Abstract
Motivation: Efficient and fast next-generation sequencing (NGS) algorithms are essential to analyze the terabytes of data generated by the NGS machines. A serious bottleneck can be the design of such algorithms, as they require sophisticated data structures and advanced hardware implementation.
Results: We propose an open-source library dedicated to genome assembly and analysis to fasten the process of developing efficient software. The library is based on a recent optimized de-Bruijn graph implementation allowing complex genomes to be processed on desktop computers using fast algorithms with low memory footprints.
Availability and implementation: The GATB library is written in C++ and is available at the following Web site http://gatb.inria.fr under the A-GPL license.
Contact: lavenier@irisa.fr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The analysis of next-generation sequencing (NGS) data remains a time- and space-consuming task. Many efforts have been made to provide efficient data structures for indexing the terabytes of data generated by the fast sequencing machines (Suffix Array, Burrows–Wheeler transform, Bloom filter, etc.). Genome assemblers such as Velvet (Zerbino and Birney, 2008), ABySS (Simpson et al., 2009), SOAPdenovo2 (Luo et al., 2012), SPAdes (Bankevich et al., 2012) or mappers such as BWA (Li and Durbin, 2009) or variant detection such as CRAC (Philippe et al., 2013) for instance make an intensive use of these data structures to keep their memory footprint as low as possible.
At the same time, parallelism has been largely investigated to reduce execution time. Many strategies such as GPU implementation (Liu et al., 2012), cloud deployment (Zhao et al., 2013), algorithm vectorization (Rizk and Lavenier, 2010), multithreading, etc., have demonstrated high potentiality on NGS processing.
The overall efficiency of NGS software depends on a smart combination of data representation and use of the available processing units. Developing such software is thus a real challenge, as it requires a large spectrum of competence from high-level data structure and algorithm concepts to tiny details of implementation.
The GATB library aims to ease the design of NGS algorithms. It offers a panel of high-level optimized building blocks to speedup the development of NGS tools related to genome assembly and/or genome analysis. The underlying data structure is a memory efficient de-Bruijn graph (Compeau et al., 2011), and the general parallelism model is multithreading. The GATB library targets standard computing resources such as current multicore processor (laptop computer, small server) with a few gigabytes of memory.
Hence, from the high-level C++ functions available in the GATB library, NGS programing designers can rapidly elaborate their own software based on state-of-the-art algorithms and data structures of the domain.
Based on the same idea, other bioinformatics libraries exist, from which domain-specific tools can be elaborated. The NGS++ library (Markovits et al., 2013) is specifically tailored for developing applications that work with genomic regions and features, such as epigenomics marks, gene features and data that are associated with BED type files. The SeqAn library (Doring et al., 2008) is a general-purpose library targeting standard sequence processing. Advanced data structures such as de-Bruijn graphs are not included in SeqAn. Khmer (Crusoe et al., 2014) is a library and toolkit for doing k-mer-based NGS dataset analysis. As with GATB, most of khmer relies on an underlying probabilistic data structure (Bloom filter). The khmer library can be used in various k-mer processing such as abundance filtering, error trimming, graph size filtering or partitioning.
2 METHODS
One of the main concerns of the GATB-core library is to provide computing modules able to run on standard machines, i.e. computers not requiring large amount of main memory.
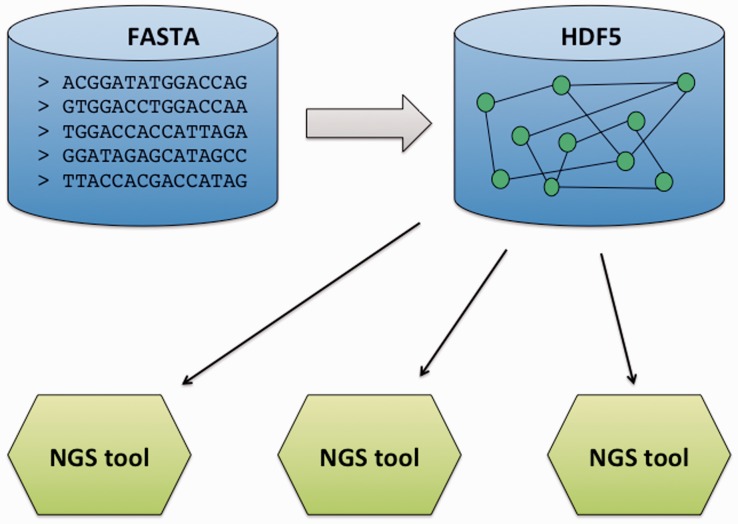
The central data structure is a de-Bruijn graph from which numerous actions can be performed as shown Figure 1: data error correction, assembly, biological motif detection [e.g. single nucleotide polymorphism (SNP)], etc. The graph is constructed by extracting and by counting all the different k-mers from one or several sequencing datasets. This time- and space-consuming task is conducted by a disk streaming algorithm, DSK (Rizk et al., 2013), which adapts its memory requirement according to the available computer memory. Trade-off between execution time and memory occupancy can be set up: the larger the computer memory, shorter the computation time (reduced disk access).
Fig. 1.
Schematic view of the GATB organization
The de-Bruijn graph memory footprint is kept low thanks to an optimized Bloom filter representation (Chikhi and Rish, 2012; Salikhov et al., 2014). Only vertices of the de-Bruijn graph are memorized. Edges are deduced by querying the Bloom filter. False positives (owing to the probabilistic behavior of the Bloom filter) are suppressed by adding an extra data structure enumerating critical vertices. This efficient de-Bruijn graph representation fits, for example, a complete mammal genome in ∼4 GB.
3 IMPLEMENTATION
The GATB library is composed of five main packages: system, tools, bank, kmer and de-Bruijn packages.
The system package holds all the operations related to the operating system (OS): file management, memory management and thread management. Using such an abstraction allows client code to be independent from the OS, thus suppressing compilation directive inside the code or improving some OS accesses by hiding specific OS optimization. The supported operating systems are Linux, Mac and Windows.
The tools package offers generic operations used throughout the user application but not specific to genomics area. For example, this package includes design pattern tools (such as iterators, observers, smart pointers, etc.) and object collections (such as containers, bags, iterables, etc.). It also optimizes the way GATB data structures are saved. The HDF5 file format is currently used (HDF5, 2012). This powerful technology is extremely well suited for large and complex data collection such as those handled in the GATB library.
The bank package provides operations related to standard genomic sequence dataset management. All the main sequence file formats are supported, and high-level interfaces allow sequences to be easily iterated regardless of the input format. In other words, algorithms are written independently of the input formats.
The kmer package is dedicated to fine-grained manipulation of k-mers. Optimized routines are provided to perform k-mer counting from large sequence datasets, to find k-mer neighborhood or to select k-mers based on different criteria.
Finally, the de-Bruijn package provides high-level functions to manipulate a static de-Bruijn graph data structure: creation from a set of k-mers, iteration through different nature of nodes (simple k-mers, branching k-mers, etc.), extraction of neighbor nodes, etc. Additional information (e.g. k-mer coverage, markers of visited nodes) is stored in the graph branching nodes. From this abstraction level, developing new tools based on de-Buijn graphs is fast, and does not require programmers to delve into low-level details.
The GATB library takes benefit of the parallel nature of today’s multicore architecture of microprocessors. When possible, time-consuming parts of the code are multithreaded to provide fast runtime execution.
The GATB library is developed in C++ under the A-GPL license and is available from the following Web site: http://gatb.inria.fr. An extensive documentation with tutorials is available to guide designers in the process of developing new NGS tools from the GATB building blocks: http://gatb-core.gforge.inria.fr (see also Supplementary File 2 for technical implementation details).
4 RESULTS
To demonstrate the efficiency of the GATB library, a few software implemented from GATB are briefly presented. The idea is to give a quick overview of the application spectrum of the GATB library and some performance numbers.
Minia (Chikhi and Rish, 2012) is a short-read de-Bruijn assembler capable of assembling large and complex genomes into contigs on a desktop computer. The assembler produces contigs of similar length and accuracy to other de-Bruijn assemblers––e.g. Velvet (Zerbino and Birney, 2008). As an example, a Boa constrictor constrictor (1.6 Gb) dataset (Illumina 2 × 120 bp reads, 125 × coverage) from Assemblathon 2 (Bradnam et al., 2013) can be processed in ∼45 h and 3 GB of memory on a standard computer (3.4 GHz 8-core processor) using a single core, yielding a contig N50 of 3.6 kb (prior to scaffolding and gap-filling).
Bloocoo is a k-mer spectrum-based read error corrector, designed to correct large datasets with low memory footprints. It uses the disk streaming k-mer counting algorithm contained in the GATB library and inserts solid k-mers in a Bloom filter. The correction procedure is similar to the Musket multistage approach (Liu et al., 2013). Bloocoo yields similar results while requiring far less memory: for example, it can correct whole human genome re-sequencing reads at 70× coverage with <4 GB of memory (see Supplementary file 1 for extra information on Bloocoo).
DiscoSNP aims to discover Single Nucleotide Polymorphism from non-assembled reads and without a reference genome. From one or several datasets a global de-Bruijn graph is constructed, then scanned to locate specific SNP graph patterns (Uricaru et al., 2014). A coverage analysis on these particular locations can finally be performed to validate and assign scores to detected biological elements. Applied on a mouse dataset (2.88 Gb, 100 bp Illumina reads), DiscoSnp takes 34 h and requires 4.5 GB RAM. In the same spirit, the TakeABreak software discovers inversion variants from non-assembled reads. It directly finds particular patterns in the de-Bruijn graph and provides execution performances similar to DiscoSNP (Lemaitre et al., 2014).
Funding: ANR (French National Research Agency) (ANR-12-EMMA- 0019-01).
Conflict of interest: none declared.
Supplementary Material
REFERENCES
- Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KR, et al. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience. 2013;2:10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi R, Risk G. Space-efficient and exact de-Bruijn graph representation based on a Bloom filter. Algorithms Bioinform. 2012;8:236–248. doi: 10.1186/1748-7188-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compeau P, et al. How to apply de Bruijn graphs to genome assembly. Nat. Biotechnol. 2011;29:987–991. doi: 10.1038/nbt.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring A, et al. SeqAn:an efficient generic C++ loibrary for sequence analysis. BMC Bioinformatics. 2008;9:11. doi: 10.1186/1471-2105-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HDF5 group help desk. File format specification v2.0. 2012 http://www.hdfgroup.org/HDF5/doc/H5.format.html. [Google Scholar]
- Crusoe MR, et al. The khmer software package: enabling efficient sequence analysis. 2014 doi: 10.12688/f1000research.6924.1. [Epub ahead of print, doi: 10.6084/m9.figshare.979190] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre C, et al. First International Conference on Algorithms for Computational Biology (AlCoB 2014) Tarragona, Spain: 2014. Mapping-free and assembly-free discovery of inversion breakpoints from raw NGS reads. [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. Musket: a multistage k-mer spectrum-based error corrector for Illumina sequence data. Bioinformatics. 2013;29:308–315. doi: 10.1093/bioinformatics/bts690. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. CUSHAW: a CUDA compatible short read aligner to large genomes based on the Burrows–Wheeler transform. Bioinformatics. 2012;28:1830–1837. doi: 10.1093/bioinformatics/bts276. [DOI] [PubMed] [Google Scholar]
- Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovits A, et al. NGS++: a library for rapid prototyping of epigenomics software tools. Bioinformatics. 2013;29:1893–1894. doi: 10.1093/bioinformatics/btt312. [DOI] [PubMed] [Google Scholar]
- Philippe N, et al. CRAC: an integrated approach to the analysis of RNA-seq reads. Genome Biol. 2013;14:R30. doi: 10.1186/gb-2013-14-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk G, Lavenier D. GASSST: global alignment short sequence search tool. Bioinformatics. 2010;26:2534–2540. doi: 10.1093/bioinformatics/btq485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk G, et al. DSK: k-mer counting with very low memory usage. Bioinformatics. 2013;29:652–653. doi: 10.1093/bioinformatics/btt020. [DOI] [PubMed] [Google Scholar]
- Salikhov K, et al. Using cascading bloom filters to improve the memory usage for de-Bruijn graph. Algorithms Mol Biol. 2014;9:2. doi: 10.1186/1748-7188-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uricaru R, et al. Reference-ree detection of genotypable SNPs, in revision to NAR. 2014 [Epub ahead of print] [Google Scholar]
- Zhao S, et al. Rainbow: a tool for large-scale whole-genome sequencing data analysis using cloud computing. BMC Genomics. 2013;14:425. doi: 10.1186/1471-2164-14-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de-Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.