Abstract
RecR, together with RecF and RecO, facilitates RecA loading in the RecF pathway of homologous recombinational DNA repair in procaryotes . The human Rad52 protein is a functional counterpart of RecFOR. We present here the crystal structure of RecR from Deinococcus radiodurans (DR RecR). A monomer of DR RecR has a two-domain structure: the N-terminal domain with a helix–hairpin–helix (HhH) motif and the C-terminal domain with a Cys4 zinc-finger motif, a Toprim domain and a Walker B motif. Four such monomers form a ring-shaped tetramer of 222 symmetry with a central hole of 30−35 Å diameter. In the crystal, two tetramers are concatenated, implying that the RecR tetramer is capable of opening and closing. We also show that DR RecR binds to both dsDNA and ssDNA, and that its HhH motif is essential for DNA binding.
Keywords: DNA clamp, DNA repair, homologous recombination, RecF pathway, RecR
Introduction
In all forms of life, the repair of damaged DNA is one of the most fundamental cellular functions. Several different mechanisms for DNA repair have been characterized such as homologous recombination, nonhomologous end joining, base excision repair, nucleotide excision repair, mismatch repair and reversion repair (Christmann et al, 2003; West, 2003). The progress of replication fork is halted by encounters with DNA damages such as double-strand breaks (DSBs) or single-strand gaps (Kuzminov, 1999; Cox et al, 2000; Marians, 2000; Cox, 2001a, 2001b). For DSB repair, homologous recombination is the main pathway in simple eucaryotes like yeast, whereas the nonhomologous end-joining pathway predominates in mammals. Mutagenic translesion synthesis by a special class of DNA polymerases can also move a replication fork past the damage. In bacteria, the major role of homologous recombination is recombinational repair associated with the replication of a damaged template DNA (Cox et al, 2000). It was estimated that at least 10−30% of all replication forks originating at the bacterial origin of replication are halted by DNA damage and must undergo recombinational DNA repair (Cox, 1999). Recombinational DNA repair is both the most complex and the least understood of bacterial DNA repair processes. In Escherichia coli, two major pathways (or ‘recombination machines') of homologous recombination operate: RecBCD and RecF pathways. The RecBCD pathway is the main route when a DNA nick is encountered and the RecF pathway seems to act when a DNA base lesion is encountered (Cox, 2001a). These recombination machines consist of at least three broad classes of activities—helicases, nucleases and synapsis proteins.
In E. coli, the RecBCD and RecF recombination machines use distinct sets of enzymes to produce an ssDNA molecule coated with RecA proteins. In the RecBCD pathway, all of the required functions reside in one machine: the RecBCD enzyme alone provides the helicase, nuclease and RecA-loading activities. In comparison, the RecF recombination pathway requires several separate proteins (RecQ helicase, RecJ nuclease, RecF, RecO and RecR) to process the DNA into a presynaptic intermediate. DNA is unwound by RecQ helicase and the 5′ end is digested by RecJ, leaving the 3′-tailed ssDNA coated with single-stranded DNA binding proteins (SSBs). Then a concerted action of the RecFOR complex directs the loading of RecA protein specifically onto gapped DNA that is coated with SSB (Morimatsu and Kowalczykowski, 2003). Recently, parts of the RecBCD and RecF recombination machines were shown to be interchangeable in promoting recombination under special circumstances (Amundsen and Smith, 2003).
The Rad52 proteins in human and yeast and UvsY in bacteriophage T4 are functional analogs of RecFOR (Kreuzer, 2000). These proteins have been termed ‘recombination mediator proteins' (Beernink and Morrical, 1999). Despite the importance of these recombination mediator proteins, the details about their roles in homologous recombination remain to be characterized. Recently, the RecFOR complex was shown to be a structure-specific mediator that targets recombinational repair to ssDNA–dsDNA junctions and it was proposed that the behavior of the RecFOR protein is mimicked by the functional counterparts in all organisms (Morimatsu and Kowalczykowski, 2003). The RecR protein plays a critical role by forming complexes with either RecF or RecO, or possibly both (Umezu and Kolodner, 1994; Hegde et al, 1996; Shan et al, 1997; Webb et al, 1997). However, no direct interaction between RecF and RecO has been observed (Morimatsu and Kowalczykowski, 2003). The RecF protein binds to DNA (Griffin and Kolodner, 1990; Madiraju and Clark, 1992) and has a weak ATPase activity (Webb et al, 1995, 1999). The RecFR complex attenuates the extension of RecA filaments beyond ssDNA gaps (Webb et al, 1997). The RecO protein promotes renaturation of complementary ssDNA, catalyzes assimilation of ssDNA into superhelical dsDNA (Luisi-DeLuca and Kolodner, 1994; Luisi-DeLuca, 1995), and anneals ssDNA with its cognate ssDNA like the human Rad52 protein (Kantake et al, 2002). RecOR helps RecA to overcome the inhibitory effect of SSB during homologous pairing of ssDNA with dsDNA (Umezu and Kolodner, 1994), prevents an end-dependent dissociation of RecA filaments from ssDNA (Shan et al, 1997), and modulates the RecA protein function at 5′ ends of ssDNA (Bork et al, 2001). An alternative, nonrecombinative role for RecFOR was also suggested (Courcelle et al, 1997, 2001). In this model, at least two genes in the recF pathway, recF and recR, are required to reassemble a replication holoenzyme at the site of a DNA replication fork when replication is disrupted.
Although crystallization of RecO from Thermus thermophilus has been reported (Aono et al, 2003), no structural information is currently available for RecF, RecO or RecR. In order to provide a structural framework for understanding the role of the RecR protein in homologous recombinational DNA repair, we have determined the crystal structure of RecR from Deinococcus radiodurans (DR RecR), a 220-residue protein. It was found to play an important role in the resistance to interstrand crosslinks in this bacterium (Kitayama et al, 2000). DR RecR, as a homotetramer, displays a ring-shaped architecture with a central hole. This structural feature, together with the observation of concatenated octamers in the crystal, provides insights into the role of RecR in homologous recombination.
Results and discussion
Structure determination and overall structures
The structure of DR RecR was solved at 2.5 Å resolution using two sets of multiwavelength anomalous diffraction (MAD) data collected from a mercury-soaked crystal of the selenomethionine (SeMet)-substituted protein and a mercury derivative crystal of the native protein (Table I). The model has been refined using the mercury-derivatized native data to crystallographic Rwork and Rfree values of 24.8 and 31.5%, respectively, for reflections with no sigma cutoff in the resolution range 43.4−2.5 Å. R values remain a little higher than usual due to the limited quality of the data, which was caused by the inherent limitation of the crystals. After repeated trials, this data set was found to be the best that we could collect. The refined model contains 791 residues of the four independent monomers in the asymmetric unit of the crystal (residues 1–199, 2–199, 2–197 and 2–199 for monomers A, B, C and D, respectively), 65 water molecules, four zinc ions and five imidazole molecules. Residues 200–220 as well as the C-terminal eight-residue tag are disordered in the crystal and are not visible in the electron density map. An alignment of RecR amino-acid sequences shows that the disordered C-terminal residues are not conserved among RecR proteins (Figure 1) and thus are not critical for the function of RecR proteins. Four monomers of DR RecR in the asymmetric unit adopt similar conformations. The r.m.s. differences are 0.43, 1.17, 1.24, 1.55, 1.13 and 0.62 Å for 196 Cα atoms for comparing the chains A–B, A–C, A–D, B–C, B–D and C–D, respectively. A and B subunits (or C and D subunits) are more similar to each other than other pairs.
Table 1.
Statistics on MAD data collection, phasing and refinement
| Crystal | Data set | X-ray wavelength (Å) | Resolution (Å) | Total/unique reflections | Completeness (%)a | I/σ(I) | Rmerge (%)b |
|---|---|---|---|---|---|---|---|
| I | SeMet-1 (remote) | 0.97182 | 50–3.1 (3.2–3.1) | 149 335/18 551 | 98.1 (99.5) | 7.1 (1.7) | 8.7 (24.7) |
| SeMet-2 (edge) | 0.97950 | 50–3.1 (3.2–3.1) | 167 793/18 623 | 98.5 (100) | 6.7 (1.5) | 9.2 (26.8) | |
| SeMet-3 (peak) | 0.97936 | 50–3.1 (3.2–3.1) | 215 277/18 188 | 99.4 (100) | 6.5 (1.6) | 9.3 (27.4) | |
| II | Hg-1 (remote) | 0.99999 | 50–2.4 (2.5–2.4) | 239 055/38 694 | 97.7 (100) | 6.3 (2.0) | 9.2 (39.2) |
| Hg-2 (edge 1) | 1.00921 | 50–2.4 (2.5–2.4) | 323 573/38 306 | 96.3 (100) | 5.7 (1.9) | 11.1 (38.6) | |
| Hg-3 (edge 2) | 1.00883 | 50–2.6 (2.7–2.6) | 316 760/30 578 | 97.3 (100) | 7.0 (2.3) | 10.6 (34.3) | |
| Hg-4 (peak) | 1.00812 | 50–2.6 (2.7–2.6) | 252 982/30 299 | 96.8 (100) | 6.5 (2.2) | 12.6 (42.0) | |
| Figure of meritc for MAD phasing: 0.46 (before RESOLVE)/0.53 (after RESOLVE) for 30–2.5 Å | |||||||
| Refinement (Hg-1 data set) | |||||||
| Resolution range (Å) |
No. of reflectionsd |
No. of atomse |
Rwork/Rfree (%)f |
R.m.s. deviationg |
Average B factorh |
Ramachandran ploti |
|
| 43.4–2.5 |
34 134 (5148) |
6036/65/4/25 |
24.8/31.5 |
0.007/1.44 |
46.3(48.0/49.2/47.8/45.1)/38.5/30.6/55.6 |
85.5/14.5/0.0/0.0 |
|
| a Completeness for I/σ(I)>1.0, high-resolution shell in parentheses. | |||||||
| b Rmerge=∑hkl∑i∣I(hkl)i−〈I(hkl)〉∣/∑hkl〈I(hkl)〉, where I(hkl)i is the ith measurement of the intensity of reflection hkl and 〈I(hkl)〉 is the average intensity. | |||||||
| c Figure of merit=〈∣∑P(α)eiα/∑P(α)∣〉, where α is the phase and P(α) is the phase probability distribution. | |||||||
| d Numbers reflect the reflections with F/σ(F)>0; numbers in parentheses reflect statistics for the last shell (2.66–2.5 Å). | |||||||
| e Protein/water/zinc ion/imidazole. | |||||||
| f R=∑∣∣Fobs∣−∣Fcalc∥/∑∣Fobs∣, where Rfree (Brünger, 1992) is calculated without a sigma cutoff for a randomly chosen 10% of reflections, which were not used for structure refinement, and Rwork is calculated for the remaining reflections. | |||||||
| g Deviations from ideal bond lengths/angles. | |||||||
| h Tetramer(A/B/C/D subunits)/water/zinc ion/imidazole. | |||||||
| i Percentage of non-glycine residues in the most favored regions/additional allowed regions/generously allowed regions/disallowed regions. | |||||||
Figure 1.
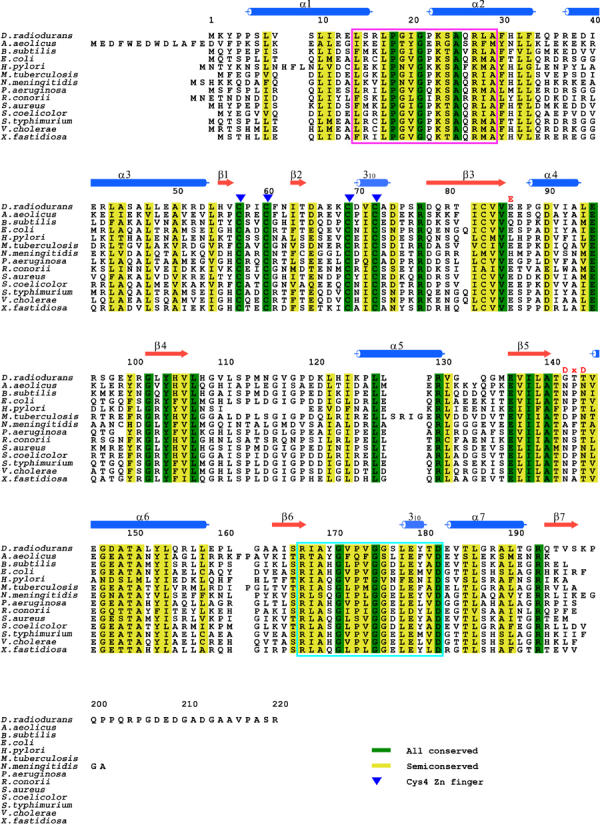
Amino-acid sequence alignment of 14 RecR proteins. The signature sequence of the HhH motif (hxxhxGhGxxxAxxhh) is marked in a pink box and the Walker B motif (R/KxxxGxxxL/VhhhhD) in a cyan box. Cysteine residues of the zinc finger are marked by blue triangles above the sequence. The conserved acidic residues found in Toprim domains are denoted as ‘E' and ‘DxD' above the sequence.
Each monomer of DR RecR has approximate dimensions of 70 Å × 50 Å × 40 Å and it can be roughly divided into two domains (Figure 2). The N-terminal domain (N-domain; residues 1–53) contains three α helices: the first two helices (α1 and α2) are folded into a helix–hairpin–helix (HhH) motif and the third helix (α3) serves as a linker between the two domains. The C-terminal domain (C-domain; residues 54–199) consists of seven β strands, four α helices and two 310 helices, and is folded into three distinct motifs (the Cys4 zinc-finger motif, the Toprim domain and the Walker B motif) and a C-terminal tail consisting of helix α7 and the strand β7 (Figure 2). Comparisons with the structural database in Protein Data Bank using the program DALI (Holm and Sander, 1995) found structural similarity between DR RecR and E. coli topoisomerase I (PDB code 1ECL, Z score=7.5, r.m.s. deviation=3.9 Å for 89 structurally aligned residues of DR RecR between residues 79 and 187) and E. coli DNA primase (PDB code 1DD9, Z score=6.2, r.m.s. deviation=2.6 Å for 82 structurally aligned residues of DR RecR between residues 55 and 162). These residues encompass roughly the Toprim domain of DR RecR.
Figure 2.
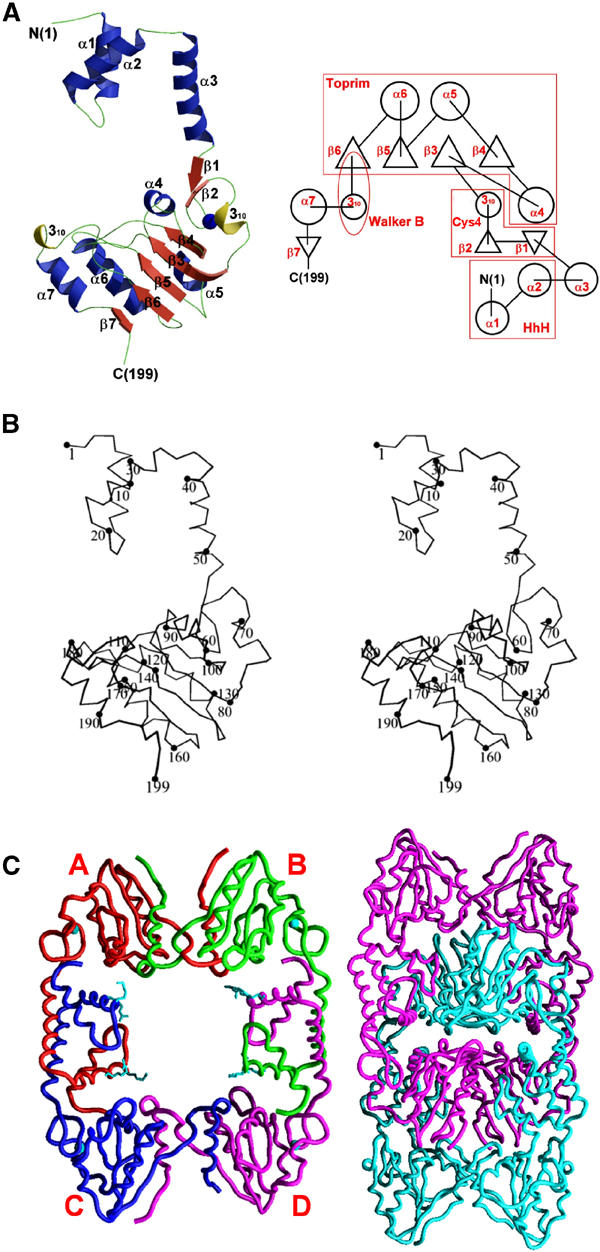
Overall structure of DR RecR. (A) Ribbon diagram and topology diagram showing the overall fold of a monomer. (B) Cα stereo view of a monomer. (C) DR RecR tetramer and octamer. A/B/C/D subunits are drawn in red/green/blue/pink, and zinc ions and the side chains of Lys23 and Arg27 are drawn in cyan in the tetramer model (left). Two tetramers (pink and cyan) are concatenated to form an octamer (right).
Four DR RecR monomers in the asymmetric unit of the crystal form a ring-shaped tetramer of 222 symmetry (Figure 2C), with dimensions of 90 Å × 70 Å × 30 Å, and the tetramer has a central hole with approximate dimensions of 30 Å × 35 Å in the narrowest section and with approximately 30 Å thickness. In each tetramer, the HhH motif (α1 and α2) of the N-domain is domain-swapped with the same motif of an adjacent subunit (Figure 3A). Furthermore, the C-terminal region including the Walker B motif (residues 167–182), helix α7 and strand β7 of the C-domain is also domain-swapped with the equivalent region of another neighboring subunit (Figure 3D). Surprisingly, two such tetramers are concatenated in the crystal and form an octamer with approximate dimensions of 120 Å × 80 Å × 60 Å (Figure 2C). To the best of our knowledge, this kind of concatenation of two identical closed circular oligomers has not been observed in proteins before. We have found by dynamic light scattering analyses that DR RecR and Helicobacter pylori (HP) RecR exist in solution as tetramers at a protein concentration of 1 mg ml−1 and as octamers at 5 mg ml−1, respectively. Analytical ultracentrifugation indicates that both RecRs are tetramers at 1 mg ml−1 (Figure 4A). We have also discovered that octamers of both RecRs dissociate into tetramers by the addition of 0.2–1.0% (v/v) Tween 20 (polyoxyethylenesorbitan monolaurate) or by diluting the protein. The conversion from octamers to tetramers was not influenced by magnesium ions, ATP or ssDNA, as analyzed by dynamic light scattering. And the purified octamers and tetramers did not re-equilibrate spontaneously in the absence of detergent. We have also grown crystals in the presence of 80 mM MgCl2 and 2 mM ATP; the structure refined at 3.4 Å is again concatenated octamers. As the intracellular concentration of RecR would be very low, the quaternary structure of RecR inside the bacterial cell would be the ring-shaped tetramer (Figure 2C). We suspect that the concatenated octamers of DR RecR in the crystal have resulted from a high protein concentration required for crystallization and the ability of DR RecR tetramer to open and close. However, it should be emphasized that the nature of the octamers in solution remains an open question. This is because many ring proteins often form the ‘back-to-back' rings in solution and there is no reason to assume that the concatenated octamers of DR RecR seen in our crystal are the same form as the octamers in solution.
Figure 3.
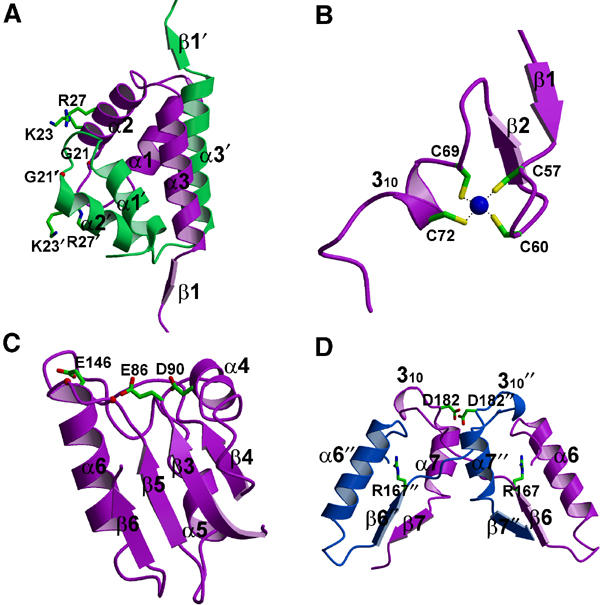
Detailed views of domains/motifs in DR RecR. (A) HhH motifs from two monomers are swapped. Gly21, Arg23 and Lys27 are shown. Primed residues belong to the second subunit B in Figure 2C. (B) Cys4 zinc-finger motif. (C) Toprim domain with residues Glu86, Asp90 and Glu146. Corresponding positions (142 and 144) of the ‘DxD' sequence are indicated by red balls. (D) The C-terminal regions from two monomers are swapped. Arg167 and Asp182 are the fingerprint residues of the canonical Walker B motif. Double-primed residues belong to the third subunit C. The orientations of domains/motifs are roughly similar to those in subunits B (colored in white green), D (purple) and C (white blue) in Figure 2C.
Figure 4.
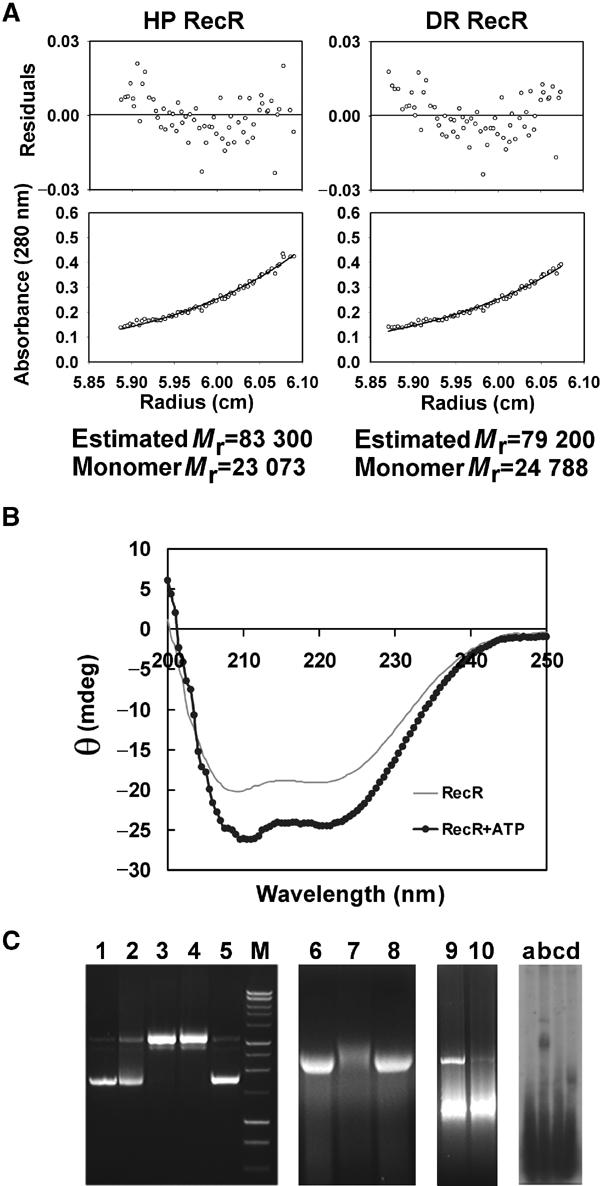
Biochemical data of RecR. (A) Sedimentation equilibrium of HP RecR and DR RecR at 1 mg ml−1 concentration. (B) Effect of ATP on the circular dichroism spectrum of HP RecR. (C) DNA-binding assay of RecR. Lane 1, supercoiled pUC19; lane 2, pUC19+DR RecR+4 mM ATP; lane 3, pUC19+DR RecR+40 mM MgCl2; lane 4, pUC19+DR RecR+4 mM ATP+40 mM MgCl2; lane 5, pUC19+DR RecR34–220+4 mM ATP+40 mM MgCl2; lane M, DNA size marker; lane 6, φX174 virion DNA (circular ssDNA)+ DR RecR; lane 7, φX174 virion DNA+DR RecR+40 mM MgCl2; lane 8, φX174 virion DNA+DR RecR+1 mM ATP; lane 9, pUC19+DR RecR+1 mM ATP+40 mM MgCl2; lane 10, pUC19+DR RecR(K23A/R27A)+1 mM ATP+40 mM MgCl2; lane a, ssDNA (30 mer; 5′-AAGGAGGAGAAGGAGAAGAAGGAGGAGAAG-3′); lane b, ssDNA+HP RecR+4 mM ATP+80 mM MgCl2; lane c, ssDNA+HP RecR39–193+4 mM ATP+80 mM MgCl2; lane d, ssDNA+HP RecR1–164+4 mM ATP+80 mM MgCl2. For lanes 9 and 10, the wild type and the double mutant K23A/R27A of DR RecR were present at the same concentration, respectively.
Motif/domain structures of a monomer
RecR protein sequences contain an HhH motif, a Cys4 zinc finger, a Toprim domain and a Walker B motif (Peláez et al, 2001). We describe the structural details of these motifs or domains below (Figure 3).
HhH motif: The distinctive sequence of the HhH motif, hxxhxGhGxxxAxxhh (where h is any hydrophobic residue, VILMWFYA), is conserved in the N-terminal part (residues 14–29) of DR RecR corresponding to the helices α1 and α2 (Figure 1). HhH motifs are present in many DNA replication and repair proteins (Doherty et al, 1996; Aravind et al, 1999), including endonuclease III from E. coli (Thayer et al, 1995), AlkA from E. coli (Labahn et al, 1996), MutY from E. coli (Guan et al, 1998) and human DNA polymerase β (Mullen and Wilson, 1997). The r.m.s. differences between HhH motifs of DR RecR and endonuclease III/AlkA/MutY/DNA polymerase β are 0.71/0.72/0.48/1.02 Å for 18 Cα atom pairs, respectively. The presence of HhH motifs has been implicated in sequence-nonspecific DNA binding (Thayer et al, 1995; Doherty et al, 1996). In the DR RecR tetramer, two HhH motifs from two adjacent monomers (subunits A and C, or subunits B and D) are domain-swapped and form a compact domain (Figures 2C and 3A). Interestingly, the conserved basic residues Lys23 and Arg27 in the second helix of the HhH motif line the central hole of the RecR tetramer, with their side chains pointing toward the hole (Figure 2C). The conserved Gly21 also lines the surface of the central hole. We suspected that the HhH motif containing these conserved residues might play an important role in DNA binding. Indeed, we were able to show by making deletion and point mutant proteins (DR RecR34–220, HP RecR39–193, DR RecR(K23A/R27A)) that the HhH motif is essential for DNA binding, as discussed further below.
Cys4zinc finger: The side chains of four strictly conserved cysteine residues (Cys57, Cys60, Cys69 and Cys72) coordinate a very strong electron density, which was interpreted as a zinc ion, and these residues are part of a Cys4 zinc-finger motif (Figure 1). The zinc-finger motifs lie on the outside of the DR RecR ring (Figure 2C). Cys4 zinc fingers are frequently present in many DNA-binding proteins such as the DNA-binding domain of hormone receptors (Schmiedeskamp and Klevit, 1994; Klug and Schwabe, 1995; Mackay and Crossley, 1998), NAD+-dependent DNA ligase (Lee et al, 2000) and the human DNA repair protein XPA (Ikegami et al, 1998). An E. coli strain carrying a mutant zinc finger of RecR displayed decreased survival, suggesting the importance of the zinc finger in the function of E. coli RecR (Clark, 1991). Plausible roles of the RecR zinc-finger motif are a structural role in protein folding and/or a direct role in DNA binding. The recombinant zinc-finger deletion mutants of both DR RecR and HP RecR (DR RecR73–220 and HP RecR77–193) were produced as inclusion bodies in E. coli. We have also prepared several point mutants. DR RecR(C60S) and HP RecR(C61S) were expressed at very low levels and as inclusion bodies in E. coli. DR RecR(C69S), DR RecR(C72S) and HP RecR(C76S) were expressed at high levels and as inclusion bodies. Thus we could not examine the role of the zinc-finger motif in DNA binding with these deletion and point mutants.
Toprim domain: The overall fold of the Toprim domain of DR RecR (residues 78–166) resembles the Rossman-like nucleotide-binding fold, with a parallel four-stranded central β sheet flanked by one α helix (α4) on one side and two α helices (α5 and α6) on the other side in each monomer (Figure 2). In the ring-shaped tetramer, the C-terminal β strand (β7) from an adjacent monomer lies antiparallel to the central β sheet and the C-terminal helix (α7) from an adjacent monomer flanks the α4-covered side of the central β sheet (Figure 2C). Similar Toprim domains are also present in topoisomerases (I, II and VI), primases and members of the OLD nuclease family (Lima et al, 1994; Berger et al, 1996; Nichols et al, 1999; Keck et al, 2000; Podobnik et al, 2000; Kato et al, 2003). In the Toprim domain structures of E. coli topoisomerase I and yeast topoisomerase II, and the modeled structure of the DnaG-type primases, a conserved glutamate and two conserved aspartate residues in the DxD sequence, that is, the fingerprint acidic residues, cluster together to form a highly acidic surface patch (Aravind et al, 1998; Podobnik et al, 2000; Kato et al, 2003). It was suggested that the DxD motif might coordinate a magnesium ion (Aravind et al, 1998). Indeed, a magnesium ion was found to bind to the predicted site in the structures of Methanococcus jannaschii topoisomerase VI (Nichols et al, 1999), E. coli DnaG primase (Keck et al, 2000; Podobnik et al, 2000) and T7 primase (Kato et al, 2003). In RecR sequences, the glutamate residue in the first turn of the Toprim domain (at position 86 of DR RecR, denoted as ‘E' in Figure 1) is semiconserved, whereas the DxD sequence (at corresponding positions 142–144 of DR RecR, denoted as ‘DxD' in Figure 1) is not conserved. However, several other acidic residues are highly conserved in RecR sequences (Figure 1): Asp90, Glu95, Glu136, Glu146 and Asp148 in DR RecR. Glu86, Asp90 and Glu146 line the central hole; Glu95, Glu136 and Asp148 are located outside the central hole. Glu86, Asp90 and the carbonyl group of Gly142 make a hydrogen bond to a water molecule in three of the four subunits of the tetramer.
Walker B motif: A highly diverged version of the Walker B motif signature sequence (R/KxxxGxxxL/VhhhhD, where x is any residue and h is a hydrophobic residue; Walker et al, 1982) is present in RecR sequences (Figure 1). The Walker B motif of DR RecR between positions 167 and 182 forms a long loop containing a 310 helix at its C-terminus between β6 and α7. It is involved in forming the dimer interface between subunits A and B (or C and D) by motif swapping. The C-terminal deletion mutant proteins DR RecR1–167 and HP RecR1–164 lacking the Walker B motif showed a tendency to aggregate upon purification. This result may have been due to a failure to form ring-shaped tetramers. The conserved aspartate residue in Walker B motifs was proposed to be important for magnesium binding in adenylate kinase and phosphofructokinase (Walker et al, 1982). Circular dichroism spectra indicated that the α helical content of DR RecR and HP RecR increased in the presence of ATP (Figure 4B), whereas spectra were not affected by the addition of magnesium ions. This suggests that the Walker B motif of DR RecR is likely to interact with ATP and that there is a conformational change in the Walker B motif upon binding ATP. We could not detect ATPase activity for both DR RecR and HP RecR. Similarly, Streptomyces coelicolor RecR did not show ATPase activity (Peláez et al, 2001). However, ATP enhanced the DNA-binding activity of Bacillus subtilis RecR (Alonso et al, 1993). Although it has not been clearly established how ATP influences the RecR function, we may imagine a possible regulatory role of ATP binding to RecR. The Walker A motif signature sequence (GxxxxGKT/S) is not present in RecR sequences. Both Walker A and B motifs are present in RecF sequences (Ayora and Alonso, 1997), being consistent with a weak ATPase activity of RecF. In contrast, neither Walker A nor Walker B motif is present in the RecO sequences. It is also conceivable that the ATPase activity of RecR is cryptic and its interaction with another protein, likely RecO, switches on its ATPase.
Functional implications for RecR
The overall shape of DR RecR tetramer is strikingly similar to the ring-shaped structures of DNA polymerase processivity factors including the β subunit of E. coli DNA polymerase III (Kong et al, 1992), gp45 of T4 DNA polymerase (Moarefi et al, 2000) and PCNA of yeast and human DNA polymerases δ (Krishna et al, 1994; Gulbis et al, 1996) (Figure 5A). Among DR RecR, the β subunit of E. coli DNA polymerase III, gp45 of T4 DNA polymerase and PCNA of eucaryotic DNA polymerases δ, there is little sequence similarity. They also differ in their quaternary structures: DR RecR is a homotetramer and E. coli β clamp is a homodimer, whereas T4 gp45 and eucaryotic PCNAs are homotrimers. A large number of other proteins involved in DNA metabolism also adopt a toroidal (or ring-shaped) structure, even though they have completely unrelated functions (Hingorani and O'Donnell, 2000). They include NAD+-dependent DNA ligase (Lee et al, 2000), λ exonuclease (Kovall and Matthews, 1997), hexameric helicases, topoisomerases, the trp RNA-binding attenuation protein, the bacteriophage head-to-tail connector and translin (Hingorani and O'Donnell, 2000, and references therein).
Figure 5.
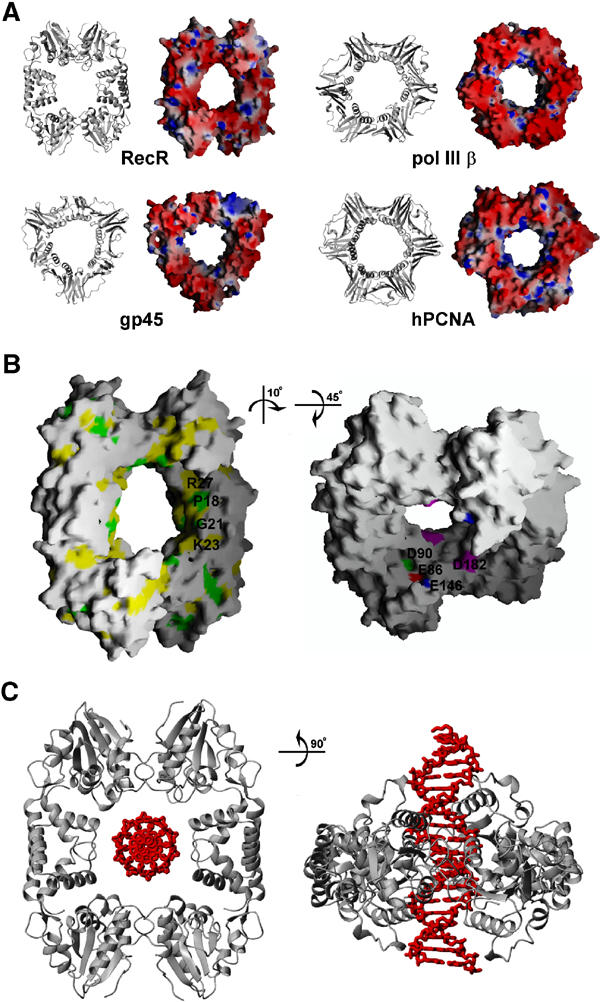
A possible DNA clamp model for RecR. (A) Comparison of DR RecR with DNA clamp proteins. Ribbon diagrams and electrostatic potential at the molecular surface are shown for DR RecR, E. coli DNA polymerase III β subunit, T4 gp45 and human PCNA. The diameter of the central hole is about 30–35 Å for DR RecR and about 35 Å for other clamp proteins. The molecular surface was colored according to the electrostatic potential: blue, 10 kT; white, 0 kT; red, −10 kT. (B) Conserved residues of DR RecR located in the putative DNA-binding region of the central hole (left). Strictly conserved residues are colored in green and semiconserved residues in yellow, as deduced by aligning 14 RecR sequences in Figure 1. Negatively charged residues of the Toprim domain and the Walker B motif that may play a role in Mg2+-enhanced DNA binding (right). (C) Model for dsDNA binding by DR RecR.
Since RecR proteins show no sequence similarity to any of the toroidal proteins that act on DNA, it is difficult to predict with confidence a possible function of RecR solely on the basis of its shape and sequence homology. However, the ring-shaped architecture of the tetrameric DR RecR with a central hole and the presence of concatenated octamers in the crystal raise an intriguing possibility that DR RecR might function as a kind of DNA holding clamp that is capable of opening and closing. This possibility does not necessarily assume that RecR slides along dsDNA like the processivity factors. Instead, we speculate that RecR, in complex with RecF or RecO, or both, might function as a nonsliding clamp that recognizes the structural features of the ssDNA–dsDNA junctions (Morimatsu and Kowalczykowski, 2003). It is also possible that RecR plays other functional roles, which may not be obvious from its ring-shaped structure.
The diameter of the central hole of tetrameric RecR is ∼30 Å in its shortest dimension and ∼35 Å in its longest dimension, just large enough to accommodate dsDNA (Figure 5). And the conserved basic residues (Lys23 and Arg27) of the HhH motif are located on the surface of the central hole in the tetrameric ring (Figures 2C and 5B). The HhH motif-truncated deletion mutant proteins of RecR (DR RecR34–220 and HP RecR39–193) exist as stable dimers in solution and they do not bind to either dsDNA or ssDNA. The DNA-binding affinity of the double mutant DR RecR(K23A/R27A) was also considerably reduced (Figure 4C, lane 10). Therefore, it seems reasonable to suggest that dsDNA would bind within the central hole of tetrameric RecR (Figure 5C) and that the HhH motifs play an essential role in dsDNA binding. We suspect that conserved acidic residues Glu86, Asp90 and Glu146 of DR RecR that line the central hole (Figure 5B) may play a role in the observed Mg2+-enhanced DNA binding (discussed below), since they might bind magnesium ions and thus reduce the electrostatic repulsion between the negatively charged side chains and the phosphate backbone of DNA bound within the central hole. Asp182 of DR RecR lies on the surface of the central hole (Figure 5B). This residue may also be partly responsible for the observed Mg2+-enhanced DNA binding by DR RecR.
If we assume that the RecR proteins play a clamp-like role in homologous recombinational DNA repair and that dsDNA is bound within their central hole, the tetrameric ring might be capable of opening and closing in a manner analogous to that proposed for T4 gp45 processivity factor (Alley et al, 1999) and E. coli DNA polymerase III β clamp (Jeruzalmi et al, 2001). T4 gp45 forms a closed trimeric ring in the crystal, but it was speculated that it might be in equilibrium between an open and closed form in solution (Alley et al, 1999). In the case of E. coli β clamp, it was suggested that the dimeric ring opens spontaneously once one of the two dimer interfaces is perturbed by the δ wrench (Jeruzalmi et al, 2001). We observed by dynamic light scattering analyses that DR RecR and HP RecR exist as either tetramers or octamers in solution and a conversion between tetramers and octamers is possible. We also observed that two tetrameric rings of DR RecR are concatenated to form an octamer in the crystal (Figure 2C). These observations raise an intriguing possibility of opening and closing of the DR RecR tetrameric ring.
Within the DR RecR tetramer, there are two types of interface between the neighboring subunits and each monomer makes two types of intersubunit interaction with the neighboring monomers. The two types of interface have similar areas of buried surface. The interface area between A and C subunits (or between B and D subunits) is ∼2420 Å2 and that between A and B subunits (or between C and D subunits) is ∼2350 Å2. However, the interaction between the HhH motif-swapped subunit A–C (or B–D) pair appears to be weaker than that between the Walker B motif-swapped subunit A–B (or C–D) pair. The A–C (or B–D) interface involves five hydrogen bonds and one (or no) salt bridge, whereas the A–B (or C–D) interface involves 36 (or 29) hydrogen bonds and two salt bridges. The polar/hydrophobic interactions are 32.0/68.0% (or 31.3/68.7%) of the total interactions in A–C (or B–D) pair and 36.9/63.1% (or 34.8/65.2%) for A–B (or C–D) pair, respectively (protein–protein interaction server, http://www.biochem.ucl.ac.kr/bsm/PP/server). Therefore, it would be easier for the RecR ring to open at the weaker A–C (or B–D) subunit interface. It is conceivable that protein–protein interactions, possibly involving RecF, RecO, or both, could induce opening of only one of the two equivalent interfaces in the tetrameric ring.
HhH motif is essential for DNA binding by RecR
There have been conflicting results regarding the DNA-binding properties of RecR proteins and there has been no report on which regions of RecR are responsible for DNA binding. B. subtilis RecR binds to both ssDNA and dsDNA, and the RecR–DNA complex formation is markedly stimulated by ATP and divalent cations (Alonso et al, 1993; Ayora et al, 1997). In contrast, E. coli RecR does not bind to either ssDNA (Umezu and Kolodner, 1994) or dsDNA (Webb et al, 1995). The E. coli RecR protein, however, greatly facilitates RecF binding to dsDNA in the presence of ATP (Webb et al, 1995). Since our structure suggests a possible DNA clamp model for RecR, we examined DNA-binding properties of both DR RecR and HP RecR by electrophoretic mobility shift assay (EMSA). We have found that the wild-type DR RecR and HP RecR bind to supercoiled circular pUC19 dsDNA at high (40 or 80 mM) magnesium ion concentrations (Figure 4C, lane 9) but they do not bind to the same dsDNA at low (2 or 5 mM) magnesium ion concentrations. Addition of 4 mM ATP did not affect dsDNA binding. We have also found that HP RecR binds to a 30-nucleotide ssDNA in the presence of 80 mM magnesium chloride (Figure 4C, lane b). With 40 mM magnesium chloride, DR RecR binds to φX174 virion DNA (a closed circular ssDNA) but it does not bind to the same ssDNA with 1 mM ATP (Figure 4C, lanes 6–8). At 0–2 mM magnesium chloride, DR RecR does not bind to φX174 virion DNA. Our results are in general agreement with the observations with B. subtilis RecR (Alonso et al, 1993; Ayora et al, 1997). RecR binding to dsDNA and ssDNA is likely to be considerably facilitated by its physical interaction with RecF and RecO. B. subtilis RecF binds to both ssDNA and dsDNA, and its interaction with ssDNA is markedly stimulated by divalent cations (Ayora and Alonso, 1997). E. coli RecF binds to the ssDNA–dsDNA junction (Morimatsu and Kowalczykowski, 2003). E. coli RecO binds to both ssDNA and dsDNA, and promotes renaturation of complementary ssDNA molecules (Luisi-DeLuca and Kolodner, 1994; Luisi-DeLuca, 1995). It is conceivable that DR RecR and HP RecR could bind to ssDNA and dsDNA at physiological magnesium ion concentrations, when they form a complex with corresponding RecF or RecO, or both. To be optimally active, RecA protein-mediated DNA strand exchange requires 6–8 mM Mg2+ in excess of that required to form complexes with ATP (Lusetti et al, 2003). It has been proposed that the C-terminus of the RecA protein acts as a regulatory switch, inhibiting homologous DNA pairing and strand exchange at low magnesium concentrations (Lusetti et al, 2003).
In order to determine which parts of RecR are crucial for DNA binding, we have tried to express several deletion and point mutant proteins: DR RecR34–220, DR RecR73–220, DR RecR1–167, HP RecR39–193, HP RecR77–193, HP RecR1–164, DR RecR(K23A/R27A), HP RecR(C61S), HP RecR(C76S), DR RecR(C60S), DR RecR(C69S) and DR RecR(C72S). DR RecR73–220, HP RecR77–193, HP RecR(C61S), HP RecR(C76S), DR RecR(C60S), DR RecR(C69S) and DR RecR(C72S) were expressed as inclusion bodies and we could not test their DNA-binding activity. The C-terminal Walker B motif-truncated deletion mutants DR RecR1–167 and HP RecR1–164 showed a tendency to aggregate upon purification and we could not determine their oligomeric state. However, these mutants appear to bind to ssDNA (Figure 4C) and dsDNA. The HhH motif-truncated deletion mutants DR RecR34–220 and HP RecR39–193 exist as stable dimers in solution according to the dynamic light scattering measurements and they do not bind to either dsDNA or ssDNA. This finding indicates that the HhH motif of RecR is essential for DNA binding in two ways. Indirectly, the lack of DNA binding by the RecR mutants without the HhH motif could be due to the failure of tetramer formation. In a direct way, the conserved Gly21, Lys23 and Arg27 of the HhH motif may play important roles in sequence-nonspecific binding to DNA (Figure 5B). The importance of two of these residues was demonstrated by showing that the double mutant DR RecR(K23A/R27A) exhibited much weaker DNA-binding affinity than the wild type (Figure 4C, lanes 9 and 10).
Conclusions
We determined the crystal structure of DR RecR, and carried out mutational and DNA-binding studies to elucidate its functional role in the recombinational DNA repair. DR RecR forms a ring-shaped tetramer with a central hole of 30−35 Å diameter, displaying a striking overall structural resemblance to DNA polymerase processivity factors. This architecture, together with the observation of concatenated octamers in the crystal, raises an intriguing possibility that the tetrameric ring of DR RecR might be capable of opening and closing, and RecR in complex with RecF or RecO, or both, might function as a kind of structure-specific, nonsliding DNA clamp. We also showed that the HhH motif is essential for DNA binding by RecR. We summarize a working model for the formation of RecA filaments onto gapped DNA in the RecF pathway in Supplementary data.
Materials and methods
Protein preparation, crystallization and data collection
Recombinant DR RecR with a C-terminal eight-residue tag was overexpressed and crystallized as described elsewhere (Lee et al, 2004). The C-terminal His6-tagged HP RecR was purified by the same procedure. Genes encoding six deletion mutant proteins DR RecR34–220, DR RecR73–220, DR RecR1–167, HP RecR39–193, HP RecR77–193 and HP RecR1–164 were cloned into the pET-28b(+) vector in the N- and C-terminal His-tagged form. Without the N-terminal tag, these proteins were not expressed in E. coli. Recombinant DR RecR73–220 and HP RecR77–193 were produced as inclusion bodies in E. coli. DR RecR34–220, DR RecR1–167, HP RecR39–193 and HP RecR1–164 were purified using a Hi-trap chelating column and a Superdex 200 prep grade column (Amersham Pharmacia Inc.). Point mutants were prepared using the QuikChange Site-Directed Mutagenesis kit (Stratagene). The mutations were confirmed by sequencing. We obtained crystals of both the full-length DR RecR and HP RecR; only the crystals of DR RecR were suitable for structure determination.
The best crystals of DR RecR grew when the reservoir solution for the hanging-drop vapor diffusion crystallization consisted of 100 mM imidazole (pH 8.0), 16–20% (w/v) polyethylene glycol 1000, 200 mM calcium acetate and 20% (v/v) (±)1,3-butanediol. The native enzyme crystallized into the C2221 space group with unit cell parameters of a=106.22 Å, b=121.39 Å and c=150.00 Å. The asymmetric unit contains four monomers. A set of Se MAD data was collected from an ethylmercurythiosalicylate (EMTS)-soaked crystal of the SeMet-substituted DR RecR and another set of Hg MAD data was collected from an EMTS-soaked crystal of the native protein using the Bruker CCD detector at beamline BL-6B of Pohang Light Source, Korea. Diffraction data were processed and scaled using the programs SMART, SAINTPLUS and PROSCALE (Bruker AXS Inc.). Dynamic light scattering was measured on the model DynaPro-801 instrument (Protein Solutions).
Structure solution and refinement
Heavy atom sites were located with SOLVE (Terwilliger and Berendzen, 1999): nine Se sites from the EMTS-soaked SeMet-substituted protein crystal and three Hg sites from the EMTS-soaked native protein crystal. Initial phases were improved using the program RESOLVE (Terwilliger and Berendzen, 1999), yielding an interpretable electron density map. Noncrystallographic symmetry (NCS) matrices were found from a partial model built from the initial electron density map and phases were further improved by the four-fold NCS averaging, solvent flattening and histogram matching with the program DM (Collaborative Computational Project, Number 4, 1994). The model was built with O (Jones et al, 1991). The protein model was refined using the CNS program (Brünger et al, 1998) against the remote data from the EMTS-soaked native crystal, including the bulk solvent correction and relaxing the four-fold NCS restraints at the last stage of the refinement.
Measurements of biochemical activities and circular dichroism spectra
We checked the DNA-binding properties of RecR by EMSA using both dsDNA and ssDNA. For dsDNA, we used the supercoiled circular plasmid pUC19. In all, 1 μl DR or HP RecR (∼2 μg) and 1 μl dsDNA (∼1 μg) were mixed with 25 μl buffer containing 20 mM HEPES (pH 7.5), 10 mM ammonium sulfate, 0.2% (v/v) Tween 20, 30 mM potassium chloride, 40 or 80 mM magnesium chloride and 0 or 4 mM ATP; the reaction mixtures were incubated for 1 h. RecR–DNA complexes were analyzed by 1% (w/v) agarose gel electrophoresis in 1 × TAE buffer (45 mM Tris-acetate, 1 mM EDTA). DNA bands were visualized by ethidium bromide staining. For closed circular ssDNA, we used φX174 virion DNA and the experimental conditions are the same as above, except that 1 mM ATP was used. To compare the DNA-binding affinity of the wild-type and the double mutant K23A/R27A of DR RecR, we used pUC19 and ∼0.2 μg of each protein. For linear ssDNA, we used a 30-nucleotide oligomer (5′-AAG GAG GAG AAG GAG AAG AAG GAG GAG AAG-3′), which was labeled using [γ-32P]ATP (Amersham, UK) and T7 polynucleotide kinase (Promega, UK). In all, 2 μl HP RecR (∼0.7 μg) and 1 μl ssDNA (∼50 000 counts per min) were mixed with 17 μl buffer containing 10 mM Tris–HCl (pH 7.5), 50 mM sodium chloride, 10% (v/v) glycerol, 0.5 mM EDTA, 0.5 mM dithiothreitol, 80 mM magnesium chloride and 4 mM ATP; the reaction mixtures were incubated for 30 min. Then, the mixtures were loaded on a 6% (w/v) polyacrylamide gel in 0.5 × TBE buffer (45 mM Tris-borate, 1 mM EDTA), and subjected to electrophoresis for 120 min. The dried gel was exposed for autoradiography on X-ray films at −70°C for 18 h.
ATPase activity of both DR and HP RecR was measured by detection of free phosphates released from ATP using the PhosFree™ phosphate assay kit (Cytoskeleton Inc.). The malachite green specifically binds to free phosphate ions and results in an increase in absorbance at 650 nm.
Sedimentation equilibrium experiments were performed in a Beckman Optima XL-A instrument equipped with a four-hole rotor (An60Ti) with six-channel standard cells at a rotor speed of 10 000 rpm for 24 h at 20°C. Each of DR RecR and HP RecR was dissolved at 1 mg ml−1 concentration in a buffer solution consisting of 50 mM Tris–HCl (pH 7.5) and 100 mM NaCl. For the molecular mass analysis, data were fit to an ideal, single-component model using partial specific volumes of 0.7387 and 0.7405 cm3 g−1 for DR RecR and HP RecR, respectively, and a solution density of 1.05 g cm−3.
Circular dichroism spectra were recorded on a JASCO J-715 spectropolarimeter using a 0.2 cm path length with 2.0 nm bandwidth and 8 s response time. HP RecR was dissolved at 0.1 mg ml−1 concentration in a buffer solution consisting of 20 mM HEPES (pH 7.5), 10 mM ammonium sulfate, 30 mM potassium chloride, 0.2% (v/v) Tween 20, 2 mM MgCl2 and 0–0.2 mM ATP. The standard far-UV spectra were collected at 20°C with a scan speed of 50 nm min−1 and 0.2 nm scan resolution. A total of 10 individual scans were averaged, followed by subtraction of the solvent signal.
Accession numbers
Atomic coordinates and the structure factor data have been deposited in the Protein Data Bank under ID code 1VDD.
Supplementary Material
Supplementary Figure 1
Acknowledgments
We are grateful to SM Shim, S-H Baek, KS Ha (SNU), SY Jeong (KJIST), Prof. KJ Hong and Mr JH Kang (Catholic University) for assistance during this work. We thank the Inter-University Center for Natural Science Research Facilities, Seoul National University, for providing the X-ray equipment. We also thank Dr HS Lee and his staff at beamline BL-6B of Pohang Light Source, Korea and Prof. Sakabe and his staff at beamline BL-18B of Photon Factory, Japan for assistance during synchrotron data collection. This work was supported by the Korea Ministry of Science and Technology (NRL-2001, grant no. M10318000132) and the Center for Functional Analysis of Human Genome (21st Century Frontier Program). BIL and KHK were supported by the BK21 Fellowship.
References
- Alley SC, Shier VK, Abel-Santos E, Sexton DJ, Soumillion P, Benkovic SJ (1999) Sliding clamp of the bacteriophage T4 polymerase has open and closed subunit interfaces in solution. Biochemistry 38: 7696–7709 [DOI] [PubMed] [Google Scholar]
- Alonso JC, Stiege AC, Dobrinski B, Lurz R (1993) Purification and properties of the RecR protein from Bacillus subtilis 168. J Biol Chem 268: 1424–1429 [PubMed] [Google Scholar]
- Amundsen SK, Smith GR (2003) Interchangeable parts of the Escherichia coli recombination machinery. Cell 112: 741–744 [DOI] [PubMed] [Google Scholar]
- Aono S, Hartsch T, Schulze-Gahmen U (2003) Crystallization of a member of the recFOR DNA repair pathway, RecO, with and without bound oligonucleotide. Acta Crystallogr D 59: 576–579 [DOI] [PubMed] [Google Scholar]
- Aravind L, Leipe DD, Koonin EV (1998) Toprim—a conserved catalytic domain in type IA and II topoisomerase, DnaG-type primases, OLD family nuclease and RecR proteins. Nucleic Acids Res 26: 4205–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res 27: 1223–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayora S, Alonso JC (1997) Purification and characterization of the RecF protein from Bacillus subtilis 168. Nucleic Acids Res 25: 2766–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayora S, Stiege AC, Lurz R, Alonso JC (1997) Bacillus subtilis 168 RecR protein–DNA complexes visualized as looped structures. Mol Gen Genet 254: 54–62 [DOI] [PubMed] [Google Scholar]
- Beernink HT, Morrical SW (1999) RMPs: recombination/replication mediator proteins. Trends Biochem Sci 24: 385–389 [DOI] [PubMed] [Google Scholar]
- Berger JM, Gamblin SJ, Harrison SC, Wang JC (1996) Structure and mechanism of DNA topoisomerases II. Nature 379: 225–232 [DOI] [PubMed] [Google Scholar]
- Bork JM, Cox MM, Inman RB (2001) The RecOR proteins modulate RecA protein function at 5′ ends of single-stranded DNA. EMBO J 20: 7313–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT (1992) The free R-value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355: 472–474 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Christmann M, Tomicic MT, Roos WP, Kaina B (2003) Mechanisms of human DNA repair: an update. Toxicology 193: 3–34 [DOI] [PubMed] [Google Scholar]
- Clark AJ (1991) Rec genes and homologous recombination proteins in Escherichia coli. Biochimie 73: 523–532 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Courcelle J, Carswell-Crumpton C, Hanawalt PC (1997) recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci USA 94: 3714–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Ganesan AK, Hanawalt PC (2001) Therefore, what are recombination proteins there for? BioEssays 23: 463–470 [DOI] [PubMed] [Google Scholar]
- Cox MM (1999) Recombinational DNA repair in bacteria and the RecA protein. Prog Nucleic Acid Res Mol Biol 63: 311–366 [DOI] [PubMed] [Google Scholar]
- Cox MM (2001a) Historical overview: searching for replication help in all of the rec places. Proc Natl Acad Sci USA 98: 8173–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM (2001b) Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu Rev Genet 35: 53–82 [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Shndler SJ, Marians KJ (2000) The importance of repairing stalled replication forks. Nature 404: 37–41 [DOI] [PubMed] [Google Scholar]
- Doherty AJ, Serpell LC, Ponting CP (1996) The helix–hairpin–helix DNA-binding motif: a structural basis for non-specific–specific recognition of DNA. Nucleic Acids Res 24: 2488–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin TJ IV, Kolodner RD (1990) Purification and preliminary characterization of the Escherichia coli K-12 recF protein. J Bacteriol 172: 6291–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, Lloyd S, Tainer JA (1998) MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol 5: 1058–1064 [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J (1996) Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell 87: 297–306 [DOI] [PubMed] [Google Scholar]
- Hegde SP, Qin M-H, Li X-H, Atkinson MA, Clark AJ, Rajagopalan M, Madiraju MV (1996) Interactions of RecF protein with RecO, RecR, and single-stranded DNA binding proteins reveal roles for the RecF–RecO–RecR complex in DNA repair and recombination. Proc Natl Acad Sci USA 93: 14468–14473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani MM, O'Donnell M (2000) A tale of toroids in DNA metabolism. Nat Rev Mol Cell Biol 1: 22–30 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1995) Dali: a network tool for protein structure comparison. Trends Biochem Sci 20: 478–480 [DOI] [PubMed] [Google Scholar]
- Ikegami T, Kuraoka I, Saijo M, Kodo N, Kyogoku Y, Morikawa K, Tanaka K, Shirakawa M (1998) Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA. Nat Struct Biol 5: 701–706 [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, O'Donnell M, Kuriyan J (2001) Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 106: 417–428 [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kantake N, Madiraju MV, Sugiyama T, Kowalczykowski SC (2002) Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: a common step in genetic recombination. Proc Natl Acad Sci USA 99: 15327–15332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Ito T, Wagner G, Richardson CC, Ellenberger T (2003) Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol Cell 11: 1349–1360 [DOI] [PubMed] [Google Scholar]
- Keck JL, Roche DD, Lynch AS, Berger JM (2000) Structure of the RNA polymerase domain of E. coli primase. Science 287: 2482–2486 [DOI] [PubMed] [Google Scholar]
- Kitayama S, Narumi I, Kikuchi M, Watanabe H (2000) Mutation in recR gene of Deinococcus radiodurans and possible involvement of its product in the repair of DNA interstrand cross-links. Mutat Res 461: 179–187 [DOI] [PubMed] [Google Scholar]
- Klug A, Schwabe JW (1995) Protein motifs 5. Zinc fingers. FASEB J 9: 597–604 [PubMed] [Google Scholar]
- Kong XP, Onrust R, O'Donnell M, Kuriyan J (1992) Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69: 425–437 [DOI] [PubMed] [Google Scholar]
- Kovall R, Matthews BW (1997) Toroidal structure of λ-exonuclease. Science 277: 1824–1827 [DOI] [PubMed] [Google Scholar]
- Kreuzer KN (2000) Recombination-dependent DNA replication in phage T4. Trends Biochem Sci 25: 165–173 [DOI] [PubMed] [Google Scholar]
- Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J (1994) Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79: 1233–1243 [DOI] [PubMed] [Google Scholar]
- Kuzminov A (1999) Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol Mol Biol Rev 63: 751–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labahn J, Scharer OD, Long A, Ezaz-Nikpay K, Verdine GL, Ellenberger TE (1996) Structural basis for the excision repair of alkylation-damaged DNA. Cell 86: 321–329 [DOI] [PubMed] [Google Scholar]
- Lee BI, Kim KH, Shim SM, Ha KS, Yang JK, Yoon H-J, Ha JY, Suh SW (2004) Crystallization and preliminary X-ray crystallographic analysis of the RecR protein from Deinococcus radiodurans, a member of the RecFOR DNA-repair pathway. Acta Crystallogr D 60: 379–381 [DOI] [PubMed] [Google Scholar]
- Lee JY, Chang C, Song HK, Moon J, Yang JK, Kim H-K, Kwon S-T, Suh SW (2000) Crystal structure of NAD+-dependent DNA ligase: molecular architecture and functional implications. EMBO J 19: 1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima CD, Wang JC, Mondragon A (1994) Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature 367: 138–146 [DOI] [PubMed] [Google Scholar]
- Luisi-DeLuca C (1995) Homologous pairing of single-stranded DNA and superhelical double-stranded DNA by RecO protein from Escherichia coli. J Bacteriol 177: 566–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi-DeLuca C, Kolodner R (1994) Purification and characterization of the Escherichia coli RecO protein. Renaturation of complementary single-stranded DNA molecules catalyzed by the RecO protein. J Mol Biol 236: 124–138 [DOI] [PubMed] [Google Scholar]
- Lusetti SL, Shaw JJ, Cox MM (2003) Magnesium ion-dependent activation of the RecA protein involves the C terminus. J Biol Chem 278: 16381–16388 [DOI] [PubMed] [Google Scholar]
- Mackay JP, Crossley M (1998) Zinc fingers are sticking together. Trends Biochem Sci 23: 1–4 [DOI] [PubMed] [Google Scholar]
- Madiraju MV, Clark AJ (1992) Evidence for ATP binding and double-stranded DNA binding by Escherichia coli RecF protein. J Bacteriol 174: 7705–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marians KJ (2000) PriA-directed replication fork restart in Escherichia coli. Trends Biochem Sci 25: 185–189 [DOI] [PubMed] [Google Scholar]
- Moarefi I, Jeruzalmi D, Turner J, O'Donnell M, Kuriyan J (2000) Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J Mol Biol 296: 1215–1223 [DOI] [PubMed] [Google Scholar]
- Morimatsu K, Kowalczykowski SC (2003) RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell 11: 1337–1347 [DOI] [PubMed] [Google Scholar]
- Mullen GP, Wilson SH (1997) DNA polymerase β in abasic site repair: a structurally conserved helix–hairpin–helix motif in lesion detection by base excision repair enzymes. Biochemistry 36: 4713–4717 [DOI] [PubMed] [Google Scholar]
- Nichols MD, DeAngelis K, Keck JL, Berger JM (1999) Structure and function of an archaeal topoisomerase IV subunit with homology to the meiotic recombination factor Spo11. EMBO J 18: 6177–6188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peláez AI, Ribas-Aparicio RM, Gómez A, Rodicio MR (2001) Structural and functional characterization of the recR gene of Streptomyces. Mol Genet Genomics 265: 663–672 [DOI] [PubMed] [Google Scholar]
- Podobnik M, Mclnerney P, O'Donnell M, Kuriyan J (2000) A Toprim domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J Mol Biol 300: 353–362 [DOI] [PubMed] [Google Scholar]
- Schmiedeskamp M, Klevit RE (1994) Zinc finger diversity. Curr Opin Struct Biol 4: 28–35 [Google Scholar]
- Shan Q, Bork JM, Webb BL, Inman RB, Cox MM (1997) RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J Mol Biol 265: 519–540 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA (1995) Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J 14: 4108–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K, Kolodner RD (1994) Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J Biol Chem 269: 30005–30013 [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1: 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb BL, Cox MM, Inman RB (1995) An interaction between the Escherichia coli RecF and RecR proteins dependent on ATP and double-stranded DNA. J Biol Chem 270: 31397–31404 [DOI] [PubMed] [Google Scholar]
- Webb BL, Cox MM, Inman RB (1997) Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell 91: 347–356 [DOI] [PubMed] [Google Scholar]
- Webb BL, Cox MM, Inman RB (1999) ATP hydrolysis and DNA binding by the Escherichia coli RecF protein. J Biol Chem 274: 15367–15374 [DOI] [PubMed] [Google Scholar]
- West SC (2003) Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 4: 435–445 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1


