Abstract
Postzygotic single-nucleotide mutations (pSNMs) have been studied in cancer and a few other overgrowth human disorders at whole-genome scale and found to play critical roles. However, in clinically unremarkable individuals, pSNMs have never been identified at whole-genome scale largely due to technical difficulties and lack of matched control tissue samples, and thus the genome-wide characteristics of pSNMs remain unknown. We developed a new Bayesian-based mosaic genotyper and a series of effective error filters, using which we were able to identify 17 SNM sites from ∼80× whole-genome sequencing of peripheral blood DNAs from three clinically unremarkable adults. The pSNMs were thoroughly validated using pyrosequencing, Sanger sequencing of individual cloned fragments, and multiplex ligation-dependent probe amplification. The mutant allele fraction ranged from 5%-31%. We found that C→T and C→A were the predominant types of postzygotic mutations, similar to the somatic mutation profile in tumor tissues. Simulation data showed that the overall mutation rate was an order of magnitude lower than that in cancer. We detected varied allele fractions of the pSNMs among multiple samples obtained from the same individuals, including blood, saliva, hair follicle, buccal mucosa, urine, and semen samples, indicating that pSNMs could affect multiple sources of somatic cells as well as germ cells. Two of the adults have children who were diagnosed with Dravet syndrome. We identified two non-synonymous pSNMs in SCN1A, a causal gene for Dravet syndrome, from these two unrelated adults and found that the mutant alleles were transmitted to their children, highlighting the clinical importance of detecting pSNMs in genetic counseling.
Keywords: single-nucleotide mutation, postzygotic mosaicism, Dravet syndrome, next-generation sequencing, Bayesian model
Introduction
Genomic mosaicism is a biological phenomenon in which genetic alterations occurring during development or aging give rise to two or more cell populations with distinct genome sequences within one individual1,2. The DNA alterations in a fraction of somatic and/or germ cells can occur at different genomic scales, varying from chromosomal abnormalities and copy number variations (CNVs) to small indels and single-nucleotide substitutions1,3. Comparisons of the whole-genome or whole-exome sequencing data from affected vs normal control tissues in the same person have discovered the role of mosaicism in multiple types of cancer4,5,6 as well as several overgrowth disorders including Proteus syndrome7, Ollier disease and Maffucci syndrome8, CLOVES syndrome9, Schimmelpenning syndrome10, Sturge-Weber syndrome11, and several types of brain malformations12,13,14.
In theory every person is a mosaic. Indeed, many sporadic cases of mosaicism have been reported in clinically unremarkable persons15, sometimes parents of children with a genetic disease, highlighting the clinical importance of mosaicism in genetic counseling. Unfortunately, at the whole-genome scale, only relatively large mosaicisms have been identified in clinically unremarkable individuals16. These include structural variations and CNVs by analyses of array comparative genomic hybridization or SNP microarray17,18,19,20 and neuronal somatic retrotransposition events using transposon-specific targeted sequencing21,22.
The mosaicism caused by postzygotic single-nucleotide mutations (pSNMs), on the other hand, have not been identified in clinically unremarkable persons at genome scale. Existing algorithms for identifying pSNMs from whole-genome or targeted resequencing data require a matched control sample, such as JointSNVMix23, Varscan 224, Strelka25, EBCall26, muTect27, Mutascope28, and LoFreq29. As a result, fundamental patterns of the pSNMs in whole genomes of clinically unremarkable individuals remain largely unknown, such as the prevalence, allele fractions, mutation characteristics, tissue variations, and transmissions to offspring. The study of these patterns is the goal of our research, starting with the development of a new detection method based on next-generation sequencing, a Bayesian genotyper, and stringent error filters.
Results
A Bayesian model and error filters for the detection of pSNM
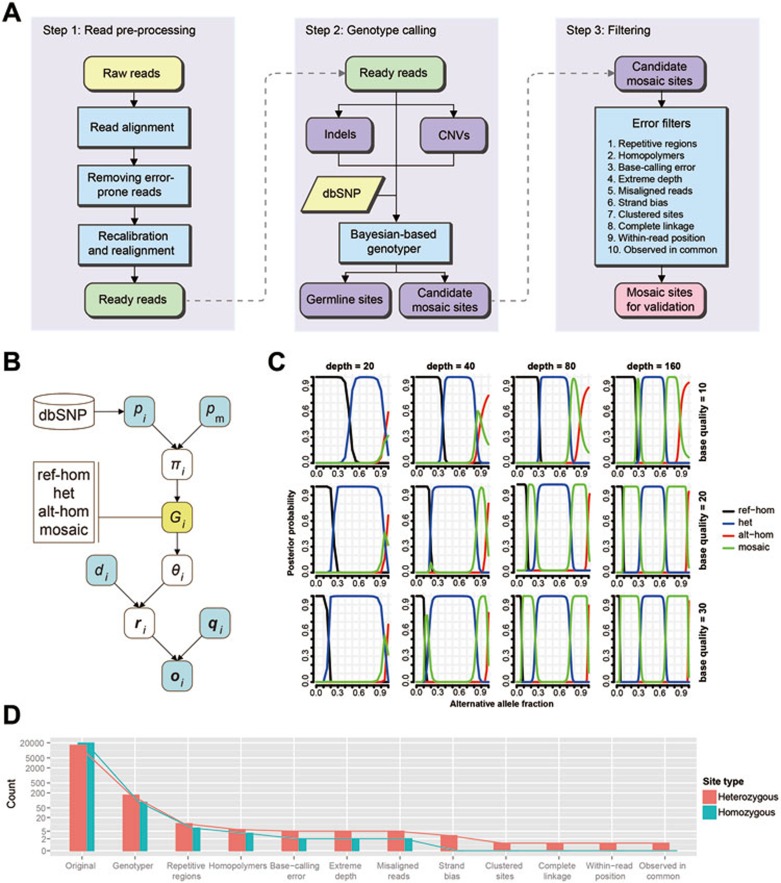
We detected the mosaic sites led by pSNMs and quantified their allele fractions in the peripheral blood of three clinically unremarkable adults using whole-genome sequences produced by Illumina Hiseq platform. Pre-processing of reads was done with standard protocols (see Materials and Methods). As we focused only on pSNMs here, we used CNVnator30 and GATK31 to mask CNVs and indels, respectively. The challenges of detecting pSNMs in non-overgrowth individuals without matched control samples involve distinguishing true mosaic sites from germline heterozygous and homozygous sites, and base-calling and alignment errors32. To address these challenges, we developed a new Bayesian-based genotyper and a series of stringent error filters, summarized in Figure 1A and below and detailed in Materials and Methods.
Figure 1.
A new computational pipeline for genome-wide identification of pSNMs without matched control tissue samples. (A) Overall framework of the pipeline including read pre-processing, genotyping and filtering. The processes of mosaic identification and filtering were implemented in our scripts. (B) The Bayesian-based genotyper demonstrated as a probabilistic graphical model. Four genotypes were defined: ref-hom for “homozygous for the reference allele”, het for “heterozygous”, alt-hom for “homozygous for the alternative allele”, and mosaic for “mosaic”. The posterior probabilities were inferred from prior and conditional probabilities that were calculated or simulated from known population genetics data and next-generation sequencing data (see Materials and Methods). (C) Simulated behavior of the Bayesian genotyper when the sequencing depth and base quality varied. The X axis denotes the alternative allele fraction. The Y axis denotes the posterior probability of the four genotypes. Columns 1 to 4 represent sequencing depths of 20, 40, 80, and 160, respectively, and rows 1 to 3 represent base qualities of 10, 20, and 30, respectively. It showed that increasing sequencing depth could improve the power to distinguish between mosaic and heterozygous sites, whereas increasing base quality could be helpful in distinguishing between mosaic and homozygous sites. (D) The power to distinguish mosaic sites from the simulated ∼20 000 homozygous and ∼20 000 heterozygous sites by sequentially applying the Bayesian genotyper and each of the ten error filters. This result demonstrates the high specificity of our pipeline in excluding germline sites and the relative contribution of the genotyper and filters.
Bayesian probabilistic models are commonly used to distinguish three germline genotypes: homozygous for the reference allele (ref-hom), heterozygous, and homozygous for the alternative allele (alt-hom)33. To distinguish the mosaic sites from germline sites, we introduced a new genotype state, named “mosaic”, into the Bayesian model. Our new model aimed at identifying and measuring the departure of observed allele fractions from germline expectations (0, 0.5 and 1), which we formulated as:
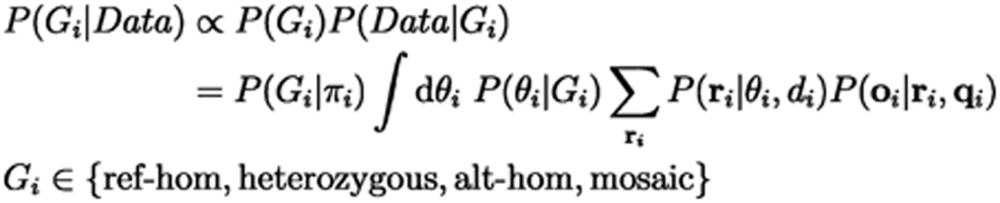 |
This model was able to incorporate known population genetics information and sequencing data characteristics to aid mosaic detection (Figure 1B). Specifically, Data was the bases, base qualities, and total sequencing depth from the aligned sequencing reads at a position. P(Gi) was the prior probability of each genotype, estimated using population genetic data from dbSNP and theoretically estimated somatic mutation rate34. Non-pseudoautosomal regions of sex chromosomes in males were modeled as a haploid. P(Data|Gi) captures sequencing data characteristics such as base-calling errors and read depth biases with a likelihood estimation based on Bernoulli sampling and binomial distribution. Sites where the posterior probability of mosaic genotype was greater than 0.05 were considered candidate sites for the next step. As shown in Figure 1C, our genotyper was able to detect a mosaic site whose alternative allele fraction was around 0.05-0.35 and 0.65-0.95 when the sequencing depth reached 80. Increasing sequencing depth could improve the power to distinguish between mosaic and heterozygous sites, whereas increasing base quality could be helpful in distinguishing between mosaic and homozygous sites.
However, the Bayesian probabilistic model could not remove the large number of false positives caused by systematic errors in read alignment and base calling. We implemented a series of empirical error filters (Table 1). Genomic regions known to cause frequent errors were removed, including repetitive regions and homopolymers. Abnormal patterns of alignment were filtered out, such as extremely high or low read depth, high percentage of misaligned reads, high strand bias of the alternative allele, and skewed alignment position of the alternative allele. Abnormal local patterns were filtered out including clustered sites which were most likely to be within heterochromatin or missed CNV regions (see Materials and Methods), and sites with complete linkage with an adjacent polymorphic site which were most likely due to misalignment of paralogous regions. Finally, we filtered out sites whose allele fractions showed large deviations from germline expectations in two or more individuals because the likelihood of hotspot pSNMs in healthy individuals was presumed to be orders of magnitude smaller than the likelihood of recurrent systematic sequencing bias.
Table 1. Error filters used in the computational pipeline.
| Filter name | Definition |
|---|---|
| Repetitive regions | We rejected nucleotide positions (“sites”) located in annotated repetitive DNA elements and self-alignment regions with similarity score > 80. |
| Homopolymers | We rejected sites located in or near homopolymers which were defined as four or more continuous identical nucleotides, and their flanking regions which were defined as 2 bp from homopolymers shorter than 6 nt or 3 bp from longer homopolymers. |
| Base-calling error | We rejected sites for which the minor allele could be explained by random base-calling errors according to LoFreq29. |
| Extreme depth | We rejected sites with sequencing depth that was either too low (< 25) or too high (> 150), compared to the average sequencing depth of ∼80. |
| Misaligned reads | We rejected sites where > 50% of the reads supporting the major or minor alleles had high risk of being misaligned, defined as when the BWA and BLAT alignments were inconsistent or when the site fell within 15 bp of the start or end of the aligned read or within 5 bp from a gap in the alignment. |
| Strand bias | We rejected sites where the majority of reads supporting the alternative allele were found in only one strand direction. The Fisher's exact test was performed to compare the ratio of the reads supporting the reference and alternative alleles between two strand directions, and sites with a P-value < 0.05 were rejected. |
| Clustered sites | We rejected sites located in or within 20 kb from the genomic regions clustered with three or more sites with minor allele fractions between 10% and 35% and maximal distance between two adjacent sites < 10 kb. |
| Complete linkage | We rejected sites for which one allele showed complete coincidence with an adjacent polymorphic site within the same read-pair. The Fisher's exact test was performed by counting the number of read-pairs supporting the four types of allele combinations, and sites with a P-value < 0.01 and no more than one disagreeing read-pair were filtered. |
| Within-read position | We rejected sites where the majority of sites supporting the alternative alleles were clustered at one end of the reads. The Wilcoxon rank-sum test was performed to compare the positions of the site along the reads between those supporting the reference and alternative alleles, and sites with a P-value < 0.05 were rejected. |
| Observed in common | We rejected sites whose allele fractions showed large deviations from germline expectations in two or more individuals. |
We demonstrated that applying the filters could dramatically decrease the discrepancies between the observed and expected distributions of allele fractions, suggesting that the filters successfully removed the majority of technical artifacts (Supplementary information, Figure S1). To further evaluate the efficacy of the error filters, we generated a benchmark dataset of simulated homozygous and heterozygous sites by in silico mixing of actual Hiseq sequencing reads from two well-genotyped individuals (see Materials and Methods). Figure 1D showed the effectiveness of the filters in removing false positives. Only 1 of 15 842 simulated heterozygous sites and none of 19 624 simulated homozygous sites were misclassified as mosaic.
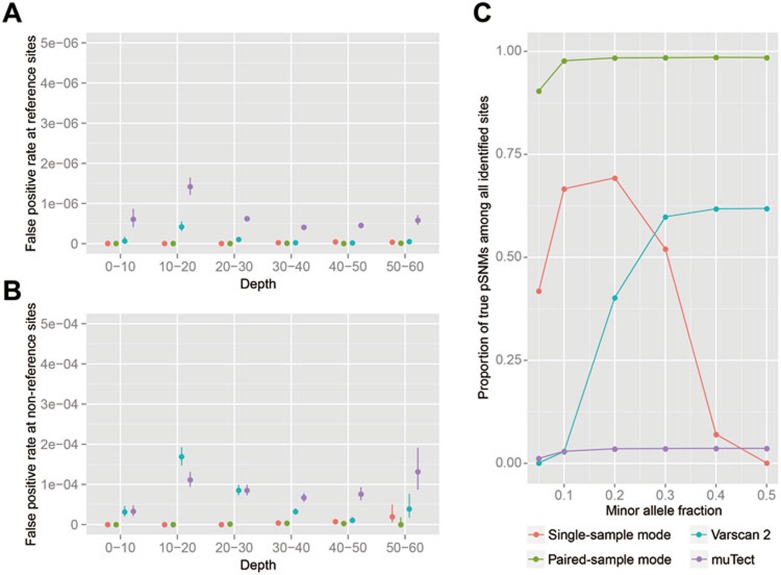
In cases where matched control tissues are available, utilizing data from the matched controls may increase detection accuracy for pSNMs. We implemented a paired-sample mode of our pipeline to utilize sequencing information in control sample (see Materials and Methods). We used two sets of simulation data to evaluate the specificity and precision, and compared the performance of the single-sample mode and paired-sample mode of our pipeline against Varscan 224 and muTect27. Both the single-sample mode and the paired-sample mode of our pipeline achieved higher specificity than Varscan 2 and muTect in true reference sites (Figure 2A) and true non-reference sites (Figure 2B), suggesting that our pipeline can effectively remove false positives. As shown in Figure 2C, without the need of matched control sample, the precision of the single-sample mode of our pipeline was above 50% to identify pSNMs in healthy individuals when the allele fractions were 0.1-0.3. With matched control sample, the paired-sample mode of our pipeline achieved over 90% precision for all the pSNMs whose allele fraction were greater than 0.1, and outperformed both Varscan 2 and muTect (Figure 2C).
Figure 2.
Specificity and precision of identifying pSNMs using our pipeline, Varscan 2, and muTect. (A-B) False positive rate for true reference sites (A) and true non-reference sites (B). Error bars: 95% confidence intervals. (C) Proportion of true pSNMs among all identified sites. The postzygotic mutation rate was set to 4.4 × 10−7 per base, based on estimates in this study. The X axis denotes the minor allele fraction of the pSNM sites.
pSNMs in the peripheral blood of three clinically unremarkable individuals
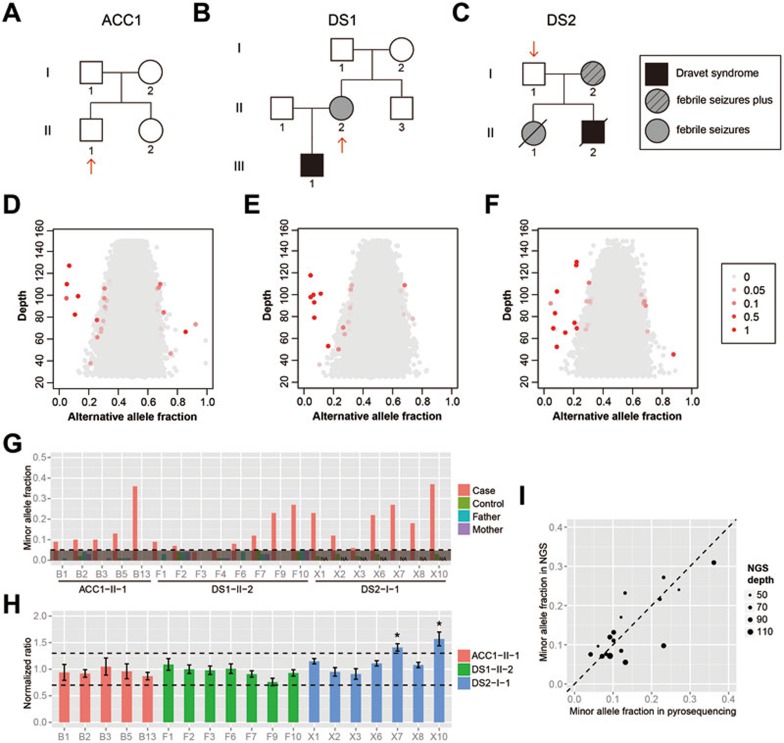
We sequenced the whole genomes of the unamplified peripheral blood DNA samples from three unrelated adults, ACC1-II-1, DS1-II-2 and DS2-I-1, with an average sequencing depth of 76-90× (Supplementary information, Table S1). ACC1-II-1 and DS2-I-1 had no diagnosable symptoms at present and no clinical history. DS1-II-2 had two episodes of mild seizures at age 4-5 but had been subsequently seizure-free with normal cognitive function and no diagnosable symptoms at present. DS1-II-2 and DS2-I-1 were the mother and the father of two unrelated children diagnosed with Dravet syndrome (Figure 3A-3C). Applying our Bayesian model and error filters, we identified 38 candidate pSNMs in the three individuals (Supplementary information, Table S2). These sites showed a distinguishable pattern of allele fractions from the backgrounds of germline polymorphic sites (Figure 3D-3F).
Figure 3.
Identification and validation of pSNMs in the whole-genome sequences from peripheral blood samples of three individuals. (A-C) Pedigree structures of the three participating families. Red arrows point to the three individuals selected for whole-genome sequencing. (D-F) Alternative allele fractions and sequencing depth of the pSNMs identified in the individuals ACC1-II-1 (D), DS1-II-2 (E), and DS2-I-1 (F) using our pipeline. The candidate pSNM sites are shown in red along with the germline sites shown in gray. The sites with extreme depth or allele fraction are not shown. Different shades of red represent mosaic posterior probabilities. (G) Validation by pyrosequencing. The X axis shows the pSNMs identified and validated in the three individuals. pSNM site IDs correspond to Table 2. The Y axis shows the alternative allele fractions of the pSNM sites in the case, unrelated control, and parents, detected by pyrosequencing. The dashed line represents allele fraction of 0.05, which is the detection threshold of pyrosequencing. (H) Copy number abnormalities are ruled out for all but two of the pSNM sites using MLPA. Seventeen sites showed normal copy numbers with normalized signal ratios between 0.7 and 1.3. Two sites with extra DNA copies are marked by asterisks. Error bars represent the SD of three replications of MLPA. (I) Correlation of the minor allele fractions estimated by whole-genome sequencing and pyrosequencing of the validated sites. The sequencing depth is represented by the size of the dots.
Validation of the candidate pSNMs were performed by pyrosequencing, Sanger sequencing of individual cloned fragments, and multiplex ligation-dependent probe amplification (MLPA). The validated pSNMs were listed in Table 2.
Table 2. Validated pSNMs in the three individuals.
| Individual | ID | Position | Ref base | Alt base | Whole-genome sequencing |
Pyrosequencing |
Sanger sequening |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Ref read # (%) | Alt read # (%) | Ref % | Alt % | Ref clone # | Alt clone # | |||||
| ACC1-II-1 | B1 | 15:80878576 | C | T | 118 (93%) | 9 (7%) | 91% | 9% | 41 | 4 |
| ACC1-II-1 | B2 | 4:165099831 | C | T | 73 (89%) | 9 (11%) | 90% | 10% | 105 | 5 |
| ACC1-II-1 | B3 | 6:85605629 | A | T | 86 (87%) | 13 (13%) | 90% | 10% | 75 | 11 |
| ACC1-II-1 | B5 | 6:154692299 | G | C | 104 (95%) | 6 (5%) | 87% | 13% | 91 | 8 |
| ACC1-II-1 | B13 | 18:70512197 | T | G | 67 (69%) | 30 (31%) | 64% | 36% | 29 | 15 |
| DS1-II-2 | F1 | 17:52287119 | G | T | 89 (88%) | 12 (12%) | 91% | 9% | 33 | 3 |
| DS1-II-2 | F2 | 19:55529705 | C | T | 93 (93%) | 7 (7%) | 93% | 7% | 167 | 2 |
| DS1-II-2 | F3 | 19:56343191 | T | C | 86 (92%) | 7 (8%) | 96% | 4% | 82 | 4 |
| DS1-II-2 | F6 | 14:71380614 | T | G | 73 (92%) | 6 (8%) | 92% | 8% | 113 | 3 |
| DS1-II-2 | F7 | 16:84073033 | A | C | 44 (83%) | 9 (17%) | 88% | 12% | 36 | 4 |
| DS1-II-2 | F9 | 13:79932615 | C | A | 51 (73%) | 19 (27%) | 77% | 23% | 25 | 9 |
| DS1-II-2 | F10 | 2:166848782 | G | C | 38 (76%) | 12 (24%) | 73% | 27% | 19 | 3 |
| DS2-I-1 | X1 | 1:72008400 | A | G | 93 (90%) | 10 (10%) | 77% | 23% | 20 | 3 |
| DS2-I-1 | X2 | 16:64312186 | C | T | 76 (92%) | 7 (8%) | 88% | 12% | 24 | 5 |
| DS2-I-1 | X3 | X:68611473 | C | A | 47 (90%) | 5 (10%) | 94% | 6% | 166 | 3 |
| DS2-I-1 | X6 | 2:166854673 | G | T | 58 (78%) | 16 (22%) | 78% | 22% | 30 | 8 |
| DS2-I-1 | X8 | 13:89662020 | A | C | 53 (77%) | 16 (23%) | 82% | 18% | 45 | 9 |
First, eighteen of the 38 sites were confirmed by pyrosequencing above the allele fraction threshold of 5% which was the detection threshold of the pyrosequencing technology35,36. The alternative alleles were present in the corresponding sample, and absent in the control sample obtained from an unrelated individual (Figure 3G). In addition, two pSNMs whose alternative allele fractions did not exceed 5% by pyrosequencing showed a statistically significant difference between the case and three negative control samples (one-tailed Z-test, P-value < 2.2 × 10−16), and were thus also included in subsequent validation.
Second, for these 20 sites, we further confirmed the presence (not the allele fraction) of the alternative allele by Sanger sequencing of individual clones after TA-cloning the amplicons. The presence of reference and alternative alleles was confirmed for 19 sites with at least two, and in most cases three or more, independent clones (Table 2 and Supplementary information, Table S3).
Third, as CNVnator may have missed some DNA copy number alterations which could cause abnormal allele fraction37, we further performed MLPA on the 19 pSNMs which were validated by both pyrosequencing and clonal Sanger sequencing. DNA copy number gain was detected for two sites in DS2-I-1. These two sites had alternative allele fractions of ∼1/3 in both whole-genome sequencing and pyrosequencing data (Figure 3G), consistent with the expected allele fraction when three copies of DNA were present. The other 17 candidate pSNMs showed normal copy numbers (Figure 3H). These 17 pSNMs were validated by all three technologies and considered bona fide pSNMs.
In summary, we identified 17 pSNMs from the peripheral blood samples of three clinically unremarkable individuals and validated them using pyrosequencing (to validate the presence and fraction of the mosaic mutant allele), clonal Sanger sequencing (to validate the presence of the mosaic mutant allele), and MLPA (to rule out copy number alterations). Because the vast majority of genomic positions were not mosaic, some false positives were inevitable despite our stringent pipeline. The current validation rate was 45% (17/38). It could be increased to 70% (14/20) if we increased the Bayesian posterior probability threshold from 0.05 to 0.5.
By counting the allele numbers in the whole-genome sequencing data, we calculated the alternative allele fractions of the validated pSNMs, which range from 5% to 31% (Table 2). The quantification accuracy was justified by the significant correlation between the allele fractions estimated by whole-genome sequencing and pyrosequencing (Pearson's r = 0.79 and P-value = 0.0001, Figure 3I).
For ACC1-II-1 and DS1-II-2 whose parents' blood samples were available, pyrosequencing confirmed the absence of the alternative alleles in their parents (Figure 3G), which suggested postzygotic but not inherited origin of the mutant pSNM alleles. We also tried to assess the candidate pSNMs by direct Sanger sequencing of the PCR products, but found that mosaic could be unequivocally detected in only eight sites (Supplementary information, Table S3), which gave a warning that direct Sanger sequencing had low sensitivity in mosaic detection.
Characteristics of pSNMs in healthy individuals
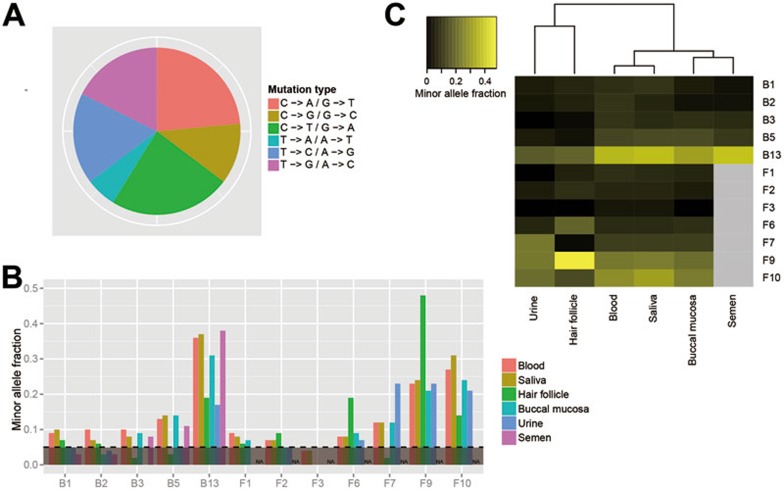
The validated pSNMs enabled us to take a first look at the mutational spectrum of pSNMs in healthy individuals. Because it was unlikely that postzygotic mutations affected both alleles at a single genomic position, the allele generated by postzygotic mutation was expected to be the minor allele in pSNM and, therefore, distinguished from the ancestral allele. Among the validated sites, C→T and C→A were the two most common mutation types at 24% each (Figure 4A), followed by T→G and T→C mutations. These two predominant mutation types were also reported in previous cancer studies4, suggesting possible shared mechanisms of somatic mutation between cancer and non-cancer samples.
Figure 4.
Characteristics of the validated pSNMs. (A) The mutation spectrum of validated pSNMs. The C→T and C→A mutations accounted for half of the mutations identified at the mosaic sites. (B) Allele fractions of the pSNM sites in different samples within the same individuals. Three pSNMs were detected in both somatic and semen samples. (C) Similarity of the allele fractions of the pSNMs between different samples. The blood and saliva samples showed the highest similarities in the six samples.
A question of interest was, “what would be the allele fraction of the pSNMs in other samples collected non-invasively from the same healthy individuals?” In addition to the peripheral blood samples, we were able to collect saliva, hair follicle, buccal mucosa, and urine samples from ACC1-II-1 and DS1-II-2, as well as semen sample from ACC1-II-1. We performed pyrosequencing of the validated pSNMs in these samples. As shown in Figure 4B, for a few pSNMs, the mutant alleles were not detected in some samples, indicating the presence of lineage-specific pSNMs where the postzygotic mutation might have occurred after the differentiation of specific cell lineages. Most of the pSNMs, however, could be detected in multiple samples. Of particular interest, in three of the five pSNMs of ACC1-II-1, the mutant alleles were observed in both the blood and semen samples, suggesting that postzygotic mutations could affect both the somatic and germ cells (Figure 4B).
The inter-sample variations in allele fractions differed widely among the pSNM sites, with the coefficients of variation ranging from 25%-137%. We further demonstrated that the inter-sample variation was not caused by technical variation of DNA extraction and pyrosequencing: we extracted the genomic DNA from the blood sample of ACC1-II-1 three times, and each DNA sample was pyrosequenced three times. Among five mosaic sites, the inter-sample coefficients of variation (25%-137%) was an order of magnitude higher than the average coefficients of variation between different pyrosequencing runs (4.6%) and between different DNA extractions (5.2%) (Supplementary information, Figure S2). Hierarchical clustering of the minor allele fractions in the six types of samples showed that the peripheral blood and saliva samples were the most similar, followed by the buccal mucosa and semen samples (Figure 4C). This finding was consistent with previous reports that 74% of the DNA extracted from saliva samples and 21% from the buccal swab samples were from leukocytes38. In addition, our results showed that the hair follicle and urine samples were the least similar to the other samples, indicating that these samples consisted of cell populations with more distant lineages compared to blood samples.
We showed that our detection pipeline had good specificity and was able to detect the small number of pSNMs among millions of germline polymorphic sites and sequencing errors. To estimate the sensitivity of the detection pipeline and infer the total number of pSNMs in clinically unremarkable individuals, we performed a computer simulation to generate ∼20 000 simulated mosaic sites each at varied minor allele fractions by mixing the real sequencing data (see Materials and Methods). The sensitivity of pSNM detection in non-repetitive genomic regions depended on the alternative allele fraction. The estimated sensitivity ranged from as high as 30% for pSNMs with alternative allele fraction 0.2 to as low as 1% for pSNMs with alternative allele fraction 0.4 (Supplementary information, Figure S3). Because we detected an average of 5.7 validated pSNMs in each individual, we propose that, based on the estimated sensitivity, a clinically unremarkable person might harbor ∼19-570 pSNMs in non-repetitive regions with minor allele fractions from 5%-40%, which corresponded to ∼1.5 × 10−8-4.4 × 10−7 per nucleotide per individual. This was significantly lower than the somatic mutation rate in tumors, where non-silent somatic mutation rate had been estimated to be varied between 1 × 10−7 and 1 × 10−4 across different cancer types, with an average of 4 × 10−64.
Clinical implications of detecting pSNMs
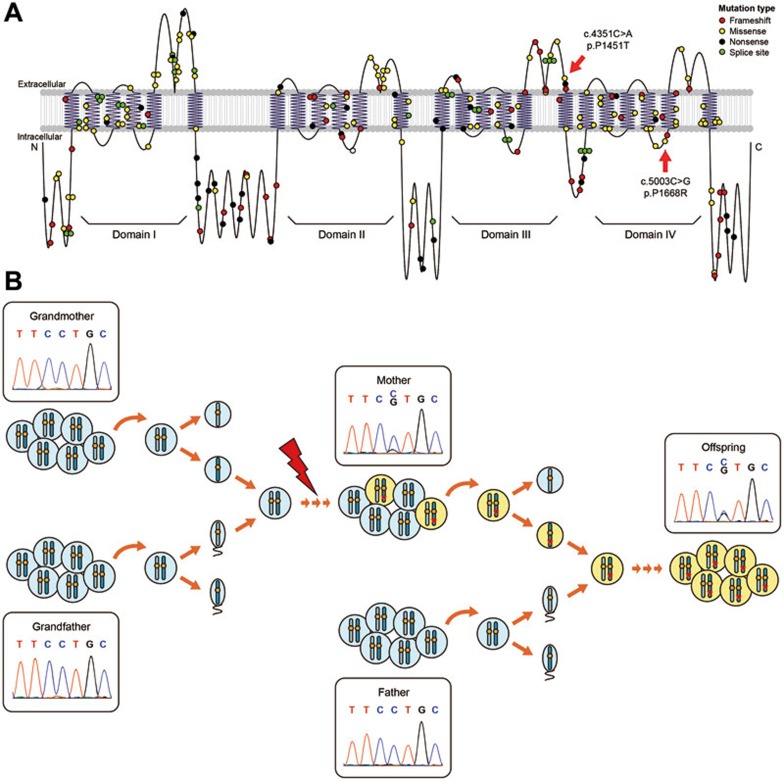
We found a non-synonymous c.5003C→G pSNM with 27% allele fraction and a non-synonymous c.4351C→A pSNM with 22% allele fraction in the SCN1A gene of DS1-II-2 and DS2-I-1, respectively. DS1-II-2 and DS2-I-1 each had a son with Dravet syndrome (DS1-III-1 and DS2-II-2). DS1-III-1 was heterozygous with c.5003C→G, and DS2-II-2 was heterozygous with c.4351C→A, in the SCN1A gene. Dravet syndrome is a rare and catastrophic form of intractable epilepsy that begins in infancy, and SCN1A is well established as the major causal gene for this disease39,40. We ruled out other possible causal mutations by sequencing all the exons of SCN1A and five other rare causal genes including PCDH1941, GABRG242, SCN1B43, GABRA144, and STXBP144 in these two boys. After filtering out the silent and common variants present in dbSNP, only one SCN1A non-synonymous mutation remained in each boy, which was exactly the same alternative allele at exactly the same positions as the two pSNMs identified in their respective parents. Among all the identified pSNMs sites in their parents, these two sites in SCN1A were the only sites where the mutant alleles were inherited. The two non-synonymous sites were located in the third and fourth domains of SCN1A, respectively, adjacent to previously identified pathogenic mutations of Dravet syndrome (Figure 5A). They were predicted to be deleterious by PolyPhen2 (score = 0.793 and 0.679)45, SIFT (score = 0.001 and 0.000)46, and SAPRED (likelihood = 0.878 and 0.867)47. Using both pyrosequencing and Sanger sequencing, we confirmed that the two boys' other parents, DS1-II-1 and DS2-I-2, did not carry the mutant allele, and thus the causal SCN1A variants were inherited from the mosaic parents (Supplementary information, Table S4).
Figure 5.
pSNMs detected in DS1-II-2 and DS2-I-1 in the gene SCN1A and transmitted to their respective child with Dravet syndrome as a heterozygous mutation. (A) The two non-synonymous mutations are highlighted by red arrows on transmembrane structure of the sodium channel encoded by SCN1A. These mutations alter residues located at the ends of the loop structures in domains III and IV, adjacent to previously known pathogenic mutations in Dravet syndrome which are shown here as small circles with different colors representing different mutation type. (B) The parent-to-offspring transmission model is illustrated for c.5003C→G pSNM. In the mother, the mutant allele generated by postzygotic mutations is present in a proportion of the cell population and identified by our pipeline as a pSNM. The mosaicism apparently affected germ cells, and thus the offspring had a chance to inherit the mutation during gametogenesis and fertilization, leading to the heterozygous genotype.
Our results highlighted the importance of accurate detection of pSNMs in genetic counseling. Consistent with previous studies48,49, our results showed that a clinically unremarkable carrier of a deleterious postzygotic mutation might transmit it to offspring and generate a heterozygous genotype that may cause serious genetic diseases (Figure 5B). Considering cell proliferation in embryogenesis, an pSNM with relatively higher mutant allele fraction might be more likely generated by postzygotic mutations at the early developmental stage and, therefore, be more likely shared between somatic and germ cells. As our results showed, simple Sanger sequencing was unable to detect more than half of the pSNMs. Furthermore, applying conventional genotyper GATK31 to our dataset resulted in 35% of the validated pSNMs being mistakenly genotyped as homozygous for the reference allele. Thus, some of the pathogenic genetic mutations currently believed to emerge de novo in affected children might be caused by transmitted parental mosaicism that were missed by Sanger sequencing or conventional genotypers16,50.
Discussion
Postzygotic single-nucleotide mutations had not been previously studied at genome scale in clinically unremarkable individuals, largely due to technical challenges caused by sequencing errors and the lack of matched control tissue. Our Bayesian model and error filters allowed us to detect pSNMs in all three clinically unremarkable individuals and enabled us to take a first look at the characteristics of pSNMs. Many factors may be involved in the generation of mutations, including external mutagens and spontaneous cellular processes51. Interestingly, we observed similar mutational spectra as in cancer samples, but at lower mutation rate. The higher mutation rate in cancer may result from the occurrence of accelerated mutagenesis after the dysfunction of the DNA replication and repair systems that is common in many types of cancer51.
The parent-to-offspring transmission of mutant SCN1A alleles highlighted the clinical implications of genome-wide identification of pSNMs in genetic counseling. In addition, mosaicisms have also previously been reported to cause diseases, often with milder symptoms than homozygous or heterozygous mutations3,52. DS1-II-2, who had a pSNM at SCN1A at allele fraction of 27% in her peripheral blood, had normal cognitive function and no diagnosable symptoms at present, but she had two episodes of mild seizure at four years old. In her whole genome sequence we identified non-synonymous mutations in three other genes that were in the epilepsy gene database CarpeDB (http://www.carpedb.ua.edu/), CASQ2, ALDH7A1, and CACNA1H, which were not present in the individuals of 1000 Genomes Project or dbSNP. Among them the mutation in ALDH7A1 was predicted to be damaging by PolyPhen245, SIFT46, and SAPRED47. It would be interesting to investigate whether this mutation or her pSNM at SCN1A was the cause of her childhood seizure, but that was beyond the scope of this study.
Fetal cells had been found to remain in the circulatory system of some mothers long after birth and vice versa, a phenomenon called feto-maternal mosaicism53. This is unlikely to be the case with the pSNMs that we identified. First, the proportion of extrinsic cells in feto-maternal mosaicism is < 0.5%54, which is at least one order of magnitude lower than what we observed for the validated pSNMs. Second, pyrosequencing of the mothers of ACC1-II-1 and DS1-II-2 found no mutant alleles at the pSNM sites (Figure 3G). Finally, except for one site in SCN1A, the mutant alleles of all the other pSNMs identified in DS1-II-2 were found absent in her offspring. Therefore, the observed pSNMs were not likely to have resulted from feto-maternal mosaicism.
Clonal dominance led by proliferative or selection advantages was observed in peripheral blood cell population55. Although we cannot completely exclude the possibility that the identified pSNMs were subjected to proliferative or selection advantages in blood cells, several lines of evidence did not support this hypothesis. First, except two non-synonymous pSNMs in SCN1A, 15 of the 17 pSNMs were located outside of the exonic regions and not likely to alter gene function. Second, SCN1A gene encodes a subunit of sodium channel which is critical for neuron functionality; however, there is no evidence about its roles in the proliferation of blood cell. Third, all the pSNMs present in blood samples were confirmed in the other non-blood samples from the same individuals, which suggested that the pSNMs are not limited to blood cells.
Single-cell sequencing is another possible approach to study pSNMs. However, because the mutant alleles of pSNMs are often present in only a small subpopulation of cells, a large number of cells would need to be sequenced to identify them. Furthermore, even larger number of cells need to be sequenced to quantify the allele fraction. In addition, the current whole-genome amplification step of single-cell sequencing might introduce unexpected locus or allele dropouts and thus cause false positives56. Thus, bulk sequencing of a population of cells is more effective and less expensive for identifying and quantifying pSNMs.
The short read length and high error rate of next-generation sequencing make it difficult to remove false positive artifacts due to genomic variations or technical errors57. Our pipeline implements a series of filters to reduce the false positives led by such artifacts, which might have potential values in other next-generation sequencing applications. Our current method could identify pSNMs with minor allele fraction of 5%-40% at sequencing depth of ∼80×. In next-generation sequencing, the observed minor allele fractions in two-allele sites are influenced by the random variation of binomial sampling. Thus, increasing sequencing depth could improve the sensitivity to distinguish pSNMs from inherited homozygous and heterozygous sites especially when their allele fractions are close to 0 and 0.5 (Figure 1C). Our simulation demonstrated that increasing the depth to 200× or increasing the base quality to 60 enabled the detection of pSNMs with minor allele fractions as low as 1%-2% (Supplementary information, Figure S4). The Bayesian model of our mosaic genotyper provides the opportunity to integrate more prior knowledge for better detection of mosaic sites, such as the genotyping information of the parents and the site-specific mutation rate which might be correlated to mutational spectrum, mRNA expression and DNA replication4,58. With next-generation sequencing technologies generating longer reads with higher sequencing depth and quality, the sensitivity and specificity of our pipeline will continue to be improved.
Previous cancer studies focused on postzygotic mutations that were restricted to be observed in only one tissue or even one clonal cell population, which might originate during later development and aging51,59. Our findings that most of the pSNMs that we identified were shared in multiple samples suggested the widespread nature of postzygotic mutations during embryogenesis and early development. This highlighted the importance of a control-free method to identify pSNMs. Indeed, when we applied conventional somatic mutation callers, Varscan 2 and muTect, to compare the whole-genome sequencing data of the blood (as case) and saliva (as control) samples of ACC1-II-1, none of the validated pSNMs in blood could be identified.
A recent paper reported that the number of substitution mutations per cell division in mouse small-bowel stem cells was estimated as ∼1.1 using organoid technology60. The pSNMs might contribute to disease risks by either interrupting biological functions of the carriers or transmitting the mutant allele to the offspring1,16,50. The accurate identification of pSNMs will reveal new avenues for studies on the mechanisms and functional consequences of postzygotic mutations and provide new insights into this previously overlooked genetic factor in applications such as finding the “missing heritability” and genetic counseling.
Materials and Methods
Sample collection and DNA processing
This study was officially approved by the Institutional Review Boards of Peking University, and informed consent was obtained from all participants or legal guardians. Blood and other samples were obtained from ACC1-II-1, DS1-II-2, and DS2-I-1 and their families whose pedigree structures were illustrated in Figure 3A-3C. ACC1-II-1 was a healthy adult with no clinical symptoms at present and no clinical history. DS1-II-2 had two episodes of mild seizures between the ages of 4 and 5 years but was subsequently seizure-free with normal cognitive function and no other symptoms. DS2-I-1 was a healthy adult with no clinical symptoms at present and no clinical history. In particular he had no seizures or epilepsy. DS1-II-2 and DS2-I-1 each had a child diagnosed with Dravet syndrome (DS1-III-1 and DS2-II-2). DS2-II-2 suffered sudden unexpected death at five years old. DS2-I-2 had several episodes of FS and DS2-II-1 had FS at an early stage and died of purulent meningitis eight months after birth. The clinical histories of all three families showed no symptoms of cancer or other known overgrowth disorders.
The genomic DNA from peripheral blood lymphocytes was extracted by the QIAamp DNA Blood Maxi Kit (Qiagen, Hilden, Germany) for family ACC1 and by a salting-out procedure61 for family DS1 and DS2. The TIANamp Micro DNA Kit (Tiangen Biotech, Beijing, China) was used to isolate genomic DNA from the hair follicle, buccal mucosa, urine, and semen samples, whereas the genomic DNA of the saliva samples was isolated using the Oragene DNA Kit (OG-500; DNA Genotek, Kanata, Canada), according to the manufacturer's instructions. Each sample of genomic DNA was divided into two parts with one part for whole-genome sequencing and the other part for low-throughput validations.
To screen for pathogenic variations in SCN1A in DS1-III-1 and DS2-II-2, 26 exons were PCR amplified and Sanger sequenced using primers as previously described62. In addition, the canonical exons of five other rare causal genes of Dravet syndrome including PCDH1941, GABRG242, SCN1B43, GABRA144, and STXBP144 were also screened by Sanger sequencing. The exonic variations that were synonymous or present in dbSNP with minor allele fraction ≥ 5% were filtered out. Information about known SCN1A variants associated with Dravet syndrome was extracted from the SCN1A Variant Database63.
Whole-genome sequencing and data analysis
Genomic DNA extracted from the peripheral blood samples of ACC1-II-1, DS1-II-2 and DS2-I-1 was selected for whole-genome sequencing. Sequencing libraries were constructed according to the manufacturer's protocol (Illumina, San Diego, CA, USA), with an average insert size of 400-500 bp. The libraries were sequenced by the Illumina HiSeq2000 platform using 100-bp paired-end reads. The reads were aligned against the GRCh37 human reference genome by BWA (version 0.6.1)64 in a paired-end mode, allowing for a maximum edit distance of four. The duplicate reads were then removed using Picard (http://picard.sourceforge.net/). To exclude ambiguous alignments, the reads flagged as improperly paired or those mapping to multiple positions were filtered out. In addition, we removed the reads with more than three mismatches, which were potentially error-prone in base calling or mapping. The remaining reads were processed by GATK (version 1.6-9)31 for indel realignment and base quality score recalibration, and piled-up by SAMtools65. The average depth of the clean reads was ∼80× for the three peripheral blood samples (Supplementary information, Table S1). To further reduce errors in base calling and alignment, all the bases with base quality or mapping quality less than 20 were excluded from subsequent analyses. CNVs and indels were identified by CNVnator30 and GATK31, respectively. The bin size of CNVnator was set to 100 bp, and the candidate lists of CNVs and indels were further filtered according to the developers' guidelines.
A new Bayesian genotyper for identifying pSNMs
We developed a new Bayesian-based genotyper, illustrated as a probabilistic graphical model in Figure 1B.
Four genotype states were considered in the probabilistic model: ref-hom, heterozygous, alt-hom, and mosaic. For each genomic position i, the genotype Gi was inferred under Bayes' rule as described below:
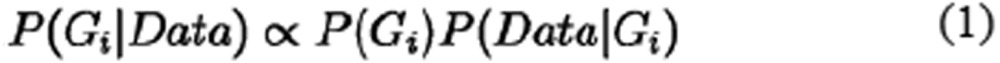 |
where the priors of each genotype, P(Gi), were estimated based on population genetics information, and the sequencing profiles incorporating read depth, allele counts and base qualities were modeled using the likelihood P(Data|Gi).
To generate the genotype priors, Gi was considered as a random variable taken from a multinomial distribution with parameter πi, where πi was determined by the probability of observing a germline alternative allele at a given site, pi, and the probability of a site becoming mosaic by postzygotic mutation, pm. We assumed the Hardy-Weinberg equilibrium in calculating the genotypic prior probabilities of ref-hom, heterozygous and alt-hom according to pi. For the haploid regions, only the ref-hom and alt-hom genotypes were considered, and their prior probabilities were set to pi and 1−pi, respectively. To obtain the prior estimation of pi, the annotations from dbSNP v137 were extracted, and pi was set to be the allele frequency of the corresponding substitution. For the substitutions present in dbSNP that lacked allele frequency, pi was set to be 0.002, because the allele frequencies were estimated from 692 individuals (http://www.ncbi.nlm.nih.gov/projects/SNP/). If a substitution was completely unannotated in dbSNP, we set pi = 1/10 000, a relatively small probability of missing such a polymorphism in the existing data. We further set pm = 10−7, according to the estimated somatic mutation rate34.
Conditional probability distributions in this model were shown as follows:
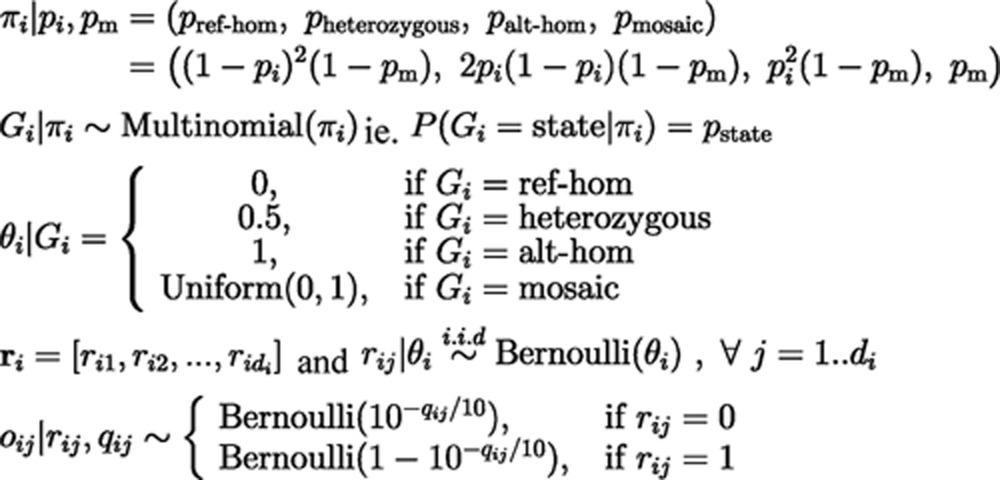 |
At each position, we observed a pile of bases oi with base qualities qi and a total sequencing depth di from the alignments of sequencing data. Because sequencing errors may have occurred, we set ri to be a vector of the real base states, showing whether a base is the reference or alternative, which could not be directly observed. ri was regarded as a series of independent random variables sampled from identical Bernoulli distributions with parameter θi, where θi is a determined variable depending only on the genotype state Gi. Therefore, the calculation of the likelihood P(Data|Gi) could be separated into two parts as follows:
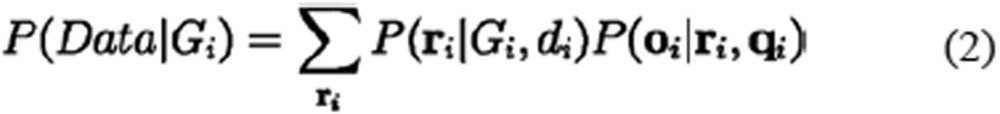 |
Because the relationship between Gi and θi was determined by definition, we expected θi = 0, 0.5, and 1 when Gi is ref-hom, heterozygous and alt-hom, respectively. P(ri|Gi, di) was calculated based on the Bernoulli trial series ri with the success (alternative base state) count ri and probability θi, which can be shown as follows:
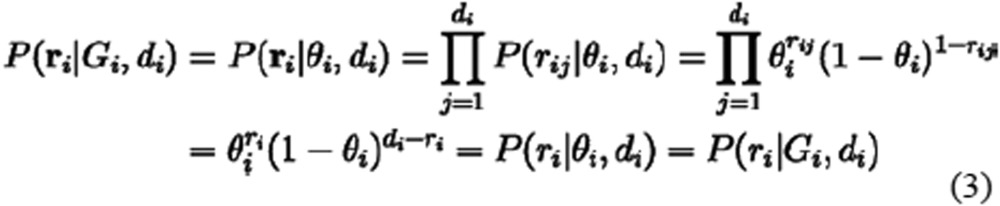 |
where,
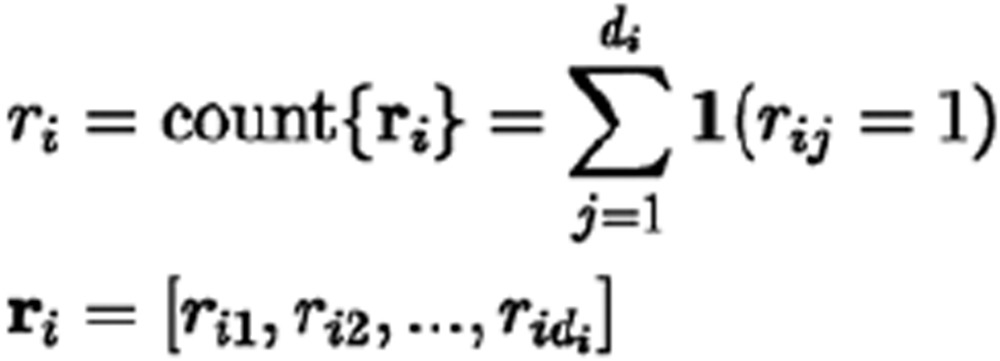 |
Specifically, the parameter θi was considered to be a uniform random variable between 0 and 1 when Gi is mosaic, because we assumed no special distribution of allele fractions in mosaic sites. The corresponding likelihood was computed by a beta function according to equation (4):
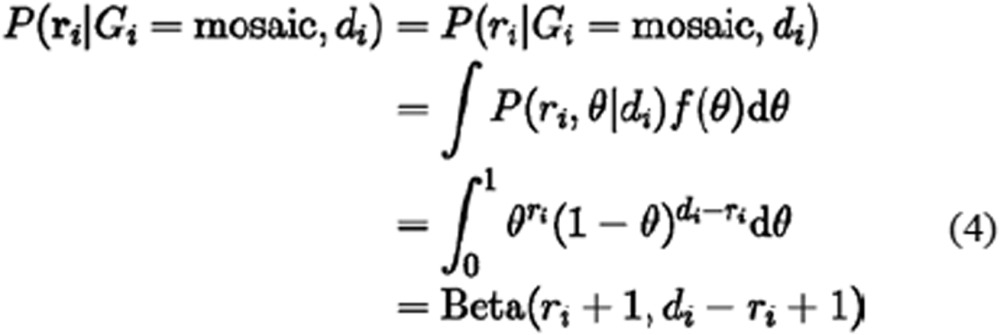 |
where 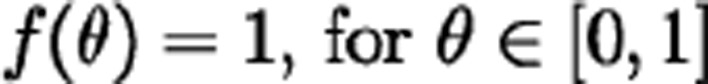 when
when 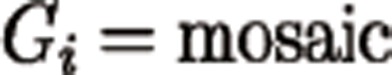
The second part P(oi|ri, qi) would be very easy to calculate if the real states ri were known. However, because ri was unknown, we traversed every possible Boolean vector ri, multiplied it by the corresponding first part P(ri|Gi, di), and added the values to obtain the final likelihood P(Data|Gi). Because P(ri|Gi, di) is constant when the success count ri is fixed, the calculation of P(Data|Gi) in equation (2) can be further simplified as follows:
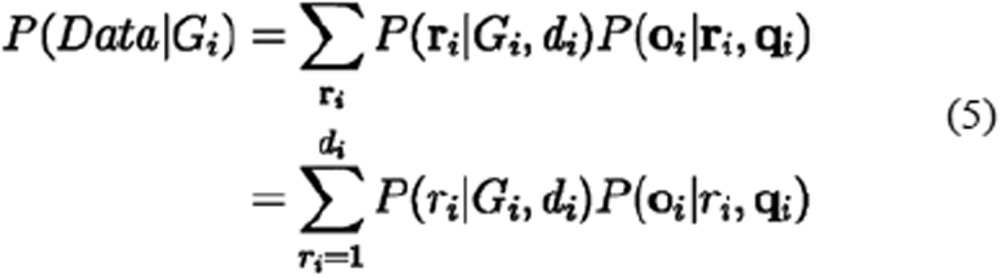 |
where, P(oi|ri, qi) denotes the sum of the probabilities P(oi|ri, qi) for all Boolean vectors ri with the same ri, as shown in equation (6):
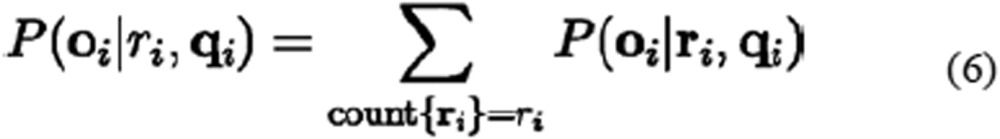 |
To compute P(oi|ri, qi), an iterative algorithm traveling over every base state was used. Each base state rij in the Boolean vector ri was assumed to be independent of the others, with a sequencing error probability perror derived from its Phred-scaled quality score qij. Therefore,
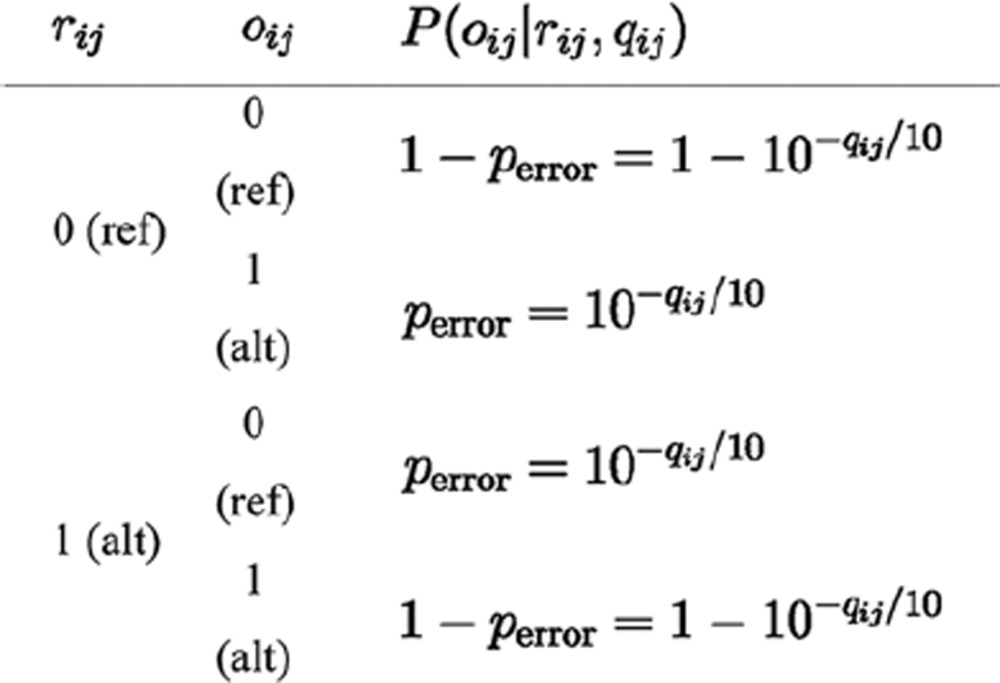 |
First, the initial P(oi1|ri1, qi1) for ri1 = 0 or 1 was set using the corresponding perror or 1-perror, according to the match between the observed and read base states, oi1 and ri1, respectively. Then, the iterative formula was employed according to equation (7) to calculate P(oi|ri, qi) by traversing all di bases:
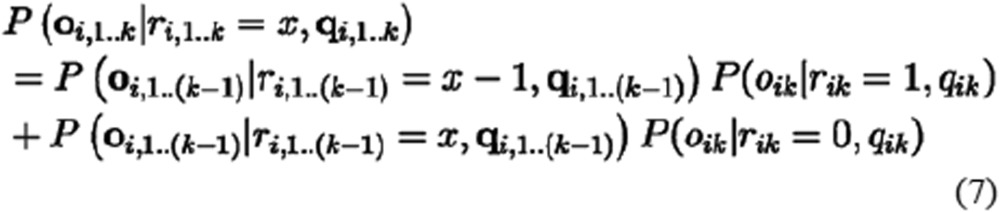 |
where P(oi,1..k|ri,1..k, qi,1..k) is the summed probability for the first k bases, as summarized in the count ri,1..k = x, which can be taken from 0 to k.
As a result, the P(Data|Gi) could be easily calculated according to equation (5), and then P(Gi|Data) for each genotype state was calculated by further multiplying P(Gi) as shown in equation (1) and normalized to set the summed probability equal to 1. The sites with < 3 reads, or < 5% reads supporting the minor allele were skipped for quality control. To achieve a high sensitivity of pSNM detection at this genotyping step, a relatively low threshold (Pmosaic > 0.05) was applied for the posterior probability of the mosaic genotype. As expected, the specificity could be improved when the threshold was increased (Pmosaic > 0.5) (Supplementary information, Table S2). To avoid potential computational underflow, our calculations were generally performed in the log-probability space.
Filtering of candidate pSNMs
There were artifacts caused by systematic errors in sequencing, base calling, and read alignment that the probabilistic model could not remove. We developed a series of error filters and integrated them into the identification pipeline. The descriptions of the filters that we implemented were summarized in Table 1.
First, we excluded the sites that were located near repetitive DNA elements and homopolymers which were known to be prone to errors from existing experimental methods57. The annotations of repetitive regions were downloaded from the UCSC genome browser66, including transposons, microsatellites, simple tandem repeats, interrupted repeats, segmental duplications, self-alignment regions with similarity score > 80, and other repeats masked by RepeatMasker (http://www.repeatmasker.org). Sites within 2 bp from homopolymers of 4-6 nt or within 3 bp from longer homopolymers were also filtered out. Theoretically pSNMs should be scattered, rather than clustered, along the chromosomes. We found that clustered sites were enriched in heterochromatic regions, including the centromeres and telomeres, and regions with copy number alterations (Supplementary information, Figure S5). Thus we filtered out clustered sites with abnormal allele fractions.
Next, we implemented several filters to remove artifacts caused by alignment errors. Reads with discordant alignment between BWA64 and BLAT67 were removed. All contigs in the hg19 assembly were added to the GRCh37 human genome sequence and applied as the reference genome in BLAT to minimize potential misalignment due to the incompleteness of the human reference genome. The sites meeting either of the following criteria were also excluded: (1) predominantly supported by alignment near the ends of reads or near gaps which were known to be error-prone; (2) one allele showing complete co-occurrence with an adjacent polymorphic site within the same sequencing read-pair. To further exclude the reads that were misaligned due to unexpected structural variations, we rejected the sites with significant bias in strand distribution of the reads or sites with skewed within-read position between the reference and alternative alleles. These criteria are known to be efficient for removing misalignment artifacts27,31.
To exclude the artifacts caused by base-calling errors, a statistical test was performed for each site following the algorithm developed in LoFreq29 to distinguish the true alternative allele from sequencing errors, and the sites with P-value > 0.05 after Bonferroni correction were filtered out. The sites with extreme depth (< 25 or > 150, the 10th and 90th percentile among all genomic positions) were also excluded because they were often caused by sequencing gaps, CNVs or alignment errors.
Simulating benchmark datasets to estimate sensitivity and specificity
To evaluate the performance of our pipeline, we generated a benchmark data set of simulated mosaic and polymorphic sites in silico by mixing the whole-genome sequencing data from two individuals (the “in silico mixture dataset”), according to Cibulskis et al.27. We selected two individuals, NA12878 and NA12891, for whom the sequencing depth was similar to our samples. The source of the sequence data and high-quality genotyping files were shown in Supplementary information, Table S5. By comparing the genotypes of the two individuals, we identified positions that were heterozygous in NA12878 and homozygous for the reference allele in NA12891. Because the genders of NA12878 and NA12891 were different, the sites located in X and Y chromosomes were excluded. For each position with enough depth to be sampled, the paired-end reads overlapping with the candidate site were extracted for both individuals. Then some of the NA12891 reads were randomly replaced with the corresponding reads of NA12891 following a binomial sampling with given alternative allele fraction and read depth. We generated ∼20 000 simulated sites for each of seven expected alternative allele fractions, or more specifically, 19 989, 19 986, 19 985, 19 968, 19 883, 19 365, and 16 224 sites with expected allele fractions of 0, 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5, respectively. Supplementary information, Figure S6 showed that the distributions of the simulated polymorphic sites mimicked the distributions of stochastic sampling of real sequenced reads at the polymorphic sites.
Sites with alternative allele fractions 0 and 0.5 were considered homozygous for the reference allele and heterozygous, respectively. Specificity of our pipeline was calculated as the proportion of reference-homozygous and heterozygous sites, respectively, that were correctly rejected as “not mosaic”. Sensitivity was calculated as the proportion of simulated pSNMs correctly identified as “mosaic” for each of the alternative allele fractions ranging from 0.05, 0.1, 0.2, 0.3, to 0.4 in non-repetitive regions.
The performance of our pSNM identification pipeline was compared against two conventional somatic mutation callers, Varscan 224 and muTect27. The latest versions of Varscan 2 (version 2.2.11) and muTect (version 1.1.4) were run under their default parameters, and the candidate lists were filtered following their instructions. Since the sequencing data from the matched control samples were required for both of the tools, we implemented a paired-sample mode of our pipeline: we extracted the candidate sites which were predicted as not homozygous for the reference allele in the case sample (Pref-hom < 0.05) and homozygous for the reference allele in the corresponding control sample (Pref-hom > 0.5) by our Bayesian genotyper, and our error filters were then applied for the candidates in the case sample. The reads of libraries Solexa-18483 and Solexa-18484 of the same individual, NA12878, were treated as the case and control datasets, respectively, following the strategy described in27. All the identified postzygotic mutations were considered as false positives and the false positive rates were reported. The depth-dependent specificities were calculated and subsequently used.
We next estimated the identification precision from paired samples. We used the original NA12891 sequencing data as the control to compare against the in silico mixture data set described above, and calculated sensitivity as the fraction of identified simulated sites in non-repetitive regions. Precision was calculated from this sensitivity and the depth-dependent specificities. The proportions of reference sites and non-reference sites were set based on estimates from previous population-based study67, which were several orders of magnitude larger than the mosaic mutation rate estimated in this work.
In addition, we compared the performance between our pipeline and the pooled-sample model of GATK when the matched control tissue was unavailable. We set the haplotype number to be 20 for practical reasons. Because GATK pooled-sample model only estimated the proportion of haplotypes carrying the alternative allele without reporting the genotype of each position, all the sites with alternative allele proportions differing from 0 and 0.5 were reported as mosaic. Only about 1 out of 10 000 candidate mosaic sites reported by the GATK pooled-sample model were expected to be real, whereas our pipeline achieved order of magnitude higher specificity (Supplementary information, Figure S7). We further showed here that all the false positives in homozygous sites identified by GATK pooled-sample model could be filtered by our stringent filters, which suggested the power of our filters to remove technical artifacts, but a large number of false positives in heterozygous sites still remained even after we combined GATK pooled-sample model with our filters (Supplementary information, Figure S7).
Validation of pSNMs by pyrosequencing
To validate the presence and allele fraction of the candidate pSNMs detected by our pipeline, pyrosequencing was performed on the genomic DNA obtained from all available samples and family members. The PCR and sequencing primers were designed using PyroMark Assay Design (2.0; Qiagen, Venlo, the Netherlands) and listed in Supplementary information, Table S6. The PCR amplification, product processing and pyrosequencing were performed using the PyroMark Q96 ID System (Qiagen) with the corresponding reagents. The raw data were analyzed using the PyroMark Q96 ID Software (Qiagen) for allele quantification. Pyrosequencing has a detection limit of 5% allele fraction35,36, and any sites with an alternative signal < 5% were usually considered technical noise. The differences in allele fraction between different samples within the same individuals were assessed using the Euclidean distance of the minor allele fractions for all the validated sites, and the six samples were further clustered using Ward's method.
Validation of pSNMs by Sanger sequencing of TA clones
To further confirm the presence of the alternative alleles in pSNMs by another independent validation platform, all the pyrosequencing-validated sites were Sanger sequenced in individual clones selected from TA-cloned PCR amplicons. The genomic DNA was amplified using primers flanking these sites (Supplementary information, Table S7) and the PCR products were purified. The amplicons were cloned into the Trans1-T1 phage resistant chemically competent cells using the pEASY-T1 Simple Cloning Kit (Transgen Biotech, Beijing, China). The DNA from the positive colonies was PCR amplified using the M13 universal primers, and then the purified products were sequenced using the Applied Biosystems 3730 DNA Analyzer (Life Technologies, Carlsbad, CA, USA). All the pSNMs were confirmed by the independent validation of at least two reference and mutation calls each by Sanger sequencing. We also sequenced the original PCR amplicons in both directions in the Applied Biosystems 3730 DNA Analyzer (Life Technologies).
MLPA
To rule out potential copy number abnormalities at the candidate pSNM sites, MLPA was performed on the case sample and a reference control sample obtained from an unrelated individual in whom no mosaicism was observed at the corresponding site. A pair of custom synthetic probes was designed for each validated pSNM to target its flanking regions; the distance to the pSNMs varied from 5 to 1 267 bp (Supplementary information, Table S8). The steps of probe preparation, ligation, and PCR amplification were performed using the EK1-FAM Probe Kit and the P300-100R Reference Probemix (MRC-Holland, Amsterdam, the Netherlands), following the manufacturer's protocol. The PCR products were analyzed on the Applied Biosystems 3730 DNA Analyzer (Life Technologies), and the signal processing, normalization and comparison were performed using Coffalyser.NET software (MRC-Holland). Each MLPA experiment on the reference sample was repeated three times. The genomic copy number analysis was reported as normal when the ratio of the normalized peak areas between the case and reference samples was 0.7-1.3, which were the default parameters on Coffalyser.NET.
Data availability
The raw whole-genome sequencing data from this study have been deposited in the Short Reads Archive of NCBI (http://www.ncbi.nlm.nih.gov/sra/) under accession number SRP028833.
We made the scripts which implemented the Bayesian-based mosaic genotyper and error filters publicly available at https://github.com/AugustHuang/MosaicHunter. The users can change the running order and the parameters of running the genotyper and the filters.
Acknowledgments
We are grateful to Drs Manyuan Long, Cheng Li, Jian Lu, and Jinzhu Jia for their valuable comments and suggestions. We thank Ms Sasha Sa for manuscript editing. We thank Xianing Zheng and Zhe Yu for assistance with validation. This work was supported by the National Natural Science Foundation of China (31025014 and 81171221) and the Ministry of Science and Technology of China (2012CB837600). Publication charges were paid by the 111 project of the Ministry of Education of China.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Distribution of the expected (pink) and observed (blue) minor allele fractions of the non-dbSNP heterozygous sites in DS1-II-2.
The minor allele fractions in ACC1-II-1 postzygotic single-nucleotide mutations among biological samples and technical replicates.
The sensitivity of detecting postzygotic single-nucleotide mutations (pSNMs) with varied alternative allele fractions.
The theoretical sensitivity to identify postzygotic single-nucleotide mutations (pSNMs) with varied minor allele fraction, sequencing depth, and base quality.
Examples of clustered sites with abnormal alternative allele fractions.
Distributions of the minor allele fractions of the simulated sites with varied expected allele fractions.
Proportion of true postzygotic single-nucleotide mutations (pSNMs) among all identified sites.
Overview of the whole-genome sequencing data.
The complete list of candidate postzygotic single-nucleotide mutations identified by our computational pipeline.
Sanger sequencing results of TA-clones and direct sequencing for the validated postzygotic single-nucleotide mutations.
Results of pyrosequencing and Sanger sequencing for the two SCN1A variants
The data source for generating simulated sequencing data
The primers of pyrosequencing for validation
The primers of Sanger sequencing for validation
The primers of MLPA for CNV detection
References
- Lupski JR. Genome mosaicism — one human, multiple genomes. Science. 2013;341:358–359. doi: 10.1126/science.1239503. [DOI] [PubMed] [Google Scholar]
- De S. Somatic mosaicism in healthy human tissues. Trends Genet. 2011;27:217–223. doi: 10.1016/j.tig.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:43. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhurst MJ, Sapp JC, Teer JK, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amary MF, Damato S, Halai D, et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet. 2011;43:1262–1265. doi: 10.1038/ng.994. [DOI] [PubMed] [Google Scholar]
- Kurek KC, Luks VL, Ayturk UM, et al. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am J Hum Genet. 2012;90:1108–1115. doi: 10.1016/j.ajhg.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groesser L, Herschberger E, Ruetten A, et al. Postzygotic HRAS and KRAS mutations cause nevus sebaceous and Schimmelpenning syndrome. Nat Genet. 2012;44:783–787. doi: 10.1038/ng.2316. [DOI] [PubMed] [Google Scholar]
- Shirley MD, Tang H, Gallione CJ, et al. Sturge-Weber syndrome and Port-Wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013;368:1971–1979. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière J-B, Mirzaa GM, O'Roak BJ, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74:41–48. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Huynh M, Silhavy JL, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H, Pyeritz RE. Mechanisms and consequences of somatic mosaicism in humans. Nat Rev Genet. 2002;3:748–758. doi: 10.1038/nrg906. [DOI] [PubMed] [Google Scholar]
- Biesecker LG, Spinner NB. A genomic view of mosaicism and human disease. Nat Rev Genet. 2013;14:307–320. doi: 10.1038/nrg3424. [DOI] [PubMed] [Google Scholar]
- Ballif BC, Rorem EA, Sundin K, et al. Detection of low-level mosaicism by array CGH in routine diagnostic specimens. Am J Med Genet A. 2006;140:2757–2767. doi: 10.1002/ajmg.a.31539. [DOI] [PubMed] [Google Scholar]
- Laurie CC, Laurie CA, Rice K, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs KB, Yeager M, Zhou W, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Huallachain M, Karczewski KJ, Weissman SM, Urban AE, Snyder MP. Extensive genetic variation in somatic human tissues. Proc Natl Acad Sci USA. 2012;109:18018–18023. doi: 10.1073/pnas.1213736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie JK, Barnett MW, Upton KR, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Ding J, Morin R, et al. JointSNVMix: A probabilistic model for accurate detection of somatic mutations in normal/tumour paired next generation sequencing data. Bioinformatics. 2012;28:907–913. doi: 10.1093/bioinformatics/bts053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- Shiraishi Y, Sato Y, Chiba K, et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 2013;41:e89. doi: 10.1093/nar/gkt126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost SE, Alakus H, Matsui H, et al. Mutascope: sensitive detection of somatic mutations from deep amplicon sequencing. Bioinformatics. 2013;29:1908–1909. doi: 10.1093/bioinformatics/btt305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm A, Aw PPK, Bertrand D, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Wendl MC, Koboldt DC, Mardis ER. Analysis of next-generation genomic data in cancer: accomplishments and challenges. Hum Mol Genet. 2010;19:R188–R196. doi: 10.1093/hmg/ddq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 2011;12:443–451. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatis AC, Norris-Kirby A, Rich RG, et al. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations. J Mol Diagn. 2010;12:425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querings S, Altmuller J, Ansen S, et al. Benchmarking of mutation diagnostics in clinical lung cancer specimens. PLoS One. 2011;6:e19601. doi: 10.1371/journal.pone.0019601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiede C, Prange-Krex G, Freiberg-Richter J, Bornhäuser M, Ehninger G. Buccal swabs but not mouthwash samples can be used to obtain pretransplant DNA fingerprints from recipients of allogeneic bone marrow transplants. Bone Marrow Transplant. 2000;25:575–577. doi: 10.1038/sj.bmt.1702170. [DOI] [PubMed] [Google Scholar]
- Ohmori I, Ouchida M, Ohtsuka Y, Oka E, Shimizu K. Significant correlation of the SCN1A mutations and severe myoclonic epilepsy in infancy. Biochem Biophys Res Commun. 2002;295:17–23. doi: 10.1016/s0006-291x(02)00617-4. [DOI] [PubMed] [Google Scholar]
- Marini C, Scheffer IE, Nabbout R, et al. The genetics of Dravet syndrome. Epilepsia. 2011;52:24–29. doi: 10.1111/j.1528-1167.2011.02997.x. [DOI] [PubMed] [Google Scholar]
- Depienne C, Bouteiller D, Keren B, et al. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet. 2009;5:e1000381. doi: 10.1371/journal.pgen.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkin LA, Bowser DN, Dibbens LM, et al. Truncation of the GABAA-receptor γ2 subunit in a family with generalized epilepsy with febrile seizures plus. Am J Hum Genet. 2002;70:530–536. doi: 10.1086/338710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino GA, Claes LRF, Lopez-Santiago LF, et al. A functional null mutation of SCN1B in a patient with Dravet syndrome. J Neurosci. 2009;29:10764–10778. doi: 10.1523/JNEUROSCI.2475-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvill GL, Weckhuysen S, McMahon JM, et al. GABRA1 and STXBP1: novel genetic causes of Dravet syndrome. Neurology. 2014;82:1245–1253. doi: 10.1212/WNL.0000000000000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Ye ZQ, Zhao SQ, Gao G, et al. Finding new structural and sequence attributes to predict possible disease association of single amino acid polymorphism (SAP) Bioinformatics. 2007;23:1444–1450. doi: 10.1093/bioinformatics/btm119. [DOI] [PubMed] [Google Scholar]
- Rump A, Hildebrand L, Tzschach A, Ullmann R, Schrock E, Mitter D. A mosaic maternal splice donor mutation in the EHMT1 gene leads to aberrant transcripts and to Kleefstra syndrome in the offspring. Eur J Hum Genet. 2013;21:887–890. doi: 10.1038/ejhg.2012.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriely A, Lord H, Lim J, et al. Germline and somatic mosaicism for FGFR2 mutation in the mother of a child with Crouzon syndrome: implications for genetic testing in “paternal age-effect” syndromes. Am J Med Genet A. 2010;152A:2067–2073. doi: 10.1002/ajmg.a.33513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nat Rev Genet. 2013;14:703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson RP. Somatic gene mutation and human disease other than cancer: An update. Mutat Res. 2010;705:96–106. doi: 10.1016/j.mrrev.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Loubiere LS, Lambert NC, Flinn LJ, et al. Maternal microchimerism in healthy adults in lymphocytes, monocyte/macrophages and NK cells. Lab Invest. 2006;86:1185–1192. doi: 10.1038/labinvest.3700471. [DOI] [PubMed] [Google Scholar]
- Nelson JL, Gillespie KM, Lambert NC, et al. Maternal microchimerism in peripheral blood in type 1 diabetes and pancreatic islet beta cell microchimerism. Proc Natl Acad Sci USA. 2007;104:1637–1642. doi: 10.1073/pnas.0606169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehse B, Roeder I. Insertional mutagenesis and clonal dominance: biological and statistical considerations. Gene Ther. 2008;15:143–153. doi: 10.1038/sj.gt.3303052. [DOI] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- Reumers J, Rijk PD, Zhao H, et al. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nat Biotechnol. 2012;30:61–68. doi: 10.1038/nbt.2053. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Mustonen V, Reva B, et al. Computational approaches to identify functional genetic variants in cancer genomes. Nat Methods. 2013;10:723–729. doi: 10.1038/nmeth.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S, Huch M, Boxtel Rv, et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes Nature 2014. Jun 29. doi: 10.1038/nature13448 [DOI] [PMC free article] [PubMed]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Zhang Y, Liu X, et al. Analysis of SCN1A mutation and parental origin in patients with Dravet syndrome. J Hum Genet. 2010;55:421–427. doi: 10.1038/jhg.2010.39. [DOI] [PubMed] [Google Scholar]
- Claes LR, Deprez L, Suls A, et al. The SCN1A variant database: a novel research and diagnostic tool. Hum Mutat. 2009;30:E904–E920. doi: 10.1002/humu.21083. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreszer TR, Karolchik D, Zweig AS, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;2012:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT — the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TGP. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of the expected (pink) and observed (blue) minor allele fractions of the non-dbSNP heterozygous sites in DS1-II-2.
The minor allele fractions in ACC1-II-1 postzygotic single-nucleotide mutations among biological samples and technical replicates.
The sensitivity of detecting postzygotic single-nucleotide mutations (pSNMs) with varied alternative allele fractions.
The theoretical sensitivity to identify postzygotic single-nucleotide mutations (pSNMs) with varied minor allele fraction, sequencing depth, and base quality.
Examples of clustered sites with abnormal alternative allele fractions.
Distributions of the minor allele fractions of the simulated sites with varied expected allele fractions.
Proportion of true postzygotic single-nucleotide mutations (pSNMs) among all identified sites.
Overview of the whole-genome sequencing data.
The complete list of candidate postzygotic single-nucleotide mutations identified by our computational pipeline.
Sanger sequencing results of TA-clones and direct sequencing for the validated postzygotic single-nucleotide mutations.
Results of pyrosequencing and Sanger sequencing for the two SCN1A variants
The data source for generating simulated sequencing data
The primers of pyrosequencing for validation
The primers of Sanger sequencing for validation
The primers of MLPA for CNV detection
Data Availability Statement
The raw whole-genome sequencing data from this study have been deposited in the Short Reads Archive of NCBI (http://www.ncbi.nlm.nih.gov/sra/) under accession number SRP028833.
We made the scripts which implemented the Bayesian-based mosaic genotyper and error filters publicly available at https://github.com/AugustHuang/MosaicHunter. The users can change the running order and the parameters of running the genotyper and the filters.