Abstract
We evaluated the importance of tumor cell selection for generating gene signatures in non–small cell lung cancer. Tumor and nontumor tissue from macroscopically dissected (Macro) surgical specimens (31 pairs from 32 subjects) was homogenized, extracted, amplified, and hybridized to microarrays. Adjacent scout sections were histologically mapped; sets of approximately 1000 tumor cells and nontumor cells (alveolar or bronchial) were procured by laser capture microdissection (LCM). Within histological strata, LCM and Macro specimens exhibited approximately 67% to 80% nonoverlap in differentially expressed (DE) genes. In a representative subset, LCM uniquely identified 300 DE genes in tumor versus nontumor specimens, largely attributable to cell selection; 382 DE genes were common to Macro, Macro with preamplification, and LCM platforms. RT-qPCR validation in a 33-gene subset was confirmatory (ρ = 0.789 to 0.964, P = 0.0013 to 0.0028). Pathway analysis of LCM data suggested alterations in known cancer pathways (cell growth, death, movement, cycle, and signaling components), among others (eg, immune, inflammatory). A unique nine-gene LCM signature had higher tumor–nontumor discriminatory accuracy (100%) than the corresponding Macro signature (87%). Comparison with Cancer Genome Atlas data sets (based on homogenized Macro tissue) revealed both substantial overlap and important differences from LCM specimen results. Thus, cell selection via LCM enhances expression profiling precision, and confirms both known and under-appreciated lung cancer genes and pathways.
Cancers are complex, even within traditional histopathological strata. Most lung tumors have supporting stromal cells, including infiltrating inflammatory cells, fibroblasts, and neovasculature. In nonmalignant lung, perhaps an even higher degree of cellular heterogeneity is present; normal lung has more than 42 identifiable cell types. The nonepithelial components, in both carcinomas and paired adjacent nonmalignant tissues, almost certainly contribute to the transcriptomes generated with traditional tissue-block homogenization. Thus, depending on the individual tumor characteristics, these other nonmalignant and nonepithelial cell types might contribute a very substantial fraction of the transcriptome that has been reported from studies with tissue-mincing homogenization. Of the handful of transcriptome studies of refined cell capture including microdissection for procurement of a verifiably enriched malignant cell fraction in lung cancers1, 2, 3 or focused on coupled platforms,4 most are limited by quite small sample size and/or nonpaired tumor and nontumor samples.
We hypothesized that lung cancer signature precision could be augmented by enhanced cell selection using laser capture microdissection (LCM) of morphologically malignant cells. Reduced contamination of tumor cells by admixed supporting stroma, combined with paired nonmalignant [ie, nontumor (NT)] lung parenchymal epithelium, alveolar (NTa) or bronchial (NTb), should help refine the features of the transcriptome unique to lung cancer. We therefore microdissected pairs of non–small cell lung cancers (T) and their far-adjacent NT tissue. In parallel, we also performed conventional macroscopic tissue-block homogenization (hereafter referred to as Macro) on the same tissue samples for comparison purposes. In addition, microaliquots of Macro T and NT lung sample extracts in a subset were preamplified using a LCM sample preamplification procedure, to control for bias inherent to the preamplification process itself.
Materials and Methods
Patient Recruitment and Sample Collection
The study population comprised 32 consenting individuals undergoing resectional surgery for clinically suspected non–small cell lung cancer (NSCLC) under a protocol5 approved by the local institutional review boards.
Lung tissue resection samples were visually divided into T and NT (far-adjacent to remote) in a room adjacent to the operating room and used for preparing frozen sections. The samples were snap-frozen in liquid isopentane within 15 minutes of surgical resection and were stored in a tissue bank (in liquid nitrogen at −180°C) until analysis. The assigned clinical surgical pathologist confirmed the diagnosis of lung cancer in all cases, per clinical routine, and classified the samples as adenocarcinoma, squamous cell carcinoma, or mixed adenosquamous NSCLC, according to the 1999 World Health Organization histological classification of lung and pleural tumors6 and recent updates.7, 8 In addition, all 32 selected cases were independently reviewed again by two other pathologists (J.L. and C.Z.), each masked to prior histological diagnosis, clinical, and transcriptome data.
Tissue Accession Sets
A set of 31 paired and 2 individual unpaired samples of macroscopic homogenized NSCLC T and NT (alveolar predominant) tissue were selected for bulk macroscopic tissue sampling and microarray analyses: 19 T–NT adenocarcinoma pairs and 1 nonpaired adenocarcinoma sample; 10 T–NT squamous cell carcinoma pairs and 1 nonpaired squamous cell carcinoma sample; and 2 T–NT pairs of adenocarcinoma–squamous cell carcinoma. Of these, 17 T–NT pairs (with alveolar cells selected for NT) and 9 unpaired samples that underwent LCM met RNA quality criteria for microarray analyses: 7 pairs and 7 nonpaired samples of adenocarcinomas; 8 pairs and 1 nonpaired sample of squamous-cell carcinoma; and 2 pairs and 1 nonpaired sample of mixed adenosquamous carcinoma. Additionally, NTb samples were microdissected from six specimens.
RNA Isolation from Macro Tissue Samples
Approximately 50 to 80 mg of snap-frozen lung tissue was added to a tube containing 1 mL of extraction buffer and was completely homogenized. Further total RNA extraction procedures were performed using an RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer’s recommendations, including an optional 30 minutes DNase I treatment. Total RNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and quality was confirmed on an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA). An RNA integrity number (RIN) of 7 was considered the threshold for proceeding.
Preparation of Macro Homogenized Samples for Microarray
Approximately 100 ng total RNA was amplified using an Ambion WT expression kit (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions; 5.5 μg of amplification product was labeled with an Affymetrix (Santa Clara, CA) WT terminal labeling kit and then hybridized to Affymetrix HG Gene 1.0ST chips using Affymetrix hybridization and wash and stain kits according to the manufacturer’s instructions.
LCM
For every paired (T and NT) tissue block obtained from each resection subject, a pathologist (J.L. and C.Z.) assessed a digitized hematoxylin and eosin–stained (scout) slide to determine and outline tumor nests, thereby mapping the section and directing the laser capture microscopist toward irrefutably high tumor cell content on the adjacent frozen sections. The same procedure was used for the NT block. Distinguishing tumor cells from two different types of NT lung tissue compartments [ie, NTa or NTb epithelium] could be achieved with confidence in these snap-frozen samples.
Once the hematoxylin and eosin–stained scout section was mapped, we accessed corresponding adjacent frozen lung tissue (T or NT) and re-embedded it in cold Tissue-Tek OCT optimal cutting temperature medium (Sakura Finetek USA, Torrance, CA). Three sections (12 μmol/L thick) from each frozen T and NT block were cut in a cryostat at −25°C and placed on nuclease-free and human nucleic acid–free membrane slides (Leica Microsystems, Wetzlar, Germany). The sections were stored at −80°C until use or were immediately stained with a rapid hematoxylin and eosin ethanol-based staining protocol.5, 9 Within 30 minutes of air drying, the slide was placed on the LCM stage of a Leica AS LMD instrument for microdissection. The desired cells (either alveolar or bronchial, separately) were microdissected into the cap of 200-μL PCR tubes filled with 20 μL RLT/β-mercaptoethanol buffer (Qiagen). This procedure was repeated on the next slide until a total of 1000 cells of interest had been captured for that case. The number of (adjacent) slides used per case to reach this 1000-cell threshold varied from 1 to 3, because tumor proportion varied across samples and donors.
RNA Extraction of LCM Samples and Preamplification for Microarray
LCM-derived RNA samples for preamplification were isolated from 1000 cells using an RNeasy micro kit (Qiagen), including the optional DNase treatment, according to the manufacturer’s instructions. Total RNA was quantified and quality was determined using the same techniques as for the macroscopic samples. Two-thirds of LCM samples met the RIN = 7 threshold for proceeding. Because of the low initial RNA template inherent to LCM-scale specimens, RNA preamplifications were performed. We used an Ovation Pico WTA whole-transcriptome amplification (NuGEN, San Carlos, CA) system according to the manufacturer’s recommendations, followed by an additional amplification step using a NuGEN exon module. One nanogram of total RNA was used as input for Pico amplification; the resulting 4 μg of amplified cDNA was then used for NuGEN Pico exon module amplification. cDNA amplification products were fragmented and labeled with biotin using a NuGEN FL module and hybridized to Affymetrix HG Gene 1.0 ST arrays using an Affymetrix hybridization control kit and were washed and stained using an Affymetrix wash and stain kit, identically to the macroscopic homogenized samples, according to the manufacturer’s recommendations.
Microaliquots of homogenized macroscopic lung T and NT sample extracts were preamplified in a subset of eight T–NT paired samples, using the same procedure as for LCM of the same samples. Because the preamplification was requisite to LCM interrogation with expression microarrays (because of low initial template concentrations typical of these samples), its use for small aliquots of homogenized materials served as a control for preamplification. This allowed direct comparison of T–NT transcript differences attributable to LCM-related cell selection versus the preamplification process itself.
Quality Control of Microarray Results Using Affymetrix Expression Console Software
For the entire set of microarray samples, the raw array data were imported into Affymetrix Expression Console version 1.1, a software package that permits visualization, quality control, and normalization of the data. Quality control results included array images, line graphs of labeling and hybridization controls, signal box plots, and histograms before and after normalization for all hybridizations, as well as a heat map for Spearman rank correlation between all pairs of hybridizations. Problematic arrays were visually inspected using images, graphs, and plots as above. The gene expression measures for all arrays that passed quality control were normalized by the robust multiarray analysis (RMA) approach in the Affymetrix Expression Console software package.
Microarray Analysis
Ranking of genes by degree of differential expression was performed using the Bioconductor limma package version 2.14 (http://www.bioconductor.org) and in-house code developed in the R statistical language (http://www.r-project.org). Selection of significantly different gene expression profiles between the two experimental conditions was based on the empirical Bayes moderated t-statistic. The Benjamini–Hochberg method was applied to correct for multiple testing.10 Significant genes were identified by adjusted P ≤ 0.05 and fold change (FC) in mean expression of FC ≥ |2|.
Power
For LCM samples (all three histologies), overall T versus NT tests were performed on 47 microarray chips (of which 6 were for NTb), leaving 41 chips treated as paired sample T–NTa comparisons (with one sample missing). The sample size provided 99% power to detect a difference of 1.0 log2 unit, which is the mean difference in log2 intensity of mean differential (ie, twofold) mRNA expression between T and NT for each pair. This permitted distinguishing between the null hypothesis (mean of 0.0) and the alternative hypothesis (mean of 1.0) with an estimated standard deviation of 1.0 and with a significance level of α = 0.05. If we enlarge the effect size (the difference) to 1.5 log2 units, the power increases to 100%. For the adenocarcinoma paired T–NT subsets, the 11 LCM pairs yielded 85% power to detect the same difference (1.0 log2 units = twofold), and 99% if the effect size is increased to 1.5 log2 units (threefold). For squamous cell carcinoma, the nine LCM pairs yielded 75% power for effect size of 1.0, and 97% for effect size of 1.5. These calculations assume paired samples (T–NT) and a P value of 0.05.
Confirmation of Microarray Results by RT-qPCR
Quantitative real-time RT-PCR (RT-qPCR) for the most dysregulated macroscopic block-derived genes was performed with Power SYBR Green PCR master mix in a 96-well optical plate using an ABI 7500 real-time PCR system (all from Life Technologies, Carlsbad, CA). A primer pair for each gene was designed with online Primer3 software version 3.0 (https://www.broadinstitute.org/genome_software/other/primer3.html) based on the published sequences (http://www.ncbi.nlm.nih.gov/genbank). The RT-qPCR was performed in technical duplicates for each sample. Additional double-distilled water blank and RNA without reverse transcriptase samples served as negative controls for each transcript run. Melting analysis for one additional cycle was performed. Where necessary, an RNA-specific strategy that avoids contaminating genomic DNA amplification and false positives was used.11 After the reaction, all PCR products underwent additional confirmatory electrophoresis on an agarose–ethidium bromide gel and were visualized under UV light. Spearman correlation was performed with the same most dysregulated transcripts as detected by expression microarray.
Tumor Classification Signature
To evaluate the performance of the respective gene signature for correct classification of the T versus NT status, a 1:1 split to training and test samples was effected, individually for Macro samples, and then for LCM samples, separately. An initial training set signature, selected on the basis of misclassification error and the number of probe sets in the model during leave-one-out cross-validation, was derived from the microarray expression patterns. A three-step strategy was performed, similar to that reported previously.12 First, probe sets were ranked by degrees of differential expression using the Bioconductor limma package and in-house R code. Second, the Pearson correlation coefficient of the expression for each of the top 3000 probe sets with T versus NT status was calculated. In all, 174 probe sets were found to be significantly associated with T versus NT status [P < 0.01, adjusted for false discovery rate (FDR)]. Third, all 174 probe sets were used to fit a Lasso regularized generalized linear model, using Bioconductor lmnet. Training and test sets were matched on all relevant clinical variables (ie, age, sex, cumulative smoking dose, smoking status, and histology); the training set therefore derived from a split sample of subjects (training and test), which were statistically indistinguishable. Data were expressed as the area under curve (AUC), with sensitivity plotted against 1 − specificity, from null (0.5) to perfect (1.0) discrimination.
Comparison with the TCGA Reference Data Set
We compared our LCM and Macro transcriptome findings with those derived from the Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov/tcga, last accessed April 9, 2014) for lung adenocarcinoma T versus NT comparisons. TCGA describes only macroscopic (ie, non-LCM) findings. Because there were no microarray-based data on T–NT pairs available in TCGA adenocarcinoma studies, TCGA macroscopic pairs interrogated by next-generation sequencing (RNA sequencing or RNA-seq) were used for this comparison. To avoid batch-to-batch variability, batch 144 alone was used, because it was the largest (n = 15 adenocarcinoma T pairs from 15 donors, all stage I or II) from a single institution and sequencing platform. The normalized RNA-seq gene level data for batch 144 was downloaded from the TCGA data portal. A total of 15 pairs of matched T–NT samples were included into the analysis. Any gene with more than 10% zero readings across these samples was excluded from analysis. Among the remaining 16,159 genes and 30 samples, the smallest nonzero normalized count was 0.7184. We then used half of that (0.7184/2 = 0.3592) as the limit of detection, to replace the few zero read counts and to allow computational transformation more easily. After log transformation, moderated t-testing was performed for to the RNA-seq data, using the Bioconductor limma package and in-house R code. The genes were ranked by Benjamini–Hochberg adjusted P values and FC.
Pathway Analysis
Gene ontology, canonical pathway, and functional network analyses were performed by using Ingenuity Pathways Analysis (IPA) software tools (Qiagen Silicon Valley, Redwood City, CA). The lists of significantly differentially expressed genes identified in gene expression analysis (adjusted P < 0.05), including Affymetrix probe set identifiers and the corresponding FC values, were uploaded into the IPA analysis tool. These genes were further filtered in IPA by the criterion of FC ≥ 2 before use for pathway and network analyses. Each probe set identifier was then mapped to its corresponding gene in the Ingenuity Pathways Knowledge Base database. A ratio of the number of genes from the gene list that map to a certain pathway divided by the total number of genes that map to that pathway was calculated. Fisher’s exact test was used to estimate the probability that the association between the gene list and a certain pathway is due to random chance. In the case of functional analysis, where a set of N molecules is queried as to whether there is enrichment of molecules with a particular (pathway) annotation, Fisher’s exact test is computationally less expensive than other plausible approaches, such as permutation strategies. IPA analyses are reported using the Benjamini–Hochberg multiple testing adjusted P value.
Results
Donor and Tissue Characteristics
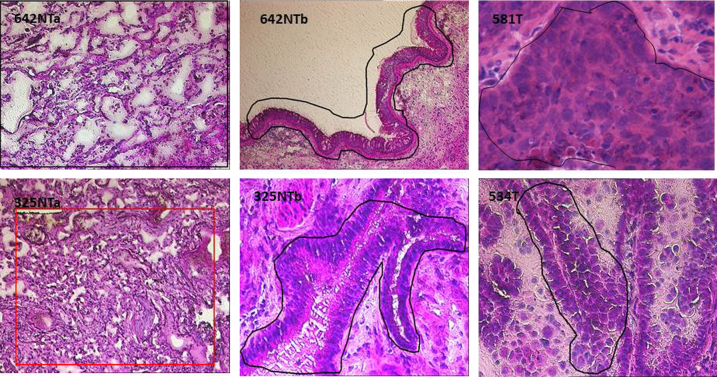
Clinicodemographic characteristics of the 32 donors for this lung cancer study are summarized in Supplemental Table S1. All subjects had a lung cancer diagnosis; by virtue of surgical candidacy and lobe resections, the diagnoses were confirmed pathologically as clinical stages I to II. Typical scout slide images, including representative margins outlined for microdissection, are shown in Figure 1.
Figure 1.
Representative images of frozen-section specimens undergoing laser capture microdissection (LCM). The target areas are outlined with black or red laser track marks. Tumors (T) for LCM were histologically characterized by consensus of two pathologists. The precise areas to be laser-captured from each tissue section were designated by a pathologist (J.L. and C.Z.) on each scout image, and laboratory personnel followed standard protocols for LCM. For tissue scans and microscopy of nontumor (NT) alveolar areas (NTa; left column), parenchymal inflammation was common. Typical nonmalignant bronchial mucosa (NTb; middle column) is outlined for microdissection. The two tumor specimens (T; right column) are squamous cell carcinoma (top row) and adenocarcinoma (bottom row).
Microarray Analysis from Macro Homogenized Tissues
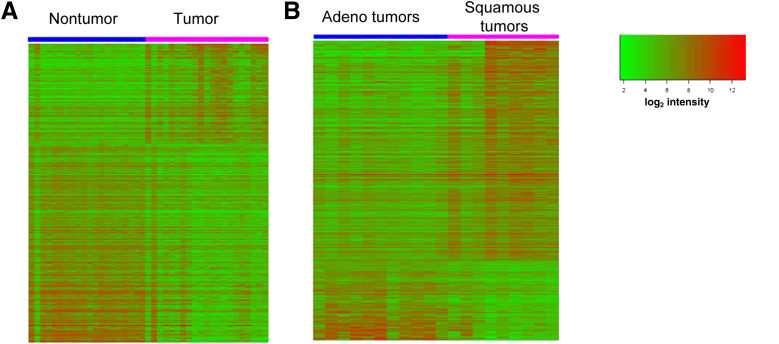
Representative heat maps for LCM samples are presented in Figure 2. Gene expression analysis of Macro samples identified a large number of DE genes (adjusted P > 0.05, FC > 2) for both adenocarcinoma and squamous cell carcinomas, compared with the paired NT lung tissue. The 20 most up-regulated and the 20 most down-regulated genes (in terms of FC, given adjusted P < 0.05) in all NSCLC histologies combined are listed in Table 1. The full data set is available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo, accession number GSE31552). The 20 most up-regulated and the 20 most down-regulated genes for adenocarcinoma are listed in Table 2; those for squamous cell carcinoma are listed in Table 3. The five most up-regulated genes for the Macro T specimens, all three histologies (Table 1), were SPP1, TMPRSS4, MMP12, GPX2, and MMP1; the five most down-regulated genes were FABP4, SLC6A4, AGER, TMEM100, and GKN2.
Figure 2.
Heat maps demonstrate technical feasibility of microarray-based transcriptome analyses on LCM samples. A: Heat map of differentially expressed (DE) gene transcripts from LCM T versus NT lung specimens. A total of 850 genes with adjusted P value < 0.05 and log2 fold change (FC) of ≥1.0 were used for this comparison. B: A similar heat map differentiates LCM adenocarcinomas from LCM squamous cell carcinomas. A total of 651 genes with adjusted P < 0.05 and log2 FC ≥ 1.0 were used for this comparison.
Table 1.
Genes Differentially Expressed (T versus NT) in Macro and LCM Samples of NSCLC, All Three Histologies Combined
| Macro |
LCM |
||||
|---|---|---|---|---|---|
| Gene | FC∗ | Padjusted† | Gene | FC∗ | Padjusted† |
| Up-regulated (all histologies) | |||||
| SPP1 | 9.1 | 7.82 × 10−10 | SPINK1‡ | 7.0 | 1.97 × 10−2 |
| TMPRSS4 | 6 | 2.31 × 10−10 | AKR1C2‡ | 5.8 | 7.14 × 10−3 |
| MMP12 | 5.7 | 2.25 × 10−7 | CENPF | 5.0 | 2.20 × 10−4 |
| GPX2‡ | 4.7 | 6.98 × 10−6 | TOP2A‡ | 4.6 | 2.93 × 10−4 |
| MMP1 | 4.4 | 2.87 × 10−6 | CP | 4.4 | 4.42 × 10−2 |
| AFAP1 | 4.1 | 8.77 × 10−8 | DSG2 | 4.2 | 1.34 × 10−5 |
| SPINK1‡ | 3.8 | 3.24 × 10−4 | SNORA73A | 4.2 | 1.80 × 10−3 |
| AKR1B10 | 3.5 | 1.64 × 10−4 | BIRC5 | 4.0 | 1.21 × 10−3 |
| CST1 | 3.5 | 2.23 × 10−6 | TP63 | 4.0 | 1.09 × 10−2 |
| TOP2A‡ | 3.5 | 9.62 × 10−9 | ANLN‡ | 3.9 | 2.35 × 10−4 |
| AKR1C2‡ | 3.4 | 3.59 × 10−4 | DSP | 3.9 | 3.86 × 10−3 |
| CYP24A1 | 3.4 | 1.75 × 10−4 | OCIAD2 | 3.9 | 2.19 × 10−4 |
| CKMT1A | 3.3 | 3.14 × 10−7 | NQO1 | 3.8 | 3.61 × 10−3 |
| CKMT1A | 3.3 | 3.14 × 10−7 | TPX2 | 3.5 | 5.10 × 10−4 |
| MMP13 | 3.3 | 1.15 × 10−4 | CKS1B | 3.5 | 1.56 × 10−4 |
| ANLN‡ | 3.2 | 1.69 × 10−7 | DSC3 | 3.5 | 1.49 × 10−2 |
| FERMT1 | 3.2 | 2.07 × 10−9 | BAIAP2L1 | 3.4 | 2.47 × 10−5 |
| SLC2A1 | 3.1 | 2.73 × 10−7 | GPX2‡ | 3.4 | 2.70 × 10−2 |
| SLC7A11 | 3.1 | 1.72 × 10−6 | LRIG3 | 3.4 | 1.31 × 10−3 |
| ANKRD22 | 3 | 4.06 × 10−8 | DSG3 | 3.4 | 3.67 × 10−2 |
| Down-regulated (all histologies) | |||||
| FABP4‡ | 0.12 | 4.10 × 10−10 | SFTPC‡ | 0.02 | 2.21 × 10−9 |
| SLC6A4 | 0.16 | 5.56 × 10−8 | HBB | 0.05 | 1.62 × 10−8 |
| AGER | 0.17 | 2.99 × 10−9 | MRC1 | 0.06 | 4.02 × 10−8 |
| AGER | 0.17 | 2.59 × 10−9 | MRC1 | 0.06 | 4.02 × 10−8 |
| AGER | 0.18 | 1.93 × 10−9 | FABP4‡ | 0.07 | 5.11 × 10−8 |
| TMEM100 | 0.19 | 2.07 × 10−9 | PECAM1 | 0.08 | 4.97 × 10−8 |
| GKN2 | 0.20 | 8.62 × 10−7 | GMFG | 0.09 | 6.40 × 10−8 |
| CPB2 | 0.21 | 2.53 × 10−7 | A2M | 0.10 | 5.10 × 10−7 |
| FCN3 | 0.22 | 1.74 × 10−8 | FMO2§ | 0.11 | 5.11 × 10−8 |
| FIGF | 0.22 | 1.08 × 10−6 | LDB2 | 0.11 | 2.21 × 10−9 |
| WIF1 | 0.22 | 1.70 × 10−6 | ABI3BP | 0.11 | 4.02 × 10−8 |
| SFTPC‡ | 0.22 | 3.18 × 10−5 | AQP4 | 0.12 | 4.21 × 10−6 |
| ANKRD1 | 0.23 | 1.24 × 10−7 | CALCRL | 0.12 | 1.14 × 10−6 |
| ADH1B | 0.23 | 3.50 × 10−6 | ANKRD1 | 0.13 | 5.90 × 10−7 |
| RTKN2 | 0.24 | 3.42 × 10−10 | LRRK2 | 0.13 | 1.94 × 10−4 |
| CLDN18 | 0.24 | 8.95 × 10−7 | PTPRB | 0.13 | 5.17 × 10−7 |
| ST8SIA6 | 0.25 | 1.22 × 10−10 | HBA1 | 0.14 | 6.08 × 10−8 |
| FHL1 | 0.25 | 3.20 × 10−9 | HBA1 | 0.14 | 6.08 × 10−8 |
| TCF21 | 0.26 | 1.17 × 10−8 | MSR1 | 0.14 | 1.39 × 10−6 |
| PKHD1L1 | 0.26 | 4.10 × 10−10 | EPAS1 | 0.14 | 4.97 × 10−8 |
Macro: n = 32 T, n = 32 NT, n = 31 pairs. LCM: n = 21 T, n = 20 NT, n = 17 pairs. Repeated gene names indicate redundant probe sets.
Fold change (FC) reflects the ratio of transcript levels in T versus NT alveolar, by microarray.
Benjamini–Hochberg false-discovery rate (FDR)–adjusted significance.
Genes common to both Macro and LCM signatures.
FMO2 for a functional protein is reclassified as FMO4 (flavin containing monooxygenase 4).
Table 2.
Genes Differentially Expressed in Adenocarcinoma (T versus NT) in Macro and LCM Samples
| Macro |
LCM |
||||
|---|---|---|---|---|---|
| Gene | FC∗ | Padjusted† | Gene | FC∗ | Padjusted† |
| Up-regulated (adenocarcinoma) | |||||
| SPP1‡ | 11.4 | 7.76 × 10−9 | SPINK1‡ | 38.3 | 3.25 × 10−3 |
| SPINK1‡ | 6.7 | 2.59 × 10−4 | CP | 15.9 | 4.31 × 10−3 |
| AFAP1‡ | 6.4 | 5.41 × 10−7 | OCIAD2 | 6.9 | 4.64 × 10−6 |
| TMPRSS4 | 5.3 | 1.51 × 10−6 | SNORA73A | 6.8 | 5.97 × 10−4 |
| MUC21 | 4.5 | 2.66 × 10−5 | AGR2‡ | 6.6 | 4.44 × 10−4 |
| MUC21 | 4.4 | 3.02 × 10−5 | GOLM1 | 4.5 | 4.88 × 10−3 |
| MUC21 | 4.2 | 1.94 × 10−5 | LRIG3 | 4.5 | 2.58 × 10−2 |
| MMP12 | 4.2 | 2.41 × 10−4 | SPP1‡ | 4.2 | 4.78 × 10−2 |
| CEACAM5 | 3.8 | 1.65 × 10−3 | NQO1 | 4.2 | 8.22 × 10−3 |
| ANKRD22 | 3.5 | 6.85 × 10−7 | BAIAP2L1 | 3.8 | 6.67 × 10−4 |
| CST1 | 3.5 | 2.28 × 10−4 | EHF | 3.8 | 3.81 × 10−5 |
| MMP1 | 3.4 | 3.13 × 10−3 | FRK | 3.4 | 2.78 × 10−3 |
| CXCL13 | 3.3 | 1.31 × 10−4 | SLC44A4 | 3.4 | 9.27 × 10−4 |
| AGR2‡ | 3.1 | 1.04 × 10−5 | SLC44A4 | 3.4 | 9.27 × 10−4 |
| GREM1 | 3.1 | 6.71 × 10−4 | SLC44A4 | 3.4 | 9.27 × 10−4 |
| COL10A1 | 3.1 | 2.81 × 10−5 | SLC41A2 | 3.4 | 2.14 × 10−3 |
| MUC5B | 3.1 | 2.54 × 10−3 | MUC1 | 3.3 | 8.90 × 10−4 |
| ABCC3 | 3.0 | 2.66 × 10−9 | LOC151009 | 3.3 | 8.78 × 10−3 |
| GCNT3 | 2.9 | 7.86 × 10−6 | AFAP1‡ | 3.3 | 2.63 × 10−2 |
| GLB1L3 | 2.9 | 2.53 × 10−3 | HOOK1 | 3.2 | 4.50 × 10−5 |
| Down-regulated (adenocarcinoma) | |||||
| SLC6A4 | 0.10 | 4.54 × 10−8 | SFTPC | 0.02 | 2.11 × 10−9 |
| FABP4‡ | 0.11 | 1.09 × 10−8 | HBB | 0.05 | 1.89 × 10−6 |
| TMEM100‡ | 0.14 | 1.59 × 10−9 | FABP4‡ | 0.05 | 2.52 × 10−8 |
| ANKRD1‡ | 0.18 | 4.27 × 10−7 | PECAM1 | 0.07 | 1.26 × 10−7 |
| AGER | 0.18 | 8.08 × 10−8 | CAV1 | 0.07 | 2.11 × 10−9 |
| AGER | 0.19 | 5.85 × 10−8 | ANKRD1‡ | 0.07 | 4.14 × 10−9 |
| AGER | 0.19 | 5.06 × 10−8 | MRC1 | 0.07 | 1.47 × 10−4 |
| RTKN2‡ | 0.19 | 5.13 × 10−9 | MRC1 | 0.07 | 1.47 × 10−4 |
| FCN3 | 0.20 | 7.74 × 10−8 | CALCRL | 0.07 | 1.05 × 10−5 |
| GKN2 | 0.21 | 7.97 × 10−5 | LDB2 | 0.09 | 8.70 × 10−9 |
| CPB2 | 0.22 | 2.69 × 10−5 | FMO2 | 0.09 | 2.21 × 10−5 |
| WIF1 | 0.23 | 7.71 × 10−5 | CRYAB | 0.09 | 1.47 × 10−6 |
| NCKAP5 (alias NAP5) | 0.23 | 7.79 × 10−10 | GMFG | 0.10 | 9.69 × 10−5 |
| PKHD1L1 | 0.24 | 4.25 × 10−9 | LPHN2 | 0.10 | 3.66 × 10−6 |
| ACADL | 0.24 | 3.67 × 10−8 | ABI3BP | 0.10 | 2.74 × 10−8 |
| CD36 | 0.25 | 1.80 × 10−6 | RTKN2‡ | 0.10 | 1.18 × 10−6 |
| FIGF | 0.25 | 7.59 × 10−6 | CD36 | 0.10 | 6.40 × 10−4 |
| IGSF10 | 0.25 | 3.29 × 10−9 | TMEM100‡ | 0.11 | 1.25 × 10−7 |
| GPM6A | 0.25 | 4.72 × 10−10 | PTPRB | 0.11 | 1.54 × 10−7 |
| EDNRB | 0.25 | 2.75 × 10−9 | DCN | 0.12 | 1.31 × 10−4 |
Macro: n = 17 T, n = 18 NT, n = 17 pairs. LCM: n = 11 T, n = 10 NT, n = 7 pairs. Repeated gene names indicate redundant probe sets.
Fold change (FC) reflects the ratio of transcript levels in T versus NT alveolar, by microarray.
Benjamini–Hochberg FDR-adjusted significance.
Genes common to both Macro and LCM signatures.
Table 3.
Genes Differentially Expressed in Squamous Cell Carcinoma (T versus NT) in Macro and LCM Samples
| Macro |
LCM |
||||
|---|---|---|---|---|---|
| Gene | FC∗ | Padjusted† | Gene | FC∗ | Padjusted† |
| Up-regulated (squamous cell carcinoma) | |||||
| DSG3‡ | 11.2 | 2.12 × 10−2 | TP63‡ | 12.2 | 2.75 × 10−2 |
| GPX2 | 10.2 | 8.36 × 10−3 | DSG3‡ | 12.0 | 3.46 × 10−2 |
| AKR1C2 | 9.7 | 1.35 × 10−3 | DSC3‡ | 11.0 | 3.01 × 10−2 |
| SERPINB5 | 9.3 | 8.99 × 10−3 | BIRC5 | 8.7 | 2.75 × 10−2 |
| DSC3‡ | 9.0 | 3.09 × 10−2 | CENPF | 8.7 | 3.96 × 10−2 |
| AKR1B10 | 7.9 | 1.83 × 10−2 | TOP2A | 8.6 | 3.39 × 10−2 |
| MMP12 | 7.6 | 2.42 × 10−2 | DSP | 8.5 | 2.64 × 10−2 |
| KRT5‡ | 7.6 | 3.21 × 10−2 | KRT6A‡ | 8.1 | 4.14 × 10−2 |
| KRT6A‡ | 7.2 | 2.42 × 10−2 | KRT5‡ | 7.6 | 3.15 × 10−2 |
| SPRR2A | 6.9 | 4.10 × 10−2 | ANLN | 7.3 | 2.75 × 10−2 |
| KRT6C‡ | 6.4 | 2.48 × 10−2 | KRT6C‡ | 6.6 | 4.14 × 10−2 |
| SPP1 | 6.3 | 3.65 × 10−2 | TPX2 | 6.4 | 3.02 × 10−2 |
| TMPRSS4 | 6.2 | 1.56 × 10−2 | ASPM | 6.3 | 3.13 × 10−2 |
| KRT6B | 6.1 | 2.73 × 10−2 | SNORD26 | 6.1 | 4.86 × 10−2 |
| ADH7 | 6.0 | 2.34 × 10−2 | FAM83B | 6.1 | 2.75 × 10−2 |
| CKMT1A | 5.7 | 8.36 × 10−3 | GPR87 | 5.8 | 3.95 × 10−2 |
| CKMT1A | 5.7 | 8.36 × 10−3 | S100A2 | 5.7 | 4.59 × 10−2 |
| UCHL1 | 5.5 | 8.36 × 10−3 | VSNL1 | 5.6 | 4.19 × 10−2 |
| TP63‡ | 5.3 | 2.60 × 10−2 | CDC6 | 5.6 | 2.71 × 10−2 |
| PTPRZ1 | 5.3 | 1.94 × 10−2 | DAPL1 | 5.6 | 4.12 × 10−2 |
| Down-regulated (squamous cell carcinoma) | |||||
| FABP4 | 0.18 | 3.47 × 10−2 | SFTPC | 0.03 | 3.57 × 10−2 |
| AGER | 0.19 | 3.3 × 10−2 | C4BPA | 0.05 | 7.07 × 10−3 |
| AGER | 0.20 | 3.3 × 10−2 | SLC34A2 | 0.06 | 3.42 × 10−2 |
| AGER | 0.21 | 3.21 × 10−2 | IGJ | 0.06 | 2.92 × 10−2 |
| CLDN18 | 0.23 | 3.65 × 10−2 | HBB | 0.06 | 3.02 × 10−2 |
| VEPH1 | 0.23 | 2.11 × 10−2 | MRC1 | 0.07 | 2.49 × 10−2 |
| TCF21 | 0.23 | 3.05 × 10−2 | MRC1 | 0.07 | 2.49 × 10−2 |
| MAMDC2 | 0.24 | 3.58 × 10−2 | SFTPA2 | 0.07 | 2.16 × 10−2 |
| PEBP4 | 0.25 | 4.44 × 10−2 | SFTPA2 | 0.07 | 2.16 × 10−2 |
| CLIC5 | 0.26 | 2.76 × 10−2 | LRRK2 | 0.08 | 4.20 × 10−2 |
| ST8SIA6 | 0.26 | 1.83 × 10−2 | STEAP4 | 0.08 | 1.78 × 10−2 |
| FHL1 | 0.26 | 3.70 × 10−2 | A2M | 0.08 | 3.03 × 10−2 |
| ITGA8 | 0.27 | 4.07 × 10−2 | CYP2B7P (previously CYP2B7P1) | 0.08 | 1.78 × 10−2 |
| CCDC141 | 0.28 | 2.40 × 10−2 | SFTPB | 0.10 | 2.38 × 10−2 |
| ANGPTL1 | 0.29 | 1.75 × 10−2 | GPR116 | 0.11 | 2.92 × 10−2 |
| BMP5 | 0.29 | 3.01 × 10−2 | GMFG | 0.11 | 2.75 × 10−2 |
| EMCN | 0.29 | 2.86 × 10−2 | SCGB3A2 | 0.11 | 3.16 × 10−2 |
| CHRDL1 | 0.30 | 3.07 × 10−2 | PDK4 | 0.11 | 2.10 × 10−2 |
| NECAB1 | 0.30 | 3.31 × 10−2 | HBA1 | 0.11 | 3.61 × 10−2 |
| HSD17B6 | 0.30 | 3.60 × 10−2 | HBA1 | 0.11 | 3.61 × 10−2 |
Macro: n = 13 T, n = 12 NT, n = 12 pairs. LCM: n = 9 T, n = 8 NT, n = 8 pairs. Repeated gene names indicate redundant probe sets.
Fold change (FC) reflects the ratio of transcript levels in T versus NT alveolar, by microarray.
Benjamini–Hochberg FDR-adjusted significance.
Genes common to both Macro and LCM signatures.
Microarray Analysis from LCM Epithelia
The 20 most up-regulated and the 20 most down-regulated genes for the 41 LCM samples are listed in Tables 1, 2, and 3 (for combined histologies, adenocarcinoma, and squamous cell carcinoma, respectively). Gene expression analysis from LCM samples identified many unique T–NT DE genes (approximately 20% to 33% nonoverlap), compared with Macro specimens (Tables 1, 2, and 3), for both the adenocarcinoma and squamous cell carcinoma histological subgroups (Figures 3 and 4).
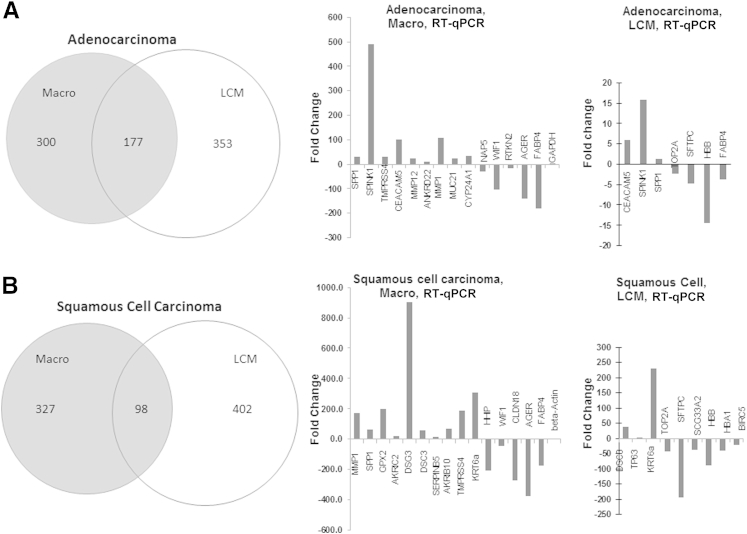
Figure 3.
Validation of adenocarcinoma-specific (A) and squamous cell carcinoma–specific (B) DE transcripts in Macro versus LCM specimens. A: The analysis revealed 177 T versus NT DE transcripts in common between the Macro and LCM platforms (approximately 27%), shown in the Venn diagram. Validation of T versus NT microarray-based differential expression by quantitative real-time RT-PCR (RT-qPCR) in Macro and LCM specimens. PCR-based assays of top microarray hits were evaluated using RNA-specific RT-qPCR on the same adenocarcinoma sample, as described under Materials and Methods, are shown in the graphs. The values for Macro tissue sets were concordant (ie, in the same direction, up-regulated or down-regulated) with LCM microarray values (Table 2) for the selected top microarray hits. Such concordance was also found with RT-qPCR validation, although of course the actual lists of the most dysregulated genes differed between Macro and LCM specimens. B: The analysis revealed 98 T–NT DE transcripts in common (approximately 23%) between the Macro and LCM platforms (shown in Venn diagram). Validation of squamous cell carcinoma T versus NT microarray-based differential expression by RT-qPCR in Macro and LCM specimens is shown in the graphs. Assays of top microarray hits were evaluated using an RNA-specific RT-qPCR on the same sample, as described under Materials and Methods. The values for Macro tissue sets were concordant with LCM microarray values (Table 1, Table 2, Table 3) for the selected top microarray hits. Such concordance was largely true of the LCM RT-qPCR validation for LCM microarray top hits, the two exceptions being TOP2A and BIRC5, which were up-regulated on the microarray but down-regulated in the RT-qPCR. Also, TP63 was up-regulated only 1.6-fold, albeit in the same qualitative direction as the microarray data. Again, the actual lists of the most dysregulated genes differed between Macro and LCM specimens. Data are expressed as mean FC (T versus NT) values, scaled to RNA-specific amplification of a housekeeping gene (GAPDH) (A) or to parallel RNA-specific amplification of a housekeeping transcript (β-actin) (B) not confounded by pseudogenes.
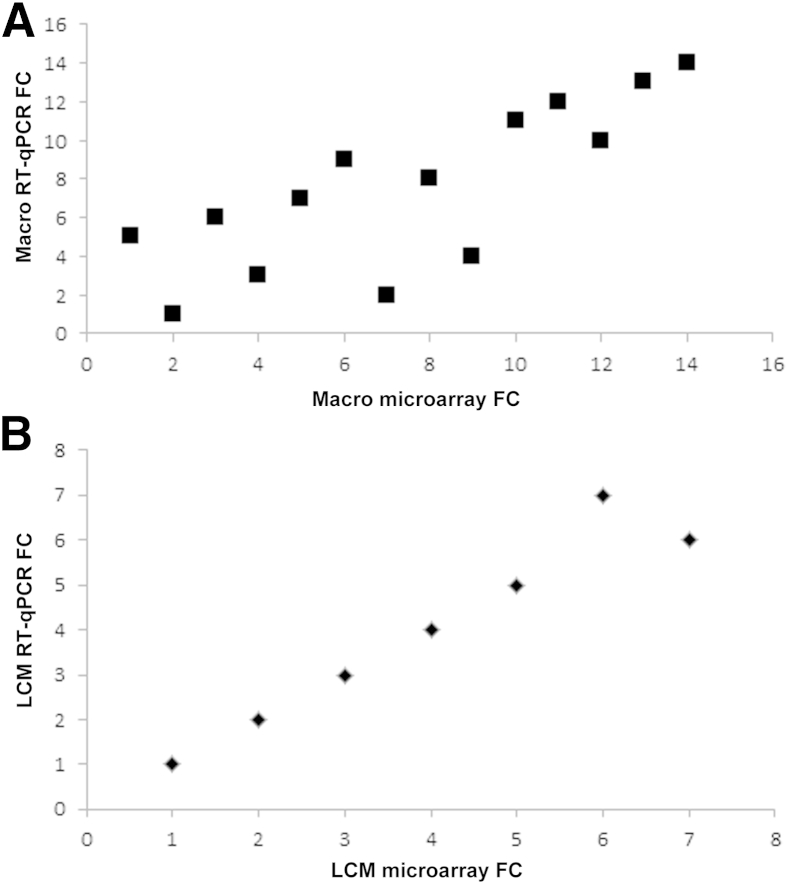
Figure 4.
Correlation of differential gene expression comparing RT-qPCR with expression cDNA microarray for Macro (A) and LCM (B) samples. Spearman correlation indicates a strong relationship between the two gene-expression platforms for both types of tissue (Macro: ρ = 0.789, P = 0.0013; LCM: ρ = 0.964, P = 0.0028). Each point represents one representative gene in replicate queried by one platform in the Macro (A) or LCM (B) tissue setting.
The five most up-regulated genes for the LCM specimens (with preamplification), all tumor histologies combined, were SPINK1, AKR1C2, CENPF, TOP2A, and CP; the five most down-regulated were SFTPC, HBB, MRC1, FABP4, and PECAM1. There was no overlap for the five most up-regulated or down-regulated (and only modest overlap for the full set of 40 dysregulated genes) between the two tissue-sampling platforms (ie, Macro homogenized and LCM) (Table 1). Comparisons of T and NT to the six LCM NTb samples are listed in Supplemental Tables S2 and S3.
The genes most up-regulated genes in LCM, prioritized by FC and for adenocarcinoma and squamous cell carcinoma histologies combined, included CENPF (mechanics of centrosome migration in mitosis), TOP2A (chromatin condensation and mitosis), and DSG2 (desmosomal protein). Genes up-regulated in both LCM and Macro homogenized samples, all three histologies, were SPINK1 (alias TATI; encoding a protease inhibitor), AKR1C2 (encoding an aldo-keto reductase), and ANLN (putatively involved in PI3K/AKT signaling). The genes most up-regulated only in adenocarcinoma LCM specimens (Table 2) included CP (metal-binding function), OCIAD2 (function unknown); SNORA73A (function unknown), GOLM1 (encoding a Golgi apparatus protein), LRIG3 (involved in EGFR signaling); and GPX2 (antioxidant enzyme glutathione peroxidone). Both Macro homogenized and LCM adenocarcinoma specimens exhibited up-regulation of SPINK1, AFAP1 (actin filament associated protein 1), AGR2 (function unknown), and SPP1 (osteopontin; tumor invasion).
Down-regulated genes unique to LCM, for all three histologies combined and prioritized by FC, included MRC1 (involved in DNA replication, S phase checkpoint); AQP4 (water channel involved in tumor progression); PTPRB (protein phosphatase with cancer association); and MSR1 (macrophage tumor microenvironment). For both platforms and both histologies, the down-regulated genes with some literature available were SFTPC (alveolar type II cell marker, involved in adenocarcinoma progression), FABP4 (fatty acid binding; relation to cancer unclear), and ANKRD1 (function unclear; not previously studied in lung cancer). Uniquely in LCM adenocarcinoma specimens, we observed tumor down-regulation in SFTPC, PECAM1 (suspected angiogenesis), CAV1 (scaffolding, links integrins to Ras–ERK), and MRC1. The following genes were down-regulated in both LCM and Macro homogenized NSCLC tumors: FABP4, ANKRD1, CD36 (linked to thrombospondin and angiogenesis), TMEM100 (member of the TGFB superfamily, downstream of BMP/ALK1), and RTKN2 (rhotekin 2; regulates NFκB pathway).
Controlling for Preamplification Bias Inherent to Small Samples, such as the LCM Platform
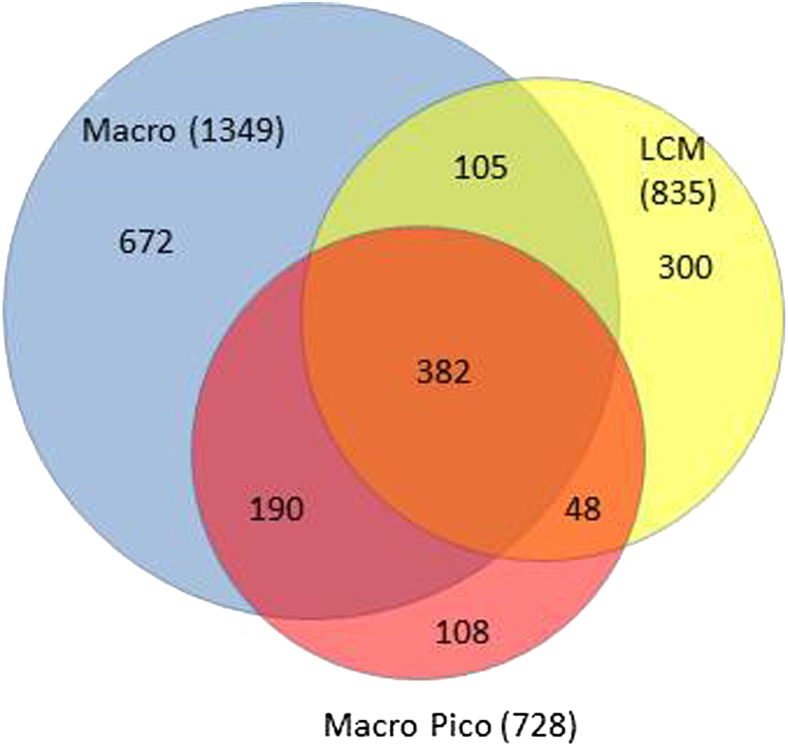
A subgroup of eight representative sample pairs was selected for comparison of tissue-sampling platform versus amplification platform (Figure 5). We identified 1349 DE genes overall for Macro samples and 728 DE genes overall for Macro–Pico control samples. The overlap of 572 DE genes between conventional Macro (without amplification) and Macro–Pico (with amplification) represents 78.6% concordance; as a corollary, there exists 21.4% discordance between these two tissue-sampling platforms that is attributable to the preamplification procedure itself. Similarly, 430/728 (59.1%) of Macro–Pico DE genes were also found in LCM samples (which were also subject to Pico preamplification); this implies that 40.9% of the DE genes are attributable to the LCM histological selection process itself. Thus, the percentage of unique T–NT DE genes attributable to amplification (21.4%) is significantly less than those attributable to LCM cell selection (40.9%, P < 0.001). Direct comparison of the gene lists for the three tissue-sampling platforms identified an overlap of 382 genes that were differentially expressed regardless of sampling or amplification technique (Figure 5), and 672 additional genes were found by Macro sampling only. The full list of 382 genes from these eight samples is available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo, accession number GSE31552).
Figure 5.
Differentially expressed genes for different sample preparation procedures, from a representative subset of eight tumor–nontumor pairs for which all three tissue-sampling platform types were available: macroscopic–conventional homogenized (Macro), macroscopic–conventional homogenized small aliquot undergoing Pico preamplification (Macro–Pico) controls, and LCM samples also undergoing Pico preamplification (LCM). The corresponding gene lists, including overlap lists, are available from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo; accession number GSE31552).
We also noted that the preamplification protocol microarrays (LCM and Macro–Pico) tended, with some exceptions, to be associated with a wider dynamic FC range, whether considering non–microdissected or microdissected material (Supplemental Table S4). LCM–Pico material tended to exhibit the widest dynamic range on these expression microarrays for down-regulated genes. The Macro–Pico data generally indicated that the preamplification was a minor contributor to the range expansion of T–NT differential expression. Rather, cell selection and/or microdissection itself appeared to be the major contributor to the differences in tissue signatures between Macro conventional and LCM samples; this was particularly notable in down-regulated T genes.
RT-qPCR Validation
In general, the RNA-specific RT-qPCR data from 33 selected transcripts (Tables 4 and 5) support the microarray data for the most up-regulated or down-regulated genes, from both the Macro sample sets (13 adenocarcinoma and 11 squamous cell carcinoma) and the LCM sample sets (6 adenocarcinoma and 8 squamous cell carcinoma). The transcripts identified as up-regulated in the microarray were identified as up-regulated by RT-qPCR (Figure 3). Two of the 16 tested gene transcripts in LCM samples (12.5%) were exceptions; in squamous cell carcinoma samples uniquely, both TOP2A and BIRC5 were up-regulated in microarrays, but down-regulated in the RT-qPCR assays. A strong correlation of differential gene expression between RT-qPCR and cDNA microarray data was observed in both Macro and LCM samples (Figure 4). Spearman correlation indicated a strong relationship between the two gene-expression platforms (RT-qPCR and microarray) for both types of tissue sampling (Macro: ρ = 0.789, P = 0.0013; LCM: ρ = 0.964, P = 0.0028, respectively) (Figure 4).
Table 4.
PCR Primers for Confirmation of the Most Dysregulated Genes from Macro Samples
| Primer name | Sequence |
|---|---|
| AGER | Forward: 5′-GATGGTGTGCCCTTGCCCCTTCC-3′ |
| Reverse: 5′-GATCCTCCCACAGAGCCTGCAGTTG-3′ | |
| AKR1B10-UR | Forward: 5′-TGTTCAACTAGGATCAGAATATCACA-3′ |
| AKR1C2-UR | Forward: 5′-TGTGGATGGTGACACAGAGG-3′ |
| ANKRD22 | Forward: 5′-GCTGCCTGAGCTGCTGGAAA-3′ |
| Reverse: 5′-GCTGCTTGGCAGATGGGCTCA-3′ | |
| AQP4l | Forward: 5′-GGAATTTCTGGCCATGCTTA-3′ |
| Reverse: 5′-AGACTTGGCGATGCTGATCT-3′ | |
| β-Actin | Forward: 5′-ACAGAGCCTCGCCTTTGCCG-3′ |
| Reverse: 5′-CATCACGCCCTGGTGCCTGG-3′ | |
| β-Actin UR | Forward: 5′-TATTTTGAATGATGAGCCTTCGT-3′ |
| CEACAM5 | Forward: 5′-GGTGCATCCCCTGGCAGA-3′ |
| Reverse: 5′-GGTTGCCATCCACTCTTTCA-3′ | |
| CYP24A | Forward: 5′-ACCCAGGTGTTGGGATCCAGTGA-3′ |
| Reverse: 5′-AGCTCTGCTAATCGGCGACCA-3′ | |
| CYP24A1 | Forward: 5′-ACCCAAAGGAACAGTGCTCATGCT-3′ |
| Reverse: 5′-TGCGGACAATCCAACAAAGAGCCA-3′ | |
| DSC3-UR | Forward: 5′-GCCAGAATGCTTGGATATGAA-3′ |
| DSG3 | Forward: 5′-CGACCGGGAGGAAACTCCAAGC-3′ |
| Forward-2: 5′-AGCTGCTAGCTTGCTCTCGGTCA-3′ | |
| Reverse: 5′-GTGCCTCAAACTCACTGGTGATGA-3′ | |
| Reverse-2: 5′-TCATCACCAGTGAGTTTGAGGCAC-3′ | |
| FABP4 | Forward: 5′-GAAGTAGGAGTGGGCTTTGCCACC-3′ |
| Reverse: 5′-GCACATGTACCAGGACACCCCC-3′ | |
| GAPDH-U | Forward: 5′-AGCCCCAGCAAGAGCACAA-3′ |
| HHIP | Forward: 5′-GCGGATGAGTTTTGCTTTTA-3′ |
| Reverse: 5′-AGCCATCCCCACTATGC-3′ | |
| KRT6a | Forward: 5′-CCTTCATCGACAAGGTGCGGT-3′ |
| Reverse: 5′-CACCAGGTCCTGCATGCCTCTG-3′ | |
| MMP1 | Forward: 5′-GGGAGATCATCGGGACAACTC-3′ |
| Reverse: 5′-GGGCCTGGTTGAAAAGCAT-3′ | |
| MMP12 | Forward: 5′-TGCTGATGACATACGTGGCA-3′ |
| Reverse: 5′-AGGATTTGGCAAGCGTTGG-3′ | |
| Muc21 | Forward: 5′-CTTCCCATAGTGCATCTACTGC-3′ |
| Reverse: 5′-AGGGACAGGCTGTTTCTCAC-3′ | |
| NAP5 | Forward: 5′-GGGAAGAACCTCAGGAAAAGGCAAT-3′ |
| Reverse: 5′-TGGGGGTTCAAGTAACACTTTTCGC-3′ | |
| RTKN2 | Forward: 5′-GCCGACTAGTTGCCCAGCCA-3′ |
| Reverse: 5′-TGCCCGGATTCTGGTTTCCTTGT-3′ | |
| RTKN2-UR | Forward: 5′-GGAGAAAATACTAATGCTGACACG-3′ |
| SERPINB5-UR | Forward: 5′-GTGTTGCAGGTTCATGGATT-3′ |
| SPINK1 | Forward: 5′-ACTCCCTGGGAAGAGAGGCCAAAT-3′ |
| Reverse: 5′-GGCCTCGCGGTGACCTGATG-3′ | |
| SPP1 | Forward: 5′-ACAGCCAGGACTCCATTGA-3′ |
| Reverse: 5′-TCAGGTCTGCGAAACTTCTTAG-3′ | |
| TMPRSS4 | Forward: 5′-CCGATGTGTTCAACTGGAAG-3′ |
| Reverse: 5′-GAGAAAGTGAGTGGGAACTG-3′ | |
| UR60 | Reverse: 5′-AACGAGACGACGACGGACTTT-3′ |
| WI | Forward: 5′-CAGTGCCTCACAAGGCATCAGTTGT-3′ |
| WIF1 | Reverse: 5′-CTCCATTTCGGCACCCGCCT-3′ |
Universal reverse PCR primer (UR), amplification with the Spivack Laboratory’s RNA-specific RT-qPCR strategy with tagged primer to avoid inadvertent genomic DNA pseudogene amplification.13
Table 5.
Primers for Confirmation of the Most Dysregulated Genes from LCM Samples
| Gene | Primer sequence |
|---|---|
| Adenocarcinoma | |
| CEACAM5 | Forward: 5′-GGTGCATCCCCTGGCAGA-3′ |
| Reverse: 5′-GGTTGCCATCCACTCTTTCA-3′ | |
| FABP4∗ | Forward: 5′-AGAGCATAAGCCAAGGGAC-3′ |
| HBB∗ | Forward: 5′-GCTGCCTATCAGAAAGTGG-3′ |
| SFTPC∗ | Forward: 5′-CCAACGGGAAAGGAAACG-3′ |
| SPINK1 | Forward: 5′-ACTCCCTGGGAAGAGAGGCCAAAT-3′ |
| Reverse: 5′-GGCCTCGCGGTGACCTGATG-3′ | |
| SPP1 | Forward: 5′-ACAGCCAGGACTCCATTGA-3′ |
| Reverse: 5′-TCAGGTCTGCGAAACTTCTTAG-3′ | |
| TOP2A∗ | Forward: 5′-TCTGAGTCTGAATCTCCCAAAG-3′ |
| Squamous cell carcinoma | |
| BIRC5∗ | Forward: 5′-GTTGGAGTGGAGTCTGGGA-3′ |
| HBA1∗ | Forward: 5′-TTTCAGACAGCAGCAGAGCAA-3′ |
| HBB∗ | Forward: 5′-GCTGCCTATCAGAAAGTGG-3′ |
| SCGB3A2∗ | Forward: 5′-GAAGAAACTGCTGGAGGC-3′ |
| SFTPC∗ | Forward: 5′-CCAACGGGAAAGGAAACG-3′ |
| TOP2A∗ | Forward: 5′-TCTGAGTCTGAATCTCCCAAAG-3′ |
| TP63∗ | Forward: 5′-GGGAGCCAGAAGCCAATC-3′ |
Amplified with reverse primer UR60 (Table 4).
LCM NT Alveolar versus NT Bronchial Microarray Comparison
Both the T versus NTa and T versus NTb comparisons, as well as the direct comparison of microdissected NTa with microdissected NTb, revealed marked differences between the two normal lung compartments (Supplemental Tables S2 and S3). This was reflected in the overall comparisons [histologies combined T versus NT (alveolar versus bronchial)] (Supplemental Table S2). Under stratification by tumor histology (such as squamous cell carcinoma compared with NTb, because squamous cell carcinoma is thought to arise from the bronchi) (Supplemental Table S2), marked T versus NT differences in DE persisted, depending on the nature of the comparison tissue (alveolar or bronchial). Direct NTa versus NTb comparisons were also performed (Supplemental Table S3).
Tumor Classification Signature
To evaluate the performance of the respective gene signature for correct classification of the T versus NT status of a given lung tissue, the expression patterns from an initial training set of T–NT pairs (all three histologies) were generated. The best models for Macro homogenized and for LCM samples are listed (Tables 6 and 7, respectively). In the Macro samples, the cross-validation misclassification rate in the best training set (entailing eight genes) was approximately 10%. For the LCM samples, a minimum of nine genes were necessary to attain the lowest misclassification rate (<5%) during initial cross-validation, establishing a training set. Subsequently, the training signatures for LCM samples obtained from 14 donors yielding 20 LCM samples (10 paired T and 10 NT alveolar tissues) were used to predict the T versus NT status of the test set of 20 independent LCM samples (10 T and 10 NT) from a separate set of 13 subjects with clinically matched characteristics. All relevant clinical variables (ie, age, sex, cumulative smoking dose, smoking status, and histology) were statistically similar between training and test sets for both Macro homogenized and LCM specimen donors (P > 0.05 for each variable).
Table 6.
Eight-Gene Signature for Macro Specimens
| Gene | Name or description (Homo sapiens genes) |
|---|---|
| ITGA5 | Integrin, alpha 5 (fibronectin receptor, alpha polypeptide), mRNA |
| FUT2 | Fucosyltransferase 2 (secretor status included), transcript variant 1, mRNA |
| CNRIP1 | Cannabinoid receptor interacting protein 1, transcript variant CRIP1a, mRNA |
| TMEM74B (previously C20orf46) | Transmembrane protein 74B; Transmembrane protein C20orf46 gene:ENSG00000125895 |
| CBR3 | Carbonyl reductase 3, mRNA |
| JAKMIP2 | Janus kinase and microtubule interacting protein 2, mRNA |
| NUPL2 | Nucleoporin like 2, mRNA |
| FRMD3 | FERM domain containing 3, mRNA |
Table 7.
Nine-Gene Signature for LCM Lung Specimens, All Three Histologies Combined
| Gene | Name or description (Homo sapiens genes) |
|---|---|
| DDR2 | Discoidin domain receptor tyrosine kinase 2; Migration-inducing gene 16 protein mRNA, complete cds |
| MMRN2 | Multimerin 2, mRNA |
| C11orf80 | Chromosome 11 open reading frame 80, mRNA |
| SLCO2B1 | Solute carrier organic anion transporter family, member 2B1, transcript variant 2, mRNA |
| RPS20P27 | Ribosomal protein S20 pseudogene 27, mRNA (cDNA clone IMAGE:5549882) |
| OVCH1 | Ovochymase 1, mRNA |
| IRF8 | Interferon regulatory factor 8, mRNA |
| SLC26A2 | Solute carrier family 26 (sulfate transporter), member 2, mRNA |
| ACKR4 (previously CCRL1) | Atypical chemokine receptor 4; chemokine (C-C motif) receptor-like 1 (CCRL1), transcript variant 1, mRNA |
We then tested the performance of the respective signatures in the test set classification (receiver operating characteristics) analyses. The predictive value of an eight-gene Macro homogenized training set signature (Table 6) in identifying tissue as a tumor in the separate test set of Macro homogenized sample pairs was very good (AUC = 0.871). Applied to a separate LCM test set of matched donor sample pairs, the predictive value of the LCM training set-derived nine-gene signature identifying tissue as a tumor (Table 7) was outstanding (AUC = 1.00), suggesting that a microdissected expression microarray signature from clinically matched subjects is more reproducible than that for Macro samples.
We also obtained an eight-gene LCM signature specific to the adenocarcinoma subtype on a training set, by similar methods (Supplemental Table S5). However, there were too few adenocarcinoma LCM cases to validate this discriminant signature in a separate test set.
Comparison with the TCGA Reference Data Set
The TCGA RNA-seq downloaded raw data from batch 144 were analyzed to rank gene transcripts for adenocarcinoma T–NT paired tissue (Macro) from the same donor (Supplemental Table S6). These RNA-seq data were queried for T versus NT discrimination of individual transcripts and were ranked by FC (given Benjamini–Hochberg adjusted P value of <0.05), yielding approximately 7000 significant genes that were tumor discriminant. Some concordance of our microarray-based data with this set of TCGA-defined genes was apparent. Of the most up-regulated genes in the present study (18 Macro and 18 LCM, excluding redundant probe sets), 17 Macro and 16 LCM transcripts exhibited >90% nonzero readings also in the TCGA RNA-seq batch 144 data set; of these, 13 Macro and 10 LCM tumor differential transcripts remained statistically significant (adjusted P < 0.05). Within these 13 and 10 significantly up-regulated overlapping genes (detected by both RNA-seq and cDNA microarray), 5/13 (38.5%) Macro genes and 2/10 (20%) LCM genes also ranked in the top 100 in RNA-seq data. For the most down-regulated genes, 18 Macro and 19 LCM transcripts and 17 Macro and 19 LCM genes exhibited more than 90% nonzero readings and remained statistically significant in TCGA RNA-seq batch 144 data. Within these TCGA significant genes, 14/17 (82.4%) Macro genes and 6/19 (31.6%) LCM genes ranked in the top 100 in RNA-seq data. The concordance in the qualitative direction of FC (up- or down-regulated) between the present data and data from TCGA is further highlighted by the finding that 68/69 such overlapping genes (ie, the most highly differentially expressed genes found also in RNA-seq data and showing more than 90% nonzero readings) were altered in the same direction; the one exception was SLC44A4.
Ingenuity Pathway Analysis of DE Genes
Genes with significant difference in expression level between T and paired NT samples were classified into key pathways, based on their biological role into key pathways, using IPA. In this fashion, we analyzed the entire set of significantly dysregulated genes (adjusted P < 0.05 and either FC > 2.0 for up-regulated genes or FC < 0.5 for down-regulated genes), from LCM and Macro data sets stratified by tumor histology. The most significant pathways represented in the gene signatures established by comparing paired T–NT tissue are listed in Supplemental Table S7. Both LCM and Macro data sets were found to be enriched significantly in genes belonging to similar pathways, although individual genes in each list variously differed or overlapped (Supplemental Table S7).
For Macro homogenized T (both NSCLC histologies) versus NT (alveolar) samples, the cancer disease associations were strong. The component pathways (Benjamini–Hochberg adjusted) included cell movement (adjusted P = 3.55 × 10−9); cell growth and proliferation (adjusted P = 5.22 × 10−8); and cell cycle progression (adjusted P = 1.70 × 10−2) (Supplemental Table S7).
For LCM T (both NSCLC histologies) versus NTa samples, the cancer disease associations were stronger. The component pathways included cell growth and proliferation (adjusted P = 2.20 × 10−17), cell cycle progression (adjusted P = 2.59 × 10−11), and apoptosis (adjusted P = 7.53 × 10−13) (Supplemental Table S7), as well as cell-to-cell signaling and interaction, among others (data not shown). The LCM data set also uniquely revealed many pathways and genes not previously well recognized in lung cancer biology, including immune functions active in allograft rejection, cytotoxic T-lymphocyte–mediated apoptosis of target cells, antigen presentation pathway, hematological system development, and tissue morphology (data not shown). A local cancer network from T–NT DE comparisons is presented for Macro specimens in Supplemental Figure S1A and for LCM specimens in Supplemental Figure S1B. The results differ substantially. Additionally, although recognizable cancer elements and nodes such as cyclin D, CDK, MSH6, eNO1, RAR, RXR, FOS, VDR, and VEGF are apparent, the role of many other elements (eg, immunological function) (Supplemental Figure S2) that exhibit differential expression and network connections have not been well described, particularly not in the lung cancer literature.
Effects of Smoking Status
In exploratory analysis, we found that, among LCM samples, T–NT DE genes varied somewhat across smoking status categories (current, former, never), (Supplemental Table S8). Within the power limits of this pilot exploratory subgroup analysis, where n varied from approximately 8 T–NT pairs for current smokers, to approximately 10 T–NT pairs for former smokers, and approximately 3 T–NT pairs for never smokers, it appears that current and former smokers differ in the vast majority of genes implicated in the tumors. However, these smoking data are unstable; numbers were too low in these smoking strata to discern the true effect of smoking on T-unique expression signatures.
Discussion
Homogenization of Cell Types Mixes Transcriptomes
We have shown that the transcriptomes of morphologically defined, selected lung T and NT cells differ substantially from those of the corresponding homogenized lung tissue sets. This finding holds even with control for the preamplification required to interrogate the transcriptome of these sets of approximately 1000 microdissected cell. A major implication of this finding is that many or most of the published lung cancer–derived transcriptomes likely contain the confounding factor of cellular heterogeneity. Thus, it seems that many previous studies are necessarily nonreplicable, because the cellular admixing differs from sample to sample.14, 15, 16, 17
To determine the effect of cell selection by LCM on transcriptomes, distinct from that of the preamplification step, we performed a separate set of expression microarray experiments. In a common and representative subset of eight T–NT pairs of samples, comparison of LCM with Macro–Pico preamplified samples revealed the T–NT DE differences attributable to the LCM cell selection process itself (approximately two-thirds), rather than to the preamplification process (approximately one third) inherent to small-sample amplification, as measured by differences between Macro (conventional homogenized tissue coupled with a conventional Affymetrix cRNA amplification protocol) to Macro–Pico preamplified samples. Thus, although the comparison of LCM with Macro in the entire sample set entails components of both cell selection–specific and preamplification-specific factors, cell selection appears to be the predominant factor.
Both the most dysregulated LCM sample–derived genes in a given histological stratum (eg, adenocarcinoma) and the composite tumor classifying signatures in the present study are quite different from those reported in the literature. We believe that this is likely attributable to the virtues of cell selection. If the LCM is accurate, one would expect a resemblance to the few microdissected lung cancer transcriptomes procured in the past. Among available studies, the most directly comparable intrasubject T versus NT transcriptome comparisons have been much smaller in scale and have used unmatched T versus NT tissues and/or have used an older microarray platform.1, 2, 3, 18, 19 Our present results are consistent with the findings of Klee et al,1 who evaluated the effect of preamplification in a highly detailed manner and demonstrated that in their study the preamplification step had a relatively modest effect on most components of the T versus NT DE signatures. Consistently, cell selection appeared to be crucial for determining these differential transcriptomes of cancer. Consonant with our lists of the 20 most up-regulated and the 20 most down-regulated genes in adenocarcinomas, they also picked out SPINK1, SPP1, and NQO1, as the most over-expressed genes.1 The consistency and reproducibility of the results in both studies is further reassuring.
Although a larger-scale study of airway transcriptomes has been performed, the epithelial bronchial brushings were limited to nonmalignant bronchial epithelium among lung cancer cases versus noncancer controls, and no transcriptomes from tumors themselves were assessed.20 Also, the LCM cell selection process procures both bronchial and alveolar cells, within the confines of some LCM imprecision, as opposed to the >90% purity of the bronchial brushings. Finally, the preamplification process required for the 103 cells procurable by LCM is not required for bronchial brushings, which typically yield 105 to 106 cells per sample.
Although there has not been a previous definitive study of expression in LCM tissues directly comparing paired T and NT samples, a large study by Selamat et al4 used manually microdissected adenocarcinoma T and adjacent NT tissue, albeit in a highly integrative fashion across clinical and mutational subtypes. Rather than providing pure transcriptome reporting, the authors reported largely integrative genomic-transcriptomic, findings filtered mostly by DNA methylation changes.4 Similarly, although genomic alterations such as copy number variations in adenocarcinomas21 and mutational profiling22, 23, 24 have been reported in macroscopic tissues, few devoted modern-platform transcriptome studies have been conducted,25 and certainly none with the precision conferred by LCM in the present study.
In terms of comparison with larger public consortium databases, we have been unable to find a primary TCGA analysis devoted to T versus NT discriminant expression for LCM; as a rule, there are no consistent LCM T–NT paired specimens interrogated in TCGA. We therefore took the initiative of analyzing a representative RNA-seq data set (batch 144, adenocarcinoma, macroscopic) ourselves (Supplemental Table S8). The TCGA data based on macroscopic T–NT tissue indicated significant overlap of T–NT discriminant RNA-seq–detected transcripts with the microarray-based data on macroscopic tissue, but less of an overlap with the LCM results. For the 20 most up-regulated transcripts, 13 Macro and 10 LCM were significantly overlapping, and of these approximately 5/13 (38.5%) Macro and 2/10 (20%) LCM ranked among the top 100 T–NT discriminant transcripts in RNA-seq data. For the down-regulated transcripts, 14/17 (82.4%) from microarray expression interrogation of Macro samples overlapped with those from comparable RNA-seq TCGA macroscopic samples; this overlap was again less evident for the LCM samples, where 6/19 (31.6%) ranked among the top 100 in TCGA RNA-seq data. These findings highlight the differences between macroscopic and LCM specimen–based signatures, as well as the RNA-seq–based detection inherent to most of the TCGA data. Overall, these macroscopic versus LCM-based differences are based on both the preamplification steps required for LCM-based transcriptome assay and especially on cell selection itself.
Because paired Macro and LMC squamous cell carcinoma samples were available from only six donors, we cannot adequately comment on the comparability of findings to those of the TCGA, but other impressively comprehensive squamous cell carcinoma data are again notably based on homogenized tumor tissue.26, 27 This TCGA effort suggested several expression and somatic alteration subtypes, characterized by 3q24 localization, gene expression (KEAP1, NFE2L2, PTEN, RB1, NF1, CDKN2A expression, which were not apparent in our limited data set and chromosome number instability (as well as mutation or copy number variation which we did not study).
In terms of practicality, LCM is a labor-intensive procedure,28 especially when capturing 1000 cells per lepidic growth pattern, as well as replicate samples per donor-tissue type across many donors. LCM is therefore not likely to be used in routine clinical practice, nor in most research protocols as manually executed. One potential alternative approach is the use of automated microdissection formats28 that might allow more routine use of this precision-enhancing procedure in a variety of settings by permitting procurement of perhaps up to 105 cells by LCM and thus avoiding the need for preamplification (which simply is not feasible manually). It should be noted that some researchers4 have used manual microdissection (microscope plus scalpel) quite successfully and with greater temporal efficiency and throughput for tissue separation, although precision in this setting may not be as good as for LCM.
The dynamic range of the up- and down-regulated genes was compressed in the Macro samples, compared with the LCM samples, particularly for genes down-regulated in the present study (as well as in the most comparable study from the literature1). This finding is not a major surprise, because of admixture dilution of the malignant cell transcriptomes with those of nonmalignant cells in homogenized specimens, but the degree of quantitative difference between Macro homogenized and LCM samples may not be fully accounted for solely by the degree of expected cellular admixture (estimated as two- to threefold). This implies that the preamplification procedure for LCM microarray assays has distorted the FC range for T–NT DE genes. This preamplification effect is evident for some genes, but not for others. Although the qualitative identity of the genes on our LCM lists at Gene Expression Omnibus (http://www.nih.ncbi.nih.gov/geo, accession number GSE31552) is largely accurate, we cannot be certain that the quantitative FCs in the LCM data reflect the precise level of altered regulation, apart from the 33 genes that we have directly validated by RNA-specific RT-qPCR. In a representative set of transcripts, the microarray–RT-qPCR correlation was strong (ρ = 0.789 to 0.964).
The range of FC values differs for RT-qPCR between Macro and LCM adenocarcinoma specimens (Figure 3) for some genes verified in common (SPP, SPINK1, CEACAM, FABP4), and certainly those entailed distinct gene transcripts among the most dysregulated T–NT genes from the same resection samples. In summary, we speculate that these qualitative and quantitative differences within a given histological stratum (eg, adenocarcinoma) depend on cell-selection technique (homogenization versus microdissection), and possibly also on unmeasured differences in surrounding NT tissues.
That the list of T–NT DE genes for the adenocarcinoma transcriptome differs markedly from that for squamous cell carcinoma is not a new finding.29 The present study provides a somewhat more refined list of DE genes in adenocarcinoma. For the most part, the most dysregulated DE genes differed between the two histologies. We compared the FC values of genes commonly detected by RT-qPCR in both squamous cell carcinoma and adenocarcinoma tissues. For Macro samples, six genes detected by RT-qPCR were common to both squamous cell carcinoma and adenocarcinoma. The differential expression of five of these genes had a FC value of <2.5 for the mRNA level in squamous cell carcinoma (FCscc) versus adenocarcinoma (FCad): SPP1 (FCscc/FCad = 64/31.7 = 2.0)30, 31; MMP1 (FCscc/FCad = 170.5/109 = 1.5)32; WIF1 (FCscc/FCad = −46/−102 = 0.45)33; AGER (FCscc/FCad = −378/−149 = 2.5; not previously reported with lung cancer); FABP4 (FCscc/FCad = −175/−181 = 0.96)34; and TMPRSS4 (FCscc/FCad = 186/30.8 = 6.1).35 For LCM samples, three genes were detected in common by RT-qPCR in squamous cell carcinoma and adenocarcinoma: TOP2A (FCscc/FCad = −41/−2.4 = 17.1); SFTCP (FCscc/FCad = −194/−4.8 = 40.4); and HBB (FCscc/FCad = −89/−14.5 = 6.1). These three genes were down-regulated in both squamous cell carcinoma and adenocarcinoma, but substantially more so in squamous cell carcinoma, which is consistent with recent reports.31, 32, 36, 37 We therefore speculate that the qualitative and quantitative differences in gene signatures between adenocarcinoma versus squamous cell carcinomas, within any one tissue-sampling platform (ie, within Macro or LCM sample sets) identify truly different biologies.
In exploratory pilot analysis, the effect of smoking was tentative but suggestive for differential expression (T versus NT) between current and former smokers. This is in agreement with previous reports on smoking-sensitive transcriptomes.20, 38 Although in the present study there was certainly inadequate power to address this issue other than as an exploratory, secondary endpoint, one of the strengths of our study is that the effects of other inherited and acquired donor factors were minimized by use of intraindividual T–NT comparisons.
The transcriptomes of LCM NT tissues of predominantly NTa versus NTb were also notably different. This was certainly not a surprise, for these are anatomically and morphologically different epithelia. Because putative epithelia of origin for adenocarcinoma and squamous cell carcinoma differ, T versus NT DE comparisons indicated different genes up-regulated or down-regulated, depending on the NT epithelium (alveolar or bronchial) chosen.
Genes and Pathways
In the LCM and Macro homogenized data sets, the collection of perturbed pathways included previously implicated pathways, of which the most prominent were cell growth and proliferation, cell movement, cell death, cell cycle progression, and cell-to-cell signaling. The IPA analysis of the LCM data set also uniquely revealed pathways and genes not previously well recognized in lung cancer biology, including immune functions. The identification of such pathways and genes simply emphasizes the principle that the precision of our knowledge pivots on the precision of our techniques, including cell-selection efforts.
Notably, the eight- and nine-gene T–NT discriminant signatures are not made up of the same genes as those on the top hits lists; this is, we speculate, largely because the latter are based on FC, not necessarily on integrated tumor discriminant function in a cohesive statistical sense. As is known from other discovery settings, the genes with the most individually pronounced tumor expression or repression, when placed in combination with other gene transcripts, may not be the most informative for the (cancer) phenotype in question, perhaps because redundancy and colinearity or other factors are sifted out in generating the most informative, multigene cohesive signatures.
Translational Implications
The existence of a gene signature for lung cancer, derived from samples substantially enriched for cancer cells, has translational implications because it allows us to focus on salient gene signatures that could, for example, be used diagnostically in ambiguous cases (eg, fine-needle aspirates of a solitary pulmonary nodule, in cases where cell morphology is unclear) or as an early detection tool, perhaps for targeting measurement of key transcripts or their underlying regulatory features in noninvasive specimens,39, 40, 41 in either clinical or broader population settings. Therapeutically, if verified in animal heterotopic and other tumor models, the most up-regulated genes could be envisioned as potential targets for treatment. Because the lists of genes with greatest differential expression in adenocarcinomas versus squamous cell carcinomas were generally quite robust and distinct, those differences may offer insight into the different pathways underlying the etiologies, morphologies, phenotypic tumor biologies, and therapeutic responses of each histological type.42
Temporal Considerations in Carcinogenesis Studies
The human lung cancer transcriptome likely encompasses genes that are important for tumor maintenance, as well as others that may be important for various carcinogenesis processes leading up to tumor manifestation. Nonetheless, because procurement of human lung tissue cannot generally be performed serially over time, but rather is generally a one-time collection for any given donor, the implications for identification of carcinogenic pathways cannot readily be inferred with certainty from these limited and essentially cross-sectional data. However, hypotheses can be generated. Further explorations of tumors, premalignant lesions, and adjacent NT tissue are likely to offer further clues to etiological pathways.41
Considerable work is involved in microdissection, preamplification, and standardization strategies, and this is the largest lung LCM study reported to date, albeit with sample size constraints. Sample size for the subset of donors providing both LCM and Macro specimens limited analysis (ie, the squamous cell carcinoma and never smoker analyses) of the microdissected tumor transcriptome. For subanalyses, including other factors (smoking dose, NT histology specifics) and interactions (smoking status × histology), a stable estimate of these factors and signature performance was precluded. An additional independent data set would be needed to validate the classifier signature.
Summary
We have shown that cell selection (along with known factors such as tumor histology and amplification strategy) plays a role in even the simplest T–NT comparisons at a transcriptome-wide level. Robustness on replication should be helpful in sifting for valid gene and pathway hits among those initially identified in the present study. For tumor biological inferences, the LCM data reveal both known and previously unrecognized genes and pathways. Although LCM data are somewhat concordant with the TCGA data and with our own data set derived from Macro specimens, the differences are notable. This suggests the need for studies exploring next-generation mRNA sequencing assessment of LCM transcriptomes, at present a technical challenge at these template levels. Additional validation steps should also include exploration of premalignant lesions to allow more direct inferences on human lung carcinogenesis.
Acknowledgments
We thank Katherine Mokhiber, RN, and M. Katherine Fernandez, RN, for excellent human subject study coordination in Albany and Bronx, NY, respectively; Rani Sellers and Tom Harris for services at the Einstein Histopathology core and laser capture microdissection station; David Reynolds (Einstein Genomics Core) for sage advice and processing of the Affymetrix microarrays; Brent Calder and Bin Ye (Einstein Computational Genomics) for initial Affymetrix quality assurance assistance; and Dawn Bowen-Jenkins for excellent secretarial aid.
Footnotes
Supported by NIH grants 1RC1-CA145422-01 (S.D.S.), 1K24-CA139054-01 (S.D.S.), 1R01-CA180126-01 (S.D.S.), and P30-CA013330 (Albert Einstein Cancer Center).
J.L. and G.M. contributed equally to this work.
Disclosures: None declared.
Current address of J.L., Department of Pathology, University of Pittsburgh School of Medicine, Pittsburgh, PA.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2014.06.028.
Supplemental Data
Network of the elements and nodes differentially expressed in Macro (A) and LCM (B) NSCLC T (adenocarcinoma and squamous cell carcinoma histologies combined) versus nontumor (NT) tissue. Recognizable cancer elements and nodes that occur in the Macro specimens (elements common between Macro and LCM specimens are in bold) include Smad, Wnt, Akt, PPARγ, cyclin A/D/E, VEGF, CAV1, KLF4, EPAS1, SOX2, ESP90, HSP7, histone H4, PKA, CDC, Rb, WIF1, DACH1, PEPCK, RAR, RXR, JINK 1/2, and E21. A different list for the LCM specimens is apparent (including cyclin D, CDK, MSH6, eNO1, RAR, RXR, FOS, VDR, VEGF, CCNB1, and ERG), but the role of many other detected elements are not well described, particularly in lung cancer. The networks were generated through the use of Qiagen’s Ingenuity Pathway Analysis (IPA).
Pathways identified by IPA include immunological pathways distinctive for tumor tissue. P values (y axis) are not adjusted for repeated measures. The networks were generated through the use of Qiagen’s Ingenuity Pathway Analysis (IPA).
References
- 1.Klee E.W., Erdogan S., Tillmans L., Kosari F., Sun Z., Wigle D.A., Yang P., Aubry M.C., Vasmatzis G. Impact of sample acquisition and linear amplification on gene expression profiling of lung adenocarcinoma: laser capture micro-dissection cell-sampling versus bulk tissue-sampling. BMC Med Genomics. 2009;2:13. doi: 10.1186/1755-8794-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohrbeck A., Neukirchen J., Rosskopf M., Pardillos G.G., Geddert H., Schwalen A., Gabbert H.E., von Haeseler A., Pitschke G., Schott M., Kronenwett R., Haas R., Rohr U.P. Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers. J Transl Med. 2008;6:69. doi: 10.1186/1479-5876-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi K., Nishioka M., Kohno T., Nakamoto M., Maeshima A., Aoyagi K., Sasaki H., Takenoshita S., Sugimura H., Yokota J. Identification of genes whose expression is upregulated in lung adenocarcinoma cells in comparison with type II alveolar cells and bronchiolar epithelial cells in vivo. Oncogene. 2004;23:3089–3096. doi: 10.1038/sj.onc.1207433. [DOI] [PubMed] [Google Scholar]
- 4.Selamat S.A., Chung B.S., Girard L., Zhang W., Zhang Y., Campan M., Siegmund K.D., Koss M.N., Hagen J.A., Lam W.L., Lam S., Gazdar A.F., Laird-Offringa I.A. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan X.T., Wang T., Xiong S., Kumar S.V., Han W., Spivack S.D. Smoking-related gene expression in laser capture microdissected human lung. Clin Cancer Res. 2009;15:7562–7570. doi: 10.1158/1078-0432.CCR-09-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Travis WD, Colby TV, Corrin B, Shimosato Y, Brambilla E: I Histological Typing of Lung and Pleural Tumors, ed 3. World Health Organization International Histological Classification of Tumours. Geneva: World Health Organization, 1999.
- 7.Travis W.D., Brambilla E., Noguchi M., Nicholson A.G., Geisinger K.R., Yatabe Y. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Travis W.T. Pathology of lung cancer. Clin Chest Med. 2011;32:669–692. doi: 10.1016/j.ccm.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Simone N.L., Bonner R.F., Gillespie J.W., Emmert-Buck M.R., Liotta L.A. Laser capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet. 1998;14:272–276. doi: 10.1016/s0168-9525(98)01489-9. [DOI] [PubMed] [Google Scholar]
- 10.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 11.Hurteau G.J., Spivack S.D. mRNA-specific RT-PCR from human tissue extracts. Anal Biochem. 2002;307:304–315. doi: 10.1016/s0003-2697(02)00058-1. [DOI] [PubMed] [Google Scholar]
- 12.van ’t Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., Schreiber G.J., Kerkhoven R.M., Roberts C., Linsley P.S., Bernards R., Friend S.H. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 13.Hurteau G.J., Spivack S.D. mRNA-specific reverse transcription-polymerase chain reaction from human tissue extracts. Anal Biochem. 2002;307:304–315. doi: 10.1016/s0003-2697(02)00058-1. [DOI] [PubMed] [Google Scholar]
- 14.Van Schaeybroeck S., Allen W.L., Turkington R.C., Johnston P.G. Implementing prognostic and predictive biomarkers in CRC clinical trials. Nat Rev Clin Oncol. 2011;8:222–232. doi: 10.1038/nrclinonc.2011.15. [DOI] [PubMed] [Google Scholar]
- 15.Kim K., Zakharkin S.O., Allison D.B. Expectations, validity, and reality in gene expression profiling. J Clin Epidemiol. 2010;63:950–959. doi: 10.1016/j.jclinepi.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi L., Perkins R.G., Fang H., Tong W. Reproducible and reliable microarray results through quality control: good laboratory proficiency and appropriate data analysis practices are essential. Curr Opin Biotechnol. 2008;19:10–18. doi: 10.1016/j.copbio.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Walker M.S., Hughes T.A. Messenger RNA expression profiling using DNA microarray technology: diagnostic tool, scientific analysis or un-interpretable data? Int J Mol Med. 2008;21:13–17. [PubMed] [Google Scholar]
- 18.Polacek D.C., Passerini A.G., Shi C., Francesco N.M., Manduchi E., Grant G.R., Powell S., Bischof H., Winkler H., Stoeckert C.J., Jr., Davies P.F. Fidelity and enhanced sensitivity of differential transcription profiles following linear amplification of nanogram amounts of endothelial mRNA. Physiol Genomics. 2003;13:147–156. doi: 10.1152/physiolgenomics.00173.2002. [DOI] [PubMed] [Google Scholar]
- 19.MAQC Consortium. Shi L., Reid L.H., Jones W.D., Shippy R., Warrington J.A. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spira A., Beane J.E., Shah V., Steiling K. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 21.Thu K.L., Vucic E.A., Chari R., Zhang W., Lockwood W.W., English J.C., Fu R., Wang P., Feng Z., MacAuley C.E., Gazdar A.F., Lam S., Lam W.L. Lung adenocarcinoma of never smokers and smokers harbor differential regions of genetic alteration and exhibit different levels of genomic instability. PLoS One. 2012;7:e33003. doi: 10.1371/journal.pone.0033003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lockwood W.W., Wilson I.M., Coe B.P., Chari R., Pikor L.A., Thu K.L., Solis L.M., Nunex M.I., Behrens C., Yee J., English J., Murray N., Tsao M.-S., Minna J.D., Gazdar A.F., Wistuba I.I., MacAulay C.E., Lam S., Lam W.L. Divergent genomic and epigenomic landscapes of lung cancer subtypes underscore the selection of different oncogenic pathways during tumor development. PLoS One. 2012;7:e37775. doi: 10.1371/journal.pone.0037775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo J.S., Ju Y.S., Lee W.C., Shin J.Y., Lee J.K., Jung Y.J., Kim J.O., Shin J.Y., Yu S.B., Kim J., Lee E.R., Kang C.H., Park I.K., Rhee H., Lee S.H., Kim J.I., Kang J.H., Kim Y.T. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S.C., Jung Y., Park J., Cho S., Seo C., Kim J., Kim P., Park J., Seo J., Kim J., Park S., Jang I., Kim N., Yang J.O., Lee B., Rho K., Jung Y., Keum J., Lee J., Han J., Kang S., Bae S., Choi S.J., Kim S., Lee J.E., Kim W., Kim J., Lee S. A high-dimensional, deep-sequencing study of lung adenocarcinoma in female never-smokers. PLoS One. 2013;8:e55596. doi: 10.1371/journal.pone.0055596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toh S.H., Prathipati P., Motakis E., Keong K.C., Yenamandra S.P., Kuznetsov V.A. A robust tool for discriminative analysis and feature selection in paired samples impacts the identification of the genes essential for reprogramming lung tissue to adenocarcinoma. BMC Genomics. 2011;12(Suppl 3):S24. doi: 10.1186/1471-2164-12-S3-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammerman P.S., Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonergan K.M., Chari R., Coe B.P., Wilson I.M., Tsao M.-S., Ng R.T., MacAulay C., Lam S., Lam W.L. Transcriptome profiles of carcinoma-in-situ and invasive non-small cell lung cancer as revealred by SAGE. PLoS One. 2010;5:e9162. doi: 10.1371/journal.pone.0009162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallagher R.I., Blakely S.R., Liotta L.A., Espina V. Laser capture microdissection: arcturus(XT) infrared capture and UV cutting methods. Methods Mol Biol. 2012;823:157–178. doi: 10.1007/978-1-60327-216-2_11. [DOI] [PubMed] [Google Scholar]
- 29.Garber M.E., Troyanskaya O.G., Schluens K., Petersen S., Thaesler Z., Pacyna-Gengelbach M., van de Rijn M., Rosen G.D., Perou C.M., Whyte R.I., Altman R.B., Brown P.O., Botstein D., Petersen I. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [Erratum appeared in Proc Natl Acad Sci USA 2002, 99:1098] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isa S., Kawaguchi T., Teramukai S., Minato K., Ohsaki Y., Shibata K., Yonei T., Hayashibara K., Fukushima M., Kawahara M., Furuse K., Mack P.C. Serum osteopontin levels are highly prognostic for survival in advanced non-small cell lung cancer: results from JMTO LC 0004. J Thorac Oncol. 2009;4:1104–1110. doi: 10.1097/JTO.0b013e3181ae2844. [DOI] [PubMed] [Google Scholar]
- 31.Blasberg J.D., Pass H.I., Goparaju C.M., Flores R.M., Lee S., Donington J.S. Reduction of elevated plasma osteopontin levels with resection of non-small-cell lung cancer. J Clin Oncol. 2010;28:936–941. doi: 10.1200/JCO.2009.25.5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauter W., Rosenberger A., Beckmann L., Kropp S., Mittelstrass K., Timofeeva M., Wölke G., Steinwachs A., Scheiner D., Meese E., Sybrecht G., Kronenberg F., Dienemann H., LUCY-Consortium. Chang-Claude J., Illig T., Wichmann H.E., Bickeböller H., Risch A. Matrix metalloproteinase 1 (MMP1) is associated with early-onset lung cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:1127–1135. doi: 10.1158/1055-9965.EPI-07-2840. [DOI] [PubMed] [Google Scholar]
- 33.Rubin E.M., Guo Y., Tu K., Xie J., Zi X., Hoang B.H. Wnt inhibitory factor 1 decreases tumorigenesis and metastasis in osteosarcoma. Mol Cancer Ther. 2010;9:731–741. doi: 10.1158/1535-7163.MCT-09-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G.1, Ye Y., Zheng W., Ma W. [Identification of candidate genes for lung adenocarcinoma using Toppgene] Zhongguo Fei Ai Za Zhi. 2010;13:282–286. doi: 10.3779/j.issn.1009-3419.2010.04.02. Chinese; abstract in English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen T.H., Weber W., Havari E., Connors T., Bagley R.G., McLaren R., Nambiar P.R., Madden S.L., Teicher B.A., Roberts B., Kaplan J., Shankara S. Expression of TMPRSS4 in non-small cell lung cancer and its modulation by hypoxia. Int J Oncol. 2012;41:829–838. doi: 10.3892/ijo.2012.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han Y., Li G., Su C., Ren H., Chu X., Zhao Q., Zhu Y., Wang Z., Hu B., An G., Kang J., Wang W., Yu D., Song X., Xiao N., Li Y., Li X., Yang H., Yu G., Liu Z. Exploratory study on the correlation between 14 lung cancer-related gene expression and specific clinical characteristics of NSCLC patients. Mol Clin Oncol. 2013;1:887–893. doi: 10.3892/mco.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nordgård O., Singh G., Solberg S., Jørgensen L., Halvorsen A.R., Smaaland R., Brustugun O.T., Helland Å. Novel molecular tumor cell markers in regional lymph nodes and blood samples from patients undergoing surgery for non-small cell lung cancer. PLoS One. 2013;8:e62153. doi: 10.1371/journal.pone.0062153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spira A., Beane J., Shah V., Liu G., Schembri F., Yang X., Palma J., Brody J.S. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA. 2004;101:10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boeri M., Verri C., Conte D., Roz L., Modena P., Facchinetti F., Calabrò E., Croce C.M., Pastorino U., Sozzi G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han W., Wang T., Reilly A.A., Keller S., Spivack S.D. Gene promoter methylation assayed in exhaled breath, with differences in smokers and lung cancer patients. Respir Res. 2009;10:86. doi: 10.1186/1465-9921-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belinsky S.A., Liechty K.C., Gentry F.D., Wolf H.J., Rogers J., Vu K., Haney J., Kennedy T.C., Hirsch F.R., Miller Y., Franklin W.A., Herman J.G., Baylin S.B., Bunn P.A., Byers T. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–3344. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 42.Sato M., Shames D.S., Gazdar A.F., Minna J.D. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2:327–343. doi: 10.1097/01.JTO.0000263718.69320.4c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Network of the elements and nodes differentially expressed in Macro (A) and LCM (B) NSCLC T (adenocarcinoma and squamous cell carcinoma histologies combined) versus nontumor (NT) tissue. Recognizable cancer elements and nodes that occur in the Macro specimens (elements common between Macro and LCM specimens are in bold) include Smad, Wnt, Akt, PPARγ, cyclin A/D/E, VEGF, CAV1, KLF4, EPAS1, SOX2, ESP90, HSP7, histone H4, PKA, CDC, Rb, WIF1, DACH1, PEPCK, RAR, RXR, JINK 1/2, and E21. A different list for the LCM specimens is apparent (including cyclin D, CDK, MSH6, eNO1, RAR, RXR, FOS, VDR, VEGF, CCNB1, and ERG), but the role of many other detected elements are not well described, particularly in lung cancer. The networks were generated through the use of Qiagen’s Ingenuity Pathway Analysis (IPA).
Pathways identified by IPA include immunological pathways distinctive for tumor tissue. P values (y axis) are not adjusted for repeated measures. The networks were generated through the use of Qiagen’s Ingenuity Pathway Analysis (IPA).