Abstract
Schizosaccharomyces pombe Rad16 is the ortholog of the XPF structure-specific endonuclease, which is required for nucleotide excision repair and implicated in the single strand annealing mechanism of recombination. We show that Rad16 is important for proper completion of meiosis. In its absence, cells suffer reduced spore viability and abnormal chromosome segregation with evidence for fragmentation. Recombination between homologous chromosomes is increased, while recombination within sister chromatids is reduced, suggesting that Rad16 is not required for typical homolog crossovers but influences the balance of recombination between the homolog and the sister. In vegetative cells, rad16 mutants show evidence for genome instability. Similar phenotypes are associated with mutants affecting Rhp14XPA but are independent of other nucleotide excision repair proteins such as Rad13XPG. Thus, the XPF/XPA module of the nucleotide excision repair pathway is incorporated into multiple aspects of genome maintenance even in the absence of external DNA damage.
Keywords: meiosis, Rad16, XPF, homologous recombination, NER
THE XPF/ERCC1 protein complex is one of several structure-specific endonucleases that function broadly as resolvases in the repair of damaged DNA (Schwartz and Heyer 2011). Rad16/Swi9 is the fission yeast ortholog of endonuclease XPF, which forms a complex with Swi10/Rad23 (ERCC1) (Carr et al. 1994). XPF has a conserved role in nucleotide excision repair (NER) to remove UV-induced lesions in the DNA (Camenisch et al. 2006; Gregg et al. 2011). In humans, mutation of XPF is associated with xeroderma pigmentosum (XP), which causes sun sensitivity and high rates of skin cancer, as well as premature aging and neurological disorders (Camenisch et al. 2006; Gregg et al. 2011). Recent studies suggest XPF mutations are also associated with Fanconi anemia (FA), which requires repair of interstrand crosslinks (ICLs) (Bogliolo et al. 2013; Kashiyama et al. 2013). The canonical model for NER suggests that upon recognition of helix-distorting lesions, the DNA is unwound to form a bubble around the damage. XPA (SpRhp14) loads XPF (SpRad16) and ERCC1 (SpSwi10); XPF cleaves at the 5′ end of the bubble while XPG (SpRad13), another endonuclease, cleaves at the 3′ end to remove the offending segment (Fagbemi et al. 2011; Schwartz and Heyer 2011). Thus, Schizosaccharomyces pombe rad16 mutants show a decrease in (6-4) photoproduct excision (McCready et al. 1993; Carr et al. 1994).
Consistent with these clinical effects, XPF is implicated in multiple mechanisms of genome maintenance (Paques and Haber 1997; Gregg et al. 2011; Schwartz and Heyer 2011). XPF is required for single strand annealing (SSA), a form of double strand break (DSB) repair distinct from typical homologous recombination (HR) (Ma et al. 2003; Kass and Jasin 2010). This occurs when short regions of homology exposed by resection are able to pair, leaving nonhomologous 3′ overhangs as substrates for XPF cleavage. In budding yeast, recruitment of ScRad1XPF and ScRad10ERCC1 in this pathway depends on interactions with other proteins including the recombination mediator Rad52, and a scaffold provided by Saw1 and Slx4 (reviewed in Lyndaker and Alani 2009). Recent studies have also implicated ScRad1XPF in recombination between dispersed repeats (Symington et al. 2000; Mazon et al. 2012) and in sister chromatid recombination to repair replication-induced double strand breaks (Muñoz-Galván et al. 2012; Pardo and Aguilera 2012). There is evidence that XPF is recruited to the replisome even in undamaged DNA, suggesting an intimate role in genome maintenance during DNA replication (Gilljam et al. 2012). Despite this, mammalian XPF is not essential for viability (Brookman et al. 1996; Tian et al. 2004), possibly because it overlaps with another structure-specific endonuclease, Mus81. In chicken DT40 cells that lack Mus81, XPF is essential for viability and inactivation leads to chromosome breakage and failure at a late stage of HR (Kikuchi et al. 2013). Mammalian XPF is also implicated in telomere protection (Zhu et al. 2003; Muñoz et al. 2005).
These observations are consistent with the phenotypes of S. pombe rad16 mutants, which are defective for gene conversion associated with mating type switching and for repair of radiation-induced DNA damage (Egel et al. 1984; Schmidt et al. 1989; Prudden et al. 2003). There is increased instability between direct repeats, leading to increased gene conversion as well as deletions (Osman et al. 2000). S. pombe Rad16 promotes recombination repair of broken replication forks using ectopic donor sequences, in contrast to Mus81, which promotes repair via sister chromatids (Roseaulin et al. 2008).
Evidence is mixed regarding XPF function in meiosis. In Drosophila, the DmMei9XPF homolog functions as a Holliday junction resolvase during meiosis; flies deficient in mei9 show reduced meiotic recombination and loss of viabile progeny (Yildiz et al. 2002). In Caenorhabditis elegans, XPF functions redundantly with other structure-specific endonucleases MUS-81 and SLX-1 in resolution of crossovers, and suppresses formation of abnormal structures (Agostinho et al. 2013; O’Neil et al. 2013; Saito et al. 2013). In humans and in budding yeast, the structure-specific endonucleases SLX1/Slx1, MUS81/Mus81, and GEN1/Yen1, but not XPF, are linked to crossover resolution of Holliday junctions (Kaliraman et al. 2001; Fricke and Brill 2003; Wyatt et al. 2013). Thus, Sc rad1∆ mutants show no decrease in spore viability, leading to the conclusion that there is no general meiotic function for ScRad1XPF (Higgins et al. 1983), although it is implicated in resolving insertions within heteroduplex DNA (Kirkpatrick et al. 2000; Kearney et al. 2001).
In fission yeast, Mus81 functions as the primary Holliday junction resolvase during meiosis (Boddy et al. 2000, 2001; Smith et al. 2003), and there is no obvious GEN1/Yen1 ortholog (Ip et al. 2008). Rad16 is required for short-patch repair of C/C mismatches in meiotic recombination intermediates, consistent with its role in NER (Fleck et al. 1999). Loss of rad16 reduces gene conversion at an ade6 hotspot that additionally contains unpaired heteroduplex DNA (Farah et al. 2005, 2009). However, a detailed analysis of its function in typical meiotic recombination has not been carried out.
In this study, we examine the effect of rad16 mutation on fission yeast meiosis. We show that rad16 mutants have reduced spore viability accompanied by chromosome mis-segregation and apparent chromosome fragmentation at both meiotic divisions. While the gross dynamics of DNA double strand break repair appear intact, there is a modest increase in the rate of meiotic interhomolog crossover (CO) exchange, accompanied by a reduction in events that use the sister chromatid. These phenotypes are shared with rhp14∆ mutants that disrupt the XPA loading factor, but not in rad13∆ mutants that disrupt the XPG endonuclease. Importantly, rad16 mutants also cause genome instability during vegetative growth, accompanied by chronic activation of the DNA damage checkpoint in the absence of external stress. Synthetic interactions with DNA repair and replication mutants suggest that Rad16 contributes to genome stability even in an unperturbed cell cycle.
Materials and Methods
Cell growth and culture
Strains used in this study are in (Table S1). General culture conditions and media are described in Sabatinos and Forsburg (2010). Cells were grown from single colonies in 5-ml cultures at 32° overnight to midlog phase for RPA and Rad52 focus imaging and serial dilution plating assays to determine drug sensitivities. Drug plates were incubated at 32° for 2–4 days before being imaged using a flatbed scanner. For imaging, cells were concentrated at 6000 rpm in a microfuge and spread on pombe minimal glutamate (PMG) agar on glass slides for imaging (Sabatinos et al. 2012). Heterothallic strains were grown independently for meiotic movies in PMG with appropriate supplements as 32° until culture was in late log phase (OD595 = ∼0.8). Cells were pelleted and washed in EMM-N and resuspended in Malt extract (ME) and incubated 12–20 hr in a 25° airshaker. Cells were concentrated using a microfuge and spread on SPAS agar pads on glass slides. Imaging was performed at 25°.
Spore viability and recombination
Spore viability and recombination were performed by mating strains on SPAS agar for 2–3 days at which point the mating patch was scraped from the plate and diluted in 1 ml 0.5% glusulase. This was digested for 16–20 hr rotating at room temperature. Spores were plated on Yeast Extract with supplements (YES) media and grown at 32° for 3–5 days before counting and replica plating colonies onto PMG media with appropriate supplements. Phloxin B was included to identify any diploids; for rad16-249, no diploids or dyad asci were observed. We also assayed diploid spore formation in rad16-249 and wild type (WT) in recombination assays using a lys4 deletion marked with kanMX in combination with the his4-239 point mutation. In this way when His+ Lys+ colonies were recovered, any colonies that were also diploid would be resistant to G418. We found no colonies that were His+ Lys+ and G418 resistant. His+ Lys+, Leu+ Ura+, or His+ Leu+ progeny were identified and genetic distance was calculated by (2(His+ Lys+)/total colonies) × 100. Within the same strains intragenic recombination was assessed using the ade6-M26 and ade6-52 alleles and scoring for the restoration of the Ade+ phenotypes (Gutz 1971; Ponticelli et al. 1988). The experiment was repeated nine times, plating 2000 spores for each genotype each trial. For tetrad analysis genetic distance was calculated using Perkin’s formula as described (Smith 2009). Sister recombination was determined as in an assay described previously (Catlett and Forsburg 2003). Significance was calculated for genetic distances using a two-tailed t-test. To test mitotic recombination in the sister recombination assay, sectored colonies were counted after germination of spores on YE and EMM low ade media. For tetrad dissection spore viability, asci were dissected after mating on SPAS media and germinated on YES for 3–5 days. Significance was determined using the χ2 test.
Imaging
Images were acquired with a DeltaVision Core widefield deconvolution microscope (Applied Precision, Issaquah, WA) using an Olympus 60×/1.40, PlanApo, NA = 1.40 objective lens and a 12-bit Photometrics CoolSnap HQII CCD, deep-cooled, Sony ICX-285 chip. The system x-y pixel size is 0.1092 µm x-y. softWoRx v4.1 (Applied Precision) software was used at acquisition electronic gain = 1.0 and pixel binning 1 × 1. Excitation illumination was from a solid-state illuminator (seven-color version); GFP was excited and detected with an (ex)475/28, (em)525/50 filter set and a 0.2-sec exposure; red fluorescent protein (RFP) was excited and detected with an (ex)575/25, (em)632/60 filter set and a 0.2-sec exposure; cerulean fluorescent protein (CFP) was excited and detected with an (ex)438/24, (em)470/24 filter set and a 0.15-sec exposure; and yellow fluorescent protein (YFP) was excited and detected with an (ex)513/17, (em)559/38 filter set and a 0.15-sec exposure for meiotic time courses; for mitotic still imaging, CFP was excited and detected with an (ex)438/24, (em)470/24 filter set and a 0.5-sec exposure; and YFP was excited and detected with an (ex)513/17, (em)559/38 filter set and a 0.5-sec exposure. Suitable polychroic mirrors was used, GFP/mCherry Chroma ET C125705 roughly: 520/50–630/80 and Semrock CFP/YFP/DsRed 61008 bs roughly: 415/20–462/32–535/50–635/74. Thirteen z sections at 0.5 µm were acquired. Three-dimensional stacks were deconvolved with manufacturer-provided optical transfer function using a constrained iterative algorithm and images were maximum-intensity projected. For live cell imaging, time points were taken 10 min apart for the length of the experiment. Images were contrast adjusted using a histogram stretch with an equivalent scale and gamma for comparability. Brightfield images were acquired with DIC. Whole cell SytoxGreen flow cytometry (FACS) was performed as described in Sabatinos and Forsburg (2009).
Western blot
Western blot analysis of cell extracts was taken from cultures grown to early midlog phase (OD595 ∼0.3) in YES media at 32°. Cultures were split in equal volumes and treated with 0.01% MMS and untreated and grown for 4 hr at 32° at which point 10× stop buffer was added. Extracts were prepared using trichloric acid (TCA) (Foiani et al. 1994). Protein extracts were quantified using Pierce bicinchoninic acid (BCA) and 100 μg protein was run on a 8% acrylamide with 1.25% crosslinker SDS/PAGE gel and probed with 16B12 α-HA (Covance) or 12CA5 α-HA (Abcam) at 1:1500 or 1:1000 dilution, respectively, in PBSt antibody.
Pulse field gel electrophoresis
Synchronous diploid meiosis was achieved using the mat2-102 and pat1-114 alleles as in Catlett and Forsburg (2003) to create stable diploids using ade6-M210/M216 complementation. Pulse field gel plugs were created by digesting the cell wall with 0.2 mg/ml 100T zymolase and 0.45 mg/ml Sigma lysing enzymes titrated to 50 and 25% of original strength for time points 1–2 and 3–6, respectively, as in Cervantes et al. (2000). Pulse field gel using a BioRad Chef II Pulse Field Machine was run for 48 hr using 2 V/cm, 1800-sec switch time, and a 106° angle. DNA was visualized via ethidium bromide. DSB quantification was done by quantification using BioRad Quant One software representing the DSB breaks as a ratio to total chromosome signal with the local background subtracted as in Borde et al. (2000).
Results
rad16 mutation reduces spore viability and perturbs meiotic chromosome segregation
The rad16-249 allele was originally identified in a screen for mutants sensitive to alkylation damage (Dolan et al. 2010). This allele carries a truncation of the 877-amino-acid protein at residue 118, and eliminates all conserved domains. The rad16-249 allele is recessive and behaves identically to a disruption allele that removes aa 313–798 (Supporting Information, Figure S1).
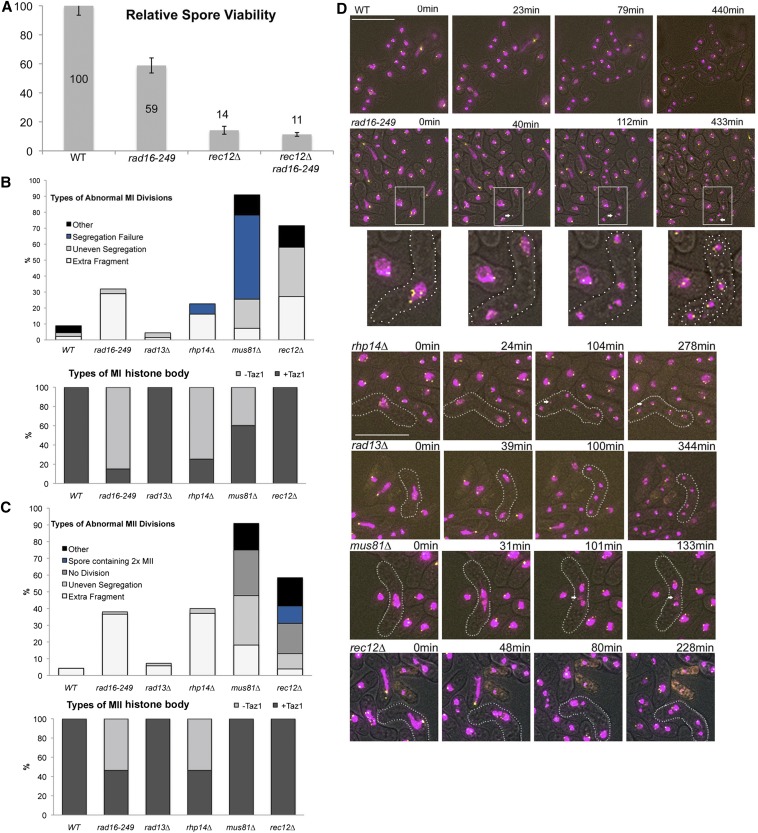
In a cross between rad16-249 strains, we observed relative spore viability dropped to 59% compared to wild type (assayed by random spore analysis; Figure 1A). This is approximately fourfold higher viability than observed for rec12∆ cells, which lack the meiosis-specific endonuclease Rec12Spo11 and cannot generate meiosis-specific DSBs (Farah et al. 2005). Relative viability of the double mutant rec12∆ rad16-249 is not significantly different from that of rec12∆ (11.35% ± 4.27 and 14.19% ± 9.06, respectively). This suggests that Rad16 operates in a Rec12-dependent pathway.
Figure 1.
Spore viability and chromosome segregation. (A) Bulk spore germination of homozygous h+/h− meiosis from homolog recombination data. Error bars represent standard error of the mean. At least nine trials for each genotype for a total of 24,600 and 38,600 spores plated for wild type and rad16-249, respectively, and 156,000 for rec12∆ and rad16-249 rec12∆. (B and C). Quantification of MI and MII segregations defects, respectively. −Taz1 indicates no Taz1-GFP signal on histone body; +Taz1 indicates at least one Taz1-GFP signal associated with histone body. The 2× MII category refers to a single spore encapsulating both daughter nuclei of an MII division. (D) Representative images selected from live cell analysis of meiosis in homozygous h+/h− meiosis for wild type, rad16-249, rhp14∆, rad13∆, mus81∆, and rec12∆ homozygous h+/h− meiosis. Still image frames are taken from live cell movies at indicated times relative to the first image panel in the series labeled 0 min. White box indicates portion of image at higher magnification in bottom row showing fragmentation in rad16-249. White arrows indicate fragments. White dots outline cell wall and spore walls. Magenta is signal from H3-mRFP and yellow from Taz1-GFP. Bar, 15 µm. Live cell analysis was performed on at least 30 cells from three different biological replicates (WT: MI = 46, MII = 47; rad16-249: MI = 69, MII = 71; rad13∆: MI = 68, MII = 69; rhp14∆: MI = 31, MII = 36; rec12∆: MI = 81, MII = 77; and mus81∆: MI = 50, MII = 44).
Spore viability determined using tetrad dissection was greater than that of a random spore analysis, likely because normal-appearing four-spored asci are preferentially selected in tetrad analysis (Table S2). However, even with this bias, only 39% of rad16-249 asci from a homozygous cross had four viable germinating spores, and some had no viable spores at all, indicating an important role for normal meiotic progression.
We examined the dynamics of meiosis in live rad16-249 cells in a cross between h+ and h− parents compared to wild type (Figure 1, B–D, File S1, and File S2). We visualized two fluorescent markers: mRFP-labeled histone H3 to label the chromatin and the telomere-associated Taz1-GFP to identify any defects in telomere clustering and bouquet formation, which can lead to disruptions in meiosis and recombination (Cooper et al. 1997, 1998; Chikashige and Hiraoka 2001; Tomita and Cooper 2007). We saw no obvious abnormalities in telomere clustering or in the characteristic horse-tailing movement in the rad16-249 mutant, suggesting that telomere organization is normal.
However, there are striking defects apparent during both MI and MII divisions, in which fragments of H3-mRFP separate from the bulk of nuclear signal (29% in MI and 37% in MII; Figure 1, B–D). In some cases, these fragments were enclosed as extra-nuclear spots within the spores (included), but at other times, they remained outside of the spore wall (excluded, 50% in MI and 61% in MII) (Figure 1, B and C). Approximately 15% MI and 46% MII fragments contained one or two Taz1-GFP signals, as would be expected if they contain full-length chromosomes (Figure 1, B and C). The absence of Taz1 in the remainder suggests they result from some form of chromosome breakage.
Defects in meiotic chromosome segregation observed in other DNA repair mutants
The fragmentation phenotype is visually similar to the abnormal segregation and more than four nuclear spots that we reported previously for rad54∆ rdh54∆ double mutants, which are completely deficient in DSB repair. However, in that case, all the spores were inviable, consistent with a catastrophic failure of DNA repair (Catlett and Forsburg 2003). We investigated the formation of fragments in other mutants.
During NER, the XPF endonuclease is recruited to the DNA by the XPA protein, encoded by rhp14+ in fission yeast (Hohl et al. 2001; Croteau et al. 2008). Similar to rad16-249, we observed fragments in MI and MII divisions in rhp14∆ (Figure 1, B–D and File S3), suggesting that XPA and XPF also act together during meiosis. In contrast, we observed no disruptions in meiosis in rad13∆ mutants, which lack the downstream NER endonuclease XPG (Figure 1, B–D and File S4).
We compared this to the meiotic phenotype of mus81∆ cells, which lack the Holliday junction resolvase and cannot resolve chiasmata (Figure 1, B–D, File S5, and File S6) (Boddy et al. 2000, 2001; Smith et al. 2003). Previous immunofluorescence analysis of fixed mus81∆ cells showed evidence for entangled chromosomes and dramatic disruption of divisions (Boddy et al. 2001; Gaillard et al. 2003; Osman et al. 2003b). Consistent with this, we observed extensively disordered MI divisions in live mus81∆ cells, with a failure of nuclear division in 52% of cells. By following live cells through the time course, we were able to observe them as they entered MII based on timing, regardless of the MI outcome. In 27% of the cells in MII, we observed no nuclear division. In those that did divide, segregation was highly unequal with multiple defects. We observed fragments, which we defined as extra spots of histone-mRFP apart from the main body of the nuclei. These were observed in 18% of MI and 32% of MII divisions. About half of MI fragments and nearly all of the MII fragments contained a Taz1 signal, indicating the presence of telomeres.
Finally, we examined rec12∆ mutants, which fail to create meiosis-specific DSBs. In most cells, both meiotic divisions occurred with irregularities, generating additional histone signals in 38% of cells (Figure 1, B–D and File S7). As expected, these extra spots appeared to be intact chromosomes, as they always contained at least one Taz1 signal, with the majority containing two or four Taz1 signals (Figure 1, B and C). About half of these fragments were not encapsulated into spores. We also observed a background level of dyad asci (39%), likely diploids as reported previously (Davis and Smith 2003). In ∼73% of these dyads we observed encapsulation of two distinct histone signals inside a single spore. Only 23% of dyads resulted from an apparent failure to undergo an MII division, whereas the remainder underwent an MII division but both products were incorporated into a single spore.
Meiotic repair dynamics in rad16-249
We reasoned that the defect in meiotic progression in rad16-249 reflects defects in repair of programmed meiotic DSBs defining a function that is important, but not essential. We therefore examined the formation of programmed DSBs using pulsed field gel electrophoresis (PFGE). In these experiments, programmed breaks are typically observed as a smear below the three chromosome pairs (Young et al. 2002; Catlett and Forsburg 2003). We used temperature shift to induce a synchronous meiosis in h−/mat2-102 pat1/pat1 diploids, which maintain ploidy and mating type signaling, but allow temperature-dependent synchronous meiotic progression (Kohli et al. 1977; Yamamoto and Hiraoka 2003; Pankratz and Forsburg 2005).
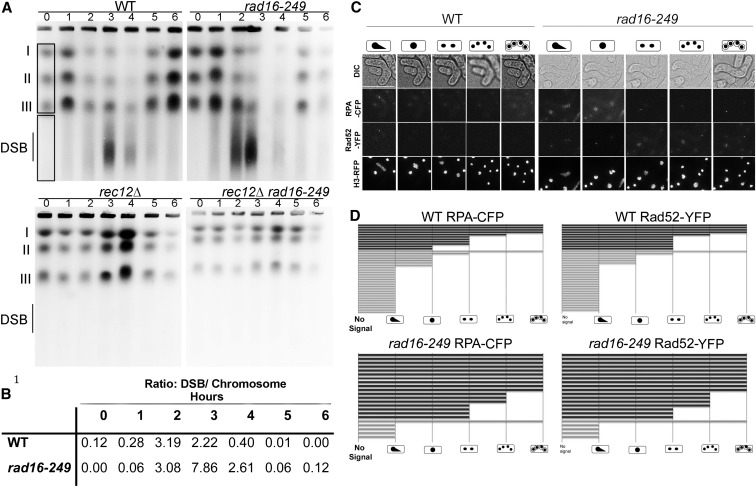
Consistent with previous studies, we observed the induction of meiotic DSBs in wild-type cells beginning 3 hr after temperature shift with the majority being repaired by 4 hr (Figure 2A). To quantify the amount of DNA in the breaks, we determined the signal intensity in the DSB smear relative to the total signal observed in the whole chromosomes that enter the gel, indicated by the boxes in lane 0 (Borde et al. 2000). In the characteristic wild-type pattern, there is a transient reduction of the signal corresponding to whole chromosomes, which is caused by DSBs and also by chromosomes with unresolved recombination intermediates that are retained in the well. Repair of the DSBs and resolution of recombination leads to restored migration of the intact chromosomes by 5 hr.
Figure 2.
Meiotic DSBs in diploids. Data from representative experiment selected from three independent trials. (A) Representative image of three pulse field gel experiments of synchronous mat2-102 pat1-114 diploid meiosis indicating chromosomes I, II, III, and DSBs for 0, 1, 2, 3, 4, 5, and 6 hr. (B) Quantification of gel in A showing the ratio of DSBs/total chromosome signal for each time point once the local background has been subtracted. (C) Representative panels of live cell imaging for RPA-CFP and Rad52-YFP. Bar, 15 μm. (D) Quantification of RPA-CFP and Rad52-YFP focus persistence in meiotic cells. Each bar is a single cell. Bars with dark shading indicate cells in which there was an abnormal segregation event; bars with light shading represent apparently normal meiotic progression.
The overall pattern in rad16-249 cells is roughly similar to wild type. The bulk of the population shows restored chromosome entry, although the DSB:whole chromosome ratio remains slightly elevated even at 6 hr (Figure 2, A and B). The DSBs we see in rad16-249 depend upon Rec12 (Figure 2A), and the timing of meiS phase, MI and MII divisions in rad16-249 are similar to wild type (Figure S2), suggesting this is a post-Rec12 effect. This result contrasts with PFGE performed with repair-defective mutants such as mus81∆, rad54∆, or rad54∆ rdh54∆, in which the DSB smear persists throughout the entire time course, consistent with their catastrophic failure to repair or resolve the broken DNA (Catlett and Forsburg 2003; Young et al. 2004).
As an independent assay to see whether damage persists in rad16-249 cells, we examined the formation and resolution of DNA damage markers during meiosis in wild-type and rad16-249 diploids. Cells with DNA damage show increased numbers of foci of fluorescently tagged RPA and Rad52, so that these markers provide a metric for unresolved DSBs (e.g., Lisby et al. 2004; Sabatinos et al. 2012). In wild-type cells, the RPA and Rad52 signals were resolved by the MI division in the majority of cells (71 and 67%, respectively). In rad16-249, we observed resolution in only 22% of cells. The majority of the rad16-249 cells (77%) had RPA and Rad52 signals that persisted through nuclear divisions and as far as spore formation (Figure 2D). The presence of fragments during meiotic divisions largely correlated with the persistence of both RPA and Rad52 signals, suggesting failure to resolve recombination structures leads to the abnormal segregation.
rad16-249 alters recombination frequencies
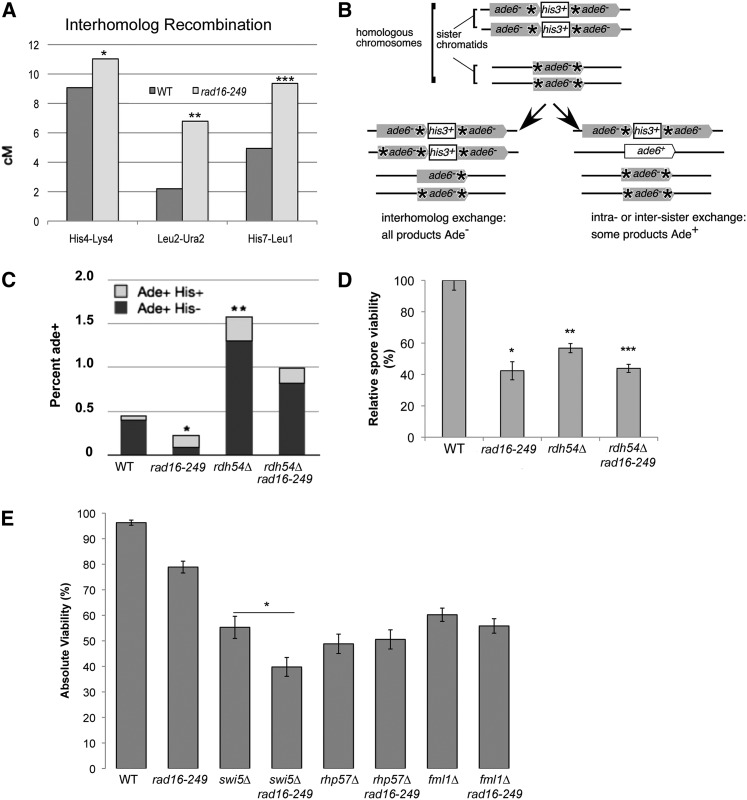
We investigated recombination outcomes in rad16-249 by examining three different intergenic regions: his4+–lys4+ and his7+–leu1+ on chromosome II and ura1+–leu2+ on chromosome I. Surprisingly, we observed a modest but statistically significant increase in crossovers among the surviving spores in rad16-249 when compared to wild type in all intervals tested (Figure 3A and Table S3). The wild-type genetic distance was 9.05 cM, 2.02 cM, and 4.94 cM for the his4–lys4, leu2–ura2, and his7–leu1 intervals, respectively, while rad16-249 was 11.03 cM (P-value = 0.0024), 6.79 cM (P-value = 0.027), and 9.36 cM (P-value = 0.018). The increased recombination in the his4–lys4 interval was also verified by tetrad dissections. Again, there was a statistically significant difference (χ2 = 5.898, alpha between 0.01 and 0.02) between WT (7.18 cM) and rad16-249 (11.73 cM; Table S2). Importantly, we saw no evidence for dyad formation or diploid offspring that might affect these ratios. We observed no striking difference in gene conversion events (3:1 or 1:3 segregation ratios) in rad16-249 (3%) compared to wild type (1%, similar to other reports, Rudolph et al. 1999).
Figure 3.
Recombination of intergenic and intragenic intervals and spore viability. Significance established by two-tailed t-test. (A) Recombination frequencies for homolog intergenic recombination. At least four trials were done for each interval for a total of at least 5000 spores analyzed. *P = 0.0023, **P = 0.027, and ***P = 0.018, respectively, for rad16-249 compared to WT for each interval. (B) Diagram indicating types of repair between the sister and the homolog using the ade-his-ade allele. (C) *P = 0.028 and **P = 0.016, for rad16-249 and rdh54∆ compared to WT, respectively. (D) Spore viability determined via random spore analysis. (E) Absolute viability assayed via tetrads. Error bars represented as standard error. *χ2 = 6.01, alpha = 0.02 for swi5∆ rad16-249 double mutant compared to swi5∆ single mutant.
Although a modest increase in crossovers was apparent in several genetic intervals, we did not see the same effects when we measured gene conversion at a single locus induced by the ade6-M26 hotspot allele (Gutz 1971; Goldman and Smallets 1979; Ponticelli et al. 1988). We observed a modest decrease in the recovery of Ade+ recombinants in rad16-249 relative to wild type (Table S3).
Typically, meiotic DSBs can be repaired using either the homologous chromosome or the sister chromatid, a distinction referred to as “partner choice.” Only recombination with the homologous chromosome has the potential to generate a genetic CO event, although these chiasmata can also be resolved as noncrossovers (NCOs). With the increased rate of homologous CO events in rad16-249, we reasoned that the balance of repair between the homologous chromosome and the sister chromatid might be disrupted. To test this, we employed a diploid in which one copy of chromosome 3 contains a double point mutant ade6-M375-M210, while the other copy contains tandem heteroallelic ade6-L469/pUC8/his3+/ade6-M375 (as described in Catlett and Forsburg 2003; Pankratz and Forsburg 2005). In this configuration, an ade6+ allele can typically only be recovered via intersister or intrasister exchange (because M210 and M469 are only 2 bp apart (Szankasi et al. 1988; R. MacFarlane and W. Wahls, personal communication), so this can be used as a rough metric for intrachromatid or intrasister events (Figure 3B). We found that rad16-249 has a 1.9-fold decrease in sister exchanges, measured by the recovery of Ade+ spore clones (Figure 3C). We examined the types of sister chromatid exchanges (SCE) events recovered by scoring the presence of the his3+ marker. In mitotic cells, conversion events that keep his3+ represent short tracts of recombination between the repeats, while deletion events are thought to result from SSA, nonconservative one-sided invasion, replication slippage, intrachromatid crossing over, or unequal sister chromatid crossing over (Osman et al. 2000). There was a modest increase in the proportion of conversion types (Ade+ His+) in rad16-249 cells compared to wild type (Ellermeier et al. 2004), although this was not statistically significant (Table S4, Figure 3C).
Previous studies have examined the effect of rad16 in haploids containing a similar ade6-his+-ade6 allele and have observed a modest increase in recombination in rad16 cells compared to wild type during vegetative growth (Osman et al. 2003a). Since we see the opposite effect in meiosis of what was observed in mitosis, we infer that this is unlikely to represent rearrangements during mitotic expansion of the spore clones. To test this, we repeated the experiment, plating the spores on low adenine media where Ade+ colonies are white and Ade− colonies are pink (Table S8). We reasoned that rearrangements that occur during meiosis should generate a clonally pure Ade+ colony, which should appear completely white, while rearrangements that occur during mitotic expansion should generate a colony with white and pink sectors. We observed sectors in ∼10% of wild-type Ade+ colonies, and close to 50% of rad16 Ade+ colonies. The frequency of nonsectored Ade+ colonies (generated by rearrangement during meiosis) is just under 0.5% in wild type, and ∼0.17% in rad16, reflecting the trend we observed in the larger experiment. Further, consistent with the report of Osman et al. (2003a), the frequency of sectored Ade+ colonies reflecting rearrangements during mitosis was higher in rad16 (0.11%) than wild type (0.06%).
In a previous study, we showed that the meiosis-specific homologous recombination mutant rdh54∆ causes an increase in sister exchanges using this assay, with no mitotic phenotype observed (Catlett and Forsburg 2003). Since this is the opposite of rad16-249, we constructed a double mutant and found that rad16-249 reduces the frequency of intrahomolog exchanges in rdh54∆, though it still remains elevated over wild type. Spore viability is not changed, with both single mutants and the double mutant each showing 50% spore viability compared to wild type (Figure 3D).
Interhomolog events, both CO and NCO, depend primarily on the Swi5/Sfr1 complex to mediate Rad51 filament formation, particularly at hotspots (Akamatsu et al. 2003; Hyppa and Smith 2010). Rad55 and Rad57 play a minor role in both interhomolog and intrasister exchanges at DSB-poor regions, while Rad52/Rti1 are implicated primarily in intrasister events (Akamatsu et al. 2003; Octobre et al. 2008; Hyppa and Smith 2010). We constructed double mutants to investigate whether rad16-249 could be placed genetically in either of these pathways. We performed tetrad analysis and found that spore viability in swi5∆ rad16-249 mutants was significantly reduced compared to either single mutant, while rad57∆ rad16-249 did not show a significant change (Figure 3E). This synthetic phenotype suggests that Rad16 functions in a pathway separate from the Swi5-mediated interhomolog events, consistent with a role in intersister exchanges.
The choice between NCO and CO for resolution of chiasmata in the homologous chromsomes depends upon the Fml1 (FANCM) translocase, which limits CO in favor of NCO (Lorenz et al. 2012). Thus, fml1∆ mutants also show evidence of increased homologous exchange. We constructed a double mutant between fml1∆ and rad16-249 and performed tetrad analysis. We see a modest decrease in spore viability in fml1∆ that is unchanged in the fml1∆ rad16-249 double mutant (Figure 3E).
rad16-249 has genetic interactions with other DNA damage repair mutants
In Drosophila, the XPF ortholog Mei9 functions as a Holliday junction resolvase in meiosis (Yildiz et al. 2002). In fission yeast, the primary meiotic resolvase is Mus81 (Boddy et al. 2001; Doe et al. 2002; Gaillard et al. 2003; Smith et al. 2003; Gaskell et al. 2007), and in its absence, cells are unable to complete meiosis (Boddy et al. 2001; Osman et al. 2003b; Smith et al. 2003) (Figure 2). We were unable to construct a double mutant between rad16-249 and mus81∆ (Table 1); this synthetic lethality suggests that lesions produced in mus81∆ mutants in vegetative growth absolutely depend upon Rad16 for resolution and vice versa. This is consistent with data suggesting that Rad16 and Mus81 overlap to maintain genome stability in metazoans (Mazon et al. 2012; Muñoz-Galván et al. 2012; Kikuchi et al. 2013; Saito et al. 2013). In vegetative fission yeast cells, Rad16 functions as a template specific resolvase during repair of replication forks, along with Mus81 (Roseaulin et al. 2008).
Table 1. Genetic interactions: phenotype of double mutants with rad16-249.
| Mutant | Viability | UV sensitivity | MMS sensitivity | Comments |
|---|---|---|---|---|
| rhp51∆ | Lethal | — | — | |
| mus81∆ | Lethal | — | — | |
| rad3∆ | Lethal | — | — | |
| rqh1∆ | Lethal | — | — | |
| pcn1-K164R | Lethal | — | — | |
| rhp18∆ | Lethal | — | — | |
| rad2∆ | Viable | Enhanced | Enhanced | c |
| mms2∆ | Viable | Enhanced | Enhanced | b,c |
| ubc13∆ | Viable | Enhanced | Enhanced | b,c |
| eso1 | Viable | Enhanced | Like rad16-249 | c |
| kpa1∆ | Viable | Enhanced | Enhanced | |
| apn2∆ | Viable | Enhanced | Enhanced | b,c |
| nth1∆ | Viable | Enhanced | Enhanced | c |
| saw1∆ | Viable | Like rad16-249a | Like rad16-249a | |
| slx4∆ | Viable | Like rad16-249a | Like rad16-24a |
One parent is not noticeably sensitive to drug.
Colony size smaller than either parent, indicating poor growth.
Colonies darker pink than either single mutant on PhloxinB, indicating poor health.
The Mus81 endonuclease is essential for viability in rqh1∆ mutants, which lack the RecQ helicase that restrains recombination in mitotic cells (Doe et al. 2002). If Rad16 and Mus81 overlap, we reasoned that rad16-249 rqh1∆ should also be lethal, and this was observed (Table 1). Similarly, rad16-249 and rad51∆ double mutants are synthetically lethal. These data indicate that Rad16 plays an important role to preserve genome stability even in unperturbed cells.
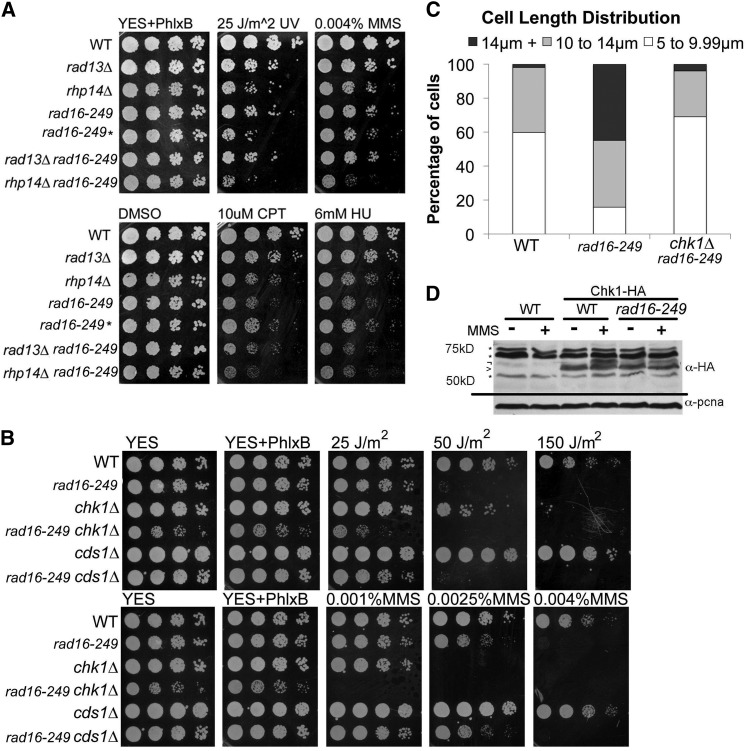
Next, we investigated the spectrum of damage sensitivity associated with mutation of Rad16XPF, Rhp14XPA, or Rad13XPG. Consistent with their role in NER, we observed similar sensitivity to UV and MMS in rad16-249, rhp14∆, and rad13∆ (Figure 4A). We also observed sensitivity to CPT in rad16-249 and rhp14∆ mutants, but not in the rad13∆ mutant. This is consistent with previous work in Saccharomyces cerevisiae that identified a role for Rad1XPF in resolution of topoisomerase-bound intermediates caused by camptothecin (CPT) treatment (Vance and Wilson 2002). Finally, we observed sensitivity to hydroxyurea (HU) in rad16-249 and rhp14∆ mutants, but again not in rad13∆. HU causes fork stalling due to nucleotide depletion, and restart occurs via recombination-based mechanisms (Meister et al. 2007; Lambert et al. 2010; Sabatinos et al. 2012).
Figure 4.
Checkpoint activation and DNA damaging drug sensitivity. (A) Representative image of long-term viability and sensitivity. Equal concentrations of exponentially growing cells in YES plated in 5× serial dilutions from left to right. *, indicates rad16-249 mutation in a background comparable to the background of rhp14∆ mutant while no * is rad16-249 mutation in background comparable to rad13∆. (B) Representative image of long-term viability and sensitivity. Equal concentrations of exponentially growing cells in YES plated in 5× serial dilutions from left to right. (C) Cell length distribution of exponentially gorwing cells in YES. (D) Western blot of Chk1-HA using 12CA5 anti-HA antibody with and without treatment of 0.01% MMS. *, indicates nonspecific bands; >, is Chk1-HA specific band; ¬, indicates modified Chk1-HA band.
Slx4 and Saw1 are proposed to function as scaffolds that assemble XPF and other structure-specific endonucleases (Lyndaker and Alani 2009; Kashiyama et al. 2013; Li et al. 2013; Wan et al. 2013). In contrast to budding yeast (Flott et al. 2007; Li et al. 2008), neither slx4∆ nor saw1∆ is sensitive to UV or MMS in fission yeast (Figure S3 and (Coulon et al. 2006). We observed no synthetic growth defects in double mutants between rad16-249 and either slx4∆ or saw1∆, and little if any effect on MMS or UV sensitivity compared to rad16-249 alone (Table 1 and Figure S3).
Finally, we examined interactions with components of postreplication repair (PRR) pathways. We found that rad16-249 is synthetically lethal with rhp18∆ or pcn1-K164R (Table 1), which affect both error-prone (translesion synthesis) and error-free postreplication repair pathways (Frampton et al. 2006). We found synthetically lethal interactions similar to that of rad16-249 in double mutants with rhp14∆, but not rad13∆.
When rad16-249 is combined with error-free PRR pathway mutants mms2∆ and ubc13∆ (Brown et al. 2002), we do not observe synthetic lethality, but rather increased sensitivity to UV and MMS (Table 1 and Figure S3). When rad16-249 is combined with kpa1∆ (error-prone DNA polymerase kappa; Kai and Wang 2003) we also observe increased sensitivity to damage caused by UV and MMS. There was an enhancement of UV sensitivity when rad16-249 was combined with an eso1 allele that deletes the polymerase eta domain, but no change in MMS sensitivity. There were modest synthetic growth defects and increased damage sensitivity in double mutants between rad16-249 and other repair mutants including the flap endonuclease rad2∆ (Yonemasu et al. 1997), the base excision repair mutant nth1∆ (Osman et al. 2003a), and the base excision repair mutant apn2∆ (Fraser et al. 2003), all consistent with linked repair functions.
The DNA damage checkpoint is constitutively active in rad16-249
The rad16-249 mutants have an elongated cell morphology, which is typically evidence of checkpoint activation from intrinsic DNA damage (Figure 4C and Table S5; reviewed in Gomez and Forsburg 2004). We examined nuclear morphology during the vegetative cell cycle using RFP-histone and observed fragmented or lagging histone signals in both rad16-249 and rhp14∆, although less frequently than in meiosis (9.62 and 8.25% of cells for rad16-249 and rhp14∆, respectively; Table S6 and File S8). This, along with the double mutant analysis, suggests Rad16 is required for chromosome stability even in the absence of external perturbations. However, despite this, rad16-249 cells maintain a high level of viability, with plating efficiency of 93.5% (SD ± 7%) relative to wild type.
We constructed double mutants between rad16-249 and cds1∆, which disrupts the intra-S phase checkpoint (Lindsay et al. 1998; Rhind and Russell 2000): chk1∆, required for the DNA damage checkpoint pathway (Walworth and Bernards 1996; Rhind and Russell 2000), and rad3∆, the ATR homolog at the apex of both pathways, which is required for other damage responses as well (Bentley et al. 1996; Edwards et al. 1999; Rhind and Russell 2000; Du et al. 2003; Rozenzhak et al. 2010). We observed no additional growth defect in the rad16-249 cds1∆ double mutants compared to either single mutant (Figure 4B). In contrast, the rad3∆ rad16-249 double mutant is synthetically lethal (Table 1), again consistent with chronic DNA damage caused by rad16-249.
The rad16-249 chk1∆ double mutants were viable, but slow growing, with reduced cell size. These cells also showed heightened sensitivity to UV and MMS sensitivity (Figure 4B). Chk1 is activated by phosphorylation, which causes a characteristic mobility shift on SDS/PAGE (Walworth and Bernards 1996). We observed a slower-migrating Chk1-HA in rad16-249 asynchronously growing cultures in both the presence and absence of MMS, consistent with intrinsic damage (Figure 4D). Similar results were observed for rhp14∆ (Figure S4). We did not see Chk1 activation in rad13∆, consistent with previously reported results (Herrero et al. 2006).
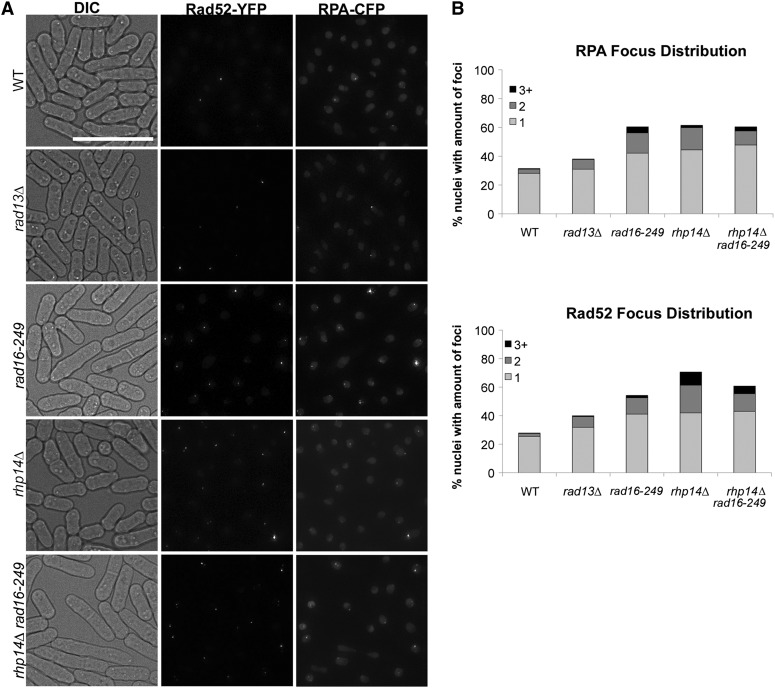
To determine whether Chk1 activation in rad16-249 reflects increased DNA damage, we examined live vegetative cells containing fluorescently tagged damage markers. We observed single stranded DNA (ssDNA) by RPA-CFP and repair foci marked by Rad52-YFP (e.g., (Lisby et al. 2004; Sabatinos et al. 2012). In wild-type cells, we found 28% contained a single RPA focus and 25% contained a single Rad52 focus (Figure 5, e.g., Sabatinos et al. 2012). However, we saw an increased number of cells with a single RPA (42%) and Rad52 (41%) focus in rad16-249. Similar results were observed for rhp14∆, but not rad13∆.
Figure 5.
Visualization of DNA damage via Rad52 and RPA foci. (A) Representative images of Rad52-YFP and RPA-CFP foci in exponentially growing cultures in YES media for designated genotypes. Number of nuclei analyzed for wild type, rad13∆, rad16-249, rhp14∆, and rhp14∆ rad16-249 is 1816, 689, 1131, 854, and 843, respectively. See Table S7 for standard error and confidence interval. (B) Distribution of nuclei containing 1, 2, or 3 + Rad52-YFP and/or RPA-CFP foci.
Discussion
XPF is a conserved, structure-specific endonuclease with multiple functions in genome maintenance (Schwartz and Heyer 2011). Originally linked to nucleotide excision repair, XPF, its binding partner ERCC1, and its loading factor XPA are also implicated in repair of ICLs, in SSA, homologous recombination, and telomere maintenance (Bogliolo et al. 2013; Kashiyama et al. 2013). XPF appears to be particularly important to trim unpaired ssDNA at the boundaries of limited homology domains and cleaves the 3′ end of nonhomologous flaps (Paques and Haber 1997; Hollingsworth and Brill 2004; Fagbemi et al. 2011; Schwartz and Heyer 2011; Mazon et al. 2012). The role of XPF in meiosis varies considerably in different species. In Drosophila, Mei9XPF is essential for resolution of meiotic chiasmata (Yildiz et al. 2002), while in C. elegans, XPF overlaps with two other nucleases, Mus81 and SLX-1, in meiosis (Agostinho et al. 2013; O’Neil et al. 2013; Saito et al. 2013). In budding yeast, Rad1XPF appears to have no function in meiosis (Higgins et al. 1983). Another structure-specific endonuclease, Yen1, functionally overlaps with ScMus81 and ScRad1XPF in budding yeast mitosis (Blanco et al. 2010; Muñoz-Galván et al. 2012) and functions in meiosis to resolve late COs (Matos et al. 2011), but there is no obvious Yen1 ortholog in fission yeast (Ip et al. 2008).
S. pombe Rad16XPF is required for normal meiosis
We investigated the role of the S. pombe XPF nuclease Rad16 in meiosis, using a newly characterized truncation allele that eliminates all the conserved domains of the protein. We find that rad16-249 mutants undergo meiosis with normal timing, including formation and repair of Rec12-dependent DSBs, but nevertheless show a reduction in spore viability to about half of wild-type levels. Loss of viability was also reported in a previous study, using a different allele (Lorenz et al. 2012). In tetrad dissection, only about one-third of four-spored tetrads are capable of germinating all four spores (4:0 viable), while the remainder range from 3:1 to 0:4 viability. This indicates a role in meiosis that is important, but not absolutely essential.
Using live-cell imaging, we observed that rad16-249 mutants suffer chromosome segregation abnormalities during both MI and MII (Figure 1). These are apparent as extra spots of histone-RFP or DAPI, which are smaller and less bright than a full nucleus. Many, but not all of these apparent fragments lack the telomere marker Taz1, suggesting they are chromosome fragments, rather than full chromosomes. They often are left outside of the spore wall, consistent with being disconnected chromosome fragments that are not attached to a kinetochore. We see similar fragmentation phenotypes for rhp14∆XPA, but not for rad13∆XPG. This agrees with data showing that XPA and XPF have functions independent of XPG and other NER proteins (Paques and Haber 1999; Lyndaker and Alani 2009) and implicates XPA and XPF in a distinct meiotic function.
The cells that produce fragments are more likely to display persistent RPA and Rad52 foci during meiotic divisions, whereas these signals are generally resolved prior to divisions in wild-type cells, or cells without fragments (Figure 2). This suggests that the fragmentation phenotype is associated with a failure to properly resolve DNA damage, either due to intrinsic stress or defects in resolution of a subset of recombination structures. The progression through meiotic divisions despite the presence of damage signals is consistent with previous observations suggesting that the damage checkpoint is not triggered in meiosis (Pankratz and Forsburg 2005).
We compared the rad16 phenotype to rec12∆ mutants (Figure 1), which do not create programmed double strand breaks (Sharif et al. 2002). In contrast to rad16, the rec12∆ fragments always contain at least one Taz1 signal, consistent with aberrant segregation of intact chromosomes. We infer therefore that Taz1-minus chromosome fragments in rad16 result from unrepaired breaks or from damage that occurs due to aberrant segregation of unresolved recombination intermediates. This is consistent with our previous report of chromosome fragments during meiosis in rad54∆ rdh54∆ double mutants, which are completely deficient in meiotic DSB repair and produce no viable spores (Catlett and Forsburg 2003). We examined the phenotype of mus81∆ cells, which lack the main Holliday junction resolvase in fission yeast and fail to segregate their chromosomes due to unresolved entanglements (Boddy et al. 2000, 2001; Osman et al. 2003b; Smith et al. 2003). Using live cell analysis, we confirmed that the majority of Mus81 cells fail to undergo chromosome segregation particularly in MI, with only a small percentage showing evidence for Taz1-minus chromosome fragments. Thus, the rad16-249 phenotype is clearly distinct from that of mutants that fail to form DSBs (rec12∆, which segregates whole chromosomes) or fail to resolve crossovers (mus81∆, which remains largely entangled) and more closely resembles the phenotype of rad54∆ rdh54∆, which is deficient in repair, although the phenotype in rad16 is much less penetrant.
The majority of breaks are repaired in rad16 mutants
We used a PFGE assay to examine DSB formation and resolution more closely (Figure 2). During DSB formation and resolution, the three chromosomes show reduced migration; replaced by a smear of low molecular weight DNA represents breakage and joint molecules that are retained in the well. We observe that the rad16-249 mutants have roughly normal timing for the formation and disappearance of the majority of programmed double strand breaks and repair, as measured by PFGE. However, at later time points, there is a modest but persistent background of low molecular weight DSB signal (Figure 2). By comparison, in mus81∆, failure to resolve joint molecules reduces the whole chromosome signal and strikingly increases in the smear of DSBs throughout the entire timecourse (Young et al. 2004). Similar observations of reduced whole chromsomes and persistent DSBs were made in rad54∆ rdh54∆, which is also completely deficient in DSB repair (e.g., Catlett and Forsburg 2003). We conclude that most DSBs are repaired in rad16∆, but a subset remains.
One possibility is that rad16-249 simply has more DSBs due to intrinsic genome instability in this strain (see below). If this were the case, there would be a fraction of DSBs occurring that are independent of Rec12. Mutations that cause breaks due to genome instability can partially rescue the viability defect associated with rec12∆ mutants (Farah et al. 2005; Pankratz and Forsburg 2005), but we see no evidence of that in rec12∆ rad16 double mutants. Additionally, we observed no DSB smear in rad16-249 rec12∆, indicating that the breaks observed are Rec12 dependent.
rad16 increases crossovers between homologous chromosomes
We observe a statistically significant increase in the rate of COs between homologous chromosomes in several genetic intervals in rad16 compared to wild type (Figure 3). This indicates that rad16 cells are competent for some form of DSB repair, consistent with the PFGE result. An increased rate could represent a shift in the balance between crossover and noncrossover resolution of chiasmata between homologous chromosomes. Alternatively, or in addition, it could indicate a shift in partner choice from the sister chromatid to the homologous chromosomes.
In contrast to the interhomolog recombination data, we observe different results for gene conversion involving the ade6-M26 hotspot, in which rad16-249 reduces the frequency of Ade+ spores recovered. A similar modest reduction at the ade6 hotspot was also observed by Lorenz et al. (2012). Biased conversion and marker effects at the ade6 hotspot have linked to defects in mismatch repair (Schuchert and Kohli 1988; Szankasi and Smith 1995; Fleck et al. 1999), which remains a possible explanation for the results with the ade6 hotspot. Previously, rad16 was reported to reduce gene conversion at an ade6 hotspot that also contained unpaired heteroduplex DNA (Farah et al. 2005, 2009); this would be consistent with XPF function at unpaired flaps (Schwartz and Heyer 2011). Why rad16 has different effects at normal intervals than at the hotspot is unclear.
To examine whether the increase in homologous CO reflects a change in partner choice, we investigated sister chromatid events using a substrate in which only intra- or interchromatid events can give an Ade+ colony (Catlett and Forsburg 2003; Pankratz and Forsburg 2005). We observed reduced frequency of Ade+ colonies in rad16-249 mutants compared to wild type (Figure 3). Although this reduction in recombination might reflect the role of Rad16 in processing heterologous flap structures, and thus be an artifact of the tandem allele construct we used, we consider this unlikely. In vegetative haploids, mutation of rad16 actually increases recovery of Ade+ at the ade6-L469/pUC8/his3+/ade6-M375 locus (Osman et al. 2003a); therefore, there is no intrinsic impediment to resolution in the absence of rad16. In agreement with this, we observe an increase in Ade+ sectoring during mitotic growth in rad16 haploids compared to wild type (Table S8). We suggest that the rad16 mutant is impaired in sister chromatid recombination during meiosis.
Previously, we observed several situations in which recombination using this sister construct was increased rather than decreased. In rdh54∆ mutants, a modest increase in use of the sister is accompanied by a modest decrease in recombination with the homolog, consistent with a role in partner choice (Catlett and Forsburg 2003). We observe that the rad16-249 mutant partly reverses the rdh54∆ effect without rescuing its spore viability (Figure 3). This also suggests that rad16-249 is deficient in the resolution of sister recombination events. Increased recovery of Ade+ offspring is also seen in DNA checkpoint mutants, which we inferred is due to sister-mediated repair of genome instability, similar to that occurring in vegetative cells (Pankratz and Forsburg 2005). However, in that case, this leads to Rec12-independent DNA damage and rescue of rec12∆ viability. Despite the genome instability associated with rad16, we see no evidence for rad16-induced DNA damage during meiosis.
A substantial fraction of the joint molecules in fission yeast are formed between sister chromatids, not between homologs (Cromie and Smith 2008), and this is particularly true for areas of efficient DSB formation (Hyppa and Smith 2010). The Swi5/Sfr1 recombination mediator appears to be particularly important for interhomolog exchanges in regions of DSB hotspots, while Rad22/Rti1 are proposed to function at intersister exchanges (Akamatsu et al. 2007; Hyppa and Smith 2010). The Rad55/Rad57 mediator, which is distinct from Swi5/Sfr1, may be more important for exchanges in cold regions, affecting both interhomolog and intersister events (Khasanov et al. 1999, 2008; Akamatsu et al. 2003; Hyppa and Smith 2010). We find that rad16-249 swi5∆ double mutants have reduced spore viability compared to the single mutants, which suggests that Rad16 operates in a pathway that is separate from Swi5. We see little additive effect in rad16-249 rad57∆ double mutants. Thus we suggest that Rad16 may play a role in resolution of events mediated by Rad55/57, particularly those involving the sister.
Consistent with this, mutations that reduce sister chromatid exchanges in C. elegans without affecting crossovers between homologs generate DNA fragments (Bickel et al. 2010), leading to the suggestion that homolog-independent recombination is important to preserve genome stability in meiosis. We conclude that events involving the sister chromatid, rather than the homologous chromosome, are similarly important for meiotic genome stability in fission yeast.
Genome instability in vegetative rad16 cells
Rad16 is clearly important for genome stability even in otherwise unperturbed vegetative fission yeast cells. The rad16-249 mutants suffer disordered segregation, with increased damage foci from markers Rad52-YFP and RPA-CFP and constitutive activation of the DNA damage checkpoint kinase Chk1. Rad16 contributes to replication fork recovery in response to different replication stress conditions (Roseaulin et al. 2008; Muñoz-Galván et al. 2012). This implies that replication stresses are intrinsic to normal cell cycle progression and require Rad16 for effective management. The synthetic lethality of rad16-249 rad3∆ indicates that in addition to Chk1 activation, other repair activities initiated by Rad3 are also important for rad16-249 viability. These could include histone H2A(X) phosphorylation, upstream checkpoint proteins, or chromatin effectors (Edwards et al. 1999; Du et al. 2003; Rozenzhak et al. 2010).
We observe CPT and HU sensitivity in both rhp14∆ and rad16-249 mutants. This contrasts with budding yeast in which Rad14XPA, the XPF loading factor, is not required for CPT resistance (Vance and Wilson 2002). Instead, Sc Slx4 and Saw1 are proposed to provide an alternative XPF loading complex, and Slx4 with Slx1 forms another structure-specific endonuclease (Lyndaker and Alani 2009; Schwartz and Heyer 2011). In fission yeast, Slx4-Slx1 endonuclease is linked to rDNA maintenance via a Rad51-independent recombination mechanism (Coulon et al. 2006), but so far there is no evidence that Slx4 and Saw1 affect Rad16XPF in S. pombe. Significantly we and others observe no damage sensitivity in slx4∆ or saw1∆ mutants (Figure S3 and Coulon et al. 2006) and no change in nucleolar morphology in rad16-249 (data not shown), suggesting they operate independently.
The instability of rad16-249 mutants even in an unperturbed mitotic cell cycle suggests that its role in recombination may be an important component to normal genome maintenance. A potential collaborator may be PCNA, the replication clamp that ensures processive replication. Ubiquitylation of PCNA on K164 by SpRhp18 (Sc Rad18) is required for postreplication repair. Polyubiquitylation is required for error-free PRR (Frampton et al. 2006). We observed synthetic lethality between rad16-249 and rhp18∆ or pcn1-K164R, but not with genes that affect its polyubiquitylation, which suggests that PCNA monoubiquitylation is essential for viability in the absence of Rad16. Interestingly, several studies implicate PCNA modification in maintenance of repeat stability (Motegi et al. 2006; Daee et al. 2007; Putnam et al. 2010) and in Exo-1-mediated resection (Chen et al. 2013). The XPF loading factor XPA binds PCNA, and XPA and XPF are found as constituents of the replication fork in unperturbed cells (Gilljam et al. 2012). Indeed, the decreased stability of the ade6 heteroallele in mitosis may reflect a rad16-related instability of the replication fork in repetitive sequences.
DNA replication is intrinsically a source of DNA damage (reviewed in Lehmann and Fuchs 2006). Structure-specific endonucleases such as Mus81 and XPF actively contribute to recombination events that rescue damaged replication forks or other structures, thus promoting genome stability (e.g., Roseaulin et al. 2008; Willis and Rhind 2009; Muñoz-Galván et al. 2012). The synthetic lethality we observe between rad16-249 and mus81∆ is consistent with data in metazoans that argues for redundancy between these two enzymes, with deficiency leading to increased double strand DNA breaks (Kikuchi et al. 2013). Yet these same proteins contribute actively to gross chromosome rearrangements, including translocations, which are typical of cancer (Mazon et al. 2012; Pardo and Aguilera 2012).
The choice of a helpful or harmful pathway may reflect access to repetitive sequences that facilitate SSA forms of repair. Typically, regions closest to the DSB are used preferentially for intrachromatid repair by the SSA pathway (Ray et al. 1988; Nickoloff et al. 1989; Sugawara and Haber 1992; Frankenberg-Schwager et al. 2009), and evidence suggests that the extent of resection in meiosis influences choice of repair pathways (Neale et al. 2002). There is likely to be a closely regulated interplay between resection, helicase-driven resolution of recombination structures, and the activity of structure-specific endonucleases that determines the outcome of these events.
Supplementary Material
Acknowledgments
We thank JiPing Yuan for help with pulsed field gels, Marc Green for assistance with imaging, and Sarah Sabatinos for advice on meiosis. We thank Julie Cooper and Henning Schmidt for strains. We thank Matt Michael, Oscar Aparicio, and members of the Forsburg lab for helpful comments on the manuscript. This work was supported by National Institutes of Health grant R01-GM81418 to S.L.F.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.171355/-/DC1.
Communicating editor: S. K. Sharan
Literature Cited
- Agostinho A., Meier B., Sonneville R., Jagut M., Woglar A., et al. , 2013. Combinatorial regulation of meiotic holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 9: e1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu Y., Dziadkowiec D., Ikeguchi M., Shinagawa H., Iwasaki H., 2003. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl. Acad. Sci. USA 100: 15770–15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu Y., Tsutsui Y., Morishita T., Siddique M. S., Kurokawa Y., et al. , 2007. Fission yeast Swi5/Sfr1 and Rhp55/Rhp57 differentially regulate Rhp51-dependent recombination outcomes. EMBO J. 26: 1352–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley N. J., Holtzman D. A., Flaggs G., Keegan K. S., DeMaggio A., et al. , 1996. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 15: 6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Bickel J. S., Chen L., Hayward J., Yeap S. L., Alkers A. E., et al. , 2010. Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability. PLoS Genet. 6: e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M. G., Matos J., Rass U., Ip S. C., West S. C., 2010. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair (Amst.) 9: 394–402. [DOI] [PubMed] [Google Scholar]
- Boddy M. N., Lopez-Girona A., Shanahan P., Interthal H., Heyer W. D., et al. , 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20: 8758–8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M. N., Gaillard P. H., McDonald W. H., Shanahan P., Yates J. R., 3rd, et al. , 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548. [DOI] [PubMed] [Google Scholar]
- Bogliolo M., Schuster B., Stoepker C., Derkunt B., Su Y., et al. , 2013. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am. J. Hum. Genet. 92: 800–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V., Goldman A. S., Lichten M., 2000. Direct coupling between meiotic DNA replication and recombination initiation. Science 290: 806–809. [DOI] [PubMed] [Google Scholar]
- Brookman K. W., Lamerdin J. E., Thelen M. P., Hwang M., Reardon J. T., et al. , 1996. ERCC4 (XPF) encodes a human nucleotide excision repair protein with eukaryotic recombination homologs. Mol. Cell. Biol. 16: 6553–6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M., Zhu Y., Hemmingsen S. M., Xiao W., 2002. Structural and functional conservation of error-free DNA postreplication repair in Schizosaccharomyces pombe. DNA Repair (Amst.) 1: 869–880. [DOI] [PubMed] [Google Scholar]
- Camenisch U., Dip R., Schumacher S. B., Schuler B., Naegeli H., 2006. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat. Struct. Mol. Biol. 13: 278–284. [DOI] [PubMed] [Google Scholar]
- Carr A. M., Schmidt H., Kirchhoff S., Muriel W. J., Sheldrick K. S., et al. , 1994. The rad16 gene of Schizosaccharomyces pombe: a homolog of the RAD1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 2029–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett M. G., Forsburg S. L., 2003. Schizosaccharomyces pombe Rdh54 (TID1) acts with Rhp54 (RAD54) to repair meiotic double-strand breaks. Mol. Biol. Cell 14: 4707–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes M. D., Farah J. A., Smith G. R., 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5: 883–888. [DOI] [PubMed] [Google Scholar]
- Chen X., Paudyal S. C., Chin R. I., You Z., 2013. PCNA promotes processive DNA end resection by Exo1. Nucleic Acids Res. 41: 9325–9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y., Hiraoka Y., 2001. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr. Biol. 11: 1618–1623. [DOI] [PubMed] [Google Scholar]
- Cooper J. P., Nimmo E. R., Allshire R. C., Cech T. R., 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747. [DOI] [PubMed] [Google Scholar]
- Cooper J. P., Watanabe Y., Nurse P., 1998. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392: 828–831. [DOI] [PubMed] [Google Scholar]
- Coulon S., Noguchi E., Noguchi C., Du L. L., Nakamura T. M., et al. , 2006. Rad22Rad52-dependent repair of ribosomal DNA repeats cleaved by Slx1-Slx4 endonuclease. Mol. Biol. Cell 17: 2081–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie G., Smith G. R., 2008. Meiotic recombination in Schizosaccharomyces pombe: a paradigm for genetic and molecular analysis. Genome Dyn Stab 3: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau D. L., Peng Y., Van Houten B., 2008. DNA repair gets physical: mapping an XPA-binding site on ERCC1. DNA Repair (Amst.) 7: 819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daee D. L., Mertz T., Lahue R. S., 2007. Postreplication repair inhibits CAG.CTG repeat expansions in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L., Smith G. R., 2003. Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics 163: 857–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C. L., Ahn J. S., Dixon J., Whitby M. C., 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277: 32753–32759. [DOI] [PubMed] [Google Scholar]
- Dolan W. P., Le A. H., Schmidt H., Yuan J. P., Green M., et al. , 2010. Fission yeast Hsk1 (Cdc7) kinase is required after replication initiation for induced mutagenesis and proper response to DNA alkylation damage. Genetics 185: 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L. L., Nakamura T. M., Moser B. A., Russell P., 2003. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell. Biol. 23: 6150–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R. J., Bentley N. J., Carr A. M., 1999. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1: 393–398. [DOI] [PubMed] [Google Scholar]
- Egel R., Beach D. H., Klar A. J., 1984. Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc. Natl. Acad. Sci. USA 81: 3481–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C., Schmidt H., Smith G. R., 2004. Swi5 acts in meiotic DNA joint molecule formation in Schizosaccharomyces pombe. Genetics 168: 1891–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagbemi A. F., Orelli B., Scharer O. D., 2011. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair (Amst.) 10: 722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah J. A., Cromie G., Steiner W. W., Smith G. R., 2005. A novel recombination pathway initiated by the Mre11/Rad50/Nbs1 complex eliminates palindromes during meiosis in Schizosaccharomyces pombe. Genetics 169: 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah J. A., Cromie G. A., Smith G. R., 2009. Ctp1 and Exonuclease 1, alternative nucleases regulated by the MRN complex, are required for efficient meiotic recombination. Proc. Natl. Acad. Sci. USA 106: 9356–9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck O., Lehmann E., Schar P., Kohli J., 1999. Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nat. Genet. 21: 314–317. [DOI] [PubMed] [Google Scholar]
- Flott S., Alabert C., Toh G. W., Toth R., Sugawara N., et al. , 2007. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol. Cell. Biol. 27: 6433–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Marini F., Gamba D., Lucchini G., Plevani P., 1994. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14: 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton J., Irmisch A., Green C. M., Neiss A., Trickey M., et al. , 2006. Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol. Biol. Cell 17: 2976–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg-Schwager M., Gebauer A., Koppe C., Wolf H., Pralle E., et al. , 2009. Single-strand annealing, conservative homologous recombination, nonhomologous DNA end joining, and the cell cycle-dependent repair of DNA double-strand breaks induced by sparsely or densely ionizing radiation. Radiat. Res. 171: 265–273. [DOI] [PubMed] [Google Scholar]
- Fraser J. L., Neill E., Davey S., 2003. Fission yeast Uve1 and Apn2 function in distinct oxidative damage repair pathways in vivo. DNA Repair (Amst.) 2: 1253–1267. [DOI] [PubMed] [Google Scholar]
- Fricke W. M., Brill S. J., 2003. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 17: 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard P. H., Noguchi E., Shanahan P., Russell P., 2003. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell 12: 747–759. [DOI] [PubMed] [Google Scholar]
- Gaskell L. J., Osman F., Gilbert R. J., Whitby M. C., 2007. Mus81 cleavage of Holliday junctions: A failsafe for processing meiotic recombination intermediates? EMBO J. 26: 1891–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilljam K. M., Muller R., Liabakk N. B., Otterlei M., 2012. Nucleotide excision repair is associated with the replisome and its efficiency depends on a direct interaction between XPA and PCNA. PLoS ONE 7: e49199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S. L., Smallets S., 1979. Site specific induction of gene conversion: the effects of homozygosity of the ade6 mutant M26 of Schizosaccharomyces pombe on meiotic gene conversion. Mol. Gen. Genet. 173: 221–225. [DOI] [PubMed] [Google Scholar]
- Gomez E. B., Forsburg S. L., 2004. Analysis of the fission yeast Schizosaccharomyces pombe cell cycle. Methods Mol. Biol. 241: 93–111. [DOI] [PubMed] [Google Scholar]
- Gregg S. Q., Robinson A. R., Niedernhofer L. J., 2011. Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease. DNA Repair (Amst.) 10: 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H., 1971. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics 69: 317–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A. B., Martin-Castellanos C., Marco E., Gago F., Moreno S., 2006. Cross-talk between nucleotide excision and homologous recombination DNA repair pathways in the mechanism of action of antitumor trabectedin. Cancer Res. 66: 8155–8162. [DOI] [PubMed] [Google Scholar]
- Higgins D. R., Prakash S., Reynolds P., Prakash L., 1983. Molecular cloning and characterization of the RAD1 gene of Saccharomyces cerevisiae. Gene 26: 119–126. [DOI] [PubMed] [Google Scholar]
- Hohl M., Christensen O., Kunz C., Naegeli H., Fleck O., 2001. Binding and repair of mismatched DNA mediated by Rhp14, the fission yeast homologue of human XPA. J. Biol. Chem. 276: 30766–30772. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N. M., Brill S. J., 2004. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 18: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppa R. W., Smith G. R., 2010. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell 142: 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip S. C., Rass U., Blanco M. G., Flynn H. R., Skehel J. M., et al. , 2008. Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361. [DOI] [PubMed] [Google Scholar]
- Kai M., Wang T. S., 2003. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 17: 64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman V., Mullen J. R., Fricke W. M., Bastin-Shanower S. A., Brill S. J., 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15: 2730–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiyama K., Nakazawa Y., Pilz D. T., Guo C., Shimada M., et al. , 2013. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am. J. Hum. Genet. 92: 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass E. M., Jasin M., 2010. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 584: 3703–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney H. M., Kirkpatrick D. T., Gerton J. L., Petes T. D., 2001. Meiotic recombination involving heterozygous large insertions in Saccharomyces cerevisiae: formation and repair of large, unpaired DNA loops. Genetics 158: 1457–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khasanov F. K., Savchenko G. V., Bashkirova E. V., Korolev V. G., Heyer W. D., et al. , 1999. A new recombinational DNA repair gene from Schizosaccharomyces pombe with homology to Escherichia coli RecA. Genetics 152: 1557–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khasanov F. K., Salakhova A. F., Khasanova O. S., Grishchuk A. L., Chepurnaja O. V., et al. , 2008. Genetic analysis reveals different roles of Schizosaccharomyces pombe sfr1/dds20 in meiotic and mitotic DNA recombination and repair. Curr. Genet. 54: 197–211. [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Narita T., Pham V. T., Iijima J., Hirota K., et al. , 2013. Structure-specific endonucleases xpf and mus81 play overlapping but essential roles in DNA repair by homologous recombination. Cancer Res. 73: 4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick D. T., Ferguson J. R., Petes T. D., Symington L. S., 2000. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics 156: 1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli J., Hottinger H., Munz P., Strauss A., Thuriaux P., 1977. Genetic mapping in Schizosaccharomyces pombe by mitotic and meiotic analysis and induced haploidization. Genetics 87: 471–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S., Mizuno K., Blaisonneau J., Martineau S., Chanet R., et al. , 2010. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol. Cell 39: 346–359. [DOI] [PubMed] [Google Scholar]
- Lehmann A. R., Fuchs R. P., 2006. Gaps and forks in DNA replication: rediscovering old models. DNA Repair (Amst.) 5: 1495–1498. [DOI] [PubMed] [Google Scholar]
- Li F., Dong J., Pan X., Oum J. H., Boeke J. D., et al. , 2008. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol. Cell 30: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Dong J., Eichmiller R., Holland C., Minca E., et al. , 2013. Role of Saw1 in Rad1/Rad10 complex assembly at recombination intermediates in budding yeast. EMBO J. 32: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay H. D., Griffiths D. J., Edwards R. J., Christensen P. U., Murray J. M., et al. , 1998. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12: 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Barlow J. H., Burgess R. C., Rothstein R., 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713. [DOI] [PubMed] [Google Scholar]
- Lorenz A., Osman F., Sun W., Nandi S., Steinacher R., et al. , 2012. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science 336: 1585–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyndaker A. M., Alani E., 2009. A tale of tails: insights into the coordination of 3′ end processing during homologous recombination. BioEssays 31: 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. L., Kim E. M., Haber J. E., Lee S. E., 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23: 8820–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J., Blanco M. G., Maslen S., Skehel J. M., West S. C., 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazon G., Lam A. F., Ho C. K., Kupiec M., Symington L. S., 2012. The Rad1-Rad10 nuclease promotes chromosome translocations between dispersed repeats. Nat. Struct. Mol. Biol. 19: 964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCready S., Carr A. M., Lehmann A. R., 1993. Repair of cyclobutane pyrimidine dimers and 6–4 photoproducts in the fission yeast Schizosaccharomyces pombe. Mol. Microbiol. 10: 885–890. [DOI] [PubMed] [Google Scholar]
- Meister P., Taddei A., Ponti A., Baldacci G., Gasser S. M., 2007. Replication foci dynamics: replication patterns are modulated by S-phase checkpoint kinases in fission yeast. EMBO J. 26: 1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi A., Kuntz K., Majeed A., Smith S., Myung K., 2006. Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz P., Blanco R., Flores J. M., Blasco M. A., 2005. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 37: 1063–1071. [DOI] [PubMed] [Google Scholar]
- Muñoz-Galván S., Tous C., Blanco M. G., Schwartz E. K., Ehmsen K. T., et al. , 2012. Distinct roles of Mus81, Yen1, Slx1-Slx4, and Rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol. Cell. Biol. 32: 1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M. J., Ramachandran M., Trelles-Sticken E., Scherthan H., Goldman A. S., 2002. Wild-type levels of Spo11-induced DSBs are required for normal single-strand resection during meiosis. Mol. Cell 9: 835–846. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A., Singer J. D., Hoekstra M. F., Heffron F., 1989. Double-strand breaks stimulate alternative mechanisms of recombination repair. J. Mol. Biol. 207: 527–541. [DOI] [PubMed] [Google Scholar]
- O’Neil N. J., Martin J. S., Youds J. L., Ward J. D., Petalcorin M. I., et al. , 2013. Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during Caenorhabditis elegans meiosis. PLoS Genet. 9: e1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Octobre G., Lorenz A., Loidl J., Kohli J., 2008. The Rad52 homologs Rad22 and Rti1 of Schizosaccharomyces pombe are not essential for meiotic interhomolog recombination, but are required for meiotic intrachromosomal recombination and mating-type-related DNA repair. Genetics 178: 2399–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F., Adriance M., McCready S., 2000. The genetic control of spontaneous and UV-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Curr. Genet. 38: 113–125. [DOI] [PubMed] [Google Scholar]
- Osman F., Bjoras M., Alseth I., Morland I., McCready S., et al. , 2003a A new Schizosaccharomyces pombe base excision repair mutant, nth1, reveals overlapping pathways for repair of DNA base damage. Mol. Microbiol. 48: 465–480. [DOI] [PubMed] [Google Scholar]
- Osman F., Dixon J., Doe C. L., Whitby M. C., 2003b Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12: 761–774. [DOI] [PubMed] [Google Scholar]
- Pankratz D. G., Forsburg S. L., 2005. Meiotic S-phase damage activates recombination without checkpoint arrest. Mol. Biol. Cell 16: 1651–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Haber J. E., 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Haber J. E., 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B., Aguilera A., 2012. Complex chromosomal rearrangements mediated by break-induced replication involve structure-selective endonucleases. PLoS Genet. 8: e1002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli A. S., Sena E. P., Smith G. R., 1988. Genetic and physical analysis of the M26 recombination hotspot of Schizosaccharomyces pombe. Genetics 119: 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J., Evans J. S., Hussey S. P., Deans B., O’Neill P., et al. , 2003. Pathway utilization in response to a site-specific DNA double-strand break in fission yeast. EMBO J. 22: 1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Hayes T. K., Kolodner R. D., 2010. Post-replication repair suppresses duplication-mediated genome instability. PLoS Genet. 6: e1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Siddiqi I., Kolodkin A. L., Stahl F. W., 1988. Intra-chromosomal gene conversion induced by a DNA double-strand break in Saccharomyces cerevisiae. J. Mol. Biol. 201: 247–260. [DOI] [PubMed] [Google Scholar]
- Rhind N., Russell P., 2000. Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways. J. Cell Sci. 113(Pt 22): 3889–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseaulin L., Yamada Y., Tsutsui Y., Russell P., Iwasaki H., et al. , 2008. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 27: 1378–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenzhak S., Mejia-Ramirez E., Williams J. S., Schaffer L., Hammond J. A., et al. , 2010. Rad3 decorates critical chromosomal domains with gammaH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet. 6: e1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph C., Kunz C., Parisi S., Lehmann E., Hartsuiker E., et al. , 1999. The msh2 gene of Schizosaccharomyces pombe is involved in mismatch repair, mating-type switching, and meiotic chromosome organization. Mol. Cell. Biol. 19: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatinos S. A., Forsburg S. L., 2009. Measuring DNA content by flow cytometry in fission yeast. Methods Mol. Biol. 521: 449–461. [DOI] [PubMed] [Google Scholar]
- Sabatinos S. A., Forsburg S. L., 2010. Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol. 470: 759–795. [DOI] [PubMed] [Google Scholar]
- Sabatinos S. A., Green M. D., Forsburg S. L., 2012. Continued DNA synthesis in replication checkpoint mutants leads to fork collapse. Mol. Cell. Biol. 32: 4986–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. T., Lui D. Y., Kim H. M., Meyer K., Colaiacovo M. P., 2013. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet. 9: e1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Kapitza-Fecke P., Stephen E. R., Gutz H., 1989. Some of the swi genes of Schizosaccharomyces pombe also have a function in the repair of radiation damage. Curr. Genet. 16: 89–94. [DOI] [PubMed] [Google Scholar]
- Schuchert P., Kohli J., 1988. The Ade6–M26 mutation of Schizosaccharomyces pombe increases the frequency of crossing over. Genetics 119: 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E. K., Heyer W. D., 2011. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120: 109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif W. D., Glick G. G., Davidson M. K., Wahls W. P., 2002. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome 1: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R., 2009. Genetic analysis of meiotic recombination in Schizosaccharomyces pombe. Methods Mol. Biol. 557: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R., Boddy M. N., Shanahan P., Russell P., 2003. Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics 165: 2289–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Haber J. E., 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12: 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Kang L. E., Moreau S., 2000. Alteration of gene conversion tract length and associated crossing over during plasmid gap repair in nuclease-deficient strains of Saccharomyces cerevisiae. Nucleic Acids Res. 28: 4649–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi P., Heyer W. D., Schuchert P., Kohli J., 1988. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination host spot allele ade6–M26. J. Mol. Biol. 204: 917–925. [DOI] [PubMed] [Google Scholar]
- Szankasi P., Smith G. R., 1995. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science 267: 1166–1169. [DOI] [PubMed] [Google Scholar]
- Tian M., Shinkura R., Shinkura N., Alt F. W., 2004. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol. Cell. Biol. 24: 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K., Cooper J. P., 2007. The telomere bouquet controls the meiotic spindle. Cell 130: 113–126. [DOI] [PubMed] [Google Scholar]
- Vance J. R., Wilson T. E., 2002. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. USA 99: 13669–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth N. C., Bernards R., 1996. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science 271: 353–356. [DOI] [PubMed] [Google Scholar]
- Wan B., Yin J., Horvath K., Sarkar J., Chen Y., et al. , 2013. SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell Reports 4: 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis N., Rhind N., 2009. Mus81, Rhp51(Rad51), and Rqh1 form an epistatic pathway required for the S-phase DNA damage checkpoint. Mol. Biol. Cell 20: 819–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt H. D., Sarbajna S., Matos J., West S. C., 2013. Coordinated actions of SLX1–SLX4 and MUS81–EME1 for Holliday junction resolution in human cells. Mol. Cell 52: 234–247. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Hiraoka Y., 2003. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 22: 2284–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz O., Majumder S., Kramer B., Sekelsky J. J., 2002. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol. Cell 10: 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemasu R., McCready S. J., Murray J. M., Osman F., Takao M., et al. , 1997. Characterization of the alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe. Nucleic Acids Res. 25: 1553–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. A., Schreckhise R. W., Steiner W. W., Smith G. R., 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9: 253–263. [DOI] [PubMed] [Google Scholar]
- Young J. A., Hyppa R. W., Smith G. R., 2004. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. D., Niedernhofer L., Kuster B., Mann M., Hoeijmakers J. H., et al. , 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12: 1489–1498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.