Abstract
Some species exhibit very high levels of DNA sequence variability; there is also evidence for the existence of heritable epigenetic variants that experience state changes at a much higher rate than sequence variants. In both cases, the resulting high diversity levels within a population (hyperdiversity) mean that standard population genetics methods are not trustworthy. We analyze a population genetics model that incorporates purifying selection, reversible mutations, and genetic drift, assuming a stationary population size. We derive analytical results for both population parameters and sample statistics and discuss their implications for studies of natural genetic and epigenetic variation. In particular, we find that (1) many more intermediate-frequency variants are expected than under standard models, even with moderately strong purifying selection, and (2) rates of evolution under purifying selection may be close to, or even exceed, neutral rates. These findings are related to empirical studies of sequence and epigenetic variation.
Keywords: hyperdiversity, epigenetic variation, site frequency spectrum, purifying selection, rate of substitution
THE infinite sites model, originally proposed by Fisher (1922, 1930) and developed in detail by Kimura (1971), has been the workhorse of molecular population genetics for four decades. Its core assumption is that any nucleotide site segregates for at most two variants and that the mutation rate scaled by effective population size (Ne) is so low that new mutations arise only at sites that are fixed within the population (see also Charlesworth and Charlesworth 2010, p. 207). This assumption facilitates calculations of the theoretical values of some key observable quantities, such as the expected level of pairwise nucleotide site diversity or the expected number of segregating sites in a sample (Kimura 1971; Watterson 1975; Ewens 2004). In the framework of coalescent theory, this implies a linear relation between the genealogical distance between two sequences and the neutral sequence divergence between them, greatly simplifying methods of inference and statistical testing (Hudson 1990; Wakeley 2008).
There has recently been some discussion of how to go beyond the infinite sites assumption of a low scaled mutation rate, which breaks down for species with very large effective population sizes, including some species of virus and bacteria, and even eukaryotes such as the sea squirt and outbreeding nematode worms, resulting in “hyperdiversity” of DNA sequence variability within a population (Cutter et al. 2013). It is important to note, however, that this problem can arise even when the scaled mutation rate is relatively low, since then the proportion of neutral nucleotide sites that are currently segregating in a population (which depends on the scaled mutation rate) can be substantial when the population size is sufficiently large. For example, with a neutral mutation rate of u per site in a population of N breeding adults, the expected fraction of sites that are segregating in a randomly mating population is fs = θ [ln(2N) + 0.6775], where θ = 4Neu (Ewens 2004, p. 298). Thus, with θ = 0.01, a reasonable value for many species (Leffler et al. 2012), we have fs = 0.15 even when N has the implausibly low value of 1 million. This implies that ∼15% of new mutations are expected to arise at sites that are already segregating, suggesting a significant departure from the assumptions of the infinite sites model. (An alternative way of looking at this is to determine the expected number of new mutations that occur at a site while a preexisting mutation is segregating, which is of a similar magnitude to fs—see Appendix, Equation A1.)
In addition, it has been known for nearly 20 years that sufficiently high scaled mutation rates at some or all sites in a sequence can lead to substantial departures from the infinite sites expectations for statistics such as Tajima’s D, which are commonly used to detect deviations from neutral equilibrium caused by population size changes or selection (Bertorelle and Slatkin 1995; Aris-Brosou and Excoffier 1996; Tajima 1996; Yang 1996; Mizawa and Tajima 1997). This is because the occurrence of mutations at sites that are already segregating increases the pairwise diversity among sequences, but does not increase the number of segregating sites (Bertorelle and Slatkin 1995). The analysis of data on DNA sequence variation in hyperdiverse species thus requires methods that deal with this problem, and a number of population genetics models that contribute to this have already been developed (Desai and Plotkin 2008; Jenkins and Song 2011; Cutter et al. 2012; Jenkins et al. 2014; Sargsyan 2014).
Finally, analyses of the inheritance of epigenetic markers, such as methylated cytosines, have suggested that these can sometimes be transmitted across several sexual generations, but with rates of origination or reversion that are several orders of magnitude higher than the mutation rates of DNA sequences (Johannes et al. 2009; Becker et al. 2011; Schmitz et al. 2011; Lauria et al. 2014). In view of the current interest in the possible functional and evolutionary significance of epigenetic variation (Richards 2006; Schmitz and Ecker 2012; Grossniklaus et al. 2013; Klironomos et al. 2013), it seems important to develop models that can shed light on their population genetics, to understand the evolutionary forces acting on them.
The purpose of this article is to develop a relatively simple analytical framework for examining the consequences of high scaled mutation rates, in the framework of the classical random mating, finite population size model with forward and backward mutations in the presence of selection and genetic drift (Wright 1931, 1937). The approach is similar in spirit to the biallelic model used by Desai and Plotkin (2008), but with a focus on sample statistics that summarize properties of the site frequency spectrum, as well as on the expected rate of substitutions along a lineage. As has been found in previous coalescent-based treatments with neutrality (Bertorelle and Slatkin 1995; Aris-Brosou and Excoffier 1996; Cutter et al. 2012), the results derived below show that very large departures from the infinite sites model occur when the scaled mutation rate is sufficiently high, even when fairly strong purifying selection is acting, resulting in features of the data such as a large excess of intermediate-frequency variants. In addition, the signal of purifying selection on substitutions along a lineage can be obscured or even converted into a signal of positive selection. The findings have significant implications for the interpretation of the results of studies of both epigenetic variability and DNA sequence variability in species with large effective population sizes. Readers who are interested primarily in the main biological conclusions may wish to skip over the details of the derivations.
Analysis of the Model of Purifying Selection, Drift, and Mutation
Basic assumptions
We assume a randomly mating, diploid, discrete generation population with N breeding adults, and effective population size Ne. Over a long sequence of m nucleotide sites, each site has two alternative types, A1 and A2, with mutation rates u and v from A2 to A1 and vice versa. (With diploidy, this means that only three genotypes are present at a site: A1A1, A1A2 and A2A2.) A1 and A2 might correspond to AT versus GC base pairs, unpreferred versus preferred synonymous codons, or selectively favored versus disfavored nonsynonymous variants. If epigenetic variation is being considered, then A1 and A2 could be regarded as the methylated or unmethylated states of a nucleotide site or a differentially methylated region (or vice-versa). This approach, while undoubtedly oversimplified, avoids the problem of modeling mutation among all four basepairs, which is difficult to deal with except by making the unrealistic assumption of equal mutation rates in all directions (Ewens 2004).
If selection is acting, we assume semidominance, with A2 having a selective advantage s over A1 when homozygous and with the fitness of A1A2 being exactly intermediate, although our general conclusions are probably not strongly dependent on this assumption. There is complete independence among sites (i.e., recombination is sufficiently frequent that linkage disequilibrium is negligible), and all evolutionary forces are weak, so that the standard results of diffusion approximations can be employed.
When the population is at statistical equilibrium, the probability distribution of variant frequencies over sites remains stationary and the mean numbers of sites in each possible state are constant over time, despite continual changes at individual sites (Charlesworth and Charlesworth 2010, pp. 270–272). At any given time, some sites are fixed for the A1 type, some are fixed for A2, and others segregate for both. Let the equilibrium proportion of sites that are fixed for A1 and A2 be f1f and f2f, respectively. The proportion of sites that are segregating is fs = 1 – f1f – f2f.
Results for some important population parameters
These assumptions allow the use of Wright’s stationary distribution formula (Wright 1931, 1937) to describe the probability density of the frequency q of A2 at a site,
| (1) |
where p = 1 – q, α = 4Neu, β = 4Nev, γ = 2Nes, and the constant C is such that the integral of φ(q) between q = 0 and q = 1 is equal to 1. It is convenient to write u in terms of the mutational bias parameter, κ; i.e., u = κv, so that α = κβ.
Properties of the distribution
An explicit expression for C can be obtained by noting that the integral of the other terms on the right-hand side with respect to q is equal to the product of Γ(α) Γ(β)/ Γ(α + β) and the confluent hypergeometric function 1F1(a, b, z) (Abramowitz and Stegun 1965, p. 503), with parameters a = β, b = α + β, z = γ, where
| (2) |
and (Pochhammer’s symbol).
This can be seen by expanding the exponential term in Equation 1 in powers of γq and integrating over the range 0 to 1 (Kimura et al. 1963).
Integrating Equation 1, we have
| (3) |
Furthermore, the jth moment of q around zero, obtained from the integral of qjφ(q) between 0 and 1, is given by
| (4) |
In particular, the mean frequency of A2 is
| (5a) |
and the mean frequency of A1 is
| (5b) |
Approximations for small β:
Approximations to these expressions for the case when β << 1 and κ is of order 1 are derived in the Appendix. Equations A3a and A3b imply that
| (6) |
The left-hand side of Equation 6 is equivalent to the fraction of sites that carry A2 in a random sequence sampled from the population; if A1 and A2 correspond to unpreferred and preferred codons, respectively, this measures the frequency of preferred codons, Fop (McVean and Charlesworth 1999). With epigenetic variation, if A1 and A2 correspond to methylated and unmethylated states, respectively, measures the fraction of unmethylated sites or regions in a random genome.
The leading term on the right-hand side of Equation 6 is identical to the Li–Bulmer equation commonly used in analyses of selection on codon usage (Li 1987; Bulmer 1991). This result is, however, often derived by assuming that nearly all sites are fixed and calculating the rate of flux between sites fixed for A1 and A2; is then taken to be the frequency of sites that are fixed for A2, with 1 – representing the frequency of sites fixed for A2 (Bulmer 1991). This raises the question of how good an approximation we obtain by neglecting the term of order β, when the infinite sites assumption is violated, so that a significant fraction of sites are in fact segregating for A1 and A2.
First, we note that it is immediately obvious from Equation 1 and Equations 5a and 5b that with γ = 0 is equal to 1/(1 + κ), so that Equation 6 for this case is exact, as has long been known (Wright 1931). We can also obtain a first-order approximation to Equations A3a and A3b when γ ≠ 0 by expanding in powers of β, which will be accurate when β is sufficiently small. Neglecting second-order and higher terms in β, as is also done in Equations 8a–8c, we obtain
| (7a) |
where
| (7b) |
and ai is the harmonic series 1 + 1/2 + 1/3 + … + 1/(i –1), with i ≥ 2.
As shown after Equation A3b of the Appendix, the leading term in Equation 6 should provide a good approximation when βκ ≤0.1.
Approximations for large γ:
For examining what happens when γ becomes very large, it is useful to note that the Taylor’s series expansion of Equation 5b for small β yields the expression
| (8a) |
For large γ, this gives
| (8b) |
i.e.,
| (8c) |
The first term on the right-hand site of Equation 8c is equivalent to the asymptotic expression for with large γ given by Kimura et al. (1963). This implies that, for sufficiently large γ compared with β, the mean frequency of the disfavored variant is equal to its equilibrium frequency under mutation–selection balance with s >> u in an infinite population, where p = 2u/s = 2vκ/s (Haldane 1927), as expected intuitively. Numerical studies show that Equation 8b performs well for γ > 1 if β << 1, when it can give a good approximation when neither the leading term in Equation 6 nor the Kimura et al. (1963) large γ approximation is accurate (results not shown). Equation 8b implies that the leading term in Equation 6 is accurate when γ << –ln(β).
Frequencies of fixed sites:
Second, the approximate frequencies of sites that are fixed for A1 and A2, f1f and f2f, can be found from the integrals of φ(q) between 0 and 1/(2N) and 1– 1/(2N) and 1, respectively (Ewens 2004, p. 178). For large N, such that γN–1 << 1, when q is close to zero we have φ(q) = C[qβ–1 + O(γN–1) + O(βN–1)], and so
| (9a) |
| (9b) |
where the terms in O(γN–1) and O(βN–1) can be neglected when N is sufficiently large (cf. Kimura 1981). Approximations for these expressions for small β can readily be obtained (see Appendix).
Nucleotide site diversity:
Third, the expected pairwise nucleotide site diversity, π, can be obtained from the expectation of 2pq, given by 2E{q – q2}. From Equation 4, we have
| (10a) |
The expectation of E{q – q2} is given by subtracting Equation 10a from Equation 5b. Using Equation 2 and simplifying, we obtain
| (10b) |
This is equal to one-half of the expected pairwise diversity per site, π. Using the same approach as for Equations 7 and 8, keeping only terms of order β we obtain
| (11) |
As expected, this is identical to equation 15 of McVean and Charlesworth (1999) for the infinite sites model at statistical equilibrium, where new mutations arise only at sites that are fixed either for A1 or for A2. When γ >> β, this term converges on the deterministic value under mutation–selection balance, 2βκ/γ, which corresponds to the diversity expected at deterministic mutation–selection balance with p = 2u/s (see above).
In the case of neutrality, Equation 10b reduces to the following expression (Charlesworth and Charlesworth 2010, p. 237):
| (12) |
As expected intuitively, the neutral diversity is always less than for the infinite sites model with a given value of β and κ, where Equation 11 with γ = 0 gives π = 2βκ/(1 + κ), because some new mutations arise at sites that are already polymorphic; π approaches 2κ/(1 + κ)2 for large β, which is the value for an infinite population at equilibrium under reversible mutation between A1 and A2.
Rate of substitution along a lineage
Analytic results:
The rate of substitution of new mutations along a lineage can be modeled as follows. Conditioning on a frequency q of the A2 variant at a site in a given generation, there is an expected number of 2Nvκq mutations per site from A2 to A1 and 2Nvp from A1 to A2. The corresponding probability that A1 becomes fixed, conditional on p, is Q1(p) = [exp(γp) – 1]/[exp(γ) – 1] (Kimura 1962). Conditioning on this fixation event, the probability that it is a new A1 mutation that has been fixed is 1/(2Np). The expected number of new A1 mutations that become fixed is thus equal to vκp–1qQ1(p). Similarly, the conditional probability that A2 eventually becomes fixed is Q2(q) = [1 – exp(–γq)]/[1 – exp(–γ)]; the net expected number of new A2 mutations that become fixed is vpq–1Q2(q). (At first sight, it would seem that this procedure cannot be applied to mutations arising in the fixed classes and that these should be treated separately, but the argument given in the Appendix shows that it provides an accurate approximation for this situation as well.)
Integrating over all values of q, the net rate at which new mutations enter the population and become fixed is thus
| (13) |
The terms involving functions of p and q in the integrand are
| (14a) |
| (14b) |
The corresponding integrals are
| (14c) |
and
| (14d) |
Note that 1F1(β – 1, α + β, γ) – 1 has a factor of β – 1, so that the term in β – 1 in the denominator of Equation 14d cancels. At first sight, Equation 14c appears to have a singularity at α = 1. However, by using the relation 1F1(a, b, z) = 1F1(b – a, b, z)exp(z), we find that 1F1(β + 1, α + β, γ) = 1F1(α – 1, α + β, γ)exp(γ), so that the numerator of Equation 14c contains a factor of α – 1, which cancels the term in the denominator.
The net rate of substitution is given by
| (15) |
As γ approaches zero, Equations 14 and 15 imply that λ tends to 2vκ/(1 + κ); this is independent of the population size and is identical to the infinite sites expression with neutrality at statistical equilibrium under reverse mutation (Charlesworth and Charlesworth 2010, p. 274), as expected from the fact that the equilibrium neutral substitution rate is equal to the net mutation rate for any class of mutational model (Kimura 1968).
When α and β are sufficiently small, the main contributions to λ come from the two fixed classes, so that the initial frequencies of the new mutations can be equated to 1/(2N), when O(β2) terms in I1 and I2 are neglected. Using the above result that the frequencies of the fixed classes are equal to the infinite sites values multiplied by a factor 1 – O(β), the infinite sites expression for the case of selection is recovered, neglecting higher-order terms in β (equation 6.11 of Charlesworth and Charlesworth 2010, p. 275). Again, this implies that, as expected, the infinite sites model provides a good approximation for the rate of substitution with sufficiently small β.
Scaling relative to the neutral rate:
There are two different ways in which we can determine the ratio of the value of λ with γ > 0 to that for a neutral standard, thereby removing the dependence on the mutation rate term in Equation 15. First, λ with selection can be compared to its value at statistical equilibrium with the same value of α and β. This would be appropriate for comparing rates of evolution at putatively neutral sites in a given genomic region with those at sites that are potentially under purifying selection, without making any corrections for differences in base composition; this is often done when comparing nonsynonymous and synonymous rates of substitution across different genes by statistics such as KA/KS. Second, λ with selection can be compared with the neutral rate conditioned on the same mean frequencies of A1 and A2 along the sequence as for the selected sites; this corresponds to methods that compare probabilities of substitution between the same pairs of nucleotides in contexts when these are putatively selected vs. putatively neutral (Halligan et al. 2004; Eory et al. 2010).
Numerical results for the population parameters
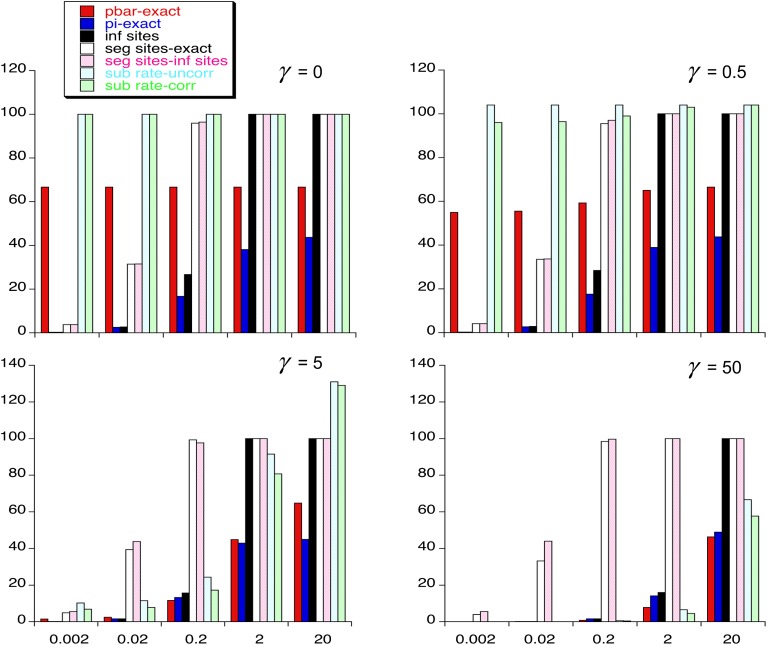
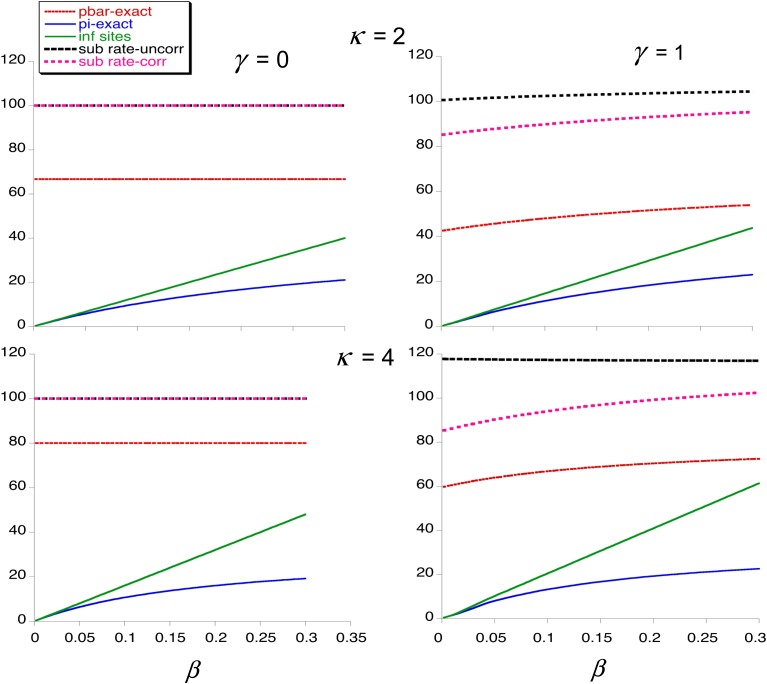
Numerical results generated from the above formulas are presented in Figure 1 and Figure 2. Figure 1 illustrates the dependence of the following variables on the scaled mutation rate (β) and the scaled intensity of selection (γ), assuming a mutational bias (κ) of 2 toward the deleterious variant at a site: the mean frequency per site of the deleterious variant A1 (), the expected diversity (π), the expected proportion of sites that segregate for variants (fs), and the above two measures of the rate of substitution relative to neutral expectation. Figure 2 illustrates the dependence of and π on β at a finer scale, for different values of κ and γ. For clarity, the infinite sites values for and the relative rates of substitution are not shown; with selection, the infinite sites values for these parameters are close to their values when β = 0.002.
Figure 1.
The vertical bars are the values (in percentages) of the mean frequency of A1, (red), π from Equation 10b (blue), π as given by the infinite sites model (black), the proportion of segregating sites from Equation 9 (white), the proportion of segregating sites under the infinite sites model (pink), the uncorrected rate of substitution relative to neutrality (light blue), and the corrected rate of substitution relative to neutrality (green).
Figure 2.
The curves are the values (in percentages) as functions of β for the mean frequency of A1, (red, dashed curve), π from Equation 10b (blue, solid curve), π as given by the infinite sites model (green, solid curve), the uncorrected rate of substitution relative to neutrality (black, dashed curve), and the corrected rate of substitution relative to neutrality (pink, dashed curve).
With neutrality, the exact value of is always equal to the infinite sites value and is independent of β for a given value of κ. With selection and low β (0.002 or 0.02), it can be seen that agreement with the infinite sites predictions is fairly good for both these values despite the fact that the proportion of sites that are segregating can be quite substantial with β = 0.02; the second-order approximation of Equations 7a and 7b gives very close agreement even for β = 0.2 with weak selection, but diverges for β > 0.2 when γ > 0.5 (results not shown), whereas the value of departs quite seriously from the infinite sites values at β = 0.2 when γ > 5. A similar pattern of departure from the infinite sites value holds for π, except when selection is strong (γ = 5 or 50), when agreement is still good at β = 2; this is because the exact diversity and the infinite sites value both approach the deterministic value under mutation–selection balance when 1 << γ and β << γ (see Equation 11). Somewhat surprisingly, the infinite sites and exact values of the proportion of sites that are segregating always agree well.
Perhaps the most interesting result to emerge is that, with κ > 1, the rate of substitution relative to neutral expectation can exceed one when there is moderate selection and mutational bias toward the deleterious variants. This has long been known to apply to the infinite sites model when the “uncorrected” relative rate is used and when there is mutational bias (Eyre-Walker 1992; McVean and Charlesworth 1999), which can cause serious problems for phylogenetic inferences concerning selective constraints (Lawrie et al. 2011). As shown in the Appendix, the “corrected” relative rate is always expected to be less than one under the infinite sites assumption (see Equation A8). But with sufficiently high β, the corrected relative rate can exceed one, even for γ = 5, and can be only just below one for lesser values of β. The reason for this seemingly paradoxical result is presumably the fact that nearly all sites are segregating if β is high; when is sufficiently high because mutation and drift are overcoming selection, there is a substantial chance that a new mutation to the favorable variant A2 can arise at a segregating site, which has a higher chance of fixation than a neutral variant and hence contributes to an elevated substitution rate. With sufficiently strong mutational bias, can be >>1/2, so that the contribution from the enhanced fixation probability of favorable mutations outweighs the lower contribution from the fixation of deleterious mutations.
As was previously shown by McVean and Charlesworth (1999) for the infinite sites model, the equilibrium diversity with selection can also considerably exceed the neutral equilibrium value with the same mutational parameters, when there is a mutational bias toward deleterious alleles (see also Kondrashov et al. 2006). For example, in Figure 1, with γ = 5 and β = 2, π = 0.43 but is 0.38 for the neutral case; with β = 20 and γ = 50 the values are 0.49 and 0.44, respectively. In this case, there is no meaningful way of correcting for differences in base composition between the neutral and selected sites when there are substantial departures from the infinite sites assumption, since the diversity in the neutral case is not related to the mean allele frequency in a simple way.
Properties of a Sample from a Population
Analytic results
This raises the question of the extent to which the properties of a sample of alleles from a population are affected by deviations from the infinite sites assumption. With the above model, the probability that a sample of n alleles segregates for k A2 variants at a site and n – k copies of A1 can be obtained from the corresponding binomial distribution with parameter q, integrated over φ(q), takes the form
| (16a) |
where C is given by Equation 3 (McVean and Charlesworth 1999; Desai and Plotkin 2008).
Using the properties of the confluent hypergeometric function, this yields
| (16b) |
| (16c) |
| (16d) |
The proportion of sites that are observed to be segregating is
| (16e) |
The conditional site frequency spectrum (SFS) for segregating sites can be obtained by dividing Equation 16b by Equation 16e. The folded SFS for segregating sites [which describes the numbers of variants of either type at frequencies 1–0.5n + 1 (n odd) or 0.5n (n even)] can also easily be obtained.
Equations 16a–16e can readily be used to obtain the theoretical values of standard sample statistics, such as the diversity per site (π) (Tajima 1983), Watterson’s θw = pseg/an (Watterson 1975), and Tajima’s D (Tajima 1989b), using the standard formulas for these quantities. A well-known problem with Tajima’s D is the fact that its magnitude is strongly dependent on both the level of variability in the population and the length of sequence used to estimate it (Tajima 1989b). Langley et al. (2014) proposed the use of the summary statistic Δπ = (π – θw)/θw for measuring the extent of departure of the SFS from the infinite sites neutral equilibrium expectation, which should not suffer from these problems. Another summary statistic for this purpose is provided by the proportion of singleton variants among segregating sites, given by
| (16f) |
(This is closely related to the widely used D statistic of Fu and Li 1993.)
Numerical results
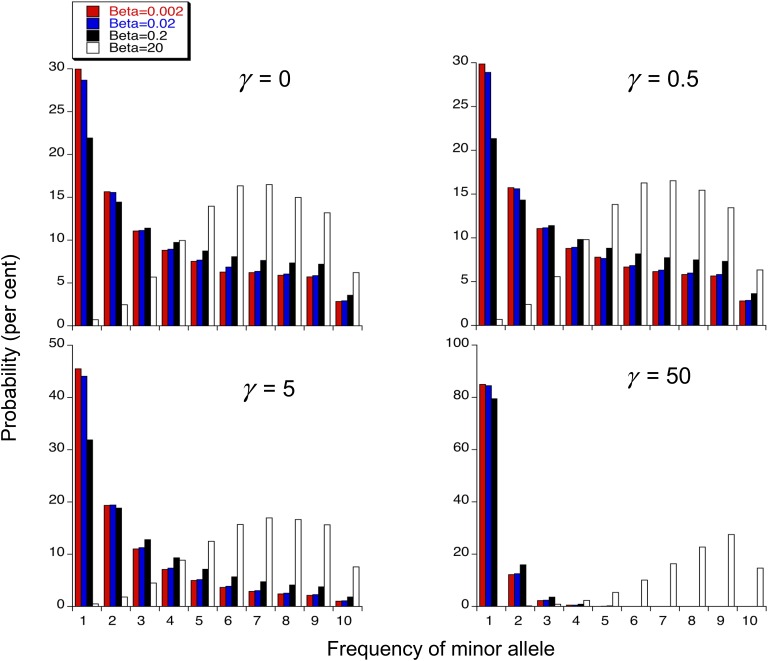
Use of the series expression for the confluent hypergeometric function allows rapid computation of all relevant statistics; to avoid overflow when γ is large, however, it is necessary to use logarithms of the individual terms and partial sums of the series (which can be done, since the selection model is defined such that γ > 0). A FORTRAN program is available on request to B. Charlesworth.
Table 1 displays some examples of such computations, for the case of a mutational bias of 2 toward deleterious mutations, for a subset of the parameter values used in Figure 1. The expected π values are not shown, since these are the same as the population diversities given in Figure 1. Figure 3 show the folded SFSs for some chosen examples, using a sample size of 20 alleles. It can be seen that a high β value (20) means that the proportion of sites that are found to be segregating (pseg) is effectively 100%, even for γ as high as 50 and a sample size (n) of 20. A moderate β value (0.2) behaves similarly in the neutral case with a sample size of 200, but otherwise is associated with a pseg of <80% (pseg is as low as 13% for n = 20 and γ = 50). With neutrality or weak selection (γ ≤ 5), moderate or high values of β cause a distortion of the SFS toward a much lower proportion of singletons (psn) and higher Tajima’s D and Δπ than is expected with the infinite sites model. Even for γ = 50, a very low psn and a positive D are found when β = 20. This reflects the tendency of high β values to push the distribution of q toward intermediate frequencies, which has long been known (Wright 1931). Some analytical approximations for pseg are derived in supporting information, File S1.
Table 1. Sample statistics for the reversible mutation model (κ = 2).
|
n = 20 |
n = 200 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| β | pseg | psn | DT | Δπ | pseg | psn | DT | Δπ | |
| γ = 0 | 0.02 | 0.088 | 0.287 | 0.038 | 0.016 | 0.142 | 0.159 | 0.089 | 0.040 |
| 0.2 | 0.533 | 0.219 | 0.322 | 0.109 | 0.728 | 0.086 | 0.745 | 0.326 | |
| 2.0 | 0.966 | 0.062 | 1.181 | 0.399 | 1.000 | 0.001 | 1.252 | 1.226 | |
| 20 | 0.999 | 0.007 | 1.637 | 0.553 | 1.000 | 0.000 | 3.570 | 1.567 | |
| γ = 0.5 | 0.02 | 0.094 | 0.289 | 0.029 | 0.009 | 0.152 | 0.159 | 0.076 | 0.034 |
| 0.2 | 0.559 | 0.213 | 0.348 | 0.117 | 0.753 | 0.080 | 0.848 | 0.373 | |
| 2.0 | 0.970 | 0.055 | 1.248 | 0.421 | 1.000 | 0.001 | 2.924 | 1.284 | |
| 20 | 0.999 | 0.007 | 1.649 | 0.556 | 1.000 | 0.000 | 3.586 | 1.574 | |
| γ = 5.0 | 0.02 | 0.071 | 0.441 | −0.654 | −0.226 | 0.146 | 0.228 | −0.843 | −0.380 |
| 0.2 | 0.511 | 0.319 | −0.241 | −0.081 | 0.787 | 0.104 | −0.032 | −0.014 | |
| 2.0 | 0.990 | 0.025 | 0.950 | 0.532 | 1.000 | 0.000 | 3.443 | 1.511 | |
| 20 | 0.999 | 0.004 | 1.755 | 0.592 | 1.000 | 0.000 | 3.722 | 1.634 | |
| γ = 50 | 0.02 | 0.014 | 0.845 | −1.539 | −0.586 | 0.063 | 0.478 | −1.820 | −0.851 |
| 0.2 | 0.129 | 0.795 | −1.650 | −0.564 | 0.478 | 0.351 | −1.826 | −0.806 | |
| 2.0 | 0.744 | 0.408 | −0.967 | −0.327 | 0.998 | 0.005 | −0.171 | −0.169 | |
| 20 | 1.000 | 0.000 | 1.329 | 0.736 | 1.000 | 0.000 | 4.270 | 1.875 | |
pseg is the proportion of sites that are segregating, psn is the proportion of singletons among segregating sites in a sample of size n, DT is the mean of Tajima’s D for a sequence of 450 bp, and Δπ = (π – θw)/θw, where θw = pseg/an and an = 1 + 1/2 + … + 1/(n – 1). All statistics were calculated from Equations 16a–16f.
Figure 3.
The vertical bars are the values (in percentages) of the probabilities of finding the minor allele in a sample of 20 at the frequencies indicated on the x-axis, for different values of β and γ.
Discussion
The results described above have some important implications for the interpretation of data on DNA sequence variation and evolution when there is hyperdiversity; i.e., the scaled mutational parameter (β in the notation used here) is sufficiently large that the infinite sites model does not accurately describe patterns of variation within populations. Recent surveys of DNA sequence polymorphisms show that that such hyperdiversity is more common than previously thought, even in multicellular organisms (Cutter et al. 2013). In addition, given the evidence from studies of organisms like Arabidopsis thaliana and maize that epigenetic variants such as methylated cytosines can be transmitted fairly stably through meiosis, but have origination and disappearance rates that are several orders of magnitude higher than those of nucleotide variants (Johannes et al. 2009; Becker et al. 2011; Schmitz et al. 2011; Lauria et al. 2014), the patterns described above are relevant to population-level studies of some classes of epigenetic variants.
Distortion of the SFS with hyperdiversity
As was pointed out ∼20 years ago in the context of human mitochondrial DNA sequence variability (Bertorelle and Slatkin 1995; Aris-Brosou and Excoffier 1996; Tajima 1996; Yang 1996), a major effect of a high scaled mutation rate (β in the notation used here) is that more intermediate-frequency variants will be present at polymorphic sites in a sample from a population than under the equilibrium infinite sites model. In particular, for a stationary population at equilibrium between drift and the input of neutral or nearly neutral mutations, the expected values of Tajima’s D statistic (DT) and the Δπ statistic proposed by Langley et al. (2014) are positive rather than slightly negative or zero, respectively, as expected under the infinite sites model (Tajima 1989b)—see Figure 1 and Table 1. This reflects the fact that the expected value of the pairwise diversity per site (π) is greater than the expected value of the measure of diversity based on the number of segregating sites at a locus (θw). As can be seen from Table 1, this effect is quite noticeable even for β as low as 0.02 when selection is absent or weak, and small positive values of DT and Δπ are found with neutrality even when β = 0.002 (of the order of 1% with n = 200).
With very high values of β, a positive Tajima’s D can occur even with quite strong purifying selection (a scaled selection parameter γ of 50), as shown in Table 1. A SFS with an excess of intermediate-frequency variants at loci across the genome is usually interpreted as indicating a recent population bottleneck or a subdivided population (e.g., Staedler et al. 2009). False positive results for tests for bottlenecks and/or subdivision may thus be obtained if infinite sites rather than finite sites models are applied to hyperdiverse populations or epigenetic variation, even when moderately strong purifying selection is acting. Given that very small positive mean values across sites of statistics such Δπ can be statistically significant with genomic-scale data and large sample sizes, caution should be used when applying infinite sites predictions to such data sets. The suggested criterion for hyperdiversity of π or θw of 5% for using finite sites models rather than the infinite sites model (Cutter et al. 2013) may be too high for such data.
Relation to the data
This raises the question of whether there is indeed evidence for the expected pattern of a skew of the SFS spectrum toward intermediate-frequency variants. In the study of Caenorhabditis sp. 5 by Cutter et al. (2012), where the within-population diversity at synonymous sites is ∼0.08, Tajima’s D values for “scattered” samples [where one allele per locus was sampled from each of 13 locations, to minimize departure from the standard coalescent process (Wakeley 2000)] were nearly all positive, with a mean of 0.28. This is consistent with the coalescent simulations of Cutter et al. (2012), who used the SIMCOAL2 program of Laval and Excoffier (2004) with a finite sites model with equal mutation rates among all four possible nucleotides (A. Cutter and L. Excoffier, personal communication). The model used here gives an expected value of Tajima’s D of ∼0.10 with γ = 0 or 0.5 and a mutational bias of 2, assuming a sample size of 13 and 150 bp per locus (corresponding approximately to the numbers of synonymous sites in the study). At least qualitatively, this species thus fits the expectation under hyperdiversity for DNA sequence variability.
In contrast, the synonymous SFS in the much more hyperdiverse species Caenorhabditis brenneri is biased toward low-frequency variants, with a mean Tajima’s D of –0.56 over 23 loci with an average of ∼150 bp per locus (Dey et al. 2103, table S3), again using scattered sampling. Similarly, in the only detailed survey of epigenomic variation published to date, that of ∼200 northern European accessions of A. thaliana (Schmitz et al. 2013, supplementary table 9), the SFS for single methylated vs. nonmethylated cystosines is also highly skewed toward low-frequency variants. The lack of linkage disequilibrium between this class of variants and SNPs suggests that these epigenetic variants are not caused by nucleotide site variants associated with methylation status, but represent true heritable epialellic variation (Schmitz et al. 2013).
There are several possible reasons for this sharp disagreement between the theoretical predictions and these observations. One is that demographic effects, such as a recent population expansion, mean that predictions based on the assumption of a stationary population are overwhelmed by the well-known excess of rare variants associated with expansion (Tajima 1989a; Slatkin and Hudson 1991). This is ruled out for the case of epigenetic variation in A. thaliana, because the SFS for SNPs is far less biased toward rare variants (Schmitz et al. 2013), but remains possible for C. brenneri. The second possibility is that purifying selection is sufficiently strong to skew the SFS toward rare variants. This seems unlikely in the case of C. brenneri, where the estimates of the overall γ for synonymous sites suggest a value close to 0.5 (Dey et al. 2103), which is insufficient to cause a skew toward rare variants (see Table 1). This explanation is more plausible for the A. thaliana example, since high levels of methylation of cytosines are nonrandomly distributed across the genome and are especially prevalent in transposable element sequences where methylation is important for their silencing (Schmitz et al. 2013). It is therefore very likely that the methylated states in such sequences are favored by selection. Another possibility is that methylation is selectively neutral, and the differences between genomic regions simply reflect different levels of mutational bias, either toward or against methylation. Calculations using the biallelic model show that extreme mutational bias at neutral or nearly neutral sites can overcome the skew of the SFS toward intermediate-frequency variants (results not shown). The published results of mutation accumulation experiments in A. thaliana (Becker et al. 2011; Schmitz et al. 2011) do not shed much light on the question of the extent of the direction and magnitude of mutational bias, since the experimental design ascertains sites for which at least one of the mutation accumulation lines contains a methylated cytosine at the site in question. It is thus strongly biased toward detecting variants at which the original state was methylation, making it hard to determine the rate of mutation toward methylation. Distinguishing between these possible interpretations is a challenging task and will require the use of numerical models that incorporate past population size changes and population structure.
Limitations of the biallelic model
It is important to note that the biallelic model used here, which is similar to that used by Bertorelle and Slatkin (1995) and Desai and Plotkin (2008), is likely to underestimate the effect of hyperdiversity on the SFS, since the presence of more than two variants at a segregating site will result in higher π but not θw. On the other hand, the infinite alleles assumption, apparently used by Aris-Brosou and Excoffier (1996), means that the upper limit to π is 1, whereas in reality there is a maximum of four segregating variants per site, leading to an upper limit to π of 3/4 (when all four variants are present at equal frequencies), as opposed to 1/2 for the biallelic model used here. Given the almost universal existence of mutational biases toward transitions vs. transversions and for GC to AT vs. AT to GC mutations, the upper limit is in practice likely to be considerably <1, so that the biallelic model with modest mutational bias probably provides a reasonably good guide to the values of measures of skew in the SFS. There has been much discussion of the question of why silent nucleotide site diversity does not a span a much wider range than is commonly seen (see Leffler et al. 2012 for a recent account). The saturation of diversity at a level considerably <3/4 when there is mutational bias may well be a contributing factor.
An intermediate situation is provided by assuming a K-allele model (Ewens 2004, pp. 192–200) with K = 4, corresponding to equal mutation rates among all four nucleotide states at a site (Tajima 1996; Yang 1996; Desai and Plotkin 2008). Under neutrality the exchangeability of the different nucleotides under this model means that the probability density φ(qi) for the frequency qi of a variant of type i (i = 1–4) is proportional to where θ is the net mutation rate per site; i.e., φ(qi) follows a β-distribution with parameters θ and θ/3 (Tajima 1996). With semidominant selection with type i having a selective advantage s over all other variants, which are assumed to be selectively equivalent to each other, this expression is simply multiplied by exp(γqi).
Following Tajima (1996), these assumptions allow simple analytical formulas for the sample statistics used above to be obtained for the case of neutrality: and where These can be compared with the statistics obtained from the biallelic model in Table 1, setting θ to the equilibrium infinite sites neutral diversity with reverse mutation 2βκ/(1 + κ) = 4β/3 (with κ = 2) to obtain comparable net scaled mutation rates per site. As expected, for very low θ, the two models yield similar results, but even with β = 0.02 the four-allele model gives noticeably higher expected values of Tajima’s D and Δπ; e.g., with a sample size of 20 and β = 0.02, the values of Tajima’s D and Δπ are 0.069 and 0.022, respectively, vs. 0.038 and 0.016 for the biallelic model. With a sample size of 20 and β = 0.2, the values of Tajima’s D and Δπ for the four-allele model are 0.61 and 0.20, respectively, compared with 0.32 and 0.11 for the biallelic model; values of D and Δπ much greater than twice the biallelic values can be generated by the four-allele model when β is large, reaching 4.7 and 1.6, respectively, with β = 20. The proportion of singletons behaves rather differently under the four-allele model; it can even increase with β up to some upper limit, after which it declines and is always higher than for the biallelic model (e.g., 0.36 vs. 0.22, respectively, for β = 0.2 and n = 20; and 0.17 vs. 0.01 for β = 20). This behavior presumably reflects the fact that there are four possible variants at each site that can behave as singletons in the case of the four-allele model, and the above formula simply sums over the probabilities that each one of these is a singleton, regardless of the status of the other three possible variants at the same site. A statistic such as Δπ is thus probably a better summary of the skew of the SFS than the proportion of singletons when a substantial fraction of polymorphic sites segregate for more than two variants, unless variants are collapsed into biallelic alternatives such as GC vs. AT base pairs (for example, Evans et al. 2014).
For studying situations with multiple alleles per nucleotide and nonequilibrium demography, numerical methods such as that of Zeng (2010) will be needed.
Some other implications
One difficulty with interpreting the results of population surveys of epiallelic variation is that it is impossible to know whether sites that lack epigenetic marks in all individuals sampled are potentially capable of acquiring them. This means that the denominator in per-site statistics such as π and θw is unknown, making it hard to apply standard population genetics methods to these kinds of data. Fortunately, however, with high β values (>0.2), nearly all sites capable of mutation will be found to be segregating in a large sample, even with a scaled selection strength as high as γ = 5; thus, the majority of sites capable of epimutations can be identified from population surveys, unless strong purifying selection is acting. Population surveys could, therefore, be a valuable tool for the characterization of the epigenome.
Another finding that is relevant for both hyperdiverse DNA sequence variation and hypermutable epigenetic variation is the fact that substitution rates for sites under purifying selection may be close to or even greater than rates at neutral sites with high β values. As described above, this may occur even after correcting for the effects of differences in base composition between neutral and selected sites (Figure 1 and Figure 2). This lack of sensitivity of substitution rates to the strength of purifying selection is consistent with the patterns described by Cutter et al. (2013), where there is only a weak relation between codon usage bias and a measure of synonymous site divergence in the hyperdiverse species Ciona savigni. Similarly, diversity at sites subject to weak purifying selection is expected to show a nonlinear pattern of relationship with γ, such that π increases with γ when sites are close to neutral and then declines again as γ approaches or exceeds 1; the range of γ values over which there is an increase is broader for large β (Figure 2). Synonymous diversity of genes in C. brenneri does indeed show a quadratic relation with the frequency of optimal codons, such that genes with ∼50% optimal codons have the highest diversity values (A. Cutter, personal communication).
With γ values typical of those reported from studies of selection on codon usage (γ ≤1), the standard Li–Bulmer equation (Li 1987; Bulmer 1991) tends to overestimate the expected level of codon bias, as measured by the mean frequency of the favored allelic type (), when β > 0.02. For example, with γ = 1 and β = 0.2, the exact value of from Equation 5a is 0.49 compared with the Li–Bulmer infinite sites prediction of 0.58, while the second-order approximation from Equations 7a and 7b gives 0.44. Analyses of codon usage in hyperdiverse species that use codon usage data to estimate γ (see Sharp et al. 2010) should probably use the exact expression. It is interesting in this context to note that there is only a small difference in the mean level of codon usage bias between C. brenneri and C. remanei, despite an approximately threefold difference in synonymous site diversity (A. Cutter, personal communication). This raises the question of whether the purifying selection model used here is appropriate for codon usage or whether a model of stabilizing selection (Kimura 1981) is more realistic, since the latter means that γ is insensitive to Ne over a wide range of parameter values, provided that there is mutational bias (Charlesworth 2013). The behavior of this model with hyperdiversity would, therefore, be worth studying.
Supplementary Material
Acknowledgments
We thank Deborah Charlesworth, Asher Cutter, Kai Zeng, and two anonymous reviewers for their comments on the manuscript and Asher Cutter and Laurent Excoffier for valuable discussions.
Appendix
The Expected Number of New Mutations That Arise at a Segregating Site
Assume that we have a nucleotide site that is segregating for a neutral mutation that arose at an initial frequency of 1/(2N). Let the probability that this variant mutates to an alternative nucleotide be u per generation (this includes the possibility that it reverts to the ancestral state); let the probability that the ancestral variant mutates to another state be v (this includes the possibility that the mutation is identical in state to the variant that is already segregating). If the frequency of the mutation in the population in a given generation is x, the expected total number of mutational events is 2N[ux + v(1 – x)]. The expected time that the original mutation spends in the frequency interval x to x + dx is given approximately by 4Ne/(1 – x) for 0 < x ≤ 1/(2N) and 2Ne/(Nx) for 1/(2N) < x ≤ 1 (Ewens 2004, p. 160). The total expected number of new mutations that arise during the sojourn of the mutation in the population is thus
| (A1) |
Approximations to Equations 5a and 5b with Small α and β
Equation 5a is equivalent to
| (A2) |
We can write terms of the form (β + i – j)/(α + β + i – j) as 1 – [βκ/(i – j)] + O(β2); keeping only O(β) terms, we have
| (A3a) |
or
| (A3b) |
where ai = 1 + 1/2 + 1/3 + … 1/(i –1) (i ≥ 2). The exponential terms in the numerator and denominator of Equation A3b can thus be replaced by 1 + O(β), yielding Equation 6 of the main text.
In Equations 7a and 7b, ai+1 < i for i > 1, the first summation in the numerator of g is less than the sum of γ i(i –1)!, so that the sum is <γ exp(γ). Similarly, ai+1 – ai = 1/i, so that the second summation is <exp(γ) – (1 + γ). It follows that g is positive and <κγ + exp(γ) – 1. This is multiplied by exp(–γ) in the numerator of Equation 5a, to obtain the multiplicand of βκ, yielding κγ exp(–γ) + 1 – exp(–γ) < κ + 1 – exp(–γ). The contribution of –βκg exp(–γ) to the numerator of Equation 7a is thus negative and smaller in magnitude than βκ (1 + κ). The leading term in Equation 6 should provide a good approximation when βκ is ≤0.1.
Approximations for the Frequencies of the Fixed Classes
Assuming that α << 1 and β << 1, and employing the approximations used in Equation A3b, we find that
| (A4a) |
Similarly,
| (A4b) |
We can use the fact that Γ(1 + x) = xΓ(x) to write Γ(x) = Γ(1 + x)/x. For small x, the representation of the gamma function as an infinite product (Abramowitz and Stegun 1965) implies that Γ(1 + x) = (1 – cx) + O(x 2), where c ≈ 0.577 is Euler’s constant, and similarly for the other gamma integrals. We can thus approximate Γ(x) for small x by 1/x, and the term involving gamma functions in Equations 9a and 9b is then κβ/(1 + κ)[1 + O(β)]. Using a similar approximation to that used in Equations 7a and 7b, and neglecting higher-order terms in β, we obtain
| (A4c) |
| (A4d) |
where
| (A4e) |
The higher-order terms in β vanish when γ = 0, suggesting that these expressions are good approximations when β and γ are both small. More rigorously, for finite i, ai is less than some constant A, which is approximately equal to ln(i – 1). If terms in i > k can be neglected in the sum that defines h, h < ln(k)exp(γ), so that h exp(–γ) < βκ ln(k), where ln(k) is a small multiple of one unless γ is very large.
In addition, for arbitrary γ, the terms involving (2N)–β in Equations A4c and A4d are equal to 1– β ln(2N) + O(β 2) and 1– βκ ln(2N) + O(β 2), respectively. Provided that ln(2N) is of order one, ff1 and ff2 are each equal to their respective infinite sites value, multiplied by a factor 1 – O(β), implying that the infinite sites values provide a good approximation unless β >> 0.
Fixations of Mutations
Consider first the case of A2 to A1 mutations that arise at a site that was initially fixed for A2. We approximate the frequency of this fixed class, f2f, by the integral in Equation 9b. The fixation probability, Q1, of an A1 mutation with initial frequency 1/(2N) when N is large is γ (2N)−1[exp(γ) – 1]−1 + O[γ 2(2N)–2], so that the net number of new A2 mutations that arise in a given generation and are expected to become fixed is 2Nκvf2f{γ/(2N)[exp(γ) – 1]−1 + O[γ 2(2N)–2]} = κvf2f{γ [exp(γ) – 1]−1 + 2N O[γ 2(2N)–2]}. Using the same approximation for Q1 and the fact that q is close to one in Equation 9b, the corresponding formula from Equations 13 and 14a is
| (A5) |
Provided that 2N is sufficiently large in relation to γ, so that the higher-order terms in γ(2N)–1 can be ignored, the two results are equivalent.
The following argument can be used for the other end of the frequency range. In this case, there is no contribution from the class fixed for A1 mutations, whose frequency is f1f as given by Equation 9a, to the fixation of new A1 mutations. The corresponding formula from Equations 13 and 14a is
| (A6) |
Again, provided that 2N is sufficiently large in relation to γ, the two results are equivalent. Parallel arguments can be used for the fixation of new A2 mutations.
The Relative Rate of Substitution Under the Infinite Sites Assumption
At equilibrium between mutation, drift, and selection, the frequencies of sites fixed for A1 and A2 under the infinite sites model are approximated by κ exp(–γ)/[1 + κ exp(–γ)] and 1/[1 + κ exp(–γ)], respectively (Li 1987; Bulmer 1991; McVean and Charlesworth 1999). Averaging over the contributions from mutations arising at each class of fixed sites, taking into account their respective fixation probabilities, the equilibrium rate of nucleotide substitution is then
| (A7a) |
(Charlesworth and Charlesworth 2010, p. 275).
If we consider neutral mutations arising at fixed sites with the same frequencies of A1 and A2 variants as the selected sites (i.e., with the same base composition), the substitution rate is
| (A7b) |
The ratio R(γ) = λ(γ)/λ(0) gives the rate of substitution of selected mutations relative to neutral expectation, conditioning on the same base composition; we have
| (A8) |
It is easily seen that R = 1 at γ = 0 and decreases as γ increases.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.167973/-/DC1.
Communicating editor: M. A. Beaumont
Literature Cited
- Abramowitz M., Stegun I. A., 1965. Handbook of Mathematical Functions. Dover, New York. [Google Scholar]
- Aris-Brosou S., Excoffier L., 1996. The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism. Mol. Biol. Evol. 13: 494–504. [DOI] [PubMed] [Google Scholar]
- Becker C., Hagmann J., Mueller J., Koenig D., Stegle O., et al. , 2011. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249. [DOI] [PubMed] [Google Scholar]
- Bertorelle G., Slatkin M., 1995. The number of segregating sites in expanding human populations with implications for estimates of demographic parameters. Mol. Biol. Evol. 12: 887–892. [DOI] [PubMed] [Google Scholar]
- Bulmer M. G., 1991. The selection-mutation-drift theory of synonomous codon usage. Genetics 129: 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 2013. Stabilizing selection, purifying selection and mutational bias in finite populations. Genetics 194: 955–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 2010. Elements of Evolutionary Genetics. Roberts & Co., Greenwood Village, CO. [Google Scholar]
- Cutter A. D., Wang G.-X., Ai H., Peng Y., 2012. Influence of finite-sites mutations, population subdivision and sampling schemes on patterns of nucleotide polymorphism for species with molecular hyperdiversity. Mol. Ecol. 21: 1345–1359. [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Jovelin R., Dey D., 2013. Molecular hyperdiversity and evolution in very large populations. Mol. Ecol. 22: 2074–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M., Plotkin J. B., 2008. The polymorphism frequency spectrum of finitely many sites under selection. Genetics 180: 2175–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A., Chan C. K. W., Thomas C. G., Cutter A. D., 2013. Molecular hyperdiversity defines populations of the nematode Caenorhabditis brenneri. Proc. Natl. Acad. Sci. USA 110: 11056–11060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eory L., Halligan D. L., Keightley P. D., 2010. Distributions of selectively constrained sites and deleterious mutation rates in the hominid and murid genomes. Mol. Biol. Evol. 27: 177–192. [DOI] [PubMed] [Google Scholar]
- Evans B. J., Zeng K., Esselstyn J. A., Charlesworth B., Melnick D. J., 2014. Reduced representation genome sequencing suggests low diversity on the sex chromosomes of Tonkean macaque monkeys. Mol. Biol. Evol. 31: 2425–2440. [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 2004. Mathematical Population Genetics. 1. Theoretical Introduction. Springer-Verlag, New York. [Google Scholar]
- Eyre-Walker A., 1992. The effect of constraint on the rate of evolution in neutral models with biased mutation. Genetics 131: 233–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1922. On the dominance ratio. Proc. R. Soc. Edinb. 42: 321–341. [Google Scholar]
- Fisher R. A., 1930. The distribution of gene ratios for rare mutations. Proc. R. Soc. Edinb. 50: 205–220. [Google Scholar]
- Fu Y.-X., Li W.-H., 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U., Kelly B., Ferguson-Smith A., Pembry M., Lindquist S., 2013. Transgenerational epigenetic inheritance: How important is it? Nat. Rev. Genet. 14: 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S., 1927. A mathematical theory of natural and artificial selection. Part V. Selection and mutation. Proc. Camb. Philos. Soc. 23: 838–844. [Google Scholar]
- Halligan D. L., Eyre-Walker A., Andolfatto P., Keightley P. D., 2004. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 14: 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 1990. Gene genealogies and the coalescent process. Oxf. Surv. Evol. Biol. 7: 1–45. [Google Scholar]
- Jenkins P. A., Song Y. S., 2011. The effect of recurrent mutation on the frequency spectrum of a segregating site and the age of an allele. Theor. Popul. Biol. 80: 158–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins P. A., Mueller J. W., Song Y. S., 2014. General triallelic frequency spectrum under demographic models with variable population size. Genetics 196: 295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes F., Porcher E., Texeira F., Saliba-Colombani V., Simon M., et al. , 2009. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 6: e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1962. On the probability of fixation of a mutant gene in a population. Genetics 47: 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1968. Evolutionary rate at the molecular level. Nature 217: 624–626. [DOI] [PubMed] [Google Scholar]
- Kimura M., 1971. Theoretical foundations of population genetics at the molecular level. Theor. Popul. Biol. 2: 174–208. [DOI] [PubMed] [Google Scholar]
- Kimura M., 1981. Possibility of extensive neutral evolution under stabilizing selection with special reference to non-random usage of synonymous codons. Proc. Natl. Acad. Sci. USA 78: 454–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Maruyama T., Crow J. F., 1963. The mutation load in small populations. Evolution 48: 1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klironomos F., Berg J., Collins S., 2013. How epigenetic mutations can affect genetic evolution: model and mechanism. BioEssays 35: 571–578. [DOI] [PubMed] [Google Scholar]
- Kondrashov F. A., Ogurtsov A. Y., Kondrashov A. S., 2006. Selection in favor of nucleotides G and C diversifies evolution rates and levels of polymorphism at mammalian synonymous sites. J. Theor. Biol. 240: 616–626. [DOI] [PubMed] [Google Scholar]
- Langley S. A., Karpen G. H., Langley C. H., 2014. Nucleosomes shape DNA polymorphism and divergence. PLoS Genet. 10: e1004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauria M., Piccinni S., Pirona R., Lund G., Viotti A., et al. , 2014. Epigenetic variation, inheritance, and parent-of-origin effects of cytosine methylation in maize (Zea mays). Genetics 196: 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval G., Excoffier L., 2004. SIMCOAL 2.0: a program to simulate genomic diversity over large recombining regions in a subdivided population with a complex history. Bioinformatics 20: 2485–2487. [DOI] [PubMed] [Google Scholar]
- Lawrie D. S., Petrov D. A., Messer P. W., 2011. Faster than neutral evolution of constrained sequences: the complex interplay of mutational biases and weak selection. Genome Biol. Evol. 3: 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler E. M., Bullaughey K., Matute D. R., Meyer W. K., Ségurel L., et al. , 2012. Revisiting an old riddle: What determines genetic diversity levels within a species? PLoS Biol. 10: e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.-H., 1987. Models of nearly neutral mutations with particular implications for non-random usage of synonymous codons. J. Mol. Evol. 24: 337–345. [DOI] [PubMed] [Google Scholar]
- McVean G. A. T., Charlesworth B., 1999. A population genetic model for the evolution of synonymous codon usage: patterns and predictions. Genet. Res. 74: 145–158. [Google Scholar]
- Mizawa K., Tajima F., 1997. Estimation of the amount of genetic variation when the mutation rate varies among sites. Genetics 147: 1959–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E. J., 2006. Inherited epigenetic variation: revisiting soft inheritance. Nat. Rev. Genet. 7: 395–401. [DOI] [PubMed] [Google Scholar]
- Sargsyan O., 2014. A framework including recombination for analyzing the dynamics of within-host HIV genetic diversity. PLoS Genet. 9: e87655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R. J., Ecker J. R., 2012. Epigenetic and epigenomic variation in Arabidopsis thaliana. Trends Plant Sci. 17: 149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R. J., Schultz M. D., Lewsey M. G., O’Malley R. C., Urich M. A., et al. , 2011. Transgenerational epigenetic instability is a source of novel methylation variants. Science 334: 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R. J., Schultz M. D., Urich M. A., Nery J. P., Pelizola M., et al. , 2013. Patterns of population epigenomic diversity. Nature 495: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Emery L. B., Zeng K., 2010. Forces that influence the evolution of codon bias. Philos. Trans. R. Soc. B 365: 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., Hudson R. R., 1991. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 143: 579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staedler T., Haubold B., Merino C., Stephan W., Pfaffelhuber P., 2009. The impact of sampling schemes on the site frequency spectrum in nonequilibrium subdivided populations. Genetics 182: 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1983. Evolutionary relationship of DNA sequences in a finite population. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989a The effect of change in population size on DNA polymorphism. Genetics 123: 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989b Statistical method for testing the neutral mutation hypothesis. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1996. The amount of DNA polymorphism maintained in a finite population when the neutral mutation rate varies among sites. Genetics 143: 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., 2000. The effects of subdivision on the genetic divergence of populations and species. Evolution 54: 1092–1101. [DOI] [PubMed] [Google Scholar]
- Wakeley J., 2008. Coalescent Theory. An Introduction. Roberts & Co., Greenwood Village, CO. [Google Scholar]
- Watterson G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Wright S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1937. The distribution of gene frequencies in populations. Proc. Natl. Acad. Sci. USA 23: 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 1996. Statistical properties of a DNA sample under the finite-sites model. Genetics 144: 1941–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K., 2010. A simple multiallele model and its application to identifying preferred–unpreferred codons using polymorphism data. Mol. Biol. Evol. 27: 1327–1337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.