A family of drugs reduces the growth response, the lipid modifications, and the pattern of gene expression triggered by inorganic phosphate starvation.
Abstract
Inorganic phosphate (Pi) is present in most soils at suboptimal concentrations, strongly limiting plant development. Plants have the ability to sense and adapt to the surrounding ionic environment, and several genes involved in the response to Pi starvation have been identified. However, a global understanding of the regulatory mechanisms involved in this process is still elusive. Here, we have initiated a chemical genetics approach and isolated compounds that inhibit the response to Pi starvation in Arabidopsis (Arabidopsis thaliana). Molecules were screened for their ability to inhibit the expression of a Pi starvation marker gene (the high-affinity Pi transporter PHT1;4). A drug family named Phosphatin (PTN; Pi starvation inhibitor), whose members act as partial suppressors of Pi starvation responses, was thus identified. PTN addition also reduced various traits of Pi starvation, such as phospholipid/glycolipid conversion, and the accumulation of starch and anthocyanins. A transcriptomic assay revealed a broad impact of PTN on the expression of many genes regulated by low Pi availability. Despite the reduced amount of Pi transporters and resulting reduced Pi uptake capacity, no reduction of Pi content was observed. In addition, PTN improved plant growth; this reveals that the developmental restrictions induced by Pi starvation are not a consequence of metabolic limitation but a result of genetic regulation. This highlights the existence of signal transduction pathway(s) that limit plant development under the Pi starvation condition.
Inorganic phosphate (Pi) is a crucial plant macronutrient. In most soils, this element is present in limiting amounts. Several factors contribute to restrict its availability to plants: assimilation by microbes and poor mobility due to its strong interaction with many cations (Shen et al., 2011). As a consequence, Pi distribution is very heterogeneous in soils and can thus be considered one of the least available plant macronutrients (Raghothama, 1999). To cope with this situation, plants have developed various mechanisms to improve Pi recovery and reduce its consumption.
Pi starvation limits plants development and modifies their architecture. Pi starvation also leads to the reduction of primary root development and to an increase in lateral roots development (Linkohr et al., 2002; López-Bucio et al., 2002; Ticconi and Abel, 2004a; Reymond et al., 2006; Sánchez-Calderón et al., 2006; Svistoonoff et al., 2007). This, in turn, promotes an increase in the root-to-shoot ratio due to modifications of the root architecture that favor exploring superficial soil layers, e.g., the soil layers that contain more Pi (for recent review, see Abel, 2011; Péret et al., 2011). Several of the genes and quantitative trait loci that control these modifications have been characterized in Arabidopsis (Arabidopsis thaliana), including low Pi root1 (Reymond et al., 2006; Svistoonoff et al., 2007), Pi deficiency response2 (PDR2; Ticconi et al., 2004b), and Low phosphorus insensitive (LPI; Sánchez-Calderón et al., 2006). This phenomenon appears to be mainly locally regulated by the external Pi concentration (Drew, 1975; Linkohr et al., 2002; Svistoonoff et al., 2007; Thibaud et al., 2010).
Systemic regulation also occurs (Thibaud et al., 2010) and affects metabolic adaptations that result from Pi starvation, such as the secretion of phosphatases (Duff et al., 1994; Tran et al., 2010a, 2010b), organic acids (Narang et al., 2000), or ribonucleases to solubilize soil-associated Pi (either located in the organic Pi pool or chelated by cations; Misson et al., 2005). Pi starvation also triggers many metabolic changes in planta to conserve and recycle Pi, such as the remobilization of membrane phospholipids (Andersson et al., 2003, 2005; Jouhet et al., 2004; Nussaume et al., 2011b; Nakamura, 2013). The conservation of ATP through the use of alternative enzymatic reactions within the glycolytic and respiratory pathways has also been suggested (Misson et al., 2005).
Due to their diversity, these metabolic modifications can be expected to be controlled by several elements. In particular, genetic screens have identified Myb transcription factors from the Phosphate Starvation Response1 (PHR1) family, which are major regulators of all of these responses (Rubio et al., 2001; Bustos et al., 2010; Thibaud et al., 2010). Although several key genes involved in this regulatory cascade have been identified (Chiou and Lin, 2011), a clearer global view of these processes is still required.
Part of the developmental limitation phenotype observed during Pi starvation results from genetic control, and not only from metabolic limitation as previously assumed (Svistoonoff et al., 2007). This hypothesis is supported by the phenotype of mutants that disconnect the external Pi concentration from root growth (lpr1, lpi, and pdr2; for review, see Abel, 2011; Péret et al., 2011) or the internal Pi concentration from shoot growth (such as the Phosphate1 [PHO1] underexpressor; Rouached et al., 2011). Nevertheless, the molecular mechanisms of this control remain largely unknown.
In this study, a chemical genetic approach was used to improve our knowledge on this pathway. A chemical screen for regulators of PHT1;4, expression, a high-affinity Pi transporter used as an early marker of Pi starvation (Karthikeyan et al., 2002; Misson et al., 2004; Hirsch et al., 2011) identified a small family of compounds that were called Phosphatins (PTNs; Pi starvation inhibitor). PTN molecules were shown to alleviate numerous Pi starvation responses and provide a measure of the importance of genetic versus metabolic impacts of Pi starvation. Moreover, due to the general impact of PTN on both morphological and biochemical traits associated with Pi starvation, this work reveals putative links between local and systemic regulation.
RESULTS
Identification of Molecules That Inhibit the Low-Pi-Induced PHT1;4 Gene
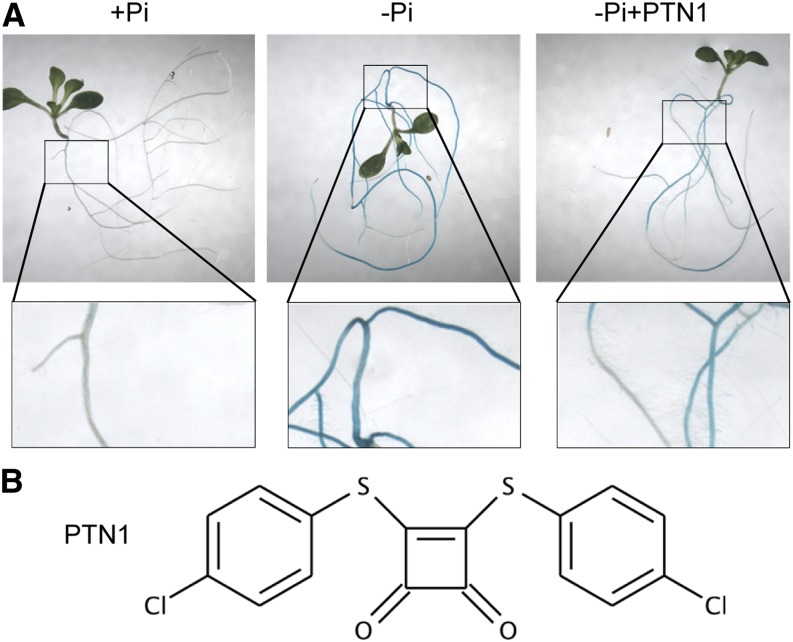
The commercially available Library of Active Compounds on Arabidopsis (University of California, Riverside) containing 3,580 biologically active compounds (Zhao et al., 2007) was screened at 25 µm to identify drugs inhibiting the expression of PHT1;4, a gene strongly induced by Pi starvation (for a recent review, see Nussaume et al., 2011a). The GeneTrap line used for the screen, pht1;4-1, contains a GUS transcriptional fusion with the endogenous PHT1;4 gene, which allows monitoring of its expression (Misson et al., 2004; Hirsch et al., 2011). For the screen, seedlings were grown on Pi-rich medium (+Pi, 500 µm) for the first 5 d. Following this, seedlings were transferred on to Pi-limiting medium (–Pi, 15 µm) with or without the compounds for an additional 5 d. GUS staining was subsequently performed to monitor the expression of PHT1.4.
This screen identified 21 compounds that partially or totally inhibited PHT1;4::GUS expression. In most cases, the reduced expression of the marker was due to the presumably toxic effects of the drugs (assessed by the growth arrest of the plantlets). Two compounds were screened in which the reduced expression of the reporter gene was not accompanied by toxicity (see below), and these were selected for further study. These compounds are commercially available from the Maybridge library (MWP00917 and KM03772) and were respectively named Phosphatin1 (PTN1) and PTN2 (Fig. 1; Supplemental Table S1). Both of these compounds, when assayed with PHT1;4 expression, acted in a dose-dependent manner. PTN1 was observed to be twice as potent as PTN2 (Supplemental Fig. S1A). Interestingly, the IC50 (defined as one-half of the maximal inhibitory concentration of a substance reducing PHT1;4 Pi starvation marker expression) of PTN1 was similar to that of PTN2 (Supplemental Table S1). This can be explained by a maximum effect of PTN2, which turned to be 50% reduced when compared with PTN1 maximal impact (Supplemental Fig. S1B). Although PTN reduced PHT1;4 expression, it stimulated root growth of plantlets in Pi-deficient medium. But maximal PTN effects on both phenotypes were observed with similar concentrations of drugs (Supplemental Fig. S1, A and B).
Figure 1.
Identification of drugs altering PHT1;4 expression. A, Screen used to identify PTN molecules. GUS staining of pht1;4-1 plantlets germinated and grown 5 d on +Pi (500 µm) and transferred for 5 d on +Pi, –Pi (15 µm), or –Pi plus PTN1 (25 µm). B, Chemical structure of PTN1.
Importance of the 4-Chlorothiophenol Motif to the Drug Effect
Several commercially available structural analogs of PTN1 were tested to narrow down the active chemical motif present in the structure of this compound. Both PTN1 and PTN2 exhibit a common structure composed of a benzene aromatic ring with sulfur and chlorine residues in para positions (Supplemental Table S1). The 4-chlorothiophenol compound (named PTN3 here) was therefore assayed (Supplemental Table S1). Three other compounds including this motif were also examined, and their characteristics are summarized in Supplemental Table S1. All of these compounds have a low Mr, smaller than 500 g mol–1. Each compound also respects Lipinski’s rule of five, which can be used to determine whether a substance is a drug (Lipinski, 2004; Supplemental Table S1). PTN3 and PTN4 repressed PHT1;4 expression, although they require different concentrations (100 and 10 µm, respectively) to promote effects similar to those observed with PTN1 and PTN2 application (data not shown). PTN4 contains two 4-chlorothiophenol motifs, just like PTN1. This could explain why PTN4 was observed to be active at a lower concentration. The IC50 of these compounds provides an indication of their gradual efficiency (Supplemental Table S1). Altogether, this analysis revealed that 4-chlorothiophenol is the minimal active motif. The importance of the sulfur environment is also underscored by PTN5 and PTN6, which are inactive and in which the S residue links to a methyl or oxygen group (Supplemental Table S1). PTN1 (the first identified compound) and PTN4 (commercially available) both triggered significant effects on plant growth. Therefore, both PTN1 and PTN4 were selected for the remainder of this study. It can be noticed that maximal impact of all active drugs (PTN1–PTN4) measured by length of primary root with optimal drug addition was highly significant; a similar higher effect was obtained with PTN1 and PTN3, whereas a 40% to 50% milder phenotype was observed with PTN4 and PTN2 (data not shown).
PTN Alters the Pi Transporter Regulations Triggered by Pi Starvation
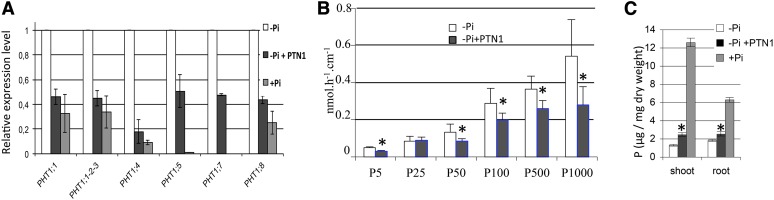
The reduced level of PHT1;4 expression with PTN1 as assayed by the GUS reporter was confirmed by quantitative PCR (qPCR) experiments (Fig. 2A) and by global transcriptomic analysis (Supplemental Table S2). We next investigated the impact of PTN1 on the expression of several additional PHT1 genes. As shown in Figure 2A, PTN1 strongly reduced the impact of Pi starvation on the induction level of all transcripts tested (by at least 50%).
Figure 2.
PTN1 modulates Pi uptake. A, Impact of PTN1 treatment (40 µm) on various PHT1 transcript levels. Measurements are from qPCR experiments on root mRNA. B, Uptake experiments performed with different Pi concentrations. C, Impact of PTN1 on total phosphorus content. Measurements were performed using ICP assays. Asterisks represent significant difference between –Pi and –Pi plus PTN1 40 µm (Student’s t test, P < 0.01). Seven-day-old (A and B) or 10-d-old (C) plantlets were used. White bars indicate –Pi, black bars indicate –Pi plus PTN1, and gray bars indicate +Pi. [See online article for color version of this figure.]
The characterization of Pi uptake capacity was then investigated by measuring total Pi influx to evaluate putative impact of these transcriptional regulations resulting from PTN addition. Consistent with PHT1 transporters being induced by Pi starvation (Misson et al., 2005; Kanno et al., 2012), a higher Pi influx capacity was observed in Pi-starved seedlings (compare –Pi with +Pi in Fig. 2B). Interestingly, PTN1 reduced the Pi uptake by 25% in –Pi, highlighting a putative impact of the observed transcriptomic modifications in term of PHT1 transporter net activity. To investigate further PTN impact on the PHT1 family, we measured the Vmax and the Km associated with Pi uptake to access the affinity and the maximum transport rate of Pi transport (Fig. 2C; Table I). The kinetics of low- and high-affinity Pi transport systems were characterized (for review, see Nussaume et al., 2011b). Neither PTN1 (nor PTN4) drugs modified both transport systems, which were still present in treated plants (similar Km value; Table I). Only the Vmax appeared affected by the drug treatments. This indicated that reduction of Pi uptake observed after PTN addition resulted from a decrease of Pi transporter amount per root length and not from the modification of affinity.
Table I. Effect of PTN1 and PTN4 on Pi absorption is similar in a wide range of Pi concentration.
Plants were grown for 5 d in complete medium (full) then transferred for 5 d in low Pi with or without (Control) drugs (at optimal concentration). Pi influx was measured during 2 h in 33P supplemented with 5, 25, 50, 100, 500, and 1,000 μm Pi for the measure of Vmax and Km. At least 10 plants were used for each point.
| Transporter | Control |
PTN1 |
PTN4 |
|||
|---|---|---|---|---|---|---|
| Vmax | Km | Vmax | Km | Vmax | Km | |
| High affinity | 0.14 | 9.4 | 0.12 | 13.3 | 0.09 | 8.2 |
| Low affinity | 0.37 | 104.3 | 0.27 | 91.6 | 0.3 | 108.6 |
Surprisingly, seedlings treated with PTN1 contained significantly higher amounts of phosphorus than the untreated –Pi control (Fig. 2C), although this is much less than the amount accumulated in the +Pi control.
We tested whether PTN1 affects acid phosphatase (PAP) secretion, as this could explain its increased phosphorus content through mobilization of any organic Pi present in the growth medium. However, PTN treatment only slightly increased the phosphatase activity (Supplemental Fig. S1C), suggesting phosphatases are not involved in the process.
PTN1 Reduces the Gene Expression Response to Pi Starvation
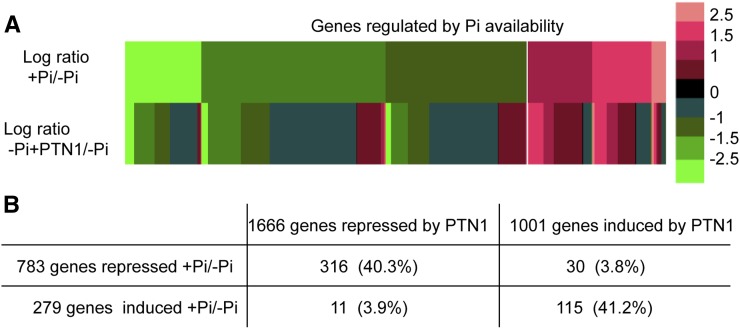
PTN1 reduced the transcription of all high-affinity PHT1 transporters tested, indicating that PTN1 effect is not restricted to the PHT1;4 marker used for the screening. To more broadly investigate PTN1 action, a transcriptional analysis of the global Arabidopsis genome was performed using the Complete Arabidopsis Transcriptome Microarray arrays (http://www.catma.org/; Unité de Recherche en Genomique Végétale). We compared transcript expression in roots of 14-d-old plantlets grown in –Pi, –Pi plus PTN1, or +Pi (Supplemental Table S2). Firstly, this approach confirmed the reduced level of PHT1 gene expression. Global analysis showed that when compared with the –Pi condition, the +Pi condition modulated the expression of 1,062 genes: 783 were repressed and 279 significantly induced (Fig. 3). By contrast, PTN1 modulated 2,667 genes: 1,001 were induced and 1,666 repressed (based on statistical analysis, Bonferroni test <0.05, with a greater than 2-fold change; Supplemental Table S2; Fig. 3). Interestingly, many of the genes whose expression was modulated by +Pi are also modified in a similar way by PTN1 in the –Pi condition (with 40.3% of transcripts repressed and 41.2% of transcripts induced; Fig. 3B). Conversely, very few genes (approximately 4%) were significantly deregulated by PTN1 when compared with the +Pi treatment (Fig. 3B). These results were confirmed by reverse transcription (RT)-qPCR experiments, using a few selected genes that are transcriptionally regulated by Pi starvation (Supplemental Fig. S2). This transcriptomic result indicates that PTN1 mimics the addition of Pi in the growth medium.
Figure 3.
PTN1 reduces the transcriptional regulation that occurs during Pi starvation. A and B, Comparison of PTN1 treatment or Pi supply on global transcriptomic analysis. For this analysis, 14-d-old wild-type root samples of plants grown on –Pi, –Pi plus PTN1 (40 µm), or +Pi were used. Data are expressed as ratio values between +Pi and –Pi or –Pi plus PTN1 and –Pi, respectively. A, Relative expression of genes induced and repressed by +Pi versus the –Pi plus PTN1 condition. B, Number of genes regulated by Pi and PTN1 addition and the percentage of genes regulated by +Pi as well as PTN1. Selected genes exhibited at least a 2-fold change difference with –Pi control, based on statistical analysis (Bonferroni test, P < 0.05).
Our transcriptomic analysis was refined by examining whether PTN1 modulated the expression of particular classes of genes, as defined by Thibaud et al. (2010), according to their local or long-distance regulation by Pi. Among the systemic genes that were repressed or induced by Pi in our conditions, respectively 65% and 55% were similarly regulated by PTN1, thus mimicking a limited Pi supply (Supplemental Table S3). Many of these genes have a predicted function in the recycling, recovery, or transport of Pi (Supplemental Table S3). Among the local genes that were repressed by Pi, 45% were also repressed by PTN1. The predicted functions of these genes are related to transcription, metal homeostasis, hormones, stress responses, or development (Thibaud et al., 2010).
Altogether, this analysis revealed that PTN1 mimics many of the effects of Pi supply on the transcriptomic regulation of different metabolic pathways; furthermore, there was no bias toward local or long-distance regulation of Pi starvation. To further investigate the effects of PTN on well-known Pi starvation features, we next performed more detailed physiological experiments.
PTN Affects Multiple Systemic Responses Triggered by Pi Starvation
Pi deficiency stimulates Pi import mechanisms (mainly through the regulation of the high-affinity PHT1 Pi transporters). Pi deficiency also triggers major remodelling of phospholipids (an important pool of organic Pi for plant), the accumulation of starch (to store excess carbon), and anthocyanin.
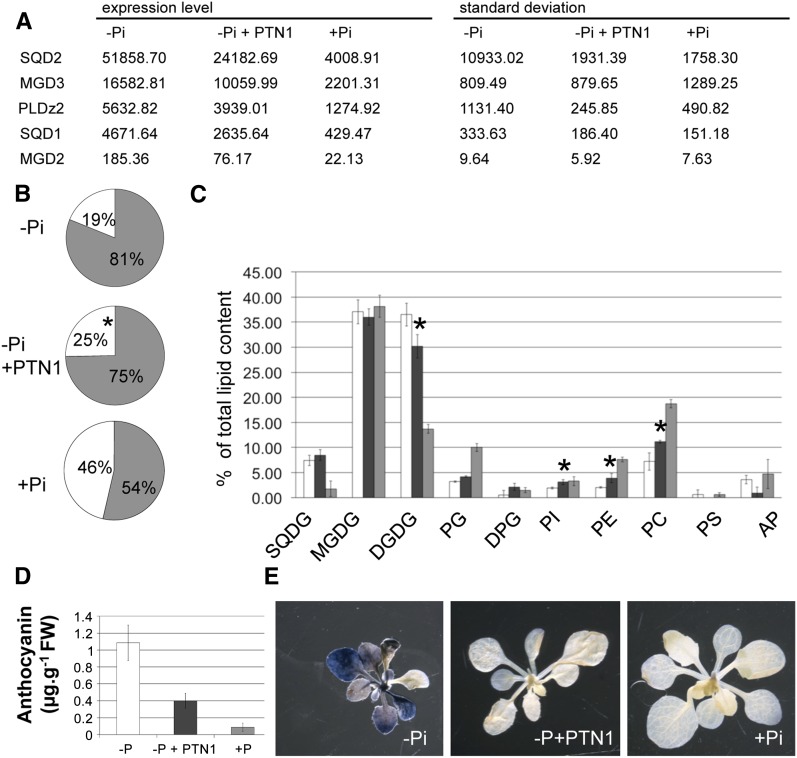
The conversion of phospholipids to galactolipids and sulfolipids is a main feature of the response triggered by Pi starvation (Dörmann and Benning, 2002; Nakamura et al., 2009; Moellering and Benning, 2011; Nussaume et al., 2011b). This lipid conversion relies on several enzymes such as sulfoquinovosyldiacylglycerol2 (SQD2), SQD1, galactosyl-1,2-diacylglycerol2 (MGD2), MGD3, digalactosyl-diacylglycerol2 (DGD2), and phospholipase DZ2 (PLDz2), whose gene expression is induced during Pi starvation (Dörmann and Benning, 2002; Nakamura et al., 2009; Moellering and Benning, 2011). Consistent with these results, our global transcriptomic analysis reveals that PTN modulates the expression of most of these genes (Supplemental Table S3). This result was confirmed by RT-qPCR analysis, indicating that PTN1 represses the expression of all tested genes in low Pi (SQD2, MGD3, PLDz2, SQD1, and MGD2; Fig. 4A). We then examined whether any of these transcriptional modifications affect lipid composition in seedlings. Figure 4B illustrates that the partitioning between phospholipids and glycolipids (galactolipids and sulfolipids) in Pi-rich and Pi-starved seedlings was 19%:81% and 46%:54%, respectively. When seedlings were grown on a –Pi medium containing PTN1, this ratio (25%:75%) was in between the two controls, indicating that PTN1 significantly reduced the effect of Pi starvation on the relative abundance of glycolipids.
Figure 4.
PTN1 reduces metabolic stress during Pi starvation. A, Transcript level measurements (by qPCR) of genes involved in lipid synthesis for plants grown on –Pi, –Pi plus PTN1 (40 µm), or +Pi for 14 d. B, Proportion of phospholipids (white) and glycolipids (gray) for plants grown on –Pi, –Pi plus PTN1, or +Pi media. Asterisks represent significant differences between –Pi and –Pi plus PTN1 (40 µm; Student’s t test, P ≤ 0.01). C, Lipid composition of wild-type plants grown on low Pi (white bars), low Pi plus PTN1 (dark gray bars), or high Pi (light gray bars). Total lipids were extracted from the aerial part of 10-d-old plants. D, Anthocyanin content for plants grown on –Pi, –Pi plus PTN1 (40 µm), or +Pi. E, Starch content revealed by staining of shoots with Lugol. FW, Fresh weight; DPG, diphosphatidylglycerol; PC, phosphatidylethanolamine; PE, phosphatidylinositol; PG, phosphatidylglycerol; PI, phosphatidylinositol; DGDG, digalactosyl-diacylglycerol.
As shown in Figure 4C, –Pi seedlings contained much less PC and PE than +Pi seedlings, and PTN1 partially reversed the effect of –Pi. Conversely, PTN1 reduced DGDG content by 27% compared with its level in Pi-starved plants, although +Pi resulted in a much greater reduction in DGDG content (Fig. 4C). This biochemical analysis demonstrates that PTN1 diminishes the conversion of phospholipids to glycolipids in Pi-starved plants, in accordance with the effect on gene expression.
PTN1 also strongly reduced the accumulation of anthocyanins and starch (Fig. 4, D and E). The amount of anthocyanin accumulated in PTN1-treated plants is reduced by approximately 60% when compared with –Pi plants (Fig. 4D). The starch accumulation was also severely attenuated by PTN1 addition, as determined by Lugol staining (Fig. 4E).
Altogether, these results indicate that PTN1 and PTN2 significantly reduce several typical plant responses to Pi starvation. To confirm the specificity of the drug, we tested its impact on the two other macronutrients for plants (potassium or nitrate). Plants growing under potassium or nitrate starvation also exhibited reduction of their biomass when compared with plants grown with optimal nutrients. Supplemental Figure S3A presents one of the assays conducted with various levels of nitrogen or potassium; they all indicate that PTN1 or PTN4 do not modulate this response. Such results reinforce the transcriptomic data, which did not reveal any impact of PTN drugs on markers of potassium or nitrogen deficiency. We also tested impact of these drugs on iron deficiency and also did not observe any modification resulting from PTN addition. The impact of PTN on ion content was also analyzed using inductively coupled plasma (ICP) analysis but did not reveal any significant modification (Supplemental Fig. S3B).
PTN1 Modulates Root Growth in Pi-Starved Plants
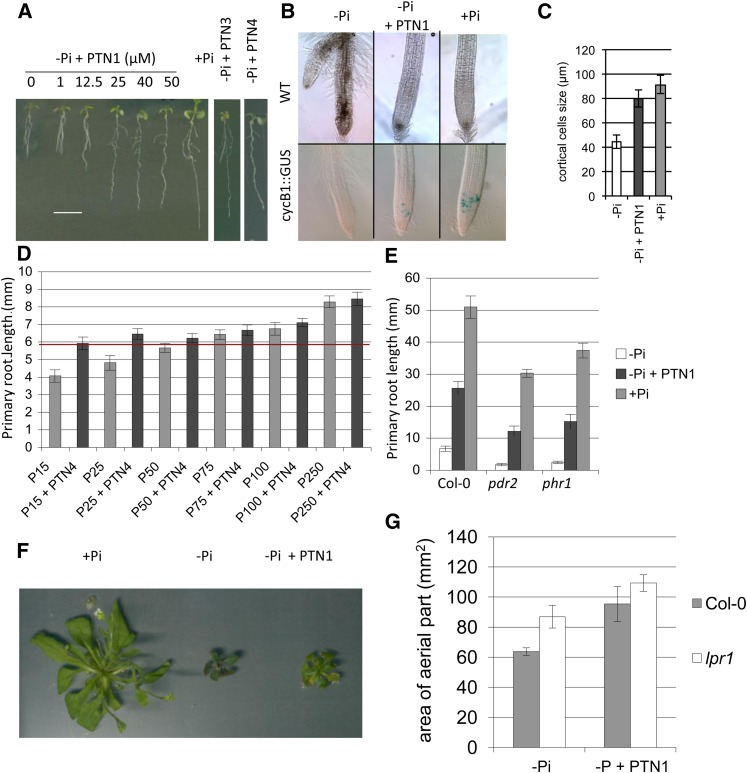
Our transcriptomic study additionally identified a global effect of PTN1 on Pi starvation responses associated with local regulation (compare with Thibaud et al., 2010). We investigated the impact of several PTN analogs on the primary root growth inhibition by low Pi, a local response conditioned by the external Pi concentration (Abel, 2011; Péret et al., 2011). Strikingly, PTN1 suppressed the primary root growth inhibition in a dose-dependent manner (Fig. 5A). As for the inhibition of the PHT1;4 transcript, a maximum effect for PTN1 was observed at 40 µm. A similar effect was observed for PTN3 and PTN4, two active PTN1 structural analogs (Supplemental Table S1) that are active at different doses (100 and 10 µm, respectively).
Figure 5.
PTNs reduce the Pi starvation-induced growth limitation. A, Identification of the optimal PTN1 level to suppress the Pi starvation symptom on root growth. A similar effect can also be observed with the PTN1 analog PTN3 and PTN4 applying 100 or 10 µm, respectively. B, Analysis of the cell cycle using the cycB1::GUS marker. Pictures of root tips from wild-type (WT; top) and cycB1::GUS (bottom) plants grown on –Pi, –Pi plus PTN1 (40 µm), or +Pi conditions after GUS staining. C, Measures of cortical cell size in wild-type plantlets grown on –Pi, –Pi plus PTN1 (40 µm), and +Pi. In A to C, 7-d-old plantlets were used for the various experiments. D, Impact of Pi concentration on PTN effect on the primary root growth. For the measures of the primary root length, plants were grown during 5 d on Pi-rich medium and transferred on various conditions tested during 24 h (PTN4, 10 µm). The red line shows the maximum gain of growth conferred by PTN1 presence. E, Effect of PTN treatment on mutants affected in the root architecture response to Pi starvation. Measurements of the primary root growth of wild-type plants, and pdr2 and phr1 mutants, were taken 9 d after germination on –Pi or –Pi plus PTN1 (40 µm) and +Pi. F, Effect of PTN1 (40 µm) addition on rosette development. G, Effect of PTN1 addition on aerial development of the wild type and the lpr1 mutant. Col-0, Ecotype Columbia.
To better understand how PTN promotes the primary root growth of –Pi seedlings, we investigated the root meristematic activity and the cell elongation rate. This was tested using the cell cycle marker Cyclin B1 (CYCB1)::GUS. CYCB1 is expressed predominantly during the G2/M phase of the cell cycle (Colón-Carmona et al., 1999). It was previously described that this marker is expressed in the root meristematic zone of seedlings grown on +Pi medium, but not in –Pi seedlings (Sánchez-Calderón et al., 2005). Expression of CYCB1 was sustained in the root tip of –Pi seedlings treated with 40 µm of PTN1, although fewer cells were marked as compared with the +Pi control (Fig. 5B).
In agreement with previously published results (Reymond et al., 2006), Pi starvation strongly reduces root cell elongation (50% in the cortical cells; Fig. 5C). Root cell elongation of –Pi seedlings was restored by PTN1 to a level close to control plants grown on +Pi medium (Fig. 5C). Similar results were observed with PTN3 and PTN4; this indicates that PTNs act on both meristematic activity and cell elongation to sustain root growth on –Pi medium.
To explore how PTN could mimic Pi, we examined the external Pi concentration necessary to promote a primary root growth similar to the one observed in the presence of PTN4 without added Pi (agar Pi content is approximately 15 µm). Throughout the tested range of Pi concentrations (15–250 µm), there was a linear correlation between primary root length and external Pi concentration (gray bars in Fig. 5D). Adding PTN4 (Fig. 5D) or PTN1 (data not shown) to the –Pi medium improved root growth, mimicking the effects of medium containing 50 to 75 µm Pi. The sustained growth promoted by PTNs reached a threshold and appears to be independent of external Pi for Pi concentrations between 15 and 75 µm. No significant differences were observed between plants grown with or without PTNs beyond 75 µm Pi.
The role of PTNs on the regulation of primary root growth was further investigated by analyzing available mutants showing altered root growth in response to Pi starvation. The lpi mutants (Sánchez-Calderón et al., 2006) as well as lpr1 and lpr2 mutants (Svistoonoff et al., 2007) were previously identified on the basis of their reduced root growth sensitivity to Pi starvation. When grown on –Pi medium, they all display a longer primary root than the wild-type control. No additive effects of PTN1 were found to affect the primary root growth of lpr1 or the three lpi mutants (Supplemental Fig. S4). By contrast, the lpr2 mutant, which exhibits a moderate root elongation phenotype compared with lpr1 or lpi, responded to PTNs with a 50% increase in the primary root length (Supplemental Fig. S4).
Unlike the lpr and lpi mutants, pdr2 mutant seedlings are hypersensitive to Pi starvation and display a very short primary root when grown on low Pi (Ticconi et al., 2004b). Interestingly, PTN1 substantially reverted the pdr2 root growth defect (Fig. 5E).
PHR1 has been described as a major regulator of the transcriptional response to Pi starvation (Franco-Zorrilla et al., 2004), and we therefore also investigated the response of the phr1 mutant. Like pdr2 and lpr2, the phr1 response to PTN1 partially alleviated the root growth restriction imposed by –Pi (Fig. 5E). These results demonstrate that the LPR2, PDR2, and PHR1 functions are not essential for PTN1 to restore root growth in low Pi.
As PTN sustained root growth in Pi starvation conditions, we next investigated whether it can also affect the development of the aerial part. Although plants cultivated for 4 weeks on –Pi medium plus PTN1 were not as big as those grown on +Pi, the drug significantly improved leaf growth (approximately +30% rosette area; Fig. 5, F and G). Similar results were obtained with PTN4 (data not shown). Together with the results from the roots (Fig. 5A), these observations show that the PTN compounds, when applied at an appropriate concentration, can substantially improve whole plant growth in low-Pi conditions, suggesting that these drugs could unlock a mechanism restricting growth. Nevertheless if PTN can partially restore growth on –Pi medium, the size of the plants remains far from that of plants grown on Pi-rich medium (500 µm). In fact, drugs only mimic the addition of 30 to 50 µm Pi. It may be noticed that we also could observe a growth stimulation of Medicago truncatula shoots maintained on this medium for several weeks with PTN4 (10 µm), suggesting that PTN effects can probably be extended to other plant species (data not shown).
PTN Uncouples Iron Accumulation and Root Growth Arrest
The primary root growth response to Pi starvation is dependent on the iron concentration in the medium (Svistoonoff et al., 2007; Ward et al., 2008). To test whether PTNs form a molecular complex with iron (and thereby impede the effect of iron on root growth), we measured the absorption spectrum of a solution containing PTN1 and Fe2+ (FeCl2). As shown in Supplemental Figure S5A, iron does not modify the absorption spectrum of PTN1. This strongly suggests that PTN1 effect on growth is not mediated by chelating iron.
Under Pi starvation conditions, roots and shoots accumulate iron (Hirsch et al., 2006; Supplemental Fig. S5). We show that PTN1 does not modify this iron accumulation (Supplemental Fig. S5B). According to these chemical results, PTN1 does not appear to affect iron bioavailability.
PTN Reduces the Growth Restriction
In some ways, the addition of PTN phenocopies the growth and development of the lpr1 and lpr2 mutants. lpr1 plants display a more developed root system in a Pi-starved medium (–Pi; Reymond et al., 2006; Svistoonoff et al., 2007), along with a phosphorus content greater than the wild type (Supplemental Fig. S6A), and a larger rosette (Fig. 5G). If PTN targets only LPR1 (to inhibit its activity or expression), then we should expect that the drug does not further improve the growth of lpr1. When grown in the –Pi condition, lpr1 plants displayed a rosette 30% larger than the wild-type control (Fig. 5G; Supplemental Fig. S6B). PTN1 and PTN4 further increased the growth of the lpr1 rosette (+25%; Fig. 5G; Supplemental Fig. S6B), despite their absence of effect on root growth (Supplemental Fig. S4). We also observed that the accumulation of anthocyanins in leaves of lpr1 grown in –Pi was substantially suppressed when Pi (+Pi condition) or PTN1 are provided (as illustrated by the paler green leaves in Supplemental Fig. S6B). Therefore, although PTN did not improve root growth in lpr1, the aerial part still responded to the drug and was found less affected than in low-Pi conditions.
We also investigated the molecular responses of lpr1 seedlings to PTN1 treatment. Firstly, we observed that in low Pi, the level of induction of genes involved in Pi uptake (PHT1;4 and PHT1;8), Pi recycling (MGD3, PLDζ2, and SQD2), and Pi signaling (SPX1, for SYG1 [suppressor of yeast gpa1], Pho81 [CDK inhibitor in yeast PHO pathway], and XPR1 [xenotropic and polytropic retrovirus receptor]) were similar between ecotype Columbia and the mutant (Supplemental Fig. S7). Secondly, PTN1 repressed the expression of these genes in lpr1 to a level similar to that observed in the wild type (Supplemental Fig. S7).
Altogether, the growth and molecular results reveal that the plant responses to PTN1 were not altered in the lpr1 mutant, suggesting that LPR1 is not a target of PTN. However, we cannot exclude the hypothesis that LPR1 is involved in the PTN1-mediated suppression of low-Pi-induced root growth inhibition.
DISCUSSION
PTNs Partially Overcome the Plant Growth Restriction Observed during Pi Starvation
PTNs were isolated due to their ability to inhibit expression of the Pi transporter PHT1;4 in low-Pi conditions (Fig. 1). We have also shown that this family of drugs inhibits the expression of other PHT1 genes (Fig. 2A) and modulates the expression of a vast number of genes associated with Pi starvation adaptation. Many of these genes are involved in Pi recovery from the external medium and physiological adaptations triggered to conserve metabolized Pi (Fig. 4; Supplemental Table S3). All of these metabolic adaptations to Pi starvation appeared systemically regulated (Thibaud et al., 2010). These observations suggest that PTNs act on the global systemic pathway to reduce the expression level of these genes. This repression was around 40% of their expression level in low Pi, reducing the metabolic adaptation usually observed in low Pi. PTN addition affected transcripts related to lipid homeostasis, Pi uptake, and Pi recovery. Physiological experiments confirmed modifications of lipid profiles and Pi absorption, but phosphatase activity remained unaffected. Discrepancy between transcript regulation and activity of phosphatases could be due to modification of root tip development or to posttranscriptional regulation, two important parameters affecting AtPAP expression (Wang et al., 2011).
Although PTNs significantly reduce the expression of many genes that function to adapt the whole-plant physiology to low-Pi conditions, their addition unexpectedly did not result in further growth inhibition. Instead, we observed an increase in root growth and a slight increase in shoot growth compared with Pi-starved plants (Fig. 5). These data support the fact that plant growth limitation in the low-Pi condition is not only due to a metabolic limitation that results from a lack of Pi. On the contrary, the root growth promoted by PTN was independent of the Pi content in the growth medium, because a plateau was observed between 15 and 75 µm Pi supplemented with PTN4 (Fig. 5D). This result supports the hypothesis that PTNs partially suppress the plant response to the low-Pi condition, independently of the available Pi (i.e., PTN partially uncouples plant growth from Pi homeostasis).
This parallels recent results obtained with the Arabidopsis PHO1 underexpressors (Rouached et al., 2011). Only a very low level of Pi accumulates in the shoots of PHO1 underexpressors (as in the pho1 null mutant), but they still grow as much as the wild-type control, suggesting that the strong growth reduction in pho1 shoots is not directly related to the level of Pi accumulated in leaves. The authors explained these results by positing the existence of an unknown growth regulation-signaling pathway.
In these experiments, genetic manipulation did not improve the growth of plants in the low-Pi condition (when compared with the wild-type control), in contrast to what was observed with PTN1 addition. The PHO1 underexpressor plants also exhibited a reduced modification of the transcriptomic response to the Pi starvation affecting only the aerial parts of the plants. Addition of PTN elicited clearly distinct responses, including a specific modification at the transcriptomic level affecting both roots and leaves, improved growth in low Pi, and no modification of Pi flux between roots and shoots, providing strong evidence for an activity distinct from PHO1.
PTN Treatment Does Not Significantly Alter Homeostasis of Ions Other Than Pi
Cross talk between plant nutrients has been already described (for review, see Ohkama-Ohtsu and Wasaki, 2010; Rouached et al., 2010). Pi anion can chelate in soil many cations including in particular metals. It therefore plays a crucial role in controlling the availability of micronutrients such as zinc, iron, and aluminum (Misson et al., 2005; Hirsch et al., 2006). Interestingly, many genes involved in metal homeostasis (such as transporters) are directly regulated by the master transcriptional regulator of Pi starvation belonging to the PHR1 family. This has been revealed by several transcriptomic or promoter analyses (Misson et al., 2005; Bournier et al., 2013). A link between main macronutrients can be illustrated by discovery of nitrogen limitation adaptation (Kant et al., 2011). This gene was identified, as it is involved in adaptive responses to low-nitrogen conditions in Arabidopsis. It turned out also to regulate PHT1 family turnover (Lin et al., 2013; Park et al., 2014). Nevertheless, here, the experiments performed to evaluate the effects of PTN on other nutrients’ deficiencies or homeostasis showed an impact restricted to Pi starvation. These results are consistent with transcriptomic analysis, which revealed a clear modulation of genes specifically associated with Pi deficiency. In addition, it is worth notice that PTN also affected many stress-related genes, which might result from the modification of plant development observed. So, PTNs do not appear to be a general plant-promoting substance, as their effects are restricted to Pi starvation.
PTN Uncouples Iron Accumulation and Root Growth Arrest in –Pi
Together, iron and Pi interfere with root growth. In low-Pi conditions, the growth of the primary root is arrested when the growth medium contains 10 µm Fe (Svistoonoff et al., 2007), but not when iron is omitted or chelated with ferrozine or deferoxamine. Ward et al. (2008) proposed that the root growth arrest in low Pi results from iron toxicity. Here, we have shown that PTN treatment uncouples high iron accumulation from root growth arrest (Supplemental Fig. S5), suggesting that such a phenotype does not directly result from toxic iron accumulation per se but possibly acts as a component of a signaling cascade requested by such a process.
Interestingly, PTN treatment mimicked +Pi treatment regarding the expression of major genes involved in iron homeostasis (for review, see Jeong and Guerinot, 2009); among these genes ferric chelate reductase2, ferric chelate reductase3, and iron-regulated transporter1 (IRT1) were induced, whereas ferritin1 and ferric reductase defective3 were repressed (Supplemental Table S2). Only IRT2 (a minor player in iron uptake; Vert et al., 2009) avoided this regulation. This pattern of regulation could be associated with iron limitation, but the iron content in PTN-treated seedlings remained similar to the –Pi control (i.e., 2- to 3-fold more iron than in +Pi seedlings). This regulation was therefore not induced by an internal iron deficiency associated with high Pi content. These observations strongly suggest that iron and Pi have common regulatory signaling steps (Misson et al., 2005; Thibaud et al., 2010) that were modulated by PTN. This is corroborated by other metal homeostasis-related genes (high-affinity copper transport protein2, zinc transporter2, nicotianamine synthase1, and nicotianamine synthase2) regulated by PTN.
PTN Overcomes the Low-Pi Growth Restriction Independently of Known Pi Pathway Signaling and Growth Regulators
Curiously, addition of PTN in our study concurrently reduced the Pi absorption capacity and resulted in improved growth. The measures of Km and Vmax confirmed that PTN addition promoted a reduction of the amount of transporters but did not change the affinity of PHT1 transporters for their substrate.
Nevertheless, it has to be noticed that the Pi concentration in –Pi medium remains extremely limiting, far below the potential Pi uptake capacity of the PHT1 transporters (even reduced by PTN addition).
One possible explanation is that the increased root length of PTN-treated seedlings improved Pi uptake and consequently reduced plant Pi starvation traits. This has been tested using lpr1 seedlings that already display a long root in –Pi. In fact, like in our plants treated with PTN1, this mutant presents a greater phosphorus content than the wild type in –Pi condition. It suggests that a longer root leads to higher phosphorus content. This view is reinforced by the recent identification of a quantitative trait loci in rice (Oryza sativa) named phosphorus uptake1 (Gamuyao et al., 2012), which encodes the protein kinase phosphorus-starvation tolerance1 that improves early root growth and subsequently resistance to low-Pi conditions.
Although lpr1 shoot growth is significantly higher than in the wild type, PTN further improves this growth. PTN alleviates also several other Pi starvation traits still present in lpr1 (accumulation of anthocyanins, modulation of low-Pi-related genes, etc.). Therefore, the increased root growth in –Pi probably accounts for the biomass gain induced by PTN.
In addition, the root growth of the pdr2 mutant responds to PTN, showing that PTN activity does not rely on the only P5-type adenylpyrophosphatase in Arabidopsis encoded by PDR2. Because both PDR2 and LPR1 act in the same functional pathway (Ticconi et al., 2009), our results corroborate the view that PTN improves root growth independently of this pathway.
Root growth improvement by PTN is independent of PHR1, because the loss-of-function phr1 mutant responds to the drug like the wild type. As this transcriptional factor belongs to a multigenic family, we cannot exclude the hypothesis that PTN modulates the expression of one or more PHR1 paralog. However, PHR1 is a major regulator of the low-Pi pathway (Bustos et al., 2010; Thibaud et al., 2010). So, it is probably not a direct target of these drugs, as its mutation would have reduced the response to PTN. This also corroborates previous results (Thibaud et al., 2010) disconnecting the local response to Pi starvation (primary root response) from the systemic pathway involving PHR1. Nevertheless, as discussed above, all phenotypes (anthocyanin suppression, modulation of low-Pi-related gene environment, etc.) reported are not observed in lpr1 mutant, which is also insensitive to root growth arrest in low Pi.
So, the singular feature of PTNs is their capacity to modulate a range of traits triggered by Pi starvation. This suggests the existence of common regulatory elements targeted by PTN affecting local and systemic Pi response. PTNs are therefore a promising tool to further dissect the Pi starvation responses.
PTN, a Tool to Dissect the Cross Talk between Morphological and Metabolic Adaptations
Previous genetic studies have demonstrated that mutants such as LPR1 (Svistoonoff et al., 2007), LPI (Sánchez-Calderón et al., 2006), and PDR2 (Ticconi et al., 2004b) were essentially altered in their root growth, whereas mutants such as PHR1 (Rubio et al., 2001), PHO1 (Hamburger et al., 2002), and PHO2 (Delhaize and Randall, 1995) had an altered Pi homeostasis without altered root growth. Strikingly, PTN modified both root growth and Pi homeostasis in our study. As commonly accepted, these two responses are sensitive to different pools of Pi: whereas root growth is correlated to Pi concentration in the growth medium (for review, see Abel, 2011; Péret et al., 2011), mechanisms that control Pi homeostasis depend on the plant internal Pi content (Thibaud et al., 2010). We cannot eliminate the possibility that the modification of Pi homeostasis is mainly a consequence of the promotion of the root growth resulting from PTN addition. This should contribute to the increase of Pi absorption by maximizing exploration of the soil. Long-term experiments of PTN addition will be very useful to study in detail the impact on plant response to Pi starvation. Unfortunately, PTN molecules are extremely insoluble in water conditions, limiting our experiments to in vitro conditions. We are currently trying to solve problems with chemists. This would offer opportunities to investigate the effect of PTNs on more adult stages of plants (grown in soils or in hydroponic solution).
MATERIALS AND METHODS
Plant Materials and Growing Conditions
For the screening, an Arabidopsis (Arabidopsis thaliana) transfer DNA (T-DNA) insertion mutant derived from the Institut National de la Recherche Agronomique collection (Ortega et al., 2002) was selected. This pht1;4-1 mutant (Wassilewskija background) contains a T-DNA construct inserted in the Pht1;4 gene, creating a transcriptional fusion with the GUS reporter gene included in the T-DNA (Misson et al., 2004). For phenotypic analyses, Arabidopsis Columbia and various lines derived from this ecotype were used. Some of these were transgenic lines that express reporter genes such as cycB1::GUS (Colón-Carmona et al., 1999), and others were various mutant lines affecting root response to low Pi, such as pdr2 (Ticconi et al., 2004b), lpr1, lpr2 (Svistoonoff et al., 2007), lpi1, lpi2, and lpi3 (Sánchez-Calderón et al., 2006).
For in vitro analyses, seeds were surface sterilized and sown in vitro on square petri plates with solid modified one-tenth-strength Murashige and Skoog (MS/10) medium (0.15 mm MgSO4, 2.1 mm NH4NO3, 1.9 mm KNO3, 0.5 or 0.005 mm NaH2PO4, 0.34 mm CaCl2, 0.5 µm KI, 10 µm FeCl2, 10 µm H3BO3, 10 µm MnSO4, 3 µm ZnSO4, 0.1 µm CuSO4, 0.1 µm CoCl2, 1 µm Na2MoO4, 5.9 µm thiamine, 4.9 µm pyridoxine, 8.1 µm nicotinic acid, 55 µm inositol, and 3.4 mm MES [pH 5.7]), 0.5% (w/v) Suc, and 0.8% (w/v) agar. In the Pi-deficient medium, NaCl (0.5 mm) was used to replace the equivalent amount of sodium provided by NaH2PO4. As a control for the effects of the molecules, 1% dimethyl sulfoxide (DMSO), the solvent for the drugs, was added. After 2 d at 4°C, the plates were placed in a vertical position, and plants were grown in a culture chamber with a 16-h-light/8-h-dark regime (24°C/21°C, 150 µE m–2 s–1) as previously described (Sarrobert et al., 2000). For the 4-week culture, large square plates (24 cm) were placed in a horizontal position. Root architecture and aerial part area were quantified using the ImageJ software with the NeuronJ plugin (Meijering et al., 2004).
Chemical Genetics Screening
For the screening, the Library of Active Compounds on Arabidopsis collection of 3,600 biologically active compounds (Zhao et al., 2007) was used at a final concentration of 25 µm in 1% DMSO. Four to five Arabidopsis seeds from the pht1;4-1 reporter line (Misson et al., 2004) were sown in 96-well plates containing 100 µL of liquid MS/10 medium with 500 µm Pi at pH 5.7. After 2 d at 4°C, plants were grown for 5 d in a culture chamber with a 16-h-light/8-h-dark regime (24°C/21°C, 150 µE m–2 s–1). The medium was then replaced by MS/10 with only 5 µm Pi, and drugs were added to the 96-well plates. Positive and negative controls were included by incubating seedlings in MS/10 solutions containing 1% DMSO and supplemented with 5 or 500 µm Pi, respectively (resulting in –Pi or +Pi media). Five days later, plantlets were analyzed by GUS staining to assay the activity of the reporter gene inserted into the pht1;4 gene. One hundred microliters of GUS staining solution was added to each well, and plates were incubated 3 h at 37°C. Subsequently, plates were analyzed with an MZ16 stereomicroscope (Leica Microsystems).
GUS Staining
GUS activity was detected as previously described (Klimyuk et al., 1995) by using a GUS staining solution containing 50 mm sodium Pi buffer (pH 7.0), 0.01% Triton X-100, 1 mm K3Fe(CN)6, 1 mm K4Fe(CN)6, and 1 mg mL–1 5-bromo-4-chloro-3-indolyl β-d-glucuronide. For cycB1::GUS staining, plants were incubated 6 h at 37°C.
Transcriptome Analysis and Quantitative Real-Time PCR
Plant RNA was extracted with the RNeasy kit (Qiagen). For transcriptome data, RNA was analyzed using the Complete Arabidopsis Transcriptome Microarray technology by the Unité de Recherche en Genomique Végétale laboratory. For qPCR, poly(dT) complementary DNA was prepared from 500 ng of total RNA with Superscript III reverse transcriptase (Invitrogen) and analyzed on a LightCycler 480 apparatus (Roche Diagnostics) with the SYBR Green I Master kit (Roche Diagnostics), according to the manufacturer’s instructions. Targets were quantified with specific primer pairs, designed using Primer3 (http://frodo.wi.mit.edu/primer3/; Supplemental Table S4). Biological triplicates were performed for all experiments. Expression levels were normalized using the At2g16600, At3g04120, At1g35160, and At1g32050 genes.
Quantification of Macro- and Microelements, Anthocyanins, and Lipids
To quantify ionic content, 50 mg of dry weight of each sample were mineralized in 14% HNO3, and a MarsX microwave system (Controls Engineering Maintenance Corporation) was used to determine macro- and microelements by ICP optical emission spectrometry (Vista MPX, Varian). Biological triplicates were performed for all experiments.
Lipids and anthocyanins were analyzed as previously described (Jouhet et al., 2004; Ticconi et al., 2004b).
Pi Uptake Experiments
Pi uptake experiments were performed as previously described (Narang et al., 2000). Twenty-four seedlings per condition were incubated separately in a Pi incubation solution containing 33PO4 (5 mm MES, 0.1 mm CaCl2, 20 µm KH2PO4, and 0.15 µCi mL–1 33PO4) for 2 h. After incubation in the uptake solution, plantlets were transferred to a chilled desorption medium (5 mm MES, 0.1 mm CaCl2, and 1 mm KH2PO4) for 2 h at 4°C. The seedlings were then dried in scintillation vials at 60°C overnight, after which 2 mL of scintillation cocktail (InstaGel, PerkinElmer) was added, and the radioactivity was measured with a scintillation analyzer (Tri-Carb, Packard Instrument Company). The amount of Pi absorbed during the experiment was calculated and normalized per centimeter of root. For Vmax and Km measurements, a similar protocol was used, with a range of Pi concentration from 5 to 1,000 μm in the incubation medium.
Protein Extraction and Phosphatase Activity
Soluble proteins were extracted with 50 mm Tris and 2 mm 1,4-dithioerythritol (Sigma, 6892–68–8). The protein concentration was determined by the Bradford method using bovine serum albumin (Sigma, 9048–46–8) as a standard. The phosphatase activity in solution was determined as previously described (Kolari and Sarjala, 1995) by measuring the release of nitrophenol from 0.6 mm p-nitrophenylphosphate in 50 mm sodium acetate (pH 5). Reactions were incubated at 30°C for 30, 60, and 90 min and were stopped with 10% Na2CO3, after which the A405 was measured.
Starch Staining
Starch staining was performed according to Zakhleniuk et al. (2001). The aerial parts of plants were placed for 6 h in 90% ethanol before iodine staining for 30 min in Lugol solution (Sigma). Afterward, samples were washed several times in water.
Statistical Analyses
Data were analyzed by Student’s t test. Asterisks indicate the means that were statistically different at P < 0.05.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Impact of PTN treatments on PHT1;4 expression and phosphatase activity.
Supplemental Figure S2. PTN1 represses several genes regulated by Pi starvation.
Supplemental Figure S3. PTN impact on other macro- and micronutrients.
Supplemental Figure S4. PTN1 addition does not impact the primary root growth of Pi-insensitive mutants.
Supplemental Figure S5. PTN1 effects are independent of Fe signaling.
Supplemental Figure S6. The lpr1 mutant responds to PTN1 addition.
Supplemental Figure S7. Molecular responses of lpr1 seedlings to PTN1 treatment.
Supplemental Table S1. Characteristics of PTN.
Supplemental Table S2. Transcriptomic data of the Pi starvation response in the wild type in the presence of PTN1.
Supplemental Table S3. PTN impact on other macro- and micronutrients.
Supplemental Table S4. List of primers used for qPCR assay.
Supplementary Material
Acknowledgments
We thank Dr. Benjamin Péret for the critical reading of this article. We also thank Dr. Luis Herrera-Estrella for providing lpi mutants.
Glossary
- Pi
phosphate
- qPCR
quantitative PCR
- RT
reverse transcription
- ICP
inductively coupled plasma
- T-DNA
transfer DNA
- DMSO
dimethyl sulfoxide
Footnotes
This work was supported by Agence Nationale de la Recherche (ANR) Bioadapt REGLISSE (grant no. ANR–13–ADAP–0008) and ANR Blanc CHEMIGENA (grant no. ANR–09–BLAN–0118).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abel S. (2011) Phosphate sensing in root development. Curr Opin Plant Biol 14: 303–309 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Larsson KE, Tjellström H, Liljenberg C, Sandelius AS. (2005) Phosphate-limited oat. The plasma membrane and the tonoplast as major targets for phospholipid-to-glycolipid replacement and stimulation of phospholipases in the plasma membrane. J Biol Chem 280: 27578–27586 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Stridh MH, Larsson KE, Liljenberg C, Sandelius AS. (2003) Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett 537: 128–132 [DOI] [PubMed] [Google Scholar]
- Bayle V, Arrighi JF, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz-Ares J, Nussaume L. (2011) Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournier M, Tissot N, Mari S, Boucherez J, Lacombe E, Briat JF, Gaymard F. (2013) Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J Biol Chem 288: 22670–22680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. (1999) Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ. (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P, Benning C. (2002) Galactolipids rule in seed plants. Trends Plant Sci 7: 112–118 [DOI] [PubMed] [Google Scholar]
- Drew MC. (1975) Comparison of the effects of a localized supply of phosphate, nitrate, ammonium and potassium on the growth of the seminal root system, and the shoot, in barley. New Phytol 75: 479–490 [Google Scholar]
- Duff SMG, Sarath G, Plaxton WC. (1994) The role of acid phosphatases in plant phosphorus metabolism. Physiol Plant 90: 791–800 [Google Scholar]
- Franco-Zorrilla JM, González E, Bustos R, Linhares F, Leyva A, Paz-Ares J. (2004) The transcriptional control of plant responses to phosphate limitation. J Exp Bot 55: 285–293 [DOI] [PubMed] [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S. (2012) The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488: 535–539 [DOI] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y. (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J, Marin E, Floriani M, Chiarenza S, Richaud P, Nussaume L, Thibaud MC. (2006) Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88: 1767–1771 [DOI] [PubMed] [Google Scholar]
- Hirsch J, Misson J, Crisp PA, David P, Bayle V, Estavillo GM, Javot H, Chiarenza S, Mallory AC, Maizel A, et al. (2011) A novel fry1 allele reveals the existence of a mutant phenotype unrelated to 5′→3′ exoribonuclease (XRN) activities in Arabidopsis thaliana roots. PLoS ONE 6: e16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Guerinot ML. (2009) Homing in on iron homeostasis in plants. Trends Plant Sci 14: 280–285 [DOI] [PubMed] [Google Scholar]
- Jouhet J, Maréchal E, Baldan B, Bligny R, Joyard J, Block MA. (2004) Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J Cell Biol 167: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno S, Yamawaki M, Ishibashi H, Kobayashi NI, Hirose A, Tanoi K, Nussaume L, Nakanishi TM. (2012) Development of real-time radioisotope imaging systems for plant nutrient uptake studies. Philos Trans R Soc Lond B Biol Sci 367: 1501–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Peng M, Rothstein SJ. (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Mukatira UT, D’Urzo MP, Damsz B, Raghothama KG. (2002) Regulated expression of Arabidopsis phosphate transporters. Plant Physiol 130: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolari KK, Sarjala T. (1995) Acid phosphatase activity and phosphorus nutrition in Scots pine needles. Tree Physiol 15: 747–752 [DOI] [PubMed] [Google Scholar]
- Klimyuk VI, Nussaume L, Harrison K, Jones JD. (1995) Novel GUS expression patterns following transposition of an enhancer trap Ds element in Arabidopsis. Mol Gen Genet 249: 357–365 [DOI] [PubMed] [Google Scholar]
- Lin WY, Huang TK, Chiou TJ. (2013) Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25: 4061–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HM. (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29: 751–760 [DOI] [PubMed] [Google Scholar]
- Lipinski CA. (2004) Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1: 337–341 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JCF, Steiner P, Hirling H, Unser M. (2004) Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58: 167–176 [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Thibaud MC, Bechtold N, Raghothama K, Nussaume L. (2004) Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Moellering ER, Benning C. (2011) Galactoglycerolipid metabolism under stress: a time for remodeling. Trends Plant Sci 16: 98–107 [DOI] [PubMed] [Google Scholar]
- Nakamura Y. (2013) Phosphate starvation and membrane lipid remodeling in seed plants. Prog Lipid Res 52: 43–50 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Koizumi R, Shui G, Shimojima M, Wenk MR, Ito T, Ohta H. (2009) Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc Natl Acad Sci USA 106: 20978–20983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang RA, Bruene A, Altmann T. (2000) Analysis of phosphate acquisition efficiency in different Arabidopsis accessions. Plant Physiol 124: 1786–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC. (2011a) Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussaume L, Marechal E, Thibaud M, Block M. (2011b) Plant plasma membrane and phosphate deprivation. In AS Murphy, W Peer, B Schulz, eds, The Plant Plasma Membrane. Plant Cell Monographs Series Vol. 19. Springer, Berlin, Heidelberg, Germany, pp 237–251 [Google Scholar]
- Ohkama-Ohtsu N, Wasaki J. (2010) Recent progress in plant nutrition research: cross-talk between nutrients, plant physiology and soil microorganisms. Plant Cell Physiol 51: 1255–1264 [DOI] [PubMed] [Google Scholar]
- Ortega D, Raynal M, Laudié M, Llauro C, Cooke R, Devic M, Genestier S, Picard G, Abad P, Contard P, et al. (2002) Flanking sequence tags in Arabidopsis thaliana T-DNA insertion lines: a pilot study. C R Biol 325: 773–780 [DOI] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua NH. (2014) NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T. (2011) Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci 16: 442–450 [DOI] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Rouached H, Secco D, Arpat BA. (2010) Regulation of ion homeostasis in plants: current approaches and future challenges. Plant Signal Behav 5: 501–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H, Stefanovic A, Secco D, Bulak Arpat A, Gout E, Bligny R, Poirier Y. (2011) Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant J 65: 557–570 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Cruz-Ramírez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L. (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol 46: 174–184 [DOI] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Gutiérrez-Ortega A, Hernández-Abreu E, Herrera-Estrella L. (2006) Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol 140: 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrobert C, Thibaud MC, Contard-David P, Gineste S, Bechtold N, Robaglia C, Nussaume L. (2000) Identification of an Arabidopsis thaliana mutant accumulating threonine resulting from mutation in a new dihydrodipicolinate synthase gene. Plant J 24: 357–367 [DOI] [PubMed] [Google Scholar]
- Shen J, Yuan L, Zhang J, Li H, Bai Z, Chen X, Zhang W, Zhang F. (2011) Phosphorus dynamics: from soil to plant. Plant Physiol 156: 997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T. (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Abel S. (2004a) Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9: 548–555 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S. (2004b) Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J 37: 801–814 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Lucero RD, Sakhonwasee S, Adamson AW, Creff A, Nussaume L, Desnos T, Abel S. (2009) ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc Natl Acad Sci USA 106: 14174–14179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Hurley BA, Plaxton WC. (2010a) Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Sci 179: 14–27 [Google Scholar]
- Tran HT, Qian W, Hurley BA, She YM, Wang D, Plaxton WC. (2010b) Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ 33: 1789–1803 [DOI] [PubMed] [Google Scholar]
- Vert G, Barberon M, Zelazny E, Séguéla M, Briat JF, Curie C. (2009) Arabidopsis IRT2 cooperates with the high-affinity iron uptake system to maintain iron homeostasis in root epidermal cells. Planta 229: 1171–1179 [DOI] [PubMed] [Google Scholar]
- Wang L, Li Z, Qian W, Guo W, Gao X, Huang L, Wang H, Zhu H, Wu JW, Wang D, et al. (2011) The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol 157: 1283–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG. (2008) The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol 147: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhleniuk OV, Raines CA, Lloyd JC. (2001) pho3: a phosphorus-deficient mutant of Arabidopsis thaliana (L.) Heynh. Planta 212: 529–534 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chow TF, Puckrin RS, Alfred SE, Korir AK, Larive CK, Cutler SR. (2007) Chemical genetic interrogation of natural variation uncovers a molecule that is glycoactivated. Nat Chem Biol 3: 716–721 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.