Abstract
Nylon-3 polymers (poly-β-peptides) have been investigated as synthetic mimics of host-defense peptides in recent years. These polymers are attractive because they are much easier to synthesize than are the peptides themselves, and the polymers resist proteolysis. Here we describe in vitro analysis of selected nylon-3 copolymers against Clostridium difficile, an important nosocomial pathogen that causes highly infectious diarrheal disease. The best polymers match the human host-defense peptide LL-37 in blocking vegetative cell growth and inhibiting spore outgrowth. The polymers and LL-37 were effective against both the epidemic 027 ribotype and the 012 ribotype. In contrast, neither vancomycin nor nisin inhibited outgrowth for the 012 ribotype. The best polymer was less hemolytic than LL-37. Overall, these findings suggest that nylon-3 copolymers may be useful for combatting C. difficle.
Introduction
Clostridium difficile is a Gram-positive, endospore-forming anaerobe that causes life-threatening intestinal infections. C. difficile infections, or CDIs, lead to billions of dollars in healthcare costs and result in over 14,000 deaths per year in the United States alone.1C. difficile has been listed by the Centers for Disease Control and Prevention (CDC) as the highest level threat of antibiotic resistance in the United States.1 Because C. difficile is a strict anaerobe, the bacterium can survive outside of the host intestine only as a dormant spore.2 For C. difficile to cause disease, the spores must be ingested and germinate when exposed to bile salts in the intestine, yielding the vegetative form of the bacterium.3 Once in the vegetative form, C. difficile can produce the toxins that are responsible for disease manifestations.4,5
C. difficile infections are often preceded by the use of therapeutic antibiotics to treat unrelated bacterial infections.6 Antibiotic use disrupts the indigenous microbiota, allowing C. difficile to colonize and proliferate within the intestine.7,8 Current treatment of CDI typically consists of metronidazole, vancomycin, or, most recently, fidaxomicin. Unfortunately, these antibiotics are not able to treat all CDIs, and recurrence of disease occurs in many patients, especially when infections involve the epidemic 027 isolates.9−11 To combat this challenge, new strategies are being explored for the treatment of CDI.12
Host-defense peptides (HDPs) have demonstrated potent activity against pathogenic bacteria and are considered promising candidates for the treatment of bacterial infections.13−15 Indeed, the human HDP LL-37 is a potent inhibitor of C. difficle growth.16,17 However, stepwise solid-phase synthesis of peptides is expensive. Synthetic polymers that can mimic the antimicrobial properties of HDPs are attractive because their production should be more facile than that of sequence-specific peptides, and the polymers resist proteolytic degradation.18−31 In this study, we evaluated a series of nylon-3 copolymers for the ability to inhibit the outgrowth of C. difficile spores and the growth of vegetative cells. Although a variety of synthetic polymers have recently been examined for inhibition of bacterial growth,18−33 we are not aware of previous efforts to assess the impact of synthetic polymers on the C. difficle or more broadly on pathogenic spore outgrowth.
Results
Nylon-3 Polymers Are Active against C. difficile Vegetative Cells
In preliminary studies, a set of 22 nylon-3 cationic homopolymers and binary hydrophobic-cationic copolymers was evaluated for the ability to inhibit growth of vegetative C. difficile (strain R20291) (Supporting Information, Figures S1 and S2). All nylon-3 polymers were active against R20291 vegetative cells, with a minimum inhibitory concentration (MIC) range of 12.5–100 μg/mL. Among copolymers containing the CH hydrophobic unit, 50:50 MM:CH and 50:50 DM:CH displayed the lowest MIC values, while among copolymers containing the TM hydrophobic unit, 50:50 DM:TM displayed the lowest MIC value. These three copolymers (Figure 1) were selected for further study with two C. difficile strains, R20291 (027 ribotype) and 630Δerm (012 ribotype), as active vegetative cell cultures. These two distinct strains were chosen because the genomes of both have been sequenced, and these strains are genotypically quite different from one another. 630 is the most commonly studied laboratory strain and is easier to manipulate genetically, while R20291 represents the current epidemic isolates. For comparison, three agents of known efficacy, the clinical antibiotic vancomycin, the lantibiotic nisin, and the human host-defense peptide LL-37,34 were also evaluated. As shown in Table 1, each agent demonstrated similar efficacy against vegetative cell growth for the two C. difficile strains. The polymer MIC values were 12.5–25 μg/mL, which is inferior to the MIC values of vancomycin, comparable to the MIC values of LL-37, and superior to the MIC values of nisin.
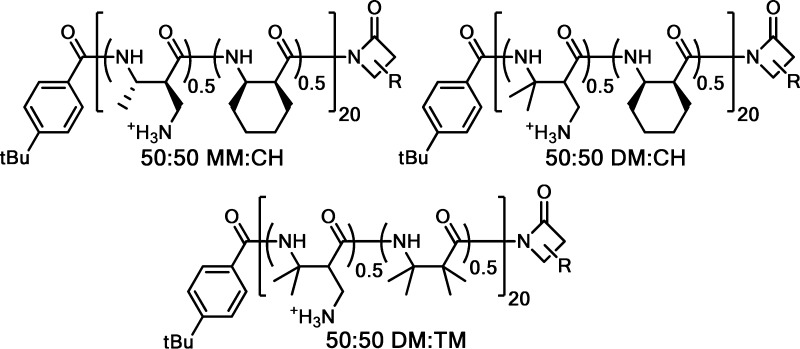
Figure 1.
Selected heterochiral nylon-3 polymers displaying the best activity in initial screening (see Supporting Information, Figures S1 and S2 for all polymers used in the initial screening). R at the C-termini of the polymer drawings represents the side chain of either of the two subunits within that polymer. (For each sample, some polymer chains will have one unit at the C-terminus, and other polymer chains will have the other unit at the C-terminus.)
Table 1. Activity of Nylon-3 Polymers and Antimicrobial Peptides toward C. difficile Vegetative Cells and Spores.
|
R20291 |
630Δerm |
|||
|---|---|---|---|---|
| antimicrobial | MICa | OICb | MICa | OICb |
| 50:50 MM:CH | 25 | 6.25 | 25 | 12.5 |
| 50:50 DM:CH | 12.5 | 6.25 | 12.5 | 12.5 |
| 50:50 DM:TM | 12.5 | 3.13 | 12.5 | 6.25 |
| LL-37 | 10 | 5 | 10 | 10 |
| nisin | 180 | 22.5 | 180 | >720c |
| vancomycin | 0.5 | 0.25 | 1 | >32c |
The miminum inhibitory concentration for vegetative cell growth (MIC; μg/mL).
The minimum inhibition concentration for spore outgrowth (OIC; μg/mL).
Higher concentrations were not examined.
Nylon-3 Polymers Reduce C. difficile Spore Outgrowth
During infection, C. difficile is present in both the vegetative and spore forms. The transition from dormant spore to vegetative bacillus is initiated by the germinant taurocholate, a bile salt found within the intestine. Once initiated, germination progresses in a pre-programmed manner that does not require active cellular metabolism (Figure 2).35−38 This transition is followed by an outgrowth phase in which metabolic processes are re-established, the cell is remodeled, and vegetative growth resumes.3,39 During the outgrowth phase the transitioning cell has limited ability of adapt to stress, creating a window of vulnerability to environmental conditions. Thus, we explored whether nylon-3 polymers can influence spore survival, germination, or the subsequent outgrowth into vegetative cells.
Figure 2.
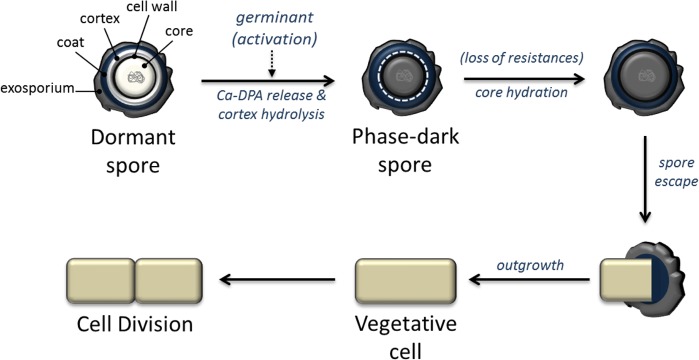
Model of spore germination and outgrowth in C. difficile. Adapted from refs (35) and (37).
To test whether polymers inhibit the outgrowth of spores into vegetative cells, we replicated MIC experiments using spores as inoculum and supplementing with 0.1% taurocholate to stimulate germination; it should be noted that this experiment does not specifically measure sporicidal activity. Table 1 summarizes the results, as manifested by outgrowth inhibitory concentration (OIC). The nylon-3 polymers displayed OIC values in the range 3.13–12.5 μg/mL, indicating that these materials are comparable to or slightly more effective at preventing the outgrowth of spores relative to their inhibition of vegetative cell growth (OIC values comparable to or slightly lower than MIC values). In addition, each polymer and each of the comparison compounds was more effective at blocking outgrowth of the clinically important 027 epidemic spores (R20291 strain) than at blocking outgrowth of the non-epidemic 630 strain. This strain-dependent variation was small for the polymers and LL-37 but large for nisin and vancomycin, with the latter two antimicrobial agents displaying no inhibition of spore outgrowth for the 630 strain. These observations suggest that strain-dependent differences in spore composition and/or outgrowth characteristics can significantly affect susceptibility to inhibition of outgrowth by conventional antibiotics but not by LL-37 or the nylon-3 polymers. In addition, the contrast between low MIC values but high OIC values for nisin and vancomycin toward the 630 strain shows that the inhibition of spore outgrowth differs in some fundamental way from inhibition of vegetative cell growth.
Antimicrobial Polymers Inhibit Spore Outgrowth but Not Germination
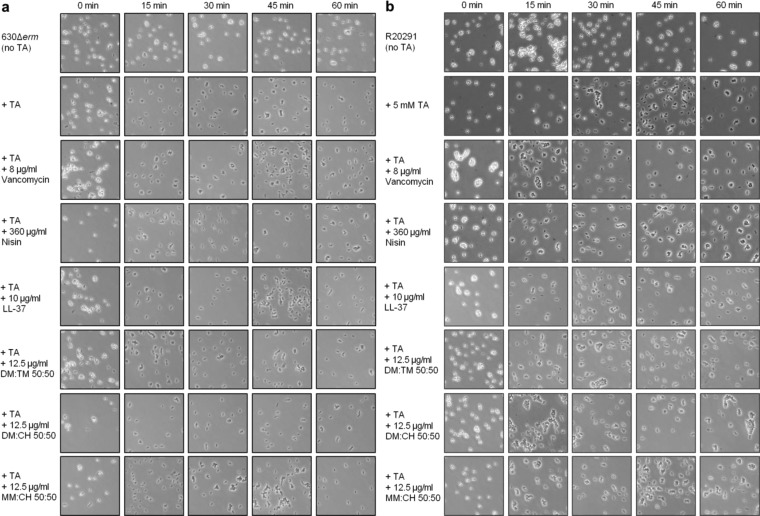
The inhibitory effects documented in Table 1 could arise in at least three different ways: (1) destruction of spore viability, (2) prevention of germination, or (3) inhibition of growth of cells transitioning from the spore to the vegetative form. In order to gain insight on the mechanism(s) of action, we conducted experiments to determine whether germination is inhibited by the polymers or reference compounds. Spores were incubated with each agent at concentrations above the OIC for 24 h in the absence of germinant. Spores were then diluted and plated onto BHIS agar medium containing the germinant taurocholate. The pre-incubation resulted in moderate decreases in colony-forming units (CFU) for all agents tested (4-fold or less; data not shown). As shown by the microscopy data in Figure 3, pre-incubation of spores with the polymers or reference compounds followed by the addition of germinant resulted in no change in the rates of spore germination for either C. difficile strain (transition from phase bright to phase dark spores). These results indicate that nylon-3 polymers and the reference compounds do not destroy spores or directly block germination, but instead they target the germinated spores. Spores are highly resistant to environmental insults, but as a spore converts into a vegetative cell during the outgrowth phase, the cell surface is completely remodeled, and there are drastic changes in gene expression necessary for growth. As a result, the spore-outgrowth period represents a point of vulnerability to antimicrobial agents, as manifested in our observations.34,40
Figure 3.
Phase-contrast microscopy of C. difficile spores incubated with antimicrobials. Purified C. difficile spores from strain 630 (a) and R20291 (b) were incubated in MH broth supplemented with 5 mM taurocholate (TA) germinant and antimicrobial compounds as indicated. Samples were taken for phase-contrast microscopy at 15 min intervals over 1 h as described in the Materials and Methods.
Evidence of Variation in Germination Propensity among Spores
We conducted experiments to determine whether any spores fail to germinate under the conditions of the OIC measurement but remain viable for subsequent germination and outgrowth. Spores were incubated for 24 h in the presence of germinant (taurocholate) and one of the polymers or other agents at multiple concentrations above the OIC. These solutions were then diluted 10–100-fold into sterile PBS, and further diluted upon plating onto BHIS agar medium containing germinant to achieve concentrations below the OIC of the relevant antimicrobial agent. Spore survival was calculated as the percentage of colony-forming units retrieved on outgrowth plates relative to the initial spore inoculum (100%). These studies were conducted with both strains of C. difficile; however, neither vancomycin nor nisin was assessed with strain 630, since these compounds did not inhibit spore outgrowth for this strain (Table 1).
The data from these experiments reveal that a small proportion of the original spores, typically in the range 0.5–3%, remain in a viable form in samples for which no spore-outgrowth activity could be detected in the OIC analysis (see Table S1). Within the limits of experimental uncertainty, this residual proportion of germination-competent spores did not change as the concentration of each nylon-3 polymer or other antibacterial agent was increased above the OIC. The small sub-population of germination-competent spores detected via these experiments presumably reflects an intrinsic heterogeneity within any sample of C. difficile spores. It is possible that the community derives a selective advantage if a small set of spores fails to undergo germination upon a given exposure to favorable conditions but nevertheless remains competent for germination at a subsequent time. Such a germination-resistant population might be functionally comparable to the small population of slow-growing “persister cells” that are proposed to occur within bacterial populations and underlie the development of antibiotic resistance.41 If our hypothesis regarding the nature of the germination-resistant spores is correct, then the nylon-3 polymers might have an advantage in vivo relative to HDPs such as LL-37 in blocking C. difficile growth: peptides are inherently susceptible to protease-mediated destruction, but previous studies with discrete β-peptide oligomers suggest that the nylon-3 polymers are not substrates for proteases.42 Thus, the polymers should retain their ability to inhibit outgrowth of slow-germinators over long periods in vivo, while a peptide would be rapidly inactivated.
Hemolytic Activities
HDPs and HDP-mimetic polymers are thought to exert antibacterial effects via disruption of membrane barrier function. Therefore, prokaryotic vs eukaryotic cell selectivity is often assessed by determining whether an agent that exerts antibacterial activity also causes disruption of human red blood cell (hRBC) membranes (“hemolysis”). As shown in Figure 4, the three nylon-3 copolymers and the three comparison compounds display varied hemolytic activities; the data for the polymers are consistent with previously reported results.43 Host-defense peptide LL-37 and polymer 50:50 DM:CH both display significant hemolytic activity. In contrast, 50:50 DM:TM causes very little hemolysis at the MIC/OIC and only mild hemolysis at high concentrations (400 μg/mL). Thus, 50:50 DM:TM manifests the most favorable activity profile among the materials we have evaluated, since this non-hemolytic polymer is quite active against both C. difficile strains in terms of inhibiting vegetative cell growth and spore outgrowth.
Figure 4.
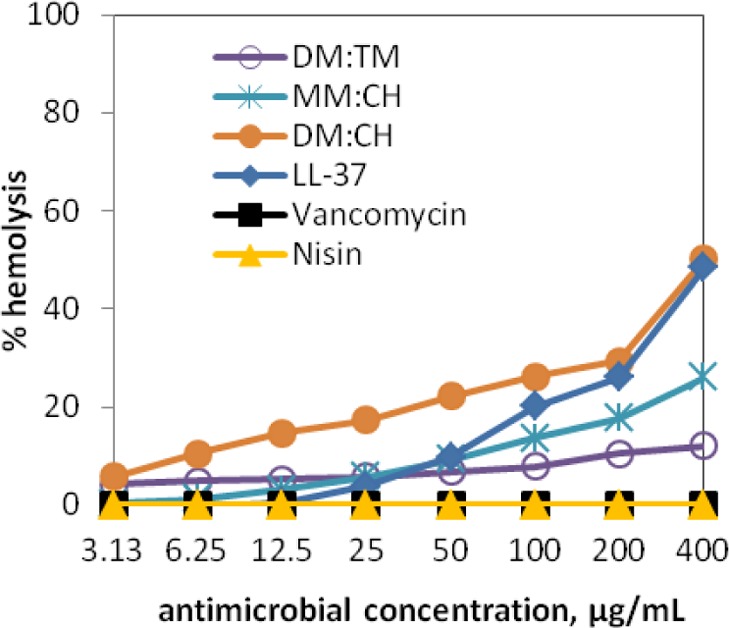
Hemolytic profiles of selected nylon-3 polymers and antimicrobials.
Discussion
C. difficile infections are often recurrent, and few effective antimicrobial treatment options are available. Because C. difficile spores are not inactivated by standard drugs or common disinfectants, patients may become re-infected by endogenous spores or spores lingering in their environment.44,45 As a result, clinical symptoms of CDI frequently return when antibiotic therapy is discontinued.46−48 With so few effective options for the treatment of CDIs and antimicrobial resistance pressure from frequent use of conventional antibiotics such as vancomycin, there is an urgent need for additional treatment strategies to combat infections by this pathogen.
In this study, we evaluated nylon-3 polymers for inhibitory activity toward C. difficile. Some of these polymers were previously shown to be active against other pathogenic bacteria including methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus faecium (VREF), Salmonella enterica LT2, Bacillus cereus ATCC14579, Pseudomonas aeruginosa PA1066, and the uropathogenic E. coli CFT073.43 Our initial survey identified three hydrophobic/cationic copolymers that are particularly effective at preventing C. difficile growth: 50:50 DM:TM, 50:50 DM:CH, and 50:50 MM:CH. These polymers inhibit growth of the vegetative form of the pathogen, and they prevent outgrowth of the spore form of C. difficile. Preventing growth of vegetative cells is key to controlling infection and preventing the production of toxins that lead to human disease. Preventing the outgrowth of spores has the added advantage of avoiding vegetative growth entirely. It should be noted that inhibition of spore outgrowth is a distinct activity relative to inhibition of vegetative cell growth, as indicated by differences between MIC and OIC for nisin and vancomycin (Table 1); the disparity between these parameters for vancomycin with the 630 strain is particularly striking. The combined effect of blocking spore outgrowth and inhibiting vegetative cell growth leads to fewer bacterial cells capable of becoming spores, thereby decreasing transmission of disease.
Antimicrobial compounds such as nisin, oritavancin, fidaxomicin, and vancomycin are known to block the outgrowth of C. difficile endospores into vegetative cells.49−52 However, our data indicate that there is considerable strain-dependent variability in the effectiveness of nisin and vancomycin at preventing spore outgrowth. For clinical applications, it is best if an agent can prevent both vegetative cell growth and spore outgrowth for multiple strains, because these two modes of action are synergistic in terms of lessening the potential for virulence by reducing the number of cells capable of producing toxins A and B.
Each nylon-3 polymer inhibits spore outgrowth for both strains of C. difficile (Table 1). The HDP LL-37 shows comparable activity, but the antibiotics vancomycin and nisin vary considerably between the R20291 and 630 strains in terms of spore outgrowth inhibition. These comparisons support our view that the nylon-3 copolymers are functional mimics of HDPs. Comparisons of hemolytic activities (Figure 4) show that a nylon-3 copolymer (specifically, 50:50 DM:TM) can be superior to a human HDP (LL-37) in terms of avoiding this undesirable property.
We were surprised to find that nisin and vancomycin manifest disparate effects on spore outgrowth for different strains of C. difficile. Nisin has been shown to kill vegetative C. difficile and to inhibit the spore outgrowth of other species, such as Bacillus anthracis and Clostridium botulinum.52 Some strains of C. botulinum exhibit higher spore resistance to nisin than others, which parallels our observations regarding nisin effects on different strains of C. difficile. C. botulinum spores are more resistant than vegetative cells to nisin.53C. difficile has mechanisms to resist killing by nisin, and spontaneous mutations that confer high nisin resistance have been described.16,34,54 Together, these factors limit the therapeutic potential of nisin and similar compounds for treatment of CDIs.
Vancomycin was previously found to prevent spore outgrowth for 027 epidemic isolates of C. difficile,51 and our findings are consistent with this precedent. However, we found that vancomycin is completely ineffective at inhibiting outgrowth of the 012 ribotype spores. This observation is striking given the low MIC measured for vancomycin against vegetative cells for the 012 ribotype. One possible explanation of these findings is that vancomycin is sequestered by 012 (strain 630) spores, which allows the cells that germinate to survive and grow in levels of vancomycin that far exceed the MIC. Because spores are always present during infections, this hypothesis could explain the failure of vancomycin treatment for some CDIs and the relapse of infections.55 Further studies are needed to determine how the efficacies of vancomycin and other antimicrobial agents are affected by the presence of spores from different strains of C. difficile. The proposed capacity of spores to sequester certain antimicrobial agents could be an important consideration in the selection of therapeutic strategies.
Altogether, our data support further evaluation of nylon-3 copolymers as potential agents to combat CDIs. The facile synthesis and ease of compositional variation of nylon-3 materials make this class of synthetic polymers attractive for clinical applications.
Materials and Methods
Bacterial Strains and Growth Conditions
Genotypically distinct C. difficile strains 630Δerm (ribotype 012)56,57 and R20291 (epidemic, ribotype 027)58 were obtained from Nigel Minton and Linc Sonenshein, respectively, and used in this study. C. difficile strains were cultured in an anaerobic chamber maintained at 37 °C (Coy Laboratory Products) with an atmosphere of 10% H2, 5% CO2, and 85% N2 as previously described.59,60 Taurocholate was added to cultures (0.1%) to induce germination of C. difficile spores as indicated (Sigma-Aldrich).61,62 Cells were routinely cultured on brain–heart infusion medium supplemented with 0.5% yeast extract and 1.5% agar (BHIS, BD Difco).63 All experiments were performed using pre-reduced Mueller–Hinton (MH) broth (BD Difco) unless otherwise specified.
Synthesis and Characterization of Nylon-3 Polymers
All nylon-3 polymers used in this study were synthesized in a moisture-controlled glovebox using either tetrahydrofuran (THF) for MM- and DM-containing polymers or dimethylacetamide (DMAc) for NM-containing polymers as the reaction solvent as previously described.64 Briefly, a mixture of β-lactam monomers and the co-initiator (tert-butylbenzoyl chloride) in a 10 mL glass vial was dissolved in THF (or DMAc), followed by the addition of co-initiator solution in THF (or DMAc). The mixture was mixed under magnetic stirring and treated with a solution of lithium bis(trimethylsilyl)amide in THF. The reaction mixture was stirred overnight at room temperature and then removed from the glovebox and quenched with a few drops of methanol. The reaction mixture was poured into a centrifuge tube containing pentane to precipitate the side-chain-protected polymer as white solid. Protected polymers were dried with N2 and then subjected to gel permeation chromatography (GPC) characterization using THF or DMAc as the mobile phase. Protected polymers were treated with trifluoroacetic acid (TFA) at room temperature for 2 h to remove the Boc groups from side chain amines. Deprotected polymers were precipitated in diethyl ether and collected after centrifugation and drying with N2 to provide TFA salts as white solids. Side-chain-protected NM-containing polymers were characterized by GPC using DMAc as the mobile phase as described previously.30 The DMAc gel permeation chromatograph (Waters) was equipped with two Waters Styragel HR 4E columns (5 μm particle) linked in series and a refractive index detector (Waters 2410). DMAc (supplemented with 10 μM LiBr) was used as the mobile phase at a flow rate of 1 mL/min at 80 °C. Side-chain-protected MM- and DM-containing polymers were characterized by GPC using THF as the mobile phase as described previously.30 The THF gel permeation chromatograph (Shimadzu) was equipped with two Waters columns (Styragel HR 4E, particle size 5 μm) linked in series, a multi-angle light scattering detector (Wyatt miniDAWN, 690 nm, 30 mW), and a refractive index detector (Wyatt Optilab-rEX, 690 nm). THF was used as the mobile phase at a flow rate of 1 mL/min at 40 °C.
Hemolysis Assays
Hemolysis assays were conducted as previously described using human red blood cells (hRBCs).65 hRBCs were obtained from the University of Wisconsin (Madison, WI) hospital blood bank, washed three times with TRIS-buffered saline (TBS; 10 mM TRIS, 150 mM NaCl, pH 7.2), and diluted 1:50 in TBS to provide a working suspension of 2% RBC relative to total RBC in whole blood. Two-fold serial dilutions of nylon-3 polymers were prepared in a 96-well plate using TBS; each well contained 100 μL of each compound in solution at concentrations ranging from 800 to 6.25 μg/mL. An aliquot of 100 μL of RBC working suspension was added to each well, followed by gentle shaking of the plate for 10 s. Wells containing TBS without polymer (blank) and wells containing Triton X-100 (positive control to give 100% hemolysis, 3.2 μg/mL in TBS) were included on the same plate. The plate was incubated at 37 °C for 1 h and then centrifuged at 3700 rpm for 5 min. An aliquot of 80 μL of the supernatant from each well was transferred to the corresponding well in a new 96-well plate, and the optical density (OD) at 405 nm was measured using a Molecular Devices Emax precision microplate reader. Measurements were performed in duplicate, and each measurement was repeated on at least two different days. The percentage of hemolysis in each well of a representative data was calculated from
and plotted against polymer concentration to give the dose–response curves of hemolysis for these polymers.
Minimal Inhibitory Concentration (MIC) Determination
Antimicrobial susceptibility tests were performed anaerobically by microdilution in MH broth (BD Difco).66,67 To determine MICs, overnight cultures of C. difficile were diluted into 10 mL of MH broth and cultures grown to an OD600 = 0.45 (∼5 × 107 CFU/mL). Cultures were then diluted 1:10 in MH broth, and 15 μL samples of diluted cultures were used to inoculate individual wells of pre-reduced 96-well round-bottom polystyrene plates containing 135 μL of MH broth or MH broth containing antimicrobials to yield a starting concentration of ∼5 × 105 CFU/mL. MH broth was supplemented with a range of concentrations (2-fold dilutions) of nisin (MP Biomedicals), LL-37 (cathelicidin; AnaSpec), vancomycin (Sigma-Aldrich), or nylon-3 polymers, as specified. Each strain and antimicrobial agent concentration was tested in duplicate for each assay. Uninoculated medium was used as a negative control to test for contamination of growth medium. The positive control was inoculated with C. difficile, but no antimicrobial compound was added. The MIC was defined as the lowest concentration of drug in which no growth was observed after 24 h at 37 °C, and the results of duplicate measurements were averaged. MIC assays were performed a minimum of three times to ensure reproducibility of results.
Spore Preparation and Quantification
C. difficile spores were prepared as described previously.100 Briefly, strains were grown in BHIS broth overnight, spread onto 70:30 agar plates and incubated for 48 h to allow spores to form.62 Following incubation, cells were scraped from the plates, washed in phosphate-buffered saline (PBS), and resuspended in 5 mL of PBS. Samples were then combined 1:1 with 95% ethanol and incubated at room temperature for 1 h to kill all vegetative cells. Spores were then pelleted, washed twice in PBS, and resuspended in 5 mL of fresh PBS. Spore suspensions were then heated to 70 °C for 20 min, followed by addition of PBS with 1% bovine serum albumin (BSA, Sigma-Aldrich) to prevent clumping. Spore preparations were serially diluted and plated onto BHIS agar containing 0.1% taurocholate to determine the number of spores present, and diluted prior to use.63
Spore Inhibition and Vegetative Outgrowth Inhibitory Concentration (OIC) Assays
Because C. difficile spores can be killed prior to or during outgrowth to the vegetative form, multiple methods were used to assess the viability of C. difficile spores exposed to antimicrobials. First, the effect of antimicrobials on the outgrowth of spores (outgrowth inhibitory concentration; OIC) was determined as follows. Spore preparations were diluted to a final concentration of 5 × 106 CFU/mL, and 15 μL aliquots of spore preps were added to individual wells of pre-reduced 96-well plates containing 135 μL of MH broth and 0.01% taurocholate, with or without antimicrobials, to allow spore germination. MH broth was supplemented with dilutions of antimicrobials as described for MIC assays. The OIC was defined as the lowest concentration of drug in which no growth was observed after 24 h at 37 °C. Assays were performed a minimum of three times to ensure reproducibility of results.
Immediately following OIC assays, samples were taken from wells in which no growth was observed to assess the viability and germination potential of spores exposed to concentrations of antimicrobials greater than or equal to the OIC values (i.e., spore outplating). Samples were diluted and plated onto BHIS medium containing 0.1% taurocholate and enumerated following incubation for at least 24 h at 37 °C. Spore survival was calculated as the percentage of CFU post-assay/initial CFU.
To assess the effects of antimicrobials on spores prior to outgrowth, OIC assays were performed in the absence of germinant. Spores were incubated for 24 h at 37 °C, followed by dilution and plating onto BHIS medium containing 0.1% taurocholate to enumerate spores that were inhibited or killed by antimicrobials. Spore survival was calculated as the percentage of CFU post-assay/initial CFU.
Phase-Contrast Microscopy of Spore Germination
C. difficile spores used for microscopy were purified as previously described with some modifications.68 First, 70:30 agar medium was used for the preparation of spores from vegetative cells. Following spore removal from plates, a 48 h incubation step at −20 °C was added prior to layering the spore prep on top of a 50% sucrose gradient. Following washes with dH2O, samples were washed three times in PBS containing 1% BSA to improve the dissociation of individual spores. Mature spores appear phase-bright under phase-contrast microscopy, while germinated spores appear phase-dark.36,69
Prior to germination assessments, purified C. difficile spores were suspended in 100 μL of MH broth. Next, each of the tested compounds was added to the broth at the indicated final concentration and incubated at room temperature for 5 min. Taurocholate was then added to a final concentration of 5 mM, and samples were incubated at 37 °C for 1 h. Positive controls containing spores and taurocholate without antimicrobials and negative controls containing spores alone were performed in parallel. After the addition of taurocholate, 5 μL samples were harvested every 15 min and placed on a solidified 0.7% agarose surface layered on a microscope slide for visualization. Phase contrast microscopy was performed using a 100X-Ph3 oil immersion objective on a Nikon Eclipse Ci-L microscope, and images were acquired using an attached DS-Fi2 camera. Each strain and condition was tested a minimum of two times. At least three fields of view were acquired for each experimental condition. A representative image was shown in Figure 3 for each experimental condition tested.
Acknowledgments
This research was supported by the U.S. National Institutes of Health through research grant GM093265 to S.H.G., and grants DK087763, DK101870 and AI109526 to S.M.M. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supporting Information Available
Bioassay results and compound characterization spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
⊥ R.L. and J.M.S. contributed equally.
The authors declare the following competing financial interest(s): B.W. and S.H.G. are co-inventors on a patent application that covers the polymers described here.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Office of the Associate Director for Communication, Digital Media Branch, Division of Public Affairs. Antibiotic Resistance Threats in the United States, 2013; Centers for Disease Control and Prevention: Atlanta, GA, Sep 16, 2013; http://www.cdc.gov/features/AntibioticResistanceThreats/.
- Deakin L. J.; Clare S.; Fagan R. P.; Dawson L. F.; Pickard D. J.; West M. R.; Wren B. W.; Fairweather N. F.; Dougan G.; Lawley T. D. Infect. Immun. 2012, 80, 2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg J. A.; Sonenshein A. L. J. Bacteriol. 2008, 190, 2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson H. E.; Price A. B. Lancet 1977, 2, 1312. [DOI] [PubMed] [Google Scholar]
- Bartlett J. G. Rev. Infect. Dis. 1979, 1, 530. [DOI] [PubMed] [Google Scholar]
- Rupnik M.; Wilcox M. H.; Gerding D. N. Nat. Rev. Microbiol. 2009, 7, 526. [DOI] [PubMed] [Google Scholar]
- Wilson K. H.; Perini F. Infect. Immun. 1988, 56, 2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C. P. Clin. Microbiol. Infect. 2012, 18Suppl 621. [DOI] [PubMed] [Google Scholar]
- Cornely O. A.; Miller M. A.; Louie T. J.; Crook D. W.; Gorbach S. L. Clin. Infect. Dis. 2012, 55Suppl 2S154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullane K. M.; Miller M. A.; Weiss K.; Lentnek A.; Golan Y.; Sears P. S.; Shue Y. K.; Louie T. J.; Gorbach S. L. Clin. Infect. Dis. 2011, 53, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie T. J.; Miller M. A.; Mullane K. M.; Weiss K.; Lentnek A.; Golan Y.; Gorbach S.; Sears P.; Shue Y. K. New Engl. J. Med. 2011, 364, 422. [DOI] [PubMed] [Google Scholar]
- Hedge D. D.; Strain J. D.; Heins J. R.; Farver D. K. Ther. Clin. Risk Manage. 2008, 4, 949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M. Nature 2002, 415, 389. [DOI] [PubMed] [Google Scholar]
- Boman H. G. J. Intern. Med. 2003, 254, 197. [DOI] [PubMed] [Google Scholar]
- Hancock R. E.; Sahl H. G. Nature Biotechnol. 2006, 24, 1551. [DOI] [PubMed] [Google Scholar]
- McBride S. M.; Sonenshein A. L. Infect. Immun. 2011, 79, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzese A.; Skerlavaj B.; Tomasinsig L.; Gennaro R.; Zanetti M. J. Antimicrob. Chemother. 2003, 52, 375. [DOI] [PubMed] [Google Scholar]
- Gelman M. A.; Weisblum B.; Lynn D. M.; Gellman S. H. Org. Lett. 2004, 6, 557. [DOI] [PubMed] [Google Scholar]
- Kuroda K.; DeGrado W. F. J. Am. Chem. Soc. 2005, 127, 4128. [DOI] [PubMed] [Google Scholar]
- Mowery B. P.; Lee S. E.; Kissounko D. A.; Epand R. F.; Epand R. M.; Weisblum B.; Stahl S. S.; Gellman S. H. J. Am. Chem. Soc. 2007, 129, 15474. [DOI] [PubMed] [Google Scholar]
- Sellenet P. H.; Allison B.; Applegate B. M.; Youngblood J. P. Biomacromolecules 2007, 8, 19. [DOI] [PubMed] [Google Scholar]
- Lienkamp K.; Madkour A. E.; Musante A.; Nelson C. F.; Nusslein K.; Tew G. N. J. Am. Chem. Soc. 2008, 130, 9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambhy V.; Peterson B. R.; Sen A. Angew. Chem., Int. Ed. 2008, 47, 1250. [DOI] [PubMed] [Google Scholar]
- Palermo E. F.; Sovadinova I.; Kuroda K. Biomacromolecules 2009, 10, 3098. [DOI] [PubMed] [Google Scholar]
- Nederberg F.; Zhang Y.; Tan J. P. K.; Xu K. J.; Wang H. Y.; Yang C.; Gao S. J.; Guo X. D.; Fukushima K.; Li L. J.; Hedrick J. L.; Yang Y. Y. Nat. Chem. 2011, 3, 409. [DOI] [PubMed] [Google Scholar]
- Song A. R.; Walker S. G.; Parker K. A.; Sampson N. S. ACS Chem. Biol. 2011, 6, 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Q.; Xu J. J.; Zhang Y. H.; Yan H. S.; Liu K. L. Macromol. Biosci. 2011, 11, 1499. [DOI] [PubMed] [Google Scholar]
- Li P.; Zhou C.; Rayatpisheh S.; Ye K.; Poon Y. F.; Hammond P. T.; Duan H. W.; Chan-Park M. B. Adv. Mater. 2012, 24, 4130. [DOI] [PubMed] [Google Scholar]
- Jiang Y. J.; Yang X.; Zhu R.; Hu K.; Lan W. W.; Wu F.; Yang L. H. Macromolecules 2013, 46, 3959. [Google Scholar]
- Liu R. H.; Chen X. Y.; Hayouka Z.; Chakraborty S.; Falk S. P.; Weisblum B.; Masters K. S.; Gellman S. H. J. Am. Chem. Soc. 2013, 135, 5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R.; Chen X.; Falk S. P.; Mowery B. P.; Karlsson A. J.; Weisblum B.; Palecek S. P.; Masters K. S.; Gellman S. H. J. Am. Chem. Soc. 2014, 136, 4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanza F.; Padhee S.; Wu H. F.; Wang Y.; Revenis J.; Cao C. H.; Li Q.; Cai J. F. RSC Adv. 2014, 4, 2089. [Google Scholar]
- Dane E. L.; Ballok A. E.; O’Toole G. A.; Grinstaff M. W. Chem. Sci. 2014, 5, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride S. M.; Sonenshein A. L. Microbiology 2011, 157, 1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. Curr. Opin. Microbiol. 2003, 6, 550. [DOI] [PubMed] [Google Scholar]
- Setlow P. J. Appl. Microbiol. 2013, 115, 1251. [DOI] [PubMed] [Google Scholar]
- Paredes-Sabja D.; Shen A.; Sorg J. A. Trends Microbiol. 2014, 22, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes-Sabja D.; Setlow P.; Sarker M. R. Trends Microbiol 2011, 19, 85. [DOI] [PubMed] [Google Scholar]
- Moir A. J. Appl. Microbiol. 2006, 101, 526. [DOI] [PubMed] [Google Scholar]
- Fisher N.; Shetron-Rama L.; Herring-Palmer A.; Heffeman B.; Bergman N.; Hanna P. J. Bacteriol. 2006, 188, 1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban N. Q.; Gerdes K.; Lewis K.; McKinney J. D. Nat. Rev. Microbiol. 2013, 11, 587. [DOI] [PubMed] [Google Scholar]
- Frackenpohl J.; Arvidsson P. I.; Schreiber J. V.; Seebach D. ChemBioChem 2001, 2, 445. [DOI] [PubMed] [Google Scholar]
- Liu R.; Chen X.; Chakraborty S.; Lemke J. J.; Hayouka Z.; Chow C.; Welch R. A.; Weisblum B.; Masters K. S.; Gellman S. H. J. Am. Chem. Soc. 2014, 136, 4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonberg R. P.; Kuijper E. J.; Wilcox M. H.; Barbut F.; Tull P.; Gastmeier P.; van den Broek P. J.; Colville A.; Coignard B.; Daha T.; Debast S.; Duerden B. I.; van den Hof S.; van der Kooi T.; Maarleveld H. J.; Nagy E.; Notermans D. W.; O’Driscoll J.; Patel B.; Stone S.; Wiuff C. Clin. Microbiol. Infect. 2008, 14Suppl 52. [DOI] [PubMed] [Google Scholar]
- Gerding D. N.; Johnson S.; Peterson L. R.; Mulligan M. E.; Silva J. Jr. Infect. Control Hosp. Epidemiol. 1995, 16, 459. [DOI] [PubMed] [Google Scholar]
- Zar F. A.; Bakkanagari S. R.; Moorthi K. M.; Davis M. B. Clin. Infect. Dis. 2007, 45, 302. [DOI] [PubMed] [Google Scholar]
- Petrella L. A.; Sambol S. P.; Cheknis A.; Nagaro K.; Kean Y.; Sears P. S.; Babakhani F.; Johnson S.; Gerding D. N. Clin. Infect. Dis. 2012, 55, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musher D. M.; Aslam S.; Logan N.; Nallacheru S.; Bhaila I.; Borchert F.; Hamill R. J. Clin. Infect. Dis. 2005, 40, 1586. [DOI] [PubMed] [Google Scholar]
- Chilton C. H.; Freeman J.; Baines S. D.; Crowther G. S.; Nicholson S.; Wilcox M. H. J. Antimicrob. Chemother. 2013, 68, 2078. [DOI] [PubMed] [Google Scholar]
- Scott V. N.; Taylor S. L. J. Food Sci. 1981, 46, 117. [Google Scholar]
- Allen C. A.; Babakhani F.; Sears P.; Nguyen L.; Sorg J. A. Antimicrob. Agents Chemother. 2013, 57, 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut I. M.; Prouty A. M.; Ballard J. D.; van der Donk W. A.; Blanke S. R. Antimicrob. Agents Chemother. 2008, 52, 4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzotta A. S.; Crandall A. D.; Montville T. J. Appl. Environ. Microbiol. 1997, 63, 2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez J. M.; Edwards A. N.; McBride S. M. J. Bacteriol. 2013, 195, 2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters B. A.; Roberts R.; Stafford R.; Seneviratne E. Gut 1983, 24, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain H. A.; Roberts A. P.; Mullany P. J. Med. Microbiol. 2005, 54, 137. [DOI] [PubMed] [Google Scholar]
- Wust J.; Hardegger U. Antimicrob. Agents Chemother. 1983, 23, 784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler R. A.; He M.; Dawson L.; Martin M.; Valiente E.; Corton C.; Lawley T. D.; Sebaihia M.; Quail M. A.; Rose G.; Gerding D. N.; Gibert M.; Popoff M. R.; Parkhill J.; Dougan G.; Wren B. W. Genome Biol. 2009, 10, R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillaut L.; McBride S. M.; Sorg J. A.. Genetic Manipulation of Clostridium difficile. Current Protocols in Microbiology; John Wiley & Sons, Inc.: New York, 2011; Chapter 9, Unit 9A.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. N.; Suarez J. M.; McBride S. M. J. Visualized Exp. 2013, 10.3791/50787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg J. A.; Dineen S. S.. Laboratory Maintenance of Clostridium difficile. Current Protocols in Microbiology; John Wiley & Sons, Inc.: New York, 2009; Chapter 9, Unit 9A.1. [DOI] [PubMed] [Google Scholar]
- Putnam E. E.; Nock A. M.; Lawley T. D.; Shen A. J. Bacteriol. 2013, 195, 1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J.; Markowitz S. M.; Macrina F. L. Antimicrob. Agents Chemother. 1981, 19, 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R. H.; Chen X. Y.; Gellman S. H.; Masters K. S. J. Am. Chem. Soc. 2013, 135, 16296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raguse T. L.; Porter E. A.; Weisblum B.; Gellman S. H. J. Am. Chem. Soc. 2002, 124, 12774. [DOI] [PubMed] [Google Scholar]
- Mueller J. H.; Hinton J. Proc. Soc. Exp. Biol. Med. 1941, 48, 330. [Google Scholar]
- EUCAST Clin. Microbiol. Infect. 2003, 9, ix. [Google Scholar]
- Edwards A. N.; Nawrocki K. L.; McBride S. M. Infect. Immun. 2014, 82, 4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg J. A.; Sonenshein A. L. J. Bacteriol. 2010, 192, 4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir A.; Smith D. A. Annu. Rev. Microbiol, 1990, 44, 531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.