Abstract
Background
Hypercapnia and elevated temperatures resulting from climate change may have adverse consequences for many marine organisms. While diverse physiological and ecological effects have been identified, changes in those molecular mechanisms, which shape the physiological phenotype of a species and limit its capacity to compensate, remain poorly understood. Here, we use global gene expression profiling through RNA-Sequencing to study the transcriptional responses to ocean acidification and warming in gills of the boreal spider crab Hyas araneus exposed medium-term (10 weeks) to intermediate (1,120 μatm) and high (1,960 μatm) PCO2 at different temperatures (5°C and 10°C).
Results
The analyses reveal shifts in steady state gene expression from control to intermediate and from intermediate to high CO2 exposures. At 5°C acid–base, energy metabolism and stress response related genes were upregulated at intermediate PCO2, whereas high PCO2 induced a relative reduction in expression to levels closer to controls. A similar pattern was found at elevated temperature (10°C). There was a strong coordination between acid–base, metabolic and stress-related processes. Hemolymph parameters at intermediate PCO2 indicate enhanced capacity in acid–base compensation potentially supported by upregulation of a V-ATPase. The likely enhanced energy demand might be met by the upregulation of the electron transport system (ETS), but may lead to increased oxidative stress reflected in upregulated antioxidant defense transcripts. These mechanisms were attenuated by high PCO2, possibly as a result of limited acid–base compensation and metabolic down-regulation.
Conclusion
Our findings indicate a PCO2 dependent threshold beyond which compensation by acclimation fails progressively. They also indicate a limited ability of this stenoecious crustacean to compensate for the effects of ocean acidification with and without concomitant warming.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-789) contains supplementary material, which is available to authorized users.
Keywords: Hyas araneus, RNA-Seq, Ocean acidification, Warming, Gene expression, Crustaceans
Background
Increasing anthropogenic emissions of CO2 induce ocean warming and acidification. These changes in environmental conditions may have adverse effects on marine organisms [1–5]. However, the responses to ocean acidification (OA) are highly variable between organisms [4, 5] based on the fact that various animals differ in their capacities to compensate for acid–base disturbances caused by elevated seawater CO2 and resulting blood hypercapnia for review see [5]. Organisms with low compensation abilities show depressed metabolism, altered energy budgets, and as a result, lower rates of growth or development [6–9]. In contrast, organisms compensating for acid–base disturbances through active ion transport, such as fish, cephalopods and some crustaceans are projected to be more tolerant towards OA [3, 10]. In parallel to these differential capacities, sensitivities within a phylum seem to be related to differences in lifestyle and associated energy turnover [5, 11]. Furthermore, species or populations from highly variable environments with natural variations in PCO2 may have evolved to be more tolerant than species from relatively stable environments. As an extreme example, the shallow living crab Cancer magister can compensate within 24 h for hypercapnia-induced acidosis, while the extracellular acidosis in the deep-sea crab Chionoecetes tanneri remains mostly uncompensated during this time [12]. However, such short-term studies have limited value if it comes to the projection of long-term ocean acidification effects.
The great spider crab Hyas araneus is an osmoconforming, slow-moving and inactive species living in relatively stable physical conditions and is thus an excellent candidate to study the medium to long-term effects of abiotic stressors. A number of physiological studies have already investigated the effects of elevated seawater PCO2 on this species: CO2 induced decreases in growth rates and fitness of larvae were demonstrated in a North Sea population, whereas an Arctic population seemed more sensitive towards thermal stress [13]. In the Spitsbergen population elevated seawater PCO2 (1,100 μatm) caused an increase in metabolic rate during larval development pointing to higher metabolic costs [14]. Adult H. araneus became more heat intolerant under elevated CO2 with potential consequences for biogeographical distribution [15]. In the Arctic population synergistic effects of increased temperature and PCO2 adversely influenced the capacities for activity associated with disturbances in acid–base status [16].
To understand organismal sensitivities and tolerance-limits to OA with and without concomitant warming it is important to identify and differentiate between the mechanisms that shape an organism’s capacity to cope with the projected changes. At the whole organism level, crustaceans are impacted by OA with and without concomitant warming with effects ranging from changes in acid–base homeostasis [12, 16, 17], metabolism [9, 18, 19], growth [7, 13, 20, 21], to development [13, 14, 22, 23] and even survival [21, 24]. These processes are highly interdependent. While active acid–base regulation is an energy-consuming process [25], eventually leading to enhanced metabolic requirements [26], uncompensated extracellular pH can elicit metabolic depression [27] via effects on transmembrane ion exchange [25]. Furthermore, low pH can trigger a decrease in protein synthesis [28]; this may result in reduction of growth under hypercapnic conditions [6]. These previous studies provide us with important insights into the mechanistic background of responses to ongoing OA and warming, but also highlight the complexity of the processes involved. To elaborate the sensitivities and potential tolerance limits further, it is important to investigate the key regulatory mechanisms shaping affected processes and the tradeoffs between them.
A transcriptomic approach makes it possible to simultaneously investigate the genetic response of a wide range of cellular processes, and thus to identify the early responses to environmental changes [29]. Gene expression analyses can be used to characterize the molecular phenotype and the cellular changes that underpin physiological responses. They can also be used to uncover molecular mechanisms that might define physiological plasticity. Transcriptomic analysis can further reveal the connections between response mechanisms to environmental changes such as OA or temperature that may otherwise be overlooked [30]. Due to technological advances in recent years, analyses of the whole transcriptome have become increasingly attractive to study non-model (marine) organisms, and their molecular responses to a variety of environmental changes such as warming [31, 32], salinity fluctuations [33], hypoxia [34, 35] or OA [29, 36, 37] in marine organisms.
In the present study, we used gene expression profiling to explore the molecular response in gills of H. araneus exposed to hypercapnia at different temperatures. In marine crustaceans, gills are the first line of defence against acid–base disturbances of body fluids and thus the most important regulatory tissue for CO2 induced acidification of the hemolymph [38, 39]. We used a quantitative transcriptomic approach based on direct cDNA sequencing using high-throughput Illumina sequencing [40]. Since the present study focuses on the mechanisms involved and the potential sensitivity of H. araneus to climate changes, we selected CO2 concentrations projected for the year 2100 and 2300 by the Intergovernmental Panel on Climate Change (IPCC) as well as two different temperatures (5°C as the summer control temperature for the Arctic population and 10°C as the median habitat temperature of the species considering the whole distribution range. This study provides comprehensive insights into the transcriptional changes involved in the responses to warming and OA.
Methods
Animals, experimental treatments and tissue sampling
Adult specimens of the Arctic spider crab H. araneus (Linnaeus 1758) were collected by scientific divers in May 2009 in Kongsfjord at the west coast of Spitsbergen at 7–12 m depth (N 78°58.635'; E 11°29.454') and transferred to the Alfred Wegener Institute, Bremerhaven, Germany. Animals were maintained at 5°C in flow through aquaria with natural seawater prior to experimentation. During this period, seawater was aerated with ambient air and animals were fed ad libitum twice per week with frozen mussels and cockles (Mytilus edulis and Cerastoderma edule).
For the medium-term experiment, male spider crabs with a carapace width of 26 to 42 mm were divided into six groups and each group was randomly assigned to the different treatments. Animals were exposed to three different CO2 concentrations (390 μatm as control, 1,120 μatm as intermediate concentration and 1,960 μatm as high CO2 treatment) and two different temperatures (5°C as control and 10°C as elevated temperature) for 10 weeks (Figure 1). For each treatment, 5–7 animals were individually placed in 2 l wide-mouth containers (Kautex, Bonn, Germany).
Figure 1.
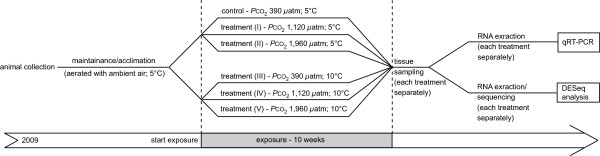
Overview of the experimental design used in the differential expression . Animals were collected in 2009 and acclimated to 5°C until the start of experimentation. Subsequently, exposure experiments were conducted for a time period of 10 weeks for all treatments. After exposure, tissue samples were taken and total RNA was extracted for analyses by quantitative real-time polymerase chain reaction (qRT-PCR) and Sequencing. Sequencing data were used for differential expression analysis by DESeq.
Experiments were carried out in recirculating seawater CO2 manipulation systems of 1 cubic meter volume each. Seawater of the storage tank was pumped to a header tank at a rate of 20 l min−1, which supplied the wide-mouth container by gravity feed at a flow rate of 200 ml min−1. Water of experimental containers was retained in a collection tank and pumped back to the storage tank at a flow rate of 20 l min−1. Ambient air temperature in the experimental rooms was thermostated to keep water temperature constant. Seawater CO2 manipulation was accomplished by constantly aerating the storage and header tanks with a defined air/CO2 mixture using an automatic mass flow controller (HTK 6 channel, HTK Hamburg GmbH, Germany). A light–dark cycle of 12:12 h was established. Water was partly changed every week by refilling the storage tank with pre-equilibrated seawater (PCO2 and temperature). Experimental animals were fed once a week ad libitum with frozen mussels (C. edule).
To monitor water physicochemistry, seawater samples were collected in airtight glass vials to prevent exchange with the atmosphere, and total dissolved carbon (DIC) concentration was immediately measured with a Seal QuAAtro SFA Analyzer (Seal Analytical, Mequon, United States of America). Temperature, salinity and pH were measured at the time of collection and, together with DIC, used to calculate the PCO2 in seawater using CO2SYS [41]. Seawater pH was measured using a pH electrode (ProfiLine pH 3310, WTW Wissenschaftlich-Technische Werkstätten GmbH, Weilheim, Germany) calibrated at the respective temperature with National Institute of Standards and Technology (NIST) standard pH buffer and salinity with a conductivity meter (ProfiLine Cond 1970i, WTW Wissenschaftlich-Technische Werkstätten GmbH, Weilheim, Germany). For CO2SYS, NBS (NIST) scale of seawater pH and constants of Mehrbach et al. [41] refitted by Dickson and Millero [41] were used. A summary of water physicochemistry data is given in Table 1.
Table 1.
Summary of the seawater physiochemical conditions during experiments with Hyas araneus
| Parameter | Control | Treatment (I) | Treatment (II) | Treatment (III) | Treatment (IV) | Treatment (V) |
|---|---|---|---|---|---|---|
| Temperature (°C) | 5.3 ± 0.2 | 4.2 ± 0.2 | 4.5 ± 0.2 | 9.9 ± 0.2 | 9.7 ± 0.3 | 9.8 ± 0.2 |
| Salinity (‰) | 32.1 ± 0.7 | 32.2 ± 0.7 | 32.2 ± 0.6 | 33.6 ± 0.4 | 33.5 ± 0.4 | 33.6 ± 0.3 |
| pH (NBS scale) | 8.15 ± 0.03 | 7.81 ± 0.04 | 7.55 ± 0.06 | 8.22 ± 0.04 | 7.85 ± 0.04 | 7.54 ± 0.05 |
| DIC (mmol kg−1) | 2,366 ± 42 | 2,436 ± 14 | 2,520 ± 39 | 2,295 ± 28 | 2,395 ± 14 | 2,488 ± 22 |
| PCO2 (μatm) | 441 ± 35 | 991 ± 96 | 1,878 ± 246 | 366 ± 30 | 942 ± 59 | 2,015 ± 147 |
| Total alkalinity (mmol kg−1) | 2,479 ± 13 | 2,479 ± 14 | 2,491 ± 14 | 2,484 ± 10 | 2,469 ± 13 | 2,473 ± 15 |
Temperature, Salinity, pH and dissolved inorganic carbon (DIC) were measured and partial pressure of CO2 (PCO2) and total alkalinity were calculated using CO2SYS [41]. Data are mean ± SD with N = 24 (5°C), N = 16–20 (10°C).
After experimental exposure, all 6 gill pairs were collected from 5–7 animals in each treatment. Tissue samples were immediately frozen in liquid nitrogen and stored at −80°C until usage.
Hemolymph sampling and measurements
Directly before tissue sampling, around 1 ml of hemolymph was extracted at the coxa of the third walking leg using a 1 ml sterile syringe (Henke-Sass, Wolf GmbH, Tuttlingen, Germany). Hemolymph was immediately transferred to a 1.5 ml tube (AG Eppendorf, Hamburg, Germany), placed in a thermostatted water bath and pH was measured at acclimation temperature using a pH microelectrode (PHM 93 Reference pH meter, Radiometer, Copenhagen, Denmark; InLab Micro, Mettler Toledo GmbH, Germany). The pH meter was calibrated at the respective temperature with NIST standard pH buffer. A hemolymph subsample was withdrawn using a gas-tight 200 μl syringe (Hamilton Company, Reno, United States of America) and total dissolved inorganic carbon (CCO2) of extracellular fluid was analysed according to the modified gas chromatographic method [42, 43]. Extracellular fluid was injected in gas tight glass vials containing 3 ml of air equilibrated 0.1 M hydrogen chloride (HCl) and analysed by gas chromatography in an Agilent 6890 N GC System (Agilent Technologies, Santa Clara, United States of America). The bicarbonate (HCO3−) concentration of the extracellular fluid was calculated from CCO2 and pH using equations derived from the Henderson-Hasselbalch equation. PCO2 was calculated as PCO2 = CCO2 * (10ph-pkIII * αCO2 + αCO2)−1 and HCO3− as HCO3− = CCO2- αCO2 * PCO2, with CCO2 being the total CO2 concentration in mM, αCO2 the physical solubility of CO2, PCO2 the partial pressure of CO2 in kPa and pK the apparent dissociation constant of the CO2/apparent HCO3− system. αCO2 and pK were calculated according to Pörtner et al. [44]. Raw data of hemolymph sampling and measurements are available at http://doi.org/10.1594/PANGAEA.833705.
RNA extraction and sequencing
Total tissue RNA of gills was extracted using the RNeasy Mini Kit according to the Purification of Total RNA from Animal Tissue protocol (QIAGEN, Hilden, Germany). RNA quantities were determined by a NanoDrop 2000c spectrometer (PeqLab, Erlangen, Germany), and RNA was analysed for quality by microfluidic electrophoresis in an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, United States of America). Total RNA from all gill pairs of 4 animals was pooled for each treatment, and used for library constructions and sequencing by GATC Biotech (Konstanz, Germany). The cDNA libraries for each treatment were constructed according to the SMART protocol for Illumina sequencing (Clontech, Mountain View, USA) and after adapter ligation pooled into two samples. To obtain appropriate deep sequencing results, samples were sequenced at least twice. Illumina sequencing was performed on a HiSeq 2000 Sequencer by GATC Biotech (Konstanz, Germany). Raw reads were quality controlled by FastQC (Babraham Institute, Cambridge, UK) and cleaned using the FastX-Toolkit (Hannon Lab - Cold Spring Harbor Laboratory, New York, USA). Quality control and trimming was performed using the following parameters: Minimum quality score of 20, minimum percentage of bases within the quality score of 90 and a minimum length of 25 bases. The cleaned raw data of the Illumina sequencing were deposited in the European Nucleotide Archive (ENA) at the European Molecular Biological Laboratory – European Bioinformatics Institute (EMBL-EBI) (http://www.ebi.ac.uk/ena/data/view/ERP002128). A summary of the cleaned sequencing results for all samples is given in Additional file 1: Table S1.
Mapping and identification of differentially expressed genes
Short reads of each sample were separately aligned against the annotated H. araneus transcriptome [40], using the Burrows-Wheeler Aligner (BWA) (version 0.5.9) with default parameters [45]. Obtained files were processed into bam files for further analysis, using SAMTools (version 0.1.18) [46]. An overview of the mapping and efficiency is described in Additional file 1: Table S1. Differential expression analysis was conducted with the R statistic software [47]. Read counts were summed up for all sequencing runs of each sample and used for the differential expression analysis without biological replicates. Differential expression of genes was evaluated using a test based on the negative binomial distribution as integrated in the Bioconductor R package DESeq [48], with a standard level of p ≤ 0.05 indicating significance. Control (control PCO2/control temperature) was compared to five treatments: (I) elevated temperature; (II) intermediate PCO2 at control temperature; (III) high PCO2 at control temperature (IV) intermediate PCO2 at elevated temperature; (V) high PCO2 at elevated temperature. The previously annotated transcriptome made a Gene Ontology enrichment analysis possible to test for particular affected terms, using Fisher’s exact test (FDR ≤ 0.05) as implemented in the Blast2GO software (version 2.6.0) [49, 50]. All subsets of significantly regulated genes identified by the binominal distribution test were tested against the full set of annotated sequences of the H. araneus transcriptome. To cut down on redundancy, GO terms were summarized into a more representative subset of terms using the web-based clustering tool REVIGO [51].
Validation by quantitative real-time polymerase chain reaction (qRT-PCR)
A set of transcript sequences known to be involved in acid–base regulation and/or transcripts that showed differential expression in one or more treatments was selected for validation of RNA-Seq results. Primers were designed using the PrimerExpress software (version 3.0) (Applied Biosystems, Darmstadt, Germany) with the Taq-Man MGB Quantification method and default parameters (Additional file 2: Table S2). Primer specificity was given by using sequences of the annotated H. araneus transcriptome [40]. All primer pairs were tested for performance and efficiency across a series of cDNA dilutions (1:20; 1:40; 1: 100; 1:200; 1:1000; 1:2000). Primers used displayed a suitable per cycle amplification rate, with an efficiency (E) of 2.0 ± 0.1 and R2 > 0.98. Efficiency was calculated as E = 10(−1/S), with s being the slope of linear regression.
Total RNA was extracted from gills as described above. Ten micrograms of total RNA per sample was treated with DNAse for DNA digestion using the Turbo DNA-free kit (Ambion, Darmstadt, Germany) and 0.4 μg DNA free RNA was transcribed into cDNA with the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Darmstadt, Germany). Real-time PCR was performed on a 7500 Real-time PCR System (Applied Biosystems, Darmstadt, Germany) and SYBR® Green PCR master mix (Applied Biosystems, Darmstadt, Germany). All genes were finally analysed in a 40-fold dilution and amplified with 300 nM of primer. To verify the amplification specificity of fragments a melting curve analysis was performed for each reaction.
Gene expression calculation was based on the CT-threshold. Absolute mRNA quantities were calculated as QX = E(CT) and normalized with the formula QN(X) = QX/QX(HK), with QX(HK) being the absolute mRNA quantity of the housekeeping gene sodium bicarbonate cotransporter (NBC). The housekeeping gene was determined using geNorm implemented in the software qbasePlus (version 2.1) (Biogazelle, Zwijnaarde, Belgium) with a relative expression stability of M ≤ 0.42 (high reference stability is given at an average geNorm of M ≤ 0.5). To ensure consistency with the differential expression results of the DESeq analysis, gene expression results of the qRT-PCR were calculated as log2 fold change (log2FC) of mean normalized quantities of treatment and control.
Statistics
To identify significant differences in the sum of all significantly up- and down-regulated transcripts between treatments, significantly changed transcripts in one or more treatment, identified by the DESeq analysis, were transformed into a matrix with 1 = significantly up-regulated, −1 = significantly down-regulated and 0 = not significantly regulated transcript. Treatments were analysed for statistical differences applying the Wilcoxon matched pairs test as implemented in SigmaPlot (Version 12.0, Systat Software Inc., San Jose, USA) with p < 0.05. Data from each treatment were tested against each other.
The correlation between the differential expression results of the DESeq analysis and the corresponding gene expression results of the qRT-PCR was determined by Pearson Correlation as implemented in SigmaPlot 12.0 (Systat Software Inc., San Jose, USA).
A one-way ANOVA was used to identify the effect of seawater PCO2 on hemolymph pH and bicarbonate (HCO3−). Data obtained under various PCO2 levels were tested against each other for each temperature separately. A Holm Sidak test for multiple comparisons was used for a posteriori analyses. Tests were performed in SigmaPlot (Version 12.0, Systat Software Inc., San Jose, United States of America) with p < 0.05 indicating significant differences.
Results and discussion
A total of 55 million reads (56%) from initial Illumina sequencing passed the quality filter and was used for the differential expression analysis. After processing, an average of 9.2 million high quality reads were produced for each sample from 2–3 sequencing runs per sample (Additional file 1: Table S1). To obtain the differential expression of each gene, high quality reads were aligned on the H. araneus transcriptome [40]. An average of 5.2 million reads for each sample produced distinct alignments. The alignment process yielded an average efficiency of 56% for the high quality reads (Additional file 1: Table S1). The achieved mapping efficiency is actually higher than in a comparable study of a non-model organism, which used an analogous approach for differential expression analysis (41% efficiency) [52]. Furthermore, 96.5% of all transcript sequences in the H. araneus transcriptome were detected in the RNA-Seq data. However, the occurrence of a large amount of unmapped reads might result from sequencing errors, repetitive sequences or inadequate quality filtering of the Illumina reads. Furthermore, it has to be considered that transcripts supported by only a small number of aligned reads (≤10) may reflect incompletely assembled transcripts in the reference. Those poorly supported transcript sequences were excluded from the subsequent analysis, and a final test-set of 16,201 transcripts sequences was used for the differential expression analysis. As there were no biological replicates, it has to be considered that the variance for genes can only be estimated by comparing the mean-variance relationship between samples/treatments, as if they were replicates, resulting in an overestimation of the variance and thus make this approach more conservative. Furthermore, expression levels of stable and highly expressed genes, based on the RNA-Seq data, were analysed by quantitative real-time PCR (qRT-PCR) confirming the RNA-Seq methodology used in this study (Additional file 3: Figure S1 and Additional file 4: Figure S2).
We could identify 864 (5.3%) out of the 16,202 tested transcripts to be differentially expressed after medium-term (10 weeks) exposure to the abiotic effectors (Additional file 5: Table S3). Out of these differentially expressed genes (DEG) 40.0% and 31.3% were differentially expressed under intermediate CO2 (treatment I) and high CO2 (treatment II; Additional file 6: Figure S3A, B), respectively. For the high temperature (treatment III), 41.0% showed significantly different expression levels (Additional file 6: Figure S3C). The combination of factors intermediate CO2 (treatment IV) and high CO2 (treatment V) with elevated temperature revealed 38.7% and 29.4% DEG, respectively (Additional file 6: Figure S3D, E). While the total amount of significant DEGs was similar in all treatments, individual genes displayed large differences in up- or down-regulation (Figure 2). Intermediate CO2 (I) led to strongly up-regulated transcript levels, while high temperature (III) caused strong down-regulation.
Figure 2.
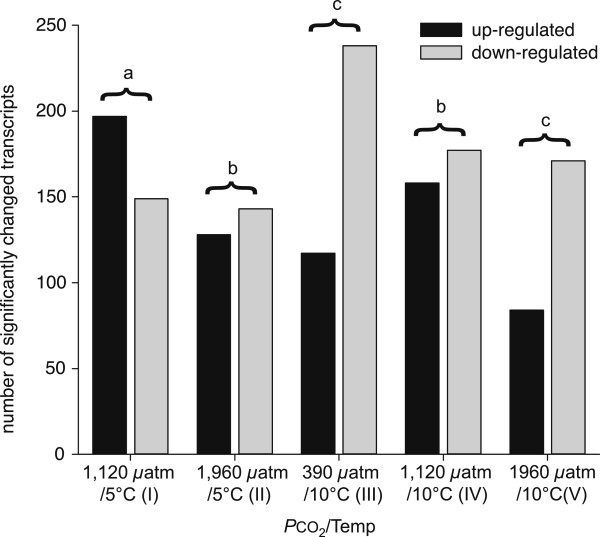
Differences in the sum of all significantly regulated transcripts of Hyas araneus after exposure experiments. For each treatment, up- and down-regulated transcripts were counted and represented as bars (black bars = up-regulated; grey bars = down-regulated). Treatments were tested for statistical differences using the Wilcoxon signed rank test (p < 0.05). Letters denote the significant differences. Differing letters indicate significant differences; identical letters indicate no significant difference.
These general results, especially the strong up-regulation of gene expression under intermediate CO2 (I), suggest that the regulatory capacity is high at moderately elevated PCO2 in adult H. araneus but reduced at higher CO2 levels. Up-regulation was also reduced at intermediate CO2 when combined with elevated temperature (IV) rather than at control temperature. Down-regulation in DEG predominated under warming alone (treatment III, high temperature) when compared to control temperature (I). 137 of the 177 genes that displayed significantly changed transcripts in the intermediate CO2/elevated temperature treatment (IV) were also differentially expressed in the high temperature treatment (III) supporting a strong temperature-dependent response, in line with the hypothesis that temperature affects most biochemical processes. Faced with a long-term temperature change, ectothermal organisms retain physiological homeostasis by several acclimation strategies, which can be of quantitative, qualitative or modifying nature [53]. The enhanced down-regulation seen in the high temperature treatments suggest that H. araneus may adopt a quantitative strategy to maintain physiological rates by down-regulating the concentrations of enzymes between 5 and 10°C [54].
To identify processes actually responding to elevated PCO2 and temperature, a first analysis was carried out using gene ontology (GO) terms. The set of GO-annotated differentially expressed genes was statistically tested for the over- and underrepresentation of GO terms to identify molecular functions, cellular components and biological processes affected most by the experimental treatments (Additional file 7: Table S4). The GO enrichment analysis revealed a variety of significantly over-represented GO terms that can primarily be summarized underneath the more generic categories ‘metabolism’ and ‘cell structure’. 23 and 25 over-represented GO terms, respectively, could be associated with these categories. It is important to mention that both intermediate CO2 treatments (I, IV) constitute 57% and 27% of the over-represented GO terms, respectively, representing compensation mechanisms mentioned above. The only over-represented GO term under high CO2, trehalose metabolic process, was evaluated for the down-regulated genes of the combined stress treatment (V). The fact that it was over-represented in all treatments indicates that enhanced expression of trehalose metabolism can be rated as a unifying response to both elevated CO2 and elevated temperature.
Within the ‘cell structure’ related GO-terms, intermediate CO2 (I, IV) led to significant up-regulation of genes concerning cell surface, extracellular matrix, structural molecule activity or brush border membrane, suggesting a structural modification of the gills. Some GO terms were found underrepresented at high temperature and in combined, intermediate CO2 and high temperature treatments (III, IV), and are assigned mainly to intracellular structures such as organelles. The over-representation of ‘cell structure’ related GO terms suggests that gill epithelial structure is adjusted in response to PCO2 disturbances. Gills are the principal organs for gas exchange and, together with the excretory organs, responsible for osmotic and ionic regulation in crustaceans [55]. As passive ion transport is influenced by the conductivity of gill epithelia [56], their structural modification might lead to a change in conductivity and would change the diffusion rate of ions. Structural changes were in fact identified in gills of Carcinus maenas during salinity exposure, with a modification of the apical plasma membrane system and an enlargement of the subcuticular compartment [57]. As environmental hypercapnia and salinity changes can cause similar mechanistic responses [39], similar transcriptomic modifications may occur. This is supported by an over-representation of the GO term response to salt stress in the intermediate CO2 treatment (I).
Although a GO enrichment analysis offers initial insights into processes affected by hypercapnia exposure and elevated temperature, a strong bias exists towards conserved and well-characterized processes, functions and cellular components in model organisms. This bias particularly applies to H. araneus, with a lack of GO annotation for about 76% of the transcripts. Additionally, many genes are grouped into more than one GO term depending on their resolution and are thus difficult to interpret. In light of these contraints, GO analysis can only provide a general overview of possibly affected processes and make a more detailed look indispensible.
For a more comprehensive understanding of the mechanisms responding to PCO2 and temperature changes, we performed a second analysis. Here, all genes included in the most affected categories ‘metabolism’ and ‘structural modification’, identified by the GO enrichment analysis, were considered. Additionally, we integrated all genes related to ‘acid–base and ion regulation’ and ‘response to stress’ into our analysis, as adjustments in these mechanisms are likely relevant in shaping resistance to hypercapnia exposure or heat stress.
For the interpretation of transcriptomic results, it has to be considered that expression profiles represent one regulatory level in the response to environmental changes and thus do not necessarily reflect the changes of other regulatory levels, e.g. protein abundances and activities. At least 25% of the proteome cannot be covered by gene expression profiling [30]. However, there was a positive correlation between transcription and translation in 87% of genes that changed ≥ twofold in living cells of yeast strains [58]. Certainly, a lower correlation has to be considered for more complex organisms, however an additional proteomics study on H. araneus revealed a comparable correlation between gene expression and protein abundance of ~70% for the intermediate treatment at 5°C (unpublished). A transcriptomic approach can provide first insights into the regulatory processes responding to environmental changes such as OA or temperature.
Response of specific groups
Acid–base regulation
The extracellular pH (pHe) measured in the hemolymph of adult H. araneus showed partial compensation under intermediate CO2 levels (I, IV) involving an increase in bicarbonate (HCO3−) concentration (Figure 3A,B). Under high CO2 (II, V), reduction in pHe was greater and the increase in 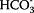 was reduced (Figure 3A,B). These findings suggest a limited capability to compensate for pHe disturbances caused by high seawater PCO2. According to a crustacean model by Freire et al. [55], proton (H+) excretion is generated by apical vacuolar-type (H+)-ATPase (V(H+)-ATPase) and/or sodium/proton exchanger (NHE), the latter dependent on sodium/potassium-ATPase (Na+/K+-ATPase). HCO3− is enriched in the hemolymph by basolateral anion exchangers. Enzymes that support active ion transport are the intracellular carbonic anhydrase (CA) and, in terms of a general support of energy consuming mechanisms, arginine kinase (AK). CA is assumed to accelerate the dissociation of carbonic acid (H2CO3) and provide the substrate for H+ and HCO3− transporters [55]. AK catalyses the reversible dephosphorylation of phosphoarginine, contributing to the restoration of adenosine triphosphate (ATP) used in energy consuming processes [59]. The expression of corresponding genes, V(H+)-ATPase, AK and partial sequences of two alpha CAs was significantly up-regulated at intermediate CO2, whereas such mRNA concentrations were only moderately increased at high CO2 (II) (Table 2). These expression levels follow the course of the hemolymph HCO3− parameters and the more effective acid–base regulation of adult H. araneus under moderately elevated CO2. However, only a few sequences encoding for Na+/K+-ATPase were up-regulated under CO2, in contrast to their response to elevated temperature treatment (III). Under the combined effect of temperature and CO2 Na+/K+-ATPase was also up-regulated. Another enzyme, DOPA decarboxylase, which catalyses the biosynthesis of dopamine by decarboxylation of L-DOPA, was found down-regulated at moderate (intermediate) CO2 elevations (I). Elevated dopamine leads to increased sodium influx and concomitantly, increased Na+/K+-ATPase activity in gills of C. maenas
[60]. In light of this finding constant mRNA levels of Na+/K+-ATPase at intermediate CO2 (I) indicate that the down-regulation of DOPA decarboxylase may even prevent an activation of Na+/K+-ATPase (Table 2). The gene expression of other transporters, supposed to be involved in acid–base regulation, such as NHE and/or bicarbonate/chloride co-transporter were not influenced at all by elevated PCO2 values or temperature. Acid–base regulation predominantly via the V(H+)-ATPase (as seen in the present transcriptome) might involve minimal disturbance to ionic composition, e.g. cellular sodium homeostasis which would be affected by strong involvement of Na+/K+-ATPase. In the sipunculid S. nudus, extracellular acidosis induced a shift in ion transporters during hypercapnia from high to low energy cost mechanisms of acid–base regulation, resulting in decreased Na+/K+-ATPase activity due to lower requirement for sodium regulation [25, 27]. In line with our present observations these results suggest a pH regulation system independent of Na+/K+-ATPase being used under hypercapnic exposure (Figure 4). No significant up-regulation of apical bicarbonate anion exchangers was observed. In C. sapidus, CO2 induced acid–base disturbances (PCO2 10,000 μatm) were mainly compensated by an uptake of HCO3− from the surrounding seawater via the gill epithelia [39]. However, our results with no up-regulation of respective transporters suggests hemolymph buffering may be achieved through the dissociation of respiratory CO2 resulting in an accumulation of HCO3− in the hemolymph. This might be exceptionally for crustaceans with low compensatory capacities. In H. araneus, also no significant up-regulation of basolateral bicarbonate anion exchangers was observed. However, expression of the anion channel bestrophin was significantly enhanced (Table 2, Figure 4). Bestrophin is activated by calcium (Ca2+) and enhances membrane permeability for anions, such as chloride and HCO3−
[61]. Even though bestrophin is commonly found in the retinal pigment epithelium, ion channels of this family have been identified in different tissues and are involved in a variety of cellular processes [62]. Its role in responses to elevated CO2 remains to be investigated.
was reduced (Figure 3A,B). These findings suggest a limited capability to compensate for pHe disturbances caused by high seawater PCO2. According to a crustacean model by Freire et al. [55], proton (H+) excretion is generated by apical vacuolar-type (H+)-ATPase (V(H+)-ATPase) and/or sodium/proton exchanger (NHE), the latter dependent on sodium/potassium-ATPase (Na+/K+-ATPase). HCO3− is enriched in the hemolymph by basolateral anion exchangers. Enzymes that support active ion transport are the intracellular carbonic anhydrase (CA) and, in terms of a general support of energy consuming mechanisms, arginine kinase (AK). CA is assumed to accelerate the dissociation of carbonic acid (H2CO3) and provide the substrate for H+ and HCO3− transporters [55]. AK catalyses the reversible dephosphorylation of phosphoarginine, contributing to the restoration of adenosine triphosphate (ATP) used in energy consuming processes [59]. The expression of corresponding genes, V(H+)-ATPase, AK and partial sequences of two alpha CAs was significantly up-regulated at intermediate CO2, whereas such mRNA concentrations were only moderately increased at high CO2 (II) (Table 2). These expression levels follow the course of the hemolymph HCO3− parameters and the more effective acid–base regulation of adult H. araneus under moderately elevated CO2. However, only a few sequences encoding for Na+/K+-ATPase were up-regulated under CO2, in contrast to their response to elevated temperature treatment (III). Under the combined effect of temperature and CO2 Na+/K+-ATPase was also up-regulated. Another enzyme, DOPA decarboxylase, which catalyses the biosynthesis of dopamine by decarboxylation of L-DOPA, was found down-regulated at moderate (intermediate) CO2 elevations (I). Elevated dopamine leads to increased sodium influx and concomitantly, increased Na+/K+-ATPase activity in gills of C. maenas
[60]. In light of this finding constant mRNA levels of Na+/K+-ATPase at intermediate CO2 (I) indicate that the down-regulation of DOPA decarboxylase may even prevent an activation of Na+/K+-ATPase (Table 2). The gene expression of other transporters, supposed to be involved in acid–base regulation, such as NHE and/or bicarbonate/chloride co-transporter were not influenced at all by elevated PCO2 values or temperature. Acid–base regulation predominantly via the V(H+)-ATPase (as seen in the present transcriptome) might involve minimal disturbance to ionic composition, e.g. cellular sodium homeostasis which would be affected by strong involvement of Na+/K+-ATPase. In the sipunculid S. nudus, extracellular acidosis induced a shift in ion transporters during hypercapnia from high to low energy cost mechanisms of acid–base regulation, resulting in decreased Na+/K+-ATPase activity due to lower requirement for sodium regulation [25, 27]. In line with our present observations these results suggest a pH regulation system independent of Na+/K+-ATPase being used under hypercapnic exposure (Figure 4). No significant up-regulation of apical bicarbonate anion exchangers was observed. In C. sapidus, CO2 induced acid–base disturbances (PCO2 10,000 μatm) were mainly compensated by an uptake of HCO3− from the surrounding seawater via the gill epithelia [39]. However, our results with no up-regulation of respective transporters suggests hemolymph buffering may be achieved through the dissociation of respiratory CO2 resulting in an accumulation of HCO3− in the hemolymph. This might be exceptionally for crustaceans with low compensatory capacities. In H. araneus, also no significant up-regulation of basolateral bicarbonate anion exchangers was observed. However, expression of the anion channel bestrophin was significantly enhanced (Table 2, Figure 4). Bestrophin is activated by calcium (Ca2+) and enhances membrane permeability for anions, such as chloride and HCO3−
[61]. Even though bestrophin is commonly found in the retinal pigment epithelium, ion channels of this family have been identified in different tissues and are involved in a variety of cellular processes [62]. Its role in responses to elevated CO2 remains to be investigated.
Figure 3.
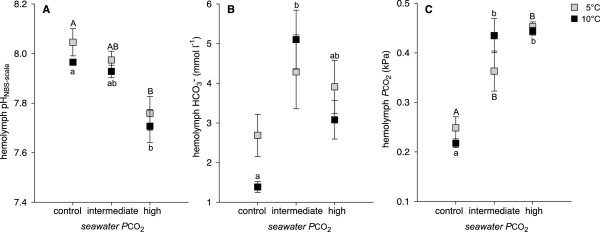
Hemolymph acid–base status Hyas araneus (A, pH-values; B, bicarbonate levels; C, P CO 2 levels) in response to different exposure experiments. Squares represent the means with error bars depicting the standard error for each treatment. Grey squares refer to treatments at 5°C and black squares to treatments at 10°C. One-way ANOVAs were used to identify the effect of seawater PCO2 concentration on hemolymph pH, bicarbonate (HCO3 −) and PCO2. A Holm-Sidak test for multiple comparisons was used for posteriori analysis (p < 0.05). Differing letters indicate significant differences; identical letters indicate no significant difference. Capital letters denote differences for 5°C treatments and lower cases for 10°C treatments.
Table 2.
Regulation of transcripts of specific interest in the hypercapnia and elevated temperature experiments on Hyas araneus
| Accession no. | Description | Rank | Treatment (I) | Treatment (II) | Treatment (III) | Treatment (IV) | Treatment (V) |
|---|---|---|---|---|---|---|---|
| HAAI01016321 | uricase | 10 | −5.24 | −4.27 | −3.68 | −7.10 | −6.20 |
| HAAI01006676 | trehalose-6-phosphate synthase 1a | 21 | −4.45 | −2.42 | −2.13 | −6.05 | −4.57 |
| HAAI01003297 | cuticle proprotein | 24 | 5.88 | 1.22 | 2.81 | 3.98 | 1.87 |
| HAAI01001762 | actin | 53 | −1.65 | 0.48 | 5.06 | −0.34 | −1.03 |
| HAAI01004150 | trehalose 6-phosphate synthase 1 | 61 | −4.35 | −1.68 | −2.73 | −4.88 | −3.99 |
| HAAI01015640 | vitellogenin like | 66 | 4.82 | 3.35 | 0.68 | 3.28 | −0.13 |
| HAAI01018061 | peroxiredoxin | 79 | 4.67 | 4.31 | 1.81 | 3.92 | 4.09 |
| HAAI01010911 | enoyl COA hydratase | 80 | −1.11 | −0.65 | 0.34 | −0.47 | −4.67 |
| HAAI01016834 | like adducin related protein | 83 | 4.62 | 3.22 | 0.81 | 3.30 | 0.29 |
| HAAI01016838 | GSH peroxidase like | 87 | 3.67 | 4.61 | 2.81 | 3.92 | 4.19 |
| HAAI01015788 | vitellogenin like | 100 | 4.48 | 3.61 | 1.22 | 3.60 | −0.71 |
| HAAI01000380 | glucose-6-phosphat dehydrogenase | 105 | −3.16 | −4.45 | −2.56 | −3.41 | −2.87 |
| HAAI01017747 | sodium glucose cotransporter | 113 | 2.94 | 4.36 | 0.64 | 1.98 | 1.29 |
| HAAI01002706 | trehalose 6-phosphate synthase 1b | 117 | −3.87 | −1.10 | −2.20 | −4.32 | −3.11 |
| HAAI01012389 | isocitrate dehydrogenase I | 128 | −1.18 | −2.36 | −0.55 | −4.19 | −2.88 |
| HAAI01000602 | dopa decarboxylase | 138 | −3.37 | −2.78 | −2.26 | −2.80 | −4.09 |
| HAAI01003033 | troponin I | 175 | −0.33 | 1.22 | 3.78 | −0.02 | 0.29 |
| HAAI01015542 | heat shock protein 90 | 192 | −3.07 | 1.07 | −3.68 | −0.09 | −3.30 |
| HAAI01002164 | alpha carbonic anhydrase | 197 | 3.67 | 1.80 | 1.64 | 3.45 | 1.87 |
| HAAI01019079 | ascorbate peroxidase | 198 | 3.67 | 2.11 | 1.17 | 2.23 | −2.52 |
| HAAI01009105 | heat shock protein 90 | 203 | 3.61 | 2.42 | 0.36 | 1.98 | −2.03 |
| HAAI01019113 | vitellogenin like | 226 | 3.52 | 2.39 | −0.36 | 2.48 | −1.56 |
| HAAI01018669 | vitellogenin like | 234 | 3.50 | 2.29 | 0.15 | 2.00 | −2.03 |
| HAAI01016527 | cytochrome p450 like | 236 | 3.23 | 2.03 | 0.07 | 3.47 | 2.61 |
| HAAI01009026 | cuticle protein like | 291 | −0.47 | 3.20 | 0.01 | 0.98 | −0.36 |
| HAAI01010727 | gelsolin precursor | 303 | 1.67 | 0.68 | 3.13 | 0.64 | 1.29 |
| HAAI01018844 | alpha tubulin | 309 | 3.11 | 1.41 | 1.46 | 2.74 | −0.20 |
| HAAI01008700 | arginine kinase | 338 | 3.01 | 2.27 | 1.43 | 2.76 | 2.14 |
| HAAI01019135 | carbohydrate phosphorylase like | 344 | 2.67 | 1.48 | 1.26 | 2.99 | 1.21 |
| HAAI01004058 | cuticle protein | 359 | 2.93 | 0.22 | −0.55 | 0.20 | −0.71 |
| HAAI01015598 | heat shock protein 90 | 367 | 2.90 | 1.47 | −0.19 | 1.85 | −0.71 |
| HAAI01019120 | bestrophin like | 368 | 2.89 | 1.8 | 2.13 | 1.56 | 1.16 |
| HAAI01000761 | heat shock protein 70 | 381 | 2.84 | 1.94 | 1.30 | 2.61 | 1.37 |
| HAAI01005842 | cuticle protein like | 386 | 2.81 | 0.48 | 1.54 | 0.63 | −0.45 |
| HAAI01001265 | beta tubulin | 403 | 2.77 | 1.77 | 0.97 | 1.98 | 1.02 |
| HAAI01018645 | V1-ATPase subunit | 427 | 2.69 | 1.80 | 1.27 | 2.54 | 1.46 |
| HAAI01018783 | cuticle protein | 435 | 2.67 | 0.71 | 1.49 | 1.50 | 1.61 |
| HAAI01004930 | heat shock protein 90 | 447 | 2.63 | 1.86 | 1.24 | 2.58 | 0.92 |
| HAAI01007246 | vitellogenin like | 455 | 2.61 | 2.15 | −1.10 | 0.98 | −1.45 |
| HAAI01003327 | actin | 461 | 2.53 | 2.26 | 1.06 | 2.60 | 1.72 |
| HAAI01000796 | alpha tubulin | 466 | 2.59 | 2.06 | 1.09 | 2.34 | 1.70 |
| HAAI01018213 | alpha-glucosidase | 477 | 1.83 | 1.26 | 1.56 | 2.55 | 2.39 |
| HAAI01005237 | alpha carbonic anhydrase like | 478 | 2.55 | 1.80 | 0.52 | 2.42 | 0.91 |
| HAAI01001455 | cuticle protein like | 498 | −1.26 | −1.86 | −0.58 | −2.44 | −2.51 |
| HAAI01014269 | cuticle protein like | 514 | −0.50 | −1.08 | −0.57 | −1.66 | −2.48 |
| HAAI01008420 | troponin I | 519 | 0.02 | −0.73 | −2.46 | −0.88 | −1.81 |
| HAAI01002591 | superoxide dismutase | 535 | −0.10 | −1.65 | −0.62 | −1.49 | −2.42 |
| HAAI01019124 | na + k + −atpase alpha subunit | 536 | 0.81 | 0.22 | 1.82 | 2.09 | 2.41 |
| HAAI01007529 | glyceraldehyde 3-phosphate dehydrogenase | 546 | 2.39 | 1.60 | 0.69 | 2.36 | 1.07 |
| HAAI01004651 | actin | 556 | 2.38 | 1.44 | 0.52 | 1.82 | 1.33 |
| HAAI01005807 | cytochrome c oxidase subunit ii | 559 | 2.23 | 1.82 | 0.96 | 2.37 | 1.33 |
| HAAI01001460 | cuticle protein like | 560 | −0.72 | −1.31 | −0.72 | −1.62 | −2.37 |
| HAAI01001217 | cuticle protein | 562 | 2.37 | 1.10 | 2.21 | 0.40 | −0.98 |
| HAAI01003904 | ankyrin related protein like | 576 | 1.33 | 1.27 | 2.33 | 0.82 | 0.59 |
| HAAI01000424 | cytochrome c oxidase subunit i | 583 | 2.32 | 1.56 | 1.10 | 2.28 | 0.84 |
| HAAI01001438 | nadh dehydrogenase subunit | 598 | 1.95 | 1.46 | 0.87 | 2.27 | 1.29 |
| HAAI01006730 | heat shock protein 90 | 608 | 2.24 | 1.44 | 0.96 | 2.13 | 1.29 |
| HAAI01005819 | cuticle protein like | 614 | 1.55 | 1.87 | 0.79 | 2.14 | 2.23 |
| HAAI01015787 | alpha tubulin | 630 | 2.20 | 0.85 | 0.47 | 1.88 | −0.27 |
| HAAI01002070 | vitellogenin like | 634 | 2.20 | 1.77 | −1.05 | 1.32 | −1.57 |
| HAAI01014788 | thioredoxin | 647 | −1.73 | −2.18 | −0.50 | −1.38 | −1.52 |
| HAAI01006091 | gelsolin precursor | 670 | 0.84 | 0.73 | 2.10 | 0.37 | −0.28 |
| HAAI01000874 | alpha tubulin | 673 | 2.03 | 2.09 | 0.90 | 1.31 | 1.69 |
| HAAI01000485 | cytochrome c oxidase subunit iii | 711 | 1.94 | 1.28 | 0.60 | 2.02 | 0.90 |
| HAAI01008219 | gelsolin precursor | 738 | 0.89 | 0.76 | 1.97 | 0.25 | −0.35 |
| HAAI01014184 | thioredoxin | 751 | −1.08 | −0.93 | −1.23 | −1.94 | −1.80 |
| HAAI01002593 | thioredoxin peroxidase | 769 | −1.89 | −0.72 | −0.98 | −1.52 | −1.52 |
| HAAI01005614 | cuticle protein like | 804 | 1.44 | 0.78 | 1.79 | 1.18 | −0.21 |
| HAAI01006347 | cuticle protein like | 817 | 1.28 | −0.22 | 1.75 | 1.37 | 1.42 |
| HAAI01018927 | spectrin like | 822 | 1.58 | 0.51 | 1.74 | 1.09 | 0.50 |
| HAAI01000352 | nesprin like | 827 | 1.70 | 0.72 | 1.73 | 1.42 | 1.58 |
Transcripts significantly regulated in response to hypercapnia and elevated temperature as identified by DESeq analysis (for details, see Methods). Accession number (accession no.) refers to the transcriptome of Hyas araneus [40] and the database ENA (EMBL). Details on transcript description and transcript length are listed for each transcript. Transcripts are sorted according to the rank in absolute regulation regardless of the treatment. Changes are given in log2-fold change for each treatment separately. Bold numbers represent significantly up-regulated transcripts; bold and italic numbers represent significantly down-regulated transcripts.
Figure 4.
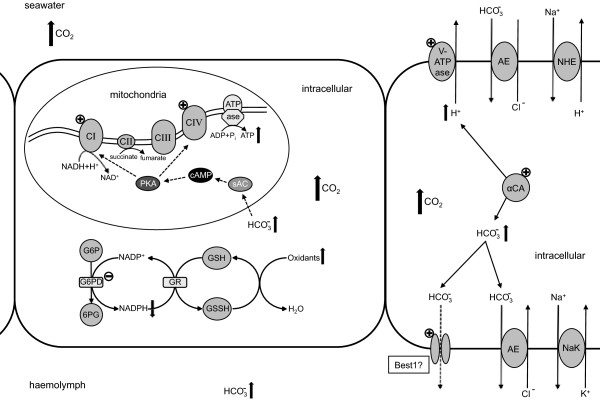
Schematic description of proposed processes in the gill epithelium of Hyas araneus in response intermediate P CO 2 exposure. Medium-term hypercapnia acclimation leads to a shift to a new acid–base equilibrium by accumulation of hemolymph bicarbonate (HCO3 −). CO2 is hydrated into H+ and HCO3 − by cytoplasmic carbonic anhydrase (CA). Protons are actively pumped out of the epithelial cell by an apical vacuolar proton ATPase (V(H+)-ATPase), followed by a transport of HCO3 − via a basolateral anion exchanger (AE) and/or ion channel, such as bestrophin (Best1). Increased energy demand is in part met by an enhanced expression of complex I (CI) and complex IV (CIV) of the electron transport system and possibly triggered by a soluble adenylyl cyclase (sAC) induced signalling pathway. sAC is stimulated by HCO3 − and increases the formation of cyclic adenosine monophosphate (cAMP), which activates protein kinase A (PKA) that subsequently leads to an induced expression of CI and CIV. While enhanced aerobic metabolic processes increase the generation of oxidants, the cellular defence against oxidative stress is adversely affected by a lower production of NADPH due to a decrease of the pentose phosphate pathway enzyme glucose-6-phosphate dehydrogenase (G6PD). NADPH acts as reducing agent for the regeneration of reduced glutathione (GSH) to oxidised glutathione (GSSH). The symbols (+) and (−) mark significantly up- and down-regulated genes, respectively modified after [55, 64].
Energy metabolism
Acid–base regulation compensating for CO2 induced disturbances likely entails a metabolic cost due to the ATP demanding movement of H+ by ion transporters [25]. Elevated metabolic rates have already been observed under hypercapnia exposure [26, 63], however, the mechanisms causing such cost increments remain unidentified. The differential expression analysis revealed an up-regulation of complex I and complex IV of the electron transport system (ETS) during exposure to intermediate CO2 levels (i,iv) (Table 2). Transcripts encoding for NADH dehydrogenase and cytochrome c oxidase subunits were significantly up-regulated above control levels possibly leading to an increase in mitochondrial density to meet the increased energy demand. Interestingly, complex II and III were not affected by hypercapnia exposure. Increased activities of complex I and IV activated by protein kinase A (PKA) resulted in an increased oxidative phosphorylation and ATP synthesis in human kidney cells [64]. The activation was triggered by a soluble adenylyl cyclase (sAC) induced signalling pathway that implies phosphorylation of PKA being stimulated by cyclic adenosine monophosphate (cAMP), which, in turn, is formed from sAC. In mammals and in elasmobranchs it is known that sAC is stimulated by HCO3− [65, 66], which would thereby support an increased activity of mitochondrial electron transport system under hypercapnia exposure. cAMP and PKA regulate key enzymes, such as complex IV, by alterating gene expression [66, 67]. Considering the higher extracellular bicarbonate levels in animals under hypercapnia (Figure 4) this signalling pathway possibly led to the increased up-regulation in ETS related genes and/or increased activity and thus might explain how the organisms meet the suggested increase in ATP demand (Figure 4).
An increased ATP production by the ETS would also lead to an elevated demand for metabolic substrates and turnover of the resulting reduction equivalents (NADH, FADH2). In the intermediate CO2 treatments (I, IV), the glycolytic pathway only experienced a significant up-regulation of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Table 2), whereas the rate-limiting enzymes phosphofructokinase or pyruvate kinase remained unaffected, suggesting no general up-regulation of glycolysis. However, the enhanced expression of sodium glucose transporter indicates increased capacity for glucose transport from the hemolymph into the gill cells (Table 2). Trehalose is the major hemolymph sugar in insects and decapod crustaceans, with higher levels than glucose [68, 69]. Enzymes for trehalose synthesis were found in crustacean tissues, including gills [70]. Trehalose and its fast transport into cells and consecutive transformation into glucose reflect its immediate availability to meet sudden bouts of energy demand. Accordingly, a 6.7 fold increase of trehalose concentration was measured in the hemolymph of C. maenas over 10 days under osmotic stress [71]. In H. araneus, however, the expression of trehalose-6-phosphate synthase was significantly down-regulated in all treatments (I-V) suggesting suppressed synthesis of the already depleted trehalose stores (Table 2). Significant up-regulation of transcripts encoding for glycogen phosphorylase (I, IV) and alpha glucosidase (IV) was found instead. Both enzymes catalyse the glucose releasing steps of glycogenolysis indicating the use of glycogen as a glucose source during long-term increased demand.
Enhanced demand for glucose is paralleled by a down-regulation of glucose-6-phosphate dehydrogenase (G6PD) in all treatments (I-V) indicating the potentially reduced production of NADPH (Table 2). G6PD is the key enzyme of the oxidative phase of the pentose phosphate pathway, the main source of NADPH for biosynthetic pathways in the cells (Figure 4). Furthermore, the cytosolic (NADP dependent) isocitrate dehydrogenase (IDH) was significantly down-regulated under intermediate CO2 levels at high temperature (IV), also suggesting lowered biosynthetic rates such as lipid biosynthesis under combined exposure (Table 2).
Oxidative stress
Besides being involved in lipid biosynthesis, NADPH is an important reducing agent in cellular antioxidative defence, e.g. by regenerating reduced glutathione, a major cellular antioxidant (Figure 4). Thus, besides a general down-regulation of anabolic reactions it seems conceivable that H. araneus encounters a reduced capacity to counteract oxidative stress under hypercapnic and thermal stress.
Significant changes in the expression of several genes involved in cellular antioxidant defence, including several peroxidases, indicate potential oxidative stress in H. araneus under intermediate CO2 exposure (I). These changes in expression level were less pronounced under high CO2 (II), suggesting a decreasing acclimation capacity of the H. araneus with increasing external PCO2(Table 2, see above). Among up-regulated genes, especially genes associated with the detoxification of hydrogen peroxide (H2O2) were affected under hypercapnia exposure. An ascorbate peroxidase (APX) was significantly up-regulated under intermediate CO2 concentrations (I). APX is a peroxidase that utilizes ascorbate as electron donor to detoxify H2O2 into water (Table 2). Additionally, a glutathione peroxidase (GPX) that reduces H2O2, using glutathione as substrate, was up-regulated under high CO2 exposure (II). This contrasts the down-regulation of two thiol-specific peroxiredoxin-1, at intermediate CO2 concentration (I), indicating a balanced response possibly by differential transcription of different splice variants. Peroxiredoxins are ubiquitous enzymes detoxifying peroxides, such as H2O2, by oxidising their active cysteine site using peroxide as substrate and are regenerated by oxidation of a thiol-containing electron donor, commonly thioredoxin [72]. However, only two sequences encoding for a thiol-containing protein (thioredoxin-1) were significantly down-regulated (II, IV).
The up-regulation of genes for anti-oxidants, such as glutathione peroxidase and peroxiredoxin, may indicate compensation for enhanced ROS (reactive oxygen species) production and concomitantly oxidative stress in the gill tissue of H. araneus under CO2 exposure. This is further supported by a significant up-regulation of a ribosomal cytochrome p450 like gene in the intermediate CO2 treatment (I). Cytochrome p450 is involved in the oxidative metabolism of a variety of organic substrates and incomplete catalytic processes can result in a continuous release of ROS [73, 74]. In contrast, a transcript encoding for urate oxidase (uricase) was significantly down-regulated in all treatments (I-V) (Table 2). Uricase is commonly located in the peroxisomes of the hepatopancreas tissue, however, uricase activity has also been detected in gill tissue of the kuruma shrimp Marsupenaeus japonicas [75]. Uricase catalyses the reaction from urate to allantoin and contributes to the generation of H2O2 by the oxidation of uric acid [76]. Consequently, a down-regulation of uricase may contribute to alleviate the generation of ROS. Interestingly, CO2 exposure also led to an increase in several vitellogenin like transcripts (Table 2). Although expression of vitellogenins is generally sex- and tissue-specific, the expression in both sexes of the mud shrimp Upogebia major [77], revealed a positive effect on oxidative stress resistance regardless of the developmental stage. Vitellogenin is also beneficial for oxidative stress resistance in honeybees, Apis mellifera [78]. Even if the function of vitellogenin in oxidative stress resistance is far from being completely understood and further investigations are needed to validate this hypothesis, the strong up-regulation of vitellogenin under CO2 (I, II, IV) may indicate that vitellogenin is an important protein in the resistance to CO2-induced oxidative stress (see below).
Hypercapnia induced enhancement of oxidative stress defence was recently demonstrated in the Eastern oyster, Crassostrea virginica [79]. In a proteomic approach, an up-regulation of several proteins, e.g. superoxide dismutase and several peroxiredoxins, was detected after exposure to high PCO2 (~3,520 μatm) for 2 weeks. The authors suggested several ways of how increased CO2 levels could directly or indirectly cause oxidative stress. On the one hand, a reaction of CO2 with peroxynitrite, a ROS formed through the reaction between superoxide anions and nitric oxide, resulting in the formation of reactive carbonate and nitrogen species, can lead to oxidative stress by oxidizing molecular compounds [80]. On the other hand, an indirect influence of elevated CO2 and/or pH could adversely affect mitochondrial functions and/or the non-enzymatic production of ROS [79]. However, our findings cannot support a direct influence of CO2 on oxidative stress generation, as one would assume an increase in the response to oxidative stress with increasing seawater CO2. Our data rather indicate an indirect influence of elevated CO2 on ROS production, likely by enhanced ROS production due to metabolic stimulation. The suggested increase in oxidative metabolic processes might cause enhanced ROS production and would also explain why the oxidative stress response was higher under intermediate CO2 than in the high CO2 treatment.
Cell structure
It is well known that the formation of ROS can damage lipids, proteins and DNA e.g. [81]. A large group of genes that belong to the functional category ‘cytoskeleton’ include several actins and tubulins which are up-regulated together with the antioxidant genes under CO2 exposure. The cytoskeleton is one major target for oxidative stress (Table 2) when the exposed cysteine component of actin forms oxidized derivates, such as intermolecular disulfide bridges [82]. This presumably has adverse effects on the interaction between actin and actin binding proteins and leads to changes in the structure of the actin cytoskeleton. The up-regulation of two transcripts encoding for actin in both intermediate CO2 treatments (I, IV) and the high CO2 treatment (II) may counter the damages caused by oxidative stress. An additional up-regulation of two actin binding proteins, nesprin and adducin, further supports the need for structural adaptation under oxidative stress triggered by CO2 exposure (Table 2). There is strong evidence that ROS induce the expression of tubulin [83], which could explain the up-regulation of β-tubulin and three transcript sequences encoding forα-tubulin in both intermediate CO2 treatments (I, IV) and the high CO2 (II) treatment (Table 2). Although the effect of oxidative stress on the cytoskeleton is still poorly understood and needs further investigation, an interaction of oxidative stress and adaptive changes in the cytoskeleton is well recognized [79, 82] and supported by our findings.
Besides enhanced antioxidative defence, a reorganisation of the cytoskeleton may occur in gill epithelia during the initial stage of hypercapnia acclimation. In C. maenas, a reorganisation of gill epithelia was observed after short-term exposure to very high PCO2 (~4,340 μatm; 7 days), but not after medium-term exposure to high PCO2 (~2,270 μatm; 11 weeks) [36]. Similarly, a reorganization of the cytoskeleton may not occur during medium-term CO2 acclimation of H. araneus (10 weeks). The response of genes that belong to the functional category ‘cytoskeleton’ was higher at intermediate CO2 (I, IV) than at high CO2 (II), in line with the findings in antioxidative defence. This also suggests that the changes observed result from oxidative stress via enhanced metabolic rate at intermediate CO2 with the cytoskeleton as a direct target of ROS formation.
Cytoskeletal genes also responded to elevated temperature (III). Several genes encoding for actin-binding proteins as well as membrane and cuticle proteins were significantly up-regulated (Table 2). Three transcripts of gelsolin, an actin-binding protein and regulator of actin filament assembly, were significantly up-regulated. The expression of an actin-binding nesprin, a membrane-binding ankyrin and an actin-related cytoskeletal structure protein spectrin was also increased. Warming affects various cellular processes, including stability and/or dynamics of the cytoskeleton for review see [84]. An increase in cytoskeletal gene expression despite the general down-regulation of expression levels indicates a requirement for the stabilization of the cytoskeleton, possibly elicited by the warm-induced stimulation of metabolic rate and associated ROS formation. Interestingly, such up-regulation of cytoskeletal gene expression could not be seen in under combined intermediate PCO2 and high temperature (IV, V), suggesting that the general down-regulation induced by high CO2 also affects cytoskeleton assembly.
Commonly, stresses such as temperature extremes, cellular energy depletion, and extreme concentrations of ions, osmolytes and gases induce the synthesis of heat-shock proteins (HSP) for review see [85]. However, HSP expression levels being unchanged in the high temperature treatment (III) suggest that 10°C is not a temperature extreme for the Arctic H. araneus population. In contrast, the expression of several HSPs was up-regulated under CO2 exposure (Table 2). One HSP70 was up-regulated in both the intermediate and the high CO2 treatments (I, II, IV). Four transcripts encoding for HSP90 were significantly up-regulated in the intermediate CO2 treatment (I) and two in the intermediate CO2 plus high temperature treatment (IV). In treatments I, III, and V one HSP90 showed decreased expression. Acting as molecular chaperones, HSPs can play an important role in maintaining proteins in a folded or unfolded state, controlling the unintended aggregation of proteins or target proteins for degradation [86]. Their induced expression, especially under intermediate CO2 concentrations, paired with increases in mRNA transcripts for proteins involved in antioxidant defence suggest an increased capacity to defend the cell against cellular damage. The up-regulation specifically of HSP90 suggests HSP90 to be of particular importance in maintaining protein homeostasis.
Conclusions
Based on a comprehensive expression analysis of genes related to acid–base regulation, metabolism, cell structure and their coupling to the stress response, this study has identified moderate, but distinct responses to ocean acidification in gills of adult H. araneus. We could demonstrate that the molecular response strongly depends on the CO2 concentration. At PCO2 values proposed for the end of the century, changes in expression suggest elevated metabolism caused by stimulated acid–base regulation and associated with increased oxidative stress. This up-regulation was attenuated at even higher PCO2 (business as usual scenario for Year 2300) with expression levels closer to control values (Figure 5), indicating down-regulation of these processes. Responses in transcripts related to acid–base regulation and energy metabolism were in line with those observed at elevated temperature. Observations under hypercapnia in the blue mussel M. edulis are in line with these conclusions. An increase in aerobic metabolism under intermediate PCO2 exposure (<2,400 μatm) was followed by a decrease at higher PCO2 [87, 88]. In earlier studies on marine invertebrates, high PCO2 exposure (~3,200-5,000 μatm) induced a decrease in metabolic rates. In the mussel Mytilus galloprovincialis and the decapod crab Necora puber, a decrease in respiration rates was seen in response to long-term exposure at a PCO2 of approx. 5,000 and 3,200 μatm, respectively [6, 9]. Our findings suggest a PCO2 dependent threshold where metabolic stimulation might turn into metabolic depression. In H. araneus this threshold may be reached at a PCO2 of approximately 2,000 μatm. Although further examinations besides transcriptomics covering a broader range of PCO2 concentrations are certainly needed to confirm a possible threshold, a recent meta-analysis revealed that 50% of all marine crustacean species were negatively affected at this PCO2 [5].
Figure 5.
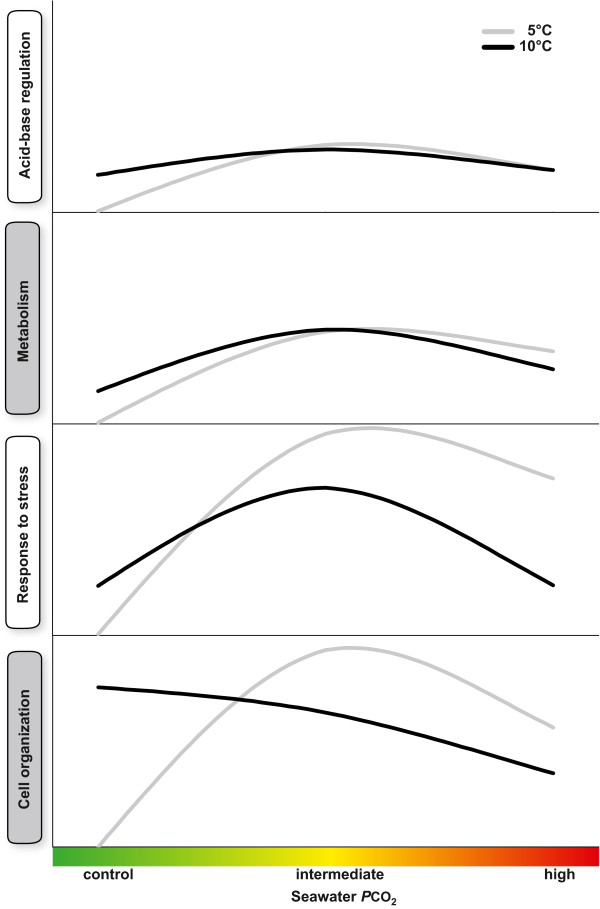
Schematic description of the response of Hyas araneus to different exposure experiments for specific categories. For each treatment, log2-fold changes of all up-regulated transcripts within one specific category were summed and plotted as a line (gray line = 5°C; black line = 10°C). Scales are harmonized for all categories. The figure shows how acclimation to warming reduces the transcriptomic response of ‘cell organization’ and ‘response to stress’ to elevated CO2 tensions.
Changes in the expression of genes related to energy metabolism paralleled the expression of those related to acid–base regulation and both paralleled the changes in extracellular pH (pHe), indicating a feedback regulation between pH and metabolic rate (Figure 5). In the marine worm S. nudus and the mussel M. galloprovincialis, metabolic depression under CO2 was caused by a drop in pHe [6, 27, 89]. H. araneus was able to partially compensate for the acidosis via bicarbonate (HCO3−) accumulation, but capacities were not sufficient to fully compensate. At both experimental temperatures (5°C and 10°C) hemolymph acidosis was compensated to the same extent. At high CO2 concentrations, the capacity to fully compensate may be even more limited related to decreased ion transporter expression and activities. Since acid–base regulation is an ATP-consuming process, the decreased expression of metabolism related genes might at least in part be explained by a pHe-triggered inhibition.
These shifts cause changes in energy balance which can therefore reflect the limits in stress tolerance [90]. A bioenergetic framework [90] builds on the physiological concepts of oxygen- and capacity-limited thermal tolerance (OCLTT) [91–93] and the dynamic energy budget (DEB) [94]. Two cases distinguished between physiological responses to moderate and extreme environmental stress. Moderate stress induces additional ATP turnover along with a higher metabolic rate, in H. araneus exemplified in enhanced acid–base regulation and cellular damage repair from oxidative stress at intermediate PCO2. These mechanisms support long-term persistence at the expense of a shift in energy budget (pejus range) [95]. Even if the enhanced energy demand of stress resistance can be met by increased feeding, it may still result in a reallocation of energy from fitness-related functions such as reproduction and growth to maintenance and damage repair [90]. More extreme stress exacerbates the disturbances in homeostasis, exemplified in stronger disturbances in acid–base status as seen in H. araneus favouring a suppression of metabolic rate [27] which preserve energy resources and lessen the generation of detrimental metabolic by-products [90]. In H. araneus the respective response to high PCO2 was paralleled by a lower expression of transcripts associated with energy metabolism, stress response and cellular organization. Depressed metabolism would result in insufficient energy balance and time-limited tolerance to environmental stress making long-term survival impossible (pessimum range) [90, 93].
To our knowledge, we report the first gene expression profiling study analysing the responses to a combination of two drivers in an osmoconforming crustacean. The study confirms the interdependence of physiological processes affected by elevated seawater PCO2 and temperature. Furthermore, the study demonstrates the importance of considering projected climate scenarios in experimental work, as responses to increasing seawater CO2 concentrations are not necessarily linear, as presented here.
Electronic supplementary material
Additional file 1: Table S1: Summary of sequencing and mapping results for Hyas araneus. Details on treatment, filename and location of sequencing raw data, used sequencer, read count, read length and total size in base pairs (bp), used alignment tool, aligned reads and percentage of aligned reads are listed separately. (XLSX 57 KB)
Additional file 2: Table S2: Details on primers for quantitative real-time polymerase chain reaction (qRT-PCR) to validate RNASeq data of Hyas araneus. Forward and backward primer sequences and descriptions of target genes used in the qRT-PCR. Accession number (accession no.) refer to the transcriptome of Hyas araneus [40] and the database ENA (EMBL). R2 and efficiency was tested in a qRT-PCR dilution series (for details, see Methods). (XLSX 40 KB)
Additional file 3: Figure S1: Changes of expression levels of transcripts in gills of Hyas araneus responding to medium-term exposure (10 weeks) at intermediate PCO2 (≈1,000 μatm) and high temperature (10°C), analysed by DESeq (gray bars) and quantitative real-time polymerase chain reaction (qRT-PCR) (black bars). Bars represent the mean log2-fold change and standard error (error bars) of the respective gene. Transcripts correspond to primers and genes used in the qRT-PCR (see Additional file 5: Table S3). (PDF 78 KB)
Additional file 4: Figure S2: Linear regression between expression levels of transcripts in gills of Hyas araneus responding to medium-term exposure (10 weeks) at intermediate PCO2 (≈1,000 μatm) and high temperature (10°C), analysed by DESeq and quantitative real-time polymerase chain reaction (qRT-PCR). Black dots represent the mean log2-fold change of transcripts analysed by DESeq plotted against the corresponding mean log2-fold change analysed by qRT-PCR. r was determined by Pearson Correlation using SigmaPlot 12.0 (Systat Software Inc., San Jose, USA). (PDF 53 KB)
Additional file 5: Table S3: Transcript levels changing significantly in gills of Hyas araneus responding to hypercapnia and elevated temperature. Transcripts regulated significantly in response to hypercapnia and elevated temperature as identified by DESeq analysis (for details, see Methods). ID and accession number (accession no.) refer to the transcriptome of Hyas araneus [40] and the database ENA (EMBL). Details on transcript description and transcript length are listed for each transcript. Transcripts are sorted according to the rank in absolute regulation regardless of the treatment. Changes are given in log2-fold change for each treatment separately. For details on treatments, see Methods). Bold numbers represent significantly up-regulated transcripts and bold and underlined numbers significantly down-regulated transcripts. (XLSX 100 KB)
Additional file 6: Figure S3: Smearplot of differentially expressed transcripts in gills of Hyas araneus. All transcripts changed in response to hypercapnia and elevated temperature. Log2-fold changes are plotted against mean readcount (log10). Blue dots represent transcripts with non-significant changes, red dots depict transcripts significantly regulated as identified by DESeq analysis (p < 0.05) and green triangles are transcripts changed significantly and identified by annotation Numbers refer to the total number of significantly up-/down-regulated transcripts. A) treatment (I) = 1,120 μatm PCO2 5°C; B) treatment (II) = 1,960 μatm PCO2 5°C; C) treatment (III) = 390 μatm PCO2 10°C; D) treatment (IV) = 1,120 μatm PCO2 10°C; E) treatment (V) = 1,960 μatm PCO2 10°C. (PDF 492 KB)
Additional file 7: Table S4: Enrichment analysis in the RNASeq study on Hyas araneus. Results of the enrichment analysis (Fisher’s Exact Test; FDR < 0.05) as implemented in Blast2GO [49, 50] and reduced by web-based clustering tool REVIGO [51]. Tested were subsets of all significantly regulated transcripts as identified by DESeq (separated by up- and down-regulated transcripts). Reference-set was the full set of annotated sequences of the H. araneus transcriptome [40]. Listed is the Gene Ontology term (GO-term), the name of the functional group (description), the category (molecular function, biological process or cellular component), the false discovery rate (FDR) and whether GO-terms are over- or under-represented (a GO-term is considered over-/under-represented if it appears significantly more often/less often in the test-set than in the reference-set. (XLSX 33 KB)
Acknowledgements
The authors would like to thank Max Schwanitz for animal collection and Laura Stapp for useful discussions. Financial support was provided by Federal Ministry of Education and Research (BMBF), within phase I of the BIOACID research programme (FKZ 03F0608B, subproject 2.2.1).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LH and MS conducted lab experiments. LH and SF performed bioinformatics analysis. LH, FCM, DS, CH, HOP and ML conceived the study and coordinated research activities. LH wrote the manuscript; all authors revised and approved the final manuscript.
Contributor Information
Lars Harms, Email: Lars.Harms@awi.de.
Stephan Frickenhaus, Email: Stephan.Frickenhaus@awi.de.
Melanie Schiffer, Email: Melanie.Schiffer@awi.de.
Felix Christopher Mark, Email: Felix.Christopher.Mark@awi.de.
Daniela Storch, Email: Daniela.Storch@awi.de.
Christoph Held, Email: Christoph.Held@awi.de.
Hans-Otto Pörtner, Email: Hans.Poertner@awi.de.
Magnus Lucassen, Email: Magnus.Lucassen@awi.de.
References
- 1.Pörtner HO, Langenbuch M, Michaelidis B. Synergistic effects of temperature extremes, hypoxia, and increases in CO2 on marine animals: from Earth history to global change. J Geophys Res. 2005;110(C9):1–15. [Google Scholar]
- 2.Fabry VJ, Seibel BA, Feely RA, Orr JC. Impacts of ocean acidification on marine fauna and ecosystem processes. ICES J Mar Sci. 2008;65:414–432. doi: 10.1093/icesjms/fsn048. [DOI] [Google Scholar]
- 3.Melzner F, Gutowska MA, Langenbuch M, Dupont S, Lucassen M, Thorndyke MC, Bleich M, Pörtner H-O. Physiological basis for high CO2 tolerance in marine ectothermic animals: pre-adaption through lifestyle and ontogeny? Biogeosciences. 2009;6:1–19. doi: 10.5194/bg-6-2313-2009. [DOI] [Google Scholar]
- 4.Kroeker KJ, Kordas RL, Crim R, Hendriks IE, Ramajo L, Singh GS, Duarte CM, Gattuso JP. Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob Chang Biol. 2013;19(6):1884–1896. doi: 10.1111/gcb.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wittmann A, Pörtner H-O. Nat Clim Chang. 2013. Sensitivities of extant animal taxa to ocean acidification. [Google Scholar]
- 6.Michaelidis B, Ouzounis C, Paleras A, Pörtner H-O. Effects of long-term moderate hypercapnia on acid–base balance and growth rate in marine mussels Mytilus galloprovincialis. Mar Ecol Prog Ser. 2005;293:109–118. doi: 10.3354/meps293109. [DOI] [Google Scholar]
- 7.Kurihara H. Effects of CO2-driven ocean acidification on the early developmental stages of invertebrates. Mar Ecol Prog Ser. 2008;373:275–284. doi: 10.3354/meps07802. [DOI] [Google Scholar]
- 8.Wood HL, Spicer JI, Widdicombe S. Ocean acidification may increase calcification rates, but at a cost. Proc Biol Sci/ Royal Soc. 2008;275(1644):1767–1773. doi: 10.1098/rspb.2008.0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Small D, Calosi P, White D, Spicer JI, Widdicombe S. Impact of medium-term exposure to CO2 enriched seawater on the physiological functions of the velvet swimming crab Necora puber. Aquat Biol. 2010;10(1):11–21. doi: 10.3354/ab00266. [DOI] [Google Scholar]
- 10.Gutowska MA, Pörtner HO, Melzner F. Growth and calcification in the cephalopod Sepia officinalis under elevated seawater pCO2. Mar Ecol Prog Ser. 2008;373:303–309. doi: 10.3354/meps07782. [DOI] [Google Scholar]
- 11.Whiteley NM. Physiological and ecological responses of crustaceans to ocean acidification. Mar Ecol Prog Ser. 2011;430:257–271. doi: 10.3354/meps09185. [DOI] [Google Scholar]
- 12.Pane EF, Barry JP. Extracellular acid–base regulation during short-term hypercapnia is effective in a shallow-water crab, but ineffective in a deep-sea crab. Mar Ecol Prog Ser. 2007;334:1–9. doi: 10.3354/meps334001. [DOI] [Google Scholar]
- 13.Walther K, Anger K, Pörtner H-O. Effects of ocean acidification and warming on the larval development of the spider crab Hyas araneus from different latitudes (54° vs. 79°N) Mar Ecol Prog Ser. 2010;417:159–170. doi: 10.3354/meps08807. [DOI] [Google Scholar]
- 14.Schiffer M, Harms L, Pörtner HO, Lucassen M, Mark FC, Storch D. Tolerance of Hyas araneus zoea I larvae to elevated seawater PCO2 despite elevated metabolic costs. Mar Biol. 2012;160(8):1943–1953. doi: 10.1007/s00227-012-2036-0. [DOI] [Google Scholar]
- 15.Walther K, Sartoris F-J, Bock C, Pörtner H-O. Impact of anthropogenic ocean acidification on thermal tolerance of the spider crab Hyas araneus. Biogeosciences. 2009;6:2207–2215. doi: 10.5194/bg-6-2207-2009. [DOI] [Google Scholar]
- 16.Zittier ZMC, Hirse T, Pörtner H-O. The synergistic effects of increasing temperature and CO2 levels on activity capacity and acid–base balance in the spider crab, Hyas araneus. Mar Biol. 2013;160(8):2049–2062. doi: 10.1007/s00227-012-2073-8. [DOI] [Google Scholar]
- 17.Spicer JI, Raffo A, Widdicombe S. Influence of CO2-related seawater acidification on extracellular acid–base balance in the velvet swimming crab Necora puber. Mar Biol. 2006;151(3):1117–1125. doi: 10.1007/s00227-006-0551-6. [DOI] [Google Scholar]
- 18.Dissanayake A, Ishimatsu A. Synergistic effects of elevated CO2 and temperature on the metabolic scope and activity in a shallow-water coastal decapod (Metapenaeus joyneri; Crustacea: Penaeidae) ICES J Mar Sci. 2011;68(6):1147–1154. doi: 10.1093/icesjms/fsq188. [DOI] [Google Scholar]
- 19.Carter HA, Ceballos-Osuna L, Miller NA, Stillman JH. Impact of ocean acidification on metabolism and energetics during early life stages of the intertidal porcelain crab Petrolisthes cinctipes. J Exp Biol. 2013;216(Pt 8):1412–1422. doi: 10.1242/jeb.078162. [DOI] [PubMed] [Google Scholar]
- 20.Wickins JF. The effect of hypercapnia sea water on growth and mineralization in Penaid prawns. Aquaculture. 1984;41:37–48. doi: 10.1016/0044-8486(84)90388-0. [DOI] [Google Scholar]
- 21.Long WC, Swiney KM, Harris C, Page HN, Foy RJ. Effects of Ocean Acidification on Juvenile Red King Crab (Paralithodes camtschaticus) and Tanner Crab (Chionoecetes bairdi) growth, condition, calcification, and survival. PLoS One. 2013;8(4):e60959. doi: 10.1371/journal.pone.0060959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurihara H, Shimode S, Shirayama Y. Effects of raised CO2 concentration on the egg production rate and early development of two marine copepods (Acartia steueri and Acartia erythraea) Mar Pollut Bull. 2004;49(9–10):721–727. doi: 10.1016/j.marpolbul.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Findlay HS, Kendall MA, Spicer JI, Widdicombe S. Future high CO2 in the intertidal may compromise adult barnacle Semibalanus balanoides survival and embryonic development rate. Mar Ecol Prog Ser. 2009;389:193–202. doi: 10.3354/meps08141. [DOI] [Google Scholar]
- 24.Watanabe Y, Yamaguchi A, Ishida H, Harimoto T, Suzuki S, Sekido Y, Ikeda T, Shirayama Y, Mac Takahashi M, Ohsumi T, Ishizaka J. Lethality of increasing CO2 levels on deep-sea copepods in the Western North Pacific. J Oceanogr. 2006;62:185–196. doi: 10.1007/s10872-006-0043-9. [DOI] [Google Scholar]
- 25.Pörtner H-O, Bock C, Reipschläger A. Modulation of the cost of pHi regulation during metabolic depression: a 31P-NMR study in invertebrate (Sipunculus nudus) isolated muscle. J Exp Biol. 2000;203:2417–2428. doi: 10.1242/jeb.203.16.2417. [DOI] [PubMed] [Google Scholar]
- 26.Stumpp M, Wren J, Melzner F, Thorndyke MC, Dupont ST. CO2 induced seawater acidification impacts sea urchin larval development I: elevated metabolic rates decrease scope for growth and induce developmental delay. Comp Biochem Physiol A Mol Integr Physiol. 2011;160(3):331–340. doi: 10.1016/j.cbpa.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 27.Reipschläger A, Pörtner H-O. Metabolic depression during environmental stress: the role of extracellular versus intracellular pH in Sipunculus nudus. J Exp Biol. 1996;199:1801–1807. doi: 10.1242/jeb.199.8.1801. [DOI] [PubMed] [Google Scholar]
- 28.Langenbuch M, Pörtner H-O. Changes in metabolic rate and N excreation in marine invertebrate Sipunculus nudus under conditions of environmental hypercapnia: identifying effective acid–base variables. J Exp Biol. 2002;205:1153–1160. doi: 10.1242/jeb.205.8.1153. [DOI] [PubMed] [Google Scholar]
- 29.Todgham AE, Hofmann GE. Transcriptomic response of sea urchin larvae Strongylocentrotus purpuratus to CO2-driven seawater acidification. J Exp Biol. 2009;212(Pt 16):2579–2594. doi: 10.1242/jeb.032540. [DOI] [PubMed] [Google Scholar]
- 30.Gracey AY. Interpreting physiological responses to environmental change through gene expression profiling. J Exp Biol. 2007;210(Pt 9):1584–1592. doi: 10.1242/jeb.004333. [DOI] [PubMed] [Google Scholar]
- 31.Stillman JH, Tagmount A. Seasonal and latitudinal acclimatization of cardiac transcriptome responses to thermal stress in porcelain crabs, Petrolisthes cinctipes. Mol Ecol. 2009;18(20):4206–4226. doi: 10.1111/j.1365-294X.2009.04354.x. [DOI] [PubMed] [Google Scholar]
- 32.Lockwood BL, Sanders JG, Somero GN. Transcriptomic responses to heat stress in invasive and native blue mussels (genus Mytilus): molecular correlates of invasive success. J Exp Biol. 2010;213(Pt 20):3548–3558. doi: 10.1242/jeb.046094. [DOI] [PubMed] [Google Scholar]
- 33.Lockwood BL, Somero G. Transcriptomic responses to salinity stress in invasive and native blue mussels (genus Mytilus) Mol Ecol. 2010;20(3):517–529. doi: 10.1111/j.1365-294X.2010.04973.x. [DOI] [PubMed] [Google Scholar]
- 34.Gracey AY, Troll JV, Somero G. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. PNAS. 2001;98(4):1993–1998. doi: 10.1073/pnas.98.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sussarellu R, Fabiouxa C, Le Moullacb G, Fleuryc E, Moragaa D. Transcriptomic response of the Pacific oyster Crassostrea gigas to hypoxia. Mar Genomics. 2010;3(3–4):133–143. doi: 10.1016/j.margen.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Fehsenfeld S, Kiko R, Appelhans Y, Towle DW, Zimmer M, Melzner F. Effects of elevated seawater pCO(2) on gene expression patterns in the gills of the green crab, Carcinus maenas. BMC Genomics. 2011;12:488. doi: 10.1186/1471-2164-12-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans TG, Chan F, Menge BA, Hofmann GE. Transcriptomic responses to ocean acidification in larval sea urchins from a naturally variable pH environment. Mol Ecol. 2013;22(6):1609–1625. doi: 10.1111/mec.12188. [DOI] [PubMed] [Google Scholar]
- 38.Henry RP, Cameron JN. The role of carbonic anhydrase in respiration, ion regulation and acid–base balance in the aquatic crab Callinectes sapidus and the terrestrial crab Gecarcinus lateralis. J Exp Biol. 1983;103:205–223. [Google Scholar]
- 39.Henry RP, Wheatly MG. Interaction of respiration, ion regulation, and acid–base balance in the everyday life of aquatic crustaceans. Am Zool. 1992;32:407–416. [Google Scholar]
- 40.Harms L, Frickenhaus S, Schiffer M, Mark FC, Storch D, Pörtner H-O, Held C, Lucassen M. Characterization and analysis of a transcriptome from the boreal spider crab Hyas araneus. Comp Biochem Physiol Part D Genomics Proteomics. 2013;8(4):344–351. doi: 10.1016/j.cbd.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Lewis E, Wallace D. ORNL/CDIAC-105a. Oak Ridge, Tennesse: Carbon Dioxide Information Analysis Center, Oak Ridge National Laboratory, U.S. Department of Energy; 1998. Program Developed for CO2 System Calculations. [Google Scholar]
- 42.Lenfant C, Aucutt C. Measurement of blood gasses by gas chromatography. Respir Physiol. 1966;1:398–407. doi: 10.1016/0034-5687(66)90007-7. [DOI] [PubMed] [Google Scholar]
- 43.Pörtner H-O, Boutillier RG, Tang Y, Toews DP. Determination of intracellular pH and PCO2 after metabolic inhibition by fluoride and nitrilotracetic acid. Respir Physiol. 1990;81:255–274. doi: 10.1016/0034-5687(90)90050-9. [DOI] [PubMed] [Google Scholar]
- 44.Pörtner H-O, Bickmeyer U, Bleich M, Bock C, Brownlee C, Melzner F, Michaelidis B, Sartoris F-J, Storch D. Studies of acid–base status and regulation. In: Riebesell U, Fabry VJ, Hansson L, Gattuso J-P, editors. Guide of Best Practise for Ocean Acidification Research and Data Reporting. Luxembourg: Publications Office of the European Union; 2010. pp. 137–166. [Google Scholar]
- 45.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 48.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 50.Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawahara-Miki R, Wada K, Azuma N, Chiba S. Expression profiling without genome sequence information in a non-model species, pandalid shrimp (Pandalus latirotris), by next-generation sequencing. PLoS One. 2011;6(11):e26043. doi: 10.1371/journal.pone.0026043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Somero G, Hochachka W. Biochemical adaptation to the environment. Am Zool. 1971;11:159–167. [Google Scholar]
- 54.Peterson ME, Daniel RM, Danson MJ, Eisenthal R. The dependence of enzyme activity on temperature: determination and validation of parameters. Biochem J. 2007;402(2):331–337. doi: 10.1042/BJ20061143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freire CA, Onken H, McNamara JC. A structure-function analysis of ion transport in crustacean gills and excretory organs. Comp Biochem Physiol A Mol Integr Physiol. 2008;151(3):272–304. doi: 10.1016/j.cbpa.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 56.Henry RP, Lucu C, Onken H, Weihrauch D. Multiple functions of the crustacean gill: osmotic/ionic regulation, acid–base balance, ammonia excretion, and bioaccumulation of toxic metals. Front Physiol. 2012;3:431. doi: 10.3389/fphys.2012.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Compere P, Wanson S, Pequeux A, Gilles R, Goffinet G. Ultrastructural changes in the gill epithelium of the green crab Carcinus maenas in relation to the external salinity. Tissue Cell. 1989;21(2):299–318. doi: 10.1016/0040-8166(89)90073-6. [DOI] [PubMed] [Google Scholar]
- 58.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 59.Kotlyar S, Weihrauch D, Paulsen RS, Towle DW. Expression of arginine kinase enzymatic activity and mRNA in gills of the euryhaline crabs Carcinus maenas and Callinectes sapidus. J Exp Biol. 2000;205:2395–2404. doi: 10.1242/jeb.203.16.2395. [DOI] [PubMed] [Google Scholar]
- 60.Sommer MJ, Mantel LH. Effect of dopamine, cyclic AMP, and pericardial organs on sodium uptake and Na/K-ATPase activity in gills of the green crab Carcinus maenas. J Exp Zool. 1988;248:272–277. doi: 10.1002/jez.1402480305. [DOI] [Google Scholar]
- 61.Lee S, Yoon BE, Berglund K, Oh SJ, Park H, Shin HS, Augustine GJ, Lee CJ. Channel-mediated tonic GABA release from glia. Science. 2010;330(6005):790–796. doi: 10.1126/science.1184334. [DOI] [PubMed] [Google Scholar]
- 62.Tsunenari T, Sun H, Williams J, Cahill H, Smallwood P, Yau KW, Nathans J. Structure-function analysis of the bestrophin family of anion channels. J Biol Chem. 2003;278(42):41114–41125. doi: 10.1074/jbc.M306150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beniash E, Ivanina A, Lieb NS, Kurochkin I, Sokolova IM. Elevated level of carbon dioxide affects metabolism and shell formation in oysters Crassostrea virginica (Gmelin) Mar Ecol Prog Ser. 2010;419:95–108. doi: 10.3354/meps08841. [DOI] [Google Scholar]
- 64.Acin-Perez R, Salazar E, Kamenetsky M, Buck J, Levin LR, Manfredi G. Cyclic AMP produced inside mitochondria regulates oxidative phosphorylation. Cell Metab. 2009;9(3):265–276. doi: 10.1016/j.cmet.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tresguerres M, Parks SK, Salazar E, Levin LR, Goss GG, Buck J. Bicarbonate-sensing soluble adenylyl cyclase is an essential sensor for acid/base homeostasis. Proc Natl Acad Sci U S A. 2010;107(1):442–447. doi: 10.1073/pnas.0911790107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zippin JH, Levin LR, Buck J. CO2/HCO3−responsive soluble adenylyl cyclase as a putative metabolic sensor. Endocrinol Metab. 2001;12(8):366–370. doi: 10.1016/s1043-2760(01)00454-4. [DOI] [PubMed] [Google Scholar]
- 67.Gopalakrishnan L, Scarpulla RC. Differential regulation of respiratory chain subunits by a CREB-dependent signal transduction pathway. J Biol Chem. 1993;269(1):105–113. [PubMed] [Google Scholar]
- 68.Wyatt GR, Kalf GF. The chemistry of insect hemolymph. J Gen Physiol. 1957;40(6):833–847. doi: 10.1085/jgp.40.6.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnston MA, Davies PS. Carbohydrates of the hepatopancreas and blood tissue of Carcinus. Comp Biochem Physiol. 1971;41B:433–443. [Google Scholar]
- 70.Chung JS. A trehalose 6-phosphate synthase gene of the hemocytes of the blue crab, Callinectes sapidus: cloning, the expression, its enzyme activity and relationship to hemolymph trehalose levels. Saline Syst. 2008;4:18. doi: 10.1186/1746-1448-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siebers D, Lucu C, Sperling K-R, Eberlein K. Kinetics of osmoregulation in the crab Carcinus maenas. Mar Biol. 1972;17:291–303. doi: 10.1007/BF00366739. [DOI] [Google Scholar]
- 72.Hofmann B, Hecht H-J, Flohé L. Peroxiredoxins. Biol Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 73.Premereur N, Van den Branden C, Roels F. Cytochrome P-450-dependent H2O2 production demonstrated in vivo. Funct Ecol. 1986;199(1):415–424. doi: 10.1016/0014-5793(86)81215-7. [DOI] [PubMed] [Google Scholar]
- 74.Livingstone DR, Marth PG, Michel X, Narbonne JF, O'hara S, Ribera D, Winston GW. Oxyradical production as a pollution-mediated mechanism of toxicity in the common mussel, Mytilus edulis L., and other molluscs. Funct Ecol. 1990;4(3):415–424. doi: 10.2307/2389604. [DOI] [Google Scholar]
- 75.Cheng SY, Lee WC, Chen JC. Increase of uricogenesis in the kuruma shrimp Marsupenaeus japonicus reared under hyper-osmotic conditions. Comp Biochem Physiol B Biochem Mol Biol. 2004;138(3):245–253. doi: 10.1016/j.cbpc.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 76.Boveris A, Oshino N, Chance B. The cellular production of hydrogen peroxide. Biochem J. 1972;128:617–630. doi: 10.1042/bj1280617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kang BJ, Nanri T, Lee JM, Saito H, Han CH, Hatakeyama M, Saigusa M. Vitellogenesis in both sexes of gonochoristic mud shrimp, Upogebia major (Crustacea): analyses of vitellogenin gene expression and vitellogenin processing. Comp Biochem Physiol B Biochem Mol Biol. 2008;149(4):589–598. doi: 10.1016/j.cbpb.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 78.Seehuus SC, Norberg K, Gimsa U, Krekling T, Amdam GV. Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc Natl Acad Sci U S A. 2006;103(4):962–967. doi: 10.1073/pnas.0502681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tomanek L, Zuzow MJ, Ivanina AV, Beniash E, Sokolova IM. Proteomic response to elevated PCO2 level in eastern oysters, Crassostrea virginica: evidence for oxidative stress. J Exp Biol. 2011;214(Pt 11):1836–1844. doi: 10.1242/jeb.055475. [DOI] [PubMed] [Google Scholar]
- 80.Denicola A, Freeman BA, Trujillo M, Radi R. Peroxynitrite reaction with carbon dioxide/bicarbonate: kinetics and influence on peroxynitrite-mediated oxidations. Arch Biochem Biophys. 1996;333(1):49–58. doi: 10.1006/abbi.1996.0363. [DOI] [PubMed] [Google Scholar]
- 81.Livingstone DR. Contaminant-stimulated reactive oxygen species production and oxidative damage in aquatic organisms. Mar Pollut Bull. 2001;42(8):656–666. doi: 10.1016/S0025-326X(01)00060-1. [DOI] [PubMed] [Google Scholar]
- 82.Dalle-Donne I, Rossi R, Milzani A, Simplicio PD, Colombo R. The actin cytoskeleton response to oxidants: from small heat shock protein phosphorilation to changes in the redox state of actin itself. Free Radic Biol Med. 2001;31(12):1624–1632. doi: 10.1016/S0891-5849(01)00749-3. [DOI] [PubMed] [Google Scholar]
- 83.Courgeon A-M, Maingourd M, Maisonhaute C, Montmory C, Rollet E, Tanguay RM, Best-Bellpomme M. Effect of hydrogen peroxide on cytoskeletal proteins of Drosophila cells: comparison with heat shock and other stressors. Exp Cell Res. 1993;204:30–37. doi: 10.1006/excr.1993.1005. [DOI] [PubMed] [Google Scholar]
- 84.Lindquist S. The heat-shock response. Ann Rev Biochemistry. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 85.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones and the stress response: evolutionary and ecological physiology. Ann Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 86.Wickner S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286(5446):1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 87.Thomsen J, Melzner F. Moderate seawater acidification does not elicit long-term metabolic depression in the blue mussel Mytilus edulis. Mar Biol. 2010;157(12):2667–2676. doi: 10.1007/s00227-010-1527-0. [DOI] [Google Scholar]
- 88.Hüning AK, Melzner F, Thomsen J, Gutowska MA, Krämer L, Frickenhaus S, Rosenstiel P, Pörtner H-O, Philipp EER, Lucassen M. Impacts of seawater acidification on mantle gene expression patterns of the Baltic Sea blue mussel: implications for shell formation and energy metabolism. Mar Biol. 2013;160(8):1845–1861. doi: 10.1007/s00227-012-1930-9. [DOI] [Google Scholar]
- 89.Pörtner H-O, Reipschläger A, Heisler N. Acid–base regulation, metabolism and energetics in Sipunculus nudus as a function of ambiant carbon dioxide level. J Exp Biol. 1998;201:43–55. doi: 10.1242/jeb.201.1.43. [DOI] [PubMed] [Google Scholar]
- 90.Sokolova IM, Frederich M, Bagwe R, Lannig G, Sukhotin AA. Energy homeostasis as an integrative tool for assessing limits of environmental stress tolerance in aquatic invertebrates. Mar Environ Res. 2012;79:1–15. doi: 10.1016/j.marenvres.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 91.Frederich M, Pörtner H-O. Oxygen limitation of thermal tolerance defined by cardiac and ventilatory performance in spider crab, Maja squinado. Am J Physiol Regul Integr Comp Physiol. 2000;279:R1531–R1538. doi: 10.1152/ajpregu.2000.279.5.R1531. [DOI] [PubMed] [Google Scholar]
- 92.Pörtner H-O. Climate change and temperature-dependent biogeography: oxygen limitation of thermal tolerance in animals. Naturwissenschaften. 2001;88(4):137–146. doi: 10.1007/s001140100216. [DOI] [PubMed] [Google Scholar]
- 93.Pörtner H-O. Oxygen- and capacity-limitation of thermal tolerance: a matrix for integrating climate-related stressor effects in marine ecosystems. J Exp Biol. 2010;213(6):881–893. doi: 10.1242/jeb.037523. [DOI] [PubMed] [Google Scholar]
- 94.Kooijman SALM. Dynamic Energy Budget Theory for Metabolic Organisation, Volume 3. Cambridge, United Kingdom: Cambridge University Press; 2010. [Google Scholar]
- 95.Pörtner H-O, Farrell AP. Physiology and climate change. Science. 2008;322:690–692. doi: 10.1126/science.1163156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1: Summary of sequencing and mapping results for Hyas araneus. Details on treatment, filename and location of sequencing raw data, used sequencer, read count, read length and total size in base pairs (bp), used alignment tool, aligned reads and percentage of aligned reads are listed separately. (XLSX 57 KB)
Additional file 2: Table S2: Details on primers for quantitative real-time polymerase chain reaction (qRT-PCR) to validate RNASeq data of Hyas araneus. Forward and backward primer sequences and descriptions of target genes used in the qRT-PCR. Accession number (accession no.) refer to the transcriptome of Hyas araneus [40] and the database ENA (EMBL). R2 and efficiency was tested in a qRT-PCR dilution series (for details, see Methods). (XLSX 40 KB)
Additional file 3: Figure S1: Changes of expression levels of transcripts in gills of Hyas araneus responding to medium-term exposure (10 weeks) at intermediate PCO2 (≈1,000 μatm) and high temperature (10°C), analysed by DESeq (gray bars) and quantitative real-time polymerase chain reaction (qRT-PCR) (black bars). Bars represent the mean log2-fold change and standard error (error bars) of the respective gene. Transcripts correspond to primers and genes used in the qRT-PCR (see Additional file 5: Table S3). (PDF 78 KB)
Additional file 4: Figure S2: Linear regression between expression levels of transcripts in gills of Hyas araneus responding to medium-term exposure (10 weeks) at intermediate PCO2 (≈1,000 μatm) and high temperature (10°C), analysed by DESeq and quantitative real-time polymerase chain reaction (qRT-PCR). Black dots represent the mean log2-fold change of transcripts analysed by DESeq plotted against the corresponding mean log2-fold change analysed by qRT-PCR. r was determined by Pearson Correlation using SigmaPlot 12.0 (Systat Software Inc., San Jose, USA). (PDF 53 KB)
Additional file 5: Table S3: Transcript levels changing significantly in gills of Hyas araneus responding to hypercapnia and elevated temperature. Transcripts regulated significantly in response to hypercapnia and elevated temperature as identified by DESeq analysis (for details, see Methods). ID and accession number (accession no.) refer to the transcriptome of Hyas araneus [40] and the database ENA (EMBL). Details on transcript description and transcript length are listed for each transcript. Transcripts are sorted according to the rank in absolute regulation regardless of the treatment. Changes are given in log2-fold change for each treatment separately. For details on treatments, see Methods). Bold numbers represent significantly up-regulated transcripts and bold and underlined numbers significantly down-regulated transcripts. (XLSX 100 KB)
Additional file 6: Figure S3: Smearplot of differentially expressed transcripts in gills of Hyas araneus. All transcripts changed in response to hypercapnia and elevated temperature. Log2-fold changes are plotted against mean readcount (log10). Blue dots represent transcripts with non-significant changes, red dots depict transcripts significantly regulated as identified by DESeq analysis (p < 0.05) and green triangles are transcripts changed significantly and identified by annotation Numbers refer to the total number of significantly up-/down-regulated transcripts. A) treatment (I) = 1,120 μatm PCO2 5°C; B) treatment (II) = 1,960 μatm PCO2 5°C; C) treatment (III) = 390 μatm PCO2 10°C; D) treatment (IV) = 1,120 μatm PCO2 10°C; E) treatment (V) = 1,960 μatm PCO2 10°C. (PDF 492 KB)
Additional file 7: Table S4: Enrichment analysis in the RNASeq study on Hyas araneus. Results of the enrichment analysis (Fisher’s Exact Test; FDR < 0.05) as implemented in Blast2GO [49, 50] and reduced by web-based clustering tool REVIGO [51]. Tested were subsets of all significantly regulated transcripts as identified by DESeq (separated by up- and down-regulated transcripts). Reference-set was the full set of annotated sequences of the H. araneus transcriptome [40]. Listed is the Gene Ontology term (GO-term), the name of the functional group (description), the category (molecular function, biological process or cellular component), the false discovery rate (FDR) and whether GO-terms are over- or under-represented (a GO-term is considered over-/under-represented if it appears significantly more often/less often in the test-set than in the reference-set. (XLSX 33 KB)


