Abstract
Living systems produce ordered structures and nanomachines that inspire the development of biomimetic nanodevices such as chips, MEMS, actuators, sensors, sorters, and apparatuses for single-pore DNA sequencing, disease diagnosis, drug or therapeutic RNA delivery. Determination of the copy numbers of subunits that build these machines is challenging due to small size. Here we report a simple mathematical method to determine the stoichiometry, using phi29 DNA-packaging nanomotor as a model to elucidate the application of a formula , where p and q are the percentage of wild-type and inactive mutant in the empirical assay; M is the copy numbers of mutant and Z is the stoichiometry in question. Variable ratios of mutants and wild-type were mixed to inhibit motor function. Empirical data were plotted over the theoretical curves to determine the stoichiometry and the value of K, which is the number of mutant needed in each machine to block the function, all based on the condition that wild-type and mutant are equal in binding affinity. Both Z and K from 1–12 were investigated. The data precisely confirmed that phi29 motor contains six copies (Z) of the motor ATPase gp16, and K = 1.
Keywords: phi29, Viral DNA packaging, ATPase, Nanobiotechnology, Copy number quantification
Background
Living beings produce a wide variety of nanomachines, ordered structures, and patterned arrays at the nanometer scale, i.e. biomotors,1–3 exosomes,4 pumps,5 arrays,6,7 valves,8–10 or membrane pores.11–13 The complexity and intricacies of these natural nanoparticles have inspired the development of biomimetic nanodevices.14,15 These nanomachines or patterned structures can be manipulated to build sophisticated nanodevices or parts for arrays, chips, MEMS,16 actuators,17 molecular sensors,18–20 molecular sorters,21 single pore DNA sequencing apparatus11–13 or other revolutionary electronic and optical devices22,23 ex vivo. Medical applications of these or their derivatives could be used for pathogen detection, disease diagnosis, and drug and therapeutic RNA delivery.19,20,24,25 Also common to living system is the transportation of dsDNA. In the ASCE (additional strand catalytic E) family including the AAA+ (ATPases Associated with diverse cellular Activities) and the FtsK-HerA superfamily, there are nanomotors that facilitate a wide range of functions involved in dsDNA riding, tracking, packaging, and translocation.26,27 These functions are critical to DNA replication, DNA repair, replication, recombination, chromosome segregation, DNA/RNA transportation, membrane sorting, cellular reorganization, cell division, and bacterial binary fission.28
The members of ASCE family usually assemble as multimeric, often hexameric,26,29 ring-shaped particles and act in coordination to perform cellular functions that involve multiple components in multi-step reactions. The hexameric geometrical shape facilitates bottom-up assembly in nanomachine manufacturing and can produce stable patterned arrays as parts for nanodevices. Quantitative analysis of copy number in these systems is difficult since the biological reactions that they catalyze are rapid and the intermediates are difficult to isolate and characterize.
Viral DNA packaging motors are intriguing molecular motors since they can package the lengthy genomic DNA to near-crystalline density into a pre-formed protein shell known as the procapsid.3 The pivotal component of the DNA packaging motor is the ATPase, which forms a ring structure and converts chemical energy from ATP hydrolysis into mechanical force to physically move the genomic DNA into the procapsid. The bacteriophage phi29 is an excellent model system for studying viral DNA packaging motors.
Four decades of extensive study have provided many details about the phi29 DNA packaging motor,8,30–32 which is composed of three essential co-axial rings: a dodecameric connector ring located at the vertex of the procapsid, a hexameric pRNA ring bound31 to the N-terminus of the connector,33 and a hexameric ring of ATPase gp16 that surrounds the helical region of pRNA26,30 (for review, see9). Sequence alignment has revealed that phi29 ATPase gp16 belongs to the FtsK/HerA family of dsDNA translocases that is a member of the FtsK/Her superfamily.34 The proteins of this family usually form hexameric rings and function in coordination. Native gel shift assays, capillary electrophoresis (CE), Hill constant determination, and titration of mutant subunits using computational binomial distribution have determined gp16 to be hexameric following a monomer → dimer → tetramer → hexamer pathway.26 The hexameric motor uses a revolution mechanism without rotation.9,26,35–37
A variety of methods have been attempted to quantify the reaction in viral DNA packaging motors.38,39 The stoichiometry of pRNA has been determined utilizing binomial distribution to predict the number of pRNA bound on the procapsid, taking advantage of the highly sensitive in vitro phi29 phage assembly system with 108 magnitude of sensitivity.40,41 This system can produce 108 virions (PFU, plaque form unit) per ml without any background. The goal of this study is to develop a simple mathematical method for determining the copy number of protein subunit in a biological complex. A Walker B mutant ATPase gp16 (gp16/ED) was constructed that dramatically inhibited the virion assembly activity of wild type gp16. The Walker B mutant gp16 could assemble into a ring on the motor together with wild type gp16. The probability of various combinations of mutant and wild type gp16 on the motor was predicted with binomial distribution. The production of the virion could be predicted under the assumption that Z gp16 copies are needed to drive the packaging motor and K copies of inhibitive mutant gp16 are required to block the motor. The pre-requisition of this method in the stoichiometry determination is that the mutant and the wild-type ATPase have an equal affinity in substrate binding. Since we can construct different mutant gp16 ATPase with only single mutation and inactive the entire system with only one amino acid modification. This trivial change will not significantly change the system, making it a feasible approach for the usage of the binomial distribution equation. This method could serve as a way to study the stoichiometry and mechanism of many other biological complexes.
Methods
Construction of mCherry-gp16 and mutant eGFP-gp16 plasmids
The strategy for construction of mCherry-gp16 was the same as that used for the construction of the eGFP-gp16 clone, as previously described.42 The mCherry gene was amplified by PCR (Polymerase Chain Reaction) with appropriate primers and digested with XbaI and KpnI. The mCherry-gp16 gene was then inserted into the previously re-engineered His-gp16 on pET32 Xa/Lic vector43 to replace the thioredoxin gene. The Walker B mutant, eGFP-gp16/ED, was cloned with the Stratagene Quick Change site-directed mutagenesis kit. The amino acid residues D255 and E256 of gp16 were mutated to E and D, respectively, with the appropriate primers. For Walker A mutant, the G27 amino acid residue was mutated to D.
Expression and purification of eGFP-gp16, mutant eGFP-gp16 and mCherry-gp16
The expression and purification of eGFP-gp16 were described previously.42 The walker B mutant eGFP-gp16/ED, walker A mutant eGFP-gp16/G27D and wildtype mCherry-gp16 protein were expressed and purified by the same protocol. Briefly, the expression of the proteins was induced with 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) in BL21 (DE3) Escherichia coli. The harvested cells were resuspended in buffer A (20 mM Tris–HCl, pH 7.9, 500 mM NaCl, 15% glycerol, (2-carboxyethyl) phosphine (TCEP) and 0.1% Tween-20) containing 5 mM imidazole. The cells were then passed through a French press and cell debris was removed by centrifugation. 0.1% Polyethylenimine (PEI) was added to remove the nucleic acids and other proteins. Lysate was loaded onto a Ni-resin column for further purification.
In vitro virion assembly assay
The components used in the in vitro phage phi29 assembly assay, including the procapsid, pRNA, DNA-gp3, gp9, and gp11-12-13-14 were prepared, as previously described.41,44 The in vitro phi29 assembly assay was also carried out as previously described.43,45 Briefly, 10 µl of purified procapsid (1 mg/ml) was mixed with 1 µl of 100 ng/µl pRNA and 3 µl of reaction buffer (10 mM ATP, 6 mM 2-mercaptoethanol, and 3 mM spermidine in TMS) at ambient temperature for 30 min. Then, DNA-gp3 and re-engineered gp16 diluted in PEG buffer (15% PEG 8000, 5% glycerin, 100 mM NaCl, 20 mM Tris–HCl, pH 7.8) were added at room temperature for 1 h to initiate the DNA packaging.
Finally, 10 µl of gp8.5-9 extract from E. coli HMS174 containing plasmid pARgp8.5-9 and 20 µl of gp11-12-13-14 extract from E. coli HMS174 containing plasmid pARgp11-12-13-14 were incubated with the DNA packaging mixture for 2 h at room temperature. The virions produced (PFU/ml) were quantified by plating Bacillus subtilis Su44+ infected with serial dilutions of the mixture on half LB plates.
Electrophoretic mobility shift assay
EMSA was carried out as previously described.46 The samples for EMSA assay were prepared in 20 µl of buffer A (20 mMTris-HCl, 50 mM NaCl, 1.5% glycerol, 1 mM Mg2+). Gp16 protein (1 µM), either wild type mCherry-gp16 (1 µM), walker A mutant eGFP-gp16/G27D (1 µM) or walker B mutant eGFP-gp16/ED (1 µM), or a mixture of wildtype mCherry-gp16 (1 µM) and mutant eGFP-gp16 (1 µM) was mixed with 0.67 µM (40 bp) Cy5-DNA with in the presence of 2.5 mM γ-S-ATP. The samples were incubated at ambient temperature for 20 min and then loaded onto a 1% agarose gel in 0.5 × TB (44.5 mM Tris, 44.5 mM boric acid) as running buffer for electrophoresis for 2 h at 80 V at 4 °C. The fluorescence produced by eGFP-gp16, mCherry-gp16 and Cy5-DNA in the gel was analyzed with a Typhoon FLA 7000 using 488 nm, 540 nm and 635 nm excitation for GFP, mCherry and Cy5, respectively.
Determination of the number of gp16 subunits in an active phi29 motor
Inhibition of the phi29 assembly by mutant eGFP-gp16s was performed to determine the gp16 subunit numbers in an active phi29 motor. Briefly, incubate the procapsid with pRNA in reaction buffer for 30 min at ambient temperature, followed by the addition of gp3-DNA and a mixture of mutant and wild type eGFP-gp16 that were pre-mixed in various ratios in the storage buffer and diluted in PEG buffer.40 They were incubated with pRNA enriched procapsid at room temperature for 30 min, then the tail protein gp9 and gp11-14 were added and incubated for another 1 h to complete the phage assembly. Assuming that wild type and mutant eGFP-gp16s have the same ability to bind together to form a hexameric ring, the probability of gp16 multimer (Z) ring formation with certain number of mutant gp16 copies (M) and wild type gp16 copies (Z-M) can be modeled with the following binomial distribution formula:
| (A) |
where p and q are the percentages of wild type mCherry-gp16 and mutant eGFP-gp16/ED, respectively and p + q = 100%, Z represents the total number of monomeric gp16 that bound to one procapsid-RNA complex as one DNA packaging motor, M and Z-M represent the number of mutant and wild type gp16, respectively. We assumed that at least K mutant copies of gp16 are needed to deactivate the phi29 packaging motor. Therefore, we can predict the theoretical activity of the phi29 DNA packaging motor with the following formula:
| (B) |
Results
Rationale and background for the method of quantification
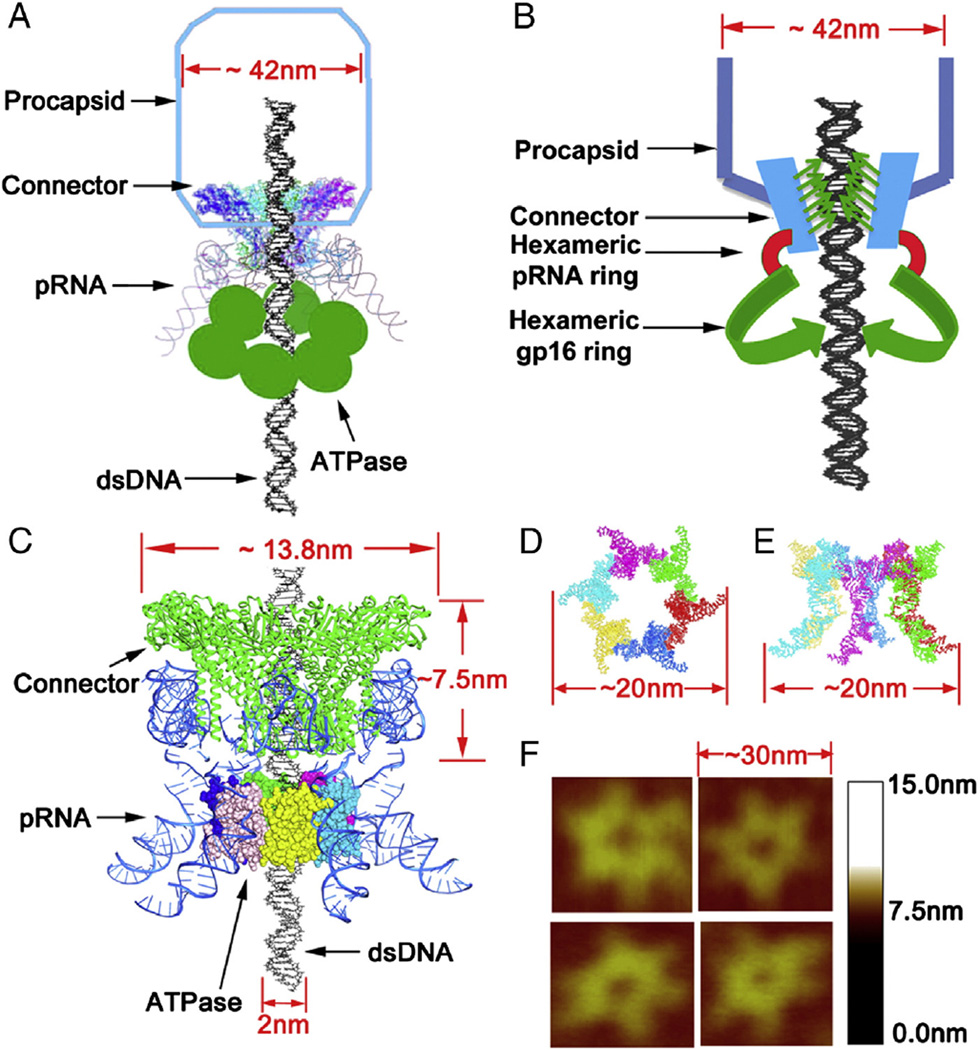
The approach for quantifying the gp16 subunits described here is based on the results and molecular details of the phi29 DNA packaging motor (Figure 1), revealed by extensive studies over four decades. pRNA assembles as a hexamer ring on the phi29 DNA packaging motor, as confirmed by AFM (Atomic Force Microscopy) imaging and an RNA crystal structure at 3.05 Å resolution.26,47 A hexameric structure is supported by the finding that pRNA binding to the dodecameric connector results in a 6-fold symmetry; but pentamer supporters proposed that pRNA binds the 5-fold procapsid shell. Cross-linking studies, though, have shown that pRNA binds to the connector protein gp10, not the procapsid protein.48
Figure 1.
Schematic illumination of the structure of bacteriophage phi29 DNA packaging motor. (A) Cartoon depiction of whole motor components with procapsid. (B) Three dimensional model of connector, pRNA, and hexameric gp16 ring assembled around DNA gp16 with DNA. (C) Ball-and-stick model. (D) Top view (E) and side view of the pRNA hexamer model. (F) AFM images of hexameric re-engineered pRNA ring.
The ATPase gp16 is the central part of the bacteriophage phi29 DNA packaging motor. Gp16 exists as a monomer in solution and assembles randomly into a strong and tight oligomeric ring on the phi29 DNA packaging motor. It binds to the motor pRNA, which also forms a ring-like structure surrounding the connector. It initiates and powers the motor mechanism of viral DNA packaging by hydrolyzing ATP.26,46
The ATPase gp16 contains two highly conserved motifs called Walker A and Walker B motifs. Walker A motif is responsible for ATP binding and Walker B motif is a very important component of ATP hydrolysis.30,35 Any alteration of the key amino acid residues in the Walker B motif will abolish ATP hydrolysis to ADP and Pi. 26 The mutation of the amino acid residues D255 and E256 in the Walker B motif of gp16 to E and D, respectively (eGFP-gp16/ED), abolished only the ATPase hydrolyzing activity, but did not affect other key functions such as DNA binding and inter-subunit interactions to form an ATPase oligomer ring. Thus we tested eGFP-gp16/ED inhibition phi29 virion assembly in vitro using the highly sensitive in vitro phi29 virion assembly system40,41 to quantify the subunit gp16 number in an active phi29 DNA packaging motor.
Walker B mutant gp16 interacted with the wild type gp16 to assemble into a protein ring
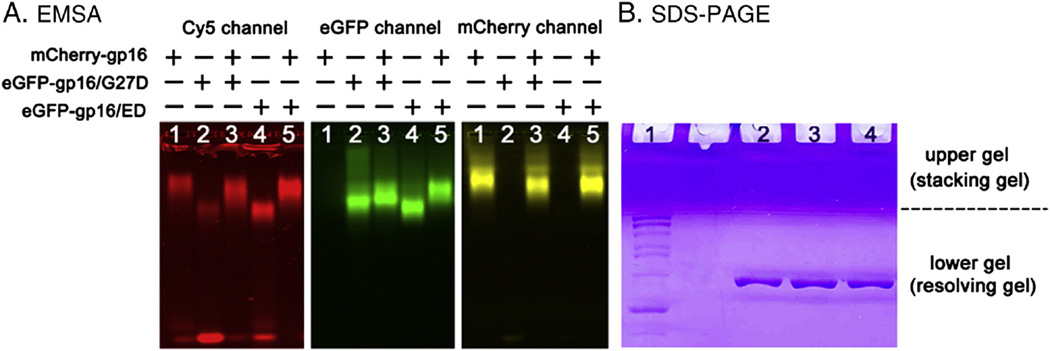
There were discrepancies regarding the stoichiometry of the motor component gp16. Binomial distribution of wild type and mutant gp16 in an inhibition assay was utilized to clarify the discrepancy and to elucidate the stoichiometry of the motor ATPase gp16.26 To inhibit the DNA packaging activity of wild type gp16, the Walker B mutant gp16 (eGFP-gp16/ED) is incorporated into the gp16 ring with wild type gp16 as competitors. To investigate the interaction between the wild type gp16 and Walker B mutant gp16, a new fluorescent fusion gp16 mCherry-gp16 was constructed. mCherry-gp16 has a different excitation and emission wavelength than eGFP-gp16, as such, the two proteins can be distinguished from each other by fluorescent scanning in the gel shift assay. Furthermore, the mCherry-gp16 protein migrated slower than eGFP-gp16 or its mutants in the EMSA (Electron Mobility Shift Assay) (Figure 2, A). If mutant eGFP-gp16/ED interacts with wild type mCherry-gp16, the mutant eGFP-gp16/ED will also migrate to a different position in a slower fashion. Since the eGFP-gp16 and mCherry-gp16 are only soluble in the presence of ATP (or its analog γ-S-ATP) and DNA, the interaction of the two proteins was examined under this condition. The eGFP-gp16/ED migrated much slower in a 1% agarose gel in the presence of wild type mCherry-gp16 in lane 5 than by itself as shown in lane 4 (Figure 2, A). This result indicated that the Walker B mutant eGFP-gp16/ED indeed interacted with wild type mCherry-gp16 to form a ring complex. The data showing the purity and homogeneity of purified gp16 and its derivatives can be found in Figure 2, B.
Figure 2.
EMSA gel shift assay showing the inter-subunit interaction between wild type gp16 and its derivatives and SDS-PAGE showing the purity and homogeneity of the protein. (A) Red (left panel) represents dsDNA labeled with Cy5, green (center panel) represents Walker B mutant gp16 labeled with GFP, and yellow (right panel) represents wild type gp16 labeled with mCherry. All the three panels were from the same gel, but produced different wavelengths for the emission of Cy5, GFP, and mCherry, respectively. The wild type mCherry-gp16 (Lane 1) migrated slower than Walker B mutant eGFP-gp16/ED (Lane 4) when they stayed alone, but their migration rate changed after mixing (Lane 5). The migration rate of the Walker A mutant eGFP-gp16/G27D, which cannot bind ATP and served as a control for Walker B mutant, changed very little in the presence of mCherry-gp16 (Lane 3). (B) SDS-PAGE showing the purity and homogeneity of purified gp16 and its derivatives. Lane 1: PageRuler protein marker; lane 2: wild-type eGFP-gp16; lane 3: eGFP-gp16/G27D; lane 4: eGFP-gp16/ED.
Determination of the distribution of gp16 in functional packaging motors
The stoichiometry of protein subunit number (denoted as Z) in the ring was determined. As explained in the Methods Section, Walker B mutant was assumed to have the same ability to form the ring complex as the wild type. The protein multimer ring composed with same number of mutant and wild type gp16 protein will appear with the same chance. For instance, two hexameric rings, WWMMWW and WMWWMW, had two mutants out of a total of six gp16 (W stands for wide-type and M for mutations), which were generated with the same frequency. This gave the binomial equation A as discussed in the Methods Section where p and q are the percentages of wild type eGFP-gp16 and mutant eGFP-gp16/ED, respectively (p + q = 100%). To determine which packaging motors were non-functional, K was denoted as the minimum number of mutant copies of gp16 needed to deactivate the phi29 packaging motor. Therefore, the probability of generating functional protein rings was calculated as equation B. The goal was to determine Z, gp16 stoichiometry, and K, the number of mutant gp16 copies sufficient to block DNA packaging based upon the above posited mathematical model. Through the detailed statistical analysis below, experimental data suggested that Z = 6 and K = 1, consistent with previous findings.26,35
Statistical analysis of phi29 assembly yields to determine the number of inactive mutant gp16 required to block motor function
Six independent experiments were conducted to determine the minimum number of inactive mutant gp16 required for blocking motor function. In each experiment 9 points of different percentages of mutant gp16 that range from 0% to 80% at 10% increments were tested, 54 experiments in total. Xi, where i = 1, 2, …, 54 was used to denote the percentages of mutant gp16. Correspondingly, Yi, where I = 1, 2, …, 54 was used to denote the yields of phi29 virion at these conditions. Therefore, the yield of phi29 virion Yi at condition of A(Xi,Z,K) was analyzed, where A was the percentage of functional motors with all wild type subunits in the ring. µ was used to denote the expectation of yields when there was no mutant, hence µA(Xi,Z,K) could be considered the theoretical yield of phi29 virion at the condition Xi. log10(Yi)~N(log10(µA(Xi,Z,K)),2 was assumed. Thus, measurement errors for Yi increased exponentially as the percentage of functional motors, A(Xi,Z,K), increased. The best parameters Z and K, as well as µ and σ2, were determined through maximum likelihood estimation. The log likelihood function was written as
and for fixed Z and K,
to maximize l(Z,K,µ,σ2).
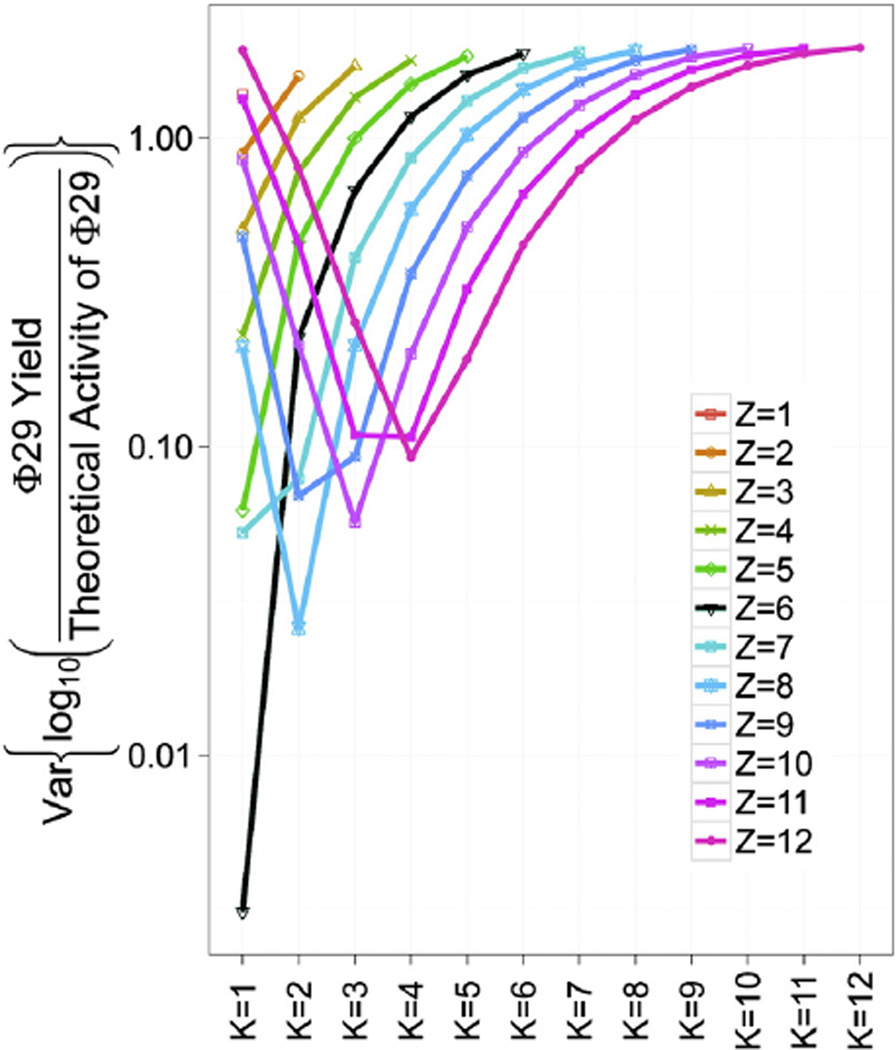
The most likely Z and K will maximize l(Z,K,µ̂(Z,K),σ̂2(Z,K)) as a function of Z and K. A grid search was used to find the optimal Z and K for all possible combinations of Z = 1, 2, …, 12 and K = 1, 2, …, 12. Figure 3 shows the profile log likelihood function l(Z,K,µ̂(Z,K),σ̂2(Z,K)) evaluated for all Z and K combinations. One can clearly see that (Z,K) = (6,1) maximizes the log likelihood function at −18.34, which is the maximum likelihood estimator. Notice that (Z,K) = (8,2) gives the second largest log likelihood function at −22.86. (Z,K) = (5,1), (Z,K) = (10,3), and (Z,K) = (7,1) are close to the third likeliest with log likelihood −27.58, −27.91, and −27.95, respectively. Our data suggest that (Z,K) = (6,1) fits experimental data better than (Z,K) = (5,1). Also (Z,K) = (8,2) has not been reported anywhere, giving a higher log likelihood than (Z,K) = (5,1), but still worse than (Z,K) = (6,1), as we hypothesized. These results strongly support the hexameric stoichiometry of gp16 and the sequential action of the motor ATPase with the fact that one inactive subunit in the hexamer abolishes motor activity.
Figure 3.
Statistical analysis used to evaluate the liability of the quantification method using binomial distribution. Log likelihood function under various combinations of Z = 1 to 12 and K = 1 to 12. Phi29 yields were the empirical virus assembly activities with various ratios of wild type eGFP-gp16 to Walker B mutant eGFP-gp16/ED. The theoretical activity of phi29 was calculated with equation A (see materials and methods). Z = 6, K = 1 (black line) gives maximum likelihood estimate).
Once maximum likelihood estimates Ẑ = 6 and K̂ = 1 were found, maximum likelihood estimates of µ and σ2 were obtained as:
The empirical normalized motor activity, when the percentage of mutant gp16 is q, was calculated as
where I(·) was the indicator function that took the value one for Xi = q; zero otherwise.
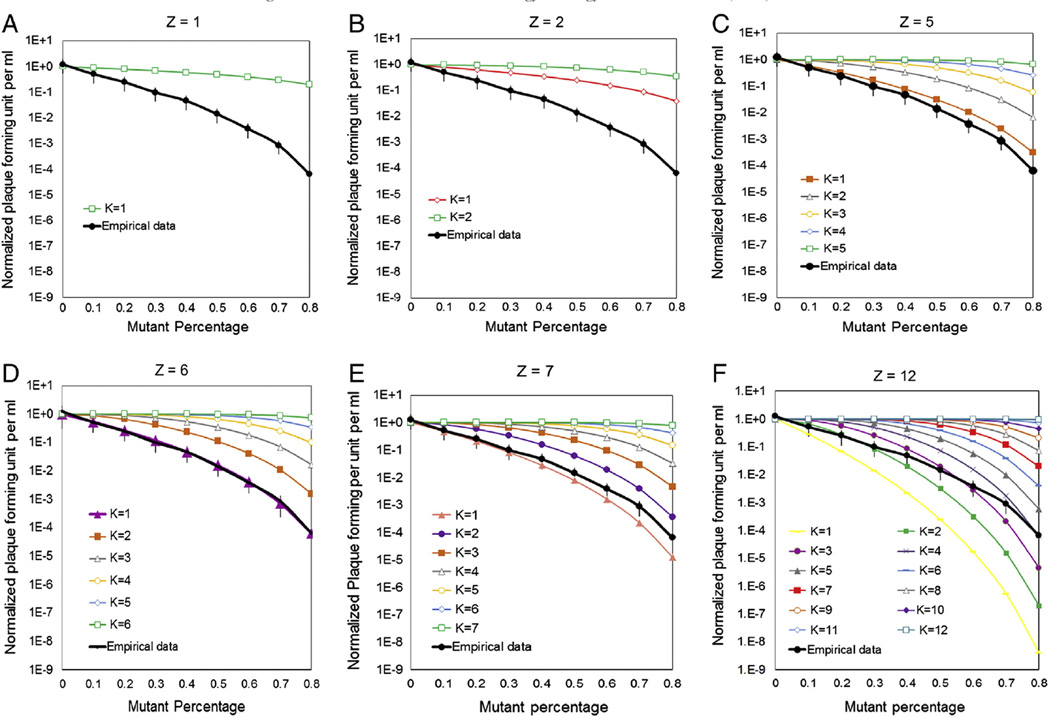
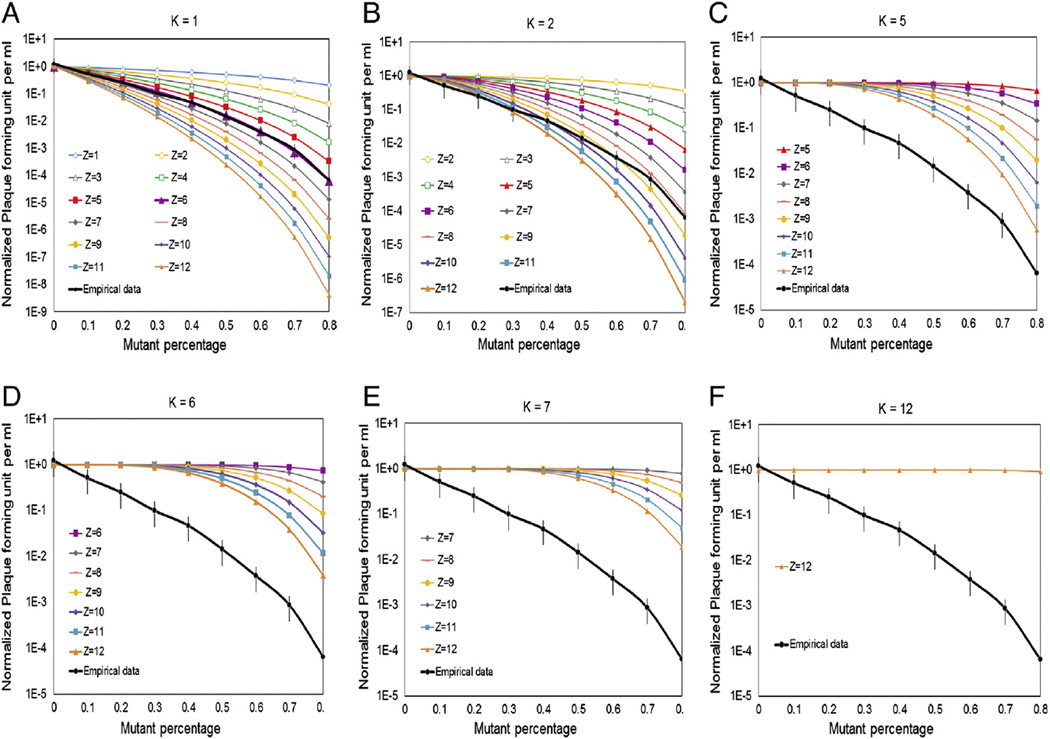
To show that the proposed model with (Z,K) = 6,1 provided an adequate fit of experimental data, the normalized empirical motor activity was plotted with the theoretical motor activity at different Z and K. The theoretical motor activities was demonstrated as shown in Figure 4 (A-F) when fix Z to be 1,2,5,6,7,12 respectively, and vary K at all possible values. The black line represents the empirical normalized motor activity with error bar corresponding to their 95% confidence intervals. The empirical data fits only to the theoretical curve for Z = 6 and K = 1 as shown in Figure 4D. We further tested this by fitting the empirical data into theoretical curve when fix K numbers to be 1, 2, 5, 6, 7 and 12 respectively and vary Z number at all possible values in Figure 5 (A-F). The empirical data can only fit into the theoretical curve for K = 1 and Z = 6 very well as shown in Figure 5A. Z denotes the total number of eGFP-gp16 per oligomer ring in the motor, and K is the minimum copy number of inactive Walker B mutant eGFP-gp16/ED required in the oligomer ring to block the motor function of wild type gp16. These data clearly show that (Z,K) = (6,1) adequately explains the variability in the experimental data.
Figure 4.
(A) Theoretical and empirical curves in logarithmic scale of minimum copy number (K) of inactive Walker B mutant eGFP-gp16/ED required in the oligomer ring to block the motor function of wild type gp16. Predictions were made using a binomial distribution with equation A where p is percent of wild type eGFP-gp16, q is percent of eGFP-gp16/ED, Z is the total number of eGFP-gp16 per oligomer ring in the motor, and M is the number of mutant eGFP-gp16 in the phi29 DNA packaging motor. The detailed equation calculation can be found in the Supplementary Material. Theoretical curves for viral assembly efficiency were predicted when assume Z equals to 1 in (A), 2 in (B) ,5 in (C), 6 in (D) , 7 in (E) and 12 in (F), with K at all possible values.
Figure 5.
Theoretical curve and empirical data to test the K value.
Discussion
We have developed a concise and simple method for the determination of the number of subunits in biological complex and nanobiomachines. The feasibility of applying this method depends on the following three conditions. First, a sensitive system for the assay of the outcome should be available. The main reason we have such elegant and precise data is due to the fact that we have developed a highly sensitive system with a range of 0 to 108 for the assay.41 Second, the experiment was based on the condition that the inactive mutants should have the binding or oligomer incorporation probability or affinity equal to the wild type. This is very important since multi-components are involved in the system. In this report, the Walker B mutant incorporated into the ring equally well as the wild type, since there was only one amino acid mutation in Walker B mutant. The change of gp16 folding after single mutation was very trivial. Third, all mutations should be 100% inactive; otherwise, the outcome of the computation would not be accurate. In the current report, all Walker B mutants were 100% inactive, since the amino acid E was well conserved in the Walter B motif, and one single E mutation rendered the ATP hydrolysis impossible.26
Supplementary Material
Acknowledgments
Statements of funding: The work was supported by National Institutes of Health grants EB012135 and EB003730 and funding to Peixuan Guo’s Endowed Chair in Nanobiotechnology position from the William Fairish Endowment Fund. The content is solely the responsibility of the authors and does not necessarily represent the official views of National Institutes of Health. The authors would like to thank Jeannie Haak for proofreading this manuscript.
Footnotes
Conflict of interests: PG is a co-founder of Kylin Therapeutics, Inc., and Biomotor and RNA Nanotechnology Development Corp. Ltd.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nano.2014.03.005.
References
- 1.Hess H, Bachand GD, Vogel V. Powering nanodevices with biomolecular motors. Chemistry. 2004;10:2110–2116. doi: 10.1002/chem.200305712. [DOI] [PubMed] [Google Scholar]
- 2.Jankowsky E, Fairman ME, Yang Q. RNA helicases: versatile ATP-driven nanomotors. J Nanosci Nanotechnol. 2005;5:1983–1989. doi: 10.1166/jnn.2005.508. [DOI] [PubMed] [Google Scholar]
- 3.Guo PX, Lee TJ. Viral nanomotors for packaging of dsDNA and dsRNA. Mol Microbiol. 2007;64:886–903. doi: 10.1111/j.1365-2958.2007.05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee BS, Lee SC, Holliday LS. Biochemistry of mechanoenzymes: biological motors for nanotechnology. Biomed Microdevices. 2003;5:269–280. [Google Scholar]
- 6.Shu D, Moll WD, Deng Z, Mao C, Guo P. Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett. 2004;4:1717–1723. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moll D, Huber C, Schlegel B, Pum D, Sleytr UB, Sara M. S-layer-streptavidin fusion proteins as template for nanopatterned molecular arrays. Proc Natl Acad Sci U S A. 2002;99:14646–14651. doi: 10.1073/pnas.232299399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang H, Jing P, Haque F, Guo P. Role of channel lysines and “push through a one-way valve” mechanism of viral DNA packaging motor. Biophys J. 2012;102:127–135. doi: 10.1016/j.bpj.2011.11.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Schwartz C, De Donatis GM, Guo P. “Push through one-way valve” mechanism of viral DNA packaging. Adv Virus Res. 2012;83:415–465. doi: 10.1016/B978-0-12-394438-2.00009-8. [DOI] [PubMed] [Google Scholar]
- 10.Jing P, Haque F, Shu D, Montemagno C, Guo P. One-way traffic of a viral motor channel for double-stranded DNA translocation. Nano Lett. 2010;10:3620–3627. doi: 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc Natl Acad Sci U S A. 2008;105:20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, et al. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat Nanotechnol. 2009;4:765–772. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalczyk SW, Blosser TR, Dekker C. Biomimetic nanopores: learning from and about nature. Trends Biotechnol. 2011;29:607–614. doi: 10.1016/j.tibtech.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Han C, Hou X, Zhang H, Guo W, Li H, Jiang L. Enantioselective recognition in biomimetic single artificial nanochannels. J Am Chem Soc. 2011;133:7644–7647. doi: 10.1021/ja2004939. [DOI] [PubMed] [Google Scholar]
- 16.Craighead HG. Nanoelectromechanical systems. Science. 2000;290:1532–1536. doi: 10.1126/science.290.5496.1532. [DOI] [PubMed] [Google Scholar]
- 17.Fennimore AM, Yuzvinsky TD, Han WQ, Fuhrer MS, Cumings J, Zettl A. Rotational actuators based on carbon nanotubes. Nature. 2003;424:408–410. doi: 10.1038/nature01823. [DOI] [PubMed] [Google Scholar]
- 18.Haque F, Li J, Wu H-C, Liang X-J, Guo P. Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano Today. 2013;8:56–74. doi: 10.1016/j.nantod.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haque F, Lunn J, Fang H, Smithrud D, Guo P. Real-time sensing and discrimination of single chemicals using the channel of Phi29 DNA packaging nanomotor. ACS Nano. 2012;6:3251–3261. doi: 10.1021/nn3001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, Haque F, Rychahou PG, Evers BM, Guo P. Engineered nanopore of Phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum. ACS Nano. 2013;7:9814–9822. doi: 10.1021/nn404435v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerion D, Parak WJ, Williams SC, Zanchet D, Micheel CM, Alivisatos AP. Sorting fluorescent nanocrystals with DNA. J Am Chem Soc. 2002;124:7070–7074. doi: 10.1021/ja017822w. [DOI] [PubMed] [Google Scholar]
- 22.McNally B, Singer A, Yu Z, Sun Y, Weng Z, Meller A. Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays. Nano Lett. 2010;10:2237–2244. doi: 10.1021/nl1012147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandler EL, Smith AL, Burden LM, Kasianowicz JJ, Burden DL. Membrane surface dynamics of DNA-threaded nanopores revealed by simultaneous single-molecule optical and ensemble electrical recording. Langmuir. 2004;20:898–905. doi: 10.1021/la035728i. [DOI] [PubMed] [Google Scholar]
- 24.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nat Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, et al. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today. 2012;7:245–257. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz C, De Donatis GM, Fang H, Guo P. The ATPase of the phi29 DNA-packaging motor is a member of the hexameric AAA + super-family. Virology. 2013;443:20–27. doi: 10.1016/j.virol.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crozat E, Grainge I. FtsK DNA translocase: the fast motor that knows where it's going. Chembiochem. 2010;11:2232–2243. doi: 10.1002/cbic.201000347. [DOI] [PubMed] [Google Scholar]
- 28.Hanson PI, Whiteheart SW. AAA + proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 29.Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Lowe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 30.Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage ϕ29. J Mol Biol. 1987;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 31.Guo P, Zhang C, Chen C, Trottier M, Garver K. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 32.Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, et al. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biology. 2007;5:558–567. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao F, Moll D, Guo S, Guo P. Binding of pRNA to the N-terminal 14 amino acids of connector protein of bacterial phage phi29. Nucleic Acids Res. 2005;33:2640–2649. doi: 10.1093/nar/gki554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz C, De Donatis GM, Zhang H, Fang H, Guo P. Revolution rather than rotation of AAA + hexameric phi29 nanomotor for viral dsDNA packaging without coiling. Virology. 2013;443:28–39. doi: 10.1016/j.virol.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Z, Khisamutdinov E, Schwartz C, Guo P. Mechanism of one-way traffic of hexameric Phi29 DNA packaging motor with four electropositive relaying layers facilitating anti-parallel revolution. ACS Nano. 2013;7:4082–4092. doi: 10.1021/nn4002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo P, Schwartz C, Haak J, Zhao Z. Discovery of a new motion mechanism of biomotors similar to the earth revolving around the sun without rotation. Virology. 2013;446:133–143. doi: 10.1016/j.virol.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrews BT, Catalano CE. Strong subunit coordination drives a powerful viral DNA packaging motor. Proc Natl Acad Sci U S A. 2013;110:5909–5914. doi: 10.1073/pnas.1222820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werbeck ND, Schlee S, Reinstein J. Coupling and dynamics of subunits in the hexameric AAA + chaperone ClpB. J Mol Biol. 2008;378:178–190. doi: 10.1016/j.jmb.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 40.Trottier M, Guo P. Approaches to determine stoichiometry of viral assembly components. J Virol. 1997;71:487–494. doi: 10.1128/jvi.71.1.487-494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee CS, Guo P. A highly sensitive system for the assay of in vitro viral assembly of bacteriophage phi29 of Bacillus subtilis. Virology. 1994;202:1039–1042. doi: 10.1006/viro.1994.1434. [DOI] [PubMed] [Google Scholar]
- 42.Lee TJ, Zhang H, Chang CL, Savran C, Guo P. Engineering of the fluorescent-energy-conversion arm of phi29 DNA packaging motor for single-molecule studies. Small. 2009;5:2453–2459. doi: 10.1002/smll.200900467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang LP, Guo P. Use of acetone to attain highly active and soluble DNA packaging protein gp16 of phi29 for ATPase assay. Virology. 2003;312:449–457. doi: 10.1016/s0042-6822(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 44.Lee CS, Guo P. In vitro assembly of infectious virions of ds-DNA phage ϕ29 from cloned gene products and synthetic nucleic acids. J Virol. 1995;69:5018–5023. doi: 10.1128/jvi.69.8.5018-5023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang LP, Guo P. Use of PEG to acquire highly soluble DNA-packaging enzyme gp16 of bacterial virus phi29 for stoichiometry quantification. J Virol Methods. 2003;109:235–244. doi: 10.1016/s0166-0934(03)00077-6. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz C, Fang H, Huang L, Guo P. Sequential action of ATPase, ATP, ADP, Pi and dsDNA in procapsid-free system to enlighten mechanism in viral dsDNA packaging. Nucleic Acids Res. 2012;40:2577–2586. doi: 10.1093/nar/gkr841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, et al. Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA. 2013;19:1226–1237. doi: 10.1261/rna.037077.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garver K, Guo P. Mapping the inter-RNA interaction of phage phi29 by site-specific photoaffinity crosslinking. J Biol Chem. 2000;275(4):2817–2824. doi: 10.1074/jbc.275.4.2817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.