Summary
Immunoglobulin G1(IgG1)-based therapies are widespread and many function through interactions with low-affinity Fc γ receptors (FcγR). N-glycosylation of the IgG1 Fc domain is required for FcγR binding, though it is unclear why. Structures of the FcγR:Fc complex fail to explain this because the FcγR polypeptide does not bind the N-glycan. Here we identify a link between motion of the N-glycan and Fc:FcγRIIIa affinity that explains the N-glycan requirement. Fc F241 and F243 mutations decreased the N-glycan/polypeptide interaction and increased N-glycan mobility. The affinity of the Fc mutants for FcγRIIIa was directly proportional to the degree of glycan restriction (R2=0.82). The IgG1 Fc K246F mutation stabilized the N-glycan and enhanced affinity for FcγRIIIa. Allosteric modulation of a protein/protein interaction represents a previously undescribed role for N-glycans in biology. Conserved features suggesting a similar N-glycan/aromatic interaction were also found in IgD, E and M, but not A.
Introduction
The fragment crystallizable (Fc) of human immunoglobulin G1 (IgG1) engages Fc γ receptors (FcγR) displayed on the surface of immune cells. In an adaptive immune response, Fc links the target-specific recognition of antigen binding fragments (Fab) to a pro-inflammatory cascade, resulting in destruction of the invading pathogen (Janeway et al., 2008).
IgG1 Fc contains a conserved asparagine-linked carbohydrate (N-glycan) that is required for productive engagement of the low-affinity FcγRs (Jefferis, 2009; Lux et al., 2013). The IgG1 Fc N-glycan is heterogeneous in nature as a result of the template-independent synthesis of carbohydrates in the Golgi (reviewed in (Varki, 2009)). Despite this source of compositional variability, a relatively small number of Fc glycoforms are observed and are predominantly of a core fucosylated, biantennary, complex-type with low levels of terminal sialic acid modification (Arnold et al., 2007).
The Fc N-glycan composition correlates strongly with rheumatoid arthritis (RA) disease state and is dominated by ungalactosylated forms in patients with advanced disease (Parekh et al., 1985). Furthermore, changes in glycan distribution can be observed years before RA symptoms arise (Ercan et al., 2010) and glycan anomalies return to normal during pregnancy-induced remission (Alavi et al., 2000; Bondt et al., 2013). The Fc N-glycan is predominantly of a biantennary, complex-type with a high level of core fucosylation (Figure 1). It was suggested that native sialic acid modification, which converts pro-inflammatory Fc to a potently anti-inflammatory form, is prevented when a galactose (Gal) residue at the non-reducing termini of the glycan is absent (Anthony et al., 2008; Kaneko et al., 2006). RA is a multifactorial disease, and though it is not known if IgG N-glycan anomalies cause RA, it is known that compositional changes to the Fc N-glycan alter FcγRIIIa affinity (Okazaki et al., 2004; Yamaguchi et al., 2006).
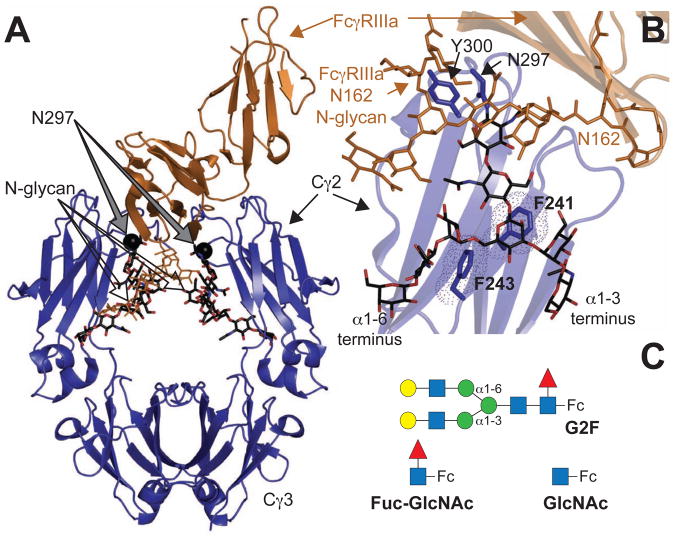
Figure 1. (A) Homodimeric IgG1 Fc.
(blue ribbon diagram) binds to monomeric FcγRIIIa (orange ribbons) (Sondermann et al., 2000). (B) Interactions between F241 and F243 amino acid sidechains stabilize the N-glycan in structures of Fc determined by x-ray crystallography. The Fc N-glycan is shown with a black stick model. An FcγRIIIa N-glycan contacts core Fc N297 glycan residues and is shown with an orange stick model (Mizushima et al., 2011) (C) Three Fc glycoforms are examined in this study: a complex-type, biantennary N-glycan containing two [13C2]-galactose (Gal) residues at the non-reducing termini (G2F-Fc), a single N-acetylglucosamine residue (GlcNAc-Fc) and Fc with a disaccharide (Fuc-GlcNAc-Fc). Carbohydrate residues are represented using the CFG convention (Varki, 2009) (GlcNAc-blue square; Gal-yellow circle; fucose (Fuc)- red triangle; mannose (Man)-green circle); α1–6 and α1–3 linkages to the branching mannose are shown that differentiate the two branches of the glycan.
Structural models of IgG1 Fc show the N-glycan interacting with the Fc polypeptide surface between the Cγ2 domains (Figure 1A; (Deisenhofer, 1981; Huber et al., 1976)). Surprisingly, the glycan termini were distal to the site of FcγRIIIa binding (Figure 1B; (Mizushima et al., 2011; Sondermann et al., 2000)) and it seems unlikely that direct interactions between the branch termini of the Fc N-glycan and the pro-inflammatory FcγRIIIa explain how composition differences at the Fc N-glycan termini affect Fc:FcγRIIIa affinity (Yamaguchi et al., 2006). A different model must be used to explain this phenomenon.
Solution nuclear magnetic resonance (NMR) spectroscopy and molecular dynamics simulations revealed significant motions of the Fc N-glycan (Barb et al., 2012; Barb and Prestegard, 2011; Frank et al., 2014), which was unexpected considering the fixed N-glycan position observed by x-ray crystallography (Huber et al., 1976). These opposing observations agreed in one key aspect: the α1–6 branch of the N-glycan interacts with amino acid residues on the Fc polypeptide surface. Motions of the Fc Cγ2 domain motion also occur (Frank et al., 2014; Krapp et al., 2003; Saphire et al., 2002) and may be related to N-glycan motion.
NMR spectroscopy provides a direct measurement of molecular motion with atom-level resolution. Though a single peak, corresponding to a resonance frequency, for each of the two Gal 13C2 nuclei was observed, further NMR analysis revealed each peak represented the population-weighted average of two distinct states (Barb and Prestegard, 2011). The (α1–6 branch)Gal residue showed the greatest effects of this interconversion and was found to exchange between a polypeptide-bound state and an unrestricted, mobile state on a μs timescale. Each resonance in each state is characterized by a distinct resonance frequency that is largely determined by covalent bonds in the Gal moiety and the immediate non-bonded chemical environment (within 5 Å). Both the rate of exchange and the difference in the resonance frequencies for each state contributed to line broadening relaxation (R2) of the glycan resonances (Barb and Prestegard, 2011; Cavanagh, 2007).
Two Fc amino acid residues likely restrict Fc N-glycan mobility (Lund et al., 1996; Voynov et al., 2009; Yu et al., 2013) and contribute to stabilizing the polypeptide-bound state of the N-glycan (Barb and Prestegard, 2011). F241 and F243 are found at the IgG1 Fc N-glycan/polypeptide interface and >7 Å from the nearest FcγRIIIa residue (Figure 1B). F241 is positioned beneath the branch point mannose residue and F243 interacts only with the glycan’s α1–6 branch (Fig 1B&C). Mutations at these positions increased the level of Gal and sialic acid incorporation at these positions (Lund et al., 1996; Voynov et al., 2009; Yu et al., 2013). Kelly and coworkers determined that aromatic residues formed higher affinity interactions with a glycan than other amino acids types (Chen et al., 2013). Though hydrophobic effects were measurable, the dispersion component of CH-π interactions contributed a greater proportion of free energy to the interaction. This observation highlights F241 and F243 as targets for mutation to disrupt the interface.
Based on recent reports (Barb et al., 2012; Barb and Prestegard, 2011; Frank et al., 2014; Krapp et al., 2003; Malhotra et al., 1995; Yamaguchi et al., 1998), we propose that conformational oscillations of the Fc Cγ2 domains and N-glycans are integral to FcγR binding and vary between naturally occurring glycoforms. Such a dynamic system endows IgG-expressing B cells with the capacity to tune FcγR affinity by changing glycan composition and would explain why serum IgG N-glycan composition changes rapidly in response to injury or disease (Lauc et al., 2013; Novokmet et al., 2014). If this model of Fc motion is accurate, one expects an Fc N-glycan with Gal on the α1–6 branch to experience less motion than an N-glycan lacking this Gal, and a Fc N-glycan with an N-acetylglucosamine residue (GlcNAc) on the α1–6 branch to experience less motion than an N-glycan lacking this GlcNAc. The restriction in both cases is due to extended glycan/polypeptide interactions. It is unclear how α1–3 branch modification might affect glycan/polypeptide interactions. This supposition is supported by the observation that Fc with longer glycans binds FcγRIIIa with higher affinity (Yamaguchi et al., 2006); the effect of sialylation, an extending modification, on receptor affinity is unresolved (Kaneko et al., 2006; Yu et al., 2013)) though recent evidence suggests sialylation does not alter either N-glycan motion or the interface character to a large extent (Barb et al., 2012; Crispin et al., 2013). To simplify analysis of the relationship between dynamics and affinity, we generated a series of Fc mutants with alterations at the N-glycan/polypeptide interface and remodeled the N-glycans in vitro to near homogeneity. Here we report the N-glycan mobility and receptor binding affinity for these Fc mutants.
Results
Fc mutations increase glycan modification during expression
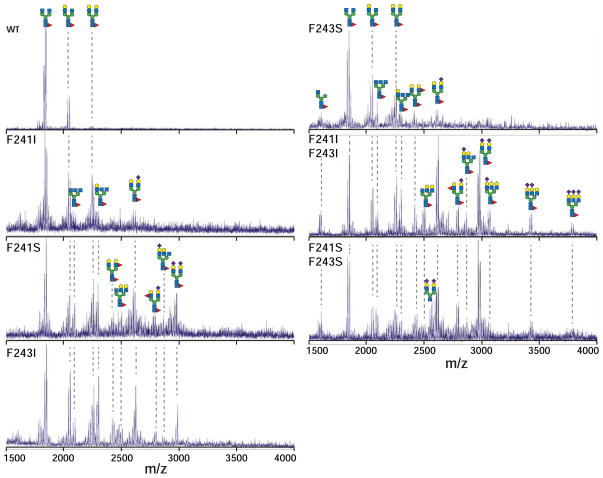
We prepared F241I, F243I and F241I/F243I Fc mutants to disrupt CH-π interactions but preserve hydrophobic contacts and F241S, F243S and F241S/F243S Fc mutants to disrupt both CH-π and hydrophobic contacts. Matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) analysis of N-glycans from purified Fc mutants showed a broader distribution of glycoforms and a greater degree of modification than those from wild-type (wt) Fc (Figure 2 & S1). Though complex-type, core fucosylated, biantennary types were the predominant glycans from all proteins, the double mutants contained triantennary glycans, extensive sialylation as well as increased fucosylation. Glycan distributions from F241S/F243S and single F241 and F243 mutants shared similarities with published reports (Lund et al., 1996; Voynov et al., 2009; Yu et al., 2013). These triantennary and branch-fucosylated glycoforms are not found on IgG1 Fc from human serum (Arnold et al., 2007). The single mutant proteins showed degrees of modification that were intermediate between wt and the two Fc double mutants. One explanation for the increased diversity of glycans on mutant Fc offered by Lund et al. (1996) is that removing aromatic sidechains reduced glycan/polypeptide interactions and decreased steric occlusion of the glycan. This would lead to greater glycan exposure and greater modification by glycosyltransferases in the Golgi during protein expression. Alternative explanations include increased retention in the Golgi or protein misfolding.
Figure 2. MALDI-Mass spectrometry of PNGaseF-released, permethylated N-glycans shows the increased diversity of glycans from Fc mutants when compared to wild type.
Cartoon diagrams highlight potential configurations that fit the observed masses and known N-glycan structures; isobaric ions were not distinguished. Peaks corresponding to Na+ and K+ adducts are observed for each species. See also Figure S1.
Fc mutants with homogenous N-glycans are well folded
The increased diversity of Fc mutant glycoforms introduces a significant challenge because N-glycan composition changes FcγRIIIa affinity and potentially Fc quaternary structure (Kaneko et al., 2006; Krapp et al., 2003; Yamaguchi et al., 2006; Yu et al., 2013). To reduce the effect of heterogeneity, N-glycans from wt and mutant Fc were enzymatically remodeled in vitro to contain 60–95% of the G2F glycoform (Figures 1C, S2–3).
All Fc proteins were judged using 1H NMR spectroscopy to be well-folded following in vitro glycan remodeling (data not shown). Furthermore, each protein bound to a Protein A affinity resin, which binds Fc through a protein-protein interaction and requires appropriately folded Fc (data not shown). Disulfide bonds that link the hinge region and stabilize the Fc dimer also formed (Figure S4).
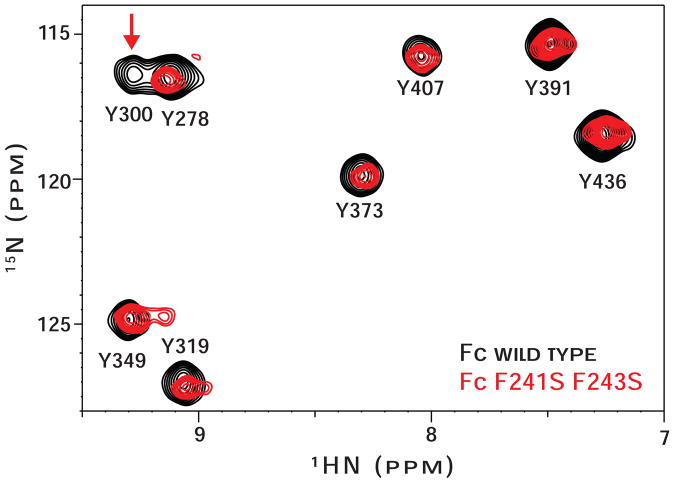
Heteronuclear single quantum coherence (HSQC) spectra provide high-resolution information regarding protein tertiary structure. We found that the 2d 1H-15N HSQC spectrum of [15N-tyrosine(Y)]-Fc F241S/F243S was nearly identical to that of [15N-Y]-Fc wt (Figure 3), indicating the Cγ2 and Cγ3 domains of both proteins are similarly folded. One difference between the spectra was disappearance of the Y300 peak in the Fc F241S/F243S spectrum. Y300 occupies the same loop as the glycan tether residue, asparagine (N)-297, and the Y300 resonance is sensitive to changes in the structure and motion of the loop (Fig. 1B;(Matsumiya et al., 2007; Yamaguchi et al., 2006)).
Figure 3. 1H-15N HSQC spectra of (15N-Tyr)-Fc wt (black contours) and (15N-Tyr)-Fc F241S/F243S (red contours) are nearly identical.
Peaks from residues Y300 and Y349 show differences in the mutant spectrum and are highlighted with a vertical arrow. Peak assignments are known (Matsumiya et al., 2007).
Fc mutants with a G2F N-glycan exhibit reduced FcγRIIIa binding
Folded Fc wt and Fc mutants, remodeled to contain a high proportion of G2F N-glycans, bound to a soluble fragment of glycosylated human FcγRIIIa. We found the Fc wt:FcγRIIIa affinity, as measured by surface plasmon resonance (SPR; 0.55±0.05 μM), isothermal titration calorimetry (ITC; 1.2±0.1 μM) and a Protein A resin-based pull down assay (0.5 μM), was similar to reported values (0.1–2 μM (Kaneko et al., 2006; Yamaguchi et al., 2006; Yu et al., 2013)). Single mutant proteins showed 3–4 fold reduced affinity by SPR when compared to Fc wt and the double mutant proteins bound with a 20 to 60-fold less affinity than wt (Table I). A previous study found a similar binding effect with single F241A and F243A mutants (Yu et al., 2013). These data are noteworthy because the effect of glycan heterogeneity has been removed by efficient in vitro glycan remodeling to a physiologically relevant form. The trend of FcγRIIIa binding by Fc mutants also mirrored the MALDI-MS data that showed greater N-glycan modification during expression for Fc double mutants and intermediate degrees observed for the single mutants (Figure 2).
Table 1.
Equilibrium FcγRIIIa dissociation constants for Fc wt and mutants from SPR experiments. Each observation represents the average of three independent measurements ± standard deviation of the mean. (n.b. = no binding detected)
| IgG1 Fc | FcγRIIIa KD (μM) |
± error |
|---|---|---|
| G2F glycan
|
||
| wt | 0.55 | 0.05 |
| F241I | 2.1 | 0.2 |
| F241S | 1.5 | 0.5 |
| F243S | 1.9 | 0.3 |
| F241I F243I | 10 | 1 |
| F241S F243S | 29 | 3 |
| K246F | 0.34 | 0.06 |
| GlcNAc glycan
|
||
| wt | 2.4 | 0.2 |
| F241I F243I | 6.3 | 0.6 |
| F241S F243S | 3.5 | 0.9 |
| No glycan
|
||
| T299A | n.b. | - |
F241/F243 Fc mutations do not affect GlcNAc-Fc:FcγRIIIa affinity
An enzymatically truncated Fc glycoform was prepared that provided a unique mechanism to test the role of the F241 and F243 mutations. It was reported that deglycosylated Fc and Fuc-GlcNAc-Fc (Figure 1C) bound FcγRIIIa below the levels of detectability (Yamaguchi et al., 2006). However, we found a similar form, GlcNAc-Fc (Figure 1C), bound to FcγRIIIa with robust affinity (2.4±0.2 μM; Table 1). This binding is likely enhanced by the absence of the core fucose residue, which reduced FcγRIIIa affinity 30–50 fold in studies using a complex-type, core fucosylated biantennary Fc N-glycan (Mizushima et al., 2011; Okazaki et al., 2004).
Equilibrium dissociation constants for the GlcNAc-Fc glycoforms of Fc wt and both Fc double mutants showed similar values (Table I), indicating the F241 and F243 mutations only affect the glycan/polypeptide interface and do not disturb other features of the Fc-FcγRIIIa interaction. A different Fc variant, containing only a T299A mutation to prevent N-glycosylation, did not bind FcγRIIIa under these conditions. Crispin and coworkers recently demonstrated that F241 and F243 mutations also affect FcγRIIIa binding of IgG1 Fc with a Man5 glycan (Yu et al., 2013). Our data confirm F241 and F243 affect FcγRIIIa affinity through the extended N-glycan and do not impact other aspects of binding.
F241/F243 Fc mutations increase N-glycan motion
The N-glycan occupies at least two distinct states that can be characterized by NMR spectroscopy. Each peak position represents the population-weighted average of a polypeptide-bound state and an unrestricted, mobile state (Barb and Prestegard 2011). Therefore, we expect to see changes in both NMR peak positions and R2 relaxation for any mutant with an altered N-glycan/polypeptide interface. Based on the distance between the F241 and F243 sidechains from the Gal residues (12 and 6 Å, respectively from (α1–6 branch)Gal 13C2), we do not expect to alter, to a large extent, the Gal chemical environment that contributes to the observed resonance frequencies of the individual states.
We measured the resonance frequencies of Gal-13C2 nuclei on Fc mutants that were remodeled to display nearly homogenous G2F N-glycans. Unlike wt Fc (Barb and Prestegard, 2011; Yamaguchi et al., 1998), 1H-13C HSQC spectra of proteolyzed Fc, degraded to remove long-range N-glycan/polypeptide interactions, and both of the double mutant proteins showed a single peak that contained contributions from both (α1–3 branch) and (α1–6 branch)Gal residues. This result indicated the contribution of the polypeptide-bound state to the observed resonance frequencies was minimal and the population was skewed towards the high mobility, unrestricted state (Figure 4A, Table 2).
Figure 4. Fc mutations impact the structure and mobility of N-glycan termini as judged by solution NMR spectroscopy.
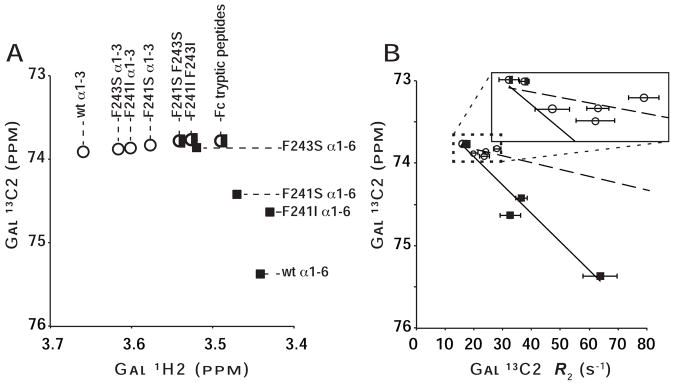
(A) Peaks from both N-glycan branches migrate incrementally away from those of the corresponding Fc wt peak and toward that of proteolyzed Fc. Spectra, from which this plot was generated, are shown in Figure S5. (B) The degree of line broadening for the (α1–6 branch)Gal 13C2 (black squares) correlates strongly with the resonance frequency (R2=0.95), indicating the frequency is sensitive to glycan mobility (R2 for α1–3=0.36; open circles). Points from (α1–3 branch)Gal and (α1–6 branch)Gal measurements are denoted with open circles or filled squares, respectively. Peaks with overlapped signals are marked with a half open circle/half filled square.
Table 2.
NMR chemical shift and relaxation parameters measured at 16.4T. (n.d. = analysis inconclusive due to likely peak overlap)
| IgG1 Fc | branch | Spin Relaxation
|
± error | Frequency
|
|||
|---|---|---|---|---|---|---|---|
| R1 (s−1) | ± error | R2 CP (s−1) | 1H (ppm) | 13C (ppm) | |||
| wt | α1–3 | 1.46 | 0.17 | 23.81 | 1.70 | 3.659 | 73.913 |
| α1–6 | 1.50 | 0.24 | 63.82 | 5.86 | 3.442 | 75.368 | |
| F241I | α1–3 | 1.22 | 0.13 | 24.02 | 1.01 | 3.601 | 73.869 |
| α1–6 | 1.10 | 0.32 | 32.65 | 3.61 | 3.430 | 74.630 | |
| F241S | α1–3 | 1.26 | 0.06 | 28.09 | 1.37 | 3.577 | 73.830 |
| α1–6 | 0.97 | 0.10 | 36.47 | 2.08 | 3.470 | 74.420 | |
| F243S | α1–3 | 1.50 | 0.10 | 19.98 | 1.55 | 3.616 | 73.873 |
| (α1–6)? | n.d. | n.d. | 3.520 | 73.856 | |||
| F241I F243I | degenerate | 1.42 | 0.20 | 16.14 | 0.87 | 3.527 | 73.766 |
| F241S F243S | degenerate | 1.59 | 0.07 | 17.51 | 0.42 | 3.541 | 73.771 |
| proteolyzed | degenerate | 1.63 | 0.04 | 10.25 | 0.44 | 3.491 | 73.770 |
Spectra of F241I, F241S and F243S proteins each contained two major resolved peaks and in that aspect was similar to wt, however, the position of these peaks was different (Fig. 4). Peaks from the α1–6 branch were found in a position between that of the wt Fc (α1–6 branch)Gal C2 and that of proteolyzed Fc and peaks from the α1–3 branch were found in a position between that of the wt Fc (α1–3 branch)Gal C2 and that of proteolyzed Fc. As mentioned previously, we anticipate that the mutations do not directly affect Gal resonance frequencies. These results indicate that N-glycans from each of the single Fc mutants exhibit reduced, but still measureable interactions with the Fc polypeptide when compared to wt Fc. The peak positions thus reflect a population-weighted average that was shifted towards the more mobile state (Barb and Prestegard, 2011).
Signal intensities for the double Fc mutants observed in 1d 13C and 2d HSQC spectra were stronger than those of the single mutants, which in turn were stronger than Fc wt despite similar protein concentrations during the experiments (data not shown). One explanation for this is that signals from mutant proteins experience less magnetization decay during the experiment. We measured the longitudinal and line broadening transverse relaxation rates (R1 and R2, respectively) that describe signal loss when the bulk magnetization vector is parallel or perpendicular to the external magnetic field, respectively. These relaxation rates report local molecular motions of Fc on fast (<20 ns (Barb and Prestegard, 2011); R1 and R2) and slow timescales (μs-ms; R2 only). Longitudinal relaxation rates did not show a clear response to the mutations (Table 2). Transverse relaxation rates from Gal-13C2 nuclei on mutant Fc were less than those of wt (Table 2) and showed a strong correlation with peak position for the (α1–6 branch)Gal 13C2 of the N-glycan (R2=0.95; Figure 4B). These values are also consistent with the observed increase in signal intensity. Furthermore, the relaxation rates are entirely consistent with measurements of the resonance frequencies that showed the decreased influence of polypeptide/N-glycan interaction.
N-glycans on Fc double mutant proteins showed transverse relaxation rates that were highly similar to those of proteolyzed Fc, indicating undetectable levels of line-broadening relaxation due to exchange between the two previously identified N-glycan states. This is likely due to increased N-glycan motions faster than the μs timescale, which were previously shown to contribute to line broadening (Barb et al., 2012; Barb and Prestegard). We do not believe that the rate of motion was reduced to a timescale of s; in this case we would expect to observe two peaks for each Gal 13C2 nucleus corresponding to each state.
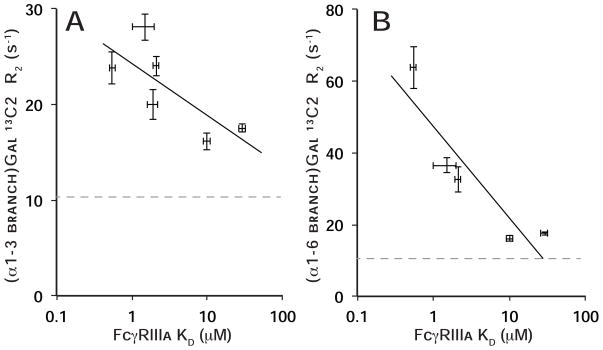
Transverse relaxation rates for both α1–3 and α1–6Man-linked Gal 13C2 nuclei were inversely correlated with FcγRIIIa affinity (R2=0.55 and 0.82, respectively) (Figure 5), indicating that reducing the N-glycan/polypeptide interactions increased glycan motion and reduced Fc:FcγRIIIa affinity. 13C2 resonance frequencies (Table 2) and N-glycan distributions (Figure 2) also showed a high correlation with FcγRIIIa affinity.
Figure 5. The degree of line broadening (R2) for the (α1–3 branch)Gal 13C2(A) or (α1–6 branch)Gal 13C2(B) nuclei correlates with FcγRIIIa binding affinity.
Standard deviations of the mean are included for both dimensions. The R2 for the same nucleus on a proteolyzed Fc fragment is shown for comparison to provide an experimental lower bound (dashed gray line). The coefficient of determination (R2) of a line fitted to each data set is 0.55 (A) and 0.82 (B).
Stabilizing the Fc N-glycan enhances affinity for FcγRIIIa
Removing N-glycan/polypeptide interactions led to increased N-glycan mobility and decreased FcγRIIIa affinity. Therefore, strengthening the glycan/polypeptide interface should enhance affinity. Previous data showed a stronger glycan/polypeptide interaction at lower temperature (Barb and Prestegard, 2011), however, temperature likely also influences other aspects of Fc binding FcγRIIIa. We again pursued an approach utilizing Fc mutation and in vitro N-glycan remodeling to test whether an increased interface would lead to higher FcγRIIIa affinity.
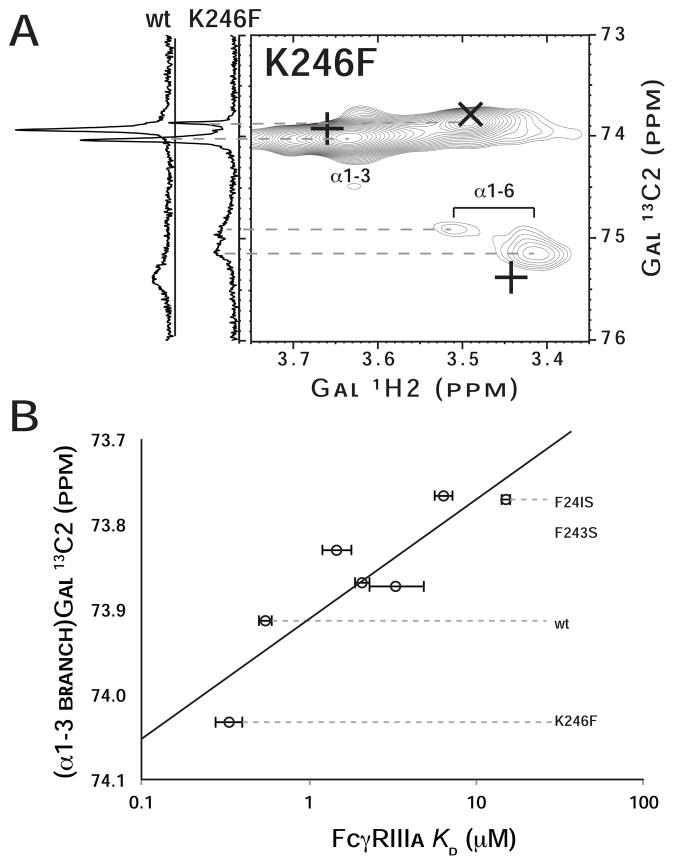
The positively charged sidechain of lysine (K)246 is positioned directly adjacent to the (α1–6 branch)Gal residue in structures of IgG1 Fc (Deisenhofer, 1981) and is thought to influence the resonance frequencies of (α1–6 branch)Gal nuclei (Barb and Prestegard, 2011). Fc K246F was prepared to determine whether a glycan stabilizing interaction could be formed at this position. Fc K246F with a G2F glycan bound FcγRIIIa with a 1.6-fold higher affinity than wt Fc (Table 1).
NMR spectroscopy of the 13C2-Gal-labeled Fc K246F showed the (α1–3 branch) Gal 13C2 nucleus had a greater chemical shift value than that of Fc wt, indicating a less mobile N-glycan (Figure 6A). Two very broad and weak peaks corresponding to the (α1–6 branch)Gal 13C2 nucleus were observed. The resonance frequency and relaxation rate of this nucleus are more difficult to interpret than those measured with F241/F243 Fc mutants because K246 is close (<4 Å) and thus mutations could be expected to alter (α1–6 branch)Gal 13C2 resonance frequency directly, unlike our expectation for the F241 and F243 mutations. However, the position of the (α1–3 branch)Gal 13C2 peak shows a strong correlation to FcγRIIIa affinity for the corresponding Fc (Figure 6B). Though the mechanism by which N-glycan mobility influences (α1–3 branch)Gal 13C2 resonance frequency is not known, this nucleus is a reliable indicator of total N-glycan mobility (R2=0.79) due to the presumed lack of direct contact between the N-glycan’s α1–3 branch and the Fc polypeptide.
Figure 6. A mutation designed to enhance the interaction between the glycan and polypeptide increases affinity for Fc.
γRIIIa.
(A) An 1H-13C HSQC spectrum of K246F mutant of Fc at 50°C containing the G2F glycan with 13C2-enriched Gal termini is shown. Positions of wild type coherences are highlighted in each spectrum with “+” signs, and the position of coherences following trypsin proteolysis are marked with “x”. To the left of the 2d HSQC spectrum, 1d 13C spectra of wt at K246F glycans are shown. (B) The resonance frequency of (α1–3 branch)Gal 13C2 nuclei correlates with FcγRIIIa affinity (R2=0.79).
Discussion
The effects of N-glycan mobility and FcγRIIIa affinity
Here we demonstrate that Fc restricts N-glycan motion through interactions with the F241 and F243 residues. We also found that the extent of Fc N-glycan/polypeptide interactions as measured by NMR correlated directly to FcγRIIIa affinity and the F241/F243 mutations only affected FcγR binding when the G2F N-glycan was present. Stabilizing N-glycan motion through the K246F mutation showed the expected increase in FcγRIIIa affinity for this mutant.
These data reveal a new role for N-glycans. N-glycans influence protein folding, stability, pharmacokinetic properties and also contain epitopes for specific recognition by many classes of receptors (reviewed in (Varki, 2009)). Here we provide an extensive characterization of a new role: an N-glycan influences a primarily protein/protein interaction through an allosteric mechanism that utilizes an extensive glycan/polypeptide interface to restrict N-glycan motion.
There are two, non-exclusive models that explain how Fc N-glycan mobility affects affinity for FcγRIIIa. First, F241 and F243 provide contacts that restrict the medial and distal N-glycan residues (relative to the N297 linkage site) and thus lock the N297-bearing Fc loop into a position that is optimal for contact with FcγRIIIa. The disappearance of the Y300 peak in the 1H-15N-HSQC spectrum of the Fc F241S/F243S mutant is consistent with this model (Figure 1A, 3). Second, it has been suggested that Fc Cγ2 domain orientation is an important component of FcγRIIIa affinity and may be sensitive to N-glycan structure or mobility (Krapp et al., 2003). Unrestricted Fc N-glycans are in a position to occupy the space between the Cγ2 domains and force these domains farther apart than is optimal for FcγRIIIa binding. Motions of the Fc Cγ2 domains relative to one another as observed in 200 ns molecular dynamics simulations, proved more extensive than previously shown by x-ray crystallography and are likewise consistent with this possibility (Frank et al., 2014).
The sensitivity of FcγRIIIa affinity to Fc N-glycan mobility suggests a potentially interesting feature of IgG: the interaction with low affinity FcγRs can be tuned in subtle ways. The advantages of this system are many, including the potential for a rapid cell-mediated response. In most cases, protein:protein interactions are only modulated over generations through genetic modification. Evidence for the existence of such a system comes from the identification of rapid changes in the IgG N-glycome in response to stress (Lauc et al., 2013; Novokmet et al., 2014), B-cell stimuli (Wang et al., 2011), and the identification of IgG N-glycan modification by ST6GalI, resulting in a potently anti-inflammatory circulating form of Fc, that occurs primarily outside the Golgi and in the serum (Jones et al., 2012). The balance point between pro- and anti-inflammatory agents must be constantly shifting to maintain sensitivity to eliminate infection but avoid autoimmune disorders.
Feature conservation in IgD, E, G and M
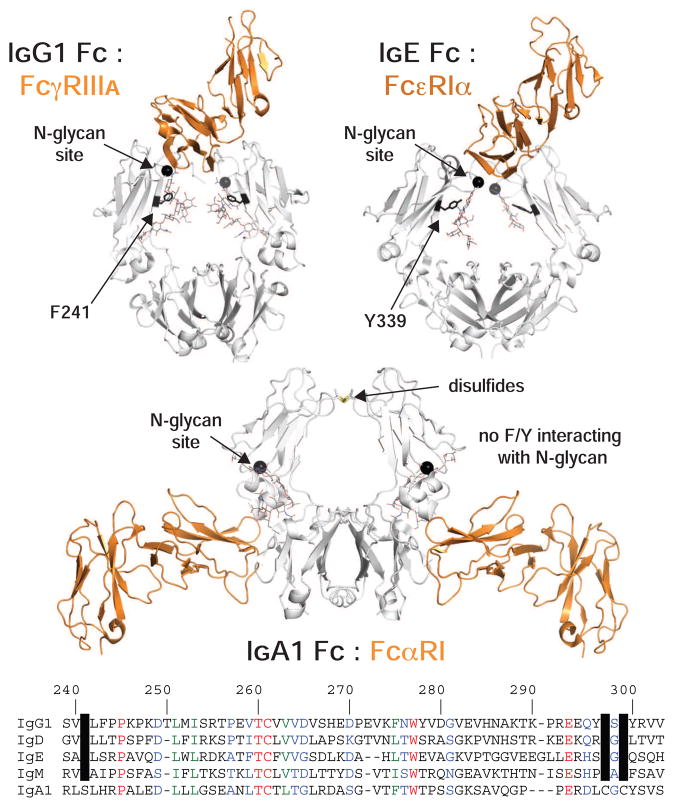
The role of the Fc N-glycan may not be a unique feature of IgG1. A strong interaction of IgG2–4 with low-affinity FcγRs also requires the N-glycan (Lux et al., 2013). IgD, E, and M also have an N-glycosylation site analogous to N297 of IgG1 and contain an aromatic residue at the same position as IgG1 F241 (Figure 7, S6). Though evidence for the role of these features is sparse for IgD, E and M, compelling evidence hints at a similar mechanism.
Figure 7. N-glycosylation and an aromatic residue at a comparable position are conserved features of IgD, E, G an M.
Of the human immunoglobulins, only IgA does not share these features, however, the IgA1:FcαRI interaction is remarkably different. The structure of receptor (orange ribbon): Fc (white ribbon) complexes is shown based (pdb: 1e4k (Sondermann et al., 2000) pdb: 1f6a (Garman et al., 2000) pdb: 1ow0(Herr et al., 2003)). At the bottom an amino acid alignment, based on IgG1 Fc numbering, is highlighted to show conserved aromatic residues and N-glycosylation sequons.
The structure of the IgE Fc:FcεRIα complex is remarkably similar to the IgG1 Fc:FcγRIIIa complex, sharing the FcεR binding which occurs asymmetrically at the top of two IgE Fc domains with 1:1 FcεR:Fc stoichiometry (Fig. 7; (Garman et al., 2000)). IgE contains an N-glycan at N394, analogous to IgG1 N297, that is predominantly of a mannose-type, representative of incomplete Golgi-mediated processing in contrast to other IgE N-glycans (Arnold et al., 2004; Dorrington and Bennich, 1978) and also required for FcεRIα binding (Nettleton and Kochan, 1995). IgD and IgM N-glycans at positions N354 and N402, respectively, corresponding to IgG1 N297, are likewise predominantly of mannose-types unlike other N-glycans on the same protein (Fig. S6; (Arnold et al., 2005)). It is unclear whether the conserved aromatic residue and glycan/polypeptide interactions contribute to restricted IgD, E and M N-glycan processing at these sites, but the degree of structural and functional conservation is remarkable. IgD, E and M also have F or Y at a position identical to F241 of IgG1 (Figure 7), though only IgG has an aromatic amino acid at the +2 position (F243). IgE, interestingly, has a F residue at the V262 position of IgG1 that is on the beta strand neighboring the F243 position of IgG1 and could potentially interact with the α1–6 branch of the N-glycan. This suggests, if the glycan-polypeptide interfaces for these immunoglobulins all form similarly, that an aromatic amino acid restricts the N-glycan in IgD, E G and M, though IgE and G have an aromatic amino acid positioned to form an additional interaction to restrict α1–6 branch residues.
Of the human Ig isotypes only IgA does not contain these features; IgA has neither an N-glycan nor aromatic residue at sites corresponding to IgG1 N297 and F241. However, the FcαRI recognizes IgA Fc by a completely different mechanism than that described above. FcαRI binds symmetrically to the IgA Cα3 domain (analogous to IgG Cγ3) with a 2:1 FcαR:Fc stoichiometry (Fig. 7; (Herr et al., 2003)). Though IgA lacks the N-glycan/aromatic residue features, inter-heavy chain disulfide bonds are present and are expected to exert conformational restrictions on the Cα2 domains.
Conclusions
This newfound connection between IgG N-glycan motion and FcγRIIIa affinity revealed why IgG Fc N-glycosylation is required. As a result we found new methods to tune antibody affinity, developed an Fc form that binds tighter than wt, and highlighted new avenues for future discovery, including how the ensemble of Fc conformations in solution are shaped by the N-glycan and investigating how glycan/polypeptide interactions influence the structure and activity of other Ig isotypes.
Experimental Procedures
Materials
All materials were purchased from Sigma-Aldrich unless otherwise noted. Structure figures were prepared using the PyMOL software package (Schrödinger).
Protein expression
Human IgG1-Fc (residues 216–447) with a 25 residues trypanosome signal peptide (Vandersall-Nairn et al., 1998) appended to the N-terminus was cloned into the NotI and HindIIII sites of the pGen2 vector (Barb et al., 2012). Plasmids encoding IgG1 Fc mutations F241I, F241S, F243I, F243S, K246F, T299A, F241I/F243I or F241S/F243S were made according to the QuikChange method (Agilent Technologies). All mutations were confirmed by DNA sequencing (ISU DNA Facility). The extracellular region of FcγRIIIa (residues 19–193, V158 allotype) containing N-terminal 8 x histidine and Green Fluorescent Protein (GFP) tags and a Tobacco etch virus (TEV) protease digestion site was cloned into the EcoRI and HindIII sites of the pGen2 vector (Barb et al., 2012).
Proteins were expressed during a transient transfection of HEK293F (Life Technologies) or HEK293S (lec1−/−) cells (Backliwal et al., 2008a; Backliwal et al., 2008b; Barb et al., 2012; Reeves et al., 2002). Cells grown in FreeStyle293™ medium (Life Technologies) on a shaker (ATR Biotech) at 125 RPM with 8% CO2 and 80% humidity at 37°C were transfected at a density of 2.5 × 106 live cells/mL with 2.5 μg/mL DNA and 7.5 – 9.0 μg/mL PEI. Cells were diluted 1:1 after a 24 h incubation with FreeStyle293™ medium supplemented with 4.4 mM valproic acid. Transfected cells were harvested after 6 days by centrifugation at 1000 g for 5 minutes. FcγRIIIa was purified using a Ni-NTA column (Qiagen). Expressed Fc fragments were purified using a Protein A-Sepharose® Column.
Fc glycoproteins expressed in human HEK293F cells to high levels (~50 mg/mL) with the exception of F243I, which expressed repeatedly at low levels. Expression medium was centrifuged to remove cells and debris, then loaded onto a Protein A-Sepharose® Column and washed with three column volumes of 25 mM 3-(N-morpholino)propanesulfonic acid (MOPS), 0.1 M sodium chloride, pH 7.2. Protein was eluted with 0.1 M glycine, pH 3.0 into collection tubes containing 1.0 M trisaminomethane (TRIS), pH 8.0 equal to one half of the elution volume and was then immediately buffer exchanged with 25 mM MOPS, pH 7.2, 0.1 M sodium chloride using 15 mL of 10 kDa molecular weigh cut-off Amicon® Ultra centrifugal filters (Millipore). Amino acid selective labeling of IgG1-Fc was achieved by supplementing custom FreeStyle293™ expression medium without lysine, phenylalanine and tyrosine with labeled [15N]-tyrosine, unlabeled l-lysine and unlabeled l-phenylalanine at 100 mg/L based on the average concentration of these amino acids in DMEM (Dulbecco and Freeman, 1959). Stock concentrations of l-lysine (40 mg/mL), l-phenylalanine (20 mg/mL) and l-tyrosine (1.5 mg/mL) were prepared in 10 mM TRIS, pH 8.0 and filter sterilized.
HEK293S cells were maintained in Freestyle 293™ expression medium supplemented with 10% ExCell medium. Transient transfection of HEK293S cells was performed only in Freestyle 293™ medium. The dilution 24 h after transfection was performed with Freestyle 293™ medium supplemented with 20% ExCell medium.
Enzymatic remodeling for 13C enrichment of IgG1-Fc N-glycans
Following purification, 13C2-labeled galactose was added to the Fc N-glycan termini as described previously (Barb and Prestegard, 2011). IgG1-Fc (3~5 mg) in a 50 mM ammonium acetate, pH 6.0 buffer was treated with neuraminidase (0.5 U/μL) and β-1,4-galactosidase (0.08 U/μL; New England Biolabs). Samples were purified as described above and exchanged into 20 mM MOPS buffer pH 7.2, 0.1 M sodium chloride and 20 mM manganese chloride. Reactions were incubated for 24–48 h following addition of β-1,4-galactosyltransferase and UDP-13C2-Galactose. Reaction completion was confirmed by MALDI MS. Samples were further washed with 5% or 100 % D2O (Cambridge Isotopes) NMR buffer (20 mM sodium phosphate, pH 7.2, 0.1 M sodium chloride, 0.5 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS)) by centrifugation using 0.5 mL of 10 kDa molecular weigh cut-off Amicon® Ultra centrifugal filters (Millipore) to remove MnCl2, UDP, and unreacted UDP-13C2-Galactose.
Preparation of Fc glycoforms consisting of only a single GlcNAc Residue
IgG1-Fc and mutants expressed in HEK293S cells were exchanged with 0.1 M potassium phosphate buffer, pH 6.0. Any higher forms of N-glycans were truncated to generate unfucosylated single N-acetylglucosamine (GlcNAc) glycoform by enzymatic cleavage with Endo-β-N-acetylglucosaminidase F1 (EndoF1) using 1:50 enzyme:protein concentration in the dark at room temperature for 16–18 h. EndoF1 treated Fc was further purified using Protein A-Sepharose® Column as described above and buffer exchanged with 20 mM MOPS pH 7.2, 0.1 M sodium chloride. The digestion reactions were judged to be complete using SDS-PAGE and MALDI-MS.
NMR Spectroscopy
NMR spectra were collected using 4 mm Shigemi NMR tubes in a spectrometer equipped with a cryogenically cooled probe and an Avance II console (Bruker) and operating at 50 °C and 16.4 T. Fc dimer concentrations were between 0.2~0.5 mM in final volume of 150 μL. 1H-15N Heteronuclear Single Quantum Coherence (HSQC) NMR spectra were collected over 64 × 2048 complex points in the t1 × t2 time-domain dimensions (1420 Hz) with approximately 768 scans per FID. The spectral digital resolution was 45 Hz. 1H-13C HSQC NMR spectra were collected over 256 × 2048 complex points in the t1 × t2 time-domain dimensions (3520 Hz) with approximately 128 scans per FID. The spectral digital resolution was 27.5 Hz.
The relaxation parameters R1 and R2(CP) for the Gal-13C2 nuclei were measured as described previously (Barb et al., 2012). Data were analyzed using Topspin (version 3.2), NMRviewJ (One Moon Scientific), NMRPipe (Delaglio et al., 1995) and the Gnuplot (v. 4.6) software packages. Chemical shifts were reference directly (1H) and indirectly (13C, 15N) to the internal DSS standard at 0.07 ppm (1H).
Glycans analysis by mass spectroscopy
IgG-Fc N-glycans were released and analyzed as described previously (Anumula and Taylor, 1992; Barb et al., 2009) using matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOFMS) on a Voyager-DE PRO (Applied Biosystems).
Binding analysis of Fc and Fc-mutants with FcγRIIIA
Surface Plasmon Resonance
GFP-FcγRIIIa was immobilized on the surface of a CM5 S Series chip using primary amine coupling in flow cells 2, 3, and 4 with a BiaCore T100 system (GE Healthcare). Flow cell 1 was left uncoupled as a reference cell. IgG1 Fc:FcγRIIIa dissociation constants were measured in triplicate by analysis of SPR response at equilibrium following subtraction of the signal from the uncoupled reference cell at 25 °C. At least five concentrations of each sample were injected onto the chip surface at a flow rate of 5 μl/min with a contact time of 420 s and a dissociation time of 420 s. Chip regeneration was achieved by applying 10 μL of 100 mM Glycine, pH 3.0 in 30 s. Dissociation constants (KD) were determined by fitting Eq 1 to equilibrium response values.
| Eq. 1 |
where RUeq is the measured response at equilibrium, [Fc] is the IgG1 Fc concentration, RUmax is the response when the chip is saturated with Fc.
Isothermal titration calorimetry
ITC measurements were performed using a MicroCal iTC200 system (GE Healthcare). All measurements were done at 25 °C in a buffer composed of 25 mM MOPS, 100 mM NaCl, pH 7.4. 166 μM IgG1 Fc (as a dimer) was titrated into 20 μM GFP-FcgRIIIa using 18 × 2 μL injections with a spacing time of 3 min. Two control experiments were performed, one by titrating IgG1 Fc into buffer, and the other by titrating buffer into GFP-FcγRIIIa. Binding isotherms were created with the MicroCal iTC200 Origin software (GE Healthcare). The data from IgG1 Fc-Buffer and Buffer-FcγRIIIa titration experiments were subtracted from the IgG1 Fc-FcγRIIIa titration data to account for heat changes resulting from non-binding interactions. The resulting binding isotherm and dissociation constant were fit using a 1:1 Fc:FcγR ratio.
Protein A-sepharose bead-based method
First, protein A sepharose beads were washed extensively with a buffer containing 25 mM MOPS, 100 mM NaCl, and 0.25 mg/mL BSA, pH 7.4. Washed beads were suspended in a volume of buffer such that the resulting slurry was 50% (v/v) beads. GFP-FcγRIIIa (0, 0.1, 0.3, 1, 3, and 6 μM) was mixed with IgG1 Fc (0, 0.1, 0.3, 1, 3, and 6 μM) and incubated at room temperature for 5 min. After incubation, protein A beads were added. The mixture was incubated at room temperature for three minutes with periodic gentle vortexing to ensure a homogenous bead distribution. Supernatant was then separated from the beads following centrifugation at 500 g. Supernatant fluorescence was measured using a Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies) (ex-484 nm em-510 nm). The fraction of fluorescence bound to the beads at each point was determined by comparing to the total amount of fluorescence in a reaction without Fc added. Dissociate constants (KD) were determined by fitting Eq 2 to fractional occupancy data.
Supplementary Material
Figure S1, related to Figure 2. Relative abundance of Fc N-glycoforms as measured from the spectra presented in Figure 2.
Figure S2, related to Figures 2 and 4. MALDI-MS of PNGaseF-released, permethylated N-glycans following in vitro remodeling to incorporate 13C2-Gal (indicated by a “*” within a yellow circle). Cartoon diagrams highlight potential configurations that fit the observed masses and known N-glycan structures; isobaric ions were not distinguished. Two peaks corresponding to Na+ and K+ adducts are observed for each species.
Figure S3, related to Figures 2,4 and S2. Relative abundance of Fc N-glycoforms following in vitro remodeling with [13C2]-Gal as measured from the spectra presented in Figure S2.
Figure S4, related to Figures 3 and 4. Hinge disulfides formed stabilize Fc dimers (expt ~55 kDa) expressed from both HEK293F and HEK293S (lec1−/−) cells. The covalently-linked Cγ2, Cγ3 domains and N-glycan that compose the Fc monomer have a combined mass of ~27.5 kDa and are observed in an SDS-PAGE gel when 1 mM beta-mercaptoethanol (β-me) is included in the sample preparation buffer.
Figure S5, related to Figure 4. 1H-13C HSQC spectra of Fc mutants collected at 50°C following enzymatic remodeling to incorporate 13C2-Gal. Positions of wild type coherences are highlighted in each spectrum with “+” signs, and the position of coherences following trypsin proteolysis are marked with “x”. Peaks labeled by “?” have not been identified, but are present in wild type. These peaks are very narrow and intense, but according to 1d 13C NMR spectroscopy only represent a small fraction of the total 13C signal.
Figure S6, related to Figure 7. Models of IgD and IgM Fc show the position of conserved N-glycosylation sites and aromatic residues. The IgD figure was generated from solution scattering data (pdb 1zvo (Sun et al., 2005)) and IgM upon x-ray scattering, crystallography and NMR data (Muller et al., 2013; Perkins et al., 1991).
Highlights.
F241 and F243 of IgG1 Fc both restrict N-glycan motion
Disrupting μs motion of the Fc N-glycan reduces affinity for FcγRIIIa
IgG1 Fc K246F stabilizes the N-glycan and enhances FcγRIIIa affinity
IgD, E, G and M share similar N-glycan restricting features; IgA does not
Acknowledgments
We thank Dr. Bruce Fulton (I.S.U.) for help with NMR instrumentation and pulse sequences. This work was financially supported by the grant K22AI099165 from the National Institutes of Health, and by funds from the Roy J. Carver Department of Biochemistry, Biophysics & Molecular Biology at Iowa State University.
Footnotes
Author Contributions
All authors performed, designed and interpreted experiments and wrote the manuscript.
The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alavi A, Arden N, Spector TD, Axford JS. Immunoglobulin G glycosylation and clinical outcome in rheumatoid arthritis during pregnancy. The Journal of rheumatology. 2000;27:1379–1385. [PubMed] [Google Scholar]
- Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science. 2008;320:373–376. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anumula KR, Taylor PB. A Comprehensive Procedure for Preparation of Partially Methylated Alditol Acetates from Glycoprotein Carbohydrates. Analytical Biochemistry. 1992;203:101–108. doi: 10.1016/0003-2697(92)90048-c. [DOI] [PubMed] [Google Scholar]
- Arnold JN, Radcliffe CM, Wormald MR, Royle L, Harvey DJ, Crispin M, Dwek RA, Sim RB, Rudd PM. The glycosylation of human serum IgD and IgE and the accessibility of identified oligomannose structures for interaction with mannan-binding lectin. J Immunol. 2004;173:6831–6840. doi: 10.4049/jimmunol.173.11.6831. [DOI] [PubMed] [Google Scholar]
- Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annual Review of Immunology. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- Arnold JN, Wormald MR, Suter DM, Radcliffe CM, Harvey DJ, Dwek RA, Rudd PM, Sim RB. Human serum IgM glycosylation: identification of glycoforms that can bind to mannan-binding lectin. J Biol Chem. 2005;280:29080–29087. doi: 10.1074/jbc.M504528200. [DOI] [PubMed] [Google Scholar]
- Backliwal G, Hildinger M, Chenuet S, Wulhfard S, De Jesus M, Wurm FM. Rational vector design and multi-pathway modulation of HEK 293E cells yield recombinant antibody titers exceeding 1 g/l by transient transfection under serum-free conditions. Nucleic Acids Res. 2008a:36. doi: 10.1093/nar/gkn423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backliwal G, Hildinger M, Hasija V, Wurm FM. High-density transfection with HEK-293 cells allows doubling of transient titers and removes need for a priori DNA complex formation with PEI. Biotechnol Bioeng. 2008b;99:721–727. doi: 10.1002/bit.21596. [DOI] [PubMed] [Google Scholar]
- Barb AW, Brady EK, Prestegard JH. Branch-specific sialylation of IgG-Fc glycans by ST6Gal-I. Biochemistry-Us. 2009;48:9705–9707. doi: 10.1021/bi901430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barb AW, Meng L, Gao Z, Johnson RW, Moremen KW, Prestegard JH. NMR characterization of immunoglobulin G Fc glycan motion on enzymatic sialylation. Biochemistry-Us. 2012;51:4618–4626. doi: 10.1021/bi300319q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barb AW, Prestegard JH. NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic. Nat Chem Biol. 7:147–153. doi: 10.1038/nchembio.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barb AW, Prestegard JH. NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic. Nat Chem Biol. 2011;7:147–153. doi: 10.1038/nchembio.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondt A, Selman MH, Deelder AM, Hazes JM, Willemsen SP, Wuhrer M, Dolhain RJ. Association between galactosylation of immunoglobulin G and improvement of rheumatoid arthritis during pregnancy is independent of sialylation. J Proteome Res. 2013;12:4522–4531. doi: 10.1021/pr400589m. [DOI] [PubMed] [Google Scholar]
- Cavanagh J. Protein NMR spectroscopy: principles and practice. 2. Amsterdam; Boston: Academic Press; 2007. [Google Scholar]
- Chen W, Enck S, Price JL, Powers DL, Powers ET, Wong CH, Dyson HJ, Kelly JW. Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins. J Am Chem Soc. 2013;135:9877–9884. doi: 10.1021/ja4040472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispin M, Yu X, Bowden TA. Crystal structure of sialylated IgG Fc: Implications for the mechanism of intravenous immunoglobulin therapy. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1310657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry-Us. 1981;20:2361–2370. [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. J Biomol Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dorrington KJ, Bennich HH. Structure-function relationships in human immunoglobulin E. Immunol Rev. 1978;41:3–25. doi: 10.1111/j.1600-065x.1978.tb01458.x. [DOI] [PubMed] [Google Scholar]
- Dulbecco R, Freeman G. Plaque production by the polyoma virus. Virology. 1959;8:396–397. doi: 10.1016/0042-6822(59)90043-1. [DOI] [PubMed] [Google Scholar]
- Ercan A, Cui J, Chatterton DE, Deane KD, Hazen MM, Brintnell W, O’Donnell CI, Derber LA, Weinblatt ME, Shadick NA, et al. Aberrant IgG galactosylation precedes disease onset, correlates with disease activity, and is prevalent in autoantibodies in rheumatoid arthritis. Arthritis and rheumatism. 2010;62:2239–2248. doi: 10.1002/art.27533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Walker RC, Lanzilotta WN, Prestegard JH, Barb AW. Immunoglobulin g1 fc domain motions: implications for fc engineering. J Mol Biol. 2014;426:1799–1811. doi: 10.1016/j.jmb.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garman SC, Wurzburg BA, Tarchevskaya SS, Kinet JP, Jardetzky TS. Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc epsilonRI alpha. Nature. 2000;406:259–266. doi: 10.1038/35018500. [DOI] [PubMed] [Google Scholar]
- Herr AB, Ballister ER, Bjorkman PJ. Insights into IgA-mediated immune responses from the crystal structures of human FcalphaRI and its complex with IgA1-Fc. Nature. 2003;423:614–620. doi: 10.1038/nature01685. [DOI] [PubMed] [Google Scholar]
- Huber R, Deisenhofer J, Colman PM, Matsushima M, Palm W. Crystallographic structure studies of an IgG molecule and an Fc fragment. Nature. 1976;264:415–420. doi: 10.1038/264415a0. [DOI] [PubMed] [Google Scholar]
- Janeway C, Murphy KP, Travers P, Walport M. Janeway’s immuno biology. 7. New York: Garland Science; 2008. [Google Scholar]
- Jefferis R. Recombinant antibody therapeutics: the impact of glycosylation on mechanisms of action. Trends in pharmacological sciences. 2009;30:356–362. doi: 10.1016/j.tips.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Jones MB, Nasirikenari M, Lugade AA, Thanavala Y, Lau JT. Anti-inflammatory IgG production requires functional P1 promoter in beta-galactoside alpha2,6-sialyltransferase 1 (ST6Gal-1) gene. J Biol Chem. 2012;287:15365–15370. doi: 10.1074/jbc.M112.345710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J Mol Biol. 2003;325:979–989. doi: 10.1016/s0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- Lauc G, Huffman JE, Pucic M, Zgaga L, Adamczyk B, Muzinic A, Novokmet M, Polasek O, Gornik O, Kristic J, et al. Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS genetics. 2013;9:e1003225. doi: 10.1371/journal.pgen.1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund J, Takahashi N, Pound JD, Goodall M, Jefferis R. Multiple interactions of IgG with its core oligosaccharide can modulate recognition by complement and human Fc gamma receptor I and influence the synthesis of its oligosaccharide chains. J Immunol. 1996;157:4963–4969. [PubMed] [Google Scholar]
- Lux A, Yu X, Scanlan CN, Nimmerjahn F. Impact of immune complex size and glycosylation on IgG binding to human FcgammaRs. J Immunol. 2013;190:4315–4323. doi: 10.4049/jimmunol.1200501. [DOI] [PubMed] [Google Scholar]
- Malhotra R, Wormald MR, Rudd PM, Fischer PB, Dwek RA, Sim RB. Glycosylation changes of IgG associated with rheumatoid arthritis can activate complement via the mannose-binding protein. Nat Med. 1995;1:237–243. doi: 10.1038/nm0395-237. [DOI] [PubMed] [Google Scholar]
- Matsumiya S, Yamaguchi Y, Saito J, Nagano M, Sasakawa H, Otaki S, Satoh M, Shitara K, Kato K. Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J Mol Biol. 2007;368:767–779. doi: 10.1016/j.jmb.2007.02.034. [DOI] [PubMed] [Google Scholar]
- Mizushima T, Yagi H, Takemoto E, Shibata-Koyama M, Isoda Y, Iida S, Masuda K, Satoh M, Kato K. Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans. Genes to cells: devoted to molecular & cellular mechanisms. 2011;16:1071–1080. doi: 10.1111/j.1365-2443.2011.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettleton MY, Kochan JP. Role of glycosylation sites in the IgE Fc molecule. International archives of allergy and immunology. 1995;107:328–329. doi: 10.1159/000237017. [DOI] [PubMed] [Google Scholar]
- Novokmet M, Lukic E, Vuckovic F, Ethuric Z, Keser T, Rajsl K, Remondini D, Castellani G, Gasparovic H, Gornik O, Lauc G. Changes in IgG and total plasma protein glycomes in acute systemic inflammation. Scientific reports. 2014;4:4347. doi: 10.1038/srep04347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki A, Shoji-Hosaka E, Nakamura K, Wakitani M, Uchida K, Kakita S, Tsumoto K, Kumagai I, Shitara K. Fucose depletion from human IgG1 oligosaccharide enhances binding enthalpy and association rate between IgG1 and FcgammaRIIIa. J Mol Biol. 2004;336:1239–1249. doi: 10.1016/j.jmb.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Parekh RB, Dwek RA, Sutton BJ, Fernandes DL, Leung A, Stanworth D, Rademacher TW, Mizuochi T, Taniguchi T, Matsuta K, et al. Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature. 1985;316:452–457. doi: 10.1038/316452a0. [DOI] [PubMed] [Google Scholar]
- Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire EO, Stanfield RL, Crispin MD, Parren PW, Rudd PM, Dwek RA, Burton DR, Wilson IA. Contrasting IgG structures reveal extreme asymmetry and flexibility. J Mol Biol. 2002;319:9–18. doi: 10.1016/S0022-2836(02)00244-9. [DOI] [PubMed] [Google Scholar]
- Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- Vandersall-Nairn AS, Merkle RK, O’Brien K, Oeltmann TN, Moremen KW. Cloning, expression, purification, and characterization of the acid α-mannosidase from Trypanosoma cruzi. Glycobiology. 1998;8:1183–1194. doi: 10.1093/glycob/8.12.1183. [DOI] [PubMed] [Google Scholar]
- Varki A. Essentials of glycobiology. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- Voynov V, Chennamsetty N, Kayser V, Helk B, Forrer K, Zhang H, Fritsch C, Heine H, Trout BL. Dynamic fluctuations of protein-carbohydrate interactions promote protein aggregation. PLoS One. 2009;4:e8425. doi: 10.1371/journal.pone.0008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Balog CI, Stavenhagen K, Koeleman CA, Scherer HU, Selman MH, Deelder AM, Huizinga TW, Toes RE, Wuhrer M. Fc-glycosylation of IgG1 is modulated by B-cell stimuli. Molecular & cellular proteomics: MCP. 2011;10:M110. doi: 10.1074/mcp.M110.004655. 004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Kato K, Shindo M, Aoki S, Furusho K, Koga K, Takahashi N, Arata Y, Shimada I. Dynamics of the carbohydrate chains attached to the Fc portion of immunoglobulin G as studied by NMR spectroscopy assisted by selective C-13 labeling of the glycans. J Biomol Nmr. 1998;12:385–394. doi: 10.1023/a:1008392229694. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Nishimura M, Nagano M, Yagi H, Sasakawa H, Uchida K, Shitara K, Kato K. Glycoform-dependent conformational alteration of the Fc region of human immunoglobulin G1 as revealed by NMR spectroscopy. Biochimica Et Biophysica Acta-General Subjects. 2006;1760:693–700. doi: 10.1016/j.bbagen.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Yu X, Baruah K, Harvey DJ, Vasiljevic S, Alonzi DS, Song BD, Higgins MK, Bowden TA, Scanlan CN, Crispin M. Engineering hydrophobic protein-carbohydrate interactions to fine-tune monoclonal antibodies. J Am Chem Soc. 2013;135:9723–9732. doi: 10.1021/ja4014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 2. Relative abundance of Fc N-glycoforms as measured from the spectra presented in Figure 2.
Figure S2, related to Figures 2 and 4. MALDI-MS of PNGaseF-released, permethylated N-glycans following in vitro remodeling to incorporate 13C2-Gal (indicated by a “*” within a yellow circle). Cartoon diagrams highlight potential configurations that fit the observed masses and known N-glycan structures; isobaric ions were not distinguished. Two peaks corresponding to Na+ and K+ adducts are observed for each species.
Figure S3, related to Figures 2,4 and S2. Relative abundance of Fc N-glycoforms following in vitro remodeling with [13C2]-Gal as measured from the spectra presented in Figure S2.
Figure S4, related to Figures 3 and 4. Hinge disulfides formed stabilize Fc dimers (expt ~55 kDa) expressed from both HEK293F and HEK293S (lec1−/−) cells. The covalently-linked Cγ2, Cγ3 domains and N-glycan that compose the Fc monomer have a combined mass of ~27.5 kDa and are observed in an SDS-PAGE gel when 1 mM beta-mercaptoethanol (β-me) is included in the sample preparation buffer.
Figure S5, related to Figure 4. 1H-13C HSQC spectra of Fc mutants collected at 50°C following enzymatic remodeling to incorporate 13C2-Gal. Positions of wild type coherences are highlighted in each spectrum with “+” signs, and the position of coherences following trypsin proteolysis are marked with “x”. Peaks labeled by “?” have not been identified, but are present in wild type. These peaks are very narrow and intense, but according to 1d 13C NMR spectroscopy only represent a small fraction of the total 13C signal.
Figure S6, related to Figure 7. Models of IgD and IgM Fc show the position of conserved N-glycosylation sites and aromatic residues. The IgD figure was generated from solution scattering data (pdb 1zvo (Sun et al., 2005)) and IgM upon x-ray scattering, crystallography and NMR data (Muller et al., 2013; Perkins et al., 1991).