Abstract
Chromatin is generally classified as euchromatin or heterochromatin, each with distinct histone modifications, compaction levels, and gene expression patterns. Although the proper formation of heterochromatin is essential for maintaining genome integrity and regulating gene expression, heterochromatin can also spread into neighboring regions in a sequence-independent manner, leading to the inactivation of genes. Because the distance of heterochromatin spreading is stochastic, the formation of boundaries, which block the spreading of heterochromatin, is critical for maintaining stable gene expression patterns. Here we review the current understanding of the mechanisms underlying heterochromatin spreading and boundary formation.
Keywords: Boundary element, Heterochromatin, Spreading, Histone modifications, Silencing
Introduction
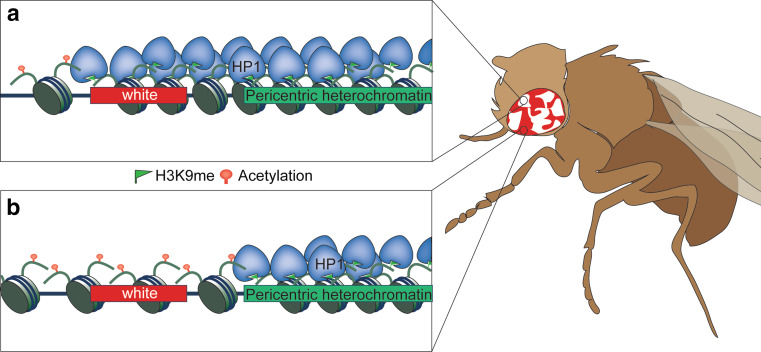
Eukaryotic genomic DNA is folded with histones and other proteins in the form of chromatin, which regulates every aspect of DNA metabolism, including transcription, replication, and DNA damage repair. Based on the level of compaction, chromatin is divided into euchromatin and heterochromatin. Euchromatin is generally gene rich, less condensed, and associated with active gene transcription, whereas heterochromatin is generally gene poor, highly condensed, and refractory to the transcription machinery. The discovery of position effect variegation (PEV) in the fruit fly Drosophila melanogaster in the 1930s paved the way to revealing the importance of chromatin in regulating gene expression [1]. The white gene, which is responsible for generating red color pigment in Drosophila eyes, normally resides in the euchromatic region. However, when the white gene is placed adjacent to pericentric heterochromatin due to chromosomal rearrangement, it is variably silenced and the different expression states are clonally inherited in different cell populations, resulting in mottled eyes [2] (Fig. 1). A similar phenomenon termed telomere position effect (TPE) was later observed in the budding yeast Saccharomyces cerevisiae, in which reporter genes placed near telomeres are also variably silenced and clonally inherited, resulting in sectored colonies [3, 4]. A general theme emerging from studies of these phenomena is that heterochromatin can spread variable distances into neighboring regions in a stochastic manner to regulate gene expression and that once established these expression states can be stably maintained through multiple cycles of cell divisions.
Fig. 1.
Position effect variegation in Drosophila. The normally euchromatic white gene is placed close to the pericentric heterochromatin due to X-ray induced chromosome inversion. During early Drosophila development, heterochromatin spreading in some progenitor cells results in the silencing of white. Such expression is clonally inherited in all the progenies of the same cell, resulting in white patches of the adult eye
The effect of heterochromatin on gene expression is not limited to reporter genes. For example, the similarities between transposon-mediated gene silencing in maize and PEV in Drosophila led Barbara McClintock to propose that transposable elements regulate the expression of neighboring genes [5]. Recent studies in Arabidopsis show that indeed transposable elements are sites of heterochromatin assembly and influence the expression of nearby genes [6]. In humans, heterochromatin domains expand to cover developmentally regulated genes during the differentiation of stem cells, resulting in large changes in the chromatin landscape [7]. But the variable nature of heterochromatin spreading can potentially lead to inappropriate expression of genes, which has been implicated in a number of serious human diseases [8]. For example, facioscapulohumeral dystrophy (FSHD) is a neuromuscular disorder predominantly affecting the skeletal muscles of the face and arms and has been correlated with deletions of D4Z4 repeats in the chromosome 4q35 subtelomeric region. Although the mechanism of this disease is still under debate, one hypothesis is that the loss of these repeats compromises heterochromatin spreading-mediated inactivation of adjacent genes [8, 9]. Therefore, the spreading of heterochromatin needs to be tightly controlled, and the discovery of boundary elements that can shield genes from position effects demonstrates their important roles in regulating gene expression. Given that the mechanisms of heterochromatin spreading and boundary formation are best studied in yeasts, in which precise genetic manipulations can be made, we will focus our review on studies conducted in yeasts and discuss their relevance to mechanisms in higher eukaryotes.
Heterochromatin spreading
It is well established that chromatin structure is regulated by both chromatin remodeling activities and the modification of histones and DNA [10]. Since many factors involved in PEV and TPE have been characterized as enzymes and proteins that regulate chromatin structure, heterochromatin assembly and spreading has been a paradigm for studying the roles of chromatin modifiers in regulating stably maintained chromatin states [2, 4]. The histones within heterochromatin regions are generally devoid of acetylation and are often methylated at H3 lysine 9 (H3K9me) [11–13]. While histone deacetylation can directly affect interactions between nucleosomes to form higher-order chromatin structures [14], histone methylation indirectly affects chromatin structure by either antagonizing acetylation at the same residue [15] or serving as a binding site for the recruitment of chromatin proteins [16]. H3K9me recruits heterochromatin protein 1 (HP1) [12, 17, 18], which acts as both a structural component and an adaptor for the recruitment of chromatin-modifying factors [19]. In addition to histone methylation, the DNA within heterochromatin regions is highly methylated in many higher eukaryotes such as mammals and plants. Although DNA methylation also contributes to heterochromatin functions, the mechanisms by which it enables gene repression are less well-understood [20].
Heterochromatin assembly can be divided into three distinct steps: establishment, spreading, and maintenance [21, 22]. Heterochromatin is established at nucleation centers through the targeting of histone-modifying activities by transcription factors or non-coding RNAs. Subsequently, heterochromatin spreads into neighboring regions, mostly via a network of interactions among chromatin proteins, resulting in the formation of large heterochromatin domains independent of the underlying DNA sequences. While the mechanisms of heterochromatin establishment and maintenance have been extensively studied, those that regulate heterochromatin spreading are less well understood. One of the most attractive models is that heterochromatin spreads by “oozing”, in which repeated cycles of histone modifications and the binding of chromatin proteins result in an “inch worm”-like spreading of heterochromatin from the nucleation center until heterochromatin-associated proteins coat the extended domain [23]. Once these domains are formed, they are maintained through interactions among chromatin proteins similar to those involved in heterochromatin spreading [24].
Heterochromatin assembly and spreading in budding yeast
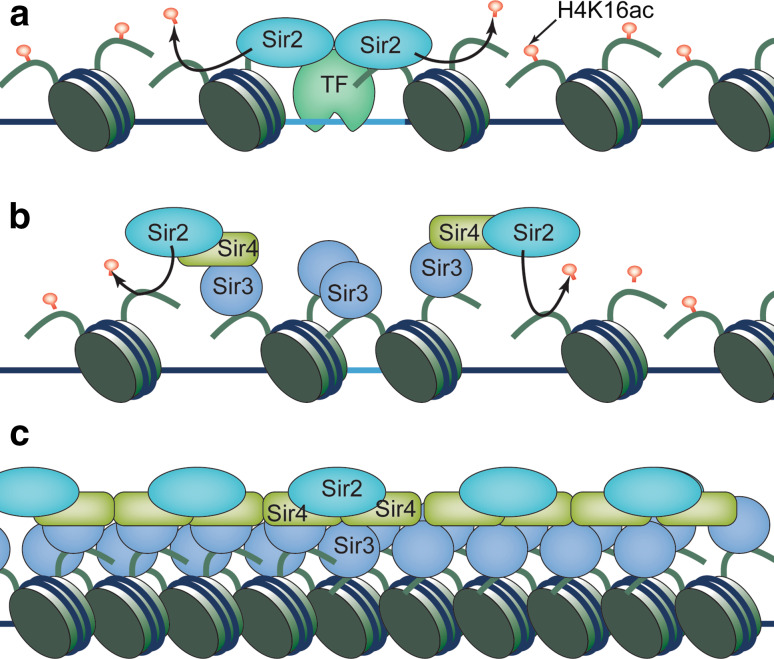
In budding yeast, heterochromatin is formed at telomeres and the silent mating type locus, mediated by the Sir (silent information regulator) protein complex, composed of Sir2, Sir3, and Sir4 [22, 25]. Sir2 is a histone deacetylase with main activity on H4K16 [26–28], the acetylation of which directly regulates higher-order chromatin folding in vitro [14] and plays a major role in heterochromatin function in vivo [13, 29]. Sir3 and Sir4 preferentially interact with histone tails devoid of H4K16ac [30–32]. At telomere regions, telomere DNA-binding protein Rap1 and the DNA end-binding complex Ku70/Ku80 recruit the Sir complex [33–36]. At the silent mating type locus, Rap1, Abf1, and the origin recognition complex (ORC) recruit the Sir complex to nucleation sites termed silencers [36–41]. In either case, Sir2 subsequently deacetylates histone H4K16, allowing Sir3 and Sir4 to bind. Sir3 oligomerizes and recruits more Sir2 to deacetylate H4K16 in the adjacent nucleosomes and thus facilitates the spread of the entire Sir complex [22, 25]. The main evidence supporting such a model include that Sir proteins cover the entire heterochromatin domain and that silencing spreads continuously through the domain [42–44] (Fig. 2). A distinct silencing mechanism operates at repressive rDNA loci, which is dependent on Sir2, but not Sir3 or Sir4, and spreads in a unidirectional manner controlled by Pol I transcription [45–48].
Fig. 2.
The stepwise assembly of heterochromatin in budding yeast. a Heterochromatin establishment is achieved by targeting of the Sir protein complex to telomeres or silencers at the silent mating type locus through DNA-binding proteins where Sir2 deacetylates H4K16. b Deacetylated histones increase the affinity of Sir3 and Sir4 for chromatin and recruit additional Sir complex. Sir2 then deacetylates adjacent nucleosomes to allow heterochromatin spreading. c The formation of an extend heterochromatin domain that is covered by Sir complex
Heterochromatin assembly and spreading in fission yeast
In fission yeast, heterochromatin forms at repetitive DNA elements in the centromeres, telomeres, and the silent mating type region [19]. Histones at these regions are not only hypoacetylated by a number of histone deacetylases (HDACs), but are also methylated on H3 Lys 9 (H3K9me) by the histone methyltransferase Clr4 [12, 49]. Similar to budding yeast, these histone-modifying enzymes can be targeted to DNA repeats by sequence-specific DNA-binding proteins to establish heterochromatin [50–53]. Interestingly, the RNA interference (RNAi) pathway is also required for heterochromatin establishment at repeat regions [54]. The DNA repeats are transcribed by RNA polymerase II during the S phase of the cell cycle [55–58]. These transcripts are converted to double-stranded RNAs by the RNA-dependent RNA polymerase complex and then processed by Dicer into small interfering RNAs (siRNAs) [59, 60]. These siRNAs are loaded into the Argonaut protein (Ago1) in the RITS (RNA-induced transcriptional gene silencing) complex, which is targeted to repeat regions through base pairing between siRNAs and nascent transcripts [61–63]. RITS directly associates with the Clr4 complex to initiate H3K9me [64], which further recruits HP1 proteins such as Swi6 and Chp2 [12, 65].
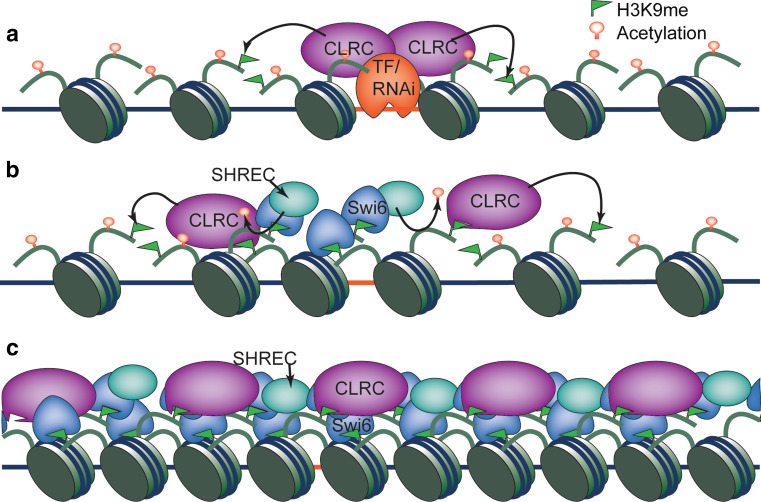
The spreading of heterochromatin from initiation sites requires Swi6, and in its absence H3K9me is restricted to heterochromatin nucleation centers [52, 66]. Because mammalian and fly HP1 interacts with histone H3K9 methyltransferases [67, 68], it was proposed that a similar interaction between Swi6 and Clr4 could result in the recruitment of additional HMTases, which in turn would modify histones of adjacent nucleosomes [66]. In addition, Clr4 contains a chromodomain that recognizes H3K9me, an interaction that could lead to heterochromatin spreading through repeated binding of H3K9me and methylation of the adjacent nucleosomes [69]. Elegant biochemical analyses demonstrate that Clr4 preferentially binds to dimethylated H3K9 while Swi6 prefers trimethylated H3K9, avoiding the potential competition between Clr4 and Swi6 and allowing efficient spreading of heterochromatin [70]. Again, the “inch worm” spreading model is supported by the fact that Swi6 and Clr4 are localized continuously throughout entire heterochromatin domains [69, 71] (Fig. 3).
Fig. 3.
The establishment and spreading of heterochromatin in fission yeast. a Heterochromatin establishment is achieved by sequence-specific DNA-binding proteins or RNAi-mediated targeting of histone methyltransferase CLRC to repetitive DNA elements, leading to local H3K9 methylation. b H3K9me recruits Swi6, which might facilitate the recruitment of additional CLRC. The chromodomain of Clr4 also recognizes H3K9me and facilitates CLRC recruitment. CLRC then methylates adjacent nucleosomes, leading to heterochromatin spreading. SHREC associates with Swi6 and deacetylate histones to promote heterochromatin spreading. c The formation of an extended heterochromatin domain that is covered by Swi6, CLRC and SHREC
In addition to the self-propagation cycles of H3K9me, heterochromatin spreading in fission yeast also requires complex crosstalk among many chromatin proteins. Like budding yeast, fission yeast also possesses Sir2, which is required for heterochromatin spreading, although functional homologues of Sir3 and Sir4 are absent [72–74]. It is possible that Sir2-mediated deacetylation of H4K16 regulates chromatin compaction, thus bringing Clr4 closer to adjacent nucleosomes [74]. Moreover, Swi6 associates with the histone deacetylase complex SHREC to deacetylate histone H3K14 and remodel chromatin to promote the spreading of H3K9me [75–78]. Meanwhile, structural and kinetic studies reveal that Swi6 undergoes a conformational change to a spreading competent state when it binds to methylated H3K9 [79].
Additional models for heterochromatin spreading have also been proposed based on studies in fission yeast. For example, spreading can be accomplished by direct or indirect coupling of CLRC to RNA polymerase II, allowing H3K9me in the wake of transcription [23]. In addition, the association between the CLRC and DNA polymerase ε suggests that heterochromatin spreads by associating with the leading strand DNA polymerase following RNAi-mediated release of Pol II that restarts stalled replication forks [80, 81].
Heterochromatin assembly and spreading in higher eukaryotes
Heterochromatin spreading in higher eukaryotes is less well defined, mostly due to the highly repetitive nature of the DNA sequences that form heterochromatin prevent precise genetic manipulations. The interactions between HP1 and H3K9 methyltransferase of the SUV39 family and the chromodomain of SUV39 are conserved, so it is possible that the inch worm spreading model functions in other systems as well [11, 68, 82]. In contrast, more is known about Polycomb protein-mediated gene silencing, which shares some similarities with heterochromatin formation and spreading and thus is often termed facultative heterochromatin.
Polycomb-silenced regions are usually characterized by the trimethylation of histone H3 lysine 27 (H3K27me3) [83]. The highly conserved Polycomb Repressive Complex 2 (PRC2) contains the SET domain-containing protein EZH2 (EZ in Drosophila) as the catalytic subunit responsible for H3K27me3 [84–87]. PRC2 also contains the histone-binding proteins RbAp46/48, the DNA-binding protein SUZ12, and EED (ESC in Drosophila). In Drosophila, PRC2 is recruited to Polycomb response elements (PREs) by sequence-specific DNA-binding proteins. In mammals, the binding sequence is less well defined and long non-coding RNAs play important roles in targeting PRC2 to specific sites [83]. The mechanism by which H3K27me3 regulates gene expression is not well understood. H3K27me3 recruits chromodomain protein Polycomb, which is part of the Polycomb Repressive Complex 1 (PRC1) [88, 89]. PRC1 also contains an E3 ubiquitin ligase that ubiquitylates K119 of H2A, which also contributes to gene silencing [90].
Importantly, PRC2 binds to H3K27me3 via the WD40 repeats of EED and stimulates methylation of nearby histone H3 on Lys 27 [91, 92], indicating that PRC2-mediated H3K27 methylation is propagated in a manner similar to that of HP1-H3K9 methylation. However, high-resolution mapping of H3K27me3 and Polycomb proteins showed that while H3K27me3 marks large chromosome domains, PRC2 is mainly concentrated at the PREs [93]. Thus it is unlikely that an inch worm spreading model applies. The exact mechanism of H3K27me3 spreading is still unknown, but it has been suggested that spreading is achieved by local diffusion of PRC2 or by the formation of chromosome loops [23, 94].
Mechanisms of boundary formation
When heterochromatin spreads into surrounding regions independently of DNA sequences, it can affect the expression of nearby genes to varying degrees depending on the extent of spreading. In certain cases, such variation of gene expression could allow for the development of new traits that help organisms adapt to new environments, facilitating evolution. However, in most cases, the disruption of normal gene expression patterns severely compromises an organism’s fitness or health, as seen in a number of human diseases linked to uncontrolled heterochromatin spreading [8]. Additionally, studies in Neurospora show that disrupting heterochromatin boundary formation leads to growth defects linked to the unchecked spreading of silenced chromatin and DNA methylation into genes outside of the normal regions, further highlighting the importance of properly restraining heterochromatin spreading for cellular fitness [95]. Thus it is essential for spreading to be tightly regulated in order to maintain stable gene expression profiles. Generally, heterochromatin regions are flanked by DNA sequences termed boundary elements, which form fixed borders accompanied by sharp transitions in histone modification profiles. Such elements result in the precise determination of epigenetic states among closely arranged chromosome loci, even when heterochromatin protein levels change. In other cases, borders are determined by the local balance of heterochromatin and euchromatin proteins, which tends to differ between cells. Such boundaries are termed negotiable borders [96].
Negotiable borders
A distinguishing feature of negotiable borders is that they are not established at a specific DNA sequence, but at a transition region defined by the balance of different proteins and histone modifications associated with heterochromatin and euchromatin [96]. For example, in budding yeast, the balance between histone acetyltransferase Sas2-mediated acetylation of H4K16 and Sir2-mediated deacetylation of the same residue defines the borders of heterochromatin at telomeric regions [29, 97]. Either loss of sas2 + or overexpression of Sir3 leads to increased heterochromatin spreading [42, 43]. In addition, loss of other histone modifications or proteins usually enriched in euchromatin may also result in increased heterochromatin spreading. For example, loss of the euchromatin-associated bromodomain protein Bdf1 or histone variant H2A.Z results in expanded heterochromatin regions [98, 99]. As a result of such competition, negotiable borders are associated with frequent changes of epigenetic states [100]. The classical example of PEV in Drosophila, where white is silenced in a portion of progenitor cells during early development, also suggests that heterochromatin spreads over varying distances rather than being constrained to a defined location. In addition, many of the factors identified as regulators of PEV affect heterochromatin spreading in a dosage-dependent manner [101], which also points to the importance of maintaining proper heterochromatin–euchromatin protein balance as a determinant of the distance of heterochromatin spreading.
Thus one important way to regulate heterochromatin spreading is by controlling the availability of heterochromatin proteins. Indeed, heterochromatin protein levels appear limiting in diverse organisms. For example, in fission yeast, ectopic heterochromatin assembly through artificial targeting of Clr4 to DNA or exogenously introduced siRNAs can only succeed when Swi6 is overexpressed or endogenous heterochromatin structures are compromised to release silencing proteins [53, 102, 103]. Moreover, overexpression of Swi6 increases the conversion rate of a less stable heterochromatin domain at the mating type region [104] and allows the cells to bypass the requirement of RNAi for pericentric heterochromatin assembly [53].
On the other hand, endogenous heterochromatin regions with negotiable borders make them ideal as “sinks” to limit the availability of heterochromatin proteins in both budding and fission yeast [53, 105–108]. Increases in heterochromatin proteins predominately localize to telomeres, leading to expansion of telomeric heterochromatin domains. In contrast, compromising heterochromatin assembly at telomeres or rDNA results in the release of heterochromatin proteins and increases the incidence of ectopic heterochromatin assembly.
Fixed borders
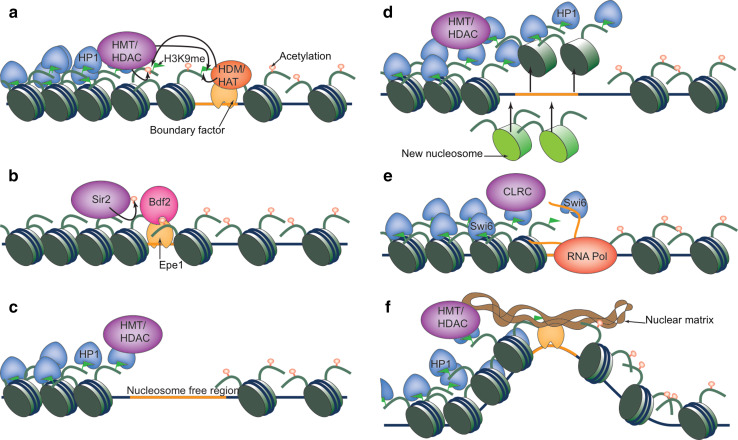
In most cases, specific DNA elements demarcate the borders of heterochromatin regions and function as boundaries to prevent spreading of heterochromatin. These boundaries precisely define chromatic regions, resulting in consistent inheritance of epigenetic states, regardless of varying heterochromatin protein levels. A general theme is that these mechanisms all converge on disrupting heterochromatin-associated histone modification cycles (Fig. 4).
Fig. 4.
Mechanism of boundary function. a Boundary elements recruit histone-modifying activities. b Boundary elements recruit proteins that protect euchromatic modifications. c Nucleosome-free regions prevent the spreading of heterochromatin modifications to establish heterochromatin boundaries. d High rate of histone turnover prevents the spreading of histone modifications. e RNA-mediated eviction of heterochromatin protein Swi6 to prevent heterochromatin spreading. f Boundary elements cluster and associate with nuclear structures to form chromatin loops
Recruitment of histone-modifying activities to directly antagonize heterochromatic histone modifications
Since heterochromatin spreading depends on repeated cycles of histone modifications, installation of incompatible histone modifications at the boundary regions can effectively block heterochromatin spreading (Fig. 4a). Indeed, in budding yeast, the boundary element at the silent mating-type locus recruits histone-modifying activities associated with euchromatic regions, and artificial tethering of Sas2 is sufficient to establish a heterochromatin boundary [109–111]. In fission yeast, the pericentric heterochromatin boundary recruits a histone demethylase complex containing Lsd1 [112]. Lsd1 was originally identified in humans as a demethylase specific for H3K4 [113], but also demethylates H3K9 in specific contexts [114]. The fission yeast Lsd1 complex localizes at the pericentric boundary regions and demethylates H3K9me to prevent heterochromatin spreading [112]. The chicken β-globin gene cluster is adjacent to a ~16 kb condensed heterochromatin region and the 5′ DNase I hypersensitive site HS4 between these two regions also has barrier activity. Transcription factors USF1 and USF2 bind to this element and recruit histone-modifying enzymes such as H3K4-specific histone methyltransferase Set1, and histone H3 acetyltransferase PCAF to block heterochromatin from spreading into the β-globin locus [115].
Protection of preexisting histone modification profiles
In addition to recruiting histone-modifying enzymes to actively counteract heterochromatin-associated histone modifications, protecting existing euchromatic modifications is also critical for establishing a heterochromatin boundary (Fig. 4b). In budding yeast, the bromodomain protein Bdf1, which protects histone H4 tail acetylation, is required for preventing heterochromatin spreading at telomeres to establish negotiable borders [98]. Another budding yeast bromodomain protein Yta7 is involved in restricting heterochromatin spreading to the silent mating type locus boundary [116, 117]. Importantly, mutations in the bromodomain lead to heterochromatin spreading outside its boundaries [118], although the acetylation events that mediate the binding of Yta7 have not been identified. In fission yeast, a double bromodomain protein Bdf2 is specifically recruited to a repeat sequence termed IRC that marks the border of pericentric heterochromatin [74]. Bdf2 is recruited to IRC by a JmjC domain-containing protein Epe1, which is highly enriched at the boundary region [74, 119, 120]. Bdf2 protects acetylated H4K16, which is essential for counteracting Sir2-mediated deacetylation to block heterochromatin spreading [74].
Nucleosome-free regions
Since heterochromatin spreading depends on cycles of histone modifications of adjacent nucleosomes, it is reasonable to expect that nucleosome-excluding sequences can function as boundaries due to the separation of substrate from histone-modifying enzymes, thus blocking the spreading of heterochromatin (Fig. 4c). Both the UAS sequence and LexA binding sites, which recruit transcription factors and exclude the formation of nucleosomes, have been shown to block the spreading of heterochromatin in budding yeast [121, 122]. Most importantly, certain DNA sequences that are known to exclude nucleosome assembly can also efficiently establish heterochromatin boundaries [121].
Regulating histone turn over rates
Heterochromatin regions are generally associated with slow turn over of histones [123, 124], which allow stable interaction between H3K9me and HP1/SUV39 to promote heterochromatin spreading. Therefore, increasing the histone turnover rate can effectively form boundary by breaking the histone modifications cycle required for heterochromatin spreading (Fig. 4d). In budding yeast, the boundary regions are indeed associated with high histone turnover rate [125]. In Drosophila, the GAGA factor directs histone H3.3 replacement that prevents heterochromatin spreading [126] and boundaries of cis-regulatory domains and GAGA binding sites are generally associated with high turn over rate of histones [127, 128].
Transcription
Transcription plays two separate roles in regulating nucleosome dynamics [129], which might contribute to boundary function. First, the transcription machinery is associated with diverse histone-modifying activities, some of which can counteract the histone modifications of heterochromatin regions. In addition, transcription increases the rate of histone turnover, which can limit the histone modification cycles required for heterochromatin spreading.
Transcription by RNA Polymerase III, presumably through the tRNA genes it transcribes, is particularly relevant to boundary function. In budding yeast, tRNA genes are required for boundary function at the silent mating type and the rDNA locus [109, 111, 130, 131], and in fission yeast, tRNA genes found at pericentric heterochromatin borders are also critical for limiting heterochromatin spreading [132, 133]. In mammals, tRNA genes also function as boundary elements, indicating an evolutionarily conserved role for tRNA genes in preventing heterochromatin encroachment [134, 135]. Mutation of the RNA pol III machinery in budding yeast, including general transcription factors TFIIIA, TFIIIC, and Pol III itself all resulted in defective boundary function, suggesting that Pol III transcription is essential for proper boundary function [109]. However, tRNA genes may play addition roles in boundary function independent of Pol III transcription. For example, at the silent mating type locus in fission yeast, the boundary region inverted repeat (IR) contains B-box sequences that recruit TFIIIC, but no Pol III was detected at this locus. TFIIIC mediates the clustering of chromosome loci at the nuclear periphery, which might contribute to boundary function through the formation of chromosome loops [136]. Similarly, in budding yeast, TFIIIC can also function as a boundary element independent of Pol III transcription [137].
In mammals, short interspersed nuclear elements (SINEs) also act as boundary elements by regulating transcription [138]. The murine growth hormone (GH) gene is regulated by the nearby SINE B2 repeat, which is transcribed by both Pol II and Pol III, though in opposite directions. During early stages of embryonic development, adjacent heterochromatin spreads past the B2 element to silence GH expression. In later stages of development, heterochromatin spreading is blocked by B2 element, allowing GH expression. Mutations of the promoters compromised boundary function, suggesting that transcription is critical for B2 boundary activity [138]. Similarly, the mouse SINE B1-X35S also has boundary activity, which is dependent on the transcription of this sequence [139].
Although the process of transcription seems to play an important role in boundary function, the transcripts themselves might also directly participate (Fig. 4e). For example, RNA directly competes with H3K9me for binding to the chromodomain of Swi6 [140]. Thus, the RNA transcripts at boundary regions may directly affect heterochromatin-mediated histone modification amplification. Consistent with such an idea, the pericentric IRC boundary of fission yeast is transcribed and mutations of the Swi6 RNA-binding residues result in heterochromatin spreading [141].
Nuclear structures
Another mechanism by which heterochromatin spreading can be blocked is through the spatial organization of chromatin. Physically separated chromatin domains can be achieved by the clustering of boundary elements or by interactions between boundary elements and nuclear structures (Fig. 4f). For example, the gypsy insulator complex in Drosophila, which was found to protect transgenes from position effects, localizes to only 20–25 sites in the nucleus despite having over 500 binding sites [142, 143]. Similarly, in fission yeast, the TFIIIC complex binding sites form clusters in the nucleus [136]. Given that the TFIIIC binding sites at the silent mating type region are critical for boundary function without local recruitment of Pol III, TFIIIC-mediated clustering may establish heterochromatin boundaries by separating chromatin domains [136]. In mammals, genome-wide analyses revealed that CTCF (CCCTC binding Factor) binding sites frequently flanked chromosome domains containing the repressive H3K27me3, often in a cell-specific manner, indicating that CTCF may regulate the spreading of facultative heterochromatin domains [144, 145]. Although CTCF is mainly known for its function as an enhancer blocker by regulating the 3D organization of the genome to control interactions between distant loci, it may perform similarly to block heterochromatin spreading [146–153]. CTCF also associates with cohesins, which have been shown to affect chromosomal architecture and organization [154, 155].
Clusters of boundary elements are often found near the nuclear periphery, suggesting that they may be tethered to the nuclear membrane. Nuclear pore proteins have been implicated in tethering DNA and may play a role in boundary activity. In an elegant “boundary trap” genetic screen, Ishii et al. screened a chimeric protein library for proteins that showed boundary activity when fused to a DNA-binding protein. One of the proteins identified, Cse1, was found to localize to the nuclear periphery, but only in the presence of the nuclear pore protein Nup2 [156]. It would be interesting to identify other nuclear membrane or nuclear matrix components that regulate the clustering of other boundary elements.
Conclusions and future directions
Since the major mechanism of heterochromatin spreading is through repeated cycles of histone modifications and binding of chromatin proteins, it is not surprising that most boundary elements function by blocking this cycle by, for example, recruiting antagonizing histone-modifying activities, protecting euchromatic modifications, creating nucleosome-free regions, altering chromatin dynamics through transcription, and tethering DNA to nuclear structures to form chromatin loops. Although each mechanism seems to be sufficient, multiple mechanisms function at each boundary. For example, the well-studied tRNA gene boundary incorporates recruitment of histone-modifying enzymes, generation of nucleosome-free regions, transcription, and TFIIIC-mediated chromatin clustering. Such redundancy might function at other heterochromatin boundary regions to ensure the efficient blocking of heterochromatin spreading.
Although the chromatin modification cycle is an attractive model to explain heterochromatin spreading, there are exceptions that suggest addition mechanisms [23]. For example, in Drosophila, H3K27me3 domains are much broader than that of PRC2 and PRC1 [93] and in budding yeast, rDNA silencing requires Sir2, but not Sir3 or Sir4 [45–47]. Moreover, in some cases, heterochromatin spreading is not continuous. For example, at native budding yeast telomeres, the spreading of heterochromatin skips reporter genes flanked by boundary elements [157, 158]. Therefore, a better understanding of the mechanism of heterochromatin spreading will provide further insights into how boundaries are formed.
References
- 1.Muller HJ. Types of visble variations induced by X-rays in Drosophila. J Genet. 1930;22:299–334. [Google Scholar]
- 2.Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor: Cold Spring Harbor Press; 2007. pp. 81–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 4.Grunstein M, Gasser SM. Epigenetics in Saccharomyces cerevisiae. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor: Cold Spring Harbor Press; 2007. pp. 63–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClintock B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 7.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleinjan DA, Lettice LA. Long-range gene control and genetic disease. Adv Genet. 2008;61:339–388. doi: 10.1016/S0065-2660(07)00013-2. [DOI] [PubMed] [Google Scholar]
- 9.Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 10.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 11.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 13.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 14.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 15.Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010;38:4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel DJ, Wang Z. Readout of epigenetic modifications. Annu Rev Biochem. 2013;82:81–118. doi: 10.1146/annurev-biochem-072711-165700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 18.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 19.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 20.Baubec T, Schubeler D. Genomic patterns and context specific interpretation of DNA methylation. Curr Opin Genet Dev. 2014;25C:85–92. doi: 10.1016/j.gde.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 21.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 22.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae . Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 23.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 24.Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011;146:510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kueng S, Oppikofer M, Gasser SM. SIR proteins and the assembly of silent chromatin in budding yeast. Annu Rev Genet. 2013;47:275–306. doi: 10.1146/annurev-genet-021313-173730. [DOI] [PubMed] [Google Scholar]
- 26.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 27.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci USA. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 30.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 31.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 32.Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Luo K, Vega-Palas MA, Grunstein M. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002;16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- 35.Mishra K, Shore D. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr Biol. 1999;9:1123–1126. doi: 10.1016/s0960-9822(99)80483-7. [DOI] [PubMed] [Google Scholar]
- 36.Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brand AH, Breeden L, Abraham J, Sternglanz R, Nasmyth K. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985;41:41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- 38.Brand AH, Micklem G, Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987;51:709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- 39.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 40.Bell SP, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 41.Foss M, McNally FJ, Laurenson P, Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae . Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- 42.Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 43.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 44.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 45.Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 46.Fritze CE, Verschueren K, Strich R, Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 48.Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 49.Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 50.Jia S, Noma K, Grewal SI. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 51.Kim HS, Choi ES, Shin JA, Jang YK, Park SD. Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6. J Biol Chem. 2004;279:42850–42859. doi: 10.1074/jbc.M407259200. [DOI] [PubMed] [Google Scholar]
- 52.Kanoh J, Sadaie M, Urano T, Ishikawa F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol. 2005;15:1808–1819. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 53.Tadeo X, Wang J, Kallgren SP, Liu J, Reddy BD, Qiao F, Jia S. Elimination of shelterin components bypasses RNAi for pericentric heterochromatin assembly. Genes Dev. 2013;27:2489–2499. doi: 10.1101/gad.226118.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 55.Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 57.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 58.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 60.Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 63.Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- 64.Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140:666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 67.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol. 2005;25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 70.Al-Sady B, Madhani HD, Narlikar GJ. Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Mol Cell. 2013;51:80–91. doi: 10.1016/j.molcel.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 72.Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol. 2003;13:1240–1246. doi: 10.1016/s0960-9822(03)00489-5. [DOI] [PubMed] [Google Scholar]
- 73.Buscaino A, Lejeune E, Audergon P, Hamilton G, Pidoux A, Allshire RC. Distinct roles for Sir2 and RNAi in centromeric heterochromatin nucleation, spreading and maintenance. EMBO J. 2013;32:1250–1264. doi: 10.1038/emboj.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J, Tadeo X, Hou H, Tu PG, Thompson J, Yates JR, 3rd, Jia S. Epe1 recruits BET family bromodomain protein Bdf2 to establish heterochromatin boundaries. Genes Dev. 2013;27:1886–1902. doi: 10.1101/gad.221010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 76.Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 77.Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SI. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci USA. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Canzio D, Liao M, Naber N, Pate E, Larson A, Wu S, Marina DB, Garcia JF, Madhani HD, Cooke R, et al. A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature. 2013;496:377–381. doi: 10.1038/nature12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475:244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marin L, Chang AY, Goto D, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479:135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schotta G, Ebert A, Dorn R, Reuter G. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol. 2003;14:67–75. doi: 10.1016/s1084-9521(02)00138-6. [DOI] [PubMed] [Google Scholar]
- 83.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 84.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 85.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 86.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 88.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 91.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 92.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 94.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 95.Honda S, Lewis ZA, Huarte M, Cho LY, David LL, Shi Y, Selker EU. The DMM complex prevents spreading of DNA methylation from transposons to nearby genes in Neurospora crassa . Genes Dev. 2010;24:443–454. doi: 10.1101/gad.1893210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kimura A, Horikoshi M. Partition of distinct chromosomal regions: negotiable border and fixed border. Genes Cells. 2004;9:499–508. doi: 10.1111/j.1356-9597.2004.00740.x. [DOI] [PubMed] [Google Scholar]
- 97.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 98.Ladurner AG, Inouye C, Jain R, Tjian R. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol Cell. 2003;11:365–376. doi: 10.1016/s1097-2765(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 99.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 100.Mano Y, Kobayashi TJ, Nakayama J, Uchida H, Oki M. Single cell visualization of yeast gene expression shows correlation of epigenetic switching between multiple heterochromatic regions through multiple generations. PLoS Biol. 2013;11:e1001601. doi: 10.1371/journal.pbio.1001601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iida T, Nakayama J, Moazed D. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol Cell. 2008;31:178–189. doi: 10.1016/j.molcel.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324:1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakayama J, Klar AJ, Grewal SI. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- 105.Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- 106.Marcand S, Buck SW, Moretti P, Gilson E, Shore D. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes Dev. 1996;10:1297–1309. doi: 10.1101/gad.10.11.1297. [DOI] [PubMed] [Google Scholar]
- 107.Michel AH, Kornmann B, Dubrana K, Shore D. Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev. 2005;19:1199–1210. doi: 10.1101/gad.340205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Taddei A, Van Houwe G, Nagai S, Erb I, van Nimwegen E, Gasser SM. The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion. Genome Res. 2009;19:611–625. doi: 10.1101/gr.083881.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae . EMBO J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oki M, Valenzuela L, Chiba T, Ito T, Kamakaka RT. Barrier proteins remodel and modify chromatin to restrict silenced domains. Mol Cell Biol. 2004;24:1956–1967. doi: 10.1128/MCB.24.5.1956-1967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oki M, Kamakaka RT. Barrier function at HMR. Mol Cell. 2005;19:707–716. doi: 10.1016/j.molcel.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 112.Lan F, Zaratiegui M, Villen J, Vaughn MW, Verdel A, Huarte M, Shi Y, Gygi SP, Moazed D, Martienssen RA. S. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Mol Cell. 2007;26:89–101. doi: 10.1016/j.molcel.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 113.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 114.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 115.West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol Cell. 2004;16:453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 116.Tackett AJ, Dilworth DJ, Davey MJ, O’Donnell M, Aitchison JD, Rout MP, Chait BT. Proteomic and genomic characterization of chromatin complexes at a boundary. J Cell Biol. 2005;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jambunathan N, Martinez AW, Robert EC, Agochukwu NB, Ibos ME, Dugas SL, Donze D. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics. 2005;171:913–922. doi: 10.1534/genetics.105.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gradolatto A, Smart SK, Byrum S, Blair LP, Rogers RS, Kolar EA, Lavender H, Larson SK, Aitchison JD, Taverna SD, et al. A noncanonical bromodomain in the AAA ATPase protein Yta7 directs chromosomal positioning and barrier chromatin activity. Mol Cell Biol. 2009;29:4604–4611. doi: 10.1128/MCB.00160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 120.Braun S, Garcia JF, Rowley M, Rougemaille M, Shankar S, Madhani HD. The Cul4-Ddb1(Cdt)(2) ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell. 2011;144:41–54. doi: 10.1016/j.cell.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bi X, Yu Q, Sandmeier JJ, Zou Y. Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol Cell Biol. 2004;24:2118–2131. doi: 10.1128/MCB.24.5.2118-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bi X, Broach JR. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 1999;13:1089–1101. doi: 10.1101/gad.13.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Choi ES, Shin JA, Kim HS, Jang YK. Dynamic regulation of replication independent deposition of histone H3 in fission yeast. Nucleic Acids Res. 2005;33:7102–7110. doi: 10.1093/nar/gki1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Aygun O, Mehta S, Grewal SI. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat Struct Mol Biol. 2013;20:547–554. doi: 10.1038/nsmb.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 126.Nakayama T, Nishioka K, Dong YX, Shimojima T, Hirose S. Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev. 2007;21:552–561. doi: 10.1101/gad.1503407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 128.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 130.Buck SW, Sandmeier JJ, Smith JS. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell. 2002;111:1003–1014. doi: 10.1016/s0092-8674(02)01193-5. [DOI] [PubMed] [Google Scholar]
- 131.Biswas M, Maqani N, Rai R, Kumaran SP, Iyer KR, Sendinc E, Smith JS, Laloraya S. Limiting the extent of the RDN1 heterochromatin domain by a silencing barrier and Sir2 protein levels in Saccharomyces cerevisiae . Mol Cell Biol. 2009;29:2889–2898. doi: 10.1128/MCB.00728-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 133.Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 134.Ebersole T, Kim JH, Samoshkin A, Kouprina N, Pavlicek A, White RJ, Larionov V. tRNA genes protect a reporter gene from epigenetic silencing in mouse cells. Cell Cycle. 2011;10:2779–2791. doi: 10.4161/cc.10.16.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Raab JR, Chiu J, Zhu J, Katzman S, Kurukuti S, Wade PA, Haussler D, Kamakaka RT. Human tRNA genes function as chromatin insulators. EMBO J. 2012;31:330–350. doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 137.Simms TA, Dugas SL, Gremillion JC, Ibos ME, Dandurand MN, Toliver TT, Edwards DJ, Donze D. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae . Eukaryot Cell. 2008;7:2078–2086. doi: 10.1128/EC.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, Garcia-Diaz A, Zhu X, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 139.Roman AC, Gonzalez-Rico FJ, Molto E, Hernando H, Neto A, Vicente-Garcia C, Ballestar E, Gomez-Skarmeta JL, Vavrova-Anderson J, White RJ, et al. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011;21:422–432. doi: 10.1101/gr.111203.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Keller C, Adaixo R, Stunnenberg R, Woolcock KJ, Hiller S, Buhler M. HP1(Swi6) mediates the recognition and destruction of heterochromatic RNA transcripts. Mol Cell. 2012;47:215–227. doi: 10.1016/j.molcel.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 141.Keller C, Kulasegaran-Shylini R, Shimada Y, Hotz HR, Buhler M. Noncoding RNAs prevent spreading of a repressive histone mark. Nat Struct Mol Biol. 2013;20:994–1000. doi: 10.1038/nsmb.2619. [DOI] [PubMed] [Google Scholar]
- 142.Gdula DA, Gerasimova TI, Corces VG. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci USA. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol. 2003;162:565–574. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 145.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 147.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci USA. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 152.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 153.Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- 154.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 155.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 157.Fourel G, Revardel E, Koering CE, Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 1999;18:2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. EMBO J. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]