Significance
Tyrosyl-DNA phosphodiesterase 1 (TDP1) is a novel repair enzyme that removes 3′-DNA blocking groups generated by topoisomerase cleavage complexes, oxidative DNA lesions, and base alkylation during normal transcription and replication, and by widely used anticancer drugs that induce DNA strand breaks including topoisomerase inhibitors (topotecan, irinotecan, etoposide), bleomycin, and alkylating agents (temozolomide). This report demonstrates that the Drosophila gene glaikit (gkt) is the ortholog of TDP1. It also demonstrates the critical importance of TDP1 (gkt) for normal neurological development and lifespan, potentially linking TDP1 and mitochondria to neurodegenerative diseases.
Keywords: aging, neuroscience, DNA repair, topoisomerase, fly
Abstract
Tyrosyl-DNA phosphodiesterase (TDP1) is a phylogenetically conserved enzyme critical for the removal of blocking lesions at the 3′ ends of DNA or RNA. This study analyzes the Drosophila TDP1 gene ortholog glaikit (gkt) and its possible role(s) in the repair of endogenous DNA lesions and neuroprotection. To do so, we studied a homozygous PiggyBac insertion (c03958) that disrupts the 5′ UTR of gkt. Protein extracts of c03958 flies were defective in hydrolyzing 3′-DNA–tyrosyl residues, demonstrating that gkt is the Drosophila TDP1. Although the mutant is generally healthy and fertile, females exhibit reduced lifespan and diminished climbing ability. This phenotype was rescued by neuronal expression of TDP1. In addition, when c03958 larvae were exposed to bleomycin, an agent that produces oxidative DNA damage, or topoisomerase I-targeted drugs (camptothecin and a noncamptothecin indenoisoquinoline derivative, LMP-776), survivors displayed rough eye patches, which were rescued by neuronal expression of TDP1. Our study establishes that gkt is the Drosophila TDP1 gene, and that it is critical for neuroprotection, normal longevity, and repair of damaged DNA.
DNA repair is indispensable to maintaining genomic integrity against the various endogenous and exogenous agents and enzymes that react with DNA, including reactive oxygen species (1–3), base-damaging agents (4, 5), and chain-terminating nucleosides (6), which yield nonligatable DNA ends. In addition, topoisomerase I (Top1) (7–9) is capable of generating strand breaks with protein-blocked 3′ ends. To ligate and extend the ends of broken DNA, the modified 3′ end must be removed. Tyrosyl-DNA phosphodiesterase (TDP1), first identified in yeast, catalyzes the hydrolysis of 3′-phosphotyrosyl, 3′-phosphoglycolate, and 3′-nucleoside bonds (8, 10–14). The irreversible Top1–DNA complexes and 3′-blocking nucleoside lesions are cleaved by TDP1, leaving 3′-terminal phosphates, which are further processed by polynucleotide kinase phosphatase before ligation and/or extension by polymerases.
Deleterious TDP1 mutants in yeast, mice, and humans are viable but have decreased capacity to repair oxidative and topoisomerase-induced damage (15–17). In humans, a homozygous mutation in TDP1 (A1478G) causes spinocerebellar ataxia with axonal neuropathy (SCAN1) (18). Cells from SCAN1 patients contain enhanced levels of Top1 cleavage complexes (Top1cc) and have defective repair of Top1cc (19, 20) and oxidative damage (21). In mice, TDP1 was recently shown to play a critical role in removing endogenous Top1cc and preventing neurodevelopmental defects (22).
A Drosophila TDP1 ortholog, encoded by the glaikit gene (gtk), has been reported, but its functions in DNA repair have not been studied because the knockout flies were found to be nonviable, with defects in epithelial polarity formation and neuronal development (23). This finding was surprising because TDP1 is not essential for viability in other eukaryotes, including yeast (10, 11), chicken and human cells (4, 24), and mice (16, 25). Here we use an insertional mutant (Exelixis line c03958) to elucidate the role of TDP1 in flies and its critical activity for the repair of 3′-blocked DNA termini and neuroprotection.
Results
The Drosophila Gene for TDP1, gkt, Is Strongly Expressed in Fly Heads.
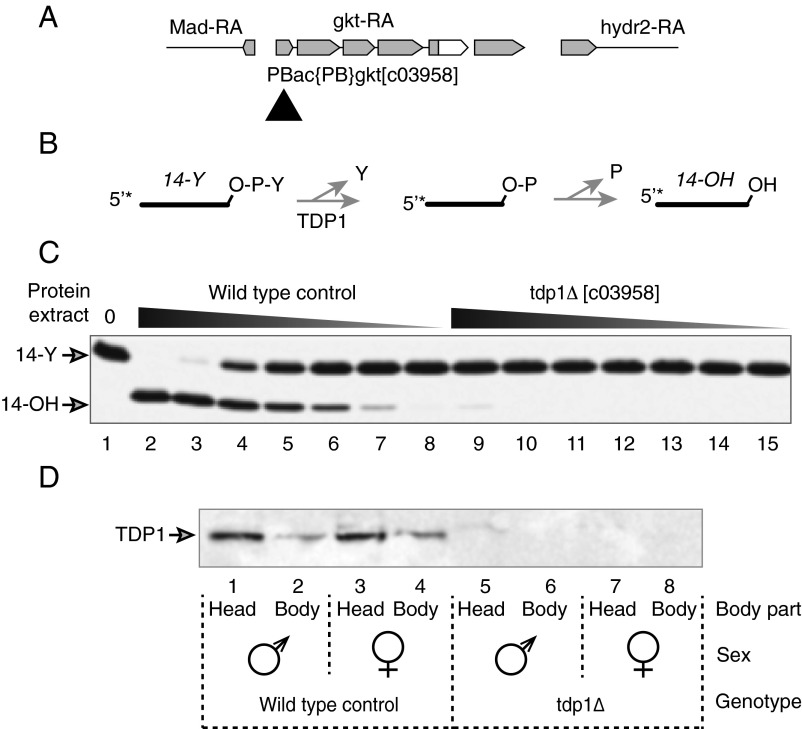
Like other species, Drosophila has only one ortholog of human TDP1 (23). The gene consists of five exons and four introns and encodes a 580-aa protein with catalytic residues identical to those found in the yeast, human, and mouse TDP1 proteins (Fig. S1). An Exelixis PiggyBac insertion, c03958, is located 38 nt upstream of the start codon of TDP1 (Fig. 1A) (23, 26). Flies carrying the insertion are seemingly healthy and fertile without any gross defect. This is in surprising contrast to the phenotypes ascribed to loss of function of the gkt/TDP1 gene in a previous report (23). To determine whether TDP1 function was indeed impaired in c03958 flies, we tested the phosphodiesterase activity of TDP1 in these animals, using a synthesized nucleopeptide that mimics the Top1–DNA covalent complex, with a phosphodiester linkage between the side-chain hydroxyl group of a tyrosine residue and the 3′ end of an oligonucleotide chain (10, 27, 28) (Fig. 1B). Different amounts of protein extracts (243-fold range) from control or c03958 tdp1 mutant flies were used in the assays (Fig. 1C). The 14-mer nucleopeptide was completely converted into the corresponding 3′-hydroxyl oligonucleotides after incubation with 6 μg of control protein extract, and phosphodiesterase activity remained detectable at 81-fold dilution of the control protein extract. However, extract from the TDP1 mutant exhibited little to no significant phosphodiesterase activity even with 6 μg of protein extract. The weak TDP1 activity observed at this high concentration of extract (Fig. 1C, lane 9) is likely caused by other (much less effective and less specific) phosphodiesterase activities. For instance, TDP2 can act as a backup pathway in the absence of TDP1 (29), and overexpression of TDP2 is sufficient to complement the hypersensitivity of TDP1-deficient budding yeast to camptothecin (CPT) (30). Mre11 has also been shown to cleave 3′-phosphotyrosyl–DNA bonds (31) and repair Top1cc in parallel with TDP1 (32–34). These results provide strong evidence that gkt encodes TDP1 in Drosophila.
Fig. 1.
Gkt is the Drosophila TDP1, and disruption of the 5′ UTR of gkt by a PiggyBac insertion (c03958) inactivates 3′-tyrosyl activity. (A) Construct showing the TDP1 gene structure (exons are in gray and insertion co3958 is indicated by a black triangle). RA, transcripts. (B) Scheme for TDP1 assay (4). (C) Extracts from wild-type Drosophila adults (lanes 2–8) hydrolyze the 3′-DNA–tyrosyl residue, and this activity is defective in extracts from c03958 flies (lanes 9–15). Proteins per reaction were serially diluted threefold in consecutive lanes, starting at 6 µg in lanes 2 and 9. (D) TDP1 protein is mainly expressed in fly heads and absent in c03958 flies. Twenty micrograms of protein extract was loaded per SDS/PAGE lane, and TDP1 was detected by Western blotting.
To directly examine the effects of the c03958 mutation on TDP1 expression, we generated an antibody against an 18-mer peptide antigen (LTPYAPDDKPFLMDYLQG) corresponding to the TDP1 C terminus (Fig. S1). Consistent with the distribution of TDP1 mRNA, which is enriched in fly brain, thoracicoabdominal ganglion, and ovary (flyatlas.org), we found that TDP1 protein is expressed in the fly head and body, with the head exhibiting higher levels (Fig. 1D). In contrast, no TDP1 protein was detected in the c03958 mutant (tdp1Δ), consistent with the conclusion that gkt exclusively encodes Drosophila TDP1. Together, our results indicate that the insertion c03958 disrupts the expression of TDP1 in Drosophila melanogaster, and establish that gkt is the Drosophila TDP1 ortholog.
TDP1 Mutant Females Have Short Lifespans and Defective Climbing Ability.
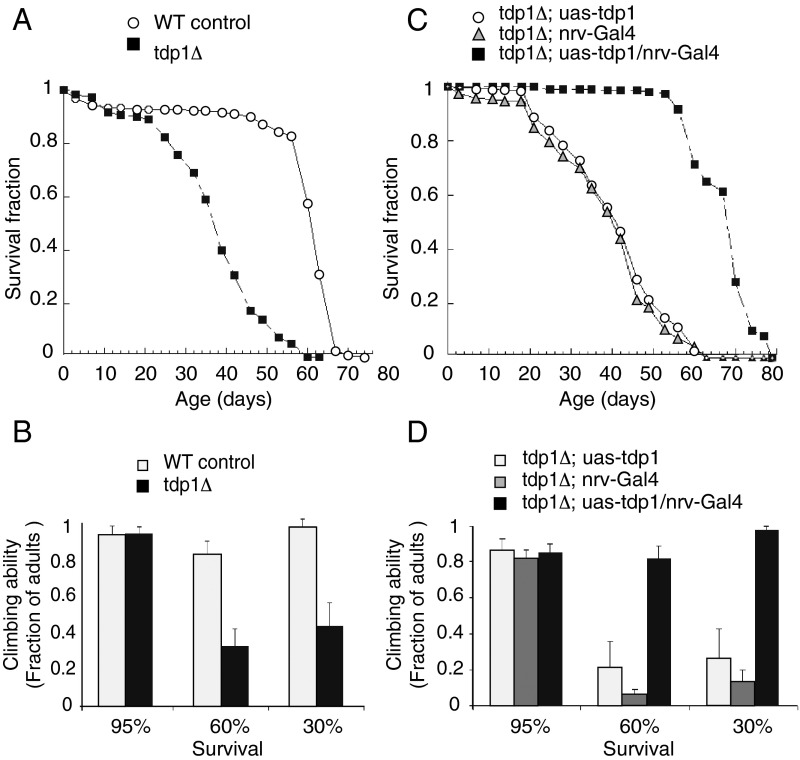
Top1 cleavage complexes accumulate at damaged DNA sites (8, 35) under normal conditions (22, 36), and TDP1 repairs not only Top1-mediated DNA damage but also endogenous oxidative DNA lesions (2, 8, 12). Both of these types of damage can result in increased mortality, and we therefore tested whether TDP1 influences the lifespan of tdp1Δ flies. Fig. 2A shows that tdp1Δ female flies have significantly reduced lifespan.
Fig. 2.
TDP1 disruption in the nervous system shortens Drosophila lifespan and reduces climbing ability. (A and B) TDP1 mutant females have reduced lifespan and impaired climbing ability. (C and D) Expression of TDP1 in the nervous system restores lifespan (C) and climbing ability (D) to normal levels. Error bars represent standard deviations.
Tdp1Δ females also exhibited decreased climbing ability (Fig. 2B). At the age where 95% of the mutant fly population survived, tdp1Δ females had similar climbing ability as their control counterparts, but at ages corresponding to both 60% and 30% survival levels, tdp1Δ females showed reduced climbing ability (around 40% of controls; Fig. 2B).
Next, we determined whether the reduced lifespan and climbing phenotypes could be rescued by TDP1/gkt expression in tdp1Δ flies. We applied the UAS/Gal4 system to express TDP1 in the nervous system. Expression of TDP1 driven by NirvanaGal4, which is known to be expressed exclusively in the nervous system of Drosophila (37), not only restored the mean lifespan of mutant flies (Fig. 2C) but also restored their climbing ability at both 60% and 30% survival levels (Fig. 2D). These results indicate that TDP1 plays a housekeeping role for the maintenance of normal functions in the nervous system.
Late Retinal Development Is Altered by Bleomycin and Top1 Inhibitors in tdp1Δ Flies and Is Partially Rescued by Expression of gkt in the Nervous System.
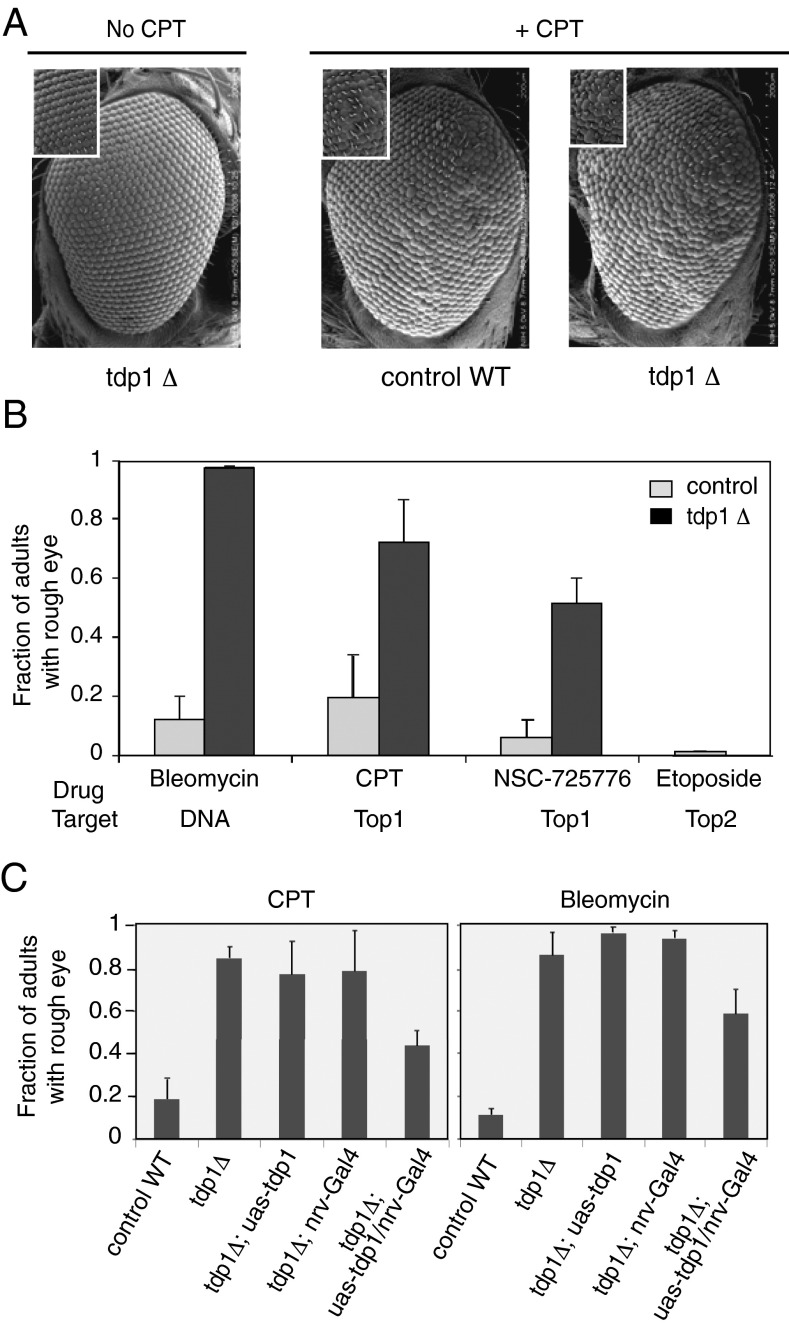
The compound eye of D. melanogaster contains 800 ommatidia, or individual units, each containing 8 photoreceptor cells, support cells, pigment cells, and a cornea. Defects in eye development, ommatidial assembly, or ommatidial spacing cause a rough eye phenotype (38). It has been reported that disruption of DNA repair genes results in rough eyes in Drosophila due to apoptosis (39). Eyes of tdp1Δ flies were mostly normal in the absence of treatment (Fig. 3A, Left), but after feeding larvae 25 µM CPT, both control and tdp1Δ flies displayed rough eyes (Fig. 3A, Right). However, the rough eye phenotype of CPT-treated tdp1Δ flies was more severe than that of the corresponding controls (Fig. 3 A and B).
Fig. 3.
Eye alterations in TDP1 mutants treated with bleomycin and Top1 inhibitors. (A) Representative SEM images of fly eyes treated with or without CPT. (B) Bleomycin (11 µM) and the Top1 poisons CPT and NSC-725776 (LMP-776) (25 µM) induce the rough eye phenotype, whereas the Top2 poison etoposide (25 µM) does not. (C) Expression of TDP1 cDNA in the nervous system partially rescues the rough eye phenotype of TDP1 mutant adult flies treated with CPT or bleomycin. Error bars represent standard deviations.
We expanded the studies to two other DNA-damaging agents known to depend on TDP1 for repair, bleomycin (15, 21) and the noncamptothecin Top1 inhibitor NSC-725776 (LMP-776), which is in clinical trials as an anticancer drug (40). Adult tdp1Δ flies, which as larvae were fed bleomycin, CPT, or NSC-725776, exhibited a more pronounced rough eye phenotype than wild-type control flies (Fig. 3B). On the contrary, there was no detectable rough eye phenotype in TDP1 adults fed as larvae with etoposide, a Top2 poison (40, 41), for which DNA repair is primarily carried out by TDP2 (8, 30, 42). Our results demonstrate that TDP1 contributes significantly to epithelial cell maintenance in the face of Top1-mediated and oxidative DNA damage.
To investigate whether expression of TDP1 in the nervous system can rescue the rough eye phenotype induced by CPT and bleomycin, we examined tdp1Δ and TDP1-complemented flies after feeding larvae CPT or bleomycin. Expression of TDP1 in the nervous system of tdp1Δ mutant flies partially ameliorated the rough eye phenotype of larvae fed either CPT or bleomycin, with the fraction of rough-eyed adults decreasing by 33% and 48% when larvae were fed 11 µM bleomycin and 25 µM CPT, respectively (Fig. 3 C and D). Expression of TDP1 in the nervous system did not bring the fraction of adults with rough eyes to that of controls treated either with CPT (44 ± 9% vs. 19 ± 8%) or bleomycin (58 ± 12% vs. 11 ± 4%). These results indicate that expression of TDP1 in the nervous system rescues the rough eye phenotype, but only partially. Because of the cellular complexity of the Drosophila eye, it is likely that the partial correction of the rough eye phenotype is due to TDP1 inactivation outside of neurons, such as in the accessory cells.
Discussion
The present study demonstrates that gkt is the tyrosyl-DNA phosphodiesterase 1 of D. melanogaster and that it is not essential for viability but is important for nervous system maintenance during development and for response to DNA damage. We show that Drosophila TDP1 hydrolyzes tyrosyl–DNA covalent bonds, just as previously reported for TDP1 in other species (2, 11, 16, 25, 43), which is consistent with the high conservation of TDP1 active site residues across species (Fig. S1). No TDP1 protein and no TDPl activity were detected in gkt/TDP1 mutant flies by Western blotting and biochemical assays, demonstrating that gkt/TDP1 repairs Top1-induced and oxidative DNA damages in flies.
Dunlop and coworkers reported that loss of gkt during neuronal development results in severe CNS architecture disruption and embryonic lethality (23). However, we find that TDP1/gktΔ flies are viable and without detectable developmental defect. Although we are unable to explain their results, which were based on the study of other mutants in which TDP1 activity was not assayed, we note that our results are consistent with those obtained with two TDP1−/− mouse models. In one, the mouse phenotype is indistinguishable from wild-type, physically, histologically, behaviorally, and electrophysiologically (25). In the other, mice display age-dependent progressive cerebellar atrophy (16), which is aggravated upon ATM inactivation (22). Both TDP1−/− mouse models show hypersensitivity to CPT as well as to bleomycin, which is consistent with our results in flies where, in addition, we observed hypersensitivity to the noncamptothecin Top1 inhibitor NSC-725776. Top1 inhibitors and bleomycin produce different forms of 3′-end DNA damage. CPT selectively traps the catalytic intermediates of the Top1–DNA reaction, the cleavage complexes (40, 44), whereas bleomycin causes strand breaks with 3′-phosphoglycolates and abasic sites due to hydrogen abstraction from the C-4′ position of deoxyribose sugars in DNA (45).
TDP1 mutant females, but not TDP1 mutant males, showed a shortened lifespan. This notable difference is not due to differential expression of TDP1 in males and females (Fig. 1D). Different lifespans for the two sexes are not uncommon. One combination of D. melanogaster insulin receptor (inr) alleles, InRp5545/InRE19, extends lifespan for females by 85% but only by 43% for males (46). Mutations in CHICO, the gene encoding the Drosophila insulin receptor substrate homolog (47), extend the lifespan of adult females by 48% but not that of males (48). The mechanisms underlying these phenotypes are not fully understood, but may be related to the higher metabolism, selective hormone-dependent gene expression, and larger size of females. Lifespan in all organisms is determined genetically and influenced by the environment. The free radical theory of aging (49, 50) proposes that accumulation of oxidative damage to cellular components leads to the progressive dysfunction and death of cells, organs, and ultimately the organism. More recent studies in diverse organisms including nematodes, flies, and rodents suggest that the central nervous system controls lifespan (51, 52). The high metabolic activity of mature neurons generates large amounts of reactive oxygen species with DNA-damaging capacity. Defects in DNA damage repair in the nervous system may lead to a shorter lifespan. The nervous system coordinates the defenses of the whole body to endogenous and exogenous insults by modulating the activities of neuroendocrine pathways and the autonomic nervous system (53). Hydrolysis of the 3′-terminal glycolate-DNA phosphodiester by TDP1 repairs oxidative damage (2, 4, 5, 15, 21). Restoration of the lifespan of TDP1 mutant females by expression of TDP1 with nrv2-Gal4 in the central nervous system suggests that resistance to oxidative stress in the nervous system is a prerequisite for normal lifespan. Our conclusions converge with those from a recent study demonstrating the critical importance of TDP1 in mice in preserving genomic integrity and preventing disease in the nervous system (22).
Materials and Methods
Fly Strains.
Flies were grown on cornmeal/molasses agar [10% (wt/vol) sucrose, 2% (wt/vol) yeast, 3.3% (wt/vol) cornmeal, 1% agar] at 25 °C and 50% humidity under a 12-h/12-h light/dark cycle using standard techniques. Line c03958 and the nrv2-Gal4 driver line were obtained from the Bloomington Drosophila Stock Center at Indiana University.
Larval Feeding.
Eggs from Drosophila were collected during a 6-h period from culture bottles containing standard medium. After 72 h, groups of 100 larvae were washed with 20% sucrose solution and transferred to standard plastic vials containing 1.5 g of Drosophila Instant Medium (Carolina Biological Supply) rehydrated with 5 mL of the respective test solutions. The larvae were kept on this medium until the emergence of adult flies (54).
Transgenic Flies.
TDP1 cDNA was obtained from GDRC (Drosophila Genomics Resource Center) (LD37277) and cloned into pUAST with PCR primers Tdp1F: 5′-CGGCAGATCTCGCGCTTTGTGTTTTTTATTTGC-3′ and Tdp1R: 5′-AGTGGGCGGCCGCCGATTTAGGTGACACTATAGAACTCGAG-3′. The following genetic crossing schemes were followed to generate stocks for the Tdp1 rescue experiments. Sp/CyO;Dr/TM3Sb virgin females were crossed to a single male carrying a UAS-TDP1 transgene or nrv2-Gal4 on the third chromosome. To double balance, male or virgin female CyO, Dr progeny flies were crossed to male or virgin female Sp, Dr flies. The resulting progeny that carried both Sp and Cyo was crossed to each other to generate Sp/CyO;UAS-TDP1 or Sp/CyO;nrv2-Gal4 stocks that were maintained. Males from these stocks were then crossed to gktc03958;Dr/TM3Sb virgin females. Non-Sp, CyO, non-Dr progeny were self-crossed to yield gktc03958;UAS-TDP1 or gktc03958;nrv2-Gal4 stocks to be used for rescue assays.
Protein Extraction, Western Blotting, and TDP1 Activity Assays.
Two to 20 flies were collected under brief CO2 anesthesia in a 1.5-mL Eppendorf tube. These flies were homogenized in 20–200 μL of 0.02 M Hepes (pH 7.5) containing 0.1 M KCl, 5% (vol/vol) glycerol, 10 mM EDTA, 0.1% Triton X-100, and a mixture of protease inhibitors (Roche). The homogenate was centrifuged at 1,000 × g for 10 min at 4 °C. Supernatant was transferred to a clean tube. The protein concentration was measured by a Bio-Rad protein assay kit (500-0006) on an Ultrospec 2100 pro spectrophotometer (Amersham Biosciences). Twenty micrograms of protein from the final supernatant was mixed with Novex Tris⋅glycine SDS sample buffer or NuPage LDS (lithium dodecyl sulfate) sample buffer (Invitrogen), heated at 75–85 °C for 5 min, and subjected to electrophoresis. After transfer to a nitrocellulose membrane, blots were probed with rabbit anti-TDP1 polyclonal antibody at 1:3,000 dilution. The secondary antibody was peroxidase-linked donkey anti-IgG, obtained from Jackson ImmunoResearch Laboratories and used according to the supplier’s instructions. Blots were developed with the ECL detection system (GE Healthcare). TDP1 activity assays were performed using a 5′-32P end-labeled 14-mer oligonucleotide with a 3′-phosphotyrosine (Midland) (27). Reactions were performed in 80 mM KCl, 2 mM EDTA, 1 mM DTT, 40 mg/mL BSA, 50 mM Tris⋅HCl (pH 7.5), and 0.01% Tween 20. After a 15-min incubation at room temperature, the reactions were terminated by adding one volume of gel loading buffer [96% (vol/vol) formamide, 10 mM EDTA, 1% (wt/vol) bromophenol blue]. Double-stranded substrates were heated at 95 °C for 3 min before loading. Samples were subjected to 16% polyacrylamide gel electrophoresis and dried and exposed on PhosphoImager screens. Imaging and quantitations were done using a Typhoon 8600 and ImageQuant software (GE Healthcare).
Longevity and Climbing Ability.
The longevity assay was performed as described (55). One hundred eighty 1- to 2-d-old TDP1 mutants or control flies were collected under brief CO2 anesthesia. The collected flies were separated by sex and kept in groups of 30 individuals in vials with standard food. The flies were transferred to new vials every 2 or 3 d and the number of dead flies in each vial was scored. This process was continued until all the flies died, and the percentage of flies alive at each time point was graphed. The climbing ability assay was performed as described (56). Aged flies at certain time points were put in an empty Drosophila vial (Genesee Scientific; 32-110) and gently tapped to the bottom of the vial. A line 3 inches from the bottom of the vial was drawn. The number of flies climbing over the line was counted after 30 s of climbing. Statistical analysis was performed with SPSS software (IBM).
Scanning Electron Microscopy.
For SEM studies, adult flies of the desired genotypes were decapitated. Heads were fixed in 2.5% glutaraldehyde overnight at 4 °C, washed three times for 30 min with 0.1 M PBS, dehydrated in ascending acetone grades, and then critical point-dried. They were then mounted on studs in the desired orientation under a stereobinocular microscope and coated with gold (thickness 30–35 nm). Scanning was done in SEM mode on an Amray 1820D electron microscope at 5 kV.
Supplementary Material
Acknowledgments
The authors thank Dr. Alena Naumova and Dr. Christophe Marchand (Laboratory of Molecular Pharmacology, Developmental Therapeutics Branch) for help in running the TDP1 activity experiments. They also thank Dr. Shar-yin N. Huang (Laboratory of Molecular Pharmacology, Developmental Therapeutics Branch) and Benjamin H. White (Neural Function Section, National Institute of Mental Health) for critical reading and editing of the manuscript and for constructive suggestions. This work was supported by the NIH Intramural Program, National Institute of Diabetes and Digestive and Kidney Diseases, and the Center for Cancer Research of the National Cancer Institute (Z01 BC 006161).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
4Deceased June 12, 2011.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415011111/-/DCSupplemental.
References
- 1.McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- 2.Inamdar KV, et al. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J Biol Chem. 2002;277(30):27162–27168. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 3.El-Khamisy SF, Hartsuiker E, Caldecott KW. TDP1 facilitates repair of ionizing radiation-induced DNA single-strand breaks. DNA Repair (Amst) 2007;6(10):1485–1495. doi: 10.1016/j.dnarep.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 4.Murai J, et al. Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells. J Biol Chem. 2012;287(16):12848–12857. doi: 10.1074/jbc.M111.333963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alagoz M, Wells OS, El-Khamisy SF. TDP1 deficiency sensitizes human cells to base damage via distinct topoisomerase I and PARP mechanisms with potential applications for cancer therapy. Nucleic Acids Res. 2014;42(5):3089–3103. doi: 10.1093/nar/gkt1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang SY, et al. TDP1 repairs nuclear and mitochondrial DNA damage induced by chain-terminating anticancer and antiviral nucleoside analogs. Nucleic Acids Res. 2013;41(16):7793–7803. doi: 10.1093/nar/gkt483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 8.Pommier Y, et al. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2) DNA Repair (Amst) 2014;19:114–129. doi: 10.1016/j.dnarep.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vance JR, Wilson TE. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc Natl Acad Sci USA. 2002;99(21):13669–13674. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang S-W, et al. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc Natl Acad Sci USA. 1996;93(21):11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286(5439):552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 12.Interthal H, Chen HJ, Champoux JJ. Human Tdp1 cleaves a broad spectrum of substrates including phosphoamide linkages. J Biol Chem. 2005;280(43):36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das BB, Dexheimer TS, Maddali K, Pommier Y. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci USA. 2010;107(46):19790–19795. doi: 10.1073/pnas.1009814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das BB, et al. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 2014;42(7):4435–4449. doi: 10.1093/nar/gku088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Pouliot JJ, Nash HA. The role of TDP1 from budding yeast in the repair of DNA damage. DNA Repair (Amst) 2004;3(6):593–601. doi: 10.1016/j.dnarep.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 16.Katyal S, et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26(22):4720–4731. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc Natl Acad Sci USA. 2001;98(21):12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takashima H, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32(2):267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 19.Miao ZH, et al. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair (Amst) 2006;5(12):1489–1494. doi: 10.1016/j.dnarep.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 20.El-Khamisy SF, et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434(7029):108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 21.Zhou T, et al. Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1) Nucleic Acids Res. 2005;33(1):289–297. doi: 10.1093/nar/gki170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katyal S, et al. Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes. Nat Neurosci. 2014;17(6):813–821. doi: 10.1038/nn.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunlop J, Morin X, Corominas M, Serras F, Tear G. glaikit is essential for the formation of epithelial polarity and neuronal development. Curr Biol. 2004;14(22):2039–2045. doi: 10.1016/j.cub.2004.10.048. [DOI] [PubMed] [Google Scholar]
- 24.Gao R, et al. Epigenetic and genetic inactivation of tyrosyl-DNA-phosphodiesterase 1 (TDP1) in human lung cancer cells from the NCI-60 panel. DNA Repair (Amst) 2014;13:1–9. doi: 10.1016/j.dnarep.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano R, et al. Spinocerebellar ataxia with axonal neuropathy: Consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007;26(22):4732–4743. doi: 10.1038/sj.emboj.7601885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thibault ST, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36(3):283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 27.Plo I, et al. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair (Amst) 2003;2(10):1087–1100. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 28.Dexheimer TS, Stephen AG, Fivash MJ, Fisher RJ, Pommier Y. The DNA binding and 3′-end preferential activity of human tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 2010;38(7):2444–2452. doi: 10.1093/nar/gkp1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng Z, et al. TDP2 promotes repair of topoisomerase I-mediated DNA damage in the absence of TDP1. Nucleic Acids Res. 2012;40(17):8371–8380. doi: 10.1093/nar/gks622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461(7264):674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 31.Sacho EJ, Maizels N. DNA repair factor MRE11/RAD50 cleaves 3′-phosphotyrosyl bonds and resects DNA to repair damage caused by topoisomerase 1 poisons. J Biol Chem. 2011;286(52):44945–44951. doi: 10.1074/jbc.M111.299347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu C, Pouliot JJ, Nash HA. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc Natl Acad Sci USA. 2002;99(23):14970–14975. doi: 10.1073/pnas.182557199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng C, Brown JA, You D, Brown JM. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics. 2005;170(2):591–600. doi: 10.1534/genetics.104.028795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malik M, Nitiss JL. DNA repair functions that control sensitivity to topoisomerase-targeting drugs. Eukaryot Cell. 2004;3(1):82–90. doi: 10.1128/EC.3.1.82-90.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pourquier P, Pommier Y. Topoisomerase I-mediated DNA damage. Adv Cancer Res. 2001;80:189–216. doi: 10.1016/s0065-230x(01)80016-6. [DOI] [PubMed] [Google Scholar]
- 36.Krawczyk C, Dion V, Schär P, Fritsch O. Reversible Top1 cleavage complexes are stabilized strand-specifically at the ribosomal replication fork barrier and contribute to ribosomal DNA stability. Nucleic Acids Res. 2014;42(8):4985–4995. doi: 10.1093/nar/gku148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun B, Xu P, Salvaterra PM. Dynamic visualization of nervous system in live Drosophila. Proc Natl Acad Sci USA. 1999;96(18):10438–10443. doi: 10.1073/pnas.96.18.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolff T, Ready DF. Cell death in normal and rough eye mutants of Drosophila. Development. 1991;113(3):825–839. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- 39.Brodsky MH, et al. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol. 2004;24(3):1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17(5):421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9(5):338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maede Y, et al. Differential and common DNA repair pathways for topoisomerase I- and II-targeted drugs in a genetic DT40 repair cell screen panel. Mol Cancer Ther. 2014;13(1):214–220. doi: 10.1158/1535-7163.MCT-13-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macovei A, Balestrazzi A, Confalonieri M, Carbonera D. The tyrosyl-DNA phosphodiesterase gene family in Medicago truncatula Gaertn.: Bioinformatic investigation and expression profiles in response to copper- and PEG-mediated stress. Planta. 2010;232(2):393–407. doi: 10.1007/s00425-010-1179-9. [DOI] [PubMed] [Google Scholar]
- 44.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260(27):14873–14878. [PubMed] [Google Scholar]
- 45.Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: Bleomycin, neocarzinostatin and other enediynes. Mutat Res. 1996;355(1-2):71–89. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- 46.Tatar M, et al. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292(5514):107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 47.Böhni R, et al. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97(7):865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- 48.Clancy DJ, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292(5514):104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 49.Harman D. Aging: A theory based on free radical and radiation chemistry. J Gerontol. 1956;11(3):298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 50.Harman D. Free radical theory of aging. Mutat Res. 1992;275(3-6):257–266. doi: 10.1016/0921-8734(92)90030-s. [DOI] [PubMed] [Google Scholar]
- 51.Mattson MP, Duan W, Maswood N. How does the brain control lifespan? Ageing Res Rev. 2002;1(2):155–165. doi: 10.1016/s1568-1637(01)00003-4. [DOI] [PubMed] [Google Scholar]
- 52.Warner HR. Longevity genes: From primitive organisms to humans. Mech Ageing Dev. 2005;126(2):235–242. doi: 10.1016/j.mad.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 53.Buijs RM, Van Eden CG. The integration of stress by the hypothalamus, amygdala and prefrontal cortex: Balance between the autonomic nervous system and the neuroendocrine system. Prog Brain Res. 2000;126:117–132. doi: 10.1016/S0079-6123(00)26011-1. [DOI] [PubMed] [Google Scholar]
- 54.Torres C, Creus A, Marcos R. Genotoxic activity of four inhibitors of DNA topoisomerases in larval cells of Drosophila melanogaster as measured in the wing spot assay. Mutat Res. 1998;413(2):191–203. doi: 10.1016/s1383-5718(98)00031-x. [DOI] [PubMed] [Google Scholar]
- 55.Lavara-Culebras E, Muñoz-Soriano V, Gómez-Pastor R, Matallana E, Paricio N. Effects of pharmacological agents on the lifespan phenotype of Drosophila DJ-1beta mutants. Gene. 2010;462(1-2):26–33. doi: 10.1016/j.gene.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 56.Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404(6776):394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.