Abstract
Biofilms formed by dissimilatory metal reducers are of interest to develop permeable biobarriers for the immobilization of soluble contaminants such as uranium. Here we show that biofilms of the model uranium-reducing bacterium Geobacter sulfurreducens immobilized substantially more U(VI) than planktonic cells and did so for longer periods of time, reductively precipitating it to a mononuclear U(IV) phase involving carbon ligands. The biofilms also tolerated high and otherwise toxic concentrations (up to 5 mM) of uranium, consistent with a respiratory strategy that also protected the cells from uranium toxicity. The enhanced ability of the biofilms to immobilize uranium correlated only partially with the biofilm biomass and thickness and depended greatly on the area of the biofilm exposed to the soluble contaminant. In contrast, uranium reduction depended on the expression of Geobacter conductive pili and, to a lesser extent, on the presence of the c cytochrome OmcZ in the biofilm matrix. The results support a model in which the electroactive biofilm matrix immobilizes and reduces the uranium in the top stratum. This mechanism prevents the permeation and mineralization of uranium in the cell envelope, thereby preserving essential cellular functions and enhancing the catalytic capacity of Geobacter cells to reduce uranium. Hence, the biofilms provide cells with a physically and chemically protected environment for the sustained immobilization and reduction of uranium that is of interest for the development of improved strategies for the in situ bioremediation of environments impacted by uranium contamination.
INTRODUCTION
In their natural environment microorganisms are most often found as surface-attached communities or biofilms (1, 2). Biofilm cells are encased in a matrix of exopolymeric substances (EPSs), such as polysaccharides, nucleic acids, lipids, and proteins, which promote adhesion to surfaces and mechanically stabilize the multilayered communities (3). In addition, the EPS matrix functions as a hydrated, catalytic microenvironment for the cells that promotes the adsorption of nutrients and, in some cases, their extracellular processing to facilitate their assimilation (3). Although the biofilm structure is dynamic and can change to minimize mass transfer limitations (4), gradients of nutrients and waste products may form within the biofilms, and as a result, the chemical composition of the matrix and the physiology of the biofilm cells are heterogeneous (5). This unique physical and chemical microenvironment also makes the physiology of biofilm cells substantially different from that of planktonic cells and leads to some general, biofilm-specific traits, such as increased resistance to antimicrobials (6), greater catalytic rates (7), and increased metabolic productivity (8).
Multimetal resistance is also a common biofilm trait (9), and for this reason, biofilms have attracted interest for applications in metal bioremediation (10). The polyionic nature of the biofilm matrix can provide, for example, 20- to 30-fold more charged groups for metal sorption than planktonic cells (11). This limits the diffusion of the metals within the biofilm and their permeation inside the cells, thus reducing the susceptibility of the biofilm cells to metal toxicity compared to that of their planktonic counterparts (9). The chemical and physiological heterogeneity of the biofilms also establishes pH and redox gradients across the biofilm matrix, which may influence metal speciation (9). The chemistry of some metals is such that their speciation affects their solubility and, therefore, their potential for spread and their bioavailability. This is particularly important for uranium (U), which often persists in contaminated groundwater and sediments as the soluble uranyl cation (UO22+) containing the oxidized U(VI) species. Its solubility facilitates the spread of the contaminant plume far away from the source (12) and results in volumes of contaminated groundwater and sediments too large to permit standard excavation-and-removal and pump-and-treat remediation approaches (13). Hence, a promising bioremediation approach would be to immobilize the U(VI) contaminant and reduce it to the U(IV) species, which is less soluble and can precipitate out of solution under the right redox conditions and pH (12). This prevents the migration of the contaminant, reduces its bioavailability, and minimizes the potential for exposure to humans and other living components of the ecosystem.
One way to reduce U(VI) to U(IV) is by the action of some dissimilatory metal-reducing bacteria, which can gain energy for growth by coupling the oxidation of various electron donors to the reduction of the uranyl cation (14–16). This metabolic ability can be stimulated in situ with field-scale additions of electron donors and results in the removal of the uranyl cation from the groundwater and its accumulation in the sediments as sparingly soluble, less mobile U(IV) minerals (17–21). Field-scale additions of electron donors often stimulate the growth and activity of dissimilatory metal-reducing microorganisms within the family Geobacteraceae (17, 19, 20, 22). Studies with the model representative Geobacter sulfurreducens indicate that the reduction of U by these organisms is extracellular; this prevents the permeation and reductive precipitation of the radionuclide inside the cell envelope, thereby protecting the cell from its toxicity (23). The extracellular reduction of U by G. sulfurreducens is primarily mediated by hair-like protein filaments or pili and, to a lesser extent, by c cytochromes of the outer membrane (23). The pili of Geobacter are conductive (24), and their expression levels are linearly proportional to the amount of U reductively precipitated by planktonic cells (23). Pilus expression in G. sulfurreducens is also required to form an electroactive biofilm (25, 26), where the cells are encased in an EPS matrix containing not only the pili but also exopolysaccharide and c cytochromes (27). This suggests that Geobacter biofilms may be particularly suitable to catalyze the speciation of U while providing a microenvironment that also protects cells from its toxicity.
Biofilms are particularly prevalent at the matrix-well screen interface and within rock fractures in subsurface environments contaminated with U, thereby influencing its mobility and speciation (13). However, the contribution of Geobacter biofilms to these processes and their potential use as bioremediation tools have not been fully investigated. This contrasts with the availability of studies about U transformations mediated by Geobacter planktonic cells (14, 15, 23, 28, 29) or by biofilms formed by other metal-reducing bacteria, such as Desulfovibrio (30, 31) and Shewanella (32, 33). Hence, we investigated U transformations mediated by biofilms of G. sulfurreducens. The results indicate that the biofilms have an enhanced capacity to immobilize and reduce U compared to planktonic cells and also tolerate exposure to higher concentrations of the contaminant for prolonged periods of time. Furthermore, microscopic, genetic, and spectroscopic studies of the U-reducing biofilms indicated that the radionuclide was reduced extracellularly in a catalytic process influenced by the biofilm structure and the presence of redox components of the biofilm matrix, most significantly, the conductive pili. These findings support the notion that Geobacter biofilms contribute to the immobilization and reduction of U in the subsurface and highlight their potential use as permeable biobarriers for the in situ bioremediation of U.
MATERIALS AND METHODS
Strains and culture conditions.
Wild-type (WT) Geobacter sulfurreducens PCA (ATCC 51573), a pilin-deficient mutant (24) (herein designated the pilA mutant), and its genetically complemented strain, pRG5::pilA (24) (herein designated the pilA+ strain), were routinely cultured in freshwater (FW) medium (34) modified as previously described (23) and supplemented with 15 mM acetate and 40 mM fumarate (FWAF). The medium was dispensed into tubes or serum bottles, sparged with N2-CO2 (80:20), sealed with butyl rubber stoppers (Bellco) and aluminum tear-off seals (Wheaton), and autoclaved for 30 min. Unless otherwise indicated, incubations were at 30°C.
Biofilm cultures.
Biofilms were grown either on 6-well, cell culture-treated polystyrene plates (Corning) or on glass coverslips assembled vertically, four at a time, onto rubber stoppers, as described previously (26). Briefly, the glass coverslips used for the stopper assemblies were first acid washed overnight in a bath of 50/50 (vol/vol) HCl-NO3− or 15% (vol/vol) HCl-H2O and rinsed thoroughly with double-distilled H2O before they were inserted into slits cut on inverted stoppers. Each coverslip assembly was placed upside down (coverslips down) in a sterile 50-ml conical tube (Corning) and submerged in FW medium lacking vitamins, minerals, acetate, and fumarate before it was sterilized by autoclaving, as previously described (26). The assembly was then transferred to a clean conical tube. The biofilm chambers (either wells of plates or conical tubes with coverslip assemblies) were filled with 6 or 20 ml of FWAF medium, respectively, and inoculated to a final optical density at 600 nm (OD600) of 0.04 with an early-stationary-phase FWAF culture. The chambers were incubated at 30°C for 24, 48, or 72 h, as specified below, inside a vinyl glove bag (Coy Labs) with an H2-CO2-N2 (7:10:83) atmosphere. At the end of the incubation period, the culture supernatants were decanted from the biofilm vessels and the biofilms were washed once with sterile, anaerobically prepared wash buffer (29).
When indicated, the biofilm biomass in the samples was estimated as the total amount of cell protein. To do this, the biofilm biomass was scraped off from the surface with a sterile plastic spatula and harvested by centrifugation (5 min, 12,000 × g), and the biofilm cells were then lysed in 2 M NaOH at 100°C for 1 h. The lysate solution was allowed to cool before neutralizing it with an equal volume of 2 M HCl. After another cycle of centrifugation to remove cellular debris, the supernatant, which contained the soluble protein released after lysing the cells, was analyzed for protein content using a Pierce Microplate BCA protein assay kit (reducing reagent compatible; Thermo Scientific) with bovine serum albumin standards, according to the manufacturer's specifications. Protein was measured as the OD562 on a Tecan Sunrise plate reader (Tecan, Inc.).
U immobilization assays with resting planktonic cell suspensions and biofilms.
The ability of planktonic or biofilm cells to immobilize U was assayed by monitoring the removal of U(VI), provided as uranyl acetate (1 mM or as indicated below), at 30°C by resting planktonic cells and biofilms, using protocols adapted from those described previously (23, 29). Planktonic cells were harvested from mid-exponential-phase cultures (OD600, 0.3 to 0.5) grown for 48 h under pilin-inducing conditions (25°C) and suspended in 10 ml of reaction buffer (OD600, 0.1) as described before (23). Resting biofilms were prepared by first growing the surface-attached communities in FWAF medium for 24, 48, or 72 h on the coverslip-stopper assemblies described above. The culture broth of the biofilm culture was then decanted, and the coverslip assembly was gently rinsed with sterile, anaerobically prepared wash buffer (29). To initiate the assay, the assemblies were submerged upside down in 50-ml conical tubes containing 20 ml of reaction buffer (23, 29) supplemented with 20 mM sodium acetate and 1 mM uranyl acetate (Electron Microscopy Sciences), which had been prepared from stock solutions in 30 mM bicarbonate buffer. Heat-killed planktonic cells (23) and uninoculated biofilm controls were also included to rule out any abiotic removal activity and/or absorption. Resting planktonic cells, resting biofilms, and the controls were incubated for up to 24 h at 30°C. Supernatant samples (500 μl) were periodically removed during incubation, filtered (pore size, 0.22 μm; Millex-GS filter; Millipore), acidified in 2% nitric acid (500 μl), and stored at −20°C. All procedures were performed inside an anaerobic glove bag, as described above. The concentration of U(VI) in the acidified samples was measured using an inductively coupled plasma mass spectrometer (ICP-MS; Micromass; Thermo Scientific) or a kinetic phosphorescence analyzer (KPA; Chemchek) and was used to estimate the total amount of U immobilized in each sample. When indicated, the amount of U immobilized by the planktonic and biofilm cells was normalized by the cell biomass, which was calculated as the amount of total cell protein released from the samples after lysis with 2 M NaOH (100°C for 1 h) and neutralization with 2 M HCl, as described above.
U valence and speciation by XAS analyses.
The valence and speciation of the U immobilized by resting biofilms were estimated by X-ray absorption near edge spectroscopy (XANES) and extended X-ray absorption fine structure (EXAFS) analyses, respectively. For these analyses, biofilms grown on coverslip-stopper assemblies and exposed to U for 24 h, as described above, were gently rinsed with wash buffer, scraped off the coverslips, and suspended in 2 ml of reaction buffer. The biofilm biomass was then harvested by centrifugation (12,000 × g, 10 min), loaded into custom-made plastic holders, and stored at −80°C (23). All procedures were carried out in an anaerobic chamber, and samples were kept frozen during X-ray absorption spectroscopy (XAS) measurements. The XAS measurements were performed with a multielement Ge detector in fluorescence mode using the PNC-CAT beamline 20-BM at the Advanced Photon Source (Argonne National Laboratory) and standard beamline parameters, as described elsewhere (35). XANES measurements were used to calculate the fraction of the immobilized U that had been reduced to U(IV) by linear combination fitting of the spectrum with U(VI) and U(IV) standards. The spectra were energy aligned using a simultaneously measured uranyl nitrate standard. The U LIII-edge EXAFS spectrum was also collected for the biofilm samples and modeled to determine the atomic coordination about U, as previously described (23). The number of axial oxygen (Oax) atoms calculated in the EXAFS spectral analyses was also used to estimate the amount of U(VI) and U(IV) in the samples, as there are two double-bonded Oax atoms for each U(VI) atom in the uranyl cation (Oax=U=Oax) and none for U(IV) (36).
Vitality fluorescent assays.
The respiratory activity of 48-h-old biofilms after exposure to various concentrations (1, 2.5, and 5 mM) of uranyl acetate for 24 h and that of biofilms not exposed to uranyl acetate (referred to as the 0 mM biofilm controls) were assayed using a fluorescent RedoxSensor vitality kit (Invitrogen), with the protocols being adapted from those described previously (23). The biofilms were first grown for 48 h on coverslip assemblies, rinsed gently with reaction buffer, and incubated in reaction buffer in the presence or absence of the uranyl acetate. The experiment also included negative controls (referred to as the 0* mM controls) in which the 0 mM biofilms were first treated for 5 min with electron transport chain uncouplers (a mix of sodium azide [10 mM] and carbonyl cyanide 3-chlorophenylhydrazone [CCCP; 10 μM]) provided in the RedoxSensor vitality kit (Invitrogen). After 24 h of incubation, the reaction buffer was decanted from the tubes and the assemblies were washed once with wash buffer. The biofilm biomass was scraped from the coverslips and suspended in 1 ml of reaction buffer, vortexed briefly, mixed 1:1 with the redox dye solution, and incubated at room temperature for 10 min before measuring the dye's fluorescence (490-nm excitation, 520-nm emission) using a SpectraMax M5 plate reader (Molecular Devices). Separate aliquots of the biofilm samples were stained with Syto 9 (Invitrogen) following the manufacturer's recommendations to determine the total biofilm biomass. The respiratory activity of the biofilms was estimated as the vitality index, i.e., the relative fluorescence emission from the RedoxSensor dye normalized by the fluorescence emission from the Syto 9 dye for six replicate samples (three biological samples and two technical replicates for each). Statistically significant changes in vitality indexes were determined in pairwise comparisons to the 0 mM control biofilms using the t-test function of Microsoft Excel software.
Microscopy.
A scanning electron microscope (SEM) was used to image 48-h-old WT biofilms, which were grown on coverslip assemblies as described above, except that round (12-mm-diameter) rather than square glass coverslips were used. The biofilms were incubated at 30°C in reaction buffer in the presence or absence of 1 mM uranyl acetate for 24 h and washed once in wash buffer before fixing the cells at 4°C for 1 to 2 h in 4% glutaraldehyde. After this, the biofilms were briefly rinsed in 0.1 M sodium phosphate buffer and then dehydrated in a series of ethanol washes (25%, 50%, 75%, 95%, 10 min each), followed by three 10-min washes in 100% ethanol. The samples were dried to the critical point using a Blazers 010 critical point dryer (Blazers Union Ltd.) with liquid CO2 as the transitional fluid. Once they were dry, the coverslips containing the biofilm samples were mounted on aluminum stubs using epoxy glue and coated with ∼10 nm of osmium using a NEOC-AT osmium coater (Meiwafosis Co., Ltd.). Samples were imaged with a JEOL JSM-7500F SEM equipped with an energy-dispersive spectroscopy (EDS) 30-mm2 detector crystal, which was used for elemental analyses.
Confocal laser scanning microscopy (CLSM) was also used to examine the biofilms and generate three-dimensional images for structural analyses. The WT biofilms were grown for 24, 48, and 72 h on 6-well plates, and the pilA and pilA+ biofilms were grown for 48 h. At the end of the incubation period, the culture supernatant from each well was carefully decanted, and the biofilms were stained for 15 min with LIVE/DEAD BacLight bacterial viability kit (Invitrogen) dye solution, following the manufacturer's recommendations. After staining, the biofilms were washed once in phosphate-buffered saline (PBS) and imaged on a Zeiss Pascal LSM microscope (Carl Zeiss Microscopy, LLC) equipped with an Achroplan ×40/0.80W submersible objective. Images were collected every 1.14 μm, and side and top projections of the biofilms were created using Zeiss LSM Image Browser software (Carl Zeiss Microscopy, LLC). Approximately 6 to 10 distinct fields of view (1,024 by 1,024 pixels, 0.22 μm/pixel) were imaged for each of three independent biofilm replicates, and the micrographs were analyzed with COMSTAT image analysis software using connected-volume filtration to remove noise in the data, as described previously (4). The biofilm structural parameters quantified with the COMSTAT analyses are listed in Table S2 in the supplemental material.
SDS-PAGE and heme staining of proteins of the biofilm EPS matrix.
The EPS matrix of 48-h-old biofilms grown on 6-well plates was extracted using a modification of previously described protocols (27, 37). Briefly, biofilms were scraped off the wells and collected in reaction buffer, and the solution was centrifuged for 10 min at 13,000 × g. After decanting the supernatant, the biofilm pellet was suspended in 1/5 volume of TNE (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM EDTA) and vortexed for 1 min. SDS was then added to a final concentration of 0.1% (wt/vol), and the solution was mixed at room temperature for 5 min. The samples were then passed 10 times through an 18-gauge needle and centrifuged at 15,500 × g for 20 min to collect the insoluble sheared biological fraction. The resulting pellet, which predominantly contained the biofilm exopolysaccharide and associated proteins, was washed 5 times to remove any SDS before suspending it in 10 mM Tris-HCl buffer (pH 7.5).
Heme-containing proteins in the EPS biofilm matrix were also analyzed, as previously described (27). The protein content in the matrix sample was determined with the bicinchoninic acid (BCA) protein assay, as described above, and the equivalent of 20 μg of protein of the EPS sample was boiled in SDS sample buffer for 10 min before electrophoretic analysis in a 12% Mini-Protean TGX gel (Bio-Rad) at 250 V for 30 min. Novex Sharp markers (Invitrogen) were used as molecular weight standards. Heme-containing proteins were visualized on the gel after N,N,N′,N′-tetramethylbenzidine staining, as described previously (23, 38). A duplicate gel was run in parallel and stained for total protein using Coomassie brilliant blue G-250 (Bio-Rad) according to the manufacturer's recommendations.
RESULTS AND DISCUSSION
Enhanced U immobilization and tolerance by biofilms.
The kinetics of U immobilization were investigated by measuring the amount of U, provided as uranyl acetate, removed from solution by resting 48-h-old biofilms in reference to the amount removed by pilus-expressing planktonic cells (Fig. 1A). Heat-killed planktonic cell controls and uninoculated biofilm vessel controls were also included to rule out any abiotic precipitation of U due to either components of the medium (such as phosphate) or adsorption to the biofilm coverslip assemblies and vessels used for the assays (Fig. 1A). The kinetics of U removal by the biofilms was linear throughout the 24-h-long assay (∼10 μM/h; R2 = 0.967). In contrast, the planktonic cells stopped removing U from solution after 12 h, and all the U immobilized by the planktonic cell biomass was solubilized again after 24 h of incubation. The enhanced capacity of the biofilms to immobilize U compared to that of planktonic cells (Fig. 1A) cannot be explained by differences in cell biomass because the rates of U immobilization per h, once normalized by the total amount of cell protein, were 6 times higher in the biofilms (∼2.8 μmol U immobilized per mg protein per h) than in planktonic cells (∼0.45 μmol/mg/h). Furthermore, the biofilms sustained the rates of U immobilization 2 times longer (24 h) than their planktonic counterparts. As a result, the yields of U immobilization by the biofilms at the end of the 24-h assay were 12 times greater (∼66 μmol of U immobilized per mg of biofilm biomass protein) than the maximum yields measured in the planktonic cells after 12 h (∼5.4 μmol of U per mg of protein).
FIG 1.
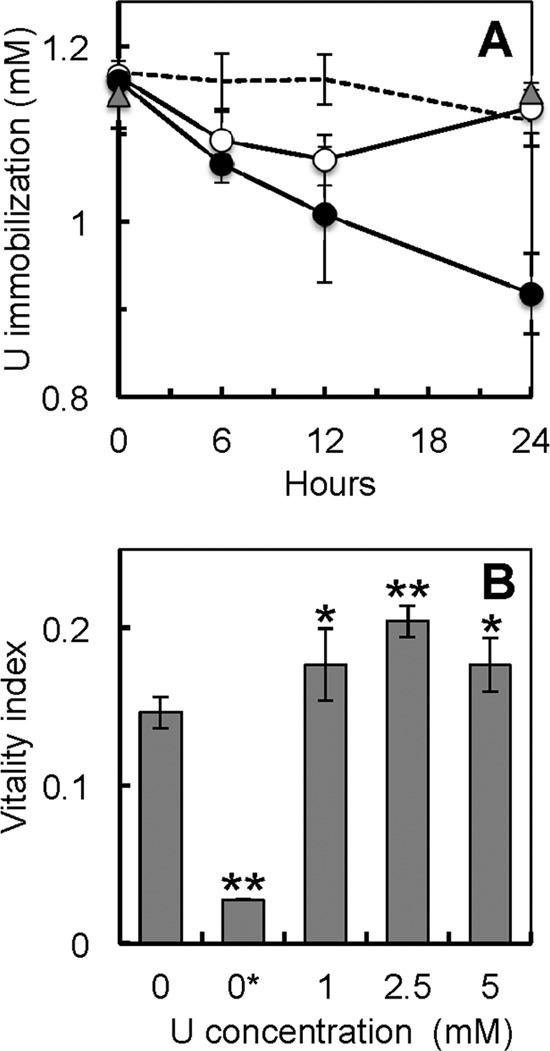
(A) Kinetics of U immobilization by 48-h-old biofilms of G. sulfurreducens (solid symbols) in reference to those of controls consisting of pilin-expressing planktonic cells (open symbols), heat-killed cells (gray triangles), and uninoculated biofilm assembly controls (dashed line). (B) Respiratory activity (measured as the vitality index) of 48-h-old biofilms exposed to increasing concentrations of uranyl acetate (0, 1, 2.5, and 5 mM) for 24 h in reference to that of a negative biofilm control chemically poisoned to uncouple the respiratory chain (0* mM controls). Shown are averages and standard deviations of 6 replicates (3 replicates for the poisoned control). Significant differences in t-test pairwise comparisons with the unexposed (0 mM) control are shown: *, P < 0.05; **, P < 0.005.
The sustained immobilization of U by 48-h-old biofilms for 24 h suggests that the biofilm cells remained viable and metabolically active throughout the assay. To investigate this, we used the fluorogenic RedoxSensor dye to measure the respiratory activities of the biofilms after exposure to increasing concentrations of U (1, 2.5, and 5 mM) for 24 h and in reference to those of biofilms incubated similarly but without U (0 mM) (Fig. 1B). Interestingly, the vitality index of the biofilms exposed to 1 mM U was greater than that of the unexposed 0 mM biofilm controls. Furthermore, increasing the concentration of U (2.5 and 5 mM) did not decrease the respiratory activities of the biofilms to or below the levels for the unexposed biofilm. The measured vitality correlated well with the respiratory activity of the biofilms because pretreatment of the biofilms with sodium azide and CCCP (0* mM controls), which are chemicals that uncouple the electron transport chain, reduced the vitality index by 80%. Furthermore, the dye used in these experiments yields green fluorescence when modified by the bacterial reductases, which are mostly located in the electron transport system of the cell envelope (39, 40). It is also unlikely that the increases in the biofilm's respiratory rate with U were due to a cellular response of the cells to U toxicity, because the vitality index serves as a proxy of viability and overall cell health in G. sulfurreducens cells after U exposure (23). For example, exposure of pilus-expressing planktonic cells to 1 mM U for only 6 h reduced the respiratory activity of the cells by 70%, because of the toxic effects of the U that permeates and mineralizes in the cell envelope (23), yet when the cells were encased in the protective biofilm microenvironment, they tolerated high, otherwise toxic levels of U, and their metabolic activities were stimulated as well.
U reduction by the biofilms.
As the reaction of interest during U bioremediation is the reduction of the soluble U(VI) species to the less mobile U(IV), we used XANES to investigate if any of the U immobilized by the biofilms had been reduced to U(IV). For these experiments, the 48-h-old biofilms for which the results are presented in Fig. 1A were collected at the end of the 24-h U challenge, and the valence of the biofilm-associated U was analyzed by XANES in reference to the valences of U(VI) and U(IV) standards. Approximately 65% ± 5% of the U immobilized by the biofilms was in the reduced state, U(IV) (see Table S1 in the supplemental material). Consistent with the reductive precipitation of U by the biofilms, SEM-EDS analyses revealed needle-like, extracellular precipitates coating the microcolonies (Fig. 2A) that contained U (see Fig. S1 in the supplemental material). The preferential localization of U minerals on the top regions of the biofilm is consistent with the metabolic stratification of cells within electroactive biofilms of G. sulfurreducens (41). As the biofilms grow, their thickness increases and a gradient of acetate is formed across the biofilm. This concentrates the metabolically active cells in the top regions of the biofilm, where acetate is more available. As a result of this metabolic stratification, U is preferentially reduced in the metabolically active, top biofilm stratum (41).
FIG 2.
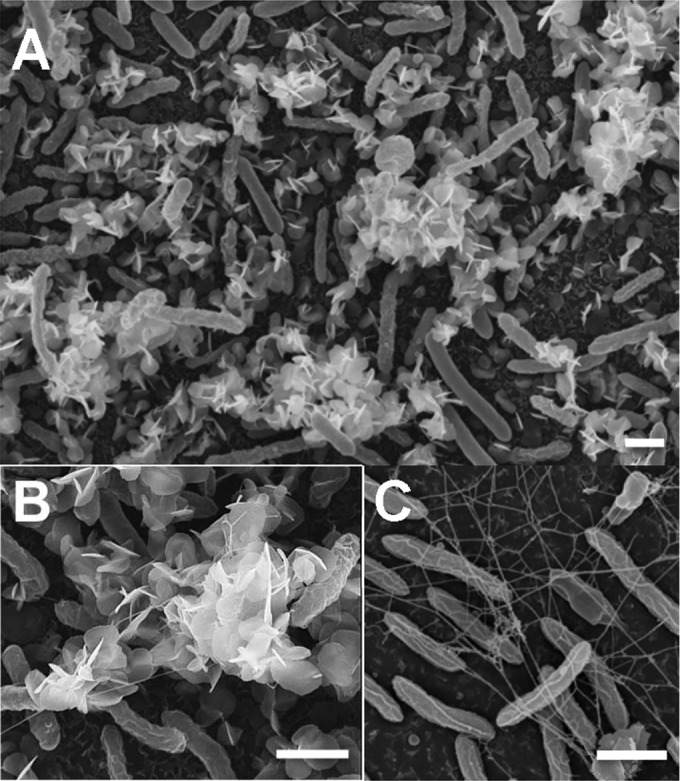
SEM micrographs of 48-h-old biofilms exposed to 1 mM uranyl acetate for 24 h (A and B) showing the extracellular needle-like, white precipitates of uranium associated with the biofilm microcolonies. (C) Control biofilms not exposed to U are also shown, allowing visualization of the network of extracellular filaments that connect the biofilm cells. Bars, 1 μm.
SEM micrographs also revealed the presence of extracellular filaments interspersed with the U precipitates (Fig. 2B). The dense filament network was more clearly visible in biofilm controls not exposed to U (Fig. 2C). Some filaments had diameters (<4 nm) that closely matched the diameters reported for the conductive pili of G. sulfurreducens (24), whereas others had larger diameters (ca. 15 to 20 nm) within the ranges reported for dehydrated EPS fibers (27). The conductive pili of G. sulfurreducens are the primary sites for extracellular immobilization and the reduction of U by pilin-expressing planktonic cells (23) and are also required for the formation of electroactive biofilms (25, 26). In addition to containing the conductive pili, the EPS matrix of G. sulfurreducens also anchors several c cytochromes involved in metal reduction (27). Thus, the Geobacter matrix not only provides an extended matrix for U immobilization but also contains redox-active components which could mediate its reduction.
U immobilization and reduction during biofilm development.
As the biofilms develop, their thickness increases and more electroactive matrix is available to immobilize and reduce U. Hence, we hypothesized that older and thicker biofilms would immobilize and reduce more U than younger and thinner biofilms. To test this hypothesis, we studied U immobilization and reduction as a function of biofilm age (24 h, 48 h, and 72 h) (see Table S1 in the supplemental material). The biofilms were also imaged by confocal microscopy (see Fig. 4), and the confocal micrographs were analyzed with COMSTAT software (4) to estimate the thickness and other structural parameters of the biofilms (see Table S2 in the supplemental material).
FIG 4.
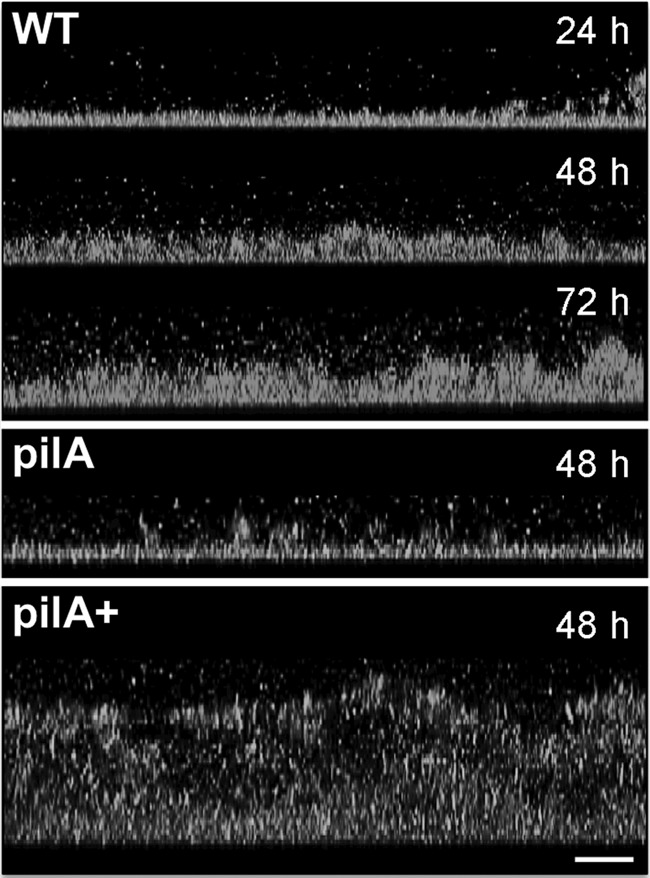
CLSM micrographs showing side-view projections of WT, pilA, and pilA+ biofilms grown for 24, 48, and/or 72 h. Bar, 20 μm. The biofilm cells were stained with dye solution from the BacLight viability kit. Color top-view projections for these images are shown in Fig. S2 in the supplemental material.
We first studied U immobilization during biofilm development. Although the capacity of the biofilms to immobilize U increased steadily as they aged (Fig. 3A) and their thickness increased (Fig. 4), the amount of U immobilized per biomass unit did not change significantly during biofilm development (see Fig. S3A in the supplemental material). For example, young (24-h-old) biofilms had less biofilm biomass, thickness, and surface area (number of cells per biofilm layer) than older (48-h- and 72-h-old) biofilms (see Fig. S4A to C in the supplemental material). Nevertheless, despite being less dense, they immobilized more U per biofilm biomass unit than older biofilms (see Fig. S3A in the supplemental material). In contrast, the surface coverage by all of the biofilms was similar and neared confluence (81 to 92%) (see Fig. S4D in the supplemental material). The surface-to-volume ratio also remained relatively constant during biofilm development (see Fig. S4E in the supplemental material), indicating that the area of the biofilm exposed to the liquid milieu relative to the biofilm volume is the same. Furthermore, the biofilm topography was relatively constant during biofilm development as well, as indicated by the similar roughness coefficients measured in all the biofilms (see Fig. S4F in the supplemental material). Taken together, the results support a model in which achieving biofilm confluence early on in biofilm development is critical to promote U immobilization. As the biofilms age, they adapt their structure to maintain a relatively constant topography that maximizes exposure to the liquid milieu. As a result, U is preferentially immobilized in the biofilm regions exposed to the liquid milieu, which is further supported by the preferential localization of U on the outer layers of the biofilms, as revealed in SEM micrographs (Fig. 2).
FIG 3.
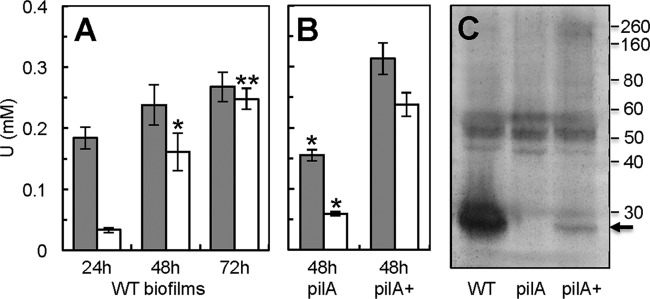
(A and B) Total U immobilized (gray bars) and fraction reduced to U(IV) (open bars) by WT biofilms grown for 24, 48, and 72 h (A) and by 48-h-old biofilms of the pilin-deficient pilA mutant and the hyperpiliated pilA+ strain (B). The biofilms were exposed to 1 mM uranyl acetate for 24 h. Shown are averages and standard deviations for triplicate biofilm samples (WT) and averages and standard errors for duplicate biofilm samples (pilA and pilA+ strain). Significant differences in t-test pairwise comparisons with the 24-h-old (A) or 48-h-old (B) WT biofilms are indicated: *, P < 0.05; **, P < 0.005. (C) Heme-stained SDS-PAGE of protein extracted from the EPS matrix of 48-h-old biofilms of the WT, pilA, and pilA+ strains. Numbers at the right are the relative molecular masses of protein markers (in kDa). The arrow points to the migration of the small, processed form of the OmcZ c cytochrome (OmcZS).
The fraction of the immobilized U that was reduced to U(IV) was also estimated by XANES (see Table S1 in the supplemental material). Interestingly, the yields of U reduction increased linearly with biofilm age (Fig. 3A), even when normalized by the biofilm biomass (see Fig. S3A in the supplemental material). None of the biofilm structural parameters determined in the COMSTAT analyses fit the linear trend of U reduction (see Fig. S4 in the supplemental material). As observed for U immobilization, surface coverage, surface-to-volume ratio, and roughness coefficient were unaffected by biofilm age. The biofilm parameters that changed were those related to biofilm biomass (biofilm biomass, thickness, and surface area), but they all stabilized in the 48-h- and 72-h-old biofilms rather than increase linearly. Only the total protein content of the biofilms, which accounts for the protein contributed by cells and by proteinaceous components of the biofilm matrix, increased linearly as the biofilms aged (see Fig. S5 in the supplemental material). Biofilm formation is a developmental process consisting of specific stages, such as attachment, microcolony formation, and biofilm maturation (42). As biofilms transition from one stage of development to the next, the biofilms increase their thickness and biomass but also undergo extensive gene reprogramming so that specific biofilm components and activities are expressed (9, 43). Pilus expression in G. sulfurreducens is, for example, required to transition from young, monolayered biofilms to thicker biofilms composed of several layers of cells (26), and this, in turn, is required to maintain the electroactivity of the biofilms (25). Hence, as the Geobacter biofilms age, they express more redox-active components in the biofilm matrix, and this, in turn, may increase their capacity to reduce the immobilized U to U(IV).
Role of the redox components of the biofilm matrix in U immobilization and reduction.
As the conductive pili of G. sulfurreducens are the primary site for U immobilization and reduction in planktonic cells (23) and are also required for biofilm formation (25, 26), we compared the capacity of 48-h-old biofilms of the WT, a pilin-deficient mutant (the pilA mutant), and its genetically complemented strain, the pilA+ strain, to immobilize and reduce U (Fig. 3B). The pilA biofilms had a diminished capacity to immobilize and reduce U (P = 0.07) compared to that of WT biofilms of the same age. The defect was due to the pilin deficiency because complementing the mutation in trans (in the pilA+ strain) restored the phenotypes and promoted U immobilization and reduction to levels comparable to those for the WT biofilms.
The defect in pilin production in the pilA mutant biofilms also prevented the biofilms from growing in thickness (Fig. 4) and led to ∼50% decreases in the values of other biomass-related structural parameters, such as biomass volume and surface area (number of cells per biofilm layer), compared with those for the WT biofilms (see Table S2 and Fig. S4A to C in the supplemental material). These defects are consistent with the reported role of the pili at promoting cell-cell aggregation during biofilm formation and providing the structural support required to build multilayered biofilms (26). However, despite the biomass and pilus defects, the pilA mutant biofilms immobilized more U per biomass unit than the WT biofilms. Furthermore, genetic complementation of the pilA mutation in the pilA+ strain resulted in denser biofilms (Fig. 4), consistent with the increased ability of the pilA+ hyperpiliated strains to aggregate (26). Nevertheless, despite the increased biofilm biomass, the pilA+ biofilms immobilized less U per biofilm biomass unit than any other biofilm (see Fig. S3B in the supplemental material). The lack of a clear correlation between biofilm biomass and U immobilization in the biofilms of the pilA, WT, and pilA+ strains may reflect differences in the rates of diffusion of electron donors and acceptors, as more diffusional constraints are expected in thicker biofilms (pilA+ > WT) than in thinner biofilms (i.e., pilA biofilms). Consistent with this, the pilA+ biofilms had lower surface-to-volume ratios than any other biofilms (see Fig. S4E and Table S2 in the supplemental material), indicating that less surface area of the biofilm volume is exposed to the liquid medium and available for electron donor and acceptor diffusion. Furthermore, the roughness coefficient, which provides a measure of the spatial heterogeneity of the biofilms as variations in biofilm thickness, was also the lowest in the pilA+ biofilms (see Fig. S4F in the supplemental material). The very dense pilA+ biofilms have, for example, a relatively uniform topography (indicated by a very low roughness coefficient of ∼0.06), whereas the thin pilA biofilms formed the most heterogeneous biofilms of all (indicated by the highest roughness coefficient of ∼0.32, which is almost twice the roughness coefficients for the WT) (see Table S2 in the supplemental material). Hence, the more heterogeneous structure of the pilA and WT biofilms also maximizes the biofilm surface area available for the diffusion of electron donors and acceptors.
Although the pilin-deficient pilA biofilms immobilized more U per biomass unit than any other strain, they reduced a lower fraction (∼38%) of the immobilized U to U(IV) per biomass unit than the WT (∼63%) and pilA+ (∼76%) biofilms (see Fig. S3 in the supplemental material). The capacity of the biofilms to reduce U (pilA mutant < WT strain < pilA+ strain) correlated linearly with the levels of piliation of the strains but only exponentially with the biofilm biomass (see Fig. S6 in the supplemental material). Hence, pilus expression in the strains had a more significant impact on their ability to reduce the immobilized U than the biofilm biomass itself. As defects in pilin expression can also affect cytochrome expression in planktonic cells (23), we investigated the potential contribution of defects in the expression of matrix-associated cytochromes to U reduction by WT, pilin-deficient pilA mutant, and hyperpiliated pilA+ biofilms. Although planktonic cells of the pilA mutant strain also have defects in outer membrane c cytochromes, such as OmcS (23), which is required for the optimal reduction of insoluble metal oxides (44), a protein with a relative molecular mass matching the mass of OmcS (∼47 kDa) (45) was expressed at similar levels in both the WT and pilA biofilms (Fig. 3C). The only notable difference in the profile of heme-stained proteins from the biofilm matrices was a heme-containing protein of about 30 kDa, which was particularly abundant in the WT biofilms but absent in the pilA biofilm matrix and expressed at low levels in the pilA+ matrix (Fig. 3C). The relative molecular mass of this band is within the ranges reported for OmcZS, a processed isoform of the outer membrane OmcZ c cytochrome that is secreted to the biofilm matrix (27) and is required for optimal current production by anode biofilms in microbial fuel cells (46). In vitro studies show that the purified OmcZS protein can reduce U (46), raising the possibility that this cytochrome participates in U reduction within the biofilm matrix. However, the contribution of OmcZS to U reduction is likely secondary to that of the conductive pili, as suggested by the fact that OmcZS expression was low in the biofilm matrix of the genetically complemented pilA+ strain (Fig. 3C), although this mutant removed and reduced significantly more U than any other biofilm (Fig. 3B) and did so in proportion to its piliation levels (see Fig. S6 in the supplemental material). Furthermore, pilin expression may be greater in the top regions of the biofilms (47), which are also the regions where U mineralization was more prominent (Fig. 2), whereas OmcZS preferentially localizes to the biofilm layers closer to the supporting surface (48), where mass transport limitations may reduce the availability of both acetate and U.
Taken together, the results support a model in which the expression of the conductive pili allows the biofilms to grow in biomass and thickness, adjusting their structure to maximize the diffusion of the electron donor and acceptor. Once bound to the biofilm matrix, the redox activity of the matrix-associated pili and, to a lesser extent, the cytochromes mediate the reduction of the immobilized U coupled to the oxidation of the electron donor, acetate. Because of this, pilus expression can be used as a predictor of the amount of U that can be reductively precipitated by the biofilms.
EXAFS analyses reveal pilin-specific U mineral signatures in the biofilms.
Knowledge of the chemical state and local atomic structure of the U mineral associated with the biofilms is critical to predict the long-term stability of the radionuclide during in situ bioremediation and can provide valuable insights into the biofilm components involved in the immobilization and reduction of U as well. Information on the average local atomic structure around U and the U valence can be obtained by least-squares modeling of the U LIII-edge extended X-ray absorption fine structure (EXAFS) spectrum, which corresponds to the oscillating part of the spectrum above the LIII absorption edge (starting approximately ∼50 eV above the edge). EXAFS analysis determines the type and average number and distance of atoms neighboring U. On the basis of the number of axial oxygen atoms (Oax), determined from the EXAFS modeling, we estimated the U valence state (VI or IV) independently of standards. This is possible because U(VI) has 2 Oax atoms whereas U(IV) has none. For example, an EXAFS result of an average of 1 Oax is interpreted as 50% U(VI) and 50% U(IV) (36). Hence, we collected the U LIII-edge EXAFS spectra for the 48-h-old WT and pilA biofilms, to investigate how the presence and absence of pili in the biofilm matrix affected the valence and atomic coordination of the immobilized U. The spectra, in both cases, were best described by a mixture of U(VI) and U(IV) atoms coordinated by carbon atoms. The magnitudes of the Fourier-transformed spectra and the models are shown in Fig. 5A, with the spectra being offset for clarity. A molecular moiety that is consistent with the models for both the WT and pilA biofilms is shown in the inset. Figures 5B and C show the contribution of each path in the model in the real part of the Fourier transform for the WT and pilA samples, respectively. In both the WT and pilA biofilms, the model includes two types of carbon (C) ligands. One of the C ligands is bound to two oxygen atoms of U in a bidentate fashion (C-2) and is followed by a distant carbon (C-3) atom. The other C ligand is bound to one of the oxygen atoms of U in a monodentate fashion (C-1) and is attached to a distant oxygen (Odist) atom. Multiple scattering paths from distant C-3 and Odist atoms were included in the model, which was then simultaneously refined to both spectra. The distances and σ2 values used to model the spectra are listed in Table S3 in the supplemental material, and the coordination numbers are listed in Table S4 in the supplemental material. The coordination numbers are consistent with 1 to 2 bidentate C ligands and 1 to 2 monodentate C ligands per U atom and closely matched the U-to-C distances determined for pilus-expressing planktonic cells, which use pili and, to a lesser extent, c cytochromes to immobilize and reduce the U extracellularly (23). From the number of Oax atoms, we estimated the amount of U(VI) and U(IV) in the biofilms. The WT and pilA biofilms contained approximately 48% and 25% U(IV), respectively (with an estimated uncertainty of 10%). These values are within the ranges estimated for the 48-h-old WT and pilA biofilms by bulk XANES (Fig. 3A and B), thereby providing additional, independent evidence that the pilA biofilms had a diminished U reduction capacity compared to the WT biofilms.
FIG 5.
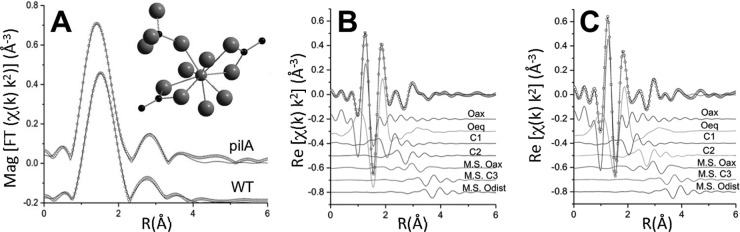
U LIII-edge XAFS analyses of 48-h-old WT and pilA biofilms. (A) Magnitude (Mag) of Fourier transform (FT) spectra (symbols) and models (lines). The spectra are offset for clarity. (Inset) U(IV) moiety consistent with the measured EXAFS spectra for both WT and pilA biofilms [small gray sphere, U(IV); large gray spheres, oxygen; black spheres, carbon]. (B and C) Real part of Fourier transform of WT (B) and pilA (C) biofilms. The components of the model are shown offset beneath the total model and the measured spectrum. χ, fine structure function; k, wave number of the photoelectron; Re, real part of FT; R, bond distance; Oeq, equatorial oxygens; M.S., multiple scattering paths.
Interestingly, the model that fit the EXAFS spectra of the WT and pilA biofilms did not require a phosphorus (P) ligand (Fig. 5A). Previous studies with planktonic cells of the pilA mutant (23) identified a correlation between the pilA mineral signature and the permeation and reduction of U inside the periplasmic space of the mutant cells. Once in the periplasmic space, U can bind C ligands in periplasmic proteins and peptidoglycan as well as membrane phospholipids, and as a result, planktonic cells of the pilA mutant required the addition of a P ligand to model the U moiety by EXAFS (23). The absence of a P ligand in the U moiety of the pilA biofilms therefore suggests that U may not have permeated inside the cells but, rather, was immobilized and reduced in the biofilm matrix, as in the WT biofilms. Hence, even without the conductive pili and the OmcZS c cytochrome, the pilA biofilm matrix immobilizes the uranyl cation, likely limiting its diffusion and permeation inside the cells. Furthermore, the pilA biofilms reduced 25% ± 10% of the immobilized U (determined by EXAFS analyses). Several heme-containing bands were detected in the pilA biofilm matrix (Fig. 3C), which could provide the redox activity needed to reduce some of the U immobilized by the pilA biofilms. The biofilm matrix encases the cells in a unique physical and chemical microenvironment, which reprograms the physiology of the biofilm very differently from that of planktonic cells (3). We showed, for example, that OmcS expression, while defective in planktonic cells of the pilA strain (23), was not affected in the pilA biofilms (Fig. 3C). Hence, other, yet to be identified components of the biofilm matrix may have contributed to the redox activities of the pilA biofilms.
Implications for U bioremediation.
U is often found in complex mixtures with toxic inorganic and organic cocontaminants (49), which can compromise the viability of planktonic cells. However, the reduced susceptibility of bacterial biofilms to toxicity by inorganic and organic pollutants makes them particularly suitable for U bioremediation applications. The ability of Geobacter biofilms to reductively precipitate U, even when it is provided at high concentrations, effectively immobilizes the contaminant as a biofilm-associated U(IV) mineral and prevents its mobility. Interestingly, providing sufficient biofilm surface coverage rather than increasing the biofilm biomass and thickness was critical for maximum U immobilization. This is advantageous for the application of biofilm-based approaches for the in situ bioremediation of U, as there is no need to stimulate the growth of very thick biofilms to create effective biopermeable barriers. On the other hand, the dependence of U reduction on the presence of specific redox components of the biofilm matrix, particularly the conductive pili, suggests that targeted approaches aimed at stimulating their synthesis in situ could significantly improve the performance of bioremediation schemes and long-term immobilization of the contaminant. Critical to these applications is a deep understanding of the genetic basis of biofilm formation and dispersal, which could provide genetic targets to monitor biofilm development and performance in situ (50). Hence, future work should be aimed at understanding the environmental parameters that regulate biofilm development and the regulatory circuits that control the biofilm structure and biofilm redox activities. Also important are insights into the ecology of Geobacter biofilms in U-impacted sites. This information could be used to elucidate the contributions of other microorganisms to the U immobilization and reduction capacity of Geobacter cells in multispecies biofilms to tailor bioremediation schemes in ways that maximize the synergistic interactions of the biofilm members and the long-term stabilization of U minerals within the biofilm.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants R01 ES017052-03 (from the National Institute of Environmental Health Sciences' Superfund program) and DE-SC0000795 (from the Biological and Environmental Research program, Office of Science, U.S. Department of Energy [DOE]) to G.R. Support from a Hensley Fellowship (College of Natural Sciences, Michigan State University) and a Biogeochemistry Environmental Research Initiative Fellowship to D.L.C. is also acknowledged. Pacific Northwest Consortium X-Ray Sciences Division facilities and research at the Advance Photon Source (APS) are supported by the DOE Basic Energy Sciences, a Major Resources Support grant from the Natural Sciences and Engineering Research Council, the University of Washington, Simon Fraser University, and the APS. APS is an Office of Science User Facility operated for the DOE's Office of Science by the Argonne National Laboratory and is supported by DOE contract DE-AC02-06CH11357.
We thank Melinda Frame, Carol Flegler, John Rotondo, Anne Otwell, APS, and Sector 20 staff at Argonne National Laboratory for assistance during different phases of this work.
Footnotes
Published ahead of print 15 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02289-14.
REFERENCES
- 1.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435–464. 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 2.Wimpenny J, Manz W, Szewzyk U. 2000. Heterogeneity in biofilms. FEMS Microbiol. Rev. 24:661–671. 10.1111/j.1574-6976.2000.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 3.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633. 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 4.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Pt 10):2395–2407. [DOI] [PubMed] [Google Scholar]
- 5.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6:199–210. 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 6.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39. 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 7.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–108. 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 8.Halan B, Buehler K, Schmid A. 2012. Biofilms as living catalysts in continuous chemical syntheses. Trends Biotechnol. 30:453–465. 10.1016/j.tibtech.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Harrison JJ, Ceri H, Turner RJ. 2007. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 5:928–938. 10.1038/nrmicro1774. [DOI] [PubMed] [Google Scholar]
- 10.Singh R, Paul D, Jain RK. 2006. Biofilms: implications in bioremediation. Trends Microbiol. 14:389–397. 10.1016/j.tim.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Fang HH. 2002. Characterization of electrostatic binding sites of extracellular polymers by linear programming analysis of titration data. Biotechnol. Bioeng. 80:806–811. 10.1002/bit.10432. [DOI] [PubMed] [Google Scholar]
- 12.O'Loughlin EJ, Boyanov MI, Antonopoulos DA, Kemner KM. 2011. Redox processes affecting the speciation of technetium, uranium, neptunium, and plutonium in aquatic and terrestrial environments. ACS Symp. Ser. 1071:477–517. 10.1021/bk-2011-1071.ch022. [DOI] [Google Scholar]
- 13.Cao B, Ahmed B, Beyenal H. 2010. Immobilization of uranium in groundwater using biofilms, p 1–37 In Shah V. (ed), Emerging environmental technologies, vol II Springer, Dordrecht, Netherlands. [Google Scholar]
- 14.Gorby YA, Lovley DR. 1992. Enzymatic uranium precipitation. Environ. Sci. Technol. 26:205–207. 10.1021/es00025a026. [DOI] [Google Scholar]
- 15.Lovley DR, Phillips EJP, Gorby YA, Landa ER. 1991. Microbial reduction of uranium. Nature 350:413–416. 10.1038/350413a0. [DOI] [Google Scholar]
- 16.Sanford RA, Wu Q, Sung Y, Thomas SH, Amos BK, Prince EK, Loffler FE. 2007. Hexavalent uranium supports growth of Anaeromyxobacter dehalogenans and Geobacter spp. with lower than predicted biomass yields. Environ. Microbiol. 9:2885–2893. 10.1111/j.1462-2920.2007.01405.x. [DOI] [PubMed] [Google Scholar]
- 17.Anderson RT, Vrionis HA, Ortiz-Bernad I, Resch CT, Long PE, Dayvault R, Karp K, Marutzky S, Metzler DR, Peacock A, White DC, Lowe M, Lovley DR. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884–5891. 10.1128/AEM.69.10.5884-5891.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Istok JD, Senko JM, Krumholz LR, Watson D, Bogle MA, Peacock A, Chang YJ, White DC. 2004. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ. Sci. Technol. 38:468–475. 10.1021/es034639p. [DOI] [PubMed] [Google Scholar]
- 19.Chang YJ, Long PE, Geyer R, Peacock AD, Resch CT, Sublette K, Pfiffner S, Smithgall A, Anderson RT, Vrionis HA, Stephen JR, Dayvault R, Ortiz-Bernad I, Lovley DR, White DC. 2005. Microbial incorporation of 13C-labeled acetate at the field scale: detection of microbes responsible for reduction of U(VI). Environ. Sci. Technol. 39:9039–9048. 10.1021/es051218u. [DOI] [PubMed] [Google Scholar]
- 20.North NN, Dollhopf SL, Petrie L, Istok JD, Balkwill DL, Kostka JE. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911–4920. 10.1128/AEM.70.8.4911-4920.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu WM, Carley J, Gentry T, Ginder-Vogel MA, Fienen M, Mehlhorn T, Yan H, Caroll S, Pace MN, Nyman J, Luo J, Gentile ME, Fields MW, Hickey RF, Gu B, Watson D, Cirpka OA, Zhou J, Fendorf S, Kitanidis PK, Jardine PM, Criddle CS. 2006. Pilot-scale in situ bioremedation of uranium in a highly contaminated aquifer. 2. Reduction of U(VI) and geochemical control of U(VI) bioavailability. Environ. Sci. Technol. 40:3986–3995. 10.1021/es051960u. [DOI] [PubMed] [Google Scholar]
- 22.Wilkins MJ, Verberkmoes NC, Williams KH, Callister SJ, Mouser PJ, Elifantz H, N′Guessan LA, Thomas BC, Nicora CD, Shah MB, Abraham P, Lipton MS, Lovley DR, Hettich RL, Long PE, Banfield JF. 2009. Proteogenomic monitoring of Geobacter physiology during stimulated uranium bioremediation. Appl. Environ. Microbiol. 75:6591–6599. 10.1128/AEM.01064-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cologgi DL, Lampa-Pastirk S, Speers AM, Kelly SD, Reguera G. 2011. Extracellular reduction of uranium via Geobacter conductive pili as a protective cellular mechanism. Proc. Natl. Acad. Sci. U. S. A. 108:15248–15252. 10.1073/pnas.1108616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101. 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 25.Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, Lovley DR. 2006. Biofilm and nanowire production lead to increased current in microbial fuel cells. Appl. Environ. Microbiol. 72:7345–7348. 10.1128/AEM.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reguera G, Pollina RB, Nicoll JS, Lovley DR. 2007. Possible nonconductive role of Geobacter sulfurreducens pilus nanowires in biofilm formation. J. Bacteriol. 189:2125–2127. 10.1128/JB.01284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rollefson JB, Stephen CS, Tien M, Bond DR. 2011. Identification of an extracellular polysaccharide network essential for cytochrome anchoring and biofilm formation in Geobacter sulfurreducens. J. Bacteriol. 193:1023–1033. 10.1128/JB.01092-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd JR, Chesnes J, Glasauer S, Bunker DJ, Livens FR, Lovley DR. 2002. Reduction of actinides and fission products by Fe(III)-reducing bacteria. Geomicrobiol. J. 19:103–120. 10.1080/014904502317246200. [DOI] [Google Scholar]
- 29.Shelobolina ES, Coppi MV, Korenevsky AA, DiDonato LN, Sullivan SA, Konishi H, Xu H, Leang C, Butler JE, Kim BC, Lovley DR. 2007. Importance of c-type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol. 7:16. 10.1186/1471-2180-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beyenal H, Sani RK, Peyton BM, Dohnalkova AC, Amonette JE, Lewandowski Z. 2004. Uranium immobilization by sulfate-reducing biofilms. Environ. Sci. Technol. 38:2067–2074. 10.1021/es0348703. [DOI] [PubMed] [Google Scholar]
- 31.Marsili E, Beyenal H, Di Palma L, Merli C, Dohnalkova A, Amonette JE, Lewandowski Z. 2007. Uranium immobilization by sulfate-reducing biofilms grown on hematite, dolomite, and calcite. Environ. Sci. Technol. 41:8349–8354. 10.1021/es071335k. [DOI] [PubMed] [Google Scholar]
- 32.Cao B, Ahmed B, Kennedy DW, Wang Z, Shi L, Marshall MJ, Fredrickson JK, Isern NG, Majors PD, Beyenal H. 2011. Contribution of extracellular polymeric substances from Shewanella sp. HRCR-1 biofilms to U(VI) immobilization. Environ. Sci. Technol. 45:5483–5490. 10.1021/es200095j. [DOI] [PubMed] [Google Scholar]
- 33.Sani RK, Peyton BM, Dohnalkova A. 2008. Comparison of uranium(VI) removal by Shewanella oneidensis MR-1 in flow and batch reactors. Water Res. 42:2993–3002. 10.1016/j.watres.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Lovley DR, Phillips EJP. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly SD, Kemner KM, Carley J, Criddle C, Jardine PM, Marsh TL, Phillips D, Watson D, Wu WM. 2008. Speciation of uranium in sediments before and after in situ biostimulation. Environ. Sci. Technol. 42:1558–1564. 10.1021/es071764i. [DOI] [PubMed] [Google Scholar]
- 36.Kelly SD, Kemner KM, Boyanov MI, O'Loughlin EJ, Jeon BH, Barnett MO, Burgos WD, Dempsey BA, Roden EE. 2005. Comparison of U valence state ratio determined from U L3-edge XANES to EXAFS measurements. In Advanced photon source activity report 2003 Argonne National Laboratory, Argonne, IL. [Google Scholar]
- 37.Chang BY, Dworkin M. 1994. Isolated fibrils rescue cohesion and development in the Dsp mutant of Myxococcus xanthus. J. Bacteriol. 176:7190–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas PE, Ryan D, Levin W. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168–176. 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 39.Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. 2008. Real-time detection of actively metabolizing microbes by redox sensing as applied to methylotroph populations in Lake Washington. ISME J. 2:696–706. 10.1038/ismej.2008.32. [DOI] [PubMed] [Google Scholar]
- 40.Gray D, Yue RS, Chueng CY, Godfrey W. 2005. Bacterial vitality detected by a novel fluorogenic redox dye using flow cytometry. Abstr. 105th Meet. Am. Soc. Microbiol. [Google Scholar]
- 41.Renslow RS, Babauta JT, Dohnalkova AC, Boyanov MI, Kemner KM, Majors PD, Fredrickson JK, Beyenal H. 2013. Metabolic spatial variability in electrode-respiring Geobacter sulfurreducens biofilms. Energy Environ. Sci. 6:1827–1836. 10.1039/c3ee40203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49–79. 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 43.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 44.Mehta T, Coppi MV, Childers SE, Lovley DR. 2005. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 71:8634–8641. 10.1128/AEM.71.12.8634-8641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qian X, Mester T, Morgado L, Arakawa T, Sharma ML, Inoue K, Joseph C, Salgueiro CA, Maroney MJ, Lovley DR. 2011. Biochemical characterization of purified OmcS, a c-type cytochrome required for insoluble Fe(III) reduction in Geobacter sulfurreducens. Biochim. Biophys. Acta 1807:404–412. 10.1016/j.bbabio.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 46.Inoue K, Qian X, Morgado L, Kim BC, Mester T, Izallalen M, Salgueiro CA, Lovley DR. 2010. Purification and characterization of OmcZ, an outer-surface, octaheme c-type cytochrome essential for optimal current production by Geobacter sulfurreducens. Appl. Environ. Microbiol. 76:3999–4007. 10.1128/AEM.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franks AE, Glaven RH, Lovley DR. 2012. Real-time spatial gene expression analysis within current-producing biofilms. ChemSusChem 5:1092–1098. 10.1002/cssc.201100714. [DOI] [PubMed] [Google Scholar]
- 48.Inoue K, Leang C, Franks AE, Woodard TL, Nevin KP, Lovley DR. 2011. Specific localization of the c-type cytochrome OmcZ at the anode surface in current-producing biofilms of Geobacter sulfurreducens. Environ. Microbiol. Rep. 3:211–217. 10.1111/j.1758-2229.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 49.Riley RG, Zachara JM, Wobber FJ. 1992. Chemical contaminants on DOE lands and selection of contaminant mixtures for subsurface science research. Subsurface Science Program, Office of Energy Research, U.S. Department of Energy, Washington, DC. [Google Scholar]
- 50.Wood TK, Hong SH, Ma Q. 2011. Engineering biofilm formation and dispersal. Trends Biotechnol. 29:87–94. 10.1016/j.tibtech.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


