Abstract
Motivation: A large number of experimental studies on ageing focus on the effects of genetic perturbations of the insulin/insulin-like growth factor signalling pathway (IIS) on lifespan. Short-lived invertebrate laboratory model organisms are extensively used to quickly identify ageing-related genes and pathways. It is important to extrapolate this knowledge to longer lived mammalian organisms, such as mouse and eventually human, where such analyses are difficult or impossible to perform. Computational tools are needed to integrate and manipulate pathway knowledge in different species.
Results: We performed a literature review and curation of the IIS and target of rapamycin signalling pathways in Mus Musculus. We compare this pathway model to the equivalent models in Drosophila melanogaster and Caenorhabtitis elegans. Although generally well-conserved, they exhibit important differences. In general, the worm and mouse pathways include a larger number of feedback loops and interactions than the fly. We identify ‘functional orthologues’ that share similar molecular interactions, but have moderate sequence similarity. Finally, we incorporate the mouse model into the web-service NetEffects and perform in silico gene perturbations of IIS components and analyses of experimental results. We identify sub-paths that, given a mutation in an IIS component, could potentially antagonize the primary effects on ageing via FOXO in mouse and via SKN-1 in worm. Finally, we explore the effects of FOXO knockouts in three different mouse tissues.
Availability and implementation: http://www.ebi.ac.uk/thornton-srv/software/NetEffects
Contact: ip8@sanger.ac.uk or thornton@ebi.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The insulin/insulin-like growth factor signalling pathway (IIS) and the target of rapamycin (TOR) signalling pathway have been shown to be important regulators of ageing across species via the transcription factor FOXO (Kenyon, 2011). The mechanisms by which ‘FOXO increased activity’ leads to lifespan extension are still unclear. However, it is thought that lifespan extension is achieved through cell-cycle arrest by FOXO in the absence of insulin signalling (van der Horst and Burgering, 2007). In addition, identification of FOXO transcriptional targets has revealed a second tier of transcription factors regulating a variety of downstream responses (Alic et al., 2011). Ageing via the IIS pathway has been intensively studied at the level of invertebrate model laboratory organisms. With their short lifespans, well-described genomes and a variety of mutants already available, Drosophila melanogaster (Clancy et al., 2001) and Caenorhabtitis elegans (Kenyon et al., 1993) provide excellent frameworks for fast identification of genetic determinants of ageing. Relating results from fly and worm to a longer lived mammalian model, such as Mus musculus (Blüher et al., 2003), is critical for the understanding of the ageing processes in human, but it is often difficult owing to the large evolutionary distance between invertebrates and mammals.
The general flow of the pathway is as follows. The insulin and insulin growth factor receptors can be activated by two different insulin molecules or two insulin-like growth factor molecules. On activation, the two receptors can activate the insulin receptor substrates (IRS1-4) by tyrosine phosphorylation. The role of IRS1 especially has been well examined and found to propagate the signal further downstream, via the PI3K complex. The phospholipid products of PI3K [phosphatidylinositol-3,4,5-triphosphate (PIP3)], once produced, can activate phosphoinositide-dependent kinase-1 (PDK1) that leads to the activation of AKT/protein kinase B-like proteins (AKT1-3, with AKT1 being well studied) and serum and glucocorticoid-inducible kinases (SGK1-3). AKT1 and the SGK1-3 kinases inhibit the activity of the Forkhead transcription factors FOXO by retaining them in the cytoplasm.
The IIS pathway is generally well conserved, with the main building blocks (INSR, PI3K, PDK1, AKT, FOXO) present in both mammals and invertebrates. The TOR pathway is also well conserved with its main building blocks present (TOR complexes 1 and 2, RHEB, S6 kinase). Important differences also exist. There are seven known insulin molecules in the fly, as opposed to 40 in the worm. In the mouse, there are two insulin and two insulin-like growth factors. Flies and worms possess a single insulin receptor each, whereas mice possess two insulin-like growth factor receptors in addition to the insulin receptor. In mice, we observe more copies of certain proteins, such as four copies of AKT (Akt1-4) instead of one in the fly and two in the worm and four genes encoding FOXO (Foxo1, Foxo2, Foxo4 and Foxo6) in contrast to a single FOXO in flies and worms (foxo and daf-16). The TOR pathway is remarkably similar between flies and mice, but exhibits differences with the worm (Ivanov et al., 2013; Riesen et al., 2014).
Previously, we developed the web-service NetEffects to solve the problem of relating gene expression results to their effects at the protein signalling level of the IIS pathway and the ageing phenotype (Papatheodorou et al., 2012). This is a common problem of many studies on ageing that use whole organism mutants to study changes in lifespan and uncover protein interactions within the signalling pathways by use of transcriptomic datasets. NetEffects uses Answer Set Programming, a logic-based method for inference that uses manually curated maps of the IIS pathway and its relationship to lifespan as prior knowledge. Given a gene mutation and the resulting genome-wide differential gene expression, NetEffects will deduce signalling effects and how they can influence lifespan according to the prior knowledge. Our applications on fly and worm datasets (Ivanov et al., 2013; Papatheodorou et al., 2012) have revealed consistent homeostatic mechanisms across both long- and short-lived mutants.
2 METHODS
The signalling network model of the IIS and TOR pathways was built using GraphML, using the editor yEd (http://www.yworks.com). This enabled a computationally readable representation of the pathways, as well as a graphical visualization that relates molecular topology to cellular components. During the curation process, we used the insulin pathway available in KEGG (Kanehisa et al., 2012) as a starting point and the rest of the connections were built by literature review. Literature searches were performed using PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) by querying mouse gene names and identifiers. Only connections with experimental support, rather than just suggestive or hypothetical, were used. Protein–protein interactions from yeast two-hybrid system or other screens were omitted. Supporting literature for each relationship is available by clicking on the pathway connections within NetEffects.
All graphs from the comparative analyses across the fly, worm and mouse pathways were produced by custom-made R (http://www.r-project.org/) scripts. These cross-species analyses were based on genomic sequence-based orthologous relationships, downloaded from Ensembl Compara (http://www.ensembl.org). Orthology relationships that were not predicted by Compara but were suggested by the topology and connectivity of the genes in the pathway models, were sought in TreeFam (http://www.treefam.org/), OrthoDB (http://cegg.unige.ch/orthodb7) and Phylome (http://phylomedb.org/).
The mouse pathway was incorporated to the web-service NetEffects, built using PHP (PHP: Hypertext Preprocessor), JavaScript and Perl. Proteins on the pathway have been annotated with Ensembl identifiers to enable the import and analysis of experimental datasets. The ‘theoretical perturbations’ option of NetEffects was used to query the pathway and produce inferences on the possible paths to longevity from different mutations. The ‘Experimental Results Analysis’ option was used to analyse the gene expression datasets. Raw files of the expression datasets in Paik et al. (2007) were analysed using the Limma package in R (Smyth, 2005).
3 RESULTS AND DISCUSSION
3.1 A model of the insulin and TOR signalling pathways for M.musculus
We curated a model of the IIS, TOR and neighbouring pathways in the mouse, providing access to the underlying literature as clickable connections on the pathway within the web-service.
We found the IIS pathway to be well connected and to include several points of cross-talks with neighbouring pathways. This enables it to respond to signals from a variety of sources rather than just extracellular insulin molecules. IRS and SHC1 bind to GRB2, which then activates the MAPK/ERK pathway. In addition to propagating insulin signals, IRS also receives feedback from other pathways, such as TOR by inhibition from S6 kinase (gene name Rps6kb1), JNK by inhibition from JNK1 and Wnt signalling through an inhibition by GSK3-beta. SHC1 is phosphorylated by the activated Insulin receptor, propagating the signal to MAPK/ERK via GRB2, SOS1 and SOS2. AKT1 provides another point of cross-talk with the TOR pathway, by directly inhibiting PRAS40, which then inhibits RPTOR, inhibiting TSC2. AKT1 can also be activated by TOR complex 2.
Previous work on mouse mutants of the IIS and TOR signalling pathways have clearly shown a role for the insulin pathway in the regulation of lifespan, with null S6K (Selman et al., 2009) mice and null IRS-1 (Selman et al., 2008) mice showing significant lifespan extension when compared with wild-type. Although there is so far no experimental confirmation of lifespan regulation by mammalian FOXOs, results from mice lacking one or more of FoxO1, FoxO3 and FoxO4 have revealed ageing-related phenotypes, such as reduced bone mass (Ambrogini et al., 2010) and the development of ageing-related diseases like thymic lymphomas, hemangiomas (Paik et al., 2007). These results suggest that mammalian FoxOs play a protective role against age-related diseases. In addition, Willcox et al. (2008) provide evidence for FoxO3A genetic variation being associated with lifespan in a large, well-phenotyped cohort of humans through a case–control study of five candidate genes.
3.2 Comparison of insulin and TOR signalling in fly, worm and mouse
With the availability of thoroughly curated pathway models for each of the three species, we are now able to make comparisons of their components and connections. Supplementary Table S2 summarizes the similarities and differences between the IIS and TOR pathway molecules across the three different species. Figure 1 presents on the mouse IIS and TOR pathway model the occurrence of fly and/or worm orthologues, also present in the species-specific models.
Fig. 1.
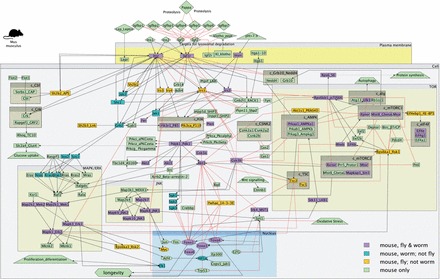
The model of the IIS and TOR pathways in M.musculus. Colours indicate the existence of orthologues in flies, worms, both or none. A larger version of this model is available in Supplementary Figure S1
‘Functional orthologues’ were also identified, where sequence similarity across species is moderate but molecular interactions and experimental evidence suggest that these pairs are indeed orthologues. Such cases include ist-1, a worm orthologue to the IRS (OrthoDB); drr-2, a worm orthologue to eukaryotic translation initiation factors 4H (Ching et al., 2010) and 4B (Phylome Orthology); unc-51, worm orthologue to Ulk1/Atg1; let-363, worm orthologue to Mtor (TreeFam) and age-1, worm orthologue to Pik3ca (TreeFam). Gene Deptor encodes a protein associated with the mammalian TORC1 that is absent from the fly and worm genomes. Glatter et al. (2011) hypothesized that the gene appeared later in vertebrate evolution.
In general, the IIS and TOR pathways in the mouse appear considerably larger and with more cross-talk (see Supplementary Section S3). This effect is partly because of the fact that the IIS and TOR pathways in the mouse have been more thoroughly studied, as well as to the different extents of the curation of the neighbouring pathways within each organism. Being used as a model organism for studies on human diseases such as cancer and diabetes and with the availability of a large number of murine cell lines, more interactions within and between the IIS, TOR and their neighbouring pathways have been discovered.
Cross-talk points between the IIS and TOR pathway include interactions between AKT1 and TORC2 in all three organisms, as well as RPS6KBA (S6 kinase) and IRS1 in mouse and fly. In the mouse, both AKT and IRS involve several paralogous copies. The IRS genes in the mouse are also involved in the cross-talk between JNK and IIS pathways, an interaction that is conserved across species. In some cases, the interactions between neighbouring pathways are not conserved due to the lack of orthologues in worms or flies or both. For example, there is no orthologue for TSC2 in the worm which is inhibited by AKT in flies and mice. There are also cases where orthologues in the invertebrates exist, but the interactions have not been observed experimentally as exemplified by the interaction of IIS and MAPK/ERK pathways via GRB2 and SOS1. Finally in the case of MAPK/ERK to TOR cross-talk, facilitated by the activation of RPS6KA1 by MAPK1 in the mouse, we found orthologues in both other organisms but no interaction in the fly. In the worm, there is evidence for a protein–protein interaction between them (see Supplementary Section S3D for the complete table of cross-talk points).
3.3 Paths to FOXO-mediated longevity with NetEffects
We incorporated the mouse pathway model into the web service NetEffects (Papatheodorou et al., 2012) to enable computational analyses. We can now compare the effects on FOXO-mediated ageing across the three species. Using the ‘theoretical perturbations’ functionality, we tested the paths to FOXO-mediated longevity in the mouse and worm from already known mutants in flies (see Supplementary Section S4 for full results). Knocking out Ins1 or Ins2 in mouse results in a path consistent with those obtained when doing the equivalent test in the fly and worm models (Table 1). This leads to increased lifespan through inhibition of AKT, which then allows translocation of FOXO into the nucleus. However, in mouse and worm, we also encounter paths that reduce longevity. In the mouse, this path involves the IGF1-receptor and RACK1 (gene Gnb2l) that leads to enhanced AKT phosphorylation and activation. This effect, however, appears to be cell-type-specific and probably also context-specific, as described in Kiely et al. (2005). In the worm, there is also an effect that might be antagonizing the FOXO-mediated lifespan increase, but is mediated by LET-60 (Kras orthologue) and the transcription factor SKN-1. Similar effects were produced when Igf1r was mutated. Mutation of Insr had similar results to the equivalent manipulations in the fly and worm.
Table 1.
Shortest paths to FOXO-mediated longevity, derived from NetEffects, where DAF-16 is the FOXO orthologue in C.elegans
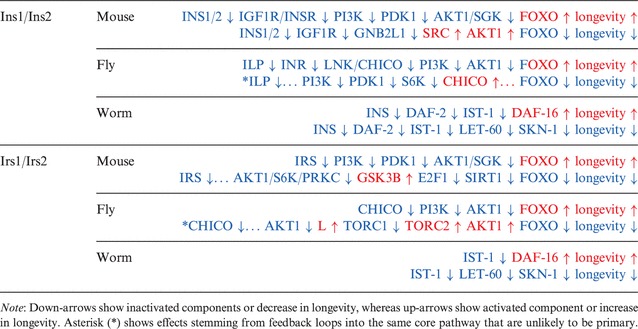 |
We also tested known mouse mutants in the IIS pathway that affect lifespan. The Irs1−/− mutant (Selman et al., 2008) results in a long-lived phenotype, as expected from previous knockout experiments on the fly orthologue chico (Clancy et al., 2001). In contrast, the Irs2−/− mutant is short-lived (Selman et al., 2008). NetEffects infers similar paths for both Irs1−/− and Irs2−/− mutants, as they exhibit similar connections to other components of the IIS and TOR pathways (Table 1). The shortest path that leads to increase in lifespan involves inhibition of AKT. The shortest paths leading to a reduction of lifespan require increased activity of GSK3B leading to inhibition of FOXO via SIRT1 and E2F1. Functional experiments, as well as genome-wide gene expression in the two mutants could show whether Irs2−/− mutant mice reduce their lifespan via a different route, and whether the activity of GSK3B plays a role. We also tested ist-1, the functional worm orthologue for Irs1 and Irs2, where a knockout experiment with lifespan analysis has not been performed. In addition to the FOXO-mediated lifespan extension, we obtained a sub-path of the same antagonistic effect via SKN-1 as in the INS1/2 tests shown above. Partial support for the opposing effect of LET-60 (RAS) via SKN-1 to FOXO-mediated lifespan extension comes from a study on long-lived age-1 (PI3K) worms, where downregulation of let-60 and skn-1 genes was observed (Tazearslan et al., 2009).
Finally, we analysed the expression datasets in cells derived from three different tissues of null and conditional alleles in the three main Foxo genes (FoxO1−/+; FoxO3−/−; FoxO4−/−). The datasets were generated by Paik et al. (2007). We analysed the Foxo mutants in liver and lung endothelial cells and thymus cells. According to the authors, liver cells presented cancer-related phenotypes, whereas lung cells did not present a detectable phenotype. With NetEffects we can show that 16 genes within the pathway model are differentially expressed in liver, one in lung and three in thymus (excluding the three Foxo genes that have been knocked out). Almost all of the shortest paths starting from these differentially expressed genes correspond to negative feedback to the mutation of Foxo genes, as shown by the predicted impact on lifespan (Supplementary Section S5). This suggests that the function of the three Foxo genes plays a greater part in the liver and thymus rather than in lung endothelial cells. Similar negative feedback was identified in our previous analyses of Foxo null mutants in the fly Papatheodorou et al. (2012).
4 CONCLUSION
The molecular basis of nutrient signalling is largely comparable across a large evolutionary space, despite striking differences in the presence or number of copies of certain components between species. The pathways, in all organisms except worm focus only on FOXO-mediated lifespan, thereby ignoring any effects through different transcription factors. However, using the mouse pathway, we can identify neighbouring signalling pathways with the potential to influence the signal transduction of the IIS through the identified cross-talk points. Comparison of the pathways in a systematic and qualitative way has the potential to explain differences in effects on lifespan across species and help design experiments that will evaluate the pathway flux. The richness and detail of the mammalian model can inform the interpretation of results in invertebrates, where molecular interactions have not been so extensively studied. Being able to compare the pathways side by side, we recorded all differences and identified ‘functional orthologues’. By use of NetEffects, we were able to suggest possible paths affecting FOXO-mediated lifespan given a single mutation and how these differ across species, thus generating hypotheses for further investigation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dobril Ivanov and Matthias Ziehm for useful discussions.
Funding: This work was funded by the Wellcome Trust Strategic Award WT081394MA (I.P., J.M.T.) and by the European Molecular Biology Laboratory (EMBL) (R.P., J.M.T.).
Conflict of interest: none declared.
REFERENCES
- Alic N, et al. Genome-wide dfoxo targets and topology of the transcriptomic response to stress and insulin signalling. Mol. Syst. Biol. 2011;7:502. doi: 10.1038/msb.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrogini E, et al. Foxo-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metab. 2010;11:136–146. doi: 10.1016/j.cmet.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher M, et al. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Ching T-T, et al. drr-2 encodes an eiF4H that acts downstream of TOR in diet-restriction-induced longevity of C. elegans. Aging Cell. 2010;9:545–557. doi: 10.1111/j.1474-9726.2010.00580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, et al. Extension of life-span by loss of chico, a drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Glatter T, et al. Modularity and hormone sensitivity of the Drosophila melanogaster insulin receptor/target of rapamycin interaction proteome. Mol. Syst. Biol. 2011;7:547. doi: 10.1038/msb.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov DK, et al. Transcriptional feedback in the insulin signalling pathway modulates ageing in both Caenorhabditis elegans and Drosophila melanogaster. Mol. BioSyst. 2013;9:1756–1764. doi: 10.1039/c3mb25485b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The first long-lived mutants: discovery of the insulin/igf-1 pathway for ageing. Philos. Trans. R. Soc. B. 2011;366:9–16. doi: 10.1098/rstb.2010.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kiely PA, et al. Rack1-mediated integration of adhesion and insulin-like growth factor i (igf-i) signaling and cell migration are defective in cells expressing an igf-i receptor mutated at tyrosines 1250 and 1251. J. Biol. Chem. 2005;280:7624–7633. doi: 10.1074/jbc.M412889200. [DOI] [PubMed] [Google Scholar]
- Paik J-H, et al. Foxos are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatheodorou I, et al. Using answer set programming to integrate RNA expression with signalling pathway information to infer how mutations affect ageing. PLoS One. 2012;7:e50881. doi: 10.1371/journal.pone.0050881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesen M, et al. MDL-1, a growth- and tumor-suppressor, slows aging and prevents germline hyperplasia and hypertrophy in C. elegans. Aging. 2014;6:98–117. doi: 10.18632/aging.100638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, et al. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 2008;22:807–818. doi: 10.1096/fj.07-9261com. [DOI] [PubMed] [Google Scholar]
- Selman C, et al. Ribosomal protein s6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman R, et al., editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Tazearslan C, et al. Positive feedback between transcriptional and kinase suppression in nematodes with extraordinary longevity and stress resistance. PLoS Genet. 2009;5:e1000452. doi: 10.1371/journal.pgen.1000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat. Rev. Mol. Cell Biol. 2007;8:440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- Willcox BJ, et al. FOXO3A genotype is strongly associated with human longevity. Proc. Natl Acad. Sci. USA. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


