Abstract
The evolution of the ability to synthesize specialized metabolites is likely to have been key for survival and diversification of different plant species. Oats (Avena spp.) produce antimicrobial triterpenoids (avenacins) that protect against disease. The oat β-amyrin synthase gene AsbAS1, which encodes the first committed enzyme in the avenacin biosynthetic pathway, is clearly distinct from other plant β-amyrin synthases. Here we show that AsbAS1 has arisen by duplication and divergence of a cycloartenol synthase-like gene, and that its properties have been refined since the divergence of oats and wheat. Strikingly, we have also found that AsbAS1 is clustered with other genes required for distinct steps in avenacin biosynthesis in a region of the genome that is not conserved in other cereals. Because the components of this gene cluster are required for at least four clearly distinct enzymatic processes (2,3-oxidosqualene cyclization, β-amyrin oxidation, glycosylation, and acylation), it is unlikely that the cluster has arisen as a consequence of duplication of a common ancestor. Although clusters of paralogous genes are common in plants (e.g., gene clusters for rRNA and specific disease resistance), reports of clusters of genes that do not share sequence relatedness and whose products contribute to a single selectable function are rare [Gierl, A. & Frey, M. (2001) Planta 213, 493–498]. Taken together, our evidence has important implications for the generation of metabolic diversity in plants.
Triterpene saponins are an important group of plant secondary metabolites that are produced by >1,000 dicotyledonous species (1). In contrast, cereals and grasses are generally deficient in these secondary metabolites with the exception of oats, which accumulate antimicrobial triterpenoid saponins (avenacins) in the roots (1–3). Saponin-deficient (sad) mutants of diploid oat (Avena strigosa) are compromised in disease resistance, indicating that avenacins protect against microbial attack (4). Metabolic engineering of this pathway into other major cereal crops such as wheat, barley, maize, and rice has clear potential for novel strategies for improved disease resistance. This represents a substantial technical challenge, however, because saponin biosynthesis is a multistep process that is not well understood for any plant species, and the genes for complete pathways have not been cloned (5).
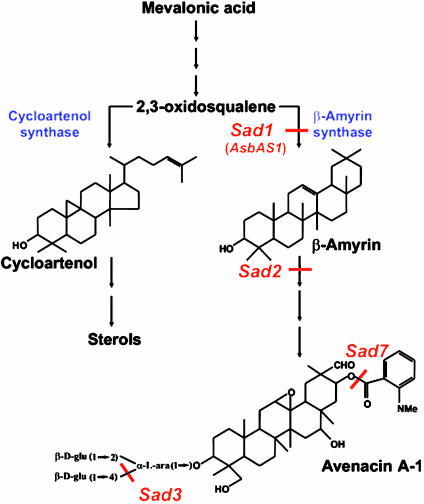
Triterpenoid saponins, like sterols, are synthesized from mevalonic acid via the isoprenoid pathway, the two pathways diverging after 2,3-oxidosqualene (Fig. 1) (1, 5). Synthesis of sterols in plants involves cyclization of 2,3-oxidosqualene to cycloartenol, mediated by the oxidosqualene cyclase enzyme cycloartenol synthase. For triterpenoid saponin synthesis, 2,3-oxidosqualene is cyclized to one of a number of different potential products, the most common being β-amyrin (1). The first committed step in avenacin synthesis is the cyclization of 2,3-oxidosqualene to β-amyrin, catalyzed by the oxidosqualene cyclase enzyme β-amyrin synthase (5–7). We have recently cloned the gene encoding oat β-amyrin synthase (AsbAS1) (7) and have shown that this corresponds to Sad1, a locus we had previously identified by mutation (4). The AsbAS1gene product is an unusual oxidosqualene cyclase that is clearly distinct from other triterpene synthases that have been characterized from plants (7).
Fig. 1.
Synthesis of sterols and triterpenoid saponins in oat. Cyclization of 2,3-oxidosqualene to cycloartenol (the committed precursor for sterol biosynthesis) or to β-amyrin (the first committed step in the avenacin biosynthetic pathway) is catalyzed by the oxidosqualene cyclase enzymes cycloartenol synthase and β-amyrin synthase, respectively. The biochemical defects of characterized sad mutants are indicated.
The saponin-deficient mutants that we originally identified define seven other genetic loci in addition to AsbAS1 (4, 7). These loci are as yet uncharacterized at the molecular genetic level. β-Amyrin, the precursor for synthesis of avenacins, is not antifungal. The conversion of β-amyrin into antifungal saponins will require cytochrome P450-dependent monooxygenases, acyltransferases, glycosyltransferases, and other enzymes (5, 7). Our data indicate that sad2 mutants accumulate β-amyrin and are likely to be blocked in a cytochrome P450-mediated step early in the pathway (8), and that sad3 and sad4 mutants are defective in saponin glucosylation (4). The sad7 mutant accumulates a metabolite that has the same sugar conjugate as avenacin A-1 but lacks the N-methyl anthranilate group (C.M., unpublished data) (Fig. 1). The biochemical defects in the other mutants (sad5, sad6, and sad8) are as yet unknown.
Here we show that AsbAS1, which maps to a region of the oat genome not conserved in other cereals, has arisen by duplication and divergence of a cycloartenol synthase-like gene, and that the properties of AsbAS1 have been refined since the divergence of oats and wheat. Intriguingly, five of the seven other Sad loci we have defined by mutation (4) are genetically linked to AsbAS1. Sad3 is 3.6 centimorgans (cM) from AsbAS1, whereas the four other linked loci (Sad2, 6, 7, and 8) showed complete cosegregation with AsbAS1 in our analyses. Because these loci are required for at least four clearly distinct enzymatic processes [AsbAS1 (Sad1), 2,3-oxidosqualene cyclization; Sad2, β-amyrin oxidation; Sad3, glycosylation; and Sad7, acylation], it is unlikely this gene cluster has arisen as a consequence of duplication of a common ancestor. The significance of these data for the evolution of metabolic diversity in plants and other eukaryotic organisms is discussed.
Methods
Nucleic Acid Extraction and Southern and Northern Blot Analysis. Seeds of A. strigosa, Avena wiestii, and other cereals were surface sterilized with 5% sodium hypochlorite and then germinated at 24°C in the dark on damp filter paper. Genomic DNA for polymorphism analysis and mapping was isolated from 7-day-old seedlings by using the DNeasy Plant Mini Kit (Qiagen, Chatsworth, CA). Total RNA was extracted by using TRI-REAGENT (Sigma). Genomic DNA for Southern blot analysis was extracted from 10 g of leaf tissue of 2-week-old seedlings, frozen in liquid nitrogen, and extracted by using the CTAB protocol (9). Southern and Northern blot analyses were carried out by using standard methods (10). Hybridizations were performed at different stringencies (55, 60, and 65°C) with washes as described by Church and Gilbert (11) by using 32P-labeled cDNA probes.
DNA Markers and Mapping. AsbAS1 was mapped in the A. strigosa CI3815 × A. wiestii CI1994 recombinant inbred lines (12) by using a single-nucleotide polymorphism in intron 17 that conferred presence or absence of a PacI restriction site. PCR fragments for digestion were amplified with the primer pair: tgggcaatgttggctttaattt/tgatgacatcggtaggaa. The PCR primer pairs for analysis of the Oisu441-derived sequence-tagged site marker (13) and the AsCS1 gene (7) (both of which detected insertion/deletion polymorphisms) were catgcctgtaccattctagctt/gcacactaacattttctatatcgtttca and tgttcacaattaccttgtgta/tgtaactgcctagaacggctt, respectively. Markers derived from other oat maps and from rice were obtained by sequence analysis and from databases [refs. 14 and 15; Rice Genome Research Program (2003) http://rgp.dna.affrc.go.jp]. Maps were constructed by using joinmap 2.0 (16), and the Kosambi map function (17) was used in the analysis.
Assessment of Sequence Divergence. Oat accessions used for comparison of sequence divergence of AsbAS1 and AsCS1 were Avena prostrata (Cc7060), Avena clauda (CAV5566/1), Avena ventricosa (CAV2835), and Avena longiglumis (Cc4719, Cc7250) (all from The Institute of Grassland and Environmental Research) and A. strigosa S75 (4) and CI3815 (12). Total RNA was isolated from 4- to 6-day-old oat roots by using TRI-REAGENT (Sigma) and products amplified from first-strand cDNA by using gene-specific primers. PCR products were gel purified (Qiagen) and sequenced by using the ABI PRISM Big-Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems). Predicted protein coding sequences were aligned by using clustal x, Version 1.8 (www-igbmc.u-strasbg.fr/bioinfo), with visual adjustments. Gaps in the alignment were not used in the analysis (complete-deletion option). Rates of synonymous and nonsynonymous nucleotide substitutions were estimated by the modified Nei–Gojobori method (18). mega2 software (19) was used for estimation of the nonsynomous (dN) to synomous (dS) ratio, sequence diversity, and Tajima's relative rate test (20).
Results
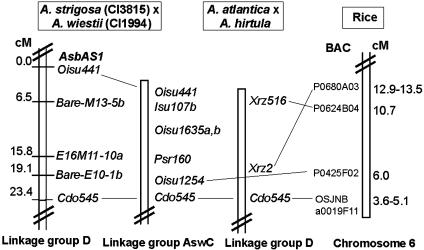
AsbAS1 Maps to a Region of the Oat Genome That Is Not Conserved in Other Cereals. We used recombinant inbred lines derived from A. strigosa CI3815 × A. wiestii CI1994 (12) to map AsbAS1 to linkage group D (Fig. 2). Linkage group D has been further resolved as linkage group AswC in a second diploid oat map constructed by Kremer et al. (13) by using the same parents. The nearest restriction fragment length polymorphism (RFLP) probe to AsbAS1 on the distal region of linkage group AswC (Oisu441; Fig. 2) was sequenced and converted to a PCR-based sequence-tagged site (STS) marker that detected a deletion/insertion polymorphism between CI3815 and CI1994. A local map was reconstructed based on a subset of markers in linkage group D plus AsbAS1 and Oisu441. AsbAS1 cosegregated with the STS marker derived from Oisu441 in the recombinant inbred population and was positioned 23.4 cM from the nearest grass RFLP anchor marker Cdo545. This region is generally colinear with part of rice chromosome 6 (15). However, sequences closely related (>80% identity) to AsbAS1 and Oisu441 are not represented in the syntenic region in rice or elsewhere in the rice genome.
Fig. 2.
Mapping of the β-amyrin synthase gene AsbAS1. AsbAS1 maps to linkage group D of a diploid oat map derived from A. strigosa CI3815 × A. wiestii CI1994 (12). This region corresponds to linkage group AswC in a second map constructed using the same parents (13). The A. atlantica × A. hirtula RFLP map was constructed by Van Deynze et al. (14). The rice chromosome 6 map was derived from the Rice Genome Research Program web site (http://rgp.dna.affrc.go.jp).
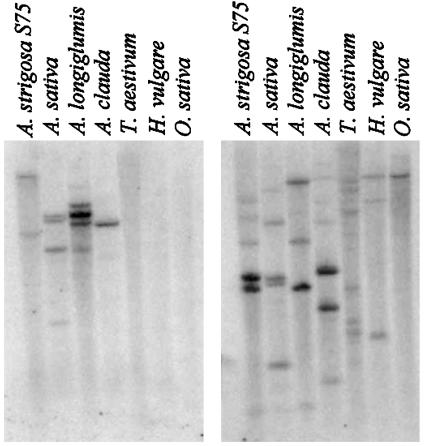
Previously, we showed that AsbAS1 is expressed strongly in oat roots but that related transcripts were not detectable in other cereals by Northern blot analysis (7). Southern blot analysis provided additional confirmation that sequences that hybridize strongly to the AsbAS1 cDNA probe are conserved in Avena species but not in other cultivated cereals (Fig. 3 Left). In contrast, a cDNA probe for the A. strigosa cycloartenol synthase gene (7) hybridized with genomic DNA from all cereals tested (Fig. 3 Right).
Fig. 3.
Sequences closely related to AsbAS1 are conserved in Avena species but not in other cereals. Southern blot analysis of XbaI-digested genomic DNA from Avena spp. and other cereals using AsbAS1(Left) and AsCS1 (Right) cDNA probes. The same high-stringency conditions were used for both blots.
Sad Genes Are Clustered. We crossed all sad mutants (wild-type S75) with A. strigosa CI3815 for further genetic analysis. First, we confirmed that the sad mutations behaved as recessive alleles of single genes within these F2 populations, as they do in crosses with the wild-type parent S75 (4). F2 populations were scored for the saponin-deficient phenotype and then genotyped for AsbAS1 alleles by single-nucleotide polymorphism analysis. Remarkably, these experiments indicated that five of the seven other Sad loci we had defined by mutation are genetically linked to AsbAS1 (Table 1). No AsbAS1/Sad2 recombinants were found in >2,000 F2 individuals; three other Sad loci (Sad6, 7, and 8) cosegregated with AsbAS1 in populations of between 150 and 170 F2 individuals; Sad3 was less closely linked, resolving this gene cluster to ≈3.6 cM around the AsbAS1 locus. Along with our biochemical data, these results indicate that genes required for at least four distinct biochemical processes in the synthesis of saponins [cyclization of 2,3-oxidosqualene to β-amyrin (AsbAS1), cytochrome P450-mediated modification of β-amyrin (Sad2), acylation (Sad7), and glucosylation (Sad3)] are clustered within linkage group D of the diploid oat genome. Because the biochemical processes affected are quite different, it is highly unlikely that this cluster of Sad loci has arisen in situ from a common ancestor by gene duplication. The probability of five loci for saponin biosynthesis being within 3.6 cM of AsbAS1, assuming random distribution of genes, is extremely low (≈10–12 to 10–15), based on estimates of total genetic distances for A. strigosa (12, 13).
Table 1. Linkage of other Sad loci to AsbASI.
| Locus | Represented by mutant number* | Number of F2 progeny assessed | Genetic distance, cM |
|---|---|---|---|
| Sad2 | 1027 | 2040 | 0.0 |
| Sad3 | 1139 | 141 | 3.6 |
| Sad4 | 9 | 43† | Unlinked |
| Sad5 | 616 | n.a.† | |
| Sad6 | 825 | 150 | 0.0 |
| Sad7 | 376 | 162 | 0.0 |
| Sad8 | 1243 | 168 | 0.0 |
See ref. 4.
Not available.
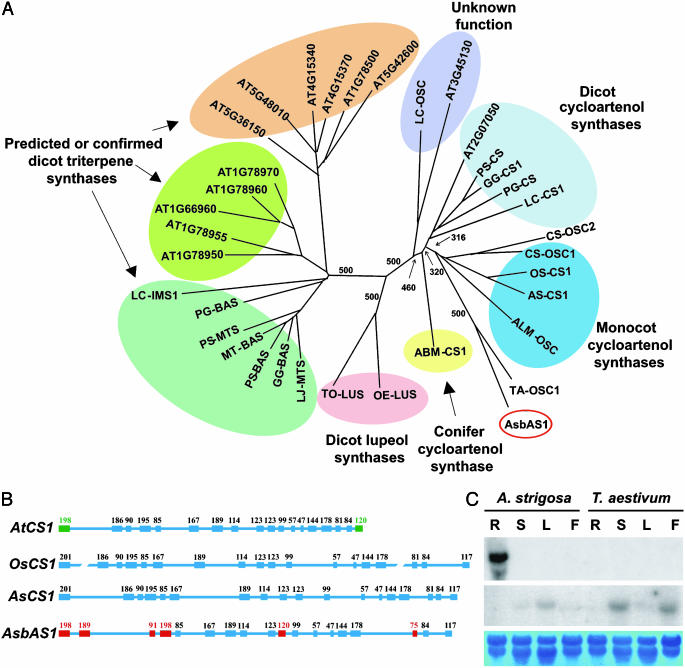
AsbAS1 Has Arisen from a Cycloartenol Synthase-Like Gene by Duplication and Rapid Sequence Divergence. The AsbAS1 gene product does not share close sequence similarity with other functionally characterized triterpene synthases from plants (7). AsbAS1 may have arisen through the divergence and vertical transmission of existing components of the plant genome or alternatively may have been introduced into oat by horizontal gene transfer. We conducted a series of experiments to distinguish between these possibilities. First, we extended our comparisons of plant oxidosqualene cyclase sequences to include all 13 predicted sequences represented in the Arabidopsis thaliana genome. These include a cycloartenol synthase gene and four triterpene synthase genes that have been functionally characterized by expression in yeast (5, 21, 22) and eight genes of unknown function. Sequences for predicted oxidosqualene cyclases from a number of other plant species were also included. Our analysis indicated that AsbAS1 is more closely related to cycloartenol synthases than to other plant β-amyrin synthases (Fig. 4A). This is surprising, given the structural differences between sterols and triterpenes (23, 24). Interestingly, a multifunctional triterpene synthase from the monocot Costus speciosus that also groups with cycloartenol synthases has recently been described (25). The most similar sequence to AsbAS1 was a predicted oxidosqualene cyclase (Ta-OSC1) from Triticum aestivum (26). Although oats are the only cereals known to produce triterpenoid saponins, β-amyrin esters have been detected in the leaf tissue and surface waxes of a variety of grass species (27–29). It is possible that Ta-OSC1 synthesizes β-amyrin in the leaves, although attempts to express this cDNA in yeast were unsuccessful.
Fig. 4.
AsbAS1 has arisen from a cycloartenol synthase-like gene. (A) Phylogenetic analysis of the coding sequences of AsbAS1and other members of the oxidosqualene cyclase superfamily from plants (see Table 2, which is published as supporting information on the PNAS web site, for further details and GenBank accession nos.). Sequences were analyzed by using the dnadist-fitch program of the phylip package (Version 6.2a) (http://evolution.genetics.washington.edu/phylip.html). The numbers indicate the numbers of bootstrap replications (of 500) in which the given branching was observed. (B) Exon–intron structures of AsbAS1and of cycloartenol synthase genes from A. strigosa S75 (AsCS1), rice (OsCS1), and A. thaliana (AtCS1). Exons are indicated by boxes; the numbers above the boxes are the exon sizes. Exons that differ in size are indicated in green for AtCS1 and in red for AsbAS1.(C) Northern blot analysis of RNA from roots of A. strigosa (GenBank accession no. S75) and T. aestivum. (Chinese Spring). The cDNA probes were AsbAS1(Top) and TaOSC1 (Middle). R, root; S, stem; L, leaf; F, flower. Twenty micrograms of RNA was loaded per lane. RNA levels were monitored with methylene blue (MB) dye (Bottom).
Our phylogenetic analysis implies that AsbAS1 has arisen from an ancestral cycloartenol synthase-like gene (Fig. 4A). Tajima's relative rate test (20) using Abies magnifica cycloartenol synthase (ABM-CS1) as an outgroup indicates that the branch for AsbAS1 is significantly longer than that for AsCS1 (χ2 = 52.74, P < 0.0001). Sequence comparisons for AsbAS1 and AsCS1 homologues from seven different oat accessions representing five different species (see Methods) revealed greater mean diversity for AsbAS1 than for AsCS1 (0.030 ± 0.003 vs. 0.017 ± 0.002, respectively). The average ratio of nonsynonymous (dN) to synonymous (dS) nucleotide substitutions per site (dN/dS) was 0.21 for AsbAS1 and 0.16 for AsCS1, indicating that both genes have been subjected to purifying selection during the evolution of oats. Collectively, these data are consistent with accelerated evolution of AsbAS1 from an ancestral cycloartenol synthase-like gene. We found general conservation of exon–intron structure between AsbAS1and the cycloartenol synthase genes from A. strigosa, rice, and A. thaliana (Fig. 4B), although six of the AsbAS1 exons differed in size from their monocot cycloartenol synthase counterparts. The other predicted/confirmed genes for triterpene synthases in A. thaliana have fewer exons (between 13 and 17) (21). Overall, the structural similarities among AsbAS1and the cycloartenol genes from oat, rice, and A. thaliana suggest these genes share a common origin. AsbAS1 may have arisen directly or indirectly by duplication of AsCS1. However, the two genes are not linked, and we have mapped AsCS1 to a different linkage group (A) (12) by using the recombinant inbred lines derived from A. strigosa CI3815 × A. wiestii CI1994.
The Patterns of Expression of AsbAS1 and Ta-OSC1 Are Different. Avenacin biosynthesis is highly tissue-specific and is restricted to the epidermal cell layer of the root tips (30). In situ hybridization experiments indicate that AsbAS1 expression is also restricted to this cell layer (7). Comparison of the expression of AsbAS1 and Ta-OSC1 [the predicted oxidosqualene cyclase from wheat (Fig. 4A)] in different plant tissues by high-stringency Northern blot analysis indicates the expression patterns of these two genes are clearly different. AsbAS1 expression was detectable in the roots of oat but not in the aerial tissues [as previously reported (7)], and this probe did not give a hybridization signal with RNA from wheat roots, shoots, flowers, or leaves (Fig. 4C). In contrast, Ta-OSC1 transcripts were detectable in the aerial tissues but not the roots of wheat (Fig. 4C). The Ta-OSC1 probe also hybridized with RNA from the aerial tissues (but not the roots) of oat (Fig. 4C). These findings are compatible with our EST-based analysis of gene expression in oat roots, in which the only two predicted oxidosqualene cyclase sequences that we identified among ≈16,000 oat-root-derived EST sequences were AsbAS1 and AsCS1 (ref. 7 and unpublished data). The expression data for Ta-OSC1 are consistent with a potential role of the gene product in the synthesis of foliar surface waxes.
Discussion
Previously, we have cloned the gene encoding oat β-amyrin synthase AsbAS1 (7). This enzyme catalyzes the cyclization of 2,3-oxidosqualene to β-amyrin, which is the first committed step in the synthesis of avenacins. This conversion forms a branch point with the sterol biosynthetic pathway in which 2,3-oxidosqualene is converted to cycloartenol by cycloartenol synthase (Fig. 1). Strikingly, our evidence indicates that AsbAS1 is more closely related to cycloartenol synthases than to other plant triterpene synthases, and that AsbAS1 has arisen by rapid evolution from an ancestral cycloartenol synthase-like gene. The close relatedness of AsbAS1 to cycoartenol synthases is surprising because, although cycloartenol synthase and β-amyrin synthase both use 2,3-oxidosqualene as a substrate, the structures of the cyclization products they generate are quite different (5, 7, 23, 24). We have also shown that the expression patterns of AsbAS1 and that of the closest database match from cereals (Ta-OSC1) are different, and that the properties of AsbAS1 (root epidermis-specific expression and probably also kinetic properties) are likely to have been refined since the divergence of oats and wheat. Sequence analysis of AsCS1 and AsbAS1 homologues from a range of different oat accessions indicates that both genes have been subjected to purifying selection. This is consistent with the critical roles of the gene products in sterol metabolism and synthesis of defense-related secondary metabolites (7), respectively.
AsbAS1 maps to a region of the oat genome not conserved in other cereals. Our genetic and biochemical analyses indicate that genes required for at least three other distinct biochemical processes in avenacin synthesis are clustered with AsbAS1 in linkage group D of the diploid oat genome. Clusters of paralogous genes of high nucleotide sequence identity are common in plants. Examples include gene clusters for rRNA and also for specific disease resistance that have arisen by gene duplication and unequal crossing over. In contrast, reports of clusters of plant genes that do not share sequence relatedness and whose products contribute to a single selectable function are rare. To our knowledge, the only other well characterized example of clustered genes for a secondary metabolic pathway in plants is that of the benzoxazinoids in maize (31–33), although surveys of the A. thaliana genome hint this may be a more widespread phenomenon (34, 35). Because benzoxazinoids are produced by a range of cereals and also by some dicots, it has been postulated that the pathway may have evolved before the divergence of monocots and dicots, and that failure to produce these compounds may be due to loss of pathway components (32, 36). In contrast, within the Gramineae, the ability to synthesize triterpene saponins appears to be restricted to Avena species (1).
Mechanisms that act to disperse genes (translocation, inversion, and unequal crossing over) are well known in eukaryotes, and genes associated with common metabolic pathways [including genes for some secondary metabolic pathways such as anthocyanin biosynthesis (37)] are generally unlinked. This raises the question of how and why gene clusters of the kind that we have observed are maintained in the genome. Although gene clusters for secondary metabolism are not well documented in plants, they are common in fungi (38–41). Fungal gene clusters for secondary metabolism include not only the genes for the biosynthetic components but also genes for specific pathway regulators and for autoresistance to the end product (38). Transmission of these self-contained “gene cassettes” by horizontal gene transfer has been suggested as an explanation for the persistence of clustering in fungi (38), although recent phylogenomics-based analyses indicate that the significance of vertical transmission has been underestimated (41, 42). Horizontal gene transfer cannot be the sole explanation for clustering of avenacin biosynthetic genes in oat, because our evidence indicates that the gene encoding the first committed enzyme in the pathway has arisen by gene duplication and sequence divergence. We believe there may be other explanations for the maintenance of gene clusters for secondary metabolism in plants, fungi, and other eukaryotic organisms. The oat avenacins and the maize benzoxazinoids both confer resistance to pests and pathogens (4, 7, 32), and so the ability to produce these compounds has obvious selective advantages. Clustering will facilitate the inheritance of the genes that confer these selective advantages as a functional unit. Disruption of the gene cluster may lead to failure to produce protective chemicals and could also result in the accumulation of deleterious intermediates (32). Our observation that oat mutants defective in avenacin glycosylation (sad3 and sad4 mutants) have aberrant root morphology (4) is consistent with this latter line of reasoning. Because many secondary metabolites and their pathway intermediates are potentially phytotoxic, there is likely to be a requirement for tight coordinate regulation of synthesis and for intimate coadaptation of individual pathway enzymes. Synthesis of avenacins, like many other plant secondary metabolites, is highly tissue specific and under strict developmental control. Avenacins are localized in the epidermal cells of the root tip (30), and AsbAS1 is expressed specifically in this cell layer (7). Clustering has the potential to facilitate coordinate regulation of expression of genes at the chromatin level. It may also confer other as-yet-undefined selective advantages associated with physical proximity and position effects. Intimate coadaptation of individual pathway enzymes is likely to be important as an additional mechanism for strict control and containment of secondary metabolites and their pathway intermediates during synthesis. This coadaptation may extend to physical interactions among pathway components, which would aid the channeling of metabolic intermediates within multienzyme complexes (43).
Although it is relatively easy to speculate about factors that may contribute to the maintenance of gene clusters, the issue of the origin of the clusters is rather more difficult to address. It is generally believed, at least for primary metabolism in plants, that new genes commonly arise by gene duplication and divergence (44, 45). Genes for secondary metabolism may in turn be derived from genes for primary metabolism by gene duplication and divergence or possibly also by allelic divergence (45). Domain swapping represents another mechanism for the creation of new composite genes (45, 46). Approximately one-quarter of the genes in the A. thaliana genome (≈5,000 genes) are predicted to be involved in secondary metabolism, and many of these are likely to have been recruited directly or indirectly from primary metabolism (47). In maize, the enzymes BX1 and IGL, which are required for the synthesis of secondary metabolites (benzoxazinoids and volicitin, respectively), have evolved from a tryptophan synthase α-subunit required for primary metabolism (32). Similarly, the plant terpene synthases (which collectively produce a diverse class of natural products) are predicted to be derived from genes for primary metabolism by duplication and consequent divergence in structural and functional specialization (48). Other examples of the recruitment of genes from primary metabolism are also known (e.g., refs. 49 and 50). Our finding that AsbAS1 has arisen from a cycloartenol synthase-like gene by duplication and rapid sequence divergence provides further evidence that genes for secondary metabolism can be recruited from primary metabolism in this way (45). The fact that other Sad loci required for distinct biochemical processes in the avenacin biosynthetic pathway are linked to AsbAS1 raises the intriguing possibility that gene clusters for the synthesis of elaborate secondary metabolites may arise de novo within plant genomes by shuffling and accelerated evolution of existing genetic components. The evolution of complex functions can be simulated in computer-based experiments with digital organisms (51). We need a more substantial body of biochemical, genetic, and genomic information for a range of secondary metabolic pathways from diverse plant species to piece together the evolutionary events and processes underlying the generation of metabolic diversity in the plant kingdom and to establish whether clustered genes for secondary metabolism may be more common in plants than first anticipated. To quote from Trowsdale (52), “Until we understand the processes that have determined the ordering and clustering of genes in genomes, our understanding of gene regulation and evolution will be incomplete.”
Supplementary Material
Acknowledgments
We thank Roger Wise (Iowa State University, Ames) for providing seed of recombinant inbred lines and for information and advice, Mike Lee (Iowa State University) for restriction fragment length polymorphism probes, Steve Reader (John Innes Centre) for seed of grass species, and Enno Krebbers (DuPont Agricultural Products) for constructive comments on the manuscript. X.Q. and S.B. were funded by the Biotechnology and Biological Sciences Research Council, United Kingdom. The Sainsbury Laboratory is supported by the Gatsby Charitable Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: cM, centimorgan.
References
- 1.Hostettmann, K. & Marston, A. (1995) Saponins (Cambridge Univ. Press, Cambridge, U.K.).
- 2.Crombie, W. M. L. & Crombie, L. (1986) Phytochemistry 25, 2069–2073. [Google Scholar]
- 3.Crombie, L., Crombie, W. M. L. & Whiting, D. A. (1986) J. Chem. Soc. Perkin Trans. 1, 1917–1922. [Google Scholar]
- 4.Papadopoulou, K., Melton, R. E., Leggett, M. Daniels, M. J. & Osbourn, A. E. (1999) Proc. Natl. Acad. Sci. USA 96, 12923–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haralampidis, K., Trojanowska, M. & Osbourn, A. E. (2001) Adv. Biochem. Eng./Biotechnol. 75, 31–49. [DOI] [PubMed] [Google Scholar]
- 6.Trojanowska, M. R., Osbourn, A. E., Daniels, M. J. & Threlfall, D. R. (2000) Phytochemistry 54, 153–164. [DOI] [PubMed] [Google Scholar]
- 7.Haralampidis, K., Bryan, G., Qi, X., Papadopoulou, K., Bakht, S., Melton, R. & Osbourn, A. E. (2001) Proc. Natl. Acad. Sci. USA 98, 13431–13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trojanowska, M. R., Osbourn, A. E., Daniels, M. J.& Threlfall, D. R. (2001) Phytochemistry 56, 121–129. [DOI] [PubMed] [Google Scholar]
- 9.Van der Beek, J. G., Verkerk, R., Zabel, P. & Lindhout, P. (1992) Theor. Appl. Genet. 84, 106–112. [DOI] [PubMed] [Google Scholar]
- 10.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 11.Church, G. M. & Gilbert, W. (1984) Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu, G. X. & Wise, R. P. (2000) Genome 43, 736–749. [PubMed] [Google Scholar]
- 13.Kremer, C. A., Lee, M. & Holland, J. B. (2001) Genome 44, 192–204. [PubMed] [Google Scholar]
- 14.Van Deynze, A. E., Nelson, J. C., O'Donoughue, L. S. Ahn., S. N., Siripoonwiwat, W., Harrington, S. E., Yglesias, E. S., Braga, D. P., McCouch, S. R. & Sorrells, M. E. (1995) Mol. Gen. Genet. 249, 349–356. [DOI] [PubMed] [Google Scholar]
- 15.Gale, M. D. & Devos, K. M. (1998) Science 282, 656–659. [DOI] [PubMed] [Google Scholar]
- 16.Stam, P. (1993) Plant J. 3, 739–744. [Google Scholar]
- 17.Kosambi, D. D. (1944) Ann. Eugen. 12, 172–175. [Google Scholar]
- 18.Zhang, J., Rosenberg, H. F. & Nei, M. (1998) Proc. Natl. Acad. Sci. USA 95, 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M., Eds. (2001) mega2: Molecular Evolutionary Genetics Analysis Software (Arizona State Univ., Tempe). [DOI] [PubMed]
- 20.Tajima, F. (1993) Genetics 135, 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Husselstein-Muller, T., Schaller, H. & Benveniste, P. (2001) Plant Mol. Biol. 45, 75–92. [DOI] [PubMed] [Google Scholar]
- 22.Ebizuka, Y., Katsube, Y., Tsutsumi, T., Kushiro, T. & Shibuya, M. (2003) Pure Appl. Chem. 75, 369–374. [Google Scholar]
- 23.Abe, I., Rohmer, M. & Prestwich, G. D. (1993) Chem. Rev. 93, 2189–2206. [Google Scholar]
- 24.Nes, W. D. (1994) Isopentenoids and Other Natural Products, ACS Symposium Series 562 (Am. Chem. Soc., Washington, DC).
- 25.Kawano, N., Ichinose, K. & Ebizuka, Y. (2002) Biol. Pharm. Bull. 25, 477–482. [DOI] [PubMed] [Google Scholar]
- 26.Bryan, G. T., Mcgonigle, V, Maxwell, C. A., Potter, S. M. & Hwang, D. C. (2001) WO 0166773-a.
- 27.Ohmoto, T. & Ikuse, T. (1970) Phytochemistry 9, 2137–2148. [Google Scholar]
- 28.Heupel, R. C. (1985) Phytochemistry 24, 2929–2937. [Google Scholar]
- 29.Casabuono, A. C. & Pomilio, A. B. (1997) Lipids 32, 205–210. [DOI] [PubMed] [Google Scholar]
- 30.Osbourn, A. E., Clarke, B. R., Lunness, P., Scott, P. R. & Daniels, M. J. (1994) Physiol. Mol. Plant Pathol. 45, 457–467. [Google Scholar]
- 31.Frey, M., Chomet, P., Glawischnig, E., Stettner, C., Grün, S, Winklmair, A., Eisenreich, W., Bacher, A., Meeley, R. B., Briggs, S. P, et al.(1997) Science 277, 696–699. [DOI] [PubMed] [Google Scholar]
- 32.Gierl, A. & Frey, M. (2001) Planta 213, 493–498. [DOI] [PubMed] [Google Scholar]
- 33.Frey, M., Huber, K., Park, W. J., Sicker, D., Lindberg, P., Meeley, R. B., Simmons, C. R., Yalpani, N. & Gierl, A. (2003) Phytochemistry 62, 371–376. [DOI] [PubMed] [Google Scholar]
- 34.Aubourg, S., Lecharny, A. & Bohlmann, J. (2002) Mol. Gen. Genom. 267, 730–745. [DOI] [PubMed] [Google Scholar]
- 35.Lange, B. M. & Ghassemian, M. (2003) Plant Mol. Biol. 51, 925–948. [DOI] [PubMed] [Google Scholar]
- 36.Nomura, T., Ishihara, A., Imaishi, H., Ohkawa, H., Endo, T. R. & Iwamura, H. (2003) Planta 217, 776–782. [DOI] [PubMed] [Google Scholar]
- 37.Neuffer, M. G., Coe, E. H. & Wessler, S. R. (1997) Mutants of Maize (Cold Spring Harbor Lab. Press, Plainview, NY).
- 38.Walton, J. D. (2000) Fung. Genet. Biol. 30, 167–171. [DOI] [PubMed] [Google Scholar]
- 39.Lawrence, J. G. & Roth, J. R. (1996) Genetics 143, 1843–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedley, K. F. & Walton, J. D. (2001) Proc. Natl. Acad. Sci. USA 98, 14174–14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward, T. J., Bielawski, J. P., Kistler, H. C., Sullivan, E. & O'Donnell, K. (2002) Proc. Natl. Acad. Sci. USA 99, 9278–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kroken, S., Glass, N. L., Taylor, J. W., Yoder, O. C. & Turgeon, B. G. (2003) Proc. Natl. Acad. Sci. USA 100, 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burbulis, I. E. & Winkel-Shirley, B. (1999) Proc. Natl. Acad. Sci. USA 96, 12929–12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohno, S. (1970) Evolution by Gene Duplication (Springer, Berlin).
- 45.Pichersky, E. & Gang, D. R. (2000) Trends Plant Sci. 5, 439–445. [DOI] [PubMed] [Google Scholar]
- 46.Doolittle, R. F. (1995) Annu. Rev. Biochem. 64, 287–314. [DOI] [PubMed] [Google Scholar]
- 47.The Arabidopsis Genome Initiative (2000) Nature 408, 796–814. [DOI] [PubMed] [Google Scholar]
- 48.Trapp, S. C. & Croteau, R. B. (2001) Genetics 158, 811–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ober, D. & Hartmann, T. (1999) Proc. Natl. Acad. Sci. USA 96, 14777–14782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lehfeldt, C., Shirley, A. M., Meyer, K., Ruegger, M. O., Cusumano, J. C., Viitanen, P. V., Strack, D. & Chapple, C. (2000) Plant Cell 12, 1295–12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lenski, R. E., Ofria, C., Pennock, R. T. & Adami, C. (2003) Nature 423, 139–144. [DOI] [PubMed] [Google Scholar]
- 52.Trowsdale, J. (2002) Genome Biol. 3, comment2002.1–2002.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.