Abstract
This chapter summarizes what is currently known of the structures, physiological roles, involvement in pathogenicity and biogenesis of a variety of non-covalently bound cell envelope lipids and glycoconjugates of Mycobacterium tuberculosis and other Mycobacterium species. Topics addressed in this chapter include phospholipids; phosphatidylinositol mannosides; triglycerides; isoprenoids and related compounds (polyprenyl phosphate, menaquinones, carotenoids, non-carotenoid cyclic isoprenoids); acyltrehaloses (lipooligosaccharides, trehalose mono- and di-mycolates, sulfolipids, di- and poly-acyltrehaloses); mannosyl-beta-1-phosphomycoketides; glycopeptidolipids; phthiocerol dimycocerosates, para-hydroxybenzoic acids and phenolic glycolipids; mycobactins; mycolactones; and capsular polysaccharides.
I. Global structure and composition of the mycobacterial cell envelope
The compositional and architectural complexity of the mycobacterial cell envelope is probably the most distinctive feature of the Mycobacterium genus. It is the basis of many of the physiological and pathogenic features of these bacteria and the site of susceptibility and resistance to many anti-mycobacterial drugs [1, 2]. In the context of the increasing incidence of multidrug-resistant strains of Mycobacterium tuberculosis (Mtb), elucidating the complex pathways allowing mycobacteria to synthesize and assemble this complex structure represents a crucial area of research.
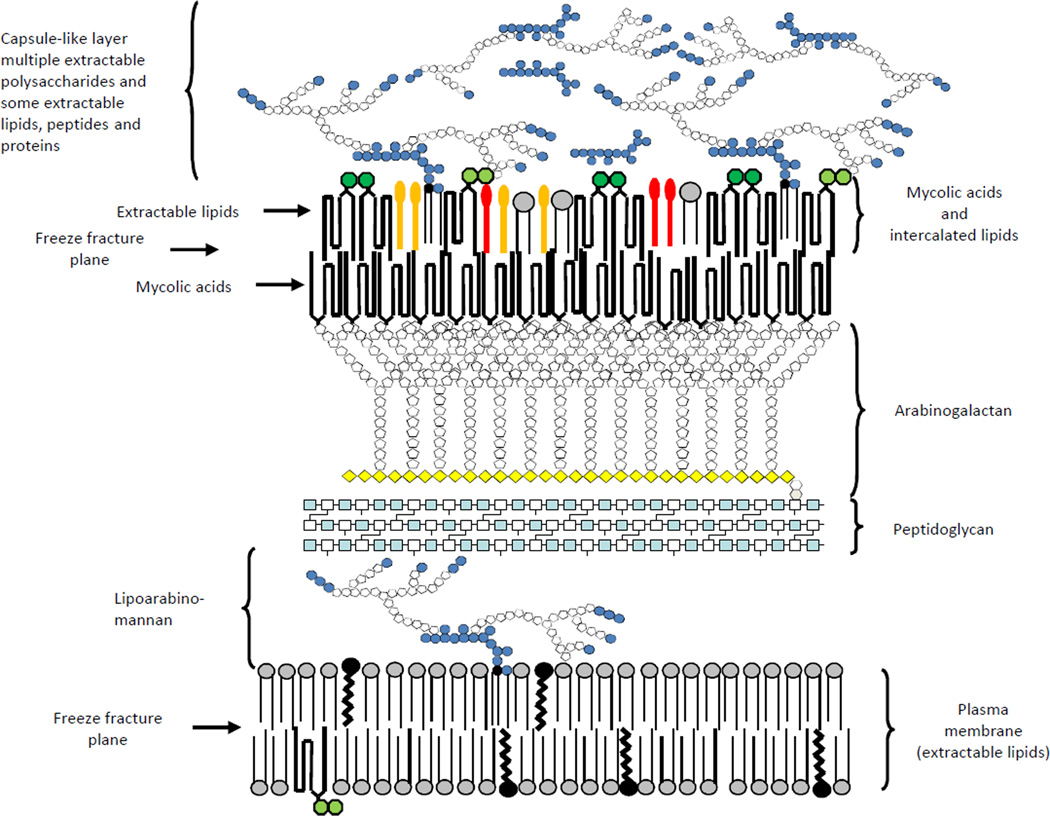
The mycobacterial cell envelope is made up of three major segments: the plasma membrane, the cell wall core and the outermost layer. The cell wall core consists of peptidoglycan (PG) in covalent attachment via phosphoryl-N-acetylglucosaminosyl-rhamnosyl linkage units with the heteropolysaccharide arabinogalactan (AG), which is in turn esterified at its non-reducing ends to α-alkyl, β-hydroxy long-chain (C70–C90) mycolic acids. The latter form the bulk of the inner leaflet of the outer membrane with the outer layer consisting of a variety of non-covalently attached (glyco)lipids, polysaccharides, lipoglycans and proteins [1, 3, 4] (Fig. 1). Only recently have developments in cryo-electron microscopy techniques allowed the different layers of the mycobacterial cell envelope to be visualized in their native state [3–5]. These studies provided direct evidence of the existence of an outer bilayer and periplasmic space in Mtb, M. bovis BCG, M. marinum, M. smegmatis and closely related Corynebacterium glutamicum (Fig. 1) [3–5]. Together with classical subfractionation and biochemical approaches, they also provided significant insights into the compositional diversity of the outermost layers of the cell envelopes of mycobacteria [5–9]. All Mycobacterium species studied to date elaborate more or less abundant ‘capsule’-like structures both in vitro and during host infection that primarily consist of polysaccharides and proteins with generally minor amounts of lipids [7, 9]. In some cases however (e.g., M. lepraemurium, M. leprae, M. avium), abundant quantities of species-specific glycolipids may be found (glycopeptidolipids and phenolic glycolipids in particular). Many of the proteins and lipids typically found in the capsules of mycobacteria also occur in the outer membrane and periplasm and their relative distribution between these three compartments seems to be species-dependent [8, 10]. This diversity in terms of surface composition most likely reflects differences in the cell envelope organization of mycobacteria and is likely to significantly impact the way that Mycobacterium species interact with the host [11, 12].
Figure 1. Schematic representation of the Mtb cell envelope.
Many of the classes of lipids and glycolipids discussed in the text are represented schematically and are shown in probable locations in the cell envelope. The structures with light and dark green hexagons represent trehalose mono- and dimycolates, respectively, the red lollipops represent phthiocerol dimycocerosates, while the gold ones represent sulfolipids, diacyltrehaloses and polyacyltrehaloses. Grey circles represent phospholipid headgroups, black circles, isoprenoids, light blue squares GlcNAc, white squares MurNAc, white pentagons arabinofuranose, yellow diamonds galactofuranose, and blue hexagons mannose. The overall schematic and individual structures are not drawn to scale and the numbers of carbohydrate residues shown are not representative of the actual molecules. Proteins and peptides are not shown for the sake of clarity.
Developments in the genetic manipulation of mycobacteria in the 1990s and the publication of the complete genome sequence of Mtb in 1998, followed later by that of several other rapidly-growing and slow-growing mycobacteria, have provided a major impetus to the study of cell envelope biosynthesis in various Mycobacterium species with the result that many of the enzymes involved in their synthesis have now been identified. The molecular genetics of the cell wall core proper (PG, AG, mycolic acids) is reviewed in other chapters of this book. This chapter focuses on what is known of the biosynthesis and translocation of the major non-covalently bound (extractable) lipid and glycoconjugate constituents populating the inner and outer membranes and capsule-like structures of mycobacteria. For those constituents ubiquitously distributed in mycobacteria, the gene nomenclature used is that of Mtb H37Rv.
II. Phospholipids, phosphatidylinositol mannosides and triglycerides
Phospholipids and triacylglycerols of mycobacteria
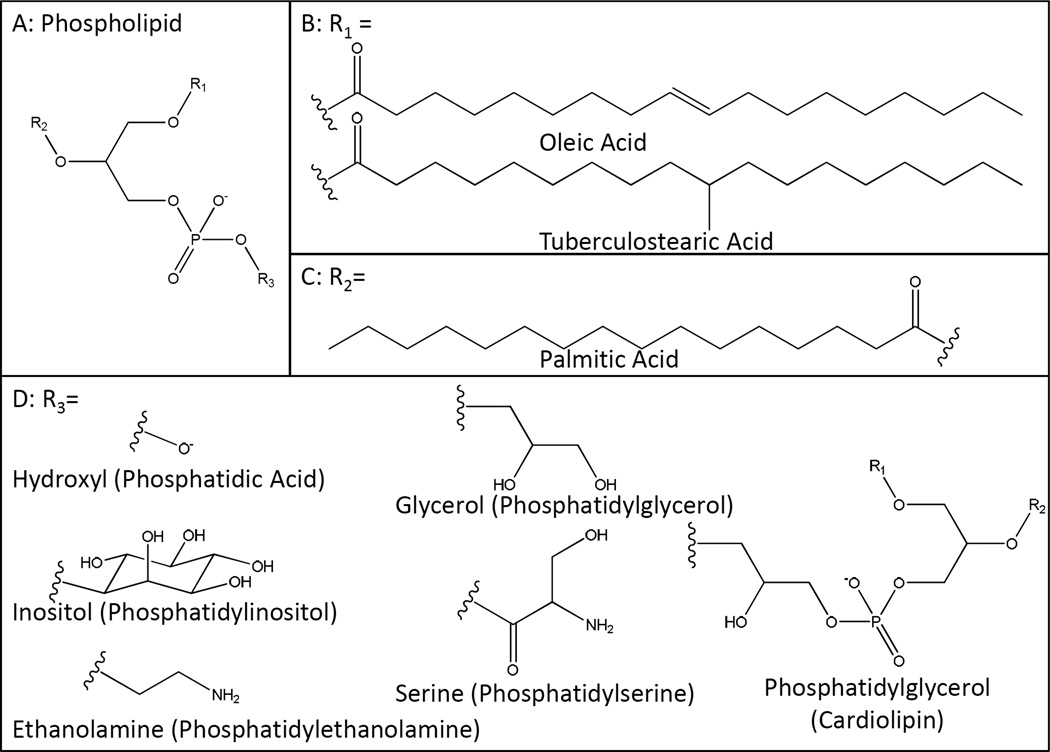
The mycobacterial phospholipids include phosphatidylglycerol (PG), diphosphatidylglycerol (i.e, cardiolipin) (CL), phosphatidylethanolamine (PE), phosphatidylinositol (PI) and mannosylated forms of PI known collectively as the phosphatidylinositol mannosides (PIM) (Fig. 2). Phosphatidylserine also occurs in limited amounts (Fig. 2) but phosphatidylcholine is apparently not produced by mycobacteria [13]. Phospholipids represent the main structural amphipathic polar lipids of the mycobacterial inner membrane and also populate the outer membrane (Fig. 1). PE and PIMs in particular were identified in the surface-exposed lipids of all Mycobacterium species investigated (Mtb, M. avium, M. kansasii, M. gastri, M. smegmatis and M. aurum) [8]. Palmitic (C16:0), oleic (C18:1) and tuberculostearic (C19) acids appear to be the major fatty acid substituents in the phospholipids of mycobacteria, with the unsaturated or branched C18:1 and C19 fatty acids principally esterifying position 1 of glycerol and C16:0 preferentially occupying position 2.
Figure 2. Structures of mycobacterial phospholipids.
Triacylglycerols, triglycerides, (TAG) have similarly been isolated from all mycobacterial species examined and represent the main apolar lipids when glycerol is the major carbon source in the culture medium [14]. Mycobacteria grown in vitro or recovered from human samples essentially accumulate TAG in the form of intracellular lipid droplets but TAG have also been identified in the surface-exposed lipids of M. smegmatis and M. avium [8]. They are proposed to act as a source of energy for actively replicating bacteria as well as a means by which free fatty acids are detoxified. TAG are also proposed to serve as an energy reserve for the long-term survival of Mtb during the persistence phase of infection [14, 15]. In M. bovis BCG and M. smegmatis, position 1 of TAG is occupied principally by stearic (C18:0), C18:1 and C19 fatty acids; position 2 is mostly esterified with C16 fatty acids, whereas the third position predominantly bears fatty acids with greater than 20 carbons (C20 to C33) [16]. The fatty acids acylating phospholipids and triglycerides in axenically-grown bacteria are thought to be synthesized by the Fatty Acid Synthase I (FAS-I) (Rv2524c) [17, 18].
Phosphatidic acid synthesis
Phosphatidic acid (Fig. 2) is a common intermediate in the biosynthesis of both TAG and phospholipids. The pathway begins with glycerol-3-phosphate which is formed by reduction of dihydroxyacetone phosphate by the glycerol-3-phosphate synthase GpsA. Two genes candidates were annotated for this function in the genome of Mtb H37Rv, gpdA1 (Rv0564c) and gpdA2 (Rv2982c), but neither of them has been confirmed biochemically. Glycerol-3-phosphate is first acylated by acyl-CoA, acyl-ACP or acyl-phosphate to form lysophosphatidate and then acylated again by acyl-CoA or acyl-ACP to yield phosphatidate [19]. Again, based on sequence similarities, two putative glycerol-3-phosphate acyltransferase genes, plsB1 (Rv1551) and plsB2 (Rv2482c), and one putative lysophosphatidate acyltransferase gene, plsC (Rv2483c), have been proposed to be involved in those acyl transfer reactions, but they have not yet been biochemically validated (Table 1).
Table 1.
Mtb H37Rv genes involved or thought to be involved in the biogenesis of phospholipids, triglycerides, isoprenoids and related lipidsa.
| Rv number |
Gene name |
Function | Evidence | Reference |
|---|---|---|---|---|
| Rv0221 | - | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv0308 | - | Putative phosphatidic acid phosphatase | H | [20] |
| Rv0436c | pssA | Putative phosphatidylserine synthase | P,H | [25] |
| Rv0437c | psd | Putative phosphatidylserine decarboxylase | H | - |
| Rv0534c | menA | Demethylmenaquinone synthase | E | [114] |
| Rv0542c | menE | o-succinylbenzoyl-CoA synthase | E | [111– 113] |
| Rv0548c | menB | 1,4-dihydroxy2-napthoic acid synthase | E | [108– 110] |
| Rv0562 | - | ω,E,E,E-geranylgeranyldiphosphate synthase | E | [87] |
| Rv0564c | gpdA1 | Putative glycerol-3-phosphate synthase | H | - |
| Rv0654 | - | Carotenoid oxygenase | E | [132] |
| Rv0895 | - | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv0989c | - | Geranyldiphosphate synthase | E | [85] |
| Rv1011 | ispE | 4-Diphosphocytidyl-2C-methyl-D-erythritol kinase | E | [61, 62] |
| Rv1086 | - | ω,E,Z-farnesyldiphosphate synthase | E | [79, 80] |
| Rv1159 | pimE | Polyprenol phosphomannose-dependent α-1,2-mannosyltransferase | E,P | [34] |
| Rv1411c | lprG | Lipoprotein; putative PIM, LM and LAM transporter | P | [38] |
| Rv1425 | - | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv1551 | plsB1 | Putative glycerol-3-phosphate acyltransferase | H | - |
| Rv1760 | - | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv1822 | pgsA2 | Putative cardiolipin synthase | H | - |
| Rv2188c | pimB’ | GDP-Man-dependent α-1,6-phosphatidylinositol mannosyltransferase | E,P | [31] |
| Rv2267c | stf3 | Putative sulfotransferase | P,H | [117] |
| Rv2285 | - | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv2361c | - | ω,E,poly-Z-decaprenyldiphosphate synthase | E | [83] |
| Rv2482c | plsB2 | Putative glycerol-3-phosphate acyltransferase | H | - |
| Rv2483c | plsC | Putative lysophosphatidate acyltransferase | H | - |
| Rv2484c | - | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv2524c | fas | Fatty acid synthetase type I | E,P | [17, 18] |
| Rv2610c | pimA | GDP-Man-dependent α-1,2-phosphatidylinositol mannosyltransferase | E,P | [28, 30] |
| Rv2611c | - | Acyltransferase involved in the 6-O-acylation of the Manp residue linked to the 2-position of myo-inositol in PIM1 and PIM2 | E,P | [32] |
| Rv2612c | pgsA1 | Phosphatidyl-myo-inositol synthase and/or phosphatidyl-myo-inositol phosphate synthase | E,P | [25, 26] |
| Rv2682c | dxs | 1-deoxy-D-xylulose-5-phosphate synthase | E | [49] |
| Rv2746c | pgsA3 | Phosphatidylglycerophosphate synthase | P,H | [25] |
| Rv2868c | ispG | 1-hydroxy-2-methyl-2(E)-butenyl 4-diphosphate synthase | H | [50] |
| Rv2870c | ispC | 1-Deoxy-D-xylulose 5-phosphate reductoisomerase | E | [54, 55] |
| Rv2881c | cdsA | Putative CDP-diacylglycerol synthase | H | [23] |
| Rv2982c | gpdA2 | Putative glycerol-3-phosphate synthase | H | - |
| Rv3087 | - | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv3088 | tgs4 | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv3130c | tgs1 | Acyl-CoA:diacylglycerol acyltransferase | E,P | [15, 21] |
| Rv3233c | - | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv3234c | tgs3 | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv3371 | - | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv3377c | - | Tuberculosinyldiphosphate synthase | E | [138] |
| Rv3378c | - | Isotuberculosinol synthase | E | [135, 139, 140] |
| Rv3383c | - | ω,E,E,E-geranylgeranyldiphosphate synthase | E | [87] |
| Rv3398c | - | ω,E,E-farnesyldiphosphate synthase | E | [86] |
| Rv3480c | - | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv3581c | ispF | 2C-methyl-D-erythritol 2,4-cyclodiphosphate | E | [63] |
| Rv3582c | ispD | 4-diphosphocytidyl-2C-methyl-D-erythritol | E | [58] |
| Rv3734c | tgs2 | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv3740c | - | Putative acyl-CoA:diacylglycerol acyltransferase | E | [15] |
| Rv3804c | fbpA | Acyl-CoA:diacylglycerol acyltransferase | E,P | [22] |
The experimental evidence for the annotation of a gene may either be ‘enzymatic’ (E) (i.e., an enzymatic activity was associated to the gene’s product in vitro) or ‘phenotypic’ (P) (i.e., the annotation results from the biochemical analysis of mycobacterial recombinant strains – e.g., knock-out/knock-down mutants, complemented mutant strains, overexpressors - or from the functional complementation of defined E. coli mutants). In some cases, the function of a gene is exclusively based on its homology to other known (myco)bacterial genes (H).
TAG synthesis
In the synthesis of TAG, phosphatidate is hydrolyzed by a specific phosphatase to yield diacylglycerol (DAG). This intermediate is then acylated to TAG in a reaction catalyzed by diglyceride acyltransferases (or triglyceride synthases). Although no phosphatidic acid phosphatases have yet been identified in mycobacteria, two proteins displaying this activity were recently characterized in Streptomyces coelicolor [20], one of which (SCO1102) displays sequence similarity with Rv0308 of Mtb H37Rv (Dr. Hugo Gramajo, personal communication). Fifteen genes were identified in the genome of Mtb H37Rv whose protein products display triglyceride synthase activity in vitro, generating triolein from diolein and oleyl-CoA [15, 21]. Interestingly, Ag85A (FbpA, Rv3804c) is also endowed with a similar acyltransferase activity, transferring long-chain acyl-CoA onto diacylglycerol [22] (Table 1).
Phospholipid biosynthesis
CDP-DAG appears to be the common precursor for the biosynthesis of phospholipids in mycobacteria and is synthesized from phosphatidic acid and CTP by the CDP-DAG synthase (CTP:phosphatidate cytidylyltransferase). Such enzymatic activity was detected in M. smegmatis and found to be membrane-associated [23]. The structural gene for CDP-DAG synthase in the genome of Mtb H37Rv is predicted to be cdsA (Rv2881c). Phosphatidyl-myo-inositol (PI) is made de novo from CDP-diacylglycerol (CDP-DAG) and myo-inositol [24] in a reaction catalyzed by the PI synthase, PgsA1 (Rv2612c) [25]. However, an alternative pathway for PI synthesis has been suggested wherein myo-Inositol is first phosphorylated to form myo-Inositol 3-phosphate which then reacts with CDP-DAG to form PI 3-phosphate (PI3P). It was proposed that pgsA1 encodes a PI3P synthase rather than a PI synthase, and that PI3P is subsequently dephosphorylated (by an as yet unknown enzyme) to yield PI [26]. Evidence based on sequence homology or changes in the phospholipid composition of M. smegmatis upon gene overexpression strongly suggest that the pgsA3 (Rv2746c) and pssA (Rv0436c) genes of Mtb encode the phosphatidylglycerophosphate synthase and phosphatidylserine synthase involved, respectively, in the formation of phosphatidylglycerol and phosphatidylserine [25]. As in other bacteria, PE is likely to arise from the decarboxylation of phosphatidylserine in a reaction catalyzed by the product of psd (Rv0437c). Cardiolipin may be formed from the condensation of two PG molecules by a cardiolipin synthase as in most prokaryotes, or through the transfer of a phosphatidyl group from CDP-DAG onto PG like in yeast and as recently shown in Streptomyces coelicolor [27]. Mtb H37Rv possesses a eukaryotic-type cardiolipin synthase bearing sequence similarity to the Streptomyces enzyme (PgsA2; Rv1822) whereas proteins displaying the characteristic phospholipase D-type features of classical prokaryotic cardiolipin synthases are missing, suggesting that the second pathway may be the one used by mycobacteria [25, 27]. Whether PgsA2 is endowed with such enzymatic activity remains however to be established (Table 1).
Phosphatidylinositol mannosides
The phosphatidylinositol dimannosides (PIM2) are considered both metabolic end products and intermediates in the biosynthesis of polar PIM (PIM5, PIM6), lipomannan (LM) and lipoarabinomannan (LAM) (for more details about these molecules and their biosynthetic pathways, see further section of this book). We will only briefly describe here the initial steps of PIM synthesis leading to the formation of PIM2 and PIM6, the two most abundant forms of PIM found in mycobacteria. The first step in PIM synthesis involves the transfer of a mannose residue from GDP-Manp to the 2-position of the myo-Inositol ring of PI to form phosphatidylinositol monomannoside, PIM1. We have identified PimA (Rv2610c) as the α-mannosyltransferase responsible for this catalytic step and found it to be an essential enzyme [28–30]. The second step involves the action of another essential α-mannosyltransferase, PimB’ (Rv2188c), which transfers a Manp residue from GDP-Manp to the 6-position of the myo-Inositol ring of PIM1 [31]. Both PIM1 and PIM2 can be acylated with palmitate at position 6 of the Manp residue transferred by PimA by the acyltransferase Rv2611c to form Ac1PIM1 and Ac1PIM2, respectively [32]. The acyltransferase responsible for the transfer of a fourth acyl group to position 3 of the myo-Inositol ring has not yet been identified. Likewise, the identity of the enzymes involved in the mannosylation of the dimannosylated forms of PIM to form PIM3 and PIM4 is at present unclear [33]. PimE (Rv1159) has been identified as the α-1,2-mannosyltransferase involved in the biosynthesis of PIM5 from PIM4 [34]. PimE belongs to the GT-C superfamily of glycosyltransferases which comprises integral membrane proteins that use polyprenyl-linked sugars as donors [33, 35]. Whether PimE also catalyzes the transfer of the second α-1,2-linked Man residue onto PIM5 to yield PIM6 or whether the formation of PIM6 results from the action of an independent mannosyltransferase is at present not known.
Translocation of phospholipids, PIM and TAG to the outer membrane and cell surface
Phospholipids and TAG are synthesized in the cytoplasm or at the periphery of the inner leaflet of the plasma membrane. Likewise, the early steps of PIM biosynthesis take place on the cytosolic face of the plasma membrane until PIM intermediates, believed to be PIM2 or PIM3, are translocated across the plasma membrane by an as yet unknown flippase to serve as substrates for further mannosylation reactions catalyzed by PimE and other GT-C polyprenyl-phosphate mannose-dependent glycosyltransferases [33, 35, 36]. Beyond their translocation across the plasma membrane, the further export of phospholipids, TAG and PIM to the outer membrane and cell surface most likely requires dedicated translocation machineries. Thus far, none of the flippases and transporters involved have been formally identified. Evidence based on physical interactions and co-crystallography suggests that the lipoprotein LprG (Rv1411c) which shares structural resemblance to LppX, a lipoprotein thought to carry phthiocerol dimycocerosates (PDIM) across the periplasm [37], may participate in the transport of PIM, LM and LAM to the cell surface [38]. This exciting hypothesis awaits further genetic and biochemical validation.
III. Isoprenoids and related lipids
Biosynthesis of isoprenoid precursors
A number of isoprenoids have been observed and characterized in Mycobacterium spp. including polyprenyl diphosphates, polyprenyl phosphates, lipid I and lipid II, carotenoids, menaquinones, sulfomenaquinones, and cyclic isoprenoids. These molecules have diverse and in some cases multiple functions. For example, polyisoprenyl phosphate (Pol-P) is involved in the biosynthesis of the arabinan portion of arabinogalactan, arabinomannan, and lipoarabinomannan, [39] and lipid I and lipid II of peptidoglycan biosynthesis [40, 41] as a lipid carrier of the activated saccharide subunits. Pol-P is also involved in the biosynthesis of the “linker unit” between two essential cell wall components, arabinogalactan and peptidoglycan [42].
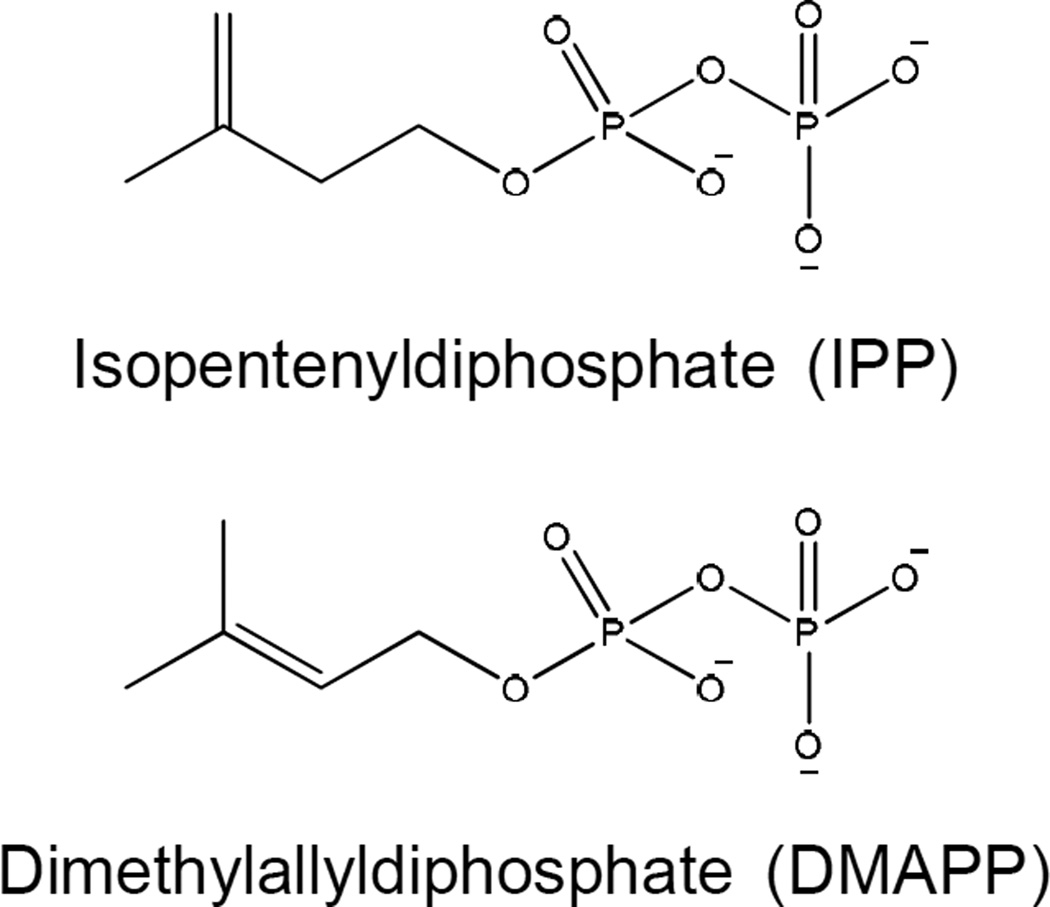
All isoprenoids are derived from the repetitive condensation of isopentenyl diphosphate (IPP) and allylic diphosphates [43] catalyzed by enzymes known as prenyldiphosphate synthases or prenyltransferases. To date, two distinct pathways for the biosynthesis of the IPP and dimethylallyl diphosphate (DMAPP, the smallest allylic diphosphate) have been identified: the mevalonate (MVA) pathway and the 2C-methyl-D-erythritol 4-phosphate (MEP) pathway. In mycobacteria, IPP and DMAPP (Fig. 3) are biosynthesized exclusively via the MEP pathway.
Figure 3. Structures of isopentenyldiphosphate and dimethylallyldiphosphate.
These molecules are precursors of all isoprenoid compounds.
(a) The 2C-methyl-D-erythritol 4-phosphate pathway of Mtb
The initial enzyme in the MEP pathway, 1-deoxy-D-xylulose-5-phosphate synthase (DXS), catalyzes the condensation of glyceraldehyde 3-phosphate (GAP) and pyruvate forming 1-deoxy-D-xylulose-5-phosphate (DXP) [44]. The product of DXS is used as not only a biosynthetic intermediate of IPP but also the precursors of thiamin (vitamin B1) and of pyridoxol (vitamin B6) in E. coli [45–47]; thus, DXS is not a committed step in the MEP pathway.
The dxs gene was first identified in E. coli [45, 46]. Sequence alignment with E. coli DXS demonstrated that Rv2682c has approximately 38% identity with a conserved DRAG motif and a key amino acid (His49) required for catalytic activity [48] (Table 1). The function of Rv2682c was demonstrated empirically as the purified recombinant enzyme is capable of producing DXS by condensation of pyruvate and GAP in the presence of thiamine pyrophosphate [49]. Interestingly, Mtb contains a second ortholog of E. coli DXS, Rv3379c. However, an alignment with E. coli DXS indicated that Rv3379c, despite a relatively high level of identity (38%), was truncated due to the positioning of an insertion element (IS6110) and, more importantly, the His49 residue is missing and the recombinant protein showed no DXS activity [49]. This, and the fact that Rv2682c is essential for bacterial survival [50] suggest that it encodes the only functional Mtb DXS.
1-Deoxy-D-xylulose 5-phosphate reductoisomerase (IspC), the second enzyme in the MEP biosynthetic pathway, catalyzes the rearrangement and reduction of DXP in the presence of NADPH to generate 2C-methyl-D-erythritol 4-phosphate (MEP) [51]. As mentioned above, DXP is a precursor not only of IPP and DMAPP but also of thiamine and pyridoxol; therefore, IspC catalyzes the first committed step for biosynthesis of IPP and DMAPP [52].
Alignments with E. coli IspC indicated that the primary structure of Rv2870c of Mtb is 25% identical to that of the E. coli IspC with conserved amino acid residues [53, 54]. Recombinant Rv2870c efficiently catalyzes the conversion of DXP to MEP in the presence of NADPH and the reverse reaction in the presence of NADP+ [54–56].
Incubation of MEP with crude, cell free extracts of E. coli in the presence of cytidine 5’-triphosphate (CTP) produces 4-diphosphocytidyl-2C-methyl-D-erythritol (CDP-ME) and the gene encoding the activity was identified as ygbP [57], which was later renamed ispD. The Rv3582c gene product has approximately 31% identity with E. coli IspD and recombinant Rv3582c protein was shown to be a functional IspD in Mtb [58].
The fourth step in the MEP pathway involves the conversion of CDP-ME to 4-diphosphocytidyl-2C-methyl-D-erythritol 2-phosphate (CDP-ME2P) in the presence of ATP catalyzed by IspE, which was initially identified in E. coli and tomatoes [59, 60]. Alignment of E. coli IspE with genes of the Mtb genome indicated that Rv1011 encodes a protein which harbors around 22% identity with conserved amino acids involved in forming the CDP-ME and ATP binding and crucial active sites and catalyzes CDP-ME phosphorylation in an ATP-dependent manner [61, 62].
The fifth step of the MEP pathway involves the formation of a metabolite containing a cyclodiphosphate moiety. The product of IspE, CDP-ME2P, is converted into 2C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) with corresponding release of cytidine 5’-monophosphate (CMP) by the ispF gene product [59]. Rv3581c encodes Mtb IspF [63] and is essential for bacterial survival [64]. The crystal structure of M. smegmatis IspF, harboring around 73% amino acid sequence identity with Mtb IspF, has been solved [64].
Recombinant E. coli ispC, ispD, ispE, ispF, and ispG were shown to catalyze the conversion of 1-deoxy-D-xylulose (DX) into 1-hydroxy-2-methyl-2(E)-butenyl 4-diphosphate (HMBPP) [65] and the ispH gene product is responsible for the conversion of HMBPP into IPP and DMAPP [66, 67]. Recombinant IspG catalyzes the reduction of MECDP resulting in opening of the cyclodiphosphate ring structure using a photoreduced deazaflavin derivative as an artificial electron donor [68, 69]. Interestingly, in vivo experiments using an E. coli strain overexpressing ispH resulted in the formation of IPP and DMAPP from HMBPP in a molar ratio of 5:1[70]. Blast searches of E. coli IspG or IspH with the Mtb genome indicates that Rv2868c, an essential gene [50], is the likely Mtb IspG and either Rv1110 or Rv3382c is the candidate gene encoding Mtb IspH.
(b) Isopentenyl diphosphate isomerase
Upon biosynthesis of IPP and DMAPP by IspH, IPP isomerase (Idi) catalyzes the interconversion of the two isoforms [71], however, the equilibrium favors the forward reaction, from IPP to DMAPP [72]. In organisms capable of synthesizing isoprenoid units by the MVA pathway, Idi is reported to be essential [73] as pyrophosphomevalonate decarboxylase in the MVA pathway produces only IPP and both DMAPP and IPP (Fig. 3) are required for further biosyntheses of isoprenoids. Orthologs of idi are also found in many organisms that utilize the MEP pathway, most of which are reported to encode non-essential enzymes [74], presumably because IspH of the MEP pathway produces both IPP and DMAPP. Two forms of bacterial Idi have been discovered to date; Type I, which includes Idi from E. coli and Type II which was identified in Streptomyces sp. strain CL190 [75]; Mtb has an ortholog of a type I Idi while M. smegmatis has an ortholog of the type II Idi.
(c) Prenyldiphosphate synthases
As mentioned above, the universal precursors of all isoprenoid compounds are synthesized from IPP, DMAPP or linear isopentenyl diphosphates that are synthesized by sequential 1’-4 condensations of IPP with DMAPP. The enzymes catalyzing this sequential process are known as prenyltransferases or prenyldiphosphate synthases. These enzymes can be divided into two families depending on the stereochemistry of the double bonds formed during product formation and the chain length of the final product. Thus, prenyldiphosphate synthases can be categorized as E-prenyldiphosphate synthases or Z-prenyldiphosphate synthases and there is no similarity between the two in terms of amino acid sequence. In the case of the E-prenyldiphosphate synthases, they can be further characterized as short-chain, having a product containing 10 to 25 carbons, medium-chain, 30 to 35 carbons, and long-chain, 40 to 50 carbons [76]. Similarly, the Z-prenyldiphosphate synthases can be characterized as short-chain, medium-chain and long-chain [77]. Both the E- and Z-prenyldiphosphate synthase families generate products with the correct chain-lengths via a molecular ruler mechanism, where one or two bulky amino acids occupy the bottom of each of the enzyme active sites to block extra chain elongation of the products, thereby determining the ultimate chain lengths [78]. Both E- and Z-prenyldiphosphate synthases have been identified and characterized in Mycobacterium species.
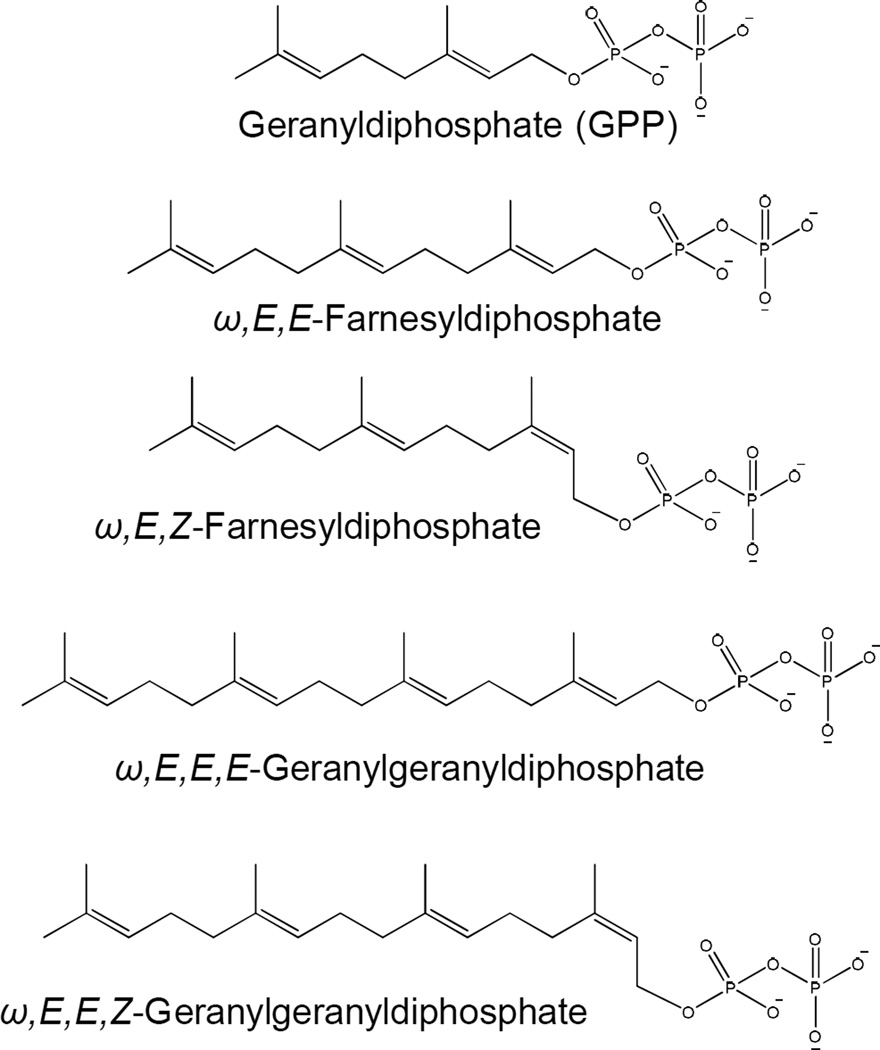
Mycobacterium spp are unusual in that they harbor two or three Z-prenyldiphosphate synthases, whereas most bacteria only have one of these enzymes. In Mtb, Rv1086 encodes a short-chain Z-prenyldiphosphate synthase that generates ω,E,Z-farnesyldiphosphate (Fig. 4, Table 1). This gene has been cloned, expressed and the enzyme activity characterized [79, 80] and was the first representative of this class of enzyme described. The crystal structure and mechanism of chain length determination has been solved [81, 82]. Rv2361c has been identified as a long-chain Z-prenyldiphosphate that synthesizes ω,E,poly-Z-decaprenyldiphosphate [79, 83] (Fig. 5). In Mycobacterium vanbaalenii, three Z-prenyldiphosphate synthases were identified and characterized [84]. Mvan_4662 accepts only geranyldiphosphate as the allylic primer producing only ω,E,Z-farnesyldiphosphate indicating a function similar to Rv1086. Mvan_1705 accepts only ω,E,E-farnesyldiphosphate synthesizing ω,Z,E,E-geranylgeranyl diphosphate, whereas, Mvan_3822 is a bifunctional Z-prenyldiphosphate synthase that preferentially synthesizes C35 or C50 products depending on the allylic reaction primer.
Figure 4. Structures of representative short-chain isopentenyldiphosphates synthesized by mycobacteria.
The sterochemical conformation is shown.
Figure 5. Structures of isoprenylphosphates reported from Mtb.
A number of E-prenyldiphosphate synthases have also been identified in mycobacteria, which synthesize E-prenyldiphosphates of various chain-lengths (Fig. 4, Table 1). These include Rv0989c, which is reported to synthesize geranyldiphosphate [85]; Rv3398c encoding an ω,E,E-farnesyldiphosphate synthase [86]; and Rv0562 and Rv3383c both of which are reported to encode ω,E,E,E-geranylgeranyldiphosphate synthases [87]. It should be noted that stereochemistry of the products of the E-prenyldiphosphate synthases is assumed based on the amino acid sequence of the enzyme not on empirical observation.
Polyprenyl phosphate
(a) Structures of mycobacterial polyprenyl phosphates
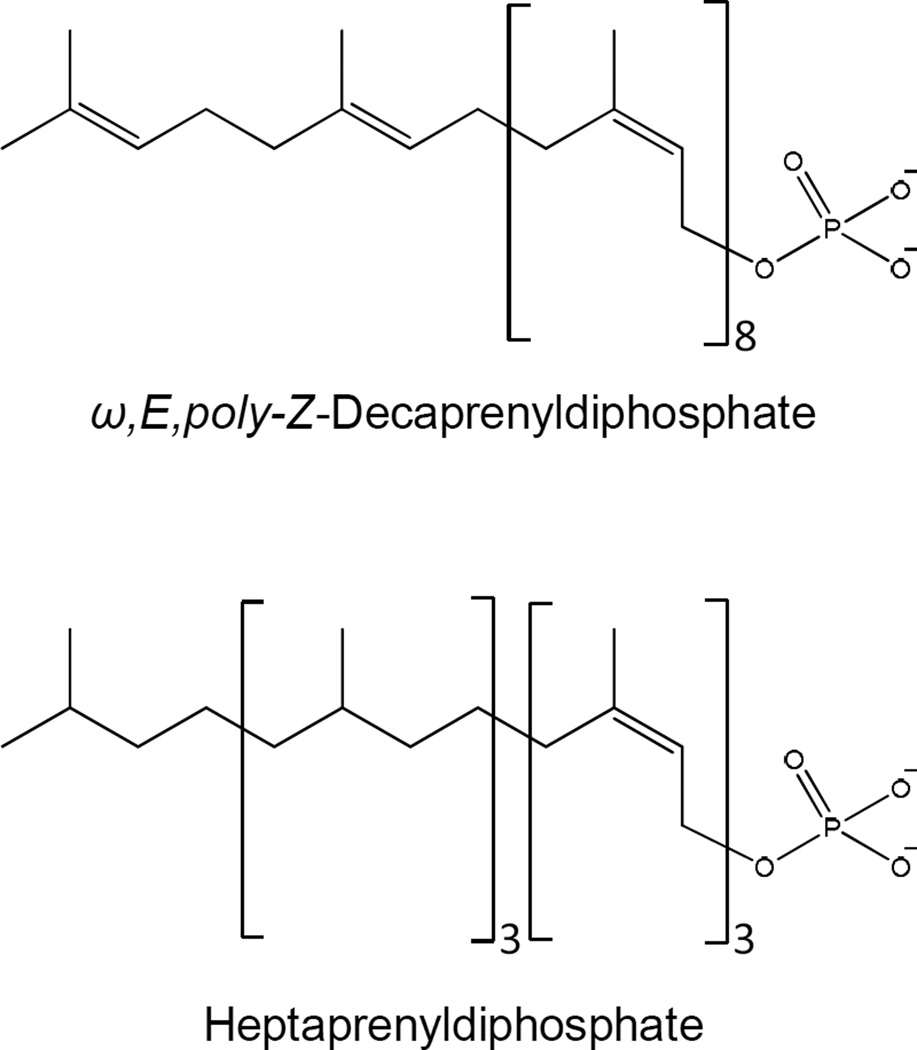
The most common structures of polyisoprenol (and therefore Pol-P) found in nature tend to be confined to four main groups: (i) ω,E-polyisoprenol, (ii) ω,di-E, poly-Z-polyisoprenol, (iii) ω,tri-E,poly-Z-polyisoprenol, and (iv) ω,Z-polyisoprenol [88]. Most bacteria utilize undecaprenylphosphate (or bactoprenylphosphate), a ω,di-E,octa-Z-prenylphosphate, as a carrier of activated sugars primarily for synthesis of oligo- and polysaccharides on the outside of the plasma membrane as is seen in peptidoglycan synthesis. However, mycobacteria synthesize and utilize at least two and perhaps three forms of Pol-P. In M. smegmatis two forms of Pol-P, decaprenyl phosphate (Dec-P) containing one ω, one E- and eight Z-isoprene units (ω,E,poly-Z) [39] and a heptaprenyl phosphate [89] containing four saturated isoprene units on the omega end of the molecule and two E- and one Z-isoprene units [90] or four saturated and three Z-isoprene units [91] have been reported (Fig 4). Mtb, on the other hand, appears to utilize a single predominant Pol-P (Dec-P). To date, the stereochemistry of the individual isoprene units of Dec-P from Mtb have not been determined [92]; however, it is likely that they are the same as those of the M. smegmatis Dec-P. In all three cases described above, the mycobacterial Pol-P molecules are structurally unusual.
(b) Polyisoprenyl phosphate biosynthesis
In general, all Pol-P molecules are synthesized via sequential condensation of IPP with allylic diphosphates catalyzed by prenyldiphosphate synthases described above forming polyisoprenyldiphosphates (Pol-PP) that are subsequently dephosphorylated. In mycobacteria, Rv1086 and Rv2361c (Table 1) can catalyze the addition of IPP to ω,E-GPP; however, kinetic analyses [80, 83] suggest that Rv1086 and Rv2361c act sequentially in the synthesis of ω,E,polyZ decaprenyl diphosphate (Dec-PP) the precursor of the ω,E,polyZ Dec-P found in mycobacteria [39, 89, 91–93], with Rv2361c adding seven isoprene units to the ω,E,Z-FPP synthesized by Rv1086. Thus, it seems likely that Rv0989c, Rv1086 and Rv2361c act in concert to generate decaprenyldiphosphate (Dec-PP), with isoprene units of the required stereochemistry. Once the Dec-PP has been synthesized, it must be dephosphorylated to form Dec-P (Fig. 5). Currently, there is little information regarding this biosynthetic transformation in mycobacteria; however, an ortholog of BacA, a phosphatase reported to be involved in dephosphorylation of Pol-PP in E. coli [94], may be involved.
Menaquinones
(a) Structure of mycobacterial lipoquinones
The lipoquinones involved in the respiratory chains of bacteria consist of menaquinones and ubiquinones [95], while mammals have only ubiquinone. Menaquinones (2-methyl-3-polyprenyl-1,4-naphthoquinones) are the predominant isoprenoid lipoquinones of mycobacteria and many Gram-positive bacteria, whereas Gram-negative bacteria typically utilize both menaquinone and ubiquinone or ubiquinone (which has a benzoquinone ring rather than a napthoquinone ring) solely [96–100].
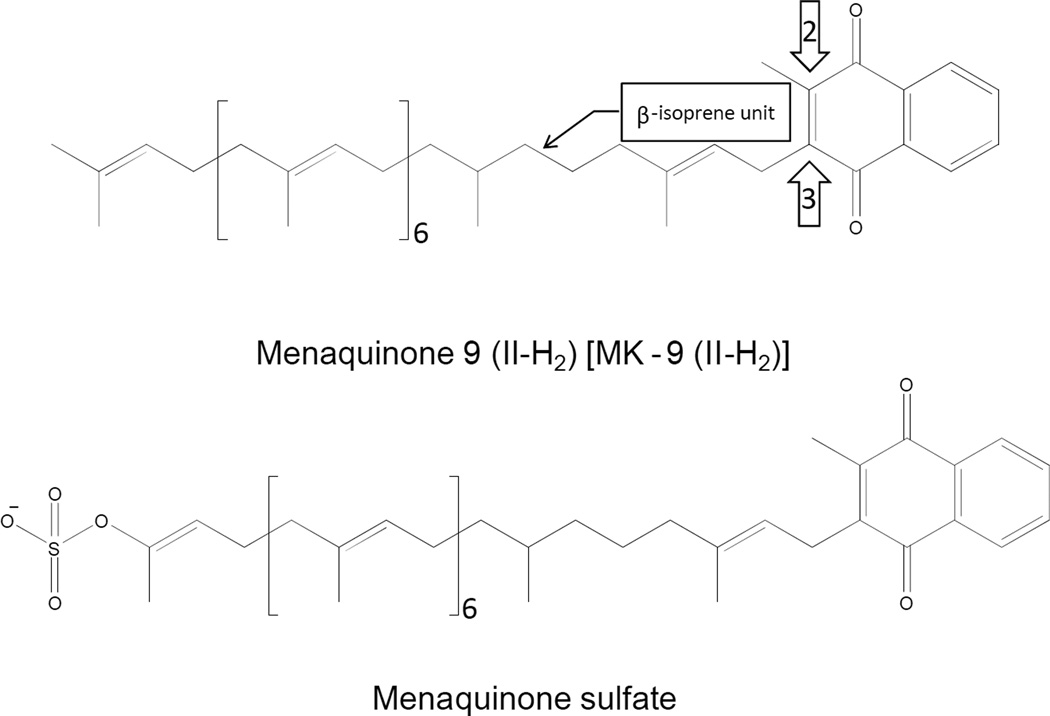
Menaquinones are identified by the variable portions of the molecules. Generally, the only variation seen in the naphthyl ring structure is whether or not the C2 position is methylated (Fig. 6). The most variant portion of the molecule is the polyisoprenyl side chain found at the C3 position. Menaquinones (and ubiquinones) are identified by the length and chemical structure of this side chain. For example, a menaquinone with a side chain of 8 isoprene units as seen in E. coli is identified as MK-8. The predominant form of menaquinone in mycobacteria has 9 isoprene units with the β position being saturated [96] (Fig. 6). Hence, this menaquinone is identified as MK-9 (II-H2).
Figure 6. Structures of the predominant menaquinone and menaquinone sulfate reported from Mtb.
Carbon positions 2, 3 and the β-isoprene unit are indicated by the arrows and call out.
(b) Functional significance of the menaquinone structure
Historically, respiratory quinones have been utilized for taxonomic purposes as the length and degree of saturation of the isoprenoid chain often reflect phylogenetic affiliation of bacteria [101]. The taxonomic distribution of structural features suggests that they are both functional and evolutionarily conserved. A great deal of effort was put into defining the significance of the various structural variations in the 1960s but this area of research has been largely ignored since. In 1970, Brodie et al. summarized the state of the knowledge [102]. Thus, it is known that the substitution at C2 of the napthyl ring is required for both oxidation and phosphorylation and must be a methyl group as conversion to a hydroxyl permits oxidation but not phosphorylation. The C3 position must be substituted with an isoprenoid chain to function as a membrane bound electron transporter. The double bond in the α-isoprene unit must be in the E-configuration, the Z-isomer does not allow phosphorylation. Thus, it appears that menaquinone in the electron transport is more than a simple electron transporter as structural modifications allow uncoupling of oxidation and phosphorylation suggesting that the menaquinone structure may regulate ATP synthesis. The single bond in the β-isoprene unit is conserved in many Gram-positive bacteria. However, the function of this modification is unknown. Recently, a novel sulfated menaquinone was isolated from Mtb, which appears to regulate virulence in mouse infection studies [103] but the precise function of this molecule is also unknown.
(c) Menaquinone biosynthesis
The biosynthesis of menaquinone takes place via the intersection of two separate pathways. 1,4-Dihydroxy-2-naphthoate is synthesized via the shikimate pathway. The naphthoate ring is then prenylated with a prenyldiphosphate, derived from a series of prenyl transferase reactions, to form demethylmenaquinone and, subsequently, the C2 position of the ring structure is methylated. The details of the biosynthesis of menaquinone studied in species other than Mycobacterium spp. have been reviewed [96, 104–106]. In mycobacteria, the β-isoprene unit of the prenyl group is reduced to form MK-9 (II-H2) after the formation of menaquinone [107].
In E. coli, the synthesis of menaquinone is accomplished by seven enzymes (menA–menG). These enzymes are encoded by two clusters of genes. The men cluster consisting of the menB,C,D,E,F and a separate cluster containing menA and menG. Menaquinone synthesis in Gram-positives in general has largely been ignored; however, the general pathway in Mtb appears to be similar. In Mtb, the menA–E genes appear to be found in a single cluster, whereas, the gene with the most homology to menF in E. coli is Rv3215 annotated as entC (isochorismate synthase).
Although menaquinone synthesis has been relatively extensively studied in E. coli (due in part to the availability of the men mutants, which can easily be generated in this organism, as it can utilize ubiquinone as an electron carrier in aerobic conditions), the synthesis of this compound in other organisms has received relatively little attention; however, MenB (1,4-dihydroxy2-napthoic acid synthase, Rv0548c) [108–110] (Table 1), MenE (o-succinylbenzoyl-CoA synthase, Rv0542c) [111–113], and MenA (Rv0534c) [114, 115] from mycobacteria have been studied as potential drug targets.
The isoprenoid tail of the menaquinone must be generated by a prenyldiphosphate synthase as described above and together with 1,4-dihydroxy-2-napthoic acid are the substrates for MenA (Rv0534c). However, the specific prenyldiphosphate synthase generating this prenyldiphosphate has yet to be identified. As noted above, other functions have been assigned to the potential candidates suggesting that additional study is required.
In addition, the saturation of the second isoprene unit from the head group of menaquinone in mycobacteria (Fig. 6) is not seen in E. coli or B. subtilis. However, this modification is seen in many Gram-positive bacteria [100, 116]. Based on the chemical mechanism of the prenyl diphosphate synthases it is likely that this modification is introduced after the mature prenyldiphosphate is synthesized and potentially after the formation of demethylmenaquinone or menaquinone. There is a single report that cell free extracts of Mycobacterium phlei are capable of reducing MK-9 to MK-9 (II-H2) [107]. The reduction required either NADH or NADPH but nothing further has been reported regarding the nature of this enzyme and it is, as yet, unknown whether this modification is required for function in mycobacteria.
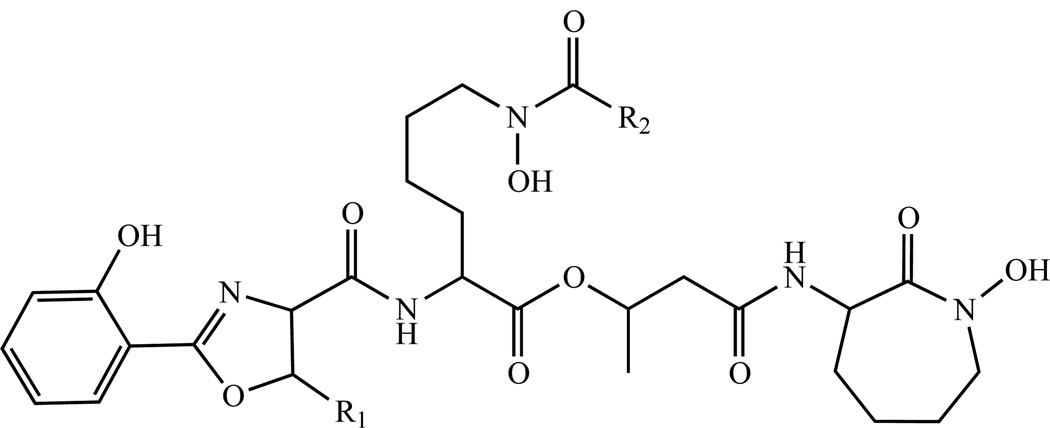
(d) Sulfated menaquinone
Sulfated menaquinone, where the sulfate is found on the ω-end of the isoprenoid tail (Fig. 6), have been isolated from Mtb [103]. The function of this unique lipid is, as yet, unknown. However, it has been reported that sulfated menaquinone, previously known as S881, negatively regulates the virulence of the organism in mouse infection models [117]. It has been postulated that this molecule is synthesized from MK-9 (II-H2) in at least two steps: 1) oxidation of the terminal position of the isoprenoid tail. 2) sulfation of the resulting hydroxyl residue. It has been shown that the putative sulfotransferase encoded by stf3, Rv2267c (Table 1), is required for the production of S881 [117] and hypothesized that Cyp128, encoded by Rv2268c, hydroxylates the MK-9 (II-H2). However this remains to be definitively demonstrated and Cyp124, encoded by Rv2266, has been shown to have appropriate ω-hydroxylase activity and a marked preference for lipids containing methyl branching such as isoprenoid compounds [118].
Carotenoids
(a) The carotenoids of mycobacteria
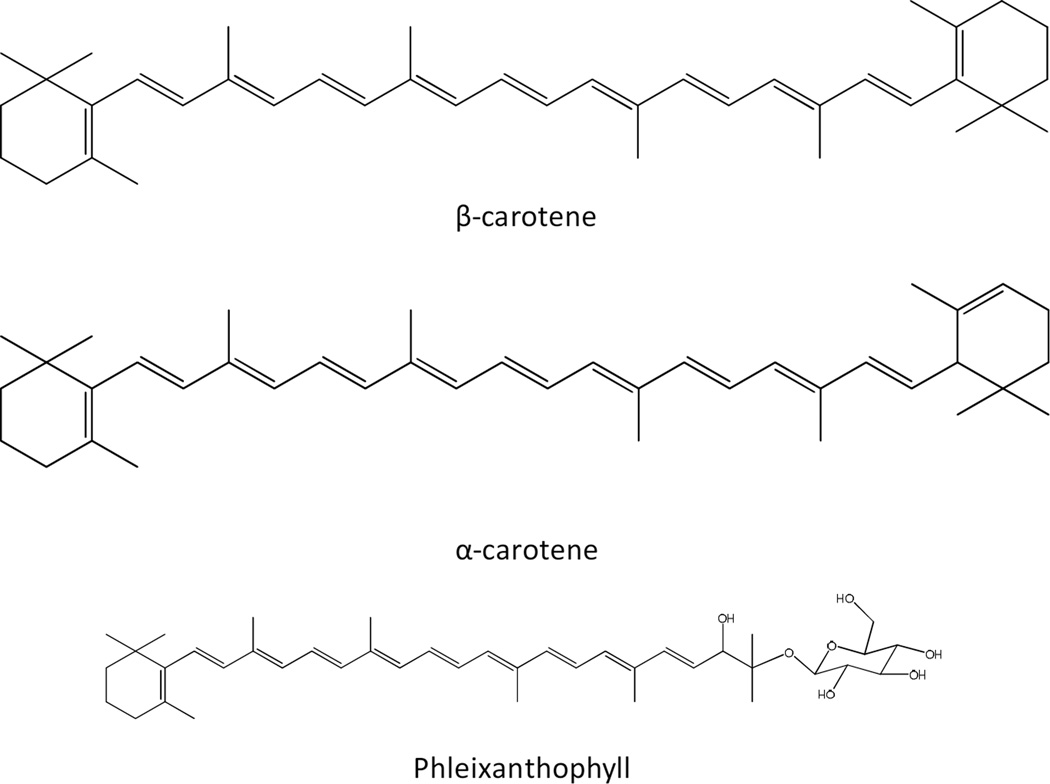
Carotenoids are a diverse family of isoprenoids that typically have 6 to 8 isoprene units. These molecules are structurally divers but are similar in general structure having a long chain of conjugated double bonds. More than 700 carotenoids have been identified and are widespread amongst bacteria including mycobacteria (Fig. 7). These, often pigmented, compounds play significant roles in protecting the organisms from oxidative damage and modify membrane fluidity [119, 120]. The carotenoids can be divided into two classes based on the presence or absence of oxygen atoms. Carotenoids without oxygen atoms in the molecule are known as carotenes, whereas, those with oxygen atoms in their structure are known as xanthophylls.
Figure 7. Structures of representative carotenoids found in mycobacteria.
Many Mycobacterium spp. produce yellow, orange or pink pigments in the dark (scotochromogens) or in the light (photochromogens) although these pigments may not be visible in culture. Very early on, mycobacteria were shown to contain carotenoid pigments [see [121] for a review]. Chargaff reported the presence of carotenoid pigments in M. phlei in 1930 and subsequent analysis showed that the major carotenoid in M. phei was leprotene (or isoneriatene) a carotene that was first isolated from an organism mistakenly identified as M. leprae [96]. In addition, many bacteria, including mycobacteria, produce carotenoid glycosides, which act as surfactants, stabilize membranes, and possibly contribute to regulating the permeability of membranes to oxygen [122–125]. The first complete structure of glycosylated carotenoids, phleixanthophyll and 4-keto-phleixanthophyll isolated from M. phlei, was determined in 1967 [126].
(b) Carotenoid biosynthesis
Carotenoid synthesis is well understood in many microorganisms [reviewed in [127]], but has received limited attention in mycobacteria; however the generally accepted pathway for carotenoid synthesis in mycobacteria, reviewed by Minnikin [96], appears to be similar to that of most non-photosynthetic microbes [127]. That is, the pathway consists of a geranylgeranyldiphosphate synthase, phytoene synthase, phytoene dehydrogenase and lycopene cyclase. In the carotenoid literature these enzymes are designated CrtE, CrtB, CrtI and CrtY, respectively. It should be noted that in non-photosynthetic bacteria, CrtI catalyzes multiple dehydrogenations (usually 2–4) that generates the conjugated double bond system and that there are multiple CrtY type cyclases with multiple designations [127]. Once lycopene has been generated in mycobacteria, the pathway splits to form α- and β-carotene [96] one of which is presumably the precursor of leprotene.
As described above orthologs of Rv0562 and Rv3383c, both of which are reported to encode E,E,E-geranylgeranyldiphosphate synthases [87], have the potential to provide the CrtE functionality in mycobacteria. Studies, aimed primarily at the development of genetic tools for manipulating mycobacteria, have provided information about other genes and enzymes involved in carotenoid synthesis in mycobacteria as well. Thus, orthologs of CrtB, CrtI and CrtY have been identified in M. marinum [128, 129] and M. aurum [130, 131]. In addition, an ortholog of CrtU, a β-carotene desaturase, has been reported in M. aurum [131] and a carotenoid oxygenase, Rv0654, has been identified in Mtb [132]. In terms of regulation of carotenoid synthesis in mycobacteria, orthologs of crtR and crtP encode a putative repressor and a positive regulator, respectively, in M. marinum and Mtb [128] and SigF controls carotenoid production in M. smegmatis [133]. Details regarding carotenoid synthesis in Mtb are not clear. The Mtb H37Rv genome encodes an ortholog of CrtB (PhyA), which may be non-functional [129].
Non-carotenoid cyclic isoprenoids
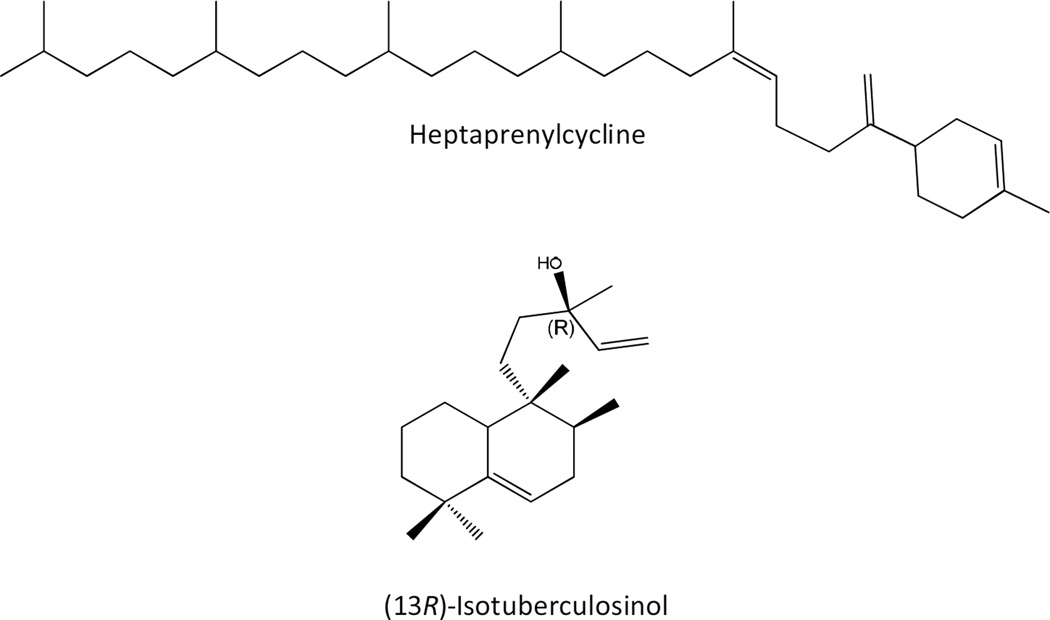
A novel class of cyclic C35 terpenes isolated from non-pathogenic Mycobacterium aichiense, Mycobacterium chlorophenolicum, Mycobacterium parafortuitum, M. smegmatis, Mycobacterium thermoresistible and Mycobacterium vanbaalenii has been described [84, 134]. These compounds, designated heptaprenylcyclines (Fig. 8), are synthesized via the cyclization of ω,E,polyZ-heptaprenyldiphosphate or ω,E,E,polyZ-heptaprenyl-diphosphate thus, the prenyldiphosphate synthases described in these species are likely involved in the production of these molecules but little else is currently known about their synthesis or function.
Figure 8. Structures of representative non-carotenoid cyclic isoprenoids found in mycobacteria.
A labdane-related diterpenoid compound, isotuberculosinol (Fig. 8), is produced by Mtb. This molecule appears to be immunomodulatory as it has been shown to block phagosome maturation in macrophages [135, 136], indeed this role was first suggested when genes encoding enzymes involved in isotuberculosinol synthesis, Rv3377c and Rv3378c, were identified in a screen for mutants defective in arresting phagosome maturation [137]. Rv3377c (Table 1) was demonstrated to be a class II diterpene cyclase, catalyzing bicyclization and rearrangement of geranylgeranyldiphosphate to form halimadienyl/tuberculosinyldiphosphate [138]. It was then shown that halimadienyl/tuberculosinyldiphosphate was hydrolyzed to tuberculosiol and isotuberculosinol by Rv3378c [135, 136, 139, 140].
IV. Acyltrehaloses
The outer membrane of mycobacteria contains a number of trehalose esters. Among them, trehalose monomycolates (TMM) and trehalose dimycolates (TDM; cord factor) are ubiquitously found across the Mycobacterium genus. Species-specific trehalose esters include di-, tri-, and poly-acyltrehaloses (DAT, TAT and PAT), sulfolipids (SL) and lipooligosaccharides (LOS). Species-specific trehalose esters are found in the outermost capsule in addition to the outer membrane [8]. TMM and TDM in contrast were identified in the surface-exposed capsular materials of M. avium and M. smegmatis but not in those of Mtb, M. kansasii and M. gastri indicating that they may be more deeply buried in the cell envelope of some Mycobacterium spp. [8]. Interest in trehalose esters stems from their demonstrated or postulated roles in host-pathogen interactions and from their potential as diagnostic tools (for a review, [1, 141]). The presence and abundance of species-specific acyltrehaloses (SL, DAT, TAT and PAT) and phthiocerol dimycocerosates (PDIM; see section VI) in the cell envelope of Mtb impact on the ability of the bacilli to stain with the cationic dye neutral red [142, 143], a property known since Dubos and Middlebrook’s early studies in the 1940s to correlate with virulence [144].
The biosynthesis of trehalose is reviewed in chapter xxx. Therefore, we will here only focus on the subsequent steps of the formation of acyltrehaloses, including the biosynthesis of the fatty acyl substituents, their transfer onto trehalose and what is known of the translocation of biosynthetic intermediates and end products across the cell envelope.
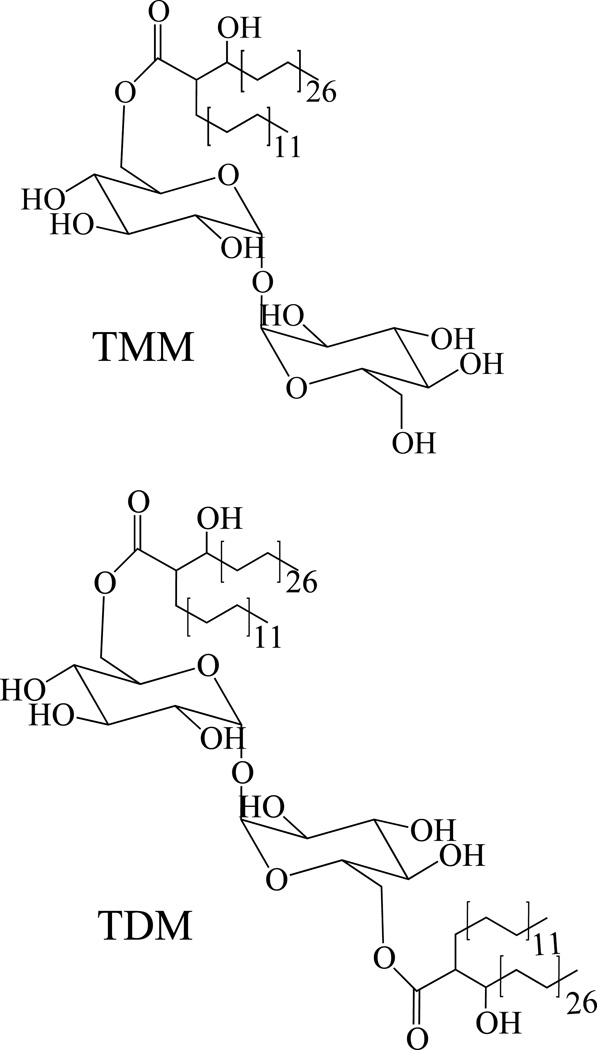
Trehalose monomycolates (TMM) and trehalose dimycolates (TDM; cord factor)
In TMM and TDM, trehalose is esterified with long-chain α-branched β-hydroxy fatty acids known as the mycolic acids. The structure and biosynthesis of mycolic acids is reviewed in chapter xxx. Any structural type of mycolic acids may esterify the positions 6 and 6’ of TDM and the position 6 of TMM (Fig. 9). The biosynthesis of mycolic acids occurs in the cytoplasm and so does that of trehalose. We recently identified MmpL3 (Rv0206c) (Table 2) as an inner membrane transporter required for the translocation of TMM to the periplasm where TMM can then serve as a mycolic acid donor for the mycolylation of arabinogalactan and the formation of TDM (Fig. 1) [145, 146]. This finding indicates that TMM is most likely the form under which mycolic acids are exported to the cell wall and outer membrane and, therefore, that TMM is probably made on the cytosolic side of the plasma membrane. The catalytic process underlying the cytoplasmic formation of TMM from fully elongated and functionalized mycolic acid chains and trehalose has not yet been elucidated. The subsequent synthesis of TDM from two TMM molecules and the transfer of mycolates to the non-reducing ends of arabinogalactan have been shown to involve the antigens 85A (Rv3804c; FbpA), 85B (Rv1886c; FbpB), and 85C (Rv0129c; FbpC) (Table 2) [147–149]. In vitro, these three mycolyltransferases display apparent redundant catalytic activities [147]. Consistent with this finding, none of the fbpA, B or C genes are individually required for the growth of Mtb. Their combined inactivation or chemical inhibition, however, leads to cell death [147, 150] (our unpublished data). Although the phenotypic characterization of fbpA, B or C null mutants of Mtb and M. smegmatis indicates that the function of these genes may in fact only partially overlap in whole cells, to this date, the precise contribution of each of the three paralogs to the transfer of mycolic acids to their cell wall and outer membrane glycolipid acceptors remains unclear. FbpC appears to be essentially involved in the transfer of mycolic acids to arabinogalactan, and FbpA in the formation of TDM [148, 149, 151–153].
Figure 9. Structures trehalose monomycolates (TMM) and trehalose dimycolates (TDM).
Table 2.
Mtb H37Rv genes involved in the biogenesis of trehalose mono- and di-mycolates, sulfolipids, di- and poly-acyltrehaloses and mannosyl-β-1-phosphomycoketides.
| Gene number |
Gene name |
Function | Evidence | Reference |
|---|---|---|---|---|
| Rv0129c | fbpC | Mycolyltransferase (antigen 85C) | E,P | [147–149] |
| Rv0206c | mmpL3 | Inner membrane transporter of the RND superfamily involved in the translocation of TMM | P | [145, 146] |
| Rv0295c | Sft0 | Sulfotransferase responsible for the formation of the trehalose-2-sulfate moiety of sulfolipids | E,P | [164] |
| Rv0757-Rv0758 | phoP-phoR | Two-component transcriptional regulator involved in the regulation of SL, DAT and PAT | P | [143, 176] |
| Rv1180-Rv1181 | pks3-pks4 | Polyketide synthase responsible for the elongation of the methyl-branched mycosanoic and mycolipenic acids found in DAT, TAT and PAT | P | [196, 197] |
| Rv1182 | papA3 | Acyltransferase catalyzing the sequential transfer of the first straight-chain saturated fatty acyl chain followed by the first mycolipenoyl group onto the 2- and 3-positions of trehalose, respectively, in the biosynthesis of DAT and PAT | E,P | [199] |
| Rv1183 | mmpL10 | Inner membrane transporter of the RND superfamily thought to be involved in the translocation of DAT and PAT | H | - |
| Rv1184c | chp2 | Acyltransferase thought to catalyze the last three acylations leading to the formation of PAT from DAT | H | - |
| Rv1185c | fadD21 | Putative fatty acid AMP ligase providing Pks3/4 with activated long-chain fatty acid starter units | H | - |
| Rv1662-Rv1663 | pks8-pks17 | Polyketide synthase responsible for the elongation of the monomethyl-branched unsaturated C16 to C20 fatty acids found in DAT and PAT | P | [162] |
| Rv1886c | fbpB | Mycolyltransferase (antigen 85B) | E | [147, 149] |
| Rv2048c | pks12 | Polyketide synthase involved in the elongation of the alkyl backbone of mycoketides | E,P | [218, 220] |
| Rv3416 | whiB3 | Regulator of SL, DAT and PAT synthesis | P | [174] |
| Rv3804c | fbpA | Mycolyltransferase (antigen 85A) | E,P | [147, 149, 152] |
| Rv3820c | papA2 | Acyltransferase catalyzing the transfer of the first straight-chain saturated fatty acyl chain onto trehalose-2-sulfate in the biosynthesis of sulfolipids | E,P | [165] |
| Rv3821 | sap | Integral membrane protein thought to facilitate the translocation of SL-1 to the cell surface | P | [168] |
| Rv3822 | chp1 | Acyltransferase catalyzing the acylation at the 6- and 6’-positions of sulfolipids | E,P | [168] |
| Rv3823c | mmpL8 | Inner membrane transporter of the RND superfamily involved in the translocation of sulfolipids | P | [168, 170, 171] |
| Rv3824c | papA1 | Acyltransferase catalyzing the transfer of the first (hydroxy)phthioceranoyl group at the 3’-position of the product of PapA2 | E,P | [165] |
| Rv3825c | pks2 | Polyketide synthase responsible for the elongation of the methyl-branched phthioceranic and hydroxyphthioceranic acids found in sulfolipids | P | [166] |
| Rv3826 | fadD23 | Putative fatty acid AMP ligase providing Pks2 with activated long-chain fatty acid starter units | H | - |
E,P,H: see Table 1.
Numerous biological activities have been associated with the TDM from tuberculous and non-tuberculous mycobacteria both in vitro and in vivo (for a review, [1, 154–157]). In fact, TDM seems to be a major contributor to the inflammation seen in mycobacterial infections. TDM contributes to protecting Mtb from killing by macrophages, is a potent modulator of the activation of macrophages, stimulates the formation of lung granulomas and enhances the resistance of mycobacteria to antibiotics [152, 154, 156, 158, 159]. The binding of TDM from Mtb to the C-type lectin Mincle is required for activation of macrophages and granuloma formation [158, 160]. Importantly, the biological activities of TDM are much dependent on the fine structure of their mycolyl substituents [156, 161].
Sulfolipids (SL)
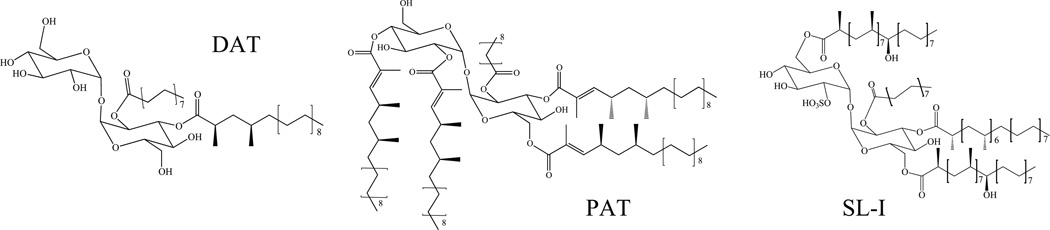
Sulfolipids (SL), also known as sulfatides and sulfoglycolipids, are sulfated trehalose esters that are acylated with three or four acyl groups consisting of one middle-chain saturated fatty acid (palmitic or stearic acid) at the 2-position and different combinations of the hepta- and octa-methyl-branched phthioceranic and hydroxyphthioceranic acids (C31-C46) at the 3-, 6-, and 6’-positions. Monomethyl-branched unsaturated C16 to C20 fatty acids have also been found as minor constituents of SL [162]. Sulfolipid-1 (SL-1), whose structure is shown on Fig. 10 is the most abundant form of sulfolipid produced by Mtb [163]. This family of lipids is exclusively found in the human pathogen Mtb.
Figure 10. Structures sulfolipids (SL), diacyltrehaloses (DAT) and polyacyltrehaloses (PAT) and biosynthetic gene clusters.
The major sulfolipid SL-I (2,3,6,6’-tetraacyl α-α’-trehalose-2’-sulfate) is represented. In SL-I, trehalose is sulfated at the 2’ position and esterified with palmitic acid and the multimethyl-branched phthioceranic and hydroxyphthioceranic acids. In DAT (2,3-di-O-acyltrehalose), trehalose is esterified with palmitic acid and the multimethylbranched mycosanoic acid. In PAT, trehalose is esterified with palmitic acid and the multimethyl-branched mycolipenic acids.
The genes involved in the biogenesis of SL-1 have been for the most part identified and, with the exception of the sulfotransferase Sft0, found to cluster on the chromosome of Mtb (Table 2). The sulfotransferase Sft0 (Rv0295c) catalyzes the first committed step in the pathway by sulfating trehalose to form trehalose-2-sulfate [164]. The acyltransferase PapA2 (Rv3820c) then catalyzes the esterification of trehalose-2-sulfate with a straight-chain saturated fatty acid (e.g., palmitic acid) at the 2’-position to generate a monoacyl intermediate, SL659 [165]. The polyketide synthase Pks2 (Rv3825c) synthesizes the methyl-branched phthioceranic and hydroxyphthioceranic acids [166] most likely using an activated long-chain fatty acid starter unit (an acyl-adenylate) provided by the fatty acid AMP ligase FadD23 (Rv3826) [167]. The polyketide-associated protein-1 (PapA1; Rv3824c) catalyzes the transfer of the first (hydroxy)phthioceranoyl group at the 3’-position of the product of PapA2 yielding a diacylated form of SL known as SL1278 [165]. The additional two acylations at the 6- and 6’-positions of SL1278 are catalyzed by the acyltransferase Chp1 (Rv3822) [168]. PapA1 and PapA2 are related to the acyltransferase PapA5 which esterifies phthiocerol with mycocerosic acids in the biosynthesis of PDIM (see section VI). Chp1 (cutinase-like hydrolase protein-1), in contrast, more closely resembles cutinase-like proteins [168]. All three acyltransferases are essential for the synthesis of SL-1 as demonstrated by the absence of fully elaborated SL-1 from the corresponding knock-out mutants [165, 168, 169].
Evidence for the involvement of MmpL8 (Rv3823c), an inner membrane transporter of the RND (Resistance, Nodulation and Division) superfamily, in the translocation of SL-1 to the cell surface was provided in 2003–2004 by two independent research groups [170, 171]. Mtb mmpL8 knock-out mutants fail to produce SL-1 and instead accumulate the diacylated SL1278 intracellularly. A possible interpretation of this finding was that the two first acylation steps catalyzed by PapA2 and PapA1 occurred on the cytoplasmic side of the plasma membrane whereas the two subsequent acylations catalyzed by Chp1 and yielding SL-1 required the prior MmpL8-mediated translocation of the diacylated SL1278 precursor across the plasma membrane. This model was however recently revised in light of the finding that the catalytic domain of the membrane-associated acyltransferase Chp1 is cytosolic and that its activity is potentiated by MmpL8 [168]. These observations are consistent with a model similar to that proposed for PDIM (see section VI) wherein the biosynthesis and transport of SL-1 is coupled and MmpL8 acts as scaffold for a cytoplasmically-oriented macromolecular complex consisting of the SL biosynthetic machinery. Further support for this assumption was recently obtained by Zheng et al. [172] in identifying MmpL8 among the component of a membrane-associated protein complex containing Pks2, PapA1 and FadD23 in M. bovis BCG. Sap (sulfolipid-1-addressing protein) (Rv3821) is an integral membrane protein that appears to facilitate the translocation of SL-1 to the cell surface. Its disruption in Mtb causes the intracellular build-up of SL1278 similar to that observed in mmpL8 knock-outs although the sap mutant retains the ability to synthesize small amounts of SL-1 [168]. Beyond MmpL8 and Sap, it is likely that the translocation of SL-1 to the cell surface requires additional periplasmic and/or outer membrane transporters but their identity is at present not known.
SL production appears to be regulated in Mtb but the environmental factors governing the synthesis of these glycolipids are still poorly understood. Supporting a role for SL during host infection, the expression of the pks2 gene was found to be strongly upregulated upon phagocytosis of Mtb by human primary macrophages [173]. It appears that one of the roles of methyl-branched fatty acid-containing lipids such as PDIM, SL, DAT and PAT during infection is to alleviate the propionate-mediated stress undergone by Mtb when the bacterium switches to host cholesterol as a major carbon source [174, 175]. The propionyl-CoA generated upon β-oxidation of cholesterol is converted to methylmalonyl-CoA by the propionyl-CoA carboxylase which is then used by dedicated polyketide synthases such as Pks2, Mas and Pks3/4 (see further) in the elongation of the methyl-branched fatty acids found in PDIM, SL, DAT and PAT. The regulator facilitating this metabolic switching to fatty acids was identified as WhiB3 (Rv3416). WhiB3 binds the promoter region of pks2 [174]. Another important regulator of SL production is the two-component transcriptional regulator PhoP-PhoR (Rv0757−Rv0758). PhoP-PhoR positively regulates the synthesis of SL and Mtb mutants deficient in the expression of this regulator are totally deficient in SL-1 production [143, 176]. It was shown that a mutation in the phoP gene of Mtb H37Ra accounts for the inability of this avirulent strain to produce SL-1 [177]. PhoP binds the promoter region of pks2 in vitro [178, 179].
The restriction of SL-1 to the human pathogen Mtb together with the observation some 50 years ago of a positive correlation between the levels of SL-1 produced by Mtb clinical isolates and their virulence in animal models has prompted extensive research aimed at elucidating the biological functions of sulfolipids during host infection (for a review, [141, 163, 180, 181]). Numerous and sometimes controversial activities were associated with purified SL-I molecules. Among these, the ability of SL-1 to potentiate the toxicity of TDM in mice, to inhibit mitochondrial oxidative phosphorylation, to prevent phagosome-lysosome fusion in cultured macrophages and to modulate the oxidative and cytokine responses of human monocytes and neutrophils are probably the ones that have received the most attention. In more recent years, the diacylated SL biosynthetic precursor SL1278 was shown to stimulate CD1b-restricted T cells through mechanisms dependent on the number of C-methyl substituents on the fatty acyl chains, the configuration of the chiral centers, and the length and respective localization of the two acyl chains on the sugar moiety [182, 183]. In the last decade, the elucidation of the biosynthetic pathway of SL finally allowed the generation of isogenic mutants of Mtb specifically deficient in their synthesis and an evaluation of the roles of these glycolipids during infection when carried by whole bacilli. Unexpectedly, pks2, papA1 and papA2 knock-out mutants, which all lack fully elaborated SL-1 while retaining in some cases the ability to synthesize sulfated trehalose, and mono- and/or di-acylated forms of SL, were found to be undistinguishable from their wild-type parent in their ability to replicate and persist in mice or guinea pigs [165, 184] In contrast, three independent studies indicated that mmpL8 KO mutants which accumulate diacylated SL1278 at the periphery of the plasma membrane display some level of attenuation in mice although the attenuation phenotypes considerably differed between studies, possibly as a result of the different Mtb strains and models of infection that were used [170, 185, 186]. Recently, Gilmore et al. [187] provided evidence that a sft0 null mutant of Mtb survives better than its wild-type parent in human but not in murine macrophages, possibly as a result of the increased resistance of this strain to human antimicrobial peptides. These results suggest that SL may only have a detectable impact on infection in the human host.
Di-acyltrehaloses (DAT) and poly-acyltrehaloses (PAT)
The 2,3-di-O-acyltrehaloses (DAT) consist of trehalose acylated at the 2-position with one middle-chain saturated fatty acid (C16–C19) and at the 3-position with the di-methyl-branched mycosanoic acids (C21–C25) (Fig. 10). In other less common forms of DAT, the tri-methyl-branched C25–C27 mycolipenic (phthienoic) or mono-hydroxylated tri-methyl-branched C24-C28 mycolipanolic acids replace the mycosanoic acids [188–190]. 2,3,6-triacyltrehaloses (TAT) harboring stearic, palmitic and mycolipenic acyl substituents have also been reported in Mtb [191]. Polyacyltrehaloses (PAT) are trehalose esters acylated with five acyl groups consisting of one middle-chain saturated fatty acid (C16-C19) at the 2-position and different combinations of the tri-methyl-branched C27-mycolipenic and C27-mycolipanolic acids at the 2’, 3’, 4 and 6’-positions (Fig. 10) [188, 192]. Monomethyl-branched unsaturated C16 to C20 fatty acids have also been found as minor constituents esterifying PAT and DAT [162]. So far, the mycolipenic acyl substituents found in DAT, TAT and PAT have only been isolated from virulent isolates of the Mtb complex species Mtb, M. bovis, and M. africanum but were not found in the avirulent laboratory strain Mtb H37Ra or in the vaccine strain M. bovis BCG. While 2,3-diacyltrehaloses and 2,3,4- and 2,3,6-triacyltrehaloses may be found in non-pathogenic species of mycobacteria such as M. fortuitum, the fatty acyl substituents identified in this species consist of straight-chain (C14–C18) and mono-methyl-branched unsaturated C16–C20 fatty acids [193, 194].
As their relative distribution to pathogenic species of the Mtb complex may suggest, DAT, TAT and PAT are biologically active molecules capable of modulating a number of host immune responses in vitro [141, 195]. Their precise role during host infection remains, however, poorly understood. Phenotypic observations made on a mutant of Mtb deficient in the biosynthesis of DAT and PAT indicated a role for these lipids in the retention of the capsular material at the cell surface [196, 197]. The modification of the surface properties of the mutant affected its binding and uptake by phagocytic and non-phagocytic cells but preliminary infection studies indicated that the mutant did not significantly differ from its wild-type parent in its ability to replicate and persist in cultured macrophages and in mice [197]. Interestingly, increased binding to phagocytic cells was also reported in the case of a SL-deficient mutant of Mtb [198]. It is thus likely that the different families of acyltrehaloses produced by Mtb have partially redundant activities in whole cells hampering the clear delineation of their individual contribution to virulence and other physiological functions. Independent from their binding or immunomodulatory properties and as noted above, methyl-branched fatty acid-containing lipids such as PDIM, SL, DAT and PAT appear to play an important role in alleviating the propionate-mediated stress undergone by Mtb when the bacterium utilizes host cholesterol as a major carbon source during infection [174, 175]. Consistently, WhiB3 acts as a positive transcriptional regulator of pks3/4 in addition to pks2 [174].
Gene knock-out studies indicated that the polyketide synthase encoded by pks3/4 (Rv1180/Rv1181) is responsible for the elongation of mycosanoic and mycolipenic acids while pks8+pks17 (Rv1662+Rv1663) encode together the polyketide synthase producing monomethyl-branched unsaturated C16 to C20 fatty acids [162, 196, 197] (Table 2). A Mtb mutant deficient in the expression of pks3/4 failed to produce PAT and DAT [196, 197]. In some Mtb strains, an intervening stop codon in pks3/4 results in two separate open reading frames (annotated as pks3 and pks4). Strains containing this mutation do not synthesize PAT [186]. Striking resemblance in the genetic organization of the regions encompassing the polyketide synthase gene pks3/4 and that involved in SL (Fig. 10) and, to a lesser extent, PDIM biosynthesis (see section VI) are suggestive of the involvement of fadD21 (Rv1185c), mmpL10 (Rv1183) and Rv1184c (chp2) in the assembly and export of DAT and PAT (Table 2). To date, however, only papA3 (Rv1182) has been characterized [199]. It encodes the acyltransferase responsible for the sequential transfer of a palmitoyl group at the 2-position of DAT/PAT followed by a mycolipenoyl group at the 3-position (Fig. 10). As is the case for SL, the two-component transcriptional regulator PhoP-PhoR (Rv0757−Rv0758) positively regulates the synthesis of DAT and PAT, and Mtb mutants deficient in the expression of this regulator are totally deficient in DAT and PAT production [143, 176]. The same mutation in the phoP gene of Mtb H37Ra which accounts for the inability of this strain to produce SL also accounts for the absence of DAT and PAT from this avirulent Mtb isolate [177]. PhoP was shown to bind the promoter regions of pks3/4 and fadD21 [178, 179].
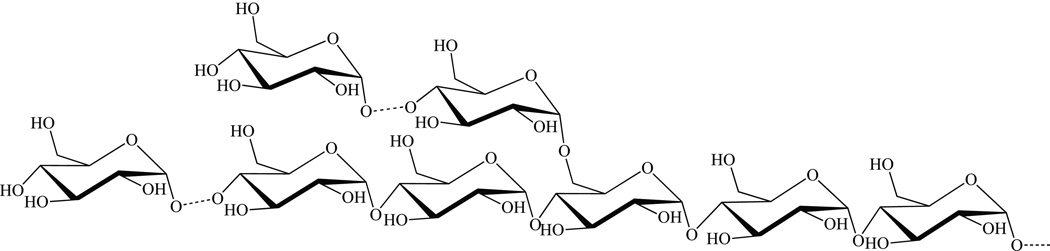
Lipooligosaccharides (LOS)
Lipooligosaccharides (LOS) are surface-exposed glycolipids [8] produced by a number of Mycobacterium species [200]. They were first found in M. kansasii [201] and M. smegmatis [202], then in nine other mycobacterial species [200], including “M. canettii” and related strains of the M. tuberculosis complex [203]. LOS are otherwise virtually absent from Mtb strains sensu stricto such as H37Rv [203].
LOS (Fig. 11 A–B) share a poly-O-acylated trehalose core further glycosylated by a mono- or, more frequently, a oligosaccharidyl unit [200]. Similar to the situation in other trehalose-based mycobacterial glycolipids such as sulfolipids and di- or tri-acyltrehaloses, the trehalose moiety of LOS is invariably acylated by polymethyl-branched fatty acids that can be either saturated, e. g. in “M. canettii”, or unsaturated, e. g. in M. smegmatis.
Figure 11. Structures of major lipooligosaccharide (LOS-A) of M. smegmatis ATCC 356.
(R1 and/or R2 : octanoic acid and tetra- or hexa-decanoic acid) (A) and Mtb Canettii (B); R = Ac. LOS biosynthetic gene cluster of M. smegmatis mc2155 (C). Shown is the 25.15 kb-region spanning MSMEG_4727 (pks5) to MSMEG_4741 (mmpL). ORF are depicted as arrows. Black arrows indicate genes encoding biosynthetic enzymes; grey arrows indicate putative transporter genes; white arrows, hypothetical genes of unknown function. Pks5, Mas-like polyketide synthase; Pap, putative acyltransferase; MSMEG_4729 and MSMEG_4730, putative acyltransferases; FadD, putative acyl-CoA synthase; Gtf (MSMEG_4732), putative glycosyltransferase; Gap2, putative transmembrane protein involved in glycolipid translocation; MSMEG_4734, hypothetical PE/PPE-like protein; Gtf (MSMEG_4735), putative glycosyltransferase; MSMEG_4736 and MSMEG_4737, putative pyrruvylyl transferases; MSMEG_4738, hypothetical protein; Mtf, possible O-methyltransferase; Gtf (MSMEG_4740), putative glycosyltransferase; MmpL, putative inner membrane transporter.
The biosynthesis of LOS molecules is still poorly understood with only a few genes experimentally demonstrated to be involved in their elongation and assembly [204–206]. The synthesis of polymethyl-branched fatty acids invariably requires a polyketide synthase (Pks) which uses methylmalonyl-CoA instead of malonyl-CoA as the elongation unit, resulting in the formation of a polymethyl branched aliphatic chain. The MSMEG_4727 (pks5) gene, whose sequence is 65.6 % identical to that of the Mtb Mas-like gene Rv1527c, was involved in the biosynthesis of LOS in M. smegmatis [207]. The genomic surroundings of pks5 from M. smegmatis reminds those described earlier for other acyltrehaloses (see SL, DAT and PAT in sections IV and Fig 9) in that pap- and fadD-like genes likely to be required for the activation and transfer of the acyl groups of LOS [206], and an mmpL gene putatively involved in the translocation of these lipids are found (Fig. 11C). In addition, genes whose products were tentatively annotated as polysaccharide pyruvyltransferases are found in the biosynthetic cluster of pyruvylated LOS-producing species such as M. smegmatis [202, 208]. Consistent with the finding of various methylated glucosyl residues in LOS (Fig. 11A), genes encoding putative glycosyltranferases and O-methyltransferases also map in the vicinity of pks5 (Fig. 11C). In M. marinum, several of these have been characterized [204, 205, 209]. It is noteworthy that orthologs of five of the M. smegmatis LOS-related genes (pks, pap, fadD, mmpL and gap) are conserved in the corresponding biosynthetic gene clusters of M. marinum and Mtb [205]. Interestingly, homologous genes are also found in the GPL biosynthetic gene clusters of M. smegmatis [210], M. abscessus [211], M. chelonae [211] and M. avium [212] (see section VII). This conserved set of genes may delineate the minimum biosynthetic machinery required for the synthesis and export of GPL- and LOS-type glycolipids in mycobacteria. The remaining ORFs identified in the confirmed or putative mycobacterial LOS biosynthetic clusters are less conserved, an observation consistent with the fact that LOS differ from other mycobacterial glycolipids in terms of the number and nature of their sugar constituents [200]. Recently, the regulatory protein WhiB4 from M. marinum was associated with LOS biosynthesis but its precise function is not known [213].
LOS are highly antigenic molecules [203]. Recent observations suggest that they play an important role in retaining proteins at the cell surface of some Mycobacterium species such as M. marinum [213]. Their precise role in the colony morphology of mycobacteria is still a matter of debate and seems to be species-specific [205, 214, 215]. In M. marinum for instance, LOS have clearly been associated with colony morphology, sliding motility, biofilm formation, and the ability of this Mycobacterium to enter macrophages [205]. The M. marinum LOS are also endowed with immunomodulatory activities [216] and modulate virulence in the zebrafish embryo model of infection [213].
V. Mannosyl-β-1-phosphomycoketides
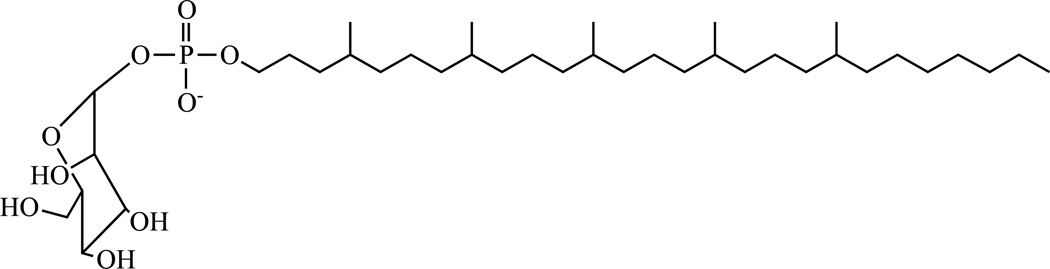
Mannosyl-β-1-phosphomycoketides consist of a mannosyl-β-1-phosphate moiety reminiscent of polyprenol phosphomannose (the lipid-linked mannose donor) and an alkyl chain of varying length (C30–C34) made of a fully saturated 4, 8, 12, 16, 20-pentamethylpentacosyl unit (Fig. 12). Mycoketides were first isolated from M. avium based on their ability to activate human CD1c-restricted T-cells [217]. This family of lipids was later identified in the slow-growing pathogenic species Mtb and M. bovis BCG but not in the rapidly growing saprophytes, M. phlei, M. fallax and M. smegmatis [218]. Under standard liquid culture conditions, mycoketides are produced in minute amounts and are found both inside the cells and released in the culture medium [219].
Figure 12. Structure of the predominant mannosyl-β-1-phosphomycoketide from Mtb H37Rv.
(See text for details)
Their restricted distribution to pathogenic slow-growing Mycobacterium spp. are suggestive of an involvement in pathogenicity and several studies aimed at comparing the virulence of mycoketide-deficient mutants of Mtb, M. avium and M. marinum to that of their wild-type parent in animal models of infection have provided support for this assumption [219]. In addition to their potential role in modulating the host immune response, mycoketides were proposed to be mycobacterial secondary metabolites acting as signaling factors to regulate cell division and virulence and to contribute to the suppression of phagosomal acidification [219]. The alkyl backbone of mycoketides is elongated by the polyketide synthase Pks12 (Rv2048c) (Table 2) [218, 220]. Pks12 is the largest predicted protein of Mtb (430 KDa) and consists of two complete sets of fatty acid synthase (FAS)-like catalytic domains capable together of using alternating C2 (malonyl-CoA) and C3 (methylmalonyl-CoA) units to elongate the alkyl backbone of mycoketides. After 5 cycles of C3 and C2 chain elongation, it is believed that the alkyl chain is released from the polyketide synthase upon hydrolysis yielding mycoketidic acid which is further reduced to the corresponding long-chain alcohol, mycoketide, and finally phosphorylated and mannosylated to generate mannosyl-β-1-phosphomycoketides [219]. The enzymes catalyzing the hydrolysis, reduction, phosphorylation and mannosylation steps have not yet been identified. The finding of orthologs of pks12 in M. marinum, M. ulcerans, M. avium paratuberculosis, and several species of the Mtb complex suggests that the production mannosyl-β-1-phosphomycoketides may be a common feature of slow-growing mycobacterial pathogens.
VI. Phthiocerol dimycocerosates, phenolic glycolipids and related compounds
Phthiocerol diesters and related compounds: structures, distribution and cell localization
Phthiocerol DiMycocerosates (PDIM) and DiPhthioceranates (PDIP) are of a family of long-chain C33–C41 β-diols (phthiocerols) esterified by two moles of polymethyl-branched (C27–C34) fatty acids. When the configuration of the asymmetric centers bearing the methyl branches are of the D series, the fatty acids are called mycocerosic acids whereas whose of the L series are known as phthioceranic acids [14] (Fig. 13). The major β-diols (phthiocerol A) are usually accompanied by structural variants of these alcohols containing either a keto group in place of the methoxy group (phthiodiolone A) or a methyl group rather than an ethyl group at the terminus of the molecules and near the methoxyl group (phthiocerol B) (Fig. 13). To date, PDIM have been found in Mtb, M. bovis, M. leprae, M. microti, M. kansasii, M. gastri and M. haemophilum, whereas PDIP have been found in M. ulcerans and M. marinum [221].
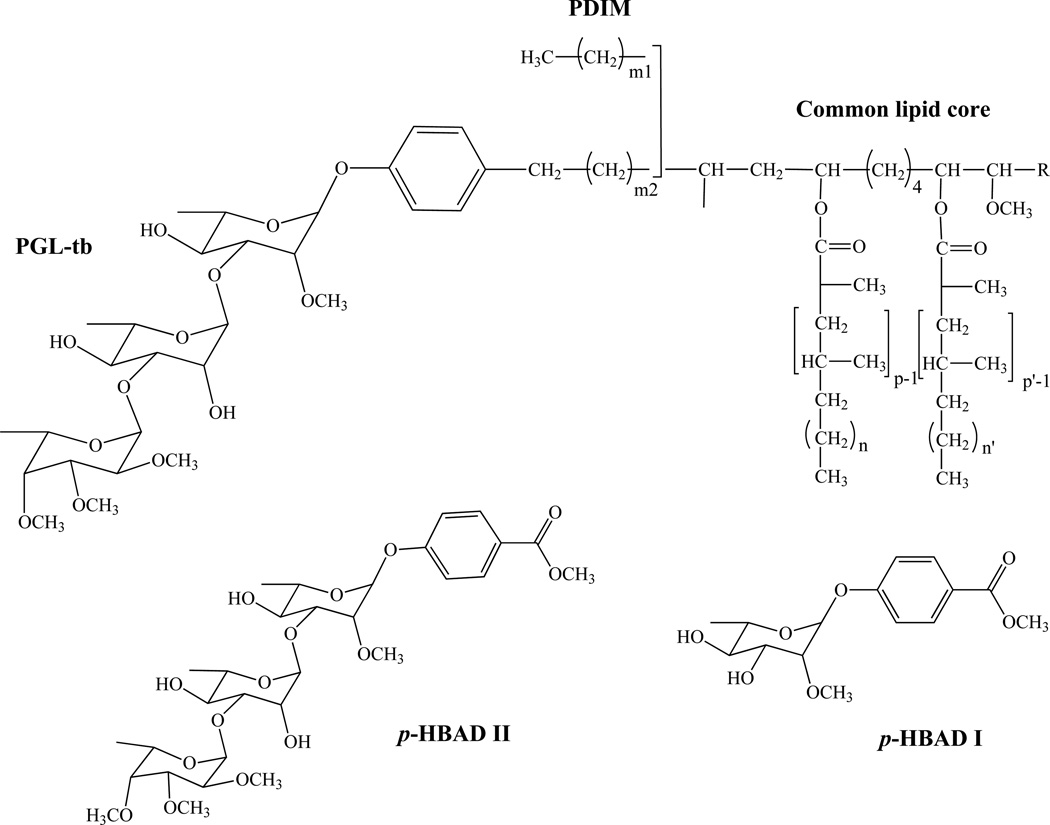
Figure 13. Structures of the phthiocerol dimycocerosates (PDIM), phenolic glycolipids (PGL) and p-hydroxybenzoic acid derivatives (p-HBADs) of Mtb.
In Mtb, p, p’=3–5; n, n’=16–18; m2=15–17; m1= 20–22; R= CH2–CH3 or CH3.
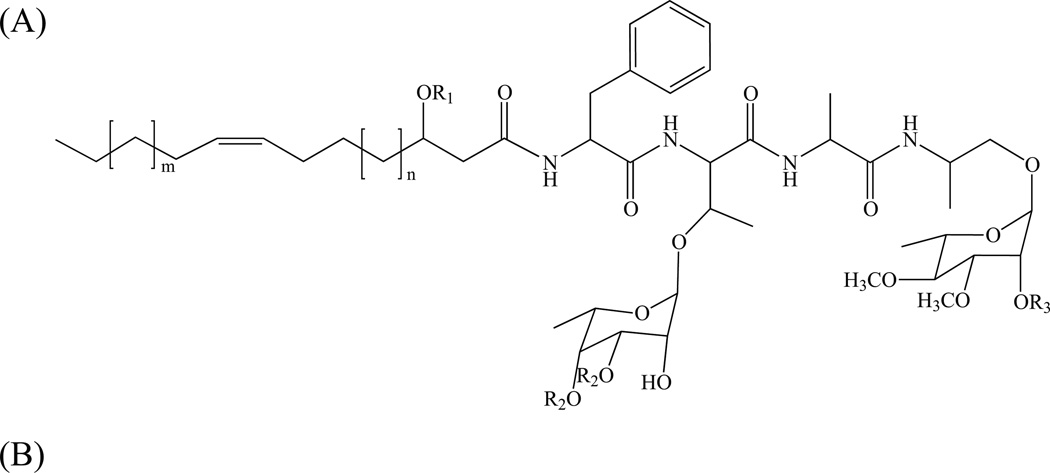
Glycosylated phenolic derivatives of PDIM and DIP, called phenolic glycolipids (PGL) are found in the same species, although they may not be present in all strains; for instance, the PGL from Mtb (PGL-tb) has only been identified in the ‘canettii’ strain [222, 223] and in some East-Asian/Beijing isolates [224, 225]. In PGL, the β-diols (phenolphthiocerols) are esterified by two moles of polymethyl-branched (C27–C34) fatty acids, except in Beijing strains where a palmitic acid is found esterifying the additional hydroxyl group occurring in the aliphatic core of phenolphthiotriol [225]. The glycosyl moiety of PGL is composed of 1 to 4 sugar residues depending on the species, most of which are O-methylated deoxysugars [14, 200]. Identical PGL structures may be found in phylogenetically-related mycobacterial species, for instance in species of the Mtb complex (M. bovis, M. microti, M. pinnipeddii and M. africanum), M. kansasii and M. gastri, and M. marinum and M. ulcerans [200, 226]. The glycosyl moiety of PGL was also found attached to p-hydroxybenzoic acid, i.e. as methyl esters, to form p-hydroxybenzoic acid derivatives (p-HBADs) in Mtb and M. bovis BCG (Fig. 13) [223]. In an attempt to correlate the lipid content with the virulence of Mtb isolates, Goren and collaborators characterized a methoxylated phenolphthiocerol, the so-called ‘attenuation indicator lipid’ [227]. The correlation between the occurrence of this lipid and reduced virulence remains, however, unclear. This lipid and its unmethylated form were detected in East-Asian/Beijing strains and accumulated in all of the Indo-Oceanic strains of Mtb examined [228].
PDIM and PGL are found in the capsules of Mtb and other pathogenic mycobacteria [8]. PDIM are otherwise abundant components of the outer membrane of Mtb where they contribute to its well-known impermeability [229]. p-HBADs, in contrast, are released in culture filtrates and tend not to remain associated with the cell envelope [223].
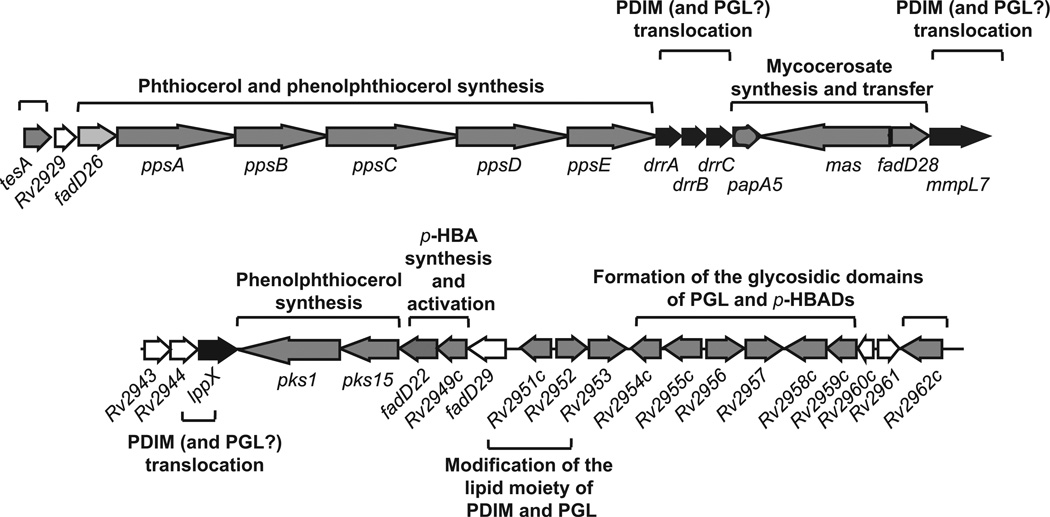
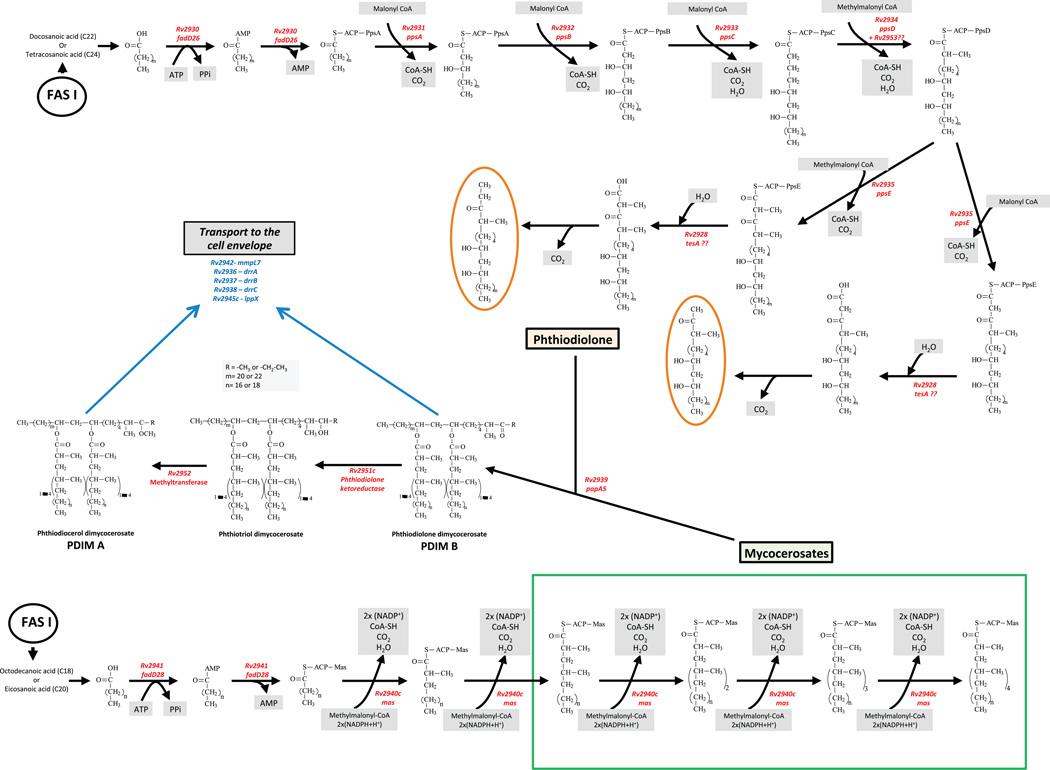
Biosynthesis of PDIM and PGL
(a) Biosynthesis of phthiocerol and related compounds
Common enzymes participate in the biosynthesis of the lipid core of PDIM and PGL (Fig. 13) where n-C22–C24 fatty-acyl chains and p-hydroxyphenylalkanoates, respectively, are elongated to form the long-chain β-diols, phthiocerol or phenolphthiocerol. Coupled genetic and biochemical strategies have allowed much of the biosynthetic pathways of PDIM and PGL to be elucidated. On the basis of mutants phenotypes, genes such as pks11 [230], pks12 [231], pks10 [232], mb0100 [233] and pks7 [234] have been associated with the biosynthesis of PDIM; however, in the absence of genetic complementation, definitive proof for their involvement in the pathway is lacking and no clear biosynthetic roles have yet been assigned to these genes. The genes unambiguously demonstrated to participate in PDIM and PGL biosynthesis are shown in Fig. 14, and their specific function in the pathway is detailed in Fig. 15 and Table 3. They are clustered on a 73-kb fragment of the Mtb chromosome (Fig. 14) and the organization of this locus is apparently highly conserved in all PDIM/PGL-producing mycobacteria, with the exception of M. leprae in which this locus is split into two loci.
Figure 14. Genetic organization of the PDIM and PGL locus of Mtb H37Rv.
ORF are depicted as arrows. Black arrows indicate genes encoding biosynthetic enzymes; grey arrows indicate putative transporter genes; white arrows, hypothetical genes of unknown function. More details about the function of each gene are provided in Table 3 and Fig. 15. Adapted from [260].
Figure 15. The PDIM biosynthetic pathway.
(See text for details)
Table 3.
Mtb H37Rv genes involved in the biogenesis of phthiocerol dimycocerosates, phenolic glycolipids and p-hydroxybenzoic acids.
| Gene number |
Gene name |
Function | Evidence | Reference |
|---|---|---|---|---|
| Rv2928 | tesA | Type II thioesterase thought to be involved in the release of phthiocerol and phenolphthiocerol from PpsE in the biosynthesis of PDIM and PGL | P | [230, 238, 239] |
| Rv2930 | fadD26 | Long-chain fatty acyl-AMP ligase providing PpsA with activated long-chain fatty acid starter units for the generation of phthiocerol in the biosynthesis of PDIM | P | [167, 229, 236, 244] |
| Rv2931 | ppsA | Type 1 polyketide synthase responsible with PpsB-C-D-E for the elongation of C22–C24 fatty acids and p-hydroxyphenylalkanoates with malonyl-CoA and methylmalonyl-CoA to yield phthiocerol and phenolphthiocerol derivatives, respectively | E,P | [235, 244] |
| Rv2932 | ppsB | Type 1 polyketide synthase responsible with PpsA-C-D-E for the elongation of C22–C24 fatty acids and p-hydroxyphenylalkanoates with malonyl-CoA and methylmalonyl-CoA to yield phthiocerol and phenolphthiocerol derivatives, respectively | E,P | [235, 244, 349] |
| Rv2933 | ppsC | Type 1 polyketide synthase responsible with PpsA-B-D-E for the elongation of C22–C24 fatty acids and p-hydroxyphenylalkanoates with malonyl-CoA and methylmalonyl-CoA to yield phthiocerol and phenolphthiocerol derivatives, respectively | P | [235, 349] |
| Rv2934 | ppsD | Type 1 polyketide synthase responsible with PpsA-B-C-E for the elongation of C22–C24 fatty acids and p-hydroxyphenylalkanoates with malonyl-CoA and methylmalonyl-CoA to yield phthiocerol and phenolphthiocerol derivatives, respectively | P | [235] |
| Rv2935 | ppsE | Type 1 polyketide synthase responsible with PpsA-B-C-D for the elongation of C22–C24 fatty acids and p-hydroxyphenylalkanoates with malonyl-CoA and methylmalonyl-CoA to yield phthiocerol and phenolphthiocerol derivatives, respectively | E,P | [235, 244] |
| Rv2936 | drrA | Component of the DrrABC ABC-transporter involved in the translocation of PDIM (and PGL?) across the plasma membrane; ATP-binding protein | P | [350] |
| Rv2937 | drrB | Component of the DrrABC ABC-transporter involved in the translocation of PDIM (and PGL?) across the plasma membrane; integral membrane protein | P | [230, 350] |
| Rv2938 | drrC | Component of the DrrABC ABC-transporter involved in the translocation of PDIM (and PGL?) across the plasma membrane; integral membrane protein | P | [229] |
| Rv2939 | papA5 | Acyltransferase responsible for the transfer of mycocerosic acids to phthiocerol to form PDIM | E,P | [244, 351, 352] |
| Rv2940c | mas | Mycocerosic acid synthase (type I polyketide synthase) involved in the biosynthesis of PDIM and PGL | E,P | [240–244] |
| Rv2941 | fadD28 | Long chain fatty acyl-AMP ligase responsible for providing the long-chain fatty acid starter unit to Mas for the generation of mycocerosic acids | E,P | [167, 229, 235, 245, 246] |
| Rv2942 | mmpL7 | RND superfamily inner membrane transporter involved in PDIM translocation to the periplasm (and PGL?) | P | [229, 235, 252] |
| Rv2945 | lppX | Lipoprotein involved in the transport of PDIM (and PGL?) to the cell surface | P | [253] |
| Rv2946c | pks1 | Together with Pks15, type I polyketide synthase involved in the elongation of p-hydroxybenzoic acid derivatives with malonyl-CoA to form p-hydroxyphenylalkanoates (precursors of PGL) | P | [223] |
| Rv2947c | pks15 | Together with Pks1, type I polyketide synthase involved in the elongation of p-hydroxybenzoic acid derivatives with malonyl-CoA to form p-hydroxyphenylalkanoates (precursors of PGL) | P | [223] |
| Rv2948c | fadD22 | p-hydroxybenzoyl-AMP ligase involved in the biosynthesis of PGL; catalyzes the activation of p-hydroxybenzoic acid and its subsequent transfer onto Pks15/1 for the production of p-hydroxyphenylalkanoates | E,P | [236, 353, 354] |
| Rv2949c | p-hydroxybenzoic acid synthase | E,P | [237] | |
| Rv2950c | fadD29 | Fatty acyl-AMP ligase involved in the biosynthesis of PGL; catalyzes the activation of hydroxyphenylalkanoates which are then transferred onto PpsA to yield phenolphthiocerol | E,P | [236] |
| Rv2951c | Ketoreductase catalyzing the reduction of (phenol)phthiodiolone to yield (phenol)phthiotriol in the biosynthesis of PDIM and PGL | P | [355, 356] | |
| Rv2952 | SAM-dependent O-methyltransferase involved in the formation of (phenol)phthiocerol dimycocerosates from (phenol)phthiodiolone dimycocerosates. Catalyzes the transfer of a methyl group to the third hydroxyl group of (phenol)phthiotriol in PDIM and glycosylated phenolphthiotriol dimycocerosates | P | [247, 356] | |
| Rv2953 | Enoyl-reductase acting in concert with PpsD in the biosynthesis of the (phenol)phthiocerol moiety of PDIM and PGL | P | [357] | |
| Rv2954c | SAM-dependent methyltransferase responsible for the O-methylation of the hydroxyl group at position 3 of the fucosyl residue of PGL (and possibly p-HBAD) | P | [249] | |
| Rv2955c | SAM-dependent methyltransferase responsible for the O-methylation of the hydroxyl group at position 4 of the fucosyl residue of PGL (and possibly p-HBAD) | P | [249] | |
| Rv2956 | SAM-dependent methyltransferase responsible for the O-methylation of the hydroxyl group at position 2 of the fucosyl residue of PGL and (and possibly p-HBAD) | P | [249] | |
| Rv2957 | Fucosyltransferase responsible for the transfer of the third glycosyl residue of the triglycosyl appendage of PGL and p-HBAD | P | [248] | |
| Rv2958c | Rhamnosyltransferase responsible for the transfer of the second rhamnosyl residue of the triglycosyl appendage of PGL and p-HBAD | P | [248] | |
| Rv2959c | SAM-dependent methyltransferase responsible for the O-methylation of the hydroxyl group at position 2 of the rhamnosyl residue linked to the phenolic group of PGL and p-HBAD | P | [247] | |
| Rv2962c | Rhamnosyltransferase responsible for the transfer of a rhamnosyl residue onto p-hydroxybenzoic ethyl ester and/or phenolphthiocerol dimycocerosates | P | [248] |
E,P,H: see Table 1 legend.
The Mtb genome encodes 36 FadD proteins with homology to acyl-CoA synthases. As noted under section IV, some of them map in the vicinity of pks genes. FadD26 is required for the synthesis of PDIM but not that of PGL in Mtb and M. bovis [229, 235, 236]. The role of the FadD proteins in activating Pks substrates was demonstrated by Trivedi et al. [167] who established that FadD26 and FadD28 belonged to a large family of fatty-acyl AMP ligases responsible for the activation of long-chain n-C22–C24 fatty acids as acyl-adenylates. They showed that FadD26 loads the activated substrates directly onto PpsA. These substrates are then elongated with malonyl-CoA and methylmalonyl-CoA by PpsA-PpsE to yield phthiocerol (Table 3; Fig. 14–15).
The enzyme encoded by Rv2949c catalyzes the formation of p-hydroxybenzoic acid from chorismate [237]. p-hydroxybenzoic acid is activated by FadD22, which displays p-hydroxybenzoyl-AMP ligase activity [236], and subsequently elongated by the type-1 polyketide synthase Pks15/1 to form p-hydroxyphenylalkanoates; the reaction may involve 8 or 9 elongation cycles using malonyl-CoA as the extender unit. A frameshift mutation within the pks15/1 gene accounts for the lack of production of PGL by the Mtb reference strains H37Rv, Erdman and CDC1551 [223]. The fatty acyl-AMP ligase FadD29 activates p-hydroxyphenylalkanoates that are then transferred onto PpsA and finally elongated with malonyl-CoA and methylmalonyl-CoA by PpsA-PpsE to yield phenolphthiocerol [236] (Table 3). The type II thioesterase TesA is thought to be involved in the release of the phthiocerol and phenolphthiocerol moieties of PDIM and PGL, respectively, from the polyketide synthase PpsE [230, 238]. The demonstrated interaction of TesA with the C-terminal half of PpsE tends to support this assumption [239].
(b) Biosynthesis and transfer of mycocerosates
The mycocerosic acids that esterify the β-diols of phthiocerol and phenolphthiocerols are elongated from C16 and C20 fatty acids with 3 or 4 propionate units by a dedicated type I polyketide synthase known as Mas (for Mycocerosic Acid Synthase) [240–244] (Fig. 15; Table 3). Mas preferentially uses methyl-malonyl-CoA instead of malonyl-CoA for fatty acid elongation, thereby introducing methyl branches into the mycocerosic acid chain. In addition, the ketoreductase and enoylreductase activities of this enzyme require NADPH as a cofactor [241]. FadD28 is the fatty-acyl AMP ligase responsible for the activation of the C16 and C20 fatty acid starter units of Mas as acyl-AMP and their transfer onto the polyketide synthase [167, 229, 235, 245, 246] (Fig. 15; Table 3). The synthesized mycocerosates are not released from Mas by a conventional thioesterase but rather directly transferred by PapA5 onto their phthiocerol or phenolphthiocerol acceptors through an interaction with Mas to catalyze the final esterification step [244] (Fig. 14–15; Table 3).
(c) Synthesis of the saccharide moiety of PGL-tb and p-HBADs
Consistent with their conserved structures (Fig. 13), the biosynthesis of the glycosyl moiety of PGL-tb and p-HBADs involves the same set of enzymes (Table 3). In the case of Mtb ‘canetti’, four genes (Rv2962c, Rv2957, Rv2958c and Rv2959c) encoding three glycosyltransferases and one methyltransferase are involved in the formation of this structure [247, 248] (Fig. 14; Table 3). The glycosyltransferase encoded by Rv2962c is involved in the transfer of the first rhamnosyl residue onto p-hydroxy-phenolphthiocerol dimycocerosates and p-hydroxy-phenolmethylester. A single nucleotide polymorphism (SNP) at position 880 of Rv2962c in the Indo-Oceanic isolates of Mtb results in a truncated open-reading frame accounting for the accumulation of phenolphthiocerol dimycocerosates and related ‘attenuation lipid’ observed in this lineage [228]. Rv2958c encodes the rhamnosyltransferase responsible for the transfer of the second rhamnosyl residue onto the mono-rhamnosylated PGL or p-hydroxy-phenolmethylester [248], and a frameshift mutation within this gene explains the lack of production triglycosylated PGL by M. bovis, M. microti, M. pinnipeddii and M. africanum [226]. Rv2957 encodes the fucosyltransferase responsible for the transfer of the third glycosyl residue of the triglycosyl appendage of PGL and p-HBAD-II (Fig. 13) [248]. Rv2959c encodes a methyltransferase involved in the methylation of position 2 of the first rhamnosyl residue of PGL-tb and p-HBADs [247]. Rv2954c, Rv2955c and Rv2956 encode the methyltransferases that catalyze the O-methylation of the hydroxyl groups located, respectively, at positions 3, 4 and 2 of the terminal fucosyl residue of PGL-tb in a sequential process, starting with methylation at position 2, followed by those of positions 4 and 3 [249]. The genes involved in the production of the glycosyl moiety of the PGL of M. leprae were identified through genetic complementation of M. bovis BCG, leading to the synthesis of M. leprae-specific PGL-1 by the vaccine strain [250].
Translocation of PDIM and related molecules
PDIM and PGL-tb are found in the outermost layers of the Mtb cell envelope [8] and p-HBADs are secreted in the culture medium [223]. Since at least some the enzymes involved for the biosynthesis of PDIM and PGL are cytosolic (e.g., polyketide synthases and FadD enzymes), the presence of these two lipids at the surface implies the existence of a translocation machinery. All the published work on this topic to date has focused on PDIM because the Mtb strains used to generate knock-out mutants were naturally deficient in the production of PGL-tb.
The mmpL7 or drrC genes, both in the PDIM and PGL locus (Fig. 14), have been involved in the translocation of PDIM. drrC and mmpL7 null mutants synthesized PDIM structurally identical to those of the wild-type strain but failed to translocate these compounds to the cell surface [229, 235]. PDIM in these mutants were apparently retained in deeper layers of the cell envelope. DrrC is an integral membrane protein belonging to an ABC transporter involving two other subunits encoded by drrA and drrB (Fig. 14; Table 3). The MmpL7 protein belongs to the RND (Resistance, Nodulation and cell Division) superfamily of transporters [251]. Like other members of this family, MmpL7 is predicted to consist of twelve transmembrane domains and two large soluble periplasmic loops. Using a yeast two-hybrid system, Jain and Cox [252] showed that the loop between the seventh and eighth transmembrane domains interacts with the polyketide synthase PpsE involved in PDIM and PGL synthesis. Based on this finding, a model was proposed wherein the synthesis and transport of PDIM are coupled [252]. Another gene, lppX, encoding a lipoprotein has been found to be required for PDIM to reach the cell surface [253]. Interestingly, LppX shares a similar fold with the periplasmic chaperone LolA and the outer membrane lipoprotein LolB which, in Gram negative bacteria, are involved in the localization of lipoproteins to the outer membrane. The crystal structure of LppX revealed a large hydrophobic cavity suitable to accommodate a single PDIM molecule [253]. It is possible that LppX acts downstream from MmpL7 and DrrABC, carrying PDIM across the periplasm to the outer membrane once the two membrane transporters have translocated the fully synthesized lipid products across the plasma membrane. The exact role of each of these transporters in the translocation process remains, however, to be determined. Given the nature of the enzymes involved in the biosynthesis of PGL and p-HBADs, it is likely that most if not all of their biosynthesis takes place in the cytoplasm or at the periphery of the plasma membrane. The same transporters as those involved in the export of PDIM may be involved in their translocation to the cell surface.
Roles of PDIM, p-HBADs and PGL-tb in the organization of the cell envelope and virulence
As glycosylated capsular or secreted components [8, 223], p-HBADs and PGLs are serologically active. Accordingly, several studies have explored the potential of PGLs as serodiagnostic tools for the detection of tuberculosis and leprosy [14, 254–258]. As most clinical isolates of Mtb do not produce PGL-tb [221–223], it is likely that the antibodies detected in patients were in fact directed against p-HBADs [223].
PDIM have been found in all Mtb clinical isolates tested [221, 227]. Their non-amphipathic character and abundance in the cell envelope have long suggested that they play a structural role, providing a hydrophobic barrier around Mtb cells and possibly a platform for anchoring other components of the cell envelope [259]. The roles of PDIM in the permeability barrier, intracellular survival and virulence of Mtb have been extensively discussed in previous reviews and will therefore not be detailed here [141, 260]. Likewise, the reader is referred to earlier reviews for details on the roles of PGL-tb and p-HBADs in the modulation of the host immune response and pathogenicity of Mtb [260].
Numerous biological activities have also been associated with the PGLs of non-tuberculous mycobacteria in general. The PGLs from M. leprae and M. kansasii, like that of M. bovis BCG, seem to non-specifically inhibit lymphoproliferative responses to various stimuli, including several antigens and mitogens [261]. Other biological activities are dependent on the nature of the carbohydrate moiety. For instance, specific suppression of cell-mediated immunity is a feature of lepromatous leprosy and the PGL-1 from M. leprae has been involved in many aspects of this process. In contrast to the PGLs from M. microti and M. kansasii, PGL-1 from M. leprae has the ability to suppress the ‘oxidative response’ of human macrophages [262, 263], probably explaining why this response is abnormally low in leprosy patients. PGL-1, but not the PGLs from M. bovis and M. kansasii, also has been reported to be active in an indirect test of specific immunosuppression, inhibiting the concanavalin A-stimulation of lymphocytes from patients with lepromatous leprosy [264]. PGL-1 also can neutralize hydroxyl- and superoxide radicals in vitro, a property shared by deacylated PGL-1 and to some extent by the carbohydrate moiety of the molecule [265].
VII. Glycopeptidolipids (GPL)
Structure and subcellular location
Mycobacteria synthesize type- or species-specific glycopeptidolipids (GPLs) that differ from one another by their sensitivity to alkali. Alkali-stable GPLs, also known as C-mycosides [266], are produced by a number of both rapid- and slow-growing mycobacterial species, including M. avium, M. abscessus, and M. smegmatis (for detailed reviews, see [200, 267, 268]). Alkali-labile GPLs have thus far only been described in M. xenopi [269, 270]. Their structures greatly differ from those of the C-type GPLs. The C-type GPLs share a common lipopeptidyl core that consists of a mixture of 3-hydroxylated and 3-methoxylated long-chain (C26–C34) fatty acids [271], amidated by a tripeptide (D-Phe-D-alloThr-D-Ala) and terminated by an aminoalcohol (L-alaninol) (Fig. 16A). The position of hydroxyl/methoxyl group A on the fatty acyl chain has been recently questioned and position 5 has been proposed [272]. C-type GPLs differ by the number and the nature of the glycosyl residues that substitute the lipopeptidyl core. In the most abundant molecular species, the apolar C-type GPLs (Fig. 16A) [also called non-specific GPLs (nsGPLs)], the alaninol is glycosylated by a mono- or di-O-methylated rhamnosyl residue while a di-O-acetylated 6-deoxytalosyl unit is attached to the alloThr residue. In the polar C-type GPLs, also known as serospecific GPLs (ssGPLs), additional sugar units are attached to the 6-deoxytalosyl residue; at least 14 out of the 28 described ssGPLs from the M. avium-intracellulare complex (MAC) have been structurally characterized [200, 268]. The serological variance among the members of the M. avium-intracellulare complex is due to subtle differences in the structure of the oligosaccharide chain that substitutes the communal C-type nsGPL core. The oligosaccharide haptens from several polar C-type GPLs contain unusual sugars: glucuronic acid and variants, acetamido-dideoxy-hexosyl residues and other branched sugars.
Figure 16.
(A) Structure of the non-specific glycopeptidolipids of M. smegmatis. R1 = -H or –CH3; R2 = -H or –Ac; R3, -CH3, -succinyl, -rhamnosyl or -2-O-succinylrhamnosyl; m = 12–14; n = 6–10. (B) GPL biosynthetic gene cluster of M. smegmatis mc2155. Shown is the 64.97 kb-region spanning MSMEG_0380 (mmpS4) to MSMEG_0413. ORF are depicted as arrows. Black arrows indicate genes encoding biosynthetic enzymes; grey arrows indicate putative transporter genes; white arrows, putative regulatory genes. Chp, putative acyltransferase; FadE, putative acyl-CoA dehydrogenase; PapA, putative acyltransferase. Other genes are described in the text.
The correlation between the presence of C-type GPLs, smooth colony morphotype and staining of the outer membrane with Ruthenium Red [273–275], and the fact that polar GPLs correspond to “Schaefer typing antigens” used in the identification of isolates of the MAC complex [276] suggested that GPLs were present at the cell surface. Consistently, the capsular materials of M. lepraemurium [277] and M. avium [278] have been shown to consist of C-mycosides.
Biological properties of GPLs
The antigenic properties of ssGPLs and the relationships between GPL production and colonial morphotype or drug resistance have been abundantly reviewed [268, 279]. Other properties associated with GPL production are as follows. Although mycobacteria are non-flagellated, M. smegmatis and the slow growing M. avium can spread on the surface of solid media by a sliding mechanism [280]. Rough strains lacking GPLs appear to be devoid of such sliding motility [210, 275]. Consistently, all of the non-sliding mutants isolated and analyzed by Recht et al. [281] had a rough morphotype and showed no detectable levels of GPLs. These non-sliding mutants were also defective for attachment and biofilm formation on PVC plastic [281]. This observation emphasizes the importance of GPLs in determining the cell surface properties of M. smegmatis. Suggestive of the important role played by GPLs in the permeability of the cell envelope, the absence of nsGPLs from the cell envelope of a defined knock-out mutant of M. smegmatis was shown to have a profound effect on the uptake of chenodeoxycholate [275], a hydrophobic molecule that diffuses through lipid domains of the mycobacterial cell envelope.
The species M. avium-intracellulare has received attention as a major opportunistic pathogen in AIDS patients. Although the specific mechanisms that define its pathogenicity have not been entirely clarified, it is becoming apparent that GPL antigens have a variety of biological activities that could influence host responses. During infection, M. avium synthesizes GPLs that accumulate in macrophages [278, 282–284]. Early studies have shown that M. avium ssGPLs suppress the mitogen-induced proliferative responses of murine splenic cells [285–287] but not that of human peripheral blood mononuclear cells (PBM) [288]. More recently, GPLs from M. avium serovar 4 have been proposed to participate in the ability of M. avium to invade human macrophages and escape bactericidal responses [289]. GPLs are also thought to impact adaptive immunity. Pre-treatment of human PBM with serovar specific GPLs, for instance, suppresses the production of Th1 cytokines including interleukin 2 (IL-2) and interferon γ (IFNγ) [290]. In contrast, ssGPLs induce the production of two important immunomodulatory substances, tumor necrosis factor α (TNF-α) and prostaglandin E2 (PGE2) [288, 291]. A group of polar GPLs from M. chelonae (pGPL-Mc) has also been reported to increase the resistance of mice to disseminated candidiasis [292] and to enhance the immune response to influenza vaccination [293]. Moreover, pGPL-Mc molecules exhibit the properties of haematopoietic growth factor [294–296].
Biosynthesis of GPLs
(a) nsGPL biosynthesis in M. smegmatis
The GPL biosynthetic gene cluster of M. smegmatis (Fig. 16B) is currently thought to encompass 24 genes. Twelve of them have been experimentally characterized using a combination of genetic approaches [210, 274, 281, 297–301]. The non-ribosomal peptide synthase genes, mps1 and msp2 encode the enzymes responsible for the synthesis of the peptidic moiety of D-Phe-D-allo-Thr-D-Ala-L-alaninol [210, 274]. Mps1 and Mps2 are each composed of two modules catalyzing the incorporation of an amino acid. The last module of Mps2, however, is devoid of a racemase domain [210] suggesting that it is responsible for the synthesis of the distal alaninol-containing moiety of the pseudo-tetrapeptide. The putative non-ribosomal peptide synthases of M. avium share the same genetic organization consisting of two ORFs [302]. The mbtH gene has been shown to be required for GPL production [303], but no exact function has yet been attributed to its protein product.
The main acyl residue of the M. smegmatis GPLs is a monounsaturated hydroxylated C30 fatty acid [271, 304] whose synthesis is thought to involve the polyketide synthase product of pks1 [210] (Fig. 16B). Three glycosyltransferase genes are also found in the GPL cluster, a number in agreement with the structure of the M. smegmatis GPLs (Fig. 16A–B) [299, 301, 305]. Two other genes of the cluster, namely rmlA (aka rfbA) encoding a putative glucose-1-phosphate thymidylyltransferase, and rmlB encoding a putative dTDP-glucose-4,6-dehydrogenase, are likely to be involved in the synthesis of the deoxyhexoses, rhamnose and 6-deoxytalose, which would subsequently be incorporated into nsGPLs [305].
The mtf1 (aka rmt3) gene [300] encodes a S-adenosylmethionine-dependent rhamnosyl-3-O-methyltransferase [297]. This enzyme is required for the O-methylation of position 3 of the rhamnosyl unit that glycosylates the alaninol. Disruption of rmt3 virtually abolishes the further methylation of the rhamnosyl unit suggesting that this enzme is the first methyltransferase to act on the GPL precursors [297]. Three other methyltransferase genes are found in the GPL gene cluster [305]. The fmt gene encodes a fatty acid O-methyltransferase that modifies the hydroxyl group of the GPL fatty acid [299], whereas rmt4 encodes a rhamnosyl-4-O-methyltransferase and rmt2 a rhamnosyl-2-O-methyltransferase [300]; all of these methyltransferases have orthologs in M. avium. The gene atf1 is predicted to encode a 6-deoxytalose acetyltransferase [306]. The methylation of the rhamnosyl residue occurs independently of the acetylation of the 6-deoxytalose residue, since the GPLs from an atf1 knock-out mutant are normally methylated.
(b) ssGPL biosynthesis in M. avium
Limited information is currently available about GPL biosynthesis in M. avium, except for the serotype 2 (ser2) ssGPLs recently reviewed by Billman-Jacobe [305] and Chatterjee and Khoo [268]. The rhamnosyltransferase gene rtfA is the first gene whose function was determined experimentally [307, 308]. When produced in M. smegmatis, RtfA catalyzed the addition of a rhamnosyl unit to the 6-deoxytalosyl residue of the GPL core [308], thus showing that the simpler nsGPLs can serve as biosynthetic precursors in the synthesis of ssGPLs. This result was confirmed by showing that the targeted disruption of rtfA in M. avium led to the loss of ser2-specific GPL [307]. Complementing M. smegmatis methyltransferase mutants with M. avium genes from the ser2 gene cluster, Jeevarajah et al. provided evidence of the 4-O-methyltransferase activity of the M. avium MtfC and MtfB proteins [300]. In addition, they showed that MtfD displays 3-O-methyltransferase activity on the rhamnosyl residue of the M. smegmatis GPLs [300]. The specificity of this methyltransferase was recently confirmed via the construction of a mtfD knock-out mutant of M. avium [309, 310]. Interestingly, the virulence of this mutant was attenuated in mice [309].
On the basis of an altered colony morphotype, Laurent et al. identified other genes likely to be involved in the biosynthesis of GPLs in M. avium [302]. Two non-ribosomal peptide synthase genes (pstA and pstB) and a probable polyketide synthase gene located downstream the ser2 cluster are orthologous to the mps and pks genes found in the GPL biosynthetic gene cluster of M. smegmatis mc2155, but a direct involvement of these genes in the mutants phenotypes remains to be established. The ser2 gene clusters of two ser2 strains of M. avium were also sequenced and compared with the homologous regions of M. avium ser1 strain 104, M. avium subsp. paratuberculosis and M. avium subsp. silvaticum [311]. Fifteen ORFs were identified and their putative functions in GPL biosynthesis determined: 5 encode glycosyltransferases (including RtfA), 6 encode O-methyltransferases (including MtfB,C,D), 1 encodes an O-acetyltransferase and 3 encode hexose synthetases (D-glucose dehydrogenase, mannose dehydratase and 6-deoxy-4-keto-D-mannose reductase/epimerase). A biosynthetic model in which ser2-specific GPLs are synthesized from a serovar-1-specific GPL intermediate, itself derived from a non-specific GPL precursor was proposed [311].
Regulation and transport of GPLs
The C-type GPL biosynthetic gene cluster begins with a triplet of transmembrane protein encoding genes possibly forming an operon, tmtp A, B and C (now named mmpS4, mmpL4a and mmpL4b). TmtpABC belong to the MmpL and MmpS families of mycobacterial proteins [210] (Fig. 16B). Both MmpL4a and MmpL4b have 12 putative transmembrane domains whereas the smaller MmpS4 protein displays only one. mmpL4a and mmpL4b transposon mutants have been reported to have a rough colony morphology, to lack sliding motility and to be devoid of GPLs [210, 281]; the precise role of the MmpL4 proteins in this phenotype, however, has yet to be determined. Interestingly, the biochemical characterization of a mmpS4 mutant of M. smegmatis established that this protein is required for the production and export of large amounts of GPLs but is dispensable for biosynthesis per se. Cross-complementation experiments demonstrated that the MmpS4 proteins from M. smegmatis, M. avium, M. tuberculosis and M. abscessus are exchangeable and thus not specific for a particular GPL species [312]. MmpS4 requires the formation of a protein complex at the pole of the bacillus to function. It was suggested that MmpS proteins facilitate lipid biosynthesis by acting as a scaffold for a coupled biosynthetic and transport machinery [312]. A similar mechanism has also been proposed for the transport of PDIM and SL in Mtb (see sections IV and VI), and is thus emerging as a common trait in the biogenesis of mycobacterial complex lipids.
While screening an M. smegmatis transposon mutant library for mutants with changes in cell surface properties, strains that failed to stain with ruthenium red were isolated [210]. All of these mutants harbored a transposon insertion in the gap gene (Fig. 16B) and produced GPLs chemically identical to those of the wild type strain. gap mutants, however, had much less GPLs at their surface suggestive of a role for Gap in the export of these lipids. The precise role of Gap in the biogenesis of GPLs – particularly in relation with the MmpL4 and MmpS4 proteins - remains to be determined. Gap may be required for the transport of GPLs across the periplasmic space upon their translocation across the plasma membrane in a process involving the MmpL4–MmpS4 proteins.
Nutrient starvation was reported to induce the production of triglycosylated C-type GPLs in M. smegmatis mc2155 [313]. The accumulation of polar GPLs in M. smegmatis mc2155 seems to be dependent on SigB, as the overexpression of sigB induces the production of these lipids while the disruption of this gene leads on the contrary to the abolition of their production [314]. The expression of gtf3, the glycosyltransferase responsible for the addition of the last sugar moiety of triglycosylated GPLs [301] is directly or indirectly controlled by sigB, at least during certain stages of growth [314]. Another gene potentially coding for an extracytoplasmic sigma factor, ecf, is present in the GPL biosynthetic cluster (Fig. 16B), but evidence for the involvement of this gene in the regulation of GPL biosynthesis is lacking. ecf is located upstream a gene encoding a putative sigma factor-associated protein. M. smegmatis displays a low frequency of spontaneous morphological variation that correlates with the production of larger amounts of GPLs [315]. The transposition of insertion elements into two GPL loci accounts for these morphological changes. One locus is the promoter region of the mps operon. The other locus is the lsr2 gene which encodes a small basic protein that likely plays a regulatory role.
VIII. Capsular polysaccharides
As mentioned above and with a few exceptions, the ‘capsule’-like structures produced by Mycobacterium species primarily consist of polysaccharides and proteins with generally minor amounts of lipids. The ratio of protein to polysaccharide varies according to the species. While in Mtb, M. kansasii and M. gastri, the major surface capsular constituents consist of polysaccharides, they mainly are proteins in M. phlei and M. smegmatis [7, 9]. Capsular polysaccharides, like other capsular components are not covalently bound to the rest of the cell envelope. The three types of capsular polysaccharides identified in the capsular material of tuberculous and non-tuberculous mycobacteria are: a high molecular weight (>1,000,000 Da) α-D-glucan composed of a →4-α-D-Glc-1→ core branched every 5 or 6 residues by oligoglucosides; a D-arabino-D-mannan (AM) similar in structure to lipoarabinomannan (LAM); and a D-mannan composed of a →6-α-D-Man-1→ core substituted at some of the 2 positions with an α-D-Man residue [6, 7, 316, 317]. All are neutral compounds, devoid of acyl substituents.
The structure of AM appears to be identical to that of LAM except for the loss of the phosphatidyl-myo-inositol anchor, suggesting that it may be formed from LAM by a specific hydrolytic enzyme. Likewise, the structure of D-mannan appears to be identical to that of the mannan domain of LM and LAM. It is therefore reasonable to assume that the same enzymes participate in the biosynthesis of LM/LAM and in that of the two extracellular polysaccharides. The reader is referred to the dedicated chapter on LAM for details about this biosynthetic pathway. d-Mannan and AM are expected to share with LM and LAM common properties in their interactions with the host.
α-d-Glucan is structurally very similar to the intracellular glycogen of Mtb and M. bovis BCG although its 3D-structure appears to be more compact and its molecular mass slightly higher (13 × 106 versus 7.5 × 106 Da). Capsular α-D-glucan was shown to be a ligand of the C-type lectin DC-SIGN of dendritic cells (DCs), to modulate the effector functions of monocyte-derived DCs and to mediate the nonopsonic binding of Mtb to complement receptor 3 (CR3) [318–320]. It was also postulated to contribute to the anti-phagocytic properties of the capsule of Mtb, thereby possibly controlling the interactions of Mtb with macrophages and promoting uptake via CR3 [321]. Altogether, these biological properties may contribute to the survival of Mtb in the host. The structural similarity between α-D-glucan and glycogen has allowed some of the genes involved in the biosynthesis of the capsular polysaccharide to be identified, among them the α-1,4-glucosyltransferases Rv3032 and GlgA (Rv1212c), the ADP-glucose pyrophosphorylase GlgC (Rv1213) and the branching enzyme GlgB (Rv1326c) responsible for introducing α-1,6-linked branches into linear α-1,4-glucans [322] (Table 4) (Fig. 17). The phenotypic analysis of Mtb recombinant strains affected or totally deficient in the expression of these genes confirmed their involvement in the elongation and branching of the capsular α-D-glucan and a partial redundancy between the two α-1,4-glucosyltransferases Rv3032 and GlgA. These analyses further revealed the participation of GlgC, GlgB and Rv3032 in the biosynthesis of other intracellular Mtb α-1,4-glucans, namely glycogen and the methylglucose lipopolysaccharides (MGLPs) [322, 323]. Attempts to knock-out both glgA and Rv3032 in Mtb mutant were unsuccessful indicating that a functional copy of at least one of the two α-1,4-glucosyltransferases is required for growth. The apparent essentiality of glgB [322] in contrast is believed to be related to the toxic accumulation of maltose-1-phosphate that follows the inactivation of this gene [324] (Fig. 17). Importantly, mycobacterial α-1,4-glucans can also be synthesized from trehalose by a four-step pathway comprising the trehalose synthase TreS, the maltokinase Pep2, the maltose-1-phosphate maltosyltransferase GlgE, and GlgB (Fig. 17; Table 4) [324, 325]. Disruption of glgE, like that of glgB, is lethal because of the toxic accumulation of maltose-1-phosphate that ensues.
Table 4.
Mtb H37Rv genes involved in the biogenesis of capsular α-D-glucan.
| Rv number |
Gene name |
Function | Evidence | Reference |
|---|---|---|---|---|
| Rv0126 | treS | Trehalose synthase; displays maltose <-> trehalose interconverting activity and glycogen amylase activity | E,P | [324, 358–360] |
| Rv0127 | pep2 | Maltokinase | E,P | [324, 361] |
| Rv1212c | glgA | α-1,4-glucosyltransferase | P | [322] |
| Rv1213 | glgC | ADP-glucose pyrophosphorylase | P | [322] |
| Rv1326c | glgB | α-1,4-glucan branching enzyme | E,P | [322, 362] |
| Rv1327c | glgE | Maltose-1-phosphate maltosyltransferase | E,P | [324, 325] |
| Rv1328 | glgP | Putative glycogen phosphorylase | H | - |
| Rv3032 | - | α-1,4-glucosyltransferase | E,P | [322, 323] |
E,P,H: see Table 1 legend.
Figure 17. Structure and biosynthesis of α-D-glucans in Mtb.
(See text for details)
As evidenced by their Rv numbers, the genes involved in the metabolism of glycogen, capsular α-D-glucan and MGLP are clustered in four major locations on the chromosome of Mtb H37Rv (Table 4). The first cluster [Rv3030−Rv3037c] encompasses the α-1,4-glucosyltransferase gene Rv3032 and other genes likely to be involved in the modifications of MGLPs [326]. The second cluster [Rv1208−Rv1213] carries glgA, glgC, and the glucosyl-3-phosphoglycerate synthase gene, Rv1208, required for the initiation of MGLPs [33]. The third cluster [Rv1326c−Rv1328] carries glgB, glgE and the probable glycogen phosphorylase gene glgP [35]. The fourth region [Rv0126−Rv0127] harbors pep2 and treS.
Consistent with the intracellular localization of glycogen and MGLPs, GlgA and Rv3032 are nucleotide sugar-utilizing glucosyltransferases predicted to catalyze the elongation of polysaccharides on the cytosolic face of the plasma membrane. GlgB, GlgC, GlgE, Pep2 and TreS are also predicted to be cytosolic enzymes. It is therefore reasonable to assume that capsular α-D-glucan, like glycogen and MGLPs, is synthesized in the cytoplasm. Nothing is known of the translocation machinery responsible for its export to the cell surface.
IX. Other lipophilic compounds
Other lipophilic compounds found in the cell envelope of some mycobacterial species include siderophores known as mycobactins and the M. ulcerans toxin, mycolactone. Their structure is shown on Figures 18 and 19. The reader is referred to recent reviews for a complete detail of their biosynthetic pathways [327, 328].
Figure 18. Representative structures of mycobactins and carboxymycobactins from Mtb.
(See text for details). Mycobactins: R1 = H; R2 = (CH2)nCH3, n = 16–19;
(CH2)xCH=CH(CH2)yCH3, x+y = 14–17.
Carboxymycobactins: R1 = H, CH3; R2 = (CH2)nCOOCH3/COOH, n = 1–7;
(CH2)xCH=CH(CH2)yCOOCH3/COOH, x+y = 1–5.
Figure 19. Representative structure of a mycolactone from M. ulcerans.
The genes involved in the biosynthesis of the various constituents of mycolactone are indicated on the structure.
Mycobactins
Pathogenic and non-pathogenic mycobacteria rely on siderophores with high affinity for the ferric ion as the primary mechanism for iron acquisition (for a review, [327]). Two classes of siderophores are produced by mycobacteria: the exochelins and the (carboxy)mycobactins. The exochelins are water-soluble peptidic molecules and are secreted into the medium. Mycobactins and carboxymycobactins are both salicyl-capped peptide polyketide-based molecules but vary in the length of an alkyl substitution and hence in polarity and solubility (Fig. 18). The lipid-soluble mycobactins have long-chain acyl chains on the first lysine residue, whereas the water-soluble carboxymycobactins have a shorter side chain that terminates with a carboxylic acid or methyl ester. Mycobactins tend to remain cell-associated while carboxymycobactins are secreted in the culture medium. Mycobacteria fall into four groups based upon the production of these molecules. Mtb produces only (carboxy)mycobactins which are essential for its virulence [327, 329]; M. vaccae produces only the exochelin type; M. smegmatis produces both types and M. leprae produces none. The biosynthesis of mycobacterial siderophores has been reviewed recently [327] and will therefore not be detailed here. Briefly, a cluster of 10 genes (annotated mbtA through mbtJ; Rv2377c−Rv2386c - the mbt locus) encompassing approximately 24-kb, another cluster of 6 genes referred to as mbt-2, the phosphopantetheinyl gene pptT (Rv2794c), the esx3 cluster (Rv0282−Rv0292) and the transport genes mmpS4/mmpL4 (Rv0450c−Rv0451c) and mmpS5/mmpL5 (Rv0676c−Rv0677c) encode the proteins required for the synthesis, export, utilization and uptake of (carboxy)mycobactins [327, 329]. The regulator IdeR (Rv2711) represses the expression of the mbtA-N genes.
Mycolactones
Mycolactones are a family of lipophilic macrocyclic polyketide molecules that is the primary virulence factor produced by M. ulcerans, the etiologic agent of Buruli ulcer in humans, and some closely related aquatic mycobacteria (Fig. 19). Mycolactones display cytotoxic, analgesic and immunosuppressive activities [328]. They are found in abundant quantities in the extracellular matrix surrounding M. ulcerans under certain in vitro growth conditions and during host infection [330]. A 174-kb megaplasmid named pMUM001 in the M. ulcerans strain Agy99 carries all of the genes required for mycolactone synthesis [331]. These include two very large type I polyketide synthase genes, mlsA1 (51 kb), mlsA2 (7 kb), responsible for the synthesis of the upper side chain and macrolactone core; another giant type I polyketide synthase gene, mlsB (42 kb), involved in the elongation of the acyl side chain; mup_045 thought to encode the transferase linking the acyl side chain and the macrolactone core; mup_053c, a P450 monooxygenase gene most likely responsible for hydroxylation of the mycolactone acyl side chain at C12’, and mup_038, a type II thioesterase gene predicted to play a role in maintaining the fidelity of the polyketide synthases by removing acyl chains from modules where synthesis has stalled [328].
Conclusions and Future Prospects
As illustrated in this chapter, knowledge of cell envelope biosynthesis in Mtb has greatly benefited from the publication of the complete genome sequence of this bacterium and developments in the genetic manipulation of mycobacteria in the late in 1990s. As more genomes from slow- and rapidly-growing mycobacteria are being sequenced, this impetus is progressively extending to other tuberculous and nontuberculous mycobacterial species with the result that the processes leading to the biosynthesis of more and more species-specific cell envelope constituents are now being elucidated. Beyond the opportunities offered by some of these pathways for drug development, interest in the biosynthesis of species-specific cell envelope constituents stems from their antigenicity and potential for serodiagnosis and as biomarkers. GPL, DAT, LOS, PGL, pHBAD and TDM in particular are potent B-cell antigens and have been the object of extensive studies aimed at assessing their potential for the diagnosis of TB, leprosy or other mycobacterial diseases [14, 223, 256, 257, 332–334]. Key to their widespread application for therapeutic or diagnostic purposes, however, will be a precise understanding of their role in the physiology and virulence of the bacterium and regulatory processes governing their synthesis during the various stages of the lifecycle of the producing mycobacterium. While it is now well established that mycobacteria adjust the composition of their cell envelope in response to the nutrients available in the environment (e.g., carbon and nitrogen source, iron concentration, …), physical conditions to which they are exposed (pH, oxygen tension) and age of the culture, knowledge of the regulatory processes involved is more limited. Yet, these changes affect all major cell envelope constituents including phospholipids, PIM, triglycerides, capsular polysaccharides, lipoglycans, peptidoglycan, arabinogalactan and mycolic acids [14, 15, 141, 180, 335–341]. Regulation appears to occur both at the transcriptional and at the post-translational levels. The two-component transcriptional regulator PhoP-PhoR, for instance, stands out as a major regulator of polymethyl-branched acyltrehalose production in Mtb (SL, DAT and PAT) [143, 177]. Other important regulators controlling cell division and cell envelope biogenesis are found among the serine/threonine kinase family (for a review, [342]). These enzymes regulate through phosphorylation the activity of multiple enzymes and transporters involved in the biosynthesis of mycolic acids, peptidoglycan, arabinogalactan, lipoarabinomannan and PDIM.
Despite the considerable advances made in deciphering the metabolic pathways of mycobacterial cell envelope constituents, most of them are not yet complete. The clustered genetic organization of many of these pathways (e.g., lipoglycans, acyltrehaloses, GPLs, LOS, PDIM, …) raises the hope that some of the missing genes will be found on the basis of their co-localization with known clusters. In the case of a few other minor or species-specific cell envelope constituents, however, biosynthetic pathways have hardly started to be explored. This is for instance the case for the glycosyl diacylglycerols described by Hunter et al. [343] and the mycobacterial carotenoids whose structural definition extends back to the work of E. Chargaff in 1930 [259]. Likewise, although there is at present no chemical evidence of LOS in Mtb, preliminary data indicate that some of the unannotated glycosyltransferases of the GT-A or -B or -C classes may participate in the synthesis of chemically undetectable amounts of these products [204, 207].
Beyond the identification of the missing biosynthetic and regulatory proteins will be the identification of the transporters required for the translocation of biosynthetic precursors or end products from their site of production, for the most part cytoplasmic, to their final periplasmic, outer membrane or capsular locations. More than 148 transport associated proteins belonging to 33 major transporter families were identified in the genome of Mtb H37Rv (http://www.membranetransport.org/). More transporters are typically found in environmental Mycobacterium species. The latest Mtb genome annotation was also updated with 134 bioinformatically-predicted outer membrane proteins [344]. The transporters required for the building of the cell envelope are thus most likely to be found in this long and diverse list of candidate genes with or without any homologs in other prokaryotes. Indeed, searches for mycobacterial transporters sharing sequence similarity with known (lipo)polysaccharide or glycolipid transporters from Gram positive or Gram negative bacteria typically yields limited if any meaningful candidates. Yet, it is becoming increasingly evident that, similar to the biogenesis of other prokaryotic (lipo)polysaccharides, the biosynthesis and translocation of many mycobacterial cell envelope constituents (e.g., mycolic acids, PDIM, acyltrehaloses, mycobactins, lipoglycans and arabinogalactan) are temporally and spatially coupled by multiprotein complexes that possibly span the cell envelope. Reasons for their elusive nature may be found in the unusual structure and composition of the mycobacterial cell envelope which may have driven mycobacteria to evolve specialized transport systems mechanistically-related but structurally divergent from that of other prokaryotes. Examples of these include the type VII secretion system ESX-3 for the uptake of mycobactins [345], the Mce4 proteins for the import of cholesterol [346], the RND-like (Resistance, Nodulation and Division) inner membrane transporters of the MmpL family involved in the export of complex lipids and mycobactins [145, 170, 171, 210, 229, 235, 252, 281, 329, 347], the periplasmic Lol-like lipoprotein carriers LppX and LprG for the export of PDIM and PIM/lipoglycans [38, 253] and the SMR-like (Small Multidrug Resistance) lipid-linked arabinose translocase Rv3789 [348].
Finally, another area where much remains to be done is in understanding the genetic bases underlying the cell envelope’s constant remodeling (including degradation and recycling) which accompanies cell division or any changes in the metabolism of the bacterium following host infection or exposure to various environmental stresses.
Acknowledgments
The authors’ research on cell envelope biogenesis is supported through NIH/NIAID grants # AI064798, AI063054, AI049151 and AI097550. We thank Dr. Hugo Gramajo (Universidad Nacional de Rosario, Rosario, Argentina) for sharing unpublished results, and Victoria Jones (CSU), Dr. Michael McNeil (CSU), Dr. Gilles Etienne, Dr. Anne Lemassu, and Lucie Spina (IPBS-CNRS, Toulouse) for critical reading of the manuscript and their help with the preparation of the figures.
References
- 1.Daffé M, Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microb Physiol. 1998;39:131–203. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- 2.Jackson M, McNeil MR, Brennan PJ. Progress in targeting cell envelope biogenesis in Mycobacterium tuberculosis. Future Microbiol. 2013 doi: 10.2217/fmb.13.52. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. USA. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffe M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 2008;190:5672–5680. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sani M, Houben ENG, Geurtsen J, Pierson J, de Punder K, van Zon M, Wever B, Piersma SR, Jimenez CR, Daffe M, Appelmelk BJ, Bitter W, van der Wel N, Peters PJ. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010;6:e1000794. doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemassu A, Daffé M. Structural features of the exocellular polysaccharides of Mycobacterium tuberculosis. Biochem. J. 1994;297:351–357. doi: 10.1042/bj2970351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortalo-Magné A, Dupont MA, Lemassu A, Andersen AB, Gounon P, Daffé M. Molecular composition of the outermost capsular material of the tubercle bacillus. Microbiology. 1995;141:1609–1620. doi: 10.1099/13500872-141-7-1609. [DOI] [PubMed] [Google Scholar]
- 8.Ortalo-Magné A, Lemassu A, Lanéelle MA, Bardou F, Silve G, Gounon P, Marchal G, Daffé M. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J. Bacteriol. 1996;178:456–461. doi: 10.1128/jb.178.2.456-461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemassu A, Ortalo-Magne A, Bardou F, Silve G, Laneelle M-A, Daffe M. Extracellular and surface-exposed polysaccharides of non-tuberculous mycobacteria. Microbiology. 1996;142:1513–1520. doi: 10.1099/13500872-142-6-1513. [DOI] [PubMed] [Google Scholar]
- 10.Raynaud C, Etienne G, Peyron P, Laneelle MA, Daffe M. Extracellular enzyme activities potentially involved in the pathogenicity of Mycobacterium tuberculosis. Microbiology. 1998;144(Pt 2):577–587. doi: 10.1099/00221287-144-2-577. [DOI] [PubMed] [Google Scholar]
- 11.Ehlers MRW, Daffé M. Interactions between Mycobacterium tuberculosis and host cells: are mycobacterial sugars the key? Trends in Microbiology. 1998;6:328–335. doi: 10.1016/s0966-842x(98)01301-8. [DOI] [PubMed] [Google Scholar]
- 12.Daffé M, Etienne G. The capsule of Mycobacterium tuberculosis and its implications for pathogenicity. Tuber Lung Dis. 1999;79:153–169. doi: 10.1054/tuld.1998.0200. [DOI] [PubMed] [Google Scholar]
- 13.Goren MB. Biosynthesis and structures of phospholipids and sulfatides. In: Kubica GP, Wayne LG, editors. The mycobacteria. A sourcebook. Vol. 1. New York and Basel: Marcel Dekker, Inc.; 1984. pp. 379–415. [Google Scholar]
- 14.Brennan PJ. Mycobacterium and other actinomycetes. In: Ratledge C, Wilkinson SG, editors. Microbial lipids. Vol. 1. London: Academic Press Ltd.; 1988. pp. 203–298. [Google Scholar]
- 15.Daniel J, Deb C, Dubey VS, Sirakova TD, Abomoelak B, Morbidoni HR, Kolattukudy PE. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol. 2004;186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker RW, Barakat H, Hung JGC. The positional distribution of fatty acids in the phospholipids and triglycerides of Mycobacterium smegmatis and M. bovis BCG. Lipids. 1970;5:684–691. doi: 10.1007/BF02531435. [DOI] [PubMed] [Google Scholar]
- 17.Fernandes ND, Kolattukudy PE. Cloning, sequencing and characterization of a fatty acid synthase-encoding gene from Mycobacterium tuberculosis var. bovis BCG. Gene. 1996;170:95–99. doi: 10.1016/0378-1119(95)00842-x. [DOI] [PubMed] [Google Scholar]
- 18.Zimhony O, Vilcheze C, Jacobs WR., Jr Characterization of Mycobacterium smegmatis expressing the Mycobacterium tuberculosis fatty acid synthase I (fas1) gene. Journal of bacteriology. 2004;186:4051–4055. doi: 10.1128/JB.186.13.4051-4055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao J, Rock CO. Phosphatidic acid synthesis in bacteria. Biochimica et biophysica acta. 2013;1831:495–502. doi: 10.1016/j.bbalip.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comba S, Menendez-Bravo S, Arabolaza A, Gramajo H. Identification and physiological characterization of phosphatidic acid phosphatase enzymes involved in triacylglycerol biosynthesis in Streptomyces coelicolor. Microbial cell factories. 2013;12:9. doi: 10.1186/1475-2859-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirakova TD, Dubey VS, Deb C, Daniel J, Korotkova TA, Abomoelak B, Kolattukudy PE. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology. 2006;152:2717–2725. doi: 10.1099/mic.0.28993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elamin AA, Stehr M, Spallek R, Rohde M, Singh M. The Mycobacterium tuberculosis Ag85A is a novel diacylglycerol acyltransferase involved in lipid body formation. Molecular microbiology. 2011;81:1577–1592. doi: 10.1111/j.1365-2958.2011.07792.x. [DOI] [PubMed] [Google Scholar]
- 23.Nigou J, Besra GS. Cytidine diphosphate-diacylglycerol synthesis in Mycobacterium smegmatis. The Biochemical journal. 2002;367:157–162. doi: 10.1042/BJ20020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salman M, Lonsdale JT, Besra GS, Brennan PJ. Phosphatidylinositol synthesis in mycobacteria. Biochim. Biophys. Acta. 1999;1436:437–450. doi: 10.1016/s0005-2760(98)00151-9. [DOI] [PubMed] [Google Scholar]
- 25.Jackson M, Crick DC, Brennan PJ. Phosphatidylinositol is an essential phospholipid of mycobacteria. J. Biol. Chem. 2000;275:30092–30099. doi: 10.1074/jbc.M004658200. [DOI] [PubMed] [Google Scholar]
- 26.Morii H, Ogawa M, Fukuda K, Taniguchi H, Koga Y. A revised biosynthetic pathway for phosphatidylinositol in Mycobacteria. Journal of biochemistry. 2010;148:593–602. doi: 10.1093/jb/mvq093. [DOI] [PubMed] [Google Scholar]
- 27.Sandoval-Calderon M, Geiger O, Guan Z, Barona-Gomez F, Sohlenkamp C. A eukaryote-like cardiolipin synthase is present in Streptomyces coelicolor and in most actinobacteria. The Journal of biological chemistry. 2009;284:17383–17390. doi: 10.1074/jbc.M109.006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korduláková J, Gilleron M, Mikusova K, Puzo G, Brennan PJ, Gicquel B, Jackson M. Definition of the first mannosylation step in phosphatidylinositol synthesis: PimA is essential for growth of mycobacteria. J. Biol. Chem. 2002;277:31335–31344. doi: 10.1074/jbc.M204060200. [DOI] [PubMed] [Google Scholar]
- 29.Guerin ME, Kordulakova J, Schaeffer F, Svetlikova Z, Buschiazzo A, Giganti D, Gicquel B, Mikusova K, Jackson M, Alzari PM. Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J. Biol. Chem. 2007;282:20705–20714. doi: 10.1074/jbc.M702087200. [DOI] [PubMed] [Google Scholar]
- 30.Guerin ME, Schaeffer F, Chaffotte A, Gest P, Giganti D, Kordulakova J, van der Woerd M, Jackson M, Alzari PM. Substrate-induced conformational changes in the essential peripheral membrane-associated mannosyltransferase PimA from mycobacteria: implications for catalysis. The Journal of biological chemistry. 2009a;284:21613–21625. doi: 10.1074/jbc.M109.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerin ME, Kaur D, Somashekar BS, Gibbs S, Gest P, Chatterjee D, Brennan PJ, Jackson M. New insights into the early steps of phosphatidylinositol mannosides biosynthesis in mycobacteria. PimB' is an essential enzyme of Mycobacterium smegmatis. J. Biol. Chem. 2009b;284:25687–25696. doi: 10.1074/jbc.M109.030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korduláková J, Gilleron M, Puzo G, Brennan PJ, Gicquel B, Mikusova K, Jackson M. Identification of the required acyltransferase step in the biosynthesis of the phosphatidylinositol mannosides of Mycobacterium species. J. Biol. Chem. 2003;278:36285–36295. doi: 10.1074/jbc.M303639200. [DOI] [PubMed] [Google Scholar]
- 33.Kaur D, Guerin ME, Skovierova H, Brennan PJ, Jackson M. Chapter 2. Biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv Appl Microbiol. 2009;69:23–78. doi: 10.1016/S0065-2164(09)69002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morita YS, Sena CCB, Waller RF, Kurokawa K, Sernee MF, Nakatani F, Haites RE, Billman-Jacobe H, McConville MJ, Maeda Y, Kinoshita T. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in mycobacteria. J. Biol. Chem. 2006;281:25143–25155. doi: 10.1074/jbc.M604214200. [DOI] [PubMed] [Google Scholar]
- 35.Berg S, Kaur D, Jackson M, Brennan PJ. The glycosyltransferases of Mycobacterium tuberculosis- roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology. 2007;17:35R–56R. doi: 10.1093/glycob/cwm010. [DOI] [PubMed] [Google Scholar]
- 36.Morita YS, Velasquez R, Taig E, Waller RF, Patterson JH, Tull D, Williams SJ, Billman-Jacobe H, McConville MJ. Compartmentalization of lipid biosynthesis in mycobacteria. J. Biol. Chem. 2005;280:21645–21652. doi: 10.1074/jbc.M414181200. [DOI] [PubMed] [Google Scholar]
- 37.Sulzenbacher G, Canaan S, Bordat Y, Neyrolles O, Stadthagen G, Roig-Zamboni V, Rauzier J, Maurin D, Laval F, Daffe M, Cambillau C, Gicquel B, Bourne Y, Jackson M. LppX is a lipoprotein required for the translocation of phthiocerol dimycocerosates to the surface of Mycobacterium tuberculosis. The EMBO journal. 2006;25:1436–1444. doi: 10.1038/sj.emboj.7601048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drage MG, Tsai HC, Pecora ND, Cheng TY, Arida AR, Shukla S, Rojas RE, Seshadri C, Moody DB, Boom WH, Sacchettini JC, Harding CV. Mycobacterium tuberculosis lipoprotein LprG (Rv1411c) binds triacylated glycolipid agonists of Toll-like receptor 2. Nature structural & molecular biology. 2010;17:1088–1095. doi: 10.1038/nsmb.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolucka BA, McNeil MR, de Hoffmann E, Chojnacki T, Brennan PJ. Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J.Biol.Chem. 1994;269:23328–23335. [PubMed] [Google Scholar]
- 40.Mahapatra S, Yagi T, Belisle JT, Espinosa BJ, Hill PJ, McNeil MR, Brennan PJ, Crick DC. Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. Journal of bacteriology. 2005;187:2747–2757. doi: 10.1128/JB.187.8.2747-2757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson RG, Hussey H, Baddiley J. The mechanism of wall synthesis in bacteria. The organization of enzymes and isoprenoid phosphates in the membrane. Biochem.J. 1972;127:11–25. doi: 10.1042/bj1270011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mikusova K, Mikus M, Besra GS, Hancock I, Brennan PJ. Biosynthesis of the linkage region of the mycobacterial cell wall. J.Biol.Chem. 1996;271:7820–7828. doi: 10.1074/jbc.271.13.7820. [DOI] [PubMed] [Google Scholar]
- 43.Sacchettini JC, Poulter CD. Creating isoprenoid diversity. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 44.Schwender J, Seemann M, Lichtenthaler HK, Rohmer M. Biosynthesis of isoprenoids (carotenoids, sterols, prenyl side-chains of chlorophylls and plastoquinone) via a novel pyruvate/glyceraldehyde 3-phosphate non-mevalonate pathway in the green alga Scenedesmus obliquus. Biochem.J. 1996;316(Pt 1):73–80. doi: 10.1042/bj3160073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sprenger GA, Schorken U, Wiegert T, Grolle S, De Graaf AA, Taylor SV, Begley TP, Bringer-Meyer S, Sahm H. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc.Natl.Acad.Sci.U.S.A. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lois LM, Campos N, Putra SR, Danielsen K, Rohmer M, Boronat A. Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of D-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc.Natl.Acad.Sci.U.S.A. 1998;95:2105–2110. doi: 10.1073/pnas.95.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill RE, Himmeldirk K, Kennedy IA, Pauloski RM, Sayer BG, Wolf E, Spenser ID. The biogenetic anatomy of vitamin B-6 - A C-13NMR investigation of the biosynthesis of pyridoxol in Escherichia coli. J.Biol.Chem. 1996;271:30426–30435. doi: 10.1074/jbc.271.48.30426. [DOI] [PubMed] [Google Scholar]
- 48.Querol J, Rodriguez-Concepcion M, Boronat A, Imperial S. Essential role of residue H49 for activity of Escherichia coli 1-deoxy-D-xylulose 5-phosphate synthase, the enzyme catalyzing the first step of the 2-C-methyl-D-erythritol 4-phosphate pathway for isoprenoid synthesis. Biochem.Biophys.Res.Commun. 2001;289:155–160. doi: 10.1006/bbrc.2001.5957. [DOI] [PubMed] [Google Scholar]
- 49.Bailey AM, Mahapatra S, Brennan PJ, Crick DC. Identification, cloning, purification, and enzymatic characterization of Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate synthase. Glycobiology. 2002;12:813–820. doi: 10.1093/glycob/cwf100. [DOI] [PubMed] [Google Scholar]
- 50.Brown AC, Eberl M, Crick DC, Jomaa H, Parish T. The nonmevalonate pathway of isoprenoid biosynthesis in Mycobacterium tuberculosis is essential and transcriptionally regulated by Dxs. J.Bacteriol. 2010;192:2424–2433. doi: 10.1128/JB.01402-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi S, Kuzuyama T, Watanabe H, Seto H. A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc.Natl.Acad.Sci.U.S.A. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arigoni D, Sagner S, Latzel C, Eisenreich W, Bacher A, Zenk MH. Terpenoid biosynthesis from 1-deoxy-D-xylulose in higher plants by intramolecular skeletal rearrangement. Proc.Natl.Acad.Sci.U.S A. 1997;94:10600–10605. doi: 10.1073/pnas.94.20.10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reuter K, Sanderbrand S, Jomaa H, Wiesner J, Steinbrecher I, Beck E, Hintz M, Klebe G, Stubbs MT. Crystal structure of 1-deoxy-D-xylulose-5-phosphate reductoisomerase, a crucial enzyme in the non-mevalonate pathway of isoprenoid biosynthesis. J.Biol.Chem. 2002;277:5378–5384. doi: 10.1074/jbc.M109500200. [DOI] [PubMed] [Google Scholar]
- 54.Argyrou A, Blanchard JS. Kinetic and chemical mechanism of Mycobacterium tuberculosis 1-deoxy-D-xylulose-5-phosphate isomeroreductase. Biochemistry. 2004;43:4375–4384. doi: 10.1021/bi049974k. [DOI] [PubMed] [Google Scholar]
- 55.Dhiman RK, Schaeffer ML, Bailey AM, Testa CA, Scherman H, Crick DC. 1-Deoxy-D-xylulose 5-phosphate reductoisomerase (IspC) from Mycobacterium tuberculosis: towards understanding mycobacterial resistance to fosmidomycin. J.Bacteriol. 2005;187:8395–8402. doi: 10.1128/JB.187.24.8395-8402.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henriksson LM, Unge T, Carlsson J, Aqvist J, Mowbray SL, Jones TA. Structures of Mycobacterium tuberculosis 1-deoxy-D-xylulose-5-phosphate reductoisomerase provide new insights into catalysis. J.Biol.Chem. 2007;282:19905–19916. doi: 10.1074/jbc.M701935200. [DOI] [PubMed] [Google Scholar]
- 57.Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisenreich W, Bacher A, Zenk MH. Cytidine 5'-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:11758–11763. doi: 10.1073/pnas.96.21.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eoh H, Brown AC, Buetow L, Hunter WN, Parish T, Kaur D, Brennan PJ, Crick DC. Characterization of the Mycobacterium tuberculosis 4-diphosphocytidyl-2-C-methyl-D-erythritol synthase: potential for drug development. J.Bacteriol. 2007;189:8922–8927. doi: 10.1128/JB.00925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herz S, Wungsintaweekul J, Schuhr CA, Hecht S, Luttgen H, Sagner S, Fellermeier M, Eisenreich W, Zenk MH, Bacher A, Rohdich F. Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2C- methyl-D-erythritol 2-phosphate to 2C-methyl-D-erythritol 2,4- cyclodiphosphate. Proc.Natl.Acad.Sci.U.S.A. 2000;97:2486–2490. doi: 10.1073/pnas.040554697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rohdich F, Wungsintaweekul J, Luttgen H, Fischer M, Eisenreich W, Schuhr CA, Fellermeier M, Schramek N, Zenk MH, Bacher A. Biosynthesis of terpenoids: 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase from tomato. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8251–8256. doi: 10.1073/pnas.140209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eoh H, Narayanasamy P, Brown AC, Parish T, Brennan PJ, Crick DC. Expression and characterization of soluble 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase from bacterial pathogens. Chem. & Biol. 2009;16:1230–1239. doi: 10.1016/j.chembiol.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Narayanasamy P, Eoh H, Crick DC. Chemoenzymatic synthesis of 4-diphosphocytidyl-2-C-methyl-D-erythritol: a substrate for IspE. Tetrahedron Lett. 2008;49:4461–4463. doi: 10.1016/j.tetlet.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Narayanasamy P, Eoh H, Brennan PJ, Crick DC. Synthesis of 4-diphosphocytidyl-2-C-methyl-D-erythritol-2-phosphate and kinetic studies of Mycobacterium tuberculosis IspF. Chem. & Biol. 2010;17:117–122. doi: 10.1016/j.chembiol.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buetow L, Brown AC, Parish T, Hunter WN. The structure of mycobacteria 2C-methyl-D-erythritol-2,4-cyclodiphosphatesynthase, an essential enzyme, provides a platform for drug discovery. BMC Struct. Biol. 2007;7:68. doi: 10.1186/1472-6807-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F. Studies on the nonmevalonate pathway to terpenes: The role of the GcpE (IspG) protein. Proc. Natl. Acad. Sci. U.S A. 2001;98:14837–14842. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altincicek B, Duin EC, Reichenberg A, Hedderich R, Kollas AK, Hintz M, Wagner S, Wiesner J, Beck E, Jomaa H. LytB protein catalyzes the terminal step of the 2-C-methyl-D-erythritol-4-phosphate pathway of isoprenoid biosynthesis. FEBS Lett. 2002;532:437–440. doi: 10.1016/s0014-5793(02)03726-2. [DOI] [PubMed] [Google Scholar]
- 67.Altincicek B, Kollas A, Eberl M, Wiesner J, Sanderbrand S, Hintz M, Beck E, Jomaa H. LytB, a novel gene of the 2-C-methyl-D-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. FEBS Lett. 2001;499:37–40. doi: 10.1016/s0014-5793(01)02516-9. [DOI] [PubMed] [Google Scholar]
- 68.Seemann M, Bui BTS, Wolff M, Tritsch D, Campos N, Boronat A, Marquet A, Rohmer M. Isoprenoid biosynthesis through the methylerythritol phosphate pathway: The (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase (GcpE) is a [4Fe-4S] protein. Ang.Chemie-Int.Ed. 2002;41:4337–4339. doi: 10.1002/1521-3773(20021115)41:22<4337::AID-ANIE4337>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 69.Rohdich F, Zepeck F, Adam P, Hecht S, Kaiser J, Laupitz R, Grawert T, Amslinger S, Eisenreich W, Bacher A, Arigoni D. The deoxyxylulose phosphate pathway of isoprenoid biosynthesis: studies on the mechanisms of the reactions catalyzed by IspG and IspH protein. Proc.Natl.Acad.Sci.U.S.A. 2003;100:1586–1591. doi: 10.1073/pnas.0337742100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rohdich F, Hecht S, Gartner K, Adam P, Krieger C, Amslinger S, Arigoni D, Bacher A, Eisenreich W. Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc.Natl.Acad.Sci.U.S.A. 2002;99:1158–1163. doi: 10.1073/pnas.032658999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Agranoff BW, Eggerer H, Henning U, Lynen F. Biosynthesis of terpenes. VII. Isopentenyl pyrophosphate isomerase. J.Biol.Chem. 2013;235:326–332. [PubMed] [Google Scholar]
- 72.Phillips MA, D'Auria JC, Gershenzon J, Pichersky E. The Arabidopsis thaliana type I isopentenyl diphosphate isomerases are targeted to multiple subcellular compartments and have overlapping functions in isoprenoid biosynthesis. Plant Cell. 2008;20:677–696. doi: 10.1105/tpc.107.053926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuzuyama T, Seto H. Diversity of the biosynthesis of the isoprene units. Nat.Prod.Rep. 2003;20:171–183. doi: 10.1039/b109860h. [DOI] [PubMed] [Google Scholar]
- 74.Hahn FM, Hurlburt AP, Poulter CD. Escherichia coli Open Reading Frame 696 Is idi, a Nonessential Gene Encoding Isopentenyl Diphosphate Isomerase. J.Bacteriol. 1999;181:4499–4504. doi: 10.1128/jb.181.15.4499-4504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaneda K, Kuzuyama T, Takagi M, Hayakawa Y, Seto H. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp strain CL190. Proc.Natl.Acad.Sci.U.S A. 2001;98:932–937. doi: 10.1073/pnas.020472198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vandermoten S, Haubruge E, Cusson M. New insights into short-chain prenyltransferases: structural features, evolutionary history and potential for selective inhibition. Cell Mol.Life Sci. 2009;66:3685–3695. doi: 10.1007/s00018-009-0100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ambo T, Noike M, Kurokawa H, Koyama T. Cloning and functional analysis of novel short-chain cis-prenyltransferases. Biochem.Biophys.Res.Commun. 2008;375:536–540. doi: 10.1016/j.bbrc.2008.08.057. [DOI] [PubMed] [Google Scholar]
- 78.Liang PH. Reaction kinetics, catalytic mechanisms, conformational changes, and inhibitor design for prenyltransferases. Biochemistry. 2009;48:6562–6570. doi: 10.1021/bi900371p. [DOI] [PubMed] [Google Scholar]
- 79.Schulbach MC, Brennan PJ, Crick DC. Identification of a short (C15) chain Z-isoprenyl diphosphate synthase and a homologous long (C50) chain isoprenyl diphosphate synthase in Mycobacterium tuberculosis. J.Biol.Chem. 2000;275:22876–22881. doi: 10.1074/jbc.M003194200. [DOI] [PubMed] [Google Scholar]
- 80.Schulbach MC, Mahapatra S, Macchia M, Barontini S, Papi C, Minutolo F, Bertini S, Brennan PJ, Crick DC. Purification, enzymatic characterization, and inhibition of the Z-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J.Biol.Chem. 2001;276:11624–11630. doi: 10.1074/jbc.M007168200. [DOI] [PubMed] [Google Scholar]
- 81.Noike M, Ambo T, Kikuchi S, Suzuki T, Yamashita S, Takahashi S, Kurokawa H, Mahapatra S, Crick DC, Koyama T. Product chain-length determination mechanism of Z,E-farnesyl diphosphate synthase. Biochem.Biophys.Res.Commun. 2008;377:17–22. doi: 10.1016/j.bbrc.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang W, Dong C, McNeil M, Kaur D, Mahapatra S, Crick DC, Naismith JH. The structural basis of chain length control in Rv1086. J.Mol.Biol. 2008;381:129–140. doi: 10.1016/j.jmb.2008.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaur D, Brennan PJ, Crick DC. Decaprenyl diphosphate synthesis in Mycobacterium tuberculosis. J.Bacteriol. 2004;186:7564–7570. doi: 10.1128/JB.186.22.7564-7570.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sato T, Takizawa K, Orito Y, Kudo H, Hoshino T. Insight into C35 terpene biosyntheses by nonpathogenic Mycobacterium species: functional analyses of three Z-prenyltransferases and identification of dehydroheptaprenylcyclines. Chembiochem. 2010;11:1874–1881. doi: 10.1002/cbic.201000328. [DOI] [PubMed] [Google Scholar]
- 85.Mann FM, Thomas JA, Peters RJ. Rv0989c encodes a novel (E)-geranyl diphosphate synthase facilitating decaprenyl diphosphate biosynthesis in Mycobacterium tuberculosis. FEBS Lett. 2011;585:549–554. doi: 10.1016/j.febslet.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dhiman RK, Schulbach MC, Mahapatra S, Baulard AR, Vissa V, Brennan PJ, Crick DC. Identification of a novel class of {omega},E,E-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J.Lipid Res. 2004;45:1140–1147. doi: 10.1194/jlr.M400047-JLR200. [DOI] [PubMed] [Google Scholar]
- 87.Mann FM, Xu M, Davenport EK, Peters RJ. Functional characterization and evolution of the isotuberculosinol operon in Mycobacterium tuberculosis and related mycobacteria. Front Microbiol. 2012;3:368. doi: 10.3389/fmicb.2012.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN) Prenol nomenclature. Recommendations 1986. Eur.J.Biochem. 1987;167:181–184. [PubMed] [Google Scholar]
- 89.Takayama K, Schnoes HK, Semmler EJ. Characterization of the alkali-stable mannophospholipids of Mycobacterium smegmatis. Biochim.Biophys.Acta. 1973;316:212–221. doi: 10.1016/0005-2760(73)90011-8. [DOI] [PubMed] [Google Scholar]
- 90.Besra GS, Sievert T, Lee RE, Slayden RA, Brennan PJ, Takayama K. Identification of the apparent carrier in mycolic acid synthesis. Proc.Natl.Acad.Sci.U.S.A. 1994;91:12735–12739. doi: 10.1073/pnas.91.26.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wolucka BA, de Hoffmann E. Isolation and characterization of the major form of polyprenyl-phospho-mannose from Mycobacterium smegmatis. Glycobiology. 1998;8:955–962. doi: 10.1093/glycob/8.10.955. [DOI] [PubMed] [Google Scholar]
- 92.Takayama K, Goldman DS. Enzymatic synthesis of mannosyl-1-phosphoryl-decaprenol by a cell-free system of Mycobacterium tuberculosis. J.Biol.Chem. 1970;245:6251–6257. [PubMed] [Google Scholar]
- 93.Wolucka BA, de Hoffmann E. The presence of beta-D-ribosyl-1-monophosphodecaprenol in mycobacteria. J.Biol.Chem. 1995;270:20151–20155. doi: 10.1074/jbc.270.34.20151. [DOI] [PubMed] [Google Scholar]
- 94.El Ghachi M, Bouhss A, Blanot D, Mengin-Lecreulx D. The bacA gene of Escherichia coli encodes a undecaprenyl pyrophosphate phosphatase activity. J.Biol.Chem. 2004 doi: 10.1074/jbc.M401701200. [DOI] [PubMed] [Google Scholar]
- 95.Sherman MM, Petersen LA, Poulter CD. Isolation and characterization of isoprene mutants of Escherichia coli. J.Bacteriol. 1989;171:3619–3628. doi: 10.1128/jb.171.7.3619-3628.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Minnikin DE. Lipids: complex lipids, their chemistry, biosynthesis and roles. In: Ratledge C, Stanford J, editors. The Biology of Mycobacteria. London: Academic Press; 1982. pp. 95–184. [Google Scholar]
- 97.Embley TM, Stackebrandt E. The molecular phylogeny and systematics of the Actinomycetes. Annu.Rev.Microbiol. 1994;48:257–289. doi: 10.1146/annurev.mi.48.100194.001353. [DOI] [PubMed] [Google Scholar]
- 98.Pandya KP, King HK. Ubiquinone and menaquinone in bacteria - a comparative study of some bacterial respiratory systems. Arch.Biochem.Biophys. 1966;114 doi: 10.1016/0003-9861(66)90316-x. 154-&. [DOI] [PubMed] [Google Scholar]
- 99.Meganathan R. Biosynthesis of menaquinone (vitamin K-2) and ubiquinone (coenzyme Q): A perspective on enzymatic mechanisms. Vitamins and Hormones - Advances in Research and Applications. 2001:173–218. doi: 10.1016/s0083-6729(01)61006-9. [DOI] [PubMed] [Google Scholar]
- 100.Collins MD, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol.Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.da Costa MS, Albuquerque L, Nobre MF, Wait R. The extraction and identification of respiratory lipoquinones of prokaryotes and their use in taxonomy. Meth.Microbiol., Vol 38: Taxonomy of Prokaryotes. 2011;38:197–206. [Google Scholar]
- 102.Brodie AF, Revsin B, Kalra V, Phillips P, Bogin E, Higashi T, Murti CR, Cavari BZ, Marquez E. Biological function of terpenoid quinones. Biochem.Soc.Symp. 1970;29:119–143. [PubMed] [Google Scholar]
- 103.Holsclaw CM, Sogi KM, Gilmore SA, Schelle MW, Leavell MD, Bertozzi CR, Leary JA. Structural characterization of a novel sulfated menaquinone produced by stf3 from Mycobacterium tuberculosis. ACS Chem.Biol. 2008;3:619–624. doi: 10.1021/cb800145r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bentley R. Biosynthesis of vitamin-K and other natural naphthoquinones. Pure and Applied Chemistry. 1975;41:47–68. [Google Scholar]
- 105.Bentley R, Meganathan R. Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol.Rev. 1982;46:241–280. doi: 10.1128/mr.46.3.241-280.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meganathan R. Biosynthesis of vitamin K (menaquinone) and ubiquinone (coenzyme Q) In: Neihardt FC, editor. Escherichia coli and Salmonella. Washington, DC: ASM Press; 1996. pp. 642–656. [Google Scholar]
- 107.Azerad R, Bleiler-Hill R, Lederer E. Biosynthesis of a vitamin K2 by cell-free extracts of Mycobacterium phlei. Biochem.Biophys.Res.Commun. 1965;19:194–197. doi: 10.1016/0006-291x(65)90503-6. [DOI] [PubMed] [Google Scholar]
- 108.Li HJ, Li X, Liu N, Zhang H, Truglio JJ, Mishra S, Kisker C, Garcia-Diaz M, Tonge PJ. Mechanism of the intramolecular Claisen condensation reaction catalyzed by MenB, a crotonase superfamily member. Biochemistry. 2011;50:9532–9544. doi: 10.1021/bi200877x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li X, Liu N, Zhang H, Knudson SE, Li HJ, Lai CT, Simmerling C, Slayden RA, Tonge PJ. CoA adducts of 4-oxo-4-phenylbut-2-enoates: Inhibitors of MenB from the M. tuberculosis menaquinone biosynthesis pathway. ACS Med.Chem.Lett. 2011;2:818–823. doi: 10.1021/ml200141e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li X, Liu N, Zhang H, Knudson SE, Slayden RA, Tonge PJ. Synthesis and SAR studies of 1,4-benzoxazine MenB inhibitors: novel antibacterial agents against Mycobacterium tuberculosis. Bioorg.Med.Chem.Lett. 2010;20:6306–6309. doi: 10.1016/j.bmcl.2010.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu X, Zhou R, Sharma I, Li X, Kumar G, Swaminathan S, Tonge PJ, Tan DS. Stable analogues of OSB-AMP: potent inhibitors of MenE, the o-succinylbenzoate-CoA synthetase from bacterial menaquinone biosynthesis. Chembiochem. 2012;13:129–136. doi: 10.1002/cbic.201100585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lu X, Zhang H, Tonge PJ, Tan DS. Mechanism-based inhibitors of MenE, an acyl-CoA synthetase involved in bacterial menaquinone biosynthesis. Bioorg.Med.Chem.Lett. 2008;18:5963–5966. doi: 10.1016/j.bmcl.2008.07.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Truglio JJ, Theis K, Feng Y, Gajda R, Machutta C, Tonge PJ, Kisker C. Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis. J.Biol.Chem. 2003;278:42352–42360. doi: 10.1074/jbc.M307399200. [DOI] [PubMed] [Google Scholar]
- 114.Dhiman RK, Mahapatra S, Slayden RA, Boyne ME, Lenaerts A, Hinshaw JC, Angala SK, Chatterjee D, Biswas K, Narayanasamy P, Kurosu M, Crick DC. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol.Microbiol. 2009;72:85–97. doi: 10.1111/j.1365-2958.2009.06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Debnath J, Siricilla S, Wan B, Crick DC, Lenaerts AJ, Franzblau SG, Kurosu M. Discovery of selective menaquinone biosynthesis inhibitors against Mycobacterium tuberculosis. J.Med.Chem. 2012;55:3739–3755. doi: 10.1021/jm201608g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Collins MD, Goodfellow M, Minnikin DE, Alderson G. Menaquinone composition of mycolic acid-containing actinomycetes and some sporoactinomycetes. J.Appl.Bacteriol. 1985;58:77–86. doi: 10.1111/j.1365-2672.1985.tb01431.x. [DOI] [PubMed] [Google Scholar]
- 117.Mougous JD, Senaratne RH, Petzold CJ, Jain M, Lee DH, Schelle MW, Leavell MD, Cox JS, Leary JA, Riley LW, Bertozzi CR. A sulfated metabolite produced by stf3 negatively regulates the virulence of Mycobacterium tuberculosis. Proc.Natl.Acad.Sci.U.S A. 2006;103:4258–4263. doi: 10.1073/pnas.0510861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Johnston JB, Kells PM, Podust LM, Ortiz de Montellano PR. Biochemical and structural characterization of CYP124: a methyl-branched lipid omega-hydroxylase from Mycobacterium tuberculosis. Proc.Natl.Acad.Sci.U.S A. 2009;106:20687–20692. doi: 10.1073/pnas.0907398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vershinin A. Biological functions of carotenoids--diversity and evolution. Biofactors. 1999;10:99–104. doi: 10.1002/biof.5520100203. [DOI] [PubMed] [Google Scholar]
- 120.Mathews MM, Krinsky NI. The relationship between carotenoid pigments and resistance to radiation in non-photosynthetic bacteria. Photochem.Photobiol. 1965;4:813–817. doi: 10.1111/j.1751-1097.1965.tb07923.x. [DOI] [PubMed] [Google Scholar]
- 121.Goodwin TW. Carotenoids in fungi and non-photosynthetic bacteria. In: Hockenhull DJD, editor. Progress in Industrial Microbiology. Vol. 11. 1972. pp. 29–88. [PubMed] [Google Scholar]
- 122.Dembitsky VM. Astonishing diversity of natural surfactants: 3. Carotenoid glycosides and isoprenoid glycolipids. Lipids. 2005;40:535–557. doi: 10.1007/s11745-005-1415-z. [DOI] [PubMed] [Google Scholar]
- 123.Subczynsk WK, Markowska E, Sieiewiesiuk J. Effect of polar carotenoids on the oxygen diffusion-concentration product in lipid bilayers. An EPR spin label study. Biochim.Biophys Acta. 1991;1068:68–72. doi: 10.1016/0005-2736(91)90061-c. [DOI] [PubMed] [Google Scholar]
- 124.Woodall AA, Britton G, Jackson MJ. Carotenoids and protection of phospholipids in solution or in liposomes against oxidation by peroxyl radicals: relationship between carotenoid structure and protective ability. Biochim.Biophys Acta. 1997;1336:575–586. doi: 10.1016/s0304-4165(97)00007-x. [DOI] [PubMed] [Google Scholar]
- 125.Woodall AA, Britton G, Jackson MJ. Antioxidant activity of carotenoids in phosphatidylcholine vesicles: chemical and structural considerations. Biochem.Soc.Trans. 1995;23:133S. doi: 10.1042/bst023133s. [DOI] [PubMed] [Google Scholar]
- 126.Hertzberg S, Liaaen JS. Bacterial carotenoids XX. The carotenoids of Mycobacterium phlei strain Vera. 2. The structures of the phlei-xanthophylls--two novel tertiary glucosides. Acta Chem.Scand. 1967;21:15–41. [PubMed] [Google Scholar]
- 127.Sieiro C, Poza M, de MT, Villa TG. Genetic basis of microbial carotenogenesis. Int.Microbiol. 2003;6:11–16. doi: 10.1007/s10123-003-0097-0. [DOI] [PubMed] [Google Scholar]
- 128.Gao LY, Groger R, Cox JS, Beverley SM, Lawson EH, Brown EJ. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect.Immun. 2003;71:922–929. doi: 10.1128/IAI.71.2.922-929.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ramakrishnan L, Tran HT, Federspiel NA, Falkow S. A crtB homolog essential for photochromogenicity in Mycobacterium marinum: isolation, characterization, and gene disruption via homologous recombination. J.Bacteriol. 1997;179:5862–5868. doi: 10.1128/jb.179.18.5862-5868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Houssaini-Iraqui M, Lazraq MH, Clavel-Seres S, Rastogi N, David HL. Cloning and expression of Mycobacterium aurum carotenogenesis genes in Mycobacterium smegmatis. FEMS Microbiol.Lett. 1992;69:239–244. doi: 10.1016/0378-1097(92)90653-6. [DOI] [PubMed] [Google Scholar]
- 131.Viveiros M, Krubasik P, Sandmann G, Houssaini-Iraqui M. Structural and functional analysis of the gene cluster encoding carotenoid biosynthesis in Mycobacterium aurum A+ FEMS Microbiol.Lett. 2000;187:95–101. doi: 10.1111/j.1574-6968.2000.tb09143.x. [DOI] [PubMed] [Google Scholar]
- 132.Scherzinger D, Scheffer E, Bar C, Ernst H, Al-Babili S. The Mycobacterium tuberculosis ORF Rv0654 encodes a carotenoid oxygenase mediating central and excentric cleavage of conventional and aromatic carotenoids. FEBS J. 2010;277:4662–4673. doi: 10.1111/j.1742-4658.2010.07873.x. [DOI] [PubMed] [Google Scholar]
- 133.Provvedi R, Kocincova D, Dona V, Euphrasie D, Daffe M, Etienne G, Manganelli R, Reyrat JM. SigF controls carotenoid pigment production and affects transformation efficiency and hydrogen peroxide sensitivity in Mycobacterium smegmatis. J.Bacteriol. 2008;190:7859–7863. doi: 10.1128/JB.00714-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sato T, Kigawa A, Takagi R, Adachi T, Hoshino T. Biosynthesis of a novel cyclic C35-terpene via the cyclisation of a Z-type C35-polyprenyl diphosphate obtained from a nonpathogenic Mycobacterium species. Org.Biomol.Chem. 2008;6:3788–3794. doi: 10.1039/b808513g. [DOI] [PubMed] [Google Scholar]
- 135.Mann FM, Xu M, Chen X, Fulton DB, Russell DG, Peters RJ. Edaxadiene: a new bioactive diterpene from Mycobacterium tuberculosis. J.Am.Chem.Soc. 2009;131:17526–17527. doi: 10.1021/ja9019287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hoshino T, Nakano C, Ootsuka T, Shinohara Y, Hara T. Substrate specificity of Rv3378c, an enzyme from Mycobacterium tuberculosis and the inhibitory activity of the bicyclic diterpenoids against macrophage phagocytosis. Org.Biomol.Chem. 2011;9:2156–2165. doi: 10.1039/c0ob00884b. [DOI] [PubMed] [Google Scholar]
- 137.Pethe K, Swenson DL, Alonso S, Anderson J, Wang C, Russell DG. Isolation of Mycobacterium tuberculosis mutants defective in the arrest of phagosome maturation. Proc.Natl.Acad.Sci.U.S A. 2004;101:13642–13647. doi: 10.1073/pnas.0401657101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nakano C, Okamura T, Sato T, Dairi T, Hoshino T. Mycobacterium tuberculosis H37Rv3377c encodes the diterpene cyclase for producing the halimane skeleton. Chem.Commun.(Camb.) 2005:1016–1018. doi: 10.1039/b415346d. [DOI] [PubMed] [Google Scholar]
- 139.Maugel N, Mann FM, Hillwig ML, Peters RJ, Snider BB. Synthesis of (+/−)-nosyberkol (isotuberculosinol, revised structure of edaxadiene) and (+/−)-tuberculosinol. Org.Lett. 2010;12:2626–2629. doi: 10.1021/ol100832h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Spangler JE, Carson CA, Sorensen EJ. Synthesis enables a structural revision of the Mycobacterium tuberculosis-produced diterpene, edaxadiene. Chem.Sci. 2010;1:202–205. doi: 10.1039/C0SC00284D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jackson M, Stadthagen G, Gicquel B. Long-chain multiple methyl-branched fatty acid-containing lipids of Mycobacterium tuberculosis: biosynthesis, transport, regulation and biological activities. Tuberculosis. 2007;87:78–86. doi: 10.1016/j.tube.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 142.Cardona P-J, Soto CY, Martin C, Gicquel B, Agusti G, Guirado E, Sirakova TD, Kolattukudy PE, Julian E, Luquin M. Neutral red reaction is related to virulence and cell wall methyl-branched lipids in Mycobacterium tuberculosis. Microbes and Infection. 2006;8:183–190. doi: 10.1016/j.micinf.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 143.Gonzalo Asensio J, Maia C, Ferrer NL, Barilone N, Laval F, Soto CY, Winter N, Daffe M, Gicquel B, Martin C, Jackson M. The virulence-associated two-component PhoP-PhoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. The Journal of biological chemistry. 2006;281:1313–1316. doi: 10.1074/jbc.C500388200. [DOI] [PubMed] [Google Scholar]
- 144.Dubos RJ, Middlebrook G. Cytochemical reaction of virulent tubercle bacilli. Amer Rev. Tuberc. 1948;58:698–699. doi: 10.1164/art.1948.58.6.698. [DOI] [PubMed] [Google Scholar]
- 145.Grzegorzewicz AE, Pham H, Gundi VA, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SE, Korduláková J, Chavadi SS, Morisseau C, Lenaerts AJ, Lee RE, McNeil MR, Jackson M. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nature chemical biology. 2012;8:334–341. doi: 10.1038/nchembio.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tahlan K, Wilson R, Kastrinsky DB, Arora K, Nair V, Fischer E, Barnes SW, Walker JR, Alland D, Barry CE, 3rd, Boshoff HI. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrobial agents and chemotherapy. 2012;56:1797–1809. doi: 10.1128/AAC.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS. Role of the major antigen of Mycobacterium tuberculosis in the cell wall biogenesis. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 148.Jackson M, Raynaud C, Lanéelle MA, Guilhot C, Laurent-Winter C, Ensergueix D, Gicquel B, Daffé M. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Molecular microbiology. 1999;31:1573–1587. doi: 10.1046/j.1365-2958.1999.01310.x. [DOI] [PubMed] [Google Scholar]
- 149.Puech V, Guilhot C, Perez E, Tropis M, Armitige LY, Gicquel B, Daffe M. Evidence for a partial redundancy of the fibronectin-binding proteins for the transfer of mycoloyl residues onto the cell wall arabinogalactan termini of Mycobacterium tuberculosis. Molecular microbiology. 2002;44:1109–1122. doi: 10.1046/j.1365-2958.2002.02953.x. [DOI] [PubMed] [Google Scholar]
- 150.Harth G, Horwitz MA, Tabatadze D, Zamecnik PC. Targeting the Mycobacterium tuberculosis 30/32-kDa mycolyl transferase complex as a therapeutic strategy against tuberculosis: Proof of principle by using antisense technology. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15614–15619. doi: 10.1073/pnas.242612299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Armitige LY, Jagannath C, Wanger AR, Norris SJ. Disruption of the genes encoding antigen 85A and antigen 85B of Mycobacterium tuberculosis H37Rv: effect on growth in culture and in macrophages. Infect. Immun. 2000;68:767–678. doi: 10.1128/iai.68.2.767-778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nguyen L, Chinnapapagari S, Thompson CJ. FbpA-Dependent biosynthesis of trehalose dimycolate is required for the intrinsic multidrug resistance, cell wall structure, and colonial morphology of Mycobacterium smegmatis. Journal of bacteriology. 2005;187:6603–6611. doi: 10.1128/JB.187.19.6603-6611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Katti MK, Dai G, Armitige LY, Rivera Marrero C, Daniel S, Singh CR, Lindsey DR, Dhandayuthapani S, Hunter RL, Jagannath C. The Delta fbpA mutant derived from Mycobacterium tuberculosis H37Rv has an enhanced susceptibility to intracellular antimicrobial oxidative mechanisms, undergoes limited phagosome maturation and activates macrophages and dendritic cells. Cellular microbiology. 2008;10:1286–1303. doi: 10.1111/j.1462-5822.2008.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hunter RL, Armitige L, Jagannath C, Actor JK. TB research at UT-Houston--a review of cord factor: new approaches to drugs, vaccines and the pathogenesis of tuberculosis. Tuberculosis (Edinb) 2009;89(Suppl 1):S18–S25. doi: 10.1016/S1472-9792(09)70007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li C, Du Q, Deng W, Xie J. The biology of Mycobacterium cord factor and roles in pathogen-host interaction. Critical reviews in eukaryotic gene expression. 2012;22:289–297. doi: 10.1615/critreveukargeneexpr.v22.i4.30. [DOI] [PubMed] [Google Scholar]
- 156.Linares C, Bernabeu A, Luquin M, Valero-Guillen PL. Cord factors from atypical mycobacteria (Mycobacterium alvei, Mycobacterium brumae) stimulate the secretion of some pro-inflammatory cytokines of relevance in tuberculosis. Microbiology. 2012;158:2878–2885. doi: 10.1099/mic.0.060681-0. [DOI] [PubMed] [Google Scholar]
- 157.Glickman MS. Cording, cord factors and trehalose dimycolate. In: Daffé M, Reyrat J-M, editors. The mycobacterial cell envelope. Washington DC: ASM Press; 2008. pp. 63–73. [Google Scholar]
- 158.Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, Yamasaki S. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. The Journal of experimental medicine. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sakamoto K, Kim MJ, Rhoades ER, Allavena RE, Ehrt S, Wainwright HC, Russell DG, Rohde KH. Mycobacterial trehalose dimycolate reprograms macrophage global gene expression and activates matrix metalloproteinases. Infection and immunity. 2013;81:764–776. doi: 10.1128/IAI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lang R. Recognition of the mycobacterial cord factor by Mincle: relevance for granuloma formation and resistance to tuberculosis. Frontiers in immunology. 2013;4:5. doi: 10.3389/fimmu.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rao V, Fujiwara N, Porcelli SA, Glickman MS. Mycobacterium tuberculosis controls host innate immune activation through cyclopropane modification of a glycolipid effector molecule. J. Exp. Med. 2005;201:535–543. doi: 10.1084/jem.20041668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dubey VS, Sirakova TD, Cynamon MH, Kolattukudy PE. Biochemical function of msl5 (pks8 plus pks17) in Mycobacterium tuberculosis H37Rv: Biosynthesis of monomethyl branched unsaturated fatty acids. J. Bacteriol. 2003;185:4620–4625. doi: 10.1128/JB.185.15.4620-4625.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Goren MB. Mycobacterial fatty acid esters of sugars and sulfosugars. In: Kates M, editor. Handbook of Lipid Research. Glycolipids, phosphoglycolipids and sulfoglycolipids. Vol. 6. New York, London: Plenum Press; 1990. pp. 363–461. [Google Scholar]
- 164.Mougous JD, Petzold CJ, Seraratne RH, Lee DH, Akey DL, Lin FL, Munchel SE, Pratt MR, Riley LW, Leary JA, Berger JM, Bertozzi CR. Identification, function and structure of the mycobacterial sulfotransferase that initiates sulfolipid-1 biosynthesis. Nature structural & molecular biology. 2004;11:721–729. doi: 10.1038/nsmb802. [DOI] [PubMed] [Google Scholar]
- 165.Kumar P, Schelle MW, Jain M, Lin FL, Petzold CJ, Leavell MD, Leary JA, Cox JS, Bertozzi CR. PapA1 and PapA2 are acyltransferases essential for the biosynthesis of the Mycobacterium tuberculosis virulence factor sulfolipid-1. Proc. Natl. Acad. Sci. USA. 2007;104:11221–11226. doi: 10.1073/pnas.0611649104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sirakova TD, Thirumala AK, Dubey VS, Sprecher H, Kolattukudy PE. The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl branched fatty acids required for sulfolipid synthesis. J. Biol. Chem. 2001;276:16833–16839. doi: 10.1074/jbc.M011468200. [DOI] [PubMed] [Google Scholar]
- 167.Trivedi OA, Arora P, Sridharan V, Tickoo R, Mohanty D, Gokhale RS. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- 168.Seeliger JC, Holsclaw CM, Schelle MW, Botyanszki Z, Gilmore SA, Tully SE, Niederweis M, Cravatt BF, Leary JA, Bertozzi CR. Elucidation and chemical modulation of sulfolipid-1 biosynthesis in Mycobacterium tuberculosis. The Journal of biological chemistry. 2012;287:7990–8000. doi: 10.1074/jbc.M111.315473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bhatt K, Gurcha SS, Bhatt A, Besra GS, Jacobs WR., Jr Two polyketide-synthase-associated acyltransferases are required for sulfolipid biosynthesis in Mycobacterium tuberculosis. Microbiology. 2007;153:513–520. doi: 10.1099/mic.0.2006/003103-0. [DOI] [PubMed] [Google Scholar]
- 170.Converse SE, Mougous JD, Leavell MD, Leary JA, Bertozzi CR, Cox JS. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc. Natl. Acad. Sci. USA. 2003;100:6121–6126. doi: 10.1073/pnas.1030024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Domenech P, Reed MB, Dowd CS, Manca C, Kaplan G, Barry CE., III The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J. Biol. Chem. 2004;279:21257–21265. doi: 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- 172.Zheng J, Wei C, Zhao L, Liu L, Leng W, Li W, Jin Q. Combining blue native polyacrylamide gel electrophoresis with liquid chromatography tandem mass spectrometry as an effective strategy for analyzing potential membrane protein complexes of Mycobacterium bovis bacillus Calmette-Guerin. BMC genomics. 2011;12:40. doi: 10.1186/1471-2164-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Graham JE, Clark-Curtiss JE. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS) Proc. Natl. Acad. Sci. USA. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJ. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS pathogens. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lee W, Vanderven BC, Fahey RJ, Russell DG. Intracellular Mycobacterium tuberculosis Exploits Host-derived Fatty Acids to Limit Metabolic Stress. The Journal of biological chemistry. 2013;288:6788–6800. doi: 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffé M, Smith I. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 2006;60:312–330. doi: 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 177.Chesne-Seck M-L, Barilone N, Boudou F, Gonzalo Asensio J, Kolattukudy PE, Martin C, Cole ST, Gicquel B, Gopaul DN, Jackson M. A point mutation in the two-component regulator PhoP-PhoR accounts for the absence of polyketide-derived acyltrehaloses but not that of phthiocerol dimycocerosates in Mycobacterium tuberculosis H37Ra. J. Bact. 2008;190:1329–1334. doi: 10.1128/JB.01465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Goyal R, Das AK, Singh R, Singh PK, Korpole S, Sarkar D. Phosphorylation of PhoP protein plays direct regulatory role in lipid biosynthesis of Mycobacterium tuberculosis. The Journal of biological chemistry. 2011;286:45197–45208. doi: 10.1074/jbc.M111.307447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Cimino M, Thomas C, Namouchi A, Dubrac S, Gicquel B, Gopaul DN. Identification of DNA binding motifs of the Mycobacterium tuberculosis PhoP/PhoR two-component signal transduction system. PloS one. 2012;7:e42876. doi: 10.1371/journal.pone.0042876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Goren MB, Brennan PJ. Mycobacterial lipids: chemistry and biologic activities. In: Youmans GP, editor. Tuberculosis. Philadelphia, London, Toronto: W. B. Saunders Company; 1979. pp. 63–193. [Google Scholar]
- 181.Bertozzi CR, Schelle MW. Sulfated metabolites from Mycobacterium tuberculosis: Sulfolipid-1 and beyond. In: Daffé M, Reyrat J-M, editors. The mycobacterial cell envelope. Washington DC: ASM Press; 2008. pp. 291–304. [Google Scholar]
- 182.Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Böhmer G, Prandi J, Mori L, Puzo G, De Libero G. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J. Exp. Med. 2004;199:649–659. doi: 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Guiard J, Collmann A, Garcia-Alles LF, Mourey L, Brando T, Mori L, Gilleron M, Prandi J, De Libero G, Puzo G. Fatty acyl structures of Mycobacterium tuberculosis sulfoglycolipid govern T cell response. J Immunol. 2009;182:7030–7037. doi: 10.4049/jimmunol.0804044. [DOI] [PubMed] [Google Scholar]
- 184.Rousseau C, Turner OC, Rush E, Bordat Y, Sirakova TD, Kolattukudy PE, Ritter S, Orme IM, Gicquel B, Jackson M. Sulfolipid deficiency does not affect the virulence of Mycobacterium tuberculosis H37Rv in mice and guinea pigs. Infect. Immun. 2003a;71:4684–4690. doi: 10.1128/IAI.71.8.4684-4690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lamichhane G, Tyagi S, Bishai WR. Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infection and immunity. 2005;73:2533–2540. doi: 10.1128/IAI.73.4.2533-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Domenech P, Reed MB, Barry CE., III Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 2005;73:3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gilmore SA, Schelle MW, Holsclaw CM, Leigh CD, Jain M, Cox JS, Leary JA, Bertozzi CR. Sulfolipid-1 biosynthesis restricts Mycobacterium tuberculosis growth in human macrophages. ACS chemical biology. 2012;7:863–870. doi: 10.1021/cb200311s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Minnikin DE, Dobson G, Sesardic D, Ridell M. Mycolipenates and mycolipanolates of trehalose from Mycobacterium tuberculosis. J. Gen. Microbiol. 1985;131:1369–1374. doi: 10.1099/00221287-131-6-1369. [DOI] [PubMed] [Google Scholar]
- 189.Lemassu A, Lanéelle M-A, Daffé M. Revised structure of a trehalose-containing immunoreactive glycolipid of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 1991;78:171–176. doi: 10.1016/0378-1097(91)90153-2. [DOI] [PubMed] [Google Scholar]
- 190.Besra GS, Bolton R, McNeil MR, Ridell M, Simpson KE, Glushka J, van Halbeek H, Brennan PJ, Minnikin DE. Structure elucidation and antigenicity of a novel family of glycolipid antigens from Mycobacterium tuberculosis H37Rv. Biochemistry. 1992;31:9832–9837. doi: 10.1021/bi00155a040. [DOI] [PubMed] [Google Scholar]
- 191.Munoz M, Lanéelle M-A, Luquin M, Torrelles J, Julian E, Ausina V, Daffé M. Occurence of an antigenic triacyl trehalose in clinical isolates and reference strains of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 1997;157:251–259. doi: 10.1111/j.1574-6968.1997.tb12781.x. [DOI] [PubMed] [Google Scholar]
- 192.Daffé M, Lacave C, Lanéelle M-A, Gillois M, Lanéelle G. Polyphthienoyl trehalose, glycolipids specific for virulent strains of the tubercle bacillus. Eur. J. Biochem. 1988a;172:579–584. doi: 10.1111/j.1432-1033.1988.tb13928.x. [DOI] [PubMed] [Google Scholar]
- 193.Gautier N, Lopez Marin LM, Lanéelle M-A, Daffé M. Structure of mycoside F, a family of trehalose-containing glycolipids of Mycobacterium fortuitum. FEMS Microbiol. Lett. 1992;98:81–88. doi: 10.1016/0378-1097(92)90136-c. [DOI] [PubMed] [Google Scholar]
- 194.Ariza MA, Martin-Luengo F, Valero-Guillen PL. A family of diacyltrehaloses isolated from Mycobacterium fortuitum. Microbiology. 1994;140:1989–1994. doi: 10.1099/13500872-140-8-1989. [DOI] [PubMed] [Google Scholar]
- 195.Lee KS, Dubey VS, Kolattukudy PE, Song CH, Shin AR, Jung SB, Yang CS, Kim SY, Jo EK, Park JK, Kim HJ. Diacyltrehalose of Mycobacterium tuberculosis inhibits lipopolysaccharide- and mycobacteria-induced proinflammatory cytokine production in human monocytic cells. FEMS microbiology letters. 2007;267:121–128. doi: 10.1111/j.1574-6968.2006.00553.x. [DOI] [PubMed] [Google Scholar]
- 196.Dubey VS, Sirakova TD, Kolattukudy PE. Disruption of msl3 abolishes the synthesis of mycolipanoic and mycolipenic acids required for polyacyltrehalose synthesis in Mycobacterium tuberculosis H37Rv and causes cell aggregation. Mol. Microbiol. 2002;45:1451–1459. doi: 10.1046/j.1365-2958.2002.03119.x. [DOI] [PubMed] [Google Scholar]
- 197.Rousseau C, Neyrolles O, Bordat Y, Giroux S, Sirakova TD, Prevost M-C, Kolattukudy PE, Gicquel B, Jackson M. Deficiency in mycolipenate- and mycosanoate-derived acyltrehaloses enhances early interactions of Mycobacterium tuberculosis with host cells. Cell. Microbiol. 2003b;5:405–415. doi: 10.1046/j.1462-5822.2003.00289.x. [DOI] [PubMed] [Google Scholar]
- 198.Lynett J, Stokes RW. Selection of transposon mutants of Mycobacterium tuberculosis with increased macrophage infectivity identifies fadD23 to be involved in sulfolipid production and association with macrophages. Microbiology. 2007;153:3133–3140. doi: 10.1099/mic.0.2007/007864-0. [DOI] [PubMed] [Google Scholar]
- 199.Hatzios SK, Schelle MW, Holsclaw CM, Behrens CR, Botyanszki Z, Lin FL, Carlson BL, Kumar P, Leary JA, Bertozzi CR. PapA3 is an acyltransferase required for polyacyltrehalose biosynthesis in Mycobacterium tuberculosis. The Journal of biological chemistry. 2009;284:12745–12751. doi: 10.1074/jbc.M809088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Daffé M, Lemassu A. Glycobiology of the mycobacterial surface. Structures and biological activities of the cell envelope glycoconjugates. In: Doyle RJ, editor. Glycomicrobiology. New York, NY, USA: Kluwer Academic/Plenum Publishers; 2000. [Google Scholar]
- 201.Hunter SW, Murphy RC, Clay K, Goren MB, Brennan PJ. Trehalose-containing lipooligosaccharides. A new class of species-specific antigens from Mycobacterium. The Journal of biological chemistry. 1983;258:10481–10487. [PubMed] [Google Scholar]
- 202.Saadat S, Ballou CE. Pyruvylated glycolipids from Mycobacterium smegmatis. Structures of two oligosaccharide components. The Journal of biological chemistry. 1983;258:1813–1818. [PubMed] [Google Scholar]
- 203.Daffé M, McNeil MR, Brennan PJ. Novel type-specific lipooligosaccharides from Mycobacterium tuberculosis. Biochemistry. 1991a;30:378–388. doi: 10.1021/bi00216a011. [DOI] [PubMed] [Google Scholar]
- 204.Burguiere A, Hitchen P, Dover LG, Kremer L, Ridell M, Alexander DC, Liu J, Morris HR, Minnikin DE, Dell A, Besra GS. LosA, a key glycosyltransferase involved in the biosynthesis of a novel family of glycosylated acyltrehalose lipooligosaccharides from Mycobacterium marinum. J. Biol. Chem. 2005;280:42124–42133. doi: 10.1074/jbc.M507500200. [DOI] [PubMed] [Google Scholar]
- 205.Ren H, Dover LG, Islam ST, Alexander DC, Chen JM, Besra GS, Liu J. Identification of the lipooligosaccharide biosynthetic gene cluster from Mycobacterium marinum. Mol. Microbiol. 2007;63:1345–1359. doi: 10.1111/j.1365-2958.2007.05603.x. [DOI] [PubMed] [Google Scholar]
- 206.Rombouts Y, Alibaud L, Carrere-Kremer S, Maes E, Tokarski C, Elass E, Kremer L, Guerardel Y. Fatty acyl chains of Mycobacterium marinum lipooligosaccharides: structure, localization and acylation by PapA4 (MMAR_2343) protein. The Journal of biological chemistry. 2011;286:33678–33688. doi: 10.1074/jbc.M111.273920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Etienne G, Malaga W, Laval F, Lemassu A, Guilhot C, Daffe M. Identification of the polyketide synthase involved in the biosynthesis of the surface-exposed lipooligosaccharides in mycobacteria. Journal of bacteriology. 2009;191:2613–2621. doi: 10.1128/JB.01235-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Besra GS, Khoo KH, Belisle JT, McNeil MR, Morris HR, Dell A, Brennan PJ. New pyruvylated, glycosylated acyltrehaloses from Mycobacterium smegmatis strains, and their implications for phage resistance in mycobacteria. Carbohydrate research. 1994;251:99–114. doi: 10.1016/0008-6215(94)84279-5. [DOI] [PubMed] [Google Scholar]
- 209.Sarkar D, Sidhu M, Singh A, Chen J, Lammas DA, van der Sar AM, Besra GS, Bhatt A. Identification of a glycosyltransferase from Mycobacterium marinum involved in addition of a caryophyllose moiety in lipooligosaccharides. Journal of bacteriology. 2011;193:2336–2340. doi: 10.1128/JB.00065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sonden B, Kocincova D, Deshayes C, Euphrasie D, Rhayat L, Laval F, Frehel C, Daffe M, Etienne G, Reyrat J-M. Gap, a mycobacterial specific integral membrane protein, is required for glycolipid transport to the cell surface. Mol. Microbiol. 2005;58:426–440. doi: 10.1111/j.1365-2958.2005.04847.x. [DOI] [PubMed] [Google Scholar]
- 211.Ripoll F, Deshayes C, Pasek S, Laval F, Beretti JL, Biet F, Risler JL, Daffe M, Etienne G, Gaillard JL, Reyrat JM. Genomics of glycopeptidolipid biosynthesis in Mycobacterium abscessus and M. chelonae. BMC genomics. 2007;8:114. doi: 10.1186/1471-2164-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Deshayes C, Kocinkova D, Etienne G, Reyrat J-M. Glycopeptidolipids: a complex pathway for small pleitotropic molecules. In: Daffé M, Reyrat J-M, editors. The mycobacterial cell envelope. Washington DC, USA: ASM Press; 2008. [Google Scholar]
- 213.van der Woude AD, Sarkar D, Bhatt A, Sparrius M, Raadsen SA, Boon L, Geurtsen J, van der Sar AM, Luirink J, Houben EN, Besra GS, Bitter W. Unexpected link between lipooligosaccharide biosynthesis and surface protein release in Mycobacterium marinum. The Journal of biological chemistry. 2012;287:20417–20429. doi: 10.1074/jbc.M111.336461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Belisle JT, Brennan PJ. Chemical basis of rough and smooth variation in mycobacteria. Journal of bacteriology. 1989;171:3465–3470. doi: 10.1128/jb.171.6.3465-3470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lemassu A, Levy-Frebault VV, Laneelle MA, Daffe M. Lack of correlation between colony morphology and lipooligosaccharide content in the Mycobacterium tuberculosis complex. Journal of general microbiology. 1992;138:1535–1541. doi: 10.1099/00221287-138-7-1535. [DOI] [PubMed] [Google Scholar]
- 216.Rombouts Y, Burguiere A, Maes E, Coddeville B, Elass E, Guerardel Y, Kremer L. Mycobacterium marinum lipooligosaccharides are unique caryophyllose-containing cell wall glycolipids that inhibit tumor necrosis factor-alpha secretion in macrophages. The Journal of biological chemistry. 2009;284:20975–20988. doi: 10.1074/jbc.M109.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Moody DB, Ulrichs T, Muhlecker W, Young DC, Gurcha SS, Grant E, Rosat JP, Brenner MB, Costello CE, Besra GS, Porcelli SA. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 218.Matsunaga I, Bhatt A, Young DC, Cheng T-Y, Eyles SJ, Besra GS, Briken V, Porcelli SA, Costello CE, Jacobs WR, Jr, Moody DB. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. The Journal of experimental medicine. 2004;200:1559–1569. doi: 10.1084/jem.20041429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Matsunaga I, Sugita M. Mycoketide: a CD1c-presented antigen with important implications in mycobacterial infection. Clinical & developmental immunology. 2012;2012:981821. doi: 10.1155/2012/981821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Chopra T, Banerjee S, Gupta S, Yadav G, Anand S, Surolia A, Roy RP, Mohanty D, Gokhale RS. Novel intermolecular iterative mechanism for biosynthesis of mycoketide catalyzed by a bimodular polyketide synthase. PLoS Biology. 2008;6:1584–1598. doi: 10.1371/journal.pbio.0060163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Daffé M, Lanéelle M-A. Distribution of phthiocerol diester, phenolic mycosides and related compounds in mycobacteria. J. Gen. Microbiol. 1988b;134:2049–2055. doi: 10.1099/00221287-134-7-2049. [DOI] [PubMed] [Google Scholar]
- 222.Daffé M, Lacave C, Laneelle MA, Laneelle G. Structure of the major triglycosyl phenol-phthiocerol of Mycobacterium tuberculosis (strain Canetti) European journal of biochemistry/FEBS. 1987;167:155–160. doi: 10.1111/j.1432-1033.1987.tb13317.x. [DOI] [PubMed] [Google Scholar]
- 223.Constant P, Pérez E, Malaga W, Lanéelle M-A, Saurel O, Daffé M, Guilhot C. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex. J. Biol. Chem. 2002;277:38148–38158. doi: 10.1074/jbc.M206538200. [DOI] [PubMed] [Google Scholar]
- 224.Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, Kaplan G, Barry CE., III A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 225.Huet G, Constant P, Malaga W, Laneelle MA, Kremer K, van Soolingen D, Daffe M, Guilhot C. A lipid profile typifies the Beijing strains of Mycobacterium tuberculosis: identification of a mutation responsible for a modification of the structures of phthiocerol dimycocerosates and phenolic glycolipids. The Journal of biological chemistry. 2009;284:27101–27113. doi: 10.1074/jbc.M109.041939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Malaga W, Constant P, Euphrasie D, Cataldi A, Daffé M, Reyrat J-M, Guilhot C. Deciphering the genetic bases of the structural diversity of phenolic glycolipids in strains of the Mycobacterium tuberculosis complex. J. Biol. Chem. 2008;283:15177–15184. doi: 10.1074/jbc.M710275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Goren MB, Brokl O, Schaefer WB. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: Phthiocerol dimycocerosate and the attenuation indicator lipid. Infect. Immun. 1974;9:150–158. doi: 10.1128/iai.9.1.150-158.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Krishnan N, Malaga W, Constant P, Caws M, Tran TH, Salmons J, Nguyen TN, Nguyen DB, Daffe M, Young DB, Robertson BD, Guilhot C, Thwaites GE. Mycobacterium tuberculosis lineage influences innate immune response and virulence and is associated with distinct cell envelope lipid profiles. PloS one. 2011;6:e23870. doi: 10.1371/journal.pone.0023870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Camacho LR, Constant P, Raynaud C, Lanéelle M-A, Triccas JA, Gicquel B, Daffé M, Guilhot C. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 2001;276:19845–19854. doi: 10.1074/jbc.M100662200. [DOI] [PubMed] [Google Scholar]
- 230.Waddell SJ, Chung GA, Gibson KJ, Everett MJ, Minnikin DE, Besra GS, Butcher PD. Inactivation of polyketide synthase and related genes results in the loss of complex lipids in Mycobacterium tuberculosis H37Rv. Letters in applied microbiology. 2005;40:201–206. doi: 10.1111/j.1472-765X.2005.01659.x. [DOI] [PubMed] [Google Scholar]
- 231.Sirakova TD, Dubey VS, Kim H-J, Cynamon MH, Kolattukudy PE. The largest open reading frame (pks12) in the Mycobacterium tuberculosis genome is involved in pathogenesis and dimycocerosyl phthiocerol synthesis. Infect. Immun. 2003a;71:3794–3801. doi: 10.1128/IAI.71.7.3794-3801.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sirakova TD, Dubey VS, Cynamon MH, Kolattukudy PE. Attenuation of Mycobacterium tuberculosis by disruption of a mas-like gene or a chalcone synthase-like gene, which causes deficiency in dimycocerosyl phthiocerol synthesis. J. Bacteriol. 2003b;185:2999–3008. doi: 10.1128/JB.185.10.2999-3008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hotter GS, Wards BJ, Mouat P, Besra GS, Gomes J, Singh M, Bassett S, Kawakami P, Wheeler PR, de Lisle GW, Collins DM. Transposon mutagenesis of Mb0100 at the ppe1-nrp locus in Mycobacterium bovis disrupts phthiocerol dimycocerosate (PDIM) and glycosylphenol-PDIM biosynthesis, producing an avirulent strain with vaccine properties at least equal to those of M. bovis BCG. Journal of bacteriology. 2005;187:2267–2277. doi: 10.1128/JB.187.7.2267-2277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rousseau C, Sirakova TD, Dubey VS, Bordat Y, Kolattukudy PE, Gicquel B, Jackson M. Virulence attenuation of two Mas-like polyketide synthase mutants of Mycobacterium tuberculosis. Microbiology. 2003c;149:1837–1847. doi: 10.1099/mic.0.26278-0. [DOI] [PubMed] [Google Scholar]
- 235.Cox JS, Chen B, McNeil M, Jacobs WR., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 236.Simeone R, Leger M, Constant P, Malaga W, Marrakchi H, Daffe M, Guilhot C, Chalut C. Delineation of the roles of FadD22, FadD26 and FadD29 in the biosynthesis of phthiocerol dimycocerosates and related compounds in Mycobacterium tuberculosis. The FEBS journal. 2010;277:2715–2725. doi: 10.1111/j.1742-464X.2010.07688.x. [DOI] [PubMed] [Google Scholar]
- 237.Stadthagen G, Kordulakova J, Griffin R, Constant P, Bottova I, Barilone N, Gicquel B, Daffe M, Jackson M. p-Hydroxybenzoic acid synthesis in Mycobacterium tuberculosis. The Journal of biological chemistry. 2005;280:40699–40706. doi: 10.1074/jbc.M508332200. [DOI] [PubMed] [Google Scholar]
- 238.Chavadi SS, Edupuganti UR, Vergnolle O, Fatima I, Singh SM, Soll CE, Quadri LE. Inactivation of tesA reduces cell wall lipid production and increases drug susceptibility in mycobacteria. The Journal of biological chemistry. 2011;286:24616–24625. doi: 10.1074/jbc.M111.247601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Rao A, Ranganathan A. Interaction studies on proteins encoded by the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Molecular genetics and genomics : MGG. 2004;272:571–579. doi: 10.1007/s00438-004-1088-3. [DOI] [PubMed] [Google Scholar]
- 240.Rainwater DL, Kolattukudy PE. Synthesis of mycocerosic acids from methylmalonyl coenzyme A by cell-free extracts of Mycobacterium tuberculosis var. bovis BCG. The Journal of biological chemistry. 1983;258:2979–2985. [PubMed] [Google Scholar]
- 241.Rainwater DL, Kolattukudy PE. Fatty acid biosynthesis in Mycobacterium tuberculosis var. bovis Bacillus Calmette-Guerin. Purification and characterization of a novel fatty acid synthase, mycocerosic acid synthase, which elongates n-fatty acyl-CoA with methylmalonyl-CoA. The Journal of biological chemistry. 1985;260:616–623. [PubMed] [Google Scholar]
- 242.Mathur M, Kolattukudy PE. Molecular cloning and sequencing of the gene for mycocerosic acid synthase, a novel fatty acid elongating multifunctional enzyme, from Mycobacterium tuberculosis var. bovis Bacillus Calmette-Guerin. The Journal of biological chemistry. 1992;267:19388–19395. [PubMed] [Google Scholar]
- 243.Azad AK, Sirakova TD, Rogers LM, Kolattukudy PE. Targeted replacement of the mycocerosic acid synthase gene in Mycobacterium bovis BCG produces a mutant that lacks mycosides. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:4787–4792. doi: 10.1073/pnas.93.10.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Trivedi OA, Arora P, Vats A, Ansari MZ, Tickoo R, Sridharan V, Mohanty D, Gokhale RS. Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Molecular cell. 2005;17:631–643. doi: 10.1016/j.molcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 245.Fitzmaurice AM, Kolattukudy PE. Open reading frame 3, which is adjacent to the mycocerosic acid synthase gene, is expressed as an acyl coenzyme A synthase in Mycobacterium bovis BCG. Journal of bacteriology. 1997;179:2608–2615. doi: 10.1128/jb.179.8.2608-2615.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Fitzmaurice AM, Kolattukudy PE. An acyl-CoA synthase (acoas) gene adjacent to the mycocerosic acid synthase (mas) locus is necessary for mycocerosyl lipid synthesis in Mycobacterium tuberculosis var. bovis BCG. The Journal of biological chemistry. 1998;273:8033–8039. doi: 10.1074/jbc.273.14.8033. [DOI] [PubMed] [Google Scholar]
- 247.Perez E, Constant P, Laval F, Lemassu A, Laneelle MA, Daffe M, Guilhot C. Molecular dissection of the role of two methyltransferases in the biosynthesis of phenolglycolipids and phthiocerol dimycoserosate in the Mycobacterium tuberculosis complex. The Journal of biological chemistry. 2004a;279:42584–42592. doi: 10.1074/jbc.M406134200. [DOI] [PubMed] [Google Scholar]
- 248.Perez E, Constant P, Lemassu A, Laval F, Daffe M, Guilhot C. Characterization of three glycosyltransferases involved in the biosynthesis of the phenolic glycolipid antigens from the Mycobacterium tuberculosis complex. The Journal of biological chemistry. 2004b;279:42574–42583. doi: 10.1074/jbc.M406246200. [DOI] [PubMed] [Google Scholar]
- 249.Simeone R, Huet G, Constant P, Malaga W, Lemassu A, Laval F, Daffe M, Guilhot C, Chalut C. Functional characterisation of three O-methyltransferases involved in the biosynthesis of phenolglycolipids in Mycobacterium tuberculosis. PloS one. 2013;8:e58954. doi: 10.1371/journal.pone.0058954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tabouret G, Astarie-Dequeker C, Demangel C, Malaga W, Constant P, Ray A, Honore N, Bello NF, Perez E, Daffe M, Guilhot C. Mycobacterium leprae phenolglycolipid-1 expressed by engineered M. bovis BCG modulates early interaction with human phagocytes. PLoS pathogens. 2010;6:e1001159. doi: 10.1371/journal.ppat.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Jain M, Chow ED, Cox JS. The MmpL protein family. In: Daffe M, Reyrat J-M, editors. The mycobacterial cell envelope. Washington, DC: ASM Press; 2008. pp. 201–210. [Google Scholar]
- 252.Jain M, Cox JS. Interaction between polyketide synthase and transporter suggests coupled synthesis and export of virulence lipid in M. tuberculosis. PLoS pathogens. 2005;1:12–19. doi: 10.1371/journal.ppat.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Sulzenbacher G, Canaan S, Bordat Y, Neyrolles O, Stadthagen G, Roig-Zamboni V, Rauzier J, Maurin D, Laval F, Daffé M, Cambillau C, Gicquel B, Bourne Y, Jackson M. LppX is a lipoprotein required for the translocation of phthiocerol dimycocerosates to the surface of Mycobacterium tuberculosis. The EMBO journal. 2006;25:1436–1444. doi: 10.1038/sj.emboj.7601048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Daffé M, Cho SN, Chatterjee D, Brennan PJ. Chemical synthesis and seroreactivity of a neoantigen containing the oligosaccharide hapten of the Mycobacterium tuberculosis-specific phenolic glycolipid. The Journal of infectious diseases. 1991b;163:161–168. doi: 10.1093/infdis/163.1.161. [DOI] [PubMed] [Google Scholar]
- 255.Cho SN, Shin JS, Daffe M, Chong Y, Kim SK, Kim JD. Production of monoclonal antibody to a phenolic glycolipid of Mycobacterium tuberculosis and its use in detection of the antigen in clinical isolates. Journal of clinical microbiology. 1992;30:3065–3069. doi: 10.1128/jcm.30.12.3065-3069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Simonney N, Molina JM, Molimard M, Oksenhendler E, Lagrange PH. Comparison of A60 and three glycolipid antigens in an ELISA test for tuberculosis. Clinical Microbiology and Infection. 1996;2:214–222. doi: 10.1016/s1198-743x(14)65145-4. [DOI] [PubMed] [Google Scholar]
- 257.Simonney N, Molina JM, Molimard M, Oksenhendler E, Perronne C, Lagrange PH. Analysis of the immunological humoral response to Mycobacterium tuberculosis glycolipid antigens (DAT, PGLTb1) for diagnosis of tuberculosis in HIV-seropositive and -seronegative patients. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 1995;14:883–891. doi: 10.1007/BF01691495. [DOI] [PubMed] [Google Scholar]
- 258.Puzo G. The carbohydrate- and lipid- containing cell wall of mycobacteria, phenolic glycolipids: structure and immunological properties. Critic. Rev. Microbiol. 1990;17:305–327. doi: 10.3109/10408419009105730. [DOI] [PubMed] [Google Scholar]
- 259.Minnikin DE. Lipids: Complex lipids, their chemistry, biosynthesis and roles. In: Ratledge C, Stanford J, editors. The Biology of Mycobacteria. Vol. 1. London: Academic Press; 1982. pp. 95–184. [Google Scholar]
- 260.Guilhot C, Chalut C, Daffé M. Biosynthesis and roles of phenolic glycolipids and related molecules in Mycobacterium tuberculosis. In: Daffé M, Reyrat J-M, editors. The mycobacterial cell envelope. Washington DC: ASM Press; 2008. pp. 273–289. [Google Scholar]
- 261.Fournié JJ, Adams E, Mullins RJ, Basten A. Inhibition of human lymphoproliferative responses by mycobacterial phenolic glycolipids. Infection and immunity. 1989;57:3653–3659. doi: 10.1128/iai.57.11.3653-3659.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Vachula M, Holzer TJ, Andersen BR. Suppression of monocyte oxidative response by phenolic glycolipid I of Mycobacterium leprae. J Immunol. 1989;60:203–206. [PubMed] [Google Scholar]
- 263.Vachula M, Holzer TJ, Kizlaitis L, Andersen BR. Effect of Mycobacterium leprae's phenolic glycolipid-I on interferon-gamma augmentation of monocyte oxidative responses. Int J Lepr Other Mycobact Dis. 1990;58:342–346. [PubMed] [Google Scholar]
- 264.Mehra V, Brennan PJ, Rada E, Convit J, Bloom BR. Lymphocyte suppression in leprosy induced by unique M. leprae glycolipid. Nature. 1984;308:194–196. doi: 10.1038/308194a0. [DOI] [PubMed] [Google Scholar]
- 265.Chan J, Fujiwara T, Brennan PJ, McNeil M, Turco SJ, Sibille J-C, Snapper M, Aisen P, Bloom BR. Microbial glycolipids: Possible virulence factors that scavenge oxygen radicals. Proc. Natl. Acad. Sci. USA. 1989;86:2453–2457. doi: 10.1073/pnas.86.7.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Smith DW, Randall HM, Maclennan AP, Lederer E. Mycosides: a new class of type-specific glycolipids of Mycobacteria. Nature. 1960;186:887–888. doi: 10.1038/186887a0. [DOI] [PubMed] [Google Scholar]
- 267.Aspinall GO, Chatterjee D, Brennan PJ. The variable surface glycolipids of mycobacteria: structures, synthesis of epitopes, and biological properties. Advances in carbohydrate chemistry and biochemistry. 1995;51:169–242. doi: 10.1016/s0065-2318(08)60194-8. [DOI] [PubMed] [Google Scholar]
- 268.Chatterjee D, Khoo KH. The surface glycopeptidolipids of mycobacteria: structures and biological properties. Cellular and molecular life sciences : CMLS. 2001;58:2018–2042. doi: 10.1007/PL00000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Riviere M, Puzo G. A new type of serine-containing glycopeptidolipid from Mycobacterium xenopi. The Journal of biological chemistry. 1991;266:9057–9063. [PubMed] [Google Scholar]
- 270.Besra GS, McNeil MR, Rivoire B, Khoo KH, Morris HR, Dell A, Brennan PJ. Further structural definition of a new family of glycopeptidolipids from Mycobacterium xenopi. Biochemistry. 1993;32:347–355. doi: 10.1021/bi00052a043. [DOI] [PubMed] [Google Scholar]
- 271.Daffé M, Laneelle MA, Puzo G. Structural elucidation by field desorption and electron-impact mass spectrometry of the C-mycosides isolated from Mycobacterium smegmatis. Biochimica et biophysica acta. 1983;751:439–443. doi: 10.1016/0005-2760(83)90304-1. [DOI] [PubMed] [Google Scholar]
- 272.Vats A, Singh AK, Mukherjee R, Chopra T, Ravindran MS, Mohanty D, Chatterji D, Reyrat JM, Gokhale RS. Retrobiosynthetic approach delineates the biosynthetic pathway and the structure of the acyl chain of mycobacterial glycopeptidolipids. The Journal of biological chemistry. 2012;287:30677–30687. doi: 10.1074/jbc.M112.384966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Barrow WW, Brennan PJ. Isolation in high frequency of rough variants of Mycobacterium intracellulare lacking C-mycoside glycopeptidolipid antigens. Journal of bacteriology. 1982;150:381–384. doi: 10.1128/jb.150.1.381-384.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Billman-Jacobe H, McConville MJ, Haites RE, Kovacevic S, Coppel RL. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Molecular microbiology. 1999;33:1244–1253. doi: 10.1046/j.1365-2958.1999.01572.x. [DOI] [PubMed] [Google Scholar]
- 275.Etienne G, Villeneuve C, Billman-Jacobe H, Astarie-Dequeker C, Dupont M-A, Daffé M. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis. Microbiol. 2002;148:3089–3100. doi: 10.1099/00221287-148-10-3089. [DOI] [PubMed] [Google Scholar]
- 276.Schaefer WB. Serologic identification and classification of the atypical mycobacteria by their agglutination. The American review of respiratory disease. 1965;92:85–93. doi: 10.1164/arrd.1965.92.6P2.85. [DOI] [PubMed] [Google Scholar]
- 277.Draper P, Rees RJ. The nature of the electron-transparent zone that surrounds Mycobacterium lepraemurium inside host cells. Journal of general microbiology. 1973;77:79–87. doi: 10.1099/00221287-77-1-79. [DOI] [PubMed] [Google Scholar]
- 278.Draper P. The mycoside capsule of Mycobacterium Avium 357. Journal of general microbiology. 1974;83:431–433. doi: 10.1099/00221287-83-2-431. [DOI] [PubMed] [Google Scholar]
- 279.Vergne I, Daffe M. Interaction of mycobacterial glycolipids with host cells. Frontiers in bioscience : a journal and virtual library. 1998;3:d865–d876. doi: 10.2741/A330. [DOI] [PubMed] [Google Scholar]
- 280.Martinez A, Torello S, Kolter R. Sliding motility in mycobacteria. Journal of bacteriology. 1999;181:7331–7338. doi: 10.1128/jb.181.23.7331-7338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Recht J, Martinez A, Torello S, Kolter R. Genetic analysis of sliding motility in Mycobacterium smegmatis. Journal of bacteriology. 2000;182:4348–4351. doi: 10.1128/jb.182.15.4348-4351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Draper P, Rees RJ. Electron-transparent zone of mycobacteria may be a defence mechanism. Nature. 1970;228:860–861. doi: 10.1038/228860a0. [DOI] [PubMed] [Google Scholar]
- 283.Tereletsky MJ, Barrow WW. Postphagocytic detection of glycopeptidolipids associated with the superficial L1 layer of Mycobacterium intracellulare. Infection and immunity. 1983;41:1312–1321. doi: 10.1128/iai.41.3.1312-1321.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Rulong S, Aguas AP, da Silva PP, Silva MT. Intramacrophagic Mycobacterium avium bacilli are coated by a multiple lamellar structure: freeze fracture analysis of infected mouse liver. Infection and immunity. 1991;59:3895–3902. doi: 10.1128/iai.59.11.3895-3902.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Brownback PE, Barrow WW. Modified lymphocyte response to mitogens after intraperitoneal injection of glycopeptidolipid antigens from Mycobacterium avium complex. Infection and immunity. 1988;56:1044–1050. doi: 10.1128/iai.56.5.1044-1050.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Hooper LC, Barrow WW. Decreased mitogenic response of murine spleen cells following intraperitoneal injection of serovar-specific glycopeptidolipid antigens from the Mycobacterium avium complex. Advances in experimental medicine and biology. 1988;239:309–325. doi: 10.1007/978-1-4757-5421-6_31. [DOI] [PubMed] [Google Scholar]
- 287.Pourshafie M, Ayub Q, Barrow WW. Comparative effects of Mycobacterium avium glycopeptidolipid and lipopeptide fragment on the function and ultrastructure of mononuclear cells. Clinical and experimental immunology. 1993;93:72–79. doi: 10.1111/j.1365-2249.1993.tb06499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Barrow WW, de Sousa JP, Davis TL, Wright EL, Bachelet M, Rastogi N. Immunomodulation of human peripheral blood mononuclear cell functions by defined lipid fractions of Mycobacterium avium. Infection and immunity. 1993;61:5286–5293. doi: 10.1128/iai.61.12.5286-5293.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Kano H, Doi T, Fujita Y, Takimoto H, Yano I, Kumazawa Y. Serotype-specific modulation of human monocyte functions by glycopeptidolipid (GPL) isolated from Mycobacterium avium complex. Biological & pharmaceutical bulletin. 2005;28:335–339. doi: 10.1248/bpb.28.335. [DOI] [PubMed] [Google Scholar]
- 290.Horgen L, Barrow EL, Barrow WW, Rastogi N. Exposure of human peripheral blood mononuclear cells to total lipids and serovar-specific glycopeptidolipids from Mycobacterium avium serovars 4 and 8 results in inhibition of TH1-type responses. Microbial pathogenesis. 2000;29:9–16. doi: 10.1006/mpat.2000.0358. [DOI] [PubMed] [Google Scholar]
- 291.Barrow WW, Davis TL, Wright EL, Labrousse V, Bachelet M, Rastogi N. Immunomodulatory spectrum of lipids associated with Mycobacterium avium serovar 8. Infection and immunity. 1995;63:126–133. doi: 10.1128/iai.63.1.126-133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Lagrange PH, Fourgeaud M, Neway T, Pilet C. Enhanced resistance against lethal disseminated Candida albicans infection in mice treated with polar glycopeptidolipids from Mycobacterium chelonae (pGPL-Mc) Comptes rendus de l'Academie des sciences. Serie III, Sciences de la vie. 1994;317:1107–1113. [PubMed] [Google Scholar]
- 293.Gjata B, Hannoun C, Boulouis HJ, Neway T, Pilet C. Adjuvant activity of polar glycopeptidolipids of Mycobacterium chelonae (pGPL-Mc) on the immunogenic and protective effects of an inactivated influenza vaccine. Comptes rendus de l'Academie des sciences. Serie III, Sciences de la vie. 1994;317:257–263. [PubMed] [Google Scholar]
- 294.Vincent-Naulleau S, Neway T, Thibault D, Barrat F, Boulouis HJ, Pilet C. Effects of polar glycopeptidolipids of Mycobacterium chelonae (pGPL-Mc) on granulomacrophage progenitors. Research in immunology. 1995;146:363–371. doi: 10.1016/0923-2494(96)81040-6. [DOI] [PubMed] [Google Scholar]
- 295.Vincent-Naulleau S, Thibault D, Neway T, Pilet C. Stimulatory effects of polar glycopeptidolipids of Mycobacterium chelonae on murine haematopoietic stem cells and megakaryocyte progenitors. Research in immunology. 1997;148:127–136. doi: 10.1016/s0923-2494(97)82484-4. [DOI] [PubMed] [Google Scholar]
- 296.de Souza Matos DC, Marcovistz R, Neway T, Vieira da Silva AM, Alves EN, Pilet C. Immunostimulatory effects of polar glycopeptidolipids of Mycobacterium chelonae for inactivated rabies vaccine. Vaccine. 2000;18:2125–2131. doi: 10.1016/s0264-410x(99)00544-7. [DOI] [PubMed] [Google Scholar]
- 297.Patterson JH, McConville MJ, Haites RE, Coppel RL, Billman-Jacobe H. Identification of a methyltransferase from Mycobacterium smegmatis involved in glycopeptidolipid synthesis. The Journal of biological chemistry. 2000;275:24900–24906. doi: 10.1074/jbc.M000147200. [DOI] [PubMed] [Google Scholar]
- 298.Recht J, Kolter R. Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. Journal of bacteriology. 2001;183:5718–5724. doi: 10.1128/JB.183.19.5718-5724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Jeevarajah D, Patterson JH, McConville MJ, Billman-Jacobe H. Modification of glycopeptidolipids by an O-methyltransferase of Mycobacterium smegmatis. Microbiology. 2002;148:3079–3087. doi: 10.1099/00221287-148-10-3079. [DOI] [PubMed] [Google Scholar]
- 300.Jeevarajah D, Patterson JH, Taig E, Sargeant T, McConville MJ, Billman-Jacobe H. Methylation of GPLs in Mycobacterium smegmatis and Mycobacterium avium. Journal of bacteriology. 2004;186:6792–6799. doi: 10.1128/JB.186.20.6792-6799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Deshayes C, Laval F, Montrozier H, Daffe M, Etienne G, Reyrat JM. A glycosyltransferase involved in biosynthesis of triglycosylated glycopeptidolipids in Mycobacterium smegmatis: impact on surface properties. Journal of bacteriology. 2005;187:7283–7291. doi: 10.1128/JB.187.21.7283-7291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Laurent JP, Hauge K, Burnside K, Cangelosi G. Mutational analysis of cell wall biosynthesis in Mycobacterium avium. Journal of bacteriology. 2003;185:5003–5006. doi: 10.1128/JB.185.16.5003-5006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Tatham E, Sundaram Chavadi S, Mohandas P, Edupuganti UR, Angala SK, Chatterjee D, Quadri LE. Production of mycobacterial cell wall glycopeptidolipids requires a member of the MbtH-like protein family. BMC microbiology. 2012;12:118. doi: 10.1186/1471-2180-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Villeneuve C, Etienne G, Abadie V, Montrozier H, Bordier C, Laval F, Daffe M, Maridonneau-Parini I, Astarie-Dequeker C. Surface-exposed glycopeptidolipids of Mycobacterium smegmatis specifically inhibit the phagocytosis of mycobacteria by human macrophages. Identification of a novel family of glycopeptidolipids. The Journal of biological chemistry. 2003;278:51291–51300. doi: 10.1074/jbc.M306554200. [DOI] [PubMed] [Google Scholar]
- 305.Billman-Jacobe H. Glycopeptidolipid synthesis in mycobacteria. Curr. Sci. 2004;86:111–114. [Google Scholar]
- 306.Bull TJ, Sheridan JM, Martin H, Sumar N, Tizard M, Hermon-Taylor J. Further studies on the GS element. A novel mycobacterial insertion sequence (IS1612), inserted into an acetylase gene (mpa) in Mycobacterium avium subsp. silvaticum but not in Mycobacterium avium subsp. paratuberculosis. Veterinary microbiology. 2000;77:453–463. doi: 10.1016/s0378-1135(00)00330-8. [DOI] [PubMed] [Google Scholar]
- 307.Maslow JN, Irani VR, Lee SH, Eckstein TM, Inamine JM, Belisle JT. Biosynthetic specificity of the rhamnosyltransferase gene of Mycobacterium avium serovar 2 as determined by allelic exchange mutagenesis. Microbiology. 2003;149:3193–3202. doi: 10.1099/mic.0.26565-0. [DOI] [PubMed] [Google Scholar]
- 308.Eckstein TM, Silbaq FS, Chatterjee D, Kelly NJ, Brennan PJ, Belisle JT. Identification and recombinant expression of a Mycobacterium avium rhamnosyltransferase gene (rtfA) involved in glycopeptidolipid biosynthesis. Journal of bacteriology. 1998;180:5567–5573. doi: 10.1128/jb.180.21.5567-5573.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Krzywinska E, Bhatnagar S, Sweet L, Chatterjee D, Schorey JS. Mycobacterium avium 104 deleted of the methyltransferase D gene by allelic replacement lacks serotype-specific glycopeptidolipids and shows attenuated virulence in mice. Molecular microbiology. 2005;56:1262–1273. doi: 10.1111/j.1365-2958.2005.04608.x. [DOI] [PubMed] [Google Scholar]
- 310.Irani VR, Lee SH, Eckstein TM, Inamine JM, Belisle JT, Maslow JN. Utilization of a ts-sacB selection system for the generation of a Mycobacterium avium serovar-8 specific glycopeptidolipid allelic exchange mutant. Annals of clinical microbiology and antimicrobials. 2004;3:18. doi: 10.1186/1476-0711-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Eckstein TM, Belisle JT, Inamine JM. Proposed pathway for the biosynthesis of serovar-specific glycopeptidolipids in Mycobacterium avium serovar 2. Microbiology. 2003;149:2797–2807. doi: 10.1099/mic.0.26528-0. [DOI] [PubMed] [Google Scholar]
- 312.Deshayes C, Bach H, Euphrasie D, Attarian R, Coureuil M, Sougakoff W, Laval F, Av-Gay Y, Daffe M, Etienne G, Reyrat JM. MmpS4 promotes glycopeptidolipids biosynthesis and export in Mycobacterium smegmatis. Molecular microbiology. 2010;78:989–1003. doi: 10.1111/j.1365-2958.2010.07385.x. [DOI] [PubMed] [Google Scholar]
- 313.Ojha AK, Varma S, Chatterji D. Synthesis of an unusual polar glycopeptidolipid in glucose-limited culture of Mycobacterium smegmatis. Microbiology. 2002;148:3039–3048. doi: 10.1099/00221287-148-10-3039. [DOI] [PubMed] [Google Scholar]
- 314.Mukherjee R, Chatterji D. Evaluation of the role of sigma B in Mycobacterium smegmatis. Biochemical and biophysical research communications. 2005;338:964–972. doi: 10.1016/j.bbrc.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 315.Kocincova D, Singh AK, Beretti JL, Ren H, Euphrasie D, Liu J, Daffe M, Etienne G, Reyrat JM. Spontaneous transposition of IS1096 or ISMsm3 leads to glycopeptidolipid overproduction and affects surface properties in Mycobacterium smegmatis. Tuberculosis (Edinb) 2008;88:390–398. doi: 10.1016/j.tube.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 316.Dinadayala P, Lemassu A, Granovski P, Cérantola S, Winter N, Daffé M. Revisiting the structure of the anti-neoplastic glucans of Mycobacterium bovis bacille Calmette-Guérin. J. Biol. Chem. 2004;279:12369–12378. doi: 10.1074/jbc.M308908200. [DOI] [PubMed] [Google Scholar]
- 317.Dinadayala P, Sambou T, Daffé M, Lemassu A. Comparative structural analyses of the alpha-glucan and glycogen from Mycobacterium bovis. Glycobiology. 2008;18:502–508. doi: 10.1093/glycob/cwn031. [DOI] [PubMed] [Google Scholar]
- 318.Cywes C, Hoppe HC, Daffé M, Ehlers MRW. Nonopsonic binding of Mycobacterium tuberculosis to human complement receptor type 3 is mediated by capsular polysaccharides and is strain dependent. Infect. Immun. 1997;65:4258–4266. doi: 10.1128/iai.65.10.4258-4266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Gagliardi MC, Lemassu A, Teloni R, Mariotti S, Sargentini V, Pardini M, Daffe M, Nisini R. Cell wall-associated alpha-glucan is instrumental for Mycobacterium tuberculosis to block CD1 molecule expression and disable the fucntion of dendritic cells derived from infected monocyte. Cellular microbiology. 2007;9:2081–2092. doi: 10.1111/j.1462-5822.2007.00940.x. [DOI] [PubMed] [Google Scholar]
- 320.Geurtsen J, Chedammi S, Mesters J, Cot M, Driessen NN, Sambou T, Kakutani R, Ummels R, Maaskant J, Takata H, Baba O, Terashima T, Bovin N, Vandenbroucke-Grauls CMJE, Nigou J, Puzo G, Lemassu A, Daffe M, Appelmelk BJ. Identification of mycobacterial α-glucan as a novel ligand for DC-SIGN: Involvement of mycobacterial capsular polysaccharides in host immune modulation. J. Immunol. 2009;183:5221–5231. doi: 10.4049/jimmunol.0900768. [DOI] [PubMed] [Google Scholar]
- 321.Stokes RW, Norris-Jones R, Brooks DE, Beveridge TJ, Doxsee D, Thorson LM. The glycan-rich outer layer of the cell wall of Mycobacterium tuberculosis acts as an antiphagocytic capsule limiting the association of the bacterium with macrophages. Infect. Immun. 2004;72:5676–5686. doi: 10.1128/IAI.72.10.5676-5686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Sambou T, Dinadayala P, Stadthagen G, Barilone N, Bordat Y, Constant P, Levillain F, Neyrolles O, Gicquel B, Lemassu A, Daffé M, Jackson M. Capsular glucan and intracellular glycogen of Mycobacterium tuberculosis: biosynthesis and impact on the persistence in mice. Molecular microbiology. 2008;70:762–774. doi: 10.1111/j.1365-2958.2008.06445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Stadthagen G, Sambou T, Guerin M, Barilone N, Boudou F, Kordulakova J, Charles P, Alzari PM, Lemassu A, Daffé M, Puzo G, Gicquel B, Riviere M, Jackson M. Genetic basis for the biosynthesis of methylglucose lipopolysaccharides in Mycobacterium tuberculosis. The Journal of biological chemistry. 2007;282:27270–27276. doi: 10.1074/jbc.M702676200. [DOI] [PubMed] [Google Scholar]
- 324.Kalscheuer R, Syson K, Veeraraghavan U, Weinrick B, Biermann KE, Liu Z, Sacchettini JC, Besra GS, Bornemann S, Jacobs WR., Jr Self-poisoning of Mycobacterium tuberculosis by targeting GlgE in an alpha-glucan pathway. Nature chemical biology. 2010;6:376–384. doi: 10.1038/nchembio.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Elbein AD, Pastuszak I, Tackett AJ, Wilson T, Pan YT. The last step in the conversion of trehalose to glycogen: A mycobacterial enzyme that transfers maltose from maltose-1-phosphate to glycogen. J. Biol. Chem. 2010;285:9803–9812. doi: 10.1074/jbc.M109.033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Jackson M, Brennan PJ. Polymethylated polysaccharides from Mycobacterium species revisited. J. Biol. Chem. 2009;284:1949–1953. doi: 10.1074/jbc.R800047200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Quadri LEN. Iron uptake in mycobacteria. In: Daffé M, Reyrat J-M, editors. The Mycobacterial Cell Envelope. Washington, DC: ASM Press; 2008. –184.pp. 167 [Google Scholar]
- 328.Stinear TP, Small P. The mycolactones: Biologically active polyketides produced by Mycobacterium ulcerans and related aquatic mycobacteria. In: Daffé M, Reyrat J-M, editors. The Mycobacterial Cell Envelope. Washington, DC: ASM Press; 2008. pp. 367–377. [Google Scholar]
- 329.Wells RM, Jones CM, Xi Z, Speer A, Danilchanka O, Doornbos KS, Sun P, Wu F, Tian C, Niederweis M. Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS pathogens. 2013;9:e1003120. doi: 10.1371/journal.ppat.1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Marsollier L, Brodin P, Jackson M, Korduláková J, Tafelmeyer P, Carbonnelle E, Aubry J, Milon G, Legras P, Andre JP, Leroy C, Cottin J, Guillou ML, Reysset G, Cole ST. Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS pathogens. 2007;3:e62. doi: 10.1371/journal.ppat.0030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Stinear TP, Mve-Obiang A, Small PLC, Frigui W, Pryor MJ, Brosch R, Jenkin GA, Johnson PDR, Davies JK, Lee RE, Adusumilli S, Granier T, Haydock SF, Leadlay PF, Cole ST. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. USA. 2004;101:1345–1349. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.He H, Oka S, Han YK, Yamamura Y, Kusunose E, Kusunose M, Yano I. Rapid serodiagnosis of human mycobacteriosis by ELISA using cord factor (trehalose-6,6'-dimycolate) purified from Mycobacterium tuberculosis as antigen. FEMS microbiology immunology. 1991;3:201–204. doi: 10.1111/j.1574-6968.1991.tb04215.x. [DOI] [PubMed] [Google Scholar]
- 333.Vera-Cabrera L, Handzel V, Laszlo A. Development of an enzyme-linked immunosorbent assay (ELISA) combined with a streptavidin-biotin and enzyme amplification method to detect anti-2,3-di-O-acyltrehalose (DAT) antibodies in patients with tuberculosis. Journal of immunological methods. 1994;177:69–77. doi: 10.1016/0022-1759(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 334.Julian E, Matas L, Perez A, Alcaide J, Laneelle MA, Luquin M. Serodiagnosis of tuberculosis: comparison of immunoglobulin A (IgA) response to sulfolipid I with IgG and IgM responses to 2,3-diacyltrehalose, 2,3,6-triacyltrehalose, and cord factor antigens. Journal of clinical microbiology. 2002;40:3782–3788. doi: 10.1128/JCM.40.10.3782-3788.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Cunningham AF, Spreadbury CL. Mycobacterial stationary phase induced by low oxygen tension: Cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J. Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.de Chastellier C, Thilo L. Phagosome maturation and fusion with lysosomes in relation to surface property and size of the phagocytic particle. European journal of cell biology. 1997;74:49–62. [PubMed] [Google Scholar]
- 337.Guerin ME, Kordulakova J, Alzari PM, Brennan PJ, Jackson M. Molecular basis of phosphatidyl-myo-inositol mannoside biosynthesis and regulation in mycobacteria. The Journal of biological chemistry. 2010;285:33577–33583. doi: 10.1074/jbc.R110.168328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nature medicine. 2010;16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Ryan GJ, Hoff DR, Driver ER, Voskuil ML, Gonzalez-Juarrero M, Basaraba RJ, Crick DC, Spencer JS, Lenaerts AJ. Multiple M. tuberculosis phenotypes in mouse and guinea pig lung tissue revealed by a dual-staining approach. PloS one. 2010;5:e11108. doi: 10.1371/journal.pone.0011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Dhiman RK, Dinadayala P, Ryan GJ, Lenaerts AJ, Schenkel AR, Crick DC. Lipoarabinomannan localization and abundance during growth of Mycobacterium smegmatis. J. Bacteriol. 2011;193:5802–5809. doi: 10.1128/JB.05299-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Bhamidi S, Shi L, Chatterjee D, Belisle JT, Crick DC, McNeil MR. A bioanalytical method to determine the cell wall composition of Mycobacterium tuberculosis grown in vivo. Analytical biochemistry. 2012;421:240–249. doi: 10.1016/j.ab.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 342.Molle V, Kremer L. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Molecular microbiology. 2010;75:1064–1077. doi: 10.1111/j.1365-2958.2009.07041.x. [DOI] [PubMed] [Google Scholar]
- 343.Hunter SW, Neil MRM, Brennan PJ. Diglycosyl diacylglycerol of Mycobacterium tuberculosis. J. Bacteriol. 1986;168:917–922. doi: 10.1128/jb.168.2.917-922.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Slayden RA, Jackson M, Zucker J, Ramirez MV, Dawson CC, Crew R, Sampson NS, Thomas ST, Jamshidi N, Sisk P, Caspi R, Crick DC, McNeil MR, Pavelka MS, Niederweis M, Siroy A, Dona V, McFadden J, Boshoff H, Lew JM. Updating and curating metabolic pathways of TB. Tuberculosis (Edinb) 2012 doi: 10.1016/j.tube.2012.11.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Fortune SM, Moody DB, Rubin EJ. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. USA. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Camacho LR, Ensergueix D, Pérez E, Gicquel B, Guilhot C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 1999;34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- 348.Larrouy-Maumus G, Škovierová H, Dhouib R, Angala SK, Zuberogoitia S, Pham H, Drumond Villela A, Mikušová K, Noguera A, Gilleron M, Valentinova L, Korduláková J, Brennan PJ, Puzo G, Nigou J, Jackson M. A small multidrug resistance-like transporter involved in the arabinosylation of arabinogalactan and lipoarabinomannan in mycobacteria. The Journal of biological chemistry. 2012;287:39933–39941. doi: 10.1074/jbc.M112.400986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Azad AK, Sirakova TD, Fernandes ND, Kolattukudy PE. Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. The Journal of biological chemistry. 1997;272:16741–16745. doi: 10.1074/jbc.272.27.16741. [DOI] [PubMed] [Google Scholar]
- 350.Choudhuri BS, Bhakta S, Barik R, Basu J, Kundu M, Chakrabarti P. Overexpression and functional characterization of an ABC (ATP-binding cassette) transporter encoded by the genes drrA and drrB of Mycobacterium tuberculosis. The Biochemical journal. 2002;367:279–285. doi: 10.1042/BJ20020615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Onwueme KC, Ferreras JA, Buglino J, Lima CD, Quadri LE. Mycobacterial polyketide-associated proteins are acyltransferases: proof of principle with Mycobacterium tuberculosis PapA5. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4608–4613. doi: 10.1073/pnas.0306928101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Buglino J, Onwueme KC, Ferreras JA, Quadri LE, Lima CD. Crystal structure of PapA5, a phthiocerol dimycocerosyl transferase from Mycobacterium tuberculosis. The Journal of biological chemistry. 2004;279:30634–30642. doi: 10.1074/jbc.M404011200. [DOI] [PubMed] [Google Scholar]
- 353.Ferreras JA, Stirrett KL, Lu X, Ryu JS, Soll CE, Tan DS, Quadri LE. Mycobacterial phenolic glycolipid virulence factor biosynthesis: mechanism and small-molecule inhibition of polyketide chain initiation. Chemistry & biology. 2008;15:51–61. doi: 10.1016/j.chembiol.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.He W, Soll CE, Chavadi SS, Zhang G, Warren JD, Quadri LE. Cooperation between a coenzyme A-independent stand-alone initiation module and an iterative type I polyketide synthase during synthesis of mycobacterial phenolic glycolipids. Journal of the American Chemical Society. 2009;131:16744–16750. doi: 10.1021/ja904792q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Onwueme KC, Vos CJ, Zurita J, Soll CE, Quadri LE. Identification of phthiodiolone ketoreductase, an enzyme required for production of mycobacterial diacyl phthiocerol virulence factors. Journal of bacteriology. 2005;187:4760–4766. doi: 10.1128/JB.187.14.4760-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Simeone R, Constant P, Malaga W, Guilhot C, Daffe M, Chalut C. Molecular dissection of the biosynthetic relationship between phthiocerol and phthiodiolone dimycocerosates and their critical role in the virulence and permeability of Mycobacterium tuberculosis. The FEBS journal. 2007a;274:1957–1969. doi: 10.1111/j.1742-4658.2007.05740.x. [DOI] [PubMed] [Google Scholar]
- 357.Simeone R, Constant P, Guilhot C, Daffe M, Chalut C. Identification of the missing trans-acting enoyl reductase required for phthiocerol dimycocerosate and phenolglycolipid biosynthesis in Mycobacterium tuberculosis. Journal of bacteriology. 2007b;189:4597–4602. doi: 10.1128/JB.00169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.De Smet KAL, Weston A, Brown IN, Young DB, Robertson BD. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology. 2000;146:199–208. doi: 10.1099/00221287-146-1-199. [DOI] [PubMed] [Google Scholar]
- 359.Pan Y-T, Carroll JD, Asano N, Pastuszak I, Edavana VK, Elbein AD. Trehalose synthase converts glycogen to trehalose. FEBS Journal. 2008;275:3408–3420. doi: 10.1111/j.1742-4658.2008.06491.x. [DOI] [PubMed] [Google Scholar]
- 360.Pan YT, Koroth Edavana V, Jourdian WJ, Edmondson R, Carroll JD, Pastuszak I, Elbein AD. Trehalose synthase of Mycobacterium smegmatis: purification, cloning, expression, and properties of the enzyme. European journal of biochemistry/FEBS. 2004;271:4259–4269. doi: 10.1111/j.1432-1033.2004.04365.x. [DOI] [PubMed] [Google Scholar]
- 361.Mendes V, Maranha A, Lamosa P, da Costa MS, Empadinhas N. Biochemical characterization of the maltokinase from Mycobacterium bovis BCG. BMC biochemistry. 2010;11:21. doi: 10.1186/1471-2091-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Garg SK, Alam MS, Kishan KVR, Agrawal P. Expression and characterization of α-(1,4)-glucan branching enzyme Rv1326c of Mycobacterium tuberculosis H37Rv. Protein Expr. Purif. 2007;51:198–208. doi: 10.1016/j.pep.2006.08.005. [DOI] [PubMed] [Google Scholar]