Abstract
Morphological and functional changes of cells are important for adapting to environmental changes and associated with continuous regulation of gene expressions. Genes are regulated–in part–by epigenetic mechanisms resulting in alternating patterns of gene expressions throughout life. Epigenetic changes responding to the environmental and intercellular signals can turn on/off specific genes, but do not modify the DNA sequence. Most epigenetic mechanisms are evolutionary conserved in eukaryotic organisms, and several homologs of epigenetic factors are present in plants and animals. Moreover, in vitro studies suggest that the plant cytoplasm is able to induce a nuclear reassembly of the animal cell, whereas others suggest that the ooplasm is able to induce condensation of plant chromatin. Here, we provide an overview of the main epigenetic mechanisms regulating gene expression and discuss fundamental epigenetic mechanisms and factors functioning in both plants and animals. Finally, we hypothesize that animal genome can be reprogrammed by epigenetic factors from the plant protoplast.
Keywords: epigenetic mechanisms, environmental signals, DNA methylation, histone acetylation, gene expression, reprogramming, protoplast
Epigenetics—A Brief History
In recent years, a new research field has emerged known as epigenetics, which studies the factors that influence the function of genes. The term “epigenetics” was suggested by the developmental biologist Waddington in 1942, who used this phrase as a “study of the processes by which genotype gives rise to phenotype” without changes at the level of the gene itself.1 However, the word “epigenetics” was used by Heinemann in the 19th century. The original concept can be traced back to Aristotle,2 who proposed a new theory known as epigenesis, which means to grow upon genesis, the opposite of preformation. Today, according to the most accepted definition, epigenetics is the study of alterations in gene expression without changes in the DNA sequence, hence the name epi-(Greek: επί- over, above, outer) -genetics.3
One example of epigenetic changes in eukaryotic biology is the process of cellular differentiation by which a single totipotent egg cell develops into various pluripotent cell lines of the embryo, which in turn become fully differentiated cells. Epigenetics also examines the role of the environment in gene expression to determine how the environment can influence the expression of genes.4
This review, after describing some of the common epigenetic mechanisms of plant and animal cells, proposes potential epigenetic reprogramming mechanisms between plant and animal cells that have not been discussed in such reviews.
Epigenetic Regulation of the Genome
It has been well-established that DNA is organized by histones and non-histone proteins into chromatin.5 Approximately 146 base pairs of DNA wraps around a complex structure of eight histone proteins (octamer) to form one bead on a chain of bead-like nucleosomes connected by 80-base pair linker-DNA. Histones are responsible for protection of DNA as well as maintaining the shape and structure of a nucleosome. There are five families of histones known to date, including H1/H5, H2A, H2B, H3, and H4.6–8 H2A, H2B, H3, and H4 are the core histones, while linker DNA is associated histones H1 and H5. Nucleosomes act as a physical barrier to transcription factors that bind to certain regions of DNA. However, specific acetylation can remove the positive charge on the lysine amino group that is acetylated, so that the nucleosome becomes loosened on the DNA.9
It has long been hypothesized that the linker histones H1 and H5 are essential for chromatin condensation; however, this dogma has not been supported by studies. Knockout experiments in Tetrahymena and Aspergillus nidulans showed that H1 is not essential for nuclear assembly. Moreover, H1 was found to control gene expression through activation and repression mechanisms.10,11 Each of the histones has an N-terminal tail with a specific sequence of amino acids, but the H2A also has a C-terminal tail.12 The C-terminus forms a globular docking domain that is packaged into the core.13 Several studies have demonstrated the importance of the histone tails for nucleosome remodeling by ATP-dependent chromatin remodeling factors.14,15
Histone N-termini undergo posttranslational modifications that alter their interaction with DNA and nuclear proteins. Such modifications include methylation,16 acetylation,17 phosphorylation,18 sumoylation,19 ubiquitination,20 and ADP-ribosylation.21 These modifications determine the interaction between the histone and other proteins, which may in turn regulate chromatin structure, and transcription.
Among core histones, the H2A family exhibits the highest sequence divergence, resulting in the largest known number of variants. These variants, found in nearly all organisms, include H2A.Z and H2A.X.22 H2A.Z is associated with the promoters of actively transcribed genes and is also involved in the prevention of the spread of silent heterochromatin.23 It has also been found that the chromatin remodeling complex SWR1 catalyzes ATP-dependent exchange of H2A in the nucleosome for H2A.Z.24
In the other hand, H2AX, another histone variant contributes to the detection, signaling and repairing of DNA double-strand breaks.25 Plants exhibit a special class of H2A isoforms with an extended C-terminus comprising SPKK motifs.26,27 Histone H3 and H4 are nearly identical in plants and animals. For instance, only two amino acids of the 102 amino acids of histone H4 differ between pea and calf thymus.28 The linker histones are similarly found in all eukaryotes. The chromatin structure is therefore essential for both preventing of DNA and regulating gene expression, thereby preventing/enhancing the binding of transcription factors, activators, and chromatin remodeling complexes to DNA.29
Fundamental Epigenetic Mechanisms
Epigenetic mechanisms are responsible for several phenomena, such as X-inactivation, genomic imprinting, and reprogramming.30,31 There are several epigenetic processes, such as methylation, acetylation, and others that modify chromatin structure.32 The main epigenetic processes are summarized in Figure 1. Generally, methylation is associated with heterochromatic gene silencing, while acetylation is associated with euchromatic gene activation.33,34 A notable exception to general rule is methylation of some lysine and arginine residues of histones that leads to gene expression.35,36 DNA and histone modifications by methylation/demethylation influence gene expression by making DNA inaccessible (by adding a methyl group to the DNA or histone tail) or accessible (by removing it) for transcription factors and other proteins. DNA methylation is implicated in fundamental processes such as genomic imprinting, X-chromosome inactivation, and in some diseases.37 In fact, during ontogenesis, these processes regulate differentiation and determine which embryonic stem cell lines should differentiate from the totipotent zygote. The main epigenetic mechanisms regulating gene expression include the modification of DNA, the modification of histone proteins, and the chromatin remodeling.
Figure 1.
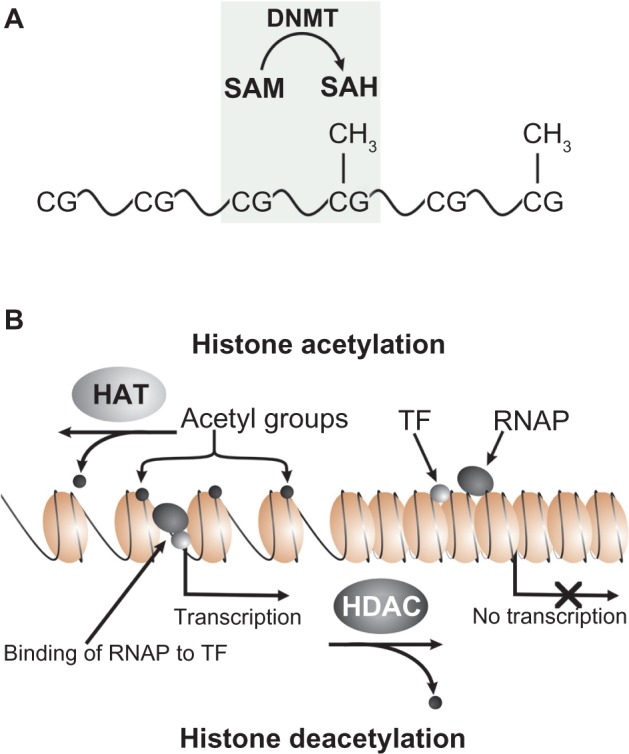
Basic epigenetic processes controlling gene expression. (A) DNA methylation, by which a methyl group (CH3; light blue) is added to DNA nucleotide, occurs at CpG sites. The reaction is catalyzed by DNA methyltransferases (DNMTs) that transfer a methyl group from the S-adenosyl methionine converting cytosine to 5-methylcytosine. DNA methylation causes gene silencing (SAH—S-adenosylehomocysteine).(B) Histone modifications include acetylation—deacetylation, methylation—demethylation processes of histones. During acetylation, histones are acetylated (light green) on lysine residues in the N-terminal tail, thereby making DNA accessible for transcription. The opposing process is deacetylation of acetylated histones, making DNA inaccessible to RNA polymerase II (RNA pol II) and thus inhibiting transcription. Histone acetylation is catalyzed by histone acetyltransferases (HATs), while histone deacetylation is catalyzed by histone deacetylases (HDACs). During histone methylation, a methyl group is transferred to the histone tail by histone methyltransferases (HMTs), turning the genes “off”, as in case of DNA methylation and histone deacetylation. Removal of methyl group from the histone tail catalyzed by histone demethylase (HDMs), turning genes “on”. The figure was drawn based on references33–37.
DNA modification
DNA methylation is a crucial epigenetic modification of the genome that involves the addition of a methyl group to the N6 position of adenine or N4 or C5 position of cytosine.38 DNA methylation is involved in regulating many cellular processes including embryonic development, chromatin structure, X-chromosome inactivation, genomic imprinting, and chromosome stability.30,31,39 This mechanism is catalyzed by DNA methyltransferases (DNMTs) that transfer a methyl group to DNA by using S-adenosyl methionine (SAM) as the methyl donor. Operation of DNMTs leads to conversion of cytosine to 5-methyl cytosine, which suppresses gene expression. DNA methylation is performed by “de novo” methyltransferases that methylate previously unmethylated cytosines, while maintenance of methylation is performed by “maintenance methyltransferases”. Maintenance of methylation refers to maintaining the methylation pattern once it is established.40,41
There are some diseases associated with aberrant DNA methylation, such as hyperhomocysteinemia, characterized by a high level of homocysteine in the blood, leading to vascular inflammation.42 Metabolic disorders, such as diabetes (and obesity), have also been linked to aberrant DNA methylation;43 moreover, it is involved in neurodegenerative disorders.44
Several data suggest that mechanisms of epigenetic regulation are general among eukaryotes, and even in prokaryotes. Enzymes that catalyze the mechanisms are highly conserved among eukaryotes.45 DNMTs catalyzing DNA methylation can be divided into three different groups, including those that generate N6-methyladenine (m6A), N4-methylcytosine (m4C), and C5-methylcytosine (m5C), each widely used by prokaryotes, fungi, plants, and animals.46 The m6A and m4C DNMTs are found primarily in prokaryotes; however, the presence of m6A in certain fungi, algae, and several ciliates has been demonstrated.47–49 The m5C DNMTs can be found in prokaryotes and eukaryotes, with 10 conserved motifs found in the catalytic domain of all DNMTs, suggesting a common origin.50,51
Based on sequence homology, DNMTs can be divided into at least 6 distinct classes, the DNMT1/methyltransferase 1 (MET1) class, DNMT2 class, DNMT3/domains rearranged methyltransferase (DRM) class, chromomethylases (CMT) class, MASC1/RID class, and MASC2/DIM2 class.52 DNMT1 was first identified in animals; however, a homolog is also found in plants. In mammals, DNMT1 is thought to be a maintenance methyltransferase, while methylation in plants is maintained by a DNMT1 homolog methyltransferase MET1.53–57 DNMT2 is the most conserved DNMT in eukaryotes58 that contains all the conserved methyltransferase motifs and is involved in methylation of tRNA.59 DNMT2 was found to have the same function in mammals and flowering plants.60 Interestingly, DNMT2 is also involved in histone deacetylation in Arabidopsis thaliana, a favorite model for plant biologists, suggesting that it participates in epigenetic regulation in plants.61 Nevertheless, very little is known regarding the function of DNMT2 in epigenetic regulation in plants and in animals. Recognition of hemimethylated DNA is catalyzed by variant in methylation (VIM) proteins in plants and ubiquitin-like, with PHD and RING finger domains 1 (UHRF1) proteins in animals.55–57
Unlike mammalian DNMT1, members of the DNMT3 subfamily (eg, DNMT3a and DNMT3b) as de novo methyltransferases are responsible for establishing cytosine methylation patterns at unmethylated DNA.62 In plants, DNA methylation is established by DRM2, which is a DNMT3 homolog.45
Demethylation of the DNA can take place through passive and active processes. Passive DNA demethylation occurs when cells fail to maintain their methylation state during DNA replication. Active DNA demethylation is primarily established by a small group of glycosylases, eg, repressor of silencing1 (ROS1), Demeter (DME) and Demeter-like3 (DML3) in plants, and 5-methylcytosine hydroxylases in animals, which introduce an abasic site.63,64
Epigenetic factors involved in DNA modification found in plants and animals are summarized in Table 1. It was reported in Arabidopsis, a regulator of DNA demethylation, IDM1, is required for preventing DNA hypermethylation, whereby binding methylated DNA at chromatin sites lacking histone methylation and acetylation protects genes from silencing.65 In animals, active DNA demethylation occurs after a sperm enters an egg.66 It was recently reported that expression of unmethylated plasmids was detected in a mouse embryo <12 h after in vitro methylated plasmid injected into the zygote. The expression of methylated plasmids was delayed until the 8 cell-stage.67 This suggests that DNA demethylation plays a critical role in regulating development both in plants and animals.
Table 1.
Summary of DNA modifications in plants and animals.
| Epigenetic mechanism | Factor
|
Function/comment | Reference | |
|---|---|---|---|---|
| Plant | Animal | |||
| Maintaining methylation | MET1 VIM |
DNMT1 UHRF family |
Transfer of a methyl group to DNA Establishment of DNA methylation patterns, recruitment of the maintenance Dnmt1/Met1 to hemimethylated DNA |
55 209 56,57,210 |
| De novo methylation | DRM2 | DNMT3 | Drm2 maintains CHH or asymmetrical methylation through a small interfering RNA (siRNA)-driven signal in a process known as RNA-directed DNA methylation |
41,211 |
| Demethylation | DME (ROS1, DML2,3) | DME Gadd45a |
Dme members have not yet been identified in animals | 210,212 |
Notes: In plants, DNA methylation commonly occurs at cytosine bases within all sequence contexts. CHH—Asymmetric CHH context, where H = A, T, or C.
Abbreviations: Met1, Maintenance DNA methyltransferase1; DNMT1, DNA methyltransferase1; VIM, Variant in methylation; UHRF, Ubiquitin-like PHD and RING finger domain; DRM2, Domains rearranged methyltransferase2; DNMT3, DNA methyltransferase3; DME, Demeter; ROS1, Repressor of silencing1; DML2,3, Demeter-like2,3; Gadd45a, growth arrest and DNA damage-inducible protein 45.
Histone modification
Acetylation is catalyzed by histone acetyltransferases (HATs). Histone acetylation enhances transcription by converting the positively charged lysine residues in the N-terminal tail into neutral residues, resulting in the loosening of the bond between DNA and the histone (Fig. 1). HATs acetylate N-terminal lysines on histones H2B and H3. Nuclear HATs are classified into several families, including the GCN5 (general control non-repressed protein5)-related N-acetyltransferase (GNAT) family. A study found that one member of the three subfamilies of GNAT is present in plants, animals, and fungi, suggesting functional conservation.
Histone deacetylases (HDACs) remove acetyl groups from an N-acetyl lysine amino acid on a histone. In plants, histone deacetylase HDA2 is a homolog of the animal HDAC11. An HDAC class, designated as class 3, has been identified within the reduced potassium dependency3/histone deacetylase 1 (RPD3/HDA1) family found only in plants and animals by phylogenetic analysis.68
Chromatin remodeling
Chromatin remodeling implies the assembly and disassembly of the nucleosomes, ATP-dependent chromatin remodeling, and modifications of histones.69 Nucleosome assembly factors, such as chromatin assembly factor1 (CAF1) and nucleosome assembly protein1 (NAP1), facilitate transcriptional regulation through deposition of histones H3 and H4 onto DNA both in plants and animals.70 Chromatin-remodeling-enzymes are ATP-dependent and responsible for conformational changes in chromatin.71,72 The subunits of chromatin-remodeling-complexes contain different domains (bromodomains, chromodomains, PHD fingers) that are involved in transcription. The members of the ATPase-dependent chromatin-remodeling-enzymes, for example the switch/sucrose nonfermentable2 (SWI2/SNF2), the imitation switch (ISWI), and the chromodomain-helicase-DNA-binding protein (CHD) groups, have ATPase domains that are responsible for their chromatin remodeling activity.73 Chromatin-remodeling-enzymes use nucleosomes as substrates and change positions of histone octamers and/or the topology of DNA that is wrapped around the nucleosome particles. The SWI/SNF family of chromatin-remodeling ATPases is conserved among the plants and animals; moreover, the SNF2 subfamily of the SWI/SNF complex is found in organisms from yeast to human.74 A summary of various epigenetic factors involved in histone modification and chromatin remodeling processes found in animal and plant cells are provided in Tables 2 and 3.
Table 2.
Summary of histone modifications in plants and animals.
| Epigenetic mechanism | Factor
|
Function/comment | Reference | |
|---|---|---|---|---|
| Plant | Animal | |||
| Acetylation | HATs (eg, GNAT, p300/CBP) | HATs (eg, GNAT/MYST, HAC1) | HATs not only specify histone modification, but also transcriptional function | 213–218 |
| Deacetylation | HDAC1 (RPD3/HDA1) | HDAC1 (RPD3/HDA1) | RPD3/HDA1-like HDACs are found in all eukaryotic genomes | 68,219–221 |
| Methylation | HMTs (SET domain proteins: CLF/SDG1, SWN/SDG1, MEA/SDG5) | HMTs (SET1, ASH1) | SET domain found from yeast to human | 222,223 |
| Demethylation | JMJC proteins (eg, KDM7B) | JMJC proteins (eg, JHDM1) | JHDM1 demethylates histone H3 at lysine 36; PKDM7B demethylates trimethyl H3K4 | 224,225 |
Abbreviations: HATs, histone acetyltransferases; GNAT, Gcn5-related N-acetyltransferase; p300/CREB, p300/CREB-binding protein; MYST, Named for the founding members MOZ (MYST3; MIM 601408), yeast YBF2 and SAS2, and TIP60 (HTATIP; MIM 601409); HAC1, Histone acetyltransferase1; HDAC1, Histone deacetylase1; RPD3, Reduced potassium dependency 3; HDA1, Histone deacetylase 1; HMTs, Histone methyltransferases; SET, [Su(var)3-9, E(z), Trx]; CLF/SDG1, Curly leaf/set domain group1; SWN/SDG1, Swinger/Set domain group1; MEA/SDG5, Medea/Set domain group5; SET1, [Su(var)3-9, E(z), Trx]1; ASH1, Discs absent, small, or homeotic-1; JMJC proteins, Jumonji domain-containing proteins; KDM7B, histone lysine (K) demethylase7B; JHDM1, JmjC domain-containing histone demethylase 1.
Table 3.
Summary of chromatin remodeling in plants and animals.
| Epigenetic mechanism | Factor
|
Function/comment | Reference | |
|---|---|---|---|---|
| Plant | Animal | |||
| Chromatin remodeling | CAF1 (eg, FAS1,2), NAP1, MSI1, HIRA | CAF1 (eg, p150, p60, p48), HIRA | Chromatin assembly, disassembly, in animals, HIRA functions as a chaperone of the variant histone H3.3 | 69,74,226,227 |
| ISWI | ISWI (SNF2H) | Facilitate sliding of histone octamers on the DNA | 228 | |
| SWI/SNF (eg, BRM, SYD), CHD (eg, PKL) | SWI/SNF (eg, BRM, BRG1, SWI2/SNF2, CDH (eg, CHD1-4,9, NURD) | SWI/SNF complexes facilitate deacetylation of histones CHDs are both transcriptional repressors and activators |
229–232 233 |
|
Abbreviations: CAF1, chromatin assembly factor1; FAS1,2, fasciata1,2; NAP1, nucleosome Assembly Protein1; MSI1, multicopy suppressor of ira1; HIRA, histone repression a factor; ISWI, imitation switch; SNF2H/L, sucrose nonfermenting 2 homolog; SWI/SNF, switch/sucrose nonfermentable; BRM, brahma; SYD, splayed; CHD, chromodomain-helicase-DNA-binding protein; PKL, pickle; BRG1, brahma-related gene1; SWI2/SNF2, switch2/sucrose nonfermentable2; CHD1-4,9, chromodomain-helicase-DNA-binding protein1-4,9; NURD, nucleosome-remodeling and histone deacetylation.
Homolog factors in plants and animals regulating gene expression
The order of the organs and body parts are determined genetically. Homeosis is the transformation of one body part into another arising from mutation of specific developmentally critical genes.75 Homeosis were first recognized in plants (Linnaeus, Goethe) and genetic studies on homeotic mutants of plants were carried out on Arabidopsis and Antirrhinum in the late 20th century.76 However, breakthrough discoveries were made in animals. Homeotic genes are conserved master genes that switch on early during ontogenesis and remain in a stable active or inactive state throughout life. These genes are involved in developmental patterns and sequences; for example, they are involved in determining when, where, and how body segments develop in flies.77 The proteins encoded by the Polycomb group (Pc-G) form structures similar to the heterochromatin and inactivate specific genes such as Hox genes.
In animals, Pc-G and trithorax group (Tr-G) proteins are responsible for continuous activity/inactivity of homeotic genes: members of the Tr-G help to fix homeotic genes into the active state, while Pc-G proteins are in the inactive state.78 Some studies reported functional similarities between plant and animal Polycomb complexes. The Polycomb repressive complex 2 (PRC2) proteins enhancer of zeste [E(z)], extra sex combs (ESC), and suppressor of variegation [Su(z)12] of mammals are involved in repression of homeotic genes. In plants, proteins such as fertilization-independent endosperm (FIE), curly leaf swinger (CLF/SWN), multi-copy suppressor of ira (MSI), and embryonic flower2 (EMF2) form PRC2-like complexes have a very similar function: to maintain the repressive condition of plant homeotic genes.69,79 A PRC2 complex carries out trimethylation of H3Lys27, which correlates with the heterochromatic gene silencing in humans and possesses the evolutionary conserved SET domain.80,81
The Su(var)3-9, E(z), Trx (SET) domain was first identified as a conserved sequence of three Drosophila proteins (Su(z), E(z), trithorax) and is highly conserved in all eukaryotes.82 The TrxG protein of SET domain catalyzes methylation of H3K4, which plays a role in transcription activation in both animals and plants.83–85
The members of the SWI/SNF family, such as ATP-dependent chromatin remodeling complexes (CRCs), are highly conserved in both animals and plants.86 The ATP-dependent CRCs function depends on their ATPase and play crucial roles in regulating transcription (activation, repression), differentiation, and ontogenesis by controlling the accessibility of DNA sequences to transcription factors.87 There is a great similarity among SWI/SNF subunits, which are homologous with those in Saccharomyces cerevisiae SWI3 in mammals and plants.88 It has been demonstrated that mammalian SWI/SNF like BRG1-associated factors (BAFs) play a crucial role in the formation of embryonic toti- and pluripotent stem cells. Embryonic stem cells express a factor distinguished as esBAF, which is defined by the presence of human BAF155 genes. Mice homozygous to the null mutation of BAF155 die during the pre-implantation stage, and heterozygous mutants develop with neural tube defects.89 Mutation of an Arabidopsis homolog of BAF155, AtSWI3 leads to inhibited development at the globular stage.90,91
The Jumonji transcription factor (Jmj) family was first identified in mouse whose members are involved in histone modification.92 It was reported that the JmjC domain containing transcription factors demethylate histones in both animals and plants. However, it was also shown that plant JmjC has both conserved and specific functions, in contrast to in mammals.93–95
Specific markers, such as sex-determining region Y-box 2 (Sox2), octamer-binding transcription factor 4 (Oct4) and Nanog, can characterize pluripotent embryonic stem cells. As in animal cells, the plant meristematic cell state is maintained by transcription factors, including WUSCHEL (Wus), CLAVATA (Clv), and PLETHORA (Plt). A HAT complex GCN5 is essential for maintaining of root stem cell niche through the attenuation of Plt.96 In animals, the proto-oncogene c-Myc has a very similar function. In addition, it regulates the expression of GCN5 via binding to the GCN5 promoter.97
Epigenetic Mechanisms Regulating Nuclear Reprogramming—Significance of Plasticity
Epigenetics is strongly related to nuclear reprogramming; in fact, it can be considered as the base of genetic reprogramming. The term nuclear reprogramming is used to describe changes in gene activity that are induced naturally (fertilization) or experimentally by introducing nuclei into a new cytoplasmic environment.98 Experimentally induced nuclear reprogramming has a close link to cloning. The term clone (ancient Greek: κλώνος [klonos], meaning “twig”) had already been used since the beginning of the 20th century in reference to plants, and later to animals.99 This can be done by removing cells from the roots of the plant and place them in a solution containing nutrients; thus, a large number of undifferentiated cells can be generated, known as the callus.100 Cells of the callus are totipotent, meaning that they have the ability to develop into any other cell type.101 Addition of plant hormones, such as auxins, cytokinins, and gibberellins to these cells results in the development of a whole plant that is genetically identical to the original plant.102
Cloning of animals is a much more complex problem because differentiated animal cells are less susceptible to dedifferentiation. However, cloning is not only observed in plants, but also several animals (aphides, fishes, lizards, frogs, etc) can reproduce without fertilization under natural conditions, cloning themselves as a part of their natural life cycle. This means that even in animals, a type of “natural cloning” is possible, which is produced by mitosis of the germ line cells of the (female) parent.102–106
The theory, originating from the great theoretician August Weismann (1834–1914) that cells forming tissues lose their reproductive ability and death becomes the natural part of their life cycle, persisted for a long time. The origin of this theory goes back to the end of the 19th century, when Weismann proposed the so called “germ plasma” (Keimplasma) theory, stating that germ line cells develop separately from somatic cells, thus indicating that acquired characteristics cannot be inherited. In many animals, there are special granules containing ooplasmic determinants in the egg which are responsible for the determination of the germ line cells.
The German embryologist Wilhelm Roux (1850–1924) stated that when the first cleavage division separated the future right half of the embryo from the future left half, there would be a separation of “right” determinants from “left” determinants in the resulting blastomeres. To test the hypothesis, in 1888 Roux used two-cell frog embryos and killed one of the cells of each embryo with a hot needle. Thus, he obtained half-embryos. Based on these and theoretical deductions, the two great scientists created a long lasting, but incorrect hypothesis: according to the Roux-Weismann theory, the diversity of the cell fates is due to the segregation of nuclear determinants during cleavage divisions, so cell nuclei become different both quantitatively and qualitatively, ascribed to the loss of genetic material.
The development of nuclear transfer experiments overthrew this thesis. In the 1930s, it was examined whether genomes of terminally differentiated cells could be reprogrammed. To answer this question, the Nobel Prize Laureate German embryologist Hans Spemann proposed an experiment: differentiated cell nuclei should be transplanted into enucleated egg cells (considered science fiction at the time of the proposal). If each cell nucleus is genetically identical to the zygote, the transplanted nucleus should be able to initiate, drive, and control the development of a new organism.107 Unfortunately, until the early 1950s, the technical background was not available to carry out these experiments and test this idea. However, in 1952, Briggs and King successfully cloned 27 tadpoles from 104 nuclear transfers in northern leopard frogs.108 In 1958, John Gurdon transferred intact nuclei from somatic cells into a Xenopus oocyte and successfully cloned a frog. He stated that the nucleus of a fully differentiated cell can return to a pluripotent state.109 In the 1960s, he also produced frogs from gut epithelial cells, yet some scientists remained skeptical, debating the fact that gut cells were differentiated and suggested that they were primordial germ cells seated in the epithelium.107 Until now, these significant experiments were praised and attributed significantly to his earning of the Noble prize in 2012, whereby many independent studies showed that fully differentiated cells can regain their totipotency.
Gurdon’s experiments were the first demonstration in animals that the nucleus of a differentiated somatic cell can regain the potential to differentiate into any cell type. Additionally, regeneration experiments showed that mature cells can be transformed into other mature cells without going through an intermediate pluripotent state.110–113 The conversion of one cell to another is termed metaplasia and includes either conversions between stem cells or direct conversion of differentiated cells, respectively.114–116 Transdifferentiation (also known as lineage reprogramming) is a type of metaplasia defined as irreversible conversion of already differentiated cell to another cell type, resulting in the loss of one phenotype and the gain of another.117 For transdifferentiation, lens regeneration, also known as Wolffian regeneration (Wolff, 1895), was demonstrated as well as metaplasia liver-to-pancreas metaplasia.118 Also in the liver, for a previous study demonstrated transdifferentiation by converting hepatocytes into bile duct cells.119 It has recently been found that using ectopic transcription factors, adult dermal fibroblasts can be converted into neural progenitor cell-like cells (iNPCs) with similar properties as primary NPCs.120
In 1996, the doctrine that the genetic material of fully differentiated cells is no longer able to produce an adult organism was finally disproved, when Scottish scientists, including the biologist Ian Wilmut, cloned a lamb successfully at the Roslin Institute using adult mammary epithelial cells incubated in depleted serum to “synchronize” to the mitotically “slow” oocyte, from a mature ewe as nucleus donor, and was transplanted to an enucleated oocyte.121 The cloning was easily verified because the phenotype and genotype of the cloned offspring could be clearly distinguished from the foster mother’s characters. The difficulty in these procedures was cleared by the fact that only 29 from the 270 nuclear transfers resulted in embryos, and just one survived. However, the only surviving sheep, Dolly, was able to produce healthy offspring, including Bonnie. Later, an entire series of mammals were successfully cloned.122
However, there are many negative aspects of cloning, including low rates and regulation problems. Cloning is very inefficient since most clones die soon after implantation.123,124 Even clones that survive often have serious abnormalities (eg, increase of body weight, kidney hypertrophy), mutations, and shortened life spans, likely a corollary of the age of somatic cells used for cloning.123,125–127
An important question arises: can a stem cell be created from a somatic cell without the need for human eggs? Two Japanese researchers, Takahashi and Yamanaka, demonstrated that ESC-like cells could be induced by some transcription factors, including Sox2, Oct4, Kruppel-like factor 4 (Klf4), and c-mycooncogene (c-Myc) pluripotency factors (also known as Yamanaka factors).128 Nuclear reprogramming using transcription factors may resolve the ethical problems related to embryonic stem cells. This method would enable development of pluripotent stem cell-based regenerative medicine. However, inducing of iPSC is very complicated and the risk of obtaining undesired cells indicates that further studies should be conducted before this method can be widely applied.129
The importance of the discovery by Gurdon130 that specialization of cells is reversible and by Shinya Yamanaka128 that intact mature cells can be reprogrammed to become immature stem cells is acclaimed by award of Nobel Prize in Physiology or Medicine in 2012. Notably, there is a great difference between the cellular plasticity of plants and animals. Cellular plasticity is the ability of cells to change their structure or function to become a different type of cell is, as we understand today, depending on the epigenetic regulation of gene expression (Fig. 2). Plasticity of plant cells to transdifferentiate into various types of cells is much higher than that of animal cells, which implies a much “looser” chromatin structure. This, however, should not be interpreted that the chromatin structure is less complex or that it is more complicated to regulate.131,132 Further in vitro epigenetic experiments and in vivo experiments, such as xeno-transplantation, may reveal this phenomenon. Based on experimental results, it may be possible to reprogram a fully differentiated animal cell nucleus using a recipient plant protoplast, a hypothesis which should be verified by future research.
Figure 2.
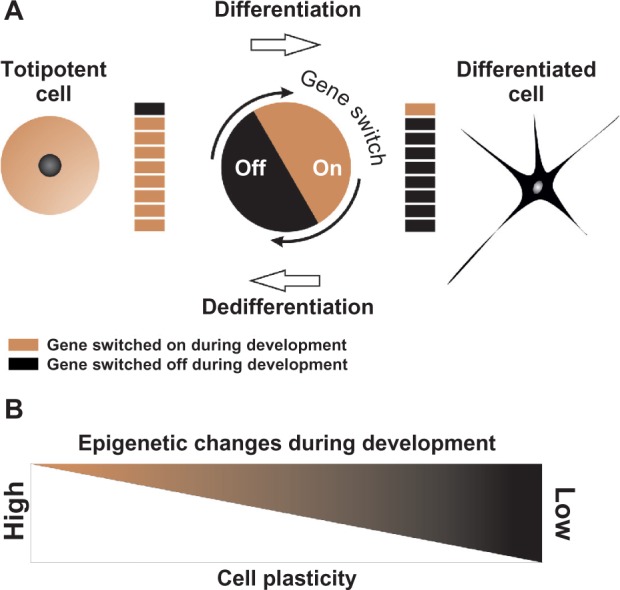
Changes in cellular plasticity. (A) Gene silencing and activation during differentiation and dedifferentiation. In a totipotent cell, such as the fertilized egg, genes responsible for segmentation and formation of pluripotent embryonic cells are switched on. Throughout differentiation, early genes are switched off, while genes needed for differentiated cell functions are switched on and others are switched off or repressed. Repressed genes can be activated reprogramming somatic cells, eg, neuron to totipotent or pluripotent states. (B) Epigenetic modifications or cell plasticity enables stem cells to differentiate into various cell types or differentiated cells to trans-differentiate to each other. During differentiation, cell plasticity is decreased. Differentiated cells have low plasticity; however, high plasticity can be increased by adding extrinsic factors that affect epigenetic processes, even in completely differentiated cells.
Conserved Epigenetic Mechanisms in Cells of Plants and Animals
The functional parallels between epigenetic elements of plant and animal development suggest that a high congruence in the epigenetic mechanisms is present in plants and animals. It should be mentioned, however, that although there is a high conservation of homeotic genes, the role of homeotic genes is dramatically different in the two groups.
In animals, the expression pattern of some ‘classical’ homeotic genes forming gene clusters is the basis for the concept “zootype”, which means that all animal phyla shared a particular pattern of gene expression. In plants, these genes are not homologous to “classical” Hox genes, suggesting that the functions of homeotic genes developed independently in the evolution of plants and animals.133,134 Comparing epigenetic patterns at the molecular level of plants and animals shows that they possess similar patterns, which are more pronounced at molecular level than at the phenomenological level. Both taxons use the same processes for epigenetic regulation, and sometimes surprising similarities are present with respect to epigenetic factors. Several examples of proteomic analysis show that a homologous transcription factor present in one group can substitute for another that is absent.93–95 Some experiments have also demonstrated that the two distant epigenetic systems may be congruent to some extent.135,136
Although several data suggest that the epigenetic regulation among plants and animals appears to be similar, an increasing amount of recent data has shown that there are many differences between the two groups, necessitating further studies.131,137
RNA Epigenetics
It has been revealed that up to 90% of eukaryotic genome is transcribed, but only 1%–2% of these transcripts encode for proteins, while the vast majority are transcribed as non-coding RNAs (ncRNAs). ncRNAs such as micro RNAs (miRNAs) are evolutionarily conserved, approximately 21 nucleotides in length, and play crucial role in development, stress responses, and chromatin states. RNA epigenetics also shows some similarities between animals and plants. In animals, such as humans, miRNAs are synthesized as single-stranded RNAs and cleaved by the RNaseIII enzymes Drosha and Dicer, producing precursor microRNAs (pre-miRNAs) and finally miRNA/miRNA duplexes.138 In plants, Dicer-like1 (DCL1) enzymes carry out these processes.139 In both plants and animals, miRNAs post-transcriptionally regulate gene expression via interactions with their target mRNAs. A major difference between plant and animal microRNA is observed for target recognition. Animal miRNAs repress gene expression by mediating translational attenuation, while nearly all plant miRNAs regulate their targets by directing mRNA cleavage at single sites in the coding regions.140 It has been demonstrated that miRNAs can also cause histone modification141 and direct DNA methylation.142,143 Interestingly, a recent study revealed that miRNAs of digested plants are present in the serum of healthy human.144
To support the cross-kingdom similarity of miRNAs with regard to epigenetic regulation of the genome, Vaucheret and Chupeau demonstrated in a recently study that ingested plant small RNAs directly influence gene expression in animals.145
Genomic imprinting
Genomic imprinting is an epigenetic process by which certain genes are expressed in a sex-dependent manner.146 It includes DNA and histone modification processes.147,148 Genomic imprinting has independently evolved in flowering plants and mammals;148,149 however, both in plants and animals, imprinting occurs in embryo-nourishing tissues, such as the placenta and the endosperm. Imprinted gene expression results from the sex-specific methylation of imprinted control regions (ICRs), such as differentially methylated regions (DMRs) in the parental germlines both in plants and mammals.150,151 Imprinting is carried out by DNA methylation and Polycomb group-mediated trimethylation of histone H3 at lysine 27 (H3K27me3) in mammals152,153 as well as in plants.154–156 However, control of imprinting differs between plants and animals.157
Role of Microenvironment in Cell Fate, Differentiation, and Dedifferentiation
In addition to epigenetic factors inside the cell, the fate of cell lineage and differentiation require continuous communications between the microenvironment of the cell, ie, extrinsic factors, extracellular matrix, and signals from neighboring cells, and the cell itself.158–160 Interactions between cells, physical conditions, and mechanical forces are also important for cell fate decision and differentiation. An interesting experiment modeled the surface geometric pattern, which affects the development of stem cells. According to this study, the shape of cells increases compressive forces in the cytoskeleton with the result that most of the flower-shaped cells form fat tissue, while star-shaped cells form bone tissue.161 In a previous experiment, the human ear was successfully grown on the back of a mouse using bovine chondrocytes with a human ear-shaped degradable polymer as a scaffold, which served as a geometric signal.162 Changes in the environment may also affect differentiated cells. When a differentiated cell nucleus is transferred into a previously activated enucleated egg cell, genetic reprogramming is taking place. It is important to note that reprogramming is not one-sided, since when a cell nucleus is located in an atypical environment, the prevailing conditions exert inductive signals. This occurs when fully differentiated plant cells undergo cell wall degradation generating protoplasts which are totipotent.163 In vitro experiments suggest that cell shape can influence cell fate determination of mesenchymal stem cells between chondrogenic and smooth muscle cell lineages through cell adhesion molecules-mediated pathways.164 Thus, extracellular stimuli, adhesion, and cell shape properties are critical determinants of cell fate and differentiation.
Epigenetic states of the cell can be modified by nutrition, behavior, stress, physical activity, and infections.165–167 It has been established that early stress effects can elicit changes in adult gene expression through epigenetic processes. Additionally, maternal stress can determine the gene expression pattern in the adult. Weaver et al found that increased pup licking and grooming (LG) and arched-back nursing (ABN) by rat mothers altered the offspring epigenome at the glucocorticoid receptor (GR) gene promoter in the hippocampus.168
In addition, a plethora of examples in plants demonstrates the importance of changes in gene expression through epigenetic regulation in response to stress adaptation.169–171 Additionally, a convincing example of adaptation to temperature stress by epigenetic regulation is the vernalization in plants growing at high altitudes.172 It has been revealed that activity of flowering locus (FLC) gene having a principal role in vernalization response state is controlled by DNA methylation, which allows the mitotically stable inheritance of the vernalized plant.173,174
In animals, dietary supplements such as vitamins can also influence epigenetic processes by affecting enzymes that regulate methylation175 or by regulating methyl-group transfer.176 These effects influence the development of diseases, such as obesity.177,178 Lack of folic acid has been shown to be associated with genomic hypomethylation179 and neural tube defects.180 Physical exercise can also influence epigenetic mechanisms, as reviewed by number of papers.181,182 It has been revealed that physical activity may affect epigenetic regulation of tumor suppressor genes contributing to cancer survival.183,184 Environmental exposure-induced abnormal epigenetic processes have been observed in many types of human185–187 and plant167 tumors. It is also important to note that miRNAs acting as tumor suppressor genes are involved in various stages of carcinogenesis.188,189 Epigenetic upregulation of suppressor miRNAs, nutritional factors, particularly vitamins, such as vitamin A,190,191 vitamin D,192 vitamin E,193 and folate194 have been shown to prevent carcinogenesis.
Interplay between Epigenetic Mechanisms of Plants and Animals— In Vitro Chromosome Condensation and Nuclear/Nucleosome Assembly
Successful xeno-transplantation or xeno-cloning, the transplantation of cells, tissues, or organs from one species to another, has been well-documented in replication and division. For instance, DNA transcription and division was observed in human nuclei, when they were injected into amphibian oocytes.195,196 The same phenomenon has not been documented between plants and animals. However, nuclear and chromatin assembly studies may deepen our understanding how to successfully conduct xeno-cloning between plant and animal.
As we described above, many transcription factors connected to the regulation of genes by rendering the chromatin state as active or silent are largely conserved in plants and animals. The ability of these transcription factors to access their binding sites depends on the structure of cellular chromatin.132,197,198 Cell shape can influence cell fate determination; therefore, cellular chromatin can be changed by the cell’s microenvironment.
In a previous study, nuclei from carrot were injected into maturing Xenopus oocyte as a recipient.199 In the control experiment, an immature oocyte was used. Prematurely condensed nuclei with premature chromosome condensation was observed after introduction of either Xenopus brain nuclei or carrot protoplast nuclei into an in vitro matured X. laevis egg immediately after germinal vesicle break down (GVBD). No chromosome condensation was observed after introduction of either Xenopus brain or carrot protoplast nuclei into Xenopus oocytes prior to GVBD.199 These findings suggest that chromosome condensation is restricted to mature oocytes that have undergone GVBD and that transplanted plant nuclei into Xenopus oocyte continues RNA synthesis, but this phenomenon was not observed when Xenopus somatic nuclei were injected into Xenopus oocytes. Breakdown of the nuclear membrane for both the plant and Xenopus nucleus by Xenopus cytoplast factors was also detected,199 suggesting that there is conservation between plants and animals regarding the enzymes involved in nuclear breakdown. However, it is possible the some nuclear damage also occurred during these procedures.
Plant Cytoplasm and Animal Chromatin
In vitro experimental evidence suggests that the plant cytoplasm is able to induce nuclear reassembly of the animal cell. For example, it was reported that carrot cytosol extract reassembled nuclear structure around a demembranated sperm chromatin from X. laevis.200 In this experiment, demembranated sperm cells and membrane vesicle purified from X. laevis was introduced into plant cell cytosol extract from carrot, which supplied an ATP-regenerating system. Immediately after introduction, the demembranated sperm chromatin was in a long, thin, and highly condensed form and strongly stained with 4′,6-diamidino-2-phenylindole (DAPI). Incubation at a specific temperature demembranated by lysolecithin sperm exhibited a series of structural and morphological changes including elongation, swelling, decondensation, changing to a round shape, and a nucleus-like structure, finally acquiring a continuous double-layered nuclear envelop with nuclear pores.200 After a long incubation, the newly assembled nuclei showed characteristics typical of normal nuclei. In the control, which contained DNase buffer, this phenomenon was not observed. Additionally, remodeling of the demembranated sperm chromatin based on the appearance of a typical DNA ladder after electrophoresis. Positive control freezing and thawing mouse liver nuclei showed a typical DNA ladder; as a negative control, lane 1 was loaded with sperm chromatin in DNase buffer. Using micrococcal nuclease did not result in DNA ladder in sperm, but a typical DNA ladder appeared in the carrot cell extract after micrococcal nuclease treatment, indicating that remodeling had occurred in this cell-free system.200 However, whether advanced structures developed, such as solenoids, remains unresolved.200
Similarly, using a cell-free system purified from Nicotiana tabaccum ovules and demembranated X. laevis sperm, chromatin decondensation and nuclear membrane assembly were observed.201 Demembranation was obtained in the same manner as in the study described above. In both cases, micrococcal nuclease digestion was used to verify nucleosome formation. Because Xenopus sperm is deficient in H1 histones, exposure to micrococcal nuclease leads to heterogeneous distribution of DNA fragment sizes.201 When Xenopus sperm nuclei were incubated with Nicotiana ovule extracts, the chromatin proteins could be replaced by histones derived from Nicotiana ovules, resulting in remodeling of the chromatin structure.201
In both cases, nuclear remodeling and nucleosome assembly were observed, suggesting that transcription factors and/or cyclin-cdk complexes originating from the plant cytoplasm may contribute to the induction of nuclear reconstitution and chromatin formation. However, complex chromatin structures, such as solenoids, were not observed and no mitosis was detected.200,201
Animal Cytoplasm and Plant Chromatin
A similar condition was applied when genetic reprogramming was carried out between an algae and an amphibian.135 In this experiment, chromosomes from the algae Crythecodinium cohnii were incubated in cytoplasmic extracts of unfertilized X. laevis oocytes or C. cohnii cell extracts. Introduction in cell-free extract from X. laevis resulted in chromosome decondensation and recondensation, nuclear membrane formation, and nuclear reconstitution. The newly assembled nuclei were morphologically different from the normal algae nuclei. Electron micrographs showed that the nuclear envelope of C. cohnii was discontinuous. However, the reconstituted nuclei possessed a normal membrane with nuclear pores which was morphologically indistinguishable from that of normal higher eukaryotic interphase nuclei. In contrast to the highly condensed chromosomes attached to the dinoflagellate C. cohnii nuclear envelope, the chromatin in the newly assembled nuclei dispersed uniformly, similar to that of typical higher eukaryotic interphase nuclei.135 In addition, there was no nuclear assembly detected when C. cohnii chromosomes were introduced into cell-free extract from C. cohnii.
These experiments clearly showed that plants and animals can influence each other through their cytoplasm and show that induction of purified DNA/chromosomes with cell-free extract from other species can lead to nuclear and nucleosome/chromatin assembly.135,200,201 However, these results do not preclude the mechanical/chemical microenvironmental effects on chromatin caused by the enucleation and nuclear transfer. In addition, each described only nuclear and nucleosome assembly as a result of purified chromosome induction with cell-free extracts, which is not extraordinary. Furthermore, in vitro nuclear assembly is independent of nucleosome/chromatin assembly.202 Early experiments demonstrated that cell-free extracts derived from species belonging to an amphibian class could induce formation of a nuclear envelope, chromatin decondensation, initiation of DNA synthesis, and chromosome condensation in sperm nuclei of X. laevis without membranes.203 The experiments described here only revealed changes in the morphology of chromatins, but not changes in DNA synthesis and mitosis.
Unicellular algae dinoflagellata C. cohnii lacks histones, which may explain why nuclear assembly did not occur when purified chromosomes from C. cohnii were introduced into cell-free extract from C. cohnii.135 In dinoflagellata, only three proteins possess similar biochemical traits as H4 in higher eukaryotes.204 Other experiments demonstrated that cytoplasm and purified chromosomes isolated from plant and from animal can induce chromatin assembly via cytoplast factors involved in histone protein synthesis.200,201 The Xenopus egg extract possesses two histone variants, histone H2A. X and histone B4, which correspond to H2A and H1 histones found in eukaryotic somatic cells.205 In the experiments, the Xenopus egg was arrested in metaphase, indicating that full components or factors are necessary for chromatin decondensation and recondensation during nuclear assembly. However, the reasons for DNA replication failure remain unclear. However, it has already been reported that in animal cells, factors involved in chromosome condensation are associated with mitosis and meiosis.206
A Novel Hypothesis
Based on the studies described above, we can hypothesize that the donor nucleus from an animal cell can reprogram the cell fate and develop into a special animal cell—“green cell”—through epigenetic mechanisms and factors of the plant protoplast. Furthermore, it can be hypothesized that external stresses, such as cell wall/membrane removal and enucleation, elicit protoplast induction/activation, resulting in the release of nuclear transcriptional regulators, thereby influencing chromatin states of the transferred nucleus. In Figure 3, a hypothetical experiment is described, in which one can transfer an animal nucleus to an enucleated plant cell, ie, protoplast, to reprogram, the donor nucleus, taking over the control of its development into differentiated animal cells. Thus, here we hypothesize that it is possible to reprogram a fully differentiated animal cell nucleus by transferring the nucleus to an enucleated protoplast (Fig. 3).
Figure 3.
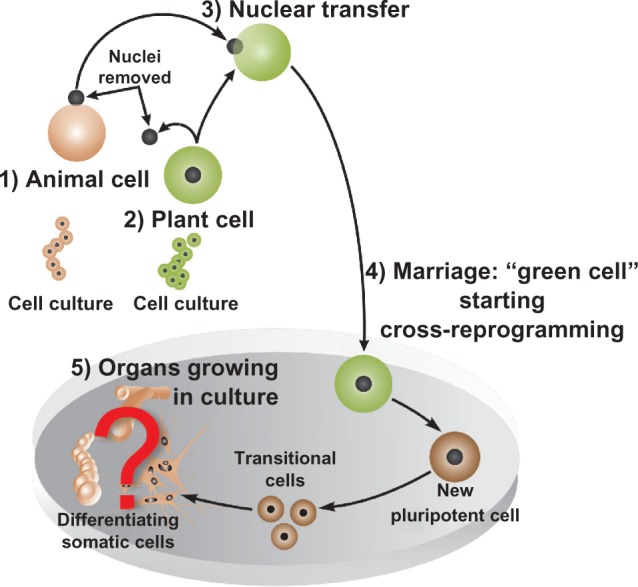
Model for reprogramming of an animal cell nucleus by transfer into enucleated protoplast. Animal cell nucleus from differentiated animal cell transferred (1) into the enucleated protoplast. In the resulting cell, nuclear reprogramming (2) takes place, resulting in totipotent cells, which under controlled conditions differentiate into a pluripotent cell known as the “green cell”. The green cell can then differentiate (3) into any cell types that are identical in genetic makeup to the donor animal cell.
Conclusions and Future Perspectives
It is known that every cell in an adult individual, either animal or plant, possess a complete set of genes with genetic and biochemical potential, and under appropriate conditions these cells are able to dedifferentiate. However, only plants have the ability to regenerate complete individuals from one single isolated somatic cell.207,208 Therefore, plants have higher dedifferentiation plasticity and capacity than animals, and utilizing these features of plants may create new avenues for research and treatment of diseases. The fully differentiated plant cells can be isolated from the original tissue by removing the cell wall, resulting in protoplasts, in which repressed genes reactivate and encode molecules needed for initiation of the developmental processes. Several studies have shown that plants and animals use conserved epigenetic mechanisms to regulate gene expression, but many different enzymes catalyze the same mechanisms.137
Thus, the main question to be answered in the future is why is complete regeneration is possible in plants, but not in animals. One possibility is that the epigenetic apparatus of plants is able to “open up”, whereas that of the animals cannot, due to its more “rigid” chromatin structure preventing the reactivation of repressed regions. However, it remains unclear whether plants have less “rigid” chromatin structure than animals or if they can utilize their epigenetic apparatus more efficiently for gene reactivation. These exciting issues need to be solved by future studies. As a further possibility for utilizing epigenetic mechanisms, more efficient methods can be designed to treat diseases that are currently incurable, such as the vast majority of cancers or neural disorders, such as Parkinson’s and Alzheimer diseases. However, diet, nutrition, and exercise also influence or control epigenetic mechanisms, and these may be used better to treat diseases such as obesity and cancer. In other areas, such as agriculture, preventing of crops from infections, or enhancing plants to adapt to stress and climate change, modification of epigenetic mechanisms could also be a target for future investigations.
In conclusion, an increased understanding of the details of epigenetic regulation of gene’s function and the use of more signaling models for detecting factors involved in regulating epigenetic processes may provide the potential to generate reprogrammed pluri- or totipotent cells without the use of cancer genes or egg cells. These developments will help to design better treatments for human diseases by using the power of epigenetic factors and mechanisms.
Acknowledgments
We thank Tibor A. Rauch for critical reading of the manuscript.
Footnotes
Author Contributions
Wrote the first draft of the manuscript: IS, ZN, AK. Contributed to the writing of the manuscript: IS, ZN, GH, RM, GS, AK. Agree with manuscript results and conclusions: IS, ZN, GH, RM, GS, AK. Jointly developed the structure and arguments for the paper: IS, ZN, GH, RM, GS, AK. Made critical revisions and approved final version: IS, ZN, GH, RM, GS, AK. All authors reviewed and approved of the final manuscript.
Funding
Supported by: SROP-4.2.2.A-11/1/KONV-2012-0024 SROP-4.2.2.A-11/1/KONV-2012-0017, Nat. Sci. Res. Fund.-OTKA K7159 and American Heart Association Founders Aff., 0555897T.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
References
- 1.Jablonka E, Lamm E. Commentary: The epigenotype—a dynamic network view of development. Int J Epidemiol. 2012;41(1):16–20. doi: 10.1093/ije/dyr185. [DOI] [PubMed] [Google Scholar]
- 2.Hurd PJ. The era of epigenetics. Brief Funct Genomics. 2010;9(5–6):425–8. doi: 10.1093/bfgp/elq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Ct, Morris JR. Genes, genetics, and epigenetics: a correspondence. Science. 2001;293(5532):1103–5. doi: 10.1126/science.293.5532.1103. [DOI] [PubMed] [Google Scholar]
- 4.Cortessis VK, Thomas DC, Levine AJ, et al. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet. 2012;131(10):1565–89. doi: 10.1007/s00439-012-1189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15(2):172–83. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 6.Behe MJ. Histone deletion mutants challenge the molecular clock hypothesis. Trends Biochem Sci. 1990;15(10):374–6. doi: 10.1016/0968-0004(90)90231-y. [DOI] [PubMed] [Google Scholar]
- 7.Wolffe A. Regulation of Chromatin Structure and Function. Austin, TX: RG Landes; 1994. [Google Scholar]
- 8.Nelson DL, Cox MM, Lehninger AL. Lehninger Principles of Biochemistry. New York, NY: Freeman; 2005. [Google Scholar]
- 9.Goodsell DS. The molecular perspective: histone deacetylase. Oncologist. 2003;8(4):389–91. doi: 10.1634/theoncologist.8-4-389. [DOI] [PubMed] [Google Scholar]
- 10.Shen X, Gorovsky MA. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86(3):475–83. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 11.Ramón A, Muro-Pastor MI, Scazzocchio C, Gonzalez R. Deletion of the unique gene encoding a typical histone H1 has no apparent phenotype in Aspergillus nidulans. Mol Microbiol. 2000;35(1):223–33. doi: 10.1046/j.1365-2958.2000.01702.x. [DOI] [PubMed] [Google Scholar]
- 12.Biswas M, Voltz K, Smith JC, Langowski J. Role of histone tails in structural stability of the nucleosome. PLoS Comput Biol. 2011;7(12):e1002279. doi: 10.1371/journal.pcbi.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang AY, Aristizabal MJ, Ryan C, Krogan NJ, Kobor MS. Key functional regions in the histone variant H2A.Z C-terminal docking domain. Mol Cell Biol. 2011;31(18):3871–84. doi: 10.1128/MCB.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgel PT, Tsukiyama T, Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16(15):4717–26. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clapier CR, Längst G, Corona DF, Becker PB, Nightingale KP. Critical role for the histone H4N terminus in nucleosome remodeling by ISWI. Mol Cell Biol. 2001;21(3):875–83. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 17.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 18.Loury R, Sassone-Corsi P. Histone phosphorylation: how to proceed. Methods. 2003;31(1):40–8. doi: 10.1016/s1046-2023(03)00086-0. [DOI] [PubMed] [Google Scholar]
- 19.Nathan D, Sterner DE, Berger SL. Histone modifications: Now summoning sumoylation. Proc Natl Acad Sci U S A. 2003;100(23):13118–20. doi: 10.1073/pnas.2436173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2012;2:26. doi: 10.3389/fonc.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Zamudio R, Ha HC. Histone ADP-ribosylation facilitates gene transcription by directly remodeling nucleosomes. Mol Cell Biol. 2012;32(13):2490–502. doi: 10.1128/MCB.06667-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talbert PB, Henikoff S. Histone variants—ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11(4):264–75. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 23.Guillemette B, Bataille AR, Gévry N, et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3(12):e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu WH, Alami S, Luk E, et al. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat Struct Mol Biol. 2005;12(12):1064–71. doi: 10.1038/nsmb1023. [DOI] [PubMed] [Google Scholar]
- 25.Chen WT, Alpert A, Leiter C, Gong F, Jackson SP, Miller KM. Systematic identification of functional residues in mammalian histone H2AX. Mol Cell Biol. 2013;33(1):111–26. doi: 10.1128/MCB.01024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigues Jde A, Brandt WF, von Holt C. The amino acid sequence of wheat histone H2A(1). A core histone with a C-terminal extension. Eur J Biochem. 1985;150(3):499–505. doi: 10.1111/j.1432-1033.1985.tb09050.x. [DOI] [PubMed] [Google Scholar]
- 27.Yi H, Sardesai N, Fujinuma T, Chan CW, Veena, Gelvin SB. Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell. 2006;18(7):1575–89. doi: 10.1105/tpc.105.039719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLange RJ, Fambrough DM, Smith EL, Bonner J. Calf and pea histone IV. 3. Complete amino acid sequence of pea seedling histone IV; comparison with the homologous calf thymus histone. J Biol Chem. 1969;244(20):5669–79. [PubMed] [Google Scholar]
- 29.Davis KL, Charney D, Coyle JT, Nemeroff C. Neuropsychopharmacology: The Fifth Generation of Progress: An Official Publication of the American College of Neuropsychopharmacology. Philadelphia, PA: Lippincott Williams & Wilkins; 2002. [Google Scholar]
- 30.Kierszenbaum AL. Genomic imprinting and epigenetic reprogramming: unearthing the garden of forking paths. Mol Reprod Dev. 2002;63(3):269–72. doi: 10.1002/mrd.90011. [DOI] [PubMed] [Google Scholar]
- 31.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303(5658):644–9. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 32.Pons D, Jukema JW. Epigenetic histone acetylation modifiers in vascular remodelling—new targets for therapy in cardiovascular disease. Neth Heart J. 2008;16(1):30–2. doi: 10.1007/BF03086114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dillon N. Heterochromatin structure and function. Biol Cell. 2004;96(8):631–7. doi: 10.1016/j.biolcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 389(6649):349–52. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 35.Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalle R, Zhang MQ. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009;10:143. doi: 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berr A, McCallum EJ, Ménard R, et al. Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell. 2010;22(10):3232–48. doi: 10.1105/tpc.110.079962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulsen M, Ferguson-Smith AC. DNA methylation in genomic imprinting, development, and disease. J Pathol. 2001;195(1):97–110. doi: 10.1002/path.890. [DOI] [PubMed] [Google Scholar]
- 38.Ponger L, Li WH. Evolutionary diversification of DNA methyltransferases in eukaryotic genomes. Mol Biol Evol. 2005;22(4):1119–28. doi: 10.1093/molbev/msi098. [DOI] [PubMed] [Google Scholar]
- 39.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 40.Giannino D, Mele G, Cozza R, et al. Isolation and characterization of a maintenance DNA-methyltransferase gene from peach (Prunus persica [L.] Batsch): transcript localization in vegetative and reproductive meristems of triple buds. J Exp Bot. 2003;54(393):2623–33. doi: 10.1093/jxb/erg292. [DOI] [PubMed] [Google Scholar]
- 41.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 42.Jamaluddin MS, Yang X, Wang H. Hyperhomocysteinemia, DNA methylation and vascular disease. Clin Chem Lab Med. 2007;45(12):1660–6. doi: 10.1515/CCLM.2007.350. [DOI] [PubMed] [Google Scholar]
- 43.Barres R, Zierath JR. DNA methylation in metabolic disorders. Am J Clin Nutr. 2011;93(4):897S–900. doi: 10.3945/ajcn.110.001933. [DOI] [PubMed] [Google Scholar]
- 44.Iraola-Guzman S, Estivill X, Rabionet R. DNA methylation in neurodegenerative disorders: a missing link between genome and environment? Clin Genet. 2011;80:1–14. doi: 10.1111/j.1399-0004.2011.01673.x. [DOI] [PubMed] [Google Scholar]
- 45.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 46.Colot V, Rossignol JL. Eukaryotic DNA methylation as an evolutionary device. Bioessays. 1999;21:402–11. doi: 10.1002/(SICI)1521-1878(199905)21:5<402::AID-BIES7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 47.Hattman S, Kenny C, Berger L, Pratt K. Comparative study of DNA methylation in three unicellular eucaryotes. J Bacteriol. 1978;135(3):1156–7. doi: 10.1128/jb.135.3.1156-1157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hattman S. DNA-[adenine] methylation in lower eukaryotes. Biochemistry (Mosc) 2005;70(5):550–8. doi: 10.1007/s10541-005-0148-6. [DOI] [PubMed] [Google Scholar]
- 49.Rogers SD, Rogers ME, Saunders G, Holt G. Isolation of mutants sensitive to 2-aminopurine and alkylating agents and evidence for the role of DNA methylation in Penicillium chrysogenum. Curr Genet. 1986;10(7):557–60. doi: 10.1007/BF00447390. [DOI] [PubMed] [Google Scholar]
- 50.Kumar S, Cheng X, Klimasauskas S, et al. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994;22(1):1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Posfai J, Bhagwat AS, Pósfai G, Roberts RJ. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989;17(7):2421–35. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen T, Li E. Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Biol. 2004;60:55–89. doi: 10.1016/S0070-2153(04)60003-2. [DOI] [PubMed] [Google Scholar]
- 53.Bartee L, Bender J. Two Arabidopsis methylation-deficiency mutations confer only partial effects on a methylated endogenous gene family. Nucleic Acids Res. 2001;29(10):2127–34. doi: 10.1093/nar/29.10.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding F, Chaillet JR. Proc Natl Acad Sci U S A; In vivo stabilization of the Dnmt1 (cytosine-5)-methyltransferase protein. ; 2002. pp. 14861–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tariq M, Paszkowski J. DNA and histone methylation in plants. Trends Genet. 20(6):244–51. doi: 10.1016/j.tig.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 56.Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 57.Bronner C, Achour M, Arima Y, Chataigneau T, Saya H, Schini-Kerth VB. The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol Ther. 2007;115(3):419–34. doi: 10.1016/j.pharmthera.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 58.Schaefer M, Lyko F. Solving the Dnmt2 enigma. Chromosoma. 2010;119(1):35–40. doi: 10.1007/s00412-009-0240-6. [DOI] [PubMed] [Google Scholar]
- 59.Schaefer M, Steringer JP, Lyko F. The Drosophila cytosine-5 methyltransferase Dnmt2 is associated with the nuclear matrix and can access DNA during mitosis. PLoS One. 2008;3(1):e1414. doi: 10.1371/journal.pone.0001414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goll MG, Kirpekar F, Maggert KA, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311(5759):395–8. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 61.Song Y, Wu K, Dhaubhadel S, An L, Tian L. Arabidopsis DNA methyltransferase AtDNMT2 associates with histone deacetylase AtHD2s activity. Biochem Biophys Res Commun. 2010;396(2):187–92. doi: 10.1016/j.bbrc.2010.03.119. [DOI] [PubMed] [Google Scholar]
- 62.Kim JK, Samaranayake M, Pradhan S. Epigenetic mechanisms in mammals. Cell Mol Life Sci. 2009;66(4):596–612. doi: 10.1007/s00018-008-8432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gong Z, Zhu JK. Active DNA demethylation by oxidation and repair. Cell Res. 2011;21(12):1649–51. doi: 10.1038/cr.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21(3):442–65. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qian W, Miki D, Zhang H, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336(6087):1445–8. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293(5532):1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 67.Zhang P, Su L, Wang Z, et al. The involvement of 5-hydroxymethylcytosine in active DNA demethylation in mice. Biol Reprod. 2012;86(4):104. doi: 10.1095/biolreprod.111.096073. [DOI] [PubMed] [Google Scholar]
- 68.Pandey R, Muller A, Napoli CA, et al. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30(23):5036–55. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen WH, Xu L. Chromatin remodeling in stem cell maintenance in Arabidopsis thaliana. Mol Plant. 2009;2(4):600–9. doi: 10.1093/mp/ssp022. [DOI] [PubMed] [Google Scholar]
- 70.Exner V, Gruissem W, Hennig L. Control of trichome branching by chromatin assembly factor-1. BMC Plant Biol. 2008;8:54. doi: 10.1186/1471-2229-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366(6453):362–5. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 72.Sudarsanam P, Winston F. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 2000;16(8):345–51. doi: 10.1016/s0168-9525(00)02060-6. [DOI] [PubMed] [Google Scholar]
- 73.Saladi SV, de la Serna IL. ATP dependent chromatin remodeling enzymes in embryonic stem cells. Stem Cell Rev. 2010;6(1):62–73. doi: 10.1007/s12015-010-9120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104(1):131–42. doi: 10.1016/s0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- 75.Akam M. Hox genes, homeosis and the evolution of segment identity: no need for hopeless monsters. Int J Dev Biol. 1998;42(3):445–51. [PubMed] [Google Scholar]
- 76.Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78(2):203–9. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 77.Gehring WJ, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–73. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- 78.Kennison JA. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 79.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 80.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 81.Rea S, Eisenhaber F, O’Carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 82.Johnson CD, Bynum TE. Hypocalcemia as a complication of jejunoileal bypass for morbid obesity. South Med J. 1976;69(5):616–8. doi: 10.1097/00007611-197605000-00041. [DOI] [PubMed] [Google Scholar]
- 83.Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol. 2003;13(8):627–37. doi: 10.1016/s0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- 84.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein super-family: protein lysine methyltransferases. Genome Biol. 2005;6(8):227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pien S, Grossniklaus U. Polycomb group and trithorax group proteins in Arabidopsis. Biochim Biophys Acta. 2007;1769(5–6):375–82. doi: 10.1016/j.bbaexp.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 86.Bezhani S, Winter C, Hershman S, et al. Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell. 2007;19(2):403–16. doi: 10.1105/tpc.106.048272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21(3):396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou C, Miki B, Wu K. CHB2, a member of the SWI3 gene family, is a global regulator in Arabidopsis. Plant Mol Biol. 2003;52(6):1125–34. doi: 10.1023/b:plan.0000004305.60407.8b. [DOI] [PubMed] [Google Scholar]
- 89.Yoo AS, Crabtree GR. ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol. 2009;19(2):120–6. doi: 10.1016/j.conb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Crosby MA, Miller C, Alon T, et al. The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol Cell Biol. 2009;19(2):1159–70. doi: 10.1128/mcb.19.2.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sarnowski TJ, Rios G, Jásik J, et al. SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell. 2005;17(9):2454–72. doi: 10.1105/tpc.105.031203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Motoyama J, Takeuchi T. The mouse embryogenesis of jumonji mutant obtained by gene-trap method. Tanpakushitsu Kakusan Koso. 1995;40(14):2152–61. [PubMed] [Google Scholar]
- 93.Lu F, Li G, Cui X, Liu C, Wang XJ, Cao X. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol. 2008;50(7):886–96. doi: 10.1111/j.1744-7909.2008.00692.x. [DOI] [PubMed] [Google Scholar]
- 94.Sun Q, Zhou DX. Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci U S A. 2008;105(36):13679–84. doi: 10.1073/pnas.0805901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 96.Kornet N, Scheres B. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell. 2009;21(4):1070–9. doi: 10.1105/tpc.108.065300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25(12):2723–34. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gurdon JB, Byrne JA, Simonsson S. Nuclear reprogramming and stem cell creation. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11819–22. doi: 10.1073/pnas.1834207100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gurdon JB, Colman A. The future of cloning. Nature. 1999;402(6763):743–6. doi: 10.1038/45429. [DOI] [PubMed] [Google Scholar]
- 100.Steward FC, Caplin SM. A tissue culture from potato tuber; the synergistic action of 2,4-D and of coconut milk. Science. 1951;113(2940):518–20. doi: 10.1126/science.113.2940.518. [DOI] [PubMed] [Google Scholar]
- 101.Schroeder CA, Kay E, Davis LH. Totipotency of Cells from Fruit Pericarp Tissue in vitro. Science. 1962;138(3540):595–6. doi: 10.1126/science.138.3540.595. [DOI] [PubMed] [Google Scholar]
- 102.Dharmawardhana P, Brunner AM, Strauss SH. Genome-wide transcriptome analysis of the transition from primary to secondary stem development in Populus trichocarpa. BMC Genomics. 2010;11:150. doi: 10.1186/1471-2164-11-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cullum AJ. Comparisons of physiological performance in sexual and asexual whiptail lizards (genus Cnemidophorus): implications for the role of heterozygosity. Am Nat. 1997;150(1):24–47. doi: 10.1086/286055. [DOI] [PubMed] [Google Scholar]
- 104.Halkett F, Plantegenest M, Prunier-Leterme N, Mieuzet L, Delmotte F, Simon JC. Admixed sexual and facultatively asexual aphid lineages at mating sites. Mol Ecol. 2005;14(1):325–36. doi: 10.1111/j.1365-294X.2004.02358.x. [DOI] [PubMed] [Google Scholar]
- 105.Smith TG, Kim B, Hong H, Desser SS. Intraerythrocytic development of species of Hepatozoon infecting ranid frogs: evidence for convergence of life cycle characteristics among apicomplexans. J Parasitol. 2000;86(3):451–8. doi: 10.1645/0022-3395(2000)086[0451:IDOSOH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 106.Schaschl H, Tobler M, Plath M, Penn DJ, Schlupp I. Polymorphic MHC loci in an asexual fish, the amazon molly (Poecilia formosa; Poeciliidae) Mol Ecol. 2008;17(24):5220–30. doi: 10.1111/j.1365-294X.2008.03997.x. [DOI] [PubMed] [Google Scholar]
- 107.Gilbert SF. Developmental Biology. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- 108.Briggs R, King TJ. Transplantation of living nuclei from blastula cells into enucleated frogs’ eggs. Proc Natl Acad Sci U S A. 1952;38(5):455–63. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182(4627):64–5. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- 110.King RS, Newmark PA. The cell biology of regeneration. J Cell Biol. 2012;196(5):553–62. doi: 10.1083/jcb.201105099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Casimir CM, Gates PB, Patient RK, Brockes JP. Evidence for dedifferentiation and metaplasia in amphibian limb regeneration from inheritance of DNA methylation. Development. 1988;104(4):657–68. doi: 10.1242/dev.104.4.657. [DOI] [PubMed] [Google Scholar]
- 112.Menke V, van Es JH, de Lau W, et al. Conversion of metaplastic Barrett’s epithelium into post-mitotic goblet cells by gamma-secretase inhibition. Dis Model Mech. 2010;3(1–2):104–10. doi: 10.1242/dmm.003012. [DOI] [PubMed] [Google Scholar]
- 113.Barros R, Freund JN, David L, Almeida R. Gastric intestinal metaplasia revisited: function and regulation of CDX2. Trends Mol Med. 2012;18(9):555–63. doi: 10.1016/j.molmed.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 114.Slack JM. Metaplasia and transdifferentiation: from pure biology to the clinic. Nat Rev Mol Cell Biol. 2007;8(5):369–78. doi: 10.1038/nrm2146. [DOI] [PubMed] [Google Scholar]
- 115.Shen CN, Burke ZD, Tosh D. Transdifferentiation, metaplasia and tissue regeneration. Organogenesis. 2004;1(2):36–44. doi: 10.4161/org.1.2.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462(7273):587–94. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 117.Eberhard D, Tosh D. Transdifferentiation and metaplasia as a paradigm for understanding development and disease. Cell Mol Life Sci. 2008;65(1):33–40. doi: 10.1007/s00018-007-7428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Horb ME, Shen CN, Tosh D, Slack JM. Experimental conversion of liver to pancreas. Curr Biol. 2003;13(2):105–15. doi: 10.1016/s0960-9822(02)01434-3. [DOI] [PubMed] [Google Scholar]
- 119.Nishikawa Y, Watanabe H, et al. Transdifferentiation of mature rat hepatocytes into bile duct-like cells in vitro. Am J Pathol. 2005;166(4):1077–88. doi: 10.1016/S0002-9440(10)62328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tian C, Ambroz RJ, Sun L, et al. Direct conversion of dermal fibroblasts into neural progenitor cells by a novel cocktail of defined factors. Curr Mol Med. 2012;12(2):126–37. doi: 10.2174/156652412798889018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380(6569):64–6. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 122.Campbell KH, Alberio R, Choi I, et al. Cloning: eight years after Dolly. Reprod Domest Anim. 2005;40(4):256–68. doi: 10.1111/j.1439-0531.2005.00591.x. [DOI] [PubMed] [Google Scholar]
- 123.Tsunoda Y, Kato Y. Recent progress and problems in animal cloning. Differentiation. 2002;69(4–5):158–61. doi: 10.1046/j.1432-0436.2002.690405.x. [DOI] [PubMed] [Google Scholar]
- 124.Wells DN. Animal cloning: problems and prospects. Rev Sci Tech. 2005;24(1):251–64. [PubMed] [Google Scholar]
- 125.Dinc L. Ethical issues regarding human cloning: a nursing perspective. Nurs Ethics. 10(3):238–54. doi: 10.1191/0969733003ne603oa. [DOI] [PubMed] [Google Scholar]
- 126.Shimozawa N, Sotomaru Y, Eguchi N, et al. Phenotypic abnormalities observed in aged cloned mice from embryonic stem cells after long-term maintenance. Reproduction. 2006;132(3):435–41. doi: 10.1530/rep.1.00745. [DOI] [PubMed] [Google Scholar]
- 127.Wang Y, Hai T, Liu Z, et al. HSPC117 deficiency in cloned embryos causes placental abnormality and fetal death. Biochem Biophys Res Commun. 2010;397(3):407–12. doi: 10.1016/j.bbrc.2010.05.105. [DOI] [PubMed] [Google Scholar]
- 128.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 129.Geoghegan E, Byrnes L. Mouse induced pluripotent stem cells. Int J Dev Biol. 2008;52(8):1015–22. doi: 10.1387/ijdb.082640eg. [DOI] [PubMed] [Google Scholar]
- 130.Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–40. [PubMed] [Google Scholar]
- 131.Avramova ZV. Heterochromatin in animals and plants. Similarities and differences. Plant Physiol. 2002;129(1):40–9. doi: 10.1104/pp.010981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Grafi G. How cells dedifferentiate: a lesson from plants. Dev Biol. 2004;268(1):1–6. doi: 10.1016/j.ydbio.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 133.Theissen G, Becker A, Di Rosa A, et al. A short history of MADS-box genes in plants. Plant Mol Biol. 2000;42(1):115–49. [PubMed] [Google Scholar]
- 134.Warren RW, Nagy L, Selegue J, Gates J, Carroll S. Evolution of homeotic gene regulation and function in flies and butterflies. Nature. 1995;373(6513):451. doi: 10.1038/373451a0. [DOI] [PubMed] [Google Scholar]
- 135.Liu XL, Shen Y, Chen EJ, Zhai ZH. Nuclear assembly of purified Crythecodinium cohnii chromosomes in cell-free extracts of Xenopus laevis eggs. Cell Res. 2000;10(2):127–37. doi: 10.1038/sj.cr.7290042. [DOI] [PubMed] [Google Scholar]
- 136.von der Haar B, Sperling K, Gregor D. Maturing Xenopus oocytes induce chromosome condensation in somatic plant nuclei. Exp Cell Res. 1981;134(2):477–81. doi: 10.1016/0014-4827(81)90450-x. [DOI] [PubMed] [Google Scholar]
- 137.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330(6004):622–7. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 139.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–87. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 140.Axtell MJ, Westholm JO, Lai EC. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12(4):221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Roccaro AM, Sacco A, Jia X, et al. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood. 2010;116(9):1506–14. doi: 10.1182/blood-2010-01-265686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wu L, Zhou H, Zhang Q, et al. DNA methylation mediated by a microRNA pathway. Mol Cell. 2010;38(3):465–75. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 143.Chavali V, Tyagi SC, Mishra PK. MicroRNA-133a regulates DNA methylation in diabetic cardiomyocytes. Biochem Biophys Res Commun. 2012;425(3):668–72. doi: 10.1016/j.bbrc.2012.07.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang L, Hou D, Chen X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22(1):107–26. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vaucheret H, Chupeau Y. Ingested plant miRNAs regulate gene expression in animals. Cell Res. 2012;22(1):3–5. doi: 10.1038/cr.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat Res. 2008;647(1–2):77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Meaney MJ, Ferguson-Smith AC. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat Neurosci. 2010;13(11):1313–8. doi: 10.1038/nn1110-1313. [DOI] [PubMed] [Google Scholar]
- 148.Feil R, Berger F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007;23(4):192–9. doi: 10.1016/j.tig.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 149.Jiang H, Kohler C. Evolution, function, and regulation of genomic imprinting in plant seed development. J Exp Bot. 2012;63(13):4713–22. doi: 10.1093/jxb/ers145. [DOI] [PubMed] [Google Scholar]
- 150.Bartolomei MS. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev. 2009;23(18):2124–33. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Macdonald WA. Epigenetic mechanisms of genomic imprinting: common themes in the regulation of imprinted regions in mammals, plants, and insects. Genet Res Int. 2012;2012:585024. doi: 10.1155/2012/585024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Umlauf D, Goto Y, Cao R, et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36(12):1296–300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- 153.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19(3):281–9. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 154.Kinoshita T, Miura A, Choi Y, et al. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303(5657):521–3. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 155.Baroux C, Gagliardini V, Page DR, Grossniklaus U. Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev. 2006;20(9):1081–6. doi: 10.1101/gad.378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xiao W, Custard KD, Brown RC, et al. DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell. 2006;18(4):805–14. doi: 10.1105/tpc.105.038836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Scott RJ, Spielman M. Genomic imprinting in plants and mammals: how life history constrains convergence. Cytogenet Genome Res. 2006;113(1–4):53–67. doi: 10.1159/000090815. [DOI] [PubMed] [Google Scholar]
- 158.Bai X, Yan Y, Song YH, et al. Both cultured and freshly isolated adipose tissue-derived stem cells enhance cardiac function after acute myocardial infarction. Eur Heart J. 2010;31(4):489–501. doi: 10.1093/eurheartj/ehp568. [DOI] [PubMed] [Google Scholar]
- 159.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5(1):17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li N, Lu X, Zhao X, et al. Endothelial nitric oxide synthase promotes bone marrow stromal cell migration to the ischemic myocardium via upregulation of stromal cell-derived factor-1alpha. Stem Cells. 2009;27(4):961–70. doi: 10.1002/stem.6. [DOI] [PubMed] [Google Scholar]
- 161.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci U S A. 2010;107(11):4872–7. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cao Y, Vacanti JP, Paige KT, Upton J, Vacanti CA. Transplantation of chondrocytes utilizing a polymer-cell construct to produce tissue-engineered cartilage in the shape of a human ear. Plast Reconstr Surg. 1997;100(2):297–302. doi: 10.1097/00006534-199708000-00001. [DOI] [PubMed] [Google Scholar]
- 163.Zhao J, Morozova N, Williams L, Libs L, Avivi Y, Grafi G. Two phases of chromatin decondensation during dedifferentiation of plant cells: distinction between competence for cell fate switch and a commitment for S phase. J Biol Chem. 2001;276(25):22772–8. doi: 10.1074/jbc.M101756200. [DOI] [PubMed] [Google Scholar]
- 164.Gao L, McBeath R, Chen CS. Stem cell shape regulates a chondrogenic versus myogenic fate through Rac1 and N-cadherin. Stem Cells. 2010;28(3):564–72. doi: 10.1002/stem.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mathers JC, Strathdee G, Relton CL. Induction of epigenetic alterations by dietary and other environmental factors. Adv Genet. 2010;71:3–39. doi: 10.1016/B978-0-12-380864-6.00001-8. [DOI] [PubMed] [Google Scholar]
- 166.Alegría-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics. 2011;3(3):267–77. doi: 10.2217/epi.11.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bierne H, Hamon M, Cossart P. Epigenetics and bacterial infections. Cold Spring Harb Perspect Med. 2012;2(12):a010272. doi: 10.1101/cshperspect.a010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–54. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 169.Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol. 2011;14(3):267–74. doi: 10.1016/j.pbi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 170.McCue AD, Nuthikattu S, Reeder SH, Slotkin RK. Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet. 8(2):e1002474. doi: 10.1371/journal.pgen.1002474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chinnusamy V, Zhu JK. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol. 2009;12(2):133–9. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sheldon CC, Finnegan EJ, Rouse DT, et al. The control of flowering by vernalization. Curr Opin Plant Biol. 2000;3(5):418–22. doi: 10.1016/s1369-5266(00)00106-0. [DOI] [PubMed] [Google Scholar]
- 173.Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC) Proc Natl Acad Sci U S A. 2000;97(7):3753–8. doi: 10.1073/pnas.060023597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sheldon CC, Burn JE, Perez PP, et al. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11(3):445–58. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Brunaud L, Alberto JM, Ayav A, et al. Effects of vitamin B12 and folate deficiencies on DNA methylation and carcinogenesis in rat liver. Clin Chem Lab Med. 2003;41(8):1012–9. doi: 10.1515/CCLM.2003.155. [DOI] [PubMed] [Google Scholar]
- 176.Niculescu MD, Zeisel SH. Diet, methyl donors and DNA methylation: interactions between dietary folate, methionine and choline. J Nutr. 2002;132(Suppl 8):2333S–5. doi: 10.1093/jn/132.8.2333S. [DOI] [PubMed] [Google Scholar]
- 177.Vucetic Z, Kimmel J, Reyes TM. Chronic high-fat diet drives postnatal epigenetic regulation of μ-opioid receptor in the brain. Neuropsychopharmacology. 2011;36(6):1199–206. doi: 10.1038/npp.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Milagro FI, Mansego ML, De Miguel C, Martínez JA. Dietary factors, epigenetic modifications and obesity outcomes: Progresses and perspectives. Mol Aspects Med. 2013;34(4):782–812. doi: 10.1016/j.mam.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 179.Pufulete M, Al-Ghnaniem R, Khushal A, et al. Effect of folic acid supplementation on genomic DNA methylation in patients with colorectal adenoma. Gut. 2005;54(5):648–53. doi: 10.1136/gut.2004.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Blom HJ. Folic acid, methylation and neural tube closure in humans. Birth Defects Res A Clin Mol Teratol. 2009;85(4):295–302. doi: 10.1002/bdra.20581. [DOI] [PubMed] [Google Scholar]
- 181.Sanchis-Gomar F, Garcia-Gimenez JL, Perez-Quilis C, Gomez-Cabrera MC, Pallardo FV, Lippi G. Physical exercise as an epigenetic modulator: Eustress, the positive stress as an effector of gene expression. J Strength Cond Res. 2012;26(12):3469–72. doi: 10.1519/JSC.0b013e31825bb594. [DOI] [PubMed] [Google Scholar]
- 182.Schwarzenbach H. Impact of physical activity and doping on epigenetic gene regulation. Drug Test Anal. 2011;3(10):682–7. doi: 10.1002/dta.294. [DOI] [PubMed] [Google Scholar]
- 183.Zeng H, Irwin ML, Lu L, et al. Physical activity and breast cancer survival: an epigenetic link through reduced methylation of a tumor suppressor gene L3MBTL1. Breast Cancer Res Treat. 2012;133(1):127–35. doi: 10.1007/s10549-011-1716-7. [DOI] [PubMed] [Google Scholar]
- 184.Coyle YM, Xie XJ, Lewis CM, Bu D, Milchgrub S, Euhus DM. Role of physical activity in modulating breast cancer risk as defined by APC and RASSF1A promoter hypermethylation in nonmalignant breast tissue. Cancer Epidemiol Biomarkers Prev. 2007;16(2):192–6. doi: 10.1158/1055-9965.EPI-06-0700. [DOI] [PubMed] [Google Scholar]
- 185.Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22(2):91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- 186.Martin FL. Epigenetic influences in the aetiology of cancers arising from breast and prostate: a hypothesised transgenerational evolution in chromatin accessibility. ISRN Oncol. 2013;624794 doi: 10.1155/2013/624794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012;41(1):79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chen X, Wang J, Shen H, et al. Epigenetics, microRNAs, and carcinogenesis: functional role of microRNA-137 in uveal melanoma. Invest Ophthalmol Vis Sci. 2011;52(3):1193–9. doi: 10.1167/iovs.10-5272. [DOI] [PubMed] [Google Scholar]
- 189.Xi S, Xu H, Shan J, et al. Cigarette smoke mediates epigenetic repression of miR-487b during pulmonary carcinogenesis. J Clin Invest. 2013;123(3):1241–61. doi: 10.1172/JCI61271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rossi A, D’Urso OF, Gatto G, et al. Non-coding RNAs change their expression profile after Retinoid induced differentiation of the promyelocytic cell line NB4. BMC Res Notes. 2010;3:24. doi: 10.1186/1756-0500-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Garzon R, Pichiorri F, Palumbo T, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26(28):4148–57. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 192.Peng X, Vaishnav A, Murillo G, Alimirah F, Torres KE, Mehta RG. Protection against cellular stress by 25-hydroxyvitamin D3 in breast epithelial cells. J Cell Biochem. 2010;110(6):1324–33. doi: 10.1002/jcb.22646. [DOI] [PubMed] [Google Scholar]
- 193.Gaedicke S, Zhang X, Schmelzer C, et al. Vitamin E dependent microRNA regulation in rat liver. FEBS Lett. 2008;582(23–4):3542–6. doi: 10.1016/j.febslet.2008.09.032. [DOI] [PubMed] [Google Scholar]
- 194.Kutay H, Bai S, Datta J, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 99(3):671–8. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Gurdon JB, De Robertis EM, Partington G. Injected nuclei in frog oocytes provide a living cell system for the study of transcriptional control. Nature. 1976;260(5547):116–20. doi: 10.1038/260116a0. [DOI] [PubMed] [Google Scholar]
- 196.Deschamps JY, Roux FA, Saï P, Gouin E. History of xenotransplantation. Xenotransplantation. 2005;12(2):91–109. doi: 10.1111/j.1399-3089.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 197.Halbach T, Scheer N, Werr W. Transcriptional activation by the PHD finger is inhibited through an adjacent leucine zipper that binds 14-3-3 proteins. Nucleic Acids Res. 2000;28(18):3542–50. doi: 10.1093/nar/28.18.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wagner D. Chromatin regulation of plant development. Curr Opin Plant Biol. 2003;6(1):20–8. doi: 10.1016/s1369526602000079. [DOI] [PubMed] [Google Scholar]
- 199.von der Haar B, Sperling K, Gregor D. Maturing Xenopus oocytes induce chromosome condensation in somatic plant nuclei. Exp Cell Res. 1981;134(2):477–81. doi: 10.1016/0014-4827(81)90450-x. [DOI] [PubMed] [Google Scholar]
- 200.Zhao Y, Liu X, Wu M, Tao W, Zhai Z. In vitro nuclear reconstitution could be induced in a plant cell-free system. FEBS Lett. 2000;480(2–3):208–12. doi: 10.1016/s0014-5793(00)01938-4. [DOI] [PubMed] [Google Scholar]
- 201.Lu P, Zhai ZH. Nuclear assembly of demembranated Xenopus sperm in plant cell-free extracts from Nicotiana ovules. Exp Cell Res. 2001;270(1):96–101. doi: 10.1006/excr.2001.5296. [DOI] [PubMed] [Google Scholar]
- 202.Jiang Z, Zhang B, Zhai Z. Nuclear reassembly in vitro is independent of nucleosome/chromatin assembly. Sci China C Life Sci. 1998;41(5):512–9. doi: 10.1007/BF02882889. [DOI] [PubMed] [Google Scholar]
- 203.Lohka MJ, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220(4598):719–21. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- 204.Vernet G, Sala-Rovira M, Maeder M, Jacques F, Herzog M. Basic nuclear proteins of the histone-less eukaryote Crypthecodinium cohnii (Pyrrhophyta): two-dimensional electrophoresis and DNA-binding properties. Biochim Biophys Acta. 1990;1048(2–3):281–9. doi: 10.1016/0167-4781(90)90068-d. [DOI] [PubMed] [Google Scholar]
- 205.Dimitrov S, Dasso MC, Wolffe AP. Remodeling sperm chromatin in Xenopus laevis egg extracts: the role of core histone phosphorylation and linker histone B4 in chromatin assembly. J Cell Biol. 1994;126(3):591–601. doi: 10.1083/jcb.126.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Berbenetz NM, Nislow C, Brown GW. Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet. 2010;6(9):e1001092. doi: 10.1371/journal.pgen.1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Vogel G. How does a single somatic cell become a whole plant? Science. 2005;309(5731):86. doi: 10.1126/science.309.5731.86. [DOI] [PubMed] [Google Scholar]
- 208.Sussex IM. The scientific roots of modern plant biotechnology. Plant Cell. 2008;20(5):1189–98. doi: 10.1105/tpc.108.058735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bronner C, Fuhrmann G, Chédin FL, Macaluso M, Dhe-Paganon S. UHRF1 links the histone code and DNA methylation to ensure faithful epigenetic memory inheritance. Genet Epigenet. 2010;2009(2):29–36. [PMC free article] [PubMed] [Google Scholar]
- 210.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 2008;4(11):e1000280. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Barreto G, Schäfer A, Marhold J, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445(7128):671–5. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 213.Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS. Plant J; In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. ; 2007. pp. 615–26. [DOI] [PubMed] [Google Scholar]
- 214.Eot-Houllier G, Fulcrand G, Magnaghi-Jaulin L, Jaulin C. Histone deacetylase inhibitors and genomic instability. Cancer Lett. 2009;274(2):169–76. doi: 10.1016/j.canlet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 215.Servet C, Conde e Silva N, Zhou DX. Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol Plant. 2010;3(4):670–7. doi: 10.1093/mp/ssq018. [DOI] [PubMed] [Google Scholar]
- 216.Jin Q, Yu LR, Wang L, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30(2):249–62. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Papaefthimiou D, Likotrafiti E, Kapazoglou A, Bladenopoulos K, Tsaftaris A. Epigenetic chromatin modifiers in barley: III. Isolation and characterization of the barley GNAT-MYST family of histone acetyltransferases and responses to exogenous ABA. Plant Physiol Biochem. 2010;48(2–3):98–107. doi: 10.1016/j.plaphy.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 218.Deng W, Liu C, Pei Y, Deng X, Niu L, Cao X. Involvement of the histone acetyltransferase AtHAC1 in the regulation of flowering time via repression of FLOWERING LOCUS C in Arabidopsis. Plant Physiol. 2007;143(4):1660–8. doi: 10.1104/pp.106.095521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9(3):206–18. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Fu W, Wu K, Duan J. Sequence and expression analysis of histone deacetylases in rice. Biochem Biophys Res Commun. 2007;356(4):843–50. doi: 10.1016/j.bbrc.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 221.Chen LT, Luo M, Wang YY, Wu K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J Exp Bot. 2010;61(12):3345–53. doi: 10.1093/jxb/erq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hallson G, Hollebakken RE, Li T, et al. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics. 2012;190(1):91–100. doi: 10.1534/genetics.111.135863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Cheung P, Lau P. Epigenetic regulation by histone methylation and histone variants. Mol Endocrinol. 2005;19(3):563–73. doi: 10.1210/me.2004-0496. [DOI] [PubMed] [Google Scholar]
- 224.Yang W, Jiang D, Jiang J, He Y. A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J. 2010;62(4):663–73. doi: 10.1111/j.1365-313X.2010.04182.x. [DOI] [PubMed] [Google Scholar]
- 225.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8(4):307–18. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 226.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14(11):997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 227.Ach RA, Taranto P, Gruissem W. A conserved family of WD-40 proteins binds to the retinoblastoma protein in both plants and animals. Plant Cell. 1997;9(9):1595–606. doi: 10.1105/tpc.9.9.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Noh YS, Amasino RM. PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell. 2003;15(7):1671–82. doi: 10.1105/tpc.012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Knizewski L, Ginalski K, Jerzmanowski A. Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci. 2008;13(10):557–65. doi: 10.1016/j.tplants.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 230.Mlynárová L, Nap JP, Bisseling T. The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J. 2007;51(5):874–85. doi: 10.1111/j.1365-313X.2007.03185.x. [DOI] [PubMed] [Google Scholar]
- 231.Farrona S, Hurtado L, Bowman JL, Reyes JC. The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development. 2004;131(20):4965–75. doi: 10.1242/dev.01363. [DOI] [PubMed] [Google Scholar]
- 232.Wagner D, Meyerowitz EM. SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr Biol. 2002;12(2):85–94. doi: 10.1016/s0960-9822(01)00651-0. [DOI] [PubMed] [Google Scholar]
- 233.Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618(1–2):30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]


