Abstract
Bifidobacteria are natural inhabitants of the human gastrointestinal tract and well known for their health-promoting effects. Tolerance to bile stress is crucial for bifidobacteria to survive in the colon and to exert their beneficial actions. In this work, RNA-Seq transcriptomic analysis complemented with proteomic analysis was used to investigate the cellular response to bile in Bifidobacterium longum BBMN68. The transcript levels of 236 genes were significantly changed (≥ threefold, p < 0.001) and 44 proteins were differentially abundant (≥1.6-fold, p < 0.01) in B. longum BBMN68 when exposed to 0.75 g l−1 ox-bile. The hemolysin-like protein and bile efflux systems were significantly over produced, which might prevent bile adsorption and exclude bile, respectively. The cell membrane composition was modified probably by an increase of cyclopropane fatty acid and a decrease of transmembrane proteins, resulting in a cell membrane more impermeable to bile salts. Our hypothesis was later confirmed by surface hydrophobicity assay. The transcription of genes related to xylose utilization and bifid shunt were up-regulated, which increased the production of ATP and reducing equivalents to cope with bile-induced damages in a xylan-rich colon environment. Bile salts signal the B. longum BBMN68 to gut entrance and enhance the expression of esterase and sortase associated with adhesion and colonization in intestinal tract, which was supported by a fivefold increased adhesion ability to HT-29 cells by BBMN68 upon bile exposure. Notably, bacterial one-hybrid and EMSA assay revealed that the two-component system senX3-regX3 controlled the expression of pstS in bifidobacteria and the role of this target gene in bile resistance was further verified by heterologous expression in Lactococcus lactis. Taken altogether, this study established a model for global response mechanisms in B. longum to bile.
Bifidobacteria are common inhabitants of the human gastrointestinal tract (GIT)1, in which they generally persist at concentrations of 109 to 1011 cells per gram of feces, constituting up to 91% of the gut microbiota in breast-fed infants (1). Although bifidobacteria account for only 3%-5% of the total fecal flora in adults (2), their presence has been associated with health-promoting effects, such as balancing of the intestinal microbiota in treatment of diarrhea and immunomodulation (3) and reducing serum cholesterol level (4). Some bifidobacteria are marketed as probiotics, and several Bifidobacterium strains have been used in functional foods, especially fermented dairy products (5, 6). Following consumption, probiotic bacteria are exposed to various physico-chemical stresses, such as low pH in the stomach or bile salts in the intestine. Typically, bifidobacteria colonize the lower GIT, where bile salts have a concentration of nearly 5 mm (7). Bile salts are detergent-like biological compounds with strong antimicrobial activities that disrupt the lipid bilayer structure of cellular membranes, induce protein misfolding and cause oxidative damage to DNA (8). Tolerance to bile stress is indeed essential to health-promoting bifidobacteria to survive in the GIT.
Many studies have been performed to explore the bile resistance factors in bifidobacteria. On one hand, it is generally considered that bile salts hydrolases (BSHs) contribute to bile tolerance in bifidobacteria by decreasing toxicity of conjugated bile salts (9, 10). On the other hand, bile efflux transporters provide protection to bile stress in bifidobacteria, such as Ctr and BetA in B. longum (11, 12) and Bbr_0838 in B. breve strains (13). In addition, the F1F0-ATPase was suggested to be involved in bile resistance by inducing proton pumping and increasing the intracellular ATP reserve in B. animalis (14). In relation to bifidobacterial adaptation to bile, several studies have shown changes in the cell envelope including fatty acids composition and membrane proteins, resulting in decreased membrane permeability to bile salts (15, 16). Furthermore, two-dimensional electrophoresis (2-DE) proteomic analyses of B. longum NCIMB 8809 and B. animalis subsp. lactis IPLA 4549 under bile stress conditions showed that differentially expressed proteins participate in various biological processes, such as general stress response, carbohydrate, amino acid and nucleotide metabolisms, and transcription and translation (17, 18). However, different mechanisms existed between these two bacterial species, for example, the carbon catabolic pathway is mainly rerouted to lactic acid production in strain NCIMB 8809, while it is displaced toward the acetate and an additional formate branch in strain IPLA 4549 (17, 18). Microarray approach also revealed the transcription level of a group of transporters was significantly up-regulated as a response to bile stress in B. breve UCC2003 (19). However, the comprehensive mechanisms of response to bile have not yet been established in bifidobacteria.
B. longum BBMN68 was isolated from a healthy centenarian in the Bama County of the Guangxi Zhuang Autonomous Region in China, which is famous for having a population with a high life-expectancy. Previous study showed that the proportion of bifidobacteria could reach up to 9.59% in the feces from the 90–109 years old population in a Bama suburb using real-time PCR and denaturing gradient gel electrophoresis (20, 21). Further study in our research group indicated that several remarkable characteristics of strain BBMN68 at the genome level, such as a higher abundance of genes associated with carbohydrate transport-metabolism category and two genes encoding bacteriocin, may be beneficial to the long-term colonization of BBMN68 in the human GIT (22). Furthermore, BBMN68 may enhance both innate and acquired immunity and improve intestinal function in mice (23, 24). The probiotic potential of BBMN68 legitimates the need to further explore the biological functions of this strain, such as bile stress response.
Nowadays, the Next-Generation Sequencing (NGS) technology, i.e. RNA-Seq, is a powerful tool for transcriptomics profiling (25, 26). Compared with microarray methods, RNA-Seq provides higher efficiency and sensitivity as it can produce more in-depth information, such as low-abundant transcripts (27). In addition, 2-DE has been widely used to investigate the proteome of lactic acid bacteria and bifidobacteria under bile stress, providing insights into how bacteria respond and tolerate to bile stress, i.e. central metabolisms variation in the cytoplasm (17, 18, 28–33). In the present study, RNA-Seq transcriptomics combined with 2-DE proteomic approach was performed to analyze the bile stress response and resistance mechanisms in B. longum BBMN68. To the best of our knowledge, this work represents the first combined functional genomic and proteomic analysis of bile response mechanisms in bifidobacteria.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
B. longum subsp. longum BBMN68 was cultured on MRS agar plate supplemented with 0.05% (w/v) l-cysteine (MRSc) at 37 °C anaerobically (5% CO2, 5% H2 and 90% N2). For the present study, B. longum was also grown in 200 ml MRSc broth supplemented with the different bile salts concentrations: 0, 0.6, 0.75, 0.9, and 1.2 g l−1 (ox-bile, Sigma, St Louis, MO), respectively. The supplemented MRSc was inoculated at 1% (v/v) with overnight cultures and then incubated anaerobically and monitored by spectrophotometry at 600 nm. Cultures of 200 ml were grown to mid-exponential phase (OD600 of 0.6), ∼2–3 × 108 CFU ml−1 and cells were harvested for both transcriptomics and proteomics experiments. Escherichia coli strains were grown aerobically at 37 °C in 2 × YT medium with shaking at 250 rpm. L. lactis was grown at 30 °C in M17 broth supplemented with 0.5% (w/v) glucose (GM17). When required, media were supplemented with the relevant antibiotics at the following concentrations: 25 μg ml−1 kanamycin, 100 μg ml−1 carbenicillin, and 10 μg ml−1 tetracycline for E. coli, and 10 μg ml−1 chloramphenicol for L. lactis.
RNA Isolation and Purification
Total RNA isolation was carried out using TRIzol reagent (Invitrogen, Carlsbad, CA) as per the manufacturer's instructions. Five milliliters of culture were centrifuged at 12,000 × g for 10 min at 4 °C, and subsequently resuspended in 2 ml of TRIzol reagent. The mixture was vortexed for 1 min and incubated for 3 min at room temperature (RT). Unbroken cells and cell debris were removed by centrifugation at 12,000 × g for 10 min at 4 °C. The supernatant was added with 400 μl of chloroform and mixed for 15 s. After incubation for 15 min at RT, the mixture was separated by centrifugation at 12,000 × g for 15 min at 4 °C. The aqueous phase was mixed with 1 ml of isopropanol and placed for 10 min at RT for precipitation of total RNA. After centrifugation at 12,000 × g for 10 min at 4 °C, the precipitate was washed by 1 ml of 75% ethanol and dissolved with 50 μl of RNase-free H2O. Total RNA was purified using a RiboMinus Kit (Invitrogen) to eliminate ribosomal RNA, and then stored at −70 °C for further use.
RNA-Seq
The RNA-Seq library was constructed using a total RNA-Seq Kit (Applied Biosystems, Foster City, CA) for SOLiD method according to manufacturer's introductions. Briefly, 1 μg of purified RNA was digested by 1 μl of RNase III (Applied Biosystems) at 37 °C for 10 min, and the reaction was terminated by incubation at 65 °C for 20 min. Digested RNA was cleaned up by a RiboMinus Concentration Module (Invitrogen) and ligated with adaptor mix (Applied Biosystems) at 65 °C for 10 min and subsequently at 16 °C for 5 min. The ligation reagents were added with the hybridization mix (Applied Biosystems) and incubated at 16 °C for 16 h. Complementary DNA (cDNA) was synthesized by incubation with RT Master Mix (Applied Biosystems) at 42 °C for 30 min. The MinElute PCR Purification Kit (Qiagen, Valencia, CA) was used to purify the cDNA preparation. cDNA was resolved by a NuPAGE Novex 4–12% Bis-Tris Gel (Invitrogen), and fragments in the size of 150–200 bp were selected for amplification with 15 emulsion PCR cycles and cleaned up with a PureLink PCR Micro Kit (Invitrogen). Then the cDNA library was sequenced using a SOLiD 4.0 sequencer (Applied Biosystems).
Data Processing
All sequenced reads were aligned to the genome of B. longum BBMN68 with accession number CP002286 (22) using SOLiD Corona Lite software (version 4.2, Applied Biosystems) available at http://waprna.big.ac.cn. We used a recursive strategy to improve the usable sequenced reads information (34). That is, 50 mers reads were firstly mapped to the genome with ≤5 color-space mismatches and unmapped reads were progressively trimmed off five bases from the 3′ end once, then the trimmed reads were mapped to the reference genome again until a match was found or unless the read had been trimmed by < 30 bases. All those uniquely mapped reads were used to calculate the RPKM values (reads per kilobase of exon per million mapped sequenced reads) of genes, which indicated the normalized gene expression level. Differentially expressed genes (DEGs) from different samples were identified according to an R package DEGseq (http://waprna.big.ac.cn/rnaseq/function/degseq.jsp) (35). The analyzed transcriptomic data has been submitted to the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE46446.
2-DE and Image Analysis
Intracellular proteins were extracted as previously described (36). Protein samples were quantified using the Bradford method (37) and 2-DE were performed as previously described (38). Briefly, 500 μg of proteins were diluted into 450 μl isoelectric focusing (IEF) buffer containing 8 m urea, 1.5% CHAPS, 0.2% DTT, 0.5% pH 4 to 7 IPG buffer, and moderate bromphenol blue. The diluted samples were used to rehydrate 24-cm pH 4 to 7 linear IPG strips (GE Healthcare, Uppsala, Sweden) for 12 h. First-dimension IEF was performed using an Ettan IPGphor II System (GE Healthcare) for a total of 100 kVh at 20 °C. Focused IPG strips were successively equilibrated for 2 × 15 min in a buffer (6 m urea, 30% glycerol [v/v], 2% SDS [w/v], 50 mm Tris-HCl, pH 8.8) supplemented with 1% DTT in the first step and with 4% iodoacetamide (Sigma) in the second one. The second dimension was performed using an Ettan DALTsix System (GE Healthcare), and proteins were resolved at 40 mA per gel for 5 h at 25 °C. The gels were stained with Coomassie Brilliant Blue G-250 (Amresco, Solon, OH) as previously described (39). Spot detection and volume quantitation were carried out with Image Master 2D Platinum software (version 5.0, GE Healthcare).
Protein Identification
The selected protein spots were cut out, and in-gel digestion using 0.05 μg/μl trypsin was performed as previously described (40). For protein identification, peptide extracts from digested proteins were redissolved in 1 μl of 0.5% trifluoroacetic acid (TFA), which was mixed with 1 μl of matrix (4-hydroxy-α-cyanocinnamic acid in 30% acetonitrile and 0.1% TFA) before spotting on the target plate. MALDI-TOF mass spectrometry and tandem TOF/TOF mass spectrometry were carried out on a 4800 Proteomics Analyzer (Applied Biosystems). In the MS mode, the generated ions were accelerated at the source (20 kV) and separated in the first TOF tube. In the MS/MS mode, the parent ion was focused into the gas cell and fragmented using CID. Combined mass and mass/mass spectra were generated through the GPS Explorer software (version 3.6, Applied Biosystems) and used to interrogate the NCBI database (NCBI nr 2011.04.09; 13,655,082 sequences) using the MASCOT database search algorithms (version 2.1, Matrix Science, Boston, MA). The search criteria were as follows: taxonomy of entries “Bacteria,” trypsin digestion with one missed cleavage allowed, fixed modification of cysteine carbamidomethylation, and variable modification of methionine oxidation. Peptide tolerance and MS/MS tolerance were both 0.2 Da. All of the automatic data analysis and database searching were fulfilled by the GPS Explorer software (version 3.6, Applied Biosystems). Protein scores > 64 were considered to be significant (p < 0.05). For unambiguous identification of proteins, more than five peptides must be matched and the sequence coverage must be greater than 15% (see supplemental Data S1).
Hydrophobicity, Autoaggregation, and Adhesion Assay
B. longum BBMN68 cells grown for 16–20 h (stationary phase) were harvested and resuspended in phosphate buffered saline (PBS, pH 7.4) and adjusted to an OD600 of 1.0. For hydrophobicity assay, 1 ml of cell suspension was added to 1 ml xylene and vortexed for 2 min. The OD600 was measured and cell surface hydrophobicity was calculated as [(1-ODaqueous phase)/ODinitial] × 100%. To test autoaggregation of bifidobacterial cells, 2 ml of cell suspension was placed in each tube and incubated anaerobically at 37 °C. Every two hours, 1 ml of the upper suspension was gently transferred and the OD600 was measured. Autoaggregation was expressed as [(1-ODupper suspension)/ODinitial] × 100%.
Human colon adenocarcinoma cell line HT29 was selected for bifidobacterial adhesion assay. Prior to adhesion, HT29 cells were routinely grown in Dulbecco's High Glucose Modified Eagles Medium (DMEM, HyClone, Thermo Scientific, Rockford, IL) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% penicillin/streptomycin (Sigma) in a humidified 5% CO2 balanced air incubator at 37 °C. Cells were subcultured every 2–3 days. For adhesion assay, cells were seeded at a density of 1 × 105 per well into 24-well plate and grown to ∼ 90% confluence (∼6 days after seeding). Bifidobacterial cultures grown for 24 h were harvested and washed twice with PBS, and resuspended in DMEM without antibiotics at a concentration of ∼ 107 CFU ml−1. Eukaryotic cells were washed twice with PBS and aliquots of 1 ml bifidobacterial suspension were added to wells. The plate was incubated for 1 h at 37 °C with 5% CO2. Afterward, the wells were gently washed three times with PBS to remove nonattached bacteria. 0.25% Trypsin (Sigma) was added to release the cells and bacterial counts were determined on MRSc plates. Adhesion ability was expressed as the percentage of bacteria adhered with respect to the bacteria added into the well.
B1H Method and Motif Searches
To determine the DNA-binding specificity of the response regulator RegX3, bacterial one-hybrid analysis was performed as previously described with slight modifications (41). The library oligonucleotide (Library-F and -R) and primers (RegX3-F and -R) for amplifying regX3 gene were listed in supplemental Table S1. The insert library DNA and vector pH3U3 were ligated after EcoRI and NotI digestion, resulting in the prey plasmid, which was then introduced into E. coli DH5α to generate a raw binding site library. Plasmids isolated from this prey library were electroporated into E. coli US0 followed by counter-selection with 2.5 mm 5-FOA, resulting in a pre-purified library composed of ∼ 5 × 107 transformants. The regX3 gene was amplified by PCR and inserted into the KpnI/XbaI sites of the bait plasmid pB1H2ω2, and the recombinant plasmid, designated as pB1H2-regX3 was transformed into E. coli US0. To confirm the expression of omega-RegX3 fusion protein, Western blotting was carried out with anti-FLAG M2 antibody (Sigma). Then, the plasmids from binding site library were introduced into E. coli US0 harboring pB1H2-regX3, and a two-step selection procedure was used to isolate preys containing recognition sequences for RegX3. 10 mm 3-AT and 3.75 mm 5-FOA were respectively employed for the first and second step. At least 10 selected “preys” were sequenced, and the 18 bp randomized cassettes were submitted to MEME algorithm (42) to identify overrepresented motif. DNA sequences encompassing 250 bp segments upstream of the putative translation start site of the genes that were up-regulated at the transcription level in bile stress condition were analyzed by Target Explorer (43) software tools for detecting target genes of RegX3.
Purification of Recombinant RegX3 and EMSA
Primers (RegX3H-F and -R) used for amplifying the regX3 gene was listed in supplemental Table S1. A His6 tag was introduced at the C-terminal of RegX3 for nickel affinity purification. The digested PCR product was ligated into pNZ8148 at the corresponding sites, resulting in the recombinant plasmid pNZregX3-His6 that was introduced into L. lactis NZ9000. The recombinant RegX3-His6 was purified with Ni Sepharose 6 Fast Flow media (GE Healthcare). The purified protein was detected by SDS-PAGE and the concentration was measured by Qubit 2.0 Fluorometer (Invitrogen).
Target DNA used for EMSA was obtained by annealing the complementary oligos (EMSA pstS-F and -R, supplemental Table S1) which were biotin-labeled using the Biotin 3′ End DNA Labeling Kit (Thermo Scientific). EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific). The binding reactions (20 μl) contained 1 × binding buffer, 50 ng μl−1 Poly dI-dC, 2.5% (v/v) glycerol, 0.05% (v/v) Nonidet P-40, 5 mm MgCl-2, 20 fmol labeled probe and 3.5 μg RegX-His6. In order to verify the protein-DNA specific interaction, a 200-fold molar excess of unlabeled probe competitor (4 pmol) was added to the reaction mixture. Samples were loaded on a 6% native polyacrylamide gel (45 mm Tris borate, 1 mm EDTA, pH 8.3) and the DNA was blotted onto a positively charged nylon membrane and fixed under UV light. Biotin-labeled DNA was detected using Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific).
Heterologous Expression and Bile Stress Survival Experiments
The pstS gene was amplified from B. longum BBMN68 chromosomal DNA using primers PstS-F and -R (supplemental Table S1) and cloned into NcoI/HindIII sites of expression vector pNZ8148. The recombinant plasmids were transformed into L. lactis NZ9000 and determined by sequencing. The recombinant strain was designed as L. lactis NZpstS. Meanwhile, the control strain L. lactis NZCK was constructed by introducing the empty vector pNZ8148 into the host.
Overnight cultures of L. lactis strains were inoculated into 10 ml of GM17 (1% inoculum). When cell density reached an OD600 of 0.2, nisin (Sigma) was added at a final concentration of 10 ng ml−1 and further incubated for 2 h at 30 °C. To assay bile stress survival, aliquots of 1 ml were collected and cells were resuspended in the same volume of Tris-Pi buffer (50 mm Tris-Cl, 25 mm Pi, 25 mm glucose, pH 7.4) supplemented with different concentrations of ox-bile (0.5%, 1.0%, and 2.0%). Samples were taken after 1 h incubation at 30 °C, and 10-fold serial dilutions were spread on GM17 plates supplemented with chloramphenicol.
RESULTS AND DISCUSSION
Global Transcriptomic and Proteomic Analysis of the Bile Stress Response in B. longum BBMN68
B. longum BBMN68 was grown in batch cultures with different ox-bile concentrations ranging from 0.6 to 1.2 g l−1. Our result showed that 1.2 g l−1 ox-bile inhibited the growth of BBMN68, whereas the growth rate was approximately reduced by 50% at 0.75 g l−1 bile (supplemental Fig. S1). Therefore, bile concentration of 0.75 g l−1 was chosen to further study the bile stress response in BBMN68.
The next generation sequencer SOLiD platform was used to investigate the transcription level changes in BBMN68 in the presence of bile salts. A total of 19,002,680 and 16,275,706 unique-mapping-reads were obtained when growing BBMN68 in the presence or not of bile salts, respectively. After filtering, the number of effective reads that were mapped to the genome of BBMN68 was 15,095,199 and 12,893,121, respectively. Further analysis showed that 1849 and 1851 out of the whole 1878 genes in the genome were covered under bile stress conditions and the control, respectively (supplemental Fig. S2). Candidate genes involved in bile stress response were chosen according to the previously described criteria with some modifications (44): 1) more than 20 unique-mapping-reads, 2) more than threefold change after normalization, and 3) statistically significant level p < 0.001. Finally, the transcription of 236 genes was detected to be associated with bile stress, including 76 genes up- and 160 genes down-regulated (supplemental Fig. S2, supplemental Table S2). Their putative functions were classified in different categories grouped by gene ontology (Fig. 1). In addition, 2-DE method was further performed to identify the differentially expressed proteins in BBMN68 between under bile stress conditions and the control. The intensity of 57 spots was significantly changed by a factor of ≥ 1.6-fold (p < 0.01), and these spots were subjected to mass spectrometry for identification. Finally, 44 spots were successfully identified (Fig. 2) and the protein functions were predicted (Table I). Among these proteins, 24 proteins were up-regulated and 20 proteins were down-regulated. Besides, four spots were detected only under bile stress conditions.
Fig. 1.
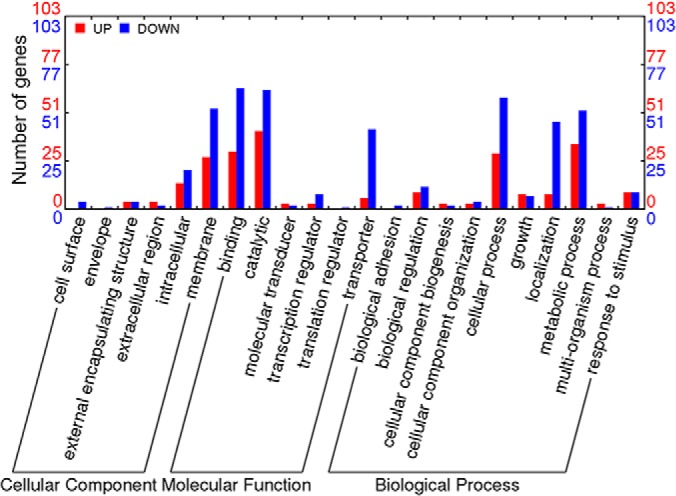
Distribution of differentially transcribed genes in B. longum BBMN68 under bile stress. Result was generated on Web Gene Ontology Annotation Plot (WEGO) (106). Red and blue bars indicate > 3-fold up- and down-regulated genes in the presence of 0.75 g l−1 ox-bile.
Fig. 2.
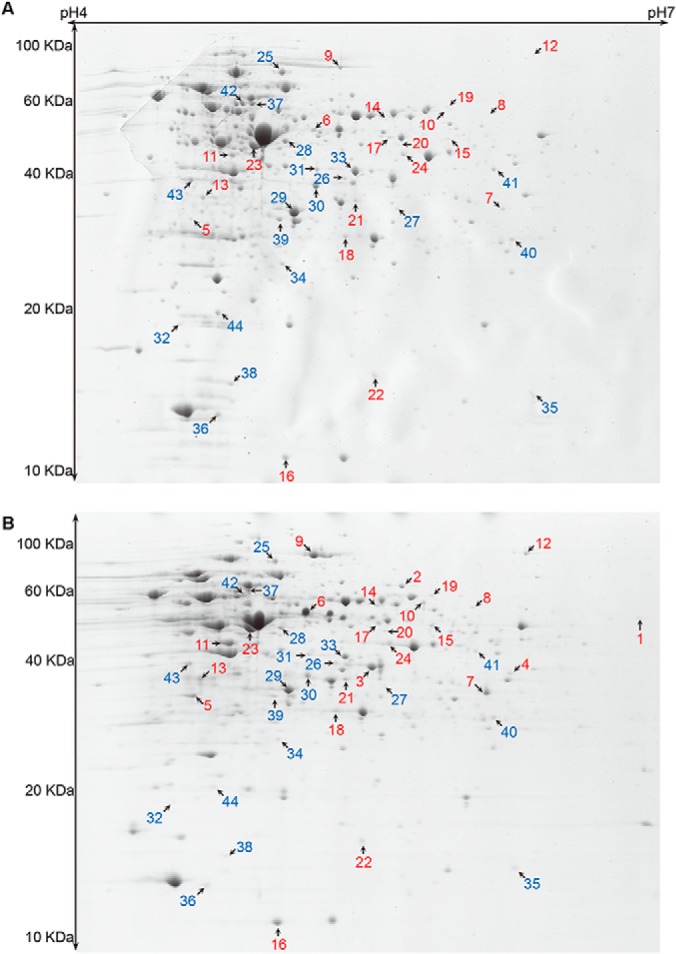
Representative 2D gels showing the intracellular soluble proteins from B. longum BBMN68. A, BBMN68 was grown in the absence of bile. B, BBMN68 was grown in the presence of 0.75 g l−1 ox-bile. Red and blue numbers indicate > 1.6-fold up-regulated and down-regulated proteins, respectively.
Table I. Identification of differentially expressed proteins in B. longum BBMN68 in the presence of bile.
| Category | Spota | Gene | Gene ID | Functionb | Mass | pI | Score | Coverage | Fold change |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Pc | Td | |||||||||
| Stress response | 13 | cbaH | BBMN68_536 | Conjugated bile acid hydrolase | 35.0 | 4.7 | 449 | 52 | 3.44 | 3.05 |
| 16 | groES | BBMN68_1589 | Co-chaperonin HSP10 | 10.6 | 5.1 | 260 | 67 | 2.66 | 2.65 | |
| 21 | uspA1 | BBMN68_51 | Universal stress protein | 34.4 | 5.2 | 333 | 72 | 2.06 | –e | |
| Carbohydrate metabolism | 1 | zwf | BBMN68_1185 | Glucose-6-phosphate 1-dehydrogenase | 57.1 | 6.2 | 325 | 57 | ++f | 3.12 |
| 4 | rbsK2 | BBMN68_1094 | Ribokinase family sugar kinase | 35.9 | 5.7 | 160 | 49 | ++ | 3.23 | |
| 6 | gapA | BBMN68_254 | Glyceraldehyde-3-phosphate dehydrogenase, type I | 37.8 | 5.2 | 469 | 63 | 12.6 | – | |
| 8 | lpd1 | BBMN68_825 | Dihydrolipoamide dehydrogenase | 52.1 | 5.6 | 210 | 58 | 6.17 | 2.55 | |
| 9 | xfp | BBMN68_708 | Xylulose-5-phosphate/fructose-6-phosphate phosphoketolase | 92.5 | 5.1 | 163 | 27 | 5.95 | – | |
| 11 | dhaT | BBMN68_1706 | 1,3-propanediol dehydrogenase/lactaldehyde reductase | 40.6 | 4.8 | 561 | 69 | 5.56 | – | |
| 12 | putA2 | BBMN68_1612 | NAD-dependent aldehyde dehydrogenase | 98.8 | 5.9 | 226 | 28 | 5.37 | 4.15 | |
| 15 | glgC | BBMN68_606 | ADP-glucose pyrophosphorylase | 45.8 | 5.5 | 253 | 60 | 2.61 | – | |
| 18 | gpmA | BBMN68_1687 | Phosphoglycerate mutase I | 28.7 | 5.8 | 334 | 87 | 2.25 | 2.41 | |
| 23 | pgk | BBMN68_399 | 3-phosphoglycerate kinase | 41.8 | 4.9 | 390 | 63 | 1.81 | – | |
| 24 | ackA | BBMN68_728 | Acetate kinase | 44.2 | 5.5 | 389 | 64 | 1.71 | – | |
| 39 | tas4 | BBMN68_1072 | Aldo/keto reductase | 31.7 | 5.0 | 360 | 64 | −2.18 | – | |
| 42 | pgm | BBMN68_1663 | Phosphoglucomutase | 60.2 | 4.9 | 686 | 59 | −3.01 | – | |
| Amino acid metabolism | 17 | BBMN68_1360 | Aspartate/tyrosine/aromatic aminotransferase | 47.2 | 5.3 | 143 | 38 | 2.62 | – | |
| 28 | argG | BBMN68_809 | Argininosuccinate synthase | 45.5 | 5.1 | 256 | 52 | −1.72 | – | |
| 30 | ilvC1 | BBMN68_1262 | Ketol-acid reductoisomerase | 38.5 | 5.1 | 641 | 67 | −1.93 | −3.81 | |
| 33 | metC3 | BBMN68_917 | Cystathionine β-lyases/cystathionine γ-synthases | 41.9 | 5.3 | 511 | 56 | −2.00 | – | |
| 40 | hisG | BBMN68_464 | ATP phosphoribosyltransferase | 30.9 | 5.7 | 121 | 52 | −2.34 | – | |
| 41 | asd | BBMN68_1227 | Aspartate-semialdehyde dehydrogenase | 40.3 | 5.7 | 449 | 74 | −2.67 | – | |
| Nucleotide metabolism | 2 | pyrG | BBMN68_614 | CTP synthase | 60.9 | 5.4 | 381 | 55 | ++ | 2.52 |
| 10 | purA | BBMN68_1276 | Adenylosuccinate synthase | 46.4 | 5.4 | 93 | 37 | 5.69 | – | |
| 19 | purH | BBMN68_424 | AICAR transformylase/IMP cyclohydrolase | 58.4 | 5.5 | 114 | 32 | 2.20 | – | |
| 26 | prsA2 | BBMN68_1158 | Phosphoribosylpyrophosphate synthetase | 36.7 | 5.3 | 200 | 43 | −1.69 | – | |
| 27 | pyrD1 | BBMN68_529 | Dihydroorotate dehydrogenase | 33.9 | 5.4 | 115 | 64 | −1.72 | −5.25 | |
| 35 | pyrI | BBMN68_533 | Aspartate carbamoyltransferase regulatory subunit | 15.5 | 5.8 | 158 | 82 | −2.07 | −3.08 | |
| 37 | guaA1 | BBMN68_709 | GMP synthase | 56.1 | 5.0 | 654 | 63 | −2.14 | – | |
| 43 | purM | BBMN68_870 | Phosphoribosylaminoimidazole (AIR) synthetase | 36.0 | 4.6 | 119 | 52 | −3.40 | – | |
| Coenzyme metabolism | 20 | npy1 | BBMN68_988 | NAD+ diphosphatase | 45.3 | 5.4 | 182 | 54 | 2.13 | 1.96 |
| 32 | mdaB | BBMN68_1782 | Putative NADPH-quinone reductase | 19.3 | 4.6 | 76 | 44 | −1.96 | – | |
| Signal transduction | 22 | luxS | BBMN68_914 | LuxS protein for autoinducer AI2 synthesis | 18.5 | 5.5 | 120 | 71 | 2.03 | 3.99 |
| Transcription | 34 | orn2 | BBMN68_1756 | Oligoribonuclease | 24.3 | 5.0 | 137 | 51 | −2.01 | 3.14 |
| 36 | greA | BBMN68_764 | Transcription elongation factor | 17.1 | 4.8 | 126 | 71 | −2.12 | 3.22 | |
| Translation | 3 | rpsB | BBMN68_371 | 30S ribosomal protein S2 | 30.8 | 5.3 | 536 | 86 | ++ | – |
| 7 | BBMN68_307 | Ribosome-associated protein Y | 24.7 | 5.7 | 288 | 48 | 7.77 | 2.23 | ||
| 25 | typA | BBMN68_271 | GTP-binding protein TypA/BipA | 70.1 | 5.0 | 380 | 54 | −1.61 | −2.09 | |
| 29 | tsf | BBMN68_372 | Translation elongation factor Ts | 30.0 | 5.1 | 904 | 79 | −1.73 | – | |
| 38 | fkpA1 | BBMN68_765 | FKBP-type peptidyl-prolyl cis-trans isomerase | 14.4 | 4.9 | 191 | 74 | −2.16 | – | |
| 44 | ppiB | BBMN68_332 | Peptidyl-prolyl cis-trans isomerase | 19.5 | 4.8 | 69 | 49 | −3.92 | 2.01 | |
| Cell wall biogenesis | 14 | murC | BBMN68_210 | UDP-N-acetylmuramate-alanine ligase | 52.6 | 5.4 | 204 | 51 | 3.32 | – |
| Unknown | 5 | BBMN68_416 | Hypothetical protein | 26.9 | 4.6 | 343 | 75 | 13.3 | – | |
| 31 | BBMN68_912 | Hypothetical protein | 49.1 | 5.8 | 70 | 35 | −1.98 | −2.22 | ||
a Spot numbers refer to the proteins labeled in Fig. 2.
b Functions were assigned from the KEGG pathways for B. longum BBMN68.
c Proteomic fold change.
d Transcriptomic fold change.
e The corresponding transcription was not influenced by bile.
f Proteins were detected only in the presence of bile.
Although transcriptomic analysis identified 236 differentially expressed genes, only 57 proteins were detected to be differentially produced. A total of 15 genes were regulated at both transcription (≥ 2-fold) and translation (≥ 1.6-fold) levels, i.e. 10 up-regulated genes and five down-regulated genes. These results illustrate a low correlation between transcriptomic and proteomic data of the bile stress response in BBMN68. Such observation constitutes another example among others where integrative “omics”-approaches proved to be a real challenge (33, 45, 46). In this study, the RNA-Seq was employed to investigate the expression profile of the whole genes at the transcription level under bile stress in BBMN68. However, the 2-DE focused on the change of cytoplasmic soluble proteins with pI of 4 to 7. The difference of coverage between these two methods may explain the low correlation between transcriptome and proteome. In addition, RNA secondary structure, Shine Dalgarno sequence differences, regulatory proteins and sRNAs, codon bias, and ribosome occupancy constitute many regulatory points that may influence the protein production (47), therefore resulting in differences between mRNA level and protein abundance of some genes. Moreover, targeted degradation of individual proteins in response to bile salts (48) may also cause the variation in protein abundances, impacting on the correlation between transcirptome and proteome in BBMN68 during the bile stress response.
In conclusion, the SOLiD combined with the 2-DE analyses showed that 236 genes and 44 proteins were affected by ox-bile and could be related with the response and resistance of B. longum BBMN68 to bile stress, which were discussed in details as follows.
Genes Involved in Bile Resistance in BBMN68
In the presence of ox-bile, the transcription or translation of eight genes related with bile resistance were significantly up-regulated in BBMN68. BBMN68 (hereafter as BN) _664, encoding a permease of the major facilitator superfamily (MFS), was the most highly up-regulated gene at the mRNA level in BBMN68 in response to bile (84.50-fold up-regulated). BN_664 shows 99% identity with BL0920 in B. longum NCC2705, which encodes a bile-inducible efflux transporter BetA conferring bile resistance (12). Meanwhile ox-bile also increased the transcription of another gene encoding MFS permease (BN_1049), which could play a similar role with BetA. In addition, the transcription level of BN_1434-1433 encoding an ABC-2 type transporter was respectively 3.59- and 4.73-fold up-regulated under bile stress conditions. Among ABC-2 type transporters, Drr and Evr have been reported to pump toxic compounds, such as daunorubicin and doxorubicin in Streptomyces lucretius (49) and viologen in Synechocystis (50). These results suggested that this transporter may possibly perform similar function of bile exclusion in BBMN68 in response to bile stress.
Remarkably, the transcription and translation of a gene encoding BSH (BN_536), which catalyzes deconjugation of glycine- or taurine-linked bile acids, was 3.05-fold and 3.44-fold up-regulated, respectively. Deconjugation of bile salts may play an important role in the bile tolerance because of the detoxification properties. Therefore, BSH activity may be a desirable feature of a probiotic, as it can increase its survival ability in gut conditions (51). However, BSH was not up-regulated in B. animalis sp. lactis IPLA 4549 (18), B. animalis sp. lactis BB-12 (52) and B. longum NCIMB 8809 (17) in response to bile, supporting the idea that different bile salts hydrolases, even within the same species, are differentially regulated in response to bile. The transcriptions of BN_167 and BN_1224, encoding hemolysin-like protein (HLP) and calcineurin-like phosphoesterase, were 10.08- and 4.23-fold up-regulated upon bile exposure, respectively. In cyanobacterium Synechocystis, HLP is located on the cell surface and functions as a barrier against the adsorption of toxic compounds (53). Another work from our research group indicated a two- to threefold increased survival of the recombinant L. lactis NZ9000 strain harboring BN_167 exposed to tauro-conjugated bile salts (TCA and TDCA) (manuscript in preparation), suggesting that HLP really conferred resistance to bile in BBMN68, especially tauro-conjugated bile salts. In addition, complete genome analysis of the most radiation-resistant bacteria Deinococcaceae reflected expansion of a calcineurin-like phosphoesterase, which appear to be related with stress resistance possibly through decomposing damage products under stress conditions (54).
Taken altogether, we proposed that these differentially expressed genes are associated with bile resistance in BBMN68 mainly by bile efflux, bile salts decomposition and prevented bile adsorption.
Bile Stress Induces General Stress Response in BBMN68
Many molecular chaperones and proteases related to the general stress response were up-regulated in BBMN68 in response to bile. The transcription of groEL (BN_44) was 3.25-fold up-regulated upon exposure to bile salts, and the expression of groES (BN_1589) was 2.65-fold and 2.66-fold up-regulated at the mRNA and protein level, respectively. GroEL/ES complex is required for the proper folding of proteins and frequently involved in common stress response in bacteria (17, 45, 55–57). At the transcription level, BN_1305 encoding HSP20 chaperone IbpA was 4.81-fold up-regulated upon bile exposure. In B. breve, the transcription of hsp20 is strongly up-regulated by heat shock and osmotic shock (58). HSP20 chaperone belongs to the small heat shock protein (sHSP) which is important for the prevention of irreversible denaturation of heat-damaged protein (59). In addition, the transcription of four genes coding for proteolysis functions were up-regulated: trypsin-like serine protease DegQ (BN_1289, 3.50-fold), proteasome assembly chaperone (BN_413, 8.17-fold), dipeptidase (BN_1184, 3.82-fold) and endopeptidase (BN_1763, 3.09-fold). DegQ has been proved to be a pH-sensitive protease that controls protein quality under mild and severe stress conditions (60). These proteasome and peptidase play a major role in degradation of damaged proteins and reutilization.
BN_86 and BN_1435 encoding nitroreductase were 4.46- and 7.03-fold up-regulated at the mRNA level upon bile salts exposure. Nitroreductase is related with oxidative stress response by regulating the activities of antioxidant-enzymes in Saccharomyces cerevisiae (61), and also protects L. lactis against oxidative stress (62). In addition, the transcription of a gene encoding cystathionine gamma-lyase (BN_1590) was 3.22-fold up-regulated in response to bile salts. This enzyme breaks down cystathionine into cysteine and 2-oxobutanoate, accompanying with the formation of H2S or NH3. Recent research has shown that H2S could mitigate oxidative stress imposed by antibiotics in bacteria (63). Meanwhile, cystathionine gamma-lyase is a component of cysteine-mediated oxidative defense in L. reuteri (64). Moreover, NH3 could capture cytoplasmic protons, thus contributing to acid stress tolerance of the host strain (65).
Other general stress-related genes up-regulated in response to bile were BN_1764 (HdeD protein, 5.74-fold at the mRNA level), BN_138 (DNA helicase, 5.04-fold at the mRNA level), and BN_51 (universal stress protein, 2.06-fold at the protein level). It has been demonstrated that HdeD is a component of the H-NS-dependent regulatory cascade of acid stress defense in E. coli (66), also involved in general stress resistance (67). DNA helicase is essential in nucleic acid metabolism involving in DNA repair, replication, recombination, and RNA processes (68). Under stress conditions, DNA helicase mainly performs repair function and deals with DNA damages caused by bile. On the other hand, a DNA-damage inducible protein D (BN_1457) was down-regulated at the transcript level, reflecting high DNA quality control under bile stress. In BBMN68, these genes play an important role in coping with other stress effects including protein misfolding, oxidative stress, low pH, and DNA damage caused by bile salts.
Bile Stress Accelerates the Xylose Catabolism and Enhances the Inhibition of Polysaccharides Utilization by Glucose
In B. longum NCIMB 8809 and B. animalis IPLA 4549, bile stress stimulated the carbohydrate catabolism, typically in the pentose phosphate pathway so-called bifid shunt (17, 18). In this study, most enzymes involved in bifid shunt were 3.02–4.15-fold up-regulated at the mRNA level (supplemental Table S2) and 1.71–12.6-fold overexpressed at the protein level (Table I), respectively, demonstrating that the bifid shunt was also enhanced in strain BBMN68 in response to bile. Meanwhile, three enzymes providing substrates for the bifid shunt were up-regulated under bile stress conditions. Glucose-6-phosphate 1-dehydrogenase (Zwf) and ribokinase (RbsK2) were only detected on the 2-DE gels of cultures treated by bile. Zwf catalyzes β-glucose-6-P to form glucono-1,5-lactone-6-P and RbsK2 catalyzes the reaction ribose + ATP → ribose-5-P. The two products can be transferred to xylulose-5-P, which feeds the key enzyme xylulose-5-phosphate/fructose-6-phosphate phosphoketolase (Xfp) in bifid shunt. Remarkably, the transcription of BN_1736 encoding xylose isomerase (XylA) was highly up-regulated (26.9-fold). XylA catalyzes the interconversion of xylose and xylulose, through which the xylose enters into the glycolysis pathway in BBMN68. In colon, xylan is abundant from plant food, which cannot be digested by the host, while it can be decomposed into xylose by gut microbes and then be used by BBMN68.
In the presence of bile, phosphoglucomutase (Pgm) was among the most highly repressed protein (3.01-fold down) in BBMN68. Pgm catalyzes the interconversion of glucose-1-P and glucose-6-P, the second step of starch and glycogen catabolism. Nine other genes involved in hydrolysis of oligo/polysaccharides were also down-regulated at the transcription level under bile stress conditions. In addition, six of the nine genes were transcribed at low levels even in sample untreated by bile salts (< 1000 mapping reads) (Table II). These results suggested that the expression of genes for polysaccharides utilization may be inhibited by glucose from the medium and such inhibition was more remarkable upon exposure to bile salts. Because glucose is more efficient for energy production, this superiority is beneficial for BBMN68 to cope with the bile stress, because ATP and reducing equivalents are essential to protect against bile stress, through pumping-out bile (12), repairing damaged protein and DNA (17), and regulating the internal pH (14).
Table II. Transcription level of genes involved in oligo/polysaccharides utilization in B. longum BBMN68.
| Gene | Reads number |
Fold change | Productiona or Domainb | Substratec | |
|---|---|---|---|---|---|
| CKd | OGe | ||||
| BN_1420 | 14909 | 5479 | −3.18 | α-galactosidase | galacto-oligosaccharides |
| BN_1446 | 11201 | 3395 | −3.85 | Endo-1,4-β-xylanase | xylan |
| BN_1018 | 86 | 23 | −4.36 | Glycosyl hydrolase family 53 | arabinogalactan |
| BN_1795 | 736 | 188 | −4.57 | β-glucosidase | β-glucan |
| BN_101 | 734 | 184 | −4.66 | α-glucosidase | α-glucan |
| BN_221 | 548 | 135 | −4.73 | β-glucosidase | β-glucan |
| BN_1363 | 158 | 34 | −5.42 | Pectinesterase | pectin |
| BN_1202 | 113338 | 19349 | −6.84 | Endo-α-N-acetylgalactosaminidase | O-glycan |
| BN_1162 | 771 | 131 | −6.89 | xylanase | xylan, araban |
a Productions were assigned through blast the amino acid sequence to the UnitProtKB database.
b Domains were identified from the KEGG SSDB database.
c Substrates were deduced from the KEGG ENZYME database.
d BBMN68 was grown in the absence of ox-bile.
e BBMN68 was grown in the presence of 0.75 g l−1 ox-bile.
Effect of Bile Stress on Amino Acids, Nucleotides, and Fatty Acids Metabolism Processes
Three enzymes related with aromatic amino acids synthesis were up-regulated in BBMN68 in response to bile salts. The transcription of BN_1136 encoding chorismate mutase was 3.98-fold up-regulated. It catalyzes the reaction chorismate → prephenate, which is the precursor of phenylalanine and tyrosine (69). BN_1775 and BN_1360 encode the same enzyme aspartate/tyrosine/aromatic aminotransferases, which were 3.90-fold and 2.62-fold up-regulated at the transcription and translation level, respectively. These results suggested that aromatic amino acids were more abundant when BBMN68 was grown in MRSc with bile. Hydrophobic amino acids, such as aromatic amino acids and branched-chain amino acids (BCAAs) could protect proteins against attack of bile by building hydrophobic areas (17). Surprisingly, the expression of a gene encoding ketol-acid reductoisomerase (BN_1262) involved in BCAA synthesis was 3.81-fold and 1.93-fold repressed at the transcription and protein levels, respectively. BCAAs played a major role in this protection mechanism in B. longum NCIMB 8809 (17), suggesting different regulation mechanism of amino acids metabolism in B. longum strains in response to bile. In addition, the transcription of a gene encoding [glutamate:ammonia-ligase] adenylyltransferase (GlnE, BN_535) was 3.22-fold up-regulated in BBMN68 in response to bile salts. GlnE controls the activity of glutamine synthetase by transferring adenylyl from ATP (70), and plays a role in nitrogen metabolism, especially under nitrogen starvation condition (71, 72). Interestingly, BN_535 and BN_536 (encoding BSH) constitute a putative operon and were simultaneously up-regulated in response to bile salts, implying that BSH may involve in amino acid metabolism because glycine or taurine released from bile salts deconjugation could potentially be used as nitrogen sources (51).
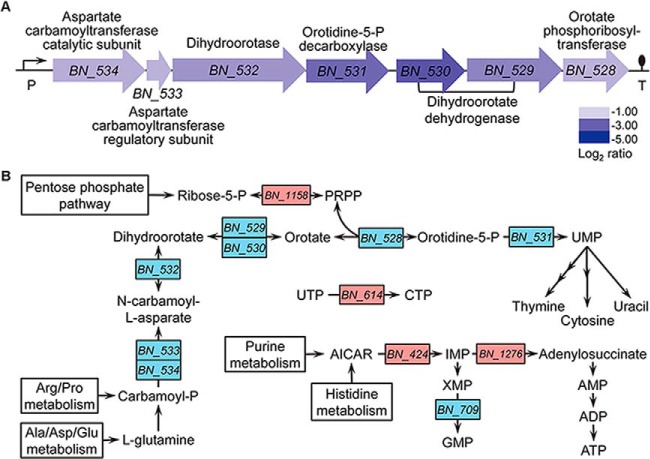
The amounts of proteins involved in purine nucleotide synthesis were altered in bile-exposed BBMN68 (AICAR transformylase, 2.20-fold; adenylosuccinate synthase, 5.69-fold; and GMP synthase, −2.14-fold). AICAR transformylase catalyzes the conversion from AICAR to IMP. Subsequently, IMP is transferred to AMP by the catalysis of adenylosuccinate synthase and adenylosuccinate lyase. On the other access, IMP is transferred to XMP, which further turns into GMP through the action of GMP synthase. As a result, more adenine nucleotides were produced in BBMN68 under bile stress conditions, probably contributing to the accumulation of ATP. In aspect of pyrimidine metabolism, CTP synthase (BN_614) involved in CTP synthesis was only detected on 2-DE gels when bile was present. CTP was an inhibitor of aspartate carbamoyltransferase (BN_534–533) (73), the first enzyme in pyrimidine biosynthesis. Clearly, a cluster containing seven genes (BN_528–534) for pyrimidine biosynthesis was 3.02- to 9.60-fold down-regulated at the transcription level. Meanwhile, the productions of BN_529 and BN_533 were also less abundant in proteome, indicating that bile stress inhibited the synthesis of pyrimidine in BBMN68 in response to bile. A schematic representation of the nucleotides metabolism regulated by bile is shown in Fig. 3.
Fig. 3.
Nucleotides metabolism influenced by bile stress in B. longum BBMN68. A, An operon composed of seven genes related with pyrimidine metabolism was repressed in the presence of bile. Changes in transcription level are represented as log2 intensity ratio values shown with the gene names. P and T indicate putative promoter and terminator, respectively. B, Schematic representation of the regulated pathways in nucleotides metabolism. Red and blue gene names indicated up- and down-regulation in the presence of bile.
BN_1705, encoding cyclopropane fatty acid (CFA) synthase, was 6.52-fold up-regulated at the transcription level upon bile exposure. CFA synthase catalyzes the cyclization of phospholipid olefinic fatty acid, producing phospholipid CFA that could modify the viscosity and permeability of cell membrane. CFAs have been previously proved to protect bacteria from stress environments, such as acid stress (74) and rifaximin (75) by decreasing membrane permeability. Moreover, CFAs is also involved in resistance to cold, osmotic and ethanol in lactic acid bacteria (76–80). On the other hand, the fatty acid biosynthesis operon (BN_1556–1558) was down-regulated, indicating a general decrease in fatty acid synthesis process in response to bile stress, which has been suggested in some lactobacilli (29, 31, 33, 81), Enterococcus faecalis (82) and B. animalis (18, 52).
Effect of Bile Stress on Transmembrane Transport in BBMN68
Transcriptomic analysis revealed that 61 annotated genes encoding transmembrane transporters were regulated in BBMN68 under bile stress conditions (supplemental Table S3). Besides three presumed bile efflux transporters mentioned above (BN_664, 1049, 1434-1433), only five transporters were more abundant, including three monosaccharide transporters or its components, encoded by BN_1738 (simple sugar ABC-type transporter permease component, 6.63-fold), BN_1664 (MFS glucose/H+ symporter, 3.84-fold), and BN_1724 (pentose ABC-type transporter ATPase component, 3.32-fold), respectively. The enhanced monosaccharide transport corresponds to the observed acceleration in glycolysis using glucose and xylose as substrates in BBMN68 upon bile exposure. In addition, the transcription of BN_1101, encoding a small-conductance mechanosensitive channel (MscS), was 3.10-fold up-regulated by bile. Mechanosensitive channels of large (MscL) and small conductance respond to the change in membrane tension and allow the passage of water and ions, preventing cell lysis (83, 84). Because bile is considered to cause membrane damage and permeability disorder leading to increased osmolality (48), increased expression of MscS may confer protection to BBMN68 from raising turgor pressure and cell lysis.
On the other side, most transporter genes were down-regulated when BBMN68 was grown in bile. Remarkably, 19 genes encoding components of eight transporters for oligo/polysaccharides utilization were strongly down-regulated (3.41- to 67.41-fold down), including six ABC-type transporters, one MFS transporters and one symporter. These results were corresponding to the repressed oligo/polysaccharides metabolisms in BBMN68 under bile stress. The repression of transporters specific for oligosaccharides has been detected by a microarray transcriptomic study in L. plantarum in response to bile (85). Meanwhile, the transcription of genes involved in transport of amino acids, peptides, inorganic ions, and other unknown substrates were also down-regulated at different levels from 3.00- to 10.14-fold by bile. These transporters are transmembrane proteins, and the down-regulated expression could result in a more hydrophobic cell surface in BBMN68, which was verified through the cell surface hydrophobicity assay. The result showed that the surface hydrophobicity was increased from 1.4% to 17.7% in BBMN68 upon bile exposure (supplemental Fig. S3). It has been suggested that bile primarily exerts its antimicrobial effects on cell membranes by causing membrane damages (48). And we hypothesized that transmembrane proteins may be the pathway through which conjugated bile acids enter into cytoplasm because of the hydrophobicity of the bilayer lipid skeleton. In this case, bifidobacteria could decrease the protein proportion of cell membrane and enhance surface hydrophobicity, thereby protecting themselves from invasion of bile salts.
Bile Salts Promote Adhesion and Adaptation of BBMN68 in the GIT
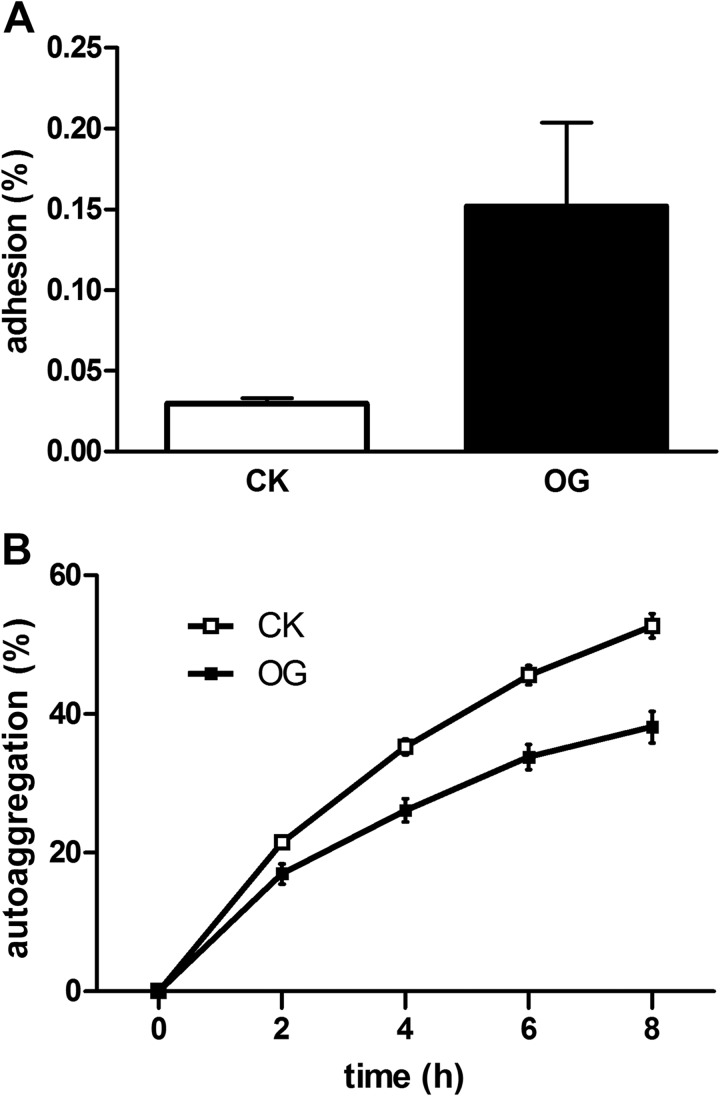
In BBMN68, the transcription of BN_119 was 7.42-fold up-regulated under bile stress conditions. BN_119 encodes an esterase that has been considered as a relevant factor with adaptation to the gastrointestinal tract, through decomposing and reorganizing peptidoglycans in cell wall (81, 86). The bile-resistant mutants of Salmonella enteria showed alteration of the cell wall lipopolysaccharide (87), which may be an adaptation mechanism to the enteric bile stress. So it was speculated that cell wall construction realignment of BBMN68 in response to bile salts could improve adaptation in the GIT. In addition, bile induced the transcription of BN_1395 (4.77-fold), encoding a housekeeping sortase related with anchoring of pili to the cell wall in Gram-positive bacteria (88, 89). Genomic analysis of B. longum NCC2705 suggested B. longum produced fimbriae for attachment in the GIT (90). To further investigate the influence of bile salts on the colonization and persistence of BBMN68 in the gut, the adhesion experiments and autoaggregation were performed in this study. As expected, the adhesion ability was increased by fivefold when BBMN68 grown with bile (Fig. 4A). Meanwhile, the autoaggregation degree was decreased from 52.67% to 38.10% after 8h incubation (Fig. 4B). In this case, once BBMN68 enters into the intestinal tract, the bifidobacterial cells tend to disaggregate because of enteric bile salts, which benefits subsequently adhere to the epithelium. Taken together, bile could act as a gut signal and facilitate the adhesion and colonization of BBMN68 in the host.
Fig. 4.
Effect of bile on adhesion to HT-29 cells A, and autoaggregation B, of B. longum BBMN68. Values are mean ± standard deviation of one representative of two independent experiments performed in triplicate. Abbreviations: CK, BBMN68 grown without bile; OG, BBMN68 grown in the presence of bile.
The transcription and translation of luxS (BN_914) encoding S-ribosylhomocysteine lyase for autoinducer-2 (AI-2) synthesis were respectively up-regulated by 3.99- and 2.03-fold, which indicated that bile also stimulates the communication between BBMN68 and other enterobacteria. Autoinducers are signal molecules that activate the quorum sensing in bacteria, and AI-2 acts as a signal which is used for interspecies cell-cell communication in both Gram-negative and Gram-positive bacterial species (91). The previous study has been proposed that B. longum NCC2705 may use AI-2 to recognize its presence within a host and enhance the expression of a series of metabolic genes required for fast propagation. Meanwhile, the secreted lactic acid and acetic acids would prevent the colonization of pathogens (92). Finally, the transcription of an IS1595 family insertion sequence (BN_1164-1166) from Streptococcus agalactiae was up-regulated (3.37–4.60-fold) in BBMN68 in response to bile. Aminoglycoside nucleotidyltransferase (BN_1166), which decomposes aminoglycosidic antibiotics, may confer resistance to the corresponding antibiotics produced by other enterobacteria or derived from diet and therapies medicine. In addition, this IS1595 family insertion sequence was only found in BBMN68 genome, which contributed to the long-term colonization of this strain in the gut of healthy centenarian.
Transcription and Translation of BBMN68 were Influenced by Bile
In the presence of bile, many transcription regulatory genes were differently expressed at the mRNA level, including three two-component systems (TCSs) and 10 transcription factors (TFs). The transcription of a gene encoding an extracytoplasmic function (ECF) sigma factor RpoE (BN_247), which belongs to the σ70 family (93), was 4.50-fold up-regulated in BBMN68 in response to bile salts stress. Upon receiving environment stimulus, ECF σ factor is released and binds to RNA polymerase to stimulate transcription of a specific group of genes (94). RpoE has been determined as an important regulator in diverse stress response, such as heat shock in E. coli (95), acid adaptation in Streptococcus mutans (96), and oxidative stress in many bacteria (97). In this study, it was for the first time reported that RpoE was involved in resistance to bile stress in bifidobacteria. Moreover, there is no clear function for the other regulatory genes except TCS senX3-regX3. But their role cannot be disregarded and the regulatory network controlling bile stress response should be deeply investigated, which may be a basis to further comprehend and examine the bile stress response in bifidobacteria.
In the aspect of translation process, ribosome-associated protein Y (BN_307) was 7.77-fold up-regulated in BBMN68 upon bile exposure. Ribosome-associated proteins are key factors that promote the folding pathways of newly synthesized proteins (98). The expression of two genes encoding putative elongation factor (BN_118) and elongation factor Ts (BN_372) were 5.12-fold down-regulated at the transcription level and 1.73-fold down-regulated at the protein level, indicating a slower translation rate in BBMN68 under bile stress. The transcription of relBE operon (BN_287–288) was 3.99-fold and 3.70-fold down-regulated under bile stress conditions, respectively. relA (BN_296), encoding GTP-pyrophosphokinase involved in (p)ppGpp formation (99), was 4.02-fold up-regulated at the transcription level. The relBE promoter is repressed by its product RelB, and RelE functions as a co-repressor (100). The decreased mRNA level of relBE suggested an accumulated amount of RelBE, which belong to toxin-antitoxin (TA) complex family. The toxin RelE can be activated by protease Lon through the polyphosphate (PolyP)-dependent signal pathway initiated by ppGpp, subsequently cleaves mRNA at the ribosomal A-site in translation process, and thus reduces the global level of translation under nutritional stress (101). These results implied that BBMN68 may employ the RelBE system to control the translation process under bile stress conditions.
The TCS senX3-regX3 Played an Important Role in Bile Stress Response in BBMN68
In BBMN68, bile salts induced the transcription of BN_1080 (5.55-fold) encoding a sensor histidine kinase, and the downstream BN_1079 encoding a response regulator was also up-regulated by 2.66-fold upon bile exposure. The two genes composed a TCS, which has been known as senX3-regX3 system in Mycobacterium tuberculosis. In M. tuberculosis, senX3-regX3 is positively autoregulated (102) and controls the expression of genes involved in inorganic phosphate (Pi) acquisition, i.e. phoA and pstS (103). However, its role in bile stress response has not been reported in bacteria.
In this study, the regulatory target genes of response regulator RegX3 were predicted by the B1H method. The recombinant bait vector pB1H2-regX3 was introduced into E. coli US0, and whole-cell lysates were prepared and subjected to Western blotting using anti-Flag antibody. A ∼ 40 kDa band was observed on the membrane, indicating the expression of RegX3 as a carboxy-terminal fusion to the ω-subunit of RNA polymerase (Fig. 5A). The DNA-binding sequence for RegX3 was identified by a two-step selection resulting in 13 sequences showed a 7-bp motif (GARRACY, where r = G/A and Y = C/T, E-value = 1.7e−6) in MEME analysis (Fig. 5B). Target Explorer analysis (cut-off score 5.00) predicted that RegX3 controlled the transcription of pstS (BN_1078), putA2 (BN_1612), hdeD (BN_1764), and six other genes (supplemental Table S4). Among these genes, pstS has been mentioned to confer resistance to some toxic compounds, such as penicillin on Streptococcus pneumonia (104) and sodium benzoate on E. coli (105). Interestingly, in the genome of bifidobacteria, senX3-regX3 is followed by a gene cluster pstSCAB, encoding the high-affinity phosphate transporter Pst. And the senX3-regX3 operon and pstS were simultaneously up-regulated at the transcription level, implying that the senX3-regX3 system enhanced the expression of pstS and then conferred bile resistance on BBMN68.
Fig. 5.
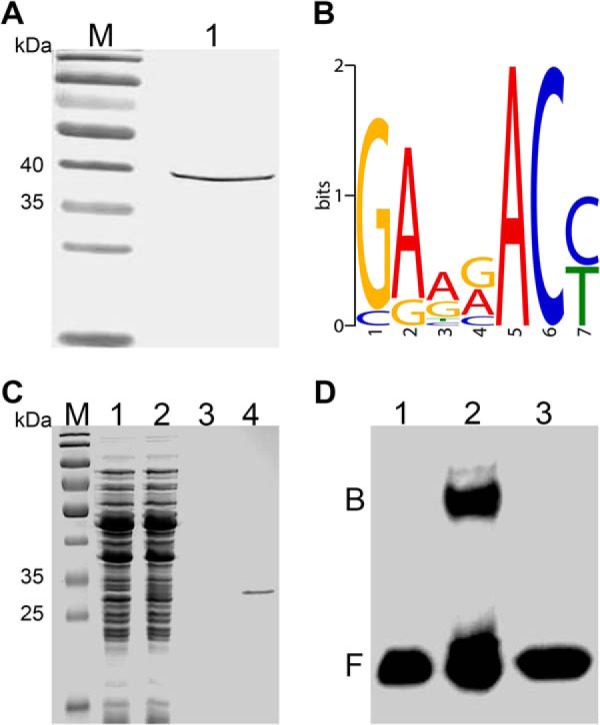
DNA binding activity of response regulator RegX3 in B. longum BBMN68. A, Western blotting was used to detect the expression of ω-RegX3 fusion protein with a theoretical molecular weight of ∼ 39 kDa in Lane 1. B, Representation of DNA binding site of RegX3 predicted by B1H. The relative height of the letters represents the frequencies of nucleotides at each position. C, SDS-PAGE analysis of the purified RegX3-His6 with a theoretical molecular weight of ∼ 27 kDa. Lane 1 and 2, L. lactis NZCK and NZregX3-His6 with 10 ng ml−1 nisin induction; lane 3 and 4, purified products of NZCK and NZregX3-His6. D, EMSA showed the specific interaction of RegX3 and predicted binding sites upstream pstS. Binding reactions consisted of: (lane 1) 20 fmol labeled probe alone, (lane 2) 20 fmol labeled probe and 3.5 μg RegX-His6, (lane 3) 20 fmol labeled probe, 4 pmol unlabeled probe, and 3.5 μg RegX-His6. Abbreviations: M, marker; B, binding complex; F, free DNA.
To verify this hypothesis, the interaction between RegX3 and the upstream sequence of pstS was further verified by EMSA. SDS-PAGE revealed a single band with an expected molecular mass of ∼ 27 kDa, suggesting that the recombinant RegX3-His6 has been successfully expressed and purified (Fig. 5C). The EMSA result indicated that the purified RegX3-His6 bounded to biotin-labeled ptsS and retarded the mobility (Fig. 5D). Assays were further performed using unlabeled probes as a specific competitor the biding site. This competitor abolished the specific shift and suggested the specific binding of RegX3His to the probe ptsS. Furthermore, the recombinant strain L. lactis NZpstS showed 31.4-fold enhanced resistance to 1.0% ox-bile compared with the control L. lactis NZCK (supplemental Fig. S4). Based on these results, we suggest that the senX3-regX3 system senses the bile stress signal in the gut and promotes the expression of pstS to maintain a high-level of Pi uptake in the sterile environment. The accumulated Pi cooperates with the enhanced glycolysis process to produce more ATP to confer bile resistance phenotype.
Concluding Remarks
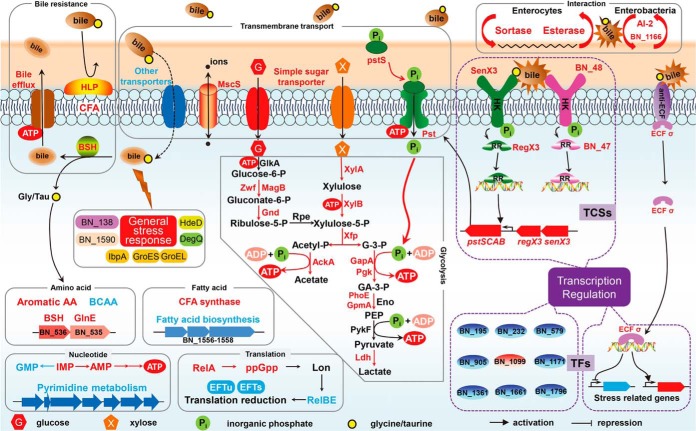
In this study, we explored the bile stress response of a potential probiotic strain B. longum BBMN68 using an RNA-Seq transcriptome profiling complemented with a 2-DE proteomic analysis. Nearly 300 genes were differentially expressed at either mRNA or protein levels in the presence of 0.75 g l−1 ox-bile. The reported changes in gene expression appeared to be associated with pathways contributing to cope with bile stress in BBMN68. First, HLP was located on the cell surface and functioned as a barrier against the adsorption of bile salts; the cell membrane composition was modified by increased CFAs and decreased transmembrane transporters, which prevent or at least reduce the influx of bile salts to the bacterial cell. When bile salts entered into cytoplasm, overexpressed BSH played a central role by catalyzing the deconjugation of glycine- or taurine-linked bile acids. The up-regulated bile efflux transporters would also pump the bile salts out from the cell. Furthermore, various genes related to general stress response, such as genes encoding DegQ, nitroreductase, and HdeD, were induced to protect cell against bile damages. On the other hand, central metabolism processes were modulated as an adaptation mechanism to bile stress. In order to effectively cope with bile stress for BBMN68 in the xylan-rich colon environment, xylose utilization and bifid shunt pathways were enhanced for producing more reducing equivalents and energies. Nucleotide metabolisms except ATP synthesis and global fatty acid biosynthesis were reduced in BBMN68 in response to bile salts. In addition, RelBE system was employed to slow down translation process under bile stress conditions. And bile functions as a gut signal and promotes interactions of BBMN68 with the host and other enterobacteria. For example, bile-induced abundance of sortases related with anchoring of pili in Gram-positive bacteria facilitated the adhesion and colonization of BBMN68 in the gut. BBMN68 was supposed to perceive the presence of competitors using the up-regulated LuxS/AI-2 system and then annihilate AI-2 produced by other bacteria, resulting in rapid propagation of BBMN68 and inhibition of pathogens' growth. Finally, 15 regulatory genes were differentially expressed in BBMN68 under bile stress conditions. Among those, an ECF sigma factor RpoE was revealed to involve in resistance to bile salts probably by stimulating the transcription of a specific group of genes related with diverse stress response. And the senX3-regX3 system was suggested to sense the bile salts signal and promote the transcription of pstS leading to accelerated Pi uptake for producing more ATP. Our present findings allowed us to propose a bile stress response mechanism model in B. longum BBMN68 and even in bifidobacteria (Fig. 6).
Fig. 6.
Proposed model for global response mechanisms of B. longum BBMN68 to bile stress. The induced and repressed gene expression changes are represented by red and blue fonts or backgrounds, respectively. Abbreviations: HLP, hemolysin-like protein; CFA, cyclopropane fatty acid; BSH, bile salt hydrolase; HK, histidine kinase; RR, response regulator; TCS, two-component system; ECF σ, extracytoplasmic function sigma factor; BCAA, branched-chain amino acid; GlkA, glucokinase; Zwf, glucose-6-phosphate-1-dehydrogenase; MagB, 6-phosphogluconolactonase; Gnd, 6-phosphogluconate dehydrogenase; Rpe, ribulose-phosphate-3-epimerase; XylA, xylose isomerase; XylB, xylulokinase; Xfp, xylulose-5-phosphate/fructose-6-phosphate phosphoketolase; AckA, acetate kinase; GapA, glyceraldehyde-3-phosphate dehydrogenase; Pgk, 3-phosphoglycerate kinase; PhoE, phosphoglycerate mutase; GpmA, 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase; PykF, pyruvate kinase; Ldh, lactate dehydrogenase; G-3-P, glyceraldehyde-3-phosphate; GA-3-P, glycerate-3-phosphate; PEP, phosphoenolpyruvate; EFTu, elongation factor Tu; EFTs, elongation factor Ts; TFs, transcription factors.
Supplementary Material
Acknowledgments
We thank Wei Zhang and Songling Liu for assistance in cell adhesion experiments, and we thank Xinghua Guo and Shangwu Chen for suggestions and Wanli Zhou and Beizhong Han for help in experiments performed at the facility.
Footnotes
Author contributions: H.A., Z.Z., J.C., F.R., and Y.H. designed research; H.A., G.W., and Z.Z. performed research; F.R., Y.L., and B.Z. contributed new reagents or analytic tools; H.A., F.P.D., J.Y., S.S., B.Z., and Y.H. analyzed data; H.A., F.P.D., and Y.H. wrote the paper.
* This work was supported by the National Natural Sciences Foundation of China [No. 31171740]; and the Chinese Universities Scientific Fund [No. 2012QJ149].
 This article contains supplemental Data S1, supplemental Tables S1–S4, and supplemental Fig. S1–S4.
This article contains supplemental Data S1, supplemental Tables S1–S4, and supplemental Fig. S1–S4.
1 The abbreviations used are:
- GIT
- gastrointestinal tract
- RNA-Seq
- RNA sequencing
- BSH
- bile salt hydrolase
- 2-DE
- two-dimensional electrophoresis
- NGS
- next-generation sequencing
- CFU
- colony forming unit
- RPKM
- reads per kilobase of exon per million mapped sequenced reads
- DEG
- differentially expressed gene
- B1H
- bacterial one-hybrid
- 5-FOA
- 5-fluoroorotic acid
- 3-AT
- 3-amino-triazole
- TF
- transcription factor
- MFS
- major facilitator superfamily
- HLP
- hemolysin-like protein
- BCAA
- branched-chain amino acid
- CFA
- cyclopropane fatty acid
- ABC
- ATP-binding cassette
- TA
- toxin-antitoxin
- AI-2
- autoinducer-2
- TCS
- two-component system
- ECF
- extracytoplasmic function
- EMSA
- electrophoretic mobility shift assay.
REFERENCES
- 1. Harmsen H. J., Wildeboer-Veloo A. C., Raangs G. C., Wagendorp A. A., Klijn N., Bindels J. G., Welling G. W. (2000) Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30, 61–67 [DOI] [PubMed] [Google Scholar]
- 2. Harmsen H. J., Raangs G. C., He T., Degener J. E., Welling G. W. (2002) Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68, 2982–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salminen S. J., Gueimonde M., Isolauri E. (2005) Probiotics that modify disease risk. J. Nutr. 135, 1294–1298 [DOI] [PubMed] [Google Scholar]
- 4. Isolauri E., Salminen S., Ouwehand A. C. (2004) Microbial-gut interactions in health and disease. Probiotics. Best Pract. Res. Clin. Gastroenterol. 18, 299–313 [DOI] [PubMed] [Google Scholar]
- 5. Masco L., Huys G., De Brandt E., Temmerman R., Swings J. (2005) Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int. J. Food Microbiol. 102, 221–230 [DOI] [PubMed] [Google Scholar]
- 6. Jayamanne V. S., Adams M. R. (2006) Determination of survival, identity, and stress resistance of probiotic bifidobacteria in bio-yoghurts. Lett. Appl. Microbiol. 42, 189–194 [DOI] [PubMed] [Google Scholar]
- 7. Hofmann A. F. (1999) The continuing importance of bile acids in liver and intestinal disease. Arch. Intern. Med. 159, 2647–2658 [DOI] [PubMed] [Google Scholar]
- 8. Bernstein H., Payne C. M., Bernstein C., Schneider J., Beard S. E., Crowley C. L. (1999) Activation of the promoters of genes associated with DNA damage, oxidative stress, ER stress, and protein malfolding by the bile salt, deoxycholate. Toxicol. Lett. 108, 37–46 [DOI] [PubMed] [Google Scholar]
- 9. Kumar R. S., Brannigan J. A., Prabhune A. A., Pundle A. V., Dodson G. G., Dodson E. J., Suresh C. G. (2006) Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J. Biol. Chem. 281, 32516–32525 [DOI] [PubMed] [Google Scholar]
- 10. Grill J. P., Perrin S., Schneider F. (2000) Bile salt toxicity to some bifidobacteria strains: role of conjugated bile salt hydrolase and pH. Can. J. Microbiol. 46, 878–884 [DOI] [PubMed] [Google Scholar]
- 11. Price C. E., Reid S. J., Driessen A. J., Abratt V. R. (2006) The Bifidobacterium longum NCIMB 702259T ctr gene codes for a novel cholate transporter. Appl. Environ. Microbiol. 72, 923–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gueimonde M., Garrigues C., van Sinderen D., de los Reyes-Gavilan C. G., Margolles A. (2009) Bile-inducible efflux transporter from Bifidobacterium longum NCC2705, conferring bile resistance. Appl. Environ. Microbiol. 75, 3153–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ruiz L., O'Connell-Motherway M., Zomer A., de los Reyes-Gavilan C. G., Margolles A., van Sinderen D. (2012) A bile-inducible membrane protein mediates bifidobacterial bile resistance. Microb. Biotechnol. 5, 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sanchez B., de los Reyes-Gavilan C. G., Margolles A. (2006) The F1F0-ATPase of Bifidobacterium animalis is involved in bile tolerance. Environ. Microbiol. 8, 1825–1833 [DOI] [PubMed] [Google Scholar]
- 15. Ruiz L., Sanchez B., Ruas-Madiedo P., de Los Reyes-Gavilan C. G., Margolles A. (2007) Cell envelope changes in Bifidobacterium animalis ssp. lactis as a response to bile. FEMS Microbiol. Lett. 274, 316–322 [DOI] [PubMed] [Google Scholar]
- 16. Ruiz L., Coute Y., Sanchez B., de los Reyes-Gavilan C. G., Sanchez J. C., Margolles A. (2009) The cell-envelope proteome of Bifidobacterium longum in an in vitro bile environment. Microbiology 155, 957–967 [DOI] [PubMed] [Google Scholar]
- 17. Sanchez B., Champomier-Verges M. C., Anglade P., Baraige F., de Los Reyes-Gavilan C. G., Margolles A., Zagorec M. (2005) Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187, 5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sanchez B., Champomier-Verges M. C., Stuer-Lauridsen B., Ruas-Madiedo P., Anglade P., Baraige F., de los Reyes-Gavilan C. G., Johansen E., Zagorec M., Margolles A. (2007) Adaptation and response of Bifidobacterium animalis subsp. lactis to bile: a proteomic and physiological approach. Appl. Environ. Microbiol. 73, 6757–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruiz L., Zomer A., O'Connell-Motherway M., van Sinderen D., Margolles A. (2012) Discovering novel bile protection systems in Bifidobacterium breve UCC2003 through functional genomics. Appl. Environ. Microbiol. 78, 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao L., Xu W., Ibrahim S. A., Jin J., Feng J., Jiang J., Meng J., Ren F. (2011) Effects of age and region on fecal microflora in elderly subjects living in Bama, Guangxi, China. Curr. Microbiol. 62, 64–70 [DOI] [PubMed] [Google Scholar]
- 21. Zhao L., Qiao X., Zhu J., Zhang X., Jiang J., Hao Y., Ren F. (2011) Correlations of fecal bacterial communities with age and living region for the elderly living in Bama, Guangxi, China. J. Microbiol. 49, 186–192 [DOI] [PubMed] [Google Scholar]
- 22. Hao Y., Huang D., Guo H., Xiao M., An H., Zhao L., Zuo F., Zhang B., Hu S., Song S., Chen S., Ren F. (2011) Complete genome sequence of Bifidobacterium longum subsp. longum BBMN68, a new strain from a healthy chinese centenarian. J. Bacteriol. 193, 787–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang H. Y., Liu S. L., Ibrahim S. A., Zhao L., Jiang J. L., Sun W. F., Ren F. Z. (2009) Oral administration of live Bifidobacterium substrains isolated from healthy centenarians enhanced immune function in BALB/c mice. Nutr. Res. 29, 281–289 [DOI] [PubMed] [Google Scholar]
- 24. Yang H., Liu A., Zhang M., Ibrahim S. A., Pang Z., Leng X., Ren F. (2009) Oral administration of live Bifidobacterium substrains isolated from centenarians enhances intestinal function in mice. Curr. Microbiol. 59, 439–445 [DOI] [PubMed] [Google Scholar]
- 25. Wang Z., Gerstein M., Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pinto A. C., Melo-Barbosa H. P., Miyoshi A., Silva A., Azevedo V. (2011) Application of RNA-seq to reveal the transcript profile in bacteria. Genet. Mol. Res. 10, 1707–1718 [DOI] [PubMed] [Google Scholar]
- 27. Mader U., Nicolas P., Richard H., Bessieres P., Aymerich S. (2011) Comprehensive identification and quantification of microbial transcriptomes by genome-wide unbiased methods. Curr. Opin. Biotechnol. 22, 32–41 [DOI] [PubMed] [Google Scholar]
- 28. Lee K., Lee H. G., Choi Y. J. (2008) Proteomic analysis of the effect of bile salts on the intestinal and probiotic bacterium Lactobacillus reuteri. J. Biotechnol. 137, 14–19 [DOI] [PubMed] [Google Scholar]
- 29. Wu R., Sun Z., Wu J., Meng H., Zhang H. (2010) Effect of bile salts stress on protein synthesis of Lactobacillus casei Zhang revealed by 2-dimensional gel electrophoresis. J. Dairy Sci. 93, 3858–3868 [DOI] [PubMed] [Google Scholar]
- 30. Bohle L. A., Faergestad E. M., Veiseth-Kent E., Steinmoen H., Nes I. F., Eijsink V. G., Mathiesen G. (2010) Identification of proteins related to the stress response in Enterococcus faecalis V583 caused by bovine bile. Proteome Sci. 8, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burns P., Sanchez B., Vinderola G., Ruas-Madiedo P., Ruiz L., Margolles A., Reinheimer J., de los Reyes-Gavilan C. G. (2010) Inside the adaptation process of Lactobacillus delbrueckii subsp. lactis to bile. Int. J. Food Microbiol. 142, 132–141 [DOI] [PubMed] [Google Scholar]
- 32. Hamon E., Horvatovich P., Izquierdo E., Bringel F., Marchioni E., Aoude-Werner D., Ennahar S. (2011) Comparative proteomic analysis of Lactobacillus plantarum for the identification of key proteins in bile tolerance. BMC Microbiol. 11, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alcantara C., Zuniga M. (2012) Proteomic and transcriptomic analysis of the response to bile stress of Lactobacillus casei BL23. Microbiology 158, 1206–1218 [DOI] [PubMed] [Google Scholar]
- 34. Cloonan N., Forrest A. R., Kolle G., Gardiner B. B., Faulkner G. J., Brown M. K., Taylor D. F., Steptoe A. L., Wani S., Bethel G., Robertson A. J., Perkins A. C., Bruce S. J., Lee C. C., Ranade S. S., Peckham H. E., Manning J. M., McKernan K. J., Grimmond S. M. (2008) Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods 5, 613–619 [DOI] [PubMed] [Google Scholar]
- 35. Zhao W., Liu W., Tian D., Tang B., Wang Y., Yu C., Li R., Ling Y., Wu J., Song S., Hu S. (2011) wapRNA: a web-based application for the processing of RNA sequences. Bioinformatics 27, 3076–3077 [DOI] [PubMed] [Google Scholar]
- 36. Zhai Z., Douillard F. P., An H., Wang G., Guo X., Luo Y., Hao Y. (2014) Proteomic characterization of the acid tolerance response in Lactobacillus delbrueckii subsp. bulgaricus CAUH1 and functional identification of a novel acid stress-related transcriptional regulator Ldb0677. Environ. Microbiol. 16, 1524–1537 [DOI] [PubMed] [Google Scholar]
- 37. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 38. Xiao M., Xu P., Zhao J., Wang Z., Zuo F., Zhang J., Ren F., Li P., Chen S., Ma H. (2011) Oxidative stress-related responses of Bifidobacterium longum subsp. longum BBMN68 at the proteomic level after exposure to oxygen. Microbiology 157, 1573–1588 [DOI] [PubMed] [Google Scholar]
- 39. Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G. M., Carnemolla B., Orecchia P., Zardi L., Righetti P. G. (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 40. Gorg A., Weiss W., Dunn M. J. (2004) Current two-dimensional electrophoresis technology for proteomics. Proteomics 4, 3665–3685 [DOI] [PubMed] [Google Scholar]
- 41. Meng X., Wolfe S. A. (2006) Identifying DNA sequences recognized by a transcription factor using a bacterial one-hybrid system. Nat. Protoc. 1, 30–45 [DOI] [PubMed] [Google Scholar]
- 42. Bailey T. L., Elkan C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 [PubMed] [Google Scholar]
- 43. Sosinsky A., Bonin C. P., Mann R. S., Honig B. (2003) Target Explorer: An automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res. 31, 3589–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qi Y., Tu J., Cui L., Guo X., Shi Z., Li S., Shi W., Shan Y., Ge Y., Shan J., Wang H., Lu Z. (2010) High-throughput sequencing of microRNAs in adenovirus type 3 infected human laryngeal epithelial cells. J. Biomed. Biotechnol. 2010:915980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koskenniemi K., Laakso K., Koponen J., Kankainen M., Greco D., Auvinen P., Savijoki K., Nyman T. A., Surakka A., Salusjarvi T., de Vos W. M., Tynkkynen S., Kalkkinen N., Varmanen P. (2011) Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol. Cell. Proteomics 10, M110.002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Qiao J., Huang S., Te R., Wang J., Chen L., Zhang W. (2013) Integrated proteomic and transcriptomic analysis reveals novel genes and regulatory mechanisms involved in salt stress responses in Synechocystis sp. PCC 6803. Appl. Microbiol. Biotechnol. 97, 8253–8264 [DOI] [PubMed] [Google Scholar]
- 47. Maier T., Guell M., Serrano L. (2009) Correlation of mRNA and protein in complex biological samples. FEBS Lett. 583, 3966–3973 [DOI] [PubMed] [Google Scholar]
- 48. Begley M., Gahan C. G., Hill C. (2005) The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651 [DOI] [PubMed] [Google Scholar]
- 49. Reizer J., Reizer A., Saier M. H., Jr. (1992) A new subfamily of bacterial ABC-type transport systems catalyzing export of drugs and carbohydrates. Protein Sci. 1, 1326–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Prosecka J., Orlov A. V., Fantin Y. S., Zinchenko V. V., Babykin M. M., Tichy M. (2009) A novel ATP-binding cassette transporter is responsible for resistance to viologen herbicides in the cyanobacterium Synechocystis sp. PCC 6803. FEBS J. 276, 4001–4011 [DOI] [PubMed] [Google Scholar]
- 51. Begley M., Hill C., Gahan C. G. (2006) Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 72, 1729–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Garrigues C., Stuer-Lauridsen B., Johansen E. (2005) Characterisation of Bifidobacterium animalis subsp. lactis BB-12 and other probiotic bacteria using genomics, transcriptomics and proteomics. Aust. J. Dairy Technol. 60, 84–92 [Google Scholar]
- 53. Sakiyama T., Araie H., Suzuki I., Shiraiwa Y. (2011) Functions of a hemolysin-like protein in the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 193, 565–571 [DOI] [PubMed] [Google Scholar]
- 54. Makarova K. S., Aravind L., Wolf Y. I., Tatusov R. L., Minton K. W., Koonin E. V., Daly M. J. (2001) Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65, 44–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Savijoki K., Suokko A., Palva A., Valmu L., Kalkkinen N., Varmanen P. (2005) Effect of heat-shock and bile salts on protein synthesis of Bifidobacterium longum revealed by [35S]methionine labelling and two-dimensional gel electrophoresis. FEMS Microbiol. Lett. 248, 207–215 [DOI] [PubMed] [Google Scholar]
- 56. Petersohn A., Brigulla M., Haas S., Hoheisel J. D., Volker U., Hecker M. (2001) Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183, 5617–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hartke A., Bouche S., Giard J. C., Benachour A., Boutibonnes P., Auffray Y. (1996) The lactic acid stress response of Lactococcus lactis subsp. lactis. Curr. Microbiol. 33, 194–199 [DOI] [PubMed] [Google Scholar]
- 58. Ventura M., Canchaya C., Zhang Z., Fitzgerald G. F., van Sinderen D. (2007) Molecular characterization of hsp20, encoding a small heat shock protein of Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73, 4695–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Narberhaus F. (1999) Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31, 1–8 [DOI] [PubMed] [Google Scholar]
- 60. Sawa J., Malet H., Krojer T., Canellas F., Ehrmann M., Clausen T. (2011) Molecular adaptation of the DegQ protease to exert protein quality control in the bacterial cell envelope. J. Biol. Chem. 286, 30680–30690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. de Oliveira I. M., Zanotto-Filho A., Moreira J. C., Bonatto D., Henriques J. A. (2010) The role of two putative nitroreductases, Frm2p and Hbn1p, in the oxidative stress response in Saccharomyces cerevisiae. Yeast 27, 89–102 [DOI] [PubMed] [Google Scholar]
- 62. Mermod M., Mourlane F., Waltersperger S., Oberholzer A. E., Baumann U., Solioz M. (2010) Structure and function of CinD (YtjD) of Lactococcus lactis, a copper-induced nitroreductase involved in defense against oxidative stress. J. Bacteriol. 192, 4172–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shatalin K., Shatalina E., Mironov A., Nudler E. (2011) H2S: a universal defense against antibiotics in bacteria. Science 334, 986–990 [DOI] [PubMed] [Google Scholar]
- 64. Lo R., Turner M. S., Barry D. G., Sreekumar R., Walsh T. P., Giffard P. M. (2009) Cystathionine gamma-lyase is a component of cystine-mediated oxidative defense in Lactobacillus reuteri BR11. J. Bacteriol. 191, 1827–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van de Guchte M., Serror P., Chervaux C., Smokvina T., Ehrlich S. D., Maguin E. (2002) Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82, 187–216 [PubMed] [Google Scholar]
- 66. Krin E., Danchin A., Soutourina O. (2010) Decrypting the H-NS-dependent regulatory cascade of acid stress resistance in Escherichia coli. BMC Microbiol. 10, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Peng C. A., Oliver M. J., Wood A. J. (2005) Is the Rehydrin TrDr3 from Tortula ruralis associated with tolerance to cold, salinity, and reduced pH? Physiological evaluation of the TrDr3-orthologue, HdeD from Escherichia coli in response to abiotic stress. Plant Biol. 7, 315–320 [DOI] [PubMed] [Google Scholar]
- 68. Abdelhaleem M. (2010) Helicases: an overview. Methods Mol. Biol. 587, 1–12 [DOI] [PubMed] [Google Scholar]
- 69. Kast P., Grisostomi C., Chen I. A., Li S., Krengel U., Xue Y., Hilvert D. (2000) A strategically positioned cation is crucial for efficient catalysis by chorismate mutase. J. Biol. Chem. 275, 36832–36838 [DOI] [PubMed] [Google Scholar]
- 70. Kingdon H. S., Shapiro B. M., Stadtman E. R. (1967) Regulation of glutamine synthetase. 8. ATP: glutamine synthetase adenylyltransferase, an enzyme that catalyzes alterations in the regulatory properties of glutamine synthetase. Proc. Natl. Acad. Sci. U. S. A. 58, 1703–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Carroll P., Pashley C. A., Parish T. (2008) Functional analysis of GlnE, an essential adenylyl transferase in Mycobacterium tuberculosis. J. Bacteriol. 190, 4894–4902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rehm N., Buchinger S., Strosser J., Dotzauer A., Walter B., Hans S., Bathe B., Schomburg D., Kramer R., Burkovski A. (2010) Impact of adenylyltransferase GlnE on nitrogen starvation response in Corynebacterium glutamicum. J. Biotechnol. 145, 244–252 [DOI] [PubMed] [Google Scholar]
- 73. Helmstaedt K., Krappmann S., Braus G. H. (2001) Allosteric regulation of catalytic activity: Escherichia coli aspartate transcarbamoylase versus yeast chorismate mutase. Microbiol. Mol. Biol. Rev. 65, 404–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shabala L., Ross T. (2008) Cyclopropane fatty acids improve Escherichia coli survival in acidified minimal media by reducing membrane permeability to H+ and enhanced ability to extrude H+. Res. Microbiol. 159, 458–461 [DOI] [PubMed] [Google Scholar]
- 75. Vitali B., Turroni S., Serina S., Sosio M., Vannini L., Candela M., Guerzoni M. E., Brigidi P. (2008) Molecular and phenotypic traits of in-vitro-selected mutants of Bifidobacterium resistant to rifaximin. Int. J. Antimicrob. Agents 31, 555–560 [DOI] [PubMed] [Google Scholar]
- 76. Montanari C., Sado Kamdem S. L., Serrazanetti D. I., Etoa F. X., Guerzoni M. E. (2010) Synthesis of cyclopropane fatty acids in Lactobacillus helveticus and Lactobacillus sanfranciscensis and their cellular fatty acids changes following short term acid and cold stresses. Food Microbiol. 27, 493–502 [DOI] [PubMed] [Google Scholar]
- 77. Gomez Zavaglia A., Disalvo E. A., De Antoni G. L. (2000) Fatty acid composition and freeze-thaw resistance in lactobacilli. J. Dairy Res. 67, 241–247 [DOI] [PubMed] [Google Scholar]
- 78. Guillot A., Obis D., Mistou M. Y. (2000) Fatty acid membrane composition and activation of glycine-betaine transport in Lactococcus lactis subjected to osmotic stress. Int. J. Food Microbiol. 55, 47–51 [DOI] [PubMed] [Google Scholar]
- 79. Beal C., Fonseca F., Corrieu G. (2001) Resistance to freezing and frozen storage of Streptococcus thermophilus is related to membrane fatty acid composition. J. Dairy Sci. 84, 2347–2356 [DOI] [PubMed] [Google Scholar]
- 80. Teixeira H., Goncalves M. G., Rozes N., Ramos A., San Romao M. V. (2002) Lactobacillic acid accumulation in the plasma membrane of Oenococcus oeni: a response to ethanol stress? Microb. Ecol. 43, 146–153 [DOI] [PubMed] [Google Scholar]
- 81. Whitehead K., Versalovic J., Roos S., Britton R. A. (2008) Genomic and genetic characterization of the bile stress response of probiotic Lactobacillus reuteri ATCC 55730. Appl. Environ. Microbiol. 74, 1812–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Solheim M., Aakra A., Vebo H., Snipen L., Nes I. F. (2007) Transcriptional responses of Enterococcus faecalis V583 to bovine bile and sodium dodecyl sulfate. Appl. Environ. Microbiol. 73, 5767–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kung C., Martinac B., Sukharev S. (2010) Mechanosensitive channels in microbes. Annu. Rev. Microbiol. 64, 313–329 [DOI] [PubMed] [Google Scholar]
- 84. Biggin P. C., Sansom M. S. (2003) Mechanosensitive channels: stress relief. Curr. Biol. 13, R183–185 [DOI] [PubMed] [Google Scholar]
- 85. Bron P. A., Molenaar D., de Vos W. M., Kleerebezem M. (2006) DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J. Appl. Microbiol. 100, 728–738 [DOI] [PubMed] [Google Scholar]
- 86. Wall T., Bath K., Britton R. A., Jonsson H., Versalovic J., Roos S. (2007) The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Appl. Environ. Microbiol. 73, 3924–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hernandez S. B., Cota I., Ducret A., Aussel L., Casadesus J. (2012) Adaptation and preadaptation of Salmonella enterica to Bile. PLoS Genet. 8, e1002459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Oh S. Y., Budzik J. M., Schneewind O. (2008) Sortases make pili from three ingredients. Proc. Natl. Acad. Sci. U. S. A. 105, 13703–13704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mandlik A., Swierczynski A., Das A., Ton-That H. (2008) Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Schell M. A., Karmirantzou M., Snel B., Vilanova D., Berger B., Pessi G., Zwahlen M. C., Desiere F., Bork P., Delley M., Pridmore R. D., Arigoni F. (2002) The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 99, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Miller M. B., Bassler B. L. (2001) Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199 [DOI] [PubMed] [Google Scholar]
- 92. Yuan J., Wang B., Sun Z., Bo X., Yuan X., He X., Zhao H., Du X., Wang F., Jiang Z., Zhang L., Jia L., Wang Y., Wei K., Wang J., Zhang X., Sun Y., Huang L., Zeng M. (2008) Analysis of host-inducing proteome changes in Bifidobacterium longum NCC2705 grown in vivo. J. Proteome Res. 7, 375–385 [DOI] [PubMed] [Google Scholar]
- 93. Rodionov D. A. (2007) Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 107, 3467–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Helmann J. D. (2002) The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46, 47–110 [DOI] [PubMed] [Google Scholar]
- 95. Raina S., Missiakas D., Georgopoulos C. (1995) The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 14, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xue X., Tomasch J., Sztajer H., Wagner-Dobler I. (2010) The delta subunit of RNA polymerase, RpoE, is a global modulator of Streptococcus mutans environmental adaptation. J. Bacteriol. 192, 5081–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. McDougald D., Gong L., Srinivasan S., Hild E., Thompson L., Takayama K., Rice S. A., Kjelleberg S. (2002) Defences against oxidative stress during starvation in bacteria. Antonie Van Leeuwenhoek 81, 3–13 [DOI] [PubMed] [Google Scholar]
- 98. Wegrzyn R. D., Deuerling E. (2005) Molecular guardians for newborn proteins: ribosome-associated chaperones and their role in protein folding. Cell. Mol. Life Sci. 62, 2727–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Srivatsan A., Wang J. D. (2008) Control of bacterial transcription, translation, and replication by (p)ppGpp. Curr. Opin. Microbiol. 11, 100–105 [DOI] [PubMed] [Google Scholar]
- 100. Overgaard M., Borch J., Gerdes K. (2009) RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. J. Mol. Biol. 394, 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Gerdes K., Christensen S. K., Lobner-Olesen A. (2005) Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 102. Himpens S., Locht C., Supply P. (2000) Molecular characterization of the mycobacterial SenX3-RegX3 two-component system: evidence for autoregulation. Microbiology 146 Pt 12, 3091–3098 [DOI] [PubMed] [Google Scholar]
- 103. Glover R. T., Kriakov J., Garforth S. J., Baughn A. D., Jacobs W. R., Jr. (2007) The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J. Bacteriol. 189, 5495–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Soualhine H., Brochu V., Menard F., Papadopoulou B., Weiss K., Bergeron M. G., Legare D., Drummelsmith J., Ouellette M. (2005) A proteomic analysis of penicillin resistance in Streptococcus pneumoniae reveals a novel role for PstS, a subunit of the phosphate ABC transporter. Mol. Microbiol. 58, 1430–1440 [DOI] [PubMed] [Google Scholar]
- 105. Critzer F. J., D'Souza D. H., Saxton A. M., Golden D. A. (2010) Increased transcription of the phosphate-specific transport system of Escherichia coli O157:H7 after exposure to sodium benzoate. J. Food Prot. 73, 819–824 [DOI] [PubMed] [Google Scholar]
- 106. Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L., Wang J. (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34, W293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.