A cytoplasmic enzyme known to synthesise chlorogenic acid in tomato has a second activity synthesising dicaffeoyl quinates at low pH and high chlorogenic acid concentrations in the vacuole.
Abstract
Tomato (Solanum lycopersicum), like other Solanaceous species, accumulates high levels of antioxidant caffeoylquinic acids, which are strong bioactive molecules and protect plants against biotic and abiotic stresses. Among these compounds, the monocaffeoylquinic acids (e.g. chlorogenic acid [CGA]) and the dicaffeoylquinic acids (diCQAs) have been found to possess marked antioxidative properties. Thus, they are of therapeutic interest both as phytonutrients in foods and as pharmaceuticals. Strategies to increase diCQA content in plants have been hampered by the modest understanding of their biosynthesis and whether the same pathway exists in different plant species. Incubation of CGA with crude extracts of tomato fruits led to the formation of two new products, which were identified by liquid chromatography-mass spectrometry as diCQAs. This chlorogenate:chlorogenate transferase activity was partially purified from ripe fruit. The final protein fraction resulted in 388-fold enrichment of activity and was subjected to trypsin digestion and mass spectrometric sequencing: a hydroxycinnamoyl-Coenzyme A:quinate hydroxycinnamoyl transferase (HQT) was selected as a candidate protein. Assay of recombinant HQT protein expressed in Escherichia coli confirmed its ability to synthesize diCQAs in vitro. This second activity (chlorogenate:chlorogenate transferase) of HQT had a low pH optimum and a high Km for its substrate, CGA. High concentrations of CGA and relatively low pH occur in the vacuoles of plant cells. Transient assays demonstrated that tomato HQT localizes to the vacuole as well as to the cytoplasm of plant cells, supporting the idea that in this species, the enzyme catalyzes different reactions in two subcellular compartments.
The importance of plant-based foods in preventing or reducing the risk of chronic disease has been widely demonstrated (Martin et al., 2011, 2013). In addition to vitamins, a large number of other nutrients in plant-based foods promote health and reduce the risk of chronic diseases; these are often referred to as phytonutrients. The presence of phytonutrients in fruit and vegetables is of significant nutritional and therapeutic importance, as many have been found to possess strong antioxidant activity (Rice-Evans et al., 1997). Phenolics are the most widespread dietary antioxidants and caffeoylquinic acids, such as chlorogenic acid (CGA), dicaffeoylquinic acids (diCQAs), and tricaffeoylquinic acids (triCQAs), play important roles in promoting health (Clifford, 1999; Niggeweg et al., 2004). CGA limits low density lipid oxidation (Meyer et al., 1998), diCQAs possess antihepatotoxic activity (Choi et al., 2005), and triCQAs reduce the blood Glc levels of diabetic rats (Islam, 2006). diCQA derivatives have been shown to protect humans from various kinds of diseases; diCQAs suppress melanogenesis effectively (Kaul and Khanduja, 1998), show anti-inflammatory activity in vitro (Peluso et al., 1995), and exhibit a selective inhibition of HIV replication (McDougall et al., 1998). The physiological effects of caffeoylquinic acid derivatives with multiple caffeoyl groups are generally greater than those of monocaffeoylquinic acids, perhaps because the antioxidant activity is largely determined by the number of hydroxyl groups present on the aromatic rings (Wang et al., 2003; Islam, 2006). Furthermore, both diCQAs and triCQAs may function as inhibitors of the activity of HIV integrase, which catalyzes the insertion of viral DNA into the genome of host cells (McDougall et al., 1998; Slanina et al., 2001; Gu et al., 2007).
CGA is the major soluble phenolic in Solanaceous crops (Clifford, 1999) and the major antioxidant in the average U.S. diet (Luo et al., 2008), while different isomers of diCQAs have been identified in many crops such as coffee (Coffea canephora), globe artichoke (Cynara cardunculus), tomato (Solanum lycopersicum), lettuce (Lactuca sativa), and sweet potato (Ipomoea batatas; Clifford, 1999; Islam, 2006; Moco et al., 2006, 2007; Moglia et al., 2008). In tomato, CGA accounts for 75% and 35% of the total phenolics in mature green and ripe fruit, respectively, amounting to 2 to 40 mg 100 g–1 dry weight (DW), although levels decline after ripening and during postharvest storage (Slimestad and Verheul, 2009). diCQAs and triCQAs also accumulate in tomato fruit (diCQAs, approximately 2 mg 100 g–1 DW; and triCQAs, 1–2 mg 100 g–1 DW; Chanforan et al., 2012).
Three pathways (Villegas and Kojima, 1986; Hoffmann et al., 2003; Niggeweg et al., 2004) have been proposed for the synthesis of CGA: (1) the direct pathway involving caffeoyl-CoA transesterification with quinic acid by hydroxycinnamoyl-Coenzyme A:quinate hydroxycinnamoyl transferase (HQT; Niggeweg et al., 2004; Comino et al., 2009; Menin et al., 2010; Sonnante et al., 2010); (2) the route by which p-coumaroyl-CoA is first transesterified with quinic acid via hydroxycinnamoyl-Coenzyme A transferase (HCT) acyltransferase (Hoffmann et al., 2003; Comino et al., 2007), followed by the hydroxylation of p-coumaroyl quinate to 5-caffeoylquinic acid, catalyzed by C3′H (p-coumaroyl-3-hydroxylase; Schoch et al., 2001; Mahesh et al., 2007; Moglia et al., 2009); and (3) the use of caffeoyl-glucoside as the acyl-donor (Villegas and Kojima, 1986). In tomato, the synthesis of CGA involves transesterification of caffeoyl-CoA with quinic acid by HQT (Niggeweg et al., 2004).
To date, it is not clear whether diCQAs are derived directly from the monocaffeoylquinic acids (such as CGA) through a second acyltransferase reaction involving an acyl-CoA or not, although their structural similarity provides good a priori evidence supporting this hypothesis. Recently the in vitro synthesis of 3,5-diCQA from CGA and CoA by HCT from coffee has been reported (Lallemand et al., 2012). By contrast, in sweet potato, an enzyme that catalyzes the transfer of the caffeoyl moiety of CGA to another molecule of CGA, leading to the synthesis of isochlorogenate (3,5-di-O-caffeoylquinate), has been described, but the corresponding gene has not been identified (Villegas and Kojima, 1986).
We report a chlorogenate:chlorogenate transferase (CCT) activity leading to the synthesis of diCQAs in tomato fruits and describe how alternative catalysis, by a single enzyme, leads to the production of both CGA and diCQA in different cellular compartments.
RESULTS
Identification of an Enzyme Activity Involved in diCQA Synthesis in Tomato
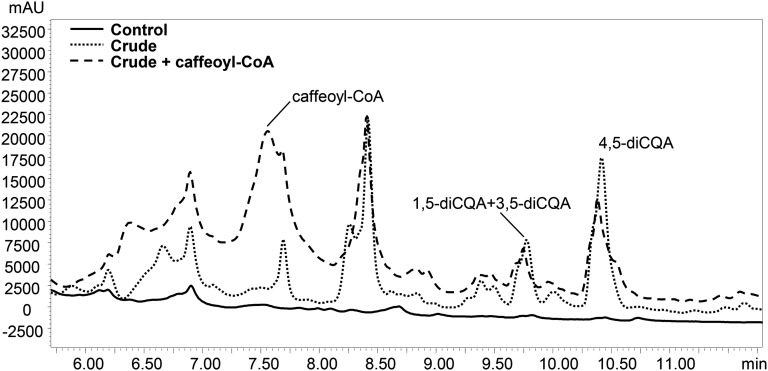
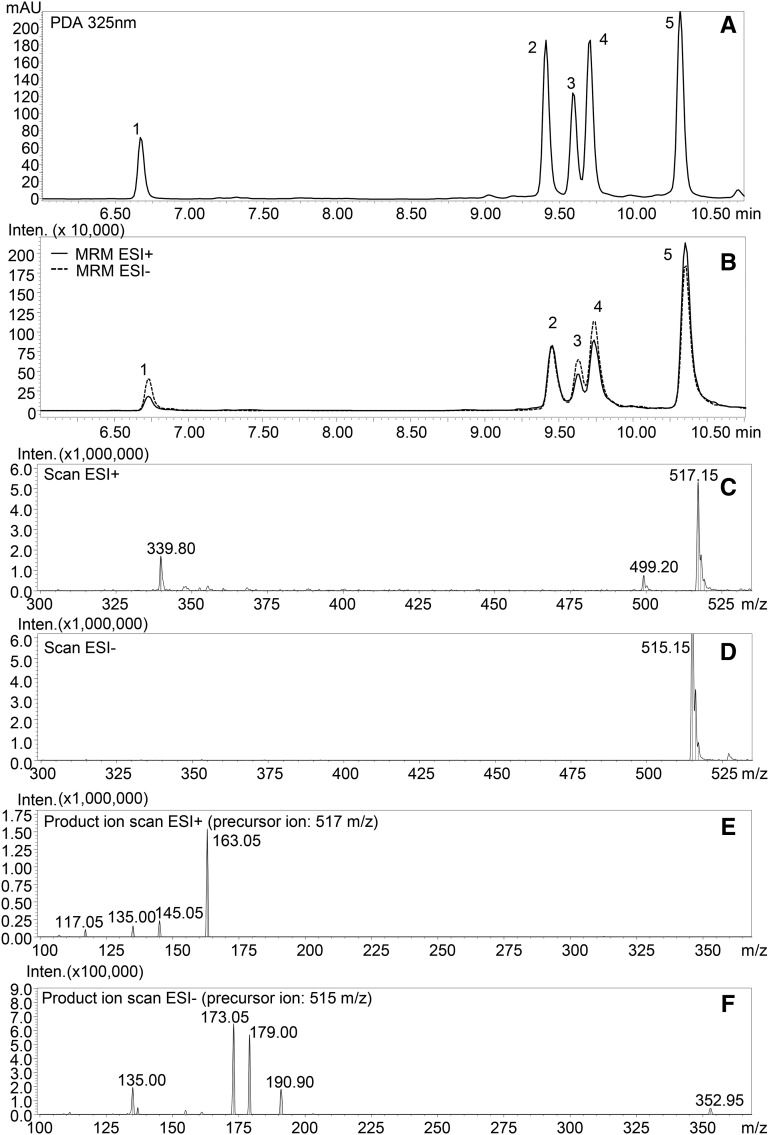
We incubated crude extracts of tomato fruit with CGA and detected the formation of three new products, absent in the negative control reactions (Fig. 1). The products were identified by liquid chromatography (LC)-photodiode array detector (PDA)-tandem mass spectrometry (MS/MS) analysis (Supplemental Fig. S1) comparing their UV profiles and mass fragmentation patterns to those obtained from the analysis of diCQA standard mixture (Fig. 2; Supplemental Fig. S2). The three new peaks (retention times: 9.60, 9.75, and 10.40 min) were identified as isomers of diCQAs; their precursor ions (mass-to-charge ratio [m/z] 515 and 517 in negative electrospray ionization mode [ESI–] and positive electrospray ionization mode [ESI+], respectively) corresponded to that of diCQA standard (Fig. 2, C and D), while in ESI–, the observed fragment ions of m/z 179 and 135 matched well with the presence of a caffeoyl moiety in the structure (Fig. 2, E and F) and the fragment ions at m/z 191 and 173 are indicative of a quinic acid moiety in the molecule.
Figure 1.
Identification of CCT enzyme activity giving synthesis of diCQAs in tomato. LC-PDA (at 325 nm) profiles of CCT activity with inactive (boiled) extract of ripe tomatoes (Control) and crude extract of ripe tomatoes (Crude). Incubations with crude enzyme extract and CGA were performed with or without caffeoyl-CoA. mAU, Milliabsorbance units.
Figure 2.
LC-PDA-MS/MS profile of diCQA standard mixture. A, PDA profile at 325 nm. B, MS profile obtained in MRM acquisition mode (for transitions, see ‟Materials and Methods”). C, ESI+ full-scan spectra of 4,5-diCQA. D, ESI– full-scan spectra of 4,5-diCQA. E, ESI+ product ion scan of 4,5-diCQA (precursor ion, 517 m/z). F, ESI– product ion scan of 4,5-diCQA (precursor ion, 515 m/z). For peak numbers, 1 indicates 1,3-diCQA, 2 indicates 3,4-diCQA, 3 indicates 1,5-diCQA, 4 indicates 3,5-diCQA, and 5 indicates 4,5-diCQA. mAU, Milliabsorbance units; Inten., ion intensity.
The three isomers were identified respectively as 1,5-diCQA, 3,5-diCQA, and 4,5-diCQA by comparison with authentic standards. This showed that the synthesis of dicaffeoyl quinates in tomato occurs through CCT activity. When tomato crude extracts were tested in the presence of CGA and caffeoyl-CoA (crude plus caffeoyl-CoA; Fig. 1) for acyl transferase activity, no increase in diCQA content was detected.
Optimization of the Enzyme Assay
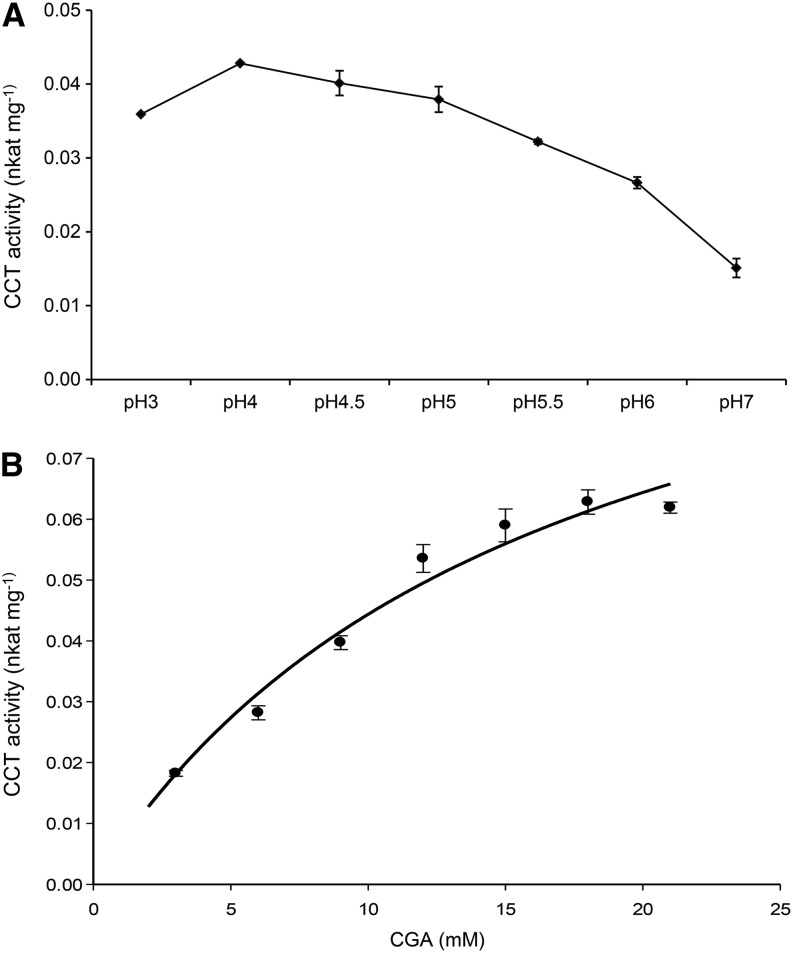
The effect of pH on the catalytic activity of the enzyme was tested from pH 3 to 7. CCT activity was highest at pH 4.0 (0.043 nkat mg–1 protein) in sodium phosphate buffer (Fig. 3A) without any cofactor requirements, although addition of MgCl2 (2.5 mm) and dithiothreitol (DTT; 1 mm) increased activity slightly. The reaction showed typical Michaelis-Menten kinetics with increasing CGA concentrations and gave a Km for CGA of 16 ± 3 mm (Fig. 3B). The activity was irreversible because when crude extracts were incubated in the presence of diCQA (3,5-, 4,5-, or 1,5-diCQA) and quinic acid, no substrate was consumed or product formed.
Figure 3.
CCT activity in red tomato fruit. A, CCT activity under different pH conditions (3–7). B, CCT activity in crude extract of ripe tomato with increasing concentrations of CGA. Values on the y axis are nkat mg–1 of total protein. The curve represents the best fit of the Michaelis-Menten equation to the data using SigmaPlot (Systat Software). Error bars indicate sd calculated from three biological replicates.
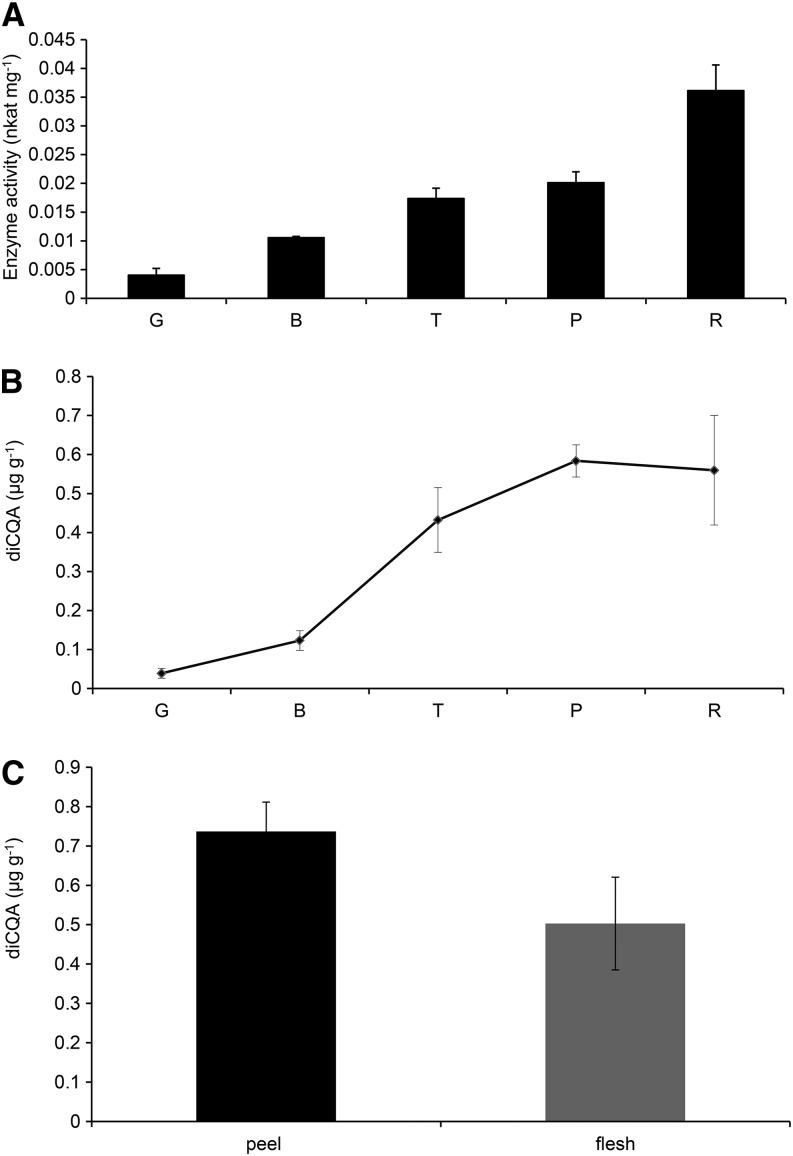
CCT activity and diCQA levels were determined at different stages during fruit ripening (green, breaker, turning, pink, and red). An increase in CCT activity was observed during ripening, reaching a maximum of 0.036 nkat mg–1 protein in the peel of red fruit (Fig. 4A). The levels of diCQAs also increased during ripening, reaching a maximum in fully ripe tomatoes (Fig. 4B). Peel tissue had slightly higher levels of CCT activity (0.034 ± 0.0034 nkat mg–1 total protein) than flesh tissue (0.027 ± 0.002 nkat mg–1 total protein) and correspondingly higher levels of diCQAs (Fig. 4C).
Figure 4.
CCT activity and levels of diCQAs during ripening and in different tissues of tomato fruit. A, Changes in CCT activity during ripening of tomato. CCT activity was measured in tissue at different stages of fruit ripening and expressed as specific activity (nkat mg–1 of total protein). Error bars indicate sd calculated from three biological replicates. G, Green; B, breaker; T, turning; P, pink; R, red stage. B, Content (µg g–1 fresh weight) of diCQAs during fruit ripening. Error bars indicate sd calculated from three biological replicates. C, Content (µg g–1 fresh weight) of diCQAs in different tissues of red fruit. Error bars indicate sd calculated from three biological replicates.
Purification of CCT Activity
Protein extracts from red fruit exhibiting high enzymatic activity were used for enzyme purification (Supplemental Table S1). A 45% to 60% ammonium sulfate-precipitated protein fraction (21.7-fold purification) was separated by ion-exchange chromatography, resulting in a fraction with a specific activity of 1.34 nkat mg–1 protein, corresponding to a purification of 388-fold (Supplemental Table S1). This protein fraction was subjected to trypsin digestion and mass spectrometric sequencing. The list of peptide masses was aligned with predicted tryptic digests of known proteins by MASCOT (Supplemental Table S2). These peptide sequences were used to interrogate the EST database of tomato to search for potential candidates for CCT. Among the peptides obtained from mass spectrometric analysis, an HQT protein (Q70G32) was selected as a good candidate for further investigation as two unique peptides from the tryptic digest (IWSSNLDLIVGR and SALDYLELQPDLSTLIR) matched exactly the predicted peptide sequence of this gene.
CCT Activity of Enzyme Expressed in Escherichia coli
As previously described, the tomato BAHD acyl transferase HQT (SlHQT) can use quinic acid as an acceptor with caffeoyl-CoA and p-coumaroyl-CoA donors for the synthesis of CGA and coumaroyl quinate, respectively (Niggeweg et al., 2004). (The acyl-CoA-dependent BAHD superfamily is defined by the names of the first four acyl-transferase enzymes of the family isolated from plant species; D'Auria, 2006.)
The crude enzyme extract of E. coli expressing SlHQT was incubated in the presence of high quantities of CGA, according to the optimum conditions determined for the CCT activity in crude extracts of tomato fruit. In the presence of SlHQT, we detected the formation of three new products, absent in control reactions. These were identified as 1,5-, 3,5-, and 4,5-diCQA acid on the basis of UV spectra, the deprotonated [M−1]– and protonated [M+1]+ molecular ions and by comparison to authentic standards (Fig. 5, wild-type protein; Supplemental Fig. S3). When SlHQT was assayed in the reverse direction (using, alternatively, 1,5-, 3,5-, 4,5-diCQA acids and quinic acid as substrates), no activity was detected. The affinity of SlHQT as CCT for CGA as an acceptor (Km = 19 ± 2 mm) was very similar to that observed earlier in crude extracts of tomato (Km = 16 ± 3 mm).
Figure 5.
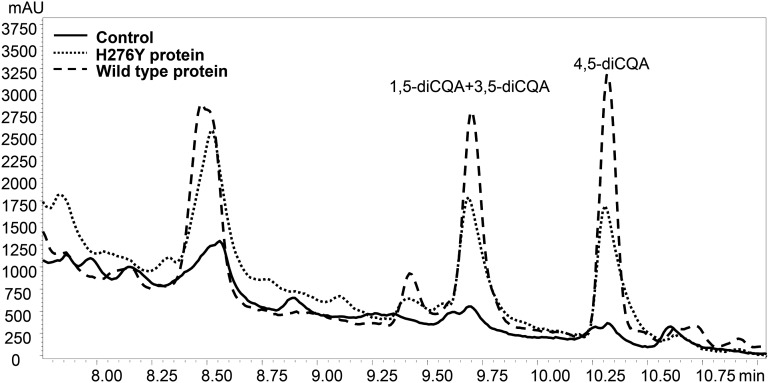
LC-PDA profiles (at 325 nm) of CCT activity with recombinant wild-type SlHQT enzyme, His-mutated SlHQT enzyme (H276Y), and inactive enzyme (Control). mAU, Milliabsorbance units.
To gain insight into the involvement of SlHQT in diCQA synthesis, the levels of transcripts were measured by means of real time quantitative PCR, using ubiquitin to normalize expression. The levels of expression were 5.9-, 8.7-, 12.0-, and 24.5-fold higher in breaker, turning, pink, and red fruits, respectively, compared with green-stage tissue, indicating an increase of gene expression levels during ripening of tomato fruits, with a peak at the red ripe stage (Supplemental Fig. S4).
Subcellular Localization of SlHQT
The subcellular localization of SlHQT was investigated using two vectors expressing the SlHQT complementary DNA (cDNA) under the control of the 35S promoter, such that the SlHQT protein was fused at its C terminus to either enhanced yellow fluorescent protein (EYFP) or monomeric red fluorescent protein (mRFP). Plasmid DNA was bombarded into onion (Allium cepa) epidermal tissue, because these cells have no chloroplasts and fluorescence could be observed free from interference from chlorophyll (Fig. 6). As a control, a vector for expression of EYFP or mRFP alone, under the control of the 35S promoter, was also bombarded into onion epidermis.
Figure 6.
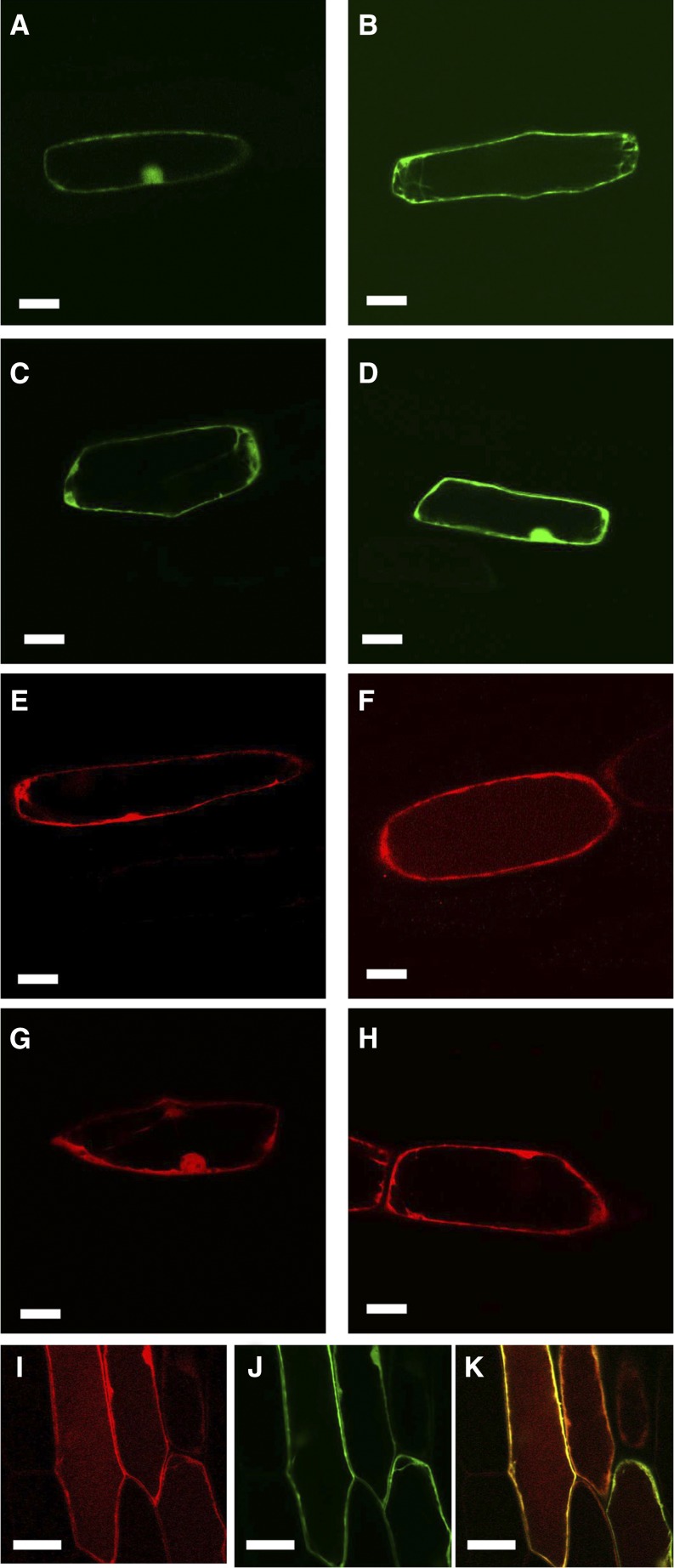
Subcellular localization of SlHQT. Onion epidermis was bombarded with 35S:SlHQT fused to EYFP or mRFP with 35S:EYFP or 35S:mRFP as controls for cytoplasmic localization. A, Fluorescence from SlHQT:EYFP after 24 h in the dark. B, Fluorescence from SlHQT:EYFP after 96 h in the dark. Fluorescence in the vacuole is faintly visible. C, Fluorescence from EYFP after 24 h in the dark. D, Fluorescence from EYFP after 96 h in the dark. E, Fluorescence from SlHQT:mRFP after 24 h in the dark. F, Fluorescence from SlHQT:mRFP after 96 h in the dark. Fluorescence in the vacuole is clearly visible. G, Fluorescence from mRFP after 24 h in the dark. H, Fluorescence from mRFP after 96 h in the dark. Fluorescence remains cytoplasmic. I, Fluorescence from SlHQT:mRFP cobombarded with EYFP after 96 h in the dark. J, Fluorescence from EYFP from the same cell. K, Merged image showing fluorescence from SlHQT:mRFP in the vacuole as well as the cytoplasm, while the EYFP remains entirely cytoplasmic. Bars = 50 μm.
Both SlHQT:EYFP- and EYFP-transformed cells showed clear cytoplasmic localization, indicated by strong fluorescence at the periphery of the cells and in cytoplasmic strands (Fig. 6, A–D). However, in contrast to 35S:EYFP, a faint fluorescence was also observed with SlHQT:EYFP in vacuolar regions, particularly in epidermal cells incubated for 96 h in the dark, implying that there might be low levels of SlHQT in vacuoles as well as in the cytoplasm (Fig. 6, B and D). Because mRFP is particularly stable at low pH (Hunter et al., 2007), transient assays were repeated with 35S:SlHQT:mRFP compared with 35S:mRFP. Again, the predominant fluorescence was cytoplasmic, as evidenced by the strong fluorescence from the zone delimiting the inner edge of the cells and the occasional cytoplasmic strands. The nucleus also fluoresced strongly, similar to 35S:mRFP. However, fainter fluorescence from SlHQT:mRFP was clearly visible in the vacuolar region of the cells, particularly in samples incubated for 96 h in the dark (Fig. 6, E and F), compared with cells bombarded with 35S:mRFP, where the fluorescence remained cytoplasmic (Fig. 6, G and H). Finally, cells were cobombarded with 35S:SlHQT:mRFP (Fig. 6I) and 35S:EYFP (Fig. 6J). Cells expressing both plasmids showed that SlHQT:mRFP was in the vacuole while EYFP was completely cytoplasmic (Fig. 6K; Supplemental Fig. S5). This demonstrated that SlHQT is localized in vacuoles as well as in the cytoplasm of plant cells.
Structural Features Determining the Duel Catalytic Activity of SlHQT/CCT
SlHQT in tomato appears to be capable of using two types of acyl donor. It uses caffeoyl-CoA or p-coumaroyl-CoA as acyl donors and quinate as the acyl acceptor in the synthesis of CGA (Niggeweg et al., 2004), but it is also able to use CGA as both acceptor and acyl donor in the synthesis of diCQAs (Fig. 7A).
Figure 7.
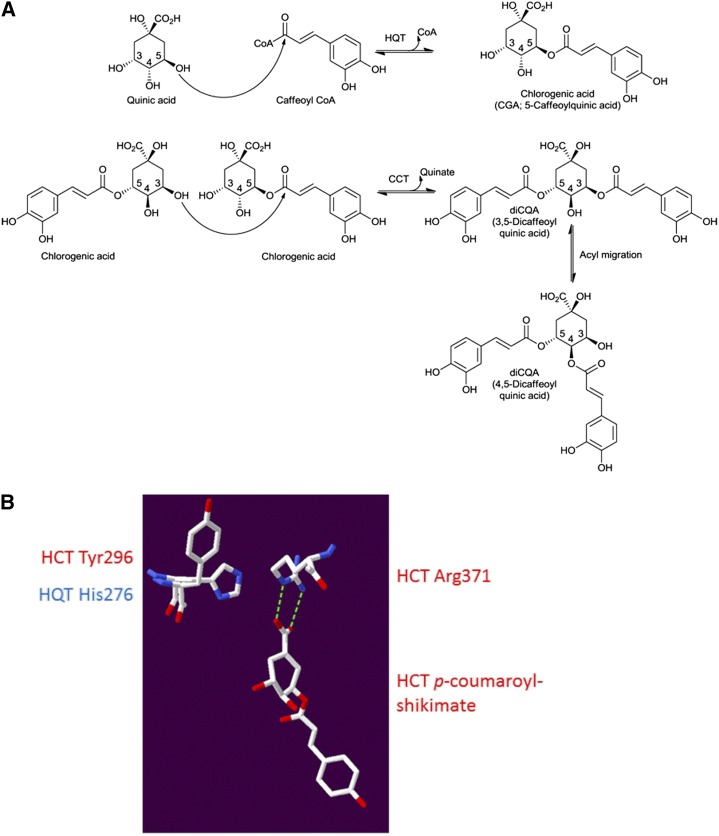
Structural analysis of SlHQT modeled on sorghum HCT (Protein Data Bank code 4KEC; Walker et al., 2013). A, Proposed reaction scheme for diCQA synthesis by CCT using CGA as the acyl donor as well as the acyl acceptor compared with the BAHD activity of HQT and HCT using caffeoyl-CoA as the acyl donor and quinate as the acyl acceptor, respectively. B, Structure of sorghum HCT (Protein Data Bank code 4KEC; Walker et al., 2013) with a superposed homology model of SlHQT based upon this structure. CGA would have to bind to SlHQT with its carboxyl group facing away from the Arg residue, allowing a favorable electrostatic interaction between His-276 and CGA that would promote CCT activity.
The 3,5-diCQA isomer could be made in one step by a CCT, but the 4,5 isomer would likely require an additional spontaneous acyl migration from the 3 to the 4 position (Fig. 7A), as suggested by Lallemand et al. (2012). We generated a homology model of HQT with Swiss model (Guex and Peitsch, 1997) using the 4KEC (a ternary complex form) structure of sorghum (Sorghum bicolor) HCT (SbHCT) bound to p-coumaroyl-shikimate described by Walker et al. (2013). The Arg-371 residue of SbHCT approaches the shikimate ring from the same face as the transferred acyl group and forms an H-bonding interaction with the carboxyl group of shikimate with a distance of 2.7 Å (Fig. 7B). In SlHQT, the equivalent Arg-352 would be expected to form a similar interaction with quinate, except that its carboxyl group is out of the plane of the ring unlike with shikimate. On the other face of the acceptor substrate and opposite the Arg residues, His-276 in SlHQT is equivalent to Tyr-296 in SbHCT. For the CCT reaction to occur, CGA would have to bind with its ring in the opposite orientation such that its carboxyl group would more likely engage with His-276 of SlHQT rather than Arg-352. The low pH optimum of the CCT reaction implies that His-276 would be positively charged and able to form a favorable interaction with CGA in this orientation, only at low pH. The absence of a potentially positively charged residue at this location in its active site would suggest that SbHCT does not have CCT activity.
To examine the role of this specific amino acid on the enzymatic activity of SlHQT, we selectively changed His-276 to Tyr-276. The activity of the H276Y mutant was compared with that of the unmutated enzyme in the CCT reaction. The SlHQT H276Y mutant showed significantly lower production of diCQA in the CCT reaction when compared with wild-type SlHQT protein (58% of wild-type activity; 0.0021 ± 0.0001 nkat mg–1 compared with 0.0036 ± 0.0001 nkat mg–1, P < 0.01 Student’s t test; Fig. 5; Supplemental Fig. S3). The H276Y mutation barely affected the activity of the enzyme as HQT in CGA synthesis, as monitored by activity with caffeoyl-CoA and quinic acid assayed using the spectrophotometric method (Niggeweg et al., 2004) on the same extracts (92% of wild-type activity; 11.16 ± 0.3 nkat mg–1 for H276Y protein compared with 12.16 ± 0.4 nkat mg–1 for wild-type SlHQT).
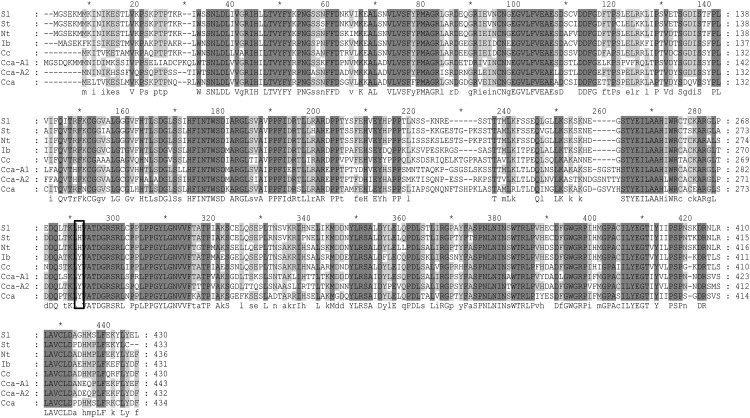
Alignment of sequences of enzymes shown to encode HCT from different plants species indicated that in all HCT proteins, the residue equivalent to Tyr-296 in SbHCT was also Tyr (Supplemental Fig. S6). Alignment of sequences of enzymes with HQT activity from Solanaceous species (tomato, tobacco (Nicotiana tabacum), and potato (Solanum tuberosum); Niggeweg et al., 2004), coffee (Lepelley et al., 2007), and globe artichoke (Comino et al., 2009; Menin et al., 2010; Sonnante et al., 2010) showed that in HQT from Solanaceous species, the residue equivalent to Tyr-296 was His (His-276 in SlHQT), but in HQT from other species, this residue was Tyr (Fig. 8). We tested whether the HQT from globe artichoke (possessing Tyr-276 instead of His-276) had CCT activity following expression in E. coli, but only traces of diCQAs were detected following incubation with CGA at pH 5 (Supplemental Fig. S7). These data confirmed our structural analysis, suggesting that the His at position 276 in SlHQT plays a key role in CCT activity and also suggesting that although CCT activity is likely responsible for diCQA formation in Solanaceous species, diCQAs may form by other routes in other plant species.
Figure 8.
Sequence alignment of sequences belonging to the plant HQT family. NP_001234850 from tomato (Sl), ABA46756 from potato (St), CAE46932 from tobacco (Nt), BAA87043 from sweet potato, ABO77957 from coffee (Cc), ABK79689 from globe artichoke (Cca), ADL62854 from globe artichoke (Cca-A1), and ADL62855 from globe artichoke (Cca-A2). Black box indicates His-276 in SlHQT and equivalent residues in HQT enzymes from other plant species.
DISCUSSION
Assays of crude extracts of tomato confirmed an early report of the synthesis of isochlorogenic acid (3,5-diCQA) in sweet potato through an enzyme reaction in which CGA acts both as acyl donor and acceptor (Villegas and Kojima, 1986; Fig. 7A). These data suggest the presence, in tomato, of a CCT, which catalyzes the formation of diCQAs using CGA as acyl donor. The increase in CCT activity correlated well with increases in levels of diCQAs with advancing fruit ripening (Fig. 4A). As previously observed by Moco et al. (2007), we detected increases in diCQA isomers in tomato epidermis upon ripening; their increase was lower in flesh (Fig. 4C), probably as a consequence of a lower CCT activity.
Partial purification of CCT activity and identification of proteins in the enriched fraction suggested HQT (an enzyme already known to bind CGA) as a candidate for CCT activity. In most cases, acyltransferases involved in hydroxycinnamate synthesis use CoA thioesters (for example, caffeoyl-CoA) as acyl donors (D’Auria, 2006). There is one reported case in which CGA is used as an acyl donor in tomato: chlorogenate:glucarate caffeoyltransferase, a lipase-like protein that catalyzes the transfer of the caffeoyl moiety of CGA to the acceptor molecule glucarate (Strack and Gross, 1990; Teutschbein et al., 2010).
Tomato CCT activity is optimal at pH 4 (Fig. 3A) and seems likely to be biologically relevant only when the HQT enzyme is located within the vacuole of tomato cells, where an acid pH prevails. Vacuolar localization of CCT activity would favor diCQA biosynthesis, because high vacuolar concentrations of CGA could promote diCQA biosynthesis despite the relatively low affinity of CCT for CGA. In the cytoplasm, SlHQT could act as a BAHD acyl transferase at pH 6 to 7 in the presence of aromatic acyl-CoA donors (Km, 0.3 ± 0.1 mm) with a high affinity for CGA (Km, 0.05 mm) and Vmax of 119 and 58 nkat mg–1 protein (forward and reverse reactions, respectively; Niggeweg et al., 2004). By contrast, it could act as a CCT in the vacuole at pH 4 to 5 in the absence of aromatic acyl-CoA donors and presence of relatively high concentrations of CGA allowing activity despite a low affinity for CGA (Km, 16 ± 3 mm) and a Vmax of 0.12 ± 0.01 nkat mg–1 protein. The low Vmax of the CCT activity could, in other circumstances, limit the biological significance of this activity, but in the context of fruit ripening, over a period of days, this level of activity is sufficient. Thus, SlHQT likely performs two distinct functions in the very different cytoplasmic and vacuolar subcellular compartments. SlHQT was consistently localized in the vacuole lumen as well as in the cytoplasm when monitored by C-terminal fluorescent tags in transient assays (Fig. 6). This dual localization was distinct from mRFP or EYFP expressed without the fusion protein, which were exclusively cytoplasmically localized. We suggest that once significant amounts of CGA have been synthesized in the cytoplasm and transported to the vacuole for storage, tomato HQT moonlights by participating in diCQA synthesis in the vacuole.
The BAHD activity of SlHQT is freely reversible (Niggeweg et al., 2004), whereas that of CCT is irreversible. Consequently, CGA can accumulate only if it is sequestered in the vacuoles of cells. There, in high concentrations, at low pH, and in the absence of acyl-CoA donors, SlHQT can form diCQAs through its CCT activity.
This possibility is supported by the close association between levels of CGA and levels of diCQAs in tomato under different cultivation conditions (Vallverdú-Queralt et al., 2012) and between HQT expression levels and CCT activity observed during fruit ripening (Fig. 4; Supplemental Fig. S4). Although our results indicate that HQT is localized in vacuoles as well as in the cytoplasm, we do not yet know whether transport to the vacuole involves an active transport mechanism. The fact that SlHQT is present in the vacuole gives significance to the CCT activity despite its high Km and low pH optimum.
In coffee, the synthesis of diCQAs has been proposed to occur via an uncharacterized activity of HCT on CGA and CoA (Lallemand et al., 2012), while in tomato, diCQAs appear to be synthesized by a different route involving the CCT activity of SlHQT.
Site-directed mutagenesis of SlHQT confirmed the role of His-276 in the CCT reaction (Fig. 5). Interestingly, HQT from globe artichoke, which has a Tyr residue rather than a His residue at the position equivalent to His-276 in SlHQT, had very low CCT activity when assayed under optimized conditions in vitro. This suggests that the ability to moonlight in the production of diCQAs may be limited to HQT enzymes from Solanaceous species that carry a His residue at an appropriate position for interaction with the carboxyl group of the quinate moiety of the CGA donor molecule, under conditions of relatively low pH (<5). Presumably, species lacking HQT or HCT enzymes with His in this position may use different mechanisms to form diCQAs, and alternative routes may have arisen by convergent evolution in different families of plants.
Our results broaden the view of one enzyme-one activity and suggest that changing subcellular environments may diversify the range of products of secondary metabolism. This concept could operate in acyl decoration of other plant natural products. It also informs the design of successful strategies to biofortify crops with diCQAs through metabolic engineering.
MATERIALS AND METHODS
Plant Material
Tomato (Solanum lycopersicum) ‘MicroTom’ was grown in the greenhouse at the John Innes Centre. Fruits were harvested at different stages of development: green, breaker, turning, pink, and red stages. From these fruit, the peel was separated from the flesh, and both tissues were frozen immediately in liquid nitrogen and stored at –80°C. Both the peel and the flesh tissues were tested for their enzyme activity as well as for their metabolite content using LC-mass spectrometry (MS).
Enzyme Assay
Tomato fruit (1 g) was ground to a fine powder in liquid nitrogen and extracted with 5 mL of extraction buffer (50 mm MOPS and 1 mm DTT). After stirring for 40 min at 4°C, the homogenate was centrifuged at 13,250g for 30 min. The clear supernatant obtained constituted the crude enzyme extract. Each 40 μL of enzyme assay reaction contained 100 mm sodium phosphate buffer (tested at pH 3–7), 1 mm DTT, 2.5 mm MgCl2, 1 mm EDTA, CGA (0–21 mm), and 10 μL of protein solution. The same enzyme reaction was also performed with caffeoyl-CoA (1 mm) combined with 1 mm CGA (provided by SIGMA). Reactions were incubated for 90 min at 30°C, extracted with 20 μL of acetonitrile, and analyzed by LC-MS. Protein concentration was quantified according to Bradford (1976) by using bovine serum albumin as internal standard.
Enzyme Extraction and Purification
Tomato fruit (60 g) was ground to a fine powder in liquid nitrogen and extracted with 300 mL of extraction buffer (50 mm MOPS and 1 mm DTT). After stirring for 40 min at 4°C, the homogenate was centrifuged at 13,250g for 30 min. The clear supernatant constituted the crude enzyme extract. The supernatant was made up to 25%, 45%, and 60% saturation with finely ground (NH4)2SO4, and the proteins were collected by centrifugation at 13,250g at 4°C. The proteins were dissolved into 20 mL of potassium phosphate buffer (0.2 m, pH 7). The protein fraction with the highest enzymatic activity was dialyzed against Tris HCl (20 m, pH 8) for 4 h and then applied to a MonoQ (Pharmacia) column (1 mL) preequilibrated with Tris HCl, pH 7.5. Proteins were eluted with a gradient of 0 to 1 m NaCl in 20 mm Tris-HCl, pH 8, at a flow rate of 0.5 mL min–1, and 0.5-mL fractions were collected. The fractions obtained were tested for their enzyme activity.
Protein MS
The fractions having the highest enzyme activities were concentrated and desalted for separation by 10% (w/v) SDS-PAGE using a Micron YM-10 filter (Millipore). Protein samples were reduced, alkylated, and digested with trypsin according to standard procedures using urea as the denaturing agent. The peptides generated by the tryptic digestion were redissolved in 0.1% (v/v) trifluoroacetic acid. For LC-MS/MS analysis, a sample aliquot was applied via a nanoAcquity UPLC system (Waters) running at a flow rate of 250 nL min–1 to an LTQ-Orbitrap mass spectrometer (Thermo Fisher). Peptides were trapped using a precolumn (Symmetry C18, 5 μm, 180 μm × 20 mm, Waters) that was then switched online to an analytical column (BEH C18, 1.7 μm, 75 μm × 250 mm, Waters) for separation. Peptides were eluted with a gradient of 5% to 40% (v/v) acetonitrile in water per 0.1% (v/v) formic acid at a rate of 0.66% min–1. The column was connected to a 10-μm SilicaTip nanospray emitter (New Objective) attached to a nanospray interface (Proxeon).
The mass spectrometer was operated in positive ion mode at a capillary temperature of 200°C. The source voltage and focusing voltages were tuned for the transmission of Met-Arg-Phe-Ala peptide (m/z 524; Sigma-Aldrich). Data-dependent analysis was carried out in Orbitrap-IT parallel mode using collision-induced dissociation fragmentation on the five most abundant ions in each cycle. The collision energy was 35, and an isolation width of 2 was used. The Orbitrap was run with a resolution of 30,000 over the MS range from m/z 350 to m/z 2,000 and an MS target of 106 and 1-s maximum scan time. The MS/MS was triggered by a minimal signal of 5,000 with an automatic gain control target of 2 × 104 ions and 150-ms scan time.
For selection of 2+ and 3+ charged precursors, charge state and monoisotopic precursor selection were used. Dynamic exclusion was set to 1 count and 40-s exclusion, with an exclusion mass window of ±0.1 D. MS scans were saved in profile mode while MS/MS scans were saved in centroid mode.
Raw data files were processed in Proteome Discoverer (Thermo Fisher) to generate an mgf file that was submitted for a database search using an in-house MASCOT 2.2.06 Server (Matrixscience). Searches were performed on the SP-trEMBL database (Sprot_sptrembl20100302.fasta) with taxonomy set to Solanum lycopersicum (Lycopersicon esculentum) using 5-ppm precursor tolerance, 0.5-D fragment tolerance, one missed cleavage, and carbamidomethylation as fixed and oxidation as variable modifications. MASCOT search results were imported and evaluated in Scaffold 3_00_01 (Proteome Software).
Enzyme Expression in Escherichia coli and CCT Enzyme Assay
His-mutated SlHQT (H276Y, His-276 replaced by Tyr-276) was obtained by Geneart (Life Technologies). The native and His-mutated SlHQT were subcloned in frame into the NotI and BamHI sites of the p-GEX 4T1 vector. The cDNA encoding the native globe artichoke (Cynara cardunculus) HQT (ABK79689) was subcloned in frame into the EcoRI and XhoI sites of the p-GEX 4T1 vector.
Recombinant plasmids p-GEX 4T1 vector (containing the cDNAs encoding the globe artichoke HQT and native and His-mutated SlHQT) were used to transform E. coli BL21(DE3)pLysE that was grown on selective medium. Transformants and control colonies were inoculated in 10 mL of Luria-Bertani medium, with 1% (w/v) Glc and selective antibiotics (ampicillin and chloramphenicol). The overnight cultures were diluted 1:25 in 50 mL of Luria-Bertani medium in the presence of selective antibiotics and incubated until the optical density at 600 nm reached 0.6. At this point, isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 1 mm, and the cultures were grown overnight at 28°C with shaking at 250 rpm. Cultures were centrifuged (3,500g, 5 min), and cells were resuspended in 1 mL of phosphate-buffered saline (pH 7.4) and lysed by three cycles of freezing (in liquid nitrogen) and thawing (at 37°C). Following sonication (five cycles, 30 s, 125 W, 20 kHz), the solution was clarified by centrifugation (11,000g, 5 min), and the supernatant was assayed for enzyme activity.
Each 100 μL of enzyme assay reaction contained 100 mm sodium phosphate buffer (pH 5), 1 mm DTT, 2.5 mm MgCl2, 1 mm EDTA, 5 mm CGA (provided by SIGMA), and 40 μL of protein solution. Reactions were incubated for 90 min at 30°C, extracted with 50 μL of acetonitrile, and analyzed by LC-MS. The reverse reactions with diCQA (3,5, 4,5, or 1,5) contained 100 mm sodium phosphate buffer (tested at pH 5–8), 1 mm DTT, 2.5 mm MgCl2, 1 mm EDTA, 1 mm diCQAs, 1 mm quinic acid, and 40 μL of bacterial extract. diCQA isomer standards were provided by Transmit. HQT activity in CGA synthesis was assayed using the spectrophotometric method as described by Niggeweg et al. (2004). Data were analyzed by Student’s t test and presented as mean ± sd, and differences were considered significant at P < 0.05.
SlHQT protein was also expressed in E. coli using the pSTAG vector (Niggeweg et al., 2004) and was used for the evaluation of kinetic parameters. The concentration of the fusion protein was determined using the S-TAG rapid assay kit (Novagen). For measuring the Km with CGA, 0.58 μg of enzyme, 1 mm DTT, 2.5 mm MgCl2, 1 mm EDTA, and 1 to 24 mm CGA were used. An HPLC calibration curve was established for each molecule for quantification purposes. Km and Vmax values were estimated in at least duplicate by fitting Michaelis-Menten curves directly using SigmaPlot.
Identification and Quantification of diCQAs
The identification and the quantification of diCQAs were performed by LC-PDA-MS/MS. The analyses were carried out on a Shimadzu Nexera X2 system equipped with a photodiode detector SPD-M20A in series to a triple quadrupole Shimadzu LCMS-8040 system provided with electrospray ionization source. An Ascentis Express C18 column (150- × 2.1-mm i.d., 2.7-μm particle size; Supelco) was used. The analysis conditions were: for the mobile phase, eluent A, 0.1% (v/v) formic acid in water; and eluent B, 0.1% (v/v) formic acid in acetonitrile; and the mobile phase gradient was as follows: 5% to 25% B in 10 min, 25% to 40% B in 5 min, 40% to 100% B in 5 min, and 100% B for 1 min. Injection volume was 5 μL, the flow rate of the mobile phase was 0.4 mL min–1, and the column was maintained at 30°C. UV spectra were acquired in the 210- to 450-nm wavelength range, and the resulting chromatograms were integrated at 325 nm.
The identification of the components was based on their UV spectra and mass spectral information in Multiple Reaction Monitoring (MRM) mode in both positive and negative ionization mode (respectively, ESI+ and ESI−). MS operative conditions were heat block temperature, 400°C; nebulizing gas (nitrogen) flow rate, 3 min–1; drying gas (nitrogen) flow rate, 15 min–1; and desolvation line temperature, 250°C. Collision gas was argon (230 kPa). Transitions monitored included ESI+, m/z 517 → 163, m/z 517 → 145, and m/z 517 → 135 (dwell time, 20 ms; collision energy, –35 V; and event time, 0.096 s), and ESI–, m/z 515 → 179, m/z 515 → 191, and m/z 515 → 135 (dwell time, 20 ms; collision energy, 35 V; and event time, 0.096 s). The MRM transitions were selected on the basis of the fragments obtained by analyzing the diCQA standards in full-scan mode in both ESI+ and ESI– in the range of 300 to 1,200 m/z, with a scan speed of 1,000 D s–1, and then in product ion scan mode in both ESI+ and ESI– in the range of 100 to 550 m/z, with a scan speed of 1,000 D s–1 and using, as precursor ions, 517 m/z protonated molecular ion ([M+H]+) for ESI+ and 515.00 m/z deprotonated molecular ion ([M–H]–) for ESI–.
The quantification of diCQAs was undertaken on the PDA profiles (at 325 nm) using the external calibration method. A three-point calibration curve was built analyzing, in triplicate, the pure standard of 4,5-diCQA in the range of 100 to 500 ppb. The determination coefficient was 0.9951.
Subcellular Localization of SlHQT
Tomato HQT was amplified from cDNA using primers for Gateway recombination (HQTF, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGGAAGTGAAA-AAATGATGAAAATTAATATC-3′; and HQTR, GGGGACCACTTTGTACAAG-AAAGCTGGGTATAATTCATATAAATATTTTTCAAATAG-3′) and recombined into entry vector pDONR207. The resulting clone was recombined into pB7YFWG2.0 for EYFP fusions and pB7RWG2.0 for mRFP fusions (Karimi et al., 2002). To construct the 35S:EYFP and 35S:mRFP control vectors, pB7YWG2.0 and pB7RWG2.0 were cut with EcoRV and SpeI (to remove the Gateway destination cassette), filled in with the Klenow fragment of DNA polymerase, and self-ligated to yield 35S:EYFP and 35S:mRFP, respectively. Plasmid DNA was prepared using a Qiagen mini-prep kit. Bombardment was conducted using a particle inflow helium gun based on the design of Vain et al. (1993). Onion (Allium cepa) epidermal tissue was prepared freshly from large onions and rinsed in sterilized and distilled water. Plasmid DNA (5 μg) was precipitated onto 2-mg gold particles (1.0-μm diameter) through the addition of 50 μL of 2.5 m CaCl2 and 20 μL of 100 mm spermidine. After precipitation, 90 μL of supernatant was discarded, and particles were washed twice in ethanol and resuspended in 50 μL of ethanol. Gold particles were prepared immediately before use. For bombardment, tissue was placed on Murashige and Skoog (1962) medium plus 8% (w/v) agar (MS medium) in a petri dish, within the desiccator in the gun range of 120 to 160 mm. Tissue was bombarded with 7 μL of gold suspension using a 50-ms burst of helium at a pressure of 7,580 kPa within a vacuum of −98 kPa. After bombardment, tissue was incubated on MS medium at 20°C in the dark for 24 or 96 h. Bombarded tissue was imaged using a Zeiss 510 Meta Confocal microscope. For EYFP, a 488-nm laser line was used, and emission at 500 to 550 nm was imaged. For mRFP, a 561-nm laser line was used, and emission at 570 to 615 nm was imaged using a ×25, 0.7 multiimmersion objective. Images were processed using Fiji (http://fiji.sc/wiki/index.php/Fiji).
Bioinformatic Analysis of Tomato HQT
Homology model of SlHQT was performed with Swiss model (Guex and Peitsch, 1997) using the 4KEC structure of SbHCT bound to p-coumaroyl-shikimate described by Walker et al. (2013). Alignment of sequences of enzymes shown to encode HCT and HQT from different plants species was performed by means of ClustalW set with standard parameters.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. CCT activity in crude extracts of tomato.
Supplemental Figure S2. MS/MS profiles of diCQA standards.
Supplemental Figure S3. CCT activity of recombinant wild-type SlHQT enzyme and H276Y-mutated SlHQT enzyme.
Supplemental Figure S4. Expression of SlHQT in tomato.
Supplemental Figure S5. Subcellular localization of SlHQT.
Supplemental Figure S6. Sequence alignment of sequences belonging to the plant hydroxycinnamoyl CoA shikimate/HCT family.
Supplemental Figure S7. CCT activity of wild-type SlHQT enzyme and globe artichoke HQT.
Supplemental Table S1. Purification scheme for chlorogenate:chlorogenate transferase activity from tomato.
Supplemental Table S2. List of peptide masses present in purified enzyme fraction.
Supplementary Material
Acknowledgments
We thank Gerhard Saalbach (John Innes Centre) for technical assistance with protein MS, Silke Robatzek (Sainsbury Laboratory) and Grant Calder (John Innes Centre) for invaluable advice and help with subcellular imaging, and Anna Maria Milani (University of Torino) for excellent technical assistance.
Glossary
- CGA
chlorogenic acid
- diCQA
dicaffeoylquinic acid
- CCT
chlorogenate:chlorogenate transferase
- HQT
hydroxycinnamoyl-Coenzyme A:quinate hydroxycinnamoyl transferase
- triCQA
tricaffeoylquinic acid
- DW
dry weight
- HCT
hydroxycinnamoyl-Coenzyme A transferase
- m/z
mass-to-charge ratio
- DTT
dithiothreitol
- cDNA
complementary DNA
- LC
liquid chromatography
- MS
mass spectrometry
- MRM
Multiple Reaction Monitoring
- PDA
photodiode array detector
- MS/MS
tandem mass spectrometry
Footnotes
This work was supported by a postdoctoral fellowship from the Piedmont Region of Italy (to A.M.), the core strategic grant of the Biological and Biotechnological Science Research Council to the John Innes Centre (to C.M.), and the Institute Strategic Program Understanding and Exploiting Plant and Microbial Secondary Metabolism from the Biological and Biotechnological Science Research Council (grant no. BB/J004596/1 to C.M., S.B., and D.K.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chanforan C, Loonis M, Mora N, Caris-Veyrat C, Dufour C (2012) The impact of industrial processing on health-beneficial tomato microconstituents. Food Chem 134: 1786–1795 [DOI] [PubMed] [Google Scholar]
- Choi J, Park JK, Lee KT, Park KK, Kim WB, Lee JH, Jung HJ, Park HJ (2005) In vivo antihepatotoxic effects of Ligularia fischeri var. spiciformis and the identification of the active component, 3,4-dicaffeoylquinic acid. J Med Food 8: 348–352 [DOI] [PubMed] [Google Scholar]
- Clifford M. (1999) Chlorogenic acids and other cinnamates: nature, occurrence and dietary burden. J Sci Food Agric 79: 362–372 [Google Scholar]
- Comino C, Hehn A, Moglia A, Menin B, Bourgaud F, Lanteri S, Portis E (2009) The isolation and mapping of a novel hydroxycinnamoyltransferase in the globe artichoke chlorogenic acid pathway. BMC Plant Biol 9: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comino C, Lanteri S, Portis E, Acquadro A, Romani A, Hehn A, Larbat R, Bourgaud F (2007) Isolation and functional characterization of a cDNA coding a hydroxycinnamoyltransferase involved in phenylpropanoid biosynthesis in Cynara cardunculus L. BMC Plant Biol 7: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria JC. (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Gu R, Dou G, Wang J, Dong J, Meng Z (2007) Simultaneous determination of 1,5-dicaffeoylquinic acid and its active metabolites in human plasma by liquid chromatography-tandem mass spectrometry for pharmacokinetic studies. J Chromatogr B Analyt Technol Biomed Life Sci 852: 85–91 [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M (2003) Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278: 95–103 [DOI] [PubMed] [Google Scholar]
- Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L (2007) Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol 145: 1371–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S. (2006) Sweetpotato (Ipomoea batatas L.) leaf: its potential effect on human health and nutrition. J Food Sci 71: R13–R21 [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kaul A, Khanduja KL (1998) Polyphenols inhibit promotional phase of tumorigenesis: relevance of superoxide radicals. Nutr Cancer 32: 81–85 [DOI] [PubMed] [Google Scholar]
- Lallemand LA, Zubieta C, Lee SG, Wang Y, Acajjaoui S, Timmins J, McSweeney S, Jez JM, McCarthy JG, McCarthy AA (2012) A structural basis for the biosynthesis of the major chlorogenic acids found in coffee. Plant Physiol 160: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepelley M, Cheminade G, Tremillon N, Simkin A, Caillet V, McCarthy J (2007) Chlorogenic acid synthesis in coffee: an analysis of CGA content and real-time RT-PCR expression of HCT, HQT, C3H1, and CCoAOMT1 genes during grain development in C. canephora. Plant Sci 172: 978–996 [Google Scholar]
- Luo J, Butelli E, Hill L, Parr A, Niggeweg R, Bailey P, Weisshaar B, Martin C (2008) AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J 56: 316–326 [DOI] [PubMed] [Google Scholar]
- Mahesh V, Million-Rousseau R, Ullmann P, Chabrillange N, Bustamante J, Mondolot L, Morant M, Noirot M, Hamon S, de Kochko A, et al. (2007) Functional characterization of two p-coumaroyl ester 3′-hydroxylase genes from coffee tree: evidence of a candidate for chlorogenic acid biosynthesis. Plant Mol Biol 64: 145–159 [DOI] [PubMed] [Google Scholar]
- Martin C, Butelli E, Petroni K, Tonelli C (2011) How can research on plants contribute to promoting human health? Plant Cell 23: 1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y, Tonelli C, Petroni K (2013) Plants, diet, and health. Annu Rev Plant Biol 64: 19–46 [DOI] [PubMed] [Google Scholar]
- McDougall B, King PJ, Wu BW, Hostomsky Z, Reinecke MG, Robinson WE Jr (1998) Dicaffeoylquinic and dicaffeoyltartaric acids are selective inhibitors of human immunodeficiency virus type 1 integrase. Antimicrob Agents Chemother 42: 140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menin B, Comino C, Moglia A, Dolzhenko Y, Portis E, Lanteri S (2010) Identification and mapping of genes related to caffeoylquinic acid synthesis in Cynara cardunculus L. Plant Sci 179: 338–347 [Google Scholar]
- Meyer A, Donovan J, Pearson D, Waterhouse A, Frankel E (1998) Fruit hydroxycinnamic acids inhibit human low-density lipoprotein oxidation in vitro. J Agric Food Chem 46: 1783–1787 [Google Scholar]
- Moco S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, Vervoort J, de Vos CH (2006) A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol 141: 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moco S, Capanoglu E, Tikunov Y, Bino RJ, Boyacioglu D, Hall RD, Vervoort J, De Vos RC (2007) Tissue specialization at the metabolite level is perceived during the development of tomato fruit. J Exp Bot 58: 4131–4146 [DOI] [PubMed] [Google Scholar]
- Moglia A, Comino C, Portis E, Acquadro A, De Vos RC, Beekwilder J, Lanteri S (2009) Isolation and mapping of a C3′H gene (CYP98A49) from globe artichoke, and its expression upon UV-C stress. Plant Cell Rep 28: 963–974 [DOI] [PubMed] [Google Scholar]
- Moglia A, Lanteri S, Comino C, Acquadro A, de Vos R, Beekwilder J (2008) Stress-induced biosynthesis of dicaffeoylquinic acids in globe artichoke. J Agric Food Chem 56: 8641–8649 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–478 [Google Scholar]
- Niggeweg R, Michael AJ, Martin C (2004) Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat Biotechnol 22: 746–754 [DOI] [PubMed] [Google Scholar]
- Peluso G, De Feo V, De Simone F, Bresciano E, Vuotto ML (1995) Studies on the inhibitory effects of caffeoylquinic acids on monocyte migration and superoxide ion production. J Nat Prod 58: 639–646 [DOI] [PubMed] [Google Scholar]
- Rice-Evans C, Miller NJ, Paganga G (1997) Antioxidant properties of phenolic compounds. Trends Plant Sci 2: 152–159 [Google Scholar]
- Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D (2001) CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem 276: 36566–36574 [DOI] [PubMed] [Google Scholar]
- Slanina J, Taborska E, Bochorakova H, Slaninova I, Humpa O, Robinson W, Schram K (2001) New and facile method of preparation of the anti-HIV-1 agent, 1,3-dicaffeoylquinic acid. Tetrahedron Lett 42: 3383–3385 [Google Scholar]
- Slimestad R, Verheul M (2009) Review of flavonoids and other phenolics from fruits of different tomato (Lycopersicon esculentum Mill.) cultivars. J Sci Food Agric 89: 1255–1270 [Google Scholar]
- Sonnante G, D’Amore R, Blanco E, Pierri CL, De Palma M, Luo J, Tucci M, Martin C (2010) Novel hydroxycinnamoyl-coenzyme A quinate transferase genes from artichoke are involved in the synthesis of chlorogenic acid. Plant Physiol 153: 1224–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack D, Gross W (1990) Properties and activity changes of chlorogenic acid:glucaric acid caffeoyltransferase from tomato (Lycopersicon esculentum). Plant Physiol 92: 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teutschbein J, Gross W, Nimtz M, Milkowski C, Hause B, Strack D (2010) Identification and localization of a lipase-like acyltransferase in phenylpropanoid metabolism of tomato (Solanum lycopersicum). J Biol Chem 285: 38374–38381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vain P, Keen N, Murillo J, Rathus C, Nemes C, Finer J (1993) Development of the particle inflow gun. Plant Cell Tissue Organ Cult 33: 237–246 [Google Scholar]
- Vallverdú-Queralt A, Odriozola-Serrano I, Oms-Oliu G, Lamuela-Raventós RM, Elez-Martínez P, Martín-Belloso O (2012) Changes in the polyphenol profile of tomato juices processed by pulsed electric fields. J Agric Food Chem 60: 9667–9672 [DOI] [PubMed] [Google Scholar]
- Villegas RJ, Kojima M (1986) Purification and characterization of hydroxycinnamoyl d-glucose. Quinate hydroxycinnamoyl transferase in the root of sweet potato, Ipomoea batatas Lam. J Biol Chem 261: 8729–8733 [PubMed] [Google Scholar]
- Walker AM, Hayes RP, Youn B, Vermerris W, Sattler SE, Kang C (2013) Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other Coenzyme A-dependent transferases and synthases. Plant Physiol 162: 640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Simon JE, Aviles IF, He K, Zheng QY, Tadmor Y (2003) Analysis of antioxidative phenolic compounds in artichoke (Cynara scolymus L.). J Agric Food Chem 51: 601–608 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.