Abstract
Membrane-derived microvesicles (MVs) shed by cells are being investigated for their role in intercellular communication and as potential biomarkers of disease, but facile and sensitive methods for their analysis do not exist. Here we demonstrate new principles for analysis of MVs that use micrometer-sized droplets of liquid crystals (LCs) to amplify MVs that are selectively captured via antibody-mediated interactions. The influence of the MVs on the micrometer-sized LC droplets is shown to be readily quantified via use of flow cytometry. The methodology was developed using MVs shed by epidermoid carcinoma A431 cells that contain epidermal growth factor receptor (EGFR) as an important and representative example of MVs containing signaling proteins that play a central role in cancer. The LC droplets were found to be sensitive to 106 MVs containing EGFR (relative to controls using isotype control antibody) and to possess a dynamic range of response across several orders of magnitude. Because the 100 nm-sized MVs captured via EGFR generate an optical response in the micrometer-sized LC droplets that can be readily detected by flow cytometry in light scattering mode, the approach possesses significant advantages over direct detection of MVs by flow cytometry. The LC droplets are also substantially more sensitive than techniques such as immunoblotting because the lipid-component of the MVs serves to amplify the antibody-mediated capture of the target proteins in the MVs. Other merits of the approach are defined and discussed in the paper.
Introduction
Microvesicles (MVs) are cell-derived membrane vesicles with sizes between 50 nm and 1 µm, and include exosomes released from multivesicular endosomes1–3 and plasma membrane-shed vesicles.4–6 MVs carry a host of cell-specific signaling proteins and nucleic acids, and have been recognized as important in cellular mechanisms underlying tumor progression, including intercellular transfer of specific biomolecules (e.g., miRNA).7–10 For example, Al-Nedawi and co-workers showed that U373 glioma cells that had been transfected with the gene for EGFRvIII, a mutant form of the epidermal growth factor receptor (EGFR) commonly associated with the glioblastoma multiforme (GBM), produced significantly more MVs than native U373 cells in vitro.11 Recent studies also suggest that elevated levels of circulating MVs correlate to the presence of diseases, including cancers such as glioblastoma.12–14 The use of MVs as potential biomarkers, in particular, has led to a growing interest in the development of analytical tools to quantify the presence of MVs in various biological fluids.15, 16 In this paper, we report new principles that employ microdroplets of thermotropic liquid crystals (LCs) for optical detection of MVs that have been specifically captured by immunobeads. A significant merit of the approach is that it can be performed using LC droplets and instrumentation that is routinely found in research and clinical laboratories.
As noted above, analysis of proteins associated with MVs (e.g., cell-surface proteins) is important because it can provide information about the state (and processes) of the cells from which the MVs originate.5, 11 Current methods for analysis of the protein content of MVs, such as immunoblotting and enzyme-linked immunosorbent assay (ELISA), require large numbers of MVs (>108 MVs).17–19 Characterization of MVs using flow cytometry (FC) across the size range of 100 nm to 1 µm is difficult in routine practice due to the weak scattering of light by the MVs and the wide variation in sizes of MVs.20–22 A light scattering technique called nanoparticle tracking analysis (NTA) has also been reported for direct visualization of MVs, but it suffers from variability of background signals generated by serum particles.16, 23 More recently, Shao and co-workers reported a technique that combines (i) immunotargeting of MVs with functionalized antibodies, (ii) covalent conjugation of superparamagnetic particles with the functionalized antibodies, (iii) a membrane filtration step for purification, and (iv) use of a minituarized nuclear magnetic resonance system to detect the labeled MVs.24 Relative to existing methods, they report a sensitivity of 105 MVs, surpassing methods such as NTA (107 MVs), FC (108 MVs) and immunoblotting (108 MVs) by at least two orders of magnitude.
While such methods are promising, techniques for molecular analysis of MVs that do not require use of covalent reactions in the presence of complex samples, do not require membrane purification steps, and use routine instrumentation found in basic biological and clinical laboratories have the potential to be quickly and widely adopted.
Herein we demonstrate principles for optical detection of MVs that utilize micrometer-sized LC droplets to report MVs that have been captured using antibodies targeted to proteins present in the MVs (Scheme 1). The LCs used in this study are nematic, thermotropic LCs and can be viewed as structured oils in which the molecules (mesogens) comprising the LC exhibit long-range orientational ordering. Our approach is enabled by two recent advances involving micrometer-sized droplets of LCs that are dispersed in aqueous solution (i.e., LC-in-water emulsions). First, the internal configurations of the LC droplets have been reported to be sensitive to the presence of lipids adsorbed to the surfaces of the droplets.25–27 Specifically, past studies have demonstrated that biological lipids and synthetic surfactants can change the orientations assumed by LCs at aqueous-LC interfaces (the so-called “anchoring” of LCs) through a mechanism that involves interactions between the aliphatic tails of the adsorbed amphiphiles and the mesogens of the LC.28–30 In the absence of an adsorbate, the mesogens confined within the droplets arrange into a so-called bipolar configuration with two surface point defects (Figure 1a–b). Upon interaction with an amphiphile, however, the LC within the droplets transforms to a radial configuration with a single defect at the cores of the droplets (Figure 1d–e). The change in the configuration of the LC within the micrometer-sized LC droplet leads to a distinct change in optical properties of the droplets (see below for additional detail). Such observations have previously led to the development of LC droplet-based sensors to detect and differentiate between types of bacteria and viruses based on their cell-wall/envelope structure.26
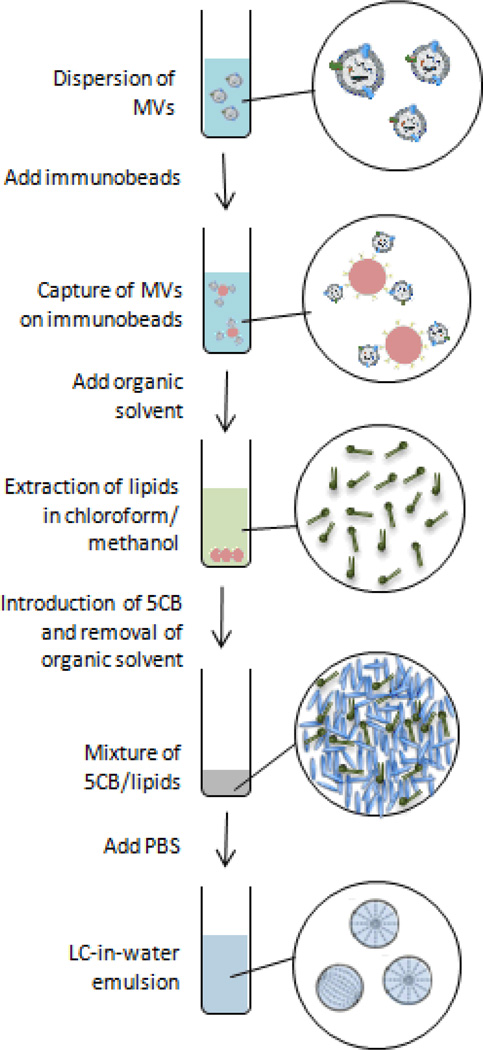
Scheme 1.
Illustration of the experimental procedure. MVs were first captured onto immunobeads via antibody-antigen recognition, and then extracted into the LC. Emulsification of the LC in PBS generated LC droplets, which were quantified using FC.
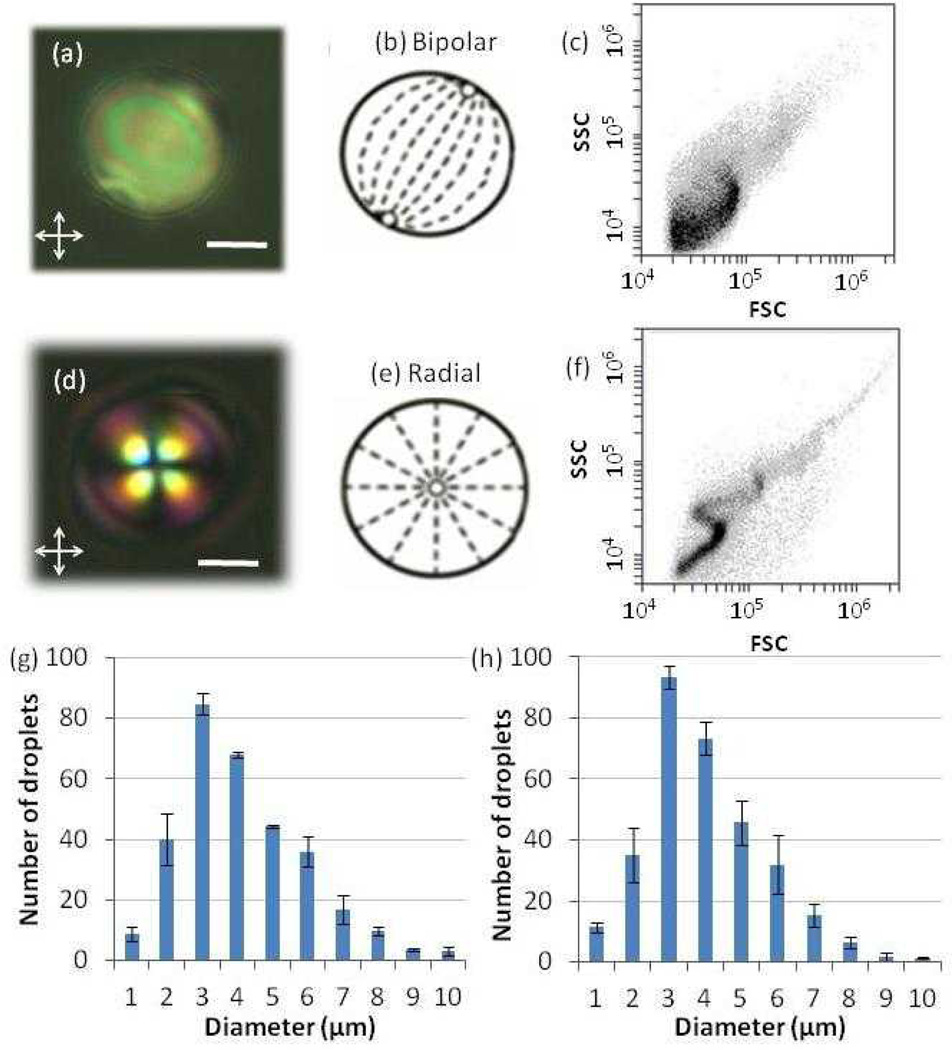
Figure 1.
(a, d) Polarized light micrographs of a LC droplet in the (a) absence (bipolar) or (d) presence (radial) of lipids extracted from MVs (scale bar: 2 µm). (b, e) Schematic illustrations of the director configurations within the LC droplets corresponding to the micrographs shown in (a) and (d), respectively. Dashed lines represent the orientations of the LC, and open circles represent defects. (c, f) Light scatter plots of the intensity of side light scattering (SSC) versus forward light scattering (FSC) obtained by flowing LC droplets in the (c) absence (bipolar) or (f) presence (radial) of lipids extracted from MVs through a flow cytometer. (g–h) Size distributions of LC droplets in the (g) absence and (h) presence of lipids from MVs, as measured by optical microscopy.
The second recent advance that is leveraged in the approach to MV analysis that is reported herein revolves around the use of FC to read-out the configurational states of LC droplets. As noted above, while FC has been used to characterize MVs, the small sizes of MVs and the presence of background signal have presented a range of challenges that have not yet been resolved for routine analysis of MVs. In contrast, because LCs possess anisotropic optical properties (birefringence), the scattering of light from micrometer-sized LC droplets is dependent on the internal configuration of the LC within the droplets and is easily characterized by FC. In particular, a recent study has demonstrated that a population of LC droplets in the radial configuration generates a light scatter plot (Figure 1f) that is distinct from that measured for bipolar droplets (Figure 1c).31 Specifically, when compared to bipolar droplets, radial droplets generate a narrower distribution of side scattering intensities (SSC, large angle light scattering) for a fixed intensity of forward light scattering (FSC, small angle light scattering). This difference in scatter plots arises from the different rotational symmetries of the refractive index profiles presented to the laser of the FC by the two configurations. Radial droplets present a spherically symmetric profile, whereas bipolar droplets do not. Furthermore, the FC-based method was demonstrated to be capable of rapidly quantifying the percentage of radial LC droplets (i.e., within a few seconds) contained in a mixture of bipolar and radial drolets with high precision, as validated by parallel analysis of polarized light micrographs of the same emulsion. Finally, we emphasize that the FC-based method described above uses a simple flow cytometer (costing approximately US$15,000) that is operated only in light scattering mode.
Whereas the prior studies reported above establish that LC droplets are a promising analytical tool, they do not address detection of membrane vesicles from mammalian cells and they do not address quantification of MVs based on the presence of a specific protein target. The principles reported in this paper were investigated to address those challenges, specifically in the context of developing methods using LC droplets that rely only on instrumentation that is routinely found in biological and clinical laboratories. Our approach integrates the use of antibody-functionalized magnetic beads that capture MVs by recognition of key signaling proteins present in the MVs along with the development of simple procedures that permit the transfer of the lipid component of the MVs into LC microdroplets. The experiments reported in this paper used MVs derived from epidermoid carcinoma A431 cells and the capture of the MVs was achieved via the presence of epidermal growth factor receptor (EGFR) in the MVs. Our focus on EGFR was motivated in part by the observation that over-expression and mutation of EGFR has been associated with a number of cancers.32 We demonstrate principles that result in facile detection of 106 MVs, which is better than NTA, FC, ELISA and immunoblotting, as well as a dynamic range of several orders of magnitude. With additional optimization of the LC-based method, higher sensitivity is envisaged.
Experimental
Materials
Dulbecco’s modified Eagle’s medium and penicillin-streptomycin solution were purchased from Mediatech (Herndon, VA). Cosmic calf serum was obtained from Hyclone (Logan, UT). Phosphate-buffered saline (PBS) (0.01 M phosphate; 0.138 M NaCl; 0.0027 M KCl; pH 7.4) and methanol was obtained from Sigma-Aldrich (St Louis, MO). Chloroform was purchased from Fisher Scientific (Pittsburgh, PA). Avidin-functionalized magnetic particles were obtained from Micromod (Rostock, Germany). Biotinylated anti-Epidermal Growth Factor Receptor (anti-EGFR) monoclonal antibody (111.6) was purchased from LifeSpan Biosciences (Seattle, WA). Biotinylated IgG1 isotype control was purchased from eBioscience (San Diego, CA). Anti-EGFR (1005, sc03) and horseradish peroxidase-conjugated secondary anti-rabbit IgG (sc-2054) were purchased from Santa Cruz Biotechnology (Dallas, TX). The nematic LC 4’-pentyl-4-cyanobiphenyl (5CB) was obtained from EMD Chemicals (Spring Valley, NY). Deionization of a distilled water source was performed with a Milli-Q system (Millipore, Bedford, MA) to give water with a resistivity of 18.2 MΩ-cm.
Cell Culture
A431 cells were cultured in Dulbecco’s modified Eagle’s medium with 10 % cosmic calf serum and 100 U/ml penicillin/streptomycin. Cells were grown for 4 days to approximately 90 % confluence in 15 mL of cell media in each T75 flask.
Isolation of MVs
Medium from the 4-day culture was collected and subjected to centrifugation at 8 000 × g for 30 min to eliminate cells and debris.11 The MV fraction was obtained after centrifugation for 4 h at 40 000 × g, washing with PBS and centrifugation again for 4 h at 40 000 × g. MVs isolated from 30 mL of cell media (2 T75 flasks) were re-suspended in 700 µL PBS for analysis. The MV-associated protein and phospholipid recovered were quantified using the Bradford assay (Thermo Scientific, Rockford, IL) and the EnzyChrom phospholipid assay kit (BioAssay Systems, Hayward, CA), respectively, according to the manufacturers’ instructions. Because the EnzyChrom phospholipid assay only measures choline-containing phospholipid, further verification of the concentration of MV-associated lipid was performed using fatty acid methylation and gas chromatography-mass spectrometry (GC-MS), according to published procedures.33 The MVs isolated through the above-described procedure are anticipated to comprise mainly of cell membrane-derived vesicles (and not exosomes) because exosomes are much smaller (< 100 nm) and thus only captured using a centrifugation speed (100 000 × g) that is higher than that employed here (40 000 × g).16
Dynamic Light Scattering
A 100 mW, 532 nm laser (Coherent Compass 315M-100) illuminated a temperature-controlled glass cell filled with a refractive-index matching fluid (decahydronaphthalene, Fisher Scientific, Pittsburgh, PA) maintained at 25 °C. The angle of the detector was set at 90°. The autocorrelation function (ACF) was obtained using a BI-9000AT digital autocorrelator (Brookhaven Instruments Corporation, Holtsville, NY) and analyzed using the CONTIN34 software package to yield a distribution of aggregate sizes of MVs.
Nanoparticle Tracking Analysis (NTA)
NTA measurements were performed with a Nanosight LM10 (Nanosight, Amesbury, U.K.) equipped with a 405 nm laser. MV dispersions were prepared as described above and diluted until the concentration was acceptable for NTA measurements (1:40 dilution ratio was found to be optimal).35 400 µL of the diluted dispersion was injected into the sample chamber using a syringe. All measurements were collected at room temperature with the camera level set to the maximal value to allow for the detection of small particles. One 60 s video was taken for each dispersion. Data were captured and analyzed using NTA version 2.3.
Immunoblotting36, 37
MV-associated proteins were separated on 10 % SDS-PAGE gels and then transferred to polyvinylidene difluoride membranes. The membrane was blocked with 5 % nonfat dried milk/ TBST (10 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH 8) overnight at 4 °C and then incubated with anti-EGFR primary antibodies (sc03, 0.4 µg/ml) in 5% nonfat dried milk/TBST for 1 h at 37 °C. Subsequently, the membrane was washed three times in TBST, incubated with HRP-conjugated goat anti-rabbit IgG secondary antibodies (sc2054, 0.08 µg/ml) for 1 h at 37 °C, and washed in TBST three times. The labeled proteins were visualized by the enhanced chemiluminescence method (Luminata Crescendo, Billerica, MA). Detection was performed using a UVP Biochemi camera system (Upland, CA).
Atomic Force Microscope (AFM)
AFM experiments were carried out with a Nanoscope III Multimode scanning probe microscope equipped with a “J” (120 µm) scanner (Veeco/Digital Instruments, Santa Barbara, CA). The images were acquired in tapping mode in solution using a commercially available liquid cell with oxide-sharpened silicon nitride V-shaped cantilevers at room temperature. The cantilever had a nominal spring constant of 0.32 N/m. Imaging was performed at a scan rate of 2 Hz and at a resolution of 256 pixels/line. Images were flattened as required.
Capture of MVs using Functionalized Magnetic Beads
To functionalize the avidin-coated magnetic beads with antibodies, 10 µL of biotinylated antibody (200 µg/mL) was mixed with 20 µL of avidin-coated magnetic particles (10 mg/mL; this corresponds to ~107 magnetic particles in 20 µL) in a glass tube and placed on a shaker for 1 h. This ratio ensured that the antibody incubated against the magnetic particles was sufficient in quantity for monolayer coverage of the magnetic particles (two-fold excess of antibody was provided). The beads were subsequently rinsed by adding 500 µL of PBS to the tube and vortexing the mixture. The beads were collected using a magnet and the PBS was removed. This cycle was repeated thrice.
For capture of MVs using functionalized beads, a 100 µL dispersion of MVs was added to the 20 µL of functionalized beads and the mixture was vortexed for 30 sec and placed on a shaker for 1 h. As above, three cycles of rinsing were performed to remove free, unbound MVs from the solution mixture. PBS was removed from the collected beads prior to extraction of lipids.
Extraction of Lipids from MVs into LC Droplets
To extract the lipids from MVs that were captured on the immunobeads, 100 µL of a chloroform:methanol (2:1) mixture was added to the beads (in a glass tube). The mixture was vortexed vigorously for 1 min and placed in a shaker for 15 min. The beads were then collected at the bottom of the vial by using a magnet. The organic overlayer was subsequently removed and transferred to a new glass tube. Next, 5CB of a prescribed volume was added to the organic phase (5CB is miscible with the organic solvent), and the solvent was evaporated under a stream of N2 gas. The dried sample containing 5CB was placed in a vacuum for 1 h to remove any residual chloroform.
LC emulsions (containing the lipids from the MVs) were formed by addition of 0.5 mL of PBS to the 5CB obtained from the above procedure, followed by sequential sonication (with a bath sonicator) and vortexing (3 000 rpm). The cycles of sonication and vortexing were performed until the emulsions started to appear slightly hazy. This procedure resulted in a polydisperse population of LC droplets with diameters ranging from 1 to 10 µm (Figure 1g–h). FC was used to establish that each emulsion contained between 7,000 and 15,000 LC droplets per µL.
Optical Characterization of LC Emulsions by Polarized Light Microscopy
Prior to characterization by FC, the LC emulsions were examined using a combination of bright field and polarized light microscopy to determine the ordering of the LC within the droplets. A 20 µL volume of the LC emulsion was dispensed onto a glass coverslip. The configurations of the LCs within the emulsion droplets were determined by observation of the droplets under an Olympus IX71 inverted microscope (Center Valley, PA) using an objective magnification power of 100x (an oil-immersion lens). Polarized light micrographs of the LC emulsions were collected with a Hamamatsu 1394 ORCAER CCD camera (Bridgewater, NJ) connected to a computer and controlled through SimplePCI imaging software (Compix, Inc., Cranberry Twp., NJ). Bright field micrographs were collected by removing the polarizer from the optical path of the microscope.
Flow Cytometry of LC Droplets
Plots of the intensity of side light scattering (SSC) versus the intensity of forward light scattering (FSC) were obtained for LC-in-water emulsions using a BD Accuri C6 flow cytometer (Ann Arbor, MI). In the experiments described below, the emulsions were pumped through the flow cytometer at a flow rate of 14 µL/min and the scatter plots were constructed from the measurement of 50 000 LC droplets (Typically, 5.1 ± 1.6 µL of the LC emulsion was flowed through to read 50 000 droplets).
Results
Characterization of MVs
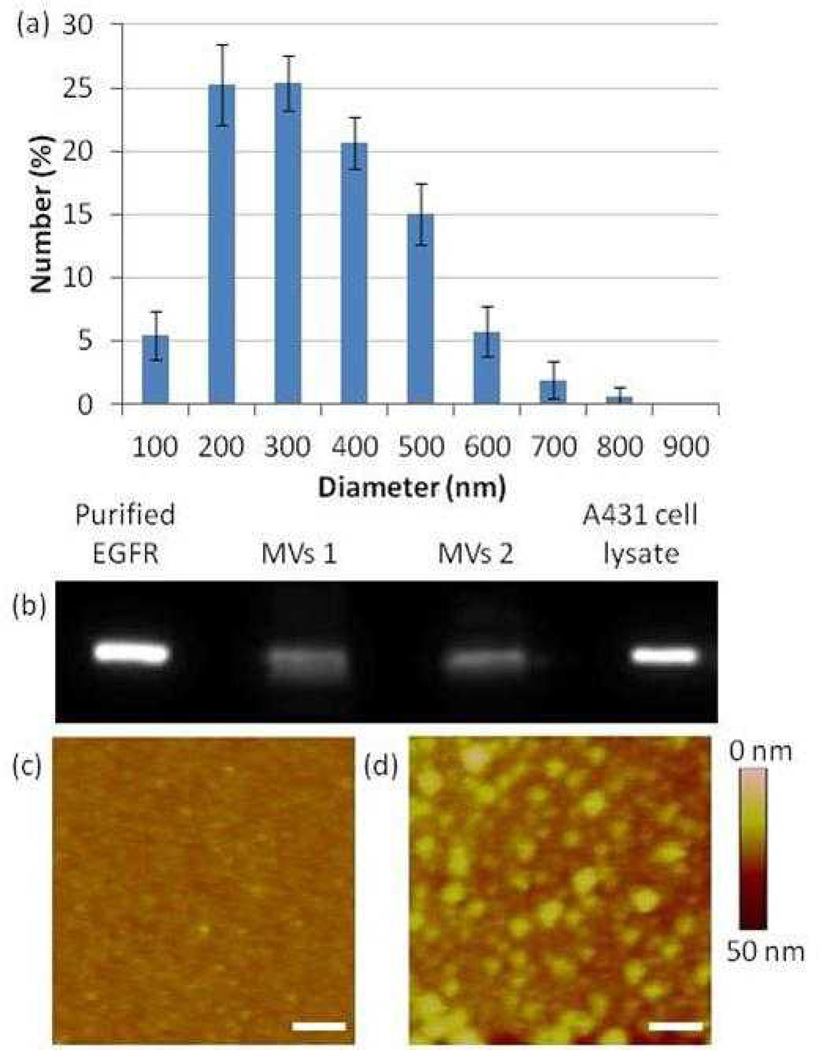
Our initial experiments were performed to characterize the MVs shed by A431 cells into culture media. The results of these experiments enabled us to design and evaluate our LC droplet-based MV detection method. MVs (from cell media containing 10% serum) were separated from cells and cell debris by centrifugation using procedures described elsewhere11 and resuspended in phosphate buffered saline (PBS) (see Experimental). DLS measurements revealed that the diameters of the MVs ranged from 100 nm to 1 µm (consistent with published literature38), with a number-average diameter of 320 ± 145 nm (Figure 2a). To verify that the isolated samples contained lipids consistent with the presence of MVs, the phospholipid content was quantified using an EnzyChrom phospholipid assay kit and gas chromatography-mass spectrometry (GC-MS). Based on the EnzyChrom assay, a phospholipid concentration of 9.8 ± 1.6 µM was measured from a dispersion of MVs which were prepared by reconstituting MVs isolated from ~ 4 mL of cell media in 100 µL of PBS. GC-MS measurements gave a phospholipid concentration (8.8 ± 1.8 µM) in close agreement with that measured using the EnzyChrom assay. GC-MS also revealed that the phospholipids of the MVs have tails that are typical of those found in mammalian cells (18-carbon aliphatic tails are the most common39; Figure S1). From the total lipid concentration and average size of the vesicles, we estimated the concentration of MVs resuspended in the PBS to be 1010 MVs/mL (or 108 MVs per mL of culture medium, which is a concentration similar to that reported by Shao et al.24). Finally, we note that the concentration of proteins in the dispersion of MVs was measured to be 63 ± 19 µg/mL by using the Bradford assay.
Figure 2.
(a) Size distribution of MVs derived from A431 cells (suspended in PBS) as measured by dynamic light scattering (DLS). (b) Immunoblot analysis of two separate MV samples derived from two different batches of cells of A431 cells. Lysates were immunoblotted for EGFR. (c–d) AFM images of (c) a surface decorated with anti-EGFR 111.6 and (d) a surface decorated with anti-EGFR 111.6 and subsequently incubated with MVs derived from A431 cells (scale bar: 500 nm).
To provide additional confirmation of the number of MVs in our samples, we used NTA. A dilution of the above-described dispersion of MVs (with original lipid concentration of 9.8 ± 1.6 µM) was required to achieve the optimal concentration required for NTA. Specifically, a 1:40 dilution of the MV dispersion was performed (using PBS), which, when measured by NTA, gave a value of 4∙108 particles/mL. Based on the earlier estimate of 1010 MVs/mL of PBS, this dilution step should result in a dispersion with a concentration of 2.5∙108 MVs/mL PBS, which is a value that is in good agreement with that experimentally obtained by NTA.
We verified the presence of EGFR in the MVs obtained from the A431 cells by using immunoblotting (Figure 2b). The immunoblots were consistent with the presence of EGFR in the MVs, as evidenced by a band obtained from the MVs that coincided with purified EGFR (MW of 170 KDa). By comparison to bands of purified EGFR, the mass of EGFR in 108 MVs was estimated to be 14 ng. From this value, we estimate there to be approximately 500 EGFR molecules/MV. Here we note that past studies have reported that A431 cells can express ~106 EGFR molecules at the cell surface40 and each cell has a typical size of approximately 40 µm.41 We calculate, therefore, that a MV of diameter of 320 nm derived from an A431 cell will theoretically contain 250 EGFR molecules, which is of the same order of magnitude as our experimental value. However, we also note that MVs have been reported to be enriched in certain components of the cell membrane, in particular those that are associated with “lipid rafts”.10, 42 Since EGFR has been suggested to localize in lipid domains43, it is plausible that the concentration of EGFR may be enriched in MVs relative to that in the cell membrane.
Finally, we explored the use of surface-immobilized anti-EGFR 111.6 to capture EGFR-containing MVs derived from A431 cells. In this experiment, clean glass surfaces were initially decorated with avidin (according to previously reported procedures44), followed by functionalization with biotinylated anti-EGFR 111.6. Images obtained by using atomic force microscopy (AFM, in tapping mode) revealed that surfaces decorated only with antibody were smooth (Figure 2c, root-mean-squared (rms) roughness of 2.1 nm measured over an area of 2 µm × 2 µm) relative to the same surfaces incubated with EGFR-containing MVs, which exhibited circular features with diameters of ~ 150 nm (Figure 2d). Figure S2 shows a measurement of the cross sectional height of the imaged surface. Notably, the size of these features (150 nm) is comparable to that measured by DLS (average at 320 nm). A control experiment performed using a surface that was functionalized with a non-specific control IgG did not lead to capture of a comparable density of MVs (Figure S3). These two results, when combined, are consistent with specific capture of MVs on surfaces presenting anti-EGFR 111.6 via antibody-antigen (EGFR) binding.
In summary, from the results presented above, we conclude that A431 cells shed membrane-bound MVs with an average diameter of 320 nm and that these MVs contain ~500 EGFR molecules. We also conclude that the A431 cells shed ~108 MVs per mL of culture media.
Interactions of Lipids from MVs with LC Droplets
Next, we performed a series of experiments to determine if lipids extracted from MVs shed by A431 cells would trigger ordering transitions in LC droplets. In this context, we note that past studies have shown that changes in the orientational ordering of LCs at aqueous interfaces (anchoring transitions) induced by amphiphiles are dependent on the structure and phase state of the amphiphiles.45, 46 For example, it was shown that amphiphiles comprised of branched acyl chains do not cause LCs to assume a perpendicular or homeotropic orientation.46 This was hypothesized to be due to frustrated packing of the branched amphiphiles, with branching preventing interdigitation (or coupling) of LC molecules with amphiphiles at the interface. Because MVs are derived from cell membranes, which are composed of a large variety of phospholipids (e.g., with different tail lengths and degree of unsaturation, see Figure S1) and other components of lipid rafts (e.g., cholesterol, sphingomyelin), we characterized the response of LC droplets as a function of the number of MVs introduced into the LC.
Our initial experiments involved incubation of dispersions of MVs shed by A431 cells with LC droplets in PBS, a procedure that was reported previously to permit transfer of lipids from model (or synthetic) vesicles, bacteria and viruses to LC droplets.25–27 When using MVs shed by the A431 cells, however, we did not observe the LC droplets to undergo configurational transitions, consistent with a barrier to lipid transfer to the LC interface associated with the phase state of the lipid and/or proteins and carbohydrates associated with the outer surface of the MV membrane. This initial observation led us to explore an alternative procedure for transfer of the MV lipids to the LC droplets that employed an organic solvent. As described in the Methods section, a simple procedure that involved extraction of MV lipids into an organic solvent and subsequent introduction of the lipids into the LC was found to facilitate rapid transfer of lipids onto the LC interfaces. By using the EnzyChrom assay, we confirmed that > 90 % of the lipid in the MVs was extracted into a 100 µL mixture of chloroform:methanol (2:1)
Following emulsification, we characterized the ordering of the LC droplets with and without lipids extracted from MVs by using optical microscopy. Figure 1a shows a representative polarized light micrograph of a nematic LC droplet (in the absence of lipids) suspended in PBS. As detailed elsewhere, Figure 1a indicates the presence of two point defects (surface defects, called boojums) located at diametrically opposite ends, or “poles”, of the LC droplet and a bright appearance of a majority of the droplet.47 From these observations and others (e.g., performed using bright-field microscopy), we conclude that the droplet is in a so-called bipolar configuration where the LC assumes a tangential orientation at the droplet surface (see Figure 1b for director profile). In contrast, following the transfer of lipids from MVs into the LC droplets, a distinct optical signature was observed. Figure 1d, with a single defect located at the droplet center and a characteristic dark cross, is consistent with a radial configuration where the LC is oriented perpendicular to the droplet surface (see Figure 1e for director profile). From these observations, we conclude that the presence of a sufficient quantity of MV-derived lipid within the LC caused the droplets to assume a radial configuration, whereas in the absence of lipid, the configuration was bipolar.
While optical microscopy permits identification of the configuration of LC droplets, as discussed in the Introduction, FC provides a facile and quantitative method to characterize the size distributions of LC microdroplets as well as the percentage of radial and bipolar droplets within an emulsion.31 Here we first address the use of FC to characterize the size distributions and more importantly changes in the size distributions due to coalescence of LC droplets during experiments. Specifically, we speculated that the presence of amphiphilic components of the MVs within the LC droplets could potentially influence the sizes and stability of the LC droplets formed by emulsification. To address this question, we used FC to measure the number of LC droplets formed by sonicating and vortexing 1 µL of 5CB (in 0.5 mL of PBS). In the absence of lipids extracted from MVs, we measured about ~107 droplets (50 000 events counted for 5.1 ± 1.6 µL of the LC emulsion) to be formed. Assuming a monodisperse LC emulsion, we calculate that 1 µL of 5CB would produce 107 LC droplets with a diameter of 5.5 µm. To test this prediction, we measured the size of the 5CB droplets using optical microscopy. Our measurements reveal that the diameter of the LC droplets ranged from 1 – 10 µm, with an average diameter of 3.8 ± 1.7 µm (Figure 1g). Given the polydispersity of the emulsion drop size, we conclude that these two measurements are in reasonable agreement. In the presence of lipids from MVs in the 5CB droplets, the size distribution of the LC droplets determined by optical microscopy was similar to that obtained in the absence of lipids, with an average diameter of 3.6 ± 1.4 µm (Figure 1h). The number of lipid-containing LC droplets counted by FC was also similar to that measured above (50 000 events counted for 4.6 ± 1.3 µL of the LC emulsion).
To assess the stability of the LC droplets, the size distributions of the 5CB droplets, both in the absence and presence of lipids, were measured after 6 h. Inspection of Figure S4 reveals that after 6 h, both distributions shifted to larger values, with an average diameter of 5.3 ± 1.6 µm and 5.2 ± 2.2 µm, respectively. This result suggests that the 5CB droplets have coalesced with time. In light of this result, as a precaution, we performed FC analysis within 1 h of preparation of the emulsions. However, we note that our current analysis of LC droplets using FC is based on a subpopulation of droplet sizes. The range of LC droplets selected for analysis (those which fall within FSC values of 30 000 to 60 000 a.u.) does not change significantly during the aging of the emulsion (Figure S5). Indeed, a significant merit of using FC for analysis of LC droplets is that subpopulations of droplets (with a given range of size) can be selected for quantification.
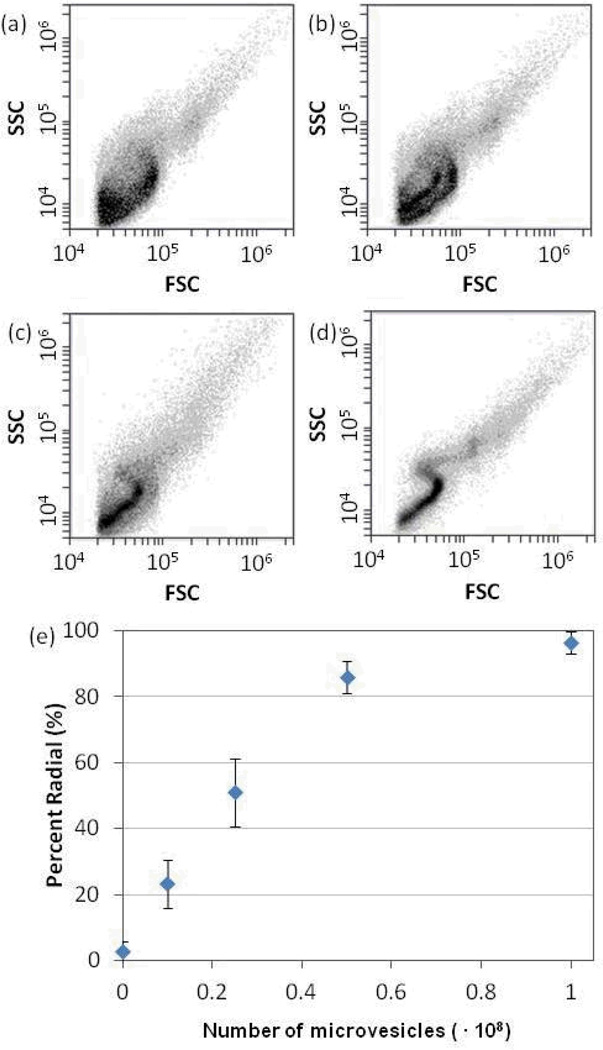
Next we examined the FC scatter plots to analyze the internal configurations of the LC droplets. Figure 1c and 1f show FC scatter plots obtained using LC droplets free of lipids and containing MV lipids, respectively. As discussed in the Introduction, the characteristic features of these scatter plots are consistent with LC droplets in bipolar (Figure 1c) and radial (Figure 1f) configurations,31 and they support our conclusion that the lipid components of MVs can induce ordering transitions in LC droplets. For FC-based methods of analysis of LC droplets to be useful for quantification of MVs, however, quantification of the number of radial droplets in a mixture of radial and bipolar droplets is needed. To this end, we varied the quantity of lipid introduced into the LC (by varying the number of MVs that were extracted into the organic solvent). Figures 3a–d show that as the number of MVs added to the LC was increased, a continuous evolution of the scatter plots from the broad distribution of SSC values at each FSC value (characteristic of emulsions containing bipolar LC droplets) to the distinct S-shape (characteristic of emulsions containing radial LC droplets) was observed. To quantify the fraction of radial droplets within a sample of LC, guided by prior studies, we analyzed FSC values between 30 000 a.u. and 60 000 a.u.31 Specifically, the number of light scattering events measured over this range of FSC values can be used to measure the percentage of radial droplets within a population of bipolar and radial droplets, a result that we found to be consistent with our measurements with MVs (Figure S6). Figure 3e shows the percentage of radial droplets in the LC emulsions plotted as a function of the number of MVs that were transferred into the LC droplets.
Figure 3.
(a–d) Scatter plots (intensity of side light scattering (SSC) versus forward light scattering (FSC)) obtained by FC-based analysis of LC emulsions obtained using LCs containing lipids extracted from A431-derived MVs. The total number of MVs extracted into the LC (1 µL 5CB) were (a) 0, (b) 1.25 ∙ 107 (c) 2.5 ∙ 107 , (d) 108. (e) Percentage of LC droplets that exhibited a radial configuration, plotted as a function of number of MVs extracted into the LC (N = 3).
A key observation made regarding Figure 3 is that the response of the LC droplets to increasing concentrations of lipids from MVs is continuous. We attribute this continuous response to the variance of the size of the droplets (Figure 1g–h), which can influence the balance of energetics that control the ordering within micrometer-sized LC droplets. Gupta and co-workers have demonstrated that the response of LC droplets to surfactants is dependent on the size of the LC droplets.26 Specifically, the concentration of surfactants required to trigger a bipolar-to-radial ordering transition in LC droplets decreases with decreasing size of the droplets. This effect is due to size-dependent contributions of elastic and surface anchoring energies to the free energy of the LC droplets. To investigate the influence of LC droplet size on the response of the LC droplets to MVs, we quantified the percentage of droplets exhibiting the radial (or near-radial) configuration using optical microscopy. Inspection of Figure S7 reveals that the percentage of LC droplets with a radial configuration decreased as the diameter of the droplets increased. This is consistent with previous observations26 and supports our conclusion that the continuous response observed in Figure 3 is likely influenced by the range of LC droplet sizes in the emulsions used in our experiments.
Anchoring Transitions Induced by MVs Captured through Antigen-Antibody Recognition
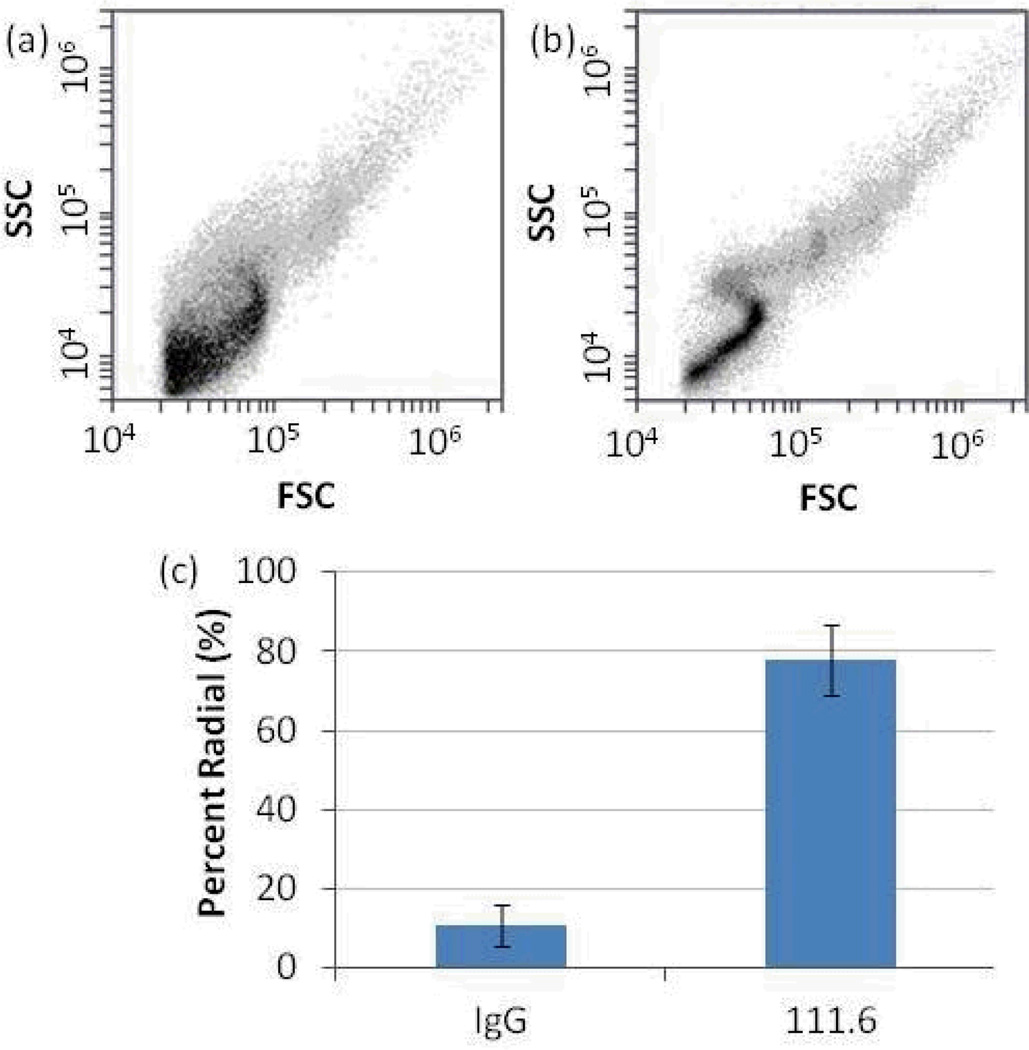
The results described above demonstrate that LC droplets in combination with FC can be utilized to quantify the presence of MVs shed by A431 cells. As discussed in the Introduction, a key goal of the study reported in this paper was to demonstrate quantification of MVs that contain key signaling proteins. To this end, we performed a series of experiments that targeted EGFR present in the MVs shed by A431 cells (as also noted in the Introduction, EGFR is used as an example of an important transmembrane signaling protein). Briefly, we incubated MVs with immunobeads presenting anti-EGFR 111.6, which were prepared using avidin-coated magnetic beads (1.5 µm) and biotinylated 111.6 (see Methods). Specificity of capture of the MVs was established using a biotinylated non-specific IgG as an isotype control to the anti-EGFR 111.6. After functionalization of the beads (107 beads) with antibodies, they were rinsed with PBS and incubated with 100 µL of PBS containing 108 MVs for 1 h. The beads were then rinsed at least three times with PBS and then introduced into 100 µL of a chloroform:methanol organic mixture to extract MV lipids from the beads. The organic solvent was separated from the beads and contacted with 1 µL of 5CB. Following evaporation of the organic solvent, an LC emulsion was formed (see Methods).
Representative FC scatter plots obtained using the above-described procedures are shown in Figure 4a and 4b. When compared to the scatter plots obtained using beads with control IgGs (Figure 4a), the LC droplets with lipids extracted from anti-EGFR 111.6-functionalized beads generated scatter plots that had qualitative features characteristic of radial LC droplets (Figure 4b). Specifically, this latter plot has a narrower distribution of SSC values at each FSC value and the characteristic S-shape discussed earlier. Figure 4c shows the percentage of radial LC droplets in each sample, clearly demonstrating that the anti-EGFR 111.6-functionalized magnetic beads captured a significantly larger number of MVs than the beads presenting IgG controls. We note that our initial efforts suffered from false positives in control experiments performed with IgG-isotype functionalized beads, which we attributed to non-specific binding. To address this issue, the beads were subjected to a more stringent washing procedure that involved contact with a 1 µM SDS solution and three cycles of PBS rinsing. This additional step was found to eliminate the false positive signals from control IgG beads.
Figure 4.
Scatter plots of LC emulsions obtained after mixing of the LC with lipids extracted from magnetic particles that had been functionalized with (a) control IgG and (b) anti-EGFR 111.6 and subsequently incubated with A431-derived MVs. (c) Percentage of LC droplets that exhibited a radial configuration from IgG- or 111.6-functionalized magnetic particles (N = 5).
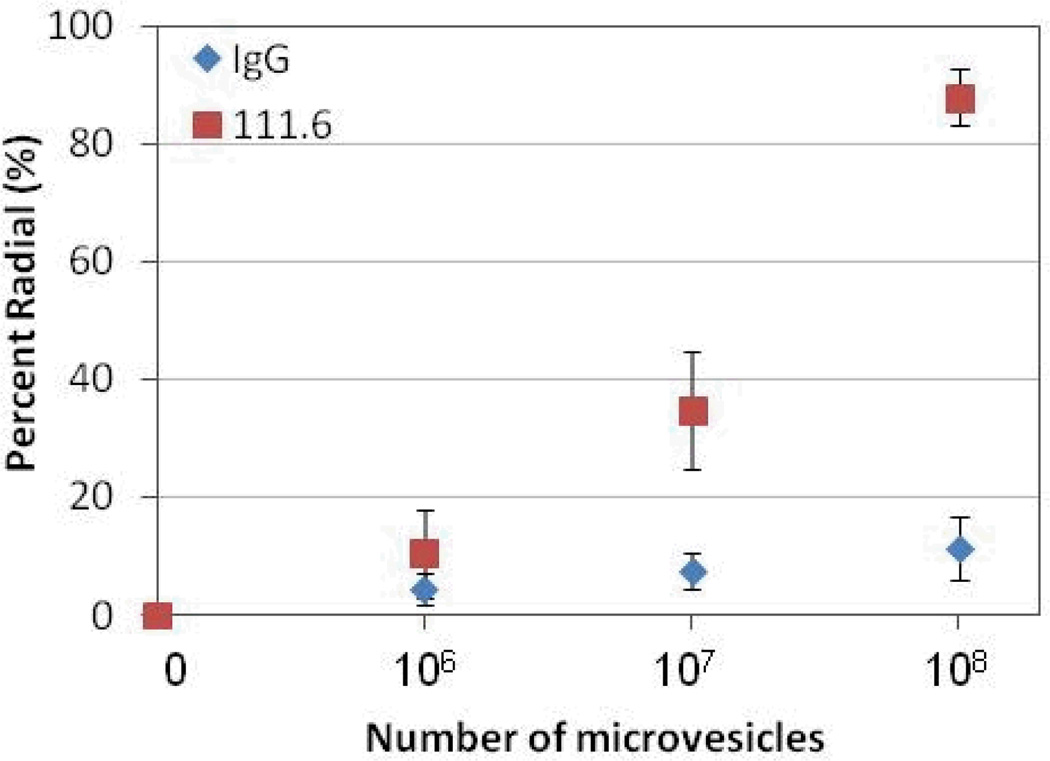
Next we quantified the response of the LC droplets as a function of the number of MVs contacted with the immunobeads. In this experiment, magnetic particles (functionalized with 111.6 or control IgG) were incubated against 100 µL dispersions that contained different numbers of MVs. Figure 5 shows a dose-response plot of the percentage of radial LC droplets as a function of the number of MVs present in the dispersion contacted with the immunobeads. The plots reveal that the percentage of radial LC droplets increased with the number of MVs that were incubated with the immunobeads. We interpret this response to reflect an increase in the number of MVs captured by the immunobeads and subsequently extracted into the LC, an interpretation that is supported by Figure 3. We also note the threshold number of MVs that gave rise to 100 % radial LC droplets was reported in Figure 3 to be 108. The experiments with immunobeads gave about 90 % radial droplets when the immunobeads had been incubated with 108 MVs, suggesting that most of the MVs were captured by the beads and transferred to the LC. This conclusion was supported by quantification of MVs in the aqueous dispersion before and after contact with immunobeads.
Figure 5.
Percentage of LC droplets that exhibited a radial configuration, plotted as a function of number of MVs shed from A431 cells that were incubated against magnetic particles functionalized with anti-EGFR (111.6) or control (isotype) IgGs (N = 3).
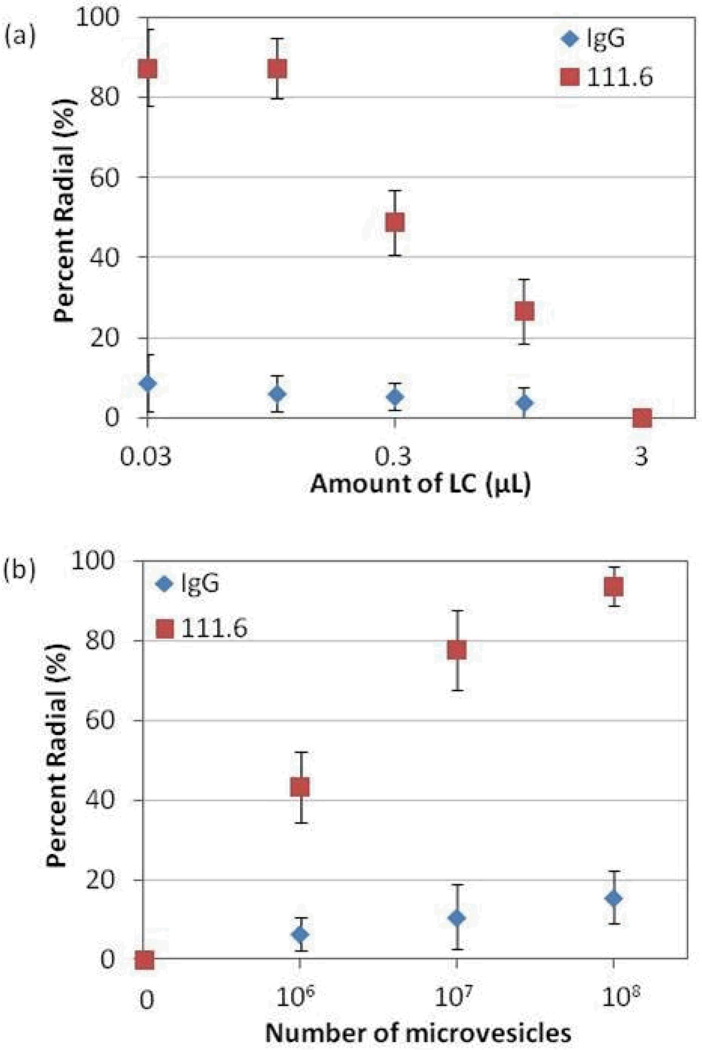
Next we determined if the results in Figure 5 could be optimized further by reducing the number of LC droplets (by emulsification of less 5CB). In this optimization step, the number of MVs incubated with the magnetic particles was kept constant at 107 MVs (a concentration of MVs that resulted in mixed radial/bipolar configurations of LC droplets when using 1 µL of 5CB; see Figure 5). The results shown in Figure 6a reveal that the percentage of radial LC droplets increased with decrease in the amount of 5CB used. This observation is consistent with our conclusion that the internal ordering of the LC droplets is dependent on the amount of lipid in each droplet and that the use of smaller amounts of 5CB led to a higher concentration of lipid within each droplet (see Table S1). Specifically, when the amount of 5CB used to prepare the LC emulsion was decreased from 1 µL to 0.1 µL, the percentage of radial LC droplets increased from 35 % (Figure 5) to 90 % (Figure 6a).
Figure 6.
(a) Percentage of LC droplets that exhibited a radial configuration, plotted as a function of the amount of 5CB into which lipids from MVs captured on antibody-functionalized beads were introduced. The beads were incubated against 107 MVs. (b) Percentage of LC droplets that exhibited a radial configuration, plotted as a function of the number of MVs that were incubated against antibody-functionalized magnetic particles. The volume of LC used to form the emulsion was 0.03 µL.
Guided by the results above, Figure 6b shows the response of the LC droplets as a function of the number of MVs incubated with the immunobeads when 0.03 µL of 5CB was used to prepare the LC emulsion. Inspection of Figure 6b reveals that the presence of 106 MVs can be readily quantified when using 0.03 µL of 5CB. As discussed above, the number of EGFR molecules per MV is estimated to be 500. This leads us to estimate that the current methodology permits detection of a concentration of EGFR in MVs in cell culture media of 5 fM.
Discussion
The key results reported in this paper are two-fold. First, we have demonstrated that LC droplets in combination with FC can be used to quantify the presence of MVs shed by mammalian cells. In particular, we found spontaneous transfer of MVs shed from mammalian cells onto the interface of LC droplets to be very slow. This contrasts to past studies of model vesicles, viruses and bacteria (see above), and suggests that either the lipid composition/phase state of the membrane of the vesicles slows spontaneous transfer, or that proteins and polysaccharides that protrude from the surfaces of mammalian cells (and thus MVs) hinder contact of the membranes and the LC droplets. To overcome this barrier to transfer, we developed and validated a simple procedure that involves extraction of the MVs into the LC phase via use of an organic solvent. We demonstrated that the procedure permits quantitative reporting of MVs shed by cells by LC droplets (using FC).
A second key accomplishment reported in this paper is the demonstration of a methodology that permits quantitation of MVs shed by cells that contain a target protein. In particular, we sought to demonstrate LC droplet-based methods that leverage protocols and instrumentation that are routinely found in biochemical laboratories. In this context, we found that capture of MVs via the use of antibody-functionalized beads provided a facile method to quantify the number of MVs in a sample that contained a targeted protein. While we have used EGFR present in MVs shed by A431 cells as a model system with which to develop and validate the methodology reported in this paper, we emphasize that EGFR is also an important target in cancer-related studies because its over-expression and mutation have been associated with some of the most aggressive forms of cancer. The methodology reported in this paper was shown to permit detection of 106 MVs that contained EGFR in a sample. Our results and others24 that have used immunoblotting to detect MVs have determined a sensitivity of 108-109 MVs, a value that is surpassed by the LC-based methods.
The high sensitivity of the LC-based detection method reported in this paper arises in part because the binding of the MV-associated transmembrane protein is amplified by the high ratio of lipid to target protein in the MVs. The amplification that is inherent in the response of the LC droplets to the lipid component of the MVs provides a significant advantage over existing assays such as ELISA and immunoblotting. In addition, due to the lipid-driven ordering of LCs, the LC-based method does not require use of secondary antibodies, thus providing a cost advantage. Furthermore, methods such as immunoblotting and ELISA detect soluble antigens present in the sample and therefore, unlike the LC-based method, are not specific to MV-associated membrane proteins.
As mentioned in the Introduction, another technique that has been employed for direct detection of MVs is FC. While FC has been successful for routine cell sorting and cell counting, most conventional FC methods are not sufficiently sensitive to support detection of particles smaller than 500 nm, including MVs.20–22 In contrast, LC-based FC reads out ordering transitions within micrometer-sized LC droplets induced by lipid-associated MVs and therefore it can readily detect MVs in the 100 nm-size range in a manner that is not strongly size-dependent. Also, the sensitivity of FC when used to probe directly for MVs was reported to be 108 MVs, a value that is two orders of magnitude higher than the sensitivity of LC-based FC. Finally, we comment that fluorescent dyes have been used to label MVs in FC-based methods, but past studies have shown that the broad range of species present in complex mixtures (e.g., free dyes, antibody aggregates, cell fragments with high levels of autofluorescence) give high background signal.48
In addition, we noted that NTA has also been used to detect MVs. Like FC, NTA analysis also suffers from false-positive signals that arise from aggregated serum proteins.16 Also, because of statistical requirements35, the limit of detection of MVs in NTA is typically in the range of 108 particles/mL, a value that is surpassed by our current method. Here we emphasize that protein aggregates do not lead to false-positive signals in LC-based methods since the LC droplets respond specifically to lipids (main component of MVs) and not protein aggregates. This unique attribute of the LC-based method arises because adsorbate-induced anchoring of LCs is strongly dependent on the structure of the adsorbate.
Other techniques employed for analysis of MVs include electron microscopy49 and AFM50, both of which require extensive sample preparation and do not provide quantitative information. One of the more sophisticated techniques developed thus far involves the use of a micro-NMR technique. In this technique, MVs are labeled with target-specific magnetic particles and introduced onto a microfluidic chip with a micro-NMR system.24 This was used to profile circulating MVs from blood samples of glioblastoma patients and is proposed as a new way to monitor and predict response to disease therapies. While this technique surpasses our current method in terms of sensitivity (105 MVs), we note the complexity and extensive time requirement for sample preparation that accompanied this technique.
As mentioned above, the analytical attributes of the methodology reported in this paper are closely tied to the response of the LC droplets to the lipid components of the MVs. We make an additional point regarding this topic in connection to the results in Figure 3. As noted above, past studies have shown that for a LC droplet to adopt a radial configuration as a result of adsorption of lipids at the droplet interface, close to saturation coverage of lipid is required at the LC interface.26 To compare our results to these past studies, we calculated the number of lipid molecules introduced into each LC droplet from the MVs. For a LC droplet with a diameter of 5 µm, we calculate that a total of 108 lipid molecules are needed to saturate the interface of the LC droplet with lipid (the density of lipid at monolayer coverage is 1.7 molecules/nm2).29 For comparison, 108 MVs, which led to radial droplets prepared from 1 µL of 5CB, contain 1014 lipid molecules. Because 1 µL of 5CB generates about 107 droplets (see above), we calculate that the average number of lipid molecules per LC droplet is 107, which corresponds to 10% of the calculated saturation coverage.
We speculate that several factors may underlie our observation that the amount of lipid required to induce the radial configuration of LC droplets in our experiments is less than a monolayer coverage. First, the cell membrane (from which MVs are derived) contains a significant amount of cholesterol and sphingomyelin (up to 30 mol %, dependent on cell type51) that is not measured by the lipid assay used to quantify the lipid content of the MVs. These components may influence the phase states of the lipid monolayer and its interactions with the mesogens at the LC interface. In addition, the influence of lipids with acyl chains of varying lengths and degrees of unsaturation on LCs (see Figure S1) has not been fully explored, and mixtures of such lipids may influence the ordering of LCs at sub-saturation coverage. We also note that our calculation above is approximate as it is based on the average droplet size, while in practice the sizes of the LC droplets in the samples used in our experiments are polydisperse (see above for size effects).
We end this discussion by noting that a wide range of protein targets have now been detected in MVs shed into biological fluids: Examples include EGFRs in MVs derived from pleural effusions of patients with non-small cell lung cancer52 and detection of HER2/neu in MVs derived from blood of gastric cancer patients.53, 54 CD147/EMMPRIN, a transmembrane protein, which is expressed at high levels by tumor cells, was found to be released from the surface of NCI-H460 cells via microvesicle shedding.18, 55 AXL receptors, which have also been reported to be overexpressed in several types of human cancers, were detected in MVs circulating in plasma of chronic lymphocytic leukemia (CLL) patients.56, 57 Overall, we note that the increased secretion of plasma membrane-derived MVs by cells in diseased states suggests that development of broadly applicable analytical platforms for MVs has the potential to impact basic biological research through to clinical diagnostics.
Conclusion
In summary, we have demonstrated principles for LC droplet-based quantification of MVs containing targeted proteins that are shed by mammalian cells. The approach uses micrometer-sized droplets of LCs to amplify MVs that are selectively captured via antibody-mediated interactions. The LC droplets are sufficiently large that they can be quantified via flow cytometry. The high lipid to protein ratio in the MVs leads to high sensitivity since LC droplets respond to the lipid component of the MVs. Specifically, we have validated the approach using MVs shed by A431 cells that contained EGFR. A significant merit of the approach is that the procedures and instrumentation that are combined with the LC droplets to create the methodology are routinely found in basic biological and clinical laboratories. In these initial proof-of-concept experiments, we demonstrate that it is straightforward to quantify 106 MVs containing EGFR, a sensitivity that is better than immunoblotting, direct FC and NTA techniques. We also demonstrate a dynamic range of several orders of magnitude. Overall this paper provides the framework for the use of LC-droplets for detection of a broad range of MVs containing targeted proteins and lipids. With additional optimization, the analytical characteristics of the methodology can likely be improved further (e.g., lower limit of detection).
Supplementary Material
Acknowledgments
This work was supported by the NSF under awards DMR-1121288 (MRSEC), by the National Institutes of Health (CA108467 and AI092004), and by the ARO (W911-NF-11-1-0251 and W911-NF-10-1-0181). We thank Dennis Yang and Professor Regina Murphy for their help with NTA measurements, and Daniel Mendez and Professor Brian Pfleger for their assistance with GC-MS measurements.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental details related to additional AFM measurements and characterization of LC droplets. See DOI: 10.1039/b000000x/
Notes and references
- 1.Kahlert C, Kalluri R. Journal of Molecular Medicine. 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koga K, Matsumoto K, Akiyoshi T, Kubo M, Yamanaka N, Tasaki A, Nakashima H, Nakamura M, Kurok S, Tanaka M, Katano M. Anticancer Res. 2005;25:3703–3707. [PubMed] [Google Scholar]
- 3.Théry C, Zitvogel L, Amigorena S. Nature Reviews Immunology. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 4.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Kidney International. 2011;78:838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 5.Cocucci E, Racchetti G, Meldolesi J. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Leukemia. 2006;20:1487–1495. doi: 10.1038/sj.leu.2404296. [DOI] [PubMed] [Google Scholar]
- 7.Ohno S-i, Ishikawa A, Kuroda M. Advanced Drug Delivery Reviews. 2012;65:398–401. doi: 10.1016/j.addr.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 8.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Nature Communications. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 10.Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3794–3799. doi: 10.1073/pnas.0804543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Nat. Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 12.Martins VR, Dias MS, Hainaut P. Current Opinion in Oncology. 2013;25:66–75. doi: 10.1097/CCO.0b013e32835b7c81. [DOI] [PubMed] [Google Scholar]
- 13.van Doormaal FF, Kleinjan A, Di Nisio M, Buller HR, Nieuwland R. Neth. J. Med. 2009;67:266–273. [PubMed] [Google Scholar]
- 14.Al-Nedawi K, Meehan B, Rak J. Cell Cycle. 2009;8:2014–2018. doi: 10.4161/cc.8.13.8988. [DOI] [PubMed] [Google Scholar]
- 15.Jayachandran M, Miller VM, Heit JA, Owen WG. Journal of Immunological Methods. 2012;375:207–214. doi: 10.1016/j.jim.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Günter M. Journal of Bioanalysis & Biomedicine. 2012;4:46–60. [Google Scholar]
- 17.Osumi K, Ozeki Y, Saito S, Nagamura Y, Ito H, Kimura Y, Ogura H, Nomura S. Thromb. Haemost. 2001;85:326–330. [PubMed] [Google Scholar]
- 18.Sidhu SS, Mengistab AT, Tauscher AN, LaVail J, Basbaum C. Oncogene. 2004;23:956–963. doi: 10.1038/sj.onc.1207070. [DOI] [PubMed] [Google Scholar]
- 19.Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. The Journal of Immunology. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 20.Kim HK, Song KS, Lee ES, Lee YJ, Park YS, Lee KR, Lee SN. Blood Coagul. Fibrinolysis. 2002;13:393–397. doi: 10.1097/00001721-200207000-00003. [DOI] [PubMed] [Google Scholar]
- 21.Gelderman MP, Simak J. Methods Mol Biol. 2008;484:79–93. doi: 10.1007/978-1-59745-398-1_6. [DOI] [PubMed] [Google Scholar]
- 22.van der Vlist EJ, Nolte EN, Stoorvogel W, Arkesteijn GJ, Wauben MH. Nature Protocols. 2012;7:1311–1326. doi: 10.1038/nprot.2012.065. [DOI] [PubMed] [Google Scholar]
- 23.Soo CY, Song Y, Zheng Y, Campbell EC, Riches AC, Gunn-Moore F, Powis SJ. Immunology. 2012;136:192–197. doi: 10.1111/j.1365-2567.2012.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Nature Medicine. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta JK, Zimmerman JS, de Pablo JJ, Caruso F, Abbott NL. Langmuir. 2009;25:9016–9024. doi: 10.1021/la900786b. [DOI] [PubMed] [Google Scholar]
- 26.Gupta JK, Sivakumar S, Caruso F, Abbott NL. Angew. Chem.-Int. Edit. 2009;48:1652–1655. doi: 10.1002/anie.200804500. [DOI] [PubMed] [Google Scholar]
- 27.Lin I-H, Miller DS, Bertics PJ, Murphy CJ, de Pablo JJ, Abbott NL. Science. 2011;332:1297–1300. doi: 10.1126/science.1195639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brake JM, Daschner MK, Luk Y-Y, Abbott NL. Science. 2003;5653:2094–2097. doi: 10.1126/science.1091749. [DOI] [PubMed] [Google Scholar]
- 29.Meli MV, Lin IH, Abbott NL. Journal of the American Chemical Society. 2008;130:4326–4333. doi: 10.1021/ja077379a. [DOI] [PubMed] [Google Scholar]
- 30.Hu Q-Z, Jang C-H. Analyst. 2012;137:567–570. doi: 10.1039/c1an15743d. [DOI] [PubMed] [Google Scholar]
- 31.Miller DS, Wang X, Buchen J, Lavrentovich OD, Abbott NL. Anal Chem. 2013;85:10296–10303. doi: 10.1021/ac4021193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholson R, Gee J, Harper M. Eur. J. Cancer. 2001;37:9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 33.Lennen RM, Braden DJ, West RM, Dumesic JA, Pfleger BF. Biotechnology and Bioengineering. 2010;106:193–202. doi: 10.1002/bit.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Provencher SW. Comput. Phys. Commun. 1982;27:213–242. [Google Scholar]
- 35.Filipe V, Hawe A, Jiskoot W. Pharmaceutical Research. 2010;27:796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bates ME, Busse WW, Bertics PJ. American Journal of Respiratory Cell and Molecular Biology. 1998;18:75–83. doi: 10.1165/ajrcmb.18.1.2766. [DOI] [PubMed] [Google Scholar]
- 37.Kim Y-N, Wiepz GJ, Guadarrama AG, Bertics PJ. Journal of Biological Chemistry. 2000;275:7481–7491. doi: 10.1074/jbc.275.11.7481. [DOI] [PubMed] [Google Scholar]
- 38.Lee TH, D’Asti E, Magnus N, Al-Nedawi K, Meehan B, Rak J. Seminars in immunopathology. 2011 doi: 10.1007/s00281-011-0250-3. [DOI] [PubMed] [Google Scholar]
- 39.Rohrer GA, Pond W, Bell A. Taylor & Francis; 2004. [Google Scholar]
- 40.Krupp MN, Connolly D, Lane M. Journal of Biological Chemistry. 1982;257:11489–11496. [PubMed] [Google Scholar]
- 41.Terakawa M, Tsunoi Y, Mitsuhashi T. International Journal of Nanomedicine. 2012;7:2653. doi: 10.2147/IJN.S31768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.del Conde I, Shrimpton CN, Thiagarajan P, Lopez JA. Blood. 2005;106:1604–1611. doi: 10.1182/blood-2004-03-1095. [DOI] [PubMed] [Google Scholar]
- 43.Couet J, Sargiacomo M, Lisanti MP. Journal of Biological Chemistry. 1997;272:30429–30438. doi: 10.1074/jbc.272.48.30429. [DOI] [PubMed] [Google Scholar]
- 44.Tan LN, Bertics PJ, Abbott NL. Langmuir. 2010;27:1419–1429. doi: 10.1021/la103975s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lockwood NA, Gupta JK, Abbott NL. Surf. Sci. Rep. 2008;63:255–293. [Google Scholar]
- 46.Lockwood NA, de Pablo JJ, Abbott NL. Langmuir. 2005;21:6805–6814. doi: 10.1021/la050231p. [DOI] [PubMed] [Google Scholar]
- 47.Lavrentovich O. Liq. Cryst. 1998;24:117–126. [Google Scholar]
- 48.Aass HCD, Øvstebø Reidun, Trøseid Anne-Marie S, Kierulf Peter, Berg Jens Petter, Henriksson Carola Elisabeth. Cytometry Part A. 2011;79:990–999. doi: 10.1002/cyto.a.21147. [DOI] [PubMed] [Google Scholar]
- 49.Heijnen HFG, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Blood. 1999;94:3791–3799. [PubMed] [Google Scholar]
- 50.Sharma S, Gillespie BM, Palanisamy V, Gimzewski JK. Langmuir. 2011;27:14394–14400. doi: 10.1021/la2038763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pike LJ, Han X, Chung K-N, Gross RW. Biochemistry. 2002;41:2075–2088. doi: 10.1021/bi0156557. [DOI] [PubMed] [Google Scholar]
- 52.Park JO, Choi DY, Choi DS, Kim HJ, Kang JW, Jung JH, Lee JH, Kim J, Freeman MR, Lee KY. Proteomics. 2013;13:2125–2134. doi: 10.1002/pmic.201200323. [DOI] [PubMed] [Google Scholar]
- 53.D'Souza-Schorey C, Clancy JW. Genes & development. 2012;26:1287–1299. doi: 10.1101/gad.192351.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baran J, Baj-Krzyworzeka M, Weglarczyk K, Szatanek R, Zembala M, Barbasz J, Czupryna A, Szczepanik A, Zembala M. Cancer Immunology Immunotherapy. 2009;59:841–850. doi: 10.1007/s00262-009-0808-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gabison EE, Hoang-Xuan T, Mauviel A, Menashi S. Biochimie. 2005;87:361–368. doi: 10.1016/j.biochi.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 56.Hong C-C, Lay J-D, Huang J-S, Cheng A-L, Tang J-L, Lin M-T, Lai G-M, Chuang S-E. Cancer letters. 2008;268:314–324. doi: 10.1016/j.canlet.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 57.Ghosh AK, Secreto CR, Knox TR, Ding W, Mukhopadhyay D, Kay NE. Blood. 2010;115:1755–1764. doi: 10.1182/blood-2009-09-242719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.