Abstract
The lysine acetylation of proteins is a reversible post-translational modification that plays a critical regulatory role in both eukaryotes and prokaryotes. Mycobacterium tuberculosis is a facultative intracellular pathogen and the causative agent of tuberculosis. Increasing evidence shows that lysine acetylation may play an important role in the pathogenesis of M. tuberculosis. However, only a few acetylated proteins of M. tuberculosis are known, presenting a major obstacle to understanding the functional roles of reversible lysine acetylation in this pathogen. We performed a global acetylome analysis of M. tuberculosis H37Ra by combining protein/peptide prefractionation, antibody enrichment, and LC-MS/MS. In total, we identified 226 acetylation sites in 137 proteins of M. tuberculosis H37Ra. The identified acetylated proteins were functionally categorized into an interaction map and shown to be involved in various biological processes. Consistent with previous reports, a large proportion of the acetylation sites were present on proteins involved in glycolysis/gluconeogenesis, the citrate cycle, and fatty acid metabolism. A NAD+-dependent deacetylase (MRA_1161) deletion mutant of M. tuberculosis H37Ra was constructed and its characterization showed a different colony morphology, reduced biofilm formation, and increased tolerance of heat stress. Interestingly, lysine acetylation was found, for the first time, to block the immunogenicity of a peptide derived from a known immunogen, HspX, suggesting that lysine acetylation plays a regulatory role in immunogenicity. Our data provide the first global survey of lysine acetylation in M. tuberculosis. The dataset should be an important resource for the functional analysis of lysine acetylation in M. tuberculosis and facilitate the clarification of the entire metabolic networks of this life-threatening pathogen.
Mycobacterium tuberculosis was responsible for 1.3 million deaths and 8.6 million new cases of tuberculosis (TB)1 worldwide in 2012 (1). This global public health crisis remains a serious problem, with the emergence of drug-resistant M. tuberculosis, especially multidrug-resistant and extensively drug-resistant M. tuberculosis, and also the emergence of coinfections of TB and human immunodeficiency virus (2, 3). To counter the increasing threat of TB, it is critical to understand fundamental aspects of TB-related biology. Such studies will not only provide new drug targets for the design of novel therapeutic agents, but also facilitate the development of novel diagnostic tools and new vaccines.
Acetylation is one of the important protein modifications and occurs both co- and post-translationally on the α-amino group at the N terminus of the protein, so-called “N-terminal acetylation,” or on the ε-amino group on the side chain of lysine (4). Lysine acetylation is one of the most common post-translational modifications to proteins in both eukaryotes and prokaryotes. As a dynamic and reversible process, protein acetylation plays important roles in many cellular physiological processes, including cell-cycle regulation and apoptosis, cell morphology (5), metabolic pathways (6–8), protein interactions (9), and enzymatic activity (8, 10). In recent years, great advances have been made in proteomic studies, and a large number of lysine-acetylated proteins have been identified in many eukaryotes, including human (5, 11, 12), rat (13), mouse (11), Drosophila (14), Arabidopsis (15, 16), Saccharomyces cerevisiae (17), and protozoans (18, 19). The global analysis of lysine acetylation has also been reported in bacteria, including Escherichia coli (20–22), Erwinia amylovora (23), Bacillus subtilis (24), and Salmonella enterica (6). These acetylome studies have generated large datasets of bacterial proteins acetylated on lysine residues and have demonstrated the diverse cellular functions of lysine acetylation in bacteria.
Increasing evidence shows that protein acetylation occurs and plays an important regulatory role in mycobacteria (8, 25–31). For example, Lange et al. reported the N-terminal acetylation of early secreted antigenic target 6 (ESAT-6) protein (31). Rv1151c is reported to be an NAD+-dependent protein deacetylase in M. tuberculosis that deacetylates and thus regulates the activity of acetyl-CoA synthase (25, 32). Two cyclic adenosine monophosphate (cAMP)-binding proteins in M. smegmatis and M. tuberculosis (MSMEG_5458 and Rv0998, respectively) show similarity to the GNAT family of acetyltransferases and could acetylate a universal stress protein (USP, MSMEG_4207) (30). Subsequent structural studies revealed the fine mechanisms of how cAMP regulates the protein lysine acetyltransferase in mycobacteria (27, 28). Very recently, reversible lysine acetylation was shown to regulate the activity of several fatty acyl-CoA synthetases in M. tuberculosis (8, 26), and also to regulate acetate and propionate metabolism in M. smegmatis (8, 26). However, to the best of our knowledge, only a few acetylated proteins in M. tuberculosis have been identified, presenting a major obstacle to further understanding the regulatory roles of reversible lysine acetylation in this life-threatening pathogen.
To fill this gap in our knowledge, we undertook a systematic study of the functional roles of lysine acetylation in M. tuberculosis. We performed an acetylomic analysis of M. tuberculosis H37Ra using high-accuracy MS combined with the identification of 226 unique lysine acetylation sites on 137 proteins. This set of M. tuberculosis proteins acetylated on lysine residues supports the emerging view that lysine acetylation is a general and fundamental regulatory process, and is not restricted to eukaryotes. It also opens the way for its detailed functional and evolutionary analysis of lysine acetylation in M. tuberculosis. The identified acetylated proteins that are involved in several important biological processes were functionally categorized into an interaction map. This is the first time that an interaction network of acetylated proteins in M. tuberculosis has been constructed, and should allow us to better understand the significance of acetylation in key cellular mechanisms in M. tuberculosis. To further explore the effects of lysine acetylation on the physiology of M. tuberculosis H37Ra, MRA_1161, the gene encoding the only known protein deacetylase in this bacterium, was deleted. The roles of MRA_1161 in the colony morphology, carbon source utilization, heat stress tolerance, and biofilm formation of M. tuberculosis were analyzed. The effect of lysine acetylation on the immunogenicity of a known immunogen, HspX, was also tested.
EXPERIMENTAL PROCEDURES
Construction of the M. tuberculosis H37Ra ΔMRA_1161 Mutant Strain
Temperature- sensitive specialized transducing plasmids (33) were constructed and used for the allelic-exchange-mediated deletion of the MRA_1161 gene (also designated cobB), which encodes an NAD+-dependent protein deacetylase in M. tuberculosis H37Ra. DNA segments of ∼1 kb flanking the gene of interest were amplified with PCR. The purified DNA segments were digested with Van91I and ligated to compatible fragments of the counter-selectable vector p0004S (a kind gift from Dr. William R. Jacobs Jr). The allelic exchange plasmids were selected and propagated following the transformation of E. coli DH5α with the hygromycin-resistance gene. The sequences of the inserted DNA fragments were verified. The allelic exchange plasmids were digested with PacI and ligated to PacI-digested phAE159 (34). The ligation products were packaged using MaxPlax™ Lambda Packaging Extracts (Epicenter Biotechnologies, Madison, WI, USA) and used to transduce E. coli HB101 for propagation as plasmids. The plasmid DNA was purified and transfected into M. smegmatis mc2155 with electroporation for phage propagation. The allelic exchange substrates were delivered to M. tuberculosis H37Ra as previously described (33). The target gene in the mutant strain was replaced with the res-hyg-sacB-res gene cassette. The γδ-resolvase-encoding vector pJH532 was then used to resolve the hygromycin-resistance cassette (which was flanked by γδ resolvase recognition sequences) in the mutant strain. The mutant strain was confirmed with PCR and DNA sequencing.
Cell Culture and Protein Extraction
Mycobacterium tuberculosis H37Ra and the ΔMRA_1161 mutant were grown at 37 °C in Middlebrook 7H9 liquid culture medium (Difco) supplemented with 10% oleic acid, albumin, dextrose, and catalase (OADC) (Difco), 0.5% glycerol, and 0.05% Tween 80 or Middlebrook 7H10 solid culture medium (Difco) supplemented with 10% OADC (Difco) and 0.5% glycerol. To extract the proteins for MS analysis, M. tuberculosis H37Ra and the ΔMRA_1161 mutant were grown under several conditions in Middlebrook 7H9 liquid culture medium supplemented with 0.5% albumin, 0.5% glycerol, 0.085% NaCl, and 0.05% Tween 80, and harvested at different growth stages. The culture extracts were obtained in: (1) log phase (200 ml), (2) late stationary phase (200 ml), (3) log phase treated with 300 μm diethylenetriamine (DETA)-NO overnight (200 ml), (4) log phase treated with 5 mm H2O2 overnight (200 ml), (5) log phase grown on 0.1% sodium acetate instead of glycerol (200 ml), (6) log phase grown on 0.5% glucose instead of glycerol (200 ml), or (7) log phase grown on 0.2% glucose with glycerol (200 ml).
The cultured cells were harvested by centrifugation at 8000 × g for 10 min at 4 °C, and the cell pellets were washed twice with ice-cold PBS (0.1 m Na2HPO4, 0.15 m NaCl, pH 7.5), suspended in 50 ml of chilled lysis buffer (50 mm Tris-HCl [pH 7.5], 100 mm NaCl, 5 mm DTT, 50 mm nicotinamide), and sonicated. Each lysate was centrifuged at 10,000 × g for 30 min at 4 °C and the supernatant was collected. The clarified lysate was precipitated with five volumes of ice-cold acetone, collected by centrifugation, and washed with 80% ice-cold acetone. The protein concentration was determined with the Bradford assay (Bio-Rad, Hercules, CA).
In-solution Trypsin Digestion and Peptide Prefractionation
Protein extracts (30 mg) were subjected to trypsin digestion, as described previously (35). Briefly, the protein disulfide bonds were reduced with 25 mm DTT (37 °C, 45 min), alkylated with 50 mm iodoacetamide (25 °C, 20 min in the dark), digested with sequencing-grade modified trypsin (1:100 w/w; Promega, Madison, WI) at 37 °C for 4 h, and further digested with trypsin (1:100 w/w) at 37 °C for 20 h. The tryptic digestion was quenched by the addition of 0.1% TFA. The solution was clarified by centrifugation at 3000 × g. The resulting peptides were then fractionated and desalted using reversed-phase C18 columns self-packed with C18 material (40 μm, 60 Å pore size; Agilent Technologies, Santa Clara, CA). After the digested peptides were loaded sequentially onto the self-packed C18 columns, the columns were washed five times with 2.0 ml of 0.1% TFA to desalt the samples, and were eluted with a series of elution buffers (2.0 ml) containing 0.1% TFA and different concentrations of acetonitrile (ACN; 10%, 15%, 20%, 25%, 30%, 35%, 40%, 50%, 60%, or 100%). Each fraction was collected, dried in a SpeedVac, and stored at −20 °C until further analysis.
In-gel Trypsin Digestion
Protein extracts (15 mg) were separated by SDS-PAGE on 12% gels (1.5 mm thick, 80 mm wide, 70 mm long), stained with Coomassie Brilliant Blue R250, which were cut into 12 gel slices. Each slice was further diced into ∼1 mm3 cubes for in-gel digestion. Each section was completely destained with 50 mm NH4HCO3 in 50% ACN at room temperature. The proteins in each fraction were disulfide reduced, alkylated, and digested with trypsin, as described above. The resulting peptides were transferred into new microcentrifuge tubes and the gels were further sonicated twice with extraction buffer (67% ACN containing 5% TFA). Finally, all peptides were combined and completely dried in a SpeedVac.
Enrichment of Acetylated Peptides
Anti-acetyl-lysine antibody (ImmuneChem Pharmaceuticals Inc. and Cell Signaling Technology) was immobilized on Protein A/G PLUS-Agarose Immunoprecipitation Reagent (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 4 °C for 5 h. The supernatant was removed, and the beads were washed three times with NETN buffer (50 mm Tris-HCl [pH 8.0], 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40). The tryptic peptides were redissolved in NETN buffer and the insoluble particles were removed by centrifugation. The agarose-conjugated anti-acetyl-lysine antibody was added to the peptide solution and incubated in Pierce Centrifuge Columns (Pierce, Rockford, IL) at 4 °C for 8 h with rotary shaking. The beads were washed three times with 1 ml of PBS and then three times with 1 ml of ice-cold water. The bound peptides were eluted with 0.1% TFA in water.
LC-MS/MS Analysis
The peptides were separated on the UltiMate 3000 Nano LC System (Dionex, Sunnyvale, CA) and analyzed online using an electrospray ion-trap mass spectrometer (HCTultra; Bruker Daltonics, Bremen, Germany). For each analysis, the sample was trapped on a C18 precolumn (Acclaim® PepMap; 300 μm i.d. × 5 mm; Dionex) and then on a C18 reversed-phase analytical column (Acclaim®; 75 μm i.d. × 150 mm; Dionex). The peptides were eluted from the column with a linear solvent gradients (A: 0.1% formic acid [FA] in water; B: 100% ACN/0.1% FA) for 120 min at a flow rate of 300 nL/min (5 min of 95% buffer A; 95 min gradient from 5–80% buffer B; 10 min of 80% buffer B to wash the column; 5 min gradient from 80–5% buffer B; and a final 5 min of 95% buffer A to equilibrate the column). This LC gradient was used for all mobile phase compositions. The mass spectrometer was operated in the positive ion mode at 2 kV and the capillary temperature was set to 180 °C. Mass spectra were recorded in stand-enhanced mode at a speed of 8100 m/z/s (as specified by the Bruker Esquire Control software). Tandem mass spectra were acquired in the ultra-scan operating mode at 26,000 m/z/s (Bruker Esquire Control software). After a survey scan (enhanced mode, 300–2000 m/z) consisting of four scans, the four most-intense peptides were selected automatically for CID MS/MS (m/z range 100–2000). All MS/MS spectra were acquired using the following parameters: normalized collision energy, 0.5 V; precursor selection threshold, 100,000 Abs; the selected ions were dynamically excluded for 0.25 min.
Data Analysis and Peptide Identification
Raw MS data files were processed using the LC/MS software DataAnalysis 4.0 (Bruker Compass™ software) and converted into the MASCOT generic format (mgf) files. All MS/MS samples were analyzed with MASCOT 2.3 (Matrix Science, London, U.K.) and pFind Studio 2.6 (36, 37) to improve the identification of the acetylated peptides. Both pFind and MASCOT were used to search a database of forward and reversed M. tuberculosis H37Ra proteins from National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) that contained 4034 protein sequences. The database search was performed with the following parameters: two missed cleavages were allowed for trypsin; carbamidomethylation (Cys) was set as a fixed modification, whereas oxidation (M) and acetylation (K) were considered variable modifications. The maximum precursor ion mass tolerance allowed was 0.4 Da (monoisotopic) and the fragment maximum mass tolerance was 0.6 Da (monoisotopic). Peptides shorter than six amino acids were excluded. For the search results, the peptide-spectrum matches (PSMs) were filtered based on the score threshold of a 1% false discovery rate (FDR), according to the formula: FDR = 2[nDecoy/(nDecoy + nTarget)] (38–40). We applied further strict criteria to the MS2 identification by requiring the peptides to have pFind E scores < 10−5 and MASCOT scores > 25, resulting in estimated FDRs that were below 1%. All fragmentation spectra of the acetylated peptides assigned to the corresponding proteins were manually evaluated using a method described by Mann et al. (41, 42). To exclude isobaric trimethylation (43), all the peptides identified based on an acetylated Lys residue were also manually inspected, using a modified method, to identify the neutral loss of ion MH+-59, as described by Mann et al. (42, 44). Finally, to pinpoint the actual acetylated lysine within the identified peptide, the localization probabilities for acetylation sites were calculated from the post-translational modification score algorithm, as described previously (35). Acetylation sites that were occupied with a probability > 0.75 were identified as class I acetylation sites.
Bioinformatic Analysis
Biological and functional analyses of our acetylome were performed using only those identified acetylation sites with high confidence (FDR < 1% and localization p value > 0.75). For their functional annotation, the identified acetylated proteins were categorized into biological process and molecular function classes according to Gene Ontology (GO) terms, with the Blast2GO tool. The subcellular localization of the acetylated proteins was analyzed with the PSORTb program, a web-based tool that predicts bacterial protein subcellular localization. To gain information on overrepresentation, a GO enrichment analysis was performed using Cytoscape plugin BiNGO (45, 46), with the previously described parameters (9). The reference GO ontology in the Cytoscape ontology format was created using the whole M. tuberculosis H37Ra proteome, extracted from UniProt-GOA Proteome (http://www.ebi.ac.uk/GOA/proteomes).
The secondary structures of all the acetylated proteins were analyzed with NetSurfP. The mean secondary structure probabilities of the modified lysine residues were compared with the mean secondary structure probabilities of a control dataset containing all the lysine residues of all the acetylated proteins identified in this study. The p values were calculated with the Wilcoxon test.
To analyze the lysine acetylation sites, the ratio of each amino acid flanking the identified acetylation site was calculated as: frequency of residues in the acetyl-13-mers [six amino acids upstream and downstream of the acetylation site]/frequency of residue in total proteome, or frequency of residues in the acetyl-13-mers/frequency of residues in the non-acetyl-13-mers. A position-specific intensity map was generated by plotting the log10 ratios of the frequencies.
A pathway analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways database (http://www.genome.jp/kegg) and the DAVID bioinformatics tools. DAVID provided the corresponding information on the p value, count, percentage (%), and fold enrichment for each KEGG pathway. In our data, KEGG pathways with p values < 0.05 were considered significantly enriched pathways. A functional interaction network analysis was performed using interaction data from the STRING database (score > 0.4) and was visualized with Cytoscape v 2.8.3 (http://www.cytoscape.org) (46).
An evolutionary conservation analysis was performed with multisequence alignments. The proteins homologous to M. tuberculosis H37Ra proteins across 231 species, from Archaea to human, were determined with two-directional BLASTP (47). Eighteen archaea, 152 bacterial, and 61 eukaryotic protein databases were retrieved from Uniprot (http://www.uniprot.org/uniprot/), NCBI (http://www.ncbi.nlm.nih.gov/), and EnsEMBL (http://www.ensembl.org), respectively. The two-directional BLAST alignments were classified as significant if the corresponding BLAST E values were lower than 1E-5.
Immunoblotting with Anti-acetyl-lysine Antibody
Mycobacterium tuberculosis H37Ra and M. tuberculosis H37Ra ΔMRA_1161 were grown, harvested, and lysed as described above. The protein concentrations were determined with the Bradford assay (Bio-Rad). The extracted proteins were separated with 10% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked overnight at 4 °C or 37 °C for 2 h with TBST (25 mm Tris-HCl [pH 8.0], 150 mm NaCl, 0.1% Tween 20) containing 5% nonfat dried milk. A primary rabbit anti-acetyl-lysine polyclonal antibody (Cell Signaling Technology) was diluted (1:1000) in TBST/1% nonfat dried milk and incubated with the membrane at 37 °C for 2 h. After the membrane was washed with TBST, it was incubated with alkaline-phosphatase-conjugated goat anti-rabbit antibody (1:5000 dilution) for 1 h at 37 °C, and then detected according to the manufacturer's instructions.
Biofilm Formation Assay
Mycobacterium tuberculosis H37Ra and M. tuberculosis H37Ra DMRA_1161 were grown in liquid culture medium to mid-log phase, and the cell density of each culture was adjusted to OD600 = 0.8. Then both strains were subcultured (1:100) in fresh 7H9 medium (10% OADC, 0.5% glycerol, 0.05% Tween 80) in 25 cm2 cell culture flasks (NEST Biotechnology) and incubated at 37 °C for 3–5 weeks without shaking. All experiments were performed in triplicate.
Carbon Source Utilization Experiments
Mycobacterium tuberculosis H37Ra and M. tuberculosis H37Ra DMRA_1161 were grown to mid-log phase in liquid culture medium, collected by centrifugation at 8000 × g for 10 min, and washed three times with 7H9 medium + 0.05% Tween 80 (7H9T) to remove the remaining medium, before they were resuspended in 7H9T and the OD600 adjusted to 0.5. Aliquots (100 μl) of the suspension culture were used to inoculate 10 ml of the following media: (1) 7H9 liquid culture medium supplemented with 10% OADC, and 0.5% glucose, 0.05% Tween 80; (2) 7H9 liquid culture medium supplemented with 0.5% albumin, 0.05% Tween 80, and different concentrations of acetate (5 mm, 10 mm, 30 mm, or 50 mm).
Heat Shock Assay
Mycobacterium tuberculosis H37Ra and M. tuberculosis H37Ra ΔMRA_1161 were grown in 7H9 medium to mid-log phase, and the cell density of each culture was adjusted to OD600 = 0.5. The cultures were then heat treated in a water bath at 53 °C. Aliquots were taken and serial dilutions were made before plating (48). The bacterial colony-forming units (cfu) were determined after 25 d at 37 °C. All the experiments were performed in triplicate.
Enzyme-linked Immunospot (ELISPOT) Tests of Two Synthetic Peptides Derived from HspX of M. tuberculosis H37Ra
Two peptides (6291 and A-6291) were synthesized (GL Biochem Ltd, Shanghai, China) based on the amino acid sequence of HspX from M. tuberculosis H37Ra. The two peptides shares the same sequence, spanning residues 62–91 of HspX (PDKDVDIMVRDGQLTIKAERTEQKDFDGRS), except that all three lysines in A-6291 are acetylated, according to our MS data. The ELISPOT test (T-SPOT TB 96 assay, catalog no. TB.200; Oxford Immunotec) was performed according to the instructions of the manufacturer. Twenty-six tuberculosis patients were recruited from Foshan Fourth People's Hospital, and the data for the patients enrolled in this experiment are listed in supplemental Table S1. The study was approved by and performed under the guidelines of the Ethical Committee of Wuhan Institute of Virology (Chinese Academy of Sciences). Written informed consent was obtained directly from each participant. Whole blood (5 ml) was drawn from each individual with venipuncture and stored at room temperature. After isolation by Ficoll-Hypaque (MD Pacific, Tianjin, China) density-gradient centrifugation, the cloudy peripheral blood mononuclear cells (PBMCs) harvested were washed twice with prewarmed RPMI 1640 medium (MD Pacific) or AIM-V medium (Invitrogen, Karlsruhe, Germany). Adequate AIM-V medium was used to resuspend the PBMCs and blood cell counting plates were used to count the cells. Four wells in a 96-well microtiter plate, precoated with monoclonal antibody directed against interferon gamma (IFN-γ; provided with the kit), were seeded with the previously prepared PBMC suspension. The positive control was stimulated with phytohemagglutinin (provided with the kit). One well was a no-peptide control, containing PBS and medium as stimulants, and was included to detect contamination. Identical concentrations (10 μg ml−1) of the acetylated peptide A-6291 or the normal peptide 6291, dissolved in PBS, were added to the other two wells. The plate was then incubated at 37 °C under 5% CO2 for 16–20 h, washed thoroughly several times with PBS, and incubated at 4 °C for 1 h with 50 μl of alkaline-phosphatase-conjugated anti-IFN-γ monoclonal antibody. The plate was then washed thoroughly again with PBS and 50 μl of substrate solution was added to each well. After incubation for 7 min at room temperature, the reaction was stopped with tap water and the plate was dried to visualize the spots. The total numbers of spots were counted and the spot-forming cells in each well was analyzed with a Bioreader 5000 (Dakewei Biotech Company, Shenzhen, China).
RESULTS AND DISCUSSION
Identification of Lysine-acetylated Proteins in M. tuberculosis H37Ra
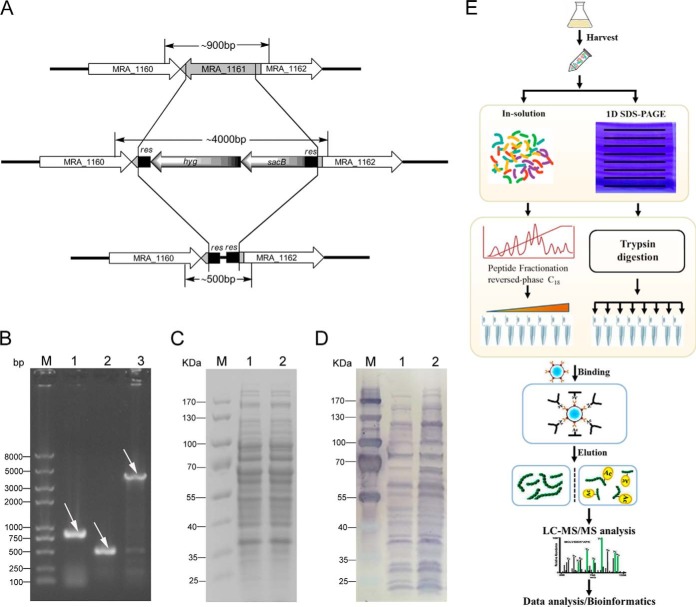
Reversible lysine acetylation is emerging as a major regulatory mechanism in metabolism and is an important post-translational modification in both prokaryotes and eukaryotes (7, 49). To evaluate the diversity and relative abundance of acetylated proteins in M. tuberculosis H37Ra, a ΔMRA_1161 (encoding lysine deacetylase) mutant was constructed (map of the MRA_1161 gene knockout and PCR verification of the knockout strain are shown in Fig. 1A and 1B) and immunoblots of the bacterial cell lysates were probed with an anti-acetyl-lysine antibody. An immunoblotting analysis of protein extracts from the wild-type and ΔMRA_1161 mutant strains detected multiple protein bands, spanning a wide mass range, with the antibody in both extracts, and many substrates showed increased acetylation levels in the ΔMRA_1161 strain (Fig. 1C and 1D). These results suggest that the antibody used is specific and that many lysine-acetylated proteins remain to be identified in M. tuberculosis. After primary verification, the protein extracts were digested both in solution and in gel with sequential rounds of trypsin treatment. To collect low-abundance modification sites, the digested peptides were fractionated and desalted on self-packed reversed-phase C18 columns. The lysine-acetylated peptides were immune-enriched with the anti-acetyl-lysine antibody and identified with nano-LC/MS/MS in an electrospray ion-trap mass spectrometer (HCTultra), as summarized in Fig. 1E. The raw data were processed with two different search algorithms (MASCOT and pFind). All spectra containing acetylation modifications were manually inspected to check for the diagnostic b-type or y-type ion series, as described by Mann et al. (see criteria under “Experimental Procedures”). All the spectra were also checked for the neutral loss of ion “MH+-59,” which can be used as a unique marker for trimethylation (43), and peptides containing this marker were removed from the analysis. In total, 226 lysine acetylation sites were identified in 137 proteins at an FDR < 0.01 for peptides with high confidence. This constitutes the first acetylation dataset obtained for the mycobacteria. Details of the modification-specific peptides, identified lysine acetylation sites and proteins are shown in supplemental Table S2. The annotated spectra of all the acetylated peptides identified are presented in supplemental Fig. S1. To understand the scope of acetylation in bacteria, we compared the M. tuberculosis H37Ra acetylome with the bacterial acetylomes reported by other groups. The results are presented in Table I. Among the highly acetylated proteins were the chaperone protein GroEL, antigen Hsp20 (HspX), and many enzymes involved in diverse metabolic pathways (supplemental Table S2). Of the acetylation sites identified in this study, 46.02% could be successfully predicted with PAIL (prediction of acetylation on internal lysines), a predictor developed from the 249 experimentally verified acetylation sites of 92 distinct proteins in the scientific literature (50) (supplemental Table S2). The raw data (including a converted MASCOT generic format file) have been uploaded onto the public database PeptideAtlas (http://www.peptideatlas.org) (51, 52) and can be accessed through the link http://www.peptideatlas.org/PASS/PASS00238.
Fig. 1.
Synopsis of lysine acetylation proteomics. A, Map of the MRA_1161 gene knockout, map of the MRA_1161 gene region in the genome and its corresponding region in the conditional mutant strain. MRA_1160 and MRA_1162 are the upstream and downstream regions of the MRA_1161 gene, respectively; res, γδ resolvase site; hyg, hygromycin-resistance gene; sacB, sucrose counterselectable gene. The primers used for PCR amplification are indicated by arrows and the expected sizes of the PCR products are indicated by straight lines. B, PCR verification of the MRA_1161 gene knockout strain. Agarose gel electrophoresis was used to verify the PCR products amplified from the genomic DNA of different mycobacterial strains. M, DNA marker; lane 1, M. tuberculosis H37Ra; lane 2, M. tuberculosis H37Ra ΔMRA_1161; lane 3, M. tuberculosis H37Ra ΔMRA_1161 (hyg-sacB). PCR products are indicated by white arrows. SDS-PAGE C, and immunoblotting D, analyses of protein lysates from the M. tuberculosis H37Ra (lane 1) and MRA_1161 knockout (lane 2) strains; M, protein marker. E, Schematic representation of the sequential steps used for the global profiling of lysine acetylation in M. tuberculosis.
Table I. Comparison of Mycobacterium tuberculosis H37Ra acetylome with other published bacterial acetylomes.
| Organism | Strain | Genome size (Mb) | Genes | Proteins | No. ac-proteins | No. ac-peptides | No. ac-sites | ref |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli | MG1655 | 4.64 | 4496 | 4146 | 91 | 138 | 138 | (21) |
| Escherichia coli | DH10 | 4.69 | 4352 | 4124 | 349 | 1034 | 1070 | (22) |
| Salmonella enterica | G2466 | 4.95 | 4732 | 4525 | 191 | 235 | 261 | (6) |
| Erwinia amylovora | Ea1189/Ea273 | 3.91 | 3712 | 3565 | 96 | 167 | 141 | (23) |
| Bacillus subtilis | 168 | 4.22 | 4422 | 4176 | 185 | 332 | 332 | (24) |
| Thermus thermophilus | HB8 | 2.12 | 2245 | 2238 | 128 | 198 | 201 | (78) |
| Geobacillus kaustophilus | HTA426 | 3.59 | 3655 | 3653 | 114 | 253 | 253 | (79) |
| Mycobacterium tuberculosis | H37Ra | 4.42 | 4084 | 4034 | 137 | 230 | 226 | This work |
Cellular Localization and Functional Annotation of Acetylated Proteins in M. tuberculosis H37Ra
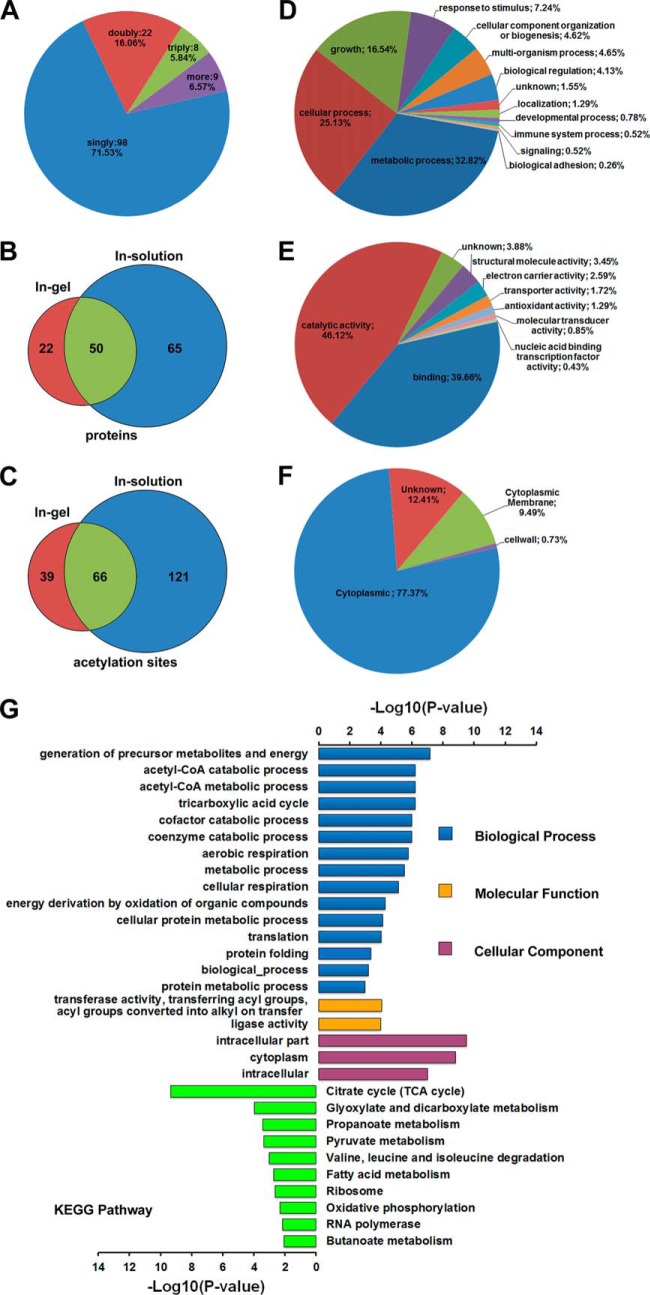
We calculated the number of modified sites per protein in the proteome of M. tuberculosis H37Ra to assess the distribution of acetylation sites. About 71.53% of the acetylated proteins contained only one acetylation site and the remaining acetylated proteins in our dataset were acetylated on multiple sites (Fig. 2A). As shown in a Venn diagram (Fig. 2B and 2C), 105 acetylation sites and 72 acetylated proteins were identified with SDS-PAGE, 187 acetylation sites and 115 acetylated proteins were identified with LC, and 66 acetylation sites and 50 acetylated proteins were detected with both methods (Fig. 2B and 2C). To further investigate, the distributions of the acetylated proteins, we classified the acetylated proteins into groups according to their cellular compartments, molecular functions, and biological processes. The biological processes and molecular functions of the acetylated proteins were assigned based on GO annotations. The 137 acetylated proteins in M. tuberculosis H37Ra are distributed broadly in terms of their biological processes: most of the detected lysine-acetylated proteins were involved in metabolic processes (127 proteins), as has been found in other bacteria: 97 were involved in cellular processes, 64 in growth, and 28 in responses to stimuli. The specific functions of the remaining six hypothetical proteins in terms of their cellular biological processes have not been identified (Fig. 2D). In the GO molecular function category, the acetylated proteins identified were assigned to several groups, including catalytic activity (46.12%), binding (39.66%), and unknown (3.88%). The remaining proteins were assigned to specific functions, such as structural molecules (3.45%), electron carrier activity (2.59), transporter activity (1.72%), antioxidant activity (1.29%), molecular transducer activity (0.85%), and nucleic-acid-binding transcription factors (0.43%) (Fig. 2E). To investigate the distribution of the acetylated proteins across the cellular compartments, we evaluated them using the PSORTb program according to the confidence values for each localization site. Our results show that the majority of acetylated proteins occurred most frequently in the cytoplasm (106); many fewer occurred on the cytoplasmic membrane (13) or the cell wall (1) (Fig. 2F; supplemental Table S3). A GO enrichment analysis was also performed to identify the biological processes and molecular functions in which the acetylated proteins were involved, and to examine the preferential subcellular localizations of the acetylated proteins. Like previous findings (6, 21), our data suggest that the acetylated proteins of M. tuberculosis H37Ra were significantly enriched for several GO biological processes, including the generation of precursor metabolites and energy, acetyl-CoA catabolic processes, metabolic processes, the citrate cycle, acetyl-CoA metabolic processes, cofactor catabolic processes, coenzyme catabolic processes, aerobic respiration, cellular respiration, energy derivation by the oxidation of organic compounds, cellular protein metabolic processes, translation, protein folding, and protein metabolic processes. From the GO enrichment analysis, we also found that the lysine-acetylated proteins were significantly enriched for two specific functions, such as ligase activity (p = 1.06 × 10−4) and transferase activity, the transfer of acyl groups, the conversion of acyl groups to alkyl groups on transfer (p = 9.11 × 10−5). In the GO cellular component categories, we found that a considerable proportion of the acetylated proteins identified were localized intracellularly (p = 3.21 × 10−10): cytoplasm (p = 1.63 × 10−9) and intracellular (p = 1.01 × 10−7) (Fig. 2G; supplemental Table S4). The identified acetyl proteins were also assigned to KEGG metabolic pathways, which indicated that lysine acetylation occurs on many enzymes involved in energy metabolism (Fig. 2G; supplemental Table S5).
Fig. 2.
Overview of the acetylated proteins identified in M. tuberculosis H37Ra. A, A pie chart illustrates the number of lysine acetylation sites identified per protein. B, Numbers of acetylated proteins and C, acetylation sites identified with SDS-PAGE and LC and their overlap are shown in the individual circles of the Venn diagram. D-F, Pie chart representations of the distributions of the acetylated proteins identified according to their biological processes, molecular functions, and cellular localization. E, Enrichment analysis. The differently colored bars indicate the corrected p values for the enrichment of the annotations.
Analysis of Lysine Acetylation Sites
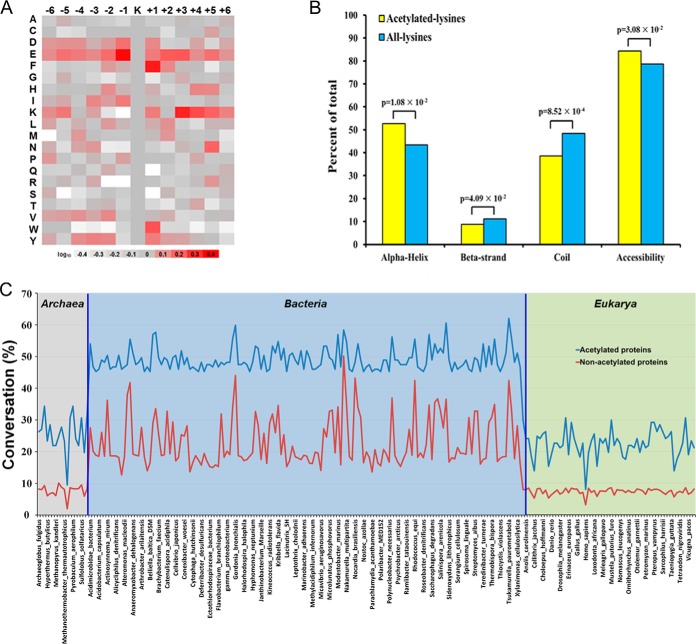
The magnitude of our dataset allowed us to identify site-specific acetylation motifs. We compared the acetylated sites to the nonacetylated lysine residues in this dataset to identify any bias that may occur in the amino acids neighboring the acetylation sites. Thus, we checked the 12 residues flanking the acetylated sites for the overrepresentation of specific amino acids relative to their expression in the proteome background distribution (Fig. 3A). An intensity map was generated as described previously (53). This analysis showed preferences for glutamic acid at the −1 position, phenylalanine at the +1 position, and lysine at the +3 position. This strong bias for a specific acetylation-site motif is quite different from the situation in other representative model organisms. No significant overall sequence recognition motif was detected among the acetylation sites of S. enterica (6) or Erwinia amylovora (23). A strong bias for the flanking amino acid residue at the +1 position was observed in other bacteria, such as the histidine or tyrosine amino acid residue preferred by E. coli (21) and the glutamic acid, aspartic acid, lysine, or proline preferred by B. subtilis (24). In contrast to the bacterial acetylome, there are consensus motifs in the Drosophila and human acetylomes. For example, glycine and glutamic acid are the preferred amino acids beyond the −1 position, whereas tyrosine, phenylalanine, and proline occur frequently at the +1 position in these organisms. Lysine residues at the-5, −6, +3, +4, +5, and +6 positions have also been observed (14). The differences in the preferred amino acid residues flanking the acetylation sites in the M. tuberculosis H37Ra proteins suggest that this modification is catalyzed by a new subset of acetyltransferases, with unique substrate preferences. In contrast to phosphorylation sites, acetylation sites frequently occur in regions with ordered secondary structures, as previously reported (54). For a more detailed investigation of the local secondary structures of the protein sequences surrounding acetylation sites, we compared the secondary structures of acetylated lysines to those of all lysines. In general, the acetylated lysines were more frequently found in structured regions (p = 1.08 × 10−2 for α-helices and p = 4.09 × 10−2 for β-strands) and less frequently in unstructured regions (p = 8.52 × 10−4 for coils) (Fig. 3B). We also evaluated the acetylated lysine sites for solvent accessibility; 84.3% of identified acetylated lysine sites were exposed to the protein surface, compared with 78.7% of all lysine residues (p = 3.08 × 10−2). This result suggests that the highly conserved acetylated sites have local structural preferences, which are quite different from those of phosphorylation sites.
Fig. 3.
Bioinformational analysis of acetylated proteins. A, The plot shows the relative abundances of the amino acids flanking the acetylated lysines; the relative abundances were calculated and are schematically represented on the intensity map. Colors were plotted using the intensity map and represent the log10 ratios of the frequencies within acetyl-13-mers versus the frequencies within non-acetyl-13-mers (red shows enrichment, gray shows depletion). B, Distribution of acetylated and nonacetylated amino acids in the protein secondary structures. The probabilities of acetylated lysines in different secondary structures (α-helices, β-strands, and coils) were compared with the secondary structure probabilities of all lysines in all the proteins identified in this study. C, Conservation of the identified M. tuberculosis H37Ra acetylated proteins relative to the whole proteome conservation among the three domains of life.
To measure acetylome conservation, we aligned the best-scoring homologous proteins from every species with all the identified acetylated proteins and all the proteomes. The orthologues that shared the highest homology with each other in both directions were counted and reported as the percentage of the total number of proteins in all the species analyzed. The percentages of M. tuberculosis H37Ra homologues of the acetylated and nonacetylated proteins in diverse organisms are shown in Fig. 3C. The acetylated proteins of M. tuberculosis H37Ra showed significantly higher conservation across the evolutionary tree, and especially among bacteria, than the nonacetylated proteins. More than half the M. tuberculosis H37Ra acetylated proteins were conserved in other bacterial species. Interestingly, 76 acetylated proteins of M. tuberculosis H37Ra were found to be conserved in other bacterial acetylomes (supplemental Table S6). Because lysine acetylation is an ancient post-translational modification and is also highly conserved throughout all species, we speculate that the functions of the detected acetylated proteins could be essential the mycobacteria.
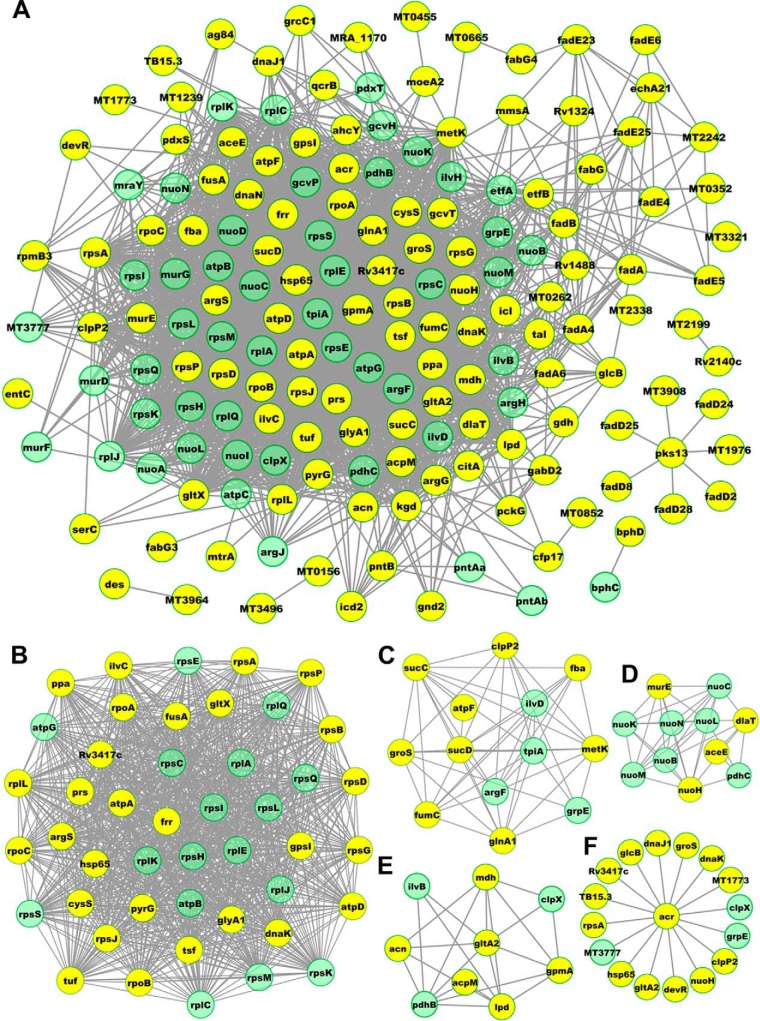
Functional Interaction Networks of Acetylated Proteins in M. tuberculosis H37Ra
Protein interaction networks were established to identify the physical and functional interactions among the identified acetylated proteins using protein interaction information from the STRING database. As shown in Fig. 4, 137 proteins in our data set were mapped to the protein interaction database of M. tuberculosis (supplemental Table S7). The interaction network presents an overview of how acetylated proteins perform various types of functions in M. tuberculosis. Five highly interconnected clusters of acetylated proteins were retrieved using the MCODE algorithm (supplemental Table S8). Interestingly, the first high-ranking complexes we identified (Cluster I) consist of ribosome-associated proteins (Fig. 4B), whereas Clusters II-IV (Fig. 4C–4E) consist of metabolism-associated proteins, especially proteins involved in the citrate cycle. These findings show that protein translation is probably regulated by lysine acetylation in M. tuberculosis, and that most of the acetylated proteins identified are involved in central metabolic pathways responsible for the control of key metabolic processes. Interestingly, one of the known immunogens of M. tuberculosis, Acr (HspX), interacts tightly with other function-associated proteins (Fig. 4F). Together with other acetylation data, these protein clusters will help to decipher the physiological roles of lysine acetylation in M. tuberculosis.
Fig. 4.
Protein interaction network of the M. tuberculosis H37Ra acetylome. A, The complete acetylation interaction network obtained with STRING (9.0) at confidence scores ≥ 0.4. B–F, The five most highly ranked and tightly connected network clusters obtained with an MCODE analysis are color-coded and then rendered as separate modules.
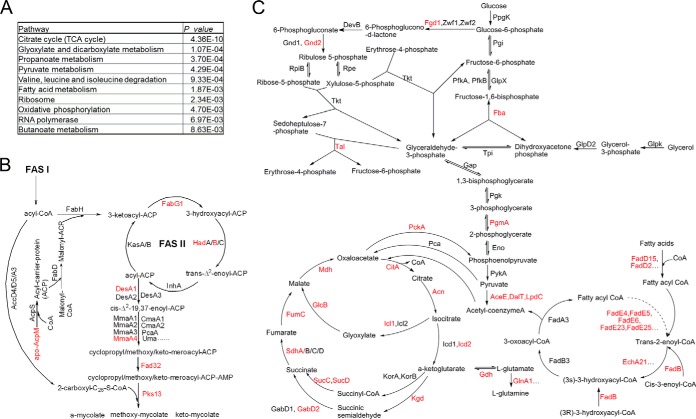
Acetylation of Enzymes Involved in the Central Metabolism and Lipid Metabolism Pathways of M. tuberculosis
It is well established that most enzymes involved in the central metabolic pathways are lysine acetylated in various organisms (6, 20–24). In this study, we found that many of the acetylated proteins identified in M. tuberculosis H37Ra are involved in central metabolic pathways. As shown in Fig. 5A, a large proportion of enzymes that are involved in glycolysis/gluconeogenesis and the citrate cycle are lysine acetylated, which is consistent with our previous understanding of lysine acetylation (6, 20–24). Therefore, lysine acetylation may regulate the central metabolic pathways at multiple sites in M. tuberculosis. Among the enzymes shown to be acetylated in the central metabolic pathways, eight are components of the citrate cycle (12, 55, 56) and two enzymes, isocitrate lyase (ICL-1) and malate synthase G (GlcB), are involved in the glyoxylate pathway. In M. tuberculosis, both copies of the isocitrate-lyase-encoding genes are required for the survival of the bacterium in vivo (57, 58). A structural analysis of ICL showed that the protein has an active-site loop region (residues 185–196), which contains the ICL signature sequence (K189KCGH193) that controls its enzymatic activity (59). The identified acetylated lysine residue (K189) occurs in this region, so the acetylation of this residue may lead to a conformational change in the protein and therefore affect its enzymatic activity. GlcB is a secreted protein and enhances the adherence of the pathogen to lung epithelial cells, so it is also related to the bacterium-host interaction (60).
Fig. 5.
Acetylation of metabolic enzymes in M. tuberculosis H37Ra. A, Central carbon metabolism network in mycobacteria. B, FAS-II fatty acid elongation pathway in mycobacteria. The acetylated metabolic enzymes identified in this proteomic survey are marked in red.
Apart from the enzymes in the citrate cycle and the glyoxylate pathway, many enzymes related to fatty acid metabolism were shown to be acetylated in M. tuberculosis, including several enzymes involved in the β-oxidation of fatty acids and several involved in fatty acid biosynthesis (Fig. 5A and 5B). The enzymes involved in fatty acid metabolism that were shown to be acetylated are significantly enriched for ligase activity and transferase activity (Fig. 2E; supplemental Table S4). In M tuberculosis, fatty acids and their metabolic derivatives are important components of the cell wall complex, which is related to the pathogenicity of the bacterium (61, 62). In fact, it has been reported that reversible acetylation can modulate the activity of several fatty-acid-CoA ligases (FadD2, FadD5, and FadD13) in M. tuberculosis (7), which play important roles in the fatty acid metabolism of the bacterium. Therefore, our proteomic data suggest that acetylation plays an important role in the fatty acid metabolism of M. tuberculosis.
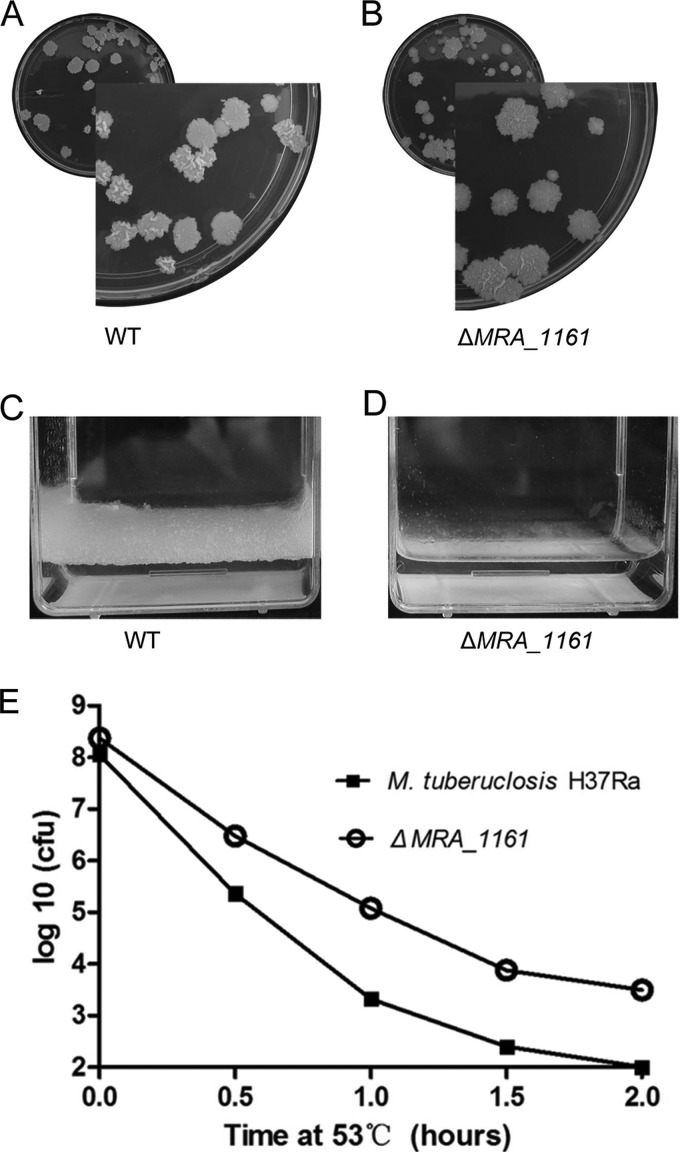
Deletion of MRA_1161 affects Colony Morphology and Biofilm Formation, and Increases Tolerance of Heat Shock Stress in M. tuberculosis H37Ra
Previous studies have shown that the lipid metabolism of mycobacteria correlates with colony morphology and biofilm formation (63–67). About 250 distinct enzymes are involved in the fatty acid metabolism of M. tuberculosis (68), and 25 of these were shown to be acetylated in this study, including two enzymes (FabG1 and HadB) essential to the type II fatty acid synthase (FAS II) system. Very recently, Nambi et al. found that acetylation affects the activity of several fatty-acid-CoA ligases in M. tuberculosis (8). Therefore, we speculate that the lysine acetylation of proteins may affect the colony morphology and biofilm formation of M. tuberculosis H37Ra. Therefore, we deleted the only known copy of the protein-deacetylase-encoding gene, MRA_1161, and showed that its deletion affected the colony morphology of M. tuberculosis H37Ra on solid 7H10 plates. The ΔMRA_1161 mutant appeared to be more granular than its parental strain (Fig. 6A and 6B) and showed defective biofilm formation after 4 weeks of growth in 7H9 medium (Fig. 6C and 6D). However, during the first 2 weeks of growth, its biofilm formation did not differ from that of the control. Our results suggest that lysine acetylation regulates fatty acid metabolism during the late growth stage, which is consistent with a previous report that the lysine acetylation of proteins is more abundant during the stationary phase of E. coli (20). Our results suggest that lysine acetylation affects the colony morphology and biofilm formation by regulating the activities of enzymes involved in the fatty acid metabolism of M. tuberculosis H37Ra.
Fig. 6.
Effects of MRA_1161 deletion in M. tuberculosis H37Ra. A, Colony morphology of M. tuberculosis H37Ra and B, the ΔMRA_1161 mutant. Bacterial cells were plated to generate single colonies on solid medium and incubated for 4 weeks at 37 °C. C, Biofilm formation of M. tuberculosis H37Ra and D, the ΔMRA_1161 mutant. Bacterial cells were grown in 7H9 medium (10% OADC + 0.5% glycerol + 0.05% Tween 80). E, Effects of MRA_1161 deletion on the heat shock response of M. tuberculosis.
Previous research has shown that the deletion of cobB in E. coli affects the stress responses of the bacterium, including its responses to oxidative stress and heat stress (69). Therefore, we tested the effects of MRA_1161 deletion on heat stress in M. tuberculosis H37Ra. In contrast to the wild-type strain, the ΔMRA_1161 mutant showed marked resistance to heat stress (53 °C) (Fig. 6E), similar to the previous observations in E. coli, suggesting that lysine acetylation affects the stress responses of bacteria. Although some stress proteins, including Dnak and GroEL, were shown to be acetylated in M. tuberculosis H37Ra, the mechanism underlying the effect of protein acetylation on the heat stress response remains unclear, and warrants further investigation.
Deletion of MRA_1161 affects Carbon Source Utilization by M. tuberculosis H37Ra
As described above, many enzymes involved in glycolysis/gluconeogenesis, the citrate cycle, and the lipid metabolism pathways of M. tuberculosis H37Ra are lysine acetylated, which is consistent with previous reports of E. coli and S. enterica (6, 20–22). In E. coli, S. enterica, M. bovis BCG, and M. smegmatis, the counterpart of MRA_1161, Pat/CobB, is responsible for the reversible acetylation of those metabolic enzymes, so the deletion of cobB and pat affects the carbon source utilization of these bacteria (6, 8, 26, 70, 71). Therefore, the effect of MRA_1161 deletion on the carbon source utilization of M. tuberculosis H37Ra was analyzed. Our results show that the ΔMRA_1161 mutant strain grew more slowly than the wild-type strain when glucose was used as the sole carbon source (supplemental Fig. S2A), which differs from previous observations of S. enterica (6). This result suggests that, although carbon source utilization is commonly regulated by reversible protein lysine acetylation, the fine mechanisms of this regulation may differ among different species. Further studies are required to determine how reversible protein lysine acetylation regulates carbon source utilization in M. tuberculosis. The effect of MRA_1161 deletion on acetate utilization also differed from that in other bacteria, including S. enterica and E. coli (70, 71). When acetate was used as the sole carbon source, the ΔMRA_1161 mutant showed no marked changes in its growth rate with different concentrations of acetate (5, 10, 30, or 50 mm) (supplemental Fig. S2B–S2E), although previous studies have shown that Rv1151c, an orthologue of MRA_1161, modulates the activity of the acetyl-CoA synthetase of M. tuberculosis in vitro by deacetylation (32). This implies either the existence of other protein deacetylases or that a different mechanism regulates acetate utilization in M. tuberculosis H37Ra.
Effect of Acetylation on the Immunogenicity of a Peptide Derived from HspX
Interestingly, the acetylome data generated in this study showed that 45 secretory proteins are acetylated in M. tuberculosis (supplemental Table S9). In many situations, these secretory proteins can stimulate the immune responses of the host (72). Therefore, we examined whether acetylation affects the immunogenicity of secretory proteins. Among the 45 secretory proteins listed in supplemental Table S9, M. tuberculosis heat-shock protein X (HspX) was shown to be acetylated at multiple sites. As a molecular chaperone, HspX is linked to the latent infection of M. tuberculosis, because its deletion results in increased bacterial growth in both mice and macrophages (73). Moreover, there is evidence that HspX of M. tuberculosis induces an effective immune response in its host, and the immunization of mice with recombinant HspX protein resulted in enhanced levels of IFN-γ. HspX also increases the expression of IFN-γ in TB patients (74–77). Therefore, HspX was selected for further investigation. Because it is difficult to prepare acetylated and deacetylated full-length HspX in vitro, two 30-mer peptides with the same sequence (spanning residues 62–91 of HspX) were synthesized, one with all three lysine residue acetylated (peptide A-6291) and the other with no acetylation (peptide 6291).
In total, 26 blood samples were collected from patients with active TB or patients with suspected TB, and ELISPOT tests were used to compare the immunogenicity of 6291 and A-6291. As shown in Fig. 7, among the 26 samples tested, three (samples 3, 13, and 25) responded positively to peptide 6291. In contrast, the responses to A-6291 were much weaker (sample 13) or no responses were observed (samples 3 and 25) for the same samples. These results suggest that acetylation affects the immunogenicity of the secretory proteins of M. tuberculosis, which warrants further investigation.
Fig. 7.
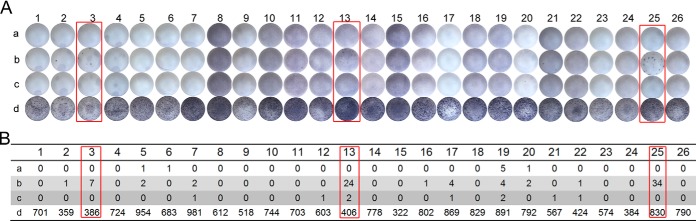
Results of ELISPOT tests. A, Images of the PVDF microfiltration membrane in each well. Each patient was assigned four reaction wells: row a, the no-peptide negative control; row b, peptide 6291; row c, A-6291; row d, the positive control. B, Total number of the spot-forming cells in each well according to A. Subjects 3, 13, and 25 (indicated in red boxes) showed different responses to peptides 6291 and A-6291, indicating the different immunogenicity of the two peptides.
CONCLUSION
In conclusion, our results provide the first extensive data on lysine acetylation in M. tuberculosis. We identified a total of 230 unique acetylated peptides from 137 M. tuberculosis proteins. Further functional studies revealed that the reversible lysine acetylation of proteins is involved in the carbon source utilization, colony morphology, biofilm formation, and stress response of M. tuberculosis, and may also affect the immunogenicity of secretory proteins. Therefore, our results provide novel insight into the range of functions regulated by lysine acetylation in M. tuberculosis. The dataset provided should help to determine the physiological functions affected by lysine acetylation and facilitate the clarification of the entire metabolic networks of M. tuberculosis.
Supplementary Material
Footnotes
Author contributions: X.Z., J.D., and F.G. designed research; F.L., M.Y., X.W., S.Y., and J.G. performed research; J.Z. contributed new reagents or analytic tools; F.L., M.Y., X.W., X.Z., J.D., and F.G. analyzed data; M.Y., J.D., and F.G. wrote the paper.
* This project was supported by the State Key Development Program for Basic Research of China (2012CB518800), the Infectious Disease Control Research Program of the Ministry of Health of China (2013ZX10003006), the Key Program of the Chinese Academy of Sciences (KJZD-EW-L02), and the Hundred Talents Program of the Chinese Academy of Sciences.
 This article contains supplemental Tables S1 to S9 and Figs. S1 and S2.
This article contains supplemental Tables S1 to S9 and Figs. S1 and S2.
1 The abbreviations used are:
- TB
- Tuberculosis
- IFN-γ
- interferon gamma
- cAMP
- cyclic adenosine monophosphate
- FA
- formic acid
- FDR
- false discovery rate
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- GO
- Gene Ontology.
REFERENCES
- 1. Team E. E. (2013) WHO publishes Global tuberculosis report 2013. Eurosurveillance 18, 39–39 [PubMed] [Google Scholar]
- 2. Corbett E. L., Watt C. J., Walker N., Maher D., Williams B. G., Raviglione M. C., Dye C. (2003) The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163, 1009–1021 [DOI] [PubMed] [Google Scholar]
- 3. Sun G., Luo T., Yang C., Dong X., Li J., Zhu Y., Zheng H., Tian W., Wang S., Barry C. E., 3rd, Mei J., Gao Q. (2012) Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J. Infect. Dis. 206, 1724–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mischerikow N., Heck A. J. (2011) Targeted large-scale analysis of protein acetylation. Proteomics 11, 571–589 [DOI] [PubMed] [Google Scholar]
- 5. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 6. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z. B., Zeng R., Xiong Y., Guan K. L., Zhao S., Zhao G. P. (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guan K. L., Xiong Y. (2011) Regulation of intermediary metabolism by protein acetylation. Trends Biochem. Sci. 36, 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nambi S., Gupta K., Bhattacharyya M., Ramakrishnan P., Ravikumar V., Siddiqui N., Thomas A. T., Visweswariah S. S. (2013) Cyclic AMP-dependent protein lysine acylation in mycobacteria regulates fatty acid and propionate metabolism. J. Biol. Chem. 288, 14114–14124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hou J., Cui Z., Xie Z., Xue P., Wu P., Chen X., Li J., Cai T., Yang F. (2010) Phosphoproteome analysis of rat L6 myotubes using reversed-phase C18 prefractionation and titanium dioxide enrichment. J. Proteome Res. 9, 777–788 [DOI] [PubMed] [Google Scholar]
- 10. Starai V. J., Escalante-Semerena J. C. (2004) Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340, 1005–1012 [DOI] [PubMed] [Google Scholar]
- 11. Kim S. C., Sprung R., Chen Y., Xu Y. D., Ball H., Pei J. M., Cheng T. L., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. M. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 12. Zhao S. M., Xu W., Jiang W. Q., Yu W., Lin Y., Zhang T. F., Yao J., Zhou L., Zeng Y. X., Li H., Li Y. X., Shi J., An W. L., Hancock S. M., He F. C., Qin L. X., Chin J., Yang P. Y., Chen X., Lei Q. Y., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lundby A., Lage K., Weinert B. T., Bekker-Jensen D. B., Secher A., Skovgaard T., Kelstrup C. D., Dmytriyev A., Choudhary C., Lundby C., Olsen J. V. (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weinert B. T., Wagner S. A., Horn H., Henriksen P., Liu W. S. R., Olsen J. V., Jensen L. J., Choudhary C. (2011) Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Science Signal. [DOI] [PubMed] [Google Scholar]
- 15. Finkemeier I., Laxa M., Miguet L., Howden A. J. M., Sweetlove L. J. (2011) Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 155, 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu X., Oh M. H., Schwarz E. M., Larue C. T., Sivaguru M., Imai B. S., Yau P. M., Ort D. R., Huber S. C. (2011) Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol. 155, 1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henriksen P., Wagner S. A., Weinert B. T., Sharma S., Bacinskaja G., Rehman M., Juffer A. H., Walther T. C., Lisby M., Choudhary C. (2012) Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol. Cell. Proteomics 11, 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miao J., Lawrence M., Jeffers V., Zhao F. Q., Parker D., Ge Y., Sullivan W. J., Cui L. W. (2013) Extensive lysine acetylation occurs in evolutionarily conserved metabolic pathways and parasite-specific functions during Plasmodium falciparum intraerythrocytic development. Mol. Microbiol. 89, 660–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeffers V., Sullivan W. J. (2012) Lysine acetylation is widespread on proteins of diverse function and localization in the protozoan parasite Toxoplasma gondii. Eukaryot. Cell 11, 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu B. J., Kim J. A., Moon J. H., Ryu S. E., Pan J. G. (2008) The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol. 18, 1529–1536 [PubMed] [Google Scholar]
- 21. Zhang J. M., Sprung R., Pei J. M., Tan X. H., Kim S., Zhu H., Liu C. F., Grishin N. V., Zhao Y. M. (2009) Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia Coli. Mol. Cell. Proteomics 8, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang K., Zheng S. Z., Yang J. S., Chen Y., Cheng Z. Y. (2013) Comprehensive profiling of protein lysine acetylation in Escherichia coli. J. Proteome Res. 12, 844–851 [DOI] [PubMed] [Google Scholar]
- 23. Wu X., Vellaichamy A., Wang D. P., Zamdborg L., Kelleher N. L., Huber S. C., Zhao Y. F. (2013) Differential lysine acetylation profiles of Erwinia amylovora strains revealed by proteomics. J. Proteomics 79, 60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim D., Yu B. J., Kim J. A., Lee Y. J., Choi S. G., Kang S., Pan J. G. (2013) The acetylproteome of Gram-positive model bacterium Bacillus subtilis. Proteomics 13, 1726–1736 [DOI] [PubMed] [Google Scholar]
- 25. Gu J., Deng J. Y., Li R., Wei H. P., Zhang Z. P., Zhou Y. F., Zhang Y., Zhang X. E. (2009) Cloning and characterization of NAD-dependent protein deacetylase (Rv1151c) from Mycobacterium tuberculosis. Biochemistry 74, 743–748 [DOI] [PubMed] [Google Scholar]
- 26. Hayden J. D., Brown L. R., Gunawardena H. P., Perkowski E. F., Chen X., Braunstein M. (2013) Reversible acetylation regulates acetate and propionate metabolism in Mycobacterium smegmatis. Microbiology 159, 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee H. J., Lang P. T., Fortune S. M., Sassetti C. M., Alber T. (2012) Cyclic AMP regulation of protein lysine acetylation in Mycobacterium tuberculosis. Nature structural & molecular biology 19, 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nambi S., Badireddy S., Visweswariah S. S., Anand G. S. (2012) Cyclic AMP-induced Conformational Changes in Mycobacterial Protein Acetyltransferases. Journal of Biological Chemistry 287, 18115–18129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu H., Hegde S. S., Blanchard J. S. (2011) Reversible Acetylation and Inactivation of Mycobacterium tuberculosis Acetyl-CoA Synthetase Is Dependent on cAMP. Biochemistry 50, 5883–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nambi S., Basu N., Visweswariah S. S. (2010) cAMP-regulated Protein Lysine Acetylases in Mycobacteria. J. Biol. Chem. 285, 24313–24323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lange S., Rosenkrands I., Stein R., Andersen P., Kaufmann S. H., Jungblut P. R. (2014) Analysis of protein species differentiation among mycobacterial low-Mr-secreted proteins by narrow pH range Immobiline gel 2-DE-MALDI-MS. J. Proteomics 97, 235–244 [DOI] [PubMed] [Google Scholar]
- 32. Li R., Gu J., Chen P., Zhang Z., Deng J., Zhang X. (2011) Purification and characterization of the acetyl-CoA synthetase from Mycobacterium tuberculosis. Acta Biochim. Biophys. Sin 43, 891–899 [DOI] [PubMed] [Google Scholar]
- 33. Bardarov S., Bardarov S., Jr., Pavelka M. S., Jr., Sambandamurthy V., Larsen M., Tufariello J., Chan J., Hatfull G., Jacobs W. R., Jr. (2002) Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148, 3007–3017 [DOI] [PubMed] [Google Scholar]
- 34. Lee S., Kriakov J., Vilcheze C., Dai Z., Hatfull G. F., Jacobs W. R., Jr. (2004) Bxz1, a new generalized transducing phage for mycobacteria. FEMS Microbiol. Lett. 241, 271–276 [DOI] [PubMed] [Google Scholar]
- 35. Yang M. K., Qiao Z. X., Zhang W. Y., Xiong Q., Zhang J., Li T., Ge F., Zhao J. D. (2013) Global phosphoproteomic analysis reveals diverse functions of serine/threonine/tyrosine phosphorylation in the model Cyanobacterium Synechococcus sp. strain PCC 7002. J. Proteome Res. [DOI] [PubMed] [Google Scholar]
- 36. Wang L. H., Li D. Q., Fu Y., Wang H. P., Zhang J. F., Yuan Z. F., Sun R. X., Zeng R., He S. M., Gao W. (2007) pFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry. Rapid Commun. Mass Spectrometry 21, 2985–2991 [DOI] [PubMed] [Google Scholar]
- 37. Li D., Fu Y., Sun R., Ling C. X., Wei Y., Zhou H., Zeng R., Yang Q., He S., Gao W. (2005) pFind: a novel database-searching software system for automated peptide and protein identification via tandem mass spectrometry. Bioinformatics 21, 3049–3050 [DOI] [PubMed] [Google Scholar]
- 38. Nesvizhskii A. I., Vitek O., Aebersold R. (2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 [DOI] [PubMed] [Google Scholar]
- 39. Elias J. E., Haas W., Faherty B. K., Gygi S. P. (2005) Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat.Methods 2, 667–675 [DOI] [PubMed] [Google Scholar]
- 40. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 41. Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., Mann M. (2008) Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7, 299–307 [DOI] [PubMed] [Google Scholar]
- 42. Macek B., Mijakovic I., Olsen J. V., Gnad F., Kumar C., Jensen P. R., Mann M. (2007) The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6, 697–707 [DOI] [PubMed] [Google Scholar]
- 43. Zhang K., Yau P. M., Chandrasekhar B., New R., Kondrat R., Imai B. S., Bradbury M. E. (2004) Differentiation between peptides containing acetylated or tri-methylated lysines by mass spectrometry: an application for determining lysine 9 acetylation and methylation of histone H3. Proteomics 4, 1–10 [DOI] [PubMed] [Google Scholar]
- 44. Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., Mann M. (2008) Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7, 299–307 [DOI] [PubMed] [Google Scholar]
- 45. Maere S., Heymans K., Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 46. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 48. Stewart G. R., Snewin V. A., Walzl G., Hussell T., Tormay P., O'Gaora P., Goyal M., Betts J., Brown I. N., Young D. B. (2001) Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat. Med. 7, 732–737 [DOI] [PubMed] [Google Scholar]
- 49. Hu L. I., Lima B. P., Wolfe A. J. (2010) Bacterial protein acetylation: the dawning of a new age. Mol. Microbiol. 77, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li A., Xue Y., Jin C., Wang M., Yao X. (2006) Prediction of Nepsilon-acetylation on internal lysines implemented in Bayesian Discriminant Method. Biochem. Biophys. Res. Commun. 350, 818–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Farrah T., Deutsch E. W., Omenn G. S., Campbell D. S., Sun Z., Bletz J. A., Mallick P., Katz J. E., Malmstrom J., Ossola R., Watts J. D., Lin B., Zhang H., Moritz R. L., Aebersold R. (2011) A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol. Cell. Proteomics 10, M110 006353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Desiere F., Deutsch E. W., King N. L., Nesvizhskii A. I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S. N., Aebersold R. (2006) The PeptideAtlas project. Nucleic Acids Res. 34, D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Treeck M., Sanders J. L., Elias J. E., Boothroyd J. C. (2011) The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites' boundaries. Cell Host Microbe 10, 410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Iakoucheva L. M., Radivojac P., Brown C. J., O'Connor T. R., Sikes J. G., Obradovic Z., Dunker A. K. (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32, 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ye H., Rouault T. A. (2010) Human iron-sulfur cluster assembly, cellular iron homeostasis, and disease. Biochemistry 49, 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim E. Y., Kim W. K., Kang H. J., Kim J. H., Chung S. J., Seo Y. S., Park S. G., Lee S. C., Bae K. H. (2012) Acetylation of malate dehydrogenase 1 promotes adipogenic differentiation via activating its enzymatic activity. J. Lipid Res. 53, 1864–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McKinney J. D., Honer zu Bentrup K., Munoz-Elias E. J., Miczak A., Chen B., Chan W. T., Swenson D., Sacchettini J. C., Jacobs W. R., Jr., Russell D. G. (2000) Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738 [DOI] [PubMed] [Google Scholar]
- 58. Honer Zu Bentrup K., Miczak A., Swenson D. L., Russell D. G. (1999) Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J. Bacteriol. 181, 7161–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sharma V., Sharma S., Hoener zu Bentrup K., McKinney J. D., Russell D. G., Jacobs W. R., Jr., Sacchettini J. C. (2000) Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis. Nat. Structural Biol. 7, 663–668 [DOI] [PubMed] [Google Scholar]
- 60. Kinhikar A. G., Vargas D., Li H., Mahaffey S. B., Hinds L., Belisle J. T., Laal S. (2006) Mycobacterium tuberculosis malate synthase is a laminin-binding adhesin. Mol. Microbiol. 60, 999–1013 [DOI] [PubMed] [Google Scholar]
- 61. Guenin-Mace L., Simeone R., Demangel C. (2009) Lipids of pathogenic Mycobacteria: contributions to virulence and host immune suppression. Transbound Emerg. Dis. 56, 255–268 [DOI] [PubMed] [Google Scholar]
- 62. Banerjee R., Vats P., Dahale S., Kasibhatla S. M., Joshi R. (2011) Comparative genomics of cell envelope components in mycobacteria. PLoS One 6, e19280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cox J. S., Chen B., McNeil M., Jacobs W. R., Jr. (1999) Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402, 79–83 [DOI] [PubMed] [Google Scholar]
- 64. Ghosh S., Indi S. S., Nagaraja V. (2013) Regulation of Lipid Biosynthesis, Sliding Motility, and Biofilm Formation by a Membrane-Anchored Nucleoid-Associated Protein of Mycobacterium tuberculosis. J. Bacteriol. 195, 1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Billman-Jacobe H., McConville M. J., Haites R. E., Kovacevic S., Coppel R. L. (1999) Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol. Microbiol. 33, 1244–1253 [DOI] [PubMed] [Google Scholar]
- 66. Recht J., Kolter R. (2001) Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J. Bacteriol. 183, 5718–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ojha A. K., Baughn A. D., Sambandan D., Hsu T., Trivelli X., Guerardel Y., Alahari A., Kremer L., Jacobs W. R., Hatfull G. F. (2008) Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol. Microbiol. 69, 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537-+. [DOI] [PubMed] [Google Scholar]
- 69. Ma Q., Wood T. K. (2011) Protein acetylation in prokaryotes increases stress resistance. Biochem. Biophys. Res. Commun. 410, 846–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Starai V. J., Takahashi H., Boeke J. D., Escalante-Semerena J. C. (2003) Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li R., Gu J., Chen Y. Y., Xiao C. L., Wang L. W., Zhang Z. P., Bi L. J., Wei H. P., Wang X. D., Deng J. Y., Zhang X. E. (2010) CobB regulates Escherichia coli chemotaxis by deacetylating the response regulator CheY. Mol. Microbiol. 76, 1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nicod L. P. (2007) Immunology of tuberculosis. Swiss Medical Weekly 137, 357–362 [DOI] [PubMed] [Google Scholar]
- 73. Hu Y. M., Movahedzadeh F., Stoker N. G., Coates A. R. M. (2006) Deletion of the Mycobacterium tuberculosis alpha-crystallin-like hspX gene causes increased bacterial growth in vivo. Infection Immunity 74, 861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Smet D. (1998) Human T-and B-Cell Reactivity to the 16 kDa α-Crystallin Protein of Mycobacterium tuberculosis. Scand. J. Immunol. 48, 403–409 [DOI] [PubMed] [Google Scholar]
- 75. Khera A., Singh R., Shakila H., Rao V., Dhar N., Narayanan P. R., Parmasivan C. N., Ramanathan V. D., Tyagi A. K. (2005) Elicitation of efficient, protective immune responses by using DNA vaccines against tuberculosis. Vaccine 23, 5655–5665 [DOI] [PubMed] [Google Scholar]
- 76. Caccamo N., Barera A., Di Sano C., Meraviglia S., Ivanyi J., Hudecz F., Bosze S., Dieli F., Salerno A. (2003) Cytokine profile, HLA restriction and TCR sequence analysis of human CD4+ T clones specific for an immunodominant epitope of Mycobacterium tuberculosis 16-kDa protein. Clin. Exp. Immunol. 133, 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Geluk A., Lin M. Y., van Meijgaarden K. E., Leyten E. M., Franken K. L., Ottenhoff T. H., Klein M. R. (2007) T-cell recognition of the HspX protein of Mycobacterium tuberculosis correlates with latent M. tuberculosis infection but not with M. bovis BCG vaccination. Infection Immunity 75, 2914–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Okanishi H., Kim K., Masui R., Kuramitsu S. (2013) Acetylome with structural mapping reveals the significance of lysine acetylation in Thermus thermophilus. J. Proteome Res. 12, 3952–3968 [DOI] [PubMed] [Google Scholar]
- 79. Lee D. W., Kim D., Lee Y. J., Kim J. A., Choi J. Y., Kang S., Pan J. G. (2013) Proteomic analysis of acetylation in thermophilic Geobacillus kaustophilus. Proteomics 13, 2278–2282 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.