Significance
Long-lived proteins in extracellular spaces (joints/tissues) or within specialized nondividing cells (eye-lens) are associated with age-related decline. However, aging also occurs in dividing stem cells. Although several hypotheses have been proposed to explain how stem cells age, none have addressed whether long-lived proteins contribute to aging, partially because of technical challenges in identifying such proteins. We developed a method to overcome these limitations in the model system Saccharomyces cerevisiae. We identified two classes of long-lived asymmetrically retained proteins (LARPs). Full-length LARPs remain intact throughout the mother cell lifespan and accumulate in abundance or become posttranslationally modified. Fragmented LARPs are original proteins that are partially degraded, yet retained by the mother cell during aging. We speculate that LARPs contribute to the aging process.
Keywords: ACD, asymmetric cell division, RITE, recombination-induced tag exchange, replicative aging
Abstract
Long-lived proteins have been implicated in age-associated decline in metazoa, but they have only been identified in extracellular matrices or postmitotic cells. However, the aging process also occurs in dividing cells undergoing repeated asymmetric divisions. It was not clear whether long-lived proteins exist in asymmetrically dividing cells or whether they are involved in aging. Here we identify long-lived proteins in dividing cells during aging using the budding yeast, Saccharomyces cerevisiae. Yeast mother cells undergo a limited number of asymmetric divisions that define replicative lifespan. We used stable-isotope pulse-chase and total proteome mass-spectrometry to identify proteins that were both long-lived and retained in aging mother cells after ∼18 cells divisions. We identified ∼135 proteins that we designate as long-lived asymmetrically retained proteins (LARPS). Surprisingly, the majority of LARPs appeared to be stable fragments of their original full-length protein. However, 15% of LARPs were full-length proteins and we confirmed several candidates to be long-lived and retained in mother cells by time-lapse microscopy. Some LARPs localized to the plasma membrane and remained robustly in the mother cell upon cell division. Other full-length LARPs were assembled into large cytoplasmic structures that had a strong bias to remain in mother cells. We identified age-associated changes to LARPs that include an increase in their levels during aging because of their continued synthesis, which is not balanced by turnover. Additionally, several LARPs were posttranslationally modified during aging. We suggest that LARPs contribute to age-associated phenotypes and likely exist in other organisms.
Long-lived proteins have a well-documented impact on age-associated decline. In mammals, extracellular proteins, such as elastin and collagen, or those within specialized nondividing cells, such as the lens crystalline proteins are turned over slowly or not at all (1–5). Consequently, these proteins are susceptible to a lifetime of damage or other changes, which contribute to reduced elasticity of tissues and vision problems. More recently, the long-lived nuclear pore proteins within metabolically active, postmitotic neurons have been implicated in nuclear “leakiness” with increasing age (6, 7). These examples indicate that long-lived proteins can contribute to the aging process, but this evidence has been limited to situations that occur in the absence of cellular division.
Many molecular studies of the aging process have been limited to nondividing cell types; however, dividing cells also age. This finding was first demonstrated in the budding yeast, Saccharomyces cerevisiae, where the number of daughter cells produced by a single mother cell is finite, typically 25–30 cell divisions (8). Since that time, limited replicative potential of cells has been observed across species; and similar to budding yeast, repeated asymmetric cell divisions have been reported to lead to dysfunction of the progenitor cell in bacteria and metazoan stem cells (reviewed in refs. 9–11).
Asymmetry is a fundamental property of cell divisions across species. It creates differentiated cells, maintains germ lineages and stem cells, generates phenotypic diversity, and is fundamental to species that propagate by polarized cell growth. Even in cells that morphologically appear to divide symmetrically, there is asymmetry in molecular constituents, for example proteins, RNA, and lipids (9, 12, 13). In budding yeast, the limited life span of mother cells has provided fundamental insights about how repeated asymmetric cell divisions may lead to cellular aging. Multiple studies have led to the idea that a “senescence factor” accumulates in a mother cell through her successive cell divisions, whereas the daughter cell is rejuvenated for a full life span (14). Candidate senescence factors include extrachromosomal ribosomal DNA circles, damaged proteins or proteins that are not renewed, dysfunctional mitochondria, and increases in vacuolar pH (reviewed in ref. 15). Each of these entities or events is reported to accumulate or change over time, preferentially in the mother cell. Although there is support for each playing a role in the determination of life span, how they become senescence factors and contribute to aging phenotypes, or whether a common mechanism or process underlies all these hypotheses, remains unclear.
The apparent links between long-lived proteins and aging in postmitotic cells or extracellular compartments (5, 16) have led us to ask whether long-lived proteins might contribute to aging in a cell that goes through repeated asymmetric divisions. It is technically challenging to explore this idea in metazoan stem cells. Therefore, we have taken a first step to explore this idea in S. cerevisiae by applying recent technology we developed (17) to identify long-lived proteins that reside and persist in budding yeast mother cells as they go through repeated asymmetric cell divisions. Our approach to identify long-lived proteins in this asymmetric dividing cell type, and our findings, are presented herein.
Results
Approach to Identify Long-Lived Asymmetrically Retained Proteins.
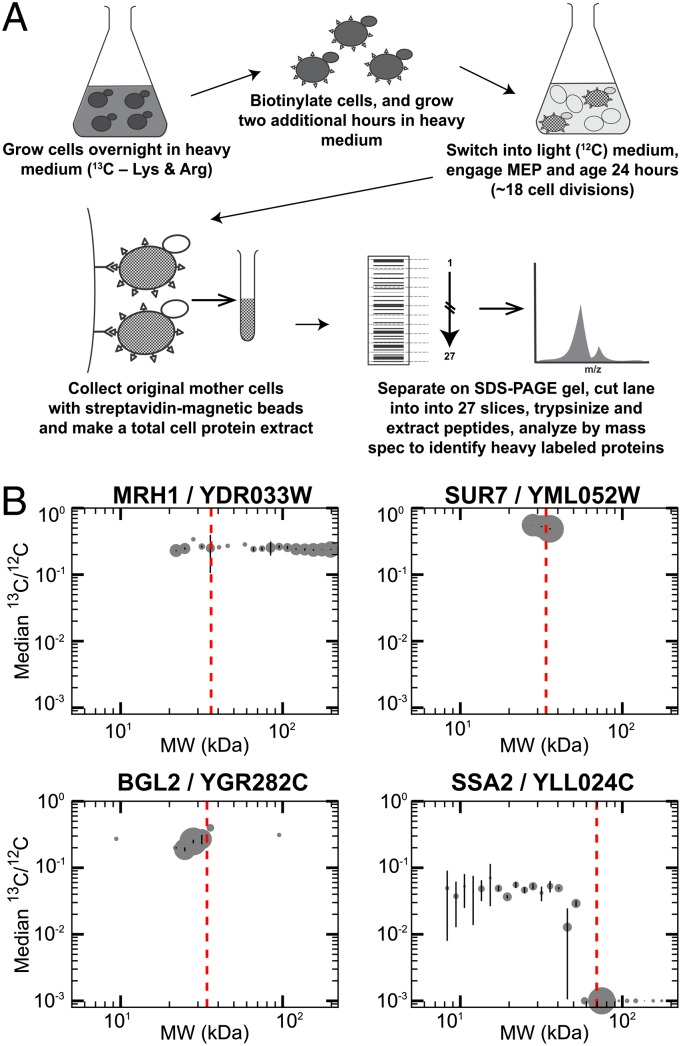
To identify long-lived proteins that are retained in yeast mother cells, we combined a method developed in our laboratory [the Mother Enrichment Program: MEP (17)] with a pulse-chase protocol using total proteome analysis (outlined in Fig. 1A). Cells were initially grown in medium with stable heavy-isotope amino acids to label proteins. The cells were then switched into media containing light-isotope amino acids and cultured for an additional ∼18 cell divisions with the MEP engaged. Finally, we determined whether any of the original heavy-labeled proteins were still present in the aged mother cells.
Fig. 1.
Heavy-isotope pulse-chase to identify long-lived proteins present in aged cells. (A) Schematic representation of experimental design to identify long-lived proteins in aged mother cells (details in Materials and Methods). (B) Representative plots of the 13C/12C ratio for peptides in given gel slices that correspond to specific proteins. The x axis is approximate MW of peptide (estimated from gel slice); y axis is median ratio 13C/12C for all peptides mapped to that protein in a gel slice. The size of dots reflects the relative number of peptide observations in that slice. Error bars are SD of the medians calculated from 1,000 bootstrapped samples. The expected MW of the full-length unmodified protein is plotted as a vertical dashed red line. Mrh1 is an integral membrane protein that appears across many gel slices, each with a similar ratio. Sur7, an integral membrane eisosome component, appears only in gel slices corresponding to its expected size with a high abundance of heavy label. Bgl2, a cell wall component, shows enrichment for heavy label. The full-length version of Ssa2, a small heat-shock protein, is primarily newly synthesized (very low 13C/12C), whereas the original 13C label is predominantly in low MW gel slices that correspond to fragmented Ssa2 protein.
We did this by purifying these aged mother cells, extracting total cellular proteins, and separating them by SDS/PAGE. The gel was cut into slices spanning apparent molecular weights of ∼200–8 kDa, and each slice was treated with trypsin and the eluted peptides were subjected to LC-MS/MS analysis.
Peptides from ∼3,200 total proteins were identified with high confidence (described in Materials and Methods). This number represents over 50% of known ORFs and afforded comparable coverage to the number of total yeast proteins identified previously by LC-MS (18). Of all of the high-confidence peptides identified, 1,581 had both heavy- and light-labeled forms (13C- and 12C-labeled, respectively) in any given gel slice; this corresponded to 465 proteins (Dataset S1). We limited further analysis to proteins in which peptide pairs with both heavy- and light-labeled events in a gel slice had an 13C/12C ratio ≥ 0.1. (This cut-off was chosen to reduce the frequency of inaccurate assignments and provide a reasonable number of candidates for further analysis described below.) For these 136 proteins, we considered all high-confidence peptide sequences and mapped their position within the SDS/PAGE gel slices. This assignment allowed us to assess the apparent molecular weight (MW) of proteins that were heavy-labeled. The median 13C/12C ratio of the peptides in each gel slice was then calculated and plotted for each protein (Fig. 1B and Datasets S1 and S2).
Two Classes of Long-Lived Asymmetrically Retained Proteins: Full-Length and Fragmented.
Analysis of these data divided the long-lived proteins into two categories: those in which the high 13C/12C ratios were present in peptides identified from full-length versions of the protein, and those in which the high 13C/12C ratio peptides migrated with apparent MW that was smaller than the expected full-length protein. For proteins in the first category, we hypothesized that the heavy-labeled peptides represent proteins synthesized very early in the lifespan of the mother cell, and that a significant fraction of each protein was still present in a full-length form after ∼18 cell divisions. Twenty-one proteins were in this category (Table 1) and were enriched in several classes. These classes included: plasma membrane proteins (Mrh1, Pma1, Snq2, and Sur7), proteins secreted into the cell wall (Bgl2, Exg1, Pho5, Pho11), and proteins involved in sulfur metabolism (Met3, Met5, Met6, Met10, Sam2, Thr1). The cell wall and proteins comprising it remain with the mother cell through her lifespan (19, 20). Thus, the presence of Bgl2, Exg1, Pho5, and Pho11 in our dataset provided support that the isotopic pulse-labeling/MS method could indeed identify proteins retained by mother cells.
Table 1.
List of prospective full-length LARPs
| Confirmed as LARP | Unable to confirm as LARP | RITE tag not successfully created |
| Mrh1 | Gcv3 | Bgl2 |
| Pma1 | Lap4 | Exg1 |
| Sur7 | Met6 | Met3 |
| Thr1 | Sam2 | Met5 |
| Hsp26 | Ynl134C | Met10 |
| Met17 | ||
| Pho5 | ||
| Pho11 | ||
| Por1 | ||
| Ser3 | ||
| Snq2 |
RITE-tagged versions of the 21 putative full-length LARPs were attempted and examined as described in the text. The proteins where then placed into three categories. (i) Confirmed as LARP: Original proteins appeared both long-lived and asymmetrically retained in the original mother cell. (ii) Unable to confirm as LARP: proteins were either not obviously long-lived or asymmetrically retained. (iii) RITE tag not successfully created: RITE tagging either caused mislocalization of the protein, was not easily visualized, or was not attempted.
The “full-length” category could be subdivided further by examining where each of the high 13C/12C ratio peptides for a particular protein migrated in the gel. Some proteins with high 13C/12C ratios had isoforms that migrated exceptionally slowly through the SDS/PAGE gel (e.g., Pho5, Pho11, Pma1, Mrh1) (Fig. 1B and Dataset S2), suggesting that they were posttranslationally modified or in an SDS-resistant aggregated form. Consistent with this idea, Pho11 and Pho5 are glycoproteins and known to run aberrantly on SDS/PAGE (21), and Pma1 and Mrh1 are integral membrane proteins and are modified or prone to aggregation (22).
Fragmented Long-Lived Asymmetrically Retained Proteins.
The majority of proteins with peptide pairs ratios ≥0.1 13C/12C contrasted with those described above. In this larger group the predicted full-length isoforms had 13C/12C ratio peptides below the 0.1 threshold. In fact, the majority of the peptides corresponding to these full-length isoforms were light-labeled (Fig. 1B and Dataset S2). Instead, the peptide pairs with ≥0.1 13C/12C ratios mapped to apparent MWs that were much smaller than the predicted MW of their respective protein. We speculate that these proteins were synthesized early in the life of the mother cell but were partially degraded into stable truncated isoforms during the next ∼18 cell divisions. These smaller fragments persist in the mother cell. The full-length version of each protein was also present; however, it was represented by few or no heavy-labeled peptides, suggesting that the full-length protein was the result of more recent synthesis that occurred as the mother cell went through successive cell divisions. Many of the proteins in this class were components of protein translation and folding (∼60%) or glycolytic enzymes (∼15%). The reason these truncated proteins were present in the mother cell after ∼18 cell divisions requires further investigation, but their detection may reflect that the full-length proteins are quite abundant (23).
Examination of Prospective Full-Length Long-Lived Asymmetrically Retained Proteins by Recombination-Induced Tag-Exchange Tagging.
To independently assess whether the full-length proteins identified above were indeed long-lived and retained in mother cells, fluorescence microscopy was combined with a protein-tagging method: the recombination-induced tag-exchange (RITE) system (24). Tagging a protein of interest with the RITE system creates a fusion protein, which initially expresses protein-GFP, then through an estradiol-inducible recombination event, expresses protein-RFP. The original (“old”) protein is labeled green, and after the switch, all subsequent (“new”) protein synthesis are labeled red. (Fig. 2A)
Fig. 2.
Full-length LARPs associated with the plasma membrane are retained in the mother cell throughout successive cell divisions. (A) Schematic representation of the RITE-tag system. The RITE-tag cassette places GFP in frame with a gene’s ORF of interest. Upon exposure to estradiol, a cre-mediated recombination event removes the GFP tag and fuses the ORF to RFP. (B) Each series of panels shows time-lapse images of microcolonies formed from single mother cells of PMA1-RITE, MRH1-RITE, and SUR7-RITE. Cre activity was induced before the first image; any newly synthesized protein was labeled with RFP and the preexisting protein was labeled with GFP. The approximate number of divisions the mother cell went through are indicated in the panels. Fig. S1 contains the unmerged channels of these images. Complete time-lapse series from which these images were taken are in Movies S1–S3. (Magnification: Pma1, 150×; Mrh1 and Sur7, 200×.)
We successfully RITE-labeled 10 of the 21 candidate proteins (Table 1) (many were mislocalized). As will be presented below, five of the full-length candidates were verified by RITE-tagging to be long-lived and asymmetrically retained in mother cells. We refer to these proteins as “long-lived asymmetrically retained proteins” (LARPs).
Plasma Membrane LARPs.
The cells containing RITE-tagged proteins were monitored by time-lapse fluorescence microscopy beginning ∼2 h after their respective RITE-tags were switched from green to red fusion proteins. The analysis of the integral plasma membrane proteins Mrh1, Pma1, and Sur7 revealed that these proteins were exceptionally long-lived and asymmetrically retained in the mother cells. For these proteins the perimeter of every original mother cell remained green for the entire period of observation (seven to eight cell divisions), with no detectable amount of the plasma membrane “old” green protein observed in daughter cells. These observations are consistent with all three of these integral plasma membrane proteins being long-lived and asymmetrically retained in the mother cell; little or none was distributed to the daughter cells (Fig. 2B and Movies S1–S3).
LARPs in Large Cytoplasmic Structures.
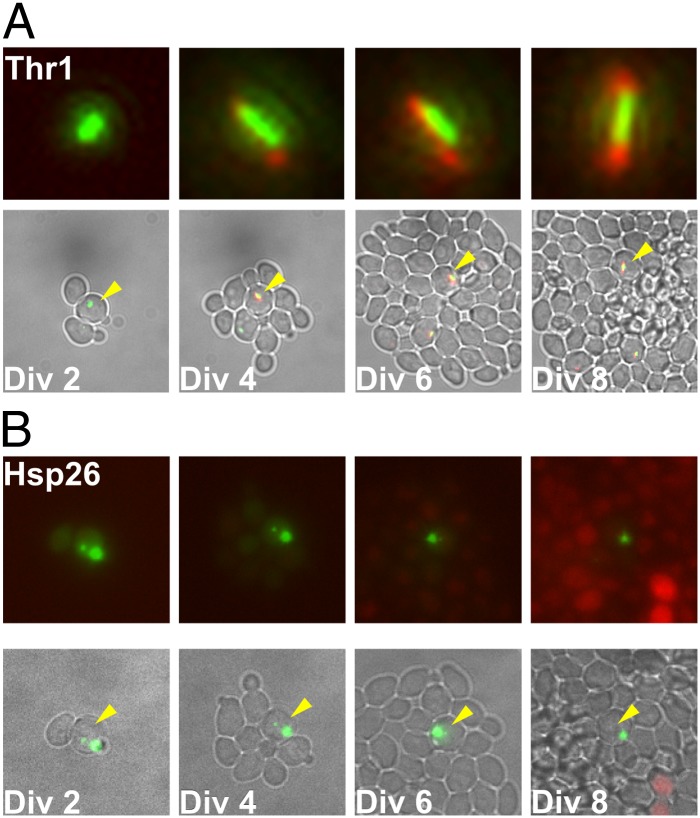
Thr1 and Hsp26 are cytoplasmic proteins that were identified as LARPs by the isotopic/MS method. These proteins also appeared to be long-lived and preferentially retained in mother cells by RITE-tagging, although differently than the plasma membrane proteins. In contrast, these cytoplasmic proteins were diffusely cytoplasmic or undetectable in most cells, but in a subpopulation of mother cells they formed distinct structures. Thr1 formed a cytoplasmic, short rod-like structure, consistent with previous reports of this metabolic enzyme forming filaments in vivo (25). However, the RITE-tagged Thr1 revealed that the original GFP-labeled protein in the filament was long-lived and had a very strong bias to remain in the mother cell. We monitored 24 cells over a total of ∼135 divisions and in only three instances (2%) did the filament transfer from the mother to the daughter. Although filaments were typically retained in mother cells, we observed small fragments of the Thr1-GFP filament break off and segregate into daughter cells during 12% of cell divisions (16 of 135). These small fragments of the original filament remained in daughter cells and appeared to serve as seeds for further filament formation once these daughters became mother cells (Fig. 3A and Movie S4). Similarly, in old mother cells the old Thr1-GFP filament appears to be a site for further deposition of newly synthesized Thr1-mRFP; the green filament was surrounded by red (Fig. 3A and Movie S4). These observations suggest that newly synthesized Thr1 was added to the old structure with increasing age.
Fig. 3.
Full-length LARPs that form cytoplasmic foci remain in the mother cell throughout successive cell divisions. (A) THR1-RITE cells were induced and imaged as described in Fig. 2. The approximate number of cell divisions the original mother cell (denoted with an arrowhead) has undergone is indicated. After the first cell division, the original Thr1-GFP containing focus remained in the mother cell. Although the original protein was present in foci throughout the course of the experiment, newly synthesized Thr1-RFP appeared to be added adjacent to existing foci (enlarged in Upper). (B) HSP26-RITE cells were induced and imaged as described in A. The Hsp26-GFP focus stayed within the original mother cell. As the colony became nutrient limited, several cells in the microcolony induced HSP26-RFP. These images were taken from Movies S4 and S5. (Magnification: A, Top, 2,000×; Bottom, 400×; B, 450×.)
Hsp26 is a small heat-shock chaperone that is involved in protein homeostasis and associates with a variety of proteins that are prone to aggregation (26, 27). One or two distinct green Hsp26 RITE-tagged foci of variable size were observed in a number of mother cells, and like Thr1, had a strong bias to remain in the mother (Fig. 3B and Movie S5). Sixty-two cells were monitored over ∼275 cell divisions, and in only 30 divisions was the Hsp26-GFP focus transferred to the daughter cell (11%). In nearly all cases, the initial Hsp26-GFP focus remained green throughout the observation of the aging mother cell. However, in one mother the focus became a composite of green and red after several cell divisions, consistent with newly synthesized Hsp26-RFP being added to the original Hsp26-GFP focus (Movie S6). Taken together, these results suggest that Hsp26 foci and Thr1 filaments are long-lived proteins with a propensity for asymmetric retention in the mother cell.
Plasma Membrane LARPs Remain, Whereas Partner Proteins Exchange.
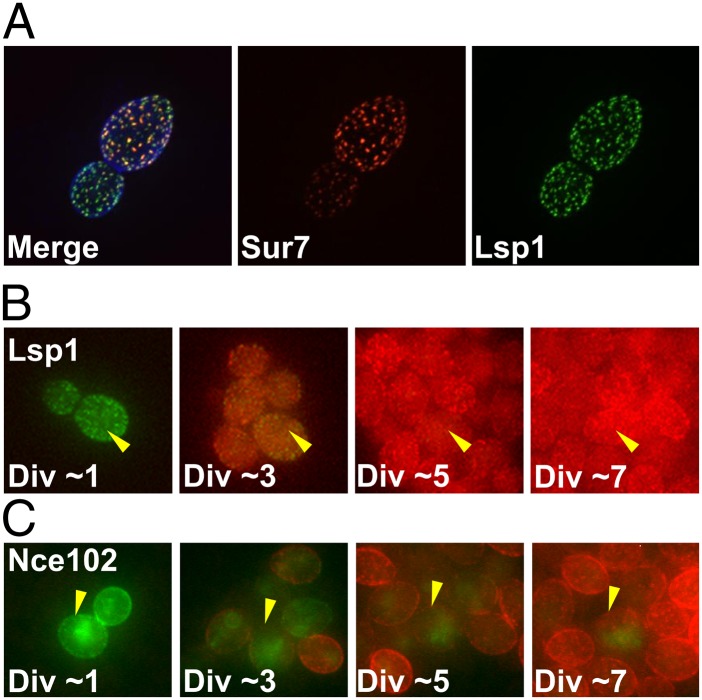
One of the plasma membrane proteins examined, Sur7, is part of a multiprotein complex: the eisosome (reviewed in refs. 28 and 29). The eisosome, whose role remains enigmatic, is defined by dozens of cortical structures distributed around the plasma membrane. In addition to the integral membrane protein Sur7, other important components of eisosomes include Lsp1. Although it is not an integral membrane protein, Lsp1 is deposited early in the bud and can assemble into a membrane scaffold (30, 31). Another important eisosome component is Nce102 which, like Sur7, is an integral membrane protein with four transmembrane domains (32, 33). We tested whether these components of the eisosome were as stable as Sur7 by examining their localization with the RITE tag system. In contrast to Sur7, Lsp1, and Nce102 were replaced with newly synthesized protein. For both of these proteins the mother’s eisosomes turned from GFP to RFP within a few cell divisions (Fig. 4 and Movies S7 and S8). This distinction between Sur7 and Lsp1 or Nce102 was further supported by the MS data; neither Lsp1 nor Nce102 had full-length proteins with a high 13C/12C ratio (Datasets S1 and S3). These results suggest that although Lsp1 and Nce102 are required for normal eisosome formation and structure, Sur7 may function as an anchor to ensure eisosome position within the plasma membrane as the cell repeatedly divides.
Fig. 4.
Not all components of the eisosome are LARPs. (A) Images of Sur7-mCherry and Lsp1-GFP, two components of the eisosome, which colocalized at puncta in young mother cells. Lsp1 appeared in daughters before Sur7, consistent with previous reports (80). Calcofluor staining in the merged image outlines cell walls in blue. (B) LSP1-RITE cells were imaged over several cell divisions, as in Fig. 2. The original Lsp1-GFP protein was distributed to both mother and daughter cells, but was replaced with newly synthesized protein (RFP-labeled) within a few cell divisions. (C) NCE102-RITE cells imaged as in B. The original Nce102-GFP at the plasma membrane was replaced with newly synthesized protein within a few cell divisions. A faint cytoplasmic haze of GFP remained in the mother cell, but it was not located at the plasma membrane. Fig. S2 contains the unmerged channels of these images. These images were taken from Movies S7 and S8. (Magnification: A, 1,000×; B and C, 600×.)
LARPs Change in the Mother Cell with Successive Cell Divisions.
A consequence of little or no protein turnover of the LARPs is that they may accumulate in the mother cell if they continue to be synthesized. In fact, this appeared to be so in some cells for the cytoplasmic LARPs (Hsp26 and Thr1) that served as seeds (see above and Fig. 3). Examination of the plasma membrane LARPs, which are expressed in all mother cells, suggested that they might also accumulate: during the time-lapse analysis of the plasma membrane-associated RITE-tagged proteins (e.g., Mrh1) mother cells appeared yellow because of expression of Mrh1-RFP, whereas Mrh1-GFP was still present in the mother cell (Movie S1).
We further characterized two plasma membrane LARPs to determine if their accumulation continued through the life of the mother cell. This possibility was explored by examining the level of fluorescence at the plasma membrane of a relatively fast-folding GFP (without a switchable RITE-tag) fused to Mrh1 or Sur7 in cells from birth through ∼20 divisions. For each of these GFP fusion proteins, there was little to no fluorescence in a small bud, but by the time the daughter had separated from the mother cell, a large increase in GFP fluorescence was evident (Fig. 5A) (e.g., Mrh1). For Sur7-GFP the total fluorescence increased a modest ∼50% over the next 15–20 divisions (Fig. 5B). Mrh1-GFP increased over the next ∼15 cell divisions as well, but it plateaued with ∼2.5-fold greater fluorescence than in a newborn mother cell (Fig. 5B). Thus, it appears that Sur7 and Mrh1 are initially deposited into the plasma membrane around the time a daughter cell matures into a mother cell, and continue to be deposited and accumulate through many cell divisions.
Fig. 5.
LARP proteins are altered with increasing age. (A) A representative image of Mrh1-GFP cells of various ages (newborn to >20 cell-divisions old) mixed together. Panels left to right show bright-field images of cells, Mrh1-GFP levels, and number of budscars (age), respectively. (Magnification: 350×.) (B) Quantification of Mrh1-GFP and Sur7-GFP levels in aging cells. Using images as shown in A, the average Mrh1-GFP and Sur7-GFP fluorescence intensity at the plasma membrane is plotted as a function of age (n = 100 cells for Mrh1-GFP, n = 39 for Sur7-GFP). (C) Western blot analysis showed altered migration of LARP proteins during aging. Samples from young and aged cells (Mrh1-GFP and Pma1-GFP) were analyzed with anti-GFP antibody by Western blot. For both Mrh1 and Pma1, protein had slower mobility relative to the expected migration (arrowhead) with increasing age. PGK1, which was not identified as a LARP, did not show this age-associated shift.
A hallmark of some long-lived proteins that are extracellular or reside in nondividing cells is that they become modified over time (5). Therefore, we examined several of the LARPs by Western analysis, comparing their migration by SDS/PAGE as a function of mother cell replicative age. For the two plasma membrane proteins examined (Pma1 and Mrh1), increasing amounts of the protein showed reduced mobility in the gel as the cell aged (Fig. 5C). The nature of this altered mobility requires further characterization, but suggests that there is an age-dependent change in both of these LARPs. Taken together, these results indicate that plasma membrane LARPs can be both modified and increase in levels with successive cell divisions.
Discussion
In this study we identified long-lived proteins that are retained within yeast mother cells as they undergo repeated asymmetric cell divisions. We designate these proteins as LARPs. Although very long-lived or mother-retained proteins have been previously identified in budding yeast (34–37), the combination of these properties in the LARPs represents a class of proteins that has not been specifically investigated. We suggest that LARPs play a role in age-associated phenotypes as a yeast mother cell transits through repeated asymmetric cell divisions.
Strengths and Limitations of the Method Used.
The ability to identify LARPs by LC-MS/MS combined several technologies, but two were uniquely important to this study. First, the MEP (17) permitted us to isolate a sufficient quantity of aged yeast mother cells for the analysis. The ability to isolate a sufficiently large population of cells that have undergone ∼18 asymmetric cell divisions is not otherwise possible. Second, the fractionation of the old cell proteins by SDS/PAGE before LC-MS/MS allowed us to discriminate between full-length and fragmented LARPs. This process allowed us to not only readily identify the best full-length candidates for testing via the RITE-tagging assay, but also to identify the fragmented class of LARPs despite the simultaneous presence of the full-length protein within the cell. The identification of this fragmented class leaves many questions to explore in the future.
We used stringent criteria in our classification of proteins as LARPs. This started with requirements of high confidence in MS sequencing calls, insistence of a 13C/12C isotope ratio > 0.1, and finally with independent testing by RITE-tagging for the full-length LARPs. Consequently, we suspect that the proteins reported here are representatives of a larger class of proteins. For example, we would not have included proteins that were very stable and retained in the mother cell, but had 10-fold more protein synthesis during the light isotope-labeling period. Additionally, our method would not be sensitive to detecting long-lived proteins that had a modest bias to remain in the mother cell. For example, ∼60% of stable nuclear pore proteins are retained in the mother each cell division (37–39). Assuming an extreme case in which these proteins are infinitely long-lived, then after 18 cell divisions the expected 13C/12C ratio would be ∼10−4, well below our cut-off threshold. A recent report using LC-MS/MS to identify long-lived proteins in nondividing mammalian tissues used a 13C/12C ratio cut-off of ∼0.05 (40). Nevertheless, our more stringent cut-off and analysis provided a view of a new spectrum of proteins. With increased experimental sensitivity and accuracy in the future, our approach may allow higher confidence in detecting more subtle mother-biased retention.
The proteins classified as LARPs had a very robust GFP signal over the course of more than eight cell divisions when analyzed by RITE-tagging. This screening tool required that the proteins be abundant and exceptionally stable (as a GFP fusion protein). Thus, less-abundant or shorter-lived proteins would be overlooked. We therefore included candidates that could not be verified by RITE-tagging in Table 1, with the expectation that more sensitive assays in the future will confirm these proteins as true LARPs. We do note, however, that several LARP candidates by MS criteria may be false-positives because of the constraints of the stable isotope-labeling protocol. For example, six proteins involved in methionine biosynthesis passed the initial MS criteria, but were not considered LARPs when analyzed by RITE-tagging. Further analysis revealed that some of these proteins were highly induced under the growth conditions of heavy-isotope labeling, but substantially repressed in the media used for the light-label chase (Fig. S3). This finding suggests that the 13C/12C ratio of these proteins was artificially high: with little light protein synthesized, the denominator of the 13C/12C ratio was quite small, even though the heavy, old protein was being distributed to the daughter cell at each division.
Our approach was also capable of identifying LARPs with variable expression in the population of cells. The plasma membrane LARPs gave a very robust MS and RITE-tagging signal, in large part because they were present in every cell and highly retained in the mother cell. In contrast, the two cytoplasmic LARPs, Hsp26 and Thr1, were present in a subset of original mother cells and had some probability of being passed onto daughter cells, albeit at a low frequency.
What Makes LARPS Long-Lived?
There are two traits that define a LARP: being long-lived and retained in the mother cell for many cell divisions. Although understanding the molecular basis of these traits will require further analysis, we can speculate about how they might be long-lived. The full-length LARPs appear to be immune from the normal pathways of protein turnover under the conditions examined. For those associated with the plasma membrane (e.g., Pma1, Mrh1, Sur7), they may lack or structurally occlude motifs necessary for the endocytic turnover of plasma membrane integral membrane proteins (reviewed in ref. 41). Similarly, the cytoplasmic LARPs may elude the ubiquitin-proteasome system, cytoplasmic proteases, and autophagy (42). It is worth noting that one of the LARPs is Hsp26, which contains the conserved α-crystallin domain (43). In eye tissues, this domain is associated with proteins that are among the most stable proteins in the human body (3).
Localization of the LARP (e.g., the plasma membrane or a filamentous structure) appears to be an important contributor to a LARP’s stability. For example, in RITE-tagged movies of Mrh1, a fraction of the protein made early in the life of the mother cell can be observed within the vacuole, whereas those located at the plasma membrane are incredibly stable (Fig. 2B and Movie S1). This finding is consistent with LARPs being sensitive to quality-control mechanisms along their transit to the plasma membrane (41), but once properly inserted into the plasma membrane they appear to be largely resistant to degradation.
The fragmented class of LARPs represents an interesting group of stable peptides in the mother cell. Proteins with long half-lives have been previously observed to progress to truncated forms with increasing age in mammals (4). In these earlier studies, the truncated proteins were found in nondividing cells, whereas the proteins described in our study were found in dividing cells. Characterization of some of the truncated proteins in mammals suggests that both enzymatic and nonenzymatic cleavage occurs. Whether similar modes of fragmented protein production can be invoked for LARPs remains to be determined.
How Are LARPs Retained?
The full-length LARPs that we were able to verify visually can be divided into those that reside in the plasma membrane (e.g., Pma1, Mrh1, Sur7) and those that are part of large cytoplasmic structures (e.g., Hsp26 and Thr1). It is likely that the mechanism of retention in mother cells is different between these two groups.
Hsp26 and Thr1 have a strong tendency to stay in the mother cell, but occasionally, part or all of the LARP structure moved into the daughter cell (Movies S4 and S5). This behavior is similar to proteins in various inclusion bodies and aggregates that form as the result of protein mis-folding in yeast cells (44–47). Each of these entities has a strong bias to remain in the mother cell, and several mechanisms have been proposed to explain their asymmetry (15, 48). However, how they are retained in the mother remains somewhat contentious and it appears that there are protein-specific distinctions between the various structures and their fate in the cell (45, 46, 49, 50). Hsp104 is associated with several of these structures and thought to facilitate disaggregation of the proteins. Interestingly, in this context, Hsp26 is hypothesized to arrive to aggregates before Hsp104 to prepare some aggregated proteins for “reactivation” by Hsp104 (26, 27). Consistent with this hypothesis, Hsp26 foci were invariably associated with Hsp104 foci; however, Hsp104 foci were not always associated with Hsp26 (Fig. S4). Thus, the long-lived retention of Hsp26 in mother cells may reflect its association with particular aggregates in cells.
As is the case for Hsp26-GFP foci, Thr1-GFP filaments are not obviously present in all mother cells. Dozens of metabolic enzymes have been discovered to form filaments, although most form filaments only under specific growth conditions, such as nutrient depletion (25, 51, 52). As is the case with Hsp26, more analysis will be needed to discover how Thr1 filaments are retained in the mother cell, but it seems likely that the narrowing at the bud neck may contribute to retention in the mother cell (53).
In contrast to the cytoplasmic proteins, the plasma membrane LARPs are present in every cell and remain within the mother. Integral plasma membrane proteins that are preferentially retained in mother cells have been previously identified (35, 36). The mechanism underlying this asymmetry remains unclear. Previously it had been suggested that septins at the bud neck serve as a diffusion barrier to the plasma membrane protein between the mother and daughter (54). We tested this idea and found no evidence that septins played a role in retaining Mrh1 in the mother cell (Fig. S5), consistent with earlier findings for another integral plasma membrane protein (35). Instead, the asymmetry in budding yeast may be explained in part by the rather slow diffusion of these proteins in the plasma membrane (55, 56). However, the plasma membrane LARPs do not appear to be completely immobile in the mother cell membrane. Careful examination of the time-lapse microscopy of Mrh1 and Pma1 fluorescent proteins reveals that the fluorescent signal of these fusion proteins is temporarily diminished near the site of budding on the mother cell as the daughter cell emerges. Mrh1 and Pma1 fusion proteins subsequently return to the bud site following cytokinesis (Movies S1 and S2).
The presence of the fragmented LARPs in the mother cell are perhaps the most enigmatic to explain. Although there are limitations to quantify peptide abundance in mass spectroscopy data (57), we can still estimate that the original fragmented 13C-labeled peptides represent ∼10−2 to 10−4 of the molar equivalents of a given protein. Given these estimates, there is still a significant bias that these fragmented peptides are retained in the mother. Assuming an infinitely long-lived protein was partitioned equally between the mother and daughter cells, and newly synthesized protein in the mother replaced the amount given to the daughter, then after 18 cell divisions the heavy-isotope form would be represented in the mother cell at (1/2)18, or ∼10−6 relative to the light-isotope form. At present we can only speculate as to where these fragmented proteins reside in the mother cell.
Consequences of LARPs.
Alterations in protein homeostasis are considered to be a major contributor to aging phenotypes, in particular the appearance and accumulation of aggregated proteins (58). We speculate changes in LARPs as a yeast mother cell matures may contribute to imbalances in protein homeostasis. For example, the truncated proteins produced by the fragmented LARPs have the potential to interfere with normal cellular functions (59). Such interference may act directly on the protein homeostasis machinery (e.g., proteasome or chaperones), or indirectly by disrupting other cellular processes, which in turn elicit a response by the protein homeostasis machinery. Furthermore, accumulation of some LARPs with successive cell divisions (e.g., Mrh1, Sur7) may result in an effective increase in dosage of protein in older cells. Such an accumulation not only has the potential to affect protein homeostasis, but may also change the phenotypic profile of the mother cell with age without impacting protein homeostasis (e.g., imbalances in metabolism, cell signaling, and so forth). Finally, accumulation of posttranslational modifications on LARPs may also alter protein activity in older cells (e.g., Mrh1, Pma1, Sur7) (Fig. 5). Taking these possibilities together, we hypothesize that LARPs may be an underlying cause of the aging process of yeast mother cells and, given the number of proteins and means by which they can change, may contribute to the complexity of age-associated phenotypes.
Finally, we speculate that LARPs are also present in metazoan cells that repeatedly divide asymmetrically (e.g., adult stem cells). There is good evidence that some stem cells have an age-associated decline, including those in muscle, the Germ-line, hematopoietic/immune systems, epithelium, and neurons (10, 11, 60, 61). Although the mechanisms underlying these age-associated changes remains a rich area of research, in Drosophila stem cells of the Germ-line and larval neuroblasts damaged proteins are retained as they go through repeated asymmetric cell divisions (62). This parallels the situation in mother cells of S. cerevisiae (63) and is consistent with the behavior of LARPs.
Materials and Methods
Plasmids and Strains.
Plasmids used in this study are presented in Table S1 and have been previously described, with the exception of pKTmCherry, which is derived from pKT127 (64), where eGFP was precisely replaced by mCherry.
Strains used in this study are presented in Table S1. Standard S. cerevisiae methods were used to generate PCR-mediated mutations, zygotes, sporulation, tetrad analysis, and selection of strains with relevant markers (65). UCC5406, which was used for the heavy isotope labeling, is derived from MEP-containing strains UCC5179 and UCC5181 (17), in which LYS1 and ARG4 were deleted to prevent arginine and lysine biosynthesis in cells (66). RITE-tagged strains were created by using BY4741 and BY4742 as parental strains (67). Into one parent, cre-EBD78 was introduced into the CYC1 locus by integrating MluI linearized pSS146 (24). Into the other parent, the gene of interest was fused to the RITE cassette as described using pKV015 as a template (24). The two strains were then mated and the resulting diploid was used for analysis.
Media and Growth Conditions.
Unless otherwise indicated, cells were grown at 30° in YEPD, Ymin, or YC media (65, 68). Stable isotope labeling by amino acids (SILAC) used [13C6]l-arginine and [13C6]l-lysine (Cambridge Isotope Laboratories), and was based on a previous protocol (66), with modifications described below. For some fluorescence-based microscopy, YEPD was treated with 30 mg/mL activated charcoal (C-3345; Sigma-Aldrich) for 20 min to reduce autofluorescence.
Pulse-Chase Labeling and Isolation of Proteins in Aged Mother Cells.
UCC5406 was grown overnight to saturation in YEPD. Cells were resuspended in heavy medium (Ymin supplemented with 0.1 mg/mL l-histidine, l-tryptophan, and uracil to support growth of auxotrophic mutations, and 0.1 mg/mL [13C6]l-arginine–HCl and [13C6]l-lysine–HCl) at a starting density of ∼4 × 104 cells/mL and grown for 24 h to a density of ∼5 × 106 cells/mL. Cells were labeled with biotin as previously described (69), and grown for an additional 2 h in fresh heavy medium. Cells were then resuspended at 2 × 104 cells/mL in light medium [YEPD supplemented with 0.1 mL/mL l-arginine–HCl and l-lysine–HCl (not heavy-isotope labeled) (Sigma-Aldrich), 200 µg/mL hygromycin (Roche) and 100 µg/mL ampicillin (Sigma-Aldich) (to discourage fungal and bacterial contamination of aging culture) and 1 µM 17β-Estradiol (Sigma-Aldrich) to engage the MEP]. Cells were aged for 24 h, completing ∼18 cell divisions. Original cells were purified with streptavidin-magnetic beads (69). Total proteins extracts were made by glass bead beating in Sample Buffer [2% (wt/vol) SDS, 10% (vol/vol) glycerol, 60 mM Tris⋅Cl pH 6.8, 5% (vol/vol) β-mercaptoethanol, Protease Inhibitors (PMSF, Leupeptin, Pepstatin, TPCK)]. The proteins were separated on an 8–16% (wt/vol) SDS/PAGE gradient gel (Bio-Rad Laboratories). The gel was then sliced in 27 fragments, ranging from an estimated molecular weight of >200 kDa to 8 kDa.
Gel Slice Digestion.
Individual gel slices in 1.5-mL tubes (Eppendorf) were subjected to consecutive 15-min washes with water, 50% (vol/vol) acetonitrile, 100% acetonitrile, 100 mM ammonium bicarbonate, and 50% acetonitrile in 50 mM ammonium bicarbonate. After removing the final wash solution, the gel slices were dried thoroughly by vacuum centrifugation. The gel slices were then cooled on ice and an ice-cold solution of 12.5-ng/µL sequencing grade trypsin (Promega) in 50 mM ammonium bicarbonate was added to the gel slices and incubated on ice for 1 h. The trypsin solution was discarded and replaced with 50 mM ammonium bicarbonate and incubated overnight at 37°. Following digestion, the supernatants were collected and the gel slices were washed with 0.1% formic acid followed by washing with 0.1% formic acid in 50% (vol/vol) acetonitrile (30 min each wash). The original digestion supernatant and the washes for a single sample were combined into a single tube and dried by vacuum centrifugation. The digestion products were desalted using Ziptips (EMD Millipore) per the manufacturer’s instructions and dried by vacuum centrifugation.
Mass Spectrometry.
Dried peptide mixtures were resuspended in 5 µL of 0.1% formic acid and analyzed by LC/ESI MS/MS with a nano2D LC (Eksigent Technologies) coupled to an LTQ-OrbiTrap mass spectrometer (Thermo Electron) using a “vented” instrument configuration, as described previously (70) and a solvent system consisting of 0.1% formic acid in water (A) and 0.1% formic acid in 100% acetonitrile (B). In-line de-salting was accomplished using an IntegraFrit trap column (100 μm × 25 mm; New Objective) packed with reverse-phase Magic C18AQ (5 μm 200 Å resin; Michrom BioResources) followed by peptide separations on a PicoFrit column (75 μm × 200 mm; New Objective) packed with reverse-phase Magic C18AQ (5-μm 100 Å resin; Michrom BioResources) directly mounted on the electrospray ion source. A 90-min nonlinear gradient was used starting at 5% B. The percentage of acetonitrile was increased to 7% B over 2 min, then 35% B over 90 min. The acetonitrile percentage was increased to 50% B over 1 min then held at 50% B for 9 min followed by ramping to 95% B over 1 min, held at 95% B for 5 min, then decreased to 5% B over 1 min. A flow rate of 400 nL/min was used for chromatographic separations and the MS capillary temperature was set to 200 °C. A spray voltage of 2,250 V was applied to the electrospray tip and the LTQ-OrbiTrap instrument was operated in the data-dependent mode, switching automatically between MS survey scans in the OrbiTrap (AGC target value 1,000,000, resolution 60,000, and ion time 500 ms) with MS/MS spectra acquisition in the linear ion trap (AGC target value of 10,000 and ion time 100 ms). The five most intense ions from the Fourier-transform full scan were selected in the linear ion trap for fragmentation by collision-induced dissociation with normalized collision energy of 35%. Selected ions were dynamically excluded for 45 s. The MS data have been deposited to the ProteomeXchange Consortium (71) via the PRIDE partner repository with the dataset identifier PXD001251.
Peptide Identification and Quantification.
Acquired LC-MS/MS spectra for all gel fractions were searched against all translated ORFs from the S. cerevisiae S288C reference assembly (yeastgenome.org). The X! Tandem search engine (72) was used with a custom scoring function (73) and parameters appropriate for SILAC labeled spectra acquired on the LTQ Orbitrap, which included trypsin cleavage, up to two missed cleavage sites, monoisotopic masses, and ±2-Da precursor mass tolerance. Variable modification of 6.02013 Da was allowed on arginine and lysine residues to accommodate dual SILAC labels, and variable modification of 15.994915 Da on methionine was allowed to accommodate a common oxidation artifact. PeptideProphet (74) was used to evaluate the likelihood of each peptide assignment.
To assess relative abundance of light- and heavy-labeled species of a given peptide, quantitation was performed with the Q3 algorithm (75) for only those peptides assigned with high confidence (PeptideProphet probability ≥ 0.9, corresponding to a false-discovery rate < 1%), and falling within 20 ppm of the measured precursor mass. Briefly, Q3 reconstructs elution curves in a 25-ppm window around major isotopes of the light and heavy peptide species in a range of precursor scans surrounding each identified MS/MS spectrum. The areas and extents of each curve are recorded, and the ratio of each pair of areas indicates the relative amounts of the SILAC labels incorporated at the two time points. Peptides species with zero area were set to a background value to avoid infinite or extreme ratios.
Custom Python scripts were developed to filter results and identify proteins of interest. We focused on proteins that had at least one peptide fragment within any single gel slice that was identified by both a heavy- and light-labeled species, and had a median 13C/12C ratio of ≥ 0.1 for all observed events. Variation in observed ratios was estimated by a bootstrapped median calculation, measuring the SD of medians repeatedly calculated from a randomly selected subset of the observations over 1,000 loops (76). A summary figure was generated for each protein of interest where the median ratio of all peptides was plotted against the gel slice (molecular weight) in which they were found (Dataset S3). These figures were analyzed manually to identify proteins that varied in the abundance of heavy-label depending on where they migrated by SDS/PAGE.
Microscopy.
Unless otherwise stated, all microscopy was performed on one of two fully automated wide-field microscopes: (i) a Nikon Eclipse Ti equipped with a 60×/1.40 objective or (ii) a Leica DMI-6000b equipped with a 63×/1.40 objective. Image processing and quantification was performed with Fiji, a distribution of ImageJ software (77).
Time-lapse microscopy of RITE-tagged strains on pads.
Cells were grown overnight in YC-Ura + 200 µg/mL hygromycin to select for cells that have not undergone spontaneous recombination and retain the GFP fusion. Cells were diluted into fresh YEPD and allowed to grow for 2 h in the presence of 1 µM 17β-Estradiol to initiate recombination of RITE tag. After 2 h, cells were placed onto YC agar pads (78). Eleven z-stack (0.6-µm steps) images in GFP, RFP, and DIC channels were taken every 30 min for 12 h. A minimum of 20 original mother cells were examined in all experiments.
Comparing young versus old mother cell fluorescence.
Cells were grown overnight to saturation and then diluted into fresh media and allowed to grow in log phase for 16 h. Cells were biotinylated, aged for 24 h, purified live with streptavidin-magnetic beads as previously described (69), and allowed to recover for 2 h in fresh YEPD. Purified cells were mixed with young cells from a log culture. Cells were stained with calcofluor white (Fluorescence Brightener 28; Sigma-Aldrich) to visualize budscars. Mean fluorescence intensity at the plasma membrane was compared between young and old mother cells within a single image.
Deltavision microscopy for Sur7 Lsp1 colocalization.
Cells were observed by high-resolution 3D deconvolution microscopy using an inverted Olympus IX71 microscope (Olympus 100×/1.40 Plan S Apo oil objective). Cells were placed on YC agar pads for imaging. The z-stacks were captured at 0.2-µm intervals using Deltavision SoftWorx software (Applied Precision). After deconvolution of the image stacks, 3D projections were made using Volocity (Perkin-Elmer).
Immunoblots.
Lysates for all samples were prepared by standard NaOH/SDS lysis (79) and analyzed as previously described (69), with a few exceptions. Sample concentrations were measured by BCA assay (Thermo Fisher Scientific). Twenty micrograms of protein was loaded per lane and run on 10% (wt/vol) polyacrylamide gels. Primary antibodies were as follows: α-PMA1 (ab4645, Abcam), α-GFP (#11814460001, Roche), and α-PGK1 (#459250, Life Technologies). Secondary antibody was α-Mouse (#715–035-150, Jackson ImmunoResearch).
Supplementary Material
Acknowledgments
We thank members of the D.E.G. laboratory for their constructive comments during the course of these studies; A. Waite (W. Shou laboratory, Fred Hutchinson Cancer Research Center) for providing reagents; and the PRoteomics IDEntifications (PRIDE) Database team for deposition of our mass spectrometry data to the ProteomeXchange Consortium. This work was supported by National Institutes of Health Grants AG037512 and AG023779 (to D.E.G.), T32GM07270 and F31AG041579 (to N.H.T.), T32CA009657 (to K.A.H.), and F30AG032808 (to J.J.H.). K.A.H. was a fellow of the Leukemia and Lymphoma Society and D.E.G. received a Glenn Award for Research in Biological Mechanisms of Aging.
Footnotes
The authors declare no conflict of interest.
See Profile on page 14007.
Data deposition: Mass spectrometry data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) with the dataset identifier PXD001251.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416079111/-/DCSupplemental.
References
- 1.Shapiro SD, Endicott SK, Province MA, Pierce JA, Campbell EJ. Marked longevity of human lung parenchymal elastic fibers deduced from prevalence of D-aspartate and nuclear weapons-related radiocarbon. J Clin Invest. 1991;87(5):1828–1834. doi: 10.1172/JCI115204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bank RA, Bayliss MT, Lafeber FP, Maroudas A, Tekoppele JM. Ageing and zonal variation in post-translational modification of collagen in normal human articular cartilage. The age-related increase in non-enzymatic glycation affects biomechanical properties of cartilage. Biochem J. 1998;330(Pt 1):345–351. doi: 10.1042/bj3300345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynnerup N, Kjeldsen H, Heegaard S, Jacobsen C, Heinemeier J. Radiocarbon dating of the human eye lens crystallines reveal proteins without carbon turnover throughout life. PLoS ONE. 2008;3:e1529. doi: 10.1371/journal.pone.0001529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Truscott RJW. Are ancient proteins responsible for the age-related decline in health and fitness? Rejuvenation Res. 2010;13(1):83–89. doi: 10.1089/rej.2009.0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyama BH, Hetzer MW. Protein homeostasis: Live long, won’t prosper. Nat Rev Mol Cell Biol. 2013;14(1):55–61. doi: 10.1038/nrm3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136(2):284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savas JN, Toyama BH, Xu T, Yates JR, 3rd, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335(6071):942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183(4677):1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 9.Kysela DT, Brown PJB, Huang KC, Brun YV. Biological consequences and advantages of asymmetric bacterial growth. Annu Rev Microbiol. 2013;67:417–435. doi: 10.1146/annurev-micro-092412-155622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones DL, Rando TA. Emerging models and paradigms for stem cell ageing. Nat Cell Biol. 2011;13(5):506–512. doi: 10.1038/ncb0511-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Signer RAJ, Morrison SJ. Mechanisms that regulate stem cell aging and life span. Cell Stem Cell. 2013;12(2):152–165. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li R. The art of choreographing asymmetric cell division. Dev Cell. 2013;25(5):439–450. doi: 10.1016/j.devcel.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Macara IG, Mili S. Polarity and differential inheritance—Universal attributes of life? Cell. 2008;135(5):801–812. doi: 10.1016/j.cell.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: The means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyström T, Liu B. The mystery of aging and rejuvenation—A budding topic. Curr Opin Microbiol. 2014;18:61–67. doi: 10.1016/j.mib.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Truscott RJW. Macromolecular deterioration as the ultimate constraint on human lifespan. Ageing Res Rev. 2011;10(4):397–403. doi: 10.1016/j.arr.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Lindstrom DL, Gottschling DE. The mother enrichment program: A genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics. 2009;183(4):413–422, 1SI–13SI. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Godoy LMF, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455(7217):1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 19.Smeal T, Claus J, Kennedy B, Cole F, Guarente L. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell. 1996;84(4):633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- 20.Orlean P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics. 2012;192(3):775–818. doi: 10.1534/genetics.112.144485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shnyreva MG, Petrova EV, Egorov SN, Hinnen A. Biochemical properties and excretion behavior of repressible acid phosphatases with altered subunit composition. Microbiol Res. 1996;151(3):291–300. doi: 10.1016/S0944-5013(96)80027-9. [DOI] [PubMed] [Google Scholar]
- 22.Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci USA. 2009;106(6):1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 24.Verzijlbergen KF, et al. Recombination-induced tag exchange to track old and new proteins. Proc Natl Acad Sci USA. 2010;107(1):64–68. doi: 10.1073/pnas.0911164107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noree C, Sato BK, Broyer RM, Wilhelm JE. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol. 2010;190(4):541–551. doi: 10.1083/jcb.201003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haslbeck M, Miess A, Stromer T, Walter S, Buchner J. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem. 2005;280(25):23861–23868. doi: 10.1074/jbc.M502697200. [DOI] [PubMed] [Google Scholar]
- 27.Cashikar AG, Duennwald M, Lindquist SL. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem. 2005;280(25):23869–23875. doi: 10.1074/jbc.M502854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merzendorfer H, Heinisch JJ. Microcompartments within the yeast plasma membrane. Biol Chem. 2013;394(2):189–202. doi: 10.1515/hsz-2012-0241. [DOI] [PubMed] [Google Scholar]
- 29.Douglas LM, Wang HX, Li L, Konopka JB. Membrane compartment occupied by Can1 (MCC) and eisosome subdomains of the fungal plasma membrane. Membranes (Basel) 2011;1(4):394–411. doi: 10.3390/membranes1040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreira KE, et al. Seg1 controls eisosome assembly and shape. J Cell Biol. 2012;198(3):405–420. doi: 10.1083/jcb.201202097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karotki L, et al. Eisosome proteins assemble into a membrane scaffold. J Cell Biol. 2011;195(5):889–902. doi: 10.1083/jcb.201104040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grossmann G, et al. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J Cell Biol. 2008;183(6):1075–1088. doi: 10.1083/jcb.200806035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fröhlich F, et al. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J Cell Biol. 2009;185(7):1227–1242. doi: 10.1083/jcb.200811081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci USA. 2006;103(35):13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eldakak A, et al. Asymmetrically inherited multidrug resistance transporters are recessive determinants in cellular replicative ageing. Nat Cell Biol. 2010;12(8):799–805. doi: 10.1038/ncb2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khmelinskii A, et al. Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat Biotechnol. 2012;30(7):708–714. doi: 10.1038/nbt.2281. [DOI] [PubMed] [Google Scholar]
- 37.Menendez-Benito V, et al. Spatiotemporal analysis of organelle and macromolecular complex inheritance. Proc Natl Acad Sci USA. 2013;110(1):175–180. doi: 10.1073/pnas.1207424110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454(7205):728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 39.Khmelinskii A, Keller PJ, Lorenz H, Schiebel E, Knop M. Segregation of yeast nuclear pores. Nature. 2010;466(7305):E1. doi: 10.1038/nature09255. [DOI] [PubMed] [Google Scholar]
- 40.Toyama BH, et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154(5):971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacGurn JA, Hsu P-C, Emr SD. Ubiquitin and membrane protein turnover: From cradle to grave. Annu Rev Biochem. 2012;81:231–259. doi: 10.1146/annurev-biochem-060210-093619. [DOI] [PubMed] [Google Scholar]
- 42.Schreiber A, Peter M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim Biophys Acta. 2014;1843(1):163–181. doi: 10.1016/j.bbamcr.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 43.Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci USA. 1992;89(21):10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454(7208):1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spokoini R, et al. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Reports. 2012;2(4):738–747. doi: 10.1016/j.celrep.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 46.Liu B, et al. Segregation of protein aggregates involves actin and the polarity machinery. Cell. 2011;147(5):959–961. doi: 10.1016/j.cell.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 47.Escusa-Toret S, Vonk WIM, Frydman J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol. 2013;15(10):1231–1243. doi: 10.1038/ncb2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denoth Lippuner A, Julou T, Barral Y. Budding yeast as a model organism to study the effects of age. FEMS Microbiol Rev. 2014;38(2):300–325. doi: 10.1111/1574-6976.12060. [DOI] [PubMed] [Google Scholar]
- 49.Liu B, et al. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140(2):257–267. doi: 10.1016/j.cell.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 50.Zhou C, et al. Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell. 2011;147(5):1186–1196. doi: 10.1016/j.cell.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Narayanaswamy R, et al. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci USA. 2009;106(25):10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Connell JD, Zhao A, Ellington AD, Marcotte EM. Dynamic reorganization of metabolic enzymes into intracellular bodies. Annu Rev Cell Dev Biol. 2012;28:89–111. doi: 10.1146/annurev-cellbio-101011-155841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16(4):493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Faty M, Fink M, Barral Y. Septins: A ring to part mother and daughter. Curr Genet. 2002;41(3):123–131. doi: 10.1007/s00294-002-0304-0. [DOI] [PubMed] [Google Scholar]
- 55.Greenberg ML, Axelrod D. Anomalously slow mobility of fluorescent lipid probes in the plasma membrane of the yeast Saccharomyces cerevisiae. J Membr Biol. 1993;131(2):115–127. doi: 10.1007/BF02791320. [DOI] [PubMed] [Google Scholar]
- 56.Valdez-Taubas J, Pelham HRB. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr Biol. 2003;13(18):1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 57.Nahnsen S, Bielow C, Reinert K, Kohlbacher O. Tools for label-free peptide quantification. Mol Cell Proteomics. 2013;12(3):549–556. doi: 10.1074/mcp.R112.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 59.Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329(6136):219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 60.Montecino-Rodriguez E, Berent-Maoz B, Dorshkind K. Causes, consequences, and reversal of immune system aging. J Clin Invest. 2013;123(3):958–965. doi: 10.1172/JCI64096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ortells MC, Keyes WM. New insights into skin stem cell aging and cancer. Biochem Soc Trans. 2014;42(3):663–669. doi: 10.1042/BST20140045. [DOI] [PubMed] [Google Scholar]
- 62.Bufalino MR, DeVeale B, van der Kooy D. The asymmetric segregation of damaged proteins is stem cell-type dependent. J Cell Biol. 2013;201(4):523–530. doi: 10.1083/jcb.201207052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299(5613):1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 64.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21(8):661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 65.Burke D, Dawson D, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab Press; 2000. [Google Scholar]
- 66.Gruhler A, et al. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4(3):310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 67.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14(2):115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 68.van Leeuwen F, Gottschling DE. Assays for gene silencing in yeast. Methods Enzymol. 2002;350:165–186. doi: 10.1016/s0076-6879(02)50962-9. [DOI] [PubMed] [Google Scholar]
- 69.Hughes AL, Gottschling DE. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 2012;492(7428):261–265. doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Licklider LJ, Thoreen CC, Peng J, Gygi SP. Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal Chem. 2002;74(13):3076–3083. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
- 71.Vizcaíno JA, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32(3):223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3(6):1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 73.MacLean B, Eng JK, Beavis RC, McIntosh M. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics. 2006;22(22):2830–2832. doi: 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
- 74.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 75.Faca V, et al. Quantitative analysis of acrylamide labeled serum proteins by LC-MS/MS. J Proteome Res. 2006;5(8):2009–2018. doi: 10.1021/pr060102+. [DOI] [PubMed] [Google Scholar]
- 76.Efron B, Tibshirani R. An Introduction to the Bootstrap. New York: Chapman and Hall; 1993. [Google Scholar]
- 77.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tran PT, Paoletti A, Chang F. Imaging green fluorescent protein fusions in living fission yeast cells. Methods. 2004;33(3):220–225. doi: 10.1016/j.ymeth.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 79.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16(9):857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 80.Walther TC, et al. Eisosomes mark static sites of endocytosis. Nature. 2006;439(7079):998–1003. doi: 10.1038/nature04472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.