Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal cancer due in part to a lack of highly robust cytotoxic or molecular-based therapies. Recent studies investigating ligand-mediated Wnt/β-catenin signaling have highlighted its importance in pancreatic cancer initiation and progression, as well as its potential as a therapeutic target in PDAC. The small molecule ICG-001 binds CREB-binding protein (CBP) to disrupt its interaction with β-catenin and inhibit CBP function as a co-activator of Wnt/β-catenin-mediated transcription. Given its ability to inhibit Wnt/β-catenin-mediated transcription in vitro and in vivo, as well as its efficacy in preclinical models of colorectal cancer and other Wnt-driven diseases, we examined ICG-001 and its potential role as a therapeutic in PDAC. ICG-001 alone significantly inhibited anchorage-dependent and -independent growth of multiple PDAC lines, and augmented in vitro growth inhibition when used in combination with gemcitabine. ICG-001 had only variable modest effects on PDAC apoptosis and instead mediated PDAC growth inhibition primarily through robust induction of G1 cell cycle arrest. These effects, however, appeared decoupled from its inhibition of Wnt/β-catenin-mediated transcription. DNA microarrays performed on PDAC cells in the context of ICG-001 treatment revealed ICG-001 altered the expression of several genes with well-established roles in DNA replication and cell cycle progression, including direct actions on SKP2 and CDKN1A. ICG-001 also significantly prolonged survival in an in vivo orthotopic xenograft model of PDAC, indicating ICG-001 or derived compounds that disrupt CBP activity are potentially useful small molecule therapeutics for pancreatic cancer.
Keywords: Pancreatic cancer, ICG-001, Wnt signaling, CREB binding protein (CBP), G1 arrest
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is among the deadliest of cancers, with an overall five-year survival rate of approximately 6%(1). PDAC is difficult to treat because it frequently presents at an advanced, non-operative stage and is highly resistant to cytotoxic or targeted molecular therapy(2). While our understanding of the molecular and cellular basis of PDAC continues to expand, present therapeutic options remain limited and offer only modest survival benefits for most patients.
Wnt/β-catenin signaling is a critical developmental signaling pathway whose dysregulation is strongly implicated in the pathogenesis of many types of cancers (3). Canonical mutations in CTNNB1, APC and AXIN that lead to constitutive hyperactivation of the pathway occur only infrequently in PDAC(4,5). Nevertheless, perturbations in the timing, context and strength of Wnt/β-catenin signaling can promote the development and progression of PDAC(4,5). Ligand-mediated Wnt/β-catenin signaling is essential for pancreatic cancer initiation and progression(6) and has been linked to aggressive tumor behavior(7). Wnt pathway activation as detected by nuclear and/or cytoplasmic accumulation of β-catenin is observed in 10-65% of pancreatic intraepithelial neoplasia (PanIN) (8,9), increasing with higher PanIN grade and invasive PDAC(10). Genetic or pharmacologic inhibition of various steps in the Wnt pathway has also been shown to prevent in vitro (7,9,10) and in vivo tumor growth (6,10–13), implicating Wnt signaling as therapeutic target in PDAC.
Although previously plagued by poor in vivo pharmacokinetics, several novel Wnt/β-catenin inhibitors have demonstrable in vivo activity and are now in various stages of preclinical or early clinical development. These include naturally occurring compounds, small molecule inhibitors, blocking antibodies and peptide antagonists (5,14,15). ICG-001 was identified in a screen of small molecules that inhibited Wnt/β-catenin transcriptional activity in a colorectal cancer cell line(16). ICG-001 selectively blocks the interaction of β-catenin with its transcriptional co-activator cyclic-AMP-response-element-binding protein (CBP) without disrupting β-catenin interaction with highly homologous p300. One significant effect ascribed to ICG-001 and its disruption of Wnt/β-catenin transcription is decreased expression of the apoptosis inhibitor BIRC5 (aka survivin protein), which leads to activation of caspase-3/7-mediated apoptosis(16–19). ICG-001 was first shown to slow colorectal cancer xenograft growth and intestinal polyp formation in the Apcmin mouse model (16) and has been subsequently shown to have in vivo efficacy in other Wnt-driven disease models, including rodent models of pulmonary fibrosis(20), renal interstitial fibrosis(21), acute lymphoblastic leukemia(22), chronic myocardial infarction(23), dermal fibrosis(24) and salivary tumorigenesis(25). The ICG-001-derived compound PRI-724 is now under investigation as a Wnt inhibitor in early phase clinical trials for advanced solid tumors (NCT01302405) and myeloid malignancies (NCT01606579) (26).
Given the importance of Wnt signaling in pancreatic carcinogenesis, we have now explored therapeutic potential and mechanism of action of ICG-001 in PDAC. ICG-001 significantly inhibited in vitro and in vivo PDAC growth by inducing G1 cell cycle arrest through effects that were largely decoupled from its activity as a Wnt inhibitor. Instead, ICG-001 appeared to more broadly impact CBP function as a co-transcriptional activator, directly and indirectly perturbing the expression of numerous genes with key roles in cell cycle progression.
Materials and Methods
Cell Lines and Reagents
All cell lines were cultured as previously described(7). AsPC-1, MiaPaCa-2 and PANC-1 were obtained in 2005 from the American Type Culture Collection (Rockville, MD). L3.6pl was obtained in 2010 from Hong Wu (UCLA). The cell lines have not been subsequently authenticated since receipt. ICG-001 was purchased from Selleck Chem and gemcitabine was kindly provided by Timothy Donahue (UCLA). Wnt3a and L-cell conditioned media were prepared as previously described(7).
Cell growth, proliferation and apoptosis assays
MTT cell viability assays (ATCC) were carried out per manufacturer's instructions at initial plating of 5,000 (AsPC-1 and L3.6pl) or 3,000 (MiaPaCa-2 and PANC-1) cells per well in 96-well plates. Soft agar assays were performed as previously described (7) in 48-well format. Media and drug were replenished once every 3-4 days. For cell cycle analysis, treated cells were stained using hypotonic DNA staining buffer: 0.1mg/ml propidium iodide (Calbiochem), 20μg/ml RNaseA, 1mg/ml sodium citrate and 0.3% Triton X-100. Flow cytometry analysis was performed in the UCLA Flow Cytometry Core using a Becton Dickinson FACSCalibur cytometer and using ModFit LT v3.1 software. Annexin V-PI flow cytometry analysis was performed with Cellquest v3.3 software using an Annexin V FITC Apoptosis Detection Kit (BD Biosciences).
Orthotopic Xenograft Tumor Assay
Animal work was performed with oversight by the UCLA Division of Laboratory Animal Medicine and approval from the UCLA Animal Research Committee. Orthotopic tumors were established by injecting 5×105 AsPC-1 cells suspended in 50% Matrigel (BD Biosciences)/RPMI into surgically-exposed pancreatic tails of 6-week old female athymic nude mice (Charles River) as previously described(27). Intraperitoneal drug injections were begun 10 days after tumor cell injections in animals randomized to four treatment arms: vehicle alone (20% PEG300, 5% solutol, 3.75% dextrose, 1% DMSO in PBS) (n=12), ICG-001 (5 mg/kg 6 days/week) (n=12), gemcitabine (25 mg/kg 2 days/week) (n=11) or ICG-001 + gemcitabine (n=12). Treatment was for 8 weeks and then discontinued for the remainder of the study (up to 120 days post-surgery). Animals were monitored daily and sacrificed per institutional guidelines when reaching either a Body Conditioning Score of 2 (28) or palpable tumor of 1.5 cm in diameter. Full necropsy was performed at the time of death with snap frozen and formalin-fixed tissue sections taken of primary and metastatic tumors, as well as normal spleen, lung, liver, small and large intestine. Histologic analysis was performed with H&E stained sections by a practicing surgical pathologist (DWD) blinded to treatment arms. Ten random high power fields per tumor were analyzed for mitotic counts. A previously described dissemination score awarding points based on tumor infiltration and metastasis(29) was used to quantify tumor spread.
Wnt reporter assays
Dual luciferase assays (Promega) were performed as previously described (7) on cell lines stably transduced with lentiviral constructs containing β-catenin-activated reporter (BAR) driving firefly luciferase expression and separate normalization control construct with constitutive EF1α promoter driving Renilla expression.
Gene Knockdown
Lipofectamine 2000 was used per manufacturer's instructions to transfect AsPC-1 cells with 20 nM control siRNA (#12935-200) or CTNNB1 s437 (#4390824) siRNA (all purchased from Life Technologies). Western blots to verify knockdown were performed using β-catenin (Sigma, C2206) and tubulin (Santa Cruz, sc-5546) antibodies.
Real-time PCR
RNA extraction, cDNA synthesis and SYBR Green-based quantitative PCR were performed as previously described(7) using primers listed in Supplemental Table 1.
Western Blots and Immunoprecipitation
Western blots were performed as previously described (7) using antibodies specific for β-catenin (Sigma C2206), PARP (Cell Signaling Technology (CST) 9542), Cleaved Caspase-3 (CST 9664), Cleaved Caspase-7 (CST 9491), SKP2 (CST 2652), MYC (Santa Cruz sc-40), p21 (CST 2947) or tubulin (Santa Cruz sc-5546). CBP co-immunoprecipitations were performed on nuclear extracts as previously described (30) with anti-CBP (Santa Cruz, sc-369) or control isotype-matched IgG (Abcam, ab46540) antibodies.
Chromatin Immunoprecipitation
AsPC-1 cells (70% confluency) were crosslinked with formalin. Chromatin immunoprecipitation (ChIP) was performed using the Pierce Agarose ChIP Kit (Thermo Scientific) on chromatin digested with 2U micrococcal nuclease. Digested chromatin extracts were precleared with 20 ug agarose beads and incubated overnight with beads pre-bound with either 5 μg IgG control or CBP antibody (Abcam, ab46540 and ab2832, respectively) blocked with 1μg fish sperm DNA. PCR was performed using primers (Supplemental Table 1) covering gene regulatory regions for CDKN1A (−231 to −117 relative to transcriptional start site, TSS), SKP2 (+1772 to +1870 relative to TSS) and CDC25A (−31 to −310 relative to TSS).
Gene expression microarray and data analysis
RNA was extracted from three biological replicates of AsPC-1 cells 6 and 24 hours post-treatment with 10 μM ICG-001 or vehicle control (DMSO) or in separate experiments from two biological replicates of AsPC1 cells transfected for 48 hours with either 20 nM control or CTNNB1 siRNA. Affymetrix U133 plus 2.0 oligonucleotide arrays were processed in the UCLA Clinical Microarray Core. Arrays are available in Gene Expression Omnibus database repository (accession code GSE57728). Data analysis was performed with dChip Analysis software package(31) using invariant set normalization and signal intensities summarized using the MBEI (model-based expression index) algorithm with mismatch probe option for background subtraction. Initial gene filtering criteria was set at >10% present call and variance across samples of 0.4<standard deviation/mean<1000. ICG-001-regulated genes were determined by comparative analysis of ICG-001 versus vehicle control (selection criteria >1.67-fold change and P<0.01 at 6 hours or >2-fold change and P<0.01 at 24 hours). β-catenin-regulated genes were determined by comparative analysis of CTNNB1 versus control siRNA tranfection (selection criteria ≥1.33-fold change and >50 absolute change). Functional enrichment was examined using default parameters in the web-based Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.7) (http://david.abcc.ncifcrf.gov) (32). Analysis was limited to the GO biological process FAT (GOTERM_BP_FAT) category terms and ranked on P-values after removal of categories with less than 6 annotated genes.
Statistics
Student t tests were used to compare continuous variables. The Cox proportional hazard model was used to estimate hazard ratios between treatment groups. The assumption of proportionality was assessed both by visual inspection of Kaplan-Meier survival curves and formal analysis of Schoenfeld residuals (p.val=0.13). Kaplan-Meier survival curves and log rank tests were performed using GraphPad Prism.
Results
ICG-001 inhibits in vitro PDAC cell line growth
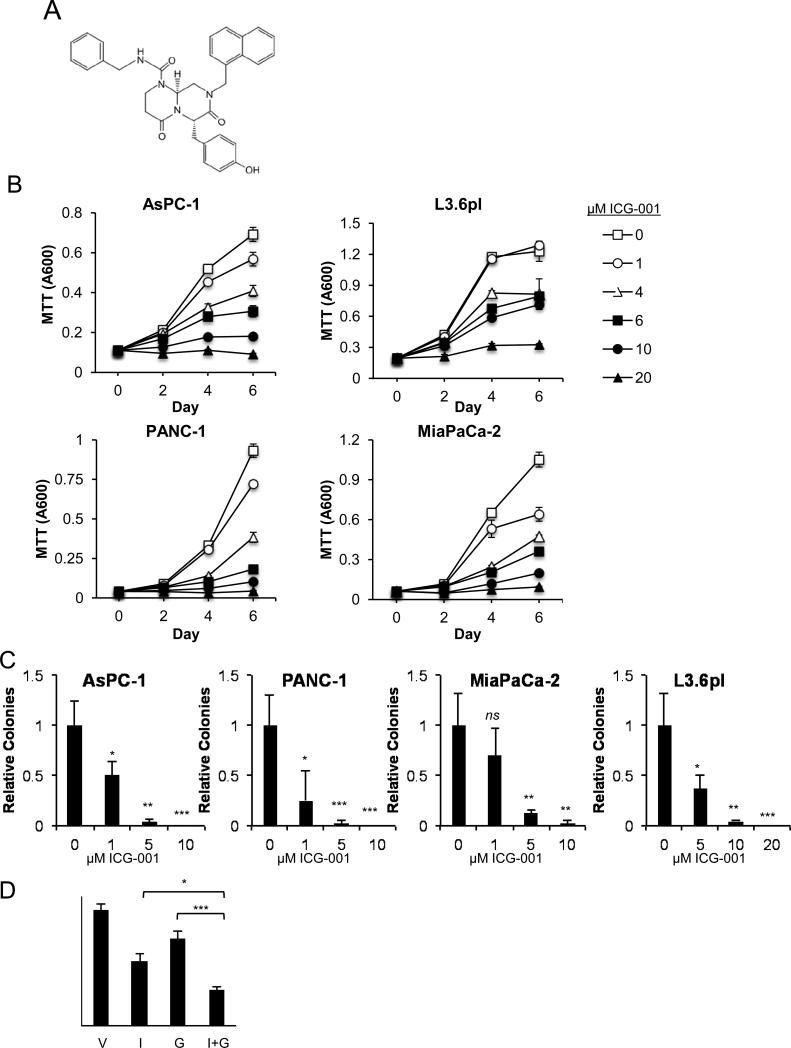
ICG-001 (Figure 1A) effects on in vitro anchorage-dependent PDAC cell growth were addressed by MTT cell viability assays. ICG-001 showed dose-dependent cell growth inhibition of AsPC-1, L3.6pl, PANC-1 and MiaPaCa-2 cell lines at IC50 values of 5.48 μM, 14.07 μM, 3.43 μM and 3.31 μM, respectively (Figure 1B, Supplemental Figure 1A-B). ICG-001 also significantly inhibited anchorage-independent growth in soft agar assays (Figure 1C). Addressing the potential for combined use of ICG-001 with cytotoxic chemotherapy, concurrent treatment with 5 μM ICG-001 and 5 nM gemcitabine further increased growth inhibition relative to individual treatments (Figure 1D).
Figure 1.
ICG-001 inhibits in vitro anchorage dependent and independent growth of pancreatic cancer lines. (A) Chemical structure of ICG-001. (B) MTT cell viability and (C) soft agar assays were performed at the indicated concentrations of ICG-001. (D) MTT assay on AsPC-1 cells treated for 6 days with vehicle (V), 5 μM ICG-001 (I), 5 nM gemcitabine (G) or 5 μM ICG-001 plus 5 nM gemcitabine (I+G). Representative experiment from at least three replicates is presented for each cell line. *P<0.05, **P<0.01, ***P<0.001, n=not significant.
ICG-001 inhibits in vivo PDAC tumor growth
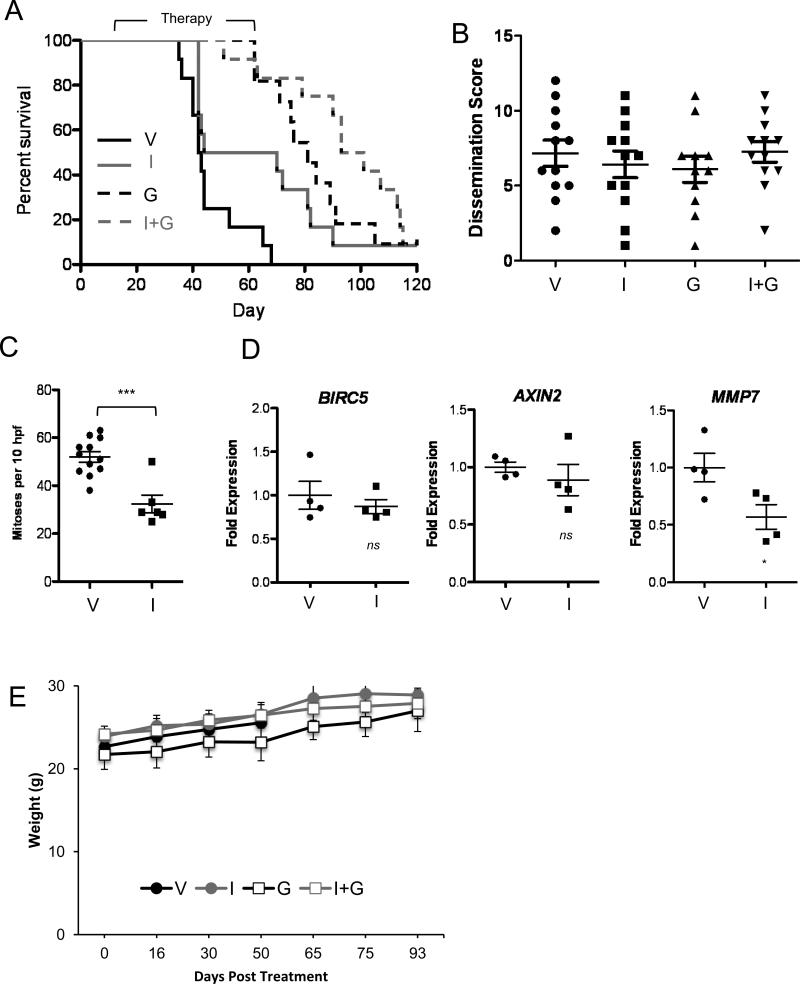
ICG-001 was next addressed in an in vivo orthotopic xenograft model of PDAC. AsPC-1 tumor cells were injected into the distal pancreas of nude mice 10 days prior to the start of treatment. Tumor-bearing mice were then randomized into four different treatment arms, including vehicle (N=12), ICG-001 (N=12), gemcitabine (N=11) or ICG-001 + gemcitabine (N=12). Treatment was continued for a total of 8 weeks, after which all surviving animals were followed until the end of the study (120 days). ICG-001 significantly improved survival compared to vehicle alone as determined by Cox proportional hazard analysis (HR=0.207, 95%CI: 0.079-0.543, P=0.001). Gemcitabine also significantly improved survival compared to vehicle (HR=0.123, 95%CI: 0.045-0.334, P< 0.001). There was no survival difference between gemcitabine alone versus ICG-001 alone (HR=0.592, 95% CI: 0.250-1.399, P=0.23). In line with in vitro results, the greatest survival benefit relative to vehicle was seen with combined gemcitabine + ICG-001 (HR= 0.074, 95%CI: 0.026, 0.212, P<0.001). Although this combination improved survival relative to ICG-001 alone (HR=0.360, 95% CI: 0.152-0.848, P=0.02), there was no significant improvement relative to gemcitabine alone (HR=0.608, 95% CI: 0.254-1.455, P=0.66). Kaplan-Meier survival curves (Figure 2A) also demonstrated improved survival relative to vehicle control (mean survival 46 ± 3.1 days) for ICG-001 (mean survival 64 ± 7.1 days, log-rank P=0.01), gemcitabine (mean survival 83 ± 5.1 days, log-rank P<0.00001) and combined ICG-001 + gemcitabine (mean survival 95 ± 6.0 days, log-rank P<0.00001) treatments.
Figure 2.
ICG-001 shows in vivo efficacy against pancreatic cancer. (A) Kaplan-Meier survival curves for mice treated with vehicle (V), ICG-001 (I), gemcitabine (G) or ICG-001 plus gemcitabine (I+G). (B) Tumor dissemination scores were determined at the time of sacrifice for all mice in the four treatment arms. (C) Mitotic counts in 10 random high power fields and (D) qPCR analysis of indicated Wnt gene targets normalized to ACTB housekeeping gene were determined in primary tumors taken from mice still receiving treatment at time of death. (E) Weight of mice at various time points following initiation of therapy. *P<0.05, **P<0.01, ***P<0.001, ns=not significant.
The in vivo biological effects of ICG-001 were further addressed by gross and histologic analysis of mice at necropsy. All tumors significantly involved the pancreas with direct extension into adjacent spleen. Metastatic tumor deposits were observed in all animals, most frequently involving the lung, small bowel and diaphragm (Supplemental Figure 2A-F and Supplemental Table 2). Using a previously described scoring system that awards points based on tumor infiltration and metastasis (29), tumor dissemination was quantified at the time of death. No differences were observed between the four treatment arms (Figure 2B), indicating that animals were reproducibly sacrificed at time points of equal tumor burden. A mitotic index calculated on tumors harvested during drug treatment revealed a significant decrease in mitotic activity in ICG-001 versus vehicle control treated tumors (Figure 2C). Quantitative real-time PCR (qPCR) for three well-established Wnt target genes in tumors harvested during drug treatment revealed a significant decrease in MMP7 expression with ICG-001 versus vehicle control treatment, but no differences in BIRC5 or AXIN2 expression (Figure 2D).
Pharmacologic inhibition of Wnt signaling could have potential deleterious effects on normal somatic stem cell maintenance and tissue homeostasis(26). However, no clinical evidence of ICG-001 cytotoxicity was observed on the basis of body weight measurements (Figure 2E). Furthermore, H&E evaluation of uninvolved liver and intestine revealed no histologic changes to suggest cytotoxicity in animals sacrificed during treatment with ICG-001 or vehicle control. Small bowel from both treatment groups revealed normal villous-crypt ratios and no changes in apoptotic or mitotic activity within crypts, while liver showed no evidence of hepatitis, cholestasis or hepatocellular injury (i.e., ballooning change or apoptosis) (Supplemental Figure 2G-2L).
ICG-001 induces G1 arrest and has variable effects on Wnt/β-catenin signaling
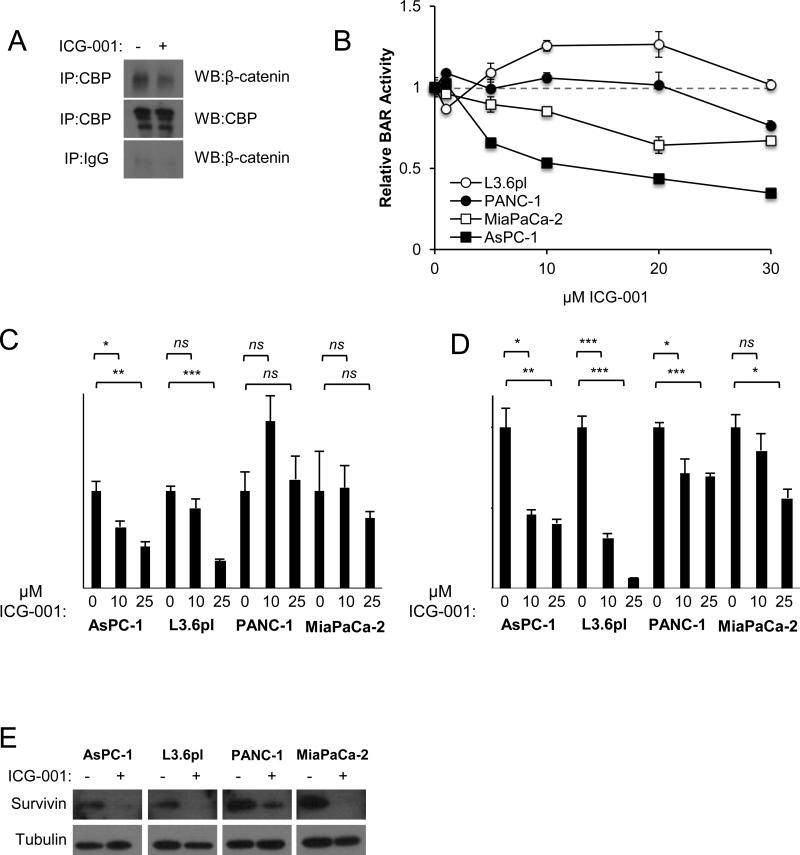
We next explored mechanisms underpinning ICG-001 pancreatic growth inhibitory effects. As previously shown in colorectal cancer(16), ICG-001 disrupted the interaction between CBP and β-catenin in AsPC1 cells as measured by co-immunoprecipitation (Figure 3A). To examine ICG-001 effects on Wnt/β-catenin signaling, PDAC cell lines stably transduced with a β-catenin-activated luciferase reporter (BAR) (7,33) were treated with increasing concentrations of ICG-001. ICG-001 exhibited strong, dose-dependent inhibition of reporter activity in AsPC-1 (Figure 3B), a cell line characterized by higher levels of autocrine Wnt/β-catenin activity(7). By comparison, high concentrations of ICG-001 only weakly inhibited reporter activity in PANC-1 and MiaPaCa-2 cells, and not at all in L3.6pl cells (Figure 3B). Parallel dose-dependent effects of ICG-001 on the expression of the endogenous Wnt/β-catenin target genes AXIN2 were also observed, with L3.6pl cells demonstrating disparate inhibition of AXIN2 expression at high ICG-001 concentration (Figure 3C). Of note, ICG-001 significantly inhibited the growth of multiple PDAC cell lines at lower concentrations that had little or no effect on Wnt/β-catenin transcriptional activity (compare Figure 1B and Figure 3B-C).
Figure 3.
ICG-001 variably influences Wnt transcriptional activity across pancreatic cancer lines. (A) Nuclear extracts from AsPC-1 cells treated with vehicle or 30 μM ICG-001 for 6 hours were immunoprecipitated with anti-CBP or control IgG antibodies followed by western blots for β-catenin and CBP. (B) PDAC cell lines carrying stably integrated Wnt/β-catenin luciferase reporter (BAR) were treated with indicated concentrations of ICG-001 and measured in dual luciferase reporter assays at 24 hours. Luciferase is normalized to Renilla and shown relative to vehicle. (C) AXIN2 and (D) BIRC5 transcript levels were determined by qPCR in PDAC cell lines treated with indicated concentrations of ICG-001. All values were normalized to ACTB and shown relative to vehicle. One representative experiment from at least two biological replicates is shown. (E) Western blot for survivin and tubulin (loading control) after 48 hours vehicle or 10 μM ICG-001. One representative experiment of three replicates is shown. *P<0.05, **P<0.01, ***P<0.001, ns=not significant.
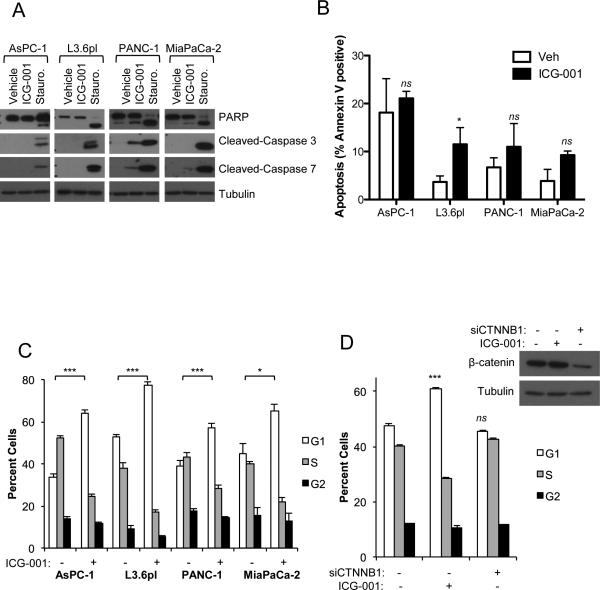
ICG-001 has been shown to induce apoptosis via downregulation of the apoptosis inhibitor BIRC5 (survivin), a known Wnt/β-catenin target gene (16–19). ICG-001 treatment decreased the expression of BIRC5 transcript and survivin protein levels in all four PDAC cell lines (Figure 3D-E). Despite this significant inhibition of survivin expression, ICG-001 had no significant effects on AsPC1 apoptosis as determined by cleaved PARP, caspase-3, caspase-7 or annexin-V-propidium iodide (PI) flow cytometry (Figures 4A-B). PANC-1 and MiaPaCa-2 cells showed mild increases in cleaved PARP, caspase-3 and caspase-7 with no appreciable differences in apoptosis by annexin-V-PI staining, while L3.6pl showed significantly increased apoptosis as measured by annexin-V-PI flow staining (Figure 4A-B). These modest changes indicated apoptosis was not the primary mechanism for the observed inhibition of PDAC growth by ICG-001. Therefore, ICG-001 effects on cell proliferation were analyzed by cell cycle analysis. ICG-001 treatment robustly induced G1 arrest in all PDAC cell lines (Figure 4C), a finding consistent with the decreased mitotic activity observed in ICG-001-treated orthotopic tumors (Figure 2C). Significant G1 arrest was observed in all four cell lines at 10μM ICG-001, a concentration for which only AsPC-1 showed appreciable inhibition of Wnt transcriptional activity.
Figure 4.
ICG-001 robustly induces G1 cell cycle arrest. (A) Cells were incubated with vehicle, 10 μM ICG-001 or 500 nM staurosporine (Stauro, positive control for apoptosis) for 24 hours and analyzed by western blot for PARP, cleaved-caspase 3, cleaved-caspase 7 and tubulin (loading control). Vehicle or 10 μM ICG-001 treated cells were analyzed by flow cytometry for (B) apoptosis by annexin V-PI staining at 48 hours or (C) cell cycle by propidium iodide staining at 24 hours. (D) Cell cycle analysis was determined for AsPC-1 cells treated simultaneously with indicated combinations of vehicle, 10 μM ICG-001, control siRNA (20 nM) or CTNNB1 siRNA (20nM) for 48 hours. Western blot confirmed >75% knockdown of β-catenin with CTNNB1 siRNA. One representative experiment of three replicates is shown for all figures. . *P<0.05, **P<0.01, ***P<0.001, ns = not significant.
If ICG-001 exerts anti-proliferative effects by inhibiting β-catenin-mediated transcription, this effect should be phenocopied by knockdown of β-catenin expression. However, siRNA-mediated CTNNB1 gene knockdown failed to induce G1 arrest in AsPC-1 cells, whereas parallel ICG-001 treatment did induce significant G1 arrest (Figure 4D). Furthermore, while exogenous Wnt3A ligand treatment fully rescued ICG-001 inhibition of Wnt reporter activity in the cell lines, it had no effect on ICG-001-mediated G1 cell cycle arrest (Supplemental Figure 3). Altogether, these data indicate that ICG-001 mediates growth arrest in PDAC cell lines through mechanisms discrete from its action as an inhibitor of Wnt signaling.
ICG-001 inhibits the expression of genes involved in cell cycle
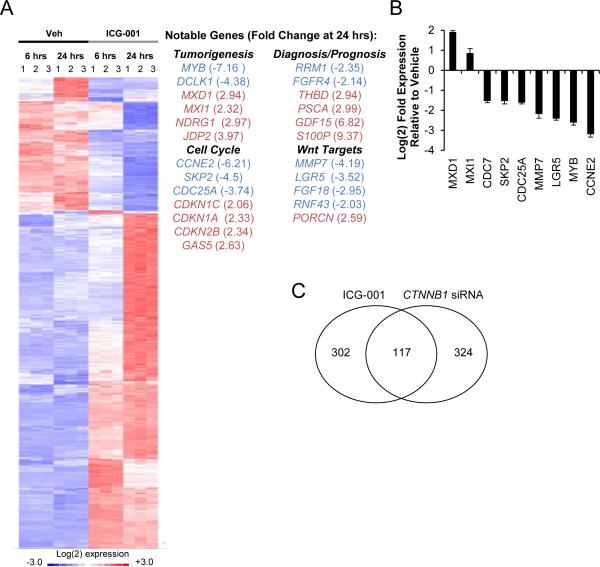
To explore mechanisms underlying ICG-001 inhibition cell cycle in PDAC, gene expression microarrays were performed on AsPC-1 cells after 6 and 24 hours of treatment with ICG-001. In total, 569 transcripts (419 upregulated and 150 downregulated) showed altered expression in response to ICG-001 treatment (P < 0.01; 1.67-fold change at 6 hours or 2-fold change at 24 hours; 0.2% median false discovery rate) (Figure 5A and Supplemental Table 3). Several of these changes were further validated by qPCR in separate ICG-001 treatment experiments (Figure 5B). To explore biological processes linked to ICG-001 treatment, we performed enrichment analysis of gene ontology (GO) functional annotations. In line with phenotypic observations, the top five GO terms for genes downregulated by ICG-001 were DNA replication, DNA metabolic processes, DNA replication initiation, DNA-dependent DNA replication and cell cycle (p < 0.00013, Supplemental Figure 4A). Top GO terms for genes upregulated by ICG-001 included multicellular organismal homeostasis, negative regulation of cell proliferation, cell cycle arrest, regulation of angiogenesis and regulation of cell proliferation (p < 0.001, Supplemental Figure 4B).
Figure 5.
ICG-001 alters the expression of cell cycle genes. (A) Gene expression was determined with Affymetrix Human U133 plus 2.0 microarrays using RNA collected from AsPC1 following 6 or 24 hour treatment with vehicle or 10 μM ICG-001. Biological replicates of significantly regulated genes are shown in the heatmap along with a partially list of genes curated based on known roles in tumorigenesis, cell cycle control, cancer diagnosis/prognosis or Wnt/β-catenin transcriptional targets. (B) qPCR validation was performed on selected genes in AsPC-1 cells separately treated with vehicle or 10 μM ICG-001 for 24 hours. (C) Venn diagram comparing AsPC-1 gene expression microarray changes observed with either 10 μM ICG-001 treatment or CTNNB1 siRNA transfection.
Although its inhibition of PDAC cell proliferation did not appear mechanistically linked to its effects on Wnt/β-catenin transcriptional activity, ICG-001 did inhibit several known Wnt transcriptional targets, including MMP7 and LGR5 that were further validated by qPCR (Figure 5B). To address ICG-001-regulated genes representing actual Wnt transcriptional targets in PDAC, we compared ICG-001 results with a second microarray study comparing gene expression changes in AsPC-1 transfected with either control or CTNNB1 siRNA. In the context of greater than 80% CTNNB1 knockdown, there were 441 altered transcripts (absolute fold change ≥ 1.33; median false discovery rate 10.4%), including 228 upregulated and 213 downregulated genes (Supplemental Table 3). Top GO terms for genes downregulated by CTNNB1 knockdown included cell fate commitment, tube morphogenesis, tube development, morphogenesis of a branching structure and angiogenesis (p < 0.0005, Supplemental Figure 5A). Top GO terms for genes upregulated by CTNNB1 siRNA included response to wounding, cell adhesion, biological adhesion, response to inorganic substance and regulation of cell migration (p < 0.0002, Supplemental Figure 5B). Cross-referencing both ICG-001 and CTNNB1 siRNA microarray results revealed 117 overlapping transcripts (Figure 5C, Supplemental Table 3). GO functional annotations were only marginally enriched in this overlapping dataset (P < 0.01), but included regulation of cell proliferation, positive regulation of developmental processes, homeostatic process, response to wounding and regulation of cell cycle.
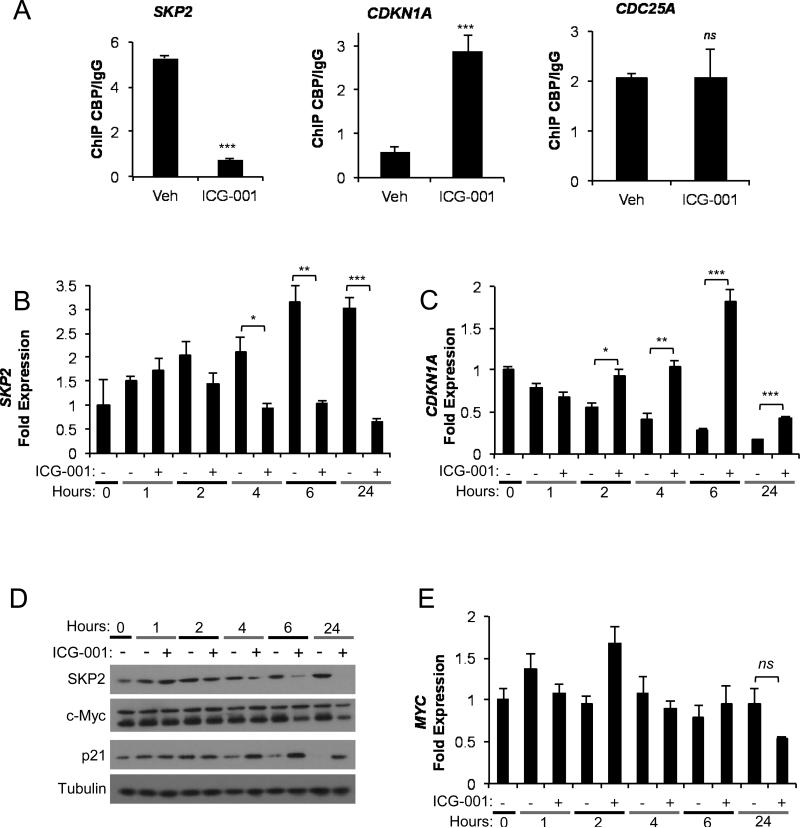
Among the most highly regulated genes identified in the ICG-001 arrays were SKP2, CDC25A and CDKN1A (p21), each regulators of cell cycle progression. We therefore explored whether these genes were direct transcriptional targets of CBP that are altered in response to ICG-001 treatment. After 1 hour ICG-001 treatment, chromatin immunoprecipitation (ChIP) revealed reduced CBP occupancy at SKP2 and increased CBP occupancy at CDKN1A (Figure 6A), corresponding respectively to aforementioned decreased SKP2 and increased CDKN1A gene expression results (Figure 5). No change in CBP occupancy was observed at CDC25A (Figure 6A) suggesting it is transcriptionally downregulated indirectly in response to ICG-001. Further detailed time-course analyses of gene and protein expression revealed SKP2 transcript levels were reduced within 4 hours while CDKN1A transcript levels were increased within 2 hours of ICG-001 treatment (Figure 6B-C). Corresponding decrease in SKP2 protein expression and increase in p21 protein expression were observed within 4-6 hours of ICG-001 treatment (Figure 6D). MYC protein levels were also decreased after 6 hours, although MYC transcript levels were unchanged (Figure 6D-E), indicating a post-transcriptional mechanism controls MYC protein levels following ICG-001 treatment.
Figure 6.
ICG-001 directly regulates change in SKP2 and CDKN1A expression. (A) Chromatin immunoprecipitation results for CBP association with SKP2, CDKN1A and CDC25A regulatory elements in AsPC-1 cells following 1 hour treatment with vehicle or 10 μM ICG-001. Data were normalized to input DNA and are presented as fold change of CBP/IgG. Representative experiments of three replicates are shown. (B-E) Time course study of AsPC-1 cells treated with vehicle or 10 μM ICG-001 measuring (B) SKP2 and (C) CDKN1A gene expression by qPCR, (D) SKP2, MYC, p21 and tubulin protein expression by western blot and (E) MYC gene expression by qPCR. All qPCR values were normalized to ACTB and shown relative to 0 hour. Representative experiments from at least two replicates are shown for all qPCR and western blots. *P<0.05, **P<0.01, ***P<0.001, ns=not significant.
Discussion
At least subsets of PDAC are dependent upon Wnt/β-catenin signaling for initiation and/or progression (6–10,34), warranting exploration of ICG-001 and other Wnt inhibitors as potential therapeutics. PRI-724, an ICG-001 derivative, is presently under investigation in combination with gemcitabine in a phase I clinical trial for patients with metastatic pancreatic cancer (NCT01764477). We have demonstrated that ICG-001 significantly inhibits in vitro PDAC growth through induction of G1 cell cycle arrest and improves survival in an in vivo orthotopic xenograft model of PDAC. Despite augmented PDAC growth inhibition in vitro, combined ICG-001 and gemcitabine did not further improve survival over gemcitabine alone in the xenograft model. It is possible that robust induction of G1 arrest with ICG-001 compromises the action of cytotoxic chemotherapy such as gemcitabine, which primarily kills cancer cells undergoing DNA synthesis in S phase. An analogous situation has been observed with CDK4 inhibitors that by inducing G1 arrest can reduce the efficacy of cytotoxic chemotherapy in certain circumstances(35). Further optimization of the dosing and delivery of ICG-001 (or PRI-724) is needed to resolve whether ICG-001 can be used effectively in combination with gemcitabine. Intriguingly, ICG-001 reduced expression of RRM1 in AsPC-1 microarrays. RRM1 expression directly correlates with gemcitabine resistance(36) and worse overall survival in PDAC(37,38). Furthermore, ICG-001 was shown to act synergistically with conventional chemotherapy to prolong survival in NOD/SCID mice engrafted with primary acute lymphoblastic leukemia(22). Thus, optimized combinations of cytotoxic regimens with ICG-001 may still offer additional survival benefit in PDAC or other tumors.
ICG-001 treatment and β-catenin knockdown regulated an overlapping set of genes in AsPC-1, including known targets of canonical Wnt signaling (i.e, MMP7, EDN1, LGR5, RNF43 and PORCN). Originally identified in a small molecule library screen of compounds that inhibited β-catenin/TCF-dependent reporter activity, ICG-001 inhibits Wnt transcriptional activity by binding to the N-terminal domain of CBP and preventing its interaction with β-catenin(16). This not only has the potential to disrupt CBP-dependent β-catenin-TCF transcription, but also may increase the pool of β-catenin available to interact with p300. Like CBP, p300 functions either as a transcriptional co-activator or co-repressor in a context-dependent manner (26,39,40). Kahn and colleagues have proposed a model of CBP/p300 differential co-activator usage whereby TCF-β-catenin-CBP-mediated transcription drives a non-differentiated proliferative state with ICG-001 inducing a switch to TCF-β-catenin-p300-mediated transcription favoring cellular differentiation(39,41). In keeping with this model, ICG-001 strongly induced G1 cell cycle arrest in PDAC cell lines and inhibited the expression of certain genes linked to pluripotency, including LGR5 that marks progenitor cells in intestinal crypts and DCLK1 which was recently shown to mark a unique tumor initiating cell population in PDAC(42). ICG-001 also regulated many genes unaltered by β-catenin knockdown, including several linked to DNA replication and cell cycle regulation. Beyond β-catenin, ICG-001 could more generally alter gene expression by disrupting CBP interaction with any of a number of other known protein binding partners that includes many transcription factors (43). As an example, ICG-001 was recently shown to block CBP binding to γ-catenin, resulting in increased p300/γ-catenin interaction and decreased BIRC5 expression (22,44).
ICG-001 can induce caspase-3/7-mediated apoptosis in colon cancer cells via downregulation of BIRC5 (16). Although ICG-001 treatment inhibited survivin protein levels, there were only variable and modest effects on apoptosis in PDAC cell lines. Rather, ICG-001 induced robust G1 cell cycle arrest, which is perhaps not unexpected given CBP/p300 histone acetyltransferase activity is essential for cell proliferation and orderly G1/S transition(45). Several groups find Wnt/β-catenin signaling promotes cellular proliferation in PDAC(4,9) and while Wnt/β-catenin transcriptional targets MYC or CCND1 are commonly linked to its regulation of cell cycle, transcript levels of these genes were not altered in PDAC cell lines in response to ICG-001 treatment. Moreover, growth inhibition and G1 cell cycle arrest was observed in multiple PDAC cell lines at ICG-001 concentrations with little or no measurable effect on Wnt reporter activity or endogenous AXIN2 expression, indicating ICG-001 inhibition of PDAC cell proliferation was decoupled from its function as a Wnt inhibitor.
ICG-001 treatment altered the expression of numerous genes linked to DNA replication and cell cycle in AsPC-1 microarray analysis, both downregulating genes that promote (i.e., SKP2, CDC25A, CDC7, CCNE and MCM2-MCM 6) and upregulating genes that inhibit (i.e., CDKN1A, CDKN1C, CDKN2B, MXD1, MXI1 and BTG1) cell cycle progression. ICG-001 induced rapid changes in CBP occupancy on SKP2 and CDKN1A regulatory regions with corresponding changes in mRNA and protein levels, implicating SKP2 and CDKN1A as direct targets of ICG-001-mediated cell cycle arrest. SKP2 is a substrate component of the SCF E3 ubiquitin-protein ligase complex that promotes cell cycle progression through ubiquitination and subsequent degradation of several proteins linked to cell cycle inhibition, including p21, p27, p57, p130 and RASSF1A among others(46). ICG-001 increased CBP occupancy at CDKN1A promoter site in AsPC-1 cells, perhaps by increasing the availability of CBP to different subsets of transcription factors mediating CDKN1A expression. CDKN1A encodes the cyclin-dependent kinase inhibitor p21 that oppose the function of multiple cyclin-CDK complexes that mediate G1 and S cell cycle progression. Increased p21 levels with ICG-001 could reflect decreased SKP2-mediated degradation of p21 and/or increased CDKN1A expression via CBP co-transcriptional activation. Although MYC transcript levels were unchanged, ICG-001 reduced MYC protein levels within 6 hours along with corresponding increases in several known targets of MYC-mediated transcriptional repression (i.e., CDKN1A, CDKN1C and CDKN2B). It is intriguing to speculate how ICG-001 might indirectly antagonize MYC through additional post-translational mechanisms linked to its protein stability or function. MXD1 and MXI1, which were both upregulated in response to ICG-001 treatment, can inhibit the transcriptional activity of MYC by competing for its important heterodimerization partner MAX (47). CBP/p300-mediated acetylation has been shown to stabilize MYC protein and function (48), which could also be perturbed by ICG-001. Finally, ICG-001 could indirectly disrupt MYC activity through its aforementioned downregulation of SKP2, which has been shown ubiquitylate MYC to license its function as a transcriptional activator(49).
In conclusion, ICG-001 appears to robustly inhibit PDAC growth through both direct and indirect mechanisms that mutually reinforce the induction of G1 cell cycle arrest. While further studies are needed to expand upon precise mechanisms underpinning its transcriptional and phenotypic activities, ICG-001 appears to more broadly disrupt CBP function beyond its effects as co-transcriptional activator of Wnt/β-catenin signaling. These future studies should not only further examine the therapeutic potential of ICG-001 and its derivatives in PDAC or other cancers, but also better establish effective drug combinations and patient selection.
Supplementary Material
Acknowledgments
Financial Support:
M.D. Arensman was supported by USHHS Ruth L. Kirschstein Institutional National Research Service Award # T32 CA009056 (UCLA Tumor Biology Training Grant). D.W. Dawson was supported by an American Cancer Society Research Scholars Grant (RSG-12-083-01-TBG), NIH (P01 CA163200) and the Hirshberg Foundation for Pancreatic Cancer Research. Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility supported by National Institutes of Health awards CA-16042 and AI-28697, and by the JCCC, UCLA AIDS Institute, and David Geffen School of Medicine at UCLA.
Abbreviations
- PDAC
pancreatic ductal adenocarcinoma
- CBP
CREB binding protein
- PanIN
pancreatic intraepithelial neoplasia
- BAR
β-catenin activated reporter
- CST
Cell Signaling Technology
- ChIP
chromatin immunoprecipitation
- MTT
(3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide)
- PI
propidium iodide
- qPCR
quantitative real-time PCR
- siRNA
small-interfering RNA
- TSS
transcriptional start site
Footnotes
Conflicts of interest: None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer Statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–20. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Morris JP, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10:683–95. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White BD, Chien AJ, Dawson DW. Dysregulation of Wnt/β-catenin Signaling in Gastrointestinal Cancers. Gastroenterology. 2011;142:219–32. doi: 10.1053/j.gastro.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Morris JP, Yan W, Schofield HK, Gurney A, Simeone DM, et al. Canonical wnt signaling is required for pancreatic carcinogenesis. Cancer Res. 2013;73:4909–22. doi: 10.1158/0008-5472.CAN-12-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arensman MD, Kovochich a N, Kulikauskas RM, Lay a R, Yang P-T, Li X, et al. WNT7B mediates autocrine Wnt/β-catenin signaling and anchorage-independent growth in pancreatic adenocarcinoma. Oncogene. 2013:1–10. doi: 10.1038/onc.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng G, Germinaro M, Micsenyi A, Monga NK, Bell A, Sood A, et al. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia. 2006;8:279–89. doi: 10.1593/neo.05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasca di Magliano M, Biankin AV, Heiser PW, Cano D a, Gutierrez PJ a, Deramaudt T, et al. Common activation of canonical Wnt signaling in pancreatic adenocarcinoma. PLoS One. 2007;2:1–9. doi: 10.1371/journal.pone.0001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Heidt DG, Lee CJ, Yang H, Logsdon CD, Zhang L, et al. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and beta-catenin stabilization. Cancer Cell. 2009;15:207–19. doi: 10.1016/j.ccr.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Froeling FEM, Feig C, Chelala C, Dobson R, Mein CE, Tuveson D a, et al. Retinoic Acid-Induced Pancreatic Stellate Cell Quiescence Reduces Paracrine Wnt-β-Catenin Signaling to Slow Tumor Progression. Gastroenterology. 2011;141:1486–97. doi: 10.1053/j.gastro.2011.06.047. [DOI] [PubMed] [Google Scholar]
- 12.Jiang X, Hao H-X, Growney JD, Woolfenden S, Bottiglio C, Ng N, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110:12649–54. doi: 10.1073/pnas.1307218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurney A, Axelrod F, Bond CJ, Cain J, Chartier C, Donigan L, et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci U S A. 2012;109:11717–22. doi: 10.1073/pnas.1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voronkov A, Krauss S. Wnt/beta-catenin signaling and small molecule inhibitors. Curr Pharm Des. 2013;19:634–64. doi: 10.2174/138161213804581837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 16.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected]. Proc Natl Acad Sci U S A. 2004;101:12682–7. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazarova DL, Chiaro C, Wong T, Drago E, Rainey A, O'Malley S, et al. CBP Activity Mediates Effects of the Histone Deacetylase Inhibitor Butyrate on WNT Activity and Apoptosis in Colon Cancer Cells. J Cancer. 2013;4:481–90. doi: 10.7150/jca.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazarova DL, Wong T, Chiaro C, Drago E, Bordonaro M. p300 Influences Butyrate-Mediated WNT Hyperactivation In Colorectal Cancer Cells. J Cancer. 2013;4:491–501. doi: 10.7150/jca.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma H, Nguyen C, Lee K-S, Kahn M. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene. 2005;24:3619–31. doi: 10.1038/sj.onc.1208433. [DOI] [PubMed] [Google Scholar]
- 20.Henderson WR, Chi EY, Ye X, Nguyen C, Tien Y, Zhou B, et al. Inhibition of Wnt/beta-catenin/CREB binding protein (CBP) signaling reverses pulmonary fibrosis. Proc Natl Acad Sci U S A. 2010;107:14309–14. doi: 10.1073/pnas.1001520107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao S, He W, Li Y, Ding H, Hou Y, Nie J, et al. Targeted inhibition of β-catenin/CBP signaling ameliorates renal interstitial fibrosis. J Am Soc Nephrol. 2011;22:1642–53. doi: 10.1681/ASN.2010101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gang EJ, Hsieh Y-T, Pham J, Zhao Y, Nguyen C, Huantes S, et al. Small-molecule inhibition of CBP/catenin interactions eliminates drug-resistant clones in acute lymphoblastic leukemia. Oncogene. 2013:1–10. doi: 10.1038/onc.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasaki T, Hwang H, Nguyen C, Kloner R a, Kahn M. The Small Molecule Wnt Signaling Modulator ICG-001 Improves Contractile Function in Chronically Infarcted Rat Myocardium. PLoS One. 2013;8:e75010. doi: 10.1371/journal.pone.0075010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beyer C, Reichert H, Akan H, Mallano T, Schramm A, Dees C, et al. Blockade of canonical Wnt signalling ameliorates experimental dermal fibrosis. Ann Rheum Dis. 2013;72:1255–8. doi: 10.1136/annrheumdis-2012-202544. [DOI] [PubMed] [Google Scholar]
- 25.Wend P, Fang L, Zhu Q, Schipper JH, Loddenkemper C, Kosel F, et al. Wnt/β-catenin signalling induces MLL to create epigenetic changes in salivary gland tumours. EMBO J. 2013;32:1977–89. doi: 10.1038/emboj.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi-Yanaga F, Kahn M. Targeting Wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res. 2010;16:3153–62. doi: 10.1158/1078-0432.CCR-09-2943. [DOI] [PubMed] [Google Scholar]
- 27.Kim MP, Evans DB, Wang H, Abbruzzese JL, Fleming JB, Gallick GE. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat Protoc. 2009;4:1670–80. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foltz C, Ullman-Cullere M. Guidelines for assessing the health and condition of mice. Lab Anim (NY) 1999;28:28–32. [PubMed] [Google Scholar]
- 29.Hotz HG, Reber HA, Hotz B, Yu T, Foitzik T, Buhr HJ, et al. An Orthotopic Nude Mouse Model for Evaluating Pathophysiology and Therapy of Pancreatic Cancer. 2003:26. doi: 10.1097/00006676-200305000-00020. [DOI] [PubMed] [Google Scholar]
- 30.Guo R, Xu D, Wang W. Identification and analysis of new proteins involved in the DNA damage response network of Fanconi anemia and Bloom syndrome. Methods. 2009;48:72–9. doi: 10.1016/j.ymeth.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Biechele TL, Moon RT. Assaying beta-catenin/TCF transcription with beta-catenin/TCF transcription-based reporter constructs. Methods Mol Biol. 2008;468:99–110. doi: 10.1007/978-1-59745-249-6_8. [DOI] [PubMed] [Google Scholar]
- 34.Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012:4–9. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickson M a. Molecular Pathways: CDK4 Inhibitors for Cancer Therapy. Clin Cancer Res. 2014:1–13. doi: 10.1158/1078-0432.CCR-13-1551. OnlineFirs. [DOI] [PubMed] [Google Scholar]
- 36.Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17:v7–12. doi: 10.1093/annonc/mdj941. [DOI] [PubMed] [Google Scholar]
- 37.Xie H, Jiang W, Jiang J, Wang Y, Kim R, Liu X, et al. Predictive and prognostic roles of ribonucleotide reductase M1 in resectable pancreatic adenocarcinoma. Cancer. 2013;119:173–81. doi: 10.1002/cncr.27715. [DOI] [PubMed] [Google Scholar]
- 38.Wei CH, Gorgan TR, Elashoff D a, Hines OJ, Farrell JJ, Donahue TR. A meta-analysis of gemcitabine biomarkers in patients with pancreaticobiliary cancers. Pancreas. 2013;42:1303–10. doi: 10.1097/MPA.0b013e3182a23ae4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMillan M, Kahn M. Investigating Wnt signaling: a chemogenomic safari. Drug Discov Today. 2005;10:1467–74. doi: 10.1016/S1359-6446(05)03613-5. [DOI] [PubMed] [Google Scholar]
- 40.Kahn M. Symmetric division versus asymmetric division: a tale of two coactivators. Future Med Chem. 2011;3:1745–63. doi: 10.4155/fmc.11.126. [DOI] [PubMed] [Google Scholar]
- 41.Miyabayashi T, Teo J, Yamamoto M, Mcmillan M, Nguyen C, Kahn M. Wnt/catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc Natl Acad Sci. 2007;104:5668–73. doi: 10.1073/pnas.0701331104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey JM, Alsina J, Rasheed Z a, McAllister FM, Fu Y-Y, Plentz R, et al. DCLK1 Marks a Morphologically Distinct Subpopulation of Cells with Stem Cell Properties in Pre-invasive Pancreatic Cancer. Gastroenterology. 2014:1–12. doi: 10.1053/j.gastro.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodman RH, Smolik S. CBP / p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–77. [PubMed] [Google Scholar]
- 44.Kim Y-M, Ma H, Oehler VG, Gang EJ, Nguyen C, Masiello D, et al. The gamma catenin/CBP complex maintains survivin transcription in β-catenin deficient/depleted cancer cells. Curr Cancer Drug Targets. 2011;11:213–25. doi: 10.2174/156800911794328420. [DOI] [PubMed] [Google Scholar]
- 45.Ait-Si-Ali S, Polesskaya a, Filleur S, Ferreira R, Duquet a, Robin P, et al. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene. 2000;19:2430–7. doi: 10.1038/sj.onc.1203562. [DOI] [PubMed] [Google Scholar]
- 46.Chan C-H, Lee S-W, Wang J, Lin H-K. Regulation of Skp2 expression and activity and its role in cancer progression. ScientificWorldJournal. 2010;10:1001–15. doi: 10.1100/tsw.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurlin PJ, Huang J. The MAX-interacting transcription factor network. Semin Cancer Biol. 2006;16:265–74. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 48.Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E, et al. Dual Regulation of MYC by p300 via Acetylation-Dependent Control of Myc Protein Turnover and Coactivation of Myc-Induced Transcription. Mol Cell Biol. 2005;25:10220–34. doi: 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin J, Harper JW. A license to kill: transcriptional activation and enhanced turnover of Myc by the SCF(kp2) ubiquitin ligase. Cancer Cell. 2003;3:517–8. doi: 10.1016/s1535-6108(03)00145-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.