Abstract
Glucose-6-phosphate dehydrogenase (G6PD) is a key enzyme in the pentose phosphate pathway (PPP) and plays an essential role in the oxidative stress response by producing NADPH, the main intracellular reductant. G6PD deficiency is the most common human enzyme defect, affecting more than 400 million people worldwide. Here, we show that G6PD is negatively regulated by acetylation on lysine 403 (K403), an evolutionarily conserved residue. The K403 acetylated G6PD is incapable of forming active dimers and displays a complete loss of activity. Knockdown of G6PD sensitizes cells to oxidative stress, and re-expression of wild-type G6PD, but not the K403 acetylation mimetic mutant, rescues cells from oxidative injury. Moreover, we show that cells sense extracellular oxidative stimuli to decrease G6PD acetylation in a SIRT2-dependent manner. The SIRT2-mediated deacetylation and activation of G6PD stimulates PPP to supply cytosolic NADPH to counteract oxidative damage and protect mouse erythrocytes. We also identified KAT9/ELP3 as a potential acetyltransferase of G6PD. Our study uncovers a previously unknown mechanism by which acetylation negatively regulates G6PD activity to maintain cellular NADPH homeostasis during oxidative stress.
Keywords: acetylation, G6PD, nicotinamide adenine dinucleotide phosphate, reactive oxygen species, SIRT2
Introduction
Nicotinamide adenine dinucleotide phosphate (NADPH) is a functionally important metabolite that is commonly used for reductive biosynthesis and maintenance of cellular redox potential. It is a required cofactor in reductive biosynthesis of fatty acids, isoprenoids, and aromatic amino acids (Turner & Turner, 1980; Graeve et al, 1994; Hauschild & von Schaewen, 2003). NADPH is also used to keep glutathione in its reduced form. Reduced glutathione (GSH) acts as a scavenger for dangerous oxidative metabolites in the cell, and it converts harmful hydrogen peroxide to water with the help of glutathione peroxidase (GSHPx) (Margis et al, 2008). Perturbed NADPH production increases sensitivity to reactive oxygen species (ROS) and provokes apoptosis (Kim et al, 2007). Despite the functional importance of NADPH, mechanisms of maintaining cellular NADPH homeostasis are not fully understood.
Numerous pathways are known to maintain cellular NADPH levels. The major NADPH-producing enzymes in the cell are glucose-6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (6PGD) in the pentose phosphate pathway (PPP), malic enzyme (ME) in the pyruvate cycling pathway, and isocitrate dehydrogenase (IDH) in the tricarboxylic acid (TCA) cycle (Salati & Amir-Ahmady, 2001). Activity of IDH1, ME1, and 6PGD remains unchanged during oxidative stress, while G6PD is the only NADPH-producing enzyme that is activated (Filosa et al, 2003). G6PD catalyzes the oxidation of glucose-6-phosphate to 6-phosphogluconate and concomitantly reduces NADP+ to NADPH, which is the rate-limiting and primary control step of the NADPH-generating portion in the PPP. Thus, G6PD acts as a guardian of cellular redox potential during oxidative stress (Filosa et al, 2003).
G6PD is highly conserved from yeast to mammalian species (Kletzien et al, 1994; Notaro et al, 2000). Yeast carries one G6PD gene, Zwf1, which when deleted causes phenotypes indicative of hypersensitivity to oxidative stress, including aerobic methionine auxotrophy and sensitivity to hydrogen peroxide (Juhnke et al, 1996; Lee et al, 1999; Blank et al, 2005). The critical function of G6PD in oxidative stress response is also conserved in mammals. Mouse embryonic stem cells with G6PD deletion display an approximately 50% reduction in the [NADPH]/[NADP+] ratio and are extremely sensitive to the lethal effects of external chemical oxidants (Pandolfi et al, 1995; Filosa et al, 2003). Furthermore, human G6PD deficiency is a common genetic abnormality found in 5–10% of the global population (Cappellini & Fiorelli, 2008). While the polymorphic mutations in G6PD affect amino acid residues throughout the enzyme and decrease the stability of the enzyme in the red blood cell, the severe mutations mostly affect residues at the dimer interface or those that interact with a structural NADP+ molecule that stabilizes the enzyme (Mason et al, 2007). The distinctive phenotype of patients with G6PD deficiency is chronic and drug- or food-induced hemolytic anemia, which is attributed to the inability to produce NADPH and withstand harmful oxidants in erythrocyte cells where other NADPH-producing enzymes are lacking (Vulliamy et al, 1993). Together, these findings further support the notion that G6PD is of central importance for NADPH homeostasis and redox regulation.
We and others have previously discovered that lysine acetylation is an evolutionarily conserved post-translational modification in the regulation of a wide range of cellular processes, particularly in nuclear transcription and cytoplasmic metabolism (Kim et al, 2006; Choudhary et al, 2009; Zhao et al, 2010). The acetylation state of a given protein results from the balanced action of lysine acetyltransferases (KATs) and deacetylases (KDACs), enzymes that catalyze the addition and removal, respectively, of an acetyl group from a lysine residue. In particular, KDACs including classical HDACs (histone deacetylases) and sirtuins (SIRTs) have received more and more attention not only for their physiological roles, but also for their involvement in disease states and, consequently, for being a therapeutic target (Haberland et al, 2009). Several recent acetylome proteomic studies have identified more than 4,500 acetylated proteins (Kim et al, 2006; Choudhary et al, 2009; Zhao et al, 2010; Lundby et al, 2012). Among these identified acetylated proteins is G6PD, implicating a novel regulatory mechanism of G6PD at the post-translational level. This study is directed toward identifying potential KAT and KDAC enzymes of G6PD and understanding how acetylation regulates G6PD activity to maintain cellular NADPH homeostasis and redox potential during oxidative stress.
Results
Lysine 403 is an important regulatory acetylation site in G6PD
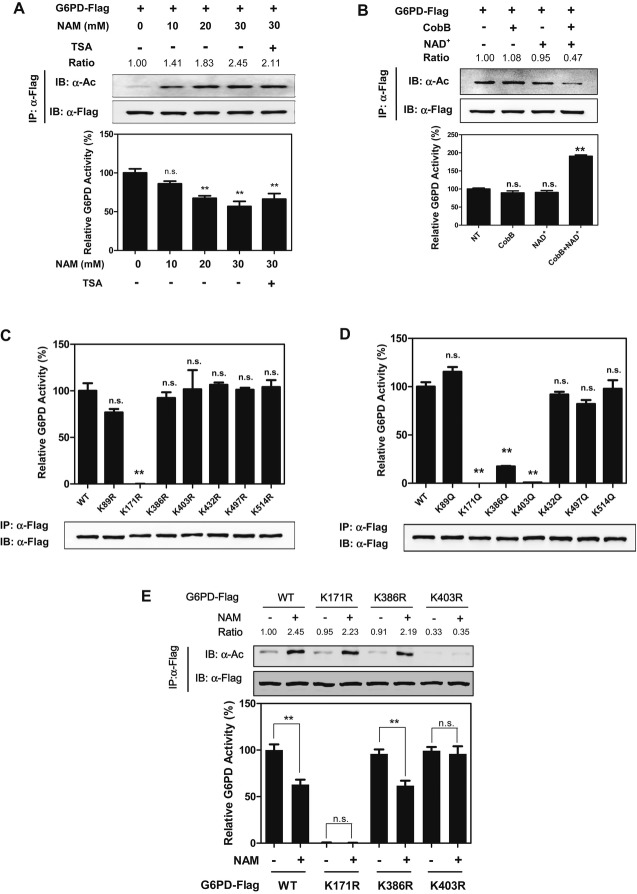
In a recent proteomic study (Choudhary et al, 2009), G6PD was identified to be acetylated on 7 lysine residues, including lysine 89 (K89), lysine 171 (K171), lysine 386 (K386), lysine 403 (K403), lysine 432 (K432), lysine 497 (K497), and lysine 514 (K514) (A Fig S1). Western blotting with a pan-anti-acetyllysine antibody demonstrated that G6PD was indeed acetylated and its acetylation was significantly elevated (up to ˜2.5-fold) in HEK293T cells after treatment with nicotinamide (NAM), an inhibitor of the SIRT family deacetylases (Bitterman et al, 2002; Avalos et al, 2005; Smith et al, 2008) (Fig1A). The effect of NAM on increasing G6PD acetylation was found to be in a dose-dependent manner, while G6PD specific activity was decreased by as much as 40% after NAM treatment (Fig1A). Treatment with trichostatin A (TSA), an inhibitor of histone deacetylase HDAC (Furumai et al, 2001), did not affect G6PD acetylation and activity (Supplementary Fig S2), and additional treatment with TSA did not further change either G6PD acetylation or its enzyme activity in cells co-treated with NAM (Fig1A). When purified G6PD protein was incubated in vitro with bacterial deacetylase CobB (Zhao et al, 2004) in the presence of NAD+, G6PD acetylation was decreased by twofold and concomitantly its enzymatic activity was increased by as much as twofold (Fig1B). These data suggest that acetylation negatively regulates G6PD activity.
Figure 1. Acetylation negatively regulates G6PD activity.
A G6PD acetylation inhibits its enzyme activity. Flag-tagged G6PD was expressed in HEK293T cells treated with NAM or NAM+TSA at the indicated concentrations. Acetylation levels and enzyme activity of Flag bead-purified G6PD were determined by Western blot analysis and enzyme assay, respectively. Acetylation levels were blotted with a pan-anti-acetyllysine antibody (α-Ac). Catalytic activity of affinity-purified G6PD was determined and normalized to protein levels. G6PD activity under no treatment condition was set as 100%. Shown are average values with standard deviation (s.d.) of triplicated experiments. IB and IP denote immunoblotting and immunoprecipitation, respectively. ** denotes P < 0.01 for cells treated with NAM/TSA for the indicated periods versus no NAM/TSA treatment; n.s. = not significant. G6PD acetylation levels were normalized against Flag.
B G6PD is activated by in vitro deacetylation. Affinity-purified Flag-tagged G6PD was incubated with recombinant CobB with or without NAD+ at 37°C for 2 h. G6PD acetylation and activity were determined. Shown are average values with standard deviation (s.d.) of triplicated experiments. ** denotes P < 0.01 for cells treated with CobB and/or NAD+ versus no treatment (NT); n.s. = not significant. G6PD acetylation levels were normalized against Flag.
C, D Mapping the major regulated sites of acetylation in G6PD. Wild-type (WT) G6PD and the indicated mutants were each expressed in HEK293T cells. Proteins were purified by IP, and specific G6PD activity was determined. Shown are average values with standard deviation (s.d.) of triplicated experiments. ** denotes P < 0.01 for cells expressing the indicated G6PD mutants versus cells expressing WT G6PD; n.s. = not significant.
E K403 is the important regulatory acetylation site in G6PD. Flag-tagged wild-type G6PD, the K171R, K386R, and K403R mutants were each expressed in HEK293T cells, followed by treatments with or without 15 mM NAM. Acetylation levels and activity of G6PD were determined. Shown are average values with standard deviation (s.d.) of triplicated experiments. ** denotes P < 0.01 for the indicated comparison; n.s. = not significant. G6PD acetylation levels were normalized against Flag.
Because G6PD is a highly conserved protein (Kletzien et al, 1994), we speculated that important regulatory sites targeted by acetylation might be also conserved. Sequence alignments from diverse species revealed that five of the acetylated lysines (K89, K386, K432, K497, and K514) are not conserved, while two lysines (K171 and K403) are invariant (Supplementary Fig S1). To determine which lysine residue(s) plays a major role in the regulation of G6PD, we mutated each of the 7 putative acetylation sites to arginine (R) or glutamine (Q) and assayed their activity individually. The K to R mutation retains a positive charge and is often used as a deacetylated mimetic, whereas the K to Q mutation abolishes the positive charge and may act as a surrogate of acetylation (Megee et al, 1990). By continuously monitoring the generation of NADPH (Tian et al, 1998), we found that substitutions at K89, K432, K497, and K514 did not significantly affect G6PD activity as compared with wild-type G6PD (Fig1C and D). Mutation of K171 to either arginine or glutamine led to a complete loss in G6PD catalytic activity (Fig1C and D). Mutation of either K386 or K403 to glutamine, but not to arginine, resulted in a significant reduction in G6PD activity (Fig1C and D). Moreover, the K386R mutant responded normally to NAM treatment regarding G6PD acetylation and enzyme activity (Fig1E). However, the K403R mutant displayed negligible response in changing acetylation and enzyme activity upon NAM treatment (Fig1E and Supplementary Fig S3). These results suggest that K403 is an important regulatory acetylation site which controls G6PD activity.
Acetylation of K403 impairs the formation of dimeric G6PD and inhibits enzyme activity
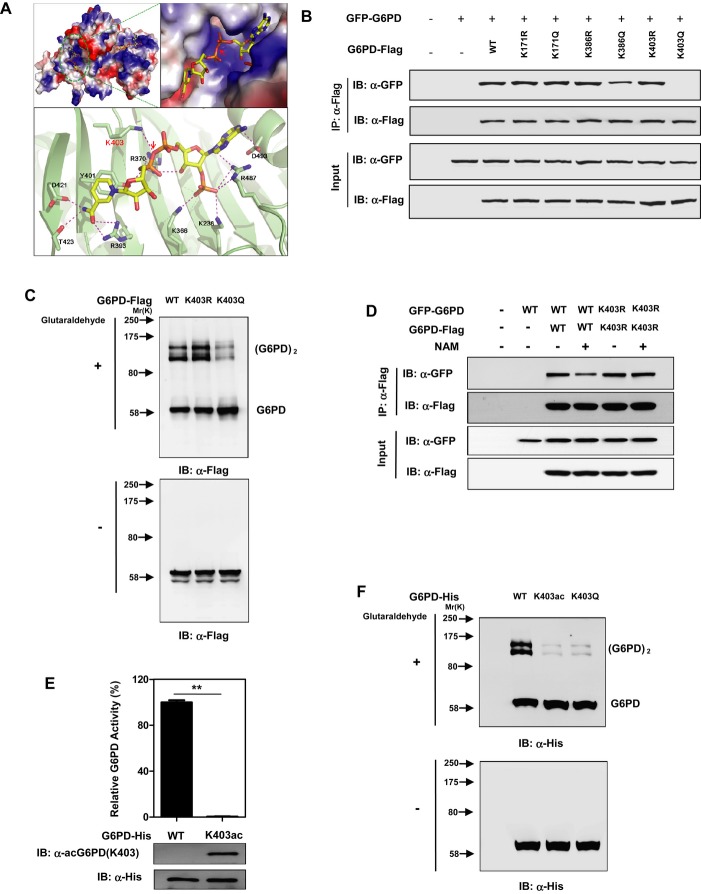
The G6PD enzyme exists as a mixture of monomer, dimer, tetramer, and hexamer, but only the dimeric and tetrameric forms are catalytic active (Cohen & Rosemeyer, 1969; Babalola et al, 1976). The structure of human G6PD reveals that each subunit contains two NADP+ binding sites, a catalytic NADP+ coenzyme-binding domain and a structural NADP+ binding domain (Au et al, 2000). The structural NADP+ binding site is distant from the catalytic site but close to the dimer interface. The interplay of the structural NADP+ and the dimer interface affects the stability and integrity of active enzyme (Au et al, 2000; Wang & Engel, 2009). A high proportion of the clinical mutations associated with severe G6PD deficiency in human clusters around the structural NADP+ site (Au et al, 2000; Vulliamy & Luzzatto, 2003), suggesting that the structural NADP+ is vital for G6PD activity and function. Notably, K171 lies in the catalysis pocket of G6PD and directly binds with both G6P and catalytic NADP+, while K386 and K403 lie close to the dimer interface (Supplementary Fig S4). In particular, K403 physically interacts with structural NADP+ (Fig2A). Therefore, it is possible that K171Q mutation would abolish G6PD activity through directly influencing the substrate recognition and/or catalysis, while K386Q and K403Q mutations would inhibit G6PD activity through impairing the integrity of dimer interface and subsequently the formation of active dimers and higher forms of G6PD.
Figure 2. K403 acetylation impairs the formation of dimeric G6PD and inhibits its enzyme activity.
A Cartoon representation of G6PD structure (PDB ID: 2BH9) (Kotaka et al, 2005) made by using Pymol (http://www.pymol.org). Upper left, electrostatic (G6PD) and stick (NADP+) representation of the crystal structure of G6PD bound to NADP+. Upper right, a closer view showing the structural NADP+ bound to a positive charged groove of G6PD. Lower panel, shown is a ribbon (G6PD)-and-stick (structural NADP+) representation of G6PD bound to the structural NADP+. The protein is colored in light green and NADP+ in yellow, with interacting residues on G6PD colored in magenta. Arrow indicates the interaction between Lys 403 and the structural NADP+.
B Acetylation at K403 blocks the protein-protein binding between wild-type G6PD. Flag-tagged G6PD, KR/KQ mutants of K171, K386, and K403 were each expressed in HEK293T cells co-expressing GFP-tagged G6PD. The interaction between Flag-tagged and GFP-tagged proteins was determined by Western blotting.
C K403Q mutation, but not K403R, inhibits the formation of dimeric G6PD. Flag-tagged G6PD, G6PDK403R, and G6PDK403Q were each expressed in HEK293T cells, followed by treatments with or without 0.025% glutaraldehyde. The formation of G6PD monomer and dimer was determined by Western blotting.
D NAM treatment impairs the protein-protein interaction between WT G6PD, but not the K403R mutant. Flag-tagged G6PD and G6PDK403R were each expressed in HEK293T cells co-expressing GFP-tagged G6PD and G6PDK403R, respectively. Cells were treated with or without 15 mM NAM, and the interaction between Flag-tagged and GFP-tagged proteins was determined by Western blotting.
E, F In vitro site-specific incorporation of acetylated K403 in G6PD. His-tagged G6PD and G6PDK403ac were recombinantly expressed and detected by Western blot with an anti-His antibody as well as a site-specific anti-acetyllysine antibody [α-acG6PD(K403)]. G6PD activity assay was performed using the purified unacetylated G6PD and G6PDK403ac proteins (E). Moreover, the purified unacetylated G6PD and G6PDK403ac proteins were treated with or without 0.025% glutaraldehyde, and the formation of G6PD monomer and dimer was tested by Western blotting (F). Purified Flag-tagged G6PDK403Q was used as a control. ** denotes P < 0.01 for the purified G6PDK403ac protein versus the unacetylated G6PD protein.
To test this hypothesis, we determined the interaction between two differentially tagged G6PD proteins, G6PD-Flag and GFP-G6PD, in HEK293T cells. We found that mutation of K171 to either R (K171R) or Q (K171Q) did not affect the interaction between G6PD subunits (Fig2B). In contrast, substitution of K386Q, but not K386R, impaired the interaction between G6PD subunits (Fig2B). Strikingly, substitution of K403Q, but not K403R, entirely disrupted the interaction between G6PD subunits (Fig2B). In addition, glutaraldehyde cross-linking assay demonstrated that the K403Q mutant displayed impaired ability to form dimers when compared to wild-type G6PD or the K403R mutant (Fig2C). Moreover, NAM treatment decreased the binding by approximately 55% between the two differentially tagged proteins of wild-type G6PD in a dose-dependent manner (Fig2D and Supplementary Fig S5). NAM treatment, however, failed to affect the interaction between Flag-tagged and GFP-tagged K403R mutant of G6PD (Fig2D), further suggesting that K403 acetylation largely hinders the interaction between G6PD subunits.
To unambiguously determine the effect of K403 acetylation on G6PD, we employed an expression system genetically encoding Nε-acetyllysine to prepare recombinant proteins in E. coli (Neumann et al, 2008, 2009). This expression system produced G6PD proteins with 100% acetylation at the targeted lysine residue. Only with Nε-acetyllysine in the growth medium was the full-length protein formed (Supplementary Fig S6). Moreover, we generated and verified an antibody specifically recognizing the K403 acetylated G6PD [α-acG6PD(K403)] (Supplementary Fig S7). The incorporated acetyllysine was confirmed by immunoblotting of the purified G6PDK403ac protein with this site-specific α-acG6PD(K403) antibody. Importantly, as compared to wild-type and K403R/Q mutants of G6PD, the recombinant G6PDK403ac protein displayed an identical pattern of proteolytic cleavage after treatments with proteases, chymotrypsin and clostripain (Supplementary Fig S8B). Moreover, the G6PDK403ac protein exhibited normal thermodynamic stability of protein folding when compared to wild-type and K403R/Q mutants of G6PD (Supplementary Fig S8C). This purified G6PDK403ac protein was catalytic inactive (Fig2E and Supplementary Fig S8A), unequivocally demonstrating that acetylation of K403 inactivates G6PD. Furthermore, our data demonstrated that the G6PDK403ac protein was defective in dimer formation (Fig2F). Together, these results clearly indicate that acetylation at K403 impairs the formation of dimeric G6PD and inhibits its enzyme activity.
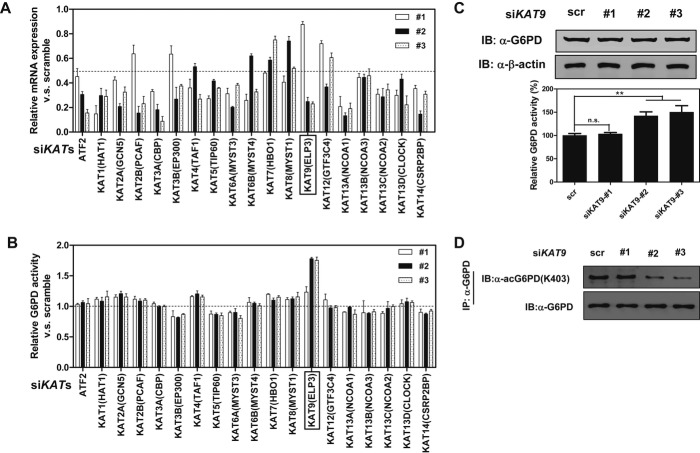
KAT9/ELP3 is involved in G6PD K403 acetylation and enzymatic inactivation
Next, we set out to search for potential KAT(s) responsible for G6PD K403 acetylation. To this end, we generated a siRNA library with three siRNAs targeting each of the 19 human KATs (Allis et al, 2007), and then determined the activity of endogenous G6PD in HEK293T cells with transient knockdown of individual KAT gene. The knockdown efficiency of each siRNA was determined by quantitative RT-PCR of its target gene (Fig3A). We found that knocking down most of the examined KAT genes did not substantially affect the enzyme activity of endogenous G6PD (Fig3B). With one exception, knocking down KAT9 (also known as ELP3), which encodes the catalytic subunit of the histone acetyltransferase elongator complex and has previously been identified as an α-tubulin acetyltransferase in mouse neurons (Creppe et al, 2009), significantly stimulated the activity of endogenous G6PD (Fig3B). Moreover, the degree of G6PD activation appeared to correlate with the KAT9 knockdown efficiency, as siRNA no. 2 and no. 3 were more potent in both KAT9 knockdown and G6PD activation than the siRNA no. 1. As expected, transient knockdown of KAT9 decreased the K403 acetylation levels of endogenous G6PD without changing its protein expression (Fig3C and D), further supporting the notion that KAT9 is the potential acetyltransferase of G6PD.
Figure 3. KAT9/ELP3 is the potential acetyltransferase of G6PD.
A A siRNA library with three siRNAs targeting each of the 19 known HAT genes was generated. Each siRNA oligonucleotide was transiently transfected into HEK293T cells, and mRNA expression of HAT genes was determined by quantitative real-time PCR at 48 h post-transfection.
B HEK293T cells were transfected as described in (A) and then were subjected to G6PD activity assay.
C, D Three different siRNAs targeting KAT9 were transiently transfected into HEK293T cells. Protein expression and enzyme activity (C) and the K403 acetylation level (D) of endogenous G6PD were determined. Shown are average values with standard deviation (s.d.) of triplicated experiments. ** denotes P < 0.01 for the indicated comparison; n.s. = not significant.
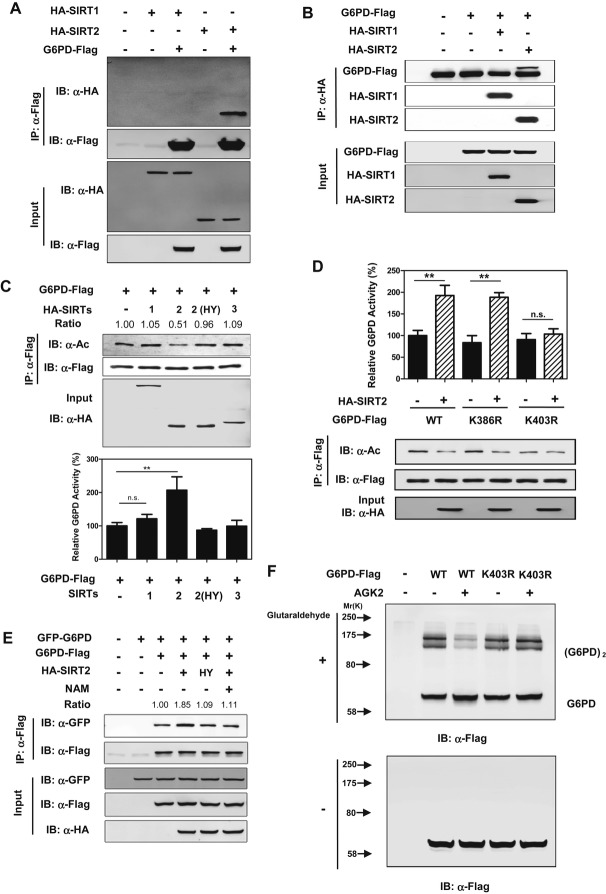
SIRT2 activates G6PD by deacetylation
Our earlier observation that NAM increases G6PD acetylation (Fig1A) led us to investigate a possible involvement of NAD+-dependent sirtuins in G6PD deacetylation. The first known sirtuin, Sir2 (silent information regulator 2) of Saccharomyces cerevisiae (Imai et al, 2000), from which the family derives its name, regulates lifespan in worms and flies (Michan & Sinclair, 2007). Mammals contain seven Sir2 homologues, SIRT1-7, each with diverse subcellular localization and protein substrates. SIRT1-3 display robust deacetylation activity, while SIRT4-7 have no detectable or very weak deacetylase activity and show diverse substrate specificity (Hirschey, 2011; Feldman et al, 2013). Given that G6PD is localized in the cytoplasm (Notaro et al, 2000), we examined whether cytosolic SIRTs, SIRT1 or SIRT2 (Michishita et al, 2005; Schwer & Verdin, 2008), could deacetylate G6PD and stimulate its enzymatic activity. We observed that G6PD directly interacted with SIRT2, but not SIRT1 (Fig4A and B). Co-expression of SIRT2 with G6PD in HEK293T cells decreased the acetylation level of G6PD by 50%, while increased G6PD enzyme activity by twofold (Fig4C). When G6PD was co-expressed with a catalytic inactive mutant of SIRT2, SIRT2H187Y (North et al, 2003), neither G6PD acetylation nor enzyme activity was changed (Fig4C), connecting G6PD deacetylation and activation directly to SIRT2 catalytic activity. Furthermore, we found that SIRT2 deacetylated and activated wild-type G6PD and the K386R mutant, but not the K403R mutant (Fig4D), confirming that K403 is an important site of acetylation in the regulation of G6PD activity by SIRT2. Co-expression of SIRT2, but not the catalytic inactive SIRT2H187Y mutant, increased the interaction between two differentially tagged G6PD, and this effect could be diminished by NAM treatment (Fig4E). In addition, AGK2, a SIRT2-specific inhibitor (Outeiro et al, 2007), inhibited the dimer formation of wild-type G6PD, while the K403R mutant displayed negligible response in the monomer-to-dimer transition upon AGK2 treatment (Fig4F). Taken together, these findings demonstrate that SIRT2 stimulates G6PD activity by deacetylating G6PD at K403 to stimulate the formation of active dimers.
Figure 4. SIRT2 decreases G6PD acetylation and increases its enzyme activity by inducing the formation of dimeric G6PD.
A, B G6PD interacts with SIRT2, but not SIRT1. Flag-tagged G6PD was expressed in HEK293T cells together with the individual HA-tagged SIRT as indicated. Proteins were purified by IP with Flag beads, followed by Western blot to detect SIRTs with a HA antibody (A). On the other hand, SIRT1 and SIRT2 proteins were purified by IP with HA beads, followed by Western blot to detect G6PD with a Flag antibody (B).
C G6PD is deacetylated and activated by SIRT2, but not SIRT1. Flag-tagged G6PD was expressed in HEK293T cells together with the individual HA-tagged SIRT as indicated. Among them, SIRT2H187Y is a catalytically inactive mutant. G6PD proteins were purified by Flag beads, and acetylation levels and enzyme activity were determined by Western blot analysis and enzyme assay, respectively. Shown are average values with standard deviation (s.d.) of triplicated experiments. G6PD acetylation levels were normalized against Flag protein levels.
D SIRT2 deacetylates G6PD mainly at the site of K403. Flag-tagged G6PD, G6PDK386R, and G6PDK403R were each expressed in HEK293T cells co-expressing HA-tagged SIRT2. G6PD proteins were purified by Flag beads, and acetylation levels were determined by Western blot. Shown are average values with standard deviation (s.d.) of triplicated experiments. ** denotes P < 0.01 for the indicated comparison; n.s. = not significant.
E SIRT2 increases the protein-protein interaction between wild-type G6PD. GFP-tagged and Flag-tagged G6PD were co-transfected along with HA-tagged SIRT2 or its catalytic inactive mutant SIRT2H187Y in HEK293T cells. Cells were treated with or without 15 mM NAM, and the interaction between Flag-tagged and GFP-tagged proteins was determined by Western blotting. GFP protein levels were normalized against Flag protein levels.
F Inhibition of SIRT2 decreases the formation of dimeric G6PD. G6PD and G6PDK403R were each expressed in HEK293T cells, followed by treatments without or with AGK2 (10 μM). Extracts of the cells were treated with or without 0.025% glutaraldehyde, and the formation of G6PD monomer and dimer was determined by Western blotting.
K403 acetylation plays a signaling role in regulating G6PD enzyme activity under oxidative stress
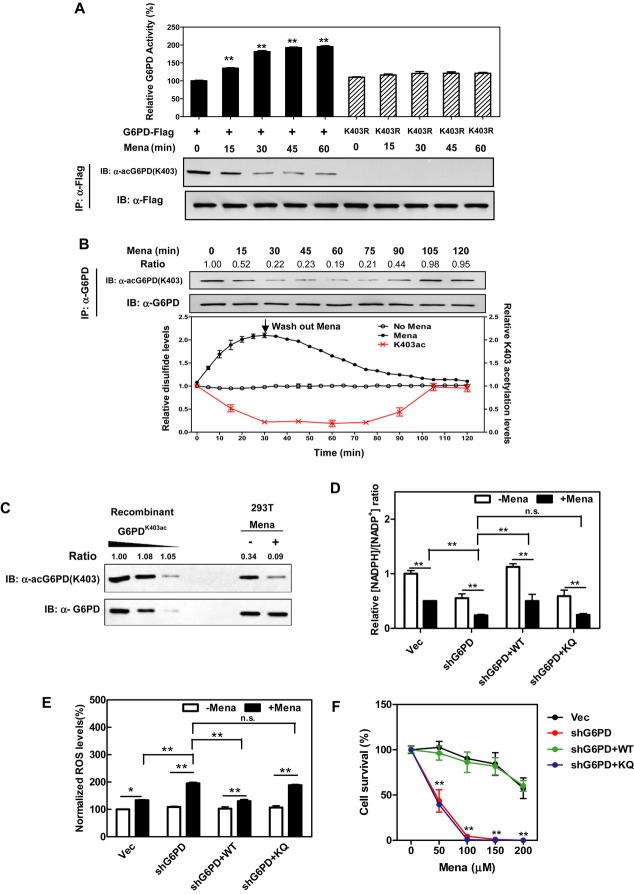
Being the key enzyme that controls the PPP, G6PD activity is tightly regulated by the [NADPH/NADP+] ratio in the cell. G6PD is inhibited by the high steady-state ratio of [NADPH] to [NADP+] in cells under non-stress conditions (Berg et al, 2006), while G6PD and the pentose phosphate pathway activity are stimulated to restore NADPH after exposure to extracellular oxidants (Cramer et al, 1995; Slekar et al, 1996; Ursini et al, 1997; Filosa et al, 2003), which are supposed to decrease the intracellular NADPH level (Xu et al, 2010). To determine whether acetylation at K403 is involved in the regulation of G6PD activity during oxidative stress, we treated HEK293T cells ectopically expressing wild-type G6PD or the K403R mutant with agents that directly induce oxidative stress, including hydrogen peroxide (H2O2) and menadione (a quinone compound that induces the production of superoxide radicals). We found that treatment with H2O2 (300 μM) led to a rapid (within 15-min) decrease in the K403 acetylation level of wild-type G6PD and thereby rapidly increased G6PD activity (Supplementary Fig S9A). In contrast, the cells expressing K403R mutant displayed a negligible response in changing enzyme activity in response to H2O2. Similar changes in G6PD K403 acetylation and enzyme activity were found in cells expressing wild-type G6PD upon treatment with menadione (50 μM), and again, the K403R-expressing cells displayed a negligible response in changing enzyme activity in response to menadione (Fig5A).
Figure 5. G6PD K403 deacetylation leads to G6PD activation in cells under oxidative stress.
- Menadione decreases G6PD K403 acetylation and activates enzyme activity of ectopically expressed G6PD. Flag-tagged WT G6PD or the K403R mutant was expressed in HEK293T cells and was then treated with 50 μM menadione for the indicated periods. The K403 acetylation levels and enzyme activity of Flag bead-purified G6PD were determined by Western blot analysis and enzyme assay, respectively. Shown are average values with standard deviation (s.d.) of triplicated experiments. ** denotes P < 0.01 for cells treated with H2O2 or menadione versus cells without oxidant treatment.
- Menadione dynamically changes the K403 acetylation level of endogenous G6PD. HEK293roGFP1 cells were treated with 25 μM menadione for the indicated periods. The relative disulfide level in the cytoplasm was monitored by using a fluorescent biosensor as described in Materials and Methods. The K403 acetylation level of endogenous G6PD was determined by Western blot analysis. Shown are average values with standard deviation (s.d.) of triplicated experiments. Relative G6PD K403 acetylation levels were normalized against G6PD protein levels.
- Determination of endogenous G6PD K403 acetylation ratio. Recombinant fully K403 acetylated G6PD was loaded onto the same gel with endogenous G6PD from HEK293T cells treated with or without 50 μM menadione for 30 min. G6PD protein and K403 acetylation were detected by Western blot. Relative K403 acetylation ratios were calculated after normalizing against G6PD protein levels.
- K403 is critical for G6PD function to produce NADPH. The ratio of reduced and oxidized forms of NADP, that is the ratio of [NADPH to NADP+], was determined by the enzymatic analysis of cell extracts from G6PD-knockdown cells re-expressing the indicated proteins after treatments with or without menadione (50 μM for 30 min). Shown are average values with standard deviation (s.d.) of triplicated experiments. ** denotes P < 0.01 for the indicated comparison; n.s. = not significant.
- K403 is critical for G6PD function to suppress cellular ROS production under oxidative stress. G6PD-knockdown cells or these cells rescued by WT G6PD or the K403Q mutant were treated with menadione (50 μM for 30 min), and ROS accumulation was determined by using a fluorescent dye as described in Supplementary Materials and Methods. Shown are average values with standard deviation (s.d.) of triplicated experiments. * denotes P < 0.05, and ** denotes P < 0.01 for the indicated comparison; n.s. = not significant.
- K403 is critical for G6PD function to protect cells from ROS-induced cell death. G6PD-knockdown cells and these cells rescued by WT G6PD or the K403Q mutant were treated with the indicated concentrations of menadione for 3 h, and cell viability was determined by counting the remaining adherent cells. Shown are average values with standard deviation (s.d.) of triplicated experiments. ** denotes P < 0.01 for knockdown and rescue cells versus knockdown cells; n.s. = not significant.
To accurately measure intracellular redox status, we established HEK293 cells stably expressing cytosolic redox-sensitive green fluorescent protein 1 (HEK293roGFP1) which allows real-time visualization of thiol-disulfide metabolic state in the cytosol of living cells (Dooley et al, 2004). We found that the disulfide level in H2O2-exposed cultures peaked at 10 min and was followed by a rapid full recovery (Supplementary Fig S9B). Notably, the K403 acetylation level of endogenous G6PD was rapidly (within 15 min) decreased in response to H2O2 followed by a period of recovery that slowly lagged after the recovery of cellular disulfide level (Supplementary Fig S9B). When cells were treated with menadione, the disulfide level was steadily increased, and a recovery occurred only after menadione was washed out from the culture (Fig5B). G6PD K403 acetylation also showed an inverse, but delayed correlation with the oxidized disulfide. To more accurately quantify these changes in endogenous G6PD K403 acetylation, we used the recombinant G6PDK403ac protein purified from E. coli as the standard and found that approximately 34% of endogenous G6PD was acetylated at K403 in HEK239T cells, which was decreased to approximately 9% after menadione treatment (Fig5C). These results indicate that G6PD K403 acetylation is regulated by cellular oxidative status and likely plays a signaling role in the dynamic regulation of G6PD activity.
To evaluate the function of G6PD K403 acetylation in regulating cellular NADPH homeostasis, we generated stable G6PD-knockdown and G6PD-rescued cells in HEK293T (Supplementary Fig S10). HEK293T cells with G6PD-knockdown displayed an approximately 45% reduction in the [NADPH]/[NADP+] ratio, reaffirming that G6PD is an important contributor to NADPH pools in the cell (Fig5D). The lower [NADPH]/[NADP+] ratio was in accord with the higher ROS production in G6PD-knockdown cells subjected to menadione (Fig5E). Importantly, re-expression of wild-type G6PD, but not the acetylated mimetic K403Q mutant, restored the [NADPH]/[NADP+] ratio and suppressed ROS production in G6PD-knockdown and rescue cells subjected to menadione (Fig5D and E). ROS has been extensively implicated in signaling cascades which function as important cell survival mechanisms in response to oxidative stress (Kregel & Zhang, 2007). As compared to control cells expressing an empty vector, when treated with menadione G6PD-knockdown cells exhibited higher levels of cleaved PARP and Caspase-3, two indicators of apoptosis (Supplementary Fig S11A), as well as higher levels of p38 MAPK phosphorylation, a stress responsive kinase (Supplementary Fig S11B). Re-introduction of wild-type G6PD, but not the acetylated mimetic K403Q mutant, reduced the levels of cleaved PARP and Caspase-3, and p38 MAPK phosphorylation in G6PD-knockdown cells when subjected to menadione (Supplementary Fig S11). Consequently, G6PD-knockdown cells exhibited a higher incidence of cell death in response to menadione, and re-expression of wild-type G6PD, but not the acetylated mimic K403Q mutant, could rescue cells from menadione-induced cell death (Fig5F). These findings clearly support an important role of G6PD K403 acetylation in controlling NADPH production and protecting cells from oxidative stress.
SIRT2 controls G6PD K403 deacetylation in response to oxidative stress
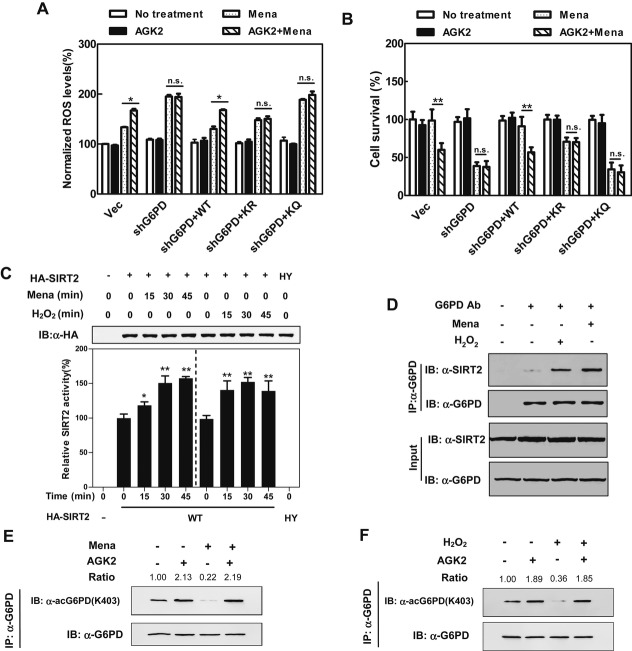
G6PD-knockdown cells were highly sensitive to menadione in both ROS production and cell death (Fig6A and B). Inhibition of SIRT2 by AGK2 did enhance the effect of menadione on ROS production and cell death in both the control and wild-type G6PD-rescued cells (Fig6A and B). Notably, AGK2 was unable to enhance the effect of menadione on ROS production and cell death in the G6PD-knockdown or K403R/K403Q-rescued cells, indicating that G6PD is required for AGK2 to synergize with menadione. Supporting this concept, transient knocking down SIRT2, but not SIRT1, increased the K403 acetylation level of endogenous G6PD in HEK293T cells (Supplementary Fig S12A and B), and SIRT2 knockdown enhanced the effect of menadione on ROS production and cell death in both the control and G6PD-rescued cells, but not in the G6PD-knockdown or K403R/K403Q-rescued cells (Supplementary Fig S12C).
Figure 6. SIRT2 controls G6PD K403 acetylation in response to oxidative stress.
A, B Inhibition of SIRT2 increases cellular susceptibility to oxidative stress. G6PD-knockdown and G6PD-rescued cells were treated with AGK2 (10 μM) for 4 h alone and/or menadione (50 μM) for 30 min. ROS accumulation was determined by using a fluorescent dye as described in Supplementary Materials and Methods (A), and cell viability was determined by counting the remaining adherent cells (B). Shown are average values with standard deviation (s.d.) of triplicated experiments. * denotes P < 0.05, and ** denotes P < 0.01 for the indicated comparison; n.s. = not significant.
C Chemical oxidants activate the deacetylase activity of SIRT2. HA-tagged SIRT2 or its catalytic inactive mutant (H187Y) was ectopically expressed in HEK293T cells and then treated with menadione (50 μM) or H2O2 (300 μM) for the indicated periods. HA-SIRT2 was purified by IP with HA beads, eluted with HA peptide, and subjected to deacetylase activity assay. Shown are average values with standard deviation (s.d.) of triplicated experiments. * denotes P < 0.05, and ** denotes P < 0.01 for cells treated with menadione or H2O2 versus cells without oxidant treatment.
D Chemical oxidants enhance the interaction between endogenous G6PD and SIRT2. HEK293T cells were treated with menadione (50 μM) or H2O2 (300 μM) for 30 min. The association of endogenous G6PD with SIRT2 was determined by Western blotting.
E, F Inhibition of SIRT2 blocks the effect of chemical oxidants on changing G6PD K403 acetylation. HEK293T cells were treated with or without 10 μM AGK2 for 4 h before treatment with menadione (50 μM) (E) or H2O2 (300 μM) (F) for 30 min, and the K403 acetylation level of endogenous G6PD was determined by Western blot analysis. Relative K403 acetylation ratios were calculated after normalizing against G6PD protein levels.
Interestingly, we found that either H2O2 or menadione did not change the transcriptional expression of SIRT2 gene (Supplementary Fig S13A and B), but significantly activated the deacetylase activity of SIRT2 (Fig6C). In vitro incubation of SIRT2 protein with redox reagents, such as either H2O2 or dithiothreitol (DTT), did not affect the deacetylase activity of SIRT2, suggesting that the observed enzymatic activation of SIRT2 by oxidants is not caused by the formation of disulfide bonds (Supplementary Fig S13C). On the other hand, the protein interaction between endogenous SIRT2 and G6PD was weak in cells under a non-stress condition, and this interaction was profoundly enhanced by treatment with either H2O2 or menadione (Fig6D). As a result, the K403 acetylation level of endogenous G6PD was decreased by > 50% by these extracellular oxidants (Fig6E and F), and such decreases were completely blocked by AGK2 treatment (Fig6E and F).
Taken together, our data indicate that K403 acetylation is crucial for the function of G6PD in maintaining cellular NADPH homeostasis and that oxidative stimuli affect G6PD K403 acetylation and activity in a SIRT2-dependent manner.
G6PD K403 deacetylation and enzyme activation are protective against oxidative stress in vivo
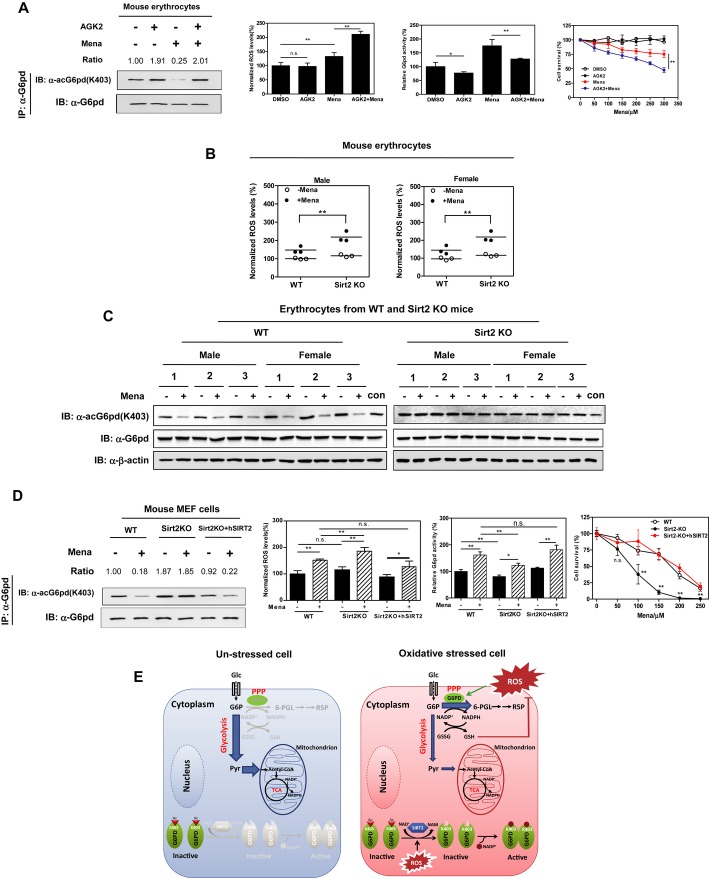
G6PD is particularly important for erythrocyte function and its deficiency leads to hemolytic anemia (Vulliamy et al, 1993). We thus investigated the functional importance of G6PD K403 acetylation in erythrocytes. We found that menadione treatment did not change the protein expression of G6pd or Sirt2 in mouse erythrocytes (Supplementary Fig S14), but did decrease the K403 acetylation level of G6pd by fourfold and consequently increased G6pd activity by 1.8-fold (Fig7A). These alterations in G6pd K403 acetylation and activity were, however, prevented by the addition of AGK2. As a result, combined exposure to AGK2 and menadione had synergistic effects on inducing ROS production and cell death in mouse erythrocytes (Fig7A). Additionally, Sirt2 deletion led to higher levels of menadione-induced ROS in erythrocytes from both female and male mice (Fig7B), further supporting a role of SIRT2 in G6PD regulation. Moreover, we found that Sirt2 deletion led to higher levels of G6pd K403 acetylation in mouse erythrocytes (Fig7C). After menadione treatment, erythrocytes from wild-type mice exhibited a dramatic reduction in G6pd K403 acetylation level, while those from Sirt2-deficient animals displayed negligible response in changing G6pd K403 acetylation (Fig7C).
Figure 7. G6PD K403 deacetylation and activation are protective against oxidative stress in vivo.
A Inhibition of SIRT2 impairs ROS scavenging in mouse erythrocytes. Mouse erythrocytes were freshly isolated and treated with AGK2 (10 μM) for 4 h alone and/or menadione (50 μM) for 30 min, and the K403 acetylation level of endogenous G6PD was determined by Western blot analysis. In addition, endogenous G6PD activity and cellular ROS accumulation were determined as described in Supplementary Materials and Methods. Moreover, mouse erythrocytes were treated with increasing concentrations of menadione as indicated along with or without 10 μM AGK2 for 6 h. Cell death assay was performed by counting floating trypan blue-positive cells. Shown are average values with standard deviation (s.d.) of triplicated experiments. * denotes P < 0.05, and ** denotes P < 0.01 for comparing cells after the indicated treatment; n.s. =not significant. Relative G6pd K403 acetylation levels were normalized against G6pd protein levels.
B, C Deletion of Sirt2 impairs ROS scavenging in mouse erythrocytes. Erythrocytes from wild-type or Sirt2-knockout mice (male n = 3 and female n = 3 per group) were isolated and treated with menadione (50 μM for 30 min). Cellular ROS accumulation was determined as described in Supplementary Materials and Methods (B) and the K403 acetylation level of endogenous G6pd was determined by Western blot analysis (C). Lysate of wild-type male mouse no. 1 was used as a loading control (con) for different panels. ** denotes P < 0.01 for the indicated comparisons; n.s. = not significant.
D Deletion of Sirt2 impairs ROS scavenging in mouse MEF cells. Wild-type, Sirt2-depleted, and Sirt2-depleted and rescued MEF cells were treated with menadione (50 μM) for 30 min. The K403 acetylation level of endogenous G6PD was determined by Western blot analysis. In addition, endogenous G6PD activity and cellular ROS accumulation were determined as described in Supplementary Materials and Methods. Moreover, mouse MEF cells were treated with increasing concentrations of menadione as indicated for 3 h. Cell survival was assessed by counting adherent MEF cells. Shown are average values with standard deviation (s.d.) of triplicated experiments. * denotes P < 0.05, and ** denotes P < 0.01 for comparing cells after the indicated treatment; n.s. = not significant. Relative G6PD K403 acetylation levels were normalized against G6PD protein levels.
E A working model for G6PD K403 acetylation plays a key role in regulating cellular NADPH homeostasis and redox balance under oxidative stress.
Finally, we examined the effects in mouse embryonic fibroblasts and again found that menadione treatment remarkably decreased K403 acetylation and increased activity of G6pd (Fig7D). To determine the function of Sirt2, Sirt2-null MEFs were tested (Supplementary Fig S15A). In these cells, Sirt2 deletion led to a higher level of K403 acetylation and impaired activity of G6pd when compared to wild-type MEFs under a non-stress condition (Fig7D). After menadione treatment, significant cell death was observed in Sirt2-null MEFs as compared to wild-type cells (Fig7D and Supplementary Fig S15B). It has to be noted that menadione treatment did not change the K403 acetylation level of G6pd in Sirt2-null MEFs, but did activate G6pd enzyme in these cells though less dramatic than the control cells (Fig7D), implying that G6PD could also be regulated by additional mechanisms besides the deacetylation by SIRT2. Nevertheless, stable expression of a human version SIRT2 in Sirt2-null MEF cells reduced the K403 acetylation of G6pd and sustainably activated G6pd to counterbalance menadione-induced ROS stress (Fig7D). As a result, human SIRT2 rescued Sirt2-null MEF cells from menadione-induced cell death (Fig7D), confirming the role of SIRT2 in G6PD activation and ROS scavenging.
Discussion
The current study uncovers a biochemical mechanism about how acetylation controls the activity/function of G6PD to modulate cellular NADPH homeostasis. SIRT2 plays a key role in G6PD deacetylation and activation, which is critical to maintain cellular redox potential and protects cells from oxidative damage.
We have identified K403 as an important regulatory acetylation site within the G6PD protein, which is supported by Western blotting of a K403 acetylation-specific antibody. K403 is located near the dimer interface and interacts with the structural NADP+, which is believed to be critical for the stability and integrity of the active form of G6PD (Wang et al, 2006; Wang & Engel, 2009). Our data show that K403Q substitution (acetylated mimic) displays a significant reduction in G6PD catalysis and is defective in dimer formation. This model is further supported by the recombinant G6PDK403ac protein, which is also inactive and fails to form dimers. Based on these findings, we hypothesize that acetylation at K403 of G6PD, partially through the ablation of the positively charged binding pocket at the dimer interface, may prevent the proper binding of the structural NADP+, thus leading to decreased dimer formation and enzyme activity.
Our study provides insights into the role of K403 acetylation in coordinating the monomer-to-dimer transition of G6PD in response to oxidative stress. It also raises a question regarding how cells sense physiologic stimuli to regulate G6PD acetylation. The NAD+ requirement of SIRTs for deacetylation suggests that these proteins may be sensors of the energy or redox state of cells. Increasing evidence indicates that SIRT2 is implicated in metabolism regulation and possibly cellular energy response (Cen et al, 2011; Guarente, 2011; Jiang et al, 2011b; Satoh et al, 2011). Other reports in yeast and mice under caloric-restricted conditions suggest that SIRT2 may also play a role in helping cells to cope with oxidative stress (Lamming et al, 2005; Wang et al, 2007; Zhu et al, 2012). To date, many substrates of SIRT2 have been identified, including α-tubulin (North et al, 2003), histone H3/H4 (Vaquero et al, 2006; Das et al, 2009), FOXO3A (forkhead box O transcription factor 3a) (Wang et al, 2007), and PEPCK1 (phosphoenolpyruvate carboxykinase 1) (Jiang et al, 2011b). Among them, only FOXO3A can be indirectly linked to ROS scavenging: SIRT2-dependent deacetylation of FOXO3A stimulates the expression of FOXO-targeting genes, thereby reducing cellular ROS and decreasing cell death (Wang et al, 2007). In this study, we show for the first time that G6PD is a direct substrate of SIRT2. Our data demonstrate that oxidative stimuli enhance the SIRT2-G6PD protein interaction and meanwhile increase the deacetylation activity of SIRT2, thereby helping cells to sense oxidative stress and maintain NADPH homeostasis through regulating G6PD K403 acetylation in a SIRT2-dependent manner. The observed enzymatic activation of SIRT2 by oxidants is apparently not attributed to the formation of disulfide bonds within the SIRT2 protein, and the underlying mechanism still requires further investigation.
In unstressed cells, PPP is greatly inhibited while glycolysis is the major carbon metabolic pathway. When cells get in contact with an oxidant, the glycolysis pathway is blocked, driving glucose flux into the PPP (Gruning et al, 2011). Both aspects need prompt change of cellular G6PD activity since it is the first rate-limiting enzyme in PPP. Our present study reveals that SIRT2-mediated K403 deacetylation and subsequent activation of G6PD rapidly (within 15 min) stimulates PPP to supply intracellular reductant in the form of NADPH to counteract oxidative damage. Interestingly, once extracellular oxidative stress is removed, G6PD K403 acetylation can be normalized to basal levels, leading to G6PD inhibition and PPP re-suppression. Our study reveals an intriguing mechanism that oxidative status dynamically regulates G6PD activity via SIRT2 to control cellular NADPH homeostasis and redox balance (Fig7E). Compared with a transcription-based mechanism that usually takes hours, a post-translational modification of G6PD by acetylation can occur much more quickly and thus could serve as a better way for acute anti-oxidative response in cell protection.
The clinical implication of human G6PD deficiency has been mostly focused on the associated hemolysis or on the relationship to protection from malaria (Ding et al, 2013). In addition, limited studies indicate that the incidence of cancer and coronary artery disease is inversely related to the frequency of G6PD deficiency (Beaconsfield et al, 1965; Long et al, 1967; Cocco et al, 1998; Meloni et al, 2008; Manganelli et al, 2013). Higher expression level of G6PD is associated with breast cancer metastasis and is thought to contribute to tumor cell proliferation by enhanced ribose and NADPH supply (Jiang et al, 2011a; Du et al, 2013). Moreover, G6PD deficiency, perhaps through decreasing the NADPH-dependent cholesterol synthesis, may be advantageous against the risk of heart disease in both G6PD-deficient mouse models and clinical studies (Matsui et al, 2006; Muntoni, 2008; Rawat et al, 2012). Furthermore, decreased G6PD activity may predispose to the occurrence of diabetes and aldosterone-induced endothelial dysfunction (Leopold et al, 2007; Zhang et al, 2010). Supporting this notion, highly significant decreases in G6PD activity due to hyperglycemia or diabetes were observed in various cultured cells and animal tissues (Zhang et al, 2000; Xu et al, 2005). Additionally, G6PD-deficient subjects were reported to show a higher frequency of diabetes mellitus when compared with subjects from the same population who have wild-type G6PD activity (Saeed et al, 1985; Niazi, 1991). Another example for the pathophysiological role of G6PD comes from the finding that aldosterone induces a G6pd-deficient phenotype to impair endothelial function, while aldosterone antagonism or gene transfer of G6pd improves vascular reactivity by restoring G6pd activity (Leopold et al, 2007). Therefore, the role of G6PD deficiency in the pathophysiology of various diseases may be context dependent. In the present study, we have provided comprehensive evidence showing that SIRT2 plays a key role in mediating G6PD deacetylation and activation, which is critical for cells to sense physiological oxidative stress and maintain cellular redox potential to protect against oxidative damage. Therefore, future therapeutic intervention(s) to modulate G6PD activity via SIRT2-mediated deacetylation may serve as a potential target for treating the related disease.
Materials and Methods
G6PD enzyme activity assay
G6PD enzyme activity was determined as described previously (Tian et al, 1998). To obtain endogenous G6PD activity, both PGD activity alone and total dehydrogenase activity (G6PD + PGD) were measured separately. G6PD activity was calculated by subtracting the activity of PGD from total enzyme activity. Reaction mixture consists of 50 mM Tris–HCl (pH 7.6), 0.1 mM NADP+, 0.2 mM glucose-6-phosphate (G6P), or 0.2 mM 6-phosphogluconate in a total volume of 300 μl. Reactions were initiated by adding enzyme and analyzed at 25°C. Activities were measured by the conversion of NADP+ to NADPH, which was monitored by measuring the increase in fluorescence (Ex. 350 nm, Em. 470 nm, HITACH F-4600 fluorescence spectrophotometer) for NADPH generation. Flag-tagged G6PD proteins were expressed in HEK293T cells, immunoprecipitated with Flag beads, eluted by Flag peptides (Gilson Biochemical), and subjected into activity assay with G6P and NADP+ as substrates.
Expression of the K403 site-specific acetylated G6PD
The K403 site-specific acetylated G6PD was expressed in E. coli as previously described (Neumann et al, 2008). In short, the Escherichia coli strain BL21 (DE3) was transformed with plasmids, pAcKRS-3 and pCDF PylT-1 carrying the ORF for G6PD with amber codon at the K403 site. Cells were grown overnight in LB containing 50 μg/ml kanamycin and 50 μg/ml spectinomycin (LB-KS) at 37°C till OD600 reached 0.6–0.8, and the culture was added with 20 mM NAM and 10 mM acetyllysine (AcK, Sigma). Protein expression was induced at 37°C 30 min later by the addition of 0.5 mM of isopropyl-1-thio-d-galactopyranoside (IPTG). Afterward, incubation was continued at 37°C for 3 h, and the cells were harvested, washed with PBS containing 20 mM NAM, and then stored at −80°C till further analysis.
Generation of stable G6PD-knockdown cell pools
To generate stable G6PD-knockdown cell pools in HEK293T cells, shRNA targeting G6PD was constructed, and retrovirus was produced using a two-plasmid packaging system as previously described (Christofk et al, 2008). The shRNA targeting sequence for G6PD is 5′-GGCCGTCACCAAGAACATTCA-3′. The shRNA construct was co-transfected with vectors expressing the gag and vsvg genes into HEK293T cells. Retroviral supernatant was harvested 36 h after transfection and mixed with 8 μg/ml polybrene to increase the infection efficiency. Cells were infected with the retrovirus and selected in 1 μg/ml puromycin for 1 week.
Real-time monitoring of the disulfide formation
Redox-sensitive green fluorescent proteins (roGFPs) allow real-time visualization of the oxidation state of the indicator (Dooley et al, 2004). The thiol-disulfide metabolic state was monitored in HEK293 cells stably expressing roGFP1 in the cytosol (HEK293roGFP1). Cells were harvested by trypsinization, washed, and resuspended in PBS containing 25 mM glucose. The aliquots of cells were incubated at 37°C with 150 μM H2O2 or 50 μM menadione. Menadione was removed from the culture at 30 min post-treatment by centrifuging cells at 100 g for 5 min and washed twice and resuspended in PBS containing 25 mM glucose. The disulfide formation potential was measured by a Spectra Max M5 microplate Reader (Molecular Devices) and calculated by determining the excitation ratio (400/485 nm). Fluorescence values were background-corrected by subtracting the intensity of HEK293 cell samples not expressing roGFP1. All the samples were run in triplicate.
Statistical analyses
Statistical analyses were performed with a two-tailed unpaired Student's t-test. All data shown represent the results obtained from triplicated independent experiments with standard errors of the mean (mean ± s.d.). The values of P < 0.05 were considered statistically significant.
Acknowledgments
We thank the members of the Fudan MCB laboratory for discussions and support throughout this study. In addition, we also thank the Biomedical Core Facility, Fudan University, for technical support throughout this study. This work was supported by the 973 Program (No. 2012CB910303, No. 2012CB910101, No. 2011CB910600, No. 2009CB918401) and by the Innovation Program of Shanghai Municipal Education Commission (No. 12ZZ008). This work was also supported by NIH Grants (CA163834 to YX and R01CA108941 to KLG) and James McDonnell Foundation Samuel Waxman Foundation (to YX).
Author contributions
YPW and DY conceived the general framework of this study. YPW designed experiments. YPW, LSZ, SWW, LLC, LXL, YPS, and JYZ performed experiments. YZZ and YY provided unique redox biosensor reagents. ZQL and FJH collected samples. YPW, CY, YY, YX, KLG, and DY prepared the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Au SW, Gover S, Lam VM, Adams MJ. Human glucose-6-phosphate dehydrogenase: the crystal structure reveals a structural NADP(+) molecule and provides insights into enzyme deficiency. Structure. 2000;8:293–303. doi: 10.1016/s0969-2126(00)00104-0. [DOI] [PubMed] [Google Scholar]
- Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol Cell. 2005;17:855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Babalola AO, Beetlestone JG, Luzzatto L. Genetic variants of human erythrocyte glucose-6-phosphate dehydrogenase. Kinetic and thermodynamic parameters of variants A, B, and A- in relation to quaternary structure. J Biol Chem. 1976;251:2993–3002. [PubMed] [Google Scholar]
- Beaconsfield P, Rainsbury R, Kalton G. Glucose-6-phosphate dehydrogenase deficiency and the incidence of cancer. Oncology. 1965;19:11–19. doi: 10.1159/000224280. [DOI] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L. Biochemistry. 6th edn. New York: W.H. Freeman & Co; 2006. pp. 577–589. [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Blank LM, Kuepfer L, Sauer U. Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol. 2005;6:R49. doi: 10.1186/gb-2005-6-6-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellini MD, Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008;371:64–74. doi: 10.1016/S0140-6736(08)60073-2. [DOI] [PubMed] [Google Scholar]
- Cen Y, Youn DY, Sauve AA. Advances in characterization of human sirtuin isoforms: chemistries, targets and therapeutic applications. Curr Med Chem. 2011;18:1919–1935. doi: 10.2174/092986711795590084. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Cocco P, Todde P, Fornera S, Manca MB, Manca P, Sias AR. Mortality in a cohort of men expressing the glucose-6-phosphate dehydrogenase deficiency. Blood. 1998;91:706–709. [PubMed] [Google Scholar]
- Cohen P, Rosemeyer MA. Human glucose-6-phosphate dehydrogenase: purification of the erythrocyte enzyme and the influence of ions on its activity. Eur J Biochem. 1969;8:1–7. doi: 10.1111/j.1432-1033.1969.tb00487.x. [DOI] [PubMed] [Google Scholar]
- Cramer CT, Cooke S, Ginsberg LC, Kletzien RF, Stapleton SR, Ulrich RG. Upregulation of glucose-6-phosphate dehydrogenase in response to hepatocellular oxidative stress: studies with diquat. J Biochem Toxicol. 1995;10:293–298. doi: 10.1002/jbt.2570100603. [DOI] [PubMed] [Google Scholar]
- Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S, Ormenese S, Belachew S, Malgrange B, Chapelle JP, Siebenlist U, Moonen G, Chariot A, Nguyen L. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding K, de Andrade M, Manolio TA, Crawford DC, Rasmussen-Torvik LJ, Ritchie MD, Denny JC, Masys DR, Jouni H, Pachecho JA, Kho AN, Roden DM, Chisholm R, Kullo IJ. Genetic variants that confer resistance to malaria are associated with red blood cell traits in African-Americans: an electronic medical record-based genome-wide association study. G3 (Bethesda) 2013;3:1061–1068. doi: 10.1534/g3.113.006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem. 2004;279:22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- Du W, Jiang P, Mancuso A, Stonestrom A, Brewer MD, Minn AJ, Mak TW, Wu M, Yang X. TAp73 enhances the pentose phosphate pathway and supports cell proliferation. Nat Cell Biol. 2013;15:991–1000. doi: 10.1038/ncb2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filosa S, Fico A, Paglialunga F, Balestrieri M, Crooke A, Verde P, Abrescia P, Bautista JM, Martini G. Failure to increase glucose consumption through the pentose-phosphate pathway results in the death of glucose-6-phosphate dehydrogenase gene-deleted mouse embryonic stem cells subjected to oxidative stress. Biochem J. 2003;370:935–943. doi: 10.1042/BJ20021614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumai R, Komatsu Y, Nishino N, Khochbin S, Yoshida M, Horinouchi S. Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc Natl Acad Sci USA. 2001;98:87–92. doi: 10.1073/pnas.011405598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeve K, von Schaewen A, Scheibe R. Purification, characterization, and cDNA sequence of glucose-6-phosphate dehydrogenase from potato (Solanum tuberosum L.) Plant J. 1994;5:353–361. doi: 10.1111/j.1365-313x.1994.00353.x. [DOI] [PubMed] [Google Scholar]
- Gruning NM, Rinnerthaler M, Bluemlein K, Mulleder M, Wamelink MM, Lehrach H, Jakobs C, Breitenbach M, Ralser M. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 2011;14:415–427. doi: 10.1016/j.cmet.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Franklin H Epstein Lecture: sirtuins, aging, and medicine. N Engl J Med. 2011;364:2235–2244. doi: 10.1056/NEJMra1100831. [DOI] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild R, von Schaewen A. Differential regulation of glucose-6-phosphate dehydrogenase isoenzyme activities in potato. Plant Physiol. 2003;133:47–62. doi: 10.1104/pp.103.025676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD. Old enzymes, new tricks: sirtuins are NAD(+)-dependent de-acylases. Cell Metab. 2011;14:718–719. doi: 10.1016/j.cmet.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011a;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011b;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhnke H, Krems B, Kotter P, Entian KD. Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol Gen Genet. 1996;252:456–464. doi: 10.1007/BF02173011. [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kim SY, Lee SM, Tak JK, Choi KS, Kwon TK, Park JW. Regulation of singlet oxygen-induced apoptosis by cytosolic NADP+-dependent isocitrate dehydrogenase. Mol Cell Biochem. 2007;302:27–34. doi: 10.1007/s11010-007-9421-x. [DOI] [PubMed] [Google Scholar]
- Kletzien RF, Harris PK, Foellmi LA. Glucose-6-phosphate dehydrogenase: a “housekeeping” enzyme subject to tissue-specific regulation by hormones, nutrients, and oxidant stress. FASEB J. 1994;8:174–181. doi: 10.1096/fasebj.8.2.8119488. [DOI] [PubMed] [Google Scholar]
- Kotaka M, Gover S, Vandeputte-Rutten L, Au SW, Lam VM, Adams MJ. Structural studies of glucose-6-phosphate and NADP+ binding to human glucose-6-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2005;61:495–504. doi: 10.1107/S0907444905002350. [DOI] [PubMed] [Google Scholar]
- Kregel KC, Zhang HJ. An integrated view of oxidative stress in aging: basic mechanisms, functional effects, and pathological considerations. Am J Physiol Regul Integr Comp Physiol. 2007;292:R18–R36. doi: 10.1152/ajpregu.00327.2006. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Lee J, Godon C, Lagniel G, Spector D, Garin J, Labarre J, Toledano MB. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J Biol Chem. 1999;274:16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- Leopold JA, Dam A, Maron BA, Scribner AW, Liao R, Handy DE, Stanton RC, Pitt B, Loscalzo J. Aldosterone impairs vascular reactivity by decreasing glucose-6-phosphate dehydrogenase activity. Nat Med. 2007;13:189–197. doi: 10.1038/nm1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long WK, Wilson SW, Frenkel EP. Associations between red cell glucose-6-phosphate dehydrogenase variants and vascular diseases. Am J Hum Genet. 1967;19:35–53. [PMC free article] [PubMed] [Google Scholar]
- Lundby A, Lage K, Weinert BT, Bekker-Jensen DB, Secher A, Skovgaard T, Kelstrup CD, Dmytriyev A, Choudhary C, Lundby C, Olsen JV. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012;2:419–431. doi: 10.1016/j.celrep.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manganelli G, Masullo U, Passarelli S, Filosa S. Glucose-6-phosphate dehydrogenase deficiency: disadvantages and possible benefits. Cardiovasc Hematol Disord Drug Targets. 2013;13:73–82. doi: 10.2174/1871529x11313010008. [DOI] [PubMed] [Google Scholar]
- Margis R, Dunand C, Teixeira FK, Margis-Pinheiro M. Glutathione peroxidase family - an evolutionary overview. FEBS J. 2008;275:3959–3970. doi: 10.1111/j.1742-4658.2008.06542.x. [DOI] [PubMed] [Google Scholar]
- Mason PJ, Bautista JM, Gilsanz F. G6PD deficiency: the genotype-phenotype association. Blood Rev. 2007;21:267–283. doi: 10.1016/j.blre.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Matsui R, Xu S, Maitland KA, Mastroianni R, Leopold JA, Handy DE, Loscalzo J, Cohen RA. Glucose-6-phosphate dehydrogenase deficiency decreases vascular superoxide and atherosclerotic lesions in apolipoprotein E(−/−) mice. Arterioscler Thromb Vasc Biol. 2006;26:910–916. doi: 10.1161/01.ATV.0000205850.49390.3b. [DOI] [PubMed] [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, Smith MM. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- Meloni L, Manca MR, Loddo I, Cioglia G, Cocco P, Schwartz A, Muntoni S. Glucose-6-phosphate dehydrogenase deficiency protects against coronary heart disease. J Inherit Metab Dis. 2008;31:412–417. doi: 10.1007/s10545-008-0704-5. [DOI] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni S. Gene-nutrient interactions in G6PD-deficient subjects–implications for cardiovascular disease susceptibility. J Nutrigenet Nutrigenomics. 2008;1:49–54. doi: 10.1159/000109874. [DOI] [PubMed] [Google Scholar]
- Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niazi GA. Glucose-6-phosphate dehydrogenase deficiency and diabetes mellitus. Int J Hematol. 1991;54:295–298. [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Notaro R, Afolayan A, Luzzatto L. Human mutations in glucose 6-phosphate dehydrogenase reflect evolutionary history. FASEB J. 2000;14:485–494. doi: 10.1096/fasebj.14.3.485. [DOI] [PubMed] [Google Scholar]
- Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, Young AB, Abagyan R, Feany MB, Hyman BT, Kazantsev AG. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- Pandolfi PP, Sonati F, Rivi R, Mason P, Grosveld F, Luzzatto L. Targeted disruption of the housekeeping gene encoding glucose 6-phosphate dehydrogenase (G6PD): G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. EMBO J. 1995;14:5209–5215. doi: 10.1002/j.1460-2075.1995.tb00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat DK, Hecker P, Watanabe M, Chettimada S, Levy RJ, Okada T, Edwards JG, Gupte SA. Glucose-6-phosphate dehydrogenase and NADPH redox regulates cardiac myocyte L-type calcium channel activity and myocardial contractile function. PLoS ONE. 2012;7:e45365. doi: 10.1371/journal.pone.0045365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed TK, Hamamy HA, Alwan AA. Association of glucose-6-phosphate dehydrogenase deficiency with diabetes mellitus. Diabet Med. 1985;2:110–112. doi: 10.1111/j.1464-5491.1985.tb00611.x. [DOI] [PubMed] [Google Scholar]
- Salati LM, Amir-Ahmady B. Dietary regulation of expression of glucose-6-phosphate dehydrogenase. Annu Rev Nutr. 2001;21:121–140. doi: 10.1146/annurev.nutr.21.1.121. [DOI] [PubMed] [Google Scholar]
- Satoh A, Stein L, Imai S. The role of mammalian sirtuins in the regulation of metabolism, aging, and longevity. Handb Exp Pharmacol. 2011;206:125–162. doi: 10.1007/978-3-642-21631-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Slekar KH, Kosman DJ, Culotta VC. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J Biol Chem. 1996;271:28831–28836. doi: 10.1074/jbc.271.46.28831. [DOI] [PubMed] [Google Scholar]
- Smith BC, Hallows WC, Denu JM. Mechanisms and molecular probes of sirtuins. Chem Biol. 2008;15:1002–1013. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian WN, Braunstein LD, Pang J, Stuhlmeier KM, Xi QC, Tian X, Stanton RC. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J Biol Chem. 1998;273:10609–10617. doi: 10.1074/jbc.273.17.10609. [DOI] [PubMed] [Google Scholar]
- Turner JF, Turner DH. The regulation of glycolysis and the pentose phosphate pathway. In: Stumpf PK, Conn EE, editors. Biochemistry of Plants. New York: Academic Press; 1980. pp. 279–316. (eds), Vol. 2. In: [Google Scholar]
- Ursini MV, Parrella A, Rosa G, Salzano S, Martini G. Enhanced expression of glucose-6-phosphate dehydrogenase in human cells sustaining oxidative stress. Biochem J. 1997;323(Pt 3):801–806. doi: 10.1042/bj3230801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliamy T, Beutler E, Luzzatto L. Variants of glucose-6-phosphate dehydrogenase are due to missense mutations spread throughout the coding region of the gene. Hum Mutat. 1993;2:159–167. doi: 10.1002/humu.1380020302. [DOI] [PubMed] [Google Scholar]
- Vulliamy TJ, Luzzatto L. Glucose-6-phosphate dehydrogenase deficiency and related disorders. In: Handin RI, et al., editors. Blood: Principles and Practice of Hematology. Philadelphia, PA: Lippincott Williams and Wilkins; 2003. pp. 1921–1950. In: (eds) [Google Scholar]
- Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- Wang XT, Lam VM, Engel PC. Functional properties of two mutants of human glucose 6-phosphate dehydrogenase, R393G and R393H, corresponding to the clinical variants G6PD Wisconsin and Nashville. Biochim Biophys Acta. 2006;1762:767–774. doi: 10.1016/j.bbadis.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Wang XT, Engel PC. Clinical mutants of human glucose 6-phosphate dehydrogenase: impairment of NADP(+) binding affects both folding and stability. Biochim Biophys Acta. 2009;1792:804–809. doi: 10.1016/j.bbadis.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Xu Y, Osborne BW, Stanton RC. Diabetes causes inhibition of glucose-6-phosphate dehydrogenase via activation of PKA, which contributes to oxidative stress in rat kidney cortex. Am J Physiol Renal Physiol. 2005;289:F1040–F1047. doi: 10.1152/ajprenal.00076.2005. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhang Z, Hu J, Stillman IE, Leopold JA, Handy DE, Loscalzo J, Stanton RC. Glucose-6-phosphate dehydrogenase-deficient mice have increased renal oxidative stress and increased albuminuria. FASEB J. 2010;24:609–616. doi: 10.1096/fj.09-135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Apse K, Pang J, Stanton RC. High glucose inhibits glucose-6-phosphate dehydrogenase via cAMP in aortic endothelial cells. J Biol Chem. 2000;275:40042–40047. doi: 10.1074/jbc.M007505200. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liew CW, Handy DE, Zhang Y, Leopold JA, Hu J, Guo L, Kulkarni RN, Loscalzo J, Stanton RC. High glucose inhibits glucose-6-phosphate dehydrogenase, leading to increased oxidative stress and beta-cell apoptosis. FASEB J. 2010;24:1497–1505. doi: 10.1096/fj.09-136572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Chai X, Marmorstein R. Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J Mol Biol. 2004;337:731–741. doi: 10.1016/j.jmb.2004.01.060. [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Zhao L, Wang E, Dimova N, Liu G, Feng Y, Cambi F. The QKI-PLP pathway controls SIRT2 abundance in CNS myelin. Glia. 2012;60:69–82. doi: 10.1002/glia.21248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.