Summary
The transcriptional activation of one out of ~2800 olfactory receptor (OR) alleles is a poorly understood process. Here, we identify a plethora of putative OR enhancers and study their in vivo activity in olfactory neurons. Distinguished by an unusual epigenetic signature, candidate OR enhancers are characterized by extensive interchromosomal interactions associated with OR transcription and share the similar pattern of transcription factor footprints. In particular, we establish the role of the transcription factor Bptf as a facilitator of both enhancer interactions and OR transcription. Our observations agree with the model whereby OR transcription occurs in the context of multiple interacting enhancers. Disruption of these interchromosomal interactions results in weak and multigenic OR expression, suggesting that the rare coincidence of numerous enhancers over a stochastically chosen OR may account for the singularity and robustness in OR transcription.
Introduction
The olfactory system has the ability to detect and distinguish among an astounding number of olfactory stimuli (Bushdid et al., 2014). This vast receptive field is afforded by the large repertoire of olfactory receptors (OR), which, in most mammals, are encoded by more than a thousand genes located in numerous genomic clusters throughout the genome (Buck and Axel, 1991; Sullivan et al., 1996; Zhang et al., 2004). ORs are expressed in olfactory sensory neurons (OSNs) in a monogenic, monoallelic, and seemingly stochastic fashion (Chess et al., 1994), in such a way that each neuron expresses only one out of the ~2800 available alleles. For each OSN, the identity of the expressed OR determines the spectrum of chemicals that it responds to and its connectivity to the brain (Wang et al., 1998). The dual role of ORs in odor detection and axon guidance makes the singularity of their expression critical for olfactory perception; were multiple ORs coexpressed in each OSN, the topographic map of OSN projections to the olfactory bulb would be perturbed, likely resulting in reduced olfactory sensitivity and resolution.
The continuous transcription of a single OR is maintained by an OR-elicited feedback signal that stabilizes the expression of the chosen OR and prevents the activation of additional ones (Ferreira et al., 2014; Lewcock and Reed, 2004; Serizawa et al., 2003; Shykind et al., 2004). In mammals, this feedback uses components of the unfolded protein response (UPR) to detect the newly translated OR in the endoplasmic reticulum and to induce transient translation of transcription factor Atf5 (Dalton et al., 2013). Atf5 orchestrates, among others, the expression of Adcy3, the major adenylyl cyclase in the OSNs that is necessary for stable OR transcription and OSN differentiation. Adcy3 expression makes OR choice permanent by signaling for the downregulation of Lsd1, a lysine demethylase with dual co-activator and co-repressor activities that regulate OR expression (Lyons et al., 2013). This feedback system is only possible because OR silencing, via the hallmarks of constitutive heterochromatin, occurs early, during OSN differentiation and before the onset of OR transcription (Magklara et al., 2011). This epigenetic silencing is reinforced by the nuclear convergence of OR loci into a few, OR-specific heterochromatic foci (Clowney et al., 2012). Active OR alleles escape these foci, supporting a role of the spatial compartmentalization between active and inactive OR alleles to the singular OR expression (Armelin-Correa et al., 2014). Indeed, disrupting the nuclear architecture of OSNs violates the “one receptor per neuron” rule, causing co-expression of multiple OR alleles per neuron, albeit at reduced levels (Clowney et al., 2012).
These observations provide the molecular underpinnings of the feedback signal and emphasize the importance of gene silencing in OR gene regulation, however, they do not explain how a single OR allele is selected for transcriptional activation at the beginning of this process. We previously hypothesized that an intergenic OR enhancer, H, could provide this singularity because it frequently associates with transcriptionally active OR alleles from the same or different chromosomes (Lomvardas et al., 2006). However, deletion of the H enhancer affects only the expression of three linked and proximal ORs (Khan et al., 2011). Redundancy for the function of H as a trans enhancer, provided by additional H-like elements, could explain why the physical association with H appears to be genetically superfluous for the transcription of most ORs (Williams et al., 2010). This model, which predicts an intricate network of genomic interactions (Bargmann, 2006), ascribes two distinct roles to each intergenic OR enhancer: a critical function as a cis regulatory element, which may open up the local chromatin architecture, orchestrating the first step of OR choice, and a redundant function as a trans enhancer which, together with other enhancers, facilitates high rates of OR transcription.
To identify elements that may provide redundancy for H as a trans enhancer, we performed a genome-wide search for intergenic OR enhancers. DNase I hypersensitivity (DHS)-seq and ChIP-seq experiments uncovered 35 predicted enhancer elements linked to OR gene clusters. Reporter assays in zebrafish embryos, followed by transgenic and knock out experiments in the mouse, support the function of at least 12 elements as OSN specific enhancers that likely regulate OR transcription. Importantly, circularized chromosome conformation capture sequencing (4C-seq) analyses of sorted OSNs, combined with two- and three-color DNA fluorescent in situ hybridization (FISH) experiments, demonstrate the convergence of multiple enhancers over the chosen OR. Hi-C analysis with chromatin from the whole MOE, along with DNA FISH experiments, revealed extensive inter- and intra-chromosomal interactions between most of these elements in olfactory neurons. Finally, two independent genetic manipulations that disrupt robust OR expression, ectopic expression of Lbr in mature OSNs and conditional deletion of Bptf, a transcription factor predicted to bind to these enhancers by in vivo footprinting experiments, result in significant decrease of trans enhancer interactions accompanying the significant reduction of OR transcription. Our experiments, which reveal the regulatory landscape of OR enhancers, propose a model of singular OR choice that depends upon the convergence of multiple enhancers in a three-dimensional nucleoprotein complex that regulates robust OR expression.
Results
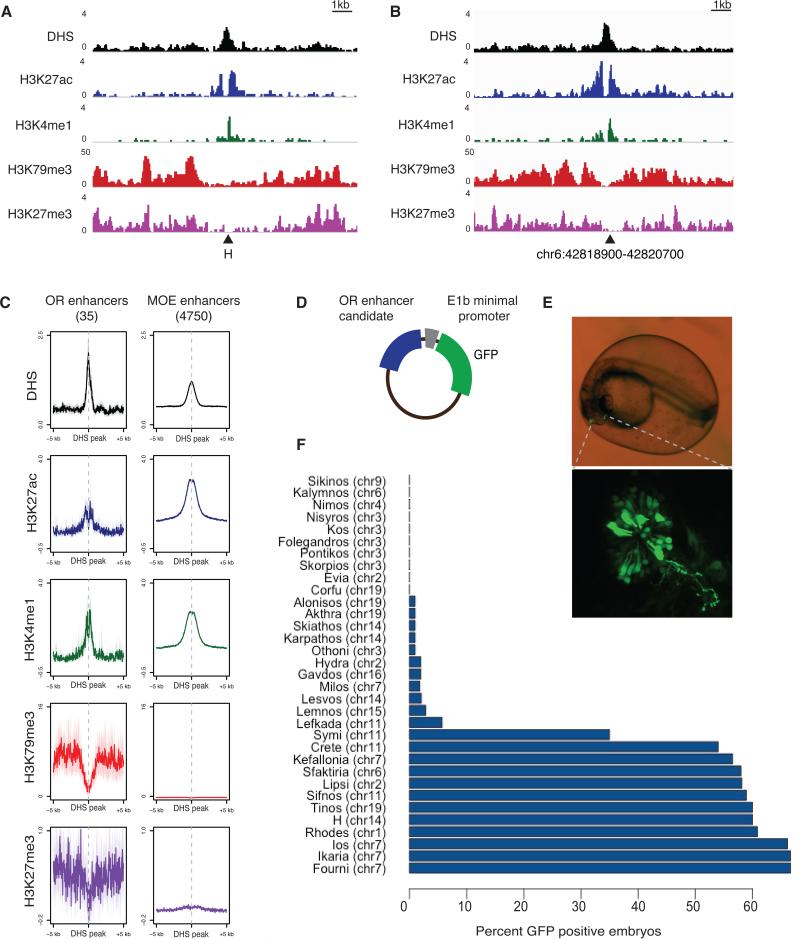
To examine the epigenetic state of the H enhancer and to use this information for the identification of novel OR enhancers, we performed DHS-seq (Thurman et al., 2012) and ChIP-seq for H3K4me1 and H3K27ac (Table S1). DHS and enhancer mark enrichment is observed at enhancers of OSN-transcribed genes, such as the protocadherin alpha enhancers (Ribich et al., 2006) (Figure S1A). The H enhancer sequence is also enriched for H3K4me1, H3K27ac, and has a well-defined DHS peak (Figure 1A), suggesting that H is transcriptionally engaged in more cells than the small fraction of neurons that express the proximal OR genes, on which we do not detect activating histone marks in whole MOE preparations. The second known OR enhancer, the P element (Khan et al., 2011), also has the same features as H.
Figure 1. An epigenetic signature for putative OR enhancers.
A. Sequencing tracks for DHS-seq and ChIP-seq. Each row displays the number of reads for each track. Triangle indicates H enhancer.
B. DHS-seq and ChIP-seq tracks over potential OR enhancer Sfaktiria (coordinates are mm9).
C. Aggregate plots of ChIP-seq and DHS-seq reads over all potential OR enhancers (left) and all potential MOE enhancers (right). Y axis is RPKM, error is bootstrapped 95% confidence intervals. X axis is centered at DHS peaks.
D. Schematic depicting the E1b-tol2 expression construct.
E. Zebrafish embryo injected with Sfaktiria-GFP at 24 hours post fertilization (hpf). Olfactory epithelium is indicated with dotted lines. Below, OSN cell bodies and axons expressing GFP.
F. Results of zebrafish enhancer screen. Percent of injected zebrafish embryos with GFP positive olfactory neurons at 48 hpf for each OR enhancer candidate.
To identify additional elements that share the chromatin pattern of the H enhancer, we performed a computational search for intergenic ChIP-seq and DHS-seq peaks using SICER (Zang et al., 2009). To remove enhancers that might generally specify neuronal cell types, we filtered out regions that overlapped with H3K4me1 and H3K27ac peaks from cerebellum ChIP-seq experiments that we performed in parallel (Figure S1B). However, even upon this filtering, there are 4750 intergenic sequences that have these characteristics in a MOE-specific fashion. To reduce this number we restricted the search to only intergenic regions residing within OR gene clusters (and 100Kb upstream and downstream of these clusters). This strategy reduced the number of positive hits to 35, with an average distance of ~35Kb from the nearest OR (Figure S1B, Table S2).
We also searched for other epigenetic marks that may be used to distinguish OR enhancers from the rest of the MOE-specific regulatory elements. Two histone modifications associated with repression, H3K79me3 and H3K27me3 (Barski et al., 2007; Ernst et al., 2011), have a unique distribution at the H locus - they are missing from the actual enhancer sequence but are enriched in the flanking sequences (Figure 1A). Visual inspection of the remaining OR enhancer candidates shows that this pattern is shared among 23 of the 35 potential enhancer elements (example in Figure 1B). Aggregate plots comparing ChIP-seq and DHS-seq reads on OR proximal elements and predicted MOE enhancers located outside OR clusters highlight the specificity of this epigenetic signature (Figure 1C). Indeed, while 65% of predicted OR enhancers overlap with regions of H3K79me3 enrichment, only 2.6% (126/4750) of all the predicted MOE enhancers show this overlap (Figure S1B). Interestingly, in drosophila embryos H3K79me3 enrichment is found on developmentally regulated enhancers (Bonn et al., 2012)
Functional analysis of predicted OR enhancers
We sought a functional assay that is appropriate for a high throughput in vivo enhancer screen. We performed transient reporter assays in zebrafish embryos and scored for MOE-specific reporter expression as previously described (Booker et al., 2013). Although the predicted OR enhancer sequences are not conserved to zebrafish, the H element supports reporter expression in zebrafish OSNs (Nishizumi et al., 2007), suggesting that this assay is appropriate for the functional identification of OR enhancers. Inspired by a previous description of enhancers as “islands” composing a “regulatory archipelago” (Montavon et al., 2011), we named the potential OR enhancers after Greek islands, a nomenclature that will be followed throughout the manuscript.We cloned the DHS peaks of 32 islands into a Tol2 retrotransposon-based reporter vector with a minimal promoter and GFP (Fig. 1D, Table S3B) (Li et al., 2010). Each construct was injected into one-cell stage zebrafish oocytes and GFP expression was monitored during embryogenesis at 24 and 48 hours post fertilization. Specific GFP expression was observed in OSNs of the olfactory epithelium for 12 of the sequences screened (Figure 1E,F, S1C,D). Empty vector controls do not support GFP expression in zebrafish OSNs.
To identify potential differences between active and inactive enhancers we summarized the levels of histone modifications around DHS peaks using principal component analysis. The first principle component (PC1) of each modification robustly represents its overall level at each enhancer locus. Ordering candidate OR enhancers by H3K79me3 PC1 grouped together enhancers that were active in zebrafish OSNs, as well as the H and P elements (Figure S1E). This set of enhancers had high levels of flanking H3K79me3. Ordering by H3K4me1 PC1 shows that enhancers that were active in zebrafish OSNs had relatively lower levels of H3K4me1 (Figure S1F). Interestingly, OR genes proximal to these 11 elements are more highly expressed than the average OR by RNA-seq of mouse OSNs (Figure S2A).
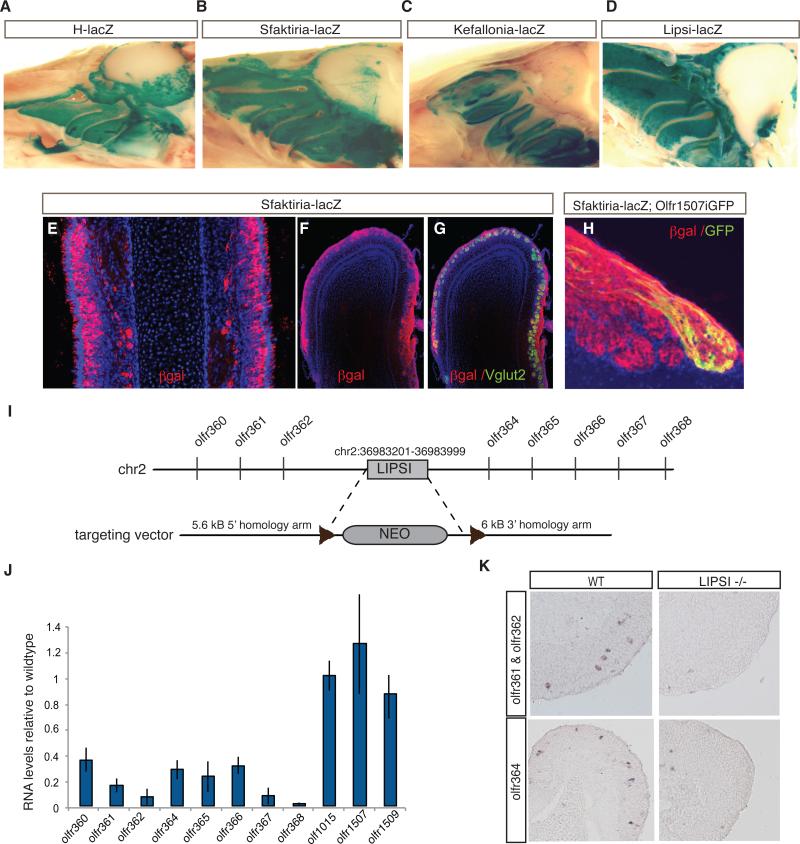
To further validate that the zebrafish reporter assay we generated transgenic beta galactosidase (βgal) reporter lines using a reporter vector driven by the hsp68 minimal promoter (Kothary et al., 1988). We tested three zebrafish-positive elements, Sfaktiria, Lipsi, and Kefallonia (Table S2), referred to as Sfaktiria-lacZ, Lipsi-lacZ and Kefallonia-lacZ in the rest of the manuscript. Whole mount x-gal staining of these transgenic mice shows widespread reporter expression specifically in the MOE similar to the H-lacZ transgenic, which we generated as positive control (Figure 2A-D, Figure S2B). Immunofluorescence (IF) for βgal in the MOE of the Sfaktiria-lacZ mouse also shows widespread expression (Figure 2E). In the olfactory bulb, βgal positive axons target multiple glomeruli and express the glutamate transporter Vglut2 (Figure 2F,G), indicating that this enhancer drives expression in mature OSNs. In contrast, there is no βgal IF signal in Neurogenin-1 positive neurons (Figure S2C), which suggests that enhancer activity is synchronous to OR expression. Similar results were obtained from Lipsi-lacZ and Kefallonia-lacZ transgenics (data not shown). In contrast βgal is coexpressed with olfactory receptor Olfr1507 in Sfaktiria-lacZ transgenics crossed to Olfr1507iresGFP knock-in mice (Barnea et al., 2004) (Figure 2H).
Figure 2. Genetic verification of enhancer function in mouse OSNs.
A-D. Whole mount x-gal staining of MOE from H-lacZ, Sfaktiria-lacZ, Kefallonia-lacZ, and Lipsi-lacZ enhancer transgenic mice, respectively.
E. IF for βgal (red) in Sfaktiria-lacZ olfactory epithelium. DAPI nuclear stain (blue)
F. IF for βgal (red) in Sfaktiria-lacZ olfactory bulb.
G. IF for βgal (red) and Vglut2 (green) in Sfaktiria-lacZ olfactory bulb.
H. IF for βgal (red) and GFP (green) in Sfaktiria-lacZ ; Olfr1507iresGFP olfactory bulb. DAPI nuclear stain (blue).
I. Targeted deletion of Lipsi allele on chromosome 2. 998 bp were replaced with a floxed neo cassette via homologous recombination. Coordinates are mm9.
J. RT-qPCR from Lipsi KO and Lipsi WT adult MOE. RT-qPCR levels for each primer set were normalized to OMP, and the results are shown as fold difference of Lipsi KO over Lipsi WT. Error bars represent SEM over duplicate experiments.
K. RNA ISH for Olfr361 and Olfr362 pooled probes and Olfr364 in Lipsi KO and WT MOE at P1. See also Figure S2 and Tables S2 and S3.
These transgenic reporter assays demonstrate an OSN-specific enhancer activity for the candidate OR enhancers. To test the requirement of these elements in OR expression we deleted one of them, Lipsi, which is located on chromosome 2 between Ofr362 and Olfr364. Our targeting strategy deleted, by homologous recombination, 1000bp of conserved sequence corresponding to the DHS peak at this location (Figure 2I). qRT-PCR analysis on RNA prepared from wild type and Lipsi KO littermates shows marked reduction in expression of the 8 genomically linked ORs that reside within this genomic cluster on chromosome 2, while ORs from a distant genomic cluster in the same or different chromosomes are unaffected (Figure 2J). RNA in situ hybridization (ISH) analysis in MOE sections from Lipsi KO and wild type littermates confirms that expression of ORs from this genomic cluster is abolished (Figure 2K, Figure S2D)
Multiple enhancers interact in trans with a transcriptionally active OR
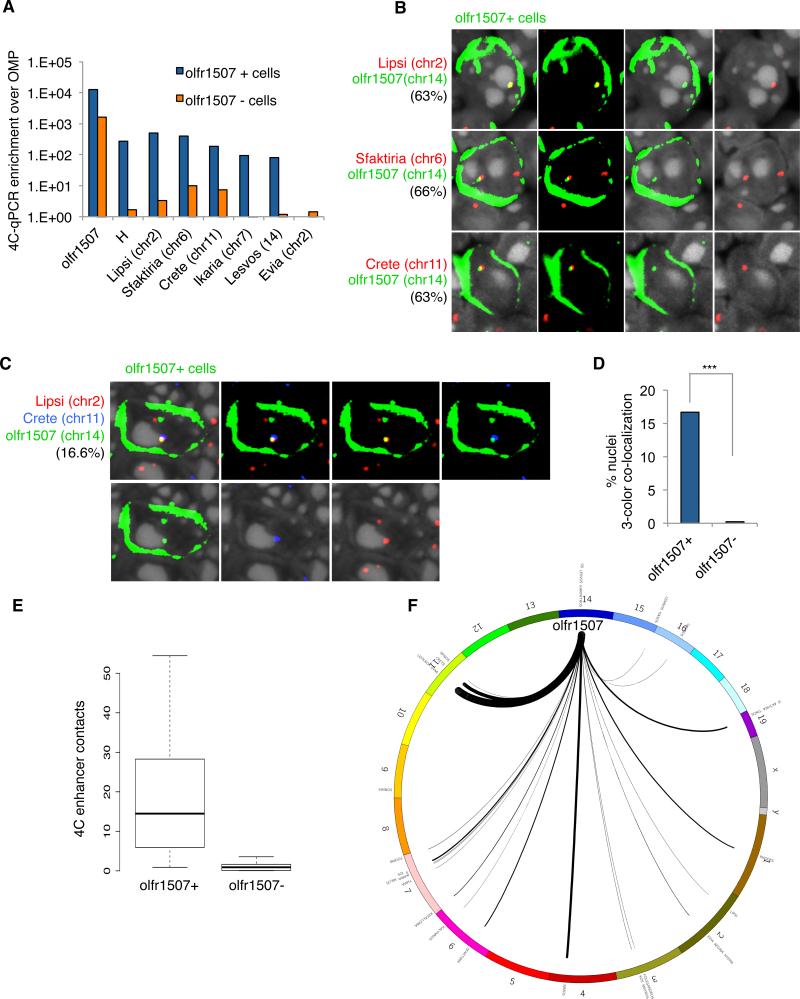
Our data thus far provide a comprehensive epigenetic and genetic characterization of intergenic DNA elements that may act as OR enhancers. To examine whether these elements associate with active OR genes in trans, like the H enhancer (Lomvardas et al., 2006), we performed 4C on an isolated population of OSNs expressing Olfr1507. We chose Olfr1507 for this experiment because its expression depends on the genomically linked H enhancer. Thus, it provides an ideal gene locus for testing the hypothesis that a cis enhancer may act in concert with trans enhancers. 4C was performed on FAC-sorted neurons from Olfr1507iresGFP knock-in mice, and libraries were amplified with inverse PCR primers anchored at the Olfr1507 promoter as previously described (Clowney et al., 2012). 4C libraries from GFP+ and GFP− cells were analyzed by qPCR to quantitate the relative enrichment of various DNA loci. Several of the newly identified sequences are enriched in the library corresponding to GFP+ cells, at levels approaching the enrichment levels of H (Figure 3A). Enrichment is significantly reduced in GFP− cells, which suggests that these associations are restricted to cells that transcribe Olfr1507. Two color DNA FISH analysis (Figure 3B) verified that the Olfr1507 locus frequently co-localizes in trans with the three most highly enriched elements, Lipsi (chr2), Sfaktiria (chr6), and Crete (chr11), in OSNs immunolabeled with an anti-Olfr1507 antibody (~63% of Olfr1507+ OSNs, n=124 for each pair). Similar results were obtained by FISH in Olfr1507iresGFP MOEs using anti-GFP immunolabeling (Figure S3A). Three-color DNA FISH (Figure 3C) revealed that Olfr1507 co-localizes with both Lipsi and Crete in 16% of Olfr1507+ OSNs, a highly significant increase over the frequency of co-localization of the three loci in Olfr1507− OSNs (0.2%, n=406, p=2E-10, chi-square test) (Figure 3D). Thus, as the network of enhancer interactions becomes more complex, it remains associated with OR transcription.
Figure 3. Multiple enhancers interact in trans with a chosen OR.
A. 4C from Olfr1507-ires-GFP FAC-sorted cells, library constructed by inverse PCR off Olfr1507 promoter. qPCR enrichment of candidate OR enhancers relative to OMP in GFP+ and GFP− cells. Error bars display SEM between triplicates.
B. IF staining for Olfr1507 (green) and two-color DNA FISH for Olfr1507 DNA (green) and Lipsi, Sfaktiria, and Crete DNA (red). DAPI is nuclear stain (gray). Percent of Olfr1507 positive cells containing co-localized red and green probes is indicated for each combination.
C. IF staining for Olfr1507 (green) and DNA FISH for Olfr1507 (green), Lipsi (red) and Crete (blue) DNA. DAPI is nuclear stain (gray). Percent of Olfr1507+cells containing three color co-localization is indicated.
D. Quantification of C. Percent Olfr1507+and Olfr1507− OSNs containing three-color co-localizations.
E. 4C-seq from Olfr1507iresGFP positive and negative cells, library constructed by inverse PCR from Olfr1507 promoter. Average number of contacts for OR enhancers are plotted. Y-axis is RPKM that span two different putative enhancers at an expected ligation site.
F. Circos plot (Krzywinski et al., 2009) of OR enhancer chromosomal locations and 4C-seq contacts with Olfr1507 promoter in Olfr1507+ cells. Lines are weighted by frequency of interaction (RPKM that span two different putative enhancers at an expected ligation site). See Table S5 for enhancer names, locations and interaction frequencies.
See also Figure S3
To explore the long-range interactions of Olfr1507 in an unbiased fashion we generated 4C-seq libraries generated from GFP+ and GFP− cells from Olfr1507iresGFP mice (Figure S3B). In agreement with our qPCR analysis, 4C-seq revealed multiple contacts between Olfr1507 and 15 candidate OR enhancers, of which 9 are functional in zebrafish OSNs (Figure S3C-E, Table S5). Interactions with OR enhancers were significantly (p<<0.01, Wilcoxon t-test) stronger in Olfr1507+ OSNs compared to the negative population (Figure 3E). The network of interchromosomal interactions between Olfr1507 and predicted OR enhancers is depicted in Figure 3F.
Extensive cis and trans interactions between potential OR enhancers in OSNs
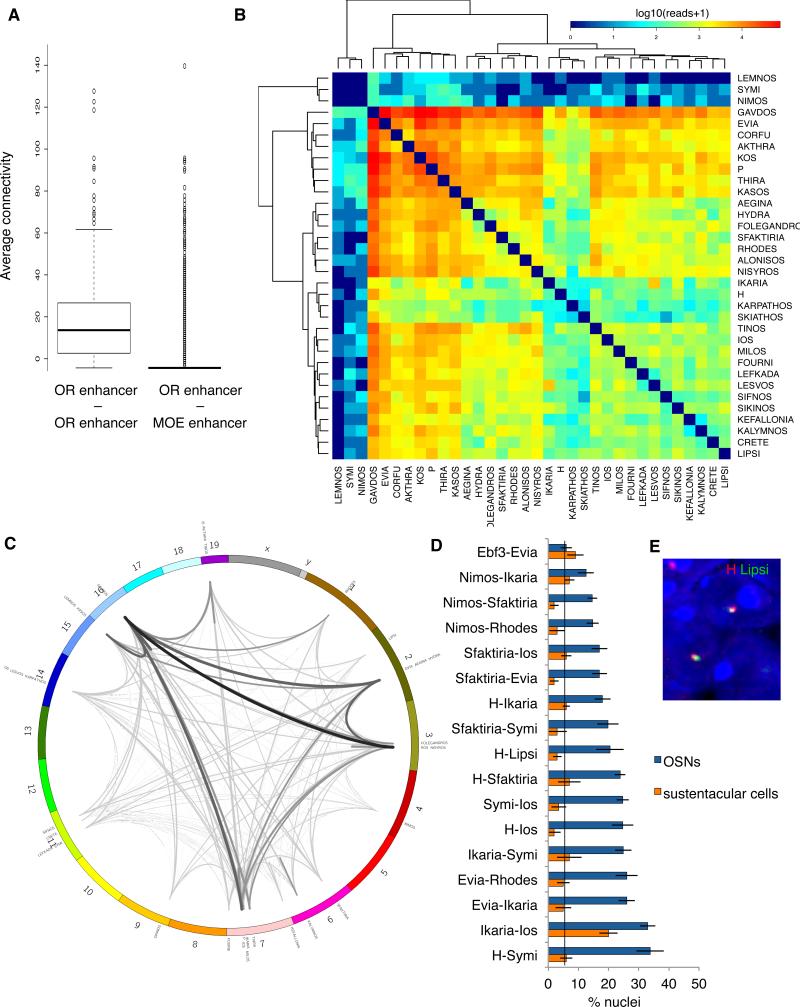
To examine the extent of putative enhancer interactions in OSNs we performed Hi-C analysis in the whole MOE. To specifically interrogate the interactions of OR enhancer candidates, we performed a modified Capture-C protocol (Hughes et al., 2014) (see extended experimental methods). Our analysis revealed that 32/35 elements associate in high frequency with other enhancers from this repertoire. These interactions appear to be highly specific; there is a significant (p<<0.01 Wilcoxon t-test) 20-fold enrichment for reads that span two different potential OR enhancers compared to reads that span an OR enhancer and one of the other MOE enhancers (Figure 4A). Within the observed repertoire there are “promiscuous” enhancers that form frequent interactions with many other elements (Evia, Gavdos), while others form fewer interactions (Nimos, Lemnos). A contact matrix depicting the pairwise frequencies of these interactions organized by hierarchical clustering reveals the existence of four clusters of potential enhancers exhibiting similar frequencies of interactions (Figure 4B). Enhancers located on chromosomes 2,3,7, and 16 make the most frequent contacts with each other and with enhancers from other chromosomes (Figure 4C).
Figure 4. An intricate network of enhancer interactions in mouse OSNs.
A. Average Hi-C connectivity of OR enhancers with other OR enhancers compared to average connectivity of OR enhancers to MOE enhancers. Y-axis is reads spanning two different genomic regions normalized to the total number of reads.
B. Contact matrix depicting interaction frequency between candidate OR enhancers (red highest, blue lowest interaction frequency). Normalized read counts spanning two enhancer regions were divided into 20 bins, with 5 bins representing each color shade (color key provided for quantitation). Interactions between enhancers are hierarchically clustered.
C. Circos plot of OR enhancer chromosomal locations and Hi-C contacts. Lines are weighted according to frequency of enhancer-enhancer interactions.
D. Results of DNA FISH screen. X-axis is percent nuclei containing two-color co-localization between OR enhancer candidates, Y-axis is enhancer candidate pairs tested. OSN and sustentacular cell nuclei indicated. Vertical line is baseline OSN co-localization frequency. Ios and Ikaria were not significantly reduced in sustentacular cells, probably due to the genomic linkage of these DNA elements which both reside on chromosome 7 at 9MB distance. Error bars are SEM from multiple sections of the MOE.
E. Representative DNA FISH for H (red) and Lipsi (green) in OSN nucleus. The blue nuclear stain is DAPI.
See also Figure S4.
To independently verify the Hi-C data, we performed two color DNA FISH analysis on sections of the MOE (Figure 4D,E, S4A). DNA sequences that interact infrequently by Hi-C, like Nimos, exhibit a low frequency of co-localization with other OR enhancers both in OSNs and in sustentacular cells, a nonneuronal cell type of the MOE that we used as an internal control (Figure 4D). In contrast, increased Hi-C interactions generally correspond to frequent co-localizations by DNA FISH that are restricted to OSNs. On average, enhancer-enhancer co-localizations are ~6 times more prevalent in OSNs than in sustentacular cells (n=3,264 nuclei, p=10−44 chi-square test). As in the Hi-C experiment, enhancerenhancer interactions are specific to OR enhancers, as co-localizations with predicted MOE enhancers are not significantly increased in OSNs compared to sustentacular cells (Figure 4D). It is worth noting that sequences engaged in interchromosomal contacts include those that were not functional in the zebrafish reporter assay, suggesting that they may have functional regulatory roles in the mouse.
The regulatory landscape of OR enhancers
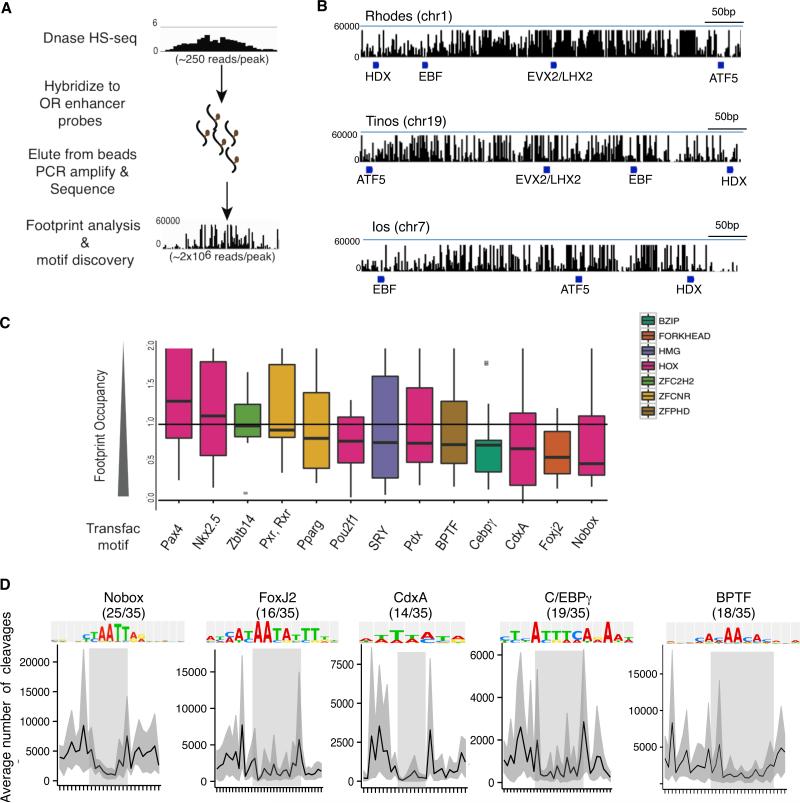
Because enhancer-bound transcription factors mediate long-range genomic interactions (Nolis et al., 2009), we searched for proteins that bind on candidate OR enhancers in order to obtain mechanistic insight into the formation of these intricate interchromosomal associations. For an unbiased search of regulatory sequences we sought to isolate DNAse I protected sequences on the candidate enhancers. In vivo footprinting by DHS-seq was recently used to identify, in a high throughput fashion, regulatory sequences and factors that control cell-type specific differentiation (Neph et al., 2012a; Neph et al., 2012b). To increase the depth of our DHS-seq reads on the 35 putative enhancers we employed a similar sequence capture strategy as done for Hi-C, using oligos tiling the OR enhancer candidates (Figure 5A), modifying a recently published enrichment strategy (Stergachis et al., 2013).
Figure 5. Shared transcription factor footprints on OR enhancers.
A. Schematic of sequence capture-based method of DNase I library construction.
B. DNase I cleavages mapped over OR enhancer candidates Rhodes, Tinos and Ios. Blue bars indicate footprints containing ATF5, HDX, EVX2/LHX2, and EBF motifs.
C. Transcription factor (TF) motifs from the TRANSFAC database that are enriched on OR enhancer candidates and average footprint score for each TF motif. Low FOS scores indicate greater footprint occupancy. TF motifs are color coded according to TF family.
D. Average DNase I cleavages for TRANSFAC motifs on 35 candidate OR enhancers. The X-axis is centered at the consensus sequence (shaded box). Fraction of OR enhancers containing each TF motif is indicated in parentheses. Error is bootstrapped 95% confidence intervals.
See also Figure S5 and Table S4 for footprint and motif locations.
After enrichment, DHS-seq read coverage of the OR enhancers increased ~10,000 fold (Figure 5A). Mapping DNase I cleavage sites across the H enhancer revealed multiple sequences that are protected from DNase I digestion (Figure S5A) and that can be computationally identified as footprints (Neph et al., 2012). We performed a motif search of footprinted sequences in the 12 enhancers with confirmed zebrafish activity using MEME (Bailey and Elkan, 1994). This analysis revealed four protected sequence motifs, which match predicted binding sites for Atf5, Evx2/Lhx2, Olf/Ebf, and Hdx (Figure 5B, S5B). Interestingly, three of these factors, Atf5, Olf/Ebf, and Lhx2, have reported roles in OR gene regulation (Dalton et al., 2013; Hirota and Mombaerts, 2004; Rothman et al., 2005). Chromatin immunoprecipitation experiments for Lhx2, for which we were able to obtain a ChIP-quality antibody, revealed enrichment on OR enhancers containing Lhx2 motifs (Figure S5C, see extended experimental methods), providing independent verification of the in vivo occupancy data.
Next we expanded our search to the complete repertoire of enhancers identified by epigenetic analysis. Across the 35 potential enhancers, 1040 footprints were identified with significant footprint occupancy scores (FOS<0.5) (Table S4A). Eleven TF motifs from the TRANSFAC database had median footprint scores less than one, indicating DNase I protection (Figure 5C). Homeodomain TF motifs comprise the majority of the footprinted motifs. Four TF motifs (Nobox, Foxj2, Cdx, and C/EBPgamma) have p-values < 0.05 and exhibit protection of the consensus sequence across all OR enhancers (Figure 5D). Among consensus sequences with reduced FOS, the predicted binding motif for transcription factor Bptf (bromo- and PHD-finger transcription factor) (Jordan-Sciutto et al., 1999) appeared as a promising candidate protein that may be involved in the formation or stabilization of long-range genomic interactions. As the histone binding component of the NURF chromatin remodeling complex, Bptf binds acetylated and methylated histone tails from different histones through the bromo and PHD-finger domains, respectively (Ruthenburg et al., 2011), which could potentially mediate the bridging of chromatin fibers from different chromosomes. Bptf has previously been shown to facilitate expression of Hox genes via chromatin remodeling at cis-regulatory sequences (Wysocka et al., 2006), but has yet to be implicated in OR expression.
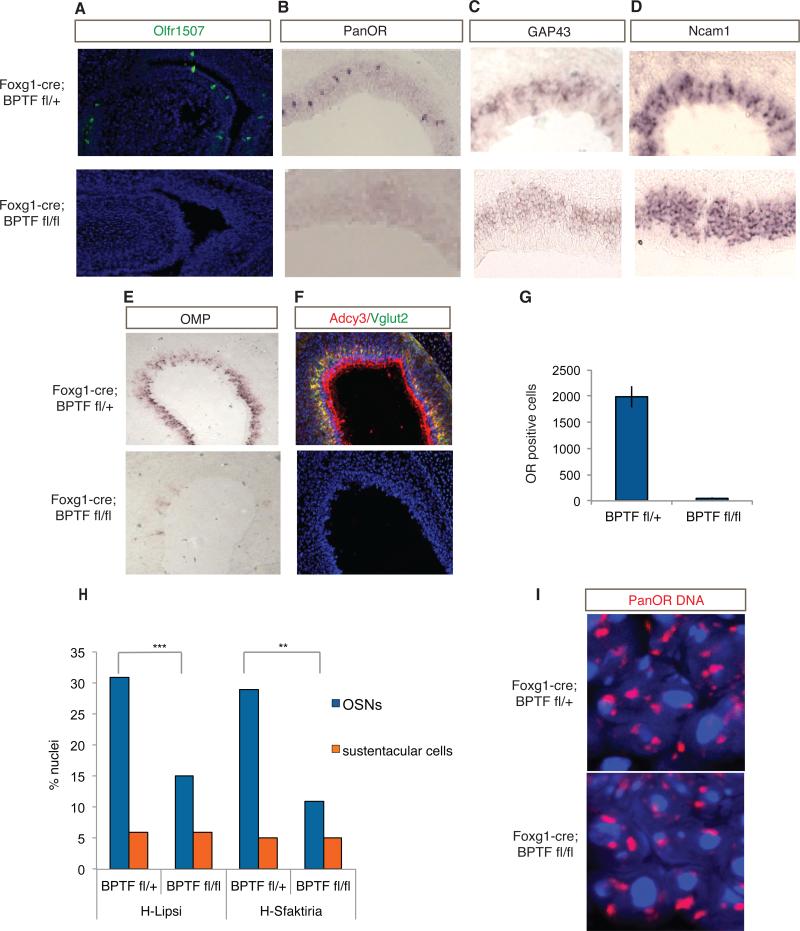
To examine the functional significance of Bptf binding on OR-proximal enhancers we conditionally deleted Bptf in the MOE by crossing a floxed Bptf allele (Landry et al., 2011) to a Foxg1-Cre driver that is expressed before the onset of OR expression (Hebert and McConnell, 2000). We refer to the Foxg1-cre, Bptf flox/flox mouse as the Bptf KO. These mice die perinatally, thus restricting our analysis to E18.5 embryos. IF for Olfr1507 in sections from Bptf KO and control MOEs showed complete loss of expression of this OR in the Bptf KO (Figure 6A). RNA ISH using a complex probe detecting several hundred ORs (see supplemental methods) also showed a dramatic reduction in OR expression (Figure 6B,G). GAP43 and Ncam1, markers of immature OSNs that are synchronous to OR expression (Lyons et al., 2013), are still expressed in Bptf KO MOEs, suggesting that the loss of OR expression is not caused by loss of the OSN lineage (Fig 6 C,D). Finally, a general deficit in OR expression is corroborated by the loss of mature OSN markers, such as Adcy3 and Vglut2, in the Bptf KO MOE (Figure 6E,F).
Fig 6. Bptf is required for OR expression and enhancer interactions.
A. Olfr1507 IF (green) in Foxg1-Cre; Bptf flox/+ (heterozygote) and Foxg1-Cre; Bptf flox/flox (KO) MOE at e18.5. DAPI is the nuclear stain (blue).
B. PanOR RNA ISH in Bptf heterozygote and KO.
C-E. ISH for developmental markers in Bptf het (top) and KO (bottom).
F. Adcy3 IF (red) and Vglut2 IF (green) in Bptf het (top) and KO (bottom). DAPI (blue) is nuclear stain.
G. Quantification of OR ISH experiment (panel B) in Bptf het and KO. Error bars represent the variance over duplicate experiments.
H. Quantification of DNA FISH co-localizations between H and Sfaktiria BAC probes and between H and Lipsi BAC probes in Bptf het and KO. The Y-axis is percent OSN and sustentacular cell nuclei containing co-localized probes.
I. DNA FISH with complex pan olfactory receptor (PanOR) probe (red) in Bptf het and KO OSNs. DAPI is nuclear stain (blue).
To test whether Bptf participates in the establishment or maintenance of associations between potential OR enhancers, we performed two-color DNA FISH in sections of control and Bptf KO MOEs. This analysis revealed a significant decrease in the frequency of interactions between H-Lipsi and HSfaktiria in Bptf KO OSNs (Figure 6H, p=0.0005, p=0.002, respectively, chi-square test n=356 nuclei). Importantly, the overall chromatin architecture of Bptf KO OSNs remains intact and the aggregation of OR foci is not impaired (Figure 6I), suggesting a rather specific role of this protein in enhancer interactions and OR gene activation.
OR gene aggregation facilitates enhancer interactions in trans
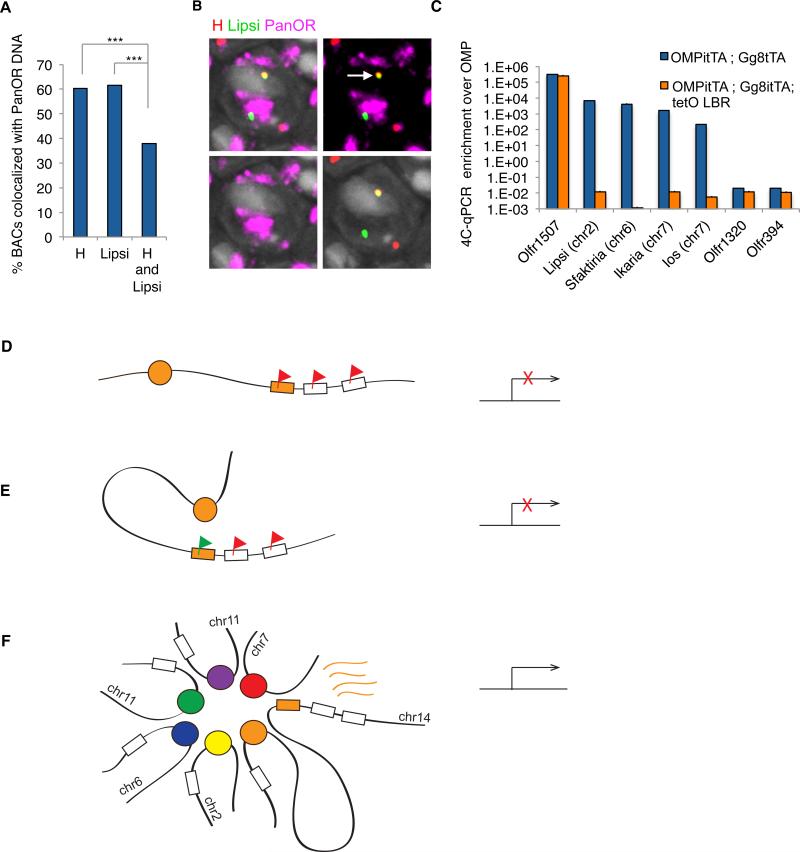
An important question emerging from our analyses is whether frequent enhancer interactions are a consequence of OR gene aggregation in OSN nuclei. The fact that we detect enhancer co-localization with Olfr1507 more frequently in Olfr1507+ cells suggests that these associations would preferentially occur outside the repressive OR foci, which is where the transcriptionally active ORs also reside (Clowney et al., 2012). Two-color DNA FISH shows that 60% of H or Lipsi alleles co-localize with a complex DNA FISH probe that recognizes most OR loci (panOR probe) (Figure 7A). However, three-color DNA FISH shows that H and Lipsi co-localization occurs preferentially outside the OR foci (p<10−12, n=1530, chi-square test), within euchromatic nuclear territories (Figure 7A,B). Thus, enhancer interactions in trans may be favored by- but are not a simple product of- the convergence of linked ORs, a result consistent with the observation that Bptf deletion impairs enhancer interactions but not OR aggregation.
Figure 7. OR gene aggregation promotes enhancer interactions.
A. Percent of OSN nuclei in which individual BAC probes for H and Lipsi are co-localized with PanOR probe (two color DNA FISH), and in which co-localized H and Lipsi BAC probes are co-localized with PanOR probe (three color DNA FISH).
B. Three-color DNA FISH with PanOR probe (magenta), H BAC (red) and Lipsi BAC (green). DAPI is nuclear stain (gray). Arrow indicates co-localized enhancer probes located outside of OR foci.
C. 4C-qPCR analysis in Lbr over-expressing mice (orange bars) and control mice (blue bars). 4C library is generated by inverse PCR from the H enhancer. Enrichment of candidate OR enhancers and OR promoter sequences on different chromosomes are normalized to control gene OMP. Error bars are SEM from duplicates.
D-F. A model for essential and redundant functions of an OR enhancer in cis and in trans respectively. Schematic representations of different states in the OSN nucleus (left) and corresponding transcriptional outputs (right). An OR gene (orange box) located proximal to an enhancer (orange circle) is repressed by H3K9me3 (red flag, D). The cis-proximal enhancer may facilitate de-repression of the OR chromatin landscape (green flag, E) but is not sufficient for OR transcription. Multiple trans-interacting enhancers (colored circles) aggregate around the transcribed cis-proximal OR (orange box, F).
It is possible that the differentiation dependent aggregation of OR loci instead facilitates enhancer interactions by bringing the elements into close proximity. To test this we analyzed the frequency of interchromosomal associations between potential OR enhancers in OSNs that express Lamin b receptor (Lbr). We previously showed that ectopic Lbr expression in OSNs disrupts the aggregation of OR genes and the interaction of the H enhancer with OR genes in trans but not in cis (Clowney et al., 2012). 4C-qPCR analysis of control and Lbr+ FAC-sorted OSNs revealed significant reduction in the frequency of trans interactions between potential OR enhancers upon Lbr expression (Figure 7C), supporting a role for the unusual nuclear architecture of OSNs, and possibly for the aggregation of OR loci, in the frequent association of predicted OR enhancers from different chromosomes. Importantly, since OR transcription is significantly reduced in Lbr+ OSNs (Clowney et al., 2012), these data provide an independent genetic manipulation whereby the specific disruption of trans enhancer interactions reduces OR transcription rates. Thus, the same process that contributes to the effective silencing of OR transcription, the aggregation of OR genes in heterochromatic foci, may also facilitate the activation of a single allele by increasing the probability of enhancer-enhancer interactions.
Discussion
Our experiments revealed 35 intergenic OR-linked sequences that share common epigenetic properties with the H element, the prototypical OR enhancer. In addition to the common enhancer features, these sequences are characterized by high flanking levels of H3K79me3. Currently, it is not clear if these epigenetic marks coexist on the same enhancer alleles or they reflect different states of these elements in the total cellular population. In any case, our reporter screen showed that 12 of the 32 elements that were tested regulate OSN-specific expression in zebrafish OSNs and revealed a positive association between flanking H3K79me3 enrichment and enhancer activity. We demonstrated the activity of three of these elements (Lipsi, Sfaktiria, Kefallonia) as OSN enhancers in the mouse, and we showed that Lipsi is necessary for the expression of proximal OR genes. Because zebrafish reporter assays for mammalian enhancers generate false negatives (Ariza-Cosano et al., 2012; Booker et al., 2013; McGaughey et al., 2008), it is likely that many of the DNA elements that did not activate transcription in zebrafish OSNs are mammalian or mouse-specific OR enhancers, especially in light of their extensive interactions with verified enhancers. It is worth noting that our screen is far from being saturated, since enhancers that are transcriptionally engaged in a smaller cell population would not meet our computational thresholds, and enhancers with a different epigenetic signature would be ignored.
Our experiments revealed an unusual network of genomic interactions occurring predominantly between putative OR-associated enhancers from different chromosomes. Interactions identified by Hi-C were verified by extensive DNA FISH experiments, which demonstrated that sequences from different chromosomes co-localize in up to 35% of OSN nuclei. 4C-seq from FAC-sorted OSNs and three-color DNA FISH experiments suggest that more than one enhancer co-localizes with an OR gene in the neurons that transcribe that OR. Previous work showed that H interacts in cis and in trans with the transcriptionally active OR allele (Lomvardas et al., 2006). Because the inactive Olfr1507 allele is epigenetically similar to other silent ORs in Olfr1507+ OSNs (Magklara et al., 2011) and likely spatially indistinguishable from them (Armelin-Correa et al., 2014), the parsimonious assumption is that multiple OR enhancers also coalesce over the transcriptionally active OR allele.
A question emerging from our observations regards the functional significance of the convergence of multiple enhancer elements over the chosen OR. Recent data revealed that many developmentally regulated genes require numerous enhancers for their proper expression. Experiments in the developing mouse embryo have shown that multiple enhancer sequences act in a coordinated fashion to activate Hox genes in various tissues, and similar observations have been made for the activation of protocadherin gene clusters (Andrey and Duboule, 2014; Delpretti et al., 2013; Guo et al., 2012; Montavon et al., 2011; Noordermeer et al., 2014). Thus, enhancers may act in an additive or even synergistic fashion to increase transcription rates. Given that ORs likely represent the most abundant protein coding mRNAs in OSNs, it is possible that the convergence of multiple enhancers contributes to the robustness of OR transcription. Coordinated action between multiple loci in trans has been also described for the activation of the human IFN β gene, which is also expressed in a stochastic but robust fashion upon virus induction (Apostolou and Thanos, 2008). Finally, genetic experiments suggest that a DNA sequence may act as a trans enhancer during the stochastic photoreceptor gene choice in drosophila ommatidia (Johnston and Desplan, 2014). Thus, while the Hox genes, with restrictive spatiotemporal expression requirements, use an “archipelago” of cis regulatory elements to achieve precise expression patterns, systems that tolerate -or even seek stochasticity- may utilize trans interactions to obtain robust but low probability transcriptional outputs.
Currently, there is no direct evidence of a role of these interactions in OR transcription. Previous work showed that deletion of H does not affect expression of more than 3 proximal OR alleles, despite the physical association of this enhancer with multiple other ORs. Similarly, deletion of P or Lipsi appears to affect the expression of linked OR alleles only, a result also observed in H,P double KO mice (Khan et al., 2011). However, with at least 15 enhancer elements interacting frequently with Olfr1507 promoter in Olfr1507+ OSNs, it is expected that deletion of a single trans enhancer does not elicit transcriptional consequences. In contrast, if each of these elements is required for a critical step in the transcriptional activation of cis-linked OR genes, such as orchestrating the de-silencing of the linked ORs, then each individual deletion may be sufficient for a detectable transcriptional effect in cis (Figure 7D-F), explaining why these enhancers are required in cis but redundant in trans.
To test this model of trans enhancement, we perturbed the ability of these elements to interact with each other in trans. Two independent experiments, deletion of Bptf and expression of Lbr in OSNs, resulted in significant downregulation of OR transcription, to the extent that ORs are not detectable by RNA ISH or IF in the mutant OSNs. Although in both cases we cannot directly attribute the downregulation of OR transcription to the reduced frequency of trans interactions, these results are consistent with our model. Recent experiments showed that an increase in the number of homeodomain and O/E sites on the promoters of transgenic ORs increases the frequency by which they are transcribed (Vassalli et al., 2011). Since the only way to increase the local concentration of binding sites for these transcription factors near an OR promoter is to recruit sites from other genomic regions, this result is consistent with the hypothesis that each enhancer on its own is required but not sufficient for the expression of proximal ORs.
There are alternative interpretations of our data that do not invoke coordinated action between distant OR enhancers. For example, enhancer convergence may reflect the existence of specialized nuclear bodies, or factories with high affinity for these enhancers and for the protein complex that supports OR transcription. Such a nuclear body is described in the regulation of vsg genes in trypanosome (Navarro and Gull, 2001). Moreover, sequestering these putative enhancers into distinct nuclear territories may primarily prevent them from interacting with their proximal ORs, essentially “decommissioning” a large number of elements in each OSN. Thus enhancer convergence may serve two functions: to eliminate the possibility of simultaneous choice of multiple ORs and to ensure robust expression of the single active OR.
In summary, our data are consistent with a model in which the robust transcription of an OR requires an enhancer in cis and numerous enhancers in trans. High levels of OR expression may be necessary for activation of the Perk pathway via ER-stress, and it is likely that only ORs expressed above a certain threshold can elicit this feedback. This prediction is consistent with the observation that transgenic ORs expressed at low levels from heterologous promoters can be coexpressed with endogenous ORs (Zhou and Belluscio, 2012). Since the vetting mechanism that stabilizes OR choice may screen for both the quality and quantity of OR protein, ORs transcribed at suboptimal levels will be turned off by sustained Lsd1 expression. If the number of enhancers associating with an OR promoter indeed determines expression levels, then stable OR expression will occur only once a sufficient number of enhancer elements associate with an OR promoter. Simple modeling of the observed experimental frequencies of pairwise enhancer interactions predicts that the co-localization of 16 different enhancers from a repertoire of 35 will occur only once in each OSN nucleus (see supplemental experimental methods). Thus, depending on the actual number of enhancers needed to achieve feedback-eliciting levels of OR transcription, the limited, or even unique generation of a nucleoprotein complex with sufficient number of enhancers may provide the elusive singularity of OR choice.
It seems counterintuitive that a sensory system critical for survival and reproduction would rely on a molecular mechanism as inefficient and probabilistic as the interchromosomal convergence of a large number of enhancer elements. However, unlike most developmental systems that are built upon tight spatiotemporal regulation, the peripheral olfactory system may be able to tolerate such a variable and often non-productive process because an efficient feedback mechanism is in place to ensure that terminal OSN differentiation occurs only upon OR choice. This mechanism is compatible with the rapid evolution of the OR gene family, which is characterized by significant copy number variations among closely related species and significant polymorphisms within species, in accordance with the essential function of this gene family in adaptation. It remains to be seen if other fast evolving gene families involved in the perception of- and the protection from- the constantly changing external environment (Clowney et al., 2011) may be employing similar radical mechanisms for stochastic and mutual exclusive gene expression.
Materials and Methods
Mouse Strains
Mice were treated in compliance with the rules and regulations of IACUC. The Lipsi enhancer knockout mouse was generated by homologous recombination in ES cells (see Extended Experimental Methods). Other mouse strains used are described in extended experimental methods.
ChIP-seq and DHS-seq
Nuclei from the olfactory epithelium of 6-8 week old widtype mice were isolated and native chromatin ChIPs were performed as described (Magklara et al., 2011). For DHS-seq, nuclei from the olfactory epithelium were isolated and digested briefly with DNase I (Ambion). 100-500bp fractions were gel extracted and prepared for Illumina sequencing. See Extended Experimental Methods for more detailed protocols.
ChIP-seq and DHS-seq analysis
Sequencing reads were mapped to the mouse genome using Bowtie2 (Langmead & Salzberg, 2012). SICER (Zang et al., 2009) was run on reads that mapped to OR clusters or the whole genome, to identify OR and MOE potential enhancers, respectively. OR clusters were defined as genomic regions containing one or more OR genes extending to the nearest non-OR Refseq gene. Peaks located within 5kB of a Refseq gene or an OR promoter (Clowney et al., 2011) were subtracted. DHS peaks that intersected H3K4me1 or H3K27ac peaks were selected, and DHS peaks that intersected with cerebellum H3K4me1 or H3K27ac peaks were subtracted.
Transgenic zebrafish and mouse assays
Enhancer candidate sequences were amplified from mouse genomic DNA using primers targeting DHS peaks (Table S3B). PCR products were cloned into the E1b-GFP-tol2 vector (Li et al., 2010)containing a minimal promoter followed by GFP and injected using standard protocols into at least 75 one-cell stage zebrafish oocytes per construct as described in (Smith et al., 2013). GFP expression was observed at 24 and 48 hpf. Embryos containing at least one GFP positive OSN were counted. Embryos with GFP expression in other tissues were excluded. For transgene reporter mice, the same PCR products were cloned into a vector containing the Hsp68 minimal promoter followed by the lacZ reporter gene (Kothary et al., 1988).
DHS-seq sequence capture
DHS-seq library was further amplified for 10 PCR cycles and 1 μg was hybridized to Seqcap EZ choice probes (NImblegen, Roche) designed to target and tile OR enhancer regions (Table S3A). See extended experimental methods
ChIP QPCR
Crosslinked ChIPs were performed on chromatin isolated from MOE of 3 week old wildtype mice using anti-Lhx2 (kind gift, Mark Roberson). See supplemental methods for detailed protocols
Supplementary Material
Acknowledgments
We would like to thank members of the Ahituv lab for their help and expertise with the zebrafish expression assay. We warmly thank Christos Papadimitriou for consultations about modeling and critical reading of the manuscript. We would like to thank Jessica Tollkuhn and members of the Lomvardas lab for critical reading of the manuscript. Raw sequencing data can be accessed at GEO GSE55174, GSE52464. This project was supported by NIH grants 5R01DA030320, 5R01DA036985, 5R01DA036894, 5R01DC013560 (for SL), and R01HD059862, 1R01NS079231 and 1R01DK090382 (for NA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
None of the authors of this manuscript have financial interests related to this work.
References Cited
- Andrey G, Duboule D. SnapShot: Hox gene regulation. Cell. 2014;156:856–856. e851. doi: 10.1016/j.cell.2014.01.060. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Thanos D. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- Ariza-Cosano A, Visel A, Pennacchio LA, Fraser HB, Gomez-Skarmeta JL, Irimia M, Bessa J. Differences in enhancer activity in mouse and zebrafish reporter assays are often associated with changes in gene expression. BMC Genomics. 2012;13:713. doi: 10.1186/1471-2164-13-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armelin-Correa LM, Gutiyama LM, Brandt DY, Malnic B. Nuclear compartmentalization of odorant receptor genes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2782–2787. doi: 10.1073/pnas.1317036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings / International Conference on Intelligent Systems for Molecular Biology ; ISMB International Conference on Intelligent Systems for Molecular Biology. 1994;2:28–36. [PubMed] [Google Scholar]
- Bargmann CI. Comparative chemosensation from receptors to ecology. Nature. 2006;444:295–301. doi: 10.1038/nature05402. [DOI] [PubMed] [Google Scholar]
- Barnea G, O'Donnell S, Mancia F, Sun X, Nemes A, Mendelsohn M, Axel R. Odorant receptors on axon termini in the brain. Science (New York, NY. 2004;304:1468. doi: 10.1126/science.1096146. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nature genetics. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Booker BM, Murphy KK, Ahituv N. Functional analysis of limb enhancers in the developing fin. Development genes and evolution. 2013;223:395–399. doi: 10.1007/s00427-013-0453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Bushdid C, Magnasco MO, Vosshall LB, Keller A. Humans can discriminate more than 1 trillion olfactory stimuli. Science (New York, NY. 2014;343:1370–1372. doi: 10.1126/science.1249168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, Magklara A, Colquitt BM, Pathak N, Lane RP, Lomvardas S. High-throughput mapping of the promoters of the mouse olfactory receptor genes reveals a new type of mammalian promoter and provides insight into olfactory receptor gene regulation. Genome research. 2011;21:1249–1259. doi: 10.1101/gr.120162.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155:321–332. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpretti S, Montavon T, Leleu M, Joye E, Tzika A, Milinkovitch M, Duboule D. Multiple enhancers regulate Hoxd genes and the Hotdog LncRNA during cecum budding. Cell reports. 2013;5:137–150. doi: 10.1016/j.celrep.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira T, Wilson SR, Choi YG, Risso D, Dudoit S, Speed TP, Ngai J. Silencing of Odorant Receptor Genes by G Protein betagamma Signaling Ensures the Expression of One Odorant Receptor per Olfactory Sensory Neuron. Neuron. 2014;81:847–859. doi: 10.1016/j.neuron.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21081–21086. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Developmental biology. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Hirota J, Mombaerts P. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8751–8755. doi: 10.1073/pnas.0400940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, De Gobbi M, Taylor S, Gibbons R, Higgs DR. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nature genetics. 2014;46:205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Jr., Desplan C. Interchromosomal communication coordinates intrinsically stochastic expression between alleles. Science (New York, NY. 2014;343:661–665. doi: 10.1126/science.1243039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan-Sciutto KL, Dragich JM, Rhodes JL, Bowser R. Fetal Alz-50 clone 1, a novel zinc finger protein, binds a specific DNA sequence and acts as a transcriptional regulator. The Journal of biological chemistry. 1999;274:35262–35268. doi: 10.1074/jbc.274.49.35262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Vaes E, Mombaerts P. Regulation of the probability of mouse odorant receptor gene choice. Cell. 2011;147:907–921. doi: 10.1016/j.cell.2011.09.049. [DOI] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Brown A, Campbell R, Peterson A, Rossant J. A transgene containing lacZ inserted into the dystonia locus is expressed in neural tube. Nature. 1988;335:435–437. doi: 10.1038/335435a0. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JW, Banerjee S, Taylor B, Aplan PD, Singer A, Wu C. Chromatin remodeling complex NURF regulates thymocyte maturation. Genes & development. 2011;25:275–286. doi: 10.1101/gad.2007311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ritter D, Yang N, Dong Z, Li H, Chuang JH, Guo S. A systematic approach to identify functional motifs within vertebrate developmental enhancers. Developmental biology. 2010;337:484–495. doi: 10.1016/j.ydbio.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154:325–336. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughey DM, Vinton RM, Huynh J, Al-Saif A, Beer MA, McCallion AS. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome research. 2008;18:252–260. doi: 10.1101/gr.6929408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012a;150:1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012b;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizumi H, Kumasaka K, Inoue N, Nakashima A, Sakano H. Deletion of the core-H region in mice abolishes the expression of three proximal odorant receptor genes in cis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20067–20072. doi: 10.1073/pnas.0706544105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D. Transcription factors mediate long-range enhancer-promoter interactions. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20222–20227. doi: 10.1073/pnas.0902454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Schorderet P, Joye E, Chabaud F, Duboule D. Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. eLife. 2014;3:e02557. doi: 10.7554/eLife.02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19719–19724. doi: 10.1073/pnas.0609445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman A, Feinstein P, Hirota J, Mombaerts P. The promoter of the mouse odorant receptor gene M71. Molecular and cellular neurosciences. 2005;28:535–546. doi: 10.1016/j.mcn.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science (New York, NY. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O'Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Stergachis AB, Haugen E, Shafer A, Fu W, Vernot B, Reynolds A, Raubitschek A, Ziegler S, LeProust EM, Akey JM, et al. Exonic transcription factor binding directs codon choice and affects protein evolution. Science (New York, NY. 2013;342:1367–1372. doi: 10.1126/science.1243490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SL, Adamson MC, Ressler KJ, Kozak CA, Buck LB. The chromosomal distribution of mouse odorant receptor genes. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:884–888. doi: 10.1073/pnas.93.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli A, Feinstein P, Mombaerts P. Homeodomain binding motifs modulate the probability of odorant receptor gene choice in transgenic mice. Molecular and cellular neurosciences. 2011;46:381–396. doi: 10.1016/j.mcn.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93:47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]
- Williams A, Spilianakis CG, Flavell RA. Interchromosomal association and gene regulation in trans. Trends Genet. 2010;26:188–197. doi: 10.1016/j.tig.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Rodriguez I, Mombaerts P, Firestein S. Odorant and vomeronasal receptor genes in two mouse genome assemblies. Genomics. 2004;83:802–811. doi: 10.1016/j.ygeno.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Belluscio L. Coding odorant concentration through activation timing between the medial and lateral olfactory bulb. Cell reports. 2012;2:1143–1150. doi: 10.1016/j.celrep.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.