Abstract
The causative agent of a recent outbreak of an atypical pneumonia, known as severe acute respiratory syndrome (SARS), has been identified as a coronavirus (CoV) not belonging to any of the previously identified groups. Fusion of coronaviruses with the host cell is mediated by the envelope spike protein. Two regions within the spike protein of SARS-CoV have been identified, showing a high degree of sequence conservation with the other CoV, which are characterized by the presence of heptad repeats (HR1 and HR2). By using synthetic and recombinant peptides corresponding to the HR1 and HR2 regions, we were able to characterize the fusion-active complex formed by this novel CoV by CD, native PAGE, proteolysis protection analysis, and size-exclusion chromatography. HR1 and HR2 of SARS-CoV associate into an antiparallel six-helix bundle, with structural features typical of the other known class I fusion proteins. We have also mapped the specific boundaries of the region, within the longer HR1 domain, making contact with the shorter HR2 domain. Notably, the inner HR1 coiled coil is a stable α-helical domain even in the absence of interaction with the HR2 region. Inhibitors binding to HR regions of fusion proteins have been shown to be efficacious against many viruses, notably HIV. Our results may help in the design of anti-SARS therapeutics.
The causative agent of the outbreak of the atypical pneumonia known as severe acute respiratory syndrome (SARS) (1) has been identified as a coronavirus (CoV) (2–9). Phylogenetic analysis of the SARS-associated CoV (SARS-CoV) shows that it is neither a mutant nor a recombinant of previously characterized CoV (10, 11), and forms a new, distinct group within the genus (3, 4, 8, 9).
The fusogenic envelope glycoprotein of CoV, the spike protein (S protein), which is displayed in ≈200 copies on the viral membrane as a trimer (12, 13), is divided into two subdomains of similar size: S1 and S2, which have distinct functions (11). S1, which forms the globular portion of the spike, mediates binding to host cell receptors (11, 14–16). For SARS-CoV, the receptor-binding domain is localized to amino acids 318–510 (15, 16), which bind angiotensin-converting enzyme 2 (15, 17). S2, which is more conserved (9, 11), forms the membrane-anchored stalk region (18) and mediates viral-host cell fusion (13, 19–21). Cleavage between S1/S2 is not an absolute requirement for the fusion of CoV (4, 11, 17, 21–24), and the available data suggest that the S protein of SARS-CoV is not cleaved into subunits (4, 9, 17).
The SARS-CoV S protein shares little amino acid similarity with other CoV (9). However, S2 contains two predicted amphipathic α-helical regions, with a 4,3 heptad repeat (HR), characteristic of coiled coils (9). The position and sequence of these HR regions are conserved among all groups of coronaviruses (18), including SARS-CoV (25).
HR regions are a common feature of Type I viral fusion proteins (26–40). These usually feature an N-terminal HR (HR1) adjacent to the fusion peptide, and a C-terminal HR (HR2) close to the transmembrane domain. In the current paradigm of virus-cell fusion mediated by these proteins, the HR form a six-helix bundle, with an inner trimeric coiled coil formed by HR1, onto which HR2 pack in antiparallel orientation. Formation of the six-helix bundle brings in close proximity the fusion peptide, which is N-terminal to HR1 and embedded in the target cell membrane, and the transmembrane domain, which is immediately downstream of HR2, thereby driving the two membranes in close contact, favoring the formation of a fusion pore (reviewed in refs. 41–43). Here we describe a systematic study of SARS-CoV HR regions, which allowed us to characterize the fusion-active complex formed by this novel CoV.
Materials and Methods
Peptides. All of the peptides in Table 1 were prepared by standard solid-phase methods, whereas the HR1 peptide spanning residues 871–972 was produced as a fusion to maltose-binding protein (MBP) by standard recombinant DNA methods. For full details see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Table 1. Synthetic SARS-CoV HR1 and HR2 peptides used in the study.
CD Spectroscopy. CD was performed on a J-710 spectropolarimeter (Jasco, Easton, MD) and at 20°C by using a rectangular quartz cell with a 0.1-cm path length. Spectra were acquired by using an 8-sec time response and a 5-nm/min scan speed; the spectra were averaged for two acquisitions. Stock solution concentration was determined by quantitative amino acid analysis. The results are reported as mean residue ellipticity [θ] having units of deg·cm2·dmol–1. Individual peptides and mixtures of HR1/HR2 peptides were analyzed at 10 μM concentration in 20 mM sodium phosphate, pH 7.3, with or without 150 mM NaCl. The percentage of α-helix was calculated according to Chen et al. (44). Thermal stability was determined at a 10 μM concentration by monitoring the change in the CD signal at 222 nm as a function of temperature, with a 10°C/h increase and 16-sec integration time.
Native PAGE. Equimolar mixtures of C1 (0.6 mM in phosphate buffer, pH 7.3) and one of the HR1 peptides N1–N7 were incubated at 25°C for 10 min (final volume of 5 μl). After the addition of an equal volume of 2× N-tris(hydroxymethyl)methylglycine (Tricine) sample buffer (0.125 M Tris·HCl, pH 6.8/10% glycerol/0.004 g of bromophenol blue) the mixture was analyzed by PAGE on 12% Tricine gel with a Tricine/glycine running buffer (pH 8.3).
Analytical Size-Exclusion Chromatography. The apparent Mr of the N0N3/C2 complex was estimated with a Superdex 75 size-exclusion column (Amersham Pharmacia), equilibrated with 10 mM sodium phosphate/150 mM NaCl, pH 7.5, (flow rate 0.5 ml/min). See also Supporting Materials and Methods.
Proteolysis Protection Experiments. Stock solutions (100 μM) of peptides C2, C3, N3, N0N3, N3N6, and HR1 [871–972]-MBP fusion protein were prepared in 20 mM Tris·HCl, pH 7.5. The peptide complexes were formed by incubating equal volumes (100 μl) of the components (final concentration of 50 μM each) for 1 h at 4°C. Proteinase K digestion was initiated by enzyme addition [1/150 proteinase K/peptide (wt/wt)], and carried out at 4°C. At selected time points an aliquot of the mixture was analyzed by HPLC-MS on a Jupiter C4 column (150 × 4.6 mm, 5 μm, 300 Å) with linear acetonitrile gradients, allowing the identification of the digested fragments. Proteolysis of the individual peptides was performed in the same conditions at 50 μM concentration.
Results
Location of HR1 and HR2. We used the programs learncoil-vmf (45) and multicoil (46) to predict the location of SARS-CoV HR1 and HR2. learncoil-vmf locates HR1 and HR2 at residues 900-1005 and 1151–1185, respectively, whereas multicoil locates HR1 and HR2 at residues 900–974 and 1148–1193, respectively (see Fig. 6, which is published as supporting information on the PNAS web site).
HR1 Peptides. We initially prepared a series of partially overlapping HR1 peptides (N1–N7) spanning the whole HR1 domain, as predicted by multicoil (Table 1). The peptides were designed to include five HR, a length sufficient to nucleate a stable coiled coil (47–51), shifted by one HR. Our peptide set was thus designed to locate the region(s) within HR1 that interacts with the HR2 domain.
The secondary structure of the HR1 peptides was analyzed by CD (Fig. 1a). Peptides N1–N4 are unordered, peptides N5 and N7 are partially helical, and, remarkably, peptide N6 is fully helical.
Fig. 1.
(a) CD spectra of peptides N1–N7. The calculated percentage of α-helix is indicated. (b) CD analysis of the interaction between peptides N3 and C2. Shown is the observed spectrum (solid trace) and the arithmetical sum of the spectra of the individual peptides (dashed trace). The calculated percentage of α-helix of the N3/C2 complex is 57%.
HR2 Peptides. We prepared three peptides: peptide C1, corresponding to the entire HR2 region, as predicted by learncoil-vmf, extended at the N terminus to residue 1148; peptide C2, corresponding to the entire HR2 region as predicted by multicoil, and peptide C3, which spans residues 1116–1185, beginning 32 residues upstream of the predicted HR2 domain (Table 1). CD analysis at neutral pH revealed an equilibrium between unordered and helical structure, higher in peptide C2: the calculated percentage of α-helix (44) was 22%, 42%, and 26% for C1, C2, and C3, respectively (Fig. 7, which is published as supporting information on the PNAS web site).
Mapping of the HR1/HR2 Interaction. We then mapped the interaction between the HR1 and HR2 peptides. We reasoned that, by analogy with other fusion proteins (26, 29, 33, 36, 39, 52) also for SARS-CoV, the interaction between the HR1 and HR2 regions should be mediated by interaction of helical domains. Among the HR1 and HR2 peptides only N6 is fully helical, whereas the others are unordered or in a conformational equilibrium between unordered–helical structure (Fig. 1a). Therefore, we expected that complex formation between one HR1/HR2 pair should be accompanied by a conformational transition toward α-helical structure.
To identify which region(s) of the longer HR1 makes direct contact with the HR2 domain, each one of peptides N1–N7 was mixed with an equimolar amount of either C1 or C2, and CD spectra were recorded. To highlight any conformational transition induced by complex formation, each spectrum of a mixture was compared with the arithmetical sum of the spectra of the individual peptides: in the absence of an interaction, the two curves should be superimposable, which is the case for all of the N peptides with the exception of N3: the combination of N3 with either C1 or C2 gives rise to a new spectrum, with a higher content of α-helix (Fig. 1b; the CD spectra of the other N peptides in complex with C1 are shown in Fig. 8, which is published as supporting information on the PNAS web site). Notably, neither N2 nor N4, which are contiguous and mostly overlapping with N3, interact with C1. This finding is not surprising, given the small size of the peptides, and the highly cooperative nature of this type of interaction.
We then independently mapped the HR1/HR2 interaction by PAGE. The results, which were similar for C1 and C2, are shown for the former (Fig. 2). No interaction is observed for peptides N5, N6, and N7, whereas the combination of equimolar amounts of N3 and C1 gives rise to the disappearance of the individual bands¶ and the formation of a new band at an intermediate position corresponding to the complex. In addition, complex formation was also observed with the pairs N1/C1, N2/C1, and N4/C1 but without complete disappearance of the individual components, which is a qualitative indication of lower affinity with respect to the fully formed N3/C1 complex. Formation of lower stability complexes was favored by the 20-fold higher concentration used for the gel-shift experiments with respect to CD. Therefore, the two sets of data, CD and PAGE, agree in locating the key region of interaction within peptide N3 (HR1 residues 914–949) and peptide C1 (HR2 1148–1185).
Fig. 2.
Analysis of the interaction between peptides N1–N7 and peptide C1 by native PAGE. The data were identical for peptide C2.
Based on the finding that (i) the core of the HR1 region interacting with HR2 is encompassed by peptide N3, (ii) the interaction might extend up to peptide N1, and (iii) peptide N6, despite its short length, is able to adopt a fully helical fold (Fig. 1a), we prepared three longer peptides: peptide N3N6, amino acids 914–970; peptide N1N3, amino acids 900–949; and peptide N0N3, amino acids 890–949, which extends beyond the predicted boundary of HR1 and toward the predicted fusion peptide (25, 53) (Table 1).
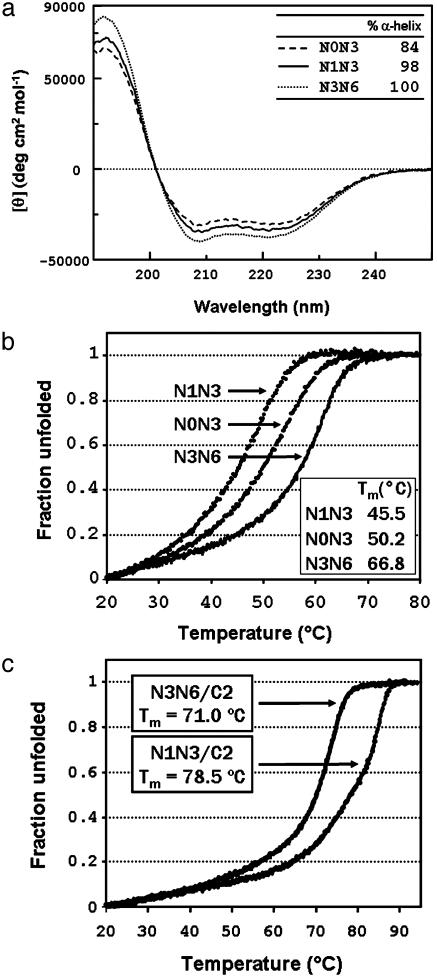
Peptides N1N3 and N3N6 were found to be fully helical, whereas the calculated percentage of α-helix was slightly lower (84%) for N0N3, in line with the predicted boundaries of the HR1 region (Fig. 3a). Thermal denaturation studies were performed at the concentration of 10 μM. All three peptides showed a reversible cooperative transition by thermal denaturation, with melting points (Tm) of 45.5°C, 50.2°C, and 66.8°C for N1N3, N0N3, and N3N6, respectively (Fig. 3b).
Fig. 3.
(a) CD spectra of peptides N0N3, N1N3, and N3N6 (Table 1). The calculated percentage of α-helix is indicated. (b) Thermal denaturation curves with indicated Tm values of peptides N0N3, N1N3, and N3N6. (c) Thermal denaturation curves with indicated Tm values of the N1N3/C2 and N3N6/C2 peptide complexes.
It appears, therefore, that the HR1 domain of SARS-CoV forms a continuous helical domain, which is fully formed in the absence of any interaction with the HR2 domain. This result is unlike HIV gp41, in which the N-peptides are unable to form a trimeric coiled coil in the absence of the C-peptide (54), and is more similar to the influenza virus hemagglutinin (26, 28, 55).
Despite being only 84% helical, N0N3 is more stable than N1N3; thus, the N-terminal boundary of HR1 should be positioned between residues 890 and 900, whereas the C-terminal boundary should at least include residue 970.
We then examined complex formation between the longer N-peptides and the HR2 peptides C1 and C2. The interaction was apparent for all of the three peptides (the CD of the complex between N3N6 and C1 is shown in Fig. 9, which is published as supporting information on the PNAS web site). Importantly, the α-helical content was increased in all of the complexes, consistent with folding of the HR2 peptides into a α-helical structure upon complex formation.
Thermal denaturation analysis shows that complex formation exerts a stabilizing effect with respect to the isolated HR1 peptides (Fig. 3c). The increase in Tm was particularly large for N0N3/C2. For this complex, complete denaturation was not yet observed at 90°C, i.e., >40°C higher than the Tm measured for N0N3. Also for the N1N3/C2 complex, we measured a Tm (78.5°C) that is 33°C higher than that measured for N1N3. The stabilization effect was much lower in the case of N3N6/C2 with a melting point of 71.0°C, only 4.2°C higher than the N3N6 alone, which however shows considerable stability by itself. The observed Tm values are comparable with those reported for the murine hepatitis coronavirus (25).
The thermal denaturation data on the complexes, showing that the stability of N0N3/C2 complex is much higher than that of the complex N1N3/C2, brings further support to the hypothesis that the N-terminal domain of HR1 might extend beyond residue 900 to include at least some residues that are comprised in N0N3 only.
Mr of the HR1/HR2 Complex in Physiologic Conditions. We performed size-exclusion chromatography experiments in physiologic conditions on the more stable N0N3/C2 complex (Fig. 4). The elution volume, relative to Mr standards, gave an apparent Mr of 39,530 Da, in good agreement with the Mr (35,360 Da) expected for a hexameric complex, formed by three molecules of N0N3 and three molecules of C2.
Fig. 4.
(a) Size-exclusion chromatography of the N0N3/C2 complex on Superdex 75. Indicated are the peaks corresponding to the complex and to excess peptide C2 and the position of isolated N0N3 run in the same conditions. (b) Mr standard calibration curve and interpolated Mr of the complex (arrow).
Mapping of HR1/HR2 Interaction by Proteolysis Protection Experiments. To identify the boundaries and the relative orientation and position of HR2 with respect to HR1 in the complex, we performed proteolysis protection experiments with proteinase K, as previously done for murine hepatitis virus (25). To this aim, we produced a recombinant peptide spanning residues 871–972 of the S protein, i.e., beginning 29 residues upstream of the predicted N terminus of the HR1 domain as a fusion with MBP.
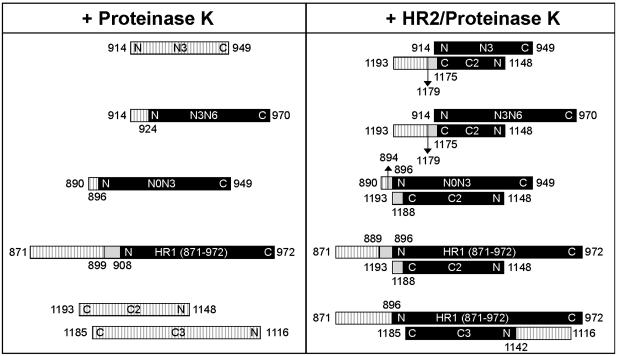
The proteolysis protection experiments are summarized in Fig. 5. Samples of individual peptides and the indicated HR1/HR2 equimolar complexes were incubated with proteinase K, and the resulting fragments were analyzed by reversed-phase HPLC-MS to unequivocally establish the sequence of each protease-resistant core peptide.
Fig. 5.
Proteolysis protection analysis of the HR1/HR2 complexes. The indicated individual peptides and mixtures were subjected to proteinase K digestion. Aliquots of the reaction mixture were analyzed by HPLC-MS. The results shown are all referred to the 5-h incubation time point. The peptides are represented by bars, with the name indicated as in Table 1. Hatched bars indicate the parts of the peptide that were readily digested by the protease, black bars indicate the protease-resistant parts, and light gray bars indicate the regions digested at a slower rate.
Digestion of isolated HR1 peptides showed the presence of a stable core in the longer peptides. Although peptide N3 is completely digested, N- and C-terminally extended peptides show increased stability. C-terminal extension up to 970 in peptide N3N6 confers stability, with cleavage only occurring at the N terminus at residue 924. Also, the N-terminal extension of N3 to amino acid 890 in peptide N0N3 confers stability, with cleavage occurring only at the N terminus at residue 896. The larger construct HR1 [871–972]-MBP was digested to fragments 899–972 and 908–972. The HR2 peptides C2 and C3, instead, were completely digested. However, when the smaller peptides N3 and C2 were complexed together, they were partially protected by degradation after 5 h. Full degradation was instead observed after 24 h. For all of the other complexes examined, the pattern at 24 h was comparable to the one at 5 h, and the latter common time point is shown in Fig. 5. For N3/C2, the protease-resistant core includes the entire N3 peptide (amino acids 914–949), whereas C2 is trimmed at the C terminus at the two positions 1179 and 1175. We used this information to align the N terminus of N3 with respect to the C terminus of C2.
The boundaries of the interaction were further probed with the longer N-peptides. The C-terminal extension of N3 to amino acid 970 in peptide N3N6 does not alter the pattern of proteolysis of C2 in the N3N6/C2 complex, with cleavages at its C terminus at positions 1179 and 1175. In the complex, N3N6 is completely protected. Conversely, the N-terminal extension of N3 to amino acid 890 in peptide N0N3 alters the pattern of proteolysis of C2 at its C terminus in the N0N3/C2 complex: complete protection is now observed up to residue 1188, and partial protection is observed up to residue 1193. The proteolysis at the N terminus of N0N3 occurs mainly at amino acid 896, as observed for the uncomplexed peptide, but with some additional protection up to amino acid 894. These results unambiguously establish the expected antiparallel orientation of the HR1/HR2 domains.
In the HR1 [871–972]-MBP/C2 complex, proteinase K hydrolyzes the N terminus of HR1 up to residue 896, with partial protection up to residue 889. HR2 is digested at the C terminus up to residue 1188, with partial protection up to residue 1193, as observed in the N0N3/C2 complex. As previously seen for the smaller HR1/HR2 complexes, the C terminus of HR1 and the N terminus of HR2 are not hydrolyzed. Finally, we examined the complex between HR1 [871–972]-MBP and the elongated HR2 peptide C3. The C terminus of C3 is contained within the previously established boundary of the protected core, and, accordingly, it remains stable. The elongated N-portion, however, is hydrolyzed by proteinase K but only up to residue 1142, which indicates that the HR1/HR2 interaction extends at least a few residues beyond the N-terminal boundary of HR2 predicted by both multicoil and learncoil-vmf (Fig. 6).
Discussion
No information is currently available on the fusion mechanism of the newly discovered coronavirus, which originates the human disease known as SARS (55–57). The only clue to how SARS-CoV fuses with the target cell is the presence in the sequence of the predicted HR regions HR1 and HR2 (9), whose position and length are conserved in SARS-CoV with respect to group I and II CoV (25), despite the low overall sequence conservation of the S protein (9). This higher degree of conservation of the HR regions that are typically associated with type I fusion proteins (27) suggests that they play a key role in the fusion of SARS-CoV. We report here a systematic study on peptides spanning the predicted sequence of the SARS-CoV HR regions, which leads to the conclusion that the HR1 and HR2 of this new virus can indeed form the stable complex typical of type I fusion proteins. More specifically, we show that:
HR1 interacts with the HR2 domain, forming a complex with an apparent molecular mass in good agreement with that expected for a six-helical bundle, as assessed by size-exclusion chromatography (Fig. 4).
Proteolysis protection experiments show that HR1 and HR2 are arranged in antiparallel fashion in the complex (Fig. 5).
The boundaries of the HR1/HR2 interaction are mapped to residues HR1 896–972 and HR2 1142–1188, possibly extending a few more residues at the N terminus of HR1 (up to residue 889) and the C terminus of HR2 (up to amino acid 1193, which is the predicted boundary of the S2 transmembrane domain); these boundaries would position the fusion peptide in the region 870–890.
The HR1 region constitutes by itself a highly stable conformational domain, even in the absence of interaction with HR2, as assessed by CD, thermal denaturation, and proteolysis protection experiments (Figs. 3 a and b, 4, and 5).
Overall, our data validate the initial expectation that the fusionactive structure of SARS-CoV features an inner trimeric coiled coil formed by HR1, onto which three HR2 peptides fold in antiparallel fashion into a six-helical bundle. Moreover, our definition of the boundaries of the interacting HR regions of SARS-CoV bears a striking resemblance with the equivalent complex of the murine hepatitis CoV (25). It would seem, therefore, that the membrane fusion and cell entry mechanisms exploited by this SARS coronavirus is similar to other CoV and more in general typical of viruses with class I fusion proteins (26, 33, 36, 39, 51, 52).
Some features of the fusion-active complex of SARS-CoV resemble those of paramyxoviruses. First, we find for SARS-CoV, as observed for the paramyxovirus SV5 F1 protein (39), that no linker region or a very short linker region is interposed between the end of the HR2 domain and the predicted beginning of the transmembrane domain (amino acid 1193). For paramyxoviruses it was suggested that the lack of a membrane-proximal domain is compensated by the presence of the large intervening domain connecting HR1 and HR2, which may provide the flexibility required for the conformational changes during fusion (39). This domain is 250 aa in length in SV5, i.e., considerably longer than in most other fusion proteins, where the region between HR1/HR2 is only 5–26 aa. It is therefore intriguing that, in SARS-CoV, the two HR are separated by a long region of 170 aa.
Second, the fusion-active structure of SF5 shows that although HR1 is fully helical, HR2 contains both a helical and an extended region, which, as our data suggest, might be the case for SARS-CoV as well. On one side, proteolysis protection of the N3/C2 complex shows a proteinase K-resistant core including the whole 36 residues of N3 (amino acids 914–949), but only the 27 N-terminal amino acid of C2 (residues 1148–1175). Cleavage of C2 at residue 1175 allows us to align this residue with HR1 residue 914. On the other side, native PAGE analysis of the interaction of C2 with the overlapping set of N1-N7 peptides indicates that the contact of C2 with N3 extends to the 942–949 region of N3, because the complex formed by C2 with peptide N3 is more stable that the one formed with either peptide N2, which is shifted seven residues along HR1 (Table 1 and Fig. 2). We know that the HR2 domain, which in isolation is in equilibrium between unordered and helical structure (Fig. 7), becomes more helical in the complex (Fig. 9). However, if we assumed C2 to be fully helical, the 27 amino acids protected in the N3/C2 complex would align perfectly well with residues 914–940, which are contained within peptide N2 (amino acids 907–942). The complex N2/C2 should then be as stable as N3, which is not the case. Overall, then, the most likely interpretation of our data is that the interaction of the 27 amino acids of C2 spans the whole of the HR1 region 914–949, which is only possible if C2 displays a mixture of helical and extended structure, like in SV5. Although proof of this hypothesis may only come from detailed structural studies, we suggest, based on the above findings, that coronaviruses and paramyxoviruses (27, 39, 59) may adopt a similar mechanism for membrane fusion.
The information provided in this study may be useful for the design of antiviral compounds. Synthetic peptides derived from the HR sequences of retroviruses (54, 60–66) and paramyxoviruses (67–70) have been shown to be potent inhibitors of viral fusion. For HIV, a peptide derived from the HR2 of gp41 has just been approved as a human therapeutic (71). Most recently, a peptide derived from the HR2 of murine hepatitis CoV was shown to inhibit viral entry and syncytia formation (25). It is thus reasonable to expect that HR-based inhibitors may be developed for SARS-CoV. Work along these lines is in progress, taking advantage of the wealth of information available on peptide fusion inhibitors (54, 60, 61, 63–69, 71).
Supplementary Material
Acknowledgments
We thank Fabio Talamo and Laura Orsatti for analytical MS; Marco Mattu for help with protein expression; Manuela Emili for the artwork; and Peter S. Kim, Gennaro Ciliberto, and Riccardo Cortese for critical revision of the manuscript.
Abbreviations: CoV, coronavirus; HR, heptad repeat; SARS, severe acute respiratory syndrome; S protein, spike protein; MBP, maltose-binding protein; Tm, melting temperature.
Note Added in Proof. Since our manuscript was submitted, two independent reports appeared on the biophysical characterization of the fusion-active core of SARS-CoV (72, 73). Both studies are in full agreement with our study on the antiparallel six-helical bundle structure of the complex. However, our study shows that the N-terminal boundary of the HR1 region involved in the complex is residue 896, i.e., ≈20 residues upstream of the boundary indicated by these other studies (in both, residue 916).
Footnotes
Although the band corresponding to the charged HR2 peptides was always visible in these conditions, for most hydrophobic N peptides the band of the isolated peptide was not visible in the gel. Care was taken to ensure that the combination experiments were performed with titrated, equimolar amounts of the two components. Observation of a residual HR2 peptide band in the presence of the complex can therefore be taken as an indication that the complex is not fully formed at equimolarity and used as a qualitative measure of the relative stability of the complexes.
References
- 1.Peiris, J. S., Yuen, K. Y., Osterhaus, A. D. & Stohr, K. (2003) N. Engl. J. Med. 349, 2431–2441. [DOI] [PubMed] [Google Scholar]
- 2.Drosten, C., Gunther, S., Preiser, W., van der Werf, S., Brodt, H.-R., Becker, S., Rabenau, H., Panning, M., Kolesnikova, L., Fouchier, R. A. M., et al. (2003) N. Engl. J. Med. 348, 1967–1976. [DOI] [PubMed] [Google Scholar]
- 3.Eickmann, M., Becker, S., Klenk, H. D., Doerr, H. W., Stadler, K., Censini, S., Guidotti, S., Masignani, V., Scarselli, M., Mora, M., et al. (2003) Science 302, 1504–1505. [DOI] [PubMed] [Google Scholar]
- 4.Stadler, K., Masignani, V., Eickman, M., Becker, S., Abrugnani, S., Klenk, H.-D. & Rappuoli, R. (2003) Nat. Rev. Microbiol. 1, 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiris, J., Lai, S., Poon, L., Guan, Y., Yam, L., Lim, W., Nicholls, J., Yee, W., Yan, W. & Cheung, M. (2003) Lancet 361, 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ksiazek, T. G., Erdman, D., Goldsmith, C. S., Zaki, S. R., Peret, T., Emery, S., Tong, S., Urbani, C., Comer, J. A., Lim, W., et al. (2003) N. Engl. J. Med. 348, 1953–1966. [DOI] [PubMed] [Google Scholar]
- 7.Kuiken, T., Fouchier, R. A. M., Schutten, M., Rimmelzwaan, G. F., van Amerongen, G., van Riel, D., Laman, J. D., de Jong, T., van Doornum, G. & Lim, W. (2003) Lancet 362, 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marra, M. A., Jones, S. J. M., Astell, C. R., Holt, R. A., Brooks-Wilson, A., Butterfield, Y. S. N., Khattra, J., Asano, J. K., Barber, S. A., Chan, S. Y., et al. (2003) Science 300, 1399–1404. [DOI] [PubMed] [Google Scholar]
- 9.Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., Penaranda, S., Bankamp, B., Maher, K., Chen, M.-H., et al. (2003) Science 300, 1394–1399. [DOI] [PubMed] [Google Scholar]
- 10.Holmes, K. V. (2001) in Fields Virology, eds. Fields, B., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B., Straus, S. E. & Knipe, D. M. (Lippincott Williams and Wilkins, New York), 4th Ed., Vol. 1, pp. 1187–1203. [Google Scholar]
- 11.Lai, M. M. C. & Holmes, K. V. (2001) Fields Virology (Lippincott Williams and Wilkins, New York), eds. Fields, B., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B., Straus, S. E. & Knipe, D. M. (Lippincott Williams and Wilkins, New York), 4th Ed., Vol. 1, pp. 1163–1185. [Google Scholar]
- 12.Delmas, B. & Laude, H. (1990) J. Virol. 64, 5367–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao, X., Chakraborti, S., Dimitrov, A. S., Gramatikoff, K. & Dimitrov, D. S. (2003) Biochem. Biophys. Res. Commun. 312, 1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taguchi, F. (1995) J. Virol. 69, 7260–7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., Somasundaran, M., Sullivan, J. L., Luzuriaga, K., Greenough, T. C., et al. (2003) Nature 426, 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong, S. K., Li, W., Moore, M. J., Choe, H. & Farzan, M. (2003) J. Biol. Chem. [DOI] [PMC free article] [PubMed]
- 17.Dimitrov, D. S. (2003) Cell 115, 652–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Groot, R. J., Luytjes, W., Horzinek, M. C., van der Zeijst, B. A., Spaan, W. J. & Lenstra, J. A. (1987) J. Mol. Biol. 196, 963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoo, D. W., Parker, M. D. & Babiuk, L. A. (1991) Virology 180, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Groot, R. J., Van Leen, R. W., Dalderup, M. J., Vennema, H., Horzinek, M. C. & Spaan, W. J. (1989) Virology 171, 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taguchi, F. (1993) J. Virol. 67, 1195–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bos, E. C. W., Heijnen, L., Luytjes, W. & Spaan, W. J. M. (1995) Virology 214, 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hingley, S. T., Leparc-Goffart, I. & Weiss, S. R. (1998) J. Virol. 72, 1606–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stauber, R., Pfleiderera, M. & Siddell, S. (1993) J. Gen. Virol. 74, 183–191. [DOI] [PubMed] [Google Scholar]
- 25.Bosch, B. J., van der Zee, R., de Haan, C. A. M. & Rottier, P. J. M. (2003) J. Virol. 77, 8801–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullough, P. A., Hughson, F. M., Skehel, J. J. & Wiley, D. C. (1994) Nature 371, 37–43. [DOI] [PubMed] [Google Scholar]
- 27.Colman, P. M. & Lawrence, M. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 309–319. [DOI] [PubMed] [Google Scholar]
- 28.Chen, J., Skehel, J. J. & Wiley, D. C. (1999) Proc. Natl. Acad. Sci. USA 96, 8967–8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. (1997) Cell 89, 263–273. [DOI] [PubMed] [Google Scholar]
- 30.Weissenhorn, W., Wharton, S. A., Calder, L. J., Earl, P. L., Moss, B., Aliprandis, E., Skehel, J. J. & Wiley, D. C. (1996) EMBO J. 15, 1507–1514. [PMC free article] [PubMed] [Google Scholar]
- 31.Malashkevich, V. N., Chan, D. C., Chutkowski, C. T. & Kim, P. S. (1998) Proc. Natl. Acad. Sci. USA 95, 9134–9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, Z. N., Mueser, T. C., Kaufman, J., Stahl, S. J., Wingfield, P. T. & Hyde, C. C. (1999) J. Struct. Biol. 126, 131–144. [DOI] [PubMed] [Google Scholar]
- 33.Fass, D., Harrison, S. C. & Kim, P. S. (1996) Nat. Struct. Biol. 3, 465–469. [DOI] [PubMed] [Google Scholar]
- 34.Malashkevich, V. N., Schneider, B. J., McNally, M. L., Milhollen, M. A., Pang, J. X. & Kim, P. S. (1999) Proc. Natl. Acad. Sci. USA 96, 2662–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weissenhorn, W., Calder, L. J., Wharton, S. A., Skehel, J. J. & Wiley, D. C. (1998) Proc. Natl. Acad. Sci. USA 95, 6032–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weissenhorn, W., Carfi, A., Lee, K. H., Skehel, J. J. & Wiley, D. C. (1998) Mol. Cell 2, 605–616. [DOI] [PubMed] [Google Scholar]
- 37.Kobe, B., Center, R. J., Kemp, B. E. & Poumbourios, P. (1999) Proc. Natl. Acad. Sci. USA 96, 4319–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malashkevich, V. N., Singh, M. & Kim, P. S. (2001) Proc. Natl. Acad. Sci. USA 98, 8502–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker, K. A., Dutch, R. E., Lamb, R. A. & Jardetzky, T. S. (1999) Mol. Cell 3, 309–319. [DOI] [PubMed] [Google Scholar]
- 40.Zhao, X., Singh, M., Malashkevich, V. N. & Kim, P. S. (2000) Proc. Natl. Acad. Sci. USA 97, 14172–14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckert, D. M. & Kim, P. S. (2001) Annu. Rev. Biochem. 70, 778–810. [DOI] [PubMed] [Google Scholar]
- 42.Skehel, J. J. & Wiley, D. C. (1998) Cell 95, 871–874. [DOI] [PubMed] [Google Scholar]
- 43.Skehel, J. J. & Wiley, D. C. (2000) Annu. Rev. Biochem. 69, 531–569. [DOI] [PubMed] [Google Scholar]
- 44.Chen, Y. H., Yang, J. T. & Chau, K. H. (1974) Biochemistry 13, 3350–3359. [DOI] [PubMed] [Google Scholar]
- 45.Singh, M., Berger, B. & Kim, P. S. (1999) J. Mol. Biol. 290, 1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolf, E., Kim, P. S. & Berger, B. (1997) Protein Sci. 6, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harbury, P. B., Kim, P. S. & Alber, T. (1994) Nature 371, 80–83. [DOI] [PubMed] [Google Scholar]
- 48.Harbury, P. B., Zhang, T., Kim, P. S. & Alber, T. (1993) Science 262, 1401–1407. [DOI] [PubMed] [Google Scholar]
- 49.Zhu, B. Y., Zhou, N. E., Semchuk, P. D., Kay, C. M. & Hodges, R. S. (1992) Int. J. Pept. Protein Res. 40, 171–179. [DOI] [PubMed] [Google Scholar]
- 50.Eckert, D. M., Malashkevich, V. N. & Kim, P. S. (1998) J. Mol. Biol. 284, 859–865. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki, K., Hiroaki, H., Kohda, D. & Tanaka, T. (1998) Protein Eng. 11, 1051–1055. [DOI] [PubMed] [Google Scholar]
- 52.Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. (1997) Nature 387, 426–430. [DOI] [PubMed] [Google Scholar]
- 53.Luo, Z. & Weiss, S. E. (1998) Virology 244, 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu, M., Blacklow, S. C. & Kim, P. S. (1995) Nat. Struct. Biol. 2, 1075–1082. [DOI] [PubMed] [Google Scholar]
- 55.Carr, C. M. & Kim, P. S. (1993) Cell 73, 823–832. [DOI] [PubMed] [Google Scholar]
- 56.Lee, N., Hui, D., Wu, A., Chan, P., Cameron, P., Joynt, G. M., Ahuja, A., Yung, M. Y., Leung, C. B., To, K. F., et al. (2003) N. Engl. J. Med. 348, 1986–1994. [DOI] [PubMed] [Google Scholar]
- 57.Tsang, K. W., Ho, P. L., Ooi, G. C., Yee, W. K., Wang, T., Chan-Yeung, M., Lam, W. K., Seto, W. H., Yam, L. Y., Cheung, T. M., et al. (2003) N. Engl. J. Med. 348, 1977–1985. [DOI] [PubMed] [Google Scholar]
- 58.Poutanen, S. M., Low, D. E., Henry, B., Finkelstein, S., Rose, D., Green, K., Tellier, R., Draker, R., Adachi, D., Ayers, M., et al. (2003) N. Engl. J. Med. 348, 1995–2005. [DOI] [PubMed] [Google Scholar]
- 59.Chen, L., Gorman, J. J., McKimm-Breschkin, J., Lawrence, L. J., Tulloch, P. A., Smith, B. J., Colman, P. M. & Lawrence, M. C. (2001) Structure (London) 9, 255–266. [DOI] [PubMed] [Google Scholar]
- 60.Root, M. J., Kay, M. S. & Kim, P. S. (2001) Science 291, 884–888. [DOI] [PubMed] [Google Scholar]
- 61.Louis, J. M., Bewley, C. A. & Clore, G. M. (2001) J. Biol. Chem. 276, 29485–29489. [DOI] [PubMed] [Google Scholar]
- 62.Bewley, C. A., Louis, J. M., Ghirlando, R. & Clore, G. M. (2002) J. Biol. Chem. 277, 14238–14245. [DOI] [PubMed] [Google Scholar]
- 63.Lombardi, S., Massi, C., Indino, E., La Rosa, C., Mazzetti, P., Falcone, M. L., Rovero, P., Fissi, A., Pieroni, O., Bandecchi, P., et al. (1996) Virology 220, 274–284. [DOI] [PubMed] [Google Scholar]
- 64.Eckert, D. M. & Kim, P. S. (2001) Proc. Natl. Acad. Sci. USA 98, 11187–11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eckert, D. M., Malashkevich, V. N., Hong, L. H., Carr, P. A. & Kim, P. S. (1999) Cell 99, 103–115. [DOI] [PubMed] [Google Scholar]
- 66.Judice, J. K., Tom, J. Y., Huang, W., Wrin, T., Vennari, J., Petropoulos, C. J. & McDowell, R. S. (1997) Proc. Natl. Acad. Sci. USA 94, 13426–13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joshi, S. B., Dutch, R. E. & Lamb, R. A. (1998) Virology 248, 20–34. [DOI] [PubMed] [Google Scholar]
- 68.Lambert, D. M., Barney, S., Lambert, A. L., Guthrie, K., Medinas, R., Davis, D. E., Bucy, T., Erickson, J., Merutka, G. & Petteway, S. R., Jr. (1996) Proc. Natl. Acad. Sci. USA 93, 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rapaport, D., Ovadia, M. & Shai, Y. (1995) EMBO J. 14, 5524–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao, Q. & Compans, R. W. (1996) Virology 223, 103–112. [DOI] [PubMed] [Google Scholar]
- 71.LaBonte, J., Lebbos, J. & Kirkpatrick, P. (2003) Nat. Rev. Drug Discovery 2, 345–346. [DOI] [PubMed] [Google Scholar]
- 72.Liu, S., Xiao, G., Chen, Y., He, Y., Niu, J., Escalante, C. R., Xiong, H., Farmar, J., Debnath, A. S., Tien, P. & Jiang, S. (2004) Lancet 363, 938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tripet, B., Howard, M. W., Jobling, M., Holmes, R. K., Holmes, K. V. & Hodges, R. S. (2004) J. Biol. Chem. 279, 20836–20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.