Abstract
With advent of several treatment options in multiple myeloma, a selection of effective regimen has become an important issue. Use of GEP is considered an important tool in predicting outcome; however, it is unclear whether such genomic analysis alone can adequately predict therapeutic response. We evaluated ability of GEP to predict complete response in MM. GEP from pre-treatment MM cells from 136 uniformly treated MM patients with response data on an IFM, France led study were analyzed. To evaluate variability in predictive power due to microarray platform or treatment types, additional datasets from three different studies (n= 511) were analyzed using same methods. We used several machine learning methods to derive a prediction model using training and test subsets of the original four datasets. Among all methods employed for GEP-based CR predictive capability, we got accuracy range of 56% to 78% in test datasets and no significant difference with regard to GEP platforms, treatment regimens or in newly-diagnosed or relapsed patients. Importantly, permuted p-value showed no statistically significant CR predictive information in GEP data. This analysis suggests that GEP-based signature has limited power to predict CR in MM, highlighting the need to develop comprehensive predictive model using integrated genomics approach.
Keywords: multiple myeloma, gene expression signature, response prediction
Introduction
With the introduction of novel agents, there has been significant improvement in outcome in multiple myeloma (MM).1–5 Although median survival has improved from 3 years to over 7 years, the clinical course is highly variable and unpredictable. Initially high-dose therapy6, 7 and subsequently novel agents8–11,12–14 have contributed to improvement in both response and survival15; however, MM remains uniformly fatal.3, 4, 14, 16, 17 Each of these treatments individually and in combination is effective in only a portion of patients, and currently predictability of response to therapy remains unreliable. A number of disease, host and therapy-specific features including age, performance status, tumor burden, tumor proliferative index, serum β-2 microglobulin, albumin, creatinine, LDH, International Staging System (ISS), and cytogenetic abnormalities have been reported to provide prognostic information.16 Although this and other systems help with prognosis for survival, they do not reliably predict probability of response and as of now, there is no pre-treatment response predictive biomarker to tailor patient-specific therapy.18
Moreover, with each new therapy attempted, patients often experience complications and toxicities requiring specialized intervention. With the improved understanding of oncogenomics in MM, a number of targeted agents have been investigated19, 20; three such novel agents, thalidomide, bortezomib and lenalidomide have already received FDA approval for use in MM, making selection of effective agents more difficult as well as economically taxing. As some of these novel agents were identified based on molecular identification of targets and the combinations were formulated based upon perturbation of expression profile by these agents, it is anticipated that integrative genomics will help provide more precise prognostic and predictive tools.
Gene expression profiling (GEP) is being widely used for tumor classification21–25 and survival risk prediction.26–31 However, it remains unclear whether the strategies used to define such prognostic genomic classifiers can be used to develop classifiers that predict response to specific therapy.32, 33 Efforts assessing potential use of GEP-based signatures in therapeutic decision making in MM have been limited. Mulligan et al. have studied pharmacogenomics in 169 relapsed multiple myeloma patients samples receiving bortezomib32 and another study by Kumar et al34 have attempted response prediction to thalidomide - dexamethasone combination using GEP based signature in newly-diagnosed patients. However, results from these studies are limited either by small sample size34 or response signature derived from GEP of relapsed and not from newly diagnosed patients with MM.32 Here, we have analyzed GEP data from one large study of 136 newly diagnosed patients with MM through a rigorous statistical approach, and have independently investigated 3 additional studies of total 349 newly diagnosed, and 162 relapsed cases to determine ability and significance of gene expression signature in predicting complete response (CR) in patients with MM.
Patients and Methods
Datasets for Response Prediction
Gene expression profiles (n=647) from four independent clinical trials were analyzed to predict treatment response. Details about these studies are listed in Table 1. HOVON35 and Mulligan et al.32 datasets are publically available, while IFM datasets are now available at NCBI GEO website with accession ID, GSE39754 (IFM I) and GSE55145(IFM II). Patients were newly diagnosed and had not received any prior therapy except those in APEX/SUMMIT trials from Mulligan et al datasets32 who were relapsed/refractory and had received 1–3 prior therapies. In two of four datasets [IFM I and HOVON], patients received three-drug regimen (VAD/PAD) followed by autologous stem cell transplantation (ASCT), and patients in the other two datasets received bortezomib with dexamethsone [IFM II and Mulligan et al.] prior to response evaluation using uniform European Group for Bone Marrow Transplantation criteria (EBMT).36
Table 1.
Characteristics and details of studies used for gene expression profile-based response prediction.
| IFM I | IFM II | HOVON | Mulligan et al. | |
|---|---|---|---|---|
| Study | IFM 2005# | IFM 2005# | HOVON 65 MM / GMMG HD4$ |
APEX / SUMMIT |
| Number of Samples | 136 | 67 | 282 | 162 |
| Platform | Affymetrix Exon 1.0 ST array | Affymetrix Exon 1.0 ST array | Affymetrix U133 Plus 2.0 array | Affymetrix U133 Plus 2.0 array |
| Patient Population | Newly-diagnosed | Newly-diagnosed | Newly-diagnosed | Relapsed |
| Treatment Protocol | VAD, ASCT | Bortezomib, ASCT | VAD/PAD, ASCT | Bortezomib |
| Time of Response Measurement | Post-transplant | Post-Induction | Post-transplant | Post Salvage therapy |
| Complete response | 44 (32%) | 24 (36 % %) | 76 (27 %) | 73 (43%)∞ |
Data Preprocessing
GEP from samples within all four datasets was obtained on Affymetrix microarray platform. The IFM I and II datasets were obtained using Affymetrix Exon 1.0 ST gene array, while HOVON and Mulligan et al datasets were obtained using HG_U133_Plus_2 gene array and gene-level signal intensities were used for subsequent analysis. Pre-processing and normalization of datasets was carried out using dChip and R package - aroma.affymetrix.
Class Prediction Methods
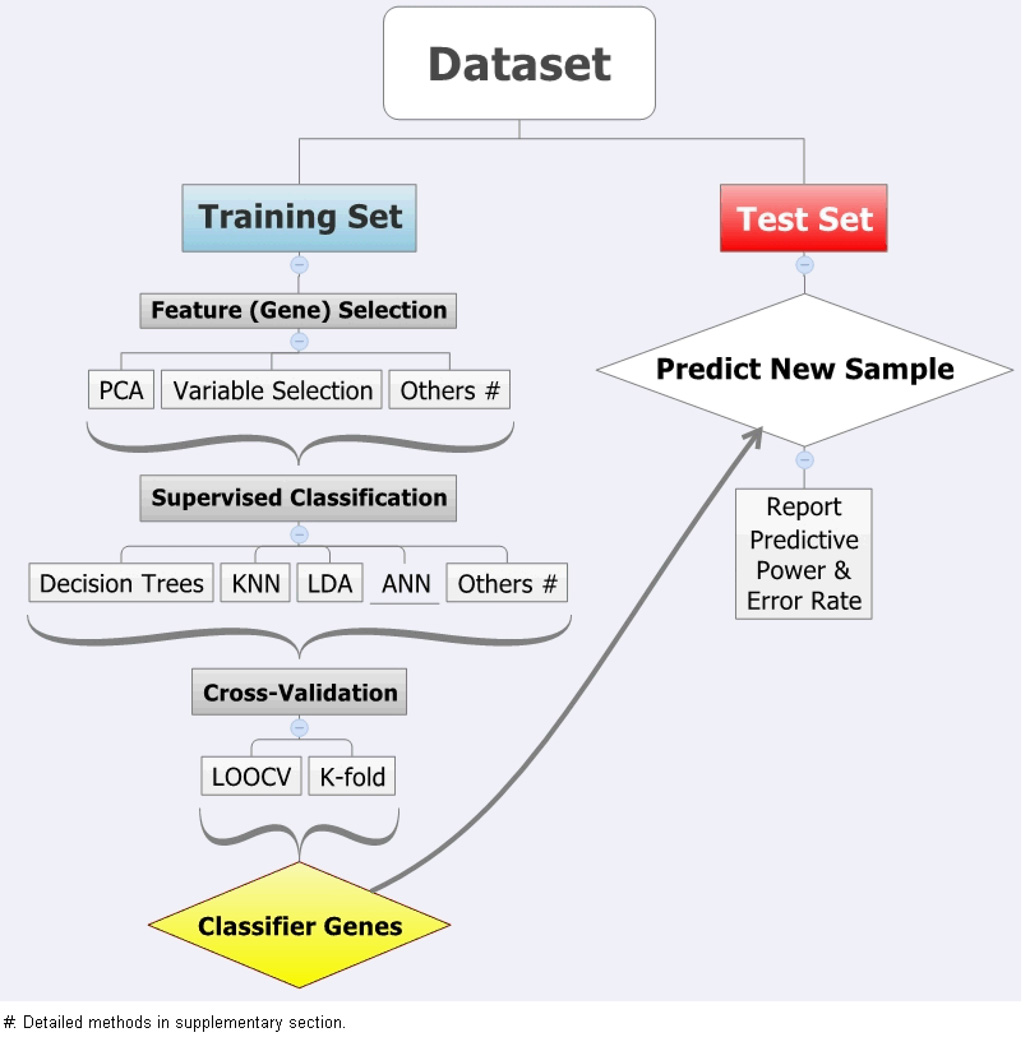
Class prediction analysis was carried out in each of four datasets using a series of steps, as summarized in Figure 1. Specifically, each dataset was split into training and test datasets, with 2/3 samples in training dataset. Various feature selection approaches were used to eliminate redundant and non-varying gene probe sets, including gene filtering by variance, variable selection using LASSO and Ridge regression, among others. [Suppl. Table 1s] Patients having CR were labeled as positive class for further analysis. Then, classifier models were built and trained within a training set based on gene expression pattern of selected genes (features). Class prediction methods from six major groups of machine learning techniques were used in building a classifier model, including decision tree, support vector machines (SVM), prediction analysis of microarray (PAM), K-nearest neighbors (KNN), Bayesian additive regression trees (BART), and artificial neural networks (ANN). [Suppl. Table 1s] Several classifier models were built using different feature sets and tuning of method-specific criteria to minimize misclassification error. Also, response labels were randomly permuted for 1000 times to build a robust model. A final classifier model was obtained following leave-one-out cross-validation (LOOCV) and/or K-fold cross-validation. The entire model-building process, including gene selection step, was repeated during each cross-validation cycle. Finally, the best-performing cross-validated model was applied to the test dataset for an unbiased assessment of response prediction, and model performance was evaluated by measuring model-specific sensitivity, specificity, positive and negative predictive value (PPV and NPV), and overall model accuracy. Additionally, receiving-operating characteristic (ROC) curves were drawn and compared among different class prediction methods. Machine learning packages in R language and BRB-ArrayTools developed by Dr. Richard Simon and BRB-ArrayTools Development Team were used for the entire analysis.37 Specific details for each of classification methods were previously described.38
Figure 1.
Flow-chart showing major steps to develop response (CR) prediction model.
Results
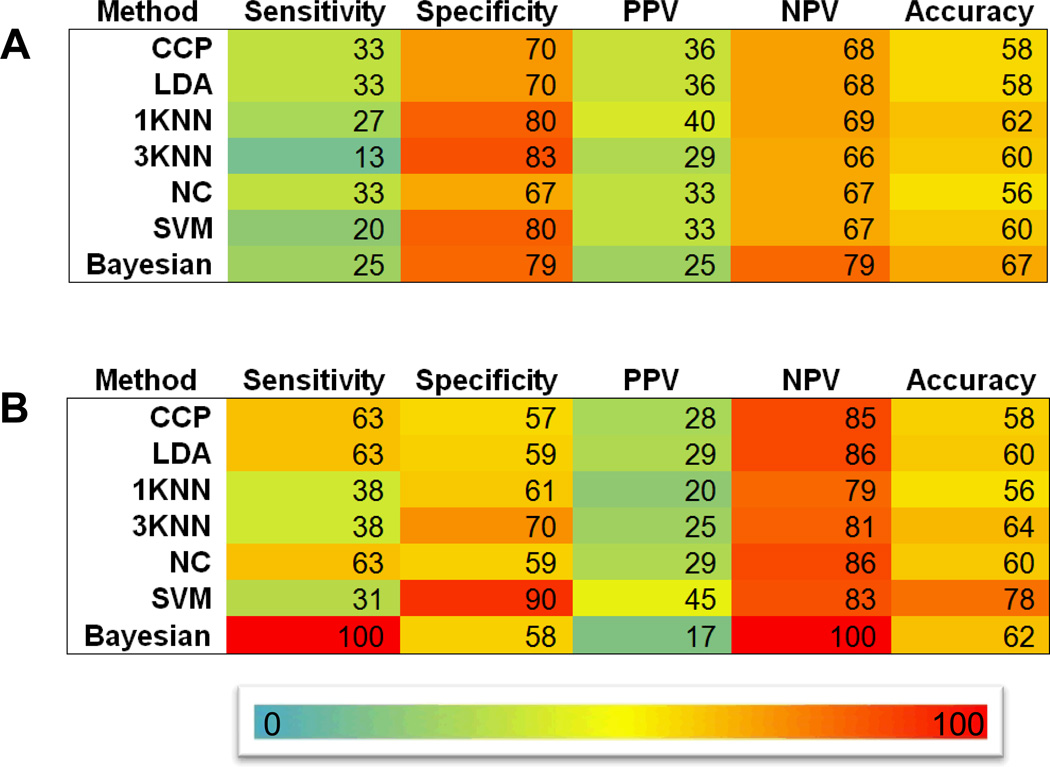
We analyzed the ability of gene expression profile to predict response to therapy in multiple myeloma. We focused on the ability to predict complete response (CR), as the majority of patients (>90%) receiving newer therapeutic modalities and/or high-dose therapy and transplant (HDT) achieve at least a partial response (PR). Moreover, attaining CR is critical to achieving a curative outcome. With this consideration in mind, we first utilized the gene expression signature as a tool to predict CR. Our primary dataset (IFM I) included 136 patients with MM receiving standard dose induction followed by HDT. Forty-four (32%) patients achieved CR in this group. Using random 2:1 sampling division, the training set of 91 patients included 30 achieving CR and the test set of the remaining 45 patients included 14 attaining CR. We applied SVM & KNN class prediction approach to the training set, as detailed in Methods and Figure 1, and observed an overall CR prediction accuracy of 60% with 24 % sensitivity, 77% specificity, 33% PPV, and 69% NPV. Applying the trained model to the test dataset gave accuracy of 62% with 27% sensitivity, 80% specificity, 40% PPV, and 69% NPV. To improve upon these results and to overcome any technical and/or statistical issues that may affect predictability, we reanalyzed the data using a number of different supervised machine learning methods. As seen in Figure 2, none of the analytical methods used significantly improved overall predictive accuracy, which remained between 56 – 78 % in both training and test sets.
Figure 2. Summary of CR prediction performance.
Performance of various class prediction methods in predicting CR in the validation dataset using IFM I dataset (a) and HOVON dataset (b). CCP: Compound covariate predictor; LDA: Linear discriminant analysis; KNN: K-nearest neighbor; NC: Nearest centroid; SVM: Support vector machine.
Different microarray platforms do not improve CR prediction accuracy
To evaluate any confounding effect of microarray platforms used on response predictability, we analyzed an independent dataset generated using the Affymetrix HG_U133_Plus_2 platform from 282 patients (HOVON trial) treated with a standard-dose induction regimen followed by HDT. Actual CR rate was 27% (76/282) in this trial. All patients within IFM I and HOVON datasets received uniform treatment, but their MM cell expression profile was performed using different microarray platforms, namely, Affymetrix Exon 1.0 ST and Affymetrix HG_U133_Plus_2 arrays, respectively. We used an identical set of prediction methods as was used for IFM I dataset described above. As noted in table 2, the maximal achievable accuracy was 78% in the HOVON test set.
Table 2. Summary of CR Prediction Analysis.
Maximum achievable performance (MAP) in both training and validation sets for all four datasets using seven different class prediction methods. MAP is defined as highest accuracy achieved by one of seven classifiers methods used for a given dataset.
| a: Class Prediction Results – Maximum Achievable Accuracy – Training Set | ||||||
|---|---|---|---|---|---|---|
| Prediction Method |
Sensitivity | Specificity | PPV | NPV | Accuracy | |
| IFM I | KNN | 24 | 77 | 33 | 69 | 60 |
| IFM II | KNN | 72 | 56 | 65 | 64 | 65 |
| HOVON | SVM | 17 | 92 | 48 | 71 | 70 |
| Mulligan et al. | KNN | 71 | 67 | 68 | 71 | 69 |
| b: Class Prediction Results – Maximum Achievable Accuracy – Test Set | ||||||
|---|---|---|---|---|---|---|
| Prediction Method |
Sensitivity | Specificity | PPV | NPV | Accuracy | |
| IFM I | KNN | 27 | 80 | 40 | 69 | 62 |
| IFM II | KNN | 67 | 56 | 25 | 88 | 58 |
| HOVON | SVM | 31 | 90 | 45 | 83 | 78 |
| Mulligan et al. | KNN | 50 | 36 | 48 | 39 | 44 |
SVM: Support Vector Machines; KNN: K-nearest neighbors (n=1)
CR prediction accuracy does not improve even if patient selection includes extremes of responses
Next, we evaluated whether gene expression signature using patients achieving CR/nCR versus those with no response (NR) or progressive disease(PD) could improve predictive power. In this case, no patients with PR or MR were included in the analysis. The IFM II dataset had 67 patients, 34 with CR/nCR and 33 with NR/PD. Using K-nearest neighbor (K=1) method, we reached maximum sensitivity of 72% with overall accuracy of 65% in the training data set. However, the same 38-gene signature failed to enrich CR prediction in the test dataset (see below), with reduced sensitivity and accuracy of 67% and 58%, respectively.
To evaluate if early-onset CR has better predictability, we performed CR modeling based on time-points when CR was achieved. Specifically, HOVON study determined CR status at the end of 3rd and 8th cycle of induction therapy (early-onset CR) and at the end of protocol (CR20). Also, we evaluated whether CR can be better predicted in those who have sustained CR, This group was classified under sustained CR group. Using similar analysis as above we did not observe significant improvement in CR prediction in these newly regrouped subsets. [Suppl. Table 2s]. Furthermore, we assessed performance of CR prediction separately in high and low GEP risk groups as defined by proliferation index (PI) and cytogenetic abnormalities.39–42 In these groups also, our results failed to show significant improvement in CR prediction. [Suppl. File 4 - Appendix]
Finally, we evaluated whether predictive accuracy changes if patients received therapy in the relapsed setting. We analyzed the Mulligan et al dataset using similar methods as above, except that we used PR as a response endpoint since not many CRs were achieved in this relapsed patient population. We achieved an accuracy of 44% in test set. Using all the additional methods described above, we did not improve upon these results.
Permutation to assess the prediction power
Finally, we compared the actual CR achieved by the patients (real CR) versus the enriched CR or positive predictive value from the classifier model giving maximum accuracy in our test set. As seen in table 3, we do not observe significant enrichment of CR compared to actual CR rate. Moreover, we performed a response permutation by randomly assigning the response labels of patients and analyzing the ability to predict. We performed 1,000 such permutations to predict CR. The permuted p-value is the proportion of permutations that give predictive ability higher than the one obtained using the actual response labels. As seen in table 3, none of the data sets have permuted p-value of < 0.05, suggesting that the data from gene expression profile is not adequately informative to predict CR outcome.
Table 3. Permuting class labels to assess the power of predicting CR.
During the training of classifiers, the treatment response labels were permuted 1000 times to measure the statistical significance of prediction performance. P-values indicate the proportion of total 1000 permutations giving better prediction performance than the original analysis using real treatment response labels. Hence, higher p-value suggests lower confidence in prediction analysis results using gene expression data.
| Real CR % | Predicted CR: Positive Predictive Value % (p-value – 1000 permutations) |
|
|---|---|---|
| IFM I | 32 (44/136) | 25 (0.36) |
| IFM II | 36 (24/67) | 27 (0.11) |
| HOVON | 27 (76/282) | 45 (0.13) |
| Mulligan et al. | 43 (73/162) | 43 (0.49) |
Predicted CR or Positive Predictive Value is derived from the test or validation dataset analysis using criteria giving the best possible overall model accuracy.
Discussion
In this study, we show that the ability of gene expression profiling (GEP) to predict CR in patients with MM is very limited. We have used uniformly treated patient population, and treatment responses were uniformly measured across all four studies using EBMT Blade Criteria.36 In our primary dataset, newly-diagnosed patients with MM in IFM I, we found the best accuracy of predicting CR at less than 67% in the test dataset. To confirm our initial observation, we have analyzed 3 different datasets using 2 different microarray platforms, as well as different treatment protocols. Among them, the Mulligan et al. study involved patients with relapsed MM who were refractory to 1–3 previous treatments. We used a set of common feature selection and supervised machine learning methods to build a robust response prediction signature in training set for each study, and evaluated the performance in a test dataset from the same study. In this thorough analysis, we have performed class prediction analysis within each of the four studies to define independent classifier gene signatures to ensure the best predictability within each dataset, and to avoid batch effects when merging different datasets. Despite these efforts, as seen in Figure 2 and Table 2, our response predictability remains low in all the analyses.
To uncover potential information that may reside in the expression data that may allow response predication, we performed permuted prediction analysis. In this approach, we randomly shuffled patients’ response labels and analyzed the ability to predict CR. If the data has some predictive power, then the prediction performance achieved with such random assignment should have significantly lower ability to predict CR than the performance achieved with the real response labels. Following 1,000 such permutations, as seen in Table 3, we observed predictive p-value to be greater than 0.05 in all the four data sets. This indicated limited inherent information in the expression profiles to predict CR.
Our study clearly shows that GEP alone has inherent limitations in CR-prediction in MM. Despite using six major classification methods with various feature selection procedures in building more than 240 models [Figure 2 and Suppl. Figure 2s], the overall prediction accuracy remained low. Furthermore, our analysis reveals inadequate response prediction using GEP regardless of the treatment type used, i.e., patients in IFM I and HOVON trials received VAD followed by ASCT therapy, while patients in IFM II and Mulligan et al. studies received bortezomib therapy. It is possible that GEP of pre and post-treatment may help in feature selection by identifying treatment-specific genes and thereby improving prediction power. However, so far such datasets are not available in public domain. Also, we do not observe significant improvement in CR prediction by stratifying data based on time of achieving CR [Suppl. Table 2s] and GEP based risk-status [Suppl. File 4 - Appenidx].
Among the number of classification methods and data sets used, the highest sensitivity was 67% in IFM II dataset, representing accurate prediction of CR amongst those achieving real CR. False negative rate (1-specificity) remained between 36% – 80%, which is the percentage of patients misclassified as non-CR who might have responded to the treatment. Finally, the sensitivity and overall accuracy dropped significantly below 60% when classifier gene signatures from one dataset was used to predict CR in a different dataset. Among various methods from six major classification groups we used, SVM and KNN performed better than the other methods. [Figure 2]
Based on our analysis, we recognize that the expression profile alone has limited ability in predicting treatment response, especially when actual response rate is high, i.e., prediction using expression profile fails to significantly enrich actual response patients using the data available from the published clinical study. [Table 3] Although we do report sensitivity of more than 70% in some prediction models (IFM II and Mulligan et al.), these values are associated with poor accuracy (65–69%); and importantly, the signature trained from the training dataset performs poorly in the validation dataset of the same study. Previous efforts at GEP-based response prediction in MM have met with very limited success. Mulligan et al.32 showed overall 71% accuracy in predicting bortezomib response using 169 patients with relapsed MM. A report from Mayo clinic researchers showed inadequate prediction power in a small number of patients receiving thalidomide-dexamethasone combination therapy34. Such limited ability of expression profiling to predict response has also been described in breast cancer, leukemia, and other cancer studies.33, 43, 44 For example, two of seven distant recurrence risk-prediction signatures in breast cancer have recently been commercialized, with test accuracy of more than 80%; however, these signatures are prognostic rather than true predictors of response. Also, risk prediction is restricted to particular subtypes of breast cancer (ER +, Node −). Similarly, a number of risk signatures have been described in myeloma, but they are developed for event-free or overall survival, not correlated with predicting response to initial therapy.27, 45, 46 In the era of personalized medicine, a response predictive signature will be required to select correct combinations of agents that provide best response with reduced toxicity.
What are the possible biological reasons for not being able to predict response using a large number of gene expression datasets? This analysis highlights the limitation of gene expression by itself to influence the eventual cellular behavior. The transcriptome and proteome modifiers play a significant role in cell growth and survival, especially following therapy.47, 48 It is well described that alternate splicing and miRNA affect the eventual translation of the expressed gene, and post-translational modifications further control cellular effects of the transcribed proteins.49–51 Variability in single nucleotide polymorphism also affects response to therapy.52, 53 Finally, intra-tumoral and inter-tumoral genetic heterogeneity54, 55 can significantly influence both, individualized response to therapy as well as a major analytical issue for development of GEP-based response signatures.
While the strength of our analysis was in having larger patient cohorts from comparable datasets and use of rigorous and comprehensive statistical classification methods, we acknowledge potential limitations in response prediction imposed by two fundamental statistical challenges in high-dimensional data analysis, e.g., curse of dimensionality (large p for genes) and curse of sparsity (small n for sample size).44, 56, 57 Nonetheless, we addressed these limits to the best of current knowledge and yet fail to significantly improve response prediction. Also, during the time of this analysis, we have not been able to assess some of GEP datasets with survival information but without response information, which is required for our analysis.
In conclusion, we report that GEP based signature have very limited ability to predict CR in MM which is in accordance with recent consensus report on risk stratification.40 This analysis highlights the need to develop more comprehensive signatures that can incorporate various genomic elements in final predictive models. Failure in predicting response as an end-point (binary) also warrants need to concurrently measure durability of response (continuous variable), which may improve prediction performance similar to available survival and prognostic GEP based signatures. Finally, integration of next generation sequencing data with available gene expression and miRNA profiling information, along with GEP, may allow development of better predictive models for potential application in clinics.
Supplementary Material
Acknowledgment
This work was supported in part by grants from the Dept. of Veterans Affairs Merit Review Awards I01-BX001584 and from the National Institutes of Health Grants RO1-124929 to NCM., RO1-050947 to KCA, and P50-100007 and PO1-78378 to NCM and KCA, PO1-155258 to NCM, KCA, HAL, SM, CL and PM and R01GM077122 to CL. KCA is an American Cancer Society Clinical Research Professor.
Footnotes
There are not any competing financial interests in relation to the work described.
Supplementary information is available at Leukemia's website.
References
- 1.Laubach J, Richardson P, Anderson K. Multiple myeloma. Annu Rev Med. 2011 Feb 18;62:249–264. doi: 10.1146/annurev-med-070209-175325. [DOI] [PubMed] [Google Scholar]
- 2.DeVita VT, Hellman S, Rosenberg SA. Cancer, principles & practice of oncology. 7th edn. Philadelphia, PA: Lippincott Williams & Wilkins; 2005. p. lxxv.p. 2898. [Google Scholar]
- 3.Kyle RA, Rajkumar SV. An overview of the progress in the treatment of multiple myeloma. Expert review of hematology. 2014 Feb;7(1):5–7. doi: 10.1586/17474086.2014.870030. [DOI] [PubMed] [Google Scholar]
- 4.Anderson KC. Therapeutic advances in relapsed or refractory multiple myeloma. Journal of the National Comprehensive Cancer Network : JNCCN. 2013 May;11(5 Suppl):676–679. doi: 10.6004/jnccn.2013.0199. [DOI] [PubMed] [Google Scholar]
- 5.Ludwig H, Miguel JS, Dimopoulos MA, Palumbo A, Garcia Sanz R, Powles R, et al. International Myeloma Working Group recommendations for global myeloma care. Leukemia. 2013 Oct 9; doi: 10.1038/leu.2013.293. [DOI] [PubMed] [Google Scholar]
- 6.Barlogie B, Shaughnessy J, Tricot G, Jacobson J, Zangari M, Anaissie E, et al. Treatment of multiple myeloma. Blood. 2004 Jan 1;103(1):20–32. doi: 10.1182/blood-2003-04-1045. [DOI] [PubMed] [Google Scholar]
- 7.Rajkumar SV., IV Initial treatment of multiple myeloma. Hematological oncology. 2013 Jun;31(Suppl 1):33–37. doi: 10.1002/hon.2064. [DOI] [PubMed] [Google Scholar]
- 8.Berenson JR, Yang HH, Vescio RA, Nassir Y, Mapes R, Lee SP, et al. Safety and efficacy of bortezomib and melphalan combination in patients with relapsed or refractory multiple myeloma: updated results of a phase 1/2 study after longer follow-up. Ann Hematol. 2008 Aug;87(8):623–631. doi: 10.1007/s00277-008-0501-0. [DOI] [PubMed] [Google Scholar]
- 9.Gay F, Hayman SR, Lacy MQ, Buadi F, Gertz MA, Kumar S, et al. Lenalidomide plus dexamethasone versus thalidomide plus dexamethasone in newly diagnosed multiple myeloma: a comparative analysis of 411 patients. Blood. 2010 Feb 18;115(7):1343–1350. doi: 10.1182/blood-2009-08-239046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jagannath S, Barlogie B, Berenson JR, Siegel DS, Irwin D, Richardson PG, et al. Updated survival analyses after prolonged follow-up of the phase 2, multicenter CREST study of bortezomib in relapsed or refractory multiple myeloma. Br J Haematol. 2008 Nov;143(4):537–540. doi: 10.1111/j.1365-2141.2008.07359.x. [DOI] [PubMed] [Google Scholar]
- 11.Munshi NC, Anderson KC. New strategies in the treatment of multiple myeloma. Clin Cancer Res. 2013 Jul 1;19(13):3337–3344. doi: 10.1158/1078-0432.CCR-12-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarthy PL, Owzar K, Hofmeister CC, Hurd DD, Hassoun H, Richardson PG, et al. Lenalidomide after stem-cell transplantation for multiple myeloma. N Engl J Med. 2012 May 10;366(19):1770–1781. doi: 10.1056/NEJMoa1114083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attal M, Lauwers-Cances V, Marit G, Caillot D, Moreau P, Facon T, et al. Lenalidomide maintenance after stem-cell transplantation for multiple myeloma. N Engl J Med. 2012 May 10;366(19):1782–1791. doi: 10.1056/NEJMoa1114138. [DOI] [PubMed] [Google Scholar]
- 14.Mateos MV, Hernandez MT, Giraldo P, de la Rubia J, de Arriba F, Lopez Corral L, et al. Lenalidomide plus dexamethasone for high-risk smoldering multiple myeloma. N Engl J Med. 2013 Aug 1;369(5):438–447. doi: 10.1056/NEJMoa1300439. [DOI] [PubMed] [Google Scholar]
- 15.Barlogie B, Kyle RA, Anderson KC, Greipp PR, Lazarus HM, Hurd DD, et al. Standard chemotherapy compared with high-dose chemoradiotherapy for multiple myeloma: final results of phase III US Intergroup Trial S9321. J Clin Oncol. 2006 Feb 20;24(6):929–936. doi: 10.1200/JCO.2005.04.5807. [DOI] [PubMed] [Google Scholar]
- 16.Kyle RA, Rajkumar SV. Treatment of multiple myeloma: a comprehensive review. Clin Lymphoma Myeloma. 2009 Aug;9(4):278–288. doi: 10.3816/CLM.2009.n.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar SK, Rajkumar SV. The current status of minimal residual disease assessment in myeloma. Leukemia. 2014 Feb;28(2):239–240. doi: 10.1038/leu.2013.306. [DOI] [PubMed] [Google Scholar]
- 18.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005 Sep 10;23(26):6333–6338. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 19.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007 Aug;7(8):585–598. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 20.Ocio EM, Richardson PG, Rajkumar SV, Palumbo A, Mateos MV, Orlowski R, et al. New drugs and novel mechanisms of action in multiple myeloma in 2013: a report from the International Myeloma Working Group (IMWG) Leukemia. 2013 Nov 20; doi: 10.1038/leu.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghadimi BM, Grade M. Cancer gene profiling for response prediction. Methods Mol Biol. 2010;576:327–339. doi: 10.1007/978-1-59745-545-9_16. [DOI] [PubMed] [Google Scholar]
- 22.Jensen EH, McLoughlin JM, Yeatman TJ. Microarrays in gastrointestinal cancer: is personalized prediction of response to chemotherapy at hand? Curr Opin Oncol. 2006 Jul;18(4):374–380. doi: 10.1097/01.cco.0000228745.56918.0f. [DOI] [PubMed] [Google Scholar]
- 23.Mariadason JM, Arango D, Shi Q, Wilson AJ, Corner GA, Nicholas C, et al. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res. 2003 Dec 15;63(24):8791–8812. [PubMed] [Google Scholar]
- 24.Nagasaki K, Miki Y. Molecular prediction of the therapeutic response to neoadjuvant chemotherapy in breast cancer. Breast Cancer. 2008;15(2):117–120. doi: 10.1007/s12282-008-0031-6. [DOI] [PubMed] [Google Scholar]
- 25.Schauer M, Janssen KP, Rimkus C, Raggi M, Feith M, Friess H, et al. Microarray-based response prediction in esophageal adenocarcinoma. Clin Cancer Res. 2010 Jan 1;16(1):330–337. doi: 10.1158/1078-0432.CCR-09-1673. [DOI] [PubMed] [Google Scholar]
- 26.Anguiano A, Tuchman SA, Acharya C, Salter K, Gasparetto C, Zhan F, et al. Gene expression profiles of tumor biology provide a novel approach to prognosis and may guide the selection of therapeutic targets in multiple myeloma. J Clin Oncol. 2009 Sep 1;27(25):4197–4203. doi: 10.1200/JCO.2008.19.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Decaux O, Lode L, Magrangeas F, Charbonnel C, Gouraud W, Jezequel P, et al. Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: a study of the Intergroupe Francophone du Myelome. J Clin Oncol. 2008 Oct 10;26(29):4798–4805. doi: 10.1200/JCO.2007.13.8545. [DOI] [PubMed] [Google Scholar]
- 28.Moreaux J, Klein B, Bataille R, Descamps G, Maiga S, Hose D, et al. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica. 2011 Apr;96(4):574–582. doi: 10.3324/haematol.2010.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006 Sep 15;108(6):2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhan F, Barlogie B, Arzoumanian V, Huang Y, Williams DR, Hollmig K, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007 Feb 15;109(4):1692–1700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhan F, Barlogie B, Mulligan G, Shaughnessy JD, Jr, Bryant B. High-risk myeloma: a gene expression based risk-stratification model for newly diagnosed multiple myeloma treated with high-dose therapy is predictive of outcome in relapsed disease treated with single-agent bortezomib or high-dose dexamethasone. Blood. 2008 Jan 15;111(2):968–969. doi: 10.1182/blood-2007-10-119321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulligan G, Mitsiades C, Bryant B, Zhan F, Chng WJ, Roels S, et al. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007 Apr 15;109(8):3177–3188. doi: 10.1182/blood-2006-09-044974. [DOI] [PubMed] [Google Scholar]
- 33.Minna JD, Girard L, Xie Y. Tumor mRNA expression profiles predict responses to chemotherapy. J Clin Oncol. 2007 Oct 1;25(28):4329–4336. doi: 10.1200/JCO.2007.12.3968. [DOI] [PubMed] [Google Scholar]
- 34.Kumar SGP, Haug J, Kline M, Chng WJ, Blood E, Bergsagel Leif, Lust John A, Gertz Morie A, Fonseca Rafael, Rajkumar SV. Gene Expression Profiling of Myeloma Cells at Diagnosis Can Predict Response to Therapy with Thalidomide and Dexamethasone Combination. Blood. (11-2005 ed) 2005:508. [Google Scholar]
- 35.Broyl A, Hose D, Lokhorst H, de Knegt Y, Peeters J, Jauch A, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010 Oct 7;116(14):2543–2553. doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- 36.Blade J, Samson D, Reece D, Apperley J, Bjorkstrand B, Gahrton G, et al. Criteria for evaluating disease response and progression in patients with multiple myeloma treated by high-dose therapy and haemopoietic stem cell transplantation. Myeloma Subcommittee of the EBMT. European Group for Blood and Marrow Transplant. Br J Haematol. 1998 Sep;102(5):1115–1123. doi: 10.1046/j.1365-2141.1998.00930.x. [DOI] [PubMed] [Google Scholar]
- 37.Dimitriadou EHK, Leisch F, Meyer D, Weingessel A. CRAN - Package e1071. Webcite. 2005 [Google Scholar]
- 38.Yip W, Amin S, Li C. A Survey of Classification Techniques for Microarray Data Analysis. Handbook of Computational Statistics: Statistical Bioinformatics. 2011 May 29; edn2011. [Google Scholar]
- 39.Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proceedings of the National Academy of Sciences of the United States of America. 1999 Aug 3;96(16):9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munshi NC, Anderson KC, Bergsagel PL, Shaughnessy J, Palumbo A, Durie B, et al. Consensus recommendations for risk stratification in multiple myeloma: report of the International Myeloma Workshop Consensus Panel 2. Blood. 2011 May 5;117(18):4696–4700. doi: 10.1182/blood-2010-10-300970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007 Apr 15;109(8):3489–3495. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 42.Dimopoulos M, Kyle R, Fermand JP, Rajkumar SV, San Miguel J, Chanan-Khan A, et al. Consensus recommendations for standard investigative workup: report of the International Myeloma Workshop Consensus Panel 3. Blood. 2011 May 5;117(18):4701–4705. doi: 10.1182/blood-2010-10-299529. [DOI] [PubMed] [Google Scholar]
- 43.Sorlie T, Perou CM, Fan C, Geisler S, Aas T, Nobel A, et al. Gene expression profiles do not consistently predict the clinical treatment response in locally advanced breast cancer. Mol Cancer Ther. 2006 Nov;5(11):2914–2918. doi: 10.1158/1535-7163.MCT-06-0126. [DOI] [PubMed] [Google Scholar]
- 44.Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003 Jan 1;95(1):14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 45.Kuiper R, Broyl A, de Knegt Y, van Vliet MH, van Beers EH, van der Holt B, et al. A gene expression signature for high-risk multiple myeloma. Leukemia. 2012 May 8; doi: 10.1038/leu.2012.127. [DOI] [PubMed] [Google Scholar]
- 46.Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007 Mar 15;109(6):2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 47.Chen Z, Yan B, Van Waes C. The Role of the NF-kappaB Transcriptome and Proteome as Biomarkers in Human Head and Neck Squamous Cell Carcinomas. Biomark Med. 2008;2(4):409–426. doi: 10.2217/17520363.2.4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng PZ, Wang KK, Zhang QY, Huang QH, Du YZ, Zhang QH, et al. Systems analysis of transcriptome and proteome in retinoic acid/arsenic trioxide-induced cell differentiation/apoptosis of promyelocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2005 May 24;102(21):7653–7658. doi: 10.1073/pnas.0502825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 50.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010 Nov 1;24(21):2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004 Nov 1;64(21):7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 52.Avet-Loiseau H, Li C, Magrangeas F, Gouraud W, Charbonnel C, Harousseau JL, et al. Prognostic significance of copy-number alterations in multiple myeloma. J Clin Oncol. 2009 Sep 20;27(27):4585–4590. doi: 10.1200/JCO.2008.20.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010 Oct 14;116(15):e56–e65. doi: 10.1182/blood-2010-04-279596. [DOI] [PubMed] [Google Scholar]
- 54.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nature communications. 2014 Jan 16;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer cell. 2014 Jan 13;25(1):91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Catchpoole DR, Kennedy P, Skillicorn DB, Simoff S. The curse of dimensionality: a blessing to personalized medicine. J Clin Oncol. 2010 Dec 1;28(34):e723–e724. doi: 10.1200/JCO.2010.30.1986. author reply e725. [DOI] [PubMed] [Google Scholar]
- 57.Michiels S, Kramar A, Koscielny S. Multidimensionality of microarrays: statistical challenges and (im)possible solutions. Mol Oncol. 2011 Apr;5(2):190–196. doi: 10.1016/j.molonc.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.