Abstract
Advanced oxidation processes are currently used for the treatment of different reactive dyes which involve use of toxic catalysts. Peroxidases are reported to be effective on such dyes and require hydrogen peroxide and/or metal ions. Cyathus bulleri laccase, expressed in Pichia pastoris, catalyzes efficient degradation (78 to 85%) of reactive azo dyes (reactive black 5, reactive orange 16, and reactive red 198) in the presence of synthetic mediator ABTS [2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)]. This laccase was engineered to degrade effectively reactive blue 21 (RB21), a phthalocyanine dye reported to be decolorized only by peroxidases. The 816-bp segment (toward the C terminus) of the lcc gene was subjected to random mutagenesis and enzyme variants (Lcc35, Lcc61, and Lcc62) were selected based on increased ABTS oxidizing ability. Around 78 to 95% decolorization of RB21 was observed with the ABTS-supplemented Lcc variants in 30 min. Analysis of the degradation products by mass spectrometry indicated the formation of several low-molecular-weight compounds. Mapping the mutations on the modeled structure implicated residues both near and far from the T1 Cu site that affected the catalytic efficiency of the mutant enzymes on ABTS and, in turn, the rate of oxidation of RB21. Several inactive clones were also mapped. The importance of geometry as well as electronic changes on the reactivity of laccases was indicated.
INTRODUCTION
Textile effluents released in water bodies are one of the major causes of pollution. The demand for color-free discharge in the effluent streams has made decolorization a top priority (1). There are several chemical classes of dyes used in the textile industry, the most common of which are the reactive dyes. These are extensively used due to their ability to bind to textile fibers through covalent bonds, resulting in enhanced fixation rates and reducing the energy consumption (2). Among these are the phthalocyanine (PC) dyes, which constitute one of the main categories of reactive dyes (3). PC's are water-soluble, predominantly copper containing metallic complexes which are resistant to bacterial degradation under aerobic as well as anaerobic conditions (4, 5). These are potentially mutagenic and implicated in causing toxicity due to their Cu content (6). Conventional physical adsorption by biomass as well as advanced oxidation processes such as TiO2, UV, Fenton, and photo-Fenton oxidations (7) is not effective for elimination of these dyes. Two reactive PC dyes, namely, reactive blue 15 (RB15) and reactive blue 38, have been reported to be decolorized by Bjerkandera adusta and Trametes versicolor (8), and this was attributed to the action of manganese peroxidase (MnP) and lignin peroxidase (LiP). Sulfophthalimides were recognized as major degradation products by comparison with chemically synthesized molecules (9). A related PC dye, reactive blue 21 (RB21), is also extensively used and has been reported to be decolorized by horseradish peroxidase (10). Although the degradation of this dye by soybean peroxidase to the extent of 95% has been reported (11), the products of degradation were recognized only recently as metabolite I (m/z = 437) and metabolite II (m/z = 524) (3). Apart from peroxidases, no other enzymatic method has been reported until now for degradation of RB21. In this report, we describe effective decolorization and degradation of RB21 using ABTS [2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)]-supplemented engineered laccase and demonstrate the versatility of this group of enzymes to act on different categories of reactive dyes.
Laccase (EC 1.10.3.2) is a blue multicopper oxidase that catalyzes the oxidation of a broad range of small organic substrates (12), primarily phenolics, with the participation of molecular oxygen. Addition of a mediator such as ABTS results in formation of ABTS·+ cation radical, which can act on nonphenolic compounds and substrates of high redox potential extending the range of laccase. LiPs and MnPs are enzymes with relatively higher redox values that use H2O2 as a primary reactant to generate a ferry iron porphyrin radical cation [Fe(IV) = O·+] which assists in substrate oxidation (13). The redox potential, in laccases, appears to be governed by the geometry at the T1 Cu center (14). Two His and one Cys have been observed to be arranged trigonally around this copper, and two noncoordinating amino acids sit within about 0.4 nm in axial positions. Of these two, one is the invariable Ile and the other can be Leu, Met, or Phe depending upon the type of laccase. The nature of this amino acid is modestly correlated with the reduction potential of the T1 copper (15). Phe at this position determines high potential, and this has been proven by site-directed mutagenesis and on the basis of several crystal structures (16). This position is occupied by Leu in laccase of Cyathus bulleri, making it a “medium redox” enzyme (17). Amino acids that line the substrate binding pocket near the T1 site also influence the redox potential (18) and, in turn, the ability to act on dyes and sterically large molecules (19). In general, the difference in the redox potential between the enzyme and the substrate is shown to affect the rate of reaction (20–22). In addition to this, the C termini of laccases, particularly from the ascomycetes, have been shown to influence the catalytic activity, azide tolerance, and thermostability (23) of the enzyme. Very little is known about the role of amino acids toward the C terminus of basidiomycete laccase on reactivity of the enzyme, except for the T. versicolor laccase Lcc2, in which modification of 11 residues at the C terminus altered the catalytic activity of the enzyme and also shifted the redox potential of the active site to a more negative value (24).
Our work on a basidiomycete laccase from C. bulleri indicated its ability to act on a variety of dyes (25) in the presence of ABTS. Pathways for degradation of simple and complex triarylmethane dyes were elucidated (26). In this study, we report on mutant variants of this laccase enzyme (WtLcc), generated through random mutagenesis of the 816-bp segment (toward the C terminus) of the wild-type gene (lcc). The variants effectively decolorized RB21, a PC high-redox dye, in the presence of ABTS. The products of degradation were monitored by electron spray ionization-mass spectrometry (ESI-MS) and a pathway for degradation was proposed. The changed amino acids in the laccase variants were determined, and this was correlated with the conformational and reactive changes in laccase. The results were integrated with hypotheses that allow us to understand the reactivity of these enzymes and role of amino acids far from the T1 site that control activity.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Cyathus bulleri (Brodie 195062) from Canadian National Collection of fungal cultures was used as a source of laccase. It belongs to the family Nidulariaceae and is commonly known as birds' nest fungus. Escherichia coli strain DH5α was used in all cloning experiments. E. coli was grown in low-salt Luria broth supplemented with 100 μg/ml of zeocin for selection when required. Pichia pastoris X33 strain was grown in YPD medium containing 10 g/liter of yeast extract (Himedia, India), 20 g/liter of peptone, and 20 g/liter of dextrose. The cDNA of C. bulleri laccase (lcc) cloned in the pPICZαB vector (17), in frame with the α-mating type factor, was used for generating the mutant library in P. pastoris. The transformants were grown in buffered complex glycerol medium (BMGY medium) followed by buffered complex methanol medium (BMMY medium) (Invitrogen). Induction of laccase was carried out by addition of 1% (vol/vol) methanol every 24 h.
Evaluation of WtLcc for dye decolorization.
Wild-type laccase enzyme (WtLcc), expressed in P. pastoris, was screened for decolorization of several reactive dyes. Decolorization assays were carried out as described by Chhabra et al. (26). The dyes used were reactive black 5 (RB5), reactive orange 16 (RO16), reactive red 198 (RR198), and reactive blue 21 (RB21). Briefly, 100 ppm of dye in 20 mM sodium citrate buffer (pH 4.0), containing 100 μM ABTS and 100 mU/ml of laccase in a final reaction volume of 20 ml, was used for decolorization experiments. The reaction mixture was incubated on a rotary shaker (100 rpm) at 37°C for up to 5 h, followed by boiling for 5 min to inactivate the enzyme and arrest reoxidation of ABTS. Heat-inactivated enzyme was used as a control. Decolorization was monitored spectrophotometrically by measuring decrease in absorbance at the λmax of the dye. Results are reported in terms of percent decolorization relative to those of the untreated dyes.
Random mutagenesis of 816-bp region of the laccase gene (lcc) corresponding to the C terminus.
A strategy was developed for creating random mutations in the region of the of the wild-type laccase gene corresponding to the C terminus (see Fig. S1 in the supplemental material). Error-prone PCR, optimized for low-frequency mutation (0 to 4.5 mutations/kb), was carried out with Mutazyme II DNA polymerase (Gene Morph kit II; Stratagene) by following the manufacturer's protocol. Each amplification reaction (50 μl) contained 0.5 μM primers (lac C-terFor, GGACATCGATGGCCACACATTTACCA, and lac Full Rev, ATTTCCCCGCGGTCAGGTGCCGGTTGG), 200 μM deoxynucleoside triphosphates (dNTPs), and 2.5 U of Mutazyme II DNA polymerase in Mutazyme II reaction buffer. (The italicized nucleotides represent the restriction sites used for cloning.) PCR was done with initial denaturation at 95°C for 5 min, 30 cycles of amplification (1 min at 95°C, 30 s at 56°C, and 50 s at 72°C), and final extension for 10 min at 72°C. The PCR product was purified using a Qiagen PCR purification kit by following the manufacturer's instructions. The purified product was digested with SacII and ClaI and ligated to similarly digested and gel-eluted pPICZαB vector containing the region of the gene corresponding to the N terminus. The recombinant plasmid library was transformed into DH5α competent cells and plated onto LB-plus-zeocin plates and incubated at 37°C. The presence of the desired PCR product was verified by colony PCR on randomly selected clones.
All the colonies (∼500) from the LB-zeocin plates were picked up and inoculated in 4 sets of 50 ml of LB containing zeocin for the isolation of plasmid. Plasmid was isolated using Qiagen miniprep kit. The recombinant plasmids were linearized with SacI and electroporated (1.5 kV for 5mS) into P. pastoris X33 strain using Bio-Rad's micropulser (Bio-Rad, CA). The transformants were selected on YPD medium containing 10% sorbitol and 100 μg/ml of zeocin.
Screening of the mutant library.
In the primary screening, the library was grown in BMMY minimal methanol plates containing 1 mM ABTS for detection of active laccase producers. The day of appearance of the green zone (on account of liberation of ABTS·+ cation) and the zone size were noted for all the clones. The colonies displaying large green zones were grown in 0.5 ml of BMMY medium (Invitrogen, CA) contained in 48-well plates. Plates were incubated at 28°C and 220 rpm for 2 days. Five microliters from each well was transferred to a 96-well plate to measure laccase activity as described under “Analytical methods” below. Equal laccase units from the microtiter plates were also transferred to another set of 96-well plates containing RB21 at 30 ppm and 100 μM ABTS. In order to monitor the time intervals carefully, lower concentrations (30 ppm) were selected. The plates were incubated at 37°C to assess the ability of the laccase variants to decolorize the dye. The plates were visualized for decolorization. Active clones (products of the lcc-35, lcc-61, and lcc-62 genes), selected on the basis of decolorization of RB21, were picked and analyzed at the genetic and biochemical levels.
Kinetic characterization of laccase variants.
For kinetic studies, the WtLcc and the mutant enzymes were purified according to the method of Garg et al. (17). Briefly, laccase expressed in the culture filtrate of recombinant P. pastoris was concentrated by ammonium sulfate precipitation and then dialyzed in 20 mM Tris-HCl (pH 8). The samples were filtered and loaded onto a DEAE anion-exchange column (GE Healthcare) preequilibrated with Tris-HCl buffer and coupled to an AKTA purifier system (GE Healthcare). The proteins were eluted with increasing concentrations of NaCl. Fractions containing laccase were pooled and concentrated by passing through a 30-kDa Centricon (Pall Lifesciences, USA). Purity of the enzymes was judged by SDS-PAGE.
Kinetic parameters were determined against three substrates (ABTS, guaiacol, and pyrogallol). Protein concentrations in the range from 0.05 to 0.3 μg were used. All assays were carried out in duplicate at 55°C in a final volume of 2 ml containing the corresponding substrate in 20 mM citrate buffer (pH 4). Substrate oxidation was followed by measurement of the absorption at 420 nm for ABTS (molar extinction coefficient [ε420], 36,000 M−1 cm−1), 470 nm for guaiacol (ε470, 6,400 M−1 cm−1) and 420 nm for pyrogallol (ε420, 4,400 M−1cm−1). The kinetic parameters were calculated using GraphPad Prism software (version 5.0b).
Characterization of metabolites by ESI-MS.
Preliminary identification of the products obtained by action of laccase variants (Lcc-35, Lcc-61, and Lcc-62) on RB21 was done by thin-layer chromatography (TLC). For this, the untreated dye and the treated dye samples were resolved using F254 silica plates and 80% MeOH–20% water as the mobile phase. Plates were dried and observed under UV at 254 nm. The metabolites from the plates were extracted and dissolved in 500 μl of acidified methanol and vortexed for 1 h. It was centrifuged and filtered with a 0.22-μm membrane filter and concentrated to 100 μl using a SpeedVac. The extracted sample (20 μl) was diluted with methanol-water (vol/vol) and injected into an ESI-MS equipped with a hybrid Q-TOF detector (AB Sciex, USA). The spectrum was monitored in positive ion mode with the following conditions: ion spray voltage, 5,500 V; nebulizer gas, 20 lb/in2; curtain gas, 25 lb/in2; declustering potential, 60 V; focusing potential, 265 V; and flow rate, 5 μl/min. The spectra were acquired using a mass range (m/z) of 100 to 5,000 atomic mass units.
Nucleotide sequencing of the laccase variants.
For sequencing of the variants of laccase gene, colony PCR was carried out using forward and reverse primers (lac C-ter For, GGACATCGATGGCCACACATTTACCA, and lac Full Rev, ATTTCCCCGCGGTCAGGTGCCGGTTGG) using proofreading Deep Vent DNA polymerase (New England BioLabs, MA). Double-stranded sequencing was done by Xcelris Labs Ltd., Gujarat, India, and the sequence was analyzed by NCBI Blast. The three-dimensional structure of the native laccase was modeled using Modeler version 9.11. The crystal structure of blue laccase from Trametes trogii complexed with p-methylbenzoate was used (PDB 2HRG) as a template for homology modeling. Lcc of C. bulleri shares 69.6% sequence identity with the laccase from T. trogii.
Chemicals and dyes.
ABTS and guaiacol were from Sigma-Aldrich (St. Louis, MO). Pyrogallol was obtained from Thermo Fisher (India). All dyes were a gift from Department of Textile Technology, IIT Delhi, and were procured from local mills. Other chemicals were of highest grade available locally.
Analytical methods.
Laccase assay was carried out at room temperature for 10 min using 100 μM ABTS in 20 mM sodium citrate buffer, pH 4. Oxidation of ABTS was followed by increase in absorbance at 420 nm, estimated using a Spectra Max M2 microplate reader (Molecular Devices, CA). Enzyme activity was expressed in U/ml as described previously (27).
Standard redox potential of RB21, WtLcc, and the laccase variants Lcc-35, Lcc-61, and Lcc-62 was measured using cyclic voltammeter as described previously (28) at 25°C. Experiments were performed using PGSTAT101 Autolabsystem (Eco-Chemie, Netherlands), controlled by Nova software. The working, counter, and reference electrodes were glassy carbon electrode (0.07 cm2), platinum wire, and an Ag/AgCl filled with saturated NaCl, respectively. Cyclic voltammograms were registered at a scan rate of 100 mV/s (for the dye) or 50 mV/s for the pure enzymes. The experiments were carried out in triplicate in sodium citrate buffer of pH 4 containing 100 ppm of the dye or 0.1 mg/ml of the purified enzyme. After each sample, the working electrode was polished and rinsed thoroughly. The results are reported against corrections made for normal hydrogen electrode (NHE).
RESULTS
Generation and screening of the mutant library.
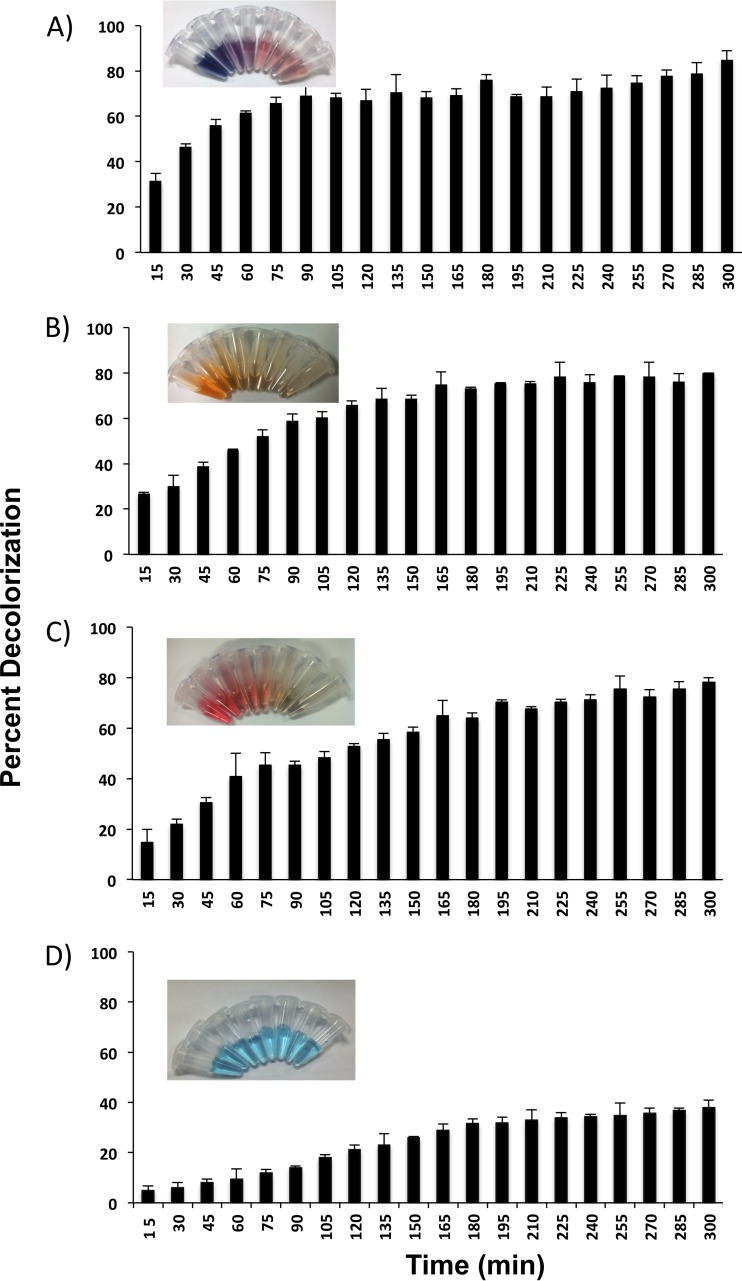
The wild-type laccase (WtLcc) from C. bulleri, expressed in P. pastoris X33, was first evaluated for decolorization of four reactive dyes, namely, RB5, RO16, RR198, and RB21, the first three being the azo dyes and the last a PC dye. All the dyes were used at a concentration of 100 ppm. Maximum decolorization of 85% was obtained in 5 h with RB5. The level of decolorization for RO16 and RR198 was about 78%. In case of RB21, a maximum decolorization of 35% was observed after 5 h (Fig. 1). The redox potential of this dye as determined by cyclic voltammeter was found to be 1.35 V, corrected for NHE (see Fig. S2 in the supplemental material). In an attempt to extend the reactivity of laccase on the phathalocyanine category of dye, mutant variants were generated. Random mutagenesis was performed in the 816-bp segment of the gene toward the region corresponding to the C terminus, as amino acids toward the C terminus have been shown to be important for activity of laccase (23). A mutation frequency of about one to two nucleotides per 816-bp segment was observed on sequencing of the various clones. The ABTS-oxidizing activity of 140 selected mutants was measured, and the distributions of residual activity were compared (see Fig. S3 in the supplemental material). Around 40 transformants (28%) displayed relative activity between 1.1- and 1.4-fold higher than that of the parent enzyme, and out of these, several (28 in number) were >1.2-fold higher. About 34 transformants (24%) displayed ABTS-oxidizing activity similar to that of the parental enzyme. A large number of transformants (28%) exhibited relative activity between 0- and 0.2-fold that of the parental enzyme. The highly ABTS-oxidizing clones (total of 28) were selected for further experiments. Each of these was tested for decolorization of RB21, and three variants, namely, Lcc-35, Lcc-61, and Lcc-62, which showed high decolorization compared to WtLcc, were selected. Around 95% decolorization was observed with Lcc-35 after 30 min of incubation with 30 ppm of RB21. About 78% decolorization was observed with the Lcc-61 and Lcc-62 variants (Fig. 2).
FIG 1.
Percent decolorization of reactive black 5 (A), reactive orange 16 (B), reactive red 198 (C), and reactive blue 21 (D) when incubated with culture filtrate of P. pastoris expressing WtLcc.
FIG 2.
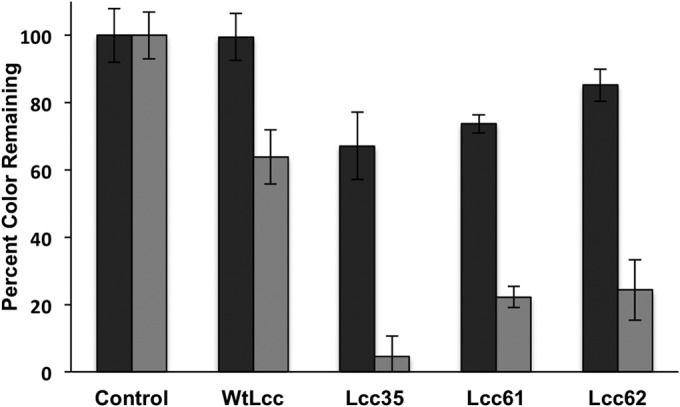
Decolorization of reactive blue 21 (30 ppm) by WtLcc and the mutant variants Lcc-35, Lcc-61, and Lcc-62 in 10 (black bars) and 30 (gray bars) min.
Characterization of highly ABTS-oxidizing mutants.
To identify the changed amino acid residues in the three mutant variants, sequencing of both the strands of the entire laccase gene was done; the results are shown in Table 1. As can be seen, the mutations in all the three genes encoding the variants were point mutations. In lcc-35, the mutation was in the codon GGA to CGA, which led to amino acid change from Gly to Arg at position 463. Based on the multiple-sequence alignment (see Fig. S4 in the supplemental material), this Gly was noted to be strongly conserved in all ascomycete, basidiomycete, plant, and bacterial laccases. In the case of lcc-61, AGU (Ser 318) was mutated to ACU (Thr). This Ser (at 318) was not so strongly conserved and was replaced by many other amino acids, such as Pro, Thr, Val, and Arg (see Fig. S4). In lcc-62, AUU (Ile 490) was changed to AUG (Met). The sequence comparison indicated the Ile at 490 to be slightly more conserved and replaced only by Val (a homologous substitution) or Thr. To determine the microenvironment around these mutations, a three-dimensional structure of WtLcc was prepared (Fig. 3) by homology modeling using theoretical values of coordinates obtained from the Protein Data Bank (PDB) database. It was found that two of the mutations, observed in Lcc-62 and Lcc-35 laccase variants, were buried. In Lcc-62, Ile at position 490 mutated to Met and is part of the helix, away from the T1 copper center. In Lcc-35, Gly at 463 was mutated to Arg, and this was localized toward the end of the beta sheet. While the two mutations appear to be localized in close proximity to each other, they were far from the T1 copper center. In the Lcc-61 variant, the mutation at position 318 was found to be on the surface and part of the loop region. This mutation was close to the substrate-binding pocket near the T1 site. The positions of these different mutations were determined from the T1 catalytic center and the T2/T3 cluster (Fig. 3 and Table 1). As shown, the buried mutations were closer to the T2/T3 cluster, while the surface mutation was close to the T1 center.
TABLE 1.
Mutations obtained in the 816-bp segment corresponding to the C. bulleri laccase (toward the C terminus)
| Laccase mutant | Nucleotide change | Amino acid substitution | Location in mature protein | Distance from T1 site (Å) | Distance from T2/T3 cluster (Å) | Secondary-structure motif |
|---|---|---|---|---|---|---|
| RB21-decolorizing clones | ||||||
| Lcc-35 | GGA-CGA (G1389C) | G463R | Buried | 23.95 | 13.99 | Beta sheet |
| Lcc-61 | AGU-ACU (G955C) | S318T | Surface | 27.97 | 33.56 | Loop |
| Lcc-62 | AUU-AUG (U1473G) | I490M | Buried | 29.01 | 18.97 | Helix |
| Low-activity clones (<0.2-fold) | ||||||
| Lcc-4 | AUC-AGC (U769G) | I256S | Buried | 19.73 | 21.08 | Loop |
| AAU-AUU (A1081U) | N360I | Surface | 15.21 | 18.78 | Loop | |
| Lcc-31 | AAC-GAT (A1311G; C 1313T) | N437D | Surface | 27.27 | 26.23 | Loop |
| AAU-AUU (A1357U) | N452I | Surface | 15.39 | 21.56 | Loop | |
| Lcc-44 | UUU-UUA (U800A) | F266L | Buried | 26.3 | 24.94 | Beta sheet |
| Lcc-49 | GCA-ACA (G915A) | A305T | Surface | 41.3 | 39.8 | Loop |
| Lcc-84 | CAG-CAT (G1125T) | Q374H | Surface | 26.86 | 18.5 | Helix |
| Lcc-85 | AUU-AAT (U1444A) | I481N | Buried | 12.86 | 10.15 | Beta sheet |
FIG 3.
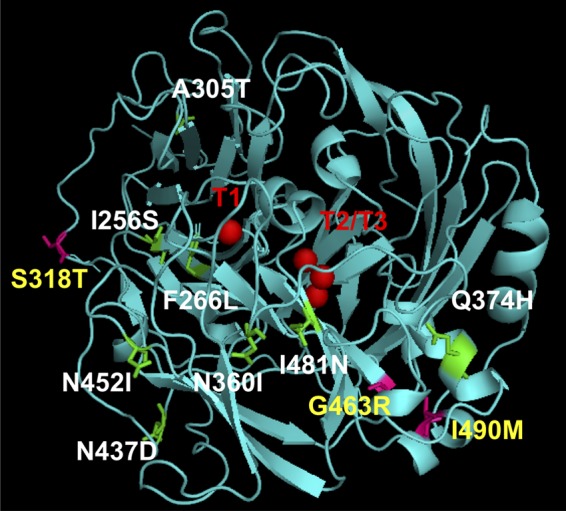
Three-dimensional structure of WtLcc generated by homology modeling using laccase from Trametes trogii (complexed with p-methylbenzoate). The T1, T2, and T3 Cu centers are marked in red. The inactive mutants are listed in white, and their positions are marked in green on the modeled structure. The mutant variants are listed in yellow, and their positions are marked in magenta.
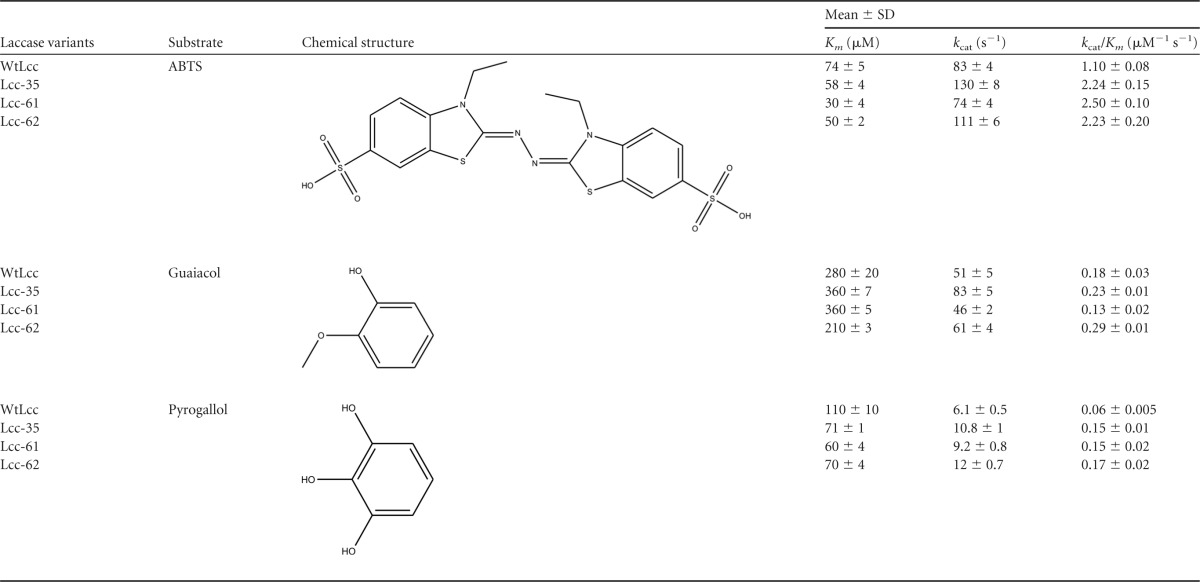
The kinetic parameters of the WtLcc and the mutants were determined on two phenolic compounds (guaiacol and pyrogallol) and the synthetic mediator ABTS. These are the most commonly used substrates for estimation of laccase activity (27, 29). The relative redox potential values of ABTS, guaiacol, and pyrogallol are 1,080 mV, 800 mV (pH 5), and 540 mV (pH 4), respectively (29, 30). The data for the kinetic parameters are shown in Table 2. All the three mutants exhibited higher catalytic efficiency on ABTS (2- to 2.3-fold) and pyrogallol (2.8- to 3-fold). This was attributed to an increase in kcat for the Lcc-35 and Lcc-62 variants and a substantial decrease in Km for ABTS and pyrogallol for the Lcc-61 variant. With guaiacol, efficiency was slightly better for the Lcc-35 and Lcc-62 variants and could again be attributed to an increase in the kcat value. The Lcc-61 variant exhibited lower catalytic efficiency on guaiacol, and this was correlated with an increase in the kcat for this variant. No difference was observed in the value of kcat compared to that of the WtLcc. The standard redox potential values were determined for RB21 and the three mutants. For RB21, it was measured as 1.35 V (see Fig. S2A in the supplemental material). The values measured for the three mutants (∼0.5 V) were found to be nearly similar to that for the WtLcc. A slight increase in the magnitude of the current was, however, observed for the mutants (see Fig. S2B).
TABLE 2.
Kinetic parameters of wild-type and mutant laccases determined at 55°C in 20 mM sodium citrate buffer (pH 4) for the oxidation of ABTS, guaiacol, and pyrogallol
Characterization of the “inactive” mutants.
To determine the positions and residues that are important for the activity of the enzyme, the inactive mutants (relative ABTS oxidizing activity <0.2-fold that of the parental enzyme) were analyzed. On sequencing of few of the inactive clones, it was found that the mutations were not localized at particular positions; instead, they were dispersed throughout the 816-bp region (Table 1). Interestingly, in the three-dimensional structure (Fig. 3), three of the inactive mutants (Lcc-31, which had the two amino acid substitutions N437D and N452I; Lcc-4, which had the two mutations I256S and N360I; and Lcc-49, with A305T) harbored mutations that were localized in the loop region. This suggested that the mutations were perturbing the flexibility of the enzyme. Moreover, two of these mutations (N452I and N360I) were also mapped close to the T1 center. In the remaining three inactive mutants, two of the mutations (in Lcc-85 and Lcc-44) were localized in the beta sheet, of which I481N in Lcc-85 was closely spaced with respect to T1 and T2/T3 cluster. The other one was F266L (Lcc-44), which was away from the catalytic center but appeared to be buried when observed in the three-dimensional structure. Lastly, at position 374 (Lcc-84), Thr was mutated to His in the helix and made the mutant inactive (Fig. 3 and Table 1). Importantly, based on the sequence alignment (see Fig. S4 in the supplemental material), all these mutations were detected in the conserved region of laccases.
Characterization of degradation products of RB21.
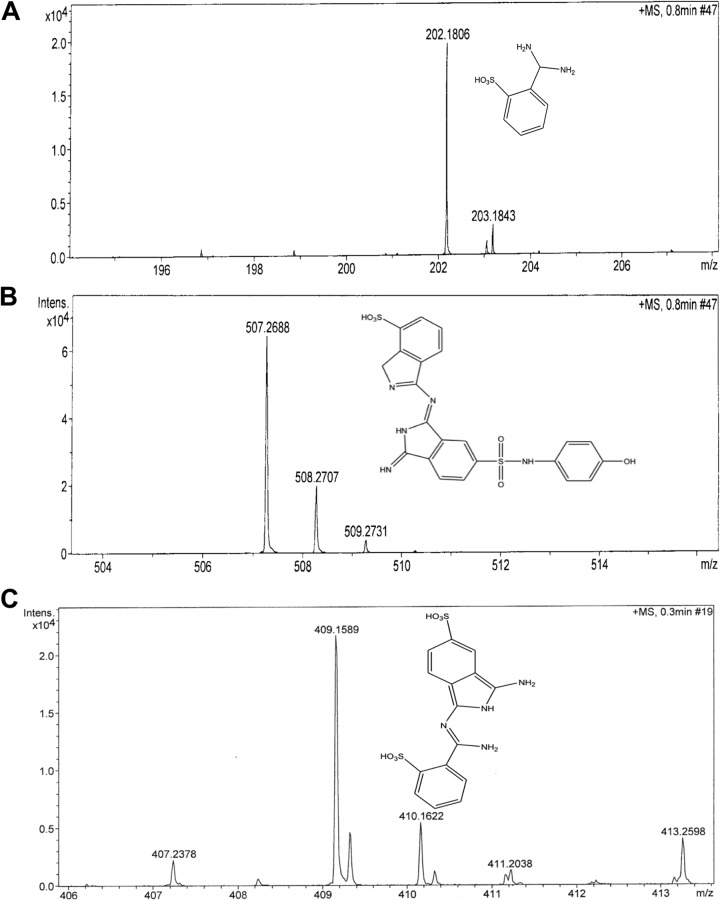
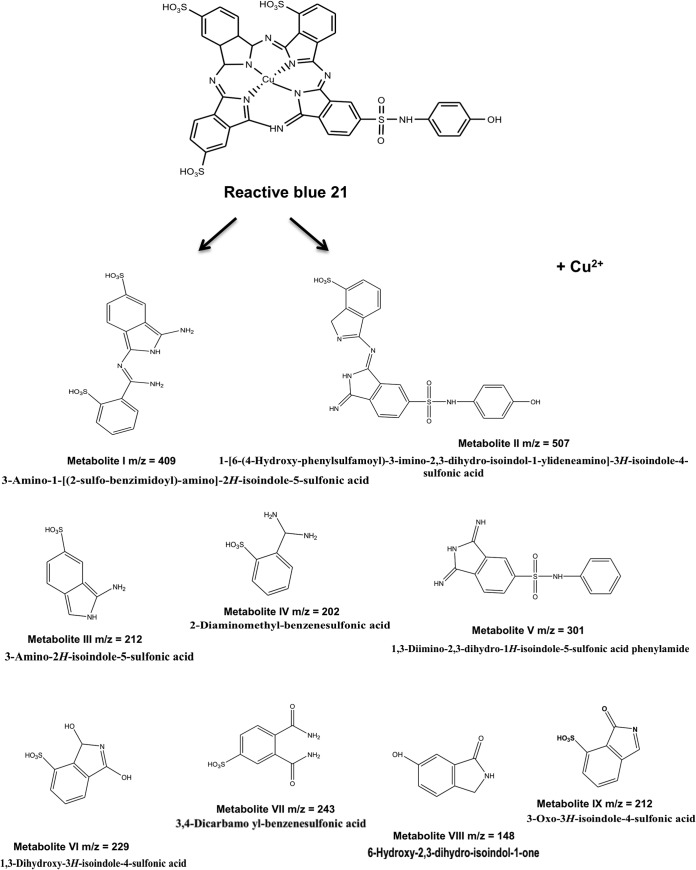
The decolorized products/metabolites formed from RB21, after treatment with the mutant laccase variant (Lcc-35), were resolved by preparative TLC followed by extraction in methanol. Three putative compounds, corresponding to m/z values of 202.18, 409.16, and 507.27, were detected in 15 min (Fig. 4 A, B, and C) samples. After 1 h of incubation, several metabolites were detected, and these had m/z values of 148.97, 202.12, 211.95, 228.96, 242.97, and 301.04 (Fig. 4D). These corresponded to derivatives of sulfophthalimide (9). Based on the products obtained and the time at which these appeared, a degradation pathway for RB21 was proposed (Fig. 5).
FIG 4.
Mass spectra of products of RB21 obtained after incubation with Lcc-35 for 15 min (A to C) or 1 h (D).
FIG 5.
Proposed degradation pathway of RB21 by Cyathus bulleri laccase.
DISCUSSION
Laccase from C. bulleri expressed in P. pastoris X33 was shown to decolorize several reactive azo dyes efficiently (78 to 85%) in the presence of the synthetic mediator ABTS. However, its activity on RB21, a high-redox PC dye, was found to be limited. An understanding of the factors that contribute to the reactivity of laccases is important, as this can lead to an increase in the range of oxidizable substrates by this enzyme and the same can be achieved through protein engineering techniques. In the present study, an attempt was made to extend the oxidizing ability of a “medium redox” laccase to the PC category of dyes. The strategy adopted to develop the library and screen for RB21-degrading variants was based on the following observations and inferences from the literature: (i) the amino acids toward the C terminus of laccase affect the redox potential of laccases (15, 18, 24) and thereby its reactivity; (ii) in general, higher catalytic activity on a laccase substrate, such as ABTS or 2,6-dimethoxy phenol, can be used as a screen for detection of altered catalytic rates or higher dye decolorization efficiencies (31–33); (iii) a high-throughput screening method would allow detection of clones that degrade the PC dye; and (iv) easy monitoring of the degradation process by qualitative TLC. Low-error-rate (0 to 4 mutations/kb) mutagenesis was used for introducing random mutations in the 816-bp segment of the lcc gene from C. bulleri. This segment was toward the C-terminal region and is proposed to contain the catalytic Cu centers and putative substrate-binding sites (34). The C-terminal region of both ascomycete and basidiomycete laccase is reported to influence the catalytic property of laccase (23, 24). The mutagenesis of this region resulted in generation of several inactive clones (∼28%) in spite of low mutation rates, indicating the importance of this region in determining catalytic activity of the enzyme. The major advantage was that with screening of a few hundred transformants, several “high activity” (40/140) variants could be obtained on ABTS. The screening procedure was, however, limited to selection of variants which were also secreted efficiently in the culture filtrate of P. pastoris.
The variant enzymes Lcc-35, Lcc-61, and Lcc-62 were selected based on their ability to decolorize RB21 effectively in the microtiter based screening method. Lcc-35 decolorized RB21 nearly completely (95%) within 30 min. The metabolites formed after treatment of RB21 with the WtLcc and the mutant laccases were characterized by ESI-MS and found to be similar, implying that the pathway of degradation was the same, but the rate was 14 times faster with the Lcc-35 variant than with WtLcc. The first two products formed due to mid-cleavage of RB21 had m/z values of 409.16 and 507.27 (metabolites I and II) (Fig. 5). These were further degraded in to a number of smaller compounds (Fig. 5). The tentative structures were assigned to the degradation products, as shown in Fig. 5, indicating an extensive breakdown of the dye in to smaller fragments. The metabolites with m/z values ranging from 331.16 to 359.17 (Fig. 5) could not be assigned any structure and may have resulted from different oxidative cleavages in the side chains of RB21. It is important to note that many of the small derivatives which were obtained by action of the engineered laccase have been either proposed to occur on degradation of the PC dyes by turnip peroxidase (3) or detected using pellet-supported, Pd-catalyzed H2 reduction of PC dyes, including RB21 (6). While the degradation products are not reported to be toxic (6), the Cu released is considered to be toxic (3) and needs to be effectively removed. This outcome is further supported by the results of Heinfling-Weidtmann et al. (9), who studied the degradation products of phathalocyanine dyes (RB15 and RB38) by white-rot fungus B. adusta. Sulfophthalamides were identified as major metabolites by comparison with synthesized reference compounds, and the release of Cu from the metal complex was proposed.
To gain insight into how the mutations were placed structurally, a three-dimensional model of WtLcc was built and all mutations were mapped on this structure. For the inactive clones, most mutations (except for Lcc-84) were localized in the lower left regions of the modeled structure. These mutations were either close to the T1 and the T2/T3 centers, as for Lcc-4, Lcc-31, and Lcc-85, or far from these, as in Lcc-44, Lcc-49, and Lcc-84 (Table 1). In Lcc-44, Phe at 266 position was mutated to Leu and has been implicated in substrate binding. In T. versicolor (LacIIIb), it is reported to be part of the substrate-binding cavity and is involved in hydrophobic interactions with the ligand (35). In Lcc-49, Ala at 305 position was mutated to Thr, which has an extra methyl and hydroxyl group. The Ala side chain is hydrophobic, whereas Thr has both hydrophobic and hydrophilic side chains. This may create steric disturbance and overall affect the conformation/folding of the enzyme, resulting in an inactive phenotype. Similarly, in Lcc-84, Glu at position 374 was mutated to His and is part of the helix. All the mutations noted here were localized in the highly conserved regions of laccases (see Fig. S4 in the supplemental material) and indicated that the activity itself is delicately balanced and point mutations can lead to drastic reduction in catalytic activity. Saturation mutagenesis at these positions can possibly predict the amino acids that can be allowed at these positions.
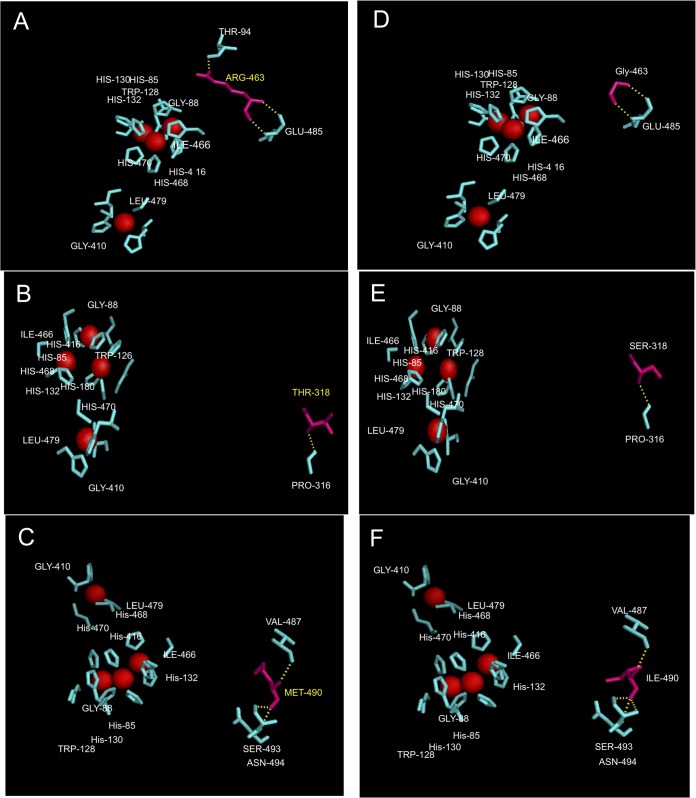
For the highly ABTS-oxidizing variants, the effect of mutations was determined at the level of additional noncovalent bond formation (if any) or interactions at the active site. In the Lcc-35 variant, Arg can form an additional hydrogen bond with neighboring Thr and possibly change the geometry of the solvent channel required for the access of dioxygen or egress of the water molecules (Fig. 6A and D). In Lcc-61, Ser 318 occurs in the loop region near the active site. Replacement with Thr can lead to a change in the steric environment of the active site, as reported for some alcohol dehydrogenases (36). This substitution maintains the hydroxyl group and thereby overall functionality but introduces an extra methyl group, which possibly changes the conformation of the enzyme at the active site (Fig. 6B and E). In Lcc-62, replacement of Ile 490 by Met does not change the microenvironment, as both these amino acids possess identical van der Waals volume of 124 Å. The only difference noted was the change in the overall conformation (Fig. 6C and F). Being in close proximity to the solvent channel, it can also affect access of oxygen or exit of the water molecule. The changed geometries in the structure correlated well with an increase in the catalytic efficiencies of the mutant enzymes, as observed from the kinetic data. The magnitude of the current in the mutant enzymes was observed to be slightly higher, suggesting an increase in electron transfer. Thus, mutations away from the T1 site (as for the Lcc-35, Lcc-61, and Lcc-62 variants) affected the reactivity of the enzymes. While the amino acid residues away from the catalytic site have been implicated in lending an additional beneficial property to laccase, such as stability in a wider pH range, in organic solvents, and thermostability (37), their role in increasing catalytic activity is not well understood.
FIG 6.
Details of the point mutations observed in Lcc-35 (G463R) (A), Lcc-61 (S318T) (B), and Lcc-62 (I490M) (C) modeled in the WtLcc structure. Corresponding positions of the native amino acids at these locations are shown in panels D, E, and F, respectively. Red spheres represent the Cu atoms. Hydrogen bonding before and after mutation is shown as yellow dashes.
There has been considerable debate on the relationship between laccase redox potential and the range of oxidizable substrates. For instance, in Thielavia arenaria laccase (TaLcc1), the redox potential of the T1 copper is reported to be slightly higher (0.51 V) than that of the recombinant Melanocarpus albomyces laccase (0.48 V) but lower than that of Trametes hirsuta laccase (0.78 V). The kinetic data of TaLcc1 thus should fit in between those of the two enzymes. However, data on kinetic parameters of this enzyme (38) showed this not to be the case, suggesting that the redox potential difference between the T1 copper and the substrate may not be the only factor that contributes to substrate oxidation. It has also been reported that a variant of the Pleurotus ostreatus laccase (1H6C) which showed a higher redox potential (by +0.12 V) relative to the wild-type enzyme exhibited catalytic efficiencies similar to those of the wild type on several substrates (39). Our studies, as reported here, also show that while the mutant laccase variants exhibited redox potentials similar to those of WtLcc, they exhibited different catalytic properties from those of the parent enzyme. This indicated the importance of factors other than the redox potential for oxidation. Clearly, altered intraprotein interactions are also important. It is very unlikely that the mutations picked up here could have been predicted by rational design, and random mutagenesis experiments allowed functionally correlation of protein regions to the activity of laccase.
Supplementary Material
ACKNOWLEDGMENTS
We thank DBT for financial support and the Council of Scientific and Industrial Research for providing a fellowship to T.K.
We thank Khushboo Bafna and Shayoni Dutta for help in the modeling of the three-dimensional structure of laccase.
Footnotes
Published ahead of print 26 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02665-14.
REFERENCES
- 1.Hao OJ, Kim H, Chiang PC. 2000. Decolorization of wastewater. Crit. Rev. Environ. Sci. Technol. 30:449–505. 10.1080/10643380091184237. [DOI] [Google Scholar]
- 2.Hrdina R. 1997. Reactive dyes for animal fibres and synthetic polyamides. Chem. Listy. 91:149–159. [Google Scholar]
- 3.Silva MC, Correa AD, Amorim MTSP, Parpot P, Torres JA, Chagas PMB. 2012. Decolorization of the phthalocyanine dye reactive blue 21 by turnip peroxidase and assessment of its oxidation products. J. Mol. Catal. B Enzym. 77:9–14. 10.1016/j.molcatb.2011.12.006. [DOI] [Google Scholar]
- 4.Pagga U, Brown D. 1986. The degradation of dyestuffs. Part II. Behaviour of dyestuffs in aerobic biodegradation tests. Chemosphere 15:479–491. 10.1016/0045-6535(86)90542-4. [DOI] [Google Scholar]
- 5.Brown D, Laboureur P. 1983. The degradation of dyestuffs. Part I. Primary biodegradation under anaerobic conditions. Chemosphere 12:397–404. 10.1016/0045-6535(83)90114-5. [DOI] [Google Scholar]
- 6.Matthews RD, Bottomley LA, Pavlostathis SG. 2009. Palladium-catalyzed hydrogen reduction and decolorization of reactive phthalocyanine dyes. Desalination 248:816–825. 10.1016/j.desal.2008.12.043. [DOI] [Google Scholar]
- 7.Arslan I, Balcioglu IA. 1999. Degradation of commercial reactive dyestuffs by heterogenous and homogenous advanced oxidation processes: a comparative study. Dyes Pigments 43:95–108. 10.1016/S0143-7208(99)00048-0. [DOI] [Google Scholar]
- 8.Heinfling A, Bergbauer M, Szewzyk U. 1997. Biodegradation of azo and phthalocyanine dyes by Trametes versicolor and Bjerkandera adusta. Appl. Microbiol. Biotechnol. 48:261–266. 10.1007/s002530051048. [DOI] [Google Scholar]
- 9.Heinfling-Weidtmann A, Reemtsma T, Storm T, Szewzyk U. 2001. Sulfophthalimide as major metabolite formed from sulfonated phthalocyanine dyes by the white-rot fungus Bjerkandera adusta. FEMS Microbiol. Lett. 203:179–183. 10.1111/j.1574-6968.2001.tb10838.x. [DOI] [PubMed] [Google Scholar]
- 10.Ulson de Souza SMAG, Forgiarini E, Ulson de Souza AA. 2007. Toxicity of textile dyes and their degradation by the enzyme horseradish peroxidase (HRP). J. Hazard. Mater. 147:1073–1078. 10.1016/j.jhazmat.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Marchis T, Avetta P, Bianco-Prevot A, Fabbri D, Viscardi G, Laurenti E. 2011. Oxidative degradation of Remazol Turquoise Blue G 133 by soybean peroxidase. J. Inorg. Biochem. 105:321–327. 10.1016/j.jinorgbio.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Messerschmidt A. 1993. Blue copper oxidases. Adv. Inorg. Chem. 40:121–185. 10.1016/S0898-8838(08)60183-X. [DOI] [Google Scholar]
- 13.Wong DWS. 2009. Structure and action mechanism of ligninolytic enzymes. Appl. Biochem. Biotechnol. 157:174–209. 10.1007/s12010-008-8279-z. [DOI] [PubMed] [Google Scholar]
- 14.Rodgers CJ, Blanford CF, Giddens SR, Skamnioti P, Armstrong FA, Gurr SJ. 2010. Designer laccases: a vogue for high-potential fungal enzymes? Trends Biotechnol. 28:63–72. 10.1016/j.tibtech.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Xu F, Berka RM, Wahleithner JA, Nelson BA, Shuster JR, Brown SH, Palmer AE, Solomon EI. 1998. Site-directed mutations in fungal laccase: effect on redox potential, activity and pH profile. Biochem. J. 334:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu F, Palmer AE, Yaver DS, Berka RM, Gambetta GA, Brown SH, Solomon EI. 1999. Targeted mutations in a Trametes villosa laccase. Axial perturbations of the T1 copper. J. Biol. Chem. 274:12372–12375. [DOI] [PubMed] [Google Scholar]
- 17.Garg N, Bieler N, Kenzom T, Chhabra M, Ansorge-Schumacher M, Mishra S. 2012. Cloning, sequence analysis, expression of Cyathus bulleri laccase in Pichia pastoris and characterization of recombinant laccase. BMC Biotechnol. 12:75. 10.1186/1472-6750-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kallio JP, Auer S, Jänis J, Andberg M, Kruus K, Rouvinen J, Koivula A, Hakulinen N. 2009. Structure-function studies of a Melanocarpus albomyces laccase suggest a pathway for oxidation of phenolic compounds. J. Mol. Biol. 392:895–909. 10.1016/j.jmb.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 19.Galli C, Gentili P, Jolivalt C, Madzak C, Vadalà R. 2011. How is the reactivity of laccase affected by single-point mutations? Engineering laccase for improved activity towards sterically demanding substrates. Appl. Microbiol. Biotechnol. 91:123–131. 10.1007/s00253-011-3240-4. [DOI] [PubMed] [Google Scholar]
- 20.Xu F. 1996. Oxidation of phenols, anilines, and benzene thiols by fungal laccases: correlation between activity and redox potentials as well as halide inhibition. Biochemistry 35:7608–7614. 10.1021/bi952971a. [DOI] [PubMed] [Google Scholar]
- 21.Xu F, Kulys JJ, Duke K, Li K, Krikstopaitis K, Deussen HJW, Abbate E, Galinyte V, Schneider P. 2000. Redox chemistry in laccase-catalyzed oxidation of N-hydroxy compounds. Appl. Environ. Microbiol. 66:2052–2056. 10.1128/AEM.66.5.2052-2056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu F, Deussen HJW, Lopez B, Lam L, Li K. 2001. Enzymatic and electrochemical oxidation of N-hydroxy compounds. Redox potential, electron-transfer kinetics, and radical stability. Eur. J. Biochem. 268:4169–4176. [DOI] [PubMed] [Google Scholar]
- 23.Andberg M, Hakulinen N, Auer S, Saloheimo M, Koivula A, Rouvinen J, Kruus K. 2009. Essential role of the C-terminus in Melanocarpus albomyces laccase for enzyme production, catalytic properties and structure. FEBS J. 276:6285–6300. 10.1111/j.1742-4658.2009.07336.x. [DOI] [PubMed] [Google Scholar]
- 24.Gelo-Pujic M, Kim HH, Butlin NG, Palmore GTR. 1999. Electrochemical studies of a truncated laccase produced in Pichia pastoris. Appl. Environ. Microbiol. 65:5515–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salony Mishra S, Bisaria VS. 2006. Production and characterization of laccase from Cyathus bulleri and its use in decolourization of recalcitrant textile dyes. Appl. Microbiol. Biotechnol. 71:646–653. 10.1007/s00253-005-0206-4. [DOI] [PubMed] [Google Scholar]
- 26.Chhabra M, Mishra S, Sreekrishnan TR. 2009. Laccase/mediator assisted degradation of triarylmethane dyes in a continuous membrane reactor. J. Biotechnol. 143:69–78. 10.1016/j.jbiotec.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Eggert C, Temp U, Eriksson KEL. 1996. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl. Environ. Microbiol. 62:1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brissos V, Pereira L, Munteanu FD, Cavaco-Paulo A, Martins LO. 2009. Expression system of CotA-laccase for directed evolution and high-throughput screenings for the oxidation of high-redox potential dyes. Biotechnol. J. 4:558–563. 10.1002/biot.200800248. [DOI] [PubMed] [Google Scholar]
- 29.Bach CE, Warnock DD, Van Horn DJ, Weintraub MN, Sinsabaugh RL, Allison SD, German DP. 2013. Measuring phenol oxidase and peroxidase activities with pyrogallol, L-DOPA, and ABTS: effect of assay conditions and soil type. Soil Biol. Biochem. 67:183–191. 10.1016/j.soilbio.2013.08.022. [DOI] [Google Scholar]
- 30.Almeida PJ, Barros AA, Rodrigues JA. 1999. Determination of guaiacol at a carbon paste electrode using cathodic stripping voltammetry. Portugaliae Electrochim. Acta 17:11–19. 10.4152/pea.199901011. [DOI] [Google Scholar]
- 31.Koschorreck K, Schmid RD, Urlacher VB. 2009. Improving the functional expression of a Bacillus licheniformis laccase by random and site-directed mutagenesis. BMC Biotechnol. 9:12. 10.1186/1472-6750-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miele A, Giardina P, Sannia G, Faraco V. 2010. Random mutants of a Pleurotus ostreatus laccase as new biocatalysts for industrial effluents bioremediation. J. Appl. Microbiol. 108:998–1006. 10.1111/j.1365-2672.2009.04505.x. [DOI] [PubMed] [Google Scholar]
- 33.Liu YH, Ye M, Lu Y, Zhang X, Li G. 2011. Improving the decolorization for textile dyes of a metagenome-derived alkaline laccase by directed evolution. Appl. Microbiol. Biotechnol. 91:667–675. 10.1007/s00253-011-3292-5. [DOI] [PubMed] [Google Scholar]
- 34.Enguita FJ, Marçal D, Martins LO, Grenha R, Henriques AO, Lindley PF, Carrondo MA. 2004. Substrate and dioxygen binding to the endospore coat laccase from Bacillus subtilis. J. Biol. Chem. 279:23472–23476. 10.1074/jbc.M314000200. [DOI] [PubMed] [Google Scholar]
- 35.Bertrand T, Jolivalt C, Briozzo P, Caminade E, Joly N, Madzak C, Mougin C. 2002. Crystal structure of a four-copper laccase complexed with an arylamine: insights into substrate recognition and correlation with kinetics. Biochemistry 41:7325–7333. 10.1021/bi0201318. [DOI] [PubMed] [Google Scholar]
- 36.Tripp AE, Burdette DS, Zeikus JG, Phillips RS. 1998. Mutation of serine-39 to threonine in thermostable secondary alcohol dehydrogenase from Thermoanaerobacter ethanolicus changes enantiospecificity. J. Am. Chem. Soc. 120:5137–5141. 10.1021/ja974129t. [DOI] [Google Scholar]
- 37.Maté D, García-Burgos C, García-Ruiz E, Ballesteros AO, Camarero S, Alcalde M. 2010. Laboratory evolution of high-redox potential laccases. Chem. Biol. 17:1030–1041. 10.1016/j.chembiol.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 38.Kallio JP, Gasparetti C, Andberg M, Boer H, Koivula A, Kruus K, Rouvinen J, Hakulinen N. 2011. Crystal structure of an ascomycete fungal laccase from Thielavia arenaria: common structural features of asco-laccases. FEBS J. 278:2283–2295. 10.1111/j.1742-4658.2011.08146.x. [DOI] [PubMed] [Google Scholar]
- 39.Macellaro G, Baratto MC, Piscitelli A, Pezzella C, Fabrizi de Biani F, Palmese A, Piumi F, Record E, Basosi R, Sannia G. 2014. Effective mutations in a high redox potential laccase from Pleurotus ostreatus. Appl. Microbiol. Biotechnol. 98:4949–4961. 10.1007/s00253-013-5491-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.