Summary
Piezo has recently been identified as a family of eukaryotic mechanosensitive channels composed of subunits containing over 2000 amino acids, without recognizable sequence similarity to other channels. Here, we present the crystal structure of a large, conserved extramembrane domain located just before the last predicted transmembrane helix of C. elegans PIEZO, which adopts a novel beta sandwich fold. The structure was also determined of a point mutation located on a conserved surface at the position equivalent to the human PIEZO1 mutation found in Dehydrated Hereditary Stomatocytosis (DHS) patients (M2225R). While the point mutation does not change the overall domain structure, it does alter the surface electrostatic potential that may perturb interactions with a yet-to-be identified ligand or protein. The lack of structural similarity between this domain and any previously characterized fold, including those of eukaryotic and bacterial channels, highlights the distinctive nature of the Piezo family of eukaryotic mechanosensitive channels.
Introduction
The ability to translate environmental cues (chemical, electrical, and mechanical) into intracellular signals is crucial to the functioning of all cells. Although membrane protein receptors for many of these environmental signals have been identified, our understanding of the receptors for mechanical signal transduction remains incomplete. The best characterized models of mechanotransduction are bacterial mechanosensitive (MS) channels that protect cells from osmotic downshock (Levina et al., 1999). In multicellular eukaryotes, mechanosensitive channels are proposed to be involved in diverse physiological and developmental processes, such as somatosensory and auditory detection, change in blood osmolarity, muscle stretch, and others (Chalfie, 2009). The identity of the MS channels involved in these physiological processes remains contentious, however.
A notable exception to the unknown molecular identities of eukaryotic mechanosensors is provided by the Piezo family of mechanosensitive channels (Coste et al., 2012). Piezo was first identified through an siRNA knock-down screen and was shown to be necessary and sufficient for mechanically-activated currents (Coste et al., 2010). Piezos have been found in both multicellular and unicellular eukaryotes with the exception of yeast (Coste et al., 2010; Prole and Taylor, 2013). A striking characteristic of Piezo is the significant size, with all characterized homologs (human, mice, and Drosophila) containing over 2000 amino acids; moreover, mouse PIEZO1 has been demonstrated to form a tetrameric complex corresponding to a total molecular weight for the channel of ~1 MDa (Coste et al., 2012). No sequence similarities have been recognized between Piezo and other types of channels, nor have internal sequence repeats within the Piezo subunit been detected. PIEZO1 can be inhibited by a general blocker of mechanosensitive channels, GsMTx4 (Bae et al., 2011), and is shown to be sensitive to ruthenium red (Coste et al., 2010; Coste et al., 2012). Piezo channels are involved in several physiological processes, such as maintaining normal cell density in zebrafish and mammalian epithelial tissue (Eisenhoffer et al., 2012), mediating mechanical nociception in Drosophila larvae (Kim et al., 2012), and light touch in Rohon-Beard neurons in zebrafish (Faucherre et al., 2013). In humans, point mutations in PIEZO1 have been identified in patients suffering from Dehydrated Hereditary Stomatocytosis (DHS), an autosomal dominant hemolytic anemia disease (Zarychanski et al., 2012; Albuisson et al., 2013; Andolfo et al., 2013). In addition, inactivation of PIEZO1 in lung epithelial cells causes integrin-independent amoeboid cell migration (McHugh et al., 2012).
Understanding the gating mechanism of Piezo will enrich our current knowledge of how mechanical cues are converted into intracellular signals. A challenge in developing a gating model for Piezo arises from the absence of any structural information on the channel. Both the large size and the eukaryotic nature of Piezo create formidable technical challenges to the direct crystallographic analysis of the intact channel. The substantial size suggests, however, that the channel likely consists of smaller domains that may be more amenable to study. Here, we present the crystal structure at 2.5 Å resolution of a Piezo channel soluble domain designated CTL2 (C-Terminal Loop 2), located just before the last transmembrane helix and the C-terminal tail. This loop is predicted to be the largest soluble domain that is conserved across Piezo homologs. Moreover, there is accumulating evidence that the C-terminal portion of Piezo, including the CTL2, is involved in the gating kinetics of the channel. One of the human PIEZO1 point mutants found in DHS patients (M2225R) maps onto this loop and is shown to exhibit a delayed activation as well as slower inactivation (Bae et al., 2013a). A combination of a naturally-occurring mutation (M2225R) and an artificial mutation (R2456K in the C-terminal loop following the last transmembrane helix) in a human PIEZO1 double mutant construct eliminates the inactivation of the channel (Bae et al., 2013b). We also present the structure of C. elegans PIEZO CTL2 domain carrying a point mutation (M31R) at the equivalent position to the mutation found in DHS patients (M2225R). These structures establish that CTL2 adopts a novel beta sandwich fold, and the absence of structural similarity between this domain and any previously characterized fold, including those of eukaryotic and bacterial channels, serves to highlight the distinctive nature of the Piezo family of eukaryotic mechanosensitive channels.
Results
Construct selection
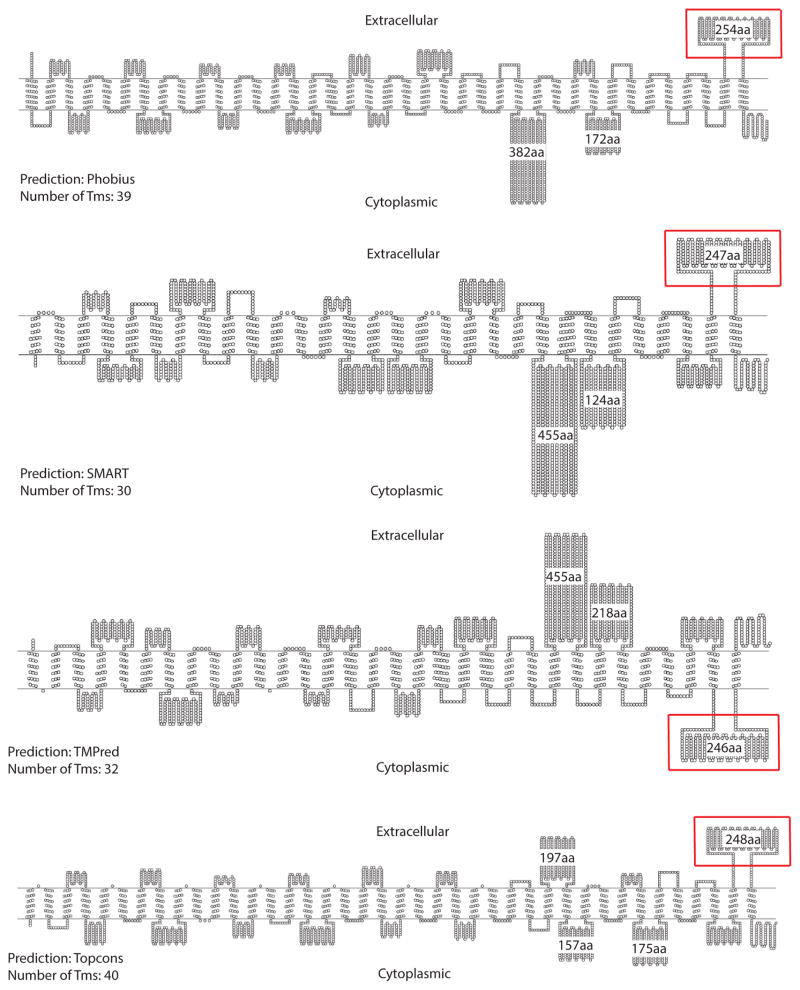
As a starting point for structural analysis of the Piezo channel, we targeted large, well-conserved extramembrane loops. To gain a broad overview of the threading of Piezo protein through the membrane, we used multiple topology prediction webservers: TMHMM2 (Sonnhammer et al., 1998), Phobius (Kall et al., 2007), TOPCONS (Bernsel et al., 2009), TMPred (Hofmann and Stoffel, 1993), and SMART (Schultz et al., 1998). Although similar trends were evident, different algorithms predicted different numbers of transmembrane helices (between 30 and 40) for all characterized Piezos and there were differences in the predicted orientation of some of the extramembrane loops (Fig. 1). For our structural study, we selected a Piezo loop that satisfied the following criteria:
Figure 1. Membrane topology of mouse PIEZO1 as predicted by various membrane topology prediction algorithms.
Depending on the membrane topology prediction algorithm, mouse PIEZO1 is predicted to have 30 to 40 transmembrane segments. The numbers of amino acids (aa) for the larger predicted loops are indicated. The loop of interest, CTL2 (red box), is predicted to be extracellular by 3 of the 4 topology prediction algorithms. Since mouse PIEZO1 is the most extensively characterized Piezo channel, it is used to illustrate the topology predictions; Diagrams are drawn using TOPO2 (Johns) http://www.sacs.ucsf.edu/TOPO2/). See also Figure S1.
it is consistently predicted by all the topology-prediction algorithms used,
it is larger than 100 amino acid residues,
it is conserved across the Piezo family.
Under these constraints, we identified a predicted soluble domain corresponding to mouse PIEZO1 residues 2210 – 2453. These residues form an extramembrane loop preceding the last predicted transmembrane helix that we designate as the C-terminal loop 2 (CTL2) domain. The orientation of CTL2 could not be conclusively defined by the bioinformatics analysis, although 3 of the 4 topology prediction webservers predict that this domain is located in the extracellular environment (Fig. 1). An alignment of Piezo homologs using MAFFT (Fig S1; (Katoh et al., 2002)) reveals that CTL2 contains over 200 amino acid residues with a calculated molecular weight between 24 – 32 kDa, making CTL2 the largest conserved loop among the predicted loops of the Piezo channel. In addition, the Phyre2 webserver (Kelley and Sternberg, 2009) predicts that CTL2 has well-defined secondary structure (Fig S2). Therefore, we selected CTL2 for crystallographic analysis.
Architecture of the C. elegans CTL2 domain
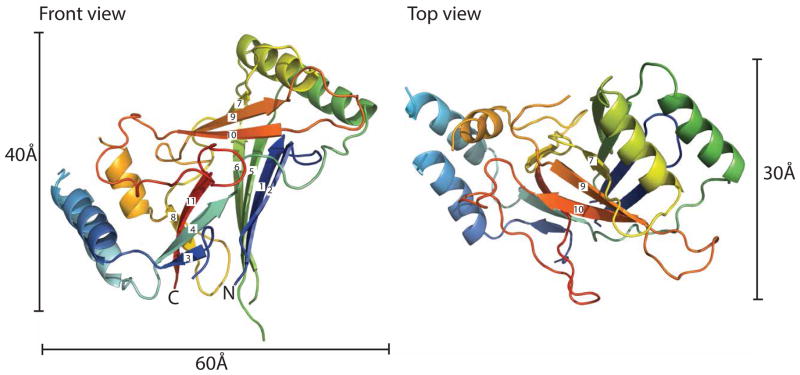
Following a screen of various Piezo orthologues (see Materials and Methods), the C. elegans CTL2 domain was found to be amenable for crystallographic analysis. The structure of wild type C. elegans CTL2 establishes that this domain is organized around a core of three beta sheets flanked by two pairs of helices with overall dimensions of ~60 Å × 40 Å × 30 Å (Fig 2). The secondary structure observed in the crystal structure generally matches that predicted by the Phyre2 webserver (Fig S2). The three beta sheets are arranged such that sheet 1 (strands 1, 2, 5, 6, with strands numbered from the N-terminus) is nearly parallel to sheet 2 (strands 3, 4, 11, 8), and perpendicular to sheet 3 (strands 7, 9, 10).
Figure 2. Structure of the wild type C. elegans CTL2 domain.
The structure of the wild type Piezo CTL2 is colored in a rainbow scheme, progressing from the N-terminus (blue) to C-terminus (red). Front view (left): The loop is oriented so that the connections to the putative transmembrane helices are positioned toward the bottom. The β-strands are numbered sequentially from the N-terminus. Top view (right): the front view, rotated 90° around the horizontal axis. See also Figures S2 and S3.
To identify whether any protein of known structure shares a similar fold with CTL2, we used SSM (Krissinel and Henrick, 2004) as well as the DALI server (Holm and Rosenstrom, 2010) to perform a structure-based alignment. None of the identified proteins exhibited close similarities to CTL2 (Max z-score: 2.8). Most of these candidate proteins possess a beta sandwich fold composed of two beta sheets; this is different than our Piezo soluble domain structure that consists of three beta sheets. However, if we consider sheet 1 and sheet 2 of CTL2 as the beta sandwich core with the rest of the protein as connectors, we obtain a two beta sheet sandwich core with a 2D connectivity diagram illustrated in (Fig S3a). A comparison between the Piezo loop structure and the identified proteins did not reveal any significant connectivity similarity (Fig S3b). Subsequently, we explored the structural classification database CATH (Sillitoe et al., 2013) for two-sheet beta sandwich containing proteins (CATH ID: 2.60). Examination of the strand order in the beta sheets of these proteins failed to identify any close matches between CTL2 and previously solved structures. Therefore, we conclude that Piezo C-terminal soluble domain exhibits a novel beta sandwich fold.
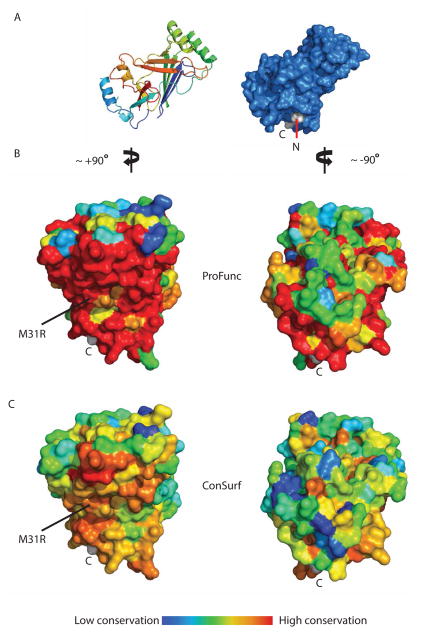
Regions of potential functional significance were identified from the patterns of residue conservation evaluated with the programs ProFunc (Laskowski et al., 2005a, b) and ConSurf (Ashkenazy et al., 2010). Both programs identified the most highly conserved region of CTL2 as consisting of the solvent exposed surface of sheet 1 (Fig 3). The higher conservation observed in the ProFunc analysis reflects the use of 50 of the closest homologs to C. elegans CTL2, while the ConSurf analysis was based on the broader set of Piezo homologues selected from those used to generate Fig S1. Consequently, while the same qualitative trends are observed with both programs, ProFunc found a higher residue conservation at this patch compared to calculation done by ConSurf.
Figure 3. Residue conservation analysis of CTL2.
(A) Surface representation of the WT CTL2 domain (marine blue) is oriented in similar fashion to the one displayed in Figure 2 (left side). The N and C terminus are labeled, with the residues colored in light gray. (B) Surface conservation representation from the closest Piezo homologs as calculated by ProFunc (Laskowski et al., 2005a, b) and displayed on the WT CTL2 domain structure. (C) Surface conservation representation from representative Piezo homologs taken from broad branches of eukaryotic kingdom (Fig S1) as calculated by ConSurf (Ashkenazy et al., 2010). The left panels of B and C show the surface of CTL2 upon ~+90° rotation along the y-axis from the view in A, dominated by the solvent exposed surface of sheet 1, while the right panel shows the surface of CTL2 upon -90° rotation from A (180° from the left panels). Surface conservation is colored in a rainbow gradient scheme in such a way that the least conserved region is colored in blue and the most conserved region is colored in red. M31R is located at the conserved sheet 1 surface of CTL2 displayed in the left panels. See also Figures S2 and S4.
Structure of M31R C. elegans PIEZO CTL2 domain variant
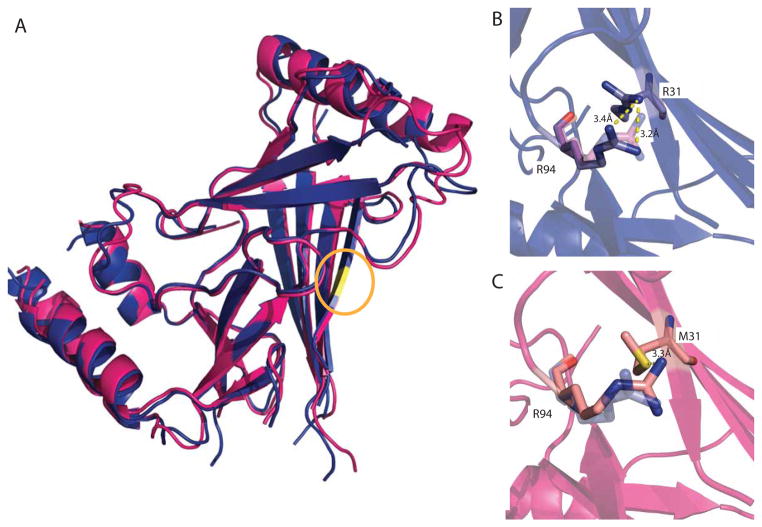
The single substitution in human PIEZO1 of methionine 2225 to arginine has been linked to the disease Dehydration Hereditary Stomatocytosis (DHS) (Zarychanski et al., 2012). From the multiple sequence alignment of Piezo homologs, we determined that M31 of the C. elegans PIEZO CTL2 construct, located in strand 2 of sheet 1, is equivalent to residue M2225 in human PIEZO1 (Fig. S4). Intriguingly, this residue is located in the conserved surface region of CTL2 (Fig. 3). The M31R C. elegans mutant CTL2 protein crystallized in a distinct crystal form from the WT domain and the structure was solved by molecular replacement. Overall, the structure of the mutant CTL2 is similar to the WT structure with some minor rearrangements (RMSD Cα = 1.3 Å overall, with 0.7 Å for residues in α-helices and β-strands (Fig. 4a)). In C. elegans PIEZO CTL2, M31 is spatially adjacent to a neighboring arginine residue, R94 (Fig. 4b). The most significant change in the M31R mutant CTL2 structure is the shift of the R94 sidechain ~3 Å away from R31, presumably due to electrostatic repulsion (Fig. 4b,c, Fig S5). However, R94 in C. elegans PIEZO loop is not conserved in the multiple sequence alignment (Fig. S4). R94 aligns to a glutamate residue that is primarily conserved in the vertebrate Piezo1 and Piezo2. Even though the C. elegans PIEZO CTL2 does not have a glutamate at this position, there are three glutamates in proximity to M31: E30, E32, and E98. This results in a net acidic local environment surrounding M31 in the C. elegans PIEZO.
Figure 4. Superposition of wild type and M31R substituted CTL2 domains.
(A) Superposition of the wild type (pink) and M31R substituted (blue) CTL2 structures. The position of residue 31 is highlighted in yellow (circled). (B, C). The M31R mutant structure is displayed in blue and the WT structure is displayed in pink; this substitution is accompanied by a modest re-arrangement of the R94 side chain. See also Figure S4 and S5.
Discussion
As a starting point for the structural analysis of Piezo, we have identified a predicted extramembrane domain designated as CTL2 (C-terminal Loop 2) as a promising target for three reasons. (1) There is a moderate degree of sequence conservation in this region through all Piezo homologs, which may imply its significance in the general protein function. (2) We were looking for a target loop with at least 100 amino acids because larger domains are more likely to have a stable core structure. With ~250 residues, CTL2 is the largest conserved loop found among Piezo homologues. Moreover, the boundaries at which this loop starts and ends as defined by different topology prediction methods are similar, simplifying the design of our constructs. (3) A point mutation in the CTL2 domain of human PIEZO1 is found in DHS patients. Removal of this loop, together with the last transmembrane segment and the C-terminal tail, severely impaired the inactivation kinetics of the channel (Bae et al., 2013a), leading to an increased ion influx that may contribute to the disease phenotype (Albuisson et al., 2013; Andolfo et al., 2013; Zarychanski et al., 2012).
The model of CTL2 establishes that the region forms a compact domain containing a beta sandwich with three sheets. The CTL2 fold is apparently unrelated to any other family of proteins, including the β sheet domains present in other eukaryotic channels, specifically the extracellular domains of the two eukaryotic ion channels of known structure (Jasti et al., 2007; Kawate et al., 2009), ASIC (CATH ID: 2.60.470.10) and P2X4 (CATH ID: 2.60.490.10). There is no evidence from either structure of the wild type or M31R mutant that this domain forms a tetramer that might help stabilize the oligomeric state of the intact channel. Mutating the conserved methionine residue into arginine (M31R), corresponding to a human DHS mutation (M2225R), does not cause a significant conformational change in C. elegans PIEZO CTL2 domain, suggesting that the effect of this mutation may be to more subtly perturb conformational energetics through changes in electrostatics, or perhaps to interfere with the ability to interact with other domains by changing the interface region.
In conclusion, we have described the first structural characterization of a domain of a eukaryotic mechanosensitive channel, the CTL2 domain of C. elegans PIEZO. We have shown that this extramembrane region forms a stable domain that adopts a previously uncharacterized beta sandwich fold. The functional role of this domain in the context of the full length channel remains unclear. In view of the monomeric nature of the CTL2 domain, it is unlikely to mediate tetramerization of the full length channel. As this domain is generally conserved across Piezo homologs, it may have a general role such as mediating proper intracellular trafficking, tethering the intact channel to one or more proteins, or binding of a ligand; the search for binding partners is currently under investigation. The lack of structural similarity between this domain and any known protein fold, including those of eukaryotic and bacterial channels, reinforces the lack of recognizable sequence homology between Piezo and other channels, and highlights the distinctive nature of this enigmatic family of eukaryotic mechanosensitive channels.
Experimental Procedures
Protein cloning and expression
The CTL2 regions from the mouse PIEZO1, mouse PIEZO2, human PIEZO1, Drosophila PIEZO and C. elegans PIEZO orthologues were cloned into a pET21 vector between the NdeI and XhoI restriction sites, generating a construct with a C-terminal 6xHis tag. The plasmid DNA was transformed into BL-21 gold E. coli cells (Stratagene). Cells were grown in Terrific Broth medium until reaching an OD600 of 1.0, when 1 mM IPTG was added. Protein induction was carried on overnight at 20 °C. The CTL2 of C. elegans PIEZO (GenBank ID: 392900032) was found to be best suited for structural analysis; the protein and nucleotide sequences of the coding region for this construct are: MSLLNQIGTISMPEKVTLRISIEGYPPLYEMEAQGSNHDNAELGMIKPDQLASLNQALTDSYTTRDTNSILRSRMSVSYLKGYTYEDILIVRFRPESEIYWPISQDSRNAMIDKLSRNTSVNFEVSLEFTRPYDPNENAALKHSKSWLVPISLDMTIRAKIQSALRGDPGHPILIPQSIPAFIQVPNQGELTLPTSIGNTIINDGNPRINTTGMEKSDEARAWFDSLTLNLEQGKSQNEKMWIATSEHPGDQNAKLWIKTANTTYSGRPYLQVVGFIDRAFPSLEHHHHHHATGTCGCTACTCAATCAAATCGGAACTATCAGTATGCCAGAAAAAGTTACGTTGAGGATTTCGATTGAAGGATATCCACCACTTTACGAGATGGAGGCACAAGGAAGTAATCACGATAACGCTGAACTTGGAATGATAAAACCTGATCAATTGGCATCTTTGAATCAAGCATTGACGGATAGTTACACGACACGAGACACTAATTCTATTTTAAGATCAAGAATGTCTGTATCATATCTGAAAGGATACACATATGAGGATATTCTGATTGTGCGATTCCGTCCAGAATCTGAAATTTATTGGCCGATTTCACAGGATTCGAGAAATGCGATGATCGATAAATTGAGCCGAAACACGTCAGTCAATTTTGAAGTATCTCTGGAATTCACTCGGCCATATGATCCAAATGAAAACGCTGCTTTAAAACATTCGAAATCGTGGCTTGTTCCAATCTCCCTGGATATGACGATTCGAGCAAAAATTCaaAGTGCTCTCCGAGGAGATCCAGGTCATCCGATTCTCATTCCACAGTCAATTCCTGCGTTTATTCAAGTTCCAAATCAAGGAGAATTGACCCTTCCAACATCAATCGGAAATACTATTATCAATGACGGAAATCCACGGATTAATACGACCGGGATGGAAAAATCAGACGAAGCTCGGGCTTGGTTCGACTCGTTAACCTTGAATCTCGAACAAGGAAAATCGCAAAACGAGAAAATGTGGATCGCCACATCCGAACATCCAGGAGACCAAAATGCGAAACTCTGGATAAAGACTGCAAATACGACGTATTCGGGAAGGCCGTACCTTCAAGTTGTCGGATTTATTGATAGAGCATTCCC ATCACTCGAGCACCACCACCACCACCACTGA
Protein residue numbers are assigned according to their position in this CTL2 construct.
Protein purification
Cells were homogenized in lysis buffer (20 mM Tris-HCl pH 8.0, 50 mM NaCl, lysozyme, DNAse, PMSF) at a ratio of 1 g cells/10 mL of buffer. Cells were lysed using a microfluidizer (Microfluidics, Model M-110L), and the lysate was centrifuged to remove insoluble material. The protein was purified in three steps. First, lysate was run through a NiNTA column, washed with 40 mM imidazole for 15 column volumes, and eluted with 250 mM imidazole. Next, the eluate from the NiNTA column was run through a Superdex-200 16/60 column (GE Healthcare), and the corresponding peak fraction was collected. Finally, the resulting protein fraction was run through an anion exchange column, HiTrap Q Sepharose FF (GE Healthcare), to obtain the final protein preparation. The identities of the purified wild type (WT) and M31R variant proteins were confirmed by mass spectrometry.
Crystallization & model refinement
WT CTL2 was crystallized by the sitting drop vapor diffusion method against a reservoir containing 0.1 M phosphate-citrate buffer pH 4.0, 26% PEG1000, and 0.2 M Li2SO4 at 20 °C in a 96-well plate, at a ratio 0.2 μL protein to 0.2 μL reservoir drop, using intelli sitting drop crystallization plates (ArtRobbins). The protein concentration was 10–12 mg/mL. WT CTL2 crystals appeared after 18 hrs and grew to maturation after 3 days to approximate dimensions of 50 μm × 50 μm × 150 μm. M31R CTL2 was crystallized against a reservoir containing 0.1 M phosphate-citrate buffer pH 4.8, 30% PEG3350, and 0.2 M Li2SO4 at 20 °C using similar procedures. These crystals grew more slowly than the WT crystals.
A heavy atom derivative of the WT CTL2 crystals was prepared by soaking in 10 mM potassium tetranitroplatinate (II) for three days, followed by back-soaking in the mother liquor for 2 hrs. Protein crystals were flash frozen in the presence of Fomblin Y 16/6 oil (Sigma), and were sent to the Stanford Synchrotron Radiation Lightsource (SSRL).
Diffraction data were collected at SSRL beamline 12-2 equipped with a PILATUS 6M PAD detector
Diffraction patterns were indexed and integrated using iMOSFLM (Battye et al., 2011) from the CCP4 software package (Potterton et al., 2003) and XDS (Kabsch, 2010). The integrated data was scaled using SCALA (Evans, 2006). Phases for the WT CTL2 were obtained experimentally by Single-wavelength Anomalous Dispersion (SAD) phasing based on the platinum derivative. Five platinum sites were located, and electron density maps were generated using Autosol (Terwilliger et al., 2009) from the PHENIX suite (Zwart et al., 2008). Phases for the M31R CTL2 structure were generated with PhaserMR (McCoy, 2007) by molecular replacement using the WT structure. Model building was done in Coot (Emsley and Cowtan, 2004), and the structure refinements were done using Refmac5 (Murshudov et al., 2011). The final molecular structures were displayed using PyMOL (Schrodinger, 2010).
Supplementary Material
Figure S1, related to Figure 1. Multiple sequence alignment of Piezo homologs.
Multiple sequence alignment was performed using representative Piezo homologs (listed below) from vertebrate Piezo1, vertebrate Piezo2, invertebrate Piezo, plant Piezo, and unicellular eukaryote Piezo. Protein sequence conservation is represented by the shading and height of the bar corresponding to each residue, with the tall yellow bar, short brown bar and no bar representing high levels, low levels and poor sequence conservation, respectively. The green bars highlight the transmembrane (TM) segment for mouse PIEZO1 as predicted by Topcons (Bernsel et al., 2009). CTL2 (shaded in light blue) is the largest conserved Piezo soluble domain. The Piezo sequence conservation is relatively higher in the C-terminal region. The diagram was generated using the MAFFT multiple sequence alignment program (Katoh et al., 2002), available at the EMBL-EBI web page.
List of Piezo homologs used:
H. sapiens 1 (NP_001136336.2), D. rerio 1 (XP_696355.4), R. norvegicus 1 (NP_001070668.2), M. lucifugus 1 (XP_006097426.1), O. garnettii 1 (XP_003800876.1), B. Taurus 1 (XP_001256011.4), S. harrisii 1 (XP_003758542.1), G. gorilla 1 (XP_004058200.1), C. porcellus 1 (XP_003460961.1), E. caballus 1 (XP_005615040.1), P. alecto 1 (XP_006927190.1), M. domestica 1 (XP_007477361.1), L. Chalumnae 1 (XP_006002160.1), T. rubripes 1 (XP_003978351.1), Anolis carolinensis 1 (XP_008120472.1), C. lupus 1 (XP_005620631.1), M. gallopavo 1 (XP_003209947.1), S. partitus 1 (XP_008279724.1), X. tropicalis 1 (XP_002933721.2), C. simum 1 (XP_004437180.1), H. glaber 1 (XP_004843264.1), O. rosmarus 1 (XP_004392217.1), M. musculus 1 (NP_001032375.1), O. orca 1 (XP_004280184.1), M. musculus 2 (NP_001034574.4), G. Gallus 2 (XP_419138.4), H. sapiens 2 (NP_071351.2), B. Taurus 2 (XP_003587835.2), C. porcelius 2 (XP_005001285.1), C. simum 2 (XP_004437180.1), H. glaber 2 (XP_004843264.1), M. lucifugus 2 (XP_006089356.1), C. millii 2 (XP_007887509.1), O. orca 2 (XP_004276120.1), O. aries 2 (XP_004020692.1), G. gorilla 2 (XP_004059237.1), O. rosmarus 2 (XP_004417033.1), M. lucifugus 2 (XP_006089356.1), X. tropicalis 2 (XP_002937522.2), C. lupus 2 (XP_005623199.1), M. gallopavo 2 (XP_003205004.1), A. platyrhynchos 2 (XP_005013163.1), T. guttata 2 (XP_002192627.2), S. harrisii 2 (XP_003760113.1), A. aegyptii (XP_001657818.1), B. impatiens (XP_003494661.1), H. saltator (EFN75267.1), C. elegans (CAA92491.3), C. sinensis (GAA51253.1), D. melanogaster (AFB77909.1), M occidentalis (XP_003747214.1), P. humanus (XP_002428649.1), N. vitripennis (XP_008202351.1), E. histolytica (XP_655549.2), P. tetraurelia (XP_001461126.1), O. trifallax (EJY84567.1), T. cruzi (EKG00857.1), S. lycopersicum (XP_004247483.1), G. max (XP_006605262.1), T. cacao (XP_007030785.1), A. thaliana (NP_182327.6), C rubella (XP_006293550.1), P. persica (XP_007200947.1), O. tauri (XP_003079754.1)
Figure S2, related to Figures 2 and 3. Comparison of the secondary structure of the WT C. elegans CTL2 from the crystal structure and from the Phyre2 prediction.
The observed secondary structure in the crystal structure of C. elegans CTL2 (drawn below the ‘Disorder’ row) generally resembles the secondary structure assignments predicted by Phyre2 (Kelley and Sternberg, 2009), (drawn below the ‘Sequence’ row). Amino acid residues that are not visible in the electron density of either the wild type or M31R mutant structures (shaded in purple) coincide with the predicted disordered region. The sequence conservation representation follows the convention used in Fig S1, with gaps in the multiple sequence alignment indicated by the magenta dotted lines.
Figure S3, related to Figure 2. Comparison of the beta strand connectivity of the Piezo CTL2 domain to potentially related domains.
Each beta strand in the connectivity diagram is drawn as an arrow. Beta strands that are parallel to the first beta strand are colored blue, while those that are antiparallel are colored yellow (A). Connectivity diagrams for each of the 14 closest matches to the Piezo CTL2 loop identified by DALI (Holm and Rosenstrom, 2010) and SSM (Krissinel and Henrick, 2004) are summarized in table (B). No exact matches in strand orientation are observed between CTL2 and these candidate structural homologs.
Figure S4, related to Figures 3 and 4. Multiple sequence alignment of Piezo homologs around the M31 mutation site.
A methionine residue that is mutated into arginine in DHS patients (colored in red) is conserved across Piezo homologs. The position of this methionine (M31 in our C. elegans CTL2 construct) is in close proximity to an arginine residue (colored in blue) that is not conserved. There are three glutamate residues (colored in purple) around M31 that provide a net negative charge at this location.
Figure S5, related to Figure 4. Substitution of M31 with Arg leads to repositioning of the R94 sidechain.
The structure of the wild type C. elegans PIEZO CTL2 structure is displayed in magenta, while the M31R mutant structure is displayed in blue. A: the electron density of WT Piezo loop is displayed in magenta. B: the electron density of the M31R mutant CTL2 domain is displayed in blue.
Table 1.
Data collection and refinement statistics of the wild type and mutant CTL2 domains.
| Data set | Wildtype CTL2 | CTL2 M31R |
|---|---|---|
| Protein Data Bank number | 4PKE | 4PKX |
| Data collection statistics | ||
| Space group | H 3 2 | P 1 21 1 |
| a, b, c (Å) | 72.59, 72.59, 241.6 | 45.18, 37.25, 73.56 |
| β, β, γ (°) | 90, 90, 120 | 90, 91.58, 90 |
| Wavelength (Å) | 1.0716 | 1.0332 |
| Resolution (Å)a | 33.09 – 2.45 (2.57 – 2.45) | 38.97 – 2.54 (2.68 – 2.54) |
| No. of unique reflection a | 9377 (1211) | 7746 (887) |
| Mean (I/sigma I) a | 17.7 (3.0) | 28.4 (11.2) |
| Completeness a | 99.9% (100%) | 93.6% (75.4%) |
| Average multiplicity a | 10.1 (10.6) | 3.1 (2.8) |
| Anom. Completeness a | 99.9% (99.9%) | |
| DelAnom. correlation between half-sets a | 0.915 (0.061) | |
| R-merge a | 0.093 (0.810) | 0.028 (0.085) |
| Matthew Coefficient | 1.951 | 1.92 |
| Refinement statistics | ||
| Resolution | 31.17 – 2.50 | 33.23 – 2.54 |
| Average B factor (Å2) | 83.9 | 38.6 |
| Rwork/Rfree (%) | 22.81/23.84 | 21.79/25.91 |
| Rmsd bonds (Å) | 0.006 | 0.004 |
| Rmsd angles (°) | 1.23 | 0.97 |
| Ramachandran plot (%) | ||
| Favored | 97.5 | 98.2 |
| Outliers | 0 | 0 |
| Rotamer outliers (%) | 5.9 | 2.9 |
| Molprobity clashscore | 13.8 | 12.8 |
| Molprobity overall score | 2.34 | 1.97 |
Values in parentheses represents the highest resolution shell
Highlights.
A conserved loop of the Piezo channel is identified that forms a stable domain.
The structure of this domain reveals a novel beta sandwich fold.
The novel fold highlights the distinctive nature of the Piezo channel family.
A disease related mutation in this domain has minimal structural consequences.
Acknowledgments
We thank S.E. Kim and A. Patapoutian for providing the C. elegans PIEZO construct, S. Hess from the Proteome Exploration Laboratory at the Beckman Institute at the California Institute of Technology for mass spectrometry analysis of the protein crystals, and P. Nikolovski from the Molecular Observatory at the Beckman Institute at the California Institute of Technology. This research was supported by National Institute of Health (NIH GM84211). We acknowledge the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi-Aventis Bioengineering Research Program and the Beckman Institute at Caltech for their generous support of the Molecular Observatory at Caltech. Operations at SSRL are supported by the US DOE and NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albuisson J, Murthy SE, Bandell M, Coste B, Louis-Dit-Picard H, Mathur J, Feneant-Thibault M, Tertian G, de Jaureguiberry JP, Syfuss PY, et al. Dehydrated hereditary stomatocytosis linked to gain-of-function mutations in mechanically activated PIEZO1 ion channels. Nat Commun. 2013;4:1884. doi: 10.1038/ncomms2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfo I, Alper SL, De Franceschi L, Auriemma C, Russo R, De Falco L, Vallefuoco F, Esposito MR, Vandorpe DH, Shmukler BE, et al. Multiple clinical forms of dehydrated hereditary stomatocytosis arise from mutations in PIEZO1. Blood. 2013;121:3925–3935. S3921–3912. doi: 10.1182/blood-2013-02-482489. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae C, Gnanasambandam R, Nicolai C, Sachs F, Gottlieb PA. Xerocytosis is caused by mutations that alter the kinetics of the mechanosensitive channel PIEZO1. Proc Natl Acad Sci U S A. 2013a;110:E1162–1168. doi: 10.1073/pnas.1219777110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae C, Gottlieb PA, Sachs F. Human PIEZO1: removing inactivation. Biophys J. 2013b;105:880–886. doi: 10.1016/j.bpj.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae C, Sachs F, Gottlieb PA. The mechanosensitive ion channel Piezo1 is inhibited by the peptide GsMTx4. Biochemistry. 2011;50:6295–6300. doi: 10.1021/bi200770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37:W465–468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M. Neurosensory mechanotransduction. Nat Rev Mol Cell Biol. 2009;10:44–52. doi: 10.1038/nrm2595. [DOI] [PubMed] [Google Scholar]
- Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE, Patapoutian A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B, Xiao B, Santos JS, Syeda R, Grandl J, Spencer KS, Kim SE, Schmidt M, Mathur J, Dubin AE, et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature. 2012;483:176–181. doi: 10.1038/nature10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhoffer GT, Loftus PD, Yoshigi M, Otsuna H, Chien CB, Morcos PA, Rosenblatt J. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 2012;484:546–549. doi: 10.1038/nature10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Faucherre A, Nargeot J, Mangoni ME, Jopling C. piezo2b Regulates Vertebrate Light Touch Response. J Neurosci. 2013;33:17089–17094. doi: 10.1523/JNEUROSCI.0522-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMBASE - A database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- Johns SJ. TOPO2, Transmembrane protein display software. [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35:W429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature. 2009;460:592–598. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kim SE, Coste B, Chadha A, Cook B, Patapoutian A. The role of Drosophila Piezo in mechanical nociception. Nature. 2012;483:209–212. doi: 10.1038/nature10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005a;33:W89–93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Watson JD, Thornton JM. Protein function prediction using local 3D templates. J Mol Biol. 2005b;351:614–626. doi: 10.1016/j.jmb.2005.05.067. [DOI] [PubMed] [Google Scholar]
- Levina N, Totemeyer S, Stokes NR, Louis P, Jones MA, Booth IR. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 1999;18:1730–1737. doi: 10.1093/emboj/18.7.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh BJ, Murdoch A, Haslett C, Sethi T. Loss of the integrin-activating transmembrane protein Fam38A (Piezo1) promotes a switch to a reduced integrin-dependent mode of cell migration. PloS one. 2012;7:e40346. doi: 10.1371/journal.pone.0040346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Prole DL, Taylor CW. Identification and analysis of putative homologues of mechanosensitive channels in pathogenic protozoa. PloS one. 2013;8:e66068. doi: 10.1371/journal.pone.0066068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillitoe I, Cuff AL, Dessailly BH, Dawson NL, Furnham N, Lee D, Lees JG, Lewis TE, Studer RA, Rentzsch R, et al. New functional families (FunFams) in CATH to improve the mapping of conserved functional sites to 3D structures. Nucleic Acids Res. 2013;41:D490–498. doi: 10.1093/nar/gks1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings/International Conference on Intelligent Systems for Molecular Biology; ISMB International Conference on Intelligent Systems for Molecular Biology. 1998;6:175–182. [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarychanski R, Schulz VP, Houston BL, Maksimova Y, Houston DS, Smith B, Rinehart J, Gallagher PG. Mutations in the mechanotransduction protein PIEZO1 are associated with hereditary xerocytosis. Blood. 2012;120:1908–1915. doi: 10.1182/blood-2012-04-422253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1. Multiple sequence alignment of Piezo homologs.
Multiple sequence alignment was performed using representative Piezo homologs (listed below) from vertebrate Piezo1, vertebrate Piezo2, invertebrate Piezo, plant Piezo, and unicellular eukaryote Piezo. Protein sequence conservation is represented by the shading and height of the bar corresponding to each residue, with the tall yellow bar, short brown bar and no bar representing high levels, low levels and poor sequence conservation, respectively. The green bars highlight the transmembrane (TM) segment for mouse PIEZO1 as predicted by Topcons (Bernsel et al., 2009). CTL2 (shaded in light blue) is the largest conserved Piezo soluble domain. The Piezo sequence conservation is relatively higher in the C-terminal region. The diagram was generated using the MAFFT multiple sequence alignment program (Katoh et al., 2002), available at the EMBL-EBI web page.
List of Piezo homologs used:
H. sapiens 1 (NP_001136336.2), D. rerio 1 (XP_696355.4), R. norvegicus 1 (NP_001070668.2), M. lucifugus 1 (XP_006097426.1), O. garnettii 1 (XP_003800876.1), B. Taurus 1 (XP_001256011.4), S. harrisii 1 (XP_003758542.1), G. gorilla 1 (XP_004058200.1), C. porcellus 1 (XP_003460961.1), E. caballus 1 (XP_005615040.1), P. alecto 1 (XP_006927190.1), M. domestica 1 (XP_007477361.1), L. Chalumnae 1 (XP_006002160.1), T. rubripes 1 (XP_003978351.1), Anolis carolinensis 1 (XP_008120472.1), C. lupus 1 (XP_005620631.1), M. gallopavo 1 (XP_003209947.1), S. partitus 1 (XP_008279724.1), X. tropicalis 1 (XP_002933721.2), C. simum 1 (XP_004437180.1), H. glaber 1 (XP_004843264.1), O. rosmarus 1 (XP_004392217.1), M. musculus 1 (NP_001032375.1), O. orca 1 (XP_004280184.1), M. musculus 2 (NP_001034574.4), G. Gallus 2 (XP_419138.4), H. sapiens 2 (NP_071351.2), B. Taurus 2 (XP_003587835.2), C. porcelius 2 (XP_005001285.1), C. simum 2 (XP_004437180.1), H. glaber 2 (XP_004843264.1), M. lucifugus 2 (XP_006089356.1), C. millii 2 (XP_007887509.1), O. orca 2 (XP_004276120.1), O. aries 2 (XP_004020692.1), G. gorilla 2 (XP_004059237.1), O. rosmarus 2 (XP_004417033.1), M. lucifugus 2 (XP_006089356.1), X. tropicalis 2 (XP_002937522.2), C. lupus 2 (XP_005623199.1), M. gallopavo 2 (XP_003205004.1), A. platyrhynchos 2 (XP_005013163.1), T. guttata 2 (XP_002192627.2), S. harrisii 2 (XP_003760113.1), A. aegyptii (XP_001657818.1), B. impatiens (XP_003494661.1), H. saltator (EFN75267.1), C. elegans (CAA92491.3), C. sinensis (GAA51253.1), D. melanogaster (AFB77909.1), M occidentalis (XP_003747214.1), P. humanus (XP_002428649.1), N. vitripennis (XP_008202351.1), E. histolytica (XP_655549.2), P. tetraurelia (XP_001461126.1), O. trifallax (EJY84567.1), T. cruzi (EKG00857.1), S. lycopersicum (XP_004247483.1), G. max (XP_006605262.1), T. cacao (XP_007030785.1), A. thaliana (NP_182327.6), C rubella (XP_006293550.1), P. persica (XP_007200947.1), O. tauri (XP_003079754.1)
Figure S2, related to Figures 2 and 3. Comparison of the secondary structure of the WT C. elegans CTL2 from the crystal structure and from the Phyre2 prediction.
The observed secondary structure in the crystal structure of C. elegans CTL2 (drawn below the ‘Disorder’ row) generally resembles the secondary structure assignments predicted by Phyre2 (Kelley and Sternberg, 2009), (drawn below the ‘Sequence’ row). Amino acid residues that are not visible in the electron density of either the wild type or M31R mutant structures (shaded in purple) coincide with the predicted disordered region. The sequence conservation representation follows the convention used in Fig S1, with gaps in the multiple sequence alignment indicated by the magenta dotted lines.
Figure S3, related to Figure 2. Comparison of the beta strand connectivity of the Piezo CTL2 domain to potentially related domains.
Each beta strand in the connectivity diagram is drawn as an arrow. Beta strands that are parallel to the first beta strand are colored blue, while those that are antiparallel are colored yellow (A). Connectivity diagrams for each of the 14 closest matches to the Piezo CTL2 loop identified by DALI (Holm and Rosenstrom, 2010) and SSM (Krissinel and Henrick, 2004) are summarized in table (B). No exact matches in strand orientation are observed between CTL2 and these candidate structural homologs.
Figure S4, related to Figures 3 and 4. Multiple sequence alignment of Piezo homologs around the M31 mutation site.
A methionine residue that is mutated into arginine in DHS patients (colored in red) is conserved across Piezo homologs. The position of this methionine (M31 in our C. elegans CTL2 construct) is in close proximity to an arginine residue (colored in blue) that is not conserved. There are three glutamate residues (colored in purple) around M31 that provide a net negative charge at this location.
Figure S5, related to Figure 4. Substitution of M31 with Arg leads to repositioning of the R94 sidechain.
The structure of the wild type C. elegans PIEZO CTL2 structure is displayed in magenta, while the M31R mutant structure is displayed in blue. A: the electron density of WT Piezo loop is displayed in magenta. B: the electron density of the M31R mutant CTL2 domain is displayed in blue.