Abstract
In this study, three phages infecting Lactobacillus delbrueckii subsp. bulgaricus, named Ld3, Ld17, and Ld25A, were isolated from whey samples obtained from various industrial fermentations. These phages were further characterized in a multifaceted approach: (i) biological and physical characterization through host range analysis and electron microscopy; (ii) genetic assessment through genome analysis; (iii) mass spectrometry analysis of the structural components of the phages; and (iv), for one phage, transcriptional analysis by Northern hybridization, reverse transcription-PCR, and primer extension. The three obtained phage genomes display high levels of sequence identity to each other and to genomes of the so-called group b L. delbrueckii phages c5, LL-Ku, and phiLdb, where some of the observed differences are believed to be responsible for host range variations.
INTRODUCTION
Lactic acid bacteria are commonly utilized in the commercial manufacture of fermented food products, where they act to reduce the pH, while also contributing to the organoleptic properties of the final product (1). Lactobacillus delbrueckii, a member of this family, contains four subspecies, two of which—L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis—are used as starter cultures in conjunction with Streptococcus thermophilus in the production of yoghurt (2) and cheeses (3). L. delbrueckii subsp. bulgaricus contributes to the flavor of these fermented products through the production of lactic acid, acetaldehyde, and exopolysaccharides (4), which all contribute to the final flavor profile of the product. Bacteriophages or phages represent a major threat to the success of such fermentation processes since these obligate parasites may cause (partial) lysis of starter cultures, thereby reducing their acidification activity (5), and thus impact negatively on the flavor of the product with consequent economic losses.
Currently, the complete genome sequence of five phages infecting two different subspecies of L. delbrueckii are available: three phages infecting L. delbrueckii subsp. lactis, namely, the temperate phage JCL1032 (6) and the virulent phages LL-Ku (6) and LL-H (7), and two virulent phages infecting L. delbrueckii subsp. bulgaricus, phage c5 (6) and phiLdb (8). Phages that infect strains of either of these two L. delbrueckii subspecies are currently classified into four distinct groups (groups a, b, c, and d) based on DNA homology (9–11). Phage JCL1032 (group c) was isolated from a dairy fermentation in Switzerland (10), while LL-H (group a) and LL-Ku (group b) were both isolated from dairy plants in Finland (6, 12), and c5 (group b) was isolated from a yoghurt production facility in France (13). The recently sequenced phiLdb phage (group b) was isolated from a Chinese yoghurt in 2010 (8). Finally, group d currently consists of just a single member, L. delbrueckii subsp. lactis phage 0235, which has not been subjected to genome sequencing. Despite the industrial significance of L. delbrueckii as a starter culture in the production of fermented foods, relatively few L. delbrueckii phages have been studied in detail, with the exception of LL-H, which was only recently reviewed (14). The LL-H genome shows little similarity to those of c5, LL-Ku, or JCL1032, demonstrating that several genetically distinct groups of phages infecting L. delbrueckii exist. Transcription of the LL-H genome appears to occur in two distinct phases, indicated as early and late transcription (7). In addition, the receptor-binding protein of this phage was identified as gp71 (15), while lipoteichoic acid has been shown to act as the LL-H host receptor (16, 17). All other functional assignments of L. delbrueckii phage genes have been based on bioinformatic predictions (6).
Here, we report on and examine the complete genomes and proteomes of three group b phages infecting L. delbrueckii subsp. bulgaricus: vB_LdbS_Ld3 (referred to here as Ld3), vB_LdbS_Ld17 (Ld17), and vB_LdbS_Ld25A (Ld25A) (18). In addition, we present transcriptional data related to the L. delbrueckii subsp. bulgaricus phage Ld17, as a typical representative of group b phages.
MATERIALS AND METHODS
Bacteriophages, bacterial strains, and growth conditions.
Phages and L. delbrueckii subsp. bulgaricus strains used as propagation hosts in the present study are listed in Table 1. Strains were grown overnight at 42°C in MRS broth (Oxoid, United Kingdom) supplemented with 1% lactose, 20 mM CaCl2, and 0.5% tryptone (MRS-LCT). Phages were isolated from a range of whey samples received from industrial yoghurt or cheese production environments involving L. bulgaricus starter cultures. Whey samples from L. delbrueckii subsp. bulgaricus fermentations were tested for phages against 68 starter cultures using the double-layer plaque assay method as single plaque isolates (19) using MRS-LCT agar supplemented with 1% glycine (20). Propagation of these phages was performed by picking an individual plaque and infecting a host at an optical density at 600 nm (OD600) of 0.15 in MRS-LCT, followed by incubation at 42°C until lysis occurred. The resulting lysate was passed through a 0.45-μm-pore-size filter to remove cell debris and stored at 4°C. Host ranges of phages were ascertained by spotting 10 μl of phage solution on MRS-LCT top agar containing 200 μl of fresh overnight culture and incubating overnight at 42°C (21, 22). Phage enumerations, expressed as PFU ml−1, were carried out by using the double-layer titration technique mentioned above (19). Phages were isolated from various whey samples, and three of these, which were found to display distinct host ranges and DNA restriction patterns, were characterized as representatives of the population.
TABLE 1.
Host range analysis of Ld3, Ld17, and Ld25Aa
| Strain | Ld3 | Ld17 | Ld25A |
|---|---|---|---|
| Ldb1 | + | – | + |
| Ldb3 | +* | – | – |
| Ldb4 | + | – | – |
| Ldb12 | – | – | + |
| Ldb13 | – | – | + |
| Ldb15 | – | – | + |
| Ldb16 | – | – | + |
| Ldb17 | – | +* | – |
| Ldb18 | – | – | + |
| Ldb19 | – | – | + |
| Ldb21 | + | – | – |
| Ldb22 | + | + | + |
| Ldb23 | – | – | + |
| Ldb24 | – | – | + |
| Ldb25 | – | – | +* |
| Ldb27 | – | – | + |
| Ldb31 | + | – | – |
| Ldb32 | + | – | – |
| Ldb33 | – | – | + |
| Ldb34 | + | + | + |
| Ldb35 | – | + | – |
| Ldb36 | + | + | + |
| Ldb37 | – | – | + |
| Ldb39 | + | – | + |
| Ldb53 | – | – | + |
| Ldb54 | – | – | + |
| Ldb55 | + | + | + |
| Ldb57 | – | – | + |
| Ldb59 | – | + | – |
| Ldb60 | – | – | + |
| Ldb61 | + | – | – |
| Ldb63 | – | + | – |
| Ldb64 | – | – | + |
| Ldb66 | – | + | – |
| Ldb68 | – | + | – |
+, the strain is susceptible to phage infection; –, the strain is resistant to phage infection; *, the strain was used as a host for the phage. All strains belong to L. delbrueckii subsp. bulgaricus.
Electron microscopic analysis.
Phage lysates were purified on a cesium chloride gradient and were dialyzed against phage buffer (20 mM Tris-HCl [pH 7.2], 10 mM NaCl, 20 mM MgSO4). Staining was performed either with 2% uranyl acetate (wt/vol) or with 1% (wt/vol) phosphotungstic acid (pH 7) on freshly prepared carbon films (Bal-Tec MED 020/EVM 030; Leica, Wetzlar, Germany). The films were picked up with 400 mesh Cu grids (Plano, Wetzlar, Germany) and analyzed in a Tecnai 10 transmission electron microscope (FEI Company, Eindhoven, The Netherlands) at an acceleration voltage of 80 kV. Micrographs were taken with a MegaView G2 charge-coupled device camera (Olympus SIS, Muenster, Germany). Measurements of phage dimensions were calculated using samples stained with uranyl acetate.
Genome sequencing.
Five μg of DNA of Ld3, Ld17 and Ld25A, as determined by NanoDrop quantification, was isolated from fresh CsCl2-purified lysates using a previously described method (23) prior to shipment to the contract sequencing facility (Macrogen, Inc., South Korea). Sequencing of the genomes was conducted using a GS-FLX Titanium sequencer. The reads generated by the 454 FLX instrument were assembled with GSassembler (454 Life Sciences, Branford, CT) to generate a consensus sequence. Quality improvement of the genome sequence involved sequencing of PCR products across the entire genome to ensure correct assembly, double stranding, and the resolution of any remaining base-conflicts occurring within homopolymer tracts.
In silico analysis.
The genomic termini were determined through homology with the cohesive ends defined for c5 and LL-Ku (6). Protein-encoding open reading frames (ORFs) were predicted using GeneMark (24). ORFs with an AUG, UUG, or GUG start codon, encoding at least 30 amino acids and preceded by a sequence resembling the consensus Shine-Dalgarno sequence (AGAAAGGAGGTG) (25) were accepted. Initial functional annotation of the ORFs was undertaken using BLASTP (26), and functions were further confirmed by querying the protein domain database PFAM (27) and the NCBI Conserved Domains Database (28) and by performing homology prediction searches using HHPred (29). The percent amino acid identity between the deduced proteomes of phages were calculated by using a CLUSTAL W alignment of concatenated coding sequences in the MegAlign program of the Lasergene software package (version 7.1.0). CLUSTAL W alignments of antireceptor genes were also performed using MegAlign software (version 7.1.0). Putative terminators in this transcript were predicted using the ARNold terminator prediction Web tool (30).
Phage structural proteome and mass spectrometry.
Purified phage proteins were extracted from CsCl2-purified phage samples by performing a single methanol-chloroform extraction (1:1:0.75 [vol/vol/vol]) and subsequently precipitated with an equal volume of methanol. Proteins were concentrated by centrifugation at 20,800 × g for 6 min and resuspended in 100 μl of TBT (100 mM NaCl, 100 mM Tris-HCl [pH 7], 10 mM MgCl2, 20 mM CaCl2). The structural protein profile was generated by standard Tris-glycine, sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis (PAGE) and visualized following staining with 0.25% Coomassie brilliant blue. Gel slices were then excised, trypsin treated, and analyzed using electrospray ionization-tandem mass spectrometry (ESI-MS/MS) as previously described (31, 32).
Transcriptome analysis.
For analysis of phage transcription, a modification of the one-step growth curve protocol of Lu et al. (33) was used for infection since it was shown to consistently yield high phage titers and helped to ensure synchronous infection. L. delbrueckii subsp. bulgaricus Ldb17 was grown in 200 ml of MRS-LCT to an OD600 of 0.15. Cells were then harvested at 5,580 × g and resuspended in 2 ml of MRS-LCT. Ld17 was added at a multiplicity of infection (MOI) of ∼0.1 and incubated at 42°C for 5 min, followed by centrifugation at 18,000 × g for 30 s to remove any unadsorbed phage, thus ensuring a synchronous infection. The resultant pellet was resuspended in 200 ml of MRS-LCT and incubated at 42°C. Then, 6-ml samples were taken at set time points (2, 5, 10, 15, 25, 35, 45, and 60 min postinfection) by centrifugation and flash freezing in a −80°C ethanol bath. Samples were maintained at −80°C until required for further analysis.
Total RNA was extracted using a High-Pure RNA isolation kit (Roche, Germany) as described previously (34). Total RNA quantity was ascertained by NanoDrop analysis (Thermo Fisher Scientific, USA) and typically yielded 100 to 230 ng μl−1.
RNA was then separated under denaturing conditions on a 1% agarose, 0.66 M formaldehyde gel. Blotting, hybridization, and washes were carried out using a NorthernMax kit (Ambion Life Technologies, USA) according to the manufacturer's instructions. Two biotinylated antisense RNA probes were generated through amplification of ∼500-bp products at the 5′ end of predicted transcripts (Table 2) with incorporation of restriction enzyme sites and subsequent cloning into pBluescript SK(−) (Invitrogen, USA) in the reverse orientation. Antisense RNA incorporating biotin-labeled nucleotides was generated using T7 polymerase and a biotin labeling kit (Roche, Germany). Signal detection was achieved using streptavidin IRD800 conjugate, and membranes were visualized using an Odyssey imager (Li-Cor Biosciences, USA).
TABLE 2.
Primers used in this study
| Analysis and primer | Sequence (5′–3′)a | Descriptionb |
|---|---|---|
| Northern hybridization | ||
| 17N1F | AAAAAATCTAGAGAGTAAATCGGAAGAGATCC | Internal ORF23 |
| 17N1R | AAAAAAGATATCCTTATCCTTCTTCATGTTC | Internal ORF23 |
| 17N6F | AAAAAATCTAGAGCACTGAAGAAGAACGGTGC | Internal TMP |
| 17N6R | AAAAAAGATATCCCGCCCACGTTTGAATGTTG | Internal TMP |
| 17N6FN | AAAAAATCTAGACCAGCTGATTACTCCAGTCGG | Internal holin |
| 17N6RN | AAAAAAGATATCCTTCATCTCCATTGCTTACAAC | Internal holin |
| Primer extension | ||
| Ter1F | GTAGCTGAGCAAATGCAAAGTG | Amplifies Ter1 region |
| Ter1R | GCCTTTAAGTTTGATCGTCC | Amplifies Ter1 region |
| Ter2F | GGTATTCAAGCAATCAGCTTC | Amplifies Ter2 region |
| Ter2R | GAACTTGCTATTTCCGTTGGC | Amplifies Ter2 region |
| Ter3F | GAAGAGTTTGAGAGAATGCAAG | Amplifies Ter3 region |
| Ter3R | GTCTTGATTAACTCTGAAGAC | Amplifies Ter3 region |
| TERLF | GACGTTATACGAATGCACTACG | Within large terminase |
| TERLR | GTCGGGTATTTCGTACTAATGATC | Within large terminase |
| MHF | GAATGACTTGATTACTCGTGCG | Within major head |
| MHR | GCGTGATGGACTTGAACGTACC | Within major head |
| TMPF | GTCGTTGCGAGAAGTAAATTCCG | Within tail tape measure |
| TMPR | GCTTGCATTATCGGATTTGTCG | Within tail tape measure |
| GDGPF | GAAGACATCATCAAGTTCGCTCG | Within glycerophosphoryl |
| GDGDR | GCTCTTCTCCGTCTTTCATACC | Diester phosphodiesterase |
| ANTF | GTTACGAGTATTCGGAGCTAC | Within antireceptor |
| ANTR | CTGATCTCTCTCAAGTTCTTCTTG | Within antireceptor |
| RT-PCR | ||
| PE2 | AGCGACTGAAACTCCAAG | IRD800 primer for E1 transcript |
| PE5 | CTTGACCGACTGGAGTAATC | IRD800 primer for L1 transcript |
| PESeqE1F | GCAACTCATATATCAAAG | Amplifies TSS region for E1 |
| PESeqE1R | TGAGGATACAACTTGAAG | Amplifies TSS region for E1 |
| PESeqL1F | CCTAAGGCGATTGTATTTGAAGC | Amplifies TSS region for L1 |
| PESeqL1R | GGTAAGCGTGCACTTTCAATC | Amplifies TSS region for L1 |
Restriction sites are underlined.
TSS, transcription start site.
Additional transcriptional profiling was performed by reverse transcription-PCR (RT-PCR). Briefly, DNA was removed from the total RNA sample (45 min postinfection) using DNase (Roche, Germany) and subsequently reverse transcribed using AMV-RT (New England BioLabs, USA) and random nonamers (MWG, Germany) according to the manufacturer's instructions. The resulting cDNA was used as a template for PCR amplifications targeting regions encompassing putative terminators in order to assess whether predicted terminators were functional. Where a PCR product was generated, the predicted terminators were deemed to be nonfunctional and to represent false-positive hits identified by the software. Identical PCRs were also undertaken on the DNase-treated RNA to confirm the removal of any residual phage DNA from the RNA samples.
Primer extension.
To define the transcriptional start sites of Ld17, 1 pmol of IRD800-labeled primer PE2 or PE5 (Table 2) was annealed to 15 to 20 μg of total RNA as previously described (35). Sequencing ladders of the predicted 5′ end of the transcripts were generated using the same primers as the primer extension; PCR products generated with the primer pairs PESeqE1F/PESeqE1R or PESeqL1F/PESeqL1R (Table 2) were used as the templates. This was achieved using a DNA cycle sequencing kit (Jena Bioscience, Germany). The sequencing reactions were run in parallel with the primer extension products on a 6.5% Li-Cor Matrix KB Plus acrylamide gel. The signal was detected, and the image was captured using a Li-Cor sequencing instrument (Li-Cor Biosciences).
Nucleotide sequence accession numbers.
The complete sequence data for the phages sequenced in the present study are available in the GenBank database under the accession numbers KJ564038 (Ld3), KJ654037 (Ld17), and KJ564036 (Ld25A).
RESULTS AND DISCUSSION
Isolation and morphological analysis of three L. delbrueckii subsp. bulgaricus infecting phages.
Three distinct phages infecting L. delbrueckii subsp. bulgaricus, Ld3, Ld17, and Ld25A were isolated by means of the standard double-layer titration method (23) from various industrial dairy fermentations: Ld3 was isolated from a natural yoghurt production facility in Jordan in 2008, Ld17 was isolated from a whey sample from a Gorgonzola cheese fermentation in Italy in 2009, and Ld25A was isolated from a Turkish yoghurt in Turkey in 2005.
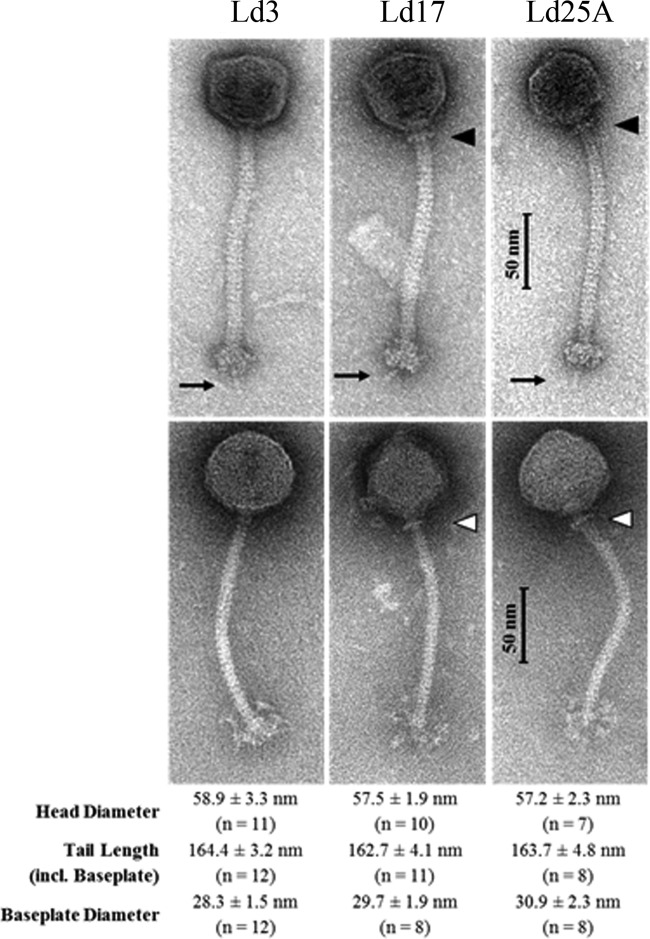
The three isolated L. delbrueckii subsp. bulgaricus phages were assessed by electron microscopy for their morphological characteristics. Ld3, Ld17, and Ld25A were all shown to be Siphoviridae phages, each possessing icosahedral heads (morphotype B1), a long noncontractile tail, including a well-defined baseplate with flexible globular appendages and a visible central tail fiber (Fig. 1). Measurements of these structural elements are provided in Fig. 1. Collar structures were evident for Ld17 and Ld25A but absent for Ld3 (Fig. 1). The phage particles are most similar to the group b family of phages infecting L. delbrueckii species which were classified as Sfi21-like Siphoviridae (9, 36), strengthening the notion that Lactobacillus species appear to be predominantly infected by Siphoviridae phages (37).
FIG 1.
Transmission electron micrographs of the L. delbrueckii subsp. bulgaricus phages stained either with uranyl acetate (UA; top three images) or with phosphotungstic acid (PTA; bottom three images). Collar-like structures of phages Ld17 and Ld25A, respectively, are indicated by triangles. The central tail fiber at the bottom of the baseplates (UA staining) is indicated by arrows. Flexible globular or fluffy appendices of baseplate structures were visualized by PTA staining.
Host range.
A total of 64 L. delbrueckii subsp. bulgaricus strains were assessed for their susceptibility to infection by Ld3, Ld17, and Ld25A. Ld3 and Ld17 were capable of infecting 12 and 10 strains, respectively, whereas Ld25A displayed a broader host range, infecting 23 strains, representing nearly 36%, of the strains assessed (Table 1).
Genome sequence analysis.
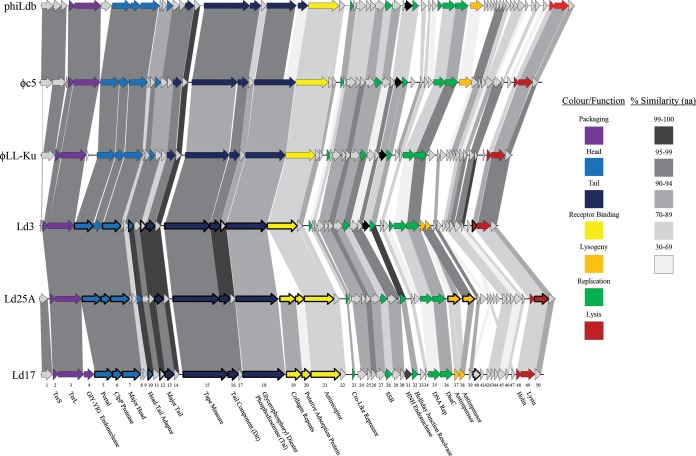
Phage genomic DNA was isolated and the respective phage genome sequences of Ld3, Ld17 and Ld25A were elucidated by 454 sequencing technology revealing genomes of 29,616 bp (39-fold coverage), 32,975 bp (45-fold coverage), and 32,799 bp (55-fold coverage) in length harboring 49, 50, and 51 ORFs, preceded by Shine-Dalgarno sequences consistent with the consensus sequence for L. delbrueckii subsp. lactis phage LL-H (see Fig. S1 in the supplemental material) (25). The general genome characteristics are summarized in Table 3. They display high levels of sequence identity both to each other, with Ld17 showing 97% identity (84% coverage) and 95% identity (87% coverage) to Ld3 and Ld25A, respectively, and also to previously published L. delbrueckii phages c5, LL-Ku (6), and phiLdb (8) (Table 3). The GC content of the phages (Table 3) is similar to phages c5 and LL-Ku (41.5 and 41.9%, respectively), for which it was noted that they exhibit a lower GC content than that of the host, leading to the possibility that they may have relatively recently evolved to infect L. delbrueckii (but originate from a different host) (6). The observed close relationship between the three newly presented and the three previously published phages clearly shows that the three newly isolated phages belong to the so-called group b phages infecting L. delbrueckii (9). Due to the high degree of sequence identity between the phages, a functional annotation of the predicted ORFs is provided for phage Ld25A only, along with the amino acid similarity to the predicted ORFs in Ld3 and Ld17 (Table 4). The organization of the genomes is highly conserved compared to c5 and LL-Ku, displaying the same gene order with all predicted functional modules in a rightward orientation (Fig. 2). The phages are organized into four functional modules: (i) the packaging module, (ii) the structural module, (iii) the replication module, and (iv) the lysis module (Fig. 2). Several insertion/deletion events are evident among the genomes; these are discussed in detail below.
TABLE 3.
General genome characteristics
| Characteristic | Phage |
||
|---|---|---|---|
| Ld3 | Ld17 | Ld25A | |
| Length (bp) | 29,616 | 32,975 | 32,799 |
| No. of ORFs | 49 | 50 | 51 |
| GC content (%) | 42.2 | 41.97 | 42.2 |
| No. of strains infected | 13 | 11 | 23 |
| % genome coding | 93 | 92.7 | 92.2 |
| Origin | Jordan | Italy | Turkey |
| Product | Yoghurt | Gorgonzola | Yoghurt |
| % identity (% coverage) | |||
| Identity to c5 | 96 (90) | 96 (87) | 95 (82) |
| Identity to LL-Ku | 95 (91) | 95 (87) | 94 (84) |
| Identity to phiLdb | 92 (92) | 93 (88) | 93 (83) |
TABLE 4.
ORFs deduced from Ld25A, Ld3, and Ld17 and their predicted proteins
| Ld25A gene | Start position | End position | Length (aa) | Size (kDa) | Strand | RBS/start codona | ORF(s) and % similarityb |
Predicted function | Representative similarity to proteins in database | % identity (no. of aa) | E-value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ld3 |
Ld17 |
|||||||||||||
| ORF | %S | ORF | %S | |||||||||||
| 1 | 90 | 515 | 142 | 15.62 | + | AAAAGCGAGGTTAAAAATG | 1 | 95 | Putative ParB nuclease | Putative ParB nuclease (Lactobacillus phage c5) | 97 (88) | 5.00E–55 | ||
| 2 | 647 | 1012 | 122 | 13.42 | + | AAGGAGTGAAAGAACATG | 2 | 97 | 2 | 97 | Putative terminase small subunit | Hypothetical protein (Lactobacillus phage c5) | 95 (115) | 4.00E–78 |
| 3 | 1002 | 2681 | 560 | 61.6 | + | ATAAATGGCGGCGAAGACAATG | 3 | 96 | 3 | 96 | Putative terminase large subunit | Putative terminase large subunit (Lactobacillus phage c5) | 98 (549) | 0 |
| 4 | 2698 | 3912 | 405 | 44.55 | + | TTAAGTTTGGGGGTGATTGATTG | 4 | 97 | 5 | 97 | Putative portal protein | Putative portal protein (Lactobacillus phage LL-Ku) | 97 (393) | 0 |
| 5 | 3902 | 4507 | 202 | 22.22 | + | AGGAGGTGAAGAAACAGATG | 5 | 98 | 6 | 98 | Putative ClpP protease | Putative ClpP protease (Lactobacillus phage c5) | 98 (196) | 4.00E–140 |
| 6 | 4510 | 5697 | 396 | 43.56 | + | TTAAGGAGGACGACTAAATATG | 6 | 96 | 7 | 95 | Putative major head protein | Putative major head protein (Lactobacillus phage c5) | 95 (376) | 0.00E + 00 |
| 7 | 5706 | 5873 | 56 | 6.16 | + | AGCCGGTTAATCAAAAAGATG | 7 | 96 | 8 | 96 | Hypothetical protein (Lactobacillus phage c5) | 96 (53) | 2.00E–30 | |
| 8 | 5866 | 6162 | 99 | 10.89 | + | AAAGGAGGTTGCTAAGAATG | 8 | 90 | 9 | 89 | Hypothetical protein (Lactobacillus phage LL-Ku) | 89 (87) | 9.00E–56 | |
| 9 | 6173 | 6526 | 118 | 12.98 | + | CTGTAAGGAGGTCATCATG | 9 | 99 | 10 | 99 | Putative head-tail adaptor | Putative head-tail adaptor (Lactobacillus phage c5) | 98 (115) | 7.00E–78 |
| 10 | 6526 | 6957 | 144 | 15.84 | + | GAAAGCGGTGATAAAGTAATG | 10 | 99 | 11 | 98 | Hypothetical protein (Lactobacillus phage c5) | 98 (141) | 2.00E–96 | |
| 11 | 6950 | 7291 | 114 | 12.54 | + | AGTGGAGGTCTATTAGGAGATG | 11 | 100 | 12 | 99 | Hypothetical protein (Lactobacillus phage c5) | 99 (112) | 3.00E–76 | |
| 12 | 7306 | 7878 | 191 | 21.01 | + | TAACGGAGGTTTTAAATATG | 12 | 100 | 13 | 98 | Putative major tail protein | Putative major tail protein (Lactobacillus phage c5) | 97 (183) | 5.00E–130 |
| 13 | 7898 | 8248 | 117 | 12.87 | + | TTGATGAGGTGAAGCACGATG | 13 | 100 | 14 | 99 | Hypothetical protein (Lactobacillus phage LL-Ku) | 100 (116) | 1.00E–76 | |
| 14 | 8493 | 11399 | 969 | 106.59 | + | AAATGGAGGTGATTTCATG | 14 | 97 | 15 | 96 | Putative tape measure protein | Putative tape measure protein (Lactobacillus phage c5) | 99 (955) | 0.00E + 00 |
| 15 | 11411 | 12115 | 235 | 25.85 | + | GATAGGAGGGCAGATAATG | 15 | 100 | 16 | 98 | Putative tail component | Putative tail component (Lactobacillus phage c5) | 99 (231) | 6.00E–166 |
| 16 | 12112 | 12450 | 113 | 12.43 | + | TATACGAGGTGGATATG | 16 | 99 | 17 | 96 | Hypothetical protein (Lactobacillus phage LL-Ku) | 95 (106) | 1.00E–71 | |
| 17 | 12464 | 15100 | 879 | 96.69 | + | TAAAAAGGTGGTTACTATG | 17 | 91 | 18 | 93 | Glycerophosphoryl diesterphosphodiesterase | Glycerophosphoryl diesterphosphodiesterase (Lactobacillus phage LL-Ku) | 93 (818) | 0.00E + 00 |
| 18 | 15248 | 16243 | 332 | 36.52 | + | GTGTGAGGAGGTAATATTTATG | 19 | 91 | Collagen repeat-containing protein | 34.8-kDa protein (Lactobacillus phage LL-K) | 81 (271) | 0 | ||
| 19 | 16248 | 16802 | 185 | 20.35 | + | CATTGAGGGGTGATTGAATG | 20 | 31 | Putative adsorption protein | Putative adsorption protein (Lactobacillus equicursoris CIP 110162) | 63 (99) | 8.00E–54 | ||
| 20 | 16804 | 18687 | 628 | 69.08 | + | AGACGTAGGAGCTTAACATG | 18 | 82 | 21 | 89 | Putative antireceptor | Putative antireceptor (Lactobacillus phage LL-Ku) | 87 (564) | 0.00E + 00 |
| 21 | 18700 | 19059 | 120 | 13.2 | + | TTAATGGAGTAAGCGAATG | 19 | 90 | 22 | 83 | Hypothetical protein (Lactobacillus phage c5) | 86 (102) | 9.00E–68 | |
| 22 | 19449 | 19652 | 68 | 7.48 | + | AAGGATGTACAAAGAAAATG | 20 | 91 | 23 | 91 | Putative Cro-like repressor | Putative Cro-like repressor (Lactobacillus phage c5) | 88 (59) | 7.00E–37 |
| 23 | 19655 | 19825 | 57 | 6.27 | + | AAGGAGGAAATGTAAAAATG | 21 | 88 | Hypothetical protein (Lactobacillus phage c5) | 96 (54) | 6.00E–27 | |||
| 24 | 19818 | 20444 | 209 | 22.99 | + | GATGGAGGAAGAAAAAAATG | 23, 22 | 87, 92 | 24 | 91 | Hypothetical protein (Lactobacillus phage c5) | 96 (199) | 6.00E–141 | |
| 25 | 20447 | 20725 | 93 | 10.23 | + | GGGAAGGACTTCTAAAAATG | 24 | 92 | 25 | 92 | Hypothetical protein (Lactobacillus phage LL-Ku) | 92 (82) | 1.00E–52 | |
| 26 | 20718 | 21035 | 106 | 11.66 | + | TAAGTGGAGCAATGACGATG | 25 | 96 | 26 | 96 | Hypothetical protein (Lactobacillus phage LL-Ku) | 96 (101) | 8.00E–66 | |
| 27 | 21040 | 21645 | 202 | 22.22 | + | GTAAAGGATTAAGCGAATG | 26 | 93 | 27 | 93 | Hypothetical protein (Lactobacillus phage LL-Ku) | 92 (185) | 1.00E–135 | |
| 28 | 21635 | 22096 | 154 | 16.94 | + | AGATTGGAGAAATAAACATG | 27 | 83 | 28 | 83 | Putative single-stranded DNA binding protein | Putative single-stranded DNA-binding protein (Lactobacillus phage LL-Ku) | 87 (133) | 2.00E–93 |
| 29 | 22107 | 22646 | 180 | 19.8 | + | CGTTTTAGGTGATGCAAGATG | 28 | 92 | 29 | 92 | Hypothetical protein (Lactobacillus phage c5) | 90 (161) | 4.00E–93 | |
| 30 | 22627 | 22908 | 94 | 10.34 | + | ATAGAGGCATTGAAAAGTG | 29 | 94 | 30 | 94 | Hypothetical protein (Lactobacillus phage LL-Ku) | 91 (85) | 6.00E–93 | |
| 31 | 22905 | 23276 | 124 | 13.64 | + | TCGGGAGGAACTGGTTATG | 31 | 100 | 32 | 91 | Putative Holliday junction resolvase | Putative Holliday junction resolvase (Lactobacillus phage LL-Ku) | 91 (107) | 8.00E–93 |
| 32 | 23588 | 23740 | 51 | 5.61 | + | GATAGGAGGTACACAAAATG | 32 | 88 | 34 | 88 | Hypothetical protein (Lactobacillus phage LL-Ku) | 88 (44) | 1.00E–92 | |
| 33 | 23795 | 24136 | 114 | 12.54 | + | TAAAGGAGTTTAAAAATG | 33 | 64 | Hypothetical protein HMPREF9465_00185 (Sutterella wadsworthensis 2_1_59BFAA) | 51 (50) | 1.00E–25 | |||
| 34 | 24164 | 24973 | 270 | 29.7 | + | CAGGAGGATCTTAAAAATG | 35 | 75 | 35 | 59 | Putative DNA replication protein | Putative DNA replication protein (Lactobacillus phage LL-Ku) | 66 (179) | 3.00E–90 |
| 35 | 24942 | 25757 | 272 | 29.92 | + | ATACGGAGGTAAAACGAGATG | 36 | 94 | 36 | 94 | Putative DnaC | Putative DnaC (Lactobacillus phage LL-Ku) | 95 (258) | 0 |
| 36 | 25915 | 26706 | 264 | 29.04 | + | AAAGGAGAAAACAAAATG | 37, 38 | 47, 92 | 37, 38 | 86, 77 | Phage antirepressor protein | Phage antirepressor protein (Lactobacillus casei A2–362) | 61 (157) | 2.00E–111 |
| 37 | 26871 | 27629 | 253 | 27.83 | + | AAAGGAAAAAACAAAATG | 37, 38 | 99, 98 | 38 | 97 | Phage antirepressor protein | Phage antirepressor (Staphylococcus prophage phiPV83) | 60 (150) | 1.00E–105 |
| 38 | 27771 | 27959 | 63 | 6.93 | + | AAAACGAGGTATTAAAAATG | 39 | 98 | 39 | 98 | Hypothetical protein (Lactobacillus phage LL-Ku) | 94 (58) | 3.00E–31 | |
| 39 | 27999 | 28400 | 134 | 14.74 | + | AAATAGAGGTAAACAAAATG | Hypothetical protein (Lactobacillus phage LL-Ku) | 88 (117) | 3.00E–75 | |||||
| 40 | 28427 | 28591 | 55 | 6.05 | + | AAAGGAGGAACTAAACATG | 46 | 63 | 47 | 63 | Hypothetical protein (Lactobacillus phage JCL1032) | 40 (17) | 8.40E + 00 | |
| 41 | 28606 | 28770 | 55 | 6.05 | + | AACTGGAGGTACAAAAATG | 39, 46 | 50, 37 | 39, 47 | 51, 37 | Hypothetical protein (Lactobacillus phage LL-Ku) | 51 (25) | 7.00E–06 | |
| 42 | 28785 | 29009 | 75 | 8.25 | + | TAAGCGGAGAAACAATCATG | Hypothetical protein (Lactobacillus phage LL-Ku) | 66 (41) | 5.00E–17 | |||||
| 43 | 28996 | 29208 | 71 | 7.81 | + | AGTGCGAGGCGGAAAATG | 41 | 74 | Hypothetical protein (Lactobacillus phage c5) | 93 (65) | 2.00E–38 | |||
| 44 | 29399 | 29665 | 89 | 9.79 | + | GATTGAGGCATTAAGCAAATG | 41 | 85 | 42 | 85 | hypothetical protein (Lactobacillus phage LL-Ku) | 97 (85) | 2.00E–54 | |
| 45 | 29662 | 29901 | 80 | 8.8 | + | TGAGGCAGGAGATCCGAAAAAATG | 42 | 85 | 43 | 85 | Hypothetical protein (Lactobacillus phage LL-Ku) | 76 (59) | 2.00E–37 | |
| 46 | 29898 | 30137 | 80 | 8.8 | + | AATGGAGGAAACGAGACCATG | 43 | 82 | 44 | 82 | Hypothetical protein (Lactobacillus phage LL-Ku) | 83 (65) | 7.00E–42 | |
| 47 | 30134 | 30622 | 163 | 17.93 | + | GAACGGGGGCTACCAAAATG | 44 | 64 | 45 | 67 | Hypothetical protein (Lactobacillus phage c5) | 64 (103) | 2.00E–60 | |
| 48 | 30628 | 30801 | 58 | 6.38 | + | CGGAAGGACTAAAAATCATG | 45 | 84 | 46 | 86 | Hypothetical protein (Lactobacillus phage LL-Ku) | 86 (49) | 2.00E–28 | |
| 49 | 31133 | 31426 | 98 | 10.78 | + | AAGGAGGAACTTAAAATATG | 47 | 89 | 48 | 89 | Putative holin | Putative holin (Lactobacillus phage c5) | 90 (87) | 5.00E–53 |
| 50 | 31419 | 32324 | 302 | 33.22 | + | TAAGGGAGACGAAAATATTG | 48 | 87 | 49 | 86 | Putative lysin | Putative lysin (Lactobacillus phage LL-Ku) | 85 (256) | 0.00E + 00 |
| 51 | 32328 | 32708 | 127 | 13.97 | + | Translationally fused | 49 | 94 | 50 | 95 | Hypothetical protein (Lactobacillus phage LL-Ku) | 97 (122) | 1.00E–82 | |
RBS, ribosome binding site. Sequences in boldface represent the putative RBS, followed by the putative start codon for each ORF.
That is, the percent identity at the indicated amino acid level to ORFs in the NCBI database (total number of amino acid matches). %S, % similarity.
FIG 2.
Genomic organization of Ld3, Ld17, and Ld25A compared to phiLdb, c5, and LL-Ku. The scale at the top of the genomes is in base pairs. Each arrow represents an ORF, with the color representing the putative function of the encoded protein that is indicated in the legend. Percent identities (amino acids) between adjacent genomes are colored as outlined on the right. ORFs confirmed as members of the structural proteome by mass spectrometric analysis are marked as bold arrows. The ORFs of Ld17 are numbered along the bottom of the genome map.
Morphogenesis module.
The group b phages are well conserved across most of the structural module from the gene encoding the portal protein (ORF4Ld25A) to the gene specifying the tail tape measure protein (TMP; ORF14Ld25A), with many predicted proteins sharing >95% amino acid (aa) identity (Table 3 and Fig. 2). Immediately downstream of the TMP-encoding gene begins the putative baseplate-encoding region. Using Ld25A as a representative, HHPred analysis of the protein encoded by ORF15Ld25A reveals similarity to the distal tail protein (Dit) of Bacillus phage SPP1 (38) and lactococcal phage TP901-1 (39), where the latter two proteins represent part of the so-called tail initiation complex upon which the baseplate is assembled (40). ORF17Ld25A encodes a protein, including a domain specifying a predicted glycerophosphoryl diester phosphodiesterase with a carbohydrate-binding domain in the C terminus. A base change from C to T at position 16255 in phiLdb has introduced a stop codon resulting in two ORFs; however, this could be an artifact of the sequencing of this phage. This ORF is found in a comparable position to the genes encoding gp-27/Tal-like protein in the baseplate model of various Gram-positive infecting phages (41). The presumed baseplate-encoding region is completed by the gene specifying the putative antireceptor, ORF20Ld25A, which shows 94 and 88% identity to the predicted antireceptor-encoding genes of LL-Ku and c5, respectively (6).
A notable region of divergence between the newly sequenced phages is located within this region represented by ORF19Ld17 and ORF20Ld17 for Ld17 and by ORF18Ld25A and ORF19Ld25A for Ld25A. This region is absent in the genomes of the other group b phages (Table 4 and Fig. 2). The GC content of this region was observed to be slightly higher than the GC content of the phages themselves (43.38% in Ld17 and 46.29% in Ld25A), suggesting a possible insertion event. This region bears resemblance to a 1.5-kb region in the group a L. delbrueckii subsp. lactis phage LL-K, termed the KIS element (LL-K insertion sequence) (42), and also to ORF36 and ORF37 and to ORF38 and ORF39 of Leuconostoc phages ΦLN03 and ΦLN12, respectively, where the presence of these genes, contained within the genes specifying the holin and lysin in the lysis module, appears to correlate with the presence of a neck passage structure (21). It is conceivable that these two ORFs were integrated into the Ld17 and Ld25A genomes as a result of acquisition from another L. delbrueckii phage or, conversely, that they were deleted from the other group b phages.
The encoded products of ORF19Ld17 and its homolog in Ld25A, designated here as Gp19Ld17 and Gp18Ld25A, respectively, contain a collagen-like triplet repeat, (G-X-Y)16, with the amino acid triplet GDK repeated seven times. This repeat sequence has been shown to facilitate binding to glycoproteins (43). ORF19Ld17 and ORF18Ld25A show 88.9 and 88.7% aa identity to ORF333LL-K of the KIS-Element, respectively. Collagen-like repeats have been found in bacteriophages, particularly in tail fiber proteins (44), and have been implicated as part of host specificity proteins in phage JCL1032 (6) and in streptococcal phages (45). The products of ORF20Ld17 (199 aa) and ORF19Ld25 (184 aa) show limited identity to each other (Table 4) and 32 and 33% identity, respectively, to ORF171LL-K. Although these proteins have no reported function they exhibit similarity to a range of phage tail proteins, including the tail fiber protein of a prophage-like element in Lactobacillus casei A2-362 and a putative adsorption protein of Lactobacillus prophage-like elements (Table 4). The low degree of identity between the two putative adsorption proteins is the only significant area of divergence in this module between the potential host recognition proteins of Ld17 and Ld25A. Notably, Ld25A has the ability to infect more than twice the number of strains compared to Ld17 (23 versus 10 strains; Table 1), suggesting that this protein may play a role in host range determination of this phage.
The KIS element is one of the main genomic differences between Ld17 and Ld25A on the one hand, and Ld3 on the other, and comparing the structures of the three phages leads us to believe that at least one of the genes in the KIS element is responsible for the presence of the collar on Ld17 and Ld25A (Fig. 1). Also, the apparent insertion/deletion of genes in this region is reminiscent of the presence or absence of the bppA gene in lactococcal phages Tuc2009 and TP901-1, respectively, where the encoded BppA acts as an accessory protein in the baseplate, which results in an increased binding affinity to the host receptor material (46). Such insertion events may thus play a crucial role in enhancing the ecological fitness of phages when the acquired genes play a role in host range expansion/diversification. Downstream of the KIS element lies the gene encoding the putative antireceptor of the group b phages, the component responsible for the binding of the phage to its receptor on the bacterial cell surface. This protein was suggested as the host recognition protein (6) based on similarities to Gp71, the confirmed antireceptor of the L. delbrueckii phage LL-H (15). A CLUSTAL W alignment of the putative antireceptors of Ld3 (Gp18), Ld17 (Gp21), and Ld25A (Gp20) with the antireceptors of two highly related phages c5, LL-Ku and phiLdb (see Fig. S2 in the supplemental material), shows a high degree of identity (93% between Ld17 and c5 from residues 1 to 438) between the putative antireceptors in their N termini. Receptor-binding proteins typically possess a conserved N terminus and variable C terminus, where the latter has been implicated to play a role in host specificity in Lactobacillus phage JCL1032 (15) and in many other lactic acid bacteria phages, such as S. thermophilus phage DT1 (45) and lactococcal 936 and P335 phages. In lactococcal phages, identity in the C-terminal portion of the antireceptors correlates with binding and subsequent infection of the same lactococcal strains (22). The group b phages show divergence in this region, especially in the C-terminal region of the putative antireceptor of Ld3 from amino acid residues 586 to 609 (see Fig. S2 in the supplemental material). This divergence may account for the different host ranges of the phages. It has previously been noted (6) that the antireceptor proteins of c5 and LL-Ku contain four and two copies, respectively, of a 20-amino-acid repeat starting at position 441. This sequence is repeated twice in both Ld3 and Ld17, whereas it is absent in the predicted antireceptor of Ld25A (see Fig. S2 in the supplemental material). Although it is tempting to implicate this repeat sequence in host recognition, its precise role, if any, in this crucial aspect of phage infection remains to be elucidated.
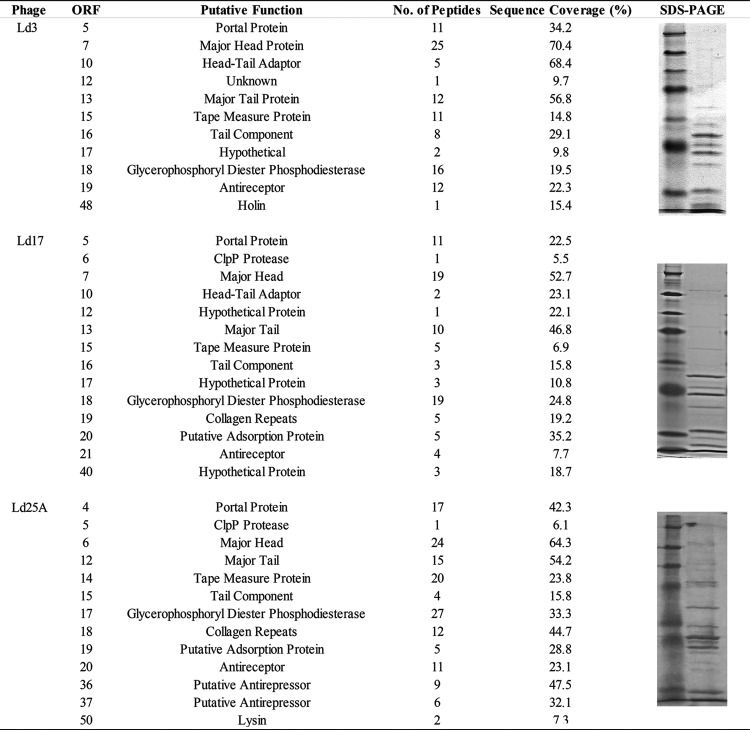
Structural protein identification.
Identified proteins associated with the phage particle of each of the three identified L. delbrueckii subsp. bulgaricus phages, as analyzed by mass spectrometry, are presented in Fig. 3, along with sequence coverage and the number of unique peptides per identified protein. Most predicted proteins in the structural module of the genomes were confirmed as structural proteins with the predicted portal, major head, major tail, tape measure, tail component, a tail fiber protein with a glycerophosphoryl diester phosphodiesterase (GDPD) motif, and the antireceptor confirmed in all phages. The putative head-tail adaptor was detected in all phages except Ld25A. This absence may be due to the small size and low relative abundance of the protein. The two ORFs of interest in the tail regions of Ld17 and Ld25A (Gp19Ld17, Gp20Ld17, Gp18Ld25A, and Gp19Ld25A [see above]), encoding a putative collagen repeat-containing protein and adsorption protein, respectively, were also identified in the corresponding phage particles. The major head protein shows a lower than predicted molecular weight, presumably due to posttranslational modification by the putative ClpP protease (47). The ClpP protease also appears as part of the virion proteome, which may be an artifact due to ClpP remaining bound to the major capsid protein following modification. Other proteins were identified in the analysis that are not considered structural elements, such as the holin of Ld3 and the lysin of Ld25A. A holin and an endolysin have previously been observed as part of the structural protein profile of the lactococcal phage Q54 (48). It is, however, unlikely that they form part of the phage particle, and it would not be inconceivable that these proteins remained in the phage environment following lysis of the cells during propagation.
FIG 3.
Structural proteins extracted from purified phage particles were identified by ESI-MS/MS after separation on a SDS–12% PAGE gel (lane 2) and broad-range protein ladder (7 to 175 kDa; New England BioLabs) (lane 1). A minimum of two independent unique peptides or 5% coverage were used as threshold values.
Replication module.
The DNA replication regions are often a common area of divergence among Lactobacillus phages (49). Some areas of divergence are evident especially in the DNA replication protein (ORF34Ld25A) (Table 4). However, in general, the gene order and functions of the replication module are conserved between all members of the group b phages (Fig. 2). The “initiator-helicase loader” type replication module, consisting of genes encoding the putative DNA replication initiator (DnaA) and the replication loader (DnaC), is conserved among the group b phages.
Also contained in this module, Ld25A carries two full-length putative antirepressor proteins—ORF36Ld25A and ORF37Ld25A—with apparently truncated antirepressors present in both Ld3 and Ld17. All three phages are obligately lytic, so the presence of these genes suggests a temperate origin of these phages. For Ld25A, these two antirepressors were identified as part of the structural proteome, a finding that is similar to that observed for the temperate lactococcal phage Tuc2009 (50).
Transcriptomic analysis of Ld17.
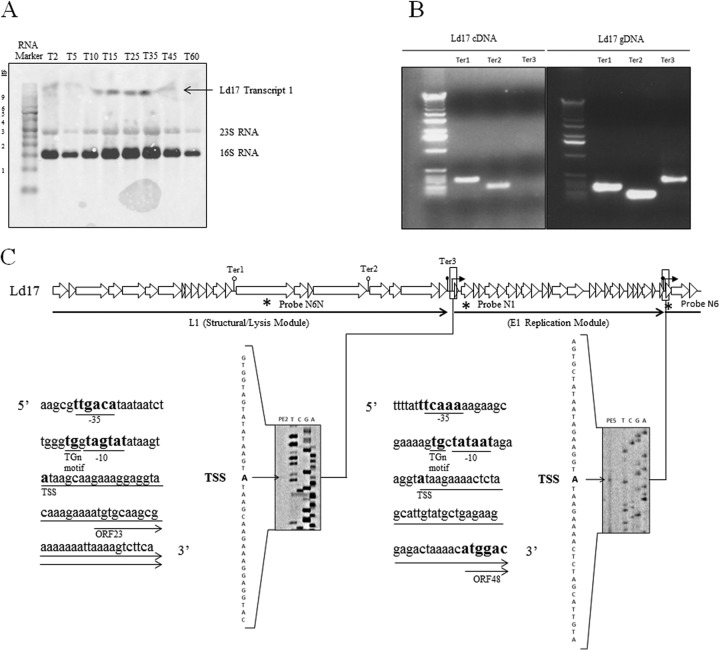
In order to obtain transcriptional information regarding a representative member of group b phages, Northern hybridizations were undertaken for Ld17. A previous in silico study (6) had predicted the transcriptome of the group b phages c5 and LL-Ku to consist of eight transcripts, and these predictions were used as a basis for probe design in the present study with the addition of a ninth predicted terminator upstream of the gene encoding the putative TMP.
Antisense RNA probes targeting the 5′ ends of these transcripts were assessed for hybridization to Ld17 total mRNA. Use of probe 17N1 (Fig. 4C) targeting the 5′ end of the replication region generated a hybridization signal corresponding to a transcript with an estimated size of >9 kb (rather than the in silico predicted 4.5-kb transcript) that was present 2 min postinfection and persisted until 45 min (Fig. 4A). The replication region of phage Ld17 therefore appears to be transcribed as a single, early transcript, designated E1, from position 20496 to the terminator at positions 31190 to 31205 (ΔG = −10.50 kcal mol−1). This suggested that the five in silico-predicted terminators in this region were nonfunctional. The transcription start site was determined by primer extension analysis and the predicted −10 and −35 promoter recognition sequences designated by visual inspection of the upstream sequences (Fig. 4C) with the −10 promoter recognition sequence exhibiting an extended TGn motif, which has been shown in other lactic acid bacteria to enhance promoter activity (51).
FIG 4.
Northern hybridization, RT-PCR, and primer extension analyses. (A) Northern hybridization of probe N1 to total RNA of Ld17 after infection with Ld17 at an MOI of 0.1. The time (in minutes) postinfection is indicated by the numbers along the top, with the sizes of the marker transcripts of the BrightStar Biotinylated Millenium Marker (Ambion) indicated down the left hand side. Bands due to nonspecific hybridization to rRNA are marked. (B) RT-PCR products for Ter1, Ter2, and Ter3. The left panel represents products amplified from cDNA; the right panel shows the products amplified from the phage genomic DNA control. (C) Primer extensions at the 5′ ends of transcripts E1 and L1. Transcription start sites are marked in boldface, along with the −35 and −10 promoter recognition sequences.
The remainder of the genome was predicted to be transcribed in two units, from the holin-encoding gene to a predicted terminator at positions 9294 to 9318 (Ter1) and from the gene encoding the putative TMP to a predicted terminator at positions 20316 to 20333 (Ter3). Probes targeting the genes encoding the predicted TMP and holin-encoding genes, 17N6N and 17N6 (Fig. 4C), respectively, failed to generate a detectable hybridization signal by Northern blotting. RT-PCR was therefore adopted to determine the boundaries of transcripts corresponding to the above-mentioned region. Total mRNA obtained at 45 min postinfection was reverse transcribed, and the generated cDNA was used as a template for PCR amplification using various primer pairs (Table 2) and targeting regions flanking the putative terminators Ter1, Ter2 (a 148-bp noncoding region upstream of the KIS element), and Ter3 (Fig. 4C). Products were amplified across both Ter1 and Ter2 (Fig. 4B) and all other targeted coding regions; therefore, these terminators were assumed to be nonfunctional, and the region is transcribed as a single transcript. Ter3 primers, however, did not generate an amplicon (Fig. 4B), demonstrating that Ter3 is a functioning transcriptional terminator. Confirmatory PCRs were also performed amplifying nonterminator regions, within coding regions (Table 2), to ensure that a PCR product was consistently amplified from cDNA in the absence of a terminator (see Fig. S3 in the supplemental material). This suggests that genes encoding the structural region are transcribed as a single long transcript, 22,059 bp in length from position 31250 to Ter3 (positions 20316 to 20333, ΔG = −12.40 kcal mol−1), with the transcription start site and promoter region defined by primer extension (Fig. 4C). The aforementioned PCRs were also carried out on cDNA from all time points used in the Northern hybridization (2, 5, 10, 15, 25, 35, 45, and 60 min postinfection) to investigate the temporal characteristics of this second transcript. Products were amplified using Ter1 and Ter2 primers for time points from 15 through to 60 min but not before (see Fig. S4 in the supplemental material). Therefore, this transcript is termed the late transcript (L1) since its transcription is initiated at a later time point than the E1 transcript. The transcriptome of Ld17 has therefore been defined as containing two transcripts, E1 and L1. Of nine predicted terminators predicted in Ld17, only two were determined to be functional, reducing the transcriptome of this group b phage from nine predicted transcripts to two transcripts in vivo.
Conclusions.
In the present study, we sequenced and analyzed the genomes of three newly identified phages infecting L. delbrueckii subsp. bulgaricus, named Ld3, Ld17, and Ld25A. Despite the differences in geographical origins and times of isolation between these phages, as well as their different host ranges, they bear a striking genetic resemblance to each other and to two previously isolated phages. To our knowledge, only four virulent L. delbrueckii phage genomes have been published to date, making it difficult to conclude whether the apparent lack of genetic diversity in these phages is due to a limited diversity of cultures being applied in such fermentations. With the addition of three phages to the complement of sequenced L. delbrueckii group b phages, more than doubling the number of sequenced members of this particular group, such phages represent the most dominant genotype infecting L. delbrueckii.
In recent years, several Siphoviridae baseplate models have emerged through structural analysis of the baseplate complexes and components of phages infecting Bacillus and Lactococcus (38, 39, 46). It appears that while sequences may not be highly conserved between these phages, their encoded structures and thus presumably their functional capacities are well conserved (reviewed in reference 41). In the present study, parallels were drawn between the baseplate structure of the L. delbrueckii group b phages and other Siphoviridae. The similarity in baseplate organization and morphology to lactococcal P335 phage members, including TP901-1 and Tuc2009, highlights the maintenance of functionally important features and structures and aids in our understanding of phage-host interactions of this industrially significant group of phages. The wide baseplate in lactococcal P335 phages has been associated with multiple binding sites to the carbohydrate receptor on the host providing high avidity and strong binding on the surface of the host. It is possible that this organelle in the group b phages of Lactobacillus provides a similar function and strengthens previous Gram-positive Siphoviridae baseplate models as broad models for phage-host interaction analyses. Functional homology is evident between phages infecting Bacillus and Lactococcus and now, once more, this baseplate structure may be responsible for the emergence of group b phages as the most dominant genotype infecting L. delbrueckii.
Supplementary Material
ACKNOWLEDGMENTS
E.C. is the recipient of a scholarship from the Irish Research Council Enterprise Partnership Scheme. D.V.S. was supported by a Principal Investigator award (reference numbers 08/IN.1/B1909 and 13/IA/1953) through Science Foundation Ireland (SFI). F.B. is supported by SFI under grants 07/CE/B1368 and SFI/12/RC/2273. A.C. is the recipient of an EMPOWER postdoctoral fellowship of the Irish Research Council. M.O.-M. is a recipient of an HRB postdoctoral fellowship (PDTM/20011/9). The Orbitrap mass spectrometric services provided in this study were supported by Hercules Foundation (Belgium) project R-3986 with technical assistance from Erik Royackers.
Footnotes
Published ahead of print 7 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01268-14.
REFERENCES
- 1.van Kranenburg R, Kleerebezem M, van Hylckama Vlieg J, Ursing BM, Boekhorst J, Smit BA, Ayad EH, Smit G, Siezen RJ. 2002. Flavour formation from amino acids by lactic acid bacteria: predictions from genome sequence analysis. Int. Dairy J. 12:111–121. 10.1016/S0958-6946(01)00132-7 [DOI] [Google Scholar]
- 2.Sieuwerts S, de Bok FA, Hugenholtz J, van Hylckama Vlieg JE. 2008. Unraveling microbial interactions in food fermentations: from classical to genomics approaches. Appl. Environ. Microbiol. 74:4997–5007. 10.1128/AEM.00113-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zago M, Suarez V, Reinheimer J, Carminati D, Giraffa G. 2007. Spread and variability of the integrase gene in Lactobacillus delbrueckii ssp. lactis strains and phages isolated from whey starter cultures. J. Appl. Microbiol. 102:344–351 [DOI] [PubMed] [Google Scholar]
- 4.Zourari A, Accolas J, Desmazeaud M. 1992. Metabolism and biochemical characteristics of yogurt bacteria: a review. Lait 72:1–34. 10.1051/lait:199211 [DOI] [Google Scholar]
- 5.Klaenhammer T, Fitzgerald G. 1994. Bacteriophages and bacteriophage resistance, p 106–168 In Gasson MJ, de Vos W. (ed), Genetics and biotechnology of lactic acid bacteria; Springer, New York, NY [Google Scholar]
- 6.Riipinen K-A, Forsman P, Alatossava T. 2011. The genomes and comparative genomics of Lactobacillus delbrueckii phages. Arch. Virol. 156:1217–1233. 10.1007/s00705-011-0980-5 [DOI] [PubMed] [Google Scholar]
- 7.Mikkonen M, Räisänen L, Alatossava T. 1996. The early gene region completes the nucleotide sequence of Lactobacillus delbrueckii subsp. lactis phage LL-H. Gene 175:49–57. 10.1016/0378-1119(96)00119-9 [DOI] [PubMed] [Google Scholar]
- 8.Wang S, Kong J, Gao C, Guo T, Liu X. 2010. Isolation and characterization of a novel virulent phage (phiLdb) of Lactobacillus delbrueckii. Int. J. Food Microbiol. 137:22–27. 10.1016/j.ijfoodmicro.2009.10.024 [DOI] [PubMed] [Google Scholar]
- 9.Lahbib-Mansais Y, Mata M, Ritzenthaler P. 1988. Molecular taxonomy of Lactobacillus phages. Biochimie 70:429–435. 10.1016/0300-9084(88)90217-9 [DOI] [PubMed] [Google Scholar]
- 10.Forsman P. 1993. Characterization of a prolate-headed bacteriophage of Lactobacillus delbrueckii subsp. lactis, and its DNA homology with isometric-headed phages. Arch. Virol. 132:321–330. 10.1007/BF01309542 [DOI] [PubMed] [Google Scholar]
- 11.Sechaud L, Cluzel P-J, Rousseau M, Baumgartner A, Accolas J-P. 1988. Bacteriophages of lactobacilli. Biochimie 70:401–410. 10.1016/0300-9084(88)90214-3 [DOI] [PubMed] [Google Scholar]
- 12.Alatossava T, Pyhtila M. 1980. Characterization of a new Lactobacillus lactis bacteriophage. IRCS Med. Sci. 8:297–298 [Google Scholar]
- 13.Accolas JP, Spillmann H. 1979. Morphology of bacteriophages of Lactobacillus bulgaricus, L. lactis, and L. helveticus. J. Appl. Microbiol. 47:309–319 [Google Scholar]
- 14.Munsch-Alatossava P, Alatossava T. 2013. The extracellular phage-host interactions involved in the bacteriophage LL-H infection of Lactobacillus delbrueckii ssp. lactis ATCC 15808. Front. Microbiol. 4:408. 10.3389/fmicb.2013.00408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravin V, Räisänen L, Alatossava T. 2002. A conserved C-terminal region in Gp71 of the small isometric-head phage LL-H and ORF474 of the prolate-head phage JCL1032 is implicated in specificity of adsorption of phage to its host, Lactobacillus delbrueckii. J. Bacteriol. 184:2455–2459. 10.1128/JB.184.9.2455-2459.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Räisänen L, Draing C, Pfitzenmaier M, Schubert K, Jaakonsaari T, von Aulock S, Hartung T, Alatossava T. 2007. Molecular interaction between lipoteichoic acids and Lactobacillus delbrueckii phages depends on d-alanyl and α-glucose substitution of poly(glycerophosphate) backbones. J. Bacteriol. 189:4135–4140. 10.1128/JB.00078-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Räisänen L, Schubert K, Jaakonsaari T, Alatossava T. 2004. Characterization of lipoteichoic acids as Lactobacillus delbrueckii phage receptor components. J. Bacteriol. 186:5529–5532. 10.1128/JB.186.16.5529-5532.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kropinski AM, Prangishvili D, Lavigne R. 2009. Position paper: the creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ. Microbiol. 11:2775–2777. 10.1111/j.1462-2920.2009.01970.x [DOI] [PubMed] [Google Scholar]
- 19.Svensson U, Christiansson A. 1991. Methods for phage monitoring. Bull. Int. Dairy Fed. 1991:29–39 [Google Scholar]
- 20.Lillehaug D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J. Appl. Microbiol. 83:85–90. 10.1046/j.1365-2672.1997.00193.x [DOI] [PubMed] [Google Scholar]
- 21.Kot W, Hansen LH, Neve H, Hammer K, Jacobsen S, Pedersen PD, Sorensen SJ, Heller KJ, Vogensen FK. 2014. Sequence and comparative analysis of Leuconostoc dairy bacteriophages. Int. J. Food Microbiol. 176:29–37. 10.1016/j.ijfoodmicro.2014.01.019 [DOI] [PubMed] [Google Scholar]
- 22.Dupont K, Vogensen FK, Neve H, Bresciani J, Josephsen J. 2004. Identification of the receptor-binding protein in 936-species lactococcal bacteriophages. Appl. Environ. Microbiol. 70:5818–5824. 10.1128/AEM.70.10.5818-5824.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24.Besemer J, Borodovsky M. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911–3920. 10.1093/nar/27.19.3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikkonen M, Vuoristo J, Alatossava T. 1994. Ribosome binding site consensus sequence of Lactobacillus delbrueckii subsp. lactis bacteriophage LL-H. FEMS Microbiol. Lett. 116:315–320. 10.1111/j.1574-6968.1994.tb06721.x [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138–D141. 10.1093/nar/gkh121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229. 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248. 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naville M, Ghuillot-Gaudeffroy A, Marchais A, Gautheret D. 2011. A web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 8:11–13. 10.4161/rna.8.1.13346 [DOI] [PubMed] [Google Scholar]
- 31.Ceyssens P-J, Mesyanzhinov V, Sykilinda N, Briers Y, Roucourt B, Lavigne R, Robben J, Domashin A, Miroshnikov K, Volckaert G. 2008. The genome and structural proteome of YuA, a new Pseudomonas aeruginosa phage resembling M6. J. Bacteriol. 190:1429–1435. 10.1128/JB.01441-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanheel A, Daniels R, Plaisance S, Baeten K, Hendriks JJ, Leprince P, Dumont D, Robben J, Brône B, Stinissen P. 2012. Identification of protein networks involved in the disease course of experimental autoimmune encephalomyelitis, an animal model of multiple sclerosis. PLoS One 7:e35544. 10.1371/journal.pone.0035544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Z, Breidt F, Jr, Fleming H, Altermann E, Klaenhammer T. 2003. Isolation and characterization of a Lactobacillus plantarum bacteriophage, ΦJL-1, from a cucumber fermentation. Int. J. Food Microbiol. 84:225–235. 10.1016/S0168-1605(03)00111-9 [DOI] [PubMed] [Google Scholar]
- 34.Van Hijum SA, De Jong A, Baerends RJ, Karsens HA, Kramer NE, Larsen R, den Hengst CD, Albers CJ, Kok J, Kuipers OP. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. 10.1186/1471-2164-6-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ventura M, Fitzgerald GF, van Sinderen D. 2005. Genetic and transcriptional organization of the clpC locus in Bifidobacterium breve UCC 2003. Appl. Environ. Microbiol. 71:6282–6291. 10.1128/AEM.71.10.6282-6291.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brüssow H, Desiere F. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213–223. 10.1046/j.1365-2958.2001.02228.x [DOI] [PubMed] [Google Scholar]
- 37.Villion M, Moineau S. 2008. Bacteriophages of Lactobacillus. Front. Biosci. 14:1661–1683. 10.2741/3332 [DOI] [PubMed] [Google Scholar]
- 38.Veesler D, Robin G, Lichière J, Auzat I, Tavares P, Bron P, Campanacci V, Cambillau C. 2010. Crystal structure of bacteriophage SPP1 distal tail protein (gp19. 1): a baseplate hub paradigm in Gram-positive infecting phages. J. Biol. Chem. 285:36666–36673. 10.1074/jbc.M110.157529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veesler D, Spinelli S, Mahony J, Lichière J, Blangy S, Bricogne G, Legrand P, Ortiz-Lombardia M, Campanacci V, van Sinderen D, Cambillau C. 2012. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc. Natl. Acad. Sci. U. S. A. 109:8954–8958. 10.1073/pnas.1200966109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vegge CS, Brøndsted L, Neve H, Mc Grath S, van Sinderen D, Vogensen FK. 2005. Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 187:4187–4197. 10.1128/JB.187.12.4187-4197.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veesler D, Cambillau C. 2011. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev. 75:423–433. 10.1128/MMBR.00014-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forsman P, Alatossava T. 1994. Repeated sequences and the sites of genome rearrangements in bacteriophages of Lactobacillus delbrueckii subsp. lactis. Arch. Virol. 137:43–54. 10.1007/BF01311172 [DOI] [PubMed] [Google Scholar]
- 43.Scarborough RM, Naughton MA, Teng W, Rose J, Phillips D, Nannizzi L, Arfsten A, Campbell A, Charo I. 1993. Design of potent and specific integrin antagonists: peptide antagonists with high specificity for glycoprotein IIb-IIIa. J. Biol. Chem. 268:1066–1073 [PubMed] [Google Scholar]
- 44.Smith M, Burns N, Sayers JR, Sorrell JA, Caskems SR, Jurgen-Engel RW. 1998. Bacteriophage collagen. Science 279:1831. [DOI] [PubMed] [Google Scholar]
- 45.Duplessis M, Moineau S. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325–336. 10.1046/j.1365-2958.2001.02521.x [DOI] [PubMed] [Google Scholar]
- 46.Collins B, Bebeacua C, Mahony J, Blangy S, Douillard FP, Veesler D, Cambillau C, van Sinderen D. 2013. Structure and functional analysis of the host recognition device of lactococcal phage Tuc2009. J. Virol. 87:8429–8440. 10.1128/JVI.00907-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dokland T. 1999. Scaffolding proteins and their role in viral assembly. Cell. Mol. Life Sci. 56:580–603. 10.1007/s000180050455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fortier L-C, Bransi A, Moineau S. 2006. Genome sequence and global gene expression of Q54, a new phage species linking the 936 and c2 phage species of Lactococcus lactis. J. Bacteriol. 188:6101–6114. 10.1128/JB.00581-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weigel C, Seitz H. 2006. Bacteriophage replication modules. FEMS Microbiol. Rev. 30:321–381. 10.1111/j.1574-6976.2006.00015.x [DOI] [PubMed] [Google Scholar]
- 50.Seegers JF, McGrath S, O'Connell-Motherway M, Arendt EK, van de Guchte M, Creaven M, Fitzgerald GF, van Sinderen D. 2004. Molecular and transcriptional analysis of the temperate lactococcal bacteriophage Tuc2009. Virology 329:40–52. 10.1016/j.virol.2004.07.003 [DOI] [PubMed] [Google Scholar]
- 51.Henkin TM, Sonenshein AL. 1987. Mutations of the Escherichia coli lacUV5 promoter resulting in increased expression in Bacillus subtilis. Mol. Gen. Genet. 209:467–474. 10.1007/BF00331151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.