Abstract
Background
The diversity of cell types and tissue types that originate throughout development derives from the differentiation potential of embryonic stem cells and somatic stem cells. While the former are pluripotent, and thus can give rise to a full differentiation spectrum, the latter have limited differentiation potential but drive tissue remodeling. Additionally cancer tissues also have a small population of self-renewing cells with stem cell properties. These cancer stem cells may arise through dedifferentiation from non-stem cells in cancer tissues, illustrating their plasticity, and may greatly contribute to the resistance of cancers to chemotherapies.
Scope of review
The capacity of the different types of stem cells for self-renewal, the establishment and maintenance of their differentiation potential, and the selection of differentiation programs are greatly defined by the interplay of signaling molecules provided by both the stem cells themselves, and their microenvironment, the niche. Here we discuss common and divergent roles of TGF-β family signaling in the regulation of embryonic, reprogrammed pluripotent, somatic, and cancer stem cells.
Major conclusions
Increasing evidence highlights the similarities between responses of normal and cancer stem cells to signaling molecules, provided or activated by their microenvironment. While TGF-β family signaling regulates stemness of normal and cancer stem cells, its effects are diverse and depend on the cell types and physiological state of the cells.
General significance
Further mechanistic studies will provide a better understanding of the roles of TGF-β family signaling in the regulation of stem cells. These basic studies may lead to the development of a new therapeutic or prognostic strategies for the treatment of cancers. This article is part of a Special Issue entitled Biochemistry of Stem Cells.
Keywords: Pluripotency, Somatic stem cell, Reprogramming, Cancer stem cell, Epithelial–mesenchymal transition, Niche
1. Introduction
Stem cells are undifferentiated cells that have an indefinite expansion potential to produce progeny through self-renewal or differentiation processes. They exist in embryonic tissues, as well as in postnatal and adult tissues. The stem cells that have received most visibility are the pluripotent embryonic stem cells (ESCs), which are derived from the inner cell mass of blastocyst stage embryos and give rise to all three germ layers (Fig. 1) [1–4]. Other pluripotent stem cells exist, such as the epiblast stem cells (EpiSCs), which are originally derived from the epiblast of mouse post implantation stage (E5.5–6.5) embryos and regarded as cells that are more similar to human than mouse ESCs [5,6]. Following early embryogenesis, most organs have resident multipotent stem cells that can give rise to a more limited set of lineages. These are called somatic or tissue stem cells (Fig. 1). These stem cells multiply through symmetric or asymmetric cell divisions to give rise to new stem cells as well as differentiated cell types, replenish dying cells and regenerate damaged tissues. Due to the inherent differentiation plasticity of stem cells, extrinsic growth and differentiation factors, or ectopically expressed key transcription factors, have the ability to direct or redirect differentiation (Fig. 1) [7–10]. Through the orchestrated balance of self-renewal and differentiation, tissues maintain their homeostasis. The properties of both ESCs and somatic stem cells are determined and maintained by their local cell environment, i.e. the stem cell niche [11–14]. For example, stem cell niches maintain somatic stem cells in quiescence, but, after tissue injury, the microenvironment signals stem cells to promote either self-renewal or differentiation to form new tissues. The niche saves stem cells from depletion, while protecting the host from abnormal stem cell proliferation. The interplay between stem cells and their niche creates a dynamic system required to sustain tissue integrity.
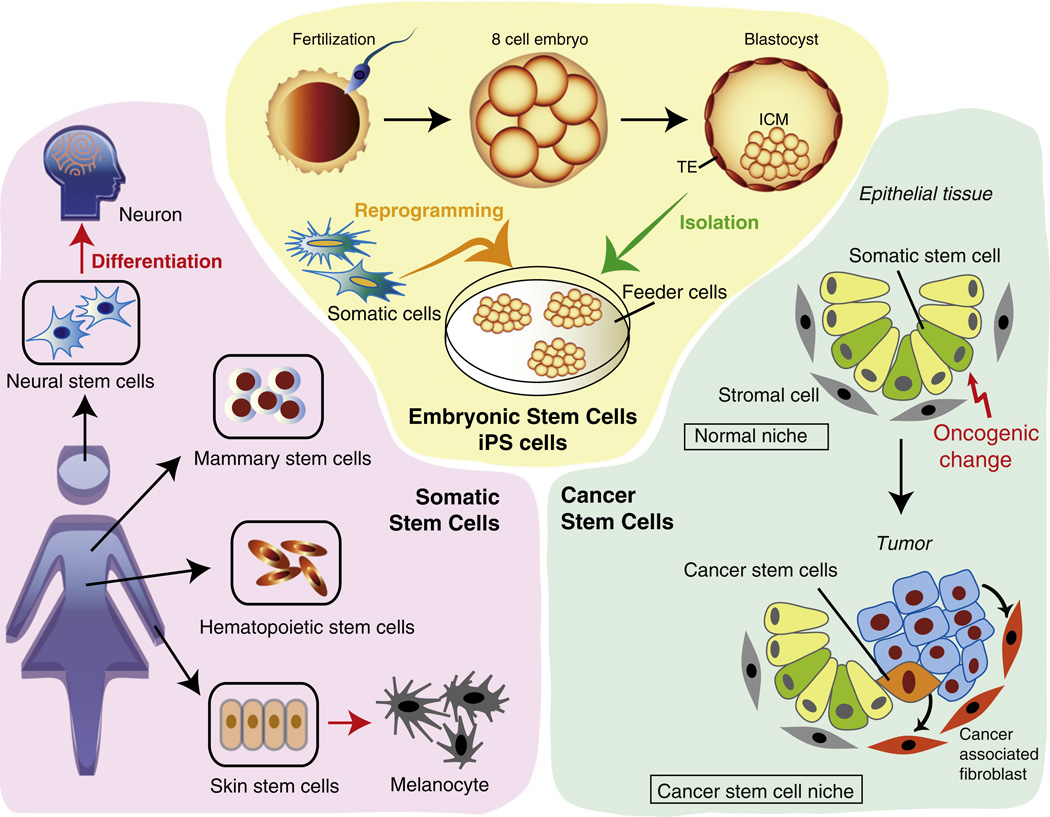
Fig. 1.
Schematic figure of four types of stem cells. Yellow area. Embryonic stem cells are derived from inner cell mass (ICM) of blastocyst stage embryos (green arrow). Induced pluripotent stem (iPS) cells are generated by reprogramming somatic cells into pluripotent cells (orange arrow). TE: trophectoderm. Pink area. Tissue stem cells, which reside in adult tissues, are the source of functionally committed cells. They generate all cell types in the same tissue through differentiation (red arrow). Some tissue stem cells are known to transdifferentiate toward other lineages (not shown). Blue area. Cancer stem cells (CSCs) represent a small subpopulation of undifferentiated cancer cells that have stem cell-like properties and are responsible for tumor expansion. The normal epithelial stem cell niche contains stromal cells with a mesenchymal phenotype that support tissue homeostasis. Oncogenic changes (red arrow) in normal somatic stem cells initiate tumorigenesis. CSCs and tumor tissues affect the properties of the stromal cells constituting the normal niche. The altered niche, where tumor stromal cells, including cancer-associated fibroblasts (CAFs), are recruited, supports the maintenance of CSCs.
In addition to these natural stem cells, induced pluripotent stem (iPS) cells have been derived from differentiated cells. By transiently expressing the transcription factors Oct4, Sox2, Klf4 and c-Myc, or some alternative ones, differentiated adult cells, such as fibroblasts or skin cells, can be reprogrammed to dedifferentiate into pluripotent cells that acquire many characteristics of ESCs [15–17] (Fig. 1). This paradigm-shifting technique of somatic cell reprogramming has facilitated the generation, by directly reprogramming somatic cells with lineage-specific master genes, of a variety of differentiated cell types with defined functions, such as pancreatic β cells, brown adipocytes, cardiac myoblasts, natural killer cells and neurons [18–22].
A rare population of cells in cancer tissues has also been shown to have stem cell properties, including self-renewal capacity and multi-potency, and is required for tumor formation and maintenance. This small fraction of cancer cells, so-called cancer stem cells (CSCs), has been identified in tumors of various organs and tissues [23,24]. Although the origin of the CSCs is subject of debate, an increasing number of studies indicate that many types of tumors initiate from normal tissue stem cells that acquired oncogenic mutations, and not from differentiated cells [25] (Fig. 1). Similarly to the normal stem cell niches, CSCs are supported by functional local microenvironments, and reciprocal interactions exist between CSCs and their microenvironment. CSCs affect the properties of the adjacent stromal cells and seem to alter the normal stem cell niche, while signals from an aberrant niche that mimics the normal stem cell niche helps maintain CSC properties [24,26] (Fig. 1).
Signaling by TGF-β family ligands plays key roles in cell differentiation and proliferation, and is important for many stem cell types. Among the TGF-β family proteins, signaling by TGF-β or activin proteins is essential for maintaining pluripotency of human ESCs [27] and mouse EpiSCs, and helps define the differentiation potential and proliferation of these cell types. Also somatic stem cells rely on TGF-β family signaling, either provided in an autocrine fashion, or by factors in their microenvironment where they reside. For example, quiescent hair follicle stem cells are activated for regeneration by TGF-β that is provided by the underlying mesenchymal dermal papillae [28]. TGF-β expressed by the follicle stem cells is also known to serve as a niche signal for melanocyte stem cells to enter quiescence [29]. Similarly to normal stem cells in development, TGF-β family signaling from the microenvironment also regulates the properties of the cancer stem cell population. One of the key events in acquiring stem cell properties of both breast cancer and normal mammary stem cells is an epithelial-mesenchymal transition (EMT) induced by TGF-β [30,31]. While TGF-β family signaling regulates stemness of normal and neoplastic stem cells, its effects are diverse and depend on the cell types, and microenvironment and physiological state of the cells. This review will focus on the roles of TGF-β family signaling in diverse functions and properties of ESCs, iPS cells, somatic stem cells, and CSCs.
2. TGF-β family signaling
The TGF-β family is encoded by 33 genes encoding structurally related polypeptides that correspond to ligand precursors. These comprise a large propeptide and the C-terminal mature polypeptide that is proteolytically cleaved from the precursor [32]. TGF-β family ligands are disulfide-linked homodimers or heterodimers of the C-terminal polypeptides, and include TGF-βs, activins, nodal, “growth and differentiation factors” (GDFs) and bone morphogenetic proteins (BMPs) [33,34]. TGF-β and myostatin are secreted as complexes with other proteins that prevent ligand binding to the receptors. These latent TGF-β complexes are activated and released by proteolytic cleavage of the propeptide [32]. TGF-β family members regulate a wide range of cellular responses, including most if not all cell differentiation events [35–38].
Signaling mediated by TGF-β ligands is transduced through cell surface receptor complexes of two distinct types of transmembrane kinases, the type I and type II receptors [34,39] (Fig. 2). In mammals, seven type I receptors (ALK-1–7) and five type II receptors exist, and a variety of heteromeric combinations have been defined. Ligand binding stabilizes the formation of a tetrameric receptor complex, consisting of a pair of type II and a pair of type I receptors, in which the type II receptors, with their constitutively active kinases, phosphorylate the type I receptors. TGF-β and activin primarily activate the type I receptors TβRI/ALK-5 and ActRIB/ALK-4, respectively, whereas BMPs activate ALK-1, ALK-2, BMPRIA/ALK-3 and BMPRIB/ALK-6. Nodal activates ActRIB/ALK-4 and ALK-7. The type I receptors are then poised to activate effector Smads and other signaling mediators that drive non-Smad signaling pathways [34,39] (Fig. 2). Activation of either Smad or non-Smad pathways by a specific ligand depends on many factors, including cell surface receptors, co-receptors, antagonists and intracellular co-factors. Differential expression of these regulators in stem cells and the niche results in the diverse effects of TGF-β family signaling on different stem cells.
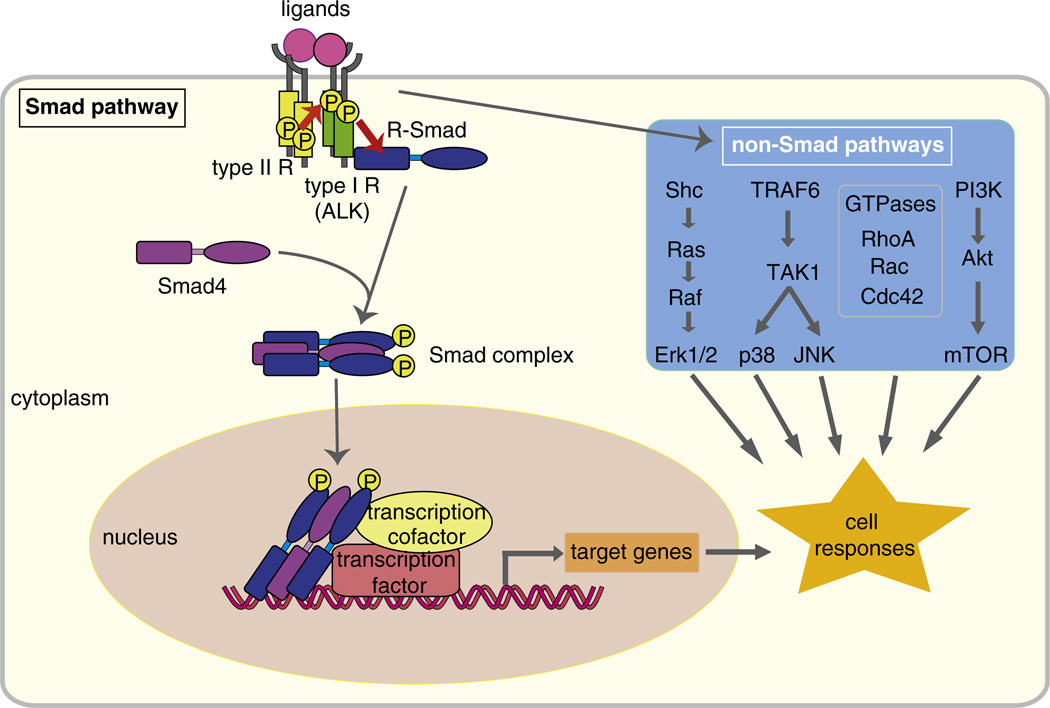
Fig. 2.
Smad-dependent and Smad-independent TGF-β family signaling. Ligands of TGF-β family members bind to type I and type II receptors. Upon ligand binding, the type II receptors phosphorylate the type I receptors, which then phosphorylate and activate effector Smads. The activated Smads form complexes with Smad4, and translocate into the nucleus. The Smad complex interacts with other transcription factors, co-activators or co-repressors to regulate transcription of target genes. TGF-β also elicits activation of other signaling cascades independent of Smad pathways. TGF-β activates the Ras–Raf–MEK–Erk MAPK pathway through tyrosine phosphorylation of ShcA, and p38 and JNK MAPK signaling through activation of TAK1 by the TRAF6. TGF-β also activates the small GTPases Rho, Rac and Cdc42, and the PI3K–Akt pathway.
2.1. Smad-mediated signaling
In vertebrates, there are eight Smads, Smad1 to Smad8 [40]. Upon ligand stimulation and subsequent activation by type II receptors, type I receptors transmit intracellular signaling through phosphorylation of downstream effector Smads [40–42]. Smad1, Smad5 and Smad8 are activated by BMP receptors, whereas Smad2 and Smad3 are activated by TGF-β/activin/nodal receptors. These receptor-activated Smads (R-Smads) form trimers with Smad4, the common Smad shared by the TGF-β/activin/nodal and BMP signaling pathways, and translocate into the nucleus (Fig. 2). The R-Smads, except for Smad2, can bind to preferred DNA sequences, yet their affinity is not sufficient to support the association of Smads with regulatory promoter sequences. Thus, Smad complexes interact and cooperate with DNA sequence-specific transcription factors, and, together, they regulate gene transcription with the help of coactivators and corepressors [43]. In the Smad complexes, Smad4 acts as coactivator of Smad-mediated transcription regulation. Many genes are activated in response to TGF-β family ligands, whereas others are transcriptionally repressed. Various families of DNA-binding transcription factors with which Smads can functionally interact have been identified. Moreover, coactivators, such as CBP and p300, or co-repressors, such as histone deacetylases, that interact with Smads, define whether the target genes are activated or repressed, and the extent of activation or repression [40,42]. Many of the responses of stem cells to TGF-β family ligands are regulated by Smad-mediated transcription activation or repression of key genes. In various cell types, and in embryonic stem cells, Smads cooperate with master regulators of cell differentiation or pluripotency [44–53]. Moreover, Smads can regulate gene expression by initially using histone marks as a platform to switch master regulator genes from the poised to the active state, and by recruiting histone demethylases [54,55].
2.2. Smad-independent signaling
TGF-β also activates, in a Smad-independent manner, signaling pathways that are generally considered as important effector pathways for tyrosine kinase receptors [41,56,57]. TGF-β induces activation of these non-Smad pathways through interactions of signaling mediators with the type I or type II receptors, either directly or through adaptor proteins. Depending on the cell system, these non-Smad pathways can also be activated indirectly as a consequence of Smad-mediated changes in gene expression. TGF-β has been shown to directly activate the Ras–Raf–MEK–Erk MAPK pathway through association of ShcA with the TGF-β receptor complex, and direct tyrosine phosphorylation of ShcA by TGF-β type I receptor in response to TGF-β, taking advantage of the dual specificity of the TGF-β receptor kinases. The phosphorylated tyrosines on ShcA then provide a docking site for the recruitment of Grb2 and Sos, and this complex initiates Ras activation leading to Erk MAPK signaling cascade [58]. TGF-β also induces p38 and JNK MAPK signaling through activation of TAK1 by the ubiquitin ligase TRAF6 that interacts with the TGF-β receptor complex [59,60]. Furthermore, TGF-β also regulates the activities of the small GTPase proteins Rho, Rac and Cdc42, which regulate the cytoskeletal organization and gene expression [61–63], but how receptor activation leads to signaling by small GTPases remains to be better defined. TGF-β-activated RhoA induces activation of its downstream targets ROCK and LIM kinase [64]. Finally, TGF-β induces Akt activation through PI3K [65,66]. Once activated, Akt initiates signaling pathways, e.g. through mTOR, that play roles in cell survival, growth, migration and invasion [67,68]. The roles of TGF-β-induced, Smad-independent signaling in stem cells are still unclear and remain to be elucidated.
3. ESCs and iPS cells
The first ESCs were isolated about 30 years ago from the inner cell mass of mouse blastocysts [1]. Nowadays, studies on mouse and human ESCs have revealed similarities, yet also differences in the properties and behavior of ESCs from both species [1,3]. ESCs can expand indefinitely in cell culture by maintaining themselves, a process named self-renewal, and produce most cell types that constitute the full organism, through various differentiation programs. These two properties, the potential to self-renew and the pluripotency, are defining characteristics of ESCs.
iPS cells are pluripotent stem cells that are artificially derived from non-pluripotent, differentiated cells [15]. By ectopically expressing certain genes, i.e. Oct4, Sox2, Klf4 and c-Myc, mouse and human somatic cells have been shown to acquire pluripotency [15–17]. iPS cells are similar to ESCs in many aspects, such as gene expression, chromatin modification patterns, cell doubling time, capacity of differentiation, chimera formation and germline transmission [15–17,69]. However, they retain an epigenetic memory of their somatic cell of origin [70–72], requiring further assessment of the full extent of their relation to ESCs [73].
3.1. Maintenance of pluripotency
Several factors are commonly important in the maintenance of pluripotency of ESCs and iPS cells, in both mouse and human cell systems. However, mouse and human cell systems also have characteristics that distinguish both species [74]. For example, their colony morphologies differ; mouse ESCs grow into compact, dome-shaped colonies, and are amenable to single-cell dissociation, whereas human ESCs form flat, two-dimensional colonies, and poorly survive single cell passaging. Most distinctly, in cell culture, mouse ESCs require leukemia inhibitory factor (LIF) (Fig. 3, orange zone) and BMP4 to maintain pluripotency, while human ESCs are maintained in the presence of basic fibroblast growth factor (bFGF) (Fig. 3, beige zone) and activin or TGF-β [27,75] (Fig. 3). In human cells, LIF signaling is dispensable for the maintenance of ESCs, whereas BMP signaling induces mesodermal and trophectodermal differentiation, and thus negatively affects cell pluripotency [5,6,76–78]. In mouse ESCs, BMP4 induces expression of Id proteins, which are DNA-binding transcription factors that inhibit differentiation into neuronal lineages [79,80]. The positive effect of BMP4 on self-renewal of mouse ESCs is additionally accomplished by inhibition of both the Erk and p38 MAPK pathways [81]. In contrast to mouse ESCs, human ESCs and mouse EpiSCs rely on FGF-induced Erk MAPK signaling to maintain pluripotency and suppress apoptosis and neuronal differentiation.
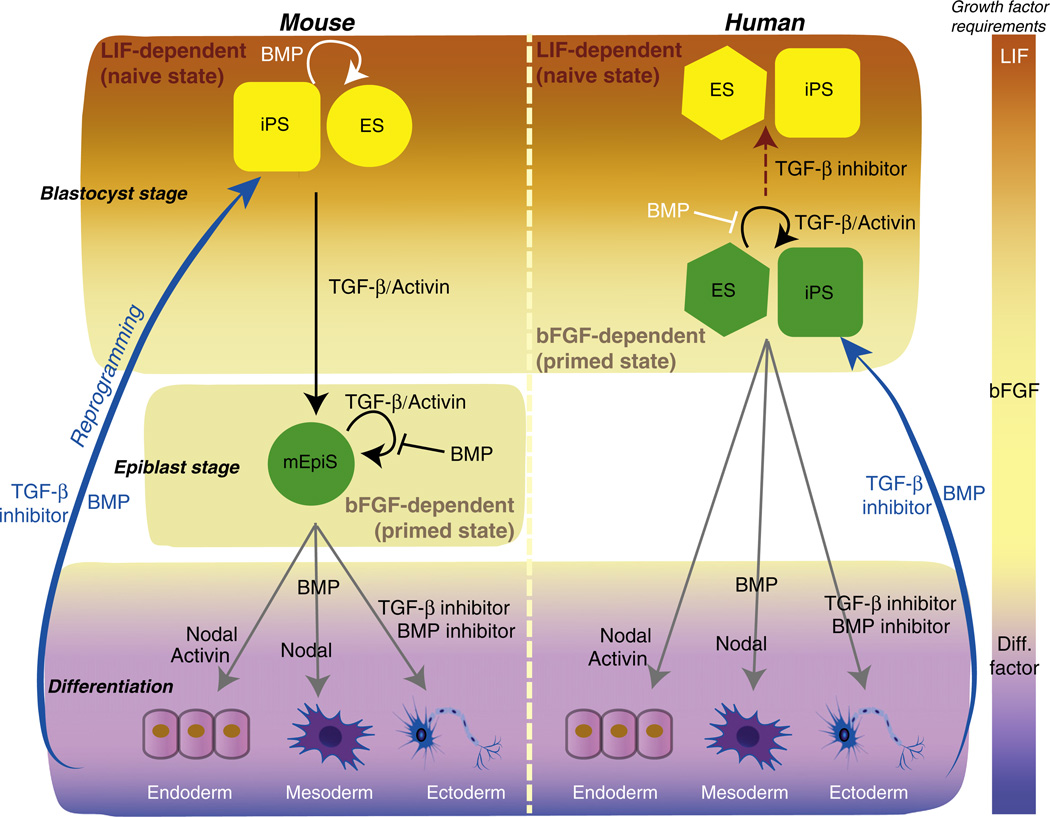
Fig. 3.
Roles of TGF-β family members in maintenance and differentiation of ESCs, and somatic cell reprogramming. ESCs derived from mouse blastocysts or human blastocysts have distinct mechanisms to maintain pluripotency. Mouse ESCs (yellow circle) rely on LIF signaling, while human ESCs (green hexagon) require bFGF signaling. Fully reprogrammed mouse and human iPS cells (yellow and green squares) require the same signaling mediators as ESCs from the same species. Low BMP levels promote pluripotency of mouse ESCs and iPS (yellow) cells in the presence of LIF (orange zone), whereas TGF-β/activin signaling induces differentiation in the absence of LIF. Pluripotent mouse EpiSCs, derived from epiblast stage embryos, require bFGF signaling, similarly to human ESCs and iPS (green) cells. These three cell types (green) require TGF-β/activin signaling together with bFGF signaling (beige zone), while BMP signaling impairs their pluripotency. Activation of TGF-β/activin signaling in mouse ESCs causes transition from the naïve ESC to the primed EpiSC state, when LIF signaling is blocked. Primed human ESCs (green hexagon) also revert to the naïve state (yellow hexagon) upon blocking TGF-β/activin signaling (dotted brown arrow). Differentiation of pluripotent cells into the three germ layers is induced by withdrawal of pluripotency factors such as LIF or bFGF (purple zone). Activin/nodal acts as an endoderm-inducing signal in the absence of bFGF. BMP and Nodal promote mesodemal differentiation, while blocking both activin/nodal and BMP signaling promotes neuroectoderm differentiation. BMP enhances somatic cell reprogramming (blue arrows), by inducing mesenchymal–epithelial transition. TGF-β type I receptor kinase inhibitors also promote iPS cell generation.
Mouse EpiSCs can give rise to multiple lineages in vitro and teratomas in vivo. However, in contrast to mouse ESCs, these EpiSCs have a limited potential to contribute to the generation of chimeras when introduced into early embryos, and have not been reported to undergo germ line transmission [6,82]. Interestingly, mouse EpiSCs rely on the same signaling pathways as human ESCs to maintain pluripotency (Fig. 3). Inhibition of TGF-β/activin signaling or activation of BMP signaling results in loss of pluripotency of both human ESCs and mouse EpiSCs, while activation of TGF-β/activin signaling induces differentiation of mouse ESCs in the absence of LIF (Fig. 3). Although the maintenance of mouse ESC pluripotency does not rely on TGF-β/activin signaling, it plays a key role in the proliferation of mouse ESCs [83].
It has been demonstrated that EpiSCs can be converted into mouse ESC-like cells in the presence of LIF without introducing an exogenous transcription factor, albeit with a lower efficiency [84]. This conversion, along with the withdrawal of TGF-β/activin and bFGF, can be enhanced by inhibiting the kinase activities of the TGF-β/activin type I receptors, thus inhibiting TGF-β/activin signaling, and/or inhibiting the Erk MAPK pathway, glycogen synthase kinase GSK3, or histone demethylase LSD1 [75,85–88]. Some reports indicate that a similar conversion can be achieved in human ESCs as well [89,90].
TGF-β/activin signaling also regulates Nanog expression and in this way acts to maintain pluripotency, in both mouse EpiSCs and human ESCs. Phosphorylated Smad2 or Smad3, which was not distinguished with the antibody used, was shown to bind to the Nanog proximal promoter region, and thus direct Nanog expression [50,51]. Upon BMP signaling, Nanog can interact with Smad1 and block BMP/Smad signaling [49], and conversely Smad1 may attenuate Nanog's function as pluripotency transcription factor. Nanog expression is heterogeneous in mouse or human pluripotent stem cells [91]. ESC markers, such as Stella, which may be involved in chromosomal organization and RNA processing, the transcription factor Oct4, and the cell surface protein SSEA3, also have revealed distinct subpopulations of mouse ESCs, EpiSCs, and substantial heterogeneity in human ESCs [92–94]. Since the response of stem cells to TGF-β/activin signaling may vary between subpopulations, the heterogeneous Nanog expression in ESCs may originate from intrinsic heterogeneity in cell signaling of the cell population. Further mechanistic studies will provide a deeper understanding of the roles of TGF-β/activin signaling, in addition to its role in Nanog gene regulation.
3.2. Differentiation
Since diverse and detailed effects of TGF-β family signaling on the differentiation of ESCs have been reported [95–100], we focus on the presumed roles of TGF-β proteins in the very early lineage decisions of ESCs. The formation of the primitive streak, which provides the origin of mesodermal and endodermal cells, represents the first step of ESC differentiation. This process, which has been well studied in mice, is controlled by BMP and activin/nodal signaling [98,101,102]. Since bFGF in undifferentiated cells suppresses BMP signaling [103], withdrawal of bFGF (Fig. 3 bottom, purple zone) potentiates the effects of the very low BMP levels in the media. Differentiation of the primitive streak population into definitive endoderm is then induced by treatment of the cells with activin or nodal [104,105]. Activation of Smad2, in response to activin/nodal signaling, results, through cooperation with other transcription factors, in the expression of definitive endodermal genes, such as Sox17, Cxcr4, Gata4, Gata6, FoxA2, Lhx1, and Gsc, and primitive streak genes, such as Eomes, Mixl1, Fgf8, and Wnt3 [106–108]. When BMP is further added to ESCs under differentiation-inducing conditions in the absence of serum, i.e. free from pluripotency growth factors such as activin and bFGF, the Brachyury-expressing mesendoderm cells are further specified toward a committed mesodermal state, characterized by expression of Flk1, the receptor for vascular endothelial growth factor (VEGF) [109]. In contrast, differentiation along the ectodermal lineage is induced in the absence of BMP and TGF-β signals. By adding inhibitors for BMP and TGF-β signaling, efficient induction of neuroectodermal differentiation can be achieved [110,111].
3.3. Reprogramming of somatic cells into iPS cells
Somatic cells can be reprogrammed into pluripotent, ESC-like cells by overexpressing defined transcription factors. This iPS cell technique was pioneered in mouse fibroblasts through ectopic expression of four transcription factors, Oct4, Sox2, Klf4 and c-Myc [15]. Depending on the reprogramming conditions, mouse somatic cells can be reprogrammed into iPS cells with either mouse ESC or EpiSC (i.e. similar to human ESC) states, i.e. naïve and primed pluripotency, respectively (Fig. 3). Naive pluripotency is maintained by LIF and low concentration of BMP, while primed pluripotency is maintained by bFGF and TGF-β/activin signaling [16,112] (Fig. 3).
Interestingly, inhibition of the TGF-β/activin/nodal type I receptor kinases enhances iPSC induction and can eliminate the requirement to introduce Sox2 expression. Furthermore, a 24–48 h treatment of intermediate, i.e. partially reprogrammed, iPS cells with this TGF-β receptor inhibitor surprisingly induces Nanog expression to promote full reprogramming [113,114]. Conversely, treating reprogramming iPS cells with TGF-β, or introducing an activated type I TGF-β receptor lowers the efficiency of iPS reprogramming [114,115]. Furthermore, expression of miRNAs that inhibits expression of the TGF-β type II receptor and blocks TGF-β-induced EMT through downstream genes, enhances iPS cell reprogramming processes [115–117]. Therefore, although TGF-β signaling is important for ES cell self-renewal, it inhibits reprogramming into iPS cells. Conversely, BMP signaling, which has been shown to induce mesenchymal-epithelial transition, and thus opposes TGF-β stimulation in some contexts, appears to promote reprogramming into iPS cells [118,119] (Fig. 3 blue arrow).
Regulation of TGF-β family signaling by miRNAs is also seen in other biological processes besides iPS reprogramming. In human undifferentiated ESCs, miR-302 is one of the most abundant microRNAs. It targets many genes such as cyclin D1 and NR2F2 [120,121], but particularly in the context of TGF-β family signaling, it targets the expression of TGF-β signaling components including Lefty, as well as BMP signaling inhibitors [122,123]. In mouse liver stem cells, miR-23b and miR-24-1 inhibit TGF-β signaling by downregulating Smad3 expression, thus contributing to stemness [124]. In human hematopoietic progenitor cells, miR-24-1 inhibits TGF-β/activin signaling by targeting the type I receptor ALK-4 [125]. miR-21 induces adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by downregulating the expression of the TGF-β type II receptor [126]. All these examples emphasize that miRNAs play a determining factor in stem cell behavior by modulating the activation of the signaling pathways. We will further discuss the role of TGF-β family signaling in somatic stem cells in the next section.
4. Somatic stem cells
As with ESCs, the stemness of somatic or tissue stem cells is regulated by intrinsic properties of the stem cells, and non-autonomous signals that are provided by their niche. Multiple external cues from soluble factors, membrane-bound molecules and extracellular matrix proteins influence the behavior, and ultimately direct the fate determination, of the somatic stem cells. To determine the key components that regulate maintenance and differentiation of somatic stem cells, the signals released in the niche need to be elucidated. TGF-β signaling by the niche cells has been implicated in the maintenance and differentiation of various types of somatic stem cells. In this section, we focus on their roles in mammary, intestinal, skin, mesenchymal, skeletal muscle, neural and hematopoietic stem cells.
4.1. Mammary stem cells
Mammary stem cells are the source of epithelial cells in the growth and development of the mammary gland during puberty and gestation. They have been identified in, and isolated from human and mouse mammary tissue, and from cell lines derived from the mammary gland [127–129]. Single mammary stem cells can give rise to both luminal and myoepithelial cells of the gland, and were shown to regenerate entire mammary gland structures in mice [130]. Whereas many signaling pathways, such as Hedgehog, EGF, Wnt, FGF, and Notch signaling, control mammary stem cell proliferation and survival, TGF-β signaling appears to play a critical role in stemness of normal mammary epithelial cells [131,132].
Increasing evidence links stemness of mammary epithelial cells with EMT. EMT is a key event in various differentiation contexts in normal embryogenesis, cancer invasion and metastasis, and fibrosis [133,134]. EMT occurs when epithelial cells lose their epithelial characteristics, and acquire mesenchymal phenotypes, resulting in increased motility. TGF-β is a potent inducer of EMT in normal mammary epithelial cells [8,135,136], and many studies have established crucial roles for TGF-β-induced EMT in tumor progression [133,137–140]. EMT in immortalized human mammary epithelial cells results in acquisition of mesenchymal markers, such as increased fibronectin and vimentin expression, increased invasiveness, and a CD44high/CD24low antigenic phenotype, which also characterizes mammary CSCs. Cells that have undergone EMT gain a higher potential to form mammospheres, a property associated with mammary epithelial stem cells [30]. Furthermore, TGF-β and Wnt signaling cooperate to induce activation of the EMT program, and function in an autocrine fashion to maintain the resulting stem cell state [31]. These findings demonstrate that differentiated epithelial cells have a potential to dedifferentiate and gain stem cell properties through EMT.
4.2. Intestinal stem cells
Intestinal stem cells divide throughout life and give rise to the epithelial cells lining the surface of the small and large intestines [141]. Intestinal stem cells reside near the base of the stem cell niche, the crypts of Lieberkühn. Genetic, inducible fate mapping studies have identified two principal epithelial stem cell pools. One pool consists of columnar Lgr5-expressing cells that proliferate rapidly and reside predominantly at the base of the crypts [142]. The other pool consists of Bmi1-expressing cells that proliferate slowly and largely reside above the crypt base [143]. Both stem cell populations were shown to give rise to differentiated epithelial cells [144,145], although, the functional relevance of having two different types of stem cell populations, and their relative contributions remain to be fully evaluated.
The proliferation and differentiation of the intestinal stem cells are regulated by factors secreted from the underlying mesenchymal layer, which include fibroblasts, enteric neurons, blood vessels, and extracellular matrix. Wnt signaling stimulates the proliferation of the stem cells and the transiently amplifying cells in the crypts [146], while BMP signaling acts as a negative regulator of stem cell proliferation in the crypts. BMP4 is highly expressed in the intravillus mesenchyme [147], and phosphorylation and nuclear localization of the BMP-specific effector Smads have been observed in differentiated villus epithelial cells and intestinal stem cells [148]. These observations suggest that BMP signaling from the mesenchyme acts on adjacent, non-proliferating epithelial cells and slowly proliferating stem cells. Mutations in the genes encoding Smad4 [149] or BMPRIA/ ALK-3 [150] have been linked to juvenile polyposis in humans. Furthermore, exogenous expression of the BMP antagonist noggin in the mouse intestine leads to de novo ectopic formation of normal crypt-villus units, and, at later stages, formation of a complex architecture of branching villi with dilated cysts, similar to the intestinal phenotype of human juvenile polyposis [147]. In addition, conditional deletion of the Bmpr1a gene in crypts in mice disturbs the regeneration of intestinal epithelial cells, with expansion of stem and transiently amplifying cells, eventually leading to a type of intestinal polyposis, reminiscent of human juvenile polyposis [148]. These data suggest that BMP signaling is important for a balanced control of stem cell self-renewal, specifically in maintaining quiescence of Bmi1-positive slowly proliferating stem cells, and in terminal differentiation of intestinal epithelial villi, possibly by opposing Wnt signaling, which stimulates proliferation of the Lgr5-positive stem cell population.
4.3. Skin stem cells
The skin epidermis and its appendages provide a protective barrier to harmful microbes and prevent dehydration. To perform their functions and maintain tissue homeostasis, skin continuously rejuvenates by replenishing old cells with new cells that differentiate from skin stem cells. Based on their localization and function, three different types of skin stem cells have been discerned, i.e. epidermal, hair follicle, and melanocyte stem cells. We focus on hair follicle stem cells (HFSCs) and melanocyte stem cells, since recent studies highlighted the role of TGF-β signaling in the maintenance of their stemness.
4.3.1. Hair follicle stem cells
Throughout adult life, HFSCs undergo dynamic, synchronized cycles of degeneration (catagen), quiescence (telogen), and regeneration (anagen) [151–153]. In the telogen phase, HFSCs are quiescent and reside in a specialized microenvironment, called the hair bulge [154]. In telogen, the base of the bulge is directly adjacent to the underlying dermal papilla, a signaling center for HFSCs.
TGF-βs are expressed by hair follicles in catagen, and their role in this state, through induction of apoptosis, has been well established [155,156]. However, targeted inactivation of the individual genes encoding the three TGF-β ligands resulted in differential effects on embryonic hair follicle development. For example, mice with a disruption of the Tgfb1 gene show slightly advanced hair follicle formation, while lack of the Tgfb3 gene does not have any effects. Tgfb2−/− mice exhibit a profound delay of hair follicle morphogenesis, with a 50% reduced number of hair follicles [157]. Thus, the roles of TGF-β ligands in HFSCs appear to be very diverse. TGF-β2 expression in the dermal papilla acts as a critical signal in promoting HFSC regeneration, and HFSCs that cannot sense TGF-β exhibit significant delays in hair follicle regeneration. Additionally, TGF-β2 antagonizes BMP signaling in HFSCs by enhancing the expression of Tmeff1, an antagonist of the BMP pathway, thus restricting and lowering BMP responsiveness in the niche [28].
While TGF-β signaling needs to be activated for the transition of the dermal papilla from the quiescent to the regeneration state, BMP signaling supports the maintenance of quiescent HFSCs, reminiscent of a similar role in intestinal stem cells. When the Bmpr1a gene is inactivated in postnatal skin epithelium, the quiescent HFSCs are activated to proliferate, leading to loss of slowly proliferating cells [158]. In line with this observation, BMP inhibitory factors from the surrounding niche cells have been implicated in the induction of anagen [159–161]. Conversely, expression of a constitutively active BMPRIA in skin promotes premature hair follicle differentiation, further supporting the notion that BMP signals are critical in maintaining the properties of quiescent stem cells [158].
4.3.2. Melanocyte stem cells
Melanocytes are the pigment-producing cells in the skin, hair and eye that determine their color. In the hair bulge, melanocyte stem cells are found where the HFSCs reside [162]. Their differentiation state is defined by the expression of genes involved in pigmentation, which is driven by the master transcription factor Mitf [163,164]. Melanocyte stem cells are usually in a quiescent, non-cycling state and are activated only at early anagen to undergo stem cell division. A precise control mechanism through Mitf and Bcl2 is important in the maintenance of stemness of melanocytes, and to avoid depletion of the stem cell source, which leads to hair graying [165].
Expression of TGF-β ligands by niche cells in the hair bulge [166] plays an important role in the physiology of the melanocyte stem cells, as they induce cell cycle arrest in melanocyte stem cells to let them enter dormancy [29]. Furthermore, TGF-β induces downregulation of Mitf expression, consequently preventing expression of melanogenic differentiation genes [29]. As a result, mice that specifically lack expression of the TGF-β type II receptor in melanocytes, exhibit mild but accelerated hair graying [29].
Interestingly, the HFSCs serve as melanocyte stem cell niche by expressing TGF-β [167]. Mice lacking collagen17α1, which exhibit hair loss due to the lack of the HFSC maintenance program, retain TGF-β1/2 expression in the bulge area of hair follicles at 5 weeks, but gradually loose TGF-β expression by 8 weeks. Correlating with defects in HFSC maintenance upon loss of TGF-β signaling, aberrant melanocyte differentiation starts at 12 weeks in col17a1−/− mice [167], suggesting that melanocyte stem cells are maintained by being adjacent to normal HFSC, which produces TGF-β as a niche signal for melanocyte stem cells. In this context, TGF-β stimulates hair regeneration by activating the cell cycle of HFSCs, while also inducing the entry to the quiescence of melanocyte stem cells to potentiate their function as a reservoir of a pigment-producing organ for longer periods.
4.4. Mesenchymal stem cells
Mesenchymal stem cells (MSCs) have been isolated from various adult tissues including connective tissue, adipose tissue, muscle, bone marrow and teeth, blood, placenta and umbilical cord [168,169]. The differentiation potential of MSCs depends on the local tissue environment from which they are isolated, and, in vivo, is dictated by their localization and microenvironment. For example, bone marrow-derived MSCs are capable of differentiating into osteoblasts, adipocytes, chondrocytes, muscle cells, and hematopoiesis-supporting stromal cells, but not into hematopoietic cells [170]. Muscle-derived MSCs are able to give rise to myogenic, osteogenic, chondrogenic, and adipogenic cells [171]. Human MSCs derived from bone marrow can be expanded more than a billion-fold in culture without losing their stem cell capacity. Owing to their potential for clinical applications, MSCs have attracted much attention [172].
Proliferation of human MSCs is stimulated by Wnt or TGF-β signaling [173–175]. TGF-β1 induces Smad3-dependent nuclear accumulation of β-catenin in MSCs, which is required for stimulation of MSC proliferation. On the other hand, BMP2 antagonizes Wnt3a signaling and inhibits proliferation of bone marrow-derived MSCs through interaction of BMP-activated Smad1/5 with Dishevelled-1, a component of the Wnt signaling pathway [176].
TGF-β signaling has also been implicated in directing the differentiation fate of MSCs [177]. BMPs induce differentiation of mesenchymal cells into chondroblasts or osteoblasts in vitro. TGF-β and activin also provide competence for chondroblast differentiation at early stages, while TGF-β inhibits osteoblast maturation at late stages in differentiation [177]. Consequently, pharmacological inhibition of TGF-β/activin signaling strongly enhances osteoblast maturation [178]. These inhibitory effects are mediated by induction of expression of inhibitory Smads, such as Smad6, which in turn represses BMP signaling [179]. In addition, BMP7 was shown to induce the generation of brown fat from MSCs in the absence of the normally required hormonal induction [180]. Since brown fat is specialized in energy expenditure and can counteract obesity, a therapeutic strategy based on the use of BMP7 may be suggested as a treatment of obesity.
While MSCs can differentiate into skeletal, smooth and cardiac muscle cells [181,182], many studies have focused on their potential to give rise to cardiomyocytes for cardiac tissue regeneration after infarction [183–189]. A correlation between in vivo transplantation of MSCs and repair of scarred myocardium after myocardial infarction in rats [190] has led to the idea that the implantation of MSCs into myocardium could be a useful clinical approach. Treatment of human MSCs with TGF-β induces the expression of the Notch ligand Jagged 1 along with cardiomyocyte markers, including α-smooth muscle actin, calponin 1, and myocardin. Increased expression of these genes depends on activation of Smad3 and Rho kinase. In addition, preventing Jagged 1 expression blocks the expression of cardiomyocyte genes shown above, suggesting that Jagged 1 plays an important role in TGF-β-induced expression of cardiomyocyte marker genes [191]. These studies implicate a role of the microenvironment in the induction of differentiation of MSCs along the cardiomyocyte lineage, and TGF-β signaling is a candidate key niche factor to promote this differentiation. Further studies in animal models will allow us to understand the underlying mechanisms of how in vivo niche factors, such as TGF-β, control MSC differentiation.
4.5. Skeletal muscle stem cells
Muscle stem cells show myogenic potential in response to environmental cues, and have been isolated from skeletal muscle [192–194]. They can be categorized into satellite cells, mesoangioblasts (vessel-associated stem cells), side population cells, muscle-derived stem cells, pericytes, and CD133-positive stem cells [194–199]. Among these stem cell types, satellite cells have been extensively studied. Since a fair amount of knowledge has accumulated in the past few decades, the role of TGF-β signaling in satellite cells has been well documented.
Satellite cells are located between the basal lamina and the sarcolemma of the muscle fiber, and become activated for proliferation and differentiation in response to trauma, mechanical injury or biological stimuli. Satellite cells are a heterogeneous cell population comprised of stem cells and committed progenitors, based on expression of Pax7 or Myf5 [200]. Once stimulated, satellite cells proliferate, differentiate into myoblasts, and fuse with existing muscle fibers or, alternatively, create fibers de novo [193,201].
In skeletal muscle, TGF-β family members have inhibitory effects on both muscle development and postnatal regeneration of skeletal muscle [202]. TGF-β1 represses skeletal muscle-specific gene expression, and regulates proliferation in satellite cells [203–208]. Myostatin is another important TGF-β family member in skeletal muscle development. Mice lacking myostatin expression exhibit increased muscle mass and fiber hypertrophy, which are indicative of the inhibitory effects of myostatin on muscle development [209–211]. Interestingly, short-term inhibition of myostatin in aged mice that normally retain less muscle regeneration potential, enhances muscle regeneration and activates satellite cell proliferation [212]. In aged human skeletal muscle, satellite cells are maintained but fail to be activated upon environmental stimuli. These cells exhibit a decline in Notch activation due to decreased Erk MAPK signaling [213]. Moreover, aged human muscle produces excessive TGF-β, but not myostatin, which induces unusually high levels of Smad3 activation in resident satellite cells and interferes with their regeneration capacity [213].
Increased TGF-β signaling is also apparent in skeletal muscle of fibrillin-1-deficient and dystrophin-deficient mice, models of human Marfan syndrome and Duchenne muscular dystrophy, respectively. Antagonizing TGF-β signaling rescues skeletal muscle regeneration in fibrillin-1-deficient mice and restores the regeneration program in these mice [214]. Furthermore, fibrillin-1-deficient mice exhibit increased satellite cell numbers when TGF-β is antagonized [215]. These findings suggest that TGF-β and myostatin inhibit satellite cell activation, and thus lead to a failure of skeletal muscle regeneration.
Signaling by TGF-β or myostatin converges in activation of Smad2 and Smad3. Several studies have elucidated roles of Smad2 and Smad3 in TGF-β1-mediated myogenic inhibition and in normal differentiation [47,216]. Smad3, but not Smad2, is the key mediator of TGF-β inhibition of myogenesis, through inhibition of MyoD and other myogenic bHLH transcription factors that drive myogenic differentiation [47]. Smad3 associates physically with MyoD, inhibiting the transactivation properties of MyoD [47], and with MEF2C, thus decreasing its transcription activity [217]. MEF2C plays a role in the late stages of myogenic differentiation, and conditional deletion in skeletal muscle affects sarcomere integrity [218].
To fully understand the molecular mechanisms of the TGF-β and myostatin signaling pathways in skeletal muscle stem cells, further work in different stem cell types will be needed. Profiling TGF-β or myostatin target genes at a genome-wide level may give further insights into common and divergent roles of these differentiation factors.
4.6. Neural stem cells
The self-renewing neural stem cells give rise to neural progenitor cells and eventually to neurons, astrocytes and oligodendrocytes [219]. Adult neurogenesis, resulting in the formation of new neurons, has been demonstrated in brains of adult mice, songbirds and primates, including humans, specifically in the subventricular zone, which lines the lateral ventricles, and the dentate gyrus of the hippocampus [220]. These observations led to the identification of neural stem cells in these structures, although the presence of true self-renewing stem cells there has been debated [221]. Under some circumstances, such as following tissue damage in ischemia, neurogenesis can be induced in other brain regions as well, including in the neocortex. Neural stem cells can be propagated in vitro as neurospheres, and differentiated into both neuronal and glia cells [219]; however, some studies suggest that this differentiation behavior is induced by culture conditions [222,223]. A better understanding of the growth factors or stimuli that control the proliferation and lineage decisions of these cells is warranted.
TGF-β signals play important roles in the maintenance and growth of neural stem cells [224]. Targeted inactivation of TGF-β type II receptor gene in the mid/hind brain enhanced the self-renewal, but not the multipotency, of mouse neural stem cells, resulting in an enlarged midbrain. Ectopic expression of FGF and Wnt ligands, such as FGF8 and Wnt1, was observed in these mutant brains, suggesting that TGF-β signaling antagonizes FGF and Wnt signaling, thus controlling the size of the midbrain by negatively regulating self-renewal of neural stem cells.
BMPs induce a variety of biological responses in embryonic neural stem cells [225]. BMP2/4 acts as a neuroepithelial proliferation signal at very early stages of embryonic central nervous system development [226]. Later in development, BMPs induce neuronal and astroglial differentiation of neural stem cells [227,228]. BMP signals have also been implicated in the maintenance of neural stem cells in adult brain. In the subventricular zone, where somatic neural stem cells reside [229], multiple cell types can be discerned, such as migrating neuroblasts, immature precursors, astrocytes, and ependymal cells. These astrocytes were shown to act as neural stem cells in both normal and regenerating brain [230]. Among the many signaling molecules that are expressed in the subventricular zone, BMPs induce the generation of astrocytes at the expense of new oligodendrocyte and neuron generation. In contrast, noggin is highly expressed in ependymal cells, which do not act as neural stem cells [231]. Local noggin expression may contribute to the formation of a neurogenic niche for stem cells in the subventricular zone, as it promotes differentiation. These findings suggest that BMPs play a role in the maintenance of neural stem cells, i.e. astrocytes, by inhibiting their commitment to neurons and oligodendrocytes.
4.7. Hematopoietic stem cells
Hematopoietic stem cells (HSCs) are found in bone marrow, and give rise to all blood cell types. Although various methods are used to purify HSCs [232], these cells are usually not defined by phenotype, but rather by their ability to reconstitute a hematopoietic cell population in bone marrow [233,234]. Accordingly, HSCs have been classified by their lineage differentiation capacity, assessed by serial transplantation of single HSCs [235]. Another classification of HSCs is based on repopulation kinetics in mice transplanted with limiting dilutions of whole bone marrow [236–238]. These findings indicate that each subtype of HSCs displays different responsiveness to the microenvironment, and thus different responses to growth factors in their niche.
TGF-β signaling has been implicated in the regulation of quiescence of HSCs [239]. Since TGF-β is produced in a latent form by a variety of cells, and multiple mechanisms can lead to TGF-β activation, the mechanism of activation of latent TGF-β in the context of HSC quiescence remains to be resolved. Non-myelinating Schwann cells were shown to be in contact with a substantial proportion of HSCs in bone marrow, and to mediate activation of TGF-β. These findings suggest that glial cells maintain HSC quiescence by limiting activation of latent TGF-β as components of a bone marrow niche [240].
In cell culture, TGF-β signaling-deficient HSCs have a higher proliferative capacity, whereas the quiescence and maintenance of HSCs in vivo may depend on TGF-β signaling [241,242]. Furthermore, the response of HSCs to TGF-β stimulation is biphasic; high concentrations of TGF-β inhibit, while low concentrations stimulate HSC proliferation [243]. Additionally, TGF-β signaling results in different effects on myeloid-biased (My-) and lymphoid-biased (Ly-) HSC subtypes in various assays, such as colony formation from single cells, cell cycle analyses, and proliferation in vivo [244]. These observations suggest a mechanism for differential regulation of distinct HSC subtypes. Differential responses between subtypes of leukemic cells, such as leukemic stem cells and non-stem cells, toward environmental cues can also be explained by cell heterogeneity.
5. Cancer stem cells
Cancer stem cells (CSCs) are a small population of self-renewing cells with the ability to initiate tumor formation and give rise to heterogenous cancer cells. Increasing evidence highlights the importance of CSCs in cancer progression [23,245]. CSCs are thought to confer resistance to chemotherapy or radiotherapy, leading to recurrence and metastases [24]. Thus, characterizing the properties of CSCs and the mechanisms of their regulation by signaling pathways is of high importance. CSCs are thought to be maintained by a cancer cell microenvironment that is conceptually similar to a normal stem cell niche [26]. Growth factors and cytokines released from the cancer microenvironment are thought to regulate CSC maintenance, differentiation and function. Recent studies have revealed critical roles of TGF-β family signaling in CSC maintenance and differentiation in several types of tumors (Table 1). Their roles are strikingly similar to those in normal somatic stem cell maintenance and differentiation.
Table 1.
Reported cancer stem cells and corresponding literature.
| Cancer type | Roles of TGF-β family signaling | References |
|---|---|---|
| Breast cancer | TGF-β-induced EMT maintains stem-like properties | [30,31] |
| TGF-β reduces the size of the SP fraction and the ability of tumor formation | [252] | |
| BMP2/7 decrease ALDH+/CD44high/CD24low stem cells | [251] | |
| Pancreas cancer | Nodal/activin signaling drives self-renewal and tumorigenicity | [255] |
| Diffuse-type gastric cancer | TGF-β decreases the CSC fraction | [269,270] |
| Colorectal cancer | BMP4 induces differentiation, apoptosis and chemosensitization of CSCs | [258] |
| Squamous cell carcinoma | TGF-β/TβRII signaling restricts self-renewal and expansion of CSCs | [268] |
| Ovarian cancer | TGF-β-induced TG2 induces EMT and a CSC phenotype | [271] |
| Prostate cancer | BMP7 induces senescence in CSCs by activating p38 MAPK and increasing the expression of p21 and NDRG1 | [272] |
| Glioma | TGF-β maintains tumorigenicity and stemness | [276,277] |
| BMP4 inhibits proliferation and induces differentiation of human glioblastoma stem cells | [274] | |
| Leukemia | TGF-β–FOXO pathway maintains stem cell-like properties | [282] |
| Smad4 negatively regulates leukemia initiation and maintenance induced by HOXA9 gene or NUP98–HOXA9 fusion oncogene | [283] |
5.1. Epithelial cancer stem cells
Epithelial cancers, also called carcinomas, are cancers formed from epithelial tissues such as skin, breast, gastrointestinal tract, kidney and bladder. Epithelial cancers make up about 85% of all the cancer types. Recent evidence suggests plasticity in epithelial cancer cells that allows non-CSCs to acquire CSC-like properties, and vice versa. Thus, the hierarchical, unidirectional model in which CSCs are often described as cells at the apex of a hierarchy is now being replaced with a model in which signals from the cancer microenvironment can create new CSCs from non-CSCs [246] (Fig. 4A). Several reports demonstrate that TGF-β-induced EMT can lead to dedifferentiation and gain of stem cell-like properties in both cancer and normal cells [247] (Table 1, Fig. 4A).
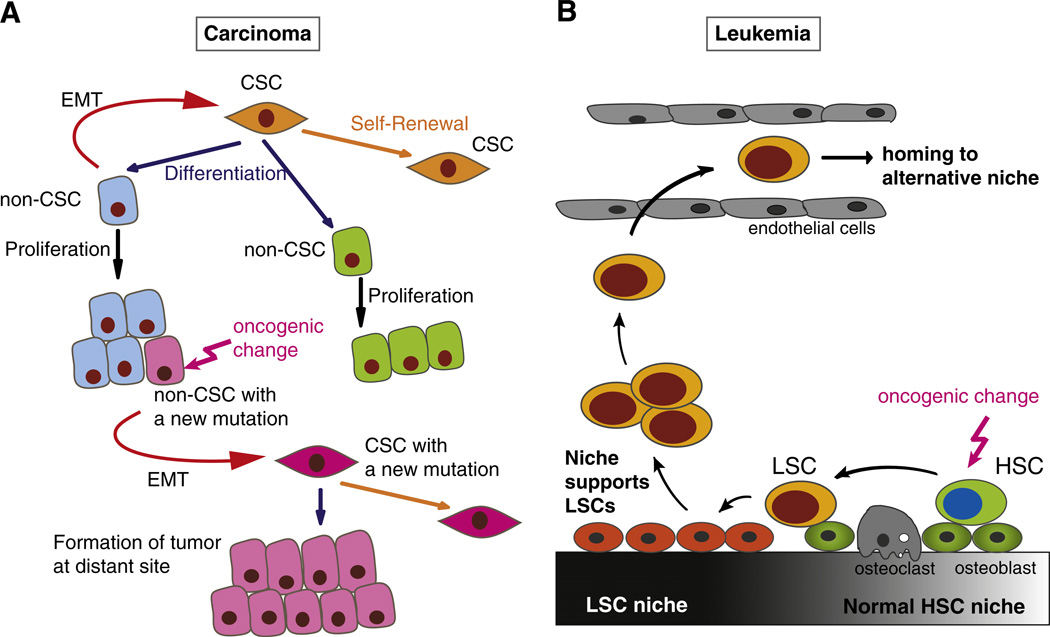
Fig. 4.
Cancer stem cells (CSCs) in tumor initiation and progression. (A) Plasticity of epithelial cancer cells through epithelial–mesenchymal transition (EMT) and its role in tumor progression. Epithelial CSCs have the ability to self-renew (orange arrow) or to differentiate (blue arrow) into different types of non-CSCs (blue, green), and acquire oncogenic changes. Non-CSCs can undergo EMT (red arrow), thus acquiring a mesenchymal phenotype and stem cell-like properties, and generating new CSCs from non-CSCs. A cancer cell in non-CSC population may acquire an additional oncogenic change (pink arrow) that promotes tumor progression. Dedifferentiation of this non-CSC (pink) through EMT generates a CSC with a second oncogenic mutation (pink), which has a more malignant phenotype and drives expansion, invasion and metastasis of the tumor. (B) Leukemic stem cells (LSCs) and their niche. The interactions between LSCs and their niche have much in common with the interactions between normal hematopoietic stem cells (HSCs) and their bone marrow niche. The normal niche contains HSCs and supporting cells, such as osteoblasts. Oncogenic changes (pink arrow) in HSCs can lead to the generation of LSCs. LSCs alter the properties of normal niche cells, and the niche adapts to LSCs. Growth factors and cytokines released from the altered niche support the LSCs to maintain their stemness. Signals from the niche also induce additional genetic or epigenetic changes in LSCs. LSCs that are no longer dependent on the original niche survive in the blood vessel and may home to an alternative niche, leading to expansion of leukemic cells.
5.1.1. Breast cancer stem cells
In human breast cancers, a small subpopulation of cancer cells with a CD44high/CD24low antigenic phenotype was shown to exhibit CSC properties [248]. High aldehyde dehydrogenase 1 (ALDH1) activity also identifies cell populations with CSC properties [249]. Interestingly, stem cell-like cells isolated from mouse or human mammary carcinomas, as well as normal mammary glands, were shown to express EMT markers. Transformed human mammary epithelial cells that have undergone EMT show an increased ability to form mammospheres, soft agar colonies and tumors, which correlates with their CSC properties [30]. Additionally, in immortalized human mammary epithelial cells, oncogenic Ras and TGF-β1 cooperate to produce CD24low stem cell-like cells from CD24high differentiated cells [250]. These observations link TGF-β-mediated EMT with acquisition of the CD24low CSC phenotype. Furthermore, a mesenchymal cell subpopulation spontaneously arises from immortalized human mammary epithelial cells through EMT. In these mesenchymal cells, autocrine TGF-β signaling is necessary for maintaining the mesenchymal state and tumorigenicity [31]. In this cell system, in which BMP antagonizes TGF-β-induced EMT, BMP signaling is decreased and TGF-β signaling is increased, when compared to the normal epithelial cell population [31]. Supporting these reports, BMP2/7 antagonizes TGF-β-mediated EMT, and decreases the frequency of ALDHhigh/CD44high/CD24low breast CSCs. Bone metastasis, formed after intra-cardiac injection of MDA-MB-231 breast cancer cells in Balb/c nu/nu mice, is also decreased with BMP2/7 treatment [251]. These reports shed light on the notion that normal mammary stem cells and breast CSCs may share similar mechanisms, which rely in part on TGF-β signaling-mediated EMT, to establish and maintain stemness. The concept of epithelial cancer plasticity through EMT may provide explanations for how multi-step cancer progression is achieved. If oncogenic changes that normally occur in non-CSCs can be introduced to CSCs by dedifferentiation through EMT, CSCs with second oncogenic mutations can drive metastasis and resistance to chemotherapies and radiotherapies. Thus, TGF-β-induced EMT may enhance cancer progression by inducing not only an invasive phenotype, but also dedifferentiation of non-CSCs into CSCs.
In contrast to reports demonstrating that TGF-β signaling induces expansion of the CSC population and promotes cancer progression, TGF-β has also been shown to decrease the number of CSCs and inhibit tumor formation. In immortalized and transformed human breast epithelial cells derived from MCF10A cells, TGF-β reduces the size of the side population cell fraction, which exhibits properties of CSCs [252]. In this model, TGF-β promotes differentiation of CSC population by decreasing the expression of Id1, which induces self-renewal and inhibits differentiation in many tissues. By enhancing differentiation and reducing the number of CSCs, TGF-β inhibits tumor formation [252]. These results suggest complex roles of TGF-β in the regulation of CSCs, and different effects of TGF-β on CSC populations from different tumors, that may result from differential expression of signaling regulators in different populations. Further studies on the roles and mechanisms of TGF-β in the regulation of CSCs using different tumor types are needed to develop treatments based on activities of TGF-β signaling.
5.1.2. Pancreas cancer stem cells
Pancreas cancers also harbor distinct subpopulations of CSCs with self-renewal capacity, abilities to differentiate and initiate tumorigenesis [253,254]. Human pancreas CSCs and stroma-derived pancreatic stellate cells express nodal and activin, important regulators of ESC pluripotency, and activated nodal/activin signaling is required for self-renewal and tumorigenicity of pancreas CSCs [255]. Pharmacological inhibition of the nodal/activin type I receptor kinases reduces self-renewal and tumorigenicity of these CSCs, and enhances their sensitivity to chemotherapy [255]. These results suggest that, to maintain their self-renewal capacity, tumorigenicity and resistance to chemotherapy, pancreas CSCs may use the same signaling pathways as those that are involved in human ESC pluripotency. Inhibition of nodal/activin signaling, combined with chemotherapy, may provide a therapeutic strategy for targeting pancreas CSCs. On the other hand, about 50% of patients with pancreas cancer bear impaired Smad4 function, which suggests other key mechanisms besides nodal/activin signaling to maintain pancreas CSCs. Further studies will be needed to define several potential therapeutic strategies that will be chosen according to the mutations.
5.1.3. Colon cancer stem cells
In human colon cancer, CSCs were initially identified as a CD133+ cell subpopulation [256,257]. These CSCs can be expanded as tumor spheres using serum-free media, but, when grown in spheres in the presence of serum, they acquire epithelial markers and lose tumorigenicity.
BMPs promote differentiation of normal colon stem cells, and inactivation of BMP signaling confers increased tumorigenesis. Inactivation of BMP signaling by impaired BMPRIA or Smad4 function is often observed in human colon cancers [147,149,150]. Consistent with these observations, BMP4, which is expressed in non-CSCs, but not in CSCs, induces differentiation, apoptosis and sensitivity to chemotherapy in the CSC population of human colorectal cancer cells that do not have impaired Smad4 function and constitutive activation of PI3K [258]. This effect of BMP4 is mediated through inhibition of the PI3K–Akt pathway, which is normally activated in CSCs, through Smad-dependent and Smad-independent mechanisms. These and other findings suggest that the same signaling pathways can enhance differentiation of both normal and cancer stem cells, and that BMP4 may be used as a therapeutic agent against CSCs in colon cancers.
5.1.4. Melanoma stem cells
Similarly to other types of CSCs, malignant melanoma stem cells have the ability to initiate new tumors and generate a heterogeneous population of cells that includes CSCs [259]. Cell-surface markers specific for melanoma stem cells, such as CD20 and CD133 have been identified [260,261], but the correlation between expression of these markers and the cells' capacities for self-renewal and tumorigenesis remains to be elucidated. More recent reports have shown that a high proportion (at least 25%) of melanoma cells isolated from patients were able to initiate tumorigenesis from a single cell [262]. Furthermore, none of the 22 heterogenously expressed markers that are commonly used to identify CSCs, can isolate tumorigenic melanoma stem cells, and any large subpopulation of melanoma cells does not lack tumorigenicity [262]. Moreover, reversible epigenetic regulatory mechanisms allow melanoma cells to readily convert between a CSC state, which is quiescent and resistant to therapy, and a non-CSC state that cannot initiate tumor formation [263]. This phenotypic plasticity in melanoma cells may represent an extreme example of functional plasticity of epithelial cancers [264]. Signals from melanoma microenvironments play a key role in regulating this fate change from a stem cell-like to a non-stem cell-like state. Nodal, a regulator of human ESC pluripotency, is expressed in human melanoma cells, and is required, at least in part, for tumor cell plasticity and aggressiveness [265–267]. Anti-nodal antibodies can inhibit nodal-induced plasticity in vitro and lung colonization in mice [265,267]. Nodal expression is not detected in normal melanocytes, suggesting deregulated nodal signaling in melanoma cells. Nodal may represent a novel prognostic and therapeutic target for human melanoma.
5.1.5. Other epithelial cancers
TGF-β family signaling also plays important roles in other types of epithelial CSCs. In squamous cell carcinoma, a subpopulation of α6β1hiCD34hi cells is thought to function as CSCs. TGF-β signaling inhibits self-renewal and expansion of α6β1hiCD34hi cells in a Tgfbr2−/− mouse model of squamous cell carcinoma [268]. In human diffuse-type gastric carcinoma, TGF-β from the tumor microenvironment decreases the side population or ALDH1+ cells, which show CSC properties, and inhibits tumor formation in immunodeficient mice [269,270]. In primary human ovarian cancer cells, TGF-β secreted in the tumor microenvironment induces expression of tissue transglutaminase 2 (TG2). TG2 induces EMT and spheroid formation, which correlate with the CSC properties, to enhance metastasis [271]. In human prostate cancer, BMP7, secreted from bone stromal cells, induces reversible senescence of CSCs. BMP7 signaling induces the CSCs to become dormant, in part by activating p38 MAPK and increasing the expression of the cell cycle inhibitor p21CIP1 and the metastasis suppressor NDRG1 (N-myc downstream-regulated gene 1) [272].
5.2. Glioma stem cells
Human gliomas, which are derived from multipotent neural stem cells, neural progenitor cells or glial progenitor cells [25], contain a small fraction of glioblastoma stem cells (GSCs), which retain many similarities to neural stem cells, including the abilities to self-renew and differentiate into neural and glial cells. GSCs express specific markers such as CD133 and nestin that are commonly expressed in normal neural stem cells [273]. Similarly to BMP-induced astrocytic differentiation of embryonic neural stem cells, BMP4 inhibits the proliferation of GSCs, and induces their differentiation into glioma cells with an astroglial-like fate [274]. Consequently, BMP4 delivered to the tumor in vivo, blocks tumor growth in mice following intracerebral grafting of human glioblastoma cells [274]. However, BMP-induced astroglial differentiation appears to be impaired in a subpopulation of GSCs, due to epigenetic silencing of the BMPR1B gene [275]. These results suggest that new oncogenic alterations that could be beneficial for the expansion of GSCs may selectively accumulate under some conditions. These deregulated signals in GSCs may reveal themselves as possible targets for therapeutic approaches.
Increased TGF-β1 expression is commonly seen in malignant glioma cells, similarly to upregulated TGF-β1 expression in carcinomas and other tumors of neuro-ectodermal origin. Recent reports have revealed key roles of TGF-β signaling in the maintenance of stem cell-like properties and the tumor initiating ability of GSCs. TGF-β maintains the tumorigenicity of human GSCs through Smad-mediated induction of LIF expression, which then, in an autocrine way, activates the JAK–STAT signaling pathway in GSCs, to increase self-renewal and decrease differentiation [276]. TGF-β signaling maintains stem cell properties of human GSCs through Smad-dependent induction of SOX4 expression and subsequent induction of SOX2 expression. SOX2 is an ESC self-renewal gene, which regulates the expression of other transcription factors that are critical for maintaining stem cell-like properties of ESCs and neural stem cells [277]. These findings show that TGF-β signaling maintains GSCs' population and promotes glioblastoma formation. Accordingly, pharmacological inhibition of the TβRI kinase can augment the response to radiation treatment in a mouse model of orthotopic, intracranially implanted human glioblastoma cells [278]. By coordinately increasing apoptosis of glioma cells and GSCs, while blocking DNA damage repair, invasion, mesenchymal transition, and angiogenesis, the TβRI kinase inhibitor blocks glioma expansion [278]. The combination of radiotherapy with inhibition of TGF-β signaling may be a therapeutic strategy for gliomas.
5.3. Leukemic stem cells
Leukemic stem cells (LSCs) were first identified as a phenotypically and functionally distinct cell population in acute myeloid leukemia (AML) [279]. Various studies examined the role of the stem cell niche in the control of LSCs. In both AML and chronic myeloid leukemia (CML), the interaction of the stem cell marker CD44, which is expressed in LSCs, with hyaluronic acid moieties at the surface of stromal cells in bone marrow, is essential for adherence of LSCs to the niche [280,281]. This mechanism resembles that of the interaction between normal HSCs and the vascular niche, again revealing parallels between normal HSCs and LSCs (Fig. 4B).
TGF-β signaling plays crucial roles in the maintenance of LSCs. In human CML, TGF-β produced by LSCs, or in the tumor microenvironment, is thought to be required for survival, proliferation, and maintenance of stem cell-like properties of LSCs, through activating Akt signaling, which induces nuclear localization of FOXO3a [282]. Regulation of Akt activation and nuclear localization of FOXO3a in response to TGF-β has also been shown in normal HSCs [239], demonstrating a common mechanism for normal HSCs and LSCs. TGF-β-induced FOXO3a nuclear localization was specific to LSC, and was not observed in the non-LSC cell population [282], suggesting that TGF-β has different effects on FOXO3a activation in LSCs and non-LSCs. Thus, inhibition of TGF-β-FOXO signaling may provide a possible therapeutic approach for the treatment of human CML.
In contrast to the stimulatory effect of TGF-β signaling on CML stem cell maintenance and proliferation, TGF-β-induced Smad signaling impairs initiation and progression of leukemia in a mouse model of AML [283]. Ina mouse model of human AML induced by HOXA9 or the fusion oncogene NUP98–HOXA9, Smad4 binds and stabilizes Hoxa9 in the cytoplasm, thus inhibiting nuclear localization of Hoxa9. Since nuclear Hoxa9 induces transformation of normal hematopoietic stem or progenitor cells to LSCs, Smad4 can negatively regulate the transformation of HSC into LSC [283]. Loss of Smad4 increases Hoxa9 levels in the nucleus, enhancing the transformation of HSC into LSC, and accelerating leukemogenesis [283]. The differences between LSCs from different leukemias or different subpopulations of LSCs may reflect complex mechanisms by which TGF-β differentially regulates the maintenance of normal HSC subpopulations [244]. Further studies on the effects of TGF-β on LSC maintenance, and their mechanisms, will yield additional prognostic methods and new therapeutic targets in the treatment of leukemia.
5.4. Tumor stromal cells
Functional interactions of CSCs with stromal cells that constitute the tumor microenvironment are important for tumor formation, maintenance and progression. Most of the tumor microenvironment consists of cancer-associated fibroblasts (CAFs), which closely resemble myofibroblasts and express α-smooth muscle actin. Although the origin of CAFs is not clear, several studies have demonstrated that bone marrow-derived MSCs and endothelial cells provide a source of CAFs [284,285]. TGF-β family signaling has important roles in the interactions between tumor cells and CAFs. In mouse models of inflammation-dependent gastric cancer, TGF-β induces differentiation of bone marrow-derived MSCs to myofibroblasts, expansion of these cells and recruitment to tumors through induced expression of the secreted chemokine SDF-1α by myofibroblasts [286]. SDF-1α functions, in an autocrine fashion, as a stimulator of proliferation of myofibroblasts, and, in a paracrine fashion, as an attractant of myofibroblasts to tumor cells, which express the SDF-1α receptor CXCR4. The myofibroblasts are recruited to tumors and support cancer cells as CAFs [286]. Additionally, inhibition of TGF-β signaling or CXCR4 signaling reduces tumor growth in vivo [286]. On the other hand, cancer-associated MSCs isolated from ovarian cancer ascites show increased expression of BMP2, 4 and 6, when compared with normal MSCs. BMPs expressed by cancer-associated MSCs increase the number of human ovarian CSCs and promote tumorigenesis [287]. TGF-β also stimulates the generation of CAFs from endothelial cells, which occurs through endothelial to mesenchymal transition [285]. Although various findings suggest the importance of TGF-β family signaling in the interactions of CSCs and CAFs, little is known about the roles of TGF-β family signaling in the regulation of tumor stromal cells. Further studies on the interaction of CSCs and tumor stromal cells may reveal additional roles of TGF-β family signaling in the maintenance of CSCs, through effects on tumor stromal cells.
6. Conclusions and future perspective
CSCs, and embryonic and somatic stem cells have striking similarities in their responses to signals from the surrounding microenvironment, yet some aspects are distinct. Since TGF-β family signaling appears to have different effects on subpopulations of certain tissue stem cells, such as intestinal stem cells or hematopoietic stem cells, it is to be expected that diverse effects of TGF-β family signaling are seen in different types of CSCs. According to their innate properties and acquired changes, several different approaches to target these sub-populations are needed. Further mechanistic studies on TGF-β family signal transduction and the cross-talk with other signaling pathways in different subpopulations of cancer and normal stem cells will provide a better understanding of the roles of TGF-β family signaling in the regulation of stem cells. These basic studies may lead to development of a new therapeutic or prognostic strategy for the treatment of cancers.
Footnotes
This article is part of a Special Issue entitled Biochemistry of Stem Cells.
References
- 1.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U. S. A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 3.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 4.Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, Powers D, Melton DA. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 5.Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 6.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 7.Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miettinen PJ, Ebner R, Lopez AR, Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 1994;127:2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulessa H, Frampton J, Graf T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 1995;9:1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- 10.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 11.Bendall SC, Stewart MH, Menendez P, George D, Vijayaragavan K, Werbowetski-Ogilvie T, Ramos-Mejia V, Rouleau A, Yang J, Bosse M, Lajoie G, Bhatia M. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature. 2007;448:1015–1021. doi: 10.1038/nature06027. [DOI] [PubMed] [Google Scholar]
- 12.Ferraro F, Celso CL, Scadden D. Adult stem cells and their niches. Adv. Exp. Med. Biol. 2010;695:155–168. doi: 10.1007/978-1-4419-7037-4_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voog J, Jones DL. Stem cells and the niche: a dynamic duo. Cell Stem Cell. 2010;6:103–115. doi: 10.1016/j.stem.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snippert HJ, Clevers H. Tracking adult stem cells. EMBO Rep. 2011;12:113–122. doi: 10.1038/embor.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BL, Dougan G, Huntly B, Gottgens B, Jenkins NA, Copeland NG, Colucci F, Liu P. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 24.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat. Rev. Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 25.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 26.Lane SW, Scadden DT, Gilliland DG. The leukemic stem cell niche: current concepts and therapeutic opportunities. Blood. 2009;114:1150–1157. doi: 10.1182/blood-2009-01-202606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 28.Oshimori N, Fuchs E. Paracrine TGF-beta signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell. 2012;10:63–75. doi: 10.1016/j.stem.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishimura EK, Suzuki M, Igras V, Du J, Lonning S, Miyachi Y, Roes J, Beermann F, Fisher DE. Key roles for transforming growth factor beta in melanocyte stem cell maintenance. Cell Stem Cell. 2010;6:130–140. doi: 10.1016/j.stem.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL, Weinberg RA. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J. Cell Sci. 2003;116:217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- 33.Derynck R, Miyazono K. In: TGF-beta and the TGF-beta family. Derynck R, Miyazono K, editors. New York: The TGF-beta family, Cold Spring Harbor Laboratory Press; 2008. pp. 29–44. [Google Scholar]
- 34.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 35.Wu MY, Hill CS. TGF-beta superfamily signaling in embryonic development and homeostasis. Dev. Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 36.Shen MM. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- 37.Derynck R, Akhurst RJ. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nat. Cell Biol. 2007;9:1000–1004. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- 38.Whitman M. Smads and early developmental signaling by the TGFbeta superfamily. Genes Dev. 1998;12:2445–2462. doi: 10.1101/gad.12.16.2445. [DOI] [PubMed] [Google Scholar]
- 39.Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 40.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 41.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 42.Feng XH, Derynck R. Specificity and versatility in TGF-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 43.Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 45.Labbe E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol. Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 46.Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu D, Black BL, Derynck R. TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 2001;15:2950–2966. doi: 10.1101/gad.925901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choy L, Derynck R. Transforming growth factor-beta inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J. Biol. Chem. 2003;278:9609–9619. doi: 10.1074/jbc.M212259200. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki A, Raya A, Kawakami Y, Morita M, Matsui T, Nakashima K, Gage FH, Rodriguez-Esteban C, Izpisua Belmonte JC. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10294–10299. doi: 10.1073/pnas.0506945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, Thomson JA. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MW, Cho CH, Martinez A, Rugg-Gunn P, Brons G, Pedersen RA. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoon SJ, Wills AE, Chuong E, Gupta R, Baker JC. HEB and E2A function as SMAD/FOXH1 cofactors. Genes Dev. 2011;25:1654–1661. doi: 10.1101/gad.16800511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, Studer L, Mark W, Patel DJ, Massague J. A poised chromatin platform for TGF-beta access to master regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahle O, Kumar A, Kuehn MR. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci. Signal. 2010;3:pra48. doi: 10.1126/scisignal.2000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J. Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 57.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J, Smith SM, Derynck R. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 2007;26:3957–3967. doi: 10.1038/sj.emboj.7601818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sorrentino A, Thakur N, Grimsby S, Marcusson A, von Bulow V, Schuster N, Zhang S, Heldin CH, Landstrom M. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 2008;10:1199–1207. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- 60.Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol. Cell. 2008;31:918–924. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell. 2001;12:27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edlund S, Landstrom M, Heldin CH, Aspenstrom P. Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol. Biol. Cell. 2002;13:902–914. doi: 10.1091/mbc.01-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 64.Vardouli L, Moustakas A, Stournaras C. LIM-kinase 2 and cofilin phosphorylation mediate actin cytoskeleton reorganization induced by transforming growth factor-beta. J. Biol. Chem. 2005;280:11448–11457. doi: 10.1074/jbc.M402651200. [DOI] [PubMed] [Google Scholar]
- 65.Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 2000;275:36803–36810. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- 66.Shin I, Bakin AV, Rodeck U, Brunet A, Arteaga CL. Transforming growth factor beta enhances epithelial cell survival via Akt-dependent regulation of FKHRL1. Mol. Biol. Cell. 2001;12:3328–3339. doi: 10.1091/mbc.12.11.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J. Cell Biol. 2007;178:437–451. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lamouille S, Connolly E, Smyth JW, Akhurst RJ, Derynck R. TGF-beta-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J. Cell Sci. 2012;125:1259–1273. doi: 10.1242/jcs.095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 70.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phanstiel DH, Brumbaugh J, Wenger CD, Tian S, Probasco MD, Bailey DJ, Swaney DL, Tervo MA, Bolin JM, Ruotti V, Stewart R, Thomson JA, Coon JJ. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat. Methods. 2011;8:821–827. doi: 10.1038/nmeth.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Greber B, Wu G, Bernemann C, Joo JY, Han DW, Ko K, Tapia N, Sabour D, Sterneckert J, Tesar P, Scholer HR. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 76.Vallier L, Touboul T, Chng Z, Brimpari M, Hannan N, Millan E, Smithers LE, Trotter M, Rugg-Gunn P, Weber A, Pedersen RA. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS One. 2009;4:e6082. doi: 10.1371/journal.pone.0006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 78.Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC, Bardwell L, Moffett A, Pedersen RA. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9:144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tropepe V, Hitoshi S, Sirard C, Mak TW, Rossant J, van der Kooy D. Direct neural fate specification from embryonic stem cells: a primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 80.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 81.Qi X, Li TG, Hao J, Hu J, Wang J, Simmons H, Miura S, Mishina Y, Zhao GQ. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6027–6032. doi: 10.1073/pnas.0401367101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ogawa K, Saito A, Matsui H, Suzuki H, Ohtsuka S, Shimosato D, Morishita Y, Watabe T, Niwa H, Miyazono K. Activin–Nodal signaling is involved in propagation of mouse embryonic stem cells. J. Cell Sci. 2007;120:55–65. doi: 10.1242/jcs.03296. [DOI] [PubMed] [Google Scholar]
- 84.Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 86.Zhou H, Li W, Zhu S, Joo JY, Do JT, Xiong W, Kim JB, Zhang K, Scholer HR, Ding S. Conversion of mouse epiblast stem cells to an earlier pluripotency state by small molecules. J. Biol. Chem. 2010;285:29676–29680. doi: 10.1074/jbc.C110.150599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang J, van Oosten AL, Theunissen TW, Guo G, Silva JC, Smith A. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu Y, Zhu X, Hahm HS, Wei W, Hao E, Hayek A, Ding S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 90.Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- 92.Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Han DW, Tapia N, Joo JY, Greber B, Arauzo-Bravo MJ, Bernemann C, Ko K, Wu G, Stehling M, Do JT, Scholer HR. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell. 2010;143:617–627. doi: 10.1016/j.cell.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 94.Stewart MH, Bosse M, Chadwick K, Menendez P, Bendall SC, Bhatia M. Clonal isolation of hESCs reveals heterogeneity within the pluripotent stem cell compartment. Nat. Methods. 2006;3:807–815. doi: 10.1038/nmeth939. [DOI] [PubMed] [Google Scholar]
- 95.Xu X, Browning VL, Odorico JS. Activin, BMP and FGF pathways cooperate to promote endoderm and pancreatic lineage cell differentiation from human embryonic stem cells. Mech. Dev. 2011;128:412–427. doi: 10.1016/j.mod.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 97.Sumi T, Tsuneyoshi N, Nakatsuji N, Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development. 2008;135:2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- 98.Gadue P, Huber TL, Paddison PJ, Keller GM. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 100.Yasunaga M, Tada S, Torikai-Nishikawa S, Nakano Y, Okada M, Jakt LM, Nishikawa S, Chiba T, Era T. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat. Biotechnol. 2005;23:1542–1550. doi: 10.1038/nbt1167. [DOI] [PubMed] [Google Scholar]
- 101.Gu Z, Nomura M, Simpson BB, Lei H, Feijen A, van den Eijnden-van Raaij J, Donahoe PK, Li E. The type I activin receptor ActRIB is required for egg cylinder organization and gastrulation in the mouse. Genes Dev. 1998;12:844–857. doi: 10.1101/gad.12.6.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wiles MV, Johansson BM. Embryonic stem cell development in a chemically defined medium. Exp. Cell Res. 1999;247:241–248. doi: 10.1006/excr.1998.4353. [DOI] [PubMed] [Google Scholar]
- 103.Xu RH, Peck RM, Li DS, Feng X, Ludwig T, Thomson JA. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat. Methods. 2005;2:185–190. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- 104.Kubo A, Shinozaki K, Shannon JM, Kouskoff V, Kennedy M, Woo S, Fehling HJ, Keller G. Development of definitive endoderm from embryonic stem cells in culture. Development. 2004;131:1651–1662. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- 105.Tada S, Era T, Furusawa C, Sakurai H, Nishikawa S, Kinoshita M, Nakao K, Chiba T. Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development. 2005;132:4363–4374. doi: 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- 106.Fei T, Zhu S, Xia K, Zhang J, Li Z, Han JD, Chen YG. Smad2 mediates Activin/Nodal signaling in mesendoderm differentiation of mouse embryonic stem cells. Cell Res. 2010;20:1306–1318. doi: 10.1038/cr.2010.158. [DOI] [PubMed] [Google Scholar]
- 107.Lee KL, Lim SK, Orlov YL, Yit le Y, Yang H, Ang LT, Poellinger L, Lim B. Graded Nodal/Activin signaling titrates conversion of quantitative phospho-Smad2 levels into qualitative embryonic stem cell fate decisions. PLoS Genet. 2011;7:e1002130. doi: 10.1371/journal.pgen.1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brown S, Teo A, Pauklin S, Hannan N, Cho CH, Lim B, Vardy L, Dunn NR, Trotter M, Pedersen R, Vallier L. Activin/Nodal signaling controls divergent transcriptional networks in human embryonic stem cells and in endoderm progenitors. Stem Cells. 2011;29:1176–1185. doi: 10.1002/stem.666. [DOI] [PubMed] [Google Scholar]
- 109.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev. Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 110.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou J, Su P, Li D, Tsang S, Duan E, Wang F. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells. 2010;28:1741–1750. doi: 10.1002/stem.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Han DW, Greber B, Wu G, Tapia N, Arauzo-Bravo MJ, Ko K, Bernemann C, Stehling M, Scholer HR. Direct reprogramming of fibroblasts into epiblast stem cells. Nat. Cell Biol. 2011;13:66–71. doi: 10.1038/ncb2136. [DOI] [PubMed] [Google Scholar]
- 113.Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, Huangfu D, Akutsu H, Liu DR, Rubin LL, Eggan K. A small-molecule inhibitor of TGF-beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maherali N, Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30:823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 117.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 119.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 120.Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lipchina I, Elkabetz Y, Hafner M, Sheridan R, Mihailovic A, Tuschl T, Sander C, Studer L, Betel D. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 2011;25:2173–2186. doi: 10.1101/gad.17221311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barroso-delJesus A, Lucena-Aguilar G, Sanchez L, Ligero G, Gutierrez-Aranda I, Menendez P. The Nodal inhibitor Lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. FASEB J. 2011;25:1497–1508. doi: 10.1096/fj.10-172221. [DOI] [PubMed] [Google Scholar]
- 124.Rogler CE, Levoci L, Ader T, Massimi A, Tchaikovskaya T, Norel R, Rogler LE. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50:575–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- 125.Wang Q, Huang Z, Xue H, Jin C, Ju XL, Han JD, Chen YG. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 2008;111:588–595. doi: 10.1182/blood-2007-05-092718. [DOI] [PubMed] [Google Scholar]
- 126.Kim YJ, Hwang SJ, Bae YC, Jung JS. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells. 2009;27:3093–3102. doi: 10.1002/stem.235. [DOI] [PubMed] [Google Scholar]
- 127.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 129.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 130.Liu S, Dontu G, Wicha MS. Mammary stem cells, self-renewal pathways, and carcinogenesis. Breast Cancer Res. 2005;7:86–95. doi: 10.1186/bcr1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kordon EC, McKnight RA, Jhappan C, Hennighausen L, Merlino G, Smith GH. Ectopic TGF beta 1 expression in the secretory mammary epithelium induces early senescence of the epithelial stem cell population. Dev. Biol. 1995;168:47–61. doi: 10.1006/dbio.1995.1060. [DOI] [PubMed] [Google Scholar]
- 132.Moses H, Barcellos-Hoff MH. TGF-beta biology in mammary development and breast cancer. Cold Spring Harb. Perspect. Biol. 2011;3:a003277. doi: 10.1101/cshperspect.a003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial–mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 134.Kalluri R, Weinberg RA. The basics of epithelial–mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996;10:2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 136.Piek E, Moustakas A, Kurisaki A, Heldin CH, ten Dijke P. TGF-(beta) type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J. Cell Sci. 1999;112(24):4557–4568. doi: 10.1242/jcs.112.24.4557. [DOI] [PubMed] [Google Scholar]
- 137.Moustakas A, Heldin CH. Signaling networks guiding epithelial–mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98:1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang J, Weinberg RA. Epithelial–mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 139.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 142.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 143.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 147.Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ, Clevers H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303:1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- 148.He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, Mishina Y, Li L. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt–beta-catenin signaling. Nat. Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- 149.Howe JR, Roth S, Ringold JC, Summers RW, Jarvinen HJ, Sistonen P, Tomlinson IP, Houlston RS, Bevan S, Mitros FA, Stone EM, Aaltonen LA. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science. 1998;280:1086–1088. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- 150.Howe JR, Bair JL, Sayed MG, Anderson ME, Mitros FA, Petersen GM, Velculescu VE, Traverso G, Vogelstein B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat. Genet. 2001;28:184–187. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- 151.Millar SE. Molecular mechanisms regulating hair follicle development. J. Invest. Dermatol. 2002;118:216–225. doi: 10.1046/j.0022-202x.2001.01670.x. [DOI] [PubMed] [Google Scholar]
- 152.Schmidt-Ullrich R, Paus R. Molecular principles of hair follicle induction and morphogenesis. Bioessays. 2005;27:247–261. doi: 10.1002/bies.20184. [DOI] [PubMed] [Google Scholar]
- 153.Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 155.Foitzik K, Lindner G, Mueller-Roever S, Maurer M, Botchkareva N, Botchkarev V, Handjiski B, Metz M, Hibino T, Soma T, Dotto GP, Paus R. Control of murine hair follicle regression (catagen) by TGF-beta1 in vivo. FASEB J. 2000;14:752–760. doi: 10.1096/fasebj.14.5.752. [DOI] [PubMed] [Google Scholar]
- 156.Soma T, Dohrmann CE, Hibino T, Raftery LA. Profile of transforming growth factor-beta responses during the murine hair cycle. J. Invest. Dermatol. 2003;121:969–975. doi: 10.1046/j.1523-1747.2003.12516.x. [DOI] [PubMed] [Google Scholar]
- 157.Foitzik K, Paus R, Doetschman T, Dotto GP. The TGF-beta2 isoform is both a required and sufficient inducer of murine hair follicle morphogenesis. Dev. Biol. 1999;212:278–289. doi: 10.1006/dbio.1999.9325. [DOI] [PubMed] [Google Scholar]
- 158.Kobielak K, Stokes N, de la Cruz J, Polak L, Fuchs E. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10063–10068. doi: 10.1073/pnas.0703004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kulessa H, Turk G, Hogan BL. Inhibition of Bmp signaling affects growth and differentiation in the anagen hair follicle. EMBO J. 2000;19:6664–6674. doi: 10.1093/emboj/19.24.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Botchkarev VA, Botchkareva NV, Nakamura M, Huber O, Funa K, Lauster R, Paus R, Gilchrest BA. Noggin is required for induction of the hair follicle growth phase in postnatal skin. FASEB J. 2001;15:2205–2214. doi: 10.1096/fj.01-0207com. [DOI] [PubMed] [Google Scholar]
- 161.Zhang J, He XC, Tong WG, Johnson T, Wiedemann LM, Mishina Y, Feng JQ, Li L. Bone morphogenetic protein signaling inhibits hair follicle anagen induction by restricting epithelial stem/progenitor cell activation and expansion. Stem Cells. 2006;24:2826–2839. doi: 10.1634/stemcells.2005-0544. [DOI] [PubMed] [Google Scholar]
- 162.Nishimura EK, Jordan SA, Oshima H, Yoshida H, Osawa M, Moriyama M, Jackson IJ, Barrandon Y, Miyachi Y, Nishikawa S. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416:854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- 163.Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 164.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 165.Nishimura EK, Granter SR, Fisher DE. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science. 2005;307:720–724. doi: 10.1126/science.1099593. [DOI] [PubMed] [Google Scholar]
- 166.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tanimura S, Tadokoro Y, Inomata K, Binh NT, Nishie W, Yamazaki S, Nakauchi H, Tanaka Y, McMillan JR, Sawamura D, Yancey K, Shimizu H, Nishimura EK. Hair follicle stem cells provide a functional niche for melanocyte stem cells. Cell Stem Cell. 2011;8:177–187. doi: 10.1016/j.stem.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 168.Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair—current views. Stem Cells. 2007;25:2896–2902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- 169.Shi S, Bartold PM, Miura M, Seo BM, Robey PG, Gronthos S. The efficacy of mesenchymal stem cells to regenerate and repair dental structures. Orthod. Craniofac. Res. 2005;8:191–199. doi: 10.1111/j.1601-6343.2005.00331.x. [DOI] [PubMed] [Google Scholar]
- 170.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 171.Williams JT, Southerland SS, Souza J, Calcutt AF, Cartledge RG. Cells isolated from adult human skeletal muscle capable of differentiating into multiple mesodermal phenotypes. Am. Surg. 1999;65:22–26. [PubMed] [Google Scholar]
- 172.Caplan AI. Mesenchymal stem cells and gene therapy. Clin. Orthop. Relat. Res. 2000:S67–70. doi: 10.1097/00003086-200010001-00010. [DOI] [PubMed] [Google Scholar]
- 173.Boland GM, Perkins G, Hall DJ, Tuan RS. Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J. Cell. Biochem. 2004;93:1210–1230. doi: 10.1002/jcb.20284. [DOI] [PubMed] [Google Scholar]
- 174.Jian H, Shen X, Liu I, Semenov M, He X, Wang XF. Smad3-dependent nuclear translocation of beta-catenin is required for TGF-beta1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev. 2006;20:666–674. doi: 10.1101/gad.1388806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Baksh D, Boland GM, Tuan RS. Cross-talk between Wnt signaling pathways in human mesenchymal stem cells leads to functional antagonism during osteogenic differentiation. J. Cell. Biochem. 2007;101:1109–1124. doi: 10.1002/jcb.21097. [DOI] [PubMed] [Google Scholar]
- 176.Liu Z, Tang Y, Qiu T, Cao X, Clemens TL. A dishevelled-1/Smad1 interaction couples WNT and bone morphogenetic protein signaling pathways in uncommitted bone marrow stromal cells. J. Biol. Chem. 2006;281:17156–17163. doi: 10.1074/jbc.M513812200. [DOI] [PubMed] [Google Scholar]
- 177.Roelen BA, Dijke P. Controlling mesenchymal stem cell differentiation by TGFbeta family members. J. Orthop. Sci. 2003;8:740–748. doi: 10.1007/s00776-003-0702-2. [DOI] [PubMed] [Google Scholar]
- 178.Maeda S, Hayashi M, Komiya S, Imamura T, Miyazono K. Endogenous TGF-beta signaling suppresses maturation of osteoblastic mesenchymal cells. EMBO J. 2004;23:552–563. doi: 10.1038/sj.emboj.7600067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lin X, Liang YY, Sun B, Liang M, Shi Y, Brunicardi FC, Feng XH. Smad6 recruits transcription corepressor CtBP to repress bone morphogenetic protein-induced transcription. Mol. Cell. Biol. 2003;23:9081–9093. doi: 10.1128/MCB.23.24.9081-9093.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, Ahrens MJ, Dudley AT, Norris AW, Kulkarni RN, Kahn CR. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ferrari G, Cusella-De Angelis G, Coletta M, Paolucci E, Stornaiuolo A, Cossu G, Mavilio F. Muscle regeneration by bone marrow-derived myogenic progenitors. Science. 1998;279:1528–1530. doi: 10.1126/science.279.5356.1528. [DOI] [PubMed] [Google Scholar]
- 182.Goldring K, Partridge T, Watt D. Muscle stem cells. J. Pathol. 2002;197:457–467. doi: 10.1002/path.1157. [DOI] [PubMed] [Google Scholar]
- 183.Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson SM, Li B, Pickel J, McKay R, Nadal-Ginard B, Bodine DM, Leri A, Anversa P. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410:701–705. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- 184.Toma C, Pittenger MF, Cahill KS, Byrne BJ, Kessler PD. Human mesenchymal stem cells differentiate to a cardiomyocyte phenotype in the adult murine heart. Circulation. 2002;105:93–98. doi: 10.1161/hc0102.101442. [DOI] [PubMed] [Google Scholar]
- 185.Wang JS, Shum-Tim D, Galipeau J, Chedrawy E, Eliopoulos N, Chiu RC. Marrow stromal cells for cellular cardiomyoplasty: feasibility and potential clinical advantages. J. Thorac. Cardiovasc. Surg. 2000;120:999–1005. doi: 10.1067/mtc.2000.110250. [DOI] [PubMed] [Google Scholar]
- 186.Pittenger MF, Martin BJ. Mesenchymal stem cells and their potential as cardiac therapeutics. Circ. Res. 2004;95:9–20. doi: 10.1161/01.RES.0000135902.99383.6f. [DOI] [PubMed] [Google Scholar]
- 187.Min JY, Sullivan MF, Yang Y, Zhang JP, Converso KL, Morgan JP, Xiao YF. Significant improvement of heart function by cotransplantation of human mesenchymal stem cells and fetal cardiomyocytes in postinfarcted pigs. Ann. Thorac. Surg. 2002;74:1568–1575. doi: 10.1016/s0003-4975(02)03952-8. [DOI] [PubMed] [Google Scholar]
- 188.Mathur A, Martin JF. Stem cells and repair of the heart. Lancet. 2004;364:183–192. doi: 10.1016/S0140-6736(04)16632-4. [DOI] [PubMed] [Google Scholar]
- 189.Stamm C, Westphal B, Kleine HD, Petzsch M, Kittner C, Klinge H, Schumichen C, Nienaber CA, Freund M, Steinhoff G. Autologous bone-marrow stem-cell transplantation for myocardial regeneration. Lancet. 2003;361:45–46. doi: 10.1016/S0140-6736(03)12110-1. [DOI] [PubMed] [Google Scholar]
- 190.Miyahara Y, Nagaya N, Kataoka M, Yanagawa B, Tanaka K, Hao H, Ishino K, Ishida H, Shimizu T, Kangawa K, Sano S, Okano T, Kitamura S, Mori H. Monolayered mesenchymal stem cells repair scarred myocardium after myocardial infarction. Nat. Med. 2006;12:459–465. doi: 10.1038/nm1391. [DOI] [PubMed] [Google Scholar]
- 191.Kurpinski K, Lam H, Chu J, Wang A, Kim A, Tsay E, Agrawal S, Schaffer DV, Li S. Transforming growth factor-beta and notch signaling mediate stem cell differentiation into smooth muscle cells. Stem Cells. 2010;28:734–742. doi: 10.1002/stem.319. [DOI] [PubMed] [Google Scholar]
- 192.Campion DR. The muscle satellite cell: a review. Int. Rev. Cytol. 1984;87:225–251. doi: 10.1016/s0074-7696(08)62444-4. [DOI] [PubMed] [Google Scholar]
- 193.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 194.Peault B, Rudnicki M, Torrente Y, Cossu G, Tremblay JP, Partridge T, Gussoni E, Kunkel LM, Huard J. Stem and progenitor cells in skeletal muscle development, maintenance, and therapy. Mol. Ther. 2007;15:867–877. doi: 10.1038/mt.sj.6300145. [DOI] [PubMed] [Google Scholar]
- 195.Asakura A, Seale P, Girgis-Gabardo A, Rudnicki MA. Myogenic specification of side population cells in skeletal muscle. J. Cell Biol. 2002;159:123–134. doi: 10.1083/jcb.200202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Torrente Y, Belicchi M, Sampaolesi M, Pisati F, Meregalli M, D'Antona G, Tonlorenzi R, Porretti L, Gavina M, Mamchaoui K, Pellegrino MA, Furling D, Mouly V, Butler-Browne GS, Bottinelli R, Cossu G, Bresolin N. Human circulating AC133(+) stem cells restore dystrophin expression and ameliorate function in dystrophic skeletal muscle. J. Clin. Invest. 2004;114:182–195. doi: 10.1172/JCI20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Galvez BG, Sampaolesi M, Brunelli S, Covarello D, Gavina M, Rossi B, Constantin G, Torrente Y, Cossu G. Complete repair of dystrophic skeletal muscle by mesoangioblasts with enhanced migration ability. J. Cell Biol. 2006;174:231–243. doi: 10.1083/jcb.200512085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dellavalle A, Sampaolesi M, Tonlorenzi R, Tagliafico E, Sacchetti B, Perani L, Innocenzi A, Galvez BG, Messina G, Morosetti R, Li S, Belicchi M, Peretti G, Chamberlain JS, Wright WE, Torrente Y, Ferrari S, Bianco P, Cossu G. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat. Cell Biol. 2007;9:255–267. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- 199.Negroni E, Riederer I, Chaouch S, Belicchi M, Razini P, Di Santo J, Torrente Y, Butler-Browne GS, Mouly V. In vivo myogenic potential of human CD133+ muscle-derived stem cells: a quantitative study. Mol. Ther. 2009;17:1771–1778. doi: 10.1038/mt.2009.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hawke TJ, Garry DJ. Myogenic satellite cells: physiology to molecular biology. J. Appl. Physiol. 2001;91:534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- 202.Kollias HD, McDermott JC. Transforming growth factor-beta and myostatin signaling in skeletal muscle. J. Appl. Physiol. 2008;104:579–587. doi: 10.1152/japplphysiol.01091.2007. [DOI] [PubMed] [Google Scholar]
- 203.Allen RE, Boxhorn LK. Regulation of skeletal muscle satellite cell proliferation and differentiation by transforming growth factor-beta, insulin-like growth factor I, and fibroblast growth factor. J. Cell. Physiol. 1989;138:311–315. doi: 10.1002/jcp.1041380213. [DOI] [PubMed] [Google Scholar]
- 204.Cook DR, Doumit ME, Merkel RA. Transforming growth factor-beta, basic fibroblast growth factor, and platelet-derived growth factor-BB interact to affect proliferation of clonally derived porcine satellite cells. J. Cell. Physiol. 1993;157:307–312. doi: 10.1002/jcp.1041570213. [DOI] [PubMed] [Google Scholar]
- 205.Florini JR, Roberts AB, Ewton DZ, Falen SL, Flanders KC, Sporn MB. Transforming growth factor-beta. A very potent inhibitor of myoblast differentiation, identical to the differentiation inhibitor secreted by Buffalo rat liver cells. J. Biol. Chem. 1986;261:16509–16513. [PubMed] [Google Scholar]
- 206.Greene EA, Allen RE. Growth factor regulation of bovine satellite cell growth in vitro. J. Anim. Sci. 1991;69:146–152. doi: 10.2527/1991.691146x. [DOI] [PubMed] [Google Scholar]
- 207.Massague J, Cheifetz S, Endo T, Nadal-Ginard B. Type beta transforming growth factor is an inhibitor of myogenic differentiation. Proc. Natl. Acad. Sci. U. S. A. 1986;83:8206–8210. doi: 10.1073/pnas.83.21.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Olson EN, Sternberg E, Hu JS, Spizz G, Wilcox C. Regulation of myogenic differentiation by type beta transforming growth factor. J. Cell Biol. 1986;103:1799–1805. doi: 10.1083/jcb.103.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Amthor H, Macharia R, Navarrete R, Schuelke M, Brown SC, Otto A, Voit T, Muntoni F, Vrbova G, Partridge T, Zammit P, Bunger L, Patel K. Lack of myostatin results in excessive muscle growth but impaired force generation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1835–1840. doi: 10.1073/pnas.0604893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mendias CL, Marcin JE, Calerdon DR, Faulkner JA. Contractile properties of EDL and soleus muscles of myostatin-deficient mice. J. Appl. Physiol. 2006;101:898–905. doi: 10.1152/japplphysiol.00126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Whittemore LA, Song K, Li X, Aghajanian J, Davies M, Girgenrath S, Hill JJ, Jalenak M, Kelley P, Knight A, Maylor R, O'Hara D, Pearson A, Quazi A, Ryerson S, Tan XY, Tomkinson KN, Veldman GM, Widom A, Wright JF, Wudyka S, Zhao L, Wolfman NM. Inhibition of myostatin in adult mice increases skeletal muscle mass and strength. Biochem. Biophys. Res. Commun. 2003;300:965–971. doi: 10.1016/s0006-291x(02)02953-4. [DOI] [PubMed] [Google Scholar]
- 212.Siriett V, Salerno MS, Berry C, Nicholas G, Bower R, Kambadur R, Sharma M. Antagonism of myostatin enhances muscle regeneration during sarcopenia. Mol. Ther. 2007;15:1463–1470. doi: 10.1038/sj.mt.6300182. [DOI] [PubMed] [Google Scholar]
- 213.Carlson ME, Hsu M, Conboy IM. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 2008;454:528–532. doi: 10.1038/nature07034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cohn RD, van Erp C, Habashi JP, Soleimani AA, Klein EC, Lisi MT, Gamradt M, ap Rhys CM, Holm TM, Loeys BL, Ramirez F, Judge DP, Ward CW, Dietz HC. Angiotensin II type 1 receptor blockade attenuates TGF-beta-induced failure of muscle regeneration in multiple myopathic states. Nat. Med. 2007;13:204–210. doi: 10.1038/nm1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ng CM, Cheng A, Myers LA, Martinez-Murillo F, Jie C, Bedja D, Gabrielson KL, Hausladen JM, Mecham RP, Judge DP, Dietz HC. TGF-beta-dependent pathogenesis of mitral valve prolapse in a mouse model of Marfan syndrome. J. Clin. Invest. 2004;114:1586–1592. doi: 10.1172/JCI22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Quinn ZA, Yang CC, Wrana JL, McDermott JC. Smad proteins function as co-modulators for MEF2 transcriptional regulatory proteins. Nucleic Acids Res. 2001;29:732–742. doi: 10.1093/nar/29.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Liu D, Kang JS, Derynck R. TGF-beta-activated Smad3 represses MEF2-dependent transcription in myogenic differentiation. EMBO J. 2004;23:1557–1566. doi: 10.1038/sj.emboj.7600179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Potthoff MJ, Arnold MA, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol. Cell. Biol. 2007;27:8143–8151. doi: 10.1128/MCB.01187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 220.Alvarez-Buylla A, Seri B, Doetsch F. Identification of neural stem cells in the adult vertebrate brain. Brain Res. Bull. 2002;57:751–758. doi: 10.1016/s0361-9230(01)00770-5. [DOI] [PubMed] [Google Scholar]
- 221.Bull ND, Bartlett PF. The adult mouse hippocampal progenitor is neurogenic but not a stem cell. J. Neurosci. 2005;25:10815–10821. doi: 10.1523/JNEUROSCI.3249-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002;36:1021–1034. doi: 10.1016/s0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 223.Marshall GP, 2nd, Laywell ED, Zheng T, Steindler DA, Scott EW. In vitro-derived “neural stem cells” function as neural progenitors without the capacity for self-renewal. Stem Cells. 2006;24:731–738. doi: 10.1634/stemcells.2005-0245. [DOI] [PubMed] [Google Scholar]
- 224.Falk S, Wurdak H, Ittner LM, Ille F, Sumara G, Schmid MT, Draganova K, Lang KS, Paratore C, Leveen P, Suter U, Karlsson S, Born W, Ricci R, Gotz M, L Sommer. Brain area-specific effect of TGF-beta signaling on Wnt-dependent neural stem cell expansion. Cell Stem Cell. 2008;2:472–483. doi: 10.1016/j.stem.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 225.Chen HL, Panchision DM. Concise review: bone morphogenetic protein pleiotropism in neural stem cells and their derivatives—alternative pathways, convergent signals. Stem Cells. 2007;25:63–68. doi: 10.1634/stemcells.2006-0339. [DOI] [PubMed] [Google Scholar]
- 226.Panchision DM, Pickel JM, Studer L, Lee SH, Turner PA, Hazel TG, McKay RD. Sequential actions of BMP receptors control neural precursor cell production and fate. Genes Dev. 2001;15:2094–2110. doi: 10.1101/gad.894701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Gross RE, Mehler MF, Mabie PC, Zang Z, Santschi L, Kessler JA. Bone morphogenetic proteins promote astroglial lineage commitment by mammalian subventricular zone progenitor cells. Neuron. 1996;17:595–606. doi: 10.1016/s0896-6273(00)80193-2. [DOI] [PubMed] [Google Scholar]
- 228.Li W, Cogswell CA, LoTurco JJ. Neuronal differentiation of precursors in the neocortical ventricular zone is triggered by BMP. J. Neurosci. 1998;18:8853–8862. doi: 10.1523/JNEUROSCI.18-21-08853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Alvarez-Buylla A, Lim DA. For the long run: maintaining germinal niches in the adult brain. Neuron. 2004;41:683–686. doi: 10.1016/s0896-6273(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 230.Doetsch F, Caille I, DA Lim, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 231.Lim DA, Tramontin AD, Trevejo JM, Herrera DG, Garcia-Verdugo JM, Alvarez-Buylla A. Noggin antagonizes BMP signaling to create a niche for adult neurogenesis. Neuron. 2000;28:713–726. doi: 10.1016/s0896-6273(00)00148-3. [DOI] [PubMed] [Google Scholar]
- 232.Challen GA, Boles N, Lin KK, Goodell MA. Mouse hematopoietic stem cell identification and analysis. Cytometry A. 2009;75:14–24. doi: 10.1002/cyto.a.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Szilvassy SJ, Humphries RK, Lansdorp PM, Eaves AC, Eaves CJ. Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proc. Natl. Acad. Sci. U. S. A. 1990;87:8736–8740. doi: 10.1073/pnas.87.22.8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Spangrude GJ, Brooks DM, Tumas DB. Long-term repopulation of irradiated mice with limiting numbers of purified hematopoietic stem cells: in vivo expansion of stem cell phenotype but not function. Blood. 1995;85:1006–1016. [PubMed] [Google Scholar]
- 235.Dykstra B, Kent D, Bowie M, McCaffrey L, Hamilton M, Lyons K, Lee SJ, Brinkman R, Eaves C. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 236.Muller-Sieburg CE, Cho RH, Thoman M, Adkins B, Sieburg HB. Deterministic regulation of hematopoietic stem cell self-renewal and differentiation. Blood. 2002;100:1302–1309. [PubMed] [Google Scholar]
- 237.Muller-Sieburg CE, Cho RH, Karlsson L, Huang JF, Sieburg HB. Myeloid-biased hematopoietic stem cells have extensive self-renewal capacity but generate diminished lymphoid progeny with impaired IL-7 responsiveness. Blood. 2004;103:4111–4118. doi: 10.1182/blood-2003-10-3448. [DOI] [PubMed] [Google Scholar]
- 238.Sieburg HB, Cho RH, Dykstra B, Uchida N, Eaves CJ, Muller-Sieburg CE. The hematopoietic stem compartment consists of a limited number of discrete stem cell subsets. Blood. 2006;107:2311–2316. doi: 10.1182/blood-2005-07-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Yamazaki S, Iwama A, Takayanagi S, Eto K, Ema H, Nakauchi H. TGF-beta as a candidate bone marrow niche signal to induce hematopoietic stem cell hibernation. Blood. 2009;113:1250–1256. doi: 10.1182/blood-2008-04-146480. [DOI] [PubMed] [Google Scholar]
- 240.Yamazaki S, Ema H, Karlsson G, Yamaguchi T, Miyoshi H, Shioda S, Taketo MM, Karlsson S, Iwama A, Nakauchi H. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell. 2011;147:1146–1158. doi: 10.1016/j.cell.2011.09.053. [DOI] [PubMed] [Google Scholar]
- 241.Larsson J, Blank U, Helgadottir H, Bjornsson JM, Ehinger M, Goumans MJ, Fan X, Leveen P, Karlsson S. TGF-beta signaling-deficient hematopoietic stem cells have normal self-renewal and regenerative ability in vivo despite increased proliferative capacity in vitro. Blood. 2003;102:3129–3135. doi: 10.1182/blood-2003-04-1300. [DOI] [PubMed] [Google Scholar]
- 242.Larsson J, Blank U, Klintman J, Magnusson M, Karlsson S. Quiescence of hematopoietic stem cells and maintenance of the stem cell pool is not dependent on TGF-beta signaling in vivo. Exp. Hematol. 2005;33:592–596. doi: 10.1016/j.exphem.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 243.Kale VP, Vaidya AA. Molecular mechanisms behind the dose-dependent differential activation of MAPK pathways induced by transforming growth factor-beta1 in hematopoietic cells. Stem Cells Dev. 2004;13:536–547. doi: 10.1089/scd.2004.13.536. [DOI] [PubMed] [Google Scholar]
- 244.Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 2010;6:265–278. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr. Opin. Biotechnol. 2007;18:460–466. doi: 10.1016/j.copbio.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 246.Scheel C, Weinber RA. Phenotypic plasticity and epithelial–mesenchymal transitions in cancer and normal stem cells? Int. J. Cancer. 2011;129:2310–2314. doi: 10.1002/ijc.26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 248.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial–mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Buijs JT, van der Horst G, van den Hoogen C, Cheung H, de Rooij B, Kroon J, Petersen M, van Overveld PG, Pelger RC, van der Pluijm G. The BMP2/7 heterodimer inhibits the human breast cancer stem cell subpopulation and bone metastases formation. Oncogene. 2012;31:2164–2174. doi: 10.1038/onc.2011.400. [DOI] [PubMed] [Google Scholar]
- 252.Tang B, Yoo N, Vu M, Mamura M, Nam JS, Ooshima A, Du Z, Desprez PY, Anver MR, Michalowska AM, Shih J, Parks WT, Wakefield LM. Transforming growth factor-beta can suppress tumorigenesis through effects on the putative cancer stem or early progenitor cell and committed progeny in a breast cancer xenograft model. Cancer Res. 2007;67:8643–8652. doi: 10.1158/0008-5472.CAN-07-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 254.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 255.Lonardo E, Hermann PC, Mueller MT, Huber S, Balic A, Miranda-Lorenzo I, Zagorac S, Alcala S, Rodriguez-Arabaolaza I, Ramirez JC, Torres-Ruiz R, Garcia E, Hidalgo M, Cebrian DA, Heuchel R, Lohr M, Berger F, Bartenstein P, Aicher A, Heeschen C. Nodal/Activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell. 2011;9:433–446. doi: 10.1016/j.stem.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 256.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 257.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 258.Lombardo Y, Scopelliti A, Cammareri P, Todaro M, Iovino F, Ricci-Vitiani L, Gulotta G, Dieli F, de Maria R, Stassi G. Bone morphogenetic protein 4 induces differentiation of colorectal cancer stem cells and increases their response to chemotherapy in mice. Gastroenterology. 2011;140:297–309. doi: 10.1053/j.gastro.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 259.Strizzi L, Hardy KM, Kirsammer GT, Gerami P, Hendrix MJ. Embryonic signaling in melanoma: potential for diagnosis and therapy. Lab. Invest. 2011;91:819–824. doi: 10.1038/labinvest.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 261.Monzani E, Facchetti F, Galmozzi E, Corsini E, Benetti A, Cavazzin C, Gritti A, Piccinini A, Porro D, Santinami M, Invernici G, Parati E, Alessandri G, La Porta CA. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur. J. Cancer. 2007;43:935–946. doi: 10.1016/j.ejca.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 262.Quintana E, Shackleton M, Foster HR, Fullen DR, Sabel MS, Johnson TM, Morrison SJ. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 2010;18:510–523. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res. 2010;23:746–759. doi: 10.1111/j.1755-148X.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- 265.Topczewska JM, Postovit LM, Margaryan NV, Sam A, Hess AR, Wheaton WW, Nickoloff BJ, Topczewski J, Hendrix MJ. Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nat. Med. 2006;12:925–932. doi: 10.1038/nm1448. [DOI] [PubMed] [Google Scholar]
- 266.Postovit LM, Margaryan NV, Seftor EA, Kirschmann DA, Lipavsky A, Wheaton WW, Abbott DE, Seftor RE, Hendrix MJ. Human embryonic stem cell microenvironment suppresses the tumorigenic phenotype of aggressive cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4329–4334. doi: 10.1073/pnas.0800467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Strizzi L, Hardy KM, Seftor EA, Costa FF, Kirschmann DA, Seftor RE, Postovit LM, Hendrix MJ. Development and cancer: at the crossroads of Nodal and Notch signaling. Cancer Res. 2009;69:7131–7134. doi: 10.1158/0008-5472.CAN-09-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Schober M, Fuchs E. Tumor-initiating stem cells of squamous cell carcinomas and their control by TGF-beta and integrin/focal adhesion kinase (FAK) signaling. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10544–10549. doi: 10.1073/pnas.1107807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Ehata S, Johansson E, Katayama R, Koike S, Watanabe A, Hoshino Y, Katsuno Y, Komuro A, Koinuma D, Kano MR, Yashiro M, Hirakawa K, Aburatani H, Fujita N, Miyazono K. Transforming growth factor-beta decreases the cancer-initiating cell population within diffuse-type gastric carcinoma cells. Oncogene. 2011;30:1693–1705. doi: 10.1038/onc.2010.546. [DOI] [PubMed] [Google Scholar]
- 270.Katsuno Y, Ehata S, Yashiro M, Yanagihara K, Hirakawa K, Miyazono K. Coordinated expression of REG4 and aldehyde dehydrogenase 1 regulating tumourigenic capacity of diffuse-type gastric carcinoma-initiating cells is inhibited by TGF-beta. J. Pathol. 2012 doi: 10.1002/path.4020. http://dx.doi.org/10.1002/path.4020. [DOI] [PubMed] [Google Scholar]
- 271.Cao L, Shao M, Schilder J, Guise T, Mohammad KS, Matei D. Tissue transglutaminase links TGF-beta, epithelial to mesenchymal transition and a stem cell phenotype in ovarian cancer. Oncogene. 2012;31:2521–2534. doi: 10.1038/onc.2011.429. [DOI] [PubMed] [Google Scholar]
- 272.Kobayashi A, Okuda H, Xing F, Pandey PR, Watabe M, Hirota S, Pai SK, Liu W, Fukuda K, Chambers C, Wilber A, Watabe K. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J. Exp. Med. 2011;208:2641–2655. doi: 10.1084/jem.20110840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 274.Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 275.Lee J, Son MJ, Woolard K, Donin NM, Li A, Cheng CH, Kotliarova S, Kotliarov Y, Walling J, Ahn S, Kim M, Totonchy M, Cusack T, Ene C, Ma H, Su Q, Zenklusen JC, Zhang W, Maric D, Fine HA. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Penuelas S, Anido J, Prieto-Sanchez RM, Folch G, Barba I, Cuartas I, Garcia-Dorado D, Poca MA, Sahuquillo J, Baselga J, Seoane J. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–327. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 277.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 278.Zhang M, Kleber S, Rohrich M, Timke C, Han N, Tuettenberg J, Martin-Villalba A, Debus J, Peschke P, Wirkner U, Lahn M, Huber PE. Blockade of TGF-beta signaling by the TGFbetaR-I kinase inhibitor LY2109761 enhances radiation response and prolongs survival in glioblastoma. Cancer Res. 2011;71:7155–7167. doi: 10.1158/0008-5472.CAN-11-1212. [DOI] [PubMed] [Google Scholar]
- 279.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 280.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat. Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 281.Krause DS, Lazarides K, von Andrian UH, Van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat. Med. 2006;12:1175–1180. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- 282.Naka K, Hoshii T, Muraguchi T, Tadokoro Y, Ooshio T, Kondo Y, Nakao S, Motoyama N, Hirao A. TGF-beta–FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 2010;463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- 283.Quere R, Karlsson G, Hertwig F, Rissler M, Lindqvist B, Fioretos T, Vandenberghe P, Slovak ML, Cammenga J, Karlsson S. Smad4 binds Hoxa9 in the cytoplasm and protects primitive hematopoietic cells against nuclear activation by Hoxa9 and leukemia transformation. Blood. 2011;117:5918–5930. doi: 10.1182/blood-2010-08-301879. [DOI] [PubMed] [Google Scholar]
- 284.Guo X, Oshima H, Kitmura T, Taketo MM, Oshima M. Stromal fibroblasts activated by tumor cells promote angiogenesis in mouse gastric cancer. J. Biol. Chem. 2008;283:19864–19871. doi: 10.1074/jbc.M800798200. [DOI] [PubMed] [Google Scholar]
- 285.Zeisberg EM, Potenta S, Xie L, Zeisberg M, Kalluri R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007;67:10123–10128. doi: 10.1158/0008-5472.CAN-07-3127. [DOI] [PubMed] [Google Scholar]
- 286.Quante M, Tu SP, Tomita H, Gonda T, Wang SS, Takashi S, Baik GH, Shibata W, Diprete B, Betz KS, Friedman R, Varro A, Tycko B, Wang TC. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.McLean K, Gong Y, Choi Y, Deng N, Yang K, Bai S, Cabrera L, Keller E, McCauley L, Cho KR, Buckanovich RJ. Human ovarian carcinoma-associated mesenchymal stem cells regulate cancer stem cells and tumorigenesis via altered BMP production. J. Clin. Invest. 2011;121:3206–3219. doi: 10.1172/JCI45273. [DOI] [PMC free article] [PubMed] [Google Scholar]