Abstract
Across taxa, female behavior and physiology change significantly following the receipt of ejaculate molecules during mating. For example, receipt of sex peptide (SP) in female Drosophila melanogaster significantly alters female receptivity, egg production, lifespan, hormone levels, immunity, sleep, and feeding patterns. These changes are underpinned by distinct tissue- and time-specific changes in diverse sets of mRNAs. However, little is yet known about the regulation of these gene expression changes, and hence the potential role of microRNAs (miRNAs), in female postmating responses. A preliminary screen of genomic responses in females to receipt of SP suggested that there were changes in the expression of several miRNAs. Here we tested directly whether females lacking four of the candidate miRNAs highlighted (miR-279, miR-317, miR-278, and miR-184) showed altered fecundity, receptivity, and lifespan responses to receipt of SP, when mated once or continually to SP null or control males. The results showed that miRNA-lacking females mated to SP null males exhibited altered receptivity, but not reproductive output, in comparison to controls. However, these effects interacted significantly with the genetic background of the miRNA-lacking females. No significant survival effects were observed in miRNA-lacking females housed continually with SP null or control males. However, continual exposure to control males that transferred SP resulted in significantly higher variation in miRNA-lacking female lifespan than did continual exposure to SP null males. The results provide the first insight into the effects and importance of miRNAs in regulating postmating responses in females.
Keywords: Acp70A, ejaculate, male–female coevolution, postmating sexual selection, sexual conflict
REPRODUCTION is a fundamental biological process and it is well established that mating itself initiates a multitude of physiological and behavioral postmating changes in females. Insights into the gene expression changes underlying mating have been gained from studies in the fruit fly Drosophila melanogaster (Lawniczak and Begun 2004; McGraw et al. 2004; Mack et al. 2006; Innocenti and Morrow 2009). D. melanogaster males transfer not only sperm in their ejaculates, but also up to ∼130 different seminal fluid peptides (Sfps) (Findlay et al. 2008; Ayroles et al. 2011). Many of the striking postmating responses of females to mating are mediated by the effects of these Sfps (Chapman 2001; Gillot 2003; Ram and Wolfner 2007a; Wolfner 2009). It is therefore of significant, fundamental interest to understand the detailed mechanisms underlying the profound reprogramming in gene expression that occurs in females due to Sfp receipt. Through this understanding, it will be possible to determine (i) how the extensive changes required to effectively coordinate reproduction are regulated and (ii) to what extent these processes are shaped by sexual selection and sexual conflict.
Our knowledge of the phenotypes and functions of individual Sfps in D. melanogaster is rapidly increasing (Herndon and Wolfner 1995; Neubaum and Wolfner 1999; Tram and Wolfner 1999; Mueller et al. 2007; Ram and Wolfner 2007a; LaFlamme et al. 2012). One well-characterized Sfp, on which we focus in this study, is the so-called “sex peptide” (SP). SP has significant effects on a range of important fitness-related traits in females. It significantly increases egg production and decreases female receptivity to remating (Chapman et al. 2003b; Liu and Kubli 2003), increases food uptake (Carvalho et al. 2006), alters nutrient balancing (Ribeiro and Dickson 2010), and increases the expression of antimicrobial peptides (Peng et al. 2005). It also inhibits female siesta sleep (Isaac et al. 2010), alters water balance (Cognigni et al. 2011), and is involved in regulating sperm release from the storage organs (Avila et al. 2010). The sex peptide receptor has been identified, and it is expressed in the female genital tract and central nervous system (Soller et al. 2006; Yapici et al. 2008; Rezaval et al. 2012).
The fitness effects of SP in males and females appear to represent hallmarks of interlocus sexual conflict (Rice 1998; Chapman et al. 2003a; Arnqvist and Rowe 2005), in that repeated receipt of SP exacerbates the survival cost of mating in females (Wigby and Chapman 2005), while simultaneously increasing a male’s “per mating” offspring production (Fricke et al. 2009). It has been hypothesized that males and females are locked into a cycle of antagonistic coevolution over the phenotypic effects of SP and the female’s physiological responses to it. Understanding the mechanisms underlying female genomic responses to Sfps such as SP is therefore of great interest in revealing the sophisticated chemical communication between the sexes at mating. It also represents an excellent starting point to understand whole genome responses to sexual conflict and the potential role of Sfps in driving sexually antagonistic coevolution between the sexes.
Transcriptome studies have provided significant insights into the extensive genome-wide changes in gene expression that together form a synchronized response to mating and to Sfp receipt (McGraw et al. 2004; Mack et al. 2006; Domanitskaya et al. 2007 Innocenti and Morrow 2009). Such studies have also sought to characterize potential genomic signatures of sexual conflict (Gioti et al. 2012). To date (McGraw et al. 2004; Mack et al. 2006; Gioti et al. 2012) expression profiles have been conducted at different levels of resolution—from whole organisms (McGraw et al. 2004) to tissue-specific profiles (Mack et al. 2006)—and from total responses to mating (Mack et al. 2006), to courtship, to ejaculate receipt, and to specific Sfps (McGraw et al. 2008; Gioti et al. 2012). For example, McGraw et al. (2004) found that many genes were differentially expressed after mating. The fold changes involved were generally quite modest, which led the authors to suggest that females are “poised” to mount rapid responses to mating and hence maintain a set of mRNAs to facilitate this. McGraw et al. (2004) showed that distinct subsets of genes alter their expression in response to receipt of sperm vs. seminal fluid proteins. However, despite this, the post-transcriptional regulatory mechanisms that modulate these gene expression changes and ultimately result in the observed phenotypic changes have not been considered in any detail. In this study, we focused on the role of such post-transcriptional gene regulation in females in mediating postmating responses.
As noted above, initial insights into the underlying genomic signatures of sexual conflict mediated by SP in particular have also been gained from gene expression studies (Gioti et al. 2012). Genome-wide responses in females revealed widespread tissue and time-specific changes in many categories of genes (Gioti et al. 2012). Gene expression changes in response to SP in the head + thorax were more varied and dynamic than in the abdomen, in which genes were mainly down-regulated. In the head + thorax, genes involved in a number of biological processes (e.g., the TOR pathway regulating nutrient sensing and genes involved in phototransduction) were differentially regulated, while in the abdomen, egg and early embryo development genes were overrepresented. This study supported the idea that substantial changes to female physiology can occur after mating due to receipt of a single Sfp and that these changes are spatially and temporally dynamic (McGraw et al. 2008). The potential for manipulation of females by males via SP is therefore widespread and thus hard for females to sidestep or “ignore,” even if the effects of SP are costly to females (Wigby and Chapman 2005).
The effective interpretation and comparison of the results of gene expression studies such as those described above requires careful consideration of biological and technical differences as well as of general biases in sequencing methodologies (Van Dijk et al. 2014). One important factor can be tissue specificity. For example, Mack et al. (2006) compared their postmating gene expression profiles in the D. melanogaster lower female genital tract to McGraw et al. (2004)’s whole organism expression patterns and found little overlap. At least part of the explanation is likely to be that signatures of local tissue-specific changes are often swamped by whole organismal responses (Chintapalli et al. 2007). Another factor is the timing of sampling, which can drive divergence in transcript levels either due to mRNA expression per se or because of differences in the level of post-transcriptional control over time. Gioti et al. (2012) found genes to be mainly down-regulated in the abdomen following receipt of SP, whereas Mack et al. (2006) observed mostly gene activation in the lower genital tract following mating. Mack et al. (2006)’s parallel measures of the associated protein expression changes revealed a general pattern of down-regulation in 84 proteins, and, though mRNA transcripts were found for the majority of the differentially expressed proteins, there was little correspondence between up-/down-regulation of mRNA vs. its protein. This discrepancy between gene and protein expression is important, as it suggests that there is significant post-transcriptional regulation in the coordination of female postmating responses. It is the existence of such post-transcriptional regulation that we investigated in this study.
Our knowledge of the nature of post-transcriptional regulation has been revolutionized over the last two decades. It has been realized that there is a huge influence of small noncoding RNAs on the regulation of transcription and translation. MicroRNAS (miRNAs) are perhaps the best studied class of noncoding RNAs to date and have been identified as post-transcriptional master regulators of gene expression, typically for at least one-third of genes (Filipowicz et al. 2008). miRNAs are ∼22 nt in length and mediate translational repression and/or mRNA degradation. They show deep evolutionary conservation and are involved in many, if not all, biological processes (Filipowicz et al. 2008). miRNAs have been identified in many insects and their importance in regulating developmental processes, cell growth, and proliferation, ageing as well as host–pathogen interactions is increasingly realized (Asgari 2013; Lucas and Raikhel 2013). miRNAs are also important in reproduction and are involved in regulating functionality of the ovaries, e.g., in the maintenance and differentiation of germline stem cells (Park et al. 2007). This suggests they are promising candidates for involvement in the regulation of female postmating responses.
In the context of sexual conflict theory, the manipulation by one sex of gene regulatory mechanisms in the other, for example by miRNAs, could be highly significant. Male ejaculate components could alter the expression of miRNAs in females in a way that alters female physiological processes to provide maximum fitness benefits for males. In this way, males could increase female reproductive output to maximize the paternity gained before the female remates. Alternatively, variation in the expression of miRNAs might represent female responses to minimize male manipulation. miRNA expression in females could, for example, be used to dampen down potentially costly oscillations in gene expression caused by responses to Sfps (Kim et al. 2013). In effect, this could be a mechanism to reduce noise and stabilize expression to a more benign level in terms of female fitness outcomes. Our aim in this study was to start an investigation into the potential importance of post-transcriptional regulation in sexual conflict. We did this by experimentally testing the involvement of a set of candidate miRNAs in responses to SP.
An initial screen of miRNA expression changes following receipt of SP from males highlighted several candidate miRNAs. Among these, miR-279, miR-317, and miR-184 showed down-regulation in female head + thorax samples. In female abdomens, miR-279 was down-regulated and miR-278 up-regulated (T. Rathjen, H. Pais, S. Moxon, C. J. Pennington, T. Dalmay, and T. Chapman, unpublished data; Supporting Information, File S1; Table S1; Figure S1; and Figure S2). Here, we tested directly the SP responses of females lacking these candidate miRNAs. However, this set of four miRNAs is only a subset of the miRNAs likely to be involved in SP responses. First, the miRNA count data in this preliminary screen were obtained prior to the finding that there can be significant RNA ligase-dependent bias in small RNA cloning. Some small RNA sequences, including miRNAs, can be preferred over others due to their ability to anneal to adapter molecules used for library generation, which leads to a higher chance for ligation and therefore sequencing (Sorefan et al. 2012). Second, we chose this set of four miRNAs for further testing not only on the basis of their validated, altered expression in response to SP (File S1; Table S1; Figure S1; Figure S2), but also on the availability of loss-of-function mutations (Materials and Methods, below).
We investigated the direct influence of miRNAs on female phenotypic responses to receipt of SP using both hypomorph and knockout (ko) mutants. We tested the effect of miRNA mutants in different genetic backgrounds and recorded the reproductive output and sexual receptivity of miRNA mutant females after single matings to SP-lacking or control males. We also measured the survival of miRNA-lacking females following continual exposure to SP-lacking or control males. Our prediction was that, as SP predominantly led to down-regulation of the four candidate miRNAs, we would see SP-like phenotypic responses in miRNA mutant females mated to SP-lacking males.
Materials and Methods
Culturing methods
Stocks were maintained at 25° on a 12:12 light:dark cycle in either large overlapping cage cultures (wild-type populations) or in bottle cultures (mutant stocks). miRNA stocks were cultured in glass bottles (189 ml) containing 70 ml of standard sugar-yeast (SY) food [100 g brewer's yeast powder, 100 g sucrose, 20 g agar, 30 ml Nipagin (10% w/v solution), 3 ml propionic acid, and 1 liter of water]. All experiments were conducted at 25° and in a humidified constant temperature room (∼50% relative humidity, RH), using glass vials (75-mm height × 25-mm diameter) containing 7 ml of SY food with ad libitum live yeast granules or live yeast paste added. To collect adults for the experiments, females were allowed to oviposit on agar–grape juice plates [50 g agar, 600 ml red grape juice, 42.5 ml nipagin (10% w/v solution), and 1.1 liters of water] with a blob of yeast paste unless stated otherwise. First instar larvae were collected the following day and groups of 100 transferred to vials with SY medium. Vials were incubated at standard conditions for 10 days. Virgin adults were ice anesthetized upon eclosion, sexed, and held in groups of 10 per sex.
Fly stocks
Wild-type flies:
The Dahomey wild-type stock was used throughout these experiments to provide experimental males for remating opportunities. The Dahomey stock was collected in the 1970s in Benin, Africa and held under the above conditions since then.
Sex peptide-lacking males:
SP gene knockout males (Liu and Kubli 2003) were used to generate males that do not transfer SP during mating. These males were produced by crossing SP0/TM3,Sb,ry males to ∆130/TM3,Sb,ry females. The resulting SP0/∆130 (SP0) males produce no SP (Liu and Kubli 2003). Control males were generated by crossing SP0,SP+/TM3,Sb,ry males to ∆130/TM3,Sb,ry females to generate SP producing SP0,SP+/∆130 (SP+) males. The strains were previously backcrossed into the Dahomey wild type to increase vigor. The ∆130/TM3,Sb,ry stock was backcrossed for three generations, and chromosomes 1, 2, and 4 of the SP0/TM3,Sb,ry and SP0,SP+/TM3,Sb,ry stocks were backcrossed for four generations. To generate SP-lacking and control males, three parental males and females for each cross were housed together in vials and transferred onto fresh food every day. Ten days later, male offspring of the correct genotype were collected and housed in groups of 10 in vials until used in the experiments.
miR-mutant females:
Lines that were hypomorphic for miR-279 and miR-317 were gratefully received from A. Yamamoto (North Carolina State University). These mutants carry a single autosomal P[GT1] transposon insertion in the respective miRNA genes in a w1118; Canton-S (w[CS]) wild-type genetic background (Yamamoto et al. 2008). We backcrossed both these hypomorphs four times into the white Dahomey (w[Dah]) genetic background (this is the wild-type Dahomey genetic background into which the w1118 allele had previously been backcrossed multiple times) (Broughton et al. 2005). We performed phenotypic tests (see below) using the two hypomorphs in both the Canton-S and Dahomey genetic backgrounds, using w[CS] or w[Dah] as the appropriate controls. We refer to hypomorphs as mir-279C and mir-317C in the w[CS] genetic background and mir-279D and mir-317D in the w[Dah] genetic background.
We used two miRNA knockout lines—one lacking miR-278 and one lacking miR-184. The mir-278 knockout strain was a kind gift from S. Cohen (National University of Singapore) (Teleman and Cohen 2006). The original line had been derived in a w1118 background. We backcrossed this line four times into the w[Dah] genetic background and refer to this backcrossed line as mir-278D. w[Dah] females were therefore an appropriate control. We received the mir-184 knockout (∆mir-184/Kr-GFP, CyO;TM2/TM6B) as a kind gift from N. Iovino (University of Munich) (Iovino et al. 2009). We used females homozygous for the mir-184 deletion in our phenotypic assays and the w1118 stock (no. 60000 from the Vienna Drosophila Stock Centre) as a control. The mir-278 and mir-184 knockouts are reported to span the two mir genes in question, leaving nearby genes unaffected (Teleman and Cohen 2006; Iovino et al. 2009). To obtain virgin females homozygous for the mir-184 knockout, we allowed three males and three females from the parental generation to interact and oviposit in vials for up to 4 days. After removing the adult flies, vials were incubated and virgin females of the correct genotype were collected shortly after eclosion and held in groups of 10 until used in the experiments.
Verification of miR-mutant lines
Northern blots for miRNA knock out verification:
Loss of miRNA expression in the knockout lines used was verified by Northern blotting using the protocol by Pall and Hamilton (2008). We extracted RNA as above from two samples and loaded 10 μg total RNA mixed with Ambion gel loading buffer II on a 15% polyacrylamide gel with urea. The gel was run at 120 V for 2 hr in 0.5× TBE. We then transferred the RNA to a Hybond-NX membrane using semidry transfer conditions at 250 mA for 45 min. We cross-linked the RNA in the membrane by adding 5 ml cross-linking solution (12 ml H2O, 122.5 μl 12.5 M 1-methylimidazole, 10 µl 1 M hydrochloric acid pH 8, and 0.373 g of EDC) and incubating at 60° for 1 hr in saran wrap. For each probe we prehybridized the membrane with Ultra-hyb-oligo buffer (Ambion) at 37° for 1 hr. We then incubated mixture of 10 μl H2O, 4 μl 5× polynucleotide kinase (PNK) forward buffer, 2 μl 10 μM oligo probe, 1 μl T4 PNK and 3 μl γ-ATP at 37° for 1 hr. The mixture was run through a Sephadex column to elute unbound isotope. We incubated the membrane in this buffer overnight at 37° and then washed it three times in 0.2× SSC:0.1% SDS before exposing it on a phosphorimaging screen in a radioactive cassette (Fujifilm) followed by imaging on a FX Pro Plus molecular imager (Bio-Rad). Antisense DNA oligonucleotide probe sequences used (Sigma-Aldrich) were as follows: miR-184 (5′-GCCCTTATCAGTTCTCCGTCCA-3′) and miR-278 (5′-AAACGGACGAAAGTCCCACCGA-3′). We used U6 as a loading control for all samples.
Quantitative real-time PCR for miRNA hypomorph verification:
To validate reduced or absent expression of the candidate miRNAs in the hypomorph lines used, we performed quantitative real-time PCR (qRT-PCR). We extracted total RNA using the mirVana miRNA isolation kit (Life Technologies) following the manufacturer’s protocol with minor modifications to the sample homogenization stage. Ten virgin females aged for 4 days from each line were homogenized on liquid nitrogen in a 2-ml microcentrifuge tube. We used the TaqMan MicroRNA assay (Life Technologies) for the qRT-PCRs with probes for mir-279 and mir-317, following the manufacturer’s protocol. We used miR-2S as a reference gene. Each 20-µl reaction was placed onto a MicroAmp (Life Technologies) plate and qRT-PCR was performed on the 7500 Fast Real-Time PCR system (Life Technologies). For each probe set, we produced a standard curve using a 1:5 serial dilution of a sample independent of any of those used in our assays. All standard curves had an R2 > 0.98 and slopes of between −2.40 and −3.24 (efficiency of 161.0 and 103.5%, respectively). We analyzed between five and six samples per stock tested. w[CS] and w[Dah] samples served as controls for the mutant stocks in respective genetic background and were reared in the same way.
Single mating reproductive output and receptivity assays
We used 4-day-old individuals to test reproductive output (number of offspring or estimated number of offspring) and sexual receptivity responses to receipt of SP in the mir-279C, mir-317C and mir-279D, mir-317D hypomorphs and the mir-278D and mir-184 knockout lines, vs. their wild-type controls. miRNA mutant and control females were mated to SP-lacking or control males or kept as virgins (to control for intrinsic differences between miR-mutant females). A total of 180 mutant or control females each were divided randomly across the three treatment groups by aspirating them into individual vials the morning of the experiment. The day before the beginning of the experiment, we introduced individual males of the appropriate genotype into vials for the “mated” treatments. Virgin females were similarly maintained but were not given a male on this day. For the females encountering a male, we recorded the time of introduction and the start and end of mating. After a successful mating, the male was removed from the vial and the female was allowed to oviposit. Females that had not mated within 2 hr after introducing a male were discarded. We then divided females from all three treatments into two groups (n = 30 per treatment combination) and allowed half of the females to remate after 24 hr, while the other half had an opportunity to remate after 48 hr. For the remating tests, females were given a 1-hr opportunity to remate with a Dahomey wild-type male and the females from the virgin treatment were simultaneously allowed to mate for the first time. We again recorded time of introduction, the start and end of remating, as well as the overall proportion of females that remated. For females assigned to the 24-hr remating treatment, we counted the number of eggs laid by females following the initial matings to either SP-lacking or control males and the number of offspring emerging from those eggs, to calculate egg–adult survival and reproductive output. We did not directly count the number of eggs laid during the 48-hr intermating period; thus we used offspring number as our measure of reproductive output. To compare this with the reproductive output of the virgin treatment (where no offspring were produced) we multiplied the number of eggs laid by virgin females with the strain-specific egg–adult survival rate.
Throughout the experiments, reproductive output was therefore a measure of offspring number. It represented the actual number of offspring for the mated females and an estimate of offspring number for the virgin female treatments (given by the number of eggs multiplied by the relevant strain-specific egg–adult survival rate).
Female reproductive output, receptivity, and survival following continual exposure to SP-lacking or control males
We tested the reproductive output (number of offspring), receptivity, and survival of miRNA-mutant and control females when continuously exposed to SP-lacking or control males throughout life. We also maintained groups of virgin females of each genotype for comparison. The day after eclosion, females were assigned to treatments at random and held in groups of three females (virgin female treatment) or three females together with three males of the appropriate genotype (mated female treatment) in vials supplemented with live yeast granules. There were 15 vials per treatment (n = 45 females, with the exception of the mir-278D virgin female treatment, for which there were n = 30 females). mir-184 knockouts were not included in this assay because we had not backcrossed them into the wild-type Dahomey genetic background. Furthermore, mir-184 knockout females do not produce eggs, which would potentially have complicated fitness comparisons with that of other treatments, including the controls, which are fertile.
We scored female survival daily until all females were dead. Every other day, groups were transferred onto fresh food during which males and females were shuffled within treatments to form new groups of three males and three females (or three females only for the virgin treatments) to minimize vial-to-vial differences and to prevent differences in density occurring over time as the females started to die. We maintained constant sex ratio (in the mated treatments) and density by combining vials. Each vial contained at least two females or two pairs. For the mated female treatments, males were replaced each week with 2- to 4-day-old fresh males of the appropriate genotype. This minimized effects on females of any age-dependent decline in male reproductive performance.
For 10 days over the first 2 weeks of the experiments we scored mating rate in the mated female treatments by performing spot checks of behavior every 20 min for 3 hr after lights on. We counted the number of mating pairs in each treatment at each time point. Twice each week, we also scored reproductive output in the mated female treatments. We scored virgin reproductive output once per week (estimated number of offspring given by virgin female fecundity multiplied by the strain-specific egg-to-adult survival). A total of 21 randomly chosen females per treatment were allowed to oviposit in vials with 7 ml of standard SYA food with added charcoal (4 g charcoal per liter) to facilitate egg counting. Females were separated and allowed to oviposit in individual vials for 6 hr before being returned to their experimental groups once again. We recorded reproductive output for each treatment until the time when there were <15 mated females of each genotype remaining alive.
Statistical analysis
General:
Mean (±SE) values are reported throughout unless otherwise specified. We calculated standard errors for all proportion data according to the following formula: SE = square root [p*(1 − p)/n], where p is the proportion of females remating and n is the number of trials in that particular test. All analyses were conducted in Rv2.15.1 (Ihaka and Gentleman 1996). For generalized linear models, we used the appropriate error structure and conducted an analysis of deviance. In this, the significance of factors was tested by individually subtracting each factor in turn from the full model and comparing models by estimating the difference in deviance (G2) between the two models. When using the quasiextension to correct for overdispersion of the data, we used an F-distribution to test for significance; otherwise the deviance values were compared against a χ2 distribution (Crawley 2007). We removed three-way interactions from models if they did not significantly affect the fit of the model.
To display the effect of receipt of SP on mutant females, we calculated effect sizes, as the standardized mean difference (Cohen’s d) over a pooled estimator of the standard deviation as the denominator. We calculated 95% confidence intervals (CIs) for effect sizes. In the single mating assay, we calculated the effect size for the number of offspring produced (using baseline corrected values, see below). In each case, we show the increase in reproductive output (baseline corrected value) relative to that of the relevant virgin female reproductive output (estimated number of offspring). To calculate effect sizes for the proportion of females that remated after 24 and 48 hr, we used the “failes” command in the “compute.es” package in R. This calculates Cohen’s d from binary data using the number of nonremating females in each group (the number of “failures”) relative to the total sample size. For the survival data, we calculated mean lifespan and used the associated pooled standard deviation to calculate the standardized mean difference plus 95% confidence intervals.
Statistical analysis of single mating reproductive output and receptivity assay:
The different female genotypes varied in intrinsic egg-laying capacity; therefore, to compare them, we used estimates of virgin female reproductive output (virgin fecundity scaled by the appropriate egg–adult survival, calculated as described above) of the appropriate genotype in each case as a baseline for standardization (for egg-to-adult survival data and analysis see File S2; Figure S3). This allowed us to directly compare reproductive output of mated females across the different genotypes. We standardized all 24- and 48-hr offspring counts by subtracting the mean corrected virgin output. This allowed us to test by how much a mating to a SP0 or SP+ male elevated female offspring production above the virgin baseline. The resulting data were then analyzed using analysis of variance.
Statistical analysis of reproductive output, receptivity, and survival following continual exposure to SP-lacking or control males:
Survival data were analyzed with a parametric Kaplan–Meier regression with a Weibull distribution, as described in Crawley (2007). For mating rate and reproductive output data, we used generalized linear models with appropriate error structures. The spot check data for mating rate were modeled using a binomial error distribution as the number of mating opportunities (e.g., the number of females in each treatment × the number of spot checks) taken vs. not taken. Reproductive output checks were analyzed using a Poisson error distribution with the quasiextension to account for overdispersion in the data (Crawley 2007).
Results
Verification of lines
All miRNA knockouts and hypomorphs had significantly lower expression of the appropriate miRNA as compared to the control lines as shown by the Northern blotting and qRT-PCR (Figure S4; Figure S5). Hence all the mutant lines in the different genetic backgrounds used in the experiments described here were successfully validated.
Effect of miRNAs on female reproductive output and sexual receptivity responses to SP after a single mating
Female reproductive output responses to SP:
mir-279D and mir-317D hypomorphs:
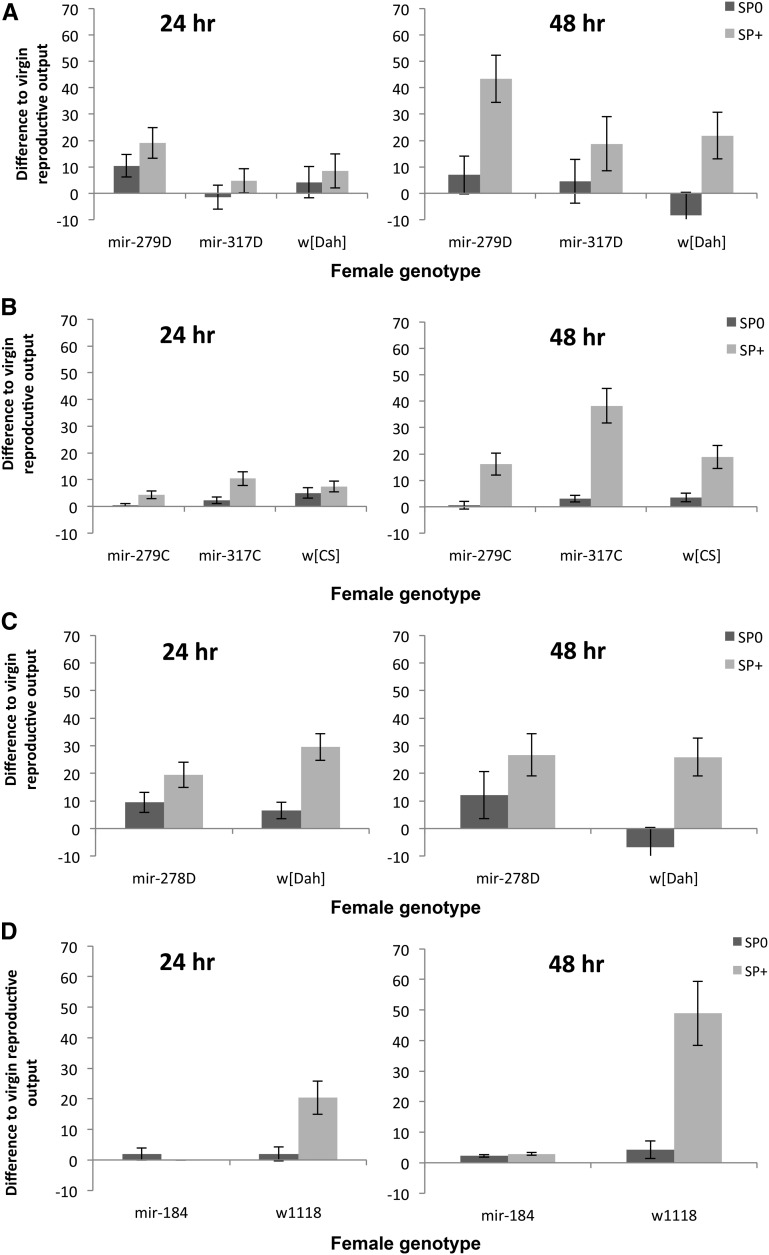
Virgin miRNA-lacking females differed significantly in fecundity [G2 = 209.46, F1,176 = 7.61, P = 0.0007 (dispersion parameter = 13.92)] with mir-279D females laying fewer eggs than the other two groups. Overall, the number of eggs laid increased from 24 to 48 hr (G2 = 697.63, F1,175 = 50.72, P < 0.0001) and this was similar for all female genotypes (interaction = ns). Therefore, to ascertain more clearly to what extent receipt of SP increased female reproductive output, we analyzed, as described above, the difference in reproductive output relative to the appropriate virgin female genotype, using the baseline corrected reproductive output data. This showed that reproductive output relative to the virgin level was significantly higher in females that received SP (F1,318 = 16.07, P < 0.0001; Figure 1A), and female genotypes also differed significantly in their reproductive output (F2,318 = 4.52, P = 0.012). However, there was no evidence that miRNA-lacking females differed in the extent to which they responded to SP. In mir-279D females, mating induced a steeper increase in reproductive output above the baseline virgin level than it did in mir-317D or control females (Figure 1A). There was also a nonsignificant trend for reproductive output to increase with time (F1,318 = 2.97, P = 0.086). Reproductive output increased significantly with time for all female genotypes mated to SP+ control males (F1,318 = 5.98, P = 0.015, Figure 1A). This pattern was also mirrored in the effect size calculation, where the standardized mean difference for the SP+ vs. SP0 treatment was larger for the 48-hr compared to the 24-hr treatment (Figure 1E).
Figure 1.
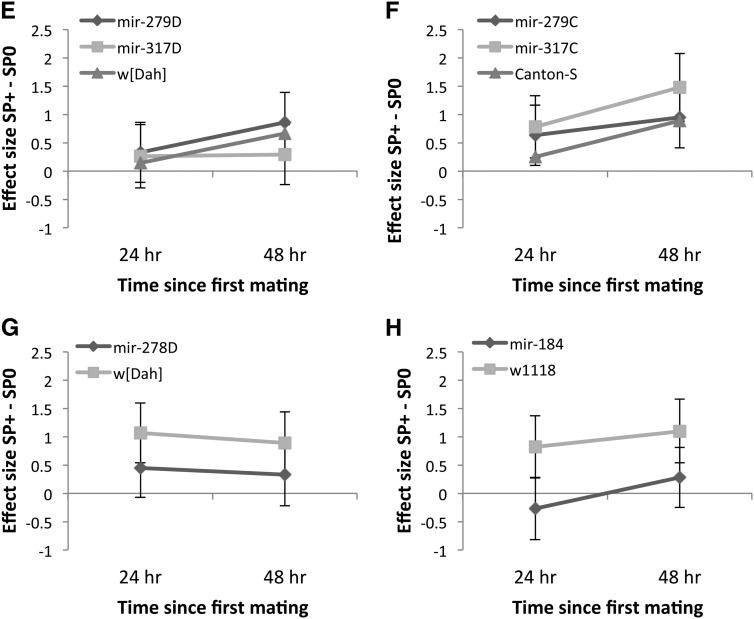
(A–D) Mean (±SE) reproductive output of miRNA mutant females and their controls, relative to the estimate for reproductive output of virgin females of the same genotype. Reproductive output was scored as offspring counts either 24 or 48 hr after a single mating to SP-lacking (SP0, bars with dark shading) or SP-transferring (SP+, bars with light shading) males. Females were either hypomorphic for mir-279 or mir-317 in two different genetic backgrounds (A) w[Dah] or (B) w[CS]. (C) Results for knockout mir-278 in the w[Dah] genetic background or (D) mir-184 in the w1118 background. (E–H) Effect sizes and 95% CI for reproductive output scored for the same female genotypes as in A–D 24 (diamonds with dark shading) or 48 hr (squares with light shading) after a single mating to either a SP+ or a SP0 male.
mir-279C and mir-317C hypomorphs:
Virgin female genotypes again differed significantly in fecundity (G2 = 2308.2, F2,177 = 73.85, P < 0.0001), but the significant increase in fecundity over time from 24 to 48 hr (G2 = 515.65, F1,176 = 33.00, P < 0.0001) was similar across female genotypes [G2 = 24.19, F2,175 = 0.78, P = 0.461 (dispersion parameter = 15.56)]. There were significant differences between female genotypes, however; all mated females, as expected, produced significantly more offspring when mated to a SP+ male. There was no evidence that the female genotypes showed altered reproductive output responses to SP. Overall, mir-317C females had higher reproductive output after receipt of SP compared to the other two groups (Figure 1B). The magnitude of this effect increased with time, and the slope increased more steeply for females mated to SP+ in comparison to SP0 males (Table 1A; Figure 1B). This was also reflected in the larger effect size for all female genotypes for the 48-hr treatment (Figure 1F).
Table 1. The results of an analysis of variance on the virgin baseline-corrected reproductive output.
| A Source | d.f. | MS | F | P |
|---|---|---|---|---|
| Female genotype (FG) | 2 | 1660 | 7.28 | <0.001 |
| Male genotype (MG) | 1 | 14,608 | 64.06 | <0.001 |
| Time period (time) | 1 | 5768 | 25.29 | <0.001 |
| FG × MG | 2 | 1322 | 5.80 | 0.003 |
| FG × time | 2 | 632 | 2.77 | 0.064 |
| MG × time | 1 | 6085 | 26.68 | <0.001 |
| Error | 319 | 228 | ||
| B Source | ||||
| Female genotype (FG) | 1 | 15,334 | 26.57 | <0.0001 |
| Male genotype (MG) | 1 | 14,404 | 24.96 | <0.001 |
| Time period (time) | 1 | 4218 | 7.31 | 0.007 |
| FG × MG | 1 | 13,878 | 24.05 | <0.0001 |
| FG × time | 1 | 2495 | 4.32 | 0.039 |
| MG × time | 1 | 3075 | 5.33 | 0.022 |
| FG × MG × time | 1 | 1872 | 3.24 | 0.073 |
| Error | 209 | 577 |
(A) mir-279C and mir-317C hypomorphic females in the w[CS] genetic background and (B) mir-184 knockout females, either 24 or 48 hr after single matings to SP-lacking (SP0) or control (SP+) males.
mir-278D knock out (ko):
Virgin mutant and control females had similar fecundity [G2 = 3.66, F1,118 = 0.38, P = 0.541 (dispersion parameter = 9.79)] and the fecundity of both groups increased significantly from 24 to 48 hr (G2 = 1601.8, F1,118 = 164.36, P < 0.0001, interaction = ns). Mutant and control females did not differ in their virgin baseline-corrected reproductive output (F1,223 = 0.53, P = 0.469), but females mated to SP0 males produced significantly fewer offspring (F1,223 = 21.92, P < 0.0001). There was, however, a nonsignificant tendency for control females to respond more strongly to receipt of SP (male × female genotype: F1,223 = 3.40, P = 0.066; Figure 1, C and G). This effect was stable over time (P = ns, as were all interactions).
mir-184 ko:
Virgin mir-184 ko females laid very few eggs [G2 = 906.61, F1,117 = 36.53, P < 0.0001 (Dispersion parameter = 18.58)] as expected based on previous work (Iovino et al. 2009), and the low level of fecundity did not increase over time, in contrast to the observation for control females (time: G2 = 340.44, F1,117 = 13.72, P = 0.0003; female genotype × time: G2 = 54.92, F1,116 = 2.96, P = 0.088; File S2). mir-184 ko females did not increase their reproductive output above the virgin level following mating to either SP+ or SP0 males (Figure 1D). This pattern was stable across 24–48 hr. Control females had a higher reproductive output following receipt of SP and this increased from 24 to 48 hr (Table 1B; Figure 1, D and H).
Overall the results showed no evidence that mir-279, mir-317 hypomorphs in either genetic background, or mir-184 ko females, showed altered reproductive output responses to SP in comparison to the appropriate controls. However, mir-278D ko females showed a nonsignificant tendency for weaker responses to SP in comparison to controls.
Sexual receptivity responses to SP:
mir-279D and mir-317D hypomorphs:
Virgin mutant and control females did not differ in their willingness to mate in their first mating (G22 = 1.67, P = 0.434) and this did not change over time (G21 = 2.84, P = 0.092; interaction = ns). There was a marginally nonsignificant difference in mating rate among the mated females (G22 = 5.33, P = 0.070) and the effect of time since the first mating on willingness to remate was similarly marginally nonsignificant (G21 = 3.15, P = 0.076). Receipt of SP significantly suppressed remating rate as expected (G21 = 84.26, P < 0.0001). There was a marginally nonsignificant interaction between female and male genotype (G22 = 5.68, P = 0.059, all other interactions = ns). This suggests that the effect of SP receipt on female sexual receptivity varied across the female genotypes tested. Across genotypes, females that received no SP remated at a rate similar to virgin females. However, SP was less efficient in suppressing remating in mir-317D females, whereas in mir-279D females, SP was equally effective in suppressing remating after 24 and 48 hr (Figure 2, A and E).
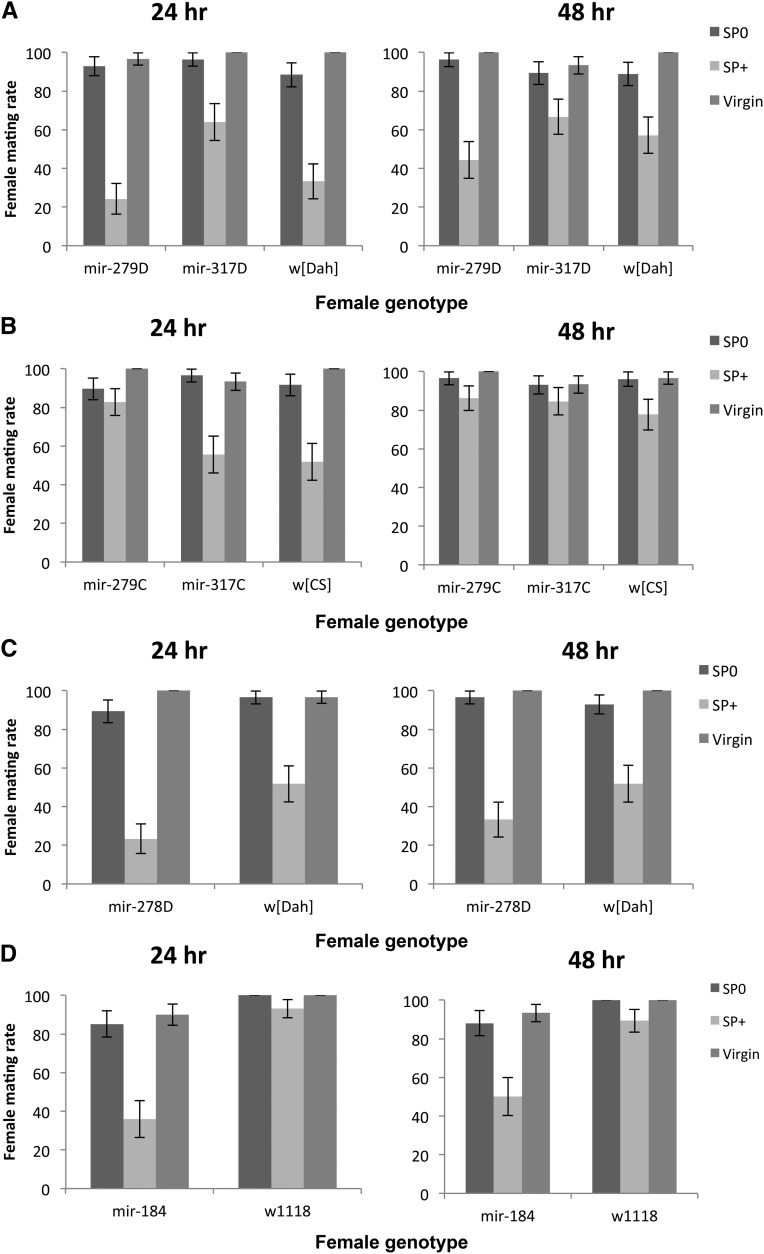
Figure 2.
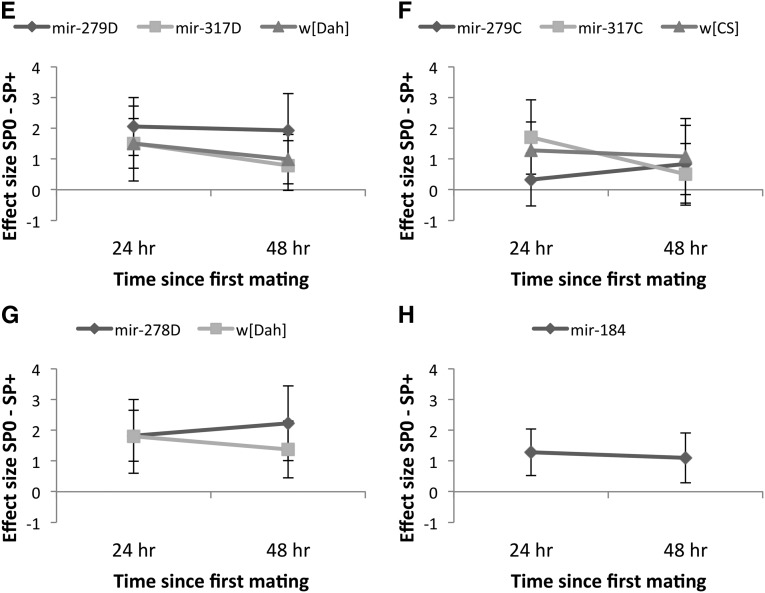
(A–D) Remating rate (number of females remating in 1 hr) at 24 or 48 hr following initial matings with either SP-transferring (SP+, bars with light shading) or SP-lacking (SP0, bars with dark shading) males. Simultaneously, the mating rate of age- and genotype-matched virgin females (bars with intermediate shading) was measured for comparison. (A and B) Remating in females hypomorphic for mir-279 and mir-317 in the w[Dah] or w[CS] genetic backgrounds, respectively. (C) Receptivity of mir-278 knockout females in the w[Dah] genetic background and (D) receptivity of mir-184 knockout females in the w1118 background. (E–H) Effect size (SP0 − SP+ treatment) and 95% CI for female remating rate for the different female genotypes mated first to a SP+ or SP0 male and provided with an opportunity to remate with a Dahomey wild-type male either 24 (diamonds with dark shading) or 48 hr (squares with light shading) after a first mating. Note that in the tests of the mir-184 knockout, (Figure 2H) all the control w1118 females remated; therefore, Cohen’s d could not be calculated. Hence only the effect size for mir-184 is shown.
mir-279C and mir-317C hypomorphs:
Virgin females differed significantly in their willingness to mate in their first mating (G22 = 6.12, P = 0.047). mir-317C females were slightly less willing to engage in mating, but this did not change significantly over time (G21 = 0.21, P = 0.650, interaction = ns). Fewer females first mated to a SP+ control male remated (G21 = 28.00, P < 0.0001), but this effect diminished with time (G21 = 7.93, P = 0.005; Figure 2B). The nonsignificant interaction term indicates that females of the different genotypes showed a similar response to receipt of SP across time (G22 = 4.43, P = 0.109, all interactions = ns). However, the standardized mean difference revealed that after 24 hr, SP was not efficient in suppressing remating in mir-279C mutant females (Figure 2F).
mir-278D ko:
Virgin females of this genotype did not differ in willingness to mate from virgin control w[Dah] females and this effect was constant across both time periods examined (all = ns). However, SP receipt suppressed female willingness to remate more strongly in mir-278D ko females than in controls (female genotype: G21 = 6.24, P = 0.012; male genotype: G21 = 85.29, P < 0.001; Figure 2, C and G), a pattern that was constant across time (G21 = 0.48, P = 0.49, all interactions = ns).
mir-184 ko:
Virgin mir-184 ko females were less willing to mate than controls (G21 = 7.05, P = 0.008) and this was constant across the two time points tested (G21 = 0.22, P = 0.639, interaction = ns). Receipt of SP was more efficient in suppressing remating in mir-184 ko mutant females in comparison to controls, which showed high receptivity following their first matings independent of SP receipt (female genotype: G21 = 41.11, P < 0.0001; male genotype: G21 = 28.89, P < 0.0001; Figure 2, D and H). This pattern did not alter from 24 to 48 hr (time: G21 = 0.46, P = 0.496, all interactions = ns).
Overall the results showed that females lacking miRNAs showed either significantly reduced or strengthened sexual receptivity responses to SP. There were also significant interactions with genetic background. mir-317D hypomorphs showed reduced receptivity responses to SP (Figure 2A) (but not when in the w[CS]genetic background; Figure 2B). Similarly, mir-279C hypomorphs also showed reduced SP receptivity responses (Figure 2B) (but not in the w[Dah] genetic background; Figure 2A). SP receipt decreased female sexual receptivity more strongly in mir-278D ko (Figure 2C) and in mir-184 ko females (Figure 2D) than in their respective controls.
Effect of mir-278D, mir-279D, and mir-317D mutations on female reproductive output, sexual receptivity, and survival responses to SP after continuous exposure to males
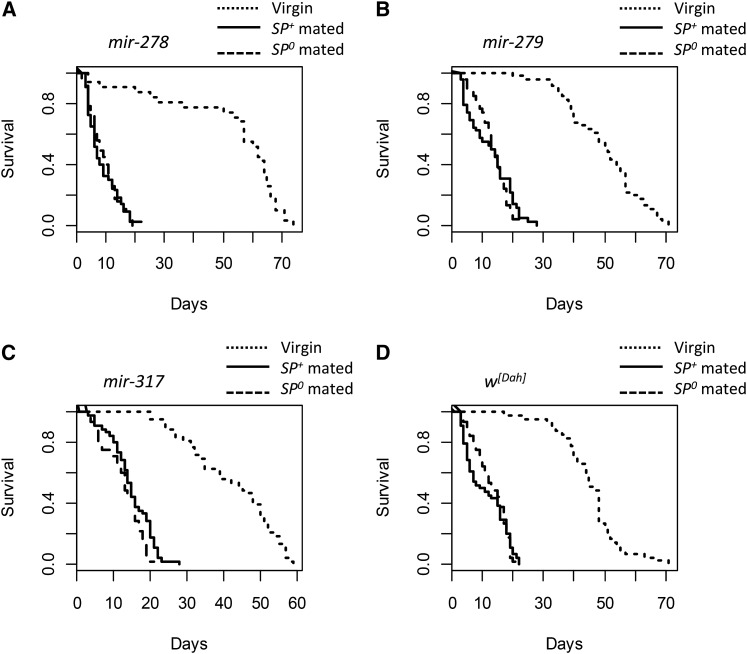
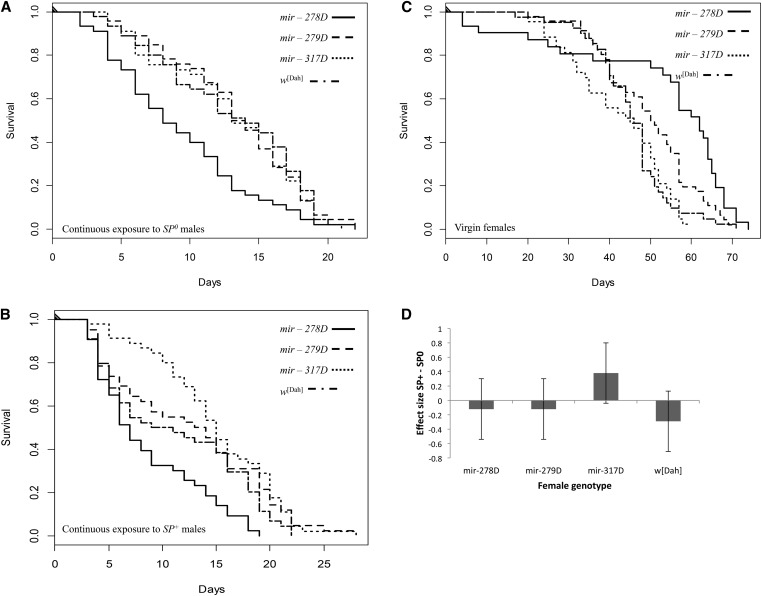
Virgin female genotypes differed significantly in survival (G23 = 23.36, P < 0.0001; Figure 3; Figure 4). Virgin mir-278D ko females had low early-life but higher late-life survival (Figure 3A). Virgin mir-317D females showed a steady decline in survival from day 20 onward, resulting in the lowest maximum lifespan (∼60 days compared to ∼70 days for the other female genotypes; Figure 3C). Continuous exposure to males significantly reduced female lifespan in comparison to virgin females in all groups (mating treatment: G22 = 623.48, P < 0.0001). However, this effect depended on female genotype (interaction: G26 = 38.53, P < 0.0001; female genotype: G23 = 11.92, P = 0.008; Table 2). Among mated females, female genotypes differed significantly in their responses to mating (G23 = 31.81, P < 0.0001; Figure 4, A and B), though SP receipt had no effect on female lifespan (male genotype: G21 = 0.12, P = 0.730; male × female genotype: G23 = 2.89, P = 0.408). Thus females differed in their susceptibility to male exposure, with mir-278D ko female lifespan (Figure 3A) being markedly reduced in comparison to the other genotypes tested (Figure 3; Figure 4); however, these effects were independent of SP receipt.
Figure 3.
Survival curves for the miRNA mutant females (backcrossed into the w[Dah] genetic background) kept as virgins or continuously exposed to SP-lacking (SP0) or SP-transferring (SP+) control males throughout their lifetimes. Shown are the survival curves for the (A) mir-278D knockouts, (B) mir-279D, (C) mir-317D hypomorphic mutant females, and (D) w[Dah] controls. For each panel, the virgins are shown by the dotted lines, females mated with SP+ males by the solid lines, and females mated with SP0 males by the dashed lines.
Figure 4.
Survival of all the females shown in Figure 3 redrawn to illustrate survival following exposure to either (A) SP0 or (B) SP+ males in comparison to the survival of (C) virgin females. (D) Mean survival effect sizes and 95% CI for the different female genotypes held continuously with SP+ vs. SP0 males.
Table 2. Lifespan data for (A) virgin females or (B) females held continuously with SP-lacking (SP0) or control (SP+) males.
| A Female genotype | Median lifespan | Upper confidence limit | Lower confidence limit | Mean lifespan | |||
|---|---|---|---|---|---|---|---|
| mir - 278D | 62.0 | 65 | 57 | 52.65 | |||
| mir - 279D | 50.5 | 57 | 46 | 49.35 | |||
| mir - 317D | 45.0 | 51 | 35 | 42.21 | |||
| w[Dah] | 46.0 | 48 | 44 | 45.37 |
| B |
SP0 male exposure |
SP+ male exposure |
||||||
|---|---|---|---|---|---|---|---|---|
| Female genotype | Median lifespan | Upper CL | Lower CL | Mean lifespan | Median lifespan | Upper CL | Lower CL | Mean lifespan |
| mir-278D | 8.0 | 12 | 6 | 9.22 | 7.0 | 11 | 6 | 8.60 |
| mir-279D | 13.5 | 16 | 13 | 13.28 | 13.5 | 16 | 8 | 12.57 |
| mir-317D | 13.0 | 16 | 12 | 12.98 | 15.0 | 19 | 13 | 15.00 |
| w[Dah] | 14.0 | 17 | 11 | 13.00 | 10.0 | 16 | 6 | 11.27 |
Lifespan data for females hypomorphic for mir-279D and mir-317D are shown together with mir-278D knockout females, all backcrossed into the w[Dah] genetic background.
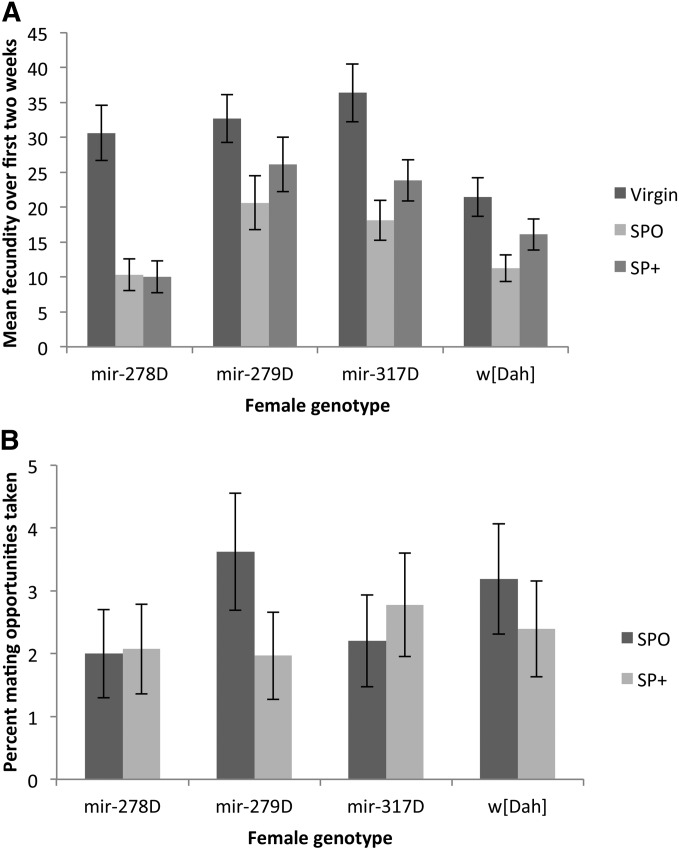
Females significantly differed in the number of eggs laid within the 6-hr tests over the first 2 weeks of the assays [female genotype: G2 = 278.32, F3,249 = 9.42, P < 0.0001 (dispersion parameter = 9.64)]. While virgin females of all four genotypes laid more eggs than mated females, females of all genotypes mated to SP+ males laid significantly more eggs than those held with SP0 males (mating treatment: G2 = 472.44, F2,248 = 23.97, P < 0.0001; mating treatment × female genotype: G2 = 110.11, F6,246 = 1.90, P = 0.081; Figure 5A). As in the single mating tests, there was therefore no evidence for altered fecundity responses to SP in the miRNA-lacking females. The results remained the same even when virgin data were excluded and data for the mated females only over the first 2 weeks of the experiment were analyzed.
Figure 5.
(A) Mean (± SE) reproductive output of miRNA mutant females shown in Figure 3. The reproductive output of 21 randomly sampled females per treatment was recorded for a period of 6 hr every week over the first 2 weeks of the survival experiment. (B) The percentage of mating opportunities taken by the females in A when continuously held with males lacking SP (SP0, bars with dark shading) or control males (SP+, bars with light shading).
miRNA-mutant females in the Dahomey genetic background differed significantly in the number of mating opportunities accepted (G23 = 14.05, P = 0.003; Figure 5B). However, this effect was driven by the mir-278D ko females accepting fewer matings, and exclusion of this line from the analysis rendered the female effect nonsignificant (G22 = 2.09, P = 0.351). However, independent of the inclusion of mir-278D, mating rate differed significantly upon male genotype (model including mir-278D: G21 = 7.22, P = 0.007), and the number of mating opportunities taken was also dependent upon an interaction between male × female genotype (model including mir-278D: G23 = 23.36, P < 0.0001; Figure 5B). While SP transfer significantly reduced mating rate in the w[Dah] control as well as in mir-279D mutant females, mir-278D ko females were unaffected and in general showed low acceptance of matings. In contrast, mir-317D hypomorph females showed a reversal from the expected mating pattern and mated more frequently when held with SP+ control as compared to SP-lacking males. Thus, while mir-278D ko females showed the lowest mating rate they showed the greatest reduction in lifespan due to continuous male exposure. In contrast while receipt of SP was less effective in suppressing mating rate in mir-317D females, these females tended to survive longer when continuously held with SP+ control in comparison to SP0 males (Figure 3C; Figure 4B).
Overall the results of the continual exposure experiment showed that female lifespan varied widely upon exposure to males, but this effect was independent of SP receipt. Consistent with the single mating tests we observed altered receptivity responses, measured as mating frequency, in mir-278D ko and mir-317D hypomorph females.
The results of all the phenotypic tests described above are summarized in Table 3.
Table 3. Summary of the single mating and continual exposure assays.
| MicroRNA | Single mating assay |
Continual exposure assay |
|||
|---|---|---|---|---|---|
| Reproductive output | Receptivity | Survival | Reproductive output | Receptivity | |
| mir-184 | Reduced output with no response to receipt of SP | SP more effective in suppressing remating | Not tested | Not tested | Not tested |
| mir-278D | No altered response to SP, tendency for a weaker response (P = 0.066) than in control | SP more effective in suppressing remating | Strong reduction in survival with continual male exposure, independent of SP receipt | No altered response to SP | Unresponsive to SP receipt, low number of matings overall |
| mir-279D | No altered response to SP, though a tendency for higher output following SP receipt than in controls | No altered response to SP | Reduced survival with continual male exposure, independent of SP receipt | No altered response to SP | No altered response to SP |
| mir-317D | No altered response to SP | SP less efficient in suppressing remating | Reduced survival with continual male exposure, independent of SP receipt | No altered response to SP | Reversal of pattern, more matings when held with SP+ males |
| mir-279C | No altered response to SP | No altered response to SP, but SP less efficient in suppressing remating after 24 hr | Not tested | Not tested | Not tested |
| mir-317C | No altered response to SP, though a tendency for higher output following SP receipt than in controls | No altered response to SP | Not tested | Not tested | Not tested |
Females either lacked the microRNAs of interest (mir-184 and mir-278D knock outs) or had reduced mir expression (mir-279 and mir-317 hypomorphs) in the wild-type Canton-S (C) or Dahomey (D) genetic background. Females were either mated singly to, or held continuously with, SP-lacking (SP0) or SP-transferring males (SP+). Female reproductive output and receptivity were recorded in both assays, and female survival was measured in the continual exposure assay.
Discussion
Overall, we found significant but subtle changes in miRNA-lacking females in response to SP receipt. There was little evidence that females lacking miRNAs exhibited grossly altered SP responses, such as SP-like phenotypes in the absence of SP. Instead, the results revealed a pattern of modulations to the strength of SP responses (Table 3). Females that had reduced or ablated expression for the four candidate miRNAs tested showed reduced or strengthened sexual receptivity responses following SP receipt, in comparison to relevant controls. There was little evidence, however, of consistent SP effects on reproductive output or survival. These key female responses to receipt of SP—i.e., egg output and receptivity—are mediated through different neurons and are the result of the activation of different regulatory pathways (Haussmann et al. 2013). The four miRNAs tested may regulate different facets of known SP responses within these distinct pathways. Our data suggest that the behavioral responses to SP, specifically, may be subject to post-transcriptional regulation by miRNAs. Below we describe the known and putative functions of the different candidate miRNAs tested and highlight how they could influence female SP-mediated phenotypic responses. However, we note that a potentially complex interplay between several miRNAs might be required to regulate biological processes. As we tested only a subset of the microRNAs differentially expressed in response to SP receipt, there is the potential for others to have stronger effects than the candidates tested here or to interact with the candidate microRNAs tested to exert even stronger combined phenotypic responses in females.
Roles and functions of the miRNAs tested
The candidate miRNAs tested mainly modulated behavioral processes (see details below), which is consistent with their signatures of differential expression in response to receipt of SP, with miR-279, miR-317, and miR-184 being down-regulated in the head + thorax. miR-278 was up-regulated in the abdomen, while miR-279 was down-regulated in both body parts, perhaps suggesting some additional reproductive functions (Table S1; Figure S1; Figure S2; and see below).
Turning to the known functions of the miRNAs tested, little is yet known about the function of miR-317, apart from its involvement in brain development and, along with miR-279, in locomotion (Yamamoto et al. 2008). miR-279, in contrast, has been relatively well studied. A role for miR-279 in regulating neuron development has been shown and nerfin-1 verified as a target (Cayirlioglu et al. 2008). Nerfin-1 contains several predicted seed sites for different miRNAs, and multiple miRNAs are needed to act cooperatively to regulate its spatial and temporal expression (Kuzin et al. 2007). In adults, miR-279 is involved in maintaining circadian rhythms and acts downstream of period neurons. As SP is known to inhibit female day-time rest and activity patterns (Isaac et al. 2010), this effect might be mediated through decreased miR-279 expression and subsequent modulation of period neuron signals to the downstream cascade. We found an effect of miR-279 on remating rate, suggesting that the effects of miR-279 on neurological function do influence fly mating behavior. However, the pattern was strongly dependent on genetic background (no effect in Canton-S and large effect in Dahomey genetic background). In addition to regulating neuronal processes, miR-279 is also active in the ovary. Here miR-279 is involved in a regulatory circuit to regulate expression of the signal transducer and activator of transcription (STAT) in follicle cells (Yoon et al. 2011). A subset of these cells change cell fate to become migratory border cells and form the micropyle (Montell 2003). Decreased miR-279 results in abnormal invasion of follicle cells (Yoon et al. 2011). However we show here that this disruption of oogenesis seems not to result in reduced fertilization success or egg-to-adult survival in mutant mir-279 females (Figure S3, A and B).
miR-278 is reported to control energy balance, principally by regulating insulin responsiveness (Teleman and Cohen 2006). Through repression of translation of the gene expanded, miR-278 regulates insulin sensitivity and flies lacking miR-278 are lean and store less triglyceride (Teleman and Cohen 2006). miR-278 is also involved in regulating germline stem cell division by regulating the cyclin-dependent kinase inhibitor dacapo (Yu et al. 2009). Thus miR-278 has multiple functions, potentially including mediating links between nutrient availability and egg development. As SP receipt increases female feeding activity (Carvalho et al. 2006), miR-278 could be an important regulatory element between nutrient uptake, storage, and mobilization toward allocation into reproductive output. The availability of nutrition significantly affects the effect of SP on female lifespan (Fricke et al. 2010) and we found that mir-278D ko females were extremely sensitive to continuous male exposure, suffering greatly reduced lifespans (Figure 3A). SP up-regulated miR-278 expression in the abdomen, which could potentially initiate enhanced fat accumulation in the fat body to direct toward increased egg production. While SP receipt still resulted in increased reproductive output in mir-278 knockout females, this increase was modest and smaller than in the control females (Figure 1, C and G). Elevated miR-278 expression might therefore allow males to reap the full reproductive benefits of SP transfer (Fricke et al. 2009, 2013).
miR-184 functions in the ovary and regulates processes during oogenesis and early embryogenesis (Iovino et al. 2009). Loss of miR-184 results in complete loss of egg production. These defects are so severe that we could not measure any effect of SP receipt on reproductive output in mir-184 knockout females. However, our initial screen showed postmating expression changes in miR-184 after SP receipt in the head + thorax only, indicating that postmating regulation of miR-184 expression may alter functions in the brain/nervous system rather than the ovary. During embryogenesis, miR-184 is expressed in the developing central nervous system and in the head and eye disc in larvae (Li et al. 2011), consistent with accumulating evidence that miR-184 is involved in regulating neuronal processes (Greenberg et al. 2010; Liu et al. 2012). For example, miR-184 is highly expressed in honey bee Apis mellifera heads (Liu et al. 2012) and is part of the regulatory processes that confer the behavioral switch from nursing to foraging (Greenberg et al. 2010). In mouse, miR-184 also regulates the balance of adult neural stem cell proliferation vs. differentiation (Liu et al. 2010). Collectively these data suggest a potential role for miR-184 in neurological processes.
miRNAs as buffers in the context of sexual conflict
We initiated in this study an investigation of the extent to which candidate microRNAs can regulate postmating female physiological processes in response to a male ejaculate signal subject to sexual conflict. We were interested in whether such processes have been coopted by males to potentially manipulate female physiological and behavioral responses for their own benefits. Alternatively, receipt of SP in females could result in alterations to miRNA expression to dampen down potentially deleterious oscillations in mRNAs in females (Kim et al. 2013). We expected that the reduced or absent levels of miR-279, miR-317, and miR-184 in the mutant females would, to some extent, mimic the down-regulation that we had previously observed following receipt of SP. Our observation of modest alterations to SP responses in the miRNA mutant females suggests that the miRNAs may function to buffer noise or stabilize responses to mating. Further work is now required to ascertain which sex is most sensitive to the relative expression level of the miRNAs involved.
miRNAs can have striking effects on the phenotype in developing individuals during ontogeny, but in adults miRNAs may have more subtle effects. The degree of protein repression by miRNAs can be modest, despite the observation that single microRNAs can suppress the translation of hundreds of proteins (Baek et al. 2008; Selbach et al. 2008). Thus it has been suggested that miRNAs might primarily perform modulatory functions, provide stability, and minimize expression noise in biological systems (Herranz and Cohen 2010). Little is known about how miRNAs confer robustness in systems characterized by gradual responses to external signals. miRNAs might act as “switches,” by effectively repressing protein expression below a threshold level of the target mRNA, but might also act to fine tune gene expression when close to the threshold (Mukherji et al. 2011). This type of phenomenon could explain the results we obtained in the two hypomorph mutant strains, mir-279 and mir-317, when expressed in two different genetic backgrounds. We found a reversal in remating behavior in response to SP in mir-279 mutant females, and for both mir-279 and mir-317 mutant females the effect of SP on reproductive output was stronger/more pronounced in one genetic background than the other (Figure 1, E and F; Figure 2, E and F). This suggests the existence of epistatic effects due to different expression of either the level of target mRNA or of the regulators of expression of the miRNAs in the two genetic backgrounds. This could alter the balance of miRNA to target mRNA and, depending on closeness to threshold levels, alter the function of the miRNA from an effective suppressor to a modulator.
Our results provide an initial investigation of some of the miRNA loci potentially involved in the regulation of postmating changes in the female transcriptome. We focused on the effects of a particular male sexual antagonistic signal—the sex peptide—as this seminal fluid protein elicits diverse female phenotypic responses and is a major contributor to the cost of mating in females. We showed that post-transcriptional regulation by miRNAs is an element of the molecular cross-talk between the sexes and mainly regulates behavioral responses in the specific miRNAs tested. Mutant female responses to receipt of SP were altered in a relatively modest manner. We suggest that females use miRNA regulators to buffer their physiological processes against transcriptional noise introduced by mating and potentially to minimize the negative fitness effects of male manipulation.
It is important to note that ours is a study of a small number of candidate miRNAs that change expression in response to receipt of a single ejaculate protein, SP. It is likely that other microRNAs are involved in mediating responses to SP and to Sfps that interact with SP. For example, even in this study there were candidate miRNAs that were not tested. In addition, the ability of SP to participate in sustained postmating responses requires the actions of a proteolytic cascade of different Sfps (Avila et al. 2010; LaFlamme et al. 2012). Therefore the actions of SP are interdependent with those of multiple other Sfps and the regulation of such networks is likely to involve as yet unknown interacting regulatory mechanisms. There are also ∼130 Sfps (Findlay et al. 2008; Ayroles et al. 2011) with diverse functions in different facets of postmating responses (e.g., Chapman 2001; Gillot 2003; Ram and Wolfner 2007a; Wolfner 2009; Avila et al. 2011). For example, postmating responses such as egg production, receptivity to remating, and sperm storage require multiple Sfps in addition to SP (e.g., Ram and Wolfner 2007b; Avila et al. 2011). It should also be noted that many Sfps have as yet unknown functions. Future investigations of the interactions between different Sfps and their regulators in maintaining reproductive functions, buffering against perturbation, and in balancing the interests of males and females are likely to yield important results.
Supplementary Material
Acknowledgments
C.F. was supported by an Emmy-Noether fellowship of the Deutsche Forschungsgemeinschaft. We thank the Biotechnology and Biological Sciences Research Council for funding (research grant to T.C.) and Amanda Bretman and Dominic Edward for help with the mating experiments. We thank two anonymous reviewers for their constructive and useful comments.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.167320/-/DC1.
Communicating editor: S. E. Bickel
Literature Cited
- Arnqvist G., Rowe L., 2005. Sexual Conflict. Princeton University Press, Princeton, NJ. [Google Scholar]
- Asgari S., 2013. MicroRNA functions in insects. Insect Biochem. Mol. Biol. 43: 388–397. [DOI] [PubMed] [Google Scholar]
- Avila F. W., Ravi Ram K., Bloch Qazi M. C., Wolfner M. F., 2010. Sex peptide is required for the efficient release of stored sperm in mated Drosophila females. Genetics 186: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila F. W., Sirot L. K., LaFlamme B. A., Rubinstein D., Wolfner M. F., 2011. Insect seminal fluid proteins: identification and function. Annu. Rev. Entomol. 56: 21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., LaFlamme B. A., Stone E. A., Wolfner M. F., Mackay T. F. C., 2011. Functional genome annotation of Drosophila seminal fluid proteins using transcriptional genetic networks. Genet. Res. 93: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Villen J., Shin C., Camargo F. D., Gygi S. P., et al. , 2008. The impact of microRNAs on protein output. Nature 455: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton S. J., Piper M. D. W., Ikeya T., Bass T. M., Jacobson J., et al. , 2005. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. USA 102: 3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho G. B., Kapahi P., Anderson D. J., Benzer S., 2006. Allocrine modulation of feeding behavior by the sex peptide of Drosophila. Curr. Biol. 16: 692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu P., Grunwald Kadow I., Zhan X., Okamura K., Suh G. S. B., et al. , 2008. Hybrid neurons in a MicroRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science 319: 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T., 2001. Seminal fluid-mediated fitness traits in Drosophila. Heredity 87: 511–521. [DOI] [PubMed] [Google Scholar]
- Chapman T., Arnqvist G., Bangham J., Rowe L., 2003a Sexual conflict. Trends Ecol. Evol. 18: 41–47. [Google Scholar]
- Chapman T., Bangham J., Vinti G., Seifried B., Lung O., et al. , 2003b The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc. Natl. Acad. Sci. USA 100: 9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A. T., 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720. [DOI] [PubMed] [Google Scholar]
- Cognigni P., Bailey A. P., Miguel-Aliaga I., 2011. Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell Metab. 13: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley M. J., 2007. The R Book. John Wiley & Sons, Chichester, UK. [Google Scholar]
- Domanitskaya E. V., Liu H., Chen S., Kubli E., 2007. The hydroxyproline motif of male sex peptide elicits the innate immune response in Drosophila females. FEBS J. 274: 5659–5668. [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N., Sonenberg N., 2008. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 9: 102–114. [DOI] [PubMed] [Google Scholar]
- Findlay G. D., Yi X. H., MacCoss M. J., Swanson W. J., 2008. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 6: 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke C., Wigby S., Hobbs R., Chapman T., 2009. The benefits of male ejaculate sex peptide transfer in Drosophila melanogaster. J. Evol. Biol. 22: 275–286. [DOI] [PubMed] [Google Scholar]
- Fricke C., Bretman A., Chapman T., 2010. Female nutritional status determines the magnitude and sign of responses to a male ejaculate signal in Drosophila melanogaster. J. Evol. Biol. 23: 157–165. [DOI] [PubMed] [Google Scholar]
- Fricke C., Green D., Mills W. E., Chapman T., 2013. Age-dependent female responses to a male ejaculate signal alter demographic opportunities for selection. Proc. Biol. Sci. 280: 20130428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillot C., 2003. Male accessory gland secretions: modulators of female reproductive physiology and behavior. Annu. Rev. Entomol. 48: 163–184. [DOI] [PubMed] [Google Scholar]
- Gioti A., Wigby S., Wertheim B., Schuster E., Martinez P., et al. , 2012. Sex peptide of Drosophila melanogaster males is a global regulator of reproductive processes in females. Proc. Biol. Sci. 279: 4423–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg J. K., Xia J., Zhou X., Thatcher S. R., Gu X., et al. , 2010. Behavioral plasticity in honey bees is associated with differences in brain microRNA transcriptome. Genes Brain Behav. 11: 660–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussmann I. U., Hemani Y., Wijesekera T., Dauwalder B., Soller M., 2013. Multiple pathways mediate the sex-peptide-regulated switch in female Drosophila reproductive behaviours. Proc. Biol. Sci. 280: 20131938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon L. A., Wolfner M. F., 1995. A Drosophila seminal fluid protein, Acp26Aa, stimulates egg-laying in females for 1 day after mating. Proc. Natl. Acad. Sci. USA 92: 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H., Cohen S. M., 2010. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 24: 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R., Gentleman R., 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5: 299–314. [Google Scholar]
- Innocenti P., Morrow E. H., 2009. Immnunogenic males: a genome-wide analysis of reproduction and the cost of mating in Drosophila melanogaster females. J. Evol. Biol. 22: 964–973. [DOI] [PubMed] [Google Scholar]
- Iovino N., Pane A., Gaul U., 2009. miR-184 has multiple roles in Drosophila female germline development. Dev. Cell 17: 123–133. [DOI] [PubMed] [Google Scholar]
- Isaac R. E., Li C., Leedale A. E., Shirras A. D., 2010. Drosophila male sex peptide inhibits siesta sleep and promotes locomotor activity in the post-mated female. Proc. Biol. Sci. 277: 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Grün D., Van Oudenaarden A., 2013. Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nat. Genet. 45: 1337–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzin A., Kundu M., Brody T., Odenwald W. F., 2007. The Drosophila nerfin-1 mRNA requires multiple microRNAs to regulate its spatial and temporal translation dynamics in the developing nervous system. Dev. Biol. 310: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFlamme B. A., Ravi Ram K., Wolfner M. F., 2012. The Drosophila melanogaster seminal fluid protease “seminase” regulates proteolytic and post-mating reproductive processes. PLoS Genet. 8: e1002435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak M. K. N., Begun D. J., 2004. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome 47: 900–910. [DOI] [PubMed] [Google Scholar]
- Li P., Peng J., Hu J., Xu Z., Xie W., et al. , 2011. Localized expression pattern of miR-184 in Drosophila. Mol. Biol. Rep. 38: 355–358. [DOI] [PubMed] [Google Scholar]
- Liu C., Teng Z.-Q., Santistevan N. J., Szulwach K. E., Guo W., et al. , 2010. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 6: 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Peng W., Li Z., Li W., Li L., et al. , 2012. Next-generation small RNA sequencing for microRNAs profiling in Apis mellifera: comparison between nurses and foragers. Insect Mol. Biol. 21: 297–303. [DOI] [PubMed] [Google Scholar]
- Liu H., Kubli E., 2003. Sex peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100: 9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Shen Y., Wu Q., He B., Shi S., et al. , 2008. The birth and death of microRNA genes in Drosophila. Nat. Genet. 40: 351–355. [DOI] [PubMed] [Google Scholar]
- Lucas K., Raikhel A. S., 2013. Insect MicroRNAs: biogenesis, expression profiling and biological functions. Insect Biochem. Mol. Biol. 43: 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack P. D., Kapelnikov A., Heifetz Y., Bender M., 2006. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103: 10358–10363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A., Hui J. H. L., Ronshaugen M., Griffiths-Jones S., 2010. Functional shifts in insect microRNA evolution. Genome Biol. Evol. 2: 686–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw L. A., Gibson G., Clark A. G., Wolfner M. F., 2004. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr. Biol. 14: 1509–1514. [DOI] [PubMed] [Google Scholar]
- McGraw L. A., Clark A. G., Wolfner M. F., 2008. Post-mating gene expression profiles of female Drosophila melanogaster in response to time and to four male accessory gland proteins. Genetics 179: 1395–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell D. J., 2003. Border-cell migration: the race is on. Nat. Rev. Mol. Cell Biol. 4: 13–24. [DOI] [PubMed] [Google Scholar]
- Mueller J. L., Page J. L., Wolfner M. F., 2007. An ectopic expression screen reveals the protective and toxic effects of Drosophila seminal fluid proteins. Genetics 175: 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S., Ebert M. S., Zheng G. X. Y., Tsang J. S., Sharp P. A., et al. , 2011. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 43: 854–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubaum D. M., Wolfner M. F., 1999. Mated female Drosophila melanogaster females require a seminal fluid protein, Acp 36DE, to store sperm efficiently. Genetics 153: 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall G. S., Hamilton A. J., 2008. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 3: 1077–1084. [DOI] [PubMed] [Google Scholar]
- Park J. K., Liu X., Strauss T. J., McKearin D. M., Liu Q., 2007. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 17: 533–538. [DOI] [PubMed] [Google Scholar]
- Peng J., Zipperlen P., Kubli E., 2005. Drosophila sex-peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Curr. Biol. 15: 1690–1694. [DOI] [PubMed] [Google Scholar]
- Price N., Cartwright R. A., Sabath N., Graur D., Azevedo R. B. R., 2011. Neutral evolution of robustness in Drosophila microRNA precursors. Mol. Biol. Evol. 28: 2115–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram K. R., Wolfner M. F., 2007a. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction Integr. Comp. Biol. 47: 427–445. [DOI] [PubMed] [Google Scholar]
- Ram K. R., Wolfner M. F., 2007b. Sustained post-mating response in Drosophila melanogaster requires multiple seminal fluid proteins. PLoS Genet. 3: 2428–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaval C., Pavlou H. J., Doman A. J., Chan Y.-B., Kravitz E. A., et al. , 2012. Neural circuitry underlying Drosophila female postmating behavioral responses. Curr. Biol. 22: 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C., Dickson B. J., 2010. Sex peptide receptor and neuronal TOR/S6K signaling modulate nutrient balancing in Drosophila. Curr. Biol. 20: 1000–1005. [DOI] [PubMed] [Google Scholar]
- Rice W. R., 1998. Intergenomic conflict, interlocus antagonistic coevolution, and the evolution of reproductive isolation, pp. 261–270 in Endless Forms: Species and Speciation, edited by Howard D. J., Berlocher S. H. Oxford University Press, Oxford. [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., et al. , 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63. [DOI] [PubMed] [Google Scholar]
- Soller M., Haussmann I. U., Hollmann M., Choffat Y., White K., et al. , 2006. Sex-peptide-regulated female sexual behavior requires a subset of ascending ventral nerve cord neurons. Curr. Biol. 16: 1771–1782. [DOI] [PubMed] [Google Scholar]
- Sorefan K., Pais H., Hall A. E., Kozomara A., Griffiths-Jones S., et al. , 2012. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman A. A., Cohen S. M., 2006. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 20: 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tram U., Wolfner M. F., 1999. Male seminal fluid proteins are essential for sperm storage in Drosophila melanogaster. Genetics 153: 837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E. L., Jaszczyszyn Y., Thermes C., 2014. Library preparation methods for next-generation sequencing: tone down the bias. Exp. Cell Res. 322: 12–20. [DOI] [PubMed] [Google Scholar]
- Wigby S., Chapman T., 2005. Sex peptide causes mating costs in female Drosophila melanogaster. Curr. Biol. 15: 316–321. [DOI] [PubMed] [Google Scholar]
- Wolfner M. F., 2009. Battle and ballet: molecular interactions between the sexes in Drosophila. J. Hered. 100: 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Zwarts L., Callaerts P., Norga K., Mackay T. F. C., et al. , 2008. Neurogenetic networks for startle-induced locomotion in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 105: 12393–12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yapici N., Kim Y.-J., Ribeiro C., Dickson B. J., 2008. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature 451: 33–37. [DOI] [PubMed] [Google Scholar]
- Yoon W. H., Meinhardt H., Montell D. J., 2011. miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nat. Cell Biol. 13: 1062–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.-Y., Reynolds S. H., Hatfield S. D., Shcherbata H. R., Fischer K. A., et al. , 2009. Dicer-1-dependent Dacapo suppression acts downstream of Insulin receptor in regulating cell division of Drosophila germline stem cells. Development 136: 1497–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.