Abstract
Immunophilins are defined as receptors for immunosuppressive drugs including cyclosporin A, FK506, and rapamycin. The cyclosporin A receptors are referred to as cyclophilins (CYPs) and FK506- and rapamycin-binding proteins are abbreviated as FKBPs. These two groups of proteins (collectively called immunophilins) share little sequence homology, but both have peptidyl prolyl cis/trans isomerase (PPIase) activity that is involved in protein folding processes. Studies have identified immunophilins in all organisms examined including bacteria, fungi, animals, and plants. Nevertheless, the physiological function of immunophilins is poorly understood in any organism. In this study, we have surveyed the genes encoding immunophilins in Arabidopsis genome. A total of 52 genes have been found to encode putative immunophilins, among which 23 are putative FKBPs and 29 are putative CYPs. This is by far the largest immunophilin family identified in any organism. Both FKBPs and CYPs can be classified into single domain and multiple domain members. The single domain members contain a basic catalytic domain and some of them have signal sequences for targeting to a specific organelle. The multiple domain members contain not only the catalytic domain but also defined modules that are involved in protein-protein interaction or other functions. A striking feature of immunophilins in Arabidopsis is that a large fraction of FKBPs and CYPs are localized in the chloroplast, a possible explanation for why plants have a larger immunophilin family than animals. Parvulins represent another family of PPIases that are unrelated to immunophilins in protein sequences and drug binding properties. Three parvulin genes were found in Arabidopsis genome. The expression of many immunophilin and parvulin genes is ubiquitous except for those encoding chloroplast members that are often detected only in the green tissues. The large number of genes and diversity of structure domains and cellular localization make PPIases a versatile superfamily of proteins that clearly function in many cellular processes in plants.
Immunosuppressive drugs cyclosporin A (CsA), FK506, and rapamycin are used clinically in transplantation to prevent graft rejection. During the course to understand the molecular mechanisms of immunosuppression by CsA, FK506, and rapamycin, the cellular receptors of these drugs have been purified and characterized (Schreiber, 1991; Fruman et al., 1994). CsA binds to a family of receptors named cyclophilins (CyPs), and FK506 and rapamycin bind to a distinct set of receptors called FKBPs (FK506 and rapamycin-binding proteins). Cyclophilins and FKBPs are collectively referred to as immunophilins (Schreiber, 1991).
The complexes formed by immunophilins and their cognate ligands are the functional modules for immunosuppression. The FKBP12-FK506 and CyP-CsA complexes, but not their separate components, bind to and inhibit the activity of calcineurin, a Ca2+, calmodulin-dependent protein phosphatase (Liu et al., 1991). Studies have demonstrated that inhibition of calcineurin activity is necessary for the immunosuppressive effect of CsA and FK506 (Clipstone and Crabtree, 1992; Liu et al., 1992; O'Keefe et al., 1992; Rao et al., 1997). The complex formed by FKBP12 and rapamycin targets FRAP (Brown et al., 1994), RAFT1 (Sabatini et al., 1994), or mTOR (Sabers et al., 1995), a mammalian homolog of yeast TOR1 and TOR2 that is required for G1-S progression in the cell cycle (Heitman et al., 1991; Kunz and Hall, 1993). FRAP belongs to a novel family of phosphatidylinositol kinase (PIK)-related kinases that are involved in a range of essential cellular functions, including cell cycle progression, DNA repair, and DNA recombination (Keith and Schreiber, 1995; Abraham, 1998).
Identification of the functional targets (calcineurin and mTOR) have elucidated the molecular basis for immunosuppression by immunosupressants CsA, FK506, and rapamycin. Meanwhile, it also demonstrates a unique gain-of-function mechanism for immunophilins in the presence of their drug ligands. However, these studies have not addressed the cellular functions of immunophilins in the absence of immunosuppressive drugs. During the past decade, a growing number of immunophilins have been characterized not only from mammalian sources but also from other organisms ranging from bacteria to yeast to higher plants (for reviews, see Schreiber, 1991; Fruman et al., 1994; Luan, 1998). The high level of conservation and ubiquitous distribution of immunophilins among divergent organisms and in almost all the subcellular compartments suggest that these proteins participate in important cellular processes.
Their attendant peptidyl prolyl cis/transisomerase (PPIase, EC 5.2.1.8) activity led to the suggestion that immunophilins facilitate protein folding in vivo. Evidence for this hypothesis is accumulating (Fruman et al., 1994; Brenner and Wainberg, 2001). Several studies suggest that immunophilins play a role in protein trafficking (Patterson et al., 2000). A genetic analysis shows that cyclophilin homolog NinaA in fruitflies (Drosophila melanogaster) is required for the transit of specific isoforms of rhodopsin from the endoplasmic reticulum (Stamnes et al., 1991). It is not known if foldase/rotamase activity is essential for these trafficking processes. Other findings suggest that each member of immunophilins may have specific functions that may or may not be related to their function in protein folding process. For example, FKBP12 associates with and modifies the activity of Ca2+-releasing ryanodine receptor in the striated muscle cells (Brillantes et al., 1994). As a result, mutant mice without a functional FKBP12 develop cardiac defect (Shou et al., 1998). Fkbp52 associates with interferon regulatory factor-4 (IRF-4) and the FKBP52-IRF-4 association inhibited IRF4-PU.1 binding to the immunoglobulin light chain enhancer as well as IRF-4-PU.1 transactivation in a PPIase activity dependent manner (Mamane et al., 2000), providing a novel posttranslational mechanism of transcriptional control.
The FKBP family also includes a member called trigger factor that is found in all eubacteria analyzed and in the chloroplast of higher plants. Trigger factor is the major protein that cross-links to virtually all nascent chains of secretory and cytosolic proteins tested (Valent et al., 1995; Hesterkamp and Bukau, 1996a). Sequence analysis and substrate specificity suggests that trigger factor contains a central domain belonging to the FKBP family (Callebaut and Mornon, 1995; Hesterkamp and Bukau, 1996b). Besides the central PPIase domain, trigger factor has a ribosome binding domain at the N-terminal domain and its C-terminal domain may strengthen the binding (Hesterkamp et al., 1997).
Another group of proteins that also possess PPIase activity but are otherwise unrelated with immunophilins are parvulins. A parvulin protein was originally found in Escherichia coli as a novel PPIase that consists of only 92 amino acids and is hence named parvulin (Latin: parvulus, very small; Rahfeld et al., 1994). Later on, parvulin homologs were found in both prokaryotic and eukaryotic organisms like SurA from E. coli (Eisenstark et al., 1992), Ess1/Ptf1 from yeast (Saccharomyces cerevisiae; Hanes et al., 1989; Hani et al., 1995), and Pin1 from human (Lu et al., 1996). Besides PPIase activity, the prokaryotic parvulins exhibit chaperone-like activities. SurA is located in the periplasm of E. coli and participates in the early stage maturation of outer membrane proteins (Lazar and Kolter, 1996; Rouviere and Gross, 1996). Eukaryotic parvulins can be subdivided into two groups according to their substrate specificity: phospho-(Ser/Thr)-Pro moieties-specific and nonspecific type. PIN1 from human belongs to the first group and is essential for cell cycle regulation (Ranganathan et al., 1997; Yaffe et al., 1997). PIN1-like parvulins have been identified as ESS1 in yeast (Hanes et al., 1989), Dodo in Drosophila (Maleszka et al., 1996), and AtPIN1 in Arabidopsis (Landrieu et al., 2000a). Unlike its homologs, the plant PIN1 like protein does not have the conserved WW-domain that is believed to be required for the recognition of phosphoprotein substrates (Lu et al., 1999). However, it still retains the phospho-substrate specificity and can complement yeast mutant lacking ESS1 (Landrieu et al., 2000; Metzner et al., 2001; Yao et al., 2001). The activity of the other human parvulin, hPar14, is independent from substrate phosphorylation (Rulten et al., 1999; Uchida et al., 1999). It was shown that phosphorylation of the N-terminal domain of hPar14 regulates its subcellular localization and DNA binding (Reimer et al., 2003). The function of Arabidopsis parvulins is unknown, but a recent study indicated that it may be involved in auxin signaling (Dharmasiri et al., 2003).
In an attempt to isolate meristem-specific genes, Gasser et al. (1990) isolated a highly abundant transcript from Arabidopsis encoding a cyclophilin. Presence of immunophilins was also supported by identification of CsA- and FK506-sensitive rotamase activity in plant organelles (Breiman et al., 1992). Luan et al. (1993) unexpectedly found a CsA- and FK506-sensitive process in plant cells, suggesting presence of both CYPs and FKBPs in higher plants. Using immunosupressants as an affinity tool, multiple CYPs and FKBPs have been purified from various subcellular compartments in a higher plant (Luan et al., 1993; Luan et al., 1994a). The most unique members of plant immunophilins are those that are localized in the chloroplast (Luan et al., 1994a). Along the way to characterizing plant immunophilins, a number of cyclophilin genes have been characterized (Gasser et al., 1990; Lippuner et al., 1994; Luan et al., 1994b; Marivet et al., 1995; Chou and Gasser, 1997). Among them, pCyPB/ROC4 encodes a chloroplast cyclophilin (Lippuner et al., 1994; Luan et al., 1994b). The first plant FKBP-type immunophilin (FKBP15) was cloned from both Arabidopsis and fava bean (Vicia faba) and was shown to be located in endoplasmic reticulum (Luan et al., 1996; J. Ting and S. Luan, unpublished data). There are at least two isoforms of FKBP15 in Arabidopsis and they are responsive to heat shock, consistent with the finding on the ER form of FKBP in yeast (Partaledis and Berlin, 1993; Sykes et al., 1993). A cytosolic FKBP12 has been characterized from fava bean and Arabidopsis (Faure et al., 1998; Xu et al., 1998). High Mr FKBP members have been identified from wheat (Triticum aestivum) and Arabidopsis (Blecher et al., 1996; Vucich and Gasser, 1996; Kurek et al., 1999). These large FKBPs contain the putative domains for interaction with Hsp90, as reported for FKBP59 and CyP 40 in animals (Pratt, 1998). Recent studies have begun to address the function of immunophilins in plants. An Arabidopsis mutant, pas1, displays abnormal developmental pattern. PAS1 gene was shown to encode a high Mr FKBP (Vittorioso et al., 1998). A cyclophilin 40 homolog has been shown to regulate development of leaf shape in Arabidopsis (Berardini et al., 2001). Several plant cyclophilins interact with an endonuclease involved in T-DNA transfer from agrobacterium to host plant cells (Deng et al., 1998). Disruption of AtFKBP42 gene function caused developmental defect (Kamphausen et al., 2002). A chloroplast FKBP has been shown to interact with a photosynthetic electron carrier and affects the accumulation of the protein (Gupta et al., 2002). As in animals, different members of immunophilins appear to play different roles in plants.
Recent completion of an Arabidopsis genome sequencing project revealed at least 52 genes encoding putative immunophilins and 3 genes encoding parvulin. Some of FKBP genes were surveyed before the completion of the genome project (Harrar et al., 2001). Based on the complete genome sequence of Arabidopsis, we made a comprehensive analysis of all immunophilin and parvulin genes. Domain analysis categorized these proteins into single and multiple domain immunophilins. Expression pattern and subcellular localization of some representative members were analyzed and implicated in a complex spectrum of functions in plant physiology and development. This study provides crucial information on the structure, genomic organization, and expression pattern of the PPIase superfamily in Arabidopsis and sets the stage for functional analysis.
RESULTS AND DISCUSSION
Immunophilin Genes in the Arabidopsis Genome
Using the BLAST programs to survey the Arabidopsis genome sequence database, we identified 52 genes encoding putative immunophilins. Among them, 23 genes encode FKBPs and 29 encode cyclophilins. Table I lists these genes with their names, accession numbers, molecular weight, potential subcellular localization, and other useful information. Arabidopsis encodes the largest immunophilin super family among organisms whose genomes have been completely sequenced. For example, yeast genome contains 4 genes for FKBPs and 8 for cyclophilins. The genome of Caenorhabditis elegans contains 8 genes for FKBPs and 16 genes for cyclophilins. Human genome contains 18 FKBPs and 24 cyclophilins (SMART). The large number of genes for immunophilins in plants indicates a diverse array of functions served by these proteins and this may also reflect significant degree of functional redundancy among the genes.
Table I.
Arabidopsis immunophilin and parvulin families
| Gene Name | Other Namea | AGI IDb | Amino Acid No.c | GeneBank Accession | Exons | Mrd | pIe | Subcellular Locationf | ESTl |
|---|---|---|---|---|---|---|---|---|---|
| kD | |||||||||
| AtFKBP Family | |||||||||
| AtFKBP121,2 | At5g64350 | 112 | AB008268 | 5 | 12 | 5.83 | S | 20 | |
| AtFKBP133 | FKBP22–14 | At5g45680 | 208/129 | AB012245 | 5 | 22/13.6 | 8.96/9.59 | TPgi | 5 |
| AtFKBP15–15 | At3g25220 | 153/128 | U52046 | 6 | 16.4/13.8 | 8.67/9.41 | ER | 1 | |
| AtFKBP15–25 | At5g48580 | 163/138 | U52047 | 6 | 17.7/14.8 | 5.05/4.73 | ER | 5 | |
| AtFKBP15–3 | At5g05420 | 143 | AB010692 | 5 | 15.3 | 9.02 | N | Y | |
| AtFKBP16–1 | At4g26555 | 207/136 | AF439821 | 8 | 22.7/15.2 | 7.67/5.32 | TP | 5 | |
| AtFKBP16–2 | FKBP22–24 | At4g39710 | 217/144 | AJ242481 | 5 | 19.7/15.5 | 8.09/9.22 | TPh | Y |
| AtFKBP16–3 | At2g43560 | 223/147 | AY113988 | 7 | 23.5/15.7 | 8.45/4.73 | TPgh | 5 | |
| AtFKBP16–4 | FKBP24–24 | At3g10060 | 230/148 | C010927 | 10 | 24.5/15.8 | 10.17/9.9 | TPh | 4 |
| AtFKBP17–1 | FKBP24–14 | At4g19830 | 229/151 | AJ242482 | 4 | 24.2/16.7 | 9.59/6.24 | TP | 2 |
| AtFKBP17–2 | At1g18170 | 247/153 | AC069551 | 3 | 26.5/16.5 | 8.97/7.32 | TP | 4 | |
| AtFKBP17–3 | At1g73655 | 234/152 | AC012679 | 3 | 25.7/16.8 | 6.68/5.12 | TP | 5 | |
| AtFKBP18 | At1g20810 | 232/161 | AF349528 | 6 | 25.5/17.9 | 9.8/9.65 | TPg | 5 | |
| AtFKBP19 | FKBP274 | At5g13410 | 256/169 | AL163572 | 10 | 27.8/18.7 | 7.55/9.17 | TPgh | 4 |
| AtFKBP20–1 | FKBP204 | At3g55520 | 190 | AL132975 | 5 | 20.3 | 8.66 | N | 1 |
| AtFKBP20–2 | At3g60370 | 242/175 | AL138646 | 7 | 27.2/19.9 | 8.65/9.53 | TPgh | 1 | |
| AtFKBP426 | TWD6 | At3g21640 | 356 | AB019232 | 7 | 40.6 | 6.02 | S | Y |
| AtFKBP43 | At3g12340 | 378 | AC069474 | 9 | 41.9 | 9.33 | N | Y | |
| AtFKBP53 | FKBP534 | At4g25340 | 477 | AL079350 | 11 | 52.2 | 4.66 | N | 2 |
| AtFKBP62 | ROF17 | At3g25230 | 555 | U49453 | 12 | 61.5 | 4.95 | S | 5 |
| AtFKBP65 | ROF24 | At5g48570 | 578 | AB015468 | 13 | 65.2 | 5.16 | S | 1 |
| AtFKBP72 | PAS18 | At3g54010 | 635 | U77366 | 20 | 71.8 | 5.12 | Nj | 1 |
| AtTIG | At5g55220 | 547/520 | AY074845 | 13 | 61.7/58.77 | 5.26/5.16 | CSP | 8 | |
| AtCYP Family | |||||||||
| AtCYP18–1 | At1g01940 | 160 | AC020622 | 5 | 17.5 | 8.49 | S | 2 | |
| AtCYP18–2 | At2g36130 | 164 | AC007135 | 6 | 18.2 | 8.69 | S | 2 | |
| AtCYP18–3 | ROC19 | At4g38740 | 172 | AL035656 | 1 | 18.4 | 7.97 | S | 31 |
| AtCYP18–4 | AtCYP1/ROC510 | At4g34870 | 172 | AL079347 | 1 | 18.4 | 8.98 | S | 40 |
| AtCYP19–1 | ROC39 | At2g16600 | 173 | AC005825 | 1 | 18.5 | 8.63 | S | 12 |
| AtCYP19–2 | AtCYP2/ROC69,11 | At2g21130 | 174 | AF020434 | 1 | 18.5 | 8.32 | S | 3 |
| AtCYP19–3 | ROC29 | At3g56070 | 176 | AL163763 | 1 | 18.9 | 7.97 | S | 1 |
| AtCYP19–4 | CyP512 | At2g29960 | 201/178 | AF020433 | 6 | 21.5/19.0 | 9.06/9.5 | SPP | Y |
| AtCYP20–1 | ROC713,14 | At5g58710 | 204/181 | AF192490 | 6 | 21.9/19.6 | 9.62/9.62 | SPP | 7 |
| AtCYP20–2 | At5g13120 | 259/183 | AL391711 | 6 | 28.3/20 | 9.82/7.99 | TPgh | 10 | |
| AtCYP20–3 | ROC49,15 | At3g62030 | 260/181 | AL138642 | 7 | 28.2/19.8 | 8.78/5.32 | CSPgk | 11 |
| AtCYP21–1 | At4g34960 | 224/197 | AL022023 | 6 | 24.6/21.4 | 6.89/5.89 | SSP | 1 | |
| AtCYP21–2 | At3g55920 | 228/197 | AF192490 | 8 | 24.5/21.1 | 7.07/5.85 | SSP | Y | |
| AtCYP21–3 | At2g47320 | 230/183 | AC002337 | 6 | 26/20.7 | 7.67/4.71 | MP | 6 | |
| AtCYP21–4 | At3g66654 | 236/189 | AC036106 | 7 | 26.4/21.3 | 9.41/5.74 | MP | 7 | |
| AtCYP22 | At2g38730 | 199 | AC005499 | 6 | 21.5 | 8.29 | S | 1 | |
| AtCYP23 | At1g26940 | 226/204 | AY085848 | 8 | 25.5/23.3 | 8.45/7.36 | SSP | 2 | |
| AtCYP26–1 | At3g22920 | 232 | AP001300 | 1 | 26 | 4.63 | S | Y | |
| AtCYP26–2 | At1g74070 | 317/244 | AY062660 | 3 | 34.4/26.2 | 10.04/9.65 | TP | 1 | |
| AtCYP28 | At5g35100 | 290/257 | AY074520 | 2 | 31.4/27.9 | 7.34/5.76 | TPg | 7 | |
| AtCYP37 | At3g15520 | 461/347 | P82869 | 12 | 45.1/37.3 | 9.5/5.08 | TPgh | 2 | |
| AtCYP38 | At3g01480 | 437/345 | AC009325 | 7 | 48/38.3 | 5.06/4.43 | TPgh | 7 | |
| AtCYP40 | Cyp4016 | At2g15790 | 361 | AC018722 | 8 | 40.5 | 5.69 | S | 1 |
| AtCYP57 | At4g33060 | 504 | AL031804 | 9 | 57.1 | 7.65 | S | 3 | |
| AtCYP59 | At1g53720 | 506 | AC024260 | 11 | 58.8 | 6.2 | N | 1 | |
| AtCYP63 | At3g63400 | 570 | AL163816 | 11 | 63.5 | 11.56 | N | 4 | |
| AtCYP65 | At5g67530 | 595 | AB013390 | 11 | 65 | 8.34 | S | 5 | |
| AtCYP71 | At3g44600 | 631 | AL353818 | 13 | 70.7 | 6.1 | S | 4 | |
| AtCYP95 | At4g32420 | 837 | AL034567 | 14 | 94.6 | 12.44 | N | 7 | |
| AtPIN Family | |||||||||
| AtPIN1 | PIN1At17 | At2g18040 | 119 | AF360318 | 2 | 13 | 9.71 | S | 5 |
| AtPIN2 | At1g26550 | 142 | AF332414 | 4 | 15 | 10.16 | S | 6 | |
| AtPIN3 | At5g19370 | 299/219 | AL035656 | 8 | 32.98/24.42 | 8.62/5.03 | CSP | 5 | |
Names used in the literature and the references are: 1, Xu, et al. 1998; 2, Faure, et al. 1998; 3, Gupta et al., 2002; 4, Harrar et al., 2001; 5, Luan, et al., 1996; 6, Kamphausen et al., 2002; 7, Vucich and Gasser, 1996; 8, Vittorioso et al., 1998; 9, Chou and Gasser, 1997; 10, Hayman and Miernyk, 1994; 11, Deng et al., 1998; 12, Saito et al., 1999; 13, Jackson and Soll, 1999; 14, Berardini et al, 2001; 15, Lippuner et al., 1994; 16, Berardini et al., 2001; 17, Landrieu et al., 2000.
Arabidopsis Genome Initiative nomenclature.
Amino acid number of the full length protein/amino acid number of predicted mature protein.
The Mr and isoelectronic point of the full-length/mature protein were predicted by protein composition (http://www.up.univ-mrs.fr).
Subcellular location: S, cytosol; ER, endoplasmic reticulum; SPP, secretory pathway protein without ER-retention signal; CSP, chloroplast stroma protein; TP, thylakoid lumenal protein; MP, mitochondrion protein; and N, nucleus. The location of some proteins were verified by other experimental methods.
Arabidopsis thylakoid lumen proteomics, Peltier, et al., 2002.
Arabidopsis thylakoid lumen proteomics, Schubert; et al., 2001.
Chloroplast import; Gupta et al. 2002.
Green fluorescent protein-fusion; Carol et al., 2001.
Chloroplast import; Lippuner et al., 1994.
EST numbers found in the database, or no EST found but our data confirmed the expression indicated as Y (Yes).
For the FKBP nomenclature, we adopted previously published rules that are rather consistent in studies on both animal and plant FKBPs. Generally the proteins are named FKBP with prefix letters to indicate species of origin (e.g. At for Arabidopsis) and a suffix number to indicate Mr. In this study, we retained the names of published FKBP members in plants. These include regular names such as AtFKBP12 (Faure et al., 1998; Xu et al., 1998), AtFKBP15-1, AtFKBP15-2 (Luan et al., 1996), and AtFKBP13 (Gupta et al., 2002), and irregular names such as PAS1 and TWD1, which are named according to other circumstances such as phenotype of the mutants (Vittorioso et al., 1998; Kamphausen et al., 2002). For newly identified genes in this study, they were named after the calculated Mr of mature proteins predicted from cDNA-deduced sequences. The potential processing/cleavage sites were predicted using several programs as specified in “Materials and Methods” or were determined according to information from earlier proteomics analysis of thylakoid lumen proteins (Peltier et al., 2002; Schubert et al., 2002). For proteins with similar Mr, an extension number was added to the Mr to distinguish them (e.g. AtFKBP15-1, AtFKBP15-2, and AtFKBP15-3). One gene encoding trigger factor-like protein was found in the Arabidopsis genome and named as AtTIG. Trigger factors are distantly related to the FKBP family (Callebaut and Mornon, 1995; Hesterkamp and Bukau, 1996b).
Nomenclature for cyclophilins has been less consistent in previous studies. Earlier literature in animal systems used several different rules to name a cyclophilin. One way is to use CyP with prefix letters to indicate species of origin and suffix number to indicate Mr (e.g. CyP40). Another way is to name a protein CyP with suffix letter to indicate the numbering system (e.g. CyPA, CyPB, CyPC, CyPD…). Studies in plants also adopted several naming strategies including those inherited from animal literature with some modifications (AtCyP40, AtCYP5, pCyPB; Luan et al., 1994b; Saito et al., 1999; Berardini et al., 2001) and a new strategy that names cyclophilins rotamase cyclophilins (ROCs; Lippuner et al., 1994; Chou and Gasser, 1997). To streamline nomenclature of cyclophilins, we suggest here that cyclophilins, like FKBPs, are named after the abbreviation CYP with prefix letters to indicate species of origin and suffix numbers to indicate Mr. Nevertheless, we retained the previously published names for comparison as shown in Table I.
Immunophilins consist of two families of proteins, FKBPs and cyclophilins, that show little sequence similarity between the members of the two families. In addition, the overall sequence similarity between the members within each family can range widely. The pair-wise analyses with the full-length protein sequences indicated that amino acid identity was between 10% (lowest) to 68.4% (highest) for FKBPs and 10% to 90.1% for CYPs. However, all members were characterized by the FKBP or CYP signature residues that form the binding pocket for CsA or FK506/rapamycin (Schreiber, 1991). The FKBP signature is derived from the available structures of FKBP12 protein (Michnick et al., 1991; Van Duyne et al., 1991) and mutagenesis analysis (Aldape et al., 1992; DeCenzo et al., 1996). It was found that the residues required for drug binding are highly conserved and most of these residues are essential for the rotamase activity (DeCenzo et al., 1996). The conserved binding site is therefore used as the signature for FKBP protein identification. In a similar manner, the residues necessary for cyclophilin activity are highly conserved (Kallen et al., 1991; Zydowsky et al., 1992) and serve as CYP signature for identifying putative CYP members. The sequence alignment for AtFKBP and AtCYP family is shown in Figure 1 and Figure 2, respectively. For comparison of the structural property, the human FKBP12 (hFKBP12) and CyPA (hCyPA) were included in the sequence alignments. The residues important for the drug binding and PPIase activity were marked. As AtTIG has only weak similarity to FKBPs, we did not include it in the sequence alignment.
Figure 1.
Multiple sequence alignment of Arabidopsis FKBPs. Human FKBP12 (hFKBP12, GenBank accession no. A35780) was included for comparison. Amino acids of full length protein of single domain AtFKBPs were used for the alignment. Only the amino acids spanning the most conserved FKBP domain were used for the multiple domain AtFKBPs. The sequences used for the alignment were: amino acids 1 to 160 for AtFKBP42, 62, and 65; amino acids 237 to 378 for AtFKBP43; amino acids 350 to 477 for AtFKBP53; and amino acids 263 to 393 for AtFKBP72. The sequences were aligned using MegAlign followed by manual refinement. Consensus sequences (threshold 65%) were boxed and similar residues were shaded. Gaps (marked by dashes) were introduced to achieve maximum similarity. The amino acids necessary for rapamycin or FK506 binding as determined for hFKBP12 were marked by asterisks (*). The putative chloroplast/thylakoid targeting signal was manually adjusted around the double Arg or Lys-Arg residues (boxed). The hydrophobic stretches following the double Args were underlined. The α-helix and β-sheets derived from the hFKBP12 were shown.
Figure 2.
Multiple sequence alignment of Arabidopsis CYPs. Human CyPA (hCyPA, GenBank accession no. A35780) was included for comparison. Amino acids of full-length protein of single domain AtCYPs were used for the alignment. Only the amino acids spanning the CYP domain of the multiple domain AtCYPs were used. The amino sequences used for the alignment were: amino acids 1 to 200 for AtCYP40, 57, 59, 63, and 95; amino acids 201 to 461 for AtCYP37; amino acids 201 to 437 for AtCYP38; amino acids 301 to 595 for AtCYP65, and amino acids 401 to 631 for AtCYP71. The sequences were aligned using MegAlign followed by manual refinement. Consensus sequences (threshold 65%) were boxed and similar residues were shaded. Gaps (marked by dashes) were introduced to achieve maximum similarity. The amino acids necessary for CsA binding as determined for hCyPA were marked by asterisk (*). The α-helix and β-sheets derived from the hCyPA were shown.
Among the FKBPs from Arabidopsis, the highest sequence identity is found between FKBP15-1 and FKBP15-2 (68.4%), and between AtFKBP62 and AtFKBP65 (68.1%). The residues shown to be critical for rotamase activity are very conserved among some of the AtFKBPs including the first six sequences aligned in Figure 1. In most of FKBPs, some conserved residues are substituted by other residues. These substitutions may affect the drug binding and PPIase activity of FKBPs. In the AtFKBP42 sequence, 11 of the 13 conserved residues are changed, resulting in the loss of PPIase activity (Kamphausen et al., 2002). Among the AtCYP members, the first 15 sequences aligned in Figure 2 are highly conserved. They are characterized by a seven-amino acid insertion between α2 and β3 compared with the archetype human CyPA except AtCYP20-2 and AtCYP20-3, two chloroplast located cyclophilins, which were referred as divergent-loop cyclophilin (Dornan et al., 1999). In this 15-member group, proteins are highly conserved and have retained almost all the 13 conserved residues known to be critical for CYP activity (Pflugl et al., 1994). In contrast, AtCYP26-2, AtCYP28, AtCYP37, and AtCYP38 are the most divergent cyclophilins that contain only a few (1–5) of the 13 conserved residues.
Genomic Organization and Phylogenetic Relationship of Arabidopsis Immunophilins
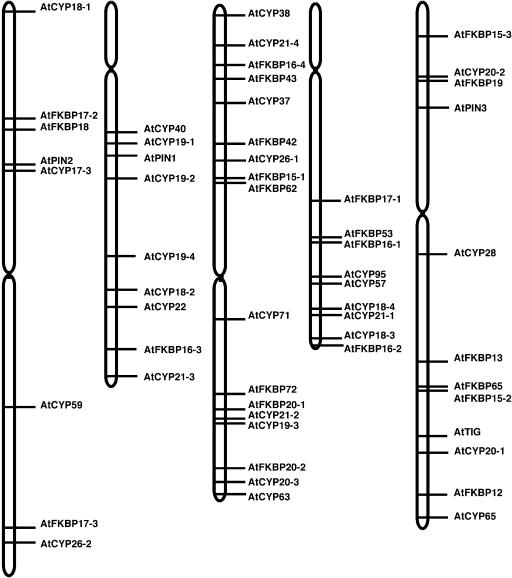
FKBP and cyclophilin genes are distributed on all five chromosomes in Arabidopsis (Fig. 3). Chromosome III has the largest number of immunophilin genes (17) and chromosomes I, II, IV, and V contain 7, 8, 9, and 11 genes, respectively. The short arms of chromosomes II and IV do not have immunophilin genes. The FKBP and cyclophilin genes are rather equally distributed on each chromosome except for chromosome II that has seven cyclophilins but only one FKBP. As discussed earlier, immunophilins consist of a very diverse group of proteins and may have very different origins during genome evolution. The only genes that appear to be produced by genomic duplication of AtFKBPs are AtFKBP15-1 and AtFKBP15-2, and AtFKBP62 and AtFKBP65. Interestingly, AtFKBP15-1 and AtFKBP62 are organized in a tandem on chromosome III and AtFKBP15-2 and AtFKBP65 are organized in a similar manner on chromosome V. Because AtFKBP15-1/AtFKBP15-2 show high sequence similarity and AtFKBP62/AtFKBP65 also resemble each other, we suggest that AtFKBP15-1/AtFKBP62 and AtFKBP15-2/AtFKBP65 are regional duplicates.
Figure 3.
Chromosomal distribution of Arabidopsis immunophilins and parvulins on the five chromosomes. The chromosome number was indicated above each chromosome. Immunophilin and parvulin genes were located along the five chromosomes by Chromosome Map Tool and redrawn by Canvas 5.
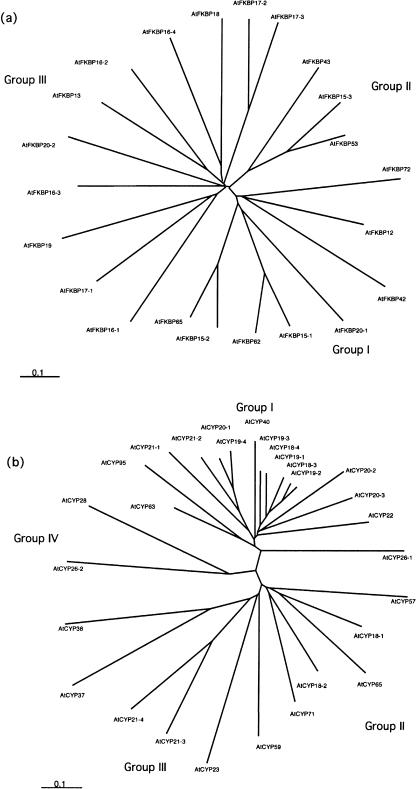
To further analyze phylogenetic relationships among Arabidopsis immunophilins, phylogenetic trees were constructed based on the alignments of AtFKBP and AtCYP protein sequences (Fig. 4, a and b). In this analysis, full sequences were used for the single domain immunophilins and only the immunophilin domains were used for the multiple domain immunophilins. For FKBP members with multiple FKBP domains, only the most conserved FKBP domain was used for the analysis as shown in Figures 1 and 2.
Figure 4.
Phylogenetic relationships of Arabidopsis FKBP (a) and CYP (b) proteins. The phylogenic analysis was based on the sequence alignments by ClustalX as described in “Materials and Methods.”
The AtFKBPs can be classified into three groups (Fig. 4a). Group I contains eight FKBPs with diverse subcellular locations, including ER located AtFKBP15-1 and AtFKBP15-2, nuclear AtFKBP20-1 and AtFKBP72, cytosolic AtFKBP12, AtFKBP62, and AtFKBP65, and membrane anchored AtFKBP42. Among them four are multidomain FKBPs that contain TPR repeats at their C-terminal region (see Fig. 5 and later discussion). Group II contains three nuclear FKBPs, including one single domain FKBP (AtFKBP15-3), two multiple domain FKBPs (AtFKBP42 and AtFKBP53) that both contain a C-terminal FKBP domain and a highly charged N-terminal domain. The last group contains the 11 thylakoid lumen FKBPs, including AtFKBP13, 16-1, 16-2, 16-3, 16-4, 17-1, 17-2, 17-3, 18-1, 18-2, 19, and 20. The thylakoid FKBPs constitute one-half of the Arabidopsis FKBP proteins.
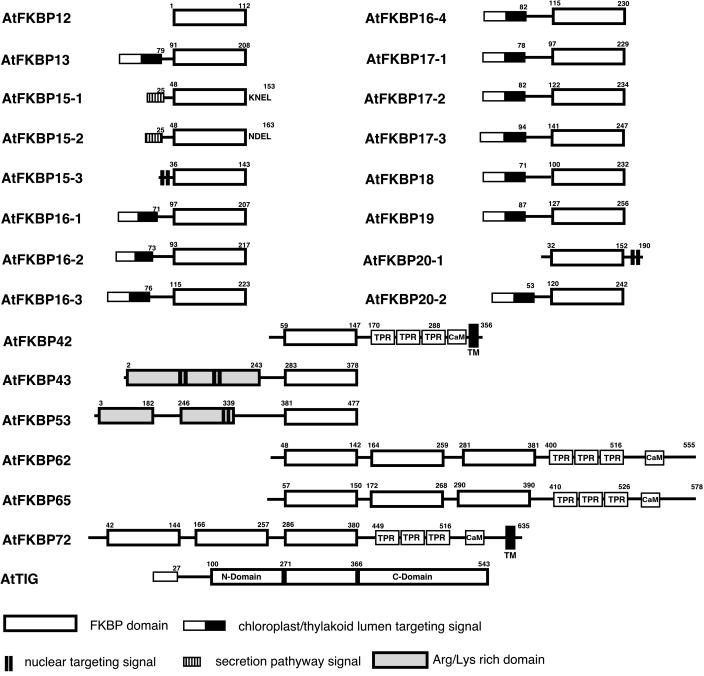
Figure 5.
Domain architecture of the Arabidopsis FKBP family immunophilins. The beginning and ending amino acid numbers of each protein are shown at each end of the diagram. FKBP domains are represented by white box. The other functional domains such as tetratricopeptide repeats (TPR), charged domains, and Calmodulin-binding motif (CaM) are indicated separately. The transmembrane domains (TM) that are not part of the signal peptide are labeled. The bipartite chloroplast/thylakoid targeting signal, ER targeting signal, and the NLS are shown. The ER retention signals of AtFKBP15-1 (KNEL) and AtFKBP15-2 (NDEL) are noted. The numbers above the boxes denote the amino acid positions of the function domains.
The AtCYP can be clustered into four groups. Group I is the most complex and includes 16 members. This group can be further divided into subgroups. The subgroup I contains five cytosolic single domain CYPs without any significant N- or C-terminal extensions. Furthermore, these five members have similar genomic structures that do not contain introns. The high similarity in both protein sequences and genomic structures indicates that they may have originated from duplication. The only other Arabidopsis immunophilin gene without any intron is AtCYP26-1 that also belongs to Group I but has a unique C-terminal transmembrane domain. Subgroup II contains two chloroplast CYPs, one located in stroma (AtCYP20-3) and the other in the thylakoid lumen (AtCYP20-2). Subgroup III contains four secretary pathway CYPs, including AtCYP19-4, 20-1, 21-1, and 21-2. Three multiple domain CYPs, AtCYP40, 63, and 95, are predicted to be nucleus localized and form subgroup IV. The remaining two members of Group I are AtCYP22 (with an N-terminal extension) and AtCYP26-1 (with a C-terminal extension) that may be in the cytosol.
Group II of the AtCYP family includes five cytosolic cyclophilins, two single domain cyclophilins AtCYP18-1 and AtCYP18-2, and three multiple-domain cyclophilins, AtCYP57, AtCYP65, and AtCYP71. The genes encoding the two single domain cyclophilins contain five and six extrons respectively, in contrast to those in the subgroup I of group I (with no intron). Group III contains six members with different subcellular locations. AtCYP21-3 and AtCYP21-4 are the only two Arabidopsis immunophilins that are predicted to be located in the mitochondria. The high similarity between both the precursors and the mature proteins of these two CYPs indicates that they may be generated by gene duplication. AtCYP37 and AtCYP38 are thylakoid lumen cyclophilins and are highly divergent AtCYPs. AtCYP37 can only be identified by the BLAST program using AtCYP38 as query. AtCYP23 (a secretary cyclophilin) and AtCYP59 (a nuclear multidomain cyclophilin) also belong to this group. Group IV contains two thylakoid lumen cyclophilins, AtCYP26-2 and AtCYP28. They are the most divergent AtCYPs. Only 1 to 3 out of the 13 residues important for PPIase and drug binding are conserved.
Subdomains and Subcellular Locations
The immunophilins, both cyclophilins and FKBPs, can be classified into single-domain members and multiple-domain members. The single-domain members contain a FKBP or CYP catalytic domain and other regions that do not show identifiable domain structures except for targeting sequences for different compartments. The multiple-domain members have other functional domains in addition to a single or duplicated CYP or FKBP domain(s). Of the 23 FKBPs, 16 are characterized as single domain members. These proteins all contain a single FKBP domain and some of them harbor a targeting presequence that determines subcellular localization of the mature protein. For example, 11 AtFKBPs contain putative chloroplast targeting sequences at the N-terminal region and 2 AtFKBPs contain putative ER targeting sequences. It is interesting to note that all the predicted chloroplast FKBPs have typical thylakoid lumen targeting signal (see Fig. 1). Except for AtFKBP16-2, the presequence of all other chloroplast FKBPs is characterized by double Arg residues followed by a hydrophobic region common to lumen proteins that are translocated via the ΔpH pathway. The thylakoid lumen signal peptide of AtFKBP16-2 has Lys-Arg instead of double Arg and most probably go through the Sec-dependent import pathway. This suggests that all the chloroplast FKBPs are located in the thylakoid lumen. Several of these FKBPs were analyzed previously by chloroplast import and proteomics procedures. The localization of AtFKBP13 was determined by both import and chloroplast fraction followed by western blot. It was also shown that AtFKBP13 was imported into thylakoid lumen through the ΔpH pathway (Gupta et al., 2002). Proteomics procedures designed to sequence all thylakoid lumen proteins uncovered several FKBPs including AtFKBP13, 16-2, 16-3, 16-4, 18, 19, and 22 (Table I and references therein). As transit peptides from AtFKBP16-1, 17-1, 17-2, and 17-3 have similar structure to that of verified lumenal FKBPs, these FKBPs may also be located in the lumen. The predicted Mrs of the mature proteins of the chloroplast FKBPs range from 13 kD to 20 kD. The chloroplast/thylakoid lumen targeting sequences in these FKBPs consist of 71 to 94 amino acid residues. The position of the last amino acid of the signal peptides is marked in the domain structure schemes (Fig. 5). All the chloroplast FKBPs have their FKBP domain at the C terminus and possess a short N-terminal extension that are highly divergent among these FKBPs.
Besides chloroplast FKBPs that constitute a large fraction (50%) of FKBP family in Arabidopsis, other FKBPs were predicted to be located in the ER, nucleus, and cytosol. None of the FKBPs is predicted to be in the mitochondria although early studies suggested that FKBP-type rotamase activity and proteins were detected in the mitochondria (Breiman et al., 1992; Luan et al., 1994a). It is interesting to note that the human FKBP38 was shown to be located in the mitochondria (Shirane and Nakayama, 2003) although such a feature is not predicted by its sequence. Thus, it is possible that the mitochondrial AtFKBP(s) exist but were simply not predicted by any of the programs used in this study. The ER-located proteins include AtFKBP15-1 and AtFKBP15-2 that possess a 25-amino acid signal peptide at the N terminus and an ER retention signal (KNEL and NDEL for AtFKBP15-1 and AtFKBP15-2, respectively) at the C terminus (Fig. 5). The nuclear FKBPs may include AtFKBP15-3, AtFKBP20-1, AtFKBP43, and AtFKBP53 that contain putative nuclear localization signal(s). The FKBPs without an identifiable targeting signal may be located in the cytosol. However, precise location of FKBPs cannot be confirmed without vigorous experimental work.
Seven of the AtFKBPs belong to multiple-domain members. Four of them including AtFKB42, AtFKBP62, AtFKBP65, and AtFKBP72, are characterized by a single (for AtFKBP42) or triple (for AtFKBP62, 65, and 72) FKBP-domain(s), the TPR domain, and putative calmodulin-binding domain. The domain structure of these four AtFKBPs resembles that of mammalian FKBP51/FKBP52 except that the mammalian FKBPs contain two FKBP domains (Callebaut et al., 1992). The human FKBP51/52 are found in the steroid hormone receptor complex that also contains hsp90 and p23 (Smith et al., 1993). The TPRs are degenerate sequences of 34 amino acids and are believed to serve as protein-protein interaction modules (Goebl and Yanagida, 1991). It is shown that mammalian hsp90 has a universal TPR binding region that can bind TPR containing proteins such as immunophilins, protein phosphatase 5, and p60 (Owens-Grillo et al., 1996; Silverstein et al., 1997; Barent et al., 1998). The human CyP40, a TPR-containing cyclophilin, also interacts with hsp90 (Ratajczak and Carrello, 1996). The homolog of human CyP40, AtCYP40, is the only AtCYP that contains TPR domain and may bind hsp90. It has been shown that wheat FKBP73/77 with TPR domains can replace animal FKBP52 in forming a complex with human hsp90 (Pratt et al., 2001). Because hsp90 is highly conserved among eukaryotes, wheat FKBP73/77, AtFKBP62, AtFKBP65, and possibly AtFKBP72 may function by interacting with hsp90 in plant cells. Two of the multiple domain AtFKBPs are AtFKBP43 and AtFKBP53 that contain highly charged N-terminal domains in addition to a single FKBP domain in the C-terminal region. The highly charged domain may be involved in DNA binding and protein-protein interaction. Both proteins have predicted nuclear localization signals, suggesting a nuclear localization.
The distantly related member of the Arabidopsis multiple-domain FKBP group is the trigger factor AtTIG. Like its homologs from eubacteria, AtTIG contains a central FKBP domain (271 amino acids–366 amino acids) located between the N-terminal ribosome binding domain and C-terminal domain that both help bind ribosome (Hesterkamp et al., 1997). AtTIG was predicted to be located in the chloroplast stroma, consistent with the cyanobacterial origin of the chloroplast. It is noteworthy that trigger factor is only found in bacteria and in the chloroplast so far. The putative chloroplast trigger factor is encoded by the nuclear genome. Trigger factor is a ribosome-associated chaperone and is the first chaperone that encounters nascent peptides, as the trigger factor can be cross-linked with polypeptide chains just emerging from the peptide exit tunnel of the ribosome (Hesterkamp et al., 1996c). Trigger factor and DnaK cooperate to promote proper folding of a variety of E. coli proteins. Deletion of either gene has no effect on viability at normal growth temperatures, but inactivation of both genes is lethal (Deuerling et al., 1999; Teter et al., 1999). The importance of trigger factor is also emphasized by the fact that it is the only PPIase in Mycoplasma species (Bang et al., 2000). The function of trigger factor in the chloroplast is unknown. But it may also function like its bacterial homologs in the protein synthesis and targeting processes.
Among the 29 cyclophilins, 21 are single domain members and 8 are characterized as multi-domain proteins (Fig. 6). Unlike the FKBPs, none of the cyclophilins contains multiple catalytic domains. In addition, the functional domains in CYPs are more divergent. Besides the TPR domain that is also found in some of the AtFKBPs, some CYP members contain other functional domains such as WD-40 repeat, U-box domain, Zinc finger, and Leu zipper domains each of which is involved in protein-protein or protein-DNA interactions. A unique RNA recognition motif (RRM) that may interact with RNA was also found in AtCYP59. It is noteworthy that most of the multiple domain cyclophilins have Arg- or Lys-rich domains with unknown function.
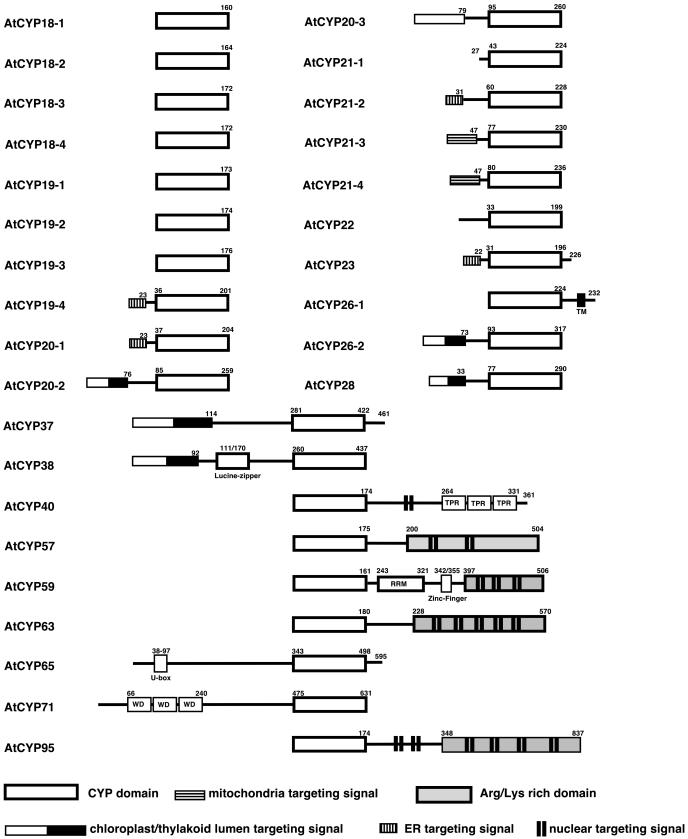
Figure 6.
Domain architecture of the Arabidopsis CYP family immunophilins. The beginning and ending amino acid numbers of each protein is shown at teach end of the diagram. CYP domains are represented by open box. The other functional domains such as tetratricopeptide repeats (TPR), U-box, WD repeats (WW), RRM, Leu-zipper, and zinc-finger are indicated. The transmembrane domains (TM) that are not part of the signal peptide are labeled. The bipartite chloroplast/thylakoid targeting signal, ER targeting signal, and the nucleus localization signal (NLS) are shown. The numbers above the boxes denote the amino acid positions of the function domains.
The single-domain cyclophilins contain the cyclophilin domain and some of them have a signal peptide for sorting to different organelles. Interestingly, a number of CYPs were again predicted to be chloroplast proteins. These CYPs (total of six) all contain the typical N-terminal transit peptide for chloroplast import. Five of them including AtCYP20-2, 26-3, 28, 37, and 38 may be translocated into the thylakoid lumen as they contain the bipartite signal sequences for crossing both envelope and thylakoid membrane (Fig. 6). AtCYP20-3 (ROC4) is predicted and experimentally confirmed by import assay to be located in the chloroplast stroma (Lippuner et al., 1994). The homolog of AtCYP38 from pea (Pisum sativum), TLP40, has been shown to be located in the thylakoid lumen and partition between thylakoid membrane and soluble lumenal fraction (Fulgosi et al., 1998). It was shown that the association/dissociation of TLP40 from thylakoid membrane regulates the phosphorylation of photosystem II components (Vener et al., 1999). The chloroplast (lumenal or stromal) localization of some of the predicted CYPs were also verified by chloroplast import, immunodetection and peptide sequencing efforts (Table I and references therein).
Two of the CYPs, AtCYP21-3 and AtCYP21-4, are predicted to target mitochondria. A mitochondrial cyclophilin (CyP-D) in animal cells modulates the mitochondrial permeability transition pore (MPTP) and plays a critical role in apoptotic and necrotic cell death (Connern and Halestrap, 1994; Crompton, 1999). A similar process may exist in plants and the plant mitochondrial cyclophilins may be involved (Arpagaus et al., 2002). The presence of mitochondrial cyclophilins was also indicated by the identification of CsA-sensitive rotamase activity and purification using CsA affinity column (Breiman et al., 1992; Luan et al., 1994a). AtCYP21-3 and AtCYP21-4 contain very similar signal peptides of 47 amino acids and have highly homologous C-terminal cyclophilin domains.
There are five cyclophilins predicted to go through the secretory pathway. These include AtCYP19-4, AtCYP20-1, AtCYP21-1, AtCYP21-2, and AtCYP23. Unlike the ER-located AtFKBPs, these cyclophilins do not have a C-terminal ER-retention signal. Their exact location needs to be determined by further experiments. The AtCYP19-4 (CyP5) has been shown to be located in the ER by N-terminal green fluorescent protein fusion (Saito et al., 1999). This protein displayed PPIase and protein refolding activities that were sensitive to cyclosporin A (Grebe et al., 2000). Furthermore, AtCYP19-4 was suggested to modulate the function of an Arabidopsis protein GNOM, a guanine nucleotide exchange factor essential for vesicle trafficking in many organisms (Grebe et al., 2000). Another secretory pathway cyclophilin is AtROC7 (Jackson and Soll, 1999), which may regulate PP2A activity.
Several single domain cyclophilins are predicted to be cytosolic proteins. These include AtCYP18-1, AtCYP18-2, AtCYP18-3, AtCYP18-4, AtCYP19-1, AtCYP19-2, and AtCYP19-3. These proteins are highly conserved and may have similar functions. AtCYP18-3 was shown to interact with Agrobacterium VirD2, an endonuclease covalently bound to the 5′ end of the T-DNA. Because T-DNA transfer is inhibited by CsA, CYP interaction with VirD2 may be important for agrobacterial infection (Deng et al., 1998). AtCYP26-1 is another CYP predicted to be cytosolic. However, there is a potential transmembrane domain at the C-terminal that may serve as a membrane anchor.
Eight Arabidopsis cyclophilins have other functional domains in addition to the CYP domain and are characterized as multiple domain cyclophilins. AtCYP59 contains a N-terminal cyclophilin domain (amino acids 1–161) followed by an RRM region (amino acids 243–321), a putative zinc-finger motif (amino acids 342–355), and a highly charged C-terminal domain (amino acids 397–506) with three putative nucleus localization signals (NLSs) located within the highly charged domain (Fig. 6). The RRM domain is a 90-amino acid structural module found in a variety of RNA binding proteins, including heterogeneous nuclear ribonucleoproteins, proteins implicated in the regulation of alternative splicing, and protein components of small nuclear ribonucleoproteins (Birney et al., 1993). Homologues of AtCYP59 are present in the genomes of Paramecium tetraurelia, Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens. These homologs differ mainly in the C-terminal region (Krzywicka et al., 2001). For example, the zinc-finger domain is present only in the AtCYP59, but not in the other homologs. The zinc-finger domain is involved in RNA-binding or single strand DNA binding even protein-protein interaction (Matthews and Sunde, 2002). The AtCYP59 homolog in Paramecium tetraurelia, KIN241, has been shown to be localized in the nucleus and involved in cell morphogenesis, cortical organization, and nuclear reorganization (Krzywicka et al., 2001). AtCYP65 is a 595 amino acid long polypeptide with a cyclophilin domain near the C terminus (amino acids 343–498) and a U-box domain near the N terminus (amino acids 38–97). U-box is a modified RING finger domain without the full complement of Zn2+-binding ligands. The U-box is a highly conserved domain present in some ubiquitin ligases. Ubiquitin ligases determine protein stability by coordinating the addition of polyubiquitin chains to proteins that are targeted to the proteasome for degradation (Hatakeyama and Nakayama, 2003). There is only one U-box-containing protein, UFD2, in yeast. Two UFD2 homologs and several other U-box domain proteins have been found in humans. Surprisingly, Arabidopsis genome encodes at least 75 proteins with a putative U-box domain (SMART). The plant U-box proteins are categorized into five distinct subclasses, suggesting that they play diverse roles (Azevedo et al., 2001). The ARC1 gene from Brassica is required for self-incompatibility and represents the only plant U-box gene with a known function (Stone et al., 1999).
AtCYP71 contains 631 amino acid residues with its cyclophilin domain located at the C terminus (amino acids 475–631) and three WD-40 repeats near the N terminus (amino acids 66–240; Fig. 6). There are 367 WD-40 proteins encoded by the Arabidopsis genome (SMART), but only a few of them have been functionally characterized. WD-40 repeat is a protein-protein interaction module that consists of about 40 amino acid residues with a central Trp-Asp that are generally present as tandem repeats. The WD-40 repeat domain of a photomorphogenesis repressor, COP1, mediates its interaction with HY5 transcription factor and thereby leads to the targeted degradation of HY5 by the 26S proteasome (Holm et al., 2001).
AtCYP95 contains 837 amino acids and is the largest immunophilin in Arabidopsis. It has a cyclophilin domain (amino acids 1–174) at the N-terminal end and a highly charged domain (amino acids 317–838) at the C terminus. Seven NLSs were identified in AtCYP95 with four located in the highly charged domain and two outside of it (Fig. 6), suggesting a possible nuclear localization. The highly charged domain is also rich in Ser-Arg (SR repeats), a feature of many RNA splicing factors (Zahler et al., 1992; Neugebauer et al., 1995). Matrin cyclophilin, the mammalian homolog of AtCYP95, is shown to associate with the nuclear matrix and splicing factors (Mortillaro and Berezney, 1998). Another Arabidopsis multiple domain cyclophilin, AtCYP63, has a domain structure similar to AtCYP95. AtCYP63 has a cyclophilin domain (amino acids 1–180), a highly charged, SR-rich domain (amino acids 228–570), and five predicted NLSs. Both AtCYP63 and AtCYP95 may be located in the nucleus and possibly play a role in RNA metabolism. AtCYP57 has two predicted NLSs, an N-terminal cyclophilin domain (amino acids 1–175), and a highly chargedC-terminal region. However, the highly charged domain, unlike those in AtCYP95 and ATCYP63, lacks an obvious SR-rich repeats; instead, it has alternated acidic and basic patches that may be involved in the interaction with the highly charged matrix proteins in the nucleus.
Parvulin Gene Family of Arabidopsis
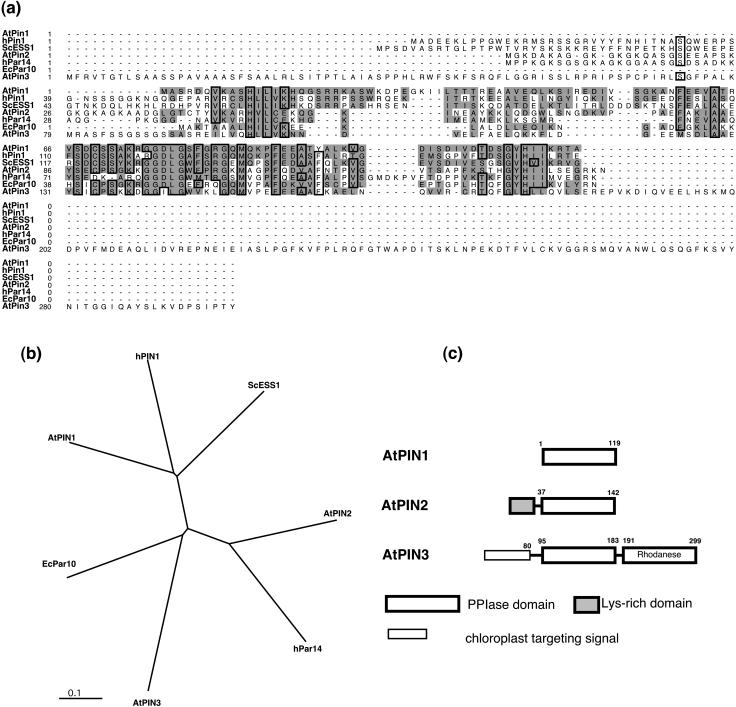
Three parvulin genes were identified in the Arabidopsis genome and were named AtPIN1, AtPIN2, and AtPIN3, respectively. Sequence alignment (Fig. 7a) showed that AtPIN1 is more similar to the eukaryotic PIN1 type parvulin with 52% and 51% identity to hPIN1 and ScESS1, respectively. AtPIN2 is more similar to hPar14 with 53% sequence identity. As a result, AtPIN1 and AtPIN2 belong to different types and share only 32% identity. AtPIN3 is most similar to the bacterial parvulin EcPar10, with a 39% identity. Indeed, studies have shown that AtPIN1 is functionally equivalent to eukaryotic PIN1 type parvulins that are characterized by specificity to phosphorylated substrates (Ranganathan et al., 1997; Landrieu et al., 2000a). AtPIN2 may have similar properties with hPar14, as they share a similar PPIase domain and a similar N-terminal domain enriched in basic amino acids (Fig. 7a). The basic N-terminal domain is responsible for the phosphorylation regulated partition between cytosol and nucleus and its affinity for DNA binding (Reimer et al., 2003). From the unrooted phylogenetic tree (Fig. 7b), the three AtPIN parvulins can also be classified into three groups: eukaryotic PIN1 type, hPar14 type, and prokaryotic type.
Figure 7.
a, Multiple sequence alignment of Arabidopsis parvulins. Some members of the parvulin family from other organisms were used for comparison. They were: hPIN1 (human, Q13526), hPar14 (human, AB009690), ScESS1 (yeast, P22696), and EcPar10 (E. coli, P39159). Amino acids of full-length protein were used for the alignment. The sequences were aligned using MegAlign followed by manual refinement. Consensus sequences (threshold 65%) were boxed and similar residues were shaded. Gaps (marked by dashes) were introduced to achieve maximum similarity. The amino acids necessary for rapamycin or FK506 binding as determined for hFKBP12 were marked by asterisks. The α-helix and β-sheets derived from the hFKBP12 were shown. b, Phylogenetic relationships of Arabidopsis parvulin proteins and selected members of parvulin from other organisms. The phylogenic analysis was based on the sequence alignments by ClustalX as described in “Materials and Methods.” c, Domain architecture of the Arabidopsis parvulins. The beginning and ending amino acid numbers of each protein are shown at each end of the diagram. Parvulin PPIase domains are represented by white box. The Rhodanese domain of AtPIN3 is shaded and labeled. The chloroplast targeting signal is shown. The numbers above the boxes denote the amino acid positions of the function domains.
Among the three parvulins, AtPIN1 is a single-domain protein with only the PPIase domain. AtPIN2 and AtPIN3 are multiple-domain proteins (Fig. 7c). AtPIN2 has a C-terminal PPIase domain and anN-terminal Lys-rich domain that may have regulatory functions as that in hPar14. AtPIN3 has a PPIase domain located in the middle, a putative C-terminal rhodanese domain that may be involved in protein-protein interaction (Bordo and Bork, 2002), and a predicted N-terminal chloroplast targeting signal (Fig. 7c). It was shown recently that another rhodanese domain containing protein (SIR1) is involved in the auxin signal transduction pathway (Zhao et al., 2003). Parvulin(s) may also participate in auxin action as an inhibitor of parvulin PPIase blocked auxin function in a cell-free system (Dharmasiri et al., 2003).
Expression Patterns of Plant Immunophilins
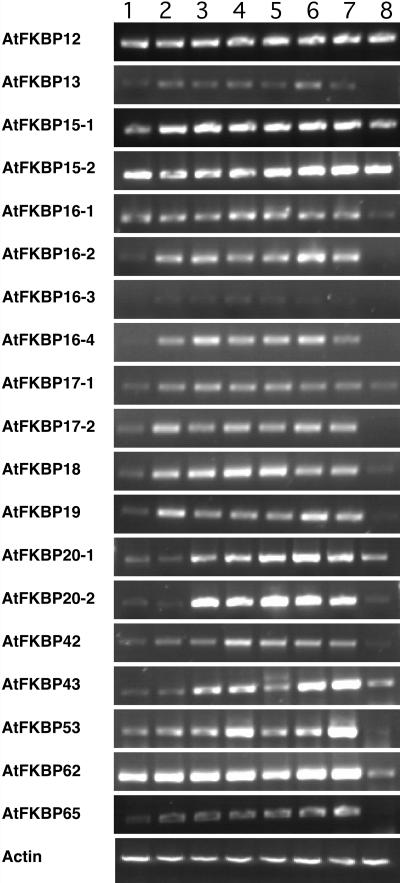
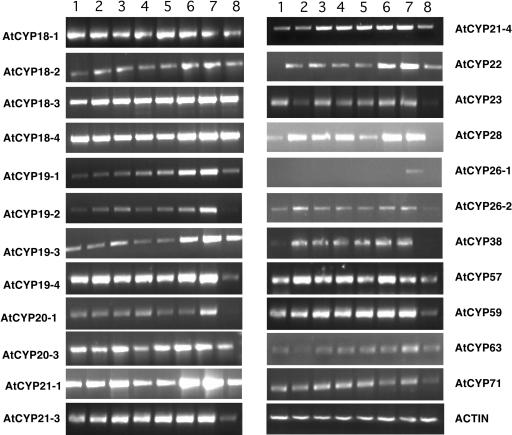
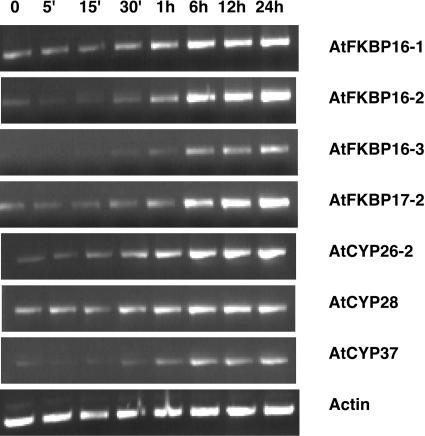
The available data from the EST database, cDNA cloning and sequencing, and our reverse transcription (RT)-PCR analysis indicate that all 51 immunophilin genes are expressed in plants (Table I). The expression patterns of some of the genes are shown in Figure 8 and Figure 9. Most of the genes are expressed in all tissues studied, including etiolated seedlings, roots, stems, leaves, and flowers. The expression pattern of some of the AtCYP genes were studied by northern blot (Chou and Gasser, 1997) and our RT-PCR results are consistent with these results. One cyclophilin, AtCYP26-1, appeared to be specifically expressed in flowers (Fig. 9), implicating CYP in further specific functions. As expected, the genes encoding chloroplast immunophilins are mainly expressed in green tissues and are hardly detectable in roots. Like other genes for chloroplast proteins, some genes for chloroplast immunophilins are also expressed in etiolated seedlings, but the transcript levels were much lower as compared to those in the green tissues. The expression levels increased with greening of the eliolated seedlings (Fig. 10). We speculate that their protein products may accumulate in etiolated seedlings should they be required for chloroplast development. At least in the case of pea TLP40 protein (homologue of AtCYP38), it was shown to accumulate in etiolated seedlings (Fulgosi et al., 1998).
Figure 8.
Expression patterns of Arabidopsis FKBP genes in different tissues. Semiquantitative RT-PCR was performed with gene specific primers using cDNAs synthesized from RNA samples isolated from different tissues. The tissues used were 3-d dark growing seedling (1), 3-d light growing seedling (2), 2-week seedling (3), leaves (4), caulin leaves (5), stems (6), flowers (7), and roots (8).
Figure 9.
Expression patterns of Arabidopsis CYP proteins in different tissues. Semiquantitative RT-PCR was performed with gene specific primers using cDNAs synthesized from RNA samples isolated from different tissues. The tissues used were described in Figure 8.
Figure 10.
Light response of the expression level of representative immunophilin genes as determined by semiquantitative RT-PCR. Six-day dark growing seedlings were exposed to light (100 μmol m−2 s−1) and tissues were harvested at the indicated time point. RNA isolation and RT-PCR was performed as detailed in “Materials and Methods.”
To examine the expression of immunophilin genes under various growth conditions, we took advantage of the available data on transcriptional profiling (Stanford Microarray Database). Analysis of microarray data in the database indicated that some of the immunophilin genes are regulated by environmental conditions (Table II). The most notable is that many genes for the chloroplast immunophilins are regulated by light-induction with greening process and decrease in the dark, consistent with our RT-PCR analysis in Figure 10.
Table II.
Expression regulation by different conditions
| Gene Names | Experiment ID | Experimental Treatment | Expression Fold Change |
|---|---|---|---|
| AtCYP18–2 | 21124/22172/22171 | Pathogen | −2.44 |
| AtCYP18–3 | 17573 | Cytokinin | −3.6 |
| 2476 | Salt | +2.43 | |
| AtCYP18–4 | 2475/2474/2476 | Salt | −9.08 |
| 2464/2463/2465 | Cytokinin | −2.74 | |
| AtCYP19–1 | 11895 | ABA insensitive mutant | −3.36 |
| AtCYP19–4 | 2471/2474/2473 | Salt | +2.1 |
| 2464/2465/2525 | Cytokinin | +2.2 | |
| AtCYP21–4 | 23184/23182/23185 | Dark | −10.6 |
| AtCYP20–2 | 11775/11770/11766 | Pathogen | −3.03 |
| 7341/14779 | Light | +3.57 | |
| AtCYP26–2 | 23548 | Sucrose | −14.3 |
| AtCYP28 | 23176/23184 | Dark | −6.5 |
| 7561 | High CO2 | −3.3 | |
| AtCYP37 | 23176 | Dark | −13.4 |
| AtCYP38 | 23176 | Dark | −29.0 |
| 7341/14779 | Light | +3.7 | |
| AtCYP71 | 11755 | Knox overexpression | −7.87 |
| 3743 | Auxin | −5.9 | |
| AtFKBP15–2 | 21124/22171 | Pathogen | −5.5 |
| AtFKBP13 | 23548 | Sucrose | −3.4 |
| 23176 | Dark | −3.0 | |
| AtFKBP16–3 | 3710/10847 | 700 ppm CO2 | −3.4 |
| AtFKBP17–3 | 23548 | Sucrose | −3.8 |
| AtFKBP19 | 7125/7114 | Iron deficiency | +3.1 |
| AtFKBP53 | 23180/23182 | Dark | −2.7 |
The expression data were retrieved from Stanford Microarray Database (http://genome-www5.stanford.edu/MicroArray/SMD/). Experiment ID was specified in the database. Detail of the experiment treatments can be found in the database under the same Experiment ID and the fold changes of the gene expression level upon treatment are indicated. +, Up-regulation; −, down-regulation.
In summary, studies in both animal and plant systems have revealed a diverse array of functions for individual immunophilin members. Such functions can be a result of their protein foldase activity, chaperone activity, scaffolding activity, and other unknown activities. A very clear distinction between immunophilins and other types of protein foldases and molecular chaperones is that each member of immunophilin family appears to have specific targets and function in the cell. This is consistent with the fact that the sequence and the structure of immunophilins are rather divergent although a conserved core for drug-binding is present in all members. The specific sequence motif in each member may present the structural basis for interacting with specific targets. Another general rule for immunophilin function is their association with super-molecular complexes in both animal and plant cells. We speculate that each immunophilin member may function in the maintenance of these complexes.
MATERIAL AND METHODS
Plant Materials and RNA Analysis
Arabidopsis (Columbia-0) plants were grown in the soil with a 16 h light/8 h dark cycle at 22°C in the greenhouse conditions. For RNA extraction, different parts of the plant (rosette leaves, caulin leaves, stems, flowers, and roots) were collected and frozen in liquid nitrogen. For plants growing on plates, surface sterilized Arabidopsis seeds were plated on one-half strength MS medium solidified by 0.8% agar. Total RNA was isolated from various tissues using TRIZOL Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instruction manual. First-strand cDNA was produced using 2 μg total RNA, oligo(dT) primer, and Superscript II RNase H− Reverse Transcriptase (Gibco-BRL) in a 20-μL reaction and the resulting cDNA was used as templates in 20 μL of PCR reaction (4 min 94°C; 28 to 32 cycles at 1 min 94°C, 1 min 52°C, and 1 min 72°C) using gene specific primers. The gene specific primers used for AtFKBPs are as follows: AtFKBP12, 5′-ctcgagctctagacatgggtgtggagaagcaag-3′ and 5′-tcaagatctctgcacgctcagtacttcg-3′; AtFKBP13, 5′-cggatccatggaaacaacttcttgtgaa-3′ and 5′-gagctcgagtcaagctttacctatgtac-3′; AtFKBP15-1, 5′-agatctagacaccatgatgagctctggatccgcc-3′ and 5′-tcaggatccaagctcattctttgatttcg-3′; AtFKBP15-2, 5′-gagctcgctagccatggcgagcaagatgagtc-3′ and 5′-tctggatcctagctcgtcatttcc-3′; AtFKBP15-3, 5′-gagctcgctagccatgtccccatctgaatctg-3′ and 5′-tcaggatcctttaacattaagcagctc-3′; AtFKBP16-1, 5′-gctctagaccagatgaaggaacctgaagtgatc-3′ and 5′-gcgggatcctaatactttcaagagctggat-3′; AtFKBP16-2, 5′-agtactagtaatgacgaggattgactactac-3′ and 5′-ccgctcgagtcatcttgtattacttcctg-3′; AtFKBP16-3, 5′-agatctagatgctggtttacctccagaag-3′ and 5′-ccgctcgagtcactcttcctctgaatcgag-3′; AtFKBP16-4, 5′-agcgctagccatggtaagcaccagaagaagag-3′and 5′-ccgctcgagtcaggatccgccttcaactatcttcactg-3′; AtFKBP17-1, 5′-ctcgagctctagagatgaagactaaatcaaagagtcc-3′ and 5′-tcaggatccttaactgcttgtgacatctg-3′; AtFKBP17-2, 5′-agatctagagcagatcaagactcg-3′ and 5′-ccgctcgagctcaggatccagctggtgcaattgagactc-3′; AtFKBP17-3, 5′-gaagatctagaaatggcgactctcttcactg-3′ and 5′-tcaggatcctacaatagtttggaaaca-3′; AtFKBP18, 5′-agatctagaatcttccgaagctagagag-3′ and 5′-gactagtcgactaggatccccttctcttctggtggagga-3′; AtFKBP19, 5′-gctctagagtctcaatttgctgacatgc-3′and 5′-ctcgagctctagatctattgggtacaattttgagg-3′; AtFKBP20-1, 5′-gagctcgctagccatgggtgatgcaatcgatttg-3′ and 5′-tcaggatcctttagctttgcctttgc-3′; AtFKBP20-2, 5′-agatctagatatgtcttccgtcgtctcctcc-3′ and 5′-ccgctcgagctcaggatcctcggtgaaggcgtctggtgctgacaagt-3′; AtFKBP42, 5′-agatctagacaccatggatgaatctctggagcatc-3′ and 5′-ttaggatccatctgctttaactctgtggcgtcg-3′; AtFKBP43, 5′-agatctagacaccatggagaaaggtagcagttatg-3′ and 5′-gacctcgtaacccttgaaggcagaggac-3′; AtFKBP53, 5′-agatctagacatgggattctggggacttgaag-3′ and 5′-cttcatcatgctcataatcatcttcatc-3′; AtFKBP62, 5′-agatctagaagccatggatgctaatttcgag-3′ and 5′-cccatatgctagctccgcaggtatagtg-3′; AtFKBP65, 5′-agcgctagccatggaagacgatttcgacac-3′and 5′-cccaagtcccatcctttaatgacatgtc-3′; and AtFKBP72, 5′-ggactagtcaccatggcggtaggcgatcagacgga-3′ and 5′-tcaggatcctgtaaatttggcgctcacaaactg-3′.
The gene specific primers used for AtCYP are as follows: AtCYP18-1, 5′-gagctcgctagccatggtgagtcgtttgtcc-3′ and 5′-tcaggatccaccagcaagtggattggc-3′; AtCYP18-2, 5′-gagctctagacatgtcggcaagacctcaag-3′ and 5′-tcaagatctatcgatcactttggtccttag-3′; AtCYP18-3, 5′-agcgctagccatggcgttccctaaggtatac-3′ and 5′-tcaagatctagagagctgaccacaatcg-3′; AtCYP18-4, 5′-gagctctagacatgtcgaaccctagagttttc-3′ and 5′-tcaggatccagaaaagctgaccacaatc-3′; AtCYP19-1, 5′-gagctcgctagccatggcaacaaaccctaaag-3′ and 5′-tcaggatccagaaatctgaccacaatc-3′; AtCYP19-2, 5′-agcgctagccatggcgtcgcatcctaaag-3′ and 5′-tcaggatccagaagagatctcaccacaatc-3′; AtCYP19-3, 5′-agcgctagccatggcgaatcctaaagtc-3′ and 5′-tcaggatcctgaacttgggttcttgagc-3′; AtCYP19-4, 5′-gagctcgctagccatggcgaaagcaagctttattc-3′ and 5′-tcaggatccgagaggaagttctccactgtc-3′; AtCYP20-1, 5′-gctagccatggcgagctcagtgacg-3′ and 5′-tcaggatcccaggggaaggctcaccactg-3′; AtCYP20-2, 5′-agatctagaaatggttactgaaccgcaatcg-3′ and 5′-tcagagctcgagttaagcttcagacattgg-3′; AtCYP20-3, 5′-agatctagacatggcttcttcgtcttctatg-3′ and 5′-gcgggatccagcatctaacgggagctctcc-3′; AtCYP21-1, 5′-gagctctagacatgcgtagagagatctcg-3′ and 5′-tcaggatcctctctcttcatcccatttgtc-3′; AtCYP21-2, 5′-gagctctagacatgggaatcacgagaaacttg-3′ and 5′-tcaggatcccgggaagcacttcattctctgtctttc-3′; AtCYP21-3, 5′-gagctcgctagccatggcgaagatcaaacctcaag-3′ and 5′-tcaggatcccatgtcttgaagcaaagtg-3′; AtCYP21-4, 5′-ggactagtaatgagtcagagatttgaggac-3′ and 5′-tcagagctcgagtcatgtctctagtttcag-3′; AtCYP22, 5′-tctagacatgaattcaggaggtgga-3′ and 5′-tcaggatcccatctccccacactcagta-3′; AtCYP26-1, 5′-agatctagacatggctaaccctaaagttttc-3′and 5′-tcaggatccaacgaaccaagaccaaaaac-3′; AtCYP26-2, 5′-agatctagatacaattgctaaccct-3′ and 5′-tcaggatcctagagtttgtgactcgatcaaacc-3′; AtCYP28, 5′-gctctagaaatgtccgcgacactctcctcc-3′ and 5′-ctaggatccaggcaaagttggagagag-3′; AtCYP37,5′-ctcgagctctagacatggttctttcatctccggacac-3′ and 5′-gacgtcgactaggatccagtggattcgttgatgttg-3′; AtCYP38, 5′-agtactagtggcgaatccagtgattc-3′ and 5′-ccgctcgagctcaggatccaccggcgattttgtaactc-3′; AtCYP40, 5′-gagctcgctagccatgggtaggtcaaagtgtttc-3′ and 5′-tcaggatcctacgaacattttgcggtac-3′; AtCYP57, 5′-gagctcgctagccatgtcagtctctattgtgac-3′ and 5′-tctctccaccactggccattgcaacag-3′; AtCYP59, 5′-agatctagacatgtcgacggtgtacgtgct-3′ and 5′-gagctcgcattagccatggcaacaatc-3′; AtCYP63, 5′-agatctagacatgactaaaaagaagaatcc-3′ and 5′-ccattgaaaggactccagctccatcatg-3′; AtCYP65, 5′-agatctagacatggggaagaaacaacacag-3′ and 5′-ttcagtgaagactttgtttagaacaggac-3′; AtCYP71, 5′-gagctctagacatggaggaagaatctaagaa-3′ and 5′-gagctctagacatggaggaagaatctaagaa-3′; and AtCYP95,5′-agatctagacatggcaaaaaagaagaatccac-3′ and 5′-ttcagtgaagactttgtttagaacaggac-3′. A total of 10 μL of PCR samples was separated by agarose gel electrophoresis and visualized with ethidium bromide staining. Primers used for the amplification were listed in Table II. The expression of an actin gene was used as an internal control for determining the RT-PCR amplification efficiency among different reactions. The RT-PCR reactions were repeated three times and representative results from one experiment were shown.
Sequence Analysis
Genes for the two families of immunophilins and parvulins were identified by sequence comparison of previously identified FKBPs, CYPs, and parvulins with entries in the public databases at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and the Arabidopsis Information Resource (http://www.arabidopsis.org/Blast/). The MEGALIGN (DNASTAR) was used to produce multi-alignments and to determine the homology scores. To produce the phylogenetic trees, ClustalX was used for the alignment of the PPIase domain sequences. Neighbor-joining method was used for tree reconstruction and confirmation of the tree topology by boostrap analysis (1,000 replicates) was done by ClustalX using default settings. The trees were drawn by Treeview (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
To search for functional domains within the immunophilin and parvulin protein sequences, SMART (http://smart.embl-heidelberg.de/) and PROSITE (http://us.expasy.org/tools/scanprosite/) were used.
Chromosome mapping of the immunophilin and parvulin genes was performed by Chromosome Map Tool (http://www.arabidopsis.org/jsp/ChromosomeMap/tool.jsp) and redrawn by Canvas 5. The protein targeting signal and subcellular location of the immunophilins and parvulins were predicted by several programs available in the World Wide Web-based resources including PSORT (http://psort.ims.u-tokyo.ac.jp/), TargetP (http://www.cbs.dtu.dk/services/TargetP/), SignalP (http://www.cbs.dtu.dk/services/SignalP/), and ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) for cross confirmation. The Mr and PI of the full-length proteins and predicted mature forms were calculated by a program from website (http://www.up.univ-mrs.fr/wabim/d_abim/compo-p.html). The microarray data of the selected gene expression were retrieved from SDM database (http://genome-www5.stanford.edu) and analyzed manually.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.031005.
References
- Abraham RT (1998) Mammalian target of rapamycin: immunosuppressive drugs uncover a novel pathway of cytokine receptor signaling. Curr Opin Immunol 10: 330–336 [DOI] [PubMed] [Google Scholar]
- Aldape RA, Futer O, DeCenzo MT, Jarrett BP, Murcko MA, Livingston DJ (1992) Charged surface residues of FKBP12 participate in formation of the FKBP12-FK506-calcineurin complex. J Biol Chem 267: 16029–16032 [PubMed] [Google Scholar]
- Arpagaus S, Rawyler A, Braendle R (2002) Occurrence and characteristics of the mitochondrial permeability transition in plants. J Biol Chem 277: 1780–1787 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Santos-Rosa MJ, Shirasu K (2001) The U-box protein family in plants. Trends Plant Sci 6: 354–358 [DOI] [PubMed] [Google Scholar]
- Bang H, Pecht A, Raddatz G, Scior T, Solbach W, Brune K, Pahl A (2000) Prolyl isomerases in a minimal cell. Catalysis of protein folding by trigger factor from Mycoplasma genitalium. Eur J Biochem 267: 3270–3280 [DOI] [PubMed] [Google Scholar]
- Barent RL, Nair SC, Carr DC, Ruan Y, Rimerman RA, Fulton J, Zhang Y, Smith DF (1998) Analysis of FKBP51/FKBP52 chimeras and mutants for Hsp90 binding and association with progesterone receptor complexes. Mol Endocrinol 12: 342–354 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Bollman K, Sun H, Poethig RS (2001) Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science 291: 2405–2407 [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res 21: 5803–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blecher O, Erel N, Callebaut I, Aviezer K, Breiman A (1996) A novel plant peptidyl-prolyl-cis-trans-isomerase (PPIase): cDNA cloning, structural analysis, enzymatic activity and expression. Plant Mol Biol 32: 493–504 [DOI] [PubMed] [Google Scholar]
- Bordo D, Bork P (2002) The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Rep 3: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman A, Fawcett TW, Ghirardi ML, Mattoo AK (1992) Plant organelles contain distinct peptidylprolyl cis,trans-isomerases. J Biol Chem 267: 21293–21296 [PubMed] [Google Scholar]
- Brenner BG, Wainberg Z (2001) Heat shock proteins: novel therapeutic tools for HIV-infection? Expert Opin Biol Ther 1: 67–77 [DOI] [PubMed] [Google Scholar]
- Brillantes AB, Ondrias K, Scott A, Kobrinsky E, Ondriasova E, Moschella MC, Jayaraman T, Landers M, Ehrlich BE, Marks AR (1994) Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell 77: 513–523 [DOI] [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL (1994) A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369: 756–758 [DOI] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP (1995) Trigger factor, one of the Escherichia coli chaperone proteins, is an original member of the FKBP family. FEBS Lett 374: 211–215 [DOI] [PubMed] [Google Scholar]
- Callebaut I, Renoir JM, Lebeau MC, Massol N, Burny A, Baulieu EE, Mornon JP (1992) An immunophilin that binds M(r) 90,000 heat shock protein: main structural features of a mammalian p59 protein. Proc Natl Acad Sci USA 89: 6270–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou IT, Gasser CS (1997) Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol Biol 35: 873–892 [DOI] [PubMed] [Google Scholar]
- Clipstone NA, Crabtree GR (1992) Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 357: 695–697 [DOI] [PubMed] [Google Scholar]
- Connern CP, Halestrap AP (1994) Recruitment of mitochondrial cyclophilin to the mitochondrial inner membrane under conditions of oxidative stress that enhance the opening of a calcium-sensitive non-specific channel. Biochem J 302: 321–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton M (1999) The mitochondrial permeability transition pore and its role in cell death. Biochem J 341: 233–249 [PMC free article] [PubMed] [Google Scholar]
- DeCenzo MT, Park ST, Jarrett BP, Aldape RA, Futer O, Murcko MA, Livingston DJ (1996) FK506-binding protein mutational analysis: defining the active-site residue contributions to catalysis and the stability of ligand complexes. Protein Eng 9: 173–180 [DOI] [PubMed] [Google Scholar]
- Deng W, Chen L, Wood DW, Metcalfe T, Liang X, Gordon MP, Comai L, Nester EW (1998) Agrobacterium VirD2 protein interacts with plant host cyclophilins. Proc Natl Acad Sci USA 95: 7040–7045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400: 693–696 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Jones AM, Estelle M (2003) Auxin action in a cell-free system. Curr Biol 13: 1418–1422 [DOI] [PubMed] [Google Scholar]
- Dornan J, Page AP, Taylor P, Wu S, Winter AD, Husi H, Walkinshaw MD (1999) Biochemical and structural characterization of a divergent loop cyclophilin from Caenorhabditis elegans. J Biol Chem 274: 34877–34883 [DOI] [PubMed] [Google Scholar]
- Eisenstark A, Miller C, Jones J, Leven S (1992) Escherichia coli genes involved in cell survival during dormancy: role of oxidative stress. Biochem Biophys Res Commun 188: 1054–1059 [DOI] [PubMed] [Google Scholar]
- Faure JD, Gingerich D, Howell SH (1998) An Arabidopsis immunophilin, AtFKBP12, binds to AtFIP37 (FKBP interacting protein) in an interaction that is disrupted by FK506. Plant J 15: 783–789 [DOI] [PubMed] [Google Scholar]
- Fruman DA, Burakoff SJ, Bierer BE (1994) Immunophilins in protein folding and immunosuppression. FASEB J 8: 391–400 [DOI] [PubMed] [Google Scholar]
- Fulgosi H, Vener AV, Altschmied L, Herrmann RG, Andersson B (1998) A novel multi-functional chloroplast protein: identification of a 40 kDa immunophilin-like protein located in the thylakoid lumen. EMBO J 17: 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser CS, Gunning DA, Budelier KA, Brown SM (1990) Structure and expression of cytosolic cyclophilin/peptidyl-prolyl cis-trans isomerase of higher plants and production of active tomato cyclophilin in Escherichia coli. Proc Natl Acad Sci USA 87: 9519–9523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M, Yanagida M (1991) The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci 16: 173–177 [DOI] [PubMed] [Google Scholar]
- Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld JU, Salchert K, Koncz C, Jurgens G (2000) A conserved domain of the arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 12: 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Mould RM, He Z, Luan S (2002) A chloroplast FKBP interacts with and affects the accumulation of Rieske subunit of cytochrome bf complex. Proc Natl Acad Sci USA 99: 15806–15811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes SD, Shank PR, Bostian KA (1989) Sequence and mutational analysis of ESS1, a gene essential for growth in Saccharomyces cerevisiae. Yeast 5: 55–72 [DOI] [PubMed] [Google Scholar]
- Hani J, Stumpf G, Domdey H (1995) PTF1 encodes an essential protein in Saccharomyces cerevisiae, which shows strong homology with a new putative family of PPIases. FEBS Lett 365: 198–202 [DOI] [PubMed] [Google Scholar]
- Harrar Y, Bellini C, Faure JD (2001) FKBPs: at the crossroads of folding and transduction. Trends Plant Sci 6: 426–431 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Nakayama KI (2003) U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun 302: 635–645 [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909 [DOI] [PubMed] [Google Scholar]
- Hesterkamp T, Bukau B (1996. a) The Escherichia coli trigger factor. FEBS Lett 389: 32–34 [DOI] [PubMed] [Google Scholar]
- Hesterkamp T, Bukau B (1996. b) Identification of the prolyl isomerase domain of Escherichia coli trigger factor. FEBS Lett 385: 67–71 [DOI] [PubMed] [Google Scholar]
- Hesterkamp T, Deuerling E, Bukau B (1997) The amino-terminal 118 amino acids of Escherichia coli trigger factor constitute a domain that is necessary and sufficient for binding to ribosomes. J Biol Chem 272: 21865–21871 [DOI] [PubMed] [Google Scholar]
- Hesterkamp T, Hauser S, Lutcke H, Bukau B (1996. c) Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains. Proc Natl Acad Sci USA 93: 4437–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW (2001) Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J 20: 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson K, Soll D (1999) Mutations in a new Arabidopsis cyclophilin disrupt its interaction with protein phosphatase 2A. Mol Gen Genet 262: 830–838 [DOI] [PubMed] [Google Scholar]
- Kallen J, Spitzfaden C, Zurini MG, Wider G, Widmer H, Wuthrich K, Walkinshaw MD (1991) Structure of human cyclophilin and its binding site for cyclosporin A determined by X-ray crystallography and NMR spectroscopy. Nature 353: 276–279 [DOI] [PubMed] [Google Scholar]
- Kamphausen T, Fanghanel J, Neumann D, Schulz B, Rahfeld JU (2002) Characterization of Arabidopsis thaliana AtFKBP42 that is membrane-bound and interacts with Hsp90. Plant J 32: 263–276 [DOI] [PubMed] [Google Scholar]
- Keith CT, Schreiber SL (1995) PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science 270: 50–51 [DOI] [PubMed] [Google Scholar]
- Krzywicka A, Beisson J, Keller AM, Cohen J, Jerka-Dziadosz M, Klotz C (2001) KIN241: A gene involved in cell morphogenesis in Paramecium tetraurelia reveals a novel protein family of cyclophilin-RNA interacting proteins (CRIPs) conserved from fission yeast to man. Mol Microbiol 42: 257–267 [DOI] [PubMed] [Google Scholar]
- Kunz J, Hall MN (1993) Cyclosporin A, FK506 and rapamycin: more than just immunosuppression. Trends Biochem Sci 18: 334–338 [DOI] [PubMed] [Google Scholar]
- Kurek I, Aviezer K, Erel N, Herman E, Breiman A (1999) The wheat peptidyl prolyl cis-trans-isomerase FKBP77 is heat induced and developmentally regulated. Plant Physiol 119: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrieu I, De Veylder L, Fruchart JS, Odaert B, Casteels P, Portetelle D, Van Montagu M, Inze D, Lippens G (2000) The Arabidopsis thaliana PIN1At gene encodes a single-domain phosphorylation-dependent peptidyl prolyl cis/trans isomerase. J Biol Chem 275: 10577–10581 [DOI] [PubMed] [Google Scholar]
- Lazar SW, Kolter R (1996) SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol 178: 1770–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner V, Chou IT, Scott SV, Ettinger WF, Theg SM, Gasser CS (1994) Cloning and characterization of chloroplast and cytosolic forms of cyclophilin from Arabidopsis thaliana. J Biol Chem 269: 7863–7868 [PubMed] [Google Scholar]
- Liu J, Albers MW, Wandless TJ, Luan S, Alberg DG, Belshaw PJ, Cohen P, MacKintosh C, Klee CB, Schreiber SL (1992) Inhibition of T cell signaling by immunophilin-ligand complexes correlates with loss of calcineurin phosphatase activity. Biochemistry 31: 3896–3901 [DOI] [PubMed] [Google Scholar]
- Liu J, Farmer JD Jr, Lane WS, Friedman J, Weissman I, Schreiber SL (1991) Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66: 807–815 [DOI] [PubMed] [Google Scholar]
- Lu KP, Hanes SD, Hunter T (1996) A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380: 544–547 [DOI] [PubMed] [Google Scholar]
- Lu PJ, Zhou XZ, Shen M, Lu KP (1999) Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283: 1325–1328 [DOI] [PubMed] [Google Scholar]
- Luan S (1998) Immunophilins in animals and higher plants. Bot Bull Acad Sin 39: 217–223 [Google Scholar]
- Luan S, Albers MW, Schreiber SL (1994. a) Light-regulated, tissue-specific immunophilins in a higher plant. Proc Natl Acad Sci USA 91: 984–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Kudla J, Gruissem W, Schreiber SL (1996) Molecular characterization of a FKBP-type immunophilin from higher plants. Proc Natl Acad Sci USA 93: 6964–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Lane WS, Schreiber SL (1994. b) pCyP B: a chloroplast-localized, heat shock-responsive cyclophilin from fava bean. Plant Cell 6: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Li W, Rusnak F, Assmann SM, Schreiber SL (1993) Immunosuppressants implicate protein phosphatase regulation of K+ channels in guard cells. Proc Natl Acad Sci USA 90: 2202–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleszka R, Hanes SD, Hackett RL, de Couet HG, Miklos GL (1996) The Drosophila melanogaster dodo (dod) gene, conserved in humans, is functionally interchangeable with the ESS1 cell division gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 93: 447–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane Y, Sharma S, Petropoulos L, Lin R, Hiscott J (2000) Posttranslational regulation of IRF-4 activity by the immunophilin FKBP52. Immunity 12: 129–140 [DOI] [PubMed] [Google Scholar]
- Marivet J, Frendo P, Burkard G (1995) DNA sequence analysis of a cyclophilin gene from maize: developmental expression and regulation by salicylic acid. Mol Gen Genet 247: 222–228 [DOI] [PubMed] [Google Scholar]
- Matthews JM, Sunde M (2002) Zinc fingers—folds for many occasions. IUBMB Life 54: 351–355 [DOI] [PubMed] [Google Scholar]
- Metzner M, Stoller G, Rucknagel KP, Lu KP, Fischer G, Luckner M, Kullertz G (2001) Functional replacement of the essential ESS1 in yeast by the plant parvulin DlPar13. J Biol Chem 276: 13524–13529 [DOI] [PubMed] [Google Scholar]
- Michnick SW, Rosen MK, Wandless TJ, Karplus M, Schreiber SL (1991) Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin. Science 252: 836–839 [DOI] [PubMed] [Google Scholar]
- Mortillaro MJ, Berezney R (1998) Matrin CYP, an SR-rich cyclophilin that associates with the nuclear matrix and splicing factors. J Biol Chem 273: 8183–8192 [DOI] [PubMed] [Google Scholar]
- Neugebauer KM, Stolk JA, Roth MB (1995) A conserved epitope on a subset of SR proteins defines a larger family of Pre-mRNA splicing factors. J Cell Biol 129: 899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe SJ, Tamura J, Kincaid RL, Tocci MJ, O'Neill EA (1992) FK-506- and CsA-sensitive activation of the interleukin-2 promoter by calcineurin. Nature 357: 692–694 [DOI] [PubMed] [Google Scholar]
- Owens-Grillo JK, Stancato LF, Hoffmann K, Pratt WB, Krishna P (1996) Binding of immunophilins to the 90 kDa heat shock protein (hsp90) via a tetratricopeptide repeat domain is a conserved protein interaction in plants. Biochemistry 35: 15249–15255 [DOI] [PubMed] [Google Scholar]
- Partaledis JA, Berlin V (1993) The FKB2 gene of Saccharomyces cerevisiae, encoding the immunosuppressant-binding protein FKBP-13, is regulated in response to accumulation of unfolded proteins in the endoplasmic reticulum. Proc Natl Acad Sci USA 90: 5450–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson CE, Schaub T, Coleman EJ, Davis EC (2000) Developmental regulation of FKBP65. An ER-localized extracellular matrix binding-protein. Mol Biol Cell 11: 3925–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Soderberg L, Roepstorff P, von Heijne G, et al. (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14: 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugl GM, Kallen J, Jansonius JN, Walkinshaw MD (1994) The molecular replacement solution and X-ray refinement to 2.8 A of a decameric complex of human cyclophilin A with the immunosuppressive drug cyclosporin A. J Mol Biol 244: 385–409 [DOI] [PubMed] [Google Scholar]
- Pratt WB (1998) The hsp90-based chaperone system: involvement in signal transduction from a variety of hormone and growth factor receptors. Proc Soc Exp Biol Med 217: 420–434 [DOI] [PubMed] [Google Scholar]
- Pratt WB, Krishna P, Olsen LJ (2001) Hsp90-binding immunophilins in plants: the protein movers. Trends Plant Sci 6: 54–58 [DOI] [PubMed] [Google Scholar]
- Rahfeld JU, Rucknagel KP, Schelbert B, Ludwig B, Hacker J, Mann K, Fischer G (1994) Confirmation of the existence of a third family among peptidyl-prolyl cis/trans isomerases. Amino acid sequence and recombinant production of parvulin. FEBS Lett 352: 180–184 [DOI] [PubMed] [Google Scholar]
- Ranganathan R, Lu KP, Hunter T, Noel JP (1997) Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell 89: 875–886 [DOI] [PubMed] [Google Scholar]
- Rao A, Luo C, Hogan PG (1997) Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol 15: 707–747 [DOI] [PubMed] [Google Scholar]
- Ratajczak T, Carrello A (1996) Cyclophilin 40 (CyP-40), mapping of its hsp90 binding domain and evidence that FKBP52 competes with CyP-40 for hsp90 binding. J Biol Chem 271: 2961–2965 [DOI] [PubMed] [Google Scholar]
- Reimer T, Weiwad M, Schierhorn A, Ruecknagel PK, Rahfeld JU, Bayer P, Fischer G (2003) Phosphorylation of the N-terminal domain regulates subcellular localization and DNA binding properties of the peptidyl-prolyl cis/trans isomerase hPar14. J Mol Biol 330: 955–966 [DOI] [PubMed] [Google Scholar]
- Rouviere PE, Gross CA (1996) SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev 10: 3170–3182 [DOI] [PubMed] [Google Scholar]
- Rulten S, Thorpe J, Kay J (1999) Identification of eukaryotic parvulin homologues: a new subfamily of peptidylprolyl cis-trans isomerases. Biochem Biophys Res Commun 259: 557–562 [DOI] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH (1994) RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78: 35–43 [DOI] [PubMed] [Google Scholar]
- Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT (1995) Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem 270: 815–822 [DOI] [PubMed] [Google Scholar]
- Saito T, Niwa Y, Ashida H, Tanaka K, Kawamukai M, Matsuda H, Nakagawa T (1999) Expression of a gene for cyclophilin which contains an amino-terminal endoplasmic reticulum-targeting signal. Plant Cell Physiol 40: 77–87 [DOI] [PubMed] [Google Scholar]
- Schreiber SL (1991) Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 251: 283–287 [DOI] [PubMed] [Google Scholar]
- Schubert M, Petersson UA, Haas BJ, Funk C, Schroder WP, Kieselbach T (2002) Proteome map of the chloroplast lumen of Arabidopsis thaliana. J Biol Chem 277: 8354–8365 [DOI] [PubMed] [Google Scholar]
- Shirane M, Nakayama KI (2003) Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat Cell Biol 5: 28–37 [DOI] [PubMed] [Google Scholar]
- Shou W, Aghdasi B, Armstrong DL, Guo Q, Bao S, Charng MJ, Mathews LM, Schneider MD, Hamilton SL, Matzuk MM (1998) Cardiac defects and altered ryanodine receptor function in mice lacking FKBP12. Nature 391: 489–492 [DOI] [PubMed] [Google Scholar]
- Silverstein AM, Galigniana MD, Chen MS, Owens-Grillo JK, Chinkers M, Pratt WB (1997) Protein phosphatase 5 is a major component of glucocorticoid receptor.hsp90 complexes with properties of an FK506-binding immunophilin. J Biol Chem 272: 16224–16230 [DOI] [PubMed] [Google Scholar]
- Smith DF, Baggenstoss BA, Marion TN, Rimerman RA (1993) Two FKBP-related proteins are associated with progesterone receptor complexes. J Biol Chem 268: 18365–18371 [PubMed] [Google Scholar]
- Stamnes MA, Shieh BH, Chuman L, Harris GL, Zuker CS (1991) The cyclophilin homolog ninaA is a tissue-specific integral membrane protein required for the proper synthesis of a subset of Drosophila rhodopsins. Cell 65: 219–227 [DOI] [PubMed] [Google Scholar]
- Stone SL, Arnoldo M, Goring DR (1999) A breakdown of Brassicaself-incompatibility in ARC1 antisense transgenic plants. Science 286: 1729–1731 [DOI] [PubMed] [Google Scholar]
- Sykes K, Gething MJ, Sambrook J (1993) Proline isomerases function during heat shock. Proc Natl Acad Sci USA 90: 5853–5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU (1999) Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97: 755–765 [DOI] [PubMed] [Google Scholar]
- Uchida T, Fujimori F, Tradler T, Fischer G, Rahfeld JU (1999) Identification and characterization of a 14 kDa human protein as a novel parvulin-like peptidyl prolyl cis/trans isomerase. FEBS Lett 446: 278–282 [DOI] [PubMed] [Google Scholar]
- Valent QA, Kendall DA, High S, Kusters R, Oudega B, Luirink J (1995) Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J 14: 5494–5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J (1991) Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science 252: 839–842 [DOI] [PubMed] [Google Scholar]
- Vener AV, Rokka A, Fulgosi H, Andersson B, Herrmann RG (1999) A cyclophilin-regulated PP2A-like protein phosphatase in thylakoid membranes of plant chloroplasts. Biochemistry 38: 14955–14965 [DOI] [PubMed] [Google Scholar]
- Vittorioso P, Cowling R, Faure JD, Caboche M, Bellini C (1998) Mutation in the Arabidopsis PASTICCINO1 gene, which encodes a new FK506-binding protein-like protein, has a dramatic effect on plant development. Mol Cell Biol 18: 3034–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucich VA, Gasser CS (1996) Novel structure of a high molecular weight FK506 binding protein from Arabidopsis thaliana. Mol Gen Genet 252: 510–517 [DOI] [PubMed] [Google Scholar]
- Xu Q, Liang S, Kudla J, Luan S (1998) Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. Plant J 15: 511–519 [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, et al. (1997) Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278: 1957–1960 [DOI] [PubMed] [Google Scholar]
- Yao JL, Kops O, Lu PJ, Lu KP (2001) Functional conservation of phosphorylation-specific prolyl isomerases in plants. J Biol Chem 276: 13517–13523 [DOI] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB (1992) SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev 6: 837–847 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J (2003) SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301: 1107–1110 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J, Grozinger CM, Chao ED, Moazed D (2003) SIR1, an upstream component in auxin signaling identified by chemical genetics. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. Science 301: 1107–1110 [DOI] [PubMed] [Google Scholar]
- Zydowsky LD, Etzkorn FA, Chang HY, Ferguson SB, Stolz LA, Ho SI, Walsh CT (1992) Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci 1: 1092–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]