Abstract
Fission yeast Rec12 (Spo11 homolog) initiates meiotic recombination by forming developmentally programmed DNA double-strand breaks (DSBs). DSB distributions influence patterns of heredity and genome evolution, but the basis of the highly nonrandom choice of Rec12 cleavage sites is poorly understood, largely because available maps are of relatively low resolution and sensitivity. Here, we determined DSBs genome-wide at near-nucleotide resolution by sequencing the oligonucleotides attached to Rec12 following DNA cleavage. The single oligonucleotide size class allowed us to deeply sample all break events. We find strong evidence across the genome for differential DSB repair accounting for crossover invariance (constant cM/kb in spite of DSB hotspots). Surprisingly, about half of all crossovers occur in regions where DSBs occur at low frequency and are widely dispersed in location from cell to cell. These previously undetected, low-level DSBs thus play an outsized and crucial role in meiosis. We further find that the influence of underlying nucleotide sequence and chromosomal architecture differs in multiple ways from that in budding yeast. DSBs are not strongly restricted to nucleosome-depleted regions, as they are in budding yeast, but are nevertheless spatially influenced by chromatin structure. Our analyses demonstrate that evolutionarily fluid factors contribute to crossover initiation and regulation.
Via homologous recombination, a broken chromosome uses its intact sister chromatid or homologous chromosome (homolog) as a repair template. In most sexually reproducing organisms, recombination is critical during meiosis because it facilitates pairing of homologs and forms physical connections between them in the form of crossovers, which, in combination with sister chromatid cohesion, are necessary for proper homolog segregation (Petronczki et al. 2003). Crossovers also reassort haplotypes, yielding progeny with increased genetic diversity (Kauppi et al. 2004). Failure to recombine can have disastrous consequences, often resulting in aneuploid, nonviable gametes. Yet despite this nearly universal role, the mechanisms and machinery of meiotic recombination are highly diverse among species.
Developmentally programmed DNA double-strand breaks (DSBs) initiate meiotic recombination in the organisms studied most thoroughly at the molecular level, the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe, and likely nearly all species (Keeney 2007; Cromie and Smith 2008). These recombinogenic lesions are generated by Spo11 (Rec12 in fission yeast), a topoisomerase-related protein found in essentially all sexually reproducing organisms. The formation of DSBs is influenced by numerous factors acting in concert, some of which are inherent to the underlying genome sequence, resulting in nonrandom distribution of breaks (Petes 2001; Cromie and Smith 2008). DSB distribution is tightly regulated, as recombination near centromeres or within repetitive DNA can lead to catastrophe (Sasaki et al. 2010). Because of the influence of recombination on genetic inheritance and genome stability and evolution, understanding the determinants of DSB distribution and repair is of great interest.
Regions where break formation, recombination, or both occur especially frequently are called hotspots. In budding yeast, DSB hotspots occur most often at most transcriptional promoters and associated nucleosome-depleted regions (NDRs) (Pan et al. 2011), while in fission yeast and mice, DSB hotspots are generally much farther apart and are infrequently promoter associated (Cromie et al. 2007; Smagulova et al. 2011). Hotspots in fission yeast, and perhaps other organisms, also appear to delineate domains of differential repair: The sister chromatid is the template of choice for breaks in hotspots while the homolog is preferred outside hotspots (Hyppa and Smith 2010). This results in a nearly uniform (interhomolog) crossover frequency along chromosomes despite the presence of strong DSB hotspots (Young et al. 2002). This phenomenon, called crossover invariance, has been observed at two well-studied hotspots (Hyppa and Smith 2010), but whether it extends to other loci is unknown. Furthermore, the provenance of the many crossovers that form outside of DSB hotspots has been unclear, as prior methods did not reveal sufficient break numbers to account for the recombination events, possibly because of insufficient signal:noise discrimination (Young et al. 2002; Cromie et al. 2007).
Several factors affecting DSB distributions have been described, particularly those governing strong DSB hotspots. Three S. pombe proteins (Rec25, Rec27, and Mug20), likely acting as a complex, bind essentially all hotspots and are required for most DSB formation there, but how they act and the chromosome features determining their binding remain unknown (Fowler et al. 2013). In a few cases, hotspots depend on transcription factors (TFs) binding a particular DNA sequence, e.g., at the ade6-M26 mutation in S. pombe and the HIS4 locus in S. cerevisiae (Schuchert et al. 1991; White et al. 1991; Kon et al. 1997; Steiner et al. 2002). However, transplacement of binding sites (Ponticelli and Smith 1992) revealed that TF-directed hotspot formation is context-dependent. These and other studies, both within and between species (Steiner and Steiner 2012), show that TFs have complex but poorly defined roles in determining break locations. DSB formation also depends on chromatin structure; for example, certain histone modifications are strongly associated with hotspots (Hirota et al. 2008; Borde et al. 2009; Acquaviva et al. 2013; Sommermeyer et al. 2013), though it remains unclear whether such modifications play a conserved role between species (Brick et al. 2012; Tischfield and Keeney 2012; Yamada et al. 2013). Thus, while DSB hotspots seem to be a widely conserved feature of meiotic recombination, aspects of their specification remain elusive. Moreover, hotspots are just one layer of organization, accounting for only a portion of the DSB landscape (Pan et al. 2011).
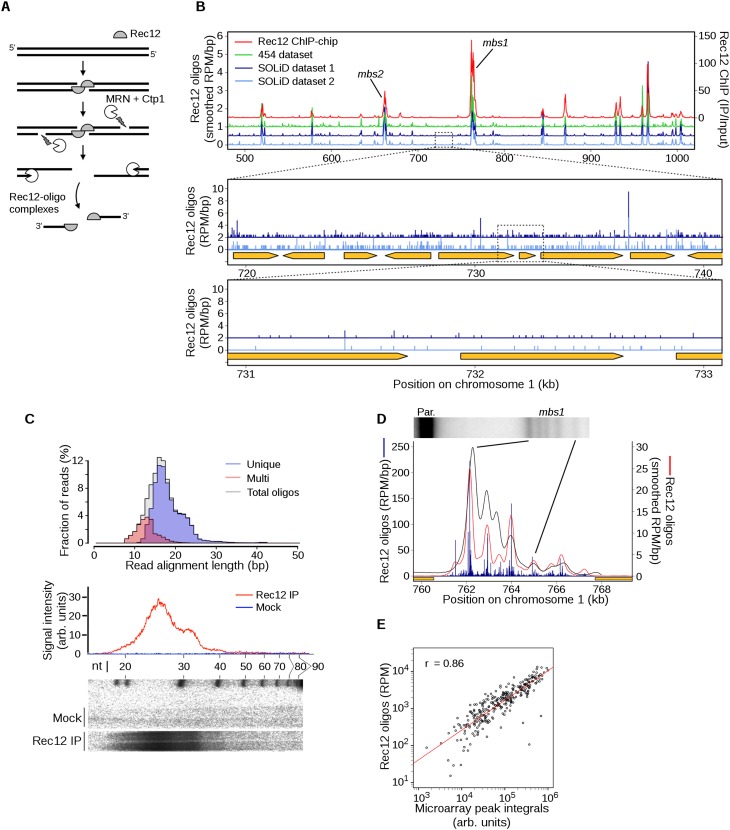
In this study, we adapted a method recently used to map DSBs at near single-nucleotide resolution in budding yeast (Pan et al. 2011), taking advantage of the mechanism of DSB formation and processing. Cleavage of duplex DNA by two molecules of Spo11 (or Rec12) acting in concert leaves the proteins covalently bound via tyrosine phosphodiester bonds to the DSB 5′ ends (Fig. 1A). Subsequent endonucleolytic cleavage releases Rec12 with an attached oligonucleotide, and the exact position of individual DSBs is revealed by isolating and sequencing these oligos. In fission yeast, most Rec12 oligos are ∼13–29 nucleotides long (Milman et al. 2009; Rothenberg et al. 2009), in contrast to the two distinct size classes in budding yeast and mice (Neale et al. 2005; Lange et al. 2011). The significance of these size classes is unclear, but the presence of a single class in fission yeast allowed us to sample deeply from all DSB-derived oligos, not just the larger subclass as was done previously in budding yeast (Pan et al. 2011). The Rec12-oligo map matches previously published microarray-based maps (Cromie et al. 2007; Hyppa et al. 2008) but has higher resolution with a higher dynamic range and lower background, thereby providing unprecedented insight into the DSB landscape and its relation to crossover distribution.
Figure 1.
Generation of DSB maps at nearly single nucleotide resolution by Rec12-oligo mapping. (A) Pathway of meiotic DSB and Rec12-oligo formation. (B) Agreement between methods for mapping DSBs. Microarray hybridization of Rec12-DNA complexes (right axis, median-normalized IP/Input; data from Fowler et al. 2013), and uniquely mapped Rec12 oligos (left axis; offset in steps of 0.5 reads per million [RPM]/bp, smoothed with a 1-kb Hann window) are shown as an example region. An artifactual sequence pile-up at a single base pair at ∼710 kb in the 454 data set is not shown. Zoomed plots are provided in the bottom panels to show dispersed Rec12 oligos across the indicated subintervals of a DSB “cold” region. Orange arrows indicate the open reading frames of protein-coding genes. (C) Length distributions of Rec12 oligos. Oligo sequences were mapped to the genome and the distribution of alignment lengths was plotted according to mapping status (top). The sequence length distribution correlates with the sizes of DNA molecules purified from meiotic cultures and resolved by denaturing PAGE (bottom autoradiograph of duplicate lanes and trace). Note that the oligos migrate more slowly on the gel than expected due to residual amino acid(s) remaining on the 5′ end of each oligo after proteolysis and the addition of labeled nucleotides. (D) Agreement between high-resolution comparisons of breakage at the mbs1 hotspot (Southern blot of MluI-digested meiotic DNA from a rad50S induction at top; black line is the signal density trace) and Rec12-oligos (smoothed with a 501-bp Hann window). Orange boxes indicate 5′ portions of divergently transcribed flanking genes. (E) Quantitative correlation between Rec12 oligos and Rec12 ChIP-chip. Rec12 oligos were summed at hotspots determined by ChIP-chip (n = 288) and compared with the integrated microarray signal (Fowler et al. 2013). The trend is slightly nonlinear, likely because of higher microarray background. See Supplemental Figure S1 for other analyses and validation of Rec12-oligo distributions.
Consonant with the phylogenetic diversity of meiotic recombination mechanisms, the high-resolution fission yeast DSB map reveals similarities but also many differences from that in budding yeast. These divergent patterns highlight the importance of determining the rules governing DSB formation in multiple organisms to better understand the many factors that are involved in homologous recombination.
Results and Discussion
Faithful map of meiotic DSBs at single-nucleotide resolution
We immunopurified Rec12-oligo complexes from synchronous meiotic cultures at the time of their maximal abundance (Milman et al. 2009). Rec12 was digested with a protease, oligos were recovered, and DNA adaptors added (Supplemental Material; Supplemental Table S1). Amplification yielded products of expected size that were not recovered from mock immunoprecipitates or water controls (Supplemental Fig. S1A). We sequenced on the Roche 454 platform one sample from a haploid strain, which in the pat1-114 background initiates meiosis (Young et al. 2002), and on the ABI SOLiD platform two samples from one culture of a diploid strain. Sequence reads were mapped to the S. pombe genome; we obtained 0.2 million mapped reads from 454 and 5.2 million total from SOLiD (Supplemental Table S2). The maps were highly reproducible between both biological and technical replicates (Figs. 1B; Supplemental Fig. S1B). The analyses below used the pooled SOLiD data because of the greater sequencing depth, but the 454 data were used to confirm findings where appropriate. Conclusions were further validated by comparison with an independently generated map (P Schlögelhofer, pers. comm.).
Mapped reads displayed a unimodal distribution of lengths (Fig. 1C) (mean ±SD of 17.2 ± 4.3 nt, median of 17 nt, mode of 16–17 nt, with a prominent shoulder at 20–23 nt). This distribution parallels that seen for denaturing polyacrylamide gel electrophoresis of proteinase-treated, 3′ end-labeled Rec12 oligos (Fig. 1C). (Note that the oligos migrate on gels significantly more slowly than expected for their original size because of residual amino acids from Rec12 attached to their 5′ ends and the addition of multiple nucleotides to their 3′ ends by terminal transferase.)
Most reads mapped uniquely in the genome, but many mapped to multiple positions (21.5% “multi-mappers”). This fraction is substantially higher than that in S. cerevisiae (1.8% multi-mappers) (Pan et al. 2011), but this difference is largely attributable to the shorter S. pombe oligos having lower mapping efficiency (mapped oligos in budding yeast were 29.5 ± 6.4-nt long). Indeed, when reads were partitioned by length, multi-mappers were highly enriched among the shortest oligos, making up about half of the 13- to 14-nt-long reads and nearly all reads under 12 nt (Fig. 1C). In contrast, >97.5% of reads longer than 18 nt mapped uniquely, comparable to totals in S. cerevisiae. We infer that many of the longer multi-mappers reflect Rec12 oligos from truly repetitive DNA elements (e.g., rDNA, pericentric repeats, and transposable elements), whereas most of the shorter reads, although mapped ambiguously, came primarily from nonrepetitive genomic regions (i.e., repeated sequences longer than the oligo read but less than 50 bp long). In support of this conclusion, multi-mappers were enriched in previously defined DSB hotspots (Fowler et al. 2013) in proportion to the unique oligo map (Supplemental Fig. S1C), and multi-mappers and unique reads showed similar fine-scale distributions within hotspots (Supplemental Fig. S1D). Moreover, for many individual multi-map reads, we observed that one map position often fell within a region with many uniquely mapped reads on both strands (i.e., a hotspot), while the other map position(s) usually fell within a region(s) with few or no uniquely mapped reads nearby (Supplemental Fig. S1E; data not shown).
We therefore reasoned that uniquely mapped reads could be used to disambiguate multi-mappers: A Rec12 oligo mapping to multiple positions is more likely to have come from the position(s) that also generated more unique mappers nearby (Supplemental Fig. S1F). Therefore, we compiled an imputed map by fractionally assigning each multi-mapper to each of its mapped positions in proportion to the density of uniquely mapped oligos within ±250 bp. This approach is similar to one developed independently to evaluate ChIP-seq data from repetitive genomic regions (Chung et al. 2011). We estimate that only 2.2% of the nonrepetitive nuclear genome lacks a unique oligo within 500 bp, which results in equal assignment between sites in those rare regions. The unique map and imputed multi-map had similar fine-scale patterns (discussed further below), validating this approach; thus, these maps were combined for subsequent analyses. However, nearly identical results were obtained if only unique mappers were used (data not shown).
Several lines of evidence affirm that we mapped bona fide Rec12 oligos. First, mock immunoprecipitates yielded only negligible amounts of nucleic acid in the size range of the sequenced oligos (Fig. 1C) and little if any PCR amplification products of the expected size after adaptor ligation (Supplemental Fig. S1A). Previous work showed that immunoprecipitated Rec12-oligo complexes yield little detectable signal on a polyacrylamide sequencing gel unless treated with protease (Milman et al. 2009). Thus, the vast majority of the purified DNA used to construct sequencing libraries was from bona fide Rec12 oligos. We cannot exclude the possibility of contaminating high molecular weight DNA, but even if present this would be irrelevant for our analyses as such material clearly has not contributed to our sequencing results.
Second, sequence reads were infrequent in regions that have little detectable crossing-over and are thus thought to be largely devoid of DSBs. In many organisms, rDNA repeats are recombinationally suppressed to prevent chromosome rearrangements (Vader et al. 2011). In our laboratory strains of S. pombe, rDNA resides in ∼65 tandem arrays on the ends of chromosome III, occupying ∼710 kb (M Eickbush, S Zanders, and GR Smith, unpubl.), and accounts for only 0.04% of reads despite making up ∼5% of nuclear DNA. Likewise, very few reads were recovered from other DSB-suppressed regions (e.g., heterochromatic centromeres) (Supplemental Fig. S1G; Ellermeier et al. 2010). Even if we conservatively assume that none of the rDNA reads are true Rec12 oligos, then the Rec12-independent background is negligible, at more than 110-fold below the genome mean: 0.7 reads per million mapped (RPM) per kb in the rDNA vs. 79.5 RPM/kb genome-wide. This value is even lower than that reported for a Spo11-oligo map in S. cerevisiae (1.1 RPM/kb in the rDNA, 75-fold below genome average) (Pan et al. 2011), which has negligible rDNA recombination during meiosis (Petes and Botstein 1977).
Third, the map agreed spatially and quantitatively with DSBs detected directly by Southern blot hybridization of genomic DNA isolated from meiotically induced cells. For example, our map faithfully recapitulated the DSB pattern at the well-studied mbs1 hotspot (Cromie et al. 2005), including the marked substructure comprising clusters of break sites within this ∼6-kb-wide region (Fig. 1D). Of the eight peaks at mbs1 determined by Southern blot analysis, six agree with Rec12-oligo peaks in position and rank order of intensity. Only one peak (at ∼764 kb in Fig. 1D) shows substantially different relative signal (but matched position); the other disagreement is between weak subsidiary peaks at the edge of the hotspot. Moreover, read counts scaled linearly with DSB frequency over at least a 10-fold range for a collection of hotspots whose activity has been measured by Southern blotting (Supplemental Fig. S1H).
Fourth, our map agreed well genome-wide with microarray hybridization of immunoprecipitated Rec12–DNA complexes (ChIP-chip) that accumulate in a rad50S mutant, whose DSB landscape appears indistinguishable from that in rad50+ (Hyppa et al. 2008). As expected for Rec12 oligos, sequence reads were highly enriched in previously defined ChIP-chip hotspots and occurred at low frequency between hotspots (Fig. 1B), and read counts were highly correlated with integrated microarray signal intensity in the ChIP-chip hotspots (Fig. 1E). Importantly, however, Rec12-oligo sequencing provided much greater spatial precision that more accurately reflected DSB distributions (see below). Moreover, the specificity of Rec12-oligo purification (see above) yielded a higher signal-to-noise ratio and thus greater sensitivity, particularly for weaker hotspots or in regions between hotspots (Fig. 1B).
We conclude that our sequence map faithfully reflects the genome-wide distribution of Rec12 oligos, and thus DSBs, with unsurpassed sensitivity and spatial precision, and with extremely low background. Below, we use this map to investigate genomic features that shape the DSB landscape.
Hotspot-centric analysis of DSBs
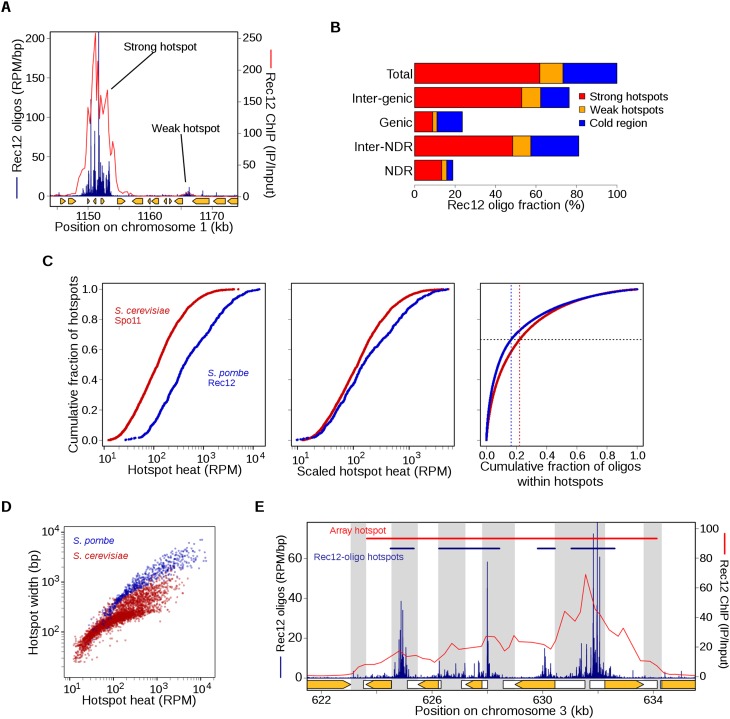
Most S. pombe DSB hotspots are in large intergenic regions (IGRs), and most protein-coding gene promoters lack associated hotspots (Cromie et al. 2007); this pattern is similar to that in mice (Brick et al. 2012) but unlike that in S. cerevisiae (Pan et al. 2011). As a result, S. pombe has fewer, more widely spaced hotspots than S. cerevisiae. We previously defined 288 DSB hotspots as regions with ChIP-chip hybridization signal at or above the level expected for a DSB frequency of 0.3%, based on linear regression of ChIP-chip signal vs. direct DSB Southern blot assays at 25 hotspots (Cromie et al. 2007; Fowler et al. 2013). This value (0.3%) represents the approximate threshold for detection on the Southern blots (Cromie et al. 2007).
In this study, we defined 603 Rec12-oligo hotspots as sites with significantly higher read densities than surrounding regions (Fig. 2A; Supplemental Material; Supplemental Table S3). Rec12-oligo densities in these hotspots ranged from 173 to 5021 RPM/kb. The density in the weakest hotspot scored was thus 247-fold higher than the density in the rDNA, and 2.2-fold higher than the genome average. This list accounted for 98% of the 288 previously defined hotspots (Fowler et al. 2013), plus 322 additional hotspots uncovered because of the greater sensitivity of Rec12-oligo sequencing. Consistent with previous findings, most Rec12-oligo hotspots overlapped with IGRs (Fig. 2B). Importantly, significant spatial enrichment of ChIP-chip signal for Rec12 and, albeit weakly, for hotspot-determinant proteins Rec25, Rec27, and Mug20 was seen even at the weak Rec12-oligo hotspots that would have been below detection limits by other means (Supplemental Fig. S2A), confirming that these are bona fide sites of preferential DSB formation.
Figure 2.
Features of DSB hotspots and comparisons with S. cerevisiae. (A) An example of adjacent strong and weak hotspots. “Strong” hotspots refer to those with an inferred break frequency of ≥0.3% of DNA, i.e., the limit of detection in prior studies. Rec12-oligo frequencies (blue) were compared with Rec12 ChIP-chip signal (red). For comparison, there are eight strong and 13 weak hotspots in Figure 1B (top). Orange arrows indicate open reading frames of protein-coding genes. (B) Rec12 oligos arise primarily from intergenic DNA and hotspots, although about a quarter arise from “cold regions,” i.e., outside the 603 hotspots defined here. NDR and Inter-NDR indicate the fraction of oligos that arise within or outside nucleosome depleted regions, respectively. (C) Rec12-oligo counts in hotspots follow a smooth continuum. (Left) Hotspots were rank-ordered by oligo counts (603 hotspots from S. pombe; 3604 hotspots from S. cerevisiae) (Pan et al. 2011). (Middle) To account for the greater number of meiotic DSBs per cell in S. cerevisiae, the rank-ordered S. pombe hotspots were scaled by a factor of 58/160, the approximate ratio of the number of DSBs per cell in S. pombe and S. cerevisiae. (Right) Plot of the cumulative fraction of hotspot oligos among the ranked hotspots. (D) Hotter hotspots tend to be wider than colder hotspots; S. pombe hotspots tend to be much wider than in S. cerevisiae. (E) Hotspot boundaries correlate poorly if at all with IGR boundaries, positions of protein-coding genes (orange arrows), or promotors (white boxes indicate transcribed regions). Gray bands indicate IGRs. Top bars show the defined hotspot regions, blue from this study and red from microarray hybridizations (Fowler et al. 2013). See Supplemental Figure S2 for further analyses of Rec12-oligo hotspots.
When hotspots were ranked by oligo count, they followed a smooth continuum with no obvious discontinuity to serve as a dividing line between quantitative classes (Fig. 2C, left). In particular, we emphasize that there was no clear distinction between previously discovered hotspots and the additional weak ones discovered here (i.e., those with an inferred DSB frequency of <0.3% of DNA). This pattern reinforces the view that the cutoff is arbitrary between sites that are hotspots and those that are not (Pan et al. 2011). Interestingly, the distribution of breaks among hotspots differed in the two species. For example, hotspots in general were hotter as a fraction of total breaks than those in budding yeast, as judged by comparing the cumulative curves in Figure 2C, left. Looked at a different way, if we scale hotspot counts to reflect the greater number of total DSBs in S. cerevisiae than in S. pombe (∼160 vs. ∼58, respectively, see below), S. pombe hotspots also tend to be hotter in absolute terms (Fig. 2C, middle). Furthermore, the weakest two-thirds of hotspots accounted for 22% of hotspot-associated oligos in S. cerevisiae, but only 16.4% in S. pombe (Fig. 2C, right); and 20% of all hotspot-associated oligos were contained in the 87 hottest hotspots in S. cerevisiae, but in only the 16 hottest hotspots in S. pombe. Thus, while there are fewer locations that score as hotspots in fission yeast, a larger fraction of these are exceptionally hot and account for a disproportionate share of breakage.
The more precise spatial information afforded by Rec12-oligo mapping allowed us to explore hotspot features in greater detail than previously possible. For example, the width of Rec12-oligo hotspots correlated strongly with heat (i.e., read count) (Fig. 2D). This relationship was not linear, however, reflecting an additional correlation of hotspot width with oligo density (Supplemental Fig. S2B). Thus, the wider break sites are broken disproportionately more frequently. Compared with budding yeast, S. pombe hotspots displayed more highly variable widths (∼50 bp to 7 kb; the widest S. cerevisiae hotspot is 2 kb) and were wider on average: The mean width (1.4 kb) was comparable to that of the widest hotspots in budding yeast (Fig. 2D).
Many wide hotspots contained clusters of closely spaced break sites, as exemplified by mbs1 (Fig. 1D). This pattern reinforces the points that the genome is not simply divided into alternating domains that favor or disfavor break formation, and that hotspot definitions are inherently arbitrary. From ChIP-chip data, it appeared that most hotspots cover the majority or entirety of the IGRs where they reside (Cromie et al. 2007). At finer scale, however, Rec12 oligos generally arose from only a portion of each IGR (Fig. 2E; Supplemental Fig. S2C). On average, intergenic hotspots occupied only 57% of their IGRs, with hotter hotspots tending to encompass more, and in only 21% of IGRs with a hotspot did oligos extend over >90% of the IGR (Supplemental Fig. S2C). Prominent clusters of largely intergenic Rec12 oligos often extended into adjacent coding regions (Fig. 2E) and spatially correlated with some sites of noncoding RNA (ncRNA) generation (Rhind et al. 2011; Chen et al. 2012). For intergenic ncRNAs, Rec12 oligos were enriched within the ncRNA region but not within their flanking regions (Supplemental Fig. S2D). Thus, DSBs are not as constrained by coding regions as it previously appeared (Cromie et al. 2007).
Susceptibility of IGRs to DSB formation did not correlate with the presence of promoters or adjacent gene expression (data not shown). Within IGRs, breakage was rarely promoter associated: Only 19% of IGR oligos were within 200 bp of coding gene transcription start sites (TSSs), and this number dropped to 5% for IGRs wider than 2.5 kb. Separating IGRs by the orientation of their flanking genes showed that oligos were most dense between divergent genes and least dense between convergent (Supplemental Fig. S2E). This overall pattern is similar to S. cerevisiae IGRs, but S. pombe IGRs had overall lower oligo densities (because many DSBs arise outside of IGR-associated hotspots; see below), and S. pombe showed a much smaller quantitative distinction between promoter-containing (divergent and tandem) and promoter-lacking (convergent) IGRs. Thus, unlike in S. cerevisiae, promoters of protein-coding genes are only rarely prominent targets for DSB formation. At least superficially, this pattern is reminiscent of that observed in mice, where DSB hotspots, measured as RAD51- or DMC1-bound DNA, are generally intergenic but not promoter associated (Smagulova et al. 2011; Brick et al. 2012).
Abundant non-hotspot DSBs that were previously undetectable
A hotspot-centric view is biased toward the idea that hotspots produce all or the vast majority of DSBs and crossovers. However, 28% of all Rec12 oligos mapped outside of definable hotspots (Figs. 1B, 2A,B). This fraction is greater than that in S. cerevisiae (11.4%) despite similar minimum thresholds for calling hotspots (approximately twofold over genome average oligo density in both cases). While the mean density of these “cold-region” Rec12 oligos was low (22.6 RPM/kb), it was 32-fold higher than that within the rDNA (Supplemental Fig. S1G). Significantly, these oligos share a hallmark local base composition bias with their hotspot counterparts (discussed further below), indicating that cold-region and hotspot oligos are formed by similar or identical mechanisms. Similar findings were obtained from both sequencing platforms used here and from an independent study (data not shown; P Schlögelhofer, pers. comm.). Thus, we conclude that the overwhelming majority of cold-region oligos represent bona fide Rec12-generated breaks. The low, nearly uniform distribution of these breaks (Fig. 1B, bottom) would render them invisible to lower sensitivity methods such as ChIP-chip or Southern blot hybridizations.
Non-hotspot DSBs account for crossovers in “DSB-cold” regions
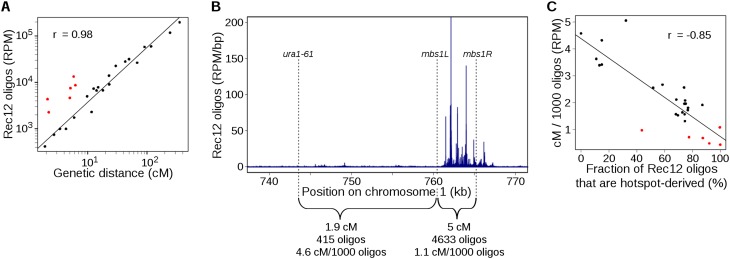
Meiotic crossing-over occurs frequently at great distances from DSB hotspots in S. pombe; these crossovers are Rec12 dependent, but prior studies were unable to detect enough DSBs to account for them (Young et al. 2002). The large number of Rec12 oligos arising from regions outside of hotspots strongly argues that this recombination originates from DSBs outside hotspots, and that such DSBs make up much of the total chromosome breakage and crossovers (see below). In support of this hypothesis, genetic intervals that had a substantial fraction of cold-region oligos and small intervals without hotspots displayed a linear relationship, over a >100-fold range, between the number of Rec12 oligos and the genetic distance (cM) (Fig. 3A; Supplemental Fig. S3A; Supplemental Table S4), consistent with the interpretation that each DSB in these intervals yields a crossover with roughly equal probability. As expected for the relatively uniform distribution of cold-region DSBs, intervals with few or no hotspot-associated oligos showed a linear relation between genetic distance and DNA length, but very short intervals (<5 kb) with the hotspots mbs1 and ade6-3049 were outliers (Supplemental Fig. S3B).
Figure 3.
DSBs in hotspots are much less likely to give rise to crossovers than are DSBs in cold regions. (A) Genetic map length is proportional to Rec12-oligo count, but regions containing strong hotspots behave differently from regions lacking hotspots. Rec12 oligos were summed across intervals with known genetic distances; replicate map lengths were averaged. Sources of map length data are in Supplemental Table S4. The Pearson correlation coefficient r is based on black points; red points are short intervals containing strong DSB hotspots. (B) Different ratios of crossing over to DSB frequency (cM/1000 oligos) at the mbs1 hotspot and an adjacent DSB “cold region.” Genetic distances are from Cromie et al. (2005). (C) DSBs from “cold regions” are more likely to yield a crossover than DSBs in “hot” regions, and the degree of bias in crossover fate correlates with the region’s heat. For each genetic interval in A, the fraction of Rec12 oligos that originated from hotspots was determined (see B for an example). Red points are the outliers in A. See Supplemental Figure S3 for other correlations and estimates of the contribution of different types of intervals to crossover numbers.
Unequal recombination potential and crossover invariance
Further examination of quantitative relationships between Rec12 oligos and crossing over provided new insight into the phenomenon of crossover invariance, in which numerous intervals display a nearly uniform crossover density (i.e., nearly constant cM/kb) despite some intervals having a prominent DSB hotspot(s) and others not (Hyppa and Smith 2010). These investigators showed that an important component of crossover invariance is locus-dependent differences in partner choice for DSB repair. DSBs at the strong hotspots mbs1 and ade6-3049 are biased toward repair by formation of a Holliday junction with the sister chromatid and less frequently produce an interhomolog crossover; distinct genetic requirements for recombination in hotspots vs. cold regions suggest that DSBs in cold regions are more likely to be repaired with the homolog and often produce a crossover (Hyppa and Smith 2010).
In agreement with prior findings (Cromie et al. 2005; Hyppa and Smith 2010), crossover frequencies between markers closely flanking the mbs1 and ade6-3049 hotspots were much lower than in other intervals with similar numbers of oligos (Fig. 3A). We now find similar behavior at other DSB hotspots in the short ade3–ura3 (55-kb) and ura2–leu2 (13-kb) intervals, where most Rec12 oligos (88% and 92%, respectively) arise from sites that score as hotspots (Fig. 3A; Supplemental Fig. S3A). We infer that this is a general pattern: Unlike small intervals without hotspots (see above), small intervals with a strong DSB hotspot experience much less crossing over than predicted from Rec12-oligo frequency. We therefore estimated how much each DSB contributes to the genetic map (cM per oligo). In the ura1–mbs1L interval, which is directly adjacent to mbs1 but does not contain a hotspot, we observed 4.6 cM/1000 oligos, whereas the nearby mbs1L–mbs1R interval containing the mbs1 hotspot displayed only 1.1 cM/1000 oligos (Fig. 3B). Similar results were obtained from the newly implicated hotspots: The ade3–ura3 and ura2–leu2 intervals, which contain DSB hotspots, had 0.7 and 0.5 cM/1000 oligos, respectively.
We considered that partner choice might vary continuously across the genome, such that crossover invariance is stronger at more intense hotspots, i.e., sites more frequently broken tend to be repaired more frequently using the sister chromatid. This view fits with the notion that hotspots exist as a continuum with the largely DSB-cold genome. As predicted by this hypothesis, we found for a large collection of genetic intervals a strong anticorrelation between cM/oligo values and the fraction of Rec12 oligos that were hotspot derived (Fig. 3C). There was a four- to fivefold difference in cM/oligo values between the coldest and hottest intervals, which is similar to the ratio of intersister:interhomolog Holliday junctions at the strong DSB hotspots mbs1 and ade6-3049 (4:1 and 3:1, respectively) (Cromie et al. 2006; Hyppa and Smith 2010). This result implies that the degree of repair template bias is proportional to the frequency of hotspot DSBs in the region and suggests that this pattern is pervasive.
One way to rationalize these patterns is to hypothesize that the frequency of DSB repair with the sister vs. with the homolog depends on the fraction of DSBs in the interval that depend on the Rec25–Rec27–Mug20 complex. This complex binds loci to variable extents and, in hotspots, in proportion to DSB frequency (Fowler et al. 2013). Loci with a high population-average level of Rec25–Rec27–Mug20 bound also have a high frequency of DSBs (hotspots) and are repaired primarily with the sister (IS repair) (Cromie et al. 2006; Hyppa and Smith 2010). Loci rarely bound by Rec25–Rec27–Mug20 have less frequent DSBs (cold regions) and, we infer from genetic requirements (Hyppa and Smith 2010), are repaired primarily with the homolog (IH repair). In this view, since the population average amount of Rec25–Rec27–Mug20 bound is a continuous variable, so is the population average IS:IH ratio of partner choice for repair, a consequence of the hypothesized dual functions of Rec25–Rec27–Mug20.
We conclude that crossovers are often formed outside hotspots from low-level, widely distributed DSBs. Indeed, we estimate below that about half of all crossovers come from intervals outside the previously identified (i.e., stronger) hotspots. These previously undetectable DSBs are repaired preferentially with the homolog to give more crossovers per DSB than do hotspot DSBs, which are repaired preferentially with the sister chromatid (Cromie et al. 2006; Hyppa and Smith 2010). Importantly, these results show that crossover invariance is not limited to the two previously tested intervals but likely extends to the entire genome.
Number of DSBs and crossovers per meiotic cell: Low-level DSBs account for approximately half of all crossovers
The nearly linear relation between Rec12-oligo count and percent DNA broken at a range of hotspots (Supplemental Fig. S1H) allowed us to estimate that the average number of DSBs formed per cell is ∼58 (95% CI = 48–68) (Supplemental Material). This is fewer than half the breaks in S. cerevisiae (∼160 on average) (Pan et al. 2011) despite similarly sized genomes.
We wished to estimate the total number of crossovers per cell, but doing so from DSB numbers is complicated by crossover invariance, particularly variability in partner choice (see above). Motivated by the analysis above (Fig. 3C), we assumed that the likelihood of interhomolog (rather than intersister) repair is a linear function of the DSB site’s break frequency (Supplemental Fig. S3C). We then calculated the partner choice bias for each hotspot, ranging from 20% interhomolog repair at the hottest hotspot to 100% at the coldest. We further assumed that 75% of interhomolog repair events result in a crossover, the approximate frequency observed among gene convertants in both hotspots and cold regions (Grimm et al. 1994; Cromie et al. 2005). We thus estimated ∼36 crossovers per meiosis, which agrees well with prior estimates of total genetic map length: 45 crossovers (Munz 1994) or 34 crossovers (Egel 2003). Importantly, our estimate was relatively robust to changes in the shape of the function used to model partner choice bias (linear, sigmoidal, exponential, and step-function variations ranged from ∼27 to 38 crossovers per meiosis; Supplemental Fig. S3C). The concordance of our estimates with experimental values reinforces our conclusion that crossover invariance is based on differences in partner choice for DSB repair genome-wide.
We estimated the fraction of all crossovers arising from DSBs outside the strong hotspots that were previously detectable (i.e., those with more than ∼0.3% breakage) by using the relation between hotspot intensity and frequency of repair with the homolog, as above. Using the linear relation (Supplemental Fig. S3C), we calculated that 46% of all crossovers arise from these DSB-poor regions, of which the majority (32% of total) came from DSBs that did not map to a scoreable hotspot at all. The sigmoidal, exponential, and step functions yielded similar estimates of 43%, 59%, and 44%, respectively. These results show that previously undetected DSBs account for nearly half of all crossovers and underscore the importance of sensitive assays when assessing genome-wide features. They also show that spatially dispersed meiotic DSBs are more important than previously suspected.
Relationship of DSBs to local chromatin structure
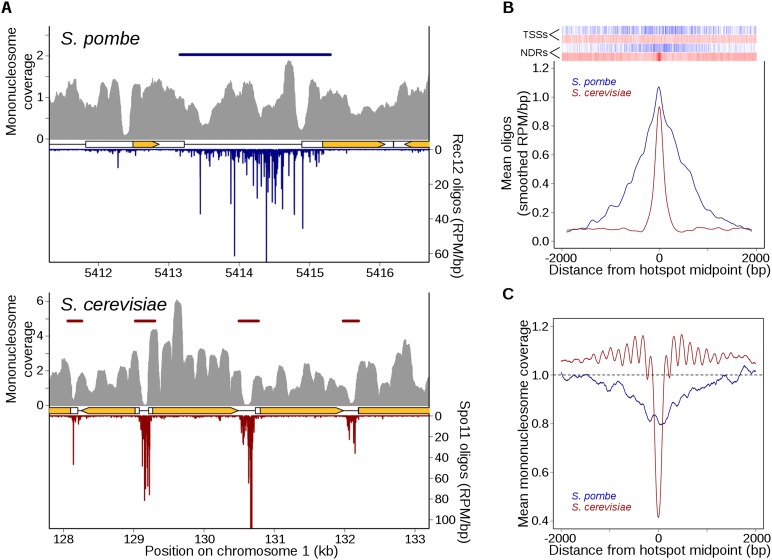
Chromatin is known or hypothesized to shape DSB distributions in many organisms (Lichten 2008), but precisely how it does so and the degree of evolutionary conservation are unknown. Until now, the only available high-resolution genome-wide data in meiosis for both chromatin structure and DSB distributions were from S. cerevisiae (Jiang and Pugh 2009; Pan et al. 2011; Zhang et al. 2011). In this yeast, most DSB hotspots correspond to places where the chromatin is hypersensitive to digestion with exogenous nucleases (DNase I or micrococcal nuclease [MNase]) (Ohta et al. 1994; Wu and Lichten 1994), and a large majority of Spo11 oligos map to clear NDRs, mostly in promoters (Pan et al. 2011). Thus, Spo11 takes advantage of constitutively accessible regions in chromatin and targets these preferentially for cleavage, further suggesting that Spo11 only rarely if ever cuts DNA wrapped into nucleosomes (Pan et al. 2011). Importantly, although the degree of nucleosome occupancy within or adjacent to an NDR is not a good predictor of hotspot heat, nucleosome occupancy is nevertheless an excellent predictor of local DSB spatial patterns (Pan et al. 2011). As a result, it is often easy to guess where DSB hotspots might be simply by looking at a nucleosome map, or conversely to guess where NDRs are by looking at a Spo11-oligo map (Fig. 4A, bottom). It has been argued that S. pombe hotspots are similarly lacking in nucleosomes and that DSBs are largely restricted to places that are nucleosome-depleted in most cells in a population (Hirota et al. 2007; de Castro et al. 2012; Yamada and Ohta 2013). However, prior genome-wide data had insufficient resolution to fully test this hypothesis. We therefore explored this question using recent maps of MNase-resistant mononucleosomal DNA generated from S. pombe meiotic chromatin (Soriano et al. 2013). As detailed below, relationships between population-average chromatin structure and DSB distributions differ substantially between fission and budding yeasts.
Figure 4.
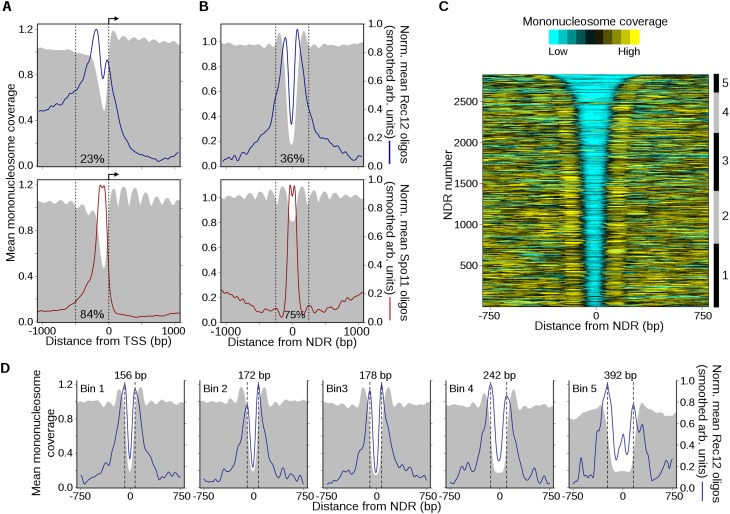
Markedly different relationship between DSBs and chromatin structure in S. pombe and S. cerevisiae. (A) Rec12 oligos are not especially enriched in NDRs, unlike Spo11 oligos. High-resolution mononucleosome coverage maps (MNase-seq) from meiotic S. pombe (Soriano et al. 2013) and S. cerevisiae (Pan et al. 2011) cells were compared with their respective oligo maps for representative regions on each organism’s chromosome 1. Nucleosome maps were normalized by taking the number of times each base pair was sequenced and dividing it by the mean genome coverage (i.e., the mean genome coverage is 1). Horizontal blue and red bars indicate hotspots. Orange arrows mark protein-coding genes and white boxes indicate transcripts. (B) Fission yeast hotspots are much wider than budding yeast hotspots and not generally centered on TSSs or NDRs. Rec12- and Spo11-oligo hotspot midpoints were aligned and the mean oligo distributions were smoothed with a 201-bp Hann window. Relative positions of annotated meiotic NDRs (Soriano et al. 2013) and TSSs (Lantermann et al. 2010) in S. pombe are indicated by blue ticks at top; S. cerevisiae NDRs and TSSs (Jiang and Pugh 2009) are in red. (C) Unlike Spo11-oligo hotspots, Rec12-oligo hotspots tend not to be strongly MNase sensitive in meiotic chromatin. Hotspots were aligned as in B and the mean mononucleosome coverage was determined at each position.
Focusing first on hotspots, we found that Rec12 oligos were not strongly associated with NDRs. Figure 4A (top) illustrates a typical hotspot region: At this site, Rec12 oligos were not restricted to NDRs but instead arose frequently from positions whose nucleosome occupancies varied over a wide range. Thus, in stark contrast to S. cerevisiae, it would not be possible to guess the locations of DSB hotspots from the chromatin map or to guess where NDRs are from the Rec12-oligo map. When hotspots were superimposed, there was little if any tendency toward local clustering of either annotated meiotic NDRs (Soriano et al. 2013) or TSSs (Lantermann et al. 2010), again very different from S. cerevisiae (Fig. 4B). Indeed, 16% of IGRs with a hotspot lacked an NDR entirely. When averaged over all hotspots, nucleosome coverage tended to be modestly lower than in flanking regions (Fig. 4C), but this pattern was weak compared with that in S. cerevisiae, where hotspots tightly overlap on average with a deep and narrow NDR flanked by arrays of relatively well-positioned nucleosomes (Fig. 4C). Thus, the strong tendency for S. cerevisiae DSBs to prefer NDRs is not true for S. pombe DSBs. As these are population-average measures, it is not possible to distinguish whether Rec12 often cuts DNA bound to nucleosomes or only cuts in subsets of cells where nucleosomes are absent from these positions. Interestingly, S. pombe hotspots do resemble a small number of exceptionally wide (>1.5 kb) S. cerevisiae hotspots, which tend to have low overall occupancy of relatively disordered nucleosomes (Pan et al. 2011).
To further investigate chromatin-DSB connections, we examined Rec12-oligo distributions from the perspective of specific chromatin contexts (i.e., without regard for the arbitrary designation of hotspots). Aligning TSSs or meiotic NDRs revealed previously described stereotypical nucleosome patterning around these features, such as the tendency for nucleosome depletion in promoters and positioned arrays of nucleosomes in transcription units and upstream flanking regions (Fig. 5A,B; Lantermann et al. 2010; Soriano et al. 2013). Many of these patterns are similar to those in S. cerevisiae, but overall, chromatin structure appears more “irregular’” in fission yeast, i.e., lower amplitude of occupancy signal for positioned nucleosomes, implying more variability from location to location.
Figure 5.
DSBs have complex relationships with local chromatin features. (A) Spatial pattern of Rec12- and Spo11-oligo formation adjacent to TSSs. Annotated TSSs in S. pombe (top) and S. cerevisiae (bottom) were aligned and oriented with transcription to the right (arrow). The mean mononucleosome coverage (Pan et al. 2011; Soriano et al. 2013) around the TSSs was determined as in Figure 4C. To determine the average spatial relationship between DSBs and TSSs, oligo counts around individual TSSs were divided by the total oligos in the 2-kb region plotted, then these normalized profiles were averaged across all TSSs and smoothed with a 101-bp Hann window for clarity. Normalization prevents sites with many oligos from dominating the average profile. Note that this procedure reveals the average shape of the Rec12- and Spo11-oligo distributions, not the average intensity. The fraction of total Rec12 or Spo11 oligos found within 500 bp upstream of TSSs (dashed lines) is indicated. (B) Rec12-oligo profiles are strongly patterned around NDRs. The midpoints of NDRs in S. pombe (constitutive and meiosis-specific) (Soriano et al. 2013) and S. cerevisiae (Jiang and Pugh 2009) were aligned and the mean normalized oligo profiles were determined as in A. The fraction of total Rec12 or Spo11 oligos found within 500 bp of NDRs (dashed lines) is indicated. Additional data are in Supplemental Figure S4. (C) Heatmap of mononucleosome occupancy around S. pombe NDRs. The NDRs from B (top) were ranked by width and the degree of mononucleosome coverage was determined around each site. The coverage profiles were smoothed for clarity and plotted on a color scale (top). NDRs were binned based upon their width for subsequent analysis (right ruler; bin 1, ≤174 bp; bin 2, 175–199 bp; bin 3, 200–249 bp; bin 4, 250–349 bp; bin 5, ≥350 bp). (D) Rec12 oligos are biased toward NDR boundaries and the flanking nucleosomes, not NDR centers. The NDRs within each bin in C were aligned and mean normalized profiles were calculated as in A. The distance between the maxima in the Rec12-oligo peaks in each bin (dashed lines) is indicated at the top. The mean width of NDRs in bin 1 is 162 bp; bin 2, 187 bp; bin 3, 221 bp; bin 4, 287 bp; bin 5, 458 bp.
To examine the spatial distribution of DSBs around these sites, we divided the oligo count at each base pair by the total number of oligos in the 2-kb window centered on an individual NDR or TSS. These normalized maps were then averaged to obtain a single Rec12-oligo profile. This normalization prevents profiles being dominated by the subset of sites with the highest numbers of oligos nearby and allows us to examine the global tendencies of local spatial patterns separate from variation in DSB frequency from one region to another. Importantly, unlike in budding yeast where TSS- or NDR-proximal regions account for a large majority of all Spo11 oligos, these regions in S. pombe account for only a minority fraction of Rec12 oligos (Fig. 5A,B).
Rec12-oligo distributions were spatially correlated with patterns of nucleosome coverage at TSSs and NDRs, with broad zones of enrichment overlapping with the regions with lowest nucleosome coverage (Fig. 5A,B). However, oligos tended to be most highly enriched along the NDR-proximal sides of the nucleosomes flanking TSSs (Fig. 5A) and to an even greater extent at meiotic NDRs (Fig. 5B). Oligos also frequently spilled over into the regions covered by the flanking nucleosomes, such as the typically well-positioned first nucleosomes of transcriptional units. (The particularly high-level enrichment to the left of TSS-associated NDRs in Fig. 5A is likely due to averaging over many large IGRs that include very wide hotspots.) NDR centers do not exhibit biased nucleotide composition relative to the genome, and sequences from them can be mapped as well as or better than random S. pombe sequences (Supplemental Fig. S4); thus, the observed central depletion of Rec12 oligos is not a mapping artifact.
By comparison, normalized Spo11-oligo distributions around S. cerevisiae TSSs and NDRs correlated much more tightly with the regions of MNase-sensitivity and appeared more strongly restricted by the flanking nucleosomes (Fig. 5A,B). Notably, Spo11 oligos within NDRs exhibited a modest central depletion akin to the strong depletion of Rec12 oligos in NDR middles, possibly indicating a shared spatial patterning that is much more pronounced in S. pombe. To better assess how the genome-wide average reflects specific sites, we rank-ordered NDRs by width and divided them into groups of similar-sized NDRs (Fig. 5C,D). Strikingly, differences in NDR widths between groups were matched by a concomitant shift in the spatial pattern of Rec12-oligo density, confirming that Rec12 oligos tend to be strongly enriched across and into the well-positioned nucleosomes flanking the NDRs, with a paucity in the NDR center itself. The group with the narrowest NDRs had widths comparable to those in budding yeast; this group displayed the same spatial bias as for wider NDRs (Fig. 5D), demonstrating that differences between the species are intrinsic to the DSB–chromatin relationship and not simply a consequence of different average NDR widths.
These findings illustrate themes in Rec12-oligo formation with regard to chromatin structure. First, loci largely depleted of nucleosomes are not especially favorable for break formation. Although IGRs containing hotspots often contain an NDR (de Castro et al. 2012), only a minority (∼20%) of Rec12 oligos originate from within NDRs themselves (Fig. 2B) and hotspots can extend several kilobases from an NDR. Indeed, most NDR cores are actually depleted locally for oligos (Fig. 5B,D). Thus, while the transplacement of large DNA fragments containing an NDR can generate an ectopic DSB hotspot (de Castro et al. 2012), the spatial patterning observed here indicates a more complex relationship. Second, Rec12 oligos formed within NDRs tended to be adjacent to the NDR boundary and often arose from the NDR-flanking regions where well-positioned nucleosomes lie. This pattern agrees well with direct comparison of DSBs and MNase cleavage at two hotspots (mbs1 and cds1) previously analyzed by Southern blotting (Hirota et al. 2007). This striking spatial positioning could reflect Rec12 preferentially cleaving DNA near, or exiting, a nucleosome. However, these analyses rely on population averages, so we do not yet know whether a mechanistic relationship exists between Rec12 and histone proteins. Nucleosomes may be absent at the subset of sites or chromatids where the breaks are made, or Rec12 or one of its partner proteins may have a DNA-binding preference similar to that of nucleosomes.
Influence of transcription factor binding on DSBs
Several sequence-specific TFs influence hotspot activity and DSB formation, perhaps through effects on local chromatin organization and/or by direct interaction with the break-forming machinery (Kon et al. 1997; Petes 2001; Steiner and Smith 2005). Although it has been proposed that TFs play a dominant role in shaping and evolving DSB landscapes (Wahls and Davidson 2010; Steiner et al. 2011), it was not previously possible to test this hypothesis in S. pombe. Rec12 oligos allowed us to investigate the genome-wide relationship between TF binding and DSBs.
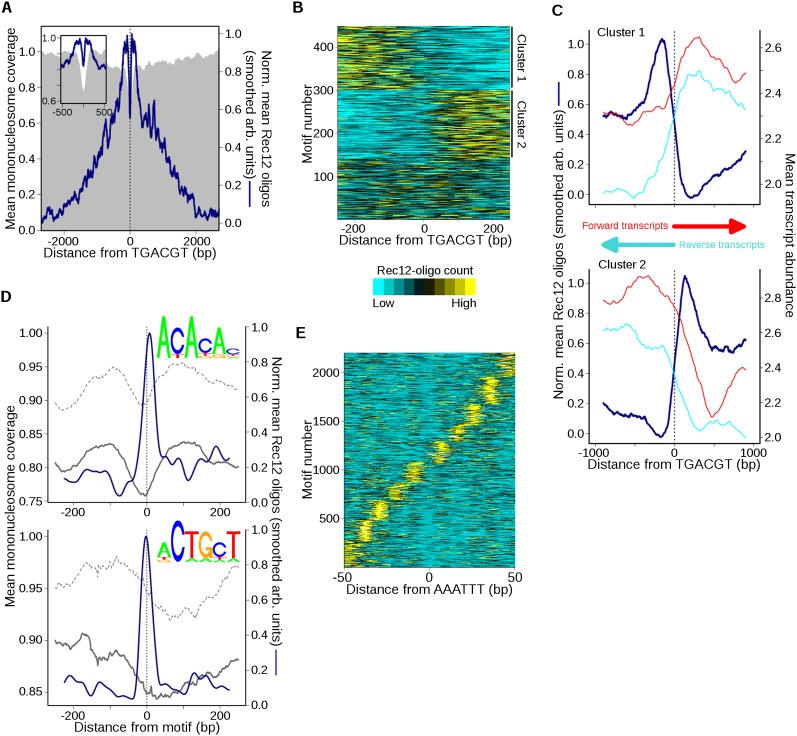
In S. pombe, DNA binding by the CREB-family TF Atf1-Pcr1 alters local chromatin structure and generates DSB hotspots in a context-dependent manner: Transplacement of the ade6-M26 allele, a binding site for Atf1-Pcr1, on 3- to 6-kb DNA fragments generates a hotspot in only a few chromosomal contexts (Ponticelli and Smith 1992; Virgin et al. 1995; Kon et al. 1997; Steiner et al. 2002; Hirota et al. 2008). We searched for the Atf1-Pcr1 core binding motif (5′-TGACGT) (Steiner and Smith 2005) and analyzed Rec12 oligos nearby; similar results were obtained with the more extended sequence 5′-ATGACGT (Schuchert et al. 1991; data not shown). For the 448 motifs (of 2141 total) with at least as many Rec12 oligos nearby as in the weakest hotspot (i.e., ≥140 RPM within ±1 kb), oligos were enriched on average for ∼2 kb on either side and were depleted in the ∼50-bp region centered on the motif (Fig. 6A). (Averages of locally normalized Rec12-oligo profiles are depicted in this and subsequent figures of sequence motifs to separate local spatial patterns from site-to-site variation in total oligo number.) This pattern was more pronounced if we considered only sites within IGRs (Supplemental Fig. S5A). Local depletion over the motif agrees with results of direct DSB detection by Southern blot hybridizations (Steiner et al. 2002) and resembles average patterns around binding sites of the S. cerevisiae TFs Abf1 and Reb1 (Pan et al. 2011).
Figure 6.
Atf1-Pcr1 and select short DNA sequence motifs are spatially correlated with DSBs. (A) Rec12-oligo distribution around Atf1-Pcr1 motifs. Sites with the motif 5′-TGACGT and ≥140 RPM total within 2 kb were oriented and aligned (n = 448). Mean normalized oligo profiles were determined as in Figure 5A and smoothed with a 51-bp Hann window for clarity (inset shows central 1 kb). (B) Atf1-Pcr1 associated oligo distributions are often asymmetric at individual sites. Oligo counts around each site in A were binned, clustered according to oligo distribution (cluster 1 loci have ≥50% more oligos to the left than to the right of the motif; cluster 2 loci have ≥50% more oligos to the right), and plotted as a heatmap. Clustering by k-means reveals a similar pattern (data not shown). (C) Transcription and Rec12 oligos predominate on opposite sides of asymmetric Atf1-Pcr1 motifs. Oligo counts in the clusters from B were averaged after normalization as in Figure 5A and compared with mean meiotic transcript abundance (red and cyan lines) (Chen et al. 2012). Oligos were smoothed with a 201-bp Hann window for clarity. (D) Rec12 oligos around simple sequence motifs. A PWM was constructed for 5′-ACACAC- and 5′-ACTGCT-like motifs (top logos), and genomic positions scoring ≥95% match to a PWM and having ≥50 oligos within the 500-bp window (n = 1021 and 1024, respectively) were identified. Averages of normalized Rec12-oligo distributions at these sites were determined as in Figure 5A and smoothed with a 51-bp Hann window (blue lines). The mean mononucleosome coverage around the aligned motifs during meiotic (solid gray lines) and mitotic (dashed gray lines) growth was then determined (Soriano et al. 2013). (E) Spatial pattern of Rec12 oligos around 5′-AAATTT motifs (n = 2200). Sites with the motif and ≥10 RPM total within 100 bp were aligned, and normalized Rec12-oligo profiles were determined as in Figure 5A. Oligo profiles were clustered by k-means (k = 10) and plotted as in B. The distance between adjacent oligo clusters is ∼10 bp; see Supplemental Figure S6F for the composite mean profile. See Supplemental Figures S5 and S6 for further analyses of sequence motifs associated with Rec12 oligos.
These average Rec12-oligo profiles seemed to show that DSBs tend to be symmetrically arrayed around this TF’s binding sites, and indeed such a pattern was seen at the Atf1-Pcr1-dependent ade6-3049 hotspot (Supplemental Fig. S5B). However, closer examination showed this was often not the case. Instead, about half of motif-containing sites had higher oligo frequencies to one side or the other (Fig. 6B). This asymmetry correlated strongly with nearby transcription such that lowest oligo density occurred in transcribed regions (Fig. 6C), but did not correlate with local chromatin structure (measured by MNase resistance; data not shown). Atf1-Pcr1 binding at some locations may stimulate local DSB formation and transcription in opposite directions, and/or transcription may inhibit DSB formation in the transcribed region. The remaining motif-containing sites had more symmetric oligo distributions and no bias in transcript abundance (data not shown).
The spatial correlation of oligos with Atf1-Pcr1 motifs is tempered by the fact that most motifs were not associated with high Rec12-oligo density: Only 19% were above the genome average, a nonstringent threshold. Even motifs in large IGRs (which generally contain a hotspot) had oligo densities above the large-IGR mean only 27% of the time. Although 28% of all hotspots contained a motif, only 12% of motifs were in hotspots (∼6% of the genome is in hotspots). Thus, the motif alone is a poor predictor of break location or frequency.
Because DNA sequence often does not predict TF binding in vivo, including for Atf1-Pcr1 (Eshaghi et al. 2010), we examined experimentally defined Atf1-Pcr1 binding sites. Atf1-Pcr1 responds to oxidative stress in addition to pre-meiotic nitrogen starvation (Shiozaki and Russell 1996). Since binding in meiosis has not been reported, we analyzed the 250 sites bound by Atf1 during oxidative stress as a proxy (Eshaghi et al. 2010). These sites—but not those bound by Atf1 only prior to oxidative stress—displayed Rec12-oligo profiles similar to that around the Atf1-Pcr1 motif, i.e., central depletion flanked by zones of enrichment (Supplemental Fig. S5C). We found that 49% of the bound sites overlapped a hotspot, but only a minority (19%) of hotspots was bound by Atf1. Note that the number of detectably bound sites is about 10 times fewer than the number of motifs.
Taken together, our findings imply that Atf1-Pcr1 can stimulate DSB formation over a region of ∼4 kb at select sites, with further spatial patterning conferred by other features of the region. In certain contexts, such as the 2.6-kb-wide ade6-3049 hotspot, breaks are induced to both sides. Importantly, most Atf1-Pcr1 motifs are not strong DSB sites, and the vast majority genome-wide are not bound by Atf1-Pcr1 in vivo and/or do not give rise to appreciable DSB activity. These findings fail to support the hypothesis that the Atf1-Pcr1 motif alone, or even the more stringent criterion of Atf1 binding, is sufficient to target high levels of DSB formation nearby (Wahls and Davidson 2010).
Other DSB-associated motifs
Two additional TF-associated motifs were more recently demonstrated to activate DSB hotspots at particular DNA consensus sequences in S. pombe: oligo-C hotspots depend on Rst2, and CCAAT hotspots depend on the Php2–Php3–Php5 complex (Steiner et al. 2011). We therefore assessed Rec12-oligo patterns near positions of their core motifs, 5′-CCCCGC (n = 456) and 5′-CCAATCA (n = 1408). Only 20% of oligo-C and 9% of CCAAT motifs fell within hotspots (Supplemental Fig. S5D); these fractions are higher than expected by chance but represent a small minority of all motif occurrences. Conversely, only 10% and 16% of hotspots contained the respective motif. Thus, as for Atf1-Pcr1 motifs, their presence was a poor hotspot predictor. However, when we aligned these motif-containing sites, Rec12 oligos were spatially correlated: oligos tended to be locally depleted directly at the motif with higher Rec12-oligo frequency in flanking regions (Supplemental Fig. S5E,F). As at Atf1-Pcr1 motif sites (Fig. 6C), Rec12-oligo frequency was anticorrelated spatially with meiotic transcript abundance (data not shown). These findings suggest that, although these TFs probably contribute fairly little to the overall hotspot map, the subset of sites they activate are also subject to spatial patterning conferred by the TF.
Simple sequence motifs associated with nonrandom DSB patterns
To uncover additional DNA motifs associated with nonrandom DSB distributions, we examined Rec12 oligos around each of the 4096 possible DNA hexamers. As expected, most sequences showed no obvious relationship with local oligo distributions (data not shown). However, several motifs were associated with clear spatial patterning of nearby Rec12 oligos, so these were analyzed further.
Two motifs, 5′-ACACAC and 5′-ACTGCT, showed elevated mean Rec12-oligo levels in a ∼60-bp region centered on the motifs (Supplemental Fig. S6A,B). We combined each with single-bp variants that had similar oligo patterns to generate position-weight matrices (PWMs). Like the original motifs, sites matching the PWMs exhibited strong spatial correlations with Rec12 oligos (Fig. 6D). The ACACAC-like (n = 3426) and ACTGCT-like (n = 6565) motifs were found in 36% and 46% of hotspots, respectively; 60% of hotspots contained at least one match. A larger fraction of ACACAC motifs were in hotspots than expected by chance, but the motif was also significantly enriched in IGRs (Supplemental Fig. S6C). Nonetheless, as with other sequence motifs, their presence was a poor predictor of hotspots: Only 15% and 6% of ACACAC and ACTGCT-like sites, respectively, were in hotspots (Supplemental Fig. S6C).
Rec12 oligos were relatively symmetrically distributed around individual ACACAC motifs (data not shown). However, oligos near a subset of ACTGCT sites were asymmetric in a manner anticorrelated with meiotic transcript abundance, analogous to Atf1 motifs (Fig. 6C; Supplemental Fig. S6D,E). Both the ACACAC and ACTGCT motifs exhibited greater MNase sensitivity than genome median, possibly indicating general nucleosome depletion, and even greater sensitivity in meiosis (Fig. 6D, insets). These motifs might represent binding sites for TFs or might simply reflect a DNA context that is preferentially cleaved by Rec12. Databases of known TF binding motifs (e.g., TOMTOM) (Gupta et al. 2007) revealed few significant matches, though the ACACAC motif is similar to the sequence bound by S. cerevisiae Rap1, a contributor to activity of the S. cerevisiae HIS4 DSB hotspot (White et al. 1991).
A more complex relationship with Rec12 oligos was seen for 5′-AAATTT and its subsequence 5′-AATT. When genomic sites with either motif were aligned, the average Rec12-oligo profile exhibited a narrow minimum at the motif center flanked for ≥100 bp on either side by a striking oscillating pattern with ∼10-bp periodicity (Supplemental Fig. S6F), the approximate number of base pairs per turn of B-form DNA. However, individual motif positions did not show the same oscillation seen for the average: Instead, each motif position was associated with a strong Rec12-oligo cluster lying to one side or the other at a distance that was a multiple of ∼10 bp (Fig. 6E). The AAATTT sequence was enriched in hotspots and IGRs (Supplemental Fig. S6C) but is abundant in the genome (n = 19,202) and is not a good predictor of hotspots (only 8% of the motifs are in a hotspot). The DNA around these motifs is more resistant to MNase during vegetative growth, suggesting the presence of a nucleosome and remodeling in meiosis (Supplemental Fig. S6F, bottom). Furthermore, the average nucleosome profile exhibits symmetric features that correlate with the region of periodic breakage (± ∼125 bp). This break pattern is largely absent at sites containing motif variants in which a G or C is located at any position within the motif (data not shown).
Certain DNA sequences are more amenable to wrapping around histone octamers (Segal et al. 2006). In vivo work in budding yeast revealed that nucleosome position stability correlates with AA, AT, TA, and TT dinucleotide frequencies (Brogaard et al. 2012). Notably, recent work in fission yeast found A/T enriched around nucleosome dyads relative to S. cerevisiae and that A/T frequency correlated with increased occupancy (Moyle-Heyrman et al. 2013). To assess a possible correlation between break formation and nucleosome binding around the AAATTT motif, we determined the mean MNase protection of each break cluster in Figure 6E. In each cluster we found that the region of MNase protection remained centered on the motif, indistinguishable from the overall mean profiles in Supplemental Figure S6F (data not shown). Thus, at an individual AAATTT site, Rec12 oligos form at a discrete distance from the motif. When considering all sites as a group, oligos arise from a set of sites that occur at regular intervals extending across the entire MNase-protected region, which itself is centered at each motif. Thus, we hypothesize that this motif is frequently found at nucleosome dyads (perhaps because it can stabilize nucleosome binding), and at individual nucleosome-bound motifs Rec12 preferentially cleaves a particular exposed DNA surface located a multiple of ∼10 bp from the nucleosome dyad. If this is the case, we might expect Rec12 oligos to exhibit base pair preferences similar to that of nucleosomes. A nucleotide-scale analysis of the individual base frequencies around Rec12 oligos revealed a strongly periodic bias with a period of 10 bp and extending for ≥100 bp from the cleavage site (Supplemental Fig. S7; Supplemental Material). Importantly, similar bias was seen for both hotspot and non-hotspot oligos (Supplemental Fig. S7C). This pattern likely reflects Rec12 acting in register with another protein that is influenced by the underlying DNA sequence, such as a nucleosome. Alternatively, this pattern may reflect a complex DNA composition preference shared by Rec12, or its partner proteins, and nucleosomes.
Conclusions
Our results highlight the fluid and probabilistic nature of meiotic DSB formation and recombination. The impression that break formation is a function of numerous hierarchical factors predicts that the majority of the phosphodiester bonds in the genome, and perhaps all, are broken at some frequency (Pan et al. 2011). Our finding of substantial breakage outside hotspots corroborates this view: The probability of a DSB at any one of these sites is low, but combined these DSBs fully account for the recombination that frequently occurs at a distance from hotspots. Our ability to assay exceptionally low-level DSBs solves the paradox of the origin of crossovers in regions without previously detected DSBs (Young et al. 2002); by accounting for nearly half of all crossovers, these low-level DSBs play a much more important role than previously thought. The phenomenon of crossover invariance, related to biased partner choice for DSB repair (Hyppa and Smith 2010), is also affirmed throughout the genome.
Our findings emphasize the importance of studying the mechanisms of meiotic recombination in varied species. Factors acting in concert to determine the break landscape in other species are also observed in S. pombe, reinforcing the fact that DSB formation in fission yeast is one of many variations on a broader theme. For example, DSBs in S. cerevisiae tend to be found near promoters, most often in the associated nucleosome-free regions; in S. pombe breaks can occur close to NDRs but are not preferentially formed there. Our data also suggest that DSBs in S. pombe are biased toward nucleosome boundaries, or at least where nucleosomes are positioned in the population. One possibility is that nucleosomal DNA is freed for Rec12 access by active remodeling of chromatin at DSB sites prior to break formation. Alternatively, nucleosomes may not be as strong a barrier to Rec12 access as previously thought from studies in budding yeast, with Rec12 itself interacting directly with nucleosomes. Such a direct link might in turn suggest that histone modifications can affect Spo11/Rec12 interactions with chromatin more directly, in addition to the known indirect connections via Spo11-accessory proteins (Acquaviva et al. 2013; Sommermeyer et al. 2013). The near-nucleotide resolution mapping of DSB formation in recombination-altered mutants will further clarify underlying molecular mechanisms, while using closely related or hybrid species could illuminate the impact of higher-order genome architecture and the evolution of meiotic DSB hotspots.
Methods
Induction of meiosis and oligo preparation
Haploid and diploid pat1-114 strains were thermally induced for meiosis and monitored for replication by flow cytometry (Cervantes et al. 2000) and for Rec12-FLAG-oligo complexes by Western blotting (Milman et al. 2009). Rec12-FLAG is nearly fully functional for DSB formation and meiotic recombination (Cromie et al. 2007). Cultures were harvested after 4.5 h; cells were resuspended in trichloroacetic acid and broken using glass beads. Cell pellets were solubilized by boiling in the presence of SDS, and Rec12-FLAG was immunoprecipitated using magnetic beads. Eluates were protease digested and DNA was recovered (Milman et al. 2009). Samples were prepared for massively parallel sequencing according to Pan et al. (2011). Two different sequencing platforms were used, and sequencing was carried out according to the manufacturer’s instructions. Rec12-oligo sequence reads were mapped to the S. pombe genome accessed August 2010. Detailed methods are in the Supplemental Material.
Assignment of multiply mapping sequences
The multiply mapping sequences from the SOLiD data set were enriched for very short read lengths (Fig. 1C), indicating that this population contained many oligos that mapped to multiple locations because they were too short rather than because they arose from bona fide repetitive DNA elements. A map incorporating these reads was generated as follows: All potential map sites were considered for each read and the number of uniquely mapping oligos within ±250 bp of each site was determined; for each ambiguous read, a fractional value was assigned to each of its map positions proportional to the number of unique oligos in the vicinity. For example, a read that mapped to two sites, one with 90 unique reads nearby and the other with 10, would be assigned to these sites as 0.9 and 0.1 oligos, respectively; see Supplemental Figure S1E for an actual example. This map, in which we have imputed each read’s originating locus, was added to the unique data to generate a combined map. The three maps (unique, multiple-mappers, and combined) were analyzed as described in the text. Sequences that mapped to two or more positions from the 454 data set were not analyzed further.
Comparison with recombination frequencies
To further validate our map and to explore the relationship between DSB formation and recombination, we used previously published recombinant frequencies (Supplemental Table S4). Most of the genetic map lengths (cM) were calculated from random spore analyses using standard auxotrophic markers, while recombination in the ade3–ura3 interval was determined by tetrad analysis and in the ade6L–ade6R by a physical measure of crossover DNA. Genetic distances were calculated from random spore analysis as cM = −50 ln(1 – 2R), where R is the fraction of recombinant spore colonies among total spore colonies analyzed; the distance for tetrad analysis was cM = 100 (T + 6NPD)/2 (PD + NPD + T), where T, NPD, and PD are the fractions of tetratype, nonparental ditype, and parental ditype asci among total asci analyzed. Crossover DNA at the ade6–3049 hotspot was measured using diploids with heterozygous restriction sites flanking the hotspot and Southern blotting to probe for the appearance of recombinant DNA fragments (Hyppa and Smith 2010). Since only one of the two recombinant DNA molecules (R2) was assayed, genetic distance was calculated as cM = 2 × (R2/total DNA), where R2 is the mean of the accumulated recombinant DNA at 6 and 7 h in a meiotic induction (R1 could arise by partial restriction digestion).
Chromatin structure analysis
Transcription start sites (TSS) in S. pombe were from previously published studies (Lantermann et al. 2010). Only TSSs with annotated start and end positions were used. Positions previously defined as depleted of nucleosomes (NDR) during meiosis in S. pombe (constitutive and meiosis-specific NDRs) were compiled from Soriano et al. (2013). The spatial distribution of Rec12 oligos around TSSs and NDRs was determined as in the analysis of sequence motifs described above. Comparisons with Spo11 oligos in S. cerevisiae used previously defined TSSs and NDRs (Jiang and Pugh 2009), and the curated S288C Spo11-oligo map and “N2” 4-h nucleosome map from Pan et al. (2011).
To allow direct comparisons between nucleosomes and Rec12 and Spo11 oligos, we used the MNase-seq data from Soriano et al. (2013) to derive mononucleosome coverage maps. Our primary interest was to determine where Rec12 oligos arise relative to DNA that resides in nucleosomes, so we wished to use nucleosome coverage maps (as in Pan et al. 2011) rather than the maps of nucleosome midpoints used by Soriano et al. (2013). We therefore mapped their raw sequence reads (GEO accession GSE41772) to the same S. pombe reference genome used above in mapping Rec12 oligos. For meiotic mononucleosome coverage we used the data set from 3 h after a synchronous pat1-114 meiotic induction (accession number GSM1024001); our analyses of mitotic coverage used data derived from asynchronous cultures grown in rich media (GSM1024003).
Since in the preparation of these libraries DNA was size selected to enrich for mononucleosome-sized fragments, sequence tags should map to the boundaries of nucleosomes on opposite strands. Since the tag positions are a function of nucleosome position and the degree of MNase digestion, we first analyzed the pairwise distances between sequence tags on the plus and minus DNA strands to estimate the degree of MNase digestion. Under ideal conditions the most frequent distance should be ∼147 bp, the length of DNA typically protected from MNase by the nucleosome. However, we found that the most frequent tag distance was only ∼127 bp (data not shown). We assume this is due to ∼10 bp being “nibbled” by MNase from each end of nucleosomal DNA, so we generated coverage maps by computationally extending the 5′ end of each sequence read 10 bp to its 5′ side and 137 bp to its 3′ side (147 bp total), representing the inferred nucleosome footprint, and then calculating the number of occurrences of each base in the genome. These coverage maps agreed well with coverage maps generated by computationally extending the nucleosome midpoint maps of Soriano et al. (2013) by 73 bp in each direction (data not shown). Since the maps of Soriano and colleagues have been subjected to extensive smoothing, we elected to use our remapped version of their data instead. Note that using coverage maps instead of midpoint maps results in more rounded peaks and narrower internucleosomal linker regions in profiles of individual sites (e.g., Fig. 4A) or averages over many sites (e.g., Fig. 5A). Importantly, however, this method of analysis makes the S. pombe data directly comparable to the S. cerevisiae data. The maps were normalized by dividing the coverage at each position by the average coverage genome-wide (the genome average ≡ 1).
Other software
Data analysis utilized R (http://www.r-project.org/) and Bioconductor (http://www.bioconductor.org/). Further details are shown in the Supplemental Material.
Data access
The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE49977.
Supplementary Material
Acknowledgments
We are especially grateful to Peter Schlögelhofer (University of Vienna) and Michael Eickbush and Sarah Zanders (FHCRC) for sharing unpublished data. We thank Agnes Viale (MSKCC Genomics Core Laboratory) for sequencing; Nick Socci (MSKCC Bioinformatics Core Facility) for assistance with sequencing data processing and mapping; Sam Tischfield (MSKCC) for assistance with data analysis; and Sarah Zanders, Naina Phadnis, Sue Amundsen (FHCRC), and Shintaro Yamada (MSKCC) for helpful comments on the manuscript. This work was supported in part by NIH grants GM031693 and GM032194 (G.R.S.) and GM058673 (S.K.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.172122.114.
References
- Acquaviva L, Szekvolgyi L, Dichtl B, Dichtl BS, de La Roche Saint Andre C, Nicolas A, Geli V. 2013. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339: 215–218 [DOI] [PubMed] [Google Scholar]
- Borde V, Robine N, Lin W, Bonfils S, Geli V, Nicolas A. 2009. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J 28: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485: 642–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogaard K, Xi L, Wang JP, Widom J. 2012. A map of nucleosome positions in yeast at base-pair resolution. Nature 486: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Farah JA, Smith GR. 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell 5: 883–888 [DOI] [PubMed] [Google Scholar]
- Chen HM, Rosebrock AP, Khan SR, Futcher B, Leatherwood JK. 2012. Repression of meiotic genes by antisense transcription and by Fkh2 transcription factor in Schizosaccharomyces pombe. PLoS ONE 7: e29917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D, Kuan PF, Li B, Sanalkumar R, Liang K, Bresnick EH, Dewey C, Keles S. 2011. Discovering transcription factor binding sites in highly repetitive regions of genomes with multi-read analysis of ChIP-Seq data. PLoS Comput Biol 7: e1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Smith GR. 2008. Meiotic recombination in Schizosaccharomyces pombe: a paradigm for genetic and molecular analysis. In Recombination and meiosis: models, means, and evolution (ed. Egel R, Lankenau D-H), pp. 195–230. Springer-Verlag, Berlin, Germany [Google Scholar]
- Cromie GA, Rubio CA, Hyppa RW, Smith GR. 2005. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics 169: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. 2006. Single Holliday junctions are intermediates of meiotic recombination. Cell 127: 1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Cam HE, Farah JA, Grewal SHIS, Smith GR. 2007. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet 3: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E, Soriano I, Marin L, Serrano R, Quintales L, Antequera F. 2012. Nucleosomal organization of replication origins and meiotic recombination hotspots in fission yeast. EMBO J 31: 124–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R. 2003. Fission yeast in general genetics. In The molecular biology of Schizosaccharomyces pombe: genetics, genomics and beyond (ed. Egel R), pp. 1–12. Springer, Berlin, Germany [Google Scholar]
- Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G, Smith GR. 2010. RNAi and heterochromatin repress centromeric meiotic recombination. Proc Natl Acad Sci 107: 8701–8705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi M, Lee JH, Zhu L, Poon SY, Li J, Cho KH, Chu Z, Karuturi RK, Liu J. 2010. Genomic binding profiling of the fission yeast stress-activated MAPK Sty1 and the bZIP transcriptional activator Atf1 in response to H2O2. PLoS ONE 5: e11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KR, Gutiérrez-Velasco S, Martín-Castellanos C, Smith GR. 2013. Protein determinants of meiotic DNA break hotspots. Mol Cell 49: 983–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Bahler J, Kohli J. 1994. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics 135: 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopolous JA, Bailey TL, Noble WS. 2007. Quantifying similarity between motifs. Genome Biol 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Steiner WW, Shibata T, Ohta K. 2007. Multiple modes of chromatin configuration at natural meiotic recombination hot spots in fission yeast. Eukaryot Cell 6: 2072–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Mizuno K, Shibata T, Ohta K. 2008. Distinct chromatin modulators regulate the formation of accessible and repressive chromatin at the fission yeast recombination hotspot ade6-M26. Mol Biol Cell 19: 1162–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppa RW, Smith GR. 2010. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell 142: 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppa RW, Cromie GA, Smith GR. 2008. Indistinguishable landscapes of meiotic DNA breaks in rad50+ and rad50S strains of fission yeast revealed by a novel rad50+ recombination intermediate. PLoS Genet 4: e1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Jeffreys AJ, Keeney S. 2004. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet 5: 413–424 [DOI] [PubMed] [Google Scholar]
- Keeney S. 2007. Spo11 and the formation of DNA double-strand breaks in meiosis. In Recombination and meiosis: crossing-over and disjunction (ed. Egel R, Lankenau D-H), pp. 81–123. Springer-Verlag, Berlin, Germany [Google Scholar]
- Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP. 1997. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc Natl Acad Sci 94: 13756–13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S. 2011. ATM controls meiotic double-strand-break formation. Nature 479: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. 2010. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol 17: 251–257 [DOI] [PubMed] [Google Scholar]
- Lichten M. 2008. Meiotic chromatin: The substrate for recombination initiation. In Recombination and meiosis: models, means, and evolution (ed. Egel R, Lankenau D-H), pp. 165–193. Springer-Verlag, Berlin, Germany [Google Scholar]
- Milman N, Higuchi E, Smith GR. 2009. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol Cell Biol 29: 5998–6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle-Heyrman G, Zaichuk T, Xi L, Zhang Q, Uhlenbeck OC, Holmgren R, Widom J, Wang JP. 2013. Chemical map of Schizosaccharomyces pombe reveals species-specific features in nucleosome positioning. Proc Natl Acad Sci 110: 20158–20163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz P. 1994. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics 137: 701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K, Shibata T, Nicolas A. 1994. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J 13: 5754–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, et al. 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD. 2001. Meiotic recombination hot spots and cold spots. Nat Rev Genet 2: 360–370 [DOI] [PubMed] [Google Scholar]
- Petes TD, Botstein D. 1977. Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc Natl Acad Sci 74: 5091–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF, Nasmyth K. 2003. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112: 423–440 [DOI] [PubMed] [Google Scholar]
- Ponticelli AS, Smith GR. 1992. Chromosomal context dependence of a eukaryotic recombinational hot spot. Proc Natl Acad Sci 89: 227–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg M, Kohli J, Ludin K. 2009. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet 5: e1000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S. 2010. Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol 11: 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert P, Langsford M, Käslin E, Kohli J. 1991. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J 10: 2157–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. 2006. A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K, Russell P. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev 10: 2276–2288 [DOI] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermeyer V, Beneut C, Chaplais E, Serrentino ME, Borde V. 2013. Spp1, a member of the Set1 complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell 49: 43–54 [DOI] [PubMed] [Google Scholar]
- Soriano I, Quintales L, Antequera F. 2013. Clustered regulatory elements at nucleosome-depleted regions punctuate a constant nucleosomal landscape in Schizosaccharomyces pombe. BMC Genomics 14: 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner WW, Smith GR. 2005. Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics 169: 1973–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner WW, Steiner EM. 2012. Fission yeast hotspot sequence motifs are also active in budding yeast. PLoS ONE 7: e53090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner WW, Schreckhise RW, Smith GR. 2002. Meiotic DNA breaks at the S. pombe recombination hotspot M26. Mol Cell 9: 847–855 [DOI] [PubMed] [Google Scholar]
- Steiner WW, Davidow PA, Bagshaw AT. 2011. Important characteristics of sequence-specific recombination hotspots in Schizosaccharomyces pombe. Genetics 187: 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield SE, Keeney S. 2012. Scale matters: the spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell Cycle 11: 1496–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G, Blitzblau HG, Tame MA, Falk JE, Curtin L, Hochwagen A. 2011. Protection of repetitive DNA borders from self-induced meiotic instability. Nature 477: 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin JB, Metzger J, Smith GR. 1995. Active and inactive transplacement of the M26 recombination hotspot in Schizosaccharomyces pombe. Genetics 141: 33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP, Davidson MK. 2010. Discrete DNA sites regulate global distribution of meiotic recombination. Trends Genet 26: 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Wierdl M, Detloff P, Petes TD. 1991. DNA-binding protein RAP1 stimulates meiotic recombination at the HIS4 locus in yeast. Proc Natl Acad Sci 88: 9755–9759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TC, Lichten M. 1994. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263: 515–518 [DOI] [PubMed] [Google Scholar]
- Yamada T, Ohta K. 2013. Initiation of meiotic recombination in chromatin structure. J Biochem 154: 107–114 [DOI] [PubMed] [Google Scholar]
- Yamada S, Ohta K, Yamada T. 2013. Acetylated histone H3K9 is associated with meiotic recombination hotspots, and plays a role in recombination redundantly with other factors including the H3K4 methylase Set1 in fission yeast. Nucleic Acids Res 41: 3504–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Schreckhise RW, Steiner WW, Smith GR. 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell 9: 253–263 [DOI] [PubMed] [Google Scholar]
- Zhang L, Ma H, Pugh BF. 2011. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res 21: 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.