Abstract
The organization of the pre-mRNA splicing machinery has been extensively studied in mammalian and yeast cells and far less is known in living plant cells and different cell types of an intact organism. Here, we report on the expression, organization, and dynamics of pre-mRNA splicing factors (SR33, SR1/atSRp34, and atSRp30) under control of their endogenous promoters in Arabidopsis. Distinct tissue-specific expression patterns were observed, and differences in the distribution of these proteins within nuclei of different cell types were identified. These factors localized in a cell type-dependent speckled pattern as well as being diffusely distributed throughout the nucleoplasm. Electron microscopic analysis has revealed that these speckles correspond to interchromatin granule clusters. Time-lapse microscopy revealed that speckles move within a constrained nuclear space, and their organization is altered during the cell cycle. Fluorescence recovery after photobleaching analysis revealed a rapid exchange rate of splicing factors in nuclear speckles. The dynamic organization of plant speckles is closely related to the transcriptional activity of the cells. The organization and dynamic behavior of speckles in Arabidopsis cell nuclei provides significant insight into understanding the functional compartmentalization of the nucleus and its relationship to chromatin organization within various cell types of a single organism.
INTRODUCTION
Pre-mRNA (pre-mRNA) splicing, a process of excision of introns and ligation of exons, is essential for generating mature transcripts and enhancing mRNA export from the nucleus to the cytoplasm. Splicing occurs within a large macromolecular complex called the spliceosome, which is composed of U1, U2, U4/U6, and U5 small nuclear ribonucleoprotein particles and a large number of non-small nuclear ribonucleoprotein particle proteins (Zhou et al., 2002). The latter include members of the SR (Ser-Arg) family of pre-mRNA splicing factors characterized by one or two N-terminal RNA recognition motifs that interact with the pre-mRNAs and a C-terminal variable-length Arg/Ser(RS)-rich domain that influences its subcellular localization and splicing activity depending upon its phosphorylation levels (for review, see Graveley, 2000).
In mammalian cells, pre-mRNA splicing factors are concentrated in 20–50 distinct nuclear domains called speckles as well as being diffusely distributed throughout the nucleoplasm, and this distribution corresponds to interchromatin granule clusters (IGCs) and perichromatin fibrils (PFs). PFs often extend from the periphery of IGCs or are present in the nucleoplasm at some distance from an IGCs (for review, see Lamond and Spector, 2003). It has been proposed that IGCs are storage and/or modification/reassembly sites for pre-mRNA splicing factors and that splicing factors are recruited from these compartments to the sites of active transcription (PFs) (Jiménez-García and Spector, 1993). Disassembly of IGCs by overexpression of an SR protein kinase Clk/STY disrupts the coordination of transcription and pre-mRNA splicing, demonstrating the functional importance of the speckled organization of splicing factors in nuclei of mammalian cells (Sacco-Bubulya and Spector, 2002). It is not clear whether the observed organization and dynamics of SR proteins in mammalian cultured cells is representative to that found in different cell types of a single organism. To address the question, we studied the organization and dynamics of member of SR splicing factors within different cell types of the flowering plant Arabidopsis thaliana.
Interestingly, the Arabidopsis genome encodes 18 SR proteins, in comparison with 10 in human and two in Schizosaccharomyces pombe (Lopato et al., 2002; Lorkovic and Barta, 2002). Two SF2/ASF-like splicing factors (SR1/atSRp34 and atSRp30) and a novel family of SC35-like splicing factors (atSCL28, atSCL30, atSCL30a, SR33/atSCL33, and atSC35), as well as a family of zinc knuckle-containing SR proteins (atRSZp21, atRSZp22, and atRSZp22a) similar in their activities to human 9G8 and SRp20 have been characterized with regard to their splicing and RNA binding activities (Lazar et al., 1995; Golovkin and Reddy, 1998, 1999; Lopato et al., 1999a,b, 2002; Lorkovic and Barta, 2002). AtSRZ21, atSRZ22, SR33, and SR45 have also been shown to interact with a plant U1-70K protein (Golovkin and Reddy, 1998, 1999). Furthermore, members of a plant-specific family of SR proteins termed the atRSp31 family (atRSp31, atRSp40, and atRSp41) were isolated (Lopato et al., 1996). More recently, a plant-specific SR protein atRSZ33 was shown to interact with SR1/atSRp34, atRSZp21, atSRZp22 and SC35-like splicing factors (Lopato et al., 2002).
In plants, spliceosomal components were found to localize in Cajal bodies (Beven et al., 1995; Boudonck et al., 1998, 1999; Acevedo et al., 2002; Cui and Moreno Díaz de la Espina, 2003; Docquier et al., 2004) and in nuclear speckles (Lopato et al., 2002; Ali et al., 2003; Docquier et al., 2004). However, only the plant-specific SR proteins were studied in living cells from a limited number of cell types, and the expression of green fluorescent protein-tagged SR proteins were under the control of a highly active constitutive 35S promoter (Lopato et al., 2002; Ali et al., 2003; Docquier et al., 2004). To gain more insight into the expression and dynamic organization of SR splicing factors in a single organism, we visualized the expression and dynamic organization of SF2/ASF and SC35-like splicing factors under the control of their endogenous promoters in Arabidopsis by deconvolution and confocal microscopy. Distinct tissue-specific expression patterns were observed, and in addition, differences in the distribution of the respective proteins within the nuclei of different cell types were identified. Fluorescence recovery after photobleaching (FRAP) analysis revealed that these factors have a rapid exchange rate in nuclear speckles. Furthermore, we found that both the organization and dynamics of SR splicing factors are related to the transcriptional activity of the cells.
MATERIALS AND METHODS
Constructs
Upstream regulatory sequence of atSRp30 (At1g09140) (Lopato et al., 1999b), SR1/atSRp34 (At1g02840) (Lazar et al., 1995; Lopato et al., 1999b), and SR33 (At1g55310) (Golovkin and Reddy, 1999) plus their complete 5′ untranslated regions was amplified by polymerase chain reaction (PCR) from genomic DNA of A. thaliana (ecotype, Columbia); primers for SR33 promoter were 5′-NNNAAGCTTCATTATAAGATAAACCTTCTAG-3′ and 5′-NNNGGATCCTGAGTCAAGCTTCAATCTCTC-3′; primers for SR1/atSRp34 promoter were 5′-NNNAAGCTTAAATATTGAACCGGCCTCGGTTC-3′ and 5′-NNNGGATCCTCTTCCTTTATCAAATCC-3′; and primers for atSRp30 promoter were 5′-NNNAAGCTTTAATTCTTAGATTCTACAATC-3′ and 5′-NNNGGATCCCTGATACCTCAGAGCAGAAA-3′. The amplified promoter fragments containing a HindIII site at their 5′ end and a BamHI site at their 3′ end were digested with HindIII and BamHI. For the SR33 promoter region with an extra HindIII site, partial HindIII digestion and complete BamHI digestion were applied to clone the intact promoter sequence.
SR33 coding sequence was amplified from a SR33 cDNA clone (Golovkin and Reddy, 1999) by using primers 5′-NNNGGATCCATGAGGGGAAGGAGCTACACT-3′ and 5′-NNNACCGGTCGCTGGCTTGGTGAACGGTCTTC-3′; SR1/atSRp34 coding sequence was amplified from pUC35S-SR1nos (Lazar et al., 1995) by using primers 5′-NNNGGATCCATGAGCA GTCGTTCGAGTAGA-3′ and 5′-NNNACCGGTCGCCTCGATGGACTCATAGTGTG-3′; atSRp30 coding sequence was amplified from pC30 (Lopato et al., 1999b) by using primers 5′-NNNGGATCCATGAGTAGCCGATGGAATCGT-3′ and 5′-NNNACCGGTCGACCAGATATCACAGGTGAAAC-3′. The amplified coding sequences containing a BamHI site at their 5′ end and an AgeI site at their 3′ end were digested with BamHI and AgeI.
The coding sequence of yellow fluorescence protein (YFP) was amplified from pYFP-N1 (BD Biosciences Clontech, Palo Alto, CA) by using primers 5′-NNNACCGGTCGGAGGTGGAGGTGGAGCTGTGAGCAAGGGCGAGGAGCTG-3′ and 5′-NNNGAGCTCTTACTTGTACAGCTCGTCCAT-3′; a flexible linker Gly-Gly-Gly-Gly-Gly-Ala was added at the N terminus of YFP. The terminator used was nopaline synthase (Nos) terminator, which was cut from pBI121 (Jefferson et al., 1987) by restriction enzymes SacI and EcoRI.
These proof-reading–amplified fragments were confirmed by sequencing and then directionally subcloned into the HindIII/EcoRI digested vector pCambia2300 (CAMBIA, Canberra, Australia) with the direction of Hin-dIII-SR protein promoter-BamHI-SR protein coding sequence-AgeI-Gly5-Ala linker-YFP-SacI-Nos terminator-EcoRI to obtain the final binary vectors. A control protein YFP-tetR-NLS (tetR, tetracycline repressor protein; NLS, nuclear localization signal) was placed under the control of pSR1/atSRp34, and then subcloned into the pCambia2300 vector.
Plant Transformation
The constructs described above were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. A. thaliana (ecotype Columbia) plants were transformed by the floral dip method (Clough and Bent, 1998). Independent T1 transgenic lines were selected on MS medium (Murashige and Skoog, 1962) containing 50 mg/l kanamycin. Transgenic lines carrying one copy of transgene were used for imaging. Plants were grown in a growth chamber under 16-h light/8-h dark conditions at 23°C.
Live Cell Imaging
For time-lapse imaging of root, transgenic T2 seeds were sterilized for 15 min in 15% sodium hypochlorite. Five to eight seeds were sown onto a LabTek II one-well chambered coverglass system (Nalge Nunc International, Naperville, IL) containing MS medium supplemented with 2% sucrose and 0.3% Gelrite (Sigma-Aldrich, St. Louis, MO). After the primary root grows down-ward and then along the No. 1.5 borosilicate coverglass, the roots that tightly contacted the coverglass were visualized on the stage of an inverted microscope.
Inflorescence stems, anthers, and rosette leaves (∼0.5 × 0.5 cm2) were cut and mounted in water between two 50 × 24-mm No. 1.5 coverslips and immediately used for imaging.
Confocal Laser Scanning Microscopy (CLSM)
An LSM510 confocal laser scanning microscope and a 10× apochromat objective (Carl Zeiss, Thornwood, NY) was used to visualize the expression of SR protein-yellow fluorescent protein fusion proteins in primary roots, inflorescence stems, leaves, flowers, and young siliques from transgenic T2 Arabidopsis plants. YFP fluorescence was monitored using a 505- to 550-nm band pass emission filter and 488-nm excitation line of an Ar/Kr laser. Eighteen optical sections with a z step of 2.0 μm for inflorescence stems, and 16 optical sections with a z step of 1.5 μm for anthers, roots, and leaves were collected and projected as final images.
Transmission Electron Microscopy (TEM)
Five-day old Arabidopsis seedlings were fixed in 2% formaldehyde/0.2% glutaraldehyde in phosphate-buffered saline (pH 7.4). The samples were dehydrated and embedded in LR White resin (Electron Microscopy Sciences, Fort Washington, PA). Embedded roots were thin sectioned (100 nm). Immunogold labeling was performed for SR splicing factors by incubating sections in monoclonal antibody 3C5 (Turner and Franchi, 1987), rinsing off unbound antibody before incubating the sections in goat anti-mouse IgM conjugated to 5-nm colloidal gold (Amersham Biosciences, Piscataway, NJ). After a rinse in distilled water, the sections were air dried and counterstained with uranyl acetate. Sections were examined using a Hitachi H-7000 TEM operated at 75 kV.
Fluorescence Deconvolution Microscopy
Three-dimensional image stacks of nuclei were acquired at room temperature with a DeltaVision restoration microscope system (Applied Precision, Issaquah, WA) consisting of an IX70 inverted microscope (Olympus, Tokyo, Japan) equipped with an UPLANAPO water immersion objective lens (60×, numerical aperture 1.20; Olympus), and a Photometrics (Roper Scientific, Trenton, NJ) cooled charged-coupled device camera configured at 0.1122 μm/pixel. Filters used for YFP were exciter, 495/20 nm/nm; emitter: 535/30 nm/nm; and 86006bs beamspitter (Chroma Technology, Brattleboro, VT). The image stacks of nuclei with a Z step size of 0.15 μm were subjected to constrained iterative (15 iterations) deconvolution (Chen et al., 1996) by using softWoRx software (Applied Precision) on an Octane Workstation (SGI, Mountain View, CA). The maximum projection or the middle optical section of the deconvolved sections were exported to TIF format and then processed by Adobe Photoshop 6.0 for the final images. The signal intensities, nuclear domain sizes or volumes were measured using softWoRx software.
FRAP of pre-mRNA Splicing Factors
A region of leaf (∼ 0.5 × 0.5 cm2) was mounted in water and observed using an LSM 510 confocal CLSM with a C-Apochromat 40×/1.20 objective lens (Carl Zeiss). The nuclei were excited with a laser at 488 nm, and emission was collected using a 505-to 530-nm-long-pass filter. After five single scans were acquired at a resolution of 128 by 128 pixels, a selected region of fixed size (1.7 × 1.7 μm2) in the nucleus embracing a speckle was bleached at a set laser power of 15 mW for 40 iterations. A series of images were collected immediately after photobleaching. Signal intensity was quantified using LSM software (Carl Zeiss) or softWorx (Applied Precision). The average background signal intensity was measured from images of wild-type plants (Kato et al., 2002). The relative fluorescence intensity (RFI) at each time point was calculated as described previously (Phair and Misteli, 2000) from the background-subtracted images. RFI = (FIBt/FINt)/(FIB0/FIN0), where FIBt is the average fluorescence intensity of the photobleached region at time points after photobleaching, FINt is the average fluorescence intensity of the entire nucleus at the corresponding time point, FIB0 is the average fluorescence intensity of the photobleached region before photobleaching, and FIN0 is the average fluorescence intensity of the entire nucleus before photobleaching.
Drug Treatment
Ten-day-old transgenic T2 seedlings or rosette leaves were immersed in 50 μg/ml 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) for 5 h at 22°C. After treatment, the leaves or seedlings were washed with water and visualized using the DeltaVision restoration microscope system described above.
RESULTS
Expression Patterns of the SR Proteins in Arabidopsis Plants
To visualize the expression patterns of SR proteins in various cell types of Arabidopsis plants, we fused two SF2/ASF-like proteins (atSRp30 and SR1/atSRp34) and the SC35-like protein SR33 with YFP. The expression of each protein was placed under the control of its endogenous upstream regulatory sequence, including the promoter and 5′ untranslated region. A Gly5-Ala flexible linker was added between the splicing factors and YFP to improve their stability and folding. Transgenic plants expressing these fusions grow normally without any obvious phenotypic changes. Using this approach we were able to detect the expression of these SR proteins in living Arabidopsis plants with high spatial sensitivity and were able to follow the same plant at different developmental stages by using CLSM and fluorescence deconvolution microscopy. Signal intensities in various nuclear regions were measured from images collected under the same microscopic settings and normalized to the same exposure time for comparison of expression levels. The nuclei in tissues can be easily distinguished irrespective of cell density or expression levels among different cell types. Several independent transgenic lines expressing each construct were analyzed to exclude the influences of the transgene locations within the genome on the promoter activity.
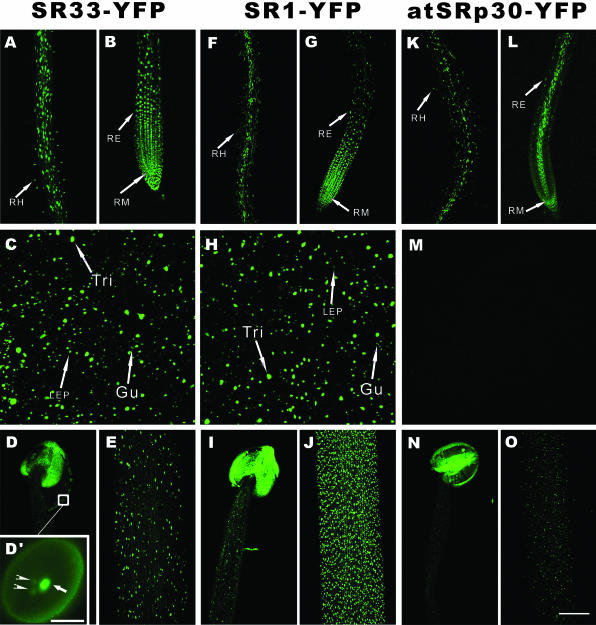
Distinct but also overlapping expression and nuclear localization patterns were observed among the three splicing factors (Figure 1). In the section of primary root near the root tip, SR33-YFP (Figure 1B) and SR1-YFP (Figure 1G) showed signals in the vascular bundle, endodermis, cortex, epidermis, and lateral root cap, whereas atSRp30-YFP was expressed mainly in the vascular bundle, lateral root cap and some epidermal cells (Figure 1L). In the differentiated basal section of the primary root, SR33-YFP (Figure 1A) and atSRp30-YFP (Figure 1K) were distributed within all cell types with the expression level of SR33-YFP higher than atSRp30-YFP, whereas SR1-YFP was expressed more highly in vascular bundles (Figure 1F). The nuclei of three distinct cell types in root, root meristematic cells (RM), root epidermal cells (RE), and root hairs (RH), are highlighted with arrows (Figure 1, A and B, F and G, K and L). In leaves, SR33-YFP (Figure 1C) shows a similar expression pattern to SR1-YFP (Figure 1H). However, the expression of atSRp30-YFP in leaves is weak and below the detection sensitivity of CLSM at these equalized settings (Figure 1M). The nuclei of three distinct cell types in leaf epidermis, guard cells (Gu), leaf epidermal pavement cells (LEP), and trichomes (Tri), are highlighted with arrows (Figure 1, C and H) (Melaragno et al., 1993).
Figure 1.
Expression and nuclear localization patterns of SR33-YFP, SR1/atSRp34-YFP, and atSRp30-YFP in Arabidopsis. Projections of a series of confocal optical sections of inflorescence stems, anthers, roots, and leaves are shown. (A, F, and K) In the basal section of primary root. (B, G, and L) In the tip section of primary root. (C, H, and M) In leaf. (D, I, and N) In anther (D′, enlarged view of a pollen grain, arrow highlights the vegetative nucleus and arrowheads highlight the nuclei of sperm cells; bar, 10 μm). (E, J, and O) In inflorescence stem. Different cell types are highlighted with arrows: Gu, guard cells; LEP, larger (C) and small (H) leaf epidermal pavement cells; RE, root epidermal cells; RH, root hairs; RM, root meristematic cells; Tri, trichomes. Bar, 100 μm.
In inflorescence stems, the strongest signals were observed in SR1-YFP–expressing plants (Figure 1J), whereas SR33 (Figure 1E) is stronger than that of atSRp30 (Figure 1O). In flowers, all three SR proteins have high expression in the pollen grains (Figure 1, D, I, and N) with signals in vegetative nuclei (Figure 1D′, arrow) much stronger than that observed in nuclei of sperm cells (Figure 1D′, arrowheads). In sepals, petals, and young siliques, SR33 and SR1 are much more highly expressed than atSRp30 (our unpublished data). After flowering, the expression level of both SR33-YFP and SR1-YFP decreases gradually; however, SR1 decreases faster than SR33, so at the later flowering stage, the expression of SR33-YFP is stronger than that of SR1-YFP. The differences of promoter activities of SR33 and SR1/atSRp34 were also shown by tobacco infiltration experiments. Strong transient expression activity of the SR33 promoter and weak activity of the SR1/atSRp34 promoter in tobacco leaves were observed (our unpublished data).
Organization of the SR Proteins in Arabidopsis Nuclei
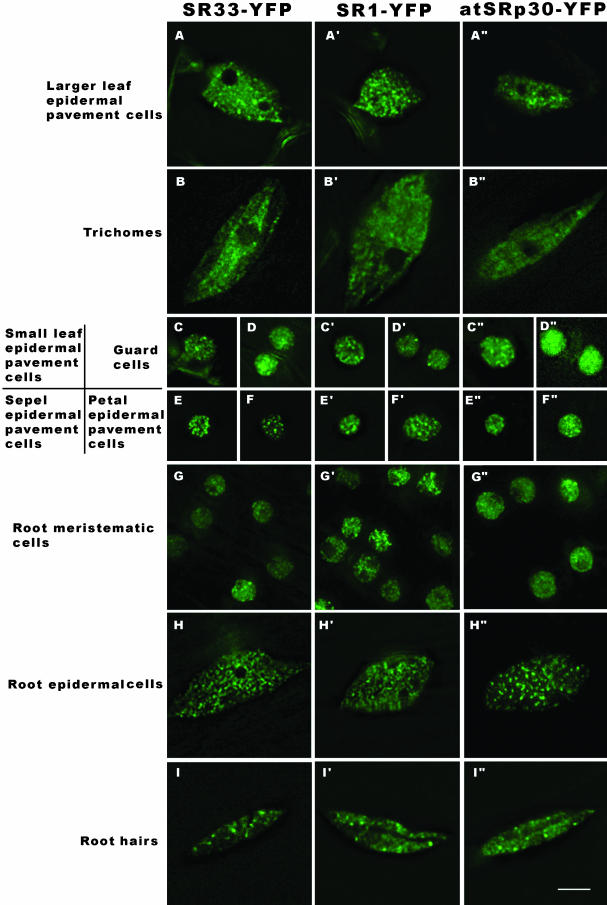
We generated transgenic Arabidopsis plants carrying individual YFP-tagged SR proteins under the control of their endogenous promoters such that the observed distribution patterns were reflection of the respective tissue and cell types. In addition, fluorescence deconvolution microscopy was used to resolve the detailed organization of SR proteins in living Arabidopsis cell types with various ploidy levels and functions. Maximum projection images of z-stacks are shown in Figure 2. Although we have not directly evaluated whether the three SR proteins colocalize in nuclei, similar localization patterns were observed in most of the cell types. The SR protein-YFP fusions localize in brightly labeled nuclear speckles varying in size, from 0.4 to 1.5 μm in diameter, in addition to a diffuse distribution throughout the nucleoplasm. Differences exist among cell types with regard to speckle number, size, density, and ratio of signal intensities between speckles and the nucleoplasm (Figure 2). We also observed a positive relationship between cellular size, nuclear volume, and ploidy level, similar to previous findings (Baluska, 1990; Melaragno et al., 1993; Kato and Lam, 2003).
Figure 2.
Localization of SR33-YFP, SR1/atSRp34-YFP, and atSRp30-YFP in different Arabidopsis cell types. Maximum projections of deconvolved optical sections are shown. The localization patterns of the three SR-YFP fusions are different among cell types with variable nuclear sizes and shapes. Bar, 5 μm.
Small LEP (Figure 2, C, C′, and C″), sepal (Figure 2, E, E′, and E″), and petal (Figure 2, F, F′, and F″) epidermal pavement cells have the same 2C ploidy level as guard cells (Figure 2, D, D′, and D″) and RM (Figure 2, G, G′, and G″) (Melaragno et al., 1993), but the latter have more diffuse signals of the expressed splicing factors. The localization patterns are also different in certain cell types for the three splicing factors; for example, in small leaf epidermal, sepal, and petal epidermal pavement cells, atSRp30 (Figure 2, C″, E″, and F″) is more diffuse than SR1/atSRp34 (Figure 2, C′, E′, and F′) and SR33(Figure 2, C, E, and F). The nuclei of larger LEP are enlarged and elongated with an irregular oval shape deformed by vacuoles or chloroplasts (Figure 2A), and their ploidy levels vary from 4C to 8C (Melaragno et al., 1993). Trichomes, the largest cell type, have the largest nuclei with irregular oval shapes (Figure 2B) and ploidy levels up to 64C (Melaragno et al., 1993). The nuclei of root epidermal cells in the differentiated zone of the root are elongated or oval shaped (Figure 2H) with ploidy levels more than 2C (Kato and Lam, 2003). The positions and shapes of root hair nuclei are highly dynamic (Figure 2I; our unpublished data). Among these endoreduplicated cell types, the distribution of SR protein in trichomes (Figure 2, B, B′, and B″) is more diffuse and the speckles are smaller than that observed in root hairs (Figure 2, I, I′, and I″), larger leaf epidermal pavement cells (Figure 2, A, A′, and A″), or root epidermal cells (Figure 2, H, H′, and H″). Compared with mammalian cell speckles, plant speckles are smaller in average size and greater in number within the same nuclear volume.
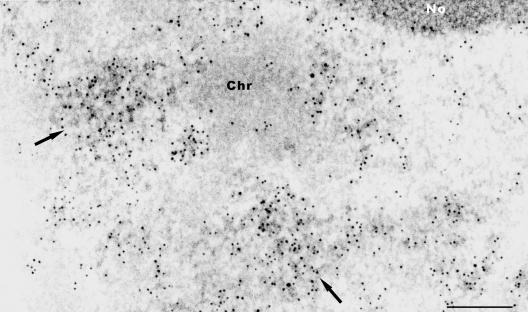
We further examined the organization of SR proteins by TEM. Ultrathin sections of the differentiated zone of Arabidopsis primary root were processed for TEM and SR proteins within IGCs were identified using 3C5 antibody (Turner and Franchi, 1987), which recognizes the SR family of pre-mRNA splicing factors. The SR proteins localize in interchromatin regions, and accumulate in speckles (arrows in Figure 3), as well as being diffusely distributed throughout the nucleoplasm (Figure 3). The regions of more concentrated antibody labeling are composed of clusters of 20- to 30-nm granules that are connected in places by thin fibrils forming a loosely associated meshwork. These clusters of granules are IGCs. Labeled fibrillar regions seem to extend between the periphery of condensed chromatin and IGCs (Figure 3). In some cases, immunolabeled fibrillar regions, possibly corresponding to perichromatin fibrils, seem directly in association with the periphery of condensed chromatin (Figure 3). This distribution is similar to that observed in mammalian cells or tissues (for reviews, see Fakan and Puvion, 1980; Thiry, 1995; Lamond and Spector, 2003).
Figure 3.
Immunoelectron microscopic localization of SR proteins in Arabidopsis. Root sections are immunolabeled with 3C5 antibody, which recognizes the SR family of pre-mRNA splicing factors in IGCs (arrows). Chr, chromatin; No, nucleolus. Bar, 200 nm.
Dynamics of Splicing Factors In Vivo
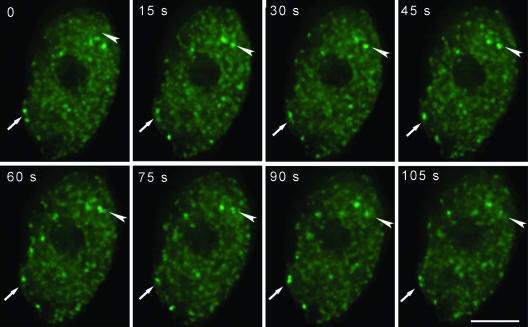
Leaf epidermal pavement cell nuclei, which have an obvious speckled pattern of pre-mRNA splicing factors, were then used to visualize the dynamics of splicing factors in vivo by time-lapse three-dimensional microscopy. A region of rosette leaf (∼0.5 cm2) was cut and mounted in water, and a 60×/1.20 numerical aperture water immersion objective lens was used to follow the dynamics of nuclear speckles at room temperature. Data sets were collected at 15-s intervals over a 30-min time period. Stacks of images at each time point were deconvolved, and the projections at eight successive time points are shown in Figure 4 (see online Supplementary Video 1). During periods of up to 30 min, most speckles were observed to be moving within a constrained volume of ∼1 μm3 with respect to the total nuclear volume of ∼280 μm3. Some speckles were observed to fuse or bud from each other during the imaging period (arrows in Figure 4). A few speckles moved into another nearby area and then resumed their constrained movement. Other speckles disassembled into the nucleoplasm or formed at new sites. Some adjacent speckles are connected by less intensely labeled fluorescent signals. The pre-mRNA splicing factors seem to be dissociating or associating with speckles or to be exchanging between nearby speckles, resulting in changes in the shape of these speckles (Figure 4, arrowhead). In ∼5% of the leaf epidermal pavement cells, one to three speckles entered into the nucleoli and moved rapidly within this nuclear domain (see online Supplementary Video 2). Identical dynamic events of speckles were observed in SR1/atSRp34-YFP, SR33-YFP, and atSRp30-YFP transgenic plants.
Figure 4.
Time-lapse deconvolution microscopy of SR1/atSRp34-YFP in a living leaf epidermal pavement cell. The projections of deconvolved optical sections are shown at each time point. The arrows highlight the fusion and disassociation of two speckles, and the arrowheads highlight the splicing factors being released from speckles. The time is indicated in seconds. Bar, 5 μm.
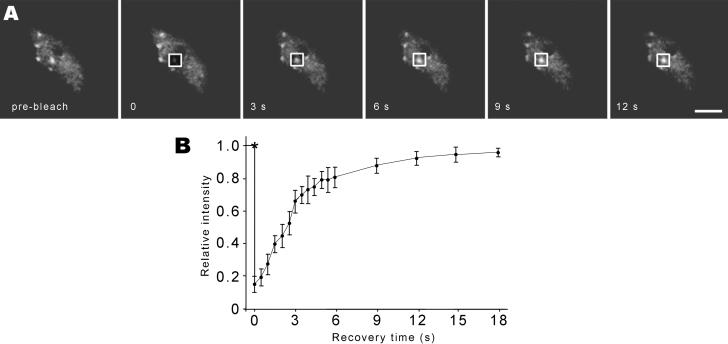
To further determine the dynamics of SR proteins in speckles, we used FRAP analysis. A region of leaf (∼0.5 cm2) from SR1/atSRp34-YFP or SR33-YFP transgenic plants was mounted in water. After five single scans were acquired, a 1.7 × 1.7-μm2 nuclear region that contains a speckle was photobleached, and a series of images were acquired immediately after bleaching (Figure 5A). Subsequently, the relative intensity of fluorescence within the photobleached area was calculated as described previously (Phair and Misteli, 2000). In addition, the level and rate of the fluorescence recovery, as fluorescent molecules from outside the photobleached zone migrated into the bleached area, was determined (Figure 5B). Consistent results were obtained from leaves of plants containing transgene SR1/atSRp34- or SR33-YFP, the SR proteins exchange from the speckle with a half time of recovery of ∼2.5 s.
Figure 5.
FRAP of SR proteins. (A) Leaf epidermal pavement cells expressing SR1/atSRp34-YFP were imaged before and during recovery from photobleaching. The white square frames represent the photobleached region. Bar, 5 μm. (B) Kinetics of recovery after bleaching of SR1/atSRp34-YFP. * indicates the photobleach point.
Organization of the SR Proteins during the Cell Cycle
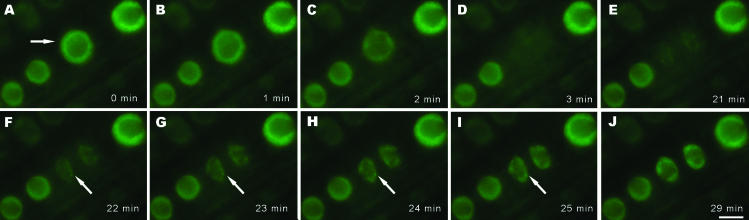
Time-lapse fluorescence microscopy was applied to study the in vivo dynamics of splicing factors during mitosis in root epidermal cells. G2 cells (Figure 6A, arrow) have larger nuclei (6.6 ± 0.25 μm in diameter) with large nucleoli (3.9 ± 0.12 μm in diameter) in comparison to G1 cell nuclei (4.8 ± 0.18 μm in diameter) with nucleoli measuring 3.15 ± 0.16 μm in diameter. On entry into mitosis, as the nuclear envelope breaks down, the splicing factor labeling intensity is gradually reduced as these factors become distributed throughout the cell (Figure 6, A–C). In metaphase, the splicing factors are diffusely distributed throughout the cytoplasm, represented as a rectangular shape, typical of the cell wall shape of root epidermal cells (Figure 6D). In later telophase, as the nuclear envelope is reformed, the SR proteins reenter into the daughter nuclei. Several speckles are observed in the telophase nuclei (Figure 6, F–I, arrows). The daughter nuclei acquire a typical interphase spherical shape and the nucleolus gradually forms (Figure 6, E–J). Mitosis in these root epidermal cells lasts ∼30 min (see online Supplementary Video 3).
Figure 6.
Dynamics of SR1/atSRp34-YFP during mitosis in living root epidermal cells. (A) A cell (arrow) just before nuclear envelope breakdown. (B and C) In prophase, as the nuclear envelope breaks down, the splicing factors enter the cytoplasm. (D) In metaphase, splicing factors are diffusely distributed in the cytoplasm. (E–J) Splicing factors are reentering into daughter nuclei. Newly forming speckles are observed in telophase nuclei (arrows in F–I). Bar, 5 μm.
Effect of Transcriptional Inhibition on the Organization and Dynamics of SR Proteins
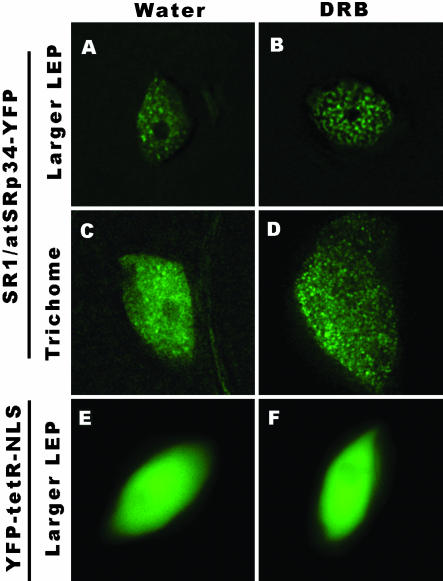
As splicing of most pre-mRNAs occurs cotranscriptionally (for review, see Proudfoot et al., 2002), we were interested in examining how the organization and dynamics we observed are related to RNA polymerase II transcription in living plant cells. Seedlings were incubated for 5 h with 50 μg/ml DRB, an inhibitor of transcription (Sehgal et al., 1976). This drug affects phosphorylation of the C-terminal domain of the large subunit of RNA polymerase II and prevents elongation of nascent transcripts (Yamaguchi et al., 1998). After drug treatment the splicing factors in leaf epidermal pavement cells further accumulated into speckles with less diffuse nucleoplasmic signals (Figure 7B), whereas the splicing factors in trichome nuclei reorganized into thousands of “microspeckles” (Figure 7D).
Figure 7.
Effect of transcription inhibition on the organization of SR proteins. The transgenic leaves were treated with 50 μg/ml DRB for 5 h at 22°C. In leaf epidermal pavement cells (B), after transcriptional inhibition, the splicing factors further accumulated in speckles with less labeling in the nucleoplasm. In trichomes (D), after transcriptional inhibition, the splicing factors organized into thousands of microspeckles. DRB (F) has no obvious effect on nuclear morphology and the diffuse distribution of a control protein YFP-tetR-NLS. In control samples, water treatment for 5 hat 22°C had no obvious effect on the distribution of SR-YFP or YFP-tetR-NLS (A, C, and E). Bar, 5 μm.
In addition to the reorganization of splicing factors upon transcriptional inhibition, we found that the most striking effect of transcriptional inhibition is the abolition of speckle dynamics, these enriched speckles or microspeckles became essentially stationary, neither the fast speckle movement nor association/disassociation of fluorescent signals from or into the speckles were observed (see online Supplementary Video 4). The organization and dynamics of SR proteins in leaves or roots immersed in water at 22°C for 5 h have no obvious changes (Figure 7, A and C, in comparison with the same cell type in Figure 2, A and B). DRB had no detectable effect on overall nuclear morphology or the diffuse distribution of a YFP-tetR-NLS fusion protein under the identical experimental conditions (Figure 7, E and F).
DISCUSSION
Spatially and Temporally Regulated Splicing Factor Expression Levels
We observed differences in SR protein expression levels in different cell types, tissues, and developmental stages. For example, in the vascular bundle of the primary root tip section, atSRp30 is highly expressed whereas SR33 and SR1/atSRp34 are expressed at significantly lower levels. In contrast, SR33 is expressed throughout the root meristem, elongation zone, and specialization zone, whereas SR1 expression drops off significantly in the specialization zone. In leaves, we observed high expression of SR33 and SR1/atSRp34 and minimal expression levels of atSRp30, whereas in pollen grains, all three proteins were expressed at similar levels. At later flowering stages, SR1/atSRp34 expression decreased to a lower level than SR33. Finally, in the inflorescence stems, SR1 is expressed at extremely high levels throughout the stem, whereas SR33 and atSRp30 are expressed at progressively lower levels. Tissue-specific promoter activities of SR proteins were also observed by histochemical analyses with lower resolution and sensitivity (Lopato et al., 1999b, 2002).
It has been proposed that the ratio of various splicing factors in the spliceosome may regulate splice site choice (for review, see Smith and Valcarcel, 2000). For example, increasing the ratio of SF2/ASF to hnRNP A1 has been shown to alter splice site selection both in vitro and in vivo in mammalian cells (Mayeda and Krainer, 1992; Cáceres et al., 1994). Similar to vertebrate introns, most plant introns have canonical GU and AG dinucleotides at their 5′ and 3′ ends, suggesting the presence of a conserved splicing mechanism. However, plant cells have special requirements for intron recognition; for example, the necessity of AU-rich sequences in the intron, a relaxed criteria for branch site selection and the unessential requirement for a polypyrimidine tract (for reviews, see Lorkovic et al., 2000; Reddy, 2001). Given that SR proteins play important roles in both constitutive and alternative pre-mRNA splicing, the larger number of SR proteins in Arabidopsis may be a reflection of specific roles in both constitutive and alternative pre-mRNA splicing among different cell types and developmental stages. This is supported by reports that overexpression of atSRp30 delays the transition from the vegetative to the flowering stage by changing the alternative splicing pattern of specific endogenous genes (Lopato et al., 1999b). In addition, ectopic expression of atRSZ33, a plant-specific SR protein, causes alteration of splicing patterns of several genes and pleiotropic changes in Arabidopsis development (Kalyna et al., 2003). In pSR33:SR33-YFP transgenic plants, we observed fluorescence in nearly all tissues or organs and at all developmental stages. This implies that SR33, a SC35-like splicing factor, is active in the processing of most pre-mRNAs in Arabidopsis, whereas atSRp30, a tissue-specific SF2/ASF-like splicing factor, may be involved in generating tissue-specific transcripts within cells of the vascular system. In contrast, the high level of expression of SR1 in the inflorescence stems and leaves may be indicative of a more global role in constitutive pre-mRNA splicing as well as a potential role in alternative pre-mRNA splicing. Further studies are necessary to elucidate the roles of specific splicing factors in regulating different aspects of plant development.
Plant Cell Nuclei Contain IGCs
Both the mammalian and plant cell nucleus are membranebound organelles that contain well-organized chromatin and the machinery to regulate gene expression. Nuclear compartmentalization is thought to facilitate the regulation of transcription and pre-mRNA splicing to generate mature transcripts (Lamond and Earnshaw, 1998). However, the presence of IGCs in plant nuclei has not been previously identified (Testillano et al., 1993; Acevedo et al., 2002; Cui and Moreno Díaz de la Espina, 2003; Docquier et al., 2004). Our observations demonstrated that two SF2/ASF and one SC35-like splicing factor localize to speckles that correspond to IGCs in plant nuclei upon TEM analysis. These IGCs were similar to those observed in mammalian nuclei because they were composed of clusters of particles measuring 20–30 nm in diameter that are connected in places by thin fibrils (Thiry, 1995). This finding demonstrates the conservation of this nuclear organelle from metazoan to plant nuclei. However, as compared with their mammalian counterparts, plant IGCs are characterized by their smaller size and larger number in the same nuclear volume, implying differences in nuclear architecture and/or organization of the splicing machinery between metazoan and plant cells.
The Organization and Dynamics of Plant SR Proteins Are Dependent upon Ongoing Transcription
We examined plant nuclear speckles by several different approaches in live plants and found them to exhibit several dynamic properties. Whereas speckles exhibited limited overall movements within a constrained volume, their protein components were shown to rapidly exchange with the nucleoplasmic population. Similar to previous studies on SR45 (Ali et al., 2003) and atRSp31 (Docquier et al., 2004), a subset of speckles, at any given time, was observed to merge, bud, disassemble, or assemble at new sites. However, after blocking transcription, we found that almost all of the above-mentioned dynamic events ceased, indicating that these dynamic events are also related to transcriptional activities in the plant nuclei. Similar dynamic events were also observed in mammalian cell nuclei (Misteli et al., 1997; Kruhlak et al., 2000; Phair and Misteli, 2000).
It is well documented that splicing of most pre-mRNAs occurs cotranscriptionally in mammalian cells (for review, see Proudfoot et al., 2002). As transcription inhibition does not disassemble the speckles in plant nuclei, and the distribution of splicing factors in highly metabolic cells such as meristematic cells is more diffuse, we propose that the plant speckles are storage or modification/assembly sites of splicing components, from which splicing factors or preassembled spliceosome subcomplexes are recruited to transcription sites similar to that suggested for their mammalian counterparts (for review, see Lamond and Spector, 2003).
In light of the observed dynamic events of pre-mRNA splicing factors, it can be speculated that regions of active chromatin around the speckle might form “transcription/splicing hot spots.” Such a model would be indicative of a speckle serving a cluster of genes. Support for this was shown in live mammalian nuclei where one or more speckles was shown to provide splicing factors to the inducible BK virus and cytomegalovirus immediate early transcripts stably integrated into the baby hamster or rat genomes, respectively (Misteli et al., 1997). More recently, Shopland et al. (2003) reported that multiple specific genes and gene-rich R-bands cluster around SC35 domains (speckles) and proposed that the speckles are functional centers for a multitude of clustered genes or local euchromatic neighborhoods.
Why is the Distribution of SR Proteins Cell-Type Specific?
We observed different distribution patterns of members of the SR family of pre-mRNA splicing factors in nuclei of different Arabidopsis cell types. These differences may be a reflection of the organization of chromatin and the amount of interchromatin space within the respective cell types. In addition, the metabolic state of the cells, including the distribution of active genes and their transcriptional levels, may also affect the distribution of SR proteins. Because a higher metabolic level correlates with more active chromatin and/or more transcriptional sites, a larger amount of SR proteins will therefore be recruited to these transcriptional sites from speckles, resulting in a lower ratio of splicing factors in speckles to that in the diffuse nucleoplasmic population.
Among cells with a 2C ploidy level, we propose that there is likely to be more decondensed or active chromatin in meristematic cells than in small leaf, petal, and sepal epidermal cells. Higher gene activity would result in the recruitment of more splicing factors to assembling spliceosomes, and therefore a more diffuse distribution pattern of splicing factors would be expected in meristematic cells than that in small leaf, petal, and sepal epidermal cells. In addition, the larger nucleoli in meristematic cells will result in less interchromatin spaces for other nuclear organelles. This is further supported by our observation that the splicing factors are more concentrated in speckles in aging leaves (our unpublished data). It will be of interest to investigate whether the observed difference in the localization of the three factors in certain cell types is a reflection of difference in the ratio of these factors between storage/assembly sites and sites of function of the three splicing factors.
Trichomes have the largest nuclei with a high degree of endoreduplication up to 64C. However, compared with other endoreduplicated cell types, the distribution of SR proteins in trichomes is essentially diffuse (Figures 2B and 7C). After transcriptional inhibition, the splicing factors accumulated into thousands of microspeckles (Figure 7D), suggesting that the chromatin organization of these cells may result in a smaller interchromatin space in the nuclei of this unique epidermal cell type. Future studies will directly examine the relationship of the compaction state of chromatin to the organization of the pre-mRNA splicing machinery in different Arabidopsis cell types.
Apart from genome and proteome, the tiny flowering plant A. thaliana might further serve as a model plant for “in situ cell biology,” i.e., to study the organization and dynamics of proteins and their related functions within cells of a living organism. The dynamic cell type specific organization of SR proteins provides insight into the organizational plans of nuclei from cell types exhibiting different ploidy levels. Moreover, the cell type-dependent expression levels and distribution patterns of pre-mRNA splicing factors implicates both chromatin organization as well as the metabolic state of the cells in the organization of the pre-mRNA splicing machinery. The observed cell type dependent nuclear organization may be a general feature of nuclei from metazoa to plants and is likely to be important for the regulation of gene expression or epigenetic functions in the nuclei of different cell types.
Supplementary Material
Acknowledgments
We acknowledge Andrea Barta (atSRp30), Anireddy S.N. Reddy (SR33), and Howard M. Goodman (SR1/atSRp34) for providing cDNA clones and Eric Lam for providing the YFP-tetR-NLS fusion. We thank Rob Martienssen, David Jackson, and members of the Spector laboratory for insightful discussions. This work was supported by Plant Genome Research Program grant 0077617 from the National Science Foundation.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-02-0100. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-02-0100.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Acevedo, R., Samaniego, R., and Moreno Díaz de la Espina, S. (2002). Coiled bodies in nuclei from plant cells evolving from dormancy to proliferation. Chromosoma 110, 559-569. [DOI] [PubMed] [Google Scholar]
- Ali, G.S., Golovkin, M., and Reddy, A.S. (2003). Nuclear localization and in vivo dynamics of a plant-specific serine/arginine-rich protein. Plant J. 36, 883-893. [DOI] [PubMed] [Google Scholar]
- Baluska, F. (1990). Nuclear size, DNA content, and chromatin condensation are different in individual tissues of the maize apex. Protoplasma 158, 1-2. [Google Scholar]
- Beven, A.F., Simpson, G.G., Brown, J.W.S., and Shaw, P.J. (1995). The organization of splicesomal components in the nuclei of higher plants. J. Cell Sci. 108, 509-518. [DOI] [PubMed] [Google Scholar]
- Boudonck, K., Dolan, L., and Shaw, P.J. (1998). Coiled body numbers in the Arabidopsis root epidermis are regulated by cell type, developmental stage and cycle parameters. J. Cell Sci. 111, 3687-3694. [DOI] [PubMed] [Google Scholar]
- Boudonck, K., Dolan, L., and Shaw, P.J. (1999). The movement of coiled bodies visualized in living plant cells by green fluorescent protein. Mol. Biol. Cell 10, 2297-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres, J.F., Stamm, S., Helfman, D.M., and Krainer, A.R. (1994). Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265, 1076-1079. [DOI] [PubMed] [Google Scholar]
- Chen, H., Hughes, D.D., Chan, T.-A., Sedat, J.W., and Agard, D.A. (1996). IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J. Struct. Biol. 116, 56-60. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- Cui, P., and Moreno Díaz de la Espina, S. (2003). Sm and U2B“proteins redistribute to different nuclear domains in dormant and proliferating onion cells. Planta 217, 21-31. [DOI] [PubMed] [Google Scholar]
- Docquier, S., Tillemans, V., Deltour, R., and Motte, P. (2004). Nuclear bodies and compartmentalization of pre-mRNA splicing factors in higher plants. Chromosoma 112, 255-266. [DOI] [PubMed] [Google Scholar]
- Fakan, S., and Puvion, E. (1980). The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. Int. Rev. Cytol. 65, 255-299. [DOI] [PubMed] [Google Scholar]
- Golovkin, M., and Reddy, A.S. (1998). The plant U1 Small Nuclear Ribonucleoprotein Particle 70K Protein Interacts with two novel serine/arginine-rich proteins. Plant Cell 10, 1637-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin, M., and Reddy, A.S. (1999). An SC35-like protein and a novel serine/arginine-rich protein interact with Arabidopsis U1–70K protein. J. Biol. Chem. 274, 36428-36438. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. (2000). Sorting out the complexity of SR protein functions. RNA 6, 1179-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: betaglucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-García, L.F., and Spector, D.L. (1993). In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell 73, 47-59. [DOI] [PubMed] [Google Scholar]
- Kalyna, M., Lopato, S., and Barta, A. (2003). Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol. Biol. Cell 14, 3565-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, N., and Lam, E. (2003). Chromatin of endoreduplicated pavement cells has greater range of movement than that of diploid guard cells in Arabidopsis thaliana. J. Cell Sci. 116, 2195-2201. [DOI] [PubMed] [Google Scholar]
- Kato, N., Pontier, D., and Lam, E. (2002). Spectral profiling for the simultaneous observation of four distinct fluorescent proteins and detection of protein-protein interaction via fluorescence resonance energy transfer in tobacco leaf nuclei. Plant Physiol. 129, 931-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak, M.J., Lever, M.A., Fischle, W., Verdin, E., Bazett-Jones, D.P., and Hendzel, M.J. (2000). Reduced mobility of the alternate splicing factor (ASF) through the nucleoplasm and steady state speckle compartments. J. Cell Biol. 150, 41-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond, A.I., and Earnshaw, W.C. (1998). Structure and function in the nucleus. Science 280, 547-553. [DOI] [PubMed] [Google Scholar]
- Lamond, A.I., and Spector, D.L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 4, 605-612. [DOI] [PubMed] [Google Scholar]
- Lazar, G., Schaal, T., Maniatis, T., and Goodman, H.M. (1995). Identification of a plant serine-arginine-rich protein similar to the mammalian splicing factor SF2/ASF. Proc. Natl. Acad. Sci. USA 92, 7672-7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato, S., Forstner, C., Kalyna, M., Hilscher, J., Langhammer, U., Indrapichate, K., Lorkovic, Z.J., and Barta, A. (2002). Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with at SC35-like splicing factors. J. Biol. Chem. 277, 39989-39998. [DOI] [PubMed] [Google Scholar]
- Lopato, S., Gattoni, R., Fabini, G., Stevenin, J., and Barta, A. (1999a). A novel family of plant splicing factors with a Zn knuckle motif: examination of RNA binding and splicing activities. Plant Mol. Biol. 39, 761-773. [DOI] [PubMed] [Google Scholar]
- Lopato, S., Kalyna, M., Dorner, S., Kobayashi, R., Krainer, A.R., and Barta, A. (1999b). AtSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 13, 987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato, S., Waigmann, E., and Barta, A. (1996). Characterization of a novel arginine/serine-rich splicing factors in Arabidopsis. Plant Cell 8, 2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic, Z.J., and Barta, A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic, Z.J., Wieczorek Kirk, D.A., Lambermon, M.H.L., and Filipowicz, W. (2000). Pre-mRNA splicing in higher plants. Trends Plant Sci. 5, 160-167. [DOI] [PubMed] [Google Scholar]
- Mayeda, A., and Krainer, A.R. (1992). Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68, 365-375. [DOI] [PubMed] [Google Scholar]
- Melaragno, J.E., Mehrotra, B., and Coleman, A.W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5, 1661-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., Cáceres, J.F., and Spector, D.L. (1997). The dynamics of a pre-mRNA splicing factor in living cells. Nature 387, 523-527. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant 15, 473-497. [Google Scholar]
- Phair, R.D., and Misteli, T. (2000). High mobility of proteins in the mammalian cell nucleus. Nature 404, 604-609. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. (2002). Integrating mRNA processing with transcription. Cell 108, 501-512. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S.N. (2001). Nuclear pre-mRNA splicing in plants. Crit. Rev. Plant Sci. 20, 523-571. [Google Scholar]
- Sacco-Bubulya, P., and Spector, D.L. (2002). Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156, 425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal, P.B., Darnell, J.E., and Tamm, I. (1976). The inhibition by DRB (5,6-Dichloro-1-B-D-ribofuranosylbenzimidazole) of hnRNA and mRNA production in HeLa cells. Cell 9, 473-480. [DOI] [PubMed] [Google Scholar]
- Shopland, L.S., Johnson, C.V., Byron, M., McNeil, J., and Lawrence, J.B. (2003). Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J. Cell Biol. 162, 981-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.W., and Valcarcel, J. (2000). Alternative pre-mRNA splicing: the logic of combinatorial control. Trend Biochem. Sci. 25, 381-388. [DOI] [PubMed] [Google Scholar]
- Testillano, P.S., Sanchez-Pina, M.A., Olmedilla, A., Fuchs, J.P., and Risueno, M.C. (1993). Characterization of the interchromatin region as the nuclear domain containing snRNPs in plant cells. A cytochemical and immunoelectron microscopy study. Eur. J. Cell Biol. 61, 349-361. [PubMed] [Google Scholar]
- Thiry, M. (1995). The interchromatin granules. Histol. Histopathol. 10, 1035-1045. [PubMed] [Google Scholar]
- Turner, B.M., and Franchi, L. (1987). Identification of protein antigens associated with the nuclear matrix and with clusters of interchromatin granules in both interphase and mitotic cells. J. Cell Sci. 87, 269-282. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, Y., Wada, T., and Handa, H. (1998). Interplay between positive and negative elongation factors: drawing a new view of DRB. Genes Cells 3, 9-15. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., and Reed, R. (2002). Comprehensive proteomic analysis of the human splicesome. Nature 419, 182-185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.