Abstract
Background
Although Cryptococcus laurentii has been considered saprophytic and its taxonomy is still being described, several cases of human infections have already reported. This study aimed to evaluate molecular aspects of C. laurentii isolates from Brazil, Botswana, Canada, and the United States.
Methods
In this study, 100 phenotypically identified C. laurentii isolates were evaluated by sequencing the 18S nuclear ribosomal small subunit rRNA gene (18S-SSU), D1/D2 region of 28S nuclear ribosomal large subunit rRNA gene (28S-LSU), and the internal transcribed spacer (ITS) of the ribosomal region.
Results
BLAST searches using 550-bp, 650-bp, and 550-bp sequenced amplicons obtained from the 18S-SSU, 28S-LSU, and the ITS region led to the identification of 75 C. laurentii strains that shared 99–100% identity with C. laurentii CBS 139. A total of nine isolates shared 99% identity with both Bullera sp. VY-68 and C. laurentii RY1. One isolate shared 99% identity with Cryptococcus rajasthanensis CBS 10406, and eight isolates shared 100% identity with Cryptococcus sp. APSS 862 according to the 28S-LSU and ITS regions and designated as Cryptococcus aspenensis sp. nov. (CBS 13867). While 16 isolates shared 99% identity with Cryptococcus flavescens CBS 942 according to the 18S-SSU sequence, only six were confirmed using the 28S-LSU and ITS region sequences. The remaining 10 shared 99% identity with Cryptococcus terrestris CBS 10810, which was recently described in Brazil. Through concatenated sequence analyses, seven sequence types in C. laurentii, three in C. flavescens, one in C. terrestris, and one in the C. aspenensis sp. nov. were identified.
Conclusions
Sequencing permitted the characterization of 75% of the environmental C. laurentii isolates from different geographical areas and the identification of seven haplotypes of this species. Among sequenced regions, the increased variability of the ITS region in comparison to the 18S-SSU and 28S-LSU regions reinforces its applicability as a DNA barcode.
Introduction
The Cryptococcus genus includes more than 100 species of which most are considered non-pathogenic, with the exceptions of Cryptococcus neoformans and Cryptococcus gattii. During the last decade Cryptococcus laurentii has occasionally been described to infect severely immunocompromised hosts [1]–[3]. In most of these reports from which isolates were obtained, the blood and the cerebrospinal fluid (CSF) were the predominant sources [2]–[5].
C. laurentii was first identified from palm wine in the Congo by Kufferath in 1920 as Torula laurentii [6]. This isolate was then reclassified as Torulopsis laurentii [7] and renamed in 1950 as Cryptococcus laurentii (CBS 139) [8]. Later in Japan, an isolate with identical phenotypic characteristics was described as Torula flavescens [9], reclassified in 1922 as Torulopsis flavescens [7], and then renamed as Cryptococcus flavescens (CBS 942) [8].
Cryptococcus aureus, Cryptococcus carnescens, and Cryptococcus peneaus, in addition to C. flavescens, were also considered to be synonymous of C. laurentii until phylogenetic analysis of the internal transcribed spacer (ITS) and D1/D2 region of 28S nuclear ribosomal large subunit rRNA gene (28S-LSU) demonstrated that they are different species [10], [11].
In 2005, Cryptococcus rajasthanensis (CBS 10406) was described and differentiated from C. laurentii due to 1.6% and 7.5% divergence of the nucleotide identity of the 28-LSU and ITS regions, respectively [12]. More recently, Cryptococcus terrestris (CBS 10810), the cryptic species of C. flavescens, was isolated and described from soil in Brazil [13].
Currently, most C. laurentii isolates described around the world have been identified by morphological criteria, which can miss subtle differences and misidentify cryptic species [1], [14], [15]. Unlike C. neoformans and C. gattii, few studies have applied DNA sequencing to describe the molecular phylogeny of C. laurentii [11], [16], [17]. Thus, considering the potential pathogenicity of this species, this study aimed to evaluate the molecular phylogeny of clinical and environmental C. laurentii isolates through the sequencing of multiple ribosomal DNA regions.
Methods
Identification and fungal strains
We evaluated 100 environmental isolates of C. laurentii that were identified by classical mycological methods, such as India ink test, urease and phenoloxidase activity, thermotolerance at 37°C on Sabouraud dextrose agar, nitrate and carbon assimilation assays, carbohydrate fermentation, and microculture on cornmeal with Tween 80 [18], [19]. Of the 56 Brazilian isolates, 26 were obtained from peri-hospital areas, 5 from unidentified trees species, 7 from captive bird droppings in Uberaba, Minas Gerais State, and 18 from various environmental sources (bird droppings, trees, and air samples) from São Paulo State. The 18 isolates from Botswana were isolated from Mopane trees (Colophospermum mopane), the 14 isolates from New York State from Norway spruce (Picea abies) and trembling aspen (Populus tremuloides), and the 12 isolates from Vancouver, Canada from Douglas fir (Pseudotsuga menziesii) and other unidentified trees (Table 1). Isolates from Canada and the United States were isolated from swab samples collected in 2010 using single-swab BD CultureSwab Liquid Amies (Becton, Dickinson and Company, Sparks, Maryland, USA). The swabs were streaked onto yeast peptone dextrose agar (YPD, Becton, Dickinson and Company, Sparks, Maryland, USA) or Niger seed (NGS) agar containing chloramphenicol (0.5 g/L, Sigma-Aldrich, St. Louis, MO, USA), and yeast colonies were selected and colony purified [20].
Table 1. Isolate, species, source, and GenBank accession numbers of Cryptococcus spp. environmental isolates.
| Isolate | Species | Source | Country | GenBank | Hap | ||
| 18S-SSU | 28S-LSU | ITS | |||||
| CL01 | C. laurentii | Peri-hospital | Brazil | JX393937 | JN626983 | JQ968462 | 1 |
| CL02 | C. laurentii | Peri-hospital | Brazil | JX393938 | JN626984 | JQ968463 | 1 |
| CL03 | C. laurentii | Peri-hospital | Brazil | JX393939 | JN626985 | JQ968464 | 1 |
| CL04 | C. laurentii | Peri-hospital | Brazil | JX393940 | JN626986 | JQ968465 | 1 |
| CL05 | C. laurentii | Peri-hospital | Brazil | JX393941 | JN626987 | JQ968466 | 1 |
| CL06 | C. laurentii | Peri-hospital | Brazil | JX393942 | JN626988 | JQ968467 | 1 |
| CL07 | C. laurentii | Peri-hospital | Brazil | JX393943 | JN626989 | JQ968468 | 1 |
| CL08 | C. laurentii | Peri-hospital | Brazil | JX393944 | JN626990 | JQ968469 | 1 |
| CL09 | C. laurentii | Peri-hospital | Brazil | JX393945 | JN626991 | JQ968470 | 1 |
| CL10 | C. laurentii | Peri-hospital | Brazil | JX393946 | JN626992 | JQ968471 | 1 |
| CL11 | C. laurentii | Trees | Brazil | JX393947 | JN626993 | JQ968472 | 1 |
| CL12 | C. laurentii | Peri-hospital | Brazil | JX393948 | JN626994 | JQ968473 | 1 |
| CL13 | C. laurentii | Peri-hospital | Brazil | JX393949 | JN626995 | JQ968474 | 1 |
| CL14 | C. laurentii | Peri-hospital | Brazil | JX393950 | JN626996 | JQ968475 | 1 |
| CL15 | C. laurentii | Peri-hospital | Brazil | JX393951 | JN626997 | JQ968476 | 1 |
| CL16 | C. laurentii | Peri-hospital | Brazil | JX393952 | JN626998 | JQ968477 | 1 |
| CL17 | C. laurentii | Peri-hospital | Brazil | JX393953 | JN626999 | JQ968478 | 1 |
| CL18 | C. laurentii | Trees | Brazil | JX393954 | JN627000 | JQ968479 | 1 |
| CL19 | C. laurentii | Peri-hospital | Brazil | JX393955 | JN627001 | JQ968480 | 1 |
| CL20 | C. laurentii | Peri-hospital | Brazil | JX393956 | JN627002 | JQ968481 | 1 |
| CL21 | C. laurentii | Peri-hospital | Brazil | JX393957 | JN627003 | JQ968482 | 1 |
| CL22 | C. laurentii | Trees | Brazil | JX393958 | JN627004 | JQ968483 | 1 |
| CL23 | C. laurentii | Peri-hospital | Brazil | JX393959 | JN627005 | JQ968484 | 1 |
| CL24 | C. laurentii | Peri-hospital | Brazil | JX393960 | JN627006 | JQ968485 | 1 |
| CL25 | C. laurentii | Trees | Brazil | JX393961 | JN627007 | JQ968486 | 1 |
| CL26 | C. laurentii | Peri-hospital | Brazil | JX393962 | JN627008 | JQ968487 | 2 |
| CL27 | C. laurentii | Peri-hospital | Brazil | JX393963 | JN627009 | JQ968488 | 1 |
| CL28 | C. laurentii | Peri-hospital | Brazil | JX393964 | JN627010 | JQ968489 | 1 |
| CL29 | C. laurentii | Peri-hospital | Brazil | JX393965 | JN627011 | JQ968490 | 1 |
| CL30 | C. laurentii | Peri-hospital | Brazil | JX393966 | JN627012 | JQ968491 | 1 |
| CL32 | C. laurentii | Peri-hospital | Brazil | JX393967 | JN627013 | JQ968492 | 1 |
| CL33 | C. laurentii | Pets shops | Brazil | JX393968 | JN627014 | JQ968493 | 1 |
| CL34 | C. laurentii | Pets shops | Brazil | JX393969 | JN627015 | JQ968494 | 1 |
| CL35 | C. laurentii | Pets shops | Brazil | JX393970 | JN627016 | JQ968495 | 1 |
| CL36 | C. laurentii | Pets shops | Brazil | JX393971 | JN627017 | JQ968496 | 1 |
| CL37 | C. laurentii | Pets shops | Brazil | JX393972 | JN627018 | JQ968497 | 1 |
| CL38 | C. laurentii | Pets shops | Brazil | JX393973 | JN627019 | JQ968498 | 1 |
| CL39 | C. laurentii | Pets shops | Brazil | JX393974 | JN627020 | JQ968499 | 1 |
| E4 | C. laurentii | Pigeon dropping | Brazil | JX393977 | JX393999 | JQ968502 | 3 |
| E5 | C. laurentii | Pigeon dropping | Brazil | JX393978 | JX394000 | JQ968503 | 3 |
| E6 | C. laurentii | Pigeon dropping | Brazil | JX393979 | JX394001 | JQ968504 | 3 |
| E7 | C. laurentii | Pigeon dropping | Brazil | JX393980 | JX394002 | JQ968505 | 1 |
| E11 | C. laurentii | Pigeon dropping | Brazil | JX393981 | JX394003 | JQ968506 | 3 |
| E12 | C. laurentii | Pigeon dropping | Brazil | JX393982 | JX394004 | JQ968507 | 3 |
| E14 | C. laurentii | Pigeon dropping | Brazil | JX393983 | JX394005 | JQ968508 | 3 |
| DS288 | C. laurentii | Mopane tree | Botswana | KC469712 | KC485478 | KC469756 | 1 |
| DS386 | C. laurentii | Mopane tree | Botswana | KC469715 | KC485481 | KC469759 | 1 |
| DS388 | C. laurentii | Mopane tree | Botswana | KC469716 | KC485482 | KC469760 | 1 |
| DS390 | C. laurentii | Mopane tree | Botswana | KC469717 | KC485483 | KC469761 | 1 |
| DS392 | C. laurentii | Mopane tree | Botswana | KC469718 | KC485484 | KC469762 | 1 |
| DS394 | C. laurentii | Mopane tree | Botswana | KC469719 | KC485485 | KC469763 | 4 |
| DS400 | C. laurentii | Mopane tree | Botswana | KC469720 | KC485486 | KC469764 | 5 |
| DS402 | C. laurentii | Mopane tree | Botswana | KC469722 | KC485488 | KC469766 | 6 |
| DS403 | C. laurentii | Mopane tree | Botswana | KC469723 | KC485489 | KC469767 | 5 |
| DS444 | C. laurentii | Mopane tree | Botswana | KC469724 | KC485490 | KC469768 | 6 |
| DS447 | C. laurentii | Mopane tree | Botswana | KC469726 | KC485492 | KC469770 | 4 |
| DS455 | C. laurentii | Mopane tree | Botswana | KC469727 | KC485493 | KC469771 | 1 |
| DS529 | C. laurentii | Norway spruce | USA | KC469728 | KC485494 | KC469772 | 3 |
| DS530 | C. laurentii | Norway spruce | USA | KC469729 | KC485495 | KC469773 | 3 |
| DS531 | C. laurentii | Norway spruce | USA | KC469730 | KC485496 | KC469774 | 3 |
| DS619 | C. laurentii | Norway spruce | USA | KC469735 | KC485501 | KC469779 | 3 |
| DS620 | C. laurentii | Norway spruce | USA | KC469736 | KC485502 | KC469780 | 3 |
| DS621 | C. laurentii | Norway spruce | USA | KC469737 | KC485503 | KC469781 | 3 |
| DS744 | C. laurentii | Douglas Fir tree | Canada | KC469742 | KC485508 | KC469786 | 3 |
| DS746 | C. laurentii | Douglas Fir tree | Canada | KC469743 | KC485509 | KC469787 | 3 |
| DS748 | C. laurentii | Douglas Fir tree | Canada | KC469744 | KC485510 | KC469788 | 3 |
| DS778 | C. laurentii | Tree | Canada | KC469745 | KC485511 | KC469789 | 5 |
| DS782 | C. laurentii | Tree | Canada | KC469746 | KC485512 | KC469790 | 5 |
| DS783 | C. laurentii | Tree | Canada | KC469747 | KC485513 | KC469791 | 5 |
| DS784 | C. laurentii | Tree | Canada | KC469748 | KC485514 | KC469792 | 5 |
| DS785 | C. laurentii | Tree | Canada | KC469749 | KC485515 | KC469793 | 5 |
| DS797 | C. laurentii | Tree | Canada | KC469750 | KC485516 | KC469794 | 3 |
| DS798 | C. laurentii | Tree | Canada | KC469751 | KC485517 | KC469795 | 3 |
| DS802 | C. laurentii | Tree | Canada | KC469752 | KC485518 | KC469796 | 3 |
| DS806 | C. laurentii | Tree | Canada | KC469753 | KC485519 | KC469797 | 5 |
| CBS 139T | C. laurentii | Palm wine | Congo | AB032640 | AF075469 | AB035043 | 7 |
| P482A | C. rajasthanensis | Tree | Brazil | JX393990 | JX394017 | JQ968514 | 8 |
| CBS 10406T | C. rajasthanensis | Flowers | India | NA | AM262324 | AM262325 | NA |
| DS569 | C. aspenensis | Trembling aspen | USA | KC469731 | KC485497 | KC469775 | 9 |
| DS570 | C. aspenensis | Trembling aspen | USA | KC469732 | KC485498 | KC469776 | 9 |
| DS572 | C. aspenensis | Trembling aspen | USA | KC469733 | KC485499 | KC469777 | 9 |
| DS573T | C. aspenensis # | Trembling aspen | USA | KC469734 | KC485500 | KC469778 | 9 |
| DS712 | C. aspenensis | Trembling aspen | USA | KC469738 | KC485504 | KC469782 | 9 |
| DS713 | C. aspenensis | Trembling aspen | USA | KC469739 | KC485505 | KC469783 | 9 |
| DS715 | C. aspenensis | Trembling aspen | USA | KC469740 | KC485506 | KC469784 | 9 |
| DS716 | C. aspenensis | Trembling aspen | USA | KC469741 | KC485507 | KC469785 | 9 |
| O242A | C. flavescens | Tree | Brazil | JX393984 | JX394006 | JQ968509 | 10 |
| I113A | C. flavescens | Air | Brazil | JX393985 | JX394007 | JQ968510 | 11 |
| I332A | C. flavescens | Tree | Brazil | JX393986 | JX394008 | JQ968511 | 10 |
| I382A | C. flavescens | Tree | Brazil | JX393987 | JX394009 | KC469798 | 11 |
| I243A | C. flavescens | Air | Brazil | JX393988 | JX394010 | JQ968512 | 11 |
| I283A | C. flavescens | Air | Brazil | JX393989 | JX394011 | JQ968513 | 12 |
| CBS 942T | C. flavescens | Air | Japan | AB085796 | AB035042 | AB035046 | 12 |
| I572B | C. terrestris | Tree | Brazil | JX393991 | JX394012 | JQ968515 | 13 |
| I573B | C. terrestris | Tree | Brazil | JX393992 | JX394013 | JQ968516 | 13 |
| 1B2011 | C. terrestris | Pigeon dropping | Brazil | JX393993 | JX394014 | JQ968517 | 13 |
| 1C2011 | C. terrestris | Pigeon dropping | Brazil | JX393994 | JX394015 | JQ968518 | 13 |
| DS233 | C. terrestris | Mopane tree | Botswana | KC469710 | KC485476 | KC469754 | 13 |
| DS234 | C. terrestris | Mopane tree | Botswana | KC469711 | KC485477 | KC469755 | 13 |
| DS290 | C. terrestris | Mopane tree | Botswana | KC469713 | KC485479 | KC469757 | 13 |
| DS291 | C. terrestris | Mopane tree | Botswana | KC469714 | KC485480 | KC469758 | 13 |
| DS401 | C. terrestris | Mopane tree | Botswana | KC469721 | KC485487 | KC469765 | 13 |
| DS446 | C. terrestris | Mopane tree | Botswana | KC469725 | KC485491 | KC469769 | 13 |
| CBS 10810T | C. terrestris | Soil | USA | NA | EF370393 | EU200782 | NA |
| CBS 142T | C. albidus | Air | Japan | AB032616 | AF075474 | AF145321 | 14 |
NA: Not applicable.
: Type strain. Hap: Haplotype number.
: C. aspenensis sp. nov. (CBS 13867).
Mopane trees (Colophospermum mopane). Douglas fir (Pseudotsuga menziesii). Norway spruce (Picea abies). Trembling aspen (Populus tremuloides). CBS: Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands. 18S-SSU: Small subunit rDNA. 28S-LSU: Large subunit rDNA. ITS: Internal transcribed spacer region.
All isolates were stored at −20°C in 70% YPD broth with 30% glycerol in 2-mL eppendorf tubes at the Mycology Laboratory at the Triangulo Mineiro Federal University (UFTM) for further analyses.
DNA sequencing
Genomic DNA was extracted from yeast cells in accordance with previously described methodology [21]. The 5′ end of the 18S nuclear ribosomal small subunit rRNA gene (18S-SSU) (AFToL project available at http://aftol.org/primers.php), internal transcribed spacer (ITS) region [10], [22], and D1/D2 region of 28S-LSU [10], [23] were amplified from genomic DNA by PCR using the primers and conditions denoted in Table 2.
Table 2. PCR conditions and primers used for the amplification of the ribosomal loci.
| Region | Forward | Reverse | Concentration | PCR Protocol |
| 18S-SSU | NS-1: 5′-GTAGTCATATGCTTGTCTC-3′ | NS-2: 5′-GGCTGCTGGCACCAGACTTGC-3′ | 50 pmol/each | 94°C for 2 min; 36 cycles of 94°C for 1 min; 57°C for 1 min; 72°C for 2 min; 72°C for 15 min; and 4°C on hold |
| 28S-LSU | NL-1: 5′-GCATATCAATAAGCGGAGGAAAAG-3′ | NL-4: 5′-GGTCCGTGTTTCAAGACGG-3′ | 70 pmol/each | 94°C for 2 min; 35 cycles of 94°C for 1 min, 57°C for 1 min; 72°C for 2 min; 72°C for 15 min; and 4°C on hold |
| ITS | ITS-1: 5′-GTCGTAACAAGGTTAACCTGCGG-3′ | ITS-4: 5′-TCCTCCGCTTATTGATATGC-3′ | 70 pmol/each | 94°C for 3 min; 29 cycles of 94°C for 30 s, 57°C for 30 s; 72°C for 45 s; 72°C for 10 min and 4°C on hold |
PCR of the 18S-SSU, 28S-LSU, and ITS regions were performed using a PTC-100 Thermocycler (MJ Research Inc., Watertown, MA, USA) in a final volume of 50 µL. Each reaction contained 20 ng of genomic DNA, 1× PCR buffer (10 mmol L−1, Tris-HCl pH 8.3, 50 mmol L−1 KCl, and 1.5 mmol L−1 MgCl2), 0.25 mmol L−1 each of dATP, dCTP, dGTP and dTTP, and 1.25 U of Taq DNA polymerase (Invitrogen, São Paulo, SP, Brazil). The amplicons were stained with 0.5 mg mL−1 of ethidium bromide and visualized under UV light after two hours of electrophoresis at 80 V [22].
Each PCR product was independently sequenced with the forward and reverse primers of each region using the BigDye terminator v. 3.1 reagent kit (Applied Biosystems, Foster City, CA, USA) including Taq DNA polymerase (Invitrogen, São Paulo, SP, Brazil) in an automated DNA sequencer (ABI PRISM 3130×L Genetic Analyzer, Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions.
Sequencing analysis
Sequences were edited using the software Sequence Scanner V. 1.0 (Applied Biosystems, USA). Only nucleotide sequences with a Phred quality score ≥20 were included in our analysis to limit the possibility of incorporating an incorrect base to ≤1 in 100 (≥99% accuracy). Bioedit software was used to obtain consensus sequences from aligned forward and reverse sequence reads. Each consensus sequence was submitted to the Basic Local Alignment Search Tool (BLAST), and identity values ≥99% were obtained to assign species. All generated sequences were deposited in GenBank (Table 1) and The Barcode of Life Database (BOLD) (http://www.barcodinglife.org) [24].
Phylogenetic relationships
Consensus sequences from newly identified isolates and those obtained from GenBank were aligned with the Clustal W2 algorithm (https://www.ebi.ac.uk/Tools/msa/clustalw2/) [25]. The phylogenetic analysis was calculated by the neighbor-joining, unweighted pair group method with arithmetic mean (UPGMA), and maximum likelihood methods in the MEGA 6.0 software [26]. For the neighbor-joining and maximum likelihood methods, the evolutionary distances were calculated in accordance with Kimura [27], while the Tamura 3-parameters method with the variation rate among sites modeled with a gamma distribution (shape parameter = 1) was used for UPGMA [28]. Phylogenetic relationships were calculated for each of the three regions and for the concatenated sequences applying a bootstrap analysis with 1,000 random resamplings. The type strain Cryptococcus albidus CBS 142 was designated as the outgroup in all phylogenetic analyses [29], [30]. Nucleotide sequences from the CBS-KNAW Fungal Biodiversity Centre (CBS) type strains were obtained from GenBank (Table 1).
To evaluate which of the three regions presented the highest variability, the intraspecific and interspecific pairwise distance was calculated by the Kimura 2-parameter model [27] in the MEGA 6.0 software [26].
Haplotype network and goeBURST analysis
Haplotype networks were generated from the three concatenated sequence regions to visualize the differences and diversity among the C. laurentii isolates. The number and diversity of each haplotype were estimated using the software DNAsp v5.10 (http://www.ub.edu/dnasp/) [31]. Median-joining networks [32] for the concatenated dataset were obtained and graphed using the software Network 4.610 (http://fluxus-engineering.com).
To confirm the haplotypes obtained by median-joining networks the analyses were replicated in MLSTest software (available at http://mlstest.codeplex.com) and graphed by goeBURST algorithm in PHILOVIZ software [33], [34]. The minimum spanning tree representing the comparison between the isolates sources and their haplotype was also calculated by goeBURST.
Coalescent species analyses
In order to estimate the time divergence between species and haplotypes, the interspecific and intraspecific net nucleotides substitutions (d) and standard error of the concatenated sequences were calculated in accordance to Kimura [27] with a bootstrap (500 replicates) as a variance method in the MEGA 6.0 software [35], [36]. The distance and standard error between closest species e. g. (C. laurentii x C. rajasthanensis 0.016±0.003; C. aspenensis x C. flavescens 0.071±0.007; C. terrestris x C. flavescens 0.009±0.002) were obtained and applied in the equation d = 2λt, where d is the number of nucleotide substitutions per site between a pair of sequences, t is the divergence time, and λ the rate of nucleotide substitution. Here, we applied the constant (λ) 10−9/bp/year previously obtained for the Eurotiomycetes lineage due to the absence of a known fossil for C. laurentii species [36]. The resulting time of divergence were used as prior parameters to calibrate the tree in the coalescent analyses.
The optimal molecular evolutionary model for the concatenated sequences was determined using the corrected Akaike Information Criterion (AICc) as executed in the software jModelTest 2 [37], [38]. The optimal molecular evolutionary model General Time Reversible (GTR+I+G) with the respective parameters: AC: 0.7675, AG: 2.3377, AT: 1.8759, CG: 0.4999, GT: 1.0, alpha (IG): 0.5430, and pinv (IG): 0.5130 were obtained and used as priors in the coalescence analyses.
The BEAST software version 1.8.0 [39] was used to calculate the mean time to the most common recent ancestor (TMRCA) by the applying the Bayesian Markov-chain Monte Carlo (MCMC) method assuming a relaxed log-normal model of molecular rate heterogeneity. The chain lengths were 107 generations with parameters sampled every 103 generations with an initial burn-in off 10%. The posterior probability for a given clade was the frequency that the clade was present among the posterior trees which means that the probability of the lineage be considered monophyletic in the used dataset. Convergence of parameter values in the MCMC were assessed by the effective sample size (ESS) in the Tracer software version 1.6 [40].
Nucleotide diversity of C. laurentii isolates
The extent of DNA polymorphisms, such as the number of polymorphic sites (S), nucleotide diversity (π), number of haplotypes (h), haplotype diversity (Hd), and average number of nucleotide differences (k), were calculated using DNAsp v5.10 [31]. In addition, Tajima's D, Fu & Li's D*, Fu & Li's F*, and Fu's Fs tests for neutrality were calculated. Negative or positive results in these tests suggest evidence of purifying or balancing selection, respectively.
Fluorescence-activated cell-sorting (FACS) analysis
The FACS protocol was modified from Tanaka et. al. [41]. Cells were grown overnight at 25°C in YPD broth, collected by centrifugation, and washed with 1× PBS. Cells were then fixed in 1 ml of 70% ethanol overnight at 4°C with mild agitation. Cell pellets were obtained by centrifugation and the supernatants were discarded. Cells were resuspended and washed with 1 mL of NS buffer (10 mM Tris-HCl pH 7.2, 0.25 M sucrose, 1 mM EDTA, 1 mM MgCl2, 0.1 mM CaCl2, 0.55 mM Phenylmethylsulfonyl floride, 0.1 mM ZnCl2, 0.049% 2-mercaptoethanol). Cells were then resuspended in 180 µl NS buffer with, 14 mL RNase A (15 mg/µl, Qiagen) and 6 µl of Propidium iodide (1.0 µg/µl, CALBIOCHEM) and incubated in the dark for 4–8 hrs at room temperature. After incubating 50 µl of the cells were mixed with 500 mL of Tris-PI mix [482 µl 1M Tris pH 7.5+18 µl Propidium Iodide (1 µg/µl)]. Flow cytometry was performed on 10,000 cells with slow laser scan, on the FL1 channel with a Becton-Dickinson FACScan.
This study was approved by the Ethical Board of Triangulo Mineiro Federal University and is registered under the protocol number 32 CBIO/UFTM.
Nomenclature
The electronic version of this article in Portable Document Format (PDF) will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants, and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone, so there is no longer any need to provide print copies. In addition, new names contained in this work have been submitted to MycoBank from where they will be made available to the Global Names Index. The unique MycoBank number can be resolved and the associated information viewed through any standard web browser by appending the MycoBank number contained in this publication to the prefix http://www.mycobank.org/MycoTaxo.aspx?Link=T&Rec=MB809723. The online version of this work is archived and available from the following digital repositories: PubMed Central and LOCKSS.
Results
All isolates produced capsule and urease but not melanin and were phenotypically characterized as C. laurentii due to their ability to assimilate arabinose, α-methyl-D-glucoside, cellobiose, D-glucose, D-mannitol, D-ribose, D-trehalose, DL-lactate, dulcitol, galactose, inositol, L-rhamnose, lactose, maltose, melizitoze, raffinose, sacarose, sorbose, xylose, and 2-keto-glutarate. However, the isolates were negative for fermentation of dextrose and assimilation of inulin and potassium nitrate. FACS analysis indicated that most of the isolates are haploid (Figure S1).
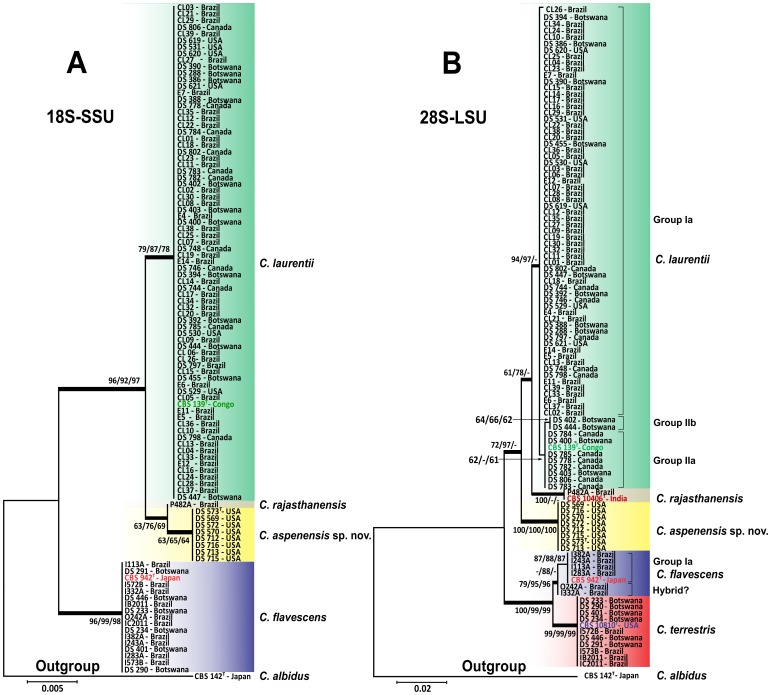
A 550-bp product was amplified from the 5′ end of 18S-SSU and sequenced with the primers NS-1 and NS-2, from which a 339-bp alignment was obtained. In this analysis, 75 isolates shared 99–100% identity with the C. laurentii CBS 139 (AB032640) type strain. Another 16 isolates shared 99% identity with C. flavescens CBS 942 (AB085796). The remaining 9 shared 99% identity with both Bullera sp. VY-68 (AB110694) from Japan and with C. laurentii RY1 from India (EF063147). High bootstrap values generated by neighbor-joining, UPGMA, and maximum likelihood analyses supported the differentiation of the following clades: C. laurentii (bootstrap values of 79, 87, and 78, respectively), C. flavescens (bootstrap values of 96, 99, and 98, respectively) and Bullera sp./C. laurentii (bootstrap values of 63, 65, and 64, respectively) (Figure 1A).
Figure 1. Phylogenetic analysis of 100 environmental Cryptococcus spp. isolates generated by the neighbor-joining, UPGMA, and maximum likelihood methods using partial nucleotide sequences of the (A) 5′end of 18S SSU-rDNA and (B) D1/D2 region of 28S LSU-rDNA.
Numbers at each branch indicate bootstrap values>50% based on 1,000 replicates (NJ/UPGMA/ML). The analysis involved 103 and 105nucleotide sequences for the 18S-SSU and 28S-LSU respectively. T: Type strain.
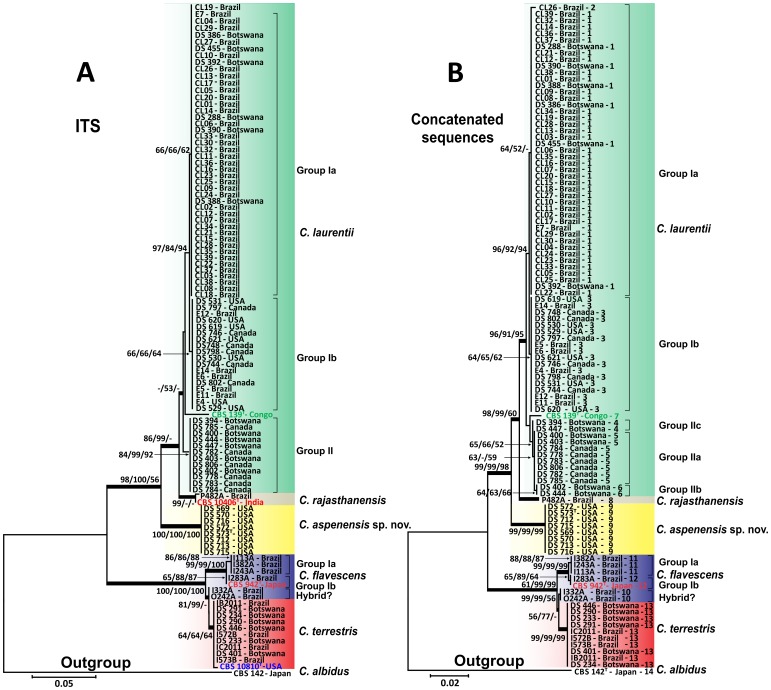
Due to the low genetic variability of the C. laurentii clade obtained at the 5′ end of the 18S-SSU gene, we sequenced two additional ribosomal loci: D1/D2 of the 28S-LSU and the ITS gene regions. The alignment and analysis of the 530-bp long amplicon of the 28S-LSU region confirmed the identification of 75 isolates as C. laurentii and showed more intraspecific variability differentiating three major groups (group Ia, IIa, and IIb) within C. laurentii isolates. Of the 16 C. flavescens isolates identified by the 18S-SSU sequencing, only 6 were confirmed in the C. flavescens clade by the 28S-LSU region with high bootstrap values of 79 (neighbor-joining), 95 (UPGMA), and 96 (maximum likelihood). The C. flavescens clade was split into two groups (Group Ia and a possible hybrid) by 28S-LSU and three groups (Groups Ia, Ib, and a possible hybrid) by ITS and analyses of the 1,328-bp amplicon of the concatenated regions. The two possible hybrid isolates I332A and O242A from Brazil were more related to C. terrestris in the ITS and concatenated sequences analyses (Figure 1B and 2). The remaining 10 isolates shared 99% identity with C. terrestris (CBS 10810), which has been recently described in Brazil and the United States (Figure 1B).
Figure 2. Phylogenetic analysis of 100 environmental Cryptococcus spp. isolates generated by the neighbor-joining, UPGMA, and maximum likelihood methods using partial nucleotide sequences of the (A) internal transcribed spacer (ITS) and (B) concatenated sequences of the three ribosomal regions.
Numbers at each branch indicate bootstrap values>50% based on 1,000 replicates (NJ/UPGMA/ML). The analysis involved 105 and 103 nucleotide sequences for ITS and concatenated sequences respectively. T: Type strain.
Among the nine Bullera sp./C. laurentii isolates identified by the 18S-SSU, isolate P482A shared 99% identity of the 28S-LSU and ITS regions with Cryptococcus rajasthanensis CBS 10406 (AM262324) from India. The eight remaining isolates (DS569, DS570, DS572, DS573, DS712, DS713, DS715, and DS716) recovered from a trembling aspen tree (Populus tremuloides) were designated as Cryptococcus aspenensis sp. nov. because they shared 100% identity with two undescribed isolates of Cryptococcus sp. APSS-862 (FM178286) and Cryptococcus sp. APSS-823 (AM931019) from India. These eight isolates exhibited a genetic distance of 3.8% and 7.1% from C. rajasthanensis and 2.3–2.7% and 6.4–7.3% from C. laurentii by 28S-LSU and ITS region analysis, respectively (Figure 1B and 2A).
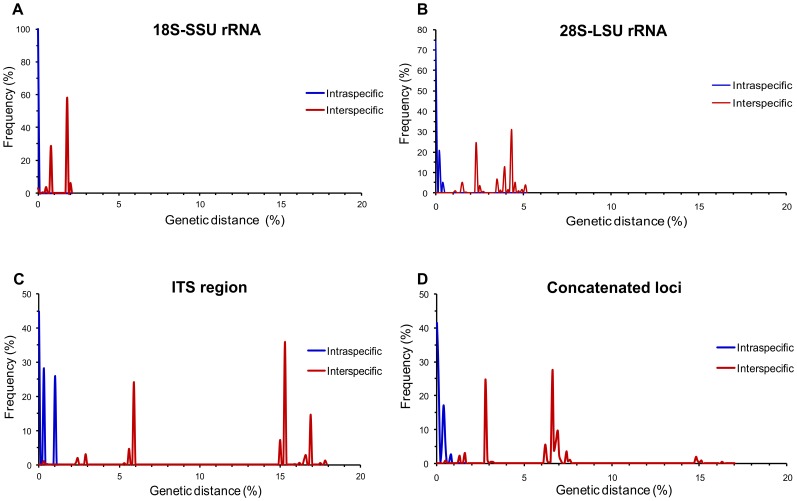
Overall, the pairwise distance of the three sequenced regions showed the highest intraspecific and interspecific variability in the ITS region (genetic distance higher than 15%) when compared with the 2.5% and 5.0% obtained with 18S-SSU and 28S-LSU, respectively (Figure 3).
Figure 3. Intraspecific and interspecific pairwise distance of the three ribosomal regions of the environmental Cryptococcus spp. calculated by the Kimura 2-parameter model revealed higher variability of the ITS region compared with the 18S-SSU and 28S-LSU regions.
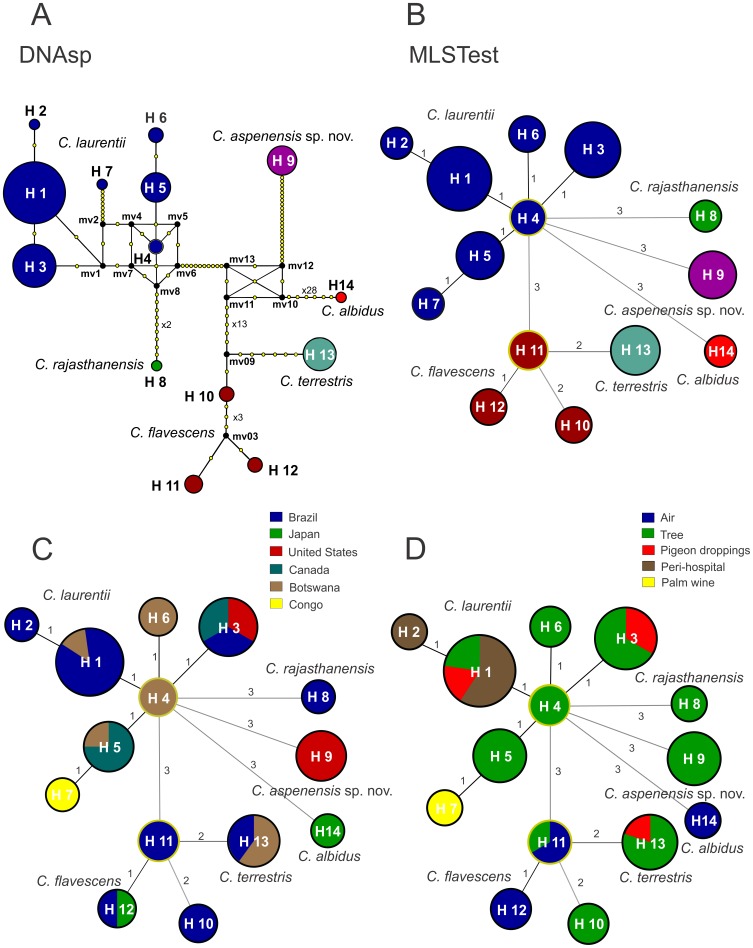
The haplotype diversity of the concatenated regions was assessed using DNAsp and MLSTest software. Multiple haplotype groups were identified within C. laurentii and C. flavescens, but not the C. aspenensis sp. nov. and C. terrestris (Figure 4A and 4B). The C. laurentii isolates were represented by seven haplotypes (H1 to H7). Haplotype 1 (H1) included 44 isolates, of which 38 (86.4%) were from Brazil, followed by the H3 composed of 6 from Brazil, 6 isolates from Canada, and 6 from the United States (Figure 4C and 4D). The highest genetic distance (12 polymorphisms) in the C. laurentii haplotypes was observed between H7 (CBS 132 type strain) and H6 (DS402 and DS444 isolates). Five of the seven C. laurentii isolates were recovered from Africa despite very limited sampling; three were unique haplotypes (H4, H6, and H7) and two were only observed in Brazil (H1) or Canada (H5). H4, which was obtained from Mopane trees in Botswana, was identified as the ancestral of C. laurentii in both Network and MLSTest analyses. H7 (C. laurentii type strain CBS 139) was restricted to the Congo and was in much closer proximity to Botswana than any other sample region. H3 was distinct from Botswana and was comprised of isolates from North and South America (Figure 4).
Figure 4. Median-joining haplotype network (A) of environmental C. laurentii isolates based on concatenated nucleotide sequences of the 5′ end of 18S-SSU, D1/D2 of 28S-LSU, and ITS regions.
The tree represents 103 Cryptococcus spp. isolates from Brazil, Botswana, Canada, Japan, India, and the United States. The seven C. laurentii and three C. flavescens haplotypes are clearly distinguished. The Botswana ancestral haplotype (H4) of C. laurentii is presented and highlighted in yellow. Each circle represents a unique haplotype (H), and the circumference is proportional to haplotype frequency (H1: 44 isolates; H2: 1; H3: 18; H4: 2; H5: 8; H6: 2; H7: 1; H8: 1; H9: 8; H10: 2; H11: 3; H12: 2; H13: 10; H14: 1; outgroup C. albidus CBS 142). Yellow dots represents the number of mutation sites, excluding gaps, between the haplotypes. Black dots (median vectors) are hypothetical missing intermediates. Minimum spanning trees (B) using the goeBURST algorithm confirm the haplotype relationships among C. laurentii isolates determined by median-joining network analysis. The size of the circle corresponds to the number of isolates within that haplotype, and the numbers between haplotypes represent the genetic distance of each haplotype, excluding the gaps. Minimum spanning trees as described in B modified to show the distribution of haplotypes according to the country of origin (C) or environmental source (D).
Three haplotypes were identified in C. flavescens isolates (H10, H11, and H12), with the ancestral haplotype H11 restricted to Brazil. H10 presented the highest genetic distance (9 polymorphisms) when compared with H11 and H12 (2 polymorphisms). H10 was also positioned closer to the C. terrestris haplotype H13 and could be a unique species, or ancestral genotype, or recombinant hybrid isolate between C. flavescens and C. terrestris. The C. aspenensis sp. nov. H9 was a completely unique genotype from New York, USA (Figure 4).
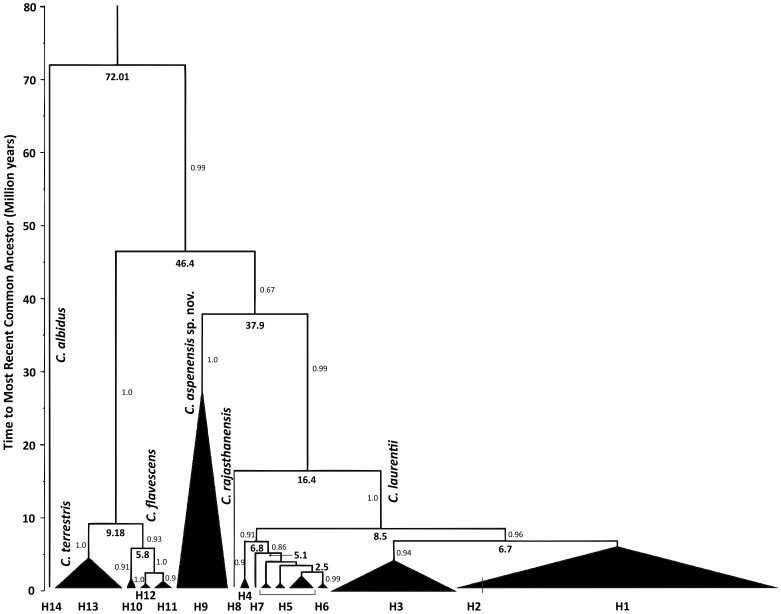
Estimates of the mean time to divergence for the C. flavescens and C. terrestris isolates were 4.02–5.4±0.87 million years (about 9 million years ago) with an effective sample size (ESS) of 1213.3 and 1006.8, respectively. For C. laurentii population, the TMRCA were 8.03±1.83 million years (about 16.4 million years ago) (ESS = 6615.0) while for the new species C. aspenensis sp. nov. 26.7±3.9 million years (about 37.9 million years ago) (ESS = 355.5). Coalescent analysis was strongly supported with (>95.0) Bayesian posterior values (Figure 5). Phylogenetic and coalescent analyses agree demonstrating additional support for the recognition of additional related haplotypes and species.
Figure 5. Species tree of the C. laurentii species complex resulting from coalescent analyses of the concatenated data set.
The speciation of C. aspenensis from C. laurentii and C. rajasthanensis took place 37.9 million years ago. The C. laurentii haplotype (H4) from Botswana was the first haplotype to be differentiated (6.8 million years ago). Numbers at branches represent the Bayesian posterior support values while the bold numbers represent the nodes ages (in millions of years).
The C. laurentii nucleotide sequences of the 18S-SSU, 28S-LSU, ITS, and the concatenated regions presented 0, 3, 11, and 14 polymorphic sites, respectively (Table 3). The highest nucleotide diversity (π) of 0.0039 was observed for ITS. Low values of haplotype (Hd = 0.604) and nucleotide diversity (π = 0.0014) of the concatenated regions may suggest clonal reproduction in this species (Table 3).
Table 3. DNA polymorphisms in the ribosomal loci of the 75 C. laurentii environmental isolates.
| Region | Length | S | π | k | h | Hd | D | FD | FF | FS |
| 18S- SSU | 399 | 0 | 0.0 | 0.0 | 1 | 0.0 | - | - | - | - |
| 28S- LSU | 530 | 3 | 0.0006 | 0.3291 | 4 | 0.280 | −0.873 | −0.521 | −0.737 | −1.537 |
| ITS | 399 | 11 | 0.0039 | 1.5881 | 4 | 0.570 | −0.977 | −2.674a | −2.472a | 2.852 |
| Concatenated | 1328 | 14 | 0.0014 | 1.9112 | 7 | 0.604 | −1.079 | −2.502a | −2.373a | 0.731 |
S: number of polymorphic sites. π: nucleotide diversity. k: average number of nucleotide differences per sequence. h: number of haplotypes. Hd: haplotype diversity. D , FD , FF and Fs: Tajima's D, Fu and Li's D*, Fu and Li's F* and Fu's Fs, respectively.
: p value<0.05.
Taxonomy
Cryptococcus aspenensis. Ferreira-Paim, K., Ferreira, T. B., Andrade-Silva, L., Mora, D. J., Springer, D. J., Heitman, J., Fonseca, F. M., Matos, D., Melhem, M. S. C., et Silva-Vergara, M. L. sp. nov. [urn:lsid:mycobank.org:names:MB809723].
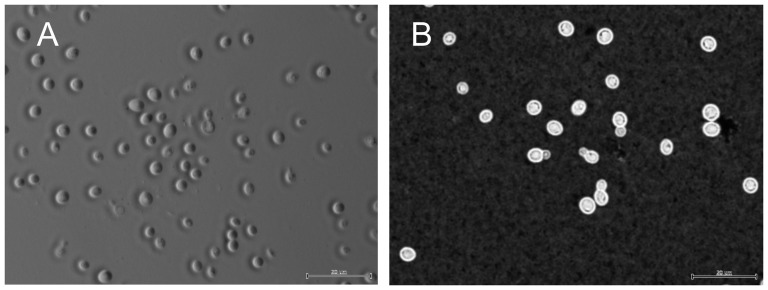
After 3 days on YPD agar at 25°C, Cryptococcus aspenensis colonies are circular, cream-colored with an entire margin, smooth, mucous to butyrous, glistening, and raised. Growth (poor) at 37°C was also observed. After 3 days at 25°C in YPD broth, the cells are ellipsoid to globose (7.5–8.7 to 5–6.2 µm), and they may be single or with one attached polar bud (Figure 6). After 15 days in slide cultures on cornmeal agar, pseudomycelium or mycelium is not formed. Fermentation ability is negative. Arabinose, α-methyl-D-glucoside, cellobiose, D-glucose, D-mannitol, D-ribose, D-trehalose, DL-lactate, dulcitol, galactose, inositol, L-rhamnose, lactose, maltose, melizitoze, raffinose, sacarose, sorbose, xylose, and 2-keto-glutarate are assimilated. Cells were haploid by FACS analysis (Figure S1).
Figure 6. Differential interference contrast (A) and India Ink staining (B) of C. aspenensis sp. nov. DS573T (CBS 13867) cells after 3 days at 25°C in YPD broth. Scale bar of 20 µm is shown.
Unambiguous identification and phylogenetic placement is based on DNA sequences of the following nuclear loci: ITS (KC469778), 18S-SSU (KC469734), D1/D2 of 28S-LSU (KC485500). The type strain DS573 was isolated from the bark of a trembling aspen (Populus tremuloides) in the New York, United States and has been deposited in the Centraalbureau voor Schimmelcultures, The Netherlands, as CBS 13867 and in the Westmead Millennium Institute, Australia, as WM 14.137. Other strains belonging to this species include DS570 (CBS 13868, WM 14.138), and DS715 (CBS 13869, WM 14.139) which were isolated from a single trembling aspen tree in New York.
Etymology: The specific epithet aspenensis L. adj. aspenensis associated with trembling aspen (Populus tremuloides), the tree substrate from which the type strain was isolated.
Discussion
Fungal identification and taxonomy has markedly improved during the last decade and as a result, several recognized species, such as Sporothrix schenckii, Paracoccidioides brasiliensis, and Coprinopsis cinerea, have been distinguished as cryptic species complexes [35], [42], [43]. In this context, the sequencing of the 18S-SSU, D1/D2 of 28S-LSU, and ITS of the ribosomal region have been useful in yeast identification for more than 10 years. However, the low variability of the 18S-SSU and 28S-LSU regions may prohibit identification of cryptic species, while the variability of the ITS region has been frequently utilized for fungal phylogenetic studies and the fungal tree of life barcoding projects (http://tolweb.org) [44], [45].
C. laurentii has classically been considered a saprophytic yeast, although 24 cases of human infection have been described, suggesting that C. laurentii is an opportunistic pathogen with potential similarities to the distantly related pathogenic C. neoformans and C. gattii species [3], [44], [46]–[48]. Cryptococcosis due to C. laurentii has been associated with severely immunocompromised patients and/or those presenting with other underlying diseases. In such cases, C. laurentii was most frequently isolated from the blood, but also from several other body sites such as the CSF, skin, and lungs [49], [50].
In this study, we evaluated 100 phenotypically identified C. laurentii isolates from several countries. Of these, 75 were confirmed to be C. laurentii by phylogenetic analysis of the 18S-SSU, 28S-LSU, and ITS regions. The obtained sequences shared 99–100% identity with sequences from Brazil, China, South Africa, and the United States, demonstrating its worldwide and overlapping geographic distribution with C. neoformans and C. gattii. Although, in North America, C. gattii has been associated with clinical infection in patients from New York, Rode Island, and other states [51]–[53]. At present, C. gattii has only been environmentally isolated from the Western United States [54] and Canada [55], while C. neoformans is broadly associated with pigeon guano in many regions of the United States, including the state of New York [56]. Hence, our study suggests that in the United States, C. laurentii appears have a much broader distribution than C. gattii as noted from its isolation in association with grasses in the USA, and goose guano and trees in New York State [57].
Within the C. laurentii clade, intraspecific variability of 0.2% (1 polymorphism), 0.2–0.4% (1–3 polymorphisms), and 0.3–2.4% (1–11 polymorphisms) was obtained for the analysis of the 18S-SSU, 28S-LSU, and ITS regions, respectively. These features are consistent with a previously published report indicating that one polymorphism exhibited in the 28S-LSU region exist up to 11 substitutions in the ITS region [58]. Through phylogenetic analysis of the 28S-LSU and ITS regions, three divergent groups were distinguishable from the 75 C. laurentii isolates. Groups IIa and II of the 28S-LSU and ITS regions contained eight isolates from Botswana and Vancouver, which differed from the remaining 67 isolates in 1–3 and 5–11 nucleotides in the 28S-LSU and ITS regions, respectively, and constituted H5 in the network and goeBURST analysis. Additional analysis of environmental and clinical samples from outside of Brazil will be valuable to determine whether H5 is distinct to Brazil. The majority of Brazilian isolates are H1 (44 isolates). The high frequency of the H1 haplotype may be related to microevolution and/or adaptation of these isolates to the environment, while the H2 haplotype may be rare.
Despite the differences in the total number of C. laurentii isolates, those from Botswana (n = 12) shared five of the seven haplotypes observed, two of them unique (H4 and H6). Interestingly, the ancestral H4 is only represented in Botswana suggesting that similar to C. neoformans var. grubii, C. laurentii may have originated from Africa [59]. The historical haplotype (H7) from palm wine is also restricted to the Congo, which is near to Botswana. Other haplotypes common in Africa are only also observed from Brazil (H1) or Canada (H5). Therefore, it is possible that C. laurentii was introduced into Brazil and Canada from Africa. To test this hypothesis, the coalescent gene analyses was performed which showed that the isolates within the haplotype 4 are the oldest in our data set (6.8 million years ago).
The remaining 25 isolates that were originally identified as C. laurentii by standard phenotypic assays were identified by ITS, 18S-SSU, and 28S-LSU analyses as C. terrestris (n = 10), the C. aspenensis sp. nov. (n = 8), C. flavescens (n = 6), and C. rajasthanensis (n = 1). C. rajasthanensis isolates are rare, and few have been previously reported in GenBank from India, Thailand, China, and Brazil. The C. rajasthanensis isolate in our study was recovered from hollow trees in São Paulo, Brazil and differed from C. laurentii by 0.4–0.6%, 1.7–2.1%, and 4.3–4.8% in the 18S-SSU, 28S-LSU, and ITS regions, respectively. In previous studies, the C. laurentii type strain (CBS 139) differed from the known Indian C. rajasthanensis reference isolate (CBS 10406) by 1.6% in the 28S-LSU region and 7.5% in the ITS region.
Despite the genetic distance observed between C. flavescens and C. laurentii (4–5.2% in 28S-LSU and 16.8–18.9% in the ITS), the species have long been considered phenotypically indistinguishable. For example, one previously identified clinical isolate of C. laurentii was later distinguished to be C. flavescens [4], [60], suggesting that opportunistic pathogen traits may have evolved more than once within this group, similar to the presence of sporadic opportunistic pathogens in Kwoniella and Cryptococcus heveanensis species groups [61].
C. flavescens has only recently been differentiated as a sibling species of C. terrestris [13] with the advancements in multi-locus sequence analysis. It is likely that the delayed recognition of C. terrestris and C. flavescens hindered the recognition of divergent phenotypic traits now recognized as important species characteristics. C. terrestris can be differentiated phenotypically from C. flavescens by delayed and/or weak assimilation of ribose and salicin [13], [44]. Our analysis supports the previous reported genetic differentiation; C. flavescens diverged from C. terrestris by 1.2–1.6% (6–10 polymorphisms) and 0.5–2.5% (2–10 polymorphisms) in the 28S-LSU and ITS regions, respectively. This difference probably occurred 9.1 million years ago as demonstrated by the coalescent analyses.
The six C. flavescens isolates recovered from Brazil were similar to isolates from China, Egypt, Italy, Japan, South Africa, and the United States, confirming the ubiquity of this species. The intraspecific variability of 0.2%, 0.4% and 0.8–2.2% observed in the 18S-SSU, 28S-LSU, and ITS regions, respectively, and the description of one haplotype in 18S-SSU, two in 28S-LSU, and three in the ITS region and concatenated analyses for the first time is relevant in the biological context of this species. Both isolates within H10 (I332A and O242A) share higher similarity with C. terrestris in 18S-SSU, the ITS region, and concatenated sequence but are more similar to C. flavescens in 28S-LSU. H10 may be a second haplotype of C. terrestris or a possible hybrid haplotype between the two species, as has been observed between C. gattii and C. neoformans [62]–[64]. Both isolates within this unique haplotype appear haploid by FACS which suggest this isolate may be a recombinant between C. flavescens and C. terrestris or a ancestral genotype. Coalescent analysis does not support the hypothesis that the two isolates in haplotype 10 are ancestral to both species and hence it is likely this haplotype arose from a productive introgression between C. flavescens and C. terrestris. Whole genome sequencing and the development of multilocus sequence primers specific to C. laurentii will be needed to support these hypotheses. Furthermore, the ancestral haplotype of C. flavescens appears to be H11 (revealed by MLSTest but not by DNAsp), which is confined to Brazil, suggesting that it may have originated in Brazil. Additional environmental and clinical isolates must be evaluated to better define the place of origin of C. flavescens.
A newly identified but distinct haploid group that we designated as C. aspenensis sp. nov. was identified consistently through phylogenetic analyses of individual and concatenated loci and confirmed by coalescent analyses. At present, this constitutes a previously unidentified species that appears to be restricted to New York, United States. All eight isolates obtained in the H9 group appear to be nearly identical/clonal and were obtained from sampling one trembling aspen tree in Long Island, New York, United States. An additional isolate was just identified from soil sample collect on July 13 in Copake, New York (personal communication D. J. Springer) and supports the recognition of this newly identified species. C. aspenensis sp. nov. appears to represent a unique ancestral lineage that diverged from the common ancestor prior to C. rajasthanensis and C. laurentii approximately 37 million years ago.
With the advent of inexpensive sequencing, alignment, and analysis, increasing numbers of sequences for bacteria, plants, viruses, animals, protozoa, and fungi are rapidly being deposited in publically accessible databases such as GenBank [65]–[67]. In fungi, several regions have been utilized for phylogenetic studies including the ITS, 28S-LSU, and 18S-SSU of the rRNA cistron regions, as well as CO1 (Penicillium), MCM7 (ascomycetes), and RBP1 (Assembling the Fungal Tree of Life, AFToL project) [45], [58], [66]. Schoch et al. recently reported that the ITS region was generally superior to the LSU in species discrimination and had a more clearly defined barcode gap, indicating that the ITS region should be designated as the universal barcode for fungi [45]. Our analyses concur with this previously published report; we found increased variability in the ITS region that resulted in better phylogenic differentiation between the highly related, globally distributed, and potentially clonal C. laurentii species group. Concatenated sequence analysis resulted in the identification of novel and distinct haplotypes within C. laurentii that appear to be associated with specific geographic regions.
Additional analysis of clinical and environmental specimens, mating type determination, sequencing of housekeeping genes, and whole genome analysis are required to further resolve potential haplotypes within C. laurentii and resolve the phylogenetic placement of the closely related species C. rajasthanensis, C. flavescens, C. terrestris, and the C. aspenensis sp. nov. described in this analysis.
Supporting Information
Representative Fluorescence-activated cell-sorting (FACS) analysis of the Cryptococcus spp. included in the study. All isolates except three C. laurentii (CL11, CL19, and E11) and one C. flavescens (I234A) appear haploid. Positive haploid (CBS10574) and diploid controls (XL143) were included in each FACS run.
(TIF)
Acknowledgments
We thank Mrs. Ângela Azôr for her technical assistance. DNA samples were sequenced at the Laboratório Multiusuário of UFTM. We would like to thank Edmond Byrnes and Laura Rusche for obtaining environmental samples from Vancouver, BC, Canada and Botswana, Africa, respectively. We thank Wieland Meyer, Catriona Halliday, and Marc Ramsperger for discussions and advice.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This work was supported by Fundação de Amparo a Pesquisa de Minas Gerais-FAPEMIG APQ-01735-10 [to M.L.S.V.]. K.F.P. is a research fellow of CAPES: Process number 9313/13-3. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Averbuch D, Boekhoutt T, Falk R, Engelhard D, Shapiro M, et al. (2002) Fungemia in a cancer patient caused by fluconazole-resistant Cryptococcus laurentii . Med Mycol 40: 479–484. [DOI] [PubMed] [Google Scholar]
- 2. Bauters TG, Swinne D, Boekhout T, Noens L, Nelis HJ (2002) Repeated isolation of Cryptococcus laurentii from the oropharynx of an immunocompromized patient. Mycopathologia 153: 133–135. [DOI] [PubMed] [Google Scholar]
- 3. Manfredi R, Fulgaro C, Sabbatani S, Legnani G, Fasulo G (2006) Emergence of amphotericin B-resistant Cryptococcus laurentii meningoencephalitis shortly after treatment for Cryptococcus neoformans meningitis in a patient with AIDS. AIDS Patient Care STDS 20: 227–232. [DOI] [PubMed] [Google Scholar]
- 4. Kordossis T, Avlami A, Velegraki A, Stefanou I, Georgakopoulos G, et al. (1998) First report of Cryptococcus laurentii meningitis and a fatal case of Cryptococcus albidus cryptococcaemia in AIDS patients. Med Mycol 36: 335–339. [PubMed] [Google Scholar]
- 5. Banerjee P, Haider M, Trehan V, Mishra B, Thakur A, et al. (2013) Cryptococcus laurentii fungemia. Indian J Med Microbiol 31: 75–77. [DOI] [PubMed] [Google Scholar]
- 6. Kufferath H (1920) Peut-on obtenir du moût de bière alcalin? Annales de la Société royale des sciences médicales et naturelles de Bruxelles 74: 16–46. [Google Scholar]
- 7. Lodder J (1934) Die anaskosporegenen Hefen, I. Hälfte. Verh K Ned Akad Wet Afd Natuurkd v. 32: 1–256. [Google Scholar]
- 8. Skinner CE (1950) Generic name for imperfect yeasts, Cryptococcus or Torulopsis ? The American Midland Naturalist Journal 43: 242–250. [Google Scholar]
- 9. Saito K (1922) Untersuchungen über die atmosphärischen Pilzkeime. Mitt Jpn J Bot 1: 1–54. [Google Scholar]
- 10. Sugita T, Takashima M, Ikeda R, Nakase T, Shinoda T (2000) Intraspecies diversity of Cryptococcus laurentii as revealed by sequences of internal transcribed spacer regions and 28S rRNA gene and taxonomic position of C. laurentii clinical isolates. J Clin Microbiol 38: 1468–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takashima M, Sugita T, Shinoda T, Nakase T (2003) Three new combinations from the Cryptococcus laurentii complex: Cryptococcus aureus, Cryptococcus carnescens and Cryptococcus peneaus . Int J Syst Evol Microbiol 53: 1187–1194. [DOI] [PubMed] [Google Scholar]
- 12. Saluja P, Prasad GS (2007) Cryptococcus rajasthanensis sp. nov., an anamorphic yeast species related to Cryptococcus laurentii, isolated from Rajasthan, India. Int J Syst Evol Microbiol 57: 414–418. [DOI] [PubMed] [Google Scholar]
- 13. Crestani J, Fontes Landell M, Faganello J, Henning Vainstein M, Simpson Vishniac H, et al. (2009) Cryptococcus terrestris sp. nov., a tremellaceous, anamorphic yeast phylogenetically related to Cryptococcus flavescens . Int J Syst Evol Microbiol 59: 631–636. [DOI] [PubMed] [Google Scholar]
- 14. Simon G, Simon G, Erdos M, Marodi L (2005) Invasive Cryptococcus laurentii disease in a nine-year-old boy with X-linked hyper-immunoglobulin M syndrome. Pediatr Infect Dis J 24: 935–937. [DOI] [PubMed] [Google Scholar]
- 15. Rosario I, Soro G, Deniz S, Ferrer O, Acosta F, et al. (2009) Presence of C. albidus, C. laurentii and C. uniguttulatus in crop and droppings of pigeon lofts (Columba livia). Mycopathologia 169: 315–319. [DOI] [PubMed] [Google Scholar]
- 16. Ferreira-Paim K, Andrade-Silva L, Mora DJ, Lages-Silva E, Pedrosa AL, et al. (2012) Antifungal susceptibility, enzymatic activity, PCR-fingerprinting and ITS sequencing of environmental Cryptococcus laurentii isolates from Uberaba, Minas Gerais, Brazil. Mycopathologia 174: 41–52. [DOI] [PubMed] [Google Scholar]
- 17. Tay ST, Na SL, Tajuddin TH (2008) Natural occurrence and growth reaction on canavanine-glycine-bromothymol blue agar of non-neoformans Cryptococcus spp. in Malaysia. Mycoses 51: 515–519. [DOI] [PubMed] [Google Scholar]
- 18. Granados DP, Castaneda E (2005) Isolation and characterization of Cryptococcus neoformans varieties recovered from natural sources in Bogota, Colombia, and study of ecological conditions in the area. Microb Ecol 49: 282–290. [DOI] [PubMed] [Google Scholar]
- 19. Staib F (1963) Membrane filtration and guizotia abyssinica culture media for the demonstration of Cryptococcus neoformans (Brown Color Effect). Z Hyg Infektionskr 149: 329–336. [PubMed] [Google Scholar]
- 20. Randhawa HS, Kowshik T, Khan ZU (2005) Efficacy of swabbing versus a conventional technique for isolation of Cryptococcus neoformans from decayed wood in tree trunk hollows. Med Mycol 43: 67–71. [DOI] [PubMed] [Google Scholar]
- 21. Ferreira-Paim K, Andrade-Silva L, Mora DJ, Pedrosa AL, Rodrigues V, et al. (2011) Genotyping of Cryptococcus neoformans isolated from captive birds in Uberaba, Minas Gerais, Brazil. Mycoses 54: e294–300. [DOI] [PubMed] [Google Scholar]
- 22. Sugita T, Nishikawa A, Ikeda R, Shinoda T (1999) Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J Clin Microbiol 37: 1985–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kurtzman CP, Robnett CJ (1997) Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol 35: 1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ratnasingham S, Hebert PD (2007) bold: The Barcode of Life Data System (http://www.barcodinglife.org) Mol Ecol Notes 7: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 28. Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol 9: 678–687. [DOI] [PubMed] [Google Scholar]
- 29. Fell JW, Roeijmans H, Boekhout T (1999) Cystofilobasidiales, a new order of basidiomycetous yeasts. Int J Syst Bacteriol 49 (Pt 2) 907–913. [DOI] [PubMed] [Google Scholar]
- 30. Scorzetti G, Petrescu I, Yarrow D, Fell JW (2000) Cryptococcus adeliensis sp. nov., a xylanase producing basidiomycetous yeast from Antarctica. Antonie Van Leeuwenhoek 77: 153–157. [DOI] [PubMed] [Google Scholar]
- 31. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 32. Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48. [DOI] [PubMed] [Google Scholar]
- 33. Francisco AP, Bugalho M, Ramirez M, Carrico JA (2009) Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, et al. (2012) PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Teixeira MM, Theodoro RC, de Carvalho MJ, Fernandes L, Paes HC, et al. (2009) Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol Phylogenet Evol 52: 273–283. [DOI] [PubMed] [Google Scholar]
- 36. Kasuga T, White TJ, Taylor JW (2002) Estimation of nucleotide substitution rates in Eurotiomycete fungi. Mol Biol Evol 19: 2318–2324. [DOI] [PubMed] [Google Scholar]
- 37. Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 39. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaocharoen S, Ngamskulrungroj P, Firacative C, Trilles L, Piyabongkarn D, et al. (2013) Molecular epidemiology reveals genetic diversity amongst isolates of the Cryptococcus neoformans/C. gattii species complex in Thailand. PLoS Negl Trop Dis 7: e2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tanaka R, Taguchi H, Takeo K, Miyaji M, Nishimura K (1996) Determination of ploidy in Cryptococcus neoformans by flow cytometry. J Med Vet Mycol 34: 299–301. [PubMed] [Google Scholar]
- 42. Marimon R, Cano J, Gene J, Sutton DA, Kawasaki M, et al. (2007) Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J Clin Microbiol 45: 3198–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nagy LG, Desjardin DE, Vagvolgyi C, Kemp R, Papp T (2013) Phylogenetic analyses of Coprinopsis sections Lanatuli and Atramentarii identify multiple species within morphologically defined taxa. Mycologia 105: 112–124. [DOI] [PubMed] [Google Scholar]
- 44. Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A (2000) Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol 50 (Pt 3) 1351–1371. [DOI] [PubMed] [Google Scholar]
- 45. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, et al. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shankar EM, Kumarasamy N, Bella D, Renuka S, Kownhar H, et al. (2006) Pneumonia and pleural effusion due to Cryptococcus laurentii in a clinically proven case of AIDS. Can Respir J 13: 275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Andrade-Silva L, Ferreira-Paim K, Silva-Vergara ML, Pedrosa AL (2010) Molecular characterization and evaluation of virulence factors of Cryptococcus laurentii and Cryptococcus neoformans strains isolated from external hospital areas. Fungal Biol 114: 438–445. [DOI] [PubMed] [Google Scholar]
- 48. Danesi P, Firacative C, Cogliati M, Otranto D, Capelli G, et al. (2014) Multilocus sequence typing (MLST) and M13 PCR fingerprinting revealed heterogeneity amongst Cryptococcus species obtained from Italian veterinary isolates. FEMS Yeast Res doi:10.1111/1567-1364.12178 [DOI] [PubMed] [Google Scholar]
- 49. Johnson LB, Bradley SF, Kauffman CA (1998) Fungaemia due to Cryptococcus laurentii and a review of non-neoformans cryptococcaemia. Mycoses 41: 277–280. [DOI] [PubMed] [Google Scholar]
- 50. Kiertiburanakul S, Sungkanuparph S, Pracharktam R (2001) Cryptococcus laurentii fungemia: A case report. J Infect Dis Antimicrob Agents 18: 112–114. [Google Scholar]
- 51. Lockhart SR, Iqbal N, Harris JR, Grossman NT, DeBess E, et al. (2013) Cryptococcus gattii in the United States: genotypic diversity of human and veterinary isolates. PLoS One 8: e74737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCulloh RJ, Phillips R, Perfect JR, Byrnes EJ 3rd, Heitman J, et al. (2011) Cryptococcus gattii genotype VGI infection in New England. Pediatr Infect Dis J 30: 1111–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Warren K, Amory C, Tobin E (2014) Meningitis Due to Cryptococcus gattii: First Reported Case of an Emerging Infectious Disease in an Immunocompetent Patient Residing in the Northeast United States. Neurology 82 Supplement P2.323. [Google Scholar]
- 54. Byrnes EJ 3rd, Li W, Lewit Y, Ma H, Voelz K, et al. (2010) Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog 6: e1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kidd SE, Chow Y, Mak S, Bach PJ, Chen H, et al. (2007) Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl Environ Microbiol 73: 1433–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Steenbergen JN, Casadevall A (2000) Prevalence of Cryptococcus neoformans var. neoformans (Serotype D) and Cryptococcus neoformans var. grubii (Serotype A) isolates in New York City. J Clin Microbiol 38: 1974–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Filion T, Kidd S, Aguirre K (2006) Isolation of Cryptococcus laurentii from Canada Goose guano in rural upstate New York. Mycopathologia 162: 363–368. [DOI] [PubMed] [Google Scholar]
- 58. Yurkov AM, Golubev WI (2013) Phylogenetic study of Cryptococcus laurentii mycocinogenic strains. Mycological Progress 12: 777–782. [Google Scholar]
- 59. Litvintseva AP, Carbone I, Rossouw J, Thakur R, Govender NP, et al. (2011) Evidence that the human pathogenic fungus Cryptococcus neoformans var. grubii may have evolved in Africa. PLoS One 6: e19688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brown JK, Hovmoller MS (2002) Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297: 537–541. [DOI] [PubMed] [Google Scholar]
- 61. Guerreiro MA, Springer DJ, Rodrigues JA, Rusche LN, Findley K, et al. (2013) Molecular and genetic evidence for a tetrapolar mating system in the basidiomycetous yeast Kwoniella mangrovensis and two novel sibling species. Eukaryot Cell 12: 746–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bovers M, Hagen F, Kuramae EE, Diaz MR, Spanjaard L, et al. (2006) Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii . FEMS Yeast Res 6: 599–607. [DOI] [PubMed] [Google Scholar]
- 63. Bovers M, Hagen F, Kuramae EE, Hoogveld HL, Dromer F, et al. (2008) AIDS patient death caused by novel Cryptococcus neoformans x C. gattii hybrid. Emerg Infect Dis 14: 1105–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Aminnejad M, Diaz M, Arabatzis M, Castaneda E, Lazera M, et al. (2012) Identification of novel hybrids between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII. Mycopathologia 173: 337–346. [DOI] [PubMed] [Google Scholar]
- 65. Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS One 6: e19254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pino-Bodas R, Martin MP, Burgaz AR, Lumbsch HT (2013) Species delimitation in Cladonia (Ascomycota): a challenge to the DNA barcoding philosophy. Mol Ecol Resour 13: 1058–1068. [DOI] [PubMed] [Google Scholar]
- 67. Scicluna SM, Tawari B, Clark CG (2006) DNA barcoding of blastocystis. Protist 157: 77–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative Fluorescence-activated cell-sorting (FACS) analysis of the Cryptococcus spp. included in the study. All isolates except three C. laurentii (CL11, CL19, and E11) and one C. flavescens (I234A) appear haploid. Positive haploid (CBS10574) and diploid controls (XL143) were included in each FACS run.
(TIF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.