Abstract
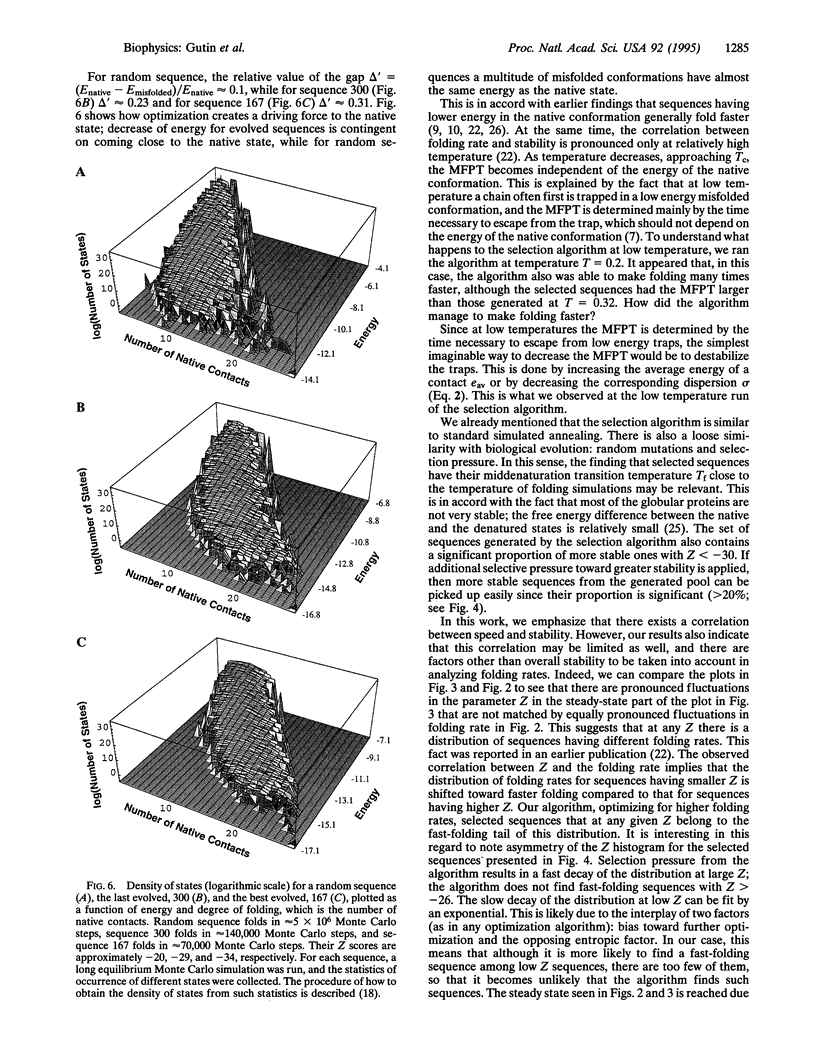
We propose an algorithm providing sequences of model proteins with rapid folding into a given target (native) conformation. This algorithm is applied to a chain of 27 residues on a cubic lattice. It generates sequences with folding 2 orders of magnitude faster than that of the practically random starting sequence. Thermodynamic analysis shows that the increase in speed is matched by an increase in stability: the evolved sequences are much more stable in their native conformation than the initial random sequence. The unfolding temperature for evolved sequences is slightly higher than the simulation temperature, bearing direct correspondence to the relatively low stability of real proteins.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abkevich V. I., Gutin A. M., Shakhnovich E. I. Specific nucleus as the transition state for protein folding: evidence from the lattice model. Biochemistry. 1994 Aug 23;33(33):10026–10036. doi: 10.1021/bi00199a029. [DOI] [PubMed] [Google Scholar]
- Bowie J. U., Lüthy R., Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991 Jul 12;253(5016):164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Camacho CJ, Thirumalai D. Minimum energy compact structures of random sequences of heteropolymers. Phys Rev Lett. 1993 Oct 11;71(15):2505–2508. doi: 10.1103/PhysRevLett.71.2505. [DOI] [PubMed] [Google Scholar]
- Goldstein R. A., Luthey-Schulten Z. A., Wolynes P. G. Protein tertiary structure recognition using optimized Hamiltonians with local interactions. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9029–9033. doi: 10.1073/pnas.89.19.9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. E., Fersht A. R. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry. 1991 Oct 29;30(43):10428–10435. doi: 10.1021/bi00107a010. [DOI] [PubMed] [Google Scholar]
- Jackson S. E., elMasry N., Fersht A. R. Structure of the hydrophobic core in the transition state for folding of chymotrypsin inhibitor 2: a critical test of the protein engineering method of analysis. Biochemistry. 1993 Oct 26;32(42):11270–11278. doi: 10.1021/bi00093a002. [DOI] [PubMed] [Google Scholar]
- Matouschek A., Serrano L., Fersht A. R. The folding of an enzyme. IV. Structure of an intermediate in the refolding of barnase analysed by a protein engineering procedure. J Mol Biol. 1992 Apr 5;224(3):819–835. doi: 10.1016/0022-2836(92)90564-z. [DOI] [PubMed] [Google Scholar]
- Privalov P. L. Thermodynamic problems of protein structure. Annu Rev Biophys Biophys Chem. 1989;18:47–69. doi: 10.1146/annurev.bb.18.060189.000403. [DOI] [PubMed] [Google Scholar]
- Ramanathan S, Shakhnovich E. Statistical mechanics of proteins with "evolutionary selected" sequences. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1994 Aug;50(2):1303–1312. doi: 10.1103/physreve.50.1303. [DOI] [PubMed] [Google Scholar]
- Sali A., Shakhnovich E., Karplus M. How does a protein fold? Nature. 1994 May 19;369(6477):248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- Sali A., Shakhnovich E., Karplus M. Kinetics of protein folding. A lattice model study of the requirements for folding to the native state. J Mol Biol. 1994 Feb 4;235(5):1614–1636. doi: 10.1006/jmbi.1994.1110. [DOI] [PubMed] [Google Scholar]
- Shakhnovich E. I., Gutin A. M. A new approach to the design of stable proteins. Protein Eng. 1993 Nov;6(8):793–800. doi: 10.1093/protein/6.8.793. [DOI] [PubMed] [Google Scholar]
- Shakhnovich E. I., Gutin A. M. Engineering of stable and fast-folding sequences of model proteins. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7195–7199. doi: 10.1073/pnas.90.15.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhnovich E. I., Gutin A. M. Implications of thermodynamics of protein folding for evolution of primary sequences. Nature. 1990 Aug 23;346(6286):773–775. doi: 10.1038/346773a0. [DOI] [PubMed] [Google Scholar]
- Shakhnovich E, Farztdinov G, Gutin AM, Karplus M. Protein folding bottlenecks: A lattice Monte Carlo simulation. Phys Rev Lett. 1991 Sep 16;67(12):1665–1668. doi: 10.1103/PhysRevLett.67.1665. [DOI] [PubMed] [Google Scholar]
- Shakhnovich EI. Proteins with selected sequences fold into unique native conformation. Phys Rev Lett. 1994 Jun 13;72(24):3907–3910. doi: 10.1103/PhysRevLett.72.3907. [DOI] [PubMed] [Google Scholar]