Abstract
Background
The Arabidopsis AtMYB80 transcription factor regulates genes involved in pollen development and controls the timing of tapetal programmed cell death (PCD). Downregulation of AtMYB80 expression precedes tapetal degradation. Inhibition of AtMYB80 expression results in complete male sterility. Full-length AtMYB80 homologs have been isolated in wheat, rice, barley and canola (C genome).
Results
The complete sequences of MYB80 genes from the Brassica. napus (A gene), B. juncea (A gene), B. oleracea (C gene) and the two orthologs from cotton (Gossypium hirsutum) were determined. The deduced amino acid sequences possess a highly conserved MYB domain, 44-amino acid region and 18-amino acid C-terminal sequence. The cotton MYB80 protein can fully restore fertility of the atmyb80 mutant, while removal of the 44 amino acid sequence abolishes its function. Two conserved MYB cis-elements in the AtMYB80 promoter are required for downregulation of MYB80 expression in anthers, apparently via negative auto-regulation. In cotton, tapetal degradation occurs at a slightly earlier stage of anther development than in Arabidopsis, consistent with an earlier increase and subsequent downregulation in GhMYB80 expression. The MYB80 homologs fused with the EAR repressor motif have been shown to induce male sterility in Arabidopsis. Constructs were designed to maximize the level of male sterility.
Conclusions
MYB80 genes are conserved in structure and function in all monocot and dicot species so far examined. Expression patterns of MYB80 in these species are also highly similar. The reversible male sterility system developed in Arabidopsis by manipulating MYB80 expression should be applicable to all major crops.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-014-0278-3) contains supplementary material, which is available to authorized users.
Keywords: Brassica, Cotton, Gossypium hirsutum, Male sterility, MYB80, Transcription factor
Background
The AtMYB80 transcription factor is involved in tapetum and pollen development and is required for the regulation of tapetal programmed cell death (PCD) in developing Arabidopsis anthers [1-3]. Using 3.2 kb of the AtMYB80 promoter fused to the GUS reporter gene and in-situ hybridization analysis, expression of AtMYB80 was found in the tapetum, middle layers and developing microspores from anther developmental stages 5 to 9 [1,4]. Functional disruption of AtMYB80 results in complete male sterility with early tapetum degeneration and collapsed pollen [2,4,5]. Three genes directly regulated by AtMYB80 have been identified using ChIP analysis, namely an A1 aspartic protease (UNDEAD), a pectin methylesterase (VANGUARD1) and a glyoxal oxidase (GLOX1). Premature tapetal PCD and degeneration were observed in the undead and atmyb80 mutants [3].
The AtMYB80 homologs from rice (Oryza sativa), wheat (Triticum aestivum), barley (Hordeum vulgare) and canola (Brassica napus) have been isolated and their protein sequences show significant conservation [6]. High similarity occurs between the R2R3 MYB domains, the 44-amino acid region immediately downstream of the MYB domain and an 18-amino acid sequence at the C-terminus [2,6]. The expression patterns driven by the OsMYB80, TaMYB80 and BnMYB80 promoters in Arabidopsis are similar to that of AtMYB80, being restricted to the tapetum and developing microspores and occurring from stages 6 to 10. When driven by the AtMYB80 or their native promoters, the full-length OsMYB80, TaMYB80 and BnMYB80 constructs are able to fully restore the fertility of the male sterile atmyb80 T-DNA mutant [6].
The two agriculturally important oilseed Brassica species, canola (B. napus, genome AACC) and brown mustard (B. juncea, genome AABB), originate from hybridisation between pairs of the diploid species B. rapa (AA), B. nigra (BB), and B. oleracea (CC) [7,8]. The full-length BnMYB80 of the C genome has been isolated [6], while the MYB80 orthologs from the A genome of B. napus and B. juncea and C genome of B. oleracea have not yet been identified. Upland cotton (Gossypium hirsutum L., genome ATDT) is the most widely cultivated allotetraploid species and originated from interspecific hybridization between G. arboreum (genome A1) and G. raimondii (genome D5) [9]. Only one MYB transcription factor, GhMYB24, has so far been found to play a role in cotton anther development [10]. GhMYB80 is the cotton homolog of AtMYB80. Two partial coding sequences of GhMYB80 were separately obtained and the deduced amino acid sequence shares high similarity with MYB80 homologs in other species [6]. However, the full-length DNA sequence of each GhMYB80 ortholog is still lacking. The expression pattern of GhMYB80 has not been determined and whether functional conservation exists between AtMYB80 and GhMYB80 is unknown.
The utilization of cytoplasmic male sterility (CMS) and nuclear encoded fertility restore genes (Rf) is an important technology for hybrid cotton and canola production [11,12]. However, the CMS-based hybridization system is difficult to develop and maintain [13]. Furthermore, the CMS phenotype is often unstable under both high and low temperatures [14-16]. Manipulation of expression of the MYB80 transcription factor provides a novel means to induce and subsequently reverse male sterility, facilitating the production of hybrid plants [2]. The experiments described here were aimed at cloning the MYB80 genes from cotton and Brassica (A and C genomes) and comparing their protein structures and promoter sequences. The expression pattern of the GhMYB80 gene in cotton anthers and its capacity to rescue the male sterile atmyb80 mutant were determined. The role of a conserved 44 amino acid sequence in MYB80 function was further assessed. The effectiveness of GhMYB80 and BnMYB80 proteins to induce male sterility in Arabidopsis was examined, when fused to the EAR sequences.
Results
Cloning of the homologous MYB80 genes from Brassica and cotton
The homologous MYB80 genes from B. napus (A gene), B. juncea (A gene), B. oleracea (C gene) and G. hirsutum were cloned and sequenced. The nucleotide sequences and the deduced amino acid sequences were compared with Arabidopsis AtMYB80 [1], B. napus MYB80 (C gene) [6] and B. rapa MYB80 (A gene) obtained from the GenBank (GI: 110797058) (Figure 1 and Additional file 1: Figure S1). The nucleotide sequences of the eight MYB80 homologs are highly conserved in their exons. The amino acid sequences are highly similar in the MYB domain (amino acids 1 – 115), a 44-amino acid region adjacent to the MYB domain (amino acids 125 – 168), and a 18 amino acid region at the end of the C-termini. A variable region of 131 to 139 amino acids is present between the 44-amino acid and the C-terminal sequences, sharing 10.7% identity (Figure 1). Among the five MYB80 homologs of the Brassica species, the amino acid sequences in the variable region of the three A genes are more similar to each other than that of the two C genes (99.1% vs. 97.8% identity). The MYB80 homolog of the Brassica B gene has not yet been cloned. The two MYB80 ortholog genes (GhMYB80-1 and 2) from G. hirsutum are highly conserved, sharing 98.4% and 99.4% identity in their nucleotide and peptide sequences, respectively (Figure 1 and Additional file 1: Figure S1). The two genes are likely to be derived from the A and D genomes.
Figure 1.

Diagram of the sequence alignment of the homologous MYB80 proteins. Sequences include AtMYB80 (A. thaliana), BnMYB80 (B. napus), BrMYB80 (B. rapa), BjMYB80 (B. juncea), BoMYB80 (B. oleracea) and GhMYB80 (G. hirsutum). Yellow highlight represents the conserved amino acids between all the homologs. Blue and green highlight represents the conserved amino acids between the Brassica and cotton MYB80 homologs, respectively. The underline indicates the MYB domains and the dash lines indicate the two conserved regions in the C-termini. CR, conserved region; C-term, C-terminus.
Deletion and mutagenesis analysis of the AtMYB80 promoter
To delineate the region of the AtMYB80 5’UTR/promoter responsible for directing expression to the tapetum and pollen, a series of four AtMYB80 promoter-GUS deletion constructs were prepared. These constructs incorporated 1651, 284, 256 or 240bp of the AtMYB80 5’UTR sequence (relative to the ATG translational start codon) into the pBI vector and were transformed into the wild-type Arabidopsis (Figure 2A). The histochemical GUS staining of florets from the transgenic lines was compared to that of the pPG construct possessing a 3200bp AtMYB80 promoter [1]. Similar GUS intensity was present in the young florets with the 3200 and 1651bp promoters. No GUS activity was detected in the 240-pBI transgenic lines. When compared with the 1651-pBI lines, very weak and weak/moderate GUS intensity was present in the 256-pBI and 284-pBI lines, respectively (Additional file 2: Table S1).
Figure 2.
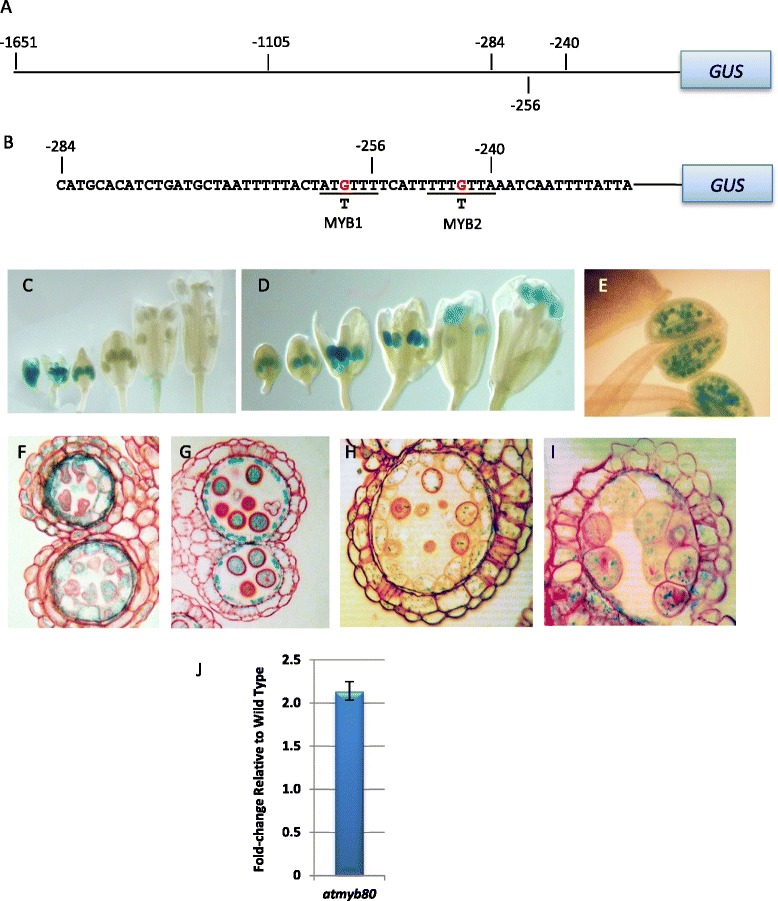
Autoregulation of the AtMYB80 promoter. A. A schematic diagram of AtMYB80 promoter-GUS deletion constructs. Numbers indicates the length of AtMYB80 promoter used for each construct. B. A schematic diagram of mutagenesis constructs within the −284 to -240bp AtMYB80 promoter region. Nucleotides that were targeted for mutagenesis are in red with the corresponding change indicated directly below. C. Floral bud line-up (stages 7 to 12) of the control line showed GUS activity was extended until stage 9. D. Floral bud line-up (stages 7 to 12) of the M2 line showed GUS activity extended to stage 12. E. GUS activity was present in the M2 anther at stage 12. F and G. Cross-sections of M2 anthers showed GUS activity in the tapetum, the outer tapetal cell wall and developing microspores at stages 9 (F) and 11 (G). H and I. GUS activities were present in the tapetum and collapsing pollen grains of the homozygous atmyb80 mutant possessing a wild-type AtMYB80 promoter-GUS construct at stage 10 (H) and 12 (I). J. Comparative qRT-PCR analysis of AtMYB80 transcript levels in the young floral buds (anther stages 5 to 9) of the atmyb80 mutant versus wild-type. The AtMYB80 transcript level is higher in the atmyb80 mutant young floral buds. The UBQ10 was used as the reference gene. Error bar represents SD.
The −284 to -240bp sequence of the AtMYB80 promoter possesses two putative cis-elements, namely MYB1 and MYB2. When the MYB1 element was mutated in a 1105bp promoter (construct M1, single base change, Figure 2B), GUS expression in the anther was unaffected (Figure 2C). However, when MYB1 and MYB2 elements were both mutated (construct M2, Figure 2B), GUS activity persisted through to stage 12 (Figure 2D) rather than being downregulated at stage 10. The activity at stage 11 was localized in the microspores or degenerating tapetal layer (Figure 2G). Pollen grains in stage 12 anthers also expressed GUS activity (Figure 2E). Both the MYB1 and MYB2 elements of the AtMYB80 promoter are conserved in the C genome of Brassica but not in the other four MYB80 genes. MYB2 is conserved in the GhMYB80 promoter and MYB1 in the BnMYB80 A gene promoter (Additional file 3: Figure S3).
To examine whether the expression of AtMYB80 is auto-regulated, a promoter-GUS construct possessing a 1105bp AtMYB80 promoter was introduced into an atmyb80 T-DNA insertion mutant (Figure 2A). Homozygous atmyb80 plants are completely male sterile whilst heterozygous plants are fully male fertile [2]. GUS activity was observed in the anthers of the heterozygous atmyb80 mutant from stages 5 to 9, the same as previously described [1,2]. GUS expression was extended to stage 13 in the two homozygous atmyb80 mutant lines (Additional file 4: Table S2). GUS activity was present in the largely vacuolated tapetal layer (stage 10) and collapsing pollen grains (stage 12) (Figure 2H and I).
Transcript levels of AtMYB80 in both wild-type and atmyb80 mutant were analysed using real-time qRT-PCR. The level of truncated AtMYB80 transcript was approximately 2.1 fold higher in the young mutant floral buds (anther developmental stages 5 to 9) than that of the wild-type (Figure 2J). Previous microarray data comparing differential gene expression in the wild-type and atmyb80 mutant anthers showed a 3.2 fold (p value 0.012) up-regulation of the truncated AtMYB80 transcript in the mutant (unpublished data) [3]. These results together suggest AtMYB80 is involved in the negative auto-regulation.
The promoters of all eight MYB80 genes possess a highly conserved sequence approximately −300 to -380bp upstream of the ATG codon. Four cis-elements are conserved in all six genes, including W-box (TTGAC), MYB (A/TACC), GTGANTG10 (TCAC) and DOFCOREZM elements (A/TAAAG) (Additional file 1: Figure S1).
GUS expression driven by the GhMYB80 promoter in Arabidopsis
To ascertain whether the GhMYB80 promoter resembles the AtMYB80 promoter in driving expression in the Arabidopsis anther, the GUS reporter gene was used. The GhMYB80-1 promoter employed was 443 bp in length (numbered from the ATG). An anther line up showed GUS activity first appeared at stage 5 and persisted to stage 9 (Figure 3C). No activity was detected at stages 10 and 11. Light and dark field microscopy of anther sections showed GUS activity in the tapetum and microspores at stages 8 and 9 (Figure 3D and E). Hence, the expression pattern driven by the GhMYB80-1 promoter in Arabidopsis resembles that of the AtMYB80 promoter.
Figure 3.
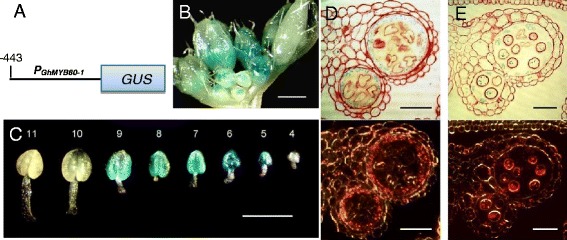
Analysis of the spatial and temporal expression pattern driven by the GhMYB80-1 promoter in Arabidopsis. A. A schematic diagram of the GhMYB80-1 promoter-GUS construct. B. GUS activity is detected in developing P GhMYB80 -GUS floral buds. C. Line-up (anther stages 4 to 11) of the P GhMYB80 :GUS anther after GUS staining. D and E. Sections of the P GhMYB80 :GUS anthers stained with safranin. Light and dark-field microscopy of stage 8 (D) and stage 9 (E) anthers. Bars = 500 μm in B and C. Bars = 25 μm in D and E.
Transcript levels of GhMYB80 in developing cotton anthers
The indicative sizes (length and width) of cotton floral buds corresponding to anther developmental stages were determined using semi-thin sections (Additional file 5: Table S3). The anther stages (from 3 to 11) were numbered in accordance with the morphological changes used for defining the stages of Arabidopsis anther development [17,18]. At stage 4, formation of the tapetum in cotton anthers was initiated (Figure 4B). At stage 6, the tapetal layer became vacuolated (Figure 4D). The tapetal cytoplasm was condensed at stage 7 (Figure 4E) and cell walls degraded at stage 8 (Figure 4F). Tapetal cell degeneration appeared to commence at stage 9 (Figure 4G) and tapetal layer was no longer visible at stage 10 (Figure 4H). The transcript levels of GhMYB80 in cotton anthers at the developmental stages 5 to 11 were analysed using real-time qPCR (Figure 4J) and RT-PCR (Additional file 6: Figure S2). The GhMYB80 transcript level was very low at early stage 5, subsequently increasing at stages 5, 6 and 7. The major increase was from stage 6 to 7 when the tapetal cytoplasm becomes condensed and tetrads appear. At late stage 8, GhMYB80 transcripts could no longer be detected.
Figure 4.
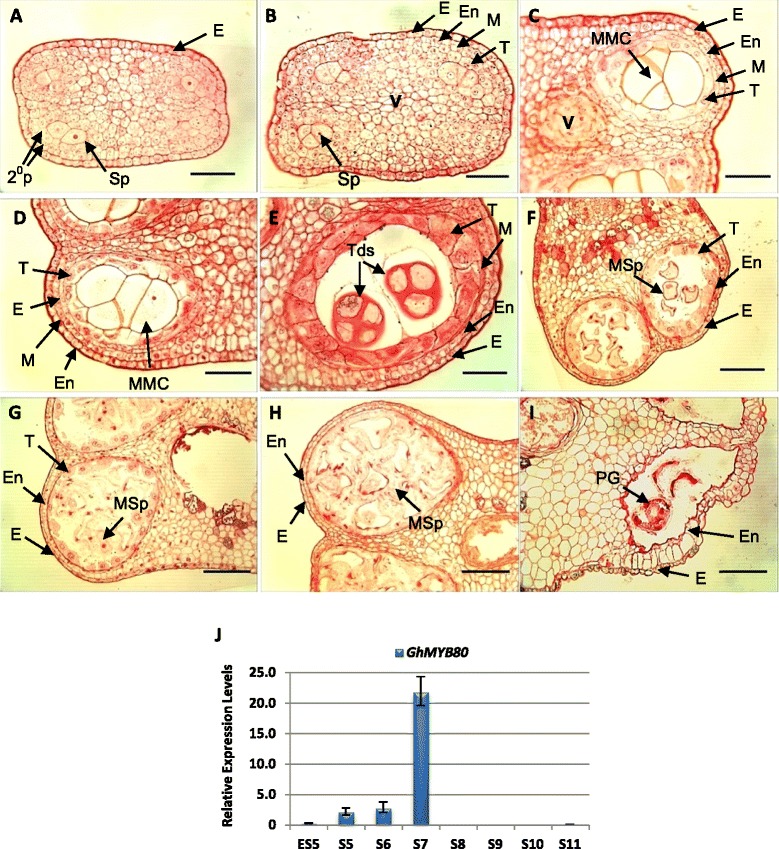
Semi-thin sections of developing G. hirsutum anthers and relative transcript levels of GhMYB80 in anthers. The indicative bud sizes for each anther developmental stage were measured (Additional file 5: Table S3). A. At stage 3, the secondary parietal layers and sporogenous cells are apparent. B. At stage 4, formation of the epidermis, endothecium, middle layer and tapetum has been initiated. C. At stage 5, the microspore mother cells appear. D. At stage 6, the microspore mother cells commence meiosis and the tapetal cells become vacuolated. E. At stage 7, the tapetal cytoplasm is condensed and tetrads appear in the anther locules. F. At late stage 8, microspores are released from the tetrads. Tapetal cell walls have been degraded. G. At stage 9, the tapetum degeneration appears to commence. Microspores are vacuolated. H. At stage 10, the tapetum has been degraded. Remnants of tapetal cells are visible. The microspores are still vacuolated. I. At stage 11, early pollen grains appear. 2°P, secondary parietal layer; E, epidermis; En, endothecium; MSp, microspores; ML, middle layer; MMC, microspore mother cell; MSp, microspore; PG, pollen grains; Sp, sporogenous cells; T, tapetum; Tds, tetrads; V, vascular. Scale bars = 50 μm in A, B, C, D and E. Scale bars = 100 μm in F, G, H and I. J. Relative expression levels of the GhMYB80 in the wild-type Gossypium hirsutum anther. The GhMYB80 transcription level was relatively low at early stage 5 (ES5), stages 5 and 6. It reached a peak level at stage 7 of anther development and was absent from late stage 8 (LS8) to stage 11. The G. hirsutum UBIQUITIN (UBI1) was used as the reference gene. S5 to S11, stages 5 to 11. Error bar represents SD.
GhMYB80 can rescue the male sterile Arabidopsis atmyb80 T-DNA mutant
To determine whether the GhMYB80 and AtMYB80 are functionally conserved, the atmyb80 mutant was transformed with the full-length GhMYB80-1 coding sequence under the control of its own promoter (443bp; PGh80:Gh80) or the AtMYB80 promoter (1100bp; PAt80:Gh80) (Figure 5A). The homozygous atmyb80 T-DNA insertion mutants possessing the transgenes were identified using PCR. Plant fertility is defined as the percentage of the elongated siliques versus the total siliques. In one of the ten PGh80:Gh80 transformed atmyb80 homozygous mutants, fertility was partially restored (20% fertility) (Figure 5B). The other nine lines were less than 10% fertile or remained completely sterile. However, fertility of the nine atmyb80 homozygous lines carrying the PAt80:Gh80 transgene was significantly or fully restored, resulting in 50-100% fertility (Figure 5C). The expression levels of the PGh80:Gh80 and PAt80:Gh80 genes in the relevant transgenic lines were determined using real-time quantitative PCR. Plant fertility was positively correlated with the relative expression levels of the transgenes (Figure 5D and E). The GhMYB80-1 promoter is apparently not as effective as the AtMYB80 promoter in Arabidopsis.
Figure 5.
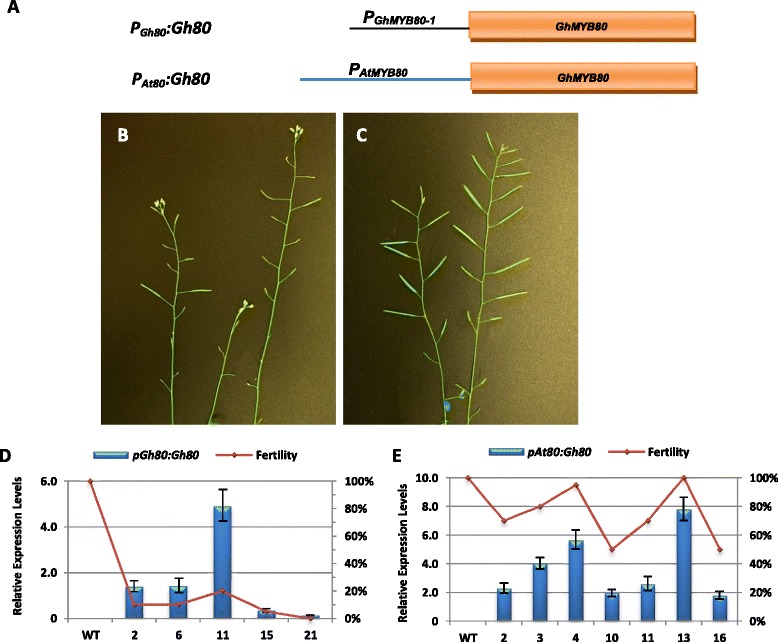
Silique phenotype and expressional analyses of transgenes in the P Gh80 :Gh80 and P At80 :Gh80 Arabidopsis lines. A. Schematic representation of the P Gh80 :Gh80 and P At80 :Gh80 complementation constructs. B. A P Gh80 :Gh80 transformed atmyb80 homozygous mutant (line 11) exhibiting 20% fertility. C. A P At80 :Gh80 transformed atmyb80 homozygous mutant exhibiting 100% fertility (line 13). D and E. The transcript levels of P Gh80 :Gh80 (D) and P At80 :Gh80 (E) relative to the UBQ10 reference gene are positively correlated with plant fertility in the selected lines. Wild type (WT) is the negative control. Error bar represents SD.
The effects of removing the 44-amino acid or the C-terminal region on MYB80 activity
To examine the functions of the 44-amino acid region and the C-terminus of MYB80 protein, two truncation constructs were created by either removing the 44-amino acid region (At80MP-LV) or the variable region and C-terminus (At80MD) from the protein (Figure 6A). The At80MP-LV construct was introduced into the atmyb80 mutant and the At80MD construct transformed into wild type Arabidopsis. Silique elongation and pollen viability were examined in the transgenic lines. All twelve atmyb80 homozygous lines transformed with the At80MP-LV transgene failed to elongate siliques (0% fertility) (Figure 6B). Hence, the 44-amino acid domain is essential for MYB80 activity and may be required for the binding of the R2R3 MYB domain to cis-elements in the promoter of target genes. A wide variation in fertility (from 5% to 95%) was found in the At80MP-LV transgenic atmyb80 heterozygous lines (Figure 6C). qRT-PCR examined expression of the At80MP-LV transgene in two atmyb80 homozygous and four heterozygous lines. Severe male sterility (5% fertility) was observed in line 8 where a high level of the At80MP-LV expression was detected (Figure 6E). In the heterozygous lines, the At80MP-LV protein may be competing for proteins that bind to the C-terminus of endogenous AtMYB80 and are required for MYB80 activity.
Figure 6.
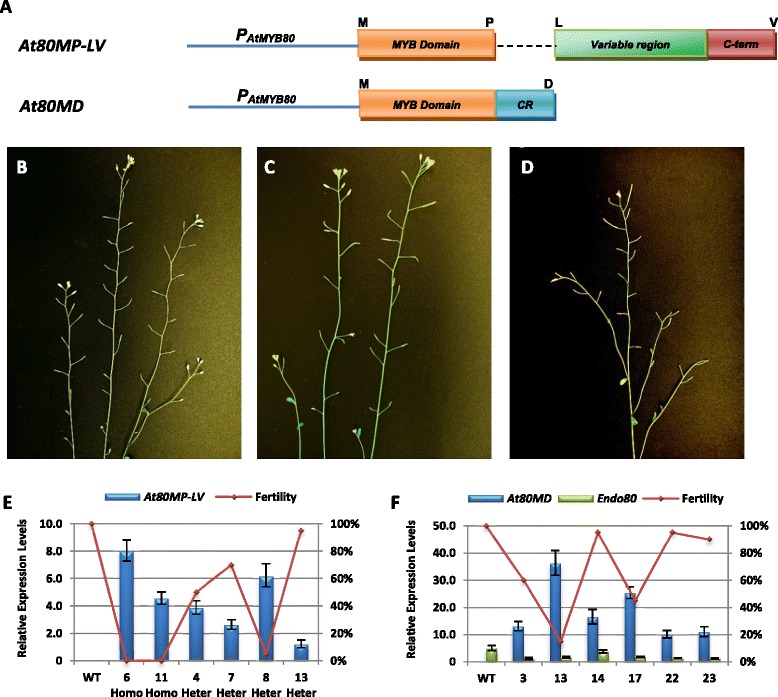
Silique elongation and expressional analyses of transgenes in the At80MP-LV and At80MD Arabidopsis. A. A schematic representation of the At80MP-LV and At80MD truncated constructs. The letters indicate amino acids at the beginning and end of domains. B. The At80MP-LV transgene was unable to rescue the atmyb80 homozygous mutant and the plant remained completely male sterile (line 6). C. Plant fertility was reduced in the heterozygous atmyb80 mutant transformed with the At80MP-LV transgene (line 8). D. The partially male sterile phenotype of the wild-type Arabidopsis transformed with the At80MD construct (line 13). E. The expression of At80MP-LV was detected in the homozygous atmyb80 mutants (line 6 and 11, 0% fertility) and the heterozygous atmyb80 mutants (line 4, 7, 8 and 13, 5% to 95% fertility). F. At80MD transcript levels and plant fertility were determined in the selected lines. The expression levels of endogenous AtMYB80 were reduced in all lines. Wild type (WT) is the negative control. Error bar represents SD.
Four out of the twenty-four wild type lines transformed with At80MD exhibited 15-50% fertility (Figure 6D). The remaining lines remained partially (90%) or fully fertile. The transcript levels of At80MD were all significantly higher than that of the endogenous AtMYB80 in all the selected lines (Figure 6F). The highest expression level of At80MD was obtained in line 13, which showed 15% fertility. The transcript levels of the endogenous AtMYB80 were reduced in all the lines when compared with the wild-type level. Tapetum and pollen development in the partially sterile At80MD lines was examined using light microscopy of anther sections. At stage 8, the tapetum cells were vacuolated and the microspores released from the tetrad were enlarged (hypertrophic) and irregularly shaped (Figure 7A). The tapetum cells became highly vacuolated and hypertrophic at stage 10. Microspore degradation had commenced and cellular debris was observed in anther locules (Figure 7B). At stage 11, a few pollen grains have developed normally in one anther locule and the tapetal layer is degenerating (Figure 7C). In a second locule, however, microspores and tapetum remained highly vacuolated and hypertrophic. Microspore debris was present and tapetal cell walls were intact. The cytoplasmic content of tapetal cells was greatly reduced. The tapetum had completely degenerated at stage 12. Pollen grains had collapsed and debris was attached to the endothecium layer (Figure 7D). The At80MD truncation protein may be able to compete with the endogenous AtMYB80 for binding the promoters of target genes, but fail to activate gene expression.
Figure 7.
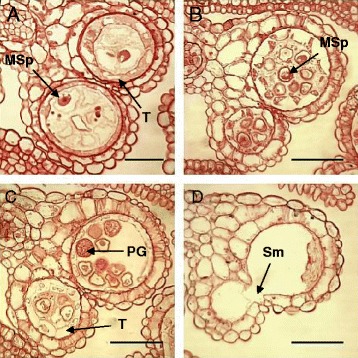
Semi-thin sections of developing Arabidopsis anthers from a transgenic At80MD plant (line 13). A. Stage 8; vacuolated tapetum cells and enlarged microspores. B. Stage 10; tapetum cells are highly vacuolated and enlarged. Microspores commence degrading. C. Stage 11; microspores remain vacuolated, enlarged tapetum with reduced cytoplasm. D. Stage 12; degenerated tapetum and collapsed pollen grains. MSp, microspores; PG, pollen grains; Sm, septum; T, tapetum. Scale bars = 25 μm in A, scale bars = 50 μm in B, C and D.
Male sterility in Arabidopsis is induced by GhMYB80/BnMYB80-EAR fusion repressors
Manipulation of AtMYB80 function has been employed to develop a reversible male sterility system in Arabidopsis. A chimeric construct of the full-length AtMYB80 with the SRDX EAR motif resulted in 60% of the transgenic lines exhibiting complete male sterility [2]. An EAR-like motif (LDLNLELRISPP), designated 32R, is a putative negative regulatory domain (NRD) found in AtMYB32 and shared by other MYB proteins in subgroup 4 [19,20]. We wished to determine if the GhMYB80 and BnMYB80 proteins are effective in inducing male sterility in Arabidopsis when the 32R motif is fused. In addition, to determine whether the effect is enhanced by truncating the MYB80 protein, adding two rather than one 32R motif or by increasing promoter strength. A full-length or a truncated GhMYB80 was fused in frame with two copies of the 32R sequence (PGh80:Gh80-32R2 and PGh80:Gh80MD-32R2). The truncated sequence consisted of the MYB domain and the 44-amino acid region. Both chimeric constructs were driven by the 443bp GhMYB80-1 promoter (Figure 8A). The full-length BnMYB80 (C gene) coding sequence was also fused with one or two copies of the 32R EAR and placed under the control of a 700bp BnMYB80 promoter (PBn80:Bn80-32R and PBn80:Bn80-32R2). The effect of double promoters was examined by using double 400 or 700bp BnMYB80 5’UTR sequences to drive the BnMYB80-32R2 chimeric constructs (PBn400x2:Bn80-32R2 and PBn700x2:Bn80-32R2) (Figure 8A).
Figure 8.
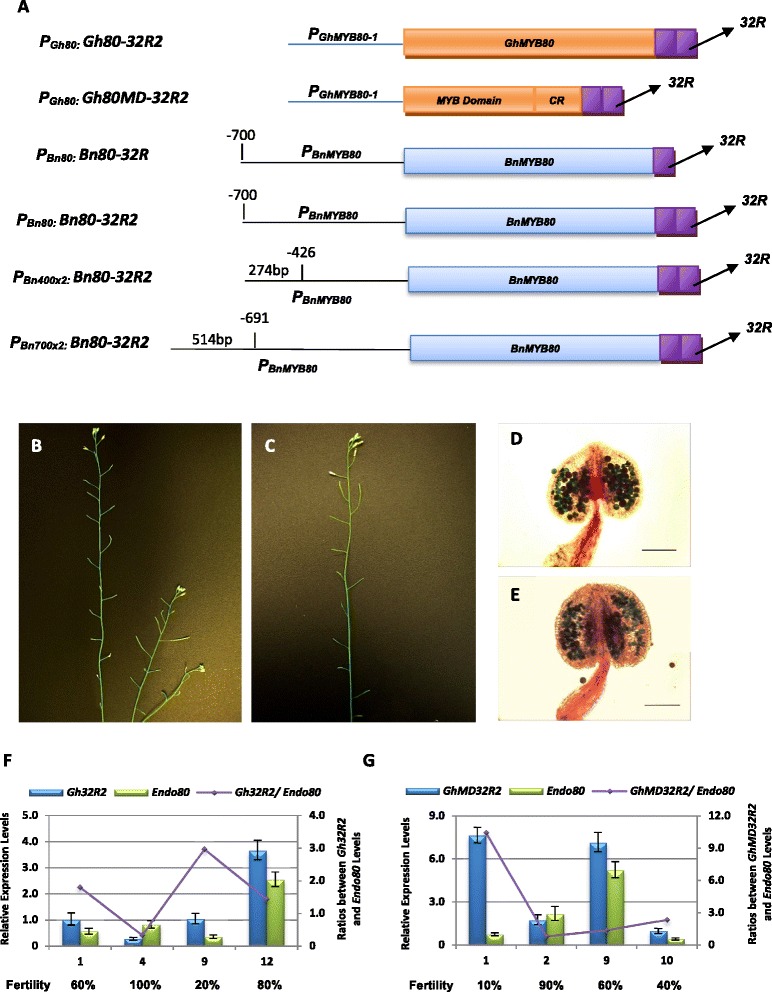
Phenotype of silique elongation and expressional analyses of transgenes in the P Gh80 :Gh80-32R2 and P Gh80 :Gh80MD-32R2 Arabidopsis. A. A schematic representation of the P Gh80 :Gh80-32R2 and P Gh80 :Gh80MD-32R2 chimeric constructs. B and C. Wild-type Arabidopsis possessing either the P Gh80 :Gh80-32R2 (B) or P Gh80 :Gh80MD-32R2 (C) transgene has a partially sterile phenotype. D and E. Alexander’s staining shows the majority of pollen grains lack cytoplasm and are aborted in the P Gh80 :Gh80-32R2 (D) and P Gh80 :Gh80MD-32R2 (E) anthers. Scale bar = 85 μm. F and G. The relative expression levels of the P Gh80 :Gh80-32R2 (F) and P Gh80 :Gh80MD-32R2 (G) transgenes in the selected lines. The higher the ratio (P Gh80 :Gh80-32R2 or P Gh80 :Gh80MD-32R2 vs. AtMYB80), the lower the plant fertility obtained. Gh32R2, P Gh80 :Gh80-32R2; GhMD32R2, P Gh80 :Gh80MD-32R2; Endo80, endogenous AtMYB80. Error bar represents SD.
PCR screening identified forty-one transgenic PGh80:Gh80-32R2 and sixty-three transgenic PGh80:Gh80MD-32R2 lines. Silique elongation in each line was examined. Approximately one-third of the transgenic PGh80:Gh80-32R2 lines and half of the PGh80:Gh80MD-32R2 lines showed less than 25% fertility (Figure 8B and C). A partially fertile phenotype (over 75% fertility) was observed in 34% of the transgenic PGh80:Gh80-32R2 lines and 3% of the transgenic PGh80:Gh80MD-32R2 lines, respectively (Additional file 7: Table. S4). Alexander’s staining of anthers from the severely sterile (less than 25% fertility) lines possessing either construct showed the majority of pollen grains lacked cytoplasmic content (Figure 8D and E). The expression levels of the PGh80:Gh80-32R2 and PGh80:Gh80MD-32R2 transgenes as well as the endogenous AtMYB80 were examined in the selected lines using qRT-PCR. Plant fertility was shown to depend on the ratio between the transcript levels of the transgenes and endogenous AtMYB80. The higher the ratio (PGh80:Gh80-32R2 or PGh80:Gh80MD-32R2 vs. AtMYB80), the lower the plant fertility obtained (Figure 8F and G). The addition of two 32R copies to the 700 bp BnMYB80 promoter driving BnMYB80 (PBn80:Bn80-32R2) was less effective than a single EAR sequence (PBn80:Bn80-32R) (Table 1). Two copies of the 700 bp BnMYB80 promoter driving the full-length BnMYB80 gene (PBn700x2:Bn80-32R2) were more effective than the two copies of the 400 bp BnMYB80 promoter (PBn400x2:Bn80-32R2). The BnMYB80-32R repressor induces male sterility more strongly in Arabidopsis than GhMYB80-32R when the two chimeric constructs were driven by their own promoters. The difference may reflect the shorter length (strength) of the GhMYB80 promoter.
Table 1.
The number of the P Bn80 :Bn80-32R , P Bn80 :Bn80-32R2 , P Bn400x2 :Bn80-32R2 and P Bn700x2 :Bn80-32R2 transgenic Arabidopsis lines obtained
| Constructs | 100% fertility | 50-75% fertility | 0% fertility | Percentage of the completely sterile lines |
|---|---|---|---|---|
| P Bn80 :Bn80-32R | 1 | 2 | 4 | 57% |
| P Bn80 :Bn80-32R2 | 2 | 5 | 3 | 30% |
| P Bn400x2 :Bn80-32R2 | 2 | 1 | 2 | 40% |
| P Bn700x2 :Bn80-32R2 | 5 | 7 | 17 | 59% |
The percentages of the completely sterile lines carrying each construct are indicated.
Discussion
Comparison of MYB80 structure and function
Among the proteins encoded by the eight MYB genes cloned from Arabidopsis, Brassica and cotton, the MYB domain, an adjacent 44 amino acid sequence and an 18 amino acid C-terminal sequence are highly conserved. The latter is extended by eight amino acids in the two cotton proteins. A variable region of 131 to 139 amino acids is located between the 44-amino acid and the C-terminal sequences, sharing 10.7% identity between the eight MYB proteins.
The sequence conservation among MYB80 proteins suggests similar functions. OsMYB80, TaMYB80, BnMYB80 (C gene) [6] and GhMYB80-1 are all able to restore male fertility of the atmyb80 mutant, implying functional conservation between monocots and dicots. The conserved 44 amino acid sequence is essential for MYB80 function as, when removed, the protein is unable to restore atmyb80 fertility.
GhMYB80 in cotton anther development
The developmental stages of cotton anther development were found to closely resemble those of Arabidopsis (Additional file 7: Table S4). In Arabidopsis AtMYB80 expression is strongest at stage 9 and tapetal cell degradation is initiated at stage 10 [1,4]. In cotton, however, tapetal cell degradation commences at anther development stage 9 and is largely completed at stage 10. These differences are consistent with the earlier increase (stage 7) and downregulation (stage 8) of MYB80 transcript levels in cotton.
Comparison of the MYB80 promoters
The promoters of all eight MYB genes share an 80 bp sequence (approximately −300 to -380 bp upstream of the ATG) which includes four cis-elements, one of which is a MYB binding site. We have not yet ascertained the importance of these elements in driving gene expression. GhMYB80 was more effective in restoring male fertility of the atmyb80 mutant when driven by the AtMYB80 (1105 bp) than the GhMYB80 promoter (443 bp), presumably reflecting the difference in promoter length. This result implies that additional cis-elements driving expression are located in the −464 to −1105 region. Alternatively, the timing of GhMYB80 promoter expression, which is perhaps slightly different from that of the AtMYB80 promoter, may also reduce the effectiveness of GhMYB80 promoter in complementing the atmyb80 mutant. The reduced autoregulation caused by the ineffective cis-elements in GhMYB80 promoter may contribute to the timing difference.
Two putative MYB-binding sites, namely MYB1 and MYB2, are situated -257bp and -246bp upstream of the transcription start site of the AtMYB80 promoter. GUS expression appeared unaffected driven by the MYB1 mutated promoter. However, when both MYB elements were mutated, GUS expression no longer ceased at anther stage 10 in Arabidopsis, persisting into stage 12 in microspores and degraded tapetal cells. Although it is not clear yet whether a mutated MYB2 element alone would affect the AtMYB80 expression, these results suggest MYB2 element plays a major role in the down-regulation of MYB80 expression at the later stages. The two MYB cis-elements in the AtMYB80 promoter are conserved in the promoter of the B. napas MYB80 C gene and the two GhMYB80 genes. However, they are absent from the wheat and rice MYB80 gene promoters, suggesting MYB80 downregulation may be regulated differently in monocots.
Disruption of the AtMYB80 gene also changes the expression pattern of its promoter. Thus the GUS expression driven by the wild-type AtMYB80 promoter was extended to stage 12 in the anthers of the homozygous atmyb80 mutant. The expression levels of the truncated AtMYB80 transcript were up-regulated in young atmyb80 anthers as shown in the microarray and qRT-PCR analyses. These results suggest that AtMYB80 protein is involved in the negative auto-regulation of its expression at the later stages of anther development. AtMYB80 positively regulates the expression of some genes but represses the expression of others [3]. The mechanism by which MYB80 changes from an activator to a repressor is not known. Three other MYB proteins, AtMYB4, 7 and 32 possess an EAR-like sequence, and have been shown to repress their own promoters [20-22].
AtMYB80 positively regulates the expression of the aspartic protease encoding gene UNDEAD. A gene that must be downregulated if the correct timing of tapetal PCD is to be achieved [3]. Thus it is critical that MYB80 expression is repressed at the appropriate stage of anther development. The downregulation of GhMYB80 at late stage 8 in cotton anthers is consistent with the earlier tapetal degradation when compared with Arabidopsis.
MYB80-EAR as an inducer of male sterility
The chimeric protein AtMYB80-EAR when introduced into Arabidopsis induces male sterility [2]. The GhMYB80 and BnMYB80 proteins fused with an EAR-like sequence, namely 32R, also resulted in male sterility in Arabidopsis. Since the sterility can be reversed [2] and MYB80 proteins from cotton, canola, wheat and rice have similar functions, the system provides a novel means to obtain hybrid vigour in crops. Important is the level of MYB80-EAR expression that can be achieved to ensure maximal levels of male sterility.
The transcript level of endogenous AtMYB80 is reduced in all lines over-expressing At80MD. The overexpressed truncated protein may compete for the AtMYB80-interacting proteins, leading to the reduced expression of the endogenous AtMYB80 gene. Whilst RNAi silencing of the endogenous AtMYB80 in At80MD lines could not be excluded as responsible for the reduction in male fertility, the silencing does not appear to significantly affect the expression of the transgene At80MD.
When the 32R sequence was fused with the truncated GhMYB80MD sequence and transformed into wild type Arabidopsis plants, the percentage of male sterile plants obtained was higher than when the full length GhMYB80 sequence was used. Fifty percent of the PGh80:Gh80MD-32R2 lines were more than 75% infertile while the figure was 30% for the full length PGh80:Gh80-32R2 lines. The At80-EAR (PAt80:At80-SRDX) construct resulted in 60% of Arabidopsis lines isolated exhibiting complete male sterility and silique abortion [2] whereas with At80MD-EAR (PAt80:At80MD-SRDX) the figure rose to 75% [6].
A strong promoter is required to drive the MYB80-EAR construct to maximize the level of male sterility obtained. The 700bp BnMYB80 promoter was more effective than the 400bp promoter, although two copies of the 700bp BnMYB80 promoter were no better than a single copy. A single EAR sequence fused to the MYB80 protein was more effective than a double sequence. The PBn700x2:Bn80-32R2 (EAR x2) construct resulted in approximately 60% of lines being completely male sterile. However, a similar percentage of lines displaying complete male sterility was obtained when Bn80-32R (single copy of EAR) was driven by a single copy of the BnMYB80 promoter (Table 1).
The results indicate that a combination of a strong promoter (driving tapetum and microspore expression) and a single copy of the EAR sequence fused to the MYB80MD protein will induce high levels of complete male sterility. In addition, the 32R EAR is less effective than the SRDX when fused to the MYB80 protein. This variability suggests the possibility of designing new EAR sequences with even greater repressive activity.
Conclusions
In this paper we extend our studies on MYB80 genes to include the Brassica A and C genomes and the two cotton orthologs. Promoter and functional analysis of the orthologs found that the expression pattern and function of a cotton ortholog are conserved and that MYB80 expression is negatively autoregulated. The developmental stages of the cotton anther were examined and GhMYB80 expression found to cease prior to the commencement of tapetal degradation.
The conservation of MYB80 genes in crops is of interest as manipulation of the gene’s expression provides a novel reversible male sterility system for obtaining hybrid vigour. We examined ways to optimize inhibition of AtMYB80 expression using a chimeric MYB80 fused with the EAR sequence from AtMYB32. A single EAR copy fused to the truncated MYB80 driven by a strong promoter (for example, B.napus MYB80) proved to be the most efficient construct for obtaining male sterility.
Methods
Plant materials and transformation
Wild type canola (B. napus cv., Westar), brown mustard (B. juncea) and cotton (G. hirsutum, Coker 315) seeds were obtained from Division of Plant Industry, Commonwealth Scientific and Industrial Research Organisation, Canberra, Australia. Wild type brussel sprout (B. oleracea) is an Australian commercial variety. Wild type Arabidopsis thaliana accession Columbia (Col-0) and the atmyb80 T-DNA insertion mutant lines were obtained from GABI-Kat (Max Planck Institute for Plant Breeding Research), the European Arabidopsis Stock Centre. Arabidopsis, canola, brown mustard and brussel sprout were grown in a plant growth room at 22°C under constant illumination. Wild type cotton was grown in a glasshouse with a temperature of 30°C/22°C (day/night). Arabidopsis transformation was performed using Agrobacterium tumefaciens strain GV3101 by dripping approximately 50 μL of the infiltration medium (2-day-grown Agrobacteria culture, 5% sucrose, 0.03% Silwet) onto each floret. The dripping procedure was repeated once a week for three weeks. Constructs were transformed into the wild type or fertile heterozygous atmyb80 plants. Genotypic and phenotypic analysis of the segregating populations was then performed in the T1 generation.
Plasmid construction
The coding sequences of the GhMYB80-1/-2, BnMYB80A, BjMYB80A, and BoMYB80C were generated by PCR amplification using primers designed from the conserved DNA sequences. The GhMYB80-1/-2 promoter sequences were obtained by the genomic walking method using the BD GenomeWalker kit (Clontech) according to the manufacturer’s protocol.
The GhMYB80 and BnMYB80 promoter fragments were cloned into pENTR/D-TOPO vector (Life Technologies) and then transferred into pKGWFS7 or pGWB533 destination vector using the LR clonase reaction. DNA fragments of the PGh80/PAt80:Gh80, GhMYB80-EAR, and AtMYB80 truncation constructs were cloned into pGWB501 destination vector. Four serial deletion of the AtMYB80 promoter fragments were amplified from the pPG construct [1] and cloned into the pBI101.1 vector using the restriction sites BamHI and HindIII. Site-specific mutagenesis was carried out using the Muta-Gene Phagemid kit (Bio-Rad) according to the manufacturer’s protocol. The two mutated promoters were cloned into the pBI101.1 vector. The double BnMYB80 promoters were created by fusing two 400 or 700 promoter repeats. The 5’ promoter repeats contain the sequence immediately upstream from TATA box (excluding the TATA box), generating a 274 plus 426 bp (double 400 promoter) and a 514 plus 691 bp (double 700 promoter) sequences. The BnMYB80-EAR fragments were fused with the single or double BnMYB80 promoter and then cloned into pCAMBIA1380 binary vector (CAMBIA). Gene specific primers are listed in Additional file 8: Table S5.
Floral buds measurement and RT/qRT-PCR Analysis
The length (from the tip of the bud to the base of the petiole) and width (the longest horizontal dimension from one side to another side) of cotton floral buds were measured under a microscope. Half of the anthers from each bud were embedded for semi-thin sectioning. The second half was used for RNA extraction. Measurements and RNA extraction were replicated for each size. Arabidopsis anther stages were determined according to the length of Arabidopsis flower bud [23].
Total RNA was extracted from the isolated anthers or floral buds using the RNeasy plant kit (Qiagen). The first strand of cDNA was synthesized using SuperScript™ III Reverse transcriptase (Life Technologies, Catalog # 18080–044) according to the original protocol. Eliminating genomic DNA contamination was then performed by DNase digestion (Life Technologies, Catalog # 18068–015). The conditions for RT-PCR amplification of cDNA were as follows: 94°C for 3 min; 26 to 28 cycles of 94°C for 30 s; 55-60°C for 30 s and 72°C for 40 s; one cycle at 72°C for 7 min. RT-qPCR was performed using the SensiFAST SYBR & Fluorescein Kit (Bioline, Catalog # BIO-96020) on the MyiQ iCycler (BIO-RAD). The PCR conditions were as follows: 94°C for 3 min; forty cycles of 94°C for 30 s; 55-60°C for 30 s; 72°C for 20 s; one cycle at 72°C for 5 min. Data was analysed using the iQ5 (BIO-RAD) software. Relative gene expression level was calculated using the primer efficiency^(−deltaCT) method. Fold change was calculated using the primer efficiency^(−delta deltaCT) method. The Arabidopsis UBIQUITIN10 (UBQ10) and G. hirsutum UBIQUITIN1 (UBI1) genes were used as reference. Gene specific primers are listed in Additional file 8: Table S5.
Sectioning of resin-embedded floral buds
Arabidopsis florets were fixed, embedded, and sectioned as described by Li [2]. Cotton anthers dissected from floral buds were fixed in FAA fixation (50% ethanol, 5% acetic acid, 3.7% formaldehyde, 41.3% water) and then embedded in LR White. Sections of cotton anther were performed in the same way as the Arabidopsis florets.
Histochemical assay of transformed arabidopsis plants
Fresh Arabidopsis floral buds were prefixed in 1% glutaraldehyde solution (made up in 50 mM sodium phosphate buffer, pH 7.4) and then covered with X-gluc solution (0.5 mg/ml X-gluc in dimethylformamide, 50 mM sodium phosphate buffer, and 0.05% Triton X-100). Samples were incubated at 37°C for 4–16 hours and washed with 95% ethanol to remove the chlorophyll. GUS activity was examined under a dissecting microscope. Arabidopsis anthers were stained with Alexander’s stain [24] and examined microscopically.
Availability of supporting data
The data set of DNA sequences supporting the results of this article is available in the GenBank repository, accession numbers KM675703 – KM675707.
Acknowledgements
We thank Edgar Sakers (La Trobe University) for providing technical support in cross-sections of Arabidopsis anther, Amila Avidic and Hanh Pham (La Trobe University) for their assistance with the plasmid construction. The first author was supported by a La Trobe University Postgraduate Research Scholarship. Part of this research was funded by an Australian Research Council Linkage Grant.
Abbreviations
- CMS
Cytoplasmic male sterility
- EAR
ERF-associated amphiphilic repression
- GUS
β-glucuronidase
- PCD
Programmed cell death
- qRT-PCR
Quantitative reverse transcription-PCR
Additional files
Nucleotide sequence alignment of MYB80 homologs from Arabidopsis (AtMYB80), canola (BnMYB80A and BnMYB80C) and cotton (GhMYB80-1).
Summary of GUS activities in the transgenic lines possessing the AtMYB80 promoter-GUS deletion constructs.
Partial promoter sequences alignment of the MYB80 homologs from Arabidopsis (pAt80), canola (pBn80A and pBn80C), cotton (pGh80), wheat (pTa80) and rice (pOs80).
GUS activities in the atmyb80 mutant lines possessing the AtMYB80 promoter-GUS construct.
Comparison of anther development stages (3 to 11) between Arabidopsis and G. hirsutum.
Expression analyses of GhMYB80 in G. hirsutum anther using semi-quantitative RT-PCR.
Plant fertility (percentage of the elongated siliques versus the total siliques) and number of the P Gh80 :Gh80-32R2 and P Gh80 :Gh80MD-32R2 transgenic lines.
Primer sequences used in this article.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YX performed the research, analyzed the data, and wrote the manuscript. SI performed the research and analyzed the data. SFL designed the research and analyzed the data. RWP wrote and edited the manuscript. All authors read and approved the final manuscript.
Contributor Information
Yue Xu, Email: yue.xu@latrobe.edu.au.
Sylvana Iacuone, Email: s.iacuone@latrobe.edu.au.
Song Feng Li, Email: s.li@latrobe.edu.au.
Roger W Parish, Email: r.parish@latrobe.edu.au.
References
- 1.Li SF, Higginson T, Parish RW. A novel MYB-related gene from Arabidopsis thaliana expressed in developing anthers. Plant Cell Physiol. 1999;40:343–347. doi: 10.1093/oxfordjournals.pcp.a029548. [DOI] [PubMed] [Google Scholar]
- 2.Li SF, Iacuone S, Parish RW. Suppression and restoration of male fertility using a transcription factor. Plant Biotech J. 2007;5:297–312. doi: 10.1111/j.1467-7652.2007.00242.x. [DOI] [PubMed] [Google Scholar]
- 3.Phan HA, Iacuone S, Li SF, Parish RW. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell. 2011;23:2209–2224. doi: 10.1105/tpc.110.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higginson T, Li SF, Parish RW. AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 2003;35:177–192. doi: 10.1046/j.1365-313X.2003.01791.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang ZB, Zhu J, Gao JF, Wang C, Li H, Li H, Zhang HQ, Zhang S, Wang DM, Wang QX, Huang H, Xia HJ, Yang ZN. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 2007;52:528–538. doi: 10.1111/j.1365-313X.2007.03254.x. [DOI] [PubMed] [Google Scholar]
- 6.Phan HA, Li SF, Parish RW. MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Mol Biol. 2012;78:171–183. doi: 10.1007/s11103-011-9855-0. [DOI] [PubMed] [Google Scholar]
- 7.Nagaharu U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. J Jap Bot. 1935;7:389–452. [Google Scholar]
- 8.Prakash S, Hinata K. Taxonomy, cytogenetics and origin of crop Brassicas, a review. Opera Bot. 1980;55:1–57. [Google Scholar]
- 9.Zhu HY, Zhang TZ, Yang LM, Guo WZ. EST-SSR sequences revealed the relationship of D-genome in diploid and tetraploid Species in Gossypium. Plant Sci. 2009;176:397–405. doi: 10.1016/j.plantsci.2008.12.007. [DOI] [Google Scholar]
- 10.Li Y, Jiang J, Du ML, Li L, Wang XL, Li XB. A cotton gene encoding MYB-like transcription factor is specifically expressed in pollen and is involved in regulation of late anther/pollen development. Plant Cell Physiol. 2013;54:893–906. doi: 10.1093/pcp/pct038. [DOI] [PubMed] [Google Scholar]
- 11.Zhang JF, Stewart JM. CMS-D8 restoration in cotton is conditioned by one dominant gene. Crop Sci. 2001;41:283–288. doi: 10.2135/cropsci2001.412283x. [DOI] [Google Scholar]
- 12.Formanová N, Li XQ, Ferrie A, DePauw M, Keller W, Landry B, Brown G. Towards positional cloning in Brassica napus: generation and analysis of doubled haploid B. rapa possessing the B. napus pol CMS and Rfp nuclear restorer gene. Plant Mol Biol. 2006;61:269–281. doi: 10.1007/s11103-006-0008-9. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Prat E, Van Lookeren Campagne MM. Hybrid seed production and the challenge of propagating male-sterile plants. Trends Plant Sci. 2002;7:199–203. doi: 10.1016/S1360-1385(02)02252-5. [DOI] [PubMed] [Google Scholar]
- 14.Fan Z, Stefansson BR. Influence of temperature on sterility of two cytoplasmic male-sterility systems in rape (Brassica napus L.) Canadian J Plant Sci. 1986;66:221–227. doi: 10.4141/cjps86-035. [DOI] [Google Scholar]
- 15.Liu WY, Gniffke PA. Stability of AVRDC's cytoplasmic male sterility (CMS) pepper lines grown under low temperatures. Capsicum Eggplant Newsletter. 2004;23:85–88. [Google Scholar]
- 16.Weider C, Stamp P, Christov N, Hüsken A, Foueillassar X, Camp KH, Munsch M. Stability of cytoplasmic male sterility in maize under different environmental conditions. Crop Sci. 2009;49:77–84. doi: 10.2135/cropsci2007.12.0694. [DOI] [Google Scholar]
- 17.Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod. 1999;11:297–322. doi: 10.1007/s004970050158. [DOI] [Google Scholar]
- 18.Parish RW, Li SF. Death of a tapetum: A programme of developmental altruism. Plant Sci. 2010;178:73–89. doi: 10.1016/j.plantsci.2009.11.001. [DOI] [Google Scholar]
- 19.Kranz H, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzaiqui A, Bevan M, Martin C, Smeekens S, Tonelli C, Paz-Ares J, Wiesshaar B. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- 20.Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 2004;40:979–995. doi: 10.1111/j.1365-313X.2004.02280.x. [DOI] [PubMed] [Google Scholar]
- 21.Hussey SG, Mizrachi E, Creux NM, Myburg AA. Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Trontiers Plant Sci. 2013;4:1–21. doi: 10.3389/fpls.2013.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peirson BN, Owen HA, Feldmann KA, Makaroff CA. Characterization of three male-sterile mutants of Arabidopsis thaliana exhibiting alterations in meiosis. Sex Plant Reprod. 1996;9:1–16. doi: 10.1007/BF00230361. [DOI] [Google Scholar]
- 24.Alexander MP. Differential staining of aborted and nonaborted pollen. Biotech Histochem. 1969;44:117–122. doi: 10.3109/10520296909063335. [DOI] [PubMed] [Google Scholar]


