done at Ghent University, authorized by biosafety permit T38-0428, activity number 13.
The nematodes were extracted using the Baermann funnel method [17]. Eighteen-day-old rice plants were inoculated with a mixed population of H. oryzae at the rate of 400 per plant, or mock-inoculated with water. This was done by inserting a 1 ml pipette tip just adjacent to the plant root system and releasing the nematode suspension (dissolved in water). One day after inoculation the plants were transferred to a hydroponic culturing system with Hoagland solution [16] to synchronize the infection process. Shoots of inoculated and non-inoculated plants were collected 3 and 7 days post inoculation (dpi). At each time point two independent biological replicates, each containing a pool of 6 plants, were collected for RNA-Sequencing. A third independent biological replicate of both inoculated and non-inoculated plants, again each containing a pool of 6 different plants, was used for qRT-PCR validation.
RNA extraction, library preparation and sequencing
RNA was extracted using the Qiagen RNeasy Plant Mini Kit (Qiagen), with an additional sonication step after addition of buffer RLT. RNA integrity was checked using the Agilent BioAnalyzer 2100 (Agilent). Approximately 2 µg of total RNA was used for mRNA-Seq library construction according to the manufacturer’s recommendations (Illumina).
We used the multiplexing sequencing adapters provided in the Multiplexing Sample Preparation Oligo Kit (Illumina). Size selection of the library was performed on a 2% agarose gel (Low Range Ultra Agarose, Biorad 161-3107). The denatured library was diluted to a final concentration of 6 pM and loaded on a paired-end read flow cell (TruSeq v5 kit, Illumina). To minimize lane effects the samples were multiplexed. Each sample was sequenced in duplicate in 2 different lanes (4 lanes total with 8 MID tags per lane). After cluster generation, the multiplexed library was sequenced on an Illumina Genome Analyzer IIx (36 cycles, paired end).
Mapping reads to genome data and annotated transcripts
Reads were mapped to the Oryza sativa subsp. japonica reference genome (build MSU7.0) in two phases using TopHat version 1.3.1 [18] and Cufflinks, version 1.0.3 [19]. The workflow and settings used in the data analysis have previously been described [4].
Calculation, normalization and profiling of gene expression
Expression was quantified per sample and per annotated or unannotated transcript as the sum of all reads mapped to the respective gene exons with a 16 base pair tolerance on either side to compensate for potential errors in the gene annotation. Expression profiles were assessed using the R-package “baySeq”, version 1.5.1. [20]. To compensate for artificial differences in read distributions, the original library sizes were multiplied by additional normalisation factors calculated using the Trimmed Median of M-values method described in [21] with standard settings as implemented in the edgeR package (version 2.0.3). For statistical significance an FDR cut-off of 0.05 was considered for the ‘general response’ analysis, where the time point after inoculation was not taken into account. For the analysis per time point (3 and 7 dpi), considering that only 2 biological replicates (although pools) per treatment were analyzed for these tissues, the statistical analysis was performed with less stringent conditions (FDR<0.1).
For all further analyses the expression level of each transcript for each condition was estimated as the fold change (FC) of mapped reads relative to the controls. The FC was calculated as follows: reads were normalized as described earlier and averaged over the biological replicates. Before calculating the base 2 log of the ratio of these averages, the number of reads was increased by 1 in each group (to avoid 0-values).
Gene ontology and enrichment analyses
Gene ontology (GO) analysis and GO enrichment were performed using agriGO [22]. Parametric Analysis of Gene Set Enrichment (PAGE) [23], based on differential gene expression levels (log2FC), was executed. Benjamin and Hochberg false discovery rate analysis (FDR) was performed to adjust the PAGE P-values.
In addition, MapMan [24] was used to visualize the expression of genes onto metabolic pathways and the WSR-test (with Benjamin and Hochberg correction) was used to test the statistical significance of differential expression of these pathways.
Analysis of Novel Transcriptionally Active Regions (nTARs)
The Cufflinks program generates a GTF file including all transcripts annotated in MSU7.0 and putative novel transcripts derived from the data. BLASTx searches were performed against Swiss-Prot and trEMBL and all predicted rice proteins (http://rice.plantbiology.msu.edu/). Homologues of the nTARs in rice ESTs (downloaded from NCBI on February 1, 2014) were searched by BLASTn. For all analyses a bitscore cut-off of 50 was used.
Validation of mRNA-Seq by qRT-PCR
Based on potential functional importance, a subset of genes was selected for validation in an independent biological sample (pool of 6 plants) by qRT-PCR, at 3 dpi. Locus numbers of these transcripts and primer sequences are presented in Table 1. PCRs were performed under the following conditions: 10 min at 95°C and 45 cycles of [25 s at 95°C, 60 s at 58°C and 20 s at 72°C]. After the PCR reaction, a melting curve was generated by gradually increasing the temperature to 95°C to test the length as an indication of PCR specificity. Normalization of the expression level of the target genes was done using two reference genes (LOC_Os03g27010, [25]; LOC_Os07g02340.1, [26]). Data was analysed as previously described [4].
Table 1. qRT-PCR validation of a selection of genes.
| Relative geneexpression level(infected versuscontrol tissue) | ||||||
| Locus numer | Annotation | primerF | primerR | RNA-Seq | qRT-PCR | Primer efficiency (%) |
| LOC_Os03g16010 | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor | TTCAGGGTGGAGCTATGTGG | TTTATAAACCGTCCCGAAGC | 0.27 | 0.85 | 74 |
| LOC_Os02g17640 | isochorismatase | TCTTTGATGCTGTAGCACTGG | AAGGTTGGTGTTTCCACTCC | 0.29 | 0.36 | 74 |
| LOC_Os01g40094 | protein phosphatase 2C, putative,expressed | GGGAGGATGAATATGCAAGG | CTCGGGGACTGGAATTATCC | 0.34 | 0.79 | 76 |
| Loc_Os01g55870 | chorismaat mutase | ATGGTAAGTTTGTGGCAGAGG | CCACCGTTTCATAGGTGAGG | 0.35 | 0.53 | 73 |
| LOC_Os03g14730 | gibberellin receptor GID1L2, putative, expressed | GTGGAGGCACTCCAGAAAGC | CTCCTTGAACAGCTCCATCG | 0.38 | 0.55 | 69 |
| LOC_Os01g08450 | ras-related protein,putative, expressed | GGAGACAAGTGCCAAGAACG | TCTTCTGGTTGACAGGTTGC | 0.45 | 0.48 | 75 |
| LOC_Os10g40570 | flavin-containing monooxygenasefamily protein, putative, expressed | GGATGGCCTAAGAGATACGC | ATTGTTCAATTGGTGGATGG | 0.49 | 0.37 | 65 |
| LOC_Os03g04920 | multidrug resistance- associated protein,putative, expressed | ACCATTGCACACCGTATCC | AGGACTTGTCCTCCAAGAGC | 0.51 | 0.61 | 73 |
| LOC_Os05g35290 | Phenylalanine ammonia-lyase,ZB8, putative, expressed | CTCGTACCCGCTCTACCG | GACGAGCACCTTGTTGAGC | 0.88 | 0.48 | 70 |
| LOC_Os01g67030 | auxin-responsive protein, putative,expressed | CCAACCTCACCTGCTTCG | CACGTACCCTCCCTTGTCC | 1.84 | 0.51 | 70 |
| LOC_Os04g41070 | zinc finger, C3HC4 type domaincontaining protein, expressed | CTGGAGGCTCAGGACCAG | CAGAAGTGGCTCCTCCAAAC | 1.91 | 1.06 | 71 |
| LOC_Os09g31450 | Polygalacturonase inhibitor 1precursor, putative, expressed | TCAACGTCAGCGACAACG | ACCTGTTGCGAGCGTAGC | 2.00 | 1.23 | 69 |
| LOC_Os04g18650 | AP2 domain containingprotein, expressed | GAGAAGGTGGAGCTTGTGTACC | GTTGTTGTTGTTGCGGTAGC | 2.38 | 1.36 | 75 |
| LOC_Os02g33840 | OsFBX52 - F-box domaincontaining protein,expressed | GAGTCCTTCGGGTGGATACC | CCTGAAGAACCCAGACATCC | 2.56 | 0.71 | 65 |
| LOC_Os03g58990 | cupin domain containingprotein, expressed | ACGTATCGTCGCCGTACAAC | GACGGCGTCGGGTATCTC | 3.29 | 1.29 | 72 |
| LOC_Os11g42290 | transferase family protein,putative, expressed | CGAGTACATCCAGTCGTTCG | CATCTCGTGGAACCTGAACC | 3.95 | 1.35 | 69 |
The table shows the relative expression levels, obtained using mRNA-Seq or qRT-PCR on an independent sample, comparing systemic tissue of H. oryzae infected plants with uninfected plants at time point 3 dpi.
Chlorophyll measurement
Chlorophyll measurements were performed in an independent experiment. Nematode collection, as well as growth conditions and inoculation of the plants were as described above. Shoots of inoculated and non-inoculated plants were harvested at 3 and 7 dpi. At each time point and for each treatment, 12 individual plants were used. At the day of sampling the length of shoot and root of each plant was measured. Then, shoots were harvested and pooled in 4 samples, each pool containing shoots of 3 individual plants. In each sample, chlorophyll content was determined after extraction in ice-cooled 100% Methanol and spectrophotometrical measurement of the absorbance (A) at 665.2 (chlorophyll a; Chla) and 652 nm (chlorophyll b; Chlb). To calculate the concentration in mg/L following formulas were applied: Chla = 16.29 * (A665.2)–8.54 * (A652); Chlb = 30.66 * (A652)–13.58 * (A665.2) [27]. After checking for normality and homoscedasticity of the data, statistical significance was evaluated using student’s t-test. This whole experiment was repeated in time, with similar results.
Results
More than 95 million RNA fragments were acquired and made publicly available in the GEO database (GSE57707). The short reads were aligned against the reference genome sequence of cv. Nipponbare (MSU7.0) and in total 81% of the sequenced reads could be mapped (Table 2). The total number of sequenced bases was nearly 7 billion, representing 18-fold the rice genome size and about 50-fold coverage of the annotated transcriptome. The expression of a total of 89,246 different rice transcripts was detected in the analyzed tissues. Comparative gene expression profiling was performed by statistical analysis of differential gene expression levels, Gene Set Enrichment, and pathway mapping. In addition, a search was performed to detect novel transcriptionally active regions (nTARs) not annotated in rice genome assembly MSU7.0.
Table 2. Overview of the obtained sequencing data and mapping of these sequences onto the rice genome.
| Sample | Total number of sequencedread pairs | Total number of mappedread pairs | Number of uniquelymapped read pairs | Number of unmapped readpairs |
| Shoots of uninfected plants,3 dpi, replicate 1 | 9407160 | 16793253 | 7475187 | 1931973 |
| Shoots of uninfected plants,3 dpi, replicate 2 | 12402694 | 22365580 | 9863309 | 2539385 |
| Shoots of uninfected plants,7 dpi, replicate 1 | 12338830 | 22597722 | 9838088 | 2500742 |
| Shoots of uninfected plants,7 dpi, replicate 2 | 11957710 | 22739886 | 9557445 | 2400265 |
| Total of shoot tissue ofuninfected plants | 46106394 | 84496441 | 36734029 | 9372365 |
| Shoots of Ho-infected plants,3 dpi, replicate 1 | 10819261 | 21665010 | 8489566 | 2329695 |
| Shoots of Ho-infected plants,3 dpi, replicate 2 | 13166886 | 29099106 | 10981032 | 2185854 |
| Shoots of Ho-infected plants,7 dpi, replicate 1 | 12601325 | 25281161 | 10705989 | 1895336 |
| Shoots of Ho-infected plants,7 dpi, replicate 2 | 13099442 | 25818811 | 10737960 | 2361482 |
| Total of shoot tissue ofinfected plants | 38867653 | 80199078 | 32424981 | 6442672 |
| Overall Total | 95793308 | 186360529 | 77648576 | 18144732 |
| sequenced bases perfragment (ie read pair) | 72 | |||
| number of sequenced bases | 6.90E+09 | |||
| length of rice genome(approximate) | 374424240 | |||
| length of rice transcriptome(approximate) | 139985298 | |||
| genome coverage | 18 | |||
| transcriptome coverage | 49 |
Systemic transcriptome changes upon rice root nematode infection
In a first analysis, the expression level in all sampled systemic tissues of infected plants (at both time points) was compared with the shoot tissues of healthy plants to look for consistent trends, regardless of the time point after inoculation.
At an FDR cut-off of 0.05, 52 annotated genes were found to be significantly differentially expressed in the shoots after root infection with H. oryzae (Table 3). Twenty one of these differentially expressed genes (DEGs) were significantly downregulated, while 31 showed higher expression in infected plant shoots. Among the downregulated genes for instance, we identified one encoding a RAS-related protein (Small monomeric GTPase), and a gene encoding 3-oxo-5-alpha-steroid 4-dehydrogenase, involved in brassinosteroid biosynthesis. Among the upregulated DEGs, 2 genes encoding AP2 domain containing proteins, and a cupin domain containing protein were found. Next to the annotated transcripts, 12 nTARs were found to be differentially expressed in shoots of inoculated vs. non-inoculated plants (Table 3).
Table 3. Differentially expressed genes (FDR<0.05), when comparing systemic tissue of H. oryzae infected plants with uninfected plants at both time points (3 and 7 dpi).
| Gene_ID | Annotation | Log2FC (shoots of infectedversus uninfected plants) | FDR |
| RNSR_1.36318 | No Description | –3.58 | 0.02 |
| LOC_Os05g34270 | inactive receptor kinase At1g27190precursor, putative, expressed | –2.00 | 0.01 |
| LOC_Os12g29670 | expressed protein | –1.93 | 0.03 |
| LOC_Os01g62950 | ras-related protein, putative, expressed | –1.85 | 0.02 |
| LOC_Os06g05130 | myristoyl-acyl carrier proteinthioesterase, chloroplast precursor,putative, expressed | –1.83 | 0.04 |
| LOC_Os05g36290 | actin, putative, expressed | –1.73 | 0.01 |
| LOC_Os10g02350 | transmembrane 9 superfamilymember, putative, expressed | –1.71 | 0.05 |
| LOC_Os01g54940 | dehydrogenase, putative, expressed | –1.69 | 0.01 |
| LOC_Os06g07630 | 26S protease regulatory subunit6A, putative, expressed | –1.68 | 0.04 |
| LOC_Os03g52040 | OsSCP19 - Putative SerineCarboxypeptidase homologue, expressed | –1.65 | 0.04 |
| LOC_Os02g36330 | RING-H2 finger proteinATL1O, putative, expressed | –1.60 | 0.02 |
| RNSR_1.55235 | No Description | –1.55 | 0.03 |
| LOC_Os04g48750 | 3-oxo-5-alpha-steroid4-dehydrogenase, putative, expressed | –1.51 | 0.05 |
| RNSR_1.61315 | No Description | –1.50 | 0.03 |
| LOC_Os02g52010 | phosphate-induced protein 1conserved region domaincontaining protein, expressed | –1.43 | 0.04 |
| LOC_Os01g07950 | OsGrx_S15.2 – glutaredoxinsubgroup II, expressed | –1.39 | 0.02 |
| RNSR_1.28714 | No Description | –1.35 | 0.01 |
| LOC_Os02g38810 | 26S proteasome non-ATPaseregulatory subunit 6,putative, expressed | –1.34 | 0.04 |
| LOC_Os04g56510 | ABC1 family domain containingprotein, putative, expressed | –1.28 | 0.03 |
| LOC_Os02g46839 | expressed protein | –1.14 | 0.04 |
| LOC_Os02g56900 | thioredoxin family protein,putative, expressed | –1.09 | 0.02 |
| LOC_Os04g33600 | hydrolase, alpha/beta foldfamily protein, putative, expressed | –1.08 | 0.02 |
| LOC_Os05g45040 | OsFBX170 - F-box domaincontaining protein, expressed | –1.01 | 0.03 |
| LOC_Os01g37700 | expressed protein | –0.88 | 0.04 |
| LOC_Os01g39310 | SEY1, putative, expressed | –0.67 | 0.04 |
| LOC_Os10g28580 | CAF1 family ribonucleasecontaining protein, expressed | 0.52 | 0.03 |
| LOC_Os10g22300 | resistance protein,putative, expressed | 0.59 | 0.03 |
| RNSR_1.47341 | No Description | 0.77 | 0.03 |
| RNSR_1.30534 | No Description | 0.82 | 0.03 |
| RNSR_1.4486 | No Description | 0.82 | 0.04 |
| RNSR_1.42503 | No Description | 0.84 | 0.01 |
| LOC_Os05g31420 | GRAS family transcriptionfactor containing protein, expressed | 0.88 | 0.04 |
| RNSR_1.27029 | No Description | 0.95 | 0.01 |
| LOC_Os06g45460 | OsFBX202 - F-box domaincontaining protein, expressed | 0.97 | 0.04 |
| LOC_Os09g31450 | polygalacturonase inhibitor 1precursor, putative, expressed | 0.97 | 0.03 |
| LOC_Os05g40570 | expressed protein | 0.99 | 0.04 |
| LOC_Os01g67030 | auxin-responsive protein,putative, expressed | 1.02 | 0.02 |
| LOC_Os03g48590 | protein transport proteinSec61 subunit alpha,putative, expressed | 1.03 | 0.05 |
| LOC_Os02g33840 | OsFBX52 - F-box domaincontaining protein, expressed | 1.08 | 0.03 |
| LOC_Os01g41140 | THION18 - Plant thioninfamily protein precursor, expressed | 1.14 | 0.01 |
| LOC_Os04g41070 | zinc finger, C3HC4 typedomain containing protein, expressed | 1.16 | 0.02 |
| LOC_Os12g09630 | expressed protein | 1.20 | 0.03 |
| LOC_Os05g10300 | expressed protein | 1.25 | 0.03 |
| LOC_Os08g04740 | expressed protein | 1.26 | 0.04 |
| LOC_Os03g42259 | hypervariable Bacillus group-specificprotein, putative, expressed | 1.30 | 0.05 |
| LOC_Os07g39450 | expressed protein | 1.32 | 0.04 |
| LOC_Os05g08580 | hypothetical protein | 1.33 | 0.02 |
| LOC_Os05g49010 | AP2 domain containingprotein, expressed | 1.35 | 0.05 |
| RNSR_1.17134 | No Description | 1.38 | 0.01 |
| LOC_Os10g23840 | conserved hypothetical protein | 1.58 | 0.02 |
| LOC_Os11g42290 | transferase family protein,putative, expressed | 1.64 | 0.04 |
| LOC_Os09g20470 | hypothetical protein | 1.71 | 0.02 |
| LOC_Os03g58990 | cupin domain containingprotein, expressed | 1.75 | 0.03 |
| LOC_Os04g02470 | expressed protein | 1.75 | 0.03 |
| LOC_Os04g18650 | AP2 domain containingprotein, expressed | 1.87 | 0.01 |
| LOC_Os12g10370 | expressed protein | 1.97 | 0.02 |
| LOC_Os03g31280 | expressed protein | 1.98 | 0.02 |
| LOC_Os08g02200 | expressed protein | 2.02 | 0.01 |
| LOC_Os06g08270 | expressed protein | 2.03 | 0.05 |
| LOC_Os02g32310 | expressed protein | 2.27 | 0.05 |
| LOC_Os12g13540 | hypothetical protein | 2.36 | 0.03 |
| RNSR_1.34363 | No Description | 2.46 | 0.03 |
| RNSR_1.11099 | No Description | 2.54 | 0.04 |
| LOC_Os12g27040 | expressed protein | 2.58 | 0.02 |
The table shows the rice locus number, the annotation (MSU7.0), the Log2 of the fold-change (FC) of infected versus uninfected shoot tissue and the FDR-value. Novel transcriptionally active regions received a gene ID starting with RNSR. FDR: false discovery rate. RNSR: Root Nematode Systemic Respons.
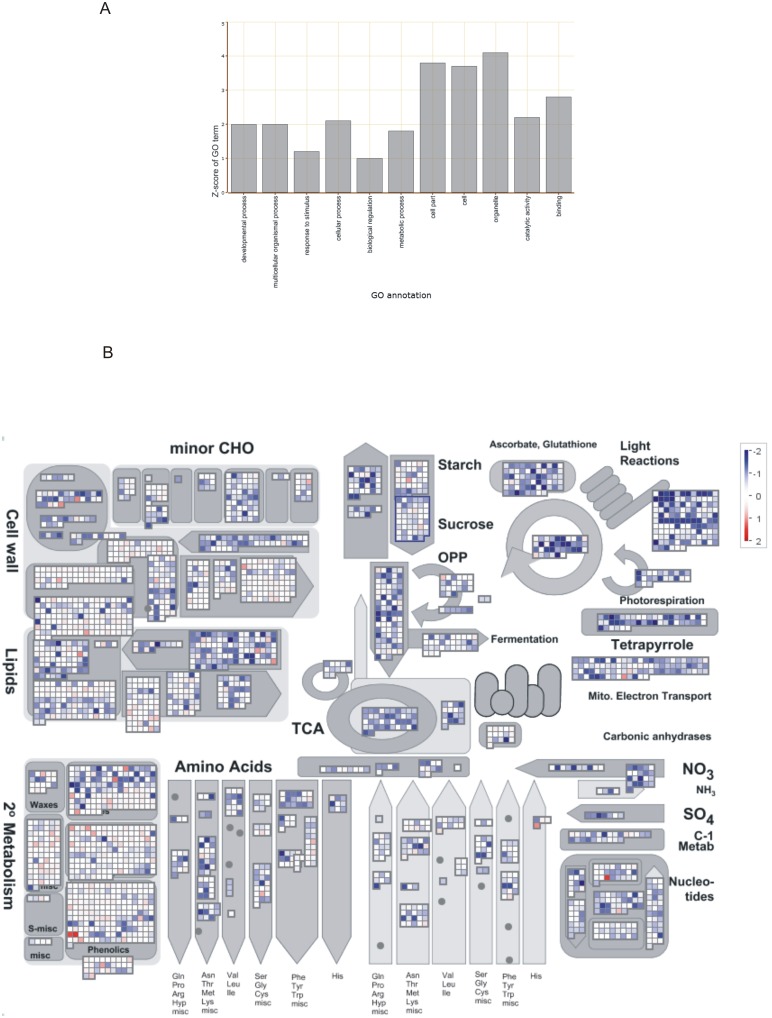
Parametric Analysis of Gene Set Enrichment of the differentially expressed genes in the infected versus uninfected plants revealed that the repressed genes were enriched for the secondary GO-groups ‘developmental process’, ‘multicellular organismal process’, ‘cellular process’, ‘metabolic process’, ‘organelle’, ‘cell’, ‘cell part’, ‘catalytic activity’ and ‘binding’ (Fig. 1A). None of the enriched gene sets showed induction in systemic tissue of infected plants in comparison with control shoot tissue.
Figure 1. Visualization of transcriptome data of shoots of rice infected with rice root nematode Hirschmanniella oryzae.
(A) Parametric Analysis of Gene Set Enrichment of the differentially expressed genes in the shoots of infected versus uninfected plants. The Z-scores of the significantly enriched secondary level GO terms are shown. (B) Mapman visualization of the expression profiles of genes involved in the general metabolism of the rice plant. The visualization shows the observed differential expression patterns, based on the Log2 fold changes of mRNA levels, in shoots of infected versus uninfected control plants. Red dots indicate that the gene is upregulated in infected plants versus the corresponding healthy control plants, while blue indicates downregulation.
When focusing on the general plant metabolism (Fig. 1B) the WSR-test of Mapman found a significant suppression of many primary metabolic processes, like ‘light reaction’, ‘fatty acid synthesis and elongation’, ‘ATP synthesis’, ‘tetrapyrrole synthesis’, ‘nucleotide metabolism’, ‘amino acid synthesis’, ‘TCA’, ‘glycolysis’, and ‘cell wall precursor synthesis’. Not only the primary metabolism of the plants was impaired, we also detected a specific suppression of a few plant defense related pathways. For instance, the biosynthesis of chorismate through the shikimate pathway, was strongly suppressed in the systemic tissue. Additionally, a significant suppression of the ‘isoprenoids’ pathway was observed.
The impact of the observed transcriptome changes in the primary metabolic pathways was analyzed by measuring the length of roots and shoots of infected versus uninfected plants. At 3 and 7 dpi, plant root and shoot length was not different from the control plants (data not shown). Because ‘light reaction’ and ‘tetrapyrrole synthesis’ were among the gene sets with the strongest levels of suppression upon infection, the chlorophyll content was measured in the leaves of H. oryzae-infected and control plants at 3 and 7 dpi (Table 4). At 3 dpi, a decreasing trend in levels of both chlorophyll a and b was detected when comparing infected with uninfected rice plants. At 7 dpi, a significant reduction in chlorophyll a was detected (18%) in the infected plants, and although not significant, the infected plants also showed 10% reduction in chlorophyll b content in comparison with uninfected control plants.
Table 4. Chlorophyll a and chlorophyll b content of shoots of plants infected with rice root nematode H. oryzae, in comparison with control (uninfected) plants of the same age.
| Chlorofyll a | Chlorofyll b | |
| Shoots of uninfected plants, 3 dpi | 704.7±150.6 | 147.9±29.2 |
| Shoots of infected plants, 3 dpi | 646.0±91.4 | 121.2±19.5* |
| Shoots of uninfected plants, 7 dpi | 809.0±76.3 | 150.6±15.0 |
| Shoots of infected plants, 7 dpi | 666.3±34.05** | 135.2±14.4 |
Values are the average ± standard deviation of four pools (n = 4) of shoot material from three plants each. Statistically significant differences in comparison with the respective control (at the same time point) are indicated with asterisks.
**P<0.05,
*P<0.1.
Systemic transcriptome changes at 3 days after rice root nematode infection
When considering the 3 dpi samples alone, only 2 biological replicates (although pools) per treatment were analyzed, and therefore the statistical analysis was performed with less stringent conditions (FDR<0.1). This resulted in 195 DEGs, with 169 repressed and 26 induced (Table S1). Next to the annotated transcripts, 25 nTARs were found to be differentially expressed in this tissue versus uninfected plant shoots (Table S1). Pathway mapping with MapMan showed a general suppression of the plant primary metabolism in the systemic tissues at 3 dpi (Figure S1).
Among the most intriguing and strongest downregulated DEGs were genes encoding (1) chalcone-flavonone isomerase (LOC_Os11g02440; Log2FC = –2.75), involved in flavonoid biosynthesis, (2) 3-beta hydroxysteroid dehydrogenase/isomerase family protein (LOC_Os03g23980; Log2FC = –2.27), involved in brassinosteroid biosynthesis, (3) the transcript encoding the brassinosteroid cell-surface leucine-rich repeat receptor-like kinase (LRR-RLK) BRASSINOSTEROID-INSENSITIVE 1 (BRI1, LOC_Os01g17250; Log2FC = –1.64), and (4) 2 other LRR-RLKs (LOC_Os02g09740, Log2FC = –2.24; LOC_Os06g47710, Log2FC = –2.84). LRR-RLKs regulate a wide variety of developmental and defense-related processes including cell proliferation, stem cell maintenance, hormone perception, host-specific as well as non-host-specific defense response, wounding response, and symbiosis [28]. Two other suppressed DEGs were encoding enzymes belonging to the shikimate pathway, chorismate mutase (Log2FC = –1.51) and isochorismatase (Log2FC = –1.78).
Among the induced DEGs, (1) a terpene synthase (LOC_Os12g30824; Log2FC = 1.10), necessary for the biosynthesis of ent-sandaracopimaradiene, a precursor of the diterpenoid phytoalexins oryzalexin A–F, (2) LOC_Os07g10240 (Log2FC = 2.05) encoding an anthocyanidin 3-O-glucosyltransferase involved in flavonoid (anthocyanin) biosynthesis, as well as many unannotated genes and 12 nTARs (Table S1).
To validate the gene expression levels as detected by RNA-seq, an independent validation was performed using qRT-PCR, comparing infected with uninfected plants at 3 dpi. In total, 14 out of the 16 tested gene expression profiles (87.5%) were confirmed (Table 1), in line with an expected FDR of 10%.
Systemic transcriptome changes at 7 days after rice root nematode infection
For the 7 dpi data, the statistical analysis was again performed with less stringent conditions (FDR<0.1), resulting in 17 DEGs, with 11 repressed and 6 induced. Next to the annotated transcripts, 10 nTARs were found to be differentially expressed in this tissue versus uninfected plant shoots (table S2). Most induced DEGs are annotated as ‘expressed protein’ or ‘hypothetical protein’, except for a gene annotated as heat shock protein DnaJ (LOC_Os01g42190; Log2FC = 0.528), and LOC_Os04g18650 (Log2FC = 1.652), coding for a pathogenesis-related transcriptional factor, with an ethylene-responsive AP2 domain (EREBP34, TSRF1, belonging to class IIIb; [29]). The tomato homolog of this AP2 gene, TSRF1 has previously been shown to enhance the osmotic and drought tolerance of rice [30]. This locus was also slightly (but not significantly) induced at 3 dpi in the systemic leaf tissue. Among the most strongly repressed DEGs are (1) a MYB family transcription factor (LOC_Os08g06110; Log2FC = –2.799), (2) a ribonuclease T2 family domain containing protein (LOC_Os08g33710; Log2FC = –2.250), (3) a CTP synthase (LOC_Os05g49770; Log2FC = –2.149), (4) monodehydroascorbate reductase (LOC_Os08g44340; Log2FC = –1.734), and (5) lactate/malate dehydrogenase (LOC_Os08g33720; Log2FC = –1.407).
Analysis of novel transcripts
A total number of 33,638 nTARs were detected in the analysed tissues (Table S3). A BLAST search against all ESTs from O. sativa resulted in 30,214 nTARs giving a significant hit (bitscore>50), while tBLASTx against all proteins of O. sativa resulted in significant hits for 19,735 of the nTARs, indicating that these nTARs are potential paralogues of known rice transcripts (Table S3). A Singular Enrichment analysis (FDR<0.05) of these rice transcripts revealed four enriched GO-terms of the category ‘Molecular function’: kinase activity, transferase activity, transferase activity - transferring phosphorus-containing groups and catalytic activity. A SwissProt search was done, and this was successful for 6,271 transcripts (Table S3), revealing for instance nTARs showing resemblance to protein domains annotated as xylanase inhibitor, MATE efflux family protein, histone-lysine N-methyltransferase, ribonuclease H protein, sugar transport protein, and ethylene-responsive transcription factor ABI4. A BLAST search against Trembl resulted in 16,134 hits, mainly to uncharacterized proteins from O. sativa.
Discussion
Gene expression analyses investigating the plant response upon nematode attack have mainly targeted the local reaction of the plant [4], [25], [31], [32], [33]. This localized approach is most appropriate to identify responses directly regulated by nematodes, while studies using systemic tissues can provide a global view of the infection response in host plants and offer a broader perspective on plant health in the context of plant-nematode interactions.
Species within the genus Hirschmanniella infest 58% of the world’s rice fields, causing 25% yield losses [1]. Hirschmanniella oryzae is one of the few plant-parasitic nematodes that can survive in anoxic environments [34] and hence this nematode is the most common plant-parasitic nematode in rice grown under constantly flooded conditions [1]. In previous studies performed by our research group we investigated (1) the systemic transcriptional responses of selected marker genes involved in hormone related defense pathways using qRT-PCR [10] and (2) the local reaction of rice roots upon H. oryzae and Meloidogyne graminicola infection using mRNA-Seq [4], [25].
In the current study, a more general overview of the systemic responses of H. oryzae-infested rice plants is provided. Using mRNA-Seq we have acquired almost 100 million reads from infected and healthy rice shoot tissues (>5 M per sample), thereby obtaining count data for more than 80,000 annotated and unannotated rice transcripts. We have used a synchronized inoculation method, in which the nematodes only had a limited period (24 hours) to infect roots. This allowed a relatively uniform infection process, so that the transcript changes detected here are not compromised by continuous entry of juveniles.
Systemic effects on defense responses
Locally, roots infected by H. oryzae exhibit a fast (3 dpi) and strong induction of genes involved in cell death as well as defense response pathways, like for instance the phenylpropanoid and jasmonate pathways [4]. The systemic shoot tissues also showed an induction of some defense-related genes at 3 dpi, including genes involved in jasmonate and ethylene production [10] and genes involved in flavonoid and phytoalexin biosynthesis (this study). However, at 7 dpi the expression of many of these genes were significantly downregulated or had returned to non-infected tissue level [10]. In contrast, shoot tissue of Arabidopsis plants infected with the cyst nematode Heterodera schachtii contained continuously higher levels of SA and JA-marker genes than corresponding control plants, from 5 till 14 dpi [8]. However, in the case of root knot nematode infection, a strong and early suppression of SA, JA, ET- biosynthesis genes, as well as PR genes has been observed already at 3 dpi [4] and 5 dpi [8], in systemic tissue of the Meloidogyne graminicola-rice [4] and Meloidogyne incognita-Arabidopsis [8] interaction, respectively. Hence, the here-observed systemic suppression of the plant defense system upon H. oryzae infection seems to be slower and less consistent than what was seen in systemic shoot tissues upon root knot nematode infection in rice and Arabidopsis [8].
In the current mRNA-Seq data we found two notable exceptions to this rule, since we detected a repression of the isoprenoid and shikimate pathway already at 3 dpi in the systemic tissue. The suppression of genes from the shikimate pathway could potentially deplete the pool of chorismate and prephenate available for the production of aromatic amino acids and salicylate. Aromatic amino acids are precursors for several secondary metabolites with proven roles in plant defense, like lignin, phenolics, flavonoids and salicylic acid, produced through the phenylpropanoid pathway [35]. Interestingly, we have recently discovered two candidate effectors of H. oryzae with homology to chorismate mutase and isochorismatase [36]. These effectors are most likely involved in the deregulation of the shikimate and subsequent phenylpropanoid pathway in the host plant. A secreted chorismate mutase from Ustilago maydis has for instance been demonstrated to lower the total salicylic acid content in infected leaves [37]. If and how these effectors can also affect the plant defence system in the above-ground tissue is an intriguing route for further research.
The isoprenoid pathway, which is also suppressed at 3 dpi in systemic shoots, is responsible for the production of a variety of both primary and secondary metabolites with very diverse functions, like tocopherol, carotenoids, mevalonate and provides precursors for brassinosteroid, gibberellin and chlorophyll biosynthesis [38].
Consistent with this view, genes involved in both chlorophyll biosynthesis as well as brassinosteroid biosynthesis and signalling were impaired in the systemic plant tissue. Brassinosteroids (BRs) are polyhydroxysteroids that offer a wide range of physiological activities starting from seed germination till flowering and senescence [39]. The strong suppression of this pathway as seen in the infected plants can potentially have dramatic effects on plant development. Although we did not observe significant effects on plant growth at 3 and 7 dpi, effects at later time points can certainly be expected.
In addition to their well-known role in plant development, BRs have recently been demonstrated to also play a role in plant responses to a broad spectrum of environmental stresses [40], [41]. For instance, BRI1, which was here found to be strongly repressed in the shoots of H. oryzae infected plants, is an important player in PAMP triggered Immunity [42].
Systemic effects on primary metabolism
The data provided in the current study reveals a remarkable suppression of the whole basic metabolic machinery in the rice shoot. The transcriptome revealed changes in the pathways needed for production of different metabolites for plant growth and development, such as carbohydrates, lipids, proteins, and nucleic acids. Similarly, Hofmann et al. [9] showed that cyst nematode (Heterodera schachtii) infection was causing dramatic changes in metabolite profiles in systemic root and shoot tissues. For instance, amino acid levels decreased in shoots of cyst-nematode infected plants in comparison with control shoots, but levels of some organic acids were increased [9]. It is known that defense metabolites such as phenylpropanoid pathway products, PR-proteins, ROS and callose represent a major flow of carbon from primary metabolism into secondary metabolism [43]. Large carbon fluxes into secondary metabolism during the defense response cannot occur without influencing reactions in primary metabolism [43]. The local strong defense induction inflicted in the H. oryzae infected root system [10], might be responsible for a depletion of carbon and energy available for the primary metabolism of the plant shoot.
Potential consequences for interactions with other pathogens and herbivores
In the case of root feeding by insects, a general induction of defense responses in the systemic shoot tissue has been observed [12], [44], [45]. For instance in cotton (Gossypium herbaceum), root-chewing by insect larvae led to increased concentrations of non-volatile terpenoid compounds in the leaves, known to deter insect feeding [12]. Recently Wondafrash et al. [15] provided a review of the systemic effects that root feeding by nematodes can have on subsequent attack by above-ground herbivores. They concluded that the effects varied depending on the feeding style of the nematode, nematode population densities and the susceptibility of the host plant, and different outcomes have been reported. For instance, soybeans with moderate and high Heterodera glycines infection levels supported greater Helicoverpa zea larval populations than observed on control or low density H. glycines infested plants. Nevertheless, very high H. glycines population densities caused severe stunting and chlorosis, reducing the suitability of these soybeans as a food source for H. zea larvae [46]. However, in Arabidopsis, a simultaneous inoculation with the sedentary endoparasitic nematode H. schachtii and the aphid Brevicoryne brassicae was performed [47] and no effects of the nematode infection were observed on the aphid performance at day 3. Again different, Agrostis capillaris and Anthoxanthum odoratum plants attacked by ectoparasitic and migratory endoparasitic nematodes, caused a reduced fecundity of the aphids Rhopalosiphum padi feeding on the shoots [48]. A lower amino acid content was observed in the phloem sap of the nematode-infected plants, and was probably one of the causes of this decreased aphid performance. Since our transcriptome data provides evidence for a reduced amino acid production in the above-ground tissues of H. oryzae infected rice plants, one could deduce that this tissue might be less attractive to phloem-feeding insects. Nevertheless, experimental evidence for this is currently lacking.
In conclusion, this transcriptome analysis of above-ground tissues upon rice root nematode infection unveils a major impairment of primary metabolite biosynthetic pathways in the shoots of infested plants. Biochemical investigation of chlorophyll levels in these tissues, corroborated the significant reduction of the plant’s photosynthetic capacity upon H. oryzae infection. Although less prominent, some subtle changes in defence-related pathways were also detected, among which mainly the isoprenoid and shikimate pathway seem to be negatively affected. If and how these changes affect the abiotic and biotic stress tolerance of the surviving rice plants remains to be further investigated.
Supporting Information
Mapman visualization of the expression profiles of genes involved in the general metabolism of the rice plant. The visualization shows the observed differential expression patterns, based on the Log2 fold changes of mRNA levels, in shoots of infected versus uninfected control plants at 3 dpi (A) and 7 dpi (B). Red dots indicate that the gene is upregulated in infected plants versus the corresponding healthy control plants, while blue indicates downregulation.
(ZIP)
Differentially expressed genes (FDR<0.1), when comparing systemic tissue of H. oryzae infected plants with uninfected plants at time point 3 dpi. The table shows the rice locus number, the annotation (MSU7.0), the Log2 of the fold-change (FC) of infected versus uninfected shoot tissue at 3 dpi and the FDR-value. Novel transcriptionally active regions received a gene ID starting with RNSR. FDR: false discovery rate. RNSR: Root Nematode Systemic Respons.
(XLSX)
Differentially expressed genes (FDR<0.1), when comparing systemic tissue of H. oryzae infected plants with uninfected plants at time point 7 dpi. The table shows the rice locus number, the annotation (MSU7.0), the Log2 of the fold-change (FC) of infected versus uninfected shoot tissue at 7 dpi and the FDR-value. Novel transcriptionally active regions received a gene ID starting with RNSR. FDR: false discovery rate. RNSR: Root Nematode Systemic Respons.
(XLSX)
List of putative novel transcriptionally active regions identified in the investigated rice shoot material, with their genomic location, BLASTn results against cDNA and genomic DNA of Oryza sativa cv. ‘Japonica’, BLASTx results against rice proteins, SwissProt/tremble. For all analyses a bitscore cut-off of 50 was used.
(XLSX)
Acknowledgments
Nematodes were originally collected and kindly provided by Zin Thu Zar Maung (Plant Protection Division, West Gyogone, Insein, Yangon, Myanmar). We thank Jean-Pierre Renard, Sarah De Keulenaer and Lien De Smet for excellent technical assistance.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The data has been submitted to the GEO repository: GSE57707, and can be freely accessed using the reviewer link provided in the paper.
Funding Statement
TK is funded by a postdoctoral fellowship of the Fund for Scientific Research Flanders (http://www.fwo.be/). SD received an IWT doctoral grant SB101371 (http://www.iwt.be/). TDM is supported by the Ghent University Multidisciplinary Research Partnership “Bioinformatics: from nucleotides to networks”, BOF_01GA0805. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bridge J, Plowright RA, Peng D (2005) Nematode parasites of rice. In: Luc M, Sikora RA, Bridge J, eds. Plant-parasitic nematodes in subtropical and tropical agriculture. CAB international, Wallingford, UK: 87–130.
- 2. Kyndt T, Fernandez D, Gheysen G (2014) Plant-Parasitic Nematode infections in rice: molecular and cellular insights. Annual Review of Phytopathology 52: 7.1–7.19. [DOI] [PubMed] [Google Scholar]
- 3. Kyndt T, Vieira P, Gheysen G, de Almeida-Engler J (2013) Nematode feeding sites: unique organs in plant roots. Planta 238: 807–818. [DOI] [PubMed] [Google Scholar]
- 4. Kyndt T, Denil S, Haegeman A, Trooskens G, Bauters L, et al. (2012) Transcriptional reprogramming by root knot and migratory nematode infection in rice. New Phytologist 196: 887–900. [DOI] [PubMed] [Google Scholar]
- 5. Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal Modulation of Plant Immunity. Annual Review of Cell and Developmental Biology, Vol 28 28: 489–521. [DOI] [PubMed] [Google Scholar]
- 6. Gheysen G, VanderEycken W, Barthels N, Karimi M, VanMontagu M (1996) The exploitation of nematode-responsive plant genes in novel nematode control methods. Pesticide Science 47: 95–101. [Google Scholar]
- 7. Bar-Or C, Kapulnik Y, Koltai H (2005) A broad characterization of the transcriptional profile of the compatible tomato response to the plant parasitic root knot nematode Meloidogyne javanica. European Journal of Plant Pathology 111: 181–192. [Google Scholar]
- 8. Hamamouch N, Li CY, Seo PJ, Park CM, Davis EL (2011) Expression of Arabidopsis pathogenesis-related genes during nematode infection. Molecular Plant Pathology 12: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hofmann J, El Ashry A, Anwar S, Erban A, Kopka J, et al. (2010) Metabolic profiling reveals local and systemic responses of host plants to nematode parasitism. Plant Journal 62: 1058–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kyndt T, Nahar K, Haegeman A, De Vleesschauwer D, Hofte M, et al. (2012) Comparing systemic defence-related gene expression changes upon migratory and sedentary nematode attack in rice. Plant Biology 14: 73–82. [DOI] [PubMed] [Google Scholar]
- 11. Manninen AM, Vuorinen M, Holopainen JK (1998) Variation in growth, chemical defense, and herbivore resistance in Scots pine provenances. Journal of Chemical Ecology 24: 1315–1331. [Google Scholar]
- 12. Bezemer TM, Wagenaar R, Van Dam NM, Van Der Putten WH, Wackers FL (2004) Above- and below-ground terpenoid aldehyde induction in cotton, Gossypium herbaceum, following root and leaf injury. Journal of Chemical Ecology 30: 53–67. [DOI] [PubMed] [Google Scholar]
- 13. Kaplan I, Sardanelli S, Denno RF (2009) Field evidence for indirect interactions between foliar-feeding insect and root-feeding nematode communities on Nicotiana tabacum. Ecological Entomology 34: 262–270. [Google Scholar]
- 14. van Dam NM (2009) Belowground Herbivory and Plant Defenses. Annual Review of Ecology Evolution and Systematics 40: 373–391. [Google Scholar]
- 15.Wondafrash M, Van Dam NM, Tytgat TOG (2013) Plant systemic induced responses mediate interactions between root parasitic nematodes and aboveground herbivorous insects. Frontiers in Plant Science 4. [DOI] [PMC free article] [PubMed]
- 16. Reversat G, Boyer J, Sannier C, Pando-Bahuon A (1999) Use of a mixture of sand and water-absorbent synthetic polymer as substrate for the xenic culturing of plant-parasitic nematodes in the laboratory. Nematology 1: 209–212. [Google Scholar]
- 17.Hooper DJ, Hallmann J, Subbotin SA. 2005. Methods for extraction, processing and detection of plant and soil nematodes. In: Luc M, Sikora RA, Bridge J, eds. Plant-parasitic nematodes in subtropical and tropical agriculture. Wallingford, UK: CAB International, 53–86.
- 18. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28: 511–U174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardcastle TJ, Kelly KA (2010) baySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. Bmc Bioinformatics 11. [DOI] [PMC free article] [PubMed]
- 21.Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology 11. [DOI] [PMC free article] [PubMed]
- 22. Du Z, Zhou X, Ling Y, Zhang ZH, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SY, Volsky DJ (2005) PAGE: Parametric analysis of gene set enrichment. Bmc Bioinformatics 6. [DOI] [PMC free article] [PubMed]
- 24. Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, et al. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant Journal 37: 914–939. [DOI] [PubMed] [Google Scholar]
- 25. Ji HL, Gheysen G, Denil S, Lindsey K, Topping JF, et al. (2013) Transcriptional analysis through RNA sequencing of giant cells induced by Meloidogyne graminicola in rice roots. Journal of Experimental Botany 64: 3885–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narsai R, Ivanova A, Ng S, Whelan J (2010) Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets. Bmc Plant Biology 10. [DOI] [PMC free article] [PubMed]
- 27. Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simulatenous equations for assaying chlorophyll A and chlorophyll B extracted with 4 different solvents – verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica Et Biophysica Acta 975: 384–394. [Google Scholar]
- 28.Torii KU (2004) Leucine-rich repeat receptor kinases in plants: Structure, function, and signal transduction pathways. International Review of Cytology - a Survey of Cell Biology, Vol 234 234: 1-+. [DOI] [PubMed]
- 29. Rashid M, Guangyuan H, Guangxiao Y, Hussain J, Xu Y (2012) AP2/ERF Transcription Factor in Rice: Genome-Wide Canvas and Syntenic Relationships between Monocots and Eudicots. Evolutionary Bioinformatics Online 8: 321–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quan RD, Hu SJ, Zhang ZL, Zhang HW, Zhang ZJ, et al. (2010) Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnology Journal 8: 476–488. [DOI] [PubMed] [Google Scholar]
- 31. Jammes F, Lecomte P, Almeida-Engler J, Bitton F, Martin-Magniette ML, et al. (2005) Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant Journal 44: 447–458. [DOI] [PubMed] [Google Scholar]
- 32. Ithal N, Recknor J, Nettleton D, Maier T, Baum TJ, et al. (2007) Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Molecular Plant-Microbe Interactions 20: 510–525. [DOI] [PubMed] [Google Scholar]
- 33. Barcala M, Garcia A, Cabrera J, Casson S, Lindsey K, et al. (2010) Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant Journal 61: 698–712. [DOI] [PubMed] [Google Scholar]
- 34. Babatola JO, Bridge J (1980) Feeding Behaviour and Histopathology of Hirschmanniella oryzae, H. imamuri, and H. spinicaudata on Rice. Journal of Nematology. 12: 48–53. [PMC free article] [PubMed] [Google Scholar]
- 35. Maeda H, Dudareva N (2012) The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annual Review of Plant Biology, Vol 63 63: 73–105. [DOI] [PubMed] [Google Scholar]
- 36. Bauters L, Haegeman A, Kyndt T, Gheysen G (2014) Analysis of the transcriptome of Hirschmanniella oryzae to explore potential survival strategies and host-nematode interactions. Molecular Plant Pathology 15: 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, et al.. (2011) Metabolic priming by a secreted fungal effector. Nature 478: 395-+. [DOI] [PubMed]
- 38. Vranova E, Coman D, Gruissem W (2012) Structure and Dynamics of the Isoprenoid Pathway Network. Molecular Plant 5: 318–333. [DOI] [PubMed] [Google Scholar]
- 39. Clouse SD, Sasse JM (1998) Brassinosteroids: Essential regulators of plant growth and development. Annual Review of Plant Physiology and Plant Molecular Biology 49: 427–451. [DOI] [PubMed] [Google Scholar]
- 40. Bajguz A, Hayat S (2009) Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiology and Biochemistry 47: 1–8. [DOI] [PubMed] [Google Scholar]
- 41. Belkhadir Y, Jaillais Y, Epple P, Balsemao-Pires E, Dangl JL, et al. (2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proceedings of the National Academy of Sciences of the United States of America 109: 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, et al. (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proceedings of the National Academy of Sciences of the United States of America 109: 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bolton MD (2009) Primary Metabolism and Plant Defense-Fuel for the Fire. Molecular Plant-Microbe Interactions 22: 487–497. [DOI] [PubMed] [Google Scholar]
- 44. Bezemer TM, van Dam NM (2005) Linking aboveground and belowground interactions via induced plant defenses. Trends in Ecology & Evolution 20: 617–624. [DOI] [PubMed] [Google Scholar]
- 45. Erb M, Flors V, Karlen D, de Lange E, Planchamp C, et al. (2009) Signal signature of aboveground-induced resistance upon belowground herbivory in maize. Plant Journal 59: 292–302. [DOI] [PubMed] [Google Scholar]
- 46. Alston DG, Bradley JR, Schmitt DP, Coble HD (1991) Response of Helicoverpa ze (Lepidoptera, Noctuidae) populations to canopy development in soybean as influenced by Heterodera glycines (Nematoda, Heteroderidae) and annual weed population densities. Journal of Economic Entomology 84: 267–276. [Google Scholar]
- 47. Kutyniok M, Muller C (2012) Crosstalk between above- and belowground herbivores is mediated by minute metabolic responses of the host Arabidopsis thaliana . Journal of Experimental Botany 63: 6199–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bezemer TM, De Deyn GB, Bossinga TM, van Dam NM, Harvey JA, et al. (2005) Soil community composition drives aboveground plant-herbivore-parasitoid interactions. Ecology Letters 8: 652–661. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mapman visualization of the expression profiles of genes involved in the general metabolism of the rice plant. The visualization shows the observed differential expression patterns, based on the Log2 fold changes of mRNA levels, in shoots of infected versus uninfected control plants at 3 dpi (A) and 7 dpi (B). Red dots indicate that the gene is upregulated in infected plants versus the corresponding healthy control plants, while blue indicates downregulation.
(ZIP)
Differentially expressed genes (FDR<0.1), when comparing systemic tissue of H. oryzae infected plants with uninfected plants at time point 3 dpi. The table shows the rice locus number, the annotation (MSU7.0), the Log2 of the fold-change (FC) of infected versus uninfected shoot tissue at 3 dpi and the FDR-value. Novel transcriptionally active regions received a gene ID starting with RNSR. FDR: false discovery rate. RNSR: Root Nematode Systemic Respons.
(XLSX)
Differentially expressed genes (FDR<0.1), when comparing systemic tissue of H. oryzae infected plants with uninfected plants at time point 7 dpi. The table shows the rice locus number, the annotation (MSU7.0), the Log2 of the fold-change (FC) of infected versus uninfected shoot tissue at 7 dpi and the FDR-value. Novel transcriptionally active regions received a gene ID starting with RNSR. FDR: false discovery rate. RNSR: Root Nematode Systemic Respons.
(XLSX)
List of putative novel transcriptionally active regions identified in the investigated rice shoot material, with their genomic location, BLASTn results against cDNA and genomic DNA of Oryza sativa cv. ‘Japonica’, BLASTx results against rice proteins, SwissProt/tremble. For all analyses a bitscore cut-off of 50 was used.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The data has been submitted to the GEO repository: GSE57707, and can be freely accessed using the reviewer link provided in the paper.