Abstract
Receptor tyrosine kinases (RTKs) are a large family of cell surface receptors that sense growth factors and hormones and regulate a variety of cell behaviours in health and disease. Contactless activation of RTKs with spatial and temporal precision is currently not feasible. Here, we generated RTKs that are insensitive to endogenous ligands but can be selectively activated by low-intensity blue light. We screened light-oxygen-voltage (LOV)-sensing domains for their ability to activate RTKs by light-activated dimerization. Incorporation of LOV domains found in aureochrome photoreceptors of stramenopiles resulted in robust activation of the fibroblast growth factor receptor 1 (FGFR1), epidermal growth factor receptor (EGFR) and rearranged during transfection (RET). In human cancer and endothelial cells, light induced cellular signalling with spatial and temporal precision. Furthermore, light faithfully mimicked complex mitogenic and morphogenic cell behaviour induced by growth factors. RTKs under optical control (Opto-RTKs) provide a powerful optogenetic approach to actuate cellular signals and manipulate cell behaviour.
Keywords: aureochrome, cell signalling, light-oxygen-voltage (LOV)-sensing domain, optogenetics, synthetic biology, synthetic physiology
Introduction
Cell surface receptors act as relays between extracellular stimuli and intracellular signalling cascades and thereby play vital and diverse roles in normal physiology and disease (Robertson et al, 2000; Ashcroft, 2006; Lagerstrom & Schioth, 2008; Casaletto & McClatchey, 2012). A wide range of techniques has been developed to manipulate receptor signalling in order to study how activation affects cell behaviour. Genetic approaches enable conditional and cell type-specific overexpression and deletion of receptor genes, and pharmacological approaches permit activation and inhibition of selected receptor families. Current evidence suggests that receptor signalling is spatially and dynamically regulated. However, it remains challenging to activate or inactivate receptors on timescales shorter than those defined by receptor biosynthesis and degradation, or by drug diffusion and pharmacokinetics. It is further difficult to manipulate receptor function locally, such as in compartments of cells or in selected cells for which no specific genetic markers are available.
Recent work in neurobiology has harnessed the power of light for the control of cellular activity with spatial and temporal precision (Szobota & Isacoff, 2010; Fenno et al, 2011). General advantages of optogenetics over pharmacology and genetics include the ease with which signals can be applied and withdrawn and the ability of rapid and local signal activation. Complementary to light-sensing ion channels and ion pumps, which are now utilized routinely, two approaches were recently developed for optogenetic activation specifically of G protein-coupled receptors (GPCRs). Chimeric proteins combine domains of light-sensing GPCRs with domains of ligand-responsive GPCRs (Airan et al, 2009), while photochromic ligands are synthetic compounds that activate GPCRs by mimicking low-molecular-weight ligands (Levitz et al, 2013). It has been argued that light-activated proteins that perform comparably to physiologically relevant signalling molecules are difficult to engineer (Fenno et al, 2011; Christie et al, 2012; Muller & Weber, 2013; Pathak et al, 2013). The challenges include avoiding background signalling in the absence of light and achieving physiological levels of pathway activation. Indeed, for receptor families other than GPCRs, activation with light has not been achieved.
Here, we utilized rational protein engineering to design receptor tyrosine kinases (RTKs) that are under optical control (Opto-RTKs). RTKs are a large and essential surface receptor family that senses growth factors and hormones. Because extracellular ligands induce and stabilize RTK dimers (Lemmon & Schlessinger, 2010; Simi & Ibanez, 2010), we reasoned that light-activated dimerization may induce receptor activation with light. We first identified protein domains that undergo homodimerization in response to blue light, and we then used these domains to induce receptor trans-phosphorylation and signal propagation to intracellular pathways. Finally, we applied Opto-RTKs to control cellular signalling and cell behaviour in human cancer and endothelial cells with spatial and temporal precision.
Results
Selection and expression of light-sensing proteins
We selected blue light-sensing protein domains that belong to the large light-oxygen-voltage (LOV)-sensing domain superfamily as candidates for light-activated dimerization of RTKs. Light-sensing LOV domains bind flavins as prosthetic groups and act as reversible light sensors in bacteria, fungi and plants. LOV domains control heterogeneous effector domains such as enzymes [for instance, in the flowering plant Arabidopsis thaliana and the green alga Chlamydomonas reinhardtii (Huang et al, 2002; Kinoshita et al, 2001)] or transcriptional regulators [for instance, in the fungus Neurospora crassa and the yellow-green alga Vaucheria frigida (Heintzen et al, 2001; Takahashi et al, 2007)]. Dimerization of LOV domains was proposed to play an important role in effector domain regulation, and a remarkable diversity in dimerization interfaces and lifetimes was observed among LOV domains (Moglich et al, 2010; Zoltowski & Gardner, 2011). As it was initially unclear whether a LOV domain would be suited to activate mammalian RTKs, we compiled an unbiased panel of diverse candidate domains (one from a proteobacterium, two from a fungus, two from algae and two from a plant; Fig1A, Supplementary Tables S1 and S2, and Materials and Methods). As the majority of these domains was never studied in mammalian cells, we first tested in two mammalian cell lines whether the domains can be expressed and whether expression causes cytotoxicity. We found that the domains were produced efficiently (as assessed by detection of a fluorescent protein fused to the domains; Fig1B and D) and with no detectable cytotoxicity (as assessed by cellular reduction of a tetrazolium dye; Fig1C and E).
Figure 1. Selection of LOV domains and expression in mammalian cells.
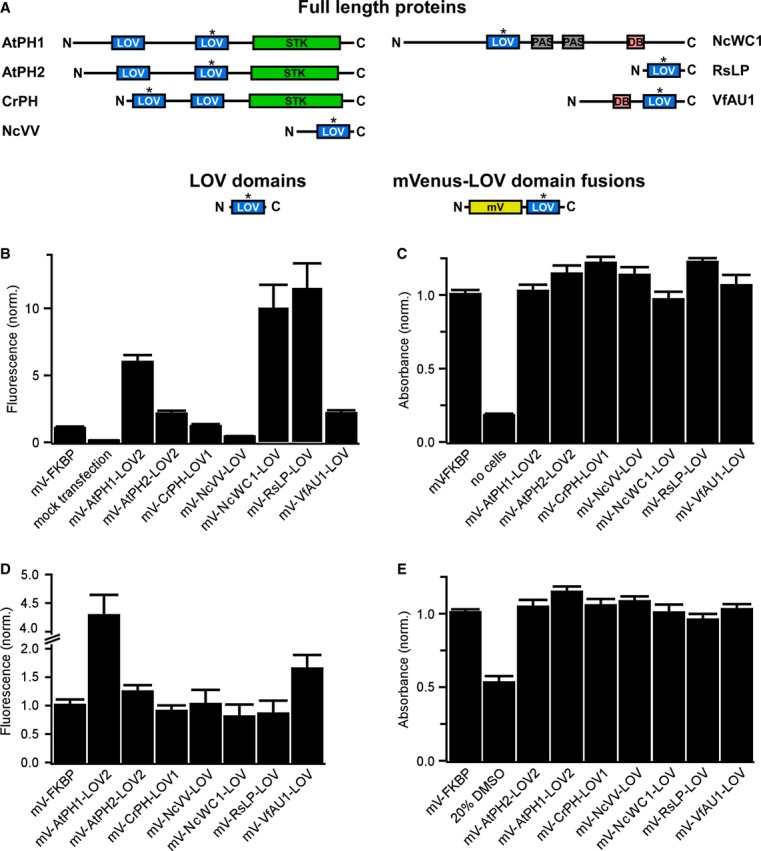
- Domain structure of light-sensing proteins from which LOV domains (highlighted with asterisk) were excised (AtPH1 and AtPH2, Arabidopsis thaliana phototropin 1 and 2; CrPH, Chlamydomonas rheinhardtii phototropin; NcVV, Neurospora crassa vivid; NcWC1, N. crassa white collar 1; RsLP, Rhodobacter sphaeroides ATCC 17025 light-sensing protein; VfAU1, Vaucheria frigida aureochrome1). In these proteins, LOV domains regulate a variety of effector domains (STK, serine/threonine kinase; DB, DNA-binding domain). To test for expression and influence on cell viability in mammalian cells, LOV domains optimized for mammalian codon usage were fused to the fluorescent protein mVenus (mV).
- Fluorescence intensity measurements of human embryonic kidney (HEK) 293 cells transfected with mVenus-LOV domain fusions.
- Viability of HEK293 cells transfected with mVenus-LOV domain fusions.
- Fluorescence intensity measurements of Chinese hamster ovary (CHO) K1 cells transfected with mVenus-LOV domain fusions.
- Viability of CHO K1 cells transfected with mVenus-LOV domain fusions.
Data information: For (B–E): fluorescence and viability were quantified 16–18 h after transfection. Data were normalized to mV fused to the small, robustly folding FK506 binding protein (FKBP). Mean values ± SD for three independent experiments each performed in quadruplicates are shown.
Engineering a light-activated fibroblast growth factor receptor
Our experiments focused on fibroblast growth factor (FGF) receptor 1, a highly conserved key regulator of cell behaviour in, for instance, embryonic development, adult neurogenesis and tumour formation (Deng et al, 1994; Zhao et al, 2007; Yang et al, 2013). We constructed chimeric receptors where each of the seven LOV domains was linked to the intracellular catalytic domain of murine FGFR1 (mFGFR1; Fig2A). The extracellular ligand-binding modules of mFGFR1 were omitted to obtain chimeras that are not responsive to native ligands. Cells expressing these chimeras should respond to blue light with activation of signalling pathways characteristic for mFGFR1. We performed cell signalling experiments in a custom-built incubator that allows illumination of cells and tissues with light of defined intensity and colour (Materials and Methods). We first examined the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) pathway, a key signalling pathway that is activated by FGFs via FGFRs and highly dynamic (Ma et al, 2009; Albeck et al, 2013). To quantify MAPK/ERK pathway activation, we utilized a reporter assay that enables simultaneous evaluation of chimeras in multi-well plates. In this assay, luciferase is produced under the control of an engineered Elk1 transcriptional activator and pathway activation is represented as raw light units (RLU). As a positive control, we employed a modified, chemically inducible mFGFR1 (imFGFR; Welm et al, 2002), which lacks the endogenous ligand-binding modules and is activated by binding of the small chemical ligand AP20187 to a single, engineered FK506 binding protein (FKBP; Clackson, 1998). We found that the chimera incorporating the LOV domain of aureochrome1 from V. frigida (mFGFR1-VfAU1-LOV) activated the MAPK/ERK pathway similarly to the positive control (Fig2B). In particular, no augmented basal pathway activation in the absence of light was observed and pathway induction by light was of comparable magnitude to that by ligand. All other chimeras either exhibited no activity or constitutive activity (Fig2B). Control experiments showed that: (i) ERK1/2 is phosphorylated upon blue light stimulation in cells transfected with mFGFR1-VfAU1-LOV (Supplementary Fig S1), (ii) blue light had no effect on cells transfected with imFGFR1 (thereby excluding reporter activation by light alone; Fig2C, left), (iii) blue light had no effect on cells transfected with mFGFR1-VfAU1-LOV with Y271F and Y272F substitutions that result in loss of autophosphorylation and kinase activity (thereby demonstrating that kinase activity of the receptor is required; Fig2C, middle) and (iv) green light or red light had no effect on cells transfected with mFGFR1-VfAU1-LOV (thereby demonstrating wavelength specificity; Fig2C, right). Collectively, these results show that mFGFR1-VfAU1-LOV, a chimeric receptor consisting of the catalytic domain of a mammalian RTK and an algal LOV domain, activates the canonical MAPK/ERK pathway in response to blue light. In addition to phosphorylation of ERK, we observed phosphorylation of AKT, the key adapter protein fibroblast growth factor receptor substrate 2 (FRS2) and phospholipase Cγ1 (PLCγ1) in response to blue light (Supplementary Fig S1). Using luciferase reporters, we also detected activation of additional pathways linked to mFGFR1 (Supplementary Fig S2). We termed the chimeric mFGFR1-VfAU1-LOV receptor ‘Opto-mFGFR1’.
Figure 2. Design and function of mFGFR1-LOV domain chimeric receptors.
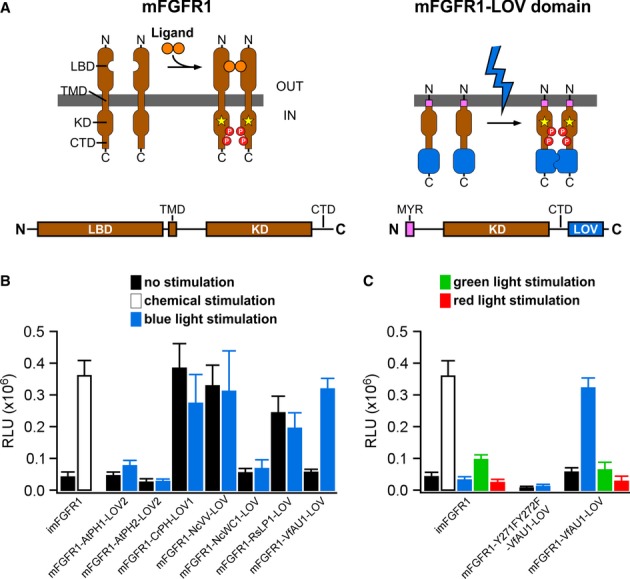
- Receptor tyrosine kinases such as mFGFR1 consist of the extracellular ligand-binding domain (LBD), single-span transmembrane domain (TMD) and intracellular domain (ICD) [kinase domain (KD) and a C-terminal tail domain (CTD)]. In mFGFR1-LOV domain chimeras, only the ICD is retained to render the protein insensitive to endogenous ligand. The ICD is attached to the membrane using a myristoylation domain (MYR) and LOV domains are incorporated at the ICD C-terminus.
- MAPK/ERK pathway activation in response to blue light for HEK293 cells that were transfected with chimeric proteins of mFGFR1-ICD and LOV domains. Activation is expressed as induction of a luciferase reporter gene. imFGFR1 is activated by the small molecule dimerizer AP20187.
- MAPK/ERK pathway activation in response to blue, green and red light for HEK293 cells that were transfected with imFGFR1, Opto-mFGFR1 (mFGFR1-VfAU1-LOV) or kinase dead Opto-mFGFR1 (Y271F, Y272F).
Data information: For (B) and (C): 24 h after transfection, cells were stimulated with light for 8 h followed by detection of luciferase. Light intensity was 1.7–2.5 μW/mm2. Mean values ± SEM for four to 16 independent experiments each performed in triplicates are shown.
Dimerization is required for Opto-mFGFR1 activation
We next investigated dimerization during activation of Opto-mFGFR1 with two independent methods. The first method utilizes a single charge inversion substitution (R195E in Opto-mFGFR1 or imFGFR1; R557E in full-length FGFR1). This substitution prevents formation of a functionally essential, asymmetric kinase domain dimer in FGFR1 (Bae et al, 2010) and inhibited MAPK/ERK pathway activation by imFGFR1 (Fig3A). We introduced this substitution into Opto-mFGFR1 as a probe for dimer formation during signalling. In HEK293 cells transfected with Opto-mFGFR1-R195E, we detected no MAPK/ERK pathway activation in response to blue light (Fig3A), indicating that receptor dimerization is required for receptor activation with light. We utilized protein cross-linking as a second method and directly observed Opto-mFGFR1 dimers in response to blue light (Fig3B). In addition, and as expected, we did not detect trans-activation of the endogenous FGFR1 in response to light stimulation of Opto-mFGFR1 in HEK293 cells that express similar levels of chimeric and endogenous receptors (Supplementary Fig S3). These results, together with the observation that VfAU1-LOV also induces dimerization of a transcription factor in response to blue light (Supplementary Fig S4), point to dimerization as the mechanism underlying Opto-mFGFR1 activation.
Figure 3. Analysis of Opto-mFGFR1 dimerization and sites required for MAPK/ERK pathway activation.
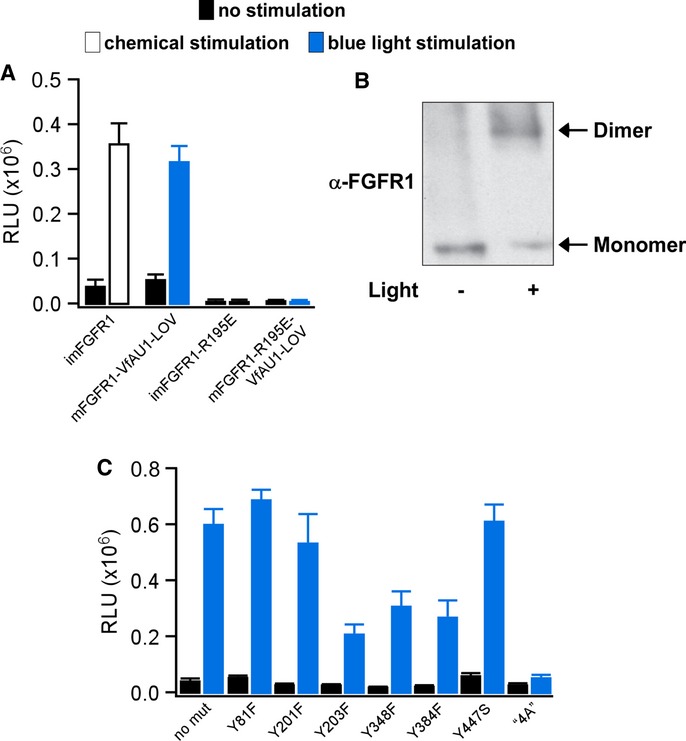
- MAPK/ERK pathway activation in response to blue light for HEK293 cells that were transfected with imFGFR1, Opto-mFGFR1 or genes with R195E substitution. This substitution prevents functional KD dimerization.
- Chemical cross-linking reveals light-induced dimerization of Opto-mFGFR1 in M38KOpto-mFGFR1 cells. Cells were stimulated with blue light for 15 min.
- Single-site or multi-site substitutions test phosphorylation site and coupling site requirements of Opto-mFGFR1. MAPK/ERK pathway activation in HEK293 cells that were transfected with Opto-mFGFR1 harbouring Tyr substitions at positions 81 (463), 201 (583), 203 (585), 348 (730), 384 (766) and 447 (3) or combined substitutions to Ala (‘4A’) at positions K37, P40, L41 and R43 (K419, P422, L423 and R425; numbering is relative to start methionine of Opto-mFGFR1; corresponding positions in mFGFR1 or VfAU1-LOV are given in parentheses). These residues were previously shown to be phosphorylated (Tyr) or required for association of FRS2 (LysProLeuArg).
Data information: For (A) and (C): 24 h after transfection, cells were stimulated with blue light for 8 h followed by detection of luciferase. Mean values ± SEM for two to 16 independent experiments each performed in triplicates are shown. For (A–C): Light intensity was ˜2.5 μW/mm2.
Coupling of Opto-mFGFR1 to cellular signalling pathways
We further investigated how Opto-mFGFR1 couples to cellular signalling pathways. Inhibition of MAPK/ERK kinase 1/2 (MEK1/2) confirmed that signals of the Elk1 reporter originate from activation of the MAPK/ERK pathway (Supplementary Fig S5). We introduced single-site substitutions into Opto-mFGFR1 to test if receptor phosphorylation is required for MAPK/ERK pathway activation. In agreement with previous work (Mohammadi et al, 1996), we found that Y271 and Y272 are required for activation of the MAPK/ERK pathway (Fig2C, middle), while Y81, Y201, Y203, Y348 and Y384 are dispensable (these residues correspond to Y653, Y654, Y463, Y583, Y585, Y730 and Y766 of mFGFR1; Fig3C). We also tested if a functionally relevant phosphorylation site has been introduced through generation of the chimera. Y447 is the sole surface-accessible Tyr in VfAU1-LOV as judged by inspecting a recent crystal structure of VfAU1-LOV (Mitra et al, 2012), and substitution at this site does not affect Opto-mFGFR1 function (Fig3C). In the juxtamembrane region of FGFR1, four residues were previously shown to be essential for association of FRS2 and coupling to the MAPK/ERK pathway (K37, P40, L41 and R43 in Opto-mFGFR1; K419, P422, L423 and R425 in mFGFR1; Ong et al, 2000). Substitutions of these residues to Ala in Opto-mFGFR1 abolished MAPK/ERK pathway activation (Fig3C). Finally, we applied confocal microscopy to detect membrane recruitment of PLCγ1. Pre- and post-stimulation images revealed that, upon blue light stimulation, PLCγ1 was recruited to the cell membrane (Supplementary Fig S6). We also note that major sites previously implicated in receptor internalization and downregulation (Sorokin et al, 1994; Persaud et al, 2011) are preserved in Opto-mFGFR1. Collectively, these results agree well with published data on mFGFR1 signalling (Sorokin et al, 1994; Mohammadi et al, 1996; Ong et al, 2000; Wang et al, 2001) and indicate that activation and coupling to downstream signalling of Opto-mFGFR1 resembles that of mFGFR1.
Long-lived VfAU1-LOV photoadduct is required for Opto-mFGFR1 activation
LOV domains respond to blue light with formation of a covalent photoadduct between a cysteine residue and the flavin prosthetic group. The photoadduct undergoes thermal reversion, and photoadduct lifetime is an important parameter in the function of light-sensing proteins (Zoltowski et al, 2009). We found that VfAU1-LOV exhibits a photoadduct lifetime that is intermediate among known LOV domains (τ = 625 s at 20°C, Supplementary Fig S7 and Supplementary Table S1; Moglich et al, 2010; Zoltowski & Gardner, 2011). We then investigated how photoadduct lifetime contributes to Opto-mFGFR1 signalling by reducing the lifetime through a point substitution. It was previously shown that substitution of a cysteinyl-facing Ile to Val results in marked decrease of photoadduct lifetime in LOV domains including VfAU1-LOV (Zoltowski et al, 2009; Mitra et al, 2012; I472V; Supplementary Fig S7 and Supplementary Table S1). Introducing this substitution into Opto-mFGFR1 reduced activation by blue light (Fig4A), and this result indicates that a long-lived VfAU1-LOV photoadduct supports Opto-mFGFR1 function. Notably, ligands establish functional FGFR1 complexes for up to 100 s (Powell et al, 2002), which is shorter than the photoadduct lifetime of wild-type VfAU1-LOV but longer than that of VfAU1-LOV after substitution.
Figure 4. Analysis of VfAU1-LOV photoadduct lifetime, additional receptor tyrosine kinases and alternative LOV domains.
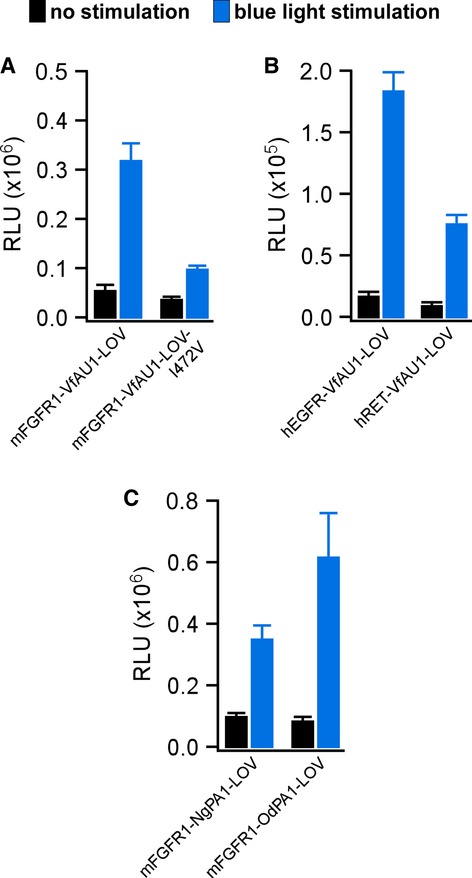
- MAPK/ERK pathway activation in response to blue light for HEK293 cells that were transfected with Opto-mFGFR1 or Opto-mFGFR1 with I472V substitution. This substitution reduces VfAU1-LOV photoadduct lifetime.
- MAPK/ERK pathway activation in response to blue light for HEK293 cells that were transfected with chimeric proteins of hEGFR1-ICD or hRET-ICD and VfAU1-LOV.
- MAPK/ERK pathway activation in response to blue light for HEK293 cells that were transfected with chimeric proteins of mFGFR1-ICD and NgPA1-LOV or OdPA1-LOV.
Data information: 24 h after transfection, cells were stimulated with blue light for 8 h followed by detection of luciferase. Light intensity was ˜2.5 μW/mm2. Mean values ± SEM for three to 16 independent experiments each performed in triplicates are shown.
Transfer of design principle to other RTKs and LOV domains
A highly desirable feature of engineered molecular systems is generalizability in which novel regulatory components are compatible with several different catalytic components. To test whether VfAU1-LOV is capable of activating other RTKs, we combined it with the catalytic domain of the human epidermal growth factor receptor (hEGFR) and human rearranged during transfection (hRET). We followed the design established in Opto-mFGFR1 and utilized membrane anchors previously shown to allow functional anchoring of these catalytic domains (Muthuswamy et al, 1999; Freche et al, 2005; Allocca et al, 2007). In line with their known signalling capability, robust activation of the MAPK/ERK pathway with blue light was observed in cells transfected with hEGFR and hRET chimeras (Fig4B). These chimeras were termed ‘Opto-hEGFR’ and ‘Opto-hRET’. We next tested whether LOV domains that resemble VfAU1-LOV can activate mFGFR1. Using database searches, we identified VfAU1-like proteins in the eustigmatophyte Nannochloropsis gaditana [N. gaditana putative aureochrome1 (NgPA1)] and in the golden algae Ochromonas danica [O. danica putative aureochrome1 (OdPA1)]. For mFGFR1 chimeras incorporating the LOV domains of NgPA1 and OdPA1 (NgPA1-LOV and OdPA1-LOV), we also observed activation of the MAPK/ERK pathway with blue light and amplitudes similar to that of the original Opto-mFGFR1 (Fig4C). We note that the LOV domain is located C-terminally to the effector domain in VfAU1, NgPA1 and OdPA1 but not in the other LOV domain-containing proteins examined here. Thus, our results allow us to propose that preserving the domain order of the naturally occurring light-sensing protein is beneficial for the function of the engineered protein.
Control of signalling in FGF2-responsive cells with spatial and temporal precision
We next tested the ability of Opto-mFGFR1 to control signalling with spatial and temporal precision in cells that are naturally responsive to FGFs. We first screened cells from different tumour entities for effects of FGF2, a prominent FGFR ligand, and we identified two cell lines derived from malignant pleural mesothelioma to be FGF2-responsive (M38K and SPC212; Schmitter et al, 1992; Kahlos et al, 1998). We also investigated human blood endothelial (hBE) cells as an additional cellular model (Schoppmann et al, 2004). We delivered Opto-mFGFR1 into these cells using virus and propagated cells with stable marker gene expression. Although expression levels of Opto-mFGFR in these stable M38K, SPC212 and hBE cells were higher than those of endogenous FGFRs (Supplementary Fig S8), cell resting states and endogenous receptor function were not compromised (see below). Immunofluorescence microscopy revealed that Opto-mFGFR1 localizes predominantly to the plasma membrane in these cells and also in HEK293 cells (Supplementary Fig S9). In M38KOpto-mFGFR1 and SPC212Opto-mFGFR1 cells, we found that 1 min of blue light was sufficient to induce phosphorylation of the receptor and ERK1/2 (Fig5A). We also observed phosphorylation of AKT and PLCγ1 in SPC212Opto-mFGFR1 cells (Fig5B). After cessation of light, phosphorylation of the receptor returned to pre-stimulation levels within 5 min, followed by phosphorylation of ERK1/2, AKT and PLCγ1 within 15–30 min (Fig5A and B). In hBEOpto-mFGFR1 cells, 5 min of blue light stimulation was sufficient to produce phosphorylation of the receptor and ERK1/2 (Fig5C). This result demonstrates control over cellular signalling in several cell types with temporal precision on the minute timescale. A remarkable diversity in the temporal signature of cell responses to this brief illumination was observed that likely reflect different activity of adaptor proteins or feedback mechanisms. To demonstrate spatially precise activation of signalling, we created monolayers of SPC212Opto-mFGFR1 and hBEOpto-mFGFR1 cells and limited illumination to small circular areas in the layers (areas contain ˜500 SPC212Opto-mFGFR1 cells in Fig6A or ˜2,500 hBEOpto-mFGFR1 cells in Fig6B; ˜50,000–250,000 surrounding cells were not illuminated). Immunodetection of ERK1/2 phosphorylation revealed activation of the MAPK/ERK pathway in illuminated but not neighbouring cells. This result and illumination of HEK293 cells in arbitrary patterns (Fig6C) demonstrates local signal activation in genetically homogenous cell layers.
Figure 5. Activation of cellular signalling with temporal precision.
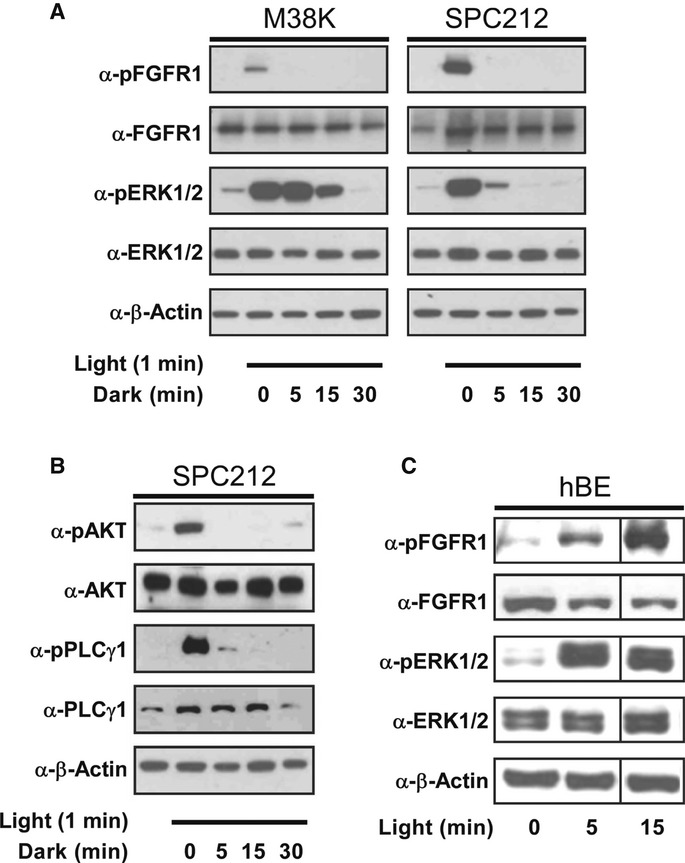
- Phosphorylation of Opto-mFGFR1 and ERK1/2 in human malignant pleural mesothelioma cells (M38KOpto-mFGFR1, SPC212Opto-mFGFR1) in response to 1 min of blue light followed by indicated duration of darkness.
- Phosphorylation of AKT and PLCγ1 in SPC212Opto-mFGFR1 in response to 1 min of blue light followed by indicated lengths of darkness.
- mFGFR1-VfAU1-LOV and ERK1/2 phosphorylation in hBEOpto-mFGFR1 cells in response to different durations of blue light.
Figure 6. Activation of cellular signalling with spatial precision.
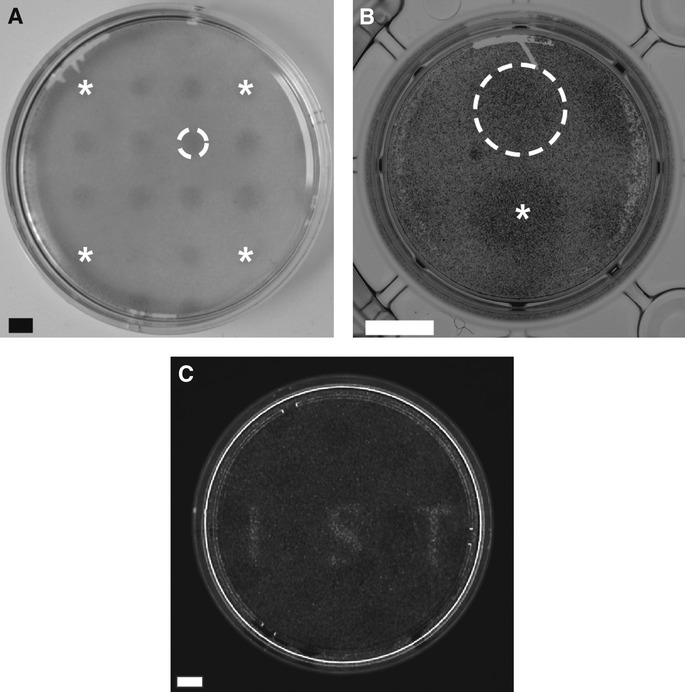
- Spatially confined ERK1/2 phosphorylation in SPC212Opto-mFGFR1 cells. A illumination pattern of 16 equidistant circles was produced using a pinhole template. Stars mark circles in corners. Dashed line marks a single circle. Cells were stimulated with blue light for 5 min followed by fixation and staining. Scale bar: 2 mm.
- Spatially confined ERK1/2 phosphorylation in hBEOpto-mFGFR1 cells. A illumination pattern of two circles was produced using a pinhole template. Star marks the center of one circle, dashed line marks the other circle. Cells were stimulated with blue light for 5 min followed by fixation and staining. Scale bar: 5 mm.
- Spatially confined MAPK-dependent gene transcription in HEK293 cells. Thirty-four hours after transfection, cells stimulated with light for 8 h followed by live-cell detection of luciferase. Scale bar: 10 mm.
Data information: All images are unprocessed raw images. Light intensity was ˜2.5 μW/mm2.
Controlling cell behaviour with light
We next analysed whether Opto-mFGFR1 could functionally substitute for ligand activation in M38K and hBE cells. In M38KOpto-mFGFR1 cells, one-h light stimulation caused an increase in cell proliferation and induced EMT-like morphological and gene expression changes (Fig7A–C, Supplementary Figs S10, S11 and S12), features characteristic of increased malignancy of cancer cells. In these experiments, changes in cell behaviour induced by light were within error of those induced by FGF2 (Fig7A and B). Blue light had no effect on control cells or cells that were pre-treated with PD166866, a selective FGFR1 kinase inhibitor (Fig7B and C). In addition to their prominent role in cancer, growth factors are important inducers of angiogenesis in physiological and pathophysiological conditions (Ferrara et al, 2003; Presta et al, 2005). The ability of several growth factors to induce angiogenesis is reflected by their ability to activate endothelial cell sprouting in vitro (Ferreras et al, 2012). Blue light illumination induced sprouting in hBEOpto-mFGFR1 spheroids to a similar extent as the major proangiogenic peptide VEGFA (Fig7D and E). These results indicate that activation of Opto-mFGFR1 can functionally substitute signalling by ligands and control the behaviour of cancer and endothelial cells.
Figure 7. Optical control of cell behaviour.
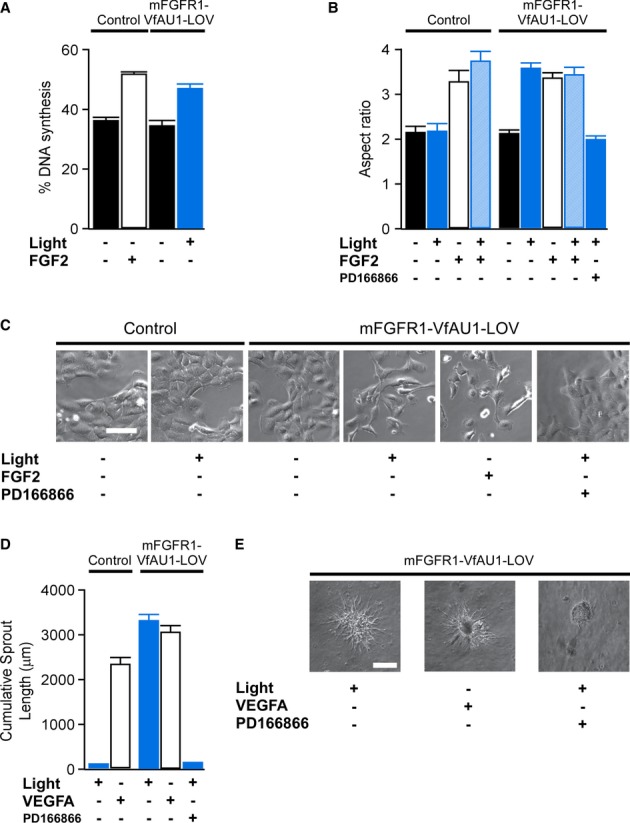
- Percentage of DNA synthesis in control and M38KOpto-mFGFR1 cells in response to blue light or FGF2 ligand. Cells were stimulated with blue light for 1 h. Proliferation was analysed after 26 h. Mean values ± SEM for 15–20 micrographs with 50–300 cells per micrograph from two independent experiments are shown.
- Morphology changes in control and M38KOpto-mFGFR1 cells in response to blue light or FGF2 ligand. Cells were stimulated with blue light for 1 h. Morphology was analysed after 24 h. Mean values ± SEM for 60 individual cells from two independent experiments are shown.
- Representative images for (B). Scale bar: 40 μm.
- Sprouting in control and hBEOpto-mFGFR1 cells in response to blue light or VEGFA ligand. Cells were stimulated with blue light for 5 min every 10 min during 10 h followed by analysis or morphology. Mean values ± SEM for 8–11 spheres per group in three independent experiments are shown.
- Representative images for (D). Scale bar: 100 μm.
Data information: Control cells were infected with mCherry. P < 0.05 for stimulated versus unstimulated cells (One-way ANOVA with Tukey's post-hoc test); n.s. for light stimulation versus FGF2/VEGFA stimulation; n.s. for unstimulated control cells versus unstimulated Opto-mFGFR1 cells. Light intensity was ˜2.5 μW/mm2.
Discussion
We demonstrate that RTKs can be light-activated through substitution of ligand-induced dimerization by light-induced dimerization. In the absence of precedence for light-activated mammalian receptor dimerization, and because of the structural and functional diversity of LOV domains (Moglich et al, 2010; Zoltowski & Gardner, 2011), we initially evaluated seven LOV domains from five different species. This evaluation resulted in the identification of VfAU1-LOV, an algal LOV domain that was never expressed in mammalian cells or incorporated into a chimeric protein. In contrast to phytochromes and LOV domains that were engineered to bind small proteins (Muller & Weber, 2013; Pathak et al, 2013), VfAU1-LOV responds to light with homodimerization (Toyooka et al, 2011). For this reason, a single transgene is sufficient to reconstitute optical activation of cellular signalling, and having to deliver only one transgene greatly facilitates application of the engineered receptors. VfAU1-LOV is capable of activating multiple, distantly related RTKs, as demonstrated here for mFGFR1, hEGFR and hRET. This observation suggests that design principles can be developed that are valid for multiple families of a large receptor superfamily and potentially beyond (see below). With the exception of a recently published gene transcription system (Wang et al, 2012), LOV domain homodimerization has not been exploited in optogenetics yet. Our work thus extends the portfolio of useful light-sensing domains with homodimerizing aureochromes and supports the notion that nature offers a large repertoire of light-sensing molecular functionalities that can be harvested by optogenetics (Chow et al, 2010).
For receptor activation, VfAU1-LOV utilizes flavin mononucleotide (FMN) as the light-sensing element. As FMN is present in animal cells, the function of VfAU1-LOV does not require addition of exogenous molecules. Furthermore, low-intensity blue light (˜2.5 μW/mm2) is sufficient for activation of all Opto-RTKs. This light intensity did not produce phototoxicity, is several orders of magnitude lower than those achieved in standard fluorescence microscopes, and is readily achieved in animal models (Janovjak et al, 2010; Ye et al, 2011). Opto-mFGFR1 enabled the precise control of receptor activation on physiologically relevant time scales and activation of cellular signalling with high spatial precision in genetically homogenous cell layers. Furthermore, activation with light triggered complex cellular programs like DNA synthesis, EMT and angiogenic sprouting in cancer cells and blood endothelial cells, which represent new cellular model systems in which behaviour is under optical control.
Beyond improved spatial and temporal precision, optical activation of RTKs will enable experiments that are difficult to realize using diffusible ligands. Light enables contactless application and complete withdrawal of stimuli without perturbing cellular environments. Furthermore, stimulation by light is limited to the receptor type that was genetically engineered, and thus light offers specificity beyond that of many ligands with promiscuous activation of receptors (Zhang et al, 2006). Work in the nervous system has demonstrated that combining optical activation with genetic targeting provides a powerful approach to manipulate cellular signalling in complex tissues. Recently, light-induced membrane localization of son of sevenless was applied to probe the dynamics of the Ras/ERK pathway (Toettcher et al, 2013). In contrast, light-activation of Opto-RTKs encoded each in a single gene enables control of multiple signalling pathways such as MAPK/ERK, AKT and PLCγ1 that all contribute to RTK-mediated biological responses. In the light of the critical involvement of RTKs in developmental biology, stem cell research and regenerative medicine, we expect that Opto-RTKs will open new avenues in these research areas. We also envision their use for cytoprotection and restoration of cell behaviours affected by disorders, particularly in those tissues for which growth factor gene therapy is already considered. Similar design principles may be developed for other cellular signals as receptor dimerization is also implicated in the function of receptor serine/threonine kinases, T-cell receptors and GPCRs (Massague, 1998; Cochran et al, 2001; Lohse, 2010).
Materials and Methods
mFGFR1 receptor constructs
pSH1/M-FGFR1-Fv-Fvls-E [D.M. Spencer, Baylor College of Medicine (Welm et al, 2002)] was obtained from Addgene (Cambridge, MA, USA). The ICD of mFGFR1 flanked by MYR, two FKBP domains and an hemagglutinin (HA)-epitope was transferred from pSH1 to pcDNA3.1(−) (Invitrogen/Life Technologies, Vienna, Austria) using NotI and EcoRI restriction enzymes. Using inverse PCR, a single or both FKBP domains were deleted to yield constructs imFGFR1 and imFGFR1-ΔFKBP. In this reaction, amplification using oligonucleotides 1 and 2 or 1 and 3 (Supplementary Table S3) produced linear DNA products in which either a single or both FKBP domain were replaced by terminal AgeI restriction sites. Products were digested with AgeI, ligated and transformed into Escherichia coli. This procedure also introduced the AgeI restriction site in imFGFR1-ΔFKBP that was used for LOV domain insertion. As an additional control, one FKBP domain was re-inserted into imFGFR1-ΔFKBP using PCR and AgeI and XmaI restriction enzymes (oligonucleotides 4 and 5, Supplementary Table S3). imFGFR1 and imFGFR1-ΔFKBP-FKBP produced similar results in MAPK/ERK pathway assays and were used interchangeably. All constructs generated in this study were verified by sequencing.
LOV domains and chimeric mFGFR1 receptors
Genes coding for the LOV domains of A. thaliana phototropin 1 (AtPH1-LOV2, residue 449–586 of Uniprot entry O48963), A. thaliana phototropin 2 (AtPH2-LOV2, residue 363–500 of Uniprot entry P93025), C. reinhardtii phototropin (CrPH-LOV1, residue 16–133 of Uniprot entry A8IXU7), N. crassa vivid (NcVV-LOV, residue 37–186 with Y50W substitution of Uniprot entry Q9C3Y6), N. crassa white collar 1 (NcWC1-LOV, residue 357–528 of Uniprot entry Q01371), Rhodobacter sphaeroides PCC 17025 light-sensing protein [RsLP-LOV, all residues of Protein Data Bank entry 4HJ4 corresponding to genomic position NC_009428:2302851–2303211; (Conrad et al, 2013)], V. frigida aureochrome1 (VfAU1-LOV, residue 204–348 of Uniprot entry A8QW55), N. gaditana hypothetical protein NGA_0015702 (NgPA1-LOV, residue 87–228 of Uniprot entry K8Z861) and O. danica aureochrome1-like protein (OdPA1-LOV, residue 180–312 of Uniprot entry C5NSW6) were synthesized with mammalian codon optimization according to the supplier's recommendation (Epoch Life Science, Inc., Missouri City, TX, USA; Supplementary Table S4). NgPA1-LOV and OdPA1-LOV were identified using database searches for proteins similar to VfAU1 in the non-redundant protein database of the National Center for Biotechnology Information. AtPH1-LOV2, AtPH2-LOV2, CrPH-LOV1, NcVV-LOV and VfAU1-LOV were inserted into imFGFR1-ΔFKBP using PCR and AgeI and XmaI restriction enzymes (oligonucleotides 6–15, Supplementary Table S3). NcWC1-LOV, RsLP-LOV, NgPA1-LOV and OdPA1-LOV were synthesized with restriction sites. Final receptor sequences are given in Supplementary Table S5.
Modified Opto-mFGFR1 receptors
Point substitutions generated were Y81F (463), R195E, Y201F (583), Y203F (585), Y271F&Y272F (653&654), Y348F (730), Y384F (766), Y447S, I472V (numbering is relative to start methionine of Opto-mFGFR1 and corresponding positions in mFGFR1 are given in parentheses where applicable). Substitutions were introduced in Opto-mFGFR1 and imFGFR1 using site-directed mutagenesis (oligonucleotides 16–33, Supplementary Table S3). Four alanine substitutions were introduced using inverse PCR and the ClaI restriction enzyme (oligonucleotides 34 and 35, Supplementary Table S3).
Opto-hEGFR and Opto-hRET
Using inverse PCR, expression plasmids were prepared based on mFGFR1-VfAU1-LOV in which the mFGFR1 ICD was replaced by a SgrAI restriction site (oligonucleotides 36 and 37, Supplementary Table S3). hEGFR ICD and hRET ICD were inserted into this plasmid using PCR and AgeI and BspEI restriction enzymes (oligonucleotides 38–41, Supplementary Table S3). The hEGFR construct was further modified by including the LBD and TMD of p75 using PCR and NotI and AscI restriction enzymes (oligonucleotides 42 and 43, Supplementary Table S3).
Light-activated VfAU1-LOV transcription factor
The plasmid GAVPO (Y. Yang, East China University of Science and Technology) contains a Gal4 DNA-binding domain, NcVV-LOV and a trans-activation domain (Wang et al, 2012; Supplementary Fig S4). Activation of GAVPO was detected with the luciferase reporter plasmid of the MAPK/ERK pathway assay (the plasmid contains multiple UAS sequences). VfAU1-LOV was amplified by PCR and oligonucleotides 44 and 45 (Supplementary Table S3) and inserted in GAVPO using BglII and EcoRI restriction enzymes. Luciferase detection was performed as described below except that mFGFR1 plasmids were replaced with 50 ng GAVPO per well.
Custom incubator for light stimulation of cells
For light stimulation of cells, an incubator (PT2499; ExoTerra/HAGEN, Holm, Germany) was equipped with 300 light-emitting diodes (JS-FS5050RGB-W30 with JS-CON-004 controller; Komerci, Ebern, Germany; λmax ≈ 630 nm (red light), λmax ≈ 530 nm (green light), λmax ≈ 470 nm (blue light), bandwidth ≈ ±5 nm). Light intensity was controlled with an analogue dimmer and measured with a digital power meter (PM120VA; Thorlabs, Munich, Germany). Intensities at maximal output were 2.3 (red light), 2.6 (green light) and 3.3 (blue light) W/m2. For stimulation over extended time periods (> 8 h), an aluminium box was equipped with the same light-emitting diodes and placed in a tissue culture incubator (also see below).
Cell culture and transfection (HEK293 and CHO K1 cells)
Cells were maintained in DMEM supplemented with 10% FBS, 100 U/ml penicillin and 0.1 mg/ml streptomycin in a humidified incubator with 5% CO2 atmosphere. After trypsination, 5 × 104 cells were seeded in each well of 96-well plates that were coated with poly-L-ornithine (Sigma, Vienna, Austria). Either transparent plates or black clear bottom plates were used. Cells were transfected using Lipofectamine 2000 (Invitrogen/Life Technologies) or polyethylenimine (Polysciences, Eppelheim, Germany).
LOV domain expression and cell viability (HEK293 and CHO K1 cells)
An expression plasmid based on pcDNA3.1(−) was prepared in which a BspEI restriction site followed the fluorescent protein mVenus (Nagai et al, 2002) and a glycine- and serine-rich linker. LOV domains were inserted into this plasmid using PCR (see above). Cells were transfected with 100 ng expression plasmid in each well of 96-well plates (three to four wells were prepared for each construct throughout the study). Expression was assessed by measuring mVenus fluorescence in a plate reader (BioTek Synergy H1, Bad Friedrichshall, Germany) 16–18 h after transfection. Transfection of pcDNA3.1(−) or a FKBP-mVenus fusion protein served as controls. Viability measurements used a tetrazolium dye following the supplier's protocol (EZ4U Cell Proliferation and Cytotoxicity Assay; Biomedica, Vienna, Austria). Absorbance measurements at 450 nm with 620 nm reference were taken after fluorescence measurements.
Stimulation and detection of MAPK/ERK pathway (HEK293 cells)
Activation of the MAPK/ERK pathway was assayed with the PathDetect Elk1 trans-Reporting System (Agilent, Vienna, Austria) consisting of an Elk1 phosphorylation-dependent trans-activator and a luciferase-based trans-reporter. Cells were transfected with 213.3 ng total DNA per well (receptor, trans-activator and trans-reporter at a ratio of 1:3:60 or 1:30:600). Six hours after transfection, medium was replaced with CO2-independent reduced serum starve medium (Gibco/Life Technologies; supplemented with 0.5% FBS, 2 mM L-glutamine, 100 U/ml penicilin and 0.1 mg/ml streptomycin) and cells were incubated for 18 h at 37°C. Cells were transferred to the stimulation incubator and for 8 h either subject to constant illumination or, for control cells throughout the study, kept in the dark. For stimulation of imFGFR1, 10 nM AP20187 (ARIAD Pharmaceuticals, Cambridge, MA, USA; Clackson, 1998) was added before transfer to the stimulation incubator. After incubation, plates were washed with PBS and processed with Luciferase 1000 Assay System (Promega, Mannheim, Germany) for cell lysis and signal development. Luminescence was detected with a microplate reader (Tecan Infinite 200 Pro, Maennedorf, Switzerland). Some samples were processed with ONE-Glo (Promega) and detected with the BioTek microplate reader. Differences in absolute intensities due to assay and plate reader were corrected using Opto-mFGFR1 as a standard if intensities were plotted in the same figure. UO126 was obtained from LC Laboratories (Woburn, MA, USA).
Stimulation and immunoblot (HEK293 cells)
2 × 106 cells were seeded in 25-cm2 dishes and transfected with 2 μg Opto-mFGFR1. After 5 h, medium was replaced with reduced serum medium. After additional 20 h, cells were illuminated for 5 min. Cells were harvested in 70 μl lysis buffer on ice and lysates were sonicated and centrifuged (12,000 g, 5 min, 4°C). Thirty μg protein per lane were separated by SDS-PAGE and electro-blotted onto PVDF membranes. Blots were incubated with primary antibodies (ERK1/2, #9102; pERK1/2, #9101; PLCγ1 #2822; pPLCγ1 #2821; AKT #9272; pAKT #4058S; pFRS2α(Tyr196), #3864; pFRS2α(Tyr436) #3861; Cell Signaling Technology, Danvers, MA, USA dilution 1:1,000; FRS2 (A-5) Santa Cruz Biotechnology, Santa Cruz, CA, USA, dilution 1:500; β-actin, Sigma, dilution 1:8,000) in blocking solution (3% BSA or 5% skim milk in TBST) overnight at 4°C. Secondary antibodies (HRP-coupled α-rabbit or α-mouse IgG; Dako, Glostrup, Denmark) were applied at a dilution of 1:10,000 for 2 h at RT. Chemiluminescence was developed with WesternC reagent (Biorad, Hercules, CA, USA) and signals recorded on X-ray film (GE Healthcare, Chalfont St. Giles, UK). For FRS2α-RFP transfection, an expression plasmid based on pcDNA3.1(−) was prepared in which a NheI restriction site precedes the red fluorescent protein and human FRS2α was inserted into this plasmid using PCR (oligonucleotides 46–47, Supplementary Table S3). For detection of transactivation, stable transfectants were selected with 500 μg/ml G418.
Detection of additional signalling pathways (HEK293 cells)
Activation of additional mFGFR1-related signalling pathways was assayed with Cignal Reporter assays (Qiagen, Hilden, Germany) consisting of mixtures of inducible pathway-luciferase reporters. Cells were transfected with 100.3 ng total DNA per well (receptor and reporter at a ratio of 1:300) and treated as described above. Cells were processed with the Dual-Glo Luciferase Assay System (Promega) and Tecan microplate reader.
Live-cell fluorescence microscopy (HEK293 cells)
An expression plasmid based on pcDNA3.1(−) was prepared in which a BspEI restriction site preceded the fluorescent protein mVenus and an in-frame glycine- and serine-rich linker. Human PLCγ1 was inserted into this plasmid using PCR and the AgeI restriction enzyme (oligonucleotides 48 and 49, Supplementary Table S3). 4 × 105 HEK293 cells were seeded in 5-cm dishes that were coated with poly-L-lysine and transfected with 200 ng expression plasmid per well (receptor and PLCγ1-mVenus at a ratio of 3:4). Medium was replaced with reduced serum medium 24 h before imaging. Stimulation with LEDs (see above) and live-cell microscopy was performed a Leica SP5 confocal microscope.
Live-cell luminescence in spatially confined illumination experiments (HEK293 cells)
3 × 106 HEK293 cells were simultaneously seeded and transfected with 24 μg expression plasmid (receptor, trans-activator and trans-reporter at a ratio of 1:3:60) in a 10-cm dish. After 16 h, cells were treated and illuminated as described above. Live cells were incubated with 0.15 mg/ml D-luciferin (PEQlab, Erlangen, Germany) in PBS for 10 min at 37°C. Luminescence was detected with a PEQLab Fusion SL imaging system (PEQLab, Erlangen, Germany).
Generation of stable Opto-mFGFR1 transgenic cell lines and virus construction (M38K, SPC212 and hBE cells)
M38K and SPC212, two malignant pleural mesothelioma cell lines, were maintained in RPMI1640 supplemented with 10% FBS. Telomerase-immortalized microvascular hBE cells were maintained in Clonetics EGM2 MV endothelial growth medium (Lonza, Wakersville, MD) supplemented with 5% FBS. For retrovirus generation, Opto-mFGFR1 or mCherry (Shaner et al, 2004) were subcloned into pQCXIP (Clontech, Mountain View, CA, USA) using NotI and EcoRI restriction enzymes. Viral particles were generated in HEK293 cells by co-transfection with the helper plasmids pVSV-G (Clontech) and p-gag-pol-gpt (Markowitz et al, 1988). Supernatants were used to transduce M38K, SPC212 or hBE cells grown to 50% confluency in 6-well plates. Cells were selected with 0.8 μg/ml puromycin for 10 days, and transgene expression was verified by immunoblotting.
Stimulation and immunoblot (M38K, SPC212 and hBE cells)
For immunoblotting, 5 × 105 cells were seeded in each well of 6-well plates. After 24 h, medium was replaced with reduced serum medium (M38K and SPC212 cells). After additional 20 h, cells were transferred to the stimulation incubator and illuminated for 1, 5 or 15 min. Cells were then either immediately or after additional 5, 15 or 30 min in the dark washed and lysed in 50 μl lysis buffer per well on ice. Lysates were sonicated and centrifuged as described above. Fifteen μg protein per lane were separated by SDS-PAGE and electro-blotted onto PVDF membranes. Blots were incubated with primary antibodies (FGFR1, #9740 Cell Signaling Technology, dilution 1:1,000; FGFRpY653/654, Thermo Scientific, Vienna, Austria, dilution 1:1,000) and secondary antibodies for processing as described above.
Immunofluorescence and confocal microscopy (HEK293, M38K, SPC212 and hBE cells)
For localization of Opto-mFGFR1, Opto-hEGFR and Opto-hRET in HEK293 cells, 3 × 105 cells per well were seeded into each well of 6-well plates and transfected with 2 μg expression plasmid. After 24 h, 1 × 105 cells were seeded into fibronectin-coated chamber slides (BD Falcon, Heidelberg, Germany). After additional 24 h, cells were fixed with 3.8% formaldehyde, blocked with 1% BSA in PBS and incubated with monoclonal rat α-HA (3F10, Roche, Mannheim, Germany, 1:100) overnight at 4°C. This was followed by Alexa Fluor 546-linked goat α-rat antibody for 1 h at RT. Slides were mounted in Vectashield mounting medium containing DAPI and images recorded on a Zeiss LSM700 confocal microscope. For localization of Opto-mFGFR1 in stable cell lines, 5 × 104 M38KOpto-mFGFR1, SPC212Opto-mFGFR1 and hBEOpto-mFGFR1 cells were seeded into chamber slides and stained and imaged as above.
Opto-mFGFR1 cross-linking (M38K cells)
For chemical cross-linking, 107 cells were collected and washed three times with PBS. Cells were stimulated for 15 min with blue light. Disuccinimidyl suberate was added to a final concentration of 5 mM and incubated at RT for 30 min in blue light or dark. The reaction was quenched with 10 mM Tris, pH 7.5 for 15 min with blue light. Cell lysis and immunoblotting were performed as described above.
ERK1/2 phosphorylation in spatially confined illumination experiments (SPC212 and hBE cells)
For detection of localized ERK1/2 phosphorylation in cell monolayers, SPC212 or hBE cells were grown to confluency in 6-cm petri dishes (SPC212) or 12-well plates (hBE cells). SPC212 were starved in medium without serum for 24 h before illumination. Templates with pinholes of 2 (SPC212) or 5 (hBE cells) mm diameter were used for localized illumination for 5 min. Afterwards, cells were washed with cold PBS and fixed with Histofix (Lactan, Graz, Austria) for 10 min. After washing with PBS and permeabilization with Triton X-100 (0.25% in PBST) and blocking in 1% BSA in PBST, dishes were incubated with pERK1/2 (#9101; Cell Signaling Technology, 1:500 for SPC212, 1:100 for hBE cells) for 1 h. Signal was developed using the UltraVision LP detection system (Thermo Scientific) and 3,3′-diaminobenzidine as chromogen. Haematoxylin was used for counterstaining of cell nuclei.
Cell proliferation (M38K cells)
2 × 104 cells were seeded in each well of 96-well plates. After 24 h, cells were stimulated for 1 h. FGF2 (Sigma, St. Louis, MO) was added to a final concentration of 10 ng/ml. After 24 h, cells were incubated with 10 μM EdU for 2 h. Subsequently, newly synthesized DNA was stained with Click-iT EdU (Life Technologies) following the manufacturer's protocol and counterstained with 5 μg/ml Hoechst dye. Cells were photographed on a Nikon Ti300 inverted microscope. To determine the percentage of cells with newly synthesized DNA, Hoechst positive nuclei and EdU positive nuclei were counted.
Cell morphology (M38K cells)
105 cells were seeded in each well of 6-well plates. After 24 h, cells were stimulated for 1 h. FGF2 (Sigma) or PD166866 (Pfizer Global Research and Development, New London, CT) were added to a final concentration of 10 ng/ml or 10 μM, respectively. After additional 24 h, cells were photographed on the Nikon Ti300 microscope. For quantification of cell morphology, all cell perimeters in randomly selected sections of phase contrast images were traced and aspect ratios (defined as length of major axis divided by length of minor axis of a fitted ellipse) calculated with ImageJ software (National Institute of Health). Sixty individual values contributed to each average. Automated analysis yielded comparable results.
Cell cycle (M38K cells)
Cell cycle distribution was analysed by flow cytometry. 5 × 105 cells were seeded in 25-cm2 tissue culture flasks. After 24 h, cells were stimulated for 1 h. After additional 24 h, cells were fixed in ethanol (70%), treated with 50 μg/ml RNase A and 50 μg/ml propidium iodide (PI). Flow cytometry was performed on a FACSCalibur (BD Biosciences, Schwechat, Austria) and cell cycle distribution calculated with ModFit LT software (Verity Software House, Topsham, ME).
Actin staining (M38K cells)
5 × 105 cells were seeded onto coverslips in 6-well plates. After 24 h, cells were illuminated with a cycle of 5 min blue light/15 min dark for 48 h. Cells were fixed (3.8% formaldehyde), permeabilized (0.5% Triton X-100 in PBS), stained with TRITC-phalloidin (1:100, 1% BSA in PBS, overnight at 4°C) and covered in mounting medium containing DAPI. Micrographs were taken on a Leica fluorescence microscope.
Gene expression analysis (M38K, SPC212 and hBE cells)
Total RNA was extracted with TRIZOL (Life Technologies) from sub-confluent flasks of M38KOpto-mFGFR1, SPC212Opto-mFGFR1 and hBEOpto-mFGFR1 cells and the respective controls. RNA was reverse transcribed with MMLV reverse transcriptase (Thermo Scientific). cDNAs corresponding to 50 ng RNA per sample were used for SYBR green qPCR on an Abi Prism 7500 Sequence Detection System. Primers recognizing both murine and human FGFR1 and GAPDH (oligonucleotides 50–53, Supplementary Table S3) were used for analysis and normalization, resp., and fold expression levels were calculated as 2−ddCt of Opto-mFGFR1 transgenic cells versus the respective control cells. For analysis of EMT marker expression, 5 × 104 M38K cells were seeded into each well of 6-well plates. After 24 h, cells were illuminated with a cycle of 5-min light and 15-min dark for 48 h. Total RNA was extracted as described above and subjected to SYBR green qPCR using published oligonucleotides (Sakuma et al, 2012). GAPDH was used for normalization, and fold change of expression level was calculated as 2−ddCt of illuminated versus non-illuminated cells.
In vitro angiogenesis (sprouting) assay (hBE cells)
For spheroid generation, cells were suspended as hanging drops (450 cells in a 25-μl drop) in M199 medium (Sigma) supplemented with 10% FBS, L-glutamine, 2.2 g/l NaHCO3 and 20% methylcellulose (Sigma) overnight in a standard tissue culture incubator. The following day, spheroids were washed in PBS containing 10% FBS, centrifuged, resuspended in Methocel/20% FBS, mixed (1:1) with neutralized rat-tail collagen and seeded into non-adhesive 24-well plates (Greiner Bio-one, Kremsmünster, Austria). After solidification of the collagen, VEGFA (30 ng/ml) or PD166866 (10 μM) were added. Plates were stimulated with blue light for 5 min every 20 min for 10 h. After stimulation, 1 ml of 8% paraformaldehyde was added to each well and spheroids were photographed on the Nikon microscope. Cumulative sprout lengths per sphere from at least 8 spheroids per group were measured (ImageJ).
Purification of VfAU1-LOV and UV-visible spectroscopy
VfAU1-LOV was subcloned into pQE80L (Qiagen) using PCR and BamHI and KpnI restriction enzymes (oligonucleotides 54 and 55, Supplementary Table S3). I28V substitution (corresponding to I472V in Opto-mFGFR1) was introduced using site-directed mutagenesis (see above). His6-tagged proteins were expressed in E. coli BL21 (DE3) and purified using Ni+ affinity columns (GE Healthcare). Purified proteins were subsequently dialysed in buffer containing 10% glycerol, 150 mM NaCl and 50 mM HEPES (pH 8.0) using PD-10 desalting columns (GE Healthcare). Work was performed in minimal light. Protein purity was visually assessed by SDS-PAGE. UV-visible spectroscopy was carried out at 20 or 37°C in a microplate reader (BioTek Synergy H1). After 5 min illumination with blue light (see above), absorption at 447 nm was recorded during 1 h. Values of OD447 as a function of time were fitted to a single exponential using Igor Pro 6 (Wavemetrics, Lake Oswego, OR) to obtain lifetimes.
Acknowledgments
We thank G. Krupitza, C. McKenzie, D. Siekhaus and M. Matveenko for discussions, E. Papusheva for help with microscopy, M. Spanova, A Kitzmann, B. Peter-Vörösmarty and D. Meindl for technical assistance, V. Baleyeava and S. zur Nedden for assistance with genetic engineering, Ariad Pharmaceuticals Inc. for AP20187, D.M. Spencer (Baylor College of Medicine) and Y. Yang (East China University of Science and Technology) for plasmids, V.L. Kinnula (University of Helsinki) and R. Stahel (University of Zurich) for mesothelioma cells, and S. Geleff (Medical University of Vienna) for hBE cells. This work was supported by grants of the European Union Seventh Framework Programme (to HJ), the Human Frontier Science Program (to HJ), funds of the Oesterreichische Nationalbank Anniversary Fund (14211 to MG) and by an Austrian Research Promotion Agency (FFG) FemTech fellowship (to ER).
Author contributions
All authors designed, performed and analysed experiments. MG, RR, AIP and HJ wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- Albeck JG, Mills GB, Brugge JS. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol Cell. 2013;49:249–261. doi: 10.1016/j.molcel.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allocca M, Di Vicino U, Petrillo M, Carlomagno F, Domenici L, Auricchio A. Constitutive and AP20187-induced Ret activation in photoreceptors does not protect from light-induced damage. Invest Ophthalmol Vis Sci. 2007;48:5199–5206. doi: 10.1167/iovs.07-0140. [DOI] [PubMed] [Google Scholar]
- Ashcroft FM. From molecule to malady. Nature. 2006;440:440–447. doi: 10.1038/nature04707. [DOI] [PubMed] [Google Scholar]
- Bae JH, Boggon TJ, Tome F, Mandiyan V, Lax I, Schlessinger J. Asymmetric receptor contact is required for tyrosine autophosphorylation of fibroblast growth factor receptor in living cells. Proc Natl Acad Sci USA. 2010;107:2866–2871. doi: 10.1073/pnas.0914157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaletto JB, McClatchey AI. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat Rev Cancer. 2012;12:387–400. doi: 10.1038/nrc3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, Boyden ES. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Gawthorne J, Young G, Fraser NJ, Roe AJ. LOV to BLUF: flavoprotein contributions to the optogenetic toolkit. Mol Plant. 2012;5:533–544. doi: 10.1093/mp/sss020. [DOI] [PubMed] [Google Scholar]
- Clackson T. Redesigning small molecule-protein interfaces. Curr Opin Struct Biol. 1998;8:451–458. doi: 10.1016/s0959-440x(98)80122-x. [DOI] [PubMed] [Google Scholar]
- Cochran JR, Aivazian D, Cameron TO, Stern LJ. Receptor clustering and transmembrane signaling in T cells. Trends Biochem Sci. 2001;26:304–310. doi: 10.1016/s0968-0004(01)01815-1. [DOI] [PubMed] [Google Scholar]
- Conrad KS, Bilwes AM, Crane BR. Light-induced subunit dissociation by a light-oxygen-voltage domain photoreceptor from Rhodobacter sphaeroides. Biochemistry. 2013;52:378–391. doi: 10.1021/bi3015373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng CX, Wynshaw-Boris A, Shen MM, Daugherty C, Ornitz DM, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- Ferreras C, Rushton G, Cole CL, Babur M, Telfer BA, van Kuppevelt TH, Gardiner JM, Williams KJ, Jayson GC, Avizienyte E. Endothelial heparan sulfate 6-O-sulfation levels regulate angiogenic responses of endothelial cells to fibroblast growth factor 2 and vascular endothelial growth factor. J Biol Chem. 2012;287:36132–36146. doi: 10.1074/jbc.M112.384875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freche B, Guillaumot P, Charmetant J, Pelletier L, Luquain C, Christiansen D, Billaud M, Manie SN. Inducible dimerization of RET reveals a specific AKT deregulation in oncogenic signaling. J Biol Chem. 2005;280:36584–36591. doi: 10.1074/jbc.M505707200. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- Huang K, Merkle T, Beck CF. Isolation and characterization of a Chlamydomonas gene that encodes a putative blue-light photoreceptor of the phototropin family. Physiol Plant. 2002;115:613–622. doi: 10.1034/j.1399-3054.2002.1150416.x. [DOI] [PubMed] [Google Scholar]
- Janovjak H, Szobota S, Wyart C, Trauner D, Isacoff EY. A light-gated, potassium-selective glutamate receptor for the optical inhibition of neuronal firing. Nat Neurosci. 2010;13:1027–1032. doi: 10.1038/nn.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlos K, Anttila S, Asikainen T, Kinnula K, Raivio KO, Mattson K, Linnainmaa K, Kinnula VL. Manganese superoxide dismutase in healthy human pleural mesothelium and in malignant pleural mesothelioma. Am J Respir Cell Mol Biol. 1998;18:570–580. doi: 10.1165/ajrcmb.18.4.2943. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature. 2001;414:656–660. doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz J, Pantoja C, Gaub B, Janovjak H, Reiner A, Hoagland A, Schoppik D, Kane B, Stawski P, Schier AF, Trauner D, Isacoff EY. Optical control of metabotropic glutamate receptors. Nat Neurosci. 2013;16:507–516. doi: 10.1038/nn.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MJ. Dimerization in GPCR mobility and signaling. Curr Opin Pharmacol. 2010;10:53–58. doi: 10.1016/j.coph.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Ma DK, Ponnusamy K, Song MR, Ming GL, Song H. Molecular genetic analysis of FGFR1 signalling reveals distinct roles of MAPK and PLCgamma1 activation for self-renewal of adult neural stem cells. Mol Brain. 2009;2:16. doi: 10.1186/1756-6606-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz D, Goff S, Bank A. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Mitra D, Yang X, Moffat K. Crystal structures of Aureochrome1 LOV suggest new design strategies for optogenetics. Structure. 2012;20:698–706. doi: 10.1016/j.str.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moglich A, Yang X, Ayers RA, Moffat K. Structure and function of plant photoreceptors. Annu Rev Plant Biol. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol Cell Biol. 1996;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K, Weber W. Optogenetic tools for mammalian systems. Mol BioSyst. 2013;9:596–608. doi: 10.1039/c3mb25590e. [DOI] [PubMed] [Google Scholar]
- Muthuswamy SK, Gilman M, Brugge JS. Controlled dimerization of ErbB receptors provides evidence for differential signaling by homo- and heterodimers. Mol Cell Biol. 1999;19:6845–6857. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Ong SH, Guy GR, Hadari YR, Laks S, Gotoh N, Schlessinger J, Lax I. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol Cell Biol. 2000;20:979–989. doi: 10.1128/mcb.20.3.979-989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak GP, Vrana JD, Tucker CL. Optogenetic control of cell function using engineered photoreceptors. Biol Cell. 2013;105:59–72. doi: 10.1111/boc.201200056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persaud A, Alberts P, Hayes M, Guettler S, Clarke I, Sicheri F, Dirks P, Ciruna B, Rotin D. Nedd4-1 binds and ubiquitylates activated FGFR1 to control its endocytosis and function. EMBO J. 2011;30:3259–3273. doi: 10.1038/emboj.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AK, Fernig DG, Turnbull JE. Fibroblast growth factor receptors 1 and 2 interact differently with heparin/heparan sulfate. Implications for dynamic assembly of a ternary signaling complex. J Biol Chem. 2002;277:28554–28563. doi: 10.1074/jbc.M111754200. [DOI] [PubMed] [Google Scholar]
- Presta M, Dell'Era P, Mitola S, Moroni E, Ronca R, Rusnati M. Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine Growth Factor Rev. 2005;16:159–178. doi: 10.1016/j.cytogfr.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Robertson SC, Tynan JA, Donoghue DJ. RTK mutations and human syndromeswhen good receptors turn bad. Trends Genet. 2000;16:265–271. doi: 10.1016/s0168-9525(00)02021-7. [DOI] [PubMed] [Google Scholar]
- Sakuma K, Aoki M, Kannagi R. Transcription factors c-Myc and CDX2 mediate E-selectin ligand expression in colon cancer cells undergoing EGF/bFGF-induced epithelial-mesenchymal transition. Proc Natl Acad Sci USA. 2012;109:7776–7781. doi: 10.1073/pnas.1111135109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitter D, Lauber B, Fagg B, Stahel RA. Hematopoietic growth factors secreted by seven human pleural mesothelioma cell lines: interleukin-6 production as a common feature. Int J Cancer. 1992;51:296–301. doi: 10.1002/ijc.2910510220. [DOI] [PubMed] [Google Scholar]
- Schoppmann SF, Soleiman A, Kalt R, Okubo Y, Benisch C, Nagavarapu U, Herron GS, Geleff S. Telomerase-immortalized lymphatic and blood vessel endothelial cells are functionally stable and retain their lineage specificity. Microcirculation. 2004;11:261–269. doi: 10.1080/10739680490425967. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Simi A, Ibanez CF. Assembly and activation of neurotrophic factor receptor complexes. Dev Neurobiol. 2010;70:323–331. doi: 10.1002/dneu.20773. [DOI] [PubMed] [Google Scholar]
- Sorokin A, Mohammadi M, Huang J, Schlessinger J. Internalization of fibroblast growth factor receptor is inhibited by a point mutation at tyrosine 766. J Biol Chem. 1994;269:17056–17061. [PubMed] [Google Scholar]
- Szobota S, Isacoff EY. Optical control of neuronal activity. Annu Rev Biophys. 2010;39:329–348. doi: 10.1146/annurev.biophys.093008.131400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F, Yamagata D, Ishikawa M, Fukamatsu Y, Ogura Y, Kasahara M, Kiyosue T, Kikuyama M, Wada M, Kataoka H. AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc Natl Acad Sci USA. 2007;104:19625–19630. doi: 10.1073/pnas.0707692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka T, Hisatomi O, Takahashi F, Kataoka H, Terazima M. Photoreactions of aureochrome-1. Biophys J. 2011;100:2801–2809. doi: 10.1016/j.bpj.2011.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Liao HJ, Chattopadhyay A, Carpenter G. EGF-dependent translocation of green fluorescent protein-tagged PLC-gamma1 to the plasma membrane and endosomes. Exp Cell Res. 2001;267:28–36. doi: 10.1006/excr.2001.5241. [DOI] [PubMed] [Google Scholar]
- Welm BE, Freeman KW, Chen M, Contreras A, Spencer DM, Rosen JM. Inducible dimerization of FGFR1: development of a mouse model to analyze progressive transformation of the mammary gland. J Cell Biol. 2002;157:703–714. doi: 10.1083/jcb.200107119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Zhang Y, Ressler SJ, Ittmann MM, Ayala GE, Dang TD, Wang F, Rowley DR. FGFR1 is essential for prostate cancer progression and metastasis. Cancer Res. 2013;73:3716–3724. doi: 10.1158/0008-5472.CAN-12-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Daoud-El Baba M, Peng RW, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–1568. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Li D, Shimazu K, Zhou YX, Lu B, Deng CX. Fibroblast growth factor receptor-1 is required for long-term potentiation, memory consolidation, and neurogenesis. Biol Psychiatry. 2007;62:381–390. doi: 10.1016/j.biopsych.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Zoltowski BD, Gardner KH. Tripping the light fantastic: blue-light photoreceptors as examples of environmentally modulated protein-protein interactions. Biochemistry. 2011;50:4–16. doi: 10.1021/bi101665s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat Chem Biol. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


