Abstract
Mitochondrial inner membrane fusion depends on the dynamin-related GTPase OPA1 and the function of OPA1 is regulated by proteolytic cleavage. The mitochondrial proteases Yme1L and OMA1 cleave OPA1 at S2 and S1 sites, respectively. Here, we show that OMA1 is cleaved to a short form (S-OMA1) by itself upon mitochondrial membrane depolarization; S-OMA1 is degraded quickly but could be stabilized by CCCP treatment or Prohibitin knockdown in cells. In addition, OMA1 processing is positively correlated with OPA1 cleavage at the S1 site and the regulation of mitochondrial morphology. Thus, our results reveal the molecular mechanism for OMA1 activation toward OPA1 processing.
Keywords: mitochondrial membrane depolarization, OMA1, OPA1
Introduction
Mitochondria in healthy cells are dynamic organelles that continually fuse and divide to maintain normal mitochondrial morphology and structure 1. Some larger dynamin-related GTPases, including mitofusins (Mfn1 and Mfn2), OPA1 and Drp1 regulate mitochondrial dynamics 2. The cytosolic protein Drp1 is essential for mitochondrial division; mitofusins locate to the mitochondrial outer membrane and are required for mitochondrial outer membrane fusion; OPA1, associated with the inner membrane in the inter-membrane space of mitochondria, regulates mitochondrial inner membrane fusion 3. In humans, point mutations in these GTPases cause severe diseases such as Charcot–Marie–Tooth type 2A for Mfn2, dominant optic atrophy for OPA1, and a severe birth defect for Drp1 4–6.
Mammalian OPA1 plays roles in several processes, including mitochondrial cristae structure regulation, apoptosis prevention and mitochondrial DNA (mtDNA) and oxidative phosphorylation maintenance 7, 8. The function of OPA1 is regulated by complex patterns of alternative mRNA splicing and proteolysis. Mitochondria in humans contain eight OPA1 isoforms that arise from alternative splicing, and different variants contain either one or two cleavage sites (S1 and S2). OPA1 is proteolytically processed by the mitochondrial proteases OMA1 and Yme1L at S1 and S2 sites to generate short forms (S-OPA1) 9–11. Uncleaved OPA1 forms are considered as OPA1 long form (L-OPA1), and the mixture of both the long and short OPA1 forms facilitate mitochondrial fusion 11.
OMA1 was identified in yeast as an ATP-independent zinc metalloprotease that locates at mitochondrial inner membrane and displays activities overlapping with a m-AAA protease 12. OMA1 is a mitochondrial quality control protease and was reported as a new key regulator of metabolic homeostasis 13. Studies in mammalian cells demonstrate that OMA1 processes OPA1 to inhibit mitochondrial fusion and facilitate apoptosis 9, 10, 13, suggesting that OMA1 may sense mitochondrial dysfunction in various states of disease; the protein level of OMA1 is decreased but OMA1’s ability of processing OPA1 is activated in response to some stress stimuli 10, so why does a decrease in OMA1 protein level lead to the activation of OMA1 protease activity toward OPA1 processing? What is the molecular mechanism of OMA1 activation? It is important to uncover the role and mechanism of the OMA1-OPA1 system.
In this study, we show that OMA1 is cleaved by itself to generate S-OMA1 in response to stress stimuli. S-OMA1 is unstable and is stabilized upon membrane depolarization; furthermore, we find that the processing of OMA1 is positively associated with its activity toward OPA1 cleavage.
Results and Discussion
OMA1 is cleaved upon mitochondrial membrane potential loss or apoptosis
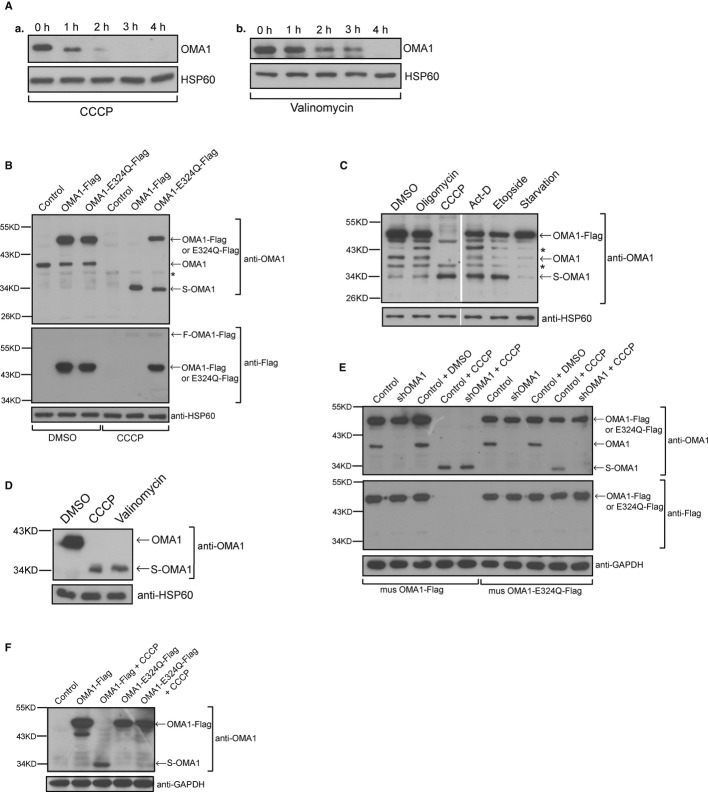
OMA1 is an ATP-independent mitochondrial protease, which cleaves OPA1 at S1 site 9, 10. In addition, the processing of OPA1 at S1 site is accelerated upon mitochondrial membrane potential loss 11. CCCP and valinomycin are effective reagents that disrupt mitochondrial membrane potential in mammalian cells; therefore, we treated HeLa cells with CCCP or valinomycin to assess the change of protein levels of OMA1. To our surprise, the protein levels of OMA1 decreased gradually upon CCCP or valinomycin treatment (Fig 1A). If the protease activity of OMA1 toward OPA1 processing is increased upon CCCP treatment 10, why does the reduction in OMA1 upon CCCP lead to activation of OMA1 protease activity toward OPA1 processing 14? We speculate that OMA1 may be cleaved rather than lost upon membrane depolarization. To test our hypothesis, HeLa cells stably expressing mouse OMA1-Flag were treated with different stimuli. As shown in Fig 1B, upon CCCP treatment for 4 h, endogenous OMA1 (~40 kDa) and exogenous overexpressed OMA1-Flag (~50 kDa) protein bands disappeared, but a new protein (~34 kDa) band appeared on the blot, suggesting that this new protein is a cleaved form of OMA1 (S-OMA1). In addition, treatment with apoptosis-inducing stimuli such as actinomycin D (Act-D) or etoposide but not starvation treatment resulted in an increase of S-OMA1 and reduction of OMA1 or OMA1-Flag (Fig 1C); upon incubation with oligomycin (an inhibitor of mitochondrial ATP synthase), endogenous OMA1 and exogenous OMA1-Flag decreased and S-OMA1 increased slightly (Fig 1C). We conclude that OMA1 is cleaved upon mitochondrial membrane depolarization or apoptosis.
Figure 1. OMA1 is processed by itself in response to membrane depolarization.
A Lysates from HeLa cells treated with 20 μM CCCP (a) or 10 μg/ml valinomycin (b) for the indicated times were analyzed by Western blot with antibodies against OMA1 and HSP60.
B HeLa cells stably expressing mouse OMA1-Flag, OMA1-E324Q-Flag or vector control were treated with DMSO or CCCP for 4 h and the cell lysates were analyzed by Western blot using the indicated antibodies. The asterisk indicates a nonspecific band.
C HeLa cells stably expressing OMA1-Flag were incubated with DMSO, oligomycin (2.5 μM, 4 h), CCCP (20 μM, 4 h), actinomycin D (Act-D, 3 μg/ml, 6 h), etoposide (100 μM, 16 h) or starved for 24 h, respectively, and were probed for OMA1 and HSP60. Asterisks indicate nonspecific bands.
D Purified mitochondrial extracts from 293T cells treated with CCCP (20 μM, 4 h) or valinomycin (10 μg/ml, 4 h) were detected by Western blot using the indicated antibodies.
E HeLa cells stably expressing mouse OMA1-Flag or OMA1-E324Q-Flag plus shRNAi against human OMA1 or vector control were incubated with DMSO or CCCP for 4 h and cell lysates were assessed for the processing of OMA1 by Western blot.
F Lysates from OMA1−/− MEFs expressing OMA1-Flag or OMA1-E324Q-Flag treated with or without CCCP (20 μM, 4 h) were probed for OMA1 and GAPDH.
To further investigate the cleavage of OMA1, we incubated 293T cells, which express high levels of endogenous OMA1, with CCCP or valinomycin and analyzed protein samples from purified mitochondrial pellets by Western blot. As expected, endogenous S-OMA1 (~34 kDa) was induced in 293T cells upon CCCP or valinomycin treatment (Fig 1D), confirming that endogenous OMA1 could be processed.
OMA1 is cleaved by itself
To examine which protease cleaves OMA1 we investigated whether the processing of OMA1 is dependent on its own activity. As the HEXXH motif of mitochondrial proteases is required for their activity, we mutated the glutamate residue of this motif in mouse OMA1 to glutamine (E324Q). 15. We then overexpressed OMA1-Flag and its mutant OMA1-E324Q-Flag in HeLa cells, incubated cells with CCCP, and analyzed OMA1 protein levels by Western blot using anti-OMA1 or anti-Flag antibody. After CCCP treatment for 4 h, endogenous OMA1 and exogenous OMA1-Flag protein bands disappeared; meanwhile, S-OMA1 accumulated, suggesting that OMA1 is cleaved completely upon CCCP treatment (Fig 1B). In contrast, only about half of OMA1-E324Q-Flag was cleaved in the presence of CCCP, suggesting that the cleavage of OMA1 is dependent on its own protease activity. To further investigate the processing of OMA1, we depleted endogenous OMA1 with shRNAi against human OMA1 (shOMA1) in HeLa cells and then overexpressed mouse OMA1-Flag or its mutant OMA1-E324Q-Flag, whose levels are not affected by shOMA1 in OMA1 knockdown HeLa cells. As shown in Fig 1E, shOMA1 resulted in the loss of endogenous OMA1 but did not lead to any change of exogenous mouse OMA1 or OMA1-E324Q. Upon CCCP treatment, OMA1 knockdown inhibited the cleavage of OMA1-E324Q-Flag since S-OMA1 is undetectable (Fig 1E), whereas it had no effect on the cleavage of OMA1-Flag in CCCP-treated HeLa cells as evidenced by the disappearance of OMA1-Flag and appearance of S-OMA1 on the blot (Fig 1E). In conclusion, OMA1 is cleaved by itself and its protease activity is required for this processing.
We further determined the processing of OMA1 in OMA1 knockout (OMA1−/−) mouse embryonic fibroblast cells (MEFs). After CCCP treatment of OMA1−/− MEFs, overexpressed OMA1-Flag was still cleaved due to self-processing (Fig 1F), while cleavage of OMA1-E324Q-Falg was completely inhibited in the presence of CCCP (Fig 1F), demonstrating that OMA1 is autocatalytically cleaved upon mitochondrial membrane depolarization.
OMA1 cleaves itself at residues 443–452 and its processing is positively correlated with its activity toward OPA1 cleavage
To determine the OMA1 processing site, the protein domains of mouse OMA1 were analyzed by SMART database, and the results show that mouse OMA1 contains a conserved domain “Peptidase_M48” (residues 232–444) (Supplementary Fig S1A); additionally, S-OMA1 is about 34 kDa in size and is undetectable by Western blot analysis with anti-Flag antibody (Fig 1E), suggesting that OMA1 is cleaved at the C-terminus. Considering that S-OMA1 may be proteolitically active, we compared the sizes of OMA1 (Δ453–521), S-OMA1 and OMA1 (Δ443–521). Supplementary Fig S1B shows that S-OMA1 is a little bit larger than OMA1 (Δ443–521) and slightly smaller than OMA1 (Δ453–521), indicating that OMA1 is processed at residues 443–452. We also inserted a Myc-tag in the OMA1 protein (between OMA1 residues 101 and 102, as shown in Supplementary Fig S1A) and examined the processing of OMA1-Myc in the presence of CCCP by Western blot analysis with anti-Myc antibody. As shown in Supplementary Fig S1C, OMA1-Myc was cleaved to a short form (S-OMA1-Myc) upon CCCP treatment, further confirming that OMA1 is cleaved at the C-terminus and S-OMA1 is an N-terminal fragment.
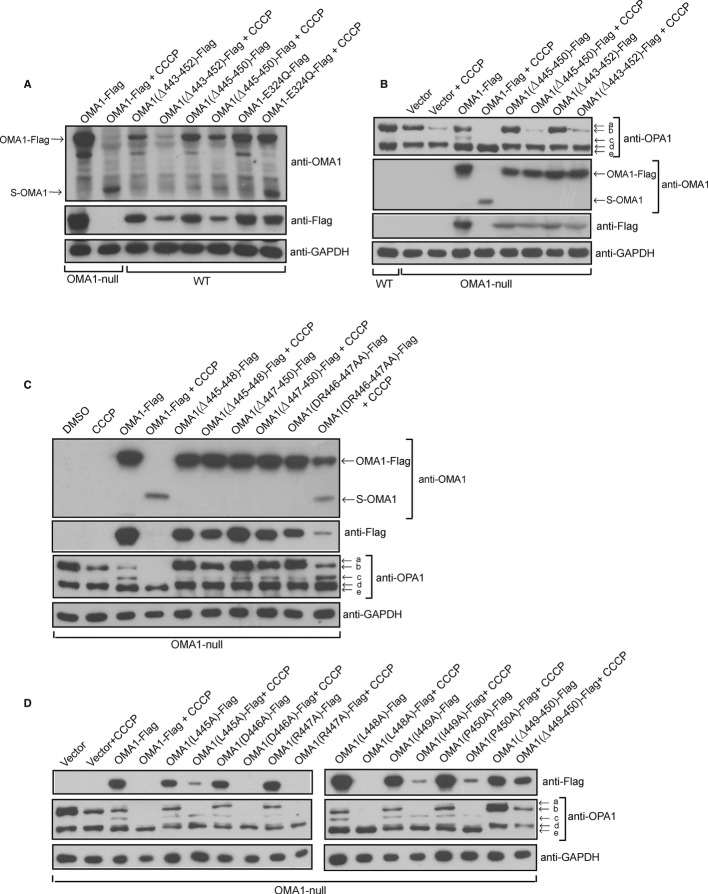
To further confirm the processing site of OMA1, we deleted ten residues 443–452 or six residues 445–450 in OMA1 and expressed OMA1 (Δ443–452)-Flag or OMA1 (Δ445–450)-Flag in WT or OMA1−/− MEFs. CCCP treatment leads to the accumulation of S-OMA1 in OMA1-Flag- or OMA1-E324Q-Flag- but not OMA1 (Δ443–452)-Flag- or OMA1 (Δ445–450)-Flag-expressing WT cells (Fig 2A); in addition, deletion of residues 443–452 or 445–450 in OMA1 could prevent the cleavage of OMA1 in OMA1−/− MEFs treated with CCCP since S-OMA1 was undetectable (Fig 2B). These results further suggest that the processing site of OMA1 is located in the region between residues 443 to 452. It should be noted that the protein levels of OMA1 (Δ443–452)-Flag- or OMA1 (Δ445–450)-Flag were reduced in response to CCCP in WT MEFs (Fig 2A), indicating that OMA1 (L-OMA1) could be degraded by itself or other mitochondrial proteases.
Figure 2. OMA1 is cleaved at residues 443 to 452 and this positively correlates with OPA1 processing.
A OMA1−/− MEFs expressing OMA1-Flag and WT MEFs expressing indicated OMA1 mutation constructs (Flag-tagged) were treated with or without CCCP (20 μM, 4 h) and whole-cell extracts were analyzed by Western blot with the indicated antibodies; “Δ” indicates deletion of residues.
B WT and OMA1−/− MEFs expressing the indicated OMA1 constructs were treated with or without CCCP (20 μM, 4 h) and the cell lysates were assessed by Western blot using the indicated antibodies.
C OMA1−/− MEFs expressing OMA1-Flag or indicated OMA1 mutants were incubated with or without CCCP (20 μM, 4 h) and the whole-cell extracts were probed with the indicated antibodies.
D Lysates from OMA1−/− MEFs expressing OMA1-Flag or OMA1 mutants as indicated treated with or without CCCP (20 μM, 4 h) were analyzed by Western blot using the indicated antibodies.
OMA1 mediates the processing of OPA1 at S1 site 9, 10 and generates the OPA1 short form bands “c” and “e” (Fig 2B). In OMA1−/− MEFs, the bands “c” and “e” disappear due to loss of cleavage at OPA1 S1 site, and exogenous expression of OMA1-Flag restores bands “c” and “e” (Fig 2C). However, exogenous expression of OMA1 (Δ443–452)-Flag or OMA1 (Δ445–450)-Flag in OMA1−/− MEFs does not restore bands “c” and “e” even in the presence of CCCP (Fig 2B), indicating that the processing of OMA1 is required for its activity toward OPA1 processing. We also expressed OMA1 (Δ445–448)-Flag, OMA1 (Δ447–450)-Flag and OMA1 (DR446–447AA)-Flag in OMA1−/− MEFs treated with or without CCCP and assessed the extent of OPA1 processing at the S1 site by Western blot analysis. As shown in Fig 2C, CCCP treatment leads to a complete loss of the OMA1-Flag long form and causes about 80% reduction in OMA1 (DR446–447AA)-Flag and a slight reduction in OMA1 (Δ445–448)-Flag and OMA1 (Δ447–450)-Flag levels. Interestingly, compared with WT OMA1-Flag, OMA1 (DR446–447AA)-Flag in OMA1−/− MEFs generates less OPA1 short form band “c” which is considered an indicator of OMA1 activity for OPA1 cleavage at S1 site here. In addition, OMA1 (Δ445–448)-Flag and OMA1 (Δ447–450)-Flag produce less OPA1 short form “c” than OMA1 (DR446–447AA)-Flag. Furthermore, OMA1 (DR446–447AA)-Flag produces more band “c” and less OPA1 long form bands “a” and “b” than OMA1 (Δ445–448)-Flag or OMA1 (Δ447–450)-Flag upon CCCP treatment in OMA1−/− MEFs (Fig 2C), suggesting that OPA1 cleavage at the S1 site is dependent on OMA1 processing. It should be noted that overexpression of OMA1-Flag in OMA1−/− MEFs results in the disappearance of protein bands “a”, “b” and “c” in response to CCCP, indicating that high expression of OMA1-Flag degrades these OPA1 protein bands but not the band “e” upon CCCP treatment. We also expressed a series of OMA1 mutant forms carrying mutations in the region between residues 443–452 (Fig 2D) in OMA1−/− MEFs and observed that increased OMA1 cleavage upon CCCP treatment was associated with increased OPA1 processing at the S1 site (Fig 2D), further confirming that OMA1 processing is positively correlated with its proteolytic activity toward OPA1 processing.
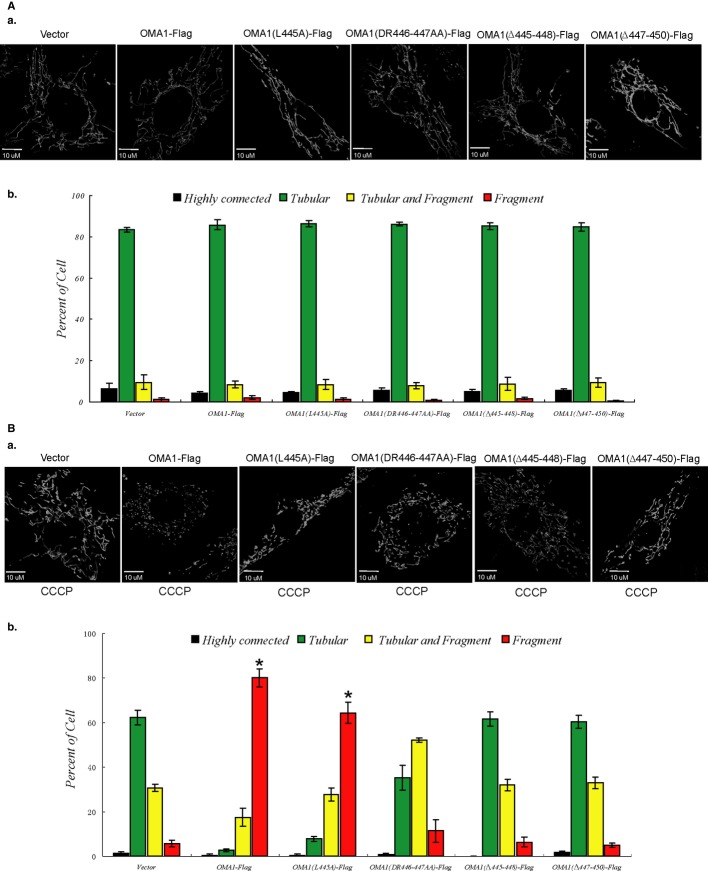
OMA1 depletion prevents CCCP-induced mitochondrial fragmentation 10, 13 and we thus examined whether the regulation of mitochondrial morphology by OMA1 is dependent on OMA1 processing. The extent of processing of OMA1-Flag, OMA1 (L445A)-Flag, OMA1 (DR446–447AA)-Flag, OMA1 (Δ445–448)-Flag or OMA1 (Δ447–450)-Flag expressed in OMA1−/− MEFs upon CCCP treatment is different (Fig 2C and D); cells expressing these proteins show similar mitochondrial morphology (tubular) under normal conditions (Fig 3A). However, compared with vector control, OMA1−/− cells expressing OMA1-Flag or OMA1 (L445A)-Flag with increased OMA1 processing induced by CCCP (Fig 2D) show significant mitochondrial fragmentation in the presence of CCCP (20 μM, 30 min) (Fig 3B), and cells expressing the mutants OMA1 (DR446–447AA)-Flag, OMA1 (Δ445–448)-Flag or OMA1 (Δ447–450)-Flag which show reduced cleavage upon CCCP (Fig 2C) show little mitochondrial fragmentation in response to CCCP treatment (Fig 3B), indicating that increased OMA1 processing upon membrane depolarization is associated with higher mitochondrial fragmentation. We conclude that mitochondrial morphology alteration upon membrane depolarization is positively correlated with OMA1 self-processing.
Figure 3. The role of OMA1 processing in regulation of mitochondrial morphology.
A, B OMA1−/− MEFs expressing OMA1-Flag or OMA1 mutation constructs were treated with DMSO (A) or CCCP (20 μM, 30 min) (B); mitochondrial morphology of cells expressing mito-GFP was visualized (a) by confocal microscope and was scored (b) according to the criteria detailed in Materials and Methods; standard deviations (s.d.) from 3 independent experiments in which 100 cell hybrids were scored. For statistical analysis, Student’s t-test was used (*P < 0.05 versus vector). A value of P < 0.05 was considered significant. Statistical significant differences are shown with asterisks.
The short form of OMA1 is less stable than the long form
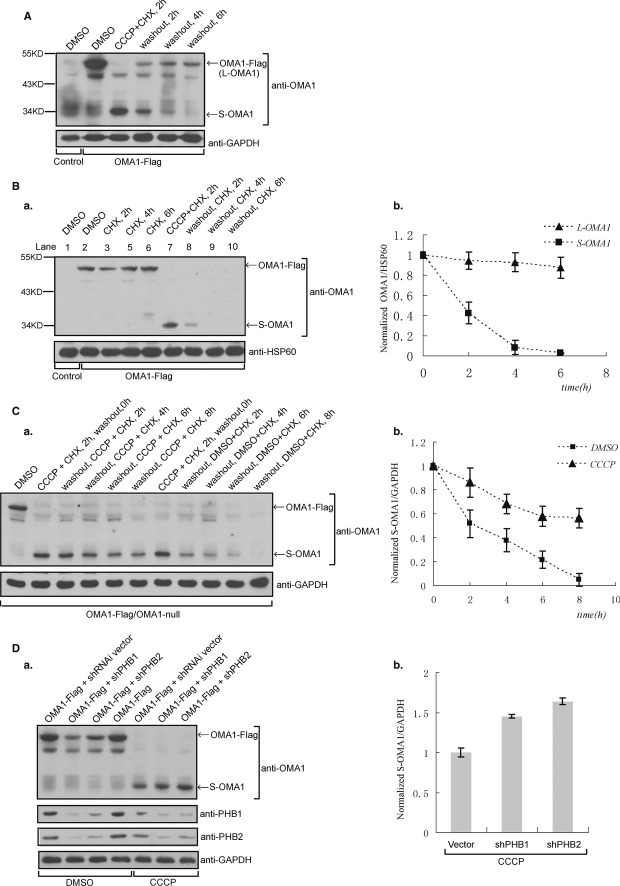
To investigate the stability of OMA1, OMA1-Flag-expressing OMA1−/− MEFs were treated with CCCP plus cycloheximide (CHX) for 2 h and then were washed to remove residual CCCP and CHX, followed by incubation with fresh medium for additional 2, 4 or 6 h. Fig 4A shows that CCCP plus CHX treatment induces complete processing of OMA1-Flag, as evidenced by the disappearance of the OMA1 long form (L-OMA1) and appearance of the OMA1 short form (S-OMA1). After washout, S-OMA1 was quickly degraded within 6 h while L-OMA1 levels recovered rapidly (Fig 4A), indicating that S-OMA1 is less stable than L-OMA1. To further test the stability of L-OMA1 and S-OMA1, we treated OMA1−/− MEFs expressing OMA1-Flag with CHX. As shown in Fig 4B, L-OMA1 protein levels slightly decreased after CHX treatment for 6 h. In contrast, S-OMA1 generated by cotreatment with CCCP plus CHX disappeared after 4 h in the presence of CHX (Fig 4B), and S-OMA1 levels decreased by 80% after CHX treatment for 2 h (Fig 4B-b), revealing that S-OMA1 is unstable and is completely degraded within 4 h while L-OMA1 remains stable under normal conditions. We also monitored the stability of the other OMA1 short form (C-terminal) generated by OMA1 cleavage. The predicted size for this short form tagged with Flag (C-OMA1-Flag) is about 17 kDa. We tried to detect C-OMA1-Flag by Western blot with anti-Flag antibody, but C-OMA1-Flag is undetectable even in response to CCCP treatment in cells (Supplementary Fig S2), indicating that C-OMA1 degrades immediately.
Figure 4. The short form of OMA1 is unstable and is stabilized by CCCP treatment or Prohibitin knockdown in cells.
A OMA1-Flag-expressing OMA1−/− MEFs were treated with CCCP (20 μM) and cycloheximide (CHX, 50 μg/ml) for 2 h; after washing and culturing them in fresh medium for additional 2, 4 or 6 h, cell lysates were analyzed by Western blot with anti-OMA1 and anti-GAPDH antibodies.
B OMA1-Flag-expressing OMA1−/− cells were incubated with CHX for 2, 4 and 6 h (a); in addition, the same cells were treated with CCCP plus CHX for 2 h to induce S-OMA1 and were then washed and cultured in the presence of CHX for additional 2, 4 and 6 h. Cell lysates were probed for OMA1 and HSP60. (b) The intensity of protein bands HSP60, OMA1 long form (L-OMA1) and short form (S-OMA1) in the blot (a) were calculated with ImageJ software and the ratios OMA1/HSP60 were normalized.
C S-OMA1 is produced by treatment of CCCP plus CHX for 2 h in OMA1-Flag-expressing OMA1−/− MEFs; cells containing S-OMA1 were then cultured in the presence of CCCP and CHX or CHX only for additional 2, 4, 6 and 8 h. The stability of S-OMA1 was analyzed by Western blot (a) with anti-OMA1 and anti-GAPDH antibodies; the ratio of S-OMA1/GAPDH protein band intensities was normalized (b).
D Control, Prohibitin 1 or 2 knockdown (shPHB1 or shPHB2) OMA1−/− MEFs expressing OMA1-Flag were treated with or without CCCP (2 h); cell lysates were assessed by Western blot (a) with indicated antibodies; the ratio of S-OMA1/GAPDH protein band intensities for CCCP-treated cell samples was normalized (b).
Data information: In (C) and (D), data are presented as mean ± s.d. of n = 3 (the values represent means and error bars indicate standard deviations from 3 independent experiments).
The degradation of S-OMA1 is dependent on the processing of OMA1 but independent of L-OMA1
The processing of OMA1 is controlled by itself (Fig 1), and S-OMA1 is an unstable protein (Fig 4B), so we then examined whether the degradation of S-OMA1 is regulated by OMA1. Since the processing site of OMA1 is located between residues 443–452, OMA1 (Δ443–521)-Flag or OMA1 (Δ453–521)-Flag which are similar to S-OMA1 in protein size were expressed in OMA1−/− MEFs or OMA1−/− MEFs expressing OMA1-Flag or OMA1-E324Q-Flag. OMA1 (Δ443–521)-Flag and OMA1 (Δ453–521)-Flag were degraded upon CCCP treatment in OMA1-Flag-expressing OMA1−/− MEFs but not in OMA1−/− MEFs expressing OMA1-E324Q-Flag (Supplementary Fig S3A), suggesting that the processing of OMA1 is required for the degradation of S-OMA1.
To determine whether L-OMA1 mediates the degradation of S-OMA1, we expressed OMA1-Flag in OMA1−/− MEFs. We observed that the cotreatment of CCCP plus CHX for 2 h results in a complete loss of L-OMA1, while S-OMA1 is still being continuously degraded even in the presence of CHX after CCCP washout (Fig 4B), suggesting that the degradation of S-OMA1 is independent of L-OMA1. Additionally, S-OMA1 is more quickly degraded than S-OMA1-E324Q in the presence of CHX (Supplementary Fig S3B), suggesting that S-OMA1 is activating its own degradation since no L-OMA1 was synthesized in the cells incubated with CHX.
S-OMA1 is stabilized upon CCCP treatment or Prohibitin knockdown
CCCP treatment increases OMA1 activity toward OPA1 processing but promotes processing of L-OMA1, indicating that OMA1’s cleaved product S-OMA1 plays an important role in OMA1 activity. To examine the effect of CCCP on the stability of S-OMA1, we treated OMA1−/−MEFs expressing OMA1-Flag with CCCP and CHX for 2 h and found that this leads to the complete cleavage of L-OMA1 and the generation of S-OMA1 (Fig 4C-a). Incubation with CHX plus DMSO for 8 h results in complete degradation of S-OMA1 (Fig 4C), while S-OMA1 levels declined by ca. 40% after incubation with CHX plus CCCP for 8 h (Fig 4C-b), suggesting that CCCP treatment stabilizes S-OMA1 in cells.
Since downregulation of the Prohibitin complex increases the processing of OPA1 at the S1 site 3, we tested whether the Prohibitin complex is associated with OMA1 processing. We depleted Prohibitin 1 (PHB1) or Prohibitin 2 (PHB2) in OMA1-Flag-expressing OMA1−/− cells, and analyzed OMA1 processing by Western blot with an anti-OMA1 antibody. We observed that PHB1 or PHB2 knockdown resulted in a marked decline of L-OMA1 (Fig 4D). To further examine whether the decrease in L-OMA1 is due to increased cleavage, we treated PHB1 or PHB2 knockdown resulted in a marked cells with CCCP and monitored S-OMA1 by Western blot. As shown in Fig 4D, in the presence of CCCP, PHB1 or PHB2 knockdown cells show increased levels of S-OMA1, indicating that Prohibitin knockdown further stabilizes S-OMA1 in the presence of CCCP, suggesting that the Prohibitin complex is associated with OMA1 processing and activity.
S-OMA1 is sufficient for OPA1 processing
To determine whether S-OMA1 alone is sufficient for OPA1 processing, the cleavage of newly synthesized L-OPA1 was tested by Western blot. As shown in Supplementary Fig S4A, L-OPA1 is completely processed after CCCP treatment, and L-OPA1 is detectable 2 h after CCCP washout in WT cells, but only 4 h after CCCP washout in OMA1-Flag-expressing cells, indicating that WT cells recovered L-OPA1 more quickly than WT cells containing S-OMA1; in addition, S-OMA1 is still detectable 2 h after CCCP washout in OMA1-Flag-expressing cells, suggesting that S-OMA1 is sufficient for L-OPA1 processing. To exclude the effect of de novo synthesized OMA1 on L-OPA1 processing, we used Retro-X Tet-On Advanced Inducible Expression System to express OMA1-Flag in OMA1−/− MEFs. Upon incubation with doxycycline (Dox; a tetracycline derivative), cells express OMA1-Flag (Supplementary Fig S4B); without Dox, the expression of OMA1-Flag is shut down but other endogenous proteins such as OPA1 are still expressed. As shown in Supplementary Fig S4C, L-OMA1 recovered slightly in lane 6 (Dox, 8 h) but not in lanes 7-14 (without Dox), indicating that no new L-OMA1 was synthesized and that only S-OMA1 was present to process newly synthesized L-OPA1. Compared with lane 2, lanes 7–10 have increased OPA1 short form band “c” (production is dependent on OMA1) (Supplementary Fig S4C), suggesting that S-OMA1 is sufficient for the processing of newly synthesized L-OPA1. Additionally, as a result of stabilization of S-OMA1 by CCCP treatment, little L-OPA1 is detectable in lanes 11–14 of Supplementary Fig S4C, indicating that de novo synthesized L-OPA1 is processed by S-OMA1, further confirming that S-OMA1 has the ability to process OPA1. It should be noted that the activity of S-OMA1 on OPA1 processing is distinct from the activity on S-OMA1 degradation because the processing of OPA1 is promoted, while S-OMA1 is stabilized in cells treated with CCCP. We also expressed S-OMA1 (OMA1_Δ443-521, OMA1_Δ453-521 or OMA1_Δ448-521 which is similar with S-OMA1 in size) in OMA1−/− MEFs cells. We did, however, not observe any effects on OPA1 processing and mitochondrial morphology, suggesting that ectopically expressed S-OMA1 alone was not sufficient to induce OPA1 processing for unknown reasons.
We performed co-IP experiments and showed that exogenously expressed OMA1-E324Q-Flag (long form) is able to precipitate endogenous OMA1 (long form) in HeLa cells (Supplementary Fig S4D-a), suggesting that OMA1 long form can interact with itself. Upon CCCP treatment, exogenous OMA1-Flag and endogenous OMA1 were degraded and S-OMA1 accumulated (lane 2 in Supplementary Fig S1C-b) due to OMA1 processing; in addition, WT OMA1-Flag (L-OMA1) failed to bind to S-OMA1 (lane 4 in Supplementary Fig S4D-b) since L-OMA1 is absent due to self-processing induced by CCCP. This suggests that S-OMA1 regulation of OPA1 processing is independent of L-OMA1 when cells undergo membrane depolarization. Additionally, in CCCP-treated HeLa cells expressing exogenous OMA1-E324Q-Flag, both L-OMA1 and S-OMA1 are present (lane 1 in Supplementary Fig S4D-b); as shown in Supplementary Fig S4D-b (lane 3), OMA1-E324Q-Flag could precipitate S-OMA1, suggesting that S-OMA1 is able to bind L-OMA1 and may degrade L-OMA1.
Taken together, our findings suggest the following mechanism for the activation of OMA1 (Supplementary Fig S5): OMA1 is self-cleaved at the C-terminus upon mitochondrial depolarization or apoptosis. The cleaved short form S-OMA1 has lower stability than L-OMA1 and is stabilized in response to membrane depolarization. Importantly, the processing of OMA1 is positively correlated with its activity toward OPA1 processing and the regulation of mitochondrial morphology (Fig 3). S-OMA1 contains an intact protease domain and may be sufficient for OPA1 processing. Additionally, compared with L-OMA1, S-OMA1 may have a different conformation or localization in mitochondria and thus may be an active form of OMA1. Our findings are consistent with a very recent report, published during revision of this work, showing that OMA1 is activated in response to various stress insults and that the autocatalyic degradation of OMA1 is associated with OMA1 activation 16. It will be interesting and important in the future to identify new OMA1 substrates and uncover the novel roles of OMA1 in mitochondrial function and related diseases.
Materials and Methods
Cell culture and transfection
MEFs, HeLa and 293T cells were maintained in Dulbecco’s Modified Eagle’s Medium containing penicillin/streptomycin, 10% fetal bovine serum and 1 mM l-glutamine. All cells were maintained, transfected and infected as described previously 17.
Antibodies and reagents
The following antibodies were used in this study: anti-OPA1 (BD Biosciences) and anti-Flag (Sigma); polyclonal anti-OMA1, anti-GAPDH and anti-HSP60 (Santa Cruz Biotechnology). The reagents used in this paper were as follows: CCCP, actinomycin D and CHX were purchased from Sigma-Aldrich, etoposide was from Enzo Life Sciences, Lipofectamine 2000 was from Invitrogen.
Acknowledgments
The authors thank Dr. David Chan (California Institute of Technology) and Dr. Alexander van der Bliek (University of California, Los Angeles) for communicating results before publication; we thank Dr. Carlos López-Otín (Universidad de Oviedo) for gift of OMA1−/− MEFs. This work was supported by National Key Basic Research Program of China (2013CB531200) and the National Natural Science Foundation of China (31171357).
Authors contributions
KZ and HL performed the research; KZ and ZS designed the study and analyzed the data; ZS supervised the research and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Ghochani M, McCaffery JM, Frey TG, Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Toothneuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- Griparic L, van der Wel NN, Orozco IJ, Peters PJ, van der Bliek AM. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J Biol Chem. 2004;279:18792–18798. doi: 10.1074/jbc.M400920200. [DOI] [PubMed] [Google Scholar]
- Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI, Langer T. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol. 2009;187:1023–1036. doi: 10.1083/jcb.200906084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol. 2009;187:959–966. doi: 10.1083/jcb.200906083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Chen H, Fiket M, Alexander C, Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser M, Kambacheld M, Kisters-Woike B, Langer T. Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J Biol Chem. 2003;278:46414–46423. doi: 10.1074/jbc.M305584200. [DOI] [PubMed] [Google Scholar]
- Quirós PM, Ramsay AJ, Sala D, Fernández-Vizarra E, Rodríguez F, Peinado JR, Fernández-García MS, Vega JA, Enríquez JA, Zorzano A, et al. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J. 2012;31:2117–2133. doi: 10.1038/emboj.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H, Soubannier V. Mitochondrial function: OMA1 and OPA1, the grandmasters of mitochondrial health. Curr Biol. 2010;20:R274–R276. doi: 10.1016/j.cub.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Truscott KN, Lowth BR, Strack PR, Dougan DA. Diverse functions of mitochondrial AAA+ proteins: protein activation, disaggregation, and degradation. Biochem Cell Biol. 2010;88:97–108. doi: 10.1139/o09-167. [DOI] [PubMed] [Google Scholar]
- Baker MJ, Lampe PA, Stojanovski D, Korwitz A, Anand R, Tatsuta T, Langer T. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 2014;33:578–593. doi: 10.1002/embj.201386474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Li H, Zhang K, Jian F, Tang J, Song Z. Loss of Yme1L perturbates mitochondrial dynamics. Cell Death Dis. 2013;4:e896. doi: 10.1038/cddis.2013.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.