Abstract
The dysregulation of protein oxidative post-translational modifications has been implicated in stress-related diseases. Trx1 is a key reductase that reduces specific disulfide bonds and other cysteine post-translational modifications. Although commonly in the cytoplasm, Trx1 can also modulate transcription in the nucleus. However, few Trx1 nuclear targets have been identified because of the low Trx1 abundance in the nucleus. Here, we report the large-scale proteomics identification of nuclear Trx1 targets in human neuroblastoma cells using an affinity capture strategy wherein a Trx1C35S mutant is expressed. The wild-type Trx1 contains a conserved C32XXC35 motif, and the C32 thiol initiates the reduction of a target disulfide bond by forming an intermolecular disulfide with one of the oxidized target cysteines, resulting in a transient Trx1–target protein complex. The reduction is rapidly consummated by the donation of a C35 proton to the target molecule, forming a Trx1 C32-C35 disulfide, and results in the concurrent release of the target protein containing reduced thiols. By introducing a point mutation (C35 to S35) in Trx1, we ablated the rapid dissociation of Trx1 from its reduction targets, thereby allowing the identification of 45 putative nuclear Trx1 targets. Unexpectedly, we found that PSIP1, also known as LEDGF, was sensitive to both oxidation and Trx1 reduction at Cys 204. LEDGF is a transcription activator that is vital for regulating cell survival during HIV-1 infection. Overall, this study suggests that Trx1 may play a broader role than previously believed that might include regulating transcription, RNA processing, and nuclear pore function in human cells.
Oxidative stress and redox signaling imbalance have been implicated in the development of neurodegenerative diseases and tissue injuries (1). One of the most common features observed in the neuronal tissues of patients with Alzheimer or Parkinson disease is the accumulation of misfolded proteins with oxidative post-translational modifications (2). Cells have evolved to utilize diverse defense mechanisms to counter the detrimental impact of oxidative post-translational modifications, including the engagement of the thioredoxin (Trx)1 family of proteins, which includes cytosolic Trx1 and mitochondrial Trx2 in mammalian cells.
Trxs are evolutionarily conserved antioxidants found in a variety of organisms, including bacteria, yeast, plants, and mammals. Trx is an oxidoreductase enzyme containing a dithiol-disulfide active site. The Trx1 reduction of oxidized protein thiol groups is coupled with the oxidation of Trx1, and the oxidized Trx1 is recycled by Trx reductase. NADPH is required as an electron donor during the reduction by Trx reductase. In mice, the loss of Trx1 results in an early stage embryonic-lethal phenotype, whereas transgenic mice overexpressing Trx1 are more resistant to a variety of oxidative stresses, including infection and inflammation (3) and focal ischemic brain damage (4). Interestingly, the median life span of the Trx transgenic mice was extended by up to 135% relative to that of the controls (5). Among redox regulatory proteins, Trx1 is a unique reducing enzyme because it regulates only specific cysteines within select target proteins. Because Trx1 itself has a limited ability to scavenge reactive oxygen species, the biological functions of Trx1 must be mediated by its target proteins. Trx1 has been shown to regulate DNA synthesis; interact with proteins related to oxidative stress, cell proliferation, and apoptosis, such as TXNIP (6), ASK1 (7), glucocorticoid receptor (8), and SENP1 (9); and facilitate nerve growth factor–mediated neurite outgrowth (10). Trx1 overexpression in rat cardiomyocytes revealed that Trx1 has a dual role as both an antioxidant and a signaling molecule involved in the development of cardiac hypertrophy (11). In E47 cells, the knockdown of Trx1 partially sensitizes the cells to oxidative stress via the ASK-1 and JNK1 signaling pathways (12). In addition to being a cytosolic disulfide reductase in the cytosol, Trx1 also translocates to the nucleus and regulates the functions of specific nuclear proteins (2), including nuclear factor κ B (NFκB) (13), activator protein 1 (13), and histone deacetylase 4 (14). In contrast to the broad understanding of Trx1 in regulating cytosolic signal transduction pathways (15–17), the significance of Trx1 in the nucleus is well documented but poorly understood (18), perhaps because of the relatively transient association of Trx1 with its targets and the relatively low levels of nuclear targets.
In this study, we identified novel human nuclear Trx1 targets using a substrate-trapping mutant of Trx1. In the past, proteomics approaches have been used to identify Trx1 targets. Fu et al. used the isotope-coded affinity tag approach and identified more than 50 cardiac Trx1 targets from a transgenic mouse model overexpressing Trx1 (15). More recently, Benhar et al. coupled the biotin switch technique with stable isotope labeling by amino acids in cell culture (SILAC) to identify 46 substrates of Trx1-mediated denitrosylation (19). However, only a small number of the Trx1 targets identified in the previous global proteomics studies have been nuclear proteins. Nuclear proteins that are sensitive to oxidative post-translational modifications and Trx1 reduction are likely to play important roles in maintaining neuronal cell functions in the presence of oxidative stress during aging and neurodegenerative diseases. The identification of novel nuclear Trx1 targets could lead to the discovery of key transcription factors, RNA processing enzymes, and nuclear structural components that are important for cell survival and an effective anti-stress response. We previously optimized a method to enrich the nuclear proteins from neuronal cells for proteome-wide studies (20). In this study, in order to obtain Trx1 target protein complexes stable enough for the proteomic identification of Trx1 targets, we expressed a Trx1C35S mutant engineered to contain an additional nuclear targeting signal in SHSY-5Y neuroblastoma cells. This enabled us to efficiently affinity capture the nuclear proteins linked to Trx1 via stable mixed disulfides. We hypothesized that more Trx1 targets would be released from the protein complexes associated with Trx1C35S rather than the wild-type Trx1 (Trx1wt) with β-mercaptoethanol. Using a subtractive proteomics approach, we identified 45 putative human nuclear Trx1 targets among the 1500+ affinity-captured proteins. Many of the putative targets are involved in the regulation of RNA transport and processing, nuclear protein import and export, nuclear pore function, and cell signaling. To validate the effectiveness of this approach, we analyzed both known and newly discovered nuclear Trx1 targets via Western blotting. Here, we report the functional validation of PSIP1 as a Trx1-regulated transcriptional regulator; Trx1 directly reduces PSIP1 at Cys 204. PSIP1 is best known as a host protein involved in HIV integration. The interaction between PSIP1 and HIV integrase is under intense study, and PSIP1 is currently being considered as a target for the development of novel anti-HIV therapies. Elevated Trx1 has also been reported in the plasma of HIV-infected patients (21). The discovery of PSIP1 as a Trx1 target in humans might provide novel insights to aid in the design of more effective anti-HIV treatments, perhaps even for neuro-AIDS symptoms. Overall, the identification and investigation of novel nuclear Trx1 targets might provide a foundation for a better understanding of the redox signal pathways involved in neurodegenerative diseases and the development of novel therapeutic approaches.
MATERIALS AND METHODS
Chemicals and Reagents
HPLC-grade acetonitrile (ACN) and water were purchased from Mallinckrodt Baker, Inc. (Phillipsburg, NJ). Triethylammonium bicarbonate, protease inhibitor mixture, and phosphatase inhibitor mixture were purchased from Sigma (St. Louis, MO). Sequencing-grade trypsin was obtained from Promega (Madison, WI). PepClean C18 spin columns were purchased from Pierce (Rockford, IL). Recombinant human PSIP1 (3468-LE, R&D Systems, Minneapolis, MN) and human Trx1 (T8690, Sigma) were used in this study. Anti-PSIP1 (ab110023, Abcam, Cambridge, MA) and anti-GFP (a11122, Invitrogen, Grand Island, NY) antibodies were used in the Western blotting.
Plasmids and Constructs
The human Trx1-containing nuclear localization sequence (PPKKKRKVEDP) was subcloned into a pcDNA3-EGFP plasmid encoding Trx1WT (nTrx1WT) (Figs. 1A and 1B). A site-directed mutagenesis was performed on the nTrx1WT construct to introduce the C35S mutation into the Trx1 redox active site to generate nTrx1C35S using the Gene Tailor site-directed mutagenesis kit (Invitrogen). GFP was used as a tag for the Western blotting, imaging, and immunoprecipitation (IP). The mammalian expression vector for human PSIP1, pCMV-SPORT6-PSIP1, was purchased from Mammalian Gene Collection (Pittsburgh, PA). A minimal promoter luciferase reporter was generated through the insertion of a minimal promoter sequence encoding the stress response element (STRE) from human vascular endothelial growth factor C (VEGF-C) into the XhoI and HindIII sites of pGL3Basic (Promega) to generate pSTRE-luc. The oligonucleotides that were used for the STRE sequence were 5′- CTAGCCTTCGGGGAAGGGGAGGGAGGAGGGGGACGAGGC-3′ and 5′- TCGAGCCTCGTCCCCCTCCTCCCTCCCCTTCCCCGAAGG-3′.
Fig. 1.
A strategy to enrich the Trx1 nuclear targets. A schematic diagram of the constructs for the nuclear-targeted Trx1 (nTrx1WT) with a C-terminal GFP tag (A) and the nTrx1C35S mutant (B). The Trx1-containing nuclear localization sequence (NLS) (PPKKKRKVEDP) was subcloned into a pcDNA3-EGFP plasmid encoding Trx1wt and Trx1C35S. C, the experimental workflow used for the identification of the Trx1 target proteins. SHSY-5Y cells were transfected with nTrx1WT or nTrx1C35S for 48 h, and the nuclear proteins were isolated for GFP IP. The eluted proteins were separated on SDS gels and identified via LC-MS/MS. D, a schematic diagram showing how the nTrx1C35S mutant traps target proteins. nTrx1WT can reduce oxidized proteins via a redox switch shown in the diagram, and nTrx1C35S binds to target proteins via mixed disulfide bonds but cannot consummate the reduction and release of the target proteins. Therefore, the disulfide-linked proteins can be isolated for proteomic identification.
Cell Culture and Transfection
The human neuroblastoma cell line SHSY-5Y was obtained from ATCC (Manassas, VA). The cells were cultured in a 1:1 mixture of Dulbecco's modified Eagle's medium and F12 medium supplemented with 0.1 mm nonessential amino acids, 1% penicillin/streptomycin, and 10% fetal bovine serum and incubated at 37 °C in 5% CO2. Exponentially growing and nearly confluent cells were harvested after several passages and washed twice with PBS. The SHSY-5Y cells were transfected with the nTrx1WT and nTrx1C35S plasmids using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Forty-eight hours after the transfection, the cells were harvested via centrifugation at 500 × g for 5 min and washed with PBS prior to the subsequent analyses. Three independent cell cultures and subsequent biochemical experiments were conducted to ensure the reproducibility of the identification of the Trx1 targets (Fig. 1C).
Nuclear Protein Extraction
Nuclear extracts were prepared from the SHSY-5Y cells using a modified procedure developed in our lab (20). Briefly, PBS-washed cells were gently suspended in a hypotonic lysis buffer consisting of 10 mm HEPES (pH 7.9), 10 mm KCl, 1.5 mm MgCl2, 0.5 mm DTT, protease inhibitors, and phosphatase inhibitors. The mixture was incubated on ice for 15 min, and then we added 0.5% Nonidet P-40. The cellular extract was then centrifuged at 800 × g for 10 min at 4 °C to separate the cytoplasmic components (supernatant) from the nuclei-enriched fraction (pellet). For the purification of the nuclear proteins, we washed the nuclear pellets two additional times with a freshly prepared hypotonic buffer and Nonidet P-40 to thoroughly remove any cytoplasmic proteins that were loosely associated with the nuclear pellets. The nuclear pellets were then suspended in a hypertonic buffer (20 mm HEPES (pH 7.9), 1.5 mm MgCl2, 420 mm NaCl, 25% v/v glycerol, 0.5 mm DTT, 0.2 mm EDTA, and protease and phosphatase inhibitor cocktails). The nuclear proteins were extracted via vigorous agitations for 15 min on ice. The solutions were further sonicated three times at 10-s intervals on ice. The resulting solutions were centrifuged at 16,000 × g at 4 °C for 15 min. The supernatants containing the solubilized nuclear proteins were stored at −80 °C until further analysis.
Immunoprecipitation and Western Blotting
Protein lysates were prepared from the cells transfected with the nTrx1WT and nTrx1C35S plasmids and then subjected to nuclear protein extraction. For the IP of the Trx1 association proteins, 500 μg of each nuclear extract was incubated with an anti-GFP antibody (A11122, Invitrogen) overnight at 4 °C; then 100 μl of protein-A agarose beads (15918–014, Invitrogen) were added and the incubation continued for 1 h with frequent mixing. The beads were then washed three times with a diluted lysis buffer (10 mm Tris, 25 mm NaCl, 0.5% Triton X-100, 25 mm EDTA), with 2 min of centrifugation at 5000 × g between each wash step. The proteins were eluted from the beads with a 2× loading buffer (100 mm Tris (pH 6.8), 4% SDS, 0.2% bromphenol blue, 20% glycerol with 5% β-mercaptoethanol (2-ME, Bio-Rad Laboratories, Hercules, CA)) and incubated on a heat block at 100 °C for 5 min. After centrifugation at 10,000 × g for 5 min, the proteins were resolved on 13.5% SDS-polyacrylamide gels and either were stained with Sypro Ruby dye (Molecular Probes, Grand Island, NY) overnight after being fixed with 40% methanol and 10% acetic acid for 30 min or were processed for Western blotting. The protein gel bands were excised for trypsin digestion and protein identification via tandem mass spectrometry, as described below. For the Western blot detection of the specific proteins, the proteins immunoprecipitated using anti-GFP were separated on a 13.5% SDS-PAGE gel and transferred onto nitrocellulose membranes. The membranes were first blocked with 5% milk and then probed with anti-Prx1 (1:5000, ab41906, Abcam), anti-P53 (9282, Cell Signaling Technology, Danvers, MA; 1:1000), anti-NF-κB (sc-372, Santa Cruz Biotechnology; 1:1000) or anti-PSIP1 (ab110023, Abcam) and visualized with a secondary antibody coupled to an enhanced chemiluminescent substrate (PerkinElmer).
In-gel Digestion and Mass Spectrometry
The Sypro Ruby–stained protein gel bands were excised, diced into 1-mm3 pieces, and washed with 30% ACN in 50 mm ammonium bicarbonate. After DTT reduction and iodoacetamide (IAA) alkylation, the gel pieces were subjected to trypsin digestion. The resulting peptides were extracted with 30% ACN in 1% trifluoroacetic acid and desalted through C18 spin columns. The LC-MS/MS was performed on an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) coupled with a Dionex Chromatography System equipped with an UltimateTM 3000 autosampler through a Proxeon nano-electrospray ion source. The desalted peptides were first trapped on a cartridge (Pepmap C18, 0.5 cm × 300 μm, Dionex) at 2% mobile phase B (mobile phase A, 2% ACN and 0.1% formic acid; mobile phase B, 85% ACN and 0.1% formic acid) at a flow rate of 30 μl/min. The peptides were resolved on a 75-μm × 150-mm capillary Acclaim PepMap100 column (C18, 3 μm, 100 Å, Dionex) using either a 75-min or a 180-min gradient (3% to 45% B) at a flow rate of 250 nL/min. The eluted peptides were ionized at 2.1 kV. The capillary temperature was 275 °C. The mass spectrometer was operated in a data-dependent mode. Full scans of the MS spectra (from m/z 300–2000) were acquired in the Orbitrap analyzer at a resolution of 60,000 at an m/z of 400, with the lock mass option enabled. The 10 most intense peptide ions with charge states of ≥2 were sequentially isolated to a target value of 10,000 and fragmented in the linear ion trap by means of collision-induced dissociation with a normalized collision energy of 30%. The ion-selection threshold was set at 3000 counts for the MS/MS. The maximum ion accumulation time was 100 ms for the full scans in the Orbitrap and 25 ms for the collision-induced dissociation measurements in the LTQ.
Database Search and Bioinformatics
Each of the raw files was analyzed using the Thermo Proteome Discoverer (v. 1.3) platform with Mascot (2.4.1) as a search engine against all of the human protein sequences (150,354 entries) in the UniRef100 protein database (downloaded on January 24, 2014; 33,613,081 entries). The following Mascot search parameters were used: trypsin; two missed cleavages; precursor mass tolerance, 10 ppm; fragment mass tolerance, 0.5 Da; dynamic modifications, methionine oxidation and carbamidomethylation of cysteines. The decoy search option for Mascot was engaged. The Mascot search results (.msf files) were further filtered and compiled into a list of non-redundant proteins with Scaffold (v. 4.2.1, Proteome Software, Portland, OR). The peptide identifications were accepted if they could be established at greater than 99.0% probability to achieve a false discovery rate of less than 1.0% with the Peptide Prophet algorithm (22), with a Scaffold delta-mass correction. The protein identifications were accepted if they could be established at a false discovery rate of less than 1.0%. The protein probabilities were assigned by the Protein Prophet algorithm (23). Proteins that contained similar peptides and could not be differentiated based on the MS/MS analysis alone were grouped to satisfy the principles of parsimony. The identified proteins were uploaded for use with Ingenuity Pathway Analysis (IPA) software to define their putative subcellular localization and function.
Fluorescence Microscopy
SHSY-5Y cells were transfected with the nTrx1WT and nTrx1C35S plasmids. Two days after the transfection, the cells were fixed in 4% paraformaldehyde in PBS for 15 min and then permeabilized with 0.1% Triton X-100 for 5 min. The cells were then incubated with either anti-PSIP1 or anti-GFP antibody overnight at 4 °C. After being washed three times with phosphate buffer saline tween (PBST), the cells were incubated with a Cy5-conjugated anti-mouse antibody (ab6563, Abcam) for 1 h. The cells were then washed in PBS and mounted with ProLong Gold anti-fade reagent with DAPI (Invitrogen). The cells were then imaged using a Nikon A1R microscope, and the images were processed using Nikon NIS Elements software.
Redox Assay of PSIP1
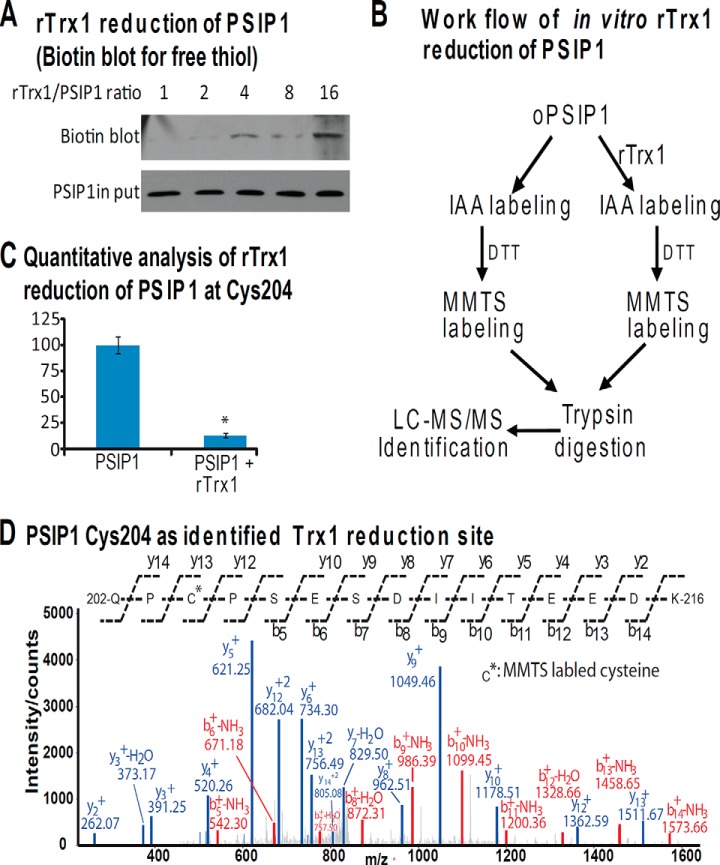
To obtain oxidized PSIP1, we oxidized recombinant human PSIP1 proteins with 100 mm H2O2 for 30 min at 37 °C. The reduced Trx1 (rTrx1) was obtained by reduction of Trx1 with 100 mm DTT for 30 min at 37 °C followed by an acetone precipitation to remove the residual DTT, and the protein pellet was further washed with 80% cold acetone three times. The solubilized PSIP1 was mixed with rTrx1 in a 1 to 0, 1, 2, 4, 8, or 16 molar ratio for 30 min at 37 °C. Subsequently, the free PSIP1 thiols were alkylated with N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)-propionamide (biotin-HPDP). The biotinylated PSIP1 was detected via Western blotting using an anti-biotin antibody (Vector Lab Inc., Burlingame, CA). For the reduced PSIP1 site identification following the rTrx1 treatment, the PSIP1 was treated with rTrx1 or buffer for 30 min at 37 °C; and this was followed by an alkylation of the free thiols with 10 mm IAA for 60 min at 37 °C. The excess IAA was quenched by the addition of 50 mm DTT, and the oxidized cysteines in PSIP1 were also reduced during the process. The residual DTT was removed from the proteins by acetone precipitation, and the protein pellets were further washed with 80% cold acetone three times. The newly freed thiols (previously oxidized) were conjugated with 50 mm methyl methanethiosulfonate. The proteins were digested with trypsin, and the resulting peptides were identified via LC-MS/MS. Trx1 reduction results in less oxidized thiols in PSIP1. The relative quantification of the methyl methanethiosulfonate–labeled peptides was performed after normalization with the top three most abundant PSIP1 peptides without cysteine modifications (n = 3 for the statistical analysis).
Luciferase Assay of PSIP1 Target Gene Expression
HeLa cells and SHSY-5Y neuroblastoma cells were transfected with a luciferase reporter construct with a PSIP1 response element derived from a minimal STRE promoter on human VEGF-C (Pierce) or co-transfected with PSIP1, Trx1, or Trx1 and PSIP1 for 48 h. The cells were washed with PBS three times prior to the luciferase activity assay (Pierce), which was performed according to the manufacturer's instructions.
Statistical Analyses
The data are expressed as means + S.E. The statistical analysis was performed using a two-tailed unpaired Student's t test with Excel. Differences with a p value < 0.05 were considered significant.
RESULTS
Identification of the Putative Nuclear Trx1 Targets in Human Neuroblastoma Cells
The overall strategy for using nTrx1C35S to trap nuclear Trx1 target proteins and to isolate them via IP is depicted in Fig. 1. In the cells expressing nTrx1WT, rTrx1 can be produced by a Trx reductase from oxidized Trx1 with the reducing equivalents derived from NADPH. rTrx1 can then form a mixed disulfide bond with one of the oxidized thiols in a target protein via the Cys32 in its catalytic site (Fig. 1D). This covalent complex can then immediately be converted to a reduced target when Cys35 donates its proton, resulting in oxidized Trx1. This process is very fast; therefore little or no mixed disulfide complex is typically observed in the cells. In order to capture the mixed disulfide complexes and identify the Trx1 targets, we constructed a Trx1 mutant in which the Cys35 was changed to a serine; nTrx1C35S cannot donate its proton to consummate the complete target reduction reaction, but Cys32 can still form mixed disulfides with the target proteins. Because there is no further target reduction by Ser35, the target proteins are trapped within the mixed disulfide complexes, providing an opportunity to isolate these complexes for proteomic analysis. We constructed nTrx1C35S with a nuclear targeting signal (for stable nuclear expression) and a GFP tag on the C terminus (Fig. 1B) for the isolation of stable nuclear Trx1-targeted protein complexes and efficient proteomic identification (Fig. 1C). As a control, we expressed nTrx1WT in separate cells. Following the successful expression of nTrx1C35S and nTrx1WT in the SHSY-5Y cells, we used a method optimized in our lab for nuclear protein enrichment in neuroblastoma cells (20) and an anti-GFP antibody to pull down the Trx1–target complexes. After the unbound proteins had been washed, the target proteins from the mixed disulfide-linked covalent complexes were eluted via a 2-ME reduction. To maximize the number of target proteins identified, we first fractionated the proteins using SDS-PAGE and analyzed the gel bands via LC-MS/MS analyses on an Orbitrap Velos tandem mass spectrometer (Fig. 1C). To ensure the reproducible identification of the nuclear Trx1 targets, we performed multiple biological-repeat experiments wherein we identified a total 1554 unique proteins. We identified 1426 proteins from only the nTrx1C35S complexes, and 784 proteins were identified from only the nTrx1WT complexes (supplemental Table S1 (A–L)). As expected, more unique proteins (Fig. 2A) and peptides (Fig. 2B) were identified in the nTrx1C35S complexes than in the nTrx1WT complexes. In one representative experiment, 1024 unique proteins were identified in the nTrx1C35S complexes, compared with 591 proteins identified in the nTrx1WT complexes, with 564 proteins associated with both (Fig. 2A). Using the proteins associated with nTrx1WT as controls to be subtracted from the specific Trx1 redox target proteins trapped by nTrx1C35S, we found 91 proteins associated with only nTrx1C35S and not with the nTrx1WT complexes in at least two out of the three repeat experiments (supplemental Table S2). Using IPA software to map the known subcellular localization of these proteins, we identified 47 proteins as canonical nuclear proteins, compared with 33 cytosolic proteins, and 9 proteins had unknown or other non-nuclear subcellular residences (supplemental Table S2). Of the 47 identified nuclear proteins, two (NUP54 and NOLC1) did not have cysteines and therefore could not be Trx1 targets; the remaining 45 were designated as putative nuclear Trx1 targets (Table I), with 1 to 87 cysteines susceptible to reduction by Trx1. Based on the number of putative Trx1 target proteins without cysteines identified in this study, the false identification rate for our approach is estimated as 4.26%.
Fig. 2.
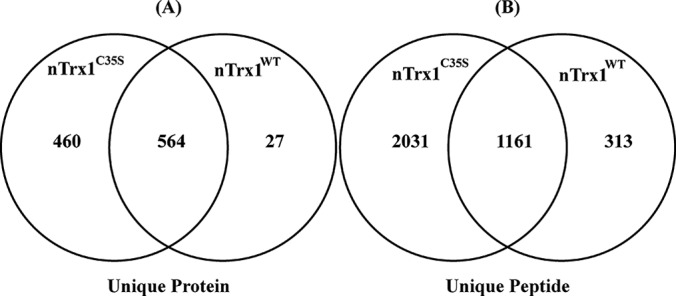
The proteins captured through IP of nTrx1C35S and nTrx1WT. The Venn diagram shows a representative dataset. Scaffold was used to filter and compare the unique proteins and peptides identified in the analysis of the nuclear extract from Experiment 2 (supplemental Tables S1 (A–L) and S2). The peptide and protein identification criteria were set with a 1% false discovery rate. As expected, more proteins and peptides were discovered through the IP of nTrx1C35S, with 460 unique proteins (A) and 2031 unique peptides (B) found only with the protein complex associated with nTrx1C35S.
Table I. Putative Trx1 targets identified in neuroblastoma cells.
| Protein name | Gene I.D. | Protein accession number | Cys number in protein | Function |
|---|---|---|---|---|
| Cell cycle | ||||
| A kinase (PRKA) anchor protein 8 | AKAP8 | O43823 | 10 | Chromatin binding |
| PIN2/TERF1 interacting, telomerase inhibitor 1 | PINX1 | Q96BK5 | 3 | Telomerase inhibitor activity |
| Structural maintenance of chromosomes 1A | SMC1A | A8K7A6 | 5 | ATP binding |
| Structural maintenance of chromosomes 3 | SMC3 | B0AZQ4 | 7 | Chromosome organization |
| Structural maintenance of chromosomes flexible hinge domain containing 1 | SMCHD1 | A6NHR9 | 30 | Chromosome organization |
| Survival of motor neuron 1, telomeric | SMN1/SMN2 | E7EQZ4 | 8 | Axonogenesis |
| DNA repair | ||||
| MutS homolog 6 | MSH6 | B4DF41 | 32 | Methylated histone binding |
| Replication factor C (activator 1) 2, 40 kDa | RFC2 | B5BUD2 | 8 | DNA repair |
| Replication factor C (activator 1) 5, 36.5 kDa | RFC5 | A8K3S0 | 5 | DNA repair |
| XPA binding protein 2 | XAB2 | Q71SV8 | 14 | DNA repair |
| Helicase | ||||
| DEAD (Asp-Glu-Ala-Asp) box helicase 24 | DDX24 | B4DKV2 | 10 | RNA helicase activity |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 | DDX10 | E9PIF2 | 6 | ATP-dependent helicase activity |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 | DDX50 | B4DW97 | 9 | ATP-dependent helicase activity |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 | DDX54 | Q8TDD1 | 5 | ATP-dependent helicase activity |
| Small nuclear ribonucleoprotein 200 kDa (U5) | SNRNP200 | Q9H8B9 | 7 | ATP-dependent helicase activity |
| RNA processing | ||||
| Fused in sarcoma | FUS | A8K4H1 | 4 | Nucleic acid binding |
| GLE1 RNA export mediator | GLE1 | A8K3B8 | 6 | mRNA export from nucleus |
| M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) | MPHOSPH10 | B3KPV5 | 2 | rRNA processing |
| Nucleolar complex associated 3 homolog (Saccharomyces cerevisiae) | NOC3L | A6NJZ9 | 8 | Poly(A) RNA binding |
| Polypyrimidine tract binding protein 1 | PTBP1 | K7EK45 | 2 | Alternative nuclear mRNA splicing |
| Ribosomal RNA processing 12 homolog (S. cerevisiae) | RRP12 | B3KMR5 | 25 | RNA processing |
| Ribosomal RNA processing 1B | RRP1B | Q14684 | 7 | rRNA processing |
| RNA binding motif protein 28 | RBM28 | Q9NW13 | 9 | mRNA processing |
| Small nuclear ribonucleoprotein polypeptide A′ | SNRPA1 | H0YMA0 | 1 | mRNA processing |
| Splicing factor 3b, subunit 3, 130 kDa | SF3B3 | A8K6V3 | 19 | mRNA processing |
| UTP18 small subunit (SSU) processome component homolog (yeast) | UTP18 | B2RAX6 | 8 | rRNA processing |
| Transcription | ||||
| Apoptosis antagonizing transcription factor | AATF | Q9NY61 | 2 | Protein binding |
| Bromodomain adjacent to zinc finger domain, 1A | BAZ1A | D3DS96 | 29 | Chromatin remodeling |
| Centromere protein B, 80 kDa | CENPB | P07199 | 7 | Centromeric DNA binding |
| Deoxynucleotidyltransferase, terminal, interacting protein 2 | DNTTIP2 | Q5QJE6 | 7 | Regulation of transcription |
| KIAA0020 | KIAA0020 | B2RDG4 | 7 | Poly(A) RNA binding |
| PC4 and SFRS1 interacting protein 1 | PSIP1 | O75475 | 2 | Activating transcription factor binding |
| PWP1 homolog (S. cerevisiae) | PWP1 | B4DNL1 | 10 | Transcription |
| Zinc finger CCCH-type containing 18 | ZC3H18 | B4DZ24 | 3 | Metal ion binding |
| Transporter | ||||
| Exportin 1 | XPO1 | B4DR01 | 13 | Transporter activity |
| Karyopherin (importin) β 1 | KPNB1 | B2RBR9 | 23 | Protein transporter activity |
| Nucleoporin 107 kDa | NUP107 | P57740 | 13 | Nucleocytoplasmic transporter |
| Nucleoporin 153 kDa | NUP153 | B4DIK2 | 28 | Protein transport |
| Nucleoporin 210 kDa | NUP210 | Q8TEM1 | 16 | Protein dimerization activity |
| Nucleoporin 214 kDa | NUP214 | P35658 | 13 | Nucleocytoplasmic transporter activity |
| RAN binding protein 2 | RANBP2 | P49792 | 77 | RNA binding |
| Transportin 1 | TNPO1 | Q92973 | 26 | Intracellular protein transport |
| Others | ||||
| DnaJ (Hsp40) homolog, subfamily C, member 9 | DNAJC9 | Q8WXX5 | 3 | Chaperone |
| Guanine nucleotide binding protein-like 2 (nucleolar) | GNL2 | Q13823 | 4 | Ribosome biogenesis |
| Nucleoporin 205 kDa | NUP205 | B4DE72 | 18 | Protein binding |
All of the proteins listed here fulfill the following requirements: (i) the protein was observed only in ntTrx1C35S and not in nTrx1WT in at least two experiments; (ii) the spectra were of high quality; (iii) the protein was designated as a nuclear protein by IPA; and (iv) the protein contains at least one cysteine. Functions annotated in the UniProt database and IPA.
Functional Analysis of the Putative Nuclear Trx1 Targets
The IPA molecular network analysis of the 45 putative nuclear Trx1 target proteins found that 23 proteins participated in a physical and functional protein–protein interaction network involved in the regulation of RNA and protein trafficking and nuclear molecular transport (Fig. 3A). Furthermore, among the nuclear Trx1 target proteins discovered in this study (Fig. 3B), six were significantly enriched with RNA trafficking functions (p value of 2.26E-9), six were enriched with protein trafficking functions (p value of 1.46E-8), and three were enriched with assembly of nuclear pore functions (p value of 4.50E-7). Interestingly, two proteins involved in HIV interactions with host cells were also observed to be regulated by Trx1. These proteins, PSIP1 and RANBP2, regulate the integration of HIV into the host genome (p value of 3.66E-04). Ten putative Trx1 target proteins are known to modulate HIV-1 infection (p value of 1.45E-05).
Fig. 3.
IPA identification of a molecular transport, RNA, and protein trafficking network within the putative novel Trx1 targets. A, IPA was used to identify a network based upon the 45 focus proteins (Table I) that were enriched in nTrx1C35S. The solid line represents a direct interaction, and the broken line represents indirect interactions between the proteins. The proteins are represented as nodes, with the node shape representing the functional class of the protein. The shaded nodes correspond to the proteins found in this study, and the uncolored nodes are additional proteins depicted based upon evidence from the IPA Knowledge Base, indicating a strong biological relevance to the network. B, the proteins enriched in various biological processes and implicated in diseases, as annotated by IPA.
Validation of Our Approach for the Identification of Nuclear Trx1 Targets
To validate the target proteins, we performed Western blotting analyses of select proteins immunoprecipitated with either nTrx1C35S or nTrx1WT. Peroxiredoxin 1 (Prx1) is one of the most studied targets of Trx1. Although Prx1 reduction by Trx1 occurs primarily in the cytosol, we detected it via both Western blotting and LC-MS/MS (supplemental Table S1 (A–L) & Fig. 4). Two known nuclear targets of Trx1 that were not observed in the MS analysis were NF-κB and P53 (18). However, the Western blot analysis indicated that significantly more P53 and NF-κB was associated with nTrx1C35S than with nTrx1WT (Fig. 4). Because the known Trx1 targets are known to associate with nTrx1C35S with greater affinity than with nTrx1WT, these observations support our hypothesis that novel nuclear Trx1 targets are identifiable via the nTrx1C35S trapping strategy.
Fig. 4.

Western blot validation of the known Trx1 targets. Representative blots showing the increased Prx1, P53, and NF-κB levels isolated from SHSY-5Y cells before and after IP with an anti-GFP antibody. The Western blot quantification was analyzed using Quantity One software (Bio-Rad). *p < 0.05 (n = 3).
Validation of PSIP1 as a Direct Trx1 Reduction Target Protein
We conducted an additional biochemical validation of PSIP1, which is commonly known as LEDGF because of its involvement in the stress response and differentiation of lens epithelial cells. PSIP1 interacts with HIV-1 integrase, thereby regulating HIV-1 integrase's nuclear localization and association with chromatin (24). The Western blot demonstrated that ∼20% more PSIP1 associated with nTrx1C35S than with the nTrx1WT complex (Fig. 5A), presumably because of the limited antibody specificity. Protein sequence alignments indicate that PSIP1 is highly conserved among human, bovine, mouse, and rat species (supplemental Fig. S1), including two conserved cysteines, Cys204 and Cys373 (the numbering is according to human PSIP1). To verify that PSIP1 is indeed associated with Trx1 in the nucleus, we performed immunofluorescence microscopy to validate the co-localization of PSIP1 with Trx1 (Fig. 5B), and we observed a large number of cells with overlapping GFP-tagged Trx1 signals, Cy5-labeled PSIP1 signals, and DAPI luminescence in the nucleus. This result suggests that both nTrx1WT and nTrx1C35S largely co-localized with PSIP1 in the nucleus of human cells. Furthermore, we performed in vitro analysis and determined that PSIP1 can be directly reduced by Trx1 (Fig. 6A). A downstream MS analysis identified PSIP1 Cys204 as the site of Trx1 reduction (Figs. 6B and 6C). Finally, using a luciferase reporter assay construct containing a known PSIP1 response element derived from VEGF-C, we confirmed that Trx1 can indeed modulate the transcriptional regulation function of PSIP1 in both HeLa and SHSY-5Y cells (Fig. 7 and supplemental Fig. S2). Interestingly, whether PSIP1 functions as a transcriptional activator or repressor appears to be cell dependent. In SHSY-5Y cells, both PSIP1 and nTrx1WT suppressed pSTRE-luc expression, but in combination, nTrx1WT was able to significantly enhance the ability of PSIP1 to suppress pSTRE-luc expression (supplemental Fig. S2). However, in HeLa cells, PSIP1 behaved as a transcriptional activator, and Trx1 reduction appeared to attenuate PSIP1's ability to activate target gene expression (Fig. 7).
Fig. 5.
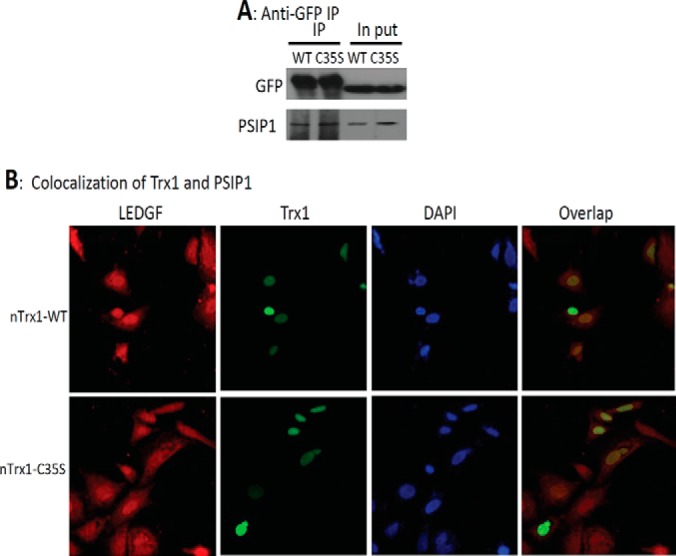
The binding and subcellular co-localization of Trx1 and PSIP1. SHSY-5Y cells were transfected with nTrx1WT and nTrx1C35S and then subjected to nuclear protein extraction. A, the nTrx1-binding proteins were pulled down using an anti-GFP antibody and separated via SDS-PAGE. After the transfer of the proteins onto nitrocellulose membranes, PSIP1 was probed using an anti-PSIP1 antibody. B, SHSY-5Y cells were transfected with nTrx1WT or nTrx1C35S. The fixed cells were stained with anti-PSIP1 and analyzed with a Zeiss confocal laser scanning microscope. The GFP-tagged Trx1 is shown in green, the antibody Cy5-labeled PSIP1 is shown in red, the DAPI-stained nuclei are shown in blue, and the Trx1 and co-localized PSIP1 are shown in yellow.
Fig. 6.
The validation of PSIP1 as a direct Trx1 reduction target. A, oxidized human PSIP1 was prepared by incubating 10 μg of PSIP1 with 100 mm H2O2. The rTrx1 was prepared by incubating 100 μg of PSIP1 with 100 mm DTT. The PSIP1 was mixed with rTrx1 at a 1 to 0, 1, 2, 4, 8, or 16 molar ratio for 30 min at 37 °C. The free thiols in PSIP1 were labeled with biotin-HPDP. The biotinylated PSIP1 was detected via Western blotting using an anti-biotin antibody. B, the work flow for the in vitro analysis of PSIP1 via rTrx1 reduction. The PSIP1 was treated with a buffer or rTrx1 for 30 min at 37 °C and then underwent 10 mm IAA alkylation for 60 min at 37 °C. The excess IAA was quenched by 50 mm DTT, and the oxidized thiols were also reduced with DTT. The residual DTT was removed via acetone precipitation. The DTT-reduced free thiols in PSIP1 (previously oxidized) were blocked with 50 mm methyl methanethiosulfonate. The proteins were trypsin digested and identified via LC-MS/MS. C, the quantitative analysis of the rTrx1 reduction of PSIP1 at Cys204. The relative quantification was calculated as the percentage of methyl methanethiosulfonate–labeled peptides normalized to the top three most abundant non-cysteine-containing peptides identified in all six LC-MS/MS runs (three controls and three Trx1 reductions). D, the MS/MS spectrum of PSIP1202–216, indicating Cys204 as the Trx1 reduction site. *p < 0.001 (n = 3).
Fig. 7.
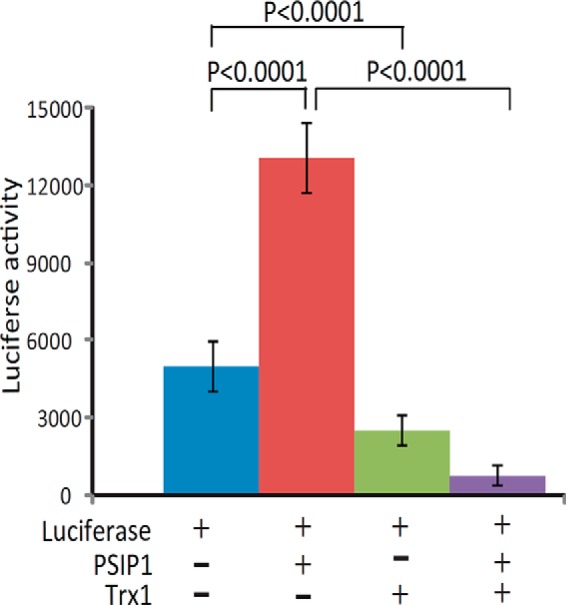
The Trx1 modulation of PSIP1-regulated target gene expression. HeLa cells were co-transfected with luciferase alone or in combination with PSIP1, Trx1, or both for 48 h. The cells were washed three times with PBS and collected for a luciferase activity assay (n = 4).
DISCUSSION
Trx1 is a distinct reductase that restores oxidized cysteine post-translational modifications back to free thiols, thereby modulating the key functions of certain target proteins. Nuclear Trx1 has unique nuclear targets that differ from its cytoplasmic ones. Therefore, the identification of Trx1 targets in the nucleus of human cells might lead to the discovery of novel stress response proteins, including transcription factors and RNA processing enzymes, that might be important in understanding the cellular-nuclear response during the development of some neurodegenerative diseases. Until now, most nuclear targets of Trx1 have been identified using traditional molecular biology approaches that have not included modern proteomics technologies. By continuing to adapt and utilize state-of-the-art mass spectrometers, we can identify more nuclear Trx1 target proteins more rapidly and more accurately.
Proteomics approaches have been widely used to identify Trx targets in non-mammalian cells and tissues (supplemental Table S3), including in plants and microbial cells. As early as 1999, Verdoucq et al. identified a Trx target protein, YLR109, in yeast using His-tagged Trx affinity (25). Yano et al. used monobromobimane to label free thiols in proteins reduced by Trx that were resolved via two-dimensional gel electrophoresis and identified five Trx targets in peanut seeds (26). The Trx affinity technique was subsequently developed to identify Trx targets in diverse cell types, including spinach (27–31), Arabidopsis (32–35), Escherichia coli (36, 37), wheat seeds (38, 39), Synechocystis (40–42), and HeLa cells (16, 43). The most successful of these studies identified up to 80 putative Trx targets (37). In 2008, Hagglund et al. used a quantitative proteomics approach based on the isotope-coded affinity tag method to identify 40 putative Trx targets in the seeds of malting barley (44). More recently, we used the isotope-coded affinity tag approach to identify over 70 targets of Trx1 transnitrosylation and 50 targets of Trx1-mediated denitrosylation (43). Although a Trx mutant has been used to identify Trx targets in non-mammalian cells (25–42, 44), its application in the identification of mammalian targets has been rare, presumably because of the presence of highly abundant cytosolic targets in the total cell lysates, such as Prx1, that compete for a limited number of binding sites on the Trx1C35S mutant. This is the first report demonstrating that an nTrx1C35S mutant can be used to identify novel Trx1 targets in the nucleus of mammalian cells. Although we report the identification of a select number of Trx1 targets here, our continued work involves refining this method to increase the speed, specificity, and sensitivity of this process. For example, increasing the salt concentration to eliminate more non-covalently bound proteins prior to the 2-ME elution of the proteins linked to nTrx1C35S via disulfides might reduce the interference of nonspecific proteins in LC-MS/MS. Therefore, transcription factors such as P53 and NF-κB might be further identified. Additionally, tandem affinity purification can be used to increase the purity of the protein complexes. Finally, following the protein elution by 2-ME, isotope-coded affinity tag or thiol-based tandem mass tag labels could be used for a more accurate quantification of only the thiol-containing peptides, allowing the identification of specific reduction sites and the quantification of their enrichment using nTrx1C35S.
PSIP1/LEDGF is a stress-regulated protein reported to be important for cell survival (45) and implicated as having oncogenic properties (46). It is also known as Dense Fine Speckles 70-kDa protein (DFS 70) or transcriptional coactivator p75/p52 and is a transcriptional co-activator and a host factor that assists HIV integration (47, 48). It has been found in both the cytosol and the nucleus of epithelial cells (49). Its ability to bind to heat or stress response elements is attenuated in cells with high levels of oxidative stress (50), suggesting that PSIP1/LEDGF itself might be sensitive to high levels of oxidative stress. Our discovery of PSIP1 as a Trx1 reduction target suggests a means for cells to preserve the PSIP1 stress modulatory function. The knockdown of PSIP1/LEDGF can significantly decrease HIV integration into the host genome (51, 52). PSIP1/LEDGF activates HIV-1 integration by interacting with HIV-1 integrase, and genetic variants of PSIP1 were reported to affect the host's susceptibility to HIV-1 infection and disease progression (53). Given PSIP1's established role in regulating HIV integrase function, increasing efforts have been made to identify drugs that can block LEDGF binding to HIV integrase (54). HIV-infected patients have elevated levels of plasma Trx1, which might impair the patient's survival by blocking chemotaxis (21). Whether oxidative stress or Trx1 plays a role in regulating the interaction between PSIP1 and HIV integrase in human cells is currently unknown. According to the National Institute of Neurological Disorders and Stroke, HIV-induced inflammation may damage the brain and spinal cord and lead to confusion, memory loss, and other behavioral and neuro-AIDS conditions. Our finding that PSIP1 is a novel target of Trx1 in the nucleus of human neuronal cells might lead to the development of a more effective therapy for neuro-AIDS patients. For example, Trx1 inhibitors are under development as cancer treatments (55); whether the combination of a Trx1 inhibitor and a drug targeting PSIP1/integrase binding would be effective in reducing HIV-associated inflammation remains to be explored.
A select number of putative nuclear Trx1 targets identified in this study have been suggested to play critical roles in diverse nuclear activities. Among the putative targets of Trx1 are several nucleoporins, such as NUP107, NUP153, NUP210, and NUP214 (Table I). Nucleoporins form the nuclear pore complex and are involved in nuclear-cytoplasmic transport. D'Angelo et al. demonstrated that the nuclear pore is leakier in 28-month-old mice than in 3-month-old mice (56). They observed that the age-related increase in nuclear permeability was accelerated by oxidative stress, and as the mice aged, many of the nucleoporins were affected by oxidation. Recent studies have also indicated that the intermolecular disulfide bonds in nucleoporins directly regulate cargo traffic within the nuclear pore channels (57). Furthermore, the E3 SUMO-protein ligase RanBP2, also known as NUP358, regulates the nuclear pore transportation of proteins. Among the 77 cysteines, the mutation of Cys2659 to Ser or Ala resulted in reduced E3 ligase activity (58). These observations suggest that Trx1, a key redox regulator, may directly control nuclear transport via the disulfide-bond-mediated functional regulation of nucleoporins. Trx1 might therefore counter the detrimental effects of oxidative stress and aging on the nuclear and cytoplasmic transport of proteins and nucleic acids to ensure proper cellular functions.
CONCLUSIONS
In this study we identified a large number of novel human nuclear Trx1 targets. These target proteins play important and diverse roles in RNA processing, RNA and protein transport, nuclear structure, and gene regulation. Until now, many of these targets were not known to be redox regulated. Our results implicate Trx1 as playing a broader role in the nucleus than previously thought, such as by directly coordinating the concerted cellular responses to oxidative stress. These results might lead to a better understanding of oxidative stress and help further elucidate the role of Trx1 in modulating nuclear processes. For example, the discovery of PSIP1/LEDGF as a direct target of Trx1 establishes a sound rationale for future studies of this interaction in regulating cell survival in models of neurodegenerative disease, as well as other diseases. In cancer and infectious diseases, the interactions between Trx1 and PSIP1/LEDGF will likely be exploited to develop therapies to attenuate the PSIP1/Trx1 interaction, thereby leading to the death of cancer cells or the attenuation of HIV infection in host cells. Our continued optimization of the proteomics technique described in this study will enable the identification of more nuclear Trx1 targets in mammalian cells and thus will allow us to continue to establish a more complete picture of how Trx1 coordinates anti-oxidative stress responses.
Supplementary Material
Acknowledgments
We appreciate the help of Luke Fritzky at NJMS Confocal Imaging Facility in the preparation of this manuscript.
Footnotes
Author contributions: C.W., M.R.J., Q.L., J.S., and H.L. designed research; C.W., M.R.J., Q.L., S.O., W.L., A.T.K., N.N., J.S., and H.L. performed research; C.W., M.R.J., Q.L., S.O., W.L., and H.L. analyzed data; C.W., M.R.J., Q.L., J.S., W.J.S., and H.L. wrote the paper.
* The project described was supported in part by Grant No. P30NS046593 from NINDS, National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- Trx
- thioredoxin
- ACN
- acetonitrile
- biotin-HPDP
- N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)-propionamide
- IAA
- iodoacetamide
- IP
- immunoprecipitation
- IPA
- Ingenuity Pathway Analysis
- LEDGF
- lens epithelium-derived growth factor
- NF-κB
- nuclear factor κ B
- nTrx1
- nuclear-targeted thioredoxin 1
- Prx1
- peroxiredoxin 1
- PSIP1
- PC4 and SFRS1 interacting protein 1
- rTrx1
- reduced thioredoxin 1
- STRE
- stress response element
- Trx1C35S
- Cys35 mutated thioredoxin 1
- WT
- wild type.
REFERENCES
- 1. Federico A., Cardaioli E., Da Pozzo P., Formichi P., Gallus G. N., Radi E. (2012) Mitochondria, oxidative stress and neurodegeneration. J. Neurol. Sci. 322, 254–262 [DOI] [PubMed] [Google Scholar]
- 2. Nakamura T., Cho D. H., Lipton S. A. (2012) Redox regulation of protein misfolding, mitochondrial dysfunction, synaptic damage, and cell death in neurodegenerative diseases. Exp. Neurol. 238, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nakamura T., Nakamura H., Hoshino T., Ueda S., Wada H., Yodoi J. (2005) Redox regulation of lung inflammation by thioredoxin. Antioxid. Redox Signal. 7, 60–71 [DOI] [PubMed] [Google Scholar]
- 4. Takagi Y., Mitsui A., Nishiyama A., Nozaki K., Sono H., Gon Y., Hashimoto N., Yodoi J. (1999) Overexpression of thioredoxin in transgenic mice attenuates focal ischemic brain damage. Proc. Natl. Acad. Sci. U.S.A. 96, 4131–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoshida T., Nakamura H., Masutani H., Yodoi J. (2005) The involvement of thioredoxin and thioredoxin binding protein-2 on cellular proliferation and aging process. Ann. N. Y. Acad. Sci. 1055, 1–12 [DOI] [PubMed] [Google Scholar]
- 6. Nishiyama A., Matsui M., Iwata S., Hirota K., Masutani H., Nakamura H., Takagi Y., Sono H., Gon Y., Yodoi J. (1999) Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J. Biol. Chem. 274, 21645–21650 [DOI] [PubMed] [Google Scholar]
- 7. Saitoh M., Nishitoh H., Fujii M., Takeda K., Tobiume K., Sawada Y., Kawabata M., Miyazono K., Ichijo H. (1998) Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 17, 2596–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Makino Y., Yoshikawa N., Okamoto K., Hirota K., Yodoi J., Makino I., Tanaka H. (1999) Direct association with thioredoxin allows redox regulation of glucocorticoid receptor function. J. Biol. Chem. 274, 3182–3188 [DOI] [PubMed] [Google Scholar]
- 9. Li X., Luo Y., Yu L., Lin Y., Luo D., Zhang H., He Y., Kim Y. O., Kim Y., Tang S., Min W. (2008) SENP1 mediates TNF-induced desumoylation and cytoplasmic translocation of HIPK1 to enhance ASK1-dependent apoptosis. Cell Death Differ. 15, 739–750 [DOI] [PubMed] [Google Scholar]
- 10. Masutani H., Bai J., Kim Y. C., Yodoi J. (2004) Thioredoxin as a neurotrophic cofactor and an important regulator of neuroprotection. Mol. Neurobiol. 29, 229–242 [DOI] [PubMed] [Google Scholar]
- 11. Yoshioka J., Schulze P. C., Cupesi M., Sylvan J. D., MacGillivray C., Gannon J., Huang H., Lee R. T. (2004) Thioredoxin-interacting protein controls cardiac hypertrophy through regulation of thioredoxin activity. Circulation 109, 2581–2586 [DOI] [PubMed] [Google Scholar]
- 12. Yang L., Wu D., Wang X., Cederbaum A. I. (2011) Depletion of cytosolic or mitochondrial thioredoxin increases CYP2E1-induced oxidative stress via an ASK-1-JNK1 pathway in HepG2 cells. Free Radic. Biol. Med. 51, 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hayashi T., Ueno Y., Okamoto T. (1993) Oxidoreductive regulation of nuclear factor kappa B. Involvement of a cellular reducing catalyst thioredoxin. J. Biol. Chem. 268, 11380–11388 [PubMed] [Google Scholar]
- 14. Ago T., Liu T., Zhai P., Chen W., Li H., Molkentin J. D., Vatner S. F., Sadoshima J. (2008) A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell 133, 978–993 [DOI] [PubMed] [Google Scholar]
- 15. Fu C., Wu C., Liu T., Ago T., Zhai P., Sadoshima J., Li H. (2009) Elucidation of thioredoxin target protein networks in mouse. Mol. Cell. Proteomics 8, 1674–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu C., Liu T., Chen W., Oka S., Fu C., Jain M. R., Parrott A. M., Baykal A. T., Sadoshima J., Li H. (2010) Redox regulatory mechanism of transnitrosylation by thioredoxin. Mol. Cell. Proteomics 9, 2262–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamamoto M., Yang G., Hong C., Liu J., Holle E., Yu X., Wagner T., Vatner S. F., Sadoshima J. (2003) Inhibition of endogenous thioredoxin in the heart increases oxidative stress and cardiac hypertrophy. J. Clin. Invest. 112, 1395–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lukosz M., Jakob S., Buchner N., Zschauer T. C., Altschmied J., Haendeler J. (2010) Nuclear redox signaling. Antioxid. Redox Signal. 12, 713–742 [DOI] [PubMed] [Google Scholar]
- 19. Benhar M., Thompson J. W., Moseley M. A., Stamler J. S. (2010) Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry 49, 6963–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Q., Jain M. R., Chen W., Li H. (2013) A multidimensional approach to an in-depth proteomics analysis of transcriptional regulators in neuroblastoma cells. J. Neurosci. Methods 216, 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakamura H., De Rosa S. C., Yodoi J., Holmgren A., Ghezzi P., Herzenberg L. A., Herzenberg L. A. (2001) Chronic elevation of plasma thioredoxin: inhibition of chemotaxis and curtailment of life expectancy in AIDS. Proc. Natl. Acad. Sci. U.S.A. 98, 2688–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 23. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 24. Llano M., Delgado S., Vanegas M., Poeschla E. M. (2004) Lens epithelium-derived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J. Biol. Chem. 279, 55570–55577 [DOI] [PubMed] [Google Scholar]
- 25. Verdoucq L., Vignols F., Jacquot J. P., Chartier Y., Meyer Y. (1999) In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J. Biol. Chem. 274, 19714–19722 [DOI] [PubMed] [Google Scholar]
- 26. Yano H., Wong J. H., Lee Y. M., Cho M. J., Buchanan B. B. (2001) A strategy for the identification of proteins targeted by thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 98, 4794–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Balmer Y., Koller A., del Val G., Manieri W., Schurmann P., Buchanan B. B. (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. U.S.A. 100, 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Balmer Y., Koller A., Val G. D., Schurmann P., Buchanan B. B. (2004) Proteomics uncovers proteins interacting electrostatically with thioredoxin in chloroplasts. Photosynthesis Res. 79, 275–280 [DOI] [PubMed] [Google Scholar]
- 29. Balmer Y., Vensel W. H., Tanaka C. K., Hurkman W. J., Gelhaye E., Rouhier N., Jacquot J. P., Manieri W., Schurmann P., Droux M., Buchanan B. B. (2004) Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 101, 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goyer A., Haslekas C., Miginiac-Maslow M., Klein U., Le Marechal P., Jacquot J. P., Decottignies P. (2002) Isolation and characterization of a thioredoxin-dependent peroxidase from Chlamydomonas reinhardtii. Eur. J. Biochem. 269, 272–282 [DOI] [PubMed] [Google Scholar]
- 31. Motohashi K., Kondoh A., Stumpp M. T., Hisabori T. (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 98, 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee K., Lee J., Kim Y., Bae D., Kang K. Y., Yoon S. C., Lim D. (2004) Defining the plant disulfide proteome. Electrophoresis 25, 532–541 [DOI] [PubMed] [Google Scholar]
- 33. Marchand C., Le Marechal P., Meyer Y., Miginiac-Maslow M., Issakidis-Bourguet E., Decottignies P. (2004) New targets of Arabidopsis thioredoxins revealed by proteomic analysis. Proteomics 4, 2696–2706 [DOI] [PubMed] [Google Scholar]
- 34. Rey P., Cuine S., Eymery F., Garin J., Court M., Jacquot J. P., Rouhier N., Broin M. (2005) Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant J. 41, 31–42 [DOI] [PubMed] [Google Scholar]
- 35. Yamazaki D., Motohashi K., Kasama T., Hara Y., Hisabori T. (2004) Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol. 45, 18–27 [DOI] [PubMed] [Google Scholar]
- 36. Kadokura H., Tian H., Zander T., Bardwell J. C., Beckwith J. (2004) Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science 303, 534–537 [DOI] [PubMed] [Google Scholar]
- 37. Kumar J. K., Tabor S., Richardson C. C. (2004) Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101, 3759–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wong J. H., Balmer Y., Cai N., Tanaka C. K., Vensel W. H., Hurkman W. J., Buchanan B. B. (2003) Unraveling thioredoxin-linked metabolic processes of cereal starchy endosperm using proteomics. FEBS Lett. 547, 151–156 [DOI] [PubMed] [Google Scholar]
- 39. Wong J. H., Cai N., Tanaka C. K., Vensel W. H., Hurkman W. J., Buchanan B. B. (2004) Thioredoxin reduction alters the solubility of proteins of wheat starchy endosperm: an early event in cereal germination. Plant Cell Physiol. 45, 407–415 [DOI] [PubMed] [Google Scholar]
- 40. Hosoya-Matsuda N., Motohashi K., Yoshimura H., Nozaki A., Inoue K., Ohmori M., Hisabori T. (2005) Anti-oxidative stress system in cyanobacteria. Significance of type II peroxiredoxin and the role of 1-Cys peroxiredoxin in Synechocystis sp. strain PCC 6803. J. Biol. Chem. 280, 840–846 [DOI] [PubMed] [Google Scholar]
- 41. Lindahl M., Florencio F. J. (2003) Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc. Natl. Acad. Sci. U.S.A. 100, 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mata-Cabana A., Florencio F. J., Lindahl M. (2007) Membrane proteins from the cyanobacterium Synechocystis sp. PCC 6803 interacting with thioredoxin. Proteomics 7, 3953–3963 [DOI] [PubMed] [Google Scholar]
- 43. Wu C., Parrott A. M., Liu T., Jain M. R., Yang Y., Sadoshima J., Li H. (2011) Distinction of thioredoxin transnitrosylation and denitrosylation target proteins by the ICAT quantitative approach. J. Proteomics 74, 2498–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hagglund P., Bunkenborg J., Maeda K., Svensson B. (2008) Identification of thioredoxin disulfide targets using a quantitative proteomics approach based on isotope-coded affinity tags. J. Proteome Res. 7, 5270–5276 [DOI] [PubMed] [Google Scholar]
- 45. Chin M. S., Caruso R. C., Detrick B., Hooks J. J. (2006) Autoantibodies to p75/LEDGF, a cell survival factor, found in patients with atypical retinal degeneration. J. Autoimmun. 27, 17–27 [DOI] [PubMed] [Google Scholar]
- 46. Daugaard M., Kirkegaard-Sorensen T., Ostenfeld M. S., Aaboe M., Hoyer-Hansen M., Orntoft T. F., Rohde M., Jaattela M. (2007) Lens epithelium-derived growth factor is an Hsp70–2 regulated guardian of lysosomal stability in human cancer. Cancer Res. 67, 2559–2567 [DOI] [PubMed] [Google Scholar]
- 47. Singh D. P., Kimura A., Chylack L. T., Jr., Shinohara T. (2000) Lens epithelium-derived growth factor (LEDGF/p75) and p52 are derived from a single gene by alternative splicing. Gene 242, 265–273 [DOI] [PubMed] [Google Scholar]
- 48. Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. (2003) HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278, 372–381 [DOI] [PubMed] [Google Scholar]
- 49. Singh D. P., Ohguro N., Chylack L. T., Jr., Shinohara T. (1999) Lens epithelium-derived growth factor: increased resistance to thermal and oxidative stresses. Invest. Ophthalmol. Vis. Sci. 40, 1444–1451 [PubMed] [Google Scholar]
- 50. Fatma N., Kubo E., Sharma P., Beier D. R., Singh D. P. (2005) Impaired homeostasis and phenotypic abnormalities in Prdx6-/-mice lens epithelial cells by reactive oxygen species: increased expression and activation of TGFbeta. Cell Death Differ. 12, 734–750 [DOI] [PubMed] [Google Scholar]
- 51. Vandekerckhove L., Christ F., Van Maele B., De Rijck J., Gijsbers R., Van den Haute C., Witvrouw M., Debyser Z. (2006) Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 80, 1886–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Llano M., Saenz D. T., Meehan A., Wongthida P., Peretz M., Walker W. H., Teo W., Poeschla E. M. (2006) An essential role for LEDGF/p75 in HIV integration. Science 314, 461–464 [DOI] [PubMed] [Google Scholar]
- 53. Madlala P., Gijsbers R., Christ F., Hombrouck A., Werner L., Mlisana K., An P., Abdool Karim S. S., Winkler C. A., Debyser Z., Ndung'u T. (2011) Association of polymorphisms in the LEDGF/p75 gene (PSIP1) with susceptibility to HIV-1 infection and disease progression. AIDS 25, 1711–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Christ F., Debyser Z. (2013) The LEDGF/p75 integrase interaction, a novel target for anti-HIV therapy. Virology 435, 102–109 [DOI] [PubMed] [Google Scholar]
- 55. Baker A. F., Dragovich T., Tate W. R., Ramanathan R. K., Roe D., Hsu C. H., Kirkpatrick D. L., Powis G. (2006) The antitumor thioredoxin-1 inhibitor PX-12 (1-methylpropyl 2-imidazolyl disulfide) decreases thioredoxin-1 and VEGF levels in cancer patient plasma. J. Lab. Clin. Med. 147, 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. D'Angelo M. A., Raices M., Panowski S. H., Hetzer M. W. (2009) Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136, 284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yoshimura S. H., Otsuka S., Kumeta M., Taga M., Takeyasu K. (2013) Intermolecular disulfide bonds between nucleoporins regulate karyopherin-dependent nuclear transport. J. Cell Sci. 126, 3141–3150 [DOI] [PubMed] [Google Scholar]
- 58. Pichler A., Knipscheer P., Saitoh H., Sixma T. K., Melchior F. (2004) The RanBP2 SUMO E3 ligase is neither HECT- nor RING-type. Nat. Struct. Mol. Biol. 11, 984–991 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.